
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 24 February 2022
Sec. Vaccines and Molecular Therapeutics
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.827605
 Jinni Chen1,2
Jinni Chen1,2 Yao Deng2
Yao Deng2 Baoying Huang2
Baoying Huang2 Di Han2,3
Di Han2,3 Wen Wang2
Wen Wang2 Mengjing Huang2,3
Mengjing Huang2,3 Chengcheng Zhai2,4
Chengcheng Zhai2,4 Zhimin Zhao2
Zhimin Zhao2 Ren Yang2
Ren Yang2 Ying Zhao5
Ying Zhao5 Wenling Wang2
Wenling Wang2 Desheng Zhai1*
Desheng Zhai1* Wenjie Tan1,2*
Wenjie Tan1,2*The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a public health emergency of international concern, and an effective vaccine is urgently needed to control the pandemic. Envelope (E) and membrane (M) proteins are highly conserved structural proteins among SARS-CoV-2 and SARS-CoV and have been proposed as potential targets for the development of cross-protective vaccines. Here, synthetic DNA vaccines encoding SARS-CoV-2 E/M proteins (called p-SARS-CoV-2-E/M) were developed, and mice were immunised with three doses via intramuscular injection and electroporation. Significant cellular immune responses were elicited, whereas no robust humoral immunity was detected. In addition, novel H-2d-restricted T-cell epitopes were identified. Notably, although no drop in lung tissue virus titre was detected in DNA-vaccinated mice post-challenge with SARS-CoV-2, immunisation with either p-SARS-CoV-2-E or p-SARS-CoV-2-M provided minor protection and co-immunisation with p-SARS-CoV-2-E+M increased protection. Therefore, E/M proteins should be considered as vaccine candidates as they may be valuable in the optimisation of vaccination strategies against COVID-19.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19), is the third novel betacoronavirus belonging to highly pathogenic human coronaviruses that have caused public health crises in the past 20 years (1). It transmits more efficiently among the population compared with its predecessors, SARS-CoV and Middle East respiratory syndrome coronavirus. Globally, SARS-CoV-2 has infected more than 260 million people, resulting in more than 5 million deaths worldwide as of November 2021 (2). Therefore, the rapid development of effective therapies and vaccines against SARS-CoV-2 and emerging coronaviruses is a critical global priority (3).
Human coronaviruses are enveloped positive-sense RNA viruses. The genetic components of coronaviruses encode four major structural proteins—spike (S), nucleocapsid (N), envelope (E), and membrane (M) proteins. The S protein is the target of neutralising antibodies during infection and is important for the development of coronavirus vaccines (4). However, SARS-CoV-2 variants are emerging in different parts of the world, posing a new threat of increased virus spread and the potential to escape vaccine-induced immunity. Most of the mutations in these variants are within the S protein (5); thus, raises the concern that monovalent vaccines targeting only the S protein may not be the most optimal strategy for conferring protection against continually emerging variants (6).
The E protein is a small ion channel-forming membrane protein (75 amino acids; ∼8.4 kDa) that plays a significant role in viral morphogenesis and assembly (6). The M protein is the most abundant protein and is approximately 222 amino acid residues in length. It interacts with other structural viral proteins and plays a central organising role in coronavirus assembly (7). Both proteins are highly conserved structural proteins of SARS-CoV-2 and SARS-CoV (8). Several reports have shown that the co-synthesis of E and M proteins is sufficient for virus-like particle assembly (9–11). Therefore, the E/M proteins are potential targets for the development of SARS-CoV-2 cross-protective vaccines. However, few studies have considered these proteins as major targets for the development of SARS-CoV-2 vaccines (12).
A variety of SARS-CoV-2 vaccines are currently being developed and some have been approved, including inactivated-, subunit-, vector-, mRNA-, and DNA vaccines (13, 14). Although several vaccines against COVID-19 are promising, developing more effective and cross-protective vaccines is urgently required. Synthetic DNA vaccines are developed at an accelerated rate because of the quick design of multiple candidates for preclinical testing in comparison to other vaccines (15–17). Currently, to the best of our knowledge, the immunogenicity and protective potential of synthetic DNA vaccines encoding the SARS-CoV-2 E/M proteins have not been reported.
To explore the immune protective potential of the SARS-CoV-2 E and M proteins as vaccine targets synthetic DNA vaccines expressing the E and M proteins were developed and their immunogenicity and protective efficacy in mice were evaluated in this study.
The full-length genes encoding the SARS-CoV-2 E and M proteins (GISAID, No. EPI_ISL_402119) were synthesised using a mammalian-optimised codon with a N-terminal Kozak sequence(GCCACC) followed by initiation codon(ATG) and a C-terminal 6x His tag (GenScript Co., Nanjing, China). Subsequently, they were inserted into the eukaryotic expression vector, pcDNA3.1 (+), via HindIII and XbaI digestion and named as p-SARS-CoV-2-E or p-SARS-CoV-2-M (Figure 1A). All DNA vaccine sequences were confirmed by Sanger DNA sequencing, and vaccines were expanded using endotoxin-free Maxiprep kits (Qiagen, Beijing, China). The expression of the E/M proteins was identified by an indirect immunofluorescence assay (IFA) and western blotting.

Figure 1 Design and expression of recombinant DNA-based SARS-CoV-2 E/M proteins vaccine constructs. (A) Schematic diagram of the recombinant DNA-based vaccines encoding severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) envelope (E)/membrane (M) protein genes. (B) E/M protein expression in DNA vaccines were tested by indirect immunofluorescence staining and (C) western blot in 293T cells transfected with either of the pSARS-CoV-2-E/M plasmids.
Human embryonic kidney 293T cells were grown in Dulbecco’s modified Eagle’s medium (Hyclone, South Logan, UT, USA) containing 10% foetal bovine serum (Gibco, NY, USA) and 1% penicillin-streptomycin (Gibco, Grand Island, NY, USA) in a 5% CO2 incubator at 37°C. 293T cells were transfected with either the p-SARS-CoV-2 E/M or pcDNA3.1 (empty/mock) vector using jet PRIME transfection reagent (Polyplus, Illkirch, France). Cells were fixed in pre-cooled 4% paraformaldehyde, mobilised in 0.2% Triton X-100, and blocked by 10% goat serum in phosphate-buffered saline. Anti-6X His tag antibody (Abcam, Cambridge, UK) diluted at 1:50 was used as the primary antibody. After incubation, cells were washed and incubated with secondary antibodies [fluorescein isothiocyanate(FITC)-labelled goat anti-rabbit IgG] and 0.1% 4′,6-diamino-2-phenylindol (DAPI) at 37°C for 10 min. Fluorescent images were acquired using a Leica TCS SP8 confocal microscope with LAS software (Leica Biosystems, Wetzlar, Germany).
