- 1Institute of Medical Genetics and Reproductive Immunity, School of Medical Science and Laboratory Medicine, Jiangsu College of Nursing, Huai’an, China
- 2Reproductive Sciences Institute, Jiangsu Key Laboratory of Medical Science and Laboratory Medicine, Department of Immunology, School of Medicine, Jiangsu University, Zhenjiang, China
RNA, one of the major building blocks of the cell, participates in many essential life processes. RNA stability is well-established to be closely related to various RNA modifications. To date, hundreds of different RNA modifications have been identified. N6-methyladenosine (m6A) is one of the most important RNA modifications in mammalian cells. An increasing body of evidence from recently published studies suggests that m6A modification is a novel immune system regulator of the generation and differentiation of hematopoietic stem cells (HSCs) and immune cells. In this review, we introduce the process and relevant regulatory mechanisms of m6A modification; summarize recent findings of m6A in controlling HSC generation and self-renewal, and the development and differentiation of T and B lymphocytes from HSCs; and discuss the potential mechanisms involved.
1 Introduction
RNA is widely acknowledged to play an extremely important role in living organisms. RNA stability largely depends on its modification, one of the most important post-transcriptional regulations (1). To date, more than 100 RNA modifications have been identified in eukaryotes (2). Among them, RNA methylation is one of the predominant forms, accounting for more than 60% of RNA modifications (2). N6-methyladenosine (m6A), a modification occurring at the 6th position of adenine (A) bases, is the most prevalent internal RNA modification (3). The first report about m6A was published in 1974 (4). However, due to the lack of m6A-mapping methods, research on m6A has been in the initial stage for a long time. Until 2012, m6A-specific methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-seq) was established and an extensive study of m6A modification was conducted by transcriptome analysis (5, 6). Since then, a mounting number of studies have substantiated m6A modification in various cell types, unveiling the mystery of m6A in a wide range of physiological and pathological processes, such as stem cell differentiation (7), the maintenance of pluripotency in embryonic development (8), X chromosome inactivation (9), virus replication (10), and the generation, development, invasion, metastasis, and drug resistance of cancer cells (11).
Overwhelming evidence substantiates that RNA m6A modification represents a significant role in the immune system. Here, we introduce the process and molecular mechanism involved in RNA m6A modification and summarize recent investigations highlighting the crucial regulatory role of RNA m6A modification in hematopoietic stem cell (HSC) generation and self-renewal, T and B lymphocyte development, and differentiation from HSCs, which provides novel insights into the role of m6A as a critical regulator of the immune system.
2 The process, hub regulators, and the mechanism of m6A modification
Studies have uncovered that the motifs of m6A modification in RNA sequences are widespread and highly conserved in eukaryotes, mainly occurring on the following consensus sequence: RRACH (R∶G or A; H=A, C, or U) (12, 13). In pre-mRNA, m6A modification motifs are highly enriched near the splicing sites and the last exon, while in mature mRNA, they are mainly distributed in the translation start sites, coding sequences (CDS), and 3’-untranslated region (3’UTR), especially near the stop codon and within internal long exons (5, 6). There are three kinds of pivotal protein factors involved in m6A methylation, including methyl-transferases (writers), demethylases (erasers), and methylation binding protein (readers) (14) (Figure 1).
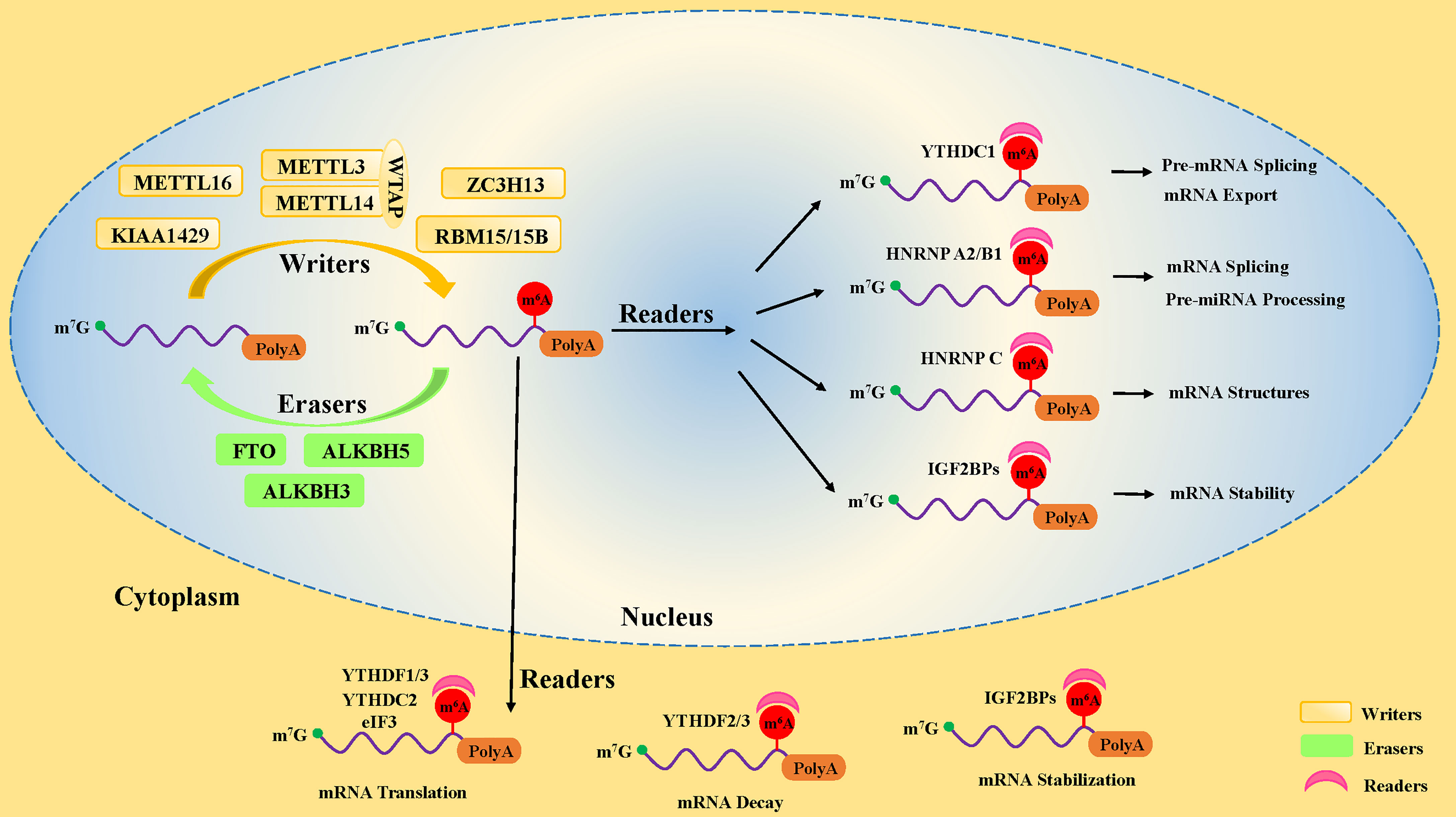
Figure 1 The dynamic process of m6A methylation in mRNA. The m6A methylation is installed by methyltransferases (“writers”), including METTL3, METTL14, WTAP, METTL16, ZC3H13, RBM15/15B, and KIAA1429. It is removed by demethylases (“Erasers”), including FTO, ALKBH5, and ALKBH3. Moreover, the fate of m6A-modification RNA is determined by RNA-binding proteins (“readers”), including YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, IGF2BPs, HNRNP A2/B1, and eIF3.
2.1 Methyl-transferases, the m6A writers, mark the RNA with the methyl group
The first protein that catalyzes the methylation of adenosine on RNA is a methyl-transferase complex (15). It has been established that methyl-transferase-like 3 (METTL3), methyl-transferase-like 14 (METTL14), and Wilms tumor 1-associated protein (WTAP) are the core components of the methyl-transferase complex that promote the incorporation of the m6A methylated group into RNA (16–18) (Figure 1). In addition to the core components, many other proteins, including METTL5 (19) and METTL16 (20), Zinc Finger CCHC-Type Containing 4 (ZCCHC4) (21), Zinc Finger CCCH-Type Containing 13 (ZC3H13) (22, 23), KIAA1429 (24), and RBM15 (and its homolog RBM15B) (9), have been found to participate in the formation of the methyl-transferase complex, playing different roles in methylation modifications of human pre-mRNAs and various non-coding RNAs (25, 26).
