- 1Research Institute of General Surgery, Affiliated Jinling Hospital, Medical School of Nanjing University, Nanjing, China
- 2Department of General Surgery, BenQ Medical Center, The Affiliated BenQ Hospital of Nanjing Medical University, Nanjing, China
Stimulator of interferon genes (STING) is an endoplasmic-reticulum resident protein, playing essential roles in immune responses against microbial infections. However, over-activation of STING is accompanied by excessive inflammation and results in various diseases, including autoinflammatory diseases and cancers. Therefore, precise regulation of STING activities is critical for adequate immune protection while limiting abnormal tissue damage. Numerous mechanisms regulate STING to maintain homeostasis, including protein-protein interaction and molecular modification. Among these, post-translational modifications (PTMs) are key to accurately orchestrating the activation and degradation of STING by temporarily changing the structure of STING. In this review, we focus on the emerging roles of PTMs that regulate activation and inhibition of STING, and provide insights into the roles of the PTMs of STING in disease pathogenesis and as potential targeted therapy.
Introduction
Innate immunity is the front line of defense, protecting the host from microbial invasion and triggering adaptive immunity to eradicate infections. When pathogen-associated molecular patterns (PAMPs), including bacterial lipopolysaccharide (LPS) and viral nucleic acids, invade the body with or without tissue damage, pattern recognition receptors (PRRs), which are located on the cell membrane or in the cytoplasm, can be activated and mediate inflammatory and antiviral pathways to deal with infection (1, 2). In addition, tissue and cell damage are accompanied by damage-associated molecular patterns (DAMPs), such as abnormal DNA and cell organelle fragments, which can also be recognized by PRRs and induce cascades of inflammatory signaling pathways (2). PRRs are protein family including Toll-like receptors (TLRs), C-type lectin receptors (CLRs), NOD-like receptors (NLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), cyclic GMP-AMP synthase (cGAS), IFN-γ-inducible protein 16 (IFI16), absent in melanoma 2 (AIM2) and multiple DNA sensors (1–3). Different PRRs can be activated by different PAMPs, which then trigger innate immune responses to combat infection (1–4).
Specifically, cGAS, a protein that can enter and exit the nucleus, is recruited to abnormal DNA, which is classified into exogenous pathogenic DNA or endogenous instable heterotopic DNA, leading to a conformational change to bind to DNA (5–7). After that, AMP and GMP are catalyzed by cGAS, releasing the second messenger 2’3’-cyclic GMP-AMP (cGAMP), which functions as the second messenger to initiate adapter protein stimulator of interferon genes (STING) on the endoplasmic reticulum (ER) membrane (7, 8). STING then relocates to the Golgi apparatus, leading to production of cytokines, including type I interferon and other cytokines (7–10). Different immune cells can also produce various cytokines through cGAS- STING pathway to perform their respective function (11).
To date, STING has been generally considered to play a necessary role in immunity and inflammation. However, the regulatory mechanism of STING remains unclear. In addition to the direct interaction between cyclic dinucleotides and STING, post-translation modifications (PTMs) and protein-protein interactions could also alter STING function (4, 9, 12, 13). PTMs of proteins have been recognized as an important regulatory switch to temporally change the functions of proteins in cells. Most cellular proteins can be decorated by a diverse range of PTMs, including phosphorylation, acetylation, methylation, glutamylation, ADP-ribosylation, SUMOylation, and ubiquitination (14–17). In this review, we review the PTMs and function of STING signaling, highlighting the potential targeted therapy afforded by PTMs of STING.
An Overview of STING Pathway
STING, namely stimulator of interferon genes, also known as TMEM173, MITA, ERIS, and MPYS (18, 19), is a ~40-kDa trans-membrane protein located on the ER membrane (20, 21). It is composed of a short N-terminal cytosolic segment, four trans-membranes (TM) located in the ER membrane, a cytosolic ligand-binding domain (LBD), and a C-terminal tail (CTT) which is responsible for binding TBK1 (19, 21). It exists widely in nature, not only humans, but also chicken (22), shrimp (23), bacteria (24), and Drosophila (25), functioning as pathogen sensors to avoid infection.
As previously mentioned, cGAMP, catalyzed by cGAS, can alter the conformation of STING, resulting in the inward rotation of the two wings of STING toward each other. This process leads to closure of the ligand binding pocket and activation of STING (7). Then STING traffics in the form of COP-II vesicles from ER to ER-Golgi intermediate compartments (ERGIC) (26, 27). Some vesicles loaded with STING serve as a membrane source for modification by the ubiquitin-like protein LC3, which is a key step in autophagosome biogenesis (26). Most STING-coated ERGIC vesicles continue to traffic through the Golgi and post-Golgi endosomes. On the ERGIC membrane, STING recruits TBK1(tank binding kinase 1) and IKK (inhibitor of kappa B kinase), then TBK1 auto-phosphorylates and STING, IRF3 and IκBα are phosphorylated (7, 27). Phosphorylated IRF3 dimerizes and translocates to activate transcription of type I IFN and interferon-stimulated genes (ISGs). Phosphorylation of IκBα results in translocation of NF-κB to the nucleus, leading to transcription of genes encoding pro-inflammatory cytokines and chemokines such as IL-6 and TNF (28).
In addition, micronuclei (29), mtDNA (30), abnormal cell cycle (31), and cytoplasmic chromatin fragments (32) can activate STING through cGAS- dependent way. Several stimuli other than cGAMP, which is catalyzed by cGAS, for example, bacterial or virus cyclic dinucleotides (CDNs) (33, 34), can also activate STING directly. Apart from the production of type I interferon and cytokines, STING can also be associated with other biological and pathological process, such as ER stress (35), oxidative stress (36), fatty acid metabolism (37), Ca2+ homeostasis (38, 39), T cell proliferation (40), senescence (32) and so on. Moreover, STING can also participate in cell death pathways (41), including autophagy (30, 42), apoptosis (43), pyroptosis (44, 45), ferroptosis (46), necroptosis (47, 48), mitotic death (49), immunogenic cell death(ICD) (50). However, sometimes STING can help herpes virus assemble viral genome to host cell’s nucleus and survive in host cells, leading to severer virus infection at the early stage of infection (51).
Now that STING plays critical roles in biochemical processes, the expression and function of STING are tightly regulated. Apart from direct interaction between cGAMP and STING, protein-protein interaction and post-translational modification (PTMs) are also important (9, 12). Hundreds of proteins may interact with STING and affect its function (52). However, the regulation of STING in cells are basically dependent on the PTMs. In a word, PTMs are the key to regulating protein function and play a critical role in modulation of STING. Thus, we review PTMs of STING and prospect related targeted therapy.
Modification and Regulation of STING
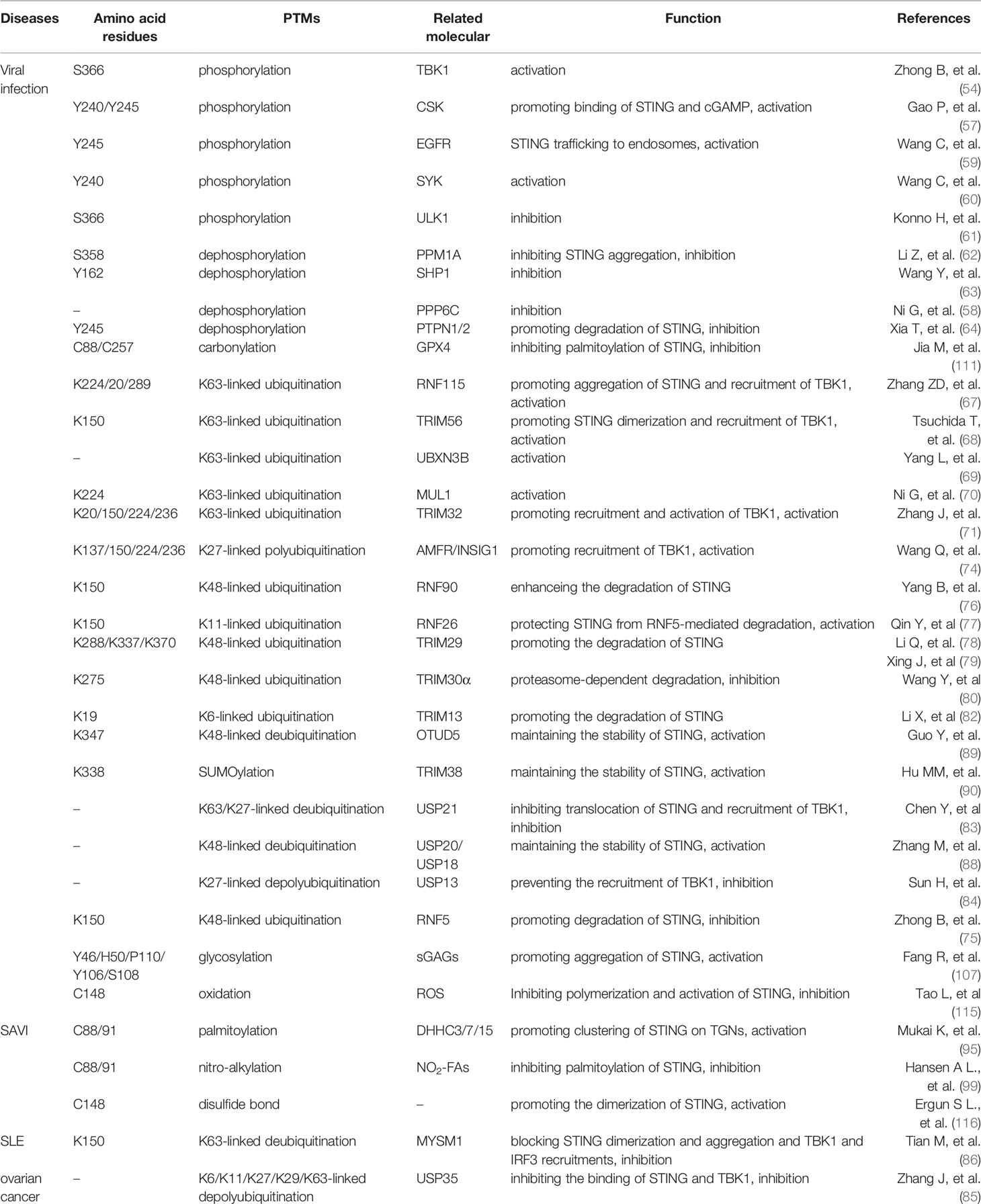
STING, a protein located in the ER, is composed of 379aa in human. Some residues of STING can be modified for its compartmentalization, dimerization, oligomerization, trafficking and degradation, which regulate immunological and other processes (Figure 1, 2). The consequence of PTMs is dependent on amino acid residue, type of modification group, thus biochemical process can be fully developed in cells. Therefore, we focus on different PTMs of STING and propose critical role of PTMs in activation or inhibition of STING. Modified residues and related proteins are shown in Figure 1. While Figure 2 shows how PTMs of STING affects STING pathway. Therefore, some PTMs in Figure 1 are not shown in Figure 2 due to the lack of literature.
