
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 22 February 2022
Sec. T Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.840457
 Motoko Yasutomi1†‡
Motoko Yasutomi1†‡ Allison F. Christiaansen2‡
Allison F. Christiaansen2‡ Naoko Imai1‡
Naoko Imai1‡ Natalia Martin-Orozco2
Natalia Martin-Orozco2 Christian V. Forst3
Christian V. Forst3 Gang Chen2*
Gang Chen2* Hideki Ueno1,4,5*
Hideki Ueno1,4,5*Costimulation pathways play an essential role in T cell activation, differentiation, and regulation. CD155 expressed on antigen-presenting cells (APCs) interacts with TIGIT, an inhibitory costimulatory molecule, and CD226, an activating costimulatory molecule, on T cells. TIGIT and CD226 are expressed at varying levels depending on the T cell subset and activation state. T follicular helper cells in germinal centers (GC-Tfh) in human tonsils express high TIGIT and low CD226. However, the biological role of the CD155/TIGIT/CD226 axis in human Tfh cell biology has not been elucidated. To address this, we analyzed tonsillar CD4+ T cell subsets cultured with artificial APCs constitutively expressing CD155. Here we show that CD226 signals promote the early phase of Tfh cell differentiation in humans. CD155 signals promoted the proliferation of naïve CD4+ T cells and Tfh precursors (pre-Tfh) isolated from human tonsils and upregulated multiple Tfh molecules and decreased IL-2, a cytokine detrimental for Tfh cell differentiation. Blocking CD226 potently inhibited their proliferation and expression of Tfh markers. By contrast, while CD155 signals promoted the proliferation of tonsillar GC-Tfh cells, their proliferation required only weak CD226 signals. Furthermore, attenuating CD226 signals rather increased the expression of CXCR5, ICOS, and IL-21 by CD155-stimulated GC-Tfh cells. Thus, the importance of CD226 signals changes according to the differentiation stage of human Tfh cells and wanes in mature GC-Tfh cells. High TIGIT expression on GC-Tfh may play a role in attenuating the detrimental CD226 signals post GC-Tfh cell maturation.
T cell activation requires a balance of stimulation and inhibition. Positive signals are required for T cells to fight against pathogens, while negative signals are required to prevent damage due to excessive inflammation. Costimulatory and coinhibitory signals serve as key regulators of this equilibrium. The most well-studied pair are CD28 and CTLA-4. Both molecules receive signals from CD80 and CD86 on antigen-presenting cells (APCs) to either induce T cell activation in the case of CD28 or inhibit T cell and APC activation in the case of CTLA-4 (1). CD155 and CD112 also play dual roles in regulating T cell activation by inducing T cell activation through the costimulatory molecule CD226 (DNAM-1) and inhibiting activation through TIGIT (T cell immunoglobulin and immunoreceptor tyrosine-based inhibitory motif ITIM domain). TIGIT deficiency or blockade results in enhanced T cell activation and exacerbated disease in experimental autoimmune encephalitis (EAE) (2–4), whereas it allows for enhanced tumor clearance in murine myeloma models and prevented metastasis (5, 6). In humans, increased TIGIT expression in cancer is associated with worse disease outcomes and TIGIT blockade and siRNA knockdown in T cells restores their effector functions (7, 8). In contrast, CD226 deficiency reduces disease susceptibility in EAE and inhibits tumor clearance (9–11). CD226 polymorphisms are also associated with a number of autoimmune diseases, including type 1 diabetes, rheumatoid arthritis, systemic lupus erythematosus, and Sjogren’s syndrome (12–15). Thus, TIGIT and CD226 play opposing roles in disease development.
TIGIT and CD226 compete for binding to their ligand CD155, however, TIGIT has a higher binding priority than CD226 (4). CD226 has been shown to signal within T cells through Vav1 to enhance TCR-mediated ERK signals and promote IL-17 production (16). By contrast, TIGIT has a cytoplasmic tail with an ITIM and Ig tail-tyrosine-like motif, which reduce TCR expression and signaling (17). Furthermore, TIGIT interferes with CD226 dimerization preventing T cell activation by its presence on the cell surface (5). Thus, TIGIT, when present, plays a dominant role in T cell regulation, while CD226 contributes to immune activation. In addition, TIGIT has been shown to introduce inhibitory signals via CD155 on dendritic cells (DCs) and macrophages that render them tolerogenic (4, 18). Thus, expression levels of either TIGIT and/or CD226 on different cell subsets contribute to the ability of CD155 to activate or inhibit cellular functions. TIGIT can be expressed on T cells and NK cells, whereas CD226 can be expressed on T cells, NK cells, B cells, monocytes, and platelets (19–21). T cell subsets express variable levels of TIGIT and CD226. CD226 is highly expressed on recently activated naïve and effector T cells, including Th1, Th2, and Th17 cells, while TIGIT is expressed on exhausted T cells and Tregs (22, 23).
Of note, T follicular helper (Tfh) cells, in particular within germinal centers (GCs), express high levels of TIGIT (22). Tfh cells play a fundamental role in generating humoral immune responses by providing B cell help in the GCs (24, 25). Human Tfh cell differentiation is initiated when CD4+ T cells interact with APCs in the presence of cytokines including IL-12, IL-23, and TGF-β, and costimulatory signals via OX40 and ICOS, which results in upregulation of the transcription factor Bcl6 (26–30). IL-2 signaling and Blimp-1 are negative regulators of Bcl6 expression and Tfh cell differentiation (31, 32). Pre-Tfh upregulate the expression of ICOS, CXCR5, and PD-1 according to their maturation stage, and accordingly, Tfh cells can be defined by low CD25 expression and high ICOS, CXCR5, and PD-1 (27, 33). Low levels of CD226 and high levels of TIGIT are observed on GC-Tfh cells (22, 34–36). TIGIT expression on circulating Tfh cells in human blood has been associated with high ICOS and IL-21 (37). However, how TIGIT and CD226 regulate the differentiation, maturation, and stability of Tfh cells in humans remains unclear.
In this study, we aimed to determine the biological importance of the CD155/TIGIT/CD226 axis in CD4+ T cell differentiation into Tfh cells and for the maintenance of mature GC-Tfh cells. To simplify the stimulatory signals, we constructed L cells constitutively co-expressing CD32 and CD155, which can act as an artificial APC when anti-CD3 is added to the culture. Here we show that CD226 signals promote Tfh cell differentiation on naïve and pre-Tfh human tonsillar cells, however, mature GC-Tfh cells become less dependent on CD226 signals for their activation and maintenance.
Naïve CD4+ T cells [CXCR5-ICOS- cell population enriched with naïve cells (38)], Tfh precursors (pre-Tfh), or GC-Tfh cells from the human tonsil were identified by the expression level of CXCR5 and ICOS as previously described (38) (Figure 1A). In these subsets expression of TIGIT was upregulated on CXCR5hi ICOShi GC-Tfh cells, as described for murine Tfh cells (36) (Figure 1B). An intermediate level of TIGIT expression was exhibited by pre-Tfh cells and low TIGIT expression by CXCR5-ICOS- cells (Figure 1B). CD226 was co-expressed by approximately half of TIGIT+ GC-Tfh cells. A majority of pre-Tfh cells and a fraction of CXCR5-ICOS- cells expressed CD226.
