- 1Aix-Marseille Université, Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), Centre d’Immunologie de Marseille-Luminy (CIML), Marseille, France
- 2Innate Pharma Research Laboratories, Marseille, France
- 3APHM, Hôpital de la Timone, Marseille-Immunopôle, Marseille, France
Natural Killer (NK) cells are innate lymphoid cells (ILCs) capable of recognizing and directly killing tumor cells. They also secrete cytokines and chemokines, which participate in the shaping of the adaptive response. NK cells identify tumor cells and are activated through a net positive signal from inhibitory and activating receptors. Several activating NK cell receptors are coupled to adaptor molecules containing an immunoreceptor tyrosine-based activation motif (ITAM). These receptors include CD16 and the natural cytotoxic receptors NKp46, NKp44, NKp30 in humans. The powerful antitumor NK cell response triggered by these activating receptors has made them attractive targets for exploitation in immunotherapy. In this review, we will discuss the different activating receptors associated with ITAM-bearing cell surface receptors expressed on NK cells, their modulations in the tumor context and the various therapeutic tools developed to boost NK cell responses in cancer patients.
NK Cells: An Overview
NK cells belong to the ILC family, which encompasses several other subsets: helper-like ILC (ILC1 to 3) and lymphoid tissue inducer cells (LTi). NK cells are endowed with cytolytic functions, whereas helper-like ILC are characterized mainly by their cytokine secretion profiles, which mirror those of helper CD4+ T cells. ILC1 produce IFN-γ type 1 cytokine, ILC2 produce type 2 cytokines, such as IL-5 and IL-13, and ILC3 secrete IL-17 and IL-22 type 3 cytokines (1).
In humans and mice, NK cells circulate in the peripheral blood and infiltrate most organs, including the spleen, liver, lymph nodes and bone marrow. They are also located in the barrier organs of the body, such as the intestines and lungs (2). The NK cell compartment is heterogeneous. Human NK cell subsets are generally defined on the basis of their cell surface expression of CD56 and CD16 (FcγRIIIA) and lack of the T cell receptor complex (CD3/TCR). Human NK cells have been split into two main subsets: CD56dim CD16+ NK cells and CD56bright CD16− NK cells, which are found in the bloodstream and most organs (3, 4). According to their transcriptomic profiles, these two subsets are found in the blood and spleen of humans and conserved in mice at homeostasis. They were defined as NK1/CD56dim-like and NK2/CD56bright-like cells (5).
CD56dim CD16+ cells are cytotoxic and express killer-cell immunoglobulin-like receptors (KIRs), whereas CD56bright CD16− NK cells are less mature, produce larger amounts of cytokines and chemokines and are more prone to proliferate under cytokine stimulation (6). Several studies have suggested that CD56dim CD16+ cells can differentiate from CD56bright CD16− cells (7, 8). We recently showed that an additional NK0/CD56bright-like subset displays specific enrichment in the expression of genes associated with the NK cell precursor (NKP) signature. This tissue-specific subset differs from NK2/CD56bright-like cells in its higher levels of expression of CD52, CD127 and a lack of CD160. A bioinformatic algorithm inferring developmental trajectories from scRNAseq data showed that NK0/CD56bright CD160−-like cells could differentiate into both NK1/CD56dim and NK2/CD56bright CD160+-like cells, but that NK1 and NK2 cells did not appear to be developmentally related under physiological conditions (9).
In addition, some individuals harbor a particular population of NK cells, “adaptive NK”, characterized by the cell surface activating receptor NKG2C and the maturation marker CD57 in humans (10). This subset results from the expansion and persistence after contraction of NK cells following cytomegalovirus (CMV) infection. Adaptive NK cells can be recovered from both humans and mice, and have been detected following the activation of NK cells following hantavirus infection (11), herpes simplex virus 2 infection (12) or influenza vaccination in humans (13).
Another population, cytokine-induced memory-like (CIML) NK cells, has also been described in both humans and mice (14–16). Following prior activation with an IL-12/IL-15/IL-18 cytokine cocktail, the capacity of these cells to produce IFN-γ and granzyme B after restimulation increases.
Thus, several subsets of NK cells have been identified in healthy donors by bulk RNA sequencing (RNAseq), single-cell RNAseq or Cytometry by time of flight (CYTOF). However, it remains to be verified whether all these NK cell populations are indeed present in the majority of individuals. Also, it is of interest to define the heterogeneity of NK cells in several cancer indications.
Role of NK Cells in Cancer
NK cells are cytolytic and exocytose cytotoxic granules containing both perforin (a membrane-disrupting protein) and granzymes (a family of proteolytic enzymes) at the immunological synapse with target cells, leading to target cell lysis. Activated human NK cells also express death-inducing ligands, such as FAS ligand and TNF-related apoptosis-inducing ligand (TRAIL). In addition to their cytotoxic activities, NK cells can produce cytokines, such as IFN-γ and TNF-α. IFN-γ has direct antitumor properties, but also activates myeloid cells and contributes to the shaping of T-cell responses. NK cells can also secrete growth factors (e.g. granulocyte–macrophage colony-stimulating factor) and chemokines (CCL3, CCL4 and CCL5), which participate in the recruitment of dendritic cells in inflamed tissues (17–19).
Many data from in vitro studies have shown that NK cells kill cancer cells of different histological origins. In vivo, depletion experiments and the use of mouse models with impaired NK cell activity have demonstrated higher rates of tumor growth and metastasis (20, 21). Unfortunately, the depleting antibodies used in these experiments were not specific for NK cells. The anti-NK1.1 antibody can also target ILC1, natural killer T cells (NKT) and activated T cells, and the anti-asialo GM1 serum can also target NKT, some T-cell populations, γδ T cells, basophils, eosinophils and macrophages (22, 23). NKp46 is mostly restricted to NK cells, but it is also expressed by ILC1, a subset of ILC3 and a minority fraction of T cells. As a result, no NKp46-depleting antibody or mouse models with modified Ncr1 gene (coding for NKp46) expression can specifically target NK cells. In this context, it is, therefore, important to bear in mind that the results obtained with these tools and attributed to NK cells might actually be due to other cell populations, such as ILC1. Along this line, investigations of role of ILC1 in tumor responses are an emerging field of research, and opposite effects have already been reported in the mouse. On the one hand, ILC1 expansion and cytotoxicity are associated with the control of tumor growth in a mouse mammary tumor model (24) and with the limitation of metastatic seeding in the liver for MC38 colon carcinoma cells and Lewis lung carcinoma cells (LLC) (25). On the other hand, NK cell differentiation towards cells with an ILC1-like gene signature has been shown to be associated with uncontrolled melanoma cell metastasis in the lung (26) and chemically induced fibrosarcoma (27, 28). In this last model, intratumoral ILC1 may limit the NK1.1+ cell-dependent antitumoral response, or even foster tumor growth (28). These results should, therefore, prompt evaluation of differential tumor infiltration and the role of ILC1 and NK cells in optimizing the harnessing of immunity in the treatment of cancer patients.
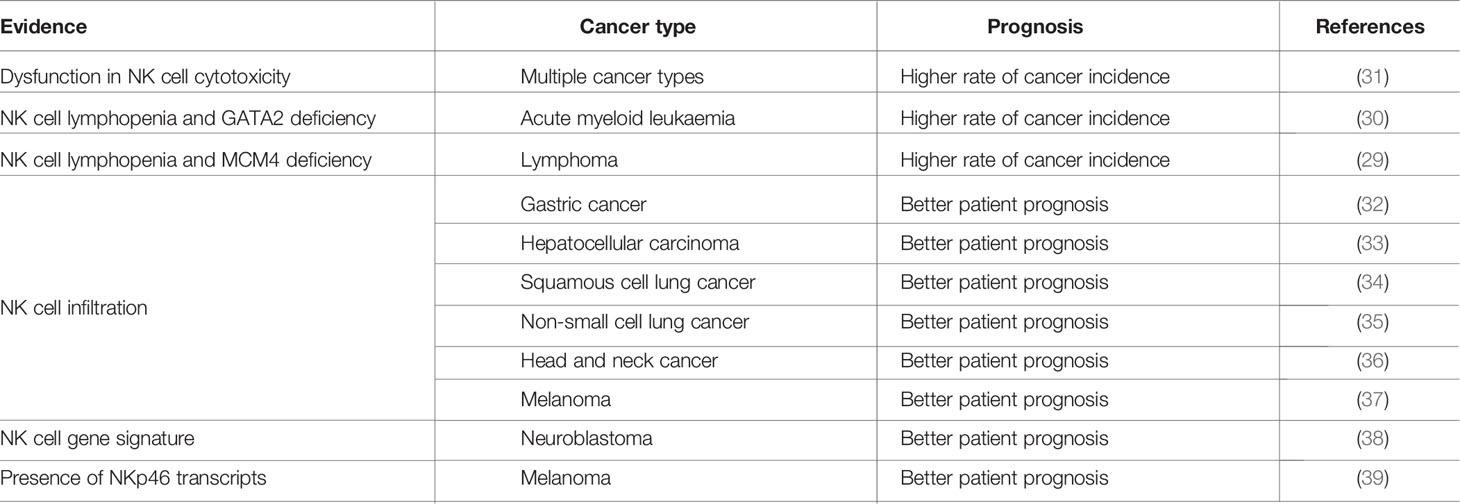
In humans, cancer incidence is higher in patients with NK cell deficiencies characterized by an absence of NK cells or poor effector functions due to mutations of the MCM4 or GATA2 genes (29, 30) (see Table 1). However, these data should be interpreted with caution as such mutations also affect immune cells other than NK cells; it is thus not possible to establish a formal link between NK cell activity and cancer control based on these findings.
A few studies have monitored the cytotoxic activity of blood NK cells and the risk of developing cancer (40, 41). In an 11-year follow-up survey conducted in the Japanese general population, the group of patients with low levels of NK cell cytotoxic activity had a higher risk of cancer (31). Along the same lines, low NK cell counts, rather than CD4+ or CD8+ T cells in peripheral blood are associated with lower overall survival in patients with follicular lymphoma (42).
Many studies have also reported defective blood NK cell function in patients with solid tumors. Our analysis of predefined NK and CD8+ T-cell signatures in multiple public transcriptome data from the TCGA database, which includes over 10,000 tumor samples corresponding to 33 different cancer types, showed that most tissues infiltrated by NK cells were also infiltrated by T cells. Contrary to the findings for ILC1, the NK signature was strongest in hematopoietic cancers (acute myeloid leukemia (AML) and B-cell lymphoma), followed by solid cancers, including kidney renal clear cell carcinoma, testicular germ cell tumors, mesothelioma (a very rare form of cancer affecting the membranes lining internal organs, such as the pleura, peritoneum and pericardium), thymoma, cervical squamous cell carcinoma, endocervical adenocarcinoma, lung cancer and gastric cancers (esophagus, stomach, pancreas) (43) (see Table 1).
Even if there is no formal proof that NK cells are involved in immunosurveillance, NK cells have been shown to have antitumor effects in clinical studies. In several xenograft models in which human cancer cells are implanted in immunodeficient mice, the injection of human NK cells controls tumor elimination (44, 45). Solid evidence for the targeting of human tumors by NK cells in patients has been provided by studies of allogeneic hematopoietic stem cell transplantation (HSCT), in which donor NK cells expressing inhibitory KIRs mismatched with the patient’s cells for MHC class I (MHC-I) molecules because they recognized recipient myeloid leukemia cells as foreign cells (46).
In solid tumors, NK cell infiltration seems to be essential for a robust immune checkpoint blockade (ICB) response (47). Interestingly, transcriptomic analysis of patient samples corresponding to gastric (32), neuroblastoma (38), hepatocellular carcinoma (33), non-small cell lung cancer (NSCLC) (48), head and neck (36) and melanoma (37) tumors have revealed a correlation between NK cell infiltration and better patient outcomes regardless of treatment. Furthermore, NK cell infiltration in solid tumors, as monitored by immunohistochemical analyses of renal cell carcinoma (49), breast cancer (50), adenocarcinoma lung cancer (51), squamous cell lung cancer (34) and NSCLC (35) patients, is associated with better overall survival.
Based on all of this evidence, the targeting of NK cells as been proposed as a means of improving immunotherapies for cancer control, at the forefront of cancer research.
