
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 26 May 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.923647
This article is part of the Research TopicCombinational Immunotherapy of Cancer: Novel Targets, Mechanisms, and StrategiesView all 85 articles
Immunotherapy has become the breakthrough strategies for treatment of cancer in recent years. The application of messenger RNA in cancer immunotherapy is gaining tremendous popularity as mRNA can function as an effective vector for the delivery of therapeutic antibodies on immune targets. The high efficacy, decreased toxicity, rapid manufacturing and safe administration of mRNA vaccines have great advantages over conventional vaccines. The unprecedent success of mRNA vaccines against infection has proved its effectiveness. However, the instability and inefficient delivery of mRNA has cast a shadow on the wide application of this approach. In the past decades, modifications on mRNA structure and delivery methods have been made to solve these questions. This review summarizes recent advancements of mRNA vaccines in cancer immunotherapy and the existing challenges for its clinical application, providing insights on the future optimization of mRNA vaccines for the successful treatment of cancer.
Cancer is one of the most lethal diseases in the world. In the recent decades, achievements in the understanding of the immune system have shed light on the treatment of cancer by immunotherapies (1). Various immune cells are able to identify antigens on the surface of cancer cells and interact with the antigenetic peptides to destroy cancer cells. Therefore, immunotherapies including immune cell-based cancer vaccines were considered as promising approaches to cure cancer. Cancer vaccines have drawn great attention since the clinical application of several novel cancer vaccines such as immune cell-based vaccines, viral vector-based vaccines and RNA or DNA-based vaccines (2, 3). Among these, mRNA-based cancer vaccines demonstrate exceptional advantages in comparison with the other types of vaccines (4). First, unlike virus-based vaccines which can be infectious in some circumstances, mRNA-based vaccines are safer and free of contamination. Second, once delivered into the cells, the genetic information of antigen carried by mRNA can be translated into protein very rapidly. Third, mRNA-based cancer vaccines can stimulate immune response and overcome vaccine resistance that is often observed in conventional chemotherapies. In addition, mRNA-based cancer vaccines encode the cancer antigens in full-length and thus can overcome the restrictions of human leukocyte antigen to activate a broader immune response. Last but not least, mRNA-based cancer vaccines are free of mutations since mRNA can’t insert into the chromosome. Given the advantages of mRNA vaccines over conventional cancer vaccines, this novel immunotherapy has become the hotspot of research for the development of new generation cancer therapies. However, the expected substantial application of mRNA-based cancer vaccines was not seen due to several problems regarding the instability of mRNA, immunogenicity and the inefficiency of in vivo delivery.
Research on mRNA-based vaccines have focused on overcoming the instability and the delivery of mRNA. One strategy is to use synthetic mRNA or modified mRNA analogs, which can enhance the stability and protein expression of mRNA. For example, circular RNA is used to avoid the detection of pathogen-associated pattern receptors; self-amplifying RNA greatly increased the expression level of proteins (5). Another strategy is to modify the in vivo delivery methods of RNA vaccines to achieve higher efficiency. For instance, in addition to electroporation, novel vectors such as polyplexes, lipid nanoparticles, peptides and cationic nanoemulsions have been used for the delivery of RNA vaccines (6). The success of RNA-based vaccines for the COVID19 pandemic has witnessed the significant progress made on the clinical application of mRNA vaccines and the increasing necessity of developing novel RNA vaccines for treatment of diseases (7).
In this review, we will focus on the most recent progress that have been made on the stability and in vivo delivery of mRNA-based vaccines for the treatment of cancer and discuss the existing challenges on the current clinical application of mRNA-based immunotherapies, hoping to accelerate the clinical application of mRNA-based cancer immunotherapy.
The first introduction of mRNA to activate immune response in vivo could be dated to the year 1993, when Martinon and co-workers constructed a liposome-mRNA expressing influenza hemagglutinin that activated CD8+ T cell responses for the detection and lysing of virus-infected cells (8). In 1995, mRNA construct expressing cancer embryonic antigen was reported to induce the generation of antibodies in mice, demonstrating the potential of mRNA vaccines in cancer therapies (9, 10). Over the past decades, mRNA vaccines quickly became the spotlight of cancer immunotherapy due to their abilities to provide safe vaccination, to improve antigen expression and to avoid gene integration. The earliest mRNA vaccines in cancer immunotherapy normally use RNA virus genomes, which have shown good efficacy against viral cancer in mouse models (11). Later, a liposome-mRNA vaccine was developed and its ability to induce cognate cytotoxic T cells has resulted in the destroy of melanoma cancer cells (12). These mRNA-based vaccines have demonstrated great advantage over conventional vaccines and thus have been established as novel strategies for cancer therapies. Since then, more and more kinds of mRNA-based vaccines are developed for treatment of cancer.
