
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 05 July 2022
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.899890
This article is part of the Research Topic Innate Immunity and Intercellular Communication View all 12 articles
Since the 2004 publication of the first study describing extracellular traps (ETs) from human neutrophils, several reports have shown the presence of ETs in a variety of different animals and plants. ETs perform two important functions of immobilizing and killing invading microbes and are considered a novel part of the phagocytosis-independent, innate immune extracellular defense system. However, several pathogens can release nucleases that degrade the DNA backbone of ETs, reducing their effectiveness and resulting in increased pathogenicity. In this review, we examined the relevant literature and summarized the results on bacterial and fungal pathogens and parasites that produce nucleases to evade the ET-mediated host antimicrobial mechanism.
Pathogens have evolved various strategies to overcome the killing mechanisms of the host immune system and establish a productive infection. The secreted proteins of pathogens, which include proteases, lipases, collagenases, hyaluronidases, chitinases, and nucleases, have been found to be essential for evading the host defense system (1). Nucleases are a diverse group of enzymes that degrade both DNA and RNA, and there are several ways to classify them (2). According to Linn formulated the criteria, the most acceptable system for classification of nucleases is to divided them into sugar specific and sugar non-specific nucleases (3). Based on their activity on ester bonds, nucleases are considered a subgroup of hydrolases and can be subdivided into exonucleases (EC 3.1.11.X to EC 3.1.16.X) and endonucleases (EC 3.1.21.X to EC 3.1.31.X). For the ability to hydrolyze DNA, sugar specific nucleases are DNases including exodeoxyribonucleases and endodeoxyribonucleases, while sugar non-specific nucleases comprise endonucleases and exonucleases (2). According to their biochemical and biological properties as well as tissue-specific production, DNases can be further divided into the DNase I and DNase II families (4). Pathogen nucleases are mainly involved in physiological processes such as genetic transformation and nutrient scavenging, DNA replication, recombination and DNA repair mechanisms (5); but, they can also act as virulence factors (5), regulate of biofilm formation, enhance fitness and bacterial shedding (6), and allow microbes to enter non-phagocytic cells or survive and replicate in phagocytic cells.
Neutrophils are the most abundant type of white blood cell, and they play an important role in inflammation and host defense through the respiratory burst, degranulation, and phagocytosis (7). In 2014, Brinkmann et al. reported that when attacked by pathogens, human neutrophils released DNA-based network structures called neutrophil extracellular traps (NETs), which immobilized and/or killed pathogens (8). This phenomenon has also been observed in different mammalian species, birds, fish, invertebrates, and plants (9). However, it is also known that certain pathogens have developed strategies to avoid the killing effects of extracellular traps (ETs) by secreting a polysaccharide capsule, forming biofilms, undergoing cell surface modifications, and inhibiting ET formation (10). The main components of ETs are granule proteins and DNA, therefore it was predicted that extracellular nucleases could directly degrade the ETs’ scaffold DNA, providing an effective way for pathogens to escape the ET defensive network structures (Figure 1). In this article, we review the latest research on the role of extracellular nucleases of bacteria, fungi, parasites, and mycoplasmas in degrading the backbone DNA of ETs, especially NETs (Table 1).
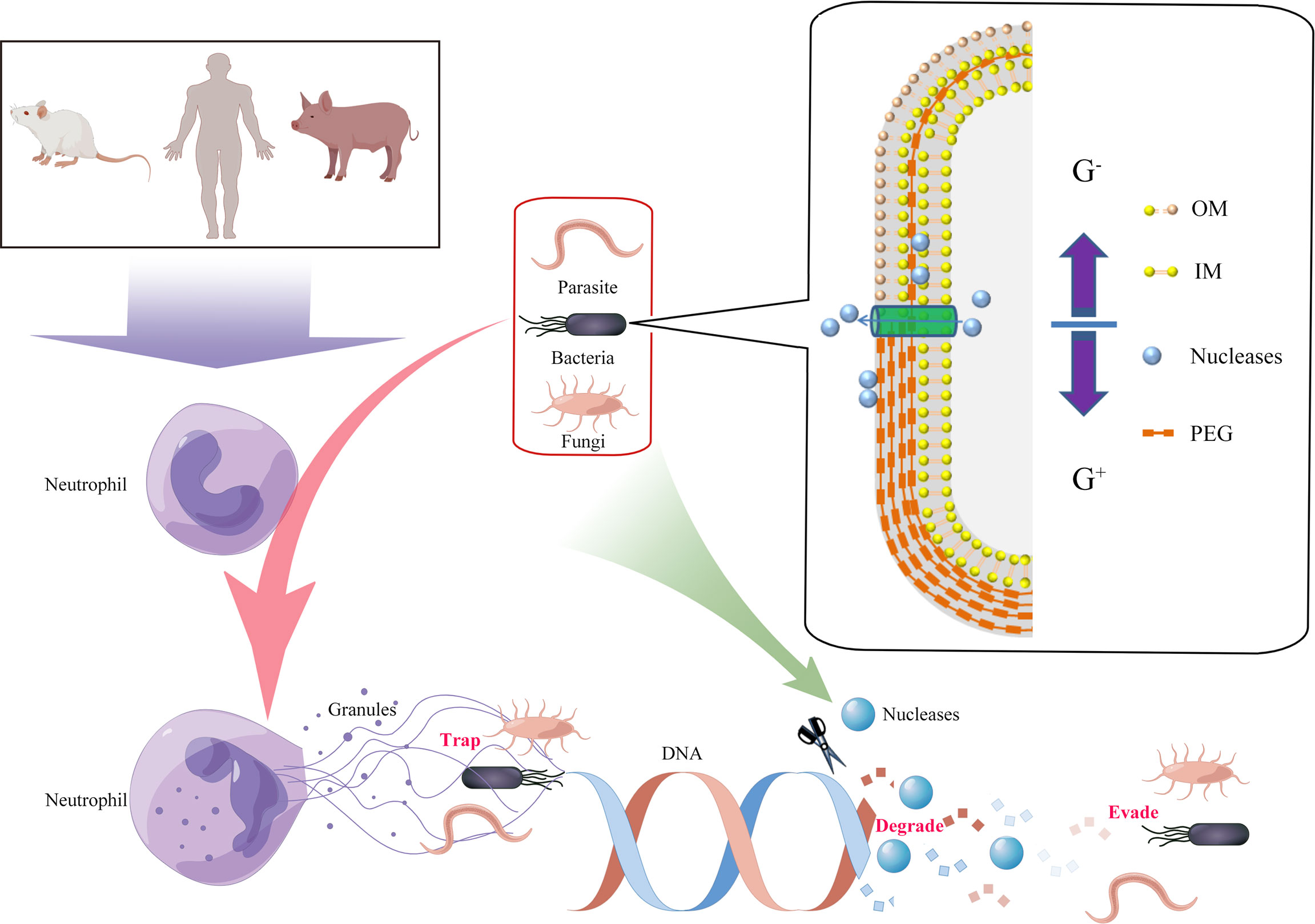
Figure 1 Degradation of extracellular traps (ETs) by the production of pathogen-derived nucleases. Innate immune cells were activated by pathogens, and DNA from nuclei or mitochondria mixed with histones, granular, and cytoplasmic proteins was released to the cellular environment (termed ETs). ETs could immobilize and/or kill the pathogens. However, secreted or cell wallanchored extracellular nucleases of pathogens were capable of destroying DNA backbones of ETs and contributing to evasion of the immune response and development of persistent infections. G+, Gram-positive bacteria; G-, Gram-negative bacteria; OM, outer membrane; IM, inner membrane; PEG, peptidoglycan.
Vibrio cholerae is a Gram-negative comma-shaped bacterium that is the causative agent of the secretory diarrheal disease cholera. Extracellular DNA is a major component of the Vibrio biofilm matrix. The two nucleases secreted by V. cholerae, Xds and Dns, are able to degrade extracellular DNA. Dns exhibits endonuclease activity, while Xds has been characterized as an exonuclease (11). Extracellular DNA degradation by the two nucleases results in accumulation of high levels of nucleotides outside the cell. OmpK was used to transport nucleotides to the outer membrane, where they were dephosphorylated by periplasmic phosphatases. Free phosphate was absorbed by the Pst/PhoU system and nucleosides were transported into cells via NupC (61). V. cholerae exposure caused human neutrophils to form NETs, while secreted Dns and Xds rapidly degraded the NETs’ DNA and released the bacteria from the network structures, thus promoting cell colonization by V. cholerae (12). Infection with a Dns/Xds deletion mutant of V. cholerae resulted in prolonged stability of the NET. The C-terminus of Xds has an exonuclease domain, and residues D787 and H837 within the exonuclease domain are the key sites of the catalytic center (13).
The endonuclease, EndA, of S. pneumoniae is localized on the membrane surface and is a key factor in DNA replication and bacterial virulence. EndA is required for the genetic transformation of S. pneumoniae and contributes to gene transfer and genetic diversification (14–16). EndA cannot bind to external DNA or degrade it when S. pneumoniae does not undergo gene conversion (17). However, in another study, it was reported that degradation of extracellular DNA by EndA did not require components of the competence regulon (18). EndA was secreted extracellularly during S. pneumoniae growth, which helped the pathogen establish an infection in the lung by degrading the DNA backbone of NETs and overcoming the host immune system (18). EndA possesses a DRGH (Asp-Arg-Gly-His) motif containing a ββα metal-finger catalytic core, and the amino acids, His160, Asn191, and Asn182 that are essential for catalytic activity. Expression of the wild-type EndA gene in E. coli was difficult due to its extreme cytotoxicity, but an EndA gene with the H160A mutation was successfully expressed. Asn182 was required for the stability of EndA (19), while Arg127/Lys128 and Arg209/Lys210 helped the enzyme bind to the substrate (20). In addition, the three mutant strains, H154A, Q186A, and Q192A, exhibited significantly decreased nuclease activities.
The twin-arginine translocase D (TatD) is a soluble cytoplasmic protein that was obtained from the extracellular secreted proteins of S. pneumoniae, but has been described in most prokaryotic and eukaryotic species (21). Although TatD functions primarily in the operation of the Tat transport system, it also exhibits endonuclease and exonuclease activity. TatD has been associated with the formation of extracellular vesicles of S. pneumoniae, and recombinant TatD (rTatD) had DNase activity. A TatD deletion mutant showed little NET degradation activity, while the addition of rTatD reduced the formation of NETs. The deletion mutant greatly reduced the bacterial load in the lung, blood, and spleen in a murine sepsis model (21). In our study, the recombinant TatD-like DNase from Listeria monocytogenes 10403s was associated with the degradation of macrophage ETs and its Mg2+-dependent nuclease activity had an optimum reaction temperature of 37°C and pH of 6.0 (62).
