
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 August 2022
Sec. Nutritional Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.913854
We previously observed the beneficial role of folic acid supplemented from maternal or offspring diet on lamb growth performance and immunity. Twenty-four Hu lambs from four groups (mother received folic acid or not, offspring received folic acid or not) were used in the current study, which was conducted consecutively to elucidate the molecular regulatory mechanisms of folic acid in lambs by analyzing blood metabolome, liver transcriptome, and muscle transcriptome. Serum metabolomics analysis showed that L-homocitrulline, hyodeoxycholic acid, 9-Hpode, palmitaldehyde, N-oleoyl glycine, hexadecanedioic acid, xylose, 1,7-dimethylxanthine, nicotinamide, acetyl-N-formyl-5-methoxykynurenamine, N6-succinyl adenosine, 11-cis-retinol, 18-hydroxycorticosterone, and 2-acetylfuran were down-regulated and methylisobutyrate was up-regulated by the feeding of folic acid from maternal and/or offspring diets. Meanwhile, folic acid increased the abundances of S100A12 and IRF6 but decreased TMEM25 in the liver. In the muscle, RBBP9, CALCR, PPP1R3D, UCP3, FBXL4, CMBL, and MTFR2 were up-regulated, CYP26B1 and MYH9 were down-regulated by the feeding of folic acid. The pathways of bile secretion, biosynthesis of unsaturated fatty acids, linoleic acid metabolism, and herpes simplex virus 1 infection were changed by folic acid in blood, liver, or muscle. Further integrated analysis revealed potential interactions among the liver, blood, and muscle, and the circulating metabolites, hub gene, and pathways, which might be the predominant acting targets of folic acid in animals. These findings provide fundamental information on the beneficial function of folic acid no matter from maternal or offspring, in regulating animal lipid metabolism and immune enhancement, providing a theoretical basis for the use of folic acid from the view of animal health care.
Folic acid (FA) is a water-soluble B vitamin involved in purine, pyrimidine and methionine metabolism as a methyl carrier (1), which plays a critical role in DNA methylation and methionine metabolism. Generally, the requirement of FA in ruminants can be met by producing the ruminal microflora (2). However, dietary supplements of FA higher than 0.5 mg/kg body weight improved animal performance and feed efficiency with increased serum folate concentrations and milk folate concentrations in most studies of ruminants (3, 4). We recently found that the supplementation of rumen-protected FA in the maternal and/or lamb diets could increase the growth performance of lambs (5) and improve slaughter performance and meat quality (6).
Furthermore, it was found that FA significantly affects nucleic acid synthesis, DNA repair, cell growth, differentiation, apoptosis, cancer prevention (7), muscle development (8), and energy metabolism (9). In addition, FA plays a crucial role in the healthy balance of the immune system and plays a double-edged sword role in offspring health via mediating DNA methylation (10, 11). Inadequate levels of FA can drastically alter immune responses (11). It was also found that maternal FA deficiency changed the methylation of genes and pathways associated with neurodevelopment and learning/memory abilities in male rat offspring (12). Similarly, FA supplementation could enhance immunity or anti-inflammation (8, 13) and stimulate hepatic gene expression in the IGF-1/PI3K/mTOR pathway (14) in ruminants.
Our previous studies showed that FA improved animal growth and meat quality (5, 6); in the meantime, it had significant effects on animal health-related characteristics in sheep, such as serum high-density lipoprotein cholesterol and very low-density lipoprotein (5) and serum immune globulins (8). FA supplementation could affect proliferation and apoptosis in bovine mammary epithelial cells by increasing the ratio of Bcl-2/Bax protein levels (15). However, few studies have investigated FA’s systematic molecular regulatory mechanism, especially for their function in immunity, in ruminant species. In addition, the gut-liver-muscle axis is associated with many diseases in patients (16, 17). Therefore, this study aimed to identify the FA’s biological function on immunological enhancement in a sheep model using multi-omics through the liver-muscle axis. We speculated that an integrative network of blood, liver, and muscle directly responds to FA’s maternal and/or offspring supplementation.
The information of rumen-protected FA has been described previously (9). The present study was conducted at Jiangsu Qianbao Animal Husbandry Co. Ltd. For the experiment, 120 Hu ewes (24 ± 4.2 months of age and 44.6 ± 5.43 kg of body weight, mean ± SD) with two parity and showing signs of estrus (based on vaginal examination) were selected and then randomly allocated into three groups (forty ewes in each group). The experimental ewes received either 0 mg (M0), 16 mg (M16), or 32 mg (M32) FA as rumen-protected FA per kg DM from mating to lambing. The added dose of FA was according to a previous study in beef steers (18). The rumen-protected FA contained 2% FA, 60% hydrogenated fat (ratio of C16:0 to C18:0 = 2:1), 16% bentonite powder, and 22% calcium stearate (rumen-protected rate 92.6%). All newborn lambs were nursed and weaned at 50 d of age. Then the twenty-two offspring were selected from each maternal group and allocated into two groups based on litter size, sex, and bodyweight to receive either 0 mg (C) or 4.0 mg/kg (F) rumen-protected FA (DM in their diets). Altogether, 66 Hu lambs (50 ± 4.3 d of age and 16.4 ± 3.84 kg of body weight) were selected and divided into a total of six groups. All the lambs were raised separately in individual pens. The feeding experiment of the weaned lambs lasted for 130 d and included a 10-d adaptation period followed by a 120-d collection period. The detailed procedures for the ewes and lambs raising were shown in our published studies (5, 6). Six lambs from each group were selected randomly for slaughtering. Due to the significant beneficial roles of M0F (only offspring received 4 mg FA), M16C (only ewe received 16 mg FA), M16F (ewe received 16 mg FA and offspring received 4 mg FA) on animal growth and health properties from our previous studies (5, 6), we selected these three groups and M0C (no FA received) for further multi-omics analysis. Three comparisons among the four groups (M0C vs. M0F, M0C vs. M16C, M0C vs. M16F) were conducted in the current study to reveal the specific function of FA in lambs.
The blood samples on d 180 before morning feeding were collected from the jugular vein, and the serum was achieved by centrifuging the blood at 4°C for 10 min at 2000 × g. After slaughtering, longissimus dorsi and liver tissues (approximately 2 g) were collected and placed in cryopreserved tubes for further transcriptome analysis. All these samples were stored under liquid nitrogen.
Serum sample extraction process: 1) The samples were thawed on ice. After thawing, vortex for 10 s to mix well; 2) Take 50 μL of the sample into an EP tube, add 150 μL of pre-cooled ice methanol (containing 1 μg/mL of 2-chlorophenylalanine as an internal standard); 3) Vortex for 3 min, 12,000 r/min, and centrifuge at 4°C for 10 min; 4) After centrifugation, pipette 200 μL of the supernatant into another new EP tube; 5) Centrifuge the supernatant with 12,000 r/min at 4°C for 5 min, and take 150 μL of the supernatant into the lining tube of the injection bottle for LC-MS/MS analysis.
The data acquisition instrument system mainly includes Ultra Performance Liquid Chromatography (UPLC) (Shim-pack UFLC SHIMADZU CBM30A, https://www.shimadzu.com/) and tandem mass spectrometry (MS/MS) (QTRAP®, https://sciex.com/). The liquid conditions mainly include Column: Waters ACQUITY UPLC HSS T3 C18 1.8 µm, 2.1 mm×100 mm; Mobile phase: ultrapure water (0.04% acetic acid) in phase A, acetonitrile (0.04% acetic acid) in phase B; Elution gradient: water/acetonitrile (95:5 V/V) at 0 min, 5:95 V/V at 11.0 min, 5:95 V/V at 12.0 min, 95:5 V/V at 12.1 min, 14.0 The min was 95:5 V/V; the flow rate was 0.4 mL/min; the column temperature was 40°C; the injection volume was 2 μL. The mass spectrometry conditions mainly include electrospray ion source temperature 500 °C, mass spectrometry voltage 5500 V (positive), -4500 V (negative), ion source gas I (GS I) 55 psi, gas II (GS II) 60 psi, curtain gas 25 psi with the Collision Induced Ionization parameter set to high. Each ion pair is scanned in triple quadrupole (Qtrap) according to the optimized declustering voltage and collision energy. Quality control samples prepared by mixing sample extracts were inserted into every 10 assay samples to monitor the repeatability of the analysis process. The high stability of the instrument provides an essential guarantee for the repeatability and reliability of the data.
