Abstract
Interferons (IFNs) are a group of cellular proteins with critical roles in the regulation of immune responses in the course of microbial infections. Moreover, expressions of IFNs are dysregulated in autoimmune disorders. IFNs are also a part of immune responses in malignant conditions. The expression of these proteins and activities of related signaling can be influenced by a number of non-coding RNAs. IFN regulatory factors (IRFs) are the most investigated molecules in the field of effects of non-coding RNAs on IFN signaling. These interactions have been best assessed in the context of cancer, revealing the importance of immune function in the pathoetiology of cancer. In addition, IFN-related non-coding RNAs may contribute to the pathogenesis of neuropsychiatric conditions, systemic sclerosis, Newcastle disease, Sjögren’s syndrome, traumatic brain injury, lupus nephritis, systemic lupus erythematosus, diabetes mellitus, and myocardial ischemia/reperfusion injury. In the current review, we describe the role of microRNAs and long non-coding RNAs in the regulation of IFN signaling.
Introduction
Being firstly recognized as antiviral factors that interfere with viral replication (1), interferons (IFNs) are a group of cellular proteins classified in three families (2). Thirteen IFN-α variants, a single IFN-β and numerous other IFNs (IFN-ϵ, -k, -ω, and -δ) are classified as Type-I IFNs (3, 4). Type II IFN family only includes IFN-γ (5), a protein that potentiates proinflammatory signals through priming macrophages for antimicrobial functions and induction of nitric oxide synthesis and inhibition of the activity of NLRP3 inflammasome (6, 7).
Secretion of IFNs from infected cells can lead to induction of innate immune response resulting in cytokine release and induction of function of natural killer cells and antigen presentation (3, 8). These proteins have critical roles in the regulation of immune responses in the course of microbial infections. Moreover, expressions of IFNs are dysregulated in autoimmune disorders. Based on these roles, the identification of cellular mechanisms of regulation of IFNs has practical significance. Regulation of IFNs expressions is accomplished by different mechanisms, including of binding of regulatory molecules to their 3′ untranslated regions (3′ UTRs). This region contains both AU-rich elements (AREs) and microRNA (miRNA) recognition elements (2). RNA-binding proteins can target AREs and either induce mRNA degradation or stabilize mRNA. Meanwhile, the binding of miRNAs with miRNA recognition elements is involved in the regulation of mRNA translation via the miRNA-induced silencing complex (2). In the current review, we describe the role of miRNAs and long non-coding RNAs (lncRNAs) in the regulation of IFNs.
Interactions Between miRNAs and IFNs
miR-301a has been found to contain a binding site in the 3′-UTR of the Interferon regulatory factor 1 (IRF-1) gene. Through the modulation of the expression of this gene, this miRNA participates in the proliferation of hepatocellular carcinoma cells. Expression of miR-301a has been increased in primary hepatocellular carcinoma tumors and cell lines, parallel with down-regulation of IRF-1. In vitro studies have shown the role of chronic hypoxia in the induction of miR-301a and down-regulation of IRF-1. Moreover, suppression of miR-301a induces cell apoptosis and reduces cell proliferation. Taken together, the regulation of miR-301a on IRF-1 expression is implicated in the pathogenesis of hepatocellular carcinoma (9). Another study in this malignancy has shown that IRF-1 can induce the expression of miR-195 to inhibit CHK1 expression. Up-regulation of IRF-1 or down-regulation of CHK1 induces cell apoptosis and increases PD-L1 expression in hepatocellular carcinoma cells (10).
In lung cancer cells, miR-19 has been shown to influence the expression of IFN-induced genes and MHC class I, signifying the impact of miR-19 in connecting inflammation and carcinogenesis (11). The IRF2-targeting miRNA miR-1290 has also been shown to be up-regulated in lung cancer. Over-expression of miR-1290 has been correlated with lymph node metastasis and advanced clinical stage. miR-1290 could enhance cell proliferation, colony formation, and invasive abilities in lung cancer cells. This miRNA could also promote the expression of cell proliferation-related proteins CDK2 and CDK4 and induce epithelial-mesenchymal transition (EMT) (12). Another study in lung cancer samples has shown up-regulation of IRF6 and down-regulation of miR-320, a miRNA that targets IRF6. IRF6 siRNA or miR-320 mimics could inhibit the growth and migration of lung cancer cells. Taken together, the miR-320/IRF6 axis has been suggested as a molecular axis involved in the pathogenesis of lung cancer (13).
Experiments in squamous cell carcinoma samples and cell liens have shown up-regulation of IRF2-targeting miRNA miR-664. This miRNA has been found to increase tumorigenic behaviors of cells both in vitro and in vivo (14).
Participation of IFN-related miRNAs has also been assessed in the pathoetiology of non-malignant conditions. For instance, the IRF2 targeting miRNA miR-221-3p has been found to be over-expressed in patients with the major depressive disorder compared with normal persons. Notably, serum miR-221-3p levels have been positively correlated with the level of depression. Mechanistically, miR-221-3p can enhance the expression of IFN-α in astrocytes through targeting IRF2. In fact, this miRNA participates in the induction of anti-neuroinflammatory signals by ketamine and paroxetine through the IRF2/IFN-α axis (15).
miR-126 and miR-139-5p are two miRNAs that participate in the dysregulation of plasmacytoid dendritic cells in systemic sclerosis. Expressions of these miRNAs have been correlated with the expression of type I IFN-responsive genes. TLR9 stimulation of plasmacytoid dendritic cells has induced expressions of miR-126 and miR-139-5p in cultures of normal cells as well as those obtained from patients with systemic sclerosis. USP24 has been identified as a target of miR-139-5p (16). Table 1 summarizes the results of investigations that assessed the effects of miRNAs on IFN signaling.
Table 1
| Type of diseases | miRNA | Sample | Cell Line | Target, Pathway | Discussion | Ref |
|---|---|---|---|---|---|---|
| Nasopharyngeal carcinoma | miR-9 | – | CNE2, 5–8F | IFI44L, PSMB8, IRF5, PSMB10, IFI27, IFIT2, TRAIL, IFIT1 | miR-9 modulates levels of IFN-induced genes and MHC class I. | (17) |
| Hepatocellular Carcinoma (HCC) | miR-301a (Up) | 20 pairs of HCC and ANTs | Huh7, Hep3B, HepG2, Hepa1-6 | IRF-1, Caspase-3 | miR-301a via down-regulating IRF-1 could induce HCC. | (9) |
| HCC | miR-195 (–) | 30 pairs of HCC and ANTs; WTB6 mice | Hepa1-6, Huh-7, Hep3B, HepG2 | IRF-1, IFN-γ, CHK1, STAT3 | IRF-1 via modulating miR-195 by down-regulating CHK1 could up-regulate apoptosis in HCC. | (10) |
| HCC | miR-146a (Up) | – | PLC/PRF/5 | INF-α, SMAD4, STAT1/2 | miR-146a could suppress the sensitivity to INF-α in HCC cells. | (18) |
