- 1Department of Pharmacy, Anhui Provincial Cancer Hospital, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Inflammation and Immune Mediated Diseases Laboratory of Anhui Province, The Key Laboratory of Anti-inflammatory of Immune Medicines, Ministry of Education, Anhui Institute of Innovative Drugs, School of Pharmacy, Anhui Medical University, Hefei, China
- 3Anhui Provincial Hospital, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 4The Third People’s Hospital of Hefei, The Third Clinical College of Anhui Medical University, Hefei, China
- 5Anhui Provincial Children’s Hospital, Children’s Hospital of Fudan University-Anhui Campus, Hefei, China
- 6Department of Pharmacy, The Second Affiliated Hospital of Anhui University of Chinese Medicine, Hefei, China
- 7School of Pharmacy, Anhui University of Chinese Medicine, Hefei, China
As the new year of 2020 approaches, an acute respiratory disease quietly caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as coronavirus disease 2019 (COVID-19) was reported in Wuhan, China. Subsequently, COVID-19 broke out on a global scale and formed a global public health emergency. To date, the destruction that has lasted for more than two years has not stopped and has caused the virus to continuously evolve new mutant strains. SARS-CoV-2 infection has been shown to cause multiple complications and lead to severe disability and death, which has dealt a heavy blow to global development, not only in the medical field but also in social security, economic development, global cooperation and communication. To date, studies on the epidemiology, pathogenic mechanism and pathological characteristics of SARS-CoV-2-induced COVID-19, as well as target confirmation, drug screening, and clinical intervention have achieved remarkable effects. With the continuous efforts of the WHO, governments of various countries, and scientific research and medical personnel, the public’s awareness of COVID-19 is gradually deepening, a variety of prevention methods and detection methods have been implemented, and multiple vaccines and drugs have been developed and urgently marketed. However, these do not appear to have completely stopped the pandemic and ravages of this virus. Meanwhile, research on SARS-CoV-2-induced COVID-19 has also seen some twists and controversies, such as potential drugs and the role of vaccines. In view of the fact that research on SARS-CoV-2 and COVID-19 has been extensive and in depth, this review will systematically update the current understanding of the epidemiology, transmission mechanism, pathological features, potential targets, promising drugs and ongoing clinical trials, which will provide important references and new directions for SARS-CoV-2 and COVID-19 research.
Introduction
To date, the 2019 coronavirus disease (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected 440 million people and caused approximately 5.97 million deaths, and these data are still growing rapidly (https://coronavirus.jhu.edu/map.html). This terrible disease not only causes a large number of casualties, but also seriously affects the world economy and peaceful development (1). Therefore, elucidating the possible mechanisms and potential targets of the disease and exploring effective therapeutic drugs and strategies are the most urgent efforts worldwide.
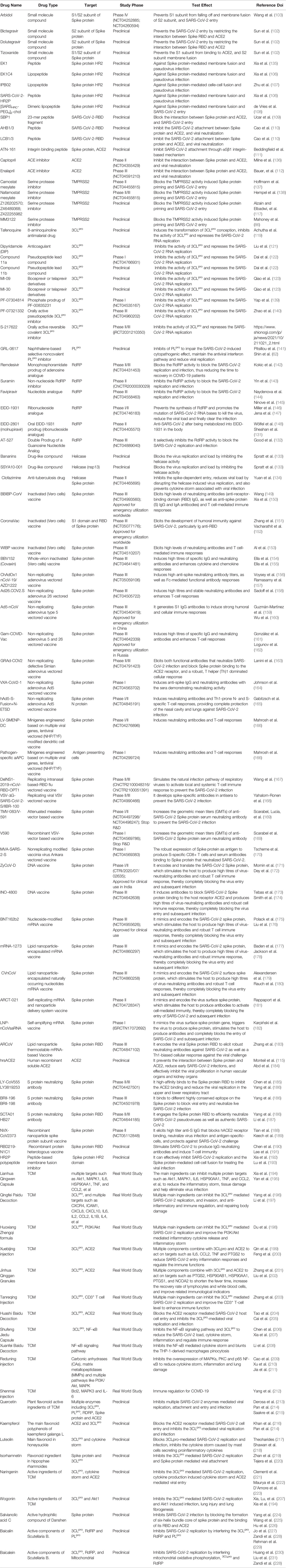
Studies have confirmed that SARS-CoV-2 is a single-stranded RNA-positive Sarbecovirus subgenus β-coronavirus (2). Homology analysis found that the genome sequence of SARS-CoV-2 is approximately 79% homologous with that of the previous SARS-CoV, and more than 50% homologous with that of MERS-CoV, which provides a certain basis and direction for its research (3). However, due to the extremely unstable genetic material of SARS-CoV-2, it is prone to mutations, producing mutant strains or promoting rapid virus evolution (Table 1), promoting the continued progress of COVID-19 and a wave of turbulence. This once again threatens the prevention and research of COVID-19 (39). Therefore, the need for targeted drugs and promising treatment strategies is urgent.
In view of this, this article will comprehensively analyze the epidemiological and pathological characteristics of SARS-CoV-2 to promote further research on COVID-19. In-depth discussion of promising therapeutic targets and possible pathogenesis during SARS-CoV-2 infection will accelerate the development of promising drugs, including small molecule drugs, vaccines and biological products, traditional Chinese medicines (TCMs) and symptomatic drugs, and the exploration of effective treatment strategies will eventually promote their clinical applications to overcome SARS-CoV-2-induced COVID-19.
Structural Information, Epidemiology and Pathology Features of SARS-CoV-2
According to statistics, there are currently two types (highly pathogenic and minimally pathogenic) of six coronaviruses (CoVs) that can cause human diseases. Among them, highly pathogenic CoVs, including SARS-CoV (Guangdong, China, 2002), Middle East respiratory syndrome coronavirus (MERS-CoV, Saudi Arabia, 2012) and the existing SARS-CoV-2 can cause severe human lung infections and multiple organ dysfunctions (40). The specific development trend of these CoVs is included in Figure 1. With the help of the latest omics, structural biology and other technologies, researchers have initially mastered the genome and structural information of SARS-CoV-2 (41). Specifically, the structure of SARS-CoV-2 is composed of the nucleocapsid (N) protein wrapped with RNA as genetic material located in the core region, accompanied by spike (S) protein, envelope (E) protein and membrane (M) protein scattered in the peripheral area, and the genome structure is mainly composed of multiple open reading frames (ORFs) (42). According to the current gene bank annotation (NC_045512.2), 2 functional ORFs (ORF1a and ORF1b) are translated into replicase complexes, and 4 functional ORFs encode S, E, M and N proteins in the 5’-3’ direction, while the remaining ORFs are distributed in the abovementioned functional genes, encoding multiple accessory proteins, including 3a/3b, 6, 7a/7b, 8a/8b and 9b (43). Further research found that the ORF1a- and ORF1b-translated viral replicase/transcriptase protein complex is cleaved to form up to 16 kinds of nonstructural proteins (nsps) by the virus/host proteolytic enzymes, including 3C-like main protease (3CLpro or Mpro) and papain-like protease (PLpro) (44). During this process, PLpro cuts the N-terminus of the polyprotein to form nsp1, nsp2 and nsp3, which are required for SARS-CoV-2 replication, while 3CLpro cleaves and separates the polyprotein pp1ab to generate nsp4-16 to form multiple active proteins, including RdRp and helicases, which are essential requirements for the life cycle of SARS-CoV-2 in host cells (45). The useful information shown in Figure 2 will help scientists better discover potential targets that interfere with the replication, spread, and pathogenicity of SARS-CoV-2 and develop promising vaccines, small molecule drugs and TCMs that can be used in the clinic.
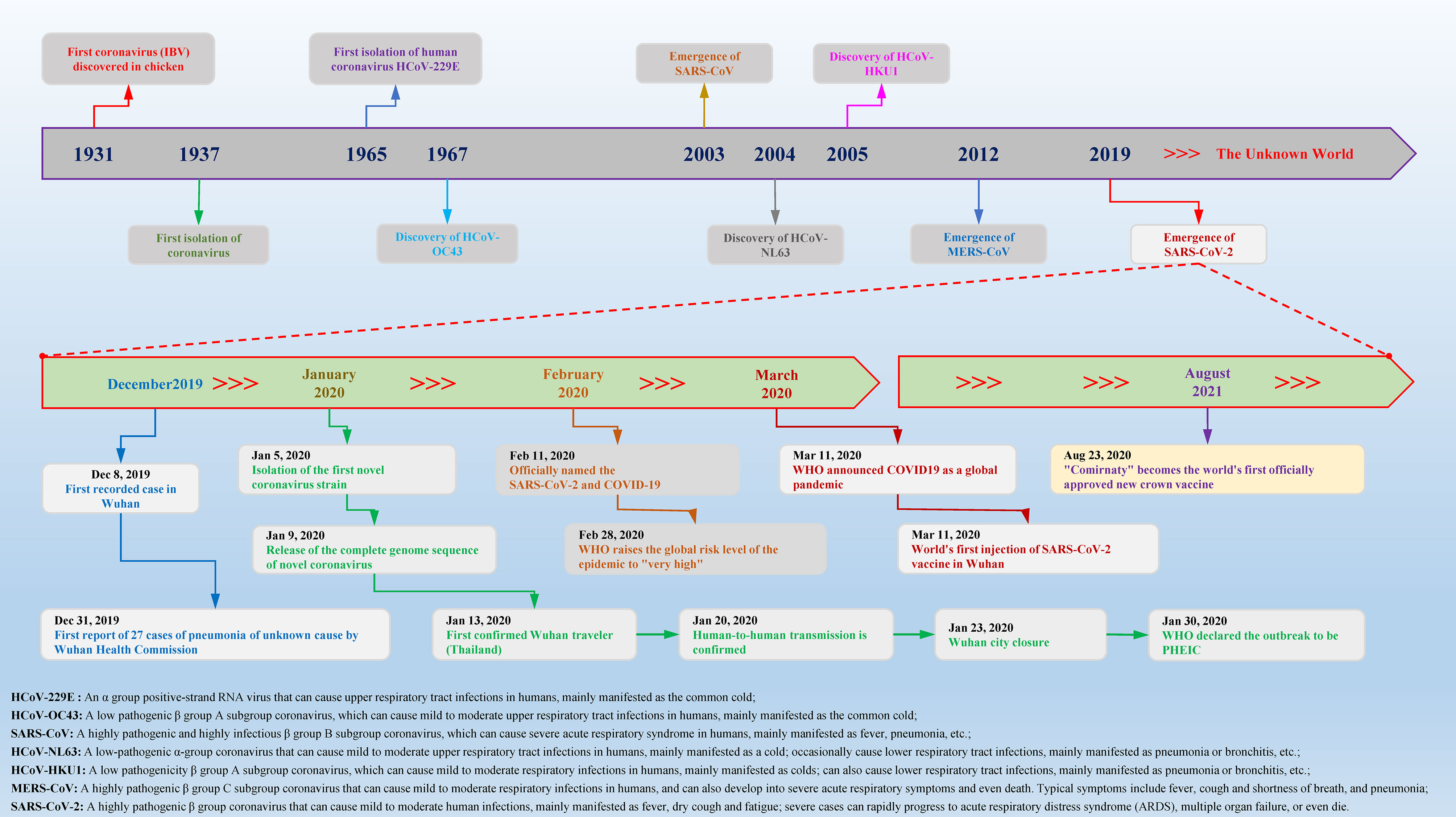
Figure 1 Timeline of key events for coronavirus discovery and research. Coronavirus was first isolated from chickens in 1937. With the passage of time and changes in the environment, in the past 84 years, a variety of different species and subgroups of coronaviruses have been discovered, identified, named, and researched. In December 2019, Wuhan, China, reported a novel coronavirus case for the first time. In a short period of time, the COVID-19 epidemic caused by SARS-CoV-2 spread to the world and caused major disasters and epidemics. In the past two years, there have been more than 440 million confirmed cases worldwide, causing approximately 5.97 million deaths, which has caused great social upheavals and dangers.
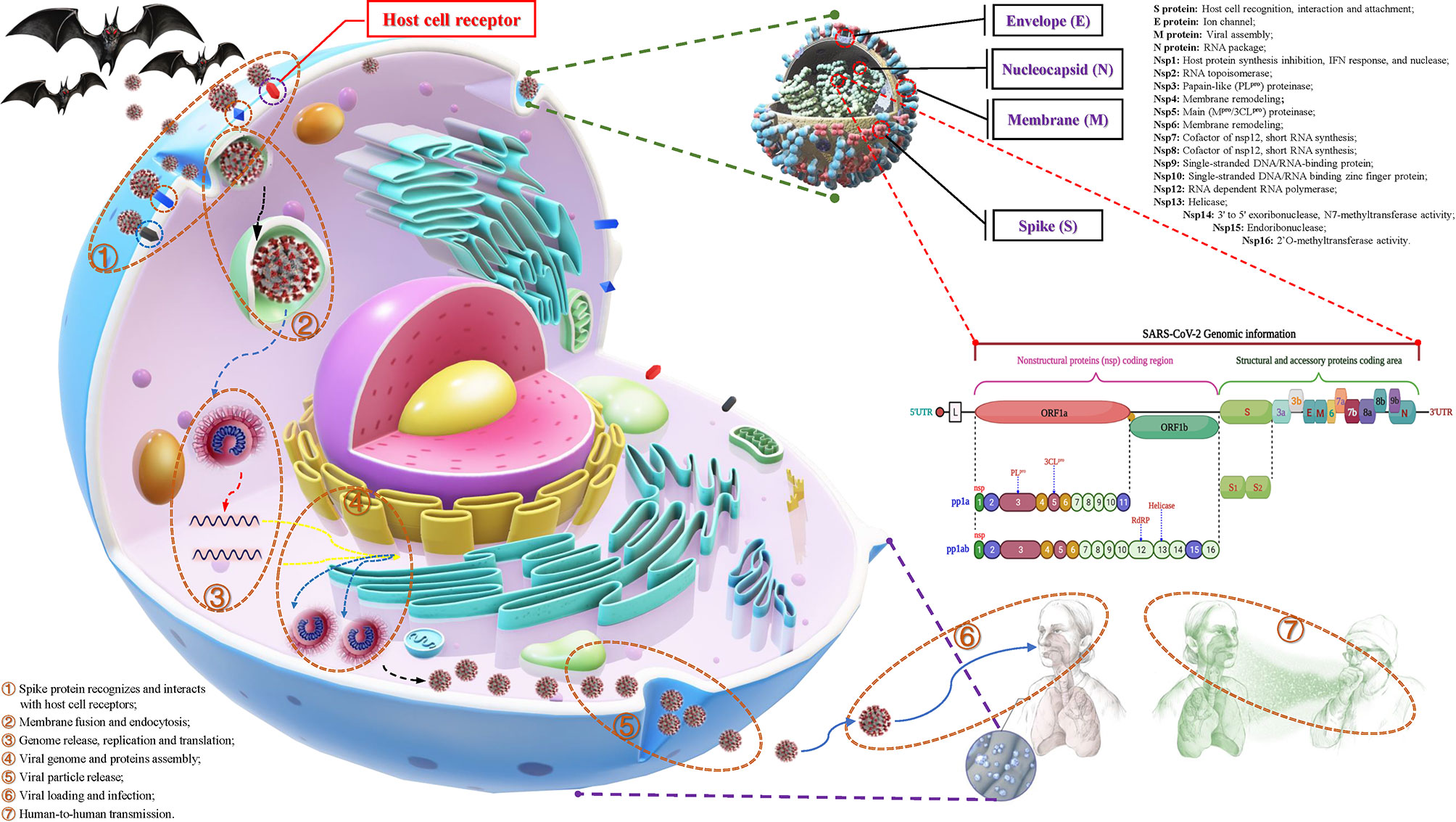
Figure 2 The structural features, potential functions and transmission process of SARS-CoV-2. Structurally, the outer side of SARS-CoV-2 is surrounded by a capsid, which is mainly composed of spike (S), membrane (M), and envelope (E) proteins, while the nucleocapsid (N) protein is accompanied by the genome. The genomic structure of SARS-CoV-2 is based on a single-stranded positive-stranded RNA, which contains a 5’-methylated cap and a 3’-polyadenylic acid tail, arranged in the following order: 5’-end; nonstructural protein (nsp) coding region [open reading frame (ORF1a/b)]; structure and accessory protein coding regions such as S, E, M, N and 3a, 3b, 6, 7a, 7b, 8a, 8b, 9b. Among them, the open reading frame (ORF) 1a/b is responsible for encoding a variety of nonstructural proteins, mainly RNA-dependent RNA polymerase (RdRP), papain-like protease (PLpro) and 3C-like protease (3CLpro). The putative functions of these proteins are mentioned in the figure. During the infection process, SARS-CoV-2 recognizes and interacts with host cell surface receptors and enters the host cell through membrane fusion and endocytosis. After entering the host cell, SARS-CoV-2 releases its genome and translates a large number of nsps, including RdRP, PLpro and 3CLpro. Under the action of these enzymes, it synthesizes the new RNA genome and assembles to form virus particles, which are then released into the extracellular space through exocytosis. Uncontrolled replication promotes SARS-CoV-2 infection, leading to immune disorders and inflammatory cytokine storms and ultimately leading to damage to multiple organs, especially the lungs.
Meanwhile, the initial epidemiological research results indicate that SARS-CoV-2 spreads from person to person mainly through the respiratory tract, droplets or aerosols (46). However, based on multiple studies, it can be seen that SARS-CoV-2 can be spread not only through the abovementioned channels but also through other means, which is mainly manifested as follows: 1. There have been cases showing that SARS-CoV-2 can spread by the placenta, but vertical transmission rarely occurs. 2. According to existing research, the virus can spread among minks and can infect humans. Meanwhile, cats and ferrets have been confirmed to be able to transmit to each other, but there are no reported cases of transmission to humans. 3. There have been studies speculating that this virus can also be spread by direct contact and pollutants, but this may be just an unusual route of transmission. 4. Although live virus has been isolated from saliva and feces, viral RNA has also been detected in semen and blood transfusions (47). There are currently no reports of sexual or blood transmission and only one report of possible fecal-respiratory transmission (48), which will provide us with important guidance for all-round protection.
Researchers conducted a systematic analysis of SARS-CoV-2-infected patients and found that almost all patients had frosted glass shadows on both sides of their lungs (49). The initial symptoms of the patient mainly included fever, cough and sputum, hemoptysis, headache and myalgia or fatigue, diarrhea, dyspnea, etc. As the disease progresses, symptoms such as inflammation, fibrosis and edema appear in the lungs, which gradually develop into acute respiratory distress syndrome (ARDS) and cause lung failure (50). Meanwhile, SARS-CoV-2 infection also causes damage to multiple organ functions, including digestive system injury, such as liver degeneration and spot necrosis, and the epithelium of the esophagus, stomach and intestine mucosa show varying degrees of degeneration, necrosis and exfoliation; brain and nervous system damage, such as cerebral congestion and edema, some neuronal degeneration and ischemic changes; cardiovascular system damage, such as increased blood pressure and arrhythmia, increases the probability of myocardial infarction, causes myocardial ischemia, necrosis, thrombosis and cardiac insufficiency; genitourinary system damage, including glomerular congestion, segmental hyperplasia or necrosis, protein exudation in the glomerular capsule, and acute kidney injury (Figure 3); and some patients still die after treatment (51). Based on this, being familiar with the pathological changes caused by SARS-CoV-2 will lay the foundation for clinical diagnosis and targeted therapy.
