
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 23 February 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.847728
This article is part of the Research Topic Role of Non-Coding RNA in Gastrointestinal Cancer Tumor Immune Microenvironment and Anticancer Therapy View all 5 articles
HCC is one of the most common malignant tumors and has an extremely poor prognosis. Accumulating studies have shown that noncoding RNA (ncRNA) plays an important role in hepatocellular carcinoma (HCC) development. However, the details of the related mechanisms remain unclear. The heterogeneity of the tumor microenvironment (TME) calls for ample research with deep molecular characterization, with the hope of developing novel biomarkers to improve prognosis, diagnosis and treatment. ncRNAs, particularly microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs), have been found to be correlated with HCC neogenesis and progression. In this review, we summarized the aberrant epigenetic and genetic alterations caused by dysregulated ncRNAs and the functional mechanism of classical ncRNAs in the regulation of gene expression. In addition, we focused on the role of ncRNAs in the TME in the regulation of tumor cell proliferation, invasion, migration, immune cell infiltration and functional activation. This may provide a foundation for the development of promising potential prognostic/predictive biomarkers and novel therapies for HCC patients.
Protein-coding genes have been well studied, but protein-coding regions account for only 1.5% of the whole human genome (1). Thousands of noncoding RNA (ncRNA) sequences that are not translated into proteins were considered “junk” of transcriptional products in the past decade. However, with advancements in high-throughput RNA sequencing technologies, increasing data have revealed that ncRNA sequences play important roles in regulating various cellular processes, including signaling pathways, transcription, posttranscriptional modifications, and chromatin remodeling (2). Multitudes of ncRNA species have been discovered, and these include microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs) (3–5).
With advancement in research, ncRNAs were found to exert biological functions at the RNA level and were identified as vital regulators in several physiological and pathological processes, especially in cancer (6–9). The regulatory function and underlying molecular mechanisms of ncRNAs in hepatocellular carcinoma (HCC) have also been explored. In recent five years, emerging role of lnRNAs in HCC has been well reviewed (10–12). Many ncRNAs have been identified as important drivers or inhibitors of HCC progression (13, 14). For instance, miRNA-15a-3p is downregulated in HCC tissues, and a low level of this miRNA is positively correlated with distant metastasis and poor prognosis (15). LncRNA MCM3AP-AS1 is highly expressed in HCC and associated with large tumor size, late stage and shorter survival in HCC patients (16). The expression of circTRIM33-12 is lower in HCC tissues and cell lines than in normal controls, and the level of circTRIM33-12 might be an independent risk factor for the overall survival of patients with HCC (17). All these studies predicted that ncRNAs play important roles in the carcinogenesis and progression of HCC and that specifically expressed ncRNAs might be developed as promising biomarkers for the prognosis and treatment strategy of cancer patients. While it was less reviewed that the genetic and epigenetic dysregulation lead to abnormal expression and dysfunction of various ncRNAs. Notably, ncRNAs have been discovered to participate in HCC cell malignant phenotypes (such as proliferation, migration, invasion, drug resistance), as well as immune cell development and function in the tumor microenvironment (TME).
In this review, we summarized the aberrant epigenetic and genetic alterations that cause aberrant ncRNA expression. We also discussed the functional mechanism of classical ncRNAs in the regulation of gene expression and provide examples of the far-reaching influence that these molecules have in affecting HCC processes. Furthermore, we summarized the effect of ncRNAs on the TME and laid the foundation for their applications in targeted therapy. In particular, we emphasized the role of ncRNAs in regulating tumor-associated macrophages (TAMs) in human HCC. Finally, we discussed the potential of ncRNAs as prognostic and diagnostic biomarkers and therapeutic targets in the future.
As documented in the previous section, various ncRNAs were aberrantly expressed in HCC. Aberrant expression of ncRNAs can arise through many different mechanisms. The section reviews mainly focused on aberrant epigenetic mechanisms (Figure 1) and genetic alterations.
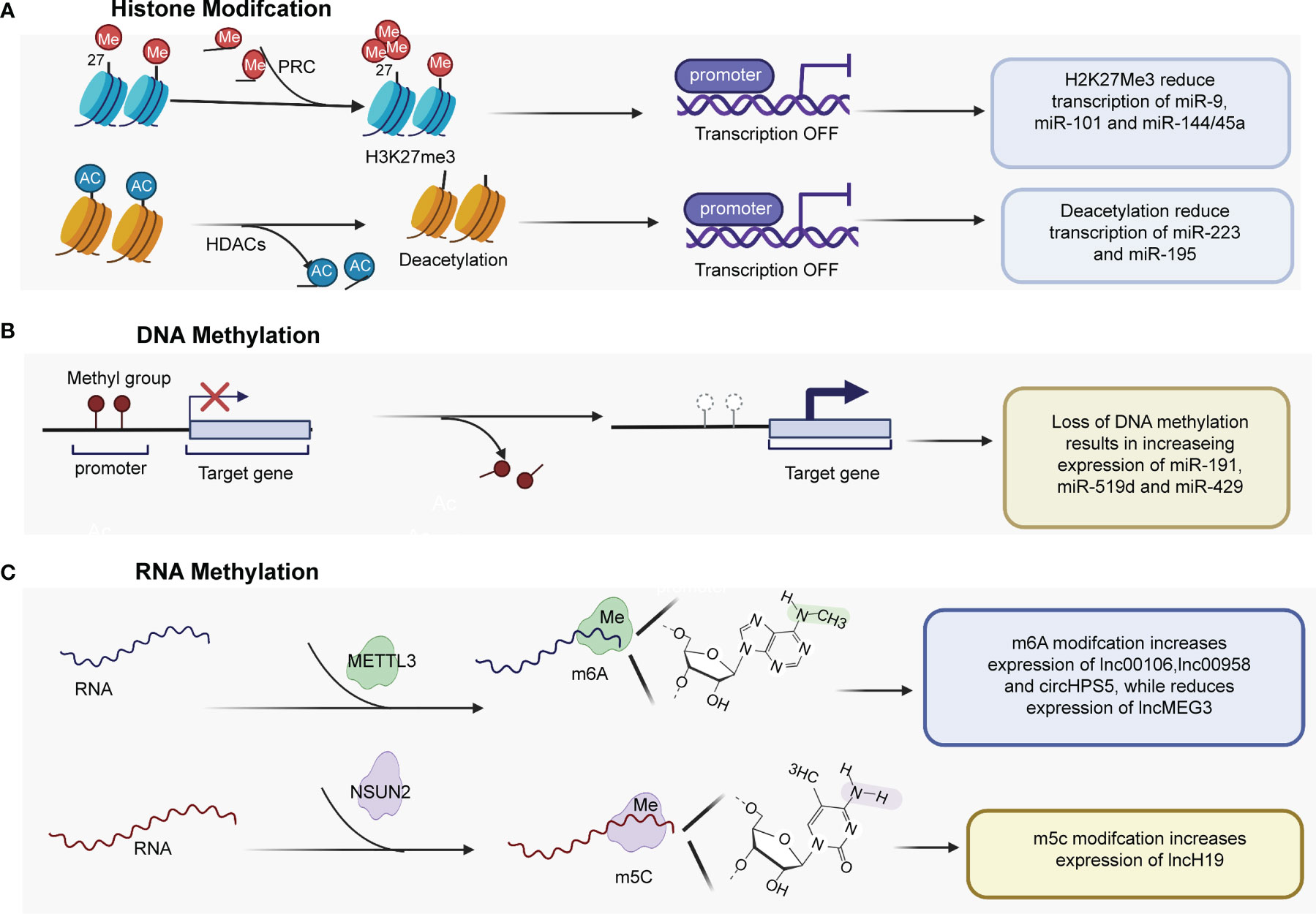
Figure 1 Epigenetic mechanisms play important roles in regulating the expression of ncRNAs. (A) H3K27me3 mediated by PRC2 regulates miRNA expression by regulating the promoters of miR-9, miR-101, and miR-144/45a. HDACs are recruited to the miR-223 and miR-195 promoter, and histone deacetylation contributes to genes downregulation. (B) Several tumor-promoting miRNAs have been found to be upregulated by promoter DNA hypomethylation. (C) m6A methyltransferase METTL3 catalyzes m6A modification of some lncRNAs and circRNA at the posttranscriptional level, promoting the expression of lnc00106, lnc00958 and cirHPS5, while reducing lncMEG3 expression level. PRC2, Polycomb repressive complex 2; HDACs, histone deacetylases; miR, microRNA; H3K27me3, trimethylation of histone H3 at lysine 27; METTL3, Methyltransferase-like 3.
CNVs are the most common DNA variations in cancer cells and are particularly common in noncoding protein regions. The whole-genome sequencing data of 49 Chinese HCC patients showed that lncRNAs were amplified in HCC tumor tissues mainly located on chromosomes 1q, 8q, 17q, and 20q, while lncRNAs deleted in the tumor tissues were mostly located on chromosomes 4q, 9q, 13q, and 16q. TaqMan copy number assay of HCC tumors and normal liver tissues from 238 patients further confirmed that lncRNAs with copy number gain in >50% of HCC samples were all upregulated in tumor tissues (18). In another study, to explore the association between HCC metastasis and lncRNAs, Yang et al. (19) identified 917 recurrently deregulated lncRNAs, and 147 of these recurrently deregulated lncRNAs were found in deleted regions. For instance, the CNV-driven lncRNA FENDRR had a pattern of decreased expression (19). Zhang et al. (20) reported that lncRNA TSLNC8 is a tumor suppressor that is located on chromosome 8p12 and is frequently deleted in HCC tissues.
