
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 10 June 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.845585
This article is part of the Research Topic Tumor-Promoting Immune Cells: Cancer Immune Escape and Beyond View all 15 articles
The AHSA1 is a main activator of ATPase of Hsp90. Hsp90 is involved in various metabolic and developmental processes of tumor cells. Although, the role of AHSA1 in tumor cells is still unrecognized. In the current research, the RNA-seq of 33 tumors were downloaded using The Cancer Genome Atlas (TCGA) database for the analysis of AHSA1 expression in tumors. The Kaplan-Meier method was used for the evaluation of the prognostic significance of AHSA1 in patients with pan-cancer. Additionally, the correlation between AHSA1 and immune cell infiltration, immune checkpoint, pyroptosis-related molecules, epithelial cell transformation-related molecules, and autophagy-related molecules were analyzed by co-expression. Furthermore, we examined the effect of AHSA1 knockdown on cell function in Huh7 and HCCLM3 cells of hepatocellular carcinoma (HCC) cell lines.
According to the finding of this study, up-regulation of AHSA1 expression was observed in numerous tumor tissues, and its over-expression in liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), and esophageal carcinoma (ESCA) could affect the overall survival and disease-specific survival of the patients. Meanwhile, as per the correlation analysis the expression of AHSA1 was greatly correlated with the expression of various immune cell infiltrates, immune checkpoint inhibitors, tumor mutation load, and microsatellite instability. Moreover, this study focused on analyzing the association of AHSA1 expression with multiple pathological stages in HCC, and confirmed that AHSA1 was an independent prognostic factor of HCC by univariate and multivariate COX regression in TCGA and The International Cancer Genome Consortium (ICGC) cohorts. At the same time, cellular experiments proved that the AHSA1 knockdown could decrease the proliferation activity, cell migration and invasion ability of HCC cells. Therefore, the results of this study indicated that AHSA1 can be used as a potential prognostic biomarker of tumors and it may have a significant role in the proliferation as well as migration of HCC cells.
In recent decades, the malignant tumor has shown an alarming incidence and mortality rate worldwide, and it is a major threat to people’s health in the 21st century (1). Although the increase of mortality rate of cancer patients in developed countries has moderated its steps in recent years, thanks to advanced prevention and treatment concepts and medical technologies, the mortality rate of cancer patients around the world still shows a prominent growth trend (2). With research development, a growing number of researchers began to pay attention to the common characteristics of various human malignant tumors, to explore the potential mechanism of tumor genesis (3). At present, pan-cancer research has been widely used to identify tumor molecular markers and signal pathways, combined with multiple omics analysis, in order to understand more comprehensively and deeply the molecular mechanism of tumor genesis and development (4–6). As a co-molecular chaperone protein, AHSA1 is currently the only ATPase activator of Hsp90 (7). AHSA1 plays a key role in regulating the molecular chaperone cycle and protein folding of Hsp90 (8). Hsp90, the acting protein of AHSA1, is a classical molecular chaperone with a highly conserved molecular structure. It can facilitate protein folding, maturation and transport of some cancer-related proteins, such as BCR-ABL, ErbB2/Neu, Akt, HIF-1α, p53 and RAF-1 (9). Lindsey B. Sheltont et al. revealed that the inhibition of AHSA1 expression in mice models of Alzheimer’s disease and in vitro cell models can eliminate the accumulation of Tau protein caused by Hsp90, which is expected to become a new therapeutic target for Alzheimer’s disease (10). According to Jianli Shao et al, AHSA1 mediates the invasion, proliferation, and apoptosis of tumor cells by regulating the Wnt/β-catenin pathway in osteosarcoma. At the same time, after the knockdown of AHSA1, the ATPase activity of Hsp90 decreased (11). However, current studies are limited to a few types of cancers, and the role of AHSA1 in various types of tumors is still not understood completely. In this study, the RNA-seq from TCGA was utilized for the examination of AHSA1 expression in a variety of human tumors. Combined with clinical follow-up data, the impact of abnormal expression of AHSA1 on the prognosis of cancer patients was analyzed. Meanwhile, combined with correlation analysis, the possible association between AHSA1 expression and tumor immune cell infiltration, tumor mutation load (TMB), microsatellite instability (MSI) and various pathway-related molecules were explored. In addition, this study verified the expression and prognostic role of AHSA1 in hepatocellular carcinoma by combining data from ICGC-LIRI-JP, and verified the expression and potential function of AHSA1 in hepatocellular carcinoma by combining cell and tissue specimens. The results of this study are helpful to understand the similarities and differences of AHSA1 expression in a variety of tumors, reveal the potential mechanism of the interaction between AHSA1 and tumor immunity, and demonstrate the potential function of AHSA1 in hepatocellular carcinoma.
The count format of 33 common tumor transcriptome data was downloaded from TCGA’s official website and converted into the TPM format. The number of tumors of each type was shown on the Table S1. The clinical follow-up survival and staging data were downloaded at the same time. The transcriptome data of hepatocellular carcinoma was provided by the ICGC database and converted into the TPM format, and corresponding clinical data were obtained. In addition, the single-nucleotide mutation data of 33 tumors were downloaded from TCGA’s official website and their tumor mutation load was calculated.
Transcriptome data from 33 tumors were filtered by “RMA” packs to remove the NA and duplicates, and log2 (TPM +1) conversion was performed afterward. Then, for comparing the expression differences of AHSA1 in the normal and tumor tissues in different tumors, the Wilcoxon rank sum test was performed. Using the “SurvMiner” and “Survival” packages, the median expression values based on AHSA1 were classified into two groups; the high risk and low-risk groups in different kinds of tumors. Their survival curves were drawn using the Kaplan-Meier method, and their statistical significance was calculated by the log-rank test.
A comprehensive website; Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) was used for the measurement of the level of tumor immune cells infiltrating degree (12). For predicting the immune cell infiltration level from tumor transcription data there are five different methods provided by TIMER website which includes TIMER, CIBERSORT, xCell, McP-counter and EPIC. The abundance of infiltrating immune cells in tumor samples from 33 tumor types was obtained from TIMER database. The single-sample gene set enrichment analysis (ssGSEA) was performed in R package GSVA. Subsequently, the relation of AHSA1 expression and the level of infiltration of these immune cells was calculated. At present, TMB and MSI are considered for use as potential biomarkers to predict the efficacy of tumor immunotherapy (13, 14). The simple nucleoside variation data of level 4 processed with MuTect2 software was acquired from the TCGA database, and the TMB was calculated (15). Microsatellite instability signature for 33 tumors were obtained from reported studies (16). Reports have indicated that Epithelial-Mesenchymal Transitions (EMT), Pyroptosis and Autophagy were all associated with tumor metastasis and malignant proliferation, and the correlation between AHSA1 and the expression of these pathway molecules was also analyzed in this study.
The GeneMANIA (http://genemania.org/) database finds out functionally identical genes on the basis of genomic and proteomic data (17). The GeneMANIA database was used to predict the genes that have functions similar to that of AHSA1. Gene oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of molecules with potential roles in AHSA1 were performed using clusterProfiler packages.
We analyzed the correlation between AHSA1 expression and clinical stage, including pathologic stage, tumor (T) stage, histologic grade, alpha-fetoprotein (AFP) level, and vascular invasion. Meanwhile, for the further evaluation of the prognostic value of AHSA1 in HCC patients, univariate and multivariate COX regression and Receiver Operating characteristic curve (ROC) analysis were performed respectively. In addition, we constructed a nomography based on the expression value of AHSA1 and pathologic stage to promote the application of AHSA1 in the evaluation of clinical prognosis of hepatocellular carcinoma, and evaluated the prediction accuracy of the nomography by calibration curve.
The American Type Culture Collection (ATCC, Manassas, VA, United States) provided the normal human liver cells LO2, HCC cell lines HepG2, Huh7, and HCCLM3 for this experiment. All cells were cultured in an incubator at 37 °C with 5% CO2. Human target gene AHSA1 short hairpin RNA (shRNA) was purchased from GenScript company (https://www.genscript.com/), the sequences are as follows:
AHSA1 shRNA-1:gGGTGAAACTTCTAAGAGAAttcaagagaTTCTCTTAGAAGTTTCACCttttt, AHSA1 shRNA-2:gGGCATGATCTTACCTACAAttcaagagaTTGTAGGTAAGATCATGCCttttt,
AHSA1 shRNA-3:gAGTCAGGAGTACAATACAAttcaagagaTTGTATTGTACTCCTGACTttttt. Huh7 and HCCLM3 cells were plated in six-well plates (4 × 105 cells/well), and cultivated in a in a 37°C, 5% CO2 incubator until they were completely adherent to the wall. Subsequently, the cells were transfected with lipo3000.
Four types of cell strains were used for total RNA was extraction. Reverse transcription was performed to convert the extracted RNA into cDNA using the reverse transcription kit provided by Beyotime, https://www.beyotime.com/. Exicycler 96 (BIONEER) was used to detect its fluorescence expression quantity. All results were processed with GAPDH for standardization. The 2-ΔΔ Ct method was used to calculate the relative expression levels of genes.
