
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 25 February 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.852702
This article is part of the Research TopicThe Function of Phagocytes in Non-Mammals - Volume IIView all 5 articles
Within the hemolymph, insect hemocytes constitute a heterogeneous population of macrophage-like cells that play important roles in innate immunity, homeostasis and development. Classification of hemocytes in different subtypes by size, morphology and biochemical or immunological markers has been difficult and only in Drosophila extensive genetic analysis allowed the construction of a coherent picture of hemocyte differentiation from pro-hemocytes to granulocytes, crystal cells and plasmatocytes. However, the advent of high-throughput single cell technologies, such as single cell RNA sequencing (scRNA-seq), is bound to have a high impact on the study of hemocytes subtypes and their phenotypes in other insects for which a sophisticated genetic toolbox is not available. Instead of averaging gene expression across all cells as occurs in bulk-RNA-seq, scRNA-seq allows high-throughput and specific visualization of the differentiation status of individual cells. With scRNA-seq, interesting cell types can be identified in heterogeneous populations and direct analysis of rare cell types is possible. Next to its ability to profile the transcriptomes of individual cells in tissue samples, scRNA-seq can be used to propose marker genes that are characteristic of different hemocyte subtypes and predict their functions. In this perspective, the identities of the different marker genes that were identified by scRNA-seq analysis to define 13 distinct cell clusters of hemocytes in larvae of the silkworm, Bombyx mori, are discussed in detail. The analysis confirms the broad division of hemocytes in granulocytes, plasmatocytes, oenocytoids and perhaps spherulocytes but also reveals considerable complexity at the molecular level and highly specialized functions. In addition, predicted hemocyte marker genes in Bombyx generally show only limited convergence with the genes that are considered characteristic for hemocyte subtypes in Drosophila.
The blood cells or hemocytes play an important role in the regulation of whole-body homeostasis in insects. While originally implicated in the cellular response against microorganisms and parasites, functions of hematocytes have continuously expanded to include tissue remodeling during development, wound repair, storage and distribution of nutrients and the humoral immune response (1, 2). In their motility and versatility to respond to various signals insect hemocytes resemble the myeloid-like blood cells in vertebrates (2, 3).
The diversity of hemocytes has been mostly studied in lepidopteran insects and in Drosophila (4–7). Despite the different nomenclature used, differentiated and functional hemocytes can be generally divided in three main types by morphological and functional criteria (8, 9).
The first category constitutes “macrophage-like cells” that are named plasmatocytes in Drosophila and correspond to granular hemocytes (granulocytes) in Lepidoptera (8). The main function of the macrophage-like cells is phagocytosis; while generally belonging to the same category, lepidopteran granulocytes differ from Drosophila plasmatocytes by the presence of specialized inclusions (granules) that are released upon activation (10, 11).
Cells that exhibit extensive spreading behavior and form capsules around large foreign bodies or nodules around bacteria and fungi are called lamellocytes in Drosophila. Confusingly, hemocytes with similar properties are called plasmatocytes in Lepidoptera (8). However, so-called plasmatocytes form a permanent part of the circulating hemocyte population in Lepidoptera (1), while lamellocytes are mainly observed in the context of parasitization in Drosophila (12), indicating functional differences.
Oenocytoids form a group of fragile cells in Lepidoptera that are characterized by the production of phenoloxidase that is responsible for melanization reactions in the hemolymph (1). In Drosophila, crystal cells show highly similar cytological and biochemical features (13) and therefore can be considered the equivalent of oenocytoids (8).
Other hemocyte types occur with lower abundance in the hemolymph. Pro-hemocytes have a round shape with a high nucleus-to-cytoplasm ratio and are considered the precursor cells of the other hemocyte types in both Lepidoptera and Drosophila (14, 15). Spherule cells or spherulocytes are recognized as another differentiated hemocyte cell type in Lepidoptera (with unknown function) but do not occur in Drosophila (16). Other rare specialized hemocyte cell types may appear in the hemolymph under specific conditions (8, 17).
The technique of single cell RNA sequencing (scRNA-seq) has emerged as a powerful technique for the characterization of cell heterogeneity and the identification of individual cell types in tissues containing multiple types of cells (18, 19). After identification of cell groups by clustering approaches, differential expression analysis can be performed in conjunction with functional profiling for the identification of biomarkers that are characteristic for the detected cell groups (20). Construction of the cell atlas and the definition of corresponding marker genes are considered a valuable resource for the execution of follow-up functional studies in various biological processes (21).
The application of scRNA-seq on blood cells has the advantage of straightforward isolation of single cells that does not require prior digestion as is the case for solid tissues (22) and was used successfully to characterize hemocyte populations in the hemolymph of Drosophila larvae (4, 5, 23). Based on differential gene expression levels, 16 clusters or subpopulations were identified, which were found to broadly correspond to the hemocyte subtypes identified by morphological and physiological criteria: 12-13 clusters of (Drosophila) plasmatocytes, 1-2 clusters of crystal cells, and 2 clusters of lamellocytes (reported in two independent studies: 4, 5). Most notably, the assignment of clusters to belong to the three hemocyte types was guided using known markers such as lozenge for crystal cells, NimC1 for plasmatocytes and atilla for lamellocytes (4, 5).
The 16 clusters/subpopulations of hemocytes in Drosophila larvae showed different abundances according to three experimental conditions: unwounded, wounded and parasitoid wasp-infected (5). As expected, the two clusters representing lamellocytes were absent in hemolymph of unwounded larvae (4). Another study employing scRNA-seq focused on the lymph gland or the larval hematopoietic organ of Drosophila and identified new hemocyte types including adipohemocytes and different subtypes of pro-hemocytes (23).
The technique of scRNA-seq was also applied to hemocytes of silkworm (Bombyx mori, Lepidoptera) (24). In this study, hemocytes were isolated from 5th instar larvae at three days after experimental manipulation which consisted either of (1) injection into the body cavity of a high infectious dose of B. mori nucleopolyhedrovirus (BmNPV) (baculovirus infection) or (2) injection of saline solution (wounding). After normalization, gene expression data from 22,286 cells were subjected to dimensionality reduction methods such as component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) (25). The R package Seurat (26) was used as a graph-based clustering method to obtain 20 hemocyte cell type clusters and to screen for marker genes in each cluster (24).
The cluster analysis of scRNA-seq of Bombyx hemocytes revealed that cell clusters associated with baculovirus infection (7 clusters: 1, 2, 3, 9, 11, 13, 18) were clearly separated from the cell clusters of the (wounded) control (9 clusters: 0, 4, 5, 7, 8, 10, 12, 16, 17), which reflects the severe impact of baculovirus infection on the composition of the hemocyte population (24). In 4 clusters (6, 14, 15, 19), the contribution from both control and BmNPV-infected groups is less polarized (10-30% contributed by cells from BmNPV-infected group) (Table 1).
The cell clusters that are predominant in the BmNPV-infected group are characterized by much lower number of differentially expressed genes (DEGs) (range: 98 to 181) compared to the control group (range: 170 to 1571) (Table 1) and their morphological features resemble those of pro-hemocytes (24). It was therefore hypothesized that the BmNPV-infected cells in clusters 1, 2, 3, 9, 11, 13 and 18 represent pro-hemocytes that were released from the hematopoietic organs in Bombyx larvae to replenish the loss of differentiated hemocytes (granulocytes, plasmatocytes, oenocytoids) in the hemolymph following baculovirus infection (24).
For the (wounded) control group of hemocyte clusters in B. mori, attempts were made to assign the clusters to the classic morphological and cytochemical categories of plasmatocytes, granulocytes and oenocytoids of lepidopteran larvae (8). Based on markers found in the literature, clusters 5, 8, 12 and 16 were identified as oenocytoids (expressing PPBP1 and PPBP2, encoding paralytic peptide-binding proteins; 16), clusters 14 and 15 were assigned as (lepidopteran) plasmatocytes (marker genes: serine protease homolog1, β-1,3-glucan recognition protein 3, paralytic peptide and integrin β3; 16, 29), and clusters 0, 4, 6, 7, 10, 17 were tentatively designated as granulocytes (based on the expression of scavenger receptor-C, cathepsin B, integrin α3, hemocyte protease-1 and peptidoglycan recognition protein precursor; 16, 30). In addition, cluster 19 was labeled as spherulocyte-like based on the preferential expression of cathepsin L-like (16).
Nevertheless, the different lineages of hemocytes and their developmental pathways are much less well understood in Bombyx than in Drosophila, and the assignment of clusters to different hemocyte types in Bombyx can only be regarded as tentative and will require further validation. Concurrently, many new marker genes were predicted based on differential gene expression and functional profiling in the designated clusters, which were not discussed in detail in the first article that presented the definition of hemocyte clusters in B. mori larvae following scRNA-seq analysis (24). Recently, lectins and monoclonal antibodies have allowed a more robust classification of hemocytes types in lepidopteran insects (7, 31) but the identity and the function of the marker molecules that are detected largely remain unknown. Sequences of marker genes assigned by scRNA-seq to specific clusters, on the other hand, contain sufficient information to allow the prediction of their function based on knowledge from better studied model organisms, most notably Drosophila, for which scRNA-seq data of hemocyte types are already available (4, 5, 23). In the analysis that follows, an attempt is made to predict the function of the hemocyte types in B. mori larvae based on the marker genes that were proposed following scRNA-seq analysis.
In the previous article (24), a likelihood-ratio test (32), performed on single cluster cells against all other cells, was used to identify DEGs in single silkworm hemocyte clusters based on differential expression. Up-regulated DEGs in each cluster were identified by the following criteria: 1) p value ≤ 0.01; 2) log2 fold change (log2FC) ≥ 0.360674 (log2FC means log fold change of the average expression between the two groups); and 3) percentage of cells in a specific cluster where the gene is detected > 25%. The top genes in each silkworm hemocyte cluster were then selected as the potential marker genes for each cluster.
To infer the function of proposed marker genes, a literature search was performed with as key word the name of the gene in association with terms such as “hemocyte” and “innate immunity” and species or taxon names such as “Drosophila”, “Bombyx mori”, “Lepidoptera” and “insects”. If no gene name was provided on the database SilkDB 3.0 (33), protein sequences were subjected to BLAST searches as well as the HHpred interactive server for remote protein homology detection and structure prediction (34). The predicted function of the 5-6 top marker genes was used to construct the main activity of the cells in the identified clusters.
The datasets analyzed in this study can be found in online repositories at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA658439.
Although in the analysis of the scRNA-seq of silkworm hemocytes 20 clusters or subpopulations were identified, 7 of these clusters consisted of cells that were heavily infected by baculovirus (> 96%; Table 1; 24). An overview of the identified clusters (UMAP plot; 24) together with their proposed subtype identity (granulocyte, oenocytoid, plasmatocyte, spherulocyte) is presented in Figure 1. Pseudo-temporal ordering and morphological staining led to the proposal that the clusters in the BmNPV-infected group could correspond to pro-hemocytes that were released from the hematopoietic organs as a response to the depletion of differentiated hemocyte cell types by baculovirus infection (clusters referred to as “others” in Figure 1; 24). However, it can be assumed that baculovirus infection has a major impact on the physiology of the cells and that the transcriptome analysis may not provide a clear picture of the characteristics of pro-hemocytes in healthy silkworms. Another transcriptome analysis revealed large changes in infected hemocytes regarding amino-acid, carbohydrate, nucleic acid and lipid metabolism, conform to the expectation that infected hemocytes are transformed into virion production factories (35). Because of this limitation, the analysis is focused on the 13 clusters that encompass the differentiated cell types of the silkworm hemocyte population (granulocytes, oenocytoids, plasmatocytes and spherulocytes).
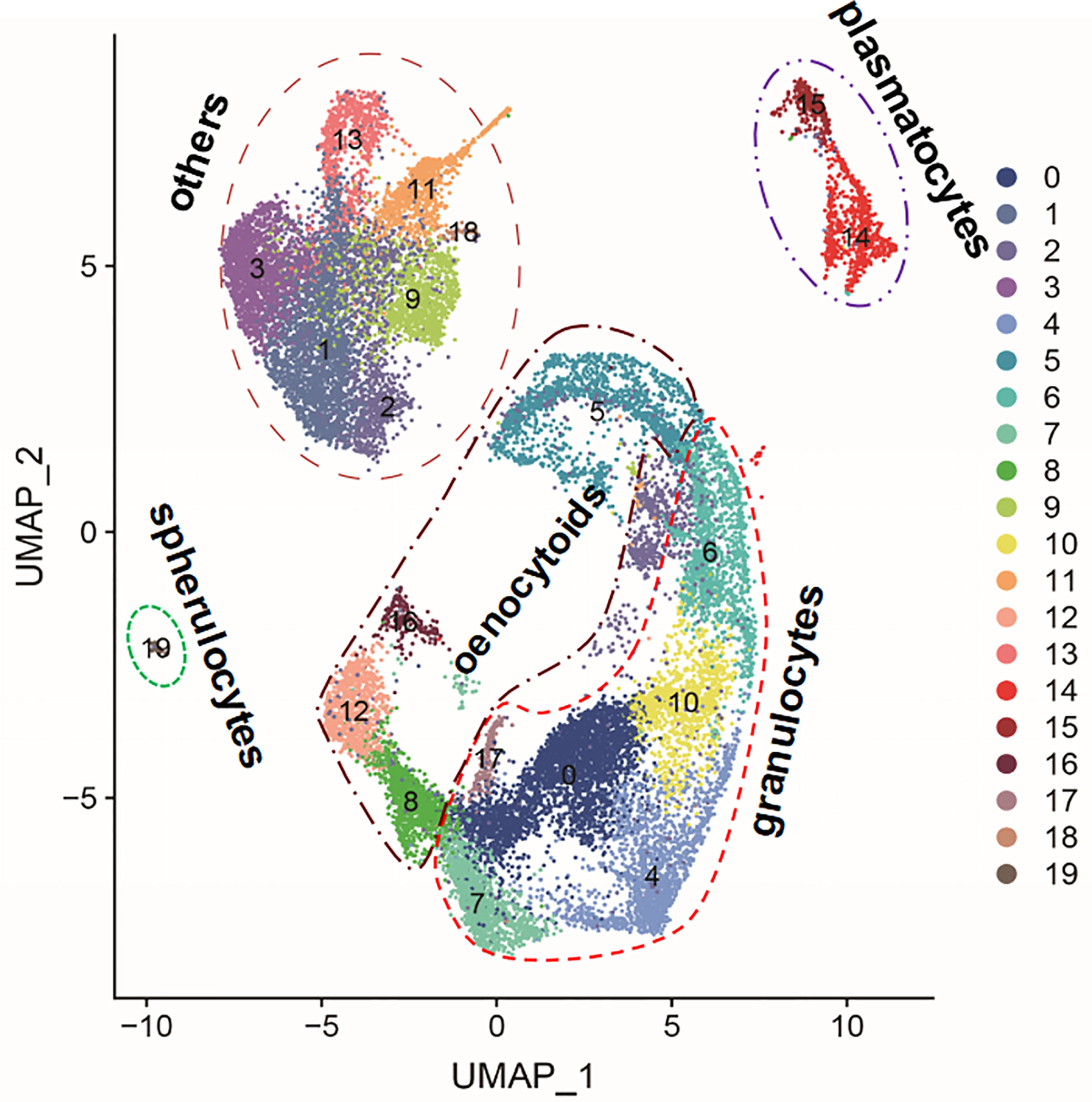
Figure 1 Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plot of the 20 hemocyte clusters identified in silkworm larvae by scRNA-seq (24). Indicated is the grouping of the clusters in the four differentiated hemocyte cell types that were proposed in the literature (granulocytes: clusters 0, 4, 6, 7, 10 and 17; plasmatocytes: clusters 14 and 15; oenocytoids: clusters 5, 8, 12 and 16; and spherulocytes: cluster 19). Clusters that were heavily infected by baculovirus (>96%; Table 1) (others: clusters 1, 2, 3, 9, 11, 13 and 18) were not included in the analysis.
Cluster 0 represents the largest group of hemocytes that was separated based on gene expression levels. The levels of differential expression are relatively mild (log2FC < 2.7) which is indicative of an intermediary cell type. The selected marker genes of cluster 0 (Table 2) reflect the function of hemocytes in tissue remodeling, phagocytosis, regulation of oxidant stress and possibly melanization.
Aliphatic nitrilase with β-ureidopropionase activity is involved in the catabolism of pyrimidine and the production of β-alanine (36) that has a role in cuticular tanning and sclerotization (37). The relatively high levels of 5-hydroxy-tryptamine (serotonin) receptor may reflect the regulation of phagocytosis by serotonin, as is observed in Drosophila (38, 39). Interestingly, another gene with high log2FC ranking is characterized with a LPS-induced TNF-activating factor (LITAF) domain that mediates targeting to intracellular membranes to regulate endosomal trafficking (40). In the malaria mosquito Anopheles gambiae, LITAF-like factors play a role in the regulation of phagocytosis (41) and are required for hemocyte differentiation into granulocytes and oenocytoids during invasion of the haemocoel by the malarial parasite (42).
Three other markers of this cluster may have a role in antioxidant signaling and stress protection. Such a role was already indicated for apolipoprotein D in the silkworm (43). Farnesyl diphosphate synthase is responsible for the synthesis of isoprenoids of which coenzyme Q may be of relevance because of its antioxidant activity and protection against reactive oxygen species (ROS) (44, 45). Finally, glutathione S-transferases (GST) of the zeta class are thought to represent an ancestral class of GST enzymes that play an important role in intermediary metabolism, i.e. the catabolism of tyrosine and phenylalanine (46, 47). Also this enzyme is thought to have a function in the protection against oxidant stress (48); moreover, by regulating tyrosine levels, it could play a role in the process of melanization (49).
All the above marker genes are also expressed in cluster 4, albeit with considerably lower ranking in log2FC values. This could indicate a developmental relationship between clusters 0 and 4. The gene encoding a LITAF-domain is also relatively highly differentially expressed in cells of cluster 15 (log2FC = 2.19).
Cells of cluster 4 contain more strongly upregulated genes (log2FC < 4.3) than several other clusters classified as granulocytes (e.g. clusters 0, 6 and 7) (Table 3). All the identified marker genes in cluster 4 are related to proliferation and mitosis, indicating their correspondence to pro-hemocytes or proliferating granulocytes.
Stathmin is involved in the regulation of microtubule dynamics and acts as a microtubule destabilizing factor (50). As such it is proposed to be involved in the regulation of mitosis in the cell cycle and the cytoskeleton (51). Similarly, mitotic spindle assembly checkpoint protein MAD2A is a key component of the spindle assembly checkpoint during mitosis which is essential to maintain genomic stability during cell division (52). Ncd is a kinesin-like protein that is required for chromosome segregation in meiosis and in early mitotic embryonic divisions in Drosophila (53). Another kinesin-like protein, KIF18A, corresponds to a microtubule depolymerase with a role in chromosome congression to form the metaphase plate during mitosis (54). Furthermore, LOC101738390 is a homolog of the mitotic spindle and nuclear protein Mink in Drosophila (55). No clear function was revealed for uncharacterized protein LOC101740936, but HHPred analysis (56) revealed the existence of a short sequence (ENTPPHSH) that is also present in the linker sequence of a peptide that interacts with the catalytic and substrate recognition sites in the CDK2/cyclin A complex, a kinase that is active in the cell cycle S phase (57). However, no substrate recruitment site (RXL) or CDK2/cyclin A consensus phosphorylation site (S/TPKK) motifs were found in the vicinity and the presence of the sequence in uncharacterized protein LOC101738390 may be fortuitous.