NK Cell Activation
NK cells gauge their environment by integrating signals delivered through ligands binding to inhibitory and activating germline-encoded receptors. The long intracellular part of inhibitory receptors contains a highly conserved signaling motif, the immunoreceptor tyrosine-based inhibitory motif (ITIM) defined by the consensus sequence S/I/V/LxYxxI/V/L. When this inhibitory receptor is triggered, the ITIM is tyrosine phosphorylated, leading to the recruitment of the SH2 domain (Src Homogy domain 2)-containing protein phosphatases SHP-1/2 or phosphatidylinositol phosphatase SHIP (52). These phosphatases dephosphorylate key signaling molecules involved in the activation pathways, thereby inhibiting cell activation. This inhibitory signal results principally from the interaction of inhibitory receptors, such as KIR in humans and Ly49 in mice, with physiological levels of MHC-I molecules. The CD94/NKG2A heterodimer is another major inhibitory receptor, expressed by around half the NK cells in human and mice; it is capable of recognizing non-classical MHC-I molecules, such as human leukocyte antigen (HLA)-E in humans and Qa-1b in mice (53, 54).
NK cell activation results from the engagement of activating receptors, such as the activating isoforms of KIRs and Ly49, the natural cytotoxicity receptors (NCRs), the SLAM (signaling lymphocyte activating molecule)-related receptors, NKG2D and CD16, leading to NK cell activation through the initiation of different signaling pathways (55). Activating receptors can be classified into three groups: receptors associating with adaptors bearing the immunoreceptor tyrosine-based activation motif (ITAM) (activating KIRs, CD16 and NCRs), NKG2D and other receptors signaling via different pathways (CD2, 2B4, DNAM-1 and NKp80). NKG2D associate with DAP10 in humans but with DAP12 (also known as TYROBP/KARAP) in mice. CD16, NKp46 and NKp30 are coupled to CD3ζ and FcRγ as homo- or heterodimers, whereas NKp44 and KIR-S signal through DAP-12 (Figure 1). CD3ζ, FcRγ and DAP12 harbor ITAM motifs defined by the consensus sequence YxxL/Ix(6-8)YxxL/I (56, 57). Cell-surface receptor aggregation induces the recruitment of Src protein tyrosine kinases, which phosphorylate the ITAM, leading to the recruitment and activation of the tandem SH2 domain–containing Syk and ZAP-70 tyrosine kinases. This cascade of events leads to the recruitment and phosphorylation of multiple signaling molecules (LAT, SLP-76, PI3K, PLCγ, Grb2, Vav, Cbl, Nck) and the expression of subsequent effector functions (52). Adaptor molecules differ in the number of ITAMs. FcRγ and DAP12 each contain only one ITAM, whereas CD3ζ has three ITAM motifs. The CD16 Fc receptor, NKp46 and NKp30 can, therefore, theoretically signal via the phosphorylation of up to six ITAMs, whereas KIR-S and NKp44 can deliver activating signals via the phosphorylation of only two ITAMs (55).
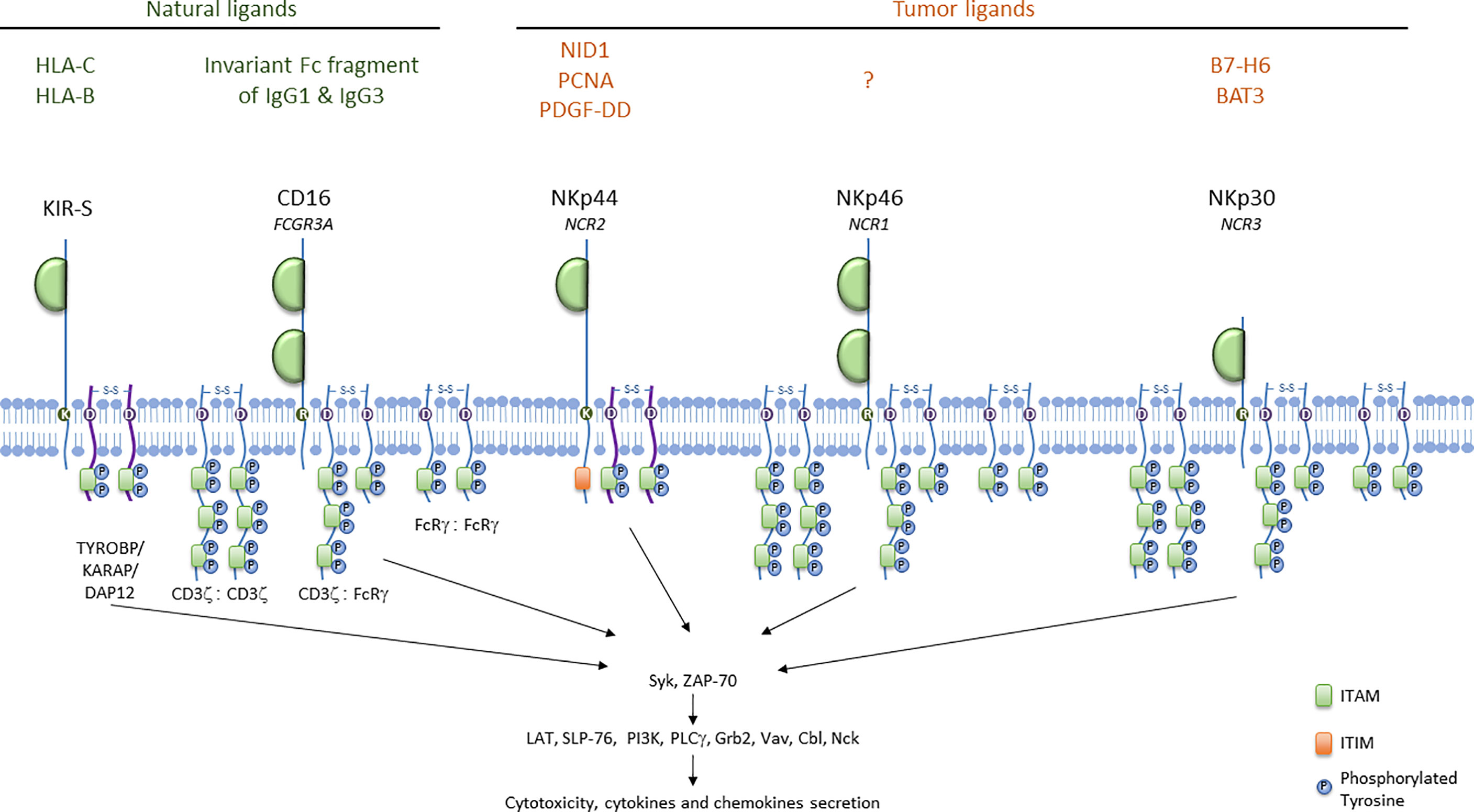
Figure 1 ITAM-bearing receptors in humans. ITAM-bearing receptors in humans are represented at the membrane associated either with CD3ζ or with FcRγ as homo- or heterodimers. ITAM motifs are represented with the proteins involved in the activation signal cascade. Natural ligands for KIR-S and CD16, and tumor ligands for NCRs are indicated.
Cell Surface Receptors Associated With ITAM-Bearing Polypeptides in Humans
Activating KIRs
KIR genes are located in the leukocyte receptor complex on chromosome 19. The KIR gene family encompasses 15 genes and 2 pseudogenes. The KIR nomenclature indicates the structure and the function of the receptor, and take into account the nucleotide sequence similarity among the different KIR family members (17). Thus, the nomenclature indicates the numbers of extracellular domains (2D or 3D), and provides information on the length of the cytoplasmic tail (long L or short S). In general, activating receptors have a short cytoplasmic fragment and possess in their transmembrane region, of a charged lysine residue that allows their association with the DAP12 bearing the ITAM motif. Only the KIR2DL4 receptor can conduct both activating and inhibitory signals. KIR genotypes are divided into A and B haplotypes: haplotype A contains only one activating receptor 2DS4 while haplotype B contain different combinations of the activating genes KIR2DS1, -S2, -S3, -S5 and KIR3DS1. Haplotype B can contains from one to five activating KIR receptors. KIR ligands are HLA class I, HLA-C and HLA-B as indicated in Figure 1. Additionally, recent work has shown that KIR2DS2+ NK cells can respond to different malignant cell lines in vitro through the interaction of KIR2DS2 with a β2-microglobulin-independent ligand (58).
Upon engagement by their ligands, activating KIRs signal via DAP12 and promote NK cell functions. Correlation between activating KIR and cancer prevalence is still unclear, but it has been shown that KIR2DS1, 2DS3, 2DS5, 3DS1 are associated with overall survival in gastric cancer (59). However, the presence of KIR2DS2, regardless of the HLA-C genotype, is associated with neuroblastoma (60).
CD16
The FCGR3A gene encodes the CD16 receptor. This receptor consists of two extracellular Ig domains, a short cytoplasmic tail and a transmembrane domain (Figure 1). It associates with FcRγ and CD3ζ homodimers or heterodimers in humans and with FcRγ homodimers in mouse NK cells (56, 57). CD16A is a low to intermediate-affinity glycoprotein transmembrane receptor for the Fc domains of IgG1 and IgG3 expressed by NK cells, whereas the isoform CD16B is mainly expressed by neutrophils. CD16 engagement initiates antibody-dependent cell-mediated cytotoxicity (ADCC), and CD16A expression levels are positively correlated with the ADCC potency of NK cells (61, 62). Upon CD16A engagement, tumor cells can be killed by the release of cytotoxic granules. Also, NK cells produce proinflammatory cytokines, such as IFN-γ, and chemokines, which lead to the recruitment and activation of tumor-infiltrating immune cells, contributing to tumor cell killing (63–71). CD16 triggering also affects NK cell survival, as CD16 ligation facilitates proliferation and sustained growth of IL-2-activated human NK cells (72). However, CD16 ligation also promotes NK cell death (73).
Two polymorphic variants of CD16A have been described resulting from a single nucleotide polymorphism (SNP). These allelic variants differ in their affinity for IgG with a KD of 0.75 µM for CD16A-158V and 5 µM for CD16A-158F, and this can affect the efficacy of tumor-targeting therapeutic mAbs (74). Patients homozygous for the higher-affinity CD16A variant with follicular lymphoma (FL) (75), non-Hodgkin lymphoma (76), CRC (77), or breast cancer (78) have a better clinical outcome after treatment with anti-tumor mAbs than patients with the low-affinity CD16 allele (see Table 2). This correlation has been observed in many, but not all studies (106–110), suggesting that other parameters may also be involved, such as variability in copy numbers of FCGR3A transcripts (61). In addition to recognizing IgG, CD16A has been shown to mediate spontaneous cytotoxicity of melanoma cells (111) by associating with CD2 in cis to recognize CD58 as a ligand (112).