Ten pairs of paraffin sections of hepatocellular carcinoma tumor tissue specimens and para-cancer specimens were provided by the Department of Pathology, North Guangdong People’s Hospital. AHSA1 antibody was purchased from Proteintech Company (Article number: 14725-1-AP). Approval for this research was given by the Ethics Committee of North Guangdong People’s Hospital. Western blotting was performed for the detection of post knockout changes in AHSA1 protein levels.
HuH7 cells transfected with shRNA-AHSA1 were digested when their level reached 90%, and then they were inoculated into 96-well culture plates with 3×103cells per well, and 5 multiple wells were designed for each group. Then, they were cultured in a 37°C, 5% CO2 incubator, and tested at 0h, 24h, 48h, 72h and 96h using the CCK-8 kit (WLA074, China).
6-well plates were used for the inoculation of these cells, and after 48 hours of transfection, a 200 μl pipette tip was used to scratch the cells. The cell surface was cleaned with serum-free medium once, and the cell fragments were removed. The cells were then observed and photographed under a 100× microscope, and their positions in the photos were recorded. Subsequently, cells in each group were placed in an incubator at 37 °C with 5% CO2 for 24h and 48h, and then photographed and recorded, and the mobility of each group was calculated.
A 24-well Transwell chamber (8 μm aperture; Corning Costar, USA) was prepared overnight at 4°C with or without 100μL matrix gel substrate provided by BD Biosciences, San Jose, CA, USA. A 200ul of cell suspension containing 1×105cells/mL was inoculated into Transwell cells with or without matrix glue, and a culture medium (800 ul) containing 10% FBS was poured into the lower chamber. After 24h culture, cell fixation was done using 4% paraformaldehyde at room temperature for 20 minutes and staining was performed for 5 minutes with 0.5% crystal violet dye. The cell count was recorded afterwards.
The mRNA expression of AHSA1 was evaluated in pan-cancer patients according to the RNA-seq data of 33 types of TCGA tumors. The results showed that AHSA1 was expressed at a relatively low level in normal bile duct tissue and a relatively high level in testicular germ cell tumor tissue. At the same time, the variance analysis indicated a relatively high expression of AHSA1 in Bladder Urothelial Carcinoma (BLCA), Breast invasive carcinoma (BRCA), Cholangiocarcinoma (CHOL), Colon adenocarcinoma (COAD), Esophageal carcinoma (ESCA), Head and Neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), LIHC, Lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), Rectum adenocarcinoma (READ) and stomach adenocarcinoma (STAD), Prostate adenocarcinoma (PRAD), and uterine corpus endometrial carcinoma (UCEC) tumor tissues as compared to the corresponding para-carcinoma tissue (Figure 1A). Furthermore, paired comparison analysis was performed and the results of this analysis also revealed the overexpression of AHSA1 in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUAD, LUSC, PRAD, READ, STAD in tumor tissues (Figure 1B). Considering that AHSA1 is an ATPase activator of HSP90AA1, we analyzed the expression of HSP90AA1 in pan-cancer. Interestingly, the similar results were observed for HSP90AA1 expression in pan-cancer (Figures S1A, B).
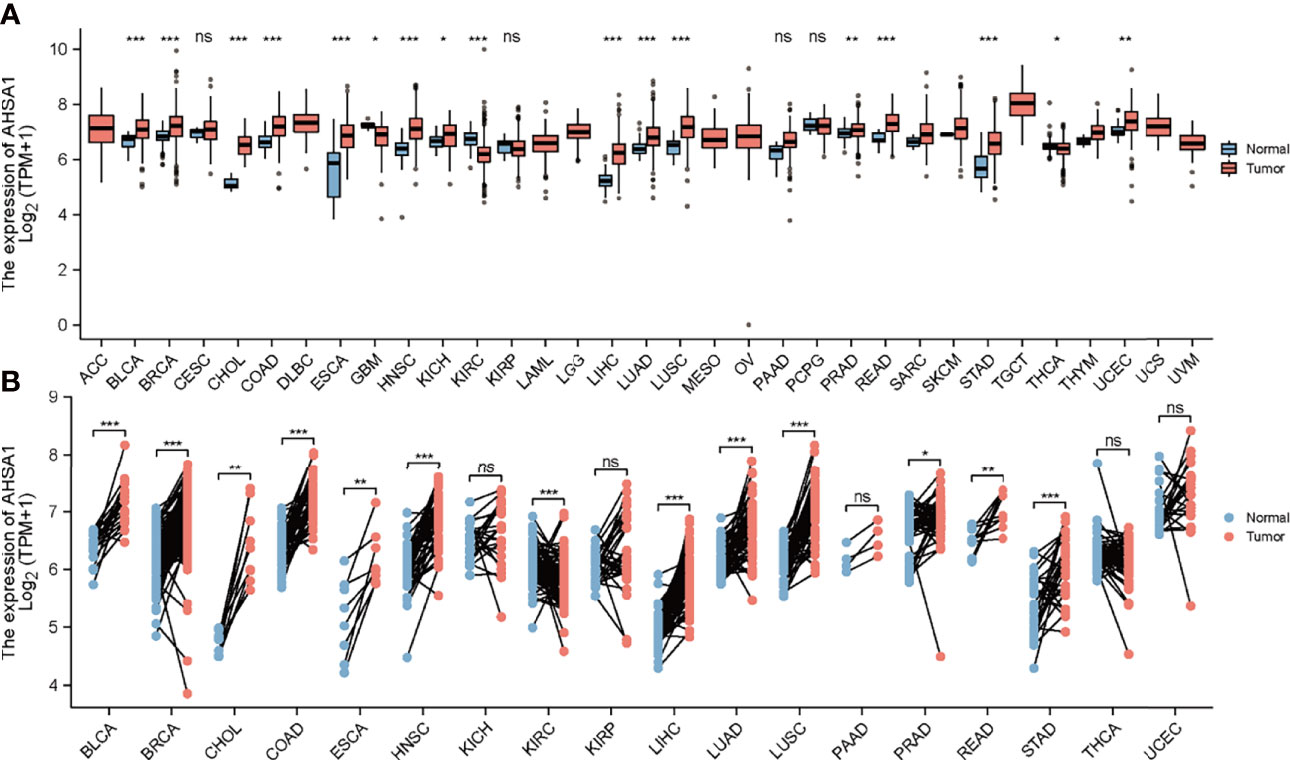
Figure 1 Differential expression analysis of AHSA1. (A) Expression of AHSA1 mRNA in pan-cancer. (B) The expression differences of AHSA1 in tumor and corresponding adjacent tissues were compared with paired analysis. Mann-Whitney U test was used for this analysis, ns, p≥0.05; *p< 0.05; **p<0.01; ***p<0.001.
Based on the TCGA database, the Kaplan-Meier curves helped in depicting the association between abnormal expression of AHSA1 and the general pan-cancer survival. The results indicated that the survival rate of patients with high expression levels of AHSA1 in LIHC, LUAD, ESCA, and KIRP was worse (Figures 2A–D). For further evaluation of the specific effect of AHSA1 on tumor survival, the influence of abnormal AHSA1 expression on disease-specific survival (DSS) of tumor patients was observed. The Kaplan-Meier plots showed that the abnormally increased AHSA1 in LIHC, LUAD, ESCA and KIRP was correlated with its poor DSS (Figures 2E–H).