During single cell RNAseq of Drosophila hemocytes, Ncd was identified as one of the marker genes of the PL-prolif cluster that are considered the mitotic precursor cells for most of the other hemocytes (4). Interestingly, both mitotic spindle and nuclear protein (mink) and stathmin (stai) were down-regulated in hemocytes following infection with Gram-positive bacteria in Drosophila, indicating that cell-cycle arrest was part of the response (58).
None of the marker genes in cluster 4 was differentially expressed in the remainder of the clusters, with the exception of cluster 14, where Ncd, KIF18A and uncharacterized protein 101740936 were marginally upregulated (log2FC value close to 1 in each case).
Log2FC values of DEGs in cells of cluster 5 range from 0.36 to 3.37, indicating an intermediate level of specialization/differentiation (Table 4).
The cells of cluster 5 are characterized by the significant induction of small heat shock protein (sHSP) genes (4 out of 5 top markers with log2FC from 3.16 to 3.37). Interestingly, the sHSP genes belong to two closely located clusters on chromosome 27, in which other sHSPs (hsp23.7; log2FC = 1.31) and HSPs (HSP68, log2FC = 2.52; HSP83, log2FC = 2.39) are also up-regulated. Additionally, expression of two other sHSP genes, hsp12.2 and α-crystallin B, that are located on chromosome 5, are increased (log2FC = 2.34 and 2.21, respectively).
The family of sHSPs is characterized by a conserved domain of about 100 residues, the α-crystallin domain, at their C-termini (59). sHSPs are abundant in conditions of stress and act as chaperone proteins. Increased production of sHSPs in hemocytes is linked to the protection against oxidative stress, e.g. after wounding or infection (60, 61). The protective function of HSPs relates to the reduction of damage from reactive oxygen species (ROS) that are produced in response to wounding and that play a role in melanization and clotting (62).
The association with ROS production is consistent with the classification of cells of cluster 5 as oenocytoids (24). Consistent with this proposition, increased expression of both prophenoloxidase genes, involved in melanization, is indeed observed (log2FC of 2.44 and 2.76).
The sequence of uncharacterized protein LOC101739676 (118 AA) corresponds to a LU domain or Ly6 module, that consists of approximately 80 amino acids and contains a conserved pattern of cysteine residues forming disulfide bridges to create the three-fingered structural motif (63, 64). The Ly6 module is found in both soluble and membrane-anchored proteins and functions as a protein-protein interaction domain that has been adopted in many different biological processes. Interestingly, a Ly6 module-containing membrane-anchored protein, Retroactive, is required for the organization of chitin fibers at the epidermal surface during cuticle assembly in larvae of Drosophila (65). Another Ly6 protein, Coiled, contributes to septate junction assembly in epithelial tissues as well as in glial cells to insulate the central nervous system from the hemolymph (66, 67). In mammals, Ly6 proteins are also known to modulate nicotinic acetylcholine receptors, voltage-gated potassium channels and Wnt/β-catenin signaling (68–70). The presence of Ly6 proteins in hemocytes is considered consistent with a role in tissue (wound) repair, encapsulation of pathogens and immune signaling.
All the marker genes of cluster 5, as well as HSP68, HSP83 and sHSP12.2 on chromosome 5, are also differentially expressed in cells of cluster 16, which are also tagged as oenocytoids (24), albeit with lower log2FC (range: 1.94 - 2.63) and lower DEG ranking (range: 34 -102). However, α-crystallin B is proposed as the top marker gene of cluster 16 (log2FC = 5.50; see further below). Two out of five marker genes of cluster 5 are also increased in clusters 6, 8 and 12 (the latter two also marked as oenocytoids; 24) but with low log2FC (range: 0.79 – 1.51). In addition, sHSP20.1 on chromosome 27 and sHSP12.2 on chromosome 5 have high log2FC values in cluster 19 (6.08 and 5.10, respectively; corresponding to relatively low respective DEG ranking of 45 and 42).
Also for cluster 6, log2FC values remained moderate and the cluster is therefore proposed to represent a precursor or intermediate cell type (Table 5).
Indeed, all identified marker genes of cluster 6 seem to be related to proliferation and growth. Most conspicuously, the second marker gene encodes Myc, a basic helix-loop-helix (bHLH) transcription factor that regulates the expression of a large number of genes mainly involved in growth and cell proliferation (71). Myc is involved in the regulation of the growth and size of the hematopoietic lymph gland in Drosophila (72) and the Myc pathway is enriched in a subset of pro-hemocytes with stem cell-like properties (PH1) that reside in the lymph gland (23).
Furthermore, the gene encoding another bHLH transcription factor, Enhancer of Split (E(spl), mbeta isoform), had the highest level of differential expression (log2FC = 3.0). bHLH proteins encoded by the E(spl) complex are target genes of Notch signaling that guide cell specification (73). Interestingly, the stem-like pro-hemocytes (PH1 subtype) in Drosophila mentioned above also preferentially express Notch, its ligand Delta, and the E(spl) target genes (23), reinforcing the idea of stem cell-like properties of cluster 6 hemocytes in Bombyx.
Notch signaling also controls crystal cell differentiation (74, 75) and the Notch target gene E(spl)m3-HLH is upregulated in immature cells of the crystal cell lineage (5). It can therefore be proposed that cluster 6 cells may represent the precursor lineage of the oenocytoid-type hemocytes in Bombyx, which are thought to have similar functions as crystal cells in Drosophila (1). However, the characteristic prophenoloxidase marker gene of crystal cells/oenocytoids is not expressed in cluster 6 cells, conform to the immature or intermediate state of the cells. Although cluster 6 hemocytes originally were categorized as granulocytes (24), more validation studies are needed with respect to the differentiation status of this hemocyte cell type.
Other genes that are highly differentially expressed can also be associated with growth and proliferation. Cleavage and polyadenylation specific factor 4 (CPSF4, SNU13) is involved in mRNA 3’-end processing and polyadenylation (76). CPSF4 plays a role in cancer cell survival and proliferation through modulation of signaling pathways (77) and regulation of gene expression (78). Interestingly, depletion of a factor of the CPSF complex could suppress the growth of the lymph gland in Drosophila, possibly through aberrant 3’-end processing of histone mRNAs (79).
Nucleolin is known as one of the most abundant (non-ribosomal) proteins in the nucleolus but can have many other functions, most notably as a histone chaperone to regulate chromatin remodeling and the facilitation of the passage of RNA polymerase II through nucleosome complexes (80, 81). The importance of nucleolar function is underlined by the enrichment of nucleolar pre-rRNA processing protein NIP7 that is involved in ribosomal large subunit biogenesis.
Another protein with high log2FC is FK506-binding protein 45 (FKBP45), which belongs to the immunophilin family of proteins that have peptidylprolyl isomerase activity and function as protein chaperones (82). In Spodoptera Sf9 cells, FKBP46 is the substrate of Sf caspase-1 during activation of apoptosis, which is inhibited by baculovirus-derived p35 protein during infection (83). While homology between Bombyx FKBP45 and Drosophila FKBP39 (closest homolog) seems to be limited to the FK506-binding domain, it was noted that FKBP39 has a central highly charged region (also present in FKBP45) that allows its quarternary organization in large multifunctional complexes (with unknown function) in the nucleolus, at nucleosomes and at microtubules (82). Whether such plasticity and flexibility in function also applies to Bombyx FKBP45, remains to be determined.
Cluster 5, 10 and 14 each have 5 marker genes of cluster 6 as DEGs but generally with much lower log2FC (range: 0.56 – 1.61). Myc, NIP7, nucleolin and FKBP46 are common as DEGs among clusters 5, 10 and 14. SNU13 as DEG is also observed clusters 10 and 14. E(spl) has a relatively high log2FC of 2.35 in cluster 5.
Also for the cells of cluster 7, DEG values are moderate (log2FC < 3.1), indicating an intermediate or possibly regulatory cell type (Table 6). Several identified marker genes (cathepsin B, metalloproteinase inhibitor, carboxylesterase) indicate a function in wound healing or tissue remodeling.
The top marker, cathepsin B, is a member of lysosomal cysteine proteases of the papain superfamily that have the unique capability to act as both endo- and exopeptidases. However, cathepsin B can also be found outside of the lysosomal compartment and be located in the cytosol, at the plasma membrane and in the pericellular environment (84). In mammals, a regulatory function in innate immunity is implicated for cathepsin B because of its role in the processing and trafficking of cytokines such as interleukin-1β and tumor necrosis factor α (85). In the flesh fly, Sarcophaga peregrina, cathepsin B is secreted by pupal hemocytes and causes the dissociation of the fat body during metamorphosis (86). Also in B. mori, expression of cathepsin B in hemocytes was proposed to play a role in the regulation of metamorphosis (87). Thus, cathepsin B produced by cluster 7 is predicted to have a role in tissue remodeling or the innate immune response.
The second marker (“uncharacterized protein LOC101739791”) corresponds to a homolog of the mitochondrial complex I intermediate-associated protein 30 (CIA30) that has been shown in Drosophila to be a chaperone for the assembly of mitochondrial complex I (88). Mitochondrial complex I (NADH:ubiquinone oxidoreductase/NADH dehydrogenase) is also involved in the regulation of oxidative stress and is considered as a main site for ROS production (89) that play a role as signaling molecules to activate various pathways including mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K) (90, 91). ROS signaling has been implicated in several homeostatic processes such as wound healing, cell differentiation and innate immunity (92). During immune defense, ROS can be released as effectors by macrophages to damage cellular structures of invading pathogens (93). The increased expression of an assembly factor for mitochondrial complex I may therefore reflect the increased importance of ROS production by mitochondria in cluster 7 hemocytes in the silkworm.
Two proposed markers are related to the function of metalloproteinases: an inducible physiological metalloproteinase inhibitor and the zinc transporter ZIP3. Metalloproteinases are well known for degrading extracellular matrix proteins (collagens, proteoglycans, laminins) but can also cleave a variety of other extracellular substrates and therefore exhibit pleiotropic roles as regulators of extracellular signaling (94, 95). In Drosophila, matrix metalloproteinases are essential for tissue remodeling during metamorphosis (96). The increased expression of inhibitor and transporter genes in cluster 7 hemocytes suggests the presence of mechanisms to regulate metalloproteinase activity either through direct interaction or controlling the availability of zinc ions, that are essential for the catalytic mechanism (97). Zinc transport has been found to be essential for remodeling of fat body tissue by matrix metalloproteases in Drosophila (98). In Lepidoptera, it was demonstrated that hemocyte behavior and function was affected by the presence of zinc in the culture medium, implicating it as a regulator of metalloprotease function (99).
Carboxylesterases form a large superfamily and have important roles in xenobiotic metabolism (including insecticide resistance), pheromone inactivation and regulation of development and neurogenesis (100). The proposed carboxylesterase marker of cluster 7 hemocytes belongs to clade H that is associated with the secreted catalytic class and has single orthologs among the silkworm, Drosophila, the honeybee, the malaria mosquito and the Colorado potato beetle (101, 102). No clear function has been assigned to clade H carboxylesterases. Microarray data show preferential expression in hemocytes and head tissue in Bombyx (101); expression of the Drosophila homolog (CG5397) in larval plasmatocytes was also reported (103). Because of its relatedness with the Drosophila glutactin clade, clade H carboxylesterases may play a role in cell segregation and adhesion.
Two marker genes of cluster 7 (ZIP3, carboxylesterase H) are also differentially expressed in clusters 0, 4 and 10; in addition, cathepsin B expression is also enriched in clusters 0 and 4. Levels of differential expression of the three genes decline gradually from clusters 7 (highest), 0, 4 and 10 (lowest). The data reinforce the developmental relationship between clusters 0 and 4.
Also the cells of cluster 8 do not seem to be highly differentiated as evidenced by the relatively low log2FC values of the DEGs (range: 0.36 to 2.30) (Table 7).
Three of the five marker genes encode 30K proteins (genes indicated as 19G1 and 21G1) and a fourth 30K protein gene (21G1, 30K protein 19) has also a high log2FC of 2.14. 30K proteins were initially identified as major proteins in the hemolymph that are synthesized by the fat body during the larval stage and become absorbed by the oocyte as yolk proteins in the pupal stage (104, 105). Altogether forty-six 30K proteins were identified in Bombyx mori that were classified as low molecular weight lipoproteins (lipoprotein 11 family), although their lipid content remains unknown (106). The four genes that are differentially expressed in cluster 8 hemocytes all encode “typical” 30K proteins (belonging to clusters III and IV; 106, 107). Interestingly, the four genes belong to two close loci on chromosome 24.
However, besides their role as nutrient proteins, 30K proteins have also been shown to be involved in inhibition of apoptosis (108) and immune defense (109). More specifically, 30K proteins can bind to fungal glucans and therefore function as pattern-recognition receptors (6G1, 19G1 and 21G1 genes; 110, 111). In addition, specific 30K proteins become induced after injection of fungal wall glucans and can promote encapsulation by hemocytes (109).
Hydroxypyruvate isomerase (HYI) catalyzes the conversion of hydroxypyruvate to 2-hydroxy-3-oxopropanoate (also known as tartronate semi-aldehyde) and therefore may be involved in carbohydrate transport and metabolism (112). HYI activity links with the glyoxylate cycle and serine metabolism. Expression of HYI was also observed in crystal cells of Drosophila (103).
Finally, HHpred analysis (56) reveals that uncharacterized protein LOC101743414 contains a short Glutamine-rich region of 87 amino-acids that is similar to subunit TRAP230 of the thyroid hormone receptor-associated protein complex. The TRAP complex mediates the interaction between specific transcriptional activators and the general transcription machinery (113). The available evidence therefore indicates that uncharacterized LOC101743414 has a role in the regulation of transcriptional activation.
Conform to its classification as oenocytoids (24), cluster 8 also displays induction of the two prophenoloxidase genes (log2FCs 2.17 and 1.75; DEG rankings 3 and 10).
HYI and the four 30K genes are up-regulated in all clusters that were identified as oenocytoids (clusters 5, 8, 12 and 16). While log2FCs in cluster 5 are below 2, higher values (range: 2.36 to 3.10) are observed in clusters 12 and 16 but these are associated with a much lower DEG ranking than in cluster 8 (range 13 to 26 for cluster 12; range 38 to 78 for cluster 16). By contrast, LOC101743414 is not a DEG in the other oenocytoid-like cells although it is up-regulated in a few other clusters that are not classified as oenocytoids [clusters 0, 4, 7 and 17 with low to moderate log2FC (0.73 to 2.38)].
With low log2FC < 1, HYI and the four 30K genes are also DEGs in cluster 7. It is also noted that another 30K gene (BMSK0011510), located on chromosome 20, is expressed in hemocytes of clusters 0, 7, 10, 14 and 15 but at DEG ranking > 100.
The 5 selected marker genes of cluster 10 have relatively high levels of differential expression (log2FC values between 2.7 and 4.7) (Table 8) while the other DEGs have moderate to low values (such as clusters 0, 6 and 7).
The highest ranked gene encodes the homolog of Imaginal Morphogenesis Protein-Late 2 (IMP-L2), a secreted factor and member of the immunoglobulin family (114). IMP-L2 binds insulin-like peptides and functions as an antagonist of insulin/insulin-like growth factor signaling (115). As such, IMP-L2 regulates developmental timing and could be involved in the regulation of aerobic glycolysis in hemocytes during the immune response (116).
Cluster 10 cells are also characterized by the increased expression of another secreted protein, the antimicrobial peptide Cecropin B (117). Actually, 6 different Cecropin genes belonging to the same cluster on chromosome 26 and with identical amino-acid sequence are upregulated in cluster 10 (with log2FC of 3.0, 2.6, 2.4, 2.1, 2.1, and 1.9). While the function of Cecropin as an inducible lytic peptide against bacterial membranes is well established (118), it is noted that Cecropins can also have a regulatory function in developmental processes such as cuticle formation via the regulation of prophenoloxidase expression (119).
The increased expression of IMP-L2 and Cecropins indicates that cluster 10 may be associated with humoral immunity or have a regulatory function by the secretion of cytokines. To support the latter, it is noted that paralytic peptide, a cytokine with diverse functions in growth and immunity (120), is also upregulated in cluster 9 (log2FC = 2.28). Another marker gene in cluster 10 encodes mannosyl-oligosaccharide alpha-1,2-mannosidase IA that participates in the maturation process of N-glycans during protein glycosylation in the Golgi complex (121). Protein glycosylation is most often associated with secreted and membrane proteins. Interestingly, expression of alpha-1,2-mannosidase I variants was associated with stress resistance in Drosophila (122).
The two other remaining marker genes correspond to unknown proteins. A region in LOC106126609 (E value 0.9 in HHpred analysis) resembles the UBA domain that is found in ubiquitin-binding proteins. The other sequence is likely incomplete and resembles a region in proteins of the major intrinsic protein (MIP) family that function as membrane channels that selectively transport water, small neutral molecules, and ions. Aquaporins belong to the MIP family and are involved in the regulation of cell shape during cellular immunity in Spodoptera exigua (123). The aquaporin Prip is one of the most enriched genes in plasmatocytes of Drosophila (58) and another aquaporin, Drip, is enriched in lamellocytes (5).
Four cecropin genes from the same cluster are also moderately upregulated in cluster 4 (log2FC between 1.7 and 2.0). Increase in expression for two cecropins of this cluster is also found in cluster 17 (log2FC of 1.1 and 2.1). One of the cecropins in this cluster (BMSK0015395) is up-regulated in clusters 0, 4, 7, 10 and 17 (log2FC of 1.32, 1.90, 1.76, 1.87 and 1.15, respectively). On the other hand, high induction of one cecropin (Cecropin A) is observed in cluster 15 (log2FC = 5.4 while ranked 30 as DEG) but this gene is located in another location in the genome on chromosome 6.
Thirteen DEGs in cluster 12 have a log2FC value of >4, which could be indicative of higher differentiation levels. (Table 9).
Mohawk is a member of the TALE class of atypical homeobox genes that functions as a potent transcriptional repressor (124). In mammals, Mohawk is a tendon-specific transcription factor that regulates collagen expression (125). It can be suggested that Mohawk may be a regulator of hemocyte-specific functions in tissue remodeling (126).