Mass spectral data were processed using the software Analyst 1.6.3. The metabolites of the samples were qualitatively and quantitatively analyzed by mass spectrometry based on the local metabolic database. Before doing the difference analysis, the principal component analysis was first performed, and then the orthogonal partial least squares discriminant analysis (OPLS-DA) was conducted. Differential metabolites were screened by combining fold change and the variable importance in projection (VIP) value of the OPLS-DA model. Screening criteria: select metabolites with VIP ≥ 1 and fold change > 1.2. Differential metabolites were annotated and displayed using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp) database.
Total RNA from liver and muscle tissue was extracted using EASYspin fibrous tissue RNA rapid extraction kit (RN44, Aidlab Biotechnologies Co., Ltd, Beijing, China), and all steps were carried out in strict accordance with the kit instructions. After the sample was qualified, magnetic beads with Oligo (dT) were used to enrich the mRNA. Then AMPure XP beads were used to purify the double-stranded cDNA, and then perform end repair, add A, and add adapters to the purified double-stranded cDNA. AMPure XP beads selected the size of the double-stranded cDNA, and finally, PCR amplification was performed to construct a cDNA library. Before sequencing, the Agilent 2100 was used to detect the size of the inserted fragment of the library. The effective concentration of the library was accurately quantified by the qPCR method (the effective concentration of the library was > 4 nM). Finally, Illumina Hiseq4000, PE 150, was used for On-board sequencing (Beijing Aoweisen Gene Technology Co., Ltd).
The raw data (raw reads) obtained by sequencing had minor contamination, and low-quality reads. To prevent the impact of subsequent analysis, they were filtered through in-house Perl scripts and used Trimmomatic software (v0.33) to remove bands. Remove reads with N (indeterminate base) content greater than 10%; remove reads with low-quality bases (Q ≤ 20) content greater than 50%. Finally, the sequencing quality Q20 > 95%, Q30 > 89% (Q20, Q30: the ratio of the number of bases with Phred quality value greater than 20, 30 to the total number of bases (clean data)) were selected to do follow-up comparison and data analysis. Alignment analysis with the reference genome of Texel sheep Oar_v3.1 (http://ftp.ensembl.org/pub/release-92/fasta/ovis_aries/dna/) was performed by Tophat2 (v2.1.0) software. The raw reads of the liver and muscle transcriptome sequencing are available at NCBI SRA (PRJNA822866 and PRJNA822707).
HTSeq (v 0.5.4 p3) software was used to analyze the gene expression level of each sample, and the model was union. Fragments per kilobase of exon model per million mapped reads (FPKM) is the number of fragments per kilobase per million reads from a gene that is used to measure gene expression levels (19). Only genes with FPKM > 1 that reached the expression threshold were analyzed in subsequent analyses. Gene differential expression analysis was performed using DESeq (1.10.1) on Reading count data obtained from gene expression level analysis (20). The standard of differential expressed gene screening was P < 0.05. KEGG was used to study the enrichment of significant pathways involved in differentially expressed genes.
Total RNA from the liver and muscle was reverse transcribed to cDNA using the TRUEscript RT (+gDNA Eraser) cDNA Synthesis Kit (Code No. PC5401, Aidlab Biotechnologies Co., Ltd, Beijing, China). Quantitative real-time PCR (qRT-PCR) was performed with the SYBR Green I (2×SYBR qPCR Mix) kit (Code No. PC3302, Aidlab Biotechnologies Co., Ltd, Beijing, China) and a LineGene 9640 instrument (BIOER Technology Co., Ltd, Hangzhou, China). The PCR conditions were set as follows: 1 cycle at 95°C for 3 min, 40 cycles of 95°C for 10 s and 60°C for 30 s, followed by a melting curve programme (60 to 95°C). The primers targeting hepatic IRF6, and muscular FBXL4, UCP3, and CYP26B1were listed in Table S1. Actin beta (ACTB) was used as a reference gene. The relative mRNA abundance of each gene was analyzed using the 2-ΔCT, ΔCT = CTtarget mRNA – CThouse keeping mRNA. Only M0C and M16F groups were used in the qPCR validation.
Spearman correlation coefficients (r) were calculated in the R program, and P < 0.01 was considered statistically significant. The network file was visualized using Cytoscape (v3.8.0) software to present the core and hub gene biological interactions.
A total of 461 compounds were identified in the serum metabolome. The OPLS-DA score plots of the serum samples from the three comparisons are shown in Figure 1A. Compared to the M0C group, 60, 128, and 121 differentially expressed metabolites were identified in the liver of the M0F, M16C, and M16F groups (Table S2). In total, 15 differentially expressed mutual metabolites were found, of which 14 differentially expressed metabolites were downregulated and 1 were upregulated in the three FA groups compared to those in the M0C group (Figure 1B). The main downregulated differentially expressed metabolites were L-homocitrulline, hyodeoxycholic acid, 9-Hpode, palmitaldehyde, N-oleoyl glycine, hexadecanedioic acid, xylose, 1,7-dimethylxanthine, nicotinamide, N6-succinyl adenosine, 11-cis-retinol, acetyl-N-formyl-5-methoxykynurenamine, 18-hydroxycorticosterone, and 2-acetylfuran, and only methylisobutyrate was upregulated by FA feeding. The metabolome map revealed 2 enriched significant KEGG metabolic pathways in comparing M0C and M0F, including synaptic vesicle cycle and neuroactive ligand-receptor interaction (Figure 1C). For comparing M0C and M16C, M0C and M16F, two same significant KEGG metabolic pathways, including biosynthesis of unsaturated fatty acids and linoleic acid metabolism, were found.
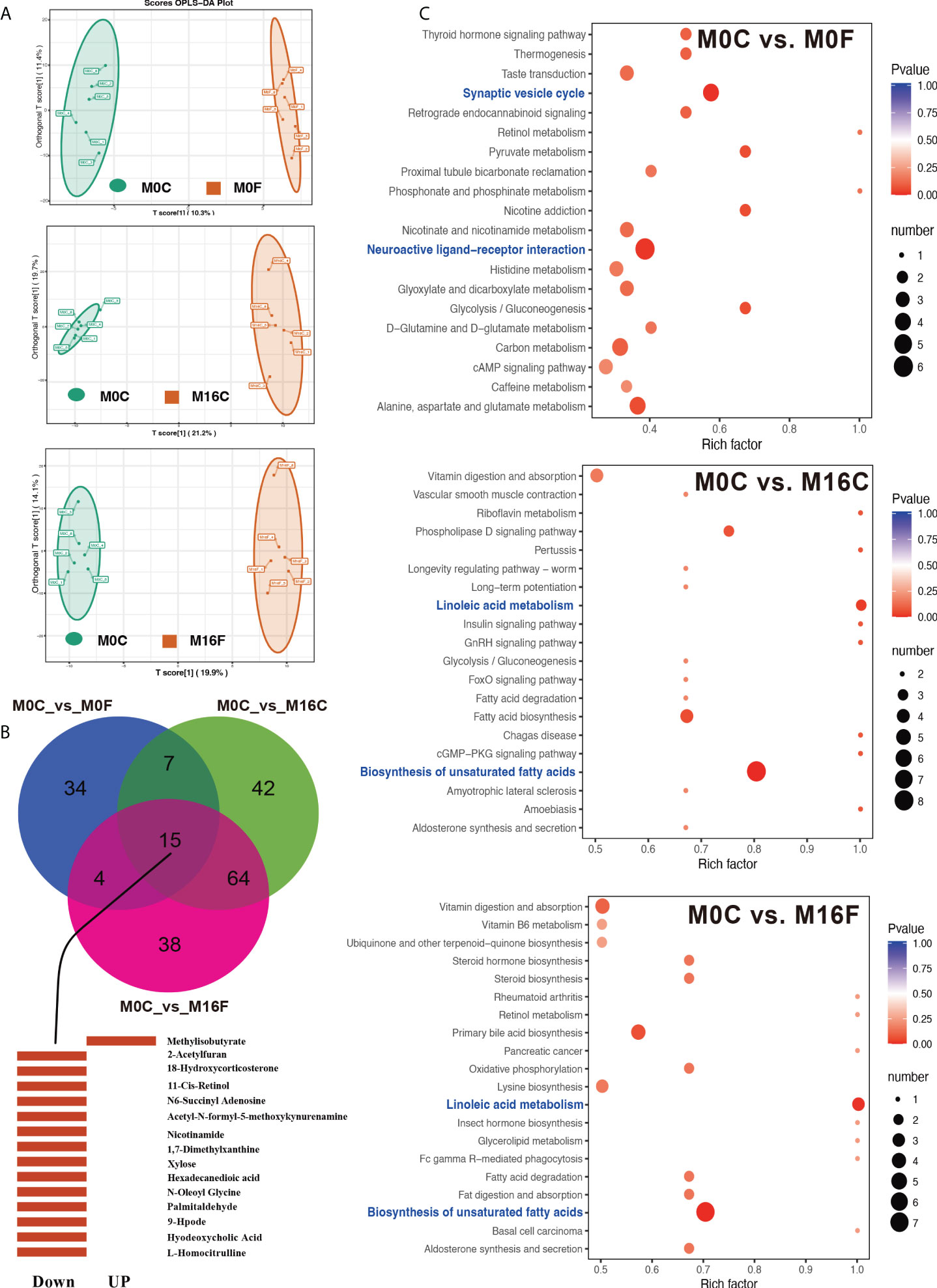
Figure 1 Folic acid changes the metabolome and pathways in the blood. OPLS-DA analyses of total blood metabolites among the comparisons of M0-OC vs. M0-OF, M0-OC vs. M16-OC, and M0-OC vs. M16-OF (A). The mutual hub different metabolites among the three comparisons (B). Top 20 enriched KEGG pathways based on the different metabolites (C). Richness factor = the ratio of different metabolites to the number of total metabolites in each pathway. The pathways highlighted with blue are the significant enriched metabolic pathways (P < 0.05).
