- 1Department of Pharmacy, The Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China
- 2Department of Clinical Laboratory, Guizhou Provincial Orthopedic Hospital, Guiyang City, China
- 3The Second Clinical Medical School of Nanjing Medical University, Nanjing Medical University, Nanjing, China
- 4The Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Henan Province Engineering Research Center of Artificial Intelligence and Internet of Things Wise Medical, Zhengzhou, China
The gut microbiota is composed of a large number of microorganisms with a complex structure. It participates in the decomposition, digestion, and absorption of nutrients; promotes the development of the immune system; inhibits the colonization of pathogens; and thus modulates human health. In particular, the relationship between gut microbiota and gastrointestinal tumor progression has attracted widespread concern. It was found that the gut microbiota can influence gastrointestinal tumor progression in independent ways. Here, we focused on the distribution of gut microbiota in gastrointestinal tumors and further elaborated on the impact of gut microbiota metabolites, especially short-chain fatty acids, on colorectal cancer progression. Additionally, the effects of gut microbiota on gastrointestinal tumor therapy are outlined. Finally, we put forward the possible problems in gut microbiota and the gastrointestinal oncology field and the efforts we need to make.
1 Introduction
The gut microbiota includes the organisms living in the gastrointestinal tract and is a large and complicated ecosystem. As a rough estimate, the total weight of adult human gut microbiota is 1.5 kg, consisting of 3.9 × 1013 microorganisms/ml of luminal content (1). The gut microbiota mainly includes bacteria, fungi, protozoa, archaea, and viruses, among which bacteria are dominant (2). The composition of the human gut microbiota is not homogeneous (3). According to the differences in intestinal pH and oxygen content, bacterial groups were distributed at different locations, and bacterial concentrations increased from stomach to rectum. During early development, the gut microbiota undergoes a systematic turnover of species until a stable adult state is reached (4). Neonates have very dynamic changes in the composition of their gut microbiota (5). Although the composition of the gut microbiota is affected by several factors, such as age, diet, and lifestyle, it is relatively stable in adults under normal physiological conditions (6).
The gut microbiota has a strong metabolic capacity and is deemed an important “metabolic organ,” which plays an important role in host digestion, nutrient absorption, metabolism, immunity, and other processes (3). Recently, with the rapid progression of sequencing technology, gut microbiota has been confirmed to be involved in the occurrence and development of various tumors. In particular, changes in gut microbial metabolites, such as short-chain fatty acids (SCFAs), tryptophan metabolites, and secondary bile acids, may have broad implications for the formation and progression of various tumors (7–9). To our knowledge of the role of the tumor microenvironment in cancer progression and treatment, the impact of the gut microbiota on tumor immunity has obtained increasing attention. Changes in gut microbiota can affect not only tumor immunotherapy but also chemotherapy treatment (10). Therefore, targeting gut the microbiota could serve as a novel therapeutic option.
The relationship between gut microbiota and tumors has become an important issue in multiple studies. In this review, we only focused on the bacteria and reviewed the potential roles of gut microbiota and microbial metabolites in gastrointestinal cancer. In addition, we discussed the therapeutic potential of gut microbiome in gastrointestinal cancer.
2 Gut Microbiota and Gastrointestinal Cancer
The carcinogenic process usually consists of three stages: initiation, progression, and metastasis. It can be influenced by the oncogenic effects of gut microbiota and their products, modulating circulatory metabolite levels that might inhibit or promote tumor growth, inducing pro-inflammatory and immunosuppressive effects (11, 12). Below, we focused on bacteria in gastric cancer (GC) and colorectal cancer (CRC).
2.1 Gastric Cancer
GC is the fifth most common cancer and the fourth leading cause of cancer-associated death worldwide (13). As illustrated by the latest WHO data, the incidence of GC exhibits significant regional differences (13). East Asia, Eastern Europe, and South America are hotspots for GC incidence and mortality (Figure 1). The risk factors for GC are associated with various etiologies, including Helicobacter pylori (H. pylori) infection, high salt intake, age, and a low fruit and vegetable diet. Chronic infection by H. pylori is the most well-described cause of noncardia GC (14).
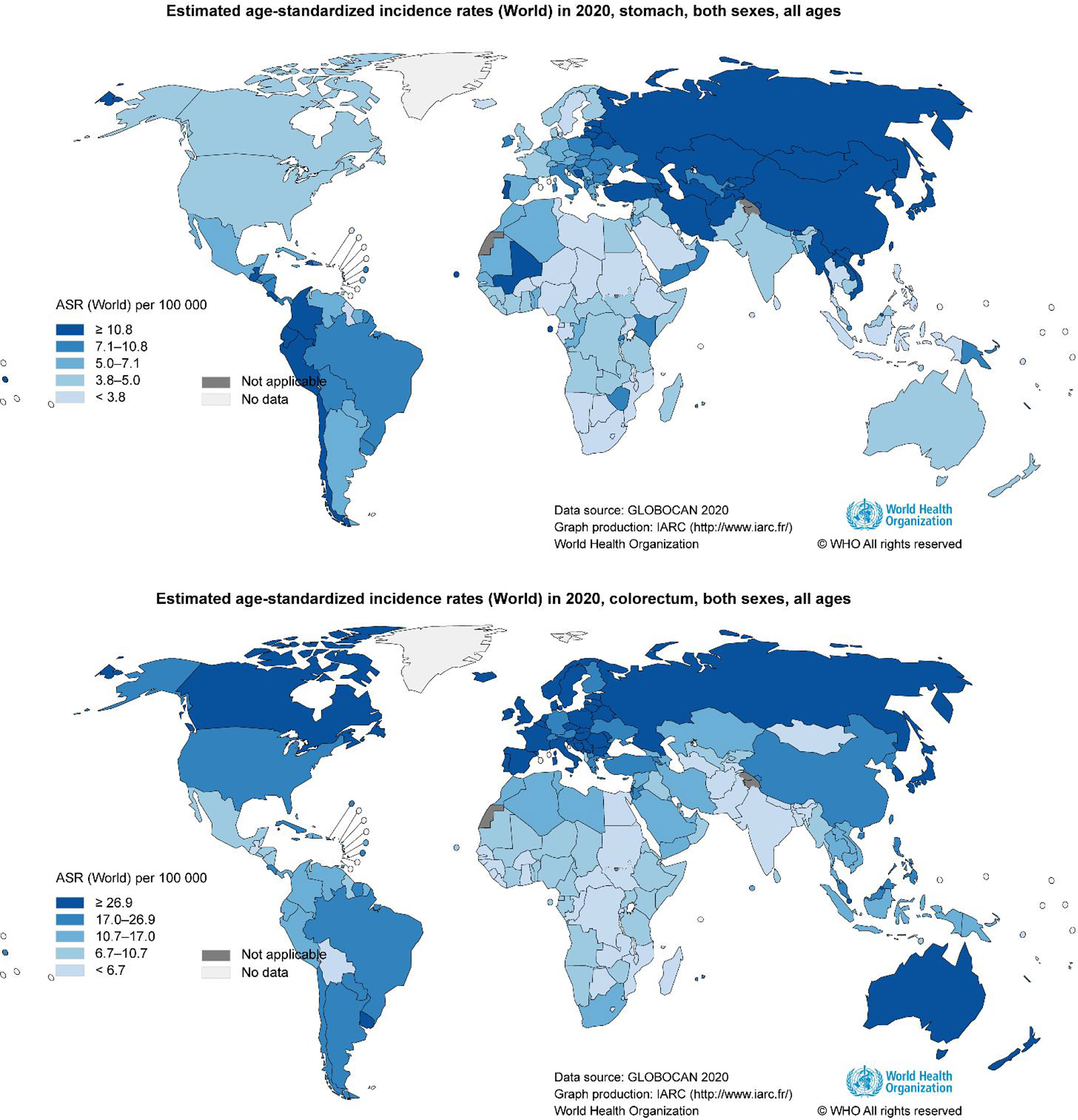
Figure 1 Geographical variation in gastrointestinal cancer rates. Data from the WHO demonstrating the high incidence of gastrointestinal cancer (13).
2.1.1 H. pylori
H. pylori is an aerobic Gram-negative bacterium, with catalase, urease, and oxidase activities (15). H. pylori has been recognized as a class I carcinogen by the World Health Organization. H. pylori infection-induced GC is associated with bacterial virulence, host genetic polymorphism, and environmental factors (14). H. pylori infection increases gastric pH, changes the composition of gastric microbiota, and then creates favorable niches for bacterial colonization.
H. pylori possibly directly affects carcinogenesis by vacuolating cytotoxin A (VacA) and cytotoxin-associated gene A (CagA) (16). On the one hand, all H. pylori strains contain a single chromosomal VacA gene, encoding a 140 kDa VacA protein, which is an intracellular-acting and channel-forming toxin. Through impairing host endolysosomal trafficking, VacA induces the accumulation of dysfunctional lysosomes and autophagosomes (17). Additionally, VacA can increase reactive oxygen species, mitochondrial damage, and inflammation (18). Furthermore, VacA may attenuate the host immune response, thereby facilitating persistent H. pylori colonization in the stomach (18). However, most H. pylori strains possess a CagA pathogenicity island encoding a 120–140 kDa CagA protein, which is a strain-specific protein and is transferred into host cells by the type IV secretion system. CagA inhibits the apoptotic pathway of epithelial cells and causes morphological aberrations, namely, loss of cell polarity and adhesion, increased cell motility, scattering, and elongation (19). Additionally, CagA is deemed a key oncogene in human chronic gastritis and gastric ulcer, MALT lymphoma, and GC (20).
Although H. pylori induces chronic gastritis and peptic and duodenal ulcers and is linked to more than 90% of GC cases, only approximately 1 to 3% of infected individuals progress to GC (21–23). During a 7.5-year follow-up randomized controlled trial, Wong et al. did not find a distinctive benefit of H. pylori eradication when compared with placebo treatment (24). However, in the subgroup of H. pylori carriers without precancerous lesions, Wong et al. found that eradication treatment of H. pylori significantly decreased GC progression (24). In a 5-year prospective study of 1,755 patients, Rugge et al. confirmed that eradication treatment of H. pylori did not abolish the risk of neoplastic progression in subjects with advanced stages (III–IV) (25). Consistently, eradication of H. pylori was shown to still be effective in a subset of early GC patients by minimizing the risk of metachronous GC (26, 27). Furthermore, Guo et al. suggested that H. pylori eradication treatment could restore the gastric microbiota to a similar status as negative subjects and may exert more beneficial effects on the gut microbiota, such as downregulation of drug-resistance and probiotic enrichment (28). Thus, H. pylori eradication treatment seems to counteract the risk of GC transformation, but the magnitude depends on the degree of preexisting damage at the time of eradication.