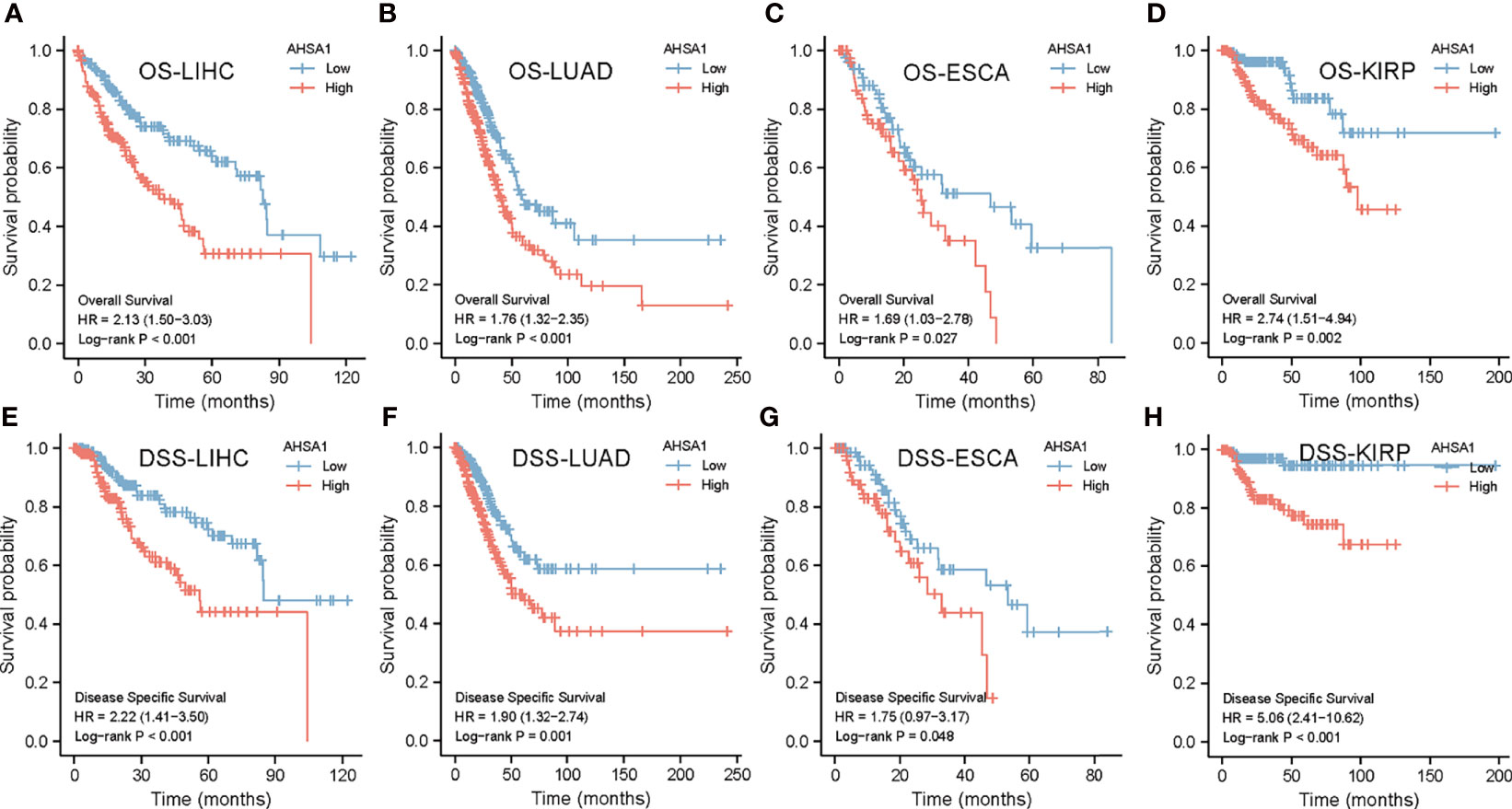
Figure 2 Survival analysis of AHSA1 in pan-cancer. The effect of AHSA1 expression on the overall survival rate (OS) of LIHC (A), LUAD (B), ESCA (C) and KIRP (D) was analyzed by Kaplan-Meier method. Meanwhile, the influence of abnormal expression of AHSA1 on disease-specific survival (DSS) of LIHC (E), LUAD (F), ESCA (G) and KIRP (H) was calculated.
Tumor microenvironment is a complicated survival environment of tumor cells, mostly comprising of immune cells, mesenchymal environment as well as related internal and external molecular composition (18). In recent years, growing evidence indicate that the immune cell infiltration level is closely associated with the survival of tumor cells. Although previous studies have suggested the prognostic importance and expression level of AHSA1 in various types of cancers, little is known if the abnormal expression of AHSA1 have an influence on immune cell infiltration. In this study, the correlation between abnormal AHSA1 expression and immune cell infiltration was evaluated using a TIMER database based on multiple immune prediction methods. The results indicated that AHSA1 was substantially positively correlated with CD4+Th1, CD4+Th2 and MDSC cells of various tumors (Figure 3A). Meanwhile, AHSA1 showed a significant positive correlation with B cells, CD4+ T cells, CD8+T cells and Myeloid dendritic cells in LIHC (Figure 3A). Moreover, we discussed the correlation between the AHSA1 expression and immune checkpoint inhibitor. The results showed that AHSA1 was significantly positively correlated with several immune checkpoint inhibitors, especially in LIHC, THCA, and THYM. Meanwhile, AHSA1 was significantly negatively correlated with several immune checkpoint in ACC, BRCA, GBM, HNSC, LUSC, LGG, and PRAD (Figure 3B). In addition, AHSA1 was significantly positively associated with CD274, CD276, and CTLA4 in LIHC, THCA, and TGCT (Figure 3B). It is understood that EMT has a significant function in tumor metastasis, which is the major cause of death. To investigate whether AHSA1 is related to tumor metastasis, the link between AHSA1 expression and the expression of EMT-related molecules was analyzed. Results show a significant positive correlation between the AHSA1 and EMT-related molecules in LIHC, KIRC, and UVM (Figure 3C). Additionally, the AHSA1 and MTHFD2, SLC3A2 and SERPINH1 showed a significant positive correlation in a wide variety of tumors. The AHSA1 and MMP2, CCL2, CFH, and CYP1B1 showed a significant negative correlation in a variety of tumors (Figure 3C). Pyroptosis is a new type of programmed inflammatory cell death that has a significant function in the development of tumor. Hence, the expression correlation between AHSA1 and pyroptosis-related molecules was observed in this study. The results showed that AHSA1 showed a significant positive correlation with pyroptosis-related molecules in BRCA, LGG, LIHC, PRAD, and THCA (Figure 4A). In addition, we also discussed the AHSA1 correlation with autophagy-related molecules, the results also showed a significant positive correlation between AHSA1 and autophagy-related molecules in BRCA, COAD, LIHC, LUAD, PAAD, PRAD and TGCT. Similar to the previous trends, AHSA1 was significantly positively correlated with HSP90AA1 and HSP90AB1 (Figure 4B). In addition, tumor mutation load (TMB) and microsatellite instability (MSI) are known as important factors that affect tumor genesis and development. Therefore, the association between TMB or MSI and AHSA1 expression was observed in 33 commonly known types of cancers. Results show that the AHSA1 presented substantial relation with TMB in CESC, COAD, HNSC, KIRC, LUAD, STAD, and UCS (Figures 4C, D).
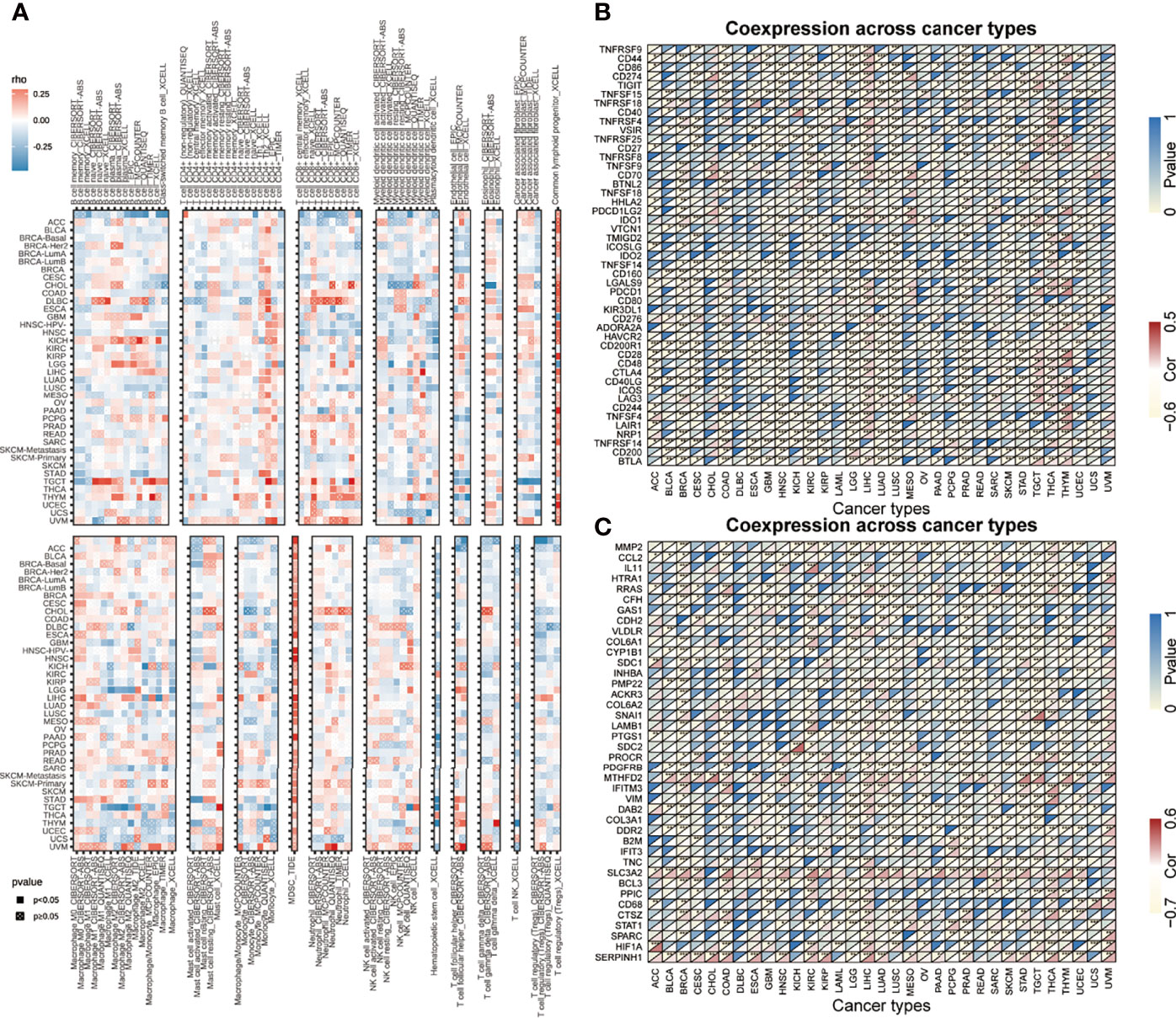
Figure 3 Correlation analysis between the expression of AHSA1 and immune cell infiltration, checkpoint inhibitors and pyroptosis-related molecules. (A) Correlation analysis between AHSA1 and immune cell infiltration in pan-cancer. (B) Correlation analysis between the expression of AHSA1 and immune checkpoint inhibitors. (C) Coexpression analysis of AHSA1 expression and epithelial-mesenchymal transition-related molecules. *p< 0.05; **p<0.01; ***p<0.001.