Group A streptococci (GAS) produce 26-30 extracellular nucleases, including Sda1, SdaD2, Spd, and Spd3. Sda1 exhibits cell wall-anchored DNase activity and highly expressed by the serotype M1 GAS strain. Sda1 contributed to enhancing the tolerance and virulence of GAS toward neutrophils in a murine model of necrotizing fasciitis. Inhibition of GAS nuclease activity with G-actin enhanced the clearance of pathogens by neutrophils in vitro and reduced virulence in vivo (63). Sda1 from invasive M1T1 GAS degraded the DNA backbone of NETs in human neutrophils. DNA degradation by Sda1 prevented GAS from inducing murine macrophages to secrete IFN-α and TNF-α, while the levels of IFN-α and TNF-α in mice were significantly decreased by Sda1-expressing GAS (64). Degradation of PMA-induced NETs by Sda1 and other phage-encoded DNases in mouse neutrophils was neutralized by antibodies from mice immunized with Sda1 (22). The prophage-encoded SdaD2 enzyme was the major DNase that contributes to GAS virulence (65). Extracellular killing of the spd3/sdaD2/spd deletion mutant of M1 GAS strain MGAS5005 in human PMNs was significantly enhanced. In addition, the virulence of the triple-mutant strain in mice was significantly reduced and bacteria were easily removed from the skin injection site (65).
The Streptococcus pyogenes nuclease A (SpnA) is an exo/endonuclease with an LPXTG motif located on the cell surface in S. pyogenes and also GAS. Two catalytic domain structures were predicted for the mature SpnA. Multiple oligonucleotide and oligosaccharide binding-fold motifs were found in the N-terminal domain at position 99-395. The C-terminal domain contained a putative endo/exonuclease domain at position 549-851. Glu592, Arg696, His716, Asp767, Asn769, Asp810, and Asp842 were necessary for SpnA activity and contributed to binding of the substrate (23). Both the full-length (99-877) and the truncated (217-877) versions of the recombinant SpnA expressed in E. coli exhibited nuclease activity. The rSpnA was capable of dismantling the NET-like structures produced by human neutrophils after stimulation with phorbol myristate acetate (PMA) (24). The nuclease activity of the spnA deletion mutant was decreased by about 70%, and antibodies against SpnA in human serum significantly inhibited nuclease activity. Although SpnA promoted bacterial survival and neutrophil killing in whole human blood, the spnA deletion mutant showed only a partial reduction in virulence in a Galleria mellonella infection model (23).
5′-nucleotidases catalyze the hydrolysis of phosphate esterified at carbon 5′ of ribonucleotides, deoxyribonucleotides, and complexnucleotides (66). 5′-nucleotidases are allocated to the enzyme commission numbers EC 3.1.3.5, and are found in all kingdoms, including plants, animals, bacteria, fungi, and parasites. The biological function of microbial 5′-nucleotidases is related to the location of the enzyme in the cell (66). Membrane-bound or cell wall-anchored 5′-nucleotidases has the ability to convert AMP into the immunomodulator adenosine, thus contributing to the evasion of the bacteria from the host immune response during infection (67). Streptococcal 5’-nucleotidase A (S5nA) is a cell wall-anchored 5′-nucleotidases and also a virulence factor of S. pyogenes N99A. Recombinant S5nA (rS5nA) produced in E. coli showed 5′-nucleotidase activity that hydrolyzed AMP, ADP, and dAMP, but not ATP, to generate adenosine and deoxyadenosine. Expression of rS5nA increased the survival of non-pathogenic Lactococcus lactis in human whole blood (68). However, the current data are still not sufficient to prove the role of S5nA in immune evasion by degrading the DNA backbone of ETs.
Streptococcal phage-encoded DNase (Spd1) is a type I extracellular DNase with 28 kDa encoded by a prophage (SF370.1) in S. pyogenes strain SF370. Spd1 existed as a monomer in solution and His121, Asn145, and Glu164 were the important conserved residues for nuclease activity (69). Although phage-encoded DNases are capable of promoting bacterial and phage particle dissemination by liquefying pus and cellular material and enhanced the survival of the phage, the crucial role of Spd1 in S. pyogenes in evading the host immune response is not fully understood.
SsnA is a secreted DNase that is anchored in the cell wall of S. suis and released into the culture medium. SsnA had strong DNase activity during exponential growth. The ssnA deletion mutant significantly reduced bacterial adherence and invasion of human laryngeal epithelial Hep-2 cells, and the virulence of the S. suis deletion mutant in CD1 mice was significantly decreased (70). SsnA was not necessary for S. suis type 2 growth and survival in human and porcine blood in vitro, but it did contribute to degradation of NETs stimulated by PMA in human and porcine neutrophils and weakened the antibacterial activity of human NETs (25).
EndAsuis is an endonuclease of S. suis with high homology and structural similarity to the membrane anchor of EndA in S. pneumoniae, but EndAsuis could not be successfully expressed in E. coli (26). The nuclease activity of a recombinant EndAsuis with a point-mutation at H165 in the DRGH motif was confirmed in the presence of Mg2+, but not Ca2+. EndAsuis was not released from S. suis into the culture medium; however, the EndAsuis deletion mutant significantly attenuated the degradation of human NETs, but had no significant effect on the NET-mediated bactericidal activity (26). High DNase activity of SsnA was found in the cerebrospinal fluid (CSF) of S. suis-infected piglets with severe suppurative meningitis. However, neither SsnA nor EndAsuis efficiently degraded NETs, and NET fibers with entrapped streptococci were observed in neutrophils from CSF (71). The up-regulated cathelicidin PR-39 in choroid plexus epithelial cells inhibited the degradation of NETs. In contrast to SsnA, host DNase 1 contributed to the enhancement of neutrophil antimicrobial activity in the CSF of S. suis-infected piglets (72).
ENuc and 5Nuc, encoded by the SESEC_RS04165 and SESEC_RS05720 genes, respectively, are two extracellular nucleases from S. equi subsp. zooepidemicus. ENuc and 5Nuc were a cell-wall anchored nucleotidase with LPXTG motif. The degradation of NETs by these nucleases reduced phagocytosis and the bactericidal capacity of macrophages, but not the intracellular killing process. An Enuc/5Nuc deletion mutant lost the ability to degrade NETs into deoxyadenosine, but it induced fewer NETs and showed greater survival in the NETs that were produced (27).
Seven genes encoding secreted nucleases from S. agalactiae have been found. NucA encoded by the gbs0661 gene displays a high degree of sequence identity with S. pneumoniae EndA and S. pyogenes Sda1. NucA contains a putative N-terminal transmembrane domain and is a major extra-cytoplasmic nuclease that required divalent cations, a stable pH, and heat stability. The nuclease activity of the H148A, R111A/H148G, K127A/H148G, K146A/H148G, and Q180A/H148G point-mutated strain was significantly decreased, while the K146R and Q183A mutant strain exhibited significantly increased activity (28). NucA was required to avoid S. agalactiae clearance from lung tissue at an early step of infection by degrading the DNA skeleton of NETs induced by PMA in mice, and contributed to dissemination in the bloodstream and persistent infection (29).
The S. mutans culture medium at 48-60 h exhibited a high level of DNase activity, which changed with the same tendency as the biofilm-released bacterial cells. The deoxyribose aldolase gene (deoC)-encoded product catalyzed the reversible aldol reaction of acetaldehyde and glyceraldehyde 3-P from the sugar phosphate, deoxyribose 5-phosphate (73) and also catabolized extracellular DNA. The nuclease DeoC was a regulator of the biofilm dispersal of S. mutans. S. mutans induced the formation of NETs in human neutrophils, and the NETs in turn enhanced the expression of the deoC gene of S. mutans. The DeoC activity of S. mutans was critical for evading killing by neutrophils through degrading NETs (30).
Morita et al. reported that oral streptococci showed extracellular DNase activity. Thirty-three cell wall-anchored proteins containing the LPXTG motif were predicted in S. sanguis. SWAN is a cell surface protein with a cell wall-sorting signal and a putative nuclease domain. The nuclease activity of a Swan deletion mutant was significantly reduced, while a strain over-expressing recombinant SWAN degraded a variety of DNAs including the DNA backbone in NETs, allowing S. sanguis to survive NETs from human neutrophils stimulated with PMA. In addition, L. lactis heterologously expressing Swan showed enhanced resistance to NET-mediated bactericidal activity (31).
The genes encoding SpnAi and S5nAi in S. iniae were similar to those for SpnA and S5nA of GAS, respectively, in terms of amino acid sequence, protein length, domain structures, and biochemical properties, as well as similar virulence mechanisms (74). The spnAi and s5nAi deletion mutants showed significant loss of DNase and nucleotidase activity, respectively. SpnAi and S5nAi supported the growth, proliferation, and dissemination of S. iniae and contributed to S. iniae virulence in zebrafish larvae. The SpnAi and S5nAi deletion mutants were still able to recruit neutrophils and macrophages to infected sites (32). NETs induced by PMA were degraded in the presence of S. iniae and also the recombinant SpnAi in neutrophils from the kidneys of adult zebrafish, but not in the presence of the spnAi deletion mutant (32).
Staphylococcal nuclease (SNase) is a nonspecific phosphodiesterase that has a strong ability to degrade a DNA scaffold to ssDNA or dsDNA, and we found that the secreted proteins of S. aureus possess DNA degradation activity (75). Similarly, Herzog et al. reported significant nuclease activity of clinical S. aureus isolates from cystic fibrosis (CF) patients (33). The nuclease activity of sequential isogenic isolates was increased in a time- and phenotype-dependent manner. Strong DNA degradation capability was observed in sputum samples from one CF patient during a long-term chronic S. aureus infection. The isolates with high nuclease activity facilitated S. aureus survival from NET-mediated killing by human neutrophils (33).
Nuc1, a thermostable nuclease, catalyzes the hydrolysis of both DNA and RNA. S. aureus eDNA maintains the structural stability of the biofilm during bacterial colonization. During the early stage of S. aureus biofilm formation, Nuc1 is expressed, which was responsible for infection persistence in a mouse subcutaneous implant model (76). Nuc1 can degrade the DNA scaffold in neutrophil NETs, thereby evading the host immune response (34). The breakdown of NET DNA by S. aureus nuclease Nuc1 contributed to the survival and dissemination of biofilm bacteria trapped in NETs and caused persistent infections (35). The ability of the nuc1 deletion mutant to degrade NETs was significantly impaired, and it was more sensitive to extracellular killing by activated neutrophils (36). Nuc2, another thermostable nuclease of S. aureus, was considered to be a cell surface-binding protein with nuclease activity. However, Nuc2 had no significant impact on NETs and was not involved in virulence and immune evasion (37).
