
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 06 April 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.856457
This article is part of the Research Topic Insect immunity and its interactions with microorganisms and parasitoids View all 31 articles
 Ulrich Aymard Ekomi Moure1,2†
Ulrich Aymard Ekomi Moure1,2† Tingshan Tan1†
Tingshan Tan1† Lin Sha1
Lin Sha1 Xiaoqin Lu1
Xiaoqin Lu1 Zhi Shao1
Zhi Shao1 Guang Yang1*
Guang Yang1* Yi Wang1,3*
Yi Wang1,3* Hongjuan Cui2,4
Hongjuan Cui2,4Insects are by far the most abundant and diverse living organisms on earth and are frequently prone to microbial attacks. In other to counteract and overcome microbial invasions, insects have in an evolutionary way conserved and developed immune defense mechanisms such as Toll, immune deficiency (Imd), and JAK/STAT signaling pathways leading to the expression of antimicrobial peptides. These pathways have accessory immune effector mechanisms, such as phagocytosis, encapsulation, melanization, nodulation, RNA interference (RNAi), lysis, autophagy, and apoptosis. However, pathogens evolved strategies that circumvent host immune response following infections, which may have helped insects further sophisticate their immune response mechanisms. The involvement of ncRNAs in insect immunity is undeniable, and several excellent studies or reviews have investigated and described their roles in various insects. However, the functional analyses of ncRNAs in insects upon pathogen attacks are not exhaustive as novel ncRNAs are being increasingly discovered in those organisms. This article gives an overview of the main insect signaling pathways and effector mechanisms activated by pathogen invaders and summarizes the latest findings of the immune modulation role of both insect- and pathogen-encoded ncRNAs, especially miRNAs and lncRNAs during insect–pathogen crosstalk.
Insects are often subjected to pathogen (bacteria, fungi, viruses, etc.) attacks, and their survival indicates advanced defense mechanisms (1). These organisms of interest endue physical barriers preventing intruders from entering their body cavity (hemocoel) (2, 3). These intruders generally reach insect body parts through the degradation of the cuticle by enzymatic digestion or by ingestion (midgut) (4), which are followed by the induction of the host’s immune defense mechanisms via the binding of their pathogen-associated molecular patterns (PAMPs) (5). Activation and coordination of insects’ innate immune defenses rely on evolutionary and highly conserved immune factors. This immunity is classified into humoral and cellular reactions (6). Humoral responses include the production of antimicrobial peptides (AMPs) (7). Studies showed that the Toll, immune deficiency (Imd), and the Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathways are the most characterized in insects, and their activation results in the production of effector molecules such as AMPs with the potential to kill insect invaders (8–12) (Section 2; Figure 1). However, it was reported that these signaling pathways could be encountered by pathogens and lead to their replication and proliferation (13–15). Cellular reactions, meanwhile, rely on insect hemocytes (primary immune cell defenses) involved in phagocytosis, nodulation, encapsulation, autophagy, apoptosis, and so on (4, 16, 17). Given all this arsenal of immune defense described above, insects lack an adaptive immune response, unlike mammals, and rely only on innate immunity (18, 19). We may speculate that insect immune defense against pathogens lacks a memory immunity that allows rapid and robust responses to neutralize and kill pathogens quickly. Nonetheless, we are far from reaching the exhaustive portrayal of how these organisms that represent the Earth’s most abundant and diverse living organisms defend themselves.
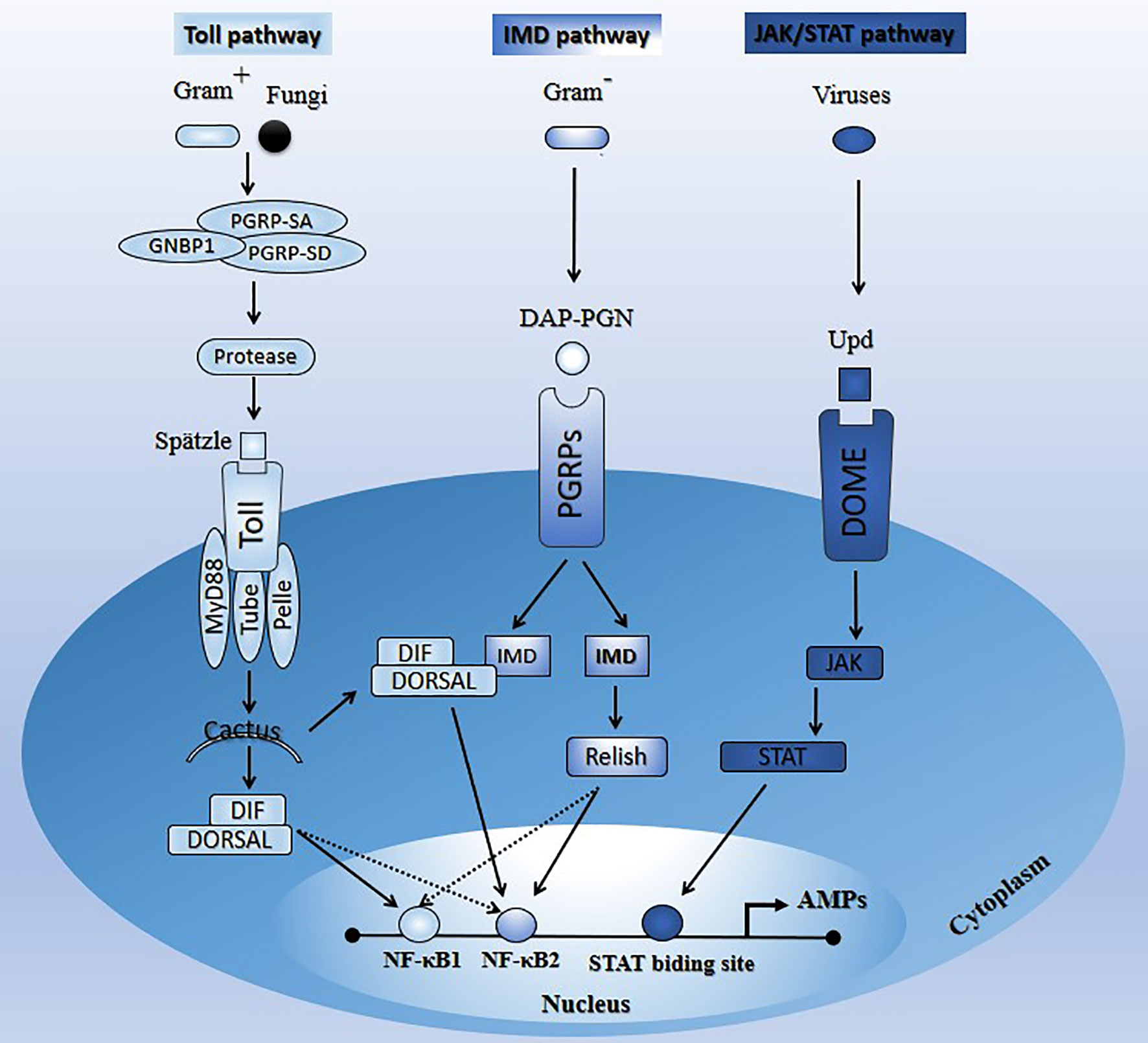
Figure 1 Schematic illustration of the main signaling pathways (Toll, Imd, and JAK/STAT) in insects upon microbial attacks. See Section 2 for the description of the paths.
Several factors such as insect gut microbiota, nutritional stress, and noncoding RNAs (ncRNAs) play an immune-modulatory role during insect–pathogen crosstalk (20–23). Noncoding RNAs (ncRNAs), also termed nonprotein-coding RNAs are RNA molecules that do not have the potentiality to encode proteins (24). In addition to ribosomal RNA (rRNA) and transfer RNA (tRNA), ncRNAs can be divided into two groups, small ncRNA (sncRNA) and long ncRNA (lncRNA), based on their size. sncRNA can be further divided into miRNA, piRNA, siRNA, etc. lncRNA can be subdivided into sense, antisense, intronic, intergenic, cis-, and trans-RNA based on their biogenesis and mechanism of action (25). This review concentrates on the involvement of miRNAs and lncRNAs in insect immune regulation upon pathogen invasions. miRNAs and lncRNAs have varied regulatory roles at the epigenetic, transcriptional, or posttranscriptional levels and participate in almost all biological processes, including immunity and host–microbial interactions. Moreover, lncRNAs and miRNAs showed differential expression levels upon insect pathogenic infections or pesticide exposure, further proving their involvement in modulating insect immune responses. For instance, infection of Galleria mellonella (G. mellonella) with both uropathogenic Escherichia coli (UPEC) and asymptomatic bacteriuria (ABU) caused significant changes in the abundance of miRNAs in the larvae. It highlighted the differential expression of 147 conserved miRNAs and 95 novel miRNA candidates. In addition, by utilizing next-generation sequencing (NGS), Dubey et al. (26) identified 126 miRNAs from Aedes aegypti (A. aegypti) cell line Aag2, and 13 of them were regulated during chikungunya virus (CHIKV) infection. Moreover and interestingly, the review written by Awais et al. (27) summarizes the diversity and multitude of differentially regulated miRNAs in Bombyx mori (B. mori) upon several viral attacks, such as B. mori cypovirus (BmCPV), B. mori nucleopolyhedrovirus (BmNPV), and B. mori Autographa californica multiple nucleopolyhedrovirus (AcMNPV). In parallel, the Drosophila melanogaster (D. melanogaster) lncRNA CR44404 (renamed lincRNA-IBIN) was highly induced (1300-fold) by Gram-positive or Gram-negative bacteria in Drosophila adults or a parasitoid wasp Leptopilina boulardi in Drosophila larvae (28). Moreover, lncRNA-155 targeted the protein tyrosine phosphatase 1B to modulate innate immunity against influenza A virus (IAV) infection in mice (29). In contrast, the lncRNA-1317 supported host antiviral defense mechanism during dengue virus serotype 2 (DENV-2) infection in A. aegypti (30). Although the fact that the expression levels of insect-encoded miRNAs and lncRNAs are altered during microbial attacks, little is known about the different mechanisms employed by these ncRNAs to regulate insects' immune defenses in response to those invaders.
This review focuses on two types of ncRNAs, lncRNAs and miRNAs. It summarizes the main immune signaling pathways and immune effector mechanisms engaged upon microbial invasions and reports the latest findings supporting the involvement of insect- and pathogen-derived miRNAs and lncRNAs in the immunological insights related to insect–pathogen interactions. It lastly provides perspectives on gaps and where future investigation must be oriented.
This insects’ pathway is responsive to Gram-positive bacteria and fungi and largely controls the expression of AMPs (28–30). The Toll pathway is a NF-κB-associated signaling pathway (8, 9). Stimulation of insects’ Toll pathway by appropriate pathogens induces proteolytic cascades activated by secreted recognition molecules, such as peptidoglycan receptor proteins (PGRP-SA, PGRP-SD, GNBP1) (11, 31–34). These recognition molecules are highly evolutionarily conserved from insects to mammals. For instance, Drosophila, mosquito, and mammals have families of 13, 7, and 4 PGRP genes, respectively (35). In Drosophila where the molecular components of the Toll signaling pathway and their functions in immune response against pathogens are well characterized (36–38), the receptor membrane Toll is activated and dimerized by the mature proteolytic product Spätzle (39–42), which subsequently causes the recruitment of three intracellular death domain-containing proteins, MyD88, Tube, and Pelle (43–45). The IκB homolog Cactus is then phosphorylated and degraded by the proteasome, leading to the release of members of the nuclear factor NF-κB family (Dif or dorsal) to translocate to the nucleus (46–48), and activate genes encoding potent antifungal and antibacterial peptides (AMPs), such as Drosomycin (Dr) (11, 38, 49). The Toll-dorsal pathway in Drosophila and the interleukin-1 receptor (IL-1R)-NF-kB pathway in mammals are homologous signal transduction pathways that mediate several different biological responses, including immunity (50).
This pathway is well characterized in Dipterans such as Drosophila; however, in Hemipterans like aphids and assassin bugs, several components of that pathway are lacking. Imd signaling components appear to be absent in Rhodnius prolixus (Reduviidae) and Acyrthosiphon pisum (Aphididae) (51–53). However, some study reported that the Toll and Imd pathways are present in the hemipteran lineages like stinkburg Plautia stali, but their functionality is blurred (54). The Imd pathway is mainly responsive to Gram-negative bacteria and controls the synthesis of several AMPs (55, 56).
During the infection, insect Imd transmembrane receptor proteins (PGRPs) recognize pathogen diaminopimelic acid-peptidoglycan (DAP-PGN). This recognition induces a cascade reaction resulting in Relish (NF-κB family member) activation (cytoplasm) and translocation (nucleus) for AMP production (Diptericin) (38, 57). Like the Toll pathway, the Imd pathway is a NF-κB-associated signaling pathway (8, 9). Although the Imd and Toll pathways have independent functions and mediate the specificity of innate immune responses towards different microorganisms, some AMP genes can be activated by both pathways. Interestingly, Tanji et al. (58) showed that a synergic interaction occurs between the Toll and Imd signaling pathways, downstream of the latter. Specifically, upon signal stimulation, Dif, dorsal, and relish factors can independently bind to NF-κB1 or NF-κB2, and relish interacts after that with Dif or dorsal to synergistically promote transcription. Alternatively, signal stimulation may enhance the formation of relish/Dif or relish/dorsal heterodimers and bind preferentially to NF-κB2 to regulate transcription synergistically with interaction with NF-κB1 (58).
The JAK/STAT pathway was originally identified as a cytokine signaling pathway in mammals (59–62). This pathway provides an antiviral protection (27, 28, 63). Unlike the Toll and Imd pathways, little is known about the transcriptional cascade induced by the JAK/STAT pathway (14, 64). Souza-Neto et al. (65) showed that the JAK-STAT pathway is part of the A. aegypti mosquito’s antidengue defense and may act independently of the Toll pathway and the RNAi-mediated antiviral defenses.
The comparative genomic analysis of A. aegypti, Anopheles gambiae, and D. melanogaster genome sequence revealed orthologs for the core JAK/STAT pathway components (dome, Hop, and STAT) (66). Activation of this pathway in Drosophila starts when the virus-induced extracellular cytokine unpaired (Upd) binds to the cellular receptor dome. The latter recruits STAT, which then dimerizes and translocates to the nucleus and activates the transcription of AMP genes (nitric oxide synthase) after binding to the STAT-binding site (12, 65). Protein inhibitor of activated STAT (PIAS) and suppressor of cytokine signaling (SOCS), are two negative regulators of the JAK-STAT pathway in D. melanogaster (67). Orthologs of these two regulators (SUMO and SOCS) have been identified, confirming the proximity and evolutionary conservation of this pathway between mammals and insects (65).
Pathogen neutralization and death are accomplished via insect immune effector mechanisms, known to be interconnected and to work synergistically (Figure 2). The primary immune cells are the hemocytes. Hemocytes are found in circulation (circulating hemocytes) and attached to tissues (sessile hemocytes), where they phagocytose, encapsulate and nodulate pathogens, and produce humoral immune factors. The fat body, the midgut, the salivary glands, and other tissues produce numerous humoral immune factors with, among other things, lytic and melanizing activity (Figure 3).