2.1.2 Non-H. pylori Gastric Bacteria
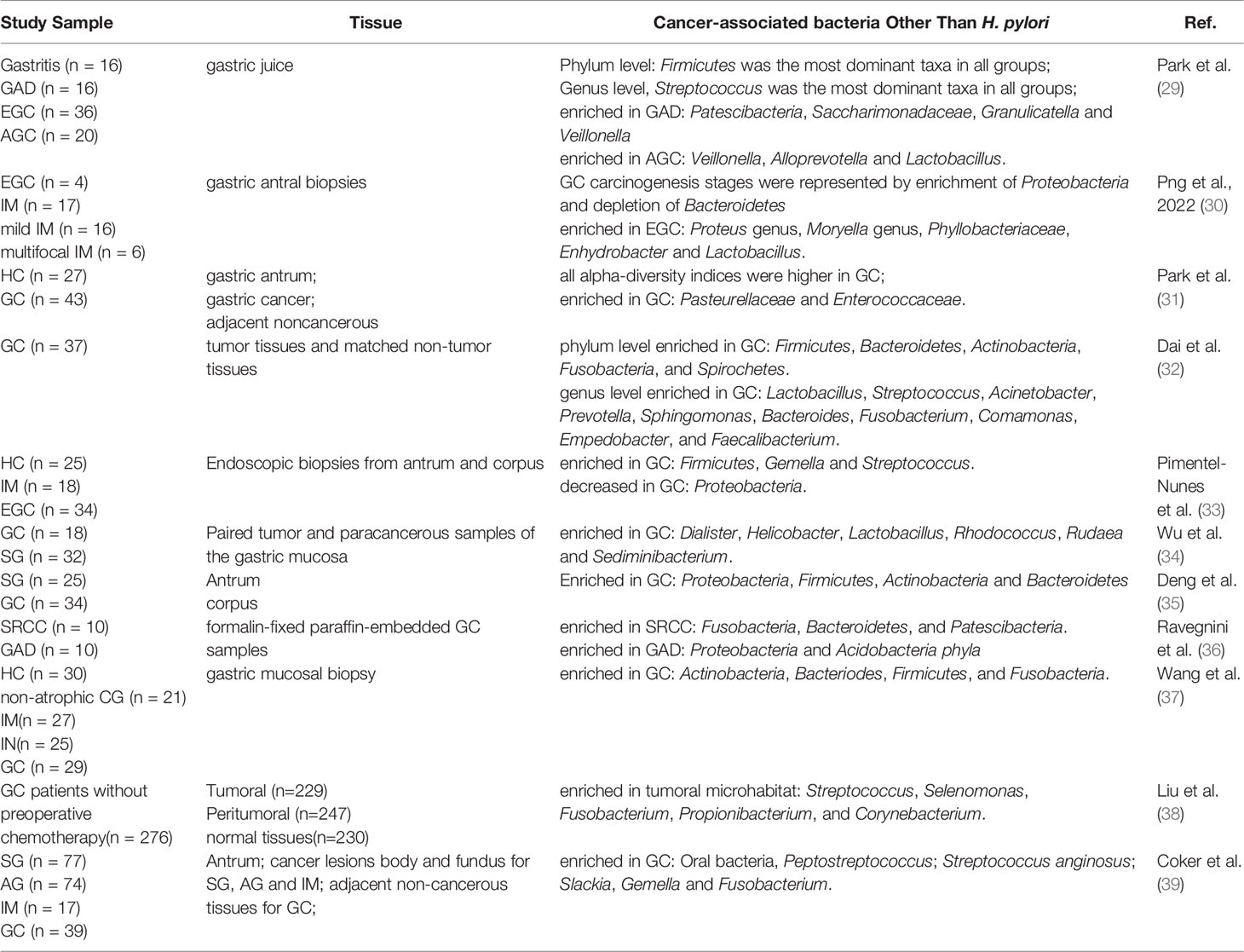
With the application of high throughput sequencing technology and metagenomics in microbiology, other acid-resistant bacteria have been found in the stomach besides H. pylori. Nasr et al. revealed the major studies in which non-H. pylori have been implicated in GC development before 2019 (20). Thus, we summarized the association between non-H. pylori and GC progression over the past five years, especially in the past three years. In summary, the limited data available to date showed that the bacterial genera that were most invariably reported to be enriched in GC patients include Lactobacillus, Streptococcus, Prevotella, Veillonella, and so on (Table 1).
Numerous studies have shown that probiotics, including lactic acid bacteria (LAB), can enhance gastrointestinal health, immune regulation, and cancer prevention (40). Even though most of the aforementioned bacteria implicated in GC are LAB, including Lactobacillus, Streptococcus, and Lactococcus. A previous study has confirmed that LAB can induce the generation of reactive oxygen species (41) which initiates cancer angiogenesis, metastasis, and survival under individual concentrations (42). In addition, LAB have been proved to form large number of N-nitroso compounds (43, 44), which promote angiogenesis, mutagenesis, protooncogene expression, and inhibit apoptosis (45–47). Furthermore, LAB can elevate lactate production, which is crucial for major carcinogenic processes, namely, cell migration, angiogenesis, metastasis, immune evasion, and cell sufficiency (48, 49).
2.2 Colorectal Cancer (CRC)
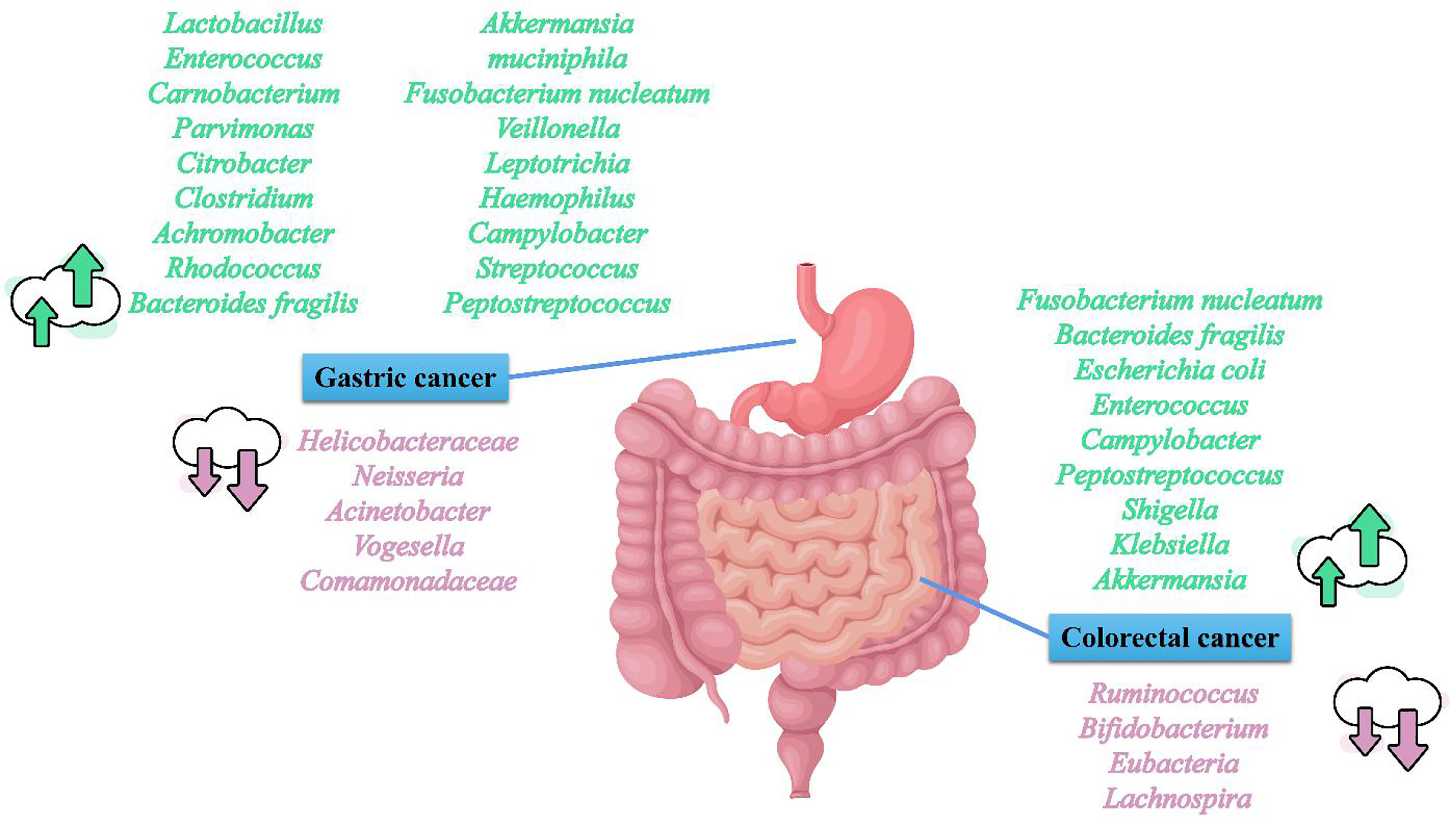
CRC is the third most common cancer and the second leading cause of cancer-associated deaths worldwide (13). Like GC, CRC incidence also displays regional differences (13). Russia, Canada, and Australia are hotspots for CRC incidence and mortality (Figure 1). CRC is constantly related to the changes in the microbial composition of the tumor and its adjacent mucosa, which is termed as dysbiosis (50–54). Dysbiosis is characterized by the expansion of the bacterial taxa to a certain extent. However, the dominant bacterial species in CRC evolution are still unknown. Several studies have reported higher proportions of Fusobacterium nucleatum (F. nucleatum), Bacteroides fragilis (B. fragilis), Escherichia coli (E. coli), Enterococcus, Campylobacter, Peptostreptococcus, Shigella, Klebsiella, and Akkermansia in CRC patients, and lower levels of Ruminococcus, Bifidobacterium, Eubacteria, and Lachnospira compared with healthy subjects (55–64) (Figure 2). Although Akkermansia has been shown to be a potential probiotic of the new generation and plays critical roles in obesity, diabetes, and atherosclerosis (65), its effects in CRC progression are still confusing. Furthermore, since F. nucleatum, B. fragilis, and E. coli are widely shown to play an important role on CRC development,we here focused on F. nucleatum, B. fragilis, and E. coli, and subsequently elucidated the correlation between gut microbiota and CRC progression.
2.2.1 F. nucleatum
F. nucleatum, a gram-negative anaerobe, which is universal in the oral cavity, is absent or rarely detected elsewhere in the body under normal conditions (66). Recently, F. nucleatum was found to be enriched in CRC and the rectal swabs of CRC patients (66–69), for example, Castellarin et al. verified F. nucleatum overabundance in tumor tissue, and observed a positive correlation with lymph node metastasis (68); Mima et al. confirmed that the amount of F. nucleatum in CRC tissues were associated with a shorter survival (70); Consistently, Yan et al. analyzed F. nucleatum level and prognostic significance in CRC patients, and found that the F. nucleatum level was abnormally high in tumor tissues and correlated with tumor invasion, lymph node metastasis status, and distant metastasis (71).
In addition, a correlation between F. nucleatum and CRC has also been found in preclinical experiments. Rubinstein et al. demonstrated that F. nucleatum promoted CRC cell progression by modulating the E-cadherin/B-catenin signaling through its unique FadA adhesin, and FadA expression was correlated with the expression of oncogenic and inflammatory genes in CRC patients (72). Kostic et al. indicated that, in the Apc (Min/+) mouse model of intestinal tumorigenesis, F. nucleatum increased tumor multiplicity and selectively recruited tumor-infiltrating myeloid cells, which can promote tumor progression (73); Yang et al. found that F. nucleatum-infected CRC cells exhibited an increased proliferation, invasive activity, and ability of tumor formation (74). Furthermore, F. nucleatum-activated Toll-like receptor 4 signaling to MYD88 activated nuclear factor-κB and thus increased miR-21 expression, and miR-21 could subsequently reduce the expression of RASA1 (74). RASA1 is a member of the RAS GTPase activating proteins (RAS-GAP) family, and mutation or loss of function of RASA1 can activate the RAS-MAPK cascade in CRC (75–77). Moreover, Brennan et al. suggested that F. nucleatum influenced intestinal immunity by shaping Th17 responses in an FFAR2-dependent manner (78). Although further studies are necessary to clarify the multifaceted roles of FFAR2, this research highlighted a conserved pathway that could be targeted to attenuate oncomicrobe-mediated CRC. These studies are consistent with the results in clinical studies that F. nucleatum is closely related to CRC, but further mechanisms between F. nucleatum and CRC still require more evidence.
2.2.2 Enterotoxigenic B. fragilis (ETBF)
B. fragilis, an anaerobic commensal that constitutes only 1–2% of the gut microbiota, can trigger diarrhea and inflammatory bowel disease by producing B. fragilis toxin (79, 80). B. fragilis phylogeny can be described in multiple ways and categorized as toxigenic or non-toxigenic (81). For example, ETBF is associated with inflammatory diseases and CRC. For example, sero-positivity of B. fragilis and E. coli was associated with CRC development, suggesting that co-infection of these bacterial species contributes to CRC tumorigenesis (82). Boleij et al. showed that the B. fragilis toxin gene was associated with CRC, especially in late-stage CRC (83). In addition, several experimental evidence has implicated enterotoxigenic ETBF in CRC development, like Liu et al. found that ETBF upregulated JMJD2B levels in a TLR4-NFAT5-dependent pathway and induced CRC stemness (84); Cao et al. confirmed that ETBF promoted intestinal inflammation and malignancy by downregulating miR-149-3p and further promoting PHF5A-mediated RNA alternative splicing of KAT2A in CRC cells (85); Guo et al. suggested that the downregulation of farnesoid X receptor promoted CRC development by facilitating ETBF colonization (86); Goodwin et al. demonstrated that B. fragilis toxin upregulated spermine oxidase, increased reactive oxygen species, and DNA damage, thereby propagating inflammation and tumorigenesis (87).