To date, replicating modified mRNA, unmodified mRNA, and virus-derived mRNA are the three main types of mRNA vaccines in cancer immunotherapy (1). The basic structure of non-replicating mRNA consists of an open reading frame flanked by a 5′-prime region and a 3′-prime untranslated region (UTR), a 5′ cap structure and a 3′ poly (A) tails (13). The ORF of mRNA encodes the sequences of the target antigen. The target antigen will be translated into proteins once mRNA is transited to the cytosol of the cell. The expressed antigen will further be post-translationally modified and fold into a full-functional protein whereas the remaining mRNA will be quickly degraded to reduce potential toxicity. However, the rapid degradation of naked mRNA also dramatically affects its stability. In addition, the intrinsic immunogenicity of mRNA further promotes the rapid degradation of mRNA, resulting in decreased expression levels of antigen. Moreover, the inefficient in vivo delivery of mRNA-based vaccines will reduce the protein expression of mRNA. These disadvantages have greatly limited the clinical application of mRNA vaccines in cancer immunotherapy. We will discuss the limitations and highlight the current state-of-the-art strategies to overcome these limitations in the next section.
Previous studies have found that the inadequate methylation of mRNA, impurity and immunogenicity could lead to low efficiency of mRNA translation (14, 15). A better understanding of the structure of mRNA has provided new strategies to improve the stability and the translation efficiency of mRNA.
mRNA consists of an ORF, a 5′-prime UTR, a 3′-prime UTR, a 5′ cap structure and a 3′ poly (A) tails (Figure 1). Firstly, modification of the 5’ cap of mRNA has been utilized to improve stability. There are mainly two types of mRNA capping methods: the in vitro post-translational capping enzymatic method that utilizes the Vaccinia virus Capping Enzyme (VCE) and the chemical capping methods (16). These capping methods largely improve mRNA translation but are limited by reverse incorporation. This limitation could be overcome by the development of anti-reverse cap analogs (ARCA), which is methylated at the C3 position. ARCA significantly increases the expression of mRNA. However, the disadvantages of ARCA including de-capping of mRNA, exogeneous motif, and relatively low capping efficiency (60-80%) in contrast to the enzymatic capping method (100%) (17, 18). To increase the capping efficiency and prevent de-capping of ARCA, a novel co-transcriptional cap analog termed CleanCap has been developed, which can generate an unmodified cap structure with high efficiency (>90%) (19).
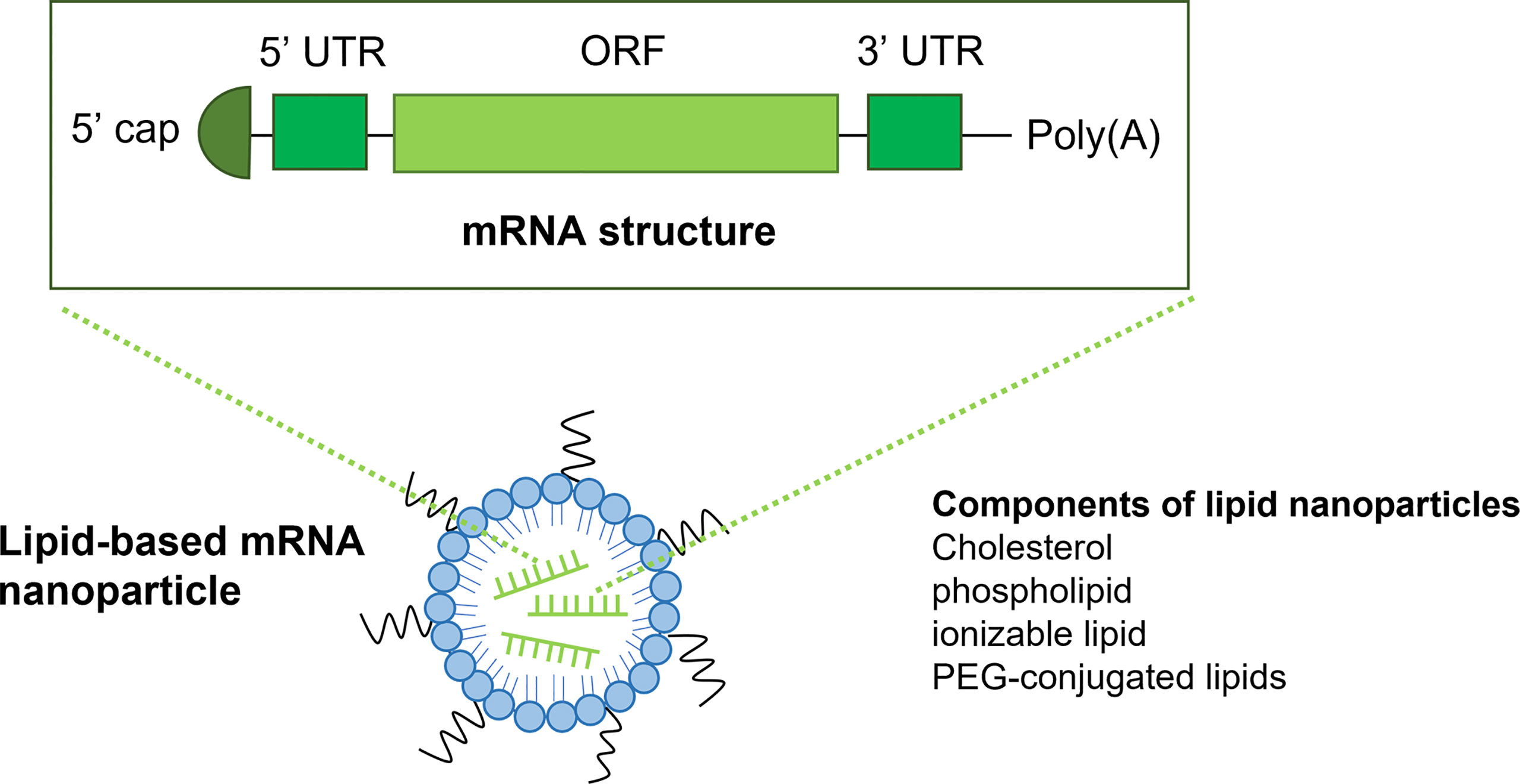
Figure 1 The structure of mRNA and lipid-based nanoparticles. The structure of mRNA (top) consists of a 5’ cap, a 5’UTR and a 3’ UTR, an open reading frame and a poly(A) tail. Lipid nanoparticles (LNPs) are the most commonly used vectors for the delivery of mRNA vaccines. LNPs often comprise of ionizable lipid, Cholesterol, phospholipid and PEG-conjugated lipids.