Cilia- and flagella-associated protein 57 (CFAP57) belongs to the WD repeat-containing proteins that function in the assembly of large protein complexes (127). More specifically, CFAP57 is localized in the axoneme of cilia in human nasal epithelial cells and mutations in the green alga Chlamydomonas cause ciliary dyskinesia (128). Interestingly, an involvement of CFAP57 (previously known as WDR65) in tissue remodeling (craniofacial development) was also implicated (129). Thus, CFAP57 is predicted to be involved in changes in hemocyte morphology, such as elongations, that are reminiscent of the function of lamellocytes to engulf large foreign bodies such as parasitoid eggs in Drosophila (130). Interestingly, in larvae of particular drosophilids, a specialized hemocyte type, the nematocyte, can be observed that have spindle-like projections with high densities of mitochondria and microtubules (131), which may be considered as cilia-like structures. However, such extreme cell types have not been described among hemocytes in Bombyx mori so far.
Also trichohyalin is predicted to have a primary role in cytoskeleton dynamics. Trichohyalin is a keratin-binding protein that mediates its assembly to keratin filaments in mammals (132). While insects lack keratins and intermediary filaments are also thought to be absent (133), it is nevertheless tempting to speculate a role for trichohyalin in tissue remodeling and the encapsulation reaction during immune defense, perhaps by interacting with other skeletal elements such as cuticular building blocks. Genes encoding cuticular proteins are indeed induced in several other clusters (but not in cluster 7), the highest in cluster 15 (log2FC = 6.75).
IAP (inhibitor of apoptosis protein) is a critical regulator of cell survival by its capacity to inhibit apoptosis (134). However, IAP proteins are also involved in the regulation of innate immune signaling pathways (135), e.g. the Imd pathway that is activated by Gram-negative bacteria (136). Increased IAP expression may reflect the activation of innate immunity and the promotion of a state of inflammation.
The fourth marker gene encodes an uncharacterized protein of 239 amino-acids (LOC114246272). HHpred analysis (56) reveals a region with a seven residue sequence repeat found in coiled coil regions that are involved in protein dimerization (137). The final marker, on the other hand, corresponds to Thioredoxin-2 (Trx-2), a small heat-stable protein that contains a redox-active disulfide with anti-oxidative function (138). Trx-2 becomes induced by various types of stress and, interestingly, provides resistance against nucleopolyhedrovirus infection in Helicoverpa armigera (53).
It is also noted that prophenoloxidase 1 and 2 are among the 25 highest ranking DEGs (log2FC of 2.65 and 2.07, respectively) in the cells of cluster 12, which therefore may also be involved in the melanization reactions that regularly follow the encapsulation of macro-pathogens (139), consistent with the classification as oenocytoids (24). Interestingly, in Drosophila, the Runt-related transcription factor Lozenge is a marker of crystal cells (140) and, similarly, Runt-related transcription factor 3 is also significantly induced (log2FC = 2.65) in cluster 12, possibly indicating conservation of hemocyte differentiation programs between Drosophila and Bombyx. However, in contrast to our analysis, the Runt-related transcription factor Lozenge was also found to be highly expressed in hemocytes of the silkworm and its role in the melanization reaction by hemocytes was indicated by over-expression and gene silencing experiments (141).
Another interesting observation is the significant upregulation of the nuclear hormone receptor HR3 (log2FC = 4.51 and 3.25 for 2 isoforms), which is a part of the regulatory cascade initiated by 20-hydroxy-ecdysone (20E) (142). 20E controls the Imd innate immune pathway in Drosophila; more specifically, RNAi of HR3 decreases the induction of antibacterial peptide (AMP) genes by peptidoglycan (143). In hemocytes of larvae of Heliothis virescens, HR3 becomes induced during baculovirus infection (144). Similar to the increased levels of IAP, stimulation by 20E signaling in the cells of cluster 12 may be related to the activation of the innate immune response.
All the marker genes, with the exception of CFAP57, are also increased in the cells of cluster 16, but with much lower log2FC (range: 0.47 - 3.04). In addition, the two prophenoloxidase genes are significantly induced in clusters 5, 8 and 16 (log2FC range: 1.75 to 3.04), consistent with the proposition that clusters 5, 8, 12 and 16 correspond to oenocytoids (24). The two prophenoloxidases also appear as DEGs in cluster 7 but with much lower log2FC (0.69 and 0.64). On the other hand, Runt-related transcription factor 3 is differentially expressed in clusters 5, 12 and 16 (log2FC values of 2.43, 2.65 and 1.21, respectively), but not in cluster 8. Runt-related transcription factor 3 also appears ad DEG in cluster 6 (log2FC = 1.79).
DEGs in cluster 14 have high log2FC values (7 genes with log 2FC > 5; 18 genes with 5 > log2FC > 4; 26 genes with 4 > log2FC > 3), which indicates a high level of differentiation (Table 10).
A gene encoding an ommochrome-binding protein is differentially expressed at high levels (log2FC = 9.72) in the cells of cluster 14. In insects, ommochrome synthesis represents the most important pathway for the removal of tryptophan metabolites, that are toxic when present in high amounts (145). In addition, ommochromes also serve as eye and integument pigments (146, 147). Secreted ommochrome-binding proteins therefore may function as ommochrome carriers in the hemolymph to sites of excretion (hindgut, Malpighian tubules) or storage in pigment granules (eye, integument). Ommochrome-binding proteins are also synthesized by the fat body and accumulate in the hemolymph at their highest levels by the end of larval life (148). On the other hand, because of the capacity of ommochromes to buffer oxidative stress (149), ommochrome-binding proteins may also be linked to oxidative metabolism and protect hemocytes from excessive production of ROS during the immune response.
The second DEG with high log2FC corresponds to antitrypsin, which belongs to the serpin family of protease inhibitors (150). The majority of serpins in insects has a role in innate immunity in which they function as negative regulators (151). Antitrypsin in Bombyx is the homolog of Serpin-1 in Manduca sexta, which has been the subject of several functional studies (150, 152). Serpin-1 isoforms were shown to block the proteinase cascades leading to the activation of the ligand of the Toll receptor, Spätzle (153), as well as the prophenoloxidase enzymes involved in melanization (154, 155).
A gene with the annotation “insulin-like peptide receptor” (BMSK0010812) is also proposed as a marker gene for cluster 14 and is highly induced. However, this gene does not correspond to the canonical “insulin receptor” (InR) gene of Bombyx mori (156; Gene ID = BMSK0002922). The amino-acid sequence is highly similar to the extracellular domain of insulin-like receptors and contains fibronectin type III and L domains (157–159). HHpred analysis (56) confirms the formation of a binding cavity that can interact with insulin-like peptides. Although no signal peptide is present, a transmembrane segment could be identified. In the absence of a tyrosine kinase domain, the “insulin-like peptide receptor” therefore represents a decoy receptor that binds insulin-like ligands efficiently but is not capable to activate a signaling pathway.
While the expression of integrin β3 in cluster 14 as a marker gene for plasmatocytes was already noted (log2FC=4.10; 24), its partner for heterodimerization, integrin αIIb is also considered as characteristic for this hemocyte type (log2FC=5.37). Interestingly, mammalian integrin αIIb has a specific function in platelets, where it joins with integrin β3 to generate a fibrinogen receptor (160). The integrin αIIb/β3 dimer is required for platelet aggregation in mammals (161) and can therefore be predicted to have a similar function in lepidopteran plasmatocytes/Drosophila lamellocytes for the encapsulation of foreign bodies and pathogens.
In Manduca sexta, specific antibodies or RNAi that inhibit integrin β1 result in the suppression of encapsulation by plasmatocytes (162). Similarly, integrin β1 plays a role in spreading and encapsulation by plasmatocytes in Ostrinia furnacalis (163, 164). In the silkworm, 11 integrin family members (6 α- and 5 β-subunits) were identified, of which the majority was preferentially expressed in the hemocytes (165). Looking at the differential expression of integrins among hemocyte clusters, it appears that integrins α3 and β1 have the widest expression although at relatively modest levels (log2FC ranges from 0.55 to 2.07). Integrin α3 can be detected as DEG in clusters 0, 4, 7, 8 and 15 while integrin β1 occurs in clusters 3, 7, 9, 11 and 15. In clusters 14 and 15, however, other integrins are expressed at high levels. As already mentioned, both αIIb and β3 integrins are uniquely and highly induced in cluster 14. Also in cluster 15, integrin αIIb (but not β3) is differentially expressed (log2FC=4.63); in addition, integrins α-PS4 and α-PS1 are induced (log2FC of 5.55 and 4.14, respectively. The high expression of unique integrins in clusters 14 and 15 may be related to their function as plasmatocytes (24). Interestingly, a recent study indicated a regulatory role for integrin β3 in the immune response because of its inhibitory effect on melanization and the expression of immune-related genes (166).
The last marker gene, encoding uncharacterized protein LOC101747033, corresponds to a fatty acyl-coA reductase (FAR) that catalyzes the synthesis fatty acyl alcohols (167). Besides a role in pheromone biosynthesis (168), FARs are engaged in the production of wax substances for the isolation of surfaces such as the inner lining of the tracheal tubes (169). The expression of FAR in plasmatocyte-like cells may be related to the function of encapsulation.
Cluster 14 cells also produce several secreted proteins such as promoting protein, that regulates the lipid composition and fluidity of the plasma-membrane (log2FC = 4.99; 170), and multifunctional peptides designated as “cytokines” that also regulate spreading behavior (growth-blocking peptide, log2FC = 4.67 and paralytic peptide, log2FC = 2.74; 171).
All proposed marker genes, with the exception of ommochrome-binding protein, are also highly induced in cluster 15 (log2FC range from 3.61 to 4.58) but with much lower DEG ranking (53 to 88). Promoting protein, growth-blocking peptide and paralytic peptide are also DEGs in cluster 15 (log2FC range from 2.28 to 4.87; DGE rank from 48 to 137). The overlap in DEGs confirms the close relation between clusters 14 and 15 as plasmatocytes (24). Regarding other clusters, paralytic peptide and promoting protein are also DEGs in cluster 10 (respective log2FC and DGE rank of 2.28 and 1.22, and 10 and 56).
Cluster 15 also represents a highly differentiated type of hemocytes with high levels of log2FC in DEGs. Forty-three and 46 DEGs have log2FC > 5 and 5 > log2FC > 3, respectively (Table 11).
The top DEG (log2FC > 12) in this cluster has a C-type lectin-like domain” (CTLD). C-type lectins were originally defined by their dependence on Ca2+ as well as by the presence of a so-called “carbohydrate-recognition domain” (CRD) but later studies indicated that such domains also occurred in many other proteins that do not bind calcium or carbohydrates (172). The designation as CTLD was subsequently introduced to solve the conundrum that CTLD proteins can bind a wide variety of ligands, including sugars, proteins, lipids and inorganic compounds (173).
The identified sequence with CTLD is small (93 amino-acids) and may be incomplete; a highly similar sequence from B. mandarina is larger (173 amino-acids) and has a signal peptide but no transmembrane domain. In B. mori, 23 genes with CTLD domains were identified (174). Several CTLD proteins from Lepidoptera can bind pathogen-associated molecular patterns in Gram-positive and -negative bacteria and fungi and are involved in the activation of the cellular immune response (175–179). Interestingly, the identified CTLD has significant homology (23% identity, 42% similarity) with mammalian DC-SIGN (CD209) that interacts with the envelope proteins of Dengue virus (180). Importantly, Dengue virus infection can be inhibited by soluble DC-SIGN proteins (181, 182). Taken into account that a significant proportion of cluster 15 cells are infected with baculovirus, it can be speculated that the increased expression of soluble CTLD may be involved in the binding of baculoviral envelope proteins as an antiviral defense mechanism. Of interest is the observation that plasmatocytes are more resistant than other hemocyte cell types against BmNPV infection in Bombyx (183).
Two other highly induced marker genes belong to the trypsin family of serine proteases (184). In the hemolymph, serine proteases are produced as zymogens that are successively activated in a cascade sequence, e.g. during clotting/coagulation and encapsulation/melanization. While factors regulating melanization have been identified earlier (185), less is known about the regulation of coagulation in insects (186). Genes belonging to both cascades are upregulated in cluster 15: serine protease snake-like is a homolog of kallikrein that is involved in clotting/coagulation while serine protease 11 corresponds to phenoloxidase-activating factor 2. In the silkworm hemolymph, both pathways become integrated in large multifunctional immune complexes following injury or pathogen infection (187). Besides the proposed marker genes, several other serine protease precursors are highly ranked DEGs: serine protease gd-like (log2FC = 7.20) and serine protease gd (log2FC = 5.20), associated with coagulation; and prophenoloxidase activating enzyme precursor (log2FC = 6.42), clip domain serine protease 11 precursor (log2FC = 6.31) and serine protease 7 (log2FC = 5.14), involved with melanization. Remarkably, β-1,3-glucan recognition protein, one of the triggers of the melanization cascade (188), is also induced (log2FC = 5.18).
The remaining marker genes encode secreted proteins (presence of signal peptide) with functions related to the extracellular matrix. The sequence of the immune-related protein (BMSK0006713) corresponds to a reeler or reelin motif (approximately 170 amino-acids) that occurs in extracellular matrix proteins that guide neuronal migration (189). KWMTBOMO16085, on the other hand, shows a short sequence that also occurs in the elastomeric protein glutenin (190). High expression of both proteins is consistent with a function in invagination or encapsulation that needs be more defined by further experimentation.
Following serine proteases are also modestly differentially expressed in cluster 14: phenoloxidase-activating factor 2 (log2FC = 1.79), serine protease gd-like (log2FC = 2.13), serine protease gd (log2FC = 3.62) and clip domain serine protease 11 precursor (log2FC = 3.08). Beta-1,3-glucan recognition protein is also a DEG in cluster 5 (log2FC = 0.92), cluster 6 (log2FC = 1.55) and cluster 14 (log2FC = 1.35).
Cells of cluster 16, which were distinguished as oenocytoids (24), have DGEs with relatively high values: two DGEs with log2FC > 5.0; six DGEs with 4.0 < log2FC < 5.0; and nineteen DGEs with 3.0 < log2FC < 4.0 (total number of DGEs = 366) (Table 12).
The top-ranked marker gene in this cluster encodes α-crystallin B, which is related to an abundant constituent in the lens of the eyes of vertebrates but functions as a chaperone (sHSP) in other tissues in other animals (191). As mentioned before, other sHSPs, located in two clusters on chromosome 27, are considered as marker genes for cluster 5. However, α-crystallin B is located on chromosome 5, as well as the sHSP12.2 gene, which is also up-regulated in cluster 16 (log2FC = 2.55), together with all the sHSP genes from chromosome 27 (see discussion regarding sHSP genes in cluster 5). Thus, in both oenocytid-like clusters 5 and 16, sHSPs are important DEGs although different sHSPs are preferentially involved: sHSPs on chromosome 27 dominate in cluster 5 while the sHSPs on chromosome 5 are more important in cluster 16.
Increased expression of sHSPs is related to the stress response (192, 193). While data in insects are limited, an increase in expression of sHSPs is observed in hemocytes and other tissues following pathogen infection in other invertebrates (194, 195).
The second highly up-regulated marker gene encodes serpin-7 that belongs to a large family of inhibitors of serine proteases called serpins (196). Serpins are referred to as “suicide inhibitors” because of their mechanism of action of irreversible trapping of the protease in a covalent complex (197). Thirty-four serpin genes have been identified in B. mori (198) and serpin-7 is similar to serpin-9 of Manduca sexta and Helicoverpa armigera (56-57% identity) that regulate serine proteases during the immune response (199 200). Interestingly, serpin-9 becomes induced during baculovirus infection in Helicoverpa to inhibit melanization and virus inactivation (200).
A glycoprotein of 27 kDa, corresponding to the third marker gene, was first described in the hemolymph of Manduca sexta (201). The glycoprotein is insect-specific and carries a domain of unknown function (DUF-1397). Also produced and secreted by the fat body, its concentration in the hemolymph ranges from 10 to 20 μg/mL, without showing obvious changes during development or between sexes (201). Interestingly, proteins with DUF-1397 are moderately (1.7 to 2.1-fold) upregulated after Flock house virus infection in Drosophila and after O’nyong–nyong virus infection in Anopheles gambiae (202, 203). A protein with DUF-1397 also appears in the venom of a centipede, possibly reflecting its recruitment from the hemolymph (204). Finally, HHpred analysis shows remote similarity with the chondroitin peptidoglycan 4 domain of the nematode C. elegans (205).
Carboxypeptidase B-like can be tracked to the gene BmMCP18 that encodes a metallocarboxypeptidase with Peptidase_M14 domain and a divalent metal (zinc) cation in its active site (206). In mammals, carboxypeptidase B can modulate the coagulation cascade in the plasma (207) but in insects it remains unknown whether a protease cascade is involved in the clotting process (208). In the crab, Scylla paramamosain, carboxypeptidase B expression in hemocytes is decreased following infection with White spot syndrome virus (WSSV) and the bacterium Vibrio alginolyticus (209). However, carboxypeptidase B may play both a positive and negative role in the immune response since its knockdown decreased WSSV replication while it increased mortality after bacterial challenge (209). In the mosquito Aedes aegypti, metallocarboxypeptidase B1 (expressed in the midgut epithelium) has antiviral activity against Dengue virus (210).
Another marker gene encodes the enzyme 24-dehydrocholesterol reductase (DHCR24) that catalyzes the last step in cholesterol biosynthesis (211). In phytophagous insects, this step corresponds to the final reaction in the conversion of plant sterols into cholesterol (212) since insects cannot synthesize sterol compounds de novo (213). High expression of DHCR24 is observed in the midgut of B. mori (211, 212). Interestingly, DHCR24 has acquired a central role in lipid metabolism in mammals and has been linked to virus infection and oxidative stress (214). Inhibition of DHCR24 in macrophages results in an anti-inflammatory phenotype that is linked to the accumulation of desmosterol (the immediate precursor to cholesterol) and the activation of the nuclear receptor LXR (215). The high expression of DHCR24 therefore indicates the importance of lipid homeostasis and invites further experimentation to investigate the role of DHCR24 in the regulation of oxidative stress and inflammation response by oenocytoid-like cluster 16 cells.
Serpin-7, 27 kDa glycoprotein, carboxypeptidase B and DHCR24 are also expressed in the other oenocytoid-like clusters 5, 8, 12 and 16 but with lower log2FC (1.18 – 3.22) and DEG rank (17 – 1420). The differential expression of sHSP genes was already discussed above and with respect to cluster 5. Regarding other serpins, serpin-4A (related to Drosophila spn77Ba) is also found as DEG in clusters 0, 4, 7, 10, 14 and 15 (log2FC = 0.47 – 2.19; DEG rank = 131 – 1156). In M. sexta, serpin-4 associates with serine proteases of the prophenoloxidase pathway (216). Serpin-12, which is not orthologous to serpins from other species (198), is highly induced in cells of cluster 15 (log2FC = 4.97; DEG rank = 44).