The expression of the E or M protein was confirmed by western blotting, as previously described (18). The anti-6× His tag antibody (Abcam, UK) diluted at 1:500 or serum antibodies (diluted at 1/10) from DNA-immunised mice were used as primary antibodies. A 1:5,000 dilution of anti-mouse horseradish peroxidase-conjugated antibody (Sigma Aldrich, St Louis, MO, USA) was used as the secondary antibody. The membranes were developed with a chemiluminescent substrate and analysed using a chemiluminescent imager.
All experiments were approved by the Committee on the Ethics of Animal Experiments of the Chinese Centre for Disease Control and Prevention where all live SARS-CoV-2 mice experiments were performed in animal biosafety level 3 containment laboratories at the National Institute for Viral Disease Control and Prevention.
For animal immunisation, 6-week-old female BALB/c mice (SPF grade) were purchased from the Beijing Vital River Laboratory Animal Technology, housed, and vaccinated at 25°C in a light-cycled facility (12 h light/12 h dark).”
Mice were divided randomly into groups (Figure 2) and immunised with pcDNA3.1 (+), p-SARS-CoV-2-E/M alone or co-immunised with p-SARS-CoV-2-E+M, on days 0, 21, and 42 via intramuscular injection plus electroporation (35μg/50μl) (19, 20). In brief, DNA vaccine were injected into the TA muscle of mice and were immediately pulsed with electricity using a two-needle array electrode (ECM830; BTX) with needles that were 5 mm apart. Their spleens were processed to measure cellular immune responses to E or M antigens, and their sera were collected and used to analyse humoral immune responses.
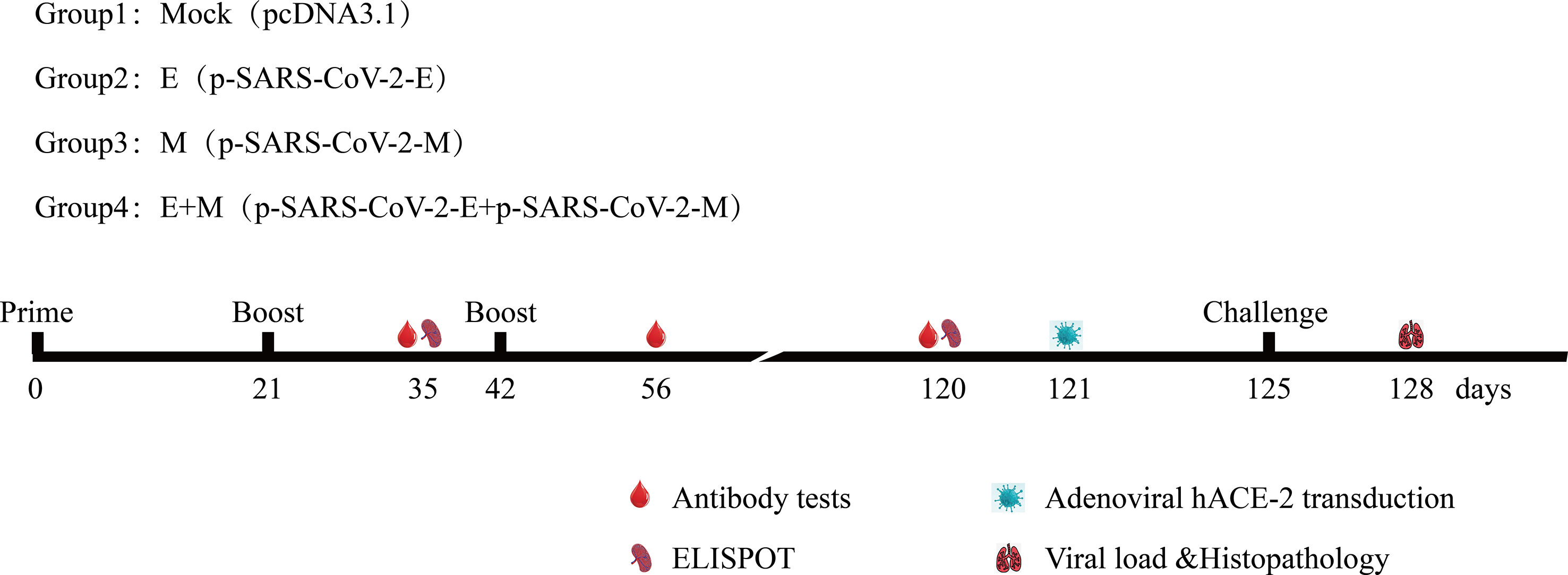
Figure 2 Immunisation and challenging schema of recombinant DNA-based SARS-CoV-2 E/M proteins coronavirus disease 2019 vaccines. Vaccination, challenging, and blood/tissue sampling time course. BALB/c mice were divided randomly into groups.
Viral challenge experiments were conducted as described previously (21). Ad5-hACE2-transduced SARS-CoV-2 mice were intranasally infected with 1×105 median tissue culture infective dose (TCID50) of SARS-CoV-2 (Wuhan/IVDC-HB-02/2019) in a total volume of 50 μL.
Either the E peptide pool or M peptide pool spanned the entire protein as consecutive 15-mers overlapping by 10 amino acids were synthesised by Scilight Biotechnology LLC. Each purified peptide of the peptide pool was at 2.5 mg per vial. The peptides were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 50 mg/ml and stored at -80°C. The experiment was conducted as described previously (22).
Synthetic extracellular peptides of the E/M proteins coupled with bovine serum albumin (synthetized by Scilight Biotechnology LLC) or E/M proteins (purchased from Unique Biotechnology LLC) diluted in carbonate buffer (0.1 M; pH 9.6) were used to coat 96-well enzyme immunoassay/radioimmunoassay plates (Thermo Fisher Scientific, Waltham, MA, USA) overnight at 4°C. ELISA was conducted as described previously (21).
The experiment was conducted in a biosafety level 3 laboratory as previously described (21).
Three days post-challenge, mice were euthanised, and necropsy was performed. Lungs of mice were harvested after sacrifice (four mice per group). Partial tissues were used for nucleic acid extraction and real-time fluorescence RT-PCR to quantify the relative amount of viral RNA in lungs as previously described (21). The TCID50 of the virus in samples was determined as previously described (21). Remaining tissue samples were fixed in a 4% formalin solution and sent to the College of Veterinary Medicine, China Agricultural University, for the preparation of haematoxylin and eosin-stained sections (four mice per group) for pathological evaluation indicated by the International Harmonisation of Nomenclature and Diagnostic Criteria (INHAND) scores.
All statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Prism Software Inc., San Diego, CA, USA). One-way ANOVA with Dunnett’s multiple comparisons test was performed to evaluate the statistical significance of differences among groups. Statistical significance was set at P<0.05.
Expression of E and M proteins in 293T cells transfected with p-SARS-CoV-2-E/M was detected by IFA (Figure 1B) and western blotting (Figure 1C). IFA showed the expression of E and M proteins in the membrane and endoplasmic area of HEK-293T cells transfected with p-SARS-CoV-2-E/M. Western blot results revealed approximately 10 kDa and 25 kDa bands that were predicted as E and M proteins based on the molecular weight in the lysates of HEK-293T cells transfected with p-SARS-CoV-2-E/M. Two bands of M proteins means that some M proteins can undergo maturation leading to N-glycosylated M proteins found in infected cells (23). This has also been observed in the case of SARS-CoV-2, revealed by two specific bands for M proteins using immunoblotting (24). No protein expression was detected in pcDNA3.1 (+)-transfected cells.
To systemically analyse the H-2d-restricted T-cell epitopes for SARS-CoV-2 E and M proteins, peptide pools consisting of 10 consecutive 15-mer overlapping peptides of the E/M proteins were used for IFN-γ ELISpot assay screening in BALB/c mice after vaccination with p-SARS-CoV-2-E/M. Individual peptide reactivity analysis showed that E-07, E-11, E-12, M-07, M-08, and M-29 contained immunodominant epitopes, the amino acid sequences of which are shown in Figure 3.
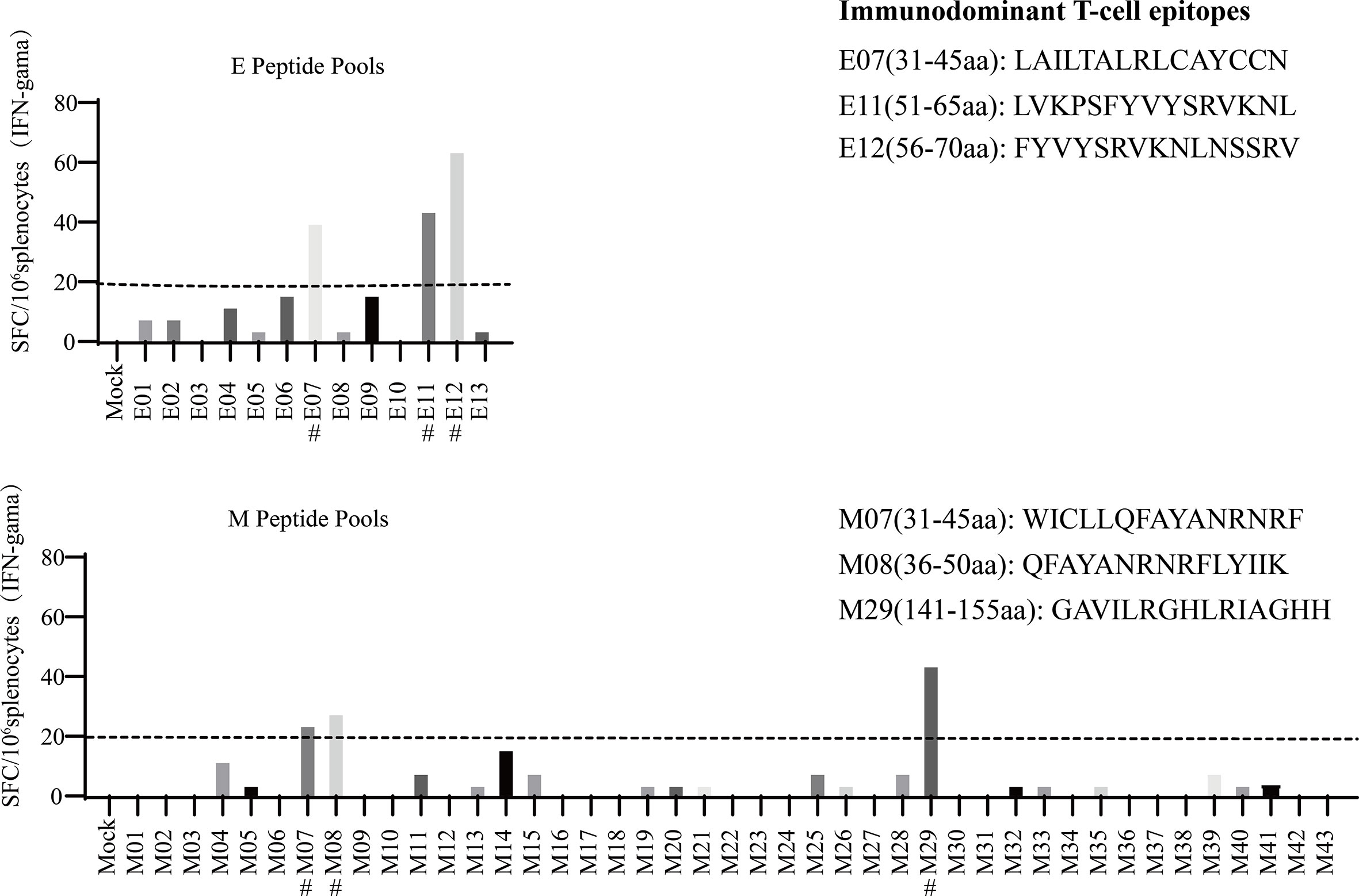
Figure 3 Mapping severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) T-cell epitopes in BALB/c mice. The fifty-six 15-mer overlapping peptides that cover the entire sequence of the SARS-CoV-2 E/M proteins were used in an enzyme-linked immune absorbent spot (ELISPOT) assay to measure the immunodominant T-cell epitopes. Candidate T-cell epitopes are labelled with #.
