- Department of Pathobiology, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, IL, United States
In recent years, fungal vaccine research emanated significant findings in the field of antifungal T-cell immunity. The generation of effector T cells is essential to combat many mucosal and systemic fungal infections. The development of antifungal memory T cells is integral for controlling or preventing fungal infections, and understanding the factors, regulators, and modifiers that dictate the generation of such T cells is necessary. Despite the deficiency in the clear understanding of antifungal memory T-cell longevity and attributes, in this review, we will compile some of the existing literature on antifungal T-cell immunity in the context of memory T-cell development against fungal infections.
Introduction
An increasing global burden of fungal diseases due to increasing immunocompromised individuals has heightened the need for effective preventive and therapeutic strategies. Fungi are one of the large biome classes, but only a handful of them are pathogenic to humans, causing a significant case fatality of up to 90%. More than 150 million severe cases and over 1.5 million succumb to fungal infections annually, despite the use of antifungal drugs (1–3). Some existing antifungals are effective but cause serious side effects and are liable to the growing drug-resistant fungal pathogens. With expanding knowledge on host–fungal pathogen interactions, there is a tremendous leap in the thrust to develop fungal vaccines. The pan-fungal vaccine is highly desirable, but the features of different fungal pathogenesis and elicitation of distinct immune responses require a clear understanding of the fungus–immune system interface, i.e., vaccine immunity and the potential to develop immunological memory. This review gives an overview of antifungal memory T cells.
Although there is a good amount of evidence of antibody-mediated immunity (4), adaptive immune cell responses against pathogenic fungi are mainly mediated by T cells, and genetic or acquired T-cell deficiency leads to a higher incidence of opportunistic infections (1, 5, 6). Antifungal defense mechanisms by CD4+ T cells, a major class of T cells, involve the secretion of proinflammatory cytokines and cell–cell interactions to activate innate immune cells, help CD8+ T cells, and provide help for the generation of antibodies from B cells (7). The antifungal CD4+ T-cell immunity involves the expression of IFNγ, TNFα, GM-CSF, and IL-17A cytokines, which are differentially produced in a fungus- and tissue-specific manner. For example, IFNγ, TNFα, and GM-CSF are predominantly induced during histoplasmosis, aspergillosis, cryptococcosis, paracoccidioidomycosis, pneumocystosis, and talaromycosis, whereas type 17 cytokines, IL-17A/F, and IL-22 are mainly induced during candidiasis, coccidioidomycosis, blastomycosis, and mucormycosis (reviewed here). Nonetheless, it is common to see both types of responses with variable degrees in most fungal infections. These secreted cytokines generate an inflammatory milieu and act on other cells for innate cell recruitment, activation, secretion of antimicrobial peptides, and killing of fungi (8–10). In contrast, the antifungal T cell-mediated immunity is compromised if their cytokine signature yields regulatory or unprotective cytokines that can lead to severe disseminated infections (11, 12). Despite the need for CD4+ T-cell help for CD8+ T-cell activation and memory maintenance in viral and bacterial infection scenarios, using mouse models of fungal infections against Pneumocystis, Histoplasma, and Blastomyces, the studies have shown that antifungal CD8+ T cells can be induced, retained as long-lasting memory, and recalled upon the challenge to provide immunity independent of the T-cell help during mouse models of Pneumocystis, Histoplasma, and Blastomyces infections (8, 13–16). Antifungal activity of CD8+ T cells involves cell cytotoxicity (17) and secretion of proinflammatory cytokines; the latter often mimics CD4+ T-cell antifungal cytokine functions.
The host’s first response to fungal invasion starts with innate immunity, which then engages the adaptive immune arm to mount antigen-specific responses to control or clear fungal infection (18). The pattern recognition receptors (PRRs) are critical for innate immune responses for initial fungal control, and their mutations are associated with higher susceptibility (6, 19–21). The activation of innate immune cells and generation of apt inflammatory milieu facilitate dendritic cell priming of naïve T cells to become effectors, which eventually differentiate to form antifungal memory T cells (22). Thus, the innate immunity dictated by fungal recognition shapes adaptive T-cell immunity and immunological memory.
Fungal recognition by the immune system: Bridging innate to adaptive immunity
Among innate cells, dendritic cells are essential for priming naïve T cells (23). The activated dendritic cells process and present the antigens to CD4+ and CD8+ T cells through MHC-II and MHC-I molecules, respectively. Along with antigen presentation, dendritic cells provide costimulatory signals for T-cell responses (24). Thus, the functions of dendritic cell maturation and activation are a critical step toward bridging innate with adaptive immunity, and such events are mainly mediated by PRR signals. PRRs are a category of host cell receptors that sense specific molecules/patterns, the pathogen-associated molecular patterns (PAMPs) such as β-glucans and mannans of pathogenic fungi, and this recognition is key for innate immune cell activation to provide a primary antifungal defense. The PRRs are mainly classified as Toll-like receptors (TLRs), C-type lectin receptors (CLRs), retinoic acid-inducible gene I-like receptors (RLRs), and NOD-like receptors (NLRs), which can directly bind to the PAMPs of fungi, whereas damage-associated molecular patterns (DAMPs) can bind to PRRs and their canonical DAMP-sensing receptors such as P2X purinoceptor 7 (P2XR7), triggering receptor expressed on myeloid cells 1/2 (TREM1/2), and receptor for advanced glycation end products (RAGE) (25–28). Several of PAMPs of fungi, including β-glucans, mannans, glycoprotein A, and glyceroglycolipids, have been identified for their functions using their PRRs in the host (29–32). There are excellent reviews on PRRs and fungal immunity elsewhere. Here, we highlight how PRRs can influence the innate immune cells to guide adaptive T-cell immunity.
Although negative signaling is noted with few PRRs, many are associated with their positive signaling to promote activation, phagocytosis, and antigen presentation by dendritic cells to T cells. The activated innate immune cells generate an inflammatory micro milieu conducive to the recruitment, activation, differentiation, and expansion of fungal-specific T cells by secreting cytokines and chemokines. Among PRRs, fungal-recognizing CLRs are instrumental in driving innate immune cell responses. Due to structural differences, different fungi show differential CLR binding properties leading to diverse host cell responses. The prototypic member of this family, the Dectin-1 receptor, expressed on innate immune cells including macrophages, neutrophils, and dendritic cells (DCs), recognizes β1-3-glucans of the fungal cell wall. The interference of Dectin-1 interaction with β-glucans by a soluble dectin-Fc fusion protein dampened the expression of inflammatory cytokines, TNFα, IL-1, IL-6, MIP-2, CCL3, G-CSF, and GM-CSF, expression in vivo, and increased fungal burden during aspergillosis (33). Ablation of Dectin-1 resulted in decreased reactive oxygen species (ROS) production by neutrophils and the ability to kill Aspergillus in vitro. Additionally, alveolar macrophages of Dectin-1−/− mice had defective production of proinflammatory cytokines and chemokines, including IL-1α, IL-1β, and TNFα. The Dectin-1 recognition of Aspergillus seems important for IL-17A production, and neutralization of IL-17A led to impaired Aspergillus fumigatus clearance and higher mortality of infected mice (34). Dectin-1 promoted the survival of antigen-specific CD4+ T cells, not the CD8+ T cells, specifically in GI-associated lymphoid tissues following systemic Candida infection, and ablation of Dectin-1 reduced the tissue-specific dendritic cells and increased activation of CD4+ T cells leading to higher susceptibility to Candida-induced colitis (35). During systemic Candida infection, the protective role of Dectin-1 was fungal strain-specific, possibly due to variable adaptation of Candida albicans strains in vivo, including the changes in the microbiota of mice due to different mouse facilities, with changes in the cell wall components and high chitin in the cell wall masks the dependability on Dectin-1 recognition (36). Further, pathogenic fungi can avoid host Dectin-1 recognition of β-(1,3)-glucan by masking with α-(1,3)-glucan, phosphatidylserine, capsule, rodlet layer/melanin, and mannans or trimming to reduce the exposure in the cell wall, thus increasing immune evasion in vivo (37–40). However, Dectin-1 was dispensable for controlling infections from Blastomyces, Cryptococcus, certain strains or species of Candida, or Candida colonization (36, 41–45), suggesting the differential requirement of CLRs for fungal immunity.
Unlike the Dectin-1 receptor, cytoplasmic domains of Dectin-2 and Mincle receptors lack their own ITAM motifs and associate with FcRγ immunoreceptor harboring cytoplasmic ITAM motif for signaling (46, 47). Dectin-2 and Mincle have been shown to be important for immunity against blastomycosis (48), aspergillosis (49), histoplasmosis (44), chromoblastomycosis (50), disseminated candidiasis (51, 52), and species-specific candidiasis (45). Dectin-2 signals through the Syk-CARD9 pathway and promote Th17 cell responses (51) by inducing the expression of IL-1 and IL-23 cytokines (53). Despite that Dectin-2 and Mincle share their signaling through FcRγ immunoreceptors, their role in activation, expansion, and differentiation of antigen-specific CD4+ T-cell responses may differ. While Dectin-2 was essential for enhancing Th17 cell differentiation, Mincle recognition suppressed Th17 polarization during chromoblastomycosis (50).
In addition to CLRs, TLRs expressed by innate cells are involved in the control of fungal infection. Myeloid differentiation primary response protein 88 (MyD88), an adaptor molecule for many TLRs signaling, has been shown to play a role in antifungal immunity against Blastomyces dermatitidis, Paracoccidioides brasiliensis, A. fumigatus, Cryptococcus neoformans, and C. albicans (54–56). TLR2 plays a significant role in conferring protective immunity against Candida infection at mucosal sites, including gastrointestinal and reproductive tracts by inducing Th17 differentiation through MyD88 signaling (57, 58). However, the role of TLR2 in controlling systemic candidiasis seems to be fungal strain specific (59, 60). Additionally, IL-1R/MyD88 signaling pathway is necessary for host resistance against Candida, and TLR4/MyD88 pathways mediate protection against Aspergillus infection by regulating Th1 and Th2 response (54, 61). TLR3 in DCs senses fungal RNA derived from dying cells and potentiates the cross-presentation to activate CD8+ T cells during aspergillosis (62). Nevertheless, compared to those of CLRs, the functions of many TLRs in the context of antifungal immunity seem to be modest or redundant.
NLRs function against fungal defense mainly involved the activation of inflammasomes, which leads to caspase-dependent production of functional IL-1β and IL-18 cytokines. Both of these cytokines have been shown to exert antifungal host defense in an NLRP3-dependent manner (63). NLRC4 negatively regulates NLRP3 inflammasome activity, suppressing early IL-1β and late IL-18-mediated antifungal CD8+ T-cell responses during pneumocystosis (64). Thus, some PRRs of the non-CLR class play a role in immunity against fungal infections (26).
Genetic polymorphisms are associated with susceptibility or resistance to infections. The genetic TLR polymorphisms related to fungal disease susceptibility in humans undergoing allogenic stem cell transplants seem to be modest or minimal (65, 66). The genetic predisposition due to PRR polymorphisms to fungal infections is variable and depends on the pathogen or the degree of inflammation. For example, TLR4 polymorphism D299G is associated with increased susceptibility to Candida bloodstream infection, possibly due to higher immunosuppressive cytokine IL-10 production. However, such susceptibility was not seen in urogenital Candida infection (67). Similarly, the TLR4 polymorphisms (D299G/T399I), despite the normal colonization of the fungus, are associated with mitigating the hyperinflammation and tissue damage during aspergillosis (66). Similarly, genetic polymorphisms of CLRs have been associated with susceptibility to fungal infections. Dectin-1 single-nucleotide polymorphisms (SNPs), rs3901533 and rs7309123, enhanced the susceptibility to invasive pulmonary aspergillosis (68). Dectin-1 polymorphism of Y238X led to decreased receptor signaling and increased susceptibility to invasive aspergillosis and recurrent vulvovaginitis caused by Candida (69, 70). Alternative splicing leading to truncated Dectin-1 seen in the C57BL/6 strain, compared to the DBA/2 mouse strain, increased the susceptibility to coccidioidomycosis (71). Thus, it is essential to decipher gene polymorphisms in humans to understand the susceptibility to fungal infections.
T cells can also express several PRRs to respond to PAMPs during fungal infections. Engagement of the cell-intrinsic PRR pathway is one of the non-classical T-cell signaling routes to enhance the activation, effector function, and memory formation of T cells as proposed originally by Janeway (72). Important PRRs on T cells that detect fungal PAMPs are TLRs, NLRs, and damage-associated molecular pattern-sensing receptors. TLRs can function as co-stimulatory receptors that complement TCR-induced signals to enhance effector T-cell proliferation, survival, and cytokine production (73). T cells expressing TLRs, including TLR2 and TLR4, can directly sense pathogens and modulate T-cell responses. Naïve CD4+ T cells do not express significant levels of TLR2/TLR4 mRNA and proteins, but activated and memory T cells express high levels of membrane-bound TLR2 and TLR4 (74, 75). TLR2 signaling in T cells can be modulated by TCR and IL-2-induced mTOR signals (76). Intrinsic MyD88 signaling can modulate the T-cell functions during fungal infections. MyD88 signaling, both extrinsic, non-CD4+ T cell-mediated (77) and intrinsic, CD8+ T cell-mediated (78), fosters fungal vaccine immunity by T cells by regulating the survival and proliferation of effector T cells. MyD88 promoted the sustained Tc17 cell proliferation by activating mTOR via Akt1, and cell-intrinsic IL-1R and TLR2 signaling, but not IL-18R, were required for MyD88-dependent Tc17 responses (78). MyD88 deletion in FoxP3+ regulatory T cells increased the fungal burden and immunopathology during oral C. albicans infection in mice, coinciding with reduced IL-17A expressing FoxP3+ T cells (Treg17) and increased dysfunctional IFNγ+/FoxP3+ cells (IFNγ+ Treg). This dysregulated IL-1β-mTOR-Treg17 axis contributes to overt inflammation during mucosal infections in elderly individuals in a model of oral candidiasis (79). NLRP3, a member of the NLR family, can indirectly sense danger signals. In a murine model of disseminated talaromycosis, compared to wild-type mice, Casp-1 and Nlrp3 global KO mice displayed higher mortality rates and fungal load, which correlated with impaired CD4+ T-cell recruitment into granulomas (80). Although this study did not look into T-cell intrinsic effect, NLRP3 signaling in CD4+ T cells has been shown to augment Th1 immunity (81). Further studies are needed to dissect the T-cell intrinsic PRR functions against fungal infections. Interestingly, PRR has been used to generate modified TCR of T cells. Dectin-1-chimeric antigen receptor (D-CAR) was bioengineered using the extracellular domain of Dectin-1 to redirect T-cell specificity toward fungal β-glucan moieties for immunity (82). D-CAR+ T cells could inhibit A. fumigatus hyphae formation in vitro and reduce pulmonary fungal burden in vivo. In this study, chimeric CD8+ T cells kill the fungi directly by pumping out cytolysins onto yeast/hyphae of Aspergillus and indirectly by secreting IFNγ that can potentiate the killing of yeasts by neutrophils (83).
Antifungal T cells
CD4+ T cells, also called helper T cells, are instrumental in controlling fungal infections, and their deficiency leads to severe disseminated infections by opportunistic fungal pathogens. As the name suggests, the helper cells bolster innate immune cell functions, aid in the generation of productive B-cell responses, help CD8+ T-cell responses, and control autoimmunity. Based on their cytokine secretion and functions during fungal infections (84, 85), T cells are classified into Th1 (IFNγ, GM-CSF, and TNFα), Th17 (IL-17A/F), Th22 (IL-22), Th2 (IL-4 and IL-13), Th9 (IL-9 and IL-10), Treg (IL-10 and TGFβ), and Tr1 (IL-10). The cytokines produced by T cells have a multifaceted role in controlling or regulating the pathogenesis during infections, including fungal infections (86). Although a mixture of different cytokine-producing T cells is often found during fungal infections, the predominant subset of T-cell responses is associated with the type of pathogen, infection, or tissue location (Table 1). Here, we will highlight some of the roles of these T cells for immunity against different pathogenic fungi with a focus on memory phenotypic cells.
Pathogen-specific antifungal T cells
Candida
Anti-Candida memory T-cell responses are studied in the context of mucosal infections, vaccine-induced responses, and commensal-specific/pre-exposed T cells in healthy donors. Candida is a commensal but opportunistic fungal pathogen that causes disseminated infection under compromised immunity. In a mouse model of oropharyngeal candidiasis (OPC), after resting for 6 weeks following primary infection, the memory CD4+ conferred immunity to secondary infection by producing antigen-specific IL-17A responses (91). However, depletion of CD4+ T cells did not cause OPC, possibly compensated by developed residual IL-17A expressing memory CD8+ T cells and CD3+CD4−CD8− cells. In a mouse model of C. albicans skin infection, IL-17+ CD4+ T cells were enriched in the skin, which transitioned into sessile CD69+/CD103+ tissue-resident memory T cells (TRM) within 90 days. This suggests that the long-lasting antifungal memory Th17 cells are generated in the non-lymphoid organ, such as the skin (105). Importantly, these TRM cells provided better immunity than migratory Th17 cells following infectious challenges. In a mouse model of vulvovaginal candidiasis, Th17 cells persisted even after the clearance of the yeast but in low numbers by day 30 (106). The vaginal washes showed the presence of IL-17A, IL-23, and β-defensins. Nevertheless, this study did not examine the long-term maintenance of effector/memory CD4+ T cells.
