- 1Center of Infectious Diseases and Pathogen Biology, Institute of Virology and AIDS Research, Key Laboratory of Organ Regeneration and Transplantation of The Ministry of Education, The First Hospital of Jilin University, Changchun, China
- 2Respiratory Department of the First Hospital of Jilin University, Changchun, China
Enterovirus infection continues to be a global health problem. The lack of specific drugs and broad-spectrum vaccines means an urgent need to develop effective strategies against enteroviruses. Host restrictive factors are a class of intrinsic host antiviral factors that have been broadly defined and investigated during HIV infections and have great significance for drug development and treatment design. In recent years, the essential role of host restrictive factors in regulating enteroviral infections has been gradually recognized and investigated. An increasing number of studies have shown that host-restrictive factors regulate multiple steps in the life cycle of enteroviruses. This mini-review discusses the restrictive factors against enteroviruses, their antiviral mechanism, and the arms race between them and enteroviruses. We also summarise the pathways that enteroviruses use to impair host antiviral signals. This mini-review characterizes the essential role of host restriction factors in enterovirus infections, which provides ideas and potential targets for antiviral drug design by regulating host restrictive factors. It also reveals potential future research on the interplay between host restrictive factors and enteroviruses.
Introduction
There are more than 100 subtypes of enteroviruses that infect humans, including the well-known enterovirus 71 (EV71), enterovirus D68 (EVD68), coxsackieviruses A and B, and poliovirus (PV) (1), and several subtypes of these induce hand-foot-and-mouth disease (HFMD) epidemics every year (2–5). Moreover, EVD68 has been the cause of an unprecedented epidemic of respiratory disease, whose symptoms are unlike its common symptoms and have been temporally associated with acute flaccid myelitis (AFM) (6–8). However, the lack of effective drugs and broad-spectrum vaccines has exacerbated severe health problems.
A series of studies have investigated the interactions between host innate immunity and enteroviruses. Host restriction factors are expressed and/or induced in response to virus infection and include proteins from interferon-stimulated genes (ISGs) (9–19). APOBEC3G (A3G), SAMHD1, and BST2 have been extensively investigated in HIV infection (13). Since the antiviral effects of restrictive factors tend to have a broad spectrum, the regulatory function of host restriction factors during enterovirus infection has been investigated in recent years and has become a rising focus of enterovirus research. For example, we identified A3G and SAMHD1 restricted multiple enteroviruses and revealed a novel antiviral mechanism (20–23). IFNs and NF-κB signals are activated by viral infection (24–30), which induces the expression of downstream host-restrictive factors to fight against viruses via their pathways. However, viruses can impair restriction through multiple strategies. Here, we discuss the host restriction factors that play essential roles in regulating enteroviruses, the underlying mechanism they suppress, and how enteroviruses break host restrictions. This mini-review provides new information that can be used to select potential drug targets against enteroviruses and enlighten the direction of future studies in antivirus research.
Host Restrictive Factors Play Antiviral Roles During Enteroviruses Infection
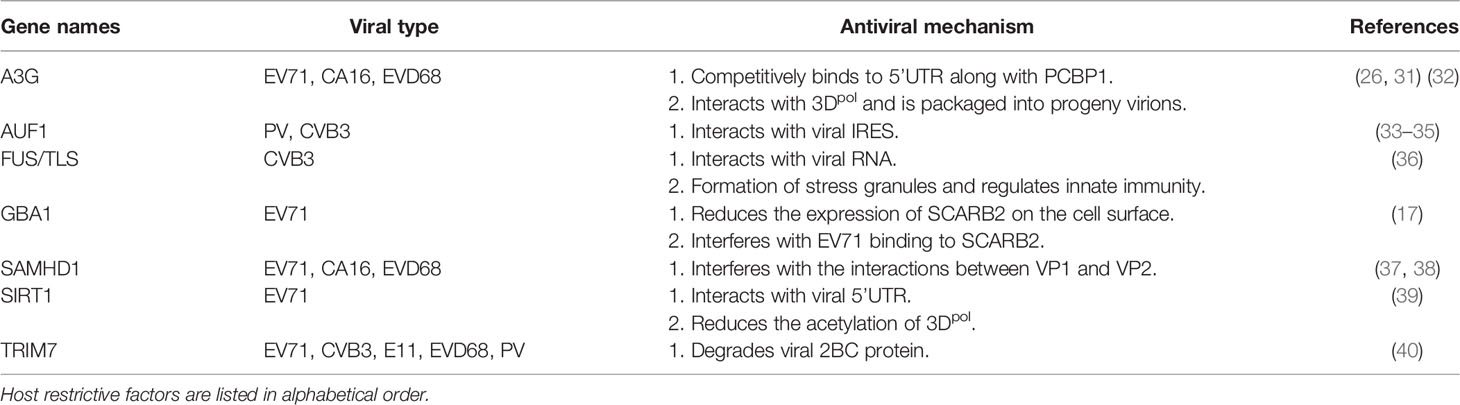
Many in-depth studies on the interaction between enteroviruses and host factors have shown that host-restrictive factors play regulatory roles in different stages of the viral life cycle. We have summarised these studies based on the lifecycles of enterovirus infections (Figure 1 and Table 1).