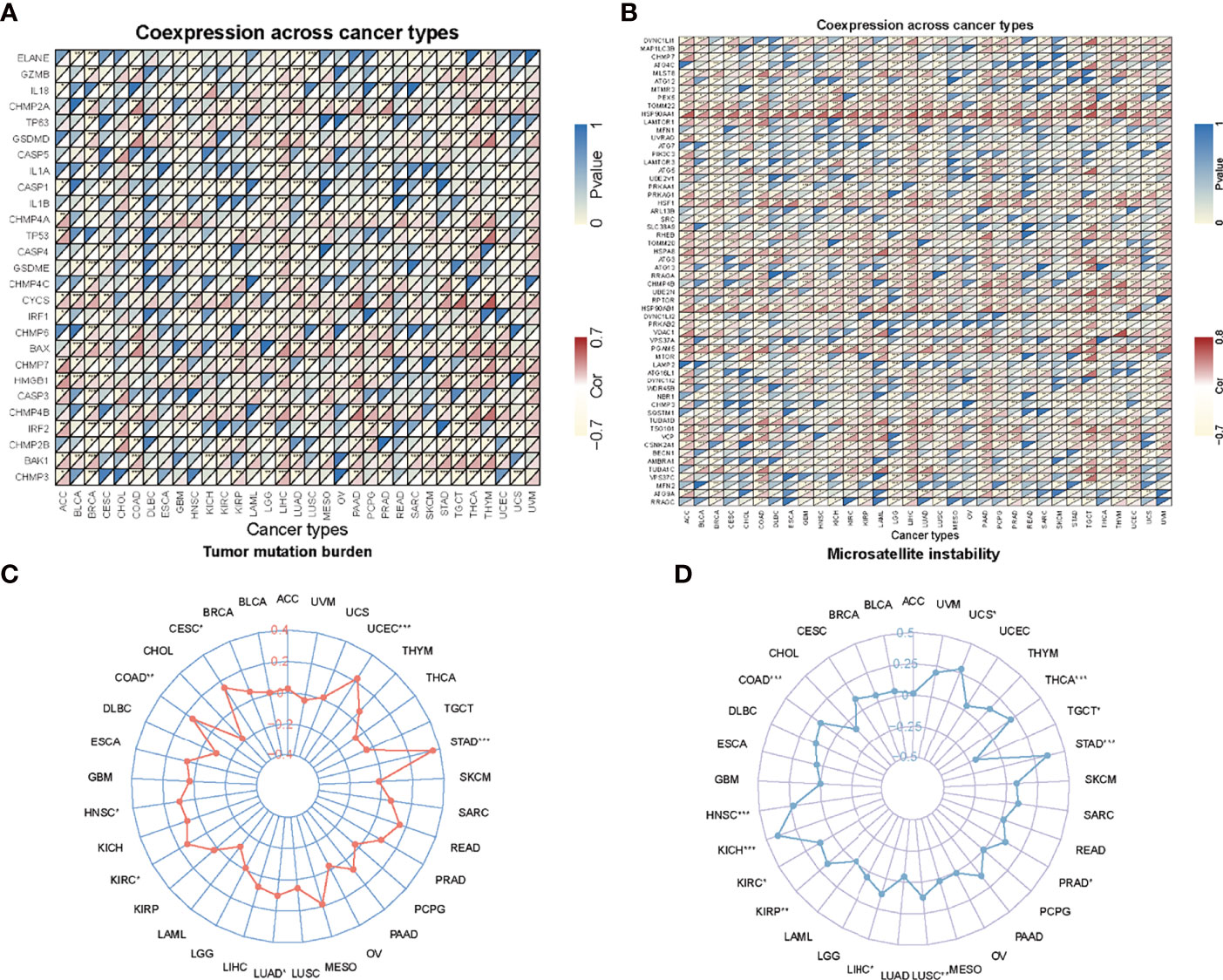
Figure 4 Correlation analysis. (A) Correlation analysis of AHSA1 expression and pyroptosis-related molecules. (B) Correlation analysis of AHSA1 expression and autophagy-related molecules. (C) Correlation between AHSA1 expression and Tumor mutation burden. (D) Correlation analysis of AHSA1 expression and Microsatellite instability. *p<0.05, **p<0.01, ***p<0.001.
To deeply understand the possible role of AHSA1, the molecules interacting with AHSA1 were analyzed by GeneMANIA and comPPI. The results from GeneMANIA showed that AHSA1 had potential interactions with AMD1, HSP90AA1, DNAJB4 and HSP90AB1. Main functions included protein folding, up-regulation of DNA biosynthetic process and regulation of the metabolic processes of reactive oxygen species (Figure 5A). Single-sample gene set enrichment analysis (ssGSEA) algorithm was introduced to predict the infiltration levels of 24 immune cell types in HCC. The results indicated that AHSA1 was positively correlated with T helper cells, Th2 cells, Macrophages, and iDC (Figure 5B). While, cytotoxic cells and Dendritic cells was negatively correlated with AHSA1 expression in HCC (Figure 5B). To further identify the possible role of AHSA1, the GO and KEGG enrichment analysis was performed on molecules that interacted with AHSA1 obtained from GeneMANIA. Cellular Component enrichment analysis showed that these molecules are mainly enriched in the cytosolic part, cell-substrate junction, cytosolic ribosome, and cytosolic small ribosomal subunit. The analysis of molecular function enrichment indicated that these molecules mostly play a role in cell adhesion molecule binding, cadherin binding, ubiquitin-protein ligase binding, stressed protein binding, heat shock protein binding, and Hsp90 protein binding (Figure 5C). Enrichment analysis of biological processes showed that these molecules are mainly involved in RNA catabolic process, mRNA catabolic process, protein targeting, protein folding, and cytoplasmic translation (Figure 5D). KEGG analysis showed that these molecules were mainly involved in ribosome, RNA transport, Spliceosome, HIF-1α signaling pathway and Legionellosis (Figure 5D). In addition, we built an interactive relationship network between GO and KEGG, as shown in Figure 5E.