The dysregulation of ncRNAs is commonly mediated by various types of histone modifications, such as histone acetylation, histone deacetylation, H3K27ac, and histone H3 lysine 27 trimethylation (H3K27me3) (Table 1). Enhancer of zeste homolog 2 (EZH2), an essential enzymatic unit of polycomb repressive complex 2 (PRC2), is the sole histone methyltransferase that promotes H3K27me3, leading to silencing genes by regulating miRNA promoters. Many studies have reported that PRC2 mediates the downregulation of tumor suppressive miRNAs, including miR-101-1 (21, 22), miR-9 (23), and miR144/451a (24), by interacting with EZH2 in HCC (Figure 1A). In addition, Zhao et al. (31) found that histone writers EP300 and WDR5 bind to the circSOD2 promoter and mediate its promoter H3K27ac and H3K4me3 modification, thereby enhancing circSOD2 expression. Additionally, several studies have reported that histone deacetylation and histone acetylation play critical roles in chromatin remodeling and ncRNA regulation (25–30, 32). For example, Dong et al. (32) reported that miR-223 was downregulated in HCC tissues and cell lines, that histone deacetylase (HDAC) 9 and HDAC10 were recruited to the miR-223 promoter and that deacetylation contributed to miR-223 downregulation. In addition, deacetylation was also found to regulate the expression of miR-195 (27) (Figure 1A). miR-122 is an important tumor suppressor and plays an essential role in the maintenance of liver homeostasis, and the expression of miR-122 is reduced in hepatic and circulating HCC patients (33–35). DNA methylation inhibitor- and HDAC inhibitor-treated HCC cells (HepG2 and Huh7) showed upregulated pri-miR-122 levels through liver-enriched transcription factors to bind the promoter region of miR-122 (30).
DNA methylation (including DNA hypomethylation and/or promoter gene CpG hypermethylation), an epigenetic modification, is a common factor that regulates the expression of ncRNAs in HCC (Table 2). DNA methylation of CpG islands within the promoter regions of tumor suppressor genes is known to suppress transcriptional initiation and thereby silence these genes. Among the different types of ncRNAs, miRNAs were the first discovered and have been the most extensively studied. Many miRNAs with tumor suppressor characteristics have been found to be silenced by promoter DNA hypermethylation (39, 40, 42, 43, 45–47). Conversely, a few tumor-promoting miRNAs have been found to be upregulated by promoter DNA hypomethylation (Figure 1B). For instance, Datta et al. (36) found that the CpG island of miR-1 was methylated in HCC cells (HepG2 and Hep3B cells) and tissues. miR-1 expression was lower in HCC cell tissues than in normal liver tissues. According to microRNA microarray analysis, the DNA hypomethylating agent 5-azacytidine restored the expression of miR-1 in HCC cells. He et al. (37) found that hsa-miR-191 was upregulated in HCC tissues and that hypomethylation of the CpG island of miR-191 led to high expression of miR-191 and enhanced epithelial-mesenchymal transition (EMT) in HCC. In addition, hypomethylation of the CpG island of miR-519d and miR-429 resulted in increasing expression of miR519d (38) and miR-429 in HCC (41, 44)
In recent years, the intriguing features of lncRNAs and circRNAs and DNA methylation have also been explored. For example, lncRNA SRHC was found to be downregulated in HCC tissues and serum samples compared with normal liver tissues and matched serum samples (48). The downregulation of SRHC was induced by DNA methylation of CpG islands in the promoter region of SRHC (48). In addition, Xu et al. (49) reported a list of dysregulated circRNAs (including circAHSA2P, circDCUN1D4, circCCNL2, and circCLEC16A, etc.) which were correlated with DNA methylation alterations based on genome-wide DNA methylation and RNA sequencing analysis of 20 pairs of HCC tissues and normal liver tissues.
Recently, RNA methylation of ncRNAs has gained much attention (50–54). For instance, M6A, a well-known type of RNA modification at the posttranscriptional level, has been reported to be involved in the expression of miRNAs, circRNAs and lncRNAs in HCC (Table 3 and Figure 1C). For example, Liang et al. (55) reported that the expression of lncRNA 00106 was higher in HCC tissues than in normal liver tissues, and the results of m6A RNA immunoprecipitation (RIP) and qRT–PCR assays showed that m6A was significantly enriched in HCC cells compared with THLE-2 cells. Further gain- and loss-of-function experiments revealed that methyltransferase-like 3 (Mettl3), an m6A methyltransferase, mediated lncRNA 00106 stability in HCC, promoting lncRNA 00106 expression by facilitating m6A modification (55). Similarly, the expression of lncRNA 00958, lncRNA MEG3, and circRNA HPS5 was also found to be regulated by m6A modification in HCC (56–58). Moreover, lncRNA H19 was abnormally increased and had a tumor-promoting effect in HCC. Mechanistically, lncRNA H19 was found to be a specific target for NSUN2, which is an RNA methyltransferase responsible for m5C modification (59).

Table 3 The expression, RNA methylation regulators and the type of RNA methylation of lncRNAs/circRNAs in HCC.
Although ncRNAs do not encode proteins, they regulate gene expression levels at various levels (epigenetic regulation, transcriptional regulation and posttranscriptional regulation, etc.) in the form of RNA. The functional mechanism of ncRNAs is complex and is not yet fully understood. Previously published studies mainly focused on the following mechanism of gene expression regulation.
One of the important functional mechanisms by which lncRNAs regulate gene transcription is via the binding and recruitment of epigenetic modifiers to specific genomic regions in HCC (60), inducing aberrant epigenetic modifications such as DNA demethylation and hypermethylation. For instance, lncRNA DDX11-AS1 directly binds with the active histone methyltransferase EZH2, which regulates H3K27me3, or DNMT1, which is a classical DNA methyltransferase that catalyzes and maintains DNA methylation, thereby epigenetically downregulating target gene expression (61). Many other lncRNAs, such as ANRIL (62), Xist (63), and HOTAIR (64), have been found to be capable of binding and recruiting EZH2 to exert their functions in the progression of HCC, indicating the close interaction between lncRNAs and EZH2-mediated hypermethylation. In addition, the demethylation of genes was also discovered in HCC progression. For example, lncRNA UPAT interacts with UHF1 and protects it from degradation, therefore inducing DNA hypomethylation to promote HCC progression (65, 66). All the evidence suggests that targeting lncRNA-induced epigenetic regulation might be a new treatment strategy for HCC.
The competing endogenous RNA (ceRNA) mechanism is a crucial mechanism by which lncRNAs indirectly regulate gene expression (67, 68). LncRNAs can sponge miRNAs, which affects the expression of miRNA-targeted genes (69). A growing body of evidence shows that lncRNAs acting as ceRNAs play a significant role in regulating tumor cell proliferation, migration, invasion, apoptosis, immune evasion and drug treatment response (70–73). For instance, lncRNA LNC00667 plays an oncogenic role by binding to miR-130a-3p, thereby attenuating the inhibition of AR expression and promoting the HCC cell malignant phenotype (71). LncRNA PICSAR is upregulated in tissues, and gain- and loss-of-function experiments indicated that lncRNA PICSAR enhances cell proliferation and cell cycle progression, inhibits apoptosis by sponging miR-194-5p and subsequently upregulates eukaryotic initiation factor 6 (EIF6) expression in HCC (74). Moreover, several ceRNA networks of lncRNA-miRNA-target genes have been screened as prognostic models for HCC, but further in vitro and in vivo experiments to verify these networks are necessary in these studies.
In the nucleus, lncRNAs can modulate transcriptional regulators that have activating or repressive activities (75). For example, the tumor-suppressive lncRNA maternally expressed gene 3 (MEG3) interacts with the p53 DNA binding domain to promote p53-mediated transcriptional activity and induce the expression of various p53 target genes in HCC cells (76).
Generally, lncRNAs indirectly regulate the stability of mRNAs. For example, UFC1 promotes the stability of β-catenin mRNA by binding with the mRNA stabilizing protein HuR, increasing the levels of β-catenin and promoting HCC progression (77). lncRNA growth-arrested DNA damage-inducible gene 7 (gadd7) mediates mRNA decay by binding to TAR DNA-binding protein (78). The same research group found that lncRNAs directly bind with mRNAs to affect mRNA stability in the cytoplasm. For instance, lncRNAs transactivate STAU1 and mediate the decay of mRNA STAU1 by binding to the 3’-untranslated regions (3’ UTRs) of STAU1 (79).
miRNAs can act as miRNA sponges, participate in the splicing of target genes, translate genes into proteins and interact with RNA binding proteins (RBPs) (80). The subcellular localization of circRNAs is essential for understanding the functional mechanism. Ample evidence has demonstrated that circRNAs are abundant in the cytoplasm and regulate miRNA function at the posttranscriptional level by sponging miRNAs. For example, circRNA 104718 promotes HCC cell proliferation, migration, invasion, and tumor growth and inhibits apoptosis through the regulation of thioredoxin domain-containing protein 5 (TXNDC5) by sponging miRNA 218-5p (81). However, a few circRNAs are distributed in the nucleus and contribute to modulating gene expression at the transcriptional and posttranscriptional levels (82–84). For example, circRNA ankrd52 is abundant in the nucleus and accumulates at transcription sites, interacting with Pol II elongation machinery and regulating local gene expression (85).
miRNAs can exert both tumor-suppressive and oncogenic effects in HCC progression (86, 87). miRNAs regulate gene expression through degradation of their target mRNAs or suppression of translation by binding the 3’ UTRs of mRNAs. Each miRNA has the capacity to repress a large number of target genes; thus, mRNAs are strong regulators of gene expression (87). For example, miR-21 acts as a tumor promoter by targeting Kruppel-like factor 5, calmodulin-regulated spectrin-associated protein family member 1, DEAD-box helicase 1, MARCKS-like 1, etc (88–90).