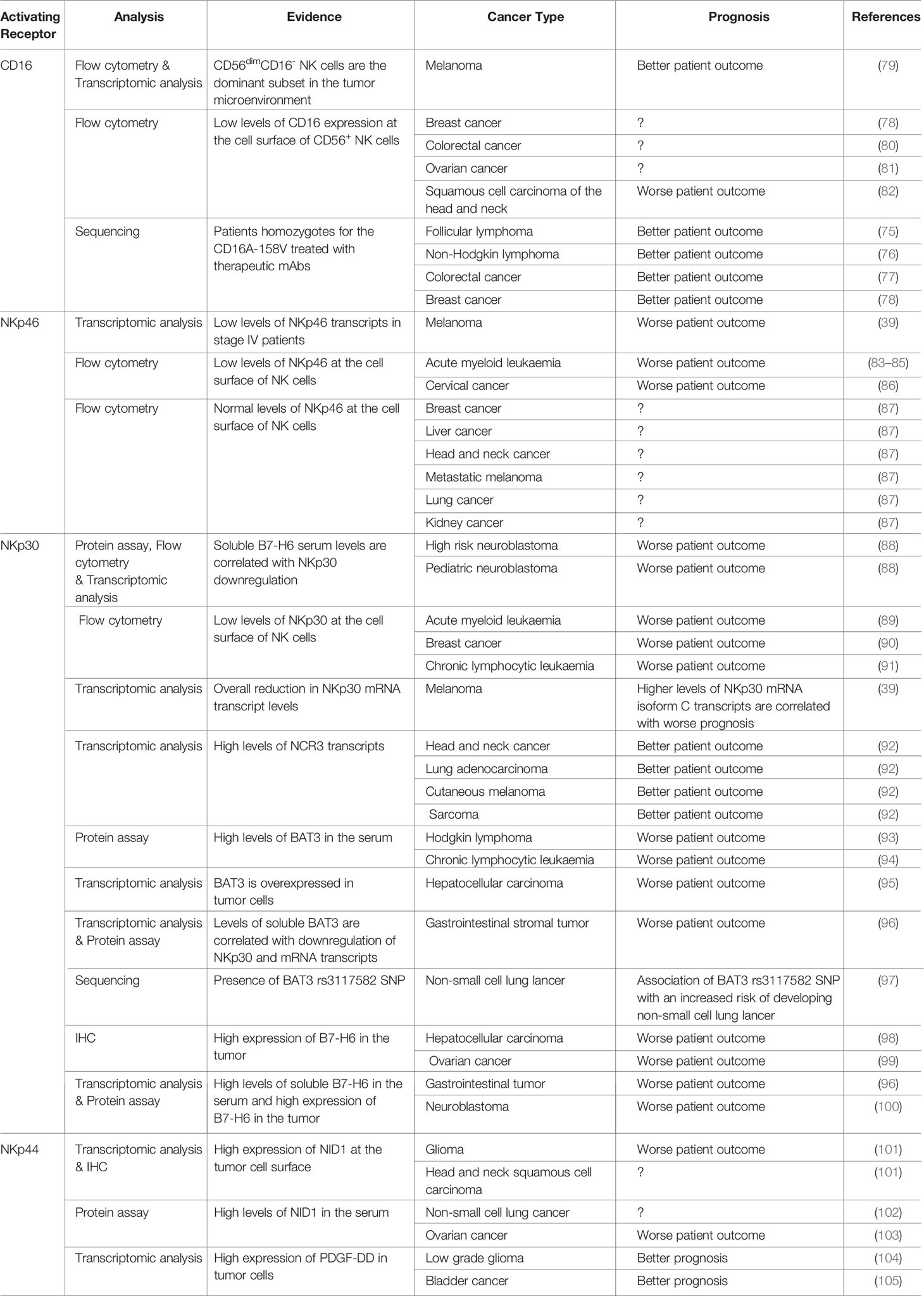
Table 2 Dysregulation of NK cell surface receptor associated with ITAM-bearing polypeptides in cancer.
Unlike other activating receptors expressed by NK cells, CD16A displays a downregulation of its surface expression after triggering, by cleavage and shedding induced by the ADAM17 metalloprotease (113–117). However, ADAM17 deficiency in humans does not affect NK cell effector activities (118). ADAM-17 is overexpressed in several human cancers, such as non-small cell lung cancer (119) and gastric carcinoma (120, 121), and this overexpression is associated with poor patient survival. CD16A levels on the surface of NK cells are downregulated in the tumor microenvironment of colorectal carcinoma (80), ovarian carcinoma (81), head and neck cancers (82), breast cancer (122) and melanoma (79), contributing to NK cell dysfunction.
NKp46
The NCR family has three members: NKp46, NKp44 and NKp30. NKp46 (or CD335, NCR1), is the only NCR conserved across mammalian species; it is expressed by all mature NK cells, ILC1 (123), a subset of ILC3, and Tγδ cells (124). NKp46, encoded by the NCR1 gene, is a 46 kDa type 1 transmembrane protein from the immunoglobulin (Ig) superfamily characterized by two immunologublin-C2-like extracellular domains connected by a stalk domain to a transmembrane domain and a short cytoplasmic tail. NKp46 is associated with CD3ζ and/or FcRγ at the cell membrane (125) (Figure 1).
NKp46 recognizes several microbial ligands, a surface protein on healthy pancreatic β cells and the soluble complement factor P (124). In vitro, NK cells can kill different types of tumor cells via NKp46, but the identity of the ligand involved remains unknown. In vivo, genetic deficiencies of NKp46 in mice impair the clearance of subcutaneous PD1.6 T lymphoma (126), RET melanoma tumors (127) and B16 melanoma metastases in the lung (127–129). Moreover, the overexpression of an NKp46 transgene has been shown to enhance the clearance of melanoma metastases in the lung (130). Enhanced NKp46 signaling has been shown to elicit IFN-γ secretion and to increase fibronectin deposition in the tumor, altering the architecture of the solid tumor and decreasing the formation of melanoma metastases (39).
NKp46 expression at the cell surface is downregulated on NK cells/ILC1 from patients with acute myeloid leukemia, this phenotype being reversible upon complete remission (83–85) (see Table 2). Patients with cervical cancer also present a decrease in NKp46 expression on the surface of NK cells/ILC1 (86). Still, the level of surface expression of NKp46 in breast, liver, head and neck, metastatic melanoma, lung and kidney cancer patients remains quite similar to the one of healthy donors and was not correlated with NK cell dysfunction (87).
NKp30
NKp30 (or CD337, NCR3) is a 30 kDa protein expressed by all mature human NK cells (131), ILC2, and tonsil-derived ILC3. This surface receptor consists of an extracellular IgV domain and a hydrophobic transmembrane domain with a charged arginine residue capable of associating with the adaptor proteins CD3ζ and/or FcRγ (132) (Figure 1).
NKp30 has six variants (NKp30a, NKp30b, NKp30c, NKp30d, NKp30e and NKp30f), due to alternative splicing. NKp30a, NKp30b and NKp30c have a V-type Ig-like extracellular domain, whereas NKp30d, NKp30e and NKp30f have a different C-type Ig extracellular domain. It has been reported that the triggering of the NKp30a and NKp30b variants promotes the production of IFN-γ, whereas cells transfected with the NKp30c variant produce high levels of immunosuppressive IL-10 (133). This difference may reflect the strong association of the NKp30a and NKp30b variants with CD3ζ and FcRγ, whereas NKp30c associates very little with these adaptors (133, 134), but the relevance of these findings remains to be understood.
NKp30 recognizes microbial and tumor-derived ligands. The tumor ligands it recognizes include nuclear HLA-B-associated transcript 3 (BAT3), a protein chaperone released from tumors (135) and reported to trigger NK cell activation. Conversely, soluble BAT3 has been detected in the serum of patients with Hodgkin lymphoma and chronic lymphocyte leukemia (93, 94), and has been shown to suppress NK cell activation. BAT3 also promotes melanoma metastasis (136, 137) and has been identified as a biomarker for poorer patient survival in hepatocellular carcinoma (95), gastrointestinal stromal tumor (96) non-small cell lung cancer (97) and lung adenocarcinoma (138) patients (see Table 2).
NKp30 binds to B7-H6, a B7 family member. Membrane expression of B7-H6 renders cancer cells sensitive to NK cell-mediated cytolysis (139). Conversely, NK cells chronically stimulated with soluble forms of B7-H6 resulting from metalloprotease-mediated shedding may display a downregulation of NKp30 expression contributing to tumor immune escape (140). Soluble B7-H6 has been detected in the serum of patients with hepatocellular carcinoma (98) or gastrointestinal tumors (96) and in peritoneal fluid from patients with ovarian cancer (99). High levels of soluble B7-H6 are also associated with bone marrow metastasis and the chemoresistance of neuroblastoma cells (88, 100).
Overall, increases in the level of NCR3 transcripts have been reported to be associated with better patient survival in the context of head and neck cancer, lung adenocarcinoma, cutaneous melanoma and sarcoma (92). NKp30a and NKp30b are associated with better survival and prognosis for gastrointestinal stromal tumors (133, 141), hepatocellular carcinoma (142) and pediatric neuroblastoma (88), whereas morbidity is higher in patients expressing predominantly the NKp30c isoform. NKp30 protein levels at the surface of NK cells are lower in patients with breast cancer (90) and chronic lymphocytic leukemia (91). The monitoring of NKp30 expression could thus be of interest to predict NK cell dysfunction. As such, NKp30 is downregulated in acute myeloid leukemia and NKp30 status has been proposed as an early prognostic biomarker identifying patients at intermediate risk with a poor prognosis (89).
NKp44
NKp44 is an activating receptor expressed by activated human NK cells, tonsil NCR+ ILC3s and plasmacytoid dendritic cells in humans, but not in mice. NKp44 is a 44 kDa protein consisting of an extracellular immunoglobulin V-like domain linked by a stalk domain to a transmembrane domain connected to an intracellular moiety (132) (Figure 1). At the membrane, NKp44 associates with the adaptor protein DAP12.Three NKp44 mRNA splice variants (NKp44-1,−2,−3) have been reported, each endowed with different signaling capabilities based on the presence (NKp44-1) or absence (NKp44-2 and−3) of an ITIM in their cytoplasmic tail. Initially described as non-functional (143), this ITIM may, in some cases, transduce an inhibitory signal (144). NKp44-1 expression has been associated with poor survival in AML patients (145).
NKp44 recognizes various tumor ligands, including the ‘proliferating cell nuclear antigen’ (PCNA) (146), platelet-derived growth factor D (PDGF-DD) (147), nidogen 1 (148).
PCNA is a nuclear protein involved in regulating DNA replication, DNA repair, and cell cycle progression; it may be overexpressed in cancer cells, promoting tumor survival and malignancy (149). The NKp44/PCNA axis inhibits NK cell-mediated cytotoxicity and IFN-γ secretion via the ITIM motif of the NKp44-1 variant (145).
NKp44 also recognizes the soluble PDGF-DD, a member of the PDGF family corresponding to the active processed form of PDGF-D (150, 151). Remarkably, NKp44/PDGF-DD interaction promotes the secretion of IFN-γ and TNF-α by IL-2-activated NK cells, but not cytotoxicity, limiting tumor cell growth in vitro and tumor spread in a transgenic NCR2 mouse model (147). PDGF-DD is secreted by various tumors and promotes cell proliferation, stromal cell recruitment, angiogenesis, epithelial-mesenchymal transition and metastasis (152, 153) (see Table 2).
Barrow and coworkers recently identified transcriptional signatures unique to PDGF-DD-activated NK cells and established the abundance of this signature in The Cancer Genome Atlas (TCGA) low-grade glioma (LGG) and bladder cancer (BLCA) datasets with CIBERSORT. They found that tumors of patients enriched in PDGF-DD-activated NK cell or memory CD8+ T-cell phenotypes were associated with a more favorable prognosis in LGG, but with a poor prognosis in BLCA (104, 105).
Nidogen 1 (NID1) is an extracellular matrix protein (148) that promotes epithelial-mesenchymal transition (EMT) and metastasis in ovarian cancer. NID1 increases the adhesion of ETV5-overexpressing endometrial cancer cells to the extracellular matrix (ECM) and promotes their proliferation and migration. The NKp44/NID1 axis does not induce classical NK cell activation, but it does inhibit the IFN-γ production induced by PDGF-DD following NKp44 engagement. Soluble NID1 levels are high in non-small cell lung cancer (102), ovarian (103), head and neck squamous cell carcinoma and glioma (101) (see Table 2).
Finally, NKp44 can also recognize a subset of HLA-DP molecules, depending on HLA-DP allotype (HLA-DP401), this interaction being further modulated by the peptide presented by HLA-DP molecules (154). The NKp44/HLA-DP401 axis induces the production of IFN-γ by NK cells. Tumor cells exposed to IFN-γ express HLA-II (155). The NKp44/HLA-DP401 axis may, therefore, contribute to the favorable prognosis of certain tumors with high levels of HLA-II expression (156).
Harnessing NK Cell Activating Receptors
The mechanisms underlying the therapeutic effect of antibodies used in clinical practice depend on the cell types infiltrating the tumor and the type of FcγR they express. Unlike myeloid cells and B cells, which also display inhibitory FcγR isoforms, NK cells express only the CD16A activating receptor for IgG, and are therefore particularly responsive to therapeutic mAb-dependent activation. Several studies have reported that increases in the numbers of intratumoral and circulating NK cells are associated with favorable outcomes of treatment with anti-HER2-mAbs, anti-EGFR-mAb (cetuximab) and anti-CD20-mAb (obinutuzumab and rituximab) for breast cancer, head and neck squamous cell carcinoma, follicular lymphoma and diffuse large B-cell lymphoma (157–162). Furthermore, the Treg-mediated suppression of ADCC is correlated with lower clinical efficacy in cetuximab-treated HNSCC patients (163).