S. aureus degrades NET DNA into deoxyadenosine to avoid its killing effect. The process triggered the caspase 3-mediated death of immune cells (38). Excessive formation of NETs contributed to the early pathological injury to the pancreas, resulting in the onset of diabetes. Lang et al. showed that a L. lactis strain expressing recombinant SNase effectively degraded PMA-induced NETs in vitro and significantly decreased circulating free DNA in the serum in non-obese diabetic (NOD) mice (39). Moreover, early treatment with SNase ameliorated the gut immune microenvironment of NOD mice by regulating the level of NETs in the intestinal mucosa (77).
Three different Yersinia enterocolitica serotypes (O:8, O:9, and O:3) were able to induce NET formation. The culture supernatants of the three serotypes also showed the capacity to degrade plasmid DNA in the presence of Ca2+/Mg2+, and NETs induced by PMA treatment of human neutrophils were degraded by the bacterial supernatants (78). Although the endonuclease-1 proteins, EndA and NucM, of Y. enterocolitica are similar to the NET-degrading V. cholerae extracellular deoxyribonuclease, their role in degrading the DNA of NETs to evade the host immune response remains unclear.
Three extracellular nucleases, EndA, ExeS, and ExeM were found in Shewanella oneidensis MR-1. ExeM was localized to the cytoplasmic membrane fraction. The activity of ExeM expressed in E. coli revealed that it was a nonspecific endonuclease requiring Ca2+ and Mg2+ or Ca2+ and Mn2+ as cofactors. ExeM was beneficial for biofilm formation under static conditions, however, externally added recombinant ExeM inhibited biofilm formation but not biofilm removal (79). Although the author hypothesized that ExeM may be related to NETs degradation, not detected in culture supernatants of S. oneidensis. Thus, the role of ExeM in NET-mediated extracellular killing is still unclear.
Sphingomyelinase catalyzes the hydrolysis of sphingomyelin into ceramide and phosphorylcholine, and is allocated to the enzyme commission numbers, EC 3.1.4.12 and EC 3.1.4.41 (80). More than six categories of sphingomyelinase have been found in a wide variety of bacteria (80, 81). The important physiological functions of sphingomyelinase are related to membrane permeability, membrane aggregation, and fusion (80). Sphingomyelinase as a virulence factor contributes to bacterial colonization, dissemination, and evasion of immune response for the establishment of persistent infection (81). Rv0888 obtained from Mycobacterium tuberculosis culture filtrate is an outer membrane protein with sphingomyelinase activity. Rv0888 increased intracellular survival, and replication of M. tuberculosis in THP-1 macrophages, but was not required for virulence of M. tuberculosis in mice (40). The C-terminal sequence of Rv0888 was highly conserved with an endonuclease-exonuclease-phosphatase domain (40). Dang et al. demonstrated that the recombinant Rv0888 expressed in E. coli required divalent metal cations (Ca2+ and Mn2+) to degrade different types of nucleic acids (41). The optimum temperature and pH for the nuclease activity were 41°C and 6.5, respectively. The activity of Rv0888 was inhibited by four traditional Chinese medicinal compounds, oleuropein, 6-gingerol, corylifolinin, and acteoside. The residues, H353, D387, D438, and H481, were essential for catalysis of Rv0888 (41). Although M. tuberculosis induced the formation of NETs and could be captured by them, the bacteria were not killed by NETs in vitro. Interestingly, the sphingomyelinase activity of recombinant Rv0888NS/MS stimulated the release of NETs from human neutrophils in vitro and in the lung tissues of mice (42).
Phosphodiesterases (EC 3.1.4.1) are a superfamily of enzymes that cleave the cyclic phosphodiester bond of cyclic adenosine and cyclic guanosine monophosphate (82). There are a total of 11 different phosphodiesterase family members (phosphodiesterase 1-11) with 21 isoforms. Phosphodiesterases are involved in diverse physiological functions and play a pivotal role in mediating the cyclic nucleotide signaling (82). In pathogens, phosphodiesterases are essential for controlling the kinetics of biofilm formation, swarming motility, survival, and dissemination (83). Pseudomonas aeruginosa has two secreted nucleases, EddA and EddB. EddA has alkaline phosphatase and phosphodiesterase activity, which can restrict the antibacterial activity of NETs (43). EddB can degrade eDNA, and the products can be taken up and used as source of nutrition by cells. The eDNA and NETs were effective inducers of the nuclease-phosphatase operon. The nucleases helped to degrade the DNA backbone of NETs and protected P. aeruginosa from NET-mediated killing. Although phosphatase did not the ability of the DNases to degrade DNA, it prevented cations from chelating phosphate from the eDNA phosphodiester backbone. Therefore, both DNase and phosphodiesterase contributed to the resistance to killing of P. aeruginosa by NETs (43).
Neisseria gonorrhoeae stimulated human neutrophils to release NETs, and the NETs showed antibacterial activity against N. gonorrhoeae. A thermostable nuclease homologue, Nuc, from N. gonorrhoeae influenced biofilm formation. Recombinant Nuc expressed in E. coli degraded DNA in PMA-induced NETs, and enhanced the survival of extracellular bacteria in human neutrophils (44).
Several studies have demonstrated that the NET-like defense structure as a mechanism is analogous to the situation in periodontitis, where bacteria colonize the periodontal region by avoiding the immune response of the host. Doke et al. reported that the periodontal pathogenic bacteria, Porphyromonas gingivalis, Prevotella intermedia, and Fusobacterium nucleatum exhibited extracellular DNA degradation activity (84). The genes, nucA and nucD, and the encoded secreted nucleases were characterized in P. intermedia (ATCC 25611). Recombinant NucA and NucD required Mg2+ and Ca2+ for optimal digestion of DNA and RNA, and their ability to degrade the DNA framework of NETs was confirmed (45). In our present study, the extracellular secreted proteins from Salmonella typhimurium, S. choleraesuis, Klebsiella pneumoniae, and Pasteurella multocida showed nuclease activity in degradation of phage λ DNA. Four proteins with nuclease activity were identified as extracellular proteins secreted from S. Typhimurium using the SDS-PAGE nuclease zymography technique combined with LC-ESI/MS/MS (85). METs -induced by Candida albicans were degraded by secreted proteins from S. choleraesuis (86). However, detailed studies are needed to assess the roles of the extracellular secreted proteins in allowing the bacteria to escape from the ETs.
In peas, corn, and cotton, extracellular DNA with histone is a component of the extracellular matrix secreted from the root cap. This structure was found to function like animal NETs to allow root border cells to capture pathogenic bacteria and fungi in the soil and immobilize them (46). Plant root border cells release DNA-containing ETs in response to the high-impact pathogenic plant bacterium, Ralstonia solanacearum. Two functional extracellular DNases, NucA and NucB, were identified in R. solanacearum strain GMI1000. The nucA/B deletion mutant was immobilized by the DNA of plant border cell traps, while the bacterial trapping was reversed by treatment with recombinant NucA and NucB (46).
3’-nucleotidase/nuclease (3′-NT/NU, EC 3.1.3.6) is a bifunctional enzyme capable of hydrolyzing 3′-monophosphorylated nucleotides and nucleic acids to generate nucleosides and inorganic phosphate. 3′-NT/NU is about 40 kDa and has five highly conserved regions. The activity of 3′-NT/NU was found in a wide variety of species, including fungi, protozoan, plants, and bacteria (87). The nuclease exhibits diverse functions in different species; however, the process of sulfate assimilation is a remarkably conserved property of the core machinery (88). The yeast nucleotide phosphatase, MET22, is closely associated with the incorporation of inorganic sulfates into amino acids, and also plays an important role in cell growth under high concentrations of Na+ and Li+. Numerous studies have reported that ETs were induced by yeast and hyphal forms of Candida spp (89). We have previously shown that METs induced by C. albicans showed the ability to trap the microbes, but with limited microbicidal activity (90). It was reported that 20% of Candida spp. showed in vitro DNase activity (91). C. albicans escape NETs through extracellular secretion of DNases that degrade the DNA backbone (92). Over recent years, several extracellular nucleotidases from fungi have been identified (93). 3′NT and 5′NT activities were determined in C. albicans and C. glabrata, and there was higher 3′NT activity at pH 4 (94). The activity was inhibited by ammonium tetrathiomolybdate (TTM), a 3′NT/NU inhibitor. NET formation and release were prevented through the 3′NT/NU activity of C. albicans and C. glabrata, and a more pronounced phagocytosis by neutrophils was observed. This feature was restored in the presence of TTM and resulted in better control and elimination of C. albicans (94).
Paracoccidioidomycosis is a fungal disease with systemic, progressive, chronic, and endemic infection. The members of Paracoccidioides spp. are the etiological agents of mycosis. P. brasiliensis induced NET formation by human PMNs in vitro, which was involved in extracellular fungicidal action (95). Recently, it was demonstrated that P. brasiliensis Pb265 and Pb18 were able to induce different patterns of NET formation (96). Pb265 is a harmless strain while Pb18 is virulent. The Pb18 strain had the ability to produce and release extracellular DNase, which degraded the DNA of NETs induced by the Pb18 strain causing them to be looser and more dispersed. Interestingly, the Pb265 strain did not consistently show DNase activity compared to the virulent strain, and its ability to induce neutrophils to form NETs was greater than that of the Pb18 strain (97).
The ET mechanism against fungal infections is also present in plants. This defensive function was abolished by the addition of DNase I, resulting in 100% incidence of root rot. The maize pathogen, C. heterostrophus, has many DNase-encoding genes and DNase activity was found in fungal culture filtrates. The deletion of the gene for secreted DNase, nuc1, significantly reduced fungal infection of leaves and roots, but virulence was restored by the addition of exogenous DNase I (98). The DNase activity of recombinant Nuc1 was dependent on Mg2+. However, the study did not directly demonstrate that C. heterostrophus used extracellular DNases as a counter mechanism against extracellular DNA in ETs from plant cells.
The class I nuclease family has been considered an important factor for several Leishmania spp. to inhibit NET-mediated killing through extracellular DNA hydrolysis (99). Leishmania spp. have a 3’-NT/NU gene with high homology to the S1 nuclease of Aspergillus oryzae and P1 of Penicillium citrinum. The 3’-NT/NU belongs to the class I nuclease family (100), and is anchored in the parasite’s cell surface membrane (101). Although the C-terminus contains the key site for the membrane anchor domain, it was not necessary for the development of enzymatic activity. The 3’-NT/NU activity did not require N-linked glycosylation, but this modification may be important for proper folding and transport of the protein to the parasite’s surface membrane (102). The 3’-NT/NU was highly constitutively expressed and helped Leishmania to infect vertebrates during the life cycle (103, 104). The 3’-NT/NU had the ability to produce extracellular adenosine through 3’-AMP hydrolysis, thus contributing to the establishment of Leishmania infection (105, 106). Inhibition of 3’-NT/NU by 3’-AMP, 5’-GMP, or the 3’-NT/NU inhibitor, TTM, decreased the survival of promastigotes in neutrophils (47). The exonuclease activity of 3’-NT/NU was directly proportional to the number of parasites, and was more efficient at alkaline pH (48). Both low phosphate (LP) and high phosphate (HP) promastigotes induced the release of NETs from human neutrophils (47), and the NET histones killed L. infantum. The L. infantum cultured in LP medium possessed higher 3’-NT/NU nuclease activity, and LP parasites showed greater resistance to neutrophil killing. The destruction of NETs can be prevented by TTM. The hydrolysis of extracellular nucleic acids by 3’-NT/NU clearly contributed to the escape and survival of parasites exposed to NETs and promoted the establishment of infection (47, 49).