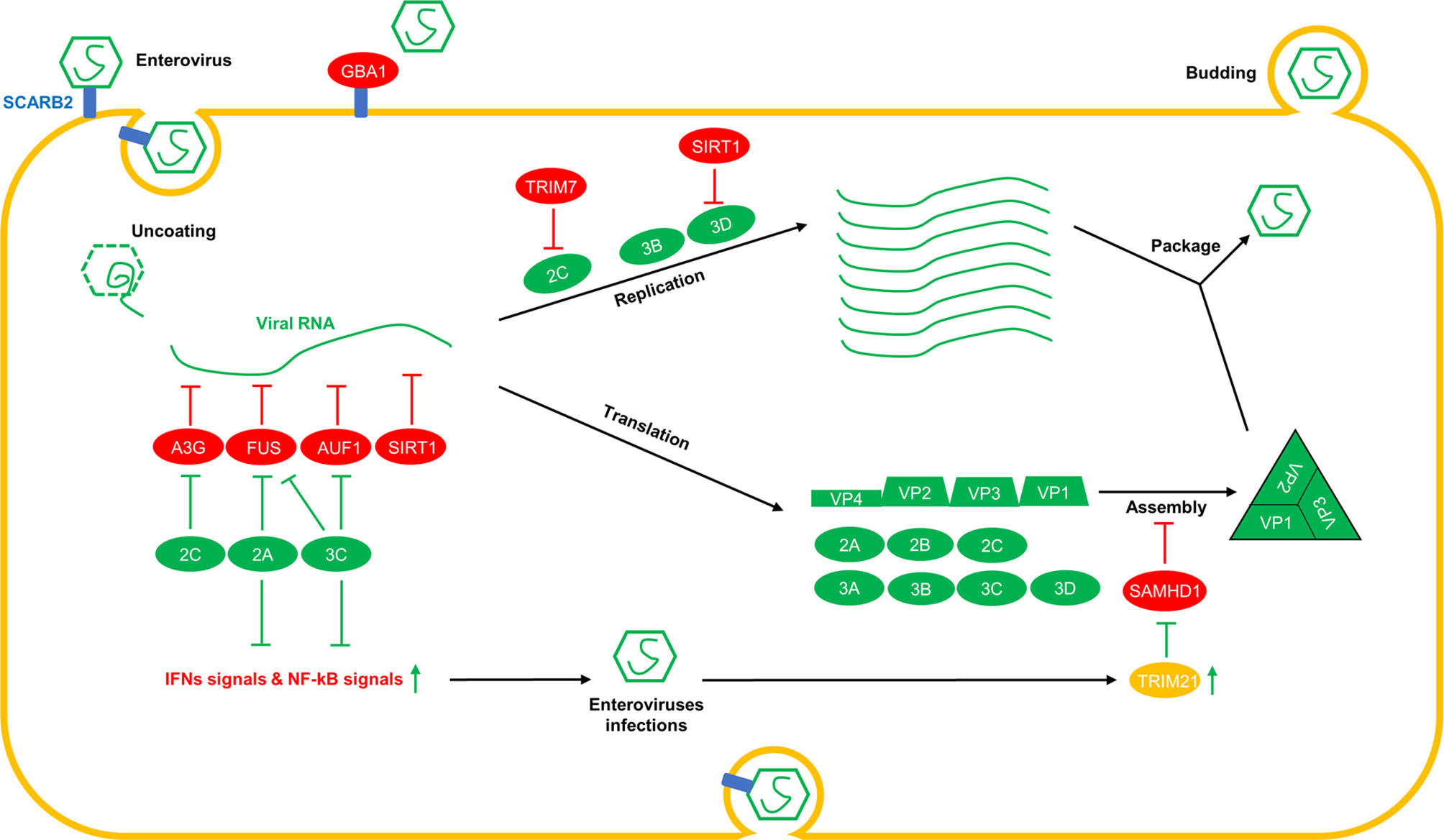
Figure 1 Overview of the interplay between host factors and enterovirus replication. Various host-restrictive factors have been shown to play regulatory roles at different stages of the virus life cycle. During the invasion phase, GBA1 interacts with SCARB2, a receptor of enteroviruses, and interferes with the binding of enteroviruses to SCARB2. After uncoating, A3G, FUS, AUF1, and SIRT1 interact with viral RNAs and reduce the replication and translation of viral RNAs. During the viral RNA replication phase, TRIM7 induces the degradation of 2C, and SIRT1 triggers the deacetylation of 3Dpol, which is required for viral RNA replication. During the assembly phase, SAMHD1 interacts with VP1 and disrupts viral capsid assembly by interfering with the interactions between the viral capsid proteins VP1 and VP2. To break through the restriction from the host, viral proteins, such as 2Apro and 3Cpro, cleave FUS, other IFNs and NF-kB signal-associated proteins, and 3Cpro cleaves AUF1. Furthermore, 2C induces the degradation of A3G. TRIM21, which is upregulated by enteroviruses, triggers polyubiquitination and the degradation of SAMHD1.
Viral Entry
Enteroviruses invade host cells by first interacting with specific receptors on the cell surface, where virions are endocytosed, or viral nucleic acids are released into the cells. For instance, scavenger receptor class B member 2 (SCARB2) was identified as a receptor for EV71 by Yamayoshi et al. (41). On this basis, Nakata et al. reported that acid beta-glucosidase 1 (GAB1) restricts EV71 infections by interacting with SCARB2 and reducing the expression of SCARB2 on the cell surface which interferes with the interactions between EV71 and SCARB2 (42). Further investigations showed that recombinant human GBA1, a molecular drug originally used to treat Gaucher’s disease (33, 40, 43), protected against EV71 infection (42), hinting that researchers could design anti-enterovirus drugs based on host restrictive factors.
Viral RNA Replication and the Protein Translation Phases
After entering host cells, RNAs from enteroviruses are replicated and translated into viral proteins under the regulation of host factors (22, 34–36, 39, 44) and viral proteins, including 2BC, 3AB, and 3D (31, 32, 45–49). During this phase, many restrictive factors are involved in inhibiting virus replication. TRIM7, an E3 ligase, has been reported to restrict the replication of multiple enteroviruses by triggering polyubiquitination of their 2BC proteins and inducing the degradation of 2BC proteins via the proteasomal pathway (50).
AU-rich element degradation factor 1 (AUF1) binds to the internal ribosome entry site (IRES) of viral RNA and restricts the replication of poliovirus and CVB3 (51–53). However, the 3Cpro component of enteroviruses can cleave AUF1 to break through the restriction (53).
