
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 20 April 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.882796
This article is part of the Research Topic Antiviral Innate Immune Sensing, Regulation, and Viral Immune Evasion View all 48 articles
 Yaru Ning1,2,3†
Yaru Ning1,2,3† Yalin Huang1,2,3†
Yalin Huang1,2,3† Mingshu Wang1,2,3†
Mingshu Wang1,2,3† Anchun Cheng1,2,3*
Anchun Cheng1,2,3* Renyong Jia1,2,3
Renyong Jia1,2,3 Mafeng Liu1,2,3
Mafeng Liu1,2,3 Dekang Zhu2,3
Dekang Zhu2,3 Shun Chen1,2,3
Shun Chen1,2,3 Xinxin Zhao1,2,3
Xinxin Zhao1,2,3 Shaqiu Zhang1,2,3
Shaqiu Zhang1,2,3 Qiao Yang1,2,3
Qiao Yang1,2,3 Ying Wu1,2,3
Ying Wu1,2,3 Juan Huang1,2,3
Juan Huang1,2,3 Bin Tian1,3
Bin Tian1,3 Xumin Ou1,2,3
Xumin Ou1,2,3 Sai Mao1,2,3
Sai Mao1,2,3 Qun Gao1,2,3
Qun Gao1,2,3 Di Sun1,2,3
Di Sun1,2,3 Yanlin Yu1,2,3
Yanlin Yu1,2,3 Ling Zhang1,2,3
Ling Zhang1,2,3Duck plague (DP) is an acute infectious disease in the duck industry. The duck plague virus (DPV) is the pathogen, a subfamily of alphaherpesvirinae. gE is a type I membrane protein that contains three parts: an extracellular domain, a transmembrane domain, and a cytoplasmic domain. gE is the major virulence determinant of α-herpesvirus. However, the functions of the gE extracellular and cytoplasmic domains have not been reported in DPV. In this study, a gE extracellular domain deletion mutant and a gE cytoplasmic domain deletion mutant were constructed from DPV. Virus replication kinetics showed that the growth titers of both the gE ectodomain-deleted mutant virus and the gE cytoplasmic domain-deleted virus in DEFs were lower than that of the parental virus CHv-50. DPV CHv-gEΔET and DPV CHv-gEΔCT were continuously passed to the 20th passage in DEFs and the 10th in ducklings. The mutant virus DNA after passage was extracted for identification. The results showed that the gE ectodomain and gE cytoplasmic domain deletion mutant viruses have good genetic stability. The ducklings in each group (n=10) were inoculated with the same titers of DPV CHv-gEΔET, DPV CHv-gEΔCT, DPV CHv-ΔgE, and parental CHv-50, respectively. Clinical symptoms and serum antibody levels were detected after inoculation. The results showed that the virulence of DPV CHv-gEΔCT to ducklings was reduced compared with parental CHv-50, while the virulence of DPV CHv-gEΔET to ducklings was significantly reduced. 105 TCID50 DPV CHv-gEΔET or DPV CHv-ΔgE can induce ducklings to produce DPV-specific antibodies, protect the ducklings from virulent CHv challenge. Therefore, DPV CHv-gEΔET may serve as a promising vaccine candidate to prevent and control duck plague.
Duck plague (DP) is an acute disease with a high fatality rate caused by DPV, which has caused significant economic losses to the duck industry around the world and has shown a trend of younger age in recent years (1–3). Studies have shown that duck herd immunization with the attenuated vaccine can prevent the occurrence of duck plague, but after immunization, it is impossible to distinguish whether it is wild virus infection or vaccine immunization produces antibodies, resulting in the lack of strong physicochemical technical support for the technical plan to purify DPV infection through immunization (4). Therefore, there is an urgent need to develop a more effective vaccine to eradicate DPV. It has been reported that many countries have successfully eradicated pseudorabies through the gene deletion DIVA (differentiating infected from vaccinated animals) vaccine. Like PRV, DPV is also a member of the alphaherpesviruses (5–7). Therefore, the emerging DPV gene-deleted attenuated mutant virus is promising as a candidate vaccine for the control and eradication of duck plague.
DPV is a member of the alphaherpesvirus subfamily, with a double-stranded DNA genome of approximately 162 kb and a capsid, tegument, and envelope. The gE protein in DPV is encoded by the US8 gene and has the characteristics of a typical type I membrane protein. The polypeptide chain transmembrane once, the N-terminus is outside the cell membrane, and the C-terminus is inside the cell membrane. Therefore, according to the transmembrane structure of gE protein, we divided it into three parts: extracellular domain, transmembrane domain and intracellular domain. gE is a non-essential structural protein that facilitates the spread of viruses from cell to cell, under certain circumstances, promotes anterograde transport of latent virions after reactivation, and is neurotoxic (8). The viral plaques of the mutant virus BAC-CHv-ΔgE on DEFs were about 60% smaller than that of the wild virus BAC-CHv (9). Electron microscopy results showed that the deletion of DPV gE caused a large number of capsids to accumulate around the vesicles, and only a few were able to bud into vesicles, which is consistent with reports in other herpesviruses such as HSV-1, HSV-2, VZV, PRV, suggesting that gE plays an important role in virion morphogenesis prior to final cytoplasmic nucleocapsid wrapping (10, 11). The gE CT is involved in the second coating of the nucleocapsid in specific parts of the Golgi apparatus, selectively distributing nascent virions to cell junctions and promoting the spread of viruses between cells. Viruses with deletions in different regions of gE CT were constructed on HSV-1 gD gene deletion strains, and it was found that after deletion of amino acids 470-495, a large number of nucleocapsids accumulated in the cytoplasm (12). DPV gE CT interacts with UL11. In the absence of gE CT, the amount of UL11 packaged into virions is reduced by 58.1 ~ 80%, and the nucleocapsid cannot complete the secondary coating to form complete virions, which inhibits the release of the virus (13, 14). gE ET mainly plays a role in the intercellular propagation between epithelial cells and polar cells such as neural tissue. Through the receptor mechanism, gE ET localizes the gE/gI complex at the extracellular junction and binds to the adjacent cell receptor, promoting the fusion of infected cells and adjacent non-infected cells. The amino acids 277, 291, and 348 of HSV gE ET were mutated to construct three mutant strains. Small plaques were formed after infecting cells, similar to the gE gene deletion strain, and the transmission of the virus from the cornea to the epithelial tissue was restricted (15). The deletion of amino acids 208-236 in the cysteine region of VZV gE ET changed the distribution of gE on the cell membrane and affected the spread of the virus between cells (16).
Notably, DPV can replicate and persist at high levels in duck tissues (17), which is associated with the ability of the virus to evade host immune defenses. gE forms a dimer with gI and participates in the immune evasion function of the virus to enhance the virulence of the virus. It is a good vaccine target protein. PRV gE has been reported to inhibit cGAS/STING-mediated IFN-β production by degrading CBP to disrupt the enhanced assembly of IRF3 and CBP (18). HSV gE binds to the Fc segment of IgG, which spatially prevents IgG or Fc-dependent effector cells from approaching the virus or virus-infected cells. Complement Clq cannot bind to the IgG Fc site, blocking the classical pathway of complement (19, 20). Protects viruses from immune processes such as antibody-dependent cytotoxicity and antibody-dependent cell-mediated phagocytosis (21–23). gE can also interact with prohibitin-1, which is conserved in all herpesviruses, for cell-to-cell transmission through the MAPK/ERK pathway (24). However, the molecular mechanism by which DPV gE plays a role in cell-to-cell transmission, evading the immune responses, and enhancing viral virulence has not been fully elucidated. The reported virulence genes mainly express non-essential envelope glycoproteins or nonstructural proteins. For example, the main virulence genes of PRV include gB, gC, TK, US3 (25–27). Therefore, studying the function of DPV gE from histopathology and in vivo colonization is crucial for the prevention and treatment of DPV infection, and there is no report on the key regions of gE virulence genes in DPV.
This study constructed CHv-gEΔET and CHv-gEΔCT deletion mutant viruses using bacterial artificial chromosome cloning of the DPV CHv-BAC-G strain. The efficacy of the mutant virus as a candidate vaccine for the control or eradication of duck plague in duck flocks was evaluated, and the safety and immunogenicity of the mutant virus were evaluated.
9-day-old Cherry Valley duck embryos and 7-day-old healthy Cherry Valley ducks were purchased from a farm operated by Sichuan Agricultural University (Sichuan, China). All experimental ducks did not contain DPV and were negative for DPV antibodies (28). The experimental animal protocol was approved by the Ethics and Animal Welfare Committee of Sichuan Agricultural University and carried out following the Chinese version of the Guide for the Care and Use of Laboratory Animals.
Parent virus CHv-50 (GenBank: JQ647509.1) (3) and DPV CHv-ΔgE deletion mutant virus were provided by our laboratory. DEFs were prepared from 9-day-old Cherry Valley duck embryos in Modified Eagle’s Medium (MEM, Gibco, Rockford, USA) supplemented with 10% neonatal bovine serum (NBS, Gibco, Rockford, USA) and 1% antibiotics (penicillin and streptavidin), cultured at 37°C in a 5% CO2 incubator.
The DPV CHv-BAC-GS1783 (29) strain is stocked in our lab, in which the entire DPV CHv genome tagged with an enhanced green fluorescent protein (EGFP) is inserted BAC for stable propagation in E. coli strain GS1783. The construction of DPV CHv-gEΔET was based on two markerless two-steps Red recombination (30). Briefly, the linear PCR product, ‘a-I-Sce I-Kan-a-b’ was amplified and electroporated into DPV CHv-BAC-GS1783 to induce the first step of Red recombination, thereby replacing the gE-ET gene with Kan, a and b are the 40 bp homology arms on the left and right sides of the gE-ET gene, respectively. Subsequently, the I-Sce I site is cleaved by the I-Sce I endonuclease, and the Kan is removed in the second step by Red recombination. Amplify the linear PCR product again, ‘a-’UL23-a’-’UL23-b’-b-I-Sce I-Kan-b-c’, electroporate it into DPV CHv-BAC-gEΔET-GS1783, induce the first step of Red recombination, a, b and c are successive homology arms downstream of the miniF sequence. Subsequently, the I-Sce I site is cleaved by the I-Sce I endonuclease, and the Kan is removed in the second step by Red recombination. The deletion of gE-ET was confirmed by identifying primers and sequencing. The plasmid DPV CHv-gEΔET-GS1783 was transfected into DEFs, and purified by spotting to obtain the DPV CHv-gEΔET deletion mutant virus. The same procedure was performed to construct and obtain DPV CHv-gEΔCT deletion mutant virus. All primers used in this study are listed in Table 1.
