
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 18 August 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.926788
This article is part of the Research Topic Immune Interactions with Pathogenic and Commensal Fungi View all 17 articles
Melioidosis is a fatal infectious disease caused by Burkholderia pseudomallei. Complications following treatment are usually due to antibiotic resistance and relapse is mainly caused by B. pseudomallei biofilm. Although the release of neutrophil extracellular traps (NETs) is crucial to capture and eliminate bacterial pathogens, to date response of NETs to B. pseudomallei biofilm is poorly understood. Here we compare the NETs produced by neutrophils in response to B. pseudomallei H777 (a biofilm-producing strain containing the bpsl0618 gene), a biofilm-defect strain lacking this gene (B. pseudomallei M10) and a bpsl0618 biofilm-complemented strain, B. pseudomallei C17, in which function of bpsl0618 was restored. Co-cultivation of these strains with healthy human neutrophils at MOI 10 with or without cytochalasin D demonstrated that H777 significantly resisted neutrophil-mediated killing and non-phagocytotic mechanisms compared to M10 (p < 0.0001). Three distinct morphotypes of NETs were seen: “aggregated”, “spiky” and “cloudy”. These were induced in different proportions by the different bacterial strains. All types of NETs were shown to confine all B. pseudomallei strains. Strains H777 and C17 could stimulate production of twice as much extracellular DNA (234.62 ng/mL and 205.43 ng/mL, respectively) as did M10 (111.87 ng/mL). Cells of H777 and C17 were better able to survive in the presence of neutrophil killing mechanisms relative to M10 (p < 0.0001) and NET formation (p < 0.0001 and 0.05). These findings suggest that NET stimulation was insufficient to eradicate B. pseudomallei H777 and C17 despite their possession of bpsl0618, a sugar-transferase gene associated with biofilm formation ability. Our findings demonstrate that B. pseudomallei biofilm phenotype may be a key factor in assisting pathogens to escape killing by neutrophils. This work provides a better understanding of how B. pseudomallei biofilm-associated infections induce and survive NET formation, resulting in bacterial persistence and increased severity of disease.
Burkholderia pseudomallei is the causative agent of fatal infectious melioidosis. The presence of this bacterial pathogen in soil and water in endemic regions, particularly in Southeast Asia and northern Australia, is correlated with high numbers of diagnosed cases (1). The global incidence of melioidosis has been increasing, with nearly 165,000 cases and close to 90,000 deaths per year (2), including 2,800 deaths in Thailand (3). In patients, B. pseudomallei infection often manifests as multiple abscesses, pneumonia, septicemia and can be chronic depending on the route of infection, host immune performance, bacterial strain and load (1, 2, 4). This bacterium can be transmitted to humans via aerosol inhalation, ingestion and inoculation through skin injuries associated with activities of daily living (5–7). The molecular and cellular basis of pathogenesis, virulence factors and mechanisms of immune evasion that allow B. pseudomallei to be such an effective opportunistic pathogen have been reviewed (2, 8). However, the ways in which bacterial interaction with the immune system result in diverse disease outcomes remain to be elucidated.
Relapsing melioidosis can be a consequence of poor patient compliance with treatment regimes, has a high mortality rate (9) and is correlated with biofilm-associated B. pseudomallei infection (10). Burkholderia pseudomallei cells growing as biofilms are sheltered as microcolonies within extracellular polymeric substances containing proteins (11), exopolysaccharide (12) and eDNA (13) both in vivo and in vitro (12, 14). As biofilm, B. pseudomallei cells are effectively protected from penetration of antimicrobial agents, resulting in high-level antibiotic resistance (15, 16). Such bacterial biofilms not only habitually evade host immune defenses (17, 18) but also alter host immune responses (19), thus changing the balance between host and pathogen and contributing to the establishment of chronic infections. Several studies have attempted to understand the role of B. pseudomallei biofilm in the melioidosis disease process. In 2005, B. pseudomallei M10, a biofilm-defective mutant produced by transposon insertion into the sugar-transferase gene bpsl0618 was constructed from the biofilm wild type H777 (20). Upregulation of bpsl0618 increases the expression of polysaccharides for development of B. pseudomallei biofilm architecture (21). It appears that bpsl0618 is crucial for B. pseudomallei biofilm formation. A third strain, derived from M10, is C17, in which the function of bpsl0618 has been restored (14). The relevance of B. pseudomallei bpsl0618 to biofilm formation, pathogenesis in alveolar epithelial cells has been established. Planktonic B. pseudomallei H777 cells are more able than those of M10 to attach to and invade host cells leading apoptosis and production of proinflammatory cytokines in the human lung epithelial cell line, A549. The biofilm complemented strain, C17 restored the biofilm formation, invasion ability, apoptosis induction and cytokines response (14). Additionally, the quantity of eDNA associated with C17 biofilm biomass was comparable to the parental strain, H777 (13). Many hypotheses regarding effects of B. pseudomallei biofilm on human innate immune cells appeared to be ill-defined.
Neutrophils are key innate immune cells for eradicating bacterial pathogens including B. pseudomallei (22). To do this, they used phagocytosis, antimicrobial peptides, enzymes, reactive oxygen species (ROS) and neutrophil extracellular traps (NETs) containing extracellular fibers with granular proteins and extracellular DNA (eDNA) (23–25). NETs have a key function in capturing and eradicating large pathogens that resist phagocytosis (26, 27). In a previous study, B. pseudomallei efficiently induced NETosis, with large amounts of NET-related components to entrap and efficiently kill B. pseudomallei but this failed to limit bacterial spreading and inflammation during B. pseudomallei-induced sepsis (28). Likewise, impaired NETs in diabetes mellitus patients were possibly correlated with susceptibility to clinical melioidosis and other forms of sepsis (29). In addition, not only host factors influenced NET formation but also bacterial components including B. pseudomallei type 3 secretory system (T3SS) and capsular polysaccharide (29). Furthermore, soluble ligands such as lipopolysaccharide (LPS) are well established to stimulate aggregated and cloudy NETs (30). The challenge of eradicating B. pseudomallei biofilm has generated considerable interest in NET activation and killing competence but to date has yielded very limited data. Therefore, we aimed to extend knowledge of the association between B. pseudomallei biofilm and NET formation. Further understanding of B. pseudomallei biofilm and NETs may provide further explanation of the complicated outcome of melioidosis pathogenesis and novel approaches to clinical therapy.
Blood was collected from healthy donors from the Blood Bank, Srinagarind hospital, Faculty of Medicine, Khon Kaen University, Thailand. This study was approved by Khon Kaen University Ethics Committee for Human Research (Reference No. HE631099).
