
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 06 April 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.882452
This article is part of the Research Topic Innovative Theranostic Approaches Towards Neuro-immunology in Gliomas View all 16 articles
Gliomas are tumors derived from mutations in glial brain cells. Gliomas cause significant morbidity and mortality and development of precision diagnostics and novel targeted immunotherapies are critically important. Radiographic imaging is the most common technique to diagnose and track response to treatment, but is an imperfect tool. Imaging does not provide molecular information, which is becoming critically important for identifying targeted immunotherapies and monitoring tumor evolution. Furthermore, immunotherapy induced inflammation can masquerade as tumor progression in images (pseudoprogression) and confound clinical decision making. More recently, circulating cell free tumor DNA (cf-tDNA) has been investigated as a promising biomarker for minimally invasive glioma diagnosis and disease monitoring. cf-tDNA is shed by gliomas into surrounding biofluids (e.g. cerebrospinal fluid and plasma) and, if precisely quantified, might provide a quantitative measure of tumor burden to help resolve pseudoprogression. cf-tDNA can also identify tumor genetic mutations to help guide targeted therapies. However, due to low concentrations of cf-tDNA, recovery and analysis remains challenging. Plasma cf-tDNA typically represents <1% of total cf-DNA due to the blood-brain barrier, limiting their usefulness in practice and motivating the development and use of highly sensitive and specific detection methods. This mini review summarizes the current and future trends of various approaches for cf-tDNA detection and analysis, including new methods that promise more rapid, lower-cost, and accessible diagnostics. We also review the most recent clinical case studies for longitudinal disease monitoring and highlight focus areas, such as novel accurate detection methodologies, as critical research priorities to enable translation to clinic.
Gliomas are a diverse set of brain tumors derived from glial brain cells. While a relatively small fraction of total cancer deaths per year, Gliomas cause significant morbidity and mortality with a five year survival rate as low as 7.2% depending on the tumor subtype (1). Current treatment options are mostly limited to surgical resection and chemoradiation, however, immunotherapy has recently been evaluated as an exciting new therapy to combat this disease (2–6).
Effective immunotherapy relies on an accurate diagnosis to guide treatment selection, and disease monitoring to identify if the glioma is responding to treatment or progressing. Because of their sensitive location in the brain, repeat biopsies are not feasible (7). Thus, radiographic imaging is commonly used for both initial diagnosis and disease monitoring. However, these images can be difficult to interpret due to various factors such as immunotherapy-induced swelling, leading to incorrect assumptions about a tumor’s response to treatment (8–12); a classic example is “pseudoprogression”, where immunotherapy-induced swelling is misinterpreted as tumor progression. Especially for immunotherapy response monitoring, which has a minimum four week iRECIST monitoring interval (13), misinterpretation can lead to unnecessarily long treatment and improper or delayed course correction (10). Additionally, imaging does not capture molecular information, which is becoming increasingly important for proper diagnosis (14, 15), identification of personalized targeted immunotherapies (16), and monitoring of tumor evolution to detect resistance mutations (17, 18).
To address these issues, liquid biopsies have emerged as a promising new diagnostic and disease monitoring approach for gliomas. Liquid biopsies work by recovering and quantifying tumor-related biomarkers shed by dying tumor cells into surrounding biofluids. Various studies have shown that biomarker levels correlate with tumor burden, and/or disease state, and may even be able to detect disease progression before it is evident in imaging (19, 20). Thus, liquid biopsies promise a minimally invasive and accurate alternative diagnostic to tissue biopsy, and a less error prone approach to quantify tumor response than radiographic imaging (13).
Circulating tumor cells from primary brain tumors have been identified in blood (21, 22), however since primary brain tumors rarely metastasize, these cells are exceptionally rare (23). In this mini-review, we focus on the use of cell-free tumor DNA (cf-tDNA) as a biomarker for gliomas and its potential to aid in development and clinical use of immunotherapies targeting gliomas. We first summarize the mechanism of glioma cf-tDNA release. We then discuss both established and novel cf-tDNA detection methods used in the literature and their strengths and weaknesses. We then discuss translational uses of cf-tDNA liquid biopsies in clinic focusing on efforts to improve immunotherapy-based treatment. Finally, we discuss current difficulties and open questions about the practical use of liquid-biopsy and new approaches to cf-tDNA detection that attempt to improve accuracy, accessibility, and cost.
As glioma cells proliferate and die via apoptosis, necrosis, or immune response, tumor DNA is immediately shed into the surrounding interstitial fluid and CSF. During apoptosis, tumor chromosomal DNA is fragmented via endonucleases around nucleosome boundaries (~140bp-180bp) resulting in a characteristic pattern of fragmentation (24). cf-tDNA fragments spread throughout the central nervous system before eventually permeating the blood-brain barrier (25). Due to the low molecular weight of post-apoptotic cf-tDNA, the molecules are more able to permeate selective filters in the body such as the blood-brain barrier and glomerulus structures in the kidneys. To date, glioma cf-tDNA has been successfully identified in CSF, plasma, and even urine (24). Figure 1 shows an overview of cf-tDNA release, recovery, current detection methods, and clinical applications.