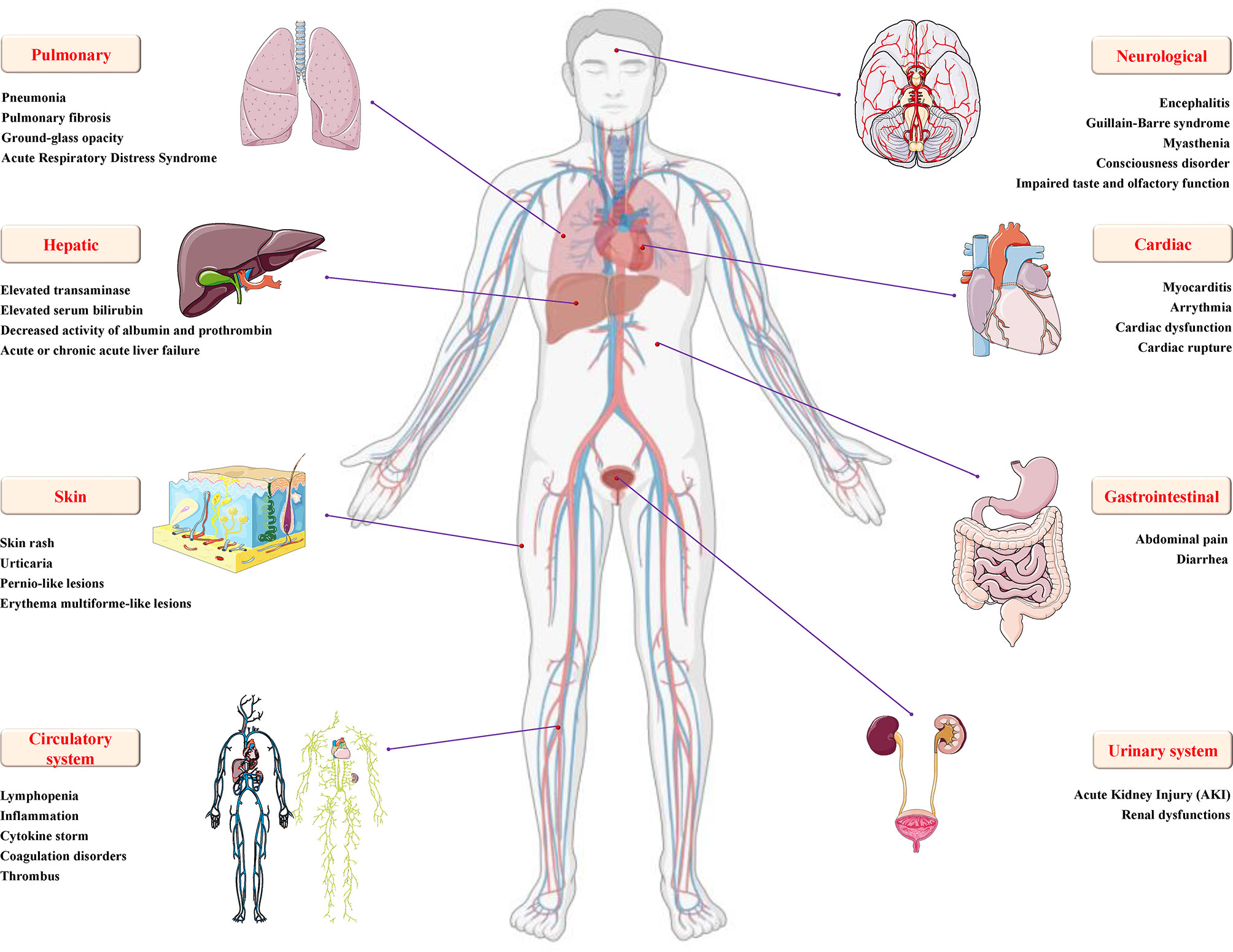
Figure 3 Details of multiple organ injury caused by SARS-CoV-2. In addition to varying degrees of pulmonary inflammation, embolism, and acute respiratory distress syndrome, COVID-19 caused by SARS-CoV-2 infection can also cause various organ dysfunctions and damages, including but not limited to encephalitis, Gillan-Barre syndrome, muscle weakness and other nervous system dysfunction; increased blood pressure, arrhythmia, myocardial ischemia, cardiac insufficiency, rupture and other cardio/cerebrovascular system damage; urogenital system damage, such as glomerular congestion and acute kidney injury; digestive system damage, such as diarrhea, increased transaminase/serum bilirubin, decreased albumin/prothrombin activity, acute or chronic acute liver failure, and skin and circulatory diseases, such as skin rash, urticaria, pernio-like lesions, inflammation, cytokine storm, coagulopathy and thrombosis.
At present, new cases of COVID-19 are caused by multiple SARS-CoV-2 variants in many countries (52). Currently, a number of major variants are rapidly growing and causing concern, including alpha (B.1.1.7), beta (B.1.351), gamma (B.1.1.28.1), delta (B.1.617.2) and omicron (B.1.1.529), and the characteristics of these variants are shown in Table 1. Meanwhile, different mutant strains have different characteristics. For example, the gamma variants increase toxicity and increase the risk of hospitalization and death, while Delta strains are highly infectious and spread quickly, especially the shortened incubation period or passage interval, which increases the risk of global epidemics (10, 53). Authoritative research shows that SARS-CoV-2 has evolved more than 800 different subtypes or branches, and its variants may have exceeded 1,000 (54). In general, the direction of the mutation and evolution of the new coronavirus is mainly to break through immunity, avoid vaccines, increase exponential replication, and be highly infectious (37). Although the mutant strains are terrible, their diversity, transmission, epidemic, and pathogenic characteristics will provide important clues for the in-depth study of virus mutation mechanisms, exploration of novel potential targets, and development of effective vaccines, drugs, and therapeutic strategies.
Potential Therapeutic Targets of SARS-CoV-2
Combining the research experience of SARS- and MERS-CoV to explore the potential therapeutic targets of SARS-CoV-2, the following aspects should be considered: enzymes and functional proteins that affect RNA synthesis and viral replication; structural proteins that affect virus entry and the self-assembly process; virulence factors that affect the host immune regulation; and host cell surface proteins and receptors (Figure 4). Correspondingly, therapeutic strategies are also divided into targeting SARS-CoV-2 and targeting host cells and the body’s immune system (55). Authoritative research shows that SARS‐CoV‐2 can encode a variety of proteins, including nsps, structural proteins, and several virulence factors (56). Moreover, multiple specific host cell surface receptors, coreceptors, and auxiliary proteases, including angiotensin converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), cluster of differentiation 147 (CD147) tyrosine-protein kinase receptor UFO (AXL) and nonmuscle myosin heavy chain IIA (MYH9) (38, 57, 58), have been identified. Obviously, these targets will be the most promising targets for fighting the COVID-19 outbreak caused by SARS-CoV-2.
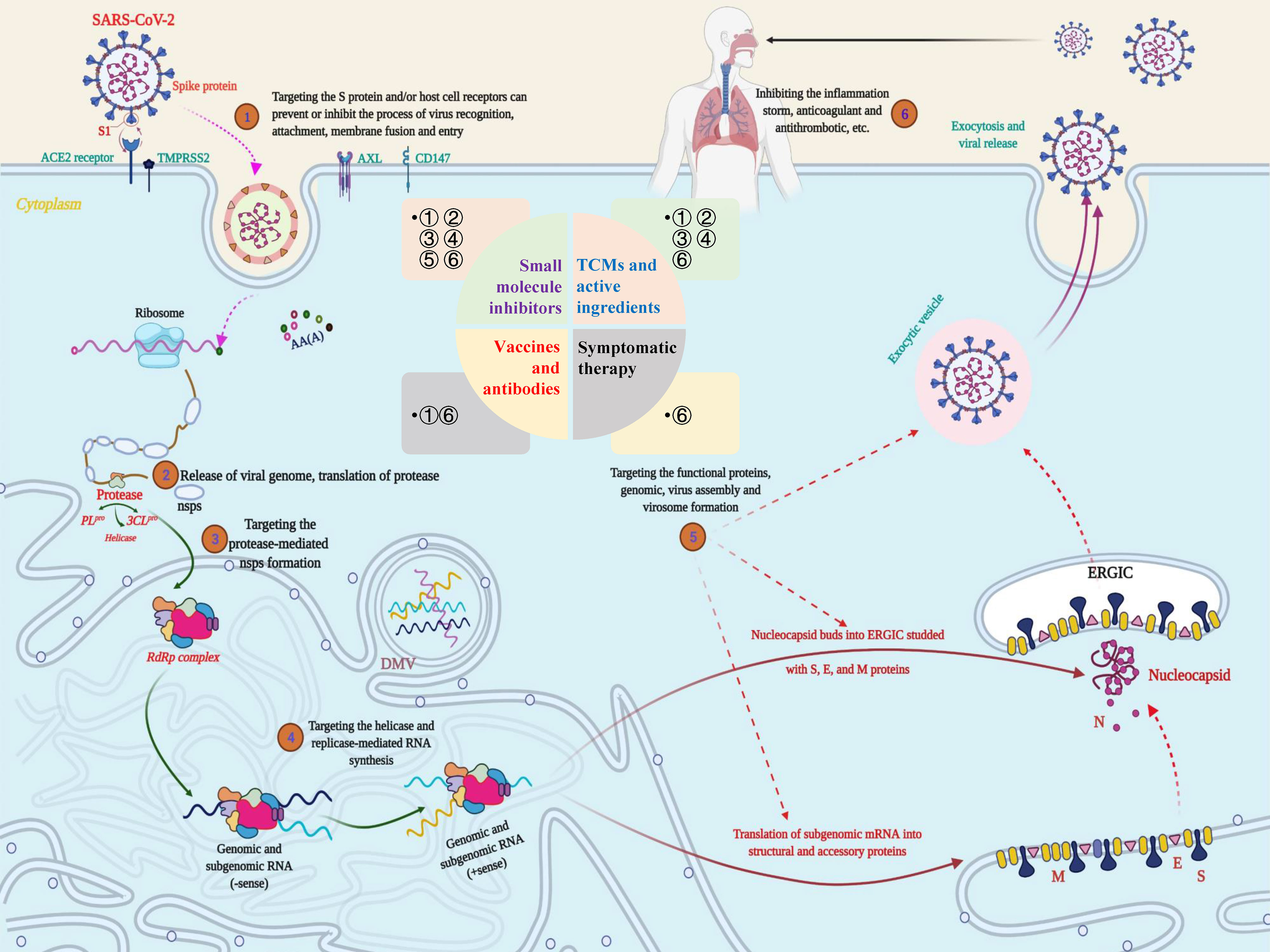
Figure 4 Potential targets and targeted therapeutic strategies for combating SARS-CoV-2-induced COVID-19. Scheme of the potential targets, intervention strategies and types of therapeutic drugs in the cycle of SARS-CoV-2 infection, replication, and transmission. During the infection stage, SARS-CoV-2 recognizes and interacts with host cell surface receptors through the spike (S) protein or transmembrane glycoprotein CD147 and enters the host cell through membrane fusion and endocytosis. After the virus enters the host cell, SARS-CoV-2 releases its nucleocapsid and genome into the cytoplasm and translates a large number of nonstructural proteins (nsps) including coding RNA-dependent RNA polymerase (RdRP), papain-like protease (PLpro) and 3C-like protease (3CLpro). Under the action of these enzymes, a full-length negative antisense genome template is synthesized to produce the new RNA genome and assembled to form virus particles, which are then released into the extracellular space through exocytosis. Uncontrolled replication promotes SARS-CoV-2 infection, leading to immune disorders and inflammatory cytokine storms and ultimately leading to damage to multiple organs, especially the lungs. The whole process exposed multiple potential targets, providing important guidance for research on anti-SARS-CoV-2 targets, drugs and treatment strategies.
RNA Synthesis and Replication Protease Targets
Nsps have proven to be widely involved in SARS-CoV-2 recognition, entry, inheritance, replication, and infection. Together with their key biological functions and relatively clear structure and active site, the main nsps, including PLpro, 3CLpro, RNA-dependent RNA polymerase (RdRP) and helicase, have become the first batch of targets to be considered for the development of small molecular inhibitors (Figure 4) (59).
3CLpro, the aforementioned nsp5, was found to cut 11 sites on the polyprotein body encoded by ORF1ab and then release mature nsp4-nsp16, which is crucial to the life cycle of SARS-CoV-2 (60). Structural analysis showed that the protease monomer mainly contains a domain I (residues 8-101) and a long loop connected domain II (residues 102-184) and a domain III (residues 185-200), and the active site is located in the gap between domains I and II (61). Mature research on the function, structure and active site of 3CLpro makes it a powerful target for anti-SARS-CoV-2 drugs such as small molecules and peptide inhibitors.
Unlike 3CLpro, PLpro mainly cuts from the N-terminus of the polyprotein to release nsp1, 2 and 3, which will affect the accuracy of SARS-CoV-2 replication. Research on MERS-/SARS-CoV suggests that it has a powerful role in antihost innate immunity (62). Moreover, homology analysis found that SARS-CoV-2 and SARS-CoV PLpro share approximately 83% of the sequence at the protein level. Combined with its indispensable role in virus replication and infection, PLpro should be a valuable target for SARS-CoV-2 inhibitor research. Meanwhile, the use of X-ray crystallography and other techniques to analyze the structure of PLpro will further facilitate the study of PLpro inhibitors against SARS-CoV-2 (63, 64).
In the RNA replication of CoVs, RdRP promotes their evolution by affecting the fidelity of replication and mutation rates to help them adapt to the environment or host cells (65). Homology analysis found that SARS-CoV and SARS-CoV-2 share approximately 82% of the homologous sequence at the genome level, while RdRP shares a sequence of more than an astonishing 96% at the protein level (66). These findings remind us that RdRP will become one of the most promising targets for the study and treatment of SARS-CoV-2. High-resolution structural analysis revealed that the functional domain of SARS-CoV-2 RdRP is located at the C-terminus of the protein, where there is a conserved Ser-Asp-Asp motif. At the RNA level, nsp8 can control the de novo synthesis of up to 6 nucleotides, which will provide primers for nsp12/RdRP RNA synthesis. Meanwhile, the nsp7-nsp8 complex can increase the activity of RdRP, which in turn affects its binding to RNA (67). All these studies provide valuable references and directions for research on anti-SARS-CoV-2 targeting RdRP.
SARS-CoV-2 helicase (nsp13) is a multifunctional nucleoside triphosphate (NTP)-dependent protein. Structural analysis revealed that helicase contains a metal binding domain (MBD) composed of 26 cysteine residues at the N-terminus and a helicase domain (Hel) consisting of a conserved motif at the C-terminus (68). Functional studies found that, helicase can unwind double-stranded (ds) DNA and RNA in an NTP-dependent manner along the 5’-3’ direction during (69). It was found that the sequence of the helicase of SARS-CoV-2 is conserved and indispensable and is an essential component of virus replication. Based on these studies, helicase is expected to become a viable target against SARS-CoV-2 infection.
Structural Protein Targets
Based on the current research results, the spike protein is one of the most critical structural proteins of SARS-CoV-2, which forms a special flower crown structure on the outer surface of the virus in the form of a trimer. Meanwhile, studies have found that the spike protein can directly affect the recognition, receptor binding, interaction, and virus entry between SARS-CoV-2 and host cells to determine the tissue or host preference in the initial stage of infection (70). In the spike (S)-mediated infection process, certain proteases in the host cell, such as TMPRSS2, can cleave the spike protein into two subtypes, the S1 subunit and the S2 subunit. The responsibility of the S1 subunit is to recognize and bind to host cell surface receptors, while the main task of the S2 subunit is to mediate the virus-cell and cell membrane fusion process (71). From the perspective of the mechanism, the structural integrity, cleavage and activation of the S protein perform crucial roles during host cell invasion and virulence. Therefore, it will have far-reaching significance to develop drugs and vaccines that affect the viral spike protein or specific receptors on the host cell surface to prevent SARS-CoV-2 from entering and infecting. Except for the outermost spike protein, the N protein is a highly immunogenic phosphoprotein and also a core and highly conserved component of SARS-CoV-2 (72). In the process of virion assembly, N protein combines with viral genomic RNA to produce a spiral nucleocapsid and is related to viral genome replication and regulation of cell signaling pathways. During this process, the N-terminal domain (NTD) and C-terminal domain (CTD) are necessary structures for effective binding to viral RNA (73). Meanwhile, studies have pointed out that the E protein mainly affects the structural integrity and virulence of SARS-CoV-2 (74). In addition, these proteins also exhibit the interferon (IFN) antagonistic properties. In particular, the M protein can prevent the formation of the MAVS-traf3-tbk1 complex and antagonize the production of IFN-I by interacting with MAVS (74, 75). Based on the above research, S (S1 and S2 subunit), N (NTD and CTD domain), E and M proteins are all have great potential to become targets for the development of anti-SARS-CoV-2 drugs and vaccines (Figure 4).
Virulence Factor Targets
Virulence factors (VFs) are molecules with virulence properties such as invasiveness and toxins produced by the metabolism of viruses and bacteria, which mainly inhibit or evade the host’s immune response when infecting the host and obtain nutrients from the host for self-proliferation (76). At present, little is known about the virulence factors of SARS-CoV-2, and there are three virulence factors, namely nsp1, nsp3c and ORF7a, which are considered to be most likely involved in interfering with the innate immunity of the host to assist in immune escape of the virus (77–79). Specifically, nsp1 induces the degradation of mRNA and inhibits the production of IFN-I by interacting with host cell 40S ribosomal subunits, while nsp3c combines with host cell ADP-ribose to resist innate immunity (77, 80). In addition, ORF7a of SARS-CoV-2 directly binds to bone marrow stromal antigen 2 (BST-2), which reduces its activity by blocking the glycosylation of BST-2 and ultimately inhibits the release of the assembled virus (79). In view of the high feasibility of virulence factors as potential targets for SARS-CoV-2 research, the development of drugs that affect the production and effects of virulence factors will be another important clue to explore the fight against SARS-CoV-2-induced COVID-19.
Hose Specific Receptor or Enzyme Targets
Authoritative studies have confirmed that host cell ACE2 is the specific receptor to which the SARS-CoV S protein receptor binding domain (RBD) binds. The latest research has found that the host receptors of SARS-CoV-2 and SARS-CoV have a high degree of consistency, which indicates that there is also an important interaction between the spike RBD of SARS-CoV-2 and ACE2 (81). During the infection stage, the RBD of the S protein S1 subunit recognizes and binds to the cell surface ACE2 receptor, which promotes the weakening or disappearance of the interaction between S1 and the S2 subunit, thereby exposing the S2 subunit (82). Subsequently, the S2 subunit changes conformation by inserting the fusion peptide (FP) into the host cell membrane, resulting in the formation of a six-helix bundle (6HB) between HR1 and HR2, which ultimately promotes fusion of the viral membrane with the host cell membrane (83). According to the receptor binding motif (RBM) analysis, a large number of amino acid residues necessary for binding to ACE2 are completely retained in the S protein of SARS-CoV-2, which is consistent with the previous discovery that the virus uses ACE2 to enter the host cell (84). Based on a number of authoritative studies, ACE2 will be the most valuable host cell target in preventing the entry and infection of SARS-CoV-2.
In addition, TMPRSS2 can cut off the spike to trigger SARS- and MERS-CoV infection. In a study of SARS-CoV-2, it was found that the virus uses TMPRSS2 instead of cathepsin B and L (CatB/L) to activate the S protein, and the spreading process may also be closely related to the activity of TMPRSS2 (85). Another study found that TMPRSS2 inhibitors can significantly inhibit the SARS-CoV-2 spike protein from entering a cell line expressing TMPRSS2, while promoting the expression of TMPRSS2 can cancel this inhibitory effect, which indicates that the initiation of the SARS-CoV-2 spike protein is dependent on TMPRSS2 (86). Furthermore, an in vitro study showed that camostat mesylate, a serine protease inhibitor, can potently stop the virus from entering Caco-2 (TMPRSS2+) cells rather than 293T (TMPRSS2-) cells by inhibiting the activity of TMPRSS2 (87). The above results suggest that inhibiting TMPRSS2 to treat patients with SARS-CoV-2 infection will be a promising and valuable therapeutic strategy.