Peripheral blood was obtained from healthy donors using lithium-heparin vacutainers (Greiner Bio-One, Chonburi, Thailand). Neutrophils were isolated as previously described (31). In brief, whole blood was mixed with HetaSep solution (STEMCELL-Technologies, Vancouver, Canada) at a ratio of 5:1 and incubated at 37°C for 30 min. Then, the upper layer containing all nucleated blood cells was taken and gently layered on top of Ficoll-Paque solution (GE Healthcare, Little Chalfont, UK) at a ratio of 1:1, followed by centrifugation at 500 g for 30 min. After peripheral blood mononuclear cells (PBMC), plasma and Ficoll-Paque solution were carefully discarded, the neutrophil pellet was resuspended in 1 mL RPMI 1640 complete medium (RPMI 1640 (Gibco, Life Technologies, Paisley, UK) with 10% fetal bovine serum (FBS) (Gibco, Life Technologies, Paisley, UK)). Hypotonic ammonium chloride lysis buffer was applied to eliminate residual erythrocytes for 4 min, followed by centrifugation at 400 g for 3 min. the neutrophil pellet was then resuspended in RPMI 1640 complete medium and checked for neutrophil purity (> 95%) and viability (> 95% required for further investigations).
Three B. pseudomallei strains, H777, a moderate biofilm producer; M10, a biofilm-defective mutant of H777 (inactivated bpsl0618) and C17, a biofilm-complemented derivative of M10 (bpsl0618 restored) (13, 14) were used in this study (Table 1). Each B. pseudomallei strain from -80°C glycerol stock was streaked on Luria-Bertani (LB) agar plate with appropriate antibiotics and incubated at 37°C for 48 h. A single colony of each strain was sub-cultured into 5 mL of LB broth and incubated at 37°C with 200 rpm for 16-18 h. The bacterial culture was adjusted to an optical density (OD) at 600 nm of 0.1 for 2% inoculums (v/v) in LB broth at 37°C at 200 rpm to obtain the mid-log phase for neutrophil co-cultivation.
Neutrophils (5 × 105 cells) in 500 µL were added into wells of a 48-well tissue-culture plate and incubated at 37°C for 90 min, 5% CO2 and pretreated or not with 10 μg/mL cytochalasin D (Sigma-Aldrich, Saint Louis, USA) for 30 min to prevent phagocytosis as previously described (29). Planktonic B. pseudomallei H777, M10 and C17 from mid-log phase cultures were added at a multiplicity of infection (MOI) of 10 and the plates further incubated for 90 min. The percentage of remaining bacteria in each well was then enumerated by serial dilutions and a drop plate technique on LB agar at 37°C for 24 h. The effect of cytochalasin D/dimethyl sulfoxide (DMSO) on neutrophil and bacterial survival was also determined in parallel assays.
NETs stimulated by B. pseudomallei were visualized as previously described (24). Briefly, neutrophils (5 × 105 cells/well) were seeded onto BSA-coated cover slips (1% BSA (Sigma-Aldrich, Saint Louis, USA) in phosphate-buffered saline (PBS), pH 7.4) in 24-well plates at 37°C, 5% CO2 for 30 min to allow for cell attachment. Planktonic B. pseudomallei H777, M10 and C17 were washed twice with PBS before being incubated with the neutrophils at MOI of 10 in parallel with 50 nM of phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich, Saint Louis, USA) (positive control) and untreated controls for 90 min. In addition, neutrophils pretreated with 1 unit/mL DNase I for 15 min prior to adding B. pseudomallei H777 were also used. After centrifugation at 1,800 g for 10 min, the cells on the cover slips were fixed with 4% paraformaldehyde for 15 min at room temperature and washed twice with PBS. Cells were permeabilized with 0.5% Triton X-100 for 1 min followed by nonspecific blocking using 1% BSA in PBST (PBS with 0.1% Tween® 20) for 30 min. The samples were stained with the primary rabbit anti-myeloperoxidase antibody (anti-MPO, ab109116, 1:100 dilution, Abcam, Cambridge, UK) for 1 h, washed 3 times with PBST followed by addition of donkey anti-rabbit IgG HL antibody (Alexa Fluor® 647, 1:500 dilution, Abcam, Cambridge, UK). In parallel, mouse anti-neutrophil elastase antibody (anti-NE, ab255935, 1:250 dilution, Abcam, Cambridge, UK) was added for 1 h followed by goat anti-mouse IgG HL antibody (Alexa Fluor® 488, 1:500 dilution, Abcam, Cambridge, UK). DNA scaffolds of NETs were observed after staining with 1 µg/mL of 4′,6-diamidino-2-phenylindole (DAPI) (Thermo scientific, Rockford, USA) for 5 min. Features of the NETs were examined on two different cover slips per treatment at 63× magnification for least 15 fields/cover slip under a confocal laser scanning microscope (LSM-500 and LSM 800; Zeiss, Germany).
To visualize B. pseudomallei entrapment, the co-cultured neutrophils and B. pseudomallei strains on cover slips were fixed with 4% paraformaldehyde, stained with Giemsa and observed at 100× magnification (Axio, Carl Zeiss, Jena, Germany). Percentage of each NET formation type stimulated by each B. pseudomallei strain were quantified (32).
Amount of eDNA from NETs was quantified as previously described (33). Briefly, purified neutrophils (5 × 105 cells) in 48-well plates were activated with either B. pseudomallei H777, M10 or C17 at MOI 10 for 90 min or with PMA. Subsequently, the mixture was treated with 1 unit/mL micrococcal nuclease (MNase) (Sigma-Aldrich, Saint Louis, USA) for 15 min at room temperature to liberate NET-DNA. After centrifugation at 1800 g for 10 min, the eDNA in the supernatant was quantified with a QuantiFluor® dsDNA system (Promega, Madison, WI, USA) (excitation: 504 nm/emission: 531 nm) using a fluorometer (Varioskan Flash Multimode Reader, Singapore) with SkanIt Software 2.4.3 RE for Varioskan Flash. A standard curve for eDNA was constructed using lambda DNA to quantify eDNA in each sample.
NETs bactericidal against B. pseudomallei H777, M10 and C17 were examined as previously (29). Neutrophils (5 × 105 cells) in 48-well plates were pretreated with either 10 μg/mL cytochalasin D for 30 min, 1 unit/mL DNase I for 15 min or both agents at 37°C prior to addition of B. pseudomallei H777, M10 or C17 at MOI of 10. After incubation for a further 90 min, the mixture in each well was 10-fold serially diluted for bacterial survival enumeration using a drop plate technique on LB agar plates. This experiment was performed in triplicate on three independent occasions.
Statistical analysis was performed using GraphPad Prism version 8.3.4 software (GraphPad, San Diego, CA). Quantities of intracellular bacterial surviving and eDNA were compared among the three experimental groups using two-way ANOVA followed by Tukey’s post hoc test for comparisons between pairs. A statistically significant difference was considered as follows: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
To determine whether the B. pseudomallei biofilm phenotype could influence bacterial survival from neutrophil mediated killing either all neutrophil eradication strategies or non-phagocytosis, we pre-treated neutrophils with the phagocytosis inhibitor, cytochalasin D before co-cultured with MOI 10 of B. pseudomallei H777 or C17 strains containing bpsl0618 compared to M10 strain with bpsl0618 defected mutant for 90 min. Significantly more B. pseudomallei H777 cells (101.36% of initial number) persisted against untreated neutrophils than was the case for M10 (78.55%). Likewise, the percentage of B. pseudomallei H777 cells surviving was significantly higher after encountering neutrophils pretreated with cytochalasin D (88.52%) than that of M10 (68.04%) (p < 0.0001) (Figures 1A, B). Meanwhile, B. pseudomallei C17, the bpsl0618-complemented derivative of M10, displayed restored bacterial survival against neutrophil functions. This evidence clearly indicated the better persistence of B. pseudomallei H777 and C17 harboring bpsl0618 against neutrophil bactericidal activities involving a non-phagocytosis mechanism.