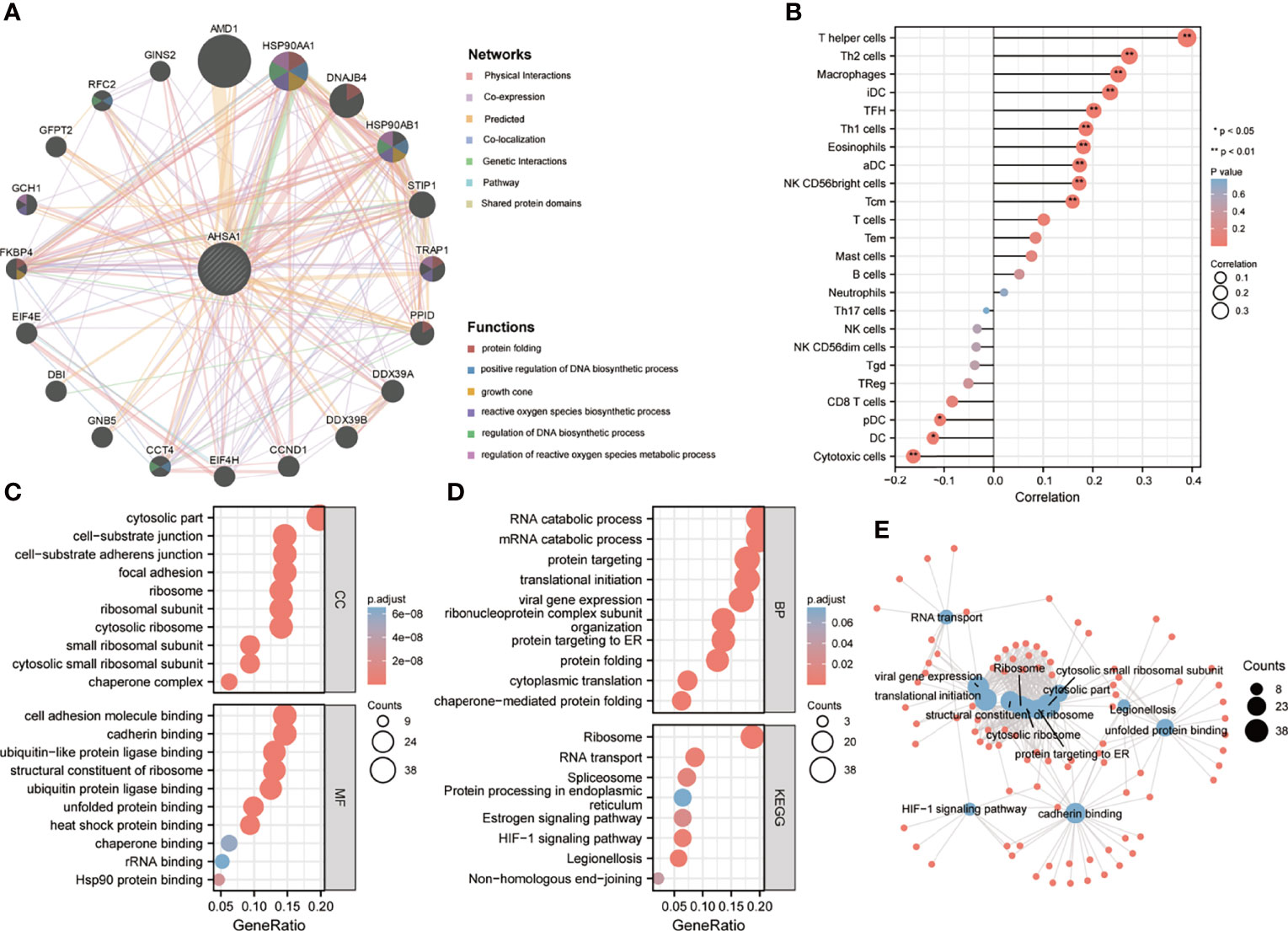
Figure 5 Potential functions of AHSA1. (A) The potential interaction molecular network of AHSA1 was created using the GeneMANIA. (B) The ssGSEA algorithm was employed to evaluate the abundance of immune cell infiltration in HCC. GO and KEGG functional enrichment analysis of the molecules interacted with AHSA1 (C, D). (E) Construction of GO and KEGG interaction networks.
The findings of this study suggests that AHSA1 may perform a key role in the development of hepatocellular carcinoma. Subsequently, the association between AHSA1 expression and clinical stage was analyzed in hepatocellular carcinoma. The results indicated that AHSA1 had a higher expression level in the higher pathologic stage (Figure 6A), T stage (Figure 6B), and histologic grade (Figure 6C). Meanwhile, AHSA1 expression was also related to the expression level of AFP and vascular invasion status (Figures 6D, E). These findings indicate that AHSA1 expression may be linked to the malignant pathological progression of hepatocellular carcinoma. Subsequently, we analyzed the prognostic evaluation efficacy of AHSA1 in the overall survival rate of HCC by ROC, and the results showed that AHSA1 showed good prognostic evaluation performance in HCC, with AUC of 0.702, 0.661 and 0.689 at 1, 3 and 5 years respectively (Figure 6F). We further evaluated the prognostic role of AHSA1 in hepatocellular carcinoma by univariate and multivariate COX regression combined with clinical data, and the results showed that AHSA1 was an independent prognostic factor of hepatocellular carcinoma (Figure 6G). Furthermore, in order to promote the application of AHSA1 in clinical evaluation, we constructed a nomography (Figure 6H) based on the expression of AHSA1 and the pathologic stage. At the same time, we evaluated the accuracy of the model for prognosis assessment of hepatocellular carcinoma patients after 1, 3 and 5 years by calibration curve, and according to the results, the nomography had very good accuracy, almost close to the ideal model (Figure 6I). For further confirmation of the value of prognosis of AHSA1 in HCC, the expression of AHSA1 in para-cancer and tumor tissues in the ICGC-LIRI-JP queue were analyzed. Both unpaired analysis (Figure 7A) and paired analysis (Figure 7B) showed that AHSA1 has higher expression in HCC tumor tissues. Meanwhile, K-M curve analysis showed that HCC patients with high AHSA1 expression in the ICGC queue had poorer survival expectations (Figure 7C). Meanwhile, the univariate and multi-variate COX analyses showed that AHSA1 was an independent prognostic risk factor for HCC (Figure 7D). For additional verification of the expression of AHSA1 in hepatocellular carcinoma, RT-PCR was performed to detect the mRNA expression levels of AHSA1 in normal hepatocyte cell line LO2 along with three hepatocellular carcinoma cell lines, HCCLM3, HepG2 and Huh7. Results showed higher mRNA expression levels of AHSA1 in hepatocellular carcinoma cells (Figure 7E). We also confirmed that the AHSA1 protein is expressed at a higher level in HCC cells compared to normal cells via western blot (Figure 7F). Subsequently, the protein expression level of AHSA1 were observed in hepatocellular tissues by immunohistochemistry, and according to the findings of this analysis, the level of protein expression of AHSA1 was greater in hepatocellular carcinoma tissues (Figure 7G).
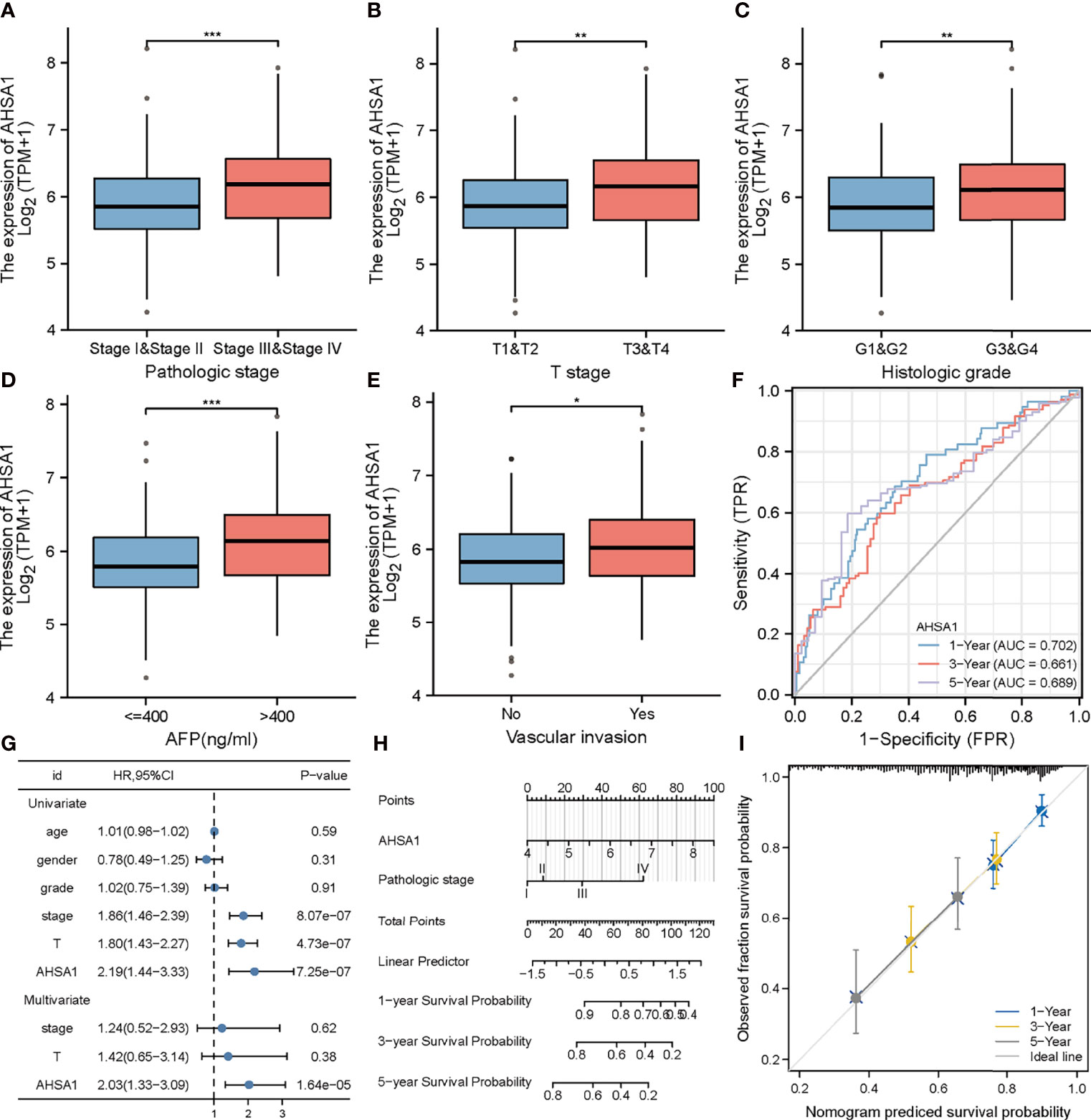
Figure 6 Clinical correlation analysis of AHSA1 in hepatocellular carcinoma. Variation analysis of AHSA1 expression in different pathological stages (A), T stage(B), Histologic grade (C), alpha-fetoprotein (AFP) (D), Vascular invasion (E). (F) Prognostic significance of AHSA1 in hepatocellular carcinoma was analyzed by COX analysis. (G) Prognostic significance of AHSA1 in hepatocellular carcinoma was analyzed by univariate and multivariate COX. (H) Nomogram based on AHSA1 expression and pathological staging. (I) Correction analysis diagram of the nomogram. *p<0.05, **p<0.01, ***p<0.001.