As shown in Figure 4A, a significant level of IFN-γ production was observed in the p-SARS-CoV-2-E/M-immunised and p-SARS-CoV-2-E+M-immunised groups, while no IFN-γ secretion was detected in the mock group. Co-immunisation with p-SARS-CoV-2-E+M induced higher levels of E protein-specific IFN-γ secretion than immunisation with p-SARS-CoV-2-E alone on days 35 (E+M: 118 spot-forming units [SFU]/106 splenocytes vs. E: 49 SFU/106 splenocytes; P<0.05) and 120 (E+M: 78 SFU/106 splenocytes vs. E: 41 SFU/106 splenocytes; P<0.05). However, co-immunisation with p-SARS-CoV-2 E+M did not induce higher levels of M-protein-specific IFN-γ secretion than immunisation with p-SARS-CoV-2 M alone on days 35 (E+M: 68 SFU/106 splenocytes vs. M: 100 SFU/106 splenocytes) and 120 (E+M: 54 SFU/106 splenocytes vs M: 93 SFU/106 splenocytes).
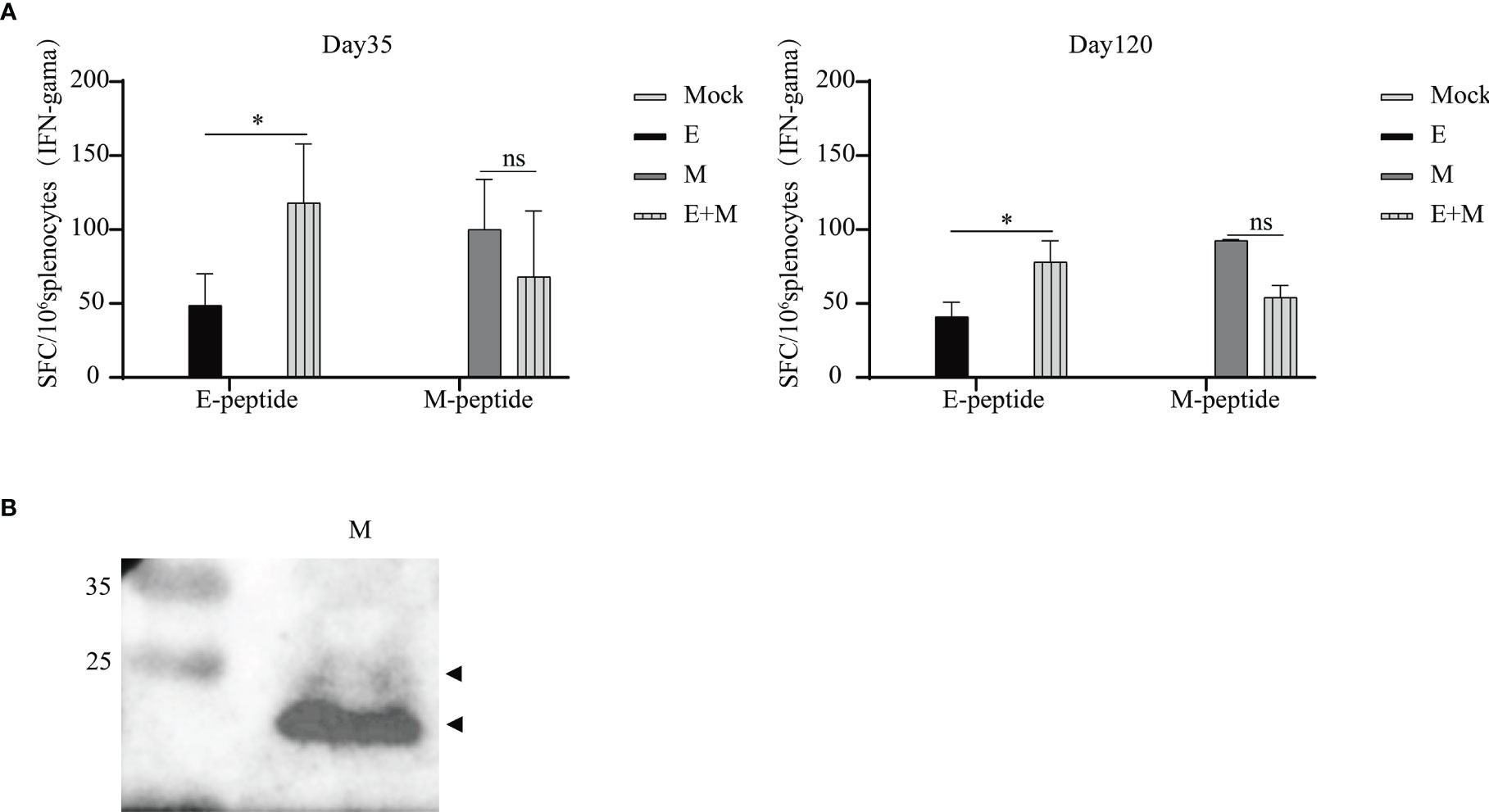
Figure 4 T and B cell immunity induced upon immunisation of mice with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) envelope (E) and/or membrane (M) proteins. (A) Splenocytes were isolated from mice (n=3 or 4 mice per group per time point) and stimulated with E/M peptides at days 35 and 120 after the 1st vaccination. (B) M protein-specific antibodies were detected by western blot analysis of purified SARS-CoV-2 particles (inactivated vaccine stock). The statistical analysis among groups was analysed by two-way ANOVA after Tukey’s multiple comparison (*P<0.05). ns, no significance.
ELISA results from plates coated with E/M proteins (AtaGenix Laboratories, Wuhan, China) or E/M peptides (Scilight Biotechnology LLC,Beijing, China) did not indicate robust E/M protein-specific antibody responses after the first or second vaccination (data not shown). Moreover, neutralising anti-E/M IgG were not detected in the sera (1:10 dilution) of immunised mice (data not shown). However, antibodies against M protein in mice could be detected by western blot (Figure 4B), and the M-specific band (as predicted by size) was observed when purified SARS-CoV-2 particles (inactivated vaccine stock) were loaded and incubated with serum (diluted at 1/10) of p-SARS-CoV-2-M-immunised mice.
We determined whether there was enhanced protection against SARS-CoV-2 challenge in p-SARS-CoV-2-E/M- and p-SARS-CoV-2-E+M-immunised mice, compared to the control group by evaluating protective effects of the DNA vaccine in mice on day 125. Tissue viral load (RNA copies and TCID50) and histopathological changes were evaluated (Figure 5). Lung viral load in DNA vaccine-immunised mice did not significantly decrease compared to that in the control group (Figure 5A). Lung histopathology demonstrated that mice in the control group had ruptured pulmonary alveoli, excessive mucus production, and immune cell infiltration. In contrast, DNA vaccine-immunized mice had milder histopathological changes (Figure 5B). INHAND scores of all DNA vaccine-immunised groups were lower than those of the control group. Notably, mice co-immunised with p-SARS-CoV-2-E+M exhibited the mildest histopathological changes and lowest INHAND scores compared to mice immunised with p-SARS-CoV-2-E/M alone. Moreover, the INHAND scores of the SARS-CoV-2-E+M vaccine group were significantly lower than those of the control group (P<0.05; Figure 5C). Taken together, both p-SARS-CoV-2-E and p-SARS-CoV-2-M immunisation provided partial protection in mice after SARS-CoV-2 challenge, and co-immunisation with p-SARS-CoV-2 E+M enhanced this protection.