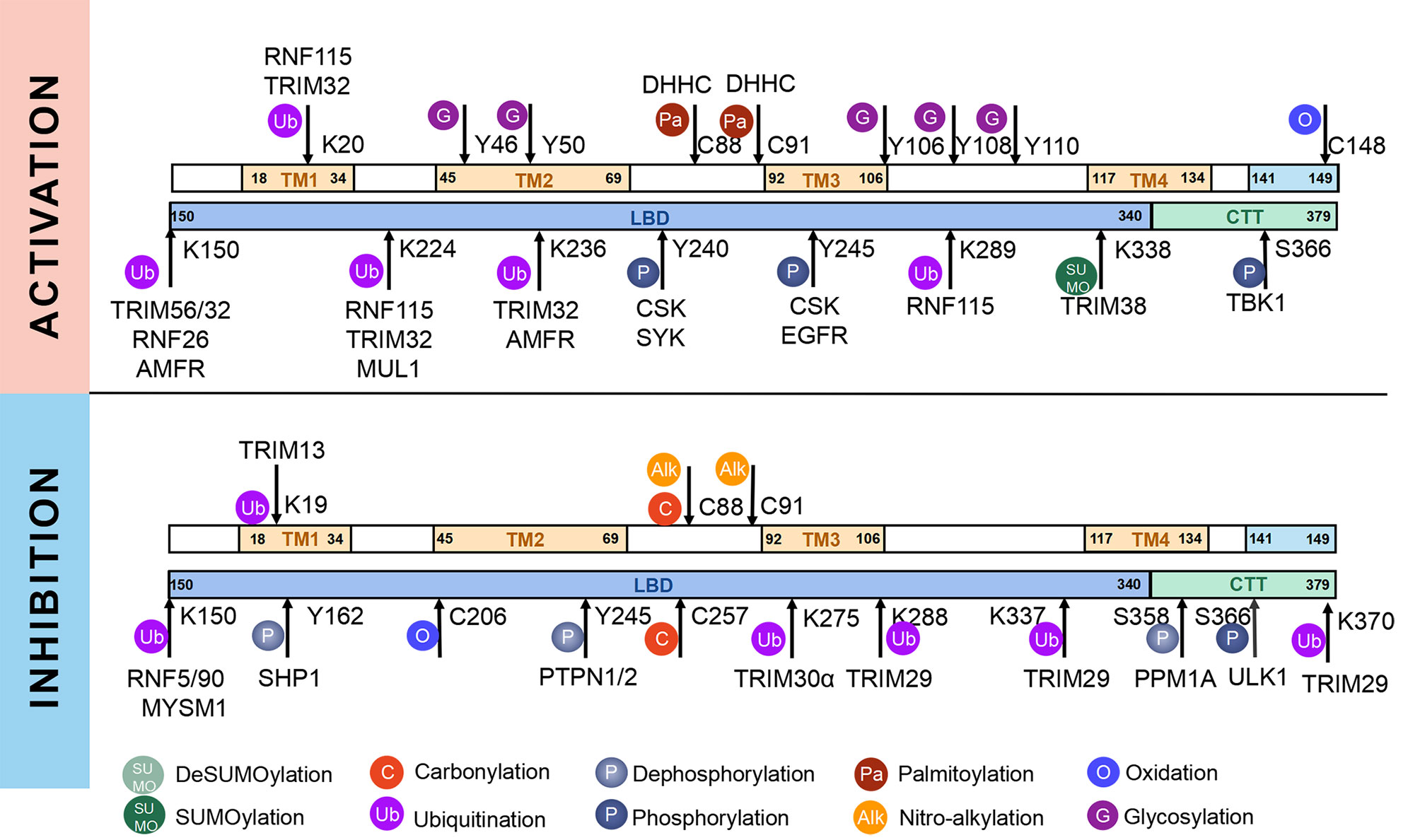
Figure 1 Post-translational modifications of STING. STING can be modified by phosphorylation and dephosphorylation, ubiquitination and deubiquitination, SUMOylation and deSUMOylation, palmitoylation, alkylation, glycosylation, carbonylation, oxidation, and disulfide bond, which are indicated according to the keys in the bottom. The PTMs that promote activation of STING are shown above, whereas those that inhibit STING activation are shown below. Related enzymes are presented around the PTMs.
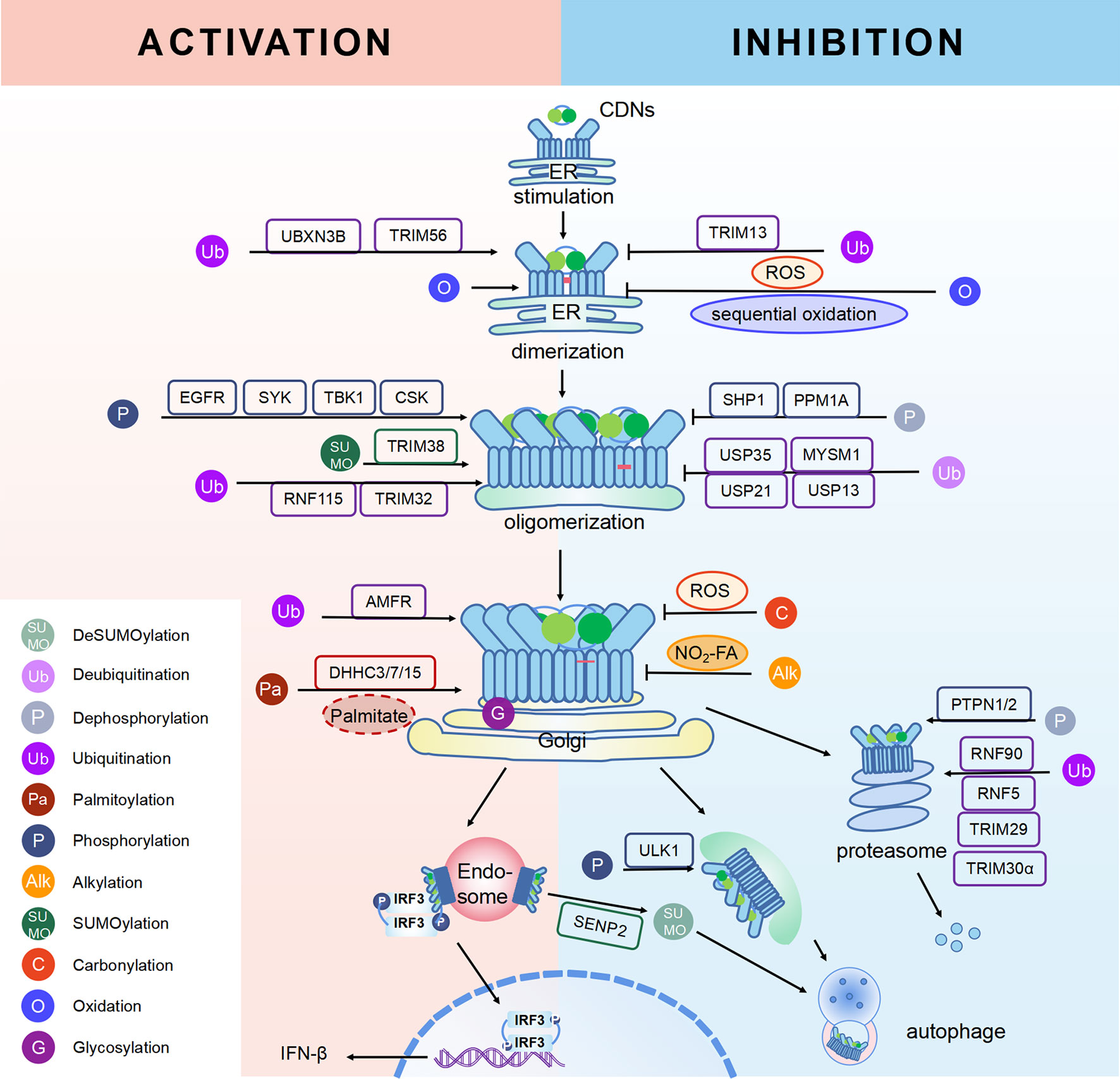
Figure 2 PTMs in STING pathway. PTMs are going along with the life of STING, from activation to degradation. When CDNs bind to STING, STING is recruited and dimerized with the help of reversible oxidation and disulfide bond. SUMOylation and ubiquitination can help STING with oligomerization, while phosphorylation is related to the activation of STING. Then STING transports from ER to Golgi. Palmitoylation and sGAGs could help STING reside on Golgi, thus continuous activation of STING leads to increasing pro-inflammatory cytokines. On the other hand, inhibition of STING happens along with activation to achieve cellular homeostasis. Sequential oxidation inhibits STING dimerization. Dephosphorylation and deubiquitination of STING decrease the oligomerization of STING. Also, carbonylation caused by ROS and alkylation caused by NO2-FAs can competitively inhibit palmitoylation and block sequential residence of STING on Golgi, thus immune responses are inhibited. In addition, dephosphorylation, ubiquitination and deSUMOylation could facilitate protein degradation to maintain proteostasis.
Phosphorylation and Dephosphorylation
Phosphorylation is the most common PTMs in mammalian cells. It means the addition of phosphate groups to proteins, especially to amino acid residues, such as Ser and Thr (53). The addition or removal of phosphate groups (dephosphorylation) acts as a biological “on/off” in many reactions, including modulation of STING (9) (Figure 2).
It is confirmed that, oligomerized STING recruited TBK1, then TBK1 phosphorylated S366 of human STING (S365 in mice), with the consequence of classic activation of STING pathway (54–56). CSK (C-terminal src kinase), another tyrosine kinase expressed broadly among mammalian cells, phosphorylates STING at Y240 and Y245 and activates immune responses via promoting aggregation of STING after HSV-1 infection (57). PPP6C, namely protein phosphatase 6 catalytic subunit, is a phosphatase. Knockdown of PPP6C greatly increased STING phosphorylation at S366 after dsDNA stimulation to enhance immune response (58). Y245 of STING can also be phosphorylated directly by EGFR (epidermal growth factor receptor) for STING trafficking to endosomes, thus type I-IFN are produced (59). SYK (spleen tyrosine kinase), another kinase, could also phosphorylate Y240 of STING and promote activation of STING (60).
However, to achieve cellular homeostasis, STING pathway can also be down-regulated via phosphorylation and dephosphorylation (Figure 2). For instance, STING is subsequently phosphorylated by UNC-51-like kinase (ULK1) at S366 so that sustained innate immune responses can be prevented (61). Dephosphorylation of S358 by Mg2+/Mn2+-dependent protein phosphatase 1A (PPM1A) can also inhibit STING aggregation and STING-dependent pathway (62). SHP1(SH2-containing protein tyrosine phosphatase), a phosphatase, can dephosphorylate STING at Y162, blocking the K63-linked ubiquitination of STING at K337 and inhibiting STING pathway (63). Tyrosine-protein phosphatase nonreceptor type (PTPN) 1 and 2 can dephosphorylate STING at Y245 with the consequence of degradation of STING via the ubiquitin-independent proteasomal pathway (64). In a word, phosphorylation and dephosphorylation of STING play important roles in modulation of STING. The imbalance between phosphorylation and dephosphorylation could be critical mechanism in development of diseases.
Ubiquitylation and Deubiquitylation
Ubiquitylation, a highly-conserved modification of protein, is the second most common PTM for proteins, after only phosphorylation (53). It can be divided into three types due to structural characteristics: mono-ubiquitination, poly-ubiquitination and branched ubiquitination (65). Ubiquitylation (Ub) is initiated by a cascade of enzymatic reactions, which is catalyzed by Ub-activating (E1), Ub-conjugating (E2) and Ub-ligating (E3) enzymes (65). Firstly, ubiquitin (Ub) is activated by E1 in an ATP-dependent manner and then is transferred to E2. Then, specific E3 catalyzes the transfer of Ub from E2 to a specific substrate protein with the assistance of E1 (66). As a result, Ub is assembled covalently to the specific protein, especially to lysine residues, resulting in the regulation of quality and quantity of proteins through degradation in various physiological and/or pathological conditions. Deubiquitylation happens along with ubiquitylation, and both of them together affect the maturation and degradation of proteins.
Specifically, STING can also be modified by Ub via E3 ubiquitin ligase (Figure 2). RNF115(RING finger protein) can catalyze K63-linked polyubiquitination of STING at K224/20/289, promoting the translocation of STING from ER to Golgi (67). TRIM56(tripartite motif protein) induces K63 linkage ubiquitination of STING at K150 due to stimulation of exogenous DNA, and promotes STING dimerization and recruitment of TBK1 (68). UBXN3B (ubiquitin regulatory X domain-containing proteins 3 b) facilitates TRIM56-dependent K63-linked ubiquitination of STING, thus leading to dimerization, trafficking, and activation STING signaling (69). MUL1 (mitochondrial E3 ubiquitin protein ligase 1) can catalyze K63-linked polyubiquitination of STING at K224 to help activation of STING (70). TRIM32 targeted STING for K63-linked ubiquitination at K20/150/224/236 and promoted the STING pathway (71). However, HSV1 VP1/2 can deubiquitinate K63-linked polyubiquitination of STING, which is mediated by TRIM32, thus help HSV escape from immune responses and promote brain infections (72). Reverse transcriptase domain of HBV can also physically bind to STING and significantly reduces the K63-linked polyubiquitination of STING, protecting HBV from innate immune responses (73). These indicate that pathogen has developed PTMs targeted strategy to evade from host immune system. Autocrine motility factor receptor (AMFR) catalyzed the K27-linked polyubiquitination of STING in an insulin-induced gene 1 (INSIG1)-dependent manner, recruiting TANK-binding kinase 1 (TBK1) and facilitating its translocation to the perinuclear microsomes (74). Therefore, K63-linked and K-27 linked polyubiquitination play important roles in activation of STING pathway.
While RNF5 and RNF90 target STING at K150 for K48-linked ubiquitination and lead to degradation of STING after viral infection (75, 76). However, RNF26 promotes K11-linked polyubiquitination of STING at K150 and protects STING from RNF5-mediated K48-linked polyubiquitination and degradation, thus enhancing quick and efficient anti-viral responses (77).TRIM29 catalyzes K48-linked polyubiquitination at K288/337/370 and promotes the degradation of STING (78, 79). TRIM30α induces K48-linked ubiquitination of STING at K275 for proteasome-dependent degradation (80). There is another interesting protein, death-associated protein kinase 3(DAPK3), which modifies STING ubiquitination differently in different situations. When cells are in basic condition, DAPK3 inhibited STING K48-linked poly-ubiquitination and proteasome-mediated degradation. By contrast, DAPK3 is required for STING K63-linked poly-ubiquitination and STING-TBK1 interaction after cGAMP stimulation (81). Recently, TRIM13 is discovered to bind to STING at resting state. After HSV-1 infection, K6-linked polyubiquitination at K19 is triggered by TRIM13, thus promoting degradation of STING (82). In a word, different types of ubiquitination precisely regulate activation or degradation of STING, including K11-linked, K27-linked, K48-linked, K63-linked, K6-linked, leading to different effects in immune responses (69). K27-linked, K11-linked and K63-linked polyubiquitination can promote transport and activation of STING, while K6-linked, and K48-linked polyubiquitination mostly lead to degradation of STING (Figures 1, 2).