2.2.3 E. coli
E. coli is a highly prevalent, but not very abundant, gram-negative facultative anaerobe of the distal gastrointestinal tract. E. coli is a vast and diverse group of bacteria. Colibactin-producing E. coli are closely related to CRC (88–91). Bonnet et al. observed an increased level of mucosa-associated and internalized E. coli in tumors compared with normal tissue, and colonization of mucosa by E. coli was associated with poor prognosis in colon cancer (tumor-node-metastasis stage) (88); E. coli from the B2 phylogenetic group is implicated in CRC as it possesses a genomic island, termed polyketide synthetase (pks), which codes for the synthesis of colibactin, a genotoxin that induces DNA damage, cell cycle arrest, mutations, and chromosomal instability in eukaryotic cells. In addition, Iyadorai found that pks+ E. coli was isolated in CRC patients (89). Furthermore, the influence of E. coli on CRC was also identified in animal studies, like E. coli-increased tumorigenesis in murine models of CRC (92, 93), and Cougnoux et al. found that colibactin-producing E. coli enhanced tumor growth in both xenograft and azoxymethane/dextran sodium sulfate models, and tumor growth was sustained by cellular senescence (92). However, note that, in addition to the tumor-facilitating effects, some E. coli are commensal and even probiotic (94).
In conclusion, the roles of F. nucleatum, B. fragilis, and E. coli in CRC susceptibility or progression are supported by preclinical studies and clinical sample-based studies. In view of the complex interactions between bacteria, these three samples are not the only microbes important for CRC, but they provide insight into targetable mechanisms of action in CRC. In the future, more research is needed to elucidate the association between gut microbiota and CRC.
3 Gut Microbiota Metabolites and Gastrointestinal Cancer
Despite various explorations into the interrelation between gut microbiota and cancer, the exact mechanisms of this interaction are still unclear. It has been reported that this interplay may be related to bacterial metabolites. Here, we focused on short-chain fatty acids (SCFAs), one of the most important gut microbiota metabolites, and discussed the current studies on the association between SCFAs and gastrointestinal cancer.
Moreover, gut microbiota can produce carbohydrate-active enzymes that ferment non-digestible carbohydrates, such as xylans, cellulose, and inulin, to generate SCFAs (95, 96). The great majority of SCFAs are the final products of bacterial fermentation, and the endogenous synthesis of the host is always trivial (97). SCFAs play multiple roles in human health and disease, such as inflammatory bowel disease, CRC, inflammatory bowel disease, diabetes, and atherosclerosis (98–100), while researchers have mainly focused on the effect of SCFAs on CRC (101–103). The gut microbiota has been revealed to produce approximately 50–100 mmol/L/day of SCFAs, mainly including acetate (C2), propionate (C3), and butyrate (C4) acid, and their ratio is 3:1:1 (104, 105). Acetate and propionate are formed by Bacteroidetes, whereas Firmicutes produce butyrate (106). Among these SCFAs, butyrate is deemed a crucial metabolite, which mediates the tumor-repressive effect of dietary fiber on CRC (105, 107, 108).
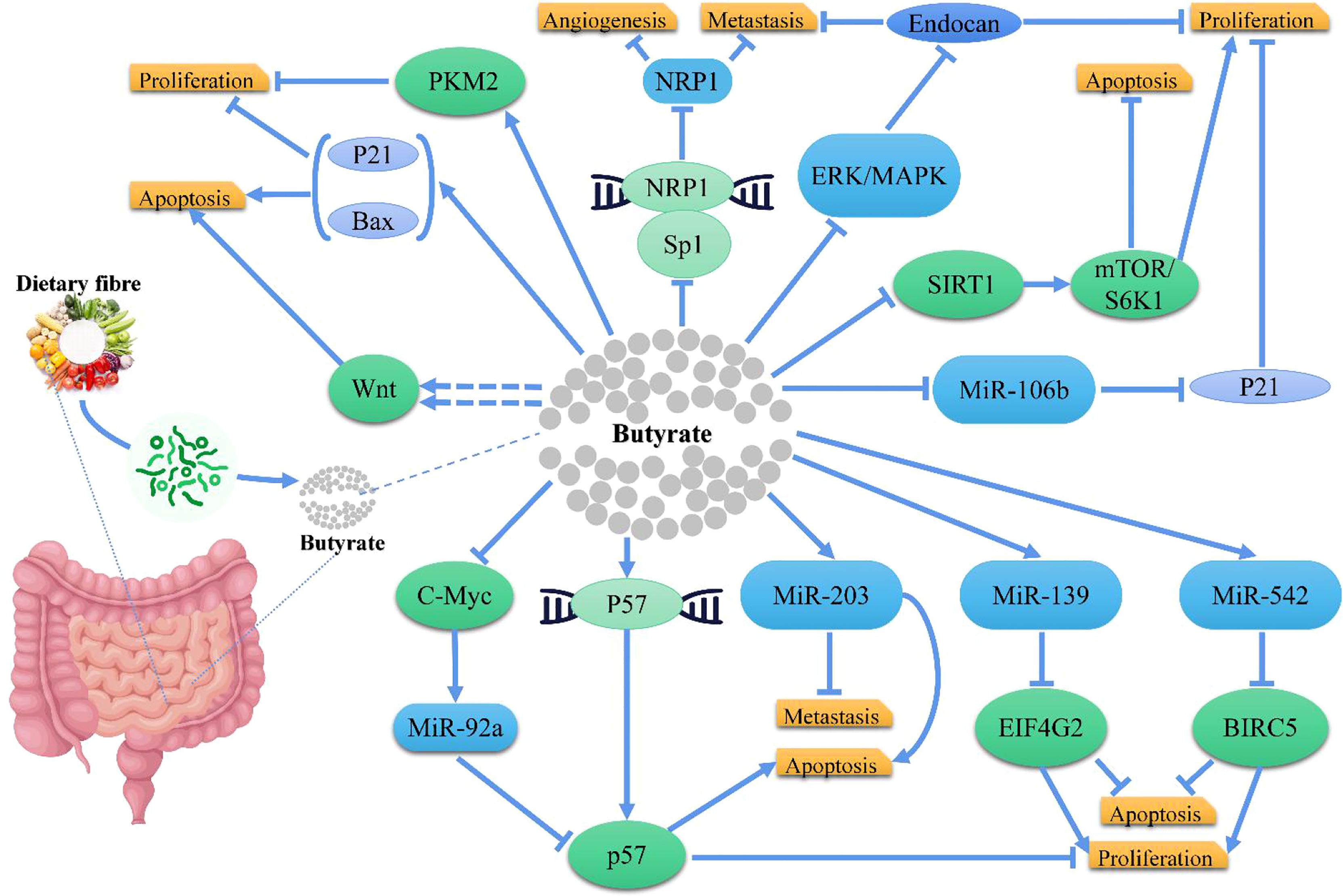
In CRC cells, butyrate inhibits histone deacetylases to increase the expression of genes that slow down the cell cycle and induce apoptosis (95, 96, 109). For example, Gamet et al. investigated the effects of SCFAs on the growth of the human adenocarcinoma cell line, HT29, and found that both butyrate and propionate inhibited the growth of HT29 cells, whereas acetate had no significant effect (110); Similarly, Hinnebusch et al. revealed that propionate, butyrate, and valerate suppressed the progression of colon carcinoma cells, while acetate and caproate had no effects (111); Zeng et al. indicated that butyrate had a greater inhibitory efficacy over propionate and acetate against CRC cell proliferation (112). Mechanistically, studies confirmed that butyrate inhibited CRC through different pathways (Figure 3), like Encarnação et al. suggested that butyrate inhibited the proliferation of CRC cells by regulating P21, and induced apoptosis by modulating BAX/BCL-2 ratio (113); Cao et al. demonstrated that butyrate treatment significantly inhibited proliferation and induced apoptosis in HCT116 cells with an increased BAX/BCL-2 ratio in CRC cells, and suggested that butyrate functioned via the deactivation of mTOR/S6K1 signaling mediated partly by SIRT1 downregulation (114); Yu et al. demonstrated that butyrate suppressed the expression of neuropilin I (NRP1) in colorectal cell lines through inhibition of Sp1 transactivation, and suppressed tumor cell migration and survival (115); Zuo et al. suggested that butyrate suppressed proliferation and migration in CRC cells through upregulating endocan expression via ERK2/MAPK signaling pathway (116); Chen et al. showed that Clostridium butyricum (one of the commonly used butyrate-producing bacteria in clinical settings) could inhibit intestinal tumor development by suppressed the Wnt/B-catenin signaling pathway, and activated G-protein coupled receptors (117); Bordonaro et al. evaluated that butyrate hyperactivated Wnt signaling, resulting in CRC cell apoptosis (118); Cucciolla et al. observed that butyrate upregulated p57 level by enhancing its transcription (119); In another research, Hu et al. identified a novel cellular mechanism that butyrate inhibited miR-92a transcription by reducing c-Myc, thus augmenting p57 level (120). Additionally, Li et al. demonstrated that butyrate suppressed the proliferation of CRC cells, through activating PKM2 via promoting its dephosphorylation and tetramerization, and thereby reprogramed the metabolism, inhibiting the Warburg effect while favoring energetic metabolism (121). Furthermore, Yoo et al. suggested that chronic exposure to butyrate induced butyrate resistance in CRC cells by triggering protective autophagy through the downregulation of AMPK/ACC and activation of Akt/mTOR signaling (122). Notably, miRNA expression is also intently related to the occurrence, development, and metastasis in CRC cells (123) and the expression of miRNA may be regulated by dietary factors, such as butyrate, like Han et al. showed that butyrate induced CRC cell apoptosis and inhibited the proliferation and invasion via upregulating miR-203 level (124); Ali et al. found that miR-139 and miR-542 acted cooperatively with butyrate to reduce CRC cell proliferation and induce apoptosis by regulating target genes, EIF4G2 and BIRC5 (125); Hu et al. concluded that butyrate regulated p21 expression via downregulating miR-106b level (126).
Although most of the available studies suggest that butyrate has an anti-tumor effect, it has also been observed to have a tumor-promoting effect in the development of CRC. This difference has been termed as the butyrate paradox and suggests that the effects of butyrate depend on its concentration, which is characterized as a high concentration of butyrate inhibiting tumorigenesis, while its low concentration promotes tumor progression (105). In more detail, on the one hand, Okumura et al. found that butyrate promoted tumorigenesis in CRC patients, and this colorectal tumorigenesis may be due to butyrate-induced senescence (127). Such effects were observed in many other studies (8, 105); however, the underlying mechanisms are still unclear. Thus, further studies are needed to understand the interplay between host genetics, microbial composition, and the presence of other gut metabolites, thereby clarifying this paradoxical result.
In conclusion, while many studies on SCFA and their association with cancer have been conducted, CRC has received the most attention. Further research is needed to explore the role of SCFA in other cancers, such as GC, pancreatic cancer, and hepatic carcinoma, to determine the exact role of SCFA in cancer development and treatment. In addition, in-depth studies are needed to investigate the effect of SCFAs on the efficacy and safety of chemotherapy or immunotherapy and their correlation with cancer prognosis.
4 Effects of Gut Microbiota on Gastrointestinal Cancer Treatment
With the increasing knowledge of gut microbiota, researchers have gained a deeper understanding of the impact of abnormal bacterial metabolism on the host. Numerous studies have shown that microbiota can metabolize, activate, and inactivate many common drugs, while their impact on cancer treatment has only received insufficient attention until recently (128, 129). Here, we have focused on the relevance of microbiota to chemotherapy, immunotherapy, and microbiota modulation.