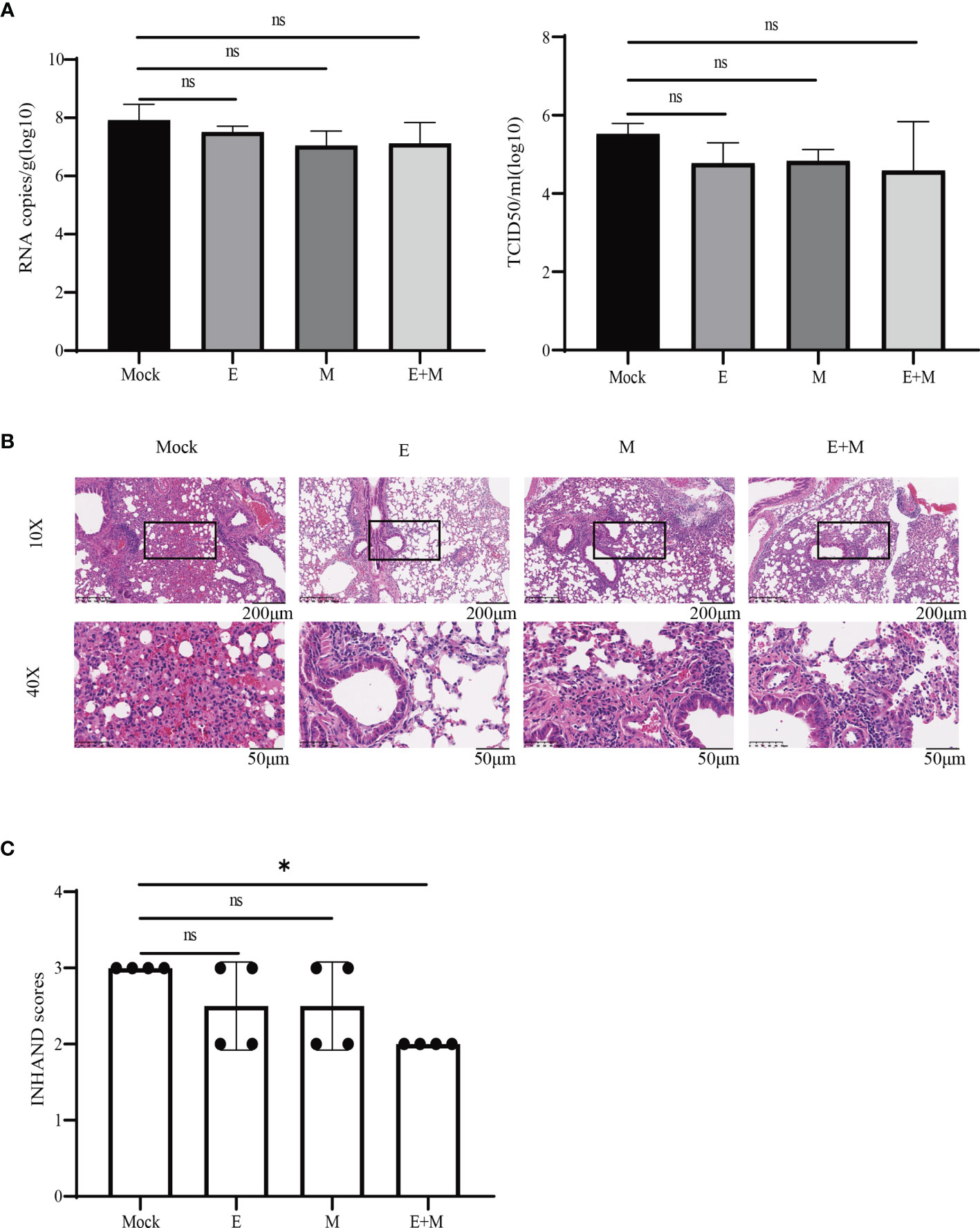
Figure 5 Immunisation protects mice from live severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) challenge. (A) Tissue viral loads and (B) histopathology analysis of SARS-CoV-2-challenged mice. (C) International Harmonisation of Nomenclature and Diagnostic Criteria (INHAND) scores of challenged mice organs, on a severity scale of 0–3 (none, mild, moderate, and severe). Statistical significance for groups of a one-way ANOVA after Dunnett’s multiple comparison correction is shown (*P < 0.05). ns, no significance.
In this study, two DNA vaccines expressing the SARS-CoV-2 E and M proteins were developed. The data showed that considerable cellular immune responses were elicited, whereas no robust humoral immunity was detected in BALB/c mice. In addition, six novel H-2d-restricted T-cell epitopes were identified in E and M proteins. Co-immunisation with two DNA vaccines expressing E and M proteins provided partial protection against SARS-CoV-2. To the best of our knowledge, this is the first study to evaluate the immune protective potential of the SARS-CoV-2 E and M proteins as vaccine targets.