RNA sequencing-based transcriptome analysis was performed to determine metabolic disorders in the liver. In total, 50.69 ± 3.69 and 51.78 ± 5.04 million clean sequence reads were obtained from the liver and muscle transcriptome, respectively. 31,565 and 30,637 genes were identified in the liver and muscle, respectively. The principal component analysis (PCA) score plots and volcano plots of the liver and muscle from the four groups were shown in Figures S1, S2, respectively. The PCA plots from both liver and muscle did not show a clear separation between the four groups. Compared to the M0C group, 238, 263, and 280 differentially expressed genes were identified in the liver of M0F, M16C, and M16F groups, and 179, 357, and 305 differentially expressed genes were identified in the muscle of M0F, M16C, and M16F groups. The mutual differentially expressed genes with annotated names in the liver among the three comparisons were S100A12, TMEM25, and IRF6 (Figure 2A), and the mutual differentially expressed genes with annotated names in muscle among the three comparisons are RBBP9, CYP26B1, CALCR, PPP1R3D, UCP3, FBXL4, CMBL, MTFR2, MYH9 (Figure 2B). The mutual differentially expressed genes between liver and muscle were CYP26B1, MFSD2B, HTATIP2, GLUL, FOXS1, BBS10, BST-2B, PRSS35, ECM1, FAM227A, HSPA6, RND2, SMPDL3A, CD274, CCDC62, and IFI44 (Figure 2C).
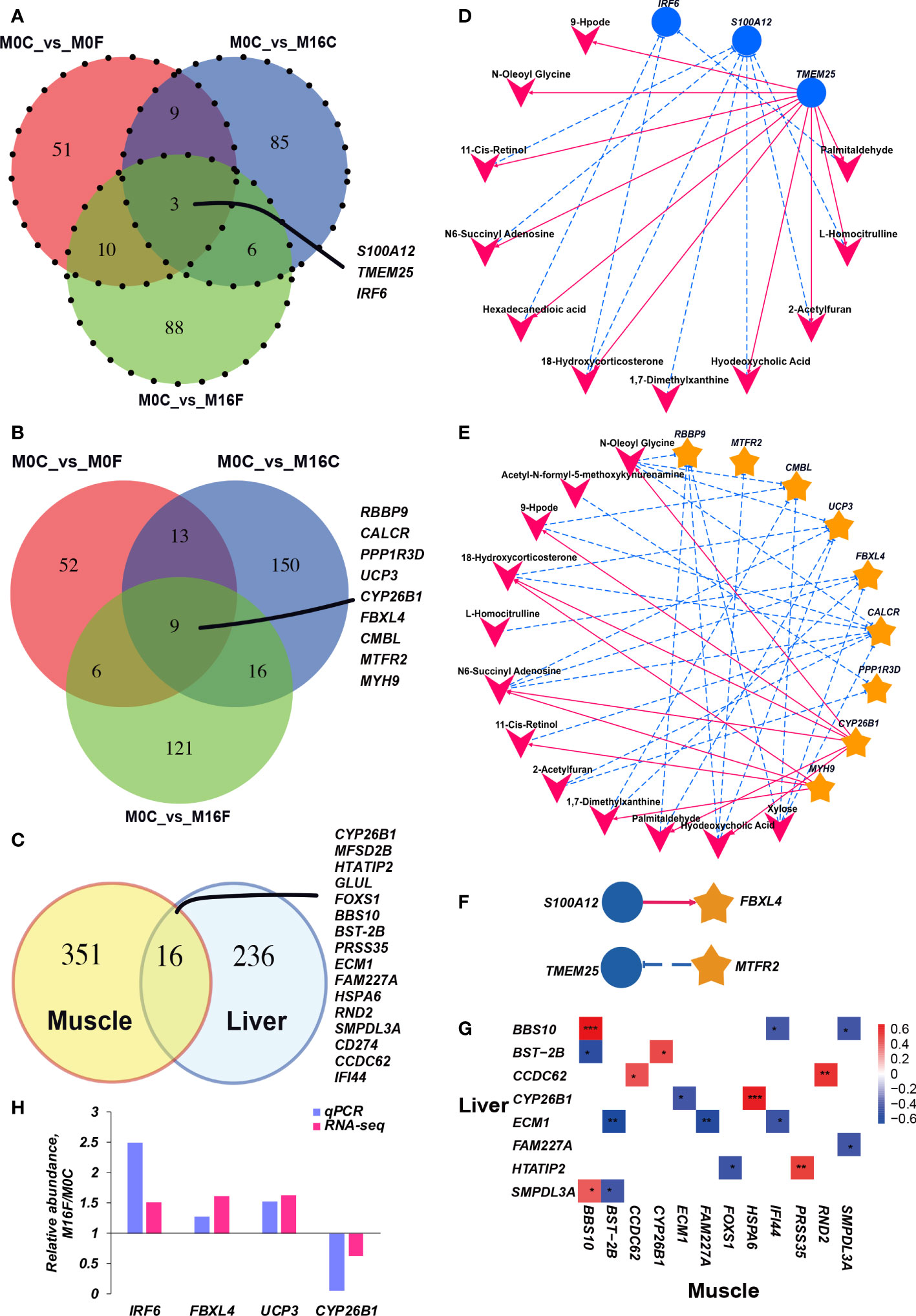
Figure 2 Cross-talk among the liver genes, muscle genes, and blood metabolites. Venn diagram illustrating the overlap of liver differential expressed genes between the three comparisons (A). Venn diagram illustrating the overlap of muscle differential expressed genes between the three comparisons (B). Venn diagram illustrating the overlap of differential expressed genes under the three comparisons between liver and muscle (C). The association between hub differentially expressed genes in the muscle and blood hub different metabolites (D). The association between hub differentially expressed genes in the liver and blood different metabolites (E). The association between hub differentially expressed genes in the liver and muscle (F). The correlation heatmap between mutual differentially expressed genes in the liver and muscle (G). Each co-occurring pair between two items had an absolute Spearman tank correlation above 0.53 (red full-line, positive correlation; blue dotted line, negative correlation) with a P-value under 0.01. The blue circle indicates differential hepatic genes, the yellow star indicates differential muscle genes, and the V-shape indicates blood metabolites. The selected core genes (IRF6 in the liver and FBXL4, UCP3, and CYP26B1 in the muscle) verified by quantitative real-time PCR (H).