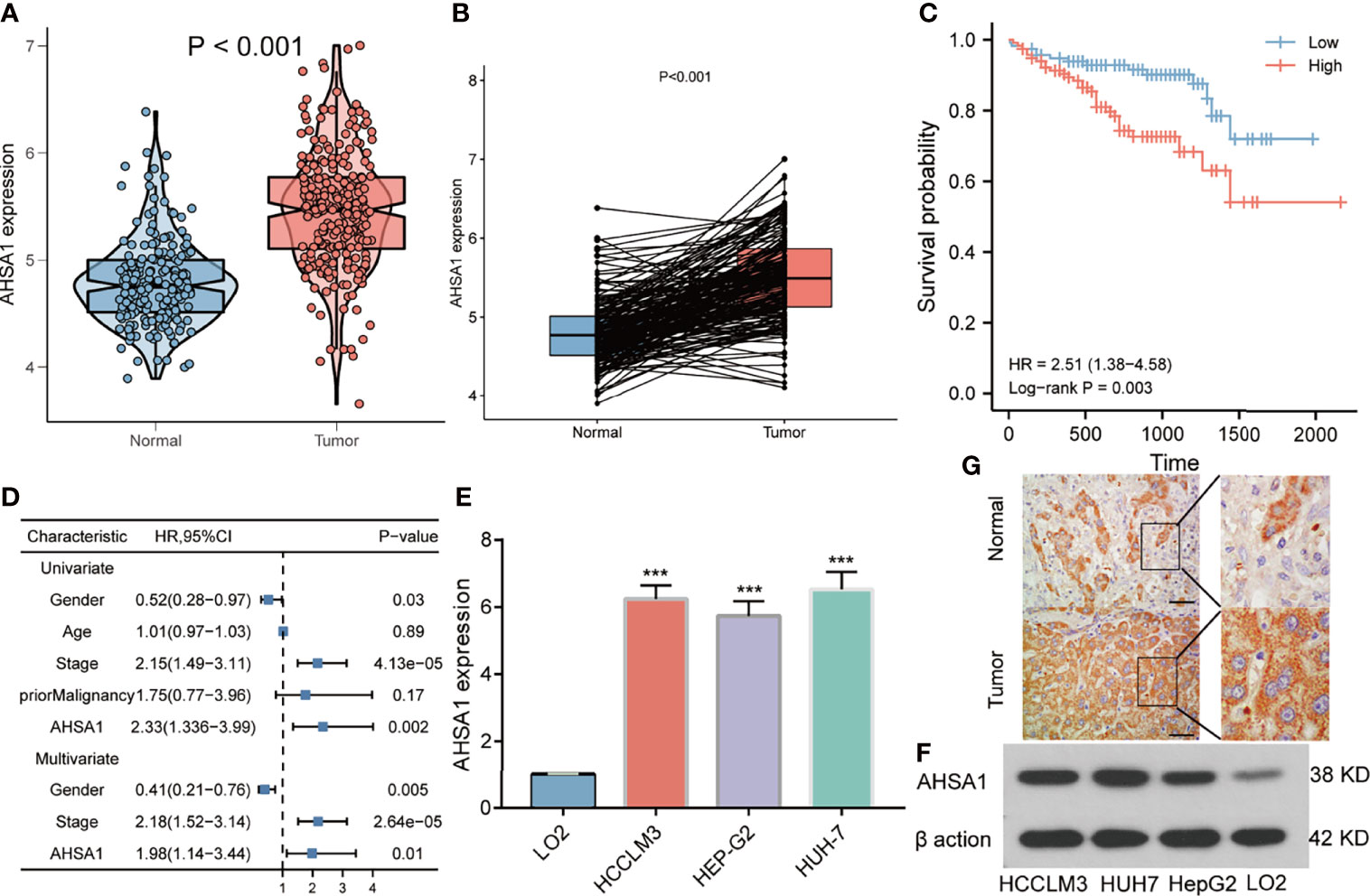
Figure 7 Validation of AHSA1 expression in hepatocellular carcinoma. (A) The mRNA expression of AHSA1 in ICGC cohort. (B) Comparing the AHSA1 expression in paired normal and tumor. (C) The relationship between AHSA1 expression and overall survival in ICGC cohort. (D) Overexpression of AHSA1 was an independent prognostic marker of HCC patients in ICGC cohort. (E) Relative mRNA expression of AHSA1 in HCC cell lines. (F) Expression levels of AHSA1 protein in HCC cell lines. (G) Representative immunohistochemical analysis of AHSA1 in HCC. ***p<0.001.
Although the previous findings indicated that AHSA1 was significantly over-expressed in hepatocellular carcinoma and patients with overexpression had poorer survival expectations, the role of AHSA1 in the occurrence of hepatocellular carcinoma needs further study. Firstly, three shRNA knockout carriers of AHSA1 were constructed and transfected into HUH7 cells. The RT-PCR and Western blot showed variations in mRNA and protein levels, respectively. According to the results, the SH2-AHSA1 had the highest knockout efficiency (Figures 8A, B), and we named the carrier as shAHSA1. Subsequently, we tested the changes of cell activity at different time points after transfection in three groups, namely normal cell group, negative vector group and shAHSA1 transfection group, using the CCK8 kit. The results showed that 48 hours after AHSA1 knockdown, the proliferation ability of cells was significantly reduced (Figure 8C). In addition, after cell scratch test, we found that the healing ability of AHSA1 knockout cells was significantly weakened (Figures 8D, E). Furthermore, the Transwell chamber experiment was performed to verify that the migration ability (Figures 8F, G) and invasion ability (Figures 8F, H) of Huh7 cells were significantly weakened after AHSA1 knockout. In additional, the knockdown efficiency of AHSA1 in HCCLM3 cells was validated by RT-PCR and Western blot (Figures 9A, B). CCK8 assays showed that knockdown of AHSA1 significantly inhibited cell proliferation after 24 hours transfection in HCCLM3 cells (Figure 9C). Furthermore, scratch assays and transwell experiment showed that knockdown of AHSA1 markedly reduced migration/invasion ability of HCCLM3 cell (Figures 9D, E). There was significant positive correlation between the mRNA expression level of AHSA1 and Hsp90AA1. In order to further explore the relationship between AHSA1 and Hsp90AA1, we knocked down AHSA1 in Huh7 cells and examined the effect on Hsp90AA1. The rt-PCR and Western blot analysis showed that the knockdown of AHSA1 significantly reduced the Hsp90AA1 mRNA and protein levels (Figures S2A, B).