DNase II belongs to a family of highly homologous DNases that can degrade DNA to produce 3′-phosphorylated and 5′-hydroxyl products. DNase II and homologs were identified in mammals, invertebrates, and non-metazoans (107). DNase II was thought to be involved in the development and homeostasis of nematodes (108). N. brasiliensis, a murine hookworm, strongly stimulated neutrophils to release NETs, but the larvae secreted Nb-DNase II which degraded the nucleic acid skeleton of NETs and reduced the neutrophil killing effect. Nb-DNase II was highly expressed at the L3 stage during early infection. The extracellular DNA fibers formed in vitro by neutrophils induced by PMA could be hydrolyzed by the recombinant Nb-DNase II, and the degradation of NETs was neutralized by antiserum against rNb-DNase II (50). DNase II activity has also been identified in the excretory/secretory products from helminths. It is noteworthy that in comparison to other nematodes, Trichinella spiralis had a more extensive expansion of the DNase II protein family with an estimated 125 genes, and almost half of those were classified as encoding secretory proteins (109). Twenty-seven developmental stage-specific genes putatively encoding DNase II homologs were isolated from a T. spiralis cDNA library by immunological screening. In our previous work, we speculated that they may play a role in digestion of host DNA to facilitate tissue migration and escape from host immune responses (110).
The TatD DNase was identified as one of the multifunctional factors in the development and transmission of malaria parasites. Chang et al. reported that a TatD-like DNase was present in every Plasmodium spp. and was highly expressed in the most virulent strains (51). The PbTatD deletion mutant of P. berghei induced J774A.1 macrophages and mouse neutrophils to generate METs and NETs in vitro, respectively; but, less ET network formation was observed in the presence of WT parasites. A recombinant PbTatD was able to restore hydrolysis of the DNA component of METs by a PbTatD deletion mutant. Interestingly, the TatD DNase activity of P. falciparum was inhibited by Mg2+, but strongly enhanced in P. knowlesi (52).
Two TatD-like DNases, TryTatD05 and TryTatD15, were identified in the supernatants of cultures of Africana trypanosome parasites, T. evansi and T. brucei, but were not released from dead parasites. T. evansi and T. brucei have been shown to induce NETs in murine neutrophils and to also be captured by NETs. However, NETs could be degraded by recombinant TryTatD05 and TryTatD15, or parasite culture supernatants (53). The degradation of NETs was significantly prevented by treatment with the DNase inhibitor, aurintricarboxylic acid. The 3’-NT/NU is a bifunctional trypanosomal enzyme, which acts as a phosphodiesterase cleaving the bond between the 3’-hydroxy and 5’-phosphoryl groups of adjacent nucleotides, or as a phosphomonoesterase to remove the 3’-terminal phosphate group of 3’-monophosphorylated nucleotides (111, 112).
Mycoplasma spp. nucleases have the ability of degradation of nucleotide substrates from host or microbial nucleic acids released in a variety of cellular processes, and several nucleases are regarded as a source of nucleotides for biosynthetic and survival purposes important and a cytotoxic factors contributing to pathogenicity (113). Intracellular, extracellularly secreted, and membrane-associated nuclease activities have been detected in most Mycoplasma spp. studied so far, and Ca2+/Mg2+ -dependent endonuclease activity has also been found in a comparatively large number of Mycoplasma spp (114). Nucleases include MGA_0676 of M. gallisepticum, MHO_0730 of M. hominis, Mhp379 and Mhp597 of M. hyopneumoniae, Mbov_rs02825 and MnuA of M. bovis, Mpn133 and Mpn491 of M. pneumoniae.
MHO_0730 is a cell-surface lipoprotein localized on the membrane of M. hominis. MHO_0730 shares significant homology with the Snase and acts as a Ca2+-dependent, sugar-nonspecific nuclease. A recombinant MHO_0730 expressed in E. coli was able to digest different nucleic acid substrates. PMA-stimulated NETs from human neutrophils were disassembled by M. hominis (54). MHO_0730 was also a potent inducer of NET formation and release through the action of the M. hominis liposoluble fraction and MHO_0730-based synthetic lipopeptides (55).
Mhp597 is a secreted nuclease of M. hyopneumoniae. The recombinant Mhp597 (rMhp597) expressed in E. coli exhibited a heat stable Ca2+- or Mg2+-dependent nuclease activity. The recombinant rMhp597 induced apoptosis and caused cytotoxicity in PK15 cells. NETs induced by PMA or M. hyopneumoniae were completely destroyed by rMhp597, as well as culture supernatants of M. hyopneumoniae (56).
MbovNase is localized in the cell membrane and a recombinant MbovNase (rMbovNase) exhibited nuclease activity at 22-65°C in the presence of Ca2+ (115). The rMbovNase Δ181-342 without the TNASE 3 domain was deficient in all biological functions. NETs were not detected in bovine neutrophils stimulated by M. bovis, and NETs induced by PMA disappeared after the addition of rMbovNase. However, the degradation of NETs and M. bovis survival were greatly reduced by the addition of EDTA (57). MnuA was the major membrane nuclease of M. bovis PG45, and NET degradation was observed in WT M. bovis. The MnuA deletion mutant lacked significant nuclease activity but still stimulated bovine neutrophils to release NETs. The generation of ROS in neutrophils was not affected by the presence or absence of mycoplasma nuclease (58).
Mpn491 is the main extracellular nuclease of M. pneumoniae and contains domains responsible for endonuclease, exonuclease, and phosphatase activities. Yamamoto et al. reported that the culture supernatants of M. pneumoniae had strong DNase activity and Mpn491 (approx. 55 kDa) was identified by zymographic analysis of the culture supernatant. The ability of the Mpn491 deletion mutant to degrade NETs from PMNs was markedly abolished (59), demonstrating that Mpn491 was essential for M. pneumoniae to evade the NET-mediated killing in vitro and in vivo.
Mycoplasma spp. have certain defects in their survival process, such as a lack of ability for de novo synthesis of nucleotides. Mycoplasma spp. secrete nucleases to obtain nucleotides from host genomic DNA to compensate for this defect. MET production induced by PMA could be observed in THP-1 cells, and was degraded by the membrane nucleases from M. hyopneumoniaea. MET degradation was inhibited by deficiency in M. hyopneumoniae nucleases. The nucleotides from MET degradation can be used for DNA synthesis (116). Therefore, nucleases are widely considered to be essential proteins for Mycoplasma, and also have been proven to be important virulence factors during infection.
It has been shown that pathogenicity and viability of Leptospira spp. were relevant factors for induction of NETs, but not for motility. A pathogenic Leptospira, L. interrogans serovar Copenhageni strain Fiocruz L1-130, induced human neutrophils to release NETs, which were capable of killing L. interrogans (60). The pathogenic Leptospira showed DNA degradation activity, but not the saprophytic Leptospira. Leptospira immunoglobulin-like proteins (Lig) are surface proteins expressed in pathogenic strains of Leptospira, including LigA, LigB, and LigC. The Lig proteins were involved in leptospiral adhesion, complement resistance, and modulation of hemostasis and play a vital role in invasion and immune evasion (117).The recombinant variable region of LigA was able to degrade DNA with both endonuclease and exonuclease activities, and could degrade PMA-induced NETs (118).
Since their discovery in 2004, ETs have been described from a wide variety of innate immune cells and are now widely regarded as an ancient and evolutionarily conserved host defense mechanism in the plant and animal kingdoms (119, 120). Several pathogens can produce one or more nucleases to directly act on the DNA skeleton of ETs (6). In this review, we summarized the current study results on nucleases expressed by pathogens to evade the ETs produced by the host immune system. The main concerns are 1) only certain pathogen-derived nucleases degrade ETs. Pathogens included V. cholerae, Streptococcus spp., S. aureus, M. tuberculosis, P. aeruginosa, N. gonorrhoeae, P. intermedius, R. solanacearum, Leishmania spp., N. brasiliensis, Plasmodium spp., Trypanosoma spp., Mycoplasma spp., and Leptospira spp. 2) types of nucleases. Thirty-four nucleases have been reported to be associated with ET degradation, and among them, eleven were exonuclease/endonuclease and twenty-three were categorized as DNase, sugar nonspecific nuclease, or nucleotidase. 3) with regard to the cellular localization of nucleases in pathogens. The formation of ETs occurs in the extracellular space of immune cells, and secreted extracellular nucleases from pathogens are the most direct way of degradating the extracellular networks. The nucleases may be anchored on the cell wall, the cell-wall surface, the membrane, or the cell-surface membrane; and parasitophorous vacuole membranes and surface proteins also have the ability to degrade ETs. 4) only ETs from certain species and cell types were used to investigate to the degradation of ETs by pathogen-derived nucleases. Neutrophils were initially used as the cells for producing and studying ETs, and considering the similar network structures of NETs and ETs from other immune cells, current research has mostly focused on the nucleases that hydrolyze the DNA of NETs. Therefore, for a number of other ETs, the role of nucleases still needs to be evaluated.
Future research will most likely focus on designing drugs to neutralize the nuclease activity of pathogens to preserve the integrity of host NETs and combat invading organisms. The protein synthesis inhibitor clindamycin and the immunoglobulins efficiently inhibited the nuclease activity of S. aureus by reducing the transcription of nuc1, resulting in enhanced NET-mediated extracellular clearance in human blood-derived neutrophils (121). The antimicrobial peptide (AMP), LL-37, a member of the cathelicidin family, facilitated the formation of NETs by human blood-derived neutrophils (122). Interestingly, LL-37 effectively prevented the nucleases of S. aureus, S. pneumoniae and GAS from degrading NETs (123), as well as the cationic AMPs, human β-defensin-3 and human neutrophil peptide-1.