Most studies have focused on the role of ncRNAs in the regulation of HCC cell functions and target genes. With advances in bioinformatics and next-generation sequencing, an increasing number of studies have shown that ncRNAs participate in cell-to-cell communication and regulate tumor immune cell activation, proliferation and cytokine secretion (91), thereby affecting tumor invasion, metastasis and immune system escape (92–95). Many miRNAs have been found to play important roles in the crosstalk between HCC cells and the TME (Table 4). Most lncRNAs and circRNAs exert their regulatory function in the TME of HCC by sponging the corresponding miRNAs (Table 5).
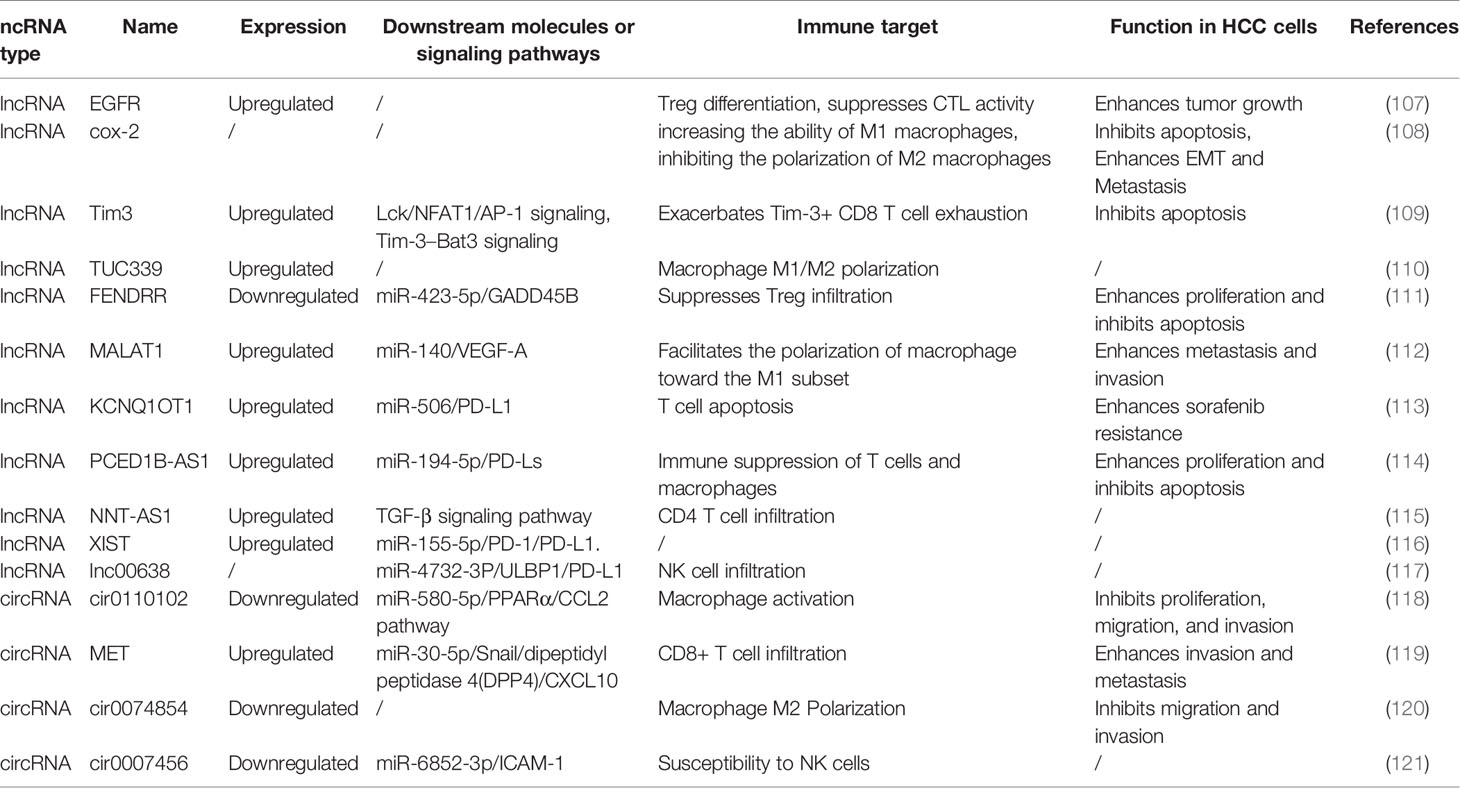
Table 5 The abnormally expressed lncRNAs and cirRNAs involved in functional regulation of immune cells.
T cells play an important role in the immune response against various pathogens and cancer cells (122). The T cell activation process is complex and involves many transcriptional and posttranscriptional regulators, including miRNAs (122). Lin et al. (96) reported that miR-570 was downregulated in HCC cell lines (including Bel-7404, Huh-7, and HepG2 cells), and the results of flow cytometry showed that the ratio of CD8+IFN-c+ T cells was markedly higher, while the ratio of CD3+CD4+ T cells was lower in the peripheral blood of nude mice injected with miR-570 mimics than that of those injected with NC-transfected SMMC7721 cells, suggesting that miR-570 might play a key role in the immune escape of HCC. Additionally, the expression of miR-26b-5 was lower in HCC tissues and cells than in normal controls and was associated with poor outcomes in HCC patients. Mechanistically, anti-miR-26b-5 mediates immunosuppression by regulating the secretion of tumor necrosis factor α (TNF-α), interferon-γ (IFN-γ), and interleukin-6 (IL-6) and IL-2 in CD4+ and CD8+ cells by targeting proviral integrations of moloney virus 2 (PIM2) in HCC (103).
Some studies have also explored the function of miRNAs in Th17 cells in the TME of HCC. During Th17 cell differentiation, treatment with the miR-132 mimic promoted the differentiation of Th17 cells, leading to a higher percentage of Th17+ cells and higher secretion of IL-17 and IL-22 (97). Furthermore, miR-132-overexpressing Th17 cells could enhance the activation of hepatic stellate cells (HSCs), inducing HCC cell EMT and migration (97).
Regulatory T cells (Tregs) normally play an immunosuppressive and tolerogenic role in the immune system and have been found to be coopted by tumor cells to escape immune surveillance (123). Yang et al. (98) reported that miR-34 exerts a tumor-suppressive function by affecting the TME by regulating the secretion of the chemokine CCL22, which facilitates the recruitment of Treg cells into the TME and immune suppression. Therefore, specific miRNAs might act as regulators of the TME during HCC progression.
TAMs play an essential role in the inflammatory microenvironment and mediate the initiation and progression of HCC according to their polarization state (M1/M2) (124). The miR-144/451a cluster was found to facilitate M1 macrophage polarization and antitumor activity in HCC (25). Mechanistically, silencing the CpG island of the miR-144/miR-451a promoter drove chromatin conformation remodeling, which promoted miR-144/miR-451a expression and further controlled TAM development and differentiated CD8+ cytotoxic T cells (25). Liu et al. (105) reported that hydrodynamic injection of miR-15a/16-1 inhibited the progression of HCC in mouse models, inhibited hepatic enrichment of Tregs and accelerated the recruitment of hepatic CD8+ cytotoxic T cells.
The expression of miR-615-5p is upregulated in NK cells of HCC patients compared with those of healthy controls. Silencing the expression of miR-615-5p impaired NK cytotoxicity and forced the expression of cytotoxic markers NKG2D, TNF-α and perforin (101). Bian et al. (106) found that downregulated miR-152 could cause epigenetic changes in HCC tumorigenesis, leading to the upregulation of human leucocyte antigen-G (HLA-G). HLA-G is a class of nonclassical MHC-I family members and is considered an immunosuppressive molecule that can bind to its receptor on NK and T cells, leading to an active immunosuppressive signaling pathway. In another study, the results of gain- and loss-of function assays in a mouse model showed that miR-561-5p was correlated with tumorigenesis and metastasis. Notably, upregulated miR-561-5p suppressed CX3CR1+ NK cell infiltration and function by targeting (C-X3-C motif) ligand 1 (CX3CL1) (104). Furthermore, miR-146a (99), miR-889 (100), and miR-506 (102) were found to regulate NK cell function and cytotoxicity against HCC cells, suggesting that modulating the expression of specific miRNAs could be a potential method to enhance NK cell-based antitumor treatments.
Increasing evidence has demonstrated that lncRNAs are involved in HCC progression and immune escape. For instance, lncRNA fetal-lethal noncoding developmental regulatory (FENDRR) acts as a miR-423-5p sponge to inhibit the immune-suppressive capacity of Tregs, therefore impairing the immune escape of HCC (111). Conversely, lncRNA epidermal growth factor receptor is highly expressed in Tregs and stimulates Treg differentiation, thus promoting HCC immune evasion and progression (107). Furthermore, lncRNA Tims and lncNNT-AS1 influence the outcome and immunotherapeutic response by decreasing tumor CD4 T cell and CD8 T cell infiltration, respectively (109, 115). Long noncoding RNA KCNQ1 overlapping transcript 1 (lncRNA KCNQ1OT1) has been shown to contribute to drug resistance and programmed death−ligand−1 (PD-L1)-mediated immune escape by sponging miR−506 in HCC cells (113). lncRNA X-inactive specific transcript (XIST) was also reported to regulate the expression of PD-1/PD-L1 by sharing a pathway between miR-194-5p and miR-155-5p in HCC (116).