CD147 is a highly glycosylated single-pass transmembrane glycoprotein that has been found to play an indelible role in tumor development, plasmodium invasion, virus infection and other processes (88). During SARS-CoV invasion of host cells, CD147 molecules can interact with cyclophilin A (CyPA) to mediate a similar mechanism of action in HIV-1 invasion, while the CD147 antagonist peptide (AP)-9 can strongly bind to HEK293 cells and exert its anti-SARS-CoV effect (89). In view of the high similarity between SARS-CoV and SARS-CoV-2, some studies have attempted to explore the possible role of CD147 in host cell invasion by SARS-CoV-2 (90). The results show that blocking host cell CD147 can significantly inhibit SARS-CoV-2 infection, suggesting that CD147 is likely to be another potential surface receptor independent of ACE2 (91). A study used the humanized anti-CD147 monoclonal antibody called meplazeumab (60 μg/ml), which can prevent virus invasion and the subsequent inflammation caused by SARS-CoV-2 and its variants, including variants α, β, γ and δ, with inhibition rates of 68.7, 75.7, 52.1, 52.1 and 62.3%, respectively (92). Furthermore, CD147 genetically modified mice are more sensitive to SARS-CoV-2 and variants such as α and β, causing the same pathological changes as COVID-19 (93). In addition, surface plasmon resonance analysis confirmed that there is an interaction between CD147 and the S protein (90). This evidence indicates that SARS-CoV-2 can also enter host cells by binding to the CD147 receptor. However, the question of whether CD147 is a coreceptor, a secondary receptor or a completely independent new receptor still needs more research to be verified. However, CD147 is a novel potential therapeutic target with further exploration value in research on fighting SARS-CoV-2 infection. While researchers have multiplied their hopes for discovering this new infection mechanism, several studies have suggested that there is no direct interaction between RBD and CD147, raising doubts about its role as a coreceptor and potential as a therapeutic target (94, 95). Science has always been developed through constant questioning. The conflicting results do not discourage us but instead provide us with new research clues. In any case, more research needs to be done to strengthen the reliability of this finding.
SARS-CoV-2 infection mainly relies on the interaction of the viral surface S protein and the well-known host cell surface receptor ACE2. However, the low expression of ACE2 in the respiratory system makes it difficult to fully explain why SARS-CoV-2 mainly infects the human respiratory system. Along with the continuous deepening of exploration, researchers proved that the AXL protein on lung cells can bind to the spike protein and show a relatively obvious colocalization phenomenon on the cell membrane through large-scale screening and a series of biochemical cellular experiments (96). Interestingly, AXL does not bind to the RBD of the S protein but instead binds to the NTD region at the N-terminus. Meanwhile, a study also found that AXL has significant retention in almost all types of airway cells, including type I/II lung epithelial cells, fibroblasts, basal cells, endothelial cells, smooth muscle cells and myeloid cells. In addition, overexpression of AXL can effectively promote the invasion of SARS-CoV-2, while knocking out AXL in human lung epithelial cells significantly reduces SARS-CoV-2 infection (97). At the same time, clinical data from patients with SARS-CoV-2 also show that the expression level of AXL is highly correlated with severe infections (98). The use of soluble AXL protein can effectively antagonize SARS-CoV-2 infection of lung cells, suggesting that AXL is another potential target during SARS-CoV-2 infection, and targeted or AXL-based drugs may be used for future clinical interventions against SARS-CoV-2 infection.
Potential Therapeutic Strategies and Promising Anti-SARS-CoV-2 Drugs
Small Molecule Inhibitors
Drawing lessons from the research and development experience of SARS-CoV and MERS therapeutic drugs and the current authoritative research about SARS-CoV-2, we need to explore small molecule inhibitors that can prevent the novel coronavirus and its epidemic from two directions (99): 1. This type of inhibitor targets viral proteins, such as the S protein, viral enzymes (PLpro, 3CLpro, RdRP and helicase) and some important structural proteins; 2. This type of inhibitor interacts with host cell surface proteins, such as receptor (ACE2 or AXL) or coreceptor (heparin sulfate), serine protease TMPRSS2, etc., to block virus invasion and some signal regulators of the human immune system, as shown in Figure 4. At the same time, the corresponding development strategies are mainly divided into three categories: 1. Virtual screening: High-throughput screening is carried out to identify possible lead compounds from existing compound databases, such as ZINC, DrugBank, or ChemDiv, on the basis of structural biology and homology modeling analysis of protein structure. 2. Experimental high-throughput screening (HTS): Identify small molecules in the active compound library, including approved drugs, clinical trial candidates, and even internal compound databases. 3. Reposition the application of clinical and preclinical drugs (100, 101). That is the so-called “new use of old medicine”. In addition, the computer-aided design and fragment-based drug exploration are also important strategies.
Under the guidance of these strategies, a variety of small molecule inhibitors targeting different stages of the SARS-CoV-2 life cycle have been discovered. When trying to block SARS-CoV-2 entry by targeting the S protein, the researchers found that Arbidol, Bictegravir, Dolutegravir, and Tizoxanide all have such a conformation that they can bind to the key sites of the S protein with a very high binding energy (102). Arbidol mainly binds to the S1 and S2 subunits of SARS-CoV-2 to promote tight subunit binding, which not only prevents the S1 subunit from falling off, but also impedes the membrane fusion function of the S2 subunit, eventually preventing virus entry. In vitro experiments showed that Arbidol has satisfactory activity against SARS-CoV-2 with IC50 and 50% cytotoxic concentration (CC50) values of 4.11 and 31.79 μmol/L, respectively, and the selectivity index (SI) was 7.73 (103). Bictegravir and Dolutegravir combine between the RBD and NTD of two adjacent S1 monomers, which can prevent SARS-CoV-2 entry by restricting the interaction between the spike RBD and ACE2 receptor (104). In addition, Tizoxanide not only affects the stability of the S1 subunit through hydrogen bonds and van der Waals forces to prevent the RBD in the metastable conformation of the S1 subunit from binding to ACE2 but also affects the membrane fusion of the S2 subunit and host cell (105). Importantly, structural optimization of these molecules produces 9 new small molecules with better anti-SARS-CoV-2 activity, which provides important references for the discovery, development and optimization of small molecule inhibitors targeting the S protein (102). According to the research experience of SARS/MERS-CoV, the design of viral fusion interference peptides based on the properties of heptad repeat 1 (HR1) and heptad repeat 2 (HR2) of the S2 subunit is also an important strategy for the research of small molecule inhibitors of SARS-CoV-2. A pancoronavirus fusion inhibitor peptide, EK1, was designed to inhibit a variety of CoVs and inhibit SARS-CoV-2 S protein-mediated membrane fusion and pseudovirus infection in a dose-dependent manner. Subsequently, its improved version, lipopeptide EK1C4 was designed to have the same inhibitory effect at IC50 values of 1.3 and 15.8 nmol/L, and these two results were 241- and 149-fold those of the former, respectively (106). In addition, another lipopeptide, IPB02, designed based on the HR2 sequence also showed a similar effect (107). Furthermore, SARS-CoV-2-HR2P, a peptide directly based on the amino acid sequence of SARS-CoV-2 HR2, showed a potent membrane fusion inhibition with an IC50 of 0.18 mmol/L (106). Unlike SARS-CoV-2-HRP2, which is designed on a single amino acid, [SARSHRC-PEG4]2-chol, as a dimeric lipopeptide has better membrane fusion inhibition and lower cytotoxicity against SARS-CoV-2 entry (108). After that, one study designed a peptide SBP1 composed of 23-mer peptides to prevent the virus from entering the host cell by disrupting the combination of SARS-CoV-2-RBD and ACE2 (109). To inhibit the combination of viral S protein and ACE2, a study designed two types of peptide inhibitors, AHB1/2 and LCB1/3, by two de novo synthesis approaches around the ACE2 helix structure and RBD motif, which have a strong SARS-CoV-2 neutralization effect with IC50 values of 35/15.5 nmol/L and 23.54/48.1 pmol/L, respectively (110). A study identified a fibronectin-derived anticancer peptide ATN-161 from existing peptides that can prevent the binding of the S protein to ACE2, thereby reducing SARS-CoV-2 infection with an IC50 of 3.16 mmol/L (111). In light of this, the design of small molecule inhibitors for the S protein should focus on the protein structure, amino acid sequence and motif characteristics of the RBD, S1 and S2 subunits. When targeting the host cell ACE2 receptor, it has recently been suggested that ACE2 inhibitors, such as captopril and enalapril, may be effective for those who have experienced SARS-CoV-2-induced pneumonia (112). Nicotinamide analogs, such as nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN), are an important class of natural vitamin derivatives. A relevant study found that it can effectively inhibit ACE2, so it is considered a potential inhibitor for the treatment of COVID-19 (113). However, these are only theoretical speculations and almost no basic or clinical research verification. It is possible that such suggestions will gradually fade out of people’s field of vision. At present, the development of anti-SARS-CoV-2 drugs targeting ACE2 is mainly focused on peptides, antibodies and other biochemical products, including ACE2 antibody, ACE2-scFv, ACE2 nanobody, and ACE2-Fc (114, 115). Although TMPRSS2 is the gateway for SARS-CoV-2 host cells to enter, there have not been many breakthroughs in the research of small molecule inhibitors against this target. The known TMPRSS2 inhibitor camostat in a clinical trial against COVID-19 shows excellent abilities to reduce the death risk and hospital stay (87). Recently, a study demonstrated that the camostat-like drug nafamostat mesylate can prevent the SARS-CoV-2 membrane fusion caused by TMPRSS2 at a concentration of its less than one tenth, suggesting that nafamostat mesylate may be a promising inhibitor against SARS-CoV-2 infection by targeting TMPRSS2 (116). Other studies identified a variety of serine protease experimental inhibitors (DB03782, DB03213, and DB04107) and potential molecules (Z126202570, Z46489368, and Z422255982) through homology modeling and molecular docking/dynamic simulation and embraced binding free energy calculations that may effectively inhibit the TMPRSS2, which all contain a positively charged warhead similar to nafamostat and camostat (117). However, these molecules need to be determined by in-depth mechanistic research. A recent study discovered a covalent small molecule ketobenzothiazole (kbt) serine protease inhibitor, MM3122, whose structure is completely different from camostat and nafamostat and is said to be effective (86). All these results indicate that the study of small molecule inhibitors targeting TMPRSS2 for SARS-CoV-2 will be a good choice. In addition to the ACE2-mediated virus entry pathway, CD147-mediated viral entry is likely to become the second pathway of SARS-CoV-2 invasion. Although still controversial, this does not affect the research of drugs targeting CD147 in the prevention of SARS-CoV-2 infection (90). At present, the drugs targeting CD147 are mainly monoclonal antibodies, and research on small molecule compounds is rarely involved, which will be a breakthrough in future research. The latest research suggests that AXL is a candidate receptor for SARS-CoV-2, which can promote the infection of lung and bronchial epithelial cells (97). As a receptor tyrosine kinase (RTK), there is currently no report on the use of small molecule compounds targeting AXL for the treatment of SARS-CoV-2 infection, but we can refer to the research of RTK small molecule inhibitors in tumors to discover potential small molecule inhibitors of AXL for preventing the entry of SARS-CoV-2.
When considering inhibiting SARS-CoV-2 replication, the study found a variety of promising small molecule inhibitors. 3CLpro (nsp5) is one of the most ideal targets for discovering inhibitors of SARS-CoV-2. The study found that the amino Cys145 residue in the catalytic pocket of 3CLpro is an effective target for exploring small molecular covalent inhibitors of SARS-CoV-2 and other coronaviruses (118). A fluorescence resonance energy transfer study found that the 8-aminoquinoline antimalarial drug tafenoquine can induce the transformation of 3CLpro to expose the hydrophobic pocket and promote the protein aggregation, ultimately reducing the activity of 3CLpro and repressing SARS-CoV-2 RNA replication with an IC50 near 2.5 μmol/L, which is appropriately 1/4 that of hydroxychloroquine (119). Although Lopinavir-Ritonavir (Kaletra) was initially used in the treatment of SARS-CoV-2 because of its ability to block the replication of SARS-CoV and MERS by inhibiting 3CLpro, the latest research results do not support its use in the treatment of COVID-19 (120). A study screened FDA-approved drug libraries and found that the anticoagulant dipyridamole (DIP) may bind to 3CLpro to inhibit more than 50% of SARS-CoV-2 replication in Vero E6 cells at 100 nmol/L. After 2 weeks of DIP treatment, 8 critically ill patients improved significantly (121). With the continuous understanding of the structure of the 3CLpro protein, more small molecule inhibitors have been discovered, and some of them have been in clinical trials. Compounds 11a and 11b have been screened and confirmed to have a strong inhibitory effect on SARS-CoV-2 3CLpro, with IC50 values of 0.053 μmol/L and 0.040 μmol/L, respectively; EC50 values of 0.53 μmol/L and 0.72 μmol/L, respectively; and have good pharmacokinetic properties (122). At present, 11a (DC402234) has submitted a clinical application registration declaration and has obtained FDA conditional clinical trial approval (Phase I: NCT04766931). After screening, the in vivo antiviral test results of the small molecule compounds MI-09 and MI-30 showed that oral or intraperitoneal injection of these compounds can significantly reduce the lung viral load and lung pathological damage in a SARS-CoV-2-infected transgenic mouse model (123). Although various research results and different inhibitors of 3CLpro have been shown in front of people one after another, the clinical entry is extremely limited; only the four inhibitors (PF-07304814 [phase III: NCT04501978] and PF-07321332 [phase III: NCT05047601] developed by Pfizer, USA; the aforementioned 11a (DC402234 made by Frontiers, China [phase I: NCT04766931]; and the code-named S-217622 produced by Shionogi Inc., Japan [phase II/III: jRCT2031210350) are in clinical trial (124). PLpro (nsp3) has also received much attention due to its important role in the replication and invasion of SARS-CoV-2. Some noncovalent small molecule inhibitors (rac3j, rac3k and rac5c) that have been effective against SARS-CoV can target SARS-CoV-2 PLpro to prevent the self-processing of nsp3 in cells, thus reducing viral-induced CPE at high concentrations (33 μmol/L) (125). Based on the crystal structure of SARS-CoV-2 PLpro, researchers obtained useful data from the FDA-approved drug database and identified 147 potential inhibitors of SARS-CoV-2 PLpro. In Vero E6 cells, dronedarone, an ion channel modifier, has good antiviral activity against SARS-CoV-2-induced CPE with an IC50 of 4.5 μmol/L (CC50 of 12.1 μmol/L) (126). The naphthalene-based inhibitor, GRL-0617, can effectively inhibit the activity of SARS-CoV-2 PLpro with an IC50 of 2.2 μmol/L, and its mechanism is not limited to occupying the substrate pocket but expands to seal the substrate binding entrance cleft, thereby preventing the binding of the substrate (62). At present, the crystal structure of PLpro has been completely resolved (PDB code: 6W9C), and more small molecule compounds will be discovered as the crystal structure is fully analyzed. To date, no small molecule inhibitors against PLpro have entered clinical studies, which suggests that there is still a long way to go in the development of PLpro-targeted small molecule inhibitors against SARS-CoV-2. RdRP (nsp 12) has become an important target for the development of anti-SARS-CoV-2 drugs because it participates in the virus replication process as a key enzyme that catalyzes the synthesis of the SARS-CoV-2 genome. A research group from Shanghai, China, successfully analyzed the three-dimensional structure of the RdRP-nsp7-nsp8 complex at near-atomic resolution (with an overall resolution of 2.9 Å) using cryo-electron microscopy, which lays a solid foundation for the design of antiviral inhibitors based on the RdRP structure (127). As research on SARS-CoV-2 RdRP continues, multiple potential drugs have been discovered and confirmed. Remdesivir [GS-5734], a nucleotide analog originally used to fight Ebola virus, was first proposed for the treatment of COVID-19 patients because it can be used as a substrate for the RdRP. In Vero E6 cells at a SARS-CoV-2 MOI of 0.05, remdesivir shows an ideal potential to fight SARS-CoV-2 with IC50 = 0.77 μmol/L, CC50 > 100 μmol/L and SI > 129.87, which also quickly promotes the quick access of RdRP small molecule inhibitors to global phase III clinical trials and their direct use in some regions (128). However, things are always dramatic. The latest clinical trial results published by the WHO do not seem to be optimistic about this small molecule inhibitor (129). As far as COVID-19 hospitalized patients are concerned, it has little or no impact on indicators such as overall mortality and duration of hospital stay. Regardless of the outcome, the emergence of remdesivir has provided an important reference and motivation for the research of small molecule inhibitors targeting RdRP. A subsequent study screened a century-old classic drug, suramin, and a variety of derivatives, which exhibited a more than 20-fold ability to fight SARS-CoV-2 infection with remdesivir by targeting RdRP (66). Another small-molecule inhibitor called favipiravir (T-705) targets RdRP to mildly resist SARS-CoV-2 infection with an IC50 of 61.88 μmol/L, CC50 > 400 μmol/L and SI > 6.46 (130). Several clinical trials (ChiCTR2000029600/200030254, etc.) have shown that favipiravir may accelerate virus clearance and alleviate the progression of COVID-19, which lays a solid foundation for its clinical application and provides a structural basis and strong evidence for the development of broad-spectrum antiviral drugs based on the strategy of “old drugs and new use”. A study reported that the oral broad-spectrum ribonucleoside analog β-D-N4-hydroxycytidine [EIDD-1931] showed good anti-SARS-CoV-2 activity in Vero cells with an IC50 of 0.3 μmol/L (131). In addition, the oral EIDD-1931 prodrug molnupiravir (MK-4482, EIDD-2801, Merck Sharp & Dohme Corp, USA), due to its ideal anti-coronavirus effect, has ended phase III clinical trials (NCT04575584, NCT04575597 and NCT04939428) ahead of schedule and is expected to be launched in the United States soon (131). The oral purine nucleotide prodrug AT-527 developed by Roche is expected to have a good phase III clinical trial (NCT04889040) result (132). These studies provide hopes and directions for the development of small molecule inhibitors targeting RdRP during SARS-CoV-2 infection. At present, there are already several small molecule inhibitors that target helicases, such as bananins, 5-hydroxychromone derivatives, and SSYA10-001, which are expected to be used in SARS-CoV-2-related experiments (133). In addition, authoritative studies suggest that the classic old drug clofazimine has the ability to inhibit the helicase activity of SARS-CoV-2, suggesting that it may play a role in controlling the current COVID-19 pandemic and the emergence of CoVs in the future (134). Although there has been hope, the greatest challenge is the relatively low selectivity of small molecule inhibitors targeting helicase, and there is no drug targeting helicase that exceeds preclinical development. However, the development of small molecule helicase inhibitors may provide another effective treatment option for the COVID-19 pandemic.
Studies are also concerned that several small molecule inhibitors can fight SARS-CoV-2 via immunoregulatory and inflammatory functions, and the specific details are introduced in the “Significant Symptomatic Therapeutic Agents” section.
At present, the continuous rapid screening of small molecule databases based on SARS-CoV-2 potential targets has found some effective lead compounds or candidate drugs, which will promote the continuation of basic research and clinical trials of small molecule inhibitors for COVID-19 (Table 2). In addition, computer-based drug design is icing on the cake for accelerating the screening and development of small molecule inhibitors, but it is conservatively estimated that new targeted interventions will still take some time. Considering the current spread of the novel coronavirus disease and the continuing case fatality rate, rapid screening of FDA-approved and clinical trial drugs is a more practical method because “old drugs and new use” may reduce development costs and shorten development time (232). To date, a large number of small molecule inhibitors against SARS-CoV-2 infection have been screened; however, many of these studies have not been fully implemented (233). Meanwhile, the safety of some confirmed promising anti-SARS-CoV-2 small molecule inhibitors or drugs is also unknown, especially reproductive toxicity, which imposes more difficulties on the clinical translation of small molecule inhibitors. Therefore, adequate research needs to be carried out to maximize safety and avoid false positive effects. The mutation of the virus and the SARS-CoV-2 epidemic have made the discovery of vaccines and drugs more uncertain. In the long run, there is still much work to be done in the screening, validation, clinical research and clinical application of specific or broad-spectrum small molecule inhibitors for SARS-CoV-2 virus entry, replication, or prevention.
Vaccines
The main therapeutic strategies for infectious diseases include controlling the source of infection, blocking the route of transmission and protecting the susceptible. Among them, vaccines, as an effective means to protect susceptible persons and block transmission, have always been the main weapon for humans to fight infectious diseases (234). Given that the current effective treatments against the new coronavirus are not fully recognized, the development of vaccines against SARS-CoV-2 is particularly important. At present, a variety of vaccine platforms against SARS-CoV-2 are rapidly being established and developed, including inactivated vaccines and live attenuated vaccines and viral vector vaccines and nucleic acid vaccines (DNA and mRNA) (Figure 5) (235). With the joint efforts of scientists from all over the world, more than 322 candidate vaccines have been developed, which are in the preclinical, Phase I, Phase II through to Phase III efficacy studies and include Phase IV registered as interventional studies (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines) (Table 2). The rapid development of vaccine research has brought dawn to the control of the epidemic, but there are many shortcomings that need to be considered and improved.