4.1 Microbiota and Chemotherapy
The gut microbiota is metabolically active, while many cancer chemotherapeutic drugs act in an anti-metabolism manner. Thus, there is a potential interaction between gut microbiota and chemotherapy. Growing evidence suggests that gut microbiota can influence the efficacy of cancer treatment. For example, certain gammaproteobacteria can metabolize gemcitabine into an inactive form, thus inducing gemcitabine resistance (130); gemcitabine resistance was abrogated by the co-treatment with ciprofloxacin in a CRC mouse model (130); and Yuan et al. also suggested that antibiotics disrupt the gut microbiota in mice and reduce 5-fluorouracil efficacy (131). Similarly, Iida et al. demonstrated that oxaliplatin and cisplatin treatment exhibited reduced antitumor efficacy and survival in various tumor-bearing mice in the presence of antibiotics (132). Additionally, Yu et al. demonstrated that Fusobacterium plays a critical role in mediating CRC chemoresistance by activating autophagy (133). Furthermore, the treatment of CRC xenograft-bearing mice with the antibiotic metronidazole reduced Fusobacterium load, cancer cell proliferation, and overall tumor growth (134). Although most of the current research is derive from preclinical experiments, we strongly believe that the gut microbiota is closely related to chemotherapy.
4.2 Microbiota and Immunotherapy
Increasing evidence has highlighted that gut microbiota is involved in the clinical response to cancer immunotherapy (135–137). These immunotherapies mainly target programmed cell death protein-1 (PD-1) and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) blockades (138). A study in 2015 confirmed that gut microbiota could modulate the effect of anti-PD-1/PD-L1 monoclonal antibodies in mice (139), and commensal Bifidobacterium enhanced the immunotherapeutic effects by activating dendritic cells for CD8+ T-cell priming and infiltration in the tumor microenvironment (139). In addition, Rizvi et al. showed that Bifidobacterium enhanced the efficacy of anti-PD-1 monoclonal antibodies in mice with melanoma by secreting the metabolite hippurate and inhibiting PD-1 expression (140); Sun et al. found that Bifidobacterium altered the composition of the gut microbiota in a manner dependent on regulatory T cells; this altered commensal community enhanced both the mitochondrial fitness and the IL-10-mediated suppressive functions of intestinal Tregs, contributing to the reduction of colitis during immune checkpoint blockade (141); and Wang et al. suggested that Bifidobacterium could mitigate intestinal immunopathology induced by CTLA-4 blockade (142). However, it remains unknown whether Bifidobacteria can reduce virulence while enhancing efficacy. Other gut bacteria besides Bifidobacterium may also modulate immunotherapy. Like Si et al. demonstrated, oral administration of live Lactobacillus rhamnosus GG augmented the antitumor activity of anti-PD-1 by increasing tumor-infiltrating dendritic and T cells (143). Mechanistically, treatment with live Lactobacillus rhamnosus GG triggered type I interferon production in dendritic cells, enhancing the cross-priming of antitumor CD8+ T cells (143); Vétizou et al. found that the antitumor effects of CTLA-4 blockade depended on distinct bacteroide species (144). Numerous studies have determined various microbial species related to the response of immune checkpoint inhibitors (135, 145–148). However, it remains unknown whether their interactions with one another and which microbes are more important in immune checkpoint inhibitors (149).
4.3 Microbiota Modulation
The microbiota modulation strategy is largely based on the regulatory role of specific gut microbiota in anti-tumor immunity. The principle of this strategy is to facilitate the effect of immune-enhancing gut microbiota while reducing the effect of immunosuppressive gut microbiota.
4.3.1 Prebiotics and Probiotics
Many clinical trials have been conducted to investigate the effect of prebiotics or probiotics on tumor therapy, some of which reported improved clinical outcomes in patients receiving probiotics, while others were not significantly different (150–153). Future studies may need to increase the number of samples and tumor types and standardize research methods to further clarify the impact of prebiotics and probiotics in tumor treatment. Notably, the limited evidence indicates that probiotic strains would not be the most suitable strains to treat tumors.The concept of prebiotics was first proposed in 1995 and redefined in December 2016 (154, 155). Prebiotics are defined by the International Scientific Association for Probiotics and Prebiotics as “substrates that are selectively utilized by host microorganisms to confer health benefits to the host” (155). The two important groups of prebiotics are fructo-oligosaccharides (FOS) and galacto-oligosaccharides (GOS) (156). FOS is naturally present in asparagus, bananas, chicory root, garlic, and onion, as well as synthesized commercially (155, 157); GOS is produced commercially from lactose by B-galactosidase (156). The effect of FOS and GOS on gut microbiota modulation has been shown previously (157). In addition, the effects of prebiotics have been shown to engage in tumor therapy, such as: Taper et al. suggested that treatment with inulin or oligofructose potentiated the effects of cancer therapy (158, 159); Dewulf et al. confirmed that treatment with inulin or oligofructose led to subtle changes in gut microbiota (160); Escherichia coli Nissle 1917 (EcN), a genetically tractable probiotic with a well-established human safety record, is emerging as a preferred chassis (161). In theory, prebiotics can selectively enrich beneficial probiotics and increase SCFA production. However, how prebiotics increase the effects of chemotherapy and immunotherapy requires further research (139, 162, 163).
Probiotics are living microorganisms, mainly including Lactobacillus and Bifidobacterium, can confer health benefits on the host at a certain concentration (164). Botta et al. suggested that Lactiplantibacillus plantarum inhibited colon cancer cell proliferation by butyrogenic capability (165). Do Carmo et al. confirmed that Propionibacterium freudenreichii alleviated mucositis induced by 5-fluorouracil chemotherapy (166); Sivan et al. suggested that Bifidobacterium promoted antitumor immunotherapy effects of anti-PD-L1 (139); Si et al. found that Lactobacillus rhamnosus GG improved response to immune checkpoint blockade (143). However, a recent study indicated an impaired-treatment response to anti-PD-1 therapy in mice receiving a low-fiber diet or probiotics, and that in the tumor microenvironment there is a lower frequency of interferon-γ-positive cytotoxic T cells (167). Compared to previous preclinical studies, this mechanistic study clarified the opposite results. Most of the current clinical studies have only shown the effect of probiotics on gut microbiota, but not clarified their impact on immunotherapy (146, 148, 168).
4.3.2 Fecal Microbiota Transplantation (FMT)
FMT is another clinical strategy for manipulating gut microbiota and has been approved by the FDA for treating Clostridium difficile infection (149). FMT can transfer entire fecal microbial community, namely, bacteria, fungi, viruses, and their metabolites, from a healthy donor to a recipient (149, 169). Wang et al. first reported a successful case series of immune checkpoint inhibitors-associated colitis treated with FMT (170), in which two patients reconstituted the gut microbiota, and a substantial reduction in CD8+ T-cell density with a concomitant increase in CD4+ FoxP3+ was observed within the colonic mucosa, offering a potential mechanism through which FMT could abrogate ICI-associated toxicity. However, certain limitations existed in this case, and more clinical trials are needed to evaluate the effect of this approach and further elucidate the underlying mechanisms. Recently, FMT has begun to be inspected in combination with checkpoint blockade therapy, as Routy et al. found that the clinical benefit of immune checkpoint inhibitors (ICIs) was attenuated in advanced cancer patients treated with antibiotics (135). Furthermore, FMT from cancer patients who responded to ICIs into antibiotic-treated or germ-free mice improved the effects of PD-1, whereas FMT from nonresponding patients failed (135). More recently, clinical trials of FMT in combination with checkpoint blockade therapy are ongoing or completed, like Matson et al. reviewed several clinical trials evaluating the potential of FMT to enhance immune checkpoint blockade therapy, primarily in patients with metastatic melanoma (171), implicating the feasibility and safety of FMT in cancer treatment. Notably, a clinical trial of FMT capsule for improving the efficacy of anti-PD-1 among patients with gastrointestinal cancer is recruiting (NCT04130763, https://clinicaltrials.gov). Additionally, Kassan et al. evaluated the suitability of stool from candidate FMT donors for clinical use and suggested that only 3% of donors would pass such quality control assessments (172). For solving such issue, more studies have been conducted and shown that a good given FMT donor should have the necessary gut microbiota composition to correct the microbiota deficiency in one patient but not another (173–178). Furthermore, unlike other microbiota modulation methods, the effects of FMT could last for more than 24 weeks, thus frequent interventions are not required (149, 169). With further studies on the mechanisms by which the gut microbiota modulates host antitumor immunity, and further confirmation of the functions of related bacteria, we believe that the selection of FMT donors for specific patients will be more convenient and faster in the future.
4.3.3 Antibiotics
Bacterial depletion by antibiotics is another strategy for manipulating the gut microbiota. Prophylactic antibiotics are always given with chemotherapy or immunotherapy to prevent potentially life-threatening infections from immunosuppression caused by chemotherapy and immunotherapy.
As previously mentioned, certain gammaproteobacteria could induce gemcitabine resistance, and antibiotics targeting gammaproteobacteria-improved gemcitabine response in patients with pancreatic ductal adenocarcinoma (130); however, Wu et al. found that antibiotic administration reduced chemotherapy efficacy and was associated with poor prognosis in patients with esophageal cancer (179); And Nenclares et al. suggested that antibiotic therapy was associated with a negative outcome in locally advanced head and neck cancer (180). Notably, Zheng et al. showed that oral or intravenous administration of irinotecan-loaded dextran nanoparticles covalently linked to azide-modified phages that inhibit the growth of F. nucleatum significantly improves the efficiency of chemotherapy in CRC mice (181). These results suggest that using antibiotics with chemotherapy is a double-edged sword. Therefore, more differentiated strategies can be applied, such as selective use of antibiotics or targeted delivery of antibiotics. With the advances in nanotechnology, targeted-delivery of antibiotics may be possible to balance the risks and benefits of prophylactic antibiotic use in cancer chemotherapeutic patients.
Researchers have observed reduced response to ICIs in patients treated with antibiotics in preclinical and clinical trials. For example, Vétizou et al. found that tumors did not respond to CTLA blockade in antibiotic-treated or germ-free mice (144); Pinato et al. suggested that gut dysbiosis caused by broad-spectrum antibiotic therapy-impaired ICIs response (182); Tinsley et al. highlighted that reduced clinical benefit from ICIs was associated with antibiotic use in advanced cancer (183). Similarly, Derosa et al. found that antibiotics were associated with worse treatment outcomes of ICIs in non-small cell lung cancer and renal cell carcinoma (184). Thus, modulation of ATB-related dysbiosis and gut microbiota composition may be a strategy to improve clinical outcomes with ICIs, as Wilson et al. confirmed that overall survival and progression-free survival are longer in patients who have not received antibiotics compared to patients with antibiotic use (185). Furthermore, antibiotic use 42 days before starting ICIs appears most harmful to the outcome, while antibiotic use in the 60 days before starting ICIs appears to have no significant difference in outcome (185). Palleja et al. found that it takes time for gut microbiota to recover in healthy adults after antibiotic exposure (186). We speculate that it takes more time for gut microbiota to recover in tumor patients after antibiotic exposure. Thus, antibiotics treatment should be avoided before ICIs (149, 187, 188). Perhaps, it is a better strategy using probiotics or FMT to modulate gut microbiota before ICIs other than using antibiotics.
5 Perspectives and Future Directions
Humans harbor trillions of resident microorganisms, which make up the microbiota. Microbiota play a vital role in various aspects of human health and disease. In recent years, an increasing number of studies have focused on the impact of gut microbiota on host metabolism and disease. Here, we reviewed the potential roles of gut microbiota and gut microbial metabolites in gastrointestinal cancer and explored the therapeutic potential of gut microbiota in gastrointestinal cancer. However, in terms of the impact of gut microbiota on tumorigenesis, development, and treatment, our knowledge may only be the tip of the iceberg, and there are still many problems to be solved.
