
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 08 July 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.875689
Aeromonas are autochthonous bacteria of aquatic environments that are considered to be emerging pathogens to humans, producing diarrhea, bacteremia, and wound infections. Genetic identification shows that 95.4% of the strains associated with clinical cases correspond to the species Aeromonas caviae (37.26%), Aeromonas dhakensis (23.49%), Aeromonas veronii (21.54%), and Aeromonas hydrophila (13.07%). However, few studies have investigated the human immune response against some Aeromonas spp. such as A. hydrophila, Aeromonas salmonicida, and A. veronii. The present study aimed to increase the knowledge about the innate human immune response against six Aeromonas species, using, for the first time, an in vitro infection model with the monocytic human cell line THP-1, and to evaluate the intracellular survival, the cell damage, and the expression of 11 immune-related genes (TLR4, TNF-α, CCL2, CCL20, JUN, RELA, BAX, TP53, CASP3, NLRP3, and IL-1β). Transcriptional analysis showed an upregulated expression of a variety of the monocytic immune-related genes, with a variable response depending upon the Aeromonas species. The species that produced the highest cell damage, independently of the strain origin, coincidentally induced a higher expression of immune-related genes and corresponded to the more prevalent clinical species A. dhakensis, A. veronii, and A. caviae. Additionally, monocytic cells showed an overexpression of the apoptotic and pyroptotic genes involved in cell death after A. dhakensis, A. caviae, and Aeromonas media infection. However, the apoptosis route seemed to be the only way of producing cell damage and death in the case of the species Aeromonas piscicola and Aeromonas jandaei, while A. veronii apparently only used the pyroptosis route.
The genus Aeromonas comprises species considered autochthonous of aquatic environments that can also cause a wide spectrum of diseases in humans, mainly gastroenteritis, bacteremia/septicemia, and wound infections. Aeromonas are considered emerging pathogens due to the increase of their isolation as well as becoming renowned as a pathogen of serious public health concern (1–5). The infection occurs more frequently in children, the elderly, and immunocompromised individuals, and cases of bacteremia with fever, jaundice, abdominal pain or septic shock, and extraintestinal infections involving meningitis, pneumonia, keratitis, and osteomyelitis among others (1, 2, 4, 6, 7). Aeromonas also were the microorganisms more isolated from wound infections after natural disasters like the tsunami in Thailand (2004), representing about 22.6% of all isolates in the survivors (8).
According to a recent review, 95.4% of the strains associated with clinical cases correspond to four species: Aeromonas caviae (37.26%), Aeromonas dhakensis (23.49%), Aeromonas veronii (21.54%), and Aeromonas hydrophila (13.07%) (5). However, other species, i.e., Aeromonas media (2.27%) and Aeromonas jandaei (0.43%), have also been involved with a lower frequency. The virulence of Aeromonas spp. has been considered multifactorial and associated with different toxins—aerolysin, hemolysin, lipases, and enterotoxins—some of them delivered by different types of secretion systems as well as produced through a signal molecules such as N-acyl homoserine-lactone (AHLs) by a phenomenon defined as quorum sensing, which results in the colonization, invasion, and proliferation of the bacteria during the infectious process (9–12). However, the development of the infection depends on the immunity of the host, which is divided into two types: innate and adaptive (13). The innate immune response, that is, the one investigated in the present study, is activated after recognition of bacteria structures, i.e., pathogen-associated molecular patterns (PAMPs) by receptors named pattern recognition receptors (PRRs), soluble and insoluble, present and expressed in a variety of immune-related cells, like neutrophils, monocytes, or macrophages (13–16). Toll-like receptors (TLRs) are membrane PRRs that induce the phagocytosis of the pathogen and activate the expression of cytokines in the host, initiating the inflammatory response (17). It has been demonstrated that TLR recognition such as TLR2, TLR3, or TLR4, depending on the organisms, induces cell death by apoptosis (18). Apoptosis is a caspase-dependent process that induces nuclear condensation and the release of cytoplasmic content from the host cell into the extracellular environment, which prevents inflammation (19). Moreover, other types of PRRs named the Nod-like receptors (NLRs) are expressed in the cytosol and induce a different type of cell death called pyroptosis, mediated by the activation of caspase-1 and by the formation of multi-protein complexes called the inflammasomes (14, 20). Pyroptosis can take place in many cell types, including macrophages, dendritic cells, neutrophils, and epithelial cells. This process is characterized by the activation of different types of caspase enzymes and contributes to the activation of pro-inflammatory cytokines, recruits more immune cells, and finally activates an inflammatory cascade in the tissue (14). In the last years, few studies have investigated the expression of several innate immune-related genes against A. hydrophila, A. veronii, and Aeromonas salmonicida in different cell lines or animal models (mice and fish) (12, 21–24). The majority of the studies has been performed with cell lines obtained from animals, with the exception of two cell lines (T-84 and WLR-68) obtained from the human colon (12, 24). However, human immune-related cell lines have, so far, not been used as cell models in the Aeromonas studies.
The upregulation of the TLR4, which is a transmembrane receptor that senses molecules such as the lipopolysaccharide (LPS) present in the cell walls of Gram-negative bacteria, has been detected in several studies after infecting a fish model with different strains of A. hydrophila (21–23, 25, 26). The TLRs also induce the activation of the nuclear factor NF-Kappa-B p65 subunit (RELA) protein, which is a protein involved with other proteins such as RELB, REL, p105/p50 NF-κB1 and NF-κB2 in the formation of the nuclear factor kappa-light-chain-enhancer (NF-κB) (17). For instance, after an A. hydrophila fish infection, the RELA protein was activated by the upregulation of the expression levels of the TLR3, TLR4-1, TLR9, and TLR22 genes and inducing the production of pro-inflammatory cytokines (27). A previous study indicated that changes in the expression of the JUN proto-oncogene, which is another transcription factor that binds with the FOS proto-oncogene, could be involved in the virulence of Aeromonas spp. in mice because pathogenic strains induced upregulation of JUN and FOS genes (28). Both genes form a complex resulting in the formation of the activator protein-1 (AP-1), which is a transcription factor family implicated in critical cell functions such as the inflammatory response (29). Moreover, after A. hydrophila and A. salmonicida infections in fish, the activation of the gene expression that encodes pro-inflammatory cytokines like TNF-α and interleukin 8 (IL-8), as well as chemokines, i.e., the C-C motif ligand 20 (CCL20) involved in the recruitment of lymphocytes and dendritic cells, was demonstrated (21, 23, 30, 31). However, the C-C motif chemokine ligand 2 (CCL2) has not been studied in Aeromonas, despite it being an interesting chemokine well studied in infections produced by other bacteria like Vibrio spp., where it plays a role in the recruitment of monocytes, T cells such as CD4+ and CD8+, and dendritic cells (32). Therefore, the chemokine CCL2 will be investigated as a novel work. In relation to apoptosis, few studies demonstrated the capacity of A. hydrophila, A. salmonicida, and A. veronii to induce this process in different cell lines, and a fish infection model has been reported (25, 26). The caspase 3 (CASP3) protein is a member of the cysteine-aspartic acid protease family related to apoptosis and is known as executioner caspase by coordinating the destruction of cellular structures such as DNA fragmentation (19). In a previous study, it was observed that this gene was expressed during the apoptosis of head kidney-derived macrophages from fish infected with A. hydrophila (33). Another gene associated with apoptosis in Aeromonas is the tumor protein P53 (TP53) as Lü et al. (34) demonstrated after infecting a fish model with A. hydrophila. Previous studies demonstrated that TP53 induces the activation of the BCL-2-associated X protein (BAX), which belongs to the BCL-2 family, which plays an important role in the apoptosis route by the intrinsic pathway (27). However, the expression of the gene encoding the BAX protein has not been studied in Aeromonas and will be also investigated in the current study. As described above, an alternative way to the cell death produced by apoptosis is pyroptosis, which involves the formation of the inflammasome (14, 35). The activation of the pyroptosis route has been demonstrated by studying two proteins associated with the inflammasome, i.e., pyrin domain containing 3 (NLRP3) and IL-1β, after infections in mice and murine macrophages with A. hydrophila and A. veronii, respectively (36, 37).
So far, in Aeromonas, the immune response studies have not yet used the human monocytic cell line (THP-1), which was used successfully in studies for the in vitro infections to investigate the host–pathogen interactions in Vibrio vulnificus (32, 35). Considering the latter and the fact that human monocytes act as the first line of defense at the beginning of the infection process (38), the THP-1 cells were selected as the host in the present study. Moreover, no information exists so far about the immune response generated against Aeromonas species, which are frequently isolated in clinical cases such as A. dhakensis and A. caviae. Additionally, whether the capacity to develop an innate-immune response in the host cells is equal to the environmental and clinical strains is another aspect that has never been explored. Therefore, this study investigates Aeromonas species that show different frequencies of occurrence in clinical cases, using genetically identified strains of clinical and environmental origin in order to clarify if there exists a species-specific immune response that could explain their differential prevalence in human infections.
The study was performed with 24 strains (10 clinical and 14 environmental) of six different species of which A. dhakensis, A. media, A. jandaei, Aeromonas piscicola, and A. caviae have not been studied until now, being the only exception A. veronii (Table 1). The clinical strains were isolated from feces, sputum, and wound human infections, while the environmental strains were isolated from sick fish and water. All strains came from a collection that was maintained in Tryptone Soya Broth (TSB) (Becton Dickinson GmbH, Heidelberg, Germany) plus glycerol (20%) at −80°C, and from there, they were grown in Tryptone Soya Agar (TSA) (Becton Dickinson GmbH, Germany) at 37°C for 24 h (similar growth rates). Their identity was previously determined based on the housekeeping gene sequencing such as rpoD (range between 498 and 596 bp) or gyrB (range between 413 and 523 bp) using primers and conditions previously described (45). Prior to infection, bacteria were regrown at 37°C in serum-free Dulbecco’s Modified Eagle’s Medium (DMEM; PAA Laboratories GmbH, Munich, Germany) under shaking conditions (100 rpm) for 18 h (32).
The presence of different virulence-associated genes such as aerolysin (aerA), hemolysin (hlyA), cytotoxic enterotoxin (act), cytotonic enterotoxins (ast and alt), flagellin A (flaA) gene, Type III secretion system genes (ascF and ascV), and shiga toxin (stx1) was evaluated by PCR using specific primers and PCR conditions described in previous studies (46, 47).
The human monocytic cell line THP-1 (48) was selected for the experiments because it is a valuable model for studying the innate immune response against bacteria. This cell line was maintained as a cell suspension in Roswell Park Memorial Institute Medium (RPMI-1640, PAA Laboratories GmbH) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories GmbH, Munich, Germany) plus 1% penicillin–streptomycin (P/S) solution (PAA Laboratories GmbH, Munich, Germany) at 37°C and 5% CO2 (32). Before the infection experiments, cells were seeded in tissue culture plates containing DMEM without FBS and P/S at a concentration of 0.5 × 106 cells/ml to obtain 1 × 106 cells/ml after 3 h (32, 35).
Cell line THP-1 was infected with each of the 24 Aeromonas strains (Table 1) using overnight cultures (18 h) in DMEM without FBS and P/S, at a multiplicity of infection (MOI) of 10 and 20, i.e., the ratio between the number of bacteria and the number of cells targeted (49). The control strain of V. vulnificus (CECT 4999) was used at MOI 5, as done in a previous study (32). The cultures were incubated at 37°C and 5% CO2.
Infected monocytes at MOI 10 and 20 (initial dose) were incubated at 37°C with 5% CO2 for 1 h, followed by gentamicin treatment (50 µg/ml) for 1 h to kill extracellular bacteria (time 0), and then the number of bacteria inside the monocytes was determined (50). Incubation of infected monocytes continued with the fresh DMEM and maintenance dose of gentamicin (2 µg/ml) for an additional 4 h. After this, the number of bacteria inside THP-1 was counted. Percent survival was calculated using the number of bacteria after 4 h of incubation and after gentamicin treatment (50 µg/ml) (50). Results were expressed as the average of the results obtained for all the strains of the same species.
After the infection at MOI 10 and 20, supernatants were obtained at different times (t = 3, 4, 5, and 6 h). Cell damage was determined by quantifying the lactate dehydrogenase (LDH) enzyme released into the culture media (supernatants), by using the Cytox 96 Non-Radioactive Cytotoxicity Assay (Promega, Madison, WI, USA), as described in the manufacturer’s instructions. To perform a standard curve, a bovine recombinant LDH (Sigma-Aldrich, St. Louis, MO, USA) was used, and the LDH levels of the samples were extrapolated from the curve (32). Results were expressed as the average of the results obtained for the clinical and environmental strains.
Eleven different genes implicated in the immune response against pathogens were selected to quantify their transcription levels by THP-1 cells in response to the infections produced with the different strains of the Aeromonas spp. in relation to the non-infected cells. The primers used to evaluate the expression of the selected genes were those from Murciano et al. (32) and Zhao et al. (51) and are listed in Table 2. The selected genes were those that encode for TLR4, cytokines, and chemokines (TNF-α, CCL2, and CCL20), apoptosis (TP53, BAX, and CASP3), and pyroptosis (NLRP3 and IL-1β) as well as genes of the transcription factors (JUN and RELA). After 4 h of infection at MOI 20, THP-1 cells were washed twice with PBS, and the RNA was isolated from the samples by using the GenElute™ Mammalian Total RNA Miniprep Kit (Sigma-Aldrich). RNA quality and integrity were confirmed spectrophotometrically using NanoDrop 2000, calculating the 260/280 and 260/230 ratios. The cDNA was transcribed from total RNA by using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). A real-time PCR was performed with cDNA for quantification by using the Power SYBR® green PCR Mastermix (Applied Biosystems®, Life Technologies, Glasgow, UK) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems®). Threshold cycle (CT) values were obtained to establish the relative RNA levels of the tested genes, using the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as a housekeeping gene of reference. The relative gene expression was determined by using the delta-delta Ct (2−ΔΔCt) method that relays the signal from the real-time PCR, as done in a previous study (32). Results corresponded to the average of the results obtained for all the strains of the same species and were expressed as fold changes in relation to the non-infected cells.
Two different genes implicated in the virulence, aerA and act, were studied after THP-1 infections with one strain of each Aeromonas species, with the exception of aerA with A. jandaei infection due to the lack of this gene in all strains. The primers used to evaluate the expression of the selected genes were those from Lee et al. (47). After 3 h of infection at MOI 20, total RNA was isolated from Aeromonas cultures using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) as previously described (52). RNA quality and integrity were confirmed using NanoDrop 2000, calculating the 260/280 and 260/230 ratios. The cDNA was transcribed from RNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s instructions. Quantitative Real-Time PCR was performed in triplicate using Real-Power SYBR® green PCR Mastermix (Applied Biosystems®, Waltham, MA, USA) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). Threshold cycle (CT) values were obtained to establish the relative RNA levels of the tested genes, using 16S rRNA gene as a housekeeping gene and then calculated with the 2−ΔΔCt method.
All the experiments were performed in triplicate, and the statistical significance was determined by using Student’s two-tailed t-test and two-way ANOVA at p < 0.05 using the GraphPad Prism 6.0 (GraphPad Software, CA, USA).
The presence of virulence-associated genes in these Aeromonas strains was screened by PCR. The distribution of the genes is summarized in Table 3. The most frequent virulence gene detected was alt (66.66%), followed by aerA, ascV, and flaA (45.83%); hlyA, ascF, and ast (41.66%); and act (37.5%). The less frequent virulence genes were aexT and aexU (20.83%). In contrast, stx gene was not detected in any of the 24 strains. In relation to the species as shown in Table 3, the results showed that A. veronii strains had a higher number of positive strains (25) for different genes, followed by A. dhakensis (23), A. piscicola (18), and A. caviae (15), while A. media (9) and A. jandaei (5) had a smaller number of positive strains.
As shown in Figure 1, independently of the MOI, the most prevalent clinical species (A. dhakensis, A. veronii, and A. caviae) showed, in general, a higher intracellular survival than the less prevalent species (A. media, A. jandaei, and A. piscicola). Additionally, at MOI 20, the intracellular survival of Aeromonas spp. was significantly higher (p < 0.05) than at MOI 10 except for the species A. jandaei and A. veronii (Figure 1).
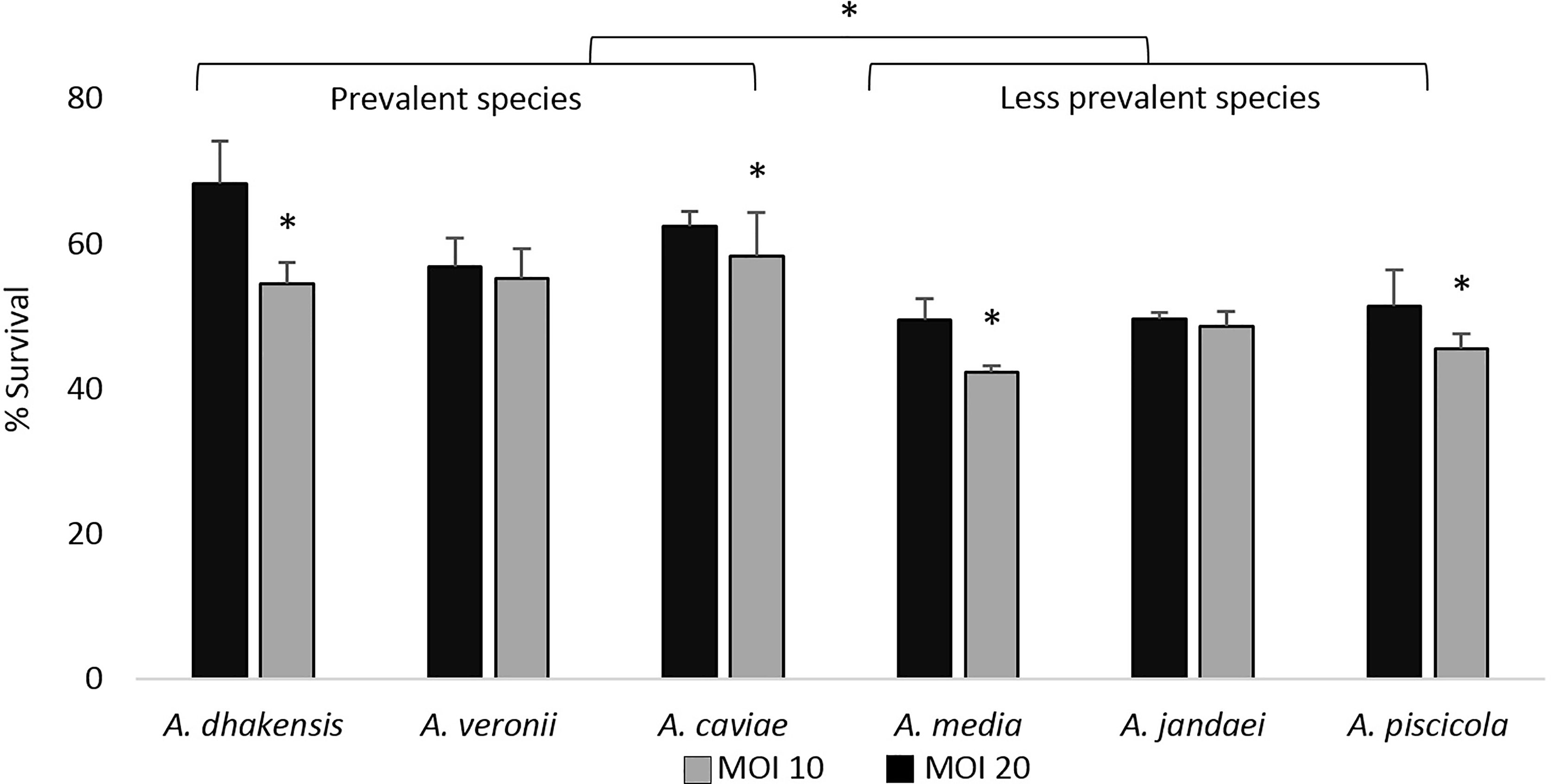
Figure 1 Intracellular survival expressed as the average of the strains of each Aeromonas species after 4 h of THP-1 monocytic cell line infection at multiplicities of infection (MOI) 10 and 20. Asterisks indicate statistical significance *p < 0.05.
The ability of Aeromonas to induce cell damage in THP-1 cells measured as the release of LDH to the cell culture supernatant is shown in Figures 2, 3. The six Aeromonas species caused significantly higher cell damage (p < 0.05) when compared with the non-infected cells, and this was higher after the infection at MOI 20 (Figure 2). All Aeromonas strains, independently of the species, were able to induce at MOI 10 and 20 significant degrees of cell damage that increased with time (p < 0.05). The clinical strains of all the species were able to induce a higher degree (p < 0.05) of THP-1 cell damage than the environmental strains (Figure 3). In addition, the more prevalent clinical species, A. dhakensis, A. caviae, and A. veronii, caused significantly higher cell damage than the rest, independently of the exposure time (p < 0.05) (Figure 2).
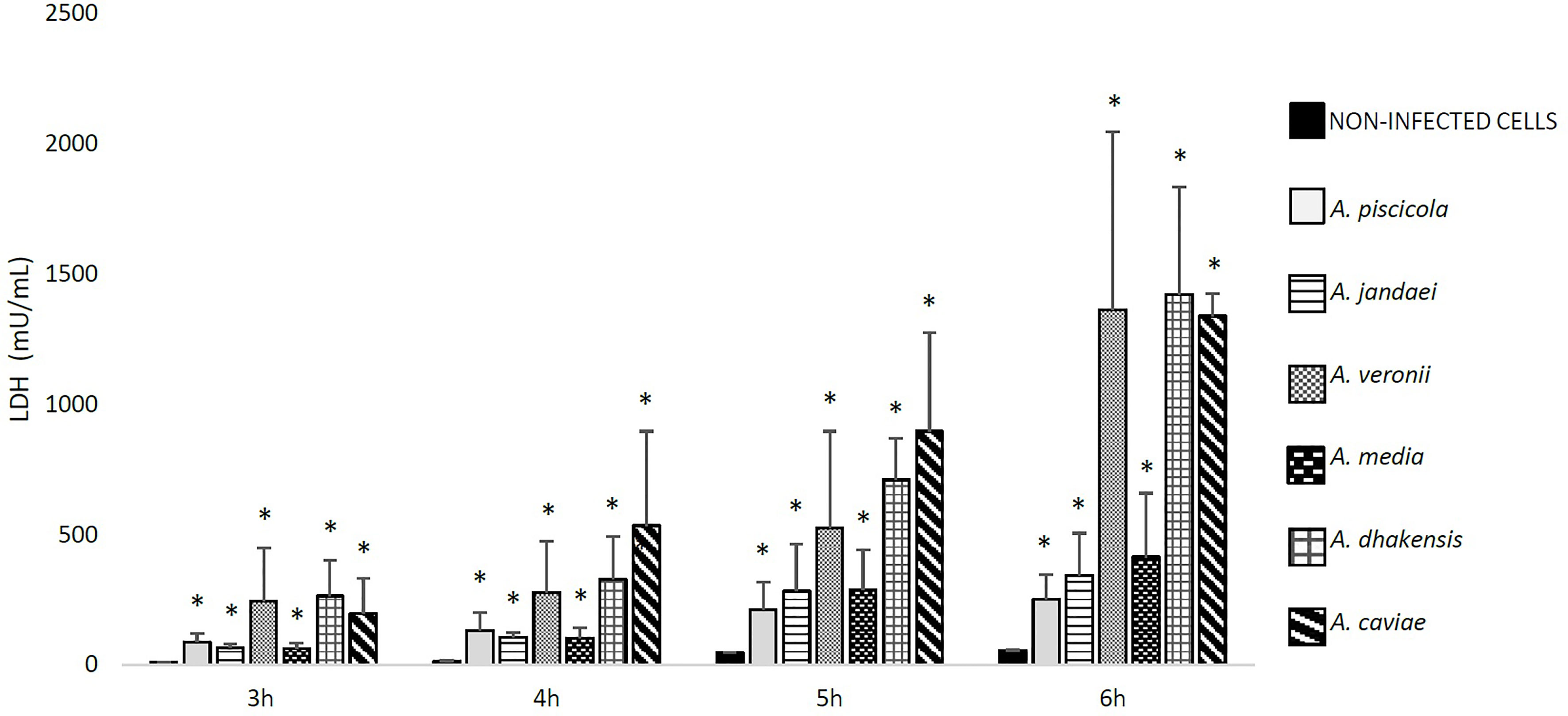
Figure 2 Detected THP-1 cell damage induced by the six different Aeromonas spp. at multiplicity of infection (MOI) 20 and at different exposure times in relation to the non-infected cells, measured by the release of lactate dehydrogenase (LDH) enzyme. Asterisks indicate statistical significance *p < 0.05.
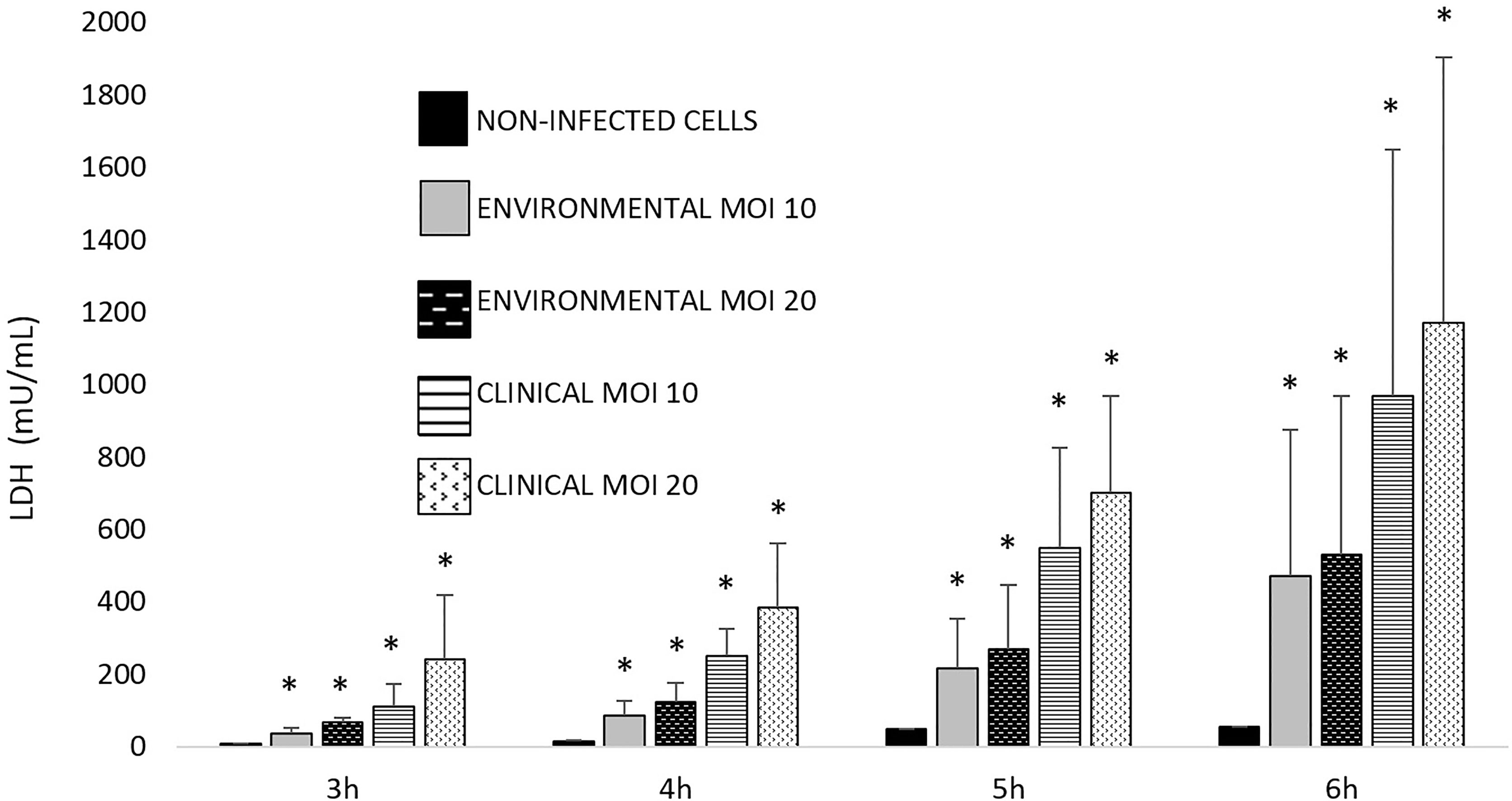
Figure 3 Observed THP-1 cell damage induced by clinical and environmental strains of Aeromonas spp. at different exposure times and multiplicities of infection (MOIs). Asterisks indicate statistical significance *p < 0.05.
After the Aeromonas infection, the transcriptional level of TLR4 gene encoding TLR4 showed significant differences (p < 0.05) in comparison with the expression of this gene in the non-infected cells, but this only occurred after infection with A. dhakensis, A. caviae, and A. veronii (Figure 4).
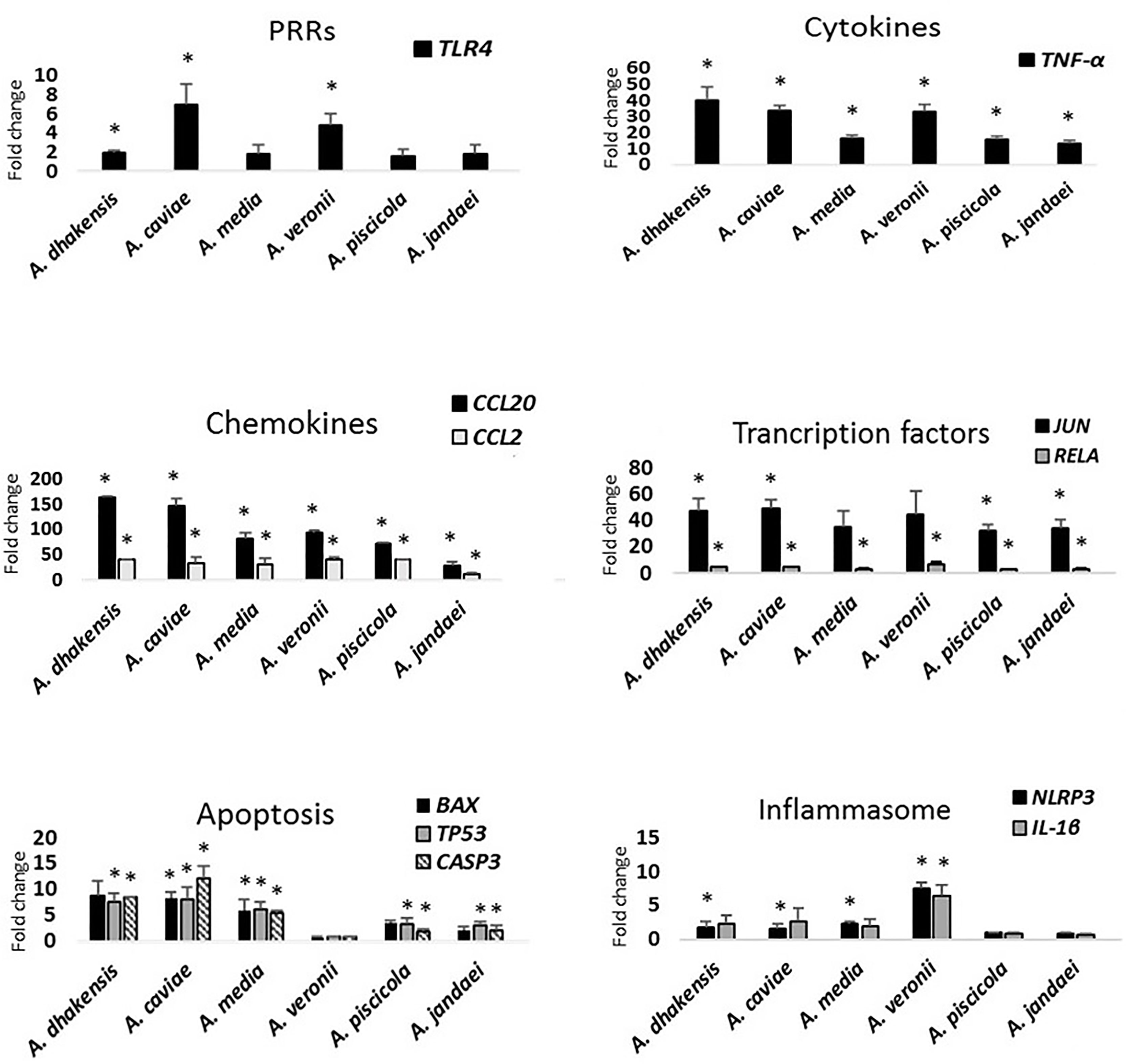
Figure 4 Gene expression profile of THP-1 cells in relation to the non-infected cells induced by the different studied Aeromonas spp. at multiplicity of infection (MOI) 20 determined by RT-qPCR. Transcript levels of the genes were normalized to the expression of GAPDH gene. Expression fold change with respect to the non-infected cells was calculated using the comparative ΔΔCt method. Asterisks indicate statistical significance *p < 0.05.
All Aeromonas strains, independently of the origin and the species, induced the THP-1 monocytic cells to express the genes that encode the cytokine TNF-α and the chemokines (CCL2 and CCL20), with a significant difference (p < 0.05) in the transcription pattern in relation to the non-infected cells (Figure 4). However, the expression levels of TNF-α gene were higher in response to the infections produced by A. dhakensis, followed by A. caviae and A. veronii (p < 0.05). The expression level of the pro-inflammatory cytokine IL-8 was below the detection limit for all species (Figure S1).
In the case of chemokines, the transcriptional levels of CCL2 and CCL20 genes were upregulated after infection with all strains in comparison with the non-infected cells, but a higher expression was observed for CCL20 gene and especially for infections produced with A. dhakensis and A. caviae than with the other species (p < 0.05) (Figure 4). The transcriptional levels of CCL2 and CCL20 genes after A. veronii infection showed no significant differences with A. media or A. piscicola. However, all species showed a significantly higher expression of chemokines (p < 0.05) than A. jandaei (Figure 4).
The THP-1 cells responded by upregulating the JUN transcription factor after infection with all the Aeromonas spp. (p < 0.05). However, the upregulation was not significantly different from the non-infected cells in the case of A. media and A. veronii (Figure 4). RELA gene was also overexpressed (p < 0.05) in relation to the non-infected cells but at a lower level than JUN gene.
The transcriptional level of BAX, TP53, and CASP3 apoptosis genes increased after infection with all Aeromonas species (p < 0.05), with the exception of A. veronii, which showed a very low expression (Figure 4). The expression of BAX gene showed no significant differences in relation to the non-infected cells after A. dhakensis, A. piscicola, and A. jandaei infections (Figure 4). The BAX, TP53, and CASP3 apoptosis genes showed a higher expression (p < 0.05) after infection with the species A. dhakensis, A. caviae, and A. media in comparison with the other species (Figure 4).
NLRP3 and IL1-β genes, which are related to pyroptosis, i.e., cell-death mediated by the formation of the inflammasome, were upregulated in THP-1 cells in response to A. dhakensis, A. caviae, A. media, and A. veronii infections (p < 0.05), while the gene expression was very low after infection with A. piscicola and A. jandaei and showed no significant differences in relation with the expression in the non-infected cells (Figure 4). The upregulation of NLRP3 and IL-1β genes showed an eightfold increase (p < 0.05) when THP-1 cells were infected with A. veronii strains. No significant differences in the level of the IL-1β gene expression were observed when comparing the infection with A. dhakensis, A. caviae, and A. media. However, significant differences in the overexpression of the NLRP3 gene were detected (p < 0.05) for the three species (Figure 4).
Gene expression of act and aerA gene is shown in Figure 5. The results showed a higher expression of both genes in A. veronii (1.01 and 0.98), followed by A. dhakensis (0.95 and 0.96), and significant differences with the other species after 3 h of monocyte infection (p < 0.05). The less transcriptional level of act gene was in A. jandaei (0.64) and of aerA gene in A. piscicola (0.69).
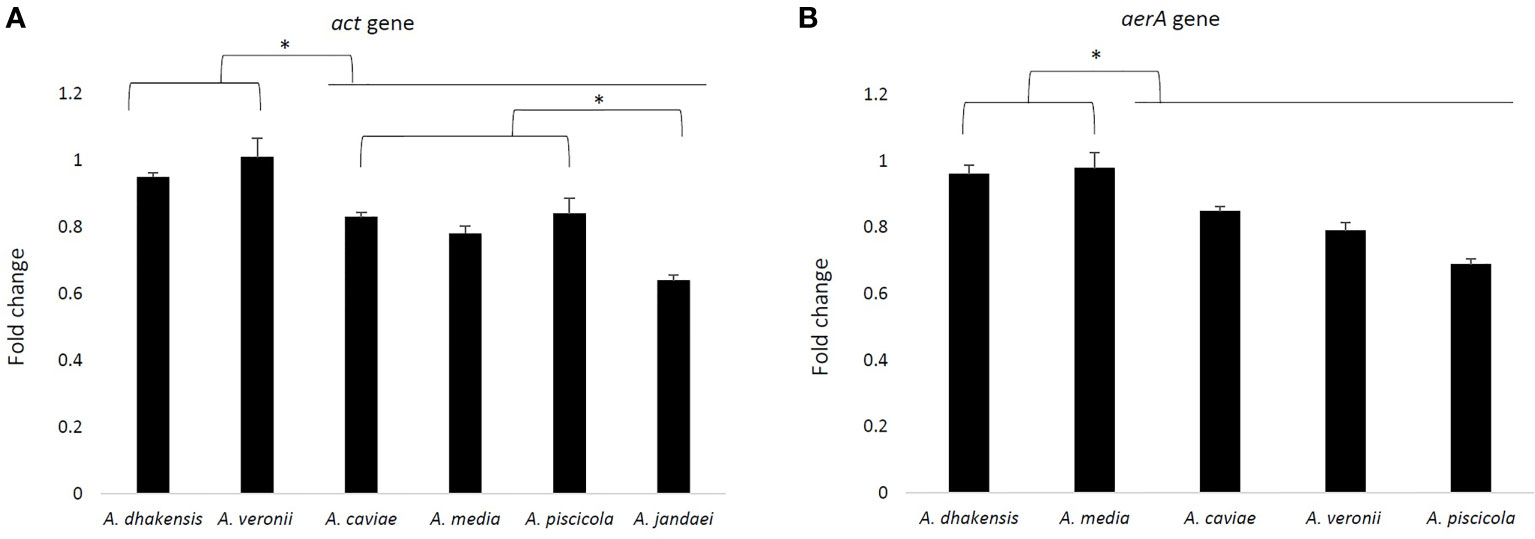
Figure 5 Fold change of the act gene expression profile of six Aeromonas strains (one strain for each species) (A) and the aerA gene with the exception of A. jandaei (B) after 3 h of THP-1 infection, analyzed by RT-qPCR and normalized to the reference gene 16S rRNA. Asterisks indicate a significant difference (*p > 0.05).
The TLRs are a class of PRR proteins that play a key role in the cell innate immune response to infection. In this study, the TLR4 gene expression in THP-1 cells in response to the infections produced by different Aeromonas strains of A. media, A. piscicola, and A. jandaei was shown to be upregulated, but the expression was not significantly higher than in the non-infected cells. The TLR4 is a transmembrane receptor that recognizes a particular type of molecules from many pathogens, in the case of Aeromonas, the LPS, and induces the inflammatory response, producing pro-inflammatory cytokines (17, 27, 53). Previous studies with other microorganisms, including SARS-CoV-2, showed that overstimulation of the TLR4 could be detrimental leading to a hyperinflammation process called cytokine storm (54). The upregulation of the TLR4 gene was demonstrated after A. hydrophila infection in catfish (27). Similarly, Srivastava et al. (55) demonstrated the activation of the TLR4 signaling pathway in zebrafish after A. hydrophila infection. These results suggest that A. hydrophila activates the TLR4 to trigger an anti-inflammatory response to facilitate their survival and pathogenesis. However, other TLR proteins such as TLR5 or TLR21 had been described after A. hydrophila infection by Zhang et al. (27) using fish as an infection model. For this reason, it is also possible that other TLRs, not evaluated in the present study, could be more related to the activation of the immune response after the Aeromonas infection with the species tested in our work. Another explanation could be related to the activation of different TLRs depending on the infected cell type studied as occurs in V. vulnificus between monocytes and endothelial cells (32).
Our results showed that the expression of the pro-inflammatory cytokine gene TNF-α was upregulated in the THP-1 monocytic cell line after infection with all the Aeromonas strains as also occurred for the THP-1 after V. vulnificus infection (32). Similar results were found after A. hydrophila infections of macrophages, yellowtail leukocytes, and grass carp intestinal cells (25, 56, 57). The TNF-α cytokine is involved in the inflammation, and in the absence of this protein, the host defense would be impaired (58). The overexpression of the TNF-α gene in the THP-1 monocytic cell line against infections produced by the six Aeromonas spp. tested confirms that this cytokine participates in the immunological response against Aeromonas infections. However, unlike results from previous studies that demonstrated the upregulated expression of the IL-8 cytokine gene, which induces chemotaxis in neutrophils and stimulate the phagocytosis, in macrophages or in a fish model after infection with Aeromonas (59–61), no upregulation of IL-8 gene in THP-1 monocytic cells could be observed in our study. An explanation of this could be associated with the varying or specific immune response of the tested infected host cell (59, 62) as it was demonstrated to occur when different cell lines were infected with V. vulnificus (32). In the case of the chemokine genes CCL2, which recruits monocytes, memory T cells, and dendritic cells, and CCL20 chemotactic for lymphocytes and neutrophils, our data showed an upregulation of their expression in the THP-1 cells after Aeromonas infection. Chemokines are involved in the chemotaxis of the cells of the immune system such as monocytes/macrophages (CCL2, CCL3, CCL5, CCL7, CCL8, CCL13, CCL17, and CCL22), neutrophils (CXC chemokines), eosinophils (CCL11, CCL24, CCL26, CCL5, CCL7, CCL13, and CCL3), and T cells (CCL2, CCL1, CCL22, and CCL17), among others, participating therefore in their recruitment and the inflammation intracellular signaling control mechanisms (63). In fact, a previous study demonstrated a higher transcriptional level of the chemokine genes (CCL2 and CCL20) after infection of THP-1 cells with V. vulnificus, confirming that monocytes played an equally important role as other immune-related cells to control the infection process (32, 35). The overexpression of CCL2 and CCL20 genes in the THP-1 monocytic cells after infection with all Aeromonas spp. suggests that monocytes play a role in inducing the recruitment of other immune-related cells.
In addition, after Aeromonas infection, the THP-1 cells showed an overexpression of RELA and JUN genes that encode for crucial proteins, i.e., RELA and JUN, for NF-κB activation, responsible for the expression of cytokines, which contribute to an effective immune response (29, 64, 65). Previous studies in a fish model of infection with A. hydrophila demonstrated the expression of RELA gene and the NF-κB pathway that could result in the production of several cytokines such as TNF-α, IL-1β, IL-12, IL-8, IL-6, and IFN (27, 60). In addition, the data obtained by Hayes et al. (28) suggested that JUN could be involved in the different mechanisms of virulence caused by Aeromonas spp. in epithelial colorectal adenocarcinoma (Caco-2) cells. Our results suggested that the overexpression of the genes that encode the transcriptional factors RELA and JUN increased the immune response against these six Aeromonas spp., inducing the production of cytokines and chemokines.
NLRP3 and IL-1β genes related to the pyroptosis cell death mediated by the inflammasome and with an important role that triggers the immune response were clearly upregulated after A. veronii infection. The results of McCoy et al. (36) demonstrated that A. hydrophila induces an inflammatory response via NLRP3 inflammasome, triggers the activation of caspase-1 (CASP1), and releases IL-1β, producing pyroptosis. Additionally, another study suggested that NLRP3 and NLRC4 inflammasomes are involved in host defense against A. veronii infection in mice, triggering the activation of CASP1 with the consequent release of IL-1β and pyroptosis, through the action of the aerolysin and the Type 3 Secretion System (T3SS) (37). However, the expression of NLRP3 and IL-1β genes after A. jandaei and A. piscicola infection of the THP-1 cells was very low and no different from the expression of the THP-1 cells without infection. An explanation for this could be the selection of a different pathway of cell death by these two species because the gene expression analysis showed an upregulation of the genes related to apoptosis (BAX, TP53, and CASP3). However, there was a very low expression of these genes after A. veronii infection. For example, in the early stage of infection, Shigella prevents caspase-4-dependent pyroptotic cell death by delivering the T3SS effector OspC3 (48). However, this affirmation could be related to the presence of virulence factors in each strain, such as T3SS, being therefore strain-dependent. Our results showed a similar expression of BAX, TP53, and CASP3 genes after infection with all A. veronii strains (Figures S1-S4). However, in future studies, it would be interesting to analyze the genome of these strains. In the case of infections with A. dhakensis, A. caviae, and A. media, the THP-1 cells overexpressed the BAX, TP53, and CASP3 genes related to the apoptosis and also those related to pyroptosis. Further studies with a higher number of strains using additional inflammasome genes, like PYCARD, IL-16, or IL-18, are necessary to confirm the hypothesis related to the different pathways of cell death.
The expression of immune-related genes in the present study showed that the selected species caused a different immune response, characterized by a species-specific activation pattern. Independently of the immune-related gene studied in this work, generally, the expression was higher in the most prevalent clinical species A. dhakensis, A. caviae, and A. veronii (5). An explanation of this could be associated with the cytokine storm, described as a systemic inflammatory response syndrome that increases the severity of the infections (66–68), and in this case, these species could induce this mechanism and the consequence could be their higher virulence and therefore prevalence in clinical cases. This is the first time that a hypothesis is provided to explain the different frequencies of occurrence of the Aeromonas species associated with clinical cases.
This study showed that all strains of Aeromonas induce the THP-1 cell damage, independently of the species (data not shown), by measuring the LDH in the supernatant. Epple et al. (69) used this technique to quantify the cell damage in epithelial colon cells, demonstrating the pathogenicity of A. hydrophila. The LDH release can be induced by lysing the cells by apoptosis or pyroptosis. Our results demonstrated that this cell damage could be related to both pathways, depending on the species, as previous studies suspect (36, 37). In addition, the most prevalent clinical species were shown to produce higher cell damage than the less prevalent, and this result agrees with their higher intracellular survival. Also, clinical strains of Aeromonas spp. produced higher cell damage than environmental strains. In the cytokine storm physiological reaction in humans, the innate immune system causes an uncontrolled and excessive release of pro-inflammatory cytokines (67). Taking into account the expression results, it could be indicated that a cytokine storm that induces a strong immune response would cause more cell damage, as well as an increase in the intracellular survival of the most prevalent clinical species A. dhakensis, A. caviae, and A. veronii in the THP-1 host cells. However, the presence/absence, as well as the expression levels of the virulence factors, is not associated with differences between species, since although the most prevalent species in the clinic such as A. veronii or A. dhakensis showed a large number of these, A. piscicola also showed in our study a large amount of virulence genes. In addition, no differences were observed between the species at the expression level of the aerA and act genes. These results coincide with those previously described in which no correlation was found between the presence/expression of virulence factors in Aeromonas and infection in humans (47, 70).
This is the first study that evaluates the immune response after Aeromonas infection using the human monocytic cell THP-1.
The data presented in the study are deposited in the NCBI GenBank repository, accession number: HQ442790.1; HQ442748.1; KJ743585.1; KJ743512.1; HQ442728.1; KR140072.1; HQ442833.1; FM999965.1; AY169338.1; HQ442709.1; JN711805.1; KJ743587.1; KJ743538.1; KP401037.1; KJ743556.1; KJ743508.1; KJ743508.1; HQ442840.1; HQ442859.1; HQ442690.1; KR140073.1; EU488664.1; HE965661.1; EF465510.1.
AF-B was involved in the performance of the assay, assay design, data analysis, and writing of the manuscript. MJF was involved in the conceptualization of assay, data analysis, and writing of the manuscript. All authors contributed to the article and approved the submitted version.
The projects JPIW2013-095-C03-03 of MINECO (Spain) and AQUAVALENS of the Seventh Framework Program (FP7/2007-2013) grant agreement 311846 from the European Union supported the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the staff in the Microbiology Unit at the University Rovira I Virgili (URV), Carme Sanmartí Solé.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.875689/full#supplementary-material
1. Janda JM, Abbott SL. The Genus Aeromonas: Taxonomy, Pathogenicity and Infection. Clin Microbiol Rev (2010) 23:35–73. doi: 10.1128/CMR.00039-09
2. Figueras MJ, Beaz-Hidalgo R. “Aeromonas Infections in Humans,”. In: Aeromonas. Norfolk, UK.: Academic Press. p. 65–108. doi: 10.21775/9781908230560.04
3. Teunis P, Figueras MJ. Reassessment of the Enteropathogenicity of Mesophilic Aeromonas Species. Front Microbiol (2016) 7:1395. doi: 10.3389/fmicb.2016.01395
4. Figueras Salvat MJ, Ashbolt N. “Aeromonas”. In: Pruden A, Ashbolt N, Miller J, editors. Part 3: Specific Excreted Pathogens: Environmental and Epidemiology Aspects - Section 2: Bacteria), Michigan State University, E. Lansing, MI, UNESCO (2019). doi: 10.14321/waterpathogens.21
5. Fernández-Bravo A, Figueras MJ. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms (2020) 8:129. doi: 10.3390/microorganisms8010129
6. Figueras MJ. Clinical Relevance of Aeromonas Sm503. Rev Med Microbiol (2005) 16:145–53. doi: 10.1097/01.revmedmi.0000184410.98677.8a
7. Igbinosa IH, Igumbor EU, Aghdasi F, Tom M, Okoh AI. Emerging Aeromonas Species Infections and Their Significance in Public Health. Sci World J (2012) 2012. doi: 10.1100/2012/625023
8. Hiransuthikul N, Tantisiriwat W, Lertutsahakul K, Vibhagool A, Boonma P. Skin and Soft-Tissue Infections Among Tsunami Survivors in Southern Thailand. Clin Infect Dis (2005) 41:e93–e96. doi: 10.1086/497372
9. Tomás JM. The Main Aeromonas Pathogenic Factors. ISRN Microbiol (2012) 2012. doi: 10.5402/2012/256261
10. Beaz-Hidalgo R, Shakèd T, Laviad S, Halpern M, Figueras MJ. Chironomid Egg Masses Harbour the Clinical Species Aeromonas taiwanensis and Aeromonas sanarellii. FEMS Microbiol Lett (2012) 337:48–54. doi: 10.1111/1574-6968.12003
11. Beaz-Hidalgo R, Figueras MJ. Aeromonas spp. Whole Genomes and Virulence Factors Implicated in Fish Disease. J Fish Dis (2013) 36:371–88. doi: 10.1111/jfd.12025
12. dos Santos PA, Pereira ACM, Ferreira AF, de Mattos Alves MA, Rosa ACP, Freitas-Almeida AC. Adhesion, Invasion, Intracellular Survival and Cytotoxic Activity of Strains of Aeromonas spp. In HEp-2, Caco-2 and T-84 Cell Lines. Antonie van Leeuwenhoek Int J Gen Mol Microbiol (2015) 107:1225–36. doi: 10.1007/s10482-015-0416-4
13. Janeway J. How the Immune System Works to Protect the Host From Infection: A Personal View. Proc Natl Acad Sci U.S.A. (2001) 3:1167–71. doi: 10.1073/pnas.131202998
14. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host Cell Death and Inflammation. Nat Rev Microbiol (2009) 7:99–109. doi: 10.1038/nrmicro2070
15. George Kerry R, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of Probiotics for Human Health: A Review. J Food Drug Anal (2018) 26:927–39. doi: 10.1016/j.jfda.2018.01.002
16. Mahla RS, Reddy MC, Prasad DVR, Kumar H. Sweeten PAMPs: Role of Sugar Complexed PAMPs in Innate Immunity and Vaccine Biology. Front Immunol (2013) 4:248. doi: 10.3389/fimmu.2013.00248
17. Kawai T, Akira S. TLR Signaling. Cell Death Differ (2006) 13:816–25. doi: 10.1038/sj.cdd.4401850
18. Salaun B, Romero P, Lebecque S. Toll-Like Receptor’s Two-Edged Sword: When Immunity Meets Apoptosis. Eur J Immunol (2007) 37:3311–18. doi: 10.1002/eji.200737744
19. Strasser A, O’Connor L, Dixit VM. Apoptosis Signaling. Annu Rev Biochem (2000) 268:10932–937. doi: 10.1146/annurev.biochem.69.1.217
20. Lopez-Castejon G, Brough D. Understanding the Mechanism of IL-1β Secretion. Cytokine Growth Factor Rev (2011) 22:189–95. doi: 10.1016/j.cytogfr.2011.10.001
21. Altmann S, Korytář T, Kaczmarzyk D, Nipkow M, Kühn C, Goldammer T, et al. Toll-Like Receptors in Maraena Whitefish: Evolutionary Relationship Among Salmonid Fishes and Patterns of Response to Aeromonas salmonicida. Fish Shellfish Immunol (2016) 54:391–401. doi: 10.1016/j.fsi.2016.04.125
22. Gong Y, Feng S, Li S, Zhang Y, Zhao Z, Hu M, et al. Genome-Wide Characterization of Toll-Like Receptor Gene Family in Common Carp (Cyprinus carpio) and Their Involvement in Host Immune Response to Aeromonas hydrophila Infection. Comp Biochem Physiol - Part D Genomics Proteomics (2017) 24:89–98. doi: 10.1016/j.cbd.2017.08.003
23. Xu T, Liao Z, Su J. Pattern Recognition Receptors in Grass Carp Ctenopharyngodon idella: II. Organization and Expression Analysis of NOD-Like Receptors. Dev Comp Immunol (2020) 110:103734. doi: 10.1016/j.dci.2020.103734
24. Moeen Abdallah M, Hadi Saleh T. Isolation and Purification of Metalloprotease Produced by Aeromonas hydrophila. Diyala J Med (2018) 80:1351–60. doi: 10.26505/djm.14023660924
25. Suarez G, Sierra JC, Kirtley ML, Chopra AK. Role of Hcp, a Type 6 Secretion System Effector, of Aeromonas hydrophila in Modulating Activation of Host Immune Cells. Microbiology (2010) 156:3678. doi: 10.1099/mic.0.041277-0
26. Shelly A, Banerjee C, Saurav GK, Ray A, Rana VS, Raman R, et al. Aeromonas hydrophila-Induced Alterations in Cytosolic Calcium Activate Pro-Apoptotic cPKC-MEK1/2-Tnfα Axis in Infected Headkidney Macrophages of Clarias gariepinus. Dev Comp Immunol (2017) 76:392–402. doi: 10.1016/j.dci.2017.07.015
27. Zhang XT, Zhang GR, Shi ZC, Yuan YJ, Zheng H, Lin L, et al. Expression Analysis of Nine Toll-Like Receptors in Yellow Catfish (Pelteobagrus fulvidraco) Responding to aeromonas hydrophila Challenge. Fish Shellfish Immunol (2017) 63:384–93. doi: 10.1016/j.fsi.2017.02.021
28. Hayes SL, Waltmann M, Donohue M, Lye DJ, Vesper SJ. Predicting Virulence of Aeromonas Isolates Based on Changes in Transcription of C-Jun and C-Fos in Human Tissue Culture Cells. J Appl Microbiol (2009) 107:964–69. doi: 10.1111/j.1365-2672.2009.04276.x
29. Lorentz A, Klopp I, Gebhardt T, Manns MP, Bischoff SC. Role of Activator Protein 1, Nuclear Factor-κb, and Nuclear Factor of Activated T Cells in IgE Receptor-Mediated Cytokine Expression in Mature Human Mast Cells. J Allergy Clin Immunol (2003) 111:1062–68. doi: 10.1067/mai.2003.1342
30. Arockiaraj J, Palanisamy R, Arasu A, Sathyamoorthi A, Kumaresan V, Bhatt P, et al. An Anti-Apoptotic B-Cell Lymphoma-2 (BCL-2) From Channa striatus: Sequence Analysis and Delayed and Advanced Gene Expression in Response to Fungal, Bacterial and Poly I: C Induction. Mol Immunol (2015) 63:586–94. doi: 10.1016/j.molimm.2014.07.018
31. Liao Z, Wan Q, Su H, Wu C, Su J. Pattern Recognition Receptors in Grass Carp Ctenopharyngodon idella: I. Organization and Expression Analysis of TLRs and RLRs. Dev Comp immunol (2017) 76:93–104. doi: 10.1016/j.dci.2017.05.019
32. Murciano C, Hor LI, Amaro C. Host-Pathogen Interactions in Vibrio vulnificus: Responses of Monocytes and Vascular Endothelial Cells to Live Bacteria. Future Microbiol (2015) 10:471–87. doi: 10.2217/fmb.14.136
33. Banerjee C, Goswami R, Verma G, Datta M, Mazumder S. Aeromonas hydrophila Induced Head Kidney Macrophage Apoptosis in Clarias batrachus Involves the Activation of Calpain and is Caspase-3 Mediated. Dev Comp Immunol (2012) 37:323–33. doi: 10.1016/j.dci.2012.02.005
34. Lü AJ, Hu XC, Wang Y, Zhu AH, Shen LL, Tian J, et al. Skin Immune Response in the Zebrafish, Danio rerio (Hamilton), to Aeromonas hydrophila Infection: A Transcriptional Profiling Approach. J Fish Dis (2015) 38:137–50. doi: 10.1111/jfd.12214
35. Murciano C, Lee CT, Fernández-Bravo A, Hsieh TH, Fouz B, Hor LI, et al. MARTX Toxin in the Zoonotic Serovar of Vibrio vulnificus Triggers an Early Cytokine Storm in Mice. Front Cell Infect Microbiol (2017) 7:1–19. doi: 10.3389/fcimb.2017.00332
36. McCoy AJ, Koizumi Y, Toma C, Higa N, Dixit V, Taniguchi S, et al. Cytotoxins of the Human Pathogen Aeromonas hydrophila Trigger, via the NLRP3 Inflammasome, Caspase-1 Activation in Macrophages. Eur J Immunol (2010) 40:2797–803. doi: 10.1002/eji.201040490
37. McCoy AJ, Koizumi Y, Higa N, Suzuki T. Differential Regulation of Caspase-1 Activation via NLRP3/NLRC4 Inflammasomes Mediated by Aerolysin and Type III Secretion System During Aeromonas veronii Infection. J Immunol (2010) 185:7077–84. doi: 10.4049/jimmunol.1002165
38. Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical Monocytes in Health and Disease. Annu Rev Immunol (2019) 37:439–56. doi: 10.1146/annurev-immunol-042617-053119
39. Schubert RHW, Hegazi M. Aeromonas eucrenophila Species Nova Aeromonas caviae a Later and Illegitimate Synonym of Aeromonas punctata. Zentralblatt fur Bakteriol Mikrobiol und Hyg - Abt 1 Orig A (1988) 268:34–39. doi: 10.1016/S0176-6724(88)80112-3
40. Vega-Sánchez V, Latif-Eugenín F, Soriano-Vargas E, Beaz-Hidalgo R, Figueras MJ, Aguilera-Arreola MG, et al. Re-Identification of Aeromonas Isolates From Rainbow Trout and Incidence of Class 1 Integron and β-Lactamase Genes. Vet Microbiol (2014) 172:528–33. doi: 10.1016/j.vetmic.2014.06.012
41. Hickman-Brenner FW, MacDonald KL, Steigerwalt AG, Fanning GR, Farmer JJ. Aeromonas veronii, a New Ornithine Decarboxylase-Positive Species That may Cause Diarrhea. J Clin Microbiol (1987) 25:900–06. doi: 10.1128/jcm.25.5.900-906.1987
42. Allen DA, Austin B, Colwell RR. Aeromonas media, a New Species Isolated From River Water. Int J Syst Bacteriol (1983) 33:599–604. doi: 10.1099/00207713-33-3-599
43. Carnahan A, Fanning GR, Joseph SW. Aeromonas jandaei (Formerly Genospecies DNA Group 9 A. sobria), a New Sucrose-Negative Species Isolated From Clinical Specimens. J Clin Microbiol (1991) 29:560–64. doi: 10.1128/jcm.29.3.560-564.1991
44. Beaz-Hidalgo R, Alperi A, Figueras MJ, Romalde JL. Aeromonas piscicola sp. nov., Isolated From Diseased Fish. Syst Appl Microbiol (2009) 32:471–9. doi: 10.1016/j.syapm.2009.06.004
45. Soler L, Yáñez MA, Chacon MR, Aguilera-Arreola MG, Catalán V, Figueras MJ, et al. Phylogenetic Analysis of the Genus Aeromonas Based on Two Housekeeping Genes. Int J Syst Evol Microbiol (2004) 54:1511–9. doi: 10.1099/IJS.0.03048-0
46. Chacón MR, Soler L, Groisman EA, Guarro J, Figueras MJ. Type III Secretion System Genes in Clinical Aeromonas Isolates. J Clin Microbiol (2004) 4:1285–87. doi: 10.1128/JCM.42.3.1285-1287.2004
47. Lee HJ, Hoel S, Lunestad BT, Lerfall J, Jakobsen AN. Aeromonas spp. Isolated From Ready-to-Eat Seafood on the Norwegian Market: Prevalence, Putative Virulence Factors and Antimicrobial Resistance. J Appl Microbiol (2021) 130:1380–93. doi: 10.1111/jam.14865
48. Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and Characterization of a Human Acute Monocytic Leukemia Cell Line (THP-1). Int J Cancer (1980) 26:171–76. doi: 10.1002/ijc.2910260208
49. Rupp M, Pilo P, Müller B, Knüsel R, von Siebenthal B, Frey J, et al. Systemic Infection in European Perch With Thermoadapted Virulent Aeromonas salmonicida (Perca fluviatilis). J Fish Dis (2019) 42:685–91. doi: 10.1111/jfd.12970
50. Fernández-Bravo A, Kilgore PB, Andersson JA, Blears E, Figueras MJ, Hasan NA, et al. T6SS and ExoA of Flesh-Eating Aeromonas hydrophila in Peritonitis and Necrotizing Fasciitis During Mono- and Polymicrobial Infections. Proc Natl Acad Sci U.S.A. (2019) 116:24084–92. doi: 10.1073/pnas.1914395116
51. Zhao X, Zhang C, Hua M, Wang R, Zhong C, Yu J, et al. NLRP3 Inflammasome Activation Plays a Carcinogenic Role Through Effector Cytokine IL-18 in Lymphoma. Oncotarget (2017) 8:108571. doi: 10.18632/oncotarget.21010
52. Fernández-Bravo A, López-Fernández L, Figueras MJ. The Metallochaperone Encoding Gene hypA is Widely Distributed Among Pathogenic Aeromonas spp. And its Expression is Increased Under Acidic pH and Within Macrophages. Microorganisms (2019) 7:415. doi: 10.3390/microorganisms7100415
53. Rosenzweig JA, Chopra AK. The Exoribonuclease Polynucleotide Phosphorylase Influences the Virulence and Stress Responses of Yersiniae and Many Other Pathogens. Front Cell Infect Microbiol (2013) 3:81. doi: 10.3389/fcimb.2013.00081
54. Aboudounya MM, Heads RJ. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators Inflammation (2021) 2021. doi: 10.1155/2021/8874339
55. Srivastava N, Shelly A, Kumar M, Pant A, Das B, Majumdar T, et al. Aeromonas hydrophila Utilizes TLR4 Topology for Synchronous Activation of MyD88 and TRIF to Orchestrate Anti-Inflammatory Responses in Zebrafish. Cell Death Discovery (2017) 7:16662–667. doi: 10.1038/cddiscovery.2017.67
56. Reyes-Becerril M, Ascencio-Valle F, Hirono I, Kondo H, Jirapongpairoj W, Esteban MA, et al. Tlr21’s Agonists in Combination With Aeromonas Antigens Synergistically Up-Regulate Functional TLR21 and Cytokine Gene Expression in Yellowtail Leucocytes. Dev Comp Immunol (2016) 61:107–15. doi: 10.1016/j.dci.2016.03.012
57. Song X, Hu X, Sun B, Bo Y, Wu K, Xiao L, et al. A Transcriptome Analysis Focusing on Inflammation-Related Genes of Grass Carp Intestines Following Infection With Aeromonas hydrophila Sci Rep (2017) 7:1–12. doi: 10.1038/srep40777
58. Pfeffer K. Biological Functions of Tumor Necrosis Factor Cytokines and Their Receptors. Cytokine Growth Factor Rev (2003) 14:185–91. doi: 10.1016/S1359-6101(03)00022-4
59. Brietzke A, Korytář T, Jaros J, Köllner B, Goldammer T, Seyfert HM, et al. Aeromonas salmonicida Infection Only Moderately Regulates Expression of Factors Contributing to Toll-Like Receptor Signaling But Massively Activates the Cellular and Humoral Branches of Innate Immunity in Rainbow Trout (Oncorhynchus mykiss). J Immunol Res (2015) 2015. doi: 10.1155/2015/901015
60. Zhang J, Li L, Kong X, Wu F, Zhou C, Nie G, et al. Expression Patterns of Toll-Like Receptors in Natural Triploid Carassius auratus After Infection With Aeromonas hydrophila. Vet Immunol Immunopathol (2015) 168:77–82. doi: 10.1016/j.vetimm.2015.08.009
61. Kong W, Huang C, Tang Y, Zhang D, Wu Z, Chen X. Effect of Bacillus subtilis on Aeromonas hydrophila-Induced Intestinal Mucosal Barrier Function Damage and Inflammation in Grass Carp (Ctenopharyngodon idella). Sci Rep (2017) 7:1–11. doi: 10.1038/s41598-017-01336-9
62. Galindo CL, Fadl AA, Sha J, Gutierrez C, Popov VL, Boldogh I, et al. Aeromonas hydrophila Cytotoxic Enterotoxin Activates Mitogen-Activated Protein Kinases and Induces Apoptosis in Murine Macrophages and Human Intestinal Epithelial Cells. J Biol Chem (2004) 279:37597–612. doi: 10.1074/jbc.M404641200
63. Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and Chemokines: At the Crossroads of Cell Signalling and Inflammatory Disease. Biochim Biophys Acta - Mol Cell Res (2014) 1843:2563–82. doi: 10.1016/j.bbamcr.2014.05.014
64. Tak PP, Firestein GS. NF-κb: A Key Role in Inflammatory Diseases. J Clin Invest (2001) 107:7–11. doi: 10.1172/JCI11830
65. Hoesel B, Schmid JA. The Complexity of NF-κb Signaling in Inflammation and Cancer. Mol Cancer (2013) 12:86. doi: 10.1186/1476-4598-12-86
66. Vidal JM, Kawabata TT, Thorpe R, Silva-Lima B, Cederbrant K, Poole S, et al. In Vitro Cytokine Release Assays for Predicting Cytokine Release Syndrome: The Current State-of-the-Science. Report of a European Medicines Agency Workshop. Cytokine (2010) 51:213–15. doi: 10.1016/j.cyto.2010.04.008
67. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the Eye of the Cytokine Storm. Microbiol Mol Biol Rev (2012) 76:16–32. doi: 10.1128/mmbr.05015-11
68. Chousterman BG, Swirski FK, Weber GF. Cytokine Storm and Sepsis Disease Pathogenesis. Semin Immunopathol (2017) 39:517–28. doi: 10.1007/s00281-017-0639-8
69. Epple HJ, Mankertz J, Ignatius R, Liesenfeld O, Fromm M, Zeitz M, et al. Aeromonas hydrophila Beta-Hemolysin Induces Active Chloride Secretion in Colon Epithelial Cells (HT-29/B6). Infect Immun (2004) 72:4848–58. doi: 10.1128/IAI.72.8.4848-4858.2004
Keywords: Aeromonas spp., immune-related genes, monocytic cells, cell damage, intracellular survival
Citation: Fernández-Bravo A and Figueras MJ (2022) Immune Response of the Monocytic Cell Line THP-1 Against Six Aeromonas spp. Front. Immunol. 13:875689. doi: 10.3389/fimmu.2022.875689
Received: 14 February 2022; Accepted: 30 May 2022;
Published: 08 July 2022.
Edited by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranReviewed by:
Inácio Mateus Assane, University Zambeze, MozambiqueCopyright © 2022 Fernández-Bravo and Figueras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Fernández-Bravo, YW5hLmZlcm5hbmRlekB1cnYuY2F0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.