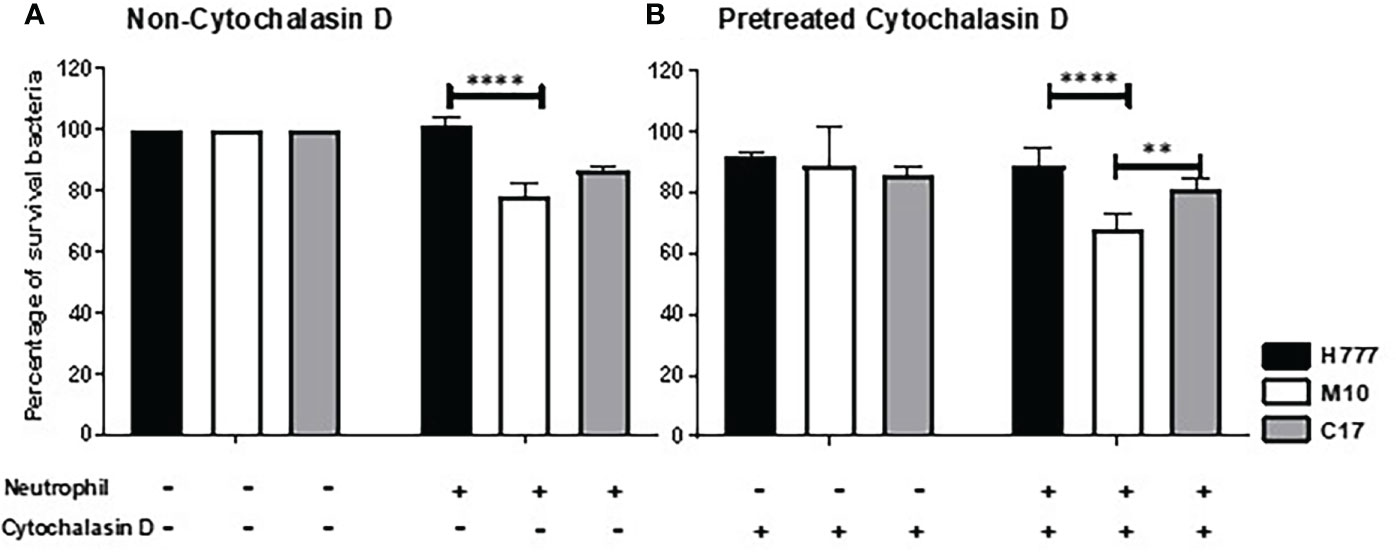
Figure 1 Burkholderia pseudomallei H777 significantly better withstood neutrophil-mediated killing with or without the presence of cytochalasin D than did M10, the strain lacking functional bpsl0618. The extent of neutrophil-mediated killing was assessed after 90 min co-cultivation of the bacteria with neutrophils untreated (A) or pretreated (B) with cytochalasin D. Percentage of B. pseudomallei cells surviving were determined from 3 independent experiments. Data are represented as mean ± standard deviation. Asterisks denote statistical significance (**p < 0.01, ****p < 0.0001).
To further investigate the ability of bpsl0618 to facilitate persistence of B. pseudomallei against non-phagocytosis mechanisms of human neutrophils, NET formation morphology was examined. Neutrophils were co-cultured with B. pseudomallei H777, M10 and C17 in company with PMA as a positive control. NET formation visualization used three NET markers; DAPI to detect released DNA backbone, anti-MPO and anti-NE antibodies. Figure 2A shows confocal fluorescence micrographs of unstimulated, intact neutrophils with nuclei, myeloperoxidase and elastase within the cytoplasm (Figure 2A). PMA-stimulated neutrophils produced mainly “cloudy” NETs (cloud-like DNA pattern) following the collapse of the nucleus and cell-membrane rupture (Figure 2B). Another appearance that NETs could adopt was “spiky” (released DNA traps with fiber-like structure) with colocalized myeloperoxidase (red) and neutrophil elastase (green) with protruding DNA (blue) after activation with PMA (Figure 2C). The presence of B. pseudomallei H777 produced mainly “aggregated” NETs (high-density aggregation of netting neutrophils) followed by cloudy and spiky NETs (Figures 2D–F; Table 2). In addition, extrusion of DNA traps from several cells appeared broad and blurry in the extracellular environment. Cells of B. pseudomallei M10 mainly led to the production of spiky NETS followed by aggregated and cloudy NETs (Figures 2G–I; Table 2), whereas B. pseudomallei C17 could elicit all types of NETs (Figures 2J–L; Table 2). Interestingly, C17 led to the appearance of aggregated (34%), cloudy (15%) at spiky NETs (51%) at proportions intermediate between those produced by H777 and M10 (Table 2). Thus, the appearance of NETs varies according to the presence or absence of functional bpsl0618 in B. pseudomallei strains stimulating them. The exceedingly high neutrophil accumulation was observed in the aggregated NETs but not in the cloudy or spiky NETs. DNase I treatment of B. pseudomallei H777-infected neutrophils resulted in diminished extracellular DNA, MPO and NE suggesting that the released materials from stimulated neutrophils were composed largely of DNA (Figure 2M).
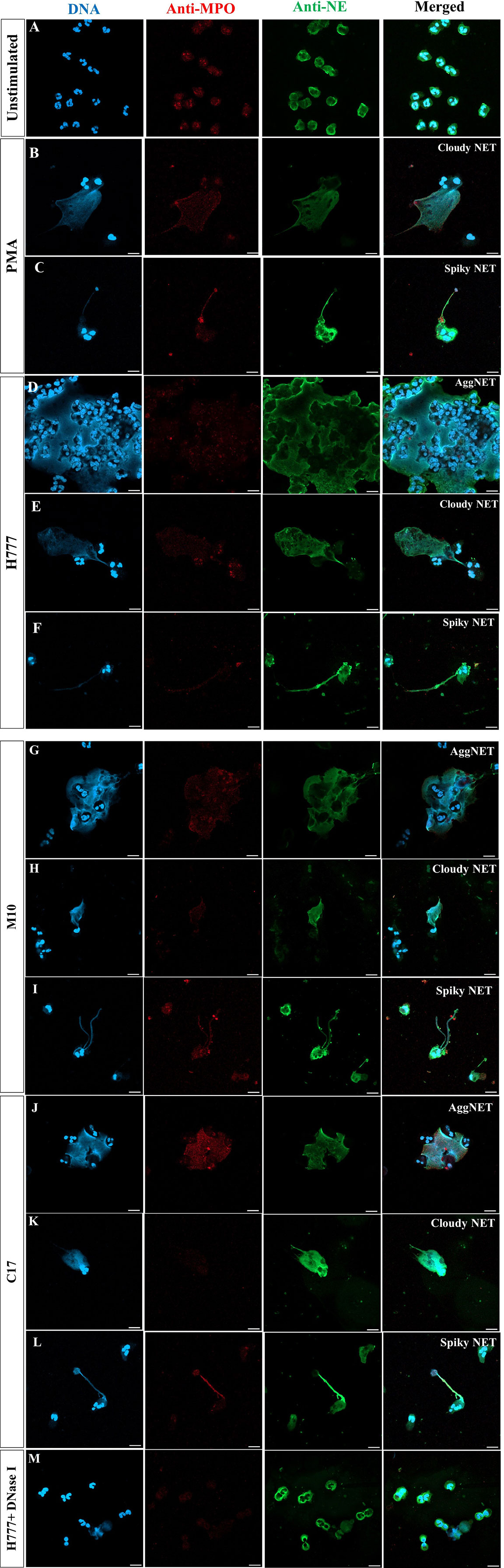
Figure 2 NET formation visualization and effect of DNase I degradation. (A) Unstimulated neutrophils; (B, C) neutrophils stimulated with PMA; (D–F) B. pseudomallei H777 (bpsl0618 wild type); (G–I) B. pseudomallei M10 (bpsl0618 mutant); (J–L) B. pseudomallei C17 (bpsl0618 complemented); (M) B. pseudomallei H777 in the presence of 1 unit/mL DNase I. Neutrophils were stained with DAPI (blue), anti-MPO (Alexa Fluor 647 conjugated, red) and anti-NE elastase (Alexa Fluor 488 conjugated, green). Magnification = 63×; scale bars = 10 µm.