Firstly, can cancer clusters be explained by microbiota? Cancer cluster is defined as the number of cancer cases occurring in a group of people over a specific period time in a specific geographic area that is higher than expected (189), for example, East Asia is a hotspot for gastric cancer incidence and mortality in this world; Linqu, Shandong Province, is a region with the highest incidence and mortality rate of GC in China (28, 190); and in sub-Saharan Africa, there is a substantial, early-onset CRC has increased significantly (191, 192). However, the specific factors contributing to the occurrence of these cancers are unclear, and further research is needed to determine whether gut microbiota is associated with increased cancer incidence in these populations. Second, the composition of gut microbiota is affected by age, diet, genetics, lifestyle, and medical conditions, are these factors also needed to be monitored? These factors are closely related to geographical location, so is it more meaningful to incorporate geographic information into microbiota studies and cancer detection? Third, what studies do researchers need to explore the relationship between gut microbiota and pathogenesis as well as treatment of gastrointestinal cancer? What is the ideal way to use gut microbes for gastrointestinal cancer treatment? Finally, in order to identify a reliable gut microbiota for predictable gastrointestinal cancer risk and patient prognosis, or for gut microbiota-based treatment strategies, we need standardized approaches and uniform designs for gut microbiota sampling in large and diverse patient populations, which requires our joint efforts.
Author Contributions
QG, HQ, XL, XZ, TQ, and LC reviewed the literature and drafted the article. ZC organized figures and tables. QG, HQ, and WZ finalized the paper and provided suggestions to improve it. All authors participated in designing the concept of this manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The study was supported by the Medical Science and Technology Research Project of Henan Province (no. SBGJ202003010), the Medical Science and Technology Research Project of Henan Province (no. LHGJ20190675), and the Doctoral Research Start-up Foundation of Henan Cancer Hospital.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviation
GC, gastric cancer; CRC, colorectal cancer; H. pylori, Helicobacter pylori; VacA, vacuolating cytotoxin A; CagA, cytotoxin-associated gene A; GAD, gastric adenoma; EGC, Early gastric cancer; AGC, Advanced gastric cancer; IM, gastritis without intestinal metaplasia; HC, healthy controls; SG, superficial gastritis; SRCC, signet-ring cell carcinoma; CG, non-atrophic chronic gastritis IN intraepithelial neoplasia; AG, atrophic gastritis; LAB, lactic acid bacteria; F. nucleatum, Fusobacterium nucleatum; B. fragilis, Bacteroides fragilis; E. coli, Escherichia coli; ETBF, Enterotoxigenic B. fragilis; SCFAs, short-chain fatty acids; PD-1, programmed cell death protein-1; CTLA-4, cytotoxic T lymphocyte-associated antigen-4; FMT, Fecal Microbiota Transplantation; ICIs, immune checkpoint inhibitors.
References
1. Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell (2016) 164(3):337–40. doi: 10.1016/j.cell.2016.01.013
2. Van Treuren W, Dodd D. Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu Rev Pathol (2020) 15:345–69. doi: 10.1146/annurev-pathol-020117-043559
3. Sędzikowska A, Szablewski L. Human Gut Microbiota in Health and Selected Cancers. Int J Mol Sci (2021) 22(24):13440. doi: 10.3390/ijms222413440
4. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature (2012) 489(7415):220–30. doi: 10.1038/nature11550
5. Imoto N, Kano C, Aoyagi Y, Morita H, Amanuma F, Maruyama H, et al. Administration of B-Lactam Antibiotics and Delivery Method Correlate With Intestinal Abundances of Bifidobacteria and Bacteroides in Early Infancy, in Japan. Sci Rep (2021) 11(1):6231. doi: 10.1038/s41598-021-85670-z
6. Koboziev I, Reinoso Webb C, Furr KL, Grisham MB. Role of the Enteric Microbiota in Intestinal Homeostasis and Inflammation. Free Radic Biol Med (2014) 68:122–33. doi: 10.1016/j.freeradbiomed.2013.11.008
7. O'Keefe SJ. Diet, Microorganisms and Their Metabolites, and Colon Cancer. Nat Rev Gastroenterol Hepatol (2016) 13(12):691–706. doi: 10.1038/nrgastro.2016.165
8. Jaye K, Li CG, Chang D, Bhuyan DJ. The Role of Key Gut Microbial Metabolites in the Development and Treatment of Cancer. Gut Microbes (2022) 14(1):2038865. doi: 10.1080/19490976.2022.2038865
9. Hanus M, Parada-Venegas D, Landskron G, Wielandt AM, Hurtado C, Alvarez K, et al. Immune System, Microbiota, and Microbial Metabolites: The Unresolved Triad in Colorectal Cancer Microenvironment. Front Immunol (2021) 12:612826. doi: 10.3389/fimmu.2021.612826
10. Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The Microbiome in Cancer Immunotherapy: Diagnostic Tools and Therapeutic Strategies. Science (2018) 359(6382):1366–70. doi: 10.1126/science.aar6918
11. Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer Effects of the Microbiome and Its Products. Nat Rev Microbiol (2017) 15(8):465–78. doi: 10.1038/nrmicro.2017.44
12. Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, et al. The Gut Microbiome Switches Mutant P53 From Tumour-Suppressive to Oncogenic. Nature (2020) 586(7827):133–8. doi: 10.1038/s41586-020-2541-0
13. Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer (2020). Available at: https://gco.iarc.fr/today. accessed [15 March 2022].
14. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric Cancer. Lancet (2020) 396(10251):635–48. doi: 10.1016/s0140-6736(20)31288-5
15. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter Pylori Infection. Clin Microbiol Rev (2006) 19(3):449–90. doi: 10.1128/cmr.00054-05
16. Brawner KM, Morrow CD, Smith PD. Gastric Microbiome and Gastric Cancer. Cancer J (2014) 20(3):211–6. doi: 10.1097/ppo.0000000000000043
17. Capurro MI, Greenfield LK, Prashar A, Xia S, Abdullah M, Wong H, et al. Vaca Generates a Protective Intracellular Reservoir for Helicobacter Pylori That Is Eliminated by Activation of the Lysosomal Calcium Channel Trpml1. Nat Microbiol (2019) 4(8):1411–23. doi: 10.1038/s41564-019-0441-6
18. Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. An Overview of Helicobacter Pylori Vaca Toxin Biology. Toxins (Basel) (2016) 8(6):173. doi: 10.3390/toxins8060173
19. Buti L, Ruiz-Puig C, Sangberg D, Leissing TM, Brewer RC, Owen RP, et al. Caga-Aspp2 Complex Mediates Loss of Cell Polarity and Favors H. Pylori Colonization of Human Gastric Organoids. Proc Natl Acad Sci USA (2020) 117(5):2645–55. doi: 10.1073/pnas.1908787117
20. Nasr R, Shamseddine A, Mukherji D, Nassar F, Temraz S. The Crosstalk Between Microbiome and Immune Response in Gastric Cancer. Int J Mol Sci (2020) 21(18):6586. doi: 10.3390/ijms21186586
21. Schulz C, Schütte K, Mayerle J, Malfertheiner P. The Role of the Gastric Bacterial Microbiome in Gastric Cancer: Helicobacter Pylori and Beyond. Therap Adv Gastroenterol (2019) 12:1756284819894062. doi: 10.1177/1756284819894062
22. Hathroubi S, Servetas SL, Windham I, Merrell DS, Ottemann KM. Helicobacter Pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol Mol Biol Rev (2018) 82(2):e00001-18. doi: 10.1128/mmbr.00001-18
23. Shah MA. Gastric Cancer: The Gastric Microbiota - Bacterial Diversity and Implications. Nat Rev Gastroenterol Hepatol (2017) 14(12):692–3. doi: 10.1038/nrgastro.2017.140
24. Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, et al. Helicobacter Pylori Eradication to Prevent Gastric Cancer in a High-Risk Region of China: A Randomized Controlled Trial. Jama (2004) 291(2):187–94. doi: 10.1001/jama.291.2.187
25. Rugge M, Meggio A, Pravadelli C, Barbareschi M, Fassan M, Gentilini M, et al. Gastritis Staging in the Endoscopic Follow-Up for the Secondary Prevention of Gastric Cancer: A 5-Year Prospective Study of 1755 Patients. Gut (2019) 68(1):11–7. doi: 10.1136/gutjnl-2017-314600
26. Choi IJ, Kook MC, Kim YI, Cho SJ, Lee JY, Kim CG, et al. Helicobacter Pylori Therapy for the Prevention of Metachronous Gastric Cancer. N Engl J Med (2018) 378(12):1085–95. doi: 10.1056/NEJMoa1708423
27. Malfertheiner P. Helicobacter Pylori Treatment for Gastric Cancer Prevention. N Engl J Med (2018) 378(12):1154–6. doi: 10.1056/NEJMe1800147
28. Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, et al. Effect of Helicobacter Pylori on Gastrointestinal Microbiota: A Population-Based Study in Linqu, a High-Risk Area of Gastric Cancer. Gut (2020) 69(9):1598–607. doi: 10.1136/gutjnl-2019-319696
29. Park JY, Seo H, Kang CS, Shin TS, Kim JW, Park JM, et al. Dysbiotic Change in Gastric Microbiome and Its Functional Implication in Gastric Carcinogenesis. Sci Rep (2022) 12(1):4285. doi: 10.1038/s41598-022-08288-9
30. Png CW, Lee WJJ, Chua SJ, Zhu F, Yeoh KG, Zhang Y. Mucosal Microbiome Associates With Progression to Gastric Cancer. Theranostics (2022) 12(1):48–58. doi: 10.7150/thno.65302
31. Park CH, Hong C, Lee AR, Sung J, Hwang TH. Multi-Omics Reveals Microbiome, Host Gene Expression, and Immune Landscape in Gastric Carcinogenesis. iScience (2022) 25(3):103956. doi: 10.1016/j.isci.2022.103956
32. Dai D, Yang Y, Yu J, Dang T, Qin W, Teng L, et al. Interactions Between Gastric Microbiota and Metabolites in Gastric Cancer. Cell Death Dis (2021) 12(12):1104. doi: 10.1038/s41419-021-04396-y
33. Pimentel-Nunes P, Barros A, Pita I, Miranda I, Conceição G, Borges-Canha M, et al. Gastric Microbiome Profile Throughout Gastric Carcinogenesis: Beyond Helicobacter. Scand J Gastroenterol (2021) 56(6):708–16. doi: 10.1080/00365521.2021.1902560
34. Wu ZF, Zou K, Wu GN, Jin ZJ, Xiang CJ, Xu S, et al. A Comparison of Tumor-Associated and Non-Tumor-Associated Gastric Microbiota in Gastric Cancer Patients. Dig Dis Sci (2021) 66(5):1673–82. doi: 10.1007/s10620-020-06415-y
35. Deng Y, Ding X, Song Q, Zhao G, Han L, Ding B, et al. Alterations in Mucosa-Associated Microbiota in the Stomach of Patients With Gastric Cancer. Cell Oncol (Dordr) (2021) 44(3):701–14. doi: 10.1007/s13402-021-00596-y
36. Ravegnini G, Fosso B, Saverio VD, Sammarini G, Zanotti F, Rossi G, et al. Gastric Adenocarcinomas and Signet-Ring Cell Carcinoma: Unraveling Gastric Cancer Complexity Through Microbiome Analysis-Deepening Heterogeneity for a Personalized Therapy. Int J Mol Sci (2020) 21(24):9735. doi: 10.3390/ijms21249735
37. Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, et al. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front Microbiol (2020) 11:997. doi: 10.3389/fmicb.2020.00997
38. Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, et al. Alterations of Gastric Mucosal Microbiota Across Different Stomach Microhabitats in a Cohort of 276 Patients With Gastric Cancer. EBioMedicine (2019) 40:336–48. doi: 10.1016/j.ebiom.2018.12.034
39. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal Microbiome Dysbiosis in Gastric Carcinogenesis. Gut (2018) 67(6):1024–32. doi: 10.1136/gutjnl-2017-314281
40. Ghosh T, Beniwal A, Semwal A, Navani NK. Mechanistic Insights Into Probiotic Properties of Lactic Acid Bacteria Associated With Ethnic Fermented Dairy Products. Front Microbiol (2019) 10:502. doi: 10.3389/fmicb.2019.00502