In the cells of cluster 17, the 5 genes that have increased expression with log2FC > 2 were selected as marker genes (Table 13). The 5 marker genes have relatively low DEG rank (47, 3, 46, 119 and 10, respectively, according to the list of genes in Table 13).
The first marker gene in this cluster encodes an enzyme that transfers glucuronic acid residues to O-glycan structures in proteins (217). Structural analysis shows that the protein possesses a transmembrane membrane at the N-terminus and that the glucuronyltransferase domain is exposed to the extracellular medium. Addition of glucuronic acid is considered as a relatively rare modification of various substrates compared to glucosidation in insects (218); on the other hand, glucuronylation is a highly prevalent elaboration of O-linked and glycosphingolipid glycans (217). Because of its negative charge, glucuronic acid has been proposed to act as an equivalent of sialic acid in vertebrates that mediates a wide variety of physiological processes (219). Consistently, mutation or knockdown of glucuronyltransferase enzymes in Drosophila leads to developmental defects (220, 221). Interestingly, expression of GlcAT-P, one of three glucuronyltransferase enzymes in Drosophila, is required in hemocytes for growth of peripheral nerve fibers (220). One of the essential functions of hemocytes is the secretion and production of extracellular matrix (222) and the glucuronyltransferase may have a function in the glycosylation of extracellular matrix components in the basal membrane or during wound repair and tissue remodeling (217, 220).
Papilin corresponds to a large extracellular matrix protein (2064 amino-acids) with multiple domains (thrombospondin type-I, Kunitz proteinase inhibitor, immunoglobulin-like, PLAC and others), of which several are present in many copies (223). In Lepidoptera, the papilin homolog is known as lacunin (224) and was identified as BmSPI58 in B, mori (196). Interestingly, in Manduca sexta, lacunin is expressed by granular hemocytes during remodeling of the basal lamina of tissues during metamorphosis (225). Migrating hemocytes are a major source for the production and secretion of papilin during development prior to the formation of basal membranes around tissues (226). In Drosophila, papilin is also proposed to play a role in the innate immune response when hemocytes synthesize a temporary extracellular matrix as a scaffold to aid the defense reaction (227). During Autographa californica multiple nucleopolyhedrovirus (AcMNPV) infection, papilin expression was suppressed in hemocytes of both Spodoptera frugiperda and Trichoplusia ni hosts (228). It can be speculated that papilin is a target for the glucuronyltransferase mentioned above because of its heavy glycosylation with O-glycans that include glucuronic acid (229).
Cluster 17 shares with cluster 10 the DGE that encodes Cecropin B albeit with lower log2FC (2.13 versus 3.04). Another cecropin B gene, that belongs to the same chromosomal location (see discussion in cluster 10) is also weakly induced (log2FC = 1.15).
Increased expression of Cecropin B may be related to the identification of Relish as a DEG in cluster 17. Relish is a transcription factor of the NF-κB family and that resemble the mammalian Nfkb1 and Nfkb2 proteins p105 and p100 (230). In Drosophila, infection by Gram-negative bacteria triggers the activation of the Imd pathway which leads to the proteolytic cleavage of Relish that is mediated by the caspase Dredd and depends on phosphorylation by the IκB kinase complex (231). In the silkworm, BmNPV infection triggers the activation of BmSTING that promotes the activation of BmRelish and the induction of AMP genes, including CecB and CecA (232). It is noted that BmRelish is also a DEG in several other hemocyte clusters with increased expression of CecB (clusters 0, 4 and 10, but not cluster 7).
Tis11 (TPA-inducible sequence 11) is a member of the tristetraprolin family of CCCH tandem zinc finger proteins that mediate posttranscriptional repression of mRNAs through interaction with AU-rich elements in their 3’-UTRs (233). In the Drosophila hemocyte-like SL2 cell line, CecA1 mRNA is a well-characterized target of TIS11 that promotes its de-adenylation and rapid degradation (234). TIS11 could have a role in the recovery phase of the immune response when mRNAs of effector molecules such as AMPs become repressed and the basal status of the hemocytes become re-established (235). The appearance of both Relish and TIS11 as DEGs in cluster 17 underlines the importance of tight regulation of immune effector molecules at both the transcriptional and posttranscriptional level.
Clusters 0, 4, 7, 10 and 17 are all clusters with increased expression of CecB genes. Strikingly, also papilin (clusters 0, 4, 7 and 17), TIS11 (clusters 0, 4, 7, 10 and 17), Relish (0, 4, 10 and 17) and glucuronyltransferase (clusters 4, 10 and 17) are DEGs in most, but not all, of these clusters (albeit with low to moderate log2FC and DEG rank). This coincidence indicates relatedness among these clusters that are qualified as granulocytes. Papilin and TIS11 are also low-ranking DEGs in granulocyte-like cluster 8; in addition, glucuronyltransferase has low log2FC in cluster 19, which was proposed to represent the spherulocyte subtype (24).
Cells of cluster 19 have many DEGs with high log2FC: 6 genes have log2FC higher than 10; 17 DEGs show log2FC between 5 and 10; and another 28 genes are induced with a log2FC between 3 and 5. High numbers of DEGs and high log2FC indicate that the cells are highly differentiated; in conjunction with their low abundance (0.2% of the total) it can be assumed they may represent a hemocyte subtype with a highly specialized function. All marker genes have log2FC higher than 11 (Table 14).
Multi-bridge factor 2 (MBF2) was originally identified as a transcriptional co-activator that connects the nuclear receptor BmFTZ-F1 with the basal transcription machinery to stimulate transcription of the fushi tarazu gene in in vitro transcription assays (236). More recently, MBF2 was shown to belong to a family of insect-specific factors that are closely related to the response to pathogens (REPAT) genes (237). Two MBF2 genes are highly induced in cluster 19 with log2FCs of 13.23 (rank 1; MBF2-3) and 11.14 (rank 6; MBF2-2). Both genes, together with MBF2-1, are closely linked on chromosome 21 and share sequence identity of 42% (237). MBF2-2 and MBF2-3 are preferentially expressed in the hemolymph and during the larval stages. The expression of MBF2-like genes is induced after bacterial and baculovirus infection and during starvation, indicating functional roles in pathogen defense and nutrient metabolism (237, 238). MBF2 proteins have a small size (100-118 amino-acids), are predicted to be glycosylated and contain a signal peptide which implies that they may be secreted.
The second marker gene emphasizes the role of the respiratory complex in the mitochondria to contribute to the phenotype of cluster 19 hemocytes. NADH:ubiquinone oxidoreductase subunit B9 (NUDFB9) is an accessory subunit of the NADH dehydrogenase complex in the inner mitochondrial membrane that contributes to the oligomerization of different respiratory chain complexes into supercomplexes (239). NUDFB9 therefore may be an important factor to regulate mitochondrial function. Mitochondria can regulate the function of hemocytes in different ways that include the activation of the immune response and the production of ROS (240). Further research is required to determine which hemocyte functions are affected by the increased expression of NUDFB9.
As already discussed for cathepsin B, that is the top marker gene for cluster 7 hemocytes (see above), the high induction of cathepsin L in cluster 19 also suggests a role in tissue remodeling (e.g. extracellular matrix degradation) and the innate immune response. Recent studies indeed have confirmed high expression of cathepsin L in the hemocytes of B. mori that could be associated with a role in innate immunity (241).
Collagenase (5th marker) could function in conjunction with cathepsin L as a tissue remodeler or in wound healing. Degradation of collagen is associated with the regulation of growth and shape of organs (242, 243). Because the collagenase has a transmembrane domain and therefore is exposed on the surface, hemocytes are expected to make close contacts with the basal membranes of epithelial cells in which collagen is the major constituent (244).
Interestingly, one of the genes with the highest differential expression in cluster 19 encodes one of the early class “CA” chorion proteins, which were thought to be exclusively expressed in the follicular epithelium of the ovary and are constituents of the eggshell (245). Early chorion proteins are proposed to form an initial scaffold during the first steps of chorion assembly and their expression in cluster 19 hemocytes may reflect a similar role in the assembly of another protective layer, the cuticle. In this respect, it should be remembered that the control group in this study was treated with PBS through injection and that the small group of cluster 19 (50 cells) therefore is involved in wound repair, as is also predicted by the markers cathepsin L and collagenase.
Of the top marker genes in cluster 19, only MBF2-2 can be found as a DEG in another hemocyte subtype, i.e. cluster 14 with moderate log2FC (1.78) and ranking (97).
By analyzing the possible function of predicted marker genes, an attempt was made to construct the phenotype of the hemocyte subtypes/clusters that are represented by them. Previously, our analysis indicated that different clusters could be allocated to the broad classes of hemocytes that prevail in the literature (24). Our current examination of the marker genes in detail largely confirms the previous assessment, especially with respect to the oenocytoid (clusters 5, 8, 12 and 16) and plasmatocyte (clusters 14 and 15) subtypes.
The four oenocytoid clusters are characterized by the expression of the prophenoloxidase enzymes that catalyze the melanization process (246). In Drosophila, the corresponding cell type is called the crystal cell of which the differentiation is controlled by the Runt-related transcription factor Lozenge (247). A role for Lozenge was also proposed for the differentiation of oenocytoids in Bombyx (141). However, in our analysis, Lozenge did not appear as a DEG in the oenocytoid-like cell clusters while Runt-related transcription factor 3 is induced in clusters 5, 12 and 16. These results invite for a re-evaluation of the type of Runt-related transcription factor that is involved in the specification of the oenocytoid lineage in the silkworm.
Beyond the common expression of the prophenoloxidase enzymes, marker genes allow the identification of oenocytoid cells with specific functions (Table 1). In cluster 5, the expression of small heat-shock proteins is predominant, which may reflect the preparedness for future possible harmful effector functions and the protection against oxidative stress; this subtype therefore may be only partially differentiated, which is consistent with the relatively low log2FC of its DEGs. In cluster 8, the emphasis is on the expression of putative pattern recognition genes that belong to the class of the 30K proteins. However, more research is needed regarding the differential binding specificity of the different 30K proteins and which pathogen-associated antigens are preferentially recognized. Cells of cluster 12 seem to be more specialized and may have a role in tissue repair with a migrating phenotype; also in this cell type induced expression of apoptosis and stress regulatory genes can be considered necessary for survival in more hazardous environments. Also cluster 16 is highly differentiated and marker genes indicate their contribution to extracellular protease cascades that have important homeostatic functions such as coagulation and melanization. Similarly to clusters 5 and 8, induction of the stress response is observed.
The two clusters (14 and 15) that are marked as plasmatocytes both represent very specialized cell types (marker genes are characterized by high log2FC). Their main functions are related to encapsulation and melanization and the subtypes therefore are reminiscent of the lamellocytes observed in Drosophila. Of both subtypes, cells of cluster 14 may have a more regulatory role because of high expression of proteins that regulate the hormone response and may provide protection against oxidative stress (Table 1).
Granulocytes are represented by six clusters and therefore cover a wider range of possible functions (Table 1). Interestingly, the marker genes in two clusters are clearly associated with cell division and proliferation, such as mitotic spindle formation (cluster 4) and protein synthesis (cluster 6).
In general, cell clusters that are grouped as granulocytes have marker genes with relatively low log2FC (Table 1), which indicate intermediate cell types that may acquire more particular effector functions in specific conditions or challenges. Although the function of phagocytosis regularly is associated with granulocytes (248), only in cluster 0 marker genes that are associated with phagocytosis are indicated. However, cluster 0 is the most abundant cluster classified as granulocyte (3145 cells or 37% among all granulocyte clusters), confirming the important role of granulocytes as dedicated phagocytes.
The remaining granulocyte clusters indicate a function in tissue repair. In cluster 7, one protease, one protease inhibitor and one esterase are indicated as marker genes; a zinc transporter protein may be required to support the function of metalloproteinases while also mitochondrial function is activated. Interestingly, in both clusters 10 and 17 cecropin B genes appear to be induced. However, the induction level of cecropin genes is lower for cluster 17 and increased cecropin B expression is noted in 5 out of 6 clusters (clusters 0, 4, 7, 10, 17) designated as granulocytes. Thus, cecropin B expression appears to be a feature of the majority of granulocytes. Although cecropins are known as AMPs, a role in the processing of cytokines for tissue repair and the regulation of melanization has recently also been proposed (119). In cluster 17, cecropin B expression may be regulated at both transcriptional and posttranscriptional level, as indicated by the induction of the transcription factor Relish and the zinc finger protein Tis11.
Spherulocytes have surfaced as an enigmatic hemocyte subtype with large inclusions (“spherules”) in lepidopteran insects for which no function could be designated (8). In our analysis, cluster 19 that consists of only 50 cells was tentatively identified as spherulocytes based on the preferential expression of cathepsin L (16, 24). Further inspection of the marker genes indicates highly specialized effector functions directed to tissue repair. A role for spherulocytes in the synthesis of extracellular matrix and cuticle has been suggested before in the literature (249, 250). Interestingly, one of the marker genes encodes an early chorion protein, of which the expression was thought to be restricted to the epithelium of ovarian follicles (251). Early chorion proteins provide an initial scaffold on which the layers of the lamellar chorion are deposited (252). By analogy, it can be imagined that wound repair is initiated by the deposition of a framework of early chorion proteins on which layers of cuticle proteins can be assembled (253).
When looking at the graphical presentation of the different hemocyte clusters in Bombyx, the clusters that represent oenocytoids and granulocytes remain associated while plasmatocytes and spherulocytes are clearly separated (Figure 1; UMAP plot). At one end of this alignment, cluster 5 (oenocytoids) borders with cluster 6 (granulocytes) and at the other end, cluster 7 (granulocytes) touches cluster 8 (oenocytoids). Linked with cluster 8 are the oenocytoid clusters that are more differentiated (cluster 12 and 16; higher log2FC values) (Figure 1). In pseudo-time trajectory construction, granulocytes and spherulocytes are grouped together and separated from oenocytoids while plasmatocytes are located at the intersection of both major groups (24). However, (baculovirus-infected) “pro-hemocytes” for which no marker genes were available were used as a starting point in this analysis. Nevertheless, the UMAP plot in Figure 1 suggests that particular clusters of oenocytoids and granulocytes are rather closely related and could function as precursors for the differentiation of plasmatocytes (clusters 14 and 15) which are more terminally differentiated. Spherulocytes, on the other hand, represent a small population which also seems to be related to both granulocytes and oenocyotoids (Figure 1).
By comparison, scRNA-seq clusters that represent crystal cells in Drosophila are more isolated in principal component analysis and, in addition, represent a much lower proportion of the total hemocyte population (5% in Drosophila versus 28% in Bombyx; Supplementary Table 1; 4, 5). This indicates that crystal cells in Drosophila correspond to a much more differentiated cell type than oenocytoids in Bombyx, where several clusters have marker genes with relatively log2FC and are closely associated with clusters designated as granulocytes (Table 1; Figure 1). In addition, lamellocytes represent a specialized cell type in Drosophila that appears as an immune response following wasp infestation following trans-differentiation from plasmatocytes (4, 5) while in Bombyx plasmatocytes (as functional equivalents of lamellocytes in Drosophila; 8) have features in common with both oenocytoids and granulocytes (pseudo-time trajectory analysis; 24).
The marker genes that are considered for the classification of hemocytes in Bombyx show little overlap with the genes that are considered important to identify hemocyte clusters in Drosophila although the analysis proposed the same broad categories (with the exception of spherulocytes that are considered Lepidoptera-specific). Categorization in Drosophila was indeed guided by the already accumulated in-depth knowledge following extensive genetic analysis of hemocyte function. However, considerable differences with Drosophila were also observed when single cell technologies, including scRNA-seq, were applied to hemocytes from malaria mosquito Anopheles gambiae (254–256). This may be expected since processes such as innate immune response and tissue repair undergo fast evolution because of the divergence of different lifestyles among insects.
To illustrate the differences between the two approaches (scRNA-seq guided by accumulated genetic analysis in Drosophila versus scRNA-seq guided by morphological/biochemical analysis in Bombyx), the differential expression of marker genes characteristic for plasmatocytes, lamellocytes and crystal cells in Drosophila (NimC1, atilla and lozenge, respectively; 5) was checked in the scRNA-seq-based hemocyte clusters of Bombyx. With respect to oenocytoids/crystal cells, the absence of lozenge as a DEG in oenocytoids of Bombyx was already mentioned together with the observation of the enrichment of Runt-related transcription factor 3 which is related to Lozenge. With respect to plasmatocytes/lamellocytes, a gene related to quiver was specifically up-regulated in clusters 14 and 15 in Bombyx, (cluster 14: log2FC=4.39, rank=19; cluster 15: log2FC=3.82, rank=74). Both Atilla and Quiver are glycosylphosphatidylinositol (GPI)-anchored proteins that are involved in the regulation of voltage-gated potassium channels (257, 258) and its expression pattern reinforces the grouping of clusters 14 and 15 as plasmatocytes.
While NimC1 (encoding a phagocytosis receptor; 259) was not found as a DEG in the scRNA-seq analysis of Bombyx hemocytes, the presence of other scavenger receptors involved in phagocytosis was also checked. Granulocyte-like cells preferentially express class B scavenger receptor ScarB1 and class C scavenger receptor Malrd1 with the highest levels in cluster 7 (log2FC of 1.79 and 2.43, respectively). In the oenocytoid-like clusters 12 and 16, the class B receptor Croquemort is more highly induced (log2FC of 4.12 and 3.49, respectively). Plasmatocyte-like cells are enriched in transmembrane serine protease 2 (TMPRSS2) which has a scavenger receptor cysteine-rich domain (260; cluster 14: log2FC=4.82; cluster 15: log2FC=4.48). Thus, while granulocyte-like clusters preferentially express particular scavenger receptors, their induction level remains relatively low compared to other scavenger receptors expressed in the putative oenocytoid and plasmatocyte cells. In Bombyx, oenocytoids are usually not considered as phagocytic while both plasmatocytes and granulocytes are engaged in the recognition and phagocytosis of foreign particles (261). The preferential expression of Croquemort in oenocytoids in Bombyx therefore is unexpected and may reflect the less differentiated nature (and closer association with granulocytes) of Bombyx oenocytoids. Low levels of phagocytosis have been reported in oenocytoids of the mosquito Aedes aegypti (262). In Drosophila, Croquemort is enriched in plasmatocytes, more specifically in a subtype characterized as “phagocytic plasmatocytes” (4, 263; see also further below).
Recently, a consensus of hematocyte subtypes in Drosophila was proposed (263) based on the integration of different scRNA-seq studies (4, 5, 264). An attempt was made to correlate the clusters identified in Bombyx with the subtypes that were proposed in Drosophila (Supplementary Table 1). With respect to Drosophila plasmatocytes, which are considered to be the equivalent of Bombyx granulocytes (8), different smaller subgroups were proposed based on putative functional properties inferred by predicted marker genes. Strikingly, the major group of plasmatocytes in Drosophila (~60%) is designated as “unidentified” since no distinctive markers were identified. Similarly, clusters with marker genes that have relatively low log2FC values in Bombyx (arbitrarily set at lower than 3.5; clusters 0, 5, 6, 7, 8 and 17) could also be considered as little differentiated and therefore characterized as “unidentified” (amounting to a proportion of ~65%, which is similar as in Drosophila) (Supplementary Table 1). However, it was also attempted to correlate the clusters with relatively low log2FC markers in Bombyx with the more specialized hemocyte subgroups in Drosophila (both scenarios are incorporated in Supplementary Table 1).