On the other hand, ubiquitin-specific protease (USP)21, a deubiquitinating enzyme, hydrolyzes K27/63-linked polyubiquitin chain on STING, resulting in negative regulation of STING and significant decrease of type I interferons (83). USP13 uncouples K27-linked polyubiquitin chains from STING and prevents the recruitment of TBK1 to inhibit STING pathway (84). USP35 directly removes K11-, K27-, K63-linked polyubiquitin of STING, thus inhibits phosphorylation and multimerization of STING to limit STING signaling (85). The MYSM1 (Myb-like, SWIRM, and MPN domains 1 protein), interacts with STING and cleaves STING K63-linked ubiquitination at K150 to inhibit cGAS-STING signaling (86). On the other hand, OTUD5 and USP20 catalyze the K48-linked deubiquitination of STING and inhibit STING degradation, thus maintain the stability of STING and promote activation of STING pathway (87–89). In conclusion, targeted ubiquitination could be potential therapeutic approach (Figures 1, 2).
SUMOylation and deSUMOylation
Small ubiquitin-related modifier (SUMO) is a widely expressed Ub-like protein, which is similar with Ub in structure and enzymatic cascade (16). There are three major SUMOs. SUMO-1 usually modifies a substrate as a monomer; while SUMO-2/3 can form poly-SUMO chains. Both SUMO-1 and poly-SUMO chains can interact with other proteins through SUMO-interactive motif (SIM) (16). Thus, SUMO modification participated mainly in enhancing protein-protein interaction and regulating proteins’ localization, stability and activity (16, 65). As for STING, TRIM38 mediates the SUMOylation of STING at K338 to inhibit STING degradation, promoting oligomerization of STING and recruitment of IRF3 (90). On the other hand, at the late stage of infection, STING is deSUMOylated by SUMO-specific protease (SENP)2 after phosphorylation at S366, eventually leading to STING degradation and dampening innate immune responses (90)(Figures 1, 2). More studies are needed to figure out whether other SUMO-enzymes play roles in regulation of STING, for example, SAE1 (SUMO-activating enzyme subunit 1), a subunit of SUMO-activating enzyme, which might interact with STING and plays a role in regulation of STING (52).
Palmitoylation
Palmitoylation, or S-palmitoylation, a type of lipidation, can make proteins bind non-polar structures more tightly, with important function for the localization, diffusion, and physical interactions of these proteins within the cell. It is namely related to palmitic acid (PA), which comes out either with intrinsic fatty acid synthesis, or fatty acid uptake from outside of cells (91). However, studies have also discovered that palmitoylation is not only associated with PA concentration, but also with the zinc finger DHHC-type containing (ZDHHC) family of palmitoyl S-acyltransferases (PATs), ZDHHC3, ZDHHC7, ZDHHC15, which are mostly localized to the ER and Golgi apparatus (92, 93). On the other hand, depalmitoylation, which means the removal of S-palmitoylation, can be catalyzed by some depalmitoylases, which belong to the serine hydrolase family, including APT1 (LYPLA1), APT2 (LYPLA2) and so on (94).
It has been confirmed that the palmitoylation of cysteines 88 and 91 of STING participates in regulation of STING, but not trafficking of STING (95)(Figures 1, 2). Palmitoylation of STING triggers composition of a multimeric complex at the lipid rafts of the trans-Golgi network, which triggers STING interacting with TBK1 (96). Suppression of palmitoylation with 2-bromopalmitate (2-BP) and hydoxylamine eliminates the transcription of downstream inflammatory cytokine genes, thus the phenotype of STING-associated vasculopathy with onset in infancy (SAVI, an auto-inflammatory disease related to gain-of-function mutations of STING) could be improved. Mutation of C88/91 inhibits palmitoylation and decreases the activation of STING-dependent host defense genes (95). However, it is still unknown whether STING can be depalmitoylated by any of the depalmitoylases. More studies are needed to investigate functions of depalmitoylated STING and related mechanisms.
Nitro-Alkylation
Nitro-alkylation, which is related to nitro-fatty acids (NO2-FAs), has been discovered as potent inhibitors of STING signaling. NO2-FAs is a recently discovered group of bioactive lipids with anti-inflammatory and tissue protective functions (97). It is produced mainly in gastrointestinal tract during digestion. Meanwhile, it can come out locally to modify specific proteins through Michael addition reactions, so that inflammation responses can be regulated accurately (98).
As mentioned above, there are two cysteine residues in the N-terminal region of STING, Cys88/91, which can be recognized by NO2-FAs to covalently interact with, resulting in nitro-alkylation of STING (92). Once STING is nitro-alkylated by NO2-FAs, palmitoylation of STING is abolished and STING pathway is inhibited (92). In addition, treatment with nitro-fatty acids is sufficient to inhibit production of type I IFN in fibroblasts derived from SAVI patients (99). In conclusion, nitro-alkylation can be a potent inhibitor of palmitoylation in cells and disease (Figures 1, 2). However, there is still no enzymes covered which is responsible for alkylation of STING, or other proteins. In addition, studies have suggested that, prostaglandin reductase-1 (PtGR-1) promotes nitroalkene transition to inactive nitroalkanes, thus decreases alkylation of protein (100). However, it is still not clear, whether PtGR-1 affects regulation of STING, and how palmitoylation could be replaced or inhibited by alkylation under physiological conditions. More studies are needed to figure out deeper mechanisms.
Glycosylation
Glycosylation is one of the most diverse post-translational modifications in eukaryotic cells (101). Proteins are glycosylated by either enzymes or interaction directly with glucose (aldehyde form) through lysine and arginine residues in proteins, and eventually leading to advanced glycation end products so that participating in biological or pathological process (101). Glycosylation of proteins are complex, indicated by the diverse types of glycans, the multiple positions of glycans, various structures of glycoproteins and different glycosyl-transferase enzymes, leading to various functions of proteins (102). Studies have discovered that, N41 of STING in mice could be a potential N-linked glycosylation site, however, no N-linked glycosylation was detected with deglycosylation experiments (43). Another experiment showed that, there are four putative N-glycosylation sites of STING in Drosophila, N84, N187, N270, N333. Among two forms of STING in drosophila (long form and short form), only long form of STING could be glycosylated thus promoting residence of STING on ER (103). However, the glycosylation of STING in human has not been studied well for unknown reasons.
Glycosaminoglycans (GAGs) are linear acidic polysaccharides, which can be divided into many groups (104). GAGs are subsequently modified by epimerization and sulfation to produce sulfated GAGs (sGAGs) (105). Researchers have discovered that sGAGs can interact with various proteins through their negatively charged sulfate groups (106). Studies have suggested that STING translocates to sGAG-containing vesicles after vaccinia virus infection and evolutionally-conserved bounds to sGAGs through its luminal, positively charged, and polar residues (Figures 1, 2). It is hypothesized that STING palmitoylation facilitates its clustering into lipid rafts on the Golgi apparatus from the STING cytosolic side, while sGAGs induce STING polymerization from the STING luminal side, and both of two methods together lead to full activation of STING and TBK1 (107). However, whether STING is glycosylated and regulated by other substances, especially glucose, are still unclear.
Carbonylation
Carbonylation, one of the most harmful irreversible oxidative protein modifications, is linked to lipid peroxidation. It is considered as a major hallmark of oxidative stress-related disorders, leading to biomolecule malfunctions and eventually cell death (108, 109). A large amount of evidence has indicated the role of carbonylated proteins in the initiation of inflammation and autoimmune responses (110). However, situation becomes different in STING. There are two carbonylation sites of STING, namely residues C88 and C257, which are conserved across species (111). It is confirmed that both HSV-1 infection and 4-hydroxynonenal (4-HNE, a type of lipid peroxidation metabolite) induce STING carbonylation through lipid peroxidation, preventing the palmitoylation and translocation of STING from the ER to the Golgi, with the consequence of down-regulating immune responses (Figure 2). While glutathione peroxidase 4 (GPX4) inhibits STING carbonylation and promote activation of STING. Although the critical carbonylation site of STING has been discovered, there are still many questions to be resolved: why does carbonylation of STING result in inhibition of inflammation, which is different from that of other proteins? Is there a deeper mechanism regulating inflammatory phenotype after carbonylation of proteins? Also, there are still doubts whether carbonylation could be a therapeutic target.
Reversible Oxidation and Others
Reversible oxidation is a type of reversible oxidative PTM, which is generally related to oxidation of cysteine (Cys), namely Cys ox-PTMs (112). Reversible Cys ox-PTMs consist of various patterns, including S-sulfenylation (Cys-SOH), S-glutathionylation (Cys-SSG), and disulfide bond (113). The characteristics of this oxidation pattern are fast and reversible, contributing to quick transition of protein function, so that cells can adapt to a complicated environment. Studies have discovered that the reversible oxidation of C148 and C206 of STING in cells participate in the contradictory regulation of STING. C148 of STING is oxidized independent on CDNs interaction in basic state. And this oxidation state of C148 makes for the binding of 2’3’-cGAMP to STING (114). However, sequential oxidation of C206 of STING in response to 2’3’-cGAMP leads to a conformational change which inhibits the phosphorylation of S366 and prevents over-activation of STING (114). In addition, excessive ROS induced by viral infection can oxidize C148 and inhibit polymerization and activation of STING, thus helping virus evade from cellular defenses (115). However, it is still unclear, whether other Cys of STING can be oxidized and play roles in regulation of STING. More researches are needed (Figures 1, 2).
It is suggested that disulfide bond is also a type of reversible oxidation, which mainly exists in secreted proteins. Disulfide bond is formed by Cys in the oxidizing environment of the cytosol and in the luminal part of proteins (lumen of mitochondria, ER, etc), with the consequence of a more stable structure of protein. There are five cysteines in the cytosolic domain of STING, only one of which (C148) are engaged in disulfide bonds (116). It is discovered that 2’,3’-cGAMP induces closing of the human STING homo-dimer and leads to the formation of disulfide bond via C148. When C148 is mutated to alanine, the affinity of STING to cGAMP is weaker (116). However, more studies are needed to figure out, why disulfide bonds of STING can be produced in the cytosol, and whether disulfide bonds are associated with phase separation. Due to the important role of disulfide bond, it could be potential therapeutic target to improve STING-related auto-inflammatory diseases (Figures 1, 2).
There are still other PTMs which are important in biological and pathological processes, such as acetylation and deacetylation, methylation, biotinylation, ribosylation, carboxylation. However, there is still no evidence favoring these PTMs on STING. Therefore, more research and efforts should be taken into to discover more detailed regulatory mechanisms of STING. Understanding and exploring the underlying network of PTMs may provide new idea of targeted STING therapy.
PTM Related Diseases and Targeted Therapy
As mentioned above, PTMs play critical roles in stabilization, activation and inhibition of STING, thus the immune responses could be accurately regulated in biological and pathological processes (9) (Figure 2). The competition of different PTMs at the same residue is important for the regulation of STING activity and can be critical factor in development of diseases, including infectious diseases (117), cancer (118), auto-inflammatory diseases (116). Therefore, targeted STING therapies have been developed on the basis of PTMs. Since phosphorylation is the classic activation PTMs form of STING, almost all of the STING-related diseases are discovered to be associated with phosphorylation of STING. Therefore, we review as follows mainly those diseases, which are covered to be related to non-phosphorylation of STING. All of STING-related diseases are presented in Figure 3.