41. Jones RM, Mercante JW, Neish AS. Reactive Oxygen Production Induced by the Gut Microbiota: Pharmacotherapeutic Implications. Curr Med Chem (2012) 19(10):1519–29. doi: 10.2174/092986712799828283
42. Kirtonia A, Sethi G, Garg M. The Multifaceted Role of Reactive Oxygen Species in Tumorigenesis. Cell Mol Life Sci (2020) 77(22):4459–83. doi: 10.1007/s00018-020-03536-5
43. Forsythe SJ, Cole JA. Nitrite Accumulation During Anaerobic Nitrate Reduction by Binary Suspensions of Bacteria Isolated From the Achlorhydric Stomach. J Gen Microbiol (1987) 133(7):1845–9. doi: 10.1099/00221287-133-7-1845
44. Calmels S, Béréziat JC, Ohshima H, Bartsch H. Bacterial Formation of N-Nitroso Compounds From Administered Precursors in the Rat Stomach After Omeprazole-Induced Achlorhydria. Carcinogenesis (1991) 12(3):435–9. doi: 10.1093/carcin/12.3.435
45. Li J, Billiar TR, Talanian RV, Kim YM. Nitric Oxide Reversibly Inhibits Seven Members of the Caspase Family Via S-Nitrosylation. Biochem Biophys Res Commun (1997) 240(2):419–24. doi: 10.1006/bbrc.1997.7672
46. Ambs S, Ogunfusika MO, Merriam WG, Bennett WP, Billiar TR, Harris CC. Up-Regulation of Inducible Nitric Oxide Synthase Expression in Cancer-Prone P53 Knockout Mice. Proc Natl Acad Sci USA (1998) 95(15):8823–8. doi: 10.1073/pnas.95.15.8823
47. Feng CW, Wang LD, Jiao LH, Liu B, Zheng S, Xie XJ. Expression of P53, Inducible Nitric Oxide Synthase and Vascular Endothelial Growth Factor in Gastric Precancerous and Cancerous Lesions: Correlation With Clinical Features. BMC Cancer (2002) 2:8. doi: 10.1186/1471-2407-2-8
48. Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial Carcinogenesis: Lactic Acid Bacteria in Gastric Cancer. Biochim Biophys Acta Rev Cancer (2019) 1872(2):188309. doi: 10.1016/j.bbcan.2019.07.004
49. San-Millán I, Brooks GA. Reexamining Cancer Metabolism: Lactate Production for Carcinogenesis Could Be the Purpose and Explanation of the Warburg Effect. Carcinogenesis (2017) 38(2):119–33. doi: 10.1093/carcin/bgw127
50. Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology (2020) 158(2):322–40. doi: 10.1053/j.gastro.2019.06.048
51. Janney A, Powrie F, Mann EH. Host-Microbiota Maladaptation in Colorectal Cancer. Nature (2020) 585(7826):509–17. doi: 10.1038/s41586-020-2729-3
52. Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell (2018) 33(6):954–64. doi: 10.1016/j.ccell.2018.03.004
53. Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res (2017) 23(8):2061–70. doi: 10.1158/1078-0432.Ccr-16-1599
54. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus Anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology (2017) 152(6):1419–33.e5. doi: 10.1053/j.gastro.2017.01.009
55. Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial Dysbiosis in Colorectal Cancer (Crc) Patients. PloS One (2011) 6(1):e16393. doi: 10.1371/journal.pone.0016393
56. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free And Conventional Mice. Gastroenterology (2017) 153(6):1621–33.e6. doi: 10.1053/j.gastro.2017.08.022
57. Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. Gut Microbiota Imbalance and Colorectal Cancer. World J Gastroenterol (2016) 22(2):501–18. doi: 10.3748/wjg.v22.i2.501
58. Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, et al. Increased Rectal Microbial Richness Is Associated With the Presence of Colorectal Adenomas in Humans. Isme J (2012) 6(10):1858–68. doi: 10.1038/ismej.2012.43
59. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-Analysis of Fecal Metagenomes Reveals Global Microbial Signatures That Are Specific for Colorectal Cancer. Nat Med (2019) 25(4):679–89. doi: 10.1038/s41591-019-0406-6
60. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic Analysis of Colorectal Cancer Datasets Identifies Cross-Cohort Microbial Diagnostic Signatures and a Link With Choline Degradation. Nat Med (2019) 25(4):667–78. doi: 10.1038/s41591-019-0405-7
61. Hu M, Chen K, Wang N, Zhao Y, Wei C, Meng L, et al. [Changes in the Structure of Intestinal Mucosal Flora in Colorectal Cancer Patients]. Nan Fang Yi Ke Da Xue Xue Bao (2022) 42(2):263–71. doi: 10.12122/j.issn.1673-4254.2022.02.14
62. Khodaverdi N, Zeighami H, Jalilvand A, Haghi F, Hesami N. High Frequency of Enterotoxigenic Bacteroides Fragilis and Enterococcus Faecalis in the Paraffin-Embedded Tissues of Iranian Colorectal Cancer Patients. BMC Cancer (2021) 21(1):1353. doi: 10.1186/s12885-021-09110-x
63. Choi S, Chung J, Cho ML, Park D, Choi SS. Analysis of Changes in Microbiome Compositions Related to the Prognosis of Colorectal Cancer Patients Based on Tissue-Derived 16s Rrna Sequences. J Transl Med (2021) 19(1):485. doi: 10.1186/s12967-021-03154-0
64. Nardelli C, Granata I, Nunziato M, Setaro M, Carbone F, Zulli C, et al. 16s Rrna of Mucosal Colon Microbiome and Ccl2 Circulating Levels Are Potential Biomarkers in Colorectal Cancer. Int J Mol Sci (2021) 22(19):10747. doi: 10.3390/ijms221910747
65. Hasani A, Ebrahimzadeh S, Hemmati F, Khabbaz A, Hasani A, Gholizadeh P. The Role of Akkermansia Muciniphila in Obesity, Diabetes and Atherosclerosis. J Med Microbiol (2021) 70(10):001435. doi: 10.1099/jmm.0.001435
66. Han YW. Fusobacterium Nucleatum: A Commensal-Turned Pathogen. Curr Opin Microbiol (2015) 23:141–7. doi: 10.1016/j.mib.2014.11.013
67. Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, et al. Towards the Human Colorectal Cancer Microbiome. PloS One (2011) 6(5):e20447. doi: 10.1371/journal.pone.0020447
68. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium Nucleatum Infection Is Prevalent in Human Colorectal Carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
69. Chen W, Liu F, Ling Z, Tong X, Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients With Colorectal Cancer. PloS One (2012) 7(6):e39743. doi: 10.1371/journal.pone.0039743
70. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut (2016) 65(12):1973–80. doi: 10.1136/gutjnl-2015-310101
71. Yan X, Liu L, Li H, Qin H, Sun Z. Clinical Significance of Fusobacterium Nucleatum, Epithelial-Mesenchymal Transition, and Cancer Stem Cell Markers in Stage Iii/Iv Colorectal Cancer Patients. Onco Targets Ther (2017) 10:5031–46. doi: 10.2147/ott.S145949
72. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/B-Catenin Signaling Via Its Fada Adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
73. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
74. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium Nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-Kb, and Up-Regulating Expression of Microrna-21. Gastroenterology (2017) 152(4):851–66.e24. doi: 10.1053/j.gastro.2016.11.018
75. Sun D, Wang C, Long S, Ma Y, Guo Y, Huang Z, et al. C/Ebp-B-Activated Microrna-223 Promotes Tumour Growth Through Targeting Rasa1 in Human Colorectal Cancer. Br J Cancer (2015) 112(9):1491–500. doi: 10.1038/bjc.2015.107
76. Kent OA, Mendell JT, Rottapel R. Transcriptional Regulation of Mir-31 by Oncogenic Kras Mediates Metastatic Phenotypes by Repressing Rasa1. Mol Cancer Res (2016) 14(3):267–77. doi: 10.1158/1541-7786.Mcr-15-0456
77. Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, et al. Microrna-31 Activates the Ras Pathway and Functions as an Oncogenic Microrna in Human Colorectal Cancer by Repressing Ras P21 Gtpase Activating Protein 1 (Rasa1). J Biol Chem (2013) 288(13):9508–18. doi: 10.1074/jbc.M112.367763
78. Brennan CA, Clay SL, Lavoie SL, Bae S, Lang JK, Fonseca-Pereira D, et al. Fusobacterium Nucleatum Drives a Pro-Inflammatory Intestinal Microenvironment Through Metabolite Receptor-Dependent Modulation of Il-17 Expression. Gut Microbes (2021) 13(1):1987780. doi: 10.1080/19490976.2021.1987780
79. Karpiński TM, Ożarowski M, Stasiewicz M. Carcinogenic Microbiota and Its Role in Colorectal Cancer Development. Semin Cancer Biol (2022). doi: 10.1016/j.semcancer.2022.01.004
80. Kaźmierczak-Siedlecka K, Skonieczna-Żydecka K, Hupp T, Duchnowska R, Marek-Trzonkowska N, Połom K. Next-Generation Probiotics - Do They Open New Therapeutic Strategies for Cancer Patients? Gut Microbes (2022) 14(1):2035659. doi: 10.1080/19490976.2022.2035659
81. Clay SL, Fonseca-Pereira D, Garrett WS. Colorectal Cancer: The Facts in the Case of the Microbiota. J Clin Invest (2022) 132(4):e155101. doi: 10.1172/jci155101
82. Butt J, Jenab M, Werner J, Fedirko V, Weiderpass E, Dahm CC, et al. Association of Pre-Diagnostic Antibody Responses to Escherichia Coli and Bacteroides Fragilis Toxin Proteins With Colorectal Cancer in a European Cohort. Gut Microbes (2021) 13(1):1–14. doi: 10.1080/19490976.2021.1903825
83. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The Bacteroides Fragilis Toxin Gene Is Prevalent in the Colon Mucosa of Colorectal Cancer Patients. Clin Infect Dis (2015) 60(2):208–15. doi: 10.1093/cid/ciu787
84. Liu QQ, Li CM, Fu LN, Wang HL, Tan J, Wang YQ, et al. Enterotoxigenic Bacteroides Fragilis Induces the Stemness in Colorectal Cancer Via Upregulating Histone Demethylase Jmjd2b. Gut Microbes (2020) 12(1):1788900. doi: 10.1080/19490976.2020.1788900
85. Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, et al. Enterotoxigenic Bacteroidesfragilis Promotes Intestinal Inflammation and Malignancy by Inhibiting Exosome-Packaged Mir-149-3p. Gastroenterology (2021) 161(5):1552–66.e12. doi: 10.1053/j.gastro.2021.08.003
86. Guo S, Peng Y, Lou Y, Cao L, Liu J, Lin N, et al. Downregulation of the Farnesoid X Receptor Promotes Colorectal Tumorigenesis by Facilitating Enterotoxigenic Bacteroides Fragilis Colonization. Pharmacol Res (2022) 177:106101. doi: 10.1016/j.phrs.2022.106101
87. Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine Catabolism Contributes to Enterotoxigenic Bacteroides Fragilis-Induced Colon Tumorigenesis. Proc Natl Acad Sci USA (2011) 108(37):15354–9. doi: 10.1073/pnas.1010203108
88. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the Human Gut by E. Coli and Colorectal Cancer Risk. Clin Cancer Res (2014) 20(4):859–67. doi: 10.1158/1078-0432.Ccr-13-1343
89. Iyadorai T, Mariappan V, Vellasamy KM, Wanyiri JW, Roslani AC, Lee GK, et al. Prevalence and Association of Pks+ Escherichia Coli With Colorectal Cancer in Patients at the University Malaya Medical Centre, Malaysia. PloS One (2020) 15(1):e0228217. doi: 10.1371/journal.pone.0228217
90. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients With Familial Adenomatous Polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science (2018) 359(6375):592–7. doi: 10.1126/science.aah3648
91. Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-Associated Fecal Microbial Markers in Colorectal Cancer Detection. Int J Cancer (2017) 141(12):2528–36. doi: 10.1002/ijc.31011
92. Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, et al. Bacterial Genotoxin Colibactin Promotes Colon Tumour Growth by Inducing a Senescence-Associated Secretory Phenotype. Gut (2014) 63(12):1932–42. doi: 10.1136/gutjnl-2013-305257
93. Tomkovich S, Yang Y, Winglee K, Gauthier J, Mühlbauer M, Sun X, et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res (2017) 77(10):2620–32. doi: 10.1158/0008-5472.Can-16-3472
94. Wassenaar TM. Insights From 100 Years of Research With Probiotic E. Coli. Eur J Microbiol Immunol (Bp) (2016) 6(3):147–61. doi: 10.1556/1886.2016.00029