The C. albicans hypha-specific surface protein antigen, agglutinin-like sequence (Als3)-based NDV-3A vaccine was used for active immunization in a mouse model that prevented Candida colonization at vein catheterization site (107), and the mechanisms involve the induction of high levels of anti-rAls3p-N antibodies. Here, the antibody titers persisted 15 days post-boost, interfered with Candida colonization at the catheter site, and reduced the fungal burdens in the kidneys. Although the elicitation of CD4+ T-cell responses or their persistence was not evaluated in this study, the blocking/inhibitory ability of the antibodies may suggest their potential. In another study, where the NDV-3 vaccine was used in a mouse model of vaccine immunity to vulvovaginal candidiasis, robust antibody responses and immunity were dependent on both T and B cells (108). However, the immunity was assessed 2 weeks following the boost, which may not give a clear understanding of long-lasting memory CD4+ T-cell development. Nevertheless, in a human study, intramuscular NDV-3 vaccination of the volunteers induced the durable serum and cervicovaginal antibody titer (anti-Als3) for up to 1 year and provided significant immunity against recurrent vulvovaginal candidiasis (109). This study found significantly higher numbers of Als3-specific cytokine (IFNγ and IL-17A) secreting peripheral blood mononuclear cells (PBMCs) even after day 90 of vaccination. Although this trial evaluated an immunotherapeutic vaccine, whether the vaccination induces the antigen-free (Candida reexposure-free) persistence of long-lasting “memory adaptive (T and B) cells” needs to be assessed.
Candida-specific memory CD4+ T cells in healthy blood donors produced IL-17A and IFNγ, but not IL-10, following restimulation (110). The human Candida-specific memory Th17 cells preferentially expressed phenotypic markers, CCR6 and CCR4 (92), which suggests the generation of long-lived anti-Candida memory T cells in humans, possibly due to stimulations from commensal microorganisms. Notably, human Candida-specific memory CD4+ T cells are heterogeneous, produce multiple cytokines, and have unique and shared clonotypes among memory subsets (111). C. albicans-specific TRM cells prevented the fungal overgrowth in human skin and oral mucosa by producing IL-17A (105, 112). Similarly, Candida-specific IL-9-producing CD4+ T cells, Th9, were found enriched in the skin of healthy donors (suggesting their memory phenotype) and have the ability to amplify IFNγ, IL-9, IL-13, and IL-17 by skin-tropic T cells (113). However, gut Th9 cells protect against Candida reinfection and mitigate associated pathology (114). Another Candida-specific subset of CD4+ T cells expressing IL-22 (Th22) is found in humans as memory cells and is increased following infection (115). Further, Th22 seems to provide defense against recurrent vulvovaginitis caused by Candida in humans (116), suggesting the formation of mucosal memory Th22 cells. In this line, defective Th22 responses are associated with chronic mucocutaneous candidiasis (117). Although little is known about the fate of pre-existing antifungal memory T cells during co-infection, a recent study suggests the impaired T-cell responses to Candida following COVID-19 infection that was associated with diminished inflammatory cytokines release (118).
In the mouse models of mucosal candidiasis, studies have shown that Tc17 cells play a role in both oral and vaginal infections (91, 119). Oral immunization of mice with C. albicans under B-cell deficiency induced systemic memory of CD8+ T cells and provided protection following the challenge (120). The screening of Candida-specific memory CD8+ T cells in healthy human blood donors showed a non-classical cytotoxic molecules expression profile, i.e., secretion of granulysin and granzyme K rather than perforin/granzyme B (121). The CD8+ T cells were reactive to C. albicans, Candida glabrata, and Sporothrix and expressed lysosomal degranulation markers, CD107a/b, and secreted IFNγ and TNFα, following ex vivo stimulation with yeast-loaded dendritic cells.
In one study, the various phylogenetically closer and distant yeast-specific T-cell responses were assessed using PBMCs of humans and found a predominant presence of IFNγ-expressing effector memory CD8+ T cells (122). Interestingly, in this study, enriched CD8+ T cells were more reactive to filamentous form than the unicellular form of Candida. In HIV+ patients, the Candida-specific activated memory CD8+ T cells were accumulated within the oropharyngeal candidiasis (OPC) lesions at the lamina propria–epithelium interface (123). Similarly, C. albicans-specific CD8+ T cells were found in the blood and nasal mucosa of chronic rhinosinusitis patients, suggesting a possible persistent T cell-mediated mucosal inflammation (124), which may be due to repeated exposure to the antigen.
Aspergillus
Aspergillus, a globally prevalent opportunistic fungal pathogen, causes pulmonary and invasive mycoses following inhalation of spores. Immunity to aspergillosis is primarily associated with the development of memory Th1 cells, while biased Th2 or regulatory responses are linked to exacerbated disease (125–127). Experimental vaccination of mice with conidia, hyphae, crude culture filtrate antigens, or adjuvanted (CpG ODN1862) cell wall glucanase Crf1 protein strongly induced Th1 (IFNγ and IL-2) responses, formed effector or memory T cells, and conferred immunity following the challenge (87, 128–131). In line with the protective role of Th1 responses, IL-4 seems to play a negative role following A. fumigatus challenge (127). In chronic rhinosinusitis with nasal polyposis patients, the mycology culture showed the presence of Aspergillus flavus, and PBMCs stimulated with aspergillus antigens showed an increased ratio of aspergillus-specific Th17 cells over Tregs, suggesting prior sensitization (132). Further, stimulated PBMC culture supernatant showed elevated levels of IL-17 and IL-10 with reduced TGFβ levels, suggesting the possible associated pathology in these patients. Prior work has shown the negative effect of IL-10 for control of experimental lethal systemic aspergillosis (133), and its overexpression, due to genetic polymorphisms, predisposes to invasive aspergillosis possibly by inhibiting TNFα secretion in hematopoietic stem cell recipients or hematological patients (134, 135). However, IL-10 seems to be protective in regulating exaggerated immune responses and inflammation in allergic bronchopulmonary aspergillosis (136), suggesting that the regulatory role of IL-10 depends on the disease context. In many healthy individuals (10%–30%), multi-Aspergillus-specific T cells were found, suggesting their memory potential and feasibility to expand, store, and be used for self-adoptive transfers following hematopoietic stem cell transplantation (137–139). Hematopoietic stem cell transplant patients undergo a period of immunocompromised state, enhancing vulnerability to many opportunistic fungal infections, including aspergillosis. Thus, rapid preventive or therapeutic reconstitution of the functional adaptive immune system is beneficial. Adoptive immunotherapy using either donor-derived (139) or partially HLA-matched antigen-specific T cells can be used to prevent or treat opportunistic fungal infections (140). In a preclinical study, in vitro expanded yeast-specific cytokine-producing Th cells have been used to reduce the severity of pulmonary and cerebral forms of aspergillus infections in mice (141).
Although the role of Th17 cells in pulmonary aspergillosis is debated, a recent study suggests the presence of aspergillus-specific Th17 cells that correlated with protective immunity (88), and possible mechanisms may include the formation of inducible bronchus-associated lymphoid tissue (iBALT) structures and development of TRM cells (142). With the use of recombinant aspergillus proteins (Asp f proteins), a study investigated the presence of yeast-specific CD4+ and CD8+ T cells in healthy non-atopic donors (143). In these individuals, the cytokine production signature suggested the presence of aspergillus-specific memory T cells expressing IFNγ, IL-17A, and to some extent IL-4, possibly due to prior exposure. Importantly, this study showed the presence of Aspergillus-reactive IFNγ+ T cells for up to 6 months of follow-up observations. However, the persistence of diverse cytokine-expressing T cells as memory in humans needs to be evaluated for their role in immunity or immunopathology following infection.
Pneumocystis
Pneumocystis is another opportunistic fungus that causes infection under immunodeficiency, especially of CD4+ T cells and B cells. It is believed that Pneumocystis may persist in individuals upon early age exposure without any apparent symptoms, akin to toxoplasma. The reactivation occurs when an individual becomes immunodeficient or severely immunosuppressed, suggesting an active role of memory or effector adaptive immunity to keep the fungus at bay. However, recent studies suggest the possibility of reinfection in immunocompromised individuals (144). Murine models have been valuable in understanding the host–Pneumocystis interface for adaptive immunity and recapitulating human primary immune disorders (145). Increasing evidence suggests that B-cell responses are important in the control of Pneumocystis. Anti-CD20 mAb therapy in humans enhanced the susceptibility to pneumocystosis, suggesting a critical role of B cells (146). Of note, the CD20 mAb therapy does not deplete mature plasma cells (147), raising questions on mature long-lived plasma cell generation against Pneumocystis. It is possible that anti-CD20 mAb therapy leads to functional impairment of antibody-secreting cells. However, in a murine model of pneumocystosis, neither the memory CD4+ T cells nor B cells are required for clearance of infection (146). Here, convalescent Pneumocystis-specific IgGs were enough to provide immunity. Interestingly, B cells are required for elicitation of antigen-specific CD4+ T-cell responses to Pneumocystis (148, 149), suggesting a potential cross-talk between these two subsets for immunity against pneumocystosis. Notably, in the simian model of vaccination with Pneumocystis jirovecii protease kexin (KEX1), once the B cells are primed, induction of CD4+ T-cell deficiency with SHIV infection did not prevent antibody-dependent control of infection, suggesting the persistence of “memory” plasma cells or threshold levels of Ab titers (150). In a murine model, memory CD4+ T cells were dispensable for pneumocystosis control, whereas memory CD8+ T cells, alveolar macrophages, and Pneumocystis-specific IgG contributed to secondary immunity (102). Here, the IgG antibody enhanced the macrophage killing of yeast, while macrophages helped CD8+ T-cell recall responses in IFNγ production. IFNγ-stimulated CD8+ T cells, in turn, can be potent antifungal cytotoxic cells (13). However, interestingly, CD8+ T cells seem to help CD4+ T cells with their IFNγ responses. Memory CD4+ T cells can indirectly potentiate NK cell functions against infection caused by Pneumocystis murina, and depletion of CD4+ T cells significantly reduced the accumulation of NK cells and NK-cell mediated immunity (151). In a mouse model of vaccine immunity, immunization with a recombinant fusion protein containing N-terminal 544-aa Pneumocystis cross-reactive antigen-1 and trigger factor (TF) induced protective and cross-reactive antibody responses that provided immunity even after memory CD4+ T cells were depleted at the time of challenge infection (152). Thus, CD4+ T cells seem to have functional duality against Pneumocystis infection, first by helping B cells and CD8+ T cells to become protective differentiated memory cells and second by secreting proinflammatory cytokines to control the primary infection.
Cryptococcus
C. neoformans is a facultative intracellular opportunistic pathogen commonly associated with AIDS patients due to severe CD4+ T-cell deficiency. The CD4+ T-cell immunity to cryptococcosis is mainly dependent on Th1 cytokines. Models of vaccination and infection suggested the role of CD4+ T cells and their Th1 cytokine profile. Cryptococcus-activated CD4+ T cells recruited other immune cells, enhanced the phagocytosis, and killed infected cells by CD8+ T cells, akin to intracellular bacterial infections (153). Striking effects of Th1-derived cytokines for immunity against cryptococcosis are noticed when genetically engineered C. neoformans strain H99 expressing IFNγ (H99-γ) was experimentally used in mice (154). Here, the “vaccinated” mice cleared the infection that was associated with a large influx of leukocytes, enhanced T-cell recruitment, and increased Th1 and decreased Th2-type cytokines following challenge infection. The use of a recombinant strain of C. neoformans (H99-γ) as a vaccine strain induced memory CD8+ T cells to mediate immunity under CD4+ T-cell deficiency, suggesting that antifungal CD8+ T cells can compensate CD4+ T cells (155). Notably, there was the development of memory T cells and enhanced secondary responses following the challenge. C. neoformans chitin deacetylase 2 peptide (Cda2-Pep1) delivered in glucan particle (GP)-based vaccination robustly protected the mice following the challenge, and the immunity was correlated with their MHC-II binding affinity (156). Similarly, multi-epitope vaccine/s may be useful in controlling cryptococcosis (157). In an experimental model, immunization with either cell wall or cytoplasmic protein preparation from Cryptococcus gattii induced vaccine immunity after challenge and protection that was associated with enhanced Th1 responses and antigen-specific serum IgG (158). Nevertheless, in such vaccination platforms, the development of memory T cells is not clear. However, Cryptococcus antigen-pulsed dendritic cell-based systemic vaccination elicited long-lived memory Th17 cells in the lungs (95). Interestingly, these cells were lung resident TRM cells, produced IL-17A but not IFNγ, and mediated protection against C. gattii challenge. Pulmonary infection with C. neoformans elicited strong CD8+ T-cell responses to control the infection independent of CD4+ T cells (15). In this line, immunization with the genetically mutant Cryptococcus strain (Δsgl1) that accumulates steryl glucosides led to induction of protective immunity that required either CD8+ or CD4+ T cells (159). However, these studies did not show memory T-cell development or persistence. In HIV-associated Cryptococcus meningitis patients, the clearance of infection was strongly correlated with Th1, not Th2 or Th17, cytokines (IFNγ or TNFα). In contrast, their defective expression led to higher mortality (160), suggesting an importance of type I immunity. Lower frequency of cytokine-producing memory CD8+ and CD4+ T cells was found in HIV-infected patients with Cryptococcus meningitis (CM), but their numbers were increased with more polyfunctional IL-2+/IL-17+ CD4+ T cells and IL-2+ CD8+ T cells following antiretroviral therapy (ART) in CM-associated immune reconstitution inflammatory syndrome (CM-IRIS) (161) patients, suggesting the pathological role of cryptococcal memory T cells under certain conditions.
Blastomyces
Mouse models of immunity to blastomycosis suggest that CD4+ T cells are essential for controlling primary pulmonary infections. An experimental mouse model of vaccination suggested that Th17 cells expressing IL-17A are the main driver for immunity against pulmonary blastomycosis by activating macrophages and neutrophils (56). A Blastomyces-specific fungal antigen, Calnexin, was found to be conserved among multiple fungal pathogens, and vaccination with Adjuplex adjuvant or encapsulated glucan mannan particles seems to induce robust CD4+ T-cell responses and immunity (162, 163). Identification of such conserved antigens may help the design of pan-fungal vaccines (164). Although homeostasis of memory CD4+ T cells was not studied, the fungal-specific CD4+ T cells persisted for 8 weeks and the adoptive transfer of vaccine-induced effector CD4+ T cells mediated the immunity following the lethal challenge even after 10 weeks of rest (162), suggesting their potential to become memory. Interestingly, intranasal delivery of vaccine-candidate Blastomyces endonuclease-2 (Bl-Eng2) induced TRM cells in the lungs but failed to provide proactive immunity, unlike systemic vaccine-induced migratory CD4+ T cells (165). Importantly, Bl-Eng2 is a glycoprotein antigen that has mannose residues that bind to Dectin-2 and a protein backbone with protective CD4 T-cell epitope/peptide (165, 166), thus a vaccine candidate with intrinsic adjuvanticity property. Admixing adjuvants, especially TLR9 (CpG55.2) and Aldeltin (formulation with alum OH) with fungal antigen (Bl-Eng2 peptide) potentiated the vaccine immunity against blastomycosis, which was dependent on type 1 and type 17 cytokine-producing migratory and lung-resident T cells (167). In the mouse model of vaccination under CD4+ T-cell deficiency, CD8+ T cells could mount sterilizing immunity to lethal pulmonary infection. Immunity was predominantly mediated by IL-17A+ CD8+ (Tc17) cells (90) with disparate dependency on the type I cytokines (by Tc1 cells), IFNγ, TNFα, and GM-CSF (168); i.e., deficiency of one Tc1 cytokine was compensated by other Tc1 cytokines. Notably, antifungal memory CD8+ T cells were long-lasting and persisted stably without plasticity in the absence of vaccine antigen or CD4+ T-cell help (16, 169). These memory CD8+ T-cell precursors portrayed stem cell-like phenotype and can be fine-tuned by MyD88-Akt-mTOR signaling (78). Additionally, targeting the negative regulator of the TCR signaling molecule, Cblb, could enhance memory CD8+ T-cell responses of both Tc1 and Tc17 responses to inactivated vaccine and potentiate immunity following lethal pulmonary challenge (170). Memory Tc17 cells predominantly expressed GM-CSF, and these co-expressing cells potentiated the fungal vaccine immunity without precipitating pathology (171).
Histoplasma
Histoplasma is an opportunistic fungal pathogen and causes disseminated infection in severely immunocompromised patients. Incidentally, most people living in the Ohio-Mississippi river valley endemic regions were reactive to Histoplasma (172), suggesting memory T-cell persistence. Memory CD4+ T cells contributed to immunity following the secondary infection, and depletion of both CD4+ and CD8+ T-cell subsets enhanced the infection and decreased survivability (173). The deficiency of IL-10 conferred salutary effects on memory T cell-mediated protection to secondary histoplasmosis (174). The immunity was dependent on T cell-derived TNFα or IFNγ, and the protection conferred by T cells generated under IL-10 deficiency was robust. Although memory responses of IL-17+ T cells are not clear, induction of their effector type and immunity has been noticed following vaccination or infection with Histoplasma (56, 90, 170, 175). In a model of histoplasmosis, CD8+ T cells could compensate for the loss of CD4+ T cells for vaccine immunity (176), and the depletion of CD8+ T cells compromised the primary immune responses (177). Immunity to histoplasmosis was perforin-dependent and perforin-independent, which included cytokine-mediated (IFNγ or TNα) mechanisms (178). The antigen cross-presentation by dendritic cells seems to be critical for the elicitation of protective antifungal CD8+ T-cell responses (14). Although the above studies do not evaluate memory homeostasis, memory responses to histoplasmosis were bolstered by IL-10 neutralization, where fewer CD8+ T cells were enough to mediate immunity (174).