Additionally, bright-field microscopic images during NET formation confirmed that B. pseudomallei were entrapped within NETs (Figure 3). Cells of B. pseudomallei H777 were entrapped within aggregated NETs and liberated NET components or extracellular DNA components (extracellular hazy-gray zone) (Figures 3A, B). Moreover, B. pseudomallei M10 cells were either confined by the sticky DNA within the boundary of complex spiky NETs (Figure 3C), hooked by single extracellular DNA fibers (Figure 3D) or entrapped with a DNA cloud of aggregated NETs (Figure 3E). Similarly, B. pseudomallei C17 were restrained by aggregated NETs (Figures 3F, G). These phenomena demonstrated that B. pseudomallei could potentially induce NET formation in different manner depending on their possession or not of bpsl0618. Extracellular NET components released from stimulated neutrophils were confirmed to capture B. pseudomallei cells.
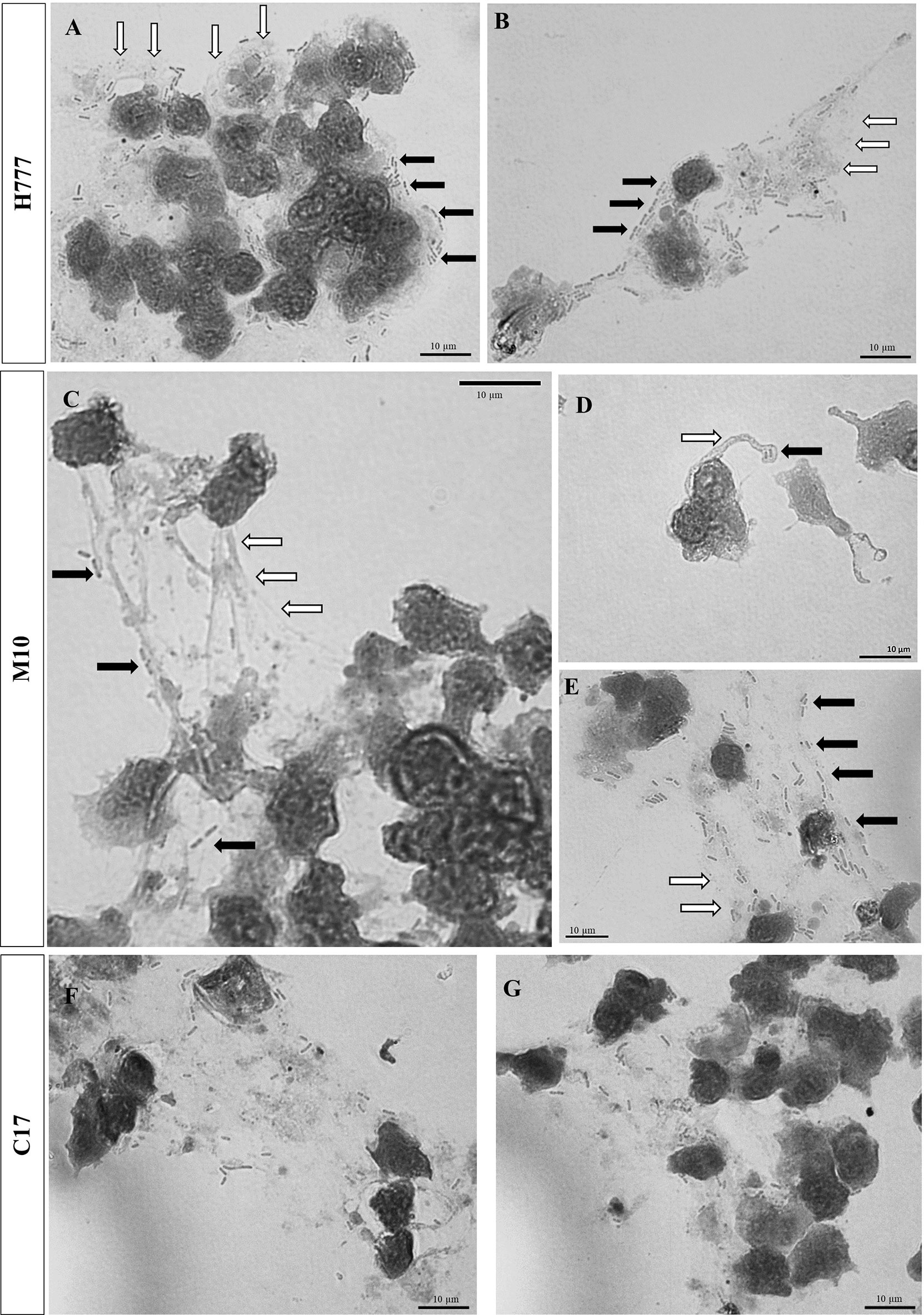
Figure 3 Bright-field images showing that B. pseudomallei cells entrapped within NETs after being encountered by neutrophils at MOI of 10 for 90 min and stained with Giemsa dye. (A, B) B. pseudomallei H777 (rod shape, black arrow) trapped by aggregated NET (gray area, white arrow); (C) B. pseudomallei M10 entrapped within NETs, (D) B. pseudomallei M10 entrapped in spiky NETs; (E) B. pseudomallei M10 trapped within aggregated NETs. (F, G) B. pseudomallei C17 entrapped within aggregated NETs. Scale bars = 10 µm.
The quantity of released DNA from NETs activated by B. pseudomallei was compared between bacterial strains with or without a functional bpsl0618 gene. DNA was measured after micrococcal nuclease (MNase) digestion. The results showed that neutrophils activated by B. pseudomallei H777 liberated significantly higher quantities of DNA in traps (234.62 ng/mL) than did B. pseudomallei M10 (111.87 ng/mL) (p ≤ 0.01) (Figure 4). While B. pseudomallei C17 elicited release of intermediate amounts of NET DNA (205.43 ng/mL). These results suggest that the B. pseudomallei bpsl0618 gene encoding a sugar transferase is associated with greater production of NETs and NET DNA.