Interestingly, “proliferative plasmatocytes” were indicated as a subgroup in Drosophila (263) which seems to be equivalent with clusters 4 and 6 in Bombyx that are also enriched in markers associated with proliferation and growth. It is also noted that 4 out of 5 marker genes in cluster 6 are also DGEs in cluster 5 (marked as oenocytoids; see also section 4.4), consistent with the close association in the UMAP plot (Figure 1).
On the other hand, other subgroups that were proposed in Drosophila such as “antimicrobial plasmatocytes”, “phagocytic plasmatocytes” and “secretory plasmatocytes” are less clearly represented as categories in Bombyx granulocytes. Regarding antimicrobial plasmatocytes in Drosophila, a variety of AMP genes is enriched in two subclusters (263) while in Bombyx granulocytes preferential expression is limited to cecropin B genes (Supplementary Table 1). On the other hand, a common theme may be the enrichment for the Imd pathway (represented by the marker gene Relish in cluster 17). Although cluster 0 in Bombyx displays some marker genes with a regulatory function associated with phagocytosis, phagocytic plasmatocytes in Drosophila show enhanced expression of effector genes directly involved with the process (263). Cluster 7 of Bombyx was also tentatively assigned as phagocytic (although a role in tissue repair was also considered, see above) because marker genes are associated with metalloproteinase function, but further research is required. Secretory plasmatocytes in Drosophila produce storage proteins that are also expressed in the fat body (263) while equivalent storage proteins such as 30K proteins are actually differentially expressed by oenocytoid-like cells (cluster 8) in Bombyx (Supplementary Table 1). 30K markers are also DEGs (with low log2FC; <1) in cluster 7 (granulocyte) which neighbors cluster 8 in the UMAP plot (Figure 1).
To complete the comparison, it is also clear that different marker genes are selected with respect to oenocytoids/crystal cells and plasmatocytes/lamellocytes in Bombyx/Drosophila although the general features of the hemocyte subtype look similar (Supplementary Table 1).
In this study, an overview and extended analysis was provided of the marker genes that were proposed for the characterization of 13 scRNA-seq clusters of hemocytes in the larvae of the silkworm (24). Because of the analysis, marker genes have now lost their anonymity and in many instances clear hypotheses can be formulated to carry out functional studies. Indeed, some marker genes stand out for their interesting features that inspire straightforward experimentation to investigate their hypothesized functions. Examples include a decoy receptor for insulin-like peptides (BMSK0010812; cluster 14), integrin αIIb that may have a conserved function in encapsulation (BMSK0005347; cluster 14) and a secreted protein with C-type lectin-like domain that could function as a decoy against baculovirus infection (BMSK0010425; cluster 15). The indicated proteins are expressed by highly differentiated plasmatocytes and therefore may be associated with specialized immune and/or repair functions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA658439.
LS and MF conceived the idea and designed the study. LS wrote the first draft of the manuscript. MF and JS critically read the manuscript and made improvements in the text and the tables. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
MF acknowledges support from the South China Agricultural University high-level talent launch project. LS acknowledges support of this work by the project ‘An Open-Access Research Infrastructure of Chemical Biology and Target-Based Screening Technologies for Human and Animal Health, Agriculture and the Environment (OPENSCREEN-GR)’ (MIS 5002691) which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational Programme ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund). JS acknowledges support from the National Natural Science Foundation of China (31872426) and the Guangdong Provincial Promotion Project on Preservation and Utilization of Local Breed of Livestock and Poultry (No.2018-143).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.852702/full#supplementary-material
1. Lavine MD, Strand MR. Insect Hemocytes and Their Role in Immunity. Insect Biochem Mol Biol (2002) 32:1295–309. doi: 10.1016/S0965-1748(02)00092-9
2. Hartenstein V. Blood Cells and Blood Cell Development in the Animal Kingdom. Annu Rev Cell Dev Biol (2006) 22:677–712. doi: 10.1146/annurev.cellbio.22.010605.093317
3. Hartenstein V. One Too Many: The Surprising Heterogeneity of Drosophila Macrophages. EMBO J (2020) 39:e105199. doi: 10.15252/embj.2020105199
4. Cattenoz PB, Sakr R, Pavlidaki A, Delaporte C, Riba A, Molina N, et al. Temporal Specificity and Heterogeneity of Drosophila Immune Cells. EMBO J (2020) 39:e104486. doi: 10.15252/embj.2020104486
5. Tattikota SG, Cho B, Liu YF, Hu Y, Barrera V, Steinbaugh MJ, et al. A Single-Cell Survey of Drosophila Blood. Elife (2020) 9:e54818. doi: 10.7554/eLife.54818
6. Eleftherianos I, Heryanto C, Bassal T, Zhang W, Tettamanti G, Mohamed A. Haemocyte-Mediated Immunity in Insects: Cells, Processes and Associated Components in the Fight Against Pathogens and Parasites. Immunology (2021) 164:401–32. doi: 10.1111/imm.13390
7. von Bredow C-R, von Bredow YM, Trenczek TE. The Larval Haematopoietic Organs of Manduca Sexta (Insecta, Lepidoptera): An Insight Into Plasmatocyte Development and Larval Haematopoiesis. Dev Comp Immunol (2021) 115:103858. doi: 10.1016/j.dci.2020.103858
8. Ribeiro C, Brehélin M. Insect Haemocytes: What Cell Type Is That? J Insect Physiol (2006) 52:417–29. doi: 10.1016/j.jinsphys.2006.01.005
9. Liu F, Xu Q, Zhang Q, Lu A, Beerntsen BT, Ling E. Hemocytes and Hematopoiesis in the Silkworm, Bombyx Mori. ISJ-Invert Surviv J (2013) 10:102–9.
10. Pech LL, Strand MR. Granular Cells are Required for Encapsulation of Foreign Targets by Insect Haemocytes. J Cell Sci (1996) 109:2053–60. doi: 10.1242/jcs.109.8.2053
11. Gillespie JP, Kanost MR, Trenczek T. Biological Mediators of Insect Immunity. Annu Rev Entomol (1997) 42:611–43. doi: 10.1146/annurev.ento.42.1.611
12. Rizki TM, Rizki RM. Lamellocyte Differentiation in Drosophila Larvae Parasitized by Leptopilina. Dev Comp Immunol (1992) 16:103–10. doi: 10.1016/0145-305X(92)90011-Z
13. Buchon N, Silverman N, Cherry S. Immunity in Drosophila Melanogaster–From Microbial Recognition to Whole-Organism Physiology. Nat Rev Immunol (2014) 14:796–810. doi: 10.1038/nri3763
14. Nakahara Y, Kanamori Y, Kiuchi M, Kamimura M. Two Hemocyte Lineages Exist in Silkworm Larval Hematopoietic Organ. PloS One (2010) 5:e11816. doi: 10.1371/journal.pone.0011816
15. Banerjee U, Girard JR, Goins LM, Spratford CM. Drosophila as a Genetic Model for Hematopoiesis. Genetics (2019) 211:367–417. doi: 10.1534/genetics.118.300223
16. Nakahara Y, Shimura S, Ueno C, Kanamori Y, Mita K, Kiuchi M, et al. Purification and Characterization of Silkworm Hemocytes by Flow Cytometry. Dev Comp Immunol (2009) 33:439–48. doi: 10.1016/j.dci.2008.09.005
17. Honti V, Csordás G, Márkus R, Kurucz E, Jankovics F, Andó I. Cell Lineage Tracing Reveals the Plasticity of the Hemocyte Lineages and of the Hematopoietic Compartments in Drosophila Melanogaster. Molec Immunol (2010) 47:1997–2004. doi: 10.1016/j.molimm.2010.04.017
18. Chen G, Ning B, Shi T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front Genet (2019) 10:317. doi: 10.3389/fgene.2019.00317
19. Kulkarni A, Anderson AG, Merullo DP, Konopka G. Beyond Bulk: A Review of Single Cell Transcriptomics Methodologies and Applications. Curr Opin Biotechnol (2019) 58:129–36. doi: 10.1016/j.copbio.2019.03.001
20. Chowdhury HA. Effective Clustering of scRNA-Seq Data to Identify Biomarkers Without User Input. Proc AAAI Conf Artif Intell (2021) 35:15710–1.
21. Hu M-W, Kim DW, Liu S, Zack DJ, Blackshaw S, Qian J. PanoView: An Iterative Clustering Method for Single-Cell RNA Sequencing Data. PloS Comput Biol (2019) 15:e1007040. doi: 10.1371/journal.pcbi.1007040
22. Luecken MD, Theis FJ. Current Best Practices in Single-Cell RNA-Seq Analysis: A Tutorial. Mol Syst Biol (2019) 15:e8746. doi: 10.15252/msb.20188746
23. Cho B, Yoon SH, Lee D, Koranteng F, Tattikota SG, Cha N, et al. Single-Cell Transcriptome Maps of Myeloid Blood Cell Lineages in Drosophila. Nat Commun (2020) 11:4483. doi: 10.1038/s41467-020-18135-y
24. Feng M, Xia J, Fei S, Peng R, Wang X, Zhou Y, et al. Identification of Silkworm Hemocyte Subsets and Analysis of Their Response to Baculovirus Infection Based on Single-Cell RNA Sequencing. Front Immunol (2021) 12:645359. doi: 10.3389/fimmu.2021.645359
25. Sun S, Zhu J, Ma Y, Zhou X. Accuracy, Robustness and Scalability of Dimensionality Reduction Methods for Single-Cell RNA-Seq Analysis. Genome Biol (2019) 20:269. doi: 10.1186/s13059-019-1898-6
26. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial Reconstruction of Single-Cell Gene Expression Data. Nat Biotechnol (2015) 33:495–502. doi: 10.1038/nbt.3192
27. Nakahara Y, Kanamori Y, Kiuchi M, Kamimura M. In Vitro Studies of Hematopoiesis in the Silkworm: Cell Proliferation in and Hemocyte Discharge From the Hematopoietic Organ. J Insect Physiol (2003) 49:907–16. doi: 10.1016/S0022-1910(03)00149-5
28. Horohov DW, Dunn PE. Changes in the Circulating Hemocyte Population of Manduca Sexta Larvae Following Injection of Bacteria. J Invertebr Pathol (1982) 40:327–39. doi: 10.1016/0022-2011(82)90171-9
29. Zhang K, Li C, Weng X, Su J, Shen L, Pan G, et al. Transgenic Characterization of Two Silkworm Tissue-Specific Promoters in the Haemocyte Plasmatocyte Cells. Insect Mol Biol (2018) 27:133–42. doi: 10.1111/imb.12360
30. Zhang K, Tan J, Xu M, Su J, Hu R, Chen Y, et al. A Novel Granulocyte-Specific Alpha Integrin is Essential for Cellular Immunity in the Silkworm Bombyx Mori. J Insect Physiol (2014) 71:61–7. doi: 10.1016/j.jinsphys.2014.10.007
31. von Bredow YM, von Bredow C-R, Trenczek TE. A Novel Site of Haematopoiesis and Appearance and Dispersal of Distinct Haemocyte Types in the Manduca Sexta Embryo (Insecta, Lepidoptera). Dev Comp Immunol (2020) 11:103722. doi: 10.1016/j.dci.2020.103722
32. McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, et al. Data Exploration, Quality Control and Testing in Single-Cell qPCR-Based Gene Expression Experiments. Bioinformatics (2013) 29:461–7. doi: 10.1093/bioinformatics/bts714
33. Lu F, Wei Z, Luo Y, Guo H, Zhang G, Xia Q, et al. SilkDB 3.0: Visualizing and Exploring Multiple Levels of Data for Silkworm. Nucleic Acids Res (2019) 48:D749–55. doi: 10.1093/nar/gkz919
34. Söding J, Biegert A, Lupas AN. The HHpred Interactive Server for Protein Homology Detection and Structure Prediction. Nucleic Acids Res (2005) 33:W244–8. doi: 10.1093/nar/gki408
35. Feng M, Fei S, Xia J, Zhang M, Wu H, Swevers L, et al. Global Metabolic Profiling of Baculovirus Infection in Silkworm Hemolymph Shows the Importance of Amino-Acid Metabolism. Viruses (2021) 13:841. doi: 10.3390/v13050841
36. Pace HC, Brenner C. The Nitrilase Superfamily: Classification, Structure and Function. Genome Biol (2001) 2:reviews0001.1. doi: 10.1186/gb-2001-2-1-reviews0001
37. Jacobs ME. Role of Beta-Alanine in Cuticular Tanning, Sclerotization, and Temperature Regulation in Drosophila Melanogaster. J Insect Physiol (1985) 31:509–15. doi: 10.1016/0022-1910(85)90099-X
38. Qi Y-x, Huang J, Li M-Q, Wu Y-S, Xia R-Y, Ye G-T. Serotonin Modulates Insect Hemocyte Phagocytosis via Two Different Serotonin Receptors. eLife (2016) 5:e12241. doi: 10.7554/eLife.12241
39. Hasan A, Yeom H-S, Ryu J, Bode HB, Kim Y. Phenylethylamides Derived From Bacterial Secondary Metabolites Specifically Inhibit an Insect Serotonin Receptor. Sci Rep (2019) 9:20358. doi: 10.1038/s41598-019-56892-z
40. Ho AK, Wagstaff JL, Manna PT, Wartosch L, Qamar S, Garman EF, et al. The Topology, Structure and PE Interaction of LITAF Underpin a Charcot-Marie-Tooth Disease Type 1C. BMC Biol (2016) 14:109. doi: 10.1186/s12915-016-0332-8
41. Lombardo F, Ghani Y, Kafatos FC, Christophides GK. Comprehensive Genetic Dissection of the Hemocyte Immune Response in the Malaria Mosquito Anopheles Gambiae. PloS Pathog (2013) 9:e1003145. doi: 10.1371/journal.ppat.1003145
42. Smith RC, Barillas-Mury C, Jacobs-Lorena M. Hemocyte Differentiation Mediates the Mosquito Late-Phase Immune Response Against Plasmodium in Anopheles Gambiae. Proc Natl Acad Sci USA (2015) 112:E3412–3420. doi: 10.1073/pnas.1420078112
43. Chen S, Liu M, Li Y, Zhou X, Hou Y, Gong J. Molecular Identification and Expression Patterns of Apolipoprotein D From Bombyx Mori (Lepidoptera: Bombycidae) in Response to Oxidative Stress and Bacterial Challenge. Ann Entomol Soc Am (2016) 109:759–67. doi: 10.1093/aesa/saw035
44. Turunen M, Olsson J, Dallner G. Metabolism and Function of Coenzyme Q. Biochim Biophys Acta (2004) 1660:171–99. doi: 10.1016/j.bbamem.2003.11.012
45. Vandermoten S, Haubruge E, Cusson M. New Insights Into Short-Chain Prenyltransferases: Structural Features, Evolutionary History and Potential for Selective Inhibition. Cell Mol Life Sci (2009) 66:3685–95. doi: 10.1007/s00018-009-0100-9
46. Thom R, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ. The Structure of a Zeta Class Glutathione S-Transferase From Arabidopsis Thaliana: Characterisation of a GST With Novel Active-Site Architecture and a Putative Role in Tyrosine Catabolism. J Mol Biol (2001) 308:949–62. doi: 10.1006/jmbi.2001.4638
47. Board PG, Baker RT, Chelvanayagam G, Jermiin LS. Zeta, a Novel Class of Glutathione Transferases in a Range of Species From Plants to Humans. Biochem J (1997) 328:929–35. doi: 10.1042/bj3280929
48. Blackburn AC, Matthaei KI, Lim C, Taylor MC, Cappello JY, Hayes JD, et al. Deficiency of Glutathione Transferase Zeta Causes Oxidative Stress and Activation of Antioxidant Response Pathways. Mol Pharmacol (2006) 69:650–7. doi: 10.1124/mol.105.018911
49. Litwack G. Metabolism of Amino-Acids. In: Litwack G, editor. Human Biochemistry. Los Angeles, CA:Elsevier (2018). p. 359–94.