| Lung Cancer (LC) | miR-19 (–) | – | CNE2, HONE1, A549, HCC827 | IRF-1/7/9, IFI-6/27/35, HLA-B/F/G | miR-19 via regulating the expression of interferon could affect the expression of IFN-induced genes and MHC class I in human lung cancer cells. | (11) |
| Non-Small Cell Lung Cancer (NSCLC) | miR-1290 (Up) | 41 pairs of NSCLC and ANTs | A549, H1299, SPC-A1, H1970, H460, BEAS-2B | IRF-2, CK2/4, E/N-cadherin | Overexpression of miR-1290 by targeting IRF2 could contribute to cell proliferation and invasion of NSCLC. | (12) |
| NSCLC | miR-320 (Down) | 21 pairs of NSCLC and ANTs | A549, NCI-H2170 | IRF-6 | miR-320 via targeting IRF-6 could affect pathogenesis of NSCLC. | (13) |
| Cutaneous Squamous Cell Carcinoma (cSCC) | miR-664 (Up) | Athymic nude mice | HSC-1, A431, HSC-5, HaCaT | IRF-2 | miR-664 via suppressing IRF-2 could function as an oncogene in cSCC. | (14) |
| Cervical Cancer (CC) | miR-587 (Up) | 41 pairs of CC and ANTs, nude mice | Ect1/E6E7, HeLa, SiHa, CaSki, C-33A | IRF-6, Cyclin-D1, CDK4 | miR-587 by repressing IRF6 could promote CC. | (19) |
| Gastric Cancer (GC) | miR-19a, miR-18a (–) | 20 pairs of GC tissues and ANTs; BALB/c nude mice | MKN45, AGS, SGC7901, GES1 | IFN-γ, IRF-1, Axin2, SMAD2, Wnt/β-catenin | IRF-6 by regulating MIR17HG-miR-18a/19a axis via Wnt/β-catenin signaling could promote GC metastasis. | (20) |
| Glioblastoma (GBM) | miR-203a (Down) | NSG mice | MT330, SJG2 | IFN-α, IFN-β, IFN-λ1, IFI-1/6, IFT20, p65, NF-κB, STAT1-3 | miR-203a via an ATM-dependent interferon response pathway could suppress GBM. | (21) |
| Osteosarcoma | miR-4295 (Up) | 15 pairs of OS and ANTs | MG-63, Saos-2, hMSC | IRF-1 | miR-4295 via targeting IRF1 could promote cell proliferation, migration and invasion. | (22) |
| Systemic Lupus Erythematosus (SLE) | miR-146 | WBCs from patients with SLE | THP-1 cells | Type I IFN inhibits miR-146a maturation via increasing expression of MCPIP1. | (23) | |
| Major Depressive Disorder (MDD) | miR-221-3p (Up) | (n=64) perioperative patients | Astrocytes | IRF-2, IFN-α, NF-κB | miR-221-3p via targeting IRF2 could up-regulate IFN-α expression in MDD patients. | (15) |
| Systemic Sclerosis | miR-126, miR-139-5p (Up) | Blood samples of SS patients (n=72) and healthy control (n=26) | pDCs | IFI-6, IFIT1, CXCL10, USP24, TLR-7/8/9 | miR-126 and miR-139-5p via TLR9-mediated response and IFN signaling could regulate the activation of plasmacytoid dendritic cells. | (16) |
| Newcastle Disease (ND) | gga-miR-455-5p (Down) | – | 293T, BHK-21 | IFN-I, SOCS3 | gga-miR-455-5p via targeting cellular suppressors of SOCS3 could suppress ND virus replication. | (24) |
| Sjögren’s Syndrome (SS) | miR-1248 (–) | – | phSG | IFN-β, IRF-1/9, IFIT1, IFI-6/44, IFIH1, MX1, JAK-1/2, STAT-1/2/3 | miR-1248 could activate IFN-β via the direct association with both AGO2 and RIG-I. | (25) |
| Traumatic Brain Injury (TBI) | miR-155 (-) | C57BL/6 mice | IFN-I, IFN-α2/4/5, IL-6, IFN-β1, IRF-1, TNF-α, SOCS1C | Up-regulation of miR-155 after brain injury promotes IFN-I to exert a neuroprotective function. | (26) | |
| Edwardsiella Tarda TX1 (E. tarda TX1) | pol-miR-194a (Up) | Fish | FG-9307, 293T | IFN-I, IRF-7 | pol-miR-194a via targeting IRF7 could participate in the regulation of flounder immune response and microbial infection. | (27) |
| – | miR-17 (-) | C57BL/6J mice | VSMCs, RAVSMCs | IRF-9 | miR-17 knockdown via up-regulating IRF-9 expression could promote vascular smooth muscle cell phenotypic modulation. | (28) |
| – | miR-155 (-) | – | EPC, BHK-21 | IFN-I, PIAS4a | Overexpression of miR-155 via targeting IFN-I could contribute to antiviral response in EPC. | (29) |
| – | miR-181a, miR-30a (-) | – | U937, 293T, monocytes, MDM, MDDC | IFN-I/II, IFN-α, IFN-β, IFN-γ, ERK, STAT-1 | Interferons via down-regulating miR-181a and miR-30a could induce the expression of SAMHD1 in monocytes. | (30) |
| – | miR-155, miR-155* (–) | – | HeLa, PDC | IFN-I, IFN-α, IFN-β, NF-κB, PI3K, AKT, p38 | miR-155 in cooperation with its star-form partner miR-155* could regulate IFN-I production. | (31) |
| – | Bta-miR-204 (Down) | bEEC | – | IFN-τ, BoLA, PD-L1/2 | IFN-τ by down-regulating bta-miR-204 could enhance the expression and function of BoLA. | (32) |
| – | miR-30c-5p (–) | Vero E6, | IFN-I/III, IFN-λ, IFIT1, ISG-15, SOCS-1 | The coronavirus PEDV via the miR-30c-5p/SOCS1 axis could evade type III interferon response. | (33) | |
| – | miR-744 (–) | – | RMCs | PTP1B, INF-I, CCL2/5, CXCL10, IL6, ERK, p38, MX1, IFIT3, TYK2, STAT1/3, JAK1, NF-κB | miR-744 by targeting PTP1B could enhance the INF-I signaling pathway in primary human renal mesangial cells (RMCs). | (34) |
| – | miR-155 (Up) | C57BL/6 mice | 293T, RAW264.7, BMMs | SOCS1, IFN-β, MITF, TRAP | INF-β-induced miR-155 by targeting SOCS1 and MITF could inhibit osteoclast differentiation. | (35) |
| – | miR-221 (–) | – | H69, HIBEpiC | ICAM-1, IFN-γ, PRRSV, p65 | miR-221 via targeting ICAM-1 translation regulating IFN-γ could induce ICAM-1 expression in human cholangiocytes. | (36) |
The effects of miRNAs on IFN signaling (ANT, adjacent normal tissue).
miR-26a is an example of miRNAs that participate in the regulation of host immune responses during viral infections. Expression of this miRNA is increased upon infection with Feline Herpes Virus 1 (FHV-1). This virus could induce the expression of miR-26a through a cGAS-dependent route since down-regulation of cellular cGAS could result in blockage of poly (dA:dT) or FHV-1-induced expression of miR-26a. Functional studies have shown the impact of miR-26a in the induction of STAT1 phosphorylation and enhancement of type I IFN signals, which inhibit viral replication. In fact, miR-26a directly targets SOCS5 mRNA. SOCS5 silencing has led to an increase in STAT1 phosphorylation and induction of antiviral responses mediated by type I IFNs (37).
Another study has shown a time-dependent down-regulation of miR-155 upon infection with the dengue virus. Exogenous up-regulation of this miRNA could limit replication of the dengue virus in vitro, indicating that down-regulation of miR-155 has a beneficial effect for replication of this virus. The results of in vivo experiments have also confirmed the impact of miR-155 in protection against the life-threatening effect of dengue virus infection. This activity of miR-155 has been shown to be exerted through targeting Bach1, and subsequent activation of the HO-1-mediated suppression of NS2B/NS3 protease activity of dengue virus. Taken together, modulation of miR-155 expression has been suggested as a therapeutic option for the management of dengue virus infection (38). miR-218 is another miRNA that can regulate host responses to viral infections since its down-regulation by porcine reproductive and respiratory syndrome virus can facilitate replication of this virus through suppression of type I IFN responses (39). Table 2 shows the effects of miRNA on IFN signaling in the context of viral infections. Figure 1 illustrates the aberrant expression of various miRNAs, which adversely affect the IFN signaling pathway triggering several kinds of human diseases and malignancies as well as their role in the context of viral infections.