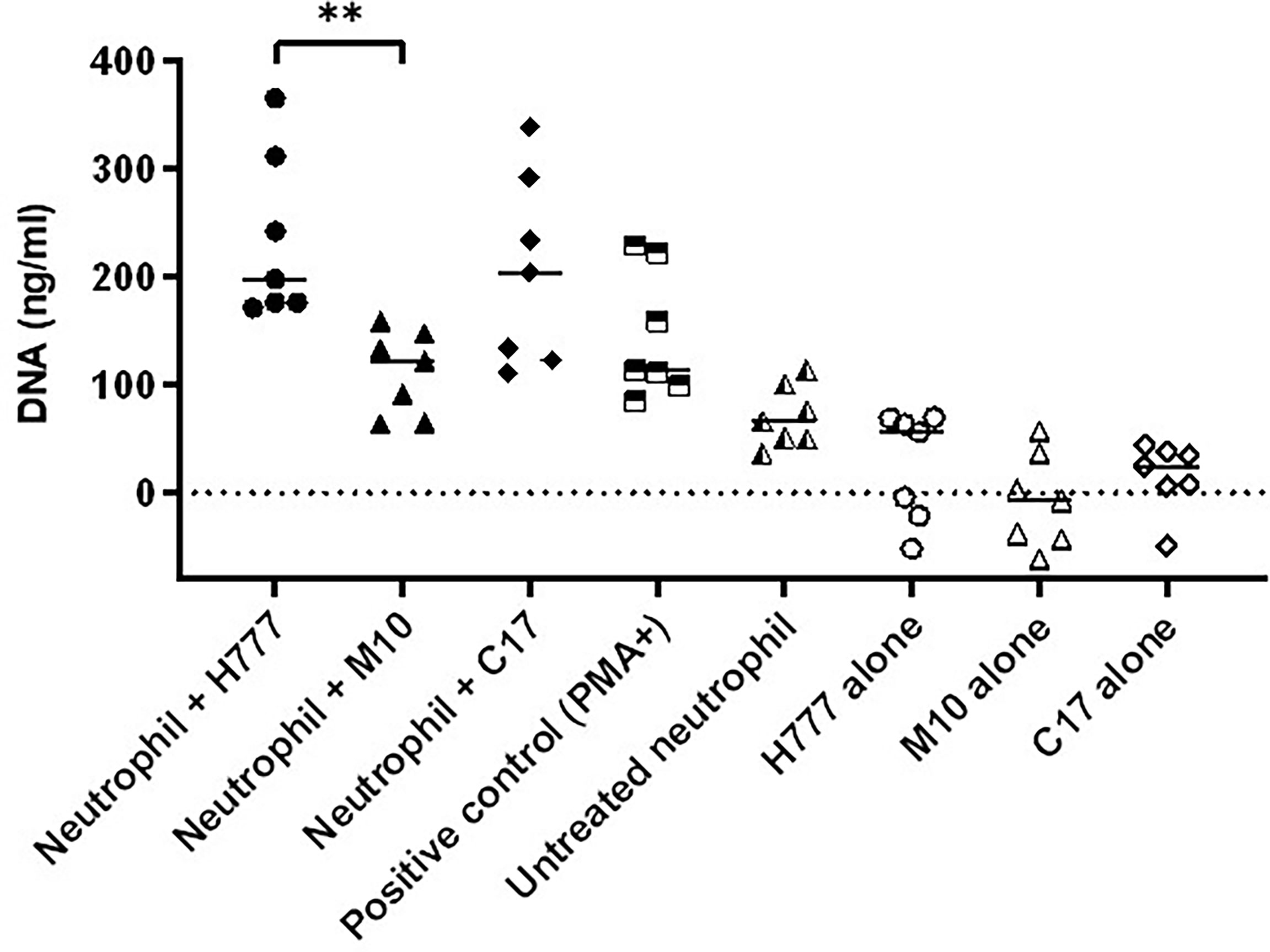
Figure 4 B. pseudomallei H777 (bpsl0618 wild type) and C17 (bpsl0618 complemented) induced greater release of extracellular DNA from neutrophils than did M10 (bpsl0618 defect mutant). Neutrophils were stimulated with either B. pseudomallei at the MOI of 10 or 50 nM PMA followed by MNase treatment in supernatant. The DNA was quantified in 3 independent experiments (n = 7). Data are represented as mean ± standard deviation. Asterisks denote statistical significance (**p < 0.01).
Following NET stimulation, the fate of each B. pseudomallei strain was monitored against neutrophil-mediated killing mechanisms: phagocytosis, NET-mediated killing and degranulation. Neutrophils were treated or not with either or both of DNase I and cytochalasin D. The results revealed that cells of B. pseudomallei H777 and C17 could persist against neutrophil-mediated killing (untreated neutrophils) (98.77 and 87.17%) but that survival was much lower in the bpsl0618-inactivated mutant, M10 (59.57%). Similar percentages of B. pseudomallei H777 and C17 could survive NETosis (pretreatment of neutrophils with cytochalasin D) (96.24 and 74.76%, respectively) while survival of M10 cells was significantly lower (59.95%) (Figure 5). However, all three strains were almost equally susceptible to phagocytic killing by neutrophils pretreated with DNase I: survival rates were 58.59% for H777, 50.58% for M10 and 54.65% for C17 cells. These data highlighted the crucial role of the bps0618 gene on B. pseudomallei biofilm formation ability that endurance against NET-mediated killing but not of phagocytosis. However, in the presence of both cytochalasin D and DNase I, leaving neutrophils only the option of degranulation-mediated killing, all three bacterial strains were equally susceptible. These results emphasized that NET stimulation by B. pseudomallei is insufficient to eradicate B. pseudomallei strains with the ability to form biofilm and that the presence of the bpsl0618 gene assists survival against NET-mediated bacterial killing.
