- 1Department of Pathophysiology, College of Basic Medical Sciences, Dalian Medical University, Dalian, China
- 2Department of Pharmacology, College of Pharmacy, Dalian Medical University, Dalian, China
Background: We aimed to characterize serine protease inhibitor Kazal type 1 (SPINK1) as a gene signature for the early diagnosis, molecular targeting, and prediction of immune checkpoint blockade (ICB) treatment response of hepatocellular carcinoma (HCC).
Methods: The transcriptomics, proteomics, and phenotypic analyses were performed separately or in combination.
Results: We obtained the following findings on SPINK1. Firstly, in the transcriptomic training dataset, which included 279 stage I and II tumor samples (out of 1,884 stage I–IV HCC specimens) and 259 normal samples, significantly higher area under curve (AUC) values and increased integrated discrimination improvement (IDI) and net reclassification improvement (NRI) were demonstrated for HCC discrimination in SPINK1-associated models compared with those of alpha-fetoprotein (AFP). The calibration of both SPINK1-related curves fitted significantly better than that of AFP. In the two independent transcriptomic validation datasets, which included 201, 103 stage I-II tumor and 192, 169 paired non-tumor specimens, respectively, the obtained results were consistent with the above-described findings. In the proteomic training dataset, which included 98 stage I and II tumor and 165 normal tissue samples, the analyses also revealed better AUCs and increased IDI and NRI in the aforementioned SPINK1-associated settings. A moderate calibration was shown for both SPINK1-associated models relative to the poor results of AFP. Secondly, in the in vitro and/or in vivo murine models, the wet-lab experiments demonstrated that SPINK1 promoted the proliferation, clonal formation, migration, chemoresistance, anti-apoptosis, tumorigenesis, and metastasis of HCC cells, while the anti-SPINK1 antibody inhibited the growth of the cells, suggesting that SPINK1 has “tumor marker” and “targetable” characteristics in the management of HCC. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that SPINK1 was engaged in immunity-related pathways, including T-cell activation. Thirdly, in the transcriptomic analyses of the 368 HCC specimens from The Cancer Genome Atlas (TCGA) cohort, the high abundance of SPINK1 was positively correlated with the high levels of activated tumor-infiltrating CD4+ and CD8+ T lymphocytes and dendritic and natural killer cells, while there were also positive correlations between SPINK1 and immune checkpoints, including PD-1, LAG-3, TIM-3, TIGIT, HAVCR2, and CTLA-4. The ESTIMATE algorithm calculated positive correlations between SPINK1 and the immune and ESTIMATE scores, suggesting a close correlation between SPINK1 and the immunogenic microenvironment within HCC tissues, which may possibly help in predicting the response of patients to ICB therapy.
Conclusions: SPINK1 could be a potential biomarker for the early detection, targeted therapy, and prediction of ICB treatment response in the management of HCC.
Introduction
With a dismal prognosis, hepatocellular carcinoma (HCC) has 5-year recurrence rates approaching 70% post-resection, and overall survival (OS) is about 3–4 months in patients with untreated HCC (1). Due to the complexity of tumor heterogeneity (2–4), the three current main obstacles in the management of HCC are mining optimal biomarkers for its early diagnosis, molecular targeted therapeutics, and prediction of the efficacy of immune checkpoint blockade (ICB) therapies. Although exploration of some biomarkers has led to new surveillance programs for the early detection of HCC, the pros and cons of these molecules need to be reevaluated and improved (2, 3), as almost half of patients are still in the advanced stage at the time of diagnosis (5). As is known, over the past decade, targeted therapy has become one systemic therapy option for patients with HCC, despite the fact that only ~25% of HCCs harbor alterations that are potentially targetable with existing drugs (6). But these treatments have shown limited efficacy, and more reliable molecular targets are required for therapeutics through further understanding of the pathogenesis of HCC (7). On the other hand, ICB therapies have become increasingly important since tumor cells and infiltrating immune cells harbor an immunosuppressive microenvironment, including the overexpression of immune checkpoint molecules (8). However, ICB monotherapy has a response rate of only about 15%–23%, while that of combination treatments increased to approximately 30% (9). In addition, ICBs can induce severe immune-related side effects (10, 11). Therefore, it is necessary to use more reliable biomarkers for the prediction of the efficacy of ICB prior to starting a treatment plan. Unfortunately, based on our literature search, there are no predictive biomarkers currently available for the prediction of ICB response (5). In addition, it is costly and inconvenient for patients to undergo gene sequencing screening before deciding on ICB and targeted therapeutics. Thus, the discovery of potentially actionable biomarkers is necessary for the early diagnosis, target therapy, and prediction of ICB response to immunotherapy of HCC.
Combining biostatistical analyses and wet-lab experiments, the aim of this study was to explore SPINK1, a potentially qualified candidate, as a better marker for the early diagnosis, molecular targeting, and the prediction of the response to ICB treatment of HCC.
Materials and methods
Online Transcriptomics Studies of Liver Cancer Cohorts
The preliminary complementary DNA (cDNA) expression of SPINK1 in HCC tissue samples was analyzed using Oncomine (12). Gene expression analyses were carried out using “Oncomine” on four cohorts of patients with HCC, including Roessler Liver 1 and 2, Chen Liver, and Wurmbach Liver. The expression levels of SPINK1 messenger RNA (mRNA) were assessed using liver tumor and precancerous tissues, and differences with p ˂ 0.05 were considered statistically significant. Oncomine is publicly available at www.oncomine.org.
UALCAN, HPA Data Sources, and Kaplan–Meier Plotter
UALCAN is an easy-to-use, interactive web portal for conducting in-depth analyses of The Cancer Genome Atlas (TCGA) gene expression data (13). HPA, the Human Protein Atlas project, is funded to generate a map of protein expression patterns in normal cells, tissues, and cancer, while the Kaplan–Meier plotter is capable of assessing the impact of genes on the survival of many cancer types.
UALCAN uses TCGA level 3 RNA sequencing (RNA-seq) and clinical data from 31 cancer types including HCC. This web portal allows analysis of the relative expression of a query gene across liver tumor and normal tissue samples, as well as in various tumor subgroups including individual cancer stages, grade, race, body weight, or other clinicopathological features. UALCAN is publicly accessible at http://ualcan.path.uab.edu. The expression levels of some specific proteins corresponding to over 50% of all human protein-encoding genes have been analyzed and can be obtained from the HPA website. The HPA-related dataset is available at https://www.proteinatlas.org. The Kaplan–Meier plotter evaluates the impact of genes on the clinical outcomes of patients with different cancer types. Sources for the databases include the Gene Expression Omnibus (GEO) and TCGA. These two databases are freely available at http://gepia.cancer-pku.cn and http://kmplot.com/analysis.
Bioinformatics Analysis of the GEO and RNA-Seq Datasets
For the early detection of HCC, out of 1,884 specimens of stage I–IV HCC, 538 stage I and II patient samples (279 tumor and 259 para-tumor samples) from GEO (GSE14520, GSE101685, and GSE62232) were selected and used as the training dataset. The two new, non-mixed cohorts of 395 (GSE36376) and 272 (OEZ005255, NODE database) stage I and II specimens (201 tumor, 192 para-tumor; 169 tumor and 103, para-tumor) from the GEO RNA-seq datasets, respectively, were used as the validation datasets (14, 15). The list of NCBI-GEO datasets employed to perform the initial biomarker screening included GSE102097, GSE107170, GSE143231, GSE14520, GSE22405, GSE29721, GSE31370, GSE39791, GSE41804, GSE45267, GSE46408, GSE51401, GSE54236, GSE57957, GSE62232, GSE69715, GSE76427, GSE84402, GSE84598, GSE89377, and GSE98383 (Supplementary Table S1). The RNA-seq data collection used for the analysis of predictive factors was from Gao et al. (15) (OEZ005255, NODE database). All of the datasets comprise 1,884 tumor and non-tumor normal tissue samples from patients with stage I–IV HCC. Out of the aforementioned sample pool, three HCC datasets were collected from the GEO database (GSE14520, GSE101685, and GSE62232). These datasets included a total of 611 samples, of which 538 stage I and II patient samples were chosen as the training dataset. The TNM staging system of the American Joint Committee on Cancer (AJCC) was used to classify the stage I and II patient specimens from the rest of the samples. The original expression profiling data were obtained from both cancerous and non-cancerous tissue specimens. For the microarray probes, the expression values for the probe-mapped genes were calculated to obtain the means of the probes if multiple probes were mapped to the same Entrez gene ID. Data batch effects among the GEO data files were examined with t-distributed stochastic neighbor embedding (tSNE) analysis and corrected using ComBat of the R package “sva.” (14)
The RNA-seq data derived from 272 liver tissue specimens and para-cancerous normal tissue samples of patients with HCC could be obtained either from downloading the files from the website (https://www.biosino.org/node/index) or directly from the authors who published it. This transcriptomics dataset from Gao et al. (15) is publicly available to researchers.
For the analyses of the prediction of response to ICB therapy, 368 stage I–IV HCC specimens were selected from TCGA RNA-seq database (https://xenabrowser.net/datapages/), as the other GEO and RNA-seq training and validation datasets lack the transcriptomic abundance information on the expression of programmed death-ligand 1 (PD-L1), which was needed for the analysis of the biomarker-related ICB response.
Proteomics Analysis
Paired 98 tumor and 165 para-tumor tissue samples from stage I and II Chinese HCC patients (CHCC) (out of 330 I–IV tumor samples) with hepatitis B virus (HBV) infection were selected for proteomics analysis (15). The published HCC proteomics CPTAC (the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium) data file was obtained directly from the web link provided by Professor Hu Zhou, University of Chinese Academy of Sciences, Beijing (https://cptac-data-portal.georgetown.edu/study-summary/S049). In the proteomics analysis, 10,783 proteins (encoded by 10,759 genes) with an average of 8,934 proteins per sample were identified using isobaric tandem mass tag (TMT)-based global proteomics (15).
Cell Culture, Vector, Antibodies, and Reagents
The HepG2, Hep3B, H22, and 293T cell lines were given as gifts by Professor Aiguo Wang (Dalian Medical University, Dalian, China), who purchased them from the Chinese Academy of Sciences, Beijing, China. The cells were grown in McCoy’s medium (Sangon Biotech, Shanghai, China), RPMI-1640, or Dulbecco’s modified Eagle’s medium (DMEM) (Sangon Biotech, Shanghai, China) with 10% fetal bovine serum (FBS) (CellMax, AusGeneX, Queensland, Australia). Human SPINK1-overexpressing (OE) and SPINK1 short hairpin RNA (shRNA) lentiviral vectors were generated by Applied Biological Materials (ABM, Nanjing, China), and the Transwell system was purchased from Nest (Southborough, MA, USA). The following commercially available antibodies were used: anti-SPINK1 (Abnova, Taipei, Taiwan) and anti-Bcl-2, anti-Bcl-XL, anti-Bax, anti-Bad, and anti-caspase-3 (all from Wanlei Biotech, Shenyang, China). The main chemicals or reagents used in this study were as follows: 5-fluoruracil (5-FU) (Shanghai Pharmaceutical Company, Shanghai, China), Lipo2000 (Invitrogen, Carlsbad, CA, USA), Cell Counting Kit-8 (CCK-8) (Xian Baiying Biotechnology Inc., Xian, China), puromycin (Solarbio, Beijing, China), and polybrene (Maokang Biotech, Shanghai, China).
Generation of Stable SPINK1-Overexpressing or SPINK1-Silencing HCC Cell Lines
The HepG1, Hep3B, and H22 HCC tumor cell lines permanently transduced with SPINK1-OE or SPINK1-silencing (shRNA) lentiviral constructs were generally constructed as previously described (16). Briefly, the pLenti-CMV-hSPINK1-2A-GFP vector or pLenti-SPINK1 shRNA was co-transfected with either ABM’s Second Generation (LV003) Packaging Mix or the mixture of pMD2.G and psPAX2 packaging plasmids (Addgene Inc., Cambridge, MA, USA) into 293T cells, while the transfected empty or scrambled vector was used as a control. The packaged lentiviral particles were harvested 24–48 h after transfection. HepG2, Hep3B, and H22 cells were infected by filtered lentiviral particles, and the transduced cells were screened under pressure of 2 µg/ml puromycin for about 3 weeks before the drug-resistant cells were collected. Subsequently, the protein expression of SPINK1 was examined in passaged HepG1, Hep3B, and H22 cells using Western blotting.
Proliferation, Colony Formation, Transwell Migration, Scratch Test (Wound Healing Assay), Chemoresistance, Anti-Apoptosis, and SPINK1 Targeting Response in HCC Cells
Experiments were carried out with respect to colony formation, Transwell assay, chemoresistance, chemically induced apoptosis resistance, and SPINK1 targeting response in SPINK1-OE and/or knockdown HCC cells. Cell proliferation was measured using CCK-8, as described in a previous publication (16). In brief, the experiments were performed in cell culture dishes or in triplicates of a 96-well plate for each designed group. The absorbance (A) for the wells was measured using a microplate reader. The proliferation rates were calculated as: inhibition rate (%) = (1 − treated group A/control A) × 100%. The colony formation assay was carried out as follows. Briefly, the corresponding HCC cells were seeded in six-well plates at about 1,000 cells per well and cultured for 14 days. The plates were then washed with phosphate-buffered saline (PBS) and stained with crystal violet. The images of each well were saved, and the number of holoclones re-grown (colonies >50 cells each) was counted to determine the efficacy of holoclone formation. The clone formation rate was equal to the number of clones/cells seeded (in percent). The experiment was performed in triplicate.
