
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 11 August 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.819574
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). COVID-19 is a complex disease which immune response can be more or less potent. In severe cases, patients might experience a cytokine storm that compromises their vital functions and impedes clearance of the infection. Gamma delta (γδ) T lymphocytes have a critical role initiating innate immunity and shaping adaptive immune responses, and they are recognized for their contribution to tumor surveillance, fighting infectious diseases, and autoimmunity. γδ T cells exist as both circulating T lymphocytes and as resident cells in different mucosal tissues, including the lungs and their critical role in other respiratory viral infections has been demonstrated. In the context of SARS-CoV-2 infection, γδ T cell responses are understudied. This review summarizes the findings on the antiviral role of γδ T cells in COVID-19, providing insight into how they may contribute to the control of infection in the mild/moderate clinical outcome.
Beginning December 2019, there was a surge of pneumonia cases of unknown etiology in Wuhan City, Hubei Province, Central China. Genomic sequencing (1) showed that this pneumonia was caused by a novel coronavirus (CoV) that belonged to the B lineage of the Beta-CoV genus along with SARS-CoV and MERS-CoV (1). Its similarity to the original SARS-CoV sequence led to the naming severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the associated illness was named coronavirus disease 19 (COVID-19). CoVs belong to the family Coronaviridae (subfamily Coronavirinae) and can infect a wide range of animal hosts. In humans, CoV-induced diseases, such as SARS, MERS, and currently COVID-19 (2, 3), can range in severity from a common cold to the fatal acute respiratory distress syndrome (ARDS). The origin of SARS-CoV-2 and the first transmission event of the disease is still unclear. However, initial group analysis pinpointed the source of the outbreak to a seafood market in Wuhan. This market is known for selling exotic animals for human consumption and it is postulated that this could be the point where the zoonotic transmission occurred (3). The route of transmission between humans is through contact with the nasopharyngeal secretions, including saliva of infected people. Transmission occurs mainly by direct contact with respiratory droplets but also hands or objects contaminated with these secretions (4, 5).
SARS-CoV-2 consists of a positive-sense single-stranded RNA genome, approximately 30 kb in length (6). The overall structure of the virus includes the viral genome contained within a nucleocapsid and encased with a glycosylated envelope. Of note, the nucleocapsid is arranged in a helical symmetry, which is not a common characteristic of positive-sense RNA viruses (7). The main structural proteins of coronaviruses are described in detail below (Figure 1A). (1) Spike (S) Glycoprotein is a multifunctional class I transmembrane protein and varies in size from 1160 to 1400 amino acids. It is located on the surface of the virion, giving the virus its characteristic crown appearance. Foe the entry of virion particles into the cell, the S glycoprotein is required through its union with different host cell receptors (8).. (2) Membrane (M) protein is the most abundant structural protein in the virion, giving a definite shape to the viral envelope. M protein is related to the shape and size of the virus (9). It binds to the nucleocapsid and plays the role of the central organizer during viral assembly. (3) Nucleocapsid (N) protein plays an important role in the packaging of viral RNA into ribonucleocapsid. This protein mediates viral assembly by interacting with the viral genome and the M protein, facilitating transcription and replication of viral RNA (10, 11). (4) Envelope (E) protein has different functions involved in the entry, assembly, and release of the virus (12, 13). It is also known to act as a viroporin which integrates into the host membrane and alters the flow of ions such as Na+ (14, 15).
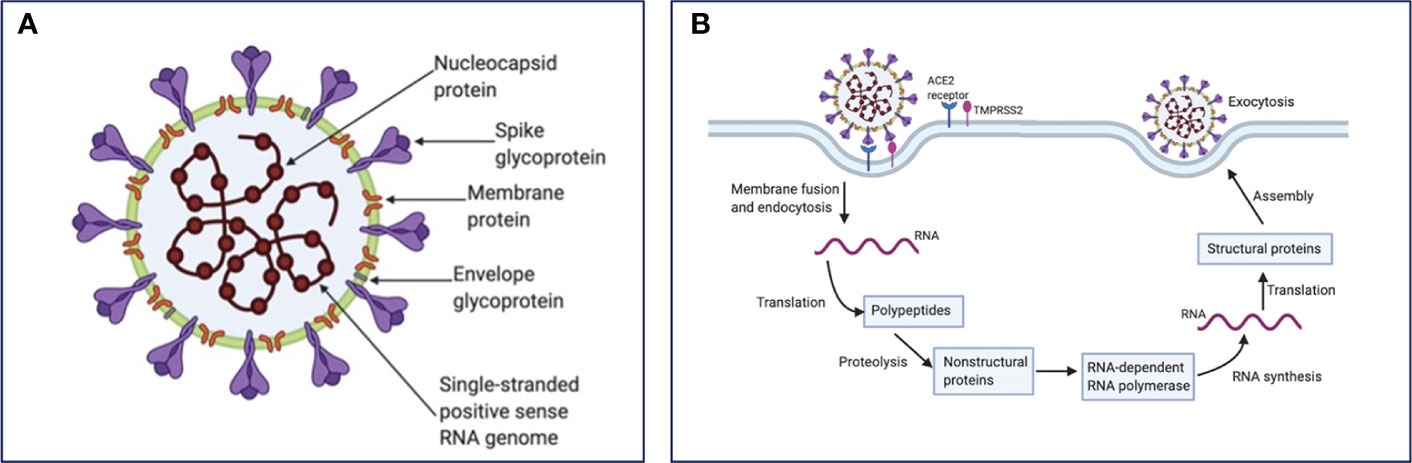
Figure 1 SARS-CoV-2 structure and life cycle. (A) SARS-CoV-2 contains four structural proteins: spike surface glycoprotein, membrane, nucleocapsid and envelope protein, as well as eight accessory proteins (not represented). (B) Coronavirus particles bind to cellular attachment factors and specific S interactions with the cellular receptors ACE2 and TMPRSS2, promoting viral uptake and membrane fusion. Positive sense single-stranded RNA is released, partially translated into SARS-CoV-2 polymerase protein, and transcribed. Structural proteins and accessory proteins (N, S, M, and E) results after RNA subgenomic translation, that are inserted into the ER–Golgi compartment for virion assembly. Subsequent positive-sense RNA genomes are incorporated into newly virions, which are secreted from the plasma membrane.
As recently reviewed, SARS-CoV-2 enters host cells by the interaction of its surface Spike (S) protein with the host’s angiotensin-converting enzyme 2 (ACE2) receptor, a membrane exopeptidase primarily expressed in the kidney, lungs, and heart (16). However, this process also requires a cellular protease that helps the cleavage of the spike protein and fusion of the cell and viral membranes. This cellular transmembrane protease is TMPRSS2 (17). Both of these proteins, TMPRSS2 and ACE2, determines viral entry (18, 19). After the virus is endocytosed, the nucleocapsid containing the RNA genome, is released into the cytoplasm of the cell. Parts of the endoplasmic reticulum are then appropriated to form double membraned vesicles which protect the viral genome to ensure replication (20). The viral genome gets translated by using the protein translation machinery of the host. Two viral proteases are used for cleavage of proteins into structural and nonstructural viral proteins. Viral particles are assembled in the endoplasmic reticulum/Golgi compartment and infectious virions are released from the cells through exocytosis (21–23). SARS-CoV-2 life cycle is summarized in Figure 1B.