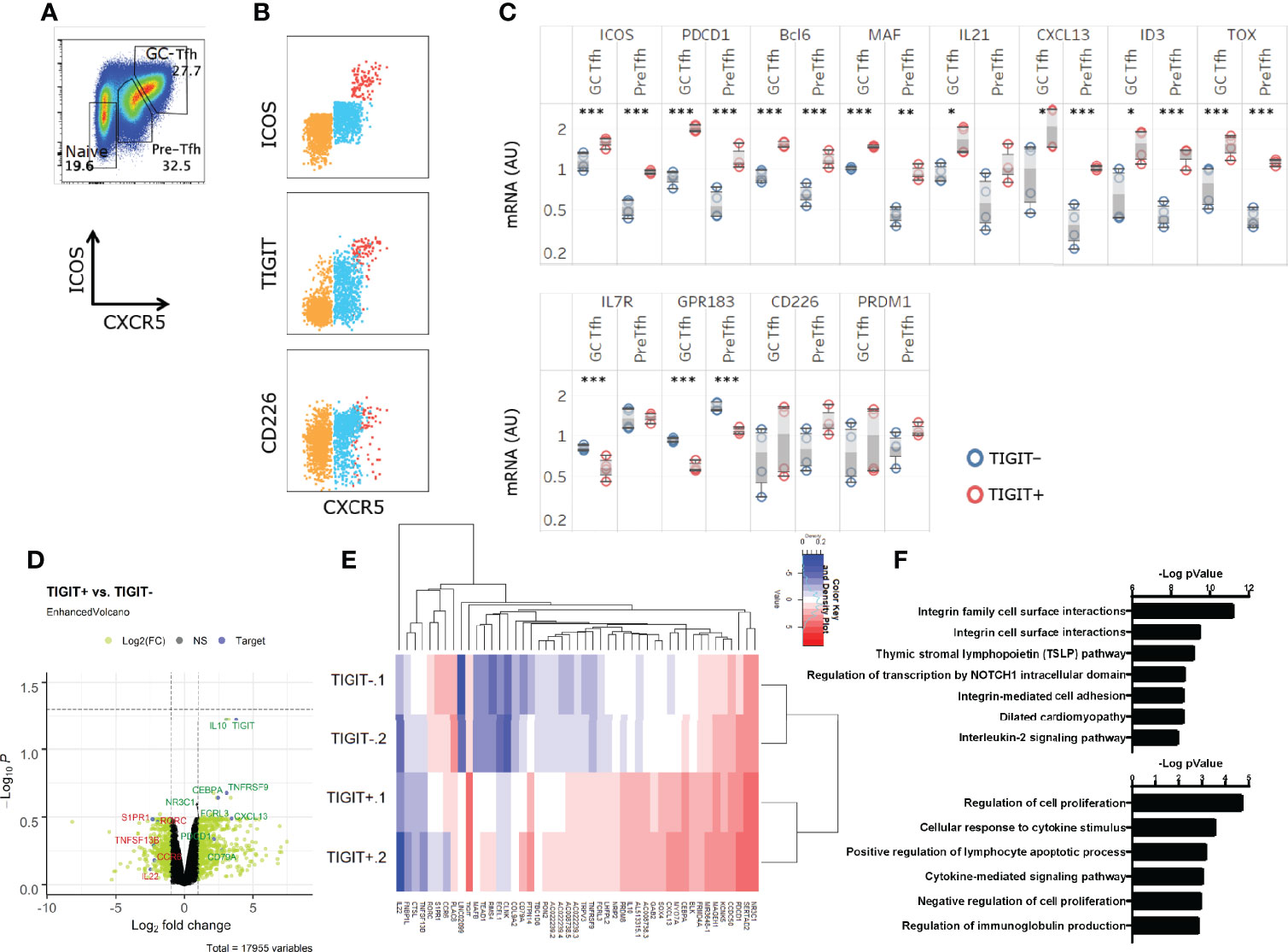
Figure 1 TIGIT+GC-Tfh cells express a strong Tfh gene signature. (A) Definition of GC-Tfh cells (CXCR5hiICOShi), pre-Tfh cells (CXCR5loICOSlo), and naïve (CXCR5-ICOS-) in human tonsils in this study. (B) TIGIT, CD226, ICOS and CXCR5 expression on tonsillar CD4+ T cell with colors depicting each pre-gated subset (Naïve – Orange, Pre-Tfh – Blue, GC-Tfh – Red). (C) GC-Tfh and Pre-Tfh subsets were each sorted according to TIGIT expression, and Tfh-associated gene expression was determined. Paired t-test. *p < 0.05; **p < 0.01; ***p < 0.001. (D, E) The transcriptome of TIGIT+ and TIGIT- pre-Tfh cells was analyzed by RNAseq. Differentially expressed genes are shown in a volcano plot with selected genes of interest highlighted in green if upregulated and downregulated genes listed in red (D) and in a heat map the 47 most significant genes are shown in (E). (F) The top differential pathways between TIGIT+ and TIGIT- pre-Tfh cells. (top) EnrichR (39) using pathway databases from BioPlanet 2019. (bottom) Biological Process 2018. Data is depicting 2 biological replicates per condition.
To determine whether TIGIT expression is associated with the maturation stage of human Tfh cells, GC-Tfh cells and pre-Tfh cells were sorted into TIGIT+ and TIGIT- subsets and analyzed for the expression of Tfh-related genes by Quantigene. We found that TIGIT+ GC-Tfh cells displayed increased ICOS, PDCD1, and IL21 and reduced IL7R, KLF2, and GPR183 than TIGIT- GC-Tfh cells (Figure 1C). Similarly, TIGIT+ pre-Tfh cells expressed stronger Tfh-gene signature than TIGIT- pre-Tfh cells (Figure 1C). Further RNA sequencing analysis on sorted cells indicated the association of TIGIT+ cells with Tfh genes, such as PDCD1 and CXCL13, and downregulation of S1PR1, RORC, and CCR6 which reaffirmed the more mature status of TIGIT+ cells (Figures 1D, E). Interestingly, IL10 was found to be substantially higher in TIGIT+ cells than TIGIT- cells, which is an inhibitory cytokine associated with suppressive functions of a human Tfh cell subset (40).
The major pathway that differed between TIGIT positive and negative cells was IL-2 signaling pathway (Figure 1F). Further analysis showed that the majority of the differentially regulated genes in the IL-2 signaling pathway (such as S1PR1, FLT3LG, and MYC) were downregulated in TIGIT+ cells, and described an inhibitory pathway for Tfh development (31), (Supplemental Table 1). TIGIT+ cells also expressed a gene signature associated with the regulation of cell proliferation (Figure 1F). These observations suggest that TIGIT expression correlates with the maturation stage of human Tfh cells and is associated with reduced IL-2 signaling.
Next, we examined proliferation of tonsil CD4 T cell subsets and whether TIGIT+ and TIGIT- cells differently respond to stimulation. Sorted GC-Tfh, pre-Tfh and naive cells were stimulated with anti-CD3 and anti-CD28, and the proliferation of these cells was measured by dilution of Cell Trace Violet with flow cytometry. In all three cell subsets, TIGIT expression was higher on cells that proliferated less (Figure 2A). To determine if cells lost TIGIT expression upon proliferation or were less proliferate cells, we sorted TIGIT+ and TIGIT- GC-Tfh and pre-Tfh cells, and stimulated with Staphylococcal enterotoxin B (SEB)-pulsed B cells. In both cell subsets, TIGIT+ cells proliferated less than TIGIT- cells (Figure 2B). This observation corresponds with the gene signature of regulation of cell proliferation in TIGIT+ cells (Figure 1F). Thus, TIGIT expression is associated with reduced proliferation in Tfh cells.
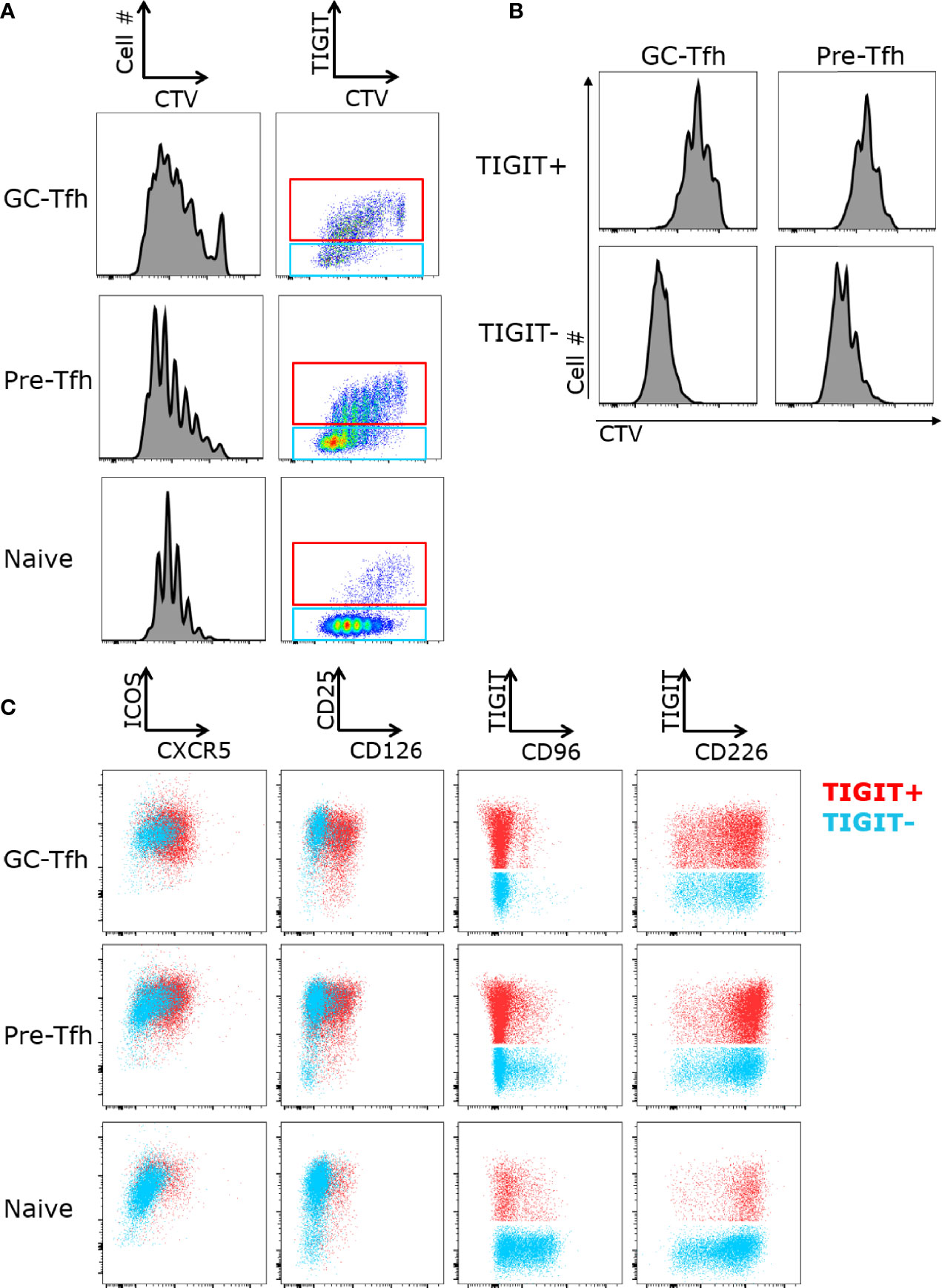
Figure 2 TIGIT+ Tfh cells proliferate less upon stimulation. (A) FACS-sorted GC-Tfh, pre-Tfh, and naïve cells from tonsils were labeled with Cell Trace Violet (CTV) and stimulated with anti-CD3 and anti-CD28 for 5 days. Cell proliferation and TIGIT expression was analyzed by flow cytometry. (B) Proliferation of TIGIT+ and TIGIT- subsets of GC-Tfh and pre-Tfh cells cultured with SEB-pulsed B cells for 5 days. (C) Expression of ICOS, CXCR5, CD226, TIGIT, CD96, CD25, and CD126 on sorted GC-Tfh, pre-Tfh and naïve T cells stimulated with anti-CD3 and anti-CD28 for 3 days. Gated to TIGIT+ (shown in red) and TIGIT- (shown in blue). Representative FACS plots shown for 3 experiments.