In vitro Transwell migration assays were performed using Nest 12-multiwell inserts with 8-µm pores. For the migration assay, 30,000 cells in 200 µl serum-free DMEM were placed into each Transwell insert with 750 µl of 10% FBS growth medium in the lower chamber. The cells migrated through the pores were processed with 3.7% formaldehyde, stained with 2.5 µg/ml Hoechst for 15 min before washing with PBS, and counted 48 h after cell seeding. The 10 fields of cells were observed per experiment.
For the scratch test, 5 × 10 (4) cells/well were seeded into six-well plates and cultured to the second day. Three parallel marks were scratched along the margin of a ruler in each well using a 200-μl pipette tip. With the supernatant removed, serum-free medium was added to the cells and the cells photographed under a microscope after 0, 24, and 48 h. The migration rate of the cells in each group was calculated with ImageJ software.
For the chemoresistance test, SPINK1-OE and/or knockdown HepG2, H22, or Hep3B cells with or without 5-FU (2.5 μM) treatment were inoculated in a 96-well plate (0.3 × 10 (4)/well) for 48 h before 10 μl of CCK-8 solution was finally added into each well; the incubation was continued for an additional 2 h prior to the analysis. The apoptosis status of SPINK1-OE or knockdown HCC cells with or without 5-FU treatment was evaluated by examining a number of the main pro- and anti-apoptotic molecules using Western blotting. After seeding with 0.3 × 10 (4) cells/well, the SPINK1 targeting experiment was carried out by assessing the targeting efficacy of the human anti-SPINK1 antibody in HCC cells at a concentration of 5 or 10 μg/ml using the CCK-8 assay after 72 h culture in a 96-well plate.
Conditioned Media and Rescue Experiment
For the collection of conditioned medium (CM), 8–10 ml of serum-free DMEM was added into each of a 10-cm culture plate seeded with HepG2 and the SPINK1-OE cell lines. The cell debris was removed and SPINK1 CM was collected 48 h later via centrifugation at 1,000 rpm for 5 min at 4°C. The supernatant was accumulated and stored at −80°C until use. The vector control CM was made for the empty vector-transfected cell strains in the same manner as described above. For the rescue experiment, permanent SPINK1-silencing Hep3B and empty vector-transduced control cells were treated with SPINK1 CM and control CM, respectively, for 48 h. Subsequently, the CCK-8 assay was performed to evaluate the proliferation rates of these two cell lines processed with the CM.
Western Blotting
The immunoblot analysis was performed as previously described (17). For this analysis, whole cell lysates were prepared using NP-40 lysis buffer, followed by measurement of the protein concentration. The proteins were separated by SDS-PAGE and transferred to a 0.2-mm PVDF membrane. SPINK1, GAPDH, and other antibodies including Bcl-2, Bcl-XL, Bid, and Bad were used to probe the targeted protein at 1:500 before the antibody–antigen complex was detected using horseradish peroxidase (HRP)-conjugated antibodies and enhanced chemiluminescence (ECL).
Experiments of Tumor Homograft, Xenograft, and Metastasis In Vivo
SPINK1-associated tumorigenesis and metastasis were analyzed in normal immunocompetent BALB/c mice using SPINK1-OE HepG2 or H22 cells. Clean-degree female BALB/c mice (6–8 weeks old, weighing around 20 g) were purchased from the Center of Experimental Animals of Dalian Medical University. The mice were reared in a room on a 12-h light/dark cycle. All procedures were approved by the Animal Care and Use Committee of Dalian Medical University. The tumor volume and metastatic lung nodules were measured based on the methods in previously published reports (18, 19). For the growth study, 3 × 10 (6) cells/0.2 ml scramble control and SPINK1–OE HepG2 or H22 cells were injected subcutaneously in the back region of the mice. Tumor growth was recorded weekly using a caliber ruler, and the tumor volume was estimated using the formula: [tumor volume = 1/2(L × W (2)]. The tumor burden or weight was scaled as soon as the mouse was sacrificed. For the metastasis study, 3 × 10 (5) cells/0.05 ml scramble control and SPINK1–OE H22 cells were injected via the tail vein, and the lungs of mice were simultaneously excised and observed grossly for metastatic lesions through counting the total volume of the number of metastatic nodules using the same method described above: tumor volume = 1/2[L × W (2)].
Co-Expression, Enrichment, and Pathway Analyses of SPINK1
LinkedOmics is a publicly available portal that includes multi-omics data from all 32 TCGA cancer types (20). The LinkFinder module of LinkedOmics was used to study the genes co-expressed with SPINK1 in the HCC cohort of TCGA. The results showed that a large number of genes had significantly positive correlations with SPINK1, whereas similar numbers of genes showed significantly negative correlations [false discovery rate (FDR) <0.01]. These data were statistically evaluated using Pearson’s correlation coefficient, followed by graphical presentation in volcano plots and heatmaps. The LinkInterpreter module of LinkedOmics carries out Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the genes differentially co-expressed with SPINK1. GO and KEGG analyses were performed with the “GSEA” tool. The LinkedOmics database is publicly available at http://linkedomics.org/login.php.
Potential Predictor of Response to ICB Therapy
In order to predict patients more likely to benefit from ICB immunotherapy (21), we presented the currently recognized immune response-associated cells or molecules and analyzed their correlation with SPINK1 using their unique expression data, assessing the possibility of SPINK1 as a predictor for precision HCC immunotherapy.
For the assessment of the association between SPINK1 and tumor-infiltrating immune cells, the transcriptomic expression of SPINK1 within each tumor sample of the HCC dataset (TCGA) was initially subcategorized as high or low level on the basis of its median value. Subsequently, the compositions and contents of the various subtypes of infiltrating immune cells in each sample were determined based on RNA expression abundance using CIBERSORT. Thirdly, the correlations between SPINK1 and the expression patterns of 28 subtypes of infiltrating immune cells were analyzed in the same “high” or “low” expression group of specimens using statistical t-test. With respect to the estimation of stromal and infiltrating immune cells in malignant tumor tissues, we chose a new algorithm that takes advantage of the unique properties of the transcriptional profiles of the HCC samples to deduce tumor cellularity, named ESTIMATE (Estimation of Stromal and Immune Cells in Malignant Tumor Tissues Using Expression Data) (22). The correlations between SPINK1 and the immune, stromal, and ESTIMATE scores were determined using the “estimate” package in R. Similarly, the expressions of several prominent immune checkpoints (ICPs) were determined as these important molecules are likely involved in the induction of ICB response (23). Tumor mutation burden (TMB), the number of mutations per million bases in tumor tissue, was used for assessing vulnerability to ICB immunotherapy, as TMB likely affects immune response (24).. The mutation data of patients with HCC were downloaded from TCGA, and TMB analysis was accomplished using the R package “maftools.” (25) The correlations among SPINK1, ICPs, and TMB were investigated using statistical t-test.
Statistical Analysis Software and Methods
Most of the bioinformatics analyses were accomplished using R statistical software, version 3.4.1 (R Core Team, Vienna, Austria). MedCalc software was used to plot the area under the curve (AUC) for AFP, SPINK1, and their combination before carrying out logistic regression to generate the receiver operating characteristic (ROC) curves. The “rms” package was used to draw the fit curves of the observed and predicted values. The R package “PredictABEL” was applied to calculate the integrated discrimination improvement (IDI) and net reclassification improvement (NRI), which represent the discrimination of a prediction pattern (26). The Akaike information criterion (AIC) and Bayesian information criterion (BIC), which reveal the calibration of the same model, were also applied (27). We performed decision curve analysis (DCA), a measurement of the clinical net benefits (28), using the “Decision Curve” package in R. The clinical impact curve (CIC) (29), a plot derived from the DCA, was used to further evaluate the clinical cost/benefit ratio of a model. Significant differences in the murine tumor volume among the groups were statistically analyzed using two-way ANOVA. Quantitative or semi-quantitative variables were counted as the mean ± SEM, and the means were compared using unpaired two-tailed Student’s t-test. A p-value <0.05 was considered statistically significant.
Results
Study Design
The flowchart shows the research project process (Figure 1). The study design included three step-by-step reasoning concerns we encountered during the investigation, the eligibility criteria for the selection of patients with HCC, and the bench-scale experiment plans during the analysis process (Figure 1). In the transcriptomics study of “HCC early detection,” the cDNA microarray expression data files were used in the analysis of the training dataset, while the GEO and RNA-seq data files were applied in two new and non-mixed validation datasets, respectively. The training dataset was used only in the proteomics analysis for the above detection. To collect more evidence of “what works and why” for the role of SPINK1 in the early detection of HCC, we carried out in vitro and in vivo wet-lab experiments including monoclonal antibody (mAb)-oriented or molecular targeting and other cellular assays (Figure 1). Interestingly, we found that SPINK1 possesses some biologically tumorigenic characteristics that make it a potential target molecule in targeted therapeutics for HCC. Thereafter, we further explored the underlying molecular signaling networks that mainly contributed to the “early detection” and “targeting” features of SPINK1. Unexpectedly, gene set enrichment analyses (GSEA) including GO and KEGG showed that the underlying molecular interactions were closely associated with immune response-related biological processes and pathways in HCC. Thus, we assumed that SPINK1 might have acted as a biomarker for the prediction of response to ICB therapy. In the “prediction” study of ICB-related therapies, the RNA-seq data of the HCC samples from the TCGA database were collected for the analyses of the associations between SPINK1, infiltrating immune cells, stromal cells, immune checkpoint molecules, and TMB (Figure 1).
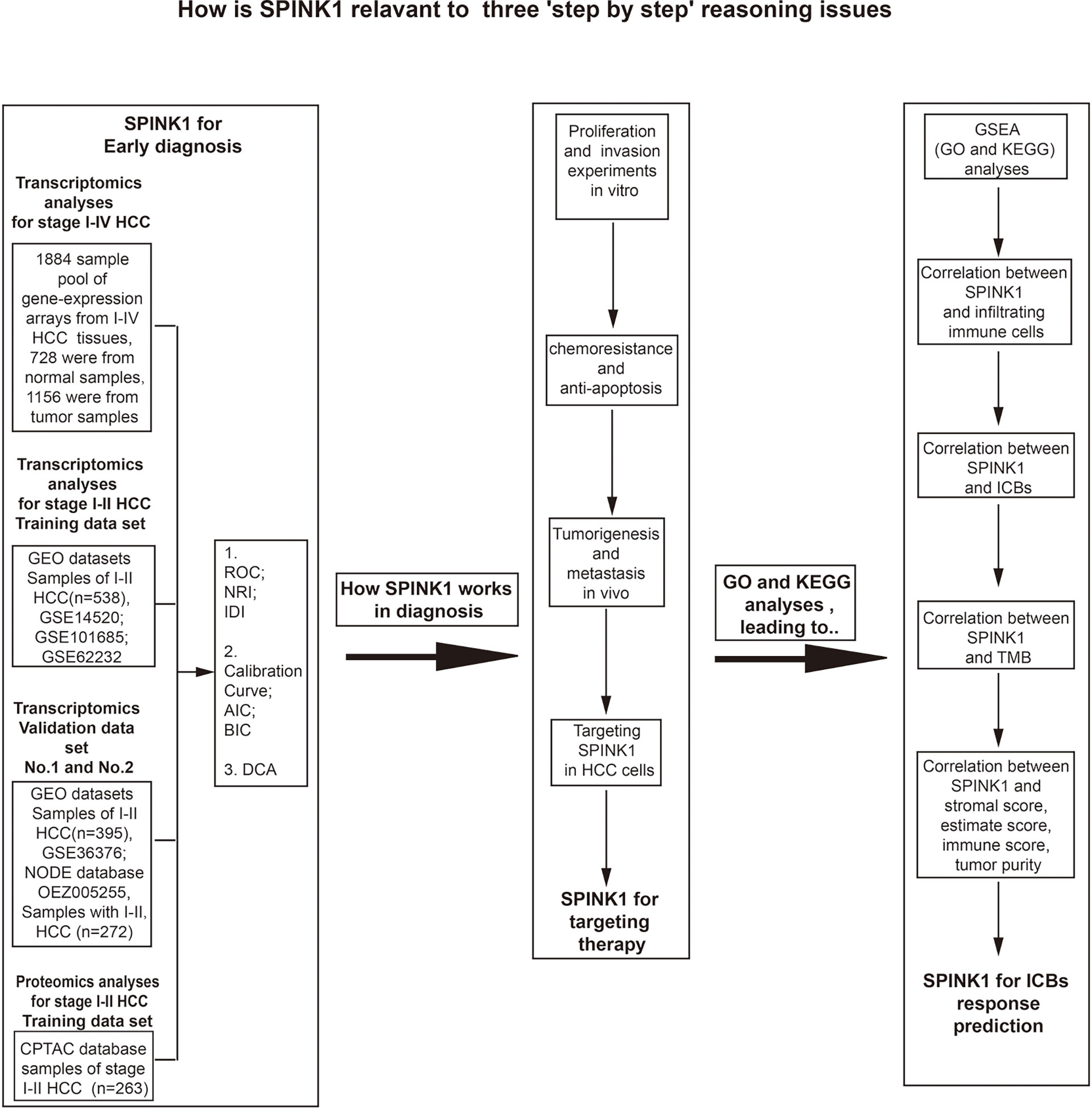
Figure 1 Flowchart of the three goals of this investigation: step-by-step bioinformatics analyses, bench-side experimental design, and eligibility criteria for the collection of tissue samples from patients with hepatocellular carcinoma (HCC) from different datasets.
