- 1Department of Pathology, School of Medicine, University of Pittsburgh, Pittsburgh, PA, United States
- 2Department of Infectious Diseases and Microbiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, United States
- 3Barrier Immunity Section, Laboratory of Viral Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States
- 4Division of Infectious Diseases, Department of Medicine, School of Medicine, University of Pittsburgh, Pittsburgh, PA, United States
The lumen of the gastrointestinal (GI) tract contains an incredibly diverse and extensive collection of microorganisms that can directly stimulate the immune system. There are significant data to demonstrate that the spatial localization of the microbiome can impact viral disease pathogenesis. Here we discuss recent studies that have investigated causes and consequences of GI tract pathologies in HIV, SIV, and SARS-CoV-2 infections with HIV and SIV initiating GI pathology from the basal side and SARS-CoV-2 from the luminal side. Both these infections result in alterations of the intestinal barrier, leading to microbial translocation, persistent inflammation, and T-cell immune activation. GI tract damage is one of the major contributors to multisystem inflammatory syndrome in SARS-CoV-2-infected individuals and to the incomplete immune restoration in HIV-infected subjects, even in those with robust viral control with antiretroviral therapy. While the causes of GI tract pathologies differ between these virus families, therapeutic interventions to reduce microbial translocation-induced inflammation and improve the integrity of the GI tract may improve the prognoses of infected individuals.
Introduction
Differently from Joni Mitchell, the Canadian-American singer-songwriter and painter who doesn’t know love at all (in spite of looking at it from both sides), we know gastrointestinal (GI) tract tissue as an immune organ very well. It contains about 80% of the total leukocytes in the body (1), and most of the human microbiota (2–4) (Figure 1A). The GI tract is constantly exposed to foreign antigens from food and this exposure is critical for normal development of the mucosal immune system and immune tolerance (5–8).
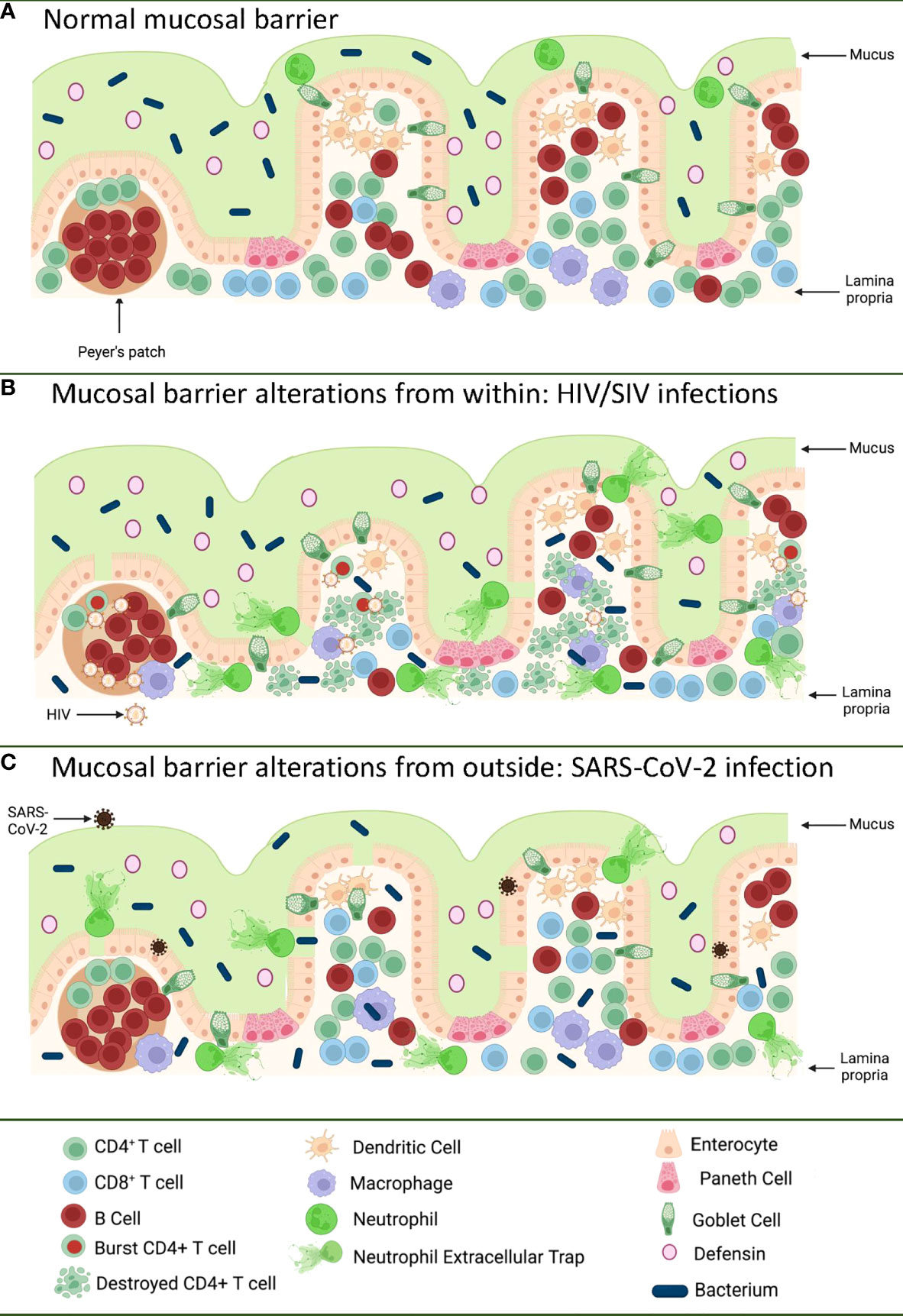
Figure 1 Pathways of the gastrointestinal tract damage in HIV/SIV and SARS-CoV-2 infections. (A) Normal GI tract is a continuous barrier which protects the internal milieu by the damage of an enormous microbiota existent in the GI lumen. This barrier is multistratified, being represented by mucus, a continuous intestinal epithelium, and immune effectors that capture translocated microbes. (B) While HIV/SIV penetrates the body at mucosal sites, GI infection occurs through systemic seeding. CD4+ T cell destruction and the inflammatory responses contribute to the destruction of the mucosal barrier from within, leading to the translocation of the intestinal flora in the lamina propria and then systemically; (C) SARS-CoV-2 infection of the enterocytes (that express high levels of the ACE-2 and TMPRSS-2 receptor) produce direct epithelial destructions also leading to translocation of the intestinal microbiota to the lamina propria and then systemically. Mucosal damage is both a major determinant of long COVID, as well as of an incomplete immune recovery even in HIV-infected individuals receiving suppressive antiretroviral therapy. Was created with BioRender.com.
The GI tract has the largest surface area exposed to the environment and the intestinal epithelia confers protection against toxic substances from food and microbes, both those normally present in the human microbiome, as well as those carried with food and water. The intestinal mucosal barrier is multilayered, with intestinal mucus, commensal bacteria, GI tract epithelium and the lamina propria immune system all contributing to host defense (9, 10) (Figure 1A). Protection is not limited to a physical barrier, but includes numerous active cell populations that exert secretory functions: goblet cells which produce mucus glycoproteins (11, 12); Paneth cells which produce antimicrobials that have the ability to specifically lyse bacteria (13); and B cells from the lamina propria which produce immunoglobulins (IgA) that capture bacteria that arriving to this gut level, preventing their successful translocation (14) (Figure 1A).
Breaches of the mucosal integrity of the GI tract are central to the pathogenesis of multiple chronic metabolic, autoimmune, and aging-related diseases (9, 10). Multiple infections can alter the integrity of the mucosal barrier including (15): human and simian immunodeficiency viruses (16–21); influenza virus infection (22); dengue (23); hepatitis B virus infection (24); hepatitis C virus infection (25); and SARS-CoV-2 (26, 27).
Furthermore, the quality of our intestinal microbiome is driving our overall morbidity (28–32). An inflammatory flora, such as the one associated with a Western diet (i.e. rich in saturated fats and sugars) drives a state of chronic inflammation, which triggers multiple systemic diseases and is roughly responsible for more than 50% of the deaths on the planet (33). Replacement with a healthy diet (i.e., Mediterranean diet rich in fiber, minerals and vitamins, and Omega 3) can alter the microbiome in as little as 3 weeks and change its phenotype to an anti-inflammatory one (34–42).
The interplay of the GI tract immune system and pathogens which disrupt this complex mucosal barrier is critically important in understanding pathogenesis, and providing targets for reducing damage. We will explore the well-studied impacts of HIV and SIV on the GI tract in addition to the parallels and distinctions that can be made in a recently emerged pandemic virus, SARS-CoV-2, and its corresponding disease, COVID-19.
Breaching the barrier from within: Mucosal pathogenesis of HIV and SIV infection
Even since the discovery of HIV, the involvement of the GI tract in the pathogenesis of AIDS was suggested by the high frequency of the gut dysfunction and wasting disease (43). Yet, the paradigm of HIV infection as a mucosal disease emerged only after the detailed characterization of the interactions between HIV and SIVs and their CD4-expressing target cells. It was reported that only the CD4+ T cell subsets that expressed high levels of CCR5 (i.e. central memory cells, transitional memory cells, and effector memory cells) are preferentially targeted by HIV and SIV (44–47) and that the main reservoir is represented by the central memory cells (48). From a functional perspective, Th-17 CD4+ T cells contribute to the maintenance of the gut integrity and are preferentially lost during progressive HIV and SIV infections (49–51). As such, since the vast majority of the effector memory cells are located at mucosal surfaces, numerous studies have shown that the first major immunologic injury inflicted by HIV/SIV to the immune system is the massive depletion of mucosal CD4+ T cells (>95%) that occurs at the mucosal sites within three weeks from infection (52–54). As memory CD4+ T cells are the preferential targets of HIV infection, their depletion is more prominent at the effector sites, such as the lamina propria, compared to inductive sites (i.e. the Peyer patches) which contain naïve CD4+ T cells (55). CD4+ T cell depletion within effector sites persists throughout chronic infection, irrespective of the virological and clinical outcome (56). Furthermore, differently from the circulating CD4+ T cells, which can be rapidly restored to preinfection levels after administration of combination antiretroviral therapy (cART), mucosal CD4+ T cell restoration is slow and incomplete (35-50% from the baseline levels) (56–58).
The severe immunologic insult produced following the interactions between HIV/SIV and their target cells within the GI tract trigger key pathogenic features of chronic SIV/HIV infection that drive disease progression (Figure 1B). Indeed, Th17 cells contribute to the maintenance of GI tract immunity through induction of mucins, claudins, and defensins, which are key components of the mucosal junctions and have antimicrobial activities; therefore, loss of Th17 cells directly compromises mucosal integrity (59). Their loss results in reduced levels of IL-17 and IL-22, which promote the recruitment of neutrophils and myeloid cells at the effector sites of the mucosa and lead to growth of epithelial cells (59–61). Alteration of the Th17/Treg ratio is associated with increased indoleamine-2,3-dioxygenase (IDO) expression by antigen-presenting cells (62–65). IDO is involved in the tryptophan metabolization (64), and IDO metabolites directly inhibit Th17 cell differentiation (66). IDO increases are also associated with decreased frequencies of CD103 antigen-presenting cells, which can induce Th17 cells (67). Altogether these features, which are specifically associated with pathogenic SIV infection and absent during the SIV infection of natural NHP hosts (in which Th17 cells are preserved) (49, 51), point to a vicious circle that leads to a continuous depletion of the Th17 population, the consequence of which is the occurrence and intensification of the mucosal damage during HIV/SIV infections.