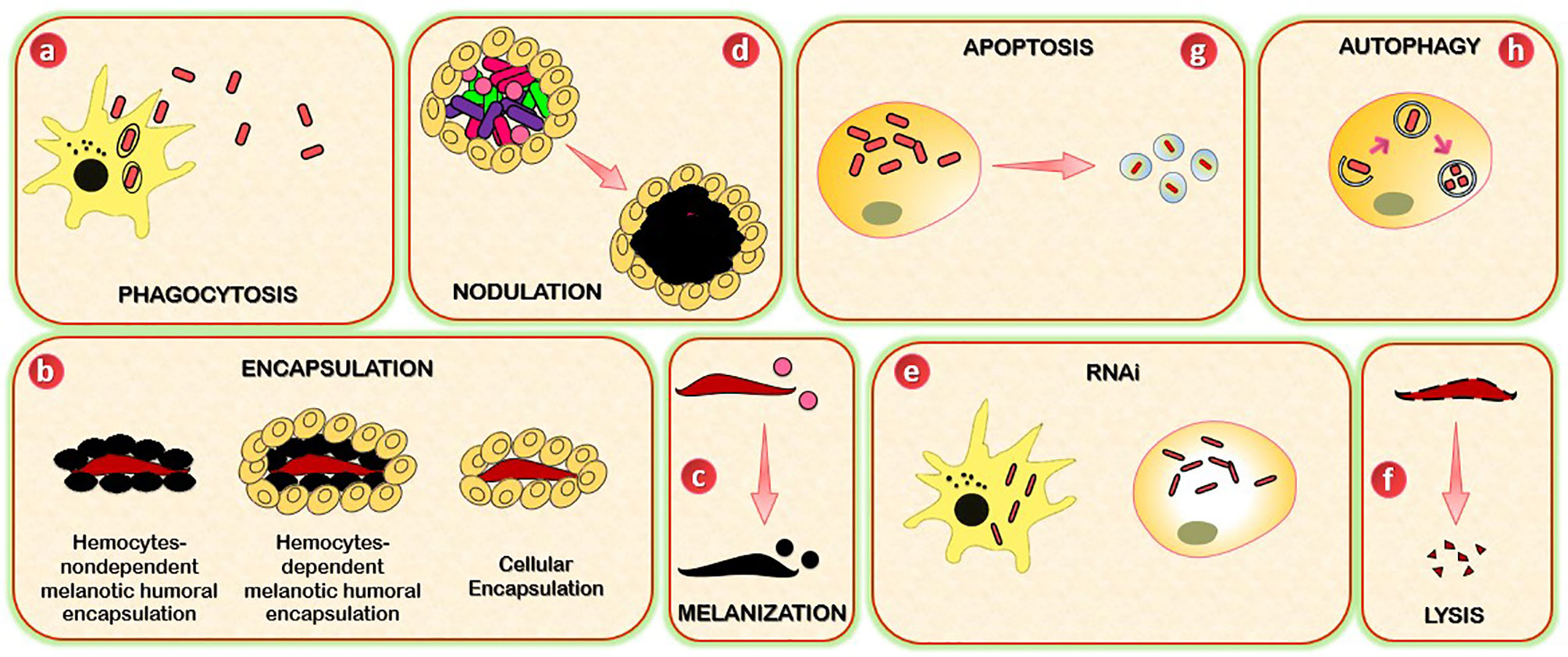
Figure 2 Schematic representation of insects’ immune effector mechanisms. (A) Insects use phagocytosis to neutralize and kill small pathogens. This process is mediated by phagocytes (hemocytes or granulocytes). (B) Encapsulation (cellular and melanotic) is a defense mechanism that insects used when pathogens are too large to be phagocytosed. Cellular encapsulation occurs without melanization, whereas melanotic humoral encapsulation is dependent on PO activity and can occur with or without the assistance of hemocytes. (C) Melanization is a process based on the conversion of PPO to PO, which leads to the formation of the melanotic capsule (melanotic enzymes) which mediates the killing of the foreign agent. (D) Nodulation is a process by which immune cells (granulocytes) adhere to each other to create layers that surround many bacteria or fungal spores. The granulocytes release their contents, which trap the bacteria in a flocculent material. This step is often followed by melanization. (E) RNAi mechanism is based on the ribonuclease cleavage of viral dsRNA and is specifically used against viruses, and can be mediated by immune circulating cells (hemocytes, macrophages). (F) Lysis is a mechanism through which insects kill pathogens by disrupting their cellular membrane. The process involves the participation of peptides and proteins with antimicrobial activity, including lysosomes. (G) Autophagy provides protection to insects against pathogens through the degradation of cytoplasmic material (bacteria or viruses). (H) Apoptosis is a programmed cell death mediated by caspases. The killing of pathogens includes the formation of apoptotic bodies.
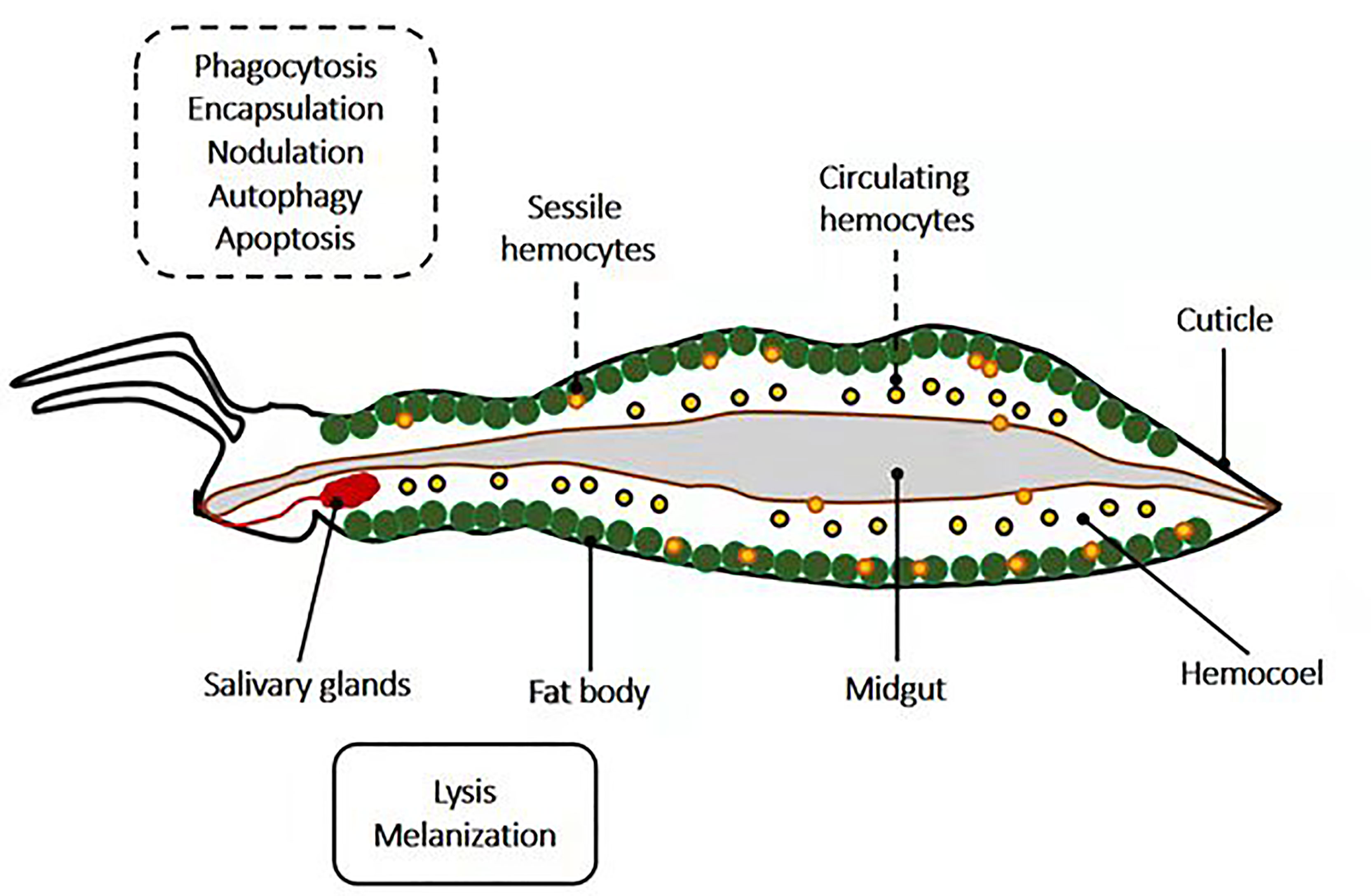
Figure 3 Schematic representation of the localization of insects’ immune effector mechanisms. Mechanisms such as phagocytosis, encapsulation, nodulation, autophagy, and apoptosis occur in insect hemocytes (sessile and circulating), which are located in the hemocoel. Meanwhile, insect parts such as salivary glands, the fat body, and midgut are potent sites for lysis and melanization.
Small pathogens that become melanized, such as bacteria, are often phagocytosed (68, 69). Phagocytosis is an evolutionarily conserved cellular immune process used by vertebrates and invertebrate animals to neutralize and kill small pathogens. Due to its incredible speed, it takes seconds to hydrolyze foreign bodies following their internalization (69–71), and hundreds of bacteria can be phagocytosed at any time (72). The phagocytes, which can be circulating and sessile hemocytes (73) or granulocytes in the cases of Lepidoptera, Hemiptera, and mosquitoes, and the plasmatocytes of fruit flies, identify the foreign body. The latter is then internalized into a membrane-delimited phagosome, which then fuses with a lysosome for enzymatic hydrolysis-mediated degradation (68, 69, 73, 74).
Although the intracellular patterns regulating phagocytosis remain poorly understood, phagocytosis initiates by binding of a cell-surface PRR or a humoral PRR on a PAMP. PRRs that have been empirically shown to be involved in phagocytosis include thioester-containing proteins, Nimrod proteins, DSCAM, β-integrins, and PGRPs (75–79). Different PRRs have different specificities. For example, D. melanogaster PGRP-LC mediates the phagocytosis of Escherichia coli (E. coli), but not Staphylococcus aureus (S. aureus) (79), and NimC1 mediates the phagocytosis of S. aureus and, to a lesser extent, E. coli (76).
Encapsulation is a cellular immune response that insects use in response to pathogens that are too large to be phagocytosed. Two types of encapsulation are described in insects: cellular encapsulation, mainly described in Lepidoptera, and melanotic humoral encapsulation, more typical in some dipterans like Drosophila (80). Cellular encapsulation can occur without any sign of melanization. In contrast, melanotic encapsulation relies on PO activity and can occur with or without the assistance of hemocytes. In cellular encapsulation, granulocytes and plasmatocytes of Lepidoptera, plasmatocytes, and lamellocytes of Drosophila are the main hemocyte types involved in encapsulation (81, 82). In Lepidoptera, an inner layer of granulocytes and an outer one of plasmatocytes surrounded encapsulated objects (83, 84). Insects such as dipteran and lepidopteran larvae commonly employed this response to infection with the eggs of parasitoid wasps. In Lepidoptera, encapsulation starts when granulocytes attach to form a layer of cells, in a process dependent on the binding of integrins to specific sites defined by an Arg-Gly-Asp (RGD) sequence (85). This layer of cells surrounding the pathogen is in turn covered by several layers of plasmatocytes, which are then circled by the adhesion of additional granulocytes. A similar process occurs in Drosophila, except that the cells involved are plasmatocytes and lamellocytes (86). Depending on the pathogen and the insect, the capsule may become melanized.
Melanization is an enzymatic process used by insects in several mechanisms, including immunity. The process involves coordinating pattern recognition receptors, serine proteases, serine protease inhibitors, and melanin production enzymes. When PRRs (β-1,3 glucan recognition proteins, C-type lectins, and Gram-negative binding proteins) identify PAMPs (87–89), the serine protease cascade is induced and leads to the conversion of the pro-phenoloxidase (PPO) to phenoloxidase (PO) (90, 91), which finally culminates in the formation of melanotic capsules. The killing of the foreign agent is mediated by two factors: the pathogen’s surrounding by the proteinaceous capsule and the oxidative stress damage or starvation (92–94). In addition, melanization also assists in the clearing of dead or dying pathogens (95, 96). Many of the enzymes and PRRs that drive the melanization response are produced by hemocytes, with oenocytoids (crystal cells) being the primary producers of PPO (68, 97). Melanization is another essential facet of the mosquito immune defense against fungi infection. It plays a crucial role in encapsulating and retarding invasive Bauveria bassiana (B. bassiana) growth and dissemination in mosquitoes (98).
Although the molecular patterns underlining this immune process are still poorly understood, nodulation relies on eicosanoid-based signaling and the extracellular matrix-like protein, Noduler (99, 100). This process starts with the adherence of granulocytes to each other to create layers that surround many bacteria or fungal spores. The granulocytes release their contents, which trap the bacteria in a flocculent material. Plasmatocytes then aggregate around the surface of the nodule. This step is often followed by melanization (101, 102).
RNA interference (RNAi) is an RNA-based mechanism for gene silencing. RNAi could be used to manage insect pests (103, 104). Nevertheless, one of the natural functions of RNAi is to protect the organism from viral infection (105–108).
The small interfering RNA (siRNA) pathway is the primary RNAi path that participates in the insect response against the virus. The process involves the ribonuclease cleavage of viral double-stranded RNA (dsRNA) by dicer-2 (Dcr2), forming a complex with its cofactor R2D2. This cleavage culminates in the production of viral-derived siRNAs (~21 nucleotides), and these siRNAs are loaded into pre-RNA induced silencing complexes (RISC) that include Argonaute-2 (Ago2). The siRNA in a RISC complex is unwound, one strand is discarded, and the other binds complementary viral RNA, which triggers its destruction by Ago2. The binding of viral RNA by Dcr2 also activates the transcription of Vago, which is a cysteine-rich polypeptide that negatively controls virus replication (109). In mosquitoes, the transcriptional activation of Vago is mediated by an NF-κB transcription factor associated with the Imd pathway (Rel2), and in turn, Vago activates the JAK/STAT pathway (110). The PIWI-associated RNA pathway (piRNA) also acts in the RNAi-mediated antiviral defense of mosquitoes but in a manner independent of dicer (105, 106).
Pathogen killing via lysis refers to death due to the immune-based disruption of the cellular membrane. Some of the earliest studied insect factors that induce pathogen death via lysis are AMPs (~2 and 20 kDa). These peptides and proteins were initially identified for their antimicrobial activity in vitro (111–113). Most AMPs can be grouped into four categories (114–116). The defensin and defensin-like peptides (defensin, drosomycin, holitricin, sapecin, and others) are rich in cysteines and contain cysteine disulfide bonds that have activity against Gram-positive bacteria. Still, others combat Gram-negative bacteria and fungi. The cecropin and cecropin-like peptides (moricins) are α-helical peptides that have activity against Gram-positive and Gram-negative bacteria and fungi. The attacins and gloverins are glycine-rich peptides and are mainly active against Gram-positive bacteria, whereas the gloverins, have activity against Gram-positive bacteria, Gram-negative bacteria, or fungi. The lebocins (lebocin, drosocin, metchnikowin, and others) are proline-rich peptides and are active against Gram-positive bacteria, Gram-negative bacteria, and some fungi. The production of antimicrobial peptides is governed by immune signaling pathways, as seen above with Toll, Imd, and JAK/STAT pathways.