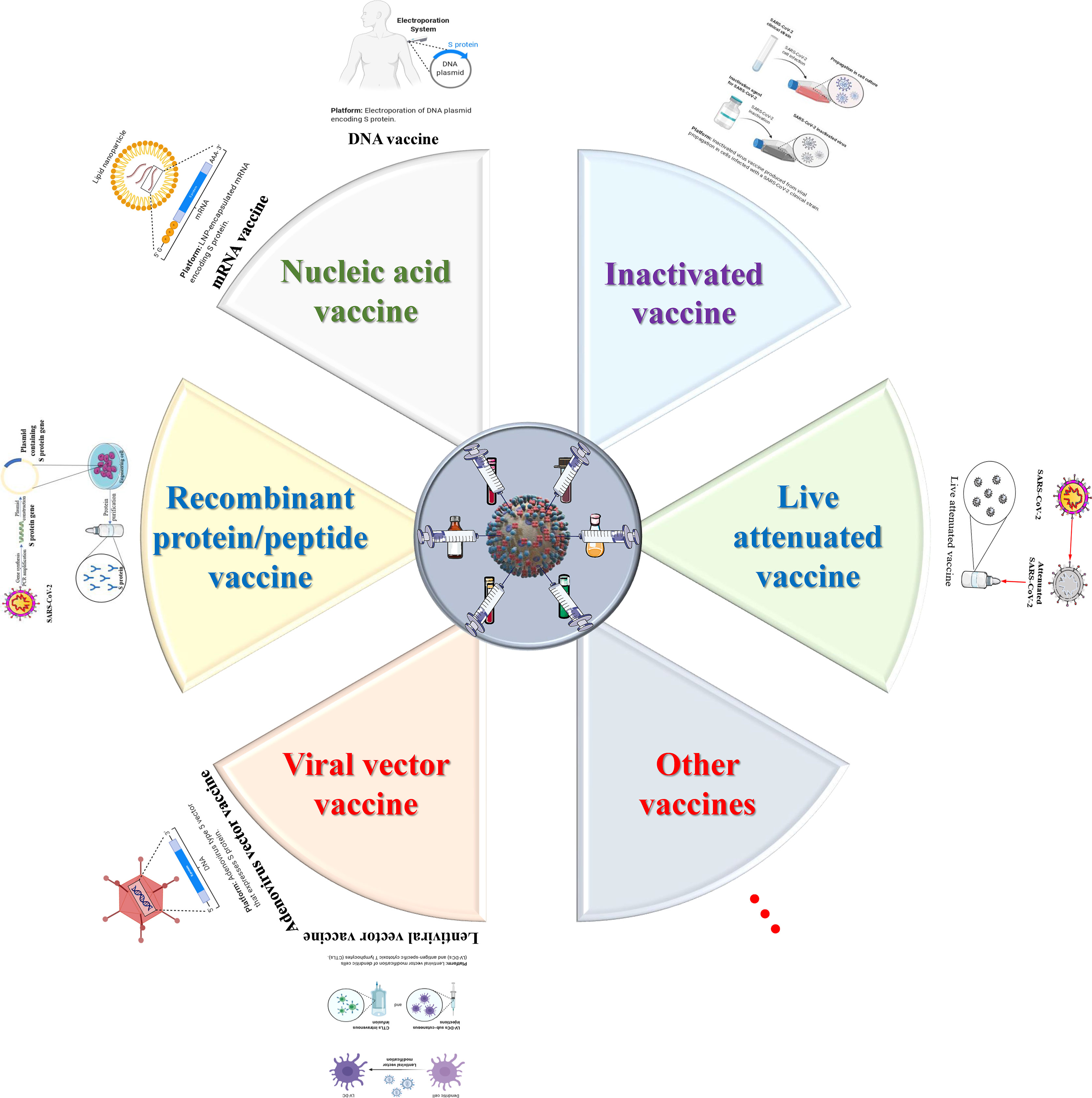
Figure 5 The design and development model of SARS-CoV-2/COVID-19 vaccines. One of the most important intervention strategies for COVID-19 is vaccine control. To date, six major types of vaccine candidates (live attenuated vaccines, recombinant protein/peptide vaccines, inactivated vaccines, viral vector vaccines, nucleic acid vaccines and other types of vaccines) are under development, clinical trials, authorized emergency use, and routine preventive use. These six types of candidate vaccines represent the direction of SARS-CoV-2 and even the entire coronavirus vaccine research.
Regarding inactivated vaccines, there are preliminary statistics of 15 such vaccines that have entered different clinical trials, including BBIBP-CorV, CoronaVac, WIBP vaccines, and Covaxin, which have entered Phase III (236). CoronaVac (Sinovac Biotech, China) can produce a wide range of neutralizing antibodies against 10 different virus strains in a variety of animals with a titer of over 90, and it has complete protection against SARS-CoV-2 infection after three immunizations (6 μg/dose) in macaques (151). Currently, CoronaVac is undergoing Phase III clinical trials in Brazil, of which 90,000 healthy participants are or will be registered. Another inactivated vaccine, covaxin, developed by an Indian Pharmaceutical company, has shown good safety and effectiveness in phase 1/2 clinical trials. At present, Covaxin is also undergoing phase III clinical trials, of which 26,000 volunteers participated (237). Other candidate inactivated vaccines are being rapidly developed in China and have been confirmed to have higher antibody titers and better safety in phase 1/2 clinical trials. Just now, a Phase III clinical trial with 15,000 participants has been launched in the United Arab Emirates (238). The development of inactivated vaccines gives us confidence in the development of vaccines against SARS-CoV-2. However, we must also recognize some of the shortcomings of inactivated vaccines and improve them. For example, the inoculation dose is large, the protection time is short, the need for multiple vaccinations, and the formation of antibody-dependent enhancement effects may aggravate viral infections.
The development of live attenuated vaccines against SARS-CoV-2 has been slow, mainly due to the limitations of the longer transformation or screening time of attenuated strains, heavy workload, and high biosafety protection standards. At present, only one attenuated influenza virus vector vaccine developed by China has entered a phase I clinical trial (ChiCTR2000037782), and three live attenuated vaccines developed by India and Turkey are undergoing preclinical evaluation (239). However, we should also realize that live attenuated vaccines can retain the complete structure of the virus and have good immunogenicity; they can simulate the natural infection process to induce humoral and cellular immunity and can produce long-lasting protection; no adjuvant is required (240). If the transformation time of attenuated strains can be optimized and biosafety is ensured, live attenuated vaccines can be an alternative direction.
At present, approximately 20 SARS-CoV-2 vaccines are being developed around the world that use the viral vector method. These vector vaccines are mainly divided into two categories: nonreplicating vector vaccines based on adenovirus and lentivirus and replicating vector vaccines based on measles, influenza, etc. The most concerning adenovirus vectors include ChAdOx1 nCoV-19/AZD1222 (Oxford University & AstraZeneca, etc., UK; D8110C00001) (241) and Ad26.COV2.S (Johnson & Johnson, USA; NCT04505722) (242), Ad5-nCoV (Academy of Military Medical Sciences & CanSino Biologics Inc, China; NCT04526990) (160) and Gam-COVID-Vac (Gamaleya, Russia; NCT04656613) (162). In addition, the 2019-nCOV candidate (Academy of Military Medical Sciences, China; ChiCTR2000031781) and defective simian adenovirus vector GRAd-COV2 (ReiThera, Italy; NCT04791423) are in phase II, with two vaccines: VXA-CoV2-1 (Vaxart, USA; NCT04563702) and hAd5-S-Fusion+N-ETSD (ImmunityBio, Inc., USA; NCT04710303) are in phase I. Lentiviral vector-based vaccines under development include LV-SMENP-DC currently in phase I/II and pathogen-specific aAPC vaccine in phase I (Shenzhen Geno-Immune Medical Institute, China; NCT04299724/NCT04276896). Phase II of the clinical trial was the intranasal influenza virus vector DelNS1-2019-nCoV-RBD-OPT1 (Beijing Wantai Biological Pharmacy, China; ChiCTR2000039715), and phase I/II of the clinical trial was the replication VSV vector rVSV-SARS-CoV-2-S/IIBR-100 vaccine (Israel Institute of Biology, Israel; NCT04608305) and three replication virus vector vaccines in Phase I: the intranasal influenza virus vector the measles vector TMV-083/V-591 (Institut Pasteur & Themis Bioscience, Austria; NCT04497298) and the VSV vector V590-001 (MSD Corp., USA; NCT04569786) and modified Ankara vector MVA-SARS-2-S (Universitätsklinikum Hamburg-Eppendorf, Germany; NCT04569383). Relying on the characteristics of few adverse reactions, good safety, and a mature production system, this type of vaccine has been developed rapidly. However, neutralizing antibodies of the vector may exist in the body, which will cause the vector to be attacked, thereby reducing the vaccine effect. Therefore, improving the effectiveness will be an important direction for the improvement of these vector vaccines.
The SARS-CoV-2 nucleic acid vaccine has quickly become the focus of vaccine research and development due to its simple development and operation, low production cost, short development and production cycle, and rapid response (243). At present, the research of such vaccines is divided into two major directions, namely, DNA vaccines and mRNA vaccines. Currently, there are 27 DNA vaccines under research in the world, 11 of which have entered the clinical trial stage, and this number will slowly increase as the technology continues to mature. ZyCoV-D is a new type of DNA vaccine candidate mainly composed of plasmid DNA loaded with the viral spike gene and signal peptide coding gene (171). The results of clinical trials (CTRI/2020/07/026352) have verified a good safety profile and induced cellular and humoral responses, which will support its further development to prevent COVID-19-related infection and death in the global population. Meanwhile, the emergency use of the ZyCoV-D vaccine in India has brought more possibilities and hopes for the development of DNA vaccines. INO-4800 is a DNA vaccine expressing S protein particles developed by Inovio Pharmaceuticals (174). Clinical trial (NCT04336410) data prove that the INO-4800 vaccine maintains one or both cells and humoral arms of the immune response for the emerging SARS-CoV-2 variant, which may be the key factor affecting the ongoing COVID-19 pandemic. Taking into account the advantages of DNA vaccines, the results of phase I clinical trials of INO-4800 (NCT04447781) and the status of entering phase II/III clinical trials (NCT04642638) once again brought great encouragement to the development of DNA vaccines. Although we have seen great hopes for DNA vaccines against the new coronavirus, we should also clearly recognize that the challenge for DNA vaccines is that they need to reach the nucleus all the way, which forces us to do more research to improve and develop a delivery system to meet the delivery efficiency of DNA vaccines (244). In addition to DNA vaccines, the development of mRNA vaccines is also in full swing. mRNA vaccines can express intracellular antigens similar to DNA vaccines, but they solve the problem of low immunogenicity of DNA vaccines and generate nonspecific immunity against the vector and delivery efficiency, so they have received more attention from researchers. Currently, two mRNA vaccines have been approved for marketing, namely BNT162b2 developed by BioNTech & Pfizer and mRNA-1273 produced by Moderna (175, 177). The results of clinical trials (NCT04368728/NCT04470427) show that the effectiveness, safety and immunogenicity of the two mRNA vaccines meet the ideal requirements. With further in-depth research on SARS-CoV-2, more mRNA vaccines have entered clinical trials, such as CVnCoV (CureVac AG; Phase II: NCT04515047), ARCT-021 (Arcturus Therapeutics, Inc.; Phase I/II: NCT04480957), LNP-nCoVsaRNA (Imperial College London; Phase I: ISRCTN17072692) and ARCoV (Academy of Military Medical Sciences; Phase I: ChiCTR2000034112), etc., and preclinical research (more than 19 candidate mRNA vaccines). Evidence from clinical trials thus far shows that mRNA vaccines are very likely to become a new platform that is fast, safe and efficient. However, to become a viable clinical alternative to traditional vaccines, mRNA vaccines must overcome two major problems related to the immunogenicity and stability of mRNA vaccines (245).
In addition to the above vaccine development strategies, recombinant protein and peptide vaccines such as human recombinant soluble ACE2 (hrsACE2), recombinant S protein nanoparticle vaccine (NVX-CoV2373), recombinant RBD protein vaccine (RBD219-N1), HR2P polypeptide and EK1C4 vaccine, etc. It can effectively induce humoral and cellular immunity to produce a wider cross-reaction, which is also an important choice for the development of SARS-CoV-2 vaccines. Each vaccine development strategy has many advantages, while at the same time, there are more or fewer shortcomings (246). The current main goal is to develop a safe and effective vaccine to curb the pandemic of SARS-CoV-2. However, we should be clearly aware that while avoiding the risks of existing vaccines, the ultimate goal of vaccine development is to develop single or mixed general vaccines for different CoVs or to establish a research and development and production platform. Only in this way can we withstand the current and future virus damage.
Traditional Chinese Medicine
Traditional Chinese medicine (TCM) has played an important role in the prevention and treatment of infectious diseases, and its theories and methods have been traced in many classic Chinese medical works (247). Meanwhile, these TCMs achieved good results in fighting against SARS-CoV infection in 2003. Moreover, in the 74187 confirmed cases of SARS-CoV-2 infection reported in China, the effective rate of receiving TCM treatment was more than 90%, and its main effect was to significantly improve and shorten the course of disease, delay disease progression, and reduce mortality (248). At the same time, traditional Chinese medicine has also been confirmed to have a low incidence of adverse reactions and often self-healing in the treatment of COVID-19 patients (249). Given that, TCM is a valuable resource for combating the epidemic of SARS-CoV-2.
Among the abundant resources of TCM, some representative drugs have shown good anti-SARS-CoV-2 activity in terms of direct anti-virus, regulation of inflammatory immunity, and organ protection, as shown in Figure 4. Analysis of cytopathic effects and plaque reduction showed that the active ingredients of Lianhua Qingwen capsule significantly inhibited the replication of SARS-CoV-2 in a dose-dependent manner through Akt signaling (194). In Vero E6 cells infected with 100 TCID50 SARS-CoV-2, the IC50 value was 411.2 μg/mL (250). In addition, Qingfei Paidu Decoction has the effect of directly inhibiting the invasion and replication of SARS-CoV-2 by acting on the host cell ACE2 and 3CLpro, respectively (251). In addition, the ingredients of Huoxiang Zhengqi capsule and Xuebijing injection are reported as potential 3CLpro inhibitors, which could inhibit SARS-CoV-2 replication by targeting PIK3CG and E2F1 through the PI3K/Akt pathway. Moreover, network pharmacology and molecular docking studies found that the active ingredients of multiple TCMs, including Jinhua Qinggan granules, Tanreqing injection and Huashi Baidu Decoction, can all act on replicating enzymes or host cell receptor proteins to inhibit the replication and invasion of SARS-CoV-2 (252). In addition to directly blocking the replication and invasion of SARS-CoV-2, several active ingredients in Qingfei Baidu Decoction, Xuanfei Baidu Decoction, Huashi Baidu Decoction, Jinhua Qinggan Granules, Huoxiang Zhengqi Capsules, Lianhua Qingwen Capsules, Shufeng Jiedu Capsules, Xuebi Jing injection, Reduning injection, Tanreqing injection and Shenmai injection have been proven to not only reduce inflammation and inflammatory storms but also regulate cytokines and immune dysfunction by regulating multiple signal pathway abnormalities in patients, thus alleviating SARS-CoV-2-induced COVID-19 (253). In the process of studying the damage of SARS-CoV-2 to organ function, clinical analysis found that Qingfei Paidu Decoction, Jinhua Qinggan Granules, Lianhua Qingwen Capsules, and Shufeng Jiedu Capsules may play a protective role in organ damage through the effects of expectorant, anti-inflammatory, antioxidant, and antifibrosis (248, 252).
The mechanisms of TCMs for anti-SARS-CoV-2 and organs protection are quite complicated. On this basis, it will be a great deal for the TCM treatment of SARS-CoV-2 if the specific active ingredients can be clarified. In this context, based on the TCM system pharmacology database and analysis platform (TCMSP) and literature, researchers have discovered that quercetin, kaempferol, luteolin, isorhamnetin, baicalein, naringenin, and wogonin (the latter three are in the same ranking) are the most promising important ingredients for anti-SARS-CoV-2 by comprehensive analysis using network pharmacology, bioinformation analysis, molecular docking, animal experiments, and clinical trials (252). In addition, as many as 401 compounds were found to have antiviral activity, and many ingredients have shown good therapeutic effects in experiments. A recent study found that salvianolic acid C, an active hydrophilic compound of Danshen, can effectively inhibit SARS-CoV-2 infection and block the formation of the S protein 6-HB core, with an IC50 value of 3.41 μmol/L (224). In a cell-based system, baicalin and baicalein, as the key active components of Scutellaria B., show strong antiviral ability by significantly inhibiting 3CLpro activity, with IC50 values of 10.27 and 1.69 μmol/L, respectively (254). The above findings suggest that TCM resources are very abundant, and many ingredients or compounds can be considered as lead compounds for the development of anti-SARS-CoV-2 drugs (Table 2). Perhaps the active ingredients of TCM can form a more promising small molecule inhibitor library in the future. Therefore, we should pay attention to and devote certain resources to screening, discovering and developing promising TCM compounds and extracts for the treatment of SARS-CoV-2.
We know that TCM prescriptions are produced in long-term exploration and practice, and their compatibility, toxicity, safety and other issues can be guaranteed. However, the abovementioned problems exist when the active ingredients and monomers of TCM are used (249, 255). We hope that TCM can be more widely used in the treatment of COVID-19, but at the same time, safety issues such as compatibility, toxicity, and adverse reactions of active ingredients and monomers of TCM should also be more studied and explored (256).
Significant Symptomatic Therapeutic Strategy
During COVID-19, aggressive inflammation and dysfunctional immune responses are the most basic, common and important pathological features that trigger cytokine storms and mediate multiple organ system damage (257). If not well controlled, the situation will worsen and even lead to death. In severe cases, most patients experience severe lung inflammation and thrombosis (258). Therefore, anti-inflammatory and anticoagulant drugs have been proposed and implemented, including the application of low molecular weight heparin to hospitalized patients as one of the standard symptomatic therapeutic strategies (Figure 4). In the serum of most COVID-19 patients, the levels of proinflammatory cytokines, including IL-1β, IL-2, IL-6, IL-8, IL-17, G/GM-CSF, MCP1, CCL3 and TNF, are significantly elevated, which is considered a cytokine storm (259). Among these cytokines, IL-6 has become a stable indicator of poor prognosis and has been used in the neutralization treatment of several inflammatory diseases. Therefore, targeting serum IL-6 levels to reduce inflammation may become an important symptomatic treatment strategy (260). One clinical study (ChiCTR2000029765) showed that tocilizumab, an IL-6 receptor-targeted monoclonal antibody, could reduce the risk of severe SARS-CoV-2 infection in patients with invasive mechanical ventilation or death (261). A randomized double-blind phase III clinical trial (NCT04320615) showed that tocilizumab (8 mg/kg, intravenous injection) can significantly shorten the intensive care unit by 5.8 days (9.8 days of standard care) and shorten the discharge time by 8 days (20 days of standard care) (262). Currently, tocilizumab has registered more than 70 SARS-CoV-2-related clinical trials. CVL218 was originally discovered through a data-driven drug reuse framework that can effectively inhibit the replication of SARS-CoV-2 with an EC50 of 5.12 μM. In-depth studies have shown that CVL218 (1 and 3 μM) treatment for 12 h can significantly reduce the production of IL-6 by 50% and 73% in peripheral blood mononuclear cells induced by CpG (microbial DNA sequence containing unmethylated CpG dinucleotides), respectively. In vivo studies have shown that CVL218 is mainly distributed in lung tissues and has no obvious toxicity (263). The above results suggest that CVL218 has a significant anti-inflammatory cytokine effect on SARS-CoV-2-induced immunopathological symptoms. Based on this, we think that targeted intervention of inflammatory cytokines is an important SARS-CoV-2 treatment strategy that can be studied in depth.