The first clinical studies of bispecific NK cell engagers (BiKEs) date back to the 1990s. A F(ab’)2 format molecule was generated to target CD16 and NK cells and CD30 as the tumor antigen (Tag), to treat end-stage Hodgkin’s disease (164–166). Unfortunately, this treatment was highly toxic. Novel BiKEs have been generated in recent years, to link scFvs against an NK-cell receptor (CD16, NKp46, NKp30 or NKG2D) and a Tag (BCMA, CD19, CD20, CD30, CD33, CD38, CD133, EPCAM and GPC3) (166–173). The engineering of the Fc portion of tumor-targeting mAbs makes specific targeting of the FcRγ subtype possible. Specific genetic mutations and glycoengineering can be used to increase the affinity of the IgG for CD16. The anti-CD20 mAb obinutuzumab differs from rituximab in having a defucosylated Fc region, increasing CD16 binding affinity (174–177). In combination with chemotherapy, the antitumor efficacy of obinutuzumab was superior to that of rituximab, with the elicitation of stronger ADCC due to its binding to both CD16A-158V and 158F, indifferently, in patients suffering from chronic lymphocytic leukemia (CLL) or follicular lymphoma (178–181)
Another way to improve the responsiveness of NK cells is to stimulate them with cytokines. The IL-12/IL-15/IL-18 inflammatory cytokine cocktail can confer memory-like features on murine and human NK cells in the absence of an antigen (14, 16, 182). Heteromeric IL-12/IL-15/IL-18/CD16scFv fusion proteins were found not to increase NK cell reactivity over that achieved with IL-12/IL-15/IL-18, and were, therefore, not developed any further. Furthermore, the reactivity of CIML NK cells against HuT-78 lymphoblasts in the presence of AFM13 (CD16/CD33) is moderately higher than that of freshly isolated NK cells (183). However, TriKEs composed of one scFv against CD16, another against CD33 and a human IL-15 crosslinker promote NK cell effector functions, and also increase NK cell expansion and persistence in vivo in mouse preclinical models of AML and ovarian cancer (184, 185). Thus, co-engagement of the ITAM and JAK/STAT pathways renders the NK cell response more potent. The humanized TriKE CD16/IL-15/CD33 is currently under evaluation in a phase I/II trial for the treatment of high-risk myelodysplastic syndromes, refractory/relapsed AML and advanced systemic mastocytosis (NCT03214666).
NKp46 appears to be a particularly interesting potential target, as it is persistently expressed on tumor-infiltrating NK cells, whereas NKp30, CD16 and NKG2D are frequently downregulated. NKp46 is also more NK-cell specific, as NKG2D is widely expressed in T cells and can cause severe T cell-associated toxicities (70). Bispecific NK cell engagers against NKp46 and tumor antigen GPC3 are being developed (186). Trispecific NK cell engagers targeting both NKp46 and CD16 on NK cells together with a Tag, named ANKET (Antibody-based NK cell engager therapeutics), have been generated. Bispecific NKp46-Tag activate NK cells, but trispecific ANKET have greater potency for the activation of NK cells in vitro and for increasing the number of NK cells at the tumor site, thereby providing a higher degree of tumor growth control in solid and invasive mouse tumor models. The co-engagement of a larger number of NK cell activating receptors therefore increases efficacy. Trispecific ANKET can control tumor development in preclinical models of B cell lymphoma (87). In pediatric B-cell acute lymphoblastic leukemia, the trispecific ANKET targeting NKp46, CD16 and CD19 has been shown to have greater cytotoxicity against BCP-ALL cell lines and to lead to an increase in the percentage of IFN-γ+ NK cells (187). Finally, tetraspecific ANKET combining NKp46/CD16/CD20 and IL-2 are now being generated and are displaying strong preclinical antitumor efficacy against CD20+ tumor cells and an ability to increase NK cell proliferation (Demaria et al., unpublished data).
Concluding Remarks
High levels of NK cell infiltration have been associated with a better prognosis, at least for some tumors. However, the tumor microenvironment may affect NK cell invasion of the tumor mass and may also have suppressive activities against NK cells. It is thus of interest to favor NK cell infiltration of tumors and to boost NK cell functions in the tumor bed. The antitumor response of NK cells is effectively stimulated by tumor-targeting mAbs, through NCRs and CD16 triggering. Current research is aiming to promote NK cell survival/proliferation, recruitment, and persistence at tumor sites. Future studies will aim to potentiate these immunotherapies by combining them with immune checkpoint inhibitors, chemotherapy or drugs targeting the immunosuppressive tumor microenvironment.
Author Contributions
All authors contributed to gathering of data, writing, editing, and revising of the manuscript.
Funding
The EV laboratory at CIML and Assistance-Publique des Hopitaux de Marseille is supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (TILC, grant agreement No. 694502 and MInfla-TILC, grant agreement No. 875102 - MInfla-Tilc), the Agence Nationale de la Recherche including the PIONEER Project (ANR-17-RHUS0007), MSDAvenir, Innate Pharma and institutional grants to the CIML (INSERM, CNRS, and Aix-Marseille University) and to Marseille Immunopole. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. HMK is recipient of a PhD grant from ‘Institut Cancer Immunologie’.
Conflict of Interest
EV is an employee of Innate Pharma.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Guia S, Narni-Mancinelli E. Helper-Like Innate Lymphoid Cells in Humans and Mice. Trends Immunol (2020) 41(5):436–52. doi: 10.1016/j.it.2020.03.002
2. Björkström NK, Ljunggren H-G, Michaëlsson J. Emerging Insights Into Natural Killer Cells in Human Peripheral Tissues. Nat Rev Immunol (2016) 16(5):310–20. doi: 10.1038/nri.2016.34
3. Hanna J, Bechtel P, Zhai Y, Youssef F, McLachlan K, Mandelboim O. Novel Insights on Human NK Cells’ Immunological Modalities Revealed by Gene Expression Profiling. J Immunol (2004) 173(11):6547–63. doi: 10.4049/jimmunol.173.11.6547
4. Yu J, Freud AG, Caligiuri MA. Location and Cellular Stages of Natural Killer Cell Development. Trends Immunol (2013) 34(12):573–82. doi: 10.1016/j.it.2013.07.005
5. Crinier A, Milpied P, Escalière B, Piperoglou C, Galluso J, Balsamo A, et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity (2018) 49(5):971–986.e5. doi: 10.1016/j.immuni.2018.09.009
6. Cooper MA, Fehniger TA, Caligiuri MA. The Biology of Human Natural Killer-Cell Subsets. Trends Immunol (2001) 22(11):633–40. doi: 10.1016/S1471-4906(01)02060-9
7. Chan A, Hong D-L, Atzberger A, Kollnberger S, Filer AD, Buckley CD, et al. CD56bright Human NK Cells Differentiate Into CD56 Dim Cells: Role of Contact With Peripheral Fibroblasts. J Immunol (2007) 179(1):89–94. doi: 10.4049/jimmunol.179.1.89
8. Dulphy N, Haas P, Busson M, Belhadj S, Peffault de Latour R, Robin M, et al. An Unusual CD56(Bright) CD16(Low) NK Cell Subset Dominates the Early Posttransplant Period Following HLA-Matched Hematopoietic Stem Cell Transplantation. J Immunol (2008) 181(3):2227–37. doi: 10.4049/jimmunol.181.3.2227
9. Crinier A, Dumas P-Y, Escaliere B, Piperoglou C, Gil L, Villacreces A, et al. Single-Cell Profiling Reveals the Trajectories of Natural Killer Cell Differentiation in Bone Marrow and a Stress Signature Induced by Acute Myeloid Leukemia. Cell Mol Immunol (2021) 18(5):1290–304. doi: 10.1038/s41423-020-00574-8
10. Smith SL, Kennedy PR, Stacey KB, Worboys JD, Yarwood A, Seo S, et al. Diversity of Peripheral Blood Human NK Cells Identified by Single-Cell RNA Sequencing. Blood Adv (2020) 4(7):1388–406. doi: 10.1182/bloodadvances.2019000699
11. Björkström NK, Lindgren T, Stoltz M, Fauriat C, Braun M, Evander M, et al. Rapid Expansion and Long-Term Persistence of Elevated NK Cell Numbers in Humans Infected With Hantavirus. J Exp Med (2011) 208(1):13–21. doi: 10.1084/jem.20100762
12. Björkström NK, Ljunggren H-G, Sandberg JK. CD56 Negative NK Cells: Origin, Function, and Role in Chronic Viral Disease. Trends Immunol (2010) 31(11):401–6. doi: 10.1016/j.it.2010.08.003
13. Dou Y, Fu B, Sun R, Li W, Hu W, Tian Z, et al. Influenza Vaccine Induces Intracellular Immune Memory of Human NK Cells. PloS One (2015) 10(3):e0121258. doi: 10.1182/blood-2004-05-2058
14. Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-Induced Memory-Like Natural Killer Cells. Proc Natl Acad Sci U S A. (2009) 106(6):1915–9. doi: 10.1126/science.1198687
15. Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine-Induced Memory-Like Natural Killer Cells Exhibit Enhanced Responses Against Myeloid Leukemia. Sci Transl Med (2016) 8(357):357ra123. doi: 10.1182/blood-2009-08-238469
16. Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, et al. Cytokine Activation Induces Human Memory-Like NK Cells. Blood. (2012) 120(24):4751–60. doi: 10.1182/blood.V80.3.670.670
17. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or Adaptive Immunity? The Example of Natural Killer Cells. Science. (2011) 331(6013):44–9. doi: 10.1002/ijc.2910300118
18. Martin-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, et al. Induced Recruitment of NK Cells to Lymph Nodes Provides IFN-Gamma for T(H)1 Priming. Nat Immunol (2004) 5(12):1260–5.
19. Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-Killer Cells and Dendritic Cells: “L’union Fait La Force”. Blood. (2005) 106(7):2252–8. doi: 10.1172/JCI8082
20. Gorelik E, Wiltrout RH, Okumura K, Habu S, Herberman RB. Role of NK Cells in the Control of Metastatic Spread and Growth of Tumor Cells in Mice. Int J Cancer. (1982) 30(1):107–12. doi: 10.1111/j.1349-7006.1996.tb00241.x
21. Talmadge JE, Meyers KM, Prieur DJ, Starkey JR. Role of Natural Killer Cells in Tumor Growth and Metastasis: C57BL/6 Normal and Beige Mice. J Natl Cancer Inst (1980) 65(5):929–35. doi: 10.1016/j.cell.2016.01.002
22. Lee U, Santa K, Habu S, Nishimura T. Murine Asialo GM1+CD8+ T Cells as Novel Interleukin-12-Responsive Killer T Cell Precursors. Jpn J Cancer Res (1996) 87(5):429–32. doi: 10.1073/pnas.2026271118
23. Trambley J, Bingaman AW, Lin A, Elwood ET, Waitze SY, Ha J, et al. Asialo GM1(+) CD8(+) T Cells Play a Critical Role in Costimulation Blockade-Resistant Allograft Rejection. J Clin Invest. (1999) 104(12):1715–22. doi: 10.1038/ni.3809
24. Dadi S, Chhangawala S, Whitlock BM, Franklin RA, Luo CT, Oh SA, et al. Cancer Immunosurveillance by Tissue-Resident Innate Lymphoid Cells and Innate-Like T Cells. Cell. (2016) 164(3):365–77. doi: 10.1038/ni.3800
25. Ducimetiere L, Lucchiari G, Litscher G, Nater M, Heeb L, Nunez NG, et al. Conventional NK Cells and Tissue-Resident ILC1s Join Forces to Control Liver Metastasis. Proc Natl Acad Sci USA (2021) 118(27). doi: 10.3389/fimmu.2021.768989
26. Cortez VS, Ulland TK, Cervantes-Barragan L, Bando JK, Robinette ML, Wang Q, et al. SMAD4 Impedes the Conversion of NK Cells Into ILC1-Like Cells by Curtailing Non-Canonical TGF-Beta Signaling. Nat Immunol (2017) 18(9):995–1003. doi: 10.1182/blood-2013-07-515528
27. Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, et al. Tumor Immunoevasion by the Conversion of Effector NK Cells Into Type 1 Innate Lymphoid Cells. Nat Immunol (2017) 18(9):1004–15. doi: 10.1172/JCI61014
28. Vienne M, Etiennot M, Escaliere B, Galluso J, Spinelli L, Guia S, et al. Type 1 Innate Lymphoid Cells Limit the Antitumoral Immune Response. Front Immunol (2021) 12:768989. doi: 10.1093/oxfordjournals.annonc.a058659
29. Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, et al. Partial MCM4 Deficiency in Patients With Growth Retardation, Adrenal Insufficiency, and Natural Killer Cell Deficiency. J Clin Invest. (2012) 122(3):821–32. doi: 10.1016/j.molimm.2004.07.037
30. Spinner MA, Sanchez LA, Hsu AP, Shaw PA, Zerbe CS, Calvo KR, et al. GATA2 Deficiency: A Protean Disorder of Hematopoiesis, Lymphatics, and Immunity. Blood. (2014) 123(6):809–21. doi: 10.18632/aging.102774
31. Imai K, Matsuyama S, Miyake S, Suga K, Nakachi K. Natural Cytotoxic Activity of Peripheral-Blood Lymphocytes and Cancer Incidence: An 11-Year Follow-Up Study of a General Population. Lancet (2000) 356(9244):1795–9. doi: 10.1016/j.coi.2017.01.003
32. Fu K, Hui B, Wang Q, Lu C, Shi W, Zhang Z, et al. Single-Cell RNA Sequencing of Immune Cells in Gastric Cancer Patients. Aging (Albany NY). (2020) 12(3):2747–63. doi: 10.1067/mtc.2001.113026
33. Wu M, Mei F, Liu W, Jiang J. Comprehensive Characterization of Tumor Infiltrating Natural Killer Cells and Clinical Significance in Hepatocellular Carcinoma Based on Gene Expression Profiles. BioMed Pharmacother. (2020) 121:109637. doi: 10.1016/j.biopha.2019.109637
34. Villegas FR, Coca S, Villarrubia VG, Jimenez R, Chillon MJ, Jareno J, et al. Prognostic Significance of Tumor Infiltrating Natural Killer Cells Subset CD57 in Patients With Squamous Cell Lung Cancer. Lung Cancer. (2002) 35(1):23–8. doi: 10.1158/0008-5472.CAN-10-4179
35. Soo RA, Chen Z, Yan Teng RS, Tan HL, Iacopetta B, Tai BC, et al. Prognostic Significance of Immune Cells in non-Small Cell Lung Cancer: Meta-Analysis. Oncotarget. (2018) 9(37):24801–20. doi: 10.1158/2159-8290.CD-13-0985
36. Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, et al. The Head and Neck Cancer Immune Landscape and Its Immunotherapeutic Implications. JCI Insight (2016) 1(17):e89829. doi: 10.4049/jimmunol.0801878
37. Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S, et al. A Gene Signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma Patients. Cancer Immunol Res (2019) 7(7):1162–74. doi: 10.1007/BF01806249
38. Melaiu O, Chierici M, Lucarini V, Jurman G, Conti LA, De Vito R, et al. Cellular and Gene Signatures of Tumor-Infiltrating Dendritic Cells and Natural-Killer Cells Predict Prognosis of Neuroblastoma. Nat Commun (2020) 11(1):5992. doi: 10.1016/S0140-6736(00)03231-
39. Messaoudene M, Fregni G, Enot D, Jacquelot N, Neves E, Germaud N, et al. NKp30 Isoforms and NKp46 Transcripts in Metastatic Melanoma Patients: Unique NKp30 Pattern in Rare Melanoma Patients With Favorable Evolution. Oncoimmunology. (2016) 5(12):e1154251. doi: 10.1016/j.leukres.2013.07.038
40. Bovbjerg DH, Valdimarsdottir H. Familial Cancer, Emotional Distress, and Low Natural Cytotoxic Activity in Healthy Women. Ann Oncol (1993) 4(9):745–52. doi: 10.1158/2159-8290.CD-20-0655
41. Strayer DR, Carter WA, Brodsky I. Familial Occurrence of Breast Cancer Is Associated With Reduced Natural Killer Cytotoxicity. Breast Cancer Res Treat (1986) 7(3):187–92. doi: 10.1016/j.bbrc.2020.01.053
42. Shafer D, Smith MR, Borghaei H, Millenson MM, Li T, Litwin S, et al. Low NK Cell Counts in Peripheral Blood Are Associated With Inferior Overall Survival in Patients With Follicular Lymphoma. Leuk Res (2013) 37(10):1213–5. doi: 10.1096/fj.202100025R
43. Cozar B, Greppi M, Carpentier S, Narni-Mancinelli E, Chiossone L, Vivier E. Tumor-Infiltrating Natural Killer Cells. Cancer Discovery (2021) 11(1):34–44. doi: 0.1038/s41467-020-19781-y
44. Cao B, Liu M, Wang L, Liang B, Feng Y, Chen X, et al. Use of Chimeric Antigen Receptor NK-92 Cells to Target Mesothelin in Ovarian Cancer. Biochem Biophys Res Commun (2020) 524(1):96–102. doi: 10.1172/jci.insight.89829
45. Mensali N, Dillard P, Fayzullin A, Koksal H, Gaudernack G, Kvalheim G, et al. “Built-In” PD-1 Blocker to Rescue NK-92 Activity From PD-L1-Mediated Tumor Escape Mechanisms. FASEB J (2021) 35(9):e21750. doi: 10.1158/2326-6066.CIR-18-0500
46. Malmberg KJ, Schaffer M, Ringden O, Remberger M, Ljunggren HG. KIR-Ligand Mismatch in Allogeneic Hematopoietic Stem Cell Transplantation. Mol Immunol (2005) 42(4):531–4. doi: 10.1158/1078-0432.CCR-12-3847
47. Melero I, Rouzaut A, Motz GT, Coukos G. T-Cell and NK-Cell Infiltration Into Solid Tumors: A Key Limiting Factor for Efficacious Cancer Immunotherapy. Cancer Discovery (2014) 4(5):522–6. doi: 10.1016/S0169-5002(01)00292-6
48. Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. Profound Coordinated Alterations of Intratumoral NK Cell Phenotype and Function in Lung Carcinoma. Cancer Res (2011) 71(16):5412–22. doi: 10.18632/oncotarget.24835
49. Remark R, Alifano M, Cremer I, Lupo A, Dieu-Nosjean MC, Riquet M, et al. Characteristics and Clinical Impacts of the Immune Environments in Colorectal and Renal Cell Carcinoma Lung Metastases: Influence of Tumor Origin. Clin Cancer Res (2013) 19(15):4079–91. doi: 10.1126/science.290.5489.84
50. Muntasell A, Ochoa MC, Cordeiro L, Berraondo P, Lopez-Diaz de Cerio A, Cabo M, et al. Targeting NK-Cell Checkpoints for Cancer Immunotherapy. Curr Opin Immunol (2017) 45:73–81. doi: 10.1128/MCB.23.17.6291-6299.2003
51. Takanami I, Takeuchi K, Giga M. The Prognostic Value of Natural Killer Cell Infiltration in Resected Pulmonary Adenocarcinoma. J Thorac Cardiovasc Surg (2001) 121(6):1058–63. doi: 10.1038/ni1581
52. Tomasello E, Blery M, Vely F, Vivier E. Signaling Pathways Engaged by NK Cell Receptors: Double Concerto for Activating Receptors, Inhibitory Receptors and NK Cells. Semin Immunol (2000) 12(2):139–47. doi: 10.1038/342803a0
54. Stebbins CC, Watzl C, Billadeau DD, Leibson PJ, Burshtyn DN, Long EO. Vav1 Dephosphorylation by the Tyrosine Phosphatase SHP-1 as a Mechanism for Inhibition of Cellular Cytotoxicity. Mol Cell Biol (2003) 23(17):6291–9. doi: 10.4049/jimmunol.1600930
55. Lanier LL. Up on the Tightrope: Natural Killer Cell Activation and Inhibition. Nat Immunol (2008) 9(5):495–502. doi: 10.29252/ibj.23.5.4
56. Lanier LL, Yu G, Phillips JH. Co-Association of CD3 Zeta With a Receptor (CD16) for IgG Fc on Human Natural Killer Cells. Nature. (1989) 342(6251):803–5. doi: 10.1111/tan.12608
57. Lanier LL, Yu G, Phillips JH. Analysis of Fc Gamma RIII (CD16) Membrane Expression and Association With CD3 Zeta and Fc Epsilon RI-Gamma by Site-Directed Mutation. J Immunol (1991) 146(5):1571–6. doi: 10.4049/jimmunol.167.3.1141
58. Thiruchelvam-Kyle L, Hoelsbrekken SE, Saether PC, Bjornsen EG, Pende D, Fossum S, et al. The Activating Human NK Cell Receptor KIR2DS2 Recognizes a Beta2-Microglobulin-Independent Ligand on Cancer Cells. J Immunol (2017) 198(7):2556–67. doi: 10.1128/JVI.02666-08
59. Yousefinejad F, Jowkar F, Barani S, Jamali E, Mahmoudi E, Ramezani A, et al. Killer Cell Immunoglobulin-Like Receptors (KIRs) Genotype and Haplotype Analysis in Iranians With Non-Melanoma Skin Cancers. Iran BioMed J (2019) 23(5):330–7. doi: 10.1182/blood-2007-01-070656
60. Keating SE, Ni Chorcora C, Dring MM, Stallings RL, O'Meara A, Gardiner CM. Increased Frequencies of the Killer Immunoglobulin-Like Receptor Genes KIR2DL2 and KIR2DS2 Are Associated With Neuroblastoma. Tissue Antigens (2015) 86(3):172–7. doi: 10.1038/nri3839
61. Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased Natural Killer Cell Expression of CD16, Augmented Binding and ADCC Activity to Rituximab Among Individuals Expressing the Fc{gamma}RIIIa-158 V/V and V/F Polymorphism. Blood. (2007) 110(7):2561–4. doi: 10.1002/eji.200324070
62. Liu Q, Sun Y, Rihn S, Nolting A, Tsoukas PN, Jost S, et al. Matrix Metalloprotease Inhibitors Restore Impaired NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Human Immunodeficiency Virus Type 1 Infection. J Virol (2009) 83(17):8705–12. doi: 10.1016/j.molimm.2004.07.034
63. Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, Smyth MJ. Increased Susceptibility to Tumor Initiation and Metastasis in TNF-Related Apoptosis-Inducing Ligand-Deficient Mice. J Immunol (2002) 168(3):1356–61. doi: 10.4049/jimmunol.168.3.1356
64. Prager I, Watzl C. Mechanisms of Natural Killer Cell-Mediated Cellular Cytotoxicity. J Leukoc Biol (2019) 105(6):1319–29. doi: 10.1002/JLB.MR0718-269R
65. Smyth MJ, Cretney E, Kelly JM, Westwood JA, Street SE, Yagita H, et al. Activation of NK Cell Cytotoxicity. Mol Immunol (2005) 42(4):501–10. doi: 10.1038/nrc.2015.5
66. Voskoboinik I, Whisstock JC, Trapani JA. Perforin and Granzymes: Function, Dysfunction and Human Pathology. Nat Rev Immunol (2015) 15(6):388–400. doi: 10.1189/jlb.5VMR0415-141R
67. Wallin RP, Screpanti V, Michaelsson J, Grandien A, Ljunggren HG. Regulation of Perforin-Independent NK Cell-Mediated Cytotoxicity. Eur J Immunol (2003) 33(10):2727–35.
68. Battella S, Cox MC, Santoni A, Palmieri G. Natural Killer (NK) Cells and Anti-Tumor Therapeutic mAb: Unexplored Interactions. J Leukoc Biol (2016) 99(1):87–96.
69. Bottcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, et al. NK Cells Stimulate Recruitment of Cdc1 Into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. (2018) 172(5):1022–37.e14. doi: 10.1084/jem.181.1.339
70. Morvan MG, Lanier LL. NK Cells and Cancer: You can Teach Innate Cells New Tricks. Nat Rev Cancer. (2016) 16(1):7–19. doi: 10.1074/jbc.M510171200
71. Trinchieri G, Valiante N. Receptors for the Fc Fragment of IgG on Natural Killer Cells. Nat Immun (1993) 12(4-5):218–34. doi: 10.1200/JCO.2003.05.013
72. Warren HS, Kinnear BF. Quantitative Analysis of the Effect of CD16 Ligation on Human NK Cell Proliferation. J Immunol (1999) 162(2):735–42. doi: 10.1182/blood-2002-02-0469
73. Ortaldo JR, Mason AT, O'Shea JJ. Receptor-Induced Death in Human Natural Killer Cells: Involvement of CD16. J Exp Med (1995) 181(1):339–44. doi: 10.1200/JCO.2006.08.8021
74. Ferrara C, Stuart F, Sondermann P, Brunker P, Umana P. The Carbohydrate at FcgammaRIIIa Asn-162An Element Required for High Affinity Binding to non-Fucosylated IgG Glycoforms. J Biol Chem (2006) 281(8):5032–6. doi: 10.1200/JCO.2007.14.8957
75. Weng WK, Levy R. Two Immunoglobulin G Fragment C Receptor Polymorphisms Independently Predict Response to Rituximab in Patients With Follicular Lymphoma. J Clin Oncol (2003) 21(21):3940–7.
76. Manches O, Lui G, Chaperot L, Gressin R, Molens JP, Jacob MC, et al. In Vitro Mechanisms of Action of Rituximab on Primary Non-Hodgkin Lymphomas. Blood. (2003) 101(3):949–54. doi: 10.1016/j.jss.2017.06.032
77. Zhang W, Gordon M, Schultheis AM, Yang DY, Nagashima F, Azuma M, et al. FCGR2A and FCGR3A Polymorphisms Associated With Clinical Outcome of Epidermal Growth Factor Receptor Expressing Metastatic Colorectal Cancer Patients Treated With Single-Agent Cetuximab. J Clin Oncol (2007) 25(24):3712–8. doi: 10.4049/jimmunol.1300313
78. Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, et al. Immunoglobulin G Fragment C Receptor Polymorphisms and Clinical Efficacy of Trastuzumab-Based Therapy in Patients With HER-2/Neu-Positive Metastatic Breast Cancer. J Clin Oncol (2008) 26(11):1789–96. doi: 10.4049/jimmunol.1301024
79. Vujanovic L, Chuckran C, Lin Y, Ding F, Sander CA, Santos PM, et al. CD56(dim) CD16(-) Natural Killer Cell Profiling in Melanoma Patients Receiving a Cancer Vaccine and Interferon-Alpha. Front Immunol (2019) 10:14. doi: 10.18632/oncotarget.23999
80. Nakagomi H, Petersson M, Magnusson I, Juhlin C, Matsuda M, Mellstedt H, et al. Decreased Expression of the Signal-Transducing Zeta Chains in Tumor-Infiltrating T-Cells and NK Cells of Patients With Colorectal Carcinoma. Cancer Res (1993) 53(23):5610–2. doi: 10.1186/1756-8722-6-1
81. Lai P, Rabinowich H, Crowley-Nowick PA, Bell MC, Mantovani G, Whiteside TL. Alterations in Expression and Function of Signal-Transducing Proteins in Tumor-Associated T and Natural Killer Cells in Patients With Ovarian Carcinoma. Clin Cancer Res (1996) 2(1):161–73. doi: 10.1172/JCI36022
82. Kuss I, Saito T, Johnson JT, Whiteside TL. Clinical Significance of Decreased Zeta Chain Expression in Peripheral Blood Lymphocytes of Patients With Head and Neck Cancer. Clin Cancer Res (1999) 5(2):329–34. doi: 10.4049/jimmunol.1102461
83. Fauriat C, Just-Landi S, Mallet F, Arnoulet C, Sainty D, Olive D, et al. Deficient Expression of NCR in NK Cells From Acute Myeloid Leukemia: Evolution During Leukemia Treatment and Impact of Leukemia Cells in NCRdull Phenotype Induction. Blood (2007) 109(1):323–30. doi: 10.1182/blood-2005-08-027979
84. Stringaris K, Sekine T, Khoder A, Alsuliman A, Razzaghi B, Sargeant R, et al. Leukemia-Induced Phenotypic and Functional Defects in Natural Killer Cells Predict Failure to Achieve Remission in Acute Myeloid Leukemia. Haematologica (2014) 99(5):836–47. doi: 10.3324/haematol.2013.087536
85. Chretien AS, Devillier R, Fauriat C, Orlanducci F, Harbi S, Le Roy A, et al. NKp46 Expression on NK Cells as a Prognostic and Predictive Biomarker for Response to Allo-SCT in Patients With AML. Oncoimmunology (2017) 6(12):e1307491. doi: 10.1080/2162402X.2017.1307491
86. Garcia-Iglesias T, Del Toro-Arreola A, Albarran-Somoza B, Del Toro-Arreola S, Sanchez-Hernandez PE, Ramirez-Duenas MG, et al. Low NKp30, NKp46 and NKG2D Expression and Reduced Cytotoxic Activity on NK Cells in Cervical Cancer and Precursor Lesions. BMC Cancer. (2009) 9:186. doi: 10.1186/1471-2407-9-186
87. Gauthier L, Morel A, Anceriz N, Rossi B, Blanchard-Alvarez A, Grondin G, et al. Multifunctional Natural Killer Cell Engagers Targeting NKp46 Trigger Protective Tumor Immunity. Cell (2019) 177(7):1701–13.e16. doi: 10.1016/j.cell.2019.04.041
88. Semeraro M, Rusakiewicz S, Zitvogel L, Kroemer G. Natural Killer Cell Mediated Immunosurveillance of Pediatric Neuroblastoma. Oncoimmunology (2015) 4(11):e1042202. doi: 10.1080/2162402X.2015.1042202
89. Chretien AS, Fauriat C, Orlanducci F, Rey J, Borg GB, Gautherot E, et al. NKp30 Expression is a Prognostic Immune Biomarker for Stratification of Patients With Intermediate-Risk Acute Myeloid Leukemia. Oncotarget (2017) 8(30):49548–63. doi: 10.18632/oncotarget.17747
90. Mamessier E, Sylvain A, Thibult ML, Houvenaeghel G, Jacquemier J, Castellano R, et al. Human Breast Cancer Cells Enhance Self Tolerance by Promoting Evasion From NK Cell Antitumor Immunity. J Clin Invest. (2011) 121(9):3609–22. doi: 10.1172/JCI45816
91. Costello RT, Knoblauch B, Sanchez C, Mercier D, Le Treut T, Sebahoun G. Expression of Natural Killer Cell Activating Receptors in Patients With Chronic Lymphocytic Leukaemia. Immunology (2012) 135(2):151–7. doi: 10.1111/j.1365-2567.2011.03521.x
92. Huntington ND, Cursons J, Rautela J. The Cancer-Natural Killer Cell Immunity Cycle. Nat Rev Cancer. (2020) 20(8):437–54. doi: 10.1038/s41568-020-0272-z
93. Reiners KS, Kessler J, Sauer M, Rothe A, Hansen HP, Reusch U, et al. Rescue of Impaired NK Cell Activity in Hodgkin Lymphoma With Bispecific Antibodies In Vitro and in Patients. Mol Ther (2013) 21(4):895–903. doi: 10.1038/mt.2013.14
94. Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M, et al. Soluble Ligands for NK Cell Receptors Promote Evasion of Chronic Lymphocytic Leukemia Cells From NK Cell Anti-Tumor Activity. Blood (2013) 121(18):3658–65. doi: 10.1182/blood-2013-01-476606
95. Zhang Y, Wang Y, Liu H, Li B. Six Genes as Potential Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma Through Data Mining. J Cell Physiol (2019) 234(6):9787–92. doi: 10.1002/jcp.27664
96. Rusakiewicz S, Perier A, Semeraro M, Pitt JM, Pogge von Strandmann E, Reiners KS, et al. NKp30 Isoforms and NKp30 Ligands are Predictive Biomarkers of Response to Imatinib Mesylate in Metastatic GIST Patients. Oncoimmunology (2017) 6(1):e1137418. doi: 10.1080/2162402X.2015.1137418
97. Etokebe GE, Zienolddiny S, Kupanovac Z, Enersen M, Balen S, Flego V, et al. Association of the FAM46A Gene VNTRs and BAG6 Rs3117582 SNP With non Small Cell Lung Cancer (NSCLC) in Croatian and Norwegian Populations. PloS One (2015) 10(4):e0122651. doi: 10.1371/journal.pone.0122651
98. Chen L, Feng J, Xu B, Zhou Y, Zheng X, Wu C, et al. B7-H6 Expression in Human Hepatocellular Carcinoma and its Clinical Significance [Corrected]. Cancer Cell Int (2018) 18:126. doi: 10.1186/s12935-018-0627-7
99. Zhou Y, Xu Y, Chen L, Xu B, Wu C, Jiang J. B7-H6 Expression Correlates With Cancer Progression and Patient's Survival in Human Ovarian Cancer. Int J Clin Exp Pathol (2015) 8(8):9428–33.
100. Semeraro M, Rusakiewicz S, Minard-Colin V, Delahaye NF, Enot D, Vely F, et al. Clinical Impact of the NKp30/B7-H6 Axis in High-Risk Neuroblastoma Patients. Sci Transl Med (2015) 7(283):283ra55. doi: 10.1126/scitranslmed.aaa2327
101. Zhang B, Xu C, Liu J, Yang J, Gao Q, Ye F. Nidogen-1 Expression is Associated With Overall Survival and Temozolomide Sensitivity in Low-Grade Glioma Patients. Aging (Albany NY). (2021) 13(6):9085–107. doi: 10.18632/aging.202789
102. Willumsen N, Bager CL, Leeming DJ, Bay-Jensen AC, Karsdal MA. Nidogen-1 Degraded by Cathepsin S can be Quantified in Serum and is Associated With Non-Small Cell Lung Cancer. Neoplasia (2017) 19(4):271–8. doi: 10.1016/j.neo.2017.01.008
103. Li L, Zhang Y, Li N, Feng L, Yao H, Zhang R, et al. Nidogen-1: A Candidate Biomarker for Ovarian Serous Cancer. Jpn J Clin Oncol (2015) 45(2):176–82. doi: 10.1093/jjco/hyu187
104. Sun Y, Sedgwick AJ, Palarasah Y, Mangiola S, Barrow AD. A Transcriptional Signature of PDGF-DD Activated Natural Killer Cells Predicts More Favorable Prognosis in Low-Grade Glioma. Front Immunol (2021) 12:668391. doi: 10.3389/fimmu.2021.668391
105. Sun Y, Sedgwick AJ, Khan MA, Palarasah Y, Mangiola S, Barrow AD. A Transcriptional Signature of IL-2 Expanded Natural Killer Cells Predicts More Favorable Prognosis in Bladder Cancer. Front Immunol (2021) 12:724107. doi: 10.3389/fimmu.2021.724107
106. Carlotti E, Palumbo GA, Oldani E, Tibullo D, Salmoiraghi S, Rossi A, et al. FcgammaRIIIA and FcgammaRIIA Polymorphisms do Not Predict Clinical Outcome of Follicular non-Hodgkin's Lymphoma Patients Treated With Sequential CHOP and Rituximab. Haematologica (2007) 92(8):1127–30. doi: 10.3324/haematol.11288
107. Ghesquieres H, Cartron G, Seymour JF, Delfau-Larue MH, Offner F, Soubeyran P, et al. Clinical Outcome of Patients With Follicular Lymphoma Receiving Chemoimmunotherapy in the PRIMA Study is Not Affected by FCGR3A and FCGR2A Polymorphisms. Blood (2012) 120(13):2650–7. doi: 10.1182/blood-2012-05-431825
108. Makanga DR, Jullien M, David G, Legrand N, Willem C, Dubreuil L, et al. Low Number of KIR Ligands in Lymphoma Patients Favors a Good Rituximab-Dependent NK Cell Response. Oncoimmunology (2021) 10(1):1936392. doi: 10.1080/2162402X.2021.1936392
109. Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A Critical Review of the Role of Fc Gamma Receptor Polymorphisms in the Response to Monoclonal Antibodies in Cancer. J Hematol Oncol (2013) 6:1. doi: 10.1186/1756-8722-6-1
110. Mitrovic Z, Aurer I, Radman I, Ajdukovic R, Sertic J, Labar B. FCgammaRIIIA and FCgammaRIIA Polymorphisms Are Not Associated With Response to Rituximab and CHOP in Patients With Diffuse Large B-Cell Lymphoma. Haematologica (2007) 92(7):998–9. doi: 10.3324/haematol.10327
111. Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL. Human CD16 as a Lysis Receptor Mediating Direct Natural Killer Cell Cytotoxicity. Proc Natl Acad Sci U S A. (1999) 96(10):5640–4. doi: 10.1073/pnas.96.10.5640
112. Grier JT, Forbes LR, Monaco-Shawver L, Oshinsky J, Atkinson TP, Moody C, et al. Human Immunodeficiency-Causing Mutation Defines CD16 in Spontaneous NK Cell Cytotoxicity. J Clin Invest. (2012) 122(10):3769–80. doi: 10.1172/JCI64837
113. Harrison D, Phillips JH, Lanier LL. Involvement of a Metalloprotease in Spontaneous and Phorbol Ester-Induced Release of Natural Killer Cell-Associated Fc Gamma RIII (CD16-Ii). J Immunol (1991) 147(10):3459–65.