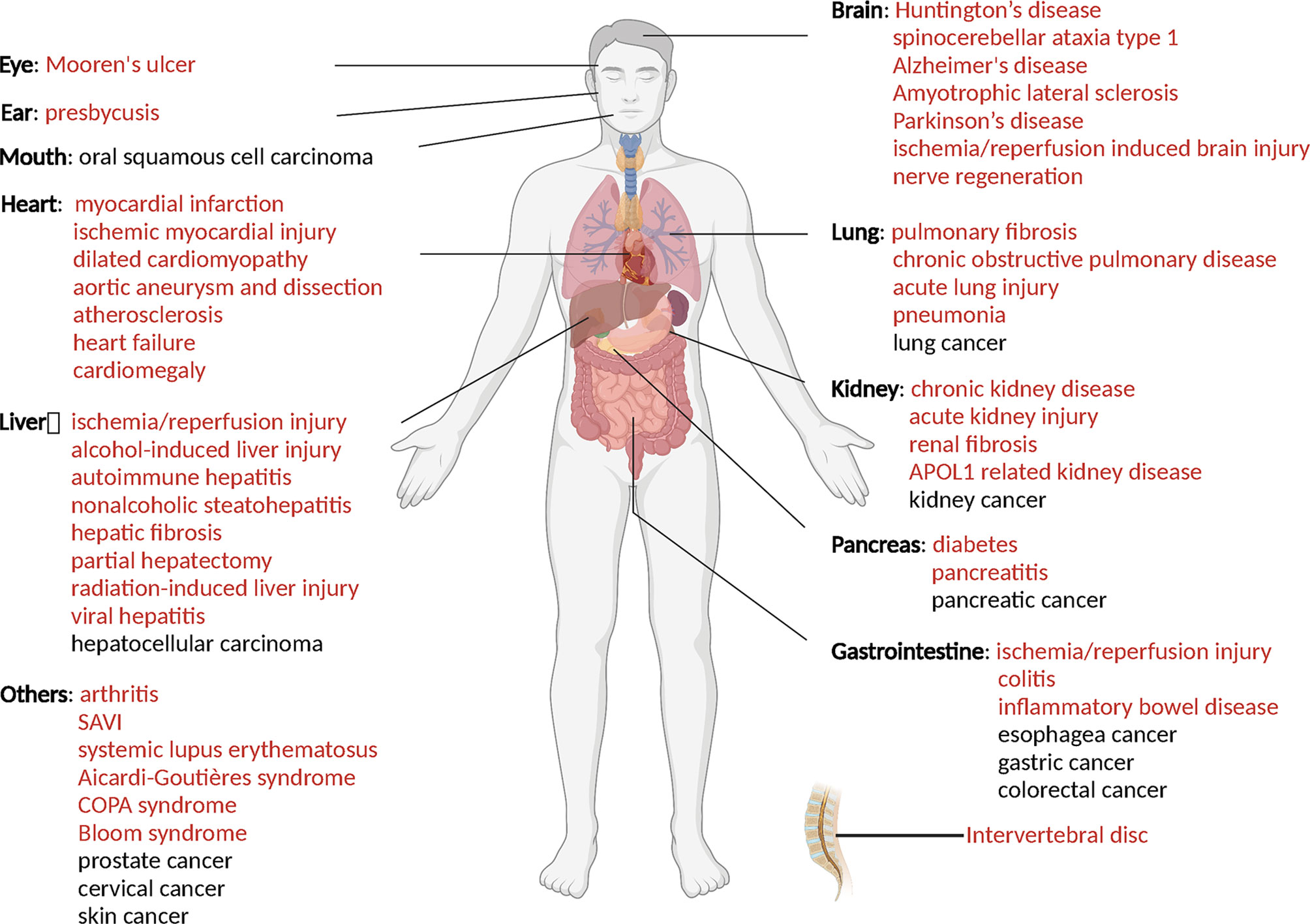
Figure 3 STING related diseases. Red indicates diseases caused by over-activation of STING, while black indicates disease with low-activation of STING. This figure is created with BioRender.com
Infectious Diseases
Infection is caused by pathogens, including bacteria, virus, by which PAMPs can be produced, thus STING pathway can be activated to deal with infection (1). In most time, activation of STING can help to eliminate pathogens (4), while sometimes STING can also help pathogens survive in cells and evade from host immune surveillance (51, 119). Virus is unique, with the simple structure consisting of nucleic acid (DNA or RNA) and protein. Most studies about STING are developed in viral infection model, for example, HSV-1. On this basis, PTMs of STING are gradually discovered, especially carbonylation, ubiquitination, SUMOylation, deubiquitination, glycosylation (Table 1), and related strategies are implemented in virus vaccination (120). However, more studies are needed to figure out in which cases STING is beneficial for clearance of virus, and how we utilize STING in treatment of viral diseases (Table 1).
Bacterial infections are major infectious diseases worldwide, leading to many diseases, including pneumonia (121), tuberculosis (122), and sepsis (123). It is confirmed that, many gram-positive and negative bacterial (117) can release not only bacterial DNA, but also cyclic diadenosine monophosphate (c-di-AMP) and virulence factors, which can trigger cGAS-STING pathway and inflammatory responses. However, not all of STING activation is beneficial for the host. Activation of the cGAS-STING pathway can promote bacterial replication and intracellular bacterial survival after staphylococcus aureus and Brucella abortus infection (124, 125). Some important physiological or pathological processes such as blood coagulation and autophagy could also be influenced by cGAS-STING pathway after bacterial infection (117). When Listeria monocytogenes enters a cell, c-di-AMP can be secreted and promote activation of STING, resulting in the reduction of protective immune responses (126). Therefore, regulation and function of cGAS-STING pathway are complicated when pathogens invade. Studies about STING potential mechanism, apart from the mediator of immune responses, are still on the way.
Cancer
To date, tumorigenesis is regarded as a process driven by inflammation (127). Previously, it is favored that the formation of tumor is related to weakened surveillance of immune system. Therefore, as an activator of immune responses, activation of STING could be potential therapy of cancer (118, 128, 129). Therapy of cGAMP or analogs into tumor-bearing mice results in substantial inhibition of tumor growth and improves the survival of the mice (130). As mentioned above, USP35 can directly deubiquitinate STING with K6/K11/K27/K29/K63-linked polyubiquitin chains and inhibit STING. Silencing USP35 potentiates cisplatin effects in ovarian cancer cells (85). Another kinase, DAPK3, is required for STING K63-linked poly-ubiquitination to activate STING pathway. However, DAPK3 loss-of-function has been discovered in several human tumor types (131), which could help tumor cells evade from host immunity and cancer immunotherapy. All of these encourage us that, cancer cells may develop anti-PTMs of STING strategy to avoid surveillance of host immune system, and targeted PTMs of STING could be potential therapy for cancer, especially those STING-sensitive cancers (Table 1).
Autoimmune and Inflammatory Disease
Mutation or abnormal activation of STING can lead to varieties of auto-immune and auto-inflammatory diseases, including systemic lupus erythematosus (SLE) and STING-associated vasculopathy with onset in infancy (SAVI). SLE is a typical auto-immune disease, in which pathogenic auto-antibodies are produced, resulting in excessive inflammation and severe tissue damage (132). It remains controversial whether STING plays necessary role in SLE (133). However, MYSM1 has been discovered as a suppressor of SLE, which actually triggers K63-linked deubiquitination of STING and inhibits STING pathway. Increased MYSM1 can decrease type I IFN, IL-6, and other inflammatory cytokines in SLE mice (86). This indicates us that targeted ubiquitination of STING therapy could help improve auto-immune and inflammatory diseases.
SAVI is an auto-inflammatory disease with the gain-of-function mutations of STING (V147L, V147M, N154S, V155M, C206Y, R281Q, R284G), characterized by early onset systemic inflammation, vasculopathy and interstitial lung disease (ILD) (134, 135). Studies have discovered that, mutants of STING in SAVI are accompanied by enhanced STING translocation, IRF3 phosphorylation, and IFN-β activity. Blocking C148-mediated disulfide bond can alleviate inflammatory responses in SAVI-modeled cells (116). In addition, treatment of NO2-FAs, which can alkylate STING and inhibit palmitoylation, can decrease production of type I IFN in fibroblasts cultured from SAVI patients. Also, 2-BP can decrease type I IFN in SAVI-mutant HEK293T cells (99). All of these indicate that inhibition of palmitoylation and conformational change of STING, or promotion of nitro-alkylation could be potential target to treat SAVI, which reveal the importance of PTMs of STING in diseases.
PTMs Targeted Therapy
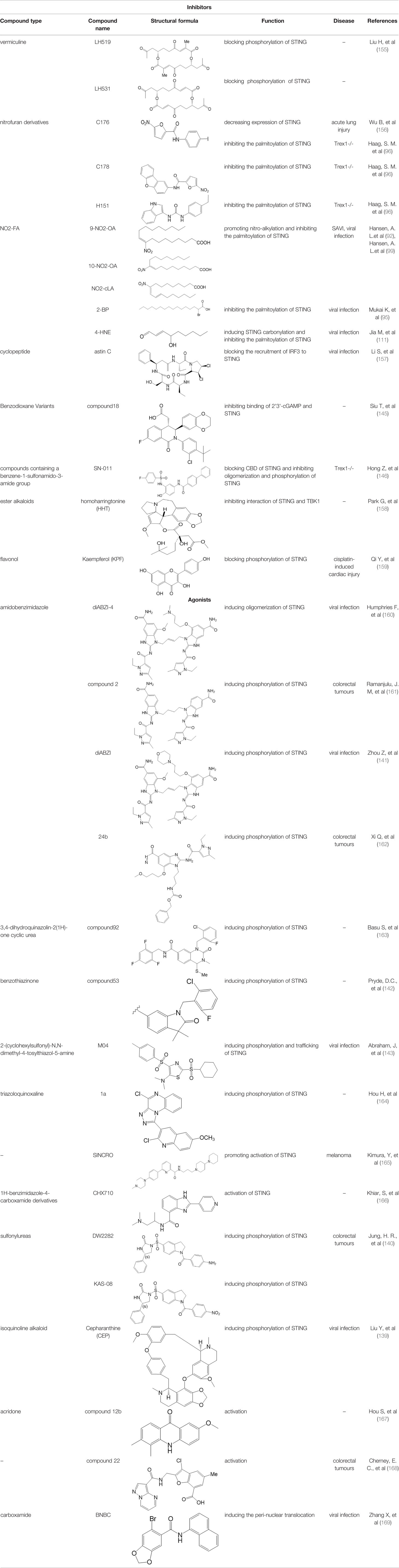
Considering the critical role of STING signaling pathway in inflammation and diseases, targeting STING may lead to novel therapeutics (27, 136). Many strategies are developed, including: production of STING, activation of STING, translocation and oligomerization of STING, degradation of STING and downstream signaling (137). Targeted these strategies develop agonists and inhibitors, some of which are related to PTMs (Table 2).
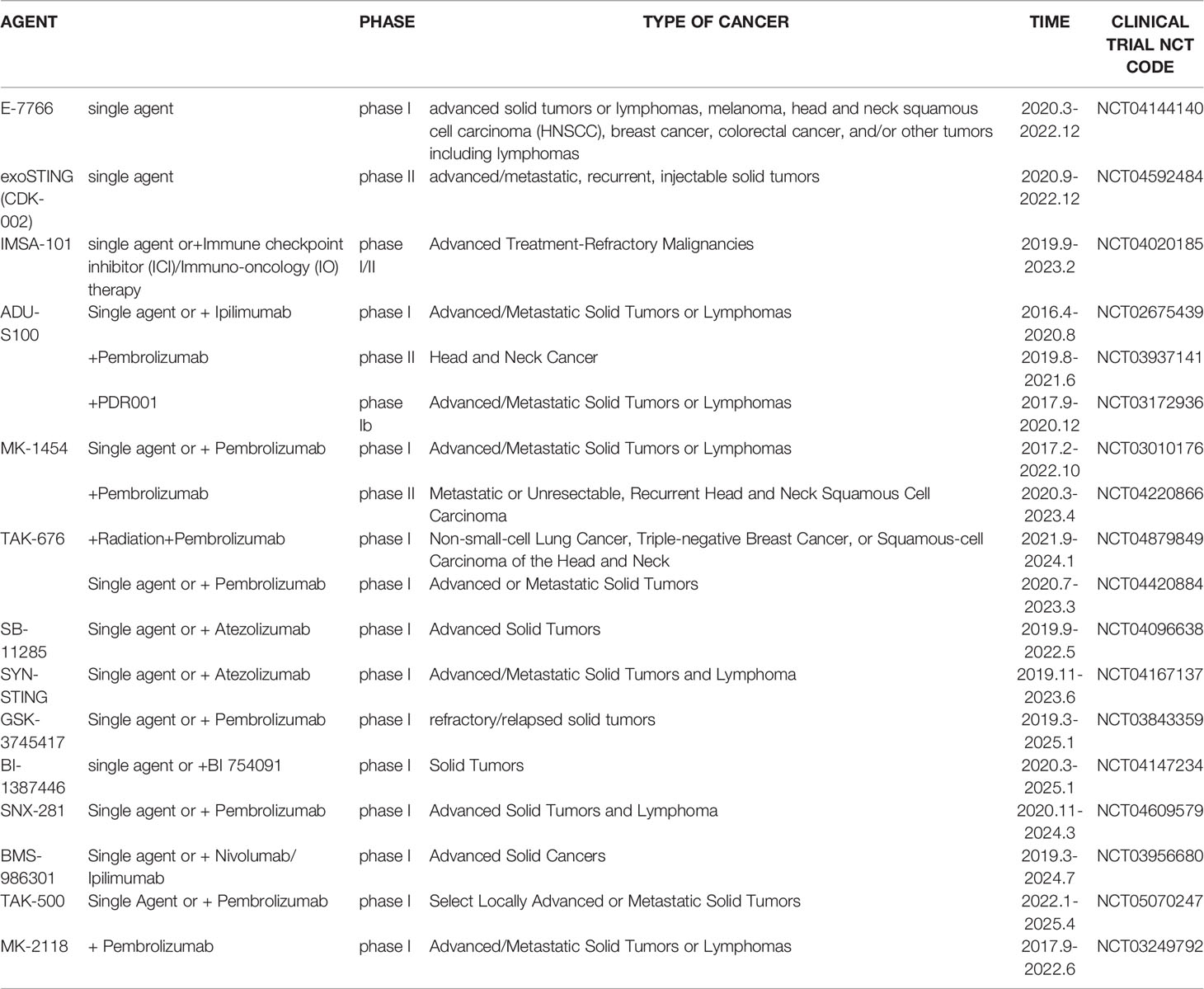
Agonists of STING have been developed for a long time. The most efficient agents are CDNs, which are fit to LBD of STING, resulting in rapid activation of STING pathway (137). Later, more agonists are reported and some of them have been applied into clinical trials to help with symptoms or enhance treatments, especially cancer (137). Novel STING agonist strategies include: bacterial vectors, CDNs, non-CDNs, nano vaccines, antibody-drug conjugate, exo-STING, etc. (160) (Table 3). In addition, other potential therapeutic agents have been explored in experiments in vivo or in vitro, such as CEP (156), KAS-08 (155), DW2282 (155), etc. These agents are mostly tightly related to phosphorylation of STING, which is considered as the marker of STING activation. It has been confirmed that diABZI (147), compound 53 (150), M04 (151), etc, regulate directly phosphorylation of STING (Table 2). While bisphenol A (BPA) can up-regulate ZDHHC1, a palmitoylase, indicating the underlying therapy of palmitoylation in regulation of STING (161). Therefore, PTMs play critical role in strategies of agonists. All of these agonists activate immune responses through cGAS-STING pathway, thus play a critical role in anti-cancer therapy.