COVID-19 has emerged as a complex disease affecting many body systems and generating a wide spectrum of clinical manifestations. Understanding SARS-CoV-2 immunopathogenesis is key to developing effective treatments. Currently, there are several drugs for the treatment of COVID approved by the FDA including several first line antivirals used to treat other infections. The most widely used are described briefly. Remdesivir, a broad-spectrum antiviral with established activity against several viruses such as respiratory syncytial virus (RSV), Ebola virus, MERS-CoV and SARS-CoV (24, 25) is also capable of inhibiting the replication of SARS-CoV-2 (26, 27). It is one of the first line drugs recommended for use in patients who require hospitalization and the support of supplemental oxygen (28). Lopinavir, an antiretroviral of the protease inhibitor class used for HIV treatment, is capable of disrupting viral replication and RNA release from host cells by inhibiting proteases such as 3CLpro, the main SARS-CoV-2 protease critical for viral replication (29). The combination of Lopinavir with IFN-1β significantly reduced the time to clinical improvement in patients with moderate symptoms (30) although this benefit did not extend to patients with severe symptoms (31).
As a result of an incredible scientific effort, we have seen the development of several highly effective vaccines in a short period of time. COVID-19 vaccines can be divided into three categories: (1) Inactivated vaccines, protein-based vaccines consisting of virus particles that generate target antigens in vitro, (2) virus-vectored, DNA or mRNA vaccines, are gene-based vaccines that encode proteins of the pathogen are delivered and (3) live-attenuated virus vaccines, it is a combination of protein-based and gene-based to produce protein antigen or antigens in vitro and in vivo (32–34). Despite the efficacy of these vaccines, we still do not fully understand the role of innate and adaptive immunity against SARS-CoV-2. Specifically, this article aims to give a comprehensive review of available literature covering the role of innate-like γδ T cells fighting SARS-CoV-2 infection.
A cytokine storm is defined by the rapid proliferation and hyperactivation of T cells, macrophages and Natural killer (NK) cells resulting in excess secretion of inflammatory cytokines and chemical mediators released by both immune or nonimmune cells (35). In older adults, having an unbalanced pro-inflammatory environment can further enhance the inflammatory responses to SARS-CoV-2 infection, leading to an exacerbated cytokine storm and could also influence ACE2 expression facilitating viral entry (36). This cytokine production leads to positive feedback on other immune cells by recruiting them to the area of inflammation causing an exponential increase in inflammation and tissue damage (37). The main cytokines involved in the cytokine storm are interleukins (IL), interferons (IFN), tumor necrosis factor (TNF), colony stimulating factors (CSF), chemokines, and growth factors (GF) (35). The induction of cytokine production typically occurs following viral entry. Target cells such as respiratory epithelial cells, alveolar cells, macrophages, and blood circulating monocytes are activated through pattern recognition receptors (PRRs) that lead to stimulation of the pro-inflammatory NF-κB cascade (38, 39). In addition, viral RNA enters endosomes and activates intracellular toll-like receptors (TLRs), mainly TLR7/8, inducing the expression of numerous inflammatory factors, cytokines, and chemokines (38, 40). One of the most detrimental effects caused by the cytokine storm is acute lung injury (35).
Recent studies have shown that patients with severe disease had high concentrations of pro-inflammatory cytokines (IL-1β, IFN-γ, interferon gamma inducible protein-10, IP-10, and monocyte chemoattractant protein-1, MCP-1) which were associated with pulmonary inflammation and extensive lung damage (41). Similarly, severe cases often display heightened activated T-helper-1 (Th1) cell responses including increased production of IFN-γ, TNF-α, and IL-2 as well as macrophage overactivation (42), both of which contribute to increased inflammation and tissue damage. The observation that patients admitted to the ICU had higher granulocyte-colony stimulating factor (G-CSF), IP-10, MCP-1, MIP-1α, and TNF-α concentrations, further suggests that the development of a cytokine storm is associated with disease severity. However, an increase in T-helper-2 (Th-2) associated cytokines IL-4 and IL-10 has also been reported in severely infected patients. These findings are consistent with other studies in which severe COVID-19 patients had mixed high levels of cytokines (IL-2, IL-6, IL-7, IL-10, IP-10, MCP-1, TNF-α) than patients with mild and moderate infections (43, 44). Therefore, an adequate Th-1/Th-2 balance plays an important role controlling the severity of the disease. However, more studies are needed to characterize the Th1 and Th2 responses, as well as when and how dysregulation starts impacting the clinical outcome of the disease.
The prolonged release of inflammatory cytokines induces a feedback loop of subsequent cytokine production that can lead to cell and organ damage. This cytokine storm can be caused by an aberrant host response to infection provoking a severe clinical syndrome known as sepsis, (45). Critically ill COVID-19 patients may develop organ dysfunction due to a dysregulated response to infection and develop sepsis (46, 47). Criteria for the diagnosis and treatment of bacterial sepsis is analogous to SARS-CoV-2 infection and has been applied to understand severe COVID-19 (48). However, there are some differences between sepsis caused by SARS-CoV-2 and bacterial sepsis. A retrospective study investigated whether there are differences in the immune system status of these two types of sepsis. A total of 64 bacterial sepsis patients and 43 patients with SARS-CoV-2 sepsis were included (49). The results of this study showed that there are key differences between the two types of sepsis, such as a milder cytokine storm in SARS-CoV-2 sepsis as well as higher immunoglobulin and complement protein levels. However, neutrophil, monocyte, infection biomarkers, individual lymphocyte subset counts (total T lymphocyte, CD4+ T cell, CD8+ T cell, B cell, and NK cell counts), and lymphocyte subset functions were similar in bacterial sepsis patients and SARS-CoV-2 sepsis patients. Treatment protocols for sepsis have a well-established history in most healthcare systems. Considering the similarities in both the immunopathogenesis and pathophysiological manifestations, our knowledge of sepsis could inform the management of severity COVID-19 (50).
The clinical course of COVID-19 is very similar to other respiratory infections such that 80% of patients present mild to moderate symptoms (Figure 2). SARS-CoV-2 primarily targets cells in the upper and lower respiratory tract, as well as pulmonary cells (51). Macrophages and NK cells belong to the innate immune system and constitute one of the first defense mechanisms (52). Pathogens are removed from circulation by phagocytosis, and processed antigens are displayed to initiate adaptive immune responses. This includes the production and secretion of inflammatory cytokines and chemokines that recruit various immune cells to the site of infection and regulate T cell responses to the pathogen (53, 54). γδ T cells, which are generally classified as innate-like T cells, secrete several cytokines including IFN-γ and TNF-α and also cytotoxic components such as perforin and granzymes (55).