After DPV CHv-ΔgE, DPV CHv-gEΔET, and DPV CHv-gEΔCT deletion mutant viruses infected DEFs to produce 80% lesions, the samples were frozen and thawed three times, and the obtained viruses were identified by PCR and Western blot. In PCR identification, primer UL30-F/R is used to identify whether the DPV UL30 gene is contained. Primers BAC-F/R are used to identify whether it contains miniF element, primer UL23-F/R is used to identify whether continuous UL23 gene is contained, Primers gE-F/R were used to identify deletions of the gE gene. DEFs were infected with DPV CHv-ΔgE, DPV CHv-gEΔET, and DPV CHv-gEΔCT deletion mutant viruses with an MOI of 0.1 to analyze the gE expression in deletion mutant viruses, and 48 h later, cells were lysed with RIPA lysate, and the protein lysate was collected. Add 1% PMSF to the lysis buffer. Western blot analysis was performed with rabbit anti-gE and goat anti-rabbit IgG antibodies.
Multistep growth kinetics of the parental strain CHv-50, DPV CHv-gEΔET, and DPV CHv-gEΔCT deletion mutant viruses were performed as previously described with minor modifications (31, 32). Briefly, DEFs cultured in 24-well cell culture plates were infected with a virus at an MOI of 0.01. After the virus was adsorbed for 2 h at 37°C and 5% CO2, the medium was discarded, the cells were washed with PBS (pH 7.4), and then the culture medium was replaced with MEM containing 2% NBS. The infected cells were collected at 12, 24, 48, and 72 h after infection, the volume of each sample was increased to 500 μL with MEM, the samples were freeze-thawed three times, and virus titers were determined by TCID50 on DEFs. All experiments were repeated 3 times.
DEFs were separately infected with the DPV CHv-ΔgE, DPV CHv-gEΔET and DPV CHv-gEΔCT deletion mutant viruses. After the cells appeared 80% lesions, the samples were frozen and thawed 3 times, and the first-generation viruses were collected. The virus was re-infected with DEFs to prepare the next generation of the virus, and the deletion mutant virus was uploaded to the 20th passage in DEFs according to this method. DNA was extracted from the 10th and 20th passages of each virus according to the instructions of the Magen kit, and PCR identification was performed. The primers used are listed in Table 1. Virus titers at passages 1, 5, 10, 15, and 20 for each virus were determined by TCID50 on DEFs. All experiments were repeated 3 times.
Forty 14-day-old ducklings were divided into 4 groups, the first 3 groups were inoculated with DPV CHv-ΔgE, DPV CHv-gEΔET, DPV CHv-gEΔCT deletion mutant virus by intramuscular injection at a dose of 106 TCID50/dose, and the last group was intramuscularly inoculated with the same volume of MEM. Seven days after inoculation, 5 ducklings in each group were randomly selected for culling, and the pathological changes of liver, spleen, and duodenum were observed. Take 1 g of liver tissue from each group for grinding, add normal saline at a ratio of 1:9 to make a homogenate, and inoculate new 14-day-old ducklings again. In this way, the mutant virus is transmitted to the 10th generation in ducklings. DNA was extracted from the 1st, 5th, and 10th passages of each virus according to the instructions of the Magen kit, and PCR identification was performed. The primers used are listed in Table 1. TaqMan-MGB probe fluorescence real-time quantitative PCR method (33) was used to detect the viral copies in liver tissue at passages 1, 5, and 10 of each virus.
130 14-day-old ducklings were divided into 13 groups. Groups 1, 2, and 3 were inoculated with CHv-50 by intramuscular injection at 104 TCID50/dose, 105 TCID50/dose, and 106 TCID50/dose, respectively. Groups 4, 5, and 6 were inoculated with DPV CHv-ΔgE by intramuscular injection at 104 TCID50/dose, 105 TCID50/dose, and 106 TCID50/dose, respectively. Groups 7, 8, and 9 were inoculated with DPV CHv-gEΔET by intramuscular injection at 104 TCID50/dose, 105 TCID50/dose, and 106 TCID50/dose, respectively. Groups 10, 11, and 12 were inoculated with DPV CHv-gEΔCT by intramuscular injection of 104 TCID50/dose, 105 TCID50/dose, and 106 TCID50/dose, respectively. The control group was intramuscularly inoculated with the same volume of MEM. Five ducks were randomly selected from each group to measure the rectal body temperature every day, and the death of each group of ducks was recorded for 10 days.
40 14-day-old ducklings were divided into 4 groups, the first group was inoculated with a dose (107.7 copies)/dose of live duck plague vaccine CVCC AV1222, and the second and third groups were inoculated with the same number of copies/dose of DPV CHv-ΔgE and DPV CHv-gEΔET deletion mutant virus, the fourth group was the control group, each inoculated with 1 mL of MEM. On the 14th day after immunization, the ducklings were injected with 100 LD50/duck virulent Chinese strains of duck plague virus (CHv) and were continuously observed for 10 days after the challenge, and the clinical symptoms and death were recorded every day.
Similar to the above grouping and inoculation, blood was collected from each group of ducklings on the 7th, 14th, 21th, and 28th days after immunization, and serum was collected for neutralizing antibody detection. The serum to be tested was filtered with a 0.22 filter and inactivated at 56°C for 30 min. The treated serum to be tested was diluted 21 ~ 28 times, and 50 µL per well was added to a 96-well plate with DEFs. Add 50 µL of 1000 TCID50 of CHv to each well, incubate at 37°C for 2 h, discard the supernatant and add 100 µL of MEM with 2% NBS. Continue to culture to observe cytopathic changes and calculate the neutralization index according to the Read-Muench method. All experiments were repeated 3 times.
The data are expressed as the means ± S.D. Statistical analysis was performed with Student’s t-test (GraphPad Prism 6); *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 indicate statistical significance compared with the control.
To explore the virulence gene functions of the ET and CT of gE, in DPV CHv-BAC-GS1783, we used two-step homologous recombination, the first homologous recombination first deleted gE-ET (Figure 1A), and constructed a DPV CHv-BAC-gEΔET-GS1783, the miniF was deleted by the second homologous recombination to construct DPV CHv-gEΔET-GS1783 (Figure 1B). DPV CHv-gEΔCT-GS1783 was constructed in the same way as described in Materials and methods. The plasmids of DPV CHv-gEΔET-GS1783 and DPV CHv-gEΔCT-GS1783 were respectively extracted and transfected into DEFs. At 48 h after transfection, small green fluorescent spots were observed in DEFs. After 144 h, green fluorescent spots were distributed in the entire field of view, and the cells produced lesions (Figure 2A). After collecting the virus solution to infect DEFs, pick the virus solution with cytopathic but no fluorescence to re-infect DEFs, resulting in stable DPV CHv-gEΔET and DPV CHv-gEΔCT deletion mutations with cytopathic and no fluorescence virus (Figure 2B).
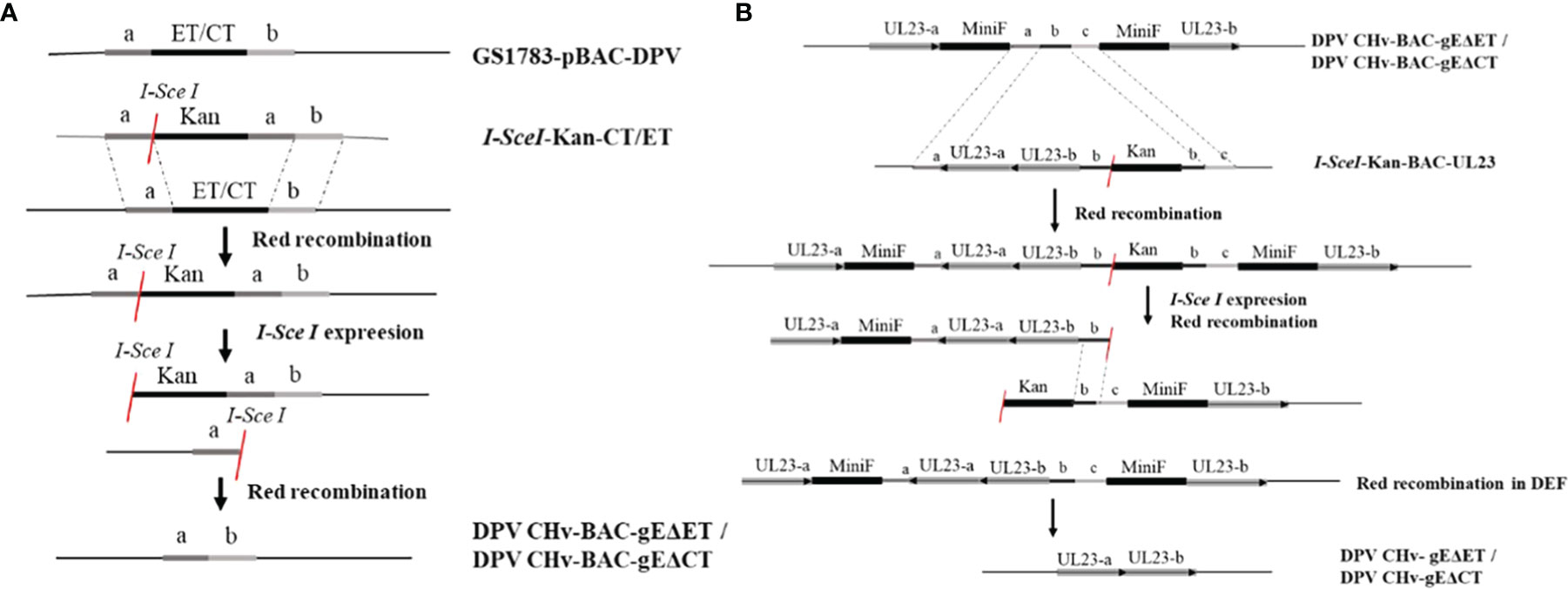
Figure 1 Homologous recombination diagram. Schematic diagram of the construction of the deletion mutant virus CHv-gEΔET and CHv-gEΔCT using the Red recombination system. (A) Schematic diagram of the construction of the deletion mutant virus CHv-BAC-gEΔET and CHv-BAC-EΔCT using the Red recombination system. (B) Schematic diagram of the construction of the deletion mutant virus CHv-gEΔET and CHvg EΔCT using the Red recombination system again.