As for inhibitors, strategies have also been developed to inhibit STING pathway: decreasing the expression of STING, blocking binding of 2’3’-cGAMP and LBD of STING, inhibiting phosphorylation, blocking traffic of STING, etc (27). Compound 18 is discovered to inhibit binding of 2’3’-cGAMP and STING, leading to inhibition of STING pathway (141). Other inhibitors are mainly associated with PTMs of STING. C176/C178/H151 have been discovered to inhibit palmitoylation of STING (96), while NO2-FAs result in nitro-alkylation to competitively inhibit palmitoylation (99). SN-011 can block phosphorylation of STING to decrease production of type I-IFN and inflammatory cytokines (142). Therefore, PTMs of STING can be important therapeutic target to regulate STING in inflammatory diseases. Targeted PTMs could be potential efficient strategy to improve related diseases.
In a word, many therapeutic targets related to STING are based on phosphorylation, palmitoylation (92), alkylation (99). There are few studies on agonists or inhibitors related to other PTMs, for example, ubiquitination, glycosylation, sumoylation, oxidation, carbonylation, which are also critical in regulation of STING. There have been developed many reviews about targeted therapy of ubiquitination (162–164), glycosylation (165), sumoylation (166), oxidation (167). More studies could focus on the role of these agonists and inhibitors in regulating STING pathway, proposing more comprehensive therapeutic strategies to improve STING-related diseases.
Summary and Future Perspective
Innate immune response is an important defender to deal with endogenous and exogenous abnormal situation in cells. As mentioned above, STING pathway participates importantly in regulation of immune responses (4). Since the discovery of the cGAS-STING pathway, a series of biochemical, structural and genetic studies have been conducted and related mechanisms have been established. It is confirmed that, PTMs of STING are important in regulation of STING pathway (9). Many types of PTMs participate in regulation of activation or degradation of STING (12), including phosphorylation and dephosphorylation, palmitoylation, nitro-alkylation, glycosylation, ubiquitylation and deubiquitylation, SUMOylation, carbonylation, oxidation and so on, so that immune responses induced by STING could be activated or inhibited efficiently.
It is inspiring that the same amino acid residues of STING could be modified by different groups, resulting in distinct, even contrary consequences. For example, C88 could be palmitoylated to activate immune responses (92), while ROS and NO2-FA could result in carbonylation and alkylation of STING to inhibit production of type I IFNs (99, 111). Add or remove phosphorylated groups on Y245 and S366 can also function differently, which indicates the importance of PTMs to precisely regulate STING (56, 57, 61, 64).
Due to important role played by STING in immune responses and inflammation, the critical mechanism of PTMs of STING is deserved to explore and transform into potential therapeutic target. Since then, targeted therapy of STING has been highlighted in treatment of many diseases (27, 137). Most agonists have been explored to improve prognosis of cancer (168). Among them, the most common mechanism is to phosphorylate STING to activate immune responses. By contrast, nitrofuran derivatives, astin C and others are inhibitors of STING (96, 169), which could potentially help with auto-immune diseases and hyper-activation of inflammation. However, the potential mechanisms of those molecular agents are not clearly clarified, which limits wider use of these drugs. In a word, due to complexity of immunity regulation, much remains to be learned about the regulation of STING and details of PTMs in STING pathway. Future therapeutic strategies could focus on optimizing PTMs on STING function in the right disease at the optimal time.
Author Contributions
JK developed and wrote this review. JW and QL revised it critically for important intellectual content. XW, YZ, and JR provide approval for publication of the content. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by National Natural Science Foundation of China (81772052, 82072149, 82072223, and 82100597), the Natural Science Foundation of Jiangsu Province (BK20201116, BK20210039), the program B for Outstanding PhD candidate of Nanjing University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author wishes to thank Hanlu Hu of the Affiliated BenQ Hospital of Nanjing Medical University for chemical structure formula figures.
References
1. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu Rev Immunol (2015) 33:257–90. doi: 10.1146/annurev-immunol-032414-112240
2. Gong T, Liu L, Jiang W, Zhou R. Damp-Sensing Receptors in Sterile Inflammation and Inflammatory Diseases. Nat Rev Immunol (2020) 20(2):95–112. doi: 10.1038/s41577-019-0215-7
3. Zhu G, Xu Y, Cen X, Nandakumar KS, Liu S, Cheng K. Targeting Pattern-Recognition Receptors to Discover New Small Molecule Immune Modulators. Eur J Med Chem (2018) 144:82–92. doi: 10.1016/j.ejmech.2017.12.026
4. Li Z, Cai S, Sun Y, Li L, Ding S, Wang X. When Sting Meets Viruses: Sensing, Trafficking and Response. Front Immunol (2020) 11:2064. doi: 10.3389/fimmu.2020.02064
5. Zhang F, Yuan Y, Ma F. Function and Regulation of Nuclear DNA Sensors During Viral Infection and Tumorigenesis. Front Immunol (2020) 11:624556. doi: 10.3389/fimmu.2020.624556
6. Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear Cgas Suppresses DNA Repair and Promotes Tumorigenesis. Nature (2018) 563(7729):131–6. doi: 10.1038/s41586-018-0629-6
7. Hopfner KP, Hornung V. Molecular Mechanisms and Cellular Functions of Cgas-Sting Signalling. Nat Rev Mol Cell Biol (2020) 21(9):501–21. doi: 10.1038/s41580-020-0244-x
8. Yum S, Li M, Chen ZJ. Old Dogs, New Trick: Classic Cancer Therapies Activate Cgas. Cell Res (2020) 30(8):639–48. doi: 10.1038/s41422-020-0346-1
9. Wang Z, Chen N, Li Z, Xu G, Zhan X, Tang J, et al. The Cytosolic DNA-Sensing Cgas-Sting Pathway in Liver Diseases. Front Cell Dev Biol (2021) 9:717610. doi: 10.3389/fcell.2021.717610
10. Balka KR, De Nardo D. Molecular and Spatial Mechanisms Governing Sting Signalling. FEBS J (2021) 288(19):5504–29. doi: 10.1111/febs.15640
11. Barrat FJ, Crow MK, Ivashkiv LB. Interferon Target-Gene Expression and Epigenomic Signatures in Health and Disease. Nat Immunol (2019) 20(12):1574–83. doi: 10.1038/s41590-019-0466-2
12. Hong Z, Ma T, Liu X, Wang C. Cgas-Sting Pathway: Post-Translational Modifications and Functions in Sterile Inflammatory Diseases. FEBS J (2021). doi: 10.1111/febs.16137. Epub ahead of print.
13. Hu T, Pan M, Yin Y, Wang C, Cui Y, Wang Q. The Regulatory Network of Cyclic Gmp-Amp Synthase-Stimulator of Interferon Genes Pathway in Viral Evasion. Front Microbiol (2021) 12:790714. doi: 10.3389/fmicb.2021.790714
14. Song B, Liu D, Greco TM, Cristea IM. Post-Translational Modification Control of Viral DNA Sensors and Innate Immune Signaling. Adv Virus Res (2021) 109:163–99. doi: 10.1016/bs.aivir.2021.03.001
15. Diskin C, Ryan TAJ, O’Neill LAJ. Modification of Proteins by Metabolites in Immunity. Immunity (2021) 54(1):19–31. doi: 10.1016/j.immuni.2020.09.014
16. Chang HM, Yeh ETH. Sumo: From Bench to Bedside. Physiol Rev (2020) 100(4):1599–619. doi: 10.1152/physrev.00025.2019
17. Snider NT, Omary MB. Post-Translational Modifications of Intermediate Filament Proteins: Mechanisms and Functions. Nat Rev Mol Cell Biol (2014) 15(3):163–77. doi: 10.1038/nrm3753
18. Cheng Z, Dai T, He X, Zhang Z, Xie F, Wang S, et al. The Interactions Between Cgas-Sting Pathway and Pathogens. Signal Transduct Target Ther (2020) 5(1):91. doi: 10.1038/s41392-020-0198-7
19. Shang G, Zhang C, Chen ZJ, Bai XC, Zhang X. Cryo-Em Structures of Sting Reveal Its Mechanism of Activation by Cyclic Gmp-Amp. Nature (2019) 567(7748):389–93. doi: 10.1038/s41586-019-0998-5
20. Ishikawa H, Barber GN. Sting Is an Endoplasmic Reticulum Adaptor That Facilitates Innate Immune Signalling. Nature (2008) 455(7213):674–8. doi: 10.1038/nature07317
21. Zhang X, Bai XC, Chen ZJ. Structures and Mechanisms in the Cgas-Sting Innate Immunity Pathway. Immunity (2020) 53(1):43–53. doi: 10.1016/j.immuni.2020.05.013
22. Li M, Raheem MA, Han C, Yu F, Dai Y, Imran M, et al. The Fowl Adenovirus Serotype 4 (Fadv-4) Induce Cellular Pathway in Chickens to Produce Interferon and Antigen-Presented Molecules (Mhci/Ii). Poult Sci (2021) 100(10):101406. doi: 10.1016/j.psj.2021.101406
23. Amparyup P, Charoensapsri W, Soponpong S, Jearaphunt M, Wongpanya R, Tassanakajon A. Stimulator of Interferon Gene (Sting) and Interferon Regulatory Factor (Irf) Are Crucial for Shrimp Antiviral Defense Against Wssv Infection. Fish Shellfish Immunol (2021) 117:240–7. doi: 10.1016/j.fsi.2021.08.016
24. Morehouse BR, Govande AA, Millman A, Keszei AFA, Lowey B, Ofir G, et al. Sting Cyclic Dinucleotide Sensing Originated in Bacteria. Nature (2020) 586(7829):429–33. doi: 10.1038/s41586-020-2719-5
25. Martin M, Hiroyasu A, Guzman RM, Roberts SA, Goodman AG. Analysis of Drosophila Sting Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep (2018) 23(12):3537–50.e6. doi: 10.1016/j.celrep.2018.05.029
26. Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy Induction Via Sting Trafficking Is a Primordial Function of the Cgas Pathway. Nature (2019) 567(7747):262–6. doi: 10.1038/s41586-019-1006-9
27. Decout A, Katz JD, Venkatraman S, Ablasser A. The Cgas-Sting Pathway as a Therapeutic Target in Inflammatory Diseases. Nat Rev Immunol (2021) 21(9):548–69. doi: 10.1038/s41577-021-00524-z
28. Motwani M, Pesiridis S, Fitzgerald KA. DNA Sensing by the Cgas-Sting Pathway in Health and Disease. Nat Rev Genet (2019) 20(11):657–74. doi: 10.1038/s41576-019-0151-1
29. Hu Y, Manasrah BK, McGregor SM, Lera RF, Norman RX, Tucker JB, et al. Paclitaxel Induces Micronucleation and Activates Pro-Inflammatory Cgas-Sting Signaling in Triple-Negative Breast Cancer. Mol Cancer Ther (2021) 20(12):2553–67. doi: 10.1158/1535-7163.MCT-21-0195
30. Liu Q, Wu J, Zhang X, Li X, Wu X, Zhao Y, et al. Circulating Mitochondrial DNA-Triggered Autophagy Dysfunction Via Sting Underlies Sepsis-Related Acute Lung Injury. Cell Death Dis (2021) 12(7):673. doi: 10.1038/s41419-021-03961-9
31. Parkes EE, Walker SM, Taggart LE, McCabe N, Knight LA, Wilkinson R, et al. Activation of STING-Dependent Innate Immune Signaling by S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst (2017) 109(1):djw199. doi: 10.1093/jnci/djw199
32. Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, et al. Cytoplasmic Chromatin Triggers Inflammation in Senescence and Cancer. Nature (2017) 550(7676):402–6. doi: 10.1038/nature24050
33. Ishikawa H, Ma Z, Barber GN. Sting Regulates Intracellular DNA-Mediated, Type I Interferon-Dependent Innate Immunity. Nature (2009) 461(7265):788–92. doi: 10.1038/nature08476
34. Nandakumar R, Tschismarov R, Meissner F, Prabakaran T, Krissanaprasit A, Farahani E, et al. Intracellular Bacteria Engage a Sting-Tbk1-Mvb12b Pathway to Enable Paracrine Cgas-Sting Signalling. Nat Microbiol (2019) 4(4):701–13. doi: 10.1038/s41564-019-0367-z
35. Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt-Jones EA, et al. Sting-Irf3 Pathway Links Endoplasmic Reticulum Stress With Hepatocyte Apoptosis in Early Alcoholic Liver Disease. Proc Natl Acad Sci USA (2013) 110(41):16544–9. doi: 10.1073/pnas.1308331110
36. Wu J, Liu Q, Zhang X, Wu X, Zhao Y, Ren J. Sting-Dependent Induction of Lipid Peroxidation Mediates Intestinal Ischemia-Reperfusion Injury. Free Radic Biol Med (2021) 163:135–40. doi: 10.1016/j.freeradbiomed.2020.12.010
37. Kanno T, Nakajima T, Yokoyama S, Asou HK, Sasamoto S, Kamii Y, et al. Scd2-Mediated Monounsaturated Fatty Acid Metabolism Regulates Cgas-Sting-Dependent Type I Ifn Responses in Cd4(+) T Cells. Commun Biol (2021) 4(1):820. doi: 10.1038/s42003-021-02310-y
38. Srikanth S, Woo JS, Wu B, El-Sherbiny YM, Leung J, Chupradit K, et al. The Ca(2+) Sensor Stim1 Regulates the Type I Interferon Response by Retaining the Signaling Adaptor Sting at the Endoplasmic Reticulum. Nat Immunol (2019) 20(2):152–62. doi: 10.1038/s41590-018-0287-8
39. Wu J, Chen YJ, Dobbs N, Sakai T, Liou J, Miner JJ, et al. Sting-Mediated Disruption of Calcium Homeostasis Chronically Activates Er Stress and Primes T Cell Death. J Exp Med (2019) 216(4):867–83. doi: 10.1084/jem.20182192
40. Cerboni S, Jeremiah N, Gentili M, Gehrmann U, Conrad C, Stolzenberg MC, et al. Intrinsic Antiproliferative Activity of the Innate Sensor Sting in T Lymphocytes. J Exp Med (2017) 214(6):1769–85. doi: 10.1084/jem.20161674
41. Zhang R, Kang R, Tang D. The Sting1 Network Regulates Autophagy and Cell Death. Signal Transduct Target Ther (2021) 6(1):208. doi: 10.1038/s41392-021-00613-4
42. Xiong R, Li N, Chen L, Wang W, Wang B, Jiang W, et al. Sting Protects Against Cardiac Dysfunction and Remodelling by Blocking Autophagy. Cell Commun Signal (2021) 19(1):109. doi: 10.1186/s12964-021-00793-0
43. Tang CH, Zundell JA, Ranatunga S, Lin C, Nefedova Y, Del Valle JR, et al. Agonist-Mediated Activation of Sting Induces Apoptosis in Malignant B Cells. Cancer Res (2016) 76(8):2137–52. doi: 10.1158/0008-5472.CAN-15-1885
44. Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, et al. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a Sting-Cell Death Program Upstream of Nlrp3. Cell (2017) 171(5):1110–24.e18. doi: 10.1016/j.cell.2017.09.039
45. Gong W, Liu P, Zhao F, Liu J, Hong Z, Ren H, et al. Sting-Mediated Syk Signaling Attenuates Tumorigenesis of Colitisassociated Colorectal Cancer Through Enhancing Intestinal Epithelium Pyroptosis. Inflammation Bowel Dis (2022) 28(4):572–85. doi: 10.1093/ibd/izab217
46. Li C, Liu J, Hou W, Kang R, Tang D. Sting1 Promotes Ferroptosis Through Mfn1/2-Dependent Mitochondrial Fusion. Front Cell Dev Biol (2021) 9:698679. doi: 10.3389/fcell.2021.698679
47. Brault M, Olsen TM, Martinez J, Stetson DB, Oberst A. Intracellular Nucleic Acid Sensing Triggers Necroptosis Through Synergistic Type I Ifn and Tnf Signaling. J Immunol (2018) 200(8):2748–56. doi: 10.4049/jimmunol.1701492
48. Zhang X, Wu J, Liu Q, Li X, Li S, Chen J, et al. Mtdna-Sting Pathway Promotes Necroptosis-Dependent Enterocyte Injury in Intestinal Ischemia Reperfusion. Cell Death Dis (2020) 11(12):1050. doi: 10.1038/s41419-020-03239-6
49. Ku JWK, Chen Y, Lim BJW, Gasser S, Crasta KC, Gan YH. Bacterial-Induced Cell Fusion Is a Danger Signal Triggering Cgas-Sting Pathway Via Micronuclei Formation. Proc Natl Acad Sci USA (2020) 117(27):15923–34. doi: 10.1073/pnas.2006908117
50. Chattopadhyay S, Liu YH, Fang ZS, Lin CL, Lin JC, Yao BY, et al. Synthetic Immunogenic Cell Death Mediated by Intracellular Delivery of Sting Agonist Nanoshells Enhances Anticancer Chemo-Immunotherapy. Nano Lett (2020) 20(4):2246–56. doi: 10.1021/acs.nanolett.9b04094
51. Hong Y, Jeong H, Park K, Lee S, Shim JY, Kim H, et al. STING Facilitates Nuclear Import of Herpesvirus Genome During Infection. Proc Natl Acad Sci USA (2021) 118(33):e2108631118. doi: 10.1073/pnas.2108631118
52. Chu TT, Tu X, Yang K, Wu J, Repa JJ, Yan N. Tonic Prime-Boost of Sting Signalling Mediates Niemann-Pick Disease Type C. Nature (2021) 596(7873):570–5. doi: 10.1038/s41586-021-03762-2
53. Deribe YL, Pawson T, Dikic I. Post-Translational Modifications in Signal Integration. Nat Struct Mol Biol (2010) 17(6):666–72. doi: 10.1038/nsmb.1842
54. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The Adaptor Protein Mita Links Virus-Sensing Receptors to Irf3 Transcription Factor Activation. Immunity (2008) 29(4):538–50. doi: 10.1016/j.immuni.2008.09.003
55. Takahashi K, Niki T, Ogawa E, Fumika K, Nishioka Y, Sawa M, et al. A Cell-Free Assay Implicates a Role of Sphingomyelin and Cholesterol in Sting Phosphorylation. Sci Rep (2021) 11(1):11996. doi: 10.1038/s41598-021-91562-z
56. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of Innate Immune Adaptor Proteins Mavs, Sting, and Trif Induces Irf3 Activation. Science (2015) 347(6227):aaa2630. doi: 10.1126/science.aaa2630
57. Gao P, Hu MM, Shu HB. Csk Promotes Innate Immune Response to DNA Virus by Phosphorylating Mita. Biochem Biophys Res Commun (2020) 526(1):199–205. doi: 10.1016/j.bbrc.2020.03.069
58. Ni G, Ma Z, Wong JP, Zhang Z, Cousins E, Major MB, et al. Ppp6c Negatively Regulates Sting-Dependent Innate Immune Responses. mBio (2020) 11(4):e01728-20. doi: 10.1128/mBio.01728-20
59. Wang C, Wang X, Veleeparambil M, Kessler PM, Willard B, Chattopadhyay S, et al. Egfr-Mediated Tyrosine Phosphorylation of Sting Determines Its Trafficking Route and Cellular Innate Immunity Functions. EMBO J (2020) 39(22):e104106. doi: 10.15252/embj.2019104106
60. Wang C, Sharma N, Veleeparambil M, Kessler PM, Willard B, Sen GC. Sting-Mediated Interferon Induction by Herpes Simplex Virus 1 Requires the Protein Tyrosine Kinase Syk. mBio (2021) 12(6):e0322821. doi: 10.1128/mbio.03228-21
61. Konno H, Konno K, Barber GN. Cyclic Dinucleotides Trigger Ulk1 (Atg1) Phosphorylation of Sting to Prevent Sustained Innate Immune Signaling. Cell (2013) 155(3):688–98. doi: 10.1016/j.cell.2013.09.049
62. Li Z, Liu G, Sun L, Teng Y, Guo X, Jia J, et al. Ppm1a Regulates Antiviral Signaling by Antagonizing Tbk1-Mediated Sting Phosphorylation and Aggregation. PloS Pathog (2015) 11(3):e1004783. doi: 10.1371/journal.ppat.1004783
63. Wang Y, Qian G, Zhu L, Zhao Z, Liu Y, Han W, et al. Hiv-1 Vif Suppresses Antiviral Immunity by Targeting Sting. Cell Mol Immunol (2022) 19(1):108–21. doi: 10.1038/s41423-021-00802-9
64. Xia T, Yi XM, Wu X, Shang J, Shu HB. Ptpn1/2-Mediated Dephosphorylation of Mita/Sting Promotes Its 20s Proteasomal Degradation and Attenuates Innate Antiviral Response. Proc Natl Acad Sci USA (2019) 116(40):20063–9. doi: 10.1073/pnas.1906431116
65. Deng L, Meng T, Chen L, Wei W, Wang P. The Role of Ubiquitination in Tumorigenesis and Targeted Drug Discovery. Signal Transduct Target Ther (2020) 5(1):11. doi: 10.1038/s41392-020-0107-0
66. Varshavsky A. The Ubiquitin System, an Immense Realm. Annu Rev Biochem (2012) 81:167–76. doi: 10.1146/annurev-biochem-051910-094049
67. Zhang ZD, Xiong TC, Yao SQ, Wei MC, Chen M, Lin D, et al. Rnf115 Plays Dual Roles in Innate Antiviral Responses by Catalyzing Distinct Ubiquitination of Mavs and Mita. Nat Commun (2020) 11(1):5536. doi: 10.1038/s41467-020-19318-3
68. Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The Ubiquitin Ligase Trim56 Regulates Innate Immune Responses to Intracellular Double-Stranded DNA. Immunity (2010) 33(5):765–76. doi: 10.1016/j.immuni.2010.10.013
69. Yang L, Wang L, Ketkar H, Ma J, Yang G, Cui S, et al. Ubxn3b Positively Regulates Sting-Mediated Antiviral Immune Responses. Nat Commun (2018) 9(1):2329. doi: 10.1038/s41467-018-04759-8
70. Ni G, Konno H, Barber GN. Ubiquitination of STING at Lysine 224 Controls Irf3 Activation. Sci Immunol (2017) 2(11):eaah7119. doi: 10.1126/sciimmunol.aah7119
71. Zhang J, Hu MM, Wang YY, Shu HB. Trim32 Protein Modulates Type I Interferon Induction and Cellular Antiviral Response by Targeting Mita/Sting Protein for K63-Linked Ubiquitination. J Biol Chem (2012) 287(34):28646–55. doi: 10.1074/jbc.M112.362608
72. Bodda C, Reinert LS, Fruhwurth S, Richardo T, Sun C, Zhang BC, et al. Hsv1 Vp1-2 Deubiquitinates Sting to Block Type I Interferon Expression and Promote Brain Infection. J Exp Med (2020) 217(7):e20191422. doi: 10.1084/jem.20191422
73. Liu Y, Li J, Chen J, Li Y, Wang W, Du X, et al. Hepatitis B Virus Polymerase Disrupts K63-Linked Ubiquitination of Sting to Block Innate Cytosolic DNA-Sensing Pathways. J Virol (2015) 89(4):2287–300. doi: 10.1128/JVI.02760-14
74. Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, et al. The E3 Ubiquitin Ligase Amfr and Insig1 Bridge the Activation of Tbk1 Kinase by Modifying the Adaptor Sting. Immunity (2014) 41(6):919–33. doi: 10.1016/j.immuni.2014.11.011
75. Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, et al. The Ubiquitin Ligase Rnf5 Regulates Antiviral Responses by Mediating Degradation of the Adaptor Protein Mita. Immunity (2009) 30(3):397–407. doi: 10.1016/j.immuni.2009.01.008
76. Yang B, Liu Y, Cui Y, Song D, Zhang G, Ma S, et al. Rnf90 Negatively Regulates Cellular Antiviral Responses by Targeting Mita for Degradation. PloS Pathog (2020) 16(3):e1008387. doi: 10.1371/journal.ppat.1008387
77. Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al. Rnf26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms. PloS Pathog (2014) 10(9):e1004358. doi: 10.1371/journal.ppat.1004358
78. Li Q, Lin L, Tong Y, Liu Y, Mou J, Wang X, et al. Trim29 Negatively Controls Antiviral Immune Response Through Targeting Sting for Degradation. Cell Discovery (2018) 4:13. doi: 10.1038/s41421-018-0010-9
79. Xing J, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, et al. Trim29 Promotes DNA Virus Infections by Inhibiting Innate Immune Response. Nat Commun (2017) 8(1):945. doi: 10.1038/s41467-017-00101-w
80. Wang Y, Lian Q, Yang B, Yan S, Zhou H, He L, et al. Trim30alpha Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting Sting. PloS Pathog (2015) 11(6):e1005012. doi: 10.1371/journal.ppat.1005012
81. Takahashi M, Lio CJ, Campeau A, Steger M, Ay F, Mann M, et al. The Tumor Suppressor Kinase Dapk3 Drives Tumor-Intrinsic Immunity Through the Sting-Ifn-Beta Pathway. Nat Immunol (2021) 22(4):485–96. doi: 10.1038/s41590-021-00896-3
82. Li X, Yu Z, Fang Q, Yang M, Huang J, Li Z, et al. The Transmembrane Endoplasmic Reticulum-Associated E3 Ubiquitin Ligase Trim13 Restrains the Pathogenic-DNA-Triggered Inflammatory Response. Sci Adv (2022) 8(4):eabh0496. doi: 10.1126/sciadv.abh0496
83. Chen Y, Wang L, Jin J, Luan Y, Chen C, Li Y, et al. P38 Inhibition Provides Anti-DNA Virus Immunity by Regulation of Usp21 Phosphorylation and Sting Activation. J Exp Med (2017) 214(4):991–1010. doi: 10.1084/jem.20161387
84. Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, et al. Usp13 Negatively Regulates Antiviral Responses by Deubiquitinating Sting. Nat Commun (2017) 8:15534. doi: 10.1038/ncomms15534
85. Zhang J, Chen Y, Chen X, Zhang W, Zhao L, Weng L, et al. Deubiquitinase Usp35 Restrains Sting-Mediated Interferon Signaling in Ovarian Cancer. Cell Death Differ (2021) 28(1):139–55. doi: 10.1038/s41418-020-0588-y