Coccidioides
Coccidioidomycosis or Valley Fever is caused by species of dimorphic fungus, Coccidioides, a major cause of mycosis endemic to the southwestern United States. Immunity to Valley Fever is primarily associated with T cells expressing both type 1 and type 17 cytokines (Th1/Th17) (179, 180). The regulatory T cells were associated with persistent coccidioidomycosis in the pediatric population that was recapitulated in resistant vs. susceptible mice (181, 182). Cytokine IL-10 plays a negative role in memory Th1 and Th17 recall responses and immunity following Coccidioides infection, but not for the development of memory T cells. Coccidioidomycosis immune donors had polyfunctional T cells composed of both effector and central memory phenotypic cells (183), suggesting their long-term immune role. In a human vaccine study using formaldehyde-killed sperules of Coccidioides immitis, no statistical differences in susceptibility to infection were found between placebo and vaccination groups (184). However, the memory T-cell development or their functions between these groups were not clear. Nevertheless, there is an active attempt to improve the vaccine efficacy using a multivalent vaccine against Coccidioides infection that required mixed Th1 and Th17 cell-mediated immunity (185, 186). In a mouse model of vaccine immunity using a temperature-sensitive, auxotrophic mutant of C. immitis, the adoptive transfers of either CD4+ or CD8+ T cells from vaccinated mice to recipient mice infected with lethal strain conferred protection, and the immunity was mediated through TNFα (97). In this study, although effector CD8+ T cells (2 weeks post-immunization) were used to show protective immunity, the mice were monitored for 50 days following the challenge, suggesting the effector cells’ persistence and possible conversion into memory cells.
Paracoccidioides
Immunity to paracoccidioidomycosis depends on the CD4+ T cells expressing Th1 cytokines, IFNγ, TNFα, and IL-2 (187). The type of T-cell response determines the nature of the disease or the susceptibility, with Th2-dominant responses being non-protective and a mix of Th17/Th22 and Th1/Th2 providing the intermediate protection (188). Experimental vaccination with P10 antigen (of gp43 protein) in Montanide ISA 720, CFA, flagellin, and DODAB adjuvants induced Th1 response and protected against intratracheal challenge infection (189, 190). Here, both effector and central memory phenotypic cells were found to be successfully recalled into the lungs after infection. Similarly, fungal-specific memory CD4+ and CD8+ T cells were seen in the patients, and low numbers of memory CD4+ T cells were associated with relapse of the disease (191). In the model of pulmonary paracoccidioidomycosis, CD8+ T cells were induced to control the fungi in the absence of CD4+ T cells by secretion of type 1 cytokines (IFNγ and IL-2), and the depletion of CD8+ T cells increased the fungal burden with a concomitant increase of non-protective IL-4 and IL-5 cytokines (101). Nitric oxide helps control Paracoccidioides infection, and deficiency of NO and CD8+ T cells seems to be detrimental to immunity. Interestingly, CD8+ T cells enhance the recruitment of TNFα/IFNγ-producing CD4+ T cells and the influx of inflammatory cells (192). However, the maintenance of such a response as memory is unclear.
Cross-reactive antifungal T cells
Cross-reactivity, the recognition of two or more peptides by the same TCR, of T cells has been documented. Cross-reactive antifungal T cells are useful, and identifying the antigens helps in the generation of a pan-fungal T-cell vaccine (193). C. albicans existence as a commensal microbe induces memory CD4+ T cells. Incidentally, these C. albicans-specific cells can cross-react with airborne fungi, like Aspergillus, and exacerbate acute inflammatory lung pathologies (194, 195). Similarly, A. fumigatus antigen-induced memory Th1 cells were cross-reactive to C. albicans (130). T cells specific to A. fumigatus were cross-reacted to induce protective immune responses against Aspergillus and Mucorales sp. infections (138). A. fumigatus-specific T cells, in culture, exhibited cross-reactivity with lysates derived from other fungi, including non-fumigatus Aspergillus, C. albicans, Penicillium spp., and Scedosporium apiospermum but not with Aspergillus terreus, C. glabrata, Fusarium spp., and Mucor spp. (196). Although such distantly related fungi have cross-reactive T cells, high cross-reactivity of T cells was noticed between phylogenetically related Scedosporium and Lomentospora species, but not with A. fumigatus (197). The cross-reactivity by adaptive immunity across kingdoms, i.e., fungus and bacteria, is noticed and exploited for vaccination against Candida and Staphylococcus aureus (198), including emerging multidrug-resistant Candida auris infection (199). Similarly, pan-fungal vaccines can be developed and used against different pathogenic fungi if the shared antigen is identified (163). Although the mechanism of cross-reactivity of fungal T cells is mainly due to shared epitope sequence (163), the broad cross-reactivity of the CD4+ T cells may be due to the nature of TCR binding with peptide-MHC class II that allows multiple anchor residues with greater flexibility of amino acid variation (200, 201). The bystander activation via TCR-independent mechanisms (202) may also be involved.
Mechanisms of T cell-mediated fungal control
T cell-mediated control of fungal infections involves mechanisms that are chiefly mediated through the effectual functions of the cytokines they secrete. Additionally, cytotoxic functions, independent of cytokines, of CD8+ T have been documented during fungal infections. Here we highlight some of the actions of cytokines/cytotoxic molecules for fungal control.
IL-17A and GM-CSF cytokines at the site of infection protected against fungal pathogens in part by enhancing the neutrophil recruitment to the site of infection (9). Further, these cytokines can activate recruited neutrophils and macrophages to bolster their ability to kill fungal cells. Activated neutrophils and macrophages may exert direct fungicidal activity via phagocytosis, degranulation, ROS production, and neutrophil extracellular trap (NET) formation (203). Different granules (primary azurophilic, secondary specific, and tertiary gelatinase) in neutrophils contain different cytolytic molecules (17). The role of the IL-17 axis and neutrophils during dimorphic fungal infections has been reviewed elsewhere (204). IL-17 and IL-22 act on cells to promote STAT3 activation, upregulate Reg proteins, and secrete antimicrobial peptides (AMPs), S100 proteins, and β-defensins from epithelial cells and keratinocytes that destroy the fungal pathogen (205). Further, IL-22 signaling helps in the regeneration of oral epithelial cells and “licenses” IL-17 signaling for resistance against oral mucosal candidiasis (206). AMPs secreted by epithelial cells induce cell wall permeabilization, mitochondrial dysfunction, and osmotic dysregulation in fungi to elicit fungicidal and fungistatic activity (207). Conversely, IL-17A may drive allergic outcomes by enhancing eosinophil recruitment following repeated exposure to A. fumigatus conidia (208).
IFNγ, GM-CSF, and TNFα enhance macrophage functions by promoting phagocyte maturation, polarization of macrophages to M1 type, and fungus-killing ability. IFNγ is known to strongly activate phagocytes and their functions against fungi (209). Both IFNγ and TNF induce ROS production from macrophages, which is fungistatic to intracellular fungal pathogens Histoplasma capsulatum and C. immitis (210, 211). Furthermore, IFNγ promotes rapid acidification of phagolysosomes in macrophages (212), upregulation of MHC-II molecules, and antigen presentation by APCs to elicit T-cell immunity (213). GM-CSF enhances macrophage ROS production and limits intracellular yeast growth by sequestration of zinc (214). GM-CSF deficiency impairs the production of TNFα and IFNγ (215), which are important for the control of intracellular fungal infections. Similarly, TNFα signaling enhances the activation, phagocytosis, and ROS-producing ability of innate immune cells for antifungal functions. Interestingly, early TNFα expression during C. neoformans infection increased the fungal burden, reduced mature dendritic cells, and increased Th2 responses (216). TNFα was needed for the maturation and recruitment of DC and the production of IL-12 and IFNγ. However, TNFα prevented biofilm development by C. albicans (217), and TNF blockade can enhance opportunistic infections.
Cytotoxic functions of T cells are well documented for CD8+ T cells. Antifungal CD8+ T cells deploy several antimicrobial granules (mainly granulysin and granzyme K) (121) for direct cytotoxic activity against the fungal pathogen or fungus-infected cells (218, 219). Granzymes can mediate cell death by induction of active caspases, generation of ROS, and mitochondrial damage, while perforin may facilitate their release from the endosomes (220). Although this review focuses on T cells and their memory, antifungal NK cell development is not well documented. NK cells, like CD8+ T cells, can produce cytotoxic molecules to kill fungi. The binding of NK cell receptors NKp46 and NCR1 to surface glycans of Candida led to degranulation and death of the yeast (221). Similar observations were made with human NK activating receptor NKp30 for the direct killing of Cryptococcus and Candida (222). Interestingly, “cytotoxic” CD4+ T cells can also produce granulysin to mediate the killing of C. neoformans (223), and this function was dysregulated in HIV-infected patients.
CD4+ T-cell help for antifungal CD8+ T cells
Most fungal infections are caused by opportunistic fungal pathogens under compromised CD4+ T cells or their functions. Unlike viral or bacterial models, effector and memory CD8+ T-cell homeostasis in fungal infections is poorly understood, and the studies have been done using animal models of fungal vaccine immunity. Attempts have been made to understand the role of antifungal CD8+ T cells in the absence of CD4+ T cells, a potential avenue to exploit residual immunity for preventive and therapeutic purposes for individuals with CD4+ T-cell lymphopenia. Because optimal programming leads to the generation of long-lasting memory CD8+ T cells (224, 225) and contributes to fungal vaccine immunity (8), we reason that a potent fungal vaccination can “license” dendritic cells for CD8+ T-cell priming independent of CD4+ T-cell help. Fungi are decorated, including some secretory, with several potential PAMPs that can bolster the dendritic cell activation and functions (226–228). Therefore, the CD8+ T-cell memory imprinting and memory homeostasis following fungal vaccination can be independent of CD4+ T-cell help (16, 169). However, evidence from viral and bacterial infections suggests that CD4+ T-cell help is essential for eliciting CD8+ T-cell responses (229). The mechanisms of CD4+ T cells help CD8+ T cells involve optimal activation of dendritic cells, enhancement of their phagocytosis, potentiation of antigen processing and presentation, upregulation of costimulatory ligands, and generation of an apt inflammatory micro milieu by dendritic cells for naïve CD8+ T-cell priming and programming (230, 231). The direct mechanisms of CD4+ T cells help involve providing IL-2 for the proliferation of differentiating CD8+ T cells and lending co-stimulation through CD40L-CD40 (7). Further, CD4+ T cells may help recruit memory CD8+ T cells into mucosal surfaces (232). Well-primed CD8+ T cells can produce IL-2 (169) and get help in an autocrine manner (233). We also found chemokine receptor-mediated recruitment of effector CD8+ T cells into the lungs, which was independent of CD4+ T cells following pulmonary challenge in a mouse model of vaccine immunity to pulmonary blastomycosis (90). Recent evidence suggests that the requirement of help from CD4+ T cells to CD8+ T cells largely depends on their distinct interactions with the dendritic cells during their activation (230, 234). Thus, well-programmed CD8+ T cells, endowed with intrinsic memory capacity, can independently bestow their recall responses and immunity (235, 236). Further studies are needed to understand the fungal antigens and the role of dendritic cells in CD8+ T-cell priming.
Memory T cells: An overview
The development of immunological memory is the hallmark of vaccination. Following infection or vaccination, the first responders, innate immune cells, exert broadly specific immunity to limit pathogen growth and initiate adaptive T-cell immunity. T-cell recognition is highly antigen-specific and binds to processed antigens/epitopes loaded onto MHC molecules, resulting in their activation, differentiation, and proliferation during the first phase of T-cell response, which is the expansion phase (237). In the ensuing contraction phase, mostly coinciding with the elimination of pathogen or antigen, ~90%–95% of effector T cells [short-lived effector cells (SLECs)] undergo attrition by apoptosis, and the remaining 5%–10% of cells [memory precursor effector cells (MPECs)] differentiate to become long-lived, quickly and robustly responding, memory cells (238) in the memory phase (Figure 1). The central paradigm of vaccination is to generate qualitatively superior threshold numbers of memory T cells (237, 239, 240).
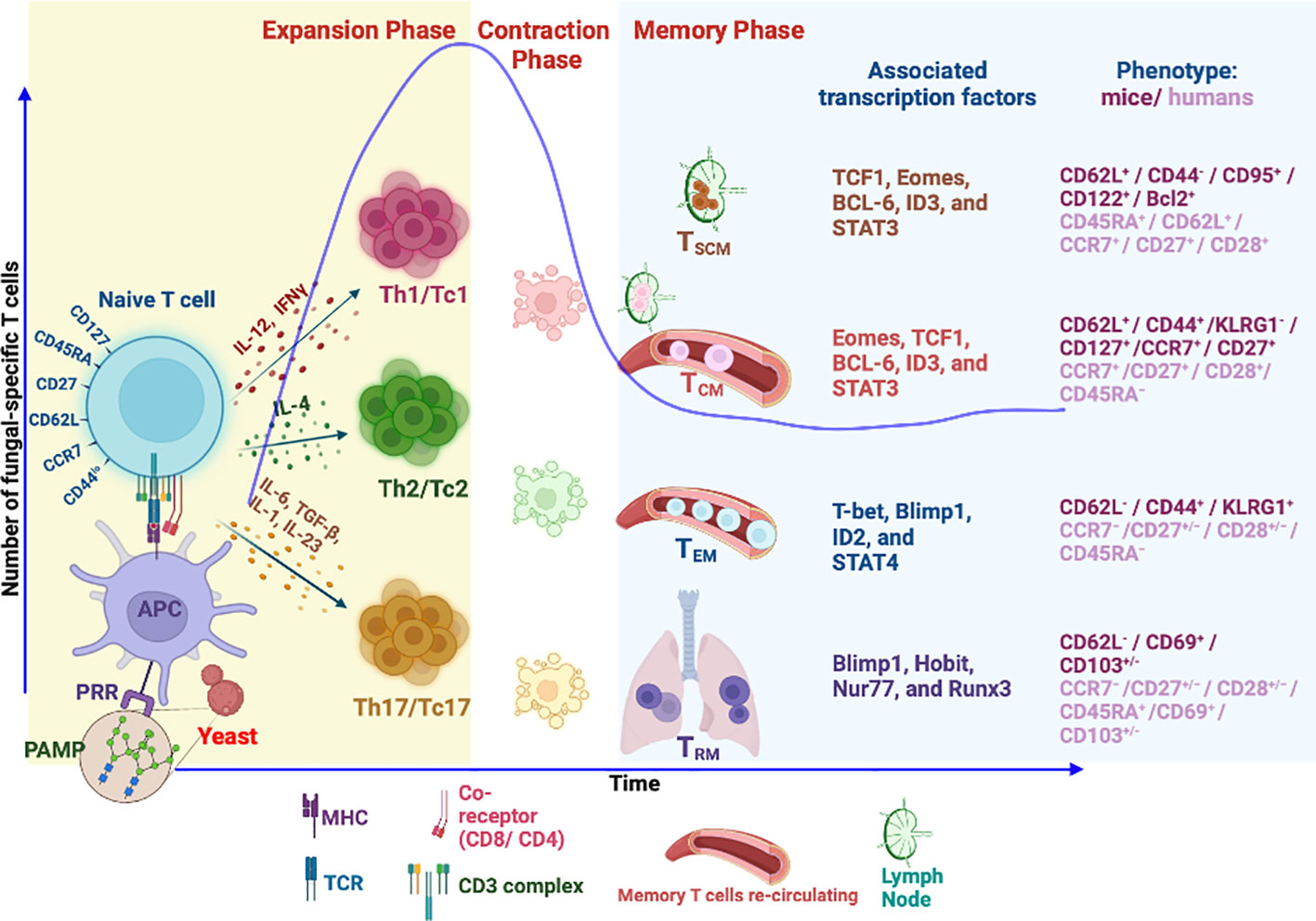
Figure 1 Differentiation and generation of memory T-cell subsets. Following recognition of fungal PAMPs by PRRs, activated antigen-presenting cells (APCs) process the antigen from phagocytosed fungus to load onto MHC molecule. MHC–peptide complex is recognized by cognate TCR of naïve T cells leading to TCR signaling, activation, and differentiation of T cells into different subsets directed by different cytokine milieus. The differentiated T cells accompanied by proliferation during expansion phase secrete inflammatory cytokines to aid in fungal killing. Fungal clearance usually coincides with initiation of T-cell contraction phase where 90% of effector cells die by apoptosis. The remaining cells differentiate to become long-lived memory cells. Memory T cells express unique phenotypic attributes and transcription factors, some of which dictate homing to lymphoid organs (CCR7/CD62L). Tissue-resident memory T cells (TRM) continue to reside in tissue of responses. The effector memory T cells (TEM) continue to invigilate the pathogen by recirculation between peripheral tissues and blood. Self-renewing central memory (TCM) and central memory stem T cells (TSCM) are preferentially home to secondary lymphoid organs and serve as “seeders” of secondary effector and memory T cells when needed. PAMPs, pathogen-associated molecular patterns; PRR, pattern recognition receptors; Th, helper T cell; Tc, cytotoxic T cell (CD8+ T cell); TSCM, stem memory T cells; TCM, central memory T cell; TEM, effector memory T cell.