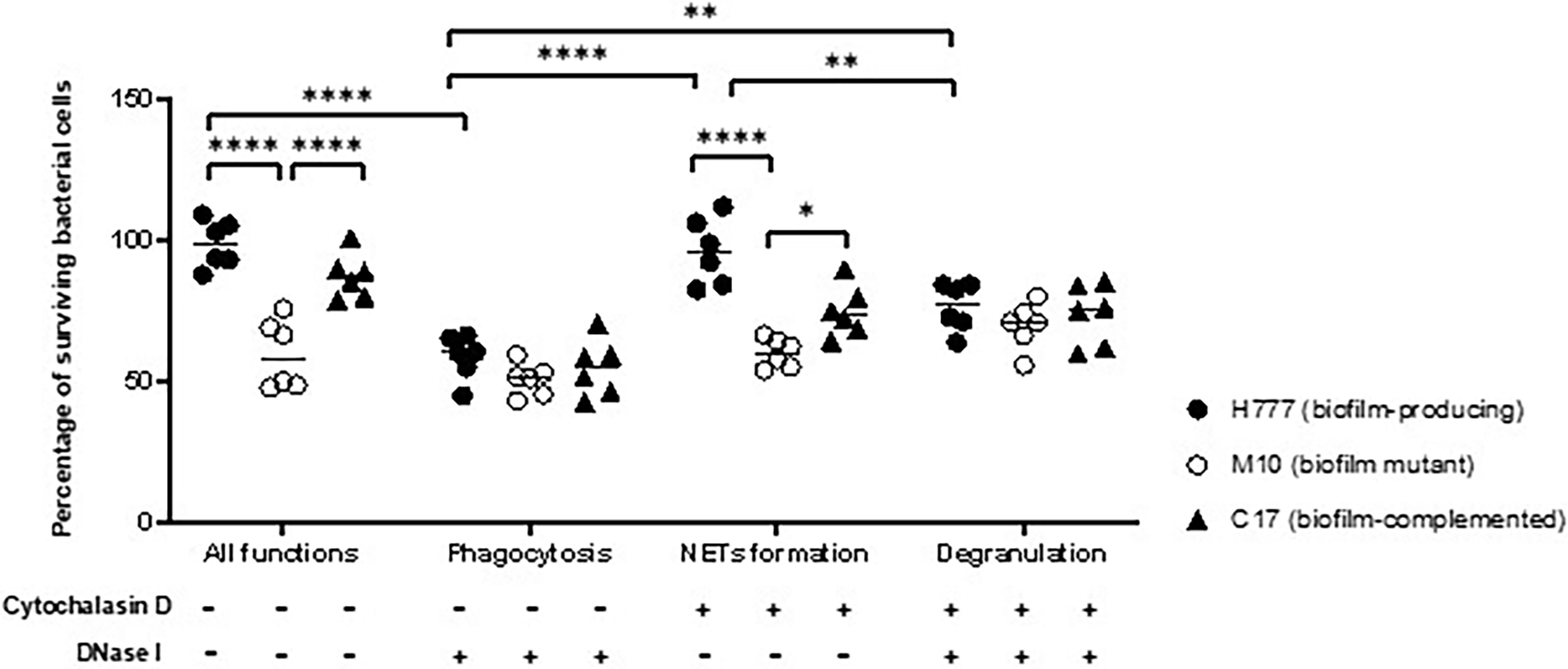
Figure 5 NET formation was insufficient to kill B. pseudomallei cells expressing bpsl0618. Co-cultivation of purified neutrophils and three strains of B. pseudomallei was done in the presence or absence of 10 μg/mL cytochalasin D and/or 1 unit/mL DNase I for 90 min. Percentage of cells surviving in each of the three strains of B. pseudomallei is shown in results from 3 independent experiments (n = 6). Data are represented as mean ± standard deviation. Asterisks denote statistical significance (*p < 0.05, **p < 0.01, ****p < 0.0001).
Biofilm-associated Burkholderia pseudomallei infections are often associated with high incidence of relapse of melioidosis (10), resulting in poor clinical outcomes (9). The possibility of bacterial biofilms skewing host immune balance remains to be elucidated (19). Production of NETs is well recognized as one of the neutrophil-mediated killing mechanisms to capture and kill pathogens capable of biofilm formation and resistant to phagocytosis. To date, a number of studies have focused on bacterial strategies to escape from NETs and influence the outcome of bacterial infections (34). Furthermore, evasion of NETs has been seen in respiratory pathogens, thus facilitating pathogen proliferation and dissemination (25). B. pseudomallei strains with the ability to produceT3SS and capsular polysaccharide were better able to evade NETs (29). The formation of NETs stimulated by presence of B. pseudomallei does not prevent bacterial dissemination and inflammation (28). To the best of our knowledge, limited information is available on the association between B. pseudomallei biofilm and NET formation and the consequences for bacterial survival. Our study offers further evidence of the effects of B. pseudomallei with the biofilm formation capability on neutrophil- and NET-mediated killing using planktonic B. pseudomallei H777, a biofilm wild type compared to that of the M10 strain, a biofilm-defective mutant of H777 (inactivated bpsl0618, a sugar transferase gene) and C17, a biofilm-complemented derivative of M10. The biofilm formation ability of these three strains was previously determined in our laboratory (13, 14). Remarkably, we have shown that B. pseudomallei biofilm phenotypes can withstand non-phagocytotic activities of neutrophils. The lowest survival rate in our experiments was seen in the B. pseudomallei biofilm-defective strain, emphasizing the susceptibility of the mutant to NETs and phagocytosis. Additionally, the distinct morphological types of NET (aggregated, cloudy and spiky) were first visualized based on presence or absence of a bpsl0618 gene in B. pseudomallei. The presence of this gene, encoding sugar transferase, was correlated with higher levels of extracellular DNA extruded from stimulated neutrophils. Notably, the twice higher level of DNA from NET stimulated by B. pseudomallei H777 and C17 than that of M10 indicated a better potential biofilm formation capability of this pathogen to stimulate NET formation. Nevertheless, NET formation failed to eradicate B. pseudomallei H777 and C17 but had a considerable impact on M10, the biofilm-defective mutant. However, all three strains were equally susceptible to phagocytotic killing by neutrophils. This study highlighted the possession of bpsl0618 that was previously demonstrated to be associated with the ability of B. pseudomallei to form biofilm (13, 14) and may allow persistence of these bacteria despite the fact that their presence stimulates production of NETs, one of the key innate eradication mechanisms employed by the body.
Our findings are consistent with previous results in which B. pseudomallei showed genuine resistance to neutrophil killing (22). Furthermore, the persistence of B. pseudomallei H777 and C17 against neutrophil- and NET-mediated killing are consistent with the findings of de Jong and colleagues (28), who demonstrated that B. pseudomallei potentially induced NET formation in humans based on the finding of NET-related components in melioidosis patients. Even so, presence of NETs hardly protects against bacterial dissemination.
Pathogenic bacteria have several approaches to protect themselves from NET capture and killing. These include using polysaccharide capsules, biofilm formation, altering electric charge of cell surface, inhibition of NETs by peptidase and DNase secretion and inhibiting ROS production (34). Both the biofilm-competent B. pseudomallei H777 harboring bpsl0618 and C17, the bpsl0618-complemented strain, efficiently induced NETs but could survive despite this. This are in line with previous findings (29), which demonstrated the crucial roles of T3SS and capsular polysaccharide produced by B. pseudomallei in evasion of NET killing mechanisms. The impact of these mutants that interrupt the bacterial cellular process represented known virulence factors of B. pseudomallei during NET formation. Similar findings have been noted for non-typeable Haemophilus influenza biofilms: despite being entrapped within NETs, these cells could survive both phagocytic and extracellular neutrophil killing resulting in increased numbers of bacterial cells. Consequently, the persistence of H. influenza correlated with chronic and recurrent otitis media infections (35). Based on our results, it is at least hypothetically possible that B. pseudomallei biofilm may stimulate NET production but escape from the bactericidal activities of NETs leading to bacterial persistence and chronic infection or relapsing melioidosis.
NET formation is an alternative defense mechanism assisting neutrophils to kill particularly large pathogens (26). NETs principally immobilize pathogens to prevent microbial spreading. The antimicrobial activity of NETs is exerted through direct contact of histones, antimicrobial peptides, neutrophil elastase, myeloperoxidase (MPO) and cathepsin G4 with the entrapped pathogens. The negatively charged DNA that traps and immobilizes pathogens manifest as sticky fibers (30). Additionally, the molecular mechanism of NET formation upon B. pseudomallei infection was indicated that NADPH oxidase was crucial but not phosphatidylinositol-3 kinase, mitogen-activated protein kinases, or Src family kinase signaling pathways (29).