Inter-omics correlation networks of all the blood metabolome variables, liver and muscle transcriptomes that were mutually different among the three comparisons were established (Figures 2D–G). In the communication between liver and blood (Figure 2D), hepatic IRF6 was negatively correlated with blood palmitaldehyde, 18-hydroxycorticosterone, hexadecanedioic acid. Hepatic S100A12 was negatively correlated with blood L-homocitrulline, 2-acetylfuran, hyodeoxycholic acid, 1,7-dimethylxanthine, 18-hydroxycorticosterone, N6-succinyl adenosine, 11-cis-retinol. TMEM25 was positively correlated with 2-acetylfuran, palmitaldehyde, L-homocitrulline, hexadecanedioic acid, N-oleoyl glycine, hyodeoxycholic acid, 18-hydroxycorticosterone, 9-Hpode, 11-cis-retinol, and N6-succinyl adenosine. In the connection between muscle and blood (Figure 2E), CALCR was negatively correlated with blood 2-acetylfuran, xylose, 11-cis-retinol, 9-Hpode, N-oleoyl glycine, 18-hydroxycorticosterone, and N6-succinyl adenosine. CMBL was negatively correlated with blood xylose, 1,7-dimethylxanthine, 9-Hpode, and N-oleoyl glycine. CYP26B1 was negatively correlated with blood N-oleoyl glycine, palmitaldehyde, N6-succinyl adenosine, hyodeoxycholic acid, 9-Hpode, and 18-hydroxycorticosterone. Only hepatic S100A12 was positively correlated with muscle FBXL4, and hepatic TMEM25 was negatively correlated with muscle MTFR2 (Figure 2F). There are 16 mutual differentially expressed genes between liver and muscle. During these 16 genes, only BBS10 and CCDC62 have a strong positive correlation between liver and muscle (Figure 2G).
To validate the core genes based on RNA-seq results, mutual differentially expressed genes, including IRF6 in the liver and FBXL4, UCP3, and CYP26B1 in the muscle, were selected for further verification using qRT-PCR (Figure 2H). As expected, the abundances of IRF6, FBXL4, and UCP3 were increased in the M16F group, and the expression level of CYP26B1 was decreased in the M16F group compared to the M0C group, which was significantly different between these two groups and overall agreed with the RNA-seq results.
The enriched KEGG pathway analysis was performed to explore the biological function of the differentially expressed genes responding to the FA supplementation. 11, 24, and 11 functional different KEGG pathways were identified in the liver tissues from the comparisons of M0C vs. M0F, M0C vs. M16C, and M0C vs. M16F respectively (P < 0.05). The mutual different KEGG pathways among the four comparisons were bile secretion, herpes simplex virus 1 infection, and ABC transporters. Linoleic acid metabolism was a mutual pathway between M0C vs. M0F and M0C vs. M16F (Figures 3A, B). In the muscle, 8, 18, and 17 functional different KEGG pathways were identified from M0C vs. M0F, M0C vs. M16C, and M0C vs. M16F, respectively (P < 0.05). The mutual different KEGG pathway was herpes simplex virus 1 infection (Figures 4A, B).
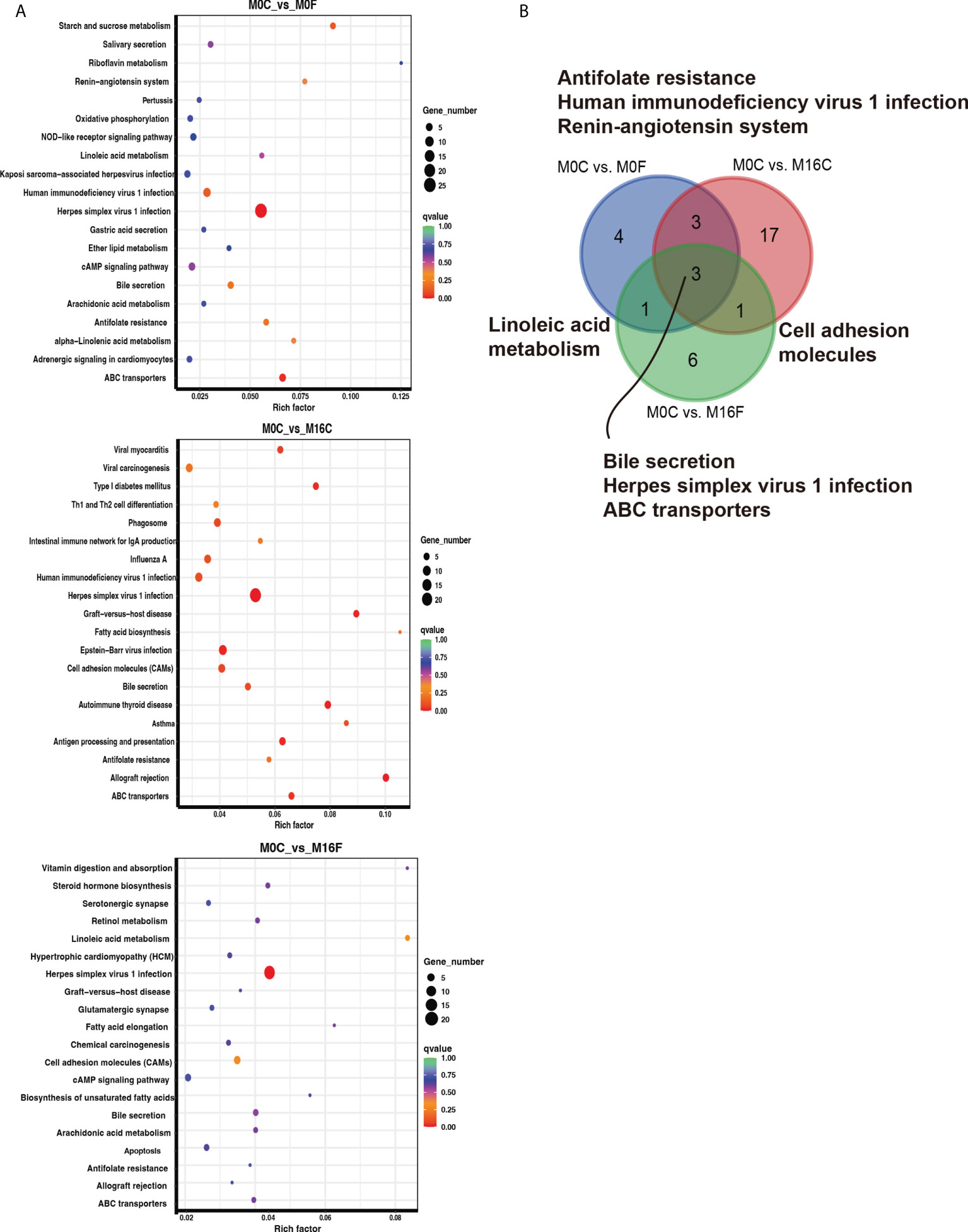
Figure 3 The hub affected KEGG pathways in the liver by folic acid supplementation. Top 20 enriched KEGG pathways based on the differentially expressed genes for the three comparisons in the liver (A). Venn diagram illustrating the mutual liver pathways between the three comparisons (B). Richness factor = the ratio of differentially expressed genes to the number of total genes in each pathway. The pathways highlighted with blue are the significant enriched metabolic pathways (P < 0.05).