Overexpression of SPINK1 in HCC Tissues
We initially examined the cDNA expression profiles of SPINK1 in the liver samples of four cohorts (Roessler Liver 1 and 2, Chen Liver, and Wurmbach Liver) of patients with HCC from Oncomine (30). The mRNA expression of SPINK1 was significantly higher in the HCC liver tissue samples of three cohorts (Roessler Liver 1 and 2 Chen Liver) compared to normal controls (p ˂ 0.01; Supplementary Figures S1A–D). In the UALCAN analysis, the transcription level of SPINK1 was ranked within the top 18 out of all the overexpressed genes in HCC tissues (Supplementary Figure S2) and was significantly higher in the HCC tissue specimens of stage I–III compared to stage IV tumor and grades I–IV (p < 0.05; Supplementary Figures S1E,F). Furthermore, the protein expression of SPINK1 was significantly increased immunohistochemically in 6 out of the 12 HCC tissue sections, as shown by the HPA data source (Supplementary Figure S1G) (31). Clinically, the Kaplan–Meier plotter (32) showed that a high SPINK1 mRNA expression was significantly associated with poor OS of patients with HCC compared with patients with low mRNA expression (data not shown). In contrast, there was no significant correlation between AFP and OS in HCC patients (data not shown). These above preliminary data suggested a possible role of SPINK1 as a biomarker for the detection and even early diagnosis of patients with HCC.
Early Detection of HCC via Analyses of cDNA Microarray Expression and RNA-Seq Profiles
We firstly mined the cDNA expression data of the 1,884 human HCC samples, which showed that the SPINK1 gene was significantly overexpressed in stage I–IV tumor samples relative to para-tumor specimens and presented better discrimination and calibration capabilities for HCC detection than AFP (Supplementary Figure S3, Table S2). We then chose the stage I and II tumor samples out of the 1,884 and used them as the training dataset for further analyses.
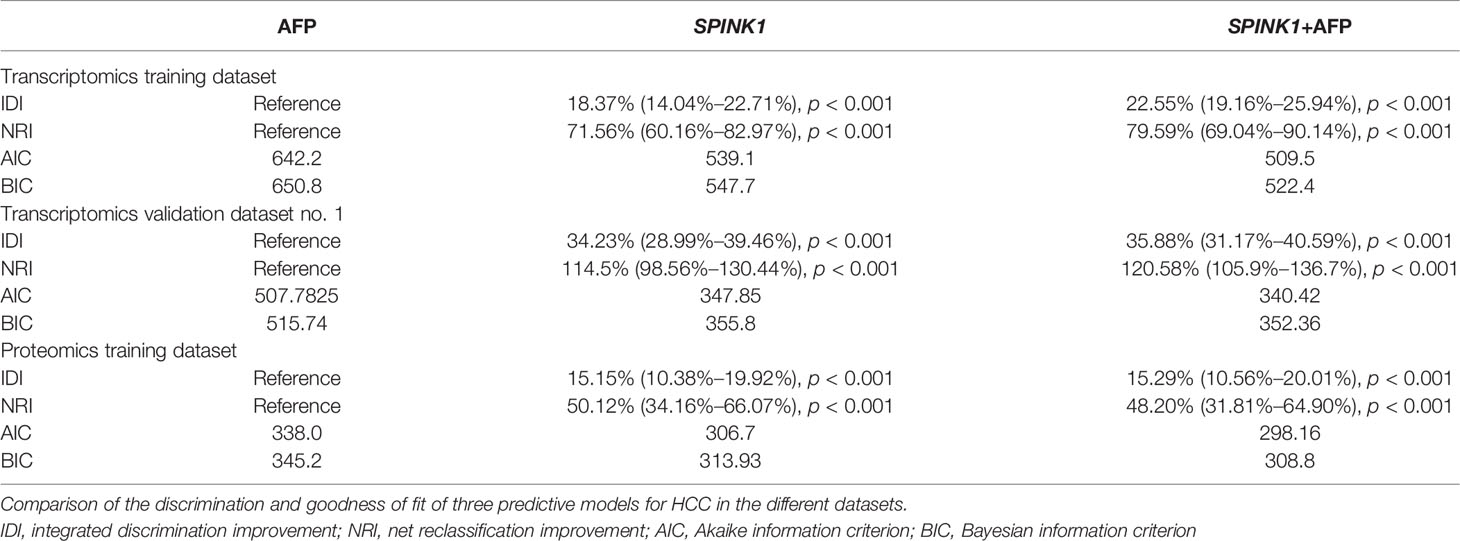
In the training dataset, we found that the ROC curves plotted for SPINK1 separated the stage I and II HCC samples from the normal adjacent to tumor controls with high discriminatory accuracy (Figure 2). Compared with AFP, which lacked sufficient sensitivity in the detection (33, 34), the SPINK1 and SPINK1+AFP models provided higher AUC values with similar specificity and a much higher sensitivity for patients with HCC (AUC = 0.83 vs. 0.84 vs. 0.66, specificity = 87.6% vs. 83.0% vs. 88.4%, sensitivity = 70% vs. 77.8.1% vs. 49.5%) (Figure 2A). Notably, the transcription level of SPINK1 was not correlated with that of AFP, suggesting SPINK1 as an independent diagnostic factor for HCC (R = 0.42; Supplementary Figure S4). To further determine how much better a new SPINK1-related model reclassified patients into the HCC and non-HCC groups, we calculated the IDI and NRI parameters. The SPINK1-related models remarkably improved the classification capacity of AFP (IDI = 118.4%:122.6%:reference, p < 0.001) (Table 1). Using the cutoff values at 0%–30% (low risk), 30%–60% (moderate risk), and 60%–100% (high risk), the NRI of the two aforementioned SPINK1 models increased by 71.6% and 79.6%, respectively, compared to that of AFP alone (all p < 0.01; Table 1), suggesting that SPINK1 improved the classification of patients with events and those without events. Thus, SPINK1 has a higher discriminatory capacity than AFP.
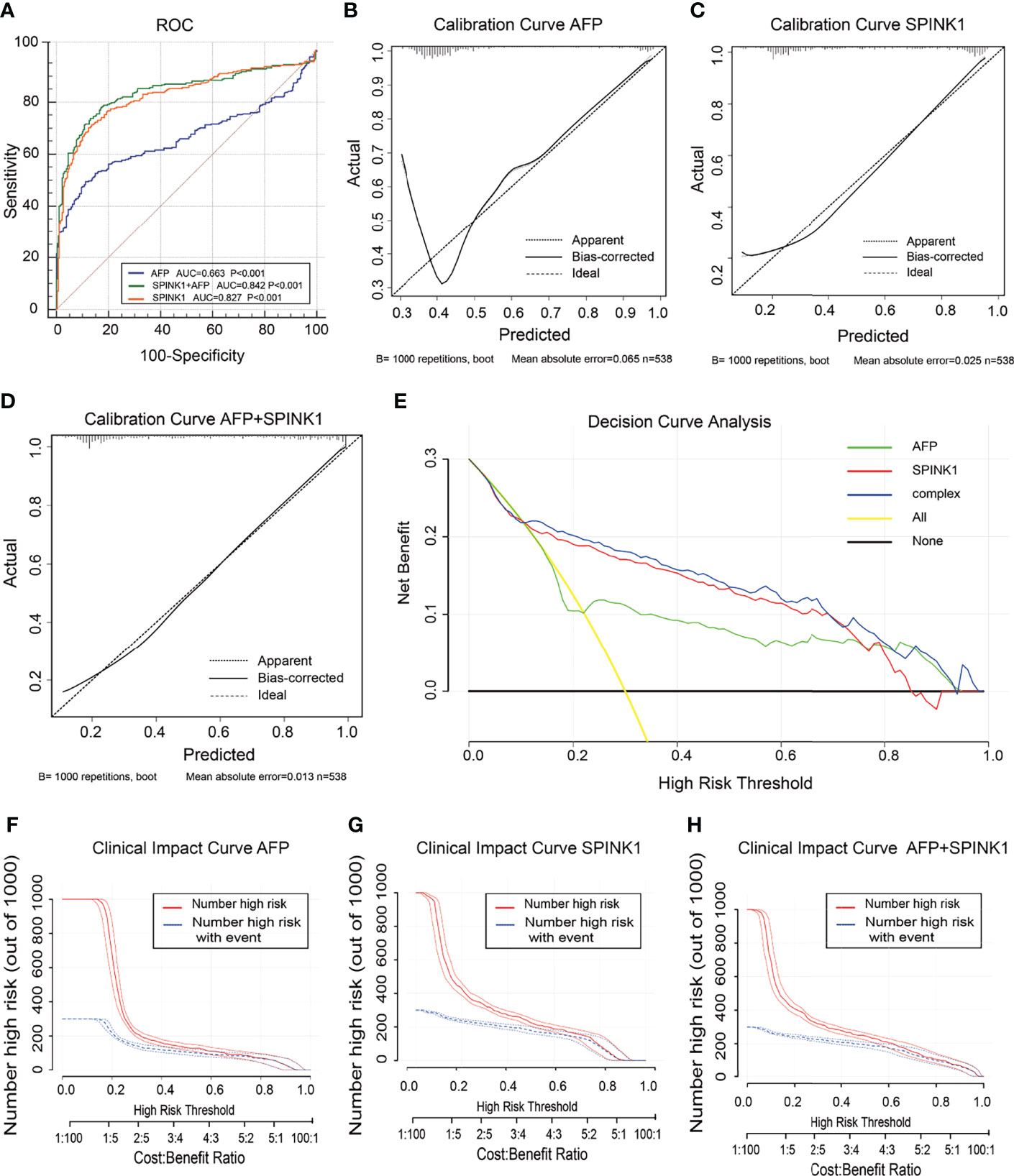
Figure 2 Discrimination and calibration of hepatocellular carcinoma (HCC) prediction models through transcriptomics analyses in the training dataset. (A) Receiver operating characteristic (ROC) curves and area under the curve (AUC) in the SPINK1, SPINK1 plus alpha-fetoprotein (AFP), and AFP models. (B–D) Calibration curves of AFP (B), SPINK1 (C), and SPINK1+AFP (D). (E) Clinical net benefits in the decision curve analysis (DCA) of SPINK1, SPINK1+AFP, and AFP. Yellow line indicates all HCC patients; middle and horizontal black line denotes non-HCC patients. (F–H) Comparison of the clinical benefits obtained from the clinical impact curve (CIC) analysis of each detection model.
Since calibration reflects the extent to which a model correctly assesses the absolute incident or risk (35), we calculated the AIC and BIC, which revealed the following: AIC = SPINK1 (539.1):AFP+SPINK1 (509.5):AFP (642.2) and BIC = 547.7:522.4:650.8. These results indicate better calibration in the SPINK1-related model compared to that in AFP (Table 1). The calibration curves between the predicted and observed values for the SPINK1-associated models generally fitted well with the predicted probability of 0.3–1.0 along the x-axis compared to the AFP model alone, in which the goodness of fit between the two curves remained better only in the range 0.7–1.0 and exhibited poor calibration particularly within the range of <0.35, where a relatively low incidence rate of predicted HCC occurred, suggesting the poor sensitivity of AFP (Figures 2B–D). To evaluate clinical significance of these markers, we performed DCA, which revealed that the SPINK1-related models both had remarkably higher net clinical benefits within the high risk threshold probability range 0.1–0.8 relative to that of AFP (Figure 2E). Moreover, the DCA-derived CICs supported the aforementioned findings (Figures 2F–H).
In the analysis of the validation dataset, we chose two new, non-mixed available GEO and RNA-seq datasets that had been created by two specific groups and platforms for primary analysis (15). In the first validation dataset, the analyses led to significantly consistent results with the findings from the training dataset (Figure 3), as the discrimination capability of the SPINK1-related curves was as good as that shown in the discovery dataset, as well as the quality of calibration in the same setting. The analyzed data were described in the following order: SPINK1 vs. SPINK1+AFP vs. AFP [AUC = 0.85:0.86:057, specificity = 91.71%:94.82%:97.93%, sensitivity = 72.77%:67.82%:39.11% (Figure3A); IDI = 1.34:1.36:reference, NRI = 214%:220%:reference) (Table 1)]. The calibration curves generally coincided with a much greater extent of the risk threshold probability at 0.2–1 in the SPINK1-related prediction models relative to that of AFP, which showed poor goodness of fit when the predicted risk probability was <0.3 (Figures 3B–D). In support of this, decreased values of AIC and BIC were calculated in these two models: AIC = 347.8:340.4:507.7 and BIC = 355.8:352.3:511.7 (Table 1). DCA and the CIC analysis also added more support to the two SPINK1 patterns (Figures 3E–H). In the second validation dataset, the analyses generally showed consistent results with those derived from the training dataset (Supplementary Figure S5,Table S2).