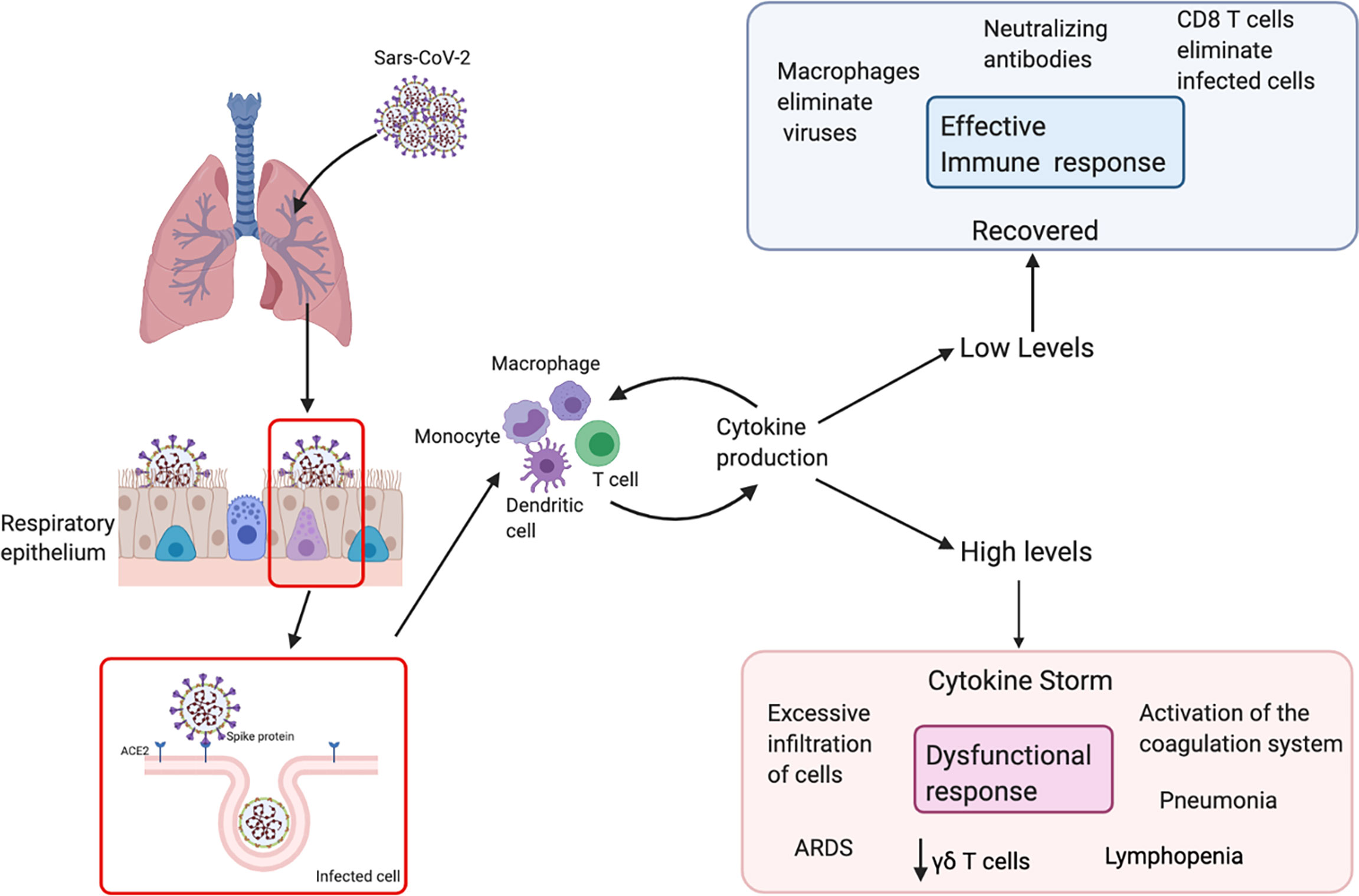
Figure 2 Schematic immune responses to SARS-CoV-2. The SARS-CoV-2 virus recognizes ACE-2 in respiratory epithelial cells, facilitating viral entry. After viral replication, the virus is released and recognized by the immune system. T cells and dendritic cells are activated through pattern recognition receptors. The virus induces the expression of numerous inflammatory factors, the synthesis of type I interferons and production of cytokines. High levels of cytokines cause a cytokine storm, which leads to a dysfunctional response with an excessive infiltration of cells, decrease in different populations of T cells in the blood such as γδ T cells and surviving T cells are immunologically exhausted, this provokes progression to acute respiratory distress syndrome (ARDS).
γδ T cell responses to coronavirus infections have been understudied, with a previous report on SARS-CoV infection showing a potent cytolytic activity against infected target monocytic cell lines (56). Although more intense research has been performed in other airway infections such as influenza, their role in SARS-CoV-2 infection remains understudied.
γδ T cells’ unique properties enable them to have a critical tole linking innate and adaptive immune responses (57, 58). These cells are key contributors to pulmonary mucosal immunity including tissue repair and rapid response against numerous respiratory pathogens (59–61), and therefore their potential role fighting SARS-CoV-2 infection is worthy of investigation.
γδ T cells are recognized for their contribution to tumor surveillance, fight against infectious diseases, and autoimmunity (57, 58, 62). In contrast to conventional αβ T cells, the T-cell receptor (TCR) of γδ T cells is comprised of variable γ and δ chains that recognize non-peptide antigens in the absence of MHC molecules. γδ T cells elicit non-redundant functions when compared to conventional αβ T cells (57, 58). Within circulating human T cells, γδ T cells account for 0.5–10% and are classified into two major peripheral blood subpopulations based on the δ chain usage, Vδ1 and Vδ2 T cells (57). The Vδ2 chain is present in the majority of circulating γδ T cells and is almost always paired with the Vγ9 chain (Vγ9Vδ2 T cells) (63–65). Vδ2 T cells specifically recognize low-weight phosphorylated metabolites of isoprenoid biosynthesis compounds, or mevalonate pathway, referred to as phosphoantigens (P-Ags) (66, 67). This pathway is essential for the synthesis of cholesterol and precursors for prenylation of proteins and as such are produced by both eukaryotes and prokaryotes and can therefore act as self (host) or non-self (pathogen) antigens. The most potent, naturally occurring P-Ag known to date is (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (HMB-PP), an intermediate of the MEP (non-mevalonate) pathway utilized by several pathogenic bacterial species (68). Another, less potent activator is isopentenyl pyrophosphate (IPP), an intermediate of the mevalonate pathway that increases in response to cellular stress and is present both in eukaryotic and procaryotic cells (69, 70). Aminobisphosphonates (N-BPs) bears structural similarity to pyrophosphates and specifically activate Vδ2 T cells. N-BPs inhibit one key enzyme of the mevalonate pathway, farnesyl pyrophosphate synthase, leading to an accumulation of IPP (71), that is then presented to Vδ2 T cells independent of major histocompatibility (MHC) molecules, but in the context of butyrophilin (BTN) molecules (72, 73). A recent study identified that in addition to BTN3A1, BTN2A1 plays an important role as a ligand that binds to the Vγ9 TCR γ chain (72). It is believed that once p-Ag binds to BTN3A1 through its intracellular domain, the BTN2A1–BTN3A1 complex engages the γδ TCR via two distinct binding sites: BTN2A1 binds to Vγ9 region, whereas another ligand (possibly BTN3A1) binds to the opposing Vγ9 and Vδ2 chains. Besides detecting p-Ags, γδ T cells can recognize cells through molecules that are expressed at the cell surface in a stress-induced manner. For example, endogenous proteins, such heat shock protein 60 (74, 75) or FI-ATPase (76), that can be ectopically expressed on the cell membrane upon transformation and recognized by Vδ2 TCRs to promote tumor cell lysis. In addition, γδ T cells recognize malignant cells through the engagement of innate-like activating receptors like NKG2D, and natural cytotoxicity receptors (NCRs) (57, 77). NKG2D acts as a costimulatory signal upon recognition of stress markers MHC class I chain related protein A and B (MICA, MICB), and UL16 binding proteins (ULBPs). Activation through this pathway provokes the secretion of proinflammatory cytokines TNF-α and IFN-γ as well as direct cytolytic activity mediated by the release of perforins and granzymes (78–80). On the contrary, the natural antigen for Vδ1 T cells has yet to be defined. These cells are mainly found within the tissue where they have a critical role in immunosurveillance (57).