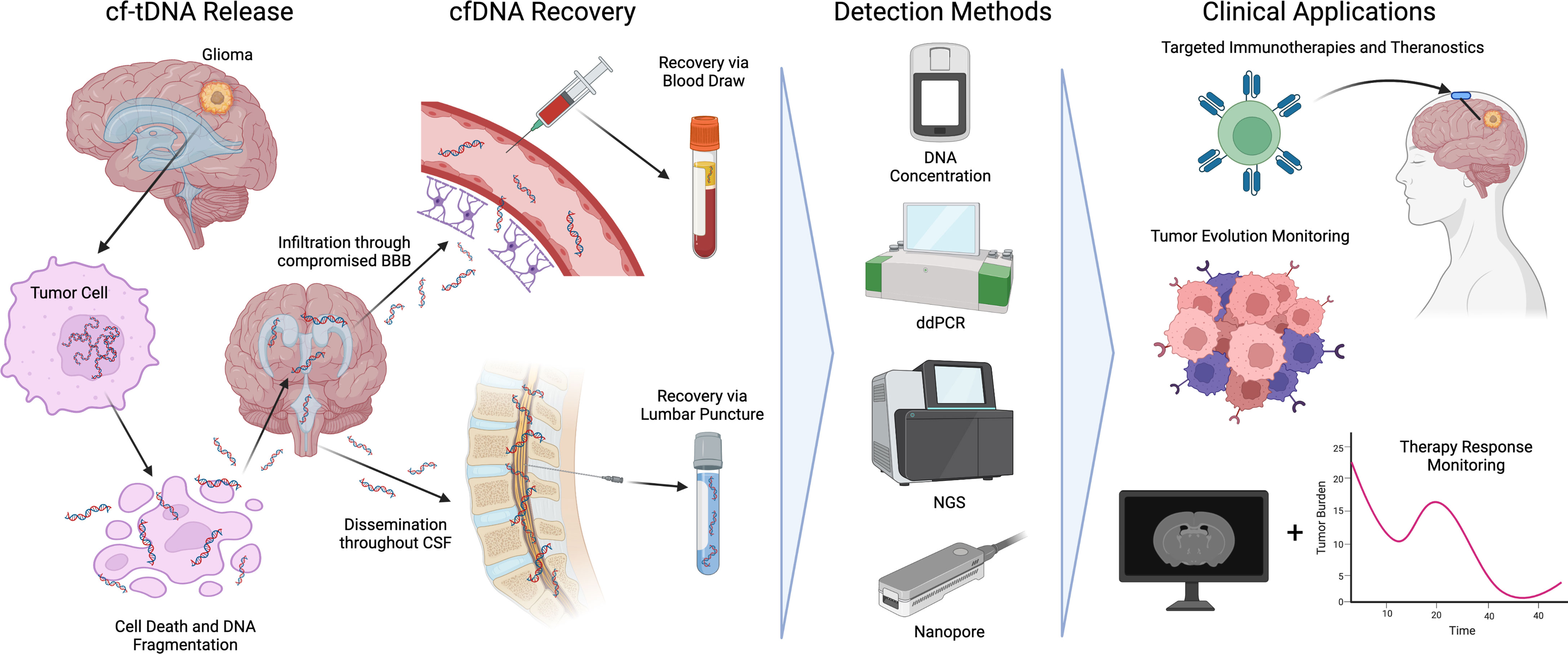
Figure 1 An overview of cf-tDNA release into biofluids, recovery, detection methods, and current clinical applications benefiting immunotherapies for glioma.
cf-tDNA signals can be distinguished from background cfDNA (from non-tumor tissue) by using either aggregate or specific detection techniques. Aggregate detection relies on biomarkers that are shared by both healthy and tumor-derived DNA but are up- or down-regulated in tumors. Prior work has detected glioma via structural variation, copy number alterations (26), methylation status of certain genomic regions (25, 27), and even cfDNA fragmentation patterns (24). However, these aggregate signals cannot uniquely discriminate tumor- vs. normal-derived cfDNA and are thus less likely to be useful when the relative amount of cf-tDNA is low (e.g., <1%). A more precise approach is to directly identify cf-tDNA by detection of genetic tumor driver mutations via probe-based quantitative PCR or cfDNA sequencing. These assays report the ratio of mutated to total cfDNA reads – i.e., the mutant or variant allele fraction (MAF/VAF) –identified in the sample.
Recovered cf-tDNA concentrations can vary widely and have been found to correlate with variables such as disease grade (28), tumor burden, tumor location relative to CSF reservoirs (29) and biofluid proximity to the tumor (30). Due to the highly-selective nature of the blood brain barrier, glioma cf-tDNA concentrations are generally several orders of magnitude higher in CSF than plasma (31) or urine (24), where typical plasma VAFs are <1% (17, 32) with suspected positives detected as low as 0.02% (32). Thus, CSF is considered the gold standard biofluid for liquid biopsy of gliomas. However, lumbar punctures to obtain CSF are significantly more invasive than blood draws and urine collection, making plasma and urine-based liquid biopsies much more desirable. This motivates ultra-sensitive detection techniques to enable accurate monitoring of both CSF-derived cf-tDNA and allow for practical use of plasma and urine-based biopsies.
Multiple cf-tDNA detection methods have been used to successfully quantify cf-tDNA related biomarkers and uncover diagnostically relevant information that might guide personalized treatment. In this section we review popular cf-tDNA detection methods and their benefits and weaknesses.
Total cfDNA concentration is the level/amount of DNA per volume of biofluid (blood, CSF, or urine). Because tumor cell turnover is higher than that of normal tissue, research has found that glioma patients tend to have higher absolute amounts of cfDNA in glioma than healthy patients (24, 33–35). However, total cfDNA can be impacted by many other factors unrelated to tumor burden (e.g. inflammation) reducing its sensitivity to detect disease without supplementary analysis (36, 37). Furthermore, concentration as a biomarker lacks molecular information that might inform targeted treatments and clinical management. Methylation of cf-DNA and recovery via methylation-specific PCR (27) or sequencing (38) has also been proposed as method of detecting disease via measurement of hypo/hyper-methylation at various genomic loci but is not discussed in this review.
qPCR (quantitative polymerase chain reaction) is a quantification method that uses sequence specific primers or fluorescent probes to detect and quantify tumor-specific somatic mutations (39). However, qPCR suffers from a variety of shortcomings that limit its sensitivity and specificity (40–42). Droplet digital PCR (ddPCR) is a modification of qPCR that improves precision and limit-of-detection (42). By dividing a typical qPCR reaction into many isolated droplets with ~1 template copy, a precise VAF can be computed from the ratio of mutant positive to wildtype droplets, with a VAF limit of detection around 0.001% (43). ddPCR has supplanted itself as a highly-accurate technique, and is considered a gold standard approach to quantify VAFs from liquid samples (36, 44). However, accurate and reproducible ddPCR assays require careful development and optimization of input template concentration, target-specific primers, and fluorescent probes and are restricted for use on a limited set of known hotspot mutations (44).
Next-generation sequencing (NGS) refers to a group of massively parallel sequencing technologies including Ion Torrent (45, 46), PacBio (47), and Illumina (48, 49), with the latter used most often for liquid biopsy due to its accuracy (50). Illumina sequencing uses synthesis of fluorescent dNTPs to clusters of template strands to recover the original template sequence (45). Unlike ddPCR, sequencing of cf-tDNA does not rely on sequence specific probes or any prior knowledge of mutations. Various library preparations enable sequencing of the whole genome (WGS), whole exome (WES), targeted hybridization capture, or amplicons from targeted panels with each technique trading genome coverage for read depth. For example, WGS can provide 20x-50x coverage over the entire genome, which enables detection of high-frequency somatic mutations and copy number variation but is too shallow to precisely measure cf-tDNA allele fractions below ~2-5%. Targeted amplicon sequencing can generate >10,000x coverage of specific genomic loci, improving VAF limit of detection but reducing the number of analyzed loci. Illumina sequencing has proven highly-accurate, with an error rate ranging between 0.5%-1% (50–52). However, this approach is relatively expensive, slow, and due to its error rate, cannot reliably detect allele fractions below the sequencer error rate without further assay modifications (53–56).