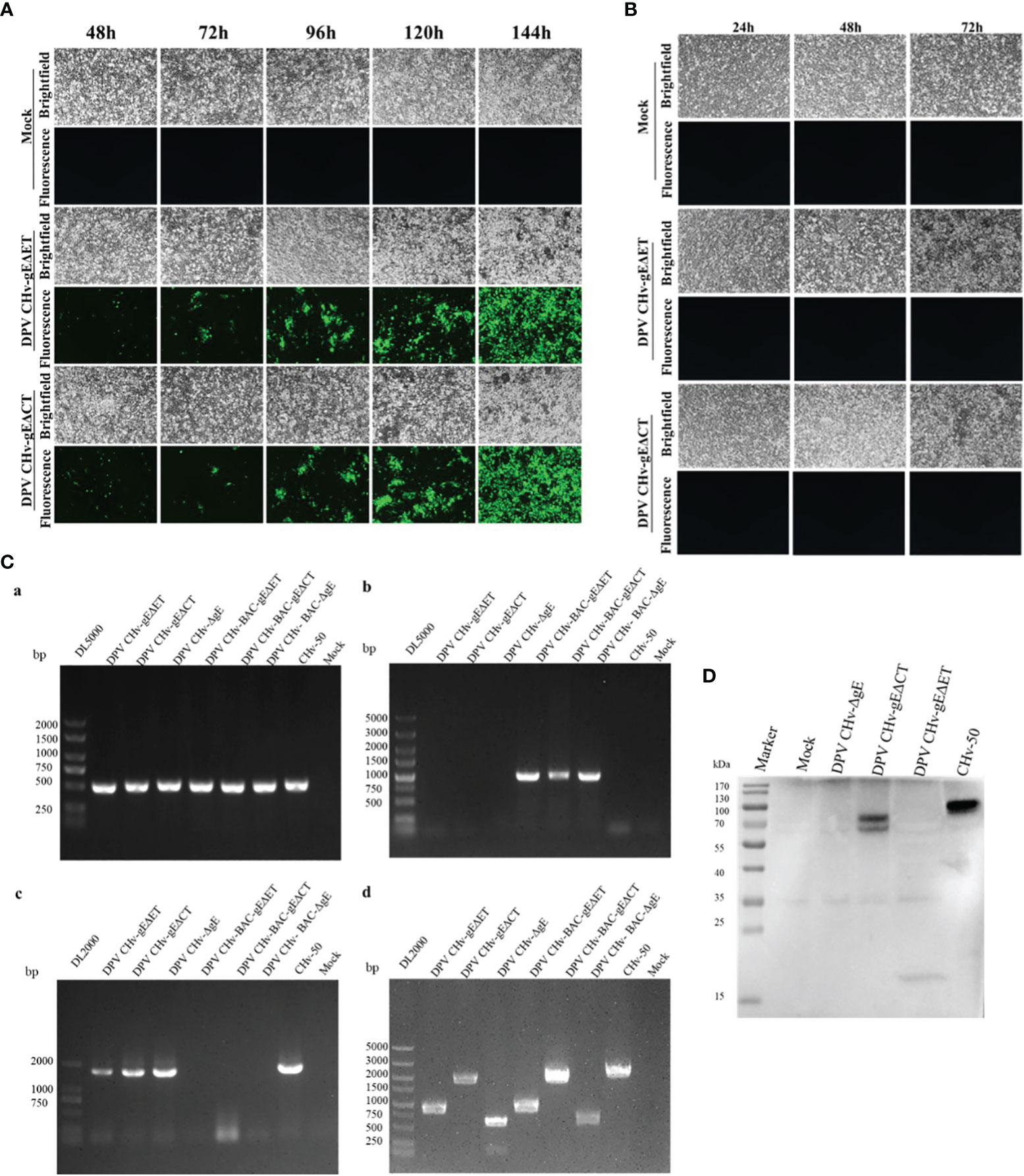
Figure 2 Generation and identification of mutant viruses for DPV CHv-gEΔET and DPV CHv-gEΔCT. (A) Transfection of the plasmids DPV CHv-gEΔET-GS1783/DPV CHv-gEΔCT-GS1783 into DEFs resulted in numerous fluorescent spots and cytopathies, the mutant virus DPV CHv-gEΔET/DPV CHv-gEΔCT were rescued. (B) Cells with cytopathic but no fluorescent spots were picked into new DEFs, resulting in the non-fluorescent mutant virus. (C) PCR identification of mutant viruses: (a) UL30 gene identification primer, (b) BAC identification primer, (c) UL23 gene identification primer, (d) gE gene deletion identification primer. (D) DEFs were infected with DPV CHv-ΔgE, DPV CHv-gEΔCT, DPV CHv-gEΔET, and DPV CHv-50, and an anti-gE polyclonal antibody was used for WB.
To identify the non-fluorescent deletion mutant virus after purification, viral DNA was extracted from DEFs infected with DPV CHv-gEΔET, DPV CHv-gEΔCT, DPV CHv-ΔgE, and CHv-50 viruses, and PCR identification analysis were performed. Using the UL30 gene identification primers can amplify the UL30 gene fragment from the above DNA, indicating that the purified deletion mutant virus is duck plague virus, using the BAC identification primers cannot amplify the miniF element from the above DNA, indicating that all the purified deletion mutant virus has completely removed the miniF element, the UL23 gene fragment can be amplified from the above DNA using the UL23 gene identification primer, indicating that the purified deletion mutant virus UL23 gene has been restored, amplification fragment of 884 bp, 1859 bp, 599 bp and 2072 bp from DNA infected with DPV CHv-gEΔET, DPV CHv-gEΔCT, DPV CHv-ΔgE and CHv-50 using gE gene deletion identification primers, respectively, indicating that the ET segment and CT segment of the gE gene have been correctly deleted, the sizes of the above target fragments were in line with expectations. DPV CHv-ΔgE and CHv-50 were used as negative and positive controls, respectively, and the Mock group was used as a control to demonstrate the specificity of the primers (Figure 2C).
The expression of gE protein in the deleted mutant virus was further analyzed by Western blot. As shown in Figure 2D, only CHv-50 can express the complete gE protein, about 20 kDa of gE protein in DPV CHv-gEΔET, DPV CHv-gEΔCT of gE protein is about 70 kDa, DPV CHv-ΔgE does not express gE protein, the results are in line with expectations, indicating that both DPV CHv-gEΔET and DPV CHv-gEΔCT recombinant viruses can be used in subsequent experiments.
The multi-step growth kinetics of DPV CHv-gEΔET and DPV CHv-gEΔCT deletion mutant viruses were evaluated after infecting DEFs with the same MOI = 0.01, collecting virus fluids at different time points, and detecting virus titers. As shown in Figure 3, the incubated virus entered the cell and started virus replication. At the initial 12 h of infection, the intracellular virus titers of the two deletion mutant viruses and the parental virus were not detected, indicating that the infectivity virus particles had not yet formed at this time. At 24 h after infection, infectious virions could be detected, indicating that a new generation of virions had been replicated in the cells. Then the virus titer gradually increased and reached a peak at 72 h, during which time the three virus strains were in the stage of replication and release. At 72 ~ 96 h after infection, the virus titers of the three virus strains all showed a downward trend. At this time, the virus titers decreased due to severe cytopathic changes. Compared with the parental virus, the viral titers of DPV CHv-gEΔET and DPV CHv-gEΔCT decreased by approximately 80 and 25 fold, respectively, at 72 h of infection. These results suggest that deletion of both gE-ET and gE-CT affects the proliferative capacity of the virus.
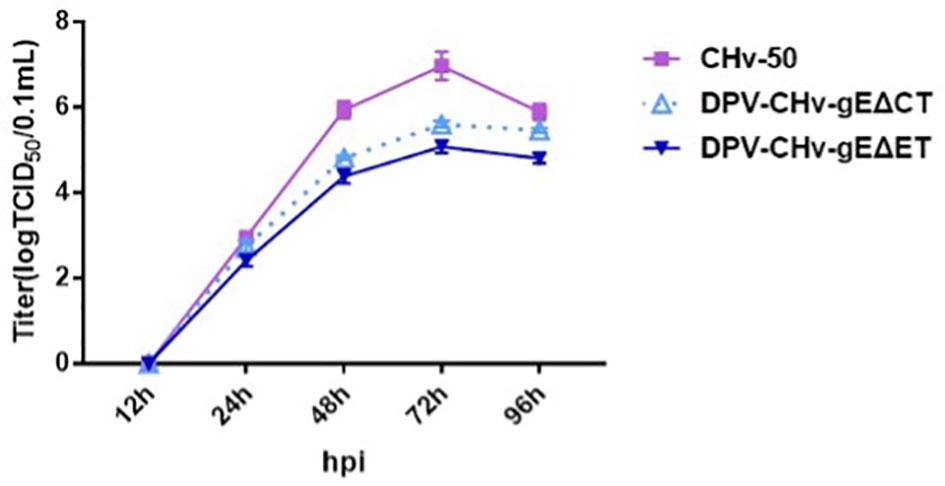
Figure 3 Determination of viral titers in growth kinetics. DEFs in 24-well plates were infected with 0.01 MOI of DPV CHv-gEΔCT, DPV CHv-gEΔET, and DPV CHv-50. Samples were collected at the indicated time points, and viral titers were determined. The data were presented as the mean ± standard deviation(SD) of three independent experiments.