We next determined the expression levels of the CD226/TIGIT family receptors after T cell activation. Naive, pre-Tfh, and GC-Tfh cells were sorted and then activated with anti-CD3 and anti-CD28. While CD226 was expressed at a low frequency on ex vivo tonsillar T cell subsets, all subsets highly expressed CD226 upon activation (Figure 2C). CD96 was also highly upregulated on naïve cells and exhibited intermediate expression on pre-Tfh cells with almost no expression on GC-Tfh cells. Additionally, CD96 was not co-expressed with TIGIT, while the majority of TIGIT expressing cells also co-expressed CD226. We also found that TIGIT+ GC-Tfh and pre-Tfh cells expressed higher levels of Tfh-associated markers CXCR5 and CD126 (Figure 2C). Collectively, while CD226 expression was strongly induced in all subsets, TIGIT expression remained correlated with Tfh markers even after activation.
We and others previously showed that the combination of IL-12 and Activin A or the combination of IL-12, IL-23, OX40L, and TGF-β induce human naïve T cells to become functional Tfh-like cells in vitro (27, 28, 30, 41). We examined whether TIGIT is expressed on human blood total CD4+ T cells cultured with these Tfh-promoting conditions. We confirmed that these culture conditions promoted the expression of ICOS and CXCR5 in association with an increase of anti-CD3 and anti-CD28-coated beads (Figure 3A). TIGIT was expressed on CXCR5+ ICOS+ cells cultured with few CD3-CD28 beads (Figure 3B, indicated by blue contour), however, upon stronger stimulation with increased CD3-CD28 beads, TIGIT expression was diminished. Thus, Tfh-promoting culture conditions did not induce TIGIT, and induction of TIGIT expression on GC-Tfh cells requires additional signals present in the GC environment in vivo.
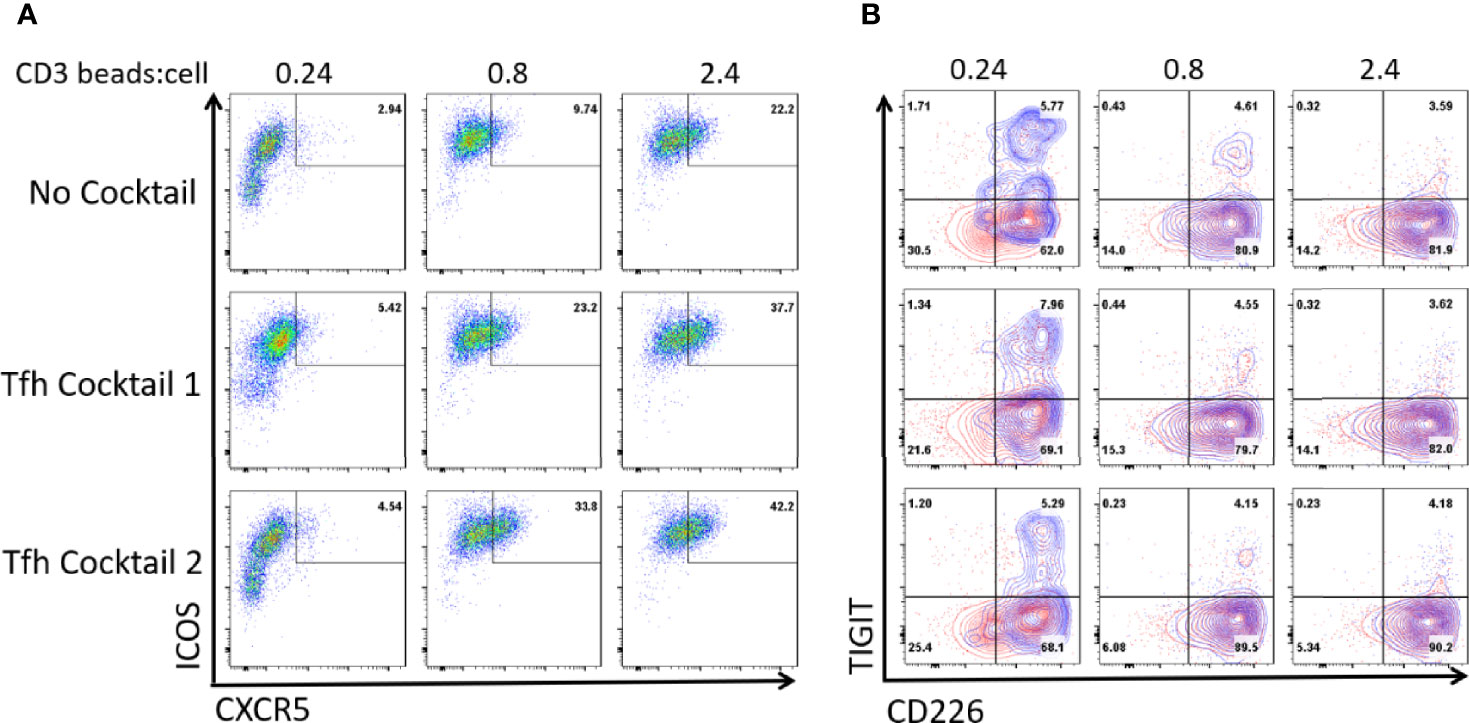
Figure 3 The culture of CD4+ T cells in Tfh-promoting conditions does not induce TIGIT. Total blood CD4+ T cells were cultured in conditions promoting Tfh cell differentiation for 3 days; using Activin A and IL-12 (Tfh cocktail 1) or IL-12, IL-23, rOX40L, and TGF-β (Tfh cocktail 2) in the presence of anti-CD3-CD28 beads at the indicated ratio. (A) ICOS and CXCR5 expression of the cultured T cells. (B) CD226 and TIGIT expression of the total cultured T cells (outlined in red) and on CXCR5+ICOS+ cells (overlayed in blue). Representative FACS plots out of 3 experiments.
To determine the cells expressing the ligands for CD226 and TIGIT in the microenvironment of human tonsils, we examined CD112 and CD155 expression on B cells, monocytes, and DCs. CD155 and the lower affinity receptor CD112 were both expressed on CD14+ monocytes and CD11c+ cDCs but not pDCs, B cells or NK cells (Figure 4). Previous studies have shown that follicular DCs (FDCs) express CD155 (34–36). The lack of CD155 and CD112 expression on B cells suggests that GC-Tfh cells do not receive signals via TIGIT and/or CD226 when they interact with B cells near and within GCs. GC response in secondary lymphoid organs is highly dependent on stromal cell network, and GC-Tfh cells and B cells migrate along FDC network in GCs (42). Therefore, within the GC environment, GC-Tfh cells likely receive signals via CD226 or TIGIT chronically from FDCs.