Previous studies have suggested that SARS-CoV-2-specific T-cells play a key role in COVID-19 resolution and modulation of disease severity (25, 26). The definition of SARS-CoV-2-specific T-cell epitopes is important for evaluating the potential influences of mutations on acquired immunity and vaccine efficacy. The immunodominant T-cell epitopes in the E and M antigen regions have only been determined in a few studies (27, 28). M protein-specific cellular immune responses have previously been reported from SARS−CoV vaccination in mice (29, 30), but there are no reports on similar responses to the E protein. Previous studies have reported that overlapping peptide pools of the E and M proteins induce SARS-CoV-2-reactive T-cell responses in humans with COVID-19 (25, 31). Immunoinformatic analyses in humans have identified SARS-CoV-2 E-specific (LVKPSFYVYSRVKNL/FYVYSRVKNLNSSRV/FLLVTLAILTALRLC) and M-specific (RGHLRIAGHHLGRCD) T-cell epitopes. These peptides elicited T-cell responses in 33%, 36%, 22%, and 72% of patients with COVID-19 and overlapped with E11, E12, E07, and M29, respectively (25, 27, 28, 32–34). Another predicted M protein-specific T-cell epitope (LLQFAYANRNRFLYI) overlapped with M07 and M08 (27, 28), which were identified in this study. However, a few predicted E/M protein-specific T-cell epitopes were not confirmed in this study, possibly due to the differences between mice and humans. Notably, one study with results inconsistent with our data reported that SARS-CoV-2 E/M-specific peptides were not able to stimulate CD4+ and CD8+ T-cells from virus replicon particle-vaccinated BALB/c mice (35). This may reflect differences in target protein expression between DNA- and virus replicon particle-based vaccines. The multiple amino-acids sequence alignment have been performed for immuno-dominant epitopes for E and M identified in our study, which shows a conservation of E07 (100% identity) between SARS-CoV and SARS-CoV-2. However, the suitability of E11, E12, M07, M08, and M29 for SARS-CoV requires further testing (36). Variant of concern (VOC) is a variant for which there is evidence of an increase in transmissibility and disease severity, significant reduction in neutralization by antibodies generated during previous infection or vaccination (37). For E protein among 5 of SARS-CoV-2 VOCs, there is only one substitution (T9I) in the omicron variant, one substitution (P71L) in the Beta variant. For M protein, there are three substitutions (D3G, Q19E, and A63T) in the omicron variant, one substitution (I82T) in the Delta variant (37, 38). Fortunately, most immuno-dominant epitopes for E and M identified in our study are conserved between SARS-CoV and VOCs of SARS-CoV-2. Notably, persistence of T-cell responses against E or M was shown by Day 120 data until weeks 11 after vaccination (Figure 4).
A few studies have tested the potential of the E protein as a vaccine target. One SARS−CoV vector vaccine study reported that none of the animals immunised with vaccines expressing E, ME, or SME proteins induced antibody responses specific to the E protein (39). Therefore, we speculated that the SARS-CoV-2 E protein has a limited ability to induce a humoral immune response. A few studies have reported that SARS-CoV M protein-based vaccines can induce antibody responses in immunised animals (29, 30, 39, 40). Of note, neutralising antibody titres specific to SARS-CoV M protein were detected in immunised animals and patients with SARS (41, 42). Previous immunoinformatic studies identified M-specific B-cell epitopes, which were confirmed by ELISA using convalescent sera from patients who previously had COVID−19 (28). However, there were no studies that identified E-specific B-cell epitopes. One study identified two linear E-specific B-cell epitopes (CoV2_E-1 and CoV2_E-1.1) by immunoinformatic prediction, but statistical analysis revealed that antibody responses against these epitopes in convalescent sera were not significantly higher than those in healthy control sera (43). Our study revealed that no significant IgG or neutralizing antibody against E/M were detected in vaccinated mice. This may be due to limited B-cell epitopes relatively smaller molecular weight of E protein, and weaker immunogenicity of DNA vaccination.
One study communicated that immunisation with bovine-human parainfluenza virus type 3 expressing the S protein provided complete and partial protection against SARS-CoV in the lower and upper respiratory tract, respectively. This was augmented slightly by co-expression with M and E. However, the expression of M, E, or M plus E in the absence of S did not confer detectable protection against SARS-CoV (39). Notably, hamsters immunised with a vaccine co-expressing the M and N proteins were protected against severe weight loss and lung pathology and had reduced viral loads in the oropharynx and lungs after SARS-CoV-2 challenge (12). Our study demonstrated that p-SARS-CoV-2-E or p-SARS-CoV-2-M immunisation provided minor protection (indicated by mild lung tissue pathology), and co-immunisation with p-SARS-CoV-2-E+M exhibited even more protection (indicated by the mildest histopathological changes and lowest INHAND scores), although no drop in lung tissue virus titre was detected in DNA-vaccinated mice after challenge with SARS-CoV-2. Furthermore, the longevity of protective immunity provided by DNA vaccines expressing SARS-CoV-2 E/M was supported here, even though the challenge study was carried out nearly 3 months post vaccination.
This study had limitations. We only observed the DNA vaccine strategy in BALB/c mice, and because of the low immunogenicity of DNA vaccines, the protection efficacy could be further explored in subunit vaccines, vector vaccines, or novel combinations of DNA and other vaccines, and future studies should estimate the immunity effect in other animal models. Importantly, additional research is needed to understand the molecular mechanisms of the E/M-mediated immune protective effect after SARS-CoV-2 challenge so as to harness this knowledge to optimise COVID-19 vaccine design.
In summary, we present a detailed immunological study of the SARS-CoV-2-specific immune response against E and M proteins after DNA vaccination. This is the first experimental report to support immune protection against SARS-CoV-2 provided by the specific cellular immune response against E and M proteins in the absence of an obvious humoral immune response. The emergence of SARS-CoV-2 variants has raised concerns about the potential loss of protection from COVID-19 vaccines targeting only the highly mutated S protein. The role of conserved structural proteins of SARS-CoV-2, including E/M protein, is worthy of attention in vaccine design and application since vaccine-induced T- cell responses against conserved epitopes will be unaffected by SARS-CoV-2 variants (39). Our results will lay a strong foundation for the development of a cross-protective COVID-19 vaccine for controlling current and emerging variants of concern, as well as for preventing future β-coronavirus pandemics.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Committee on the Ethics of Animal Experiments of the Chinese Center for Disease Control and Prevention.
WT, DZ, and YD conceived of the study. JC, DH, WW, MH, CZ, ZZ, RY, YZ, and BH performed the experiment. JC, WW and BH analysed the data. JC drafted the manuscript. DZ and WT revised the manuscript. All authors reviewed and approved the final manuscript.
This work was supported by grants from the National Natural Science Foundation of China (82041041, 82061138008). Partly supported by the Support Project of Scientific and Technological Innovation Team in Universities of Henan Province [No. 20IRTSTHN027].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
COVID-19, coronavirus disease 2019; E, envelope; ELISA, enzyme-linked immunosorbent assay; IFA, immunofluorescence assay; IFN-γ, interferon gamma; INHAND, International Harmonisation of Nomenclature and Diagnostic; M, membrane; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SFU, spot-forming unit; FITC, fluorescein isothiocyanate; DAPI, 4′,6-diamino-2-phenylindol; DMSO, dimethyl sulfoxide; VOC, variant of concern.