γδ T cells exhibit a high degree of polyfunctionality, or functional plasticity as regulatory and effector cells, as characterized by their membrane receptors and cytokine production (57). γδ T cells can secrete a range of different cytokines and can directly interact with other immune cell populations (81). Distinct subpopulations of γδ T cells are capable of producing either anti-inflammatory or pro-inflammatory signals, giving them a direct role in maintaining immunological homeostasis and response to disease (82–84). They exert different immunomodulatory and adjuvant functions on CD4 T, CD8 T, B, and dendritic cells (DC) which vary according to the infectious challenge or environment, and the subset of γδ T cells involved in the antiviral response (85). γδ T cells execute direct cytolytic activity mediated by the release of perforin and several types of granzymes (86). γδ T cells may also induce apoptosis through the expression of tumor necrosis factor receptor superfamily members TNF- related apoptosis-inducing ligand (TRAIL) or FasL (87, 88). Both of which have been demonstrated to be important for mediating the resolution of inflammation and clearing tumor cells. Effector γδ T cells also express CD16 (FcγRIII) which affords them the ability to execute antibody-dependent cellular cytotoxicity (ADCC) against virally infected cells as well as phagocytosis of opsonized cell-free pathogens (89, 90). Studies from our group have highlighted the importance of the CD16 receptor expression as a marker of cytotoxic capacity associated to Vδ2 T cell elimination of HIV-latently infected CD4 T cells (91, 92).
γδ T cells play a key role in controlling a variety of different viral infections. Their anti-HIV activity was recognized since the beginning of the pandemic (93) and although not completely yet understood, their key role fighting the infection has been documented (86, 94). Less is known about their specific involvement in HIV latency, which is the focus of our studies (86, 91, 92, 94). In respiratory infections, such as influenza or respiratory syncytial virus (RSV), lung-resident and infiltrating γδ T cells play critical roles in controlling the severity of the infection (89, 90, 95, 96). More intensive research has been performed in influenza, providing important insights on general γδ T cell responses to viral infections that cause respiratory distress. Using a murine model of influenza, it was recently reported that early production of IL-17 by γδ T cells promoted viral clearance (89) and more importantly, adoptive cell transfer of expanded γδ T cells resulted in cells trafficking to the lungs and induction of potent antiviral function that led to control of viral replication (90). In line with these results, activated γδ T cells express high levels of chemokine receptors CXCR5, CCR1 and CCR5 allowing their migration along CCL3 and CCL5 chemotactic signals towards the lungs (97, 98). Interestingly, influenza-infected macrophages and DCs alter the mevalonate pathway leading to the production of IPP and subsequent γδ T cell activation directly conferring immune protection independent of viral subtypes (99).
Lymphopenia is associated with disease progression where a significant reduction in T cell counts is often observed in severe COVID-19 patients compared to non-infected and mild cases (100, 101). SARS-CoV-2 has the ability to evade the immune system, allowing the virus to proliferate and disseminate into various tissues. Dysregulation of host immunity has been linked to the severity of the disease. This includes macrophage hyperactivation, the development of a cytokine storm, as well as the depletion of key lymphocytic populations such as NK cells and cytotoxic T cell lymphocytes (CTLs) cells (35).
γδ T cell frequency in the blood of hospitalized COVID-19 patients is lower compared to healthy controls (102–104), indicating γδ T cells as one of the most affected lymphocyte subsets (105). Carissimo et al. (105) evaluated the frequency of different T cell populations, CD8, CD4, γδ (Vδ1 and Vδ2), and mucosal-associated invariant T (MAIT) cells in the acute phase of the infection in COVID-19 patients showing a decrease in cytolytic populations CD8, MAIT and Vδ2 T cells. On the contrary, Vδ1 and CD4 T cells did not show a significant decrease during infection. In addition, as the disease severity increased, there was a gradual reduction of Vδ2 T cells in the peripheral blood, which was hypothesized to be a consequence of activation and infiltration of these cells into the lungs (105). This inverse correlation was more pronounced in Vδ2 and CD8 T cells, which suggested that there was a selective activation and infiltration of these cells in the lungs. In line with these observations, a recent work on the first lung transplant of a COVID-19 patient, highlighted a key role of γδ T cells being specifically recruited to the lungs (106). In addition to decreased circulating Vδ2 T cells, other studies reported a switch to an effector memory population at the time of hospital admission compared to healthy control (102, 107, 108). This increase in memory phenotype provides further insights into the possibility of effector-like γδ T cells recruitment from the blood to the lungs therefore, strengthening their involvement in the immune response against SARS-CoV-2 (109).
γδ T cells from COVID-19 patients with mild symptoms exhibited a strong activated phenotype based on CD25 expression, although early activation marker CD69 and exhaustion marker PD-1 levels were similarly expressed in healthy donors (108). This limited data suggests that γδ T cells were not exhausted, raising the possibility that CD69 was expressed earlier during infection followed by reversion to the quiescent state during prolonged recovery. Whether this is the case for severe cases of COVID-19 requires further investigation. In addition, patients in the acute phase of infection had elevated expression of CD38 in Vδ2 T cells compared to healthy individuals. Interestingly, Carissimo et al. also found that the immature neutrophil-to-Vδ2 T cell ratio was an excellent tool to predict patient progression to pneumonia (105).
Some γδ T cell subsets constitutively express CD8 while CD4 is generally absent (110). However, we and others previously reported that they can transiently upregulate CD4 expression upon activation (111, 112). Similarly, the proportion of γδ T cells expressing CD4 was increased in COVID-19 compared to healthy donors, while frequency of CD8 γδ T cells remained unchanged (108). In our previous study, CD4 was transiently upregulated after in vitro exposure to IL-2, without requiring TCR-antigen recognition suggesting that CD4 expression on Vδ2 cells could constitute an additional activation marker (111). Interestingly, a higher frequency of CD16 expression was observed in moderate COVID-19 disease and was almost absent in the severe cases of the disease (113, 114). Therefore, similar to HIV infection, in addition to its involvement in ADCC, CD16 could constitute a critical marker to determine the extent of the cytotoxic capacity of γδ T cells and their ability to control SARS-CoV-2 infection (115). The involvement of γδ T cell-mediated ADCC in COVID-19 responses is currently unknown, and constitute an additional area of critical investigation, similar to NK-ADCC mediated functions (116, 117).