In this study, we have illustrated the appearances of different types of NETs induced in vitro by several B. pseudomallei strains: biofilm wild type (H777), biofilm-defective mutant (M10) and biofilm-complemented (C17). Aggregated NETs were mainly found after exposure of neutrophils to B. pseudomallei biofilm wild type, less commonly for the biofilm-defective mutant but this type of NET was restored when neutrophils were exposed to the complemented strain. This is in complete agreement with the immunofluorescence images of LPS-stimulated NETs resulting in clumps of multiple neutrophils, both alive and dead, and bacteria (30). Large NETs of this type contain excess enzymes, including nuclease, which is vital for bacterial clearance. Additionally, aggregated NETs also contain both inflammatory mediators and pro-inflammatory cytokines and chemokines which support resolution of neutrophil-driven inflammation (30). Furthermore, the aggregated NETs are responsible to limit neutrophil inflammation by degrading cytokines and chemokines and disrupting neutrophil recruitment and activation in gout was demonstrated (36). Therefore, the higher proportion of aggregated NETs induced by B. pseudomallei biofilm phenotypes compared to that of the biofilm mutant, might be associated with a pathogen-survival stragegy. The bright-field images in our study have shown that B. pseudomallei H777 cells were entrapped within the boundaries of aggregated NETs in which were present myeloperoxidase and neutrophil elastase. Despite this, the NETs were unable to kill the entrapped cells. In a similar way, non-typable H. influenzae cells found within biofilm-associated chronic otitis media were not killed by NETs (37, 38).
Single “cloudy” NETs were commonly induced by PMA and LPS (30, 39). The neutrophils exhibited round nuclei and ruptured membranes with chromosomal DNA extruded giving a cloudy appearance. Moreover, NETosis-driven cloudy NETs may result in extracellular release of danger-associated molecular patterns (DAMPs) such as the necrotic cell-death pathway, leading to overwhelming inflammation and pathological immune responses (39). Cloudy NETs were most strongly induced by biofilm-producing H777, with the bpsl0618-complemented C17 strain inducing levels intermediate between H777 and the biofilm-defective strain, M10. This may imply a correlation of the biofilm phenotype with pathological immune response.
The fiber-like structure of “spiky” NETs exocytosed from intact neutrophils implied that the neutrophils remain viable (30). Spiky NETs were most frequently triggered by the M10 strain, which lacks bpsl0618. Survival of neutrophils was noted in the severe Gram-positive infection reported by Yipp and colleagues (40). They showed that the NETing neutrophils turned to anuclear cytoplasts and continued to phagocytose and prevent bacterial dissemination.
Our study has some limitations. We conducted experiments only in vitro and using only neutrophils. However, it may provide information concerning the direct consequence of B. pseudomallei biofilm on NET formation. The study of cytokines during NET formation upon encountered with B. pseudomallei would strengthen our study to disclose the impact of the bacterial biofilm on NET formation and melioidosis pathogenesis. In addition, planktonic cells of B. pseudomallei biofilm phenotype, mutant and complemented strains were used in this study to ensure comparable bacterial numbers. The role of biofilm-associated phenotypes were verified for their characters and correlated in cellular pathogenesis (14). The similar bactericidal efficacy of NETs on Streptococcus suis biofilms and planktonic cells were illustrated (27). Furthermore, the introduction of the empty vector introduced into the B. pseudomallei M10 may discard the unknown effect of the vector on the NET morphotypes.
Taken together, we have first illustrated various morphotypes of NETs (aggregated, cloudy and spiky) triggered by exposure to B. pseudomallei strains with or without the bpsl0618 gene, a sugar-transferase gene that is associated with the ability to form biofilm. The presence of this gene may transform planktonic B. pseudomallei cell surfaces, thus boosting production of NETs as double-edged swords that can shield B. pseudomallei cells rather than eradicate them. This would facilitate pathogen persistence leading to chronic infection or relapse of melioidosis. Our findings provide novel information on the encounters between biofilm-associated B. pseudomallei and NETs that extend knowledge and may inform management of chronic or relapsing melioidosis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
This study was approved by Khon Kaen University Ethics Committee for Human Research (Reference No. HE631099). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
MK, SP, KS, and SC contributed to conception and design of the study. MK performed the experiments, the statistical analysis and wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was financially supported by Faculty of Medicine, Khon Kaen University research grant (IN64129).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med (2012) 367:1035–44. doi: 10.1056/NEJMra1204699
2. Wiersinga WJ, Virk HS, Torres AG, Currie BJ, Peacock SJ, Dance DAB, et al. Melioidosis. Nat Rev Dis Primers (2018) 4:17107. doi: 10.1038/nrdp.2017.107
3. Hinjoy S, Hantrakun V, Kongyu S, Kaewrakmuk J, Wangrangsimakul T, Jitsuronk S, et al. Melioidosis in Thailand: Present and future. Trop Med Infect Dis (2018) 3:38. doi: 10.3390/tropicalmed3020038
5. Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev (2005) 18:383–416. doi: 10.1128/CMR.18.2.383-416.2005
6. Limmathurotsakul D, Kanoksil M, Wuthiekanun V, Kitphati R, Destavola B, Day NP, et al. Activities of daily living associated with acquisition of melioidosis in northeast Thailand: a matched case-control study. PLoS Negl Trop Dis (2013) 7:e2072. doi: 10.1371/journal.pntd.0002072
7. Chen PS, Chen YS, Lin HH, Liu PJ, Ni WF, Hsueh PT, et al. Airborne transmission of melioidosis to humans from environmental aerosols contaminated with B. pseudomallei. PLoS Negl Trop Dis (2015) 9:e0003834. doi: 10.1371/journal.pntd.0003834
8. Adler NR, Govan B, Cullinane M, Harper M, Adler B, Boyce JD. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol Rev (2009) 33:1079–99. doi: 10.1111/j.1574-6976.2009.00189.x
9. Chaowagul W, Suputtamongkol Y, Dance DA, Rajchanuvong A, Pattara-Arechachai J, White NJ. Relapse in melioidosis: incidence and risk factors. J Infect Dis (1993) 168:1181–5. doi: 10.1093/infdis/168.5.1181
10. Limmathurotsakul D, Paeyao A, Wongratanacheewin S, Saiprom N, Takpho N, Thaipadungpanit J, et al. Role of Burkholderia pseudomallei biofilm formation and lipopolysaccharide in relapse of melioidosis. Clin Microbiol Infect (2014) 20:O854–856. doi: 10.1111/1469-0691.12614
11. Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, et al. Bacterial biofilm and associated infections. J Chin Med Assoc (2018) 81:7–11. doi: 10.1016/j.jcma.2017.07.012
12. Vorachit M, Lam K, Jayanetra P, Costerton JW. Electron microscopy study of the mode of growth of Pseudomonas pseudomallei in vitro and in vivo. J Trop Med Hyg (1995) 98:379–91.