Macrophages respond to environmental signals in two polarized ways: classical proinflammatory activation (M1) and alternative anti-inflammatory activation (M2). Many lncRNAs have been found to participate in the regulation of macrophage M1/M2 polarization, such as lnc TUC339 (110), MALAT1 (112), cox-2 (108) and PCED1B-AS1 (114). For example, it was observed in M1 macrophages that lncRNA cox-2 expression was increased compared with that in nonpolarized macrophages and M2 macrophages. Silencing the lncRNA cox-2 decreased the expression levels of proinflammatory factors in M1 macrophages while enhancing the expression levels of anti-inflammatory cytokines in M2 macrophages, suggesting that lncRNA cox-2 suppresses HCC immune evasion and tumor progression by regulating M1/M2 macrophage polarization (108).
An increasing number of studies have indicated the important role of lncRNAs in NK cell development and function (117, 125, 126). For instance, lncRNA GAS5 deficiency in activated NK cells has been demonstrated to reduce IFN-γ secretion, NK cell killing effects, and the apoptosis rate of HepG2 and Huh7 cells (126). However, the mechanism by which lncRNAs regulate NK cells is very complicated, and there are still relatively few related studies at present.
Emerging evidence demonstrates that specific circRNAs play regulatory roles in innate and adaptive immune pathways (118, 119, 121). For example, circ0110102 was found to be upregulated in HCC tissues versus normal tissues, and this upregulation was correlated with a short survival time. Functionally, silencing circ0110102 enhanced tumor cell proliferation, migration, and invasion. Mechanistically, circ0110102 targeted miR-580-5p and inhibited CCL2 secretion into the TME by reducing the expression of peroxisome proliferator-activated receptor α (PPARα) in HCC cells and then suppressing macrophage secretion of proinflammatory cytokines by regulating the COX-2/PGE2 pathway (118). The expression of circRNA circ-0007456 is low in HCC tissues, and this miRNA functions by sponging miR-6852-3p to regulate the expression of intercellular adhesion molecule-1 (ICAM-1), resulting in a reduction in NK cytotoxicity toward HCC cells (127). Huang et al. (119) found that circ MET (circ 0082002) was upregulated in HCC tumors, and that circ MET overexpression facilitated HCC invasion, metastasis and immune suppression through the miR-30-5p/Snail/dipeptidyl peptidase 4 (DPP4)/CXCL10 axis. In addition, CXCL10 regulated CD8+ lymphocyte infiltration through the PD-1/PD-L1 pathway (119). In addition, exosomes are important messengers of intercellular communication (120, 128). Wang et al. (120) reported that hsa_circ_0074854 in exosomes could transfer from HCC cells to macrophages. Silencing hsa_circ_0074854 in exosomes inhibited macrophage M2 polarization, which impaired the migration and invasion of HCC cells (120) (Figure 2).
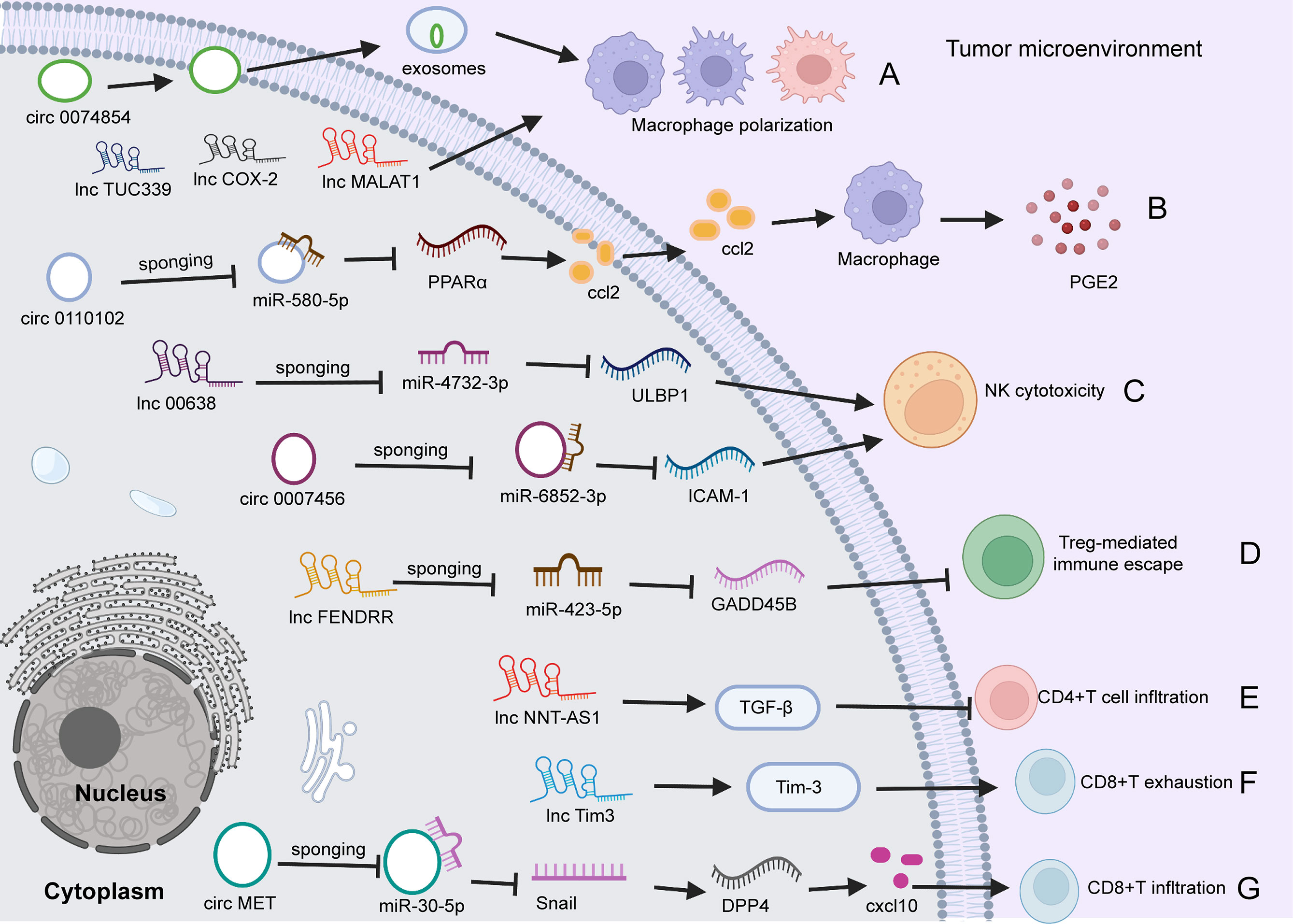
Figure 2 The role of circRNAs and lncRNAs in regulating HCC immunity. (A) circ0074854 in exosomes transfers from HCC cells to macrophages, regulating macrophage polarization. Besides, lncTUC339, lncCOX-2, and lncMALAT1 involve in macrophage polarization. (B) Upregulated circ0110102 promotes CCL2 secretion into the TME by reducing the level of PPARα by sponging miR-580-5P in HCC cells, leading to macrophage secretion of proinflammatory PGE2. (C) circ0007456 is downregulated in HCC cells and regulates NK cytotoxicity by mediating ICAM-1 expression by sponging miR-6852-3P. Similarly, lnc00638/miR-4732-3p/ULBP1 axis regulates NK cell cytotoxicity. (D) lncFENDRR acts as a miR-423-5p sponge to suppress the Treg cell mediated immune escape. (E). lncNNT-AS1 activates TGF-β signaling to decrease tumor CD4+T cell infiltration. (F) Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3. (G) circ MET is upregulated in HCC tumors, and overexpressed circMET facilitates CD8+T cells infiltration through the miR-30-5p/Snail/DPP4/CXCL10 axis. TME, tumor microenvironment; circRNA, circular RNA; PPARα, peroxisome proliferator-activated receptor α; miR, microRNA; DPP4, dipeptidyl peptidase 4; NK, natural killer; ICAM-1, intercellular adhesion molecule 1; PGE2, Prostaglandin E2; NK, natural killer; ICAM-1, intercellular adhesion molecule-1; ULBP1, UL16-binding protein 1; Treg, regulatory; TGF, Transforming growth factor; DPP4, dipeptidyl peptidase; CXCL10, CXC chemokine ligand 10.
HCC is one of the most common malignancies and ranks as the fourth leading cause of cancer-related death globally. The major reason is that most of the patients are diagnosed at an advanced stage. Therefore, exploration of diagnostic and predictive biomarkers for HCC patients in the early stage is needed. Specific ncRNAs released from cancer cells and tissues exist in exosomes, which are small membrane-derived vesicles. Interestingly, circulating exosomes can serve as vesicles loaded with ncRNAs (129–131). Previously, miRNAs have been identified in exosomes of biological fluids (including urine, serum and plasma) (132–134). More recently, various lncRNAs and circRNAs were applied as noninvasive diagnostic biomarkers for HCC due to their dysregulated expression in body fluids (135–137). Because obtaining biological fluids is noninvasive and can be repeated and because ncRNAs commonly have tissue specificity, ncRNAs in serum or exosomes are more ideal candidates for diagnostic biomarkers than those in tissues.
Furthermore, an increasing number of studies have confirmed that several ncRNAs might serve as promising prognostic biomarkers and therapeutic targets for the treatment of HCC. Numerous circRNAs, lncRNAs and miRNAs have been found to be dysregulated in tumor tissues, tumor cells, immune cells, plasma, and exosomes and to be markedly correlated with the prognosis of HCC patients, even with the activation of immune cell function involving the remodeling of TAMs. Therapeutically targeting tumor-promoting ncRNAs or restoring the function of ncRNAs with tumor-suppressive function might be promising methods. In fact, the regulatory networks of ncRNAs are extremely complicated because ncRNAs work with other biomolecules, such as coding RNAs, ncRNAs, DNAs and proteins (138). A better understanding of the regulatory mechanism of ncRNAs is necessary before ncRNAs can be tailored to therapeutic applications.