Secondly, post-translational modifications of mRNA can inhibit the immune recognition of mRNA. Studies have revealed that minimization of mRNA reorganization by the innate immune system could prevent sensing and destruction of mRNA translation, resulting in increased expression (15, 20). For example, removal of the uncapped phosphate of mRNA by phosphatases can effectively avoid immunogenicity. In addition, modifications of mRNA through substitution of natural uridine and cytidine with pseudouridine (Ψ) or 1-methylpseudouridine (m1Ψ) can also effectively avoid immunogenicity and improve stability (21, 22). Pseudouridine is an isomer of uridine in which the nitrogen-carbon glycosidic bond of uridine is replaced by a carbon-carbon bond. N1-Methylpseudouridine is the methylated derivative of pseudouridine and is usually used for mRNA vaccines. Substitution of uridine with pseudouridine (Ψ) or 1-methylpseudouridine (m1Ψ) in mRNA synthesis can reduce cytotoxicity of mRNA vaccines because Ψ/m1Ψ mRNAs trigger low-to-no immune responses to the cells and enhance the expression of tumor antigen (23, 24).
Thirdly, increased purity of RNA and removal of impurities can prevent the degradation of mRNA. During the process of transcription, double stranded RNA impurities are easily formed, which leads to the immunogenicity of mRNA cancer vaccines. Thess and co-workers have shown that the RNA impurities could be removed by optimization of the sequence and purification of RNA by high-pressure liquid chromatography (HPLC), which could result in reduced immunogenicity and improved stability of mRNA vaccines (25). However, HPLC is limited by low yield and high cost. A better way to achieve high purity of mRNA is to use synthetic mRNA (26).
Fourthly, optimization of the ORF of mRNA can improve mRNA translation. Increased GC content in the ORF can be applied to improve stability (27). Methods to increase GC content including codon optimization of ORF and depletion of uridine (28). For example, rare codons in ORF are often substituted by codons with higher tRNA abundance to achieve higher translation rate (29). However, high translation rate sometimes can be harmful, which is the major limitation of codon optimization (30). The reason underlying this observation is that the correctly folding of some proteins may require a low translation rate rather than a high translation rate. Despite the defects of codon optimization, this strategy is one of the most important methods for improving mRNA translation.
Finally, optimization of the untranslated 5’ UTR and 3’ UTR of mRNA can also significantly improve stability and protein expression (31). For example, start codon AUG or CUG at 5’ UTR can inhibit the translation of mRNA (22). Therefore, avoiding these can improve translation rate of mRNA. Increasing the 3’UTR sequence in tandem may also improve mRNA translation (32). In conclusion, modifications of mRNA structure have proven to be of great importance to improve the stability and protein expression of mRNA vaccines. Future studies will focus on overcoming the limitations of current strategies and developing novel mRNA vaccines with higher stability and translation rate.