95. Tsvetikova SA, Koshel EI. Microbiota and Cancer: Host Cellular Mechanisms Activated by Gut Microbial Metabolites. Int J Med Microbiol (2020) 310(4):151425. doi: 10.1016/j.ijmm.2020.151425
96. Mirzaei R, Afaghi A, Babakhani S, Sohrabi MR, Hosseini-Fard SR, Babolhavaeji K, et al. Role of Microbiota-Derived Short-Chain Fatty Acids in Cancer Development and Prevention. BioMed Pharmacother (2021) 139:111619. doi: 10.1016/j.biopha.2021.111619
97. Høverstad T, Midtvedt T. Short-Chain Fatty Acids in Germfree Mice and Rats. J Nutr (1986) 116(9):1772–6. doi: 10.1093/jn/116.9.1772
98. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (Scfas)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
99. Kim CH. Microbiota or Short-Chain Fatty Acids: Which Regulates Diabetes? Cell Mol Immunol (2018) 15(2):88–91. doi: 10.1038/cmi.2017.57
100. Ohira H, Tsutsui W, Fujioka Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J Atheroscler Thromb (2017) 24(7):660–72. doi: 10.5551/jat.RV17006
101. Scheppach W, Bartram HP, Richter F. Role of Short-Chain Fatty Acids in the Prevention of Colorectal Cancer. Eur J Cancer (1995) 31a(7-8):1077–80. doi: 10.1016/0959-8049(95)00165-f
102. Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, et al. Role of Scfas in Gut Microbiome and Glycolysis for Colorectal Cancer Therapy. J Cell Physiol (2019) 234(10):17023–49. doi: 10.1002/jcp.28436
103. Hou H, Chen D, Zhang K, Zhang W, Liu T, Wang S, et al. Gut Microbiota-Derived Short-Chain Fatty Acids and Colorectal Cancer: Ready for Clinical Translation? Cancer Lett (2022) 526:225–35. doi: 10.1016/j.canlet.2021.11.027
104. Duncan SH, Louis P, Thomson JM, Flint HJ. The Role of Ph in Determining the Species Composition of the Human Colonic Microbiota. Environ Microbiol (2009) 11(8):2112–22. doi: 10.1111/j.1462-2920.2009.01931.x
105. Louis P, Hold GL, Flint HJ. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
106. Levy M, Thaiss CA, Elinav E. Metabolites: Messengers Between the Microbiota and the Immune System. Genes Dev (2016) 30(14):1589–97. doi: 10.1101/gad.284091.116
107. Bultman SJ. Interplay Between Diet, Gut Microbiota, Epigenetic Events, and Colorectal Cancer. Mol Nutr Food Res (2017) 61(1):1500902. doi: 10.1002/mnfr.201500902
108. Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int J Mol Sci (2019) 20(5):1214. doi: 10.3390/ijms20051214
109. Han A, Bennett N, Ahmed B, Whelan J, Donohoe DR. Butyrate Decreases Its Own Oxidation in Colorectal Cancer Cells Through Inhibition of Histone Deacetylases. Oncotarget (2018) 9(43):27280–92. doi: 10.18632/oncotarget.25546
110. Gamet L, Daviaud D, Denis-Pouxviel C, Remesy C, Murat JC. Effects of Short-Chain Fatty Acids on Growth and Differentiation of the Human Colon-Cancer Cell Line Ht29. Int J Cancer (1992) 52(2):286–9. doi: 10.1002/ijc.2910520222
111. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The Effects of Short-Chain Fatty Acids on Human Colon Cancer Cell Phenotype Are Associated With Histone Hyperacetylation. J Nutr (2002) 132(5):1012–7. doi: 10.1093/jn/132.5.1012
112. Zeng H, Hamlin SK, Safratowich BD, Cheng WH, Johnson LK. Superior Inhibitory Efficacy of Butyrate Over Propionate and Acetate Against Human Colon Cancer Cell Proliferation Via Cell Cycle Arrest and Apoptosis: Linking Dietary Fiber to Cancer Prevention. Nutr Res (2020) 83:63–72. doi: 10.1016/j.nutres.2020.08.009
113. Encarnação JC, Pires AS, Amaral RA, Gonçalves TJ, Laranjo M, Casalta-Lopes JE, et al. Butyrate, a Dietary Fiber Derivative That Improves Irinotecan Effect in Colon Cancer Cells. J Nutr Biochem (2018) 56:183–92. doi: 10.1016/j.jnutbio.2018.02.018
114. Cao M, Zhang Z, Han S, Lu X. Butyrate Inhibits the Proliferation and Induces the Apoptosis of Colorectal Cancer Hct116 Cells Via the Deactivation of Mtor/S6k1 Signaling Mediated Partly by Sirt1 Downregulation. Mol Med Rep (2019) 19(5):3941–7. doi: 10.3892/mmr.2019.10002
115. Yu DC, Waby JS, Chirakkal H, Staton CA, Corfe BM. Butyrate Suppresses Expression of Neuropilin I in Colorectal Cell Lines Through Inhibition of Sp1 Transactivation. Mol Cancer (2010) 9:276. doi: 10.1186/1476-4598-9-276
116. Zuo L, Lu M, Zhou Q, Wei W, Wang Y. Butyrate Suppresses Proliferation and Migration of Rko Colon Cancer Cells Though Regulating Endocan Expression by Mapk Signaling Pathway. Food Chem Toxicol (2013) 62:892–900. doi: 10.1016/j.fct.2013.10.028
117. Chen D, Jin D, Huang S, Wu J, Xu M, Liu T, et al. Clostridium Butyricum, a Butyrate-Producing Probiotic, Inhibits Intestinal Tumor Development Through Modulating Wnt Signaling and Gut Microbiota. Cancer Lett (2020) 469:456–67. doi: 10.1016/j.canlet.2019.11.019
118. Bordonaro M, Lazarova D. Amlexanox and Upf1 Modulate Wnt Signaling and Apoptosis in Hct-116 Colorectal Cancer Cells. J Cancer (2019) 10(2):287–92. doi: 10.7150/jca.28331
119. Cucciolla V, Borriello A, Criscuolo M, Sinisi AA, Bencivenga D, Tramontano A, et al. Histone Deacetylase Inhibitors Upregulate P57kip2 Level by Enhancing Its Expression Through Sp1 Transcription Factor. Carcinogenesis (2008) 29(3):560–7. doi: 10.1093/carcin/bgn010
120. Hu S, Liu L, Chang EB, Wang JY, Raufman JP. Butyrate Inhibits Pro-Proliferative Mir-92a by Diminishing C-Myc-Induced Mir-17-92a Cluster Transcription in Human Colon Cancer Cells. Mol Cancer (2015) 14:180. doi: 10.1186/s12943-015-0450-x
121. Li Q, Cao L, Tian Y, Zhang P, Ding C, Lu W, et al. Butyrate Suppresses the Proliferation of Colorectal Cancer Cells Via Targeting Pyruvate Kinase M2 and Metabolic Reprogramming. Mol Cell Proteomics (2018) 17(8):1531–45. doi: 10.1074/mcp.RA118.000752
122. Yoo HY, Park SY, Chang SY, Kim SH. Regulation of Butyrate-Induced Resistance Through Ampk Signaling Pathway in Human Colon Cancer Cells. Biomedicines (2021) 9(11):1604. doi: 10.3390/biomedicines9111604
123. Michael MZ, OC SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced Accumulation of Specific Micrornas in Colorectal Neoplasia. Mol Cancer Res (2003) 1(12):882–91.
124. Han R, Sun Q, Wu J, Zheng P, Zhao G. Sodium Butyrate Upregulates Mir-203 Expression to Exert Anti-Proliferation Effect on Colorectal Cancer Cells. Cell Physiol Biochem (2016) 39(5):1919–29. doi: 10.1159/000447889
125. Ali SR, Orang A, Marri S, McKinnon RA, Meech R, Michael MZ. Integrative Transcriptomic Network Analysis of Butyrate Treated Colorectal Cancer Cells. Cancers (Basel) (2021) 13(4):636. doi: 10.3390/cancers13040636
126. Hu S, Dong TS, Dalal SR, Wu F, Bissonnette M, Kwon JH, et al. The Microbe-Derived Short Chain Fatty Acid Butyrate Targets Mirna-Dependent P21 Gene Expression in Human Colon Cancer. PloS One (2011) 6(1):e16221. doi: 10.1371/journal.pone.0016221
127. Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, et al. Gut Bacteria Identified in Colorectal Cancer Patients Promote Tumourigenesis Via Butyrate Secretion. Nat Commun (2021) 12(1):5674. doi: 10.1038/s41467-021-25965-x
128. Kim J, Lee HK. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front Immunol (2021) 12:807648. doi: 10.3389/fimmu.2021.807648
129. LaCourse KD, Johnston CD, Bullman S. The Relationship Between Gastrointestinal Cancers and the Microbiota. Lancet Gastroenterol Hepatol (2021) 6(6):498–509. doi: 10.1016/s2468-1253(20)30362-9
130. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential Role of Intratumor Bacteria in Mediating Tumor Resistance to the Chemotherapeutic Drug Gemcitabine. Science (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
131. Yuan L, Zhang S, Li H, Yang F, Mushtaq N, Ullah S, et al. The Influence of Gut Microbiota Dysbiosis to the Efficacy of 5-Fluorouracil Treatment on Colorectal Cancer. BioMed Pharmacother (2018) 108:184–93. doi: 10.1016/j.biopha.2018.08.165
132. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
133. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell (2017) 170(3):548–63.e16. doi: 10.1016/j.cell.2017.07.008
134. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science (2017) 358(6369):1443–8. doi: 10.1126/science.aal5240
135. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut Microbiome Influences Efficacy of Pd-1-Based Immunotherapy Against Epithelial Tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
136. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The Commensal Microbiome Is Associated With Anti-Pd-1 Efficacy in Metastatic Melanoma Patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
137. Hayase E, Jenq RR. Role of the Intestinal Microbiome and Microbial-Derived Metabolites in Immune Checkpoint Blockade Immunotherapy of Cancer. Genome Med (2021) 13(1):107. doi: 10.1186/s13073-021-00923-w
138. Waldman AD, Fritz JM, Lenardo MJ. A Guide to Cancer Immunotherapy: From T Cell Basic Science to Clinical Practice. Nat Rev Immunol (2020) 20(11):651–68. doi: 10.1038/s41577-020-0306-5
139. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-Pd-L1 Efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
140. Rizvi ZA, Dalal R, Sadhu S, Kumar Y, Kumar S, Gupta SK, et al. High-Salt Diet Mediates Interplay Between Nk Cells and Gut Microbiota to Induce Potent Tumor Immunity. Sci Adv (2021) 7(37):eabg5016. doi: 10.1126/sciadv.abg5016
141. Sun S, Luo L, Liang W, Yin Q, Guo J, Rush AM, et al. Bifidobacterium Alters the Gut Microbiota and Modulates the Functional Metabolism of T Regulatory Cells in the Context of Immune Checkpoint Blockade. Proc Natl Acad Sci USA (2020) 117(44):27509–15. doi: 10.1073/pnas.1921223117
142. Wang F, Yin Q, Chen L, Davis MM. Bifidobacterium Can Mitigate Intestinal Immunopathology in the Context of Ctla-4 Blockade. Proc Natl Acad Sci USA (2018) 115(1):157–61. doi: 10.1073/pnas.1712901115
143. Si W, Liang H, Bugno J, Xu Q, Ding X, Yang K, et al. Lactobacillus Rhamnosus Gg Induces Cgas/Sting- Dependent Type I Interferon and Improves Response to Immune Checkpoint Blockade. Gut (2022) 71(3):521–33. doi: 10.1136/gutjnl-2020-323426
144. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer Immunotherapy by Ctla-4 Blockade Relies on the Gut Microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
145. Limeta A, Ji B, Levin M, Gatto F, Nielsen J. Meta-Analysis of the Gut Microbiota in Predicting Response to Cancer Immunotherapy in Metastatic Melanoma. JCI Insight (2020) 5(23):e140940. doi: 10.1172/jci.insight.140940
146. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated With Ipilimumab. Ann Oncol (2017) 28(6):1368–79. doi: 10.1093/annonc/mdx108
147. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut Microbiome Modulates Response to Anti-Pd-1 Immunotherapy in Melanoma Patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
148. Peters BA, Wilson M, Moran U, Pavlick A, Izsak A, Wechter T, et al. Relating the Gut Metagenome and Metatranscriptome to Immunotherapy Responses in Melanoma Patients. Genome Med (2019) 11(1):61. doi: 10.1186/s13073-019-0672-4
149. Ting NL, Lau HC, Yu J. Cancer Pharmacomicrobiomics: Targeting Microbiota to Optimise Cancer Therapy Outcomes. Gut (2022). doi: 10.1136/gutjnl-2021-326264
150. Theodoropoulos GE, Memos NA, Peitsidou K, Karantanos T, Spyropoulos BG, Zografos G. Synbiotics and Gastrointestinal Function-Related Quality of Life After Elective Colorectal Cancer Resection. Ann Gastroenterol (2016) 29(1):56–62.