Table 2
| Virus | miRNA | Sample | Cell Line | Target | Discussion | Ref |
|---|---|---|---|---|---|---|
| Infectious stress | miR-22 | Mir22-KO mice | – | – | miR-22 enhances the IFN response to viral infections. | (40) |
| Infection with influenza virus | miR-144 | Wild-type mice | – | TRAF6-IRF7 | miR-144 diminishes host responses to the influenza virus. | (41) |
| Feline Herpes Virus 1(FHV-1) | miR-26a (Up) | – | F81, 293T | IFN-α, IFN-β, ISG-15, SOCS5, STAT-1 | miR-26a by targeting SOCS5 and promoting Type I IFN signals could inhibit FHV-1 replication. | (37) |
| Human Herpes Simplex Virus Type 1 (HSV-1) | miR-23a (–) | – | HeLa | IRF-1, RSAD2, EGFP, Myc | miR-23a via suppression of IRF-1 could facilitate the replication of HSV-1. | (42) |
| Dengue Virus (DENV) | miR-155 (Down) | Breeder ICR mice | Huh-7 | HO-1, IFN-α-2/5/17, BACH1, Nrf2, OAS-1/2/3 | miR-155 by inducing HO-1-mediated antiviral interferon responses could inhibit DENV replication. | (38) |
| Porcine Reproductive & Respiratory Syndrome Virus (PRRSV) | miR-218, miR339-5p, miR-99b, miR-365-5p, miR-378, miR-345, miR-27b-3p | SPF pig | PAMs, marc-145, Vero-E6, ST, 293T | IFN-I, IFN-β, SOCS3 | Downregulation of miR-218 by PRRSV could facilitate viral replication via repressing of type I IFN responses. | (39) |
| PRRSV | miR-30c (–) | – | PAM, Marc-145, U4A | IFN-I, IFNAR-2, ISG-15, OAS-1, JAK-1 | miR-30c via targeting IFNAR-2 could promote type 2 PRRSV infection. | (43) |
| PRRSV | miR-382-5p (Up) | – | MARC-145, 293T, BHK-21 | IFN-I, IFN-β, HSP60, MAVS, IRF-3, TBK1 | miR-382-5p by negatively regulating the induction of IFN-I could promote PRRSV replication. | (44) |
| PRRSV2 | miR-541-3p (Up) | MARC-145, MA-104, 293T | IRF7, | miR-541-3p via IRF7 could promote the replication of PRRSV2. | (45) | |
| Influenza A virus, TMEV | miR-673 (–) | Dgcr8-/- mouse, Dicer+/+ mouse | NIH3T3, ESC | IFN-β1, MAVS | During pluripotency, an interaction between MAVS (mitochondrial antiviral signaling protein) and miR-673 could act as a switch to suppress the antiviral IFN. | (46) |
| Influenza Virus A/WSN/33 (H1N1) | miR-302a (–) | C57BL/6 mice | A549, THP-1, 293T, MLE-12, H9 | IFN-β, TNF-α, IRF-5, CCL-2/5, IL-6/8, M1, NP, NF-κB | miR-302a via targeting IRF5 expression and cytokine storm induction could suppress IAV. | (47) |
| H1N1 | miR-93 (Down) | C57BL/6 mice | AT2, MLE12, A549, 293T, Murine T-cells, Murine B-cells, B-cells, NK cells | IFN-I, IFN-β, IRF-3, IL-6/8/10, NF-κB, ISG15, OAS1, RIG-I, p38/65, ERK, JAK-1/2 | Inhibition of miR-93 by up-regulating JAK-1 could promote interferon effector signaling to suppress influenza A infection. | (48) |
| Influenza A virus (IAV) H5N1 | miR-21-3p (Down) | 26 H5N1-infected patients serum samples and 13 serum samples from normal persons | A549 | IFN-I, FGF2, IFN-β, IFN-α, MxA, OAS | miR-21-3p by refraining IFN-I response could modulate FGF2 to facilitate influenza A virus H5N1 replication. | (49) |
| Foot & Mouth Disease Virus (FMDV) | miR-103, miR-107 (Down) | 20 pairs of blood samples from patients with enterovirus 71 (EV71) and normal blood samples | VERO, RD | IFN-I, IFN-α, IFN-β, SOCS3, STAT3 | miR-103/miR-107 by regulating SOCS3/STAT3 pathway could inhibit EV71 replication and facilitate IFN-I response. | (50) |
| FMDV | miR-4334-5p (Up) | – | PK-15, BHK-21 | IFN-β, TNF-α, OAS, ISG54, ID1, VP1 | miR-4334-5p by suppressing IFN pathways via direct targeting ID1 could facilitate FMDV propagation. | (51) |
| HIV-1 | miR-128 (–) | – | HeLa, 293T, THP-1, Jurkat | INF I, IFN-α, TNPO3, | IFN-I via enhancing miR-128 by targeting TNPO3 mRNA could modulate HIV-1 Replication. | (52) |
| Infectious Bursal Disease Virus (IBDV) | gga-miR-27b-3p (Up) | – | DF-1 | IFN-I, IFN-β, IRF3, NF-κB, SOCS3, SOC6, STAT-1 | gga-miR-27b-3p via targeting cellular suppressors of SOCS3 and SOCS6 could enhance type I IFN signals and inhibit replication of IBDV. | (53) |
| IBDV | gga-miR-155 (Up) | – | DF-1 | TANK, SOCS1, IFN-I, chIRF3 | gga-miR-155 via targeting SOCS1, and TANK could enhance IFN-I and suppress IBDV. | (54) |
| IBDV | gga-miR-9* (Up) | – | DF-1 | IRF-2, INF-β | gga-miR-9* by targeting IRF-2 to promote IBDV replication could inhibit IFN production in antiviral innate immunity. | (55) |
| Hepatitis C Virus (HCV) | miR-122 (–) | – | Huh7 | INF-α, INF-β, EGFP, SOCS1 | miR-122 via blocking suppressor of SOCS1 could modulate INF-I expression. | (56) |
| Human Papillomavirus 16 (HPV16) | miR-122 (–) | – | SiHa, CaSki, C33A | OAS-1, MxA, pmCherry-E6, IFN-α, IFN-β, STAT1, SOCS1 | miR-122 via blocking suppressor of cytokine signaling 1 in SiHa cells could inhibit HPV E6 gene and enhance interferon signaling. | (57) |
| Human Cytomegalovirus (HCMV) | Hcmv-miR-UL112 (–) | – | PBMCs, K562 | TNF-I, IFNAR, CD107 | Hcmv-miR-UL112 activity by inhibiting INF-I secretion could attenuate NK cells. | (58) |
The effects of miRNA on IFN signaling in the context of viral infections.
Figure 1

A schematic diagram of the interaction between several miRNAs and interferons in causing various human diseases. Mounting evidence has demonstrated that miRNAs could have an important contribution to the regulation of expression of IFN-induced genes. Aberrant expression of such ncRNAs could lead to various human diseases such as major depressive disorder, Sjögren’s Syndrome, Systemic Sclerosis as well as different kinds of cancers. As an illustration, a recent study has detected that overexpression of miR-301a could promote hepatocellular carcinoma via directly targeting IRF1 (9). Moreover, another research has figured out that miR-587 could play a key role in the progression of cervical cancer by down-regulating the expression of IRF6 (19). In addition, another finding has denoted that miR-1248 via activating the expression levels of IFN-β, IRF1/9, MX1, JAK-1/2, STAT-1/2, TYK2 as well as direct association with both AGO2 and RIG-I could have a crucial role in Sjögren’s syndrome (25). Furthermore, miR-26a could suppress feline herpesvirus 1 (FHV-1) replication via targeting SOCS5 and up-regulating the expression levels of IFN-α, IFN-β, ISG-15, STAT-1, and IFITM1 in type I IFN signaling (37). Blue lines indicate the positive regulatory effect among miRNAs and their targets, and crimson lines depict negative effects among them. All information regarding the role of these miRNAs in the modulation of the IFN signaling cascade in various types of human diseases and cancers can be seen in Tables 1 and 2.
Interactions Between lncRNAs and IFNs
LncRNAs are a group of regulatory non-coding RNAs that share several characteristics with mRNAs, but lacking open reading frames. They participate in epigenetic regulation of gene expression through modulation of histone or DNA marks as well as regulation of the stability of RNAs and interacting with regulatory proteins (59).
Several lines of evidence suggest that lncRNAs include an important subgroup of the IFN target genes. Additionally, the IFN response has been shown to be regulated by several lncRNAs encoded by host or pathogens (60). Kambara et al. have identified approximately 200 lncRNAs whose expressions are induced by IFN in primary human hepatocytes (61). Notably, among them has been lncRNA-CMPK2/NRIR which has exhibited an intense induction after IFN stimulation in various human and mouse cells. This lncRNA is located near the protein-coding IFN-stimulated gene CMPK2. Expression of this lncRNA has been shown to be induced in a JAK-STAT-dependent manner. Silencing of lncRNA-CMPK2/NRIR has resulted in a significant decrease in HCV replication in IFN-induced hepatocytes, implying the role of this lncRNA in the modulation of antiviral effects of IFN (61).
NRAV is another lncRNA whose expression is regulated by IFNs. Microarray analyses in cells overexpressing this lncRNA has shown down-regulation of several ISGs. NRAV has been shown to be able to partially preclude IFN-induced expression of its target ISGs, possibly via affecting its transcription or through epigenetic mechanisms (62).
IFNG-antisense-1 (IFNG-AS1) is another lncRNA that participates in the regulation of IFN responses. This lncRNA is located downstream of the IFNG locus. Expression of IFNG-AS1 is strongly correlated with expression of IFNG (63, 64). CD4+ and CD8+ T cells as well as NK cells express this lncRNA. IFNG-AS1 expression by CD4+ T cells depends on two transcription factors being involved in Th1 polarization, namely STAT4 and TBX21 (65).
AFAP1-AS1, lncMX1-215, linc00513, BANCR, IFITM4P, LUCAT1, NEAT1, MALAT1, DANCR, NRIR, and FIRRE are among lncRNAs whose interactions with IFN signaling have been assessed (Table 3). For instance, the up-regulated lncRNA AFAP1-AS1 can participate in the invasiveness of lung cancer cells through increased expression of IRF7 and induction of RIG-I-like receptor signals (66). On the other hand, lncMX1-215 is an IFNα-induced lncRNA that can affect the immunosuppressive responses through interfering with H3K27 acetylation (67).
Table 3
| Type of Diseases | LncRNAs | Sample | Cell Line | Target | Discussion | Ref |
|---|---|---|---|---|---|---|
| NSCLC | AFAP1-AS1 (Up) | NSCLC (n=165), banging lung tumor patients (n=118), health control (n=173) | A549, H1975, H1650, H1395 H12994 | IRF-7, IFN-γ, RIG-I, Th1/2, IL-10/12, Bcl-2, TNF-α, NF-κB | AFAP1-AS1 via upregulating IRF-7 and the RIG-I-like receptor signaling could promote migration and invasion of NSCLC. | (66) |
| Head & Neck Squamous Cell Carcinoma (HNSCC) | lncMX1-215 | 70 HNSCC and 18 normal oral mucosa tissues from patients; BALB/c nude mice | HN4, HN6, HN30, Cal27, SCC4, SCC25, Detroit 562, 293T | IFN-α, H3K27ac, H3k18ac, H3K9ac, Caspase-3 PARP, Snail, STAT-1 | IFN-α-induced lncMX1-215 by interfering with H3K27 acetylation could decrease immunosuppression in HNSCC. | (67) |
| Cryptosporidium Infection | NR_033736 | BV2 mice, | IEC4.1, HCT-8, BV2, RAW264.7 | IFN-α, IFN-β1, IFN-α12/13, IFN-I, ISGF-3, IFI-44, IFIT-1, OAS2/3, IRF-9, H3K4me3, STAT-2 | NR_033736 via regulating IFN-I-mediated gene transcription could induce intestinal epithelial anti-cryptosporidium defense. | (68) |
| Lupus Nephritis (LN) | RP11-2B6.2 (UP) | 22 LN kidney biopsies and 7 control samples, PBMC | HeLa, HK2 | IFN-I, IFI27, IFIT-1/3, ISG, Mx2, OASL, ASO1, CXCL10, JAK1, STAT-1, SOCS1 | RP112B6.2 via targeting the IFN-I by epigenetically inhibiting the expression of SOCS1 could aggravate symptoms of LN disease. | (69) |
| Systemic Lupus Erythematosus (SLE) | linc00513 (–) | 139 SLE patients | Hela, THP-1, PBMCs | IFN-I, IRF-9, OAS-1/2/3, IFI-27/44/44L, ISG-15/20, IFIT-1/3, Mx1/2, XAF1, NF-kB, STAT-1/2 | Overexpression of linc00513 via promoting IFN signaling could play a role in lupus pathogenesis. | (70) |
| Diabetes Mellitus Type 1 | Lnc10 (–) | – | EndoC-βH1 | IFN-I, IFN-γ, IFITM1, IL-1β, STAT-1 | Overexpression of lnc10 via IFN-I could enhance the immune response in pancreatic β-cells. | (71) |
| Myocardial I/R Injury | BANCR (–) | – | iPS cell-derived cardiomyocytes | IFN-β, IFNAR-1, STAT-1/2 | BANCR by targeting STAT-1 could promote IFN-β-induced cardiomyocyte apoptosis. | (72) |
| Infectious Bursal Disease Virus (IBDV) | loc107051710 (–) | – | DF-1 | IRF-8, IFI-1/6, IFN-α, IFN-β, Mx1, IFIT-5, STAT-1/2 | loc107051710 by regulating IRF-8 could promote the production of IFN-α and IFN-β, thereby modulating the antiviral activity of ISGs. | (73) |
| Influenza A Virus (IAV) | IVRPIE (Up) | – | A549, BEAS-2B, MDCK, BHK21 | IFN-β1, ISG, IRF-1, IFIT-1/3, Mx1, ISG-15, IFI44L | IVRPIE via regulating IFN-β1 and ISG expression could promote host antiviral immune responses. | (74) |
| Influenza A Virus (IAV); H1N1, IAV-PR8, IAV- CA04 | ISR (–) | C57BL/6 mice | A549, 293T, NIH/3T3, 4T1, MDCK | IFN-β, IFNAR-1, RIG-I, MxA, ISG-15, OAS2 | ISR could be regulated by RIG-I-dependent signaling; during IAV infection, it could also govern IFN-β production and inhibit viral replication. | (75) |
| Influenza Virus A/WSN/33 (H1N1) | IFITM4P (–) | – | A549, 293T, K562, HeLa, MDCK, Huh7, Mcf7, HepG2 | IFITM-1/2/3, miR-24, Mx1, RIG-I, p65, IL-6 | IFITM4P by acting as a competing endogenous RNA could regulate host antiviral responses. | (76) |
| Influenza Virus A/WSN/33 (H1N1), Sendai Virus (SeV) | Lnc-MxA (–) | – | MDCK, 293T, A549, | IFN-β, RIG-I, MAVS, IRF-3, INFAR-1, p65, ISG-15, MxA | Lnc-MxA by forming RNA-DNA triplexes could inhibit β interferon transcription. | (77) |
| Herpes Simplex Virus 1 (HSV-1), Influenza A Virus (IAV), LPS | LUCAT1 (–) | PBMCs | THP-1, THP-1 KO, hMDDC | IFN-I, IFN-α, IFN-β, IRF-3, IFI1-6, ISG, TNF-α, Mx2, JAK-1/3, STAT1 | LUCAT1 by interacting with STAT1 in the nucleus could limit the transcription of ISGs. | (78) |
| Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) | RP1-20B21.4, RP11-329L6.1, RP11-498C9.3, NEAT1, MALAT1 (–) | Dataset | – | miR-122, miR-122-5p, IRF-9, IFIT-1/2/3, MX1, OAS2/3, IFNL-1 IFNG, JAK, STAT-1 | The SARS-CoV-2 infection could lead to differential expression of lncRNAs. Also, IFN response is involved in SARS-CoV-2 infection. | (79) |
| HIV-1-BAL-HSA | NRIR, MIR3945HG, C8orf3, AC053503.1, AL359551.1 (–)f | – | CD14+ monocytes, MDMs | IFN-α, IFN-ϵ, IFN-γ, IFN-λ, Mx1, IFIT2 | Interferons could mediate the Response of lncRNAs in macrophages in HIV. | (80) |
| Vesicular Stomatitis Virus (VSV), VSV-GFP | lncLrrc55-AS (–) | C57BL/6 mice | RAW264.7, NIH/3T3, 293T, MDCK, MLE12, 3LL, Hepa | IFN-α4, IFN-β, IRF-3, IFN-I, p65, p38, ERK, JAK, STAT-1 | Interferon-inducible cytoplasmic lncLrrc55-AS by strengthening IRF3 phosphorylation could promote antiviral innate responses. | (81) |
| – | GRASLND, NEAT1 (–) | – | ASCs | IFN-II, IFN-α, IFN-β, IFN-γ, IRF-1/2/6, IFI-44/44L, IFNGR-1/2, STAT-1/2 | GRASLND via suppressing the IFN-II pathway could enhance chondrogenesis. | (82) |
| – | BANCR (–) | – | ARPE-19, | IFN-γ, IL-1β, TNF-α, JAK, STAT-1 | IFN-γ by activating the JAK-STAT1 pathway could upregulate the expression of BANCR in retinal pigment epithelial cells. | (83) |
Interactions between lncRNAs and IFNs.
LncRNAs can also affect response to protozoan parasites such as cryptosporidium. NR_033736 is a novel lncRNA that has been found to be up-regulated in intestinal epithelial cells upon infection with this protozoon. This lncRNA can suppress transcription of type I IFN-controlled genes in host cells infected with this microorganism. Notably, type I IFN signaling can trigger the expression of NR_033736. In fact, NR_033736 participation in the negative feedback regulatory mechanism of type I IFN signaling results in fine-tuning of innate defense mechanism against microorganisms in the epithelial cells (68).
Investigations in the context of lupus nephritis have shown that RP112B6.2 via targeting the IFN-I by epigenetically inhibiting the expression of SOCS1 could aggravate symptoms of this disease (69). Linc00513 is another lncRNA that participates in the pathogenesis of lupus through promoting IFN signaling (70).
The impact of lncRNAs on IFN signaling has also been assessed in the context of diabetes mellitus. Lnc10 contains a type I diabetes-associated single nucleotide polymorphism. This lncRNA can regulate the expression of the IRF7-driven inflammatory network regulating gene Ebi2 in immune cells. Expression of Lnc10 in pancreatic β-cells has been shown to be up-regulated by diabetogenic incitements, including pro-inflammatory cytokines and viral infections (71). Figure 2 represents the role of several lncRNAs in various types of human cancers and immune-related disorders as well as their impact on viral infections via regulating the IFN signaling pathway.
Figure 2
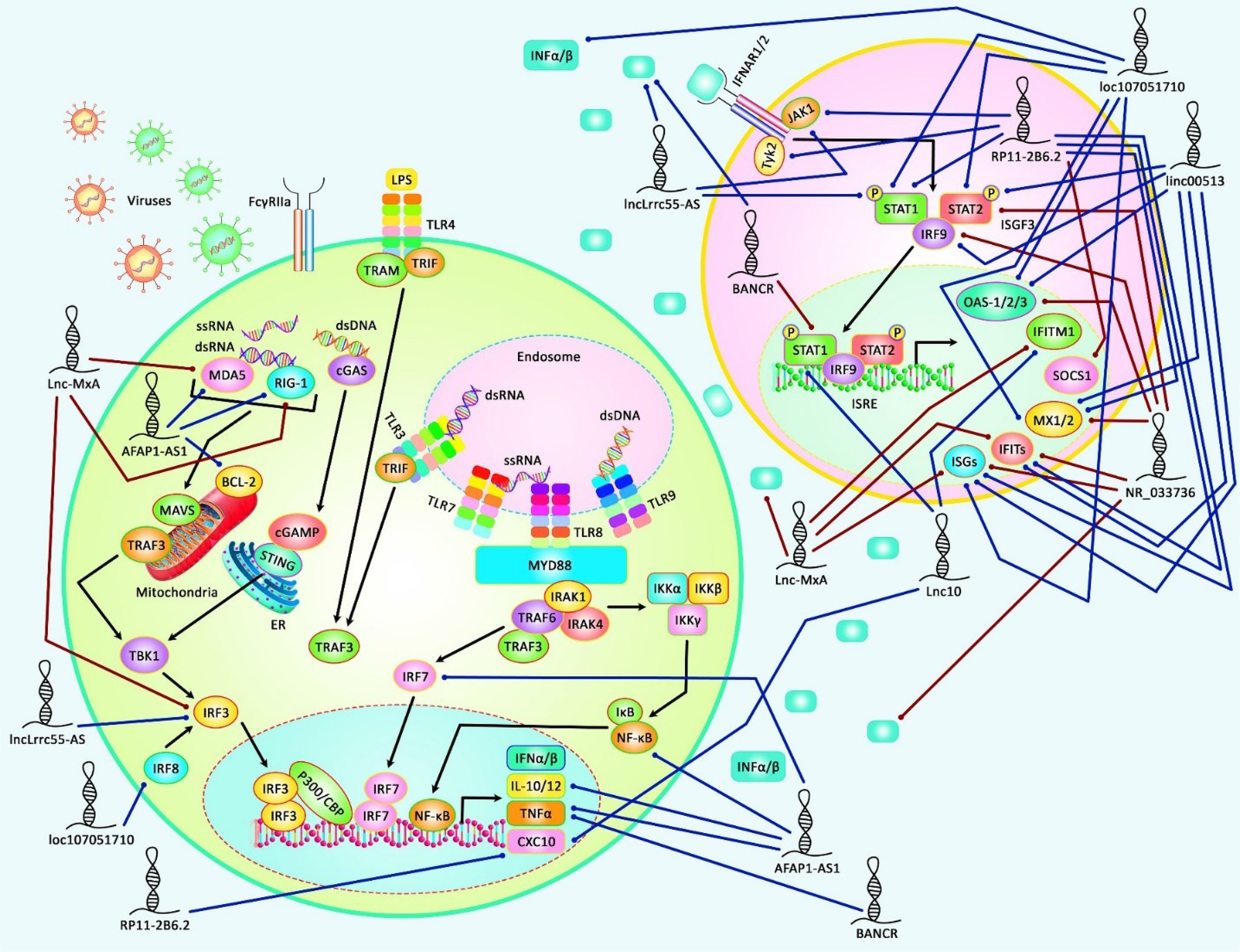
A schematic illustration of the role of several lncRNAs in regulating the IFN signaling pathway in several human diseases, including autoimmune conditions and viral infections. Accumulating evidence has illustrated that lncRNAs modulating IFN signaling cascade could participate in the pathogenesis of various kinds of human cancers as well as immune-related disorders. It has been reported that lncRNA RP11-2B6.2 could play an important role as a positive regulator of type I INF signaling pathway in Lupus Nephritis via up-regulating the expression levels of IFIT-1/3, ISG, Mx2, CXCL10, JAK1, STAT-1, TYK2, and decreasing SOCS1 expression (69). Moreover, another research has revealed that lncRNA loc107051710 could elevate the expression levels of IFN-α, IFN-β, Mx1, STAT-1/2, OAS via modulating IRF8, thereby enhancing the antiviral activity of ISGs to prevent infectious bursal disease virus (IBDV) infection (73). Blue lines indicate the positive regulatory effect among lncRNAs and their targets, and brown lines depict a negative one among them. All the information regarding the role of these lncRNAs involved in the modulation of the IFN signaling cascade in various types of immune deficiency diseases and cancers can be seen in Table 3.
Discussion
Non-coding RNAs that regulate IFN signaling have been shown to participate in the pathogenesis of different types of cancer as well as immune-related disorders. IRFs are the most investigated molecules in the field of effects of non-coding RNAs on IFN signaling. For instance, IRF-1 has been shown to have functional interactions with miR-301a, miR-195, miR-19a, miR-18a, miR-4295, miR-124, and miR-155. Meanwhile, IRF-2 has interactions with miR-1290, miR-664, and miR-221-3p. Besides, IRF-6 interacts with miR-320, miR-587, miR-19, and miR-18a. These interactions have been best assessed in the context of cancer, revealing the importance of immune function in the pathoetiology of cancer.
In addition, IFN-related non-coding RNAs may contribute to the pathogenesis of neuropsychiatric conditions through modulation of immune responses in CNS-resident cells. Major depressive disorder is an example of these conditions in which the role of IRF-targeting miRNAs has been identified. Among non-malignant conditions, are systemic sclerosis, Newcastle disease, Sjögren’s syndrome, traumatic brain injury, lupus nephritis, systemic lupus erythematosus, diabetes mellitus, and myocardial ischemia/reperfusion injury have been found to be associated with dysregulation of IFN-related non-coding RNAs. These non-coding RNAs represent a novel group of biomarkers for these conditions since their expressions are dysregulated in the biofluids of patients with these disorders.
Consistent with the important role of IFN signaling in the response of the immune system to viral infections, non-coding RNAs that regulate these signals can also participate in the pathophysiology of these conditions. The interactions between non-coding RNAs and IFN signaling have been assessed in the context of SARS-CoV-2, HIV, and influenza infections. Particularly, some miRNAs have been reported to enhance antiviral responses through modulation of IFN signaling. Identification of the impact of these miRNAs in response to viral infections could facilitate the design of efficient therapeutic modalities for these disorders. The preliminary results of in vitro and in vivo studies have suggested modulation of expression of certain miRNAs as an efficient strategy for limiting viral infections.
Notably, single nucleotide polymorphisms in the seed region of IFN-interacting miRNAs can interfere with or induce their bindings with miRNA targets. These polymorphic regions can hypothetically affect IFN responses, thus participating in the pathogenesis of autoimmune disorders, malignancies, or viral infections. Identification of these variants within the human genome might facilitate the design of specific treatment modalities for these conditions in the context of personalized medicine.
Several dysregulated IFN-related miRNAs, particularly miR-9, miR-18, miR-301a, miR-195, miR-19, miR-1290, miR-320, miR-664, miR-587, miR-203a, and miR-4295 have been shown to participate in the pathogenesis of human cancers. These miRNAs represent appropriate targets for anti-cancer therapies since they can affect immune responses against cancer. Future studies are needed to evaluate the effects of these miRNAs-targeting therapies in xenograft models of cancer.
Taken together, non-coding RNAs that regulate IFN signaling can participate in a variety of malignant and non-malignant disorders, particularly those related to abnormal immune responses.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Author contributions
SG-F wrote the draft and revised it. MT designed and supervised the study. EJ, BH, YP and FD collected the data and designed the figures and tables. All the authors read and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ESCs, Embryonic Stem Cells; FGF2, Growth Factor2; bEEC, Primary Bovine Endometrial Epithelial Cell; BoLA, Bovine Leukocyte Antigen; VSMCs, Vascular Smooth Muscle Cells; RAVSMCs, Rat Aortic VSMCs; RD, Rhabdomyosarcoma Cells; pDCs, Plasmacytoid Dendritic Cells; hMSC, Normal Human Mesenchymal Stem Cells; ATM, Ataxia-Telangiectasia Mutated; MDDCs, Monocyte-Derived Dendritic Cells; MDM, Monocyte-Derived Macrophage (MDMs); PBMC, Peripheral Blood Mononuclear Cell; GES1, Gastric Epithelial Cell Line; HSP60, Heat Shock Protein 60; MAVS, Mitochondrial Antiviral Signaling Protein; BHK, Baby Hamster Kidney; DENV, Dengue Virus; FHV-1, Feline Herpesvirus 1; CHK1, Checkpoint Kinase 1; INCR1, IFN-Stimulated Non-Coding RNA 1; ASCs, Adipose-Derived Stem Cells; MDCK, Madin-Darby Canine Kidney; mulNTEPI, Murine Intestinal Epithelial; BoLA, Bovine Leukocyte Antigen; PME-1, Phosphatase Methylesterase 1; PAMs, Porcine Alveolar Macrophages; VERO, African Green Monkey Kidney Cells; RD, Human Rhabdomyosarcoma Cells; EPC, Epithelioma Papulosum Cyprini; LPS, lipopolysacaridase; IRF-1, Interferon Regulatory Factor-1; ISGs, Interferon-Stimulated Genes; IFNAR, Heterodimeric Interferon Receptor; hMDDC, Primary Human Monocyte-Derived Dendritic Cells; MDMs, Monocyte-Derived Macrophages; ASCs, Adipose-Derived Stem Cells; LPS, Lipopolysaccharide; bEEC, Primary Bovine Endometrial Epithelial Cell; RIG-I, Retinoic Acid-Inducible Gene I; IFT, Intraflagellar Transport; IFIT1, Interferon Induced Protein with Tetratricopeptide Repeats 1; IFIH1, Interferon Induced with Helicase C Domain 1.
Glossary
| ESCs | Embryonic Stem Cells |
| FGF2 | Growth Factor2 |
| bEEC | Primary Bovine Endometrial Epithelial Cell |
| BoLA | Bovine Leukocyte Antigen |
| VSMCs | Vascular Smooth Muscle Cells |
| RAVSMCs | Rat Aortic VSMCs |
| RD | Rhabdomyosarcoma Cells |
| pDCs | Plasmacytoid Dendritic Cells |
| hMSC | Normal Human Mesenchymal Stem Cells |
| ATM | Ataxia-Telangiectasia Mutated |
| MDDCs | Monocyte-Derived Dendritic Cells |
| MDMs | MDM, Monocyte-Derived Macrophage |
| PBMC | Peripheral Blood Mononuclear Cell |
| GES1 | Gastric Epithelial Cell Line |
| HSP60 | Heat Shock Protein 60 |
| MAVS | Mitochondrial Antiviral Signaling Protein |
| BHK | Baby Hamster Kidney |
| DENV | Dengue Virus |
| FHV-1 | Feline Herpesvirus 1 |
| CHK1 | Checkpoint Kinase 1 |
| INCR1 | IFN-Stimulated Non-Coding RNA 1 |
| ASCs | Adipose-Derived Stem Cells |
| MDCK | Madin-Darby Canine Kidney |
| mulNTEPI | Murine Intestinal Epithelial |
| BoLA | Bovine Leukocyte Antigen |
| PME-1 | Phosphatase Methylesterase 1 |
| PAMs | Porcine Alveolar Macrophages |
| VERO | African Green Monkey Kidney Cells |
| RD | Human Rhabdomyosarcoma Cells |
| EPC | Epithelioma Papulosum Cyprini |
| LPS | lipopolysacaridase |
| IRF-1 | Interferon Regulatory Factor-1 |
| ISGs | Interferon-Stimulated Genes |
| IFNAR | Heterodimeric Interferon Receptor |
| hMDDC | Primary Human Monocyte-Derived Dendritic Cells |
| MDMs | Monocyte-Derived Macrophages |
| ASCs | Adipose-Derived Stem Cells |
| LPS | Lipopolysaccharide |
| bEEC | Primary Bovine Endometrial Epithelial Cell |
| RIG-I | Retinoic Acid-Inducible Gene I |
| IFT | Intraflagellar Transport |
| IFIT1 | Interferon Induced Protein with Tetratricopeptide Repeats 1 |
| IFIH1 | Interferon Induced with Helicase C Domain 1 |
References
1
IsaacsALindenmannJ. Virus Interference. I. The Interferon. Proc R Soc London Ser B-Biol Sci (1957) 147(927):258–67.
2
SavanR. Post-Transcriptional Regulation of Interferons and Their Signaling Pathways. J Interferon Cytokine Res (2014) 34(5):318–29. doi: 10.1089/jir.2013.0117
3
IvashkivLBDonlinLT. Regulation of Type I Interferon Responses. Nat Rev Immunol (2014) 14(1):36–49. doi: 10.1038/nri3581
4
De WeerdNANguyenT. The Interferons and Their Receptors—Distribution and Regulation. Immunol Cell Biol (2012) 90(5):483–91. doi: 10.1038/icb.2012.9
5
MedzhitovR. Origin and Physiological Roles of Inflammation. Nature (2008) 454(7203):428–35. doi: 10.1038/nature07201
6
MaoKChenSChenMMaYWangYHuangBet al. Nitric Oxide Suppresses NLRP3 Inflammasome Activation and Protects Against LPS-Induced Septic Shock. Cell Res (2013) 23(2):201–12. doi: 10.1038/cr.2013.6
7
MishraBBRathinamVAMartensGWMartinotAJKornfeldHFitzgeraldKAet al. Nitric Oxide Controls the Immunopathology of Tuberculosis by Inhibiting NLRP3 Inflammasome–Dependent Processing of IL-1β. Nat Immunol (2013) 14(1):52–60. doi: 10.1038/ni.2474
8
McNabFMayer-BarberKSherAWackAO’garraA. Type I Interferons in Infectious Disease. Nat Rev Immunol (2015) 15(2):87–103. doi: 10.1038/nri3787
9
DongKDuQCuiXWanPKaltenmeierCLuoJet al. MicroRNA-301a (miR-301a) Is Induced in Hepatocellular Carcinoma (HCC) and Down-Regulates the Expression of Interferon Regulatory Factor-1. Biochem Biophys Res Commun (2020) 524(2):273–9. doi: 10.1016/j.bbrc.2020.01.034
10
YanYZhengLDuQCuiXDongKGuoYet al. Interferon Regulatory Factor 1 (IRF-1) Downregulates Checkpoint Kinase 1 (CHK1) Through miR-195 to Upregulate Apoptosis and PD-L1 Expression in Hepatocellular Carcinoma (HCC) Cells. Br J Cancer (2021) 125(1):101–11. doi: 10.1038/s41416-021-01337-6
11
LiJLinT-YChenLLiuYDianM-JHaoW-Cet al. miR-19 Regulates the Expression of Interferon-Induced Genes and MHC Class I Genes in Human Cancer Cells. Int J Med Sci (2020) 17(7):953. doi: 10.7150/ijms.44377
12
JinJ-jLiuY-hSiJ-mNiRWangJ. Overexpression of miR-1290 Contributes to Cell Proliferation and Invasion of Non Small Cell Lung Cancer by Targeting Interferon Regulatory Factor 2. Int J Biochem Cell Biol (2018) 95:113–20. doi: 10.1016/j.biocel.2017.12.017
13
LiuYShaoGYangZLinXLiuXQianBet al. Interferon Regulatory Factor 6 Correlates With the Progression of Non-Small Cell Lung Cancer and Can Be Regulated by miR-320. J Pharm Pharmacol (2021) 73(5):682–91. doi: 10.1093/jpp/rgab009
14
XiaoXGuYWangGChenS. C-Myc, RMRP, and miR-34a-5p Form a Positive-Feedback Loop to Regulate Cell Proliferation and Apoptosis in Multiple Myeloma. Int J Biol Macromol (2019) 122:526–37. doi: 10.1016/j.ijbiomac.2018.10.207
15
FengJWangMLiMYangJJiaJLiuLet al. Serum miR-221-3p as a New Potential Biomarker for Depressed Mood in Perioperative Patients. Brain Res (2019) 1720:146296. doi: 10.1016/j.brainres.2019.06.015
16
ChouriEWangMHillenMRAngiolilliCSilva-CardosoSCWichersCGet al. Implication of miR-126 and miR-139-5p in Plasmacytoid Dendritic Cell Dysregulation in Systemic Sclerosis. J Clin Med (2021) 10(3):491. doi: 10.3390/jcm10030491
17
GaoFZhaoZLZhaoWTFanQRWangSCLiJet al. miR-9 Modulates the Expression of Interferon-Regulated Genes and MHC Class I Molecules in Human Nasopharyngeal Carcinoma Cells. Biochem Biophys Res Commun (2013) 431(3):610–6. doi: 10.1016/j.bbrc.2012.12.097
18
TomokuniAEguchiHTomimaruYWadaHKawamotoKKobayashiSet al. miR-146a Suppresses the Sensitivity to Interferon-α in Hepatocellular Carcinoma Cells. Biochem Biophys Res Commun (2011) 414(4):675–80. doi: 10.1016/j.bbrc.2011.09.124
19
RenYDongJHePLiangYWuLWangJet al. miR-587 Promotes Cervical Cancer by Repressing Interferon Regulatory Factor 6. J Gene Med (2020) 22(11):e3257. doi: 10.1002/jgm.3257
20
YuanJTanLYinZZhuWTaoKWangGet al. MIR17HG-miR-18a/19a Axis, Regulated by Interferon Regulatory Factor-1, Promotes Gastric Cancer Metastasis via Wnt/β-Catenin Signalling. Cell Death Dis (2019) 10(6):1–16. doi: 10.1038/s41419-019-1685-z
21
YangCHWangYSimsMCaiCHePHäckerHet al. MicroRNA203a Suppresses Glioma Tumorigenesis Through an ATM-Dependent Interferon Response Pathway. Oncotarget (2017) 8(68):112980. doi: 10.18632/oncotarget.22945
22
ChengJPHuangBDuanJHYiKJZhuangZL. Mir−4295 Promotes Cell Proliferation, Migration and Invasion of Osteosarcoma Through Targeting Interferon Regulatory Factor 1. Oncol Lett (2020) 20(5):1–. doi: 10.3892/ol.2020.12123
23
QuBCaoJZhangFCuiHTengJLiJet al. Type I Interferon Inhibition of MicroRNA-146a Maturation Through Up-Regulation of Monocyte Chemotactic Protein-Induced Protein 1 in Systemic Lupus Erythematosus. Arthritis Rheumatol (Hoboken NJ) (2015) 67(12):3209–18. doi: 10.1002/art.39398
24
WangXJiaYRenJLiuHXiaoSWangXet al. MicroRNA gga-miR-455-5p Suppresses Newcastle Disease Virus Replication via Targeting Cellular Suppressors of Cytokine Signaling 3. Vet Microbiol (2019) 239:108460. doi: 10.1016/j.vetmic.2019.108460
25
JangS-ITandonMTeosLZhengCWarnerBMAlevizosI. Dual Function of miR-1248 Links Interferon Induction and Calcium Signaling Defects in Sjögren’s Syndrome. EBioMedicine (2019) 48:526–38. doi: 10.1016/j.ebiom.2019.09.010
26
HarrisonEBEmanuelKLambertyBGMorseyBMLiMKelsoMLet al. Induction of miR-155 After Brain Injury Promotes Type 1 Interferon and Has a Neuroprotective Effect. Front Mol Neurosci (2017) 10:228. doi: 10.3389/fnmol.2017.00228
27
GuanX-lZhangB-cSunL. pol-miR-194a of Japanese Flounder (Paralichthys Olivaceus) Suppresses Type I Interferon Response and Facilitates Edwardsiella Tarda Infection. Fish Shellfish Immunol (2019) 87:220–5. doi: 10.1016/j.fsi.2019.01.017
28
LiWDengPWangJLiZZhangH. MiR-17 Knockdown Promotes Vascular Smooth Muscle Cell Phenotypic Modulation Through Upregulated Interferon Regulator Factor 9 Expression. Am J Hypertens (2020) 33(12):1119–26. doi: 10.1093/ajh/hpaa087
29
KwakJSKimKH. Effect of miR-155 on Type I Interferon Response in Epithelioma Papulosum Cyprini Cells. Fish Shellfish Immunol (2021) 111:1–5. doi: 10.1016/j.fsi.2021.01.005
30
RiessMFuchsNVIdicaAHamdorfMFloryEPedersenIMet al. Interferons Induce Expression of SAMHD1 in Monocytes Through Down-Regulation of miR-181a and miR-30a. J Biol Chem (2017) 292(1):264–77. doi: 10.1074/jbc.M116.752584
31
ZhouHHuangXCuiHLuoXTangYChenSet al. miR-155 and its Star-Form Partner miR-155* Cooperatively Regulate Type I Interferon Production by Human Plasmacytoid Dendritic Cells. Blood J Am Soc Hematol (2010) 116(26):5885–94. doi: 10.1182/blood-2010-04-280156
32
WangXYuanTYinNMaXYangYYangJet al. Interferon−τ Regulates the Expression and Function of Bovine Leukocyte Antigen by Downregulating Bta−Mir−204. Exp Ther Med (2021) 21(6):1–11. doi: 10.3892/etm.2021.10026
33
WangCShanLQuSXueMWangKFuFet al. The Coronavirus PEDV Evades Type III Interferon Response Through the miR-30c-5p/SOCS1 Axis. Front Microbiol (2020) 11:1180. doi: 10.3389/fmicb.2020.01180
34
ZhangXHanXTangYWuYQuBShenN. miR-744 Enhances Type I Interferon Signaling Pathway by Targeting PTP1B in Primary Human Renal Mesangial Cells. Sci Rep (2015) 5(1):1–11. doi: 10.1038/srep12987
35
ZhangJZhaoHChenJXiaBJinYWeiWet al. Interferon-β-Induced miR-155 Inhibits Osteoclast Differentiation by Targeting SOCS1 and MITF. FEBS Lett (2012) 586(19):3255–62. doi: 10.1016/j.febslet.2012.06.047
36
HuGGongA-YLiuJZhouRDengCChenX-M. miR-221 Suppresses ICAM-1 Translation and Regulates Interferon-γ-Induced ICAM-1 Expression in Human Cholangiocytes. Am J Physiology-Gastrointest Liver Physiol (2010) 298(4):G542–G50. doi: 10.1152/ajpgi.00490.2009
37
ZhangJLiZHuangJYinHTianJQuL. miR-26a Inhibits Feline Herpesvirus 1 Replication by Targeting SOCS5 and Promoting Type I Interferon Signaling. Viruses (2020) 12(1):2. doi: 10.3390/v12010002
38
SuYCHuangYFWuYWChenHFWuYHHsuCCet al. MicroRNA-155 Inhibits Dengue Virus Replication by Inducing Heme Oxygenase-1-Mediated Antiviral Interferon Responses. FASEB J (2020) 34(6):7283–94. doi: 10.1096/fj.201902878R
39
ZhangLZhangLPanYGaoJXuYLiXet al. Downregulation of miR-218 by Porcine Reproductive and Respiratory Syndrome Virus Facilitates Viral Replication via Inhibition of Type I Interferon Responses. J Biol Chem (2021) 296. doi: 10.1016/j.jbc.2021.100683
40
KadmonCSLandersCTLiHSWatowichSSRodriguezAKingKY. MicroRNA-22 Controls Interferon Alpha Production and Erythroid Maturation in Response to Infectious Stress in Mice. Exp Hematol (2017) 56:7–15. doi: 10.1016/j.exphem.2017.09.001
41
RosenbergerCMPodyminoginRLDiercksAHTreutingPMPeschonJJRodriguezDet al. miR-144 Attenuates the Host Response to Influenza Virus by Targeting the TRAF6-IRF7 Signaling Axis. PloS Pathogens (2017) 13(4):e1006305–e. doi: 10.1371/journal.ppat.1006305
42
RuJSunHFanHWangCLiYLiuMet al. MiR-23a Facilitates the Replication of HSV-1 Through the Suppression of Interferon Regulatory Factor 1. PloS One (2014) 9(12):e114021. doi: 10.1371/journal.pone.0114021
43
LiuFWangHDuLWeiZZhangQFengW-h. MicroRNA-30c Targets the Interferon–Alpha/Beta Receptor Beta Chain to Promote Type 2 PRRSV Infection. J Gen Virol (2018) 99(12):1671–80. doi: 10.1099/jgv.0.001166
44
ChangXShiXZhangXChenJFanXYangYet al. miR-382-5p Promotes Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Replication by Negatively Regulating the Induction of Type I Interferon. FASEB J (2020) 34(3):4497–511. doi: 10.1096/fj.201902031RRR
45
ShiXYangYZhangXChangXChenJWangCet al. miR-541-3p Promoted Porcine Reproductive and Respiratory Syndrome Virus 2 (PRRSV-2) Replication by Targeting Interferon Regulatory Factor 7. Viruses (2022) 14(1):126. doi: 10.3390/v14010126
46
WitteveldtJKnolLIMaciasS. MicroRNA-Deficient Mouse Embryonic Stem Cells Acquire a Functional Interferon Response. Elife (2019) 8:e44171. doi: 10.7554/eLife.44171
47
ChenXZhouLPengNYuHLiMCaoZet al. MicroRNA-302a Suppresses Influenza A Virus–Stimulated Interferon Regulatory Factor-5 Expression and Cytokine Storm Induction. J Biol Chem (2017) 292(52):21291–303. doi: 10.1074/jbc.M117.805937
48
GuoMLiFJiJLiuYLiuFZhaoYet al. Inhibition of miR-93 Promotes Interferon Effector Signaling to Suppress Influenza A Infection by Upregulating JAK1. Int Immunopharmacol (2020) 86:106754. doi: 10.1016/j.intimp.2020.106754
49
GaoJLiuJZhangYGuanBQuHChaiHet al. PBMCs-Derived microRNA Signature as a Prethrombotic Status Discriminator in Stable Coronary Artery Disease. Thromb Haemostasis (2020) 120(01):121–31. doi: 10.1055/s-0039-1700518
50
HuangBChenHZhengY. MiR-103/miR-107 Inhibits Enterovirus 71 Replication and Facilitates Type I Interferon Response by Regulating SOCS3/STAT3 Pathway. Biotechnol Lett (2021) 43(7):1357–69. doi: 10.1007/s10529-021-03115-z
51
WangYRenTChenHWangKZhangYLiuLet al. MiR-4334-5p Facilitates Foot and Mouth Disease Virus Propagation by Suppressing Interferon Pathways via Direct Targeting Id1. Genes (2020) 11(10):1136. doi: 10.3390/genes11101136
52
BochnakianAZhenAZisoulisDGIdicaAKewalRamaniVNNeelNet al. Interferon-Inducible MicroRNA miR-128 Modulates HIV-1 Replication by Targeting TNPO3 mRNA. J Virol (2019) 93(20):e00364-19. doi: 10.1128/JVI.00364-19
53
DuanXZhaoMLiXGaoLCaoHWangYet al. gga-miR-27b-3p Enhances Type I Interferon Expression and Suppresses Infectious Bursal Disease Virus Replication via Targeting Cellular Suppressors of Cytokine Signaling 3 and 6 (SOCS3 and 6). Virus Res (2020) 281:197910. doi: 10.1016/j.virusres.2020.197910
54
WangBFuMLiuYWangYLiXCaoHet al. gga-miR-155 Enhances Type I Interferon Expression and Suppresses Infectious Burse Disease Virus Replication via Targeting SOCS1 and TANK. Front Cell Infect Microbiol (2018) 8:55. doi: 10.3389/fcimb.2018.00055
55
OuyangWY-sWDuX-nLiuH-jZhangH-b. gga-miR-9* Inhibits IFN Production in Antiviral Innate Immunity by Targeting Interferon Regulatory Factor 2 to Promote IBDV Replication. Vet Microbiol (2015) 178(1-2):41–9. doi: 10.1016/j.vetmic.2015.04.023
56
LiASongWQianJLiYHeJZhangQet al. MiR-122 Modulates Type I Interferon Expression Through Blocking Suppressor of Cytokine Signaling 1. Int J Biochem Cell Biol (2013) 45(4):858–65. doi: 10.1016/j.biocel.2013.01.008
57
HeJJiYLiAZhangQSongWLiYet al. MiR-122 Directly Inhibits Human Papillomavirus E6 Gene and Enhances Interferon Signaling Through Blocking Suppressor of Cytokine Signaling 1 in SiHa Cells. PloS One (2014) 9(9):e108410. doi: 10.1371/journal.pone.0108410
58
HuangYChenDHeJCaiJShenKLiuXet al. Hcmv-miR-UL112 Attenuates NK Cell Activity by Inhibition Type I Interferon Secretion. Immunol Lett (2015) 163(2):151–6. doi: 10.1016/j.imlet.2014.12.003
59
ZhangXWangWZhuWDongJChengYYinZet al. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci (2019) 20(22):5573. doi: 10.3390/ijms20225573
60
ValadkhanSGunawardaneLS. lncRNA-Mediated Regulation of the Interferon Response. Virus Res (2016) 212:127–36. doi: 10.1016/j.virusres.2015.09.023
61
KambaraHNiaziFKostadinovaLMoonkaDKSiegelCTPostABet al. Negative Regulation of the Interferon Response by an Interferon-Induced Long Non-Coding RNA. Nucleic Acids Res (2014) 42(16):10668–80. doi: 10.1093/nar/gku713
62
OuyangJZhuXChenYWeiHChenQChiXet al. NRAV, a Long Noncoding RNA, Modulates Antiviral Responses Through Suppression of Interferon-Stimulated Gene Transcription. Cell Host Microbe (2014) 16(5):616–26. doi: 10.1016/j.chom.2014.10.001
63
CollierSPCollinsPLWilliamsCLBoothbyMRAuneTM. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. J Immunol (2012) 189(5):2084–8. doi: 10.4049/jimmunol.1200774
64
VigneauSRohrlichP-SBrahicMBureauJ-F. Tmevpg1, a Candidate Gene for the Control of Theiler’s Virus Persistence, Could Be Implicated in the Regulation of Gamma Interferon. J Virol (2003) 77(10):5632–8. doi: 10.1128/JVI.77.10.5632-5638.2003
65
CollierSP. TMEVPG1, a Long Noncoding RNA Within the Immune System. (2014).
66
TangX-DZhangD-DJiaLJiWZhaoY-S. lncRNA AFAP1-AS1 Promotes Migration and Invasion of Non-Small Cell Lung Cancer via Up-Regulating IRF7 and the RIG-I-Like Receptor Signaling Pathway. Cell Physiol Biochem (2018) 50(1):179–95. doi: 10.1159/000493967
67
MaHChangHYangWLuYHuJJinS. A Novel Ifnα-Induced Long Noncoding RNA Negatively Regulates Immunosuppression by Interrupting H3K27 Acetylation in Head and Neck Squamous Cell Carcinoma. Mol Cancer (2020) 19(1):1–16. doi: 10.1186/s12943-019-1123-y
68
LiJJinKLiMMathyNWGongA-YDengSet al. A Host Cell Long Noncoding RNA NR_033736 Regulates Type I Interferon-Mediated Gene Transcription and Modulates Intestinal Epithelial Anti-Cryptosporidium Defense. PloS Pathogens (2021) 17(1):e1009241. doi: 10.1371/journal.ppat.1009241
69
LiaoZYeZXueZWuLOuyangYYaoCet al. Identification of Renal Long non-Coding RNA RP11-2B6. 2 as a Positive Regulator of Type I Interferon Signaling Pathway in Lupus Nephritis. Front Immunol (2019) 10:975. doi: 10.3389/fimmu.2019.00975
70
XueZCuiCLiaoZXiaSZhangPQinJet al. Identification of LncRNA Linc00513 Containing Lupus-Associated Genetic Variants as a Novel Regulator of Interferon Signaling Pathway. Front Immunol (2018) 9:2967. doi: 10.3389/fimmu.2018.02967
71
Gonzalez-MoroISantinI eds. A T1D-Associated lncRNA Modulates the Type I Interferon Signaling and the Antiviral Response in Pancreatic Beta Cells. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (2020) p. 2914–17. IEEE.
72
WangSHeFLiZHuYHuangfuNXieD. Long Non-Coding RNA BANCR Promotes Interferon-β-Induced Cardiomyocyte Apoptosis by Targeting Signal Transducer and Activator of Transcription 1 In Vitro. Int J Clin Exp Pathol (2020) 13(11):2840.
73
HuangXXuYLinQGuoWZhaoDWangCet al. Determination of Antiviral Action of Long Non-Coding RNA Loc107051710 During Infectious Bursal Disease Virus Infection Due to Enhancement of Interferon Production. Virulence (2020) 11(1):68–79. doi: 10.1080/21505594.2019.1707957
74
ZhaoLXiaMWangKLaiCFanHGuHet al. A Long non-Coding RNA IVRPIE Promotes Host Antiviral Immune Responses Through Regulating Interferon β1 and ISG Expression. Front Microbiol (2020) 11:260. doi: 10.3389/fmicb.2020.00260
75
PanQZhaoZLiaoYChiuS-HWangSChenBet al. Identification of an Interferon-Stimulated Long Noncoding RNA (LncRNA ISR) Involved in Regulation of Influenza A Virus Replication. Int J Mol Sci (2019) 20(20):5118. doi: 10.3390/ijms20205118
76
XiaoMChenYWangSLiuSRaiKRChenBet al. Long Noncoding RNA IFITM4P Regulates Host Antiviral Responses by Acting as a Competing Endogenous RNA. J Virol (2021) 95(21):e00277-21. doi: 10.1128/JVI.00277-21
77
SuYYuanJZhangFLeiQZhangTLiKet al. MicroRNA-181a-5p and microRNA-181a-3p Cooperatively Restrict Vascular Inflammation and Atherosclerosis. Cell Death Dis (2019) 10(5):1–15. doi: 10.1038/s41419-019-1599-9
78
AgarwalSVierbuchenTGhoshSChanJJiangZKandasamyRKet al. The Long Non-Coding RNA LUCAT1 Is a Negative Feedback Regulator of Interferon Responses in Humans. Nat Commun (2020) 11(1):1–11. doi: 10.1038/s41467-020-20165-5
79
VishnubalajiRShaathHAlajezNM. Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes (2020) 11(7):760. doi: 10.3390/genes11070760
80
SchynkelTSzaniawskiMASpivakAMBosqueAPlanellesVVandekerckhoveLet al. Interferon-Mediated Long Non-Coding RNA Response in Macrophages in the Context of HIV. Int J Mol Sci (2020) 21(20):7741. doi: 10.3390/ijms21207741
81
ZhouYLiMXueYLiZWenWLiuXet al. Interferon-Inducible Cytoplasmic Lnclrrc55-AS Promotes Antiviral Innate Responses by Strengthening IRF3 Phosphorylation. Cell Res (2019) 29(8):641–54. doi: 10.1038/s41422-019-0193-0
82
HuynhNPGlossCCLorentzJTangRBrungerJMMcAlindenAet al. Long non-Coding RNA GRASLND Enhances Chondrogenesis via Suppression of the Interferon Type II Signaling Pathway. Elife (2020) 9:e49558. doi: 10.7554/eLife.49558.sa2
83
KuttyRKSamuelWDuncanTPostnikovaOJaworskiCNagineniCNet al. Proinflammatory Cytokine Interferon-γ Increases the Expression of BANCR, a Long Non-Coding RNA, in Retinal Pigment Epithelial Cells. Cytokine (2018) 104:147–50. doi: 10.1016/j.cyto.2017.10.009
Summary
Keywords
lncRNA, miRNA, interferon, expression, biomarker
Citation
Ghafouri-Fard S, Poornajaf Y, Dashti F, Hussen BM, Taheri M and Jamali E (2022) Interaction Between Non-Coding RNAs and Interferons: With an Especial Focus on Type I Interferons. Front. Immunol. 13:877243. doi: 10.3389/fimmu.2022.877243
Received
16 February 2022
Accepted
01 April 2022
Published
27 April 2022
Volume
13 - 2022
Edited by
Bertrand Kaeffer, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), France
Reviewed by
Grégoire Mignot, INRA UMREA4644 Endocrinologie Cellulaire et Moléculaire Immuno, France; Rezvan Noroozi, Jagiellonian University, Poland; Amin Safa, Complutense University of Madrid, Spain
Updates
Copyright
© 2022 Ghafouri-Fard, Poornajaf, Dashti, Hussen, Taheri and Jamali.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Taheri, mohammad.taheri@uni-jena.de; Elena Jamali, Elena.jamali@yahoo.com
This article was submitted to Cytokines and Soluble Mediators in Immunity, a section of the journal Frontiers in Immunology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.