METTL3 and METTL14 form a 1:1 stable heterodimer complex in the nucleus (27). METTL3 is a molecule with catalytic activity, while METTL14 is a binding protein that acts as a scaffold for mRNA binding (28). METTL3 could modify adenosine196 (A196) in the 5’-external transcribed spacer of 47S pre-rRNA, leading to decreased rates of pre-rRNA maturation (29). In the nucleus, the METTL3/14 heterodimer interacts with WTAP. It has been reported that WTAP has no methyl-transferase activity, regulates the binding of METTL3/14 complex to transcription sites, and recruits the heterodimers to nuclear speckles (18, 27). Knockdown of WTAP results in attenuation of the interaction between the METTL3/METTL14 complex and mRNA and reduced localization of the complex on the subcellular organelle nuclear plaques of mRNA alternative splicing and ribonucleoprotein assembly (18). WTAP also recruits other related factors, such as RBM15 and KIAA0853, to the methyl-transferase complex, regulating the methylation activity (30). These pieces of evidence indicate that WTAP is critical for m6A methylation activation.
It has been shown that METTL5 forms a heterodimeric complex with the methyl-transferase activator TRMT112 through the formation of a parallel β-zipper between the main chain atoms and is responsible for installing m6A at A1832 on 18S rRNA, thus regulating the translation process (19). METTL16 can reportedly act as a triple-stranded RNA binding protein and support the formation of lncRNA triple helices (20). In addition, under S-adenosylmethionine (SAM)-limiting conditions, METTL16 occupancy on the hairpin (hp1) of the MAT2A 3’-UTR is increased, thus promoting MAT2A splicing (25). ZCCHC4 accumulates in the nucleolus, and the methyl-transferase (MTase) domain of ZCCHC4 is packed against N-terminal GRF-type and C2H2 zinc finger domains and a C-terminal CCHC domain, creating an integrated RNA-binding surface (31). Then, ZCCHC4 deposited m6A at A4220 of human 28S rRNA, which impacts ribosome subunit distribution and global translation (19, 21). In the nucleus, ZCCHC13 was reportedly essential for nuclear localization of WATP, KIAA1429, and HAKAI to facilitate m6A methylation of mRNA (23).
2.2 M6A erasers remove the methyl groups from RNA by oxidative demethylation
The modification of m6A is a dynamic reversible process, and the m6A methylated groups on RNA can be removed by demethylase. Till now, three kinds of m6A demethylases, Fat mass and obesity-associated protein (FTO), ALKB homolog 5 (ALKBH5), and ALKB homolog 3 (ALKBH3), have been identified (32, 33) (Figure 1). All of them belong to the ferrous iron and α-ketoglutarate (αKG)-dependent dioxygenase ALKB family. However, the expression and cellular localization of FTO and ALKBH5 are different in diverse tissues, indicating that they possess many biological functions. FTO was the first identified m6A demethylase in 2011 (32). FTO knockdown with siRNA in HeLa and 293FT cells resulted in increased methylation levels in vitro, while the m6A level of mRNA was decreased by FTO overexpression using a mammalian expression vector in HeLa cells (32).
Varying subcellular localization of FTO is associated with different catalytic substrates. FTO located in the nucleus has been reported to catalyze the mRNA demethylation of m6A, while FTO located in the cytoplasm preferentially demethylates the m2 isoform m6Am (N6-2’-O-dimethyladenosine) of mRNA (34). In addition, cytoplasm FTO demethylates m6A in U6 RNA and m6Am in snRNAs. FTO mediates m1A demethylation in tRNA (34). Interestingly, FTO can reportedly oxidize m6A to form two intermediates, N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (fm6A), with a half-life of 3 h in the nucleus, which may dynamically modulate RNA–protein interactions (35).
Another demethylase ALKBH5 specifically and directly catalyzes the demethylation of m6A without any intermediate (35, 36). According to a recent study in 2013, ALKB homolog 3 (ALKBH3) is also an m6A demethylase (33). However, ALKBH3 preferentially modifies tRNA, not mRNA or rRNA, to enhance protein translation efficiency by extending tRNA’s half-life (37). Whether other ALKB protein family members possess demethylase activity remains poorly understood, warranting further clarification.
2.3 M6A readers determine the fate of m6A-modified RNA
The third most important factor of m6A modification is the RNA-binding proteins called “readers”. They properly decode the m6A RNA methylation information in cells by recognizing and binding m6A-modified RNA. Moreover, they are involved in the regulation of RNA processing and metabolism (Figure 1), such as RNA degradation (38), alternative splicing (39), and translation (40). The earliest identified readers were the YT521-B homology (YTH) domain family proteins in the mammals, consisting of the YTH domain family (YTHDF) and the YTH domain-containing protein (YTHDC) subsets, including YTHDF1, YTHDF2, YTHDF3 (38), YTHDC1, and YTHDC2 (41).
YTHDF subtypes are mainly found in the cytoplasm (42). Different YTHDF readers have different biological effects through dissimilar pathways. YTHDF1 improves the efficiency of m6A-containing mRNA translation by interacting with eukaryotic initiation factor 3 (eIF3) and contributing to ribosome occupancy of target mRNA (40), while YTHDF2 catalyzes mRNA degradation by shortening the half-life of mRNA in the cytoplasm (43). Two different research teams have confirmed that YTHDF3 interacting with YTHDF1 and YTHDF2 enhanced the YTHDF1-facilitated translation of methylated mRNA and promoted YTHDF2-mediated mRNA decay (44, 45). This finding suggests that these three YTHDF proteins work synergistically in the regulation m6A-modified mRNA modulation (44, 45). Moreover, it may also imply that YTHDF2 promotes genes expression in the short term while shortening the subsequent effects, which may benefit the cell response to the environment under stress conditions. However, Du et al. discovered that these three types of YTHDF proteins function similarly on mRNA degradation (43), probably because of their high homology. Intriguingly, the YTHDF2 protein has been reported to promote cap-independent mRNA translation in MEF cells in response to heat shock stress, since YTHDF2 transferred from cytoplasm to nucleus was found to compete with FTO in preserving 5’-UTR methylation of stress-induced mRNA (46). The 5’-UTR translation initiation codon is known to mediate the initiation of translation of eukaryotic mRNA (47). Moreover, it has been shown that YTHDF2 binds directly to 5-methylcytosine in rRNA and modulated the maturation of rRNA (48).
YTDHC1 has been established to be only located in the nucleus, regulating mRNA expression levels (39) and mRNA exporting from the nucleus by influencing alternative splicing (49). In addition, YTDHC1 is required for sufficient rRNA synthesis via regulating the scaffold function of long interspersed nuclear element-1 (LINE1) RNA (50). YTDHC2 exists in the nucleus and cytoplasm, which improves the translation efficiency of target mRNA in mice spermatogenesis (51), supporting its testicular special function. However, gene database analysis revealed that YTDHC2 is moderately expressed in immune cells, which indicates potential existence of an undocumented role in immune cells (52).
Several researchers have reported that some additional RNA-binding proteins, such as eIF3, HNRNP family proteins (including HNRNP A2/B1, HNRNP C, HNRNP G), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs, including IGF2BP1, IGF2BP2, and IGF2BP3), also function as “readers” to decode the m6A RNA methylation information. EIF3 binds to mRNA in the 5’-UTR region and mediates the initiation of mRNA translation (53). HNRNP A2/B1 was reportedly involved in alternative splicing of mRNA and processing of precursor miRNAs in the nucleus (54, 55). However, HNRNP C preferentially bound to m6A-modified sites, weakening the capability of mRNAs and long non-coding RNAs (lncRNAs) to form local secondary structures in the nucleus (56). Little is known about HNRNP G and its role in m6A modification. IGF2BPs have been reported to promote the stability and storage of their target mRNAs in an m6A-dependent manner, thus enhancing target mRNA expression in the nucleus and cytoplasm (57). In recent years, new writer and reader proteins involved in m6A modification are continued to be discovered, suggesting that the biological effects and significance in potentially regulating the m6A modification remain poorly understood, leaving a broad area of research to explore.