The impact of HIV/SIV infection on the innate immune cell populations at the mucosal sites has also been extensively investigated. Progressive HIV and SIV infections lead to a reduction of both plasmacytoid dendritic cells (pDCs) and myeloid dendritic cells (mDCs) in both the peripheral blood and spleen, and alter their homing to the gut (68). Progressive infection leads to their excessive activation, leading to increased turnover in tissues (68). Similar profiles of increased apoptosis and an altered functional profile upon HIV/SIV infections are observed for the gut-resident innate lymphocyte type III cells (69–71). As a result, instead of facilitating control of the virus through recruitment to the mucosal sites, the innate immune cells produce excess of cytokines; meanwhile their high mortality triggers release of more inflammatory cytokines by the surrounding cells, further enhancing mucosal inflammation and epithelial cell activation (72). Interestingly, mDC and macrophage recruitment to the mucosal sites also occurs during the nonprogressive SIV infections of the natural hosts or controller rhesus macaques (68). This process is, however, only transient, is not associated with excessive production of inflammatory cytokines, and does not result in their excessive death, strongly suggesting that the fate of the immune cell subsets and their functions in the GI tract is driven by the local environment (73). As such, the current view is that, being programmed to fight against the infections, the innate cells migrate to the gut in progressive, as well as in nonprogressive and controlled SIV infections. Yet, the innate cells become hyperactivated only in the pathogenic infections, due to their mucosal environment, which is altered by both the virus and translocated microbial products, and thus further fuel the inflammation, deepen the damage of the mucosal barrier, and contribute to the negative outcome of HIV/SIV infection (74, 75) (Figure 1B).
The HIV/SIV-associated immunological alterations at the mucosal sites result in structural and functional pathologies of the GI tract. Virus replication, inflammation and immune activation together with bystander apoptosis of the epithelial cells throughout the GI tract result in enterocyte loss and alterations of mucosal integrity (16). Progressive HIV and SIV infections trigger enterocyte loss through multiple mechanisms: (i) the virus itself can decrease glucose uptake by enterocytes through a Tat-mediated microtubule disruption or through GP120 binding to GPR15 on epithelial cells (76, 77); (ii) increased enterocyte apoptosis occurs through bystander effects, similar to other colitis (i.e. celiac disease) (78); (iii) excessive production of inflammatory cytokines (i.e., tumor necrosis factor-TNFα by innate and adaptive immune cells from the lamina propria) at the mucosal sites lead to increased apoptosis of the epithelial cells and perturbations of the tight junction epithelial barrier (79); and (iv) loss of IL22-producing innate lymphoid cells and Th17 cells leads to decreased proliferation of enterocytes (59, 61). Loss of epithelial GI tract integrity through any of these mechanisms in progressive HIV/SIV infection is associated with inflammation (80–83).
Enterocyte loss and subsequent intestinal alterations are associated with: (i) low levels of serum citrulline (a protein that is produced by the enterocytes); (ii) decreased ratio of the villous height/crypt depth (i.e., atrophy) (84); (iii) hyperproliferation of the crypt stem cells (resulting in malabsorption) (85); (iv) Increased plasma levels of the biomarkers of enterocyte damage, i.e., intestinal fatty acid binding protein (I-FABP) (86); (v) abnormal enterocyte differentiation through alterations of the sodium glucose transport and of the concentrations of intraepithelial calcium (87–89). GI tract dysfunction occurs as early as 14 days during progressive HIV/SIV infections and is associated with colitis, diarrhea, and malabsorption (43, 90).
These pathologies are specific to pathogenic SIV infections in macaques and absent during nonpathogenic SIV infections of the African nonhuman primates that are natural hosts of SIV (91–93). In these species, the mucosal lesions characteristic to pathogenic SIV infections do not occur during either the acute or chronic stages of infection (84, 94) due to an exquisite ability to maintain gut health throughout the SIV infection (94, 95).
Breaching the barrier from outside: Mucosal pathogenesis of SARS-CoV-2 infection
SARS-CoV-2 is the etiological agent of COVID-19, a respiratory disease characterized by severe pneumonia and a plethora of symptoms suggestive of viral pneumonia: cough and sputum production, sore throat, shortness of breath, fever, myalgia, and fatigue (96–99). However, despite SARS-CoV-2 infection’s main clinical presentation as a respiratory tract infection, it may also cause symptoms associated with multiple organs, including the GI tract (diarrhea, anorexia, nausea, vomiting, and abdominal pain), liver (abnormal enzymes levels), pancreas (pancreatitis), kidney (proteinuria and hematuria, abnormal creatinine levels), brain (strokes, seizures, confusion, and brain inflammation), heart and blood vessels (elevations of cardiac injury biomarkers, palmus, chest distress, cardiac inflammation and injury, arrhythmias, and blood clots), eyes (conjunctivitis, membrane inflammation), anosmia (loss of smell), and ageusia (loss of sense of taste) (100–124).
To enter target cells, SARS-CoV-2 engages angiotensin-converting enzyme 2 (ACE2) as the entry receptor and serine protease TMPRSS2 for the Spike (S) protein priming (125, 126). Use of ACE2 is shared with SARS (127), but not with MERS, which uses a different receptor, DPP4 (128). ACE2 is widely distributed in the body, being identified in up to 72 tissues (129), and SARS-CoV-2 infection is likewise highly disseminated (130). The ACE2 protein is expressed in enterocytes, renal tubules, gallbladder, cardiomyocytes, male reproductive cells, placental trophoblasts, ductal cells, eyes, and vasculature (131). Notably, limited ACE2 expression is observed in the respiratory system both on the protein and mRNA level (132). However, a relatively limited number of cells express high levels of both ACE2 and TMPRSS2: Type II pneumocytes, nasal secretory cells, and absorptive enterocytes (131).
ACE2 expression in the human respiratory tract is highly heterogenous, being highest within regions of the sinonasal cavity (in the nasal ciliated cells) and pulmonary alveoli; these are the sites of viral transmission and severe disease development, respectively (133–137). In the lung parenchyma, ACE2 is expressed on the apical surface of a small subset of alveolar type II cells, where it was colocalized with TMPRSS2 (133–137). Interestingly, ACE2 protein expression is not reported to be lower in children, who have a lower incidence of severe COVID-19, in some studies (133); however, other investigations have described lower levels of the protein transcript in children’s airways (138).
ACE2 expression is increased in physiologic and pathologic circumstances: smoking is correlated with increased expression of the ACE2 gene in the upper airway, but lower expression in certain lung cells (139). As such, smokers are 14 times more likely to develop a severe form of the disease (140). Interferon and influenza increase ACE2 in human nasal epithelia and lung tissue (131). Some ACE2 inhibitors (i.e. lisinopril) have the ability to raise tissue levels of ACE2 in mice (141), while other studies did not find an increase of ACE2 expression in people treated with ACE2 inhibitors (137). Severe COVID-19, which is associated with high levels of inflammatory cytokines (IL-1β and type I and type III interferons), upregulates ACE2 expression, which has the potential to increase target cell availability and, thus, viral replication (131, 134, 139, 142). Yet, the impact on the variations of ACE2 expression on disease severity it is not known, and recently, it was reported that interferon-stimulated expression of ACE2 yields a truncated isoform that cannot bind SARS-CoV-2 (143).
Different clinical conditions were also reported to modulate ACE2 expression: hypertension, hyperlipidaemia, diabetes, chronic pulmonary diseases, and aging (134) (144). All these conditions are also risk factors for more severe clinical expression of COVID-19 (145–157). Note, however, that these data regarding ACE2 are highly debated and, to date, no comorbidity has been unambiguously associated with ACE2 expression level (144).
Several molecules were reported as alternative receptors for SARS-CoV-2, such as the C-type lectins DC-SIGN and L-SIGN (158–160), and TIM1 and AXL (161, 162). However, lectins and phosphatidylserine are not classical receptors for the virus: they are nonspecific and do not function efficiently in binding SARS-CoV-2 in the absence of ACE2 (163). Therefore, it was proposed that a more correct term for these molecules would be that of ‘attachment factors’ (144). CD147 is a transmembrane glycoprotein expressed ubiquitously in epithelial and immune cells, that was proposed as a receptor for SARS-CoV-2, yet its role as a viral receptor is downplayed by the observation that CD147 cannot bind to the S protein (164–166). Neuropilin 1 (NRP1) was also reported to be a host factor for SARS-CoV-2 (167, 168). NRP1 is expressed in olfactory and respiratory epithelial cells (167), yet its expression is low in the SARS-CoV-2 target cells (ciliated cells) and high in the goblet cells, which are not susceptible to SARS-CoV-2 (134, 169). B0AT1 is a virus cofactor that is expressed in the GI tract and kidney, but not in the lung; B0AT1 expression in the small intestine depends on interaction with ACE2 (170). Additional human genes are important for SARS-CoV-2 infection of lung epithelial cells: the GTPase encoded by RAB7A is critical for endocytosis, and CTSL encoding cathepsin L contributes to SARS-CoV-2 spike cleavage; yet more genes support other viral life cycle stages (171). Integrins were also reported to mediate cell entry of SARS-CoV-2 (172, 173), although other studies did not confirm these observations (174). Reduction of human ACE2 in the epithelia of K18 transgenic mice in concert with increased CTSL did not alter the pathogenesis of SARS-CoV-2 (175), further suggesting the importance of the interplay between host factors at mucosal sites for successful viral entry and propagation.
Enterocytes express ACE2 and support viral replication that is enhanced by TMPRSS2 and 4 (176, 177), and SARS-CoV-2 virions have been directly visualized in the GI tracts of COVID-19 patients (178). SARS-CoV-2 infection rapidly induces activated CD8+ T cell infiltration to the intestinal epithelium (179) and increased effector CD4+ and CD8+ T cells in the lamina propria (180). This is in spite of a lack of gross pathological changes in histological findings on endoscopy in the same patients (179, 180), though others have reported abnormalities such as crypt hyperplasia with necrotic cell debris in the absence of inflammation following a positive SARS-CoV-2 test (181). Similar to this dichotomy, several studies have reported presence (182, 183) or absence (184) of viable virus isolation from stool, while viral RNA may be shed in feces for prolonged periods compared to respiratory tract samples (185, 186); persistence of viral antigens have also been reported in GI biopsies for approximately three months following infection while nasopharyngeal swabs were negative for SARS-CoV-2 RNA (187). Such a paucity of consensus regarding the impact of viral replication on GI inflammation and/or pathology is in stark contrast to HIV/SIV infection, in which ongoing viral replication in untreated infection is a clear determinant of mucosal and systemic inflammation, although such inflammation is reduced but not eliminated with the drastic reduction of viral replication during ART (188–190).
SARS-CoV-2 infection of an in vitro GI tract model demonstrates direct damage to tight junctions and upregulated proinflammatory cytokine transcripts (191) (Figure 1C). GI symptoms in COVID-19 have also been associated with elevated liver enzymes (192) while increased markers of inflammation such as TNFα and IL-6 have separately been associated with severe and/or fatal disease (193–195). The capability of SARS-CoV-2 to enter and replicate in GI barrier cells, with corresponding immune responses and GI symptoms, suggests GI tract damage may be a critical component of COVID-19 disease.
Microbial translocation and its role in inflammation: Are lessons learned from HIV/SIV relevant to SARS-CoV-2?
GI tract dysfunction in progressive HIV and SIV infection leads to translocation of microbial products from the lumen. However, this phenomenon is not specific to SIV/HIV infection, and occurs in multiple clinical conditions in which mucosal epithelium is altered and gut permeability is increased (17). Microbial translocation is a key determinant of systemic inflammation, which is the most important driver of progressive HIV/SIV disease progression. The intestinal flora is large and diverse (approximately 1014 bacteria, fungi, protozoans, helminths, and viruses) and is composed of numerous antigens which can directly stimulate the immune system, including: peptidoglycan and lipoteichoic acid (through TLR2), lipopolysaccharide (LPS, through TLR4), flagellin (through TLR5), CpG-containing DNA (through TLR9 and other cytoplasmic sensors), and double stranded and single stranded RNAs (through TLR 7/8 and other cytoplasmic sensors) (88). Microbial translocation also includes fungal products that have relevance for immune activation and clinical outcome independently of bacterial products (196, 197). GI tract dysfunction, therefore, leads to significant inflammation with increased production of proinflammatory cytokines IL-1β, IL-6, TNFα and interferons (88).