13. Pakkulnan R, Anutrakunchai C, Kanthawong S, Taweechaisupapong S, Chareonsudjai P, Chareonsudjai S. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One (2019) 14:e0213288. doi: 10.1371/journal.pone.0213288
14. Kunyanee C, Kamjumphol W, Taweechaisupapong S, Kanthawong S, Wongwajana S, Wongratanacheewin S, et al. Burkholderia pseudomallei biofilm promotes adhesion, internalization and stimulates proinflammatory cytokines in human epithelial A549 cells. PLoS One (2016) 11:e0160741. doi: 10.1371/journal.pone.0160741
15. Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, Wongratanacheewin S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One (2010) 5:e9196. doi: 10.1371/journal.pone.0009196
16. Pibalpakdee P, Wongratanacheewin S, Taweechaisupapong S, Niumsup PR. Diffusion and activity of antibiotics against Burkholderia pseudomallei biofilms. Int J Antimicrob Agents (2012) 39:356–9. doi: 10.1016/j.ijantimicag.2011.12.010
17. Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. How biofilms evade host defenses. Microbiol Spectr (2015) 3.3.22. doi: 10.1128/microbiolspec.MB-0012-2014
18. Arciola CR, Campoccia D, Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol (2018) 16:397–409. doi: 10.1038/s41579-018-0019-y
19. Gonzalez JF, Hahn MM, Gunn JS. Chronic biofilm-based infections: skewing of the immune response. Pathog Dis (2018) 76 fty023. doi: 10.1093/femspd/fty023
20. Taweechaisupapong S, Kaewpa C, Arunyanart C, Kanla P, Homchampa P, Sirisinha S, et al. Virulence of Burkholderia pseudomallei does not correlate with biofilm formation. Microb Pathog (2005) 39:77–85. doi: 10.1016/j.micpath.2005.06.001
21. Chin CY, Hara Y, Ghazali AK, Yap SJ, Kong C, Wong YC, et al. Global transcriptional analysis of Burkholderia pseudomallei high and low biofilm producers reveals insights into biofilm production and virulence. BMC Genomics (2015) 16:471. doi: 10.1186/s12864-015-1692-0
22. Chanchamroen S, Kewcharoenwong C, Susaengrat W, Ato M, Lertmemongkolchai G. Human polymorphonuclear neutrophil responses to Burkholderia pseudomallei in healthy and diabetic subjects. Infect Immun (2009) 77:456–63. doi: 10.1128/IAI.00503-08
23. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science (2004) 303:1532–5. doi: 10.1126/science.1092385
24. Brinkmann V, Laube B, Abed UA, Goosmann C, Zychlinsky A. Neutrophil extracellular traps: how to generate and visualize them. J Vis Exp (2010) (36):e1724. doi: 10.3791/1724
25. Storisteanu DM, Pocock JM, Cowburn AS, Juss JK, Nadesalingam A, Nizet V, et al. Evasion of neutrophil extracellular traps by respiratory pathogens. Am J Respir Cell Mol (2017) 56:423–31. doi: 10.1165/rcmb.2016-0193PS
26. Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol (2014) 15:1017–25. doi: 10.1038/ni.2987
27. Ma F, Yi L, Yu N, Wang G, Ma Z, Lin H, et al. Streptococcus suis serotype 2 biofilms inhibit the formation of neutrophil extracellular traps. Front Cell Infect Microbiol (2017) 7:86. doi: 10.3389/fcimb.2017.00086
28. De Jong HK, Koh GC, Achouiti A, van der Meer AJ, Bulder I, Stephan F, et al. Neutrophil extracellular traps in the host defense against sepsis induced by Burkholderia pseudomallei (melioidosis). Intensive Care Med Exp (2014) 2:21. doi: 10.1186/s40635-014-0021-2
29. Riyapa D, Buddhisa S, Korbsrisate S, Cuccui J, Wren BW, Stevens MP, et al. Neutrophil extracellular traps exhibit antibacterial activity against Burkholderia pseudomallei and are influenced by bacterial and host factors. Infect Immun (2012) 80:3921–9. doi: 10.1128/IAI.00806-12
30. Daniel C, Leppkes M, Muñoz LE, Schley G, Schett G, Herrmann M. Extracellular DNA traps in inflammation, injury and healing. Nat Rev Nephrol (2019) 15:559–75. doi: 10.1038/s41581-019-0163-2
31. Chokesuwattanaskul S, Phelan MM, Edwards SW, Wright HL. A robust intracellular metabolite extraction protocol for human neutrophil metabolic profiling. PLoS One (2018) 13:e0209270. doi: 10.1371/journal.pone.0209270
32. De Buhr N, Von Köckritz-Blickwede M. How neutrophil extracellular traps become visible. J Immunol Res (2016)2016 4604713. doi: 10.1155/2016/4604713
33. White PC, Chicca IJ, Ling MR, Wright HJ, Cooper PR, Milward MR, et al. Characterization, quantification, and visualization of neutrophil extracellular traps. Methods Mol Biol (2017) 1537:481–97. doi: 10.1007/978-1-4939-6685-1_29
34. Arazna M, Pruchniak MP, Demkow U. Neutrophil extracellular traps in bacterial infections: strategies for escaping from killing. Respir Physiol Neurobiol (2013) 187:74–7. doi: 10.1016/j.resp.2013.03.002
35. Hong W, Juneau RA, Pang B, Swords WE. Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J Innate Immun (2009) 1:215–24. doi: 10.1159/000205937
36. Schauer C, Janko C, Munoz LE, Zhao Y, Kienhofer D, Frey B, et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med (2014) 20:511–7. doi: 10.1038/nm.3547
37. Jones EA, Mcgillivary G, Bakaletz LO. Extracellular DNA within a nontypeable Haemophilus influenzae-induced biofilm binds human beta defensin-3 and reduces its antimicrobial activity. J Innate Immun (2013) 5:24–38. doi: 10.1159/000339961
38. Thornton RB, Wiertsema SP, Kirkham L-AS, Rigby PJ, Vijayasekaran S, Coates HL, et al. Neutrophil extracellular traps and bacterial biofilms in middle ear effusion of children with recurrent acute otitis media–a potential treatment target. PLoS One (2013) 8:e53837. doi: 10.1371/journal.pone.0053837
39. Yousefi S, Simon H-U. NETosis–does it really represent nature’s “suicide bomber”? Front Immunol (2016) 7:328. doi: 10.3389/fimmu.2016.00328
Keywords: neutrophil extracellular traps, aggregated NETs, spiky NETs, cloudy NETs, Burkholderia pseudomallei, biofilm, neutrophil, confined but survive
Citation: Khamwong M, Phanthanawiboon S, Salao K and Chareonsudjai S (2022) Burkholderia pseudomallei biofilm phenotypes confined but surviving in neutrophil extracellular traps of varying appearance. Front. Immunol. 13:926788. doi: 10.3389/fimmu.2022.926788
Received: 23 April 2022; Accepted: 26 July 2022;
Published: 18 August 2022.
Edited by:
Wenjie Fang, Shanghai Changzheng Hospital, ChinaReviewed by:
Ashwinkumar Subramenium Ganapathy, College of Medicine, The Pennsylvania State University, United StatesCopyright © 2022 Khamwong, Phanthanawiboon, Salao and Chareonsudjai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sorujsiri Chareonsudjai, c29ydWpzckBra3UuYWMudGg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.