Multiple studies suggest that excessive inflammatory production of proinflammatory cytokines such as IL-6 and TNF-α may trigger ARDS, which will accelerate disease progression and increase the risk of death in COVID-19 patients (264, 265). Therefore, controlling the development of ARDS may also be a feasible treatment strategy for COVID-19. At present, a number of clinical studies (NCT04244591/NCT04327401/NCT04476992/NCT04306393…) are using strategies such as glucocorticoids, small molecule drugs, recombinant interferon and NO inhalation to explore the effectiveness of intervening in ARDS to affect COVID-19 (266, 267). Perhaps this strategy will provide more evidence for the safety and efficacy of treating COVID-19.
Immunomodulators are an important class of substances that affect the function of the immune system. Among them, pegylated interferon-α, which is approved for the treatment of hepatitis B/C viruses (HBV/HCV), can be used to stimulate the innate antiviral response of patients infected with SARS-CoV-2 (ChiCTR2000029387) (268). A retrospective study showed that pegylated interferon-α aerosol (5 million IU, bid) and arbidol (600 mg/day) treatment can significantly reduce the upper respiratory tract viral load and shorten the time for the inflammatory response indicators (IL-6 and CRP) in blood to return to normal with no obvious adverse reaction (269). Meanwhile, some clinical trials evaluated the therapeutic effect of glucocorticoids and found that they can significantly reduce the cytokine storm and relieve the corresponding tissue damage, which is beneficial to the treatment of severe COVID-19 (reduced 1/3 of mortality rate in patients using ventilators) and may affect the clearance of the virus in mild patients (270). The above results indicate that the use of immunomodulators to affect immune function will be a symptomatic treatment strategy for COVID-19 that can be considered. Although it is not given priority, the indications, dosage and course of treatment can be strictly controlled in consideration of the patient’s situation to ensure the maximum benefit of the patient.
The use of antibodies contained in the plasma of convalescent patients to suppress viremia for passive immunotherapy is considered to be a promising option for anti-SARS-CoV-2 infection. Currently, there have been clinical trials to test the effectiveness of plasma in recovering patients, and a study showed that the mortality rate of patients receiving convalescent plasma therapy is significantly lower than that of patients not receiving plasma therapy (271). In vitro experiments showed that antibodies in the serum of SARS-CoV-2-infected patients can effectively neutralize SARS-CoV-2. Moreover, clinical trials of administering convalescent plasma to 5,000 COVID-19 hospitalized patients in the early stages have also proven to be safe because the incidence of serious adverse events is very low (272). Therefore, convalescent plasma seems to be a good symptomatic treatment strategy in the case of solving the problems of ionomer safety and whether it needs a different storage method from ordinary plasma.
In addition, blood purification, NK-cell therapy, MSC transplantation therapy and Treg cell therapy have also been mentioned and are being studied. These therapies mainly alleviate and eliminate the pathological symptoms of patients, including inflammation, immune dysfunction, organ failure, etc., by adjusting immune function, removing inflammatory cytokines from the body, and directly killing SARS-CoV-2 infections (186). Changes such as lymphopenia and increased inflammatory cytokines in COVID-19 patients can induce symptoms of inflammation, immune function, and organ system dysfunction, which can be considered potential biomarkers and intervention targets for disease progression (273). Therefore, symptomatic treatments such as improving lymphopenia, reducing inflammation, and regulating immunity will become promising treatment strategies.
Perspectives and Conclusions
The continuous outbreak of SARS-CoV-2 and the endless emergence of new mutant strains once again emphasize the urgency of continuing to explore, screen, and prevent COVID-19 globally. All this urgently requires precise target determination and mechanism elucidation in order to develop specific or broad-spectrum drugs for SARS-CoV-2 virus entry, replication, pathological changes or prevention.
While exploring and determining the effective targets for fighting against SARS-CoV-2, we should highly combine the experience of the three CoV pandemics, clarify the SARS-CoV-2 genome and structural information, and lay the foundation for screening targets; comprehensively consider the pathophysiological characteristics and mechanisms of viral entry, replication, assembly, infection and pathogenic processes to accurately analyze the crystal structure of related enzymes and proteins, and provide direct evidence for the target; and combine omics, bioinformatics, computer virtual screening and artificial intelligence and other technologies to explore, screen and confirm targets with maximum efficiency.
Furthermore, the development of vaccines and drugs needs to be carried out at multiple levels. Specifically, considering the lethality and disability of COVID-19, short-term research focuses on “old drugs and new use”, rapid screening of FDA-approved drugs and clinical trials, and cooperation with other medication considerations to speed up the treatment of patients. After multichannel experience accumulation, developing innovative drugs targeted at different populations with good activity and selectivity against viruses through virtual screening and computer drug design, candidate drug preclinical research, and corresponding protective measures are key to future prevention and treatment. Moreover, it is necessary to minimize the occurrence and impact of drug resistance to maintain the efficacy of these innovative drugs; from a long-term perspective, broad-spectrum anti-CoV drugs should be developed to provide sufficient R&D experience and test platforms for possible future outbreaks.
Currently, there are only a few clinically approved drugs, vaccines and corresponding therapeutic strategies for COVID-19, and we cannot control the long-term consequences. Therefore, through the existing vaccination prevention, contact tracing, isolation of infected persons, and effective supportive treatment of SARS-CoV-2-infected persons, the diagnosis of symptomatic and asymptomatic persons and their close contacts as soon as possible is still the key means to prevent the further spread and control the disease. Furthermore, we should also realize that focusing on international cooperation and sharing anti-epidemic experiences will provide new impetus for the dissemination and confirmation of treatment strategies.
Author Contributions
HZ and W-JN contributed to the conception or design of the review. HZ and W-JN wrote the manuscript. WH, MC, ZW, and Y-CS collected and analyzed the latest literature and intelligence on the pandemic. W-JN, MC, and ZW revised the original draft. W-JN, MC, and Y-CS critically reviewed and edited the latest version for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Science Foundation of Anhui Provincial Cancer Hospital (No. 2020YJQN008), the National Natural Science Foundation of China (No. 81803602), the Natural Science Foundation of Anhui Province (No. 1708085QH207), and the Fundamental Research Funds for the Central Universities (No. WK9110000018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the American Journal Experts (AJE) for their guidance on the grammar and language of this article. We thank Microsoft Office and Bio Render for providing materials to help us better beautify the figures.
References
1. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus From Patients With Pneumonia in China, 2019. N Engl J Med (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
2. Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV. Genomic Determinants of Pathogenicity in SARS-CoV-2 and Other Human Coronaviruses. Proc Natl Acad Sci USA (2020) 117:15193–9. doi: 10.1073/pnas.2008176117
3. Tang X, Ying R, Yao X, Li G, Wu C, Tang Y, et al. Evolutionary Analysis and Lineage Designation of SARS-CoV-2 Genomes. Sci Bull (Beijing) (2021) 66:2297–311. doi: 10.1016/j.scib.2021.02.012
4. Sabino EC, Buss LF, Carvalho M, Prete CJ, Crispim M, Fraiji NA, et al. Resurgence of COVID-19 in Manaus, Brazil, Despite High Seroprevalence. Lancet (2021) 397:452–5. doi: 10.1016/S0140-6736(21)00183-5
5. Thye AY, Law JW, Pusparajah P, Letchumanan V, Chan KG, Lee LH. Emerging SARS-CoV-2 Variants of Concern (VOCs): An Impending Global Crisis. Biomedicines (2021) 9:1303. doi: 10.3390/biomedicines9101303
6. Martin DP, Weaver S, Tegally H, San JE, Shank SD, Wilkinson E, et al. The Emergence and Ongoing Convergent Evolution of the SARS-CoV-2 N501Y Lineages. Cell (2021) 184:5189–200. doi: 10.1016/j.cell.2021.09.003
7. Benton DJ, Wrobel AG, Roustan C, Borg A, Xu P, Martin SR, et al. The Effect of the D614G Substitution on the Structure of the Spike Glycoprotein of SARS-CoV-2. Proc Natl Acad Sci USA (2021) 118:e2022586118. doi: 10.1073/pnas.2022586118
8. Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D, Mishra S, et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science (2021) 372:815–21. doi: 10.1126/science.abh2644
9. Hemmer CJ, Lobermann M, Reisinger EC. [COVID-19: Epidemiology and Mutations: An Update]. Radiologe (2021) 61:880–7. doi: 10.1007/s00117-021-00909-0
10. Mishra A, Kumar N, Bhatia S, Aasdev A, Kanniappan S, Sekhar AT, et al. SARS-CoV-2 Delta Variant Among Asiatic Lions, India. Emerg Infect Dis (2021) 27:2723–5. doi: 10.3201/eid2710.211500
11. Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, et al. Evolutionary Analysis of the Delta and Delta Plus Variants of the SARS-CoV-2 Viruses. J Autoimmun (2021) 124:102715. doi: 10.1016/j.jaut.2021.102715
12. Sapkal G, Yadav PD, Ella R, Abraham P, Patil DY, Gupta N, et al. Neutralization of VUI B.1.1.28 P2 Variant With Sera of COVID-19 Recovered Cases and Recipients of Covaxin an Inactivated COVID-19 Vaccine. J Travel Med (2021) 28:taab077. doi: 10.1093/jtm/taab077
13. Zhang L, Cui Z, Li Q, Wang B, Yu Y, Wu J, et al. Ten Emerging SARS-CoV-2 Spike Variants Exhibit Variable Infectivity, Animal Tropism, and Antibody Neutralization. Commun Biol (2021) 4:1196. doi: 10.1038/s42003-021-02728-4
14. McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, et al. SARS-CoV-2 Immune Evasion by the B.1.427/B.1.429 Variant of Concern. Science (2021) 373:648–54. doi: 10.1126/science.abi7994
15. Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant. Cell (2021) 184:3426–37. doi: 10.1016/j.cell.2021.04.025
16. Romero PE, Davila-Barclay A, Salvatierra G, Gonzalez L, Cuicapuza D, Solis L, et al. The Emergence of Sars-CoV-2 Variant Lambda (C.37) in South America. Microbiol Spectr. (2021) 9:e78921. doi: 10.1128/Spectrum.00789-21
17. Darvishi M, Rahimi F, Talebi BAA. SARS-CoV-2 Lambda (C.37): An Emerging Variant of Concern? Gene Rep (2021) 25:101378. doi: 10.1016/j.genrep.2021.101378
18. Laiton-Donato K, Franco-Munoz C, Alvarez-Diaz DA, Ruiz-Moreno HA, Usme-Ciro JA, Prada DA, et al. Characterization of the Emerging B.1.621 Variant of Interest of SARS-CoV-2. Infect Genet Evol (2021) 95:105038. doi: 10.1016/j.meegid.2021.105038
19. Uriu K, Kimura I, Shirakawa K, Takaori-Kondo A, Nakada TA, Kaneda A, et al. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. N Engl J Med (2021) 385:2397–9. doi: 10.1056/NEJMc2114706
20. Shuai H, Chan JF, Yuen TT, Yoon C, Hu JC, Wen L, et al. Emerging SARS-CoV-2 Variants Expand Species Tropism to Murines. Ebiomedicine (2021) 73:103643. doi: 10.1016/j.ebiom.2021.103643
21. Moubarak M, Kasozi KI, Hetta HF, Shaheen HM, Rauf A, Al-Kuraishy HM, et al. The Rise of SARS-CoV-2 Variants and the Role of Convalescent Plasma Therapy for Management of Infections. Life (Basel). (2021) 11:734. doi: 10.3390/life11080734
22. van der Veer BMJW, Dingemans J, van Alphen LB, Hoebe CJPA, Savelkoul PHM. A Novel B.1.1.523 SARS-CoV-2 Variant That Combines Many Spike Mutations Linked to Immune Evasion With Current Variants of Concern. bioRxiv (2021), 2021–9. doi: 10.1101/2021.09.16.460616
23. Albayat SS, Arshad S, Arshad MA, Jabbar A, Ullah I. Precautionary Measures for More Transmissible C.1.2 COVID-19 Variant: A Caution for Qatar and the Rest of the World. J Med Virol (2022) 94:842–3. doi: 10.1002/jmv.27438
24. Yang X. SARS-COV-2 C.1.2 Variant is Highly Mutated But may Possess Reduced Affinity for ACE2 Receptor. bioRxiv (2021), 2010–21. doi: 10.1101/2021.10.16.464644
25. Nagano K, Tani-Sassa C, Iwasaki Y, Takatsuki Y, Yuasa S, Takahashi Y, et al. SARS-CoV-2 R.1 Lineage Variants That Prevailed in Tokyo in March 2021. J Med Virol (2021) 93:6833–6. doi: 10.1002/jmv.27240
26. Sekizuka T, Itokawa K, Hashino M, Okubo K, Ohnishi A, Goto K, et al. A Discernable Increase in the Severe Acute Respiratory Syndrome Coronavirus 2 R.1 Lineage Carrying an E484K Spike Protein Mutation in Japan. Infect Genet Evol (2021) 94:105013. doi: 10.1016/j.meegid.2021.105013
27. Rodriguez-Maldonado AP, Vazquez-Perez JA, Cedro-Tanda A, Taboada B, Boukadida C, Wong-Arambula C, et al. Emergence and Spread of the Potential Variant of Interest (VOI) B.1.1.519 of SARS-CoV-2 Predominantly Present in Mexico. Arch Virol (2021) 166:3173–7. doi: 10.1007/s00705-021-05208-6
28. Laine P, Nihtila H, Mustanoja E, Lyyski A, Ylinen A, Hurme J, et al. SARS-CoV-2 Variant With Mutations in N Gene Affecting Detection by Widely Used PCR Primers. J Med Virol (2021) 94:1227–31. doi: 10.1002/jmv.27418
29. Manouana GP, Nzamba MM, Bikangui R, Oye BS, Ondo NG, Honkpehedji JY, et al. Emergence of B.1.1.318 SARS-CoV-2 Viral Lineage and High Incidence of Alpha B.1.1.7 Variant of Concern in the Republic of Gabon. Int J Infect Dis (2022) 114:151–4. doi: 10.1016/j.ijid.2021.10.057
30. Fibriani A, Stephanie R, Alfiantie AA, Siregar A, Pradani G, Yamahoki N, et al. Analysis of SARS-CoV-2 Genomes From West Java, Indonesia. Viruses (2021) 13:2097. doi: 10.3390/v13102097
31. Sam IC, Chong YM, Abdullah A, Fu J, Hasan MS, Jamaluddin FH, et al. Changing Predominant SARS-CoV-2 Lineages Drives Successive COVID-19 Waves in Malaysia, February 2020 to March 2021. J Med Virol (2021) 94:1146–53. doi: 10.1002/jmv.27441
32. Dudas G, Hong SL, Potter BI, Calvignac-Spencer S, Niatou-Singa FS, Tombolomako TB, et al. Emergence and Spread of SARS-CoV-2 Lineage B.1.620 With Variant of Concern-Like Mutations and Deletions. Nat Commun (2021) 12:5769. doi: 10.1038/s41467-021-26055-8
33. Zahradnik J, Marciano S, Shemesh M, Zoler E, Harari D, Chiaravalli J, et al. SARS-CoV-2 Variant Prediction and Antiviral Drug Design are Enabled by RBD. Vitro Evol Nat Microbiol (2021) 6:1188–98. doi: 10.1038/s41564-021-00954-4
34. Annavajhala MK, Mohri H, Wang P, Nair M, Zucker JE, Sheng Z, et al. Emergence and Expansion of the SARS-CoV-2 Variant B.1.526 Identified in New York. medRxiv (2021). doi: 10.1101/2021.02.23.21252259
35. Thompson CN, Hughes S, Ngai S, Baumgartner J, Wang JC, McGibbon E, et al. Rapid Emergence and Epidemiologic Characteristics of the SARS-CoV-2 B.1.526 Variant - New York City, New York, January 1-April 5, 2021. MMWR Morb Mortal Wkly Rep (2021) 70:712–6. doi: 10.15585/mmwr.mm7019e1
36. Bugembe DL, Phan M, Abias AG, Ayei J, Deng LL, Lako R, et al. SARS-CoV-2 Variants, South Sudan, January-March 2021. Emerg Infect Dis (2021) 27:3133–6. doi: 10.3201/eid2712.211488
37. Abdool KS, de Oliveira T. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications. N Engl J Med (2021) 384:1866–8. doi: 10.1056/NEJMc2100362
38. Chen J, Fan J, Chen Z, Zhang M, Peng H, Liu J, et al. Nonmuscle Myosin Heavy Chain IIA Facilitates SARS-CoV-2 Infection in Human Pulmonary Cells. Proc Natl Acad Sci U S A (2021) 118:e2111011118. doi: 10.1073/pnas.2111011118
39. Fontanet A, Autran B, Lina B, Kieny MP, Karim S, Sridhar D. SARS-CoV-2 Variants and Ending the COVID-19 Pandemic. Lancet (2021) 397:952–4. doi: 10.1016/S0140-6736(21)00370-6
40. Cui J, Li F, Shi ZL. Origin and Evolution of Pathogenic Coronaviruses. Nat Rev Microbiol (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9
41. Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, et al. The Coding Capacity of SARS-CoV-2. Nature (2021) 589:125–30. doi: 10.1038/s41586-020-2739-1
42. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
43. Jungreis I, Sealfon R, Kellis M. SARS-CoV-2 Gene Content and COVID-19 Mutation Impact by Comparing 44 Sarbecovirus Genomes. Nat Commun (2021) 12:2642. doi: 10.1038/s41467-021-22905-7
44. Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, Darby AC, et al. Generation and Transmission of Interlineage Recombinants in the SARS-CoV-2 Pandemic. Cell (2021) 184:5179–88. doi: 10.1016/j.cell.2021.08.014
45. Artese A, Svicher V, Costa G, Salpini R, Di Maio VC, Alkhatib M, et al. Current Status of Antivirals and Druggable Targets of SARS CoV-2 and Other Human Pathogenic Coronaviruses. Drug Resist Updat (2020) 53:100721. doi: 10.1016/j.drup.2020.100721