Due to the dearth of studies that specifically focus on γδ T cell responses to COVID-19, there are important caveats to consider when comparing these collective findings. Patient demographics (e.g., age, biological sex, and race) can influence γδ T cell frequencies, phenotypes, and functions during both normal health and disease (118, 119). Since age is an additional risk factor for severe COVID-19 disease and Vδ2 T cell frequencies are diminished in the elderly where general a systemic low-grade inflammation is present, the low Vδ2 T cell count could explain why the disease is more severe in older patients (120, 121). The lack of age and sex-matched cohorts within most existing studies presents a clear limitation that should be addressed in future investigations. There is also a growing body of evidence suggesting COVID-19 disease severity is associated with pre-existing comorbidities such as obesity and Type 2 diabetes. These chronic inflammatory health conditions may independently contribute to alterations in γδ T cells including their antiviral responses (122, 123). This necessitates mechanistic studies to determine the independent contribution of SARS-CoV-2 infection on γδ T cell characteristics. Lastly, the definition and stratification of disease severity is often discordant between studies. Identifying robust clinical biomarkers as well as widespread utilization of nucleic acid and rapid antigen testing may allow for more clearly defined methods for analyzing γδ T cells at different stages of COVID-19.
Few studies for the therapeutic use of Vδ2 T cells to treat infectious disease have been performed to date. A pilot study analyzed the effect of combined treatment with the N-BP zoledronate and interleukin-2 in people living with HIV without antiretroviral treatment (124). The authors observed a partial restoration of Vδ2 T cell functionality including enhancement of dendritic maturation and possibly HIV-specific CD8 lymphocyte anti-viral responses.
A therapeutic effect against influenza has been demonstrated (125), showing that human N-BP expanded Vδ2 T cells killed influenza virus-infected cells and inhibit viral replication in vitro (90). Further work demonstrated in immunodeficient mice reconstituted with human peripheral mononuclear cells, that the N-BP pamidronate reduced disease severity caused by human seasonal H1N1 and avian H5N1 influenza virus. In a more recent study, the potential of pamidronate on treating H7N9 virus-infected humanized mice was evaluated showing that intraperitoneal injection of the drug induced expansion of Vδ2 T cells that controlled both viral replication and inflammation in affected lungs (126). The therapeutic effect of Vδ2 T cells has also been demonstrated in other infections such as flavivirus infections (127, 128) and Hepatitis C virus (129, 130).
The tuberculosis vaccine, Bacille Calmette-Guerin (BCG), also affords protection against other diseases, due to immune mechanisms such as activation of non-conventional T-cells, and cross-reactive adaptive immunity (131–133). A recent study used a rhesus macaque model to explore the protective efficacy of aerosol-delivered BCG vaccination against SARS-CoV-2 (134). Immune parameters were monitored in vaccinated and unvaccinated rhesus macaques for 28 days following aerosol BCG vaccination. Interestingly, CD14+ monocytes and Vδ2 T cell frequencies increased rapidly in vaccinated animals following SARS-CoV-2 infection. Despite the lack of efficacy of the vaccination clearing infection, this study adds on the potential immunotherapeutic role of γδ T cells for SARS-CoV2.
These findings provide a strong justification for further studies aimed at translating Vδ2 T cells into the clinic for infectious diseases that currently have limited curative treatment options.
Despite the limited studies on γδ T cells, there is evidence that indicates their active participation fighting SARS-CoV-2 infection similar to their known pivotal antiviral and cytotoxic functions against other viral infections. In addition to their migration and infiltration capabilities into sites of infection, including the lungs (106), their functional plasticity is reflected in their ability to secrete numerous cytokines and crosstalk with other cells including monocytes/macrophages and DCs, to induce strong adaptive immune responses. Our capacity to manipulate the mevalonate pathway allows us to generate γδ T cells with enhanced migration and cytotoxic capabilities (135) and as such, γδ T cell immunotherapy is being pursued from several angles to treat different malignancies (136, 137), and for HIV cure (91, 92). The use of nBPs to prevent or treat COVID-19 was previously postulated (138) although a recent study showed that oral nBPs did not prevent, nor ameliorate severe COVID-19 disease (139). One possible explanation for this outcome could be related to Vδ2 T cell exhaustion after repeated dosage of the nBP (140). As recently reviewed, nBPs could be beneficial for SARS-CoV-2 acute response in the lung (141) since nBPs directly activate Vδ2 T cell homing to tissues (105, 142) and therefore, migration into the lungs in response to SARS-CoV-2 would be achieved. As we reviewed herein, Vδ2 T cells exhibit a potent adjuvant function that includes modulation of B, T, DC, and NK cells functions (143) and their specific activation could shape the full adaptive immune response.
An alternative approach to modulating the mevalonate pathway are statins. Statins are widely used for the treatment of hypercholesterolemia and they function by inhibiting HMG-CoA reductase, which is the rate-limiting enzyme in the mevalonate pathway (144, 145). SARS-CoV-2, like other enveloped viruses, depend on their lipid envelopes for entry and replication in host cells (146). Therefore, to reduce entry of coronaviruses, cholesterol can be depleted from the plasma membranes of target cells (147) In addition, statins have anti-inflammatory and immunomodulatory effects, inhibiting the secretion of proinflammatory cytokines (148) and suppressing T cell activation (149). Different observational and experimental studies suggest that treatment with statins could be associated with better prognosis in severe COVID-19 infection. A meta-analysis demonstrated that the use of statins was associated with a lower risk of mortality in COVID-19 patients (150). However, more investigation is still needed to confirm these findings.
The major adverse effect of intravenously administered nBPs is the acute phase response associated with the release of TNF-α and IL-6 (151–153). Combined administration of nBPs and statins could be a possibility for SARS-CoV-2, similar to a pilot study in patients with (bone)metastasized malignancies receiving nBP-treatment (154). Although statins reduced the production of perforin and GrzB from Vδ2 T cells, they did not prevent nBP effects increasing cell frequency, and IFN-γ and TNF-α production. An additional study showed that prior use of statins avoided the acute phase response by inhibiting nBP-induced production of IL-6 (155). The sequential use of statins and nBPs in strategic paucity to treat mild versus severe COVID-19, constitutes a valuable area of research to find the balance between immune hyperactivation and suppression.
Although SARS-CoV-2 infection research has been centered in other more abundant T cell populations, available studies demonstrate a critical involvement of γδ T cells in fighting SARS-CoV-2 infection, and in determining the severity of COVID-19. Exploiting Vδ2 T cell functional plasticity could lead to better means of prevention and mitigation of severe cases, potentially improving clinical outcomes.