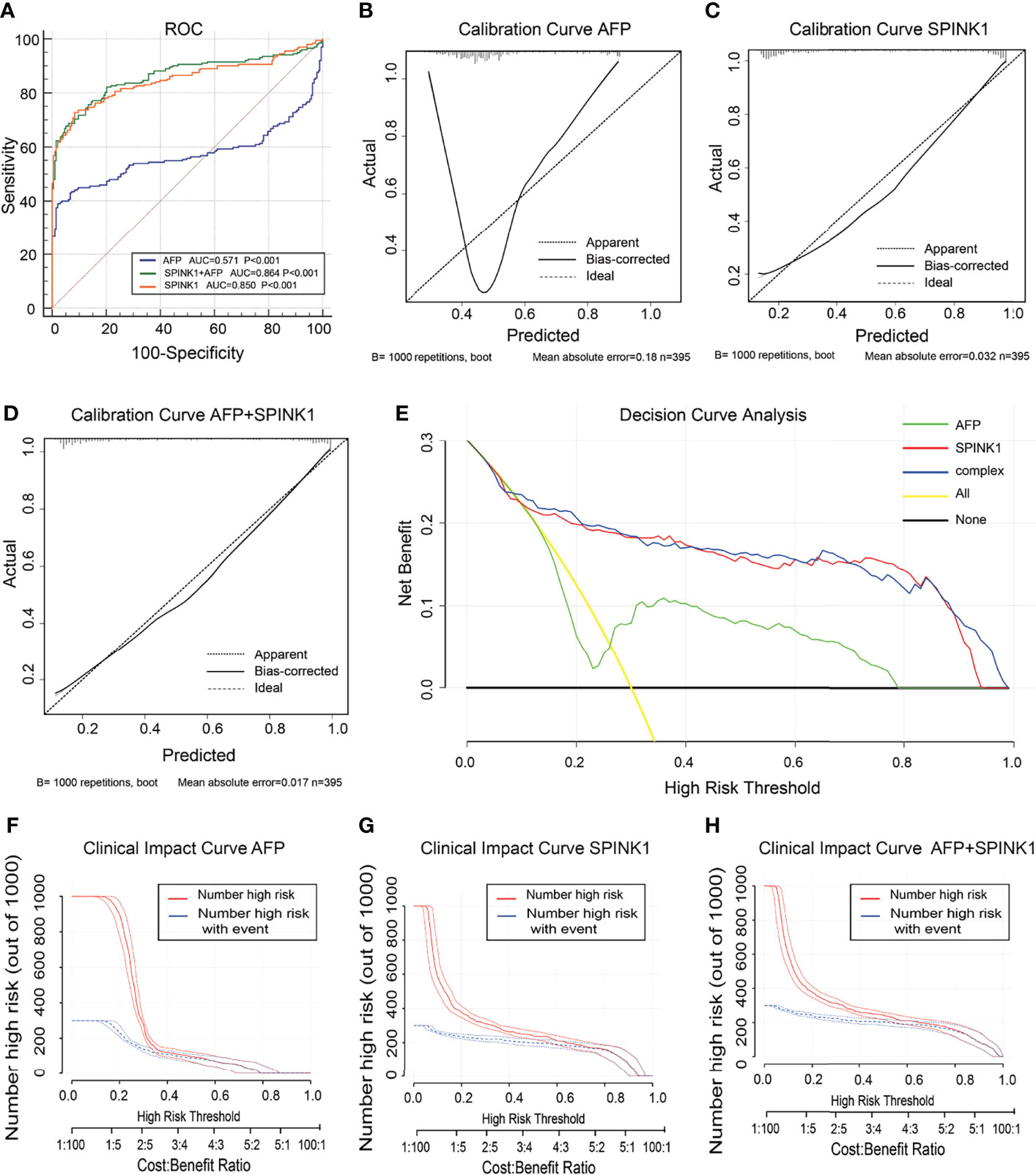
Figure 3 Discrimination and calibration of hepatocellular carcinoma (HCC) prediction models through transcriptomics analyses in the validation dataset no. 1. (A) Receiver operating characteristic (ROC) curves and area under the curve (AUC) in the SPINK1, SPINK1 plus alpha-fetoprotein (AFP), and AFP models. (B–D) Calibration curves of AFP (B), SPINK1 (C), and SPINK1+AFP (D). (E) Clinical net benefits shown by decision curve analysis (DCA) of SPINK1, SPINK1+AFP, and AFP. (F–H) Comparison of the clinical benefits obtained from the clinical impact curve (CIC) analysis of each detection model.
Proteomic Abundance of SPINK1 in the Detection of HCC
In the training dataset, the proteomics analyses revealed similar trends to those shown in the previously described transcriptomics data. The SPINK1 and SPINK1+AFP models exhibited a higher discriminatory efficacy than did AFP alone in stage I and II tumor samples: AUC = 0.72:0.72:0.53, specificity = 92.0%:92.4%:90.6%, sensitivity = 46.9%:45.7%:23.4% (Figure 4A). SPINK1 rendered an almost equal specificity and a considerably elevated sensitivity relative to the AFP control, although the sensitivity values decreased for each of the three predictive patterns. The IDI results also strengthened the above findings by showing almost the same trend in this dataset, i.e., increased by 15.15% and 15.29% (IDI = 115.15%:115.29%:reference), while the NRI values showed increases of 50.12% and 48.20% compared to the AFP reference (NRI = 150.12%:148.20%:reference) (Table 1). The calibration of the predicted and observed values was remarkably improved with the inclusion of SPINK1 compared to that of AFP alone, which displayed a poorly calibrated pattern (Figures 4B–D). Similarly, the decreased AIC and BIC values were observed correspondingly for the SPINK1-associated patterns (Table 1), reconfirming the better goodness of fit in these two SPINK1 models (AIC = 306.7:298.16:338.0, BIC = 313.93:308.8:345.2). DCA also added more positive data to the SPINK1-related groups, indicating more clinical benefits obtained with the inclusion of SPINK1 within the risk threshold probability of 0.2–0.9 (Figure 4E). The CICs clearly showed greatly improved clinical benefits in the abovementioned two settings within the range of predicted probability of 0.1–0.2, in which the incidence of HCC is quite low (Figures 4F–H). The protein level of SPINK1 was also not correlated with that of AFP, indicating SPINK1 as an independent predictive factor (R = −0.001; Supplementary Figure S6).
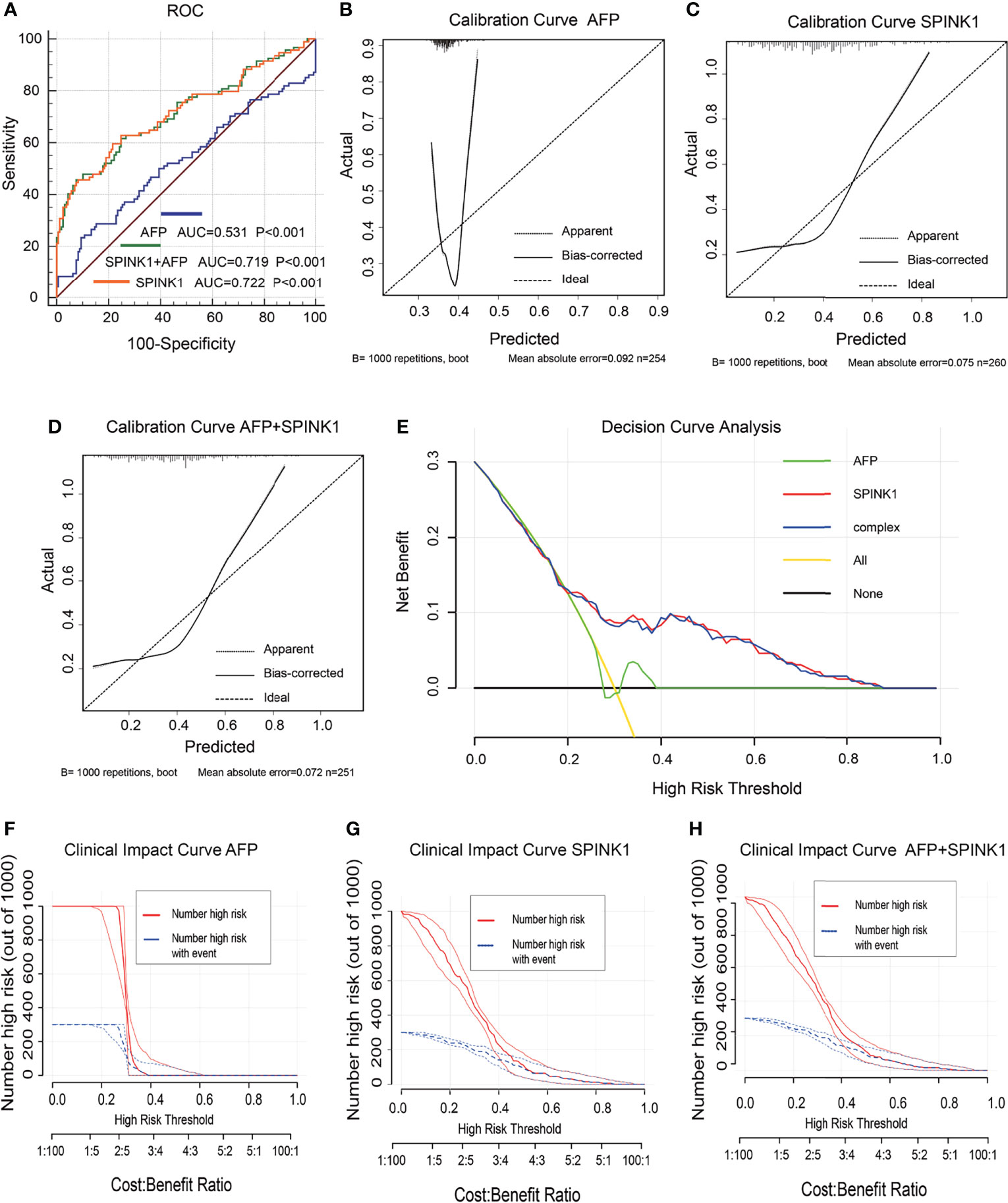
Figure 4 Discrimination and calibration of hepatocellular carcinoma (HCC) prediction models via proteomics analyses in the training dataset. (A) Receiver operating characteristic (ROC) curves and area under the curve (AUC) in the SPINK1, SPINK1 plus alpha-fetoprotein (AFP), and AFP models. (B–D) Calibration curves of AFP (B), SPINK1 (C), and SPINK1+AFP (D). (E) Comparison of the decision curve analysis (DCA) of SPINK1, SPINK1+AFP, and AFP in terms of clinical net benefits. (F–H) Clinical benefits obtained from the clinical impact curve (CIC) analysis of each detection model.
Although the transcriptomics and proteomics analyses of the expression patterns of SPINK1 in HCC tissues laid the foundation for its qualification as a promising biomarker for the early detection of the disease, there probably are other hidden properties of SPINK1 that greatly supported or contributed to its role in this scenario. Therefore, we next studied, using bench-scale experiments, some but not all of the tumorigenic phenotypes of SPINK1 that were associated with the proliferation, clonal formation, migration, metastasis, chemoresistance, and anti-apoptosis of HCC cells. The following showed that the accumulated data not only favorably supported the function of SPINK1 in the detection of HCC but also gradually led to the temporary conclusion that it may also be a possible molecular target in HCC therapies based on some of its unique biological features of cancer.
Stimulation of Cellular Proliferation and Clonal Formation by SPINK1
We determined the possible carcinogenic phenotypes of SPINK1 that were possibly responsible for its predictive capability in HCC at the cellular level. We initially created the permanently SPINK1-OE HepG2 and H22 cell strains and silenced the SPINK1 Hep3B cell line using the C2 shRNA vector, the most efficient SPINK1 knockdown construct (Supplementary Figure S7). Compared with the control, the metabolic and proliferative capabilities increased significantly in SPINK1-OE HepG2 and H22 cells and decreased considerably in SPINK1 knockdown Hep3B cells (p < 0.01; Figures 5A–C). The clonogenic assay showed that the number of colonies formed in SPINK1-OE HepG2 cells was greater than that in empty vector controls, whereas SPINK1 knockdown Hep3B cells rendered a significant inhibitory effect on the number of cell colonies formed (p < 0.01; Figures 5E, F).