3 Techniques for m6A detection in RNA
M6A modifications have been found in mRNA as early as the 1970s (4, 58). However, due to technical limitations, researchers could not detect m6A, especially quantify m6A levels, and let alone identify m6A from the single base level, which retarded the progress of scientific studies in this field for a long time. With the rapid development of next-generation sequencing (NGS) technologies and the improvement of liquid chromatography sensitivity (6), scientists have developed various m6A detection methods.
At present, to detect m6A, high-throughput sequencing is used, such as methylated RNA immunoprecipitation with next-generation sequencing (MeRIP-seq), m6A individual-nucleotide-resolution cross-linking and immunoprecipitation with sequencing (miCLIP-seq), ligation-assisted extraction and thin-layer chromatography (SCARLET), and liquid chromatography-mass spectrometry (LC-MS/MS). MeRIP-seq enables qualitative analysis of the hypermethylated mRNA region, while single-base resolution is not feasible (5). However, miCLIP-seq and SCARLET have been reported to accurately locate m6A loci in the whole transcriptome at a single-nucleotide resolution level (59, 60), while LC-MS/MS could detect the overall m6A level of mRNA (61).
4 The Role of m6A modification in the hematopoietic stem cells, and T and B lymphocytes
HSCs are multipotent cells with the lifelong ability to self-renew and can differentiate into all of the cells of the blood and immune system. In vertebrates, HSCs are derived from hematopoietic endothelial cells (HECs) through endothelial-to-hematopoietic transition (EHT) in the aorta-gonad-mesonephros (AGM) region during the embryonic development stage. Subsequently, HSCs migrate to the fetal liver for massive amplification and transfer to bone marrow (BM) after birth (62). In the BM, HSCs give rise to multipotent progenitor cells (MPPs), consisting of common myeloid progenitors (CMPs, with myeloid, erythroid and megakaryocytic potential) and common lymphoid progenitors (CLPs, with only lymphoid potential) (63). CMPs eventually differentiate into neutrophils, macrophages, eosinophils, basophils, erythroid cells, and monocytes, whereas CLPs subsequently develop and differentiate into T lymphocytes, B lymphocytes, and natural killer cells (NK cells) (63). Recent studies have shown that m6A-mediated various RNA metabolic processes are involved in the stepwise differentiation process (Figure 2). The following sub-chapters introduce more specific details of the role of the RNA m6A modification and its associated regulatory proteins in the HSC generation and self-renewal, cell development, and differentiation of T and B lymphocytes from HSCs.
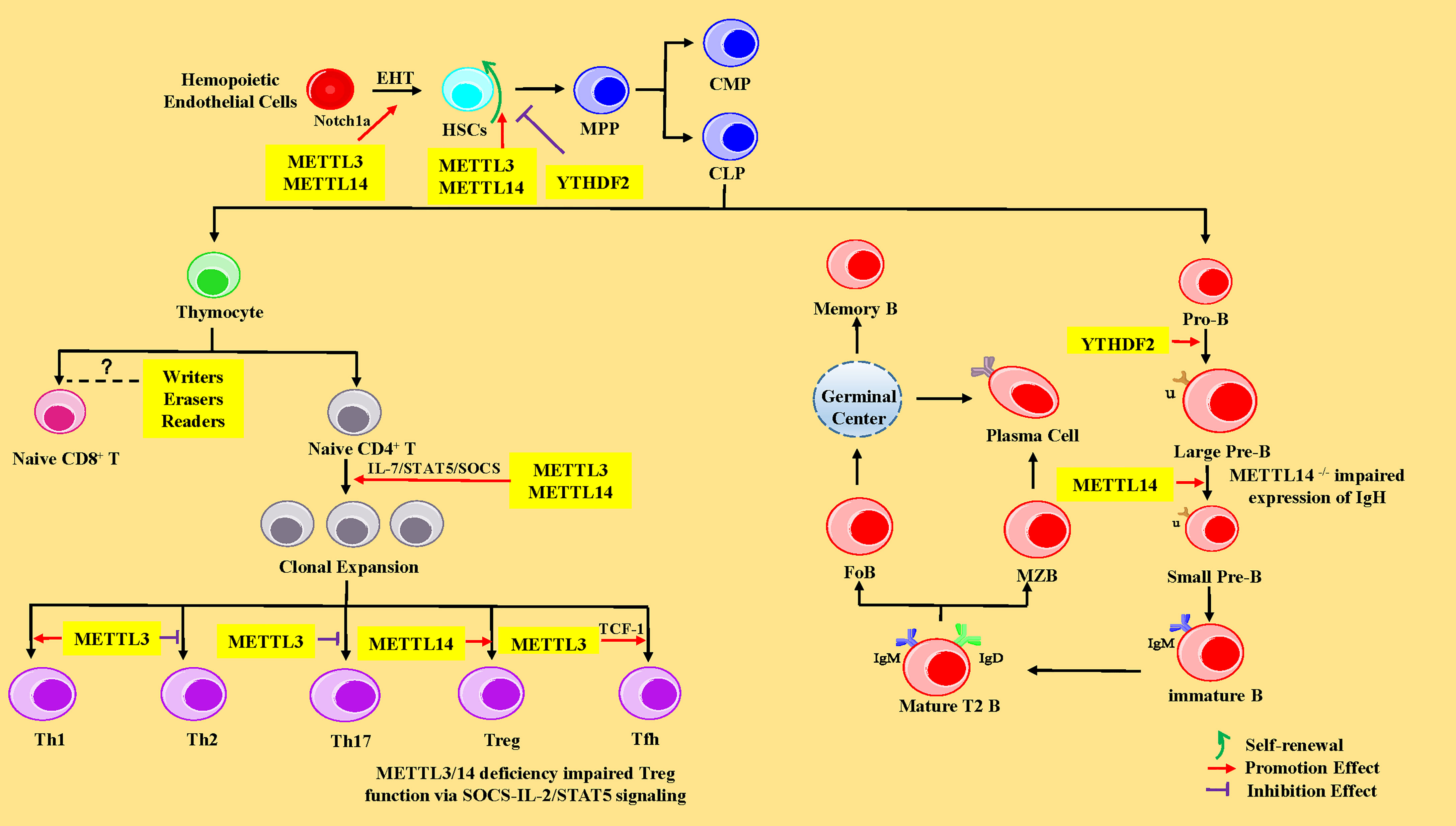
Figure 2 The regulatory role of m6A modification in HSCs, and T and B lymphocytes. In the HSCs, METTL3 and YTHDF2 promote endothelial-to-hematopoietic transition (EHT) via Notch1a leading to HSC generation. METTL3 and METTL14 enhance the ability of HSC self-renewal through MYC, whereas YTHDF2 inhibits HSC self-renewal by promoting the decay of mRNA encoding transcription factors, including TAL1, GATA2, RUNX1, and STAT5. MPP, multipotent progenitor cells; CMP, common myeloid progenitors; CLP, common lymphoid progenitors. In CD4+ T cells, METTL3 and METTL14 promote naive CD4+ T-cell proliferation by the IL-7/STAT5/SOCS pathway. Meanwhile, METTL3 promotes the differentiation of Th1 and Tfh cells, but represses Th2 and Th17 cell differentiation. METTL14 promotes Tregs differentiation, and METTL3/14 is essential for the suppressive function of Tregs. However, the exact roles of m6A modification in CD8+ T cells is unknown. In B cells, YTHDF2 and RBM15 promote pro-B-to-large-pre-B transition, and METTL14 promotes the transition from large pre-B cells to small pre-B cells via increasing chromatin accessibility of key transcription factors loci (Ikzf3, Irf4, Spib, and Bcl6). However, loss of METTL14 did not affect IgH recombination, but might impair the expression of recombined IgH. IGF2BPs promote the differentiation of MZB and FoB by enhancing the stability of B-cell regulators Pax5 and Arid3a mRNA. Pro-B, progenitor B cells; Large Pre-B, large precursor B cells; Small Pre-B, small precursor B cells; MZB, marginal zone B cells; FoB, follicular B cells.