To evaluate the genetic stability of deletion mutant viruses in vitro, the viral titers of DPV CHv-gEΔET and DPV CHv-gEΔCT deletion mutant viruses at passages 1, 5, 10, 15, and 20 after infection in DEFs were detected. As shown in Figure 4A, the titers of deletion mutant viruses at different passages were all around 105 TCID50/0.1 mL, with no significant difference (P>0.05). The virus titers of deletion mutant viruses were stable after serial passage on DEFs. DNA of passage 10 and 20 deletion mutant viruses were extracted and identified by PCR as described in Methods. As shown in Figure 4B, the UL30 gene fragments from the DPV CHv-gEΔET and DPV CHv-gEΔCT mutant viral DNAs after serial passages were identified by the UL30 primers. The target gene fragments amplified from the DPV CHv-gEΔET and DPV CHv-gEΔCT mutant viral DNAs after gE deletion primers identified serial passages, and the sizes were in line with expectations. The above results show that after the serial passage of DPV CHv-gEΔET and DPV CHv-gEΔCT mutant viruses in DEFs, the deleted genes in the viral genome will not undergo reverse mutation and stably inherited in vitro.
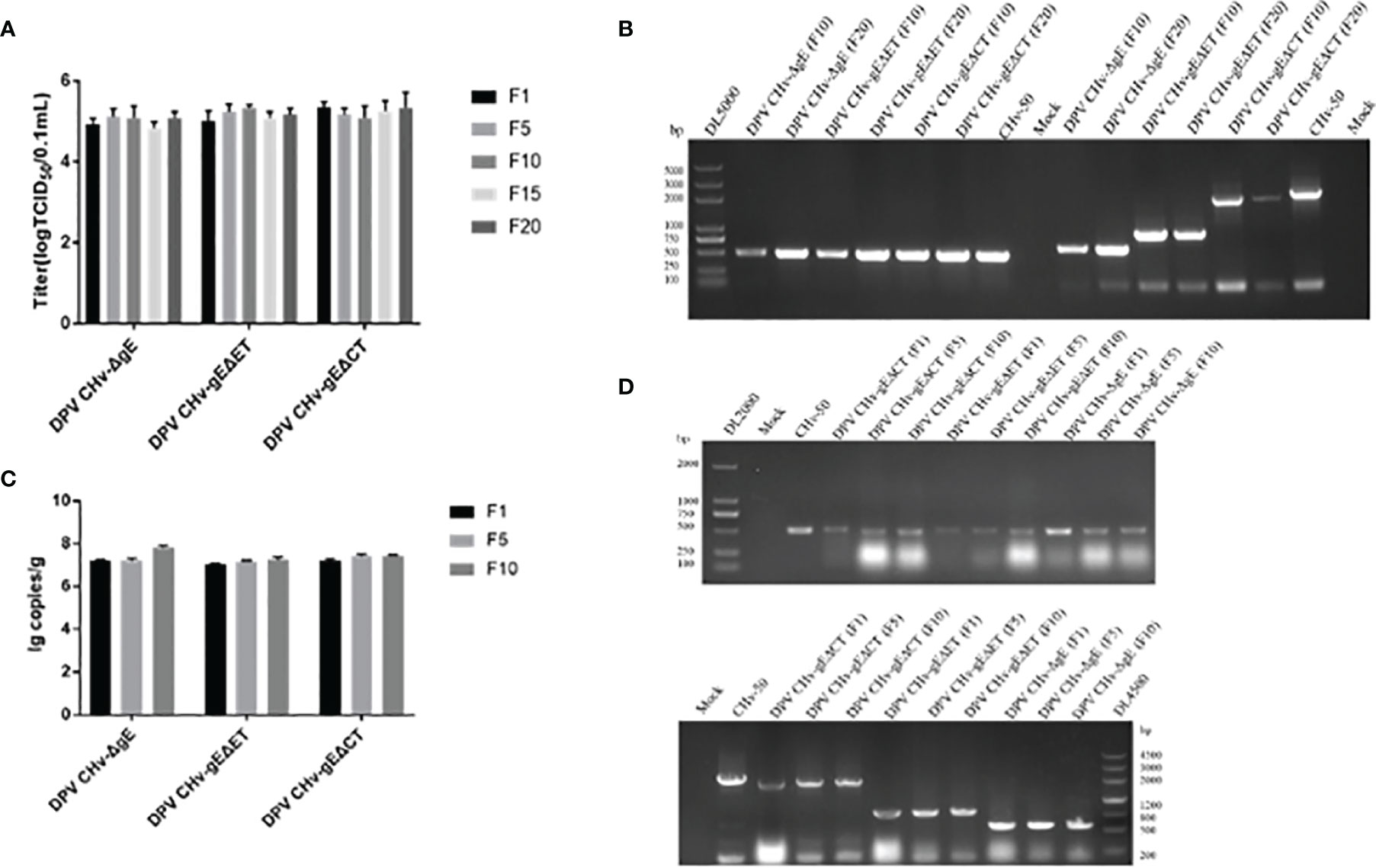
Figure 4 (A) Mutant viruses at passages 1st, 5th, 10th, 15th, and 20th were collected to infect DEFs at a 1 MOI. The cell samples were collected for virus copy number detection, P>0.05. (B) The DNA of the 10th and 20th generation mutant viruses was extracted from DEFs, respectively, and then the primers for UL30 gene identification and gE gene identification primers were used for PCR identification. (C) Mutant viruses at passages 1st, 5th, and 10th were collected to infect ducklings at a 1 MOI. After 7 days of infection, the ducklings were slaughtered, 1 g of liver tissue was taken out, and the viral genome in the liver tissue was extracted for copy number determination, P>0.05. (D) DNAs of the 1st, 5th, and 10th generation mutant viruses were extracted from liver tissue, respectively, and PCR identification was performed with UL30 gene identification primers and gE gene identification primers, respectively.
To assess the genetic stability of deletion mutant viruses in vivo, the mortality of DPV CHv-gEΔET and DPV CHv-gEΔCT deletion mutant virus in ducklings after serial passage was detected, and the disease and virus infection in different organs were detected. As shown in Table 2, after the DPV CHv-gEΔCT group was infected with ducklings, only a small number of deaths occurred in the 2nd, 5th, and 6th generations, while the DPV CHv-gEΔET group and DPV CHv-ΔgE group were the same as the Mock group, no deaths occurred. The liver, spleen, and duodenum of ducklings infected with different generations of mutant deletion virus were necropsied to observe the lesions. After the ducklings were infected with DPV CHv-gEΔCT, only the duodenal mucosa showed slight hemorrhage, and other organs showed slight hemorrhage. No lesions were observed in all organs in the DPV CHv-gEΔET group and the DPV CHv-ΔgE group, as in the Mock group (Figure 5). The viral DNA in the liver of ducks infected with 1, 5, and 10 generations of deletion mutant virus was extracted, and PCR identification and quantitative detection of the virus content were carried out. As with the in vitro detection, all the target bands were in line with expectations, and the virus in the liver of different generations mutant virus loads was stable between 107 ~ 108 copies/g (Figures 4C, D). The above results show that after the DPV CHv-gEΔET and DPV CHv-gEΔCT mutant viruses are serially passaged in ducklings, the deleted genes in the viral genome will not undergo reverse mutation and can be stably inherited in vivo.
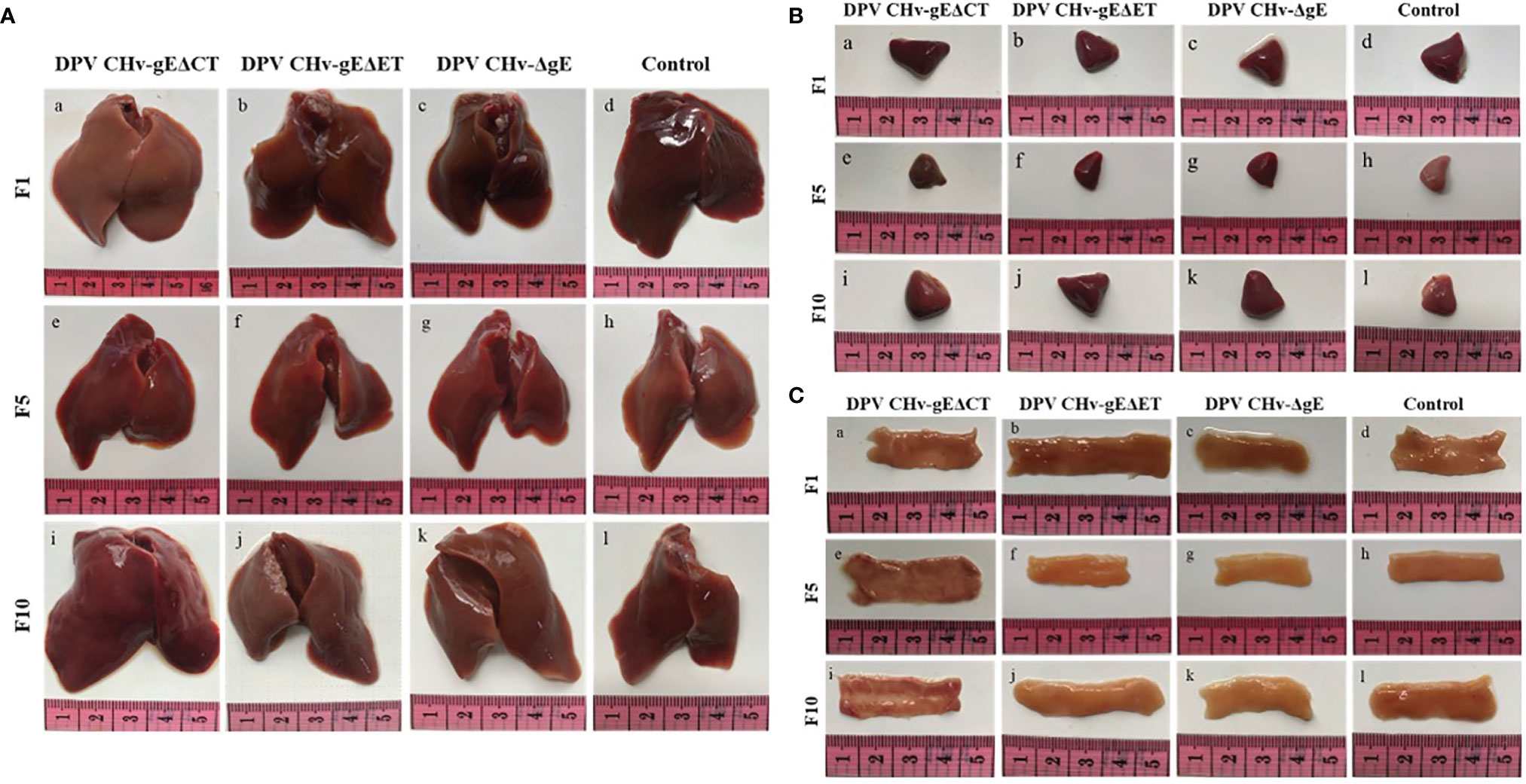
Figure 5 Pathological lesions of ducklings after challenge with the mutant virus at passages 1, 5, and 10. (A) Liver; (B) Spleen; (C) Duodenum.