46. Rolf JD. Clinical Characteristics of Covid-19 in China. N Engl J Med (2020) 382:1860. doi: 10.1056/NEJMc2005203
47. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol (2021) 19:141–54. doi: 10.1038/s41579-020-00459-7
48. Guo M, Tao W, Flavell RA, Zhu S. Potential Intestinal Infection and Faecal-Oral Transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol (2021) 18:269–83. doi: 10.1038/s41575-021-00416-6
49. Oude MB, Worp N, Nieuwenhuijse DF, Sikkema RS, Haagmans B, Fouchier R, et al. The Next Phase of SARS-CoV-2 Surveillance: Real-Time Molecular Epidemiology. Nat Med (2021) 27:1518–24. doi: 10.1038/s41591-021-01472-w
50. Yang B, Fan J, Huang J, Guo E, Fu Y, Liu S, et al. Clinical and Molecular Characteristics of COVID-19 Patients With Persistent SARS-CoV-2 Infection. Nat Commun (2021) 12:3501. doi: 10.1038/s41467-021-23621-y
51. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med (2020) 382:1708–20. doi: 10.1056/NEJMoa2002032
52. Deng H, Yan X, Yuan L. Human Genetic Basis of Coronavirus Disease 2019. Signal Transduct Target Ther (2021) 6:344. doi: 10.1038/s41392-021-00736-8
53. Abdelnabi R, Boudewijns R, Foo CS, Seldeslachts L, Sanchez-Felipe L, Zhang X, et al. Comparing Infectivity and Virulence of Emerging SARS-CoV-2 Variants in Syrian Hamsters. Ebiomedicine (2021) 68:103403. doi: 10.1016/j.ebiom.2021.103403
54. McCormick KD, Jacobs JL, Mellors JW. The Emerging Plasticity of SARS-CoV-2. Science (2021) 371:1306–8. doi: 10.1126/science.abg4493
55. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses - Drug Discovery and Therapeutic Options. Nat Rev Drug Discov (2016) 15:327–47. doi: 10.1038/nrd.2015.37
56. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat Rev Microbiol (2021) 19:155–70. doi: 10.1038/s41579-020-00468-6
57. Xiang R, Yu Z, Wang Y, Wang L, Huo S, Li Y, et al. Recent Advances in Developing Small-Molecule Inhibitors Against SARS-CoV-2. Acta Pharm Sin B (2021) 12:995–1026. doi: 10.1016/j.apsb.2021.06.016
58. Zhou H, Fang Y, Xu T, Ni WJ, Shen AZ, Meng XM. Potential Therapeutic Targets and Promising Drugs for Combating SARS-CoV-2. Br J Pharmacol (2020) 177:3147–61. doi: 10.1111/bph.15092
59. Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm Sin B (2020) 10:766–88. doi: 10.1016/j.apsb.2020.02.008
60. Kneller DW, Phillips G, O'Neill HM, Jedrzejczak R, Stols L, Langan P, et al. Structural Plasticity of SARS-CoV-2 3cl M(pro) Active Site Cavity Revealed by Room Temperature X-Ray Crystallography. Nat Commun (2020) 11:3202. doi: 10.1038/s41467-020-16954-7
61. Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved Alpha-Ketoamide Inhibitors. Science (2020) 368:409–12. doi: 10.1126/science.abb3405
62. Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, et al. Papain-Like Protease Regulates SARS-CoV-2 Viral Spread and Innate Immunity. Nature (2020) 587:657–62. doi: 10.1038/s41586-020-2601-5
63. Gao X, Qin B, Chen P, Zhu K, Hou P, Wojdyla JA, et al. Crystal Structure of SARS-CoV-2 Papain-Like Protease. Acta Pharm Sin B (2021) 11:237–45. doi: 10.1016/j.apsb.2020.08.014
64. Osipiuk J, Azizi SA, Dvorkin S, Endres M, Jedrzejczak R, Jones KA, et al. Structure of Papain-Like Protease From SARS-CoV-2 and its Complexes With non-Covalent Inhibitors. Nat Commun (2021) 12:743. doi: 10.1038/s41467-021-21060-3
65. Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, et al. Structure of the RNA-Dependent RNA Polymerase From COVID-19 Virus. Science (2020) 368:779–82. doi: 10.1126/science.abb7498
66. Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, et al. Structural Basis for Inhibition of the RNA-Dependent RNA Polymerase From SARS-CoV-2 by Remdesivir. Science (2020) 368:1499–504. doi: 10.1126/science.abc1560
67. Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P. Structure of Replicating SARS-CoV-2 Polymerase. Nature (2020) 584:154–6. doi: 10.1038/s41586-020-2368-8
68. White MA, Lin W, Cheng X. Discovery of COVID-19 Inhibitors Targeting the SARS-CoV-2 Nsp13 Helicase. J Phys Chem Lett (2020) 11:9144–51. doi: 10.1021/acs.jpclett.0c02421
69. Chen J, Malone B, Llewellyn E, Grasso M, Shelton P, Olinares P, et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell (2020) 182:1560–73. doi: 10.1016/j.cell.2020.07.033
70. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell (2020) 181:281–92. doi: 10.1016/j.cell.2020.02.058
71. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-Specific Glycan Analysis of the SARS-CoV-2 Spike. Science (2020) 369:330–3. doi: 10.1126/science.abb9983
72. Carlson CR, Asfaha JB, Ghent CM, Howard CJ, Hartooni N, Safari M, et al. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for Its Dual Functions. Mol Cell (2020) 80:1092–103. doi: 10.1016/j.molcel.2020.11.025
73. Peng Y, Du N, Lei Y, Dorje S, Qi J, Luo T, et al. Structures of the SARS-CoV-2 Nucleocapsid and Their Perspectives for Drug Design. EMBO J (2020) 39:e105938. doi: 10.15252/embj.2020105938
74. Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers. Nat Struct Mol Biol (2020) 27:1202–8. doi: 10.1038/s41594-020-00536-8
75. Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR, et al. The SARS-CoV-2 Nucleocapsid Phosphoprotein Forms Mutually Exclusive Condensates With RNA and the Membrane-Associated M Protein. Nat Commun (2021) 12:502. doi: 10.1038/s41467-020-20768-y
76. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: A Comparative Pathogenomic Platform With an Interactive Web Interface. Nucleic Acids Res (2019) 47:D687–92. doi: 10.1093/nar/gky1080
77. Schuller M, Correy GJ, Gahbauer S, Fearon D, Wu T, Diaz RE, et al. Fragment Binding to the Nsp3 Macrodomain of SARS-CoV-2 Identified Through Crystallographic Screening and Computational Docking. Sci Adv (2021) 7:eabf8711. doi: 10.1126/sciadv.abf8711
78. Yuan S, Peng L, Park JJ, Hu Y, Devarkar SC, Dong MB, et al. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery Toward Viral RNA. Mol Cell (2020) 80:1055–66. doi: 10.1016/j.molcel.2020.10.034
79. Nemudryi A, Nemudraia A, Wiegand T, Nichols J, Snyder DT, Hedges JF, et al. SARS-CoV-2 Genomic Surveillance Identifies Naturally Occurring Truncation of ORF7a That Limits Immune Suppression. Cell Rep (2021) 35:109197. doi: 10.1016/j.celrep.2021.109197
80. Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural Basis for Translational Shutdown and Immune Evasion by the Nsp1 Protein of SARS-CoV-2. Science (2020) 369:1249–55. doi: 10.1126/science.abc8665
81. Xiao T, Lu J, Zhang J, Johnson RI, McKay L, Storm N, et al. A Trimeric Human Angiotensin-Converting Enzyme 2 as an Anti-SARS-CoV-2 Agent. Nat Struct Mol Biol (2021) 28:202–9. doi: 10.1038/s41594-020-00549-3
82. Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. Receptor Binding and Priming of the Spike Protein of SARS-CoV-2 for Membrane Fusion. Nature (2020) 588:327–30. doi: 10.1038/s41586-020-2772-0
83. Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford K, Dingens AS, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell (2020) 182:1295–310. doi: 10.1016/j.cell.2020.08.012
84. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature (2020) 581:221–4. doi: 10.1038/s41586-020-2179-y
85. Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach PY. TMPRSS2 Expression Dictates the Entry Route Used by SARS-CoV-2 to Infect Host Cells. EMBO J (2021) 40:e107821. doi: 10.15252/embj.2021107821
86. Mahoney M, Damalanka VC, Tartell MA, Chung DH, Lourenco AL, Pwee D, et al. A Novel Class of TMPRSS2 Inhibitors Potently Block SARS-CoV-2 and MERS-CoV Viral Entry and Protect Human Epithelial Lung Cells. Proc Natl Acad Sci USA (2021) 118:e2108728118. doi: 10.1073/pnas.2108728118
87. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell (2020) 181:271–80. doi: 10.1016/j.cell.2020.02.052
88. Zhang MY, Zhang Y, Wu XD, Zhang K, Lin P, Bian HJ, et al. Disrupting CD147-RAP2 Interaction Abrogates Erythrocyte Invasion by Plasmodium Falciparum. Blood (2018) 131:1111–21. doi: 10.1182/blood-2017-08-802918
89. Chen Z, Mi L, Xu J, Yu J, Wang X, Jiang J, et al. Function of HAb18G/CD147 in Invasion of Host Cells by Severe Acute Respiratory Syndrome Coronavirus. J Infect Dis (2005) 191:755–60. doi: 10.1086/427811
90. Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, et al. CD147-Spike Protein is a Novel Route for SARS-CoV-2 Infection to Host Cells. Signal Transduct Target Ther (2020) 5:283. doi: 10.1038/s41392-020-00426-x
91. Chen H, Zou TH, Xuan B, Yan Y, Yan T, Shen C, et al. Single Cell Transcriptome Revealed SARS-CoV-2 Entry Genes Enriched in Colon Tissues and Associated With Coronavirus Infection and Cytokine Production. Signal Transduct Target Ther (2020) 5:121. doi: 10.1038/s41392-020-00237-0
92. Bian H, Zheng ZH, Wei D, Wen A, Zhang Z, Lian JQ, et al. Safety and Efficacy of Meplazumab in Healthy Volunteers and COVID-19 Patients: A Randomized Phase 1 and an Exploratory Phase 2 Trial. Signal Transduct Target Ther (2021) 6:194. doi: 10.1038/s41392-021-00603-6
93. Geng J, Chen L, Yuan Y, Wang K, Wang Y, Qin C, et al. CD147 Antibody Specifically and Effectively Inhibits Infection and Cytokine Storm of SARS-CoV-2 and Its Variants Delta, Alpha, Beta, and Gamma. Signal Transduct Target Ther (2021) 6:347. doi: 10.1038/s41392-021-00760-8
94. Ragotte RJ, Pulido D, Donnellan FR, Hill ML, Gorini G, Davies H, et al. Human Basigin (CD147) Does Not Directly Interact With SARS-CoV-2 Spike Glycoprotein. mSphere (2021) 6:e64721. doi: 10.1128/mSphere.00647-21
95. Shilts J, Crozier T, Greenwood E, Lehner PJ, Wright GJ. No Evidence for Basigin/CD147 as a Direct SARS-CoV-2 Spike Binding Receptor. Sci Rep (2021) 11:413. doi: 10.1038/s41598-020-80464-1
96. Morales A, Rojo RS, Cristobal H, Fiz-Lopez A, Arribas E, Mari M, et al. Growth Arrest-Specific Factor 6 (GAS6) Is Increased in COVID-19 Patients and Predicts Clinical Outcome. Biomedicines (2021) 9:335. doi: 10.3390/biomedicines9040335
97. Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, et al. AXL is a Candidate Receptor for SARS-CoV-2 That Promotes Infection of Pulmonary and Bronchial Epithelial Cells. Cell Res (2021) 31:126–40. doi: 10.1038/s41422-020-00460-y
98. Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa MM, et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell (2020) 182:685–712. doi: 10.1016/j.cell.2020.06.034
99. Beeraka NM, Sadhu SP, Madhunapantula SV, Rao PR, Svistunov AA, Nikolenko VN, et al. Strategies for Targeting SARS CoV-2: Small Molecule Inhibitors-The Current Status. Front Immunol (2020) 11:552925. doi: 10.3389/fimmu.2020.552925
100. Jan JT, Cheng TR, Juang YP, Ma HH, Wu YT, Yang WB, et al. Identification of Existing Pharmaceuticals and Herbal Medicines as Inhibitors of SARS-CoV-2 Infection. Proc Natl Acad Sci USA (2021) 118:e2021579118. doi: 10.1073/pnas.2021579118
101. Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al. Discovery of SARS-CoV-2 Antiviral Drugs Through Large-Scale Compound Repurposing. Nature (2020) 586:113–9. doi: 10.1038/s41586-020-2577-1
102. Sun C, Zhang J, Wei J, Zheng X, Zhao X, Fang Z, et al. Screening, Simulation, and Optimization Design of Small Molecule Inhibitors of the SARS-CoV-2 Spike Glycoprotein. PloS One (2021) 16:e245975. doi: 10.1371/journal.pone.0245975
103. Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, et al. The Anti-Influenza Virus Drug, Arbidol is an Efficient Inhibitor of SARS-CoV-2 In Vitro. Cell Discov (2020) 6:28. doi: 10.1038/s41421-020-0169-8
104. Khan RJ, Jha RK, Amera GM, Jain M, Singh E, Pathak A, et al. Targeting SARS-CoV-2: A Systematic Drug Repurposing Approach to Identify Promising Inhibitors Against 3C-Like Proteinase and 2'-O-Ribose Methyltransferase. J Biomol Struct Dyn (2021) 39:2679–92. doi: 10.1080/07391102.2020.1753577
105. Yamamoto KA, Blackburn K, Migowski E, Goshe MB, Brown DT, Ferreira DF, et al. Quantitative Proteomic Analysis of the Tizoxanide Effect in Vero Cells. Sci Rep (2020) 10:14733. doi: 10.1038/s41598-020-71634-2
106. Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion Mechanism of 2019-Ncov and Fusion Inhibitors Targeting HR1 Domain in Spike Protein. Cell Mol Immunol (2020) 17:765–7. doi: 10.1038/s41423-020-0374-2
107. Zhu Y, Yu D, Yan H, Chong H, He Y. Design of Potent Membrane Fusion Inhibitors Against SARS-CoV-2, an Emerging Coronavirus With High Fusogenic Activity. J Virol (2020) 94:e00635–20. doi: 10.1128/JVI.00635-20
108. de Vries RD, Schmitz KS, Bovier FT, Predella C, Khao J, Noack D, et al. Intranasal Fusion Inhibitory Lipopeptide Prevents Direct-Contact SARS-CoV-2 Transmission in Ferrets. Science (2021) 371:1379–82. doi: 10.1126/science.abf4896
109. Ucar B, Acar T, Arayici PP, Derman S. A Nanotechnological Approach in the Current Therapy of COVID-19: Model Drug Oseltamivir-Phosphate Loaded PLGA Nanoparticles Targeted With Spike Protein Binder Peptide of SARS-CoV-2. Nanotechnology (2021) 32:485601. doi: 10.1088/1361-6528/ac1c22
110. Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, et al. De Novo Design of Picomolar SARS-CoV-2 Miniprotein Inhibitors. Science (2020) 370:426–31. doi: 10.1126/science.abd9909
111. Beddingfield BJ, Iwanaga N, Chapagain PP, Zheng W, Roy CJ, Hu TY, et al. The Integrin Binding Peptide, ATN-161, as a Novel Therapy for SARS-CoV-2 Infection. JACC Basic Transl Sci (2021) 6:1–8. doi: 10.1016/j.jacbts.2020.10.003
112. Bauer A, Schreinlechner M, Sappler N, Dolejsi T, Tilg H, Aulinger BA, et al. Discontinuation Versus Continuation of Renin-Angiotensin-System Inhibitors in COVID-19 (ACEI-COVID): A Prospective, Parallel Group, Randomised, Controlled, Open-Label Trial. Lancet Respir Med (2021) 9:863–72. doi: 10.1016/S2213-2600(21)00214-9
113. Esam Z, Akhavan M, Lotfi M, Bekhradnia A. Molecular Docking and Dynamics Studies of Nicotinamide Riboside as a Potential Multi-Target Nutraceutical Against SARS-CoV-2 Entry, Replication, and Transcription: A New Insight. J Mol Struct (2022) 1247:131394. doi: 10.1016/j.molstruc.2021.131394
114. Huo J, Le Bas A, Ruza RR, Duyvesteyn H, Mikolajek H, Malinauskas T, et al. Neutralizing Nanobodies Bind SARS-CoV-2 Spike RBD and Block Interaction With ACE2. Nat Struct Mol Biol (2020) 27:846–54. doi: 10.1038/s41594-020-0469-6
115. Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human Ace2. Cell (2020) 181:905–13. doi: 10.1016/j.cell.2020.04.004
116. Zhuravel SV, Khmelnitskiy OK, Burlaka OO, Gritsan AI, Goloshchekin BM, Kim S, et al. Nafamostat in Hospitalized Patients With Moderate to Severe COVID-19 Pneumonia: A Randomised Phase II Clinical Trial. EClinicalMedicine (2021) 41:101169. doi: 10.1016/j.eclinm.2021.101169
117. Alzain AA, Elbadwi FA. Identification of Novel TMPRSS2 Inhibitors for COVID-19 Using E-Pharmacophore Modelling, Molecular Docking, Molecular Dynamics and Quantum Mechanics Studies. Inform Med Unlocked (2021) 26:100758. doi: 10.1016/j.imu.2021.100758
118. Vuong W, Khan MB, Fischer C, Arutyunova E, Lamer T, Shields J, et al. Feline Coronavirus Drug Inhibits the Main Protease of SARS-CoV-2 and Blocks Virus Replication. Nat Commun (2020) 11:4282. doi: 10.1038/s41467-020-18096-2
119. Achutha AS, Pushpa VL, Suchitra S. Theoretical Insights Into the Anti-SARS-CoV-2 Activity of Chloroquine and Its Analogs and In Silico Screening of Main Protease Inhibitors. J Proteome Res (2020) 19:4706–17. doi: 10.1021/acs.jproteome.0c00683
120. Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized With Severe Covid-19. N Engl J Med (2020) 382:1787–99. doi: 10.1056/NEJMc2008043
121. Liu X, Li Z, Liu S, Sun J, Chen Z, Jiang M, et al. Potential Therapeutic Effects of Dipyridamole in the Severely Ill Patients With COVID-19. Acta Pharm Sin B (2020) 10:1205–15. doi: 10.1016/j.apsb.2020.04.008
122. Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, et al. Structure-Based Design of Antiviral Drug Candidates Targeting the SARS-CoV-2 Main Protease. Science (2020) 368:1331–5. doi: 10.1126/science.abb4489
123. Qiao J, Li YS, Zeng R, Liu FL, Luo RH, Huang C, et al. SARS-CoV-2 M(pro) Inhibitors With Antiviral Activity in a Transgenic Mouse Model. Science (2021) 371:1374–8. doi: 10.1126/science.abf1611
124. Wu Y, Li Z, Zhao YS, Huang YY, Jiang MY, Luo HB. Therapeutic Targets and Potential Agents for the Treatment of COVID-19. Med Res Rev (2021) 41:1775–97. doi: 10.1002/med.21776
125. Huynh T, Cornell W, Luan B. In Silico Exploration of Inhibitors for SARS-CoV-2's Papain-Like Protease. Front Chem (2020) 8:624163. doi: 10.3389/fchem.2020.624163
126. Xiao X, Wang C, Chang, Wang Y, Dong X, Jiao T, et al. Identification of Potent and Safe Antiviral Therapeutic Candidates Against SARS-CoV-2. Front Immunol (2020) 11:586572. doi: 10.3389/fimmu.2020.586572
127. Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell (2020) 182:417–28. doi: 10.1016/j.cell.2020.05.034
128. Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, et al. Compassionate Use of Remdesivir for Patients With Severe Covid-19. N Engl J Med (2020) 382:2327–36. doi: 10.1056/NEJMc2015312
129. Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al. Remdesivir in Adults With Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet (2020) 395:1569–78. doi: 10.1016/S0140-6736(20)31022-9