Nanopore sequencing is a relatively new technology (57) that has been used for liquid biopsies (58, 59). Nanopore sequencers work by feeding DNA strands through small pores embedded in a membrane. As they flow through the pore, each DNA base-pair creates a unique electrical disturbance that can be measured and used to call each base. Nanopore devices are low-cost, can sequence any length DNA strand, have a small form factor, and offer a rapid time to result making them ideal devices for liquid biopsy. However, the device’s error rate has traditionally prevented it from being applied to liquid samples where allele frequencies are less than ~2% (58). Our group previously analyzed CSF samples from 12 pediatric high grade glioma patients and found that nanopore had a 85% sensitivity and 100% specificity in CSF samples (58), which compared favorably to Illumina-based targeted sequencing. More recent improvements to basecaller accuracy, and also the use of circular consensus sequencing (59–61) have improved accuracy to <0.05%, comparable with ddPCR-based approaches (59).
While not perfect, these cf-tDNA detection methods have been used to demonstrate a variety of potential uses for glioma diagnosis and monitoring. In the next section, we highlight translational research that attempts to utilize these instruments to improve disease management.
Accurate cf-tDNA-based liquid biopsies have several promising clinical applications for immunotherapy. Here, we highlight recent translational research (also summarized in Table 1) attempting to use liquid biopsy diagnostics that could help guide the use of personalized immunotherapy and monitor disease response in gliomas.
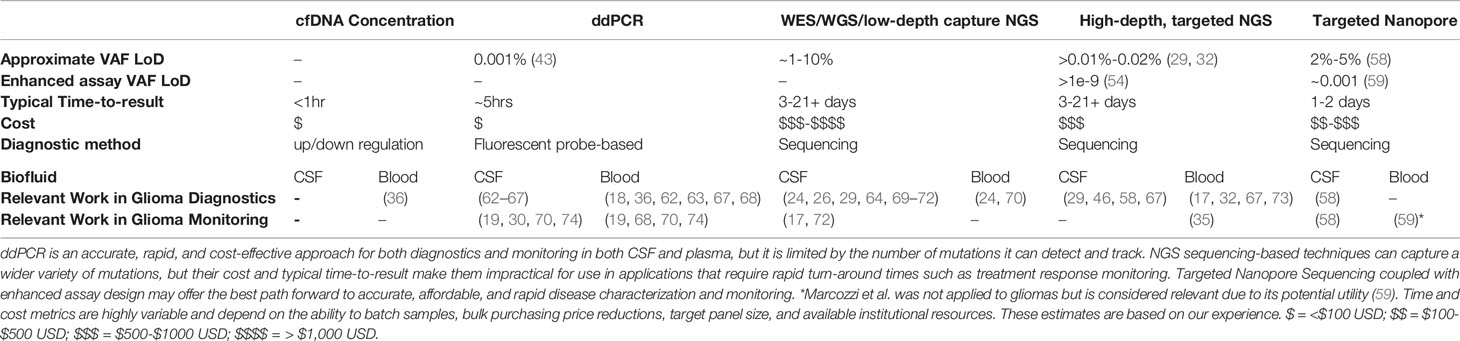
Table 1 A summary of cf-tDNA-based liquid biopsy detection methods as applied to gliomas and associated limit of detection, typical time-to-result, and cost.
Identifying tumor-specific molecular information in cf-tDNA that provides an accurate diagnosis, prognosis, and predicts response of a particular treatment is a “holy grail” clinical application for liquid biopsies. There is some work linking molecular markers (e.g. SNVs or CNVs) to the predicted response to radiation or chemotherapy in gliomas [reviewed in Birko et al. (75)]. However, minimal work has explored cf-tDNA diagnostics to personalize immunotherapy treatment. Studies looking at other solid tumors have identified several cell free DNA biomarkers as predictors of immunotherapy response (76), most notably increased tumor mutational burden (TMB) (77–79) and reduced copy number variations (80, 81). Pepe et al. showed feasibility for assessment of TMB in cytological samples from patients with NSCLC using a NGS platform (82). Studies have aimed to identify specific hotspot mutations that predict response to immune checkpoint inhibition or other immunotherapies. Guibert et al. identified that mutations in KRAS or TP53 without PTEN loss lead to increased PD-L1 expression and increased tumor mutational burden, increasing response to PD-1 immune checkpoint inhibition (77). This work was done in lung cancer, but the aforementioned mutations are also commonly present in gliomas, raising a potential opportunity to use glioma cf-tDNA to predict immunotherapy efficacy.
Another important clinical application of liquid biopsy for gliomas is the monitoring of tumor evolution. It has been shown that tumors undergo considerable evolution over the course of treatment, resulting in genetic changes that might suggest a new diagnosis and an adjustment to disease management (69). Several studies have investigated tumor evolution in glioma, with estimates ranging from 33-73% of genetic mutations at recurrence matching with alterations at biopsy (83, 84). However, as previously discussed, serial biopsies are discouraged due to the increased chance of morbidity. Miller et al. used CSF-derived cf-tDNA to monitor tumor evolution in adult gliomas (17) and was able to identify cf-tDNA in 42 of 85 patients who underwent CSF collection and NGS sequencing. In patients with hypermutated tumors, the median percentage match between the CSF derived cf-tDNA mutations and the initial tissue alterations was only 19.6%, while non-hypermutated tumors had an 81.7% match. These results indicate that CSF-based liquid biopsies can capture tumor heterogeneity and used to monitor tumor evolution over time. While this work was not applied to immunotherapy-based treatment, tumor evolution monitoring could be used to identify increased TMB and PD-1 sensitivity (77), or acquired resistance markers (85).