The body temperature of most ducklings inoculated with parental virus CHv-50 will rise to above 43°C. The body temperature of ducklings in the 104 TCID50 groups reached the peak on the 5th day, and the body temperature of the ducklings in the 105 TCID50 and 106 TCID50 groups both reached the peak on the 4th day. After reaching the peak, it will drop to the normal body temperature range of 40.5 ~ 42.5°C (Figure 6). 6 ~ 7 days is the peak period of death, and the mortality of different inoculation dose groups is distributed in a gradient. The ducklings in the 104 TCID50 group did not die, and the ducklings in the 105 TCID50 group had a mortality rate of 50%, and 1, 2, 2, died on days 5, 6, and 7, respectively. Ducklings in the106 TCID50 group had a mortality rate of 60%, 1, 1, 2, and 2 died on days 4, 5, 6, and 7, respectively (Figure 6). The DPV CHv-gEΔET group, DPV CHv-gEΔCT group, DPV CHv-ΔgE group were the same as the MEM group, the body temperature of the different dose groups after inoculation of the ducklings always fluctuated within the normal range within 10 days. Only DPV CHv -gEΔCT group died at 105 TCID50 and 106 TCID50 doses, and the mortality rate was 20% (Table 3). No deaths occurred within 10 days in the DPV CHv-gEΔET group, DPV CHv-gEΔCT group, DPV CHv-ΔgE group, and MEM group. The above results indicated that both ET and CT lacking gE would reduce the pathogenicity of the virus to ducklings, while the pathogenicity of the ET virus lacking gE was more significantly reduced to ducklings.
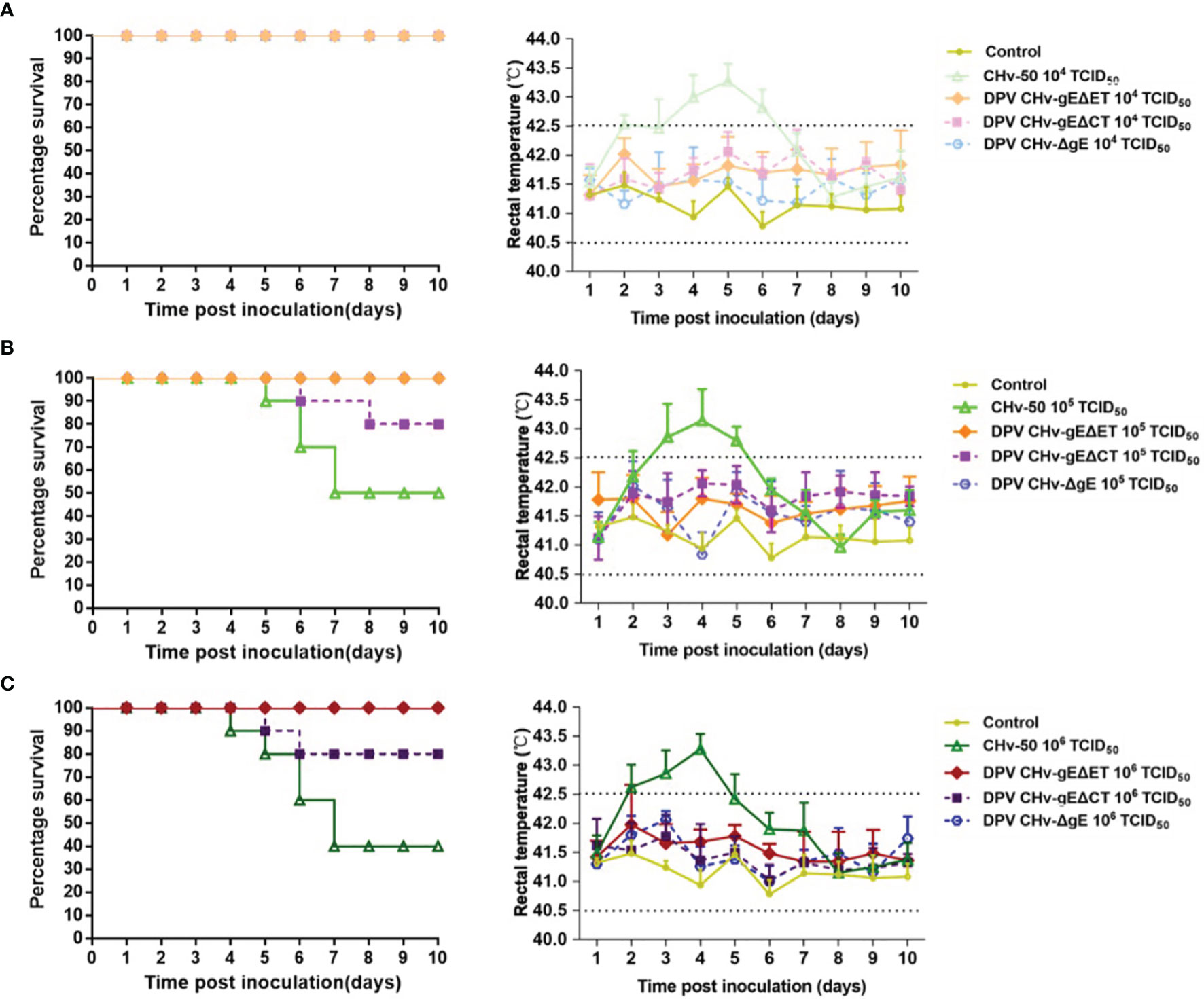
Figure 6 Survival percentage (left) and rectal temperatures (right) of ducklings. (A) Inoculated with a dose of 104 TCID50 of mutant viruses; (B) Inoculated with a dose of 105 TCID50 of mutant viruses; (C) Inoculated with a dose of 106 TCID50 of mutant viruses.
The clinical symptoms of the ducklings in different immunization groups after the challenge were observed. Only the ducklings in the control group began to be lethargic, poor in appetite, tearing, and the feathers around the eyelids formed eye circles on the 4th day after the challenge. As shown in Figure 7, the body temperature of the ducklings in different immunization groups was measured after the challenge, the body temperature of the control group increased, and the body temperature exceeded 42.5°C on the 4th day after the challenge, and then the body temperature continued to be high until all died. The body temperature of the DPV CHv-ΔgE group and DPV CHv-gEΔET group was slightly higher than that of the vaccine group, but both fluctuated within the normal range. Statistics on the death of ducklings in different immunization groups after challenge, the control group began to die on the 5th day, and the 7th day was the peak period of death, and all died within 7 days, while the DPV CHv-ΔgE group, DPV CHv-gEΔET group like the vaccine group, all the ducklings were survived, indicating that the ducklings immunized with DPV CHv-ΔgE and DPV CHv-gEΔET could resist the challenge of virulent DPV CHv by 100%.