1. Hazarika BB, Gupta D. Modelling and Forecasting of COVID-19 Spread Using Wavelet-Coupled Random Vector Functional Link Networks. Appl Soft Comput (2020) 96:106626. doi: 10.1016/j.asoc.2020.106626
2. World Health Organization. (2021). Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
3. Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 Vaccines. JAMA (2021) 325:1318–20. doi: 10.1001/jama.2021.3199
4. Vandelli A, Monti M, Milanetti E, Armaos A, Rupert J, Zacco E, et al. Structural Analysis of SARS-CoV-2 Genome and Predictions of the Human Interactome. Nucleic Acids Res (2020) 48:11270–83. doi: 10.1093/nar/gkaa864
5. Wang L, Zhou T, Zhang Y, Yang ES, Schramm CA, Shi W, et al. Ultrapotent Antibodies Against Diverse and Highly Transmissible SARS-CoV-2 Variants. Science (2021) 373. doi: 10.1126/science.abh1766
6. Li S, Yuan L, Dai G, Chen RA, Liu DX, Fung TS. Regulation of the ER Stress Response by the Ion Channel Activity of the Infectious Bronchitis Coronavirus Envelope Protein Modulates Virion Release, Apoptosis, Viral Fitness, and Pathogenesis. Front Microbiol (2019) 10:3022. doi: 10.3389/fmicb.2019.03022
7. Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus Membrane Fusion Mechanism Offers a Potential Target for Antiviral Development. Antiviral Res (2020) 178:104792. doi: 10.1016/j.antiviral.2020.104792
8. Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, et al. Insights Into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim Biophys Acta Mol Basis Dis (2020) 1866:165878. doi: 10.1016/j.bbadis.2020.165878
9. Xu R, Shi M, Li J, Song P, Li N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front Bioeng Biotechnol (2020) 8:862. doi: 10.3389/fbioe.2020.00862
10. Wang C, Zheng X, Gai W, Zhao Y, Wang H, Wang H, et al. MERS-CoV Virus-Like Particles Produced in Insect Cells Induce Specific Humoural and Cellular Imminity in Rhesus Macaques. Oncotarget (2017) 8:12686–94. doi: 10.18632/oncotarget.8475
11. Lokugamage KG, Yoshikawa-Iwata N, Ito N, Watts DM, Wyde PR, Wang N, et al. Chimeric Coronavirus-Like Particles Carrying Severe Acute Respiratory Syndrome Coronavirus (SCoV) S Protein Protect Mice Against Challenge With SCoV. Vaccine (2008) 26:797–808. doi: 10.1016/j.vaccine.2007.11.092
12. Jia Q, Bielefeldt-Ohmann H, Maison RM, Masleša-Galić S, Cooper SK, Bowen RA, et al. Replicating Bacterium-Vectored Vaccine Expressing SARS-CoV-2 Membrane and Nucleocapsid Proteins Protects Against Severe COVID-19-Like Disease in Hamsters. NPJ Vaccines (2021) 6:47. doi: 10.1038/s41541-021-00321-8
13. Günl F, Mecate-Zambrano A, Rehländer S, Hinse S, Ludwig S, Brunotte L. Shooting at a Moving Target-Effectiveness and Emerging Challenges for SARS-CoV-2 Vaccine Development. Vaccines (Basel) (2021) 9. doi: 10.3390/vaccines9101052
14. Tse LV, Meganck RM, Graham RL, Baric RS. The Current and Future State of Vaccines, Antivirals and Gene Therapies Against Emerging Coronaviruses. Front Microbiol (2020) 11:658. doi: 10.3389/fmicb.2020.00658
15. Mallapaty S. India's DNA COVID Vaccine Is a World First - More Are Coming. Nature (2021) 597:161–2. doi: 10.1038/d41586-021-02385-x
16. Tebas P, Kraynyak KA, Patel A, Maslow JN, Morrow MP, Sylvester AJ, et al. Intradermal SynCon® Ebola GP DNA Vaccine Is Temperature Stable and Safely Demonstrates Cellular and Humoral Immunogenicity Advantages in Healthy Volunteers. J Infect Dis (2019) 220:400–10. doi: 10.1093/infdis/jiz132
17. Smith TRF, Patel A, Ramos S, Elwood D, Zhu X, Yan J, et al. Immunogenicity of a DNA Vaccine Candidate for COVID-19. Nat Commun (2020) 11:2601. doi: 10.1038/s41467-020-16505-0
18. Zhan Y, Deng Y, Huang B, Song Q, Wang W, Yang Y, et al. Humoral and Cellular Immunity Against Both ZIKV and Poxvirus Is Elicited by a Two-Dose Regimen Using DNA and Non-Replicating Vaccinia Virus-Based Vaccine Candidates. Vaccine (2019) 37:2122–30. doi: 10.1016/j.vaccine.2019.02.063
19. Guan J, Deng Y, Chen H, Yin X, Yang Y, Tan W. Priming With Two DNA Vaccines Expressing Hepatitis C Virus NS3 Protein Targeting Dendritic Cells Elicits Superior Heterologous Protective Potential in Mice. Arch Virol (2015) 160:2517–24. doi: 10.1007/s00705-015-2535-7
20. Chen H, Wen B, Deng Y, Wang W, Yin X, Guan J, et al. Enhanced Effect of DNA Immunization Plus In Vivo Electroporation With a Combination of Hepatitis B Virus Core-PreS1 and S-PreS1 Plasmids. Clin Vaccine Immunol (2011) 18:1789–95. doi: 10.1128/cvi.05113-11
21. Yang R, Deng Y, Huang B, Huang L, Lin A, Li Y, et al. A Core-Shell Structured COVID-19 mRNA Vaccine With Favorable Biodistribution Pattern and Promising Immunity. Signal Transduct Target Ther (2021) 6:213. doi: 10.1038/s41392-021-00634-z
22. Zhao Z, Deng Y, Niu P, Song J, Wang W, Du Y, et al. Co-Immunization With CHIKV VLP and DNA Vaccines Induces a Promising Humoral Response in Mice. Front Immunol (2021) 12:655743. doi: 10.3389/fimmu.2021.655743
23. Nal B, Chan C, Kien F, Siu L, Tse J, Chu K, et al. Differential Maturation and Subcellular Localization of Severe Acute Respiratory Syndrome Coronavirus Surface Proteins S, M and E. J Gen Virol (2005) 86:1423–34. doi: 10.1099/vir.0.80671-0
24. Kumar B, Hawkins GM, Kicmal T, Qing E, Timm E, Gallagher T. Assembly and Entry of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2): Evaluation Using Virus-Like Particles. Cells (2021) 10. doi: 10.3390/cells10040853