Despite the many advantages of mRNA cancer vaccine described above, the difficulties of in vivo delivery of mRNA vaccine have greatly inhibited its clinical application. Therefore, it is of urgency to increase the delivery efficiency of mRNA vaccines. There are mainly three types of vectors used for in vivo delivery: viral, non-viral and cell-based vectors. Dendritic cells (DCs) are usually used as cell-based vectors for adoptive transfer of mRNA cancer vaccines. Designated ‘nature’s adjuvants’, dendritic cells (DCs) are a group of bone marrow-derived antigen-presenting cells that play essential roles in activating and mediating immune response (33, 34). DCs can transit antigen to T cells and facilitate the transduction of immunomodulatory signals via cytokines and cell interactions. The generation of mRNA-based cancer vaccines using DCs as vectors involves the use of mRNA extracted from autologous cancers (35). In this process, dendritic cells were isolated from patients, cultured ex vivo, induced to maturation via adjuvant, loaded with mRNA encoding target antigens and injected back to patients to activate efficient anti-cancer immunity (36). DC-based mRNA cancer vaccines have been examined by clinical trials (Table 1). The initial results from phase I/II have shown that these mRNA vaccines display dramatically increased survival rate and reduced toxicity (37). However, the differentiation, maturation and antigen loading of DCs impact T cell co-stimulation and lead to weak immune response (38, 39). In addition, low production and high variability of mRNA-based DC vaccines have limited the application of this novel immunotherapy. Future studies may focus on the direct injection of mRNA as an alternative approach to overcome the limitations of mRNA-based DC vectors.
Viral vectors have been extensively studied for the development of mRNA cancer vaccines, with promising preliminary results being reported (33, 40, 41). For instance, mRNA vaccines for Covid19 pandemic have used viral vectors. In fact, an adenovirus type-5 (Ad5) vector-based RNA vaccines had been on clinical trials in China as early as March 2020 (42). The most used viral vector is self-amplifying mRNA (replicons or saRNA), which encodes target antigen and self-replicates in the cytoplasm of the host cell for expression of target antigen. The viral structural protein is deleted from the sequences of the self-amplifying mRNA thus it is unable to generate infectious virus in the host cells. In the past decades, we have witnessed the utilization of self-amplifying mRNA derived from the genomes of RNA viruses including picornaviruses, alphaviruses and flaviviruses (43, 44). In comparison with non-viral vectors, viral-based mRNA vaccines demonstrate advantages such as modular design, rapid manufacturing and low dose requirement because of its self-replicative properties (45). Viral-based mRNA vaccines against colorectal tumor, urothelial carcinoma, gastroesophageal tumor and rabies have been applied in clinical trials (46). However, the infectious property of virus vectors and the difficulties for large-scale manufacturing of virus vectors have limited the applicability of this approach.
The most famous non-viral vector used for mRNA vaccine is the lipid nanoparticle-based mRNA delivery system, which has been used for the mRNA vaccine of Covid19 and has been very successful (47, 48). LNPs usually consists of an ionizable lipid-like molecule, polyethylene glycol (PEG)-conjugated lipid, cholesterol and a helper phospholipid. Lipid-based nanoparticles (LNPs) could significantly improve delivery efficiency of mRNA cancer vaccines and reduce the system toxicity. Since the ionized lipid is positively charged at low pH, it could enhance the encapsulation of the negatively charged mRNA by electrostatic interaction and promote membrane fusion and destabilization upon delivery. In addition, the neutrality of lipid at the physiological environment improves the stability of mRNA. The first-generation lipids used for LNPs including Dlin-DMA, DLin-KC2-DMA and cKK-E12, etc (49, 50). The second-generation lipids are the derivatives of DLin-KC2-DMA and cKK-E12 (51). In addition to lipid, the other components of LNPs such as cholesterol and phospholipid could facilitate membrane fusion and improve mRNA stability, whereas the diffusive PEG could inhibit particle aggregation, leading to increased stability of mRNA vaccines. Over the past decades, improvements have been made to LPNs. Current LPNs have demonstrated enhanced delivery specificity and improved capacity to be rapidly metabolized and cleared, leading to reduced toxicity of the system and increased stability of mRNA vaccines. Studies have shown that LNPs act as an effective vector for delivery of mRNA vaccines to liver cancers (52, 53). In addition, LPNs have also been reported to target tumours in other organs such as lung, spleen and bone marrow. For example, the Selective Organ Targeting approach developed by Cheng and co-workers using cationic or anionic lipid can target liver, lung and spleen (54, 55). Overall, LNPs have made great contributions to the clinical application of mRNA vaccines.
In addition to LNP vectors, peptide vectors and polymer vectors also play important roles in facilitating the delivery of mRNA cancer vaccines. Polyethylenimine (PEI) is one of the most commonly used polymer-based vectors for mRNA vaccine delivery (56, 57). The efficacy of PEI-based hemagglutinin antigen from influenza virus has been evaluated in mice models (58). In comparison with LNP-based delivery system, polymer vectors display lower purity and reduced clearance rate but higher toxicity. Peptide-based delivery system have been widely studied (59–61). For example, cationic cell-penetrating peptides (CPPs) can condense mRNA complexes and induce strong immune response upon injection (62). However, the precise mechanism of this delivery system remains to be understood.