Lysozymes are another family of proteins with lytic activity. Lysozymes hydrolyze the β-1,4-glycosidic linkage between N-acetylmuramic acid and N-acetylglucosamine of the peptidoglycan present in bacteria’s cell wall. Lysozymes are usually present in low, constitutive levels and are transcriptionally upregulated following infection. Although lysozymes are classically active in the lytic antibacterial response (117, 118), some also have antiplasmodium and antifungal activity (119, 120) and can activate the PO-based melanization pathway (121, 122).
Autophagy is an evolutionarily conserved mechanism involved in the degradation of cytoplasmic material through the lysosomal degradation pathway. It plays crucial roles in cellular homeostasis, adaptive response to nutrient deprivation, energy homeostasis, and survival during starvation (123, 124). In immunity, autophagy provides insect protective immune responses against both viral and bacterial attacks. Evidence showed that the Toll pathway induces autophagy during an antiviral response. It is excluded in the activation of autophagy during an antibacterial response and therefore needs further investigation (125). In Drosophila, the activation of autophagy by vesicular stomatitis virus (VSV) is mediated by Toll-7 recognition of VSV-G, a glycoprotein on the virion surface. This PAMP-PRR interaction leads to autophagy initiation by acting on the (PI3K)-Akt-signaling pathway (126, 127). The connection of a Toll receptor to antiviral autophagy in flies suggests an evolutionarily conserved requirement for PRR in triggering autophagy between insects and mammals. Autophagy may, in some cases, promote virus infection as shown for the Sindbis virus, where the activation of the (PI3K)-Akt-signaling pathway enhances the infection of insect cells (128). Lysteria monocytogenes invades and replicates in hemocytes and is recognized by PGRP-LE (a PRR which senses a DAP-type PGN), eliciting an autophagic response that prevents intracellular growth of the bacterium and promotes host survival (125).
Apoptosis is a form of programmed cell death. The caspase, Dronc, and the adaptor protein, Ark, form a complex at the molecular level. Dronc activates effector caspases such as Drice and Dcp1, and these caspases cleave proteins in a manner that eventually leads to programmed cell death (129). In Lepidoptera, apoptosis is involved in the response against baculoviruses (130). In mosquitoes, this process appears to be involved in the response against West Nile Virus and Sindbis virus (131, 132), and in the fruit fly, apoptosis protects against Drosophila C virus in a manner that is dependent on the phagocytosis of virus-infected apoptotic cells (133).
Using high-throughput sequencing techniques and advanced bioinformatics tools, researchers have done great work in discovering and identifying novel insect ncRNAs and their regulated transcripts, respectively (134, 135). The involvement of ncRNAs in insects’ immunity is evidenced, and changes in expression levels of these insect elements generally occur to be relatively altered upon microbial attacks. Below, we go beyond the altered differential expression of these insect elements and highlight their immune targets and influence on insect immune responses to pathogen invasions. Inversely, we will also consider the regulation of insect immunity by pathogen-encoded ncRNAs. Tables 1 and 2 enclose both conserved and novel insect- and pathogen-derived miRNAs and lncRNAs and their targets in insect-pathogen crosstalk.
miRNAs play a crucial role in host–pathogen interactions. The dynamic miRNA–mRNA is essential for immune response to pathogen attacks, including regulating critical insect signaling pathways to promote or inhibit innate immune responses and maintain homeostasis. For instance, differentially expressed miRNAs and mRNAs represented extensively dynamic changes during Drosophila with Micrococcus luteus (M. luteus) infection and enriched diverse signaling pathways, including Toll and Imd as other signaling pathways (139). Figure 4 summarizes the immune regulatory role of insect miRNAs when invaded by pathogens.
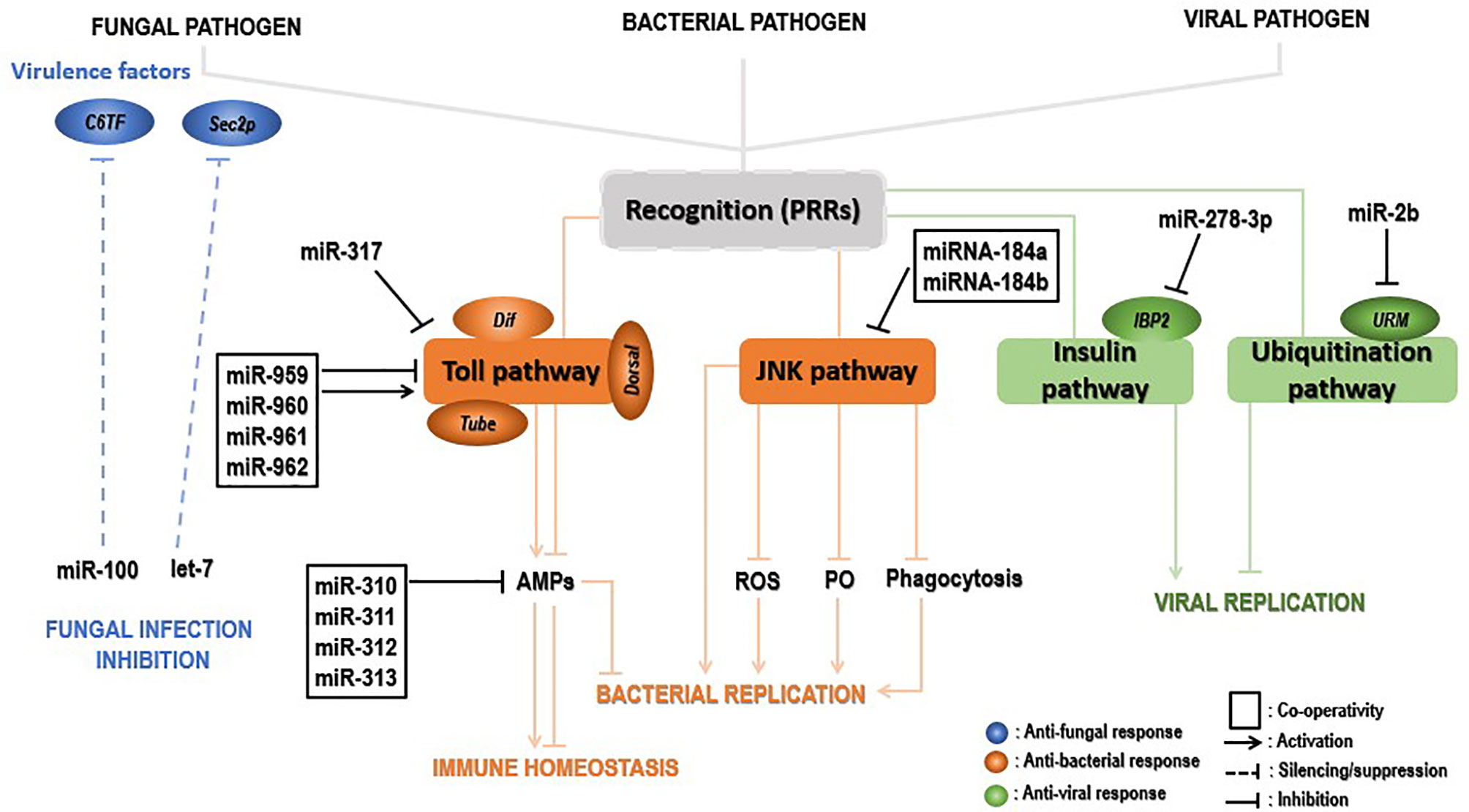
Figure 4 Insect immune defense modulation by miRNAs upon pathogenic invaders. Upon fungal invasion, insect miRNAs can silence fungal virulence genes, such as C6TF and Sec2p, and inhibits its replication (blue). Upon bacterial invasion, insect miRNAs individually or synergistically regulate the Toll pathway key components (Tube, Dorsal, Dif, etc.) positively, leading to activation of AMP gene effectors and inhibition of bacterial replication. Those elements can negatively modulate the same signaling at the late stage of the infection, decreasing the AMP gene expression for insect immune homeostasis. On the other hand, insect miRNAs can promote bacterial replication by inhibiting the JNK signaling pathway and indirectly those under its control (ROS, PO, phagocytosis) (in the pea aphid Acyrthosiphon pisum, for instance) (brown). Lastly, IBP2 is significantly upregulated upon viral invasion of insects. The latter’s expression is drastically repressed by insect miRNAs, leading to viral replication. In contrast, insect miRNAs inhibit viral replication by downregulating components (URM) of the ubiquitinylation process (green).
The expression levels of miRNAs in the insect–pathogen crosstalk used to be altered. Detection and characterization of miRNA cellular targets would therefore provide more insights to understand the immune modulatory role of these miRNAs. Insect immune mechanisms may deal directly with the pathogens by eliminating them from the host organism or disarm them by abolishing the synthesis of toxins and virulence factors that promote the invasion and destructive action of the invader within the host. Insect-encoded miRNAs seem to preferentially deal with fungal pathogens by disarming them through the suppression of invaders’ virulence factors. This silencing interferes with some translation factors associated with 5’-cap to 3’-tail structures of mRNAs (140). Recently, a study revealed mosquitoes increase the expression levels of both let-7 and miR-100 miRNAs when the fungus B. bassiana penetrates inside their hemocoel. Both miRNAs translocate into the fungal hyphae to silence the virulence-related genes sec2p and C6TF, encoding a Rab guanine nucleotide exchange factor and Zn(II)2Cys6 transcription factors, respectively (23). Suggestions state that extracellular vesicles (EVs) may transport those insect miRNAs to the fungal pathogen. To our knowledge, this is the first report describing the cross-kingdom miRNA transfer from arthropod hosts to their pathogen cells. Globally, the above evidence uncovers an insect defense mechanism whereby infection leads to the upregulation of specific miRNAs that are transferred to the invading fungus and suppress fungal virulence genes, in this way, bestowing antifungal protection. This study opens avenues to improve fungal virulence by expression of host miRNA sponges.
Another crucial role played by miRNAs is to restore insects’ immune homeostasis by negatively regulating immune signaling pathways. This role is achieved by an individual or synergistic action of miRNAs, which repress expression levels of hosts’ immune signaling key components. Dl and Toll are critical transcription and transmembrane factors, respectively, whereas the tube is an indispensable effector molecule in the Toll pathway. The synergic regulatory mechanism of miR-959-962 cluster in the Drosophila immune response to M. luteus infection could negatively regulate the Toll pathway in combination via directly targeting the 3’UTR of the tube, dl, or Toll mRNAs, leading to a reduced survival rate of flies by inhibiting the expression of AMPs at the late stage of the infection (137). The same study further demonstrated that miR-960 might modulate antibacterial defense only at the late 12-h stage upon infection. At the same time, miR-959 may constantly repress the Dr expression at 6 and 12 h, respectively. Meanwhile, miR-961 may contribute more than miR-962 to repress antibacterial defense. Furthermore, using in silico screening strategy combined with the Gal80ts-Gal4 driver system, miR-958 was identified as a candidate miRNA that could potentially regulate the Toll pathway signaling, both in vitro and in vivo, by negatively targeting Dif and Toll. The latter is specifically and significantly inhibited by miR-958 at its site 3, as the Toll 3’UTR harbors four miR-958-binding sites (141). Similarly, Drosophila miR-317 negatively regulated Drosophila Toll signaling response via suppressing only the Dif-Rc of Dif four isoforms (142). However, the same authors found in a previous study that miR-317 regulates the Drosophila Toll pathway via targeting the three other Dif isoforms (Dif-Ra/b/d) (141). A set of studies showed the involvement of miR-317 in Drosophila reproductive responses and larval ovary morphogenesis (143–145). Remarkably, flies transiently overexpressing miR-317 have poor survival. In contrast, the knockout miR-317 flies (miR-317 KO/+) have better survival than the control group, respectively, during Gram-positive bacterial infection (142), implying a new insight of miRNA involved in the crosstalk between Drosophila immune and survival. Moreover, four members of the Drosophila miR-310, namely miR-310, miR-311, miR-312, and miR-313, negatively regulated the Toll-mediated immune response by repressing the expression of Drs and directly cotargeting the 3’UTR of Drs in Drosophila upon Gram-positive bacterial infection (146). In summary, insect-encoded miRNAs can act individually or collectively to inhibit AMP expression and impair antibacterial defenses for the purpose of immune homeostasis. These mechanisms, in most cases, involve the regulation of the Toll pathway components and allow not only the identification of a new miRNAs but also enrich the repertoire of Toll-related immune-modulating miRNAs in insects.
While some insects enjoy the conservation of genes encoding immune effectors, other from the hemipteran species such as pea aphid Acyrthosiphon pisum showed decreased immune responses as they lack the genes coding for AMP, IMD, PGRPs, and other immune-related molecules (53). Fortunately, the conserved Jun N-terminal kinase (JNK) pathway has been suggested in the pea aphid immune defense. In their investigation on how this pathway regulates the pea aphid immune defense upon bacterial invasion, Ma et al. (147) found that miRNA-184a/b targeted the JNK-3’UTR and repressed its expression, hence resulting in more bacteria in the aphids and increased aphids’ mortality after infection. The expression of miRNA-184a and miRNA-184b remarkably dropped after M. luteus and Pseudomonas aeruginosa infection, with the lowest expression observed at 24 h postinfection, revealing a negative correlation with JNK expression (147). Interestingly, PO, reactive oxygen species, and phagocytosis are under the control of the JNK pathway, suggesting miRNA-184 indirectly controls these antibacterial immune response mechanisms in the pea aphid. Finally, the regulation of the JNK pathway by miRNA-184 is likely a universal mechanism in animals, as prediction using the RNAhybrid program showed that JNK is a potential target of miRNA-184 in insects, zebrafish, frogs, mice, and humans (147).
UPEC strains provoke symptomatic urinary tract infections in humans, whereas commensal-like E. coli strains in the urinary bladder cause long-term asymptomatic bacteriuria (ABU). G. mellonella is a surrogate insect model host used to study human pathogens, including UPEC (148). miRNA sequencing in G. mellonella larvae infected with UPEC strain CFT073 or ABU strain 83972 showed significant changes in the expression levels of G. mellonella miRNAs, respectively, suggesting that insect immune response-mediated miRNAs can distinguish between pathogenic and commensal E. coli invasions (136).
Host miRNAs have an essential role in defense against viral attacks, hence influencing the course of the infection. For example, during chikungunya virus (CHIKV) infection of A. aegypti, upregulated A. aegypti miR-2b binds to the ubiquitin-related modifier (Urm) 3’UTR, decreasing its translation. This finally leads to decreasing CHIKV replication within A. aegypti (26).