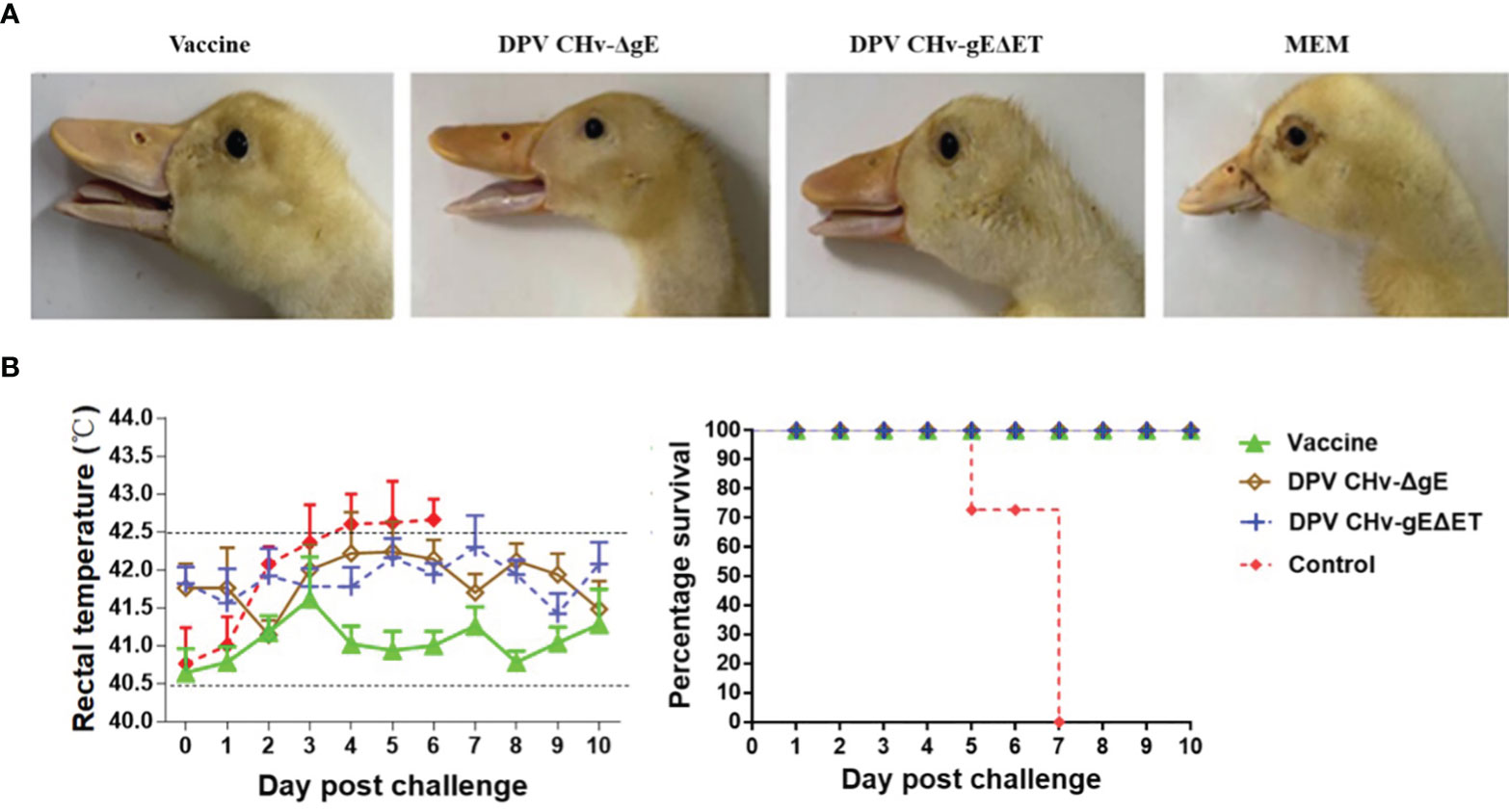
Figure 7 The ducklings were challenged after immunization, and the clinical symptoms of the ducklings after the challenge were observed. (A) Eyelid condition. (B) Rectal temperature (left) and survival rate (right).
Neutralizing antibodies were detected in the sera of ducklings in different immunization groups. Neutralizing antibodies could be detected in the DPV CHv-ΔgE group, DPV CHv-gEΔET group, and vaccine group on the 7th day after immunization, and then the antibody levels gradually increased. On the 28th day after immunization, the neutralizing titers in the sera of ducklings in different immunization groups DPV CHv-ΔgE group, DPV CHv-gEΔET group, and vaccine group reached 24.8, 24.4, and 25.6 (Figure 8), respectively, there was no significant difference in valence (P>0.05), but both were extremely significantly higher than those in the control group (P<0.001, P<0.0001), indicating that DPV CHv-ΔgE and DPV CHv-gEΔET could stimulate ducklings to produce significant neutralizing antibodies.
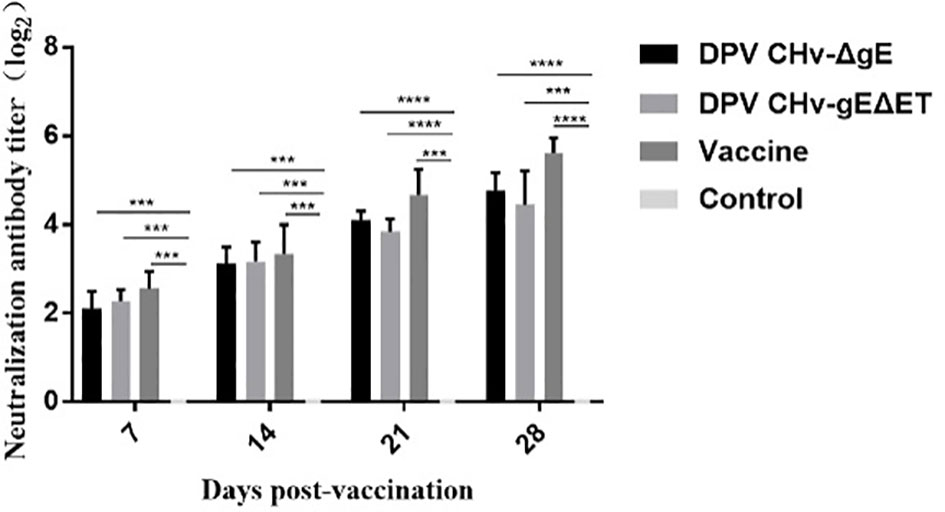
Figure 8 The determination of neutralizing antibody titer in ducks of different groups after immunization. ***P< 0.001, ****P< 0.0001.
In alphaherpesviruses, gE is an important virulence gene that affects the virus’s virulence. Selecting the deletion of the gE gene can reduce the virulence of the virus without affecting the immunogenicity and growth ability of the virus (34, 35). Virulence genes such as gE and gI have various degrees of insertion, deletion, and base substitution mutations, resulting in the emergence of highly toxic and lethal PRV mutant strains (36–39). The PRV mutant strain ZJ01, with 2 amino acid insertions in its gE sequence, caused all 14/80-day-old pigs to die (40). Two recombinant viruses, rZJ01-LA/gEI and rLA-ZJ01/gEI, were generated by exchanging the gE and gI genes of the LA strain and the ZJ01 strain. It was found that rLA-ZJ01/gEI exhibited higher virulence than its parental virus rLA, indicating that changes in the gE gene were part of the reason for the increased virulence of PRV strains in recent years (41). The bacterial artificial chromosome (BAC) of DPV is an effective tool for the study of duck plague virus, an infectious clone of a virulent strain of DPV CHv (DPV CHv-BAC-GS1783) was previously constructed in our laboratory and used in this study to generate mutant virus DPV CHv-ΔgE, DPV CHv-gEΔET and DPV CHv-gEΔCT (29, 30, 42, 43). It should be further mentioned that the above mutant virus lacks gE and different functional regions of gE and lacks miniF on its basis. Otherwise, the deletion-mutant virus cannot infect ducks or be used as a live vaccine. The gE gene of DPV has been confirmed to play an important role in the assembly of virions, with only minor damage to virus replication. In this study, the dynamic growth curve of the mutant virus after gE deletion in different regions was determined, which also confirmed this conclusion. By measuring the in vitro proliferation of DPV CHv-gEΔET and DPV CHv-gEΔCT mutant viruses on DEFs, both mutant viruses were found to reduce virus titers on host cells, but to a lesser extent. However, further research is needed on the specific functions of gE ET and gE CT.
Vaccine immunization is an important means of preventing and controlling viral diseases. Recombinant vector vaccines have become one of the hotspots in vaccine research due to their unique advantages. There are many non-essential regions in the DPV genome (1, 3), such as the gE/gI genes (44, 45), which can be used as insertion sites for foreign genes. Therefore, DPV attenuated/attenuated strains have the advantage of being used as multiplex recombinant live vaccine carriers in addition to being used as DPV vaccines. Many researchers have expressed the immunogenic genes of Avian Influenza virus, duck Tembusu virus, duck hepatitis virus, chicken infectious bronchitis virus, and Newcastle disease virus using traditional attenuated vaccine strains (46–52). DPV immunized ducks with recombinant Avian Influenza virus can produce better protective effects. Immunized chickens can also quickly produce immune protection. The virus cannot replicate efficiently in chickens, so the safety is high. Therefore, the analysis of DPV-related genes, especially the functions of virulence-related genes similar to gE, is of great significance for the prevention and control of DPV and immune purification and promoting the application of DPV in avian multivalent live vector vaccine vectors.
The genetic stability of mutant virus is one of the important contents of mutant virus research. The stability of the mutant virus is evaluated mainly through the continuous passage of the mutant virus in vitro and in vivo. We serially passaged the mutant viruses DPV CHv-gEΔET and DPV CHv-gEΔCT on DEFs and in ducklings, respectively, and measured the virus titer, whether the deleted gene was restored, and the pathogenic changes in susceptible animals. Found that the missing gene sequence of the mutant virus has not recovered, nor has the virulence returned, indicating that the mutant virus has good genetic stability, which is the premise for subsequent studies of mutant viruses as multivalent live vector vaccines or attenuated vaccine vectors. We grouped the mutant viruses DPV CHv-gEΔET and DPV CHv-gEΔCT according to the challenge dose gradient setting and conducted a pathogenicity study in 14-day-old ducklings, and preliminary analysis was made from body temperature, death, and antibody production after challenge. The results showed that the body temperature of the ducklings in the parental virus CHv-50 105 TCID50, 106 TCID50 groups increased and exceeded the normal body temperature range 3 to 4 days after the challenge, and the mortality rate showed a gradient difference according to the challenge dose. The body temperature of DPV CHv-gEΔCT fluctuated within the normal range, and the mortality rates were 20% (106 TCID50), 20% (105 TCID50), and 0% (104 TCID50), respectively. The body temperature of DPV CHv-gEΔET and DPV CHv-ΔgE in each dose group, fluctuated within the normal range, and no death occurred. DPV-specific serum antibodies could be detected in the ducklings on the 10th day after inoculation (data not shown). The above data show that gE deletion of ET or CT will reduce the pathogenicity of the virus. The pathogenicity of the mutant virus to ducklings after gE deletion of ET is significantly reduced, and the ducklings can be induced to produce specific serum antibodies. It further shows that DPV gE is also an important virulence gene, and the gE ET is an important region associated with virulence.