Various ncRNAs have been found to be aberrantly expressed in HCC and are regulated by different mechanisms. Here, we mainly focused on the dysregulation of epigenetic mechanisms and genetic alterations, such as CNVs, histone modification, DNA methylation, and RNA methylation. Although ncRNAs do not encode proteins, they regulate gene expression levels at various levels (epigenetic regulation, transcriptional regulation and posttranscriptional regulation, etc.) in the form of RNA. Additionally, ncRNAs play important roles in the TME and are involved in the regulation of tumor cell proliferation, invasion, migration, immune cell infiltration and functional activation. Because ncRNAs serve as either tumor suppressors or oncogenes in the progression of HCC, a better understanding of the role and related regulatory mechanism is necessary for the development of promising prognostic/predictive biomarkers and potential targeted treatments.
LL and JL designed and guided the study. CX and XG wrote and edited the manuscript. ZB and YS helped with reference collection. All authors read and approved the final manuscript.
This work was funded by the National Key Research and Development Program of China (2021YFC2301804, and 2021YFA1301104), and the National Nature Science Foundation of China (U20A20343).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating Non-Coding Regions of the Genome. Nat Rev Genet (2010) 11:559–71. doi: 10.1038/nrg2814
2. Anastasiadou E, Jacob LS, Slack FJ. Non-Coding RNA Networks in Cancer. Nat Rev Cancer (2018) 18:5–18. doi: 10.1038/nrc.2017.99
3. Quinn JJ, Chang HY. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat Rev Genet (2016) 17:47–62. doi: 10.1038/nrg.2015.10
4. Guttman M, Rinn JL. Modular Regulatory Principles of Large Non-Coding RNAs. Nature (2012) 482:339–46. doi: 10.1038/nature10887
5. Xue C, Li G, Lu J, Li L. Crosstalk Between circRNAs and the PI3K/AKT Signaling Pathway in Cancer Progression. Signal Transduct Target Ther (2021) 6:400. doi: 10.1038/s41392-021-00788-w
6. Xue C, Chen C, Gu X, Li L. Progress and Assessment of lncRNA DGCR5 in Malignant Phenotype and Immune Infiltration of Human Cancers. Am J Cancer Res (2021) 11:1–13.
7. Xue C, Zhao Y, Jiang J, Li L. Expression Levels of lncRNAs Are Prognostic for Hepatocellular Carcinoma Overall Survival. Am J Transl Res (2020) 12:1873–83.
8. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE Mediates Sorafenib Resistance in Hepatocellular Carcinoma by Stabilizing YBX1. Signal Transduct Target Ther (2020) 5:298. doi: 10.1038/s41392-020-00375-5
9. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 Promotes Hepatocellular Carcinoma Metastasis via Enhancing Invasiveness and Angiogenesis. J Exp Clin Cancer Res (2020) 39:20. doi: 10.1186/s13046-020-1529-9
10. Huang Z, Zhou JK, Peng Y, He W, Huang C. The Role of Long Noncoding RNAs in Hepatocellular Carcinoma. Mol Cancer (2020) 19:77. doi: 10.1186/s12943-020-01188-4
11. Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, et al. The Emerging Role of microRNAs and Long Noncoding RNAs in Drug Resistance of Hepatocellular Carcinoma. Mol Cancer (2019) 18:147. doi: 10.1186/s12943-019-1086-z
12. Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L, et al. Dysregulated Long Noncoding RNAs (lncRNAs) in Hepatocellular Carcinoma: Implications for Tumorigenesis, Disease Progression, and Liver Cancer Stem Cells. Mol Cancer (2017) 16:165. doi: 10.1186/s12943-017-0734-4
13. Wang M, Yu F, Li P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers (Basel) (2018) 10:258. doi: 10.3390/cancers10080258
14. Li D, Zhang J, Li J. Role of miRNA Sponges in Hepatocellular Carcinoma. Clin Chim Acta (2020) 500:10–9. doi: 10.1016/j.cca.2019.09.013
15. Jiang C, He ZL, Hu XH, Ma PY. MiRNA-15a-3p Inhibits the Metastasis of Hepatocellular Carcinoma by Interacting With HMOX1. Eur Rev Med Pharmacol Sci (2020) 24:12694–700. doi: 10.26355/eurrev_202012_24167
16. Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, et al. A Novel lncRNA MCM3AP-AS1 Promotes the Growth of Hepatocellular Carcinoma by Targeting miR-194-5p/FOXA1 Axis. Mol Cancer (2019) 18:28. doi: 10.1186/s12943-019-0957-7
17. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA Circtrim33-12 Acts as the Sponge of MicroRNA-191 to Suppress Hepatocellular Carcinoma Progression. Mol Cancer (2019) 18:105. doi: 10.1186/s12943-019-1031-1
18. Yin D, Hu ZQ, Luo CB, Wang XY, Xin HY, Sun RQ, et al. LINC01133 Promotes Hepatocellular Carcinoma Progression by Sponging miR-199a-5p and Activating Annexin A2. Clin Transl Med (2021) 11:e409. doi: 10.1002/ctm2.409
19. Yang Y, Chen L, Gu J, Zhang H, Yuan J, Lian Q, et al. Recurrently Deregulated lncRNAs in Hepatocellular Carcinoma. Nat Commun (2017) 8:14421. doi: 10.1038/ncomms14421
20. Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, et al. Long Noncoding RNA TSLNC8 is a Tumor Suppressor That Inactivates the Interleukin-6/STAT3 Signaling Pathway. Hepatology (2018) 67:171–87. doi: 10.1002/hep.29405
21. Wang L, Zhang X, Jia LT, Hu SJ, Zhao J, Yang JD, et al. C-Myc-Mediated Epigenetic Silencing of MicroRNA-101 Contributes to Dysregulation of Multiple Pathways in Hepatocellular Carcinoma. Hepatology (2014) 59:1850–63. doi: 10.1002/hep.26720
22. Huang D, Wang X, Zhuang C, Shi W, Liu M, Tu Q, et al. Reciprocal Negative Feedback Loop Between EZH2 and miR-101-1 Contributes to miR-101 Deregulation in Hepatocellular Carcinoma. Oncol Rep (2016) 35:1083–90. doi: 10.3892/or.2015.4467
23. Tsang DP, Wu WK, Kang W, Lee YY, Wu F, Yu Z, et al. Yin Yang 1-Mediated Epigenetic Silencing of Tumour-Suppressive microRNAs Activates Nuclear Factor-κb in Hepatocellular Carcinoma. J Pathol (2016) 238:651–64. doi: 10.1002/path.4688
24. Zhao J, Li H, Zhao S, Wang E, Zhu J, Feng D, et al. Epigenetic Silencing of miR-144/451a Cluster Contributes to HCC Progression via Paracrine HGF/MIF-Mediated TAM Remodeling. Mol Cancer (2021) 20:46. doi: 10.1186/s12943-021-01343-5
25. Dong YW, Wang R, Cai QQ, Qi B, Wu W, Zhang YH, et al. Sulfatide Epigenetically Regulates miR-223 and Promotes the Migration of Human Hepatocellular Carcinoma Cells. J Hepatol (2014) 60:792–801. doi: 10.1016/j.jhep.2013.12.004
26. Wang Y, Toh HC, Chow P, Chung AY, Meyers DJ, Cole PA, et al. MicroRNA-224 Is Up-Regulated in Hepatocellular Carcinoma Through Epigenetic Mechanisms. FASEB J (2012) 26:3032–41. doi: 10.1096/fj.11-201855
27. Zhao N, Li S, Wang R, Xiao M, Meng Y, Zeng C, et al. Expression of microRNA-195 Is Transactivated by Sp1 But Inhibited by Histone Deacetylase 3 in Hepatocellular Carcinoma Cells. Biochim Biophys Acta (2016) 1859:933–42. doi: 10.1016/j.bbagrm.2016.05.006
28. Henrici A, Montalbano R, Neureiter D, Krause M, Stiewe T, Slater EP, et al. The Pan-Deacetylase Inhibitor Panobinostat Suppresses the Expression of Oncogenic miRNAs in Hepatocellular Carcinoma Cell Lines. Mol Carcinog (2015) 54:585–97. doi: 10.1002/mc.22122
29. Yang H, Lan P, Hou Z, Guan Y, Zhang J, Xu W, et al. Histone Deacetylase Inhibitor SAHA Epigenetically Regulates miR-17-92 Cluster and MCM7 to Upregulate MICA Expression in Hepatoma. Br J Cancer (2015) 112:112–21. doi: 10.1038/bjc.2014.547
30. Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M, et al. Epigenetic Regulation of MicroRNA-122 by Peroxisome Proliferator Activated Receptor-Gamma and Hepatitis B Virus X Protein in Hepatocellular Carcinoma Cells. Hepatology (2013) 58:1681–92. doi: 10.1002/hep.26514
31. Zhao Z, Song J, Tang B, Fang S, Zhang D, Zheng L, et al. CircSOD2 Induced Epigenetic Alteration Drives Hepatocellular Carcinoma Progression Through Activating JAK2/STAT3 Signaling Pathway. J Exp Clin Cancer Res (2020) 39:259. doi: 10.1186/s13046-020-01769-7
32. Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT Promotes Proliferation and Invasion of HCC Cells via Sponging miR-214. Am J Physiol Gastrointest Liver Physiol (2018) 314:G559–g65. doi: 10.1152/ajpgi.00242.2017