Memory T cells are unique and often behave like stem cells in their homeostasis and longevity. Many models of memory T-cell generation are proposed, and one of them suggests that memory programming or imprinting can happen as early as the first antigen encounter (241, 242). Others suggested dynamic and progressive imprinting of memory from a subset of effector cells. In the absence of cognate antigen, memory T cells are quiescent and slow dividing and share many features with naïve T cells (243, 244). However, they are uniquely programmed for stem cell potential and balanced cell apoptosis and proliferation for their steady homeostasis (240, 245), chiefly controlled by cytokines IL-7 and IL-15 (246). Naïve, effector, and memory T cells are differentiated based on their expression of surface and intracellular markers (241, 247–249), which endorse their homing and functional attributes. Memory T cells are a heterogeneous pool of cells derived from multiple clones and fates of polarization and differentiation based on the “one cell multiple fate theory” (250, 251). Hence, these features are imprinted in memory T-cell heterogeneity in homeostatic turnover, effector function, location, and trafficking properties (111, 248, 252).
Classically, memory T cells were divided into two groups, TEM and TCM, based on their lymph node homing properties with distinct proliferative renewal and functional properties (240, 241). With the advancement of our understanding and discovery of new markers, memory cells are classified into many groups in both humans and mice. Based on the markers, memory cells are broadly classified as central memory T (TCM), effector memory T (TEM), tissue-resident memory T (TRM), and stem cell memory T (TSCM) cells (241, 253–255). Effector memory T cells (TEM), CD45RA−CD127+CD122+CD27−/+CD62L−/CCR7−, can migrate into peripheral tissues and exhibit immediate higher effector function with limited proliferation potential during the reinfection. TCM cells, CD45RA−CD127+CD122+CD27+CD62L+/CCR7+, are housed in secondary lymphoid organs with high proliferative potential but weaker effector properties as compared to TEM cells (255, 256). The dogma is that TCM cells are reservoirs or seeders of TEM cells in need. The property of CD62Llo/CCR7lo defines the TEM cell exclusion from lymph nodes and migration to peripheral tissues to rapidly exude effector molecules upon antigen encounter. In contrast, TCM cells are CD62Lhi/CCR7hi, allowing them to be preferentially home to lymph nodes (257). Both types of memory cells endow memory homeostasis properties where TCM mediates secondary immune responses for long-term protection and TEM offers instant protection (258). Nevertheless, as alluded earlier, the homeostatic proliferative potential of TEM and TCM cells varies (259).
A newly emerging T-cell memory subset, tissue-resident memory (TRM), is restricted to a particular tissue and is identified based on the expression of CD103+/−, CD49a, and CD69 (260). The main feature of TRM is their restriction to the tissue (261), possibly due to the expression of CD69 that suppresses the S1P receptor function for lymph node homing (262). Further, TGFβ signaling in these T cells augments the CD103 expression that facilitates latching onto epithelial cells (263, 264). Given the strategic position, TRM cells offer immediate local tissue immunity against invading pathogens (265).
The memory stem T cell (TSCM) subset, unique and often considered closest to naïve T cells, exhibits stem cell-like properties with higher self-renewal capacity. Unlike conventional memory T-cell subsets, the TSCM subset maintains a naïve cell phenotype with multipotency and serves as a reservoir of memory T cells for a lifetime (266, 267). However, the identity of such unique memory T cells that are fungal-specific is lacking despite the description of antifungal memory cells in humans (111). Further studies are required to reveal the existence of antifungal TSCM in mice and humans.
With our understanding of memory T-cell differentiation and function, the memory cells can be classified in many ways, may depend on the infection or model system, and may be largely due to graded responses during the early programming of effector cells (254, 268–270). Unlike an acute viral infection, in chronic viral infection, the central memory phenotypic T cells poorly develop (271), and high antigen levels induce an exhausted phenotype (272). It should be noted that antifungal memory T-cell homeostasis is poorly defined and not well understood.
Factors influencing generation of antifungal memory T cells
Our understanding of memory T-cell generation suggests that their attributes are bestowed by early programming during the expansion phase. However, the inflammatory milieu and the antigen persistence can affect their fate. The graded imprinting and epigenetic changes during the effector phase determine the memory T cell fate and the type. Naïve T cells must recognize cognate antigens portrayed on MHC molecules (Signal 1) of dendritic cells. Studies have shown that degree of antigen recognition by naïve or effector cells and antigen persistence (chronic and persistent infections) has a greater impact (273–275). However, this feature can be valuable during vaccine formulations where the antigen is delivered to the site of T-cell activation gradually to enhance the magnitude of their differentiation and expansion. Not surprisingly, booster doses are often given to augment adaptive immune responses, especially with subunit or inactivated vaccines. The costimulatory signal/s (Signal 2) is essential to break the TCR signaling threshold or tolerance to activate the T cells. The role of the classical costimulatory molecule, CD28, in recognizing B7 ligands on antigen-presenting cells is well defined, including during fungal vaccine immunity (276). However, other costimulatory molecules may influence memory T cells’ qualitative and quantitative traits (277), including fungal immunity (278–282). Interestingly, CD28 may play a negative role in Th17 subset differentiation (283), but in the absence of CD28, the differentiation required proinflammatory cytokine signaling (284). Nevertheless, for subunit or less potent vaccine antigen formulations, engaging multiple costimulatory molecules may help potentiate the T-cell responses and eventual memory formation (167, 226). The inflammatory milieu generated by proinflammatory cytokines (Signal 3) is instrumental for effector T-cell differentiation and memory feature imprinting (285). Immunity to different fungal infections needs distinct cytokines produced by T-cell subsets (4), and the deviation from protective T-cell subsets may lead to enhanced pathology and disseminated infections (286–289). Immunity to different fungal pathogens predominantly requires either T cell-derived type I cytokines (IFNγ, GM-CSF, and TNFα) or type 17 (IL-17A/F) responses (80, 290) mainly at systemic and mucosal surfaces, respectively. Nevertheless, following fungal vaccination or infections, both types of subsets are induced at different magnitudes and found to contribute to immunity at variable degrees (8, 22, 168, 291–294). Future studies are necessary to understand the elements of memory T-cell differentiation, homeostasis, and their recall responses for immunity following infection or vaccination.
Co-stimulatory and coinhibitory molecules influencing antifungal potential memory T cells
Co-stimulatory and coinhibitory receptor molecules present in T cells can fine-tune immune responses to fungal vaccines and infections. While co-stimulation leads to the potentiation of cell signaling, inhibitory signals deliver opposite effects during T-cell activation, thus inhibiting T-cell responses (295). Similarly, coinhibitory receptors present on memory T cells restrict recall responses but preserve memory cells by inhibiting terminal differentiation (296). Interestingly, the expression of coinhibitory molecules on resident memory CD8+ T cells (Trm) is an intrinsic property present in their core gene signature (297, 298). Compared to circulatory memory T cells, resident memory T cells expressing high amounts of coinhibitory receptors (2B4, CTLA-4, LAG3, PD-1, and Tim-3) were able to undergo local proliferation following secondary rechallenge (299). Here, we highlight a few studies where the co-stimulatory or inhibitory molecules modulate antifungal T-cell responses.
PD-1 is a coinhibitory molecule expressed in T cells associated with dysfunction, and blocking PD-1 enhances the T-cell functions. Hence, anti-PD-1 mAb administration improved the fungal clearance in a model of persistent cryptococcosis (297). Interestingly, the effect was independent of effector cell numbers and myeloid cell activation, but reduced expression of IL-5 and IL-10 by lung leukocytes and enhanced sustained expression of OX40, a costimulatory molecule, on T cells. Similarly, the blockade of PD-1 and CTLA-4 improves survival during primary and secondary fungal sepsis (300) associated with improved T-cell functions. Signaling lymphocyte activation molecule (SLAM) family members act as co-receptors for T cells fine-tuning immune homeostasis during infections (301). Mutation in SLAM-associated protein (SAP), required for SLAM signaling, results in X-linked lymphoproliferative disease (XLP). A recent study showed that SLAMF1, a member of the SLAM family, was dispensable for T-cell activation and expansion following fungal vaccination, but the fungal immunity was severely compromised (302). This study of vaccine immunity against lethal fungal pneumonia implied that SLAMF1 is mainly important for innate host control of lung fungal overgrowth as well as inflammation, recruitment, or expansion of fungal-specific effector CD4+ T cells. Another molecule, CD43, also called sialophorin, is a membrane-bound receptor that exists in two forms with one highly glycosylated on T cells (303, 304). CD43 can be co-stimulatory and inhibitory, and depending on the context, it is known to promote T-cell contraction and reduced memory (305). In the context of cell-to-cell interactions, studies showed that CD43 deficiency led to enhanced homotypic binding of T cells through ligands such as ICAM-1 and fibronectin while augmenting the T-cell proliferation (306, 307). In other studies, pre-activation of CD43 with a mAb reduced the TCR signaling threshold, enhanced the degradation of Cbl, prolonged the TCR signaling, and augmented the T-cell response (308). Our recent study showed the indispensable role of CD43 for Tc17 responses and vaccine immunity to pulmonary fungal infection (309).
T cells typically express one or more co-stimulatory receptors. Thus, understanding the role of their signaling helps design vaccines to bolster qualitatively superior antifungal memory T cells and potentiate their functions for immunotherapeutics. Further, formulations of vaccines for fungal infections should account for the type of T-cell responses desired. Nevertheless, additional studies are warranted to delineate the role of co-stimulatory molecules during fungal infections. Adjuvants in vaccines act to enhance the T-cell stimulatory signals and proinflammatory cytokine production that polarize the T-cell responses (310). Different adjuvants have different characteristics in biasing T-cell response/s. For example, Alum potentiates Th2 responses, while Monophosphoryl lipid A (MPL)/CpG1018 bolsters Th1 responses. For controlling some fungal infections, especially those that are dependent on IL-17 responses, novel adjuvants are necessary.
Conclusions and future directions
Immunological memory of fungal infections is poorly defined, but their existence and longevity are documented in preclinical and clinical studies. Most studies on T-cell memory came from preclinical model systems of fungal vaccine immunity. It is increasingly evident that T cells play a dominant role in fungal immunity, although other immune elements, including B cells and antibodies, cannot be overlooked. Although the innate cell inflammatory micro milieu is key for defining T-cell lineages and their functions for fungal immunity, the direct costimulatory signals or blockade of inhibitory signals delivered to T cells can regulate their effectors and memory cells. Non-canonical T cells such as natural T helper, MAIT, and NKT cell’s role in some fungal infections have been documented, but their persistence as memory cells is not clear (32). Harnessing non-canonical T cells for vaccine immunity can be a new avenue for controlling fungal infections.
The mechanisms of the generation of antifungal memory T cells are not well understood and need in-depth investigations. The studies will be particularly relevant for the development and application of vaccine platforms. As with many different bacterial and viral defense mechanisms, the protective T-cell effectors vary depending on the pathogenic fungi, so the memory T-cell development mechanisms. Further, fungal co-infections and the disease outcomes need thorough studies, such as mucormycosis in COVID-19 patients. Novel findings on the use of fungal PAMPs as adjuvants for vaccines call for an understanding of adjuvanticity properties and their role in the programming of immunological memory. The new concept-based emergence of chimeric antigen receptor T (CAR-T) cells for immunotherapy to treat fungal infections is attractive, and its efficiency or utility needs attention. The plasticity of antifungal T cells has not been clearly understood, and its bases for immunopathology during recall responses and immunity need further evaluation. Further, the identification of phenotypic and functional markers of protective immunological memory T cells would be useful in designing and assessing the potency and efficacy of fungal vaccines.
Author contributions
JS, SM, and SN conceived, wrote, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by NIAID-NIH 5R01AI153522 (SN).
Acknowledgment
Wee used biorender 2022 for genereation of a figure.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Brown GD, Denning DW, Gow N, Levitz SM, Netea MG, White TC. Hidden killers: Human fungal infections. Sci Transl Med (2012) 4:165rv113–165rv113. doi: 10.1126/scitranslmed.3004404
2. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel) (2017) 3(4):57. doi: 10.3390/jof3040057
3. Kainz K, Bauer MA, Madeo F, Carmona-Gutierrez D. Fungal infections in humans: the silent crisis. Microb Cell (2020) 7:143–5. doi: 10.15698/mic2020.06.718
4. Verma A, Wuthrich M, Deepe G, Klein B. Adaptive immunity to fungi. Cold Spring Harb Perspect Med (2014) 5:a019612. doi: 10.1101/cshperspect.a019612.
5. Wuthrich M, Deepe GS Jr., Klein B. Adaptive immunity to fungi. Annu Rev Immunol (2012) 30:115–48. doi: 10.1146/annurev-immunol-020711-074958
6. Lionakis MS, Netea MG, Holland SM. Mendelian genetics of human susceptibility to fungal infection. Cold Spring Harb Perspect Med (2014) 4(6):a019638. doi: 10.1101/cshperspect.a019638.
7. Castellino F, Germain RN. Cooperation between CD4+ and CD8+ T cells: When, where, and how. Annu Rev Immunol (2006) 24:519–40. doi: 10.1146/annurev.immunol.23.021704.115825
8. Nanjappa SG, Klein BS. Vaccine immunity against fungal infections. Curr Opin Immunol (2014) 28:27–33. doi: 10.1016/j.coi.2014.01.014
9. Conti HR, Gaffen SL. IL-17-Mediated immunity to the opportunistic fungal pathogen candida albicans. J Immunol (2015) 195:780–8. doi: 10.4049/jimmunol.1500909
10. Lionakis MS, Iliev ID, Hohl TM. Immunity against fungi. JCI Insight (2017) 2(11):e93156. doi: 10.1172/jci.insight.93156
11. Van De Veerdonk FL, Netea MG. T-Cell subsets and antifungal host defenses. Curr Fungal Infect Rep (2010) 4:238–43. doi: 10.1007/s12281-010-0034-6
12. Bacher P, Kniemeyer O, Schonbrunn A, Sawitzki B, Assenmacher M, Rietschel E, et al. Antigen-specific expansion of human regulatory T cells as a major tolerance mechanism against mucosal fungi. Mucosal Immunol (2014) 7:916–28. doi: 10.1038/mi.2013.107
13. Mcallister F, Steele C, Zheng M, Young E, Shellito JE, Marrero L, et al. T Cytotoxic-1 CD8+ T cells are effector cells against pneumocystis in mice. J Immunol (2004) 172:1132–8. doi: 10.4049/jimmunol.172.2.1132
14. Lin JS, Yang CW, Wang DW, Wu-Hsieh BA. Dendritic cells cross-present exogenous fungal antigens to stimulate a protective CD8 T cell response in infection by histoplasma capsulatum. J Immunol (2005) 174:6282–91. doi: 10.4049/jimmunol.174.10.6282
15. Lindell DM, Moore TA, Mcdonald RA, Toews GB, Huffnagle GB. Generation of antifungal effector CD8+ T cells in the absence of CD4+ T cells during cryptococcus neoformans infection. J Immunol (2005) 174:7920–8. doi: 10.4049/jimmunol.174.12.7920
16. Nanjappa SG, Mcdermott AJ, Fites JS, Galles K, Wuthrich M, Deepe GS Jr., et al. Antifungal Tc17 cells are durable and stable, persisting as long-lasting vaccine memory without plasticity towards IFNgamma cells. PloS Pathog (2017) 13:e1006356. doi: 10.1371/journal.ppat.1006356
17. Mok AC, Mody CH, Li SS. Immune cell degranulation in fungal host defence. J Fungi (2021) 7:484. doi: 10.3390/jof7060484
18. Cutler JE, Deepe GS Jr., Klein BS. Advances in combating fungal diseases: Vaccines on the threshold. Nat Rev Microbiol (2007) 5:13–28. doi: 10.1038/nrmicro1537
19. Jaeger M, Stappers MH, Joosten LA, Gyssens IC, Netea MG. Genetic variation in pattern recognition receptors: Functional consequences and susceptibility to infectious disease. Future Microbiol (2015) 10:989–1008. doi: 10.2217/fmb.15.37
20. Merkhofer RM, Klein BS. Advances in understanding human genetic variations that influence innate immunity to fungi. Front Cell Infect Microbiol (2020) 10:69. doi: 10.3389/fcimb.2020.00069
21. Naik B, Ahmed SMQ, Laha S, Das SP. Genetic susceptibility to fungal infections and links to human ancestry. Front Genet (2021) 12:709315. doi: 10.3389/fgene.2021.709315
22. Verma A, Wüthrich M, Deepe G, Klein B. Adaptive immunity to fungi Cold Spring Harb Perspect Med (2015) 5(3):a019612. doi: 10.1101/cshperspect.a019612
23. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature (1998) 392:245–52. doi: 10.1038/32588
24. Steinman RM. Dendritic cells and the control of immunity: Enhancing the efficiency of antigen presentation. Mt Sinai J Med (2001) 68:160–6.
25. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140:805–20. doi: 10.1016/j.cell.2010.01.022
26. Plato A, Hardison SE, Brown GD. Pattern recognition receptors in antifungal immunity. Semin Immunopathol (2015) 37:97–106. doi: 10.1007/s00281-014-0462-4
27. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol (2019) 89:24–33. doi: 10.1016/j.semcdb.2018.03.003
28. Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol (2020) 20:95–112. doi: 10.1038/s41577-019-0215-7
29. Brown GD. Innate antifungal immunity: The key role of phagocytes. Annu Rev Immunol (2011) 29:1–21. doi: 10.1146/annurev-immunol-030409-101229
30. Lionakis MS, Levitz SM. Host control of fungal infections: Lessons from basic studies and human cohorts. Annu Rev Immunol (2018) 36:157–91. doi: 10.1146/annurev-immunol-042617-053318
31. Salazar F, Brown GD. Antifungal innate immunity: A perspective from the last 10 years. J Innate Immun (2018) 10:373–97. doi: 10.1159/000488539
32. Dunne MR, Wagener J, Loeffler J, Doherty DG, Rogers TR. Unconventional T cells - new players in antifungal immunity. Clin Immunol (2021) 227:108734. doi: 10.1016/j.clim.2021.108734
33. Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of aspergillus fumigatus. PloS Pathog (2005) 1:0323–34. doi: 10.1371/journal.ppat.0010042
34. Werner JL, Metz AE, Horn D, Schoeb TR, Hewitt MM, Schwiebert LM, et al. Requisite role for the dectin-1 β-glucan receptor in pulmonary defense against aspergillus fumigatus. J Immunol (2009) 182:4938–46. doi: 10.4049/jimmunol.0804250
35. Drummond RA, Dambuza IM, Vautier S, Taylor JA, Reid DM, Bain CC, et al. CD4+ T-cell survival in the GI tract requires dectin-1 during fungal infection. Mucosal Immunol (2015) 9:492–502. doi: 10.1038/mi.2015.79
36. Marakalala MJ, Vautier S, Potrykus J, Walker LA, Shepardson KM, Hopke A, et al. Differential adaptation of candida albicans in vivo modulates immune recognition by dectin-1. PloS Pathog (2013) 9:e1003315. doi: 10.1371/journal.ppat.1003315
37. Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U.S.A. (2007) 104:1366–70. doi: 10.1073/pnas.0609848104
38. Davis SE, Hopke A, Minkin SC Jr., Montedonico AE, Wheeler RT, Reynolds TB. Masking of beta(1-3)-glucan in the cell wall of candida albicans from detection by innate immune cells depends on phosphatidylserine. Infect Immun (2014) 82:4405–13. doi: 10.1128/IAI.01612-14
39. Hopke A, Brown AJP, Hall RA, Wheeler RT. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol (2018) 26:284–95. doi: 10.1016/j.tim.2018.01.007
40. Yang M, Solis NV, Marshall M, Garleb R, Zhou T, Wang D, et al. Control of beta-glucan exposure by the endo-1,3-glucanase Eng1 in candida albicans modulates virulence. PloS Pathog (2022) 18:e1010192. doi: 10.1371/journal.ppat.1010192
41. Nakamura K, Kinjo T, Saijo S, Miyazato A, Adachi Y, Ohno N, et al. Dectin-1 is not required for the host defense to cryptococcus neoformans. Microbiol Immunol (2007) 51:1115–9. doi: 10.1111/j.1348-0421.2007.tb04007.x
42. Saijo S, Fujikado N, Furuta T, Chung S-H, Kotaki H, Seki K, et al. Dectin-1 is required for host defense against pneumocystis carinii but not against candida albicans. Nat Immunol (2007) 8:39–46. doi: 10.1038/ni1425
43. Vautier S, Drummond RA, Redelinghuys P, Murray GI, Maccallum DM, Brown GD. Dectin-1 is not required for controlling candida albicans colonization of the gastrointestinal tract. Infect Immun (2012) 80:4216–22. doi: 10.1128/IAI.00559-12
44. Wang H, Lebert V, Hung CY, Galles K, Saijo S, Lin X, et al. C-type lectin receptors differentially induce th17 cells and vaccine immunity to the endemic mycosis of north America. J Immunol (2014) 192:1107–19. doi: 10.4049/jimmunol.1302314
45. Thompson A, Da Fonseca DM, Walker L, Griffiths JS, Taylor PR, Gow N, et al. Dependence on mincle and dectin-2 varies with multiple candida species during systemic infection. Front Microbiol (2021) 12:633229. doi: 10.3389/fmicb.2021.633229
46. Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, et al. Dectin-2 is a pattern recognition receptor for fungi that couples with the fc receptor gamma chain to induce innate immune responses. J Biol Chem (2006) 281:38854–66. doi: 10.1074/jbc.M606542200
47. Graham LM, Brown GD. The dectin-2 family of c-type lectins in immunity and homeostasis. Cytokine (2009) 48:148–55. doi: 10.1016/j.cyto.2009.07.010
48. Wang H, Lee TJ, Fites SJ, Merkhofer R, Zarnowski R, Brandhorst T, et al. Ligation of dectin-2 with a novel microbial ligand promotes adjuvant activity for vaccination. PloS Pathog (2017) 13:e1006568. doi: 10.1371/journal.ppat.1006568
49. Loures FV, Rohm M, Lee CK, Santos E, Wang JP, Specht CA, et al. Recognition of aspergillus fumigatus hyphae by human plasmacytoid dendritic cells is mediated by dectin-2 and results in formation of extracellular traps. PloS Pathog (2015) 11:e1004643. doi: 10.1371/journal.ppat.1004643
50. Wuthrich M, Wang H, Li M, Lerksuthirat T, Hardison SE, Brown GD, et al. Fonsecaea pedrosoi-induced Th17-cell differentiation in mice is fostered by dectin-2 and suppressed by mincle recognition. Eur J Immunol (2015) 45:2542–52. doi: 10.1002/eji.201545591
51. Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against candida albicans. Immunity (2010) 32:681–91. doi: 10.1016/j.immuni.2010.05.001
52. Ifrim DC, Quintin J, Courjol F, Verschueren I, Van Krieken JH, Koentgen F, et al. The role of dectin-2 for host defense against disseminated candidiasis. J Interferon Cytokine Res (2016) 36:267–76. doi: 10.1089/jir.2015.0040
53. Saijo S, Iwakura Y. Dectin-1 and dectin-2 in innate immunity against fungi. Int Immunol (2011) 23:467–72. doi: 10.1093/intimm/dxr046
54. Bellocchio S, Montagnoli C, Bozza S, Gaziano R, Rossi G, Mambula SS, et al. The contribution of the toll-like/IL-1 receptor superfamily to innate and adaptive immunity to fungal pathogens in vivo. J Immunol (2004) 172:3059–69. doi: 10.4049/jimmunol.172.5.3059
55. Calich VLG, Pina A, Felonato M, Bernardino S, Costa TA, Loures FV. Toll-like receptors and fungal infections: the role of TLR2, TLR4 and MyD88 in paracoccidioidomycosis. FEMS Immunol Med Microbiol (2008) 53:1–7. doi: 10.1111/j.1574-695X.2008.00378.x
56. Wuthrich M, Gern B, Hung CY, Ersland K, Rocco N, Pick-Jacobs J, et al. Vaccine-induced protection against 3 systemic mycoses endemic to north America requires Th17 cells in mice. J Clin Invest (2011) 121:554–68. doi: 10.1172/JCI43984
57. Prieto D, Carpena N, Maneu V, Gil ML, Pla J, Gozalbo D. TLR2 modulates gut colonization and dissemination of candida albicans in a murine model. Microbes Infect (2016) 18:656–60. doi: 10.1016/j.micinf.2016.05.005
58. Miró MS, Rodríguez E, Vigezzi C, Icely PA, García LN, Painetti N, et al. Contribution of TLR2 pathway in the pathogenesis of vulvovaginal candidiasis. Pathog Dis (2017) 75(7). doi: 10.1093/femspd/ftx096
59. Netea MG, Sutmuller R, Hermann C, van der Graaf CA, van der Meer JW, Van Krieken JH, et al. Toll-like receptor 2 suppresses immunity against candida albicans through induction of IL-10 and regulatory T cells. J Immunol (2004) 172:3712–8. doi: 10.4049/jimmunol.172.6.3712
60. Villamon E, Gozalbo D, Roig P, O’connor JE, Fradelizi D, Gil ML. Toll-like receptor-2 is essential in murine defenses against candida albicans infections. Microbes Infect (2004) 6:1–7. doi: 10.1016/j.micinf.2003.09.020
61. Dubourdeau M, Athman R, Balloy V, Huerre M, Chignard M, Philpott DJ, et al. Aspergillus fumigatus induces innate immune responses in alveolar macrophages through the MAPK pathway independently of TLR2 and TLR4. J Immunol (2006) 177:3994–4001. doi: 10.4049/jimmunol.177.6.3994
62. Carvalho A, De Luca A, Bozza S, Cunha C, D’angelo C, Moretti S, et al. TLR3 essentially promotes protective class I–restricted memory CD8+ T-cell responses to aspergillus fumigatus in hematopoietic transplanted patients. Blood (2012) 119:967–77. doi: 10.1182/blood-2011-06-362582
63. Van De Veerdonk FL, Joosten LA, Netea MG. The interplay between inflammasome activation and antifungal host defense. Immunol Rev (2015) 265:172–80. doi: 10.1111/imr.12280
64. Souza COS, Ketelut-Carneiro N, Milanezi CM, Faccioli LH, Gardinassi LG, Silva JS. NLRC4 inhibits NLRP3 inflammasome and abrogates effective antifungal CD8+ T cell responses. iScience (2021) 24:102548. doi: 10.1016/j.isci.2021.102548
65. Bochud PY, Chien JW, Marr KA, Leisenring WM, Upton A, Janer M, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med (2008) 359:1766–77. doi: 10.1056/NEJMoa0802629
66. Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, et al. Polymorphisms in toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol (2009) 37:1022–9. doi: 10.1016/j.exphem.2009.06.004
67. Morre SA, Murillo LS, Spaargaren J, Fennema HS, Pena AS. Role of the toll-like receptor 4 Asp299Gly polymorphism in susceptibility to candida albicans infection. J Infect Dis (2002) 186:1377–1379; author reply 1379. doi: 10.1086/344328
68. Sainz J, Perez E, Hassan L, Moratalla A, Romero A, Collado MD, et al. Variable number of tandem repeats of TNF receptor type 2 promoter as genetic biomarker of susceptibility to develop invasive pulmonary aspergillosis. Hum Immunol (2007) 68:41–50. doi: 10.1016/j.humimm.2006.10.011
69. Plantinga TS, van der Velden WJ, Ferwerda B, Van Spriel AB, Adema G, Feuth T, et al. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis (2009) 49:724–32. doi: 10.1086/604714
70. Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, et al. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood (2010) 116:5394–402. doi: 10.1182/blood-2010-04-279307
71. Del Pilar Jimenez AM, Viriyakosol S, Walls L, Datta SK, Kirkland T, Heinsbroek SE, et al. Susceptibility to coccidioides species in C57BL/6 mice is associated with expression of a truncated splice variant of dectin-1 (Clec7a). Genes Immun (2008) 9:338–48. doi: 10.1038/gene.2008.23
72. Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol (1989) 54 Pt 1:1–13. doi: 10.1101/SQB.1989.054.01.003
73. Rahman AH, Taylor DK, Turka LA. The contribution of direct TLR signaling to T cell responses. Immunol Res (2009) 45:25–36. doi: 10.1007/s12026-009-8113-x
74. Komai-Koma M, Jones L, Ogg GS, Xu D, Liew FY. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc Natl Acad Sci (2004) 101:3029–34. doi: 10.1073/pnas.0400171101
75. Imanishi T, Hara H, Suzuki S, Suzuki N, Akira S, Saito T. Cutting edge: TLR2 directly triggers Th1 effector functions. J Immunol (2007) 178:6715–9. doi: 10.4049/jimmunol.178.11.6715
76. Imanishi T, Unno M, Kobayashi W, Yoneda N, Akira S, Saito T. mTORC1 signaling controls TLR2-mediated T-cell activation by inducing TIRAP expression. Cell Rep (2020) 32:107911. doi: 10.1016/j.celrep.2020.107911
77. Wang H, Li M, Hung CY, Sinha M, Lee LM, Wiesner DL, et al. MyD88 shapes vaccine immunity by extrinsically regulating survival of CD4+ T cells during the contraction phase. PloS Pathog (2016) 12:e1005787. doi: 10.1371/journal.ppat.1005787
78. Nanjappa SG, Hernandez-Santos N, Galles K, Wuthrich M, Suresh M, Klein BS. Intrinsic MyD88-Akt1-mTOR signaling coordinates disparate Tc17 and Tc1 responses during vaccine immunity against fungal pneumonia. PloS Pathog (2015) 11:e1005161. doi: 10.1371/journal.ppat.1005161
79. Bhaskaran N, Faddoul F, Paes Da Silva A, Jayaraman S, Schneider E, Mamileti P, et al. Il-1β-Myd88-Mtor axis promotes immune-protective IL-17a+ Foxp3+ cells during mucosal infection and is dysregulated with aging. Front Immunol (2020) 11:2818. doi: 10.3389/fimmu.2020.595936
80. Ma H, Chan JFW, Tan YP, Kui L, Tsang CC, Pei SLC, et al. NLRP3 inflammasome contributes to host defense against talaromyces marneffei infection. Front Immunol (2021) 12:760095. doi: 10.3389/fimmu.2021.760095
81. Arbore G, West EE, Spolski R, Robertson A, Klos A, Rheinheimer C, et al. T Helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4(+) T cells. Science (2016) 352:aad1210. doi: 10.1126/science.aad1210
82. Kumaresan PR, Manuri PR, Albert ND, Maiti S, Singh H, Mi T, et al. Bioengineering T cells to target carbohydrate to treat opportunistic fungal infection. Proc Natl Acad Sci U.S.A. (2014) 111:10660–5. doi: 10.1073/pnas.1312789111
83. Roilides E, Uhlig K, Venzon D, Pizzo PA, Walsh TJ. Enhancement of oxidative response and damage caused by human neutrophils to aspergillus fumigatus hyphae by granulocyte colony-stimulating factor and gamma interferon. Infect Immun (1993) 61:1185–93. doi: 10.1128/iai.61.4.1185-1193.1993
84. Borghi M, Renga G, Puccetti M, Oikonomou V, Palmieri M, Galosi C, et al. Antifungal Th immunity: Growing up in family. Front Immunol (2014) 5:506. doi: 10.3389/fimmu.2014.00506
85. Mcdermott AJ, Klein BS. Helper T-cell responses and pulmonary fungal infections. Immunology (2018) 155:155–63. doi: 10.1111/imm.12953
86. Gray JI, Westerhof LM, Macleod MKL. The roles of resident, central and effector memory CD4 T-cells in protective immunity following infection or vaccination. Immunology (2018) 154(4):574–581. doi: 10.1111/imm.12929
87. Cenci E, Mencacci A, Bacci A, Bistoni F, Kurup VP, Romani L. T Cell vaccination in mice with invasive pulmonary aspergillosis. J Immunol (2000) 165:381–8. doi: 10.4049/jimmunol.165.1.381
88. Jolink H, De Boer R, Hombrink P, Jonkers RE, Van Dissel JT, Falkenburg JH, et al. Pulmonary immune responses against aspergillus fumigatus are characterized by high frequencies of IL-17 producing T-cells. J Infect (2017) 74:81–8. doi: 10.1016/j.jinf.2016.10.010
89. Jolink H, Hagedoorn RS, Lagendijk EL, Drijfhout JW, Van Dissel JT, Falkenburg JH, et al. Induction of a. Fumigatus-specific CD4-positive T cells in patients recovering from invasive aspergillosis. Haematologica (2014) 99:1255–63. doi: 10.3324/haematol.2013.098830
90. Nanjappa SG, Heninger E, Wuthrich M, Gasper DJ, Klein BS. Tc17 cells mediate vaccine immunity against lethal fungal pneumonia in immune deficient hosts lacking CD4+ T cells. PloS Pathog (2012) 8:e1002771. doi: 10.1371/journal.ppat.1002771
91. Hernandez-Santos N, Huppler AR, Peterson AC, Khader SA, Mckenna KC, Gaffen SL. Th17 cells confer long-term adaptive immunity to oral mucosal candida albicans infections. Mucosal Immunol (2013) 6:900–10. doi: 10.1038/mi.2012.128
92. Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol (2007) 8:639–46. doi: 10.1038/ni1467
93. Iannitti RG, Carvalho A, Romani L. From memory to antifungal vaccine design. Trends Immunol (2012) 33:467–74. doi: 10.1016/j.it.2012.04.008
94. Kirchner FR, Leibundgut-Landmann S. Tissue-resident memory Th17 cells maintain stable fungal commensalism in the oral mucosa. Mucosal Immunol (2021) 14:455–67. doi: 10.1038/s41385-020-0327-1
95. Ueno K, Urai M, Sadamoto S, Shinozaki M, Takatsuka S, Abe M, et al. A dendritic cell-based systemic vaccine induces long-lived lung-resident memory Th17 cells and ameliorates pulmonary mycosis. Mucosal Immunol (2019) 12:265–76. doi: 10.1038/s41385-018-0094-4
96. Ueno K, Kinjo Y, Okubo Y, Aki K, Urai M, Kaneko Y, et al. Dendritic cell-based immunization ameliorates pulmonary infection with highly virulent cryptococcus gattii. Infect Immun (2015) 83:1577–86. doi: 10.1128/IAI.02827-14
97. Fierer J, Waters C, Walls L. Both CD4+ and CD8+ T cells can mediate vaccine-induced protection against coccidioides immitis infection in mice. J Infect Dis (2006) 193:1323–31. doi: 10.1086/502972
98. Hung CY, Castro-Lopez N, Cole GT. Vaccinated C57BL/6 mice develop protective and memory T cell responses to coccidioides posadasii infection in the absence of interleukin-10. Infect Immun (2014) 82:903–13. doi: 10.1128/IAI.01148-13
99. Diep AL, Hoyer KK. Host response to coccidioides infection: Fungal immunity. Front Cell Infect Microbiol (2020) 10:581101. doi: 10.3389/fcimb.2020.581101
100. Deepe JGS, Buesing WR, Ostroff GR, Abraham A, Specht CA, Huang H, et al. Vaccination with an alkaline extract of histoplasma capsulatum packaged in glucan particles confers protective immunity in mice. Vaccine (2018) 36:3359–67. doi: 10.1016/j.vaccine.2018.04.047
101. Chiarella AP, Arruda C, Pina A, Costa TA, Ferreira RC, Calich VL. The relative importance of CD4+ and CD8+T cells in immunity to pulmonary paracoccidioidomycosis. Microbes Infect (2007) 9:1078–88. doi: 10.1016/j.micinf.2007.04.016
102. De La Rua NM, Samuelson DR, Charles TP, Welsh DA, Shellito JE. CD4+ T-cell-independent secondary immune responses to pneumocystis pneumonia. Front Immunol (2016) 7:178.