4.1 The m6A modification in hematopoietic stem cells
4.1.1 M6A modification promotes HSC generation via degrading Notch1a
In 2017, researchers found that METTL3 was abundantly expressed in endothelial cells and HECs in zebrafish, indicating that the function of METTL3 was closely related to EHT (7). Previous studies have elucidated that the inhibition of Notch signaling in endothelial cells promotes EHT and the generation of HSCs (64, 65). Once METTL3 was deleted in the embryos of zebrafish with the CRISPR-Cas9 system or in the endothelial cells of mice AGM region using the Cre/Loxp system, the mRNA level of Notch1a was significantly increased, leading to activation of Notch signaling, thus significantly repressing the EHT process and hindering the generating of HSCs (7, 66) (Figure 2). The aforementioned phenomenon could be reversed by forcing METTL3 expression in endothelial cells (7). METTL14 is a heterologous partner of METTL3 (27), implying the importance and role of METTL14 in hematopoiesis. Furthermore, the YTHDF2 recognizes the m6A peak near the stop codon of Notch1 mRNA (7, 66) and mediates Notch1a mRNA decay (67). Interestingly, the YTHDF2 morphants in zebrafish and mice conferred a similar phenotype in METTL3-deficient embryos via mediating Notch1a mRNA decay (7). In summary, the regulation of m6A on HSC specification is evolutionally conserved during EHT in vertebrates and mainly influences the stability of Notch1a.
4.1.2 M6A modification regulates self-renewal and lineage differentiation of HSCs by affecting the half-life or translation of MYC mRNA
HSCs are well-characterized with the ability for self-renewal and multi-lineage differentiation. Recent evidence has revealed that m6A modification at the mRNA level is a new way to regulate HSC self-renewal and differentiation. The transcripts of m6A “writers” METTL3 and METTL14 were abundant in mouse HSCs and significantly downregulated in more mature committed CMPs, especially in myeloid cells (68–70). METTL3 governed the abundance of MYC in HSCs at a decision point that dictates the choice between self-renewal versus differentiation (71). The deletion of METTL3 with Mx1-cre downregulated MYC expression via regulating mRNA translation rather than changes in MYC transcript levels, contributing to an accumulation of HSCs in the BM and a marked reduction of reconstitution potential due to a symmetrical differentiation defects in vivo (70, 71). However, short hairpin RNA (shRNA)-mediated depletion of METTL3 in human umbilical cord blood (hUCB)-derived HSCs and human myeloid leukemia cell line (68) or knocking down of METTL14 in primary leukemic blasts (CD34+) resulted in myeloid differentiation with increasing phosphorylated AKT levels and the inhibition of cell growth with the reduction of colony formation in vitro (68, 69). It is well-recognized that the development and differentiation of HSCs in vivo are not only regulated by cell-intrinsic genes, but also affected by external factors, such as hematopoietic microenvironment. A study revealed that the self-renewal ability of HSCs predominantly depends on METTL3, not on METTL14 (72). In addition, METTL3 depletion in HSCs did not affect apoptosis (68), whereas knocking down of METTL14 induced acute myeloid leukemia (AML) cell apoptosis and promoted myeloid differentiation of normal HSCs via the SPI1–METTL14–MYB/MYC signaling axis (69).
RBM15 is another important component of m6A “writers”, originally found to be involved in hematopoiesis. RBM15 has been shown to exhibit high expression in HSCs. The deletion of RBM15 increased the number of HSCs in RBM15flox/null Mx1-Cre BM, and RBM15-deficent HSCs showed a shift in progenitor fate toward granulocyte differentiation or favored megakaryocyte development with abnormally small and low-ploidy megakaryocytes via downregulation of c-MYC (73, 74). Furthermore, RBM15 knockdown with RNA interference in 32DWT18 myeloid precursor cell line enhanced myeloid differentiation via a mechanism mediated by Notch signaling stimulation via altering recombination signal-binding protein for immunoglobulin kappa J region (RBP Jκ) and hairy and enhancer of split homolog-1 (HES1) promoter activity (75).
Furthermore, one research team used Mx1-Cre mice to achieve m6A “reader” YTHDF2 conditional knockout mice and observed that deletion of YTHDF2 in HSCs led to a significant increase in the absolute numbers as well as the frequency of functional HSCs with normal lineage differentiation in vivo via the activation of WNT signaling downstream targets including MYC, CCND1, and AXIN2 (76). In addition, another study revealed that lentivirus mediated-knockdown of YTHDF2 in human CD34+ HSCs resulted in expansion ex vivo. However, in molecular mechanism, YTHDF2 deficiency promotes the decay of mRNAs that encode transcription factors involved in stem cell self-renewal, including TAL1, GATA2, RUNX1, and STAT5 (76, 77). However, there was no significant difference in the absolute number of progenitors (such as MPPs, CMPs, and CLPs), as well as myeloid cells, B cells, and T cells (76, 77). In a nutshell, the above studies corroborated that MYC is a critical target of m6A modification in HSCs. M6A modification-related proteins can regulate the self-renewal and differentiation of HSCs by affecting the half-life or translation of MYC mRNA.
HSC transplantation is widely used to treat a broad spectrum of disorders, such as hematologic diseases, immune disorders, and cancer. However, the inability to expand HSCs in vitro hampers their potential clinical application. Accordingly, the expansion of HSCs remains a conundrum, warranting further investigation. Recent studies have clarified that m6A modification regulates the self-renewal of HSCs. Thus, the self-renewal and induced expansion of HSCs in vitro after treatment with specific inhibitors or activators of m6A are widely believed to provide an adequate source for the clinical application of HSCs in the future.
4.2 The m6A modification in T cells
HSCs can differentiate into CLPs in the BM, then migrate into the thymus through blood circulation in which they undergo gene rearrangement to express diverse TCR and become mature CD4+ or CD8+ naive T cells through negative and positive selections (78). After that, naive T cells are driven into peripheral immune organs where naive CD4+ T cells, activated by antigens, eventually differentiate into distinct effector T cells sub-populations (Th1, Th2, Th17, Treg, and Tfh) or memory T cells. At the same time, the naive CD8+ T cells develop to cytotoxic T cells (CTL) after recognizing the antigens by antigen-presenting cells (79).
4.2.1 M6A modification plays a dispensable role in the development of T cells and promotes T-cell proliferation via the IL-7/STAT5/SOCS pathway
Most recent studies have revealed that m6A methylation is critical in the development, proliferation, and functional performance of CD4+ T cells. The numbers of naive T cells were increased in the spleen and lymph nodes but not in the thymus in CD4-Cre conditional mice with CD4+ T cell-specific deletion of the writer protein METTL3 or METTL14 (80). However, METTL3-/- or METTL14-/- naive T cells remained in the naive “progenitor” state for more than 4 weeks (80). Similarly, the development of T cells was not affected in the thymus and peripheral lymphoid tissues of CD4+ T cell-specific eraser protein ALKBH5-deficient mice in the steady state (81), suggesting a dispensable role for m6A in the development of T cells (Figure 2).
The mRNA level of ALKBH5 was upregulated after activation of T-cell receptor (TCR) signaling, while the expression of FTO mRNA exhibited no evident change (81). Interestingly, it was puzzling that lack of ALKBH5 did not affect the activation, proliferation, apoptosis, and cytokine secretion of T cells in vivo (81). It is well established that elevated levels of IL-7 can induce proliferation of naive T cells in lymphocytopenia mice after adoptive transfer (82), and SOCS proteins can repress IL-7/JAK/STAT signaling and play an important role in T-cell proliferation and differentiation (83–87). The mRNA 3’-UTR and 5’-UTR of suppressor of cytokine signaling (SOCS) family genes (SOCS1, SOCS3, and CISH) have specific m6A peaks, with a conservative sequence of GG/AACA/U (80, 88). Loss of METTL3 increased the expression of SOCS, especially SOCS1, via increasing the mRNA half-life and decreasing m6A-mediated degradation, leading to the inactivation of the IL-7/STAT5/SOCS pathway (80). Hence, researchers found that T cells lacking METTL3 or METTL14 failed to proliferate using the adoptive transfer model in vivo (80, 89). In summary, m6A modification promotes T-cell proliferation via the IL-7/STAT5/SOCS pathway.