33. Thakral S, Ghoshal K. miR-122 Is a Unique Molecule With Great Potential in Diagnosis, Prognosis of Liver Disease, and Therapy Both as miRNA Mimic and Antimir. Curr Gene Ther (2015) 15:142–50. doi: 10.2174/1566523214666141224095610
34. Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 Expression in Liver Cancer Correlates With Suppression of the Hepatic Phenotype and Gain of Metastatic Properties. Oncogene (2009) 28:3526–36. doi: 10.1038/onc.2009.211
35. Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in Patients With Hepatocellular Carcinoma or Chronic Hepatitis. Mol Carcinog (2011) 50:136–42. doi: 10.1002/mc.20712
36. Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, et al. Methylation Mediated Silencing of MicroRNA-1 Gene and Its Role in Hepatocellular Carcinogenesis. Cancer Res (2008) 68:5049–58. doi: 10.1158/0008-5472.Can-07-6655
37. He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, et al. Hypomethylation of the hsa-miR-191 Locus Causes High Expression of Hsa-Mir-191 and Promotes the Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma. Neoplasia (2011) 13:841–53. doi: 10.1593/neo.11698
38. Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, et al. In Hepatocellular Carcinoma miR-519d Is Up-Regulated by P53 and DNA Hypomethylation and Targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol (2012) 227:275–85. doi: 10.1002/path.3995
39. Dohi O, Yasui K, Gen Y, Takada H, Endo M, Tsuji K, et al. Epigenetic Silencing of miR-335 and Its Host Gene MEST in Hepatocellular Carcinoma. Int J Oncol (2013) 42:411–8. doi: 10.3892/ijo.2012.1724
40. Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, et al. Epigenetic Repression of miR-132 Expression by the Hepatitis B Virus X Protein in Hepatitis B Virus-Related Hepatocellular Carcinoma. Cell Signal (2013) 25:1037–43. doi: 10.1016/j.cellsig.2013.01.019
41. Li L, Tang J, Zhang B, Yang W, LiuGao M, Wang R, et al. Epigenetic Modification of MiR-429 Promotes Liver Tumour-Initiating Cell Properties by Targeting Rb Binding Protein 4. Gut (2015) 64:156–67. doi: 10.1136/gutjnl-2013-305715
42. Long XR, He Y, Huang C, Li J. MicroRNA-148a Is Silenced by Hypermethylation and Interacts With DNA Methyltransferase 1 in Hepatocellular Carcinogenesis. Int J Oncol (2014) 44:1915–22. doi: 10.3892/ijo.2014.2373
43. Zhang PP, Wang XL, Zhao W, Qi B, Yang Q, Wan HY, et al. DNA Methylation-Mediated Repression of miR-941 Enhances Lysine (K)-Specific Demethylase 6B Expression in Hepatoma Cells. J Biol Chem (2014) 289:24724–35. doi: 10.1074/jbc.M114.567818
44. Tang J, Li L, Huang W, Sui C, Yang Y, Lin X, et al. MiR-429 Increases the Metastatic Capability of HCC via Regulating Classic Wnt Pathway Rather Than Epithelial-Mesenchymal Transition. Cancer Lett (2015) 364:33–43. doi: 10.1016/j.canlet.2015.04.023
45. Chen Z, Wang X, Liu R, Chen L, Yi J, Qi B, et al. KDM4B-Mediated Epigenetic Silencing of miRNA-615-5p Augments RAB24 to Facilitate Malignancy of Hepatoma Cells. Oncotarget (2017) 8:17712–25. doi: 10.18632/oncotarget.10832
46. Yu Q, Xiang L, Yin L, Liu X, Yang D, Zhou J. Loss-Of-Function of miR-142 by Hypermethylation Promotes TGF-β-Mediated Tumour Growth and Metastasis in Hepatocellular Carcinoma. Cell Prolif (2017) 50:e12384. doi: 10.1111/cpr.12384
47. Wang Y, Jiang F, Jiao K, Ju L, Liu Q, Li Y, et al. De-Methylation of miR-148a by Arsenic Trioxide Enhances Sensitivity to Chemotherapy via Inhibiting the NF-κb Pathway and CSC Like Properties. Exp Cell Res (2020) 386:111739. doi: 10.1016/j.yexcr.2019.111739
48. Zheng H, Yang S, Yang Y, Yuan SX, Wu FQ, Wang LL, et al. Epigenetically Silenced Long Noncoding-SRHC Promotes Proliferation of Hepatocellular Carcinoma. J Cancer Res Clin Oncol (2015) 141:1195–203. doi: 10.1007/s00432-014-1871-4
49. Xu T, Wang L, Jia P, Song X, Zhao Z. An Integrative Transcriptomic and Methylation Approach for Identifying Differentially Expressed Circular RNAs Associated With DNA Methylation Change. Biomedicines (2021) 9:657. doi: 10.3390/biomedicines9060657
50. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The Interplay Between M6a RNA Methylation and Noncoding RNA in Cancer. J Hematol Oncol (2019) 12:121. doi: 10.1186/s13045-019-0805-7
51. Xue C, Zhao Y, Li L. Advances in RNA Cytosine-5 Methylation: Detection, Regulatory Mechanisms, Biological Functions and Links to Cancer. Biomark Res (2020) 8:43. doi: 10.1186/s40364-020-00225-0
52. Yi YC, Chen XY, Zhang J, Zhu JS. Novel Insights Into the Interplay Between M(6)A Modification and Noncoding RNAs in Cancer. Mol Cancer (2020) 19:121. doi: 10.1186/s12943-020-01233-2
53. Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction Between N(6)-Methyladenosine (M(6)A) Modification and Noncoding RNAs in Cancer. Mol Cancer (2020) 19:94. doi: 10.1186/s12943-020-01207-4
54. Xue C, Zhao Y, Li G, Li L. Multi-Omic Analyses of the M(5)C Regulator ALYREF Reveal Its Essential Roles in Hepatocellular Carcinoma. Front Oncol (2021) 11:633415:633415. doi: 10.3389/fonc.2021.633415
55. Liang W, Wang Y, Zhang Q, Gao M, Zhou H, Wang Z. M(6)A-Mediated Upregulation of LINC00106 Promotes Stemness and Metastasis Properties of Hepatocellular Carcinoma via Sponging Let7f. Front Cell Dev Biol (2021) 9:781867:781867. doi: 10.3389/fcell.2021.781867
56. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A-Mediated Upregulation of LINC00958 Increases Lipogenesis and Acts as a Nanotherapeutic Target in Hepatocellular Carcinoma. J Hematol Oncol (2020) 13:5. doi: 10.1186/s13045-019-0839-x
57. Wu J, Pang R, Li M, Chen B, Huang J, Zhu Y. M6a-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell Through miR-544b/BTG2 Signaling. Onco Targets Ther (2021) 14:3745–55. doi: 10.2147/ott.S289198
58. Rong D, Wu F, Lu C, Sun G, Shi X, Chen X, et al. M6a Modification of Circhps5 and Hepatocellular Carcinoma Progression Through HMGA2 Expression. Mol Ther Nucleic Acids (2021) 26:637–48. doi: 10.1016/j.omtn.2021.09.001
59. Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant NSUN2-Mediated M(5)C Modification of H19 lncRNA Is Associated With Poor Differentiation of Hepatocellular Carcinoma. Oncogene (2020) 39:6906–19. doi: 10.1038/s41388-020-01475-w
60. Amicone L, Citarella F, Cicchini C. Epigenetic Regulation in Hepatocellular Carcinoma Requires Long Noncoding RNAs. BioMed Res Int (2015) 2015:473942. doi: 10.1155/2015/473942
61. Li Y, Zhuang W, Huang M, Li X. Long Noncoding RNA DDX11-AS1 Epigenetically Represses LATS2 by Interacting With EZH2 and DNMT1 in Hepatocellular Carcinoma. Biochem Biophys Res Commun (2019) 514:1051–7. doi: 10.1016/j.bbrc.2019.05.042
62. Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, et al. Long Non-Coding RNA ANRIL Is Upregulated in Hepatocellular Carcinoma and Regulates Cell Proliferation by Epigenetic Silencing of KLF2. J Hematol Oncol (2015) 8:57. doi: 10.1186/s13045-015-0153-1
63. Sado T, Hoki Y, Sasaki H. Tsix Silences Xist Through Modification of Chromatin Structure. Dev Cell (2005) 9:159–65. doi: 10.1016/j.devcel.2005.05.015
64. Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, et al. Hotair Mediates Hepatocarcinogenesis Through Suppressing miRNA-218 Expression and Activating P14 and P16 Signaling. J Hepatol (2015) 63:886–95. doi: 10.1016/j.jhep.2015.05.016
65. Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, et al. UHRF1 Overexpression Drives DNA Hypomethylation and Hepatocellular Carcinoma. Cancer Cell (2014) 25:196–209. doi: 10.1016/j.ccr.2014.01.003
66. Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K, et al. Long Noncoding RNA UPAT Promotes Colon Tumorigenesis by Inhibiting Degradation of UHRF1. Proc Natl Acad Sci USA (2016) 113:1273–8. doi: 10.1073/pnas.1500992113
67. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in Cancer: Possible Functions and Clinical Implications. J Med Genet (2015) 52:710–8. doi: 10.1136/jmedgenet-2015-103334
68. Tay Y, Rinn J, Pandolfi PP. The Multilayered Complexity of ceRNA Crosstalk and Competition. Nature (2014) 505:344–52. doi: 10.1038/nature12986