Like AUF1, fused sarcoma/translocated in liposarcoma (FUS/TLS) is a novel host antiviral factor that restricts CVB3 replication by directly inhibiting viral RNA transcription and protein translation. Moreover, FUS, which binds to viral RNA, triggers the formation of stress granules and regulates the activity of host antiviral innate immunity (54). CVB3 infection induces cytoplasmic mislocalization and cleavage of FUS through the enzymatic activity of viral proteases to evade the FUS-mediated antiviral response and innate immunity (54). In addition, as a class III NAD+-dependent histone, the deacetylase (HDAC), SIRT1, suppresses EV71 replication by repressing viral RNA transcription and attenuating viral RNA translation (55). Mechanistically, Han et al. identified the interactions between SIRT1 and viral 3Dpol, and revealed that SIRT1 inhibits 3Dpol activity by reducing the acetylation of 3Dpol. They also found that SIRT1 was able to interact with the viral 5’-UTR and interfere with viral RNA transcription and translation. Additionally, the expression of SIRT1 is upregulated by EV71 infection (55). However, Li et al. observed that EV71 infections could reduce the expression of SIRT1, and administration of the ROS inhibitor N-acetyl-L-cysteine (NAC) reduced apoptosis levels and inflammation, downregulated EV71 propagation, and increased SIRT1 expression in EV71-infected cells (56). Nevertheless, the mechanism underlying the regulation of SIRT1 expression induced by EV71 infection remains unclear.
A3G has been identified as a host-restrictive factor that suppresses HIV replication via cytosine deaminase activity (37, 38). In recent years, A3G has been shown to restrict the replication of enteroviruses, such as EV71, CA16, and EVD68, but not CA6 (20, 22, 57). These reports propose novel antiviral mechanisms independent of the cytosine deaminase activity shown by A3G. Li et al. suggested that while suppressing multiple enteroviruses, A3G competitively binds to the viral 5’UTR together with PCBP1, which is required for the transcription and translation of viral RNAs (22, 57). Further investigation showed that PCBP2, but not PCBP1, was required for CA6 replication, which would explain why A3G failed to restrict CA6 replication (22). In addition, Wang et al. suggested that A3G interacts with the 3Dpol of EV71 and packages it into progeny virions to reduce its infectivity (20). They also found that an inhibitor named IMB-Z inhibited EV71 replication by upregulating the expression of A3G (20). These studies confirmed that A3G inhibits enterovirus and that the mechanism was independent of its cytosine deaminase activity. In addition, other members of the APOBEC3 family, including A3A, A3D, and A3F, were found to possess antiviral activity against EV71 (57).
In contrast, Li et al. reported that A3G is degraded by the 2C proteins in enteroviruses, including EV71, CA6, CA16, CVB3, and EVD68 (57). In their study, viral 2C proteins triggered the polyubiquitination of A3G. Then the polyubiquitinated A3G was recognized by P62 and degraded by autolysosomes.
Viral Assembly Phase
Like A3G, SAMHD1, another anti-HIV-restrictive factor, has been extensively investigated (14, 58, 59). SAMHD1 inhibits multiple retroviruses and DNA viruses (60–62), but its antiviral mechanism is unclear. Several studies have suggested that SAMHD1 restricts viruses through its dNTPase activity (14, 60, 63–65), and other studies have argued that its nuclease activity also contributes to its antiviral activity (66–68). For enteroviruses, Li et al. reported that SAMHD1 restricted EV71 replication independently of the dNTPase and nuclease activity of SAMHD1 (21). Furthermore, Zhao et al. reported that SAMHD1 restricted the replication of enteroviruses, including EV71, EVD68, and CA16, but not CA6, by interfering with the interactions between the viral capsid proteins VP1 and VP2 (23). Zhao et al. showed that SAMHD1 interacts with the EV71-VP1 domain, which is essential for the interaction between EV71-VP1 and EV71-VP2 and attenuates the interaction between EV71 VP1 and VP2. However, the interaction between SAMHD1 and CA6-VP1 did not disrupt the interaction between VP1 and VP2 of CA6, which may explain why SAMHD1 failed to inhibit CA6 (23).
In response to the inhibition of SAMHD1, EV71 has evolved a strategy to overcome this restriction and ensure the survival of its progeny. In a study by Li et al., EV71 infection induced proteasome-associated degradation of SAMHD1 by upregulating the expression of E3 ligase TRIM21, which triggers the polyubiquitination SAMHD1. TRIM21 upregulation is interferon receptor-dependent (21). Li et al. also identified the interaction domains between SAMHD1 and TRIM21 and the ubiquitination site on SAMHD1, which may provide clues for further drug target screening and design.
Several host restrictive factors play essential regulatory roles in various stages of the enterovirus life cycle, and exploring the mechanism underlying the interplay between host restrictive factors and enteroviruses will provide an important scientific basis for strategies against strategies for enterovirus infection.
Enteroviruses Break Out of Host Restriction by Blocking Antiviral Pathways
During the long-term arms race, viruses evolve strategies to impair restriction from the host. In addition to the antagonistic strategies against the host restrictive factors mentioned above, enteroviruses also can disrupt other antiviral pathways to ensure their life cycle within the host (Figure 1). EV71 3Cpro has been reported to cleave multiple innate immune pathway-related proteins, including TRIF (26, 69), TRIM25 (70), TAK1, TAB1, TAB2, TAB3 (71), NLRP3 (72), IRF3 (73), IRF7 (74), IRF9 (75) and PMLIII and IV (76), and reduce IFN and NF-kB signals (77). As this type of research expands, the 3Cpro in EVD68 has also been reported to cleave IRF7 and affect IFN signaling (78, 79). Furthermore, the 2Apro in EV71 has been reported to cleave MAVS, MDA5, and NLPR3 (72, 80, 81) and downregulate IFN and NF-kB signaling. In addition to these viral proteases, the 2C proteins in multiple enteroviruses have been reported to suppress NF-kB and IFN signals by binding to IKKβ, P65, and MDA5 (82–86). After that, reducing the antiviral signal levels will decrease the expression of antiviral factors, which contain many host restrictive factors and are beneficial to the unscrupulous replication of viruses.