ETs are thought to be dismantled by the secreted nucleases, DNase I and DNase1-like three protein (DNase1L3, also known as DNase γ) in host (124). The fragments or remnants are removed by macrophages and dendritic cells, and the process was dependent on the cytosolic exonuclease TREX1 (also known as DNaseIII) and extracellular DNase1L3, respectively (125). It is worth noting that uncontrolled NET formation or insufficient NET removal can cause highly detrimental effects to host cells in vivo such as cell damage, delay in tissue repair, inflammation, vaso-occlusion, autoantibody production, tumor capture, tumor growth, cytokine, and chemokine degradation (126). The crucial role of NETs in the pathogenesis of metabolic diseases, sepsis, thrombosis, autoimmune and autoinflammatory diseases, cancer, and other human diseases has been extensively studied and reviewed (127). The administration of DNase to dismantle ET structures can ameliorate disease progression in mouse models of breast cancer, lung injury, and systemic lupus erythematosus (128–130). Dornase alfa (Pulmozyme®, recombinant human DNase) is currently used in the clinic to treat pulmonary disease in CF (131). Dornase alfa treatment reduced the amount of NETs in the respiratory tract, leading to less airway obstruction in severe bovine RSV infection (132). Therefore, pathogen-derived nucleases should be considered as the treatment of choice for ET-mediated diseases in the future.
On the one hand, the massive amounts of ETs degraded by nucleases in vivo are helpful for preventing the occurrence of cardiovascular, immunological, and metabolic diseases, and cancer (133). On the other hand, some pathogens such as V. cholerae, S. aureus, P. intermedia,and Actinobacillus pleuropneumoniae (A. pp) may benefit from circulating cell-free DNA, adenosine, and NAD from degraded NETs as a source of nutrients. For example, A. pp did not produce extracellular DNases, whereas porcine NETs were efficiently degraded by nucleases from S. suis and the products could be used as an external NAD source for A. pp growth when co-infecting S. suis (134). Therefore, more studies are needed in the future to unambiguously determine the relationship between ET formation and degradation by pathogen nucleases or host DNases (135, 136).
CL conceived the idea and guided the whole work, and wrote the draft. FM and MQ searched the literature and wrote the draft. MQ and XW revised the manuscript. All authors read and approved the final manuscript.
The review was supported by the National Natural Science Foundation of China (32102705 and 31802159) and the youth backbone teachers training program of Henan University of Science and Technology (13450009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Pivot Sciedit for editing the manuscript. Special thanks to Figdraw (www.figdraw.com) for providing the resources and platform to draw the figure.
1. Tam K, Torres VJ. Staphylococcus Aureus Secreted Toxins and Extracellular Enzymes. Microbiol Spectr (2019) 7(2):10.1128/microbiolspec.GPP3-0039-2018. doi: 10.1128/microbiolspec.GPP3-0039-2018.
2. Yang W. Nucleases: Diversity of Structure, Function and Mechanism. Q Rev Biophys (2011) 44(1):1–93. doi: 10.1017/S0033583510000181
3. Joshi AP, Deshmukh SS. Streptomyces Nucleases. Crit Rev Microbiol (2011) 37(3):227–36. doi: 10.3109/1040841X.2011.562173
4. Lauková L, Konečná B, Janovičová Ľ, Vlková B, Celec P. Deoxyribonucleases and Their Applications in Biomedicine. Biomolecules (2020) 10(7):1036. doi: 10.3390/biom10071036
5. Sharma P, Garg N, Sharma A, Capalash N, Singh R. Nucleases of Bacterial Pathogens as Virulence Factors, Therapeutic Targets and Diagnostic Markers. Int J Med Microbiol (2019) 309(8):151354. doi: 10.1016/j.ijmm.2019.151354
6. Garcia Gonzalez J, Hernandez FJ. Nuclease Activity: An Exploitable Biomarker in Bacterial Infections. Expert Rev Mol Diagn (2022) 22(3):265–94. doi: 10.1080/14737159.2022.2049249
7. Fine N, Tasevski N, McCulloch CA, Tenenbaum HC, Glogauer M. The Neutrophil: Constant Defender and First Responder. Front Immunol (2020) 11:571085. doi: 10.3389/fimmu.2020.571085
8. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil Extracellular Traps Kill Bacteria. Science (2004) 303(5663):1532–5. doi: 10.1126/science.1092385
9. Cubillo-Martínez AA, Pereyra MA, Garfias Y, Guluarte C, Zenteno E, Sánchez-Salgado JL. Extracellular Traps Involved in Invertebrate Immune Mechanisms. Fish Shellfish Immunol (2022) 121:380–6. doi: 10.1016/j.fsi.2022.01.024
10. Ríos-López AL, González GM, Hernández-Bello R, Sánchez-González A. Avoiding the Trap: Mechanisms Developed by Pathogens to Escape Neutrophil Extracellular Traps. Microbiol Res (2021) 243:126644. doi: 10.1016/j.micres.2020.126644
11. Seper A, Fengler VH, Roier S, Wolinski H, Kohlwein SD, Bishop AL, et al. Extracellular Nucleases and Extracellular DNA Play Important Roles in Vibrio Cholerae Biofilm Formation. Mol Microbiol (2011) 82(4):1015–37. doi: 10.1111/j.1365-2958.2011.07867.x
12. Seper A, Hosseinzadeh A, Gorkiewicz G, Lichtenegger S, Roier S, Leitner DR, et al. Vibrio Cholerae Evades Neutrophil Extracellular Traps by the Activity of Two Extracellular Nucleases. PLoS Pathog (2013) 9(9):e1003614. doi: 10.1371/journal.ppat.1003614
13. Pressler K, Mitterer F, Vorkapic D, Reidl J, Oberer M, Schild S. Characterization of Vibrio Cholerae's Extracellular Nuclease Xds. Front Microbiol (2019) 10:2057. doi: 10.3389/fmicb.2019.02057
14. Lacks S, Greenberg B, Neuberger M. Role of a Deoxyribonuclease in the Genetic Transformation of Diplococcus Pneumoniae. Proc Natl Acad Sci USA (1974) 71(6):2305–9. doi: 10.1073/pnas.71.6.2305
15. Lacks S, Greenberg B, Neuberger M. Identification of a Deoxyribonuclease Implicated in Genetic Transformation of Diplococcus Pneumoniae. J Bacteriol (1975) 123(1):222–32. doi: 10.1128/jb.123.1.222-232.1975
16. Puyet A, Greenberg B, Lacks SA. Genetic and Structural Characterization of endA. A Membrane-Bound Nuclease Required for Transformation of Streptococcus Pneumoniae. J Mol Biol (1990) 213(4):727–38. doi: 10.1016/S0022-2836(05)80259-1
17. Bergé M, Moscoso M, Prudhomme M, Martin B, Claverys JP. Uptake of Transforming DNA in Gram-Positive Bacteria: A View From Streptococcus PNeumoniae. Mol Microbiol (2002) 45(2):411–21. doi: 10.1046/j.1365-2958.2002.03013
18. Zhu L, Kuang Z, Wilson BA, Lau GW. Competence-Independent Activity of Pneumococcal EndA [Corrected] Mediates Degradation of Extracellular Dna and Nets and is Important for Virulence. PLoS One (2013) 8(7):e70363. doi: 10.1371/journal.pone.0070363
19. Midon M, Schäfer P, Pingoud A, Ghosh M, Moon AF, Cuneo MJ, et al. Mutational and Biochemical Analysis of the DNA-Entry Nuclease EndA From Streptococcus Pneumoniae. Nucleic Acids Res (2011) 39(2):623–34. doi: 10.1093/nar/gkq802
20. Moon AF, Midon M, Meiss G, Pingoud A, London RE, Pedersen LC. Structural Insights Into Catalytic and Substrate Binding Mechanisms of the Strategic EndA Nuclease From Streptococcus Pneumoniae. Nucleic Acids Res (2011) 39(7):2943–53. doi: 10.1093/nar/gkq1152
21. Jhelum H, Sori H, Sehgal D. A Novel Extracellular Vesicle-Associated Endodeoxyribonuclease Helps Streptococcus Pneumoniae Evade Neutrophil Extracellular Traps and Is Required for Full Virulence. Sci Rep (2018) 8(1):7985. doi: 10.1038/s41598-018-25865-z
22. Bi S, Wang J, Xu M, Li N, Wang B. Immunity to Sda1 Protects Against Infection by Sda1+ and Sda1- Serotypes of Group A Streptococcus. Vaccines (Basel) (2022) 10(1):102. doi: 10.3390/vaccines10010102
23. Chalmers C, Khemlani AHJ, Sohn CR, Loh JMS, Tsai CJ, Proft T. Streptococcus Pyogenes Nuclease A (SpnA) Mediated Virulence Does Not Exclusively Depend on Nuclease Activity. J Microbiol Immunol Infect (2020) 53(1):42–8. doi: 10.1016/j.jmii.2017.09.006
24. Chang A, Khemlani A, Kang H, Proft T. Functional Analysis of Streptococcus Pyogenes Nuclease A (SpnA), a Novel Group A Streptococcal Virulence Factor. Mol Microbiol (2011) 79(6):1629–42. doi: 10.1111/j.1365-2958.2011.07550.x