In contrast, in some cases, insect miRNAs can promote viral replication by reducing the expression of virus-induced host genes. That is the case for the insulin-related peptide-binding protein 2 (IBP2) which is known to be significantly upregulated in viruses- infected B. mori (149) but has been negatively regulated by miR-278-3p in vitro and vivo, leading to BmCPV replication. However, the definite mechanism of miR-278-3p and IPB2 on BmCPV replication has been ambiguous, requiring further investigation in the future (150).
The functional exploration of lncRNAs in insects is by far exhaustive, mainly upon microbial attacks. Figure 5 gives an overview of how insects use lncRNAs to defend against fungal, bacterial, or viral attacks.
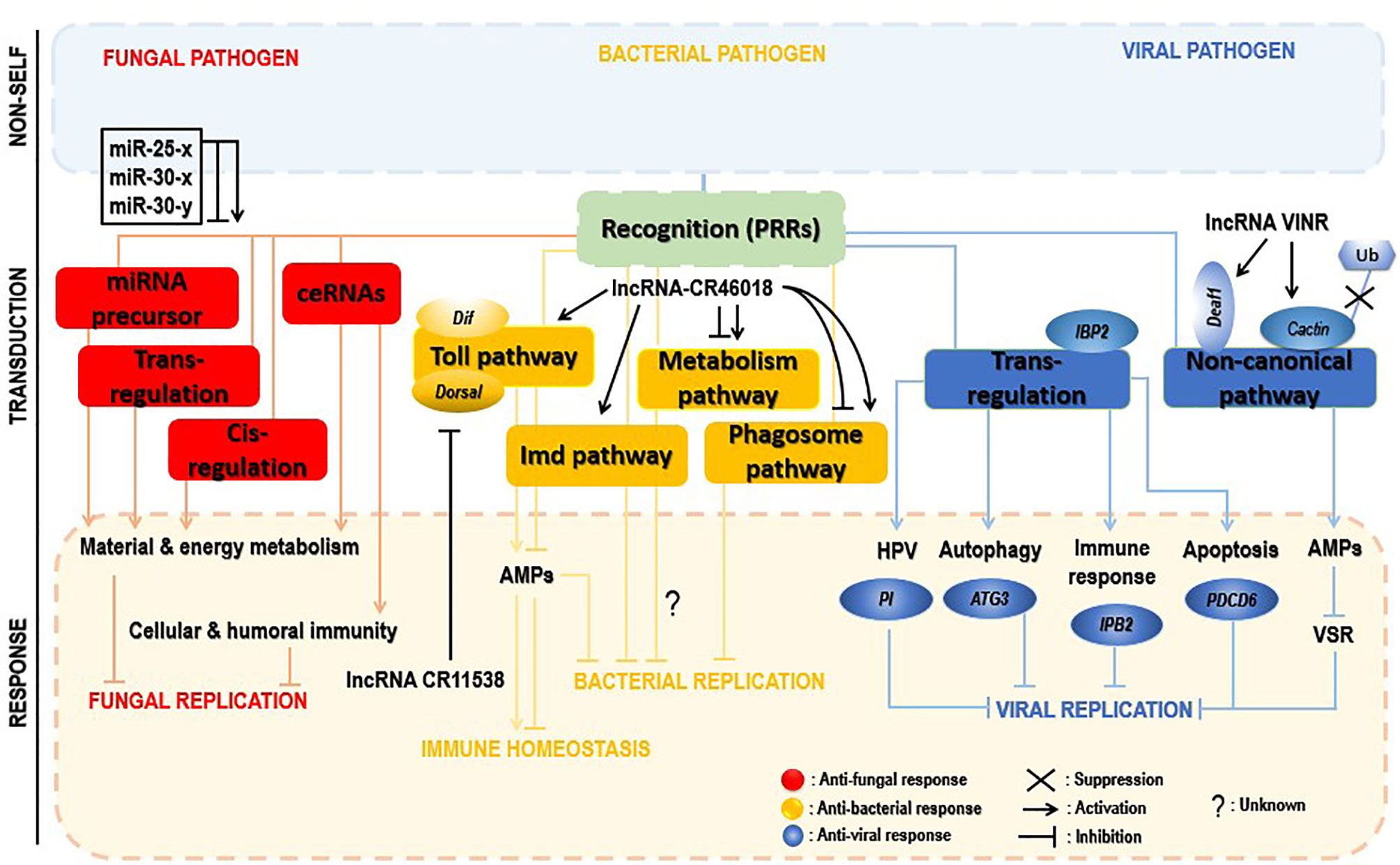
Figure 5 Insect immune defense modulation by lncRNAs upon microbial attacks. The fungal invasion of insects induced alteration of a myriad of insect lncRNAs, which regulate neighboring genes in cis and trans or act by interacting with miRNAs (sponges) or being miRNA precursors. Those trans-and cis-acting predominantly activate material and energy metabolism processes and cellular and humoral immunity, hence helping in control of the infection (red). lncRNAs inhibit bacterial replication via positive regulation of the Toll pathway, the phagosome pathway, or the metabolism process. However, the latter signaling pathway required more investigation to understand how its deactivation contributes to decreased pathogen replication. Additionally, to avoid immune overactivation after bacterial invasion, insect lncRNAs decoy the critical components of the Toll pathway, lowering the expression of AMPs, thus promoting host immune response homeostasis (yellow). During the viral invasion of insects, insect lncRNAs positively trans-regulate insect genes (PI, ATG3, IBP2, PDC6, etc.) involved in cellular and humoral immune-related pathways (HPV, autophagy, apoptosis, etc.). Viral suppression is also achieved through activation of a noncanonical pathway, as an alternative to compensate the RNAi pathway failure; deployed lncRNAs inhibit both the virulence suppressor of RNAi (VSR) and the ubiquitination of cactin in the nucleus or indirectly target the transcription factor Deaf1 and the RNA polymerase II (RNAPII) for transcription of AMPs to control the viral replication (blue).
Few reports investigated interactions between insects and fungi, mainly insect lncRNA response to fungal stress. The western honeybee Apis mellifera (A. mellifera) is domestically used worldwide for honey production and crop pollination. The spore-forming and obligate intracellular fungal pathogen, Nosema ceranae (N. ceranae), can infect a variety of insects, including honeybees (151). The expression of A. mellifera lncRNAs was drastically altered by N. ceranae infection, revealing 4,749 conserved lncRNAs and 1,604 novel lncRNAs. Some differentially expressed lncRNAs regulated gene expression in cis and trans manners or served as precursors of miRNAs, or competitive endogenous RNAs (ceRNAs) acting as miRNA sponges, leading to the activation of the host key signaling pathways and infection control (152). One of the essential suggestions was that through their sponge potential, A. mellifera differential expressed lncRNAs might suppress N. ceranae via crosstalk with miR-25-x, miR-30-x, and miR-30-y. However, the binding of these lncRNAs and miRNAs is limited to bioinformatic prediction, and further experimental study is required.
LncRNAs can promote or impair insect immune responses, especially the Toll immune response upon bacterial attacks. Dif and Dorsal genes are two crucial components of the Toll immune signaling in insects. These two effectors can activate the transcription of AMPs for pathogen eradication. The lncRNA CR46018 was approximatively tenfold overexpressed after infection of Drosophila with M. luteus. In addition, RNA-seq analysis of lncRNA CR46018-overexpressing Drosophila after infection with M. luteus revealed that upregulated genes were mainly enriched in Toll and Imd signaling pathways, supported by bioinformatics predictions and RNA-immunoprecipitation experiments which showed that CR46018 interacted with the transcription factors Dif and dorsal to enhance the Toll pathway. All this indicates lncRNA-CR46018 as a positive regulator of the Toll signaling pathway and essential for Drosophila survival. Moreover, lncRNA-CR46018 could up- and downregulate genes involved in the phagosome pathway and metabolism-related regulation, respectively. The latter pathway seems to be a promising target of insect-derived lncRNAs during insect–pathogen interactions. For example, a previous report unveiled that the Drosophila lncRNA CR44404 (lincRNA-IBIN) links immunity and metabolism in Drosophila upon infection with M. luteus (28). However, how insect-derived lncRNAs regulate this pathway to support insect immune response mechanisms needs further investigation.
Inversely, lncRNAs could negatively regulate insect immune responses by suppressing insect immune effectors to avoid abusive immune stimulation at a late stage of the infection. A recent study reports that at the later stage (24 h) of infection of Drosophila with M. luteus, the lncRNA-CR11538 inhibited the transcription of AMPs via decoying Dif/dorsal away from AMP promoter, thereby negatively modulating the Toll signaling pathway and inhibiting their transcriptions to prevent abusive immune activation in Drosophila with M. luteus infection (153).
The RNAi pathway is an essential antiviral response in insects. However, several studies suggested that the RNAi pathway is ineffective in preventing viral replication (154, 155). An obvious viral strategy to bypass insect RNAi defense mechanisms is to encode RNAi suppressors. Several RNAi cancellers have been described in plant and insect RNA viruses (156, 157). Meanwhile, no RNAi suppressors have been associated with arboviruses until the recent investigation of Zhang et al. (158). During infection of Drosophila with the Drosophila C virus (DCV), the antiviral lncRNA VINR was accumulated in the nucleus because of DCV’s viral RNAi suppressors for their inhibition. LncRNA VINR acted by binding to cactin, preventing its degradation by ubiquitin-proteasome and promoting a noncanonical antiviral and AMP defense leading to a reduced viral replication (159). This suggests a counter counter-defense strategy activated by a lncRNA in response to the viral suppression of the primary antiviral RNAi immunity in Drosophila.
LncRNAs regulate gene expression via cis- or trans-acting regulation (160, 161). In a comprehensive study of lncRNAs associated with Rice black-streaked dwarf virus (RBSDV) infection in Laodelphax striatellus midgut, all predicted and differentially expressed mRNA targets were regulated in a trans manner by 176 differentially expressed lncRNAs. In addition, although the KEGG pathway analysis revealed significantly enriched pathways such as purine metabolism, valine, leucine, and isoleucine degradation, fatty acid elongation, and so on as the most significantly enriched pathways of those trans-regulated genes, the fact remains that the Human papillomavirus infection pathway (which is pivotal for viral infection) was enriched considerably during RBSDV infection. It, therefore, might be involved in RBSDV infection of L. striatellus midgut (138). The eight differentially expressed lncRNAs and the two coexpressed targets in the Human papillomavirus pathway predicted by KEGG analysis and confirmed by RT-qPCR can be found in Table 1. Moreover, a protease inhibitor (PI), one of the lncRNAs’ targets, plays a vital role in antivirus and preventing carcinogenesis (162, 163). Interestingly, lncRNA MSTRG15394 and its target PI (15-fold) were significantly expressed, and scrutinizing their involvement in RBSDV infection of L. striatellus midgut showed that knockdown of MSTRG15394 or PI drastically increased the expression patterns of RBSDV replication-related genes, S5-1, S6, and S9-1, suggesting that MSTRG15394 and PI could inhibit the accumulation and proliferation of RBSDV in L. striatellus midgut (138). Moreover, upon B. mori cypovirus (BmCPV) infection of silkworm larvae, the expression of mRNA targets was mainly affected via trans-regulation by BmCPV-induced lncRNAs (164). Interestingly and remarkably, analysis of the differentially expressed lncRNAs and mRNAs network reveals that these differentially expressed lncRNAs simultaneously targeted some genes involved in relevant mechanisms such as apoptosis (PDCD6), autophagy (ATG3), and immune response (IPB2). Genes such as PDCD6, ATG3, IPB2, MFB1, and VPS52 could be trans-targeted by the most significantly expressed lncRNA MSTRG.20486.1 (164).
Pathogens can encounter insect immune responses and create a suitable environment for their replication. Insect pathogens have employed several strategies to escape host immune responses, including pathogen-encoded miRNAs and lncRNAs. The pathogen-encoded ncRNAs targeting insect-derived genes are listed in Table 3.
Pathogen-derived miRNAs or miRNA-like-RNAs (milRNAs) are necessary components for host–pathogen crosstalk, promoting pathogen proliferation and replication in most cases. These milRNAs can modulate both pathogen and insect host immune genes. The following concentrates on the milRNA modulation of insect key genes. Figure 6 highlights the manipulation of insect immunity by pathogen-encoded miRNAs.
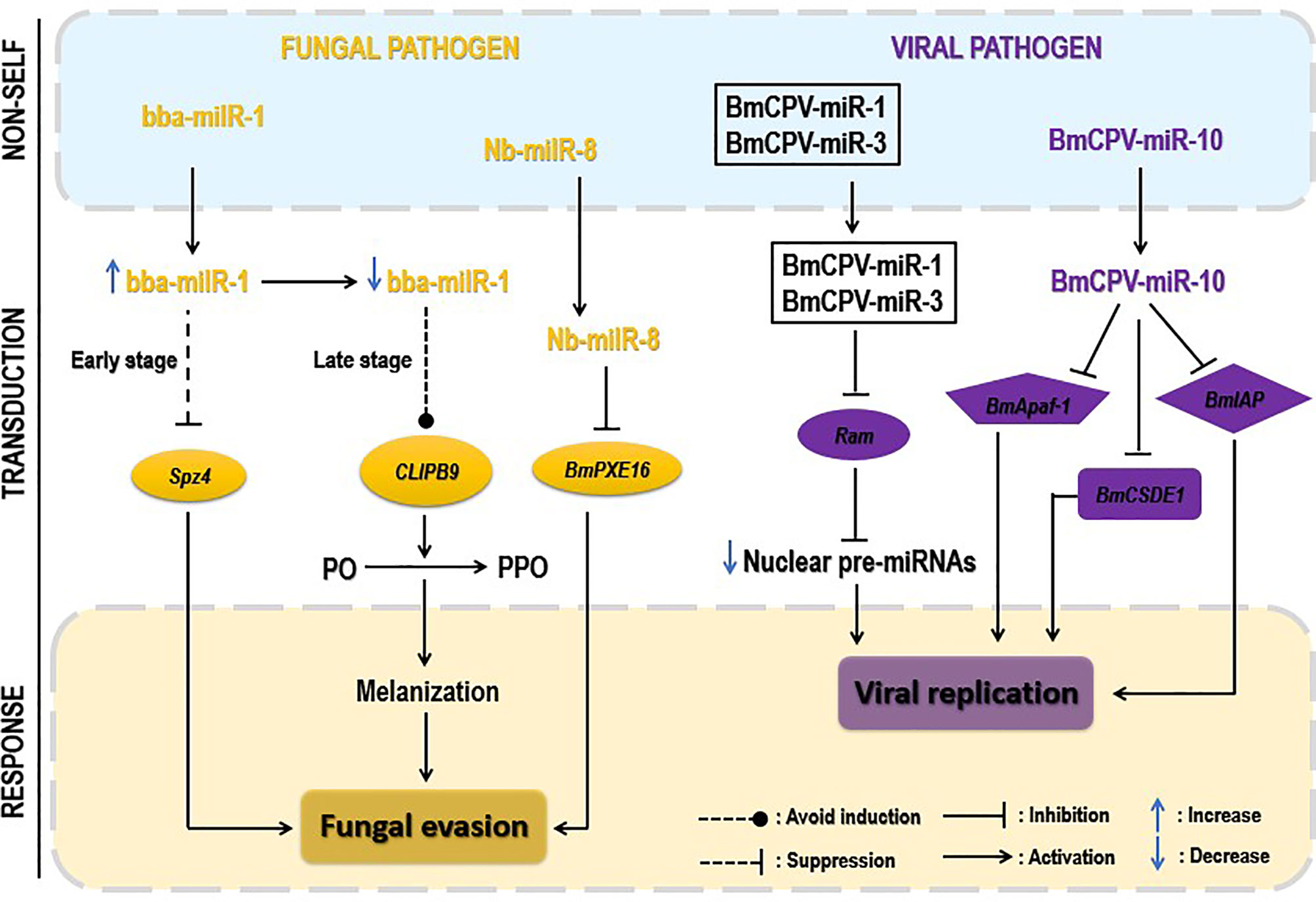
Figure 6 Pathogen-encoded miRNAs regulating insect immunity. Pathogen-derived miRNAs often translocate via extracellular vesicles (EVs) to regulate insect host immunity. Upon fungal invasion, translocated fungal miRNAs downregulate the insect Toll signaling pathway by repressing the expression of critical genes, such as Spz4, or inhibiting the peroxisome pathway, repressing the expression of the PXE16 gene. At the late stage of infection, fungi act by decreasing the expression of their miRNAs to escape the melanization process (yellow). Upon viral attacks, translocated virus-derived miRNAs act individually or synergistically to negatively regulate the expression of insect key genes (Ran, Apaf-1, etc.) by targeting their 3′UTR regions. The latter downregulation reduces host miRNAs’ production, leading to viral replication and proliferation (purple). However, the figure does not show the bacterial-derived miRNAs due to missing data on their role in insect–bacterium interaction.
milRNAs play crucial roles during fungal invasion (169). However, few fungal-derived milRNAs have been reported. Still, their active investigation remains challenging.