On the basis that the DPV CHv-gEΔET mutant virus has been proved to have stable inheritance, significantly reduce virus virulence, and induce DPV-specific serum antibodies in ducklings, further research on subsequent vaccines was carried out. In this study, the DPV CHv-gEΔET mutant virus was used to study the immune efficacy of the DPV CHv-gEΔET mutant virus by selecting the live duck plague vaccine that is widely used in China as a comparison. In the challenge protection trial, a lethal dose of CHv was administered after immunization, and no death and clinical symptoms were found in the DPV CHv-gEΔET, DPV CHv-ΔgE, and vaccine groups, and the challenge protection rate was 100%, the control group all died. In addition, judging whether the potential to develop into a vaccine is related to the challenge protection rate and observing the changes in antibody levels after immunization. Whether the body can produce high antibodies for a long time after vaccination is very important to fight viral infection. It is also one of the criteria for evaluating the quality of vaccines. Through neutralizing antibody detection, it was found that ducklings could produce certain neutralizing antibodies on the 7th day after immunizing with DPV CHv-ΔgE or DPV CHv-gEΔET mutant virus, and the antibody level on the 7th to 28th day after immunization rising. The levels of neutralizing antibodies produced after immunizing ducklings with the two mutant viruses were similar, significantly higher than those in the Mock group but slightly lower than those in the vaccine group. The above shows that the deletion of the gE gene or the deletion of gE ET in DPV reduces the virus’s virulence and stimulates the ducklings to produce a similar level of the humoral immune response, which can provide the same protection to the immunized animals. In this study, we lacked the detection of cellular immunity, and perhaps the reason for the immune protection of the DPV CHv-gEΔET mutant virus could be further explained by the study of cellular immunity.
It is well known that gE and gI function as dimers in alphaherpesviruses (10, 53). Previous studies in our laboratory have also demonstrated that gE and gI in DPV can form dimers, and amino acids 1-279 of gI can locate gE from the endoplasmic reticulum to the Golgi O-glycosylation modification (data unpublished). However, whether the complex position between gE and gI in DPV is also in the gE ET region, and the specific amino acid sites in gE that affect the complex formation with gI are currently being screened. The current research results and subsequent related research will provide a corresponding theoretical basis for the prevention and control of DPV and duck plague purification and promote the application of DPV in avian multivalent live vector vaccine vectors.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The experimental animal protocol was approved by the Ethics and Animal Welfare Committee of Sichuan Agricultural University and carried out following the Chinese version of the Guide for the Care and Use of Laboratory Animals.
Conceptualization, YN and AC. Methodology, YN and YH. Software, YN and QY. Validation, MW. Formal analysis, YN. Investigation, YN. Resources, AC, MW, SC, DZ, ML, QY, YW, XZ, SZ, JH, BT, RJ, XO, SM, QG, DS, YY, and LZ. Data curation, YN. Writing-original draft preparation, YN. Writing-review and editing, AC. Visualization, BT, RJ, ML, QY, and DS. Supervision, YN and AC. Project administration, MW and AC. Funding acquisition, MW, AC, RJ, SC, DZ, and ML. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Natural Science Foundation of China (32072894), the China Agriculture Research System of MOF and MARA, and the Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We apologize to the authors of articles reporting relevant research not cited in this manuscript due to limited space.
2. Qi X, Yang X, Cheng A, Wang M, Zhu D, Jia R, et al. Intestinal Mucosal Immune Response Against Virulent Duck Enteritis Virus Infection in Ducklings. Res Vet Sci (2009) 87(2):218–25. doi: 10.1016/j.rvsc.2009.02.009
3. Wu Y, Cheng A, Wang M, Yang Q, Zhu D, Jia R, et al. Complete Genomic Sequence of Chinese Virulent Duck Enteritis Virus. J Virol (2012) 86(10):5965. doi: 10.1128/JVI.00529-12
4. Dhama K, Kumar N, Saminathan M, Tiwari R, Karthik K, Kumar MA, et al. Duck Virus Enteritis (Duck Plague) - A Comprehensive Update. Vet Q (2017) 37(1):57–80. doi: 10.1080/01652176.2017.1298885
5. Koyanagi N, Kawaguchi Y. Evasion of the Cell-Mediated Immune Response by Alphaherpesviruses. Viruses (2020) 12(12):1354. doi: 10.3390/v12121354
6. Wang J, Song Z, Ge A, Guo R, Qiao Y, Xu M, et al. Safety and Immunogenicity of an Attenuated Chinese Pseudorabies Variant by Dual Deletion of TK&gE Genes. BMC Vet Res (2018) 14(1):287. doi: 10.1186/s12917-018-1536-7
7. Yu X, Jia R, Huang J, Shu B, Zhu D, Liu Q, et al. Attenuated Salmonella Typhimurium Delivering DNA Vaccine Encoding Duck Enteritis Virus UL24 Induced Systemic and Mucosal Immune Responses and Conferred Good Protection Against Challenge. Vet Res (2012) 43(1):56. doi: 10.1186/1297-9716-43-56
8. Dogrammatzis C, Waisner H, Kalamvoki M. "Non-Essential" Proteins of HSV-1 With Essential Roles In Vivo: A Comprehensive Review. Viruses (2020) 13(1):17. doi: 10.3390/v13010017
9. Liu T, Wang M, Cheng A, Jia R, Yang Q, Wu Y, et al. Duck Plague Virus gE Serves Essential Functions During the Virion Final Envelopment Through Influence Capsids Budding Into the Cytoplasmic Vesicles. Sci Rep (2020) 10(1):5658. doi: 10.1038/s41598-020-62604-9
10. Farnsworth A, Johnson DC. Herpes Simplex Virus Ge/gI Must Accumulate in the Trans-Golgi Network at Early Times and Then Redistribute to Cell Junctions to Promote Cell-Cell Spread. J Virol (2006) 80(7):3167–79. doi: 10.1128/JVI.80.7.3167-3179.2006
11. Brack AR, Dijkstra JM, Granzow H, Klupp BG, Mettenleiter TC. Inhibition of Virion Maturation by Simultaneous Deletion of Glycoproteins E, I, and M of Pseudorabies Virus. J Virol (1999) 73(7):5364–72. doi: 10.1128/JVI.73.7.5364-5372.1999
12. Farnsworth A, Wisner TW, Johnson DC. Cytoplasmic Residues of Herpes Simplex Virus Glycoprotein gE Required for Secondary Envelopment and Binding of Tegument Proteins VP22 and UL11 to gE and gD. J Virol (2007) 81(1):319–31. doi: 10.1128/JVI.01842-06
13. Yang L, Shen B, Wang M, Cheng A, Yang Q, Wu Y, et al. The Intracellular Domain of Duck Plague Virus Glycoprotein E Affects UL11 Protein Incorporation Into Viral Particles. Vet Microbiol (2021) 257:109078. doi: 10.1016/j.vetmic.2021.109078
14. Han J, Chadha P, Meckes DG Jr, Baird NL, Wills JW. Interaction and Interdependent Packaging of Tegument Protein UL11 and Glycoprotein E of Herpes Simplex Virus. J Virol (2011) 85(18):9437–46. doi: 10.1128/JVI.05207-11
15. Polcicova K, Goldsmith K, Rainish BL, Wisner TW, Johnson DC. The Extracellular Domain of Herpes Simplex Virus gE is Indispensable for Efficient Cell-to-Cell Spread: Evidence for Ge/gI Receptors. J Virol (2005) 79(18):11990–2001. doi: 10.1128/JVI.79.18.11990-12001.2005
16. Berarducci B, Rajamani J, Reichelt M, Sommer M, Zerboni L, Arvin AM. Deletion of the First Cysteine-Rich Region of the Varicella-Zoster Virus Glycoprotein E Ectodomain Abolishes the gE and gI Interaction and Differentially Affects Cell-Cell Spread and Viral Entry. J Virol (2009) 83(1):228–40. doi: 10.1128/JVI.00913-08
17. Chang H, Cheng A, Wang M, Jia R, Zhu D, Lou Q, et al. Immunofluorescence Analysis of Duck Plague Virus gE Protein on DPV-Infected Ducks. Virol J (2011) 8:19. doi: 10.1186/1743-422X-8-19
18. Lu M, Qiu S, Zhang L, Sun Y, Bao E, Lv Y. Pseudorabies Virus Glycoprotein gE Suppresses Interferon-β Production via CREB-Binding Protein Degradation. Virus Res (2021) 291:198220. doi: 10.1016/j.virusres.2020.198220
19. Quast I, Keller CW, Maurer MA, Giddens JP, Tackenberg B, Wang LX, et al. Sialylation of IgG Fc Domain Impairs Complement-Dependent Cytotoxicity. J Clin Invest (2015) 125(11):4160–70. doi: 10.1172/JCI82695
20. Sun H, Olsen HS, Mérigeon EY, So E, Burch E, Kinsey S, et al. Recombinant Human IgG1 Based Fc Multimers, With Limited FcR Binding Capacity, can Effectively Inhibit Complement-Mediated Disease. J Autoimmun (2017) 84:97–108. doi: 10.1016/j.jaut.2017.08.004