103. Galdino N, Loures FV, De Araújo EF, Da Costa TA, Preite NW, Calich VLG. Depletion of regulatory T cells in ongoing paracoccidioidomycosis rescues protective Th1/Th17 immunity and prevents fatal disease outcome. Sci Rep (2018) 8(1):16544. doi: 10.1038/s41598-018-35037-8
104. Pruksaphon K, Nosanchuk JD, Ratanabanangkoon K, Youngchim S. Talaromyces marneffei infection: Virulence, intracellular lifestyle and host defense mechanisms. J Fungi (2022) 8:200. doi: 10.3390/jof8020200
105. Park CO, Fu X, Jiang X, Pan Y, Teague JE, Collins N, et al. Staged development of long-lived T-cell receptor αβ TH17 resident memory T-cell population to candida albicans after skin infection. J Allergy Clin Immunol (2018) 142:647–62. doi: 10.1016/j.jaci.2017.09.042
106. Pietrella D, Rachini A, Pines M, Pandey N, Mosci P, Bistoni F, et al. Th17 cells and IL-17 in protective immunity to vaginal candidiasis. PloS One (2011) 6:e22770. doi: 10.1371/journal.pone.0022770
107. Alqarihi A, Singh S, Edwards JE Jr., Ibrahim AS, Uppuluri P. NDV-3A vaccination prevents c. albicans colonization of jugular vein catheters in mice. Sci Rep (2019) 9:6194. doi: 10.1038/s41598-019-42517-y
108. Ibrahim AS, Luo G, Gebremariam T, Lee H, Schmidt CS, Hennessey JP Jr., et al. NDV-3 protects mice from vulvovaginal candidiasis through T- and b-cell immune response. Vaccine (2013) 31:5549–56. doi: 10.1016/j.vaccine.2013.09.016
109. Edwards JE Jr., Schwartz MM, Schmidt CS, Sobel JD, Nyirjesy P, Schodel F, et al. A fungal immunotherapeutic vaccine (NDV-3A) for treatment of recurrent vulvovaginal candidiasis-a phase 2 randomized, double-blind, placebo-controlled trial. Clin Infect Dis (2018) 66:1928–36. doi: 10.1093/cid/ciy185
110. Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature (2012) 484:514–8. doi: 10.1038/nature10957
111. Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, et al. T Cell immunity. functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science (2015) 347:400–6. doi: 10.1126/science.1260668
112. Kirchner FR, Leibundgut-Landmann S. Tissue-resident memory Th17 cells maintain stable fungal commensalism in the oral mucosa. Mucosal Immunol (2020) 14:455–67. doi: 10.1038/s41385-020-0327-1
113. Schlapbach C, Gehad A, Yang C, Watanabe R, Guenova E, Teague JE, et al. Human TH9 cells are skin-tropic and have autocrine and paracrine proinflammatory capacity. Sci Transl Med (2014) 6:219ra218. doi: 10.1126/scitranslmed.3007828
114. Renga G, Moretti S, Oikonomou V, Borghi M, Zelante T, Paolicelli G, et al. IL-9 and mast cells are key players of candida albicans commensalism and pathogenesis in the gut. Cell Rep (2018) 23:1767–78. doi: 10.1016/j.celrep.2018.04.034
115. Liu Y, Yang B, Zhou M, Li L, Zhou H, Zhang J, et al. Memory IL-22-producing CD4+ T cells specific for candida albicans are present in humans. Eur J Immunol (2009) 39:1472–9. doi: 10.1002/eji.200838811
116. De Luca A, Carvalho A, Cunha C, Iannitti RG, Pitzurra L, Giovannini G, et al. IL-22 and IDO1 affect immunity and tolerance to murine and human vaginal candidiasis. PloS Pathog (2013) 9:e1003486. doi: 10.1371/journal.ppat.1003486
117. Zelante T, Iannitti R, De Luca A, Romani L. IL-22 in antifungal immunity. Eur J Immunol (2011) 41:270–5. doi: 10.1002/eji.201041246
118. Moser D, Biere K, Han B, Hoerl M, Schelling G, Chouker A, et al. COVID-19 impairs immune response to candida albicans. Front Immunol (2021) 12:640644. doi: 10.3389/fimmu.2021.640644
119. Ghaleb M, Hamad M, Abu-Elteen KH. Vaginal T lymphocyte population kinetics during experimental vaginal candidosis: evidence for a possible role of CD8+ T cells in protection against vaginal candidosis. Clin Exp Immunol (2003) 131:26–33. doi: 10.1046/j.1365-2249.2003.02032.x
120. Jones-Carson J, Vazquez-Torres FA, Balish E. B cell-independent selection of memory T cells after mucosal immunization with candida albicans. J Immunol (1997) 158:4328–35.
121. Breinig T, Scheller N, Glombitza B, Breinig F, Meyerhans A. Human yeast-specific CD8 T lymphocytes show a nonclassical effector molecule profile. Med Microbiol Immunol (2012) 201:127–36. doi: 10.1007/s00430-011-0213-2
122. Heintel T, Breinig F, Schmitt MJ, Meyerhans A. Extensive MHC class I-restricted CD8 T lymphocyte responses against various yeast genera in humans. FEMS Immunol Med Microbiol (2003) 39:279–86. doi: 10.1016/S0928-8244(03)00294-3
123. Myers TA, Leigh JE, Arribas AR, Hager S, Clark R, Lilly E, et al. Immunohistochemical evaluation of T cells in oral lesions from human immunodeficiency virus-positive persons with oropharyngeal candidiasis. Infect Immun (2003) 71:956–63. doi: 10.1128/IAI.71.2.956-963.2003
124. Ickrath P, Sprugel L, Beyersdorf N, Scherzad A, Hagen R, Hackenberg S. Detection of candida albicans-specific CD4+ and CD8+ T cells in the blood and nasal mucosa of patients with chronic rhinosinusitis. J Fungi (Basel) (2021) 7(6):403. doi: 10.3390/jof7060403
125. Roilides E, Dimitriadou A, Kadiltsoglou I, Sein T, Karpouzas J, Pizzo PA, et al. IL-10 exerts suppressive and enhancing effects on antifungal activity of mononuclear phagocytes against aspergillus fumigatus. J Immunol (1997) 158:322–9.
126. Cenci E, Mencacci A, Fe D’ostiani C, Del Sero G, Mosci P, Montagnoli C, et al. Cytokine- and T helper-dependent lung mucosal immunity in mice with invasive pulmonary aspergillosis. J Infect Dis (1998) 178:1750–60. doi: 10.1086/314493
127. Cenci E, Mencacci A, Del Sero G, Bacci A, Montagnoli C, D’ostiani CF, et al. Interleukin-4 causes susceptibility to invasive pulmonary aspergillosis through suppression of protective type I responses. J Infect Dis (1999) 180:1957–68. doi: 10.1086/315142
128. Bozza S, Gaziano R, Lipford GB, Montagnoli C, Bacci A, Di Francesco P, et al. Vaccination of mice against invasive aspergillosis with recombinant aspergillus proteins and CpG oligodeoxynucleotides as adjuvants. Microbes Infect (2002) 4:1281–90. doi: 10.1016/S1286-4579(02)00007-2
129. Ito JI, Lyons JM. Vaccination of corticosteroid immunosuppressed mice against invasive pulmonary aspergillosis. J Infect Dis (2002) 186:869–71. doi: 10.1086/342509
130. Stuehler C, Khanna N, Bozza S, Zelante T, Moretti S, Kruhm M, et al. Cross-protective TH1 immunity against aspergillus fumigatus and candida albicans. Blood (2011) 117:5881–91. doi: 10.1182/blood-2010-12-325084
131. Levitz SM. Aspergillus vaccines: Hardly worth studying or worthy of hard study? Med Mycol (2017) 55:103–8. doi: 10.1093/mmy/myw081
132. Rai G, Das S, Ansari MA, Singh PK, Dar SA, Haque S, et al. TLR-2 expression and dysregulated human Treg/Th17 phenotype in aspergillus flavus infected patients of chronic rhinosinusitis with nasal polyposis. Microb Cell Fact (2020) 19:215. doi: 10.1186/s12934-020-01481-3
133. Clemons KV, Grunig G, Sobel RA, Mirels LF, Rennick DM, Stevens DA. Role of IL-10 in invasive aspergillosis: increased resistance of IL-10 gene knockout mice to lethal systemic aspergillosis. Clin Exp Immunol (2000) 122:186–91. doi: 10.1046/j.1365-2249.2000.01382.x
134. Sainz J, Hassan L, Perez E, Romero A, Moratalla A, Lopez-Fernandez E, et al. Interleukin-10 promoter polymorphism as risk factor to develop invasive pulmonary aspergillosis. Immunol Lett (2007) 109:76–82. doi: 10.1016/j.imlet.2007.01.005
135. Cunha C, Goncalves SM, Duarte-Oliveira C, Leite L, Lagrou K, Marques A, et al. IL-10 overexpression predisposes to invasive aspergillosis by suppressing antifungal immunity. J Allergy Clin Immunol (2017) 140:867–870 e869. doi: 10.1016/j.jaci.2017.02.034
136. Grunig G, Corry DB, Leach MW, Seymour BW, Kurup VP, Rennick DM. Interleukin-10 is a natural suppressor of cytokine production and inflammation in a murine model of allergic bronchopulmonary aspergillosis. J Exp Med (1997) 185:1089–99. doi: 10.1084/jem.185.6.1089
137. Bacher P, Schink C, Teutschbein J, Kniemeyer O, Assenmacher M, Brakhage AA, et al. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J Immunol (2013) 190:3967–76. doi: 10.4049/jimmunol.1202221
138. Stuehler C, Nowakowska J, Bernardini C, Topp MS, Battegay M, Passweg J, et al. Multispecific aspergillus T cells selected by CD137 or CD154 induce protective immune responses against the most relevant mold infections. J Infect Dis (2015) 211:1251–61. doi: 10.1093/infdis/jiu607
139. Gottlieb DJ, Clancy LE, Withers B, Mcguire HM, Luciani F, Singh M, et al. Prophylactic antigen-specific T-cells targeting seven viral and fungal pathogens after allogeneic haemopoietic stem cell transplant. Clin Transl Immunol (2021) 10:e1249. doi: 10.1002/cti2.1249
140. Inam SS,G, Avdic S, Street J, Atkins E, Clany LE, Gottlieb DJ. Third-party partially HLA matched fungus-specific T-cells (FSTs) used to treat invasive fungal infection (IFI) with scedosporium aurantiacum after allogeneic hemopoietic stem cell transplant (aHSCT). Blood (2021) 138(1):2825. doi: 10.1182/blood-2021-152384
141. Castellano-Gonzalez G, Mcguire HM, Luciani F, Clancy LE, Li Z, Avdic S, et al. Rapidly expanded partially HLA DRB1-matched fungus-specific T cells mediate in vitro and in vivo antifungal activity. Blood Adv (2020) 4:3443–56. doi: 10.1182/bloodadvances.2020001565
142. Hirahara K, Kokubo K, Aoki A, Kiuchi M, Nakayama T. The role of CD4(+) resident memory T cells in local immunity in the mucosal tissue - protection versus pathology. Front Immunol (2021) 12:616309. doi: 10.3389/fimmu.2021.616309
143. Chaudhary N, Staab JF, Marr KA. Healthy human T-cell responses to aspergillus fumigatus antigens. PloS One (2010) 5:e9036. doi: 10.1371/journal.pone.0009036
144. Ma L, Cisse OH, Kovacs JA. A molecular window into the biology and epidemiology of pneumocystis spp. Clin Microbiol Rev (2018) 31(3):e00009-18. doi: 10.1128/CMR.00009-18
145. Elsegeiny W, Zheng M, Eddens T, Gallo RL, Dai G, Trevejo-Nunez G, et al. Murine models of pneumocystis infection recapitulate human primary immune disorders. JCI Insight (2018) 3(12):e91894. doi: 10.1172/jci.insight.91894
146. Elsegeiny W, Eddens T, Chen K, Kolls JK. Anti-CD20 antibody therapy and susceptibility to pneumocystis pneumonia. Infect Immun (2015) 83:2043–52. doi: 10.1128/IAI.03099-14
147. Hofmann K, Clauder AK, Manz RA. Targeting b cells and plasma cells in autoimmune diseases. Front Immunol (2018) 9. doi: 10.3389/fimmu.2018.00835
148. Lund FE, Hollifield M, Schuer K, Lines JL, Randall TD, Garvy BA. B cells are required for generation of protective effector and memory CD4 cells in response to pneumocystis lung infection. J Immunol (2006) 176:6147–54. doi: 10.4049/jimmunol.176.10.6147
149. Opata MM, Hollifield ML, Lund FE, Randall TD, Dunn R, Garvy BA, et al. B lymphocytes are required during the early priming of CD4(+) T cells for clearance of pneumocystis infection in mice. J Immunol (2015) 195:611–20. doi: 10.4049/jimmunol.1500112
150. Kling HM, Norris KA. Vaccine-induced immunogenicity and protection against pneumocystis pneumonia in a nonhuman primate model of HIV and pneumocystis coinfection. J Infect Dis (2016) 213:1586–95. doi: 10.1093/infdis/jiw032
151. Kelly MN, Zheng M, Ruan S, Kolls J, D’souza A, Shellito JE. Memory CD4+ T cells are required for optimal NK cell effector functions against the opportunistic fungal pathogen pneumocystis murina. J Immunol (2013) 190:285–95. doi: 10.4049/jimmunol.1200861
152. Tesini BL, Wright TW, Malone JE, Haidaris CG, Harber M, Sant AJ, et al. Immunization with pneumocystis cross-reactive antigen 1 (Pca1) protects mice against pneumocystis pneumonia and generates antibody to pneumocystis jirovecii. Infect Immun (2017) 85(4):e00850-16. doi: 10.1128/IAI.00850-16
153. Huffnagle GB, Yates JL, Lipscomb MF. Immunity to a pulmonary cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J Exp Med (1991) 173:793–800. doi: 10.1084/jem.173.4.793
154. Wormley FL Jr., Perfect JR, Steele C, Cox GM. Protection against cryptococcosis by using a murine gamma interferon-producing cryptococcus neoformans strain. Infect Immun (2007) 75:1453–62. doi: 10.1128/IAI.00274-06
155. Wozniak KL, Young ML, Wormley FL Jr. Protective immunity against experimental pulmonary cryptococcosis in T cell-depleted mice. Clin Vaccine Immunol (2011) 18:717–23. doi: 10.1128/CVI.00036-11
156. Specht CA, Homan EJ, Lee CK, Mou Z, Gomez CL, Hester MM, et al. Protection of mice against experimental cryptococcosis by synthesized peptides delivered in glucan particles. mBio (2022) 13(1):e0336721. doi: 10.1128/mbio.03367-21
157. Hester MM, Lee CK, Abraham A, Khoshkenar P, Ostroff GR, Levitz SM, et al. Protection of mice against experimental cryptococcosis using glucan particle-based vaccines containing novel recombinant antigens. Vaccine (2020) 38:620–6. doi: 10.1016/j.vaccine.2019.10.051
158. Chaturvedi AK, Hameed RS, Wozniak KL, Hole CR, Leopold Wager CM, Weintraub ST, et al. Vaccine-mediated immune responses to experimental pulmonary cryptococcus gattii infection in mice. PloS One (2014) 9:e104316. doi: 10.1371/journal.pone.0104316
159. Normile TG, Rella A, Del Poeta M. Cryptococcus neoformans Deltasgl1 vaccination requires either CD4(+) or CD8(+) T cells for complete host protection. Front Cell Infect Microbiol (2021) 11:739027. doi: 10.3389/fcimb.2021.739027
160. Jarvis JN, Casazza JP, Stone HH, Meintjes G, Lawn SD, Levitz SM, et al. The phenotype of the cryptococcus-specific CD4+ memory T-cell response is associated with disease severity and outcome in HIV-associated cryptococcal meningitis. J Infect Dis (2013) 207:1817–28. doi: 10.1093/infdis/jit099
161. Meya DB, Okurut S, Zziwa G, Cose S, Boulware DR, Janoff EN. HIV-Associated cryptococcal immune reconstitution inflammatory syndrome is associated with aberrant T cell function and increased cytokine responses. J Fungi (Basel) (2019) 5(2):42. doi: 10.20944/preprints201904.0086.v1
162. Wuthrich M, Hung CY, Gern BH, Pick-Jacobs JC, Galles KJ, Filutowicz HI, et al. A TCR transgenic mouse reactive with multiple systemic dimorphic fungi. J Immunol (2011) 187:1421–31. doi: 10.4049/jimmunol.1100921
163. Wuthrich M, Brandhorst TT, Sullivan TD, Filutowicz H, Sterkel A, Stewart D, et al. Calnexin induces expansion of antigen-specific CD4(+) T cells that confer immunity to fungal ascomycetes via conserved epitopes. Cell Host Microbe (2015) 17:452–65. doi: 10.1016/j.chom.2015.02.009
164. Rivera A, Hohl TM. Calnexin bridges the gap toward a pan-fungal vaccine. Cell Host Microbe (2015) 17:421–3. doi: 10.1016/j.chom.2015.03.012
165. Dobson HE, Dias LDS, Kohn EM, Fites S, Wiesner DL, Dileepan T, et al. Antigen discovery unveils resident memory and migratory cell roles in antifungal resistance. Mucosal Immunol (2020) 13:518–29. doi: 10.1038/s41385-019-0244-3
166. Dos Santos Dias L, Dobson HE, Bakke BK, Kujoth GC, Huang J, Kohn EM, et al. Structural basis of blastomyces endoglucanase-2 adjuvancy in anti-fungal and -viral immunity. PloS Pathog (2021) 17:e1009324. doi: 10.1371/journal.ppat.1009324
167. Wuthrich M, Dobson HE, Ledesma Taira C, Okaa UJ, Dos Santos Dias L, Isidoro-Ayza M, et al. Combination adjuvants enhance recombinant protein vaccine protection against fungal infection. mBio (2021) 12:e0201821. doi: 10.1128/mBio.02018-21
168. Wuthrich M, Filutowicz HI, Warner T, Deepe GS Jr., Klein BS. Vaccine immunity to pathogenic fungi overcomes the requirement for CD4 help in exogenous antigen presentation to CD8+ T cells: implications for vaccine development in immune-deficient hosts. J Exp Med (2003) 197:1405–16. doi: 10.1084/jem.20030109
169. Nanjappa SG, Heninger E, Wuthrich M, Sullivan T, Klein B. Protective antifungal memory CD8(+) T cells are maintained in the absence of CD4(+) T cell help and cognate antigen in mice. J Clin Invest (2012) 122:987–99. doi: 10.1172/JCI58762
170. Nanjappa SG, Mudalagiriyappa S, Fites JS, Suresh M, Klein BS. CBLB constrains inactivated vaccine-induced CD8(+) T cell responses and immunity against lethal fungal pneumonia. J Immunol (2018) 201:1717–26. doi: 10.4049/jimmunol.1701241
171. Mudalagiriyappa SS,J, Vieson M, Klein BS, Nanjappa SG. GM-CSF + Tc17 cells are required to bolster vaccine immunity against lethal fungal pneumonia without causing overt pathology. SSRN (2022). doi: 10.2139/ssrn.4099074
172. Edwards LB, Acquaviva FA, Livesay VT, Cross FW, Palmer CE. An atlas of sensitivity to tuberculin, PPD-b, and histoplasmin in the united states. Am Rev Respir Dis (1969) 99:Suppl:1–132.