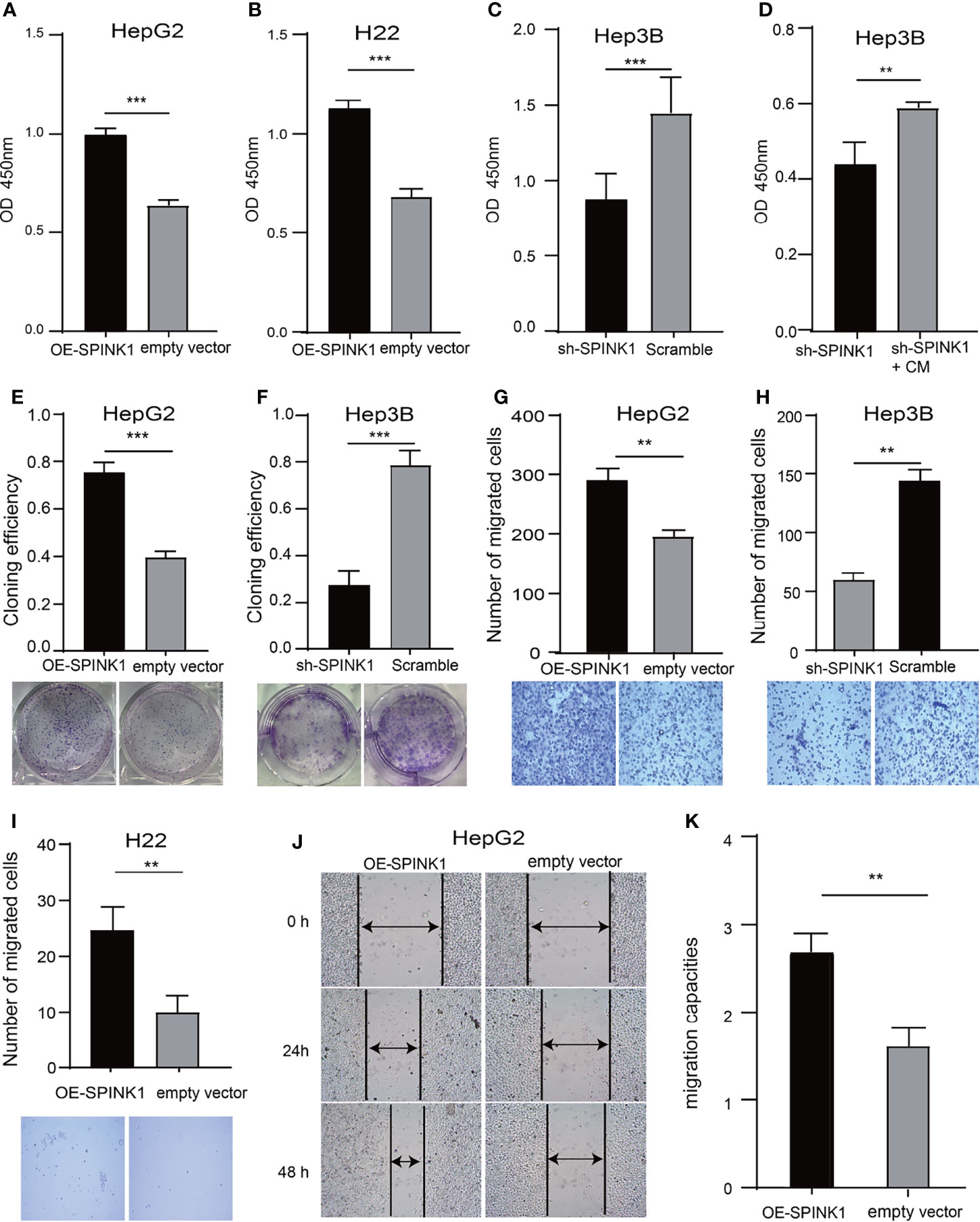
Figure 5 SPINK1’s regulation on proliferation, clonogenic formation and migration of SPINK1-OE and SPINK1-silencing HCC cells. (A, B) CCK-8 assays for metabolic and proliferative activities in stable SPINK1-OE HepG2 and SPINK1-OE H22 cells. (C, D) Proliferative capability and corresponding phenotype rescue after addition of conditioned medium (CM) for SPINK1-silencing Hep3B cells. (E, F) Clonogenic formation in SPINK1-OE HepG2 and knockdown Hep3B HCC cells, along with images of formed clones following inoculation of corresponding SPINK1-OE HepG2, SPINK1-silencing H3B and control cells for two weeks. (G–I) Transwell assays for SPINK1-OE HepG2, SPINK1-OE H22 and SPINK1 knockdown Hep3B cells, with photos that showed cells passing through well’s pores both in control and experimental groups, respectively. (J, K) Images and bar chart for measurement of migration distances of SPINK1-OE HepG2 cells 24 and 48 hrs post-scratching in Wound-healing assay (Scratch test). Error bars represent mean ± standard deviation of independent experiments or the experiments performed in triplicates either in 6-well or 96-well plates. OD, optical density; **, P < 0.01; ***, P < 0.001.
Rescue of shRNA-Mediated Phenotypes by Conditioned Medium
To further confirm the indispensable role of SPINK1 in the growth of HCC cells, we performed the “rescue” experiment in SPINK1 shRNA-transfected Hep3B cells. It was found that the survival rate of Hep3B cells was significantly decreased following transfection of the SPINK1 shRNA lentiviral construct (Figure 5C). However, the CCK-8 assay showed that the addition of exogenous SPINK1 CM could remarkably rescue Hep3B cells from the proliferation-inhibitory state, while the vector CM was unable to reach this outcome (p < 0.01; Figure 5D), suggesting the specificity of SPINK1 as a survival factor in the growth of Hep3B cells.
Migration and Invasion via SPINK1 Regulation
The results of the Transwell assays demonstrated that the number of migrated cells via the pores in the SPINK1-OE HepG2 and H22 groups were much more than those in the respective controls, whereas fewer number of migrated cells could be observed in SPINK1 knockdown Hep3B cells (p < 0.01; Figures 5G–I). In addition, the scratch test also showed similar results (p < 0.01; Figures 5J, K). Together, these data suggest that SPINK1 possesses the ability to promote the proliferation, clonal formation, migration, and invasion of HCC cells.
SPINK1-Associated Tumorigenesis and Metastasis In Vivo
We attempted to reproduce some of the aforementioned in vitro observation in in vivo experiments. To simulate the tumorigenic and metastatic capacities of SPINK1 under immunocompetent circumstances, we employed murine HCC models of implanted homograft and metastasis using SPINK1-OE H22 cells. A significant increment in the tumor size, on average, was observed weekly, and this trend gradually reached the peak at 3 weeks after implantation of SPINK1-OE H22 tumor cells in mice (Figure 6A). The tumor graft volume and the weight of SPINK1-OE H22 mice were significantly higher than those of the control (p < 0.01; Figures 6C, D). Moreover, we estimated the total volumes of the metastatic lung nodules; the results showed greater numbers in the SPINK1-OE cell-injected mice compared to the H22 control animals 4 weeks after tail vein administration of SPINK1-OE H22 cells, suggesting the pro-metastatic capability of SPINK1 (p < 0.01; Figures 6E, F). Simulating the real immunocompetent environment, we also injected human SPINK1-OE HepG2 cells subcutaneously in the flank of Balb/c mice. Despite xenograft rejection, SPINK1 overexpression also promoted the tumorigenesis and metastasis of HepG2 cells in mice under normal immunological conditions (p < 0.01; Figures 6B, G, H, I).
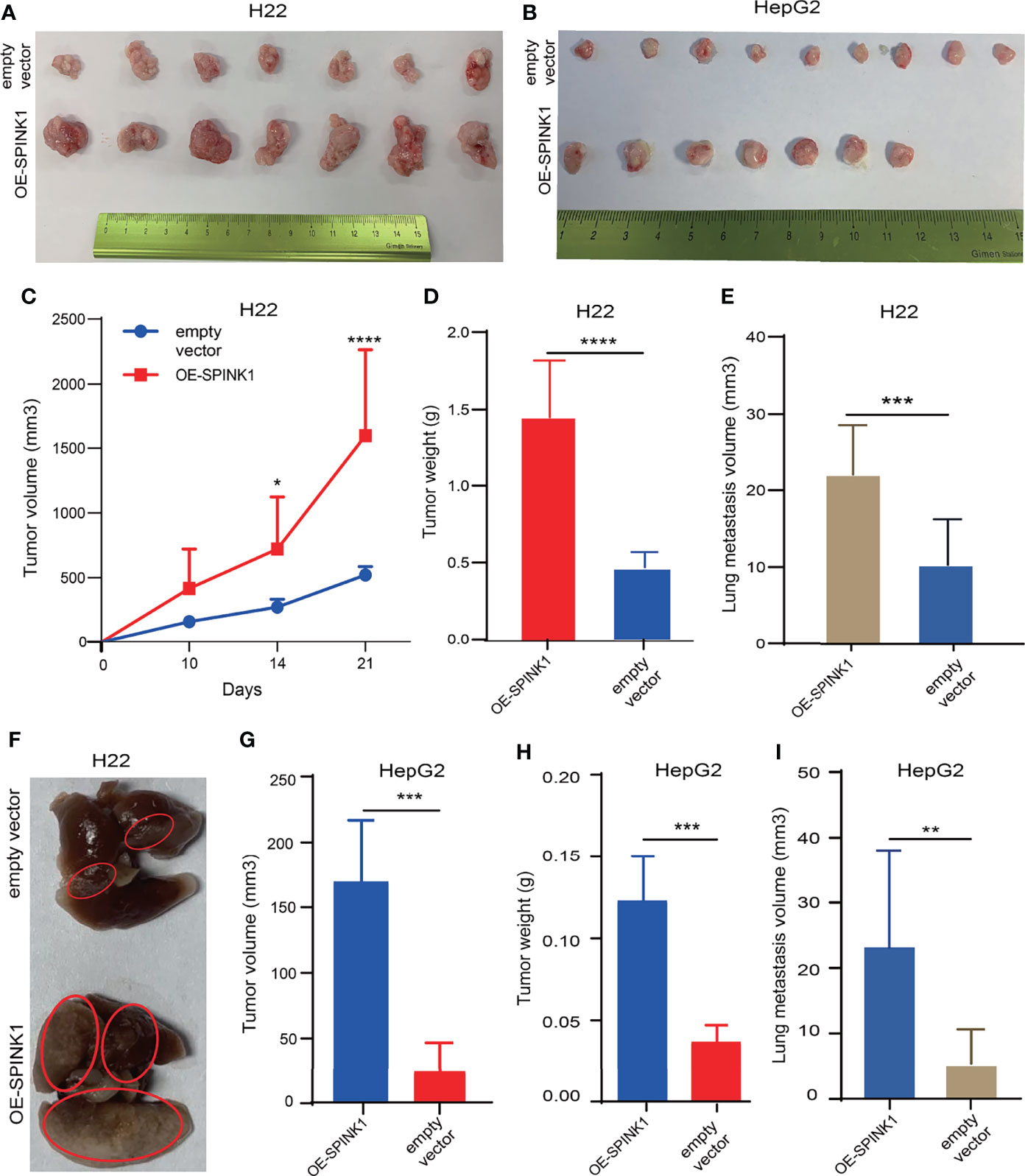
Figure 6 SPINK1-stimulated tumorigenesis , metastasis in vivo experiments. (A, B) Photos for homografts and xenografts generated from subcutaneous injection of murine SPINK1-OE H22 and human SPINK1-OE HepG2 and corresponding control cells in flank region of normal immuno-competent Balb/c mice ( H22: n1control = 7, n2 test = 7; HepG2: n1 control = 9, n2 test = 7). (C, D) Alterations of tumor volume and weight during growth period in Balb/c mice of ‘SPINK1-OE H22’ and control groups. (E, F) Estimated volume and images of metastatic lung nodules at 4 weeks post-injection of SPINK1-OE H22 and respective control H22 cells via murine tail vein. (G, H) Tumor volume and weight measurement in the process of growth in Balb/c mice (SPINK1- OE HepG2 cells). (I) Measurement of lung metastasis via estimated volume of pulmonary tumor nodules (SPINK1-OE HepG2 and control HepG2 cells). Error bars represent mean ± standard deviation of one independent experiment. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
SPINK1 Delivery of Chemoresistance and Anti-Apoptosis
Closely associated with the nature of the tumor, chemoresistance has been reported to be induced in the process of chemotherapy for HCC treatment. As previous data demonstrated the pro-survival feature of SPINK1, we then wondered whether SPINK1 confers resistance to chemotherapeutic drugs in HCC cells. We treated both SPINK1-OE HepG2 and H22 cells and SPINK1-silencing Hep3B cells with or without 2.5 µg/ml 5-fluorouracil (5-FU), an accepted dose used in in vitro chemoresistance-related experiments. The CCK-8 assay showed a significant increase in the number of SPINK1-OE HepG2 and H22 cells with 5-FU treatment relative to the vector control cells, whereas a remarkable decrement could be detected in SPINK1-silencing Hep3B cells treated with 5-FU (p < 0.01; Figures 7A–C) compared to the scramble control. Thus, despite its unknown mechanisms, SPINK1 was able to increase chemoresistance in SPINK1-OE HCC cells in vitro.