151. Cheng WY, Wu CY, Yu J. The Role of Gut Microbiota in Cancer Treatment: Friend or Foe? Gut (2020) 69(10):1867–76. doi: 10.1136/gutjnl-2020-321153
152. Mego M, Chovanec J, Vochyanova-Andrezalova I, Konkolovsky P, Mikulova M, Reckova M, et al. Prevention of Irinotecan Induced Diarrhea by Probiotics: A Randomized Double Blind, Placebo Controlled Pilot Study. Complement Ther Med (2015) 23(3):356–62. doi: 10.1016/j.ctim.2015.03.008
153. Consoli ML, da Silva RS, Nicoli JR, Bruña-Romero O, da Silva RG, de Vasconcelos Generoso S, et al. Randomized Clinical Trial: Impact of Oral Administration of Saccharomyces Boulardii on Gene Expression of Intestinal Cytokines in Patients Undergoing Colon Resection. JPEN J Parenter Enteral Nutr (2016) 40(8):1114–21. doi: 10.1177/0148607115584387
154. Gibson GR, Roberfroid MB. Dietary Modulation of the Human Colonic Microbiota: Introducing the Concept of Prebiotics. J Nutr (1995) 125(6):1401–12. doi: 10.1093/jn/125.6.1401
155. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (Isapp) Consensus Statement on the Definition and Scope of Prebiotics. Nat Rev Gastroenterol Hepatol (2017) 14(8):491–502. doi: 10.1038/nrgastro.2017.75
156. Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, et al. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods (2019) 8(3):92. doi: 10.3390/foods8030092
157. Pothuraju R, Chaudhary S, Rachagani S, Kaur S, Roy HK, Bouvet M, et al. Mucins, Gut Microbiota, and Postbiotics Role in Colorectal Cancer. Gut Microbes (2021) 13(1):1974795. doi: 10.1080/19490976.2021.1974795
158. Taper HS, Roberfroid MB. Nontoxic Potentiation of Cancer Chemotherapy by Dietary Oligofructose or Inulin. Nutr Cancer (2000) 38(1):1–5. doi: 10.1207/s15327914nc381_1
159. Taper HS, Roberfroid MB. Possible Adjuvant Cancer Therapy by Two Prebiotics–Inulin or Oligofructose. In Vivo (2005) 19(1):201–4.
160. Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, et al. Insight Into the Prebiotic Concept: Lessons From an Exploratory, Double Blind Intervention Study With Inulin-Type Fructans in Obese Women. Gut (2013) 62(8):1112–21. doi: 10.1136/gutjnl-2012-303304
161. Lynch JP, Goers L, Lesser CF. Emerging Strategies for Engineering Escherichia Coli Nissle 1917-Based Therapeutics. Trends Pharmacol Sci (2022). doi: 10.1016/j.tips.2022.02.002
162. So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, et al. Dietary Fiber Intervention on Gut Microbiota Composition in Healthy Adults: A Systematic Review and Meta-Analysis. Am J Clin Nutr (2018) 107(6):965–83. doi: 10.1093/ajcn/nqy041
163. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A Defined Commensal Consortium Elicits Cd8 T Cells and Anti-Cancer Immunity. Nature (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
164. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat Rev Gastroenterol Hepatol (2014) 11(8):506–14. doi: 10.1038/nrgastro.2014.66
165. Botta C, Spyridopoulou K, Bertolino M, Rantsiou K, Chlichlia K, Cocolin L. Lactiplantibacillus Plantarum Inhibits Colon Cancer Cell Proliferation as Function of Its Butyrogenic Capability. BioMed Pharmacother (2022) 149:112755. doi: 10.1016/j.biopha.2022.112755
166. do Carmo FLR, Rabah H, Cordeiro BF, da Silva SH, Pessoa RM, Fernandes SOA, et al. Probiotic Propionibacterium Freudenreichii Requires Slpb Protein to Mitigate Mucositis Induced by Chemotherapy. Oncotarget (2019) 10(68):7198–219. doi: 10.18632/oncotarget.27319
167. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary Fiber and Probiotics Influence the Gut Microbiome and Melanoma Immunotherapy Response. Science (2021) 374(6575):1632–40. doi: 10.1126/science.aaz7015
168. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated With Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia (2017) 19(10):848–55. doi: 10.1016/j.neo.2017.08.004
169. Borody TJ, Khoruts A. Fecal Microbiota Transplantation and Emerging Applications. Nat Rev Gastroenterol Hepatol (2011) 9(2):88–96. doi: 10.1038/nrgastro.2011.244
170. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal Microbiota Transplantation for Refractory Immune Checkpoint Inhibitor-Associated Colitis. Nat Med (2018) 24(12):1804–8. doi: 10.1038/s41591-018-0238-9
171. Matson V, Chervin CS, Gajewski TF. Cancer and the Microbiome-Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology (2021) 160(2):600–13. doi: 10.1053/j.gastro.2020.11.041
172. Kassam Z, Dubois N, Ramakrishna B, Ling K, Qazi T, Smith M, et al. Donor Screening for Fecal Microbiota Transplantation. N Engl J Med (2019) 381(21):2070–2. doi: 10.1056/NEJMc1913670
173. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated With Acquired Resistance to Pd-1 Blockade in Melanoma. N Engl J Med (2016) 375(9):819–29. doi: 10.1056/NEJMoa1604958
174. Spranger S, Gajewski TF. Tumor-Intrinsic Oncogene Pathways Mediating Immune Avoidance. Oncoimmunology (2016) 5(3):e1086862. doi: 10.1080/2162402x.2015.1086862
175. Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary Resistance to Pd-1 Blockade Mediated by Jak1/2 Mutations. Cancer Discov (2017) 7(2):188–201. doi: 10.1158/2159-8290.Cd-16-1223
176. Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of Resistance to Immune Checkpoint Inhibitors. Br J Cancer (2018) 118(1):9–16. doi: 10.1038/bjc.2017.434
177. Trujillo JA, Luke JJ, Zha Y, Segal JP, Ritterhouse LL, Spranger S, et al. Secondary Resistance to Immunotherapy Associated With B-Catenin Pathway Activation or Pten Loss in Metastatic Melanoma. J Immunother Cancer (2019) 7(1):295. doi: 10.1186/s40425-019-0780-0
178. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal Microbiota Transplant Overcomes Resistance to Anti-Pd-1 Therapy in Melanoma Patients. Science (2021) 371(6529):595–602. doi: 10.1126/science.abf3363
179. Wu C, Lai R, Li J, Zhang J, Zhao Y, Zhang X, et al. Antibiotics Modulate Chemotherapy Efficacy in Patients With Esophageal Cancer. Cancer Manag Res (2020) 12:4991–7. doi: 10.2147/cmar.S248130
180. Nenclares P, Bhide SA, Sandoval-Insausti H, Pialat P, Gunn L, Melcher A, et al. Impact of Antibiotic Use During Curative Treatment of Locally Advanced Head and Neck Cancers With Chemotherapy and Radiotherapy. Eur J Cancer (2020) 131:9–15. doi: 10.1016/j.ejca.2020.02.047
181. Zheng DW, Dong X, Pan P, Chen KW, Fan JX, Cheng SX, et al. Phage-Guided Modulation of the Gut Microbiota of Mouse Models of Colorectal Cancer Augments Their Responses to Chemotherapy. Nat BioMed Eng (2019) 3(9):717–28. doi: 10.1038/s41551-019-0423-2
182. Pinato DJ, Howlett S, Ottaviani D, Urus H, Patel A, Mineo T, et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol (2019) 5(12):1774–8. doi: 10.1001/jamaoncol.2019.2785
183. Tinsley N, Zhou C, Tan G, Rack S, Lorigan P, Blackhall F, et al. Cumulative Antibiotic Use Significantly Decreases Efficacy of Checkpoint Inhibitors in Patients With Advanced Cancer. Oncologist (2020) 25(1):55–63. doi: 10.1634/theoncologist.2019-0160
184. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative Association of Antibiotics on Clinical Activity of Immune Checkpoint Inhibitors in Patients With Advanced Renal Cell and Non-Small-Cell Lung Cancer. Ann Oncol (2018) 29(6):1437–44. doi: 10.1093/annonc/mdy103
185. Wilson BE, Routy B, Nagrial A, Chin VT. The Effect of Antibiotics on Clinical Outcomes in Immune-Checkpoint Blockade: A Systematic Review and Meta-Analysis of Observational Studies. Cancer Immunol Immunother (2020) 69(3):343–54. doi: 10.1007/s00262-019-02453-2
186. Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, et al. Recovery of Gut Microbiota of Healthy Adults Following Antibiotic Exposure. Nat Microbiol (2018) 3(11):1255–65. doi: 10.1038/s41564-018-0257-9
187. Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur Urol (2020) 78(2):195–206. doi: 10.1016/j.eururo.2020.04.044
188. Hakozaki T, Richard C, Elkrief A, Hosomi Y, Benlaïfaoui M, Mimpen I, et al. The Gut Microbiome Associates With Immune Checkpoint Inhibition Outcomes in Patients With Advanced Non-Small Cell Lung Cancer. Cancer Immunol Res (2020) 8(10):1243–50. doi: 10.1158/2326-6066.Cir-20-0196
189. Thun MJ, Sinks T. Understanding Cancer Clusters. CA Cancer J Clin (2004) 54(5):273–80. doi: 10.3322/canjclin.54.5.273
190. You WC, Blot WJ, Chang YS, Ershow AG, Yang ZT, An Q, et al. Diet and High Risk of Stomach Cancer in Shandong, China. Cancer Res (1988) 48(12):3518–23.
191. Herman AM, Hawkins AT, Misso K, Issangya C, Tarmohamed M, Mremi A, et al. Colorectal Cancer in Northern Tanzania: Increasing Trends and Late Presentation Present Major Challenges. JCO Glob Oncol (2020) 6:375–81. doi: 10.1200/jgo.19.00301
Keywords: gut microbiota, gastric cancer, colorectal cancer, SCFAs, therapy
Citation: Guo Q, Qin H, Liu X, Zhang X, Chen Z, Qin T, Chang L and Zhang W (2022) The Emerging Roles of Human Gut Microbiota in Gastrointestinal Cancer. Front. Immunol. 13:915047. doi: 10.3389/fimmu.2022.915047
Received: 07 April 2022; Accepted: 16 May 2022;
Published: 15 June 2022.
Edited by:
Le Guo, General Hospital of Ningxia Medical University, ChinaCopyright © 2022 Guo, Qin, Liu, Zhang, Chen, Qin, Chang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianqian Guo, emx5eWdxcTQyNjVAenp1LmVkdS5jbg==; Wenzhou Zhang, aG56enp3enhAc2luYS5jb20=
†These authors have contributed equally to this work
 Qianqian Guo
Qianqian Guo Hai Qin
Hai Qin Xueling Liu
Xueling Liu Xinxin Zhang
Xinxin Zhang Zelong Chen
Zelong Chen Tingting Qin
Tingting Qin Linlin Chang
Linlin Chang Wenzhou Zhang
Wenzhou Zhang