86. Tian M, Liu W, Zhang Q, Huang Y, Li W, Wang W, et al. Mysm1 Represses Innate Immunity and Autoimmunity Through Suppressing the Cgas-Sting Pathway. Cell Rep (2020) 33(3):108297. doi: 10.1016/j.celrep.2020.108297
87. Zhang MX, Cai Z, Zhang M, Wang XM, Wang Y, Zhao F, et al. Usp20 Promotes Cellular Antiviral Responses Via Deconjugating K48-Linked Ubiquitination of Mita. J Immunol (2019) 202(8):2397–406. doi: 10.4049/jimmunol.1801447
88. Zhang M, Zhang MX, Zhang Q, Zhu GF, Yuan L, Zhang DE, et al. Usp18 Recruits Usp20 to Promote Innate Antiviral Response Through Deubiquitinating Sting/Mita. Cell Res (2016) 26(12):1302–19. doi: 10.1038/cr.2016.125
89. Guo Y, Jiang F, Kong L, Wu H, Zhang H, Chen X, et al. Otud5 Promotes Innate Antiviral and Antitumor Immunity Through Deubiquitinating and Stabilizing Sting. Cell Mol Immunol (2021) 18(8):1945–55. doi: 10.1038/s41423-020-00531-5
90. Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, et al. Sumoylation Promotes the Stability of the DNA Sensor Cgas and the Adaptor Sting to Regulate the Kinetics of Response to DNA Virus. Immunity (2016) 45(3):555–69. doi: 10.1016/j.immuni.2016.08.014
91. Carta G, Murru E, Banni S, Manca C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front Physiol (2017) 8:902. doi: 10.3389/fphys.2017.00902
92. Hansen AL, Mukai K, Schopfer FJ, Taguchi T, Holm CK. Sting Palmitoylation as a Therapeutic Target. Cell Mol Immunol (2019) 16(3):236–41. doi: 10.1038/s41423-019-0205-5
93. Ko PJ, Dixon SJ. Protein Palmitoylation and Cancer. EMBO Rep (2018) 19(10):e46666. doi: 10.15252/embr.201846666. Epub 2018 Sep 19.
94. Lin H. Protein Cysteine Palmitoylation in Immunity and Inflammation. FEBS J (2021) 288(24):7043–59. doi: 10.1111/febs.15728
95. Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, et al. Activation of Sting Requires Palmitoylation at the Golgi. Nat Commun (2016) 7:11932. doi: 10.1038/ncomms11932
96. Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, et al. Targeting Sting With Covalent Small-Molecule Inhibitors. Nature (2018) 559(7713):269–73. doi: 10.1038/s41586-018-0287-8
97. Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PR, et al. Nitrated Fatty Acids: Endogenous Anti-Inflammatory Signaling Mediators. J Biol Chem (2006) 281(47):35686–98. doi: 10.1074/jbc.M603357200
98. Villacorta L, Minarrieta L, Salvatore SR, Khoo NK, Rom O, Gao Z, et al. In Situ Generation, Metabolism and Immunomodulatory Signaling Actions of Nitro-Conjugated Linoleic Acid in a Murine Model of Inflammation. Redox Biol (2018) 15:522–31. doi: 10.1016/j.redox.2018.01.005
99. Hansen AL, Buchan GJ, Ruhl M, Mukai K, Salvatore SR, Ogawa E, et al. Nitro-Fatty Acids Are Formed in Response to Virus Infection and Are Potent Inhibitors of Sting Palmitoylation and Signaling. Proc Natl Acad Sci USA (2018) 115(33):E7768–E75. doi: 10.1073/pnas.1806239115
100. Vitturi DA, Chen CS, Woodcock SR, Salvatore SR, Bonacci G, Koenitzer JR, et al. Modulation of Nitro-Fatty Acid Signaling: Prostaglandin Reductase-1 Is a Nitroalkene Reductase. J Biol Chem (2013) 288(35):25626–37. doi: 10.1074/jbc.M113.486282
101. Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global View of Human Protein Glycosylation Pathways and Functions. Nat Rev Mol Cell Biol (2020) 21(12):729–49. doi: 10.1038/s41580-020-00294-x
102. Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision Mapping of the Human O-Galnac Glycoproteome Through Simplecell Technology. EMBO J (2013) 32(10):1478–88. doi: 10.1038/emboj.2013.79
103. Goto A, Okado K, Martins N, Cai H, Barbier V, Lamiable O, et al. The Kinase Ikkbeta Regulates a Sting- and Nf-Kappab-Dependent Antiviral Response Pathway in Drosophila. Immunity (2018) 49(2):225–34.e4. doi: 10.1016/j.immuni.2018.07.013
104. Pomin VH, Mulloy B. Glycosaminoglycans and Proteoglycans. Pharmaceuticals (Basel) (2018) 11(1):27. doi: 10.3390/ph11010027
105. Soares da Costa D, Reis RL, Pashkuleva I. Sulfation of Glycosaminoglycans and Its Implications in Human Health and Disorders. Annu Rev BioMed Eng (2017) 19:1–26. doi: 10.1146/annurev-bioeng-071516-044610
106. Smock RG, Meijers R. Roles of Glycosaminoglycans as Regulators of Ligand/Receptor Complexes. Open Biol (2018) 8(10):180026. doi: 10.1098/rsob.180026
107. Fang R, Jiang Q, Guan Y, Gao P, Zhang R, Zhao Z, et al. Golgi Apparatus-Synthesized Sulfated Glycosaminoglycans Mediate Polymerization and Activation of the Cgamp Sensor Sting. Immunity (2021) 54(5):962–75.e8. doi: 10.1016/j.immuni.2021.03.011
108. Fedorova M, Bollineni RC, Hoffmann R. Protein Carbonylation as a Major Hallmark of Oxidative Damage: Update of Analytical Strategies. Mass Spectrom Rev (2014) 33(2):79–97. doi: 10.1002/mas.21381
109. Barreiro E. Protein Carbonylation and Muscle Function in Copd and Other Conditions. Mass Spectrom Rev (2014) 33(3):219–36. doi: 10.1002/mas.21394
110. Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein Carbonylation in Human Diseases. Trends Mol Med (2003) 9(4):169–76. doi: 10.1016/s1471-4914(03)00031-5
111. Jia M, Qin D, Zhao C, Chai L, Yu Z, Wang W, et al. Redox Homeostasis Maintained by Gpx4 Facilitates Sting Activation. Nat Immunol (2020) 21(7):727–35. doi: 10.1038/s41590-020-0699-0
112. Boronat S, Domenech A, Hidalgo E. Proteomic Characterization of Reversible Thiol Oxidations in Proteomes and Proteins. Antioxid Redox Signal (2017) 26(7):329–44. doi: 10.1089/ars.2016.6720
113. Paulsen CE, Carroll KS. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem Rev (2013) 113(7):4633–79. doi: 10.1021/cr300163e
114. Zamorano Cuervo N, Fortin A, Caron E, Chartier S, Grandvaux N. Pinpointing Cysteine Oxidation Sites by High-Resolution Proteomics Reveals a Mechanism of Redox-Dependent Inhibition of Human Sting. Sci Signal (2021) 14(680):eaaw4673. doi: 10.1126/scisignal.aaw4673
115. Tao L, Lemoff A, Wang G, Zarek C, Lowe A, Yan N, et al. Reactive Oxygen Species Oxidize Sting and Suppress Interferon Production. Elife (2020) 9:e57837. doi: 10.7554/eLife.57837
116. Ergun SL, Fernandez D, Weiss TM, Li L. Sting Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell (2019) 178(2):290–301.e10. doi: 10.1016/j.cell.2019.05.036
117. Liu N, Pang X, Zhang H, Ji P. The Cgas-Sting Pathway in Bacterial Infection and Bacterial Immunity. Front Immunol (2021) 12:814709. doi: 10.3389/fimmu.2021.814709
118. Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, et al. Sting-Dependent Cytosolic DNA Sensing Mediates Innate Immune Recognition of Immunogenic Tumors. Immunity (2014) 41(5):830–42. doi: 10.1016/j.immuni.2014.10.017
119. Triantafilou M, Ramanjulu J, Booty LM, Jimenez-Duran G, Keles H, Saunders K, et al. Human Rhinovirus Promotes Sting Trafficking to Replication Organelles to Promote Viral Replication. Nat Commun (2022) 13(1):1406. doi: 10.1038/s41467-022-28745-3
120. Chattopadhyay S, Hu CJ. Nanomedicinal Delivery of Stimulator of Interferon Genes Agonists: Recent Advances in Virus Vaccination. Nanomedicine (Lond) (2020) 15(29):2883–94. doi: 10.2217/nnm-2020-0269
121. van der Poll T, Opal SM. Pathogenesis, Treatment, and Prevention of Pneumococcal Pneumonia. Lancet (2009) 374(9700):1543–56. doi: 10.1016/S0140-6736(09)61114-4
122. Gagneux S. Ecology and Evolution of Mycobacterium Tuberculosis. Nat Rev Microbiol (2018) 16(4):202–13. doi: 10.1038/nrmicro.2018.8
123. Fang YL, Wang CH, Chen YS, Chien CC, Kuo FC, You HL, et al. An Integrated Microfluidic System for Early Detection of Sepsis-Inducing Bacteria. Lab Chip (2021) 21(1):113–21. doi: 10.1039/d0lc00966k
124. Guimaraes ES, Gomes MTR, Campos PC, Mansur DS, Dos Santos AA, Harms J, et al. Brucella Abortus Cyclic Dinucleotides Trigger Sting-Dependent Unfolded Protein Response That Favors Bacterial Replication. J Immunol (2019) 202(9):2671–81. doi: 10.4049/jimmunol.1801233
125. Scumpia PO, Botten GA, Norman JS, Kelly-Scumpia KM, Spreafico R, Ruccia AR, et al. Opposing Roles of Toll-Like Receptor and Cytosolic DNA-Sting Signaling Pathways for Staphylococcus Aureus Cutaneous Host Defense. PloS Pathog (2017) 13(7):e1006496. doi: 10.1371/journal.ppat.1006496
126. Archer KA, Durack J, Portnoy DA. Sting-Dependent Type I Ifn Production Inhibits Cell-Mediated Immunity to Listeria Monocytogenes. PloS Pathog (2014) 10(1):e1003861. doi: 10.1371/journal.ppat.1003861
127. Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity (2019) 51(1):27–41. doi: 10.1016/j.immuni.2019.06.025
128. Takashima K, Takeda Y, Oshiumi H, Shime H, Okabe M, Ikawa M, et al. Sting in Tumor and Host Cells Cooperatively Work for Nk Cell-Mediated Tumor Growth Retardation. Biochem Biophys Res Commun (2016) 478(4):1764–71. doi: 10.1016/j.bbrc.2016.09.021
129. Francica BJ, Ghasemzadeh A, Desbien AL, Theodros D, Sivick KE, Reiner GL, et al. Tnfalpha and Radioresistant Stromal Cells Are Essential for Therapeutic Efficacy of Cyclic Dinucleotide Sting Agonists in Nonimmunogenic Tumors. Cancer Immunol Res (2018) 6(4):422–33. doi: 10.1158/2326-6066.CIR-17-0263
130. Demaria O, De Gassart A, Coso S, Gestermann N, Di Domizio J, Flatz L, et al. Sting Activation of Tumor Endothelial Cells Initiates Spontaneous and Therapeutic Antitumor Immunity. Proc Natl Acad Sci USA (2015) 112(50):15408–13. doi: 10.1073/pnas.1512832112
131. Brognard J, Zhang YW, Puto LA, Hunter T. Cancer-Associated Loss-Of-Function Mutations Implicate Dapk3 as a Tumor-Suppressing Kinase. Cancer Res (2011) 71(8):3152–61. doi: 10.1158/0008-5472.CAN-10-3543
132. Kato Y, Park J, Takamatsu H, Konaka H, Aoki W, Aburaya S, et al. Apoptosis-Derived Membrane Vesicles Drive the Cgas-Sting Pathway and Enhance Type I Ifn Production in Systemic Lupus Erythematosus. Ann Rheum Dis (2018) 77(10):1507–15. doi: 10.1136/annrheumdis-2018-212988
133. Motwani M, McGowan J, Antonovitch J, Gao KM, Jiang Z, Sharma S, et al. Cgas-Sting Pathway Does Not Promote Autoimmunity in Murine Models of Sle. Front Immunol (2021) 12:605930. doi: 10.3389/fimmu.2021.605930
134. Fremond ML, Hadchouel A, Berteloot L, Melki I, Bresson V, Barnabei L, et al. Overview of Sting-Associated Vasculopathy With Onset in Infancy (Savi) Among 21 Patients. J Allergy Clin Immunol Pract (2021) 9(2):803–18.e11. doi: 10.1016/j.jaip.2020.11.007
135. Melki I, Rose Y, Uggenti C, Van Eyck L, Fremond ML, Kitabayashi N, et al. Disease-Associated Mutations Identify a Novel Region in Human Sting Necessary for the Control of Type I Interferon Signaling. J Allergy Clin Immunol (2017) 140(2):543–52.e5. doi: 10.1016/j.jaci.2016.10.031
136. Kwon J, Bakhoum SF. The Cytosolic DNA-Sensing Cgas-Sting Pathway in Cancer. Cancer Discovery (2020) 10(1):26–39. doi: 10.1158/2159-8290.CD-19-0761
137. Jiang M, Chen P, Wang L, Li W, Chen B, Liu Y, et al. Cgas-Sting, an Important Pathway in Cancer Immunotherapy. J Hematol Oncol (2020) 13(1):81. doi: 10.1186/s13045-020-00916-z
138. Liu H, Ottosen RN, Jennet KM, Svenningsen EB, Kristensen TF, Biltoft M, et al. Macrodiolide Diversification Reveals Broad Immunosuppressive Activity That Impairs the Cgas-Sting Pathway. Angew Chem Int Ed Engl (2021) 60(34):18734–41. doi: 10.1002/anie.202105793
139. Wu B, Xu MM, Fan C, Feng CL, Lu QK, Lu HM, et al. Sting Inhibitor Ameliorates Lps-Induced Ali by Preventing Vascular Endothelial Cells-Mediated Immune Cells Chemotaxis and Adhesion. Acta Pharmacol Sin (2021). doi: 10.1038/s41401-021-00813-2. Epub ahead of print.