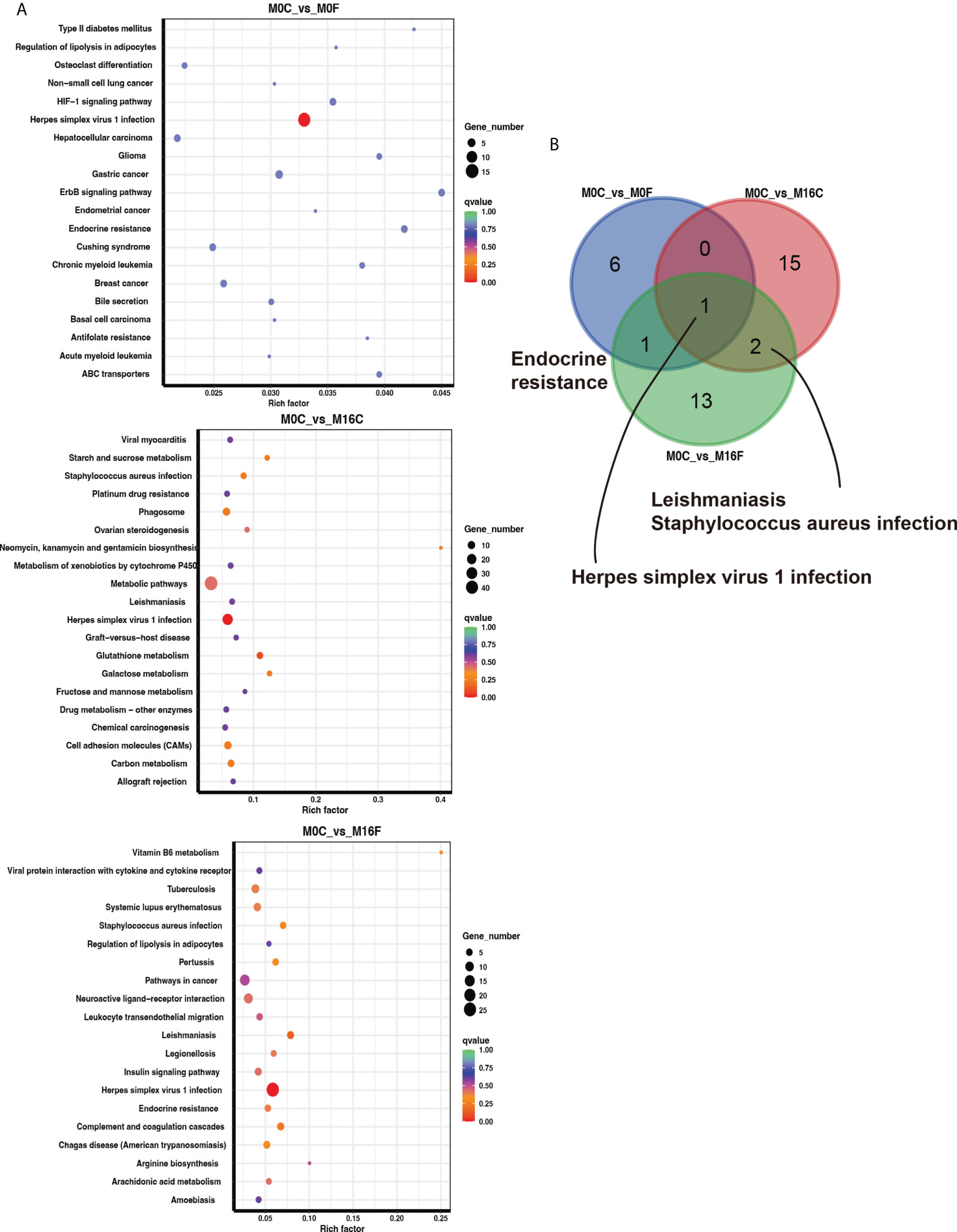
Figure 4 The hub affected KEGG pathways in the muscle by folic acid supplementation. Top 20 enriched KEGG pathways based on the differentially expressed genes for the three comparisons in the muscle (A). Venn diagram illustrating the mutual muscle pathways between the three comparisons (B). Richness factor = the ratio of differentially expressed genes to the number of total genes in each pathway. The pathways highlighted with blue are the significant enriched metabolic pathways (P < 0.05).
The present study’s findings systematically revealed the FA’s effect on the liver-muscle axis using multi-omics, which provided clues regarding the potential protective roles of FA on the immune function of lamb. To the best of our knowledge, we identified potent crosstalk between the blood, liver, and muscle in ruminants for the first time. This crosstalk may be the potential mechanism of FA-induced host transcription and metabolic responses through maternal and offspring communication (Figure 5).
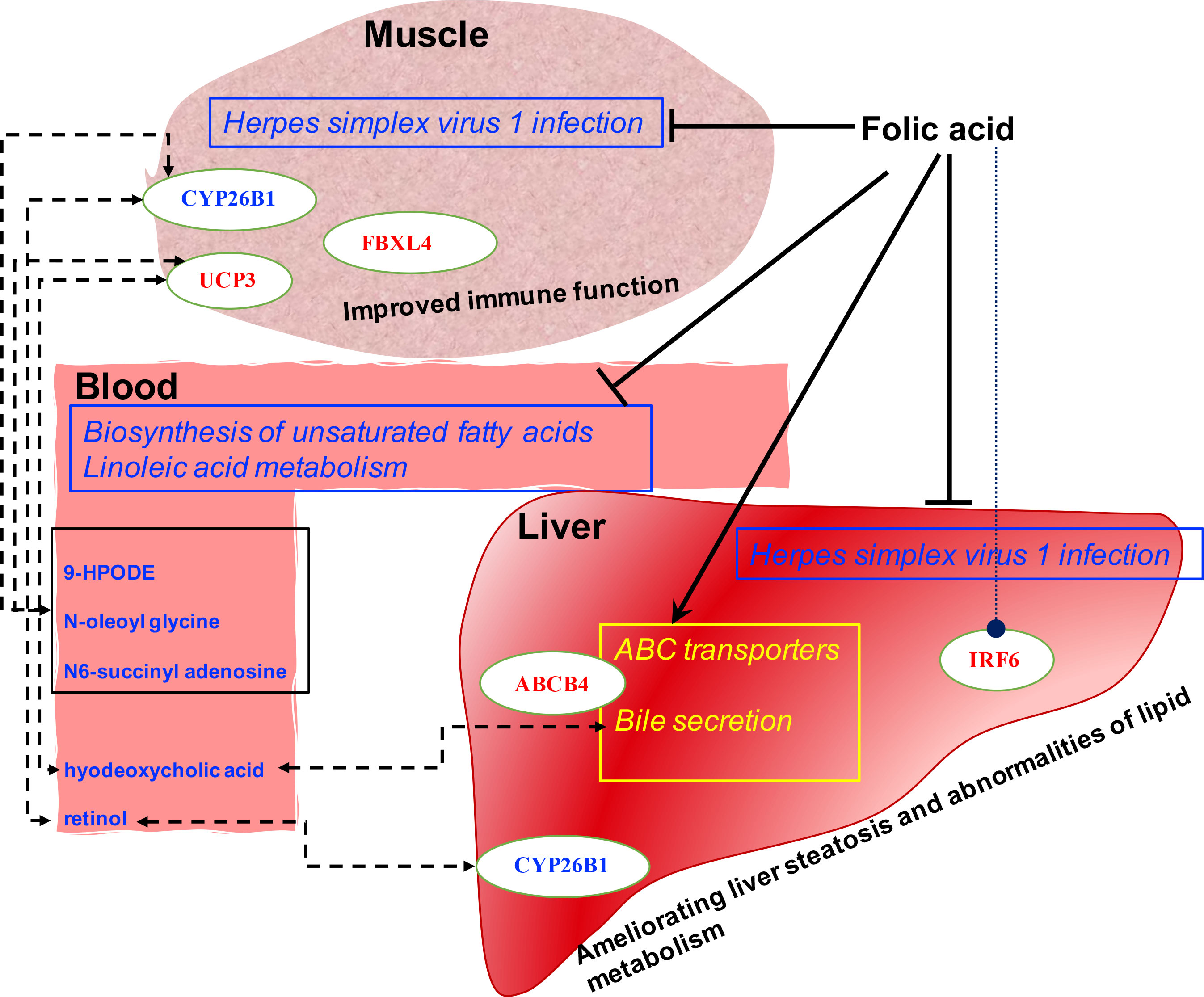
Figure 5 Proposed mechanism of folic acid’s function by the altered liver and muscle genes network and blood metabolic profiles. The upregulated and downregulated pathways are yellow and blue with squares, respectively. The upregulated and down-regulated hepatic and muscle genes and blood metabolites are labeled in red and blue, respectively. The dotted line indicates a potential biological communication.