Microbial translocation is specifically associated with progressive SIV/HIV infections and is nearly absent in African nonhuman primates that are natural hosts of SIV (198), and studies in nonhuman primates have established a direct link between microbial translocation and inflammation. Chronically SIV-infected African green monkeys (AGMs) that do not progress to AIDS maintain a healthy mucosal barrier and lack evidence of microbial translocation and systemic inflammation (91, 92, 94). However, intravenous administration of LPS, either in single dose or in prolonged administration over a three-week duration, resulted in increased levels of inflammation and coagulation markers (199). Similarly, alcohol or dextran sulphate administration to rhesus macaques increased GI tract permeability, induced microbial translocation, and resulted in increased levels of inflammation and SIV replication (200). Conversely, direct blockade of microbial translocation in progressively SIV infected Asian macaques with sevelamer, a chelator of LPS, resulted in a significant reduction of systemic inflammation and coagulation markers (201). Altogether, these studies provide direct evidence for microbial translocation as a key determinant of immune activation and associated pathologies, such as non-AIDS comorbidities, in SIV infection (202, 203).
Due to the key role of microbial translocation in the pathogenesis of HIV/SIV infection, studies have also focused on characterization of the impact of infection on the composition of the GI microbiome. Analysis of longitudinal samples from Asian macaques has shown that, while levels of enteric virus genomes increase, the bacterial microbiome is not dramatically altered (204–206). However, analyses of cross-sectional cohorts of HIV-infected and uninfected individuals routinely demonstrate the bacterial microbiomes of infected humans are altered (207–209). Recent studies have shown that one major contributor to the bacterial dysbiosis observed in HIV-infected individuals are risk factors for HIV acquisition (210, 211); when these risk factors are controlled for, significant dysbiosis is observed only in individuals with advanced HIV disease (210, 212). Moreover, while high fat diets lead to accelerated SIV disease in Asian macaques, with significantly increased inflammation (213), antibiotic-induced dysbiosis of the GI tract microbiome is insufficient to accelerate SIV disease (214).
Alteration to the GI tract virome may also play a role in disease. A significant increase in the size of the fecal virome was reported to occur in the progressive SIV infection of macaques, while no such change was detected in the nonprogressive SIV infection of AGMs (204). Furthermore, potentially pathogenic viruses, such as adenoviruses, are specifically colocalized with the areas of structural damage of the GI tract in progressively SIV-infected macaques (204). Finally, analysis of circulating microbial nucleic acids and those in tissues have demonstrated that microbes which translocate are not a representation of those present within the lumen, and the individual types of translocating organisms can be associated with prognosis (20, 205). Taken together it is clear that GI tract dysfunction, microbial translocation, and resulting inflammation play important roles in progressive HIV and SIV infections.
Alterations to the GI tract bacterial microbiome have been reported in hospitalized (215, 216) and even asymptomatic COVID-19 patients (217), though it is challenging to control for the confounding effects of diet, environment, and chronic conditions between infected and uninfected individuals to assess changes in microbial communities. K18 transgenic mice with a controlled diet and environment demonstrate dose-dependent GI tract microbiome alterations with SARS-CoV-2 infection (218), but the integrity of the intestinal barrier was not assessed. However, inflammation of the intestine itself has been implicated in SARS-CoV-2 infection, as COVID-19 patients with diarrhea demonstrated significantly higher levels of fecal calprotectin, largely produced by neutrophils and an indication of neutrophilic inflammation, which correlated with systemic IL-6 levels (219). Additionally, GI tract microbial dysbiosis and an increase in LPS-binding protein (LBP) were observed in severe COVID-19 patients over those with mild COVID-19, with LBP correlating to other inflammatory markers such as C-reactive protein (CRP) and IL-6 (220). Furthermore, bacterial proteins were found in COVID-19 patient blood plasma (220). Finally, in a comprehensive study by Giron et al., the tight junction protein zonulin was significantly elevated in COVID-19 patients with moderate or severe disease over controls, as were LBP and the product of monocyte inflammation in response to LPS, soluble CD14 (221). The levels of zonulin and LBP were correlated with a number of systemic inflammatory markers, again including IL-6 and CRP (221). Interestingly, both in Giron et al. (221) and another study from Hoel et al. investigating GI tract barrier integrity in COVID-19 patients (222), there was an increase in LBP without an increase in I-FABP indicative of enterocyte damage, suggesting that the epithelial barrier is disrupted by another means. The translocation of microbes and/or microbial products across a damaged intestinal epithelium, however, can induce systemic inflammation and contribute to the pathogenesis of SARS-CoV-2 infection (Figure 1C), as in HIV and SIV infection. Furthermore, intestinal dysbiosis in HIV infection was reported to be associated with low CD4+ T cell reconstitution, which may be relevant for COVID-19-associated lymphopenia (223).
While systemic inflammation, including that which may be induced by microbial translocation, is associated with COVID-19 mortality, there are additional mechanisms in which inflammation influences COVID-19 morbidity. Symptoms may persist or recur after primary infection, leading to the diagnosis of Post-Acute Sequalae of SARS-CoV-2 (PASC) or “long COVID-19” (https://recovercovid.org). Multisystem inflammatory syndrome can also occur in children (MIS-C) or adults (MIS-A) following COVID-19 diagnosis (https://www.cdc.gov/mis/about.html), and is manifested by severe organ system inflammation similar to Kawasaki disease that can occur in the presence or absence of viral antigen (224) and may be attributed to super-antigen-like attributes of SARS-CoV-2 spike protein (225). Notably, children and adults exhibit differential inflammatory responses during primary COVID-19, with adults demonstrating higher levels of LBP and IL-6, while healthy adult and pediatric controls were not significantly different in these markers (226). However, children with MIS had higher rates of GI symptoms than children with primary COVID-19, as well as increased zonulin, LBP, and IL-6 in the early stage of MIS-C (226, 227). Furthermore, mortality in MIS-C cases and primary severe pediatric COVID-19 is similar (228), suggesting that the high levels of inflammation in MISC-C may contribute to mortality as in adult COVID-19 cases. The impact of GI tract barrier disruption has been minimally explored in MIS-A or PASC cases, with one study reporting gut microbiome dysbiosis in adults with PASC at six months post-infection versus convalescent COVID-19 patients without PASC, who had returned to microbial communities similar to previously uninfected individuals (229). An additional study observed higher TNFα and IP-10 in the early recovery phase from primary COVID-19 in adults who would go on to experience PASC (230). Understanding the mechanisms of PASC and MIS, including GI damage, microbial translation, and resulting inflammation that may contribute to mortality, is therefore of critical importance. Insights from HIV/SIV infections that persistent immune activation and inflammation may occur with low levels or absence of viral antigen during virologically suppressive antiretroviral therapy (188–190) are the foundation upon which a more detailed knowledge of inflammation following primary COVID-19 may be built to provide prevention and treatment strategies.
Therapeutic approaches aimed at limiting the impact of gut dysfunction on the outcome of HIV and SARS-CoV-2 infections
Although ART has dramatically improved the lifespan of individuals living with HIV, with life expectancy reaching near that of uninfected individuals (231, 232), treatment neither eliminates the virus nor all inflammation (233, 234). Therapeutics to complement ART and reduce the GI tract dysfunction and inflammation experienced from early infection on have taken many forms, from microbial products to probiotics to small peptides such as an apoA-I mimetic (235). Additionally, immunomodulatory treatments for reducing GI inflammation in inflammatory bowel diseases (IBD) have been assessed, and at least one therapy was evaluated for loss of gut barrier integrity and inflammation in a MIS-C case (227). The shared mechanisms of GI tract permeability and resulting inflammation in these infectious and chronic conditions suggest that strategies to effectively address inflammation in one condition may prove beneficial in another as well.
Gut microbiota are key regulators of GI tract immunity, and promotion of anti-inflammatory functions can be attempted in many ways, including provision of prebiotics, probiotics, and microbial metabolic products. Prebiotic therapies including bacterial energy sources such as short and long chain oligosaccharides have shown modest improvements to gut-related inflammation in HIV infected individuals, with significant reductions in CRP and IL-6 (236) or sCD14 (237). However, these studies were conducted in small numbers of individuals, and only demonstrated these effects in people not receiving cART (237) or individuals who had initiated treatment but poorly reconstituted CD4+ T cell counts of <350, and without significant change to gut microbiota alpha diversity (236). Polyphenol, a key component of the Amazonian fruit Camu Camu (CC), has also been suggested as a prebiotic candidate based on its anti-inflammatory and antioxidant properties in animal models and tobacco smokers, and is under investigation for use in HIV infected individuals (238, 239).
Directly modifying the gut microbiota through administration of microbial strains as probiotics has also been trialed in HIV patients receiving cART to mixed results: men with CD4+ T cell counts <350 did not experience changes in systemic inflammation with probiotics including eight bacterial strains, and may have experienced increased T cell activation (240); two additional studies with distinct single bacterial strain probiotics observed no significant changes with treatment (241, 242); a study with multi-strain bacterial probiotics has demonstrated reductions in systemic inflammatory markers (D-dimer, IL-6, CRP), but no reductions in LPS or sCD14 (243); one study has shown improved gut barrier health with lower enterocyte apoptosis in the intestine and increased Th17 cell in GALT with high-dose, multi-strain bacterial probiotics (244). Probiotic effects (or lack thereof) may be influenced by a number of factors such as the strain(s) used, dose, and duration of treatment; in the studies detailed above, gut bacterial microbiome alterations were not assessed (241) or not observed (242) in the single bacterial strain probiotic treatments, with only multi-strain treatments demonstrating changes to the microbial communities (243, 244). Attempts to alter the complex gut microbiota may therefore require complex therapeutics, and indeed combinations of pre- and probiotics (synbiotics) have been utilized. However, like their probiotic counterparts, these studies have shown mixed results, with unaltered sCD14 and CRP levels in women (245), reduction in IL-6 in ART-naïve individuals (246), and lessened gut dysfunction in ART-treated macaques (247).
Supplementation with microbial metabolic products such as short-chain fatty acids, which are produced by GI tract microbiota through fiber fermentation and promote intestinal homeostasis (248, 249), has long been sought as a means of reducing GI tract inflammation (250). A recent study utilizing sodium propionate in conjunction with cART has shown a transient increase in circulating IL-17, but consistent decline in CD4+ Th17 and Treg cells (251), which may not promote improved gut dysfunction.
Additional microbial therapies to promote intestinal barrier integrity warrant further investigation, however: mucosaly-associated fungi promoted IL-22 and IL-17 production in the intestine of mice, promoting barrier integrity and reducing damage during infection (252). Modulating bacterial communities to specifically reduce those associated with enhanced inflammation, rather than providing beneficial bacteria as probiotics, also may be a promising alternative approach: bacteriophage mediated delivery of CRISPR-Cas9 has successfully reduced specific bacterial strains in the intestines of mice (253).
Although most therapies for reduced inflammation induced by GI tract damage target the gut microbiota, another means of modulating dysregulated gut inflammation includes apoA-I mimetics, which bind LPS and lipids. Not only has an apoA-I mimetic peptide demonstrated reduction of HDL cholesterol ex vivo (254), but the molecule and another mimetic have also reduced inflammatory cytokines such as TNFα and IL-6 in the plasma of HIV-infected humanized mice (235). These peptides do not directly interact with the virus, and have already been implicated in treatment of chronic non-infectious inflammatory GI tract conditions such as inflammatory bowel disease (IBD) (255). Investigated as a complement to ART, apoA-I mimetics could be an excellent candidate for reduction of HIV or SARS-CoV-2 induced GI tract dysregulation and inflammation.
Steroids are a clear treatment for consideration to reduce inflammation, but are not components of standard therapies for individuals living with HIV. However, in an acute infection characterized by hyperinflammatory conditions such as COVID-19, the immunosuppressive effects of corticosteroids have been beneficial: in severe COVID-19 patients, moderate doses of dexamethasone administered for a short duration reduced the duration of hospitalization and mortality (256–260).