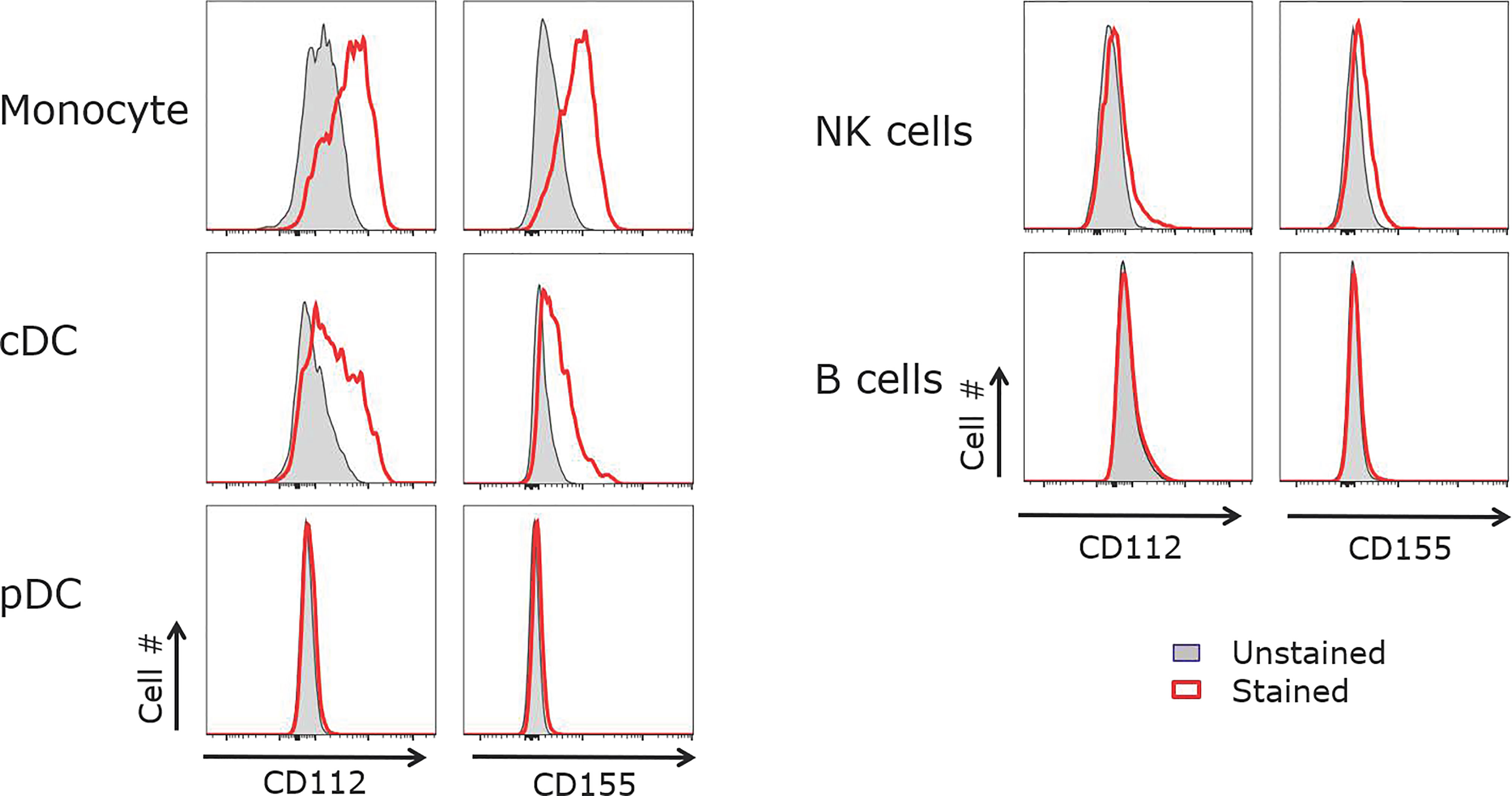
Figure 4 CD155 and CD112 are expressed by monocytes and cDCs but not B cells. Tonsillar APCs were analyzed for CD155 and CD112 expression. Monocytes were defined as CD3-CD19-CD14+, cDCs as CD3-CD19-MHCII+CD11c+, pDCs as CD3-CD19-MHCII+CD123+, NK cells as CD3-CD19-, and B cells as CD3-CD56-CD19+. Representative data out of 5 experiments.
To determine the role of CD155 stimulation on tonsillar CD4+ T cells, we generated CD155 expressing L cells. Parental mouse fibroblast L cells express human CD32, which can mount anti-CD3 upon loading. L cells were successfully transfected with a CD155 expressing lentiviral vector (Supplemental Figure 1). The culture of GC-Tfh cells with CD155+ L cells without anti-CD3 did not largely affect the survival or cell activity as compared to the culture with the parental L cells (data now shown). Therefore, anti-CD3 along with L cells or CD155+ L cells were used to activate tonsillar GC-Tfh, pre-Tfh, and naïve CD4+ T cells. CD155+ L cells induced a robust increase in proliferation in all three cell subsets as compared to parent L cells (Figure 5A). Of note, GC-Tfh cells cultured with CD155+ L cells lost the expression of TIGIT and CD226 (Supplemental Figure 2), probably due to the internalization of these molecules upon interaction with CD155. Thus, CD155 can act as a costimulatory molecule even on GC-Tfh cells and promote cell proliferation. The addition of anti-CD28 did not significantly increase the proliferation of T cells stimulated with CD155+ L cells (not shown).

Figure 5 CD155 costimulation promotes the early phase of Tfh cell differentiation. Tonsilar GC-Tfh, pre-Tfh, and naïve cells were stimulated with L cells or CD155 L cells coated with anti-CD3. (A) Cell proliferation assessed at day 5 by CTV dilution. (B) Phenotype assessed at day 5 by flow cytometry. Per stain each left panel was cultured with L cells; Right panels were cultured with CD155+ L cells as denoted in (A). P0: undivided cells, P1&P2: divided cells one and two times. (C) CXCR5, Bcl-6, CD25, ICOS, and PD-1 expression was measured in proliferated (P1, P2, P3, and P4≤) and undivided (P0) GC-Tfh, pre-Tfh, and naïve cells stimulated with L cells or CD155+ L cells. (D) Cytokine concentration in culture supernatants is shown with dotted lines indicating matching donor samples (n= 4, 4 separate experiments). Paired t-test. *p <0.05; **p < 0.01; ***p < 0.001. (E) Top 38 most significantly different genes shown between CD155+ L cell and L cell stimulated naïve and pre-Tfh cells by RNAseq analysis (n= 2 donors). Pathway analysis for genes from (F) pre-Tfh or (G) naïve cells that were found to be significantly different when stimulated with L cells or CD155+ L cells by RNAseq (enrichR (39): BioPlanet 2019). ns, not significant.
CD155 stimulation induced upregulation of CXCR5 and PD-1 on proliferating GC-Tfh cells (Figures 5B, C). Pre-Tfh cells also upregulated CXCR5. In naïve cells, CD155 stimulation reduced CD25 expression. Interestingly, we also observed a strong reduction in the production of IL-2 (Figure 5D), the cytokine that inhibits Tfh cell differentiation by activating STAT5 (33). Thus, CD155 stimulation upregulates Tfh markers CXCR5 and Bcl-6 while downregulating IL-2 and CD25 that limit Tfh cell differentiation.
Activated and dividing CD4+ T cells change the phenotype according to the number of cell divisions. To assess the effect of CD155 stimulation on the phenotype of CD4+ T cells in the early phase of activation, we analyzed the expression of Tfh markers in cells divided less than three times (Figure 5C). We found that CD155 stimulation upregulated the expression of CXCR5, Bcl6, and PD-1 on GC-Tfh cells even at undivided and the first-division phases. CD155 stimulation also upregulated CXCR5 and Bcl6 expression on pre-Tfh cells. Notably, CD155 stimulation almost abolished the expression of CD25 at an early activation phase of pre-Tfh cells and naïve T cells.
To determine the functionality of the CD155 stimulated cells, we examined the induction of Tfh associated cytokines and chemokines. In GC-Tfh cells, CD155 stimulation promoted IL-10 and CXCL13 production (Figure 5D), both of which are associated with Tfh-driven B cell responses (43, 44). CD155 stimulation on pre-Tfh cells promoted IL-10, IL-17A, and CD40L production. Additionally, CD155 promoted naïve cells to produce FasL and CD40L, which promote B cell survival and antibody production (45). Although not statistically significant, CD155 stimulation tended to increase IL-21 expression by all the subsets. Importantly, CD155 induced all three tonsillar CD4+ T cells subsets to inhibit IL-2 production (Figure 5D). These data suggest that CD155 stimulation promotes an environment to aid B cell antibody production.
Further investigation into the gene transcription changes induced by CD155 signaling was performed with RNAseq analysis. CD155+ L cell treatment induced a greater reduction in gene expression than enhancement compared to L cells (Figure 5E). Further confirmation of the role of CD155 treatment displayed in IL-2 signaling was observed by the reduction in IL2 gene expression and the significance of the IL-2 signaling pathway highlighted from the differentially expressed genes for both pre-Tfh and naïve cells (Figures 5E–G). In addition, many genes of the interferon signaling pathways were reduced by CD155 treatment resulting in both type I and type II interferon pathways being expressed within the top 7 differentially expressed pathways in both pre-Tfh and naïve cells with CD155 treatment (Figures 5F, G and Supplemental Table 1). Of note, upregulation of IL10 gene expression by CD155 stimulation was confirmed. These results indicate that CD155 signaling in tonsillar CD4+ T cells can modify cytokine expression patterns.
To determine the contribution of CD226 in CD155-mediated costimulation, a CD226 blocking antibody was utilized. Following CD155+ L cell and anti-CD3 stimulation, CD226 blockade abrogated the proliferation of naïve T cells and blocked most pre-Tfh proliferation but inhibited GC-Tfh proliferation only modestly (Figure 6A). Upon examination of the expression of Tfh markers, with anti-CD226 blocking antibody naïve and pre-Tfh cells showed a reduction in CXCR5, Bcl-6, CD25, and ICOS (Figures 6B, C). These observations show that the CD155-CD226 axis promotes the proliferation and the expression of Tfh markers on naïve and pre-Tfh cells. Interestingly, the opposite pattern was seen in GC-Tfh cells. Following CD155 stimulation, the anti-CD226 treated group exhibits a significant increase in CXCR5 and ICOS expression (Figures 6B, C). Thus, GC-Tfh cells, which express high levels of TIGIT, are less dependent on the signals via CD226 for cell proliferation and the maintenance of Tfh molecules.