The liver transcriptome depended on maternal protein and FA intakes during pregnancy (21). The coated FA had stimulatory impacts on hepatic gene expression in Holstein bulls (14), indicating the roles of FA on hepatic gene transcription. The maternal supplementation with FA had defending effects on inflammation in Holstein calves (22). FA could protect host defense against the challenge of Staphylococcus aureus through activating cytoplasmic DNA sensing and tight junction pathway, indicating the FA’s potentially biological function in improving animal immune function (23). In the current study, the Herpes simplex virus 1 infection pathway was inhibited by feeding FA, which was enriched in both liver and muscle. Herpes simplex virus type 1 is a nuclear replicating enveloped virus, usually acquired through direct contact with infected lesions or body fluids (24). The recurrence rate of herpetic keratitis was lower in patients with a higher blood level of FA, indicating FA’s vital protective role in herpes simplex keratitis (25). The folate-conjugated herpes simplex virus type 1 G207 presents a folate receptor-targeted oncolytic virus with a potential therapeutic value and an increased anti-tumor efficiency (26). A recent study reported that the FA and its derivates like tetrahydrofolic acid and 5-methyl tetrahydrofolic acid against SARS-COV-2 (27). Therefore, FA might have an essential function in immunological enhancement in sheep based on the result of the current study.
Furthermore, we found three hepatic hub genes: upregulated IRF6 and S100A12 and downregulated TMEM25. Interferon regulatory factor 6, a molecular marker associated with the serum IL10 level, plays a functional role in regulating the immune system (28). The IRF6 is significantly downregulated in high-fat diet-induced nonalcoholic fatty liver disease, a worldwide epidemic. IRF6 in hepatocytes is a protective factor in liver steatosis and abnormalities of lipid metabolism (29). In addition, an association of interaction between the IRF6 gene and maternal FA intake with non-syndromic cleft lip/palate in Mexican Mestizos was found (30). The current study found that the upregulated IRF6 by feeding FA confirms the FA’s beneficial role in hepatic immune function and metabolism and indicates that the IRF6 might be the targeted gene of FA in hepatocytes. S100 calcium binding protein A12 is a pro-inflammatory factor associated with non-infectious inflammatory diseases (31). S100A12 gene silencing decreased serum levels of anti-inflammatory/pro-inflammatory cytokines in septic rats (32). However, the down-regulation of TMEM25 is colorectal cancer-related (33), which needs further studies to clarify the molecular mechanism of TMEM25 in sheep.
CYP26B1, a mutual gene between liver and muscle transcriptome, was down-regulated by feeding FA. Similarly, CYP26B1 was down-regulated by dietary vitamin E supplementation in sheep (34), similar to the finding of this study. Retinoic acid and CYP26B1 are critical regulators of osteogenesis in the axial skeleton (35). Retinoic acid is synthesized intracellularly from its precursor, vitamin A (or retinol) (36). CYP26B1 can regulate retinoic acid-dependent signals in T cells and can be inhibited by transforming growth factor-β (TGF-β) (37). Thus, the down-regulated levels of CYP26B1 mRNA abundance in muscle and blood 11-cis-retinol are consistent with the enhanced immune function of feeding FA. MYH9 encodes the heavy chain of non-muscle myosin IIA, a cytoplasmic myosin that plays a significant role in human development and disease (38, 39). RBBP9 can suppress the TGF-β signaling required for a concomitant decrease in the integrity of tumor cell-cell junctions, indicating its protective role for body health (40). The PPP1R3D can regulate the structure of glycogen, and the deficiency of PPP1R3D preferentially inhibits neuronal and cardiac Lafora body formation in a mouse model of the fatal epilepsy Lafora disease (41). FBXL4 has a crucial function in development, metabolism, and mitochondrial dynamics and may be used to develop novel therapies for mitochondrial encephalopathy (42). Thus, the decreased abundances of CYP26B1 and MYH9 and increased abundances of RBBP9, PPP1R3D, and FBXL4 in muscle, confirmed the improved immune function of sheep by supplementation of FA.
On the other hand, FA increased the bile flow, bile acid synthesis from cholesterol, and bile acid excretion via feces (43). It was suggested that FA is an essential therapeutic intervention in dyslipidemia caused by cholestasis, protecting the liver against cholestasis (44). The enhanced bile synthesis pathway in the liver from the current study confirms the function of FA on bile acid metabolism in sheep. In addition, the blood hyodeoxycholic acid, a primary secondary bile acid, was reduced by feeding FA from the maternal or offspring diet. A significantly negative correlation between blood hyodeoxycholic acid and muscle uncoupling protein 3 (UCP3) gene was observed. Hyodeoxycholic acid can solubilize dietary fat and promote fat absorption and glucose homeostasis (45). UCP3 gene was an important candidate gene for regulating intramuscular fat content in chicken breast muscle (46). We did find the changed lipid metabolism by decreasing serum cholesterol parameters and increasing fat deposition in tail or omentum from our previous studies (5, 6). Two enriched metabolic pathways, biosynthesis of unsaturated fatty acids and linoleic acid metabolism, were down-regulated by feeding FA from the maternal diet solely and maternal and offspring diet together, indicating the crucial roles of FA on lipid metabolism. However, no information indicated the relationship between UCP3 and fat metabolism in sheep. Thus, we estimated that the FA has potential roles in alleviating fat and unsaturated fatty acids deposition in sheep by regulating the association between hyodeoxycholic acid and UCP3.
9-Hpode, N-oleoyl glycine, and N6-succinyl adenosine were positively correlated with CYP26B1 but negatively correlated with UCP3. Blood 9-Hpode is the peroxidation product of linoleic acid (47). 9-Hpode is the main product of lipid peroxidation, and the increased 9-Hpode can cause some inflammatory reactions, such as atherosclerosis (48) and rheumatoid arthritis (49). The decreased blood 9-Hpode by adding FA to the maternal or offspring diet might indicate the reduced peroxidation of linoleic acid and prevent these diseases, mainly because that FA can scavenge free radicals in the body to prevent oxidative damage (50, 51). In addition, N-oleoyl glycine and N6-succinyl adenosine were down-regulated by feeding FA. The N-oleoyl glycine can potentially increase insulin sensitivity and suppress obesity and diabetes (52). The blood concentration of N6-succinyl adenosine was a feasible predictor of chronic renal failure in rat plasma (53). Homocitrulline is a biomarker for differentiating acute from chronic renal failure (54). The increased serum homocitrulline concentration is associated with the severity of coronary artery disease (55). Thus, these reduced blood hub differential metabolites by feeding FA from either maternal or offspring diets also prove their protection roles in improved immune function in sheep.
In conclusion, a multi-omics assessment of the blood, liver, and muscle systematically revealed the molecular biological mechanisms of FA on lamb health and lipid metabolic performance. The inhibited pathway of herpes simplex virus 1 infection might play a crucial role of FA in enhancing lamb health. Moreover, the enhanced bile secretion pathway in the liver by FA from maternal or offspring diet and the inhibited blood biosynthesis of unsaturated fatty acids and linoleic acid metabolism by FA from offspring diet might contribute to the changed fat deposition and lipid metabolism in sheep. We identified the possible hub genetic biomarkers IRF6 in the liver and CYP26B1, FBXL4, and UCP3 in the muscle, and potential folic related functional biological or metabolic intermediates including blood hyodeoxycholic acid, 9-Hpode, N-oleoyl glycine, and N6-succinyl adenosine, as being responsible for FA’s function in lamb. Our results provide a systemic picture of FA’s roles in immunological enhancement in a lamb model, which enhances the understanding of FA from either maternal and offspring diet in improving animal health in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA822866 https://www.ncbi.nlm.nih.gov/, PRJNA822707.
The animal study was reviewed and approved by Animal Care and Use Committee of China Agricultural University.