4.2.2 The effect of m6A modification on the differentiation of Th1, Th2, and Th17 cells
Naive T cells can differentiate into distinct T helper effector cell subsets and perform different functions after stimulation by cytokines in vitro and various antigens in vivo. Recent studies have illustrated that m6A methylation plays a crucial regulatory role in T-cell differentiation. The deletion of METTL3 in the CD4+ T cells reinforced Th2 cell differentiation (80) but repressed Th1 and Th17 cell differentiation (80, 90). However, METTL3 deficiency did not affect Th2 cell differentiation following keyhole limpet hemocyanin (KLH) immunization in METTL3-deficient mice (90). Moreover, mice with specific deletion of METTL14 in CD4+ T cells developed spontaneous colitis, characterized by increased inflammatory cell infiltration along with a marked increase of Th1 cytokines (IFN-γ and TNF-α) and Th17 cytokines (IL-17a and IL-17c) in colonic epithelial (91). In contrast, the level of IL-25 produced by Th2 cells was dramatically reduced, while IL-13 levels did not change (91). Moreover, researchers uncovered that the expression of ALKBH5 mRNA was increased, and there were no evident changes in the expression of FTO mRNA in Th1, Th2, Th17, and regulatory T cells (Tregs) compared to naive CD4+ T cells (81). Mice with CD4+ T cell-specific deletion of ALKBH5 were resistant to autoimmune colitis and experimental autoimmune encephalomyelitis (EAE) induction with diminished recruitment of neutrophils into the central nervous system. This phenomenon was explained by the fact that ALKBH5 ablation decreased the mRNA stability of CXCL2 and IFN-γ by enhancing RNA decay and promoted CD4+ T-cell pathogenicity (81). Unlike ALKBH5, ablation of FTO did not impair T-cell development or promote EAE pathogenesis (81). One possibility can be advanced that lack of FTO may result in compensation response by other epigenetic regulation.
4.2.3 M6A modification regulates the immunosuppressive function of Tregs via SOCS-IL-2/STAT5 signaling
Tregs, the critical specialized T cell subset, are involved in reducing inflammation and immunosuppression by producing anti-inflammatory cytokines such as IL-10 and TGF-β, regulating the activity of various immune cells, and eventually suppressing the immune response and guarding against autoimmune diseases (92). The results from three research groups substantiated that METTL3 had no significant regulatory effect on directing Treg cell differentiation (80, 88, 89) (Figure 2). Similar to METTL3, loss of YTHDF1 yielded no significant difference in the number of Tregs (93). However, mice lacking METTL3 in CD4+ T cells and Tregs developed chronic intestinal inflammation and alopecia, respectively. These severe autoimmune diseases were attributed to an absolute lack of immunosuppressive function for Tregs (80, 88). The IL-2/STAT5 pathway was crucial for Tregs function and stability (94). The researchers used m6A irCLIP-Sep technology (a UV-C crosslinking and immunoprecipitation platform) and revealed that m6A modifications were indeed enriched at 3’-UTR and 5’-UTR regions of SOCS genes, with a conservative sequence of GG/AACA/U (88). It has been elicited that deletion of METTL3 increased the levels of SOCS proteins by enhancing SOCS mRNA stability (80, 88), thereby suppressing the IL-2/STAT5 signaling pathway (80, 88). Additionally, deficiency of another methyl-transferase METTL14 in mice blocked the differentiation of naive T cells into induced Tregs and resulted in Tregs dysfunction with loss of suppressive capacity, which was supported by the fact that the METTL14-deficient Tregs were unable to suppress naive T cell-induced colonic inflammation (91). However, m6A demethylase ALKBH5 deficiency in Tregs exerted no effect on immunosuppressive function (81). The role of RNA binding proteins in Tregs has not been reported, which could be future directions for investigation.
4.2.4 M6A modification promotes Tfh cell differentiation via the METTL3–TCF-1 axis
T follicular helper (Tfh) cells, a specialized subset of CD4+ T cell located within germinal centers (GC) of lymph nodes, are involved in the development of humoral immunity by controlling the GC formation and B-cell responses, especially in the differentiation of B cells into plasma cells, antibody production, and Ig class switch (95). T cell-special transcription factor 1 (TCF-1), a crucial transcription factor for Tfh cells differentiation, is encoded by the Tcf7 gene (96, 97). Tcf7 is a bona fide m6A target. METTL3 directly binds to the 3’-UTR region of TCF-1 mRNA to slow down the degradation and enhance its stability, ensuring TCF-1 protein expression (90). Therefore, the knocking down of METTL3 or METTL14, rather than ALKBH5, with shRNA in CD4+ T cells could promote Tfh cell development after lymphocytic choriomeningitis virus (LCMV) infection (89). However, conflicting findings were reported by Yao et al. that the conditional deletion of METTL3 with CD4-cre in CD4+ T cells severely impaired Tfh cell differentiation in a cell-intrinsic manner after LCMV infection (90). This inconsistency may be accounted for by the heterogeneity in experimental systems. Meanwhile, METTL3 deficiency in CD4+ T cells reduced the frequency and cell numbers of GC B cells and plasma cells and METTL3-/- mice exhibited a significantly lower concentration of the LCMV-specific IgG, indicating that the function of Tfh cells was impaired (90).
To sum up, the METTL3–TCF-1 axis functions as an important regulator to initiate and ensure the differentiation of Tfh cells post-transcriptionally. Further efforts are needed to investigate whether other m6A modified proteins are involved in Tfh cell differentiation.
4.3 The m6A modification in B cells
B-cell development represents a highly ordered process that involves sequential immunoglobulin gene recombination. In the BM, CLPs give rise to immature B cells that undergo stepwise differentiation stages from progenitor B cells (pro-B cells) to precursor B cells (pre-B cells), and to immature B cells. Subsequently, the immature B cells further mature in the spleen (98, 99). Pre-B cells can be divided into two subsets: the immature, actively dividing large pre-B cells and the more mature, quiescent small pre-B cells (100). These stages are defined by rearranging the gene’s loci encoding immunoglobulin (H and L chain) and the expression of differentiation-specific molecules on the cell surface. Pro-B cells undergo VDJ gene recombination mediated by recombination activating genes (RAG) and express surface molecules such as CD19. Once the μ heavy chain (Igμ) is expressed, the cell becomes a pre-B cell. Pre-B cells express a set of B lineage-specific genes called λ5 (CD179b) and VpreB (CD179a), which form an IgL chain-like structure known as the surrogate light chain (SLC) to pair with the Igμ heavy chain to combine to form a functional pre-B cell receptor (pre-BCR) (101, 102). Furthermore, IL7 is a pre-B cell stimulant (103), and pre-BCR enhances the reactivity of pre-B cells to IL-7 and promotes clonal expansion along with IL-7R (100). Then, the complete IgM molecule (BCR) formed by V to J rearrangements at the κ and λ light chain gene loci is expressed on the cell surface, indicating that the cells have developed into immature B cells. Lastly, immature B cells migrate into the spleen to mature and become transitional type 1 and type 2 B lymphocyte subsets (T1 and T2) (104). T2 B cells have been established to differentiate into follicular B cells (FoB) or marginal zone B cells (MZB) (104). Upon encountering antigen, mature B cells are activated and enter the GC, undergoing rapid growth and proliferation. Furthermore, GC B cells differentiate into either antibody-secreting plasma cells or long-lived memory B cells (105).