Finally, cell signaling approaches have been taken to reduce inflammation resulting from GI tract disruption. In a case of severe MIS-C, inhibiting zonulin signaling with a zonulin receptor agonist was undertaken to improve tight junctions, with tight junction loss hypothesized to lead to antigenemia and severe systemic inflammation (227). The child’s condition did improve with treatment, as evidenced by decreased CRP, D-dimer, and indeed lower SARS-CoV-2 spike protein in the blood (227). This virus-independent means of reducing GI tract disruption, which is currently approved for a clinical trial for celiac disease treatment (261), may be appropriate for HIV as well, as might anti-inflammatory treatments for other chronic immune conditions such as IBD. Although TNF antagonist and immunosuppressive thiopurine treatment was associated with risk of hospitalization or death from COVID-19, TNF antagonist treatment alone was associated with lower odds ratios of hospitalization or death (262). Treatment with anti-TNFα antibodies has proven successful at reducing inflammation in clinical trials (263) and may be a safe strategy for reducing GI tract inflammation that results from viral infection, either chronically in HIV or acutely in SARS-CoV-2; indeed anti-TNFα antibodies were successful at reducing pulmonary pathology in a case study of a COVID-19 patient (264) and in inflammation and pathology in progressive SIV infection (79). Furthermore, anti-IL-6 therapies have been investigated for HIV and SARS-CoV-2 and proposed for inflammatory gut diseases, though efficacy has been mixed for both viral infections (265–268).
In conclusion, despite the distinctions of SARS-CoV-2 and HIV infections in terms of target cells, viral persistence, and symptomatology, there are considerable parallels in the loss of gut barrier integrity and corresponding inflammation that results. These parallels suggest that therapies to address chronic HIV inflammation, as well as that of non-infectious diseases, may be appropriate for treating SARS-CoV-2. Although the infection is acute rather than chronic, MIS cases strongly suggest persistent or recrudescent damage of organ systems including the GI tract that can lead to serious and fatal inflammation. Treatment therapies to reduce GI tract damage and/or resulting inflammation may therefore not only improve acute SARS-CoV-2 infection outcomes, but also improve morbidity and mortality associated with subsequent multisystem inflammation.
Author contributions
IP KB, JB, and CA designed, wrote, and edited the manuscript. All authors contributed to literature screening, writing, and editing and approved the submitted version.
Funding
IP and CA are supported by grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases/National Heart, Lung and Blood Institute/National Institute of Allergy and Infectious Diseases: R01 DK130481 (IP), R01 DK113919 (IP/CA), R01 DK119936 (CA), R01 DK131476 (CA), RO1 HL117715 (IP), R01 HL123096 (IP), R01 HL154862 (IP), R01 AI119346 (CA). This study was funded, in part, by the Division of Intramural Research, NIAID. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgement
Figure 1 was created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wiertsema SP, van Bergenhenegouwen J, Garssen J, Knippels LMJ. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients 13 (2021). doi: 10.3390/nu13030886
2. Dickson RP. The microbiome and critical illness. Lancet Respir Med (2016) 4:59–72. doi: 10.1016/S2213-2600(15)00427-0
3. Miniet AA, Grunwell JR, Coopersmith CM. The microbiome and the immune system in critical illness. Curr Opin Crit Care (2021) 27:157–63. doi: 10.1097/MCC.0000000000000800
4. Virgin HW. The virome in mammalian physiology and disease. Cell (2014) 157:142–50. doi: 10.1016/j.cell.2014.02.032
5. Rodriguez-Sillke Y, Visekruna A, Glauben R, Siegmund B, Steinhoff U. Recognition of food antigens by the mucosal and systemic immune system: Consequences for intestinal development and homeostasis. Int J Med Microbiol (2021) 311:151493. doi: 10.1016/j.ijmm.2021.151493
6. Ku HJ, Kim YT, Lee JH. Microbiome study of initial gut microbiota from newborn infants to children reveals that diet determines its compositional development. J Microbiol Biotechnol (2020) 30:1067–71. doi: 10.4014/jmb.2002.02042
7. Mohammadkhah AI, Simpson EB, Patterson SG, Ferguson JF. Development of the gut microbiome in children, and lifetime implications for obesity and cardiometabolic disease. Children (Basel) 5 (2018). doi: 10.3390/children5120160
8. Mendez CS, Bueno SM, Kalergis AM. Contribution of gut microbiota to immune tolerance in infants. J Immunol Res (2021) 2021:7823316. doi: 10.1155/2021/7823316
9. Martel J, Chang SH, Ko YF, Hwang TL, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends Endocrinol Metab (2022). doi: 10.1016/j.tem.2022.01.002
10. Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol (2009) 9:799–809. doi: 10.1038/nri2653
11. Knoop KA, Newberry RD. Goblet cells: multifaceted players in immunity at mucosal surfaces. Mucosal Immunol (2018) 11:1551–7. doi: 10.1038/s41385-018-0039-y
12. Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut (2020) 69:2232–43. doi: 10.1136/gutjnl-2020-322260
13. Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol (2011) 9:356–68. doi: 10.1038/nrmicro2546
14. Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. The intestinal epithelium: Central coordinator of mcosal immunity. Trends Immunol (2018) 39:677–96. doi: 10.1016/j.it.2018.04.002
15. Serek P, Oleksy-Wawrzyniak M. The effect of bacterial infections, probiotics and zonulin on intestinal barrier integrity. Int J Mol Sci 22 (2021). doi: 10.3390/ijms222111359
16. Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS (2008) 3:356–61. doi: 10.1097/COH.0b013e3282f9ae9c
17. Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol (2012) 30:149–73. doi: 10.1146/annurev-immunol-020711-075001
18. Brenchley JM, Price DA, Douek DC. HIV Disease: fallout from a mucosal catastrophe? Nat Immunol (2006) 7:235–9. doi: 10.1038/ni1316
19. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med (2006) 12:1365–71. doi: 10.1038/nm1511
20. Nganou-Makamdop K, Talla A, Sharma AA, Darko S, Ransier A, Laboune F, et al. Translocated microbiome composition determines immunological outcome in treated HIV infection. Cell (2021) 184:3899–914.e16. doi: 10.1016/j.cell.2021.05.023
21. Kleinman AJ, Pandrea I, Apetrei C. So pathogenic or so what?-A brief overview of SIV pathogenesis with an emphasis on cure research. Viruses 14 (2022). doi: 10.3390/v14010135
22. Sencio V, Gallerand A, Gomes Machado M, Deruyter L, Heumel S, Soulard D, et al. Influenza virus infection impairs the gut’s barrier properties and favors secondary enteric bacterial infection through reduced production of short-chain fatty acids. Infect Immun (2021) 89:e0073420.
23. van de Weg CA, Pannuti CS, de Araujo ES, van den Ham HJ, Andeweg AC, Boas LS, et al. Microbial translocation is associated with extensive immune activation in dengue virus infected patients with severe disease. PLoS Negl Trop Dis (2013) 7:e2236.
24. Kassa Y, Million Y, Gedefie A, Moges F. Alteration of gut microbiota and its impact on immune response in patients with chronic HBV infection: A review. Infect Drug Resist (2021) 14:2571–8. doi: 10.2147/IDR.S305901
25. Preveden T, Scarpellini E, Milic N, Luzza F, Abenavoli L. Gut microbiota changes and chronic hepatitis c virus infection. Expert Rev Gastroenterol Hepatol (2017) 11:813–9. doi: 10.1080/17474124.2017.1343663
26. Devaux CA, Lagier JC, Raoult D. New insights into the physiopathology of COVID-19: SARS-CoV-2-associated gastrointestinal illness. Front Med (Lausanne) (2021) 8:640073. doi: 10.3389/fmed.2021.640073
27. Johnson SD, Olwenyi OA, Bhyravbhatla N, Thurman M, Pandey K, Klug EA, et al. Therapeutic implications of SARS-CoV-2 dysregulation of the gut-brain-lung axis. World J Gastroenterol (2021) 27:4763–83. doi: 10.3748/wjg.v27.i29.4763
28. Fromentin S, Forslund SK, Chechi K, Aron-Wisnewsky J, Chakaroun R, Nielsen T, et al. Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat Med (2022) 28:303–14. doi: 10.1038/s41591-022-01688-4
29. Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science (2016) 352:544–5. doi: 10.1126/science.aad9358
30. Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nat Rev Endocrinol (2015) 11:182–90. doi: 10.1038/nrendo.2014.210
31. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature (2020) 579:567–74. doi: 10.1038/s41586-020-2095-1
32. Jasinska AJ, Dong TS, Lagishetty V, Katzka W, Jacobs JP, Schmitt CA, et al. Shifts in microbial diversity, composition, and functionality in the gut and genital microbiome during a natural SIV infection in vervet monkeys. Microbiome (2020) 8:154. doi: 10.1186/s40168-020-00928-4
33. Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med (2019) 25:1822–32. doi: 10.1038/s41591-019-0675-0
34. Del Chierico F, Vernocchi P, Dallapiccola B, Putignani L. Mediterranean Diet and health: food effects on gut microbiota and disease control. Int J Mol Sci (2014) 15:11678–99. doi: 10.3390/ijms150711678
35. De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut (2016) 65:1812–21. doi: 10.1136/gutjnl-2015-309957
36. Mitsou EK, Kakali A, Antonopoulou S, Mountzouris KC, Yannakoulia M, Panagiotakos DB, et al. Adherence to the Mediterranean diet is associated with the gut microbiota pattern and gastrointestinal characteristics in an adult population. Br J Nutr (2017) 117:1645–55. doi: 10.1017/S0007114517001593
37. Nagpal R, Shively CA, Appt SA, Register TC, Michalson KT, Vitolins MZ, et al. Gut microbiome composition in non-human primates consuming a Western or Mediterranean diet. Front Nutr (2018) 5:28. doi: 10.3389/fnut.2018.00028
38. Podadera-Herreros A, Alcala-Diaz JF, Gutierrez-Mariscal FM, Jimenez-Torres J, Cruz-Ares S, Arenas-de Larriva AP, et al. Long-term consumption of a mediterranean diet or a low-fat diet on kidney function in coronary heart disease patients: The CORDIOPREV randomized controlled trial. Clin Nutr (2022) 41:552–9. doi: 10.1016/j.clnu.2021.12.041
39. Estruch R, Ros E, Salas-Salvado J, Covas MI, Corella D, Aros F, et al. Primary prevention of cardiovascular disease with a Mediterranean diet. N Engl J Med (2013) 368:1279–90. doi: 10.1056/NEJMoa1200303
40. Pauwels EK. The protective effect of the Mediterranean diet: focus on cancer and cardiovascular risk. Med Princ Pract (2011) 20:103–11. doi: 10.1159/000321197
41. Panagiotakos DB, Dimakopoulou K, Katsouyanni K, Bellander T, Grau M, Koenig W, et al. Mediterranean Diet and inflammatory response in myocardial infarction survivors. Int J Epidemiol (2009) 38:856–66. doi: 10.1093/ije/dyp142
42. Feart C, Samieri C, Alles B, Barberger-Gateau P. Potential benefits of adherence to the Mediterranean diet on cognitive health. Proc Nutr Soc (2013) 72:140–52. doi: 10.1017/S0029665112002959
43. Kotler DP, Gaetz HP, Lange M, Klein EB, Holt PR. Enteropathy associated with the acquired immunodeficiency syndrome. Ann Intern Med (1984) 101:421–8. doi: 10.7326/0003-4819-101-4-421
44. Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science (1998) 280:427–31. doi: 10.1126/science.280.5362.427
45. Veazey RS, Lackner AA. The gastrointestinal tract and the pathogenesis of AIDS. AIDS 12 Suppl A (1998), S35–42.
46. Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4+ T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J Virol (1998) 72:6646–56. doi: 10.1128/JVI.72.8.6646-6656.1998
47. Jasinska AJ, Pandrea I, Apetrei C. CCR5 as a coreceptor for human immunodeficiency virus and simian immunodeficiency viruses: A prototypic love-hate affair. Front Immunol (2022) 13:835994. doi: 10.3389/fimmu.2022.835994
48. Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV Reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med (2009) 15:893–900. doi: 10.1038/nm.1972
49. Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood (2008) 112:2826–35. doi: 10.1182/blood-2008-05-159301
50. Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol (2008) 1:279–88. doi: 10.1038/mi.2008.14
51. Favre D, Lederer S, Kanwar B, Ma ZM, Proll S, Kasakow Z, et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog (2009) 5:e1000295. doi: 10.1371/journal.ppat.1000295
52. Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature (2005) 434:1148–52. doi: 10.1038/nature03513
53. Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature (2005) 434:1093–7. doi: 10.1038/nature03501
54. Pandrea IV, Gautam R, Ribeiro RM, Brenchley JM, Butler IF, Pattison M, et al. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol (2007) 179:3035–46. doi: 10.4049/jimmunol.179.5.3035
55. Veazey RS, Marx PA, Lackner AA. The mucosal immune system: primary target for HIV infection and AIDS. Trends Immunol (2001) 22:626–33. doi: 10.1016/S1471-4906(01)02039-7
56. Le Hingrat Q, Sereti I, Landay AL, Pandrea I, Apetrei C. The hitchhiker guide to CD4+ T-cell depletion in lentiviral infection. A critical review of the dynamics of the CD4+ T cells in SIV and HIV infection. Front Immunol (2021) 12:695674. doi: 10.3389/fimmu.2021.695674
57. Mehandru S, Poles MA, Tenner-Racz K, Jean-Pierre P, Manuelli V, Lopez P, et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med (2006) 3:e484.