The successful application of mRNA vaccines for Covid19 has demonstrated the great potential of mRNA vaccines as novel therapies for the treatment of lethal diseases. It’s reported that mRNA-based vaccines can induce immune response in cancer cells, leading to the destruction of cancer cells and the control of tumour growth. Conventional therapies of cancer treatment such as chemotherapy and radiotherapy often suffer from multi-drug resistance and strong toxicity, whereas mRNA-based cancer vaccines have shown enhanced efficacy and reduced toxicity. Therefore, mRNA-based vaccines have gained more and more popularity for the development of novel immunotherapies. However, the instability and in vivo delivery of mRNA cancer vaccine have impaired its clinical application. Although progress has been made over the past decades to overcome these limitations, challenges still exist on the development of mRNA cancer vaccines. To promote the wide application of mRNA cancer vaccines, more strategies should be taken to improve the stability and translation rate of mRNA vaccines.
In conclusion, mRNA vaccines have the potential to significantly affect the battle against cancer. Future studies should cast more investigations on the combination of mRNA cancer vaccines and conventional cancer therapies.
X-YZ, D-XY and CZ conceived the topic, revised and proofread the manuscript. L-JD and QW drafted the paper and prepared the figure and table. All authors approved the submitted version.
This study was supported by the Major Projects in Provincial and National Union Construction of Henan Medical Science Research Plan (SBGJ202001002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Huff AL, Jaffee EM, Zaidi N. Messenger RNA Vaccines for Cancer Immunotherapy: Progress Promotes Promise. J Clin Invest (2022) 132(6):e156211. doi: 10.1172/JCI156211
2. Van Lint S, Wilgenhof S, Heirman C, Corthals J, Breckpot K, Bonehill A, et al. Optimized Dendritic Cell-Based Immunotherapy for Melanoma: The TriMix-Formula. Cancer Immunol Immunother (2014) 63(9):959–67. doi: 10.1007/s00262-014-1558-3
3. Zeng C, Zhang C, Walker PG, Dong Y. Zeng C, Zhang C, Walker PG, Dong Y. “Formulation and Delivery Technologies for mRNA Vaccines,”. In: Current Topics in Microbiology and Immunology. Berlin: Springer Press (2020) 1–40. doi: 10.1007/82_2020_217
4. Pastor F, Berraondo P, Etxeberria I, Frederick J, Sahin U, Gilboa E, et al. An RNA Toolbox for Cancer Immunotherapy. Nat Rev Drug Discov (2018) 17(10):751–67. doi: 10.1038/nrd.2018.132
5. Flemming A. Vaccines: Self-Amplifying RNA in Lipid Nanoparticles: A Next-Generation Vaccine? Nat Rev Drug Discov (2012) 11(10):748–9. doi: 10.1038/nrd3854
6. Faghfuri E, Pourfarzi F, Faghfouri AH, Abdoli Shadbad M, Hajiasgharzadeh K, Baradaran B. Recent Developments of RNA-Based Vaccines in Cancer Immunotherapy. Expert Opin Biol Ther (2021) 21(2):201–18. doi: 10.1080/14712598.2020.1815704
7. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med (2021) 384(5):403–16. doi: 10.1056/NEJMoa2035389
8. Martinon F, Krishnan S, Lenzen G, Magne R, Gomard E, Guillet JG, et al. Induction of Virus-Specific Cytotoxic T Lymphocytes In Vivo by Liposome-Entrapped mRNA. Eur J Immunol (1993) 23(7):1719–22. doi: 10.1002/eji.1830230749
9. Conry RM, LoBuglio AF, Loechel F, Moore SE, Sumerel LA, Barlow DL, et al. A Carcinoembryonic Antigen Polynucleotide Vaccine has In Vivo Antitumor Activity. Gene Ther (1995) 2(1):59–65.
10. Conry RM, LoBuglio AF, Loechel F, Moore SE, Sumerel LA, Barlow DL, et al. A Carcinoembryonic Antigen Polynucleotide Vaccine for Human Clinical Use. Cancer Gene Ther (1995) 2(1):33–8.