4.3.1 M6A modification controls early B cell development
More recently, it has been demonstrated that RNA m6A modification plays a pivotal regulatory role during early B-cell development. Two recent studies reported that there were no major defects in the BM B cells after poly I:C treatment-induced METTL3 deletion, which might be explained by two hypotheses: deletion of METTL3 was incomplete, or METTL3 was dispensable for the maintenance and survival of B cells (70, 71). However, METTL14 deficiency has been shown to block the transition from large pre-B cells to small pre-B cells in vivo and in vitro (106). Moreover, METTL14 deficiency did not affect IgH recombination but might impair the expression of recombined IgH (106). METTL14-deficient B cells showed limited ability to rearrange Igκ or downregulate the pre-BCR component VpreB (106). Meanwhile, deletion of YTHDF2 resulted in a significant block between the pro-B stage and the late large pre-B stage in vivo (106) (Figure 2). However, YTHDF1 is not essential for B-cell development (106). These developmental defects were attributed to the decreased chromatin accessibility of key transcription factors loci (Ikzf3, Irf4, Spib, and BCL6) mediating the large-pre-B-to-small-pre-B transition and BCR recombination components (Rag1 and Rag2), thus resulting in the fact that these key transcription factors could not be transcribed (106). RBM15flox/null Mx1-Cre mice treated with polyinosinic-polycytidylic acid (PIC) showed a dramatic decrease in peripheral B cells due to a block in pro/pre-B differentiation (73). However, the percentages of IgM+ IgD+ and IgM- IgD+ B cells were maintained in peripheral blood, which suggested that RBM15 is not essential for B cells in the GC (73) (Figure 2). Additionally, IGF2BP3 forced expression with transduction of FH-IGF2BP3-RV in HSCs increased the frequency of MZB and FoB by enhancing the stability of B-cell regulators Pax5 and Arid3a mRNA (107).
CD40 (also called TNFRSF5) is a member of the tumor necrosis factor receptor (TNFR) superfamily with essential roles in B-cell development, activation, GC formation, and class-switched antibodies (108, 109). Notably, the m6A writer WTAP and the m6A reader YTHDF2 are key suppressors of CD40 (110). However, little is known about the role of WTAP in B cell development.
4.3.2 The effect of m6A on B cell activation and proliferation
Furthermore, IL-7 induces the proliferation and differentiation of pro-B cells to pre-B cells (103). After IL-7 stimulation, loss of METTL14 impaired pro-B-cell proliferation and cell size enlargement in vitro and led to significantly lower proliferation rates in pro-B cells and the early large pre-B cells in vivo, consistent with the observation in the YTHDF2-deficient B cells in vitro (106). These findings strongly suggest that the mRNA m6A methylation is important for the IL-7-induced pro-B-cell proliferation by promoting the decay of a group of YTHDF2-bound transcripts (106). Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy derived from germinal center B cells with malignant proliferation (111). Cheng et al. revealed that METTL3 expression was increased both in lymph nodes from DLBCL patients and in DLBCL cell lines, including SU-DHL4, OCILy10, Farage, U2932, and HBL1. Silencing METTL3 using lentivirus-mediated shRNA in cell lines inhibited cell proliferation by abating the total mRNA level of pigment epithelium-derived factors (PEDF), which was usually regarded as a canonical WNT signaling inhibitor. However, WNT/β-catenin signaling was not activated as a result (112). These findings suggest that MEETL3 may be involved in B-cell proliferation.
Taken together, we conclude that m6A modification controls early B-cell development and IL-7-induced pro-B-cell proliferation. Impairment of m6A modification hinders B-cell proliferation, development, and maturation.
5 Conclusion
The physiological and pathological function of m6A modification has become an emerging field of investigation since discovering m6A modification on RNA. Over the past decade, ample evidence has corroborated that m6A modification is involved in the development, differentiation, and function of many immune cells. In this review, we summarize the components of m6A regulators (Figure 1) and recent findings of m6A modification from HSCs to T and B lymphocytes.
We provided a comprehensive overview that m6A modification regulates the generation, self-renewal, and lineage differentiation of HSCs; governs CD4+ T-cell development, activation and clonal proliferation, differentiation, and subsequent effector functions; and controls B cells’ early development, activation, and proliferation through different mechanisms. Moreover, we summarize the molecular mechanisms involved in m6A modification in HSCs, T and B lymphocyte development. These molecules are recognized by the RNA methylation readers and are degraded or translated subsequently. Nevertheless, it remains unknown how m6A modification functions precisely in some special T-cell and B-cell subset, such as Th9 cells, memory CD4+ T cells, Breg cells, plasma cells, and memory B cells, nor is it clear whether m6A modifications regulate the development and function of CD8+ T cells, warranting additional investigation (Figure 2).
Current studies have merely revealed m6A modifications on mRNA in immune cells and overlooked the effect of m6A modification on tRNA and rRNA. M6A modification may regulate the synthesis of proteins that play an important role in the development of immune cells by influencing ribosome occupancy and translation efficiency. Overall, m6A methylation in immune cells is a new research hotspot with more exciting discoveries expected in the future. Indeed, m6A methylation modification could be a novel therapeutic target in alleviating immune cell-related inflammatory diseases and infections and promoting cancer immunotherapy. For example, m6A small-molecule drugs are used to treat HSCs to enhance the self-renewal ability of HSCs and can yield a large number of HSCs in vitro, which provide sufficient HSCs for stem cell transplantation. Moreover, it should be borne in mind that selective deletion of m6A in tumor-infiltrated Tregs may abate the inhibitory function, thereby recovering the tumor-killing functions of CD8+ T cells. Furthermore, the deficiency of m6A in Tfh cells and the enhancement of m6A in Tregs reduce the number of plasma cells and the production of autoantibodies and increase immunosuppressive function for Tregs, representing a promising therapeutic strategy against autoimmune diseases.
Author contributions
CZ wrote the original draft. QS, GX, and XZ provided suggestions about the structure of the manuscript and the writing of molecular mechanisms of M6A modification in the immune system and summarized relevant studies. WC and QS helped to organize and revise the manuscript. QS, CZ, GX, and YY were the funding recipients. All authors contributed to the article and approved the submitted version.
Funding
Our work was supported by grants from the National Natural Science Foundation of China (Grant No. 82071738), Huai’an “Tianyixing” key Laboratory of Medical Examination (Grant No. HAP202004), Huai’an Natural Science Research Program (Grant No. HABL202114), and the Medical Leadership Program of Jiangsu College of Nursing (Grant No. 2021001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med (2020) 52(3):400–8. doi: 10.1038/s12276-020-0407-z
2. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res (2018) 46(D1):D303–D7. doi: 10.1093/nar/gkx1030
3. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
4. Desrosiers R FK, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc Natl Acad Sci USA (1974) 71):3971–5. doi: 10.1073/pnas.71.10.3971
5. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
6. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse M6a RNA methylomes revealed by M6a-seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
7. Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. M(6)A modulates haematopoietic stem and progenitor cell specification. Nature (2017) 549(7671):273–6. doi: 10.1038/nature23883
8. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. M(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell (2014) 15(6):707–19. doi: 10.1016/j.stem.2014.09.019
9. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature (2016) 537(7620):369–73. doi: 10.1038/nature19342
10. Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M, et al. N(6)-Methyladenosine-Binding proteins suppress HIV-1 infectivity and viral production. J Biol Chem (2018) 293(34):12992–3005. doi: 10.1074/jbc.RA118.004215
11. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer (2019) 18(1):176. doi: 10.1186/s12943-019-1109-9
12. Csepany T, Lin A, Baldick CJ, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem (1990) 265(33):20117–22. doi: 10.1016/s0021-9258(17)30477-5
13. Iba K, Hamada N, Sowa E, Morii H, Wada M, Shiode S., et al. A female case of kallmann’s syndrome. Endocrinol Jpn (1977) 23(4):289–93.
14. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic M(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res (2018) 28(6):616–24. doi: 10.1038/s41422-018-0040-8
15. Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. internal mRNA methylation requires a multisubunit complex. J Biol Chem (1994) 269(26):17697–704. doi: 10.1016/s0021-9258(17)32497-3
16. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA(N6-Adenosine)-Methyltransferase. RNA (1977) 3(11):1233–47.
17. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol (2014) 16(2):191–8. doi: 10.1038/ncb2902
18. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
19. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA M6a methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res (2019) 47(15):7719–33. doi: 10.1093/nar/gkz619
20. Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA (2016) 113(49):14013–8. doi: 10.1073/pnas.1614759113
21. Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol (2019) 15(1):88–94. doi: 10.1038/s41589-018-0184-3
22. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the M(6)A machinery component Wtap/Fl(2)d. Genes Dev (2018) 32(5-6):415–29. doi: 10.1101/gad.309146.117
23. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA M(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell (2018) 69(6):1028–38 e6. doi: 10.1016/j.molcel.2018.02.015
24. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential M(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov (2018) 4:10. doi: 10.1038/s41421-018-0019-0
25. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA M(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell (2017) 169(5):824–35 e14. doi: 10.1016/j.cell.2017.05.003
26. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, et al. Human METTL16 is a N(6)-methyladenosine (M(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep (2017) 18(11):2004–14. doi: 10.15252/embr.201744940
27. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
28. Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, et al. Interactions, localization, and phosphorylation of the M6a generating METTL3-METTL14-WTAP complex. RNA (2018) 24(4):499–512. doi: 10.1261/rna
29. Sergeeva O, Sergeev P, Melnikov P, Prikazchikova T, Dontsova O, Zatsepin T. Modification of Adenosine196 by Mettl3 methyltransferase in the 5'-external transcribed spacer of 47S pre-rRNA affects rRNA maturation. Cells (2020) 9(4):1061. doi: 10.3390/cells9041061
30. Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, et al. Identification of wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem (2013) 288(46):33292–302. doi: 10.1074/jbc.M113.500397
31. Ren W, Lu J, Huang M, Gao L, Li D, Wang GG, et al. Structure and regulation of ZCCHC4 in M(6)A-methylation of 28S rRNA. Nat Commun (2019) 10(1):5042. doi: 10.1038/s41467-019-12923-x
32. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
33. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
34. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential M(6)A, M(6)Am, and M(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell (2018) 71(6):973–85 e5. doi: 10.1016/j.molcel.2018.08.011
35. Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun (2013) 4:1798. doi: 10.1038/ncomms2822
36. Wu B, Li L, Huang Y, Ma J, Min J. Readers, writers and erasers of N(6)-methylated adenosine modification. Curr Opin Struct Biol (2017) 47:67–76. doi: 10.1016/j.sbi.2017.05.011
37. Ueda Y OI, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer. Sci Rep (2017) 13(7):42271. doi: 10.1038/srep42271
38. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-Methyladenosine-Dependent regulation of messenger RNA stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
39. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear M(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
40. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
41. Meyer KD, Jaffrey SR. Rethinking M(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
42. Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, et al. The YTH domain is a novel RNA binding domain. J Biol Chem (2010) 285(19):14701–10. doi: 10.1074/jbc.M110.104711
43. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes M(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
44. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-Methyladenosine-Modified RNA. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
45. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic M(6)A reader YTHDF3 promotes mRNA translation. Cell Res (2017) 27(3):444–7. doi: 10.1038/cr.2017.10
46. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic M(6)A mRNA methylation directs translational control of heat shock response. Nature (2015) 526(7574):591–4. doi: 10.1038/nature15377
47. Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem (2014) 83(1):779–812. doi: 10.1146/annurev-biochem-060713-035802
48. Dai X, Gonzalez G, Li L, Li J, You C, Miao W, et al. YTHDF2 binds to 5-methylcytosine in RNA and modulates the maturation of ribosomal RNA. Anal Chem (2020) 92(1):1346–54. doi: 10.1021/acs.analchem.9b04505
49. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife (2017) 6. doi: 10.7554/eLife.31311
50. Chen C, Liu W, Guo J, Liu Y, Liu X, Liu J, et al. Nuclear M(6)A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell (2021) 12(6):455–74. doi: 10.1007/s13238-021-00837-8
51. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
52. Shulman Z, Stern-Ginossar N. The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat Immunol (2020) 21(5):501–12. doi: 10.1038/s41590-020-0650-4
53. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR M(6)A promotes cap-independent translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
54. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of M(6)A-dependent nuclear RNA processing events. Cell (2015) 162(6):1299–308. doi: 10.1016/j.cell.2015.08.011
55. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature (2015) 519(7544):482–5. doi: 10.1038/nature14281
56. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-Dependent RNA structural switches regulate RNA-protein interactions. Nature (2015) 518(7540):560–4. doi: 10.1038/nature14234
57. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
58. Perry RP, Kelley DE. Existence of methylated messenger RNA in mouse l cells. Cell (1974) 1(1):37–42. doi: 10.1016/0092-8674(74)90153-6
59. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-Nucleotide-Resolution mapping of M6a and m6Am throughout the transcriptome. Nat Methods (2015) 12(8):767–72. doi: 10.1038/nmeth.3453
60. Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA (2013) 19(12):1848–56. doi: 10.1261/rna.041178.113
61. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible M(6)A RNA methylation. Nat Rev Genet (2014) 15(5):293–306. doi: 10.1038/nrg3724
62. Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature (2010) 464(7285):108–11. doi: 10.1038/nature08738
63. Cheng H, Zheng Z, Cheng T. New paradigms on hematopoietic stem cell differentiation. Protein Cell (2020) 11(1):34–44. doi: 10.1007/s13238-019-0633-0
64. Zhang P, He Q, Chen D, Liu W, Wang L, Zhang C, et al. G Protein-coupled receptor 183 facilitates endothelial-to-Hematopoietic transition via Notch1 inhibition. Cell Res (2015) 25(10):1093–107. doi: 10.1038/cr.2015.109
65. Lizama CO, Hawkins JS, Schmitt CE, Bos FL, Zape JP, Cautivo KM, et al. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat Commun (2015) 6:7739. doi: 10.1038/ncomms8739
66. Lv J, Zhang Y, Gao S, Zhang C, Chen Y, Li W, et al. Endothelial-specific M(6)A modulates mouse hematopoietic stem and progenitor cell development via notch signaling. Cell Res (2018) 28(2):249–52. doi: 10.1038/cr.2017.143
67. Zhang C, Liu F. RNA Methylation regulates hematopoietic Stem/Progenitor cell specification. Sci China Life Sci (2018) 61(5):610–2. doi: 10.1007/s11427-017-9193-9
68. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (M(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med (2017) 23(11):1369–76. doi: 10.1038/nm.4416
69. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic Stem/Progenitor differentiation and promotes leukemogenesis via mRNA M(6)A modification. Cell Stem Cell (2018) 22(2):191–205 e9. doi: 10.1016/j.stem.2017.11.016
70. Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, et al. Stage-specific requirement for Mettl3-dependent M(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol (2019) 21(6):700–9. doi: 10.1038/s41556-019-0318-1
71. Cheng Y, Luo H, Izzo F, Pickering BF, Nguyen D, Myers R, et al. M(6)A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep (2019) 28(7):1703–16 e6. doi: 10.1016/j.celrep.2019.07.032