21. Lubinski JM, Lazear HM, Awasthi S, Wang F, Friedman HM. The Herpes Simplex Virus 1 IgG Fc Receptor Blocks Antibody-Mediated Complement Activation and Antibody-Dependent Cellular Cytotoxicity In Vivo. J Virol (2011) 85(7):3239–49. doi: 10.1128/JVI.02509-10
22. Jenks JA, Goodwin ML, Permar SR. The Roles of Host and Viral Antibody Fc Receptors in Herpes Simplex Virus (HSV) and Human Cytomegalovirus (HCMV) Infections and Immunity. Front Immunol (2019) 10:2110. doi: 10.3389/fimmu.2019.02110
23. Bournazos S, DiLillo DJ, Ravetch JV. The Role of Fc-Fcγr Interactions in IgG-Mediated Microbial Neutralization. J Exp Med (2015) 212(9):1361–9. doi: 10.1084/jem.20151267
24. Watanabe M, Arii J, Takeshima K, Fukui A, Shimojima M, Kozuka-Hata H, et al. Prohibitin-1 Contributes to Cell-To-Cell Transmission of Herpes Simplex Virus 1 via the MAPK/ERK Signaling Pathway. J Virol (2021) 95(3):e01413–20. doi: 10.1128/JVI.01413-20
25. Tang YD, Liu JT, Wang TY, Sun MX, Tian ZJ, Cai XH. Comparison of Pathogenicity-Related Genes in the Current Pseudorabies Virus Outbreak in China. Sci Rep (2017) 7(1):7783. doi: 10.1038/s41598-017-08269-3
26. Zhou J, Li S, Wang X, Zou M, Gao S. Bartha-K61 Vaccine Protects Growing Pigs Against Challenge With an Emerging Variant Pseudorabies Virus. Vaccine (2017) 35(8):1161–6. doi: 10.1016/j.vaccine.2017.01.003
27. Wang J, Cui X, Wang X, Wang W, Gao S, Liu X, et al. Efficacy of the Bartha-K61 Vaccine and A Ge/Gi/TK Prototype Vaccine Against Variant Porcine Pseudorabies Virus (vPRV) in Piglets With Sublethal Challenge of vPRV. Res Vet Sci (2020) 128:16–23. doi: 10.1016/j.rvsc.2019.10.005
28. Wu Y, Cheng A, Wang M, Zhang S, Zhu D, Jia AB, et al. Serologic Detection of Duck Enteritis Virus Using an Indirect ELISA Based on Recombinant UL55 Protein. Avian Dis (2011) 55(4):626–32. doi: 10.1637/9766-042111-Reg.1
29. Wu Y, Li Y, Wang M, Sun K, Jia R, Chen S, et al. Preliminary Study of the UL55 Gene Based on Infectious Chinese Virulent Duck Enteritis Virus Bacterial Artificial Chromosome Clone. Virol J (2017) 14(1):78. doi: 10.1186/s12985-017-0748-y
30. Tischer BK, Smith GA, Osterrieder N. En Passant Mutagenesis: A Two Step Markerless Red Recombination System. Methods Mol Biol (Clifton NJ) (2010) 634:421–30. doi: 10.1007/978-1-60761-652-8_30
31. You Y, Liu T, Wang M, Cheng A, Jia R, Yang Q, et al. Duck Plague Virus Glycoprotein J Is Functional But Slightly Impaired in Viral Replication and Cell-to-Cell Spread. Sci Rep (2018) 8(1):4069. doi: 10.1038/s41598-018-22447-x
32. Deng L, Wang M, Cheng A, Yang Q, Wu W, Jia R, et al. The Pivotal Roles of US3 Protein in Cell-To-Cell Spread and Virion Nuclear Egress of Duck Plague Virus. Sci Rep (2020) 10(1):7181. doi: 10.1038/s41598-020-64190-2
33. Guo Y, Cheng A, Wang M, Shen C, Jia R, Chen S, et al. Development of TaqMan MGB Fluorescent Real-Time PCR Assay for the Detection of Anatid Herpesvirus 1. Virol J (2009) 6:71. doi: 10.1186/1743-422X-6-71
34. Bouma A. Determination of the Effectiveness of Pseudorabies Marker Vaccines in Experiments and Field Trials. Biologicals J Int Assoc Biol Standard (2005) 33(4):241–5. doi: 10.1016/j.biologicals.2005.08.011
35. Casal J, Planasdemunt L, Varo JA, Martín M. The Use of Different Vaccination Schedules for Sows to Protect Piglets Against Aujeszky's Disease. J Vet Med B Infect Dis Vet Public Health (2004) 51(1):8–11. doi: 10.1046/j.1439-0450.2003.00721.x
36. Sun Y, Liang W, Liu Q, Zhao T, Zhu H, Hua L, et al. Epidemiological and Genetic Characteristics of Swine Pseudorabies Virus in Mainland China Between 2012 and 2017. PeerJ (2018) 6:e5785. doi: 10.7717/peerj.5785
37. Wang Y, Qiao S, Li X, Xie W, Guo J, Li Q, et al. Molecular Epidemiology of Outbreak-Associated Pseudorabies Virus (PRV) Strains in Central China. Virus Genes (2015) 50(3):401–9. doi: 10.1007/s11262-015-1190-0
38. Tong W, Liu F, Zheng H, Liang C, Zhou YJ, Jiang YF, et al. Emergence of a Pseudorabies Virus Variant With Increased Virulence to Piglets. Vet Microbiol (2015) 181(3-4):236–40. doi: 10.1016/j.vetmic.2015.09.021
39. Wu X-M, Chen QY, Chen RJ, Che YL, Wang LB, Wang CY, et al. Pathogenicity and Whole Genome Sequence Analysis of a Pseudorabies Virus Strain FJ-2012 Isolated From Fujian, Southern China. Can J Infect Dis Med Microbiol = J Canadien Des Maladies Infectieuses la Microbiologie Medicale (2017) p:9073172. doi: 10.1155/2017/9073172
40. Gu Z, Hou C, Sun H, Yang W, Dong J, Bai J, et al. Emergence of Highly Virulent Pseudorabies Virus in Southern China. Can J Vet Res = Rev Can Recherche Veterinaire (2015) 79(3):221–8.
41. Dong J, Gu Z, Jin L, Lv L, Wang J, Sun T, et al. Polymorphisms Affecting the gE and gI Proteins Partly Contribute to the Virulence of a Newly-Emergent Highly Virulent Chinese Pseudorabies Virus. Virology (2018) 519:42–52. doi: 10.1016/j.virol.2018.03.024
42. Chai Y, Shan S, Weissman KJ, Hu S, Zhang Y, et al. Heterologous Expression and Genetic Engineering of the Tubulysin Biosynthetic Gene Cluster Using Red/ET Recombineering and Inactivation Mutagenesis. Chem Biol (2012) 19(3):361–71. doi: 10.1016/j.chembiol.2012.01.007
43. Bian X, Huang F, Stewart FA, Xia L, Zhang Y, Mϋller R. Direct Cloning, Genetic Engineering, and Heterologous Expression of the Syringolin Biosynthetic Gene Cluster in E. Coli Through Red/ET Recombineering. Chembiochem Eur J Chem Biol (2012) 13(13):1946–52. doi: 10.1002/cbic.201200310
44. Chang H, Cheng A, Wang M, Zhu D, Jia R, Liu F, et al. Cloning, Expression and Characterization of gE Protein of Duck Plague Virus. Virol J (2010) 7:120. doi: 10.1186/1743-422X-7-120
45. Zhou X, Wang M, Cheng A, Yang Q, Wu Y, Jia R, et al. Development of a Simple and Rapid Immunochromatographic Strip Test for Detecting Duck Plague Virus Antibodies Based on gI Protein. J Virol Methods (2020) 277:113803. doi: 10.1016/j.jviromet.2019.113803
46. Liu J, Chen P, Jiang Y, Deng G, Shi J, Wu L, et al. Recombinant Duck Enteritis Virus Works as a Single-Dose Vaccine in Broilers Providing Rapid Protection Against H5N1 Influenza Infection. Antiviral Res (2013) 97(3):329–33. doi: 10.1016/j.antiviral.2012.12.015
47. Zou Z, Hu Y, Liu Z, Zhong W, Cao H, Chen H, et al. Efficient Strategy for Constructing Duck Enteritis Virus-Based Live Attenuated Vaccine Against Homologous and Heterologous H5N1 Avian Influenza Virus and Duck Enteritis Virus Infection. Vet Res (2015) 46(1):42. doi: 10.1186/s13567-015-0174-3
48. Zou Z, Liu Z, Jin M. Efficient Strategy to Generate a Vectored Duck Enteritis Virus Delivering Envelope of Duck Tembusu Virus. Viruses (2014) 6(6):2428–43. doi: 10.3390/v6062428
49. He D, Zhang X, Chen L, Tang Y, Diao Y. Development of an Attenuated Live Vaccine Candidate of Duck Tembusu Virus Strain. Vet Microbiol (2019) 231:218–25. doi: 10.1016/j.vetmic.2019.03.022
50. Kang M, Roh J-H, Jang H-K. Protective Efficacy of a Bivalent Live Attenuated Vaccine Against Duck Hepatitis A Virus Types 1 and 3 in Ducklings. Vet Microbiol (2018) 214:108–12. doi: 10.1016/j.vetmic.2017.12.018
51. Li H, Wang Y, Han Z, Wang Y, Liang S, Jiang L, et al. Recombinant Duck Enteritis Viruses Expressing Major Structural Proteins of the Infectious Bronchitis Virus Provide Protection Against Infectious Bronchitis in Chickens. Antiviral Res (2016) 130:19–26. doi: 10.1016/j.antiviral.2016.03.003
52. Ding L, Chen P, Bao X, Li A, Jiang Y, Hu Y, et al. Recombinant Duck Enteritis Viruses Expressing the Newcastle Disease Virus (NDV) F Gene Protects Chickens From Lethal NDV Challenge. Vet Microbiol (2019) 232:146–50. doi: 10.1016/j.vetmic.2019.04.022
Keywords: duck plague virus, gE, extracellular domain, genetic stability, pathogenicity, vaccine
Citation: Ning Y, Huang Y, Wang M, Cheng A, Jia R, Liu M, Zhu D, Chen S, Zhao X, Zhang S, Yang Q, Wu Y, Huang J, Tian B, Ou X, Mao S, Gao Q, Sun D, Yu Y and Zhang L (2022) Evaluation of the Safety and Immunogenicity of Duck-Plague Virus gE Mutants. Front. Immunol. 13:882796. doi: 10.3389/fimmu.2022.882796
Received: 24 February 2022; Accepted: 28 March 2022;
Published: 20 April 2022.
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Xiaochuan Liu, University of California, Riverside, United StatesCopyright © 2022 Ning, Huang, Wang, Cheng, Jia, Liu, Zhu, Chen, Zhao, Zhang, Yang, Wu, Huang, Tian, Ou, Mao, Gao, Sun, Yu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.