25. de Buhr N, Neumann A, Jerjomiceva N, von Köckritz-Blickwede M, Baums CG. Streptococcus Suis DNase SsnA Contributes to Degradation of Neutrophil Extracellular Traps (NETs) and Evasion of NET-Mediated Antimicrobial Activity. Microbiol (Reading) (2014) 160(Pt 2):385–95. doi: 10.1099/mic.0.072199-0
26. de Buhr N, Stehr M, Neumann A, Naim HY, Valentin-Weigand P, von Köckritz-Blickwede M, et al. Identification of a Novel DNase of Streptococcus Suis (EndAsuis) Important for Neutrophil Extracellular Trap Degradation During Exponential Growth. Microbiol (Reading) (2015) 161(Pt 4):838–50. doi: 10.1099/mic.0.000040
27. Ma F, Guo X, Fan H. Extracellular Nucleases of Streptococcus Equi Subsp. Zooepidemicus Degrade Neutrophil Extracellular Traps and Impair Macrophage Activity of the Host. Appl Environ Microbiol (2016) 83(2):e02468–16. doi: 10.1128/AEM.02468-16
28. Moon AF, Gaudu P, Pedersen LC. Structural Characterization of the Virulence Factor Nuclease A From Streptococcus Agalactiae. Acta Crystallogr D Biol Crystallogr (2014) 70(Pt 11):2937–49. doi: 10.1107/S1399004714019725
29. Derré-Bobillot A, Cortes-Perez NG, Yamamoto Y, Kharrat P, Couvé E, Da Cunha V, et al. (Gbs0661), an Extracellular Nuclease of Streptococcus Agalactiae, Attacks the Neutrophil Extracellular Traps and Is Needed for Full Virulence. Mol Microbiol (2013) 89(3):518–31. doi: 10.1111/mmi.12295
30. Liu J, Sun L, Liu W, Guo L, Liu Z, Wei X, et al. A Nuclease From Streptococcus Mutans Facilitates Biofilm Dispersal and Escape From Killing by Neutrophil Extracellular Traps. Front Cell Infect Microbiol (2017) 7:97. doi: 10.3389/fcimb.2017.00097
31. Morita C, Sumioka R, Nakata M, Okahashi N, Wada S, Yamashiro T, et al. Cell Wall-Anchored Nuclease of Streptococcus Sanguinis Contributes to Escape From Neutrophil Extracellular Trap-Mediated Bacteriocidal Activity. PLoS One (2014) 9(8):e103125. doi: 10.1371/journal.pone.0103125
32. Soh KY, Loh JMS, Hall C, Proft T. Functional Analysis of Two Novel Streptococcus INIAE Virulence Factors Using a Zebrafish Infection Model. Microorganisms (2020) 8(9):1361. doi: 10.3390/microorganisms8091361
33. Herzog S, Dach F, de Buhr N, Niemann S, Schlagowski J, Chaves-Moreno D, et al. High Nuclease Activity of Long Persisting Staphylococcus Aureus Isolates Within the Airways of Cystic Fibrosis Patients Protects Against NET-Mediated Killing. Front Immunol (2019) 10:2552. doi: 10.3389/fimmu.2019.02552
34. Sultan AR, Hoppenbrouwers T, Lemmens-den Toom NA, Snijders SV, van Neck JW, Verbon A, et al. During the Early Stages of Staphylococcus Aureus Biofilm Formation, Induced Neutrophil Extracellular Traps Are Degraded by Autologous Thermonuclease. Infect Immun (2019) 87(12):e00605–19. doi: 10.1128/IAI.00605-19
35. Bhattacharya M, Berends ETM, Zheng X, Hill PJ, Chan R, Torres VJ, et al. Leukocidins and the Nuclease Nuc Prevent Neutrophil-Mediated Killing of Staphylococcus Aureus Biofilms. Infect Immun (2020) 88(10):e00372–20. doi: 10.1128/IAI.00372-20
36. Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. Nuclease Expression by Staphylococcus Aureus Facilitates Escape From Neutrophil Extracellular Traps. J Innate Immun (2010) 2(6):576–86. doi: 10.1159/000319909
37. Yu J, Jiang F, Zhang F, Hamushan M, Du J, Mao Y, et al. Thermonucleases Contribute to Staphylococcus Aureus Biofilm Formation in Implant-Associated Infections- A Redundant and Complementary Story. Front Microbiol (2021) 12:687888. doi: 10.3389/fmicb.2021.687888
38. Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus Aureus Degrades Neutrophil Extracellular Traps to Promote Immune Cell Death. Science (2013) 342(6160):863–6. doi: 10.1126/science.1242255
39. Lang J, Wang X, Liu K, He D, Niu P, Cao R, et al. Oral Delivery of Staphylococcal Nuclease by Lactococcus Lactis Prevents Type 1 Diabetes Mellitus in NOD Mice. Appl Microbiol Biotechnol (2017) 101(20):7653–62. doi: 10.1007/s00253-017-8480-5
40. Speer A, Sun J, Danilchanka O, Meikle V, Rowland JL, Walter K, et al. Surface Hydrolysis of Sphingomyelin by the Outer Membrane Protein Rv0888 Supports Replication of Mycobacterium Tuberculosis in Macrophages. Mol Microbiol (2015) 97(5):881–97. doi: 10.1111/mmi.13073
41. Dang G, Cao J, Cui Y, Song N, Chen L, Pang H, et al. Characterization of Rv0888, a Novel Extracellular Nuclease From Mycobacterium Tuberculosis. Sci Rep (2016) 6:19033. doi: 10.1038/srep19033
42. Ramos-Kichik V, Mondragón-Flores R, Mondragón-Castelán M, Gonzalez-Pozos S, Muñiz-Hernandez S, Rojas-Espinosa O, et al. Neutrophil Extracellular Traps Are Induced by Mycobacterium Tuberculosis. Tuberculosis (Edinb) (2009) 89(1):29–37. doi: 10.1016/j.tube.2008.09.009
43. Wilton M, Halverson TWR, Charron-Mazenod L, Parkins MD, Lewenza S. Secreted Phosphatase and Deoxyribonuclease Are Required by Pseudomonas Aeruginosa to Defend Against Neutrophil Extracellular Traps. Infect Immun (2018) 86(9):e00403–18. doi: 10.1128/IAI.00403-18
44. Juneau RA, Stevens JS, Apicella MA, Criss AK. A Thermonuclease of Neisseria Gonorrhoeae Enhances Bacterial Escape From Killing by Neutrophil Extracellular Traps. J Infect Dis (2015) 212(2):316–24. doi: 10.1093/infdis/jiv031
45. Doke M, Fukamachi H, Morisaki H, Arimoto T, Kataoka H, Kuwata H. Nucleases From Prevotella Intermedia can Degrade Neutrophil Extracellular Traps. Mol Oral Microbiol (2017) 32(4):288–300. doi: 10.1111/omi.12171
46. Tran TM, MacIntyre A, Hawes M, Allen C. Escaping Underground Nets: Extracellular DNases Degrade Plant Extracellular Traps and Contribute to Virulence of the Plant Pathogenic Bacterium Ralstonia Solanacearum. PLoS Pathog (2016) 12(6):e1005686. doi: 10.1371/journal.ppat.1005686
47. Guimarães-Costa AB, DeSouza-Vieira TS, Paletta-Silva R, Freitas-Mesquita AL, Meyer-Fernandes JR, Saraiva EM. 3'-Nucleotidase/Nuclease Activity Allows Leishmania Parasites to Escape Killing by Neutrophil Extracellular Traps. Infect Immun (2014) 82(4):1732–40. doi: 10.1128/IAI.01232-13
48. Freitas-Mesquita AL, Dick CF, Dos-Santos ALA, Nascimento MTC, Rochael NC, Saraiva EM, et al. Cloning, Expression and Purification of 3'-Nucleotidase/Nuclease, an Enzyme Responsible for the Leishmania Escape From Neutrophil Extracellular Traps. Mol Biochem Parasitol (2019) 229:6–14. doi: 10.1016/j.molbiopara.2019.02.004
49. Freitas-Mesquita AL, Meyer-Fernandes JR. 3'Nucleotidase/Nuclease in Protozoan Parasites: Molecular and Biochemical Properties and Physiological Roles. Exp Parasitol (2017) 179:1–6. doi: 10.1016/j.exppara.2017.06.001
50. Bouchery T, Moyat M, Sotillo J, Silverstein S, Volpe B, Coakley G, et al. Hookworms Evade Host Immunity by Secreting a Deoxyribonuclease to Degrade Neutrophil Extracellular Traps. Cell Host Microbe (2020) 27(2):277–89.e6. doi: 10.1016/j.chom.2020.01.011
51. Chang Z, Jiang N, Zhang Y, Lu H, Yin J, Wahlgren M, et al. The TatD-Like DNase of Plasmodium Is a Virulence Factor and a Potential Malaria Vaccine Candidate. Nat Commun (2016) 7:11537. doi: 10.1038/ncomms11537
52. Zhou Y, Xiao B, Jiang N, Sang X, Yang N, Feng Y, et al. Expression and Functional Analysis of the TatD-Like DNase of Plasmodium Knowlesi. Parasit Vectors (2018) 11(1):629. doi: 10.1186/s13071-018-3251-4
53. Zhang K, Jiang N, Chen H, Zhang N, Sang X, Feng Y, et al. TatD DNases of African Trypanosomes Confer Resistance to Host Neutrophil Extracellular Traps. Sci China Life Sci (2021) 64(4):621–32. doi: 10.1007/s11427-020-1854-2
54. Cacciotto C, Dessì D, Cubeddu T, Cocco AR, Pisano A, Tore G, et al. MHO_0730 as a Surface-Exposed Calcium-Dependent Nuclease of Mycoplasma Hominis Promoting Neutrophil Extracellular Trap Formation and Escape. J Infect Dis (2019) 220(12):1999–2008. doi: 10.1093/infdis/jiz406
55. Cacciotto C, Cubeddu T, Addis MF, Anfossi AG, Tedde V, Tore G, et al. Mycoplasma Lipoproteins Are Major Determinants of Neutrophil Extracellular Trap Formation. Cell Microbiol (2016) 18(12):1751–62. doi: 10.1111/cmi.12613
56. Li P, Zhang Y, Li X, Zhou W, Li X, Jiang F, et al. Mycoplasma Hyopneumoniae Mhp597 Is a Cytotoxicity, Inflammation and Immunosuppression Associated Nuclease. Vet Microbiol (2019) 235:53–62. doi: 10.1016/j.vetmic.2019.05.011