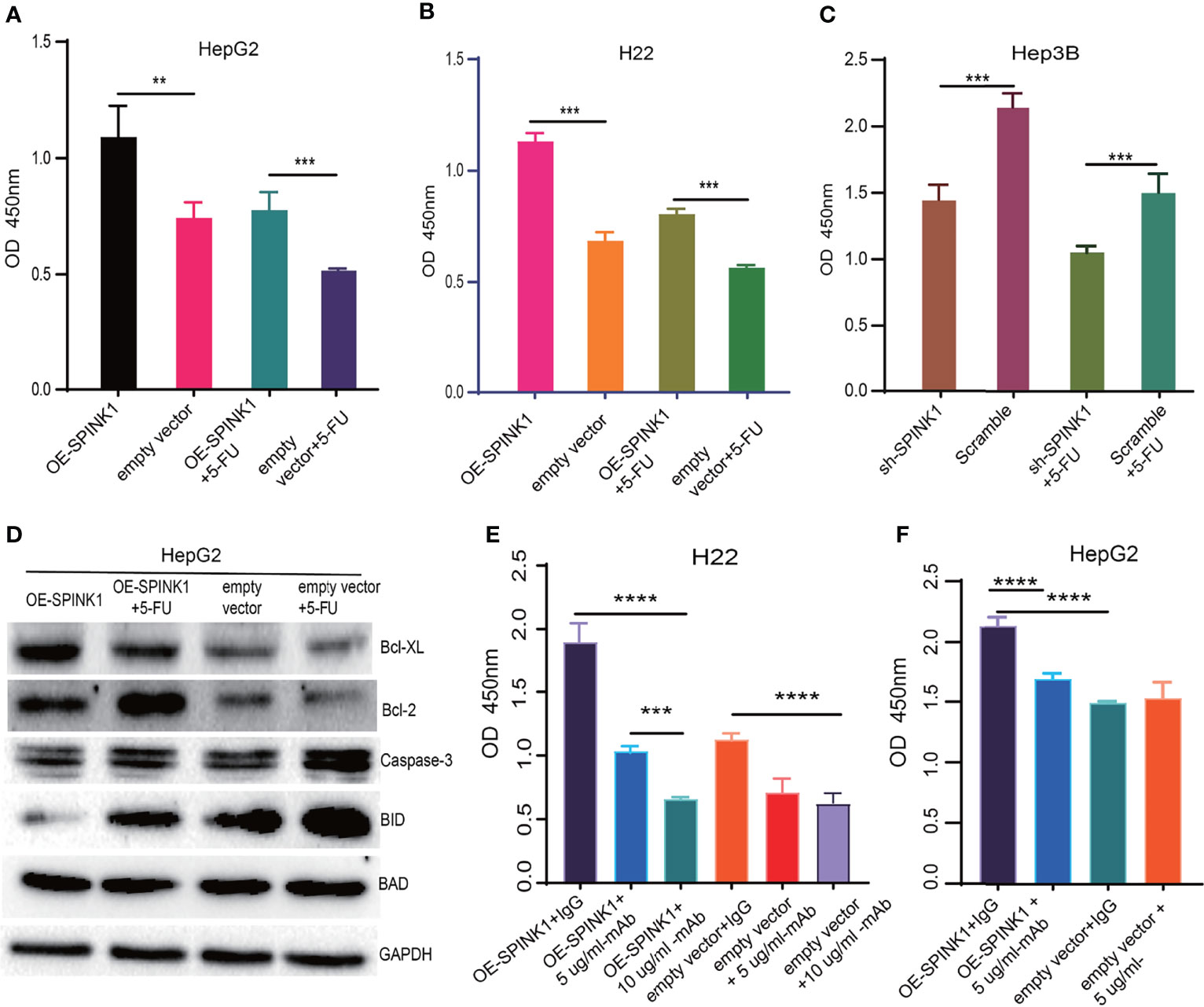
Figure 7 SPINK1-regulated chemoresistance, anti-apoptosis and SPINK1-mAb targeting experiments. (A, B) SPINK1 enhanced chemoresistance in stable SPINK1-OE HepG2 and SPINK1-OE H22 cells following 24 h of exposure to 5-FU; (C) Decreased chemoresistance to 5-FU in SPINK1 knockdown Hep3B cells 24 h post-5-FU treatment. (D) Examination of apoptosis-related molecules including main pro- (Caspase 3, Bid and Bad) and anti-apoptotic regulators (Bcl-XL and Bcl-2) using Western blot in SPINK1-OE HepG2 and empty-vector control cells with or without 5-FU addition. (E, F) Targeting SPINK1 using anti-human SPINK1- mAb in SPINK1- OE H22 and - OE HepG2 cells, with IgG as an isotype control. Error bars represent mean ± standard deviation of independent experiments or the experiments performed in triplicates either in 6-well or 96-well plates. OD, optical density; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To investigate the possible mechanisms through which SPINK1 promoted the growth of HCC and conferred chemoresistance with 5-FU administration, we examined the apoptotic status of SPINK1-OE HepG2 cells with or without 5-FU treatment. A number of apoptotic players, including pro- and anti-apoptotic molecular signals, were analyzed: caspase-3, Bid, Bad, Bcl-2, and Bcl-XL. Interestingly, the anti-apoptotic levels of Bcl-2 and BcL-XL were significantly upregulated in SPINK1-OE and 5-FU-administered HepG2 OE cells compared with the empty vector HepG2 controls, whereas the pro-apoptotic molecule Bid was remarkably downregulated in these cells (Figure 7D). The expression level of caspase-3, one of the key factors inducing apoptosis, remained decreased in SPINK1-OE HepG2 cells treated with or without 5-FU (Figure 7D). All of these results suggest that the imbalance between the anti-apoptotic and pro-apoptotic power may possibly favor the former “anti-aspect” (Figure 7D) in respective circumstances. Together, the SPINK1-associated inhibition of apoptosis may be one of the explanations for the SPINK1-induced overgrowth and chemoresistance in HCC cells.
Targeting SPINK1 in HCC Cells
Analyses of the acquired experimental data provided more understanding on SPINK1. We therefore further explored the likelihood of SPINK1 as a target molecule in targeted therapeutics for HCC, as we verified its targeting exhibited during HCC progression and development. The efficacy of HCC targeted therapy currently remains quite limited primarily due to the difficulty in finding a “precision” molecular target (5). To prove the possibility of SPINK1 as an actionable target, we performed in vitro SPINK1-targeting experiment using anti-human SPINK1 mAb. As expected, the SPINK1 mAb significantly (5 µg/ml) decreased the proliferation of SPINK1-OE HepG2 and H22 cells by 25% and 50%, respectively, as well as caused a proliferative decrement of 40% in H22 empty vector control cells, compared to a control monoclonal IgG antibody (p < 0.01; Figures 7E, F), suggesting considerable efficacy at targeting SPINK1. Interestingly, the increased concentration of the SPINK1 antibody (10 µg/ml) could even significantly inhibit the growth rate by 75% in SPINK1-OE H22 and by 45% in H22 control cells (Figure 7E). These data may indicate the potential of SPINK1 as a promising target molecule in the management of HCC.
Exploration of the Underlying Molecular Signaling Pathways
We wanted to discover the underlying molecular mechanisms that contributed to the tumorigenic features and targetable characteristics of SPINK1 in the in vitro and in vivo experiments. For this purpose, we used the LinkedOmics online tool to determine the co-expression genes that positively or negatively correlated with SPINK1 expression using the RNA-seq data of 371 HCC patients in the TCGA cohort (20). Subsequently, GSEA (GO and KEGG) was performed using this tool. The GO analysis revealed that the most SPINK1-related biological processes to the differentially expressed related genes were “T-cell activation,” “response to tumor necrosis factor,” “immune response regulating signaling pathway,” “humoral immune response,” and “leukocyte migration,” which are almost all required for delivering antitumor immunity (Figure 8A). On the other hand, KEGG signaling pathway analysis showed that these genes were mainly enriched in “cytokine–cytokine receptor interaction,” “complement cascade,” “phagosome,” and “NOD-like receptor signaling pathway,” which are also associated with the delivery of antitumor response (Figure 8B). Thus, both GO and KEGG analyses revealed the positive involvement of SPINK1 in the regulation of an active immune response, which was likely aimed at directly or indirectly resisting HCC.
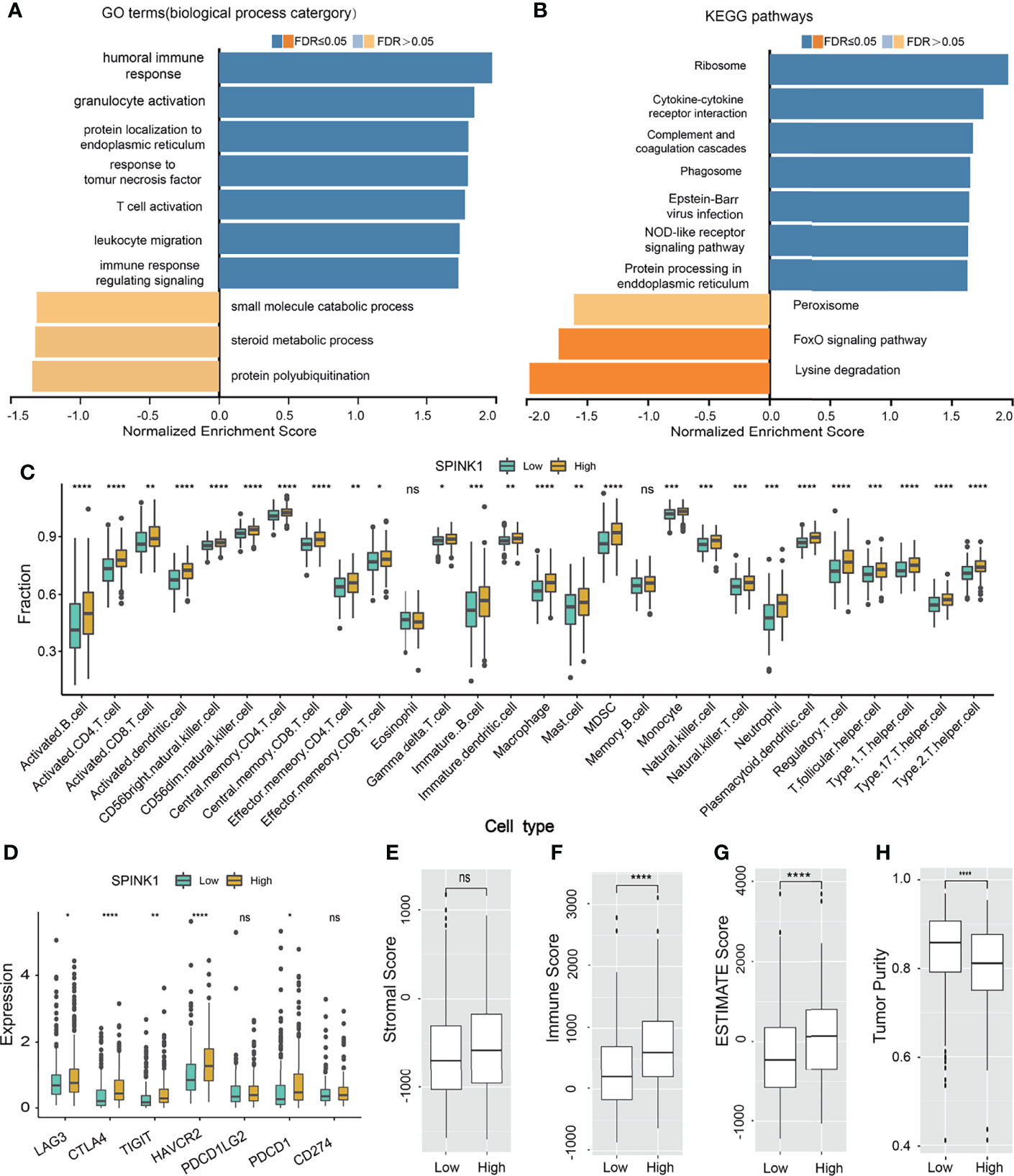
Figure 8 SPINK1-related gene set enrichment analyses (GSEA) and SPINK1, tumor infiltrating microenvironment (TIME) (A, B) SPINK1-associated gene set enrichment analyses (GSEA) of GO terms (biological process categories) and KEGG pathways using HCC RNAseq data from LinkedOmics database. (C) Relationships among SPINK1 expression abundance (low-blue vs high-yellow expression groups) and those of 28 subtypes of tumor infiltrating immune cells in terms of compositions and contents in tissue specimens of HCC patients from TCGA dataset. (D)Correlations among SPINK1 and other seven immune checkpoints molecu1es in SPINK1- low and -high abundance groups of above-mentioned HCC samples. (E–H) Correlations among SPINK1 and scores of ‘stromal’, ‘immune’, ‘estimate’ and ‘tumor purity’ in SPINK1- low and -high expression groups of same cohort of patient samples, respectively. NS, no significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The data from the GO and KEGG analyses inspired the realization that SPINK1 has been engaged in immune response-associated biological processes and signaling pathways in the development of HCC and that it might be a suitable biomarker for the prediction of a “hot” tumor immune microenvironment (TIME) and the response to ICB therapy for HCC. Currently, based on our literature search, there are no efficacious predictive biomarkers for ICB remedy in this regard (5). Thus, we decided to proceed to the last assumption of this investigation: test the possibility of SPINK1 in the prediction of response to HCC immunotherapy.
Correlation Between SPINK1 and Infiltrating Immune Cells
To probe the relationship between SPINK1 and the tumor-infiltrating immune cells in the HCC specimens from the TCGA dataset, which was currently found to be critical in predicting the effectiveness of ICB therapy (36, 37), we analyzed 28 types of infiltrating immune cells within the HCC samples using RNA-seq data. Thereafter, we examined the correlation between the expression of SPINK1 and the intrinsically infiltrating immune cell subtypes. The statistical analyses showed the relative proportions of the 28 types of immune cells in each of the HCC samples (Supplementary Figure S8). The dominantly infiltrating immune cells were activated CD4 T, CD8 T natural killer, effector memory CD8 T, dendritic, and type 1 and 2 T helper cells (Supplementary Figure S8). All these subtypes of immune cells play essential roles in the antigen-specific response to tumor immunity in the host (37). Interestingly, the transcriptomic expression of SPINK1 was positively correlated with most of the infiltrating immune cells, particularly activated CD4 T, CD8 T, effector memory CD4 T, effector CD8 T, and activated dendritic cells (Figure 8C). These immune cells are believed to be involved in CD8+ T-cell-mediated antitumor immune response (36). Collectively, SPINK1 abundance was positively associated with most of the important subtypes of infiltrating immune cells.