Delivering cell-entering effector molecules is a well-known employed strategy of plant pathogenic fungi to suppress host immunity (170, 171). No description has been documented for such effector molecules for pathogenic insect fungi until the brilliant discovery of Cui et al. (158). The pathogenic fungal B. bassiana deployed a miRNA-like RNA (bba-milR1) acting as a cell-entering effector to suppress mosquito immune response. The authors found that during the early stages of infection, B. bassiana bba-milR1 expression increased remarkably and translocated into the mosquito cells to mitigate mosquito immune responses by suppressing the expression of the critical activator gene Spz4. During this early invasion in the integument, the bba-milR1 is not accessible to circulating hemocytes, the site of CLIPB9 gene expression. In contrast, during the late stage of infection (hemocoel invasion), B. bassiana strategically decreased the expression level of bba-milR1 and avoided induction of CLIPB9 and activation of melanization. Quantification of bba-milR1 expression level during B. bassiana infection of Anopheles stephensi (A. stephensi) showed that bba-milR1 was induced by ~30-fold at 36 h postinfection and then declined to deficient levels as the fungus enters the host’s hemocoel at about 60 h after infection (158). Moreover, Nosema bombycis (N. bombycis) proliferation within B. mori is mediated by the increased expression of its miRNA-like RNA, Nb-milR8, which negatively regulates the B. mori peroxisome metabolic pathway via BmPXE16 gene expression, ultimately inhibiting the latter expression (165).
Studies have shown that one miRNA can target multiple genes and several miRNAs can also regulate a target gene. Generally, miRNA mainly binds to the 3’UTR of mRNA to repress the target gene translation, and cooperativity between two or more miRNA-binding sites can enhance the repression of the mRNA translation (172–174). In the case of viral infections, virus-encoded miRNAs generally target the 3’UTR of key insect immune genes to repress their expressional changes and reduce the generation of insect miRNAs to create a favorable environment for viral replication (175). Lin et al. (166), for example, reported that to facilitate its replication in B. mori, BmCPV released two miRNAs, BmCPV-miR-1 and BmCPV-miR-3, that co-operatively targeted the 3’UTR of B. mori GTP-biding nuclear protein Ran (BmRan) by lowering its expression level. Ran is a GTP-binding nuclear protein of 25 kDa, which plays a role of transporting small RNAs, including pre-miRNAs, from the nucleus to the cytoplasm (176), and its repression level led to a significant reduction in nuclear export of pre-miRNA. Similarly, BmCPV-miR-10 downregulated the B. mori cold shock domain E1 protein (BmCSDE1) mRNA expression level after binding to its 3’UTR in the infected larvae. BmCPV-miR-10 also inhibited the expressional changes of B. mori apoptotic protease activating factor 1 (BmApaf-1) during in vivo infection, creating suitable conditions for virus replication and proliferation (167). BmCPV-miR-1 inhibited cell apoptosis in the infected silkworm via increasing B. mori inhibitor of apoptosis protein (BmIAP) expression, promoting the virus replication (168). Obviously, the roles of virus-encoded miRNAs in the crosstalk seem to be largely explored compared with miRNA-encoded fungi or bacteria. An interesting insight in investigating this element-encoded virus is that they have the possibility to simultaneously regulate both insect innate and cellular immune responses, and further demonstrate that there is a dynamic evolution in the insect–pathogen crosstalk implicating ncRNAs, where insects evolutionally sophisticate their defense mechanisms while pathogens simultaneously elaborate strategies to encompass these immune obstacles for their own benefit.
Although new technologies and bioinformatics tools emerge for the detection of ncRNAs, most lncRNAs’ expression is low, probably limiting their identification and functional characterization (177). Several studies identified pathogen-encoded lncRNAs, their differential expression levels, and how the latter regulates pathogen’s genes (178). However, studies describing how these pathogen-encoded lncRNAs regulate insect immune responses during insect–pathogen interactions are considerably missing. For example, no virus-encoded lncRNAs were generated during silkworm larvae-BmCPV interaction, while 41 B. mori lncRNAs among 1845 were identified (164). We are tempted to speculate that pathogen-encoded lncRNAs may act similarly to miRNAs derived-pathogens, where their differential expression negatively regulates insect key genes and creates a favorable environment for pathogen replication and proliferation. However, the above suggestion needs further experimental shreds of evidence. This area could be an exciting and hotspot topic for researchers to investigate in the future.
ncRNAs are the emerging and fate-determining players of insect–pathogen interactions. The long history of host and pathogen coevolution suggests that the pathogen keeps on evolving its arsenals to succeed in infection, whereas the host continues investing in defense mechanisms. Therefore, portraying the landscape of ncRNAs in insect–pathogen crosstalk at the immunological level is of great scientific worth. In short, we briefly reviewed the main signaling pathways and accessory immune mechanisms engaged in insect defense mechanisms and the recent progress on the involvement of ncRNAs, especially miRNAs and lncRNAs, in promoting or suppressing insect immune responses upon pathogen invasion. In addition, how pathogen-encoded miRNAs and lncRNAs regulate insect immunity was a part of the scope of this study.
The Toll, Imd, and JAK/STAT pathways are evolutionarily conserved pathways playing a manifest role in insect immune defense mechanisms against pathogens. They protect a wide range of pathogens (bacteria, fungi, parasites, etc.). In addition, with the assistance of immune effector mechanisms, this protection might be strengthened. However, looking deep inside, although the conferred protection, several factors exclude the idea of total protection. The fact that these pathways are not present in all insects might influence and change the ability of insects to defend themselves upon pathogens invasions. Moreover, the inexistence of adaptive immunity, which confers immune memory, allowing organisms to mount a more rapid and potent immune response when it is re-exposed to the same pathogen or a highly related one, might probably be a situation that affects insect immune defense intensity and efficacy. However, it has been hypothesized that immune priming occurs because the insect is inherently able to activate immune responses more rapidly during a second exposure or because pathogens or pathogen components are retained within the insect, which maintains the animal in a heightened state of immune alertness (179).
Insect immunity is well known to be modulated by several factors, including microbiota and ncRNAs. The latter, mainly miRNAs and lncRNAs, hugely reinforce insect host immune defense mechanisms during their crosstalk with pathogens. Their positive and negative modulation of insects’ immune responses passes through their ability to target insect- or pathogen-encoded mRNA targets and influence the course of the infection for the benefice of insects. These elements can be viewed as alternative insects’ defense mechanisms such as those mentioned above (immune effector mechanisms). They can be put on the stage as primary insect immune defense mechanisms, when principal pathways are inefficient to secure insects’ integrity from being invaded. The RNAi immune effector mechanism, for example, is well documented to provide a robust protection to insects against viral attacks. However, pathogen VSRs were recently proved to overcome this path (180). In this present case, the insect’s survival was accountable to the immune modulation ability of ncRNAs. Therefore, it is hoped that these noncoding elements would be exploited as a mainstream player to achieve food security or avoid economic waste in the industry by tackling different insect invaders.
Although their ability to maintain and restore insect immune homeostasis or enhance host immune defenses, ncRNA protection seems to be limited as their downregulation action of insect immunity promotes, in some cases, the proliferation of the pathogen. Interestingly, in opposition to pathogen-derived miRNAs, there is a deficiency of studies describing the modulation of insect immunity by pathogen-derived lncRNAs in order to facilitate pathogen replication and proliferation. Such topics should retain researchers’ attention, as exploring this area will undoubtedly bring new insights into the role of these elements in insect–pathogen crosstalk. In addition, it seems like ncRNAs support insect immunity against pathogens by regulating insect main and evolutionarily conserved signaling pathways (Toll, Imd). Simultaneously, their action is less observed in insect accessory immune mechanisms. The regulation of these insect accessory defense mechanisms by ncRNAs has not been extensively explored. Few studies investigated the link between ncRNAs and insect accessory defense mechanisms. miRNAs of Snellenius manilae bracovirus (SmBV-miR-199b5p and SmBV-miR-2989) were found to regulate innate (domeless and toll-7) and cellular (encapsulation and phagocytosis activity of the hemocytes) immune responses of its host Spodoptera litura (181). Plutella xylostella miR-8 is downregulated following parasitization by Diadegma semiclausum, leading to significant declines in Serpin 27 transcript levels (182). The serine protease inhibitor Serpin 27 regulates activation of the Toll pathway and PPO involved in the melanization response in insects. Taken together, we are tempted to speculate that there is no direct role of ncRNAs on insect accessory immunity. Nonetheless, to a lesser extent, and based on evidence, ncRNAs undoubtedly modulate insect immune effector mechanisms indirectly by targeting insect main signaling pathway components. The Toll ligand Spätzle 3 controlled the melanization process in the stripe pattern formation of caterpillars (183).
It is now known that ncRNAs have a drastic immunomodulation influence in insect–pathogen crosstalk. All the changes, such as the differentially expressed miRNAs, lncRNAs, the co/target transcripts, and the diverse signaling pathways activated during insect–pathogen crosstalk, are proof of their crucial role. Proper understanding of insect–pathogen interaction is essential to deciphering the immune molecular patterns employed by insects against various pathogens. This understanding will positively impact the economy based on important insects, such as D. melanogaster and B. mori. Here are some perspectives: (1) improvement of methods and technologies used for ncRNA investigations; (2) promotion of more in-depth studies instead of preliminary ones; and (3) the link between pathogen-encoded lncRNAs and insect immunity represents an attracting topic to investigate and may hide tons of critical information, which will undoubtedly give new insights into the immunological landscape of insect–pathogen interaction.
UAEM and TT reviewed the literature, designed the figures, and wrote the manuscript. LS, GY, and XL designed the tables and reviewed the manuscript. ZS, YW, and HC reviewed and revised the text. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the National Natural Science Foundation of China (31802142), the Doctoral Start-up Fund of Southwest University (SWU120019, SWU020023), the Fundamental Research Funds for the Central Universities (XDJK2019C089), and the China Postdoctoral Science Foundation (2019T120801 and 2017M620408).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Satyavathi V, Ghosh R, Subramanian S. Long Non-Coding RNAs Regulating Immunity in Insects. Non-coding RNA. (2017) 3(1):14. doi: 10.3390/ncrna3010014
2. Lundgren JG, Jurat-Fuentes JL. The Physiology and Ecology of Host Defense Against Microbial Invaders. Insect Pathol (2012) 2nd edition):460–80. doi: 10.1016/B978-0-12-384984-7.00013-0
3. Siva-Jothy MT, Moret Y, Rolff J. Insect Immunity: An Evolutionary Ecology Perspective. In: Simpson SJ, editor. Advances in Insect Physiology, vol. 32 . Academic Press (2005). p. 1–48.
4. Hillyer JF. Insect Immunology and Hematopoiesis. Dev Comp Immunol (2016) 58:102–18. doi: 10.1016/j.dci.2015.12.006
5. Kingsolver MB, Huang Z, Hardy RW. Insect Antiviral Innate Immunity: Pathways, Effectors, and Connections. J Mol Biol (2013) 425(24):4921–36. doi: 10.1016/j.jmb.2013.10.006
6. Zhang W, Tettamanti G, Bassal T, Heryanto C, Eleftherianos I, Mohamed A. Regulators and Signalling in Insect Antimicrobial Innate Immunity: Functional Molecules and Cellular Pathways. Cell signalling (2021) 83:110003. doi: 10.1016/j.cellsig.2021.110003
7. Mylonakis E, Podsiadlowski L, Muhammed M, Vilcinskas A. Diversity, Evolution and Medical Applications of Insect Antimicrobial Peptides. Philos Trans R Soc London Ser B Biol Sci (2016) 371(1695):20150290. doi: 10.1098/rstb.2015.0290
8. Lemaitre B, Hoffmann J. The Host Defense of Drosophila Melanogaster. Annu Rev Immunol (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
9. Hultmark D. Drosophila Immunity: Paths and Patterns. Curr Opin Immunol (2003) 15(1):12–9. doi: 10.1016/S0952-7915(02)00005-5
10. Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila Systemic Immune Response: Sensing and Signalling During Bacterial and Fungal Infections. Nat Rev Immunol (2007) 7(11):862–74. doi: 10.1038/nri2194
11. Valanne S, Wang JH, Rämet M. The Drosophila Toll Signaling Pathway. J Immunol (Baltimore Md 1950) (2011) 186(2):649–56. doi: 10.4049/jimmunol.1002302
12. Myllymäki H, Rämet M. JAK/STAT Pathway in Drosophila Immunity. Scandinavian J Immunol (2014) 79(6):377–85. doi: 10.1111/sji.12170
13. Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to Overcome Insect Defences. Nat Rev Microbiol (2008) 6(4):302–13.
14. Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, et al. The Jak-STAT Signaling Pathway is Required But Not Sufficient for the Antiviral Response of Drosophila. Nat Immunol (2005) 6(9):946–53. doi: 10.1038/ni1237
15. Palmer WH, Joosten J, Overheul GJ, Jansen PW, Vermeulen M, Obbard DJ, et al. Induction and Suppression of NF-kB Signalling by a DNA Virus of Drosophila. bioRxiv (2018) 358176.
