- 1Department of Clinical Veterinary Medicine, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 3The Center for Animal Disease Control and Prevention in Wuhan, Wuhan Bureau of Agriculture and Rural Bureau Affairs, Wuhan, China
- 4Department of Preventive Veterinary Medicine, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
Mastitis is a common disease that hinders the development of dairy industry and animal husbandry. It leads to the abuse of antibiotics and the emergence of super drug-resistant bacteria, and poses a great threat to human food health and safety. Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) are the most common pathogens of mastitis in dairy cows and usually cause subclinical or clinical mastitis. CircRNAs and N6-methyladenosine (m6A) play important roles in immunological diseases. However, the mechanisms by which m6A modifies circRNA in bovine mammary epithelial cells remain poorly understood. The aim of our study was to investigate m6A-modified circRNAs in bovine mammary epithelial cells (MAC-T cells) injured by S. aureus and E. coli. The profile of m6A-modified circRNA showed a total of 1,599 m6A peaks within 1,035 circRNAs in the control group, 35 peaks within 32 circRNAs in the S. aureus group, and 1,016 peaks within 728 circRNAs in the E. coli group. Compared with the control group, 67 peaks within 63 circRNAs were significantly different in the S. aureus group, and 192 peaks within 137 circRNAs were significantly different in the E. coli group. Furthermore, we found the source genes of these differentially m6A-modified circRNAs in the S. aureus and E. coli groups with similar functions according to GO and KEGG analyses, which were mainly associated with cell injury, such as inflammation, apoptosis, and autophagy. CircRNA–miRNA–mRNA interaction networks predicted the potential circRNA regulation mechanism in S. aureus- and E. coli-induced cell injury. We found that the mRNAs in the networks, such as BCL2, MIF, and TNFAIP8L2, greatly participated in the MAPK, WNT, and inflammation pathways. This is the first report on m6A-modified circRNA regulation of cells under S. aureus and E. coli treatment, and sheds new light on potential mechanisms and targets from the perspective of epigenetic modification in mastitis and other inflammatory diseases.
Introduction
Mastitis is currently the most widely investigated disease in dairy cows and is usually caused by infection with specific pathogenic microorganisms (1). Currently, treatment with antibiotic causes the abuse of antibiotics and the emergence of super drug-resistant bacteria. It is a great threat to human health. For instance, methicillin-resistant Staphylococcus aureus (MRSA) from bovines is now the most serious acute infectious disease in hospital, neonatal delivery room, and community (2). S. aureus and Escherichia coli are the most common pathogens that cause mastitis, and the infection has variable clinical symptoms ranging from different pathogens (3). The characteristics of subclinical mastitis caused by S. aureus are chronic or invisible, which makes disease detection and therapy difficult (4). Conversely, mastitis caused by E. coli and some Gram-negative bacteria usually passes through acutely and is associated with a list of severe symptoms (5). Bovine mammary epithelial cells, as the first layer, have an essential defense function, and the tight junctions of the cells provide a barrier to enhance natural mammary immunity. The cells secrete inflammatory factors to fight bacterial infections when activated (6). Moreover, it will express a set of cytokine/chemokine factors and directly resist pathogens, such as bactericidal β-defensins (7), complement factors, and acute phase proteins (8, 9). This response occurs through an inflammatory-related pathway (10–13). Some studies have confirmed the immune response of mastitis in S. aureus and E. coli infection (14–18). However, the regulatory function of noncoding RNA and epigenetic modification is ignored.
Circular RNA (circRNA) has attracted considerable critical attention in recent years. As an indispensable noncoding RNA, circRNA is constituted by a highly stable and conserved covalent closed loop structure without a 3’ cap and 5’ polyA structure in organisms. Depending on the source gene of circRNA, different studies reported that the circRNA could be separated into different categories (19–21), such as exonic circRNA, intergenic circRNA, intronic circRNA, antisense circRNA, and sense overlapping circRNA. CircRNAs have been reported to bind to miRNAs as sponges and regulate target gene expression by combining with miRNAs (22). With the development of sequencing technology, many researchers have found that circRNAs widely exist in immune cells (23–25). Additionally, KEGG and GO analyses indicated that circRNAs played an essential role in many pathways associated with inflammation and immunity, such as the NF-κB, NLR, and TNF pathways (26). Therefore, circRNA has been considered a breakthrough in the innovative treatment of inflammatory disease.
M6A is a common epitranscriptome modification of RNA that is widely distributed in mammary epithelial cells (27). m6A has been found in diverse ribonucleic acids (28–31). By regulating RNA splicing (32), degradation (33), stability (34), and translation (35), the final manifestation is the impact on various pathological or physiological processes, such as the cell cycle (36) and immune function (37). Interestingly, circRNA could be regulated by m6A modification, showing a different m6A pattern from mRNA (38). Recent studies have shown that m6A-modified circRNAs have a certain impact on immune liver metastasis (39). Nevertheless, there is no study of the relationship between m6A-modified circRNAs and mastitis in dairy bovines.
To identify the potential function of m6A modification in regulating circRNA, we used RNA immunoprecipitation and high-throughput sequencing (MeRIP-seq) for the first time to profile the circRNA regulated by m6A modification in mammary epithelial cells (MAC-T) injured by inactivated S. aureus and E. coli. The features of m6A-modified circRNAs were identified first, and the potential functions were also predicted by GO enrichment analysis and KEGG pathway analysis. Moreover, a circRNA–miRNA–mRNA interaction network was constructed to predict competitive endogenous RNAs (ceRNAs). For further investigation, conjoint analysis of circRNA-seq and MeRIP-seq was performed. Together, our findings demonstrated the potential molecular function of circRNAs regulated by m6A modification caused by S. aureus- and E. coli-induced cell injury, which suggested that S. aureus and E. coli may affect the cell inflammation process through m6A-modified circRNAs, and provided us with possible diagnosis and treatment targets in mastitis.
Materials and Methods
Cell Lines and Bacteria
MAC-T cells (an immortalized bovine mammary epithelial cell line) were donated by Professor Mark Hanigan of Virginia Tech University. MAC-T cells were cultured by adapting the procedure used by Li et al. (40).
S. aureus (ATCC 29213) and E. coli (ATCC 25922) were donated by Professor Zhou Rui and Professor Wang Xiangru of Huazhong Agricultural University. S. aureus and E. coli were resuscitated, single colony purified, cultured, and heat-inactivated by adapting the procedure used by Li et al. (40).
MAC-T Cell Injury Induced by Inactivated S. aureus and E. coli
MAC-T cells were seeded in 6-well plates (Corning, US) at a density of 2×105 cells/well. After 12 h of culture, inactivated bacteria were added at an MOI of 10:1. Inactivated bacteria and cells were cocultured for 24 h. Cells in each well were washed 3 times with cold PBS (HyClone, China) before TRIzol (Invitrogen, US) was added. There were 3 replicate wells for each treatment and 3 replicate wells for the control, for a total of 9 samples.
Extraction of Total RNA and RT-qPCR
Total RNA of the above 9 samples was extracted according to the instructions of the commercial reagent manufacturer and the RNA isolation steps. A NanoDrop ND-1000 instrument (Agilent Inc. USA) was used to measure RNA concentration and purity, and the quality control index of RNA purity was based on the OD260/OD280 value between 1.8 and 2.0. Denaturing agarose gel electrophoresis was used to measure RNA integrity and gDNA contamination. The sample was stored at −80°C for later use.
The RNA samples obtained were reverse transcribed into cDNA using Vazyme HiScript II Reverse Transcriptase (+gDNA wiper) (Vazyme, Nanjing, China), and the expression of related RNA was detected by Vazyme AceQ® SYBR qPCR Master Mix (Vazyme) in a ViiA7 Real-time PCR System (Applied Biosystems Inc., Foster City, CA, United States). The 2−△△ct method was used to analyze the fluorescence quantitative data, and GraphPad Prism 7.0 was used to process the data. Related primer sequences are shown at Table S1.
Flow Cytometry
After MAC-T cells were digested, collected, and washed three times with PBS, the cells were resuspended in 100 µl of Binding Buffer. Then, 5 µl each of FITC and PI dyes (Vazyme, China) was added and left for 10 min at room temperature in the dark. After adding 400 µl of Binding Buffer, the cells were detected using CytoFLEX-LX (Beckman Coulter, Indianapolis, IN, USA).
Transmission Electron Microscopy
MAC-T cells were treated with S. aureus and E. coli and fixed with 2% glutaraldehyde fixation buffer. The cells were used for TEM slide preparation. Sections were observed via a 100 KV H7650 transmission electron microscope (HITACHI, Japan).
Establishment of circRNA Library
Nine samples under different treatments were sent to Cloud-Seq Biotech (Shanghai, China) for MeRIP-CircRNA sequencing. The operation was briefly described as follows: (1) To enrich circRNA, a circRNA Enrichment Kit (Cloud-seq, USA) was used according to the supplier’s instructions. (2) An NEBNext® Ultra™ II Directional RNA Library Prep Kit (New England Biolabs, Inc., Massachusetts, USA) was used to pretreat RNA, and a sequencing library was constructed by immunoprecipitation. (3) Quality control and quantification of the library were performed with the BioAnalyzer 2100 system (Agilent Technologies, USA), and the Illumina HiSeq instrument was used to perform 150 bp paired-end sequencing.
CircRNA Raw Data
After paired-end sequencing, raw circRNA data were obtained. According to Q30 for quality control, cutadapt software (v1.9.3) was used to obtain high-quality reads. STAR software (v2.5.1b) was used to compare high-quality reads to the reference genome/transcriptome (bosTau9), and DCC software (v0.4.4) was used for circRNA detection and identification. The circBase database and Circ2Traits were used to annotate the identified circRNA. Then, edgeR software (v3.16.5) was used for data standardization and differential expression circRNA screening (fold change ≥ 1.5, p-value ≤ 0.05). The standardized number of reads was used to calculate the differential expression of circRNA between the three groups of samples. A fold change ≥ 2.0 and p-value ≤ 0.05 were considered the thresholds for differential circRNA.
Bioinformatics Analysis and Statistical Analysis
According to the circRNA verified by sequencing, the differentially expressed circRNA was selected. Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/keg, accessed on 27 April 2020) and Gene Ontology (GO; http://www.geneontology.org, accessed on 27 April 2020) enrichment analyses for the circRNA source gene were performed using the DAVID biometric analysis tool. The downstream mRNAs were predicted using miRBase (http://mirbase.org/index.shtml).
Statistical analysis of the data was performed using GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA, United States). Data are represented as the mean with SD, and the significant differences were analyzed by Student’s t-test; p < 0.05 was considered statistically significant.
Additionally, RNAhybrid (2.1.2) was used to predict circRNA and miRNA interaction (41). Mirbase 22 (https://mirbase.org/, accessed on March 3, 2018) was used to predict the downstream mRNA of the predicted miRNA.
Results
Inflammation, Apoptosis, and Autophagy in MAC-T Cells Treated With Inactivated S. aureus and E. coli
To determine whether the stimulation of inactivated bacteria on MAC-T cells can cause cell damage, we added bacteria to stimulate the cells for 24 h. The results showed that IL-1β, IL-6, and TNF-α were significantly upregulated in the S. aureus group and E. coli group, and the expression of these three genes in the E. coli group was significantly higher than that in the S. aureus group (Figures 1A–C). Additionally, cell apoptosis was significantly increased (Figures 1D–F). It showed almost 5 times the apoptosis rate in the S. aureus group and E. coli group compared to the control group. In contrast to the trend in mRNA expression associated with inflammation, flow cytometry illustrated a similar apoptosis rate when cells were treated with S. aureus and E. coli (Figure 1G).
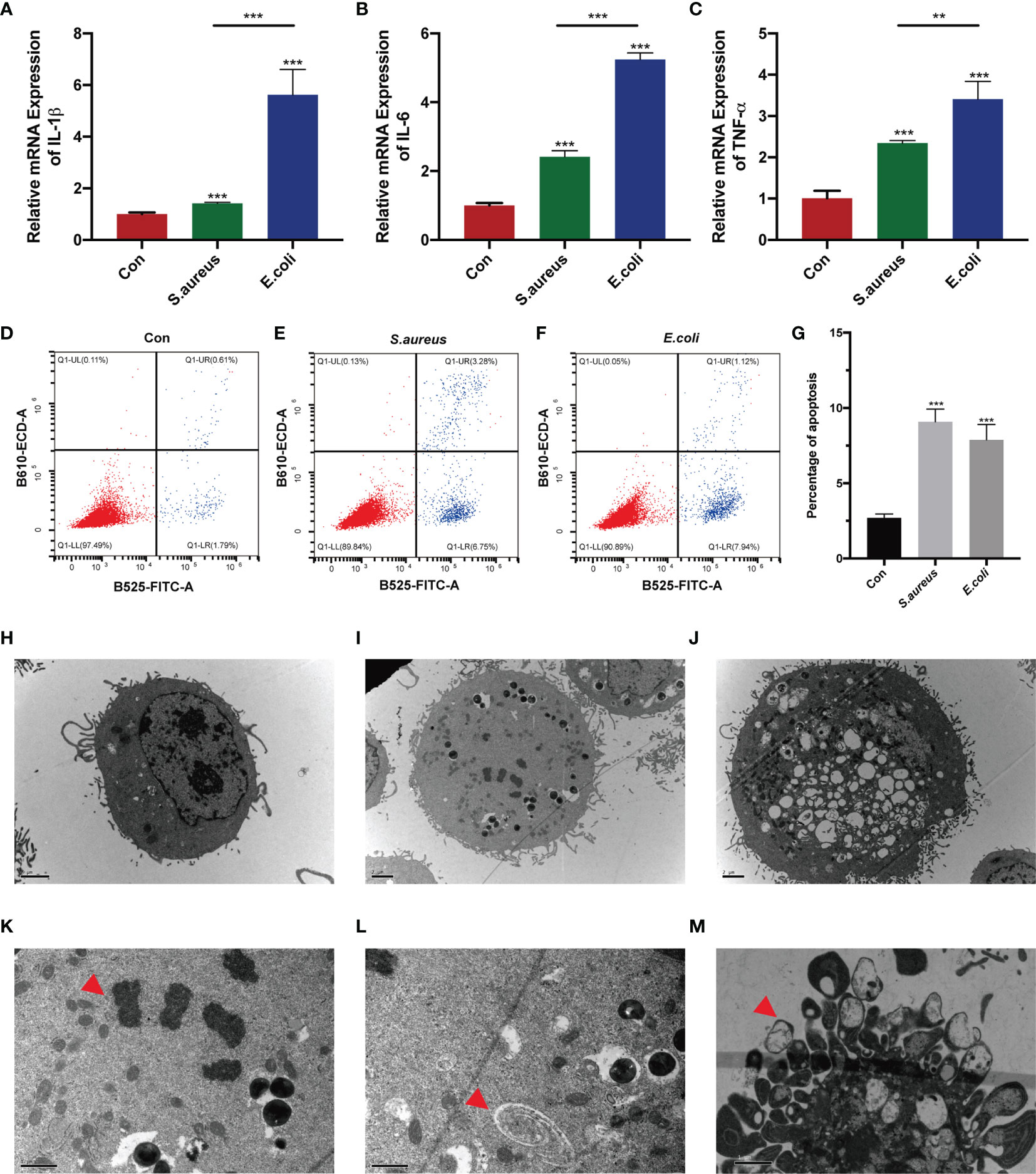
Figure 1 MAC-T cells injured by S. aureus and E. coli. (A–C) MAC-T cells were stimulated by inactivated S. aureus and E. coli and significantly higher IL-1β, IL-6, and TNF-α mRNA expression was found in the treatment groups. (D–F) Flow cytometry detected apoptosis in the S. aureus group and E. coli group. (G) The percentage of apoptotic cells in the S. aureus group and E. coli group was significantly higher than that in the control group. (H–M) Transmission electron microscopy (TEM) imaging of MAC-T cells. TEM illustrated (H) normal MAC-T cells, (I) MAC-T cells injured by S. aureus, and (J) MAC-T cells injured by E. coli. (K) Nuclear membrane disappearance, chromatin condensation (red arrow) and (L) autophagosomes (red arrow) were found in the S. aureus group. (M) Apoptotic bodies (red arrow) were found in the E. coli group (**p < 0.01, ***p < 0.001).
To more intuitively observe the cell injury in S. aureus treatment and E. coli treatment, transmission electron microscopy was used (Figures 1H–M). Interestingly, we found the nuclear membrane disappearance, chromatin condensation (Figure 1K), and autophagosomes (Figure 1L) in MAC-T cells injured by S. aureus (Figure 1I), which indicated minor damage in these cells. However, cells with acute cytosolic voids (Figure 1J) and apoptotic bodies (Figure 1M) were found in MAC-T cells injured by E. coli.
Identification of m6A-Modified circRNAs in MAC-T Cells Treated With Inactivated S. aureus and E. coli
To investigate the circRNA profile in MAC-T cells injured by heat-inactivated S. aureus or E. coli, purified cellular RNA was subjected to circRNA-seq and MeRIP-seq. The sequencing raw reads were generated from the control group, S. aureus group, and E. coli group. Bioinformatic approaches, including Cutadapt and Bowtie2 software, were performed for data filtering and quality control. We obtained clean reads and the proportions of net reads were between 99.122% and 99.995% with stringent quality control (Table S2).
To clarify the relationship between m6A modification and normal circRNA, we first analyzed the circRNA profile from circRNA-seq and found that most circRNAs were less than 2,000 bp and derived from exons (Figures S1A, B). Moreover, genes were mainly distributed on chromosome 3 (Figure S1E). When MAC-T cells were injured by S. aureus and E. coli, the source and length of the differential circRNA showed similar patterns (Figures S1C, D, F). Most circRNAs were located on chromosome 3 in the S. aureus group, while most circRNAs were located on chromosome 8 in the E. coli group (Figures S1F, G).
Further analysis of the MeRIP-seq data was performed, and the m6A methylation peaks were compared and analyzed based on the sequencing data. There were 1,599 m6A methylation peaks within 1,035 circRNAs in the control group, 35 peaks within 32 circRNAs in the S. aureus group, and 1,016 within 728 circRNAs in the E. coli group (Figures 2A, B).
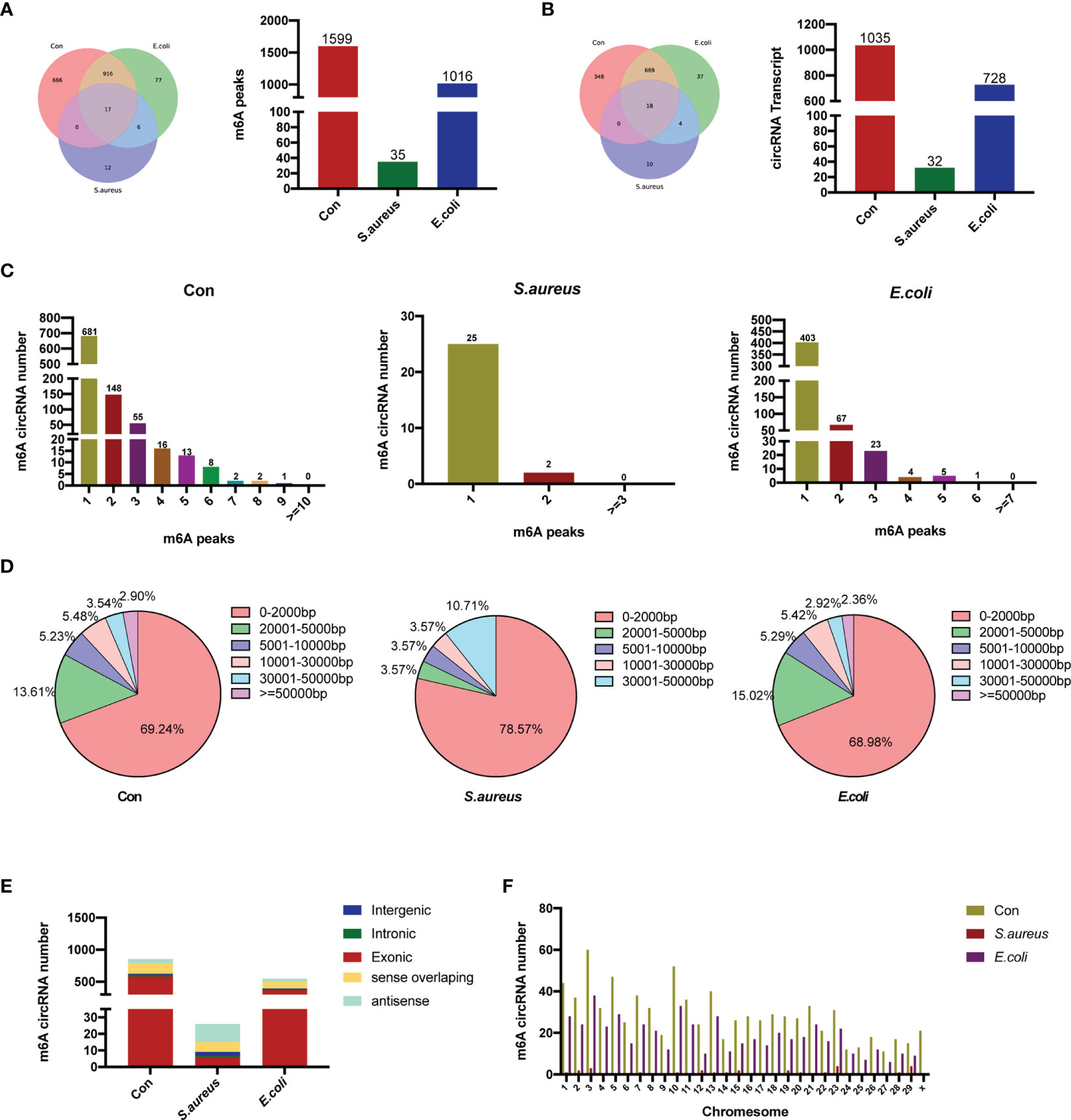
Figure 2 Overview of m6A-modified circRNAs in MAC-T cells injured by S. aureus and E. coli. (A) Venn diagram and histogram displaying the specific m6A peaks between the control group, S. aureus group, and E. coli group. (B) Venn diagram and histogram showing the specific circRNA with m6A modification between the three groups. (C) The number of m6A peaks per circRNA in the three groups. (D) The length of m6A-modified circRNAs in the three groups. (E) The source of m6A-modified circRNAs in the three groups. (F) Chromosome distribution.
Characteristics of m6A-Modified circRNAs in MAC-T Cells Treated With Inactivated S. aureus and E. coli
A total of 17 m6A peaks and 18 circRNA transcripts were observed in all three groups, whereby 933 methylation peaks and 687 circRNAs occurred in both the E. coli group and the control group. The number of m6A methylation peaks was obviously different among the three groups. There were 18 m6A peaks in the S. aureus group instead of the control group and 83 m6A peaks in the E. coli group instead of the control group.
Further analysis was performed to assess the features of m6A-modified circRNAs. The number of m6A methylation peaks in each circRNA was highly similar in the control group, S. aureus group, and E. coli group (Figure 2C). Similar to recent studies, we have found that most circRNAs have one m6A methylation peak, but there are also a large number of circRNAs with more than one m6A methylation peak, which indicates that m6A modification sites are not unique in circRNAs. Moreover, the lengths of m6A-modified circRNAs in each group were analyzed. The length of most m6A-modified circRNAs was less than 5,000 bp, and similar characteristics of length were shown in all three groups (Figure 2D). In our studies, the sources of the circRNA in the three groups were subsequently analyzed. CircRNAs in the control group and E. coli group derived from exons were the most abundant. However, in the S. aureus group, there was a certain amount of m6A-modified circRNA derived from antisense and sense overlapping (Figure 2E). Finally, chromosome distribution also revealed that m6A-methylated circRNA is more likely to be present on chromosome 3 (Figure 2F).
Comparison of m6A-Modified circRNAs in MAC-T Cells Injured by S. aureus and E. coli
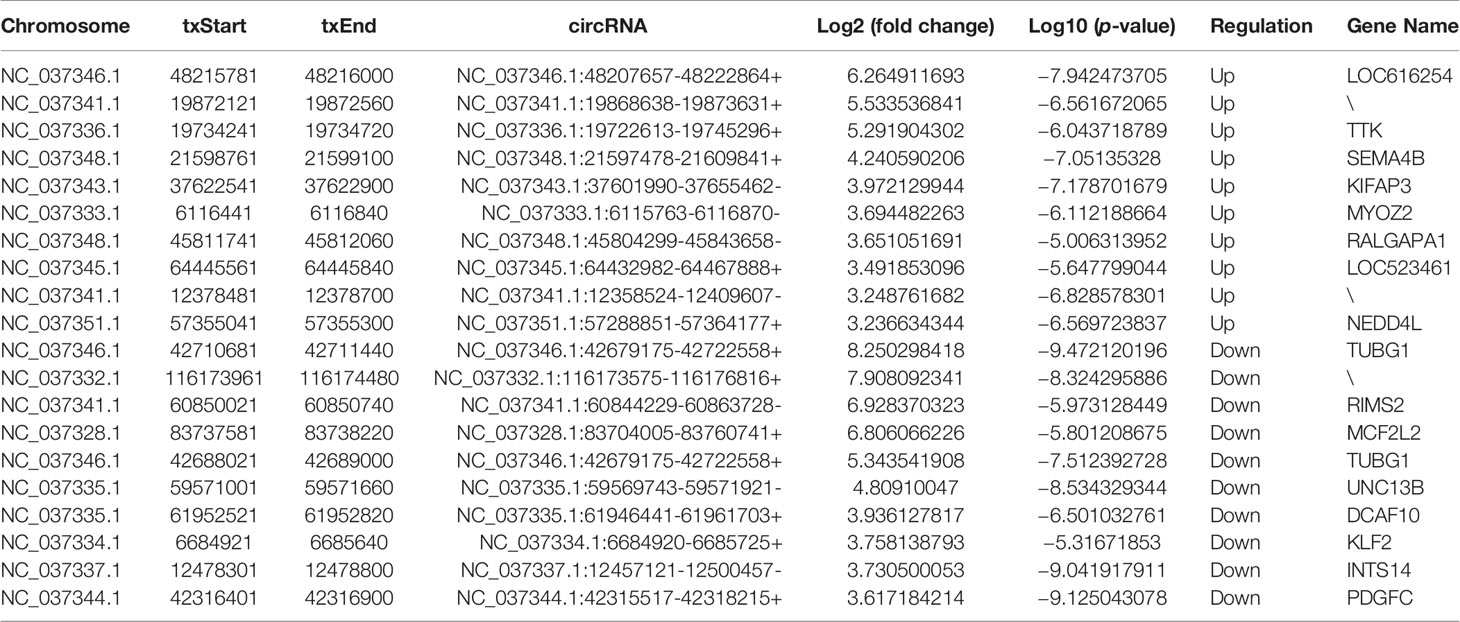
To identify the role of m6A-modified circRNA in regulating MAC-T cell injury induced by S. aureus and E. coli, the various m6A modification levels between S. aureus/E. coli and the control group were analyzed (fold change > 2, p-value < 0.05). Compared to the control group, a total of 259 differential m6A modification peaks within 200 circRNAs were found. Further analysis confirmed that 67 m6A methylation peaks within 63 circRNAs were significantly different between the S. aureus group and the control group, of which 26 hypermethylated peaks were within 25 circRNAs (e.g., circRNA_OPA3), and 41 hypomethylated peaks were within 38 circRNAs (e.g., circRNA_AKT3). Meanwhile, 192 m6A methylation peaks within 137 circRNAs were significantly different between the E. coli group and the control group, of which 75 hypermethylated peaks were within 47 circRNAs (e.g., circRNA_GRB10), and 117 hypomethylated peaks were within 90 circRNAs (e.g., circRNA_SMAD1). Data visualization analysis was performed by IGV to show the differential m6A peaks between the control group and treatment group (Figure 3A). The top 20 differentially methylated circRNAs with hypermethylation or hypomethylation in the S. aureus and E. coli group compared to the control group are shown (Tables 1, 2).
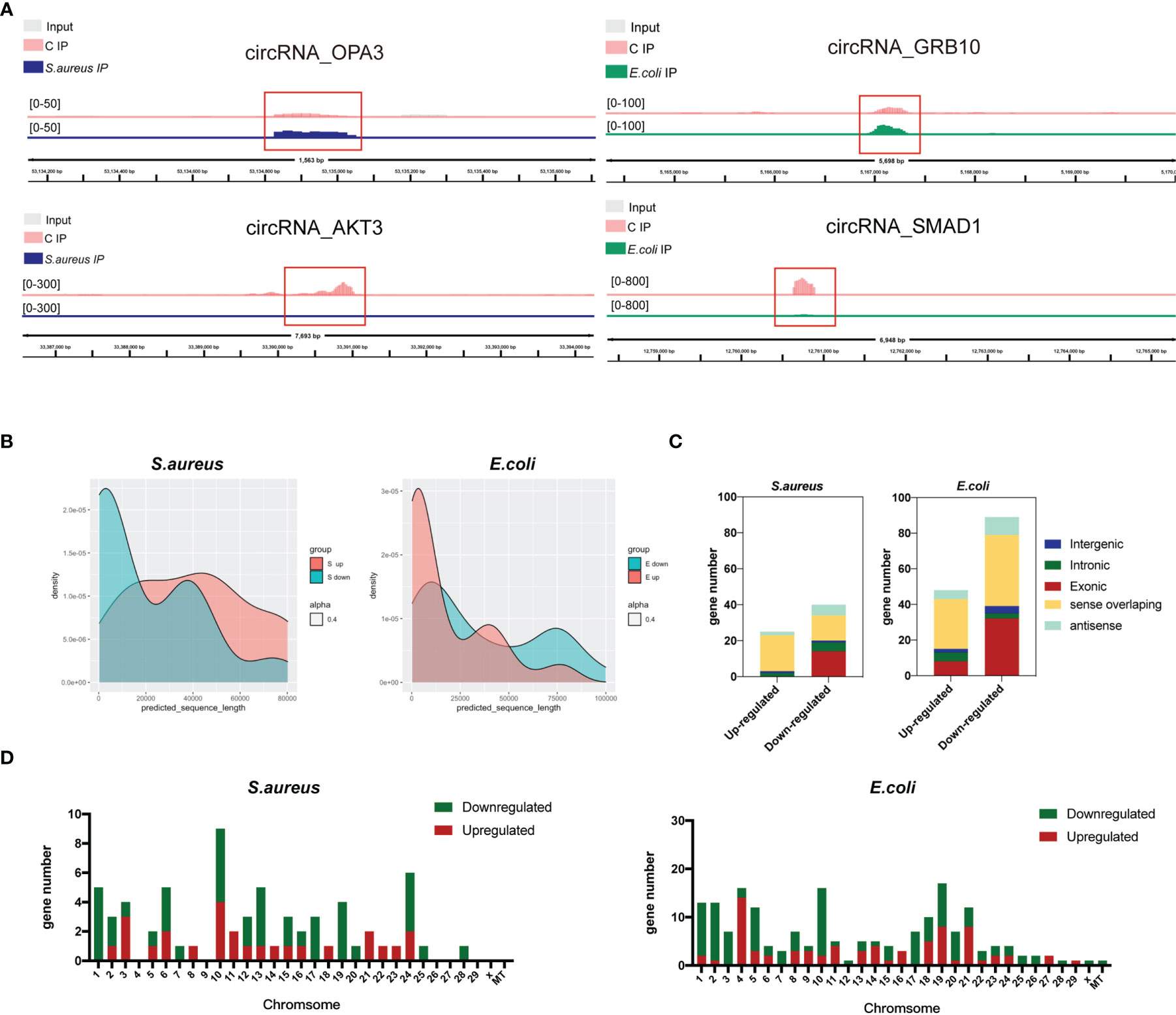
Figure 3 Distribution of significantly differentially expressed m6A-modified circRNAs in the S. aureus/E. coli group compared to the control group. (A) Data visualization analysis was performed by IGV, showing the location of differential m6A peaks in the source gene of circRNA (circRNA_OPA3, circRNA_AKT3, circRNA_GRB10, and circRNA_SMAD1) between the control group and treatment group. (B) The length, (C) source, and (D) chromosome distribution of differentially expressed m6A-modified circRNAs in the S. aureus and E. coli groups (circRNAs were named by their host genes).
Additionally, the feature of the differentially m6A-methylated circRNA in the S. aureus/E. coli group compared to the control group is also shown. Instead of being a major part of short m6A-modified circRNA derived from exon (Figure 2D), it is longer and derived more from sense overlapping when MAC-T cells are injured by S. aureus and E. coli (Figures 3B, C). Additionally, the differential circRNA with methylation showed totally reverse patterns in the S. aureus group compared to the E. coli group, which were mainly hypomethylation in the S. aureus group and hypermethylation in the E. coli group. The most differentially methylated circRNA in the S. aureus group was found on chromosome 10, while in the E. coli group, it was on chromosome 19 (Figure 3D).
GO and KEGG Pathway Enrichment Analyses of m6A-Modified circRNAs in MAC-T Cells Treated With S. aureus and E. coli
Recent studies have shown that circRNAs can affect the expression of its cis genes (42). Therefore, to clarify the essential role of m6A-modified circRNAs in MAC-T cell injury induced by S. aureus and E. coli, GO and KEGG pathway enrichment analyses for the m6A-modified circRNA source genes were performed (p-value < 0.05) (Tables S3, S4). In the S. aureus group, the GO annotation of circRNA with hypermethylation illustrated that it was mainly enriched in transmembrane, cell part morphogenesis, plasma membrane-bounded cell projection organization, P-body, and protein tyrosine kinase activator activity (Figure 4A). Furthermore, the KEGG analyses indicated that they were enriched in endocytosis, antigen processing and presentation, and ubiquitin-mediated proteolysis (Figure 5A). Likewise, the GO analysis of circRNA with hypomethylation showed that it was closely related to clathrin, SMAD binding, integrin binding, and PI3K (Figure 4B). KEGG analysis of hypomethylated circRNAs revealed enrichment in focal adhesion, the RAP1 signaling pathway, and regulation of the actin cytoskeleton (Figure 5B).
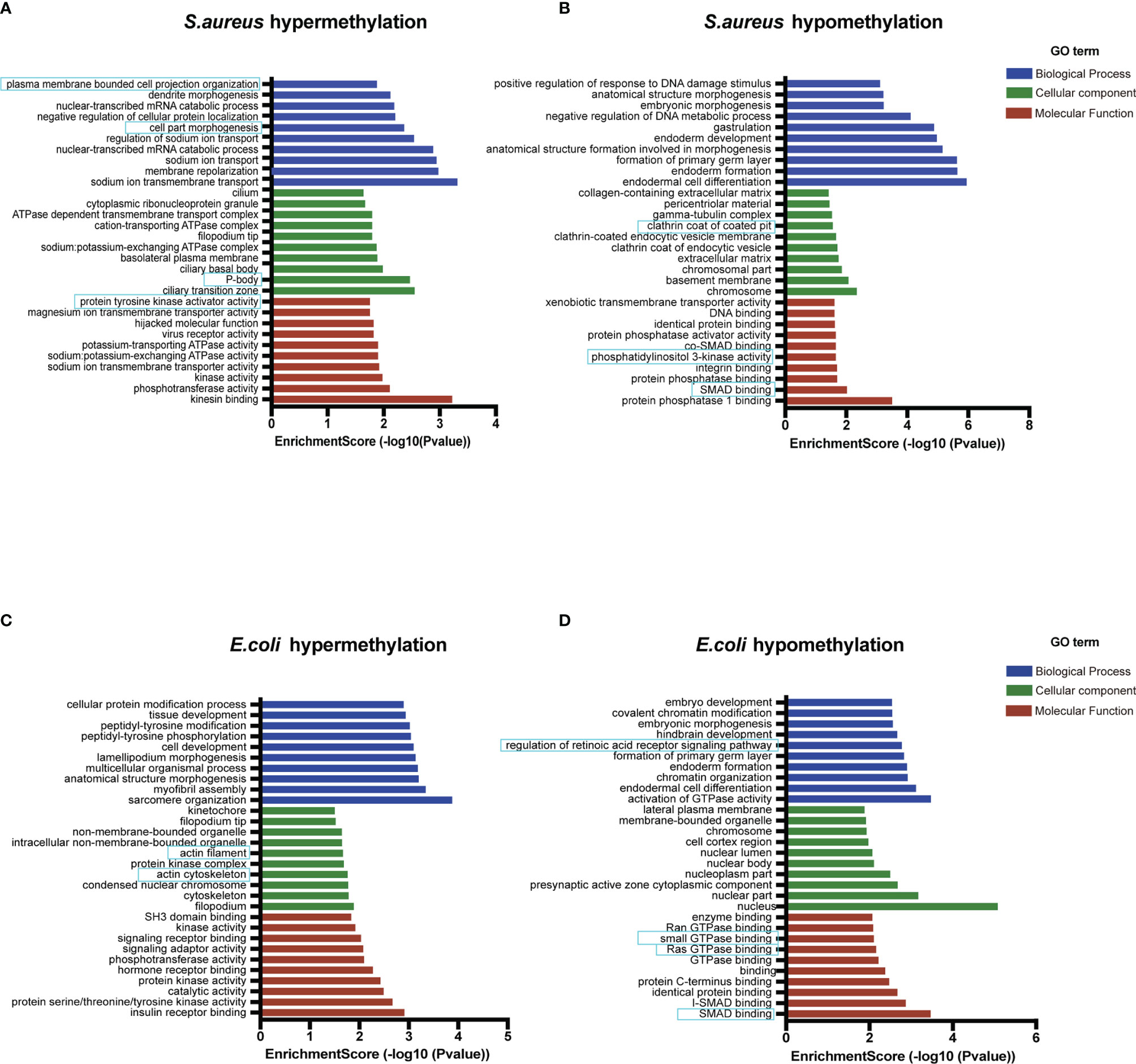
Figure 4 Go enrichment analysis. (A) The top 10 GO terms enriched for the source genes of differential circRNA with hypermethylation and (B) hypomethylation in the S. aureus group. (C) The top 10 GO terms enriched for the source genes of differential circRNA with hypermethylation and (D) hypomethylation in the E. coli group (the predicted GO pathways in the blue box were related to inflammation, apoptosis, and autophagy).
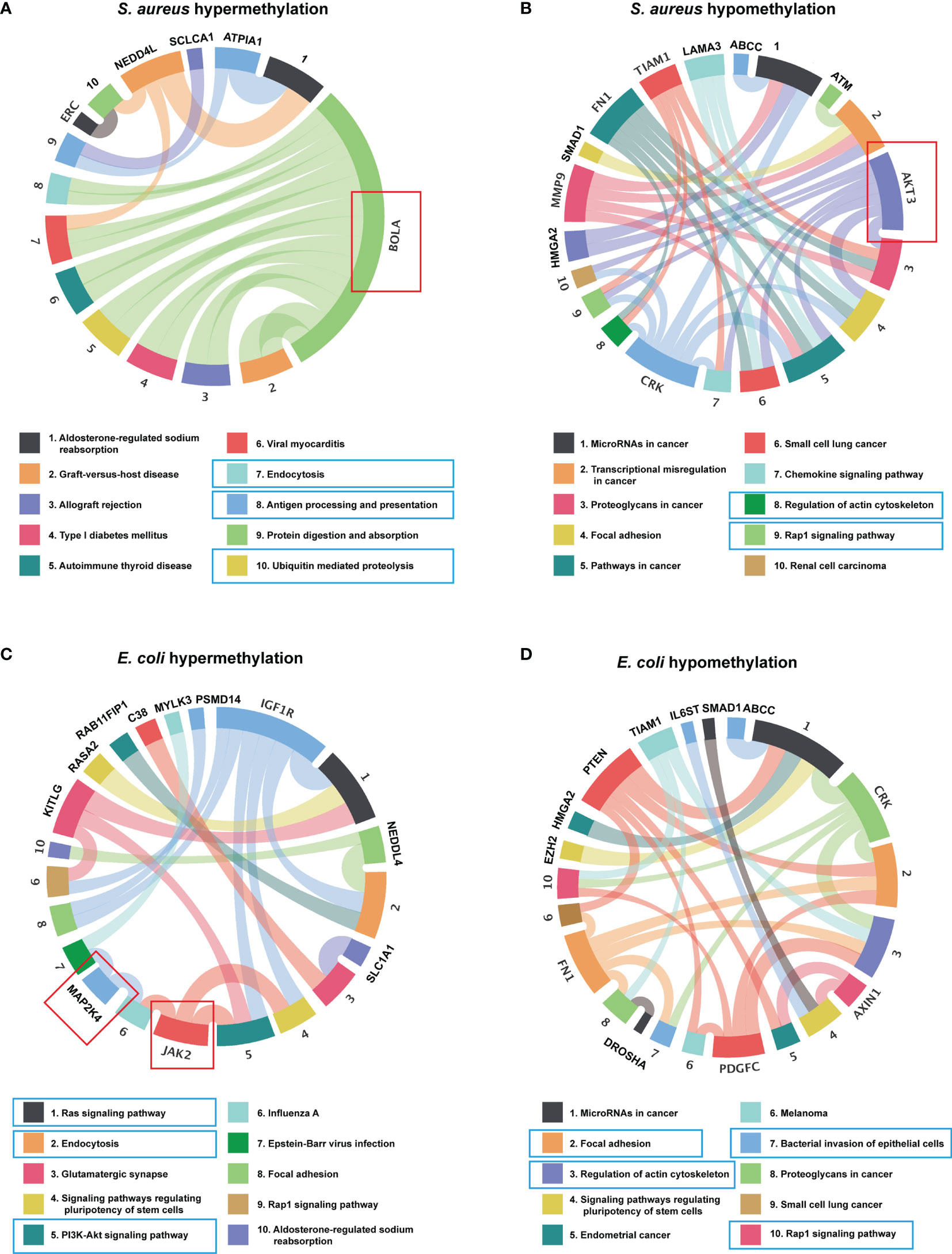
Figure 5 KEGG enrichment analysis. (A) The top 10 KEGG pathway enriched for the source genes of differential circRNA with hypermethylation and (B) hypomethylation in the S. aureus group. (C) The top 10 KEGG pathway enriched for the source genes of differential circRNA with hypermethylation and (D) hypomethylation in the E. coli group (the predicted KEGG pathways in the blue box and representative genes in the red box were related to inflammation, apoptosis, and autophagy).
In the E. coli group, the GO analysis predicted that the functions of circRNAs with hypermethylation were significantly enriched in cytoskeleton, and actin filament (Figure 4C). The KEGG pathway analysis indicated that circRNAs with hypermethylation were significantly related to the Ras signaling pathway, endocytosis, and PI3K-Akt signaling pathway (Figure 5C). Correspondingly, the GO analysis of differential m6A-modified circRNA with hypomethylation illustrated that the meaningful terms may be related to regulation of the retinoic acid receptor signaling pathway, SMAD binding, Ras GTPase binding, and small GTPase binding (Figure 4D). The KEGG analysis also revealed enrichment in focal adhesion, regulation of actin cytoskeleton, bacterial invasion of epithelial cells, and Rap1 signaling pathway (Figure 5D).
Also, some representative genes closely related to apoptosis and inflammation in the KEGG pathways analysis were shown, such as BOLA, AKT3, MAP2K4, and JAK2 (Figures 5A–D). Given the above, we found that the functions of differentially expressed m6A-modified circRNAs were highly similar when MAC-T cell injury was induced by either S. aureus or E. coli. These findings may suggest that these m6A-modified circRNAs regulated immunity, cell junctions, growth metabolism, and resistance to bacterial invasion through a similar pathway in both induced groups.
CircRNA–miRNA–mRNA Interaction Network Identified Similar Enrichment of mRNA in the S. aureus and E. coli Groups
CircRNAs are able to regulate the expression of target genes as sponges for miRNAs based on complementary base pairing. By predicting the target miRNA between circRNA and mRNA, we constructed a circRNA–miRNA–mRNA interaction network (Table S5). In this study, 10 circRNAs (5 hypermethylation and 5 hypomethylation), 10 miRNAs, and 22 mRNAs were included in the S. aureus group (Figure 6A), and 10 circRNAs (5 hypermethylation and 5 hypomethylation), 8 miRNAs, and 19 mRNAs were included in the E. coli group (Figure 6B).
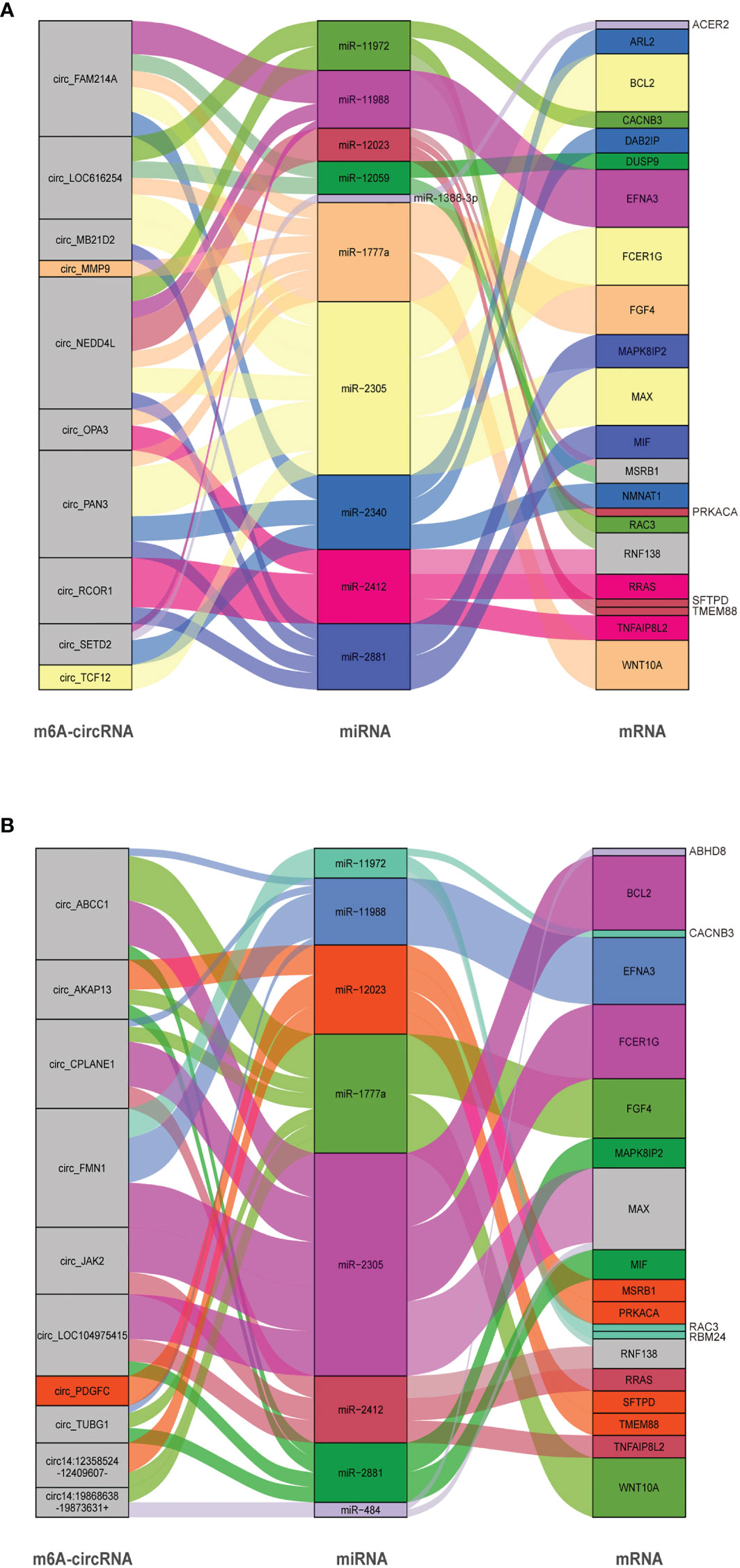
Figure 6 CircRNA–miRNA–mRNA ceRNA interaction network. The box size represents the strength of the binding. The color represents that the miRNA has a binding site with mRNA and circRNA, and if the color is gray, it represents two or more target binding sites. (A) The S. aureus group. (B) The E. coli group (circRNAs were named by their host genes or chromosome location).
In the interactive network of the S. aureus group, many inflammation-related miRNAs were predicted. For example, miR-2305 and miR-1777a, which have been considered to have an effect on immunity (43), were also shown to enrich for transcripts related to inflammation and apoptosis in our study (including key genes such as MAPK8IP2 and WNT10A). Additionally, we predicted that some miRNAs may be regulated by multiple circRNAs at the same time. For instance, miR-11988 was predicted to be regulated by circ_FAM214A, circ_NEDD4 L, and circ_SETD2 simultaneously.
Similarly, in the E. coli group, a large amount of mRNA was also predicted to be associated with inflammation, mainly in the MAPK signaling pathway and the WNT signaling pathway. In addition, BCL2, which is the key molecule in apoptosis, was predicted to be regulated by miR-2305. Some natural immune-related mRNAs such as TNFAIP8L2, FCER1G, and MSR1B were present in the network. It is predicted that the miRNAs may regulate multiple mRNAs. For example, miR-2305 could be able to interact with BCL2, FCER1G, and MAX. Interestingly, in both groups, we found a large number of similar miRNAs, suggesting that circRNAs modified with different m6A modifications may play a similar role in S. aureus- and E. coli-induced cell injury.
Conjoint Analysis of circRNA-seq and MeRIP-seq
To further explore the potential function of circRNAs with m6A modification in S. aureus- and E. coli-induced cell injury, a conjoint analysis of circRNA-seq and MeRIP-seq was performed.
In addition, the differential expression of circRNAs with significant changes in the S. aureus and the E. coli groups compared to the control group was analyzed (Figures 7A–D). We found 3,914 novel circRNAs, of which 1,532 were upregulated and 11 were downregulated in the S. aureus group, and 536 were upregulated and 11 were downregulated in the E. coli group. The top 20 most significantly differentially expressed circRNAs in the S. aureus group and E. coli group are listed separately (Tables S6, S7).
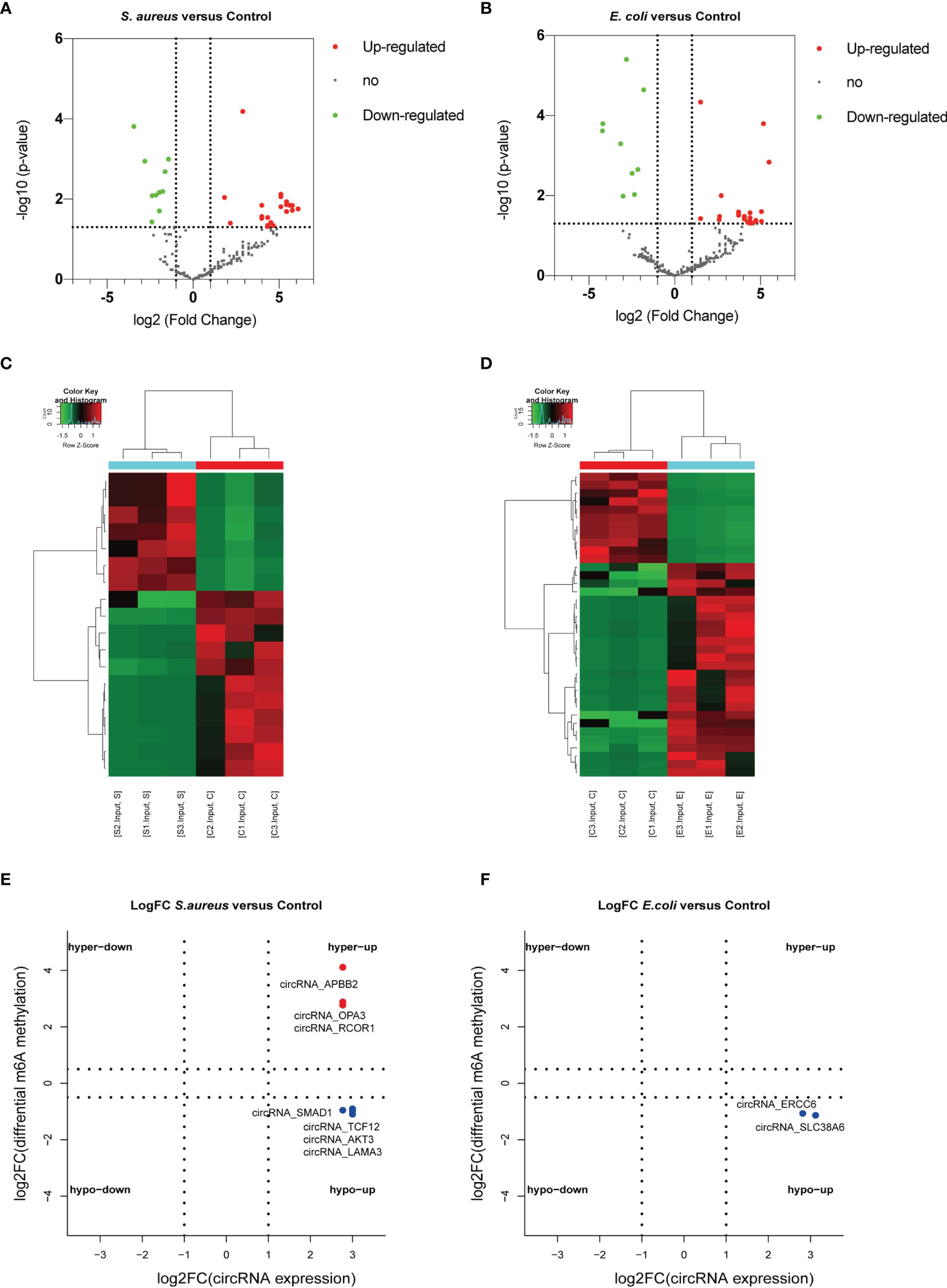
Figure 7 Conjoint analysis of circRNA with or without m6A modification. Volcano plot showing the differentially expressed circRNAs in the (A) S. aureus group and (B) E. coli group. Heatmap illustrating the differentially expressed circRNAs in the (C) S. aureus group and (D) E. coli group. Conjoint analysis between the expression level and m6A modification in the (E) S. aureus group and (F) E. coli group.
To predict the especially important circRNA in MAC-T injury and reveal the relationship between m6A modification and expression, a conjoint analysis was performed. Finally, 3 significantly upregulated circRNAs with hypermethylation and 4 upregulated circRNAs with hypomethylation were found in the S. aureus group (Figure 7E). Two upregulated circRNA with hypomethylation were found in the E. coli group (Figure 7F).
Discussion
Bovine mastitis is a common disease that seriously harms the dairy industry (44). Currently, antibiotics are still the major treatment for mastitis, which also causes some serious problems, such as drug resistance and threatening side effects. Recent studies have confirmed that circRNA plays an important role in diseases (45), and it can be modified by N6-methyladenosine and plays a more complex regulatory role (46). Nevertheless, the mechanism of m6A-modified circRNA in bovine mastitis is still undiscovered. This study is the first to reveal the relationship between m6A-modified circRNA and cell injury induced by S. aureus and E. coli, and it reveals that circRNA with m6A modification may be related to cell injury by S. aureus and E. coli treatment (Figure 8).
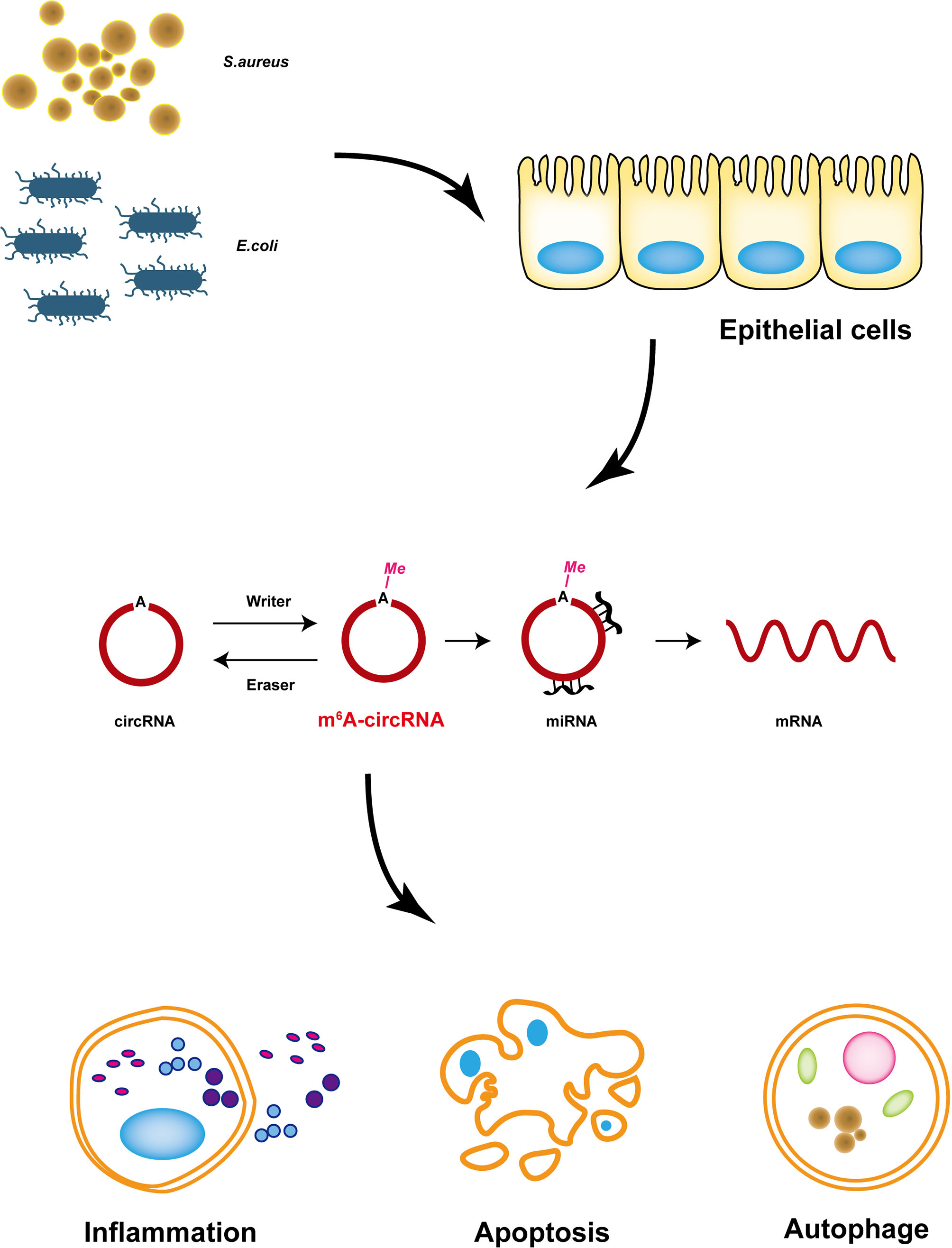
Figure 8 Proposed model of m6A-modified circRNAs that regulate MAC-T cells injured by inactivated S. aureus and E. coli. m6A-modified circRNAs regulated MAC-T cell apoptosis, inflammation, and autophagy.
It was reported that cell injury plays an important role in mastitis induced by S. aureus and E. coli (14). Perhaps the most compelling finding was that IL-1β, IL-6, and TNF-α were significantly upregulated, which indicated inflammatory changes. Additionally, apoptosis and autophagy are common cell injuries during inflammation (47). Inflammatory mRNA expression levels and TEM illustrations both demonstrated that E. coli induced more accelerated cell injury.
It is known that m6A modification of circRNA usually changes the development of disease. In our research, we found some novel m6A peaks first, and also identified the differential m6A-modified circRNA profile in the S. aureus group and E. coli group. This result suggested that this circRNA with different levels of m6A modification may play a vital role in S. aureus- and E. coli-induced cell injury. Most circRNAs upstream of mRNAs are able to regulate gene expression (48), so the altered m6A-modified circRNA could be a potential treatment target in mastitis. Furthermore, the unique structure of the conserved covalent closed loop allowed circRNA to remain stable, which makes it a better target than mRNA or other linear RNAs.
CircRNAs as a result of the diverse splicing of mRNAs, and a large number of studies have identified that the major source of circRNAs is derived from exons (49–51), which was also proven in our research. Some researchers have found that circRNAs are produced through contranscription and competition with conventional splicing (52). As a result, biogenesis of circRNA leads to a decrease in mRNA synthesis from the same location. Here, circRNA production acts as an RNA trap for mRNA production. However, we found that the features of m6A-modified circRNAs changed when MAC-T cells were injured by S. aureus and E. coli. Most of the differential m6A-modified circRNAs are longer and come from sense overlapping regions, which means that these differential and long m6A-modified circRNAs derived from sense overlapping regions play a more important function and provide us with a new direction in the m6A-modified circRNA regulatory mechanism of mastitis. In addition, the m6A-modified circRNA in both groups showed reverse density patterns of length, which may suggest a difference in cell injury in S. aureus and E. coli treatment. In fact, there are currently few studies on the m6A methylation of circRNA, and whether m6A modification can regulate mastitis in dairy cows needs further confirmation.
Since the function of circRNA in dairy cow mastitis is not fully understood, we performed GO/KEGG analysis to predict the function of altered m6A methylated circRNA. Previous studies have confirmed that PI3K-AKT and Ras-MAPK are important signaling pathways of natural immunity (53, 54). Li et al. found that ulinastatin reduced LPS-induced inflammation in mouse macrophage cells by activating the PI3K/Akt/Nrf2 pathway (55). Dong et al. found that Astragalus polysaccharides alleviate LPS-induced inflammation via the NF-κB/MAPK signaling pathway (56). When epithelial cells are injured by bacteria, the tight junctions between cells are compromised through focal adhesion and actin cytoskeletal signaling pathways (57). In our research, we predicted that the differentially expressed circRNAs with m6A modification were enriched in the Ras-MAPK, focal adhesion, and PI3K-AKT signaling pathways in MAC-T cells injured by S. aureus and E. coli, which are closely related to cell apoptosis and inflammation. The pathways and GO annotations of differentially expressed m6A-modified circRNAs in the S. aureus and E. coli groups were highly similar.
Many studies have shown that circRNA can be used as a molecular sponge to interact with miRNA to regulate mRNA (22). Recent studies have proven that specific circRNA–miRNA–mRNA axes cause apoptosis and inflammation in bovine mammary epithelial cells (58). In our ceRNA network, we found that 20 circRNAs and 26 mRNAs had a total of 10 miRNA binding sites, which were obviously related to apoptosis and inflammation. MAX, an important apoptosis gene, was upregulated by direct binding to the miR-181a promoter to increase MYCT1 expression, leading to apoptosis (59). In our study, we found that miR-2305 was able to regulate MAX and BCL2 in both ceRNA networks, and multiple circRNAs had binding sites with miR-2305 (60). In addition, some researchers have shown that downregulation could block the AKT pathway, inhibiting cell survival and proliferation, which means that MIF may be an important gene working in cell proliferation (61).
When MAC-T cells were treated with S. aureus and E. coli, we found that circ_RCOR1, circ_NEDD4 L, circ_MB21D2, and circ_PAN3 regulate MIF through miR-2881 in the S. aureus group, and circ_AKAP13, circ_ABCC1, circ_TUBG1, and circ_LOC104975415 regulate MIF through miR-2881 in the E. coli group. This ultimately had a potential effect on MAC-T cell proliferation. RAC3 is an NF-κB coactivator closely related to inflammation, whose expression is regulated by TNF, in addition to being a negative regulator of autophagy (62–64). In our research, miR-11972 was predicted to have binding sites for RAC3. Strangely, the expression of IL-1β, IL-6, and TNFα was different in the cell injury model, while the levels of apoptosis in the two groups were similar (Figures 1A–F). Through our ceRNA networks, we conjectured that the m6A-circRNA regulated the mRNA enriched in some similar pathways of apoptosis, but there may be more complicated mechanisms that were related to inflammation. Overall, circRNAs with different m6A modification levels regulated mRNAs enriched in the MAPK, WNT, and natural immune signaling pathways through circRNA–miRNA–mRNA axes and ultimately affected S. aureus- and E. coli-induced mastitis. In our research, ceRNA networks showed that mRNAs were enriched in MAPK, WNT, and natural immunology signaling pathways in both the S. aureus and E. coli groups, which may be the reason for the similar levels of apoptosis.
To further reveal the relationship between circRNA and m6A modification, we performed an analysis of circRNA-seq. We found a total of 3,914 novel circRNAs with expression differences. The length and source of altered circRNAs in the S. aureus and E. coli groups are consistent with the circRNA characteristics of previous studies, and most of them are shorter and exon-derived circRNAs. In the conjoint analysis of MeRIP-seq, we found that a total of 9 circRNAs showed a significant association between expression and m6A modification. To change circRNA expression, m6A modification has many functions, such as regulated circRNA splicing and degradation (45). For instance, Li et al. confirmed that circMETTL3 is upregulated in a m6A-dependent manner and promotes breast cancer progression (65).
This study is the first report to clarify the profile of m6A-modified circRNAs in bovine mammary epithelial cells injured by S. aureus and E. coli. Additionally, we predicted similar pathways and target genes in both groups. This finding provided us with new insights for exploring the cellular injury mechanism of mastitis. However, we did have some limitations. For the purpose of exploring the innate immune response, we used heat-inactivated bacteria instead of live bacteria, which may differ from real mastitis disorders in bovines. In our ceRNA networks, only the circRNAs with different m6A levels are shown, without a detailed description of the “up–down–up” or “down–up–down” model of differential expression. Therefore, the mechanism of cell injury needs to be revealed by further experiments, and animal models are needed to clarify the complex function of m6A-modified circRNAs in vivo.
Conclusion
Mastitis is a common disease with a complicated pathological mechanism in bovines. m6A-modified circRNA plays an important role in regulating disease, but its function in mastitis remains unknown. Our study clearly profiled the m6A methylation of circRNA in MAC-T cell injury induced by S. aureus and E. coli and made predictions on differential m6A-methylated circRNA firstly. We provided novel insight into MAC-T cell injury induced by S. aureus or E. coli via m6A-modified circRNAs, which may represent a new research direction for future studies in mastitis.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196736.
Author Contributions
HX and CL: Conceptualization, Validation, Formal analysis, Investigation, Resources, Data curation, Writing—original draft, Visualization, Writing—review and editing, and Supervision. YZ and TL: Validation, Formal analysis, and Investigation. JY and SC: Writing—review and editing. CH, JC, XC, YC, and AG: Project administration and Funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31972758; 31101874).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank State Key Laboratory of Agricultural Microbiology of Huazhong Agricultural University for providing a platform to conduct this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.873330/full#supplementary-material
Supplementary Figure 1 | Overview of the circRNA profile. (A) Novel circRNAs, mainly exonic, intronic, intergenic, sense overlapping, and antisense circRNAs. (B) The length of the novel circRNA found by circRNA-seq. (C, D) Pie charts showing the length of differentially expressed circRNAs in the S. aureus and E. coli groups. (E) The chromosome distribution of the novel circRNAs. (F) Histogram showing the source of differentially expressed circRNAs in the S. aureus and E. coli groups. (G) The chromosome distribution of differential circRNAs in the S. aureus and E. coli groups.
References
1. De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW. Invited Review: Mastitis in Dairy Heifers: Nature of the Disease, Potential Impact, Prevention, and Control. J Dairy Sci (2012) 95(3):1025–40. doi: 10.3168/jds.2010-4074
2. Holmes MA, Zadoks RN. Methicillin Resistant S. Aureus in Human and Bovine Mastitis. J Mammary Gland Biol Neoplasia (2011) 16(4):373–82. doi: 10.1007/s10911-011-9237-x
3. Zadoks RN, Middleton JR, McDougall S, Katholm J, Schukken YH. Molecular Epidemiology of Mastitis Pathogens of Dairy Cattle and Comparative Relevance to Humans. J Mammary Gland Biol Neoplasia (2011) 16(4):357–72. doi: 10.1007/s10911-011-9236-y
4. Erskine RJ, Walker RD, Bolin CA, Bartlett PC, White DG. Trends in Antibacterial Susceptibility of Mastitis Pathogens During a Seven-Year Period. Dairy Sci (2002) 85(5):1111–8. doi: 10.3168/jds.S0022-0302(02)74172-6
5. Vangroenweghe F, Lamote I, Burvenich C. Physiology of the Periparturient Period and Its Relation to Severity of Clinical Mastitis. Domest Anim Endocrinol (2005) 29(2):283–93. doi: 10.1016/j.domaniend.2005.02.016
6. Tang D, Kang R, Coyne C, Zeh H, Lotze M. Pamps and Damps: Signal 0s That Spur Autophagy and Immunity. Immunol Rev (2012) 249(1):158–75. doi: 10.1111/j.1600-065X.2012.01146.x
7. Gunther J, Petzl W, Zerbe H, Schuberth HJ, Koczan D, Goetze L, et al. Lipopolysaccharide Priming Enhances Expression of Effectors of Immune Defence While Decreasing Expression of Pro-Inflammatory Cytokines in Mammary Epithelia Cells From Cows. BMC Genomics (2012) 13:17. doi: 10.1186/1471-2164-13-17
8. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
9. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient Microrna Sponges. Nature (2013) 495(7441):384–90. doi: 10.1038/nature11993
10. Lai JL, Liu YH, Liu C, Qi MP, Liu RN, Zhu XF, et al. Indirubin Inhibits Lps-Induced Inflammation Via TLR4 Abrogation Mediated by the NF-κb and MAPK Signaling Pathways. Inflammation (2017) 40(1):1–12. doi: 10.1007/s10753-016-0447-7
11. Lai JL, Liu YH, Peng YC, Ge P, He CF, Liu C, et al. Indirubin Treatment of Lipopolysaccharide-Induced Mastitis in a Mouse Model and Activity in Mouse Mammary Epithelial Cells. Mediators Inflammation (2017) 2017:1–13. doi: 10.1155/2017/3082805
12. Liu C, Tang X, Zhang W, Li G, Chen Y, Guo A, et al. 6-Bromoindirubin-3′-Oxime Suppresses Lps-Induced Inflammation Via Inhibition of the TLR4/NF-κb and TLR4/MAPK Signaling Pathways. Inflammation (2019) 42(6):2192–204. doi: 10.1007/s10753-019-01083-1
13. Tang X, Liu C, Li T, Lin C, Hu C. Gambogic Acid Alleviates Inflammation and Apoptosis and Protects the Blood-Milk Barrier in Mastitis Induced by LPS. Int Immunopharmacol (2020) 86:106697. doi: 10.1016/j.intimp.2020.106697
14. Bannerman DD, Paape MJ, Lee JW, Zhao X, Hope JC, Rainard P. Escherichia Coli and Staphylococcus Aureus Elicit Differential Innate Immune Responses Following Intramammary Infection. Clin Diagn Lab Immunol (2004) 11(3):463–72. doi: 10.1128/CDLI.11.3.463-472.2004
15. Ibeagha-Awemu EM, Ibeagha AE, Messier S, Zhao X. Proteomics, Genomics, and Pathway Analyses of Escherichia Coli and Staphylococcus Aureus Infected Milk Whey Reveal Molecular Pathways and Networks Involved in Mastitis. J Proteome Res (2010) 9:4604–19. doi: 10.1021/pr100336e
16. Fu Y, Zhou E, Liu Z, Li F, Liang D, Liu B, et al. Staphylococcus Aureus and Escherichia Coli Elicit Different Innate Immune Responses From Bovine Mammary Epithelial Cells. Vet Immunol Immunopathol (2013) 155(4):245–52. doi: 10.1016/j.vetimm.2013.08.003
17. Gilbert FB, Cunha P, Jensen K, Glass EJ, Foucras G, Robert-Granié C, et al. Differential Response of Bovine Mammary Epithelial Cells to Staphylococcus Aureus or Escherichia Coli Agonists of the Innate Immune System. Vet Res (2013) 44(1):1–23. doi: 10.1186/1297-9716-44-40
18. Lin C, Zhu Y, Hao Z, Xu H, Li T, Yang J, et al. Genome-Wide Analysis of LncRNA in Bovine Mammary Epithelial Cell Injuries Induced by Escherichia Coli and Staphylococcus Aureus. Int J Mol Sci (2021) 22(18):9719. doi: 10.3390/ijms22189719
19. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
20. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-Intron Circular RNAs Regulate Transcription in the Nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
21. Ebbesen KK, Hansen TB, Kjems J. Insights Into Circular RNA Biology. RNA Biol (2017) 14(8):1035–45. doi: 10.1080/15476286.2016.1271524
22. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, et al. Circular RNAs in Immune Responses and Immune Diseases. Theranostics (2019) 9(2):588–607. doi: 10.7150/thno.29678
23. Ma X, Zhao X, Zhang Z, Guo J, Guan L, Li J, et al. Differentially Expressed Non-Coding RNAs Induced by Transmissible Gastroenteritis Virus Potentially Regulate Inflammation and NF-κb Pathway in Porcine Intestinal Epithelial Cell Line. BMC Genomics (2018) 19(1):747. doi: 10.1186/s12864-018-5128-5
24. Agirre X, Meydan C, Jiang Y, Garate L, Doane AS, Li Z, et al. Long Non-Coding RNAs Discriminate the Stages and Gene Regulatory States of Human Humoral Immune Response. Nat Commun (2019) 10(1):821. doi: 10.1038/s41467-019-08679-z
25. Zhang Y, Zhang Y, Li X, Zhang M, Lv K. Microarray Analysis of Circular RNA Expression Patterns in Polarized Macrophages. Int J Mol Med (2017) 39(2):373–9. doi: 10.3892/ijmm.2017.2852
26. Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RASGEF1B Circular RNA is a Positive Regulator of ICAM-1 in the TLR4/LPS Pathway. RNA Biol (2016) 13(9):861–71. doi: 10.1080/15476286.2016.1207036
27. Zhao C, Ling X, Xia Y, Yan B, Guan Q. The m6A Methyltransferase METTL3 Controls Epithelial-Mesenchymal Transition, Migration and Invasion of Breast Cancer Through the MALAT1/Mir-26b/HMGA2 Axis. Cancer Cell Int (2021) 21(1):441. doi: 10.1186/s12935-021-02113-5
28. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
29. Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B. N6-Adenosine Methylation in miRNAs. PloS One (2015) 10(2):e0118438. doi: 10.1371/journal.pone.0118438
30. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N6-Methyladenosine (m6A) Methyltransferase That Targets Pre-mRNAs and Various Non-Coding RNAs. EMBO Rep (2017) 18(11):2004–14. doi: 10.15252/embr.201744940
31. Lan Y, Liu B, Guo H. The Role of M(6)a Modification in the Regulation of Tumor-Related LncRNAs. Mol Ther Nucleic Acids (2021) 24:768–79. doi: 10.1016/j.omtn.2021.04.002
32. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-Dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and is Required For Adipogenesis. Cell Res (2014) 24(12):1403–19. doi: 10.1038/cr.2014.151
33. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. Ythdf2 Destabilizes M(6)a-Containing RNA Through Direct Recruitment of the CCR4-Not Deadenylase Complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
34. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-Methyladenosine Modification Destabilizes Developmental Regulators in Embryonic Stem Cells. Nat Cell Biol (2014) 16(2):191–8. doi: 10.1038/ncb2902
35. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-Methyladenosine Modulates Messenger RNA Translation Efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
36. Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, et al. N(6)-Methyladenosine RNA Modification Regulates Embryonic Neural Stem Cell Self-Renewal Through Histone Modifications. Nat Neurosci (2018) 21(2):195–206. doi: 10.1038/s41593-017-0057-1
37. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)a mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
38. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-Wide Maps of m6A CircRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns That Are Distinct From mRNAs. Cell Rep (2017) 20(9):2262–76. doi: 10.1016/j.celrep.2017.08.027
39. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell (2019) 76(1):96–109 e9. doi: 10.1016/j.molcel.2019.07.016
40. Li T, Lin C, Zhu Y, Xu H, Yin Y, Wang C, et al. Transcriptome Profiling of M(6)a mRNA Modification in Bovine Mammary Epithelial Cells Treated With Escherichia Coli. Int J Mol Sci (2021) 22(12):6254. doi: 10.3390/ijms22126254
41. Kruger J, Rehmsmeier M. Rnahybrid: MicroRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res (2006) 34(Web Server issue)::W451–4. doi: 10.1093/nar/gkl243
42. Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. Circrna Inhibits DNA Damage Repair by Interacting With Host Gene. Mol Cancer (2020) 19(1):128. doi: 10.1186/s12943-020-01246-x
43. Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, et al. Bovine Milk Exosomes Contain MicroRNA and mRNA and Are Taken Up by Human Macrophages. J Dairy Sci (2015) 98(5):2920–33. doi: 10.3168/jds.2014-9076
44. Zhao X, Lacasse P. Mammary Tissue Damage During Bovine Mastitis: Causes and Control. J Anim Sci (2008) 86(13 Suppl):57–65. doi: 10.2527/jas.2007-0302
45. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
46. Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, et al. The Role of N6-Methyladenosine (m6A) Modification in the Regulation of CircRNAs. Mol Cancer (2020) 19(1):105. doi: 10.1186/s12943-020-01224-3
47. Green DR, Galluzzi L, Kroemer G. Mitochondria and the Autophagy-Inflammation-Cell Death Axis in Organismal Aging. Science (2011) 333(6046):1109–12. doi: 10.1126/science.1201940
48. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell (2018) 71(3):428–42. doi: 10.1016/j.molcel.2018.06.034
49. Ge P, Zhang J, Zhou L, Lv MQ, Li YX, Wang J, et al. CircRNA Expression Profile and Functional Analysis in Testicular Tissue of Patients With Non-Obstructive Azoospermia. Reprod Biol Endocrinol (2019) 17(1):100. doi: 10.1186/s12958-019-0541-4
50. Ebermann C, Schnarr T, Muller S. Recent Advances in Understanding Circular RNAs. F1000Res (2020) 9 F1000 Faculty Rev-655. doi: 10.12688/f1000research.25060.1
51. Chen LL, Yang L. Regulation of CircRNA Biogenesis. RNA Biol (2015) 12(4):381–8. doi: 10.1080/15476286.2015.1020271
52. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. Circrna Biogenesis Competes With Pre-mRNA Splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
53. Arthur JS, Ley SC. Mitogen-Activated Protein Kinases in Innate Immunity. Nat Rev Immunol (2013) 13(9):679–92. doi: 10.1038/nri3495
54. Hawkins PT, Stephens LR. PI3K Signalling in Inflammation. Biochim Biophys Acta (2015) 1851(6):882–97. doi: 10.1016/j.bbalip.2014.12.006
55. Li S, Dai Q, Zhang S, Liu Y, Yu Q, Tan F, et al. Ulinastatin Attenuates LPS-Induced Inflammation in Mouse Macrophage Raw264.7 Cells by Inhibiting the JNK/NF-κb Signaling Pathway and Activating the PI3K/AKT/NRF2 Pathway. Acta Pharmacol Sin (2018) 39(8):1294–304. doi: 10.1038/aps.2017.143
56. Dong N, Li X, Xue C, Zhang L, Wang C, Xu X, et al. Astragalus Polysaccharides Alleviates LPS-Induced Inflammation Via the NF-κb /MAPK Signaling Pathway. J Cell Physiol (2020) 235(7-8):5525–40. doi: 10.1002/jcp.29452
57. Zihni C, Balda MS, Matter K. Signalling at Tight Junctions During Epithelial Differentiation and Microbial Pathogenesis. J Cell Sci (2014) 127(Pt 16):3401–13. doi: 10.1242/jcs.145029
58. Chen Z, Liang Y, Lu Q, Nazar M, Mao Y, Aboragah A, et al. Cadmium Promotes Apoptosis and Inflammation Via the Circ08409/Mir-133a/TGFB2 Axis in Bovine Mammary Epithelial Cells and Mouse Mammary Gland. Ecotoxicol Environ Saf (2021) 222:112477. doi: 10.1016/j.ecoenv.2021.112477
59. Wang HT, Tong X, Zhang ZX, Sun YY, Yan W, Xu ZM, et al. MYCT1 Represses Apoptosis of Laryngeal Cancerous Cells Through the MAX/Mir-181a/NPM1 Pathway. FEBS J (2019) 286(19):3892–908. doi: 10.1111/febs.14942
60. Siddiqui WA, Ahad A, Ahsan H. The Mystery of BCL2 Family: BCL-2 Proteins and Apoptosis: An Update. Arch Toxicol (2015) 89(3):289–317. doi: 10.1007/s00204-014-1448-7
61. Cao F, Xiao Z, Chen S, Zhao C, Chen D, Haisma HJ, et al. HDAC/MIF Dual Inhibitor Inhibits Nsclc Cell Survival and Proliferation by Blocking the AKT Pathway. Bioorg Chem (2021) 117:105396. doi: 10.1016/j.bioorg.2021.105396
62. Alvarado CV, Rubio MF, Fernández Larrosa PN, Panelo LC, Azurmendi PJ, Ruiz Grecco M, et al. The Levels of RAC3 Expression Are Up Regulated by TNF in the Inflammatory Response. FEBS Open Bio (2014) 4(1):450–7. doi: 10.1016/j.fob.2014.04.009
63. Zhu WL, Hossain MS, Guo DY, Liu S, Tong H, Khakpoor A, et al. A Role for RAC3 GTPase in the Regulation of Autophagy. J Biol Chem (2011) 286(40):35291–8. doi: 10.1074/jbc.M111.280990
64. Werbajh S, Nojek I, Lanz R, Costas MA. RAC-3 Is a NF-κb Coactivator. FEBS Lett (2000) 485(2-3):195–9. doi: 10.1016/S0014-5793(00)02223-7
Keywords: N6-methyladenosine, circRNA, epithelial cells, cell injury, inflammation, S. aureus, E. coli
Citation: Xu H, Lin C, Li T, Zhu Y, Yang J, Chen S, Chen J, Chen X, Chen Y, Guo A and Hu C (2022) N6-Methyladenosine-Modified circRNA in the Bovine Mammary Epithelial Cells Injured by Staphylococcus aureus and Escherichia coli. Front. Immunol. 13:873330. doi: 10.3389/fimmu.2022.873330
Received: 10 February 2022; Accepted: 11 March 2022;
Published: 04 April 2022.
Edited by:
Haitao Wang, National Cancer Institute (NIH), United StatesReviewed by:
Yaqiang Cao, National Institutes of Health (NIH), United StatesWenjie Wang, National Institutes of Health (NIH), United States
Copyright © 2022 Xu, Lin, Li, Zhu, Yang, Chen, Chen, Chen, Chen, Guo and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changmin Hu, aGNtQG1haWwuaHphdS5lZHUuY24=
 Haojun Xu
Haojun Xu Changjie Lin1,2
Changjie Lin1,2 Jinghan Yang
Jinghan Yang Jianguo Chen
Jianguo Chen Xi Chen
Xi Chen Yingyu Chen
Yingyu Chen Aizhen Guo
Aizhen Guo Changmin Hu
Changmin Hu