58. Saluzzo S, Pandey RV, Gail LM, Dingelmaier-Hovorka R, Kleissl L, Shaw L, et al. Delayed antiretroviral therapy in HIV-infected individuals leads to irreversible depletion of skin- and mucosa-resident memory T cells. Immunity (2021) 54:2842–58.e5. doi: 10.1016/j.immuni.2021.10.021
59. Stockinger B, Omenetti S. The dichotomous nature of T helper 17 cells. Nat Rev Immunol (2017) 17:535–44. doi: 10.1038/nri.2017.50
60. Neil JA, Matsuzawa-Ishimoto Y, Kernbauer-Holzl E, Schuster SL, Sota S, Venzon M, et al. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat Microbiol (2019) 4:1737–49. doi: 10.1038/s41564-019-0470-1
61. Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med (2009) 206:1465–72. doi: 10.1084/jem.20082683
62. Page EE, Greathead L, Metcalf R, Clark SA, Hart M, Fuchs D, et al. Loss of Th22 cells is associated with increased immune activation and IDO-1 activity in HIV-1 infection. J Acquir Immune Defic Syndr (2014) 67:227–35. doi: 10.1097/QAI.0000000000000294
63. Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, et al. Gut-resident lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep (2015) 13:1589–97. doi: 10.1016/j.celrep.2015.10.026
64. Byakwaga H, Boum Y, Huang 2Y, Muzoora C, Kembabazi A, Weiser SD, et al. Hunt, the kynurenine pathway of tryptophan catabolism, CD4+ T-cell recovery, and mortality among HIV-infected ugandans initiating antiretroviral therapy. J Infect Dis (2014) 210:383–91. doi: 10.1093/infdis/jiu115
65. Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med 2 (2010), 32ra36.
66. Brenchley JM, Douek DC. HIV Infection and the gastrointestinal immune system. Mucosal Immunol (2008) 1:23–30. doi: 10.1038/mi.2007.1
67. Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol (2012) 5:646–57. doi: 10.1038/mi.2012.38
68. Wijewardana V, Kristoff J, Xu C, Ma D, Haret-Richter G, Stock JL, et al. Kinetics of myeloid dendritic cell trafficking and activation: impact on progressive, nonprogressive and controlled SIV infections. PLoS Pathog (2013) 9:e1003600. doi: 10.1371/journal.ppat.1003600
69. Li H, Richert-Spuhler LE, Evans TI, Gillis J, Connole M, Estes JD, et al. Hypercytotoxicity and rapid loss of NKp44+ innate lymphoid cells during acute SIV infection. PLoS Pathog (2014) 10:e1004551. doi: 10.1371/journal.ppat.1004551
70. Mudd JC, Brenchley JM. Innate lymphoid cells: Their contributions to gastrointestinal tissue homeostasis and HIV/SIV disease pathology. Curr HIV/AIDS Rep (2019) 16:181–90. doi: 10.1007/s11904-019-00439-4
71. Mudd JC, Busman-Sahay K, DiNapoli SR, Lai S, Sheik V, Lisco A, et al. Hallmarks of primate lentiviral immunodeficiency infection recapitulate loss of innate lymphoid cells. Nat Commun (2018) 9:3967. doi: 10.1038/s41467-018-05528-3
72. Sivanandham R, Brocca-Cofano E, Krampe N, Falwell E, Venkatraman SMK, Ribeiro RM, et al. Neutrophil extracellular trap production contributes to pathogenesis in SIV-infected nonhuman primates. J Clin Invest (2018) 128:5178–83. doi: 10.1172/JCI99420
73. van Wijk F, Cheroutre H. Mucosal T cells in gut homeostasis and inflammation. Expert Rev Clin Immunol (2010) 6:559–66. doi: 10.1586/eci.10.34
74. Schechter ME, Andrade BB, He T, Richter GH, Tosh KW, Policicchio BB, et al. Inflammatory monocytes expressing tissue factor drive SIV and HIV coagulopathy. Sci Transl Med 9 (2017). doi: 10.1126/scitranslmed.aam5441
75. Reno TA, Tarnus L, Tracy R, Landay AL, Sereti I, Apetrei C, et al. Complement, and NET formation in HIV/SIV pathogenesis. Front Virol (2022).
76. de Mareuil J, Carre M, Barbier P, Campbell GR, Lancelot S, Opi S, et al. HIV-1 tat protein enhances microtubule polymerization. Retrovirology (2005) 2:5. doi: 10.1186/1742-4690-2-5
77. Lien K, Mayer W, Herrera R, Rosbe K, Tugizov SM. HIV-1 proteins gp120 and tat induce the epithelial-mesenchymal transition in oral and genital mucosal epithelial cells. PLoS One (2019) 14:e0226343. doi: 10.1371/journal.pone.0226343
78. Blander JM. Death in the intestinal epithelium-basic biology and implications for inflammatory bowel disease. FEBS J (2016) 283:2720–30. doi: 10.1111/febs.13771
79. Tabb B, Morcock DR, Trubey CM, Quinones OA, Hao XP, Smedley J, et al. Reduced inflammation and lymphoid tissue immunopathology in rhesus macaques receiving anti-tumor necrosis factor treatment during primary simian immunodeficiency virus infection. J Infect Dis (2013) 207:880–92. doi: 10.1093/infdis/jis643
80. Sim JH, Mukerji SS, Russo SC, Lo J. Gastrointestinal dysfunction and HIV comorbidities. Curr HIV/AIDS Rep (2021) 18:57–62. doi: 10.1007/s11904-020-00537-8
81. Alzahrani J, Hussain T, Simar D, Palchaudhuri R, Abdel-Mohsen M, Crowe SM, et al. Inflammatory and immunometabolic consequences of gut dysfunction in HIV: Parallels with IBD and implications for reservoir persistence and non-AIDS comorbidities. EBioMedicine (2019) 46:522–31. doi: 10.1016/j.ebiom.2019.07.027
82. Tincati C, Douek DC, Marchetti G. Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res Ther (2016) 13:19. doi: 10.1186/s12981-016-0103-1
83. Pandrea I, Xu C, Stock JL, Frank DN, Ma D, Policicchio BB, et al. Antibiotic and antiinflammatory therapy transiently reduces inflammation and hypercoagulation in acutely SIV-infected pigtailed macaques. PLoS Pathog (2016) 12:e1005384. doi: 10.1371/journal.ppat.1005384
84. Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog (2010) 6:e1001052. doi: 10.1371/journal.ppat.1001052
85. Batman PA, Kotler DP, Kapembwa MS, Booth D, Potten CS, Orenstein JM, et al. HIV Enteropathy: crypt stem and transit cell hyperproliferation induces villous atrophy in HIV/Microsporidia-infected jejunal mucosa. AIDS (2007) 21:433–9. doi: 10.1097/QAD.0b013e3280142ee8
86. He T, Brocca-Cofano E, Gillespie DG, Xu C, Stock JL, Ma D, et al. Critical role for the adenosine pathway in controlling simian immunodeficiency virus-related immune activation and inflammation in gut mucosal tissues. J Virol (2015) 89:9616–30. doi: 10.1128/JVI.01196-15
87. Das B, Okamoto K, Rabalais J, Young JA, Barrett KE, Sivagnanam M. Aberrant epithelial differentiation contributes to pathogenesis in a murine model of congenital tufting enteropathy. Cell Mol Gastroenterol Hepatol (2021) 12:1353–71. doi: 10.1016/j.jcmgh.2021.06.015
88. Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol (2012) 10:655–66. doi: 10.1038/nrmicro2848
89. Hartigan-O’Connor DJ, Hirao LA, McCune JM, Dandekar S. Th17 cells and regulatory T cells in elite control over HIV and SIV. Curr Opin HIV AIDS (2011) 6:221–7. doi: 10.1097/COH.0b013e32834577b3
90. Li Q, Estes JD, Duan L, Jessurun J, Pambuccian S, Forster C, et al. Simian immunodeficiency virus-induced intestinal cell apoptosis is the underlying mechanism of the regenerative enteropathy of early infection. J Infect Dis (2008) 197:420–9. doi: 10.1086/525046
91. Ma D, Jasinska A, Kristoff J, Grobler JP, Turner T, Jung Y, et al. SIVagm infection in wild African green monkeys from south Africa: Epidemiology, natural history, and evolutionary considerations. PLoS Pathog (2013) 9:e1003011. doi: 10.1371/journal.ppat.1003011
92. Ma D, Jasinska AJ, Feyertag F, Wijewardana V, Kristoff J, He T, et al. Factors associated with siman immunodeficiency virus transmission in a natural African nonhuman primate host in the wild. J Virol (2014) 88:5687–705. doi: 10.1128/JVI.03606-13
93. Pandrea I, Silvestri G, Onanga R, Veazey RS, Marx PA, Hirsch V, et al. Simian immunodeficiency viruses replication dynamics in African non-human primate hosts: Common patterns and species-specific differences. J Med Primatol (2006) 35:194–201. doi: 10.1111/j.1600-0684.2006.00168.x
94. Raehtz KD, Barrenas F, Xu C, Busman-Sahay K, Valentine A, Law L, et al. African Green monkeys avoid SIV disease progression by preventing intestinal dysfunction and maintaining mucosal barrier integrity. PLoS Pathog (2020) 16:e1008333. doi: 10.1371/journal.ppat.1008333
95. Barrenas F, Raehtz K, Xu C, Law L, Green RR, Silvestri G, et al. Macrophage-associated wound healing contributes to African green monkey SIV pathogenesis control. Nat Commun (2019) 10:5101. doi: 10.1038/s41467-019-12987-9
96. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
97. Huang D, Lian X, Song F, Ma H, Lian Z, Liang Y, et al. Clinical features of severe patients infected with 2019 novel coronavirus: A systematic review and meta-analysis. Ann Transl Med (2020) 8:576. doi: 10.21037/atm-20-2124
98. Li J, Gong X, Wang Z, Chen R, Li T, Zeng D, et al. Clinical features of familial clustering in patients infected with 2019 novel coronavirus in wuhan, China. Virus Res (2020) 286:198043. doi: 10.1016/j.virusres.2020.198043
99. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet (2020) 395:507–13. doi: 10.1016/S0140-6736(20)30211-7
100. Mukherjee R, Smith A, Sutton R. Covid-19-related pancreatic injury. Br J Surg (2020) 107:e190. doi: 10.1002/bjs.11645
101. Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut (2020) 69:997–1001. doi: 10.1136/gutjnl-2020-321013
102. de-Madaria E, Siau K, Cardenas-Jaen K. Increased amylase and lipase in patients with COVID-19 pneumonia: Don’t blame the pancreas just yet! Gastroenterology (2021) 160:1871. doi: 10.1053/j.gastro.2020.04.044
103. Wang D, Yin Y, Hu C, Liu X, Zhang X, Zhou S, et al. Clinical course and outcome of 107 patients infected with the novel coronavirus, SARS-CoV-2, discharged from two hospitals in wuhan, China. Crit Care (2020) 24:188. doi: 10.1186/s13054-020-02895-6
104. Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, et al. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun (2020) 87:18–22. doi: 10.1016/j.bbi.2020.03.031