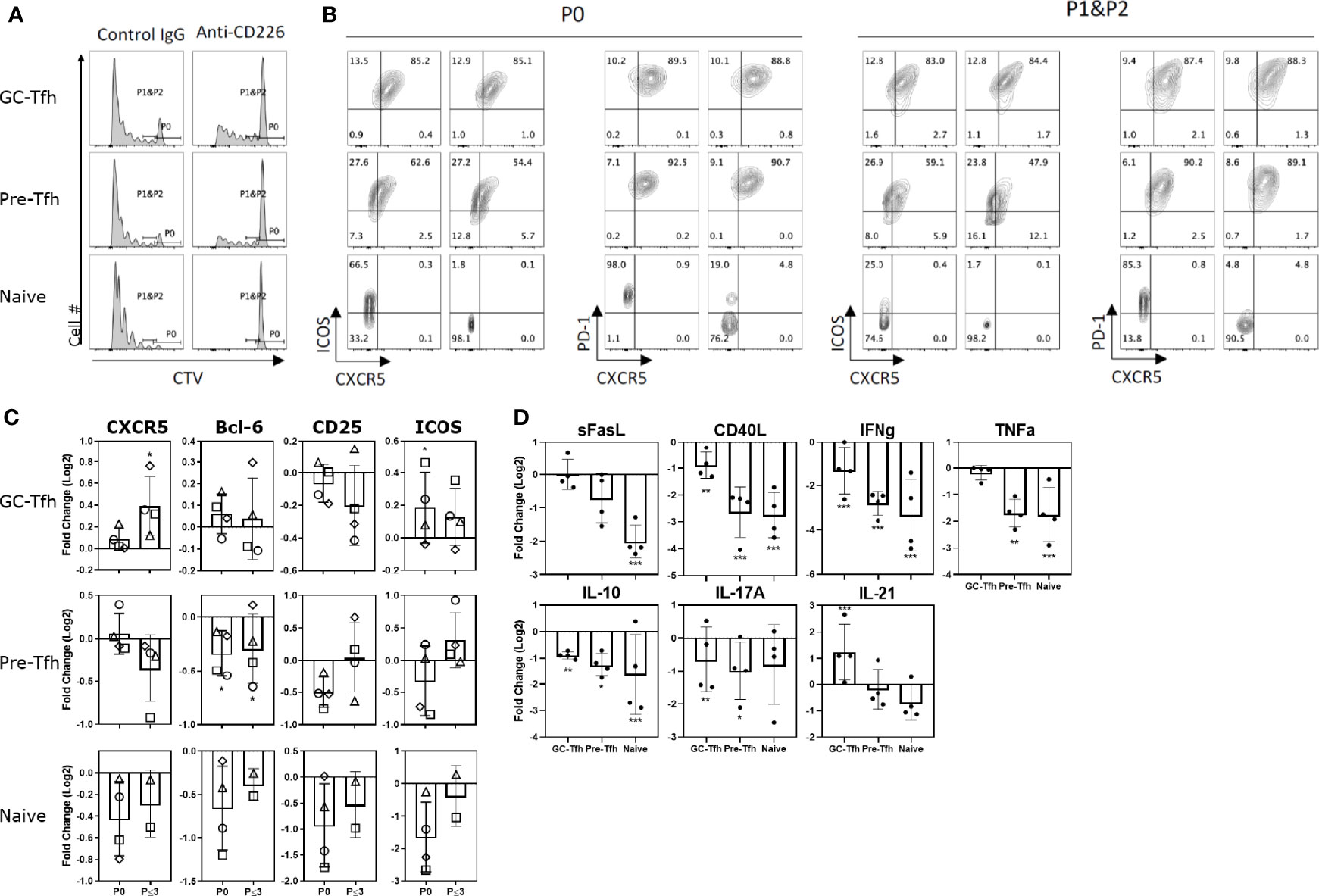
Figure 6 CD155 signaling on T cells is CD226 dependent. Tonsilar GC-Tfh, pre-Tfh, and naïve cells were stimulated with CD155+ L cells coated with anti-CD3 in the presence of control IgG or anti-CD226 blocking mAb. (A) Cell proliferation assessed at day 5 by CTV dilution. Left panels – cultured with control IgG; Right panels – cultured with anti-CD226. (B) Phenotype assessed at day 5 by flow cytometry. Per stain each left panel was cultured with control IgG; Right panels were cultured with anti-CD226 as denoted in (A). P0: undivided cells, P1&P2: divided cells one and two times. (C) The fold change in CXCR5, Bcl-6, CD25, and ICOS expression by anti-CD226 treatment compared to control IgG is shown in undivided (P0) and proliferated (P ≤ 3) GC-Tfh, pre-Tfh, and naïve cells. Paired t-test. l*p < 0.05; **p < 0.01. (D) The fold change in cytokine concentration by anti-CD226 treatment compared to control IgG is shown. Paired t-test. *p < 0.05; **p < 0.01; ***p < 0.001.
We then examined the change in cytokine production by CD226 blockade. CD226 blocking antibody substantially reduced the cytokine production by all three CD4+ T cell subsets compared to the control, with the exception of IL-21 by GC-Tfh cells (Figure 6D). Unexpectedly, IL-10 production was also reduced by CD226 blocking. Thus, CD226 has a dominant role in the expression of a wide range of cytokines by all the tonsillar CD4+ T cell subsets. Increased IL-21 production from GC-Tfh cells by CD226 blocking further supports the notion that GC-Tfh cells are less dependent on CD226 signals.
The relatively increased level of TIGIT expression on tonsillar GC-Tfh cells compared to other T effector cell subsets (22) led us to determine the association between TIGIT expression and the maturation stage of Tfh cells. As compared to TIGIT- compartment, TIGIT+ pre-Tfh cells upregulated gene and protein expression of Tfh markers, including IL-21, IL-10, CXCR5, and PD-1, an observation consistent with blood circulating TIGIT+ Tfh cells (37). TIGIT+ pre-Tfh and naïve cells also exhibited increased expression of CD126 (IL-6R) and reduced genes associated with the IL-2 signaling pathway compared to TIGIT- pre-Tfh cells. Given that IL-6 promotes while IL-2 inhibits Tfh differentiation (31, 46), TIGIT+ pre-Tfh cells appear more prone to mature in Tfh cell differentiation. Furthermore, TIGIT expression on tonsillar CD4+ T cells is associated with reduced proliferation and increased Tfh gene signature. Thus, our study shows that TIGIT expression is associated with the maturation status of human Tfh cells. Of note, TIGIT expression was not upregulated upon the culture of naïve CD4+ T cells in the conditions promoting in vitro Tfh differentiation; thus, other pathways must be operative for the upregulation of TIGIT on differentiating Tfh cells in vivo.
The lack of CD155 expression on B cells including GC-B cells suggests that TIGIT and CD226 are not directly involved in the interactions between Tfh cells and B cells in GCs. To determine the functional significance of TIGIT and CD226 signals in Tfh cell differentiation, we simplified the culture system by generating L cells constitutively expressing CD155 and CD32 and using these cells as an artificial APC. L cells are derived from murine fibroblasts and thus do not produce human cytokines. Pre-incubation of anti-CD3 with L cells permits its binding to CD32, and CD4+ T cells cultured with CD155+ L cells receive activation signals only via T cell receptor and the TIGIT-CD226 family. Using this system, we found that despite high TIGIT expression on GC-Tfh cells, CD226 plays a dominant role during an early phase of Tfh cell differentiation. Several fundamental differences between naïve and pre-Tfh cells vs. GC-Tfh cells were also noted in the context of CD226 signals.
The culture with CD155+ L cells strongly promoted the proliferation of GC-Tfh, pre-Tfh, and naïve CD4+ T cells, confirming that CD155 signals act as costimulatory signals in all tonsillar CD4+ T cell subsets. We found that CD155 stimulation promoted naïve or pre-Tfh cells to proliferate, produced B helper molecules including IL-10, IL-21, ICOS, and sCD40L, and upregulated Tfh markers including CXCR5 and Bcl6, while inhibited the IL-2-CD25 axis. These changes were largely dependent on CD226 signaling in naïve and pre-Tfh cells. Thus, the CD155-CD226 axis drives the early phase of Tfh cell differentiation. Consistent with our conclusion, murine studies have shown that CD155 or CD226 deficient mice exhibit reduced Tfh cell generation in Peyer’s patches (36, 47).
By contrast, GC-Tfh cells required only weak CD226 signals for cell proliferation. Attenuation of CD226 signals by blocking antibody rather increased the expression of Tfh markers, including CXCR5, ICOS, and Bcl-6 in GC-Tfh cells. Thus, unlike naive and pre-Tfh cells, GC-Tfh cells are less dependent on CD226 signals, and too strong CD226 signals seem detrimental to the integrity of GC-Tfh cells. Herein, high TIGIT on GC-Tfh cells likely contributes to decreasing CD226 signals. Maybe weak and tonic TIGIT signals mediated by interactions with FDCs also contribute to insulating CD226 signals in GC-Tfh cells. Studies in both CD8 T cells and Tregs have noted an association with the ratio of CD226 to TIGIT expression and cell activity (5, 48, 49). While these previous studies focused on the ability of TIGIT blockade to restore CD226 activation, we have noted that this ratio also impacts the ability of CD226 blockade to inhibit T cell activation.