72. Yao QJ, Sang L, Lin M, Yin X, Dong W, Gong Y, et al. Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res (2018) 28(9):952–4. doi: 10.1038/s41422-018-0062-2
73. Raffel GD, Mercher T, Shigematsu H, Williams IR, Cullen DE, Akashi K, et al. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc Natl Acad Sci U.S.A. (2007) 104(14):6001–6. doi: 10.1073/pnas.0609041104
74. Niu C, Zhang J, Breslin P, Onciu M, Ma Z, Morris SW. C-myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood (2009) 114(10):2087–96. doi: 10.1182/blood-2009-01-197921
75. Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW, et al. Rbm15 modulates notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol (2007) 27(8):3056–64. doi: 10.1128/MCB.01339-06
76. Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of YTHDF2-mediated M(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res (2018) 28(10):1035–8. doi: 10.1038/s41422-018-0082-y
77. Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of M(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res (2018) 28(9):904–17. doi: 10.1038/s41422-018-0072-0
78. Hou X, Chen W, Zhang X, Wang G, Chen J, Zeng P, et al. Preselection TCR repertoire predicts CD4(+) and CD8(+) T-cell differentiation state. Immunology (2020) 161(4):354–63. doi: 10.1111/imm.13256
79. Wu H, Shao Q. The role of inhibitor of binding or differentiation 2 in the development and differentiation of immune cells. Immunobiology (2019) 224(1):142–6. doi: 10.1016/j.imbio.2018.09.006
80. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
81. Zhou J ZX, Hu J, Qu R, Yu Z, Xu H, Chen H, et al. mA demethylase ALKBH5 controls CD4 T cell pathogenicity and promotes autoimmunity. Sci Adv (2021) 7(25):eabg0470. doi: 10.1126/sciadv.abg0470
82. Sprent J, Surh CD. Normal T cell homeostasis: The conversion of naive cells into memory-phenotype cells. Nat Immunol (2011) 12(6):478–84. doi: 10.1038/ni.2018
83. Chong MM CA, Darwiche R, Stanley EG, Purton JF, Godfrey DI, Hilton DJ, et al. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-Dependent CD8+ T cell differentiation. Immunity (2003) 18(4):475–87. doi: 10.1016/S1074-7613(03)00078-5
84. Cacalano NA SD, Johnston JA. Tyrosine-phosphorylated SOCS-3 inhibits STAT activation but binds to P120 RasGAP and activates ras. Nat Cell Biol (2001) 3(5):460–5. doi: 10.1038/35074525
85. Matsumoto A, Seki Y, Watanabe R, Hayashi K, Johnston JA, Harada Y, et al. A role of suppressor of cytokine signaling 3 (SOCS3/CIS3/SSI3) in CD28-mediated interleukin 2 production. J Exp Med (2003) 197(4):425–36. doi: 10.1084/jem.20020939
86. Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol (2007) 7(6):454–65. doi: 10.1038/nri2093
87. Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol (2009) 30(12):592–602. doi: 10.1016/j.it.2009.09.009
88. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. M(6)A mRNA methylation sustains treg suppressive functions. Cell Res (2018) 28(2):253–6. doi: 10.1038/cr.2018.7
89. Zhu Y, Zhao Y, Zou L, Zhang D, Aki D, Liu YC. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J Exp Med (2019) 216(7):1664–81. doi: 10.1084/jem.20190337
90. Yao Y, Yang Y, Guo W, Xu L, You M, Zhang YC, et al. METTL3-dependent M(6)A modification programs T follicular helper cell differentiation. Nat Commun (2021) 12(1):1333. doi: 10.1038/s41467-021-21594-6
91. Lu TX, Zheng Z, Zhang L, Sun HL, Bissonnette M, Huang H, et al. A new model of spontaneous colitis in mice induced by deletion of an RNA M(6)A methyltransferase component METTL14 in T cells. Cell Mol Gastroenterol Hepatol (2020) 10(4):747–61. doi: 10.1016/j.jcmgh.2020.07.001
92. Lee GR. The balance of Th17 versus treg cells in autoimmunity. Int J Mol Sci (2018) 19(3):730. doi: 10.3390/ijms19030730
93. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA M(6)A methylation and YTHDF1 in dendritic cells. Nature (2019) 566(7743):270–4. doi: 10.1038/s41586-019-0916-x
94. Li MO, Rudensky AY. T Cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol (2016) 16(4):220–33. doi: 10.1038/nri.2016.26
95. Mintz MA, Cyster JG. T Follicular helper cells in germinal center b cell selection and lymphomagenesis. Immunol Rev (2020) 296(1):48–61. doi: 10.1111/imr.12860
96. Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat Immunol (2014) 15(7):646–56. doi: 10.1038/ni.2897
97. Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol (2015) 16(9):991–9. doi: 10.1038/ni.3229
98. Rolink AG, Boekel tE, Yamagami T, Ceredig R, Andersson J, Melchers F. B cell development in the mouse from early progenitors to mature b cells. Immunol Lett (1999) 68(1):89–93. doi: 10.1016/s0165-2478(99)00035-8
99. Hardy RR LY, Allman D, Asano M, Gui M, Hayakawa K. B-cell commitment, development and selection. Immunol Rev (2000) 175:23–32. doi: 10.1111/j.1600-065X.2000.imr017517.x
100. Stein M, Dutting S, Mougiakakos D, Bosl M, Fritsch K, Reimer D, et al. A defined metabolic state in pre b cells governs b-cell development and is counterbalanced by swiprosin-2/Efhd1. Cell Death Differ (2017) 24(7):1239–52. doi: 10.1038/cdd.2017.52
101. Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating b cell lymphopoiesis through interplay of IL-7 receptor and pre-b cell receptor signalling. Nat Rev Immunol (2014) 14(2):69–80. doi: 10.1038/nri3570
102. Hendershot L BD, Kearney JF. The role of immunoglobulin heavy chain binding protein in immunoglobulin transport. Immunol Today (1987) 8(4):111–4. doi: 10.1016/0167-5699(87)90861-9
103. Takatsu K. Cytokines involved in b-cell differentiation and their sites of action. Proc Soc Exp Biol Med (1997) 215(2):121–33. doi: 10.3181/00379727-215-44119
104. Loder F MB, Ray RJ, Paige CJ, Sideras P, Torres R, Lamers MC, et al. B cell development in the spleen takes place in discrete steps and is determined by the quality of b cell receptor–derived signals. J Exp Med (1999) 190(1):75–89. doi: 10.1084/jem.190.1.75
105. Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, et al. Transcriptional profiling of antigen-dependent murine b cell differentiation and memory formation. J Immunol (2007) 179(10):6808–19. doi: 10.4049/jimmunol.179.10.6808
106. Zheng Z, Zhang L, Cui XL, Yu X, Hsu PJ, Lyu R, et al. Control of early b cell development by the RNA N(6)-methyladenosine methylation. Cell Rep (2020) 31(13):107819. doi: 10.1016/j.celrep.2020.107819
107. Wang S, Chim B, Su Y, Khil P, Wong M, Wang X, et al. Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes Dev (2019) 33(15-16):1048–68. doi: 10.1101/gad.325100.119
108. Bishop GA. The many faces of CD40: Multiple roles in normal immunity and disease. Semin Immunol (2009) 21(5):255–6. doi: 10.1016/j.smim.2009.08.002
109. Elgueta R VV, Noelle RJ. Mechanisms of CD40 signaling in the immune system. Handb Cell Signaling (2010) 1:353–8. doi: 10.1016/B978-0-12-374145-5.00051-6
110. Jiang C, Trudeau SJ, Cheong TC, Guo R, Teng M, Wang LW, et al. CRISPR/Cas9 screens reveal multiple layers of b cell CD40 regulation. Cell Rep (2019) 28(5):1307–22 e8. doi: 10.1016/j.celrep.2019.06.079
111. Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse Large b-cell lymphomas of germinal-center origin. Nat Genet (2010) 42(2):181–5. doi: 10.1038/ng.518
Keywords: N6-methyladenosine, hematopoietic stem cell, T cell, B cell, RNA
Citation: Zhao C, Xu G, Zhang X, Ye Y, Cai W and Shao Q (2022) RNA m6A modification orchestrates the rhythm of immune cell development from hematopoietic stem cells to T and B cells. Front. Immunol. 13:839291. doi: 10.3389/fimmu.2022.839291
Received: 19 December 2021; Accepted: 28 June 2022;
Published: 22 July 2022.
Edited by:
Claudia Waskow, Technical University Dresden, GermanyReviewed by:
Weifeng Gu, University of California, Riverside, United StatesChristoph Schaniel, Icahn School of Medicine at Mount Sinai, United States
Christian Kosan, Friedrich Schiller University Jena, Germany
Copyright © 2022 Zhao, Xu, Zhang, Ye, Cai and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qixiang Shao, c2hhb19xeEBqc2NuLmVkdS5jbg==
 Chuanxiang Zhao
Chuanxiang Zhao Guoying Xu
Guoying Xu Xiaoxian Zhang
Xiaoxian Zhang Yunfeng Ye1
Yunfeng Ye1 Weili Cai
Weili Cai Qixiang Shao
Qixiang Shao