105. Adao R, Guzik TJ. Inside the heart of COVID-19. Cardiovasc Res (2020) 116:e59–61. doi: 10.1093/cvr/cvaa086
106. Akhmerov A, Marban E. COVID-19 and the heart. Circ Res (2020) 126:1443–55. doi: 10.1161/CIRCRESAHA.120.317055
107. Azarkish M, Laleh Far V, Eslami M, Mollazadeh R. Transient complete heart block in a patient with critical COVID-19. Eur Heart J (2020) 41:2131. doi: 10.1093/eurheartj/ehaa307
108. Bader F, Manla Y, Atallah B, Starling RC. Heart failure and COVID-19. Heart Fail Rev (2021) 26:1–10. doi: 10.1007/s10741-020-10008-2
109. Bohm M, Frey N, Giannitsis E, Sliwa K, Zeiher AM. Coronavirus disease 2019 (COVID-19) and its implications for cardiovascular care: expert document from the German cardiac society and the world heart federation. Clin Res Cardiol (2020) 109:1446–59. doi: 10.1007/s00392-020-01656-3
110. Cheng R, Leedy D. COVID-19 and acute myocardial injury: The heart of the matter or an innocent bystander? Heart (2020) 106:1122–4. doi: 10.1136/heartjnl-2020-317025
111. Dhakal BP, Sweitzer NK, Indik JH, Acharya D, William P. SARS-CoV-2 infection and cardiovascular disease: COVID-19 heart. Heart Lung Circ (2020) 29:973–87. doi: 10.1016/j.hlc.2020.05.101
112. Goha A, Mezue K, Edwards P, Nunura F, Baugh D, Madu E. COVID-19 and the heart: An update for clinicians. Clin Cardiol (2020) 43:1216–22. doi: 10.1002/clc.23406
113. Khalid N, Chen Y, Case BC, Shlofmitz E, Wermers JP, Rogers T, et al. COVID-19 (SARS-CoV-2) and the heart - an ominous association. Cardiovasc Revasc Med (2020) 21:946–9. doi: 10.1016/j.carrev.2020.05.009
114. Khan IH, Zahra SA, Zaim S, Harky A. At The heart of COVID-19. J Card Surg (2020) 35:1287–94. doi: 10.1111/jocs.14596
115. Libby P. The heart in COVID-19: Primary target or secondary bystander? JACC Basic Transl Sci (2020) 5:537–42. doi: 10.1016/j.jacbts.2020.04.001
116. Mehra MR, Ruschitzka F. COVID-19 illness and heart failure: A missing link? JACC Heart Fail (2020) 8:512–4. doi: 10.1016/j.jchf.2020.03.004
117. Mishra AK, Sahu KK, Lal A, Sargent J. Patterns of heart injury in COVID-19 and relation to outcome. J Med Virol (2020) 92:1747. doi: 10.1002/jmv.25847
118. Otto CM. Heartbeat: Heart disease and COVID-19. Heart (2020) 106:1115–6. doi: 10.1136/heartjnl-2020-317674
120. Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, et al. Hematological findings and complications of COVID-19. Am J Hematol (2020) 95:834–47. doi: 10.1002/ajh.25829
121. Al-Namaeh M. COVID-19 and conjunctivitis: a meta-analysis. Ther Adv Ophthalmol (2021) 13:25158414211003368. doi: 10.1177/25158414211003368
122. Loffredo L, Fallarino A, Paraninfi A, Pacella F, Pacella E, Oliva A, et al. Different rates of conjunctivitis in COVID-19 eastern and western hospitalized patients: a meta-analysis. Intern Emerg Med (2021). doi: 10.1007/s11739-021-02880-z
123. Loffredo L, Pacella F, Pacella E, Tiscione G, Oliva A, Violi F. Conjunctivitis and COVID-19: A meta-analysis. J Med Virol (2020) 92:1413–4. doi: 10.1002/jmv.25938
124. Vaira LA, Salzano G, Fois AG, Piombino P, De Riu G. Potential pathogenesis of ageusia and anosmia in COVID-19 patients. Int Forum Allergy Rhinol (2020) 10:1103–4. doi: 10.1002/alr.22593
125. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell (2020) 181:271–80.e8. doi: 10.1016/j.cell.2020.02.052
126. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (2020) 367:1444–8. doi: 10.1126/science.abb2762
127. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature (2003) 426:450–4. doi: 10.1038/nature02145
128. Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res (2013) 23:986–93. doi: 10.1038/cr.2013.92
129. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step understanding SARS pathogenesis. J Pathol (2004) 203:631–7. doi: 10.1002/path.1570
130. Chertow D, Stein S, Ramelli S, Grazioli A, Chung J-Y, Singh M, et al. SARS-CoV-2 infection and persistence throughout the human body and brain (2022). Available at: https://www.researchsquare.com/article/rs-1139035/v1.
131. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al.THVuZy1uZXR3b3JrQGh1bWFuY2VsbGF0bGFzLm9yZw==and H.C.A.L.B. network, SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell (2020) 181:1016–35 e19.
132. Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol (2020) 16:e9610. doi: 10.15252/msb.20209610
133. Ortiz ME, Thurman A, Pezzulo AA, Leidinger MR, Klesney-Tait JA, Karp PH, et al. Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMedicine (2020) 60:102976. doi: 10.1016/j.ebiom.2020.102976
134. Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell (2020) 182:429–46.e14. doi: 10.1016/j.cell.2020.05.042
135. Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med (2020) 26:681–7. doi: 10.1038/s41591-020-0868-6
136. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med (2020) 14:185–92. doi: 10.1007/s11684-020-0754-0
137. Lee IT, Nakayama T, Wu CT, Goltsev Y, Jiang S, Gall PA, et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun (2020) 11:5453. doi: 10.1038/s41467-020-19145-6
138. Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA (2020) 323:2427–9. doi: 10.1001/jama.2020.8707
139. Smith JC, Sausville EL, Girish V, Yuan ML, Vasudevan A, John KM, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev Cell (2020) 53:514–29.e3. doi: 10.1016/j.devcel.2020.05.012
140. Liu W, Tao ZW, Wang L, Yuan ML, Liu K, Zhou L, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl) (2020) 133:1032–8. doi: 10.1097/CM9.0000000000000775
141. Brooks SD, Smith RL, Moreira A, Ackerman HC. Oral lisinopril raises tissue levels of ACE2, the SARS-CoV-2 receptor, in healthy male and female mice. bioRxiv (2022) 2021:10. doi: 10.3389/fphar.2022.798349
142. Zhuang MW, Cheng Y, Zhang J, Jiang XM, Wang L, Deng J, et al. Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection. J Med Virol (2020) 92:2693–701. doi: 10.1002/jmv.26139
143. Baker SA, Kwok S, Berry GJ, Montine TJ. Angiotensin-converting enzyme 2 (ACE2) expression increases with age in patients requiring mechanical ventilation. PLoS One (2021) 16:e0247060. doi: 10.1371/journal.pone.0247060
144. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol (2022) 23:3–20. doi: 10.1038/s41580-021-00418-x
145. Blair RV, Vaccari M, Doyle-Meyers LA, Roy CJ, Russell-Lodrigue K, Fahlberg M, et al. Acute respiratory distress in aged, SARS-CoV-2-Infected African green monkeys but not rhesus macaques. Am J Pathol (2021) 191:274–82. doi: 10.1016/j.ajpath.2020.10.016
146. Loske J, Rohmel J, Lukassen S, Stricker S, Magalhaes VG, Liebig J, et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol (2022) 40:319–24. doi: 10.1038/s41587-021-01037-9
147. Rossi AD, de Araujo JLF, de Almeida TB, Ribeiro-Alves M, de Almeida Velozo C, Almeida JM, et al. Association between ACE2 and TMPRSS2 nasopharyngeal expression and COVID-19 respiratory distress. Sci Rep (2021) 11:9658. doi: 10.1038/s41598-021-88944-8
148. Saheb Sharif-Askari N, Saheb Sharif-Askari F, Alabed M, Temsah MH, Al Heialy S, Hamid Q, et al. Airways expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev (2020) 18:1–6. doi: 10.1016/j.omtm.2020.05.013
149. Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell (2022) 185:881–95.e20. doi: 10.1016/j.cell.2022.01.014
150. Toh ZQ, Anderson J, Mazarakis N, Neeland M, Higgins RA, Rautenbacher K, et al. Comparison of seroconversion in children and adults with mild COVID-19. JAMA Netw Open (2022) 5:e221313. doi: 10.1001/jamanetworkopen.2022.1313
151. Weisberg SP, Connors TJ, Zhu Y, Baldwin MR, Lin WH, Wontakal S, et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat Immunol (2021) 22:25–31. doi: 10.1038/s41590-020-00826-9
152. Yoshida M, Worlock KB, Huang N, Lindeboom RGH, Butler CR, Kumasaka N, et al. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature (2022) 602:321–7. doi: 10.1038/s41586-021-04345-x
153. Prendergast H, Tyo C, Colbert C, Kelley M, Pobee R. Medical complications of obesity: heightened importance in a COVID era. Int J Emerg Med (2022) 15:29. doi: 10.1186/s12245-022-00431-7
154. Sharma P, Behl T, Sharma N, Singh S, Grewal AS, Albarrati A, et al. COVID-19 and diabetes: Association intensify risk factors for morbidity and mortality. BioMed Pharmacother (2022) 151:113089. doi: 10.1016/j.biopha.2022.113089
155. Kastora S, Patel M, Carter B, Delibegovic M, Myint PK. Impact of diabetes on COVID-19 mortality and hospital outcomes from a global perspective: An umbrella systematic review and meta-analysis. Endocrinol Diabetes Metab (2022) 5:e00338. doi: 10.1002/edm2.338
156. Boden I, Bernabeu MO, Dhillon B, Dorward DA, MacCormick I, Megaw R, et al. Pre-existing diabetic retinopathy as a prognostic factor for COVID-19 outcomes amongst people with diabetes: A systematic review. Diabetes Res Clin Pract (2022) 187:109869. doi: 10.1016/j.diabres.2022.109869
157. Karampela I, Vallianou N, Magkos F, Apovian CM, Dalamaga M. Obesity And COVID-19: The Bermuda triangle in public health. Curr Obes Rep (2022). doi: 10.1007/s13679-022-00471-3
158. Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA (2004) 101:15748–53.
159. Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, et al. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol (2004) 78:5642–50. doi: 10.1128/JVI.78.11.5642-5650.2004
160. Amraei R, Yin W, Napoleon MA, Suder EL, Berrigan J, Zhao Q, et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Cent Sci (2021) 7:1156–65. doi: 10.1021/acscentsci.0c01537
161. Mori Y, Fink C, Ichimura T, Sako K, Mori M, Lee NN, et al. KIM-1/TIM-1 is a receptor for SARS-CoV-2 in lung and kidney. medRxiv (2022).
162. Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res (2021) 31:126–40. doi: 10.1038/s41422-020-00460-y
163. Jemielity S, Wang JJ, Chan YK, Ahmed AA, Li W, Monahan S, et al. TIM-family proteins promote infection of multiple enveloped viruses through virion-associated phosphatidylserine. PLoS Pathog (2013) 9:e1003232. doi: 10.1371/journal.ppat.1003232
164. Chen Z, Mi L, Xu J, Yu J, Wang X, Jiang J, et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis (2005) 191:755–60. doi: 10.1086/427811
165. Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther (2020) 5:283. doi: 10.1038/s41392-020-00426-x
166. Shilts J, Crozier TWM, Greenwood EJD, Lehner PJ, Wright GJ. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci Rep (2021) 11:413. doi: 10.1038/s41598-020-80464-1
167. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (2020) 370:856–60. doi: 10.1126/science.abd2985
168. Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Anton-Plagaro C, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (2020) 370:861–5. doi: 10.1126/science.abd3072
169. Ahn JH, Kim J, Hong SP, Choi SY, Yang MJ, Ju YS, et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J Clin Invest 131 (2021). doi: 10.1172/JCI148517
170. Camargo SM, Singer D, Makrides V, Huggel K, Pos KM, Wagner CA, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology (2009) 136:872–82. doi: 10.1053/j.gastro.2008.10.055
171. Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M, et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell (2021) 184:92–105.e16. doi: 10.1016/j.cell.2020.10.030
172. Liu J, Lu F, Chen Y, Plow E, Qin J. Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J Biol Chem (2022) 298:101710. doi: 10.1016/j.jbc.2022.101710
173. Huntington KE, Carlsen L, So EY, Piesche M, Liang O, El-Deiry WS. Integrin/TGF-beta1 inhibitor GLPG-0187 blocks SARS-CoV-2 delta and omicron pseudovirus infection of airway epithelial cells In vitro, which could attenuate disease severity. Pharm (Basel) 15 (2022). doi: 10.1101/2022.01.02.22268641
174. Othman H, Messaoud HB, Khamessi O, Ben-Mabrouk H, Ghedira K, Bharuthram A, et al. SARS-CoV-2 spike protein unlikely to bind to integrins via the arg-Gly-Asp (RGD) motif of the receptor binding domain: Evidence from structural analysis and microscale accelerated molecular dynamics. Front Mol Biosci (2022) 9:834857. doi: 10.3389/fmolb.2022.834857
175. Alvarado DM, Son J, Thackray LB, Gomez Castro MF, Prasad S, Cui X, et al. Mesalamine reduces intestinal ACE2 expression without modifying SARS-CoV-2 infection or disease severity in mice. Inflammation Bowel Dis (2022) 28:318–21. doi: 10.1093/ibd/izab274
176. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science (2020) 369:50–4. doi: 10.1126/science.abc1669
177. Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5 (2020). doi: 10.1126/sciimmunol.abc3582
178. Qian Q, Fan L, Liu W, Li J, Yue J, Wang M, et al. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin Infect Dis (2021) 73:361–6. doi: 10.1093/cid/ciaa925
179. Lehmann M, Allers K, Heldt C, Meinhardt J, Schmidt F, Rodriguez-Sillke Y, et al. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8+ T cells. Mucosal Immunol (2021) 14:1381–92. doi: 10.1038/s41385-021-00437-z
180. Livanos AE, Jha D, Cossarini F, Gonzalez-Reiche AS, Tokuyama M, Aydillo T, et al. Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology (2021) 160:2435–50.e34. doi: 10.1053/j.gastro.2021.02.056
181. Yantiss RK, Qin L, He B, Crawford CV, Seshan S, Patel S, et al. Intestinal abnormalities in patients with SARS-CoV-2 infection: Histopathologic changes reflect mechanisms of disease. Am J Surg Pathol (2022) 46:89–96. doi: 10.1038/s41385-021-00437-z
182. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA (2020) 323:1843–4. doi: 10.1097/PAS.0000000000001755
183. Xiao F, Sun J, Xu Y, Li F, Huang X, Li H, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis (2020) 26:1920–2. doi: 10.1001/jama.2020.3786
184. Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature (2020) 581:465–9. doi: 10.3201/eid2608.200681
185. Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol (2020) 5:434–5. doi: 10.1038/s41586-020-2196-x
186. Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med (2020) 26:502–5. doi: 10.1016/S2468-1253(20)30083-2
187. Ramakrishnan RK, Kashour T, Hamid Q, Halwani R, Tleyjeh IM. Unraveling the mystery surrounding post-acute sequelae of COVID-19. Front Immunol (2021) 12:686029. doi: 10.1038/s41591-020-0817-4
188. Guadalupe M, Sankaran S, George MD, Reay E, Verhoeven D, Shacklett BL, et al. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J Virol (2006) 80:8236–47. doi: 10.3389/fimmu.2021.686029
189. Mendez-Lagares G, Romero-Sanchez MC, Ruiz-Mateos E, Genebat M, Ferrando-Martinez S, Munoz-Fernandez MA, et al. Long-term suppressive combined antiretroviral treatment does not normalize the serum level of soluble CD14. J Infect Dis (2013) 207:1221–5. doi: 10.1128/JVI.00120-06
190. Zevin AS, McKinnon L, Burgener A, Klatt NR. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS (2016) 11:182–90. doi: 10.1093/infdis/jit025
191. Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M, et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull (Beijing) (2021) 66:783–93. doi: 10.1097/COH.0000000000000234
192. Pan L, Mu M, Yang P, Sun Y, Wang R, Yan J, et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: A descriptive, cross-sectional, multicenter study. Am J Gastroenterol (2020) 115:766–73. doi: 10.1016/j.scib.2020.11.015
193. Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, et al. Prognostic value of interleukin-6, c-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol (2020) 127:104370. doi: 10.14309/ajg.0000000000000620
194. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (2020) 369:718–24. doi: 10.1016/j.jcv.2020.104370
195. Mandel M, Harari G, Gurevich M, Achiron A. Cytokine prediction of mortality in COVID19 patients. Cytokine (2020) 134:155190. doi: 10.1126/science.abc6027
196. Isnard S, Lin J, Bu S, Fombuena B, Royston L, Routy JP. Gut leakage of fungal-related products: Turning up the heat for HIV infection. Front Immunol (2021) 12:656414. doi: 10.1016/j.cyto.2020.155190
197. Mehraj V, Ramendra R, Isnard S, Dupuy FP, Ponte R, Chen J, et al. Circulating (1–>3)-beta-D-glucan is associated with immune activation during human immunodeficiency virus infection. Clin Infect Dis (2020) 70:232–41. doi: 10.3389/fimmu.2021.656414
198. Pandrea I, Apetrei C. Where the wild things are: pathogenesis of SIV infection in African nonhuman primate hosts. Curr HIV/AIDS Rep (2010) 7:28–36. doi: 10.1093/cid/ciz212
199. Pandrea I, Gaufin T, Brenchley JM, Gautam R, Monjure C, Gautam A, et al. Cutting edge: Experimentally induced immune activation in natural hosts of simian immunodeficiency virus induces significant increases in viral replication and CD4+ T cell depletion. J Immunol (2008) 181:6687–91. doi: 10.1007/s11904-009-0034-8
200. Hao XP, Lucero CM, Turkbey B, Bernardo ML, Morcock DR, Deleage C, et al. Experimental colitis in SIV-uninfected rhesus macaques recapitulates important features of pathogenic SIV infection. Nat Commun (2015) 6:8020. doi: 10.4049/jimmunol.181.10.6687
201. Kristoff J, Haret-Richter G, Ma D, Ribeiro RM, Xu C, Cornell E, et al. Early microbial translocation blockade reduces SIV-mediated inflammation and viral replication. J Clin Invest (2014) 124:2802–6. doi: 10.1038/ncomms9020
202. Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity (2013) 39:633–45. doi: 10.1172/JCI75090
203. Pandrea I, Landay A, Wilson C, Stock J, Tracy R, Apetrei C. Using the pathogenic and nonpathogenic nonhuman primate model for studying non-AIDS comorbidities. Curr HIV/AIDS Rep (2015) 12:54–67. doi: 10.1016/j.immuni.2013.10.001
204. Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell (2012) 151:253–66. doi: 10.1007/s11904-014-0245-5
205. Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol (2015) 8:1009–20. doi: 10.1016/j.cell.2012.09.024
206. Brenchley JM, Ortiz AM. Microbiome studies in non-human primates. Curr HIV/AIDS Rep (2021) 18:527–37. doi: 10.1038/mi.2014.128
207. Rosel-Pech C, Chavez-Torres M, Bekker-Mendez VC, Pinto-Cardoso S. Therapeutic avenues for restoring the gut microbiome in HIV infection. Curr Opin Pharmacol (2020) 54:188–201. doi: 10.1007/s11904-021-00584-9
208. Ashuro AA, Lobie TA, Ye DQ, Leng RX, Li BZ, Pan HF, et al. Review on the alteration of gut microbiota: The role of HIV infection and old age. AIDS Res Hum Retroviruses (2020) 36:556–65. doi: 10.1016/j.coph.2020.09.010
209. Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS (2016) 30:2737–51. doi: 10.1089/aid.2019.0282
210. Vujkovic-Cvijin I, Sortino O, Verheij E, Sklar J, Wit FW, Kootstra NA, et al. HIV-Associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat Commun (2020) 11:2448. doi: 10.1097/QAD.0000000000001289
211. Noguera-Julian M, Rocafort M, Guillen Y, Rivera J, Casadella M, Nowak P, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine (2016) 5:135–46. doi: 10.1038/s41467-020-16222-8
212. Guillen Y, Noguera-Julian M, Rivera J, Casadella M, Zevin AS, Rocafort M, et al. Low nadir CD4+ T-cell counts predict gut dysbiosis in HIV-1 infection. Mucosal Immunol (2019) 12:232–46. doi: 10.1016/j.ebiom.2016.01.032
213. He T, Xu C, Krampe N, Dillon SM, Sette P, Falwell E, et al. High-fat diet exacerbates SIV pathogenesis and accelerates disease progression. J Clin Invest (2019) 129:5474–88. doi: 10.1038/s41385-018-0083-7
214. Ortiz AM, Flynn JK, DiNapoli SR, Vujkovic-Cvijin I, Starke CE, Lai SH, et al. Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat Med (2018) 24:1313–6. doi: 10.1172/JCI121208
215. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut (2021) 70:698–706. doi: 10.1038/s41591-018-0132-5
216. Reinold J, Farahpour F, Fehring C, Dolff S, Konik M, Korth J, et al. A pro-inflammatory gut microbiome characterizes SARS-CoV-2 infected patients and a reduction in the connectivity of an anti-inflammatory bacterial network associates with severe COVID-19. Front Cell Infect Microbiol (2021) 11:747816. doi: 10.1136/gutjnl-2020-323020
217. Liu Y, Zhang H, Tang X, Jiang X, Yan X, Liu X, et al. Distinct metagenomic signatures in the SARS-CoV-2 infection. Front Cell Infect Microbiol (2021) 11:706970. doi: 10.3389/fcimb.2021.747816
218. Seibert B, Caceres CJ, Cardenas-Garcia S, Carnaccini S, Geiger G, Rajao DS, et al. Mild and severe SARS-CoV-2 infection induces respiratory and intestinal microbiome changes in the K18-hACE2 transgenic mouse model. Microbiol Spectr (2021) 9:e0053621. doi: 10.3389/fcimb.2021.706970
219. Effenberger M, Grabherr F, Mayr L, Schwaerzler J, Nairz M, Seifert M, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut (2020) 69:1543–4. doi: 10.1128/Spectrum.00536-21
220. Sun Z, Song ZG, Liu C, Tan S, Lin S, Zhu J, et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med (2022) 20:24. doi: 10.1136/gutjnl-2020-321388
221. Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front Immunol (2021) 12:686240. doi: 10.1186/s12916-021-02212-0
222. Hoel H, Heggelund L, Reikvam DH, Stiksrud B, Ueland T, Michelsen AE, et al. Elevated markers of gut leakage and inflammasome activation in COVID-19 patients with cardiac involvement. J Intern Med (2021) 289:523–31. doi: 10.3389/fimmu.2021.686240
223. Ferrari B, Da Silva AC, Liu KH, Saidakova EV, Korolevskaya LB, Shmagel KV, et al. Gut-derived bacterial toxins impair memory CD4+ T cell mitochondrial function in HIV-1 infection. J Clin Invest 132 (2022). doi: 10.1111/joim.13178
224. Patel P, DeCuir J, Abrams J, Campbell AP, Godfred-Cato S, Belay ED. Clinical characteristics of multisystem inflammatory syndrome in adults: A systematic review. JAMA Netw Open (2021) 4:e2126456. doi: 10.1172/JCI149571
225. Porritt RA, Paschold L, Rivas MN, Cheng MH, Yonker LM, Chandnani H, et al. HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J Clin Invest 131 (2021). doi: 10.1001/jamanetworkopen.2021.26456
226. Sacco K, Castagnoli R, Vakkilainen S, Liu C, Delmonte OM, Oguz C, et al. Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19. Nat Med (2022). doi: 10.1172/JCI146614
227. Yonker LM, Gilboa T, Ogata AF, Senussi Y, Lazarovits R, Boribong BP, et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J Clin Invest 131 (2021).