16. Strand MR. The Insect Cellular Immune Response. Insect Sci (2008) 15(1):1–14. doi: 10.1111/j.1744-7917.2008.00183.x
17. Browne N, Heelan M, Kavanagh K. An Analysis of the Structural and Functional Similarities of Insect Hemocytes and Mammalian Phagocytes. Virulence (2013) 4(7):597–603. doi: 10.4161/viru.25906
18. Kounatidis I, Ligoxygakis P. Drosophila as a Model System to Unravel the Layers of Innate Immunity to Infection. Open Biol (2012) 2(5):120075. doi: 10.1098/rsob.120075
19. Hoffmann JA. The Immune Response of Drosophila. Nature (2003) 426(6962):33–8. doi: 10.1038/nature02021
20. Bai L, Wang L, Vega-Rodríguez J, Wang G, Wang S. A Gut Symbiotic Bacterium Serratia Marcescens Renders Mosquito Resistance to Plasmodium Infection Through Activation of Mosquito Immune Responses. Front Microbiol (2019) 10:1580. doi: 10.3389/fmicb.2019.01580
21. Castelli L, Branchiccela B, Garrido M, Invernizzi C, Porrini M, Romero H, et al. Impact of Nutritional Stress on Honeybee Gut Microbiota, Immunity, and Nosema Ceranae Infection. Microbial Ecol (2020) 80(4):908–19. doi: 10.1007/s00248-020-01538-1
22. Ahmad P, Bensaoud C, Mekki I, Rehman MU, Kotsyfakis M. Long Non-Coding RNAs and Their Potential Roles in the Vector-Host-Pathogen Triad. Life (Basel Switzerland) (2021) 11(1):56. doi: 10.3390/life11010056
23. Wang Y, Cui C, Wang G, Li Y, Wang S. Insects Defend Against Fungal Infection by Employing microRNAs to Silence Virulence-Related Genes. Proc Natl Acad Sci USA (2021) 118(19):e2023802118. doi: 10.1073/pnas.2023802118
24. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of Transcription in Human Cells. Nature (2012) 489(7414):101–8. doi: 10.1038/nature11233
25. Choudhary C, Sharma S, Meghwanshi KK, Patel S, Mehta P, Shukla N, et al. Long Non-Coding RNAs in Insects. Anim an Open Access J MDPI (2021) 11(4):1118. doi: 10.3390/ani11041118
26. Dubey SK, Shrinet J, Jain J, Ali S, Sunil S. Aedes Aegypti microRNA miR-2b Regulates Ubiquitin-Related Modifier to Control Chikungunya Virus Replication. Sci Rep (2017) 7(1):17666. doi: 10.1038/s41598-017-18043-0
27. Awais MM, Shakeel M, Sun J. MicroRNA-Mediated Host-Pathogen Interactions Between Bombyx Mori and Viruses. Front Physiol (2021) 12:672205. doi: 10.3389/fphys.2021.672205
28. Valanne S, Salminen TS, Järvelä-Stölting M, Vesala L, Rämet M. Immune-Inducible Non-Coding RNA Molecule lincRNA-IBIN Connects Immunity and Metabolism in Drosophila Melanogaster. PloS Pathog (2019) 15(1):e1007504. doi: 10.1371/journal.ppat.1007504
29. Maarouf M, Chen B, Chen Y, Wang X, Rai KR, Zhao Z, et al. Identification of lncRNA-155 Encoded by MIR155HG as a Novel Regulator of Innate Immunity Against Influenza A Virus Infection. Cell Microbiol (2019) 21(8):e13036. doi: 10.1111/cmi.13036
30. Etebari K, Asad S, Zhang G, Asgari S. Identification of Aedes Aegypti Long Intergenic Non-Coding RNAs and Their Association With Wolbachia and Dengue Virus Infection. PloS neglected Trop Dis (2016) 10(10):e0005069. doi: 10.1371/journal.pntd.0005069
31. Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is Activated by Gram-Positive Bacteria Through a Circulating Peptidoglycan Recognition Protein. Nature (2001) 414(6865):756–9. doi: 10.1038/414756a
32. Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the Drosophila Pattern-Recognition Receptor PGRP-SD in the Detection of Gram-Positive Bacteria. Nat Immunol (2004) 5(11):1175–80. doi: 10.1038/ni1123
33. Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Sensing of Gram-Positive Bacteria in Drosophila: GNBP1 is Needed to Process and Present Peptidoglycan to PGRP-Sa. EMBO J (2006) 25(20):5005–14. doi: 10.1038/sj.emboj.7601363
34. Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, et al. Dual Detection of Fungal Infections in Drosophila via Recognition of Glucans and Sensing of Virulence Factors. Cell (2006) 127(7):1425–37. doi: 10.1016/j.cell.2006.10.046
35. Dziarski R. Peptidoglycan Recognition Proteins (PGRPs). Mol Immunol (2004) 40(12):877–86. doi: 10.1016/j.molimm.2003.10.011
36. De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-Wide Analysis of the Drosophila Immune Response by Using Oligonucleotide Microarrays. Proc Natl Acad Sci USA (2001) 98(22):12590–5. doi: 10.1073/pnas.221458698
37. De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd Pathways are the Major Regulators of the Immune Response in Drosophila. EMBO J (2002) 21(11):2568–79. doi: 10.1093/emboj/21.11.2568
38. Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila Host Defense: Differential Induction of Antimicrobial Peptide Genes After Infection by Various Classes of Microorganisms. Proc Natl Acad Sci USA (1997) 94(26):14614–9. doi: 10.1073/pnas.94.26.14614
39. Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, et al. Binding of the Drosophila Cytokine Spätzle to Toll is Direct and Establishes Signaling. Nat Immunol (2003) 4(8):794–800. doi: 10.1038/ni955
40. Hu X, Yagi Y, Tanji T, Zhou S, Ip YT. Multimerization and Interaction of Toll and Spätzle in Drosophila. Proc Natl Acad Sci USA (2004) 101(25):9369–74. doi: 10.1073/pnas.0307062101
41. Morisato D, Anderson KV. The Spätzle Gene Encodes a Component of the Extracellular Signaling Pathway Establishing the Dorsal-Ventral Pattern of the Drosophila Embryo. Cell (1994) 76(4):677–88. doi: 10.1016/0092-8674(94)90507-X
42. Schneider DS, Jin Y, Morisato D, Anderson KV. A Processed Form of the Spätzle Protein Defines Dorsal-Ventral Polarity in the Drosophila Embryo. Dev (Cambridge England) (1994) 120(5):1243–50. doi: 10.1242/dev.120.5.1243
43. Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is Required for the Response to Fungal and Gram-Positive Bacterial Infections. Nat Immunol (2002) 3(1):91–7. doi: 10.1038/ni747
44. Horng T, Medzhitov R. Drosophila MyD88 is an Adapter in the Toll Signaling Pathway. Proc Natl Acad Sci USA (2001) 98(22):12654–8. doi: 10.1073/pnas.231471798
45. Xiao T, Towb P, Wasserman SA, Sprang SR. Three-Dimensional Structure of a Complex Between the Death Domains of Pelle and Tube. Cell (1999) 99(5):545–55. doi: 10.1016/S0092-8674(00)81542-1
46. Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, González-Crespo S, et al. Dif, a Dorsal-Related Gene That Mediates an Immune Response in Drosophila. Cell (1993) 75(4):753–63. doi: 10.1016/0092-8674(93)90495-C
47. Lemaitre B, Meister M, Govind S, Georgel P, Steward R, Reichhart JM, et al. Functional Analysis and Regulation of Nuclear Import of Dorsal During the Immune Response in Drosophila. EMBO J (1995) 14(3):536–45. doi: 10.1002/j.1460-2075.1995.tb07029.x
48. Wu LP, Anderson KV. Regulated Nuclear Import of Rel Proteins in the Drosophila Immune Response. Nature (1998) 392(6671):93–7. doi: 10.1038/32195
49. Hetru C, Hoffmann JA. NF-kappaB in the Immune Response of Drosophila. Cold Spring Harbor Perspect Biol (2009) 1(6):a000232. doi: 10.1101/cshperspect.a000232
50. Belvin MP, Anderson KV. A Conserved Signaling Pathway: The Drosophila Toll-Dorsal Pathway. Annu Rev Cell Dev Biol (1996) 12:393–416. doi: 10.1146/annurev.cellbio.12.1.393
51. Ribeiro JM, Genta FA, Sorgine MH, Logullo R, Mesquita RD, Paiva-Silva GO, et al. An Insight Into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius Prolixus. PloS neglected Trop Dis (2014) 8(1):e2594. doi: 10.1371/journal.pntd.0002594
52. Mesquita RD, Vionette-Amaral RJ, Lowenberger C, Rivera-Pomar R, Monteiro FA, Minx P, et al. Genome of Rhodnius Prolixus, an Insect Vector of Chagas Disease, Reveals Unique Adaptations to Hematophagy and Parasite Infection. Proc Natl Acad Sci United States America (2015) 112(48):14936–41. doi: 10.1073/pnas.1506226112
53. Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, de Vos M, et al. Immunity and Other Defenses in Pea Aphids, Acyrthosiphon Pisum. Genome Biol (2010) 11(2):R21. doi: 10.1186/gb-2010-11-2-r21
54. Nishide Y, Kageyama D, Yokoi K, Jouraku A, Tanaka H, Futahashi R, et al. Functional Crosstalk Across IMD and Toll Pathways: Insight Into the Evolution of Incomplete Immune Cascades. Proc Biol Sci (2019) 286(1897):20182207. doi: 10.1098/rspb.2018.2207
55. Tzou P, Reichhart JM, Lemaitre B. Constitutive Expression of a Single Antimicrobial Peptide can Restore Wild-Type Resistance to Infection in Immunodeficient Drosophila Mutants. Proc Natl Acad Sci USA (2002) 99(4):2152–7. doi: 10.1073/pnas.042411999
56. Hoffmann JA, Reichhart JM. Drosophila Innate Immunity: An Evolutionary Perspective. Nat Immunol (2002) 3(2):121–6. doi: 10.1038/ni0202-121
57. Myllymäki H, Valanne S, Rämet M. The Drosophila Imd Signaling Pathway. J Immunol (Baltimore Md 1950) (2014) 192(8):3455–62. doi: 10.4049/jimmunol.1303309
58. Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila Melanogaster. Mol Cell Biol (2007) 27(12):4578–88. doi: 10.1128/MCB.01814-06
59. Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE Jr. The Proteins of ISGF-3, the Interferon Alpha-Induced Transcriptional Activator, Define a Gene Family Involved in Signal Transduction. Proc Natl Acad Sci USA (1992) 89(16):7840–3. doi: 10.1073/pnas.89.16.7840
60. Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr. Proteins of Transcription Factor ISGF-3: One Gene Encodes the 91-and 84-kDa ISGF-3 Proteins That are Activated by Interferon Alpha. Proc Natl Acad Sci USA (1992) 89(16):7836–9. doi: 10.1073/pnas.89.16.7836
61. Shuai K, Ziemiecki A, Wilks AF, Harpur AG, Sadowski HB, Gilman MZ, et al. Polypeptide Signalling to the Nucleus Through Tyrosine Phosphorylation of Jak and Stat Proteins. Nature (1993) 366(6455):580–3. doi: 10.1038/366580a0
62. Silvennoinen O, Ihle JN, Schlessinger J, Levy DE. Interferon-Induced Nuclear Signalling by Jak Protein Tyrosine Kinases. Nature (1993) 366(6455):583–5. doi: 10.1038/366583a0
63. Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, et al. Impaired Response to Interferon-Alpha/Beta and Lethal Viral Disease in Human STAT1 Deficiency. Nat Genet (2003) 33(3):388–91. doi: 10.1038/ng1097
64. Barillas-Mury C, Han YS, Seeley D, Kafatos FC. Anopheles Gambiae Ag-STAT, a New Insect Member of the STAT Family, is Activated in Response to Bacterial Infection. EMBO J (1999) 18(4):959–67. doi: 10.1093/emboj/18.4.959
65. Souza-Neto JA, Sim S, Dimopoulos G. An Evolutionary Conserved Function of the JAK-STAT Pathway in Anti-Dengue Defense. Proc Natl Acad Sci USA (2009) 106(42):17841–6. doi: 10.1073/pnas.0905006106
66. Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary Dynamics of Immune-Related Genes and Pathways in Disease-Vector Mosquitoes. Sci (New York NY) (2007) 316(5832):1738–43. doi: 10.1126/science.1139862
67. Arbouzova NI, Zeidler MP. JAK/STAT Signalling in Drosophila: Insights Into Conserved Regulatory and Cellular Functions. Dev (Cambridge England) (2006) 133(14):2605–16. doi: 10.1242/dev.02411
68. Hillyer JF, Schmidt SL, Christensen BM. Hemocyte-Mediated Phagocytosis and Melanization in the Mosquito Armigeres Subalbatus Following Immune Challenge by Bacteria. Cell Tissue Res (2003) 313(1):117–27. doi: 10.1007/s00441-003-0744-y
69. Hillyer JF, Schmidt SL, Christensen BM. Rapid Phagocytosis and Melanization of Bacteria and Plasmodium Sporozoites by Hemocytes of the Mosquito Aedes Aegypti. J parasitology (2003) 89(1):62–9. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2
70. King JG, Hillyer JF. Infection-Induced Interaction Between the Mosquito Circulatory and Immune Systems. PloS Pathog (2012) 8(11):e1003058. doi: 10.1371/journal.ppat.1003058
71. Sigle LT, Hillyer JF. Mosquito Hemocytes Preferentially Aggregate and Phagocytose Pathogens in the Periostial Regions of the Heart That Experience the Most Hemolymph Flow. Dev Comp Immunol (2016) 55:90–101. doi: 10.1016/j.dci.2015.10.018
72. Dean P, Potter U, Richards EH, Edwards JP, Charnley AK, Reynolds SE. Hyperphagocytic Haemocytes in Manduca Sexta. J Insect Physiol (2004) 50(11):1027–36. doi: 10.1016/j.jinsphys.2004.09.003
73. Hillyer JF, Strand MR. Mosquito Hemocyte-Mediated Immune Responses. Curr Opin Insect Sci (2014) 3:14–21. doi: 10.1016/j.cois.2014.07.002
74. Honti V, Csordás G, Kurucz É, Márkus R, Andó I. The Cell-Mediated Immunity of Drosophila Melanogaster: Hemocyte Lineages, Immune Compartments, Microanatomy and Regulation. Dev Comp Immunol (2014) 42(1):47–56. doi: 10.1016/j.dci.2013.06.005