BW and HLL conceived and designed the study; BW wrote the manuscript; BW, HQL, ZL, and BoW carried out the animal experiment, laboratory work, data analysis and bioinformatic analyses; BoW and ZL did the partial data analysis; HZ did the qPCR test; BZ did the Figures visualization; BW and HLL revised the manuscript. All the authors gave final approval for the publication of the manuscript and agree to be held accountable for the work performed therein.
We greatly acknowledge the grants supported by the earmarked fund for CARS-38 and the National Key Research and Development Program of China (No. 2018YFD0500402).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.913854/full#supplementary-material
1. Bailey LB, Gregory JF 3rd. Folate metabolism and requirements. J Nutr (1999) 129(4):779–82. doi: 10.1093/jn/129.4.779
2. Poe SE, Mitchell GE Jr., Ely DG. Rumen development in lambs. 3. microbial b-vitamin synthesis. J Anim Sci (1972) 34(5):826–9. doi: 10.2527/jas1972.345826x
3. Ragaller V, Huther L, Lebzien P. Folic acid in ruminant nutrition: A review. Br J Nutr (2009) 101(2):153–64. doi: 10.1017/S0007114508051556
4. Wu Q, La S, Wang C, Zhang J, Liu Q, Guo G, et al. Effects of coated copper sulphate and coated folic acid supplementation on growth, rumen fermentation and urinary excretion of purine derivatives in Holstein bulls. Anim Feed Sci Tech (2021) 276:114921. doi: 10.1016/j.anifeedsci.2021.114921
5. Li H, Wang B, Li Z, Luo H, Wang Y, Zhang C, et al. Effects of rumen-protected folic acid addition in maternal and post-weaning diets on growth performance, total tract digestibility, ruminal fermentation and blood metabolites in lambs. Anim Feed Sci Tech (2020) 260:114364. doi: 10.1016/j.anifeedsci.2019.114364
6. Li HQ, Wang B, Li Z, Luo HL, Zhang C, Jian LY, et al. Effects of maternal and post-weaned rumen-protected folic acid supplementation on slaughter performance and meat quality in offspring lambs. Br J Nutr (2021) 126(8):1140–8. doi: 10.1017/S0007114520005206
7. Trujillo E, Davis C, Milner J. Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J Am Diet Assoc (2006) 106(3):403–13. doi: 10.1016/j.jada.2005.12.002
8. Wang B, Li H, Li Z, Jian L, Gao Y, Qu Y, et al. Maternal folic acid supplementation modulates the growth performance, muscle development and immunity of hu sheep offspring of different litter size. J Nutr Biochem (2019) 70:194–201. doi: 10.1016/j.jnutbio.2019.05.011
9. Duplessis M, Girard C, Santschi D, Lefebvre D, Pellerin D. Folic acid and vitamin B12 supplement enhances energy metabolism of dairy cows in early lactation. J Dairy Sci (2012) 95(Suppl 2):118. doi: 10.3168/jds.2021-21677
10. Liu HY, Liu SM, Zhang YZ. Maternal folic acid supplementation mediates offspring health via DNA methylation. Reprod Sci (2020) 27(4):963–76. doi: 10.1007/s43032-020-00161-2
11. Mikkelsen K, Apostolopoulos V. Vitamin B12, folic acid, and the immune system. In: Nutr immun. Cham:Switzerland (2019) 103–14. doi: 10.1007/978-3-030-16073-9_6
12. Wang X, Li Z, Zhu Y, Yan J, Liu H, Huang G, et al. Maternal folic acid impacts DNA methylation profile in male rat offspring implicated in neurodevelopment and learning/memory abilities. Genes Nutr (2021) 16(1):1. doi: 10.1186/s12263-020-00681-1
13. Khan MZ, Zhang Z, Liu L, Wang D, Mi S, Liu X, et al. Folic acid supplementation regulates key immunity-associated genes and pathways during the periparturient period in dairy cows. Asian-Australas J Anim Sci (2019) 33(9):1507–19. doi: 10.5713/ajas.18.0852
14. Liu YJ, Chen JZ, Wang DH, Wu MJ, Zheng C, Wu ZZ, et al. Effects of guanidinoacetic acid and coated folic acid supplementation on growth performance, nutrient digestion and hepatic gene expression in Angus bulls. Br J Nutr (2021) 126(4):510–7. doi: 10.1017/S0007114520004341
15. Bae D, Chon JW, Kim DH, Kim H, Seo KH. Effect of folic acid supplementation on proliferation and apoptosis in bovine mammary epithelial (MAC-T) cells. Anim Biotechnol (2022) 33(1):13–21. doi: 10.1080/10495398.2020.1758123
16. Ponziani FR, Picca A, Marzetti E, Calvani R, Conta G, Del Chierico F, et al. Characterization of the gut-liver-muscle axis in cirrhotic patients with sarcopenia. Liver Int (2021) 41(6):1320–34. doi: 10.1111/liv.14876
17. De Bandt JP, Jegatheesan P, Tennoune-El-Hafaia N. Muscle loss in chronic liver diseases: The example of nonalcoholic liver disease. Nutrients (2018) 10(9):1195. doi: 10.3390/nu10091195
18. Wang C, Liu Q, Guo G, Huo W, Ma L, Zhang Y, et al. Effects of rumen-protected folic acid on ruminal fermentation, microbial enzyme activity, cellulolytic bacteria and urinary excretion of purine derivatives in growing beef steers. Anim Feed Sci Tech (2016) 221:185–94. doi: 10.1016/j.anifeedsci.2016.09.006
19. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods (2008) 5(7):621–8. doi: 10.1038/nmeth.1226
20. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol (2010) 11(10):R106. doi: 10.1186/gb-2010-11-10-r106
21. Lillycrop KA, Rodford J, Garratt ES, Slater-Jefferies JL, Godfrey KM, Gluckman PD, et al. Maternal protein restriction with or without folic acid supplementation during pregnancy alters the hepatic transcriptome in adult male rats. Br J Nutr (2010) 103(12):1711–9. doi: 10.1017/S0007114509993795
22. Lopes MG, Alharthi AS, Lopreiato V, Abdel-Hamied E, Liang Y, Coleman DN, et al. Maternal supplementation with cobalt sources, folic acid, and rumen-protected methionine and its effects on molecular and functional correlates of the immune system in neonatal Holstein calves. J Dairy Sci (2021) 104(8):9340–54. doi: 10.3168/jds.2020-19674
23. Mi S, Tang Y, Shi L, Liu X, Si J, Yao Y, et al. Protective roles of folic acid in the responses of bovine mammary epithelial cells to different virulent staphylococcus aureusstrains. Biol (Basel) (2021) 10(11):1164. doi: 10.3390/biology10111164
24. Arduino PG, Porter SR. Herpes simplex virus type 1 infection: overview on relevant clinico-pathological features. J Oral Pathol Med (2008) 37(2):107–21. doi: 10.1111/j.1600-0714.2007.00586.x
25. Savić BM, Stanojlović S, Stojković M, Mišić M, Savić B, Draganić V. Potential effect of decreased levels of folic acid and vitamin B12 on herpes simplex virus keratitis reactivation. Vojnosanit Pregl (2021) 78(1):35–9. doi: 10.2298/VSP181001037S
26. Ye ZQ, Zou CL, Chen HB, Lv QY, Wu RQ, Gu DN. Folate-conjugated herpes simplex virus for retargeting to tumor cells. J Gene Med (2020) 22(7):e3177. doi: 10.1002/jgm.3177
27. Kumar V, Kancharla S, Jena MK. In silico virtual screening-based study of nutraceuticals predicts the therapeutic potentials of folic acid and its derivatives against COVID-19. Virusdisease (2021) 32:29–37. doi: 10.1007/s13337-020-00643-6
28. Liu Y, Xu J, Fu W, Weng Z, Niu X, Liu J, et al. Tissues expression, polymorphisms of IFN regulatory factor 6 (IRF6) gene and their associated with immune traits in three pig populations. Asian-Australas J Anim Sci (2012) 25(2):163–9. doi: 10.5713/ajas.2011.11168