Associations Among SPINK1, Immune Checkpoints, and Tumor Mutation Burden
We also tested the likelihood SPINK1 being a predictor of the efficacy of ICB therapy, a burgeoning field in HCC treatment (10). We examined the correlation between the transcriptomic expression of SPINK1 and a number of critical ICPs—PD-1, LAG-3, TIM-3, TIGIT, and CTLA-4—which are the likely signatures for the response to ICB in cancer immunotherapy (21, 38). Unexpectedly, a high level of SPINK1 expression was found to be significantly and positively correlated with the high expressions of PD-1, LAG-3, TIM-3, TIGIT, and CTLA-4 in the tumor samples from the TCGA cohort (Figure 8D). The expression of PD-L1, a recommended potential predictor for immunotherapy vulnerability (39), was not positively associated with that of SPINK1 in the TCGA cohort (Figure 8D). We then analyzed the association between SPINK1 and TMB, a temporarily used marker for the prediction of ICB efficacy. The results showed no correlation between them (Supplementary Figure S9). Altogether, investigation of the RNA-seq data of the HCC samples from the TCGA database revealed a positive correlation between SPINK1 and the expression of some of the important immune checkpoint molecules, suggesting the necessity of reprogramming the immunosuppressive milieu of this category of patients for better prognosis (21).
Patterns of Connection Between SPINK1 and the ESTIMATE and Immune Cell Scores
To further evaluate the association between SPINK1 and TIME, laying the possible foundation to predict the efficacy of ICB therapy in HCC patients, we analyzed a few important parameters from different angles using ESTIMATE software: the “immune score” (proportion of immune cells), “stromal score” (proportion of stromal cells), “ESTIMATE score” (proportions of immune cells plus stromal cells), and “tumor purity” (proportions of tumor cells/the total) of the tumor specimens (40) (Figures 8E–H). Except for the expression of SPINK1 not being associated with the stromal score (non-immune cells) and being negatively correlated with tumor purity, the ESTIMATE and immune scores were positively correlated with the abundance of SPINK1 (Figures 8E–H), suggesting that SPINK1 could be considered as an indicator of the unique reflection of a hot or immune cell-infiltrating tumor microenvironment (41), with the likelihood of a positive response to ICB in HCC patients (42).
Discussion
Beginning with mining an optimal biomarker for the early detection of HCC in this study, we then discovered some of the “targetable” characteristics of SPINK1 through phenotypic analyses of its tumorigenic biological characteristics before finally determining that SPINK1 is also eligible for predicting the response to ICB treatment. The three SPINK1-associated research issues were resolved through the analyses of data derived from genomics, transcriptomics, and proteomics datasets and from real bench-scale experiments.
There have been few reports, if any, on the integration of multi-omics and phenotypic analyses of SPINK1 for the detection of early-stage (I and II) HCC, particularly compared to AFP, a routinely used marker for HCC (43, 44). Holah et al. and Zhu et al. found that the protein expression levels of SPINK1 in sera were remarkably higher in patients with liver cirrhosis and HCC (stages I–IV) than in those with chronic hepatitis (45, 46). With the small number of patient specimens, however, the previously reported data showed no or very limited discriminatory accuracy and calibrated analyses of SPINK1 in the early diagnosis of the disease (44–46).
We used ROC plots and showed that the SPINK1 transcripts displayed a much better discriminatory capability than AFP for early (stages I and II) diagnosis at the transcriptomic level (47). These findings indirectly re-confirmed previous reports of the low sensitivity of serum AFP as a diagnostic criterion for HCC (2–4, 33, 34). Impressively, the sensitivity increased greatly when AFP was replaced by SPINK1 or when SPINK1 was combined with AFP in the prediction model. Moreover, the AUC, specificity, and sensitivity of the SPINK1-related models for stage I and II HCC were better than those for stage I–IV patients of the same cohorts, implying the superiority of SPINK1 in the early diagnosis of HCC (stages I and II) than that in the late stages (III and IV).
It is worth noting that the calibration curves of the SPINK1-related models in the analysis of the first validation dataset were well fitted, indicating the generality of SPINK1 as a better diagnostic biomarker for HCC. The results of the Hosmer–Lemeshow test in the second validation dataset showed an increased p-value (>0.05), which indicated an overall better fit of the SPINK1-related models to the observations.
Our corresponding findings for SPINK1 were contrary to those of a previous report by Yan and Chen describing SPINK1 overexpression as only correlated with late-stage HCC (stages IIIB to IV) (24), possibly due to the small number of samples included in their study. Some researchers have also stated that the addition of one or more serum biomarkers to AFP, such as des-gamma-carboxy prothrombin (DCP), glypican-3 (GPC3), and Golgi protein 73 (GP73), does not significantly improve its diagnostic accuracy for HCC (48). In contrast, our data clearly demonstrated that SPINK1 and the combination of SPINK1 and AFP both showed more beneficial outcomes than AFP alone using the tissue samples, implying the possible advantages of using SPINK1 over the other currently recommended biomarkers for early detection (48).
For the first time, to our knowledge, proteomics was integrated with transcriptomics datasets for the analysis of the early detection of HCC using SPINK1. It was noteworthy that the discrimination curve of AFP was poor, possibly due to the selection of early-stage (I and II) tumor samples, for which the detection using AFP was not sensitive enough. AFP also exhibited poor calibration on the prediction curve in the proteomics dataset. Although the discriminatory ability of the SPINK1-associated models was much better than that of AFP, it was not as good as that from the transcriptomics study. One of the reasons for this phenomenon might be related to the limited number of samples from early-stage patients. Nevertheless, all the other indices including AIC, BIC, IDI, and NRI still showed the same positive trends as those of transcriptomics in the proteomics analyses.
In fact, the switch from “early detection” to “targeted therapy” naturally occurred out of the continuously obtained data during further phenotype analyses of the tumorigenic characteristics of SPINK1. We did not find reports on the targeting of SPINK1 in HCC within our PubMed search, although Ateeq et al. experimentally showed that the human anti-SPINK1 antibody against prostate cancer could reduce tumor growth (49). Currently, the efficacy of targeted therapy for HCC remains quite limited primarily due to the difficulty in finding a “precision” molecular target (5). Our in vitro and in vivo experimental data convincingly demonstrated the role of SPINK1 in tumor growth, invasion, chemoresistance, anti-apoptosis, and metastasis and the favorable effect of the anti-SPINK1 antibody on the anti-proliferation of HCC cells. These findings will likely lay the foundation for providing a new targetable molecule in order to improve the efficacy of targeted treatment for the management of HCC.
The SPINK1-related immune response biological processes and signaling routes, including T-cell activation based on GO and KEGG analyses, led us to focus on the next ICB-associated concern. Currently, the biomarkers for the identification of ICB response remain critically important and challenging in the management of HCC (50). To our knowledge, for the first time, we have shown SPINK1 as a potentially practicable and accessible biomarker that could be utilized for the prediction of the presence of a hot immunogenic environment (40) in HCC tissues and may likely be useful for starting ICB treatment. Our data clearly showed that the high abundance of SPINK1 corresponded with the high levels of CD4+ and CD8+ effector T lymphocytes, as well as activated dendritic and natural killer cells in the same tumor samples. There has been a general consensus that tumor-infiltrating CD4+ and CD8+ effector T lymphocytes are mediators of the response to ICB and that these cells are primed through tumor antigen presentation by dendritic cells, while natural killer cells play important roles in the immune surveillance of tumors (10). Specifically, the overexpression of SPINK1 in the samples suggested the possibility of a hot tumor, in which effector T cells may play the central part in the antitumor response; therefore, patients with a hot TIME are more likely to respond to immunotherapy (40). Interestingly, the high abundance of SPINK1 was also exclusively associated with the high levels of ICPs in our analyses, and these ICPs were thought to be involved in the mechanisms by which cancer cells disguise themselves as regular components of the human body (51). The assumption was that inhibitors against ICPs can reinvigorate anticancer immunity, which are currently the standard of care in a number of malignancies (52). Thus, preexisting abundant ICPs are important signals for the delivery of ICB therapy (53). Moreover, GSEA (GO) revealed that SPINK1 participated primarily in the biological process of antitumor-related immune response, including response to T-cell activation, and humoral immune response, which was associated with favorable outcomes when individuals undergo immunotherapy (54). Thus, the high expression of SPINK1 may be an accurate parameter to indicate the appropriate time point for ICB intervention.
Notably, there was no positive correlation between SPINK1 and PD-L1 in our study. Havel et al. reported that a number of lung cancer patients with PD-L1-negative tumors still responded to ICB and that, in some tumor types, the expression of PD-L1 did not correlate with response to treatment (55). El-Khoueiry et al. also showed that the tumoral expression of PD-L1 was not predictive of the response to nivolumab or pembrolizumab in HCC treatment (56). Whether PD-L1 could be considered as a marker for the prediction of ICB vulnerability is a topic for further investigation in HCC management.
In our analyses, the group with a high expression of SPINK1 did not reveal a positive correlation with TMB, which may increase the likelihood of ICB response, as described by other researchers in cancer control (55). This discrepancy was possibly due to the general low TMB in HCC and the unsuitability of TMB as a biomarker to predict ICB response in this regard (57).
The data on the immune, stromal, and ESTIMATE scores and tumor purity were generally consistent with previous results, indicating that the high abundance of SPINK1 suggests a “hot” state of accumulation of highly infiltrating immune cells, termed a “better” immune score. Although ICB therapeutics including ipilumimab and nivolumab alter the T-lymphocyte checkpoint control and may be particularly efficacious in tumors with intrinsically high levels of infiltrating leukocytes, further subclassification of the lineage features of infiltrating cells, such as discriminating between various leukocytes, may possibly represent a more actionable characteristic of clinical application (22). Specifically, more data are still needed to determine whether the ESTIMATE immune score is a more appropriate parameter for the prediction of ICB response.
Some limitations in the analyses of this investigation should be noted. Firstly, we lacked clinical data with respect to the confirmed significance of the level of SPINK1 in serum for the early detection of patients with HCC, which is the most accessible and practical test in the laboratory for cancer assessment. Secondly, the molecular targeting experiment using the anti-human SPINK1 antibody was performed only at the cellular level in vitro rather than in an in vivo rodent model. Thirdly, the preliminary data derived from the GO and KEGG analyses deciphered in part why SPINK1 could be a potential biomarker for the prediction of an immunologically hot tumor, which is possibly suitable for subsequent ICB therapy response in HCC (40). However, there were no controlled clinical data regarding the association between transcriptomic SPINK1 and the response outcomes of patients with HCC during the ICB treatment. Further experimental and clinical evidence needs to be accumulated in order to reach definite conclusions. Fourthly, all of the transcriptomics and proteomics data were collected from varied batches of patients with HCC who may even be from the same or different hospitals, meaning that we were not able to avoid selection bias and imbalances in this study, which may have resulted in the precision of the obtained results to some extent (58).
Through a number of dry and wet experiments, we have demonstrated that SPINK1 could act as a potential biomarker for the early detection, targeted therapeutics, and the prediction of ICB response in the management of HCC. Moreover, the combination of targeted therapeutics and ICB immunotherapy has become the forefront in HCC management (5). Thus, aside from being a “target,” there is a rational expectation to consider SPINK1-oriented ICB plus SPINK1-targeting therapeutics in the future. However, this temporary conclusion requires prospective validation instead of using tumor tissue specimens, a routinely accessible serum marker used in large-scale clinical trials, which should be applied in this regard before reaching definite conclusions. Hopefully, the application of SPINK1 would pave a new avenue for the diagnosis and treatment of HCC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Committee of Dalian Medical University.
Author Contributions
JJ and JW: conception and design and manuscript writing. JJ and LG: performed the experiments. JJ: collection and assembly of data. JJ, LG, YL, ZY, YW, XG, RM, RL, TL, ZT, and JW: data analysis and interpretation. All authors are accountable for all aspects of the work. All authors contributed to the article and approved the submitted version.