140. Li S, Hong Z, Wang Z, Li F, Mei J, Huang L, et al. The Cyclopeptide Astin C Specifically Inhibits the Innate Immune Cdn Sensor Sting. Cell Rep (2018) 25(12):3405–21.e7. doi: 10.1016/j.celrep.2018.11.097
141. Siu T, Altman MD, Baltus GA, Childers M, Ellis JM, Gunaydin H, et al. Discovery of a Novel Cgamp Competitive Ligand of the Inactive Form of Sting. ACS Med Chem Lett (2019) 10(1):92–7. doi: 10.1021/acsmedchemlett.8b00466
142. Hong Z, Mei J, Li C, Bai G, Maimaiti M, Hu H, et al. Sting Inhibitors Target the Cyclic Dinucleotide Binding Pocket. Proc Natl Acad Sci USA (2021) 118(24):e2105465118. doi: 10.1073/pnas.2105465118
143. Park G, Kim SY, Song YJ. Ester Alkaloids From Cephalotaxus Interfere With the 2’3’-Cgamp-Induced Type I Interferon Pathway in Vitro. PloS One (2017) 12(8):e0182701. doi: 10.1371/journal.pone.0182701
144. Qi Y, Ying Y, Zou J, Fang Q, Yuan X, Cao Y, et al. Kaempferol Attenuated Cisplatin-Induced Cardiac Injury Via Inhibiting STING/NF-κB-Mediated Inflammation. Am J Transl Res (2020) 12(12):8007–18.
145. Humphries F, Shmuel-Galia L, Jiang Z, Wilson R, Landis P, Ng SL, et al. A Diamidobenzimidazole STING Agonist Protects Against SARS-CoV-2 Infection. Sci Immunol (2021) 6(59):eabi9002. doi: 10.1126/sciimmunol.abi9002
146. Ramanjulu JM, Pesiridis GS, Yang J, Concha N, Singhaus R, Zhang SY, et al. Design of Amidobenzimidazole Sting Receptor Agonists With Systemic Activity. Nature (2018) 564(7736):439–43. doi: 10.1038/s41586-018-0705-y
147. Zhou Z, Zhang X, Lei X, Xiao X, Jiao T, Ma R, et al. Sensing of Cytoplasmic Chromatin by Cgas Activates Innate Immune Response in Sars-Cov-2 Infection. Signal Transduct Target Ther (2021) 6(1):382. doi: 10.1038/s41392-021-00800-3
148. Xi Q, Wang M, Jia W, Yang M, Hu J, Jin J, et al. Design, Synthesis, and Biological Evaluation of Amidobenzimidazole Derivatives as Stimulator of Interferon Genes (Sting) Receptor Agonists. J Med Chem (2020) 63(1):260–82. doi: 10.1021/acs.jmedchem.9b01567
149. Basu S, Middya S, Banerjee M, Ghosh R, Pryde DC, Yadav DB, et al. The Discovery of Potent Small Molecule Cyclic Urea Activators of Sting. Eur J Med Chem (2022) 229:114087. doi: 10.1016/j.ejmech.2021.114087
150. Pryde DC, Middya S, Banerjee M, Shrivastava R, Basu S, Ghosh R, et al. The Discovery of Potent Small Molecule Activators of Human Sting. Eur J Med Chem (2021) 209:112869. doi: 10.1016/j.ejmech.2020.112869
151. Abraham J, Botto S, Mizuno N, Pryke K, Gall B, Boehm D, et al. Characterization of a Novel Compound That Stimulates Sting-Mediated Innate Immune Activity in an Allele-Specific Manner. Front Immunol (2020) 11:1430. doi: 10.3389/fimmu.2020.01430
152. Hou H, Yang R, Liu X, Wu X, Zhang S, Chen K, et al. Discovery of Triazoloquinoxaline as Novel Sting Agonists Via Structure-Based Virtual Screening. Bioorg Chem (2020) 100:103958. doi: 10.1016/j.bioorg.2020.103958
153. Kimura Y, Negishi H, Matsuda A, Endo N, Hangai S, Inoue A, et al. Novel Chemical Compound Sincro With Dual Function in Sting-Type I Interferon and Tumor Cell Death Pathways. Cancer Sci (2018) 109(9):2687–96. doi: 10.1111/cas.13726
154. Khiar S, Lucas-Hourani M, Nisole S, Smith N, Helynck O, Bourgine M, et al. Identification of a Small Molecule That Primes the Type I Interferon Response to Cytosolic DNA. Sci Rep (2017) 7(1):2561. doi: 10.1038/s41598-017-02776-z
155. Jung HR, Jo S, Jeon MJ, Lee H, Chu Y, Lee J, et al. Development of Small-Molecule STING Activators for Cancer Immunotherapy. Biomedicines (2021) 10(1):33. doi: 10.3390/biomedicines10010033
156. Liu Y, Tang Q, Rao Z, Fang Y, Jiang X, Liu W, et al. Inhibition of Herpes Simplex Virus 1 by Cepharanthine Via Promoting Cellular Autophagy Through Up-Regulation of Sting/Tbk1/P62 Pathway. Antiviral Res (2021) 193:105143. doi: 10.1016/j.antiviral.2021.105143
157. Hou S, Lan XJ, Li W, Yan XL, Chang JJ, Yang XH, et al. Design, Synthesis and Biological Evaluation of Acridone Analogues as Novel Sting Receptor Agonists. Bioorg Chem (2020) 95:103556. doi: 10.1016/j.bioorg.2019.103556
158. Cherney EC, Zhang L, Lo J, Huynh T, Wei D, Ahuja V, et al. Discovery of Non-Nucleotide Small-Molecule Sting Agonists Via Chemotype Hybridization. J Med Chem (2022) 65(4):3518–38. doi: 10.1021/acs.jmedchem.1c01986
159. Zhang X, Liu B, Tang L, Su Q, Hwang N, Sehgal M, et al. Discovery and Mechanistic Study of a Novel Human-Stimulator-Of-Interferon-Genes Agonist. ACS Infect Dis (2019) 5(7):1139–49. doi: 10.1021/acsinfecdis.9b00010
160. Amouzegar A, Chelvanambi M, Filderman JN, Storkus WJ, Luke JJ. STING Agonists as Cancer Therapeutics. Cancers (Basel) (2021) 13(11):2695. doi: 10.3390/cancers13112695
161. Sowers ML, Tang H, Tian B, Goldblum R, Midoro-Horiuti T, Zhang K. Bisphenol a Activates an Innate Viral Immune Response Pathway. J Proteome Res (2020) 19(2):644–54. doi: 10.1021/acs.jproteome.9b00548
162. Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of Ubiquitin Ligases and the Proteasome in Oncogenesis: Novel Targets for Anticancer Therapies. J Clin Oncol (2013) 31(9):1231–8. doi: 10.1200/JCO.2012.44.0958
163. Senft D, Qi J, Ronai ZA. Ubiquitin Ligases in Oncogenic Transformation and Cancer Therapy. Nat Rev Cancer (2018) 18(2):69–88. doi: 10.1038/nrc.2017.105
164. Rape M. Ubiquitylation at the Crossroads of Development and Disease. Nat Rev Mol Cell Biol (2018) 19(1):59–70. doi: 10.1038/nrm.2017.83
165. Smith BAH, Bertozzi CR. The Clinical Impact of Glycobiology: Targeting Selectins, Siglecs and Mammalian Glycans. Nat Rev Drug Discovery (2021) 20(3):217–43. doi: 10.1038/s41573-020-00093-1
166. Hua D, Wu X. Small-Molecule Inhibitors Targeting Small Ubiquitin-Like Modifier Pathway for the Treatment of Cancers and Other Diseases. Eur J Med Chem (2022) 233:114227. doi: 10.1016/j.ejmech.2022.114227
167. Cuello F, Wittig I, Lorenz K, Eaton P. Oxidation of Cardiac Myofilament Proteins: Priming for Dysfunction? Mol Aspects Med (2018) 63:47–58. doi: 10.1016/j.mam.2018.08.003
168. Zhang H, You QD, Xu XL. Targeting Stimulator of Interferon Genes (Sting): A Medicinal Chemistry Perspective. J Med Chem (2020) 63(8):3785–816. doi: 10.1021/acs.jmedchem.9b01039
Keywords: STING, post translational modification, targeted therapy, diseases, phosphorylation
Citation: Kang J, Wu J, Liu Q, Wu X, Zhao Y and Ren J (2022) Post-Translational Modifications of STING: A Potential Therapeutic Target. Front. Immunol. 13:888147. doi: 10.3389/fimmu.2022.888147
Received: 02 March 2022; Accepted: 11 April 2022;
Published: 06 May 2022.
Edited by:
Junji Xing, Houston Methodist Research Institute, United StatesReviewed by:
Nathalie Grandvaux, Université de Montréal, CanadaSinisa Savic, University of Leeds, United Kingdom
Copyright © 2022 Kang, Wu, Liu, Wu, Zhao and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Zhao, emhhb3l1bjA1NjIwMTlAMTYzLmNvbQ==; Jianan Ren, amlhbmFuckBnbWFpbC5jb20=; Xiuwen Wu, d3V4aXV3ZW5Abmp1LmVkdS5jbg==
 Jiaqi Kang
Jiaqi Kang Jie Wu
Jie Wu Qinjie Liu
Qinjie Liu Xiuwen Wu1*
Xiuwen Wu1* Jianan Ren
Jianan Ren