130. Kaptein S, Jacobs S, Langendries L, Seldeslachts L, Ter Horst S, Liesenborghs L, et al. Favipiravir at High Doses has Potent Antiviral Activity in SARS-CoV-2-Infected Hamsters, Whereas Hydroxychloroquine Lacks Activity. Proc Natl Acad Sci USA (2020) 117:26955–65. doi: 10.1073/pnas.2014441117
131. Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, et al. An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci Transl Med (2020) 12:eabb5883. doi: 10.1126/scitranslmed.abb5883
132. Good SS, Westover J, Jung KH, Zhou XJ, Moussa A, La Colla P, et al. AT-527, a Double Prodrug of a Guanosine Nucleotide Analog, Is a Potent Inhibitor of SARS-CoV-2 In Vitro and a Promising Oral Antiviral for Treatment of COVID-19. Antimicrob Agents Chemother (2021) 65:e02479–20. doi: 10.1128/AAC.02479-20
133. Spratt AN, Gallazzi F, Quinn TP, Lorson CL, Sonnerborg A, Singh K. Coronavirus Helicases: Attractive and Unique Targets of Antiviral Drug-Development and Therapeutic Patents. Expert Opin Ther Pat (2021) 31:339–50. doi: 10.1080/13543776.2021.1884224
134. Yuan S, Yin X, Meng X, Chan JF, Ye ZW, Riva L, et al. Clofazimine Broadly Inhibits Coronaviruses Including SARS-CoV-2. Nature (2021) 593:418–23. doi: 10.1038/s41586-021-03431-4
135. Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, et al. A Pan-Coronavirus Fusion Inhibitor Targeting the HR1 Domain of Human Coronavirus Spike. Sci Adv (2019) 5:v4580. doi: 10.1126/sciadv.aav4580
136. Milne S, Yang CX, Timens W, Bosse Y, Sin DD. SARS-CoV-2 Receptor ACE2 Gene Expression and RAAS Inhibitors. Lancet Respir Med (2020) 8:e50–1. doi: 10.1016/S2213-2600(20)30224-1
137. Hoffmann M, Arora P, Gross R, Seidel A, Hornich BF, Hahn AS, et al. SARS-CoV-2 Variants B.1.351 and P.1 escape From Neutralizing Antibodies. Cell (2021) 184:2384–93. doi: 10.1016/j.cell.2021.03.036
138. Hempel T, Elez K, Kruger N, Raich L, Shrimp JH, Danov O, et al. Synergistic Inhibition of SARS-CoV-2 Cell Entry by Otamixaban and Covalent Protease Inhibitors: Pre-Clinical Assessment of Pharmacological and Molecular Properties. Chem Sci (2021) 12:12600–9. doi: 10.1039/d1sc01494c
139. Vandyck K, Deval J. Considerations for the Discovery and Development of 3-Chymotrypsin-Like Cysteine Protease Inhibitors Targeting SARS-CoV-2 Infection. Curr Opin Virol (2021) 49:36–40. doi: 10.1016/j.coviro.2021.04.006
140. Zhao Y, Fang C, Zhang Q, Zhang R, Zhao X, Duan Y, et al. Crystal Structure of SARS-CoV-2 Main Protease in Complex With Protease Inhibitor PF-07321332. Protein Cell (2021) 13:1–5. doi: 10.1007/s13238-021-00883-2
141. Pitsillou E, Liang J, Ververis K, Lim KW, Hung A, Karagiannis TC. Identification of Small Molecule Inhibitors of the Deubiquitinating Activity of the SARS-CoV-2 Papain-Like Protease: In Silico Molecular Docking Studies and In Vitro Enzymatic Activity Assay. Front Chem (2020) 8:623971. doi: 10.3389/fchem.2020.623971
142. Kokic G, Hillen HS, Tegunov D, Dienemann C, Seitz F, Schmitzova J, et al. Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir. Nat Commun (2021) 12:279. doi: 10.1038/s41467-020-20542-0
143. Yin W, Luan X, Li Z, Zhou Z, Wang Q, Gao M, et al. Structural Basis for Inhibition of the SARS-CoV-2 RNA Polymerase by Suramin. Nat Struct Mol Biol (2021) 28:319–25. doi: 10.1038/s41594-021-00570-0
144. Naydenova K, Muir KW, Wu LF, Zhang Z, Coscia F, Peet MJ, et al. Structure of the SARS-CoV-2 RNA-Dependent RNA Polymerase in the Presence of Favipiravir-RTP. Proc Natl Acad Sci USA (2021) 118:e2021946118. doi: 10.1073/pnas.2021946118
145. Ninove L, Nougairede A, Gazin C, Thirion L, Delogu I, Zandotti C, et al. RNA and DNA Bacteriophages as Molecular Diagnosis Controls in Clinical Virology: A Comprehensive Study of More Than 45,000Routine PCR Tests. PloS One (2011) 6:e16142. doi: 10.1371/journal.pone.0016142
146. Miller SR, McGrath ME, Zorn KM, Ekins S, Wright SH, Cherrington NJ. Remdesivir and EIDD-1931 Interact With Human Equilibrative Nucleoside Transporters 1 and 2: Implications for Reaching SARS-CoV-2 Viral Sanctuary Sites. Mol Pharmacol (2021) 100:548–57. doi: 10.1124/molpharm.121.000333
147. Jena NR. Role of Different Tautomers in the Base-Pairing Abilities of Some of the Vital Antiviral Drugs Used Against COVID-19. Phys Chem Chem Phys (2020) 22:28115–22. doi: 10.1039/d0cp05297c
148. Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological Assessment of Hospitalized Patients With COVID-2019. Nature (2020) 581:465–9. doi: 10.1038/s41586-020-2196-x
149. Wang H, Zhang Y, Huang B, Deng W, Quan Y, Wang W, et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, With Potent Protection Against SARS-CoV-2. Cell (2020) 182:713–21. doi: 10.1016/j.cell.2020.06.008
150. Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, et al. Safety and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine, BBIBP-CorV: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Trial. Lancet Infect Dis (2021) 21:39–51. doi: 10.1016/S1473-3099(20)30831-8
151. Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, et al. Safety, Tolerability, and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine in Healthy Adults Aged 18-59 Years: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Clinical Trial. Lancet Infect Dis (2021) 21:181–92. doi: 10.1016/S1473-3099(20)30843-4
152. Vacharathit V, Aiewsakun P, Manopwisedjaroen S, Srisaowakarn C, Laopanupong T, Ludowyke N, et al. CoronaVac Induces Lower Neutralising Activity Against Variants of Concern Than Natural Infection. Lancet Infect Dis (2021) 21:1352–4. doi: 10.1016/S1473-3099(21)00568-5
153. Al KN, Zhang Y, Xia S, Yang Y, Al QM, Abdulrazzaq N, et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA (2021) 326:35–45. doi: 10.1001/jama.2021.8565
154. Ella R, Reddy S, Jogdand H, Sarangi V, Ganneru B, Prasad S, et al. Safety and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine, BBV152: Interim Results From a Double-Blind, Randomised, Multicentre, Phase 2 Trial, and 3-Month Follow-Up of a Double-Blind, Randomised Phase 1 Trial. Lancet Infect Dis (2021) 21:950–61. doi: 10.1016/S1473-3099(21)00070-0
155. Ella R, Reddy S, Blackwelder W, Potdar V, Yadav P, Sarangi V, et al. Efficacy, Safety, and lot-to-lot Immunogenicity of an Inactivated SARS-CoV-2 Vaccine (BBV152): Interim Results of a Randomised, Double-Blind, Controlled, Phase 3 Trial. Lancet (2021) 398:2173–84. doi: 10.1016/S0140-6736(21)02000-6
156. Voysey M, Clemens S, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. Safety and Efficacy of the ChAdOx1 nCoV-19 Vaccine (AZD1222) Against SARS-CoV-2: An interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK. Lancet (2021) 397:99–111. doi: 10.1016/S0140-6736(20)32661-1
157. Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, et al. Safety and Immunogenicity of ChAdOx1 nCoV-19 Vaccine Administered in a Prime-Boost Regimen in Young and Old Adults (COV002): A Single-Blind, Randomised, Controlled, Phase 2/3 Trial. Lancet (2021) 396:1979–93. doi: 10.1016/S0140-6736(20)32466-1
158. Sadoff J, Gray G, Vandebosch A, Cardenas V, Shukarev G, Grinsztejn B, et al. Safety and Efficacy of Single-Dose ad26.COV2.s Vaccine Against Covid-19. N Engl J Med (2021) 384:2187–201. doi: 10.1056/NEJMoa2101544
159. Guzman-Martinez O, Guardado K, de Guevara EL, Navarro S, Hernandez C, Zenteno-Cuevas R, et al. IgG Antibodies Generation and Side Effects Caused by Ad5-nCoV Vaccine (CanSino biologics) and BNT162b2 Vaccine (Pfizer/BioNTech) Among Mexican Population. Vaccines (Basel) (2021) 9:999. doi: 10.3390/vaccines9090999
160. Wu S, Huang J, Zhang Z, Wu J, Zhang J, Hu H, et al. Safety, Tolerability, and Immunogenicity of an Aerosolised Adenovirus Type-5 Vector-Based COVID-19 Vaccine (Ad5-Ncov) in Adults: Preliminary Report of an Open-Label and Randomised Phase 1 Clinical Trial. Lancet Infect Dis (2021) 21:1654–64. doi: 10.1016/S1473-3099(21)00396-0
161. Gonzalez S, Olszevicki S, Salazar M, Calabria A, Regairaz L, Marin L, et al. Effectiveness of the First component of Gam-COVID-Vac (Sputnik V) on Reduction of SARS-CoV-2 Confirmed Infections, Hospitalisations and Mortality in Patients Aged 60-79: A Retrospective Cohort Study in Argentina. EClinicalMedicine (2021) 40:101126. doi: 10.1016/j.eclinm.2021.101126
162. Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, et al. Safety and Efficacy of an Rad26 and Rad5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine: An Interim Analysis of a Randomised Controlled Phase 3 Trial in Russia. Lancet (2021) 397:671–81. doi: 10.1016/S0140-6736(21)00234-8
163. Lanini S, Capone S, Antinori A, Milleri S, Nicastri E, Camerini R, et al. GRAd-COV2, a Gorilla Adenovirus-based Candidate Vaccine Against COVID-19, Is Safe and Immunogenic in Younger and Older Adults. Sci Transl Med (2022) 14:j1996. doi: 10.1126/scitranslmed.abj1996
164. Johnson S, Martinez CI, Tedjakusuma SN, Peinovich N, Dora EG, Birch SM, et al. Oral Vaccination Protects Against Severe Acute Respiratory Syndrome Coronavirus 2 in a Syrian Hamster Challenge Model. J Infect Dis (2022) 225:34–41. doi: 10.1093/infdis/jiab561
165. Gabitzsch E, Safrit JT, Verma M, Rice A, Sieling P, Zakin L, et al. Complete Protection of Nasal and Lung Airways Against SARS-CoV-2 Challenge by Antibody Plus th1 Dominant n- and S-Specific T-Cell Responses to Subcutaneous Prime and Thermally-Stable Oral Boost Bivalent hAd5 Vaccination in an NHP Study. bioRxiv (2021) 2012–20. doi: 10.1101/2020.12.08.416297
166. Mahrosh HS, Mustafa G. The COVID-19 Puzzle: A Global Nightmare. Environ Dev Sustain (2021) 23:12710–37. doi: 10.1007/s10668-021-01224-3
167. Wang P, Zheng M, Lau SY, Chen P, Mok BW, Liu S, et al. Generation of DelNS1 Influenza Viruses: A Strategy for Optimizing Live Attenuated Influenza Vaccines. Mbio (2019) 10:e02180–19. doi: 10.1128/mBio.02180-19
168. Yahalom-Ronen Y, Tamir H, Melamed S, Politi B, Shifman O, Achdout H, et al. A Single Dose of Recombinant VSV-G-Spike Vaccine Provides Protection Against SARS-CoV-2 Challenge. Nat Commun (2020) 11:6402. doi: 10.1038/s41467-020-20228-7
169. Scarabel L, Guardascione M, Dal Bo M, Toffoli G. Pharmacological Strategies to Prevent SARS-CoV-2 Infection and Treat the Early Phases of COVID-19. Int J Infect Dis (2021) 104:441–51. doi: 10.1016/j.ijid.2021.01.035
170. Tscherne A, Schwarz JH, Rohde C, Kupke A, Kalodimou G, Limpinsel L, et al. Immunogenicity and Efficacy of the COVID-19 Candidate Vector Vaccine MVA-SARS-2-S in Preclinical Vaccination. Proc Natl Acad Sci USA (2021) 118:e2026207118. doi: 10.1073/pnas.2026207118
171. Momin T, Kansagra K, Patel H, Sharma S, Sharma B, Patel J, et al. Safety and Immunogenicity of a DNA SARS-CoV-2 Vaccine (ZyCoV-D): Results of an Open-Label, non-Randomized Phase I Part of Phase I/II Clinical Study by Intradermal Route in Healthy Subjects in India. EClinicalMedicine (2021) 38:101020. doi: 10.1016/j.eclinm.2021.101020
172. Dey A, Chozhavel RT, Chandra H, Pericherla H, Kumar S, Choonia HS, et al. Immunogenic Potential of DNA Vaccine Candidate, ZyCoV-D Against SARS-CoV-2 in Animal Models. Vaccine (2021) 39:4108–16. doi: 10.1016/j.vaccine.2021.05.098
173. Tebas P, Yang S, Boyer JD, Reuschel EL, Patel A, Christensen-Quick A, et al. Safety and Imunogenicity of INO-4800 DNA Vaccine Against SARS-CoV-2: A Preliminary Report of an Open-Label, Phase 1 Clinical Trial. EClinicalMedicine (2021) 31:100689. doi: 10.1016/j.eclinm.2020.100689
174. Smith T, Patel A, Ramos S, Elwood D, Zhu X, Yan J, et al. Immunogenicity of a DNA Vaccine Candidate for COVID-19. Nat Commun (2020) 11:2601. doi: 10.1038/s41467-020-16505-0
175. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
176. Liu Y, Liu J, Xia H, Zhang X, Zou J, Fontes-Garfias CR, et al. BNT162b2-Elicited Neutralization Against New SARS-CoV-2 Spike Variants. N Engl J Med (2021) 385:472–4. doi: 10.1056/NEJMc2106083
177. Baden LR, El SH, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
178. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med (2020) 383:1920–31. doi: 10.1056/NEJMoa2022483
179. Alexandersen S, Chamings A, Bhatta TR. SARS-CoV-2 Genomic and Subgenomic RNAs in Diagnostic Samples Are not an Indicator of Active Replication. Nat Commun (2020) 11:6059. doi: 10.1038/s41467-020-19883-7
180. Rauch S, Roth N, Schwendt K, Fotin-Mleczek M, Mueller SO, Petsch B. MRNA-Based SARS-CoV-2 Vaccine Candidate CVnCoV Induces High Levels of Virus-Neutralising Antibodies and Mediates Protection in Rodents. NPJ Vaccine (2021) 6:57. doi: 10.1038/s41541-021-00311-w
181. Rappaport AR, Hong S, Scallan CD, Gitlin L, Akoopie A, Boucher GR, et al. A Self-Amplifying mRNA COVID-19 Vaccine Drives Potent and Broad Immune Responses at Low Doses That Protects Non-Human Primates Against SARS-CoV-2. bioRxiv (2021) 2011–21. doi: 10.1101/2021.11.08.467773
182. Karpinski TM, Ozarowski M, Seremak-Mrozikiewicz A, Wolski H, Wlodkowic D. The 2020 Race Towards SARS-CoV-2 Specific Vaccines. Theranostics (2021) 11:1690–702. doi: 10.7150/thno.53691
183. Zhang NN, Li XF, Deng YQ, Zhao H, Huang YJ, Yang G, et al. A Thermostable mRNA Vaccine Against COVID-19. Cell (2020) 182:1271–83. doi: 10.1016/j.cell.2020.07.024
184. Abd ET, Stockand JD. Recent Progress and Challenges in Drug Development Against COVID-19 Coronavirus (SARS-CoV-2) - An Update on the Status. Infect Genet Evol (2020) 83:104327. doi: 10.1016/j.meegid.2020.104327
185. Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients With Covid-19. N Engl J Med (2021) 384:229–37. doi: 10.1056/NEJMoa2029849
186. Yang L, Liu S, Liu J, Zhang Z, Wan X, Huang B, et al. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct Target Ther (2020) 5:128. doi: 10.1038/s41392-020-00243-2
187. Li Y, Qi L, Bai H, Sun C, Xu S, Wang Y, et al. Safety, Tolerability, Pharmacokinetics, and Immunogenicity of a Monoclonal Antibody (SCTA01) Targeting SARS-CoV-2 in Healthy Adults: A Randomized, Double-Blind, Placebo-Controlled, Phase I Study. Antimicrob Agents Chemother (2021) 65:e106321. doi: 10.1128/AAC.01063-21
188. Tian JH, Patel N, Haupt R, Zhou H, Weston S, Hammond H, et al. SARS-CoV-2 Spike Glycoprotein Vaccine Candidate NVX-CoV2373 Immunogenicity in Baboons and Protection in Mice. Nat Commun (2021) 12:372. doi: 10.1038/s41467-020-20653-8
189. Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med (2020) 383:2320–32. doi: 10.1056/NEJMoa2026920
190. Chen WH, Hotez PJ, Bottazzi ME. Potential for Developing a SARS-CoV Receptor-Binding Domain (RBD) Recombinant Protein as a Heterologous Human Vaccine Against Coronavirus Infectious Disease (COVID)-19. Hum Vaccin Immunother (2020) 16:1239–42. doi: 10.1080/21645515.2020.1740560
191. Lee J, Liu Z, Chen WH, Wei J, Kundu R, Adhikari R, et al. Process Development and Scale-Up Optimization of the SARS-CoV-2 Receptor Binding Domain-Based Vaccine Candidate, RBD219-N1C1. Appl Microbiol Biotechnol (2021) 105:4153–65. doi: 10.1007/s00253-021-11281-3
192. Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, et al. A Pan-Coronavirus Fusion Inhibitor Targeting the HR1 Domain of Human Coronavirus Spike. Sci Adv (2019) 5:v4580. doi: 10.1126/sciadv.aav4580
193. Lu L, Liu Q, Zhu Y, Chan KH, Qin L, Li Y, et al. Structure-Based Discovery of Middle East Respiratory Syndrome Coronavirus Fusion Inhibitor. Nat Commun (2014) 5:3067. doi: 10.1038/ncomms4067
194. Xia QD, Xun Y, Lu JL, Lu YC, Yang YY, Zhou P, et al. Network Pharmacology and Molecular Docking Analyses on Lianhua Qingwen Capsule Indicate Akt1 is a Potential Target to Treat and Prevent COVID-19. Cell Prolif (2020) 53:e12949. doi: 10.1111/cpr.12949
195. Yan H, Zou C. Mechanism and Material basis of Lianhua Qingwen Capsule for Improving Clinical Cure Rate of COVID-19: A Study Based on Network Pharmacology and Molecular Docking Technology. Nan Fang Yi Ke Da Xue Xue Bao (2021) 41:20–30. doi: 10.12122/j.issn.1673-4254.2021.01.03
196. Yang R, Liu H, Bai C, Wang Y, Zhang X, Guo R, et al. Chemical Composition and Pharmacological Mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction Against Coronavirus Disease 2019 (COVID-19): In Silico and Experimental Study. Pharmacol Res (2020) 157:104820. doi: 10.1016/j.phrs.2020.104820
197. Li X, Tang H, Tang Q, Chen W. Decoding the Mechanism of Huanglian Jiedu Decoction in Treating Pneumonia Based on Network Pharmacology and Molecular Docking. Front Cell Dev Biol (2021) 9:638366. doi: 10.3389/fcell.2021.638366
198. Du H, Wang P, Ma Q, Li N, Ding J, Sun T, et al. Preliminary Study on the Effective Components and Mechanism of Huoxiang Zhengqi Decoction in Inhibiting the Replication of Novel Coronavirus. World Sci Technol-Modern Tradit Chin Med Mater Med (2020) 22:645–51. doi: 10.11842/wst.20200221002
199. Qin F, Li S, Sun Y, Liu Y, Peng S. Molecular Mechanism of Xuebijing Treating ARDS Caused by SARS-CoV-2 Based on Network Pharmacology and Molecular Docking. Pharmacol Clinics Chin Mater Med (2020) 36:21–8. doi: 10.13412/j.cnki.zyyl.20200528.001
200. Feng Y, Xie Y, Wang Y, Lian Q, Wang Y, Luo G, et al. Molecular Mechanism of Xuebijing Injection in treatment of Sepsis According to “Drug-Target-Pathway” Network. Acta Pharm Sin (2017) 52:556–62. doi: 10.16438/j.0513-4870.2016-1048