Discussion
Enterovirus infections are prevalent worldwide. However, the lack of specific drugs and broad-spectrum multivalent vaccines poses an urgent health threat. So, considerable studies on drug design targeting viral proteins have been conducted but unsuccessful (87). The high mutagenicity of RNA viruses and the similarity between the virus enzyme active domain and the host protein present considerable obstacles to selecting drug targets (88–92). The discovery of host restriction factors against enteroviruses and their interactions with viruses has attracted attention as a new antiviral strategy. Under this strategy, we could regulate the expression of host restrictive factors and effectively inhibit viral infections. Furthermore, we have identified the ‘Achilles heel’ of enteroviruses based on studies of hosting restrictive factors against enteroviruses. For instance, Zhao et al. reported that 119-223aa in VP1 were essential for the interactions between VP1 and VP2 (23). Based on this assumption, the inhibitors targeting 119-223aa in VP1 would possess a space-occupying effect and restrict the replication of enteroviruses, which may be a promising drug against EV71 infection.
On the other hand, viruses have evolved various methods to overcome the restriction of host restrictive factors, and treatment design against enteroviruses from this perspective will kill two birds with one stone. As reported by Li et al., the 2C protein of enteroviruses interacts with A3G and triggers the degradation of A3G (22, 57). Thus, inhibitors targeting the 2C domain that binds A3G can interfere with the interaction between the 2C protein and A3G and prevent the escape of the enteroviruses from A3G. At the same time, even if the target domain of the 2C protein mutates and causes the effects of inhibitors to be off-target, the mutant 2C protein will fail to bind to A3G and break out the restriction from A3G, indicating that A3G could exert its antiviral activity and that the inhibitors targeting this domain will stably inhibit enteroviruses by inducing virus mutation to a greater extent.
Third, as the endogenous component of host cells, it is important to note that antiviral strategies that regulate the expression of host restrictive factors will greatly reduce any side effects, which will be milder and safer than those experienced after using drugs. As Wang et al. showed in their study, IMB-Z inhibits EV71 replication by upregulating the expression of A3G (20). Moreover, 80 µM IMB-Z induced adequate A3G expression and greatly inhibited the replication of EV71 in a variety of cells. At the same time, 200 µM IMB-Z did not affect cell activity in cell lines, including Vero, HeLa, HCT-8, HEK293T, and SK-N-SH. Therefore, these findings have implications for the safety of antiviral strategies against enteroviruses by regulating the expression of host-restrictive factors.
In recent years, antimicrobial peptides (93) and mRNA drugs (94) have attracted increased interest among scientists, health professionals, and pharmaceutical companies because of their therapeutic potential. With the development of polypeptide and mRNA drugs, the functional domains of host restrictive factors will rapidly develop into antiviral drugs and become the mainstay of novel antiviral therapies. Therefore, identifying human host restrictive factors and exploring the interaction mechanism between a virus and host restrictive factors will become the premise and basis for us to master important antivirus strategies in the future.
In conclusion, studies on the interactions between host restrictive factors and enteroviruses will deepen understanding of virus-host interactions, provide a theoretical basis, and reveal potential targets that are not prone to off-target effects. This information can then be used to develop anti-enterovirus drugs.
Author Contributions
ZL conceptualized the ideas. XQ performed the literature search, drafted the original manuscript, and drew the figures. CH revised the manuscript. All the authors approved the final version of the manuscript.
Funding
This work was supported in part by funding from the National Natural Science Foundation of China (81701987 to ZL and 81801994 to CH), the Science and Technology Department of Jilin Province (20210101300JC), China Postdoctoral Science Foundation (2020M670826), and the Education Department of Jilin Province (JJKH20211141KJ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Baggen J, Thibaut HJ, Strating J, van Kuppeveld FJM. The Life Cycle of Non-Polio Enteroviruses and How to Target It. Nat Rev Microbiol (2018) 16(6):368–81. doi: 10.1038/s41579-018-0005-4
2. Bian L, Wang Y, Yao X, Mao Q, Xu M, Liang Z. Coxsackievirus A6: A New Emerging Pathogen Causing Hand, Foot and Mouth Disease Outbreaks Worldwide. Expert Rev Anti Infect Ther (2015) 13(9):1061–71. doi: 10.1586/14787210.2015.1058156