114. Lajoie L, Congy-Jolivet N, Bolzec A, Gouilleux-Gruart V, Sicard E, Sung HC, et al. ADAM17-Mediated Shedding of Fc Gamma RIIIA on Human NK Cells: Identification of the Cleavage Site and Relationship With Activation. J Immunol (2014) 192(2):741–51. doi: 10.4049/jimmunol.1301024
115. Peruzzi G, Femnou L, Gil-Krzewska A, Borrego F, Weck J, Krzewski K, et al. Membrane-Type 6 Matrix Metalloproteinase Regulates the Activation-Induced Downmodulation of CD16 in Human Primary NK Cells. J Immunol (2013) 191(4):1883–94. doi: 10.4049/jimmunol.1300313
116. Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, et al. NK Cell CD16 Surface Expression and Function Is Regulated by a Disintegrin and Metalloprotease-17 (ADAM17). Blood (2013) 121(18):3599–608. doi: 10.1182/blood-2012-04-425397
117. Wang Y, Wu J, Newton R, Bahaie NS, Long C, Walcheck B. ADAM17 Cleaves CD16b (FcgammaRIIIb) in Human Neutrophils. Biochim Biophys Acta (2013) 1833(3):680–5. doi: 10.1016/j.bbamcr.2012.11.027
118. Tsukerman P, Eisenstein EM, Chavkin M, Schmiedel D, Wong E, Werner M, et al. Cytokine Secretion and NK Cell Activity in Human ADAM17 Deficiency. Oncotarget (2015) 6(42):44151–60. doi: 10.18632/oncotarget.6629
119. Ni SS, Zhang J, Zhao WL, Dong XC, Wang JL. ADAM17 is Overexpressed in non-Small Cell Lung Cancer and its Expression Correlates With Poor Patient Survival. Tumour Biol (2013) 34(3):1813–8. doi: 10.1007/s13277-013-0721-3
120. Fang W, Qian J, Wu Q, Chen Y, Yu G. ADAM-17 Expression is Enhanced by FoxM1 and is a Poor Prognostic Sign in Gastric Carcinoma. J Surg Res (2017) 220:223–33. doi: 10.1016/j.jss.2017.06.032
121. Ferretti E, Carlomagno S, Pesce S, Muccio L, Obino V, Greppi M, et al. Role of the Main Non HLA-Specific Activating NK Receptors in Pancreatic, Colorectal and Gastric Tumors Surveillance. Cancers (Basel) (2020) 12(12). doi: 10.3390/cancers12123705
122. Shen H, Li L, Zhou S, Yu D, Yang S, Chen X, et al. The Role of ADAM17 in Tumorigenesis and Progression of Breast Cancer. Tumour Biol (2016). doi: 10.1007/s13277-016-5418-y
123. Seillet C, Belz GT, Huntington ND. Development, Homeostasis, and Heterogeneity of NK Cells and ILC1. Curr Top Microbiol Immunol (2016) 395:37–61.
124. Guia S, Fenis A, Vivier E, Narni-Mancinelli E. Activating and Inhibitory Receptors Expressed on Innate Lymphoid Cells. Semin Immunopathol (2018) 40(4):331–41. doi: 10.1007/s00281-018-0685-x
125. Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, et al. Molecular Cloning of NKp46: A Novel Member of the Immunoglobulin Superfamily Involved in Triggering of Natural Cytotoxicity. J Exp Med (1998) 188(5):953–60. doi: 10.1084/jem.188.5.953
126. Halfteck GG, Elboim M, Gur C, Achdout H, Ghadially H, Mandelboim O. Enhanced In Vivo Growth of Lymphoma Tumors in the Absence of the NK-Activating Receptor Nkp46/NCR1. J Immunol (2009) 182(4):2221–30. doi: 10.4049/jimmunol.0801878
127. Lakshmikanth T, Burke S, Ali TH, Kimpfler S, Ursini F, Ruggeri L, et al. NCRs and DNAM-1 Mediate NK Cell Recognition and Lysis of Human and Mouse Melanoma Cell Lines In Vitro and In Vivo. J Clin Invest (2009) 119(5):1251–63. doi: 10.1172/JCI36022
128. Glasner A, Ghadially H, Gur C, Stanietsky N, Tsukerman P, Enk J, et al. Recognition and Prevention of Tumor Metastasis by the NK Receptor Nkp46/NCR1. J Immunol (2012) 188(6):2509–15. doi: 10.4049/jimmunol.1102461
129. Merzoug LB, Marie S, Satoh-Takayama N, Lesjean S, Albanesi M, Luche H, et al. Conditional Ablation of NKp46+ Cells Using a Novel Ncr1(greenCre) Mouse Strain: NK Cells are Essential for Protection Against Pulmonary B16 Metastases. Eur J Immunol (2014) 44(11):3380–91. doi: 10.1002/eji.201444643
130. Glasner A, Isaacson B, Viukov S, Neuman T, Friedman N, Mandelboim M, et al. Increased NK Cell Immunity in a Transgenic Mouse Model of NKp46 Overexpression. Sci Rep (2017) 7(1):13090. doi: 10.1038/s41598-017-12998-w
131. Pende D, Parolini S, Pessino A, Sivori S, Augugliaro R, Morelli L, et al. Identification and Molecular Characterization of NKp30, a Novel Triggering Receptor Involved in Natural Cytotoxicity Mediated by Human Natural Killer Cells. J Exp Med (1999) 190(10):1505–16. doi: 10.1084/jem.190.10.1505
132. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating Receptors and Coreceptors Involved in Human Natural Killer Cell-Mediated Cytolysis. Annu Rev Immunol (2001) 19:197–223. doi: 10.1146/annurev.immunol.19.1.197
133. Delahaye NF, Rusakiewicz S, Martins I, Menard C, Roux S, Lyonnet L, et al. Alternatively Spliced NKp30 Isoforms Affect the Prognosis of Gastrointestinal Stromal Tumors. Nat Med (2011) 17(6):700–7. doi: 10.1038/nm.2366
134. von Strandmann EP, Hansen HP, Reiners KS, Schnell R, Borchmann P, Merkert S, et al. A Novel Bispecific Protein (ULBP2-BB4) Targeting the NKG2D Receptor on Natural Killer (NK) Cells and CD138 Activates NK Cells and has Potent Antitumor Activity Against Human Multiple Myeloma In Vitro and In Vivo. Blood (2006) 107(5):1955–62. doi: 10.1182/blood-2005-05-2177
135. Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP, et al. Human Leukocyte Antigen-B-Associated Transcript 3 is Released From Tumor Cells and Engages the NKp30 Receptor on Natural Killer Cells. Immunity (2007) 27(6):965–74. doi: 10.1016/j.immuni.2007.10.010
136. Dassler-Plenker J, Reiners KS, van den Boorn JG, Hansen HP, Putschli B, Barnert S, et al. RIG-I Activation Induces the Release of Extracellular Vesicles With Antitumor Activity. Onco Immunol (2016) 5(10):e1219827. doi: 10.1080/2162402X.2016.1219827
137. Schuldner M, Dorsam B, Shatnyeva O, Reiners KS, Kubarenko A, Hansen HP, et al. Exosome-Dependent Immune Surveillance at the Metastatic Niche Requires BAG6 and CBP/p300-Dependent Acetylation of P53. Theranostics (2019) 9(21):6047–62. doi: 10.7150/thno.36378
138. Renieri A, Mencarelli MA, Cetta F, Baldassarri M, Mari F, Furini S, et al. Oligogenic Germline Mutations Identified in Early Non-Smokers Lung Adenocarcinoma Patients. Lung Cancer. (2014) 85(2):168–74. doi: 10.1016/j.lungcan.2014.05.020
139. Cao G, Wang J, Zheng X, Wei H, Tian Z, Sun R. Tumor Therapeutics Work as Stress Inducers to Enhance Tumor Sensitivity to Natural Killer (NK) Cell Cytolysis by Up-Regulating NKp30 Ligand B7-H6. J Biol Chem (2015) 290(50):29964–73. doi: 10.1074/jbc.M115.674010
140. Pesce S, Tabellini G, Cantoni C, Patrizi O, Coltrini D, Rampinelli F, et al. B7-H6-Mediated Downregulation of NKp30 in NK Cells Contributes to Ovarian Carcinoma Immune Escape. Oncoimmunology (2015) 4(4):e1001224. doi: 10.1080/2162402X.2014.1001224
141. Fogel LA, Sun MM, Geurs TL, Carayannopoulos LN, French AR. Markers of Nonselective and Specific NK Cell Activation. J Immunol (2013) 190(12):6269–76. doi: 10.4049/jimmunol.1202533
142. Mantovani S, Oliviero B, Lombardi A, Varchetta S, Mele D, Sangiovanni A, et al. Deficient Natural Killer Cell NKp30-Mediated Function and Altered NCR3 Splice Variants in Hepatocellular Carcinoma. Hepatology (2019) 69(3):1165–79. doi: 10.1002/hep.30235
143. Campbell KS, Yusa S, Kikuchi-Maki A, Catina TL. NKp44 Triggers NK Cell Activation Through DAP12 Association That is Not Influenced by a Putative Cytoplasmic Inhibitory Sequence. J Immunol (2004) 172(2):899–906. doi: 10.4049/jimmunol.172.2.899
144. Fuchs A, Cella M, Kondo T, Colonna M. Paradoxic Inhibition of Human Natural Interferon-Producing Cells by the Activating Receptor Nkp44. Blood (2005) 106(6):2076–82. doi: 10.1182/blood-2004-12-4802
145. Shemesh A, Brusilovsky M, Hadad U, Teltsh O, Edri A, Rubin E, et al. Survival in Acute Myeloid Leukemia Is Associated With NKp44 Splice Variants. Oncotarget (2016) 7(22):32933–45. doi: 10.18632/oncotarget.8782
146. Rosental B, Brusilovsky M, Hadad U, Oz D, Appel MY, Afergan F, et al. Proliferating Cell Nuclear Antigen Is a Novel Inhibitory Ligand for the Natural Cytotoxicity Receptor Nkp44. J Immunol (2011) 187(11):5693–702. doi: 10.4049/jimmunol.1102267
147. Barrow AD, Edeling MA, Trifonov V, Luo J, Goyal P, Bohl B, et al. Natural Killer Cells Control Tumor Growth by Sensing a Growth Factor. Cell (2018) 172(3):534–48.e19. doi: 10.1016/j.cell.2017.11.037
148. Gaggero S, Bruschi M, Petretto A, Parodi M, Del Zotto G, Lavarello C, et al. Nidogen-1 Is a Novel Extracellular Ligand for the NKp44 Activating Receptor. Oncoimmunology (2018) 7(9):e1470730. doi: 10.1080/2162402X.2018.1470730
149. Horton NC, Mathew SO, Mathew PA. Novel Interaction Between Proliferating Cell Nuclear Antigen and HLA I on the Surface of Tumor Cells Inhibits NK Cell Function Through Nkp44. PloS One (2013) 8(3):e59552. doi: 10.1371/journal.pone.0059552
150. Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, et al. PDGF-D Is a Specific, Protease-Activated Ligand for the PDGF Beta-Receptor. Nat Cell Biol (2001) 3(5):512–6. doi: 10.1038/35074588
151. Li X, Eriksson U. Novel PDGF Family Members: PDGF-C and PDGF-D. Cytokine Growth Factor Rev (2003) 14(2):91–8. doi: 10.1016/S1359-6101(02)00090-4
152. Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-Derived Growth Factor (PDGF) Autocrine Signaling Regulates Survival and Mitogenic Pathways in Glioblastoma Cells: Evidence That the Novel PDGF-C and PDGF-D Ligands may Play a Role in the Development of Brain Tumors. Cancer Res (2002) 62(13):3729–35.