In addition to tumor evolution, several studies have shown the utility of serial cf-tDNA sampling for treatment response monitoring and resolution of pseudoprogression in gliomas (19, 37, 58, 62, 68, 69, 74, 86). One of the largest studies thus far from Panditharatna et al. collected serial CSF samples and concordant MRI from 22 patients (74). They found that cf-tDNA decreased in response to radiotherapy in 83% of patients, which was corroborated by a decrease in tumor size on MRI. The first prospective high grade glioma clinical trial with serial liquid biopsy was recently published by our group (19). We collected serial CSF and plasma samples from 24 patients and found that patients with decreased H3K27M CSF and plasma cf-tDNA VAF had prolonged progression free survival (19). A similar trend was identified by Jensen et al. while tracking response to immunotherapy over a variety of cancers (20).
We also compared serial ct-DNA levels with corresponding radiographic imaging. In individual cases, they were able to identify instances of suspected pseudoprogression, where radiographic progression was accompanied by a decrease in cf-DNA VAF. In another patient, a large increase in cf-tDNA VAF (>25%) preceded radiographic progression in many patients, suggesting that cf-tDNA VAF changes may act as an earlier warning sign of tumor progression versus radiographic imaging. For immunotherapy-based response, Jensen et al. used shallow WGS (0.3x) of cf-DNA to identify copy number alterations (CNAs) and report a metric of “genome instability” over a variety of cancers (20). This study demonstrated that dynamic changes in CNAs could track immunotherapy response and were able to resolve pseudoprogression, but specific use in gliomas has yet to be demonstrated. A common theme among these studies is relative changes in cf-tDNA signals—rather than absolute values—are better indicators of tumor response. Taken together, these results reaffirm the potential clinical utility of serial liquid biopsies for improved molecular profiling and effective therapeutic monitoring for gliomas.
While exciting progress is being made developing liquid biopsies that can support immunotherapy-based treatment of gliomas, further work is required to improve understanding tumor-specific biomarker release and how it corresponds to tumor burden, improve detection accuracy of various assays, and investigate novel liquid biopsy approaches that offer improved sensitivity and specificity.
Our current understanding of glioma cf-tDNA release and dissemination to various biofluids is still limited. Research has highlighted variability depending on a tumor’s proximity to CSF reservoirs in the brain (17, 69). This raises concerns about the ability of liquid biopsies to accurately track tumor burden if disease spreads. Blood-brain barrier permeability can also vary highly case-to-case, further complicating efforts to correlate tumor burden with cf-tDNA levels in blood. It is also unclear how various treatments (e.g. radiation, chemotherapy, and immunotherapy) impact both normal cfDNA and cf-tDNA release over time, which could bias cf-tDNA levels and VAF if not properly accounted for (10, 12). Future work might incorporate variables such as tumor ventricle proximity, tumor biology, and treatment type to better understand these patterns.
Even though there is a general consensus that the sensitivity and specificity of CSF-based assays are superior to plasma (18, 28, 70, 87, 88), plasma- and urine-based liquid biopsies are still highly desirable due to the ease of sample collection. It is likely that the large disparity between CSF- and plasma-derived results are partly due to limitations of current gold standard detection methods. As an example of the difficulty of implementing precise detection methods, Li et al. measured the performance of multiple ddPCR-based assays for the detection of H3.3K27M mutations in matched tissue, CSF, and plasma samples across three independent labs using two commercially available ddPCR machines (44). Results indicated that ddPCR was capable of precisely measuring small VAFs from plasma-derived cfDNA, but discovered high-variation among replicates, and statistically significant differences across assays and ddPCR instrument vendors. Significant protocol optimizations were required to improve the sensitivity, repeatability, and reliability of the assay (44). When considering NGS detection methods, even the most accurate NGS instruments have an established raw error rate of ~0.1%, which is most likely too high to precisely resolve plasma-derived cf-tDNA levels that fluctuate between 1% and 0.05%. These results highlight the need for highly optimized and standardized versions of current approaches, as well as improved detection methods to enable proper translation to clinic.
The current limitations with ddPCR, NGS, and Nanopore-based liquid biopsy approaches are not easy to solve, but progress is being made via improved assay design and bioinformatic error correction. For example, the use of universal molecular identifiers (UMIs) during targeted amplification can help resolve sequencing errors targeted amplification, enabling detection of 1 mutant molecule in 10,000 (89), with other assay design techniques further improving detection sensitivity by several orders of magnitude (53–56). Nanopore, long-read sequencing offers the ability to sequence single-molecule tandem-repeats constructed from small cf-tDNA fragments using rolling circle amplification. Even though the native error rate for Nanopore sequencing is relatively high (58), its ability to sequence long DNA strands with multiple redundant copies of a single cf-tDNA template allows for accuracy beyond any available NGS or ddPCR approach (59–61). Some of these methods are so accurate, that they are limited by polymerase error rather than sequencer error rate (59). Furthermore, continual improvements to basecalling software, library preparation methods, and assay design are certain to further reduce false positive rates. Because of these factors we expect long-read, consensus sequencing approaches to become a gold standard liquid biopsy approach for plasma cf-tDNA in the future.
Recent work has explored the effectiveness of CAR-T cell therapy in diffuse midline gliomas, administered serially into CSF via Ommaya reservoirs (6, 90). Ommaya reservoirs are used for intra-cranial administration of immunotherapies as well as frequent, minimally invasive recovery of CSF without the need for lumbar punctures. Ommaya reservoirs would allow for more practical, and frequent use of CSF to monitor disease and apply liquid biopsy techniques more frequently. More frequent sequencing-based liquid biopsies might add undue cost to treatment. This motivates use of lower-cost techniques such as ddPCR as well as investigation of cost-effective sequencing approaches like single-use Oxford Nanopore flow-cells (91).
JW designed the original paper. JW, KR, VJ, and CB were responsible for initial manuscript generation and editing. JW was responsible for final manuscript preparation. CK provided funding and editing. All authors contributed to the article and approved the submitted version.
JW is supported by the National Institutes of Health under award number T32HL749, the University of Michigan Chad Carr Pediatric Brain Tumor Center, Catching Up With Jack, and the Pediatric Brain Tumor Foundation. CK is supported by NIH/NINDS Grant R01-NS124607 and R01-NS119231 and Department of Defense Grant CA201129P1, the University of Michigan Chad Carr Pediatric Brain Tumor Center, the ChadTough Defeat DIPG Foundation, the DIPG Collaborative, Catching Up With Jack, The Pediatric Brain Tumor Foundation, The Yuvaan Tiwari Memorial Foundation, The Morgan Behen Golf Classic, and the Michael Miller Memorial Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the editors and reviewers for their work. Figure 1 was created using BioRender.com.
1. Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro-Oncol (2020) 22(12 Suppl 2):iv1–96. doi: 10.1093/neuonc/noaa200
2. Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, et al. Regression of Glioblastoma After Chimeric Antigen Receptor T-Cell Therapy. N Engl J Med (2016) 375(26):2561–9. doi: 10.1056/NEJMoa1610497
3. Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol Off J Am Soc Clin Oncol (2016) 34(19):2206–11. doi: 10.1200/JCO.2016.66.6552
4. Friedman GK, Johnston JM, Bag AK, Bernstock JD, Li R, Aban I, et al. Oncolytic HSV-1 G207 Immunovirotherapy for Pediatric High-Grade Gliomas. N Engl J Med (2021) 384(17):1613–22. doi: 10.1056/NEJMoa2024947
5. Khasraw M, Reardon DA, Weller M, Sampson JH. PD-1 Inhibitors: Do They Have a Future in the Treatment of Glioblastoma? Clin Cancer Res Off J Am Assoc Cancer Res (2020) 26(20):5287–96. doi: 10.1158/1078-0432.CCR-20-1135
6. Majzner RG, Ramakrishna S, Yeom KW, Patel S, Chinnasamy H, Schultz LM, et al. GD2-CAR T Cell Therapy for H3K27M-Mutated Diffuse Midline Gliomas. Nature (2022) 603:1–10. doi: 10.1038/s41586-022-04489-4
7. Nishihara M, Sasayama T, Kudo H, Kohmura E. Morbidity of Stereotactic Biopsy for Intracranial Lesions. Kobe J Med Sci (2011) 56(4):E148–153.
8. Brandsma D, Stalpers L, Taal W, Sminia P, van den Bent MJ. Clinical Features, Mechanisms, and Management of Pseudoprogression in Malignant Gliomas. Lancet Oncol (2008) 9(5):453–61. doi: 10.1016/S1470-2045(08)70125-6
9. Nishino M. Tumor Response Assessment for Precision Cancer Therapy: Response Evaluation Criteria in Solid Tumors and Beyond. Am Soc Clin Oncol Educ Book (2018) 38):1019–29. doi: 10.1200/EDBK_201441
10. Tie J. Tailoring Immunotherapy With Liquid Biopsy. Nat Cancer (2020) 1(9):857–9. doi: 10.1038/s43018-020-00113-4
11. Jia W, Gao Q, Han A, Zhu H, Yu J. The Potential Mechanism, Recognition and Clinical Significance of Tumor Pseudoprogression After Immunotherapy. Cancer Biol Med (2019) 16(4):655–70. doi: 10.20892/j.issn.2095-3941.2019.0144
12. Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized Circulating Tumor DNA Analysis as a Predictive Biomarker in Solid Tumor Patients Treated With Pembrolizumab. Nat Cancer (2020) 1(9):873–81. doi: 10.1038/s43018-020-0096-5
13. Seymour L, Bogaerts J, Perrone A, Ford R, Schwartz LH, Mandrekar S, et al. iRECIST: Guidelines for Response Criteria for Use in Trials Testing Immunotherapeutics. Lancet Oncol (2017) 18(3):e143–52. doi: 10.1016/S1470-2045(17)30074-8
14. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol (Berl) (2016) 131(6):803–20. doi: 10.1007/s00401-016-1545-1
15. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncol (2021) 23(8):1231–51. doi: 10.1093/neuonc/noab106
16. Minati R, Perreault C, Thibault P. A Roadmap Toward the Definition of Actionable Tumor-Specific Antigens. Front Immunol (2020) 11:583287. doi: 10.3389/fimmu.2020.583287
17. Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, et al. Tracking Tumour Evolution in Glioma Through Liquid Biopsies of Cerebrospinal Fluid. Nature (2019) 565(7741):654–8. doi: 10.1038/s41586-019-0882-3
18. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med (2014) 6(224):224ra24. doi: 10.1126/scitranslmed.3007094
19. Cantor E, Wierzbicki K, Tarapore RS, Ravi K, Thomas C, Cartaxo R, et al. Serial H3K27M Cell-Free Tumor DNA (cf-tDNA) Tracking Predicts ONC201 Treatment Response and Progression in Diffuse Midline Glioma. Neuro-Oncol (2022), noac030. doi: 10.1093/neuonc/noac030
20. Jensen TJ, Goodman AM, Kato S, Ellison CK, Daniels GA, Kim L, et al. Genome-Wide Sequencing of Cell-Free DNA Identifies Copy-Number Alterations That Can Be Used for Monitoring Response to Immunotherapy in Cancer Patients. Mol Cancer Ther (2019) 18(2):448–58. doi: 10.1158/1535-7163.MCT-18-0535
21. Zhang H, Yuan F, Qi Y, Liu B, Chen Q. Circulating Tumor Cells for Glioma. Front Oncol (2021) 11:607150. doi: 10.3389/fonc.2021.607150
22. Krol I, Castro-Giner F, Maurer M, Gkountela S, Szczerba BM, Scherrer R, et al. Detection of Circulating Tumour Cell Clusters in Human Glioblastoma. Br J Cancer (2018) 119(4):487–91. doi: 10.1038/s41416-018-0186-7
23. Sindeeva OA, Verkhovskii RA, Sarimollaoglu M, Afanaseva GA, Fedonnikov AS, Osintsev E, et al. New Frontiers in Diagnosis and Therapy of Circulating Tumor Markers in Cerebrospinal Fluid In Vitro and In Vivo. Cells (2019) 8(10):1195. doi: 10.3390/cells8101195
24. Mouliere F, Smith CG, Heider K, Su J, van der Pol Y, Thompson M, et al. Fragmentation Patterns and Personalized Sequencing of Cell-Free DNA in Urine and Plasma of Glioma Patients. EMBO Mol Med (2021) 13(8):e12881. doi: 10.15252/emmm.202012881
25. Müller Bark J, Kulasinghe A, Chua B, Day BW, Punyadeera C. Circulating Biomarkers in Patients With Glioblastoma. Br J Cancer (2020) 122(3):295–305. doi: 10.1038/s41416-019-0603-6