11. Ying H, Zaks TZ, Wang RF, Irvine KR, Kammula US, Marincola FM, et al. Cancer Therapy Using a Self-Replicating RNA Vaccine. Nat Med (1999) 5(7):823–7. doi: 10.1038/10548
12. Zhou WZ, Hoon DS, Huang SK, Fujii S, Hashimoto K, Morishita R, et al. RNA Melanoma Vaccine: Induction of Antitumor Immunity by Human Glycoprotein 100 mRNA Immunization. Hum Gene Ther (1999) 10(16):2719–24. doi: 10.1089/10430349950016762
13. Emens LA, Adams S, Cimino-Mathews A, Disis ML, Gatti-Mays ME, Ho AY, et al. Society for Immunotherapy of Cancer (SITC) Clinical Practice Guideline on Immunotherapy for the Treatment of Breast Cancer. J Immunother Cancer (2021) 9(8):e002597. doi: 10.1136/jitc-2021-002597
14. Foster JB, Choudhari N, Perazzelli J, Storm J, Hofmann TJ, Jain P, et al. Purification of mRNA Encoding Chimeric Antigen Receptor Is Critical for Generation of a Robust T-Cell Response. Hum Gene Ther (2019) 30(2):168–78. doi: 10.1089/hum.2018.145
15. Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC, et al. Systemic RNA Delivery to Dendritic Cells Exploits Antiviral Defence for Cancer Immunotherapy. Nature (2016) 534(7607):396–401. doi: 10.1038/nature18300
16. Muttach F, Muthmann N, Rentmeister A. Synthetic mRNA Capping. Beilstein J Org Chem (2017) 13:2819–32. doi: 10.3762/bjoc.13.274
17. Rydzik AM, Kulis M, Lukaszewicz M, Kowalska J, Zuberek J, Darzynkiewicz ZM, et al. Synthesis and Properties of mRNA Cap Analogs Containing Imidodiphosphate Moiety–Fairly Mimicking Natural Cap Structure, Yet Resistant to Enzymatic Hydrolysis. Bioorg Med Chem (2012) 20(5):1699–710. doi: 10.1016/j.bmc.2012.01.013
18. Shuman S. Catalytic Activity of Vaccinia mRNA Capping Enzyme Subunits Coexpressed in Escherichia Coli. J Biol Chem (1990) 265(20):11960–6. doi: 10.1016/S0021-9258(19)38494-7
19. Vaidyanathan S, Azizian KT, Haque A, Henderson JM, Hendel A, Shore S, et al. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity Without HPLC Purification. Mol Ther Nucleic Acids (2018) 12:530–42. doi: 10.1016/j.omtn.2018.06.010
20. Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA Recognition by Toll-Like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity (2005) 23(2):165–75. doi: 10.1016/j.immuni.2005.06.008
21. Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio (2016) 7(5):e00833–16. doi: 10.1128/mBio.00833-16
22. Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada T. N(1)-Methylpseudouridine-Incorporated mRNA Outperforms Pseudouridine-Incorporated mRNA by Providing Enhanced Protein Expression and Reduced Immunogenicity in Mammalian Cell Lines and Mice. J Control Release (2015) 217:337–44. doi: 10.1016/j.jconrel.2015.08.051
23. Svitkin YV, Cheng YM, Chakraborty T, Presnyak V, John M, Sonenberg N. N1-Methyl-Pseudouridine in mRNA Enhances Translation Through Eif2alpha-Dependent and Independent Mechanisms by Increasing Ribosome Density. Nucleic Acids Res (2017) 45(10):6023–36. doi: 10.1093/nar/gkx135
24. Parr CJC, Wada S, Kotake K, Kameda S, Matsuura S, Sakashita S, et al. N 1-Methylpseudouridine Substitution Enhances the Performance of Synthetic mRNA Switches in Cells. Nucleic Acids Res (2020) 48(6):e35. doi: 10.1093/nar/gkaa070
25. Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, et al. Sequence-Engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol Ther (2015) 23(9):1456–64. doi: 10.1038/mt.2015.103
26. Nelson J, Sorensen EW, Mintri S, Rabideau AE, Zheng W, Besin G, et al. Impact of mRNA Chemistry and Manufacturing Process on Innate Immune Activation. Sci Adv (2020) 6(26):eaaz6893. doi: 10.1126/sciadv.aaz6893
27. Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, et al. A Role for Codon Order in Translation Dynamics. Cell (2010) 141(2):355–67. doi: 10.1016/j.cell.2010.02.036
28. Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, et al. mRNA Structure Regulates Protein Expression Through Changes in Functional Half-Life. Proc Natl Acad Sci U S A (2019) 116(48):24075–83. doi: 10.1073/pnas.1908052116