69. Peng Y, Croce CM. The Role of MicroRNAs in Human Cancer. Signal Transduct Target Ther (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
70. Qin Z, Liu X, Li Z, Wang G, Feng Z, Liu Y, et al. LncRNA LINC00667 Aggravates the Progression of Hepatocellular Carcinoma by Regulating Androgen Receptor Expression as a miRNA-130a-3p Sponge. Cell Death Discovery (2021) 7:387. doi: 10.1038/s41420-021-00787-4
71. Shi T, Morishita A, Kobara H, Masaki T. The Role of Long Non-Coding RNA and microRNA Networks in Hepatocellular Carcinoma and Its Tumor Microenvironment. Int J Mol Sci (2021) 22:10630. doi: 10.3390/ijms221910630
72. Xu Q, Chen S, Hu Y, Huang W. Prognostic Role of ceRNA Network in Immune Infiltration of Hepatocellular Carcinoma. Front Genet (2021) 12:739975. doi: 10.3389/fgene.2021.739975
73. Zhou Y, Li K, Dai T, Wang H, Hua Z, Bian W, et al. Long non-Coding RNA HCP5 Functions as a Sponge of miR-29b-3p and Promotes Cell Growth and Metastasis in Hepatocellular Carcinoma Through Upregulating DNMT3A. Aging (Albany NY) (2021) 13:16267–86. doi: 10.18632/aging.203155
74. Liu Z, Mo H, Sun L, Wang L, Chen T, Yao B, et al. Long Noncoding RNA PICSAR/miR-588/EIF6 Axis Regulates Tumorigenesis of Hepatocellular Carcinoma by Activating PI3K/AKT/mTOR Signaling Pathway. Cancer Sci (2020) 111:4118–28. doi: 10.1111/cas.14631
75. Long Y, Wang X, Youmans DT, Cech TR. How Do lncRNAs Regulate Transcription? Sci Adv (2017) 3:eaao2110. doi: 10.1126/sciadv.aao2110
76. Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, et al. Long Noncoding RNA MEG3 Interacts With P53 Protein and Regulates Partial P53 Target Genes in Hepatoma Cells. PloS One (2015) 10:e0139790. doi: 10.1371/journal.pone.0139790
77. Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, et al. The Long Intergenic Noncoding RNA UFC1, a Target of MicroRNA 34a, Interacts With the mRNA Stabilizing Protein HuR to Increase Levels of β-Catenin in HCC Cells. Gastroenterology (2015) 148:415–26.e18. doi: 10.1053/j.gastro.2014.10.012
78. Liu X, Li D, Zhang W, Guo M, Zhan Q. Long Non-Coding RNA Gadd7 Interacts With TDP-43 and Regulates Cdk6 mRNA Decay. EMBO J (2012) 31:4415–27. doi: 10.1038/emboj.2012.292
79. Gong C, Maquat LE. lncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing With 3’ UTRs via Alu elements. Nature (2011) 470:284–8. doi: 10.1038/nature09701
80. Zang J, Lu D, Xu A. The Interaction of circRNAs and RNA Binding Proteins: An Important Part of circRNA Maintenance and Function. J Neurosci Res (2020) 98:87–97. doi: 10.1002/jnr.24356
81. Yu J, Yang M, Zhou B, Luo J, Zhang Z, Zhang W, et al. CircRNA-104718 Acts as Competing Endogenous RNA and Promotes Hepatocellular Carcinoma Progression Through microRNA-218-5p/TXNDC5 Signaling Pathway. Clin Sci (Lond) (2019) 133:1487–503. doi: 10.1042/cs20190394
82. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs Are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
83. Yu B, Shan G. Functions of Long Noncoding RNAs in the Nucleus. Nucleus (2016) 7:155–66. doi: 10.1080/19491034.2016.1179408
84. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs Are Abundant, Conserved, and Associated With ALU Repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
85. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular Intronic Long Noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
86. Lee YS, Dutta A. MicroRNAs in Cancer. Annu Rev Pathol (2009) 4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222
87. Oura K, Morishita A, Masaki T. Molecular and Functional Roles of MicroRNAs in the Progression of Hepatocellular Carcinoma-A Review. Int J Mol Sci (2020) 21:8362. doi: 10.3390/ijms21218362
88. Wan L, Yuan X, Liu M, Xue B. miRNA-223-3p Regulates NLRP3 to Promote Apoptosis and Inhibit Proliferation of Hep3b Cells. Exp Ther Med (2018) 15:2429–35. doi: 10.3892/etm.2017.5667
89. Yang J, Xu C, Wu M, Wu Y, Jia X, Zhou C, et al. MicroRNA-124 Inhibits Hepatic Stellate Cells Inflammatory Cytokines Secretion by Targeting IQGAP1 Through NF-κb Pathway. Int Immunopharmacol (2021) 95:107520. doi: 10.1016/j.intimp.2021.107520
90. Koenig AB, Barajas JM, Guerrero MJ. Ghoshal K. A Comprehensive Analysis of Argonaute-CLIP Data Identifies Novel, Conserved and Species-Specific Targets of miR-21 in Human Liver and Hepatocellular Carcinoma. Int J Mol Sci (2018) 19:851. doi: 10.3390/ijms19030851
91. Liu Z, Ma M, Yan L, Chen S, Li S, Yang D, et al. miR-370 Regulates ISG15 Expression and Influences IFN-α Sensitivity in Hepatocellular Carcinoma Cells. Cancer biomark (2018) 22:453–66. doi: 10.3233/cbm-171075
92. Lin YH, Wu MH, Yeh CT, Lin KH. Long Non-Coding RNAs as Mediators of Tumor Microenvironment and Liver Cancer Cell Communication. Int J Mol Sci (2018) 19:3742. doi: 10.3390/ijms19123742
93. Alasaad S, Oleaga Á, Casais R, Rossi L, Min AM, Soriguer RC, et al. Temporal Stability in the Genetic Structure of Sarcoptes Scabiei Under the Host-Taxon Law: Empirical Evidences From Wildlife-Derived Sarcoptes Mite in Asturias, Spain. Parasit Vectors (2011) 4:151. doi: 10.1186/1756-3305-4-151
94. Gnoni A, Santini D, Scartozzi M, Russo A, Licchetta A, Palmieri V, et al. Hepatocellular Carcinoma Treatment Over Sorafenib: Epigenetics, microRNAs and Microenvironment. Is There a Light at the End of the Tunnel? Expert Opin Ther Targets (2015) 19:1623–35. doi: 10.1517/14728222.2015.1071354
95. Gramantieri L, Giovannini C, Piscaglia F, Fornari F. MicroRNAs as Modulators of Tumor Metabolism, Microenvironment, and Immune Response in Hepatocellular Carcinoma. J Hepatocell Carcinoma (2021) 8:369–85. doi: 10.2147/jhc.S268292
96. Lin Y, Liu S, Su L, Su Q, Lin J, Huang X, et al. miR-570 Inhibits Proliferation, Angiogenesis, and Immune Escape of Hepatocellular Carcinoma. Cancer Biother Radiopharm (2018) 33:252–7. doi: 10.1089/cbr.2017.2389
97. Feng R, Cui Z, Liu Z, Zhang Y. Upregulated microRNA-132 in T Helper 17 Cells Activates Hepatic Stellate Cells to Promote Hepatocellular Carcinoma Cell Migration In Vitro. Scand J Immunol (2021) 93:e13007. doi: 10.1111/sji.13007
98. Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 Signaling-Induced Treg Cell Recruitment Promotes Venous Metastases of HBV-Positive Hepatocellular Carcinoma. Cancer Cell (2012) 22:291–303. doi: 10.1016/j.ccr.2012.07.023
99. Sun X, Zhang J, Hou Z, Han Q, Zhang C, Tian Z. miR-146a Is Directly Regulated by STAT3 in Human Hepatocellular Carcinoma Cells and Involved in Anti-Tumor Immune Suppression. Cell Cycle (2015) 14:243–52. doi: 10.4161/15384101.2014.977112
100. Xie H, Zhang Q, Zhou H, Zhou J, Zhang J, Jiang Y, et al. microRNA-889 Is Downregulated by Histone Deacetylase Inhibitors and Confers Resistance to Natural Killer Cytotoxicity in Hepatocellular Carcinoma Cells. Cytotechnology (2018) 70:513–21. doi: 10.1007/s10616-017-0108-1
101. Rahmoon MA, Youness RA, Gomaa AI, Hamza MT, Waked I, El Tayebi HM, et al. MiR-615-5p Depresses Natural Killer Cells Cytotoxicity Through Repressing IGF-1R in Hepatocellular Carcinoma Patients. Growth Factors (2017) 35:76–87. doi: 10.1080/08977194.2017.1354859
102. Su Z, Ye X, Shang L. MiR-506 Promotes Natural Killer Cell Cytotoxicity Against Human Hepatocellular Carcinoma Cells by Targeting Stat3. Yonsei Med J (2019) 60:22–9. doi: 10.3349/ymj.2019.60.1.22
103. Han W, Li N, Liu J, Sun Y, Yang X, Wang Y. MicroRNA-26b-5p Enhances T Cell Responses by Targeting PIM-2 in Hepatocellular Carcinoma. Cell Signal (2019) 59:182–90. doi: 10.1016/j.cellsig.2018.11.011
104. Chen EB, Zhou ZJ, Xiao K, Zhu GQ, Yang Y, Wang B, et al. The miR-561-5p/CX(3)CL1 Signaling Axis Regulates Pulmonary Metastasis in Hepatocellular Carcinoma Involving CX(3)CR1(+) Natural Killer Cells Infiltration. Theranostics (2019) 9:4779–94. doi: 10.7150/thno.32543
105. Liu N, Chang CW, Steer CJ, Wang XW, Song G. MicroRNA-15a/16-1 Prevents Hepatocellular Carcinoma by Disrupting the Communication Between Kupffer Cells and Regulatory T Cells. Gastroenterology (2022) 162:575–89. doi: 10.1053/j.gastro.2021.10.015