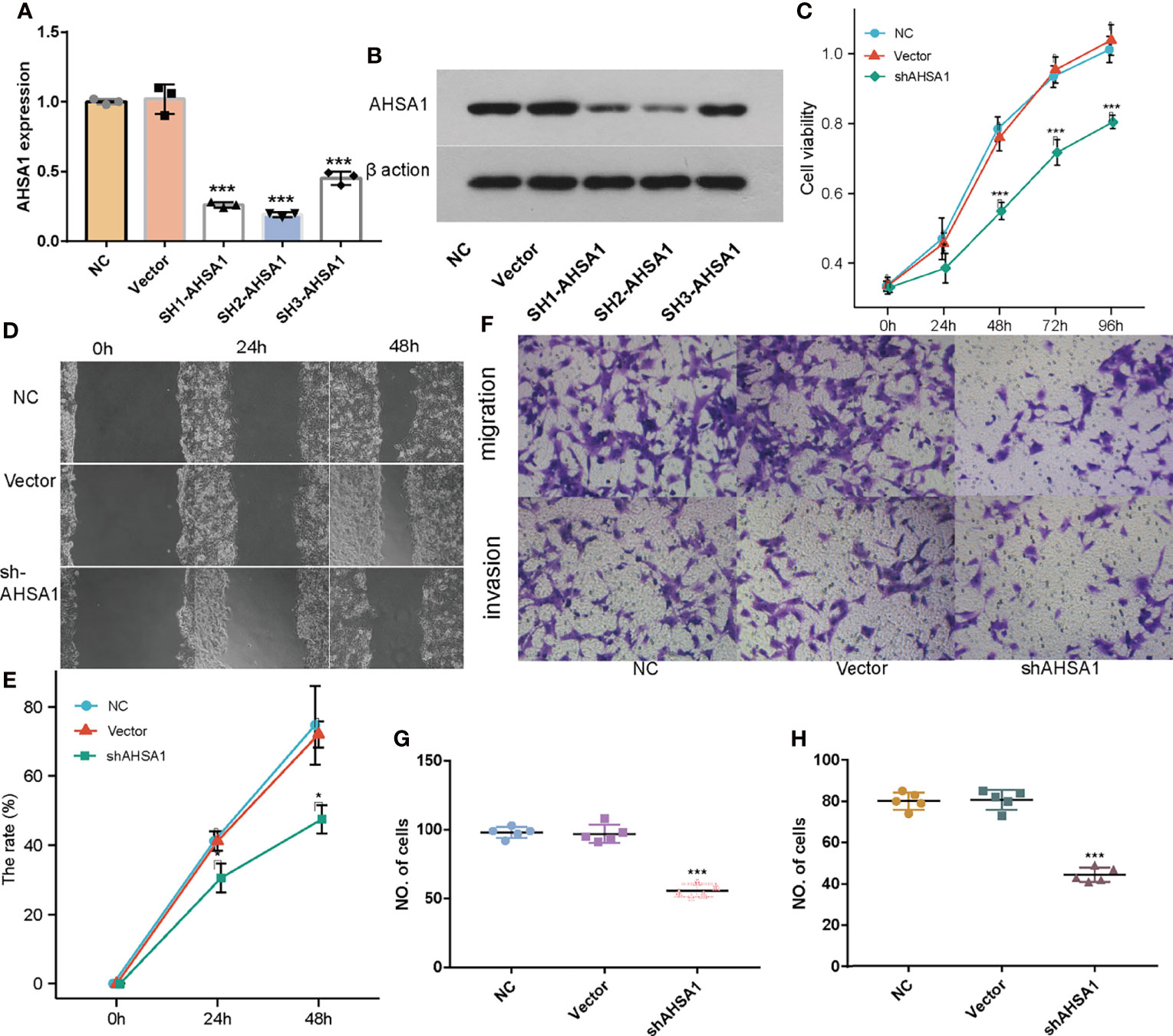
Figure 8 Effects of AHSA1 knockdown on Huh7 cell proliferation and migration. Real time PCR (A) and Weston blot (B) determine the efficiency of AHSA1 knockdown in Huh7 cells. (C) Huh7 cell proliferation was detected by CCK8 assays. (D, E). Cell migration ability was examined by cell scratch assay. (F) Tranwell analysis was preformed to determine the cell migration (G) and invasion (H). *p<0.05, ***p<0.001.

Figure 9 Effects of AHSA1 knockdown on HCCLM3 cell proliferation and migration. Real time PCR (A) and Weston blot (B) determine the efficiency of AHSA1 knockdown in Huh7 cells. (C) CCK8 assays was performed to detect the cell proliferation of HCCLM3. (D) Scratch test was used to determine the cell migration. (E) Cell migration and invasion was detected by Tranwell assay. ***p<0.001.
Heat Shock protein 90 (Hsp90) is a highly conserved, extensively exit molecular chaperone that has a role in the stabilizing and proper folding of at least 300 proteins (19). Reportedly, the Hsp90 protein is known to have a significant function in various cellular processes, signaling, tumor metastasis and tumor immunity (20). Hsp90 is a kind of molecular chaperone highly dependent on ATP activity, and its executive function also requires the cooperation of various auxiliary chaperone molecules. AHSA1 can strongly stimulate the ATPase of Hsp90 and plays a significant role in the executive function of Hsp90 (21). Moreover, AHSA1 itself is also a helper molecular chaperone involved in the maturation and stabilization of various proteins. However, systematic studies on the expression and role of AHSA1 in tumors are still inefficient.
According to this study, based on the TCGA database, AHSA1 was abnormally overexpressed in different types of tumors through unpaired and paired comparisons, but was expressed in low quantities in the KIRC tumor tissues. Meanwhile, by using the K-M survival analysis method, it was found that abnormally high expression of AHSA1 in LIHC, LUAD, ESCA, and KIRP was associated with poor OS and DSS. These findings indicate that AHSA1 may have a significant role in tumor genesis and prognosis. The tumor immune microenvironment can interact with tumor cells and play a significant function in tumor cell clearance and immune escape (22). Several reports have suggested that immune cell infiltration level in the tumor immune microenvironment is associated to tumor development and prognosis (23, 24). This study reported that the expression of AHSA1 was significantly positively related to the infiltration level of CD4+ T cells and MDSC cells in a variety of tumors. Moreover, ssGSEA analysis also confirmed that AHSA1 was substantially positively associated with infiltration levels of T helper cells, Th2 cells and Macrophages cells in LIHC. B Th2 cells are known to inhibit Th1 cells differentiation and IFN-γ-secreting but promote tumor cell proliferation via secretion of IL-4 and IL-10 (25, 26). As an important part of tumor immune microenvironment, macrophages have attracted more and more attention (27, 28). M2 macrophage can be induced to differentiate by IL-4, and promote tumorigenesis and tumor progression (29). Therefore, we preliminarily speculated that AHSA1 might promote the tumor progression through regulating the polarization of Th2 and macrophages cells. Immune checkpoint is an important immunomodulator to maintain immune homeostasis and prevent autoimmunity (30). It consists of stimulant and inhibitory pathways, which are important for maintaining autoimmune tolerance and regulating the type, intensity, and duration of immune responses (31). The results of this study showed that AHSA1 was substantially linked with immune checkpoints in BRCA, LIHC, and LUSC. According to these findings the AHSA1 may play a certain role in the regulation of tumor microenvironment. Epithelial mesenchymal cell transformation, pyroptosis, and autophagy are significantly involved in the genesis, progression, and metastasis of tumors (32–34). The results of this study also indicate that AHSA1 expression is significantly positively co-expressed with these pathway-related molecules in a variety of tumors. These results suggest that abnormal expression of AHSA1 may be associated with tumor genesis and metastasis. Moreover, molecules with possible effects of AHSA1 were also explored and the molecular interaction network was constructed. As expected, AHSA1 showed strong interaction with AMD1 and HSP90, which is consistent with previous reports (35). At the same time, by conducting enrichment analysis, we explained the molecules potentially interacting with AHSA1 are mainly involved in RNA transport, spliceosome and HIF-1α signaling pathway. The pathways above play a significant role in tumor development (36–38). This also indicated the significant involvement of AHSA1 in the progression of tumor. The current research highlighted that the abnormal expression of AHSA1 was closely related to the prognosis of patients with hepatocellular carcinoma. Therefore, further analysis of the association between the expression of AHSA1 and the clinicopathological stage was carried out, and to facilitate the application of AHSA1 in the prognosis assessment of hepatocellular carcinoma, a nomography was constructed. Previous studies have shown that AHSA1 inhibition can significantly inhibit the proliferation and viability of breast cancer cells (39). The results of this study showed that AHSA1 knockdown significantly reduced the proliferation activity, migration and invasion ability of Huh7 and HCCLM3 cells. Meanwhile, AHSA1 knockdown significantly reduced the expression of Hsp90AA1. And Hsp90AA1, as the autophagy-related genes, plays key roles in the autophagy pathway and tumorigenesis. This result suggests AHSA1 might plays a role in regulating autophagy.
AHSA1 is abnormally upregulated in different tumor tissues, and its anomalous expression is related to the prognosis of tumors. The abnormal expression of AHSA1 is related to the infiltration of immune cells, the expression of immune checkpoints, and the expression of various tumor pathway molecules in pan-cancer. Moreover, knockdown of AHSA1 can affect the proliferation and transformation of hepatocellular carcinoma cells. Therefore, AHSA1 can be used as a potential prognostic biomarker.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Yue Bei People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
JL and WL both involved in drafting the manuscript. JL and WL took part in the design and data collection process of the study and are responsible for the content of the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.845585/full#supplementary-material
Supplementary Figure 1 | Differential expression analysis of Hsp90AA1 in Pan-cancer. (A) Expression of Hsp90AA1 mRNA in pan-cancer. (B) The expression differences of Hsp90AA1 in tumor and corresponding adjacent tissues were compared with paired analysis. Mann-Whitney U test was used for this analysis, ns, p≥0.05; * p< 0.05; ** p<0.01; *** p<0.001.