29. Linares-Fernandez S, Lacroix C, Exposito JY, Verrier B. Tailoring mRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol Med (2020) 26(3):311–23. doi: 10.1016/j.molmed.2019.10.002
30. Schlake T, Thess A, Fotin-Mleczek M, Kallen KJ. Developing mRNA-Vaccine Technologies. RNA Biol (2012) 9(11):1319–30. doi: 10.4161/rna.22269
31. Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, et al. Modification of Antigen-Encoding RNA Increases Stability, Translational Efficacy, and T-Cell Stimulatory Capacity of Dendritic Cells. Blood (2006) 108(13):4009–17. doi: 10.1182/blood-2006-04-015024
32. Orlandini von Niessen AG, Poleganov MA, Rechner C, Plaschke A, Kranz LM, Fesser S, et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3' UTRs Identified by Cellular Library Screening. Mol Ther (2019) 27(4):824–36. doi: 10.1016/j.ymthe.2018.12.011
33. Palucka K, Banchereau J. Cancer Immunotherapy via Dendritic Cells. Nat Rev Cancer (2012) 12(4):265–77. doi: 10.1038/nrc3258
34. Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic Cells in Cancer Immunology and Immunotherapy. Nat Rev Immunol (2020) 20(1):7–24. doi: 10.1038/s41577-019-0210-z
35. Perez CR, De Palma M. Engineering Dendritic Cell Vaccines to Improve Cancer Immunotherapy. Nat Commun (2019) 10(1):5408. doi: 10.1038/s41467-019-13368-y
36. Sabado RL, Balan S, Bhardwaj N. Dendritic Cell-Based Immunotherapy. Cell Res (2017) 27(1):74–95. doi: 10.1038/cr.2016.157
37. Beck JD, Reidenbach D, Salomon N, Sahin U, Tureci O, Vormehr M, et al. mRNA Therapeutics in Cancer Immunotherapy. Mol Cancer (2021) 20(1):69. doi: 10.1186/s12943-021-01348-0
38. Kongsted P, Borch TH, Ellebaek E, Iversen TZ, Andersen R, Met O, et al. Dendritic Cell Vaccination in Combination With Docetaxel for Patients With Metastatic Castration-Resistant Prostate Cancer: A Randomized Phase II Study. Cytotherapy (2017) 19(4):500–13. doi: 10.1016/j.jcyt.2017.01.007
39. Gu YZ, Zhao X, Song XR. Ex Vivo Pulsed Dendritic Cell Vaccination Against Cancer. Acta Pharmacol Sin (2020) 41(7):959–69. doi: 10.1038/s41401-020-0415-5
40. Fiedler K, Lazzaro S, Lutz J, Rauch S, Heidenreich R. mRNA Cancer Vaccines. Recent Results Cancer Res (2016) 209:61–85. doi: 10.1007/978-3-319-42934-2_5
41. Diken M, Kranz LM, Kreiter S, Sahin U. mRNA: A Versatile Molecule for Cancer Vaccines. Curr Issues Mol Biol (2017) 22:113–28. doi: 10.21775/cimb.022.113
42. Cohen J. Vaccine Designers Take First Shots at COVID-19. Science (2020) 368(6486):14–6. doi: 10.1126/science.368.6486.14
43. Tews BA, Meyers G. Self-Replicating RNA. Methods Mol Biol (2017) 1499:15–35. doi: 10.1007/978-1-4939-6481-9_2
44. Bloom K, van den Berg F, Arbuthnot P. Self-Amplifying RNA Vaccines for Infectious Diseases. Gene Ther (2021) 28(3-4):117–29. doi: 10.1038/s41434-020-00204-y
45. Li Y, Teague B, Zhang Y, Su Z, Porter E, Dobosh B, et al. In Vitro Evolution of Enhanced RNA Replicons for Immunotherapy. Sci Rep (2019) 9(1):6932. doi: 10.1038/s41598-019-43422-0
46. Gebre MS, Brito LA, Tostanoski LH, Edwards DK, Carfi A, Barouch DH. Novel Approaches for Vaccine Development. Cell (2021) 184(6):1589–603. doi: 10.1016/j.cell.2021.02.030
47. Thomas SJ, Perez JL, Lockhart SP, Hariharan S, Kitchin N, Bailey R, et al. Efficacy and Safety of the BNT162b2 mRNA COVID-19 Vaccine in Participants With a History of Cancer: Subgroup Analysis of a Global Phase 3 Randomized Clinical Trial. Vaccine (2022) 40(10):1483–92. doi: 10.1016/j.vaccine.2021.12.046
48. Buschmann MD, Carrasco MJ, Alishetty S, Paige M, Alameh MG, Weissman D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines (Basel) (2021) 9(1):65. doi: 10.3390/vaccines9010065
49. Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew Chem Int Ed Engl (2012) 51(34):8529–33. doi: 10.1002/anie.201203263
50. Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, et al. Lipid-Like Materials for Low-Dose, In Vivo Gene Silencing. Proc Natl Acad Sci U S A (2010) 107(5):1864–9. doi: 10.1073/pnas.0910603106