Writing original draft, MS, AC, BTM., and NS-S. Resources, NS-S. Conceptualization, NS-S. All authors have read and agreed to the published version of the manuscript.
This work was funded by NIH Grant R01-AI125097 and R21-AI157864.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature (2020) 579(7798):265–9. doi: 10.1038/s41586-020-2008-3
2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in china, 2019. New Engl J Med (2020) 382(8):727–33. doi: 10.1056/NEJMoa2001017
3. Gralinski LE, Menachery VD. Return of the coronavirus: 2019-nCoV. Viruses (2020) 12(2):135. doi: 10.3390/v12020135
4. To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis (2020) 71(15):841–3. doi: 10.1093/cid/ciaa149
5. Hung LS. The SARS epidemic in Hong Kong: what lessons have we learned? J R Soc Med (2003) 96(8):374–8. doi: 10.1258/jrsm.96.8.374
6. Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome composition and divergence of the novel coronavirus (2019-ncov) originating in China. Cell Host Microbe (2020) 27(3):325–8. doi: 10.1016/j.chom.2020.02.001
7. Fehr AR, Perlman S. Coronaviruses: An overview of their replication and pathogenesis. New York: Springer (2015) p. 1–23.
8. Beniac DR, Andonov A, Grudeski E, Booth TF.. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol (2006) 13(8):751–2. doi: 10.1038/nsmb1123
9. Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S. A structural analysis of m protein in coronavirus assembly and morphology. J Struct Biol (2011) 174(1):11–22. doi: 10.1016/j.jsb.2010.11.021
10. Cong Y, Ulasli M, Schepers H, Mauthe M, V'kovski P, Kriegenburg F, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol (2020) 94(4). doi: 10.1128/JVI.01925-19
11. Chang C-K, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, et al. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci (2006) 13(1):59–72. doi: 10.1007/s11373-005-9035-9
12. Nieto-Torres JL, De Diego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PloS Pathog (2014) 10(5):e1004077. doi: 10.1371/journal.ppat.1004077
13. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J (2019) 16(1). doi: 10.1186/s12985-019-1182-0
14. Pervushin K, Tan E, Parthasarathy K, Lin X, Jiang FL, Yu D, et al. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PloS Pathog (2009) 5(7):e1000511. doi: 10.1371/journal.ppat.1000511
15. Cao Y, Yang R, Lee I, Zhang W, Sun J, Wang W, Meng X. Characterization of the SARS-CoV-2 e protein: Sequence, structure, viroporin, and inhibitors. Protein Sci (2021) 30(6):1114–30. doi: 10.1002/pro.4075
16. Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol (2021) 433(2):166725. doi: 10.1016/j.jmb.2020.11.024
17. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell (2020) 181(2):271–280.e8.
18. Astuti I, Ysrafil. Severe acute respiratory syndrome coronavirus 2 (sars-cov-2): an overview of viral structure and host response. Diabetes Metab Syndr (2020) 14(4):407–12.
19. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell (2020) 181(5):1016–1035.e19.
20. Al-Horani RA, Kar S, Aliter KF. Potential anti-COVID-19 therapeutics that block the early stage of the viral life cycle: structures, mechanisms, and clinical trials. Int J Mol Sci (2020) 21(15).
21. de Haan CA, et al. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol (1998) 72(8):6838–50.
22. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (2020) 395(10224):565–74. doi: 10.1016/S0140-6736(20)30251-8
23. Wu HY, Brian DA. Subgenomic messenger RNA amplification in coronaviruses. Proc Natl Acad Sci U.S.A. (2010) 107(27):12257–62. doi: 10.1073/pnas.1000378107
24. Malin JJ, et al. Remdesivir against COVID-19 and other viral diseases. Clin Microbiol Rev (2020) 34(1). doi: 10.1128/CMR.00162-20
25. Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med (2017) 9(396). doi: 10.1126/scitranslmed.aal3653
26. Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, et al. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of covid-19. ACS Cent Sci (2020) 6(5):672–83. doi: 10.1021/acscentsci.0c00489
27. Pirzada RH, et al. Remdesivir and ledipasvir among the fda-approved antiviral drugs have potential to inhibit sars-cov-2 replication. Cells (2021) 10(5). doi: 10.3390/cells10051052
28. Jorgensen SCJ, Kebriaei R, Dresser LD. Remdesivir: review of pharmacology, pre-clinical data, and emerging clinical experience for covid-19. Pharmacotherapy (2020) 40(7):659–71. doi: 10.1002/phar.2429
29. Choy KT, Wong AY, Kaewpreedee P, Sia SF, Chen D, Hui KPY, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res (2020) 178:104786. doi: 10.1016/j.antiviral.2020.104786
30. Rahmani H, Davoudi-Monfared E, Nourian A, Khalili H, Hajizadeh N, Jalalabadi NZ, et al. Interferon β-1b in treatment of severe COVID-19: A randomized clinical trial. Int Immunopharmacol (2020) 88:106903. doi: 10.1016/j.intimp.2020.106903
31. Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet (2020) 395(10238):1695–704. doi: 10.1016/S0140-6736(20)31042-4
32. Graham BS. Rapid COVID-19 vaccine development. Science (2020) 368(6494):945–6. doi: 10.1126/science.abb8923
33. Li YD, Chi WY, Su JH, Ferrall L, Hung CF, Wu TC. Coronavirus vaccine development: from SARS and MERS to COVID-19. J BioMed Sci (2020) 27(1):104. doi: 10.1186/s12929-020-00695-2
34. Marian AJ. Current state of vaccine development and targeted therapies for COVID-19: impact of basic science discoveries. Cardiovasc Pathol (2021) 50:107278. doi: 10.1016/j.carpath.2020.107278
35. Hu B, Huang S, Yin L. The cytokine storm and COVID-19. J Med Virol (2021) 93(1):250–6. doi: 10.1002/jmv.26232
36. Song P, et al. Cytokine storm induced by SARS-CoV-2. Clin Chim Acta (2020) 509:280–7. doi: 10.1016/j.cca.2020.06.017
37. Chen Y, Klein SL, Garibaldi BT, Li H, Wu C, Osevala NM, et al. Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res Rev (2021) 65:101205. doi: 10.1016/j.arr.2020.101205
38. Dosch SF, Mahajan SD, Collins AR. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res (2009) 142(1-2):19–27. doi: 10.1016/j.virusres.2009.01.005
39. Hariharan A, et al. The role and therapeutic potential of NF-kappa-B pathway in severe COVID-19 patients. Inflammopharmacology (2021) 29(1):91–100. doi: 10.1007/s10787-020-00773-9
40. Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol (2012) 66:177–96. doi: 10.1146/annurev-micro-092611-150203
41. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5
42. Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood (2008) 112(5):1557–69. doi: 10.1182/blood-2008-05-078154
43. Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine (2020) 55:102763. doi: 10.1016/j.ebiom.2020.102763
44. Wang J, et al. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: Review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol (2020) 108(1):17–41. doi: 10.1002/JLB.3COVR0520-272R
45. Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol (2017) 39(5):517–28.
46. Li H, Liu L, Zhang D, Xu J, Dai H, Tang N, et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet (2020) 395(10235):1517–20.
47. Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe (2020) 27(6):992–1000.e3.
48. Alhazzani W, Møller MH, Arabi YM, Loeb M, Gong MN, Fan E, et al. Surviving sepsis campaign: Guidelines on the management of critically Ill adults with coronavirus disease 2019 (COVID-19). Crit Care Med (2020) 48(6):e440–69.
49. Dong X, Wang C, Liu X, Gao W, Bai X, Li Z, et al. Lessons learned comparing immune system alterations of bacterial sepsis and SARS-CoV-2 sepsis. Front Immunol (2020) 11:598404.
50. Olwal CO, Nganyewo NN, Tapela K, Djomkam Zune AL, Owoicho O, Bediako Y, Duodu S, et al. Parallels in sepsis and COVID-19 conditions: Implications for managing severe COVID-19. Front Immunol (2021) 12:602848.
51. Tay MZ, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol (2020) 20(6):363–74. doi: 10.1038/s41577-020-0311-8
52. Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol (2020) 15:123–47. doi: 10.1146/annurev-pathmechdis-012418-012718
53. Mercer J, Greber UF. Virus interactions with endocytic pathways in macrophages and dendritic cells. Trends Microbiol (2013) 21(8):380–8. doi: 10.1016/j.tim.2013.06.001
54. MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol (1997) 15:323–50. doi: 10.1146/annurev.immunol.15.1.323
55. Chien YH, Meyer C, Bonneville M. γδ T cells: first line of defense and beyond. Annu Rev Immunol (2014) 32:121–55. doi: 10.1146/annurev-immunol-032713-120216
56. Poccia F, Agrati C, Castilletti C, Bordi L, Gioia C, Horejsh D, et al. Anti-severe acute respiratory syndrome coronavirus immune responses: the role played by V gamma 9V delta 2 T cells. J Infect Dis (2006) 193(9):1244–9. doi: 10.1086/502975
57. Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol (2013) 13(2):88–100. doi: 10.1038/nri3384
58. Silva-Santos B, Serre K, Norell H. γδ T cells in cancer. Nat Rev Immunol (2015) 15(11):683–91. doi: 10.1038/nri3904
59. Deschler S, Kager J, Erber J, Fricke L, Koyumdzhieva P, Georgieva A, et al. Mucosal-associated invariant T (MAIT) cells are highly activated and functionally impaired in COVID-19 patients. Viruses (2021) 13(2). doi: 10.3390/v13020241
60. Trottein F, Paget C. Natural killer t cells and mucosal-associated invariant t cells in lung infections. Front Immunol (2018) 9:1750. doi: 10.3389/fimmu.2018.01750
61. McCarthy NE, Eberl M. Human γδ t-cell control of mucosal immunity and inflammation. Front Immunol (2018) 9:985. doi: 10.3389/fimmu.2018.00985
62. Tanaka Y. Human gamma delta T cells and tumor immunotherapy. J Clin Exp Hematop (2006) 46(1):11–23. doi: 10.3960/jslrt.46.11
63. Wu D, et al. Human γδT-cell subsets and their involvement in tumor immunity. Cell Mol Immunol (2017) 14(3):245–53.
64. Takihara Y, Reimann J, Michalopoulos E, Ciccone E, Moretta L, Mak TW, et al. Diversity and structure of human T cell receptor delta chain genes in peripheral blood gamma/delta-bearing T lymphocytes. J Exp Med (1989) 169(2):393–405. doi: 10.1084/jem.169.2.393
65. Dimova T, Brouwer M, Gosselin F, Tassignon J, Leo O, Donner C, et al. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc Natl Acad Sci U.S.A. (2015) 112(6):E556–65. doi: 10.1073/pnas.1412058112
66. Green AE, Lissina A, Hutchinson SL, Hewitt RE, Temple B, James D, et al. Recognition of nonpeptide antigens by human V gamma 9V delta 2 T cells requires contact with cells of human origin. Clin Exp Immunol (2004) 136(3):472–82. doi: 10.1111/j.1365-2249.2004.02472.x
67. Jomaa H, Feurle J, Lühs K, Kunzmann V, Tony HP, Herderich M, et al. Vgamma9/Vdelta2 T cell activation induced by bacterial low molecular mass compounds depends on the 1-deoxy-D-xylulose 5-phosphate pathway of isoprenoid biosynthesis. FEMS Immunol Med Microbiol (1999) 25(4):371–8.
68. Eberl M, Hintz M, Reichenberg A, Kollas AK, Wiesner J, Jomaa H, et al. Microbial isoprenoid biosynthesis and human gammadelta T cell activation. FEBS Lett (2003) 544(1-3):4–10. doi: 10.1016/S0014-5793(03)00483-6
69. Gober HJ, Kistowska M, Angman L, Jenö P, Mori L, De Libero G, et al. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med (2003) 197(2):163–8. doi: 10.1084/jem.20021500
70. Gossman W, Oldfield E. Quantitative structure–activity relations for gammadelta T cell activation by phosphoantigens. J Med Chem (2002) 45(22):4868–74. doi: 10.1021/jm020224n
71. van Beek E, et al. Farnesyl pyrophosphate synthase is the molecular target of nitrogen-containing bisphosphonates. Biochem Biophys Res Commun (1999) 264(1):108–11. doi: 10.1006/bbrc.1999.1499
72. Rigau M, Ostrouska S, Fulford TS, Johnson DN, Woods K, Ruan Z, et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by γδ T cells. Science (2020), 367(6478). doi: 10.1126/science.aay5516
73. Harly C, Peigné CM, Scotet E. Molecules and mechanisms implicated in the peculiar antigenic activation process of human vγ9vδ2 t cells. Front Immunol (2014) 5:657.