CD226 and TIGIT are primarily considered as an analogy of CD28 and CTLA-4. CD28 and CD226 are expressed on activated naïve and pre-Tfh cells, while CTLA-4 and TIGIT are highly expressed on GC-Tfh cells (22, 50). However, their roles might be somewhat different in the context of Tfh cell differentiation. CD28 plays a role in Tfh cell differentiation until CTLA-4 is upregulated by GC-Tfh cells (51), whereas the importance of CD28 signals for GC-Tfh cells is unclear (52). Similarly, CD226 contributes to Tfh cell differentiation for naïve and pre-Tfh cells until TIGIT is upregulated by GC-Tfh cells. The main difference between CD28 and CD226 signals is that the latter eventually shuts off IL-2 production, whereas CD28 promotes IL-2 production (53). TIGIT signals also inhibit IL-2 production by human CD4+ T cells (2). Thus, CD226 and TIGIT seem to contribute to Tfh cell differentiation by inhibiting IL-2 production, at least in humans.
Collectively, our study shows that CD226 and TIGIT collaborate in Tfh cell differentiation and maturation in humans. TIGIT seems important to attenuate CD226 signals on GC-Tfh cells, which express CD226 upon TCR activation. Our study may provide novel insights into the mechanism for controlled human Tfh cell response and the alterations in this axis in the pathogenesis of human autoimmune diseases. Blocking CD226 signals might decrease the generation of Tfh cells and their precursors in patients with autoimmune diseases. However, this needs to be tested with caution, as CD226-blocking might increase the stability of the differentiated mature GC-Tfh cells, contributing to the generation of autoantibodies.
RPMI 1640 media, L-glutamine, Penicillin-Streptomycin, sodium pyruvate, MEM non-essential amino acid solution and HEPES were purchased from Thermo Fisher Scientific (Waltham, MA). Fetal bovine serum, 2-mercaptoethanol, and Mitomycin C (MMC) were purchased from Sigma-Aldrich (Munich, Germany). Staphylococcus aureus Enterotoxin B (SEB) was from Toxin Technology, Inc. (Sarasota, FL). Recombinant human IL-12, IL-23 and TGF-β were purchased from PeproTech (Rocky Hill, NJ). Recombinant human/mouse/rat activin A and soluble OX40L (sOX40L) were purchased from R&D Systems, Inc. (Minneapolis, MN). Cell lines and PBMCs were cultured in complete RPMI medium (RPMI-1640 medium containing 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 1X MEM non-essential amino acid, 25mM HEPES and 50 μM 2-mercaptoethanol supplemented with 10% fetal bovine serum) in the presence of 5% CO2 in a humidified atmosphere at 37°C.
Mouse fibroblastic L cells stably transfected with the human CD32 (CD32-L cells) was kindly provided by Dr. Yong-Jun Liu (54). For generation of L cells stably expressed human CD32 and human CD155 (CD32/CD155-L cells), human CD155 gene was transduced into CD32-L cells by using lentiviral vector. A human CD155 alpha CDS 300-1553 expressing pLenti6.3/V5-DEST was purchased from Thermo Fisher Scientific. The vector was transfected to HEK293T cells using Pantropic Lentiviral Packaging System (Cell Biolabs, Inc., San Diego, CA) and PEI max (Polysciences, Inc., Warrington, PA). The supernatants were collected after 72 hours. CD32-L cells were infected with the supernatants by spin infection for 2 hours. After 72 hours, CD155 and CD32 double expressing L cells were selected by sorting and Mitomycin C selection.
For immunophenotyping, cells were stained with LIVE/Dead dye and Fc blocked prior to staining. Then, cells were stained with fluorochrome-conjugated antibodies in FACS buffer (PBS with 2% heat inactivated FBS) or Brilliant Stain Buffer (BD). Staining of transcription factor Bcl-6 was performed with Tru-Nuclear Transcription Factor Buffer Set (BioLegend). Stained cells were acquired on LSRII (BD Biosciences, NJ) or Cytek Aurora (Cytek Biosciences Inc. CA)) and the flow data were analyzed with FlowJo (BD). The antibodies and viability dyes used are listed in the Supplemental Table 2.
To purify naïve (CD4+CD45RO-) and memory (CD4+CD45RO+) blood CD4+ T cells, thawed PBMCs were magnetically isolated by human CD4+ T cell isolation kit and human CD45RO Microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer’s protocol.
To obtain B cells, B cell subsets and CD4+ T cell subsets from tonsillar cells, thawed cells were separated into CD19+ and CD19- fractions by EasySep Human CD19 Positive Selection Kit II (Stemcell Technologies) using double-triple dose of antibody and beads in manufacturer’s protocol. To purify B cells, CD19+ fraction was stained with fluorescence conjugated antibodies and viability dye and Live/Dead-CD20+CD3- CD14-CD56- cells with FACSAria. To isolate CD4+ T cell subsets, CD19- fraction (T cell-enriched fraction) was stained with fluorescence conjugated antibodies and viability dye and sorted into CXCR5hiICOShi (GC-Tfh), CXCR5loICOSlo (pre-Tfh) and CXCR5-ICOS- (naïve) subsets from Live/Dead-CD4+CD8-CD20-CD11c-CD14- CD56-CD123- cells with FACSAria. In some experiments, T cell-enriched fraction was sorted into smaller populations according to TIGIT expression.
If required, sorted populations were labeled with 2.5μM CTV (Cell Trace Violet, Thermo Fisher Scientific) according to manufacturer’s instruction.
Blood naïve (CD45RO-) CD4+ T cells (5 x104/well) were cultured in 96-well round bottom and activated by Dynabeads Human T-Activator CD3/CD28 (Thermo Fisher Scientific) at various bead: cell ratios. After overnight, a combination of activin A (50 ng/ml) and IL-12 (100 pg/ml) (Tfh Cocktail 1); or a combination of IL-12 (100 pg/ml), sOX40L (100 ng/ml), IL-23 (25 ng/ml), and TGF-β (5 ng/ml) (Tfh Cocktail 2) was added. After three days culture, the cells were analyzed for assessing the expression of surface molecules by flow cytometry.
Tonsillar GC-Tfh, pre-Tfh or naïve cells (5 x104/well) were cultured in 96-well round bottom plates in the presence of Dynabeads Human T-Activator CD3/CD28 at a bead: cell ratio 0.16 for 3-5 days. Phenotype were analyzed by flow cytometry.
Sorted TIGIT+ or TIGIT- GC-Tfh or pre-Tfh cells were co-cultured with tonsillar B cells in 96-well round bottom plates (2 x 104 cells/well) in the presence of 0.1 ug/ml of SEB. In the presence of 0.1 μg/ml of SEB. Cells were cultured for 5 days and analyzed by flow cytometry.
On day -1, L cells and CD155-L cells were treated 4 μg/ml MMC for 3 hours. Cells were collected by Trypsin-EDTA (Thermo Fisher Scientific), washed 2 times and transferred to 96-well flat bottom plates (2.5 x 104/well). On day 0, anti-human CD3 (OKT3; prepared ‘in house’) were added at final concentration of 0.2 - 0.5 μg/ml and plates were incubated at 37°C for at least an hour. Sorted (and CTV-labeled if indicated) GC-Tfh, pre-Tfh and naïve cells were added to the plate (5.0 x 104/well) and cultured for 3-5 days for further analysis.
In some experiments, control IgG (30 μg/ml), or blocking anti-CD226 (10 μg/ml; LeoA1; Millipore Sigma) antibodies were added on day 0 (and day 2 and day 4).
CD4+ T cell subsets were co-cultured with L or CD155-L cells. On day 5, supernatant was collected for determination of cytokine levels. The following cytokines were quantified by ProcartaPlex Immunoassay BLC, CD40L, FAS-L, IFNg, IL-1b, IL-2, IL-4, IL-6, IL-9, IL-10, IL-17A, IL-21, IL-27, IL-31, TNFa (Thermo Fisher) according to the manufacturers’ instructions. Data was collected using the Luminex 200
TIGIT+ and TIGIT- GC-Tfh cells were sorted by FACSAria. The sorted cells were directly collected into the lysis mixture and prepared the cell lysates by using QuantiGene Sample Processing Kit (Thermo Fisher Scientific). The expression levels of mRNA were analyzed with using QuantiGene Plex Assay kit (Thermo Fisher Scientific) and MAGPIX system (Luminex Corporation, Austin, Texas) according to the manufactures’ protocols. Fold changes to average of three house-keeping genes were calculated after subtraction of background.
Sorted and CTV-labeled GC-Tfh, pre-Tfh and naïve were co-cultured with L cells or CD155-L cells. On day3, cells were collected, stained with APC-Cy7-CD4+ and PI and sorted live CD4+CTV+ cells by FACSAria. Total RNA was extracted from sorted cells by Qiagen RNAeasy kit and RNA sequencing was run by Genewiz Inc.