51. Maier MA, Jayaraman M, Matsuda S, Liu J, Barros S, Querbes W, et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol Ther (2013) 21(8):1570–8. doi: 10.1038/mt.2013.124
52. Lai I, Swaminathan S, Baylot V, Mosley A, Dhanasekaran R, Gabay M, et al. Lipid Nanoparticles That Deliver IL-12 Messenger RNA Suppress Tumorigenesis in MYC Oncogene-Driven Hepatocellular Carcinoma. J Immunother Cancer (2018) 6(1):125. doi: 10.1186/s40425-018-0431-x
53. Stadler CR, Bahr-Mahmud H, Celik L, Hebich B, Roth AS, Roth RP, et al. Elimination of Large Tumors in Mice by mRNA-Encoded Bispecific Antibodies. Nat Med (2017) 23(7):815–7. doi: 10.1038/nm.4356
54. Parhiz H, Shuvaev VV, Pardi N, Khoshnejad M, Kiseleva RY, Brenner JS, et al. PECAM-1 Directed Re-Targeting of Exogenous mRNA Providing Two Orders of Magnitude Enhancement of Vascular Delivery and Expression in Lungs Independent of Apolipoprotein E-Mediated Uptake. J Control Release (2018) 291:106–15. doi: 10.1016/j.jconrel.2018.10.015
55. Basha G, Novobrantseva TI, Rosin N, Tam YY, Hafez IM, Wong MK, et al. Influence of Cationic Lipid Composition on Gene Silencing Properties of Lipid Nanoparticle Formulations of siRNA in Antigen-Presenting Cells. Mol Ther (2011) 19(12):2186–200. doi: 10.1038/mt.2011.190
56. Khan OF, Kowalski PS, Doloff JC, Tsosie JK, Bakthavatchalu V, Winn CB, et al. Endothelial siRNA Delivery in Nonhuman Primates Using Ionizable Low-Molecular Weight Polymeric Nanoparticles. Sci Adv (2018) 4(6):eaar8409. doi: 10.1126/sciadv.aar8409
57. Romano E, Pascolo S, Ott P. Implications of mRNA-Based SARS-CoV-2 Vaccination for Cancer Patients. J Immunother Cancer (2021) 9(6):e002932. doi: 10.1136/jitc-2021-002932
58. Vogel AB, Lambert L, Kinnear E, Busse D, Erbar S, Reuter KC, et al. Self-Amplifying RNA Vaccines Give Equivalent Protection Against Influenza to mRNA Vaccines But at Much Lower Doses. Mol Ther (2018) 26(2):446–55. doi: 10.1016/j.ymthe.2017.11.017
59. Sebastian M, Schroder A, Scheel B, Hong HS, Muth A, von Boehmer L, et al. A Phase I/IIa Study of the mRNA-Based Cancer Immunotherapy CV9201 in Patients With Stage IIIB/IV non-Small Cell Lung Cancer. Cancer Immunol Immunother (2019) 68(5):799–812. doi: 10.1007/s00262-019-02315-x
60. Kubler H, Scheel B, Gnad-Vogt U, Miller K, Schultze-Seemann W, Vom Dorp F, et al. Self-Adjuvanted mRNA Vaccination in Advanced Prostate Cancer Patients: A First-in-Man Phase I/IIa Study. J Immunother Cancer (2015) 3:26. doi: 10.1186/s40425-015-0068-y
61. De Keersmaecker B, Claerhout S, Carrasco J, Bar I, Corthals J, Wilgenhof S, et al. TriMix and Tumor Antigen mRNA Electroporated Dendritic Cell Vaccination Plus Ipilimumab: Link Between T-Cell Activation and Clinical Responses in Advanced Melanoma. J Immunother Cancer (2020) 8(1):e000329. doi: 10.1136/jitc-2019-000329
62. Udhayakumar VK, De Beuckelaer A, McCaffrey J, McCrudden CM, Kirschman JL, Vanover D, et al. Arginine-Rich Peptide-Based mRNA Nanocomplexes Efficiently Instigate Cytotoxic T Cell Immunity Dependent on the Amphipathic Organization of the Peptide. Adv Healthc Mater (2017) 6(13):1601412. doi: 10.1002/adhm.201601412
Keywords: mRNA, cancer vaccine, immunotherapy, efficient delivery, optimization, strategies
Citation: Duan L-J, Wang Q, Zhang C, Yang D-X and Zhang X-Y (2022) Potentialities and Challenges of mRNA Vaccine in Cancer Immunotherapy. Front. Immunol. 13:923647. doi: 10.3389/fimmu.2022.923647
Received: 19 April 2022; Accepted: 02 May 2022;
Published: 26 May 2022.
Edited by:
Xian Zeng, Fudan University, ChinaReviewed by:
Yan Cao, Sun Yat-sen University Cancer Center (SYSUCC), ChinaCopyright © 2022 Duan, Wang, Zhang, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuilian Zhang, bHVja3l6Y2xAcXEuY29t; Dong-Xiao Yang, ZG9uZ3hpYW95YW5nMkBnbWFpbC5jb20=; Xu-Yao Zhang, aGhzdHV5eHpAb3V0bG9vay5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.