Supplementary Figure 2 | Real time PCR (A) and Weston blot (B) were adopted to evaluate the effects of AHSA1 knockdown on the expression of Hsp90AA1. ns, p≥0.05; * p< 0.05; ** p<0.01; *** p<0.001.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Santucci C, Carioli G, Bertuccio P, Malvezzi M, Pastorino U, Boffetta P, et al. Progress in Cancer Mortality, Incidence, and Survival: A Global Overview. Eur J Cancer Prev (2020) 29(5):367–81. doi: 10.1097/CEJ.0000000000000594
3. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat Genet (2013) 45(10):1113–20. doi: 10.1038/ng.2764
4. Saidak Z, Soudet S, Lottin M, Salle V, Sevestre M-A, Clatot F, et al. A Pan-Cancer Analysis of the Human Tumor Coagulome and its Link to the Tumor Immune Microenvironment. Cancer Immunol Immunother (2021) 70(4):923–33. doi: 10.1007/s00262-020-02739-w
5. Liu J, Zhang S, Dai W, Xie C, Li J-C. A Comprehensive Prognostic and Immune Analysis of SLC41A3 in Pan-Cancer. Front Oncol (2020) 10:586414. doi: 10.3389/fonc.2020.586414
6. Wang Z, Zhang H, Cheng Q. PDIA4: The Basic Characteristics, Functions and its Potential Connection With Cancer. Biomed Pharmacother (2020) 122:109688. doi: 10.1016/j.biopha.2019.109688
7. Wolmarans A, Lee B, Spyracopoulos L, LaPointe P. The Mechanism of Hsp90 ATPase Stimulation by Aha1. Sci Rep (2016) 6:33179. doi: 10.1038/srep33179
8. Xu W, Beebe K, Chavez JD, Boysen M, Lu Y, Zuehlke AD, et al. Hsp90 Middle Domain Phosphorylation Initiates a Complex Conformational Program to Recruit the ATPase-Stimulating Cochaperone Aha1. Nat Commun (2019) 10(1):2574. doi: 10.1038/s41467-019-10463-y
9. Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the Dynamic HSP90 Complex in Cancer. Nat Rev Cancer (2010) 10(8):537–49. doi: 10.1038/nrc2887
10. Shelton LB, Baker JD, Zheng D, Sullivan LE, Solanki PK, Webster JM, et al. Hsp90 Activator Aha1 Drives Production of Pathological Tau Aggregates. Proc Natl Acad Sci USA (2017) 114(36):9707–12. doi: 10.1073/pnas.1707039114
11. Shao J, Wang L, Zhong C, Qi R, Li Y. AHSA1 Regulates Proliferation, Apoptosis, Migration, and Invasion of Osteosarcoma. Biomed Pharmacother (2016) 77:45–51. doi: 10.1016/j.biopha.2015.11.008
12. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–e10. doi: 10.1158/0008-5472.CAN-17-0307
13. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of Tumor Mutation Burden as an Immunotherapy Biomarker: Utility for the Oncology Clinic. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495
14. van Velzen MJM, Derks S, van Grieken NCT, Haj Mohammad N, van Laarhoven HWM. MSI as a Predictive Factor for Treatment Outcome of Gastroesophageal Adenocarcinoma. Cancer Treat Rev (2020) 86:102024. doi: 10.1016/j.ctrv.2020.102024
15. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The Landscape of Somatic Copy-Number Alteration Across Human Cancers. Nature (2010) 463(7283):899–905. doi: 10.1038/nature08822
16. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol (2017) 2017:PO.17.00073. doi: 10.1200/PO.17.00073
17. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res (2010) 38(Web Server issue):W214–W20. doi: 10.1093/nar/gkq537
18. Riera-Domingo C, Audigé A, Granja S, Cheng W-C, Ho P-C, Baltazar F, et al. Immunity, Hypoxia, and Metabolism-The Ménage À Trois of Cancer: Implications for Immunotherapy. Physiol Rev (2020) 100(1):1–102. doi: 10.1152/physrev.00018.2019
19. Schopf FH, Biebl MM, Buchner J. The HSP90 Chaperone Machinery. Nat Rev Mol Cell Biol (2017) 18(6):345–60. doi: 10.1038/nrm.2017.20
20. Hoter A, El-Sabban ME, Naim HY. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int J Mol Sci (2018) 19(9):2560. doi: 10.3390/ijms19092560
21. Walton-Diaz A, Khan S, Bourboulia D, Trepel JB, Neckers L, Mollapour M. Contributions of Co-Chaperones and Post-Translational Modifications Towards Hsp90 Drug Sensitivity. Future Med Chem (2013) 5(9):1059–71. doi: 10.4155/fmc.13.88
22. Lei X, Lei Y, Li J-K, Du W-X, Li R-G, Yang J, et al. Immune Cells Within the Tumor Microenvironment: Biological Functions and Roles in Cancer Immunotherapy. Cancer Letters (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
23. Huntington ND, Cursons J, Rautela J. The Cancer-Natural Killer Cell Immunity Cycle. Nat Rev Cancer (2020) 20(8):437–54. doi: 10.1038/s41568-020-0272-z
24. Engelhard VH, Rodriguez AB, Mauldin IS, Woods AN, Peske JD, Slingluff CL. Immune Cell Infiltration and Tertiary Lymphoid Structures as Determinants of Antitumor Immunity. J Immunol (2018) 200(2):432–42. doi: 10.4049/jimmunol.1701269
25. Lee HL, Jang JW, Lee SW, Yoo SH, Kwon JH, Nam SW, et al. Inflammatory Cytokines and Change of Th1/Th2 Balance as Prognostic Indicators for Hepatocellular Carcinoma in Patients Treated With Transarterial Chemoembolization. Sci Rep (2019) 9(1):3260. doi: 10.1038/s41598-019-40078-8
26. Narsale A, Moya R, Davies JD. Human CD4(+) CD25(+) CD127(hi) Cells and the Th1/Th2 Phenotype. Clin Immunol (2018) 188:103–12. doi: 10.1016/j.clim.2018.01.003
27. Mehla K, Singh PK. Metabolic Regulation of Macrophage Polarization in Cancer. Trends Cancer (2019) 5(12):822–34. doi: 10.1016/j.trecan.2019.10.007
28. Locati M, Curtale G, Mantovani A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu Rev Pathol (2020) 15:123–47. doi: 10.1146/annurev-pathmechdis-012418-012718
29. Wu K, Lin K, Li X, Yuan X, Xu P, Ni P, et al. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front Immunol (2020) 11:1731. doi: 10.3389/fimmu.2020.01731
30. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune Checkpoint Inhibitors: Recent Progress and Potential Biomarkers. Exp Mol Med (2018) 50(12):1–11. doi: 10.1038/s12276-018-0191-1
31. Kalbasi A, Ribas A. Tumour-Intrinsic Resistance to Immune Checkpoint Blockade. Nat Rev Immunol (2020) 20(1):25–39. doi: 10.1038/s41577-019-0218-4
32. Pastushenko I, Blanpain C. EMT Transition States During Tumor Progression and Metastasis. Trends Cell Biol (2019) 29(3):212–26. doi: 10.1016/j.tcb.2018.12.001
33. Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell (2019) 176(1-2):11–42. doi: 10.1016/j.cell.2018.09.048
34. Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang Y, et al. Pyroptosis: A New Frontier in Cancer. Biomed Pharmacother (2020) 121:109595. doi: 10.1016/j.biopha.2019.109595
35. Oroz J, Blair LJ, Zweckstetter M. Dynamic Aha1 Co-Chaperone Binding to Human Hsp90. Protein Sci (2019) 28(9):1545–51. doi: 10.1002/pro.3678
36. Semenza GL. HIF-1 Mediates Metabolic Responses to Intratumoral Hypoxia and Oncogenic Mutations. J Clin Invest (2013) 123(9):3664–71. doi: 10.1172/JCI67230
37. Lee SC-W, Abdel-Wahab O. Therapeutic Targeting of Splicing in Cancer. Nat Med (2016) 22(9):976–86. doi: 10.1038/nm.4165
38. Obeng EA, Stewart C, Abdel-Wahab O. Altered RNA Processing in Cancer Pathogenesis and Therapy. Cancer Discov (2019) 9(11):1493–510. doi: 10.1158/2159-8290.CD-19-0399
Keywords: AHSA1, HCC, immune, cell migration, prognosis
Citation: Li W and Liu J (2022) The Prognostic and Immunotherapeutic Significance of AHSA1 in Pan-Cancer, and Its Relationship With the Proliferation and Metastasis of Hepatocellular Carcinoma. Front. Immunol. 13:845585. doi: 10.3389/fimmu.2022.845585
Received: 30 December 2021; Accepted: 17 May 2022;
Published: 10 June 2022.
Edited by:
Nicolas Larmonier, Université de Bordeaux, FranceReviewed by:
Pulak Ranjan Nath, National Cancer Institute (NIH), United StatesCopyright © 2022 Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Liu, liuyu8566@126.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.