Raw reads were aligned with STARaligner v2.7.0f against the human genome GRCh38.v27 with outFilterScoreMinOverLread = 0.3, outFilterMatchNminOverLread = 0.3, alignSJoverhangMin = 8, alignSJDBoverhangMin = 1. Otherwise default parameters were used. Reads were summarized and assigned to genomic features by feature Counts v2.0.1. Raw counts were further processed using the edgeR v3.30.0/limma v3.4.4.1 pipeline with R v4.0.0 to normalize counts, for quality control, and to identify differentially expressed genes. Genes were considered significantly differentially expressed with a fold change (FC) of |FC| ≥ 2 and a P-value ≤ 0.05 (adjusted for multiple testing after Benjamini & Hochberg). Genes and groups in heatmaps were clustered using their expression values (deviating from the collective median expression) together with “single linkage method” (which is closely related to the minimum spanning tree).
Significantly differentially expressed genes (see Statistical Analysis) from each comparison was entered into enrichR (39) and the top pathways from BioPlanet2019 and Biological Process 2018 databases are shown sorted by P-value rankings. EnrichR employs a modified Fisher’s Exact Test to calculate a rank score (Z-score) of enriched gene sets. The p-values are adjusted for multiple testing after Benjamini & Hochberg.
The significance of the difference between groups in the experiments was evaluated by two-tailed paired t test and one-way ANOVA with GraphPad Prism (GraphPad Software, San Diego, CA). A value of p < 0.05 was considered significant.
All human studies described here were approved by the Institutional Review Board of Icahn School of Medicine at Mount Sinai. Buffy coat from healthy donors was obtained from the New York Blood Center, and PBMCs were isolated via gradient centrifugation with Ficoll (Stemcell Technologies, Vancouver, British Columbia, Canada). Tonsil samples were obtained from the Cooperative Human Tissue Network and single cells were collected by mechanical disruption. The donors (or their parents or the guardians) have provided an informed consent. PBMCs and tonsillar cells were cryopreserved for further experiments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GEO, accession no: GSE194293.
All human studies described here were approved by the Institutional Review Board of Icahn School of Medicine at Mount Sinai. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
MY conducted experiments and acquired and analyzed data. AC conducted experiments, acquired and analyzed data, drafted the initial manuscript and revised the manuscript. NI conducted experiments, acquired and analyzed data, drafted manuscript methods and revised the manuscript. CF drafted manuscript methods and analyzed data. NM-O designed research studies and revised the manuscript. GC oversaw the research, assisted with experimental design, and revised the manuscript. HU conceived and oversaw the research, designed research studies, drafted manuscript methods, and revised the manuscript. All authors approved the final manuscript as written.
This research was supported by the collaborative fund from EMD Serono. The funder also involved in the study design, collection of RNAseq study data, and the decision to submit the manuscript for publication.
Authors AC, NM-O and GC were employed by EMD Serono Research and Development Institute Inc. during the project and have equity interest.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank all the donors who provided us blood and tonsillar samples. We thank Collaborative Human Tissue Network for the collection and shipping of human tonsil samples. We thank the Flow Cytometry Core members at Mount Sinai for their continuous support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.840457/full#supplementary-material
1. Alegre M-L, Frauwirth KA, Thompson CB. T-Cell Regulation by CD28 and CTLA-4. Nat Rev Immunol (2001) 1(3):220–8. doi: 10.1038/35105024
2. Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA. The TIGIT/CD226 Axis Regulates Human T Cell Function. J Immunol (2012) 188(8):3869–75. doi: 10.4049/jimmunol.1103627
3. Joller N, Hafler JP, Brynedal B, Kassam N, Spoerl S, Levin SD, et al. Cutting Edge: TIGIT has T Cell-Intrinsic Inhibitory Functions. J Immunol (Baltimore Md 1950) (2011) 186(3):1338–42. doi: 10.4049/jimmunol.1003081
4. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The Surface Protein TIGIT Suppresses T Cell Activation by Promoting the Generation of Mature Immunoregulatory Dendritic Cells. Nat Immunol (2009) 10(1):48–57. doi: 10.1038/ni.1674
5. Johnston Robert J, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8+ T Cell Effector Function. Cancer Cell (2014) 26(6):923–37. doi: 10.1016/j.ccell.2014.10.018
6. Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the Checkpoint Receptor TIGIT Prevents NK Cell Exhaustion and Elicits Potent Anti-Tumor Immunity. Nat Immunol (2018) 19(7):723–32. doi: 10.1038/s41590-018-0132-0
7. Kong Y, Zhu L, Schell TD, Zhang J, Claxton DF, Ehmann WC, et al. T-Cell Immunoglobulin and ITIM Domain (TIGIT) Associates With CD8+ T-Cell Exhaustion and Poor Clinical Outcome in AML Patients. Clin Cancer Res (2016) 22(12):3057–66. doi: 10.1158/1078-0432.CCR-15-2626
8. Yang Z-Z, Kim HJ, Wu H, Jalali S, Tang X, Krull JE, et al. TIGIT Expression Is Associated With T-Cell Suppression and Exhaustion and Predicts Clinical Outcome and Anti–PD-1 Response in Follicular Lymphoma. Clin Cancer Res (2020) 26(19):5217–31. doi: 10.1158/1078-0432.CCR-20-0558
9. Dardalhon V, Schubart AS, Reddy J, Meyers JH, Monney L, Sabatos CA, et al. CD226 is Specifically Expressed on the Surface of Th1 Cells and Regulates Their Expansion and Effector Functions. J Immunol (Baltimore Md 1950) (2005) 175(3):1558–65. doi: 10.4049/jimmunol.175.3.1558
10. Lozano E, Joller N, Cao Y, Kuchroo VK, Hafler DA. The CD226/CD155 Interaction Regulates the Proinflammatory (Th1/Th17)/anti-Inflammatory (Th2) Balance in Humans. J Immunol (Baltimore Md 1950) (2013) 191(7):3673–80. doi: 10.4049/jimmunol.1300945
11. Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, S-i H, et al. Accelerated Tumor Growth in Mice Deficient in DNAM-1 Receptor. J Exp Med (2008) 205(13):2959–64. doi: 10.1084/jem.20081611
12. Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, et al. CD226 Gly307Ser Association With Multiple Autoimmune Diseases. Genes Immun (2009) 10(1):5–10. doi: 10.1038/gene.2008.82
13. Reinards THCM, Albers HM, Brinkman DMC, Kamphuis SSM, van Rossum MAJ, Girschick HJ, et al. CD226 (DNAM-1) is Associated With Susceptibility to Juvenile Idiopathic Arthritis. Ann Rheum Dis (2015) 74(12):2193–8. doi: 10.1136/annrheumdis-2013-205138
14. Dieudé P, Guedj M, Truchetet ME, Wipff J, Revillod L, Riemekasten G, et al. Association of the CD226 Ser(307) Variant With Systemic Sclerosis: Evidence of a Contribution of Costimulation Pathways in Systemic Sclerosis Pathogenesis. Arthritis rheumatism (2011) 63(4):1097–105. doi: 10.1002/art.30204
15. Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JHM, et al. Shared and Distinct Genetic Variants in Type 1 Diabetes and Celiac Disease. N Engl J Med (2008) 359(26):2767–77. doi: 10.1056/NEJMoa0807917
16. Gaud G, Roncagalli R, Chaoui K, Bernard I, Familiades J, Colacios C, et al. The Costimulatory Molecule CD226 Signals Through VAV1 to Amplify TCR Signals and Promote IL-17 Production by CD4+ T Cells. Sci Signal (2018) 11(538):eaar3083. doi: 10.1126/scisignal.aar3083
17. Ge Z, Peppelenbosch MP, Sprengers D, Kwekkeboom J. TIGIT, the Next Step Towards Successful Combination Immune Checkpoint Therapy in Cancer. Front Immunol (2021) 12:699895. doi: 10.3389/fimmu.2021.699895
18. Chen X, Lu P-H, Liu L, Fang Z-M, Duan W, Liu Z-L, et al. TIGIT Negatively Regulates Inflammation by Altering Macrophage Phenotype. Immunobiology (2016) 221(1):48–55. doi: 10.1016/j.imbio.2015.08.003
19. Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, et al. DNAM-1, a Novel Adhesion Molecule Involved in the Cytolytic Function of T Lymphocytes. Immunity (1996) 4(6):573–81. doi: 10.1016/S1074-7613(00)70060-4
20. Vo AV, Takenaka E, Shibuya A, Shibuya K. Expression of DNAM-1 (CD226) on Inflammatory Monocytes. Mol Immunol (2016) 69:70–6. doi: 10.1016/j.molimm.2015.11.009