75. Dong Y, Taylor HE, Dimopoulos G. AgDscam, a Hypervariable Immunoglobulin Domain-Containing Receptor of the Anopheles Gambiae Innate Immune System. PloS Biol (2006) 4(7):e229. doi: 10.1371/journal.pbio.0040229
76. Kurucz E, Márkus R, Zsámboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a Putative Phagocytosis Receptor With EGF Repeats in Drosophila Plasmatocytes. Curr Biol CB (2007) 17(7):649–54. doi: 10.1016/j.cub.2007.02.041
77. Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved Role of a Complement-Like Protein in Phagocytosis Revealed by dsRNA Knockout in Cultured Cells of the Mosquito, Anopheles Gambiae. Cell (2001) 104(5):709–18. doi: 10.1016/S0092-8674(01)00267-7
78. Mamali I, Lamprou I, Karagiannis F, Karakantza M, Lampropoulou M, Marmaras VJ. A Beta Integrin Subunit Regulates Bacterial Phagocytosis in Medfly Haemocytes. Dev Comp Immunol (2009) 33(7):858–66. doi: 10.1016/j.dci.2009.02.004
79. Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional Genomic Analysis of Phagocytosis and Identification of a Drosophila Receptor for E. Coli. Nature (2002) 416(6881):644–8. doi: 10.1038/nature735
80. Yang L, Qiu LM, Fang Q, Stanley DW, Ye GY. Cellular and Humoral Immune Interactions Between Drosophila and its Parasitoids. Insect Sci (2021) 28(5):1208–27. doi: 10.1111/1744-7917.12863
81. Mortimer NT, Kacsoh BZ, Keebaugh ES, Schlenke TA. Mgat1-Dependent N-Glycosylation of Membrane Components Primes Drosophila Melanogaster Blood Cells for the Cellular Encapsulation Response. PloS Pathog (2012) 8(7):e1002819. doi: 10.1371/journal.ppat.1002819
82. Ono M, Arimatsu C, Kakinoki A, Matsunaga K, Yoshiga T. Comparison of Cellular Encapsulation With Nematodes in Two Lepidopteran Insects. Appl Entomol Zoology (2020) 55(3):337–44. doi: 10.1007/s13355-020-00687-6
83. Akai H, Sato S. Ultrastructure of the Larval Hemocytes of the Silkworm, Bombyx Mori L. (Lepidoptera : Bombycidae). Int J Insect Morphol Embryol (1973) 2(3):207–31. doi: 10.1016/0020-7322(73)90029-9
84. Takahashi S, Enomoto G. The Initial Phase of Encapsulation of Silicone Oil Injected in <Span Class="Genus-Species">Samia Cynthia Ricini (Lepidoptera, Saturniidae): The Innermost Structure of the Developing Capsule. Zoological Sci (1995) 12(3):303–9, 7. doi: 10.2108/zsj.12.303
85. Pech LL, Strand MR. Granular Cells are Required for Encapsulation of Foreign Targets by Insect Haemocytes. J Cell Sci (1996) 109(Pt 8):2053–60. doi: 10.1242/jcs.109.8.2053
86. Russo J, Dupas S, Frey F, Carton Y, Brehelin M. Insect Immunity: Early Events in the Encapsulation Process of Parasitoid (Leptopilina Boulardi) Eggs in Resistant and Susceptible Strains of Drosophila. Parasitology (1996) 112(Pt 1):135–42. doi: 10.1017/S0031182000065173
87. Matskevich AA, Quintin J, Ferrandon D. The Drosophila PRR GNBP3 Assembles Effector Complexes Involved in Antifungal Defenses Independently of its Toll-Pathway Activation Function. Eur J Immunol (2010) 40(5):1244–54. doi: 10.1002/eji.200940164
88. Wang JL, Zhang Q, Tang L, Chen L, Liu XS, Wang YF. Involvement of a Pattern Recognition Receptor C-Type Lectin 7 in Enhancing Cellular Encapsulation and Melanization Due to its Carboxyl-Terminal CRD Domain in the Cotton Bollworm, Helicoverpa Armigera. Dev Comp Immunol (2014) 44(1):21–9. doi: 10.1016/j.dci.2013.11.002
89. Wang X, Fuchs JF, Infanger LC, Rocheleau TA, Hillyer JF, Chen CC, et al. Mosquito Innate Immunity: Involvement of Beta 1,3-Glucan Recognition Protein in Melanotic Encapsulation Immune Responses in Armigeres Subalbatus. Mol Biochem parasitology (2005) 139(1):65–73. doi: 10.1016/j.molbiopara.2004.09.009
90. An C, Budd A, Kanost MR, Michel K. Characterization of a Regulatory Unit That Controls Melanization and Affects Longevity of Mosquitoes. Cell Mol Life Sci CMLS (2011) 68(11):1929–39. doi: 10.1007/s00018-010-0543-z
91. An C, Ishibashi J, Ragan EJ, Jiang H, Kanost MR. Functions of Manduca Sexta Hemolymph Proteinases HP6 and HP8 in Two Innate Immune Pathways. J Biol Chem (2009) 284(29):19716–26. doi: 10.1074/jbc.M109.007112
92. Cerenius L, Lee BL, Söderhäll K. The proPO-System: Pros and Cons for its Role in Invertebrate Immunity. Trends Immunol (2008) 29(6):263–71. doi: 10.1016/j.it.2008.02.009
93. Christensen BM, Huff BM, Miranpuri GS, Harris KL, Christensen LA. Hemocyte Population Changes During the Immune Response of Aedes Aegypti to Inoculated Microfilariae of Dirofilaria Immitis. J Parasitol (1989) 75(1):119–23. doi: 10.2307/3282948
94. Nappi AJ, Christensen BM. Melanogenesis and Associated Cytotoxic Reactions: Applications to Insect Innate Immunity. Insect Biochem Mol Biol (2005) 35(5):443–59. doi: 10.1016/j.ibmb.2005.01.014
95. Osta MA, Christophides GK, Kafatos FC. Effects of Mosquito Genes on Plasmodium Development. Sci (New York NY) (2004) 303(5666):2030–2. doi: 10.1126/science.1091789
96. Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. A Genetic Module Regulates the Melanization Response of Anopheles to Plasmodium. Cell Microbiol (2006) 8(9):1392–405. doi: 10.1111/j.1462-5822.2006.00718.x
97. Ashida M, Ochiai M, Niki T. Immunolocalization of Prophenoloxidase Among Hemocytes of the Silkworm, Bombyx Mori. Tissue Cell (1988) 20(4):599–610. doi: 10.1016/0040-8166(88)90061-4
98. Yassine H, Kamareddine L, Osta MA. The Mosquito Melanization Response is Implicated in Defense Against the Entomopathogenic Fungus Beauveria Bassiana. PloS Pathog (2012) 8(11):e1003029. doi: 10.1371/journal.ppat.1003029
99. Carton Y, Frey F, Stanley DW, Vass E, Nappi AJ. Dexamethasone Inhibition of the Cellular Immune Response of Drosophila Melanogaster Against a Parasitoid. J parasitology (2002) 88(2):405–7. doi: 10.1645/0022-3395(2002)088[0405:DIOTCI]2.0.CO;2
100. Gandhe AS, John SH, Nagaraju J. Noduler, a Novel Immune Up-Regulated Protein Mediates Nodulation Response in Insects. J Immunol (Baltimore Md 1950) (2007) 179(10):6943–51. doi: 10.4049/jimmunol.179.10.6943
101. Ratcliffe NA, Gagen SJ. Studies on the In Vivo Cellular Reactions of Insects: An Ultrastructural Analysis of Nodule Formation in Galleria Mellonella. Tissue Cell (1977) 9(1):73–85. doi: 10.1016/0040-8166(77)90050-7
102. Satyavathi VV, Minz A, Nagaraju J. Nodulation: An Unexplored Cellular Defense Mechanism in Insects. Cell signalling (2014) 26(8):1753–63. doi: 10.1016/j.cellsig.2014.02.024
103. Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G, et al. Towards the Elements of Successful Insect RNAi. J Insect Physiol (2013) 59(12):1212–21. doi: 10.1016/j.jinsphys.2013.08.014
104. Yu N, Christiaens O, Liu J, Niu J, Cappelle K, Caccia S, et al. Delivery of dsRNA for RNAi in Insects: An Overview and Future Directions. Insect Sci (2013) 20(1):4–14. doi: 10.1111/j.1744-7917.2012.01534.x
105. Blair CD, Olson KE. Mosquito Immune Responses to Arbovirus Infections. Curr Opin Insect Sci (2014) 3:22–9. doi: 10.1016/j.cois.2014.07.005
106. Bronkhorst AW, van Rij RP. The Long and Short of Antiviral Defense: Small RNA-Based Immunity in Insects. Curr Opin Virol (2014) 7:19–28. doi: 10.1016/j.coviro.2014.03.010
107. Karlikow M, Goic B, Saleh MC. RNAi and Antiviral Defense in Drosophila: Setting Up a Systemic Immune Response. Dev Comp Immunol (2014) 42(1):85–92. doi: 10.1016/j.dci.2013.05.004
108. Sim S, Jupatanakul N, Dimopoulos G. Mosquito Immunity Against Arboviruses. Viruses (2014) 6(11):4479–504. doi: 10.3390/v6114479
109. Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, et al. The DExD/H-Box Helicase Dicer-2 Mediates the Induction of Antiviral Activity in Drosophila. Nat Immunol (2008) 9(12):1425–32. doi: 10.1038/ni.1664
110. Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ. Secreted Vago Restricts West Nile Virus Infection in Culex Mosquito Cells by Activating the Jak-STAT Pathway. Proc Natl Acad Sci United States America (2012) 109(46):18915–20. doi: 10.1073/pnas.1205231109
111. Hoffmann JA, Hoffmann D. The Inducible Antibacterial Peptides of Dipteran Insects. Res Immunol (1990) 141(9):910–8. doi: 10.1016/0923-2494(90)90192-2
112. Steiner H, Hultmark D, Engström A, Bennich H, Boman HG. Sequence and Specificity of Two Antibacterial Proteins Involved in Insect Immunity. Nature (1981) 292(5820):246–8. doi: 10.1038/292246a0
113. Vizioli J, Bulet P, Hoffmann JA, Kafatos FC, Müller HM, Dimopoulos G. Gambicin: A Novel Immune Responsive Antimicrobial Peptide From the Malaria Vector Anopheles Gambiae. Proc Natl Acad Sci USA (2001) 98(22):12630–5. doi: 10.1073/pnas.221466798
114. He Y, Cao X, Li K, Hu Y, Chen YR, Blissard G, et al. A Genome-Wide Analysis of Antimicrobial Effector Genes and Their Transcription Patterns in Manduca Sexta. Insect Biochem Mol Biol (2015) 62:23–37. doi: 10.1016/j.ibmb.2015.01.015
115. Imler JL. Overview of Drosophila Immunity: A Historical Perspective. Dev Comp Immunol (2014) 42(1):3–15. doi: 10.1016/j.dci.2013.08.018
116. Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect Antimicrobial Peptides and Their Applications. Appl Microbiol Biotechnol (2014) 98(13):5807–22. doi: 10.1007/s00253-014-5792-6
117. Elmogy M, Bassal TT, Yousef HA, Dorrah MA, Mohamed AA, Duvic B. Isolation, Characterization, Kinetics, and Enzymatic and Nonenzymatic Microbicidal Activities of a Novel C-Type Lysozyme From Plasma of Schistocerca Gregaria (Orthoptera: Acrididae). J Insect Sci (2015) 15(1):57. doi: 10.1093/jisesa/iev038
118. Kwon H, Bang K, Lee M, Cho S. Molecular Cloning and Characterization of a Lysozyme cDNA From the Mole Cricket Gryllotalpa Orientalis (Orthoptera: Gryllotalpidae). Mol Biol Rep (2014) 41(9):5745–54. doi: 10.1007/s11033-014-3446-5
119. Kajla MK, Shi L, Li B, Luckhart S, Li J, Paskewitz SM. A New Role for an Old Antimicrobial: Lysozyme C-1 can Function to Protect Malaria Parasites in Anopheles Mosquitoes. PloS One (2011) 6(5):e19649. doi: 10.1371/journal.pone.0019649
120. Sowa-Jasiłek A, Zdybicka-Barabas A, Stączek S, Wydrych J, Mak P, Jakubowicz T, et al. Studies on the Role of Insect Hemolymph Polypeptides: Galleria Mellonella Anionic Peptide 2 and Lysozyme. Peptides (2014) 53:194–201. doi: 10.1016/j.peptides.2014.01.012
121. Li B, Paskewitz SM. A Role for Lysozyme in Melanization of Sephadex Beads in Anopheles Gambiae. J Insect Physiol (2006) 52(9):936–42. doi: 10.1016/j.jinsphys.2006.06.002
122. Rao XJ, Ling E, Yu XQ. The Role of Lysozyme in the Prophenoloxidase Activation System of Manduca Sexta: An In Vitro Approach. Dev Comp Immunol (2010) 34(3):264–71. doi: 10.1016/j.dci.2009.10.004
123. Casati B, Terova G, Cattaneo AG, Rimoldi S, Franzetti E, de Eguileor M, et al. Molecular Cloning, Characterization and Expression Analysis of ATG1 in the Silkworm, Bombyx Mori. Gene (2012) 511(2):326–37. doi: 10.1016/j.gene.2012.09.086
124. Tettamanti G, Malagoli D, Marchesini E, Congiu T, de Eguileor M, Ottaviani E. Oligomycin A Induces Autophagy in the IPLB-LdFB Insect Cell Line. Cell Tissue Res (2006) 326(1):179–86. doi: 10.1007/s00441-006-0206-4
125. Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic Control of Listeria Through Intracellular Innate Immune Recognition in Drosophila. Nat Immunol (2008) 9(8):908–16. doi: 10.1038/ni.1634
126. Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, et al. Virus Recognition by Toll-7 Activates Antiviral Autophagy in Drosophila. Immunity (2012) 36(4):658–67. doi: 10.1016/j.immuni.2012.03.003
127. Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an Essential Component of Drosophila Immunity Against Vesicular Stomatitis Virus. Immunity (2009) 30(4):588–98. doi: 10.1016/j.immuni.2009.02.009
128. Patel RK, Hardy RW. Role for the Phosphatidylinositol 3-Kinase-Akt-TOR Pathway During Sindbis Virus Replication in Arthropods. J Virol (2012) 86(7):3595–604. doi: 10.1128/JVI.06625-11
129. Cao X, He Y, Hu Y, Wang Y, Chen YR, Bryant B, et al. The Immune Signaling Pathways of Manduca Sexta. Insect Biochem Mol Biol (2015) 62:64–74. doi: 10.1016/j.ibmb.2015.03.006
130. Clem RJ. Baculoviruses and Apoptosis: A Diversity of Genes and Responses. Curr Drug Targets (2007) 8(10):1069–74. doi: 10.2174/138945007782151405