29. Tong J, Han CJ, Zhang JZ, He WZ, Zhao GJ, Cheng X, et al. Hepatic interferon regulatory factor 6 alleviates liver steatosis and metabolic disorder by transcriptionally suppressing peroxisome proliferator-activated receptor gamma in mice. Hepatology (2019) 69(6):2471–88. doi: 10.1002/hep.30559
30. Velázquez-Aragón JA, Alcántara-Ortigoza MA, Estandia-Ortega B, Reyna-Fabián ME, Cruz-Fuentes C, Villagómez S, et al. Association of interactions among the IRF6 gene, the 8q24 region, and maternal folic acid intake with non-syndromic cleft lip/palate in Mexican mestizos. Am J Med Genet A (2012) 158a(12):3207–10. doi: 10.1002/ajmg.a.35641
31. Ma D, Li X, Zhang L, Deng C, Zhang T, Wang L, et al. S100A12 expression in patients with primary biliary cirrhosis. Immunol Invest (2015) 44(1):13–22. doi: 10.3109/08820139.2014.914530
32. Wen X, Han XR, Wang YJ, Fan SH, Zhang ZF, Wu DM, et al. Effects of S100A12 gene silencing on serum levels of anti-inflammatory/pro-inflammatory cytokines in septic rats through the ERK signaling pathway. J Cell Biochem (2018) 119(5):4038–49. doi: 10.1002/jcb.26568
33. Hrašovec S, Hauptman N, Glavač D, Jelenc F, Ravnik-Glavač M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis Markers (2013) 34(2):93–104. doi: 10.3233/DMA-120948
34. Qu YH, Jian LY, Ce L, Ma Y, Xu CC, Gao YF, et al. Identification of candidate genes in regulation of spermatogenesis in sheep testis following dietary vitamin e supplementation. Anim Reprod Sci (2019) 205:52–61. doi: 10.1016/j.anireprosci.2019.04.003
35. Spoorendonk KM, Peterson-Maduro J, Renn J, Trowe T, Kranenbarg S, Winkler C, et al. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development (2008) 135(22):3765–74. doi: 10.1242/dev.024034
36. Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol Rev (2000) 80(3):1021–54. doi: 10.1152/physrev.2000.80.3.1021
37. Takeuchi H, Yokota A, Ohoka Y, Iwata M. Cyp26b1 regulates retinoic acid-dependent signals in T cells and its expression is inhibited by transforming growth factor-beta. PLoS One (2011) 6(1):e16089. doi: 10.1371/journal.pone.0016089
38. Pecci A, Ma X, Savoia A, Adelstein RS. MYH9: Structure, functions and role of non-muscle myosin IIA in human disease. Gene (2018) 664:152–67. doi: 10.1016/j.gene.2018.04.048
39. Asensio-Juarez G, Llorente-Gonzalez C, Vicente-Manzanares M. Linking the landscape of MYH9-related diseases to the molecular mechanisms that control non-muscle myosin II-a function in cells. Cells (2020) 9(6):1458. doi: 10.3390/cells9061458
40. Shields DJ, Niessen S, Murphy EA, Mielgo A, Desgrosellier JS, Lau SK, et al. RBBP9: A tumor-associated serine hydrolase activity required for pancreatic neoplasia. Proc Natl Acad Sci U S A (2010) 107(5):2189–94. doi: 10.1073/pnas.0911646107
41. Israelian L, Nitschke S, Wang P, Zhao X, Perri AM, Lee JPY, et al. Ppp1r3d deficiency preferentially inhibits neuronal and cardiac lafora body formation in a mouse model of the fatal epilepsy lafora disease. J Neurochem (2021) 157(6):1897–910. doi: 10.1111/jnc.15176
42. Haydu JEM. The roles of f-box and leucine-rich repeat protein 4 (FBXL4) in mitochondrial encephalopathy and T-cell acute lymphoblastic leukemia. (New York City:Columbia University) (2015). doi: 10.7916/D8028Q64
43. Delgado-Villa MJ, Ojeda ML, Rubio JM, Murillo ML, Sanchez OC. Beneficial role of dietary folic acid on cholesterol and bile acid metabolism in ethanol-fed rats. J Stud Alcohol Drugs (2009) 70(4):615–22. doi: 10.15288/jsad.2009.70.615
44. Mohammadian Z, Eidi A, Mortazavi P, Tavangar MM, Asghari A. Effects of folic acid on dyslipidemia and serum homocysteine in a rat model of cholestasis and hepatic fibrosis. Pol J Pathol (2015) 66(1):49–56. doi: 10.5114/pjp.2015.51153
45. Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E. Bile acids in glucose metabolism in health and disease. J Exp Med (2018) 215(2):383–96. doi: 10.1084/jem.20171965
46. Gao GL, Wang HW, Zhao XZ, Li Q, Li J, Li QR, et al. Feeding conditions and breed affect the level of DNA methylation of the mitochondrial uncoupling protein 3 gene in chicken breast muscle. J Anim Sci (2015) 93(4):1522–34. doi: 10.2527/jas.2014-8431
47. Spiteller P, Kern W, Reiner J, Spiteller G. Aldehydic lipid peroxidation products derived from linoleic acid. Biochim Biophys Acta (2001) 1531(3):188–208. doi: 10.1016/s1388-1981(01)00100-7
48. Jira W, Spiteller G. Dramatic increase of linolelic acid peroxidation products by aging, atherosclerosis, and rheumatoid arthritis. In: Eicosanoids and other bioactive lipids in cancer, inflammation, and radiation injury4. Advances in Experimental Medicine and Biology, 469. (Boston, MA: Springer) (1999). doi: 10.1007/978-1-4615-4793-8_70
49. Jira W, Spiteller G, Richter A. Increased levels of lipid oxidation products in rheumatically destructed bones of patients suffering from rheumatoid arthritis. Z für Naturforschung C (1998) 53(11-12):1061–71. doi: 10.1515/znc-1998-11-1220
50. Gursu MF, Onderci M, Gulcu F, Sahin K. Effects of vitamin c and folic acid supplementation on serum paraoxonase activity and metabolites induced by heat stress in vivo. Nutr Res (2004) 24(2):157–64. doi: 10.1016/j.nutres.2003.11.008
51. Asbaghi O, Ghanavati M, Ashtary-Larky D, Bagheri R, Rezaei Kelishadi M, Nazarian B, et al. Effects of folic acid supplementation on oxidative stress markers: a systematic review and meta-analysis of randomized controlled trials. Antioxidants (Basel) (2021) 10(6):871. doi: 10.3390/antiox10060871
52. Wang S, Xu Q, Shu G, Wang L, Gao P, Xi Q, et al. N-oleoyl glycine, a lipoamino acid, stimulates adipogenesis associated with activation of CB1 receptor and akt signaling pathway in 3T3-L1 adipocyte. Biochem Biophys Res Commun (2015) 466(3):438–43. doi: 10.1016/j.bbrc.2015.09.046
53. Kobayashi T, Matsumura Y, Ozawa T, Yanai H, Iwasawa A, Kamachi T, et al. Exploration of novel predictive markers in rat plasma of the early stages of chronic renal failure. Anal Bioanal Chem (2014) 406(5):1365–76. doi: 10.1007/s00216-013-7472-2
54. Desmons A, Jaisson S, Pietrement C, Rieu P, Wynckel A, Gillery P. Homocitrulline: A new marker for differentiating acute from chronic renal failure. Clin Chem Lab Med (2016) 54(1):73–9. doi: 10.1515/cclm-2015-0398
Keywords: folic acid, metabolomics, transcriptomics, lipid metabolism, immune function
Citation: Wang B, Li H, Li Z, Wang B, Zhang H, Zhang B and Luo H (2022) Integrative network analysis revealed the molecular function of folic acid on immunological enhancement in a sheep model. Front. Immunol. 13:913854. doi: 10.3389/fimmu.2022.913854
Received: 06 April 2022; Accepted: 26 July 2022;
Published: 12 August 2022.
Edited by:
Yangchun Cao, Northwest A&F University, ChinaReviewed by:
Yan Tu, Feed Research Institute (CAAS), ChinaCopyright © 2022 Wang, Li, Li, Wang, Zhang, Zhang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailing Luo, bHVvaGFpbGluZ0BjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.