228. Feldstein LR, Tenforde MW, Friedman KG, Newhams M, Rose EB, Dapul H, et al. Characteristics and outcomes of US children and adolescents with multisystem inflammatory syndrome in children (MIS-c) compared with severe acute COVID-19. JAMA (2021) 325:1074–87. doi: 10.1172/JCI149633
229. Liu Q, Mak JWY, Su Q, Yeoh YK, Lui GC, Ng SSS, et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut (2022) 71:544–52. doi: 10.1001/jama.2021.2091
230. Peluso MJ, Lu S, Tang AF, Durstenfeld MS, Ho HE, Goldberg SA, et al. Markers of immune activation and inflammation in individuals with postacute sequelae of severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis (2021) 224:1839–48. doi: 10.1136/gutjnl-2021-325989
231. Antiretroviral Therapy Cohort C. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: A collaborative analysis of cohort studies. Lancet HIV (2017) 4:e349–56. doi: 10.1093/infdis/jiab490
232. Marcus JL, Leyden WA, Alexeeff SE, Anderson AN, Hechter RC, Hu H, et al. Comparison of overall and comorbidity-free life expectancy between insured adults with and without HIV infection, 2000-2016. JAMA Netw Open (2020) 3:e207954.
233. Siliciano JD, Siliciano RF. Nonsuppressible HIV-1 viremia: A reflection of how the reservoir persists. J Clin Invest (2020) 130:5665–7. doi: 10.1001/jamanetworkopen.2020.7954
234. Babu H, Ambikan AT, Gabriel EE, Svensson Akusjarvi S, Palaniappan AN, Sundaraj V, et al. Systemic inflammation and the increased risk of inflamm-aging and age-associated diseases in people living with HIV on long term suppressive antiretroviral therapy. Front Immunol (2019) 10:1965. doi: 10.1172/JCI141497
235. Daskou M, Mu W, Sharma M, Vasilopoulos H, Heymans R, Ritou E, et al. ApoA-I mimetics reduce systemic and gut inflammation in chronic treated HIV. PLoS Pathog (2022) 18:e1010160. doi: 10.3389/fimmu.2019.01965
236. Serrano-Villar S, Vazquez-Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando-Martinez S, et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol (2017) 10:1279–93. doi: 10.1371/journal.ppat.1010160
237. Gori A, Rizzardini G, Van’t Land B, Amor KB, van Schaik J, Torti C, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: Results of the “COPA” pilot randomized trial. Mucosal Immunol (2011) 4:554–63. doi: 10.1038/mi.2016.122
238. Isnard S, Fombuena B, Ouyang J, Royston L, Lin J, Bu S, et al. Camu camu effects on microbial translocation and systemic immune activation in ART-treated people living with HIV: Protocol of the single-arm non-randomised camu camu prebiotic pilot study (CIHR/CTN PT032). BMJ Open (2022) 12:e053081. doi: 10.1038/mi.2011.15
239. Messaoudene M, Pidgeon R, Richard C, Ponce M, Diop K, Benlaifaoui M, et al. And circumvents anti-PD-1 resistance through effects on the gut microbiota. Cancer Discov (2022) 12:1070–87. doi: 10.1136/bmjopen-2021-053081
240. Rousseau RK, Walmsley SL, Lee T, Rosenes R, Reinhard RJ, Malazogu F, et al. Randomized, blinded, placebo-controlled trial of de Simone formulation probiotic during HIV-associated suboptimal CD4+ T cell recovery. J Acquir Immune Defic Syndr (2022) 89:199–207. doi: 10.1158/2159-8290.CD-21-0808
241. Yang OO, Kelesidis T, Cordova R, Khanlou H. Immunomodulation of antiretroviral drug-suppressed chronic HIV-1 infection in an oral probiotic double-blind placebo-controlled trial. AIDS Res Hum Retroviruses (2014) 30:988–95. doi: 10.1097/QAI.0000000000002840
242. Tenore SB, Avelino-Silva VI, Costa PR, Franco LM, Sabino EC, Kalil J, et al. Immune effects of lactobacillus casei shirota in treated HIV-infected patients with poor CD4+ T-cell recovery. AIDS (2020) 34:381–9. doi: 10.1089/aid.2014.0181
243. Stiksrud B, Nowak P, Nwosu FC, Kvale D, Thalme A, Sonnerborg A, et al. Reduced levels of d-dimer and changes in gut microbiota composition after probiotic intervention in HIV-infected individuals on stable ART. J Acquir Immune Defic Syndr (2015) 70:329–37. doi: 10.1097/QAD.0000000000002420
244. Christensen AD, Skov S, Haase C. The role of neutrophils and G-CSF in DNFB-induced contact hypersensitivity in mice. Immun Inflammation Dis (2014) 2:21–34. doi: 10.1097/QAI.0000000000000784
245. Schunter M, Chu H, Hayes TL, McConnell D, Crawford SS, Luciw PA, et al. Randomized pilot trial of a synbiotic dietary supplement in chronic HIV-1 infection. BMC Complement Altern Med (2012) 12:84. doi: 10.1002/iid3.16
246. Gonzalez-Hernandez LA, Jave-Suarez LF, Fafutis-Morris M, Montes-Salcedo KE, Valle-Gutierrez LG, Campos-Loza AE, et al. Synbiotic therapy decreases microbial translocation and inflammation and improves immunological status in HIV-infected patients: a double-blind randomized controlled pilot trial. Nutr J (2012) 11:90. doi: 10.1186/1472-6882-12-84
247. Klatt NR, Canary LA, Sun X, Vinton CL, Funderburg NT, Morcock DR, et al. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques. J Clin Invest (2013) 123:903–7. doi: 10.1186/1475-2891-11-90
248. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev (2001) 81:1031–64. doi: 10.1172/JCI66227
249. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell (2016) 165:1332–45. doi: 10.1152/physrev.2001.81.3.1031
250. Parada Venegas D, de la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.1016/j.cell.2016.05.041
251. Brauckmann V, Nambiar S, Potthoff A, Hoxtermann S, Wach J, Kayser A, et al. Influence of dietary supplementation of short-chain fatty acid sodium propionate in people (PLHIV). J Eur Acad Dermatol Venereol (2022). doi: 10.3389/fimmu.2019.00277
252. Leonardi I, Gao IH, Lin WY, Allen M, Li XV, Fiers WD, et al. Mucosal fungi promote gut barrier function and social behavior via type 17 immunity. Cell (2022) 185:831–46.e14. doi: 10.1111/jdv.18006
253. Lam KN, Spanogiannopoulos P, Soto-Perez P, Alexander M, Nalley MJ, Bisanz JE, et al. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep (2021) 37:109930. doi: 10.1016/j.cell.2022.01.017
254. Kelesidis T, Yang OO, Currier JS, Navab K, Fogelman AM, Navab M. HIV-1 infected patients with suppressed plasma viremia on treatment have pro-inflammatory HDL. Lipids Health Dis (2011) 10:35. doi: 10.1016/j.celrep.2021.109930
255. Meriwether D, Sulaiman D, Volpe C, Dorfman A, Grijalva V, Dorreh N, et al. Apolipoprotein a-I mimetics mitigate intestinal inflammation in COX2-dependent inflammatory bowel disease model. J Clin Invest (2019) 129:3670–85. doi: 10.1186/1476-511X-10-35
256. Benfield T, Bodilsen J, Brieghel C, Harboe ZB, Helleberg M, Holm C, et al. Improved survival among hospitalized patients with coronavirus disease 2019 (COVID-19) treated with remdesivir and dexamethasone. A nationwide population-based cohort study. Clin Infect Dis (2021) 73:2031–6. doi: 10.1172/JCI123700
257. Calabrese LH, Calabrese C. Baricitinib and dexamethasone for hospitalized patients with COVID-19. Cleve Clin J Med (2021). doi: 10.1093/cid/ciab536
258. Calzetta L, Aiello M, Frizzelli A, Rogliani P, Chetta A. Dexamethasone in patients hospitalized with COVID-19: Whether, when and to whom. J Clin Med 10 (2021). doi: 10.3949/ccjm.88a.ccc073
259. Group RC, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, et al. Dexamethasone in hospitalized patients with covid-19. N Engl J Med (2021) 384:693–704. doi: 10.3390/jcm10081607
260. Ranjbar K, Moghadami M, Mirahmadizadeh A, Fallahi MJ, Khaloo V, Shahriarirad R, et al. Methylprednisolone or dexamethasone, which one is superior corticosteroid in the treatment of hospitalized COVID-19 patients: A triple-blinded randomized controlled trial. BMC Infect Dis (2021) 21:337. doi: 10.1056/NEJMoa2021436
261. Leffler DA, Kelly CP, Green PH, Fedorak RN, DiMarino A, Perrow W, et al. Larazotide acetate for persistent symptoms of celiac disease despite a gluten-free diet: a randomized controlled trial. Gastroenterology (2015) 148:1311–9.e6. doi: 10.1186/s12879-021-06045-3
262. Ungaro RC, Brenner EJ, Agrawal M, Zhang X, Kappelman MD, Colombel JF. Surveillance epidemiology of coronavirus under research exclusion for inflammatory bowel disease research, impact of medications on COVID-19 outcomes in inflammatory bowel disease: Analysis of more than 6000 patients from an international registry. Gastroenterology (2022) 162:316–19.e5. doi: 10.1053/j.gastro.2015.02.008
263. Zhou HY, Guo B, Lufumpa E, Li XM, Chen LH, Meng X, et al. Comparative of the effectiveness and safety of biological agents, tofacitinib, and fecal microbiota transplantation in ulcerative colitis: Systematic review and network meta-analysis. Immunol Invest (2021) 50:323–37. doi: 10.1053/j.gastro.2021.09.011
264. Bezzio C, Manes G, Bini F, Pellegrini L, Saibeni S. Infliximab for severe ulcerative colitis and subsequent SARS-CoV-2 pneumonia: A stone for two birds. Gut (2021) 70:623–4. doi: 10.1080/08820139.2020.1714650
265. Gupta S, Leaf DE. Tocilizumab in COVID-19: some clarity amid controversy. Lancet (2021) 397:1599–601. doi: 10.1136/gutjnl-2020-321760
266. Gupta S, Padappayil RP, Bansal A, Daouk S, Brown B. Tocilizumab in patients hospitalized with COVID-19 pneumonia: Systematic review and meta-analysis of randomized controlled trials. J Investig Med (2022) 70:55–60. doi: 10.1016/S0140-6736(21)00712-1
267. Nagao A, Nakazawa S, Hanabusa H. Short-term efficacy of the IL6 receptor antibody tocilizumab in patients with HIV-associated multicentric castleman disease: report of two cases. J Hematol Oncol (2014) 7:10. doi: 10.1136/jim-2021-002001
Keywords: HIV - human immunodeficiency virus, SIV, SARS-CoV-2, AIDS - acquired immunodeficiency syndrome, COVID - 19, inflammation, microbial translocation, barrier integrity
Citation: Pandrea I, Brooks K, Desai RP, Tare M, Brenchley JM and Apetrei C (2022) I’ve looked at gut from both sides now: Gastrointestinal tract involvement in the pathogenesis of SARS-CoV-2 and HIV/SIV infections. Front. Immunol. 13:899559. doi: 10.3389/fimmu.2022.899559
Received: 18 March 2022; Accepted: 25 July 2022;
Published: 12 August 2022.
Edited by:
Pei-Hui Wang, Shandong University, ChinaReviewed by:
Jean-Pierre Routy, McGill University, CanadaBrandon Beddingfield, Tulane University, United States
Copyright © 2022 Pandrea, Brooks, Desai, Tare, Brenchley and Apetrei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristian Apetrei, YXBldHJlaWNAcGl0dC5lZHU=
†These authors have contributed equally to this work
‡These authors have contributed equally to this work
 Ivona Pandrea
Ivona Pandrea Kelsie Brooks
Kelsie Brooks Rahul P. Desai
Rahul P. Desai Minali Tare
Minali Tare Jason M. Brenchley
Jason M. Brenchley Cristian Apetrei
Cristian Apetrei