74. Chen H, He X, Wang Z, Wu D, Zhang H, Xu C, et al. Identification of human T cell receptor gammadelta-recognized epitopes/proteins via CDR3delta peptide-based immunobiochemical strategy. J Biol Chem (2008) 283(18):12528–37. doi: 10.1074/jbc.M708067200
75. Laad AD, et al. Human gamma delta T cells recognize heat shock protein-60 on oral tumor cells. Int J Cancer (1999) 80(5):709–14. doi: 10.1002/(SICI)1097-0215(19990301)80:5<709::AID-IJC14>3.0.CO;2-R
76. Scotet E, Martinez LO, Grant E, Barbaras R, Jenö P, Guiraud M, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein a-I. Immunity (2005) 22(1):71–80. doi: 10.1016/j.immuni.2004.11.012
77. Hudspeth K, Fogli M, Correia DV, Mikulak J, Roberto A, Della Bella S, et al. Engagement of NKp30 on Vδ1 T cells induces the production of CCL3, CCL4, and CCL5 and suppresses HIV-1 replication. Blood (2012) 119(17):4013–6. doi: 10.1182/blood-2011-11-390153
78. Wu J, Groh V, Spies T. T Cell antigen receptor engagement and specificity in the recognition of stress-inducible MHC class I-related chains by human epithelial gamma delta T cells. J Immunol (2002) 169(3):1236–40. doi: 10.4049/jimmunol.169.3.1236
79. Rincon-Orozco B, Kunzmann V, Wrobel P, Kabelitz D, Steinle A, Herrmann T, et al. Activation of V gamma 9V delta 2 T cells by NKG2D. J Immunol (2005) 175(4):2144–51. doi: 10.4049/jimmunol.175.4.2144
80. González S, et al. NKG2D ligands: key targets of the immune response. Trends Immunol (2008) 29(8):397–403. doi: 10.1016/j.it.2008.04.007
81. Wu YL, Ding YP, Tanaka Y, Shen LW, Wei CH, Minato N, et al. γδ T cells and their potential for immunotherapy. Int J Biol Sci (2014) 10(2):119–35. doi: 10.7150/ijbs.7823
82. Peters C, Kabelitz D, Wesch D. Regulatory functions of γδ T cells. Cell Mol Life Sci (2018) 75(12):2125–35. doi: 10.1007/s00018-018-2788-x
83. Wakita D, Sumida K, Iwakura Y, Nishikawa H, Ohkuri T, Chamoto K, et al. Tumor-infiltrating IL-17-producing gammadelta T cells support the progression of tumor by promoting angiogenesis. Eur J Immunol (2010) 40(7):1927–37. doi: 10.1002/eji.200940157
84. Ma Y, Aymeric L, Locher C, Mattarollo SR, Delahaye NF, Pereira P, et al. Contribution of IL-17-producing gamma delta T cells to the efficacy of anticancer chemotherapy. J Exp Med (2011) 208(3):491–503. doi: 10.1084/jem.20100269
85. Poccia F, Agrati C, Martini F, Capobianchi MR, Wallace M, Malkovsky M, et al. Antiviral reactivities of gammadelta T cells. Microbes Infect (2005) 7(3):518–28. doi: 10.1016/j.micinf.2004.12.009
86. Mann BT, et al. Boosting the immune system for HIV cure: A γδ T cell perspective. Front Cell Infect Microbiol (2020) 10:221. doi: 10.3389/fcimb.2020.00221
87. Dalton JE, et al. Fas-fas ligand interactions are essential for the binding to and killing of activated macrophages by gamma delta T cells. J Immunol (2004) 173(6):3660–7. doi: 10.4049/jimmunol.173.6.3660
88. Ponomarev ED, Dittel BN. Gamma delta T cells regulate the extent and duration of inflammation in the central nervous system by a fas ligand-dependent mechanism. J Immunol (2005) 174(8):4678–87. doi: 10.4049/jimmunol.174.8.4678
89. Palomino-Segura M, et al. Early production of IL-17A by γδ T cells in the trachea promotes viral clearance during influenza infection in mice. Eur J Immunol (2020) 50(1):97–109. doi: 10.1002/eji.201948157
90. Tu W, Zheng J, Liu Y, Sia SF, Liu M, Qin G, et al. The aminobisphosphonate pamidronate controls influenza pathogenesis by expanding a gammadelta T cell population in humanized mice. J Exp Med (2011) 208(7):1511–22. doi: 10.1084/jem.20110226
91. Garrido C, Clohosey ML, Whitworth CP, Hudgens M, Margolis DM, Soriano-Sarabia N, et al. γδ T cells: an immunotherapeutic approach for HIV cure strategies. JCI Insight (2018) 3(12). doi: 10.1172/jci.insight.120121
92. James KS, Trumble I, Clohosey ML, Moeser M, Roan NR, Adimora AA, et al. Measuring the contribution of γδ T cells to the persistent HIV reservoir. Aids (2020) 34(3):363–71. doi: 10.1097/QAD.0000000000002434
93. Autran B, Triebel F, Katlama C, Rozenbaum W, Hercend T, Debre P, et al. T Cell receptor gamma/delta+ lymphocyte subsets during HIV infection. Clin Exp Immunol (1989) 75(2):206–10.
94. Juno JA, Eriksson EM. γδ T-cell responses during HIV infection and antiretroviral therapy. Clin Transl Immunol (2019) 8(7):e01069.
95. Cheng M, Hu S. Lung-resident γδ T cells and their roles in lung diseases. Immunology (2017) 151(4):375–84. doi: 10.1111/imm.12764
96. Sant S, Jenkins MR, Dash P, Watson KA, Wang Z, Pizzolla A, et al. Human γδ T-cell receptor repertoire is shaped by influenza viruses, age and tissue compartmentalisation. Clin Transl Immunol (2019) 8(9):e1079. doi: 10.1002/cti2.1079
97. Qin G, Liu Y, Zheng J, Ng IH, Xiang Z, Lam KT, et al. Type 1 responses of human Vγ9Vδ2 T cells to influenza a viruses. J Virol (2011) 85(19):10109–16. doi: 10.1128/JVI.05341-11
98. Glatzel A, Wesch D, Schiemann F, Brandt E, Janssen O, Kabelitz D. Patterns of chemokine receptor expression on peripheral blood gamma delta T lymphocytes: strong expression of CCR5 is a selective feature of V delta 2/V gamma 9 gamma delta T cells. J Immunol (2002) 168(10):4920–9. doi: 10.4049/jimmunol.168.10.4920
99. Jameson JM, et al. A role for the mevalonate pathway in the induction of subtype cross-reactive immunity to influenza a virus by human gammadelta T lymphocytes. Cell Immunol (2010) 264(1):71–7. doi: 10.1016/j.cellimm.2010.04.013
100. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest (2020) 130(5):2620–9. doi: 10.1172/JCI137244
101. Jiang M, Guo Y, Luo Q, Huang Z, Zhao R, Liu S, et al. T-Cell subset counts in peripheral blood can be used as discriminatory biomarkers for diagnosis and severity prediction of coronavirus disease 2019. J Infect Dis (2020) 222(2):198–202. doi: 10.1093/infdis/jiaa252
102. Rijkers G, Vervenne T, van der Pol P. More bricks in the wall against SARS-CoV-2 infection: involvement of γ9δ2 T cells. Cell Mol Immunol (2020) 17(7):771–2. doi: 10.1038/s41423-020-0473-0
103. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med (2020) 26(7):1070–6. doi: 10.1038/s41591-020-0944-y
104. Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med (2020) 26(10):1623–35. doi: 10.1038/s41591-020-1038-6
105. Carissimo G, Xu W, Kwok I, Abdad MY, Chan YH, Fong SW, et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat Commun (2020) 11(1):5243. doi: 10.1038/s41467-020-19080-6
106. Chen XJ, Li K, Xu L, Yu YJ, Wu B, He YL, et al. Novel insight from the first lung transplant of a COVID-19 patient. Eur J Clin Invest (2021) 51(1):e13443. doi: 10.1111/eci.13443
107. Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J Exp Med (2020) 217(12). doi: 10.1084/jem.20200872
108. Lei L, Qian H, Yang X, Zhang X, Zhang D, Dai T, et al. The phenotypic changes of γδ T cells in COVID-19 patients. J Cell Mol Med (2020) 24(19):11603–6. doi: 10.1111/jcmm.15620
109. Odak I, Barros-Martins J, Bošