- 1Natural and Medical Sciences Research Center, University of Nizwa, Nizwa, Oman
- 2Biomedical Research Center, School of Science, Engineering and Environment, University of Salford, Manchester, United Kingdom
Coronavirus disease 2019 (COVID-19) is caused by SARS-CoV-2. During T-cell activation, the immune system uses different checkpoint pathways to maintain co-inhibitory and co-stimulatory signals. In COVID-19, expression of immune checkpoints (ICs) is one of the most important manifestations, in addition to lymphopenia and inflammatory cytokines, contributing to worse clinical outcomes. There is a controversy whether upregulation of ICs in COVID-19 patients might lead to T-cell exhaustion or activation. This review summarizes the available studies that investigated IC receptors and ligands in COVID-19 patients, as well as their effect on T-cell function. Several IC receptors and ligands, including CTLA-4, BTLA, TIM-3, VISTA, LAG-3, TIGIT, PD-1, CD160, 2B4, NKG2A, Galectin-9, Galectin-3, PD-L1, PD-L2, LSECtin, and CD112, were upregulated in COVID-19 patients. Based on the available studies, there is a possible relationship between disease severity and increased expression of IC receptors and ligands. Overall, the upregulation of some ICs could be used as a prognostic biomarker for disease severity.
Introduction
Coronavirus disease 2019 (COVID-19) is a pandemic disease from December 2019 (1). Since the initial wave of cases appeared in Wuhan, China, over 260 million individuals worldwide have been infected with COVID-19, resulting in about six million deaths until now. Most infected patients are without any symptoms or have mild symptoms, but some patients become severely ill and need to be admitted to the hospitals (1, 2). This unexpected outbreak has highlighted the necessity to develop new vaccinations and different therapies to combat COVID-19 (3). Importantly, there are new approved direct antiviral medications for COVID-19 patients (4). For example, remdesivir, a nucleoside analog, is incorporated into the SARS-CoV-2 viral RNA-dependent RNA polymerase (RdRp) complex and prevents its translocation (5). The US Food and Drug Administration (FDA) has authorized it for the treatment of hospitalized COVID-19 patients (5, 6). Furthermore, molnupiravir, a nucleoside analogue, is the first orally taken direct-acting antiviral drug that has been shown to be effective in the eradication of viral RNA, while maintaining high safety and tolerability profiles (4, 7).
COVID-19 could be an immune-related disorder, characterized by lymphopenia, increased proinflammatory cytokines, and abnormal T-cell responses (3, 8, 9). It can stimulate both innate and adaptive immune responses. Later, this causes severe inflammatory reactions leading to systemic cellular damaging (10). However, the transition from innate to adaptive immune responses is crucial in defining the clinical implications of COVID-19 infections. First responses are often protective, whereas later leads to a reduction in viral clearance and a low survival rate (8, 11). Tissue injury observed in acute COVID-19 infections is mediated primarily by the hyperreactivity of lymphocyte responses (8).
T Cells in COVID-19 Patients
Lymphopenia is a general characteristic of many respiratory viral diseases such as human rhinovirus and influenza (12). COVID-19-associated lymphopenia could be more severe and persistent, compared with other respiratory infections (12, 13). Although lymphopenia is not fully understood in COVID-19, the decline in T-cell numbers is a common symptom among patients with severe diseases (14). Recent studies showed a decline in the total number of T cells, as well as a negative relationship between T-cell depletion and prognosis, particularly in COVID-19 patients who require admission to the ICU (15, 16). Moreover, COVID-19 can be more severe in patients who arrive at the hospital with low CD4+ and CD8+ T cell numbers, which can lead to worse clinical outcomes (17). Clearly, these patients should be monitored for any changes in levels of T cells (18). In severe cases of COVID-19, it has been shown that CD8+ T cells and natural killer (NK) cells were reduced in numbers, but they were hyperactive (19). The number and immunological status of GrA+CD8+ T cells and NK cells were recovered after the patients’ condition improved (19). According to this study, perforin+ NK cells and GrA+CD8+ T cells could be useful for the diagnosis of COVID-19 patients. Memory T cells are essentially important to fight against SARS-CoV-2 reinfection and to determine the duration of vaccine protection (20). A study demonstrated that virus-specific T cells induced by betacoronaviruses are long-lasting, suggesting that COVID-19 patients will develop a long-term T-cell immunity, which may be able to protect against SARS-CoV-2 (21). In addition, Odak et al. found that hospitalized COVID-19 patients showed altered effector/effector memory and naïve T-cell frequencies, compared with healthy controls (22). Also, they found that T regulatory cells were significantly lower in both severe and mild COVID-19 patients, compared with healthy controls (22). Moreover, they observed increased levels of effector and memory T-cell populations in mild disease but not in severe disease (22).
Inhibitory Immune Checkpoints in COVID-19
During T-cell activation, the immune system uses checkpoint pathways to maintain co-stimulatory and co-inhibitory signals. As a result, a disturbance in the function of ICs may lead to autoimmune diseases. Some cytokines regulate the expression of immune checkpoint proteins. As an example, transforming growth factor-β1 (TGF-β1) increases the expression of the programmed cell death-1 (PD-1) receptor by enhancing antigen-driven PD-1 gene transcription through Smad3 transcriptional activation in T cells in vitro and in tumor-infiltrating lymphocytes in vivo (23). Moreover, Schlichtner et al. found that VISTA upregulation is regulated by the TGF-β1-Smad3 signaling pathway (24). However, in T cells, TGF-β regulate the expression of VISTA only on T cells lacking granzyme B expression (24). Indeed, they also reported that TGF-β may regulate galectin-9 (Gal-9) expression by the Smad3 pathway in tumor cells (24).
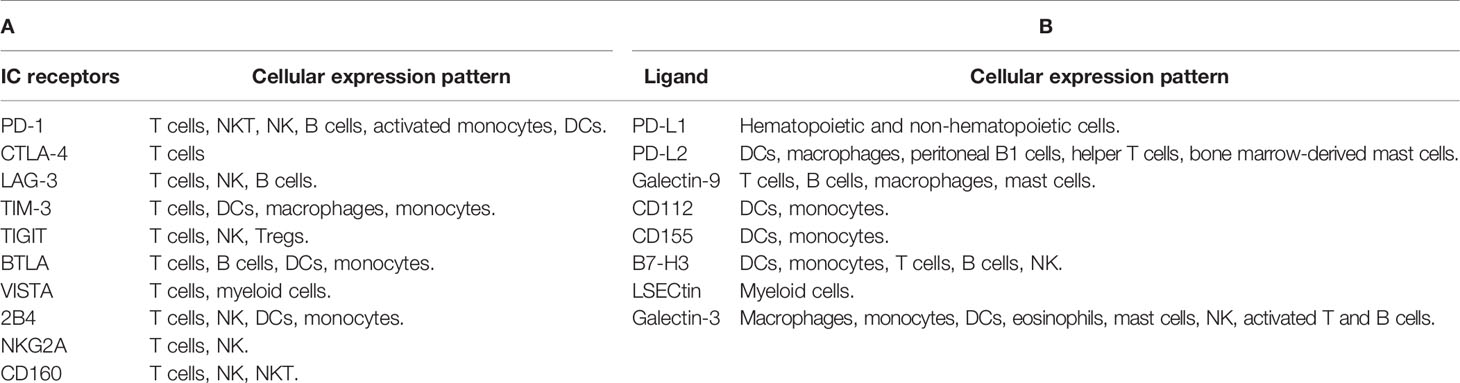
Many pathogens are able to induce overexpression of these checkpoint molecules in different immune cells, leading to increases in IC inhibitory signals and immune evasion (25, 26). As a consequence of IC expression, T cells are exhausted, leading to viral escape from immune monitoring (26, 27). Table 1 summarizes IC receptors and ligands covered in this review.
Programmed Cell Death-1
PD-1 works by inhibiting innate and adaptive immune responses (28, 29). It is expressed on B cells, T cells, activated monocytes, natural killer T (NKT) cells, natural killer cells (NK), and dendritic cells (DCs) (28, 30–33). PD-1 modulates T-cell function and tolerance, as well as immune-mediated tissue injury (34, 35). There are two known ligands for the PD-1 receptor: PD-L1 and PD-L2. In normal circumstances, the PD-1/PD-L1 pathway plays a crucial role in the modulation of immune function and preventing autoimmunity by inhibiting T-cell activation (34, 36, 37). PD-1 is elevated during acute and chronic viral diseases, such as HCV, HBV, or HIV (31, 38). T-cell depletion and disease progression are linked to PD-1 expression in HIV-specific CD4+ and CD8+ T cells (31, 32, 38, 39).
In COVID patients, PD-1 was shown to be overexpressed on both peripheral blood CD4+ and CD8+ T cells, compared with healthy controls (40). Some studies indicate that PD-1 is thought to have a role in T-cell exhaustion and disease progression (31, 39, 41, 42). The observed PD-1 expression was higher in peripheral blood CD4+ and CD8+ T lymphocytes in COVID-19 patients of all ages, compared with healthy controls (26, 39, 43). PD-1 was found to be upregulated on both peripheral blood CD8+ and CD4+ T cells in severe compared with mild and moderate diseases (Figure 1) (40, 43). Moreover, Kong et al. observed a significant increase in serum levels of soluble PD-1 (sPD-1) in severe COVID-19 patients, compared with mild disease (44). Jeannet et al. reported that the expression of PD-1 was increased exponentially with the period of illness in COVID-19 patients in the ICU, thus reducing the effectiveness of immune responses to viral infections (45). However, according to Rha et al., peripheral blood CD8+ T cells expressing PD-1 during COVID-19 infection are not exhausted but rather functional (46). In line with these findings, Shahbaz et al. found that the overexpression of PD-1 in peripheral blood was not associated with exhaustion and impairment of T-cell function (40).
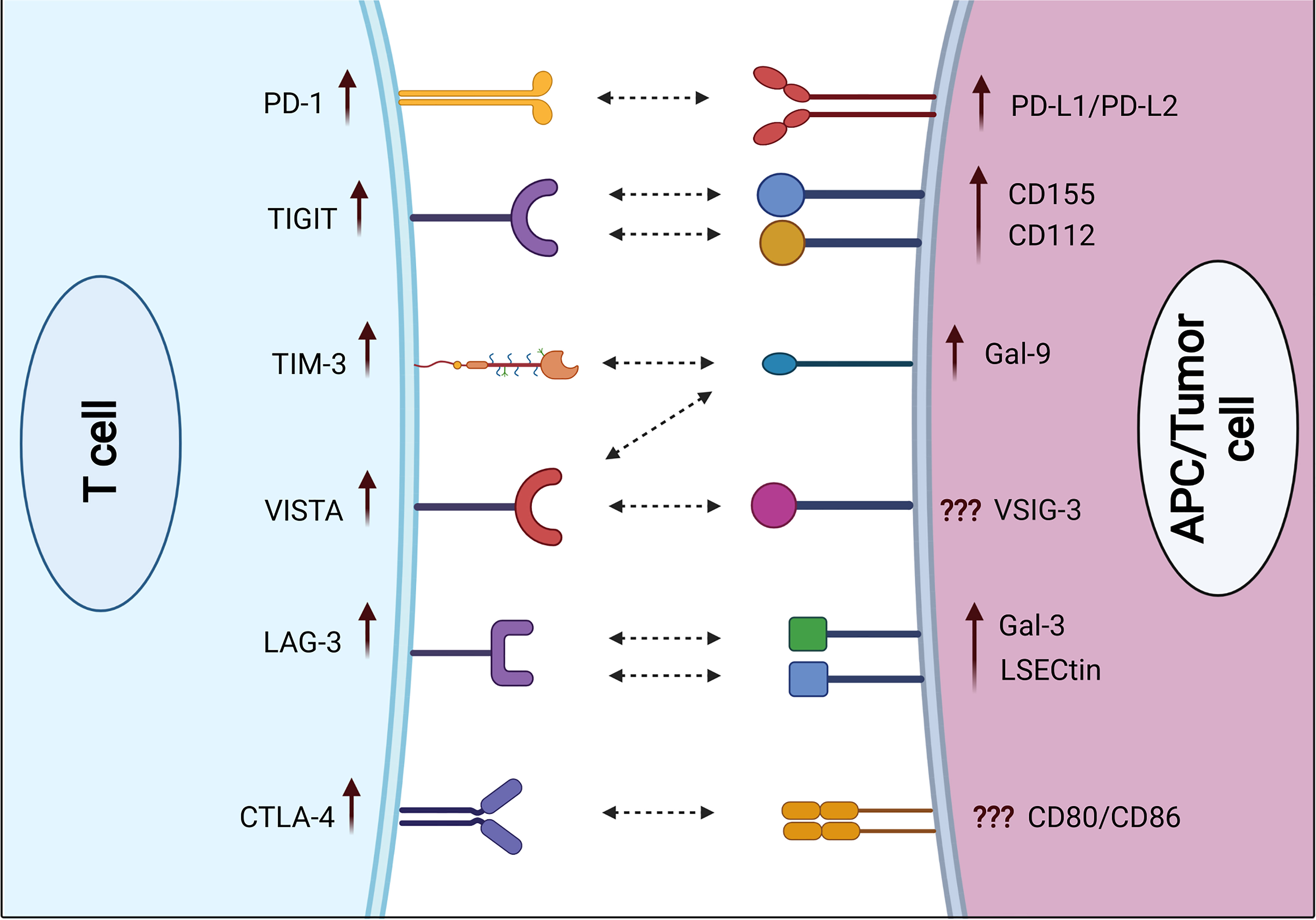
Figure 1 Expression of immune checkpoint receptors on T cells and their respective ligands on APCs and/or cancer cells in severe COVID-19 patients. Some ICs including PD-1, TIGIT, TIM-3, VISTA, LAG-3, and CTLA-4 are upregulated on both CD4+ and CD8+ T cells in severe COVID-19 patients, compared with mild/moderate patients. Various IC ligands including PD-L1, PD-L2, CD155, CD112, Gal-9, Gal-3, and LSECtin are upregulated on APCs in severe COVID-19 patients.
Cytotoxic T Lymphocyte-Associated Antigen-4
Cytotoxic T lymphocyte-associated antigen (CTLA-4) works by suppression of T-cell stimulatory signals (47–49). It binds both B7 family members (B7-1 and B7-2) with considerably higher affinity than CD28 (50). As a result, the CD28 receptor is excluded from the immunological synapse (50). This receptor also sends inhibitory signals to T cells, limiting their activation and finally leading to the depletion of its ligands via endocytosis on antigen-presenting cells (51, 52). CTLA-4 is hypothesized to control T-cell proliferation early in the immune responses, mainly in lymph nodes, while PD-1 inhibits T cells later, mainly in the peripheral tissues (53, 54).
Zheng et al. showed that the increased expression of CTLA-4 in severe symptomatic COVID-19 patients leads to CD8+ T-cell exhaustion in peripheral blood and impairs their specific immune activity (43). Moreover, Kong et al. found a significant increase in serum levels of soluble CTLA-4 (sCTLA-4) in severe COVID-19 patients compared with mild disease (44). Another study found that the upregulation of CTLA-4 in blood and bronchoalveolar lavage fluid (BALF) CD8+ and CD4+ T cells is due to viral invasion and excessive immune responses (55). Some recent studies reported that CTLA-4 is upregulated in peripheral blood CD8+ T cells in severe disease, compared with mild and moderate diseases (Figure 1) (40). Moreover, a specific upregulation of CTLA-4 was seen in BALF CD8+ T cells isolated from severe COVID-19 patients (56). Hou et al. showed that the expression of CTLA-4 on CD4+ T cells was dramatically elevated in patients with COVID-19 after 1 year of recovery (57). A study indicated that the presence of CTLA-4 and PD-1 on T cells was not associated with a T-cell inhibition, but rather with a strong activation (40). Other studies demonstrated that the presence of CTLA-4 and PD-1 on T cells may modulate the immune response and protect the vital organ from an excessive inflammatory environment in severe COVID-19 patients (51, 58).
T-Cell Immunoglobulin and Mucin Domain-Containing Protein 3 and Lymphocyte-Activation Gene 3
T-cell immunoglobulin and mucin domain-containing protein 3 (TIM-3) is expressed on CD8+ T cells and T helper 1 (Th1) cells, serving as a potent immune inhibitor (35, 38, 49, 59). It is also detected on monocytes, dendritic cells, and macrophages (38, 60, 61). Lymphocyte-activation gene 3 (LAG-3) expression is increased on activated CD4+ T cells, CD8+ T cells, B cells, and NK cells (35, 49, 62, 63). Some studies have shown that LAG-3 and TIM-3 are strongly upregulated on T cells in COVID-19 patients (62, 64, 65). Importantly, TIM-3 and LAG-3 could be utilized to identify COVID-19 patients with bad prognoses (66–68). Furthermore, Shahbaz et al. found significant upregulations of TIM-3 on both peripheral blood CD4+ and CD8+ T cells in COVID-19 patients, compared with healthy controls (40). Other studies reported significant upregulations of TIM-3 and LAG-3 on both peripheral blood CD4+ and CD8+ T cells in severe compared with mild and moderate diseases (Figure 1) (40, 62). Another study observed significant elevations of soluble TIM-3 (sTIM-3) and soluble LAG-3 (sLAG-3) in severe COVID-19 patients, compared with mild disease (44). Furthermore, Chen et al. found that the plasma level of sTIM-3 was significantly higher in severe COVID-19 patients, compared with healthy controls (69). Moreover, Diao et al. showed a significant increase in TIM-3 expression on peripheral blood CD4+ T cells in COVID-19 patients, which could contribute to the functional exhaustion of these cells (67). Also, they found a correlation between TIM-3 expression and the severity of the disease in COVID-19 patients (67). In line with these findings, Modabber et al. identified higher TIM-3 expression on peripheral blood CD4+ T cells in critical COVID-19 patients than in moderate and severe diseases (60). Furthermore, some COVID-19 inpatients from Nanjing Hospital/China were evaluated, and it was found that the majority of exhausted T cells expressed LAG-3 (70). Another study found that NK cells from the majority of COVID-19 patients appeared exhausted based on the expression of LAG-3 (71). Therefore, exhaustion of these cells could be associated with serious illness and weak antiviral immune responses.
T-Cell Immunoreceptor With Immunoglobulin and ITIM Domain
T-cell immunoreceptor with immunoglobulin and ITIM domain (TIGIT) is expressed on activated T cells, as well as NK cells, and Tregs (72–74). In severe viral diseases, the sustained expression of TIGIT in response to persistent antigen can result in T-cell exhaustion (8, 43). Shahbaz et al. found a significant upregulation of TIGIT on peripheral blood CD8+ and CD4+ T cells in COVID-19 patients, compared with controls (40). In line with these observations, TIGIT expression on peripheral blood CD8+ T cells was higher in severe compared with mild patients (Figure 1) (40, 43, 75). Conversely, Herrmann et al. observed no significant differences in TIGIT expression in COVID-19 patients but substantially lower than that of controls (62). In addition, Hsieh et al. found that higher frequencies of NK cell subsets expressing TIGIT eliminated the viruses faster than cells with lower levels of TIGIT in COVID-19 patients (76). Moreover, Shahbaz et al. indicated that overexpressions of TIGIT, TIM-3, and CTLA-4 were not associated with exhaustion and impairment of peripheral blood T-cell functions. More accurately, these expressions on activated T cells are to avoid harmful hyper-immune reactions (40).
V-Domain Ig Suppressor of T-Cell Activation
V-domain Ig suppressor of T-cell activation (VISTA) is an immune checkpoint receptor that regulates T-cell function (77). It is expressed in significant levels on T cells and myeloid cells (78). In contrast to other IC receptors that are expressed after immune-cell activation, VISTA is expressed in stable conditions on both T cells and myeloid cells (79). Overexpression of VISTA leads to increase in T-cell exhaustion and reduction in their proliferation (77, 80). Some studies found that V-set and immunoglobulin domain-containing 3 (VSIG-3) is a ligand of VISTA, and its interaction can inhibit T-cell proliferation (79, 81, 82). Moreover, another study found that VISTA interacts with Gal-9 secreted by tumor cells as a ligand in acute myeloid leukemia (83).
In COVID-19, VISTA was highly expressed on peripheral blood T cells (40, 84). Shahbaz et al. reported a significant upregulation of VISTA on peripheral blood CD8+ and CD4+ T cells in COVID-19 patients, compared with controls (40). Furthermore, VISTA expression levels on T cells were found to be considerably greater in severe COVID-19 patients versus those with mild diseases (Figure 1) (40). Another study found that overexpression of VISTA on exhausted T cells can occur in chronic viral illnesses like COVID-19 (85). As a result, viral multiplication is likely to be uncontrollable (85).
B- and T-Lymphocyte Attenuator
B- and T-lymphocyte attenuator (BTLA), a member of the CD28 Ig-superfamily, is structurally and functionally similar to CTLA-4 and PD-1 (86–89). BTLA is mostly expressed on B cells and both CD4+ and CD8+ T cells (90–93). Also, it can be expressed on DCs and monocytes (88). BTLA expression is reduced rapidly upon T-cell activation (89). BTLA differs from the rest of the Ig superfamily because it can bind to the herpesvirus entry mediator (HVEM), one of the TNFR superfamily members (35, 86–88). In COVID-19 patients, the BTLA was significantly elevated on peripheral blood CD4+ and CD8+ T cells, when compared to the normal group (90). This elevation of BTLA serves to counteract the initial activation of T cells (94). Another study observed a significant elevation of soluble BTLA (sBTLA) in severe COVID-19 patients, compared with mild disease (44). Moreover, Sharif-Askari et al. found a link between BTLA upregulation and COVID-19 severity (56). Moreover, Schultheiß et al. reported a significant upregulation of BTLA on both CD8+ and CD4+ T cells in COVID-19 patients, compared with healthy controls (90). In an in vitro study, Sumida et al. found that production of IFN-β during viral infection suppresses the expression of CD160, TIGIT, and BTLA on CD8+ and CD4+ T cells (95).
Other Immune Checkpoints
CD244 (2B4) is expressed on T cells, as well as NK cells, DCs, and monocytes (96). Also, CD160 is expressed on T cells, NK cells, and NKT cells (96, 97). In addition, NKG2A is expressed on T cells and NK cells (98). The expression of these ICs might lead to exhaustion of CD4+ and CD8+ T cells (99–101). Some studies have linked the increased expression of these inhibitory receptors to CD8+ T-cell exhaustion in chronic viral infections such as influenza, HIV, and HCV (31, 96, 100). In COVID-19 patients, Shahbaz et al. found an overexpression of 2B4 on peripheral blood CD4+ and CD8+ T cells; however, CD160 was upregulated on CD4+ T cells but not on CD8+ T cells, compared with controls (40). Additionally, NKG2A was upregulated on peripheral blood CD4+ and not on CD8+ T cells in severe COVID-19 patients, compared with mild and moderate diseases (Figure 1) (40). Despite previously reported associations between T-cell dysfunction and overexpression of these inhibitory receptors in viral infections (96, 98, 100), Shahbaz et al. showed that such overexpression was associated with functional T cells against SARS-CoV-2 (40). Furthermore, Zhang et al. showed that the expression of CD160 on NKT cells was increased significantly in moderate COVID-19 patients, compared with severe illness (97). This might imply that the presence of CD160 on NKT cells improves disease control through direct cytotoxicity (97, 102). On the other hand, Zheng et al. observed an overexpression of NKG2A on exhausted NK cells and CD8+ T cells in severe COVID-19 patients (98). Therefore, the upregulation of NKG2A could be associated with functional exhaustion of cytotoxic lymphocytes at the early stage, which could result in progression of the disease (98).
Cross Talks Between Immune Checkpoints
Some of T-cell inhibitory receptors appear to be co-expressed during exhausted T-cell differentiation. Interestingly, Yang et al. showed that PD-1 binds to the TIM-3 ligand Gal-9, which attenuates Gal-9/TIM-3-induced cell death (103). Moreover, Baitsch et al. found that naive T cells are primarily controlled by BTLA and TIM-3 receptors, whereas effector cells interact via larger amounts of inhibitory receptors (104). Furthermore, Okazaki et al. demonstrated that a synergistic effect was found between LAG-3 and PD-1 in the regulation of T-cell function (105). Indeed, Koyama et al. observed an upregulation of TIM-3 in tumor tissues following anti-PD-1 treatment. Consequently, adaptive resistance to anti-PD-1 therapy was acquired (106). It is possible that blocking several immune checkpoints with particular monoclonal antibodies may lead to improvements in the outcomes of various chronic viral infections, as well as in several types of cancer (107).
Immune Checkpoint Ligands in COVID-19
Binding of IC receptors with their ligands suppresses T-cell activity and function, helping in the regulation of immunity (108). Viral infections induce the overexpression of some IC ligands in different immune cells, resulting in a decrease of the viral clearance and increased mortality (109, 110). Herein, we present the few available studies that investigated IC ligands in COVID-19 patients.
PD-L1 and PD-L2
PD-L1 is broadly expressed on hematopoietic and non-hematopoietic cells (111). PD-L2 (also known as B7-DC) is mostly expressed in macrophages, activated DCs, Th2 cells, bone marrow-derived mast cells, and peritoneal B1 cells (112). Importantly, PD-1 and its ligands PD-L1 and PD-L2 were elevated during acute viral infections and after sustained viral infections (111). The expression of PD-L1 on basophils and eosinophils was associated with COVID-19 severity (Figure 1) (113). In COVID-19, dendritic cells and monocytes lack maturation markers and have elevated levels of PD-L1 (114). Moreover, Monaghan et al. reported a significant overexpression of PD-L1 and PD-L2 in peripheral blood of patients who died from COVID-19 (115). SARS-CoV-2 induced an overexpression of PD-L1 in epithelial cells, and it was dysregulated in a variety of immune cells including neutrophils, gamma delta T cells, monocytes, and CD4+ T cells of COVID-19 patients (116). These results indicate that PD-L1 has a prognostic role in COVID-19 patients (116). Blood levels of pro-inflammatory cytokines IL-6, IL-17, and IL-8 were markedly elevated in severe COVID-19 patients, together with elevated macrophage and neutrophil activity (117, 118). Therefore, overexpression of PD-L1 on the surface of immune cells in COVID-19 patients could be due to the presence of these pro-inflammatory cytokines (118). In other studies, it has been reported that PD-L1 was overexpressed on monocytes, and the plasma of COVID-19 patients contains higher levels of soluble PD-L1 (sPD-L1), compared with healthy controls (119, 120). Moreover, another study found that the serum level of sPD-L1, but not sPD-L2, was significantly higher in severe COVID-19 patients (44).
Galectin-9
Galectin-9 (Gal-9) is a galactoside-binding protein expressed by different types of immune cells including T cells, B cells, macrophages, and mast cells, and it is involved in the regulation of overactive immune responses (121). Gal-9 is a ligand for TIM-3, and their interactions induce apoptosis and reduce T-cell activity (121). Gal-9 is significantly expressed on immune cells in viral infections, and autoimmune and malignant diseases (109). Soluble Gal-9 (sGal-9) was increased in the plasma during chronic viral disease, and it may suppress the immune activity against the viral infection (109). A recent investigation reported that circulating Gal-9 levels were elevated in humans infected with various viruses (122). These findings imply that viral infections induce Gal-9 overexpression. Schultheiß et al. reported that sGal-9 was significantly increased in severe COVID-19 patients, compared with patients after recovery and healthy controls (90). Moreover, Bozorgmehr et al. found that plasma Gal-9 concentrations were significantly greater in patients with severe COVID-19, compared with those with mild/moderate disease (Figure 1) (123). Plasma levels of the full-length and truncated forms of Gal-9 and Osteopontin (OPN) could serve as representative inflammatory biomarkers. In severe patients, cleavage of Gal-9 and OPN was found to be related to lung function and inflammation, but not the full length of Gal-9 and OPN (124). Therefore, the cleaved forms of OPN and Gal-9 could be useful in monitoring inflammation in COVID-19 patients with pneumonia (124, 125). In another study, COVID-19 patients were shown to have higher levels of Gal-9, Gal-3, and Gal-1, compared with healthy controls (120). Another study reported that Gal-9 was overexpressed on T cells in severe patients, compared with healthy controls (40). Gal-9 has been associated with a cytokine storm in COVID-19 (123). Furthermore, it has a positive correlation with pro-inflammatory cytokines such as IL-6 and tumor necrosis factor-α (TNF-α), suggesting that Gal-9 inhibition could be a potential therapeutic approach in COVID-19 patients (123).
Galectin-3
Galectin-3 (Gal-3) is a galactoside-binding protein expressed by all types of immune cells (126). Gal-3 has been related to several inflammatory diseases (126). A recent study reported that Gal-3 levels in macrophages, monocytes, and dendritic cells were increased in patients with severe COVID-19, compared with mild diseases (Figure 1) (127). Moreover, the serum level of Gal-3 was significantly higher in severe COVID-19 patients, compared with healthy controls (69, 128). It has been reported that Gal-3 was upregulated in proliferating T cells in severe cases of COVID-19, and frequently the hyperinflammation phase involves the overexpression of Gal-3, TNF-α, and IL-6 (129). Therefore, inhibition of Gal-3 could be a helpful approach in the treatment of COVID-19 by lowering the inflammatory reaction and preventing viral adherence to host cells (126, 127, 130). Additionally, a recent study reported higher levels of Gal-3, and Gal-1 in COVID-19 patients, compared with healthy controls, implying that Gal-3 could be a useful biomarker for disease prognosis (131). Another study reported that COVID-19 patients with serum levels of Gal-3 more than 35.3 ng/ml were associated with higher mortality, ICU hospitalization, and severe acute respiratory syndrome, implying its importance as a prognostic biomarker for mortality and disease severity (132).
B7-H3 (CD276)
B7-H3 (CD276) has both co-stimulatory and co-inhibitory roles (133). It interacts with the TLT-2 receptor to enhance T-cell activation, whereas binding to unknown receptors results in co-inhibition of T cells (133, 134). It is expressed on activated DCs, NK cells, T cells, B cells, and monocytes (135). There are very limited studies investigating B7-H3 in COVID-19 patients. A recent study reported that CD276 was upregulated in the lung during COVID-19 (136).
CD155 and CD112
CD155 (PVR) and CD112 (PVRL2, nectin-2) have both co-stimulatory and co-inhibitory roles. Both are expressed on monocytes, and DCs (137, 138), and they are recognized by a different group of receptors expressed on T cells and NK cells, namely, DNAM-1 (CD226), TIGIT, and TACTILE (CD96) (139, 140). During the activation process, CD155 and CD112 interact with DNAM-1 to enhance NK- and T-cell activity (141). On the other hand, TIGIT interacts with these ligands to inhibit the activation of NK and T cells (72, 140–142). With regard to COVID-19, Hsieh et al. reported that SARS-CoV-2 induced the overexpression of CD155 on infected cells, which binds to its receptor TIGIT on NK cells, resulting in decreased immune responses and viral clearance (76). Additionally, Wilk et al. reported a significant expression of CD112 on monocytes of hospitalized COVID-19 patients, compared with mild disease and healthy controls (Figure 1) (143).
LSECtin
Lectin (LSECtin), also known as CLEC4G, is a co-inhibitor of human T-cell immunity (144). A recent study showed that LSECtin suppresses human T-cell activation and proliferation via the butyrophilin family receptor BTN3A1 (144). Lu et al. reported that analysis of pulmonary cells from COVID-19 patients showed an overexpression of different C-type lectins such as L-SIGN, LSECtin, DC-SIGN, ASGR1, and CLEC10A on myeloid cells (145). Although these receptors do not promote active multiplication of SARS-CoV-2, they generate pro-inflammatory responses in myeloid cells, which are associated with COVID-19 severity (145).
Perspective
The expression of ICs in COVID-19 patients is an important manifestation, contributing to worse clinical outcomes. Most available studies evaluated IC receptors/ligands individually or in small combinations. Comprehensive co-expression and cross talk investigations of multiple IC receptors or ligands on specific immune-cell subpopulations in COVID-19 patients are lacking. Identification of the specific immune-cell subpopulations expressing IC receptors or ligands in severe versus mild/asymptomatic COVID-19 patients is critical for prognostic purposes and therapeutic targeting. Overall, few studies investigated different receptors/ligands in the same COVID-19 patients. Based on these studies, there are some evidence supporting the use of a panel of IC receptors/ligands as prognostic biomarkers in severe COVID-19 patients; this panel could include upregulations of PD-1, CTLA-4, TIM-3, PD-L1, Gal-3, and Gal-9. Further and well-designed studies are still needed to investigate expression profiles and functions of IC receptors and ligands in severe, compared to mild and asymptomatic COVID-19 patients.
Conclusion
COVID-19 is a pandemic disease that is impacting people all over the world. The severity of the disease is determined by the signs and symptoms that individuals exhibit. An enhanced expression of immune checkpoint molecules can result in stimulation of the apoptosis of T cells, decline in the number of T cells, and lymphopenia. Some studies reported a relationship between upregulation of IC receptors on T cells and the severity of COVID-19. Specifically, when immune cells are overactivated, ICs are upregulated and inflammatory cytokines are produced in excessive amounts, which increases the disease severity. Therefore, IC overexpression in COVID-19 patients might not be due to T-cell exhaustion with impaired antiviral responses. Some studies found that the overexpression of IC receptors on T cells may modulate the immune response and protect vital organs from an excessive inflammatory response in severe COVID-19 patients. Overexpression of some of these IC receptors can be used as prognostic biomarkers for COVID severity. Clearly, targeting inhibitory ICs should be carefully considered because the efficacy and safety of blocking inhibitory ICs in COVID-19 patients have not yet been fully elucidated.
Few studies have investigated the expression level of IC ligands in COVID-19 patients. Based on the few available studies, there is a relationship between disease severity and increased expression of IC ligands. However, there are no available studies investigating the expression levels of some IC ligands including B7-H4, B7-H5, and B7-H6 in COVID-19 patients, and it would be interesting to do that.
Cancer patients receiving immune checkpoint inhibitors (ICI) may have greater immunological competence as a consequence of their reactivated T cells. However, this may lead to an increase in the risk of cytokine release syndrome (CRS), a vital manifestation in COVID-19 patients (146, 147). Few studies found a high percentage of ICI-related CRS cases following ICI administration in cancer patients (147, 148). However, other studies found that there were no associations between administration of ICI with mortality in cancer patients with COVID-19 (149, 150). Recently, ICI could be used as a potential therapeutic approach against COVID-19 in non-cancer patients (26). The majority of the concerns regarding ICI administration are related to an increase in inflammatory cytokine secretion as a consequence of reactivated of exhausted T cells, which might lead to organ damage (147). However, another study found that organ damage in COVID-19 patients is caused by virus infection itself rather than cytokine storm (151). Additionally, Yatim et al. demonstrated that ICI therapy was not associated with severe COVID-19, rather it increases specific anti–SARS-CoV-2 T-cell immunity (152). Furthermore, another study found that the PD-1 inhibitor is able to enhance the specific T-cell immune response to SARS-CoV-2 antigens (153). In addition, TGF-β and IL-6 were upregulated in COVID-19 patients, suggesting that targeting these cytokines may improve COVID-19 outcomes (154–156).
Most of the available studies on COVID-19 patients who have undertaken ICI are concentrated on PD-1 inhibition. Other ICIs in this setting should be studied as well. More studies are needed to evaluate the safety of ICI in cancer and non-cancer COVID-19 patients.
Currently, there are different COVID-19 vaccinations including BNT162b2 (BioNTech, Pfizer), AZD1222 (Oxford, AstraZeneca), Ad26.CoV2.S (Janssen), mRNA-1273 (Moderna), BBIBP-CorV (Sinopharm), Sinovac-CoronaVac, BBV152 COVAXIN (Bharat Biotech), and NVX-CoV2373 (Covovax). Unfortunately, there are no available studies investigating IC receptors and/or ligands in individuals following any of the different COVID-19 vaccinations. Comprehensive studies are required on patients receiving COVID-19 vaccines to determine any changes in the expression and function of IC receptors and ligands on different immune cells following these vaccinations.
Author Contributions
MA-M and AA wrote the article and prepared the figure/table. EE conceived the concept, acquired the funds, and supervised and performed the writing of the review and the editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
BALF, bronchoalveolar lavage fluid; BTLA, B- and T-lymphocyte attenuator; COVID-19, Coronavirus disease 2019; CTLA-4, cytotoxic T lymphocyte-associated antigen; DCs, dendritic cells; FDA, Food and Drug Administration; Gal-1, Galectin-1; Gal-3, Galectin-3; Gal-9, Galectin-9; HVEM, herpesvirus entry mediator; IC, immune checkpoint; ICU, intensive care unit; IL-6, Interleukin 6; IL8, Interleukin 8; LAG-3, lymphocyte-activation gene 3; NK, natural killer; NKT, natural killer T; OPN, Osteopontin; PD-1, programmed cell death 1; PD-L1, programmed cell death ligand 1; PD-L2, programmed death ligand 2; RdRp, RNA-dependent RNA polymerase; TGF-β1, transforming growth factor-β1; Th1, T helper; TIGIT, T-cell immunoreceptor with immunoglobulin; ITIM domain, TIM-3 T-cell immunoglobulin and mucin domain-containing protein 3; TLT-2, TREM-like transcript 2; TNFR, tumor necrosis factor receptor; TNF-α, tumor necrosis factor alpha; Treg, regulatory T cells; VISTA, V-domain Ig suppressor of T-cell activation; VSIG-3, V-set and immunoglobulin domain-containing 3.
References
1. Wang K, Qiu Z, Liu J, Fan T, Liu C, Tian P, et al. Analysis of the Clinical Characteristics of 77 Covid-19 Deaths. Sci Rep (2020) 10(1):16384. doi: 10.1038/s41598-020-73136-7
2. Zavascki AP, Falci DR. Clinical Characteristics of Covid-19 in China. N Engl J Med (2020) 382(19):1859. doi: 10.1056/NEJMc2005203
3. Paces J, Strizova Z, Smrz D, Cerny J. Covid-19 and the Immune System. Physiol Res (2020) 69(3):379–88. doi: 10.33549/physiolres.934492
4. Fischer WA, Eron JJ Jr, Holman W, Cohen MS, Fang L, Szewczyk LJ, et al. A Phase 2a Clinical Trial of Molnupiravir in Patients with Covid-19 Shows Accelerated SARS-CoV-2 RNA Clearance and Elimination of Infectious Virus. Sci Transl Med (2022) 14(628):eabl7430. doi: 10.1126/scitranslmed.abl7430
5. Frediansyah A, Nainu F, Dhama K, Mudatsir M, Harapan H. Remdesivir and Its Antiviral Activity Against Covid-19: A Systematic Review. Clin Epidemiol Global Health (2021) 9:123–7. doi: 10.1016/j.cegh.2020.07.011
6. Lamb YN. Remdesivir: First Approval. Drugs (2020) 80(13):1355–63. doi: 10.1007/s40265-020-01378-w
7. Kabinger F, Stiller C, Schmitzová J, Dienemann C, Kokic G, Hillen HS, et al. Mechanism of Molnupiravir-Induced SARS-CoV-2 Mutagenesis. Nat Struct Mol Biol (2021) 28(9):740–6. doi: 10.1038/s41594-021-00651-0
8. Toor SM, Saleh R, Sasidharan Nair V, Taha RZ, Elkord E. T-Cell Responses and Therapies Against SARS-CoV-2 Infection. Immunology (2021) 162(1):30–43. doi: 10.1111/imm.13262
9. Gambichler T, Reuther J, Scheel CH, Becker JC. On the Use of Immune Checkpoint Inhibitors in Patients With Viral Infections Including Covid-19. J Immunother Cancer (2020) 8(2):e001145. doi: 10.1136/jitc-2020-001145
10. Anka AU, Tahir MI, Abubakar SD, Alsabbagh M, Zian Z, Hamedifar H, et al. Coronavirus Disease 2019 (Covid-19): An Overview of the Immunopathology, Serological Diagnosis and Management. Scand J Immunol (2021) 93(4):e12998–e. doi: 10.1111/sji.12998
11. García LF. Immune Response, Inflammation, and the Clinical Spectrum of Covid-19. Front Immunol (2020) 11:1441. doi: 10.3389/fimmu.2020.01441
12. Chen Z, Wherry EJ. T Cell Responses in Patients With Covid-19. Nat Rev Immunol (2020) 20(9):529–36. doi: 10.1038/s41577-020-0402-6
13. McClain MT, Park LP, Nicholson B, Veldman T, Zaas AK, Turner R, et al. Longitudinal Analysis of Leukocyte Differentials in Peripheral Blood of Patients With Acute Respiratory Viral Infections. J Clin Virol (2013) 58(4):689–95. doi: 10.1016/j.jcv.2013.09.015
14. Tavakolpour S, Rakhshandehroo T, Wei EX, Rashidian M. Lymphopenia During the Covid-19 Infection: What It Shows and What Can Be Learned. Immunol Lett (2020) 225:31–2. doi: 10.1016/j.imlet.2020.06.013
15. Huang W, Berube J, McNamara M, Saksena S, Hartman M, Arshad T, et al. Lymphocyte Subset Counts in Covid-19 Patients: A Meta-Analysis. Cytometry Part A (2020) 97(8):772–6. doi: 10.1002/cyto.a.24172
16. Koblischke M, Traugott MT, Medits I, Spitzer FS, Zoufaly A, Weseslindtner L, et al. Dynamics of Cd4 T Cell and Antibody Responses in Covid-19 Patients With Different Disease Severity. Front Med (2020) 7. doi: 10.3389/fmed.2020.592629
17. Liu R, Wang Y, Li J, Han H, Xia Z, Liu F, et al. Decreased T Cell Populations Contribute to the Increased Severity of Covid-19. Clinica Chimica Acta (2020) 508:110–4. doi: 10.1016/j.cca.2020.05.019
18. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J Clin Invest (2020) 130(5):2620–9. doi: 10.1172/jci137244
19. Jiang Y, Wei X, Guan J, Qin S, Wang Z, Lu H, et al. Covid-19 Pneumonia: Cd8+ T and Nk Cells Are Decreased in Number But Compensatory Increased in Cytotoxic Potential. Clin Immunol (2020) 218:108516. doi: 10.1016/j.clim.2020.108516
20. Zhao J, Wang L, Schank M, Dang X, Lu Z, Cao D, et al. SARS-CoV-2 Specific Memory T Cell Epitopes Identified in Covid-19-Recovered Subjects. Virus Res (2021) 304:198508. doi: 10.1016/j.virusres.2021.198508
21. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-Specific T Cell Immunity in Cases of Covid-19 and Sars, and Uninfected Controls. Nature (2020) 584(7821):457–62. doi: 10.1038/s41586-020-2550-z
22. Odak I, Barros-Martins J, Bošnjak B, Stahl K, David S, Wiesner O, et al. Reappearance of Effector T Cells Is Associated With Recovery From Covid-19. EBioMedicine (2020) 57:102885. doi: 10.1016/j.ebiom.2020.102885
23. Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J, et al. Tgfβ1-Mediated Smad3 Enhances Pd-1 Expression on Antigen-Specific T Cells in Cancer. Cancer Discovery (2016) 6(12):1366–81. doi: 10.1158/2159-8290.Cd-15-1347
24. Schlichtner S, Yasinska IM, Ruggiero S, Berger SM, Aliu N, Prunk M, et al. Expression of the Immune Checkpoint Protein Vista Is Differentially Regulated by the Tgf-B1 - Smad3 Signaling Pathway in Rapidly Proliferating Human Cells and T Lymphocytes. Front Med (Lausanne) (2022) 9:790995. doi: 10.3389/fmed.2022.790995
25. Khair DO, Bax HJ, Mele S, Crescioli S, Pellizzari G, Khiabany A, et al. Combining Immune Checkpoint Inhibitors: Established and Emerging Targets and Strategies to Improve Outcomes in Melanoma. Front Immunol (2019) 10:453. doi: 10.3389/fimmu.2019.00453
26. Pezeshki PS, Rezaei N. Immune Checkpoint Inhibition in Covid-19: Risks and Benefits. Expert Opin Biol Ther (2021) 21(9):1173–9. doi: 10.1080/14712598.2021.1887131
27. McLane LM, Abdel-Hakeem MS, Wherry EJ. Cd8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol (2019) 37:457–95. doi: 10.1146/annurev-immunol-041015-055318
28. Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor Antigen-Specific Cd8 T Cells Infiltrating the Tumor Express High Levels of Pd-1 and Are Functionally Impaired. Blood (2009) 114(8):1537–44. doi: 10.1182/blood-2008-12-195792
29. Han Y, Liu D, Li L. Pd-1/Pd-L1 Pathway: Current Researches in Cancer. Am J Cancer Res (2020) 10(3):727–42.
30. Almahmoud S, Zhong HA. Molecular Modeling Studies on the Binding Mode of the Pd-1/Pd-L1 Complex Inhibitors. Int J Mol Sci (2019) 20(18):4654. doi: 10.3390/ijms20184654
31. Youngblood B, Wherry EJ, Ahmed R. Acquired Transcriptional Programming in Functional and Exhausted Virus-Specific Cd8 T Cells. Curr Opin HIV AIDS (2012) 7(1):50–7. doi: 10.1097/COH.0b013e32834ddcf2
32. Horne-Debets JM, Faleiro R, Karunarathne DS, Liu XQ, Lineburg KE, Poh CM, et al. Pd-1 Dependent Exhaustion of Cd8+ T Cells Drives Chronic Malaria. Cell Rep (2013) 5(5):1204–13. doi: 10.1016/j.celrep.2013.11.002
33. Kim PS, Ahmed R. Features of Responding T Cells in Cancer and Chronic Infection. Curr Opin Immunol (2010) 22(2):223–30. doi: 10.1016/j.coi.2010.02.005
34. Francisco LM, Sage PT, Sharpe AH. The Pd-1 Pathway in Tolerance and Autoimmunity. Immunol Rev (2010) 236:219–42. doi: 10.1111/j.1600-065X.2010.00923.x
35. Wherry EJ, Kurachi M. Molecular and Cellular Insights Into T Cell Exhaustion. Nat Rev Immunol (2015) 15(8):486–99. doi: 10.1038/nri3862
36. Cheng X, Veverka V, Radhakrishnan A, Waters LC, Muskett FW, Morgan SH, et al. Structure and Interactions of the Human Programmed Cell Death 1 Receptor. J Biol Chem (2013) 288(17):11771–85. doi: 10.1074/jbc.M112.448126
37. Chen D, Tan S, Zhang H, Wang H, He W, Shi R, et al. The Fg Loop of Pd-1 Serves as a “Hotspot” for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy. iScience (2019) 14:113–24. doi: 10.1016/j.isci.2019.03.017
38. Vali B, Jones RB, Sakhdari A, Sheth PM, Clayton K, Yue FY, et al. Hcv-Specific T Cells in Hcv/Hiv Co-Infection Show Elevated Frequencies of Dual Tim-3/Pd-1 Expression That Correlate With Liver Disease Progression. Eur J Immunol (2010) 40(9):2493–505. doi: 10.1002/eji.201040340
39. Bellesi S, Metafuni E, Hohaus S, Maiolo E, Marchionni F, D’Innocenzo S, et al. Increased Cd95 (Fas) and Pd-1 Expression in Peripheral Blood T Lymphocytes in Covid-19 Patients. Br J Haematol (2020) 191(2):207–11. doi: 10.1111/bjh.17034
40. Shahbaz S, Xu L, Sligl W, Osman M, Bozorgmehr N, Mashhouri S, et al. The Quality of SARS-CoV-2-Specific T Cell Functions Differs in Patients With Mild/Moderate Versus Severe Disease, and T Cells Expressing Coinhibitory Receptors Are Highly Activated. J Immunol (Baltimore Md: 1950) (2021) 207(4):1099–111. doi: 10.4049/jimmunol.2100446
41. Chiappelli F, Khakshooy A, Greenberg G. Covid-19 Immunopathology and Immunotherapy. Bioinformation (2020) 16(3):219. doi: 10.6026/97320630016219
42. Mahmoud FM, Zaki WK, ElSheshtawy NM. Expression of Programmed Cell Death 1 (Pd-1) as a Marker of T-Cell Exhaustion and Its Correlation With Interleukin-10 Serum Level in Patients With Covid-19. J Pure Appl Microbiol (2021) 15(2):650–8. doi: 10.22207/JPAM.15.2.08
43. Zheng HY, Zhang M, Yang CX, Zhang N, Wang XC, Yang XP, et al. Elevated Exhaustion Levels and Reduced Functional Diversity of T Cells in Peripheral Blood May Predict Severe Progression in Covid-19 Patients. Cell Mol Immunol (2020) 17(5):541–3. doi: 10.1038/s41423-020-0401-3
44. Kong Y, Wang Y, Wu X, Han J, Li G, Hua M, et al. Storm of Soluble Immune Checkpoints Associated With Disease Severity of Covid-19. Signal Transduct Target Ther (2020) 5(1):192. doi: 10.1038/s41392-020-00308-2
45. Jeannet R, Daix T, Formento R, Feuillard J, François B. Severe Covid-19 Is Associated With Deep and Sustained Multifaceted Cellular Immunosuppression. Intensive Care Med (2020) 46(9):1769–71. doi: 10.1007/s00134-020-06127-x
46. Rha MS, Jeong HW, Ko JH, Choi SJ, Seo IH, Lee JS, et al. Pd-1-Expressing SARS-CoV-2-Specific Cd8(+) T Cells Are Not Exhausted, But Functional in Patients With Covid-19. Immunity (2021) 54(1):44–52.e3. doi: 10.1016/j.immuni.2020.12.002
47. Palma M, Gentilcore G, Heimersson K, Mozaffari F, Näsman-Glaser B, Young E, et al. T Cells in Chronic Lymphocytic Leukemia Display Dysregulated Expression of Immune Checkpoints and Activation Markers. Haematologica (2017) 102(3):562. doi: 10.3324/haematol.2016.151100
48. Stephen-Victor E, Das M, Karnam A, Pitard B, Gautier JF, Bayry J. Potential of Regulatory T-Cell-Based Therapies in the Management of Severe Covid-19. Eur Respir J (2020) 56(3):2002182. doi: 10.1183/13993003.02182-2020
49. Yi JS, Cox MA, Zajac AJ. T-Cell Exhaustion: Characteristics, Causes and Conversion. Immunology (2010) 129(4):474–81. doi: 10.1111/j.1365-2567.2010.03255.x
50. Krummel MF, Allison JP. Cd28 and Ctla-4 Have Opposing Effects on the Response of T Cells to Stimulation. J Exp Med (1995) 182(2):459–65. doi: 10.1084/jem.182.2.459
51. Quezada SA, Peggs KS. Exploiting Ctla-4, Pd-1 and Pd-L1 to Reactivate the Host Immune Response Against Cancer. Br J Cancer (2013) 108(8):1560–5. doi: 10.1038/bjc.2013.117
52. Rowshanravan B, Halliday N, Sansom DM. Ctla-4: A Moving Target in Immunotherapy. Blood (2018) 131(1):58–67. doi: 10.1182/blood-2017-06-741033
53. Buchbinder EI, Desai A. Ctla-4 and Pd-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am J Clin Oncol (2016) 39(1):98–106. doi: 10.1097/coc.0000000000000239
54. Fife BT, Bluestone JA. Control of Peripheral T-Cell Tolerance and Autoimmunity Via the Ctla-4 and Pd-1 Pathways. Immunol Rev (2008) 224:166–82. doi: 10.1111/j.1600-065X.2008.00662.x
55. Ronit A, Berg RMG, Bay JT, Haugaard AK, Ahlström MG, Burgdorf KS, et al. Compartmental Immunophenotyping in Covid-19 Ards: A Case Series. J Allergy Clin Immunol (2021) 147(1):81–91. doi: 10.1016/j.jaci.2020.09.009
56. Saheb Sharif-Askari N, Saheb Sharif-Askari F, Mdkhana B, Al Heialy S, Alsafar HS, Hamoudi R, et al. Enhanced Expression of Immune Checkpoint Receptors During SARS-CoV-2 Viral Infection. Mol Ther Methods Clin Dev (2021) 20:109–21. doi: 10.1016/j.omtm.2020.11.002
57. Hou H, Zhang Y, Tang G, Luo Y, Liu W, Cheng C, et al. Immunologic Memory to SARS-CoV-2 in Convalescent Covid-19 Patients at 1 Year Postinfection. J Allergy Clin Immunol (2021) 148(6):1481–92.e2. doi: 10.1016/j.jaci.2021.09.008
58. Siddiqi HK, Neilan TG. Covid-19, Immuno-Oncology and Cardiovascular Disease: Viewpoint From the Intersection. J Cardiovasc Trans Res (2020) 13(3):347–8. doi: 10.1007/s12265-020-10013-8
59. Sakhdari A, Mujib S, Vali B, Yue FY, MacParland S, Clayton K, et al. Tim-3 Negatively Regulates Cytotoxicity in Exhausted Cd8+ T Cells in Hiv Infection. PloS One (2012) 7(7):e40146. doi: 10.1371/journal.pone.0040146
60. Modabber Z, Shahbazi M, Akbari R, Bagherzadeh M, Firouzjahi A, Mohammadnia-Afrouzi M. Tim-3 as a Potential Exhaustion Marker in Cd4(+) T Cells of Covid-19 Patients. Immunity Inflammation Dis (2021) 9(4):1707–15. doi: 10.1002/iid3.526
61. Saleh R, Toor SM, Elkord E. Targeting Tim-3 in Solid Tumors: Innovations in the Preclinical and Translational Realm and Therapeutic Potential. Expert Opin Ther Targets (2020) 24(12):1251–62. doi: 10.1080/14728222.2020.1841750
62. Herrmann M, Schulte S, Wildner NH, Wittner M, Brehm TT, Ramharter M, et al. Analysis of Co-Inhibitory Receptor Expression in Covid-19 Infection Compared to Acute Plasmodium Falciparum Malaria: Lag-3 and Tim-3 Correlate With T Cell Activation and Course of Disease. Front Immunol (2020) 11:1870. doi: 10.3389/fimmu.2020.01870
63. Kisielow M, Kisielow J, Capoferri-Sollami G, Karjalainen K. Expression of Lymphocyte Activation Gene 3 (Lag-3) on B Cells Is Induced by T Cells. Eur J Immunol (2005) 35(7):2081–8. doi: 10.1002/eji.200526090
64. Ueland T, Heggelund L, Lind A, Holten AR, Tonby K, Michelsen AE, et al. Elevated Plasma Stim-3 Levels in Patients With Severe Covid-19. J Allergy Clin Immunol (2021) 147(1):92–8. doi: 10.1016/j.jaci.2020.09.007
65. Shahbazi M, Moulana Z, Sepidarkish M, Bagherzadeh M, Rezanejad M, Mirzakhani M, et al. Pronounce Expression of Tim-3 and Cd39 But Not Pd1 Defines Cd8 T Cells in Critical Covid-19 Patients. Microbial Pathogen (2021) 153:104779. doi: 10.1016/j.micpath.2021.104779
66. Martín-Quirós A, Maroun-Eid C, Avendaño-Ortiz J, Lozano-Rodríguez R, Valentín Quiroga J, Terrón V, et al. Potential Role of the Galectin-9/Tim-3 Axis in the Disparate Progression of SARS-CoV-2 in a Married Couple: A Case Report. Biomed Hub (2021) 6(1):48–58. doi: 10.1159/000514727
67. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (Covid-19). Front Immunol (2020) 11:827. doi: 10.3389/fimmu.2020.00827
68. Xiong Q, Peng C, Yan X, Yan X, Chen L, Sun B, et al. Characteristics of SARS-CoV-2-Specific Cytotoxic T Cells Revealed by Single-Cell Immune Profiling of Longitudinal Covid-19 Blood Samples. Signal Transduct Target Ther (2020) 5(1):285. doi: 10.1038/s41392-020-00425-y
69. Chen PK, Lan JL, Huang PH, Hsu JL, Chang CK, Tien N, et al. Interleukin-18 Is a Potential Biomarker to Discriminate Active Adult-Onset Still’s Disease From Covid-19. Front Immunol (2021) 12:719544. doi: 10.3389/fimmu.2021.719544
70. Qing X, Cheng P, Xiaomin Y, Lin C, Xueqi Y, Beicheng S, et al. Single-Cell Immune Profiling of Matched Disease and Recovery Phase Blood of Covid-19 Patients Reveals Anti-Lag3 Based Immune Checkpoint Therapy as a Promising Option. Res Square (2021). doi: 10.21203/rs.3.rs-24576/v1
71. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A Single-Cell Atlas of the Peripheral Immune Response in Patients With Severe Covid-19. Nat Med (2020) 26(7):1070–6. doi: 10.1038/s41591-020-0944-y
72. Joller N, Hafler JP, Brynedal B, Kassam N, Spoerl S, Levin SD, et al. Cutting Edge: Tigit Has T Cell-Intrinsic Inhibitory Functions. J Immunol (Baltimore Md: 1950) (2011) 186(3):1338–42. doi: 10.4049/jimmunol.1003081
73. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The Surface Protein Tigit Suppresses T Cell Activation by Promoting the Generation of Mature Immunoregulatory Dendritic Cells. Nat Immunol (2009) 10(1):48–57. doi: 10.1038/ni.1674
74. Krämer B, Knoll R, Bonaguro L, ToVinh M, Raabe J, Astaburuaga-García R, et al. Early Ifn-A Signatures and Persistent Dysfunction Are Distinguishing Features of Nk Cells in Severe Covid-19. Immunity (2021) 54(11):2650–69.e14. doi: 10.1016/j.immuni.2021.09.002
75. Mazzoni A, Maggi L, Capone M, Spinicci M, Salvati L, Colao MG, et al. Cell-Mediated and Humoral Adaptive Immune Responses to SARS-CoV-2 Are Lower in Asymptomatic Than Symptomatic Covid-19 Patients. Eur J Immunol (2020) 50(12):2013–24. doi: 10.1002/eji.202048915
76. Hsieh WC, Lai EY, Liu YT, Wang YF, Tzeng YS, Cui L, et al. Nk Cell Receptor and Ligand Composition Influences the Clearance of SARS-CoV-2. J Clin Invest (2021) 131(21):e146408. doi: 10.1172/jci146408
77. Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, et al. Vista, a Novel Mouse Ig Superfamily Ligand That Negatively Regulates T Cell Responses. J Exp Med (2011) 208(3):577–92. doi: 10.1084/jem.20100619
78. Mehta N, Maddineni S, Mathews II, Andres Parra Sperberg R, PS H, Cochran JR. Structure and Functional Binding Epitope of V-Domain Ig Suppressor of T Cell Activation. Cell Rep (2019) 28(10):2509–16.e5. doi: 10.1016/j.celrep.2019.07.073
79. ElTanbouly MA, Zhao Y, Schaafsma E, Burns CM, Mabaera R, Cheng C, et al. Vista: A Target to Manage the Innate Cytokine Storm. Front Immunol (2020) 11:595950. doi: 10.3389/fimmu.2020.595950
80. Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O’Connell S, et al. Vista Is an Immune Checkpoint Molecule for Human T Cells. Cancer Res (2014) 74(7):1924–32. doi: 10.1158/0008-5472.Can-13-1504
81. Xie X, Chen C, Chen W, Jiang J, Wang L, Li T, et al. Structural Basis of Vsig3: The Ligand for Vista. Front Immunol (2021) 12:625808. doi: 10.3389/fimmu.2021.625808
82. Wang J, Wu G, Manick B, Hernandez V, Renelt M, Erickson C, et al. Vsig-3 as a Ligand of Vista Inhibits Human T-Cell Function. Immunology (2019) 156(1):74–85. doi: 10.1111/imm.13001
83. Yasinska IM, Meyer NH, Schlichtner S, Hussain R, Siligardi G, Casely-Hayford M, et al. Ligand-Receptor Interactions of Galectin-9 and Vista Suppress Human T Lymphocyte Cytotoxic Activity. Front Immunol (2020) 11:580557. doi: 10.3389/fimmu.2020.580557
84. Rendeiro AF, Casano J, Vorkas CK, Singh H, Morales A, DeSimone RA, et al. Profiling of Immune Dysfunction in Covid-19 Patients Allows Early Prediction of Disease Progression. Life Sci Alliance (2021) 4(2):e202000955. doi: 10.26508/lsa.202000955
85. Rendeiro AF, Casano J, Vorkas CK, Singh H, Morales A, DeSimone RA, et al. Longitudinal Immune Profiling of Mild and Severe Covid-19 Reveals Innate and Adaptive Immune Dysfunction and Provides an Early Prediction Tool for Clinical Progression. medRxiv (2020). doi: 10.1101/2020.09.08.20189092
86. Ning Z, Liu K, Xiong H. Roles of Btla in Immunity and Immune Disorders. Front Immunol (2021) 12:654960. doi: 10.3389/fimmu.2021.654960
87. Sedy JR, Gavrieli M, Potter KG, Hurchla MA, Lindsley RC, Hildner K, et al. B and T Lymphocyte Attenuator Regulates T Cell Activation Through Interaction With Herpesvirus Entry Mediator. Nat Immunol (2005) 6(1):90–8. doi: 10.1038/ni1144
88. Murphy TL, Murphy KM. Slow Down and Survive: Enigmatic Immunoregulation by Btla and Hvem. Annu Rev Immunol (2010) 28:389–411. doi: 10.1146/annurev-immunol-030409-101202
89. Otsuki N, Kamimura Y, Hashiguchi M, Azuma M. Expression and Function of the B and T Lymphocyte Attenuator (Btla/Cd272) on Human T Cells. Biochem Biophys Res Commun (2006) 344(4):1121–7. doi: 10.1016/j.bbrc.2006.03.242
90. Schultheiß C, Paschold L, Simnica D, Mohme M, Willscher E, von Wenserski L, et al. Next-Generation Sequencing of T and B Cell Receptor Repertoires From Covid-19 Patients Showed Signatures Associated With Severity of Disease. Immunity (2020) 53(2):442–55.e4. doi: 10.1016/j.immuni.2020.06.024
91. Adler G, Steeg C, Pfeffer K, Murphy TL, Murphy KM, Langhorne J, et al. B and T Lymphocyte Attenuator Restricts the Protective Immune Response Against Experimental Malaria. J Immunol (2011) 187(10):5310–9. doi: 10.4049/jimmunol.1101456
92. Fourcade J, Sun Z, Pagliano O, Guillaume P, Luescher IF, Sander C, et al. Cd8(+) T Cells Specific for Tumor Antigens Can Be Rendered Dysfunctional by the Tumor Microenvironment Through Upregulation of the Inhibitory Receptors Btla and Pd-1. Cancer Res (2012) 72(4):887–96. doi: 10.1158/0008-5472.Can-11-2637
93. Chen YL, Lin HW, Chien CL, Lai YL, Sun WZ, Chen CA, et al. Btla Blockade Enhances Cancer Therapy by Inhibiting Il-6/Il-10-Induced Cd19(High) B Lymphocytes. J Immunother Cancer (2019) 7(1):313. doi: 10.1186/s40425-019-0744-4
94. de Joode K, Oostvogels AAM, GeurtsvanKessel CH, de Vries RD, Mathijssen RHJ, Debets R, et al. Case Report: Adequate T and B Cell Responses in a SARS-CoV-2 Infected Patient After Immune Checkpoint Inhibition. Front Immunol (2021) 12:627186. doi: 10.3389/fimmu.2021.627186
95. Sumida TS, Dulberg S, Schupp J, Stillwell HA, Axisa PP, Comi M, et al. Type I Interferon Transcriptional Network Regulates Expression of Coinhibitory Receptors in Human T Cells. bioRxiv: Preprint Server Biol (2020). doi: 10.1101/2020.10.30.362947
96. Crawford A, Wherry EJ. The Diversity of Costimulatory and Inhibitory Receptor Pathways and the Regulation of Antiviral T Cell Responses. Curr Opin Immunol (2009) 21(2):179–86. doi: 10.1016/j.coi.2009.01.010
97. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-Cell Landscape of Immunological Responses in Patients With Covid-19. Nat Immunol (2020) 21(9):1107–18. doi: 10.1038/s41590-020-0762-x
98. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional Exhaustion of Antiviral Lymphocytes in Covid-19 Patients. Cell Mol Immunol (2020) 17(5):533–5. doi: 10.1038/s41423-020-0402-2
99. Fuertes Marraco SA, Neubert NJ, Verdeil G, Speiser DE. Inhibitory Receptors Beyond T Cell Exhaustion. Front Immunol (2015) 6:310. doi: 10.3389/fimmu.2015.00310
100. Okoye IS, Houghton M, Tyrrell L, Barakat K, Elahi S. Coinhibitory Receptor Expression and Immune Checkpoint Blockade: Maintaining a Balance in Cd8(+) T Cell Responses to Chronic Viral Infections and Cancer. Front Immunol (2017) 8:1215. doi: 10.3389/fimmu.2017.01215
101. Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, et al. T Cells From Cll Patients Exhibit Features of T-Cell Exhaustion But Retain Capacity for Cytokine Production. Blood (2013) 121(9):1612–21. doi: 10.1182/blood-2012-09-457531
102. Liu J, Yang X, Wang H, Li Z, Deng H, Liu J, et al. Analysis of the Long-Term Impact on Cellular Immunity in Covid-19-Recovered Individuals Reveals a Profound Nkt Cell Impairment. mBio (2021) 12(2):e00085. doi: 10.1128/mBio.00085-21
103. Yang R, Sun L, Li CF, Wang YH, Yao J, Li H, et al. Galectin-9 Interacts With Pd-1 and Tim-3 to Regulate T Cell Death and Is a Target for Cancer Immunotherapy. Nat Commun (2021) 12(1):832. doi: 10.1038/s41467-021-21099-2
104. Baitsch L, Legat A, Barba L, Fuertes Marraco SA, Rivals JP, Baumgaertner P, et al. Extended Co-Expression of Inhibitory Receptors by Human Cd8 T-Cells Depending on Differentiation, Antigen-Specificity and Anatomical Localization. PloS One (2012) 7(2):e30852. doi: 10.1371/journal.pone.0030852
105. Okazaki T, Okazaki IM, Wang J, Sugiura D, Nakaki F, Yoshida T, et al. Pd-1 and Lag-3 Inhibitory Co-Receptors Act Synergistically to Prevent Autoimmunity in Mice. J Exp Med (2011) 208(2):395–407. doi: 10.1084/jem.20100466
106. Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, et al. Adaptive Resistance to Therapeutic Pd-1 Blockade Is Associated With Upregulation of Alternative Immune Checkpoints. Nat Commun (2016) 7:10501. doi: 10.1038/ncomms10501
107. Nirschl CJ, Drake CG. Molecular Pathways: Coexpression of Immune Checkpoint Molecules: Signaling Pathways and Implications for Cancer Immunotherapy. Clin Cancer Res (2013) 19(18):4917–24. doi: 10.1158/1078-0432.Ccr-12-1972
108. Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel Immune Checkpoint Targets: Moving Beyond Pd-1 and Ctla-4. Mol Cancer (2019) 18(1):1–14. doi: 10.1186/s12943-019-1091-2
109. Merani S, Chen W, Elahi S. The Bitter Side of Sweet: The Role of Galectin-9 in Immunopathogenesis of Viral Infections. Rev Med Virol (2015) 25(3):175–86. doi: 10.1002/rmv.1832
110. Inaguma S, Wang Z, Lasota J, Sarlomo-Rikala M, McCue PA, Ikeda H, et al. Comprehensive Immunohistochemical Study of Programmed Cell Death Ligand 1 (Pd-L1): Analysis in 5536 Cases Revealed Consistent Expression in Trophoblastic Tumors. Am J Surg Pathol (2016) 40(8):1133–42. doi: 10.1097/pas.0000000000000653
111. Schönrich G, Raftery MJ. The Pd-1/Pd-L1 Axis and Virus Infections: A Delicate Balance. Front Cell Infect Microbiol (2019) 9:207. doi: 10.3389/fcimb.2019.00207
112. Zhang N, Tu J, Wang X, Chu Q. Programmed Cell Death-1/Programmed Cell Death Ligand-1 Checkpoint Inhibitors: Differences in Mechanism of Action. Immunotherapy (2019) 11(5):429–41. doi: 10.2217/imt-2018-0110
113. Vitte J, Diallo AB, Boumaza A, Lopez A, Michel M, Allardet-Servent J, et al. A Granulocytic Signature Identifies Covid-19 and Its Severity. J Infect Dis (2020) 222(12):1985–96. doi: 10.1093/infdis/jiaa591
114. Parackova Z, Zentsova I, Bloomfield M, Vrabcova P, Smetanova J, Klocperk A, et al. Disharmonic Inflammatory Signatures in Covid-19: Augmented Neutrophils’ But Impaired Monocytes’ and Dendritic Cells’ Responsiveness. Cells (2020) 9(10):2206. doi: 10.3390/cells9102206
115. Monaghan SF, Fredericks AM, Jentzsch MS, Cioffi WG, Cohen M, Fairbrother WG, et al. Deep Rna Sequencing of Intensive Care Unit Patients With Covid-19. medRxiv (2021). doi: 10.1101/2021.01.11.21249276
116. Sabbatino F, Conti V, Franci G, Sellitto C, Manzo V, Pagliano P, et al. Pd-L1 Dysregulation in Covid-19 Patients. Front Immunol (2021) 12:2198. doi: 10.3389/fimmu.2021.695242
117. Pandolfi L, Fossali T, Frangipane V, Bozzini S, Morosini M, D’Amato M, et al. Broncho-Alveolar Inflammation in Covid-19 Patients: A Correlation With Clinical Outcome. BMC Pulmon Med (2020) 20(1):1–10. doi: 10.1186/s12890-020-01343-z
118. Chen J, Vitetta L. Increased Pd-L1 Expression May Be Associated With the Cytokine Storm and Cd8+ T-Cell Exhaustion in Severe Covid-19. J Infect Dis (2021) 223(9):1659–60. doi: 10.1093/infdis/jiab061
119. Gibellini L, De Biasi S, Paolini A, Borella R, Boraldi F, Mattioli M, et al. Altered Bioenergetics and Mitochondrial Dysfunction of Monocytes in Patients With Covid-19 Pneumonia. EMBO Mol Med (2020) 12(12):e13001. doi: 10.15252/emmm.202013001
120. De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, et al. Marked T Cell Activation, Senescence, Exhaustion and Skewing Towards Th17 in Patients With Covid-19 Pneumonia. Nat Commun (2020) 11(1):1–17. doi: 10.1038/s41467-020-17292-4
121. Oomizu S, Arikawa T, Niki T, Kadowaki T, Ueno M, Nishi N, et al. Cell Surface Galectin-9 Expressing Th Cells Regulate Th17 and Foxp3+ Treg Development by Galectin-9 Secretion. PloS One (2012) 7(11):e48574. doi: 10.1371/journal.pone.0048574
122. Katoh S, Ikeda M, Shimizu H, Fukushima K, Oka M. Induction of Galectin-9 Production by Viral Infection in the Lung. Eur Respir Soc (2015) 46:OA1780. doi: 10.1183/13993003.congress-2015.OA1780
123. Bozorgmehr N, Mashhouri S, Perez Rosero E, Xu L, Shahbaz S, Sligl W, et al. Galectin-9, A Player in Cytokine Release Syndrome and a Surrogate Diagnostic Biomarker in SARS-CoV-2 Infection. mBio (2021) 12(3):e00384–21. doi: 10.1128/mBio.00384-21
124. Bai G, Furushima D, Niki T, Matsuba T, Maeda Y, Takahashi A, et al. High Levels of the Cleaved Form of Galectin-9 and Osteopontin in the Plasma Are Associated With Inflammatory Markers That Reflect the Severity of Covid-19 Pneumonia. Int J Mol Sci (2021) 22(9):4978. doi: 10.3390/ijms22094978
125. Padilla ST, Niki T, Furushima D, Bai G, Chagan-Yasutan H, Telan EF, et al. Plasma Levels of a Cleaved Form of Galectin-9 Are the Most Sensitive Biomarkers of Acquired Immune Deficiency Syndrome and Tuberculosis Coinfection. Biomolecules (2020) 10(11):1495. doi: 10.3390/biom10111495
126. Dong R, Zhang M, Hu Q, Zheng S, Soh A, Zheng Y, et al. Galectin-3 as a Novel Biomarker for Disease Diagnosis and a Target for Therapy. Int J Mol Med (2018) 41(2):599–614. doi: 10.3892/ijmm.2017.3311
127. Caniglia JL, Asuthkar S, Tsung AJ, Guda MR, Velpula KK. Immunopathology of Galectin-3: An Increasingly Promising Target in Covid-19. F1000Research (2020) 9:1078. doi: 10.12688/f1000research.25979.1
128. Kuśnierz-Cabala B, Maziarz B, Dumnicka P, Dembiński M, Kapusta M, Bociąga-Jasik M, et al. Diagnostic Significance of Serum Galectin-3 in Hospitalized Patients With Covid-19-a Preliminary Study. Biomolecules (2021) 11(8):1136. doi: 10.3390/biom11081136
129. Aminpour M, Cannariato M, Zucco A, Di Gregorio E, Israel S, Perioli A, et al. Computational Study of Potential Galectin-3 Inhibitors in the Treatment of Covid-19. Biomedicines (2021) 9(9):1208. doi: 10.3390/biomedicines9091208
130. Caniglia JL, Guda MR, Asuthkar S, Tsung AJ, Velpula KK. A Potential Role for Galectin-3 Inhibitors in the Treatment of Covid-19. PeerJ (2020) 8:e9392. doi: 10.7717/peerj.9392
131. Kazancioglu S, Yilmaz FM, Bastug A, Ozbay BO, Aydos O, Yücel Ç, et al. Assessment of Galectin-1, Galectin-3, and Pge2 Levels in Patients With Covid-19. Jpn J Infect Dis (2021) 74(6):530–6. doi: 10.7883/yoken.JJID.2021.020
132. Portacci A, Diaferia F, Santomasi C, Dragonieri S, Boniello E, Di Serio F, et al. Galectin-3 as Prognostic Biomarker in Patients With Covid-19 Acute Respiratory Failure. Respir Med (2021) 187:106556. doi: 10.1016/j.rmed.2021.106556
133. Li G, Quan Y, Che F, Wang L. B7-H3 in Tumors: Friend or Foe for Tumor Immunity? Cancer Chemother Pharmacol (2018) 81(2):245–53. doi: 10.1007/s00280-017-3508-1
134. Hofmeyer KA, Ray A, Zang X. The Contrasting Role of B7-H3. Proc Natl Acad Sci (2008) 105(30):10277–8. doi: 10.1073/pnas.0805458105
135. Ni L, Dong C. New B7 Family Checkpoints in Human Cancers. Mol Cancer Ther (2017) 16(7):1203–11. doi: 10.1158/1535-7163.MCT-16-0761
136. Nie X, Qian L, Sun R, Huang B, Dong X, Xiao Q, et al. Multi-Organ Proteomic Landscape of Covid-19 Autopsies. Cell (2021) 184(3):775–91. e14. doi: 10.1016/j.cell.2021.01.004
137. Yan Z. Increased Expression of Cd155 and Cd112 on Monocytes in Septic Patients (Inc6p.327). J Immunol (2015) 194(1 Supplement):192.29.
138. Pende D, Castriconi R, Romagnani P, Spaggiari GM, Marcenaro S, Dondero A, et al. Expression of the Dnam-1 Ligands, Nectin-2 (Cd112) and Poliovirus Receptor (Cd155), on Dendritic Cells: Relevance for Natural Killer-Dendritic Cell Interaction. Blood (2006) 107(5):2030–6. doi: 10.1182/blood-2005-07-2696
139. Hua B, Yang M, Xue J, Dong C, Mao Y-T, Li O, et al. Abstract 2451: A Novel Single Domain Antibody Targeting Tigit for Cancer Use in Combination Therapies. Cancer Res (2021) 81(13 Supplement):2451. doi: 10.1158/1538-7445.AM2021-2451
140. Sanchez-Correa B, Valhondo I, Hassouneh F, Lopez-Sejas N, Pera A, Bergua JM, et al. Dnam-1 and the Tigit/Pvrig/Tactile Axis: Novel Immune Checkpoints for Natural Killer Cell-Based Cancer Immunotherapy. Cancers (2019) 11(6):877. doi: 10.3390/cancers11060877
141. Nagumo Y, Iguchi-Manaka A, Yamashita-Kanemaru Y, Abe F, Bernhardt G, Shibuya A, et al. Increased Cd112 Expression in Methylcholanthrene-Induced Tumors in Cd155-Deficient Mice. PloS One (2014) 9(11):e112415. doi: 10.1371/journal.pone.0112415
142. Sfera A, Osorio C, Jafri N, Diaz EL, Campo Maldonado JE. Intoxication With Endogenous Angiotensin Ii: A Covid-19 Hypothesis. Front Immunol (2020) 11:1472. doi: 10.3389/fimmu.2020.01472
143. Wilk AJ, Lee MJ, Wei B, Parks B, Pi R, Martínez-Colón GJ, et al. Multi-Omic Profiling Reveals Widespread Dysregulation of Innate Immunity and Hematopoiesis in Covid-19. J Exp Med (2021) 218(8):e20210582. doi: 10.1101/2020.12.18.423363
144. Wang J, Manick B, Renelt M, Gerassenkov T, Bi M, Kalabokis V, et al. Lsectin (Clec4g) Inhibits T-Cell Activation and Proliferation in the Presence of Tcr and Cd28 Signaling. J Immunol (2021) 206(1 Supplement):25.06. doi: 10.1038/s41423-020-0361-7
145. Lu Q, Liu J, Zhao S, Castro MFG, Laurent-Rolle M, Dong J, et al. SARS-CoV-2 Exacerbates Proinflammatory Responses in Myeloid Cells Through C-Type Lectin Receptors and Tweety Family Member 2. Immunity (2021) 54(6):1304–19.e9. doi: 10.1016/j.immuni.2021.05.006
146. Rokni M, Hamblin MR, Rezaei N. Cytokines and Covid-19: Friends or Foes? Hum Vaccines Immunother (2020) 16(10):2363–5. doi: 10.1080/21645515.2020.1799669
147. Ceschi A, Noseda R, Palin K, Verhamme K. Immune Checkpoint Inhibitor-Related Cytokine Release Syndrome: Analysis of Who Global Pharmacovigilance Database. Front Pharmacol (2020) 11:557. doi: 10.3389/fphar.2020.00557
148. Rotz SJ, Leino D, Szabo S, Mangino JL, Turpin BK, Pressey JG. Severe Cytokine Release Syndrome in a Patient Receiving Pd-1-Directed Therapy. Pediatr Blood Cancer (2017) 64(12):e26642. doi: 10.1002/pbc.26642
149. Luo J, Rizvi H, Egger JV, Preeshagul IR, Wolchok JD, Hellmann MD. Impact of Pd-1 Blockade on Severity of Covid-19 in Patients With Lung Cancers. Cancer Discov (2020) 10(8):1121–8. doi: 10.1158/2159-8290.Cd-20-0596
150. Gonzalez-Cao M, Basa MA, Puertolas T, Muñoz E, Manzano JL, Carrera C, et al. Cancer Immunotherapy Does Not Increase the Risk of Death by Covid-19 in Melanoma Patients. medRxiv (2020):2020.05.19.20106971. doi: 10.1101/2020.05.19.20106971
151. Zheng Y, Huang Z, Yin G, Zhang X, Ye W, Hu Z, et al. Study of the Lymphocyte Change Between Covid-19 and Non-Covid-19 Pneumonia Cases Suggesting Other Factors Besides Uncontrolled Inflammation Contributed to Multi-Organ Injury. medRxiv (2020):2020.02.19.20024885. doi: 10.1101/2020.02.19.20024885
152. Yatim N, Boussier J, Tetu P, Smith N, Bruel T, Charbit B, et al. Immune Checkpoint Inhibitors Increase T Cell Immunity During SARS-CoV-2 Infection. Sci Adv (2021) 7(34):eabg4081. doi: 10.1126/sciadv.abg4081
153. Loretelli C, Abdelsalam A, D’Addio F, Ben Nasr M, Assi E, Usuelli V, et al. Pd-1 Blockade Counteracts Post-Covid-19 Immune Abnormalities and Stimulates the Anti-SARS-CoV-2 Immune Response. JCI Insight (2021) 6(24):e146701. doi: 10.1172/jci.insight.146701
154. He L, Ding Y, Zhang Q, Che X, He Y, Shen H, et al. Expression of Elevated Levels of Pro-Inflammatory Cytokines in SARS-CoV-Infected Ace2+ Cells in Sars Patients: Relation to the Acute Lung Injury and Pathogenesis of Sars. J Pathol (2006) 210(3):288–97. doi: 10.1002/path.2067
155. Al-helfawi MA. Potential Approach for Fighting Against Corona Virus Disease. Am Acad Sci Res J Engineering Technology Sci (2020) 66(1):127–44. doi: 10.1016/j.immuni.2021.05.006
Keywords: SARS-CoV-2, COVID-19, inhibitory immune checkpoints, ligands, prognostic biomarker
Citation: Al-Mterin MA, Alsalman A and Elkord E (2022) Inhibitory Immune Checkpoint Receptors and Ligands as Prognostic Biomarkers in COVID-19 Patients. Front. Immunol. 13:870283. doi: 10.3389/fimmu.2022.870283
Received: 06 February 2022; Accepted: 07 March 2022;
Published: 31 March 2022.
Edited by:
Milos Jesenak, Comenius University, SlovakiaReviewed by:
Vadim V. Sumbayev, University of Kent, United KingdomYangqiu Li, Jinan University, China
Nargess Arandi, Shiraz University of Medical Sciences, Iran
Copyright © 2022 Al-Mterin, Alsalman and Elkord. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eyad Elkord, ZS5lbGtvcmRAdW5pendhLmVkdS5vbQ==; ZS5lbGtvcmRAc2FsZm9yZC5hYy51aw==
 Mohammad A. Al-Mterin
Mohammad A. Al-Mterin Alhasan Alsalman
Alhasan Alsalman Eyad Elkord
Eyad Elkord