131. Vaidyanathan R, Scott TW. Apoptosis in Mosquito Midgut Epithelia Associated With West Nile Virus Infection. Apoptosis (2006) 11(9):1643–51. doi: 10.1007/s10495-006-8783-y
132. Wang H, Gort T, Boyle DL, Clem RJ. Effects of Manipulating Apoptosis on Sindbis Virus Infection of Aedes Aegypti Mosquitoes. J Virol (2012) 86(12):6546–54. doi: 10.1128/JVI.00125-12
133. Nainu F, Tanaka Y, Shiratsuchi A, Nakanishi Y. Protection of Insects Against Viral Infection by Apoptosis-Dependent Phagocytosis. J Immunol (Baltimore Md 1950) (2015) 195(12):5696–706. doi: 10.4049/jimmunol.1500613
134. Zhang X, Aksoy E, Girke T, Raikhel AS, Karginov FV. Transcriptome-Wide microRNA and Target Dynamics in the Fat Body During the Gonadotrophic Cycle of Aedes Aegypti. Proc Natl Acad Sci (2017) 114(10):E1895–E903. doi: 10.1111/nyas.13381
135. Yang S, Zhang J, Wang S, Zhang X, Liu Y, Xi J. Identification and Profiling of miRNAs in Overwintering Lissorhoptrus Oryzophilus via Next-Generation Sequencing. Cryobiology (2017) 74:68–76. doi: 10.1016/j.cryobiol.2016.11.013
136. Mukherjee K, Amsel D, Kalsy M, Billion A, Dobrindt U, Vilcinskas A. MicroRNAs Regulate Innate Immunity Against Uropathogenic and Commensal-Like Escherichia Coli Infections in the Surrogate Insect Model Galleria Mellonella. Sci Rep (2020) 10(1):2570. doi: 10.1038/s41598-020-59407-3
137. Li R, Yao X, Zhou H, Jin P, Ma F. The Drosophila miR-959-962 Cluster Members Repress Toll Signaling to Regulate Antibacterial Defense During Bacterial Infection. Int J Mol Sci (2021) 22(2):886. doi: 10.3390/ijms22020886
138. Zhang JH, Dong Y, Wu W, Yi DS, Wang M, Wang HT, et al. Comprehensive Identification and Characterization of Long Non-Coding RNAs Associated With Rice Black-Streaked Dwarf Virus Infection in Laodelphax Striatellus (Fallén) Midgut. Front Physiol (2020) 11:1011. doi: 10.3389/fphys.2020.01011
139. Wei G, Sun L, Li R, Li L, Xu J, Ma F. Dynamic miRNA-mRNA Regulations are Essential for Maintaining Drosophila Immune Homeostasis During Micrococcus Luteus Infection. Dev Comp Immunol (2018) 81:210–24. doi: 10.1016/j.dci.2017.11.019
140. Huntzinger E, Izaurralde E. Gene Silencing by microRNAs: Contributions of Translational Repression and mRNA Decay. Nat Rev Genet (2011) 12(2):99–110. doi: 10.1038/nrg2936
141. Li S, Li Y, Shen L, Jin P, Chen L, Ma F. miR-958 Inhibits Toll Signaling and Drosomycin Expression via Direct Targeting of Toll and Dif in Drosophila Melanogaster. Am J Physiol Cell Physiol (2017) 312(2):C103–c10. doi: 10.1152/ajpcell.00251.2016
142. Li R, Huang Y, Zhang Q, Zhou H, Jin P, Ma F. The miR-317 Functions as a Negative Regulator of Toll Immune Response and Influences Drosophila Survival. Dev Comp Immunol (2019) 95:19–27. doi: 10.1016/j.dci.2019.01.012
143. Fricke C, Green D, Smith D, Dalmay T, Chapman T. MicroRNAs Influence Reproductive Responses by Females to Male Sex Peptide in Drosophila Melanogaster. Genetics (2014) 198(4):1603–19. doi: 10.1534/genetics.114.167320
144. Pushpavalli SN, Sarkar A, Bag I, Hunt CR, Ramaiah MJ, Pandita TK, et al. Argonaute-1 Functions as a Mitotic Regulator by Controlling Cyclin B During Drosophila Early Embryogenesis. FASEB J (2014) 28(2):655–66. doi: 10.1096/fj.13-231167
145. Yang H, Li M, Hu X, Xin T, Zhang S, Zhao G, et al. MicroRNA-Dependent Roles of Drosha and Pasha in the Drosophila Larval Ovary Morphogenesis. Dev Biol (2016) 416(2):312–23. doi: 10.1016/j.ydbio.2016.06.026
146. Li Y, Li S, Li R, Xu J, Jin P, Chen L, et al. Genome-Wide miRNA Screening Reveals miR-310 Family Members Negatively Regulate the Immune Response in Drosophila Melanogaster via Co-Targeting Drosomycin. Dev Comp Immunol (2017) 68:34–45. doi: 10.1016/j.dci.2016.11.014
147. Ma L, Liu L, Zhao Y, Yang L, Chen C, Li Z, et al. JNK Pathway Plays a Key Role in the Immune System of the Pea Aphid and is Regulated by microRNA-184. PloS Pathog (2020) 16(6):e1008627. doi: 10.1371/journal.ppat.1008627
148. Heitmueller M, Billion A, Dobrindt U, Vilcinskas A, Mukherjee K. Epigenetic Mechanisms Regulate Innate Immunity Against Uropathogenic and Commensal-Like Escherichia Coli in the Surrogate Insect Model Galleria Mellonella. Infection Immun (2017) 85(10). doi: 10.1128/IAI.00336-17
149. Gao K, Deng XY, Qian HY, Wu P, Qin GX, Liu T, et al. Novel Protein of IBP From Silkworm, Bombyx Mori, Involved in Cytoplasmic Polyhedrosis Virus Infection. J invertebrate Pathol (2012) 110(1):83–91. doi: 10.1016/j.jip.2012.02.011
150. Wu P, Qin G, Qian H, Chen T, Guo X. Roles of miR-278-3p in IBP2 Regulation and Bombyx Mori Cytoplasmic Polyhedrosis Virus Replication. Gene (2016) 575(2 Pt 1):264–9. doi: 10.1016/j.gene.2015.09.009
151. Fries I, Feng F, da Silva A, Slemenda SB, Pieniazek NJ. Nosema Ceranae N. Sp.(Microspora, Nosematidae), Morphological and Molecular Characterization of a Microsporidian Parasite of the Asian Honey Bee Apis Cerana (Hymenoptera, Apidae). Eur J Protistology (1996) 32(3):356–65. doi: 10.1016/S0932-4739(96)80059-9
152. Chen D, Chen H, Du Y, Zhou D, Geng S, Wang H, et al. Genome-Wide Identification of Long Non-Coding RNAs and Their Regulatory Networks Involved in Apis Mellifera Ligustica Response to Nosema Ceranae Infection. Insects (2019) 10(8):245. doi: 10.3390/insects10080245
153. Zhou H, Ni J, Wu S, Ma F, Jin P, Li S. lncRNA-CR46018 Positively Regulates the Drosophila Toll Immune Response by Interacting With Dif/Dorsal. Dev Comp Immunol (2021) 124:104183. doi: 10.1016/j.dci.2021.104183
154. Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. Dengue Virus Type 2: Replication and Tropisms in Orally Infected Aedes Aegypti Mosquitoes. BMC Microbiol (2007) 7:9. doi: 10.1186/1471-2180-7-9
155. Richardson J, Molina-Cruz A, Salazar MI. Black Wt. Quantitative Analysis of Dengue-2 Virus RNA During the Extrinsic Incubation Period in Individual Aedes Aegypti. Am J Trop Med hygiene (2006) 74(1):132–41. doi: 10.4269/AJTMH.2006.74.132
156. Li H, Li WX, Ding SW. Induction and Suppression of RNA Silencing by an Animal Virus. Sci (New York NY) (2002) 296(5571):1319–21. doi: 10.1126/science.1070948
157. Roth BM, Pruss GJ, Vance VB. Plant Viral Suppressors of RNA Silencing. Virus Res (2004) 102(1):97–108. doi: 10.1016/j.virusres.2004.01.020
158. Cui C, Wang Y, Liu J, Zhao J, Sun P, Wang S. A Fungal Pathogen Deploys a Small Silencing RNA That Attenuates Mosquito Immunity and Facilitates Infection. Nat Commun (2019) 10(1):4298. doi: 10.1038/s41467-019-12323-1
159. Zhang L, Xu W, Gao X, Li W, Qi S, Guo D, et al. lncRNA Sensing of a Viral Suppressor of RNAi Activates Non-Canonical Innate Immune Signaling in Drosophila. Cell Host Microbe (2020) 27(1):115–28.e8. doi: 10.1016/j.chom.2019.12.006
160. Zhan S, Dong Y, Zhao W, Guo J, Zhong T, Wang L, et al. Genome-Wide Identification and Characterization of Long Non-Coding RNAs in Developmental Skeletal Muscle of Fetal Goat. BMC Genomics (2016) 17(1):666. doi: 10.1186/s12864-016-3009-3
161. Ran MX, Li Y, Zhang Y, Liang K, Ren YN, Zhang M, et al. Transcriptome Sequencing Reveals the Differentially Expressed lncRNAs and mRNAs Involved in Cryoinjuries in Frozen-Thawed Giant Panda (Ailuropoda Melanoleuca) Sperm. Int J Mol Sci (2018) 19(10):3066. doi: 10.3390/ijms19103066
162. Kennedy AR. Chemopreventive Agents: Protease Inhibitors. Pharmacol Ther (1998) 78(3):167–209. doi: 10.1016/S0163-7258(98)00010-2
163. Bruno R, Sacchi P, Maiocchi L, Patruno S, Filice G. Hepatotoxicity and Antiretroviral Therapy With Protease Inhibitors: A Review. Digestive liver Dis (2006) 38(6):363–73. doi: 10.1016/j.dld.2006.01.020
164. Zhang Z, Zhao Z, Lin S, Wu W, Tang W, Dong Y, et al. Identification of Long Noncoding RNAs in Silkworm Larvae Infected With Bombyx Mori Cypovirus. Arch Insect Biochem Physiol (2021) 106(3):1–12. doi: 10.1002/arch.21777
165. Dong Z, Zheng N, Hu C, Deng B, Fang W, Wu Q, et al. Nosema Bombycis microRNA-Like RNA 8 (Nb-Milr8) Increases Fungal Pathogenicity by Modulating BmPEX16 Gene Expression in Its Host, Bombyx Mori. Microbiol Spectr (2021) 9(2):e0104821-e. doi: 10.1128/Spectrum.01048-21
166. Lin S, Wang Y, Zhao Z, Wu W, Su Y, Zhang Z, et al. Two Putative Cypovirus-Encoded miRNAs Co-Regulate the Host Gene of GTP-Binding Nuclear Protein Ran and Facilitate Virus Replication. Front Physiol (2021) 12:663482. doi: 10.3389/fphys.2021.663482
167. Wang Y, Lin S, Zhao Z, Xu P, Gao K, Qian H, et al. Functional Analysis of a Putative Bombyx Mori Cypovirus miRNA BmCPV-miR-10 and its Effect on Virus Replication. Insect Mol Biol (2021) 30(6):552–65. doi: 10.1111/imb.12725
168. Guo JY, Wang YS, Chen T, Jiang XX, Wu P, Geng T, et al. Functional Analysis of a miRNA-Like Small RNA Derived From Bombyx Mori Cytoplasmic Polyhedrosis Virus. Insect Sci (2020) 27(3):449–62. doi: 10.1111/1744-7917.12671
169. Feng H, Xu M, Gao Y, Liang J, Guo F, Guo Y, et al. Vm-Milr37 Contributes to Pathogenicity by Regulating Glutathione Peroxidase Gene VmGP in Valsa Mali. Mol Plant Pathol (2021) 22(2):243–54. doi: 10.1111/mpp.13023
170. Uhse S, Djamei A. Effectors of Plant-Colonizing Fungi and Beyond. PloS Pathog (2018) 14(6):e1006992. doi: 10.1371/journal.ppat.1006992
171. Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, et al. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Sci (New York NY) (2013) 342(6154):118–23. doi: 10.1126/science.1239705
172. Saetrom P, Heale BS, Snøve O Jr., Aagaard L, Alluin J, Rossi JJ. Distance Constraints Between microRNA Target Sites Dictate Efficacy and Cooperativity. Nucleic Acids Res (2007) 35(7):2333–42. doi: 10.1093/nar/gkm133
173. Fang Z, Rajewsky N. The Impact of miRNA Target Sites in Coding Sequences and in 3'utrs. PloS One (2011) 6(3):e18067. doi: 10.1371/journal.pone.0018067
174. Liu G, Zhang R, Xu J, Wu CI, Lu X. Functional Conservation of Both CDS- and 3'-UTR-Located microRNA Binding Sites Between Species. Mol Biol Evol (2015) 32(3):623–8. doi: 10.1093/molbev/msu323
175. Singh CP, Singh J, Nagaraju J. A Baculovirus-Encoded MicroRNA (miRNA) Suppresses its Host miRNA Biogenesis by Regulating the Exportin-5 Cofactor Ran. J Virol (2012) 86(15):7867–79. doi: 10.1128/JVI.00064-12
176. Bischoff FR, Ponstingl H. Catalysis of Guanine Nucleotide Exchange on Ran by the Mitotic Regulator RCC1. Nature (1991) 354(6348):80–2. doi: 10.1038/354080a0
177. Bergmann JH, Spector DL. Long Non-Coding RNAs: Modulators of Nuclear Structure and Function. Curr Opin Cell Biol (2014) 26:10–8. doi: 10.1016/j.ceb.2013.08.005
178. Guo R, Chen D, Xiong C, Hou C, Zheng Y, Fu Z, et al. First Identification of Long Non-Coding RNAs in Fungal Parasite Nosema Ceranae. Apidologie (2018) 49(5):660–70. doi: 10.1007/s13592-018-0593-z
179. Masri L, Cremer S. Individual and Social Immunisation in Insects. Trends Immunol (2014) 35(10):471–82. doi: 10.1016/j.it.2014.08.005
180. Betting V, Van Rij RP. Countering Counter-Defense to Antiviral RNAi. Trends Microbiol (2020) 28(8):600–2. doi: 10.1016/j.tim.2020.05.018
181. Tang CK, Tsai CH, Wu CP, Lin YH, Wei SC, Lu YH, et al. MicroRNAs From Snellenius Manilae Bracovirus Regulate Innate and Cellular Immune Responses of its Host Spodoptera Litura. Commun Biol (2021) 4(1):52. doi: 10.1038/s42003-020-01563-3
182. Etebari K, Asgari S. Conserved microRNA miR-8 Blocks Activation of the Toll Pathway by Upregulating Serpin 27 Transcripts. RNA Biol (2013) 10(8):1356–64. doi: 10.4161/rna.25481
Keywords: insect immune pathways, insect–pathogen interaction, miRNAs and lncRNAs, mRNA targets, immune modulation
Citation: Moure UAE, Tan T, Sha L, Lu X, Shao Z, Yang G, Wang Y and Cui H (2022) Advances in the Immune Regulatory Role of Non-Coding RNAs (miRNAs and lncRNAs) in Insect-Pathogen Interactions. Front. Immunol. 13:856457. doi: 10.3389/fimmu.2022.856457
Received: 17 January 2022; Accepted: 10 March 2022;
Published: 06 April 2022.
Edited by:
Zhen Zou, Institute of Zoology (CAS), ChinaReviewed by:
Pei Liang, China Agricultural University, ChinaCopyright © 2022 Moure, Tan, Sha, Lu, Shao, Yang, Wang and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guang Yang, MTY0MDgyMzUxOEBxcS5jb20=; Yi Wang, MzA0ODk3ODYxOUBxcS5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.