106. Bian X, Si Y, Zhang M, Wei R, Yang X, Ren H, et al. Down-Expression of miR-152 Lead to Impaired Anti-Tumor Effect of NK via Upregulation of HLA-G. Tumour Biol (2016) 37:3749–56. doi: 10.1007/s13277-015-3669-7
107. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The Long Noncoding RNA lnc-EGFR Stimulates T-Regulatory Cells Differentiation Thus Promoting Hepatocellular Carcinoma Immune Evasion. Nat Commun (2017) 8:15129. doi: 10.1038/ncomms15129
108. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long non-Coding RNA Cox-2 Prevents Immune Evasion and Metastasis of Hepatocellular Carcinoma by Altering M1/M2 Macrophage Polarization. J Cell Biochem (2018) 119:2951–63. doi: 10.1002/jcb.26509
109. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long Non-Coding RNA Lnc-Tim3 Exacerbates CD8 T Cell Exhaustion via Binding to Tim-3 and Inducing Nuclear Translocation of Bat3 in HCC. Cell Death Dis (2018) 9:478. doi: 10.1038/s41419-018-0528-7
110. Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA Tuc339. Int J Mol Sci (2018) 19:2958. doi: 10.3390/ijms19102958
111. Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, et al. Long Non-Coding RNA FENDRR Acts as a miR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids (2019) 17:516–29. doi: 10.1016/j.omtn.2019.05.027
112. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-Coding RNA MALAT1 Promotes Angiogenesis and Immunosuppressive Properties of HCC Cells by Sponging miR-140. Am J Physiol Cell Physiol (2020) 318:C649–c63. doi: 10.1152/ajpcell.00510.2018
113. Zhang J, Zhao X, Ma X, Yuan Z, Hu M. KCNQ1OT1 Contributes to Sorafenib Resistance and Programmed Death−Ligand−1−Mediated Immune Escape via Sponging Mir−506 in Hepatocellular Carcinoma Cells. Int J Mol Med (2020) 46:1794–804. doi: 10.3892/ijmm.2020.4710
114. Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual Targeting of PD-L1 and PD-L2 by PCED1B-AS1 via Sponging hsa-miR-194-5p Induces Immunosuppression in Hepatocellular Carcinoma. Hepatol Int (2021) 15:444–58. doi: 10.1007/s12072-020-10101-6
115. Wang Y, Yang L, Dong X, Yang X, Zhang X, Liu Z, et al. Overexpression of NNT-AS1 Activates TGF-β Signaling to Decrease Tumor CD4 Lymphocyte Infiltration in Hepatocellular Carcinoma. BioMed Res Int (2020) 2020:8216541. doi: 10.1155/2020/8216541
116. Atwa SM, Handoussa H, Hosny KM, Odenthal M, El Tayebi HM. Pivotal Role of Long Non-Coding Ribonucleic Acid-X-Inactive Specific Transcript in Regulating Immune Checkpoint Programmed Death Ligand 1 Through a Shared Pathway Between miR-194-5p and miR-155-5p in Hepatocellular Carcinoma. World J Hepatol (2020) 12:1211–27. doi: 10.4254/wjh.v12.i12.1211
117. Qi F, Du X, Zhao Z, Zhang D, Huang M, Bai Y, et al. Tumor Mutation Burden-Associated LINC00638/miR-4732-3p/ULBP1 Axis Promotes Immune Escape via PD-L1 in Hepatocellular Carcinoma. Front Oncol (2021) 11:729340. doi: 10.3389/fonc.2021.729340
118. Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA Hsa_Circ_0110102 Inhibited Macrophage Activation and Hepatocellular Carcinoma Progression via miR-580-5p/Pparα/CCL2 Pathway. Aging (Albany NY) (2021) 13:11969–87. doi: 10.18632/aging.202900
119. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET Drives Immunosuppression and Anti-PD1 Therapy Resistance in Hepatocellular Carcinoma via the miR-30-5p/Snail/DPP4 Axis. Mol Cancer (2020) 19:92. doi: 10.1186/s12943-020-01213-6
120. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of Hsa_Circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting With HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int J Nanomed (2021) 16:2803–18. doi: 10.2147/ijn.S284560
121. Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 Regulates the Natural Killer Cell-Mediated Cytotoxicity Toward Hepatocellular Carcinoma via the miR-6852-3p/ICAM-1 Axis. Cell Death Dis (2021) 12:94. doi: 10.1038/s41419-020-03334-8
122. Diener C, Hart M, Kehl T, Rheinheimer S, Ludwig N, Krammes L, et al. Quantitative and Time-Resolved miRNA Pattern of Early Human T Cell Activation. Nucleic Acids Res (2020) 48:10164–83. doi: 10.1093/nar/gkaa788
123. Mailloux AW, Young MR. Regulatory T-Cell Trafficking: From Thymic Development to Tumor-Induced Immune Suppression. Crit Rev Immunol (2010) 30:435–47. doi: 10.1615/critrevimmunol.v30.i5.30
124. Li Z, Wu T, Zheng B, Chen L. Individualized Precision Treatment: Targeting TAM in HCC. Cancer Lett (2019) 458:86–91. doi: 10.1016/j.canlet.2019.05.019
125. Mowel WK, McCright SJ, Kotzin JJ, Collet MA, Uyar A, Chen X, et al. Group 1 Innate Lymphoid Cell Lineage Identity Is Determined by a Cis-Regulatory Element Marked by a Long Non-Coding RNA. Immunity (2017) 47:435–49.e8. doi: 10.1016/j.immuni.2017.08.012
126. Fang P, Xiang L, Chen W, Li S, Huang S, Li J, et al. LncRNA GAS5 Enhanced the Killing Effect of NK Cell on Liver Cancer Through Regulating miR-544/Runx3. Innate Immun (2019) 25:99–109. doi: 10.1177/1753425919827632
127. Weinstein CL, Griffith OW. Cysteinesulfonate and Beta-Sulfopyruvate Metabolism. Partitioning Between Decarboxylation, Transamination, and Reduction Pathways. J Biol Chem (1988) 263:3735–43. doi: 10.1016/S0021-9258(18)68986-0
128. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem (2019) 88:487–514. doi: 10.1146/annurev-biochem-013118-111902
129. Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, et al. Serum and Exosome Long Non Coding RNAs as Potential Biomarkers for Hepatocellular Carcinoma. J Cancer (2018) 9:2631–9. doi: 10.7150/jca.24978
130. Liu WH, Ren LN, Wang X, Wang T, Zhang N, Gao Y, et al. Combination of Exosomes and Circulating microRNAs May Serve as a Promising Tumor Marker Complementary to Alpha-Fetoprotein for Early-Stage Hepatocellular Carcinoma Diagnosis in Rats. J Cancer Res Clin Oncol (2015) 141:1767–78. doi: 10.1007/s00432-015-1943-0
131. Wang Y, Li Z, Xu S, Guo J. Novel Potential Tumor Biomarkers: Circular RNAs and Exosomal Circular RNAs in Gastrointestinal Malignancies. J Clin Lab Anal (2020) 34:e23359. doi: 10.1002/jcla.23359
132. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA Spectrum in 12 Body Fluids. Clin Chem (2010) 56:1733–41. doi: 10.1373/clinchem.2010.147405
133. Fiorino S, Bacchi-Reggiani ML, Visani M, Acquaviva G, Fornelli A, Masetti M, et al. MicroRNAs as Possible Biomarkers for Diagnosis and Prognosis of Hepatitis B- and C-Related-Hepatocellular-Carcinoma. World J Gastroenterol (2016) 22:3907–36. doi: 10.3748/wjg.v22.i15.3907
134. Świtlik WZ, Bielecka-Kowalska A, Karbownik MS, Kordek R, Jabłkowski M, Szemraj J. Forms of Diagnostic Material as Sources of miRNA Biomarkers in Hepatocellular Carcinoma: A Preliminary Study. Biomark Med (2019) 13:523–34. doi: 10.2217/bmm-2018-0485
135. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA Circtmem45a Acts as the Sponge of MicroRNA-665 to Promote Hepatocellular Carcinoma Progression. Mol Ther Nucleic Acids (2020) 22:285–97. doi: 10.1016/j.omtn.2020.08.011
136. Huang X, Sun L, Wen S, Deng D, Wan F, He X, et al. RNA Sequencing of Plasma Exosomes Revealed Novel Functional Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Sci (2020) 111:3338–49. doi: 10.1111/cas.14516
137. Hou Y, Yu Z, Tam NL, Huang S, Sun C, Wang R, et al. Exosome-Related lncRNAs as Predictors of HCC Patient Survival: A Prognostic Model. Am J Transl Res (2018) 10:1648–62.
Keywords: HCC, ncRNA, epigenetic modification, functional mechanisms, TME
Citation: Xue C, Gu X, Bao Z, Su Y, Lu J and Li L (2022) The Mechanism Underlying the ncRNA Dysregulation Pattern in Hepatocellular Carcinoma and Its Tumor Microenvironment. Front. Immunol. 13:847728. doi: 10.3389/fimmu.2022.847728
Received: 03 January 2022; Accepted: 04 February 2022;
Published: 23 February 2022.
Edited by:
Ximing Xu, Renmin Hospital of Wuhan University, ChinaReviewed by:
James Ahodantin, University of Maryland, Baltimore, United StatesCopyright © 2022 Xue, Gu, Bao, Su, Lu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanjuan Li, bGpsaUB6anUuZWR1LmNu; Juan Lu, bHVqdWFuemp1QHpqdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.