173. Allendorfer R, Brunner GD, Deepe GS Jr. Complex requirements for nascent and memory immunity in pulmonary histoplasmosis. J Immunol (1999) 162:7389–96.
174. Deepe GS Jr., Gibbons RS. Protective and memory immunity to histoplasma capsulatum in the absence of IL-10. J Immunol (2003) 171:5353–62. doi: 10.4049/jimmunol.171.10.5353
175. Deepe GS Jr., Gibbons RS. Interleukins 17 and 23 influence the host response to histoplasma capsulatum. J Infect Dis (2009) 200:142–51. doi: 10.1086/599333
176. Deepe GS Jr., Gibbons RS. Cellular and molecular regulation of vaccination with heat shock protein 60 from histoplasma capsulatum. Infect Immun (2002) 70:3759–67. doi: 10.1128/IAI.70.7.3759-3767.2002
177. Deepe GS. Role of Cd8(+) T-cells in host-resistance to systemic infection with histoplasma-capsulatum in mice. J Immunol (1994) 152:3491–500.
178. Zhou P, Freidag BL, Caldwell CC, Seder RA. Perforin is required for primary immunity to histoplasma capsulatum. J Immunol (2001) 166:1968–74. doi: 10.4049/jimmunol.166.3.1968
179. Hung CY, Hsu AP, Holland SM, Fierer J. A review of innate and adaptive immunity to coccidioidomycosis. Med Mycol (2019) 57:S85–92. doi: 10.1093/mmy/myy146
180. Ward RA, Thompson GR 3rd, Villani AC, Li B, Mansour MK, Wuethrich M, et al. The known unknowns of the immune response to coccidioides. J Fungi (Basel) (2021) 7(5):377. doi: 10.3390/jof7050377
181. Fierer J, Walls L, Eckmann L, Yamamoto T, Kirkland TN. Importance of interleukin-10 in genetic susceptibility of mice to coccidioides immitis. Infect Immun (1998) 66:4397–402. doi: 10.1128/IAI.66.9.4397-4402.1998
182. Davini D, Naeem F, Phong A, Al-Kuhlani M, Valentine KM, Mccarty J, et al. Elevated regulatory T cells at diagnosis of coccidioides infection associates with chronicity in pediatric patients. J Allergy Clin Immunol (2018) 142:1971–1974 e1977. doi: 10.1016/j.jaci.2018.10.022
183. Nesbit L, Johnson SM, Pappagianis D, Ampel NM. Polyfunctional T lymphocytes are in the peripheral blood of donors naturally immune to coccidioidomycosis and are not induced by dendritic cells. Infect Immun (2010) 78:309–15. doi: 10.1128/IAI.00953-09
184. Pappagianis D. Evaluation of the protective efficacy of the killed coccidioides immitis spherule vaccine in humans. the valley fever vaccine study group. Am Rev Respir Dis (1993) 148:656–60. doi: 10.1164/ajrccm/148.3.656
185. Tarcha EJ, Basrur V, Hung CY, Gardner MJ, Cole GT. Multivalent recombinant protein vaccine against coccidioidomycosis. Infect Immun (2006) 74:5802–13. doi: 10.1128/IAI.00961-06
186. Campuzano AP, Komali Devi P, Liao Y, Zhang H, Wiederhold N, Ostroff G, et al. A recombinant multivalent vaccine (rCpa1) induces protection for C57BL/6 and HLA transgenic mice against pulmonary infection with both species of coccidioides. bioRxiv (2021). doi: 10.1101/2021.10.20.465232
187. Burger E. Paracoccidioidomycosis protective immunity. J Fungi (Basel) (2021) 7(2):137. doi: 10.3390/jof7020137
188. De Castro LF, Ferreira MC, Da Silva RM, Blotta MH, Longhi LN, Mamoni RL. Characterization of the immune response in human paracoccidioidomycosis. J Infect (2013) 67:470–85. doi: 10.1016/j.jinf.2013.07.019
189. Mayorga O, Munoz JE, Lincopan N, Teixeira AF, Ferreira LC, Travassos LR, et al. The role of adjuvants in therapeutic protection against paracoccidioidomycosis after immunization with the P10 peptide. Front Microbiol (2012) 3:154. doi: 10.3389/fmicb.2012.00154
190. Holanda RA, Munoz JE, Dias LS, Silva LBR, Santos JRA, Pagliari S, et al. Recombinant vaccines of a CD4+ T-cell epitope promote efficient control of paracoccidioides brasiliensis burden by restraining primary organ infection. PloS Negl Trop Dis (2017) 11:e0005927. doi: 10.1371/journal.pntd.0005927
191. Bozzi A, Reis BS, Goulart MI, Pereira MC, Pedroso EP, Goes AM. Analysis of memory T cells in the human paracoccidioidomycosis before and during chemotherapy treatment. Immunol Lett (2007) 114:23–30. doi: 10.1016/j.imlet.2007.08.004
192. Bernardino S, Pina A, Felonato M, Costa TA, Frank De Araujo E, Feriotti C, et al. TNF-alpha and CD8+ T cells mediate the beneficial effects of nitric oxide synthase-2 deficiency in pulmonary paracoccidioidomycosis. PloS Negl Trop Dis (2013) 7:e2325.
193. Deo SS, Virassamy B, Halliday C, Clancy L, Chen S, Meyer W, et al. Stimulation with lysates of aspergillus terreus, candida krusei and rhizopus oryzae maximizes cross-reactivity of anti-fungal T cells. Cytotherapy (2016) 18:65–79. doi: 10.1016/j.jcyt.2015.09.013
194. Aggor FEY, Way SS, Gaffen SL. Fungus among us: The frenemies within. Trends Immunol (2019) 40:469–71. doi: 10.1016/j.it.2019.04.007
195. Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, et al. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against candida albicans. Cell (2019) 176:1340–55:e1315. doi: 10.1016/j.cell.2019.01.041
196. Gaundar SS, Clancy L, Blyth E, Meyer W, Gottlieb DJ. Robust polyfunctional T-helper 1 responses to multiple fungal antigens from a cell population generated using an environmental strain of aspergillus fumigatus. Cytotherapy (2012) 14:1119–30. doi: 10.3109/14653249.2012.704013
197. Buldain I, Pellon A, Zaldibar B, Antoran A, Martin-Souto L, Aparicio-Fernandez L, et al. Study of humoral responses against Lomentospora/Scedosporium spp. and aspergillus fumigatus to identify l. prolificans antigens of interest for diagnosis and treatment. Vaccines (2019) 7:212. doi: 10.3390/vaccines7040212
198. Lin L, Ibrahim AS, Xu X, Farber JM, Avanesian V, Baquir B, et al. Th1-Th17 cells mediate protective adaptive immunity against staphylococcus aureus and candida albicans infection in mice. PloS Pathog (2009) 5:e1000703. doi: 10.1371/journal.ppat.1000703
199. Singh S, Uppuluri P, Mamouei Z, Alqarihi A, Elhassan H, French S, et al. The NDV-3A vaccine protects mice from multidrug resistant candida auris infection. PloS Pathog (2019) 15:e1007460. doi: 10.1371/journal.ppat.1007460
200. Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity (2013) 38:373–83. doi: 10.1016/j.immuni.2012.10.021
201. Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell (2014) 157:1073–87. doi: 10.1016/j.cell.2014.03.047
202. Lee H-G, Lee J-U, Kim D-H, Lim S, Kang I, Choi J-M. Pathogenic function of bystander-activated memory-like CD4+ T cells in autoimmune encephalomyelitis. Nat Commun (2019) 10:1–14. doi: 10.1038/s41467-019-08482-w
203. Thompson-Souza GA, Santos GMP, Silva JC, Muniz VS, Braga Y, Figueiredo RT, et al. Histoplasma capsulatum-induced extracellular DNA trap release in human neutrophils. Cell Microbiol (2020) 22:e13195. doi: 10.1111/cmi.13195
204. Puerta-Arias JD, Mejia SP, Gonzalez A. The role of the interleukin-17 axis and neutrophils in the pathogenesis of endemic and systemic mycoses. Front Cell Infect Microbiol (2020) 10:595301. doi: 10.3389/fcimb.2020.595301
205. De Luca A, Zelante T, D’angelo C, Zagarella S, Fallarino F, Spreca A, et al. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol (2010) 3:361–73. doi: 10.1038/mi.2010.22
206. Aggor FEY, Break TJ, Trevejo-Nunez G, Whibley N, Coleman BM, Bailey RD, et al. Oral epithelial IL-22/STAT3 signaling licenses IL-17-mediated immunity to oral mucosal candidiasis. Sci Immunol (2020) 5(48):eaba0570. doi: 10.1126/sciimmunol.aba0570
207. Swidergall M, Ernst JF. Interplay between candida albicans and the antimicrobial peptide armory. Eukaryot Cell (2014) 13:950–7. doi: 10.1128/EC.00093-14
208. Murdock BJ, Falkowski NR, Shreiner AB, Sadighi Akha AA, Mcdonald RA, White ES, et al. Interleukin-17 drives pulmonary eosinophilia following repeated exposure to aspergillus fumigatus conidia. Infect Immun (2012) 80:1424–36. doi: 10.1128/IAI.05529-11
209. Netea MG, Joosten L, van der Meer JWM, Kullberg B-J, Van De Veerdonk FL. Immune defence against candida fungal infections. Nat Rev Immunol (2015) 15:630–42. doi: 10.1038/nri3897
210. Kroetz DN, Deepe GS. The role of cytokines and chemokines in histoplasma capsulatum infection. Cytokine (2012) 58:112–7. doi: 10.1016/j.cyto.2011.07.430
211. Castro-Lopez N, Hung CY. Immune response to coccidioidomycosis and the development of a vaccine. Microorganisms (2017) 5:13. doi: 10.3390/microorganisms5010013
212. Kak G, Raza M, Tiwari BK. Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol Concepts (2018) 9:64–79. doi: 10.1515/bmc-2018-0007
213. Speakman EA, Dambuza IM, Salazar F, Brown GD. T Cell antifungal immunity and the role of c-type lectin receptors. Trends Immunol (2020) 41:61–76. doi: 10.1016/j.it.2019.11.007
214. Vignesh KS, Figueroa J, Porollo A, Caruso JA, Deepe JGS. Granulocyte macrophage-colony stimulating factor induced zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity (2013) 39:697–710. doi: 10.1016/j.immuni.2013.09.006
215. Deepe GS, Gibbons R, Woodward E. Neutralization of endogenous granulocyte-macrophage colony-stimulating factor subverts the protective immune response to histoplasma capsulatum. J Immunol (1999) 163:4985–93.