25. Heide J, Schulte S, Kohsar M, Brehm TT, Herrmann M, Karsten H, et al. Broadly Directed SARS-CoV-2-Specific CD4+ T Cell Response Includes Frequently Detected Peptide Specificities Within the Membrane and Nucleoprotein in Patients With Acute and Resolved COVID-19. PLoS Pathog (2021) 17:e1009842. doi: 10.1371/journal.ppat.1009842
26. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and Strong Memory CD4(+) and CD8(+) T Cells Induced by SARS-CoV-2 in UK Convalescent Individuals Following COVID-19. Nat Immunol (2020) 21:1336–45. doi: 10.1038/s41590-020-0782-6
27. Abdelmageed MI, Abdelmoneim AH, Mustafa MI, Elfadol NM, Murshed NS, Shantier SW, et al. Design of a Multiepitope-Based Peptide Vaccine Against the E Protein of Human COVID-19: An Immunoinformatics Approach. BioMed Res Int (2020) 2020:2683286. doi: 10.1155/2020/2683286
28. Crooke SN, Ovsyannikova IG, Kennedy RB, Poland GA. Immunoinformatic Identification of B Cell and T Cell Epitopes in the SARS-CoV-2 Proteome. Sci Rep (2020) 10:14179. doi: 10.1038/s41598-020-70864-8
29. Wang Z, Yuan Z, Matsumoto M, Hengge UR, Chang YF. Immune Responses With DNA Vaccines Encoded Different Gene Fragments of Severe Acute Respiratory Syndrome Coronavirus in BALB/c Mice. Biochem Biophys Res Commun (2005) 327:130–5. doi: 10.1016/j.bbrc.2004.11.147
30. Okada M, Okuno Y, Hashimoto S, Kita Y, Kanamaru N, Nishida Y, et al. Development of Vaccines and Passive Immunotherapy Against SARS Corona Virus Using SCID-PBL/hu Mouse Models. Vaccine (2007) 25:3038–40. doi: 10.1016/j.vaccine.2007.01.032
31. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans With COVID-19 Disease and Unexposed Individuals. Cell (2020) 181:1489–501.e1415. doi: 10.1016/j.cell.2020.05.015
32. Kared H, Redd AD, Bloch EM, Bonny TS, Sumatoh H, Kairi F, et al. SARS-CoV-2-Specific CD8+ T Cell Responses in Convalescent COVID-19 Individuals. J Clin Invest (2021) 131. doi: 10.1172/jci145476
33. Snyder TM, Gittelman RM, Klinger M, May DH, Osborne EJ, Taniguchi R, et al. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv (2020). doi: 10.1101/2020.07.31.20165647
34. Akbay B, Abidi SH, Ibrahim MAA, Mukhatayev Z, Ali S. Multi-Subunit SARS-CoV-2 Vaccine Design Using Evolutionarily Conserved T- and B- Cell Epitopes. Vaccines (Basel) (2021) 9. doi: 10.3390/vaccines9070702
35. Zhuang Z, Lai X, Sun J, Chen Z, Zhang Z, Dai J, et al. Mapping and Role of T Cell Response in SARS-CoV-2-Infected Mice. J Exp Med (2021) 218. doi: 10.1084/jem.20202187
36. Bianchi M, Benvenuto D, Giovanetti M, Angeletti S, Ciccozzi M, Pascarella S. SARS-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics? BioMed Res Int (2020) 2020:4389089. doi: 10.1155/2020/4389089
37. Alkhatib M, Svicher V, Salpini R, Ambrosio FA, Bellocchi MC, Carioti L, et al. SARS-CoV-2 Variants and Their Relevant Mutational Profiles: Update Summer 2021. Microbiol Spectr (2021) 9:e0109621. doi: 10.1128/Spectrum.01096-21
38. He X, Hong W, Pan X, Lu G, Wei X. SARS-CoV-2 Omicron Variant: Characteristics and Prevention. MedComm (2020) (2021) 2:838–45. doi: 10.1002/mco2.110
39. Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, et al. Contributions of the Structural Proteins of Severe Acute Respiratory Syndrome Coronavirus to Protective Immunity. Proc Natl Acad Sci USA (2004) 101:9804–9. doi: 10.1073/pnas.0403492101
40. He Y, Zhou Y, Siddiqui P, Niu J, Jiang S. Identification of Immunodominant Epitopes on the Membrane Protein of the Severe Acute Respiratory Syndrome-Associated Coronavirus. J Clin Microbiol (2005) 43:3718–26. doi: 10.1128/jcm.43.8.3718-3726.2005
41. Li YD, Chi WY, Su JH, Ferrall L, Hung CF, Wu TC. Coronavirus Vaccine Development: From SARS and MERS to COVID-19. J BioMed Sci (2020) 27:104. doi: 10.1186/s12929-020-00695-2
42. Pang H, Liu Y, Han X, Xu Y, Jiang F, Wu D, et al. Protective Humoral Responses to Severe Acute Respiratory Syndrome-Associated Coronavirus: Implications for the Design of an Effective Protein-Based Vaccine. J Gen Virol (2004) 85:3109–13. doi: 10.1099/vir.0.80111-0
43. Polyiam K, Phoolcharoen W, Butkhot N, Srisaowakarn C, Thitithanyanont A, Auewarakul P, et al. Immunodominant Linear B Cell Epitopes in the Spike and Membrane Proteins of SARS-CoV-2 Identified by Immunoinformatics Prediction and Immunoassay. Sci Rep (2021) 11:20383. doi: 10.1038/s41598-021-99642-w
Keywords: SARS-CoV-2, DNA vaccine, envelope protein, membrane protein, humoral response, cellular response
Citation: Chen J, Deng Y, Huang B, Han D, Wang W, Huang M, Zhai C, Zhao Z, Yang R, Zhao Y, Wang W, Zhai D and Tan W (2022) DNA Vaccines Expressing the Envelope and Membrane Proteins Provide Partial Protection Against SARS-CoV-2 in Mice. Front. Immunol. 13:827605. doi: 10.3389/fimmu.2022.827605
Received: 02 December 2021; Accepted: 07 February 2022;
Published: 24 February 2022.
Edited by:
Pedro A. Reche, Complutense University of Madrid, SpainReviewed by:
Marc Paul Girard, Université Paris Diderot, FranceCopyright © 2022 Chen, Deng, Huang, Han, Wang, Huang, Zhai, Zhao, Yang, Zhao, Wang, Zhai and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjie Tan, dGFud2pAaXZkYy5jaGluYWNkYy5jbg==; Desheng Zhai, emRzQHh4bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.