3. Fu X, Wan Z, Li Y, Hu Y, Jin X, Zhang C. National Epidemiology and Evolutionary History of Four Hand, Foot and Mouth Disease-Related Enteroviruses in China From 2008 to 2016. Virol Sin (2020) 35(1):21–33. doi: 10.1007/s12250-019-00169-2
4. Zhang C, Zhu R, Yang Y, Chi Y, Yin J, Tang X, et al. Phylogenetic Analysis of the Major Causative Agents of Hand, Foot and Mouth Disease in Suzhou City, Jiangsu Province, China, in 2012-2013. Emerg Microbes Infect (2015) 4(2):e12. doi: 10.1038/emi.2015.12
5. Zhao TS, Du J, Sun DP, Zhu QR, Chen LY, Ye C, et al. A Review and Meta-Analysis of the Epidemiology and Clinical Presentation of Coxsackievirus A6 Causing Hand-Foot-Mouth Disease in China and Global Implications. Rev Med Virol (2020) 30(2):e2087. doi: 10.1002/rmv.2087
6. Dyda A, Stelzer-Braid S, Adam D, Chughtai AA, MacIntyre CR. The Association Between Acute Flaccid Myelitis (AFM) and Enterovirus D68 (EV-D68) - What Is the Evidence for Causation? Euro Surveill (2018) 23(3):17-00310. doi: 10.2807/1560-7917.ES.2018.23.3.17-00310
7. Holm-Hansen CC, Midgley SE, Fischer TK. Global Emergence of Enterovirus D68: A Systematic Review. Lancet Infect Dis (2016) 16(5):e64–75. doi: 10.1016/S1473-3099(15)00543-5
8. Hixon AM, Frost J, Rudy MJ, Messacar K, Clarke P, Tyler KL. Understanding Enterovirus D68-Induced Neurologic Disease: A Basic Science Review. Viruses (2019) 11(9):821. doi: 10.3390/v11090821
9. Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, et al. Interferon-Inducible Expression of APOBEC3 Editing Enzymes in Human Hepatocytes and Inhibition of Hepatitis B Virus Replication. Hepatology (2006) 43(6):1364–74. doi: 10.1002/hep.21187
10. Sarkis PT, Ying S, Xu R, Yu XF. STAT1-Independent Cell Type-Specific Regulation of Antiviral APOBEC3G by IFN-Alpha. J Immunol (2006) 177(7):4530–40. doi: 10.4049/jimmunol.177.7.4530
11. Ying S, Zhang X, Sarkis PT, Xu R, Yu X. Cell-Specific Regulation of APOBEC3F by Interferons. Acta Biochim Biophys Sin (Shanghai) (2007) 39(4):297–304. doi: 10.1111/j.1745-7270.2007.00275.x
12. Mohanram V, Skold AE, Bachle SM, Pathak SK, Spetz AL. IFN-Alpha Induces APOBEC3G, F, and A in Immature Dendritic Cells and Limits HIV-1 Spread to CD4+ T Cells. J Immunol (2013) 190(7):3346–53. doi: 10.4049/jimmunol.1201184
13. Simon V, Bloch N, Landau NR. Intrinsic Host Restrictions to HIV-1 and Mechanisms of Viral Escape. Nat Immunol (2015) 16(6):546–53. doi: 10.1038/ni.3156
14. Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, et al. HIV-1 Restriction Factor SAMHD1 Is a Deoxynucleoside Triphosphate Triphosphohydrolase. Nature (2011) 480(7377):379–82. doi: 10.1038/nature10623
15. Cobos Jimenez V, Booiman T, de Taeye SW, van Dort KA, Rits MA, Hamann J, et al. Differential Expression of HIV-1 Interfering Factors in Monocyte-Derived Macrophages Stimulated With Polarizing Cytokines or Interferons. Sci Rep (2012) 2:763. doi: 10.1038/srep00763
16. Nguyen TT, Hu Y, Widney DP, Mar RA, Smith JB. Murine GBP-5, a New Member of the Murine Guanylate-Binding Protein Family, Is Coordinately Regulated With Other GBPs In Vivo and In Vitro. J Interferon Cytokine Res (2002) 22(8):899–909. doi: 10.1089/107999002760274926
17. Aebi M, Fah J, Hurt N, Samuel CE, Thomis D, Bazzigher L, et al. cDNA Structures and Regulation of Two Interferon-Induced Human Mx Proteins. Mol Cell Biol (1989) 9(11):5062–72. doi: 10.1128/mcb.9.11.5062-5072.1989
18. Passos V, Zillinger T, Casartelli N, Wachs AS, Xu S, Malassa A, et al. Characterization of Endogenous SERINC5 Protein as Anti-HIV-1 Factor. J Virol (2019) 93(24):e01221-19. doi: 10.1128/JVI.01221-19
19. D'Urbano V, Bertoldi A, Re MC, De Crignis E, Tamburello M, Primavera A, et al. Restriction Factors Expression Decreases in HIV-1 Patients After cART. New Microbiol (2021) 44(2):95–103.
20. Wang H, Zhong M, Li Y, Li K, Wu S, Guo T, et al. APOBEC3G Is a Restriction Factor of EV71 and Mediator of IMB-Z Antiviral Activity. Antiviral Res (2019) 165:23–33. doi: 10.1016/j.antiviral.2019.03.005
21. Li Z, Huan C, Wang H, Liu Y, Liu X, Su X, et al. TRIM21-Mediated Proteasomal Degradation of SAMHD1 Regulates Its Antiviral Activity. EMBO Rep (2020) 21(1):e47528. doi: 10.15252/embr.201847528
22. Li Z, Yang X, Zhao Z, Liu X, Zhang W. Host Restriction Factor A3G Inhibits the Replication of Enterovirus D68 Through Competitively Binding 5' UTR With PCBP1. J Virol (2022) 96(2):e0170821. doi: 10.1128/JVI.01708-21
23. Zhao Z, Li Z, Huan C, Liu X, Zhang W. SAMHD1 Inhibits Multiple Enteroviruses by Interfering With the Interaction Between VP1 and VP2 Proteins. J Virol (2021) 95(13):e0062021. doi: 10.1128/JVI.00620-21
24. Takaoka A, Yanai H. Interferon Signalling Network in Innate Defence. Cell Microbiol (2006) 8(6):907–22. doi: 10.1111/j.1462-5822.2006.00716.x
25. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and Their Class II Cytokine Receptor IL-28r. Nat Immunol (2003) 4(1):63–8. doi: 10.1038/ni873
26. Wang C, Ji L, Yuan X, Jin Y, Cardona CJ, Xing Z. Differential Regulation of TLR Signaling on the Induction of Antiviral Interferons in Human Intestinal Epithelial Cells Infected With Enterovirus 71. PloS One (2016) 11(3):e0152177. doi: 10.1371/journal.pone.0152177