Funding
This study was partially sponsored by Dalian Medical University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Jessica Zucman-Rossi, Prof. Sandrine Imbeaud, and Dr. Jean-Charles Nault for kindly sending us the GSE62232-related clinical data files.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.923031/full#supplementary-material
References
1. Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterol (2019) 156:477–91. doi: 10.1053/j.gastro.2018.08.065
2. Choi J, Kim GA, Han S, Lee W, Chun S, Lim YS. Longitudinal Assessment of Three Serum Biomarkers to Detect Very Early-Stage Hepatocellular Carcinoma. Hepatol (2019) 69:1983–94. doi: 10.1002/hep.30233
3. De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R. Novel Biomarkers in Hepatocellular Carcinoma. Dig Liver Dis (2018) 50:1115–23. doi: 10.1016/j.dld.2018.08.019
4. Harris PS, Hansen RM, Gray ME, Massoud OI, McGuire BM, Shoreibah MG. Hepatocellular Carcinoma Surveillance: An Evidence-Based Approach. World J Gastroenterol (2019) 25:1550–9. doi: 10.3748/wjg.v25.i13.1550
5. Llovet JM, Montal R, Sia D, Finn RS. Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat Rev Clin Oncol (2018) 15:599–616. doi: 10.1038/s41571-018-0073-4
6. Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome Sequencing of Hepatocellular Carcinomas Identifies New Mutational Signatures and Potential Therapeutic Targets. Nat Genet (2015) 47:505–11. doi: 10.1038/ng.3252
7. Rebouissou S, Nault J-C. Advances in Molecular Classification and Precision Oncology in Hepatocellular Carcinoma. J Hepatol (2020) 72:215–29. doi: 10.1016/j.jhep.2019.08.017
8. Pinter M, Jain RK, Duda DG. The Current Landscape of Immune Checkpoint Blockade in Hepatocellular Carcinoma. JAMA Oncol (2021) 7:113–23. doi: 10.1001/jamaoncol.2020.3381
9. Yau T, Park J-W, Finn RS, Cheng A-L, Mathurin P, Edeline J, et al. Nivolumab Versus Sorafenib in Advanced Hepatocellular Carcinoma (CheckMate 459): A Randomised, Multicentre, Open-Label, Phase 3 Trial. Lancet Oncol (2022) 23:77–90. doi: 10.1016/s1470-2045(21)00604-5
10. Champiat S, Ferrara R, Massard C, Besse B, Marabelle A, Soria J-C, et al. Hyperprogressive Disease: Recognizing a Novel Pattern to Improve Patient Management. Nat Rev Clin Oncol (2018) 15:748–62. doi: 10.1038/s41571-018-0111-2
11. Scheiner B, Kirstein MM, Hucke F, Finkelmeier F, Schulze K, von Felden J, et al. Programmed Cell Death Protein-1 (PD-1)-Targeted Immunotherapy in Advanced Hepatocellular Carcinoma: Efficacy and Safety Data From an International Multicentre Real-World Cohort. Aliment Pharmacol Ther (2019) 49:1323–33. doi: 10.1111/apt.15245
12. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia (2004) 6:1–6. doi: 10.1016/s1476-5586(04)80047-2
13. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002
14. Gan X, Luo Y, Dai G, Lin J, Liu X, Zhang X, et al. Identification of Gene Signatures for Diagnosis and Prognosis of Hepatocellular Carcinomas Patients at Early Stage. Front Genet (2020) 11:857. doi: 10.3389/fgene.2020.00857
15. Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell (2019) 179:561–77.e22. doi: 10.1016/j.cell.2019.08.052
16. Zhang J, Wang D, Hu N, Wang Q, Yu S, Wang J. The Construction and Proliferative Effects of a Lentiviral Vector Capable of Stably Overexpressing SPINK1 Gene in Human Pancreatic Cancer AsPC-1 Cell Line. Tumor Biol (2015) 37:5847–55. doi: 10.1007/s13277-015-4405-z
17. Wang J, Ohmuraya M, Suyama K, Hirota M, Ozaki N, Baba H, et al. Relationship of Strain-Dependent Susceptibility to Experimentally Induced Acute Pancreatitis With Regulation of Prss1 and Spink3 Expression. Lab Invest (2010) 90:654–64. doi: 10.1038/labinvest.2010.44
19. Liu H, Ye SL, Xue Q. The Establishment of Rat Liver Tumor Model With Spontaneous Lung Metatasis in Nude Mice. Clin J Exp Surg (2004) 21:102.
20. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46:D956–D63. doi: 10.1093/nar/gkx1090
21. Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring Immune-Checkpoint Blockade: Response Evaluation and Biomarker Development. Nat Rev Clin Oncol (2017) 14:655–68. doi: 10.1038/nrclinonc.2017.88
22. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture From Expression Data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612
23. Zemek RM, Chin WL, Nowak AK, Millward MJ, Lake RA, Lesterhuis WJ. Sensitizing the Tumor Microenvironment to Immune Checkpoint Therapy. Front Immunol (2020) 11:223. doi: 10.3389/fimmu.2020.00223
24. Yan D, Chen Y. Tumor Mutation Burden (TMB)-Associated Signature Constructed to Predict Survival of Lung Squamous Cell Carcinoma Patients. Sci Rep (2021) 11:9020. doi: 10.1038/s41598-021-88694-7
25. Mayakonda A, Lin D-C, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res (2018) 28:1747–56. doi: 10.1101/gr.239244.118
26. Demler OV, Pencina MJ, Cook NR, D'Agostino RB. Asymptotic Distribution of ∆AUC, NRIs, and IDI Based on Theory of U-Statistics. Stat Med (2017) 36:3334–60. doi: 10.1002/sim.7333
27. Vrieze SI. Model Selection and Psychological Theory: A Discussion of the Differences Between the Akaike Information Criterion (AIC) and the Bayesian Information Criterion (BIC). Psychol Methods (2012) 17:228–43. doi: 10.1037/a0027127
28. Van Calster B, Wynants L, Verbeek JFM, Verbakel JY, Christodoulou E, Vickers AJ, et al. Reporting and Interpreting Decision Curve Analysis: A Guide for Investigators. Eur Urol (2018) 74:796–804. doi: 10.1016/j.eururo.2018.08.038
29. Kerr KF, Brown MD, Zhu K, Janes H. Assessing the Clinical Impact of Risk Prediction Models With Decision Curves: Guidance for Correct Interpretation and Appropriate Use. J Clin Oncol (2016) 34:2534–40. doi: 10.1200/jco.2015.65.5654
30. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia (2007) 9:166–80. doi: 10.1593/neo.07112
31. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-Based Map of the Human Proteome. Science (2015) 347:6220. doi: 10.1126/science.1260419
32. Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An Online Survival Analysis Tool to Rapidly Assess the Effect of 22,277 Genes on Breast Cancer Prognosis Using Microarray Data of 1,809 Patients. Breast Cancer Res Treat (2009) 123:725–31. doi: 10.1007/s10549-009-0674-9
33. Bruix J, Sherman M. Sherman: Management of Hepatocellular Carcinoma: An Update. Hepatol (2011) 53:1020–2. doi: 10.1002/hep.24199
34. Galle PR, Foerster F, Kudo M, Chan SL, Llovet JM, Qin S, et al. Biology and Significance of Alpha-Fetoprotein in Hepatocellular Carcinoma. Liver Int (2019) 39:2214–29. doi: 10.1111/liv.14223
35. Alba AC, Agoritsas T, Walsh M, Hanna S, Iorio A, Devereaux PJ, et al. Discrimination and Calibration of Clinical Prediction Models. Jama (2017) 318:1377–84. doi: 10.1001/jama.2017.12126
36. Farhood B, Najafi M, Mortezaee K. CD8+cytotoxic T Lymphocytes in Cancer Immunotherapy: A Review. J Cell Physiol (2018) 234:8509–21. doi: 10.1002/jcp.27782
37. Petitprez F, Meylan M, de Reyniès A, Sautès-Fridman C, Fridman WH. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front Immunol (2020) 11:784. doi: 10.3389/fimmu.2020.00784
38. D'Alessio A, Rimassa L, Cortellini A, Pinato DJ. PD-1 Blockade for Hepatocellular Carcinoma: Current Research and Future Prospects. J Hepatocell Carcinoma (2021) 8:887–97. doi: 10.2147/jhc.S284440
39. Zeng Z, Yang B, Liao Z. Biomarkers in Immunotherapy-Based Precision Treatments of Digestive System Tumors. Front Oncol (2021) 11:650481. doi: 10.3389/fonc.2021.650481
40. Xiang Z, Zhong C, Chang A, Ling J, Zhao H, Zhou W, et al. Immune-Related Key Gene CLDN10 Correlates With Lymph Node Metastasis But Predicts Favorable Prognosis in Papillary Thyroid Carcinoma. Aging (2020) 12:2825–39. doi: 10.18632/aging.102780
41. Galon J, Bruni D. Approaches to Treat Immune Hot, Altered and Cold Tumours With Combination Immunotherapies. Nat Rev Drug Discovery (2019) 18:197–218. doi: 10.1038/s41573-018-0007-y
42. Paijens ST, Vledder A, de Bruyn M, Nijman HW. Tumor-Infiltrating Lymphocytes in the Immunotherapy Era. Cell Mol Immunol (2020) 18:842–59. doi: 10.1038/s41423-020-00565-9
43. Hass HG, Jobst J, Scheurlen M, Vogel U, Nehls O. Gene Expression Analysis for Evaluation of Potential Biomarkers in Hepatocellular Carcinoma. Anticancer Res (2015) 35:2021–8.
44. Huang K, Xie W, Wang S, Li Q, Wei X, Chen B, et al. High SPINK1 Expression Predicts Poor Prognosis and Promotes Cell Proliferation and Metastasis of Hepatocellular Carcinoma. J Invest Surg (2020) 34:1011–20. doi: 10.1080/08941939.2020.1728443
45. Holah NS, El-Azab DS, Aiad HAES, Sweed DMM. The Diagnostic Role of SPINK1 in Differentiating Hepatocellular Carcinoma From Nonmalignant Lesions. Appl Immunohistochem Mol Morphol (2017) 25:703–11. doi: 10.1097/pai.0000000000000363
46. Zhu C, Han H, Li J, Xu L, Liu F, Wu K, et al. Hepatitis B Virus X Protein-Induced Serine Protease Inhibitor Kazal Type 1 is Associated With the Progression of HBV-Related Diseases. BioMed Res Int (2019) 2019:1–7. doi: 10.1155/2019/9321494
47. Beudeker BJB, Boonstra A. Boonstra: Circulating Biomarkers for Early Detection of Hepatocellular Carcinoma. Therap Adv Gastroenterol (2020) 13:1756284820931734. doi: 10.1177/1756284820931734
48. Wu M, Liu Z, Li X, Zhang A, Li N. Dynamic Changes in Serum Markers and Their Utility in the Early Diagnosis of All Stages of Hepatitis B-Associated Hepatocellular Carcinoma. Onco Targets Ther (2020) 13:827–40. doi: 10.2147/ott.S229835
49. Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, et al. Therapeutic Targeting of SPINK1-Positive Prostate Cancer. Sci Transl Med (2011) 3:72ra17. doi: 10.1126/scitranslmed.3001498
50. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. Atezolizumab Plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med (2020) 382:1894–905. doi: 10.1056/NEJMoa1915745
51. Li B, Chan HL, Chen P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr Med Chem (2019) 26:3009–25. doi: 10.2174/0929867324666170804143706
52. Liu X, Hogg GD, DeNardo DG. Rethinking Immune Checkpoint Blockade: ‘Beyond the T Cell’. J Immunother Cancer (2021) 9:e001460. doi: 10.1136/jitc-2020-001460
53. Topalian Suzanne L, Drake Charles G, Pardoll Drew M. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell (2015) 27:450–61. doi: 10.1016/j.ccell.2015.03.001
54. Bruno TC. New Predictors for Immunotherapy Responses Sharpen Our View of the Tumour Microenvironment. Nature (2020) 577:474–6. doi: 10.1038/d41586-019-03943-0
55. Havel JJ, Chowell D, Chan TA. The Evolving Landscape of Biomarkers for Checkpoint Inhibitor Immunotherapy. Nat Rev Cancer (2019) 19:133–50. doi: 10.1038/s41568-019-0116-x
56. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in Patients With Advanced Hepatocellular Carcinoma (CheckMate 040): An Open-Label, non-Comparative, Phase 1/2 Dose Escalation and Expansion Trial. Lancet (2017) 389:2492–502. doi: 10.1016/s0140-6736(17)31046-2
57. Ang C, Klempner SJ, Ali SM, Madison R, Ross JS, Severson EA, et al. Prevalence of Established and Emerging Biomarkers of Immune Checkpoint Inhibitor Response in Advanced Hepatocellular Carcinoma. Oncotarget (2019) 10:4018–25. doi: 10.18632/oncotarget.26998
Keywords: serine peptidase inhibitor Kazal type 1 (SPINK1), immune checkpoint blockade (ICB), hepatocellular carcinoma (HCC), tumor immune microenvironment (TIME), biomarker
Citation: Jia J, Ga L, Liu Y, Yang Z, Wang Y, Guo X, Ma R, Liu R, Li T, Tang Z and Wang J (2022) Serine Protease Inhibitor Kazal Type 1, A Potential Biomarker for the Early Detection, Targeting, and Prediction of Response to Immune Checkpoint Blockade Therapies in Hepatocellular Carcinoma. Front. Immunol. 13:923031. doi: 10.3389/fimmu.2022.923031
Received: 18 April 2022; Accepted: 01 June 2022;
Published: 18 July 2022.
Edited by:
Apostolos Zaravinos, European University Cyprus, CyprusReviewed by:
Nooran Elleboudy, Ain Shams University, EgyptRunzi Sun, University of Pittsburgh, United States
Copyright © 2022 Jia, Ga, Liu, Yang, Wang, Guo, Ma, Liu, Li, Tang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeyao Tang, MjIwNzU5NzA4OEBxcS5jb20=; Jun Wang, MTEwOTkyNDY4OUBxcS5jb20=
 Jianlong Jia1
Jianlong Jia1 Yang Liu
Yang Liu Jun Wang
Jun Wang