57. Gondaira S, Higuchi H, Nishi K, Iwano H, Nagahata H. Mycoplasma Bovis Escapes Bovine Neutrophil Extracellular Traps. Vet Microbiol (2017) 199:68–73. doi: 10.1016/j.vetmic.2016.12.022
58. Mitiku F, Hartley CA, Sansom FM, Coombe JE, Mansell PD, Beggs DS, et al. The Major Membrane Nuclease MnuA Degrades Neutrophil Extracellular Traps Induced by Mycoplasma Bovis. Vet Microbiol (2018) 218:13–9. doi: 10.1016/j.vetmic.2018.03.002
59. Yamamoto T, Kida Y, Sakamoto Y, Kuwano K. Mpn491, a Secreted Nuclease of Mycoplasma Pneumoniae, Plays a Critical Role in Evading Killing by Neutrophil Extracellular Traps. Cell Microbiol (2017) 19(3):e12666. doi: 10.1111/cmi.12666
60. Scharrig E, Carestia A, Ferrer MF, Cédola M, Pretre G, Drut R, et al. Neutrophil Extracellular Traps Are Involved in the Innate Immune Response to Infection With Leptospira. PLoS Negl Trop Dis (2015) 9(7):e0003927. doi: 10.1371/journal.pntd.0003927
61. McDonough E, Kamp H, Camilli A. Vibrio Cholerae Phosphatases Required for the Utilization of Nucleotides and Extracellular DNA as Phosphate Sources. Mol Microbiol (2016) 99(3):453–69. doi: 10.1111/mmi.13128
62. Mao FC, Zhang MK, Liao CS, Jia YY, Wang XL, Yu C, et al. Structural Characterization, Expression Purification and Activity of Recombinant Protein of Listeria Monocytogenes TatD-Like DNase. Chin Vet Sci (2020) 50(1):84–92. doi: 10.16656/j.issn.1673-4696.2020.0003
63. Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, et al. DNase Expression Allows the Pathogen Group A Streptococcus to Escape Killing in Neutrophil Extracellular Traps. Curr Biol (2006) 16(4):396–400. doi: 10.1016/j.cub.2005.12.039
64. Uchiyama S, Andreoni F, Schuepbach RA, Nizet V, Zinkernagel AS. DNase Sda1 Allows Invasive M1T1 Group A Streptococcus to Prevent TLR9-Dependent Recognition. PLoS Pathog (2012) 8(6):e1002736. doi: 10.1371/journal.ppat.1002736
65. Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, et al. Extracellular Deoxyribonuclease Made by Group A Streptococcus Assists Pathogenesis by Enhancing Evasion of the Innate Immune Response. Proc Natl Acad Sci USA (2005) 102(5):1679–84. doi: 10.1073/pnas.0406641102
66. Zimmermann H. 5'-Nucleotidase: Molecular Structure and Functional Aspects. Biochem J (1992) 285(Pt 2):345–65. doi: 10.1042/bj2850345
67. Zakataeva NP. Microbial 5'-Nucleotidases: Their Characteristics, Roles in Cellular Metabolism, and Possible Practical Applications. Appl Microbiol Biotechnol (2021) 105(20):7661–81. doi: 10.1007/s00253-021-11547-w
68. Zheng L, Khemlani A, Lorenz N, Loh JM, Langley RJ, Proft T. Streptococcal 5'-Nucleotidase A (S5nA), a Novel Streptococcus Pyogenes Virulence Factor That Facilitates Immune Evasion. J Biol Chem (2015) 290(52):31126–37. doi: 10.1074/jbc.M115.677443
69. Korczynska JE, Turkenburg JP, Taylor EJ. The Structural Characterization of a Prophage-Encoded Extracellular DNase From Streptococcus Pyogenes. Nucleic Acids Res (2012) 40(2):928–38. doi: 10.1093/nar/gkr789
70. Li M, Cai RJ, Li CL, Song S, Li Y, Jiang ZY, et al. Deletion of ssnA Attenuates the Pathogenicity of Streptococcus Suis and Confers Protection Against Serovar 2 Strain Challenge. PLoS One (2017) 12(1):e0169791. doi: 10.1371/journal.pone.0169791
71. de Buhr N, Reuner F, Neumann A, Stump-Guthier C, Tenenbaum T, Schroten H, et al. Neutrophil Extracellular Trap Formation in the Streptococcus Suis-Infected Cerebrospinal Fluid Compartment. Cell Microbiol (2017) 19(2):e12649. doi: 10.1111/cmi.12649
72. Meurer M, Öhlmann S, Bonilla MC, Valentin-Weigand P, Beineke A, Hennig-Pauka I, et al. Role of Bacterial and Host DNases on Host-Pathogen Interaction During Streptococcus Suis Meningitis. Int J Mol Sci (2020) 21(15):5289. doi: 10.3390/ijms21155289
73. Han TK, Zhu Z, Dao ML. Identification, Molecular Cloning, and Sequence Analysis of a Deoxyribose Aldolase in Streptococcus Mutans GS-5. Curr Microbiol (2004) 48(3):230–6. doi: 10.1007/s00284-003-4159-5
74. Soh KY, Loh JMS, Proft T. Orthologues of Streptococcus Pyogenes Nuclease A (SpnA) and Streptococcal 5'-Nucleotidase A (S5nA) Found in Streptococcus Iniae. J Biochem (2018) 164(2):165–71. doi: 10.1093/jb/mvy039
75. Li Q, Liao CS, Mao FC, Wang XL, Zhang MK, Zhang XJ, et al. Characterization of the Nuclease Activity of Extracellular Proteins From Staphylococcus Aureus. Chin J Prev Vet Med (2020) 42(2):175–81. doi: 10.3969/j.issn.1008-0589.201905030
76. Forson AM, Rosman CWK, van Kooten TG, van der Mei HC, Sjollema J. Micrococcal Nuclease Stimulates Staphylococcus Aureus Biofilm Formation in a Murine Implant Infection Model. Front Cell Infect Microbiol (2022) 11:799845. doi: 10.3389/fcimb.2021.799845
77. Liang Y, Wang X, He D, You Q, Zhang T, Dong W, et al. Ameliorating Gut Microenvironment Through Staphylococcal Nuclease-Mediated Intestinal NETs Degradation for Prevention of Type 1 Diabetes in NOD Mice. Life Sci (2019) 221:301–10. doi: 10.1016/j.lfs.2019.02.034
78. Möllerherm H, Neumann A, Schilcher K, Blodkamp S, Zeitouni NE, Dersch P, et al. Yersinia Enterocolitica-Mediated Degradation of Neutrophil Extracellular Traps (NETs). FEMS Microbiol Lett (2015) 362(23):fnv192. doi: 10.1093/femsle/fnv192
79. Binnenkade L, Kreienbaum M, Thormann KM. Characterization of ExeM, an Extracellular Nuclease of Shewanella Oneidensis MR-1. Front Microbiol (2018) 9:1761. doi: 10.3389/fmicb.2018.01761
80. Goñi FM, Alonso A. Sphingomyelinases: Enzymology and Membrane Activity. FEBS Lett (2002) 531(1):38–46. doi: 10.1016/s0014-5793(02)03482-8
81. Flores-Díaz M, Monturiol-Gross L, Naylor C, Alape-Girón A, Flieger A. Bacterial Sphingomyelinases and Phospholipases as Virulence Factors. Microbiol Mol Biol Rev (2016) 80(3):597–628. doi: 10.1128/MMBR.00082-15
82. Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in Targeting Cyclic Nucleotide Phosphodiesterases. Nat Rev Drug Discov (2014) 13(4):290–314. doi: 10.1038/nrd4228
83. Martínez-Méndez R, Camacho-Hernández DA, Sulvarán-Guel E, Zamorano-Sánchez D. A Trigger Phosphodiesterase Modulates the Global C-Di-GMP Pool, Motility, and Biofilm Formation in Vibrio Parahaemolyticus. J Bacteriol (2021) 203(13):e0004621. doi: 10.1128/JB.00046-21
84. Palmer LJ, Chapple IL, Wright HJ, Roberts A, Cooper PR. Extracellular Deoxyribonuclease Production by Periodontal Bacteria. J Periodontal Res (2012) 47(4):439–45. doi: 10.1111/j.1600-0765.2011.01451.x
85. Liao C, Zhang M, Cheng X, Li Q, Mao F, Wang X, et al. Identification and Characterization of the Nuclease Activity of the Extracellular Proteins From Salmonella Enterica Serovar Typhimurium. Curr Microbiol (2020) 77(11):3651–60. doi: 10.1007/s00284-020-02201-1
86. Li Q, Niu JH, Wang XL, Mao J, Quan YY, Liu GK, et al. The Nuclease Activity of Extracellular Products From Salmonella Choleraesuis and Its Effect on the Formation of Macrophages Extracellular Traps. Acta Veterinaria Zootechnica Sin (2021) 52(3):733–41. doi: 10.11843/j.issn.0366-6964.2021.03.017
87. Koval T, Dohnálek J. Characteristics and Application of S1-P1 Nucleases in Biotechnology and Medicine. Biotechnol Adv (2018) 36(3):603–12. doi: 10.1016/j.biotechadv.2017.12.007
88. Hudson BH, York JD. Roles for Nucleotide Phosphatases in Sulfate Assimilation and Skeletal Disease. Adv Biol Regul (2012) 52(1):229–38. doi: 10.1016/j.advenzreg.2011.11.002
89. Urban CF, Nett JE. Neutrophil Extracellular Traps in Fungal Infection. Semin Cell Dev Biol (2019) 89:47–57. doi: 10.1016/j.semcdb.2018.03.020
90. Liu P, Wu X, Liao C, Liu X, Du J, Shi H, et al. Escherichia Coli and Candida Albicans Induced Macrophage Extracellular Trap-Like Structures With Limited Microbicidal Activity. PLoS One (2014) 9(2):e90042. doi: 10.1371/journal.pone.0090042
91. Riceto ÉB, Menezes Rde P, Penatti MP, Pedroso Rdos S. Enzymatic and Hemolytic Activity in Different Candida Species. Rev Iberoam Micol (2015) 32(2):79–82. doi: 10.1016/j.riam.2013.11.003
92. Zhang X, Zhao S, Sun L, Li W, Wei Q, Ashman RB, et al. Different Virulence of Candida Albicans Is Attributed to the Ability of Escape From Neutrophil Extracellular Traps by Secretion of DNase. Am J Transl Res (2017) 9(1):50–62.