201. Zhang Q, Cao F, Wang Y, Xu X, Sun Y, Li J, et al. The Efficacy and Safety of Jinhua Qinggan granule (JHQG) in the Treatment of Coronavirus Disease 2019 (COVID-19): A Protocol for Systematic Review and Meta Analysis. Med (Baltimore) (2020) 99:e20531. doi: 10.1097/MD.0000000000020531
202. Liu Z, Li X, Gou C, Li L, Luo X, Zhang C, et al. Effect of Jinhua Qinggan Granules on Novel Coronavirus Pneumonia in Patients. J Tradit Chin Med (2020) 40:467–72. doi: 10.19852/j.cnki.jtcm.2020.03.016
203. Zhang X, Xue Y, Chen X, Wu JM, Su ZJ, Sun M, et al. Effects of Tanreqing Capsule on the Negative Conversion Time of Nucleic Acid in Patients With COVID-19: A Retrospective Cohort Study. J Integr Med (2021) 19:36–41. doi: 10.1016/j.joim.2020.10.002
204. Tao Q, Du J, Li X, Zeng J, Tan B, Xu J, et al. Network Pharmacology and Molecular Docking Analysis on Molecular Targets and Mechanisms of Huashi Baidu Formula in the Treatment of COVID-19. Drug Dev Ind Pharm (2020) 46:1345–53. doi: 10.1080/03639045.2020.1788070
205. Cai Y, Zeng M, Chen YZ. The Pharmacological Mechanism of Huashi Baidu Formula for the Treatment of COVID-19 by Combined Network Pharmacology and Molecular Docking. Ann Palliat Med (2021) 10:3864–95. doi: 10.21037/apm-20-1759
206. Chen X, Yin YH, Zhang MY, Liu JY, Li R, Qu YQ. Investigating the Mechanism of ShuFeng JieDu Capsule for the Treatment of Novel Coronavirus Pneumonia (COVID-19) Based on Network Pharmacology. Int J Med Sci (2020) 17:2511–30. doi: 10.7150/ijms.46378
207. Xia L, Shi Y, Su J, Friedemann T, Tao Z, Lu Y, et al. Shufeng Jiedu, a Promising Herbal Therapy for Moderate COVID-19: Antiviral and Anti-Inflammatory Properties, Pathways of Bioactive Compounds, and a Clinical Real-World Pragmatic Study. Phytomedicine (2021) 85:153390. doi: 10.1016/j.phymed.2020.153390
208. Li Y, Li B, Wang P, Wang Q. Traditional Chinese Medicine, Qingfei Paidu Decoction and Xuanfei Baidu Decoction, Inhibited Cytokine Production via NF-kappaB Signaling Pathway in Macrophages: Implications for Coronavirus Disease 2019 (COVID-19) Therapy. Front Pharmacol (2021) 12:722126. doi: 10.3389/fphar.2021.722126
209. Cao C, Zhen Z, Kuang S, Xu T. Reduning Injection Combined With Western Medicine for Pneumonia: A pProtocol for Systematic Review and Meta-Analysis. Med (Baltimore) (2020) 99:e22757. doi: 10.1097/MD.0000000000022757
210. Xu X, Zhang J, Zheng W, Yang Z, Zhao X, Wang C, et al. Efficacy and Safety of Reduning Injection in the Treatment of COVID-19: A Randomized, Multicenter Clinical Study. Ann Palliat Med (2021) 10:5146–55. doi: 10.21037/apm-20-2121
211. Jia S, Luo H, Liu X, Fan X, Huang Z, Lu S, et al. Dissecting the Novel Mechanism of Reduning Injection in Treating Coronavirus Disease 2019 (COVID-19) Based on Network Pharmacology and Experimental Verification. J Ethnopharmacol (2021) 273:113871. doi: 10.1016/j.jep.2021.113871
212. Wang Y, Lu C, Li H, Qi W, Ruan L, Bian Y, et al. Efficacy and Safety Assessment of Severe COVID-19 Patients With Chinese Medicine: A Retrospective Case Series Study at Early Stage of the COVID-19 Epidemic in Wuhan, China. J Ethnopharmacol (2021) 277:113888. doi: 10.1016/j.jep.2021.113888
213. Derosa G, Maffioli P, D'Angelo A, Di Pierro F. A Role for Quercetin in Coronavirus Disease 2019 (COVID-19). Phytother Res (2021) 35:1230–6. doi: 10.1002/ptr.6887
214. Pan B, Fang S, Zhang J, Pan Y, Liu H, Wang Y, et al. Chinese Herbal Compounds Against SARS-CoV-2: Puerarin and Quercetin Impair the Binding of Viral S-Protein to ACE2 Receptor. Comput Struct Biotechnol J (2020) 18:3518–27. doi: 10.1016/j.csbj.2020.11.010
215. Saakre M, Mathew D, Ravisankar V. Perspectives on Plant Flavonoid Quercetin-Based Drugs for Novel SARS-CoV-2. Beni Suef Univ J Basic Appl Sci (2021) 10:21. doi: 10.1186/s43088-021-00107-w
216. Khan A, Heng W, Wang Y, Qiu J, Wei X, Peng S, et al. In Silico and In Vitro Evaluation of Kaempferol as a Potential Inhibitor of the SARS-CoV-2 Main Protease (3CLpro). Phytother Res (2021) 35:2841–5. doi: 10.1002/ptr.6998
217. Theoharides TC. COVID-19, Pulmonary Mast Cells, Cytokine Storms, and Beneficial Actions of Luteolin. Biofactors (2020) 46:306–8. doi: 10.1002/biof.1633
218. Shawan M, Halder SK, Hasan MA. Luteolin and Abyssinone II as Potential Inhibitors of SARS-CoV-2: An In Silico Molecular Modeling Approach in Battling the COVID-19 Outbreak. Bull Natl Res Cent (2021) 45:27. doi: 10.1186/s42269-020-00479-6
219. Zhan Y, Ta W, Tang W, Hua R, Wang J, Wang C, et al. Potential Antiviral Activity of Isorhamnetin Against SARS-CoV-2 Spike Pseudotyped Virus In Vitro. Drug Dev Res (2021) 82:1124–30. doi: 10.1002/ddr.21815
220. Tejera E, Perez-Castillo Y, Toscano G, Noboa AL, Ochoa-Herrera V, Giampieri F, et al. Computational Modeling Predicts Potential Effects of the Herbal Infusion "Horchata" Against COVID-19. Food Chem (2022) 366:130589. doi: 10.1016/j.foodchem.2021.130589
221. Clementi N, Scagnolari C, D'Amore A, Palombi F, Criscuolo E, Frasca F, et al. Naringenin Is a Powerful Inhibitor of SARS-CoV-2 Infection In Vitro. Pharmacol Res (2021) 163:105255. doi: 10.1016/j.phrs.2020.105255
222. Maurya VK, Kumar S, Prasad AK, Bhatt M, Saxena SK. Structure-Based Drug Designing for Potential Antiviral Activity of Selected Natural Products From Ayurveda Against SARS-CoV-2 Spike Glycoprotein and Its Cellular Receptor. Virusdisease (2020) 31:179–93. doi: 10.1007/s13337-020-00598-8
223. D'Amore A, Gradogna A, Palombi F, Minicozzi V, Ceccarelli M, Carpaneto A, et al. The Discovery of Naringenin as Endolysosomal Two-Pore Channel Inhibitor and Its Emerging Role in SARS-CoV-2 Infection. Cells-Basel (2021) 10:1130. doi: 10.3390/cells10051130
224. Yang C, Pan X, Xu X, Cheng C, Huang Y, Li L, et al. Salvianolic Acid C Potently Inhibits SARS-CoV-2 Infection by Blocking the Formation of Six-Helix Bundle Core of Spike Protein. Signal Transduct Target Ther (2020) 5:220. doi: 10.1038/s41392-020-00325-1
225. Wang W, Li SS, Xu XF, Yang C, Niu XG, Yin SX, et al. Danshensu Alleviates Pseudo-typed SARS-CoV-2 Induced Mouse Acute Lung Inflammation. Acta Pharmacol Sin (2021) 0:1–10. doi: 10.1038/s41401-021-00714-4
226. Hu S, Wang J, Zhang Y, Bai H, Wang C, Wang N, et al. Three Salvianolic Acids Inhibit 2019-nCoV Spike Pseudovirus viropexis by Binding to Both Its RBD and Receptor ACE2. J Med Virol (2021) 93:3143–51. doi: 10.1002/jmv.26874
227. Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL Protease by Flavonoids. J Enzyme Inhib Med Chem (2020) 35:145–51. doi: 10.1080/14756366.2019.1690480
228. Zandi K, Musall K, Oo A, Cao D, Liang B, Hassandarvish P, et al. Baicalein and Baicalin Inhibit SARS-CoV-2 RNA-Dependent-RNA Polymerase. Microorganisms (2021) 9:893. doi: 10.3390/microorganisms9050893
229. Rehman M, Akhter S, Batool AI, Selamoglu Z, Sevindik M, Eman R, et al. Effectiveness of Natural Antioxidants Against SARS-CoV-2? Insights From the In-Silico World. Antibiotics (Basel) (2021) 10:1011. doi: 10.3390/antibiotics10081011
230. Huang S, Liu Y, Zhang Y, Zhang R, Zhu C, Fan L, et al. Baicalein Inhibits SARS-CoV-2/VSV Replication With Interfering Mitochondrial Oxidative Phosphorylation in a mPTP Dependent Manner. Signal Transduct Target Ther (2020) 5:266. doi: 10.1038/s41392-020-00353-x
231. Liu H, Ye F, Sun Q, Liang H, Li C, Li S, et al. Scutellaria Baicalensis Extract and Baicalein Inhibit Replication of SARS-CoV-2 and Its 3C-Like Protease In Vitro. J Enzyme Inhib Med Chem (2021) 36:497–503. doi: 10.1080/14756366.2021.1873977
232. Sadegh S, Matschinske J, Blumenthal DB, Galindez G, Kacprowski T, List M, et al. Exploring the SARS-CoV-2 Virus-Host-Drug Interactome for Drug Repurposing. Nat Commun (2020) 11:3518. doi: 10.1038/s41467-020-17189-2
233. Su H, Zhou F, Huang Z, Ma X, Natarajan K, Zhang M, et al. Molecular Insights Into Small-Molecule Drug Discovery for SARS-CoV-2. Angew Chem Int Ed Engl (2021) 60:9789–802. doi: 10.1002/anie.202008835
234. DeFrancesco L. Whither COVID-19 Vaccines? Nat Biotechnol (2020) 38:1132–45. doi: 10.1038/s41587-020-0697-7
235. Sempowski GD, Saunders KO, Acharya P, Wiehe KJ, Haynes BF. Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19. Cell (2020) 181:1458–63. doi: 10.1016/j.cell.2020.05.041
236. Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A Systematic Review of SARS-CoV-2 Vaccine Candidates. Signal Transduct Target Ther (2020) 5:237. doi: 10.1038/s41392-020-00352-y
237. Dash GC, Subhadra S, Turuk J, Parai D, Rath S, Sabat J, et al. Breakthrough SARS-CoV-2 Infections Among BBV-152 (COVAXIN(R)) and AZD1222 (COVISHIELD(TM) ) Recipients: Report From the Eastern State of India. J Med Virol (2021) 94:1201–5. doi: 10.1002/jmv.27382
238. Awadasseid A, Wu Y, Tanaka Y, Zhang W. Current Advances in the Development of SARS-CoV-2 Vaccines. Int J Biol Sci (2021) 17:8–19. doi: 10.7150/ijbs.52569
239. Cevik M, Grubaugh ND, Iwasaki A, Openshaw P. COVID-19 Vaccines: Keeping Pace With SARS-CoV-2 Variants. Cell (2021) 184:5077–81. doi: 10.1016/j.cell.2021.09.010
240. Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological Considerations for COVID-19 Vaccine Strategies. Nat Rev Immunol (2020) 20:615–32. doi: 10.1038/s41577-020-00434-6
241. van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, et al. ChAdOx1 Ncov-19 Vaccine Prevents SARS-CoV-2 Pneumonia in Rhesus Macaques. Nature (2020) 586:578–82. doi: 10.1038/s41586-020-2608-y
242. Yu J, Tostanoski LH, Mercado NB, McMahan K, Liu J, Jacob-Dolan C, et al. Protective Efficacy of Ad26.COV2.S Against SARS-CoV-2 B.1.351 in Macaques. Nature (2021) 596:423–7. doi: 10.1038/s41586-021-03732-8
243. Pushparajah D, Jimenez S, Wong S, Alattas H, Nafissi N, Slavcev RA. Advances in Gene-Based Vaccine Platforms to Address the COVID-19 Pandemic. Adv Drug Deliv Rev (2021) 170:113–41. doi: 10.1016/j.addr.2021.01.003
244. Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, et al. DNA Vaccine Protection Against SARS-CoV-2 in Rhesus Macaques. Science (2020) 369:806–11. doi: 10.1126/science.abc6284
245. Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA Vaccine-Elicited Antibodies to SARS-CoV-2 and Circulating Variants. Nature (2021) 592:616–22. doi: 10.1038/s41586-021-03324-6
246. Callaway E. Coronavirus Vaccines Leap Through Safety Trials - But Which Will Work is Anybody's Guess. Nature (2020) 583:669–70. doi: 10.1038/d41586-020-02174-y
247. Atanasov AG, Zotchev SB, Dirsch VM, Supuran CT. Natural Products in Drug Discovery: Advances and Opportunities. Nat Rev Drug Discov (2021) 20:200–16. doi: 10.1038/s41573-020-00114-z
248. Lyu M, Fan G, Xiao G, Wang T, Xu D, Gao J, et al. Traditional Chinese Medicine in COVID-19. Acta Pharm Sin B (2021) 11:3337–63. doi: 10.1016/j.apsb.2021.09.008
249. Xing D, Liu Z. Effectiveness and Safety of Traditional Chinese Medicine in Treating COVID-19: Clinical Evidence From China. Aging Dis (2021) 12:1850–6. doi: 10.14336/AD.2021.0906
250. Runfeng L, Yunlong H, Jicheng H, Weiqi P, Qinhai M, Yongxia S, et al. Lianhuaqingwen Exerts Anti-Viral and Anti-Inflammatory Activity Against Novel Coronavirus (SARS-CoV-2). Pharmacol Res (2020) 156:104761. doi: 10.1016/j.phrs.2020.104761
251. Yan H, Zou Y, Zou C. [Mechanism of Qingfei Paidu Decoction for Treatment of COVID-19: Analysis Based on Network Pharmacology and Molecular Docking Technology]. Nan Fang Yi Ke Da Xue Xue Bao (2020) 40:616–23. doi: 10.12122/j.issn.1673-4254.2020.05.02
252. Huang YF, Bai C, He F, Xie Y, Zhou H. Review on the Potential Action Mechanisms of Chinese Medicines in Treating Coronavirus Disease 2019 (COVID-19). Pharmacol Res (2020) 158:104939. doi: 10.1016/j.phrs.2020.104939
253. Tong T, Wu YQ, Ni WJ, Shen AZ, Liu S. The Potential Insights of Traditional Chinese Medicine on Treatment of COVID-19. Chin Med (2020) 15:51. doi: 10.1186/s13020-020-00326-w
254. Su H, Yao S, Zhao W, Li M, Liu J, Shang W, et al. Discovery of Baicalin and Baicalein as Novel, Natural Product Inhibitors of SARS-CoV-2 3CL Protease In Vitro. bioRxiv (2020), 2020–4. doi: 10.1101/2020.04.13.038687
255. Wang H, Xu B, Zhang Y, Duan Y, Gao R, He H, et al. Efficacy and Safety of Traditional Chinese Medicine in Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis. Front Pharmacol (2021) 12:609213. doi: 10.3389/fphar.2021.609213
256. Liu M, Gao Y, Yuan Y, Yang K, Shi S, Zhang J, et al. Efficacy and Safety of Integrated Traditional Chinese and Western Medicine for Corona Virus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis. Pharmacol Res (2020) 158:104896. doi: 10.1016/j.phrs.2020.104896
257. Naldi L, Cazzaniga S. More on Covid-19 in Immune-Mediated Inflammatory Diseases. N Engl J Med (2020) 383:795–6. doi: 10.1056/NEJMc2018011
258. Ramlall V, Thangaraj PM, Meydan C, Foox J, Butler D, Kim J, et al. Immune Complement and Coagulation Dysfunction in Adverse Outcomes of SARS-CoV-2 Infection. Nat Med (2020) 26:1609–15. doi: 10.1038/s41591-020-1021-2
259. Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological Findings of COVID-19 Associated With Acute Respiratory Distress Syndrome. Lancet Respir Med (2020) 8:420–2. doi: 10.1016/S2213-2600(20)30076-X
260. Kim JS, Lee JY, Yang JW, Lee KH, Effenberger M, Szpirt W, et al. Immunopathogenesis and Treatment of Cytokine Storm in COVID-19. Theranostics (2021) 11:316–29. doi: 10.7150/thno.49713
261. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature (2021) 593:130–5. doi: 10.1038/s41586-021-03398-2
262. Soin AS, Kumar K, Choudhary NS, Sharma P, Mehta Y, Kataria S, et al. Tocilizumab Plus Standard Care Versus Standard Care in Patients in India With Moderate to Severe COVID-19-Associated Cytokine Release Syndrome (COVINTOC): An Open-Label, Multicentre, Randomised, Controlled, Phase 3 Trial. Lancet Respir Med (2021) 9:511–21. doi: 10.1016/S2213-2600(21)00081-3
263. Ge Y, Tian T, Huang S, Wan F, Li J, Li S, et al. An Integrative Drug Repositioning Framework Discovered a Potential Therapeutic Agent Targeting COVID-19. Signal Transduct Target Ther (2021) 6:165. doi: 10.1038/s41392-021-00568-6
264. Perez-Fernandez XL, Sabater-Riera J, Fuset-Cabanes M. COVID-19 ARDS: Getting Ventilation Right. Lancet (2022) 399:22. doi: 10.1016/S0140-6736(21)02439-9
265. O'Donnell JS, Peyvandi F, Martin-Loeches I. Pulmonary Immuno-Thrombosis in COVID-19 ARDS Pathogenesis. Intensive Care Med (2021) 47:899–902. doi: 10.1007/s00134-021-06419-w
266. Aslan A, Aslan C, Zolbanin NM, Jafari R. Acute Respiratory Distress Syndrome in COVID-19: Possible Mechanisms and Therapeutic Management. Pneumonia (Nathan) (2021) 13:14. doi: 10.1186/s41479-021-00092-9
267. Dyavar SR, Singh R, Emani R, Pawar GP, Chaudhari VD, Podany AT, et al. Role of Toll-Like Receptor 7/8 Pathways in Regulation of Interferon Response and Inflammatory Mediators During SARS-CoV2 Infection and Potential Therapeutic Options. BioMed Pharmacother (2021) 141:111794. doi: 10.1016/j.biopha.2021.111794
268. Li G, De Clercq E. Therapeutic Options for the 2019 Novel Coronavirus (2019-Ncov). Nat Rev Drug Discov (2020) 19:149–50. doi: 10.1038/d41573-020-00016-0
269. Yin P, Meng J, Chen J, Gao J, Wang D, Liu S, et al. Antiviral Drugs Arbidol and Interferon Alpha-1b Contribute to Reducing the Severity of COVID-19 Patients: A Retrospective Cohort Study. Virol J (2021) 18:142. doi: 10.1186/s12985-021-01617-w
270. Matthay MA, Thompson BT. Dexamethasone in Hospitalised Patients With COVID-19: Addressing Uncertainties. Lancet Respir Med (2020) 8:1170–2. doi: 10.1016/S2213-2600(20)30503-8
271. Casadevall A, Joyner MJ, Pirofski LA. SARS-CoV-2 Viral Load and Antibody Responses: The Case for Convalescent Plasma Therapy. J Clin Invest (2020) 130:5112–4. doi: 10.1172/JCI139760
272. Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. Effectiveness of Convalescent Plasma Therapy in Severe COVID-19 Patients. Proc Natl Acad Sci U S A (2020) 117:9490–6. doi: 10.1073/pnas.2004168117
Keywords: severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), coronavirus disease 2019 (COVID-19), small molecular inhibitor, vaccine, traditional Chinese medicine, potential target, targeted therapeutic strategy
Citation: Zhou H, Ni W-J, Huang W, Wang Z, Cai M and Sun Y-C (2022) Advances in Pathogenesis, Progression, Potential Targets and Targeted Therapeutic Strategies in SARS-CoV-2-Induced COVID-19. Front. Immunol. 13:834942. doi: 10.3389/fimmu.2022.834942
Received: 14 December 2021; Accepted: 07 March 2022;
Published: 05 April 2022.
Edited by:
Fabio Bagnoli, GlaxoSmithKline, ItalyReviewed by:
Shetty Ravi Dyavar, Adicet Bio, Inc, United StatesWen Zhang, Zhejiang University of Technology, China
Tomohiro Tanaka, Tokyo University of Science, Japan
Copyright © 2022 Zhou, Ni, Huang, Wang, Cai and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Jian Ni, bml3ZWlqaWFuQHVzdGMuZWR1LmNu; Ming Cai, Y2FpbWluZ0BhaHRjbS5lZHUuY24=; Yan-Cai Sun, MTMzNDkyOTMzNTlAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Hong Zhou
Hong Zhou Wei-Jian Ni
Wei-Jian Ni Wei Huang
Wei Huang Zhen Wang
Zhen Wang Ming Cai
Ming Cai Yan-Cai Sun
Yan-Cai Sun