153. Wang Z, Ahmad A, Li Y, Kong D, Azmi AS, Banerjee S, et al. Emerging Roles of PDGF-D Signaling Pathway in Tumor Development and Progression. Biochim Biophys Acta (2010) 1806(1):122–30. doi: 10.1016/j.bbcan.2010.04.003
154. Niehrs A, Garcia-Beltran WF, Norman PJ, Watson GM, Holzemer A, Chapel A, et al. A Subset of HLA-DP Molecules Serve as Ligands for the Natural Cytotoxicity Receptor Nkp44. Nat Immunol (2019) 20(9):1129–37. doi: 10.1038/s41590-019-0448-4
155. de Bruin EC, van de Velde CJ, van Krieken JH, Marijnen CA, Medema JP. Epithelial Human Leukocyte Antigen-DR Expression Predicts Reduced Recurrence Rates and Prolonged Survival in Rectal Cancer Patients. Clin Cancer Res (2008) 14(4):1073–9. doi: 10.1158/1078-0432.CCR-07-1597
156. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science (2006) 313(5795):1960–4. doi: 10.1126/science.1129139
157. Arnould L, Gelly M, Penault-Llorca F, Benoit L, Bonnetain F, Migeon C, et al. Trastuzumab-Based Treatment of HER2-Positive Breast Cancer: An Antibody-Dependent Cellular Cytotoxicity Mechanism? Br J Cancer (2006) 94(2):259–67. doi: 10.1038/sj.bjc.6602930
158. Jin WJ, Erbe AK, Schwarz CN, Jaquish AA, Anderson BR, Sriramaneni RN, et al. Tumor-Specific Antibody, Cetuximab, Enhances the In Situ Vaccine Effect of Radiation in Immunologically Cold Head and Neck Squamous Cell Carcinoma. Front Immunol (2020) 11:591139. doi: 10.3389/fimmu.2020.591139
159. Klanova M, Oestergaard MZ, Trneny M, Hiddemann W, Marcus R, Sehn LH, et al. Prognostic Impact of Natural Killer Cell Count in Follicular Lymphoma and Diffuse Large B-Cell Lymphoma Patients Treated With Immunochemotherapy. Clin Cancer Res (2019) 25(15):4634–43. doi: 10.1158/1078-0432.CCR-18-3270
160. Kohrt HE, Thielens A, Marabelle A, Sagiv-Barfi I, Sola C, Chanuc F, et al. Anti-KIR Antibody Enhancement of Anti-Lymphoma Activity of Natural Killer Cells as Monotherapy and in Combination With Anti-CD20 Antibodies. Blood (2014) 123(5):678–86. doi: 10.1182/blood-2013-08-519199
161. Muntasell A, Rojo F, Servitja S, Rubio-Perez C, Cabo M, Tamborero D, et al. NK Cell Infiltrates and HLA Class I Expression in Primary HER2(+) Breast Cancer Predict and Uncouple Pathological Response and Disease-Free Survival. Clin Cancer Res (2019) 25(5):1535–45. doi: 10.1158/1078-0432.CCR-18-2365
162. Muntasell A, Servitja S, Cabo M, Bermejo B, Perez-Buira S, Rojo F, et al. High Numbers of Circulating CD57(+) NK Cells Associate With Resistance to HER2-Specific Therapeutic Antibodies in HER2(+) Primary Breast Cancer. Cancer Immunol Res (2019) 7(8):1280–92. doi: 10.1158/2326-6066.CIR-18-0896
163. Jie HB, Schuler PJ, Lee SC, Srivastava RM, Argiris A, Ferrone S, et al. CTLA-4(+) Regulatory T Cells Increased in Cetuximab-Treated Head and Neck Cancer Patients Suppress NK Cell Cytotoxicity and Correlate With Poor Prognosis. Cancer Res (2015) 75(11):2200–10. doi: 10.1158/0008-5472.CAN-14-2788
164. Hartmann F, Renner C, Jung W, da Costa L, Tembrink S, Held G, et al. Anti-CD16/CD30 Bispecific Antibody Treatment for Hodgkin's Disease: Role of Infusion Schedule and Costimulation With Cytokines. Clin Cancer Res (2001) 7(7):1873–81.
165. Hartmann F, Renner C, Jung W, Deisting C, Juwana M, Eichentopf B, et al. Treatment of Refractory Hodgkin's Disease With an Anti-CD16/CD30 Bispecific Antibody. Blood (1997) 89(6):2042–7. doi: 10.1182/blood.V89.6.2042
166. Thakur A, Huang M, Lum LG. Bispecific Antibody Based Therapeutics: Strengths and Challenges. Blood Rev (2018) 32(4):339–47. doi: 10.1016/j.blre.2018.02.004
167. Chan WK, Kang S, Youssef Y, Glankler EN, Barrett ER, Carter AM, et al. A CS1-NKG2D Bispecific Antibody Collectively Activates Cytolytic Immune Cells Against Multiple Myeloma. Cancer Immunol Res (2018) 6(7):776–87. doi: 10.1158/2326-6066.CIR-17-0649
168. Davis ZB, Vallera DA, Miller JS, Felices M. Natural Killer Cells Unleashed: Checkpoint Receptor Blockade and BiKE/TriKE Utilization in NK-Mediated Anti-Tumor Immunotherapy. Semin Immunol (2017) 31:64–75. doi: 10.1016/j.smim.2017.07.011
169. Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M. Killers 2.0: NK Cell Therapies at the Forefront of Cancer Control. J Clin Invest (2019) 129(9):3499–510.
170. Hu W, Wang G, Huang D, Sui M, Xu Y. Cancer Immunotherapy Based on Natural Killer Cells: Current Progress and New Opportunities. Front Immunol (2019) 10:1205. doi: 10.3389/fimmu.2019.01205
171. Li H, Er Saw P, Song E. Challenges and Strategies for Next-Generation Bispecific Antibody-Based Antitumor Therapeutics. Cell Mol Immunol (2020) 17(5):451–61. doi: 10.1038/s41423-020-0417-8
172. Pahl JHW, Koch J, Gotz JJ, Arnold A, Reusch U, Gantke T, et al. CD16A Activation of NK Cells Promotes NK Cell Proliferation and Memory-Like Cytotoxicity Against Cancer Cells. Cancer Immunol Res (2018) 6(5):517–27. doi: 10.1158/2326-6066.CIR-17-0550
173. Reusch U, Duell J, Ellwanger K, Herbrecht C, Knackmuss SH, Fucek I, et al. A Tetravalent Bispecific TandAb (CD19/CD3), AFM11, Efficiently Recruits T Cells for the Potent Lysis of CD19(+) Tumor Cells. MAbs (2015) 7(3):584–604. doi: 10.1080/19420862.2015.1029216
174. Mazor Y, Hansen A, Yang C, Chowdhury PS, Wang J, Stephens G, et al. Insights Into the Molecular Basis of a Bispecific Antibody's Target Selectivity. MAbs (2015) 7(3):461–9. doi: 10.1080/19420862.2015.1022695
175. Mossner E, Brunker P, Moser S, Puntener U, Schmidt C, Herter S, et al. Increasing the Efficacy of CD20 Antibody Therapy Through the Engineering of a New Type II Anti-CD20 Antibody With Enhanced Direct and Immune Effector Cell-Mediated B-Cell Cytotoxicity. Blood (2010) 115(22):4393–402. doi: 10.1182/blood-2009-06-225979
176. Niederfellner G, Lammens A, Mundigl O, Georges GJ, Schaefer W, Schwaiger M, et al. Epitope Characterization and Crystal Structure of GA101 Provide Insights Into the Molecular Basis for Type I/II Distinction of CD20 Antibodies. Blood (2011) 118(2):358–67. doi: 10.1182/blood-2010-09-305847
177. Salles G, Morschhauser F, Lamy T, Milpied N, Thieblemont C, Tilly H, et al. Phase 1 Study Results of the Type II Glycoengineered Humanized Anti-CD20 Monoclonal Antibody Obinutuzumab (GA101) in B-Cell Lymphoma Patients. Blood (2012) 119(22):5126–32. doi: 10.1182/blood-2012-01-404368
178. Goede V, Fischer K, Busch R, Engelke A, Eichhorst B, Wendtner CM, et al. Obinutuzumab Plus Chlorambucil in Patients With CLL and Coexisting Conditions. N Engl J Med (2014) 370(12):1101–10. doi: 10.1056/NEJMoa1313984
179. Luo C, Wu G, Huang X, Ma Y, Zhang Y, Song Q, et al. Efficacy and Safety of New Anti-CD20 Monoclonal Antibodies Versus Rituximab for Induction Therapy of CD20(+) B-Cell non-Hodgkin Lymphomas: A Systematic Review and Meta-Analysis. Sci Rep (2021) 11(1):3255. doi: 10.1038/s41598-021-82841-w
180. Marcus R, Davies A, Ando K, Klapper W, Opat S, Owen C, et al. Obinutuzumab for the First-Line Treatment of Follicular Lymphoma. N Engl J Med (2017) 377(14):1331–44. doi: 10.1056/NEJMoa1614598
181. Prica A, Crump M. Improving CD20 Antibody Therapy: Obinutuzumab in Lymphoproliferative Disorders. Leuk Lymphoma. (2019) 60(3):573–82. doi: 10.1080/10428194.2018.1498490
182. Ni J, Miller M, Stojanovic A, Garbi N, Cerwenka A. Sustained Effector Function of IL-12/15/18-Preactivated NK Cells Against Established Tumors. J Exp Med (2012) 209(13):2351–65. doi: 10.1084/jem.20120944
183. Kerbauy LN, Marin ND, Kaplan M, Banerjee PP, Berrien-Elliott MM, Becker-Hapak M, et al. Combining AFM13, a Bispecific CD30/CD16 Antibody, With Cytokine-Activated Blood and Cord Blood-Derived NK Cells Facilitates CAR-Like Responses Against CD30(+) Malignancies. Clin Cancer Res (2021) 27(13):3744–56. doi: 10.1158/1078-0432.CCR-21-0164
184. Vallera DA, Felices M, McElmurry R, McCullar V, Zhou X, Schmohl JU, et al. IL15 Trispecific Killer Engagers (TriKE) Make Natural Killer Cells Specific to CD33+ Targets While Also Inducing Persistence, In Vivo Expansion, and Enhanced Function. Clin Cancer Res (2016) 22(14):3440–50. doi: 10.1158/1078-0432.CCR-15-2710
185. Vallera DA, Ferrone S, Kodal B, Hinderlie P, Bendzick L, Ettestad B, et al. NK-Cell-Mediated Targeting of Various Solid Tumors Using a B7-H3 Tri-Specific Killer Engager In Vitro and In Vivo. Cancers (Basel). (2020) 12(9). doi: 10.3390/cancers12092659
186. Golay J, Choblet S, Iwaszkiewicz J, Cerutti P, Ozil A, Loisel S, et al. Design and Validation of a Novel Generic Platform for the Production of Tetravalent IgG1-Like Bispecific Antibodies. J Immunol (2016) 196(7):3199–211. doi: 10.4049/jimmunol.1501592
Keywords: NK cells, ITAM, cancer, NCR, activating receptors
Citation: Medjouel Khlifi H, Guia S, Vivier E and Narni-Mancinelli E (2022) Role of the ITAM-Bearing Receptors Expressed by Natural Killer Cells in Cancer. Front. Immunol. 13:898745. doi: 10.3389/fimmu.2022.898745
Received: 17 March 2022; Accepted: 19 April 2022;
Published: 10 June 2022.
Edited by:
Nadia Guerra, Imperial College London, United KingdomReviewed by:
Anne Caignard, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceEmanuela Marcenaro, University of Genoa, Italy
Copyright © 2022 Medjouel Khlifi, Guia, Vivier and Narni-Mancinelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emilie Narni-Mancinelli, bmFybmlAY2ltbC51bml2LW1ycy5mcg==
 Hakim Medjouel Khlifi
Hakim Medjouel Khlifi Sophie Guia
Sophie Guia Eric Vivier
Eric Vivier Emilie Narni-Mancinelli
Emilie Narni-Mancinelli