21. Scott JL, Dunn SM, Jin B, Hillam AJ, Walton S, Berndt MC, et al. Characterization of a Novel Membrane Glycoprotein Involved in Platelet Activation. J Biol Chem (1989) 264(23):13475–82. doi: 10.1016/S0021-9258(18)80021-7
22. Kunicki MA, Amaya Hernandez LC, Davis KL, Bacchetta R, Roncarolo M-G. Identity and Diversity of Human Peripheral Th and T Regulatory Cells Defined by Single-Cell Mass Cytometry. J Immunol (2017) 200(1):336–46. doi: 10.4049/jimmunol.1701025
23. Fuhrman CA, Yeh WI, Seay HR, Saikumar Lakshmi P, Chopra G, Zhang L, et al. Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226. J Immunol (Baltimore Md 1950) (2015) 195(1):145–55. doi: 10.4049/jimmunol.1402381
24. Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity (2019) 50(5):1132–48. doi: 10.1016/j.immuni.2019.04.011
25. Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T Follicular Helper Cells in Humans and Mice. Nat Immunol (2015) 16(2):142–52. doi: 10.1038/ni.3054
26. Choi Youn S, Kageyama R, Eto D, Escobar Tania C, Johnston Robert J, Monticelli L, et al. ICOS Receptor Instructs T Follicular Helper Cell Versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity (2011) 34(6):932–46. doi: 10.1016/j.immuni.2011.03.023
27. Schmitt N, Liu Y, Bentebibel SE, Ueno H. Molecular Mechanisms Regulating T Helper 1 Versus T Follicular Helper Cell Differentiation in Humans. Cell Rep (2016) 16(4):1082–95. doi: 10.1016/j.celrep.2016.06.063
28. Jacquemin C, Schmitt N, Contin-Bordes C, Liu Y, Narayanan P, Seneschal J, et al. OX40 Ligand Contributes to Human Lupus Pathogenesis by Promoting T Follicular Helper Response. Immunity (2015) 42(6):1159–70. doi: 10.1016/j.immuni.2015.05.012
29. Pattarini L, Trichot C, Bogiatzi S, Grandclaudon M, Meller S, Keuylian Z, et al. TSLP-Activated Dendritic Cells Induce Human T Follicular Helper Cell Differentiation Through OX40-Ligand. J Exp Med (2017) 214(5):1529–46. doi: 10.1084/jem.20150402
30. Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K, et al. The Cytokine TGF-Beta Co-Opts Signaling via STAT3-STAT4 to Promote the Differentiation of Human TFH Cells. Nat Immunol (2014) 15(9):856–65. doi: 10.1038/ni.2947
31. DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, et al. Differential IL-2 Expression Defines Developmental Fates of Follicular Versus Nonfollicular Helper T Cells. Science (2018) 361(6407):eaao2933. doi: 10.1126/science.aao2933
32. Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Sci (New York NY) (2009) 325(5943):1006–10. doi: 10.1126/science.1175870
33. Ballesteros-Tato A, León B, Graf Beth A, Moquin A, Adams Pamela S, Lund Frances E, et al. Interleukin-2 Inhibits Germinal Center Formation by Limiting T Follicular Helper Cell Differentiation. Immunity (2012) 36(5):847–56. doi: 10.1016/j.immuni.2012.02.012
34. Lange R, Peng X, Wimmer E, Lipp M, Bernhardt G. The Poliovirus Receptor CD155 Mediates Cell-To-Matrix Contacts by Specifically Binding to Vitronectin. Virology (2001) 285(2):218–27. doi: 10.1006/viro.2001.0943
35. Boles KS, Vermi W, Facchetti F, Fuchs A, Wilson TJ, Diacovo TG, et al. A Novel Molecular Interaction for the Adhesion of Follicular CD4 T Cells to Follicular DC. Eur J Immunol (2009) 39(3):695–703. doi: 10.1002/eji.200839116
36. Seth S, Ravens I, Kremmer E, Maier MK, Hadis U, Hardtke S, et al. Abundance of Follicular Helper T Cells in Peyer's Patches is Modulated by CD155. Eur J Immunol (2009) 39(11):3160–70. doi: 10.1002/eji.200939470
37. Godefroy E, Zhong H, Pham P, Friedman D, Yazdanbakhsh K. TIGIT-Positive Circulating Follicular Helper T Cells Display Robust B-Cell Help Functions: Potential Role in Sickle Cell Alloimmunization. Haematologica (2015) 100(11):1415–25. doi: 10.3324/haematol.2015.132738
38. Bentebibel SE, Schmitt N, Banchereau J, Ueno H. Human Tonsil B-Cell Lymphoma 6 (BCL6)-Expressing CD4+ T-Cell Subset Specialized for B-Cell Help Outside Germinal Centers. Proc Natl Acad Sci USA (2011) 108(33):E488–97. doi: 10.1073/pnas.1100898108
39. Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene Set Knowledge Discovery With Enrichr. Curr Protoc (2021) 1(3):e90. doi: 10.1002/cpz1.90
40. Canete PF, Sweet RA, Gonzalez-Figueroa P, Papa I, Ohkura N, Bolton H, et al. Regulatory Roles of IL-10-Producing Human Follicular T Cells. J Exp Med (2019) 216(8):1843–56. doi: 10.1084/jem.20190493
41. Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT, et al. Activin A Programs the Differentiation of Human TFH Cells. Nat Immunol (2016) 17(8):976–84. doi: 10.1038/ni.3494
42. Stebegg M, Kumar SD, Silva-Cayetano A, Fonseca VR, Linterman MA, Graca L. Regulation of the Germinal Center Response. Front Immunol (2018) 9:2469. doi: 10.3389/fimmu.2018.02469
43. Rasheed A-U, Rahn H-P, Sallusto F, Lipp M, Müller G. Follicular B Helper T Cell Activity is Confined to CXCR5hiICOShi CD4 T Cells and is Independent of CD57 Expression. Eur J Immunol (2006) 36(7):1892–903. doi: 10.1002/eji.200636136
44. Rousset F, Garcia E, Defrance T, Péronne C, Vezzio N, Hsu DH, et al. Interleukin 10 is a Potent Growth and Differentiation Factor for Activated Human B Lymphocytes. Proc Natl Acad Sci U S A (1992) 89(5):1890–3. doi: 10.1073/pnas.89.5.1890
45. Rathmell JC, Townsend SE, Xu JC, Flavell RA, Goodnow CC. Expansion or Elimination of B Cells In Vivo: Dual Roles for CD40- and Fas (CD95)-Ligands Modulated by the B Cell Antigen Receptor. Cell (1996) 87(2):319–29. doi: 10.1016/S0092-8674(00)81349-5
46. Chavele KM, Merry E, Ehrenstein MR. Cutting Edge: Circulating Plasmablasts Induce the Differentiation of Human T Follicular Helper Cells via IL-6 Production. J Immunol (Baltimore Md 1950) (2015) 194(6):2482–5. doi: 10.4049/jimmunol.1401190
47. Danisch S, Qiu Q, Seth S, Ravens I, Dorsch M, Shibuya A, et al. CD226 Interaction With CD155 Impacts on Retention and Negative Selection of CD8 Positive Thymocytes as Well as T Cell Differentiation to Follicular Helper Cells in Peyer's Patches. Immunobiology (2013) 218(2):152–8. doi: 10.1016/j.imbio.2012.02.010
48. Fourcade J, Sun Z, Chauvin J-M, Ka M, Davar D, Pagliano O, et al. CD226 Opposes TIGIT to Disrupt Tregs in Melanoma. JCI Insight (2018) 3(14):e121157. doi: 10.1172/jci.insight.121157
49. Lucca LE, Lerner BA, Park C, DeBartolo D, Harnett B, Kumar VP, et al. Differential Expression of the T-Cell Inhibitor TIGIT in Glioblastoma and MS. Neurol Neuroimmunol Neuroinflamm (2020) 7(3):e712. doi: 10.1212/NXI.0000000000000712
50. Sage PT, Paterson AM, Lovitch SB, Sharpe AH. The Coinhibitory Receptor CTLA-4 Controls B Cell Responses by Modulating T Follicular Helper, T Follicular Regulatory, and T Regulatory Cells. Immunity (2014) 41(6):1026–39. doi: 10.1016/j.immuni.2014.12.005
51. Webb LMC, Linterman MA. Signals That Drive T Follicular Helper Cell Formation. Immunology (2017) 152(2):185–94. doi: 10.1111/imm.12778
52. Weber JP, Fuhrmann F, Feist RK, Lahmann A, Baz MSA, Gentz L-J, et al. ICOS Maintains the T Follicular Helper Cell Phenotype by Down-Regulating Krüppel-Like Factor 2. J Exp Med (2015) 212(2):217–33. doi: 10.1084/jem.20141432
53. Jenkins MK, Taylor PS, Norton SD, Urdahl KB. CD28 Delivers a Costimulatory Signal Involved in Antigen-Specific IL-2 Production by Human T Cells. J Immunol (1991) 147(8):2461–6.
Keywords: TIGIT, CD226 (DNAM-1), T follicular helper (Tfh) cell, human, CD155
Citation: Yasutomi M, Christiaansen AF, Imai N, Martin-Orozco N, Forst CV, Chen G and Ueno H (2022) CD226 and TIGIT Cooperate in the Differentiation and Maturation of Human Tfh Cells. Front. Immunol. 13:840457. doi: 10.3389/fimmu.2022.840457
Received: 21 December 2021; Accepted: 28 January 2022;
Published: 22 February 2022.
Edited by:
Michael Loran Dustin, University of Oxford, United KingdomReviewed by:
Luis Graca, University of Lisbon, PortugalCopyright © 2022 Yasutomi, Christiaansen, Imai, Martin-Orozco, Forst, Chen and Ueno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Chen, Z2FuZy5jaGVuQGVtZHNlcm9uby5jb20=; Hideki Ueno, dWVuby5oaWRla2kuOGVAa3lvdG8tdS5hYy5qcA==
†Present address:Motoko Yasutomi, Department of Pediatrics, Faculty of Medical Sciences, University of Fukui, Fukui, Japan
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.