26. Mouliere F, Mair R, Chandrananda D, Marass F, Smith CG, Su J, et al. Detection of Cell-Free DNA Fragmentation and Copy Number Alterations in Cerebrospinal Fluid From Glioma Patients. EMBO Mol Med (2018) 10(12):e9323. doi: 10.15252/emmm.201809323
27. Wang Z, Jiang W, Wang Y, Guo Y, Cong Z, Du F, et al. MGMT Promoter Methylation in Serum and Cerebrospinal Fluid as a Tumor−Specific Biomarker of Glioma. BioMed Rep (2015) 3(4):543–8. doi: 10.3892/br.2015.462
28. Simonelli M, Dipasquale A, Orzan F, Lorenzi E, Persico P, Navarria P, et al. Cerebrospinal Fluid Tumor DNA for Liquid Biopsy in Glioma Patients’ Management: Close to the Clinic? Crit Rev Oncol Hematol (2020) 146:102879. doi: 10.1016/j.critrevonc.2020.102879
29. Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L, et al. Detection of Tumor-Derived DNA in Cerebrospinal Fluid of Patients With Primary Tumors of the Brain and Spinal Cord. Proc Natl Acad Sci (2015) 112(31):9704–9. doi: 10.1073/pnas.1511694112
30. Stallard S, Savelieff MG, Wierzbicki K, Mullan B, Miklja Z, Bruzek A, et al. CSF H3F3A K27M Circulating Tumor DNA Copy Number Quantifies Tumor Growth and In Vitro Treatment Response. Acta Neuropathol Commun (2018) 6(1):80. doi: 10.1186/s40478-018-0580-7
31. Escudero L, Llort A, Arias A, Diaz-Navarro A, Martínez-Ricarte F, Rubio-Perez C, et al. Circulating Tumour DNA From the Cerebrospinal Fluid Allows the Characterisation and Monitoring of Medulloblastoma. Nat Commun (2020) 11(1):5376. doi: 10.1038/s41467-020-19175-0
32. Piccioni DE, Achrol AS, Kiedrowski LA, Banks KC, Boucher N, Barkhoudarian G, et al. Analysis of Cell-Free Circulating Tumor DNA in 419 Patients With Glioblastoma and Other Primary Brain Tumors. CNS Oncol (2019) 8(2):CNS34. doi: 10.2217/cns-2018-0015
33. Bagley SJ, Nabavizadeh SA, Mays JJ, Till JE, Ware JB, Levy S, et al. Clinical Utility of Plasma Cell-Free DNA in Adult Patients With Newly Diagnosed Glioblastoma: A Pilot Prospective Study. Clin Cancer Res (2020) 26(2):397–407. doi: 10.1158/1078-0432.CCR-19-2533
34. Corcoran RB, Chabner BA. Application of Cell-Free DNA Analysis to Cancer Treatment. N Engl J Med (2018) 379(18):1754–65. doi: 10.1056/NEJMra1706174
35. Nabavizadeh SA, Ware JB, Guiry S, Nasrallah MP, Mays JJ, Till JE, et al. Imaging and Histopathologic Correlates of Plasma Cell-Free DNA Concentration and Circulating Tumor DNA in Adult Patients With Newly Diagnosed Glioblastoma. Neuro-Oncol Adv (2020) 2(1):vdaa016. doi: 10.1093/noajnl/vdaa016
36. Boisselier B, Pérez-Larraya JG, Rossetto M, Labussière M, Ciccarino P, Marie Y, et al. Detection of IDH1 Mutation in the Plasma of Patients With Glioma. Neurology (2012) 79(16):1693–8. doi: 10.1212/WNL.0b013e31826e9b0a
37. Nørøxe DS, Østrup O, Yde CW, Ahlborn LB, Nielsen FC, Michaelsen SR, et al. Cell-Free DNA in Newly Diagnosed Patients With Glioblastoma – A Clinical Prospective Feasibility Study. Oncotarget (2019) 10(43):4397–406. doi: 10.18632/oncotarget.27030
38. Nassiri F, Chakravarthy A, Feng S, Shen SY, Nejad R, Zuccato JA, et al. Detection and Discrimination of Intracranial Tumors Using Plasma Cell-Free DNA Methylomes. Nat Med (2020) 26(7):1044–7. doi: 10.1038/s41591-020-0932-2
39. Heid CA, Stevens J, Livak KJ, Williams PM. Real Time Quantitative PCR. Genome Res (1996) 6(10):986–94. doi: 10.1101/gr.6.10.986
40. Taylor SC, Laperriere G, Germain H. Droplet Digital PCR Versus qPCR for Gene Expression Analysis With Low Abundant Targets: From Variable Nonsense to Publication Quality Data. Sci Rep (2017) 7(1):2409. doi: 10.1038/s41598-017-02217-x
41. Sedlak RH, Kuypers J, Jerome KR. A Multiplexed Droplet Digital PCR Assay Performs Better Than qPCR on Inhibition Prone Samples. Diagn Microbiol Infect Dis (2014) 80(4):285–6. doi: 10.1016/j.diagmicrobio.2014.09.004
42. Coccaro N, Tota G, Anelli L, Zagaria A, Specchia G, Albano F. Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies. Int J Mol Sci (2020) 21(9):3141. doi: 10.3390/ijms21093141
43. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal Chem (2011) 83(22):8604–10. doi: 10.1021/ac202028g
44. Li D, Bonner ER, Wierzbicki K, Panditharatna E, Huang T, Lulla R, et al. Standardization of the Liquid Biopsy for Pediatric Diffuse Midline Glioma Using ddPCR. Sci Rep (2021) 11(1):5098. doi: 10.1038/s41598-021-84513-1
45. Buermans HPJ, den Dunnen JT. Next Generation Sequencing Technology: Advances and Applications. Biochim Biophys Acta BBA - Mol Basis Dis (2014) 1842(10):1932–41. doi: 10.1016/j.bbadis.2014.06.015
46. Zhao Z, Zhang C, Li M, Shen Y, Feng S, Liu J, et al. Applications of Cerebrospinal Fluid Circulating Tumor DNA in the Diagnosis of Gliomas. Jpn J Clin Oncol (2020) 50(3):325–32. doi: 10.1093/jjco/hyz156
47. Jovčevska I. Next Generation Sequencing and Machine Learning Technologies Are Painting the Epigenetic Portrait of Glioblastoma. Front Oncol (2020) 10:798. doi: 10.3389/fonc.2020.00798.