50. Andersen SSL. Balanced Regulation of Microtubule Dynamics During the Cell Cycle: A Contemporary View. BioEssays (1999) 21:53–60. doi: 10.1002/(SICI)1521-1878(199901)21:1<53::AID-BIES7>3.0.CO;2-L
51. Bouchet BP, Akhmanova A. Microtubules in 3D Cell Motility. J Cell Sci (2017) 130:39–50. doi: 10.1242/jcs.189431
52. Orr B, Bousbaa H, Sunkel CE. Mad2-Independent Spindle Assembly Checkpoint Activation and Controlled Metaphase–Anaphase Transition in Drosophila S2 Cells. Mol Biol Cell (2007) 18:850–63. doi: 10.1091/mbc.e06-07-0587
53. Zhang S, Li Z, Nian X, Wu F, Shen Z, Zhang B, et al. Sequence Analysis, Expression Profiles and Function of Thioredoxin 2 and Thioredoxin Reductase 1 in Resistance to Nucleopolyhedrovirus in Helicoverpa Armigera. Sci Rep (2015) 5:15531. doi: 10.1038/srep15531
54. Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, et al. The Human Kinesin Kif18A is a Motile Microtubule Depolymerase Essential for Chromosome Congression. Curr Biol (2007) 17:488–98. doi: 10.1016/j.cub.2007.02.036
55. Chen JWC, Barker AR, Wakefield JG. The Ran Pathway in Drosophila Melanogaster Mitosis. Front Cell Dev Biol (2015) 3:74. doi: 10.3389/fcell.2015.00074
56. Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, et al. A Completely Reimplemented MPI Bioinformatics Toolkit With a New HHpred Server at its Core. J Mol Biol (2018) 430:2237–43. doi: 10.1016/j.jmb.2017.12.007
57. Cheng K-Y, Noble MEM, Skamnaki V, Brown NR, Low ED, Kontogiannis L, et al. The Role of the Phospho-CDK2/Cyclin A Recruitment Site in Substrate Recognition. J Biol Chem (2006) 281:23167–79. doi: 10.1074/jbc.M600480200
58. Ramond E, Dudzic JP, Lemaitre B. Comparative RNA-Seq Analyses of Drosophila Plasmatocytes Reveal Gene Specific Signatures in Response to Clean Injury and Septic Injury. PloS One (2020) 15:e0235294. doi: 10.1371/journal.pone.0235294
59. Van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal Structure and Assembly of a Eukaryotic Small Heat Shock Protein. Nat Struct Biol (2001) 8:1025–30. doi: 10.1038/nsb722
60. Chakrabarti S, Visweswariah SS. Intramacrophage ROS Primes the Innate Immune System via JAK/STAT and Toll Activation. Cell Rep (2020) 33:108368. doi: 10.1016/j.celrep.2020.108368
61. Wrońska AK, Boguś MI. Heat Shock Proteins (HSP 90, 70, 60, and 27) in Galleria Mellonella (Lepidoptera) Hemolymph are Affected by Infection With Conidiobolus Coronatus (Entomophthorales). PloS One (2020) 15:e0228556. doi: 10.1371/journal.pone.0228556
62. Krautz R, Arefin B, Theopold U. Damage Signals in the Insect Immune Response. Front Plant Sci (2014) 5:342. doi: 10.3389/fpls.2014.00342
63. Kong HK, Park JH. Characterization and Function of Human Ly-6/uPAR Molecules. BMB Rep (2012) 45:595–603. doi: 10.5483/BMBRep.2012.45.11.210
64. Upadhyay G. Emerging Role of Lymphocyte Antigen-6 Family of Genes in Cancer and Immune Cells. Front Immunol (2019) 10:819. doi: 10.3389/fimmu.2019.00819
65. Moussian B, Soding J, Schwarz H, Nusslein-Volhard C. Retroactive, a Membrane-Anchored Extracellular Protein Related to Vertebrate Snake Neurotoxin- Like Proteins, is Required for Cuticle Organization in the Larva of Drosophila Melanogaster. Dev Dyn (2005) 233:1056–63. doi: 10.1002/dvdy.20389
66. Hijazi A, Haenlin M, Waltzer L, Roch F. The Ly6 Protein Coiled is Required for Septate Junction and Blood Brain Barrier Organization in Drosophila. PloS One (2011) 6:e17763. doi: 10.1371/journal.pone.0017763
67. Nilton A, Oshima K, Zare F, Byri S, Nannmark U, Nyberg KG, et al. Crooked, Coiled and Crimpled are Three Ly6-Like Proteins Required for Proper Localization of Septate Junction Components. Development (2010) 137:2427–37. doi: 10.1242/dev.052605
68. Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, et al. SLEEPLESS, a Ly-6/Neurotoxin Family Member, Regulates the Levels, Localization and Activity of Shaker. Nat Neurosci (2010) 13:69–75. doi: 10.1038/nn.2454
69. Ozhan G, Sezgin E, Wehner D, Pfister AS, Kuhl SJ, Kagermeier-Schenk B, et al. Lypd6 Enhances Wnt/beta-Catenin Signaling by Promoting Lrp6 Phosphorylation in Raft Plasma Membrane Domains. Dev Cell (2013) 26:331–45. doi: 10.1016/j.devcel.2013.07.020
70. Paramonov AS, Kocharovskaya MV, Tsarev AV, Kulbatskii DS, Loktyushov EV, Shulepko MA, et al. Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors. Int J Mol Sci (2020) 21:7280. doi: 10.3390/ijms21197280
71. Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila Myc Regulates Cellular Growth During Development. Cell (1999) 98:779–90. doi: 10.1016/S0092-8674(00)81512-3
72. Pennetier D, Oyallon J, Morin-Poulard I, Defean S, Vincent A, Crozatier M. Size Control of the Drosophila Hematopoietic Niche by Bone Morphogenetic Protein Signaling Reveals Parallels With Mammals. Proc Natl Acad Sci USA (2012) 109:3389–94. doi: 10.1073/pnas.1109407109
73. Dearden PK. Origin and Evolution of the Enhancer of Split Complex. BMC Genomics (2015) 16:712. doi: 10.1186/s12864-015-1926-1
74. Duvic B, Hoffmann JA, Meister M, Royet J. Notch Signaling Controls Lineage Specification During Drosophila Larval Hematopoiesis. Curr Biol (2002) 122:1923–7. doi: 10.1016/S0960-9822(02)01297-6
75. Jin LH, Choi JK, Kim B, Cho HS, Kim J, Kim-Ha J, et al. Requirement of Split Ends for Epigenetic Regulation of Notch Signal-Dependent Genes During Infection-Induced Hemocyte Differentiation. Mol Cell Biol (2009) 29:1515–25. doi: 10.1128/MCB.01239-08
76. Calvo O, Manley JL. Strange Bedfellows: Polyadenylation Factors at the Promoter. Genes Dev (2003) 17:1321–7. doi: 10.1101/gad.1093603
77. Chen W, Guo W, Li M, Shi D, Tian Y, Li Z, et al. Upregulation of Cleavage and Polyadenylation Specific Factor 4 in Lung Adenocarcinoma and its Critical Role for Cancer Cell Survival and Proliferation. PloS One (2013) 8:e82728. doi: 10.1371/journal.pone.0082728
78. Yi C, Wang Y, Zhang C, Xuan Y, Zhao S, Liu T, et al. Cleavage and Polyadenylation Specific Factor 4 Targets NF-κb/Cyclooxygenase-2 Signaling to Promote Lung Cancer Growth and Progression. Cancer Lett (2016) 381:1–13. doi: 10.1016/j.canlet.2016.07.016
79. Kurihara M, Komatsu K, Awane R, Inoue Y. Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in Mxc Mutants of Drosophila. Int. J Mol Sci (2020) 21:1586. doi: 10.3390/ijms21051586
80. Mongelard F, Bouvet P. Nucleolin: A Multifaceted Protein. Trends Cell Biol (2006) 17:80–6. doi: 10.1016/j.tcb.2006.11.010
81. Cong R, Das S, Bouvet P. The Multiple Properties and Functions of Nucleolin. In: Olson M, editor. The Nucleolus. Protein Reviews, 15. New York, NY: Springer (2011). p. 185–212.
82. Ghartey-Kwansah G, Li Z, Feng R, Wang L, Zhou X, Chen FZ, et al. Comparative Analysis of FKBP Family Protein: Evaluation, Structure, and Function in Mammals and Drosophila Melanogaster. BMC Dev Biol (2018) 18:7. doi: 10.1186/s12861-018-0167-3
83. Ahmad M, Srinivasula SM, Wang L, Litwack G, Fernandes-Alnemri T, Alnemri ES. Spodoptera Frugiperda Caspase-1, a Novel Insect Death Protease That Cleaves the Nuclear Immunophilin FKBP46, is the Target of the Baculovirus Antiapoptotic Protein P35. J Biol Chem (1997) 272:1421–4. doi: 10.1074/jbc.272.3.1421
84. Hentze H, Lin XY, Choi MSK, Porter AG. Critical Role for Cathepsin B in Mediating Caspase-1-Dependent Interleukin-18 Maturation and Caspase-1- Independent Necrosis Triggered by the Microbial Toxin Nigericin. Cell Death Differ (2003) 10:956–68. doi: 10.1038/sj.cdd.4401264
85. Yan X, Wu Z, Wang B, Yu T, Hu Y, Wang S, et al. Involvement of Cathepsins in Innate and Adaptive Immune Responses in Periodontitis. Evid Based Complement Altern Med (2020) 2020:4517587, 9 pages. Hindawi. doi: 10.1155/2020/4517587
86. Kurata S, Saito H, Natori S. The 29-kDa Hemocyte Proteinase Dissociates Fat Body at Metamorphosis of Sarcophaga. Dev Biol (1992) 153:115–21. doi: 10.1016/0012-1606(92)90096-Y
87. Xu YS, Kawasaki H. Isolation and Expression of Cathepsin B cDNA in Hemocytes During Metamorphosis of Bombyx Mori. Comp Biochem Physiol B (2001) 130:393–9. doi: 10.1016/S1096-4959(01)00448-1
88. Cho J, Hur JH, Graniel J, Benzer S, Walker DW. Expression of Yeast NDI1 Rescues a Drosophila Complex I Assembly Defect. PloS One (2012) 7:e50644. doi: 10.1371/journal.pone.0050644
89. Scialò F, Sriram A, Stefanatos R, Spriggs RV, Loh SHY, Martin LM, et al. Mitochondrial Complex I Derived ROS Regulate Stress Adaptation in Drosophila Melanogaster. Redox Biol (2020) 32:101450. doi: 10.1016/j.redox.2020.101450
90. Sharma LK, Lu J, Bai Y. Mitochondrial Respiratory Complex I: Structure, Function Andiimplication in Human Diseases. Curr Med Chem (2009) 16:1266–77. doi: 10.2174/092986709787846578
91. Sinenko SA, Starkova TY, Kuzmin AA, Tomilin AN. Physiological Signaling Functions of Reactive Oxygen Species in Stem Cells: From Flies to Man. Front Cell Dev Biol (2021) 9:714370. doi: 10.3389/fcell.2021.714370
92. Shadel GS, Horvath TL. Mitochondrial ROS Signaling in Organismal Homeostasis. Cell (2015) 163:560–9. doi: 10.1016/j.cell.2015.10.001
93. Shekhova E. Mitochondrial Reactive Oxygen Species as Major Effectors of Antimicrobial Immunity. PloS Pathog (2020) 16:e1008470. doi: 10.1371/journal.ppat.1008470
94. Knorr E, Schmidtberg H, Vilcinskas A, Altincicek B. MMPs Regulate Both Development and Immunity in the Tribolium Model Insect. PloS One (2009) 4:e4751. doi: 10.1371/journal.pone.0004751
95. LaFever KS, Wang X, Page-McCaw P, Bhave G, Page-McCaw A. Both Drosophila Matrix Metalloproteinases Have Released and Membrane-Tethered Forms But Have Different Substrates. Sci Rep (2017) 7:44560. doi: 10.1038/srep44560
96. Page-McCaw A, Serano J, Santé JM, Rubin GM. Drosophila Matrix Metalloproteinases are Required for Tissue Remodeling, But Not Embryonic Development. Dev Cell (2002) 4:95–106. doi: 10.1016/S1534-5807(02)00400-8
97. Gomis-Rüth FX. Structural Aspects of the Metzincin Clan of Metalloendopeptidases. Mol Biotechnol (2003) 24:157–202. doi: 10.1385/MB:24:2:157
98. Wei T, Ji X, Yu Q, Li G, Wu L, Gao Y, et al. Fear-Of-Intimacy-Mediated Zinc Transport Controls Fat Body Cell Dissociation Through Modulating Mmp Activity in Drosophila. Cell Death Dis (2021) 12:874. doi: 10.1038/s41419-021-04147-z
99. Willott E, Tran HQ. Zinc and Manduca Sexta Hemocyte Functions. J Insect Sci (2002) 2:1–9. doi: 10.1673/031.002.0601
100. Oakeshott JG, Claudianos C, Campbell PM, Newcomb RD, Russell RJ. Biochemical Genetics and Genomics of Insect Esterases. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science, vol. 5 . London: Elsevier (2005). p. 309–61.
101. Yu Q-Y, Lu C, Li W-L, Zhang Z-H, Zhang Z. Annotation and Expression of Carboxylesterases in the Silkworm, Bombyx Mori. BMC Genomics (2009) 10:553. doi: 10.1186/1471-2164-10-553
102. Lü F-G, Fu K-Y, Li Q, Guo W-C, Ahmat T, Li G-Q. Identification of Carboxylesterase Genes and Their Expression Profiles in the Colorado Potato Beetle Leptinotarsa Decemlineata Treated With Fipronil and Cyhalothrin. Pestic Biochem Physiol (2015) 122:86–95. doi: 10.1016/j.pestbp.2014.12.015
103. re3data.org: Berkeley Drosophila Genome Project in situ; editing status 2021-07-02. re3data.org - Registry of Research Data Repositories. doi: 10.17616/R32H0K
104. Izumi S, Fujie J, Yamada S, Tomino S. Molecular Properties and Biosynthesis of Major Plasma Proteins in Bombyx Mori. Biochim. Biophys Acta (1981) 670:222–9. doi: 10.1016/0005-2795(81)90013-1
105. Mori S, Izumi S, Tomino S. Structures and Organization of Major Plasma Protein Genes of the Silkworm Bombyx Mori. J Mol Biol (1991) 218:7–12. doi: 10.1016/0022-2836(91)90866-5
106. Zhang Y, Dong Z, Liu S, Yang Q, Zhao P, Xia Q. Identification of Novel Members Reveals the Structural and Functional Divergence of Lepidopteran-Specific Lipoprotein_11 Family. Funct Integr Genom (2012) 12:705–15. doi: 10.1007/s10142-012-0281-4
107. Sun Q, Zhao P, Lin Y, Hou Y, Xia QY, Xiang ZH. Analysis of the Structure and Expression of the 30 K Protein Genes in Silkworm, Bombyx Mori. Insect Sci (2007) 14:5–14. doi: 10.1111/j.1744-7917.2007.00121.x
108. Kim EJ, Park HJ, Park TH. Inhibition of Apoptosis by Recombinant 30K Protein Originating From Silkworm Hemolymph. Biochem Biophys Res Commun (2003) 308:523–8. doi: 10.1016/S0006-291X(03)01425-6
109. Ye L, Zhang Y, Dong Z, Guo P, Zhao D, Li H, et al. Five Silkworm 30K Proteins are Involved in the Cellular Immunity Against Fungi. Insects (2021) 12:107. doi: 10.3390/insects12020107
110. Ujita M, Katsuno Y, Kawachi I, Ueno Y, Banno Y, Fujii H, et al. Glucan-Binding Activity of Silkworm 30-kDa Apolipoprotein and its Involvement in Defense Against Fungal Infection. J Agric Chem Soc Jpn (2005) 69:1178–85. doi: 10.1271/bbb.69.1178
111. Li R, Hu C, Shi Y, Geng T, Lv D, Gao K, et al. Silkworm Storage Protein Bm30K-19G1 has a Certain Antifungal Effects on Beauveria Bassiana. J Invertebr Pathol (2019) 163:34–42. doi: 10.1016/j.jip.2019.02.008
112. Lv HG, Chen KP, Yao Q, Wang L. Cloning and Characterization of Hydroxypyruvate Isomerase (EC 5.3.1.22) Gene in Silkworm Bombyx Mori. Int J Indust Entomol (2008) 17:189–95.
113. Zhou R, Bonneaud N, Yuan CX, de Santa Barbara P, Boizet B, Schomber T, et al. SOX9 Interacts With a Component of the Human Thyroid Hormone Receptor-Associated Protein Complex. Nucleic Acids Res (2002) 30:3245–52. doi: 10.1093/nar/gkf443
114. Garbe JC, Yang E, Fristrom JW. IMP-L2: An Essential Secreted Immunoglobulin Family Member Implicated in Neural and Ectodermal Development in Drosophila. Development (1993) 119:1237–50. doi: 10.1242/dev.119.4.1237
115. Sarraf-Zadeh L, Christen S, Sauer U, Cognigni P, Miguel-Aliaga I, Stocker H, et al. Local Requirement of the Drosophila Insulin Binding Protein Imp-L2 in Coordinating Developmental Progression With Nutritional Conditions. Dev Biol (2013) 381:97–106. doi: 10.1016/j.ydbio.2013.06.008
116. Krejčová G, Danielová A, Nedbalová P, Kazek M, Strych L, Chawla G, et al. Drosophila Macrophages Switch to Aerobic Glycolysis to Mount Effective Antibacterial Defense. eLife (2019) 8:e50414. doi: 10.7554/eLife.50414
117. Kato Y, Taniai K, Hirochika H, Yamakawa M. Expression and Characterization of cDNAs for Cecropin B, an Antibacterial Protein of the Silkworm, Bombyx Mori. Insect Biochem Mol Biol (1993) 23:285–90. doi: 10.1016/0965-1748(93)90009-H
118. Brady D, Grapputo A, Romoli O, Sandrelli F. Insect Cecropins, Antimicrobial Peptides With Potential Therapeutic Applications. Int J Mol Sci (2019) 20:5862. doi: 10.3390/ijms20235862
119. Liu W-T, Tu W-C, Lin C-H, Yang U-C, Chen C-C. Involvement of Cecropin B in the Formation of the Aedes Aegypti Mosquito Cuticle. Sci Rep (2017) 7:16395. doi: 10.1038/s41598-017-16625-6
120. Matsumoto H, Tsuzuki S, Date-Ito A, Ohnishi A, Hayakawa Y. Characteristics Common to a Cytokine Family Spanning Five Orders of Insects. Insect Biochem Mol Biol (2012) 42:446–54. doi: 10.1016/j.ibmb.2012.03.001
121. Jarvis DL, Bohlmeyer DA, Liao Y-F, Lomax KK, Merkle RK, Weinkauf C, et al. Isolation and Characterization of a Class II a-Mannosidase cDNA From Lepidopteran Insect Cells. Glycobiology (1997) 7:113–27. doi: 10.1093/glycob/7.1.113
122. Liu Y-L, Lu W-C, Brummel TJ, Yuh C-H, Lin P-T, Kao T-Y, et al. Reduced Expression of Alpha-1,2-Mannosidase I Extends Lifespan in Drosophila Melanogaster and Caenorhabditis Elegans. Aging Cell (2009) 8:370–9. doi: 10.1111/j.1474-9726.2009.00471.x
123. Ahmed S, King Y. An Aquaporin Mediates Cell Shape Change Required for Cellular Immunity in the Beet Armyworm, Spodoptera Exigua. Sci Rep (2019) 9:4988. doi: 10.1038/s41598-019-41541-2
124. Anderson DM, George R, Noyes MB, Rowton M, Liu W, Jiang R, et al. Characterization of the DNA-Binding Properties of the Mohawk Homeobox Transcription Factor. J Biol Chem (2012) 287:35351–9. doi: 10.1074/jbc.M112.399386
125. Koda N, Sato T, Shinohara M, Ichinose S, Ito Y, Nakamichi R, et al. The Transcription Factor Mohawk Homeobox Regulates Homeostasis of the Periodontal Ligament. Development (2017) 144:313–20. doi: 10.1242/dev.135798
126. Beetz S, Brinkmann M, Trenczek T. Differences Between Larval and Pupal Hemocytes of the Tobacco Hornworm, Manduca Sexta, Determined by Monoclonal Antibodies and Density Centrifugation. J Insect Physiol (2004) 50:805–19. doi: 10.1016/j.jinsphys.2004.06.003
127. He S, Tong X, Han M, Hu H, Dai F. Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx Mori. Int J Mol Sci (2018) 19:527. doi: 10.3390/ijms19020527
128. Bustamante-Marin XM, Horani A, Stoyanova M, Charng W-L, Bottier M, Sears PR, et al. Mutation of CFAP57, a Protein Required for the Asymmetric Targeting of a Subset of Inner Dynein Arms in Chlamydomonas, Causes Primary Ciliary Dyskinesia. PloS Genet (2020) 16:e1008691. doi: 10.1371/journal.pgen.1008691
129. Rorick NK, Kinoshita A, Weirather J, Peyrard-Janvid M, Ferreira de Lima RLL, Dunnwald M, et al. Genomic Strategy Identifies a Missense Mutation in WD-Repeat Domain 65 (WDR65) in an Individual With Van Der Woude Syndrome. Am J Med Genet A (2011) 155A:1314–21. doi: 10.1002/ajmg.a.33980
130. Liegeois S, Ferrandon D. An Atlas for Hemocytes in an Insect. eLife (2020) 9:e59113. doi: 10.7554/eLife.59113
131. Bozler J, Kacsoh BZ, Bosco G. Nematocytes: Discovery and Characterization of a Novel Anculeate Hemocyte in Drosophila Falleni and Drosophila Phalerata. PloS One (2017) 12:e0188133. doi: 10.1371/journal.pone.0188133
132. Takase T, Hirai Y. Identification of the C-Terminal Tail Domain of AHF/trichohyalin as the Critical Site for Modulation of the Keratin Filamentous Meshwork in the Keratinocyte. J Dermatol Sci (2012) 65:141–8. doi: 10.1016/j.jdermsci.2011.12.014
133. Simons M. Flies With Skin Blisters. J Invest Dermatol (2015) 135:1944–5. doi: 10.1038/jid.2015.193
134. Zhang J-Y, Pan M-H, Sun Z-Y, Huang S-J, Yu Z-S, Liu D, et al. The Genomic Underpinnings of Apoptosis in the Silkworm, Bombyx Mori. BMC Genomic (2010) 11:611. doi: 10.1186/1471-2164-11-611
135. Damgaard RB, Gyrd-Hansen M. Inhibitor of Apoptosis (IAP) Proteins in Regulation of Inflammation and Innate Immunity. Discov Med (2011) 11:221–31.
136. Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila Inhibitor of Apoptosis Protein DIAP2 Functions in Innate Immunity and is Essential to Resist Gram-Negative Bacterial Infection. Mol Cell Biol (2006) 26:7821–31. doi: 10.1128/MCB.00548-06
137. Liu J, Xheng Q, Deng Y, Cheng C-S, Kallenbach NR, Lu M. A Seven-Helix Coiled Coil. Proc Natl Acad Sci USA (2006) 103:15457–62. doi: 10.1073/pnas.0604871103
138. Kim YJ, Lee KS, Kim BY, Choo YM, Sohn HD, Jin BR. Thioredoxin From the Silkworm, Bombyx Mori: cDNA Sequence, Expression, and Functional Characterization. Comp Biochem Physiol B (2007) 147:574–81. doi: 10.1016/j.cbpb.2007.03.012
139. Li E, Yu X-Q. Cellular Encapsulation and Melanization are Enhanced by Immulectins, Pattern Recognition Receptors From the Tobacco Hornworm Manduca Sexta. Dev Comp Immunol (2006) 30:289–99. doi: 10.1016/j.dci.2005.05.005
140. Waltzer L, Ferjoux G, Bataillé L, Haenlin M. Cooperation Between the GATA and RUNX Factors Serpent and Lozenge During Drosophila Hematopoiesis. EMBO J (2003) 22:6516–25. doi: 10.1093/emboj/cdg622
141. Xu M, Wang X, Tan J, Zhang K, Guan X, Patterson LH, et al. A Novel Lozenge Gene in Silkworm, Bombyx Mori Regulates the Melanization Response of Hemolymph. Dev Comp Immunol (2015) 53:191–8. doi: 10.1016/j.dci.2015.07.001
142. Yamanaka N, Rewitz KF, O’Connor MB. Ecdysone Control of Developmental Transitions: Lessons From Drosophila Research. Annu Rev Entomol (2013) 58:497–516. doi: 10.1146/annurev-ento-120811-153608
143. Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, et al. Ecdysone Triggered PGRP-LC Expression Controls Drosophila Innate Immunity. EMBO J (2013) 32:1626–38. doi: 10.1038/emboj.2013.100
144. Breitenbach JE, Shelby KS, Popham HJR. Baculovirus Induced Transcripts in Hemocytes From the Larvae of Heliothis Virescens. Viruses (2011) 3:2047–64. doi: 10.3390/v3112047
145. Sawada H, Iino T, Tsusué M. Properties of Ommochrome-Binding Proteins From the Pigment Granules in Epidermal Cells of the Silkworm, Bombyx Mori. J Sericult Sci Jpn (1997) 66:393–402. doi: 10.11416/kontyushigen1930.66.393
146. Sawada H, Nakagoshi M, Mase K, Yamamoto T. Occurrence of Ommochrome-Containing Pigment Granules in the Central Nervous System of the Silkworm, Bombyx Mori. Comp. Biochem Physiol (2000) 125:421–8. doi: 10.1016/S0305-0491(99)00179-0
147. Insausti TC, Casas J. Turnover of Pigment Granules: Cyclic Catabolism and Anabolism of Ommochromes Within Epidermal Cells. Tissue Cell (2009) 41:421–9. doi: 10.1016/j.tice.2009.05.002
148. Yepiz-Plascencia GM, Ho C, Martel RR, Law JH. Molecular Cloning and Sequence of a Novel Ommochrome-Binding Protein From an Insect, Manduca Sexta. J Biol Chem (1993) 268:2337–40. doi: 10.1016/S0021-9258(18)53780-7
149. Figon F, Casas J. Ommochromes in Invertebrates: Biochemistry and Cell Biology. Biol Rev (2019) 94:156–83. doi: 10.1111/brv.12441
150. Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the Serpin Superfamily: Implications of Patterns of Amino Acid Conservation for Structure and Function. Genome Res (2000) 10:1845–64. doi: 10.1101/gr.147800
151. Bao J, Pan G, Poncz M, Wei J, Ran M, Zhou Z. Serpin Functions in Host-Pathogen Interactions. Peer J (2018) 6:e4557. doi: 10.7717/peerj.4557
152. Meekins DA, Kanost MR, Michel K. Serpins in Arthropod Biology. Semin Cell Dev Biol (2017) 62:105–19. doi: 10.1016/j.semcdb.2016.09.001
153. An C, Ragan EJ, Kanost MR. Serpin-1 Splicing Isoform J Inhibits the Prospätzle-Activating Proteinase HP8 to Regulate Expression of Antimicrobial Hemolymph Proteins in Manduca Sexta. Dev Comp Immunol (2011) 35:135–41. doi: 10.1016/j.dci.2010.09.004
154. Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-Activating Proteinase-3 (PAP-3) From Manduca Sexta Hemolymph: A Clip-Domain Serine Proteinase Regulated by Serpin-1J and Serine Proteinase Homologs. Insect Biochem Mol Biol (2003) 33:1049–60. doi: 10.1016/S0965-1748(03)00123-1
155. He Y, Wang Y, Yang F, Jiang H. Manduca Sexta Hemolymph Protease-1, Activated by an Unconventional non-Proteolytic Mechanism, Mediates Immune Responses. Insect Biochem. Mol Biol (2017) 84:23–31. doi: 10.1016/j.ibmb.2017.03.008
156. Liu Y, Zhou S, Ma L, Tian L, Wang S, Sheng Z, et al. Transcriptional Regulation of the Insulin Signaling Pathway Genes by Starvation and 20-Hydroxyecdysone in the Bombyx Fat Body. J Insect Physiol (2010) 56:1436–44. doi: 10.1016/j.jinsphys.2010.02.011
157. Garrett TP, McKern NM, Lou M, Frenkel MJ, Bentley JD, Lovrecz GO, et al. Crystal Structure of the First Three Domains of the Type-1 Insulin-Like Growth Factor Receptor. Nature (1998) 394:395–9. doi: 10.1038/28668
158. Marino-Buslje C, Mizuguchi K, Siddle K, Blundell TL. A Third Fibronectin Type III Domain in the Extracellular Region of the Insulin Receptor Family. FEBS Lett (1998) 441:331–6. doi: 10.1016/S0014-5793(98)01509-9
159. Uchikawa E, Choi E, Shang G, Yu H, Bai X-C. Activation Mechanism of the Insulin Receptor Revealed by Cryo-EM Structure of the Fully Liganded Receptor–Ligand Complex. eLife (2019) 8:e48630. doi: 10.7554/eLife.48630
160. Akiyama SK, Nagata K, Yamada KM. Cell Surface Receptors for Extracellular Matrix Components. Biochim Biophys Acta Biomembr (1990) 1031:91–110. doi: 10.1016/0304-4157(90)90004-V
161. Bennett JS. Structure and Function of the Platelet Integrin αIIbβ3. J Clin Invest (2005) 115:3363–9. doi: 10.1172/JCI26989
162. Levin DM, Breuer LN, Zhuang S, Anderson SA, Nardi JB, Kanost MR. A Hemocyte-Specific Integrin Required for Hemocytic Encapsulation in the Tobacco Hornworm, Manduca Sexta. Insect Biochem. Mol Biol (2005) 35:369–80. doi: 10.1016/j.ibmb.2005.01.003
163. Hu J, Zhao H, Yu X, Liu J, Wang P, Chen J, et al. Integrin β1 Subunit From Ostrinia Furnacalis Hemocytes: Molecular Characterization, Expression, and Effects on the Spreading of Plasmatocytes. J Insect Physiol (2010) 56:1846–56. doi: 10.1016/j.jinsphys.2010.08.001
164. Xu Q, Yu X, Liu J, Zhao H, Wang P, Hu S, et al. Ostrinia Furnacalis Integrin β1may be Involved in Polymerization of Actin to Modulate Spreading and Encapsulation of Plasmatocytes. Dev Comp Immunol (2012) 37:438–45. doi: 10.1016/j.dci.2012.02.003
165. Zhang K, Xu M, Su J, Yu S, Sun Z, Li Y, et al. Characterization and Identification of the Integrin Family in Silkworm Bombyx Mori. Gene (2014) 549:149–55. doi: 10.1016/j.gene.2014.07.060
166. Zhang K, Tan J, Su J, Liang H, Shen L, Li C, et al. Integrin B3 Plays a Novel Role in Innate Immunity in Silkworm, Bombyx Mori. Dev Comp Immunol (2017) 77:307–17. doi: 10.1016/j.dci.2017.08.009
167. Teerawanichpan P, Robertson AJ, Qiu X. A Fatty Acyl-CoA Reductase Highly Expressed in the Head of Honey Bee (Apis Mellifera) Involves Biosynthesis of a Wide Range of Aliphatic Fatty Alcohols. Insect Biochem Mol Biol (2010) 40:641–9. doi: 10.1016/j.ibmb.2010.06.004
168. Moto K, Yoshiga T, Yamamoto M, Takahashi S, Okano K, Ando T, et al. Pheromone Gland-Specific Fatty-Acyl Reductase of the Silkmoth, Bombyx Mori. Proc Natl Acad Sci USA (2003) 100:9156–61. doi: 10.1073/pnas.1531993100
169. Jaspers MH, Pflanz R, Riedel D, Kawelke S, Feussner I, Schuh R. The Fatty Acyl-CoA Reductase Waterproof Mediates Airway Clearance in Drosophila. Dev Biol (2014) 385:23–31. doi: 10.1016/j.ydbio.2013.10.022
170. Iatrou K, Swevers L. Transformed Lepidopteran Cells Expressing a Protein of the Silkmoth Fat Body Display Enhanced Susceptibility to Baculovirus Infection and Produce High Titers of Budded Virus in Serum-Free Media. J Biotechnol (2005) 120:237–50. doi: 10.1016/j.jbiotec.2005.06.037
171. Strand MR, Hayakawa Y, Clark KD. Plasmatocyte Spreading Peptide (PSP1) and Growth Blocking Peptide (GBP) are Multifunctional Homologs. J Insect Physiol (2000) 46:817–24. doi: 10.1016/S0022-1910(99)00171-7
172. Zelensky AN, Gready JE. The C-Type Lectin-Like Domain Superfamily. FEBS J (2005) 272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x
173. Pees B, Yang W, Zárate-Potes A, Schulenburg H, Dierking K. High Innate Immune Specificity Through Diversified C-Type Lectin-Like Domain Proteins in Invertebrates. J Innate Immun (2016) 8:129–42. doi: 10.1159/000441475
174. Rao X-J, Shahzad T, Liu S, Wu P, He Y-T, Sun W-J, et al. Identification of C-Type Lectin-Domain Proteins (CTLDPs) in Silkworm Bombyx Mori. Dev Comp Immunol (2015) 53:328–38. doi: 10.1016/j.dci.2015.07.005
175. Koizumi N, Imamura M, Kadotani T, Yaoi K, Iwahana H, Sato R. The Lipopolysaccharide-Binding Protein Participating in Hemocyte Nodule Formation in the Silkworm Bombyx Mori is a Novel Member of the C-Type Lectin Superfamily With Two Different Tandem Carbohydrate-Recognition Domains. FEBS Lett (1999) 443:139–43. doi: 10.1016/S0014-5793(98)01701-3
176. Watanabe A, Miyazawa S, Kitami M, Tabunoki H, Ueda K, Sato R. Characterization of a Novel C-Type Lectin, Bombyx Mori Multibinding Protein, From the B. Mori Hemolymph: Mechanism of Wide-Range Microorganism Recognition and Role in Immunity. J Immunol (2006) 177:4594–604. doi: 10.4049/jimmunol.177.7.4594
177. Shahzad T, Zhan M-Y, Yang P-J, Yu X-Q, Rao X-J. Molecular Cloning and Analysis of a C-Type Lectin From Silkworm Bombyx Mori. Arch Insect Biochem Physiol (2017) 95:e21391. doi: 10.1002/arch.21391
178. Geng T, Lu F, Wu H, Wang Y, Lou D, Tu N, et al. C-Type Lectin 5, A Novel Pattern Recognition Receptor for the JAK/STAT Signaling Pathway in Bombyx Mori. J Invertebr Pathol (2021) 179:107473. doi: 10.1016/j.jip.2020.107473
179. Shen D, Tong M, Guo J, Mei X, Xia D, Qiu Z, et al. A Pattern Recognition Receptor C-Type Lectin-S6 (CTL-S6) is Involved in the Immune Response in the Silkworm (Lepidoptera: Bombycidae). J Insect Sci (2021) 21:1–8. doi: 10.1093/jisesa/ieaa146
180. Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, et al. Cryo-EM Reconstruction of Dengue Virus in Complex With the Carbohydrate Recognition Domain of DC-SIGN. Cell (2006) 124:485–93. doi: 10.1016/j.cell.2005.11.042
181. Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier J-L, et al. Dendritic- Cell-Specific ICAM3-Grabbing non-Integrin is Essential for the Productive Infection of Human Dendritic Cells by Mosquito-Cell-Derived Dengue Viruses. EMBO Rep (2003) 4:723–8. doi: 10.1038/sj.embor.embor866
182. Tassaneetrithep B, Burgess T, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, et al. DC-SIGN (CD209) Mediates Dengue Virus Infection of Human Dendritic Cells. J Exp Med (2003) 197:823–9. doi: 10.1084/jem.20021840
183. Hori T, Kiuchi, Shimada T, Nagata M, Katsuma S. Silkworm Plasmatocytes are More Resistant Than Other Hemocyte Morphotypes to Bombyx Mori Nucleopolyhedrovirus Infection. J Invertebr Pathol (2013) 112:102–4. doi: 10.1016/j.jip.2012.09.004
184. Zhao P, Wang G-H, Dong Z-M, Duan J, Xu P-Z, Cheng T-C, et al. Genome-Wide Identification and Expression Analysis of Serine Proteases and Homologs in the Silkworm Bombyx Mori. BMC Genomics (2010) 11:405. doi: 10.1186/1471-2164-11-405
185. Satoh D, Horii A, Ochiai M, Ashida M. Prophenoloxidase-Activating Enzyme of the Silkworm, Bombyx Mori: Purification, Characterization and cDNA Cloning. J Biol Chem (1999) 274:7441–53. doi: 10.1074/jbc.274.11.7441
186. Theopold U, Krautz R, Dushay MS. The Drosophila Clotting System and its Messages for Mammals. Dev Comp Immunol (2014) 42:42–6. doi: 10.1016/j.dci.2013.03.014
187. Phillips DR, Clark KD. Bombyx Mori and Aedes Aegypti Form Multi-Functional Immune Complexes That Integrate Pattern Recognition, Melanization, Coagulants, and Hemocyte Recruitment. PloS One (2017) 12:e0171447. doi: 10.1371/journal.pone.0171447
188. Ochiai M, Ashida M. A Pattern-Recognition Protein for β-1,3-Glucan. The Binding Domain and the cDNA Cloning of Beta-1,3-Glucan Recognition Protein From the Silkworm, Bombyx Mori. J Biol Chem (2000) 275:4995–5002. doi: 10.1074/jbc.275.7.4995
189. Jossin Y. Reelin Functions, Mechanisms of Action and Signaling Pathways During Brain Development and Maturation. Biomolecules (2020) 10:964. doi: 10.3390/biom10060964
190. Tatham AS, Shrewny PR. Elastomeric Proteins: Biological Roles, Structures and Mechanisms. Trends Biochem Sci (2000) 25:567–71. doi: 10.1016/S0968-0004(00)01670-4
191. Ingolia TD, Craig EA. Four Small Drosophila Heat Shock Proteins are Related to Each Other and to Mammalian Alpha-Crystallin. Proc Natl Acad Sci USA (1982) 79:2360–4. doi: 10.1073/pnas.79.7.2360
192. Li Z-W, Li X, Yu Q-Y, Xiang Z-H, Kishino H, Zhang Z. The Small Heat Shock Protein (sHSP) Genes in the Silkworm, Bombyx Mori, and Comparative Analysis With Other Insect sHSP Genes. BMC Evol Biol (2009) 9:215. doi: 10.1186/1471-2148-9-215
193. Manjunatha HB, Rajesh RK, Aparna HS. Silkworm Thermal Biology: A Review of Heat Shock Response, Heat Shock Proteins and Heat Acclimation in the Domesticated Silkworm, Bombyx Mori. J Insect Sci (2010) 10:204. doi: 10.1673/031.010.20401
194. Bao Y, Wang Q, Liu H, Lin Z. A Small HSP Gene of Bloody Clam (Tegillarca Granosa) Involved in the Immune Response Against Vibrio Parahaemolyticus and Lipopolysaccharide. Fish Shellfish Immunol (2011) 30:729–33. doi: 10.1016/j.fsi.2010.12.002
195. Li C-S, Kausar S, Gul I, Yao X-X, Li M-Y, Chen C-C, et al. Heat Shock Protein 20 From Procambarus Clarkii is Involved in the Innate Immune Responses Against Microbial Infection. Dev Comp Immunol (2020) 106:103638. doi: 10.1016/j.dci.2020.103638
196. Zhao P, Dong Z, Duan J, Wang G, Wang L, Li Y, et al. Genome-Wide Identification and Immune Response Analysis of Serine Protease Inhibitor Genes in the Silkworm, Bombyx Mori. PloS One (2012) 7:e31168. doi: 10.1371/journal.pone.0031168
197. Wright HT. The Structural Puzzle of How Serpin Serine Proteinase Inhibitors Work. Bioessays (1996) 18:453–64. doi: 10.1002/bies.950180607
198. Zou Z, Picheng Z, Weng H, Mita K, Jiang H. A Comparative Analysis of Serpin Genes in the Silkworm Genome. Genomics (2009) 93:367–75. doi: 10.1016/j.ygeno.2008.12.010
199. He Y, Wang Y, Zhao P, Rayaprolu S, Wang X, Cao X, et al. Serpin-9 and -13 Regulate Hemolymph Proteases During Immune Responses of Manduca Sexta. Insect Biochem. Mol Biol (2017) 90:71–81. doi: 10.1016/j.ibmb.2017.09.015
200. Yuan C, Xing L, Wang M, Wang X, Yin M, Wang Q, et al. Inhibition of Melanization by Serpin-5 and Serpin-9 Promotes Baculovirus Infection in Cotton Bollworm Helicoverpa Armigera. PloS Pathog (2017) 13:e1006645. doi: 10.1371/journal.ppat.1006645
201. Samaraweera P, Law JH. Isolation, Cloning and Deduced Amino Acid Sequence of a Novel Glycoprotein From the Haemolymph of the Hawkmoth Manduca Sexta. Insect Mol Biol (1995) 4:7–13. doi: 10.1111/j.1365-2583.1995.tb00002.x
202. Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, et al. Broad RNA Interference–Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. J Immunol (2013) 190:650–8. doi: 10.4049/jimmunol.1102486
203. Rider MA, Zou J, Vanlandingham D, Nuckols JT, Higgs S, Zhang Q, et al. Quantitative Proteomic Analysis of the Anopheles Gambiae (Diptera: Culicidae) Midgut Infected With O'nyong—Nyong Virus. J Med Entomol (2013) 50:1077–88. doi: 10.1603/ME12155
204. Undheim EAB, Jones A, Clauser KR, Holland JW, Pineda SS, King GF, et al. Clawing Through Evolution: Toxin Diversification and Convergence in the Ancient Lineage Chilopoda (Centipedes). Mol Biol Evol (2014) 31:2124–48. doi: 10.1093/molbev/msu162
205. Olson SK, Bishop JR, Yates JR, Oegema K, Esko JD. Identification of Novel Chondroitin Proteoglycans in Caenorhabditis Elegans: Embryonic Cell Division Depends on CPG-1 and CPG-2. J Cell Biol (2006) 173:985–94. doi: 10.1083/jcb.200603003
206. Ye J, Li Y, Liu H-W, Li J, Dong Z, Xia Q, et al. Genome-Wide Identification and Characterization of Carboxypeptidase Genes in Silkworm (Bombyx Mori). Int J Mol Sci (2016) 17:1203. doi: 10.3390/ijms17081203
207. Leung LLK, Morser J. Carboxypeptidase B2 and Carboxypeptidase N in the Crosstalk Between Coagulation, Thrombosis, Inflammation, and Innate Immunity. J Thromb Haemost (2018) 16:1474–86. doi: 10.1111/jth.14199
208. Loof TG, Schmidt O, Herwald H, Theopold U. Coagulation Systems of Invertebrates and Vertebrates and Their Roles in Innate Immunity: The Same Side of Two Coins? J Innate Immun (2011) 3:34–40. doi: 10.1159/000321641
209. Qian X, Lai Y, Zhu F. Molecular Characterization of Carboxypeptidase B-Like (CPB) in Scylla Paramamosain and its Role in White Spot Syndrome Virus and Vibrio Alginolyticus Infection. Fish Shellfish Immunol (2019) 94:434–46. doi: 10.1016/j.fsi.2019.09.036
210. Gavor E, Choong YK, Tulsian NK, Nayak D, Idris F, Sivaraman H, et al. Structure of Aedes Aegypti Procarboxypeptidase B1 and its Binding With Dengue Virus for Controlling Infection. Life Sci Alliance (2021) 5:e202101211. doi: 10.26508/lsa.202101211
211. Ciufo LF, Murray PA, Thompson A, Rigden DJ, Rees HH. Characterisation of a Desmosterol Reductase Involved in Phytosterol Dealkylation in the Silkworm, Bombyx Mori. PloS One (2011) 6:e21316. doi: 10.1371/journal.pone.0021316
212. Fujimori H, Zhou YJ, Fukumura K, Matsumoto S, Tukamoto Y, Nagata S. Specific Distribution of Expression and Enzymatic Activity of Cholesterol Biosynthetic Enzyme DHCR24 Orthologs in the Phytophagous Insect. Biosci Biotechnol Biochem (2020) 84:126–33. doi: 10.1080/09168451.2019.1667221
213. Clark AJ, Block K. The Absence of Sterol Synthesis in Insects. J Biol Chem (1959) 234:2578–82. doi: 10.1016/S0021-9258(18)69741-8
214. Körner A, Zhou E, Müller C, Mohammed Y, Herceg S, Bracher F, et al. Inhibition of Δ24-Dehydrocholesterol Reductase Activates Pro-Resolving Lipid Mediator Biosynthesis and Inflammation Resolution. Proc Natl Acad Sci USA (2019) 116:20623–34. doi: 10.1073/pnas.1911992116
215. Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, et al. Regulated Accumulation of Desmosterol Integrates Macrophage Lipid Metabolism and Inflammatory Responses. Cell (2012) 151:138–52. doi: 10.1016/j.cell.2012.06.054
216. Tong Y, Jiang H, Kanost MR. Identification of Plasma Proteases Inhibited by Manduca Sexta Serpin-4 and Serpin-5 and Their Association With Components of the Prophenol Oxidase Activation Pathway. J Biol Chem (2005) 280:14932–42. doi: 10.1074/jbc.M500532200
217. Aoki K, Tiemeyer M. The Glycomics of Glycan Glucuronylation in Drosophila Melanogaster. Methods Enzymol (2010) 480:297–321. doi: 10.1016/S0076-6879(10)80014-X
218. Huang F-F, Chai C-L, Zhang Z, Liu Z-H, Dai F-Y, Lu C, et al. The UDP-Glucosyltransferase Multigene Family in Bombyx Mori. BMC Genomics (2008) 9:563. doi: 10.1186/1471-2164-9-563
219. Walski T, De Schutter K, Van Damme EJM, Smagghe G. Diversity and Functions of Protein Glycosylation in Insects. Insect Biochem Mol Biol (2017) 83:21–34. doi: 10.1016/j.ibmb.2017.02.005
220. Pandey R, Blanco J, Udolph G. The Glucuronyltransferase GlcAT-P is Required for Stretch Growth of Peripheral Nerves in Drosophila. PloS One (2011) 6:e28106. doi: 10.1371/journal.pone.0028106
221. Yamamoto-Hino M, Yoshida H, Ichimiya T, Sakamura S, Maeda M, Kimura Y, et al. Phenotype-Based Clustering of Glycosylation-Related Genes by RNAi-Mediated Gene Silencing. Genes Cells (2015) 20:521e542. doi: 10.1111/gtc.12246
222. Mase A, Augsburger J, Brückner K. Macrophages and Their Organ Locations Shape Each Other in Development and Homeostasis – A Drosophila Perspective. Front Cell Dev Biol (2021) 9:630272. doi: 10.3389/fcell.2021.630272
223. Fessler JH, Kramerova I, Kramerov A, Chen Y, Fessler LI. Papilin, a Novel Component of Basement Membranes, in Relation to ADAMTS Metalloproteases and ECM Development. Int J Biochem Cell Biol (2004) 36:1079–84. doi: 10.1016/j.biocel.2003.12.010
224. Nardi JB, Martos R, Walden KK, Lampe DJ, Robertson HM. Expression of Lacunin, a Large Multidomain Extracellular Matrix Protein, Accompanies Morphogenesis of Epithelial Monolayers in Manduca Sexta. Insect Biochem Mol Biol (1999) 29:883–97. doi: 10.1016/s0965-1748(99)00064-8
225. Nardi JB, Gao C, Kanost MR. The Extracellular Matrix Protein Lacunin is Expressed by a Subset of Hemocytes Involved in Basal Lamina Morphogenesis. J Insect Physiol (2001) 47:997–1006. doi: 10.1016/S0022-1910(01)00074-9
226. Kramerova IA, Kramerov AA, Fessler JH. Alternative Splicing of Papilin and the Diversity of Drosophila Extracellular Matrix During Embryonic Morphogenesis. Dev Dyn (2003) 226:634–42. doi: 10.1002/dvdy.10265
227. Kramerova IA, Kawaguchi N, Fessler LI, Nelson RE, Chen Y, Kramerov AA, et al. Papilin in Development; a Pericellular Protein With a Homology to the ADAMTS Metalloproteinases. Development (2000) 127:5475–85. doi: 10.1242/dev.127.24.5475
228. Pantha P, Chalivendra S, Oh D-H, Elderd BD, Dassanayake M. A Tale of Two Transcriptomic Responses in Agricultural Pests via Host Defenses and Viral Replication. Int J Mol Sci (2021) 22:3568. doi: 10.3390/ijms22073568
229. Campbell AG, Fessler LI, Salo T, Fessler JH. Papilin: A Drosophila Proteoglycan-Like Sulfated Glycoprotein From Basement Membranes. J Biol Chem (1987) 262:17605–12. doi: 10.1016/S0021-9258(18)45424-5
230. Savinova OV, Hoffman A, Ghosh G. The Nfkb1 and Nfkb2 Proteins P105 and P100 Function as the Core of High-Molecular-Weight Heterogeneous Complexes. Mol Cell (2009) 34:591–602. doi: 10.1016/j.molcel.2009.04.033
231. Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, et al. Caspase-Mediated Processing of the Drosophila NF-κB Factor Relish. Proc Natl Acad Sci USA (2003) 100:5991–6. doi: 10.1073/pnas.1035902100
232. Hua X, Li B, Song L, Hu C, Li X, Wang D, et al. Stimulator of Interferon Genes (STING) Provides Insect Antiviral Immunity by Promoting Dredd Caspase-Mediated NF-kB Activation. J Biol Chem (2018) 293:11878–90. doi: 10.1074/jbc.RA117.000194
233. Spasic M, Friedel CC, Schott J, Kreth J, Leppek K, Hofmann S, et al. Genome-Wide Assessment of AU-Rich Elements by the AREScore Algorithm. PloS Genet (2012) 8:e1002433. doi: 10.1371/journal.pgen.1002433
234. Lauwers A, Twyffels L, Soin R, Wauquier C, Kruys V, Gueydan C. Posttranscriptional Regulation of Genes Encoding Anti-Microbial Peptides in Drosophila. J Biol Chem (2009) 284:8973–83. doi: 10.1074/jbc.M806778200
235. McCelland L, Jasper H, Biteau B. Tis11 Mediated mRNA Decay Promotes the Reacquisition of Drosophila Intestinal Stem Cell Quiescence. Dev Biol (2017) 426:8–16. doi: 10.1016/j.ydbio.2017.04.013
236. Li FQ, Ueda H, Hirose S. Mediators of Activation of Fushi Tarazu Gene Transcription by BmFTZ-F1. Mol Cell Biol (1994) 14:3013–21. doi: 10.1128/mcb.14.5.3013
237. Zhou C-Y, Zha X-F, Liu C, Han M-J, Zhang L-Y, Shi P-P, et al. Identification of MBF2 Family Genes in Bombyx Mori and Their Expression in Different Tissues and Stages and in Response to Bacillus Bombysepticus Infection and Starvation. Insect Sci (2016) 23:502–12. doi: 10.1111/1744-7917.12349
238. Herrero S, Ansems M, Van Oers MM, Vlak JM, Bakker PL, De Maagd RA. REPAT, a New Family of Proteins Induced by Bacterial Toxins and Baculovirus Infection in Spodoptera Exigua. Insect Biochem. Mol Biol (2007) 37:1109–18. doi: 10.1016/j.ibmb.2007.06.007
239. Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell (2016) 167:1598–609. doi: 10.1016/j.cell.2016.11.012
240. Angajala A, Lim S, Phillips JB, Kim J-H, Yates C, You Z, et al. Diverse Roles of Mitochondria in Immune Responses: Novel Insights Into Immuno-Metabolism. Front Immunol (2018) 9:1605. doi: 10.3389/fimmu.2018.01605
241. Pan G, Zhang K, Li C, Hu X, Kausar S, Gu H, et al. A Hemocyte-Specific Cathepsin L-Like Cysteine Protease is Involved in Response to 20-Hydroxyecdysone and Microbial Pathogens Stimulation in Silkworm, Bombyx Mori. Mol Immunol (2021) 131:78–88. doi: 10.1016/j.molimm.2020.12.013
242. Pastor-Pareja JC, Xu T. Shaping Cells and Organs in Drosophila by Opposing Roles of Fat Body-Secreted Collagen IV and Perlecan. Dev Cell (2011) 21:245–56. doi: 10.1016/j.devcel.2011.06.026
243. Ma M, Cao X, Dai J, Pastor-Pareja J. Basement Membrane Manipulation in Drosophila Wing Discs Affects Dpp Retention But Not Growth Mechanoregulation. Dev Cell (2017) 42:97–106. doi: 10.1016/j.devcel.2017.06.004
244. Hatz RA, von Jan NCS, Schildberg FW. The Role of Collagenase in Wound Healing. In: Westerhof W, Vanscheidt W, editors. Proteolytic Enzymes and Wound Healing. Berlin, Heidelberg: Springer (1994). p. 75–88.
245. Chen Z, Nohata J, Guo H, Li S, Liu J, Guo Y, et al. A Comprehensive Analysis of the Chorion Locus in Silkmoth. Sci Rep (2015) 5:16424. doi: 10.1038/srep16424
246. Lu A, Zhang Q, Zhang J, Yang B, Wu K, Xie W, et al. Insect Prophenoloxidase: The View Beyond Immunity. Front Physiol (2014) 5:252. doi: 10.3389/fphys.2014.00252
247. Koranteng F, Cha N, Shin M, Shim J. The Role of Lozenge in Drosophila Hematopoiesis. Mol Cells (2020) 43:114–20. doi: 10.14348/molcells.2019.0249
248. Akai H, Sato S. Ultrastructure of the Larval Hemocytes of the Silkworm, Bombyx Mori L. (Lepidoptera: Bombycidae). Int J Insect Morphol Embryol (1973) 2(207):231. doi: 10.1016/0020-7322(73)90029-9
249. Ashhurst DE. Histochemical Properties of the Spherulocytes of Galleria Mellonella L. (Lepidoptera: Pyralidae). Int J Insect Morphol Embryol (1982) 11:285–92. doi: 10.1016/0020-7322(82)90017-4
250. Sass M, Kiss A, Locke M. Integument and Hemocyte Peptides. J Insect Physiol (1994) 40:407–21. doi: 10.1016/0022-1910(94)90159-7
251. Papantonis A, Swevers L, Iatrou K. Chorion Genes: A Landscape of Their Evolution, Structure, and Regulation. Annu Rev Entomol (2015) 60:177–94. doi: 10.1146/annurev-ento-010814-020810
252. Swevers L, Raikhel AS, Sappington TW, Shirk P, Iatrou K. Vitellogenesis and Post-Vitellogenic Maturation of the Insect Ovarian Follicle. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science, vol. 1. Oxford, UK:Elsevier Pergamon (2005). p. 87–155.
253. Sviben S, Spaeker O, Bennet M, Alberic M, Dirks J-H, Moussian B, et al. Epidermal Cell Surface Structure and Chitin–Protein Co-Assembly Determine Fiber Architecture in the Locust Cuticle. ACS Appl Mater Interfaces (2020) 12:25581–90. doi: 10.1021/acsami.0c04572
254. Severo MS, Landry JJM, Lindquist RL, Goosmann C, Brinkmann V, Collier P, et al. Unbiased Classification of Mosquito Blood Cells by Single-Cell Genomics and High-Content Imaging. Proc Natl Acad Sci USA (2018) 115:E7568–77. doi: 10.1073/pnas.1803062115
255. Kwon H, Mohammed M, Franzén O, Ankarlev J, Smith RC. Single-Cell Analysis of Mosquito Hemocytes Identifies Signatures of Immune Cell Subtypes and Cell Differentiation. eLife (2020) 10:e66192. doi: 10.7554/eLife.66192
256. Raddi G, Barletta ABF, Efremova M, Ramirez JL, Cantera R, Teichmann SA, et al. Mosquito Cellular Immunity at Single-Cell Resolution. Science (2020) 369:1128–32. doi: 10.1126/science.abc0322
257. Honti V, Kurucz E, Csordas G, Laurinyecz B, Markus R, Ando I. In Vivo Detection of Lamellocytes in Drosophila Melanogaster. Immunol Lett (2009) 126:83–4. doi: 10.1016/j.imlet.2009.08.004
258. Ruan H, Ueda A, Xing X, Wan X, Strub B, Mukai S, et al. Generation and Characterization of New Alleles of Quiver (Qvr) That Encodes an Extracellular Modulator of the Shaker Potassium Channel. J Neurogenet (2017) 31:325–36. doi: 10.1080/01677063.2017.1393076
259. Kurucz E, Marcus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a Putative Phagocytosis Receptor With EGF Repeats in Drosophila Plasmatocytes. Curr Biol (2007) 17(7):649–54. doi: 10.1016/j.cub.2007.02.041
260. Vaarala MH, Porvari K, Kellokumpu S, Kyllonen AP, Vihko P. J. T. J. O. P. Expression of Transmembrane Serine Protease TMPRSS2 in Mouse and Human Tissues. J Pathol (2001) 193:134–40. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T
261. Mir AH, Qamar A, Qadir I, Naqvi AH, Begum R. Accumulation and Trafficking of Zinc Oxide Nanoparticles in an Invertebrate Model, Bombyx Mori, With Insights on Their Effects on Immuno-Competent Cells. Sci Rep (2020) 10:1617. doi: 10.1038/s41598-020-58526-1
262. Hillyer JF, Schmidt SL, Christensen BM. Rapid Phagocytosis and Melanization of Bacteria and Plasmodium Sporozoites by Hemocytes of the Mosquito Aedes Aegypti. J Parasitol (2003) 89:62–9. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2
263. Cattenoz PB, Monticelli S, Pavlidaki A, Giangrande A. Toward a Consensus in the Repertoire of Hemocytes Identified in Drosophila. Front Cell Dev Biol (2021) 9:643712. doi: 10.3389/fcell.2021.643712
Keywords: scRNA-seq, silkworm, hemocyte, marker gene, Bombyx mori
Citation: Feng M, Swevers L and Sun J (2022) Hemocyte Clusters Defined by scRNA-Seq in Bombyx mori: In Silico Analysis of Predicted Marker Genes and Implications for Potential Functional Roles. Front. Immunol. 13:852702. doi: 10.3389/fimmu.2022.852702
Received: 11 January 2022; Accepted: 07 February 2022;
Published: 25 February 2022.
Edited by:
Xinjiang Lu, Zhejiang University, ChinaReviewed by:
Amr A. Mohamed, Cairo University, EgyptCopyright © 2022 Feng, Swevers and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Feng, aHVuYW5mZW5nbWluQDE2My5jb20=; Luc Swevers, c3dldmVyc0BiaW8uZGVtb2tyaXRvcy5ncg==; Jingchen Sun, Y3lmekBzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.