27. Kawai T, Akira S. TLR Signaling. Cell Death Differ (2006) 13(5):816–25. doi: 10.1038/sj.cdd.4401850
28. Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, et al. The RNA Helicase Lgp2 Inhibits TLR-Independent Sensing of Viral Replication by Retinoic Acid-Inducible Gene-I. J Immunol (2005) 175(8):5260–8. doi: 10.4049/jimmunol.175.8.5260
29. Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and Unique Functions of the DExD/H-Box Helicases RIG-I, MDA5, and LGP2 in Antiviral Innate Immunity. J Immunol (2005) 175(5):2851–8. doi: 10.4049/jimmunol.175.5.2851
30. Ho HY, Cheng ML, Weng SF, Chang L, Yeh TT, Shih SR, et al. Glucose-6-Phosphate Dehydrogenase Deficiency Enhances Enterovirus 71 Infection. J Gen Virol (2008) 89(Pt 9):2080–9. doi: 10.1099/vir.0.2008/001404-0
31. Schein CH, Oezguen N, Volk DE, Garimella R, Paul A, Braun W. NMR Structure of the Viral Peptide Linked to the Genome (VPg) of Poliovirus. Peptides (2006) 27(7):1676–84. doi: 10.1016/j.peptides.2006.01.018
32. Pfister T, Jones KW, Wimmer E. A Cysteine-Rich Motif in Poliovirus Protein 2C(ATPase) Is Involved in RNA Replication and Binds Zinc In Vitro. J Virol (2000) 74(1):334–43. doi: 10.1128/JVI.74.1.334-343.2000
33. Davies EH, Erikson A, Collin-Histed T, Mengel E, Tylki-Szymanska A, Vellodi A. Outcome of Type III Gaucher Disease on Enzyme Replacement Therapy: Review of 55 Cases. J Inherit Metab Dis (2007) 30(6):935–42. doi: 10.1007/s10545-007-0577-z
34. Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. Heterogeneous Nuclear Ribonuclear Protein K Interacts With the Enterovirus 71 5' Untranslated Region and Participates in Virus Replication. J Gen Virol (2008) 89(Pt 10):2540–9. doi: 10.1099/vir.0.2008/003673-0
35. Lin JY, Shih SR, Pan M, Li C, Lue CF, Stollar V, et al. hnRNP A1 Interacts With the 5' Untranslated Regions of Enterovirus 71 and Sindbis Virus RNA and Is Required for Viral Replication. J Virol (2009) 83(12):6106–14. doi: 10.1128/JVI.02476-08
36. Luo Z, Dong X, Li Y, Zhang Q, Kim C, Song Y, et al. PolyC-Binding Protein 1 Interacts With 5'-Untranslated Region of Enterovirus 71 RNA in Membrane-Associated Complex to Facilitate Viral Replication. PloS One (2014) 9(1):e87491. doi: 10.1371/journal.pone.0087491
37. Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a Human Gene That Inhibits HIV-1 Infection and Is Suppressed by the Viral Vif Protein. Nature (2002) 418(6898):646–50. doi: 10.1038/nature00939
38. Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for Editing of Human Papillomavirus DNA by APOBEC3 in Benign and Precancerous Lesions. Science (2008) 320(5873):230–3. doi: 10.1126/science.1153201
39. Lin JY, Brewer G, Li ML. HuR and Ago2 Bind the Internal Ribosome Entry Site of Enterovirus 71 and Promote Virus Translation and Replication. PloS One (2015) 10(10):e0140291. doi: 10.1371/journal.pone.0140291
40. Weinreb NJ, Barranger JA, Charrow J, Grabowski GA, Mankin HJ, Mistry P. Guidance on the Use of Miglustat for Treating Patients With Type 1 Gaucher Disease. Am J Hematol (2005) 80(3):223–9. doi: 10.1002/ajh.20504
41. Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, et al. Scavenger Receptor B2 Is a Cellular Receptor for Enterovirus 71. Nat Med (2009) 15(7):798–801. doi: 10.1038/nm.1992
42. Nakata K, Takeda S, Tanaka A, Kwang J, Komano J. Antiviral Activity of Acid Beta-Glucosidase 1 on Enterovirus 71, a Causative Agent of Hand, Foot and Mouth Disease. J Gen Virol (2017) 98(4):643–51. doi: 10.1099/jgv.0.000723
43. Charrow J, Andersson HC, Kaplan P, Kolodny EH, Mistry P, Pastores G, et al. Enzyme Replacement Therapy and Monitoring for Children With Type 1 Gaucher Disease: Consensus Recommendations. J Pediatr (2004) 144(1):112–20. doi: 10.1016/j.jpeds.2003.10.067
44. Li Z, Liu X, Wang S, Li J, Hou M, Liu G, et al. Identification of a Nucleotide in 5' Untranslated Region Contributing to Virus Replication and Virulence of Coxsackievirus A16. Sci Rep (2016) 6:20839. doi: 10.1038/srep20839
45. Sun Y, Wang Y, Shan C, Chen C, Xu P, Song M, et al. Enterovirus 71 VPg Uridylation Uses a Two-Molecular Mechanism of 3D Polymerase. J Virol (2012) 86(24):13662–71. doi: 10.1128/JVI.01712-12
46. Liu Y, Franco D, Paul AV, Wimmer E. Tyrosine 3 of Poliovirus Terminal Peptide VPg(3B) has an Essential Function in RNA Replication in the Context of Its Precursor Protein, 3AB. J Virol (2007) 81(11):5669–84. doi: 10.1128/JVI.02350-06
47. Morasco BJ, Sharma N, Parilla J, Flanegan JB. Poliovirus Cre(2C)-Dependent Synthesis of VPgpUpU Is Required for Positive- But Not Negative-Strand RNA Synthesis. J Virol (2003) 77(9):5136–44. doi: 10.1128/JVI.77.9.5136-5144.2003
48. Tang WF, Yang SY, Wu BW, Jheng JR, Chen YL, Shih CH, et al. Reticulon 3 Binds the 2C Protein of Enterovirus 71 and Is Required for Viral Replication. J Biol Chem (2007) 282(8):5888–98. doi: 10.1074/jbc.M611145200
49. Xia H, Wang P, Wang GC, Yang J, Sun X, Wu W, et al. Human Enterovirus Nonstructural Protein 2catpase Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone. PloS Pathog (2015) 11(7):e1005067. doi: 10.1371/journal.ppat.1005067
50. Fan W, Mar KB, Sari L, Gaszek IK, Cheng Q, Evers BM, et al. TRIM7 Inhibits Enterovirus Replication and Promotes Emergence of a Viral Variant With Increased Pathogenicity. Cell (2021) 184(13):3410–25.e17. doi: 10.1016/j.cell.2021.04.047
51. Lin JY, Li ML, Brewer G. mRNA Decay Factor AUF1 Binds the Internal Ribosomal Entry Site of Enterovirus 71 and Inhibits Virus Replication. PloS One (2014) 9(7):e103827. doi: 10.1371/journal.pone.0103827
52. Wu S, Lin L, Zhao W, Li X, Wang Y, Si X, et al. AUF1 Is Recruited to the Stress Granules Induced by Coxsackievirus B3. Virus Res (2014) 192:52–61. doi: 10.1016/j.virusres.2014.08.003
53. Ullmer W, Semler BL. Direct and Indirect Effects on Viral Translation and RNA Replication Are Required for AUF1 Restriction of Enterovirus Infections in Human Cells. mBio (2018) 9(5):e01669-18. doi: 10.1128/mBio.01669-18
54. Xue YC, Ng CS, Mohamud Y, Fung G, Liu H, Bahreyni A, et al. FUS/TLS Suppresses Enterovirus Replication and Promotes Antiviral Innate Immune Responses. J Virol (2021) 95(12):e00304-21. doi: 10.1128/JVI.00304-21
55. Han Y, Wang L, Cui J, Song Y, Luo Z, Chen J, et al. SIRT1 Inhibits EV71 Genome Replication and RNA Translation by Interfering With the Viral Polymerase and 5'UTR RNA. J Cell Sci (2016) 129(24):4534–47. doi: 10.1242/jcs.193698
56. Li H, Bai Z, Li C, Sheng C, Zhao X. EV71 Infection Induces Cell Apoptosis Through ROS Generation and SIRT1 Activation. J Cell Biochem (2020) 121(10):4321–31. doi: 10.1002/jcb.29628
57. Li Z, Ning S, Su X, Liu X, Wang H, Liu Y, et al. Enterovirus 71 Antagonizes the Inhibition of the Host Intrinsic Antiviral Factor A3G. Nucleic Acids Res (2018) 46(21):11514–27. doi: 10.1093/nar/gky840
58. Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, et al. Vpx Relieves Inhibition of HIV-1 Infection of Macrophages Mediated by the SAMHD1 Protein. Nature (2011) 474(7353):658–61. doi: 10.1038/nature10195
59. Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, et al. SAMHD1 is the Dendritic- and Myeloid-Cell-Specific HIV-1 Restriction Factor Counteracted by Vpx. Nature (2011) 474(7353):654–7. doi: 10.1038/nature10117
60. Ballana E, Este JA. SAMHD1: At the Crossroads of Cell Proliferation, Immune Responses, and Virus Restriction. Trends Microbiol (2015) 23(11):680–92. doi: 10.1016/j.tim.2015.08.002
61. Hollenbaugh JA, Gee P, Baker J, Daly MB, Amie SM, Tate J, et al. Host Factor SAMHD1 Restricts DNA Viruses In non-Dividing Myeloid Cells. PloS Pathog (2013) 9(6):e1003481. doi: 10.1371/journal.ppat.1003481
62. Chen Z, Zhu M, Pan X, Zhu Y, Yan H, Jiang T, et al. Inhibition of Hepatitis B Virus Replication by SAMHD1. Biochem Biophys Res Commun (2014) 450(4):1462–8. doi: 10.1016/j.bbrc.2014.07.023
63. Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres Syndrome Gene and HIV-1 Restriction Factor SAMHD1 Is a dGTP-Regulated Deoxynucleotide Triphosphohydrolase. J Biol Chem (2011) 286(51):43596–600. doi: 10.1074/jbc.C111.317628
64. Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, et al. SAMHD1 Restricts the Replication of Human Immunodeficiency Virus Type 1 by Depleting the Intracellular Pool of Deoxynucleoside Triphosphates. Nat Immunol (2012) 13(3):223–8. doi: 10.1038/ni.2236
65. Antonucci JM, St Gelais C, de Silva S, Yount JS, Tang C, Ji X, et al. SAMHD1-Mediated HIV-1 Restriction in Cells Does Not Involve Ribonuclease Activity. Nat Med (2016) 22(10):1072–4. doi: 10.1038/nm.4163
66. Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, et al. Nuclease Activity of the Human SAMHD1 Protein Implicated in the Aicardi-Goutieres Syndrome and HIV-1 Restriction. J Biol Chem (2013) 288(12):8101–10. doi: 10.1074/jbc.M112.431148
67. Choi J, Ryoo J, Oh C, Hwang S, Ahn K. SAMHD1 Specifically Restricts Retroviruses Through its RNase Activity. Retrovirology (2015) 12:46. doi: 10.1186/s12977-015-0174-4
68. Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, et al. The Ribonuclease Activity of SAMHD1 Is Required for HIV-1 Restriction. Nat Med (2014) 20(8):936–41. doi: 10.1038/nm.3626
69. Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. Cleavage of the Adaptor Protein TRIF by Enterovirus 71 3C Inhibits Antiviral Responses Mediated by Toll-Like Receptor 3. J Virol (2011) 85(17):8811–8. doi: 10.1128/JVI.00447-11
70. Xiao H, Li J, Yang X, Li Z, Wang Y, Rui Y, et al. Ectopic Expression of TRIM25 Restores RIG-I Expression and IFN Production Reduced by Multiple Enteroviruses 3C(Pro). Virol Sin (2021) 36(6):1363–74. doi: 10.1007/s12250-021-00410-x
71. Lei X, Han N, Xiao X, Jin Q, He B, Wang J. Enterovirus 71 3C Inhibits Cytokine Expression Through Cleavage of the TAK1/TAB1/TAB2/TAB3 Complex. J Virol (2014) 88(17):9830–41. doi: 10.1128/JVI.01425-14
72. Wang H, Lei X, Xiao X, Yang C, Lu W, Huang Z, et al. Reciprocal Regulation Between Enterovirus 71 and the NLRP3 Inflammasome. Cell Rep (2015) 12(1):42–8. doi: 10.1016/j.celrep.2015.05.047
73. Lei X, Liu X, Ma Y, Sun Z, Yang Y, Jin Q, et al. The 3C Protein of Enterovirus 71 Inhibits Retinoid Acid-Inducible Gene I-Mediated Interferon Regulatory Factor 3 Activation and Type I Interferon Responses. J Virol (2010) 84(16):8051–61. doi: 10.1128/JVI.02491-09
74. Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. Cleavage of Interferon Regulatory Factor 7 by Enterovirus 71 3C Suppresses Cellular Responses. J Virol (2013) 87(3):1690–8. doi: 10.1128/JVI.01855-12
75. Hung HC, Wang HC, Shih SR, Teng IF, Tseng CP, Hsu JT. Synergistic Inhibition of Enterovirus 71 Replication by Interferon and Rupintrivir. J Infect Dis (2011) 203(12):1784–90. doi: 10.1093/infdis/jir174
76. Li Z, Wu Y, Li H, Li W, Tan J, Qiao W. 3C Protease of Enterovirus 71 Cleaves Promyelocytic Leukemia Protein and Impairs PML-NBs Production. Virol J (2021) 18(1):255. doi: 10.1186/s12985-021-01725-7
77. Wen W, Qi Z, Wang J. The Function and Mechanism of Enterovirus 71 (EV71) 3c Protease. Curr Microbiol (2020) 77(9):1968–75. doi: 10.1007/s00284-020-02082-4
78. Tan J, George S, Kusov Y, Perbandt M, Anemuller S, Mesters JR, et al. 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses. J Virol (2013) 87(8):4339–51. doi: 10.1128/JVI.01123-12
79. Xiang Z, Liu L, Lei X, Zhou Z, He B, Wang J. 3c Protease of Enterovirus D68 Inhibits Cellular Defense Mediated by Interferon Regulatory Factor 7. J Virol (2016) 90(3):1613–21. doi: 10.1128/JVI.02395-15
80. Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, et al. Enterovirus 71 Protease 2Apro Targets MAVS to Inhibit Anti-Viral Type I Interferon Responses. PloS Pathog (2013) 9(3):e1003231. doi: 10.1371/journal.ppat.1003231
81. Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K, et al. Enterovirus 2Apro Targets MDA5 and MAVS in Infected Cells. J Virol (2014) 88(6):3369–78. doi: 10.1128/JVI.02712-13
82. Wang SH, Wang K, Zhao K, Hua SC, Du J. The Structure, Function, and Mechanisms of Action of Enterovirus Non-Structural Protein 2c. Front Microbiol (2020) 11:615965. doi: 10.3389/fmicb.2020.615965
83. Zheng Z, Li H, Zhang Z, Meng J, Mao D, Bai B, et al. Enterovirus 71 2C Protein Inhibits TNF-Alpha-Mediated Activation of NF-kappaB by Suppressing IkappaB Kinase Beta Phosphorylation. J Immunol (2011) 187(5):2202–12. doi: 10.4049/jimmunol.1100285
84. Li Q, Zheng Z, Liu Y, Zhang Z, Liu Q, Meng J, et al. 2c Proteins of Enteroviruses Suppress IKKbeta Phosphorylation by Recruiting Protein Phosphatase 1. J Virol (2016) 90(10):5141–51. doi: 10.1128/JVI.03021-15
85. Du H, Yin P, Yang X, Zhang L, Jin Q, Zhu G. Enterovirus 71 2c Protein Inhibits NF-KappaB Activation by Binding to RelA(P65). Sci Rep (2015) 5:14302. doi: 10.1038/srep14302
86. Li L, Fan H, Song Z, Liu X, Bai J, Jiang P. Encephalomyocarditis Virus 2C Protein Antagonizes Interferon-Beta Signaling Pathway Through Interaction With MDA5. Antiviral Res (2019) 161:70–84. doi: 10.1016/j.antiviral.2018.10.010
87. Wang L, Wang J, Wang L, Ma S, Liu Y. Anti-Enterovirus 71 Agents of Natural Products. Molecules (2015) 20(9):16320–33. doi: 10.3390/molecules200916320
88. Caligiuri P, Cerruti R, Icardi G, Bruzzone B. Overview of Hepatitis B Virus Mutations and Their Implications in the Management of Infection. World J Gastroenterol (2016) 22(1):145–54. doi: 10.3748/wjg.v22.i1.145
89. Liang B, Jiang J, Pan P, Chen R, Zhuang D, Zhao F, et al. Morphine Increases Lamivudine- and Nevirapine-Induced Human Immunodeficiency Virus-1 Drug-Resistant Mutations In Vitro. Microb Drug Resist (2017) 23(3):285–93. doi: 10.1089/mdr.2015.0347
90. Ma J, Zhang Y, Chen X, Jin Y, Chen D, Wu Y, et al. Association of Preexisting Drug-Resistance Mutations and Treatment Failure in Hepatitis B Patients. PloS One (2013) 8(7):e67606. doi: 10.1371/journal.pone.0067606
91. Razonable RR. Drug-Resistant Cytomegalovirus: Clinical Implications of Specific Mutations. Curr Opin Organ Transplant (2018) 23(4):388–94. doi: 10.1097/MOT.0000000000000541
92. Yin F, Wu Z, Fang W, Wu C, Rayner S, Han M, et al. Resistant Mutations and Quasispecies Complexity of Hepatitis B Virus During Telbivudine Treatment. J Gen Virol (2015) 96(11):3302–12. doi: 10.1099/jgv.0.000285
93. Boparai JK, Sharma PK. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept Lett (2020) 27(1):4 16. doi: 10.2174/0929866526666190822165812
Keywords: host restrictive factors, enteroviruses, virus-host interplay, antiviral, arms race
Citation: Huan C, Qu X and Li Z (2022) Host Restrictive Factors Are the Emerging Storm Troopers Against Enterovirus: A Mini-Review. Front. Immunol. 13:910780. doi: 10.3389/fimmu.2022.910780
Received: 01 April 2022; Accepted: 12 April 2022;
Published: 04 May 2022.
Edited by:
Junji Xing, Houston Methodist Research Institute, United StatesReviewed by:
Longhuan Ma, University of Florida, United StatesCopyright © 2022 Huan, Qu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaolong Li, bGl6aGFvbG9uZ0BqbHUuZWR1LmNu
 Chen Huan
Chen Huan Xinglong Qu
Xinglong Qu Zhaolong Li
Zhaolong Li