48. Slatko BE, Gardner AF, Ausubel FM. Overview of Next Generation Sequencing Technologies. Curr Protoc Mol Biol (2018) 122(1):e59. doi: 10.1002/cpmb.59
49. Hughes GM, Gang L, Murphy WJ, Higgins DG, Teeling EC. Using Illumina Next Generation Sequencing Technologies to Sequence Multigene Families in De Novo Species. Mol Ecol Resour (2013) 13(3):510–21. doi: 10.1111/1755-0998.12087
50. Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A Tale of Three Next Generation Sequencing Platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq Sequencers. BMC Genomics (2012) 13(1):341. doi: 10.1186/1471-2164-13-341
51. Thys K, Verhasselt P, Reumers J, Verbist BMP, Maes B, Aerssens J. Performance Assessment of the Illumina Massively Parallel Sequencing Platform for Deep Sequencing Analysis of Viral Minority Variants. J Virol Methods (2015), 221:29–38. doi: 10.1016/j.jviromet.2015.04.022
52. Liang J, Zhao W, Lu C, Liu D, Li P, Ye X, et al. Next-Generation Sequencing Analysis of ctDNA for the Detection of Glioma and Metastatic Brain Tumors in Adults. Front Neurol (2020) 11:544. doi: 10.3389/fneur.2020.00544.
53. Dai P, Wu LR, Chen SX, Wang MX, Cheng LY, Zhang JX, et al. Calibration-Free NGS Quantitation of Mutations Below 0.01% VAF. Nat Commun (2021) 12(1):6123. doi: 10.3389/fneur.2020.00544
54. Abascal F, Harvey LMR, Mitchell E, Lawson ARJ, Lensing SV, Ellis P, et al. Somatic Mutation Landscapes at Single-Molecule Resolution. Nature (2021) 593(7859):405–10. doi: 10.1038/s41586-021-03477-4
55. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and Quantification of Rare Mutations With Massively Parallel Sequencing. Proc Natl Acad Sci USA (2011) 108(23):9530–5. doi: 10.1073/pnas.1105422108
56. Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting Ultralow-Frequency Mutations by Duplex Sequencing. Nat Protoc (2014) 9(11):2586–606. doi: 10.1038/nprot.2014.170
57. Wang Y, Yang Q, Wang Z. The Evolution of Nanopore Sequencing. Front Genet (2015) 5:449. doi: 10.3389/fgene.2014.00449
58. Bruzek AK, Ravi K, Muruganand A, Wadden J, Babila CM, Cantor E, et al. Electronic DNA Analysis of CSF Cell-Free Tumor DNA to Quantify Multi-Gene Molecular Response in Pediatric High-Grade Glioma. Clin Cancer Res (2020) 26(23):6266–76. doi: 10.1158/1078-0432.CCR-20-2066
59. Marcozzi A, Jager M, Elferink M, Straver R, van Ginkel JH, Peltenburg B, et al. Accurate Detection of Circulating Tumor DNA Using Nanopore Consensus Sequencing. NPJ Genomic Med (2021) 6(1):106. doi: 10.1038/s41525-021-00272-y
60. Volden R, Palmer T, Byrne A, Cole C, Schmitz RJ, Green RE, et al. Improving Nanopore Read Accuracy With the R2C2 Method Enables the Sequencing of Highly Multiplexed Full-Length Single-Cell cDNA. Proc Natl Acad Sci USA (2018) 115(39):9726–31. doi: 10.1073/pnas.1806447115
61. Wilson BD, Eisenstein M, Soh HT. High-Fidelity Nanopore Sequencing of Ultra-Short DNA Targets. Anal Chem (2019) 91(10):6783–9. doi: 10.1021/acs.analchem.9b00856
62. Izquierdo E, Proszek P, Pericoli G, Temelso S, Clarke M, Carvalho DM, et al. Droplet Digital PCR-Based Detection of Circulating Tumor DNA From Pediatric High Grade and Diffuse Midline Glioma Patients. Neuro-Oncol Adv (2021) 3(1):vdab013. doi: 10.1093/noajnl/vdab013
63. Juratli TA, Stasik S, Zolal A, Schuster C, Richter S, Daubner D, et al. TERT Promoter Mutation Detection in Cell-Free Tumor-Derived DNA in Patients With IDH Wild-Type Glioblastomas: A Pilot Prospective Study. Clin Cancer Res (2018) 24(21):5282–91. doi: 10.1158/1078-0432.CCR-17-3717
64. Martínez-Ricarte F, Mayor R, Martínez-Sáez E, Rubio-Pérez C, Pineda E, Cordero E, et al. Molecular Diagnosis of Diffuse Gliomas Through Sequencing of Cell-Free Circulating Tumor DNA From Cerebrospinal Fluid. Clin Cancer Res Off J Am Assoc Cancer Res (2018) 24(12):2812–9. doi: 10.1158/1078-0432.CCR-17-3800
65. Ye Z, Chatterton Z, Pflueger J, Damiano JA, McQuillan L, Simon Harvey A, et al. Cerebrospinal Fluid Liquid Biopsy for Detecting Somatic Mosaicism in Brain. Brain Commun (2021) 3(1):fcaa235. doi: 10.1093/braincomms/fcaa235
66. Fujioka Y, Hata N, Akagi Y, Kuga D, Hatae R, Sangatsuda Y, et al. Molecular Diagnosis of Diffuse Glioma Using a Chip-Based Digital PCR System to Analyze IDH, TERT, and H3 Mutations in the Cerebrospinal Fluid. J Neurooncol (2021) 152(1):47–54. doi: 10.1007/s11060-020-03682-7
67. Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain Tumor Mutations Detected in Cerebral Spinal Fluid. Clin Chem (2015) 61(3):514–22. doi: 10.1373/clinchem.2014.235457
68. Muralidharan K, Yekula A, Small J, Rosh Z, Kang K, Wang L, et al. TERT Promoter Mutation Analysis for Blood-Based Diagnosis and Monitoring of Gliomas. Clin Cancer Res (2020) 27:clincanres.3083.2020. doi: 10.1101/2020.09.03.20135236
69. Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S, et al. Evaluating Cancer of the Central Nervous System Through Next-Generation Sequencing of Cerebrospinal Fluid. J Clin Oncol (2016) 34(20):2404–15. doi: 10.1200/JCO.2016.66.6487
70. De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal Fluid-Derived Circulating Tumour DNA Better Represents the Genomic Alterations of Brain Tumours Than Plasma. Nat Commun (2015) 6:8839. doi: 10.1038/ncomms9839
71. Duan H, Hu J-L, Chen Z-H, Li J-H, He Z-Q, Wang Z-N, et al. Assessment of Circulating Tumor DNA in Cerebrospinal Fluid by Whole Exome Sequencing to Detect Genomic Alterations of Glioblastoma. Chin Med J (Engl) (2020) 133(12):1415–21. doi: 10.1097/CM9.0000000000000843
72. Miller AM, Szalontay L, Bouvier N, Hill K, Ahmad H, Rafailov J, et al. Next-Generation Sequencing of Cerebrospinal Fluid for Clinical Molecular Diagnostics in Pediatric, Adolescent and Young Adult (AYA) Brain Tumor Patients. Neuro-Oncol (2022). doi: 10.1093/neuonc/noac035/6527231
73. Schwaederle M, Husain H, Fanta PT, Piccioni DE, Kesari S, Schwab RB, et al. Detection Rate of Actionable Mutations in Diverse Cancers Using a Biopsy-Free (Blood) Circulating Tumor Cell DNA Assay. Oncotarget (2016) 7(9):9707–17. doi: 10.18632/oncotarget.7110
74. Panditharatna E, Kilburn LB, Aboian MS, Kambhampati M, Gordish-Dressman H, Magge SN, et al. Clinically Relevant and Minimally Invasive Tumor Surveillance of Pediatric Diffuse Midline Gliomas Using Patient Derived Liquid Biopsy. Clin Cancer Res Off J Am Assoc Cancer Res (2018) 24(23):5850–9. doi: 10.1158/1078-0432.CCR-18-1345
75. Birkó Z, Nagy B, Klekner Á, Virga J. Novel Molecular Markers in Glioblastoma—Benefits of Liquid Biopsy. Int J Mol Sci (2020) 21(20):7522. doi: 10.3390/ijms21207522
76. Cabel L, Proudhon C, Romano E, Girard N, Lantz O, Stern M-H, et al. Clinical Potential of Circulating Tumour DNA in Patients Receiving Anticancer Immunotherapy. Nat Rev Clin Oncol (2018) 15(10):639–50. doi: 10.1038/s41571-018-0074-3
77. Guibert N, Jones G, Beeler JF, Plagnol V, Morris C, Mourlanette J, et al. Targeted Sequencing of Plasma Cell-Free DNA to Predict Response to PD1 Inhibitors in Advanced Non-Small Cell Lung Cancer. Lung Cancer (2019) 137:1–6. doi: 10.1016/j.lungcan.2019.09.005
78. Wang Z, Duan J, Cai S, Han M, Dong H, Zhao J, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non–Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol (2019) 5(5):696–702. doi: 10.1001/jamaoncol.2018.7098
79. Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-Based Tumor Mutational Burden as a Predictor of Clinical Benefit in Non-Small-Cell Lung Cancer Patients Treated With Atezolizumab. Nat Med (2018) 24(9):1441–8. doi: 10.1038/s41591-018-0134-3
80. Yang X, Hu Y, Yang K, Wang D, Lin J, Long J, et al. Cell-Free DNA Copy Number Variations Predict Efficacy of Immune Checkpoint Inhibitor-Based Therapy in Hepatobiliary Cancers. J Immunother Cancer (2021) 9(5):e001942. doi: 10.1136/jitc-2020-001942
81. Weiss GJ, Beck J, Braun DP, Bornemann-Kolatzki K, Barilla H, Cubello R, et al. Tumor Cell-Free DNA Copy Number Instability Predicts Therapeutic Response to Immunotherapy. Clin Cancer Res Off J Am Assoc Cancer Res (2017) 23(17):5074–81. doi: 10.1158/1078-0432.CCR-17-0231
82. Pepe F, Pisapia P, Gristina V, Rocco D, Micheli M, Micheli P, et al. Tumor Mutational Burden on Cytological Samples: A Pilot Study. Cancer Cytopathol (2021) 129(6):460–7. doi: 10.1002/cncy.22400
83. Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science (2014) 343(6167):189–93. doi: 10.1126/science.1239947
84. Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DIS, Zairis S, et al. Clonal Evolution of Glioblastoma Under Therapy. Nat Genet (2016) 48(7):768–76. doi: 10.1038/ng.3590
85. Blons H, Garinet S, Laurent-Puig P, Oudart J-B. Molecular Markers and Prediction of Response to Immunotherapy in Non-Small Cell Lung Cancer, an Update. J Thorac Dis (2019) 11(Suppl 1):S25–36. doi: 10.21037/jtd.2018.12.48
86. Wierzbicki K, Ravi K, Franson A, Bruzek A, Cantor E, Harris M, et al. Targeting and Therapeutic Monitoring of H3K27M-Mutant Glioma. Curr Oncol Rep (2020) 22(2):19. doi: 10.1007/s11912-020-0877-0
87. Saenz-Antoñanzas A, Auzmendi-Iriarte J, Carrasco-Garcia E, Moreno-Cugnon L, Ruiz I, Villanua J, et al. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers (2019) 11(7):950. doi: 10.3390/cancers11070950
88. McEwen AE, Leary SES, Lockwood CM. Beyond the Blood: CSF-Derived cfDNA for Diagnosis and Characterization of CNS Tumors. Front Cell Dev Biol (2020) 8:45. doi: 10.3389/fcell.2020.00045
89. Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of Ultra-Rare Mutations by Next-Generation Sequencing. Proc Natl Acad Sci USA (2012) 109(36):14508–13. doi: 10.1073/pnas.1208715109
90. Vitanza NA, Johnson AJ, Wilson AL, Brown C, Yokoyama JK, Künkele A, et al. Locoregional Infusion of HER2-Specific CAR T Cells in Children and Young Adults With Recurrent or Refractory CNS Tumors: An Interim Analysis. Nat Med (2021) 27(9):1544–52. doi: 10.1038/s41591-021-01404-8
Keywords: liquid biopsy, glioma, immunotherapy, cell-free tumor DNA (cf-tDNA), Csf, plasma
Citation: Wadden J, Ravi K, John V, Babila CM and Koschmann C (2022) Cell-Free Tumor DNA (cf-tDNA) Liquid Biopsy: Current Methods and Use in Brain Tumor Immunotherapy. Front. Immunol. 13:882452. doi: 10.3389/fimmu.2022.882452
Received: 23 February 2022; Accepted: 14 March 2022;
Published: 06 April 2022.
Edited by:
Lijie Zhai, Northwestern Medicine, United StatesReviewed by:
Albino Eccher, Integrated University Hospital Verona, ItalyCopyright © 2022 Wadden, Ravi, John, Babila and Koschmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jack Wadden, d2FkZGVuQHVtaWNoLmVkdQ==; Carl Koschmann, Y2tvc2NobWFAdW1pY2guZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.