216. Herring AC, Falkowski NR, Chen G-H, Mcdonald RA, Toews GB, Huffnagle GB. Transient neutralization of tumor necrosis factor alpha can produce a chronic fungal infection in an immunocompetent host: potential role of immature dendritic cells. Infect Immun (2005) 73:39–49. doi: 10.1128/IAI.73.1.39-49.2005
217. Rocha F, Alves A, Rocha MFG, Cordeiro RA, Brilhante RSN, Pinto A, et al. Tumor necrosis factor prevents candida albicans biofilm formation. Sci Rep (2017) 7:1206. doi: 10.1038/s41598-017-01400-4
218. Ma LL, Spurrell JC, Wang JF, Neely GG, Epelman S, Krensky AM, et al. CD8 T cell-mediated killing of cryptococcus neoformans requires granulysin and is dependent on CD4 T cells and IL-15. J Immunol (2002) 169:5787–95. doi: 10.4049/jimmunol.169.10.5787
219. Dotiwala F, Lieberman J. Granulysin: killer lymphocyte safeguard against microbes. Curr Opin Immunol (2019) 60:19–29. doi: 10.1016/j.coi.2019.04.013
220. Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol (2015) 15:388–400. doi: 10.1038/nri3839
221. Vitenshtein A, Charpak-Amikam Y, Yamin R, Bauman Y, Isaacson B, Stein N, et al. NK cell recognition of candida glabrata through binding of NKp46 and NCR1 to fungal ligands Epa1, Epa6, and Epa7. Cell Host Microbe (2016) 20:527–34. doi: 10.1016/j.chom.2016.09.008
222. Li SS, Ogbomo H, Mansour MK, Xiang RF, Szabo L, Munro F, et al. Identification of the fungal ligand triggering cytotoxic PRR-mediated NK cell killing of cryptococcus and candida. Nat Commun (2018) 9:751. doi: 10.1038/s41467-018-03014-4
223. Zheng CF, Ma LL, Jones GJ, Gill MJ, Krensky AM, Kubes P, et al. Cytotoxic CD4+ T cells use granulysin to kill cryptococcus neoformans, and activation of this pathway is defective in HIV patients. Blood (2007) 109:2049–57. doi: 10.1182/blood-2006-03-009720
224. Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol (2001) 2:415–22. doi: 10.1038/87720
225. Chung HK, Mcdonald B, Kaech SM. The architectural design of CD8+ T cell responses in acute and chronic infection: Parallel structures with divergent fates. J Exp Med (2021) 218(4):e20201730. doi: 10.1084/jem.20201730
226. Portuondo DL, Ferreira LS, Urbaczek AC, Batista-Duharte A, Carlos IZ. Adjuvants and delivery systems for antifungal vaccines: current state and future developments. Med Mycol (2015) 53:69–89. doi: 10.1093/mmy/myu045
227. Oliveira LVN, Wang R, Specht CA, Levitz SM. Vaccines for human fungal diseases: close but still a long way to go. NPJ Vaccines (2021) 6:33. doi: 10.1038/s41541-021-00294-8
228. Borriello F, Poli V, Shrock E, Spreafico R, Liu X, Pishesha N, et al. An adjuvant strategy enabled by modulation of the physical properties of microbial ligands expands antigen immunogenicity. Cell (2022) 185:614–629 e621. doi: 10.1016/j.cell.2022.01.009
229. Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science (2003) 300:339–42. doi: 10.1126/science.1083317
230. Hor JL, Whitney PG, Zaid A, Brooks AG, Heath WR, Mueller SN. Spatiotemporally distinct interactions with dendritic cell subsets facilitates CD4+ and CD8+ T cell activation to localized viral infection. Immunity (2015) 43:554–65. doi: 10.1016/j.immuni.2015.07.020
231. Ahrends T, Spanjaard A, Pilzecker B, Babala N, Bovens A, Xiao Y, et al. CD4(+) T cell help confers a cytotoxic T cell effector program including coinhibitory receptor downregulation and increased tissue invasiveness. Immunity (2017) 47:848–861 e845. doi: 10.1016/j.immuni.2017.10.009
232. Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature (2009) 462:510–3. doi: 10.1038/nature08511
233. Redeker A, Welten SP, Baert MR, Vloemans SA, Tiemessen MM, Staal FJ, et al. The quantity of autocrine IL-2 governs the expansion potential of CD8+ T cells. J Immunol (2015) 195:4792–801. doi: 10.4049/jimmunol.1501083
234. Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H, et al. Robust anti-viral immunity requires multiple distinct T cell-dendritic cell interactions. Cell (2015) 162:1322–37. doi: 10.1016/j.cell.2015.08.004
235. Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol (2003) 4:1191–8. doi: 10.1038/ni1009
236. Ahrends T, Busselaar J, Severson TM, Babala N, De Vries E, Bovens A, et al. CD4(+) T cell help creates memory CD8(+) T cells with innate and help-independent recall capacities. Nat Commun (2019) 10:5531. doi: 10.1038/s41467-019-13438-1
237. Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol (2009) 9:153–61. doi: 10.1038/nri2496
238. Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity (2007) 27:393–405. doi: 10.1016/j.immuni.2007.08.007
239. Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol (2003) 4:835–42. doi: 10.1038/ni969
240. Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol (2004) 22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702
241. Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol (2002) 2:251–62. doi: 10.1038/nri778
242. Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science (2007) 315:1687–91. doi: 10.1126/science.1139393
243. Stemberger C, Neuenhahn M, Gebhardt FE, Schiemann M, Buchholz VR, Busch DH. Stem cell-like plasticity of naive and distinct memory CD8+ T cell subsets. Semin Immunol (2009) 21:62–8. doi: 10.1016/j.smim.2009.02.004
244. Fuertes Marraco SA, Soneson C, Cagnon L, Gannon PO, Allard M, Abed Maillard S, et al. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci Transl Med (2015) 7:282ra248. doi: 10.1126/scitranslmed.aaa3700
245. Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity (2008) 29:848–62. doi: 10.1016/j.immuni.2008.11.002
246. Raeber ME, Zurbuchen Y, Impellizzieri D, Boyman O. The role of cytokines in T-cell memory in health and disease. Immunol Rev (2018) 283:176–93. doi: 10.1111/imr.12644
247. Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol (2004) 16:217–25. doi: 10.1016/j.coi.2004.02.005
248. Martin MD, Badovinac VP. Defining memory CD8 T cell. Front Immunol (2018) 9:2692. doi: 10.3389/fimmu.2018.02692
249. Muroyama Y, Wherry EJ. Memory T-cell heterogeneity and terminology. Cold Spring Harb Perspect Biol (2021) 13(10):a037929. doi: 10.1101/cshperspect.a037929
250. Reiner SL, Sallusto F, Lanzavecchia A. Division of labor with a workforce of one: Challenges in specifying effector and memory T cell fate. Science (2007) 317:622–5. doi: 10.1126/science.1143775
251. Gerlach C, Van Heijst JWJ, Swart E, Sie D, Armstrong N, Kerkhoven RM, et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med (2010) 207:1235–46. doi: 10.1084/jem.20091175
252. Bresser K, Kok L, Swain AC, King LA, Jacobs L, Weber TS, et al. Replicative history marks transcriptional and functional disparity in the CD8(+) T cell memory pool. Nat Immunol (2022) 23(5):791–801. doi: 10.1038/s41590-022-01171-9
253. Ahmed R, Bevan MJ, Reiner SL, Fearon DT. The precursors of memory: models and controversies. Nat Rev Immunol (2009) 9:662–8. doi: 10.1038/nri2619
254. Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol (2012) 12:749–61. doi: 10.1038/nri3307
255. Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol (2014) 15:1104–15. doi: 10.1038/ni.3031
256. Hope JL, Stairiker CJ, Bae EA, Otero DC, Bradley LM. Striking a balance-cellular and molecular drivers of memory T cell development and responses to chronic stimulation. Front Immunol (2019) 10:1595. doi: 10.3389/fimmu.2019.01595
257. Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature (1999) 401:708–12. doi: 10.1038/44385
258. Lanzavecchia A, Sallusto F. Understanding the generation and function of memory T cell subsets. Curr Opin Immunol (2005) 17:326–32. doi: 10.1016/j.coi.2005.04.010
259. Geginat J, Lanzavecchia A, Sallusto F. Proliferation and differentiation potential of human CD8+ memory T-cell subsets in response to antigen or homeostatic cytokines. Blood (2003) 101:4260–6. doi: 10.1182/blood-2002-11-3577
260. Leibundgut-Landmann S. Tissue-resident memory T cells in antifungal immunity. Front Immunol (2021) 0:1976. doi: 10.3389/fimmu.2021.693055
261. Szabo PA, Miron M, Farber DL. Location, location, location: Tissue resident memory T cells in mice and humans. Sci Immunol (2019) 4(34):eaas9673. doi: 10.1126/sciimmunol.aas9673
262. Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol (2015) 194:2059–63. doi: 10.4049/jimmunol.1402256
263. Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol (2012) 188:4866–75. doi: 10.4049/jimmunol.1200402
264. Hirai T, Yang Y, Zenke Y, Li H, Chaudhri VK, de la Cruz Diaz JS, et al. Competition for active TGFbeta cytokine allows for selective retention of antigen-specific tissue- resident memory T cells in the epidermal niche. Immunity (2021) 54:84–98 e85. doi: 10.1016/j.immuni.2020.10.022
265. Sasson SC, Gordon CL, Christo SN, Klenerman P, Mackay LK. Local heroes or villains: tissue-resident memory T cells in human health and disease. Cell Mol Immunol (2020) 17:113–22. doi: 10.1038/s41423-019-0359-1
266. Lugli E, Dominguez MH, Gattinoni L, Chattopadhyay PK, Bolton DL, Song K, et al. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest (2013) 123:594. doi: 10.1172/JCI66327
267. Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, et al. Serial transfer of single-Cell-Derived immunocompetence reveals stemness of CD8+ central memory T cells. Immunity (2014) 41:116–26. doi: 10.1016/j.immuni.2014.05.018
268. Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity (2007) 27:281–95. doi: 10.1016/j.immuni.2007.07.010
269. Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, et al. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol (2014) 192:5881–93. doi: 10.4049/jimmunol.1303187
270. Jameson SC, Masopust D. Understanding subset diversity in T cell memory. Immunity (2018) 48:214–26. doi: 10.1016/j.immuni.2018.02.010
271. Wherry EJ, Barber DL, Kaech SM, Blattman JN, Ahmed R. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc Natl Acad Sci U.S.A. (2004) 101:16004–9.
272. Utzschneider DT, Alfei F, Roelli P, Barras D, Chennupati V, Darbre S, et al. High antigen levels induce an exhausted phenotype in a chronic infection without impairing T cell expansion and survival. J Exp Med (2016) 213:1819–34. doi: 10.1084/jem.20150598
273. Wirth TC, Xue HH, Rai D, Sabel JT, Bair T, Harty JT, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity (2010) 33:128–40. doi: 10.1016/j.immuni.2010.06.014
274. Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol (2012) 86:8161–70. doi: 10.1128/JVI.00889-12
275. Daniels MA, Teixeiro E. TCR signaling in T cell memory. Front Immunol (2015) 6:617. doi: 10.3389/fimmu.2015.00617
276. Wuthrich M, Warner T, Klein BS. CD28 is required for optimal induction, but not maintenance, of vaccine-induced immunity to blastomyces dermatitidis. Infect Immun (2005) 73:7436–41. doi: 10.1128/IAI.73.11.7436-7441.2005
277. Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol (2013) 13:227–42. doi: 10.1038/nri3405
278. Humphreys IR, Edwards L, Walzl G, Rae AJ, Dougan G, Hill S, et al. OX40 ligation on activated T cells enhances the control of cryptococcus neoformans and reduces pulmonary eosinophilia. J Immunol (2003) 170:6125–32. doi: 10.4049/jimmunol.170.12.6125
279. Wuthrich M, Fisette PL, Filutowicz HI, Klein BS. Differential requirements of T cell subsets for CD40 costimulation in immunity to blastomyces dermatitidis. J Immunol (2006) 176:5538–47. doi: 10.4049/jimmunol.176.9.5538
280. Zhou Q, Gault RA, Kozel TR, Murphy WJ. Immunomodulation with CD40 stimulation and interleukin-2 protects mice from disseminated cryptococcosis. Infect Immun (2006) 74:2161–8. doi: 10.1128/IAI.74.4.2161-2168.2006
281. Chen GH, Osterholzer JJ, Choe MY, Mcdonald RA, Olszewski MA, Huffnagle GB, et al. Dual roles of CD40 on microbial containment and the development of immunopathology in response to persistent fungal infection in the lung. Am J Pathol (2010) 177:2459–71. doi: 10.2353/ajpath.2010.100141
282. Felonato M, Pina A, Bernardino S, Loures FV, De Araujo EF, Calich VL. CD28 exerts protective and detrimental effects in a pulmonary model of paracoccidioidomycosis. Infect Immun (2010) 78:4922–35. doi: 10.1128/IAI.00297-10
283. Bouguermouh S, Fortin G, Baba N, Rubio M, Sarfati M. CD28 co-stimulation down regulates Th17 development. PloS One (2009) 4:e5087. doi: 10.1371/journal.pone.0005087
284. Revu S, Wu J, Henkel M, Rittenhouse N, Menk A, Delgoffe GM, et al. IL-23 and IL-1beta drive human Th17 cell differentiation and metabolic reprogramming in absence of CD28 costimulation. Cell Rep (2018) 22:2642–53. doi: 10.1016/j.celrep.2018.02.044
285. Dong C. Cytokine regulation and function in T cells. Annu Rev Immunol (2021) 39:51–76. doi: 10.1146/annurev-immunol-061020-053702
286. Cavassani KA, Campanelli AP, Moreira AP, Vancim JO, Vitali LH, Mamede RC, et al. Systemic and local characterization of regulatory T cells in a chronic fungal infection in humans. J Immunol (2006) 177:5811–8. doi: 10.4049/jimmunol.177.9.5811
287. Romani L. Immunity to fungal infections. Nat Rev Immunol (2011) 11:275–88. doi: 10.1038/nri2939
288. Murdock BJ, Teitz-Tennenbaum S, Chen GH, Dils AJ, Malachowski AN, Curtis JL, et al. Early or late IL-10 blockade enhances Th1 and Th17 effector responses and promotes fungal clearance in mice with cryptococcal lung infection. J Immunol (2014) 193:4107–16. doi: 10.4049/jimmunol.1400650
289. Break TJ, Oikonomou V, Dutzan N, Desai JV, Swidergall M, Freiwald T, et al. Aberrant type 1 immunity drives susceptibility to mucosal fungal infections. Science (2021) 371(6526):eaay5731. doi: 10.1126/science.aay5731
290. Montano DE, Voigt K. Host immune defense upon fungal infections with mucorales: Pathogen-immune cell interactions as drivers of inflammatory responses. J Fungi (2020) 6(3):173. doi: 10.3390/jof6030173
291. Deepe GS Jr., Gibbons RS. T Cells require tumor necrosis factor-alpha to provide protective immunity in mice infected with histoplasma capsulatum. J Infect Dis (2006) 193:322–30. doi: 10.1086/498981
292. Romani L. Innate and acquired cellular immunity to fungi. Mol Principles Fungal Pathogene (2006), 471–86. doi: 10.1128/9781555815776.ch32
293. Romani L. Cell mediated immunity to fungi: a reassessment. Med Mycol (2008) 46:515–29. doi: 10.1080/13693780801971450
294. Leibundgut-Landmann S, Wuthrich M, Hohl TM. Immunity to fungi. Curr Opin Immunol (2012) 24:449–58. doi: 10.1016/j.coi.2012.04.007
295. Azuma M. Co-Signal molecules in T-cell activation: Historical overview and perspective. Adv Exp Med Biol (2019) 1189:3–23. doi: 10.1007/978-981-32-9717-3_1
296. Morris AB, Adams LE, Ford ML. Influence of T cell coinhibitory molecules on CD8+ recall responses. Front Immunol (2018) 1810. doi: 10.3389/fimmu.2018.01810
297. Roussey JA, Viglianti SP, Teitz-Tennenbaum S, Olszewski MA, Osterholzer JJ. Anti–PD-1 antibody treatment promotes clearance of persistent cryptococcal lung infection in mice. J Immunol (2017) 199:3535–46. doi: 10.4049/jimmunol.1700840
298. Shwetank, Abdelsamed HA, Frost EL, Schmitz HM, Mockus TE, Youngblood BA, et al. Maintenance of PD-1 on brain-resident memory CD8 T cells is antigen independent. Immunol Cell Biol (2017) 95:953–9. doi: 10.1038/icb.2017.62
299. Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol (2018) 19:183–91. doi: 10.1038/s41590-017-0027-5
300. Chang KC, Burnham CA, Compton SM, Rasche DP, Mazuski RJ, Mcdonough JS, et al. Blockade of the negative co-stimulatory molecules PD-1 and CTLA-4 improves survival in primary and secondary fungal sepsis. Crit Care (2013) 17:R85. doi: 10.1186/cc12711
301. Gartshteyn Y, Askanase AD, Mor A. SLAM associated protein signaling in T cells: Tilting the balance toward autoimmunity. Front Immunol (2021) 12:654839. doi: 10.3389/fimmu.2021.654839
302. Kohn EM, Dos Santos Dias L, Dobson HE, He X, Wang H, Klein BS, et al. SLAMF1 is dispensable for vaccine-induced T cell development but required for resistance to fungal infection. J Immunol (2022) 208:1417–23. doi: 10.4049/jimmunol.2100819
303. Cyster JG, Shotton DM, Williams AF. The dimensions of the T lymphocyte glycoprotein leukosialin and identification of linear protein epitopes that can be modified by glycosylation. EMBO J (1991) 10:893–902. doi: 10.1002/j.1460-2075.1991.tb08022.x
304. Ostberg JR, Barth RK, Frelinger JG. The Roman god janus: A paradigm for the function of CD43. Immunol Today (1998) 19:546–50. doi: 10.1016/S0167-5699(98)01343-7
305. Onami TM, Harrington LE, Williams MA, Galvan M, Larsen CP, Pearson TC, et al. Dynamic regulation of T cell immunity by CD43. J Immunol (2002) 168:6022–31. doi: 10.4049/jimmunol.168.12.6022
306. Manjunath N, Johnson RS, Staunton DE, Pasqualini R, Ardman B. Targeted disruption of CD43 gene enhances T lymphocyte adhesion. J Immunol (1993) 151:1528–34.
307. Manjunath N, Correa M, Ardman M, Ardman B. Negative regulation of T-cell adhesion and activation by CD43. Nature (1995) 377:535–8. doi: 10.1038/377535a0
308. Pedraza-Alva G, Merida LB, Del Rio R, Fierro NA, Cruz-Munoz ME, Olivares N, et al. CD43 regulates the threshold for T cell activation by targeting cbl functions. IUBMB Life (2011) 63:940–8. doi: 10.1002/iub.554
309. Mudalagiriyappa SD, George S D Jr, Nanjappa SG. Sialophorin is an essential host element for vaccine immunity against pulmonary fungal infections. bioRxiv (2022). doi: 10.1101/2022.03.31.486552
Keywords: CD4+ T cells, CD8+ T cells, memory, fungal, vaccination, infection, immunity
Citation: Sharma J, Mudalagiriyappa S and Nanjappa SG (2022) T cell responses to control fungal infection in an immunological memory lens. Front. Immunol. 13:905867. doi: 10.3389/fimmu.2022.905867
Received: 28 March 2022; Accepted: 22 August 2022;
Published: 13 September 2022.
Edited by:
Eui Ho Kim, Institut Pasteur Korea, South KoreaReviewed by:
Fabian Salazar, University of Exeter, United KingdomNa Cui, Peking Union Medical College Hospital (CAMS), China
Copyright © 2022 Sharma, Mudalagiriyappa and Nanjappa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Som Gowda Nanjappa, nanjappa@illinois.edu
†These authors have contributed equally to this work
 Jaishree Sharma
Jaishree Sharma Srinivasu Mudalagiriyappa
Srinivasu Mudalagiriyappa Som Gowda Nanjappa
Som Gowda Nanjappa