93. Rodrigues L, Russo-Abrahão T, Cunha RA, Gonçalves T, Meyer-Fernandes JR. Characterization of Extracellular Nucleotide Metabolism in Candida Albicans. FEMS Microbiol Lett (2016) 363(1):fnv212. doi: 10.1093/femsle/fnv212
94. Afonso M, Mestre AR, Silva G, Almeida AC, Cunha RA, Meyer-Fernandes JR, et al. Candida Extracellular Nucleotide Metabolism Promotes Neutrophils Extracellular Traps Escape. Front Cell Infect Microbiol (2021) 11:678568. doi: 10.3389/fcimb.2021.678568
95. Bachiega TF, Dias-Melicio LA, Fernandes RK, de Almeida Balderramas H, Rodrigues DR, Ximenes VF, et al. Participation of Dectin-1 Receptor on NETs Release Against Paracoccidioides Brasiliensis: Role on Extracellular Killing. Immunobiology (2016) 221(2):228–35. doi: 10.1016/j.imbio.2015.09.003
96. Della Coletta AM, Bachiega TF, de Quaglia e Silva JC, Soares ÂM, De Faveri J, Marques SA, et al. Neutrophil Extracellular Traps Identification in Tegumentary Lesions of Patients With Paracoccidioidomycosis and Different Patterns of NETs Generation In Vitro. PLoS Negl Trop Dis (2015) 9(9):e0004037. doi: 10.1371/journal.pntd.0004037
97. Zonta YR, Dezen ALO, Della Coletta AM, Yu KST, Carvalho L, Dos Santos LA, et al. Paracoccidioides Brasiliensis Releases a DNase-Like Protein That Degrades NETs and Allows for Fungal Escape. Front Cell Infect Microbiol (2021) 10:592022. doi: 10.3389/fcimb.2020.592022
98. Park HJ, Wang W, Curlango-Rivera G, Xiong Z, Lin Z, Huskey DA, et al. A DNase From a Fungal Phytopathogen Is a Virulence Factor Likely Deployed as Counter Defense Against Host-Secreted Extracellular DNA. mBio (2019) 10(2):e02805–18. doi: 10.1128/mBio.02805-18
99. Freitas-Mesquita AL, Meyer-Fernandes JR. Stage-Specific Class I Nucleases of Leishmania Play Important Roles in Parasite Infection and Survival. Front Cell Infect Microbiol (2021) 11:769933. doi: 10.3389/fcimb.2021.769933
100. Debrabant A, Gottlieb M, Dwyer DM. Isolation and Characterization of the Gene Encoding the Surface Membrane 3'-Nucleotidase/Nuclease of Leishmania Donovani. Mol Biochem Parasitol (1995) 71(1):51–63. doi: 10.1016/0166-6851(95)00035-y
101. Debrabant A, Ghedin E, Dwyer DM. Dissection of the Functional Domains of the Leishmania Surface Membrane 3'-Nucleotidase/Nuclease, a Unique Member of the Class I Nuclease Family. J Biol Chem (2000) 275(21):16366–72. doi: 10.1074/jbc.M908725199
102. Yamage M, Debrabant A, Dwyer DM. Molecular Characterization of a Hyperinducible, Surface Membrane-Anchored, Class I Nuclease of a Trypanosomatid Parasite. J Biol Chem (2000) 275(46):36369–79. doi: 10.1074/jbc.M004036200
103. Gottlieb M, Dwyer DM. Evidence for Distinct 5'- and 3'-Nucleotidase Activities in the Surface Membrane Fraction of Leishmania Donovani Promastigotes. Mol Biochem Parasitol (1983) 7(4):303–17. doi: 10.1016/0166-6851(83)90013-0
104. Kaye P, Scott P. Leishmaniasis: Complexity at the Host-Pathogen Interface. Nat Rev Microbiol (2011) 9(8):604–15. doi: 10.1038/nrmicro2608
105. Paletta-Silva R, Vieira DP, Vieira-Bernardo R, Majerowicz D, Gondim KC, Vannier-Santos MA, et al. Leishmania Amazonensis: Characterization of an Ecto-3'-Nucleotidase Activity and Its Possible Role in Virulence. Exp Parasitol (2011) 129(3):277–83. doi: 10.1016/j.exppara.2011.07.014
106. Vieira DP, Paletta-Silva R, Saraiva EM, Lopes AH, Meyer-Fernandes JR. Leishmania Chagasi: An Ecto-3'-Nucleotidase Activity Modulated by Inorganic Phosphate and Its Possible Involvement in Parasite-Macrophage Interaction. Exp Parasitol (2011) 127(3):702–7. doi: 10.1016/j.exppara.2010.11.003
107. Evans CJ, Aguilera RJ. DNase II: Genes, Enzymes and Function. Gene (2003) 322:1–15. doi: 10.1016/j.gene.2003.08.022
108. Liu MF, Wu XP, Wang XL, Yu YL, Wang WF, Chen QJ, et al. The Functions of Deoxyribonuclease II in Immunity and Development. DNA Cell Biol (2008) 27(5):223–8. doi: 10.1089/dna.2007.0691
109. Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, et al. The Draft Genome of the Parasitic Nematode Trichinella Spiralis. Nat Genet (2011) 43(3):228–35. doi: 10.1038/ng.769
110. Liao C, Liu M, Bai X, Liu P, Wang X, Li T, et al. Characterisation of a Plancitoxin-1-Like DNase II Gene in Trichinella Spiralis. PLoS Negl Trop Dis (2014) 8(8):e3097. doi: 10.1371/journal.pntd.0003097
111. Gottlieb M. The Surface Membrane 3'-Nucleotidase/Nuclease of Trypanosomatid Protozoa. Parasitol Today (1989) 5(8):257–60. doi: 10.1016/0169-4758(89)90259-7
112. Volbeda A, Lahm A, Sakiyama F, Suck D. Crystal Structure of Penicillium Citrinum P1 Nuclease at 2.8 A Resolution. EMBO J (1991) 10(7):1607–18. doi: 0.1002/j.1460-2075.1991.tb07683.x
113. Bendjennat M, Blanchard A, Loutfi M, Montagnier L, Bahraoui E. Purification and Characterization of Mycoplasma Penetrans Ca2+/Mg2+-Dependent Endonuclease. J Bacteriol (1997) 179(7):2210–20. doi: 10.1128/jb.179.7.2210-2220.1997
114. Li L, Krishnan M, Baseman JB, Kannan TR. Molecular Cloning, Expression, and Characterization of a Ca2+-Dependent, Membrane-Associated Nuclease of Mycoplasma Genitalium. J Bacteriol (2010) 192(19):4876–84. doi: 10.1128/JB.00401-10
115. Zhang H, Zhao G, Guo Y, Menghwar H, Chen Y, Chen H, et al. Mycoplasma Bovis MBOV_RS02825 Encodes a Secretory Nuclease Associated With Cytotoxicity. Int J Mol Sci (2016) 17(5):628. doi: 10.3390/ijms17050628
116. Henthorn CR, Chris Minion F, Sahin O. Utilization of Macrophage Extracellular Trap Nucleotides by Mycoplasma Hyopneumoniae. Microbiol (Reading) (2018) 164(11):1394–404. doi: 10.1099/mic.0.000717
117. Haake DA, Matsunaga J. Leptospiral Immunoglobulin-Like Domain Proteins: Roles in Virulence and Immunity. Front Immunol (2021) 11:579907. doi: 10.3389/fimmu.2020.579907
118. Kumar A, Varma VP, Sridhar K, Abdullah M, Vyas P, Ashiq Thalappil M, et al. Deciphering the Role of Leptospira Surface Protein LigA in Modulating the Host Innate Immune Response. Front Immunol (2021) 12:807775. doi: 10.3389/fimmu.2021.807775
119. Neumann A, Brogden G, von Köckritz-Blickwede M. Extracellular Traps: An Ancient Weapon of Multiple Kingdoms. Biol (Basel) (2020) 9(2):34. doi: 10.3390/biology9020034
120. Schultz BM, Acevedo OA, Kalergis AM, Bueno SM. Role of Extracellular Trap Release During Bacterial and Viral Infection. Front Microbiol (2022) 13:798853. doi: 10.3389/fmicb.2022.798853
121. Schilcher K, Andreoni F, Uchiyama S, Ogawa T, Schuepbach RA, Zinkernagel AS. Increased Neutrophil Extracellular Trap-Mediated Staphylococcus Aureus Clearance Through Inhibition of Nuclease Activity by Clindamycin and Immunoglobulin. J Infect Dis (2014) 210(3):473–82. doi: 10.1093/infdis/jiu091
122. Neumann A, Berends ET, Nerlich A, Molhoek EM, Gallo RL, Meerloo T, et al. The Antimicrobial Peptide LL-37 Facilitates the Formation of Neutrophil Extracellular Traps. Biochem J (2014) 464(1):3–11. doi: 10.1042/BJ20140778
123. Neumann A, Völlger L, Berends ET, Molhoek EM, Stapels DA, Midon M, et al. Novel Role of the Antimicrobial Peptide LL-37 in the Protection of Neutrophil Extracellular Traps Against Degradation by Bacterial Nucleases. J Innate Immun (2014) 6(6):860–8. doi: 10.1159/000363699
124. Jiménez-Alcázar M, Rangaswamy C, Panda R, Bitterling J, Simsek YJ, Long AT, et al. Host DNases Prevent Vascular Occlusion by Neutrophil Extracellular Traps. Science (2017) 358(6367):1202–6. doi: 10.1126/science.aam8897
125. Lazzaretto B, Fadeel B. Intra- and Extracellular Degradation of Neutrophil Extracellular Traps by Macrophages and Dendritic Cells. J Immunol (2019) 203(8):2276–90. doi: 10.4049/jimmunol.1800159
126. Papayannopoulos V. Neutrophil Extracellular Traps in Immunity and Disease. Nat Rev Immunol (2018) 18(2):134–47. doi: 10.1038/nri.2017.105
127. Daniel C, Leppkes M, Muñoz LE, Schley G, Schett G, Herrmann M. Extracellular DNA Traps in Inflammation, Injury and Healing. Nat Rev Nephrol (2019) 15(9):559–75. doi: 10.1038/s41581-019-0163-2
128. Macanovic M, Sinicropi D, Shak S, Baughman S, Thiru S, Lachmann PJ. The Treatment of Systemic Lupus Erythematosus (SLE) in NZB/W F1 Hybrid Mice; Studies With Recombinant Murine DNase and With Dexamethasone. Clin Exp Immunol (1996) 106(2):243–52. doi: 10.1046/j.1365-2249.1996.d01-839.x
129. Meng W, Paunel-Görgülü A, Flohé S, Witte I, Schädel-Höpfner M, Windolf J, et al. Deoxyribonuclease is a Potential Counter Regulator of Aberrant Neutrophil Extracellular Traps Formation After Major Trauma. Mediators Inflammation (2012) 2012:149560. doi: 10.1155/2012/149560
130. Jorch SK, Kubes P. An Emerging Role for Neutrophil Extracellular Traps in Noninfectious Disease. Nat Med (2017) 23(3):279–87. doi: 10.1038/nm.4294
131. Yang C, Montgomery M. Dornase Alfa for Cystic Fibrosis. Cochrane Database Syst Rev (2021) 3(3):CD001127. doi: 10.1002/14651858.CD001127.pub5
132. Cortjens B, de Jong R, Bonsing JG, van Woensel JBM, Antonis AFG, Bem RA. Local Dornase Alfa Treatment Reduces NETs-Induced Airway Obstruction During Severe RSV Infection. Thorax (2018) 73(6):578–80. doi: 10.1136/thoraxjnl-2017-210289
133. Brinkmann V. Neutrophil Extracellular Traps in the Second Decade. J Innate Immun (2018) 10(5-6):414–21. doi: 10.1159/000489829
134. de Buhr N, Bonilla MC, Pfeiffer J, Akhdar S, Schwennen C, Kahl BC, et al. Degraded Neutrophil Extracellular Traps Promote the Growth of Actinobacillus Pleuropneumoniae. Cell Death Dis (2019) 10(9):657. doi: 10.1038/s41419-019-1895-4
135. de Buhr N, von Köckritz-Blickwede M. The Balance of Neutrophil Extracellular Trap Formation and Nuclease Degradation: An Unknown Role of Bacterial Coinfections in COVID-19 Patients? mBio (2021) 12(1):e03304–20. doi: 10.1128/mBio.03304-20
Keywords: immune evasion, innate immune cells, extracellular traps, pathogens, nucleases
Citation: Liao C, Mao F, Qian M and Wang X (2022) Pathogen-Derived Nucleases: An Effective Weapon for Escaping Extracellular Traps. Front. Immunol. 13:899890. doi: 10.3389/fimmu.2022.899890
Received: 19 March 2022; Accepted: 08 June 2022;
Published: 05 July 2022.
Edited by:
Hang Yin, Tsinghua University, ChinaReviewed by:
Werner Solbach, University of Lübeck, GermanyCopyright © 2022 Liao, Mao, Qian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengshui Liao, bGlhb2NoZW5nc2h1aTMzQDE2My5jb20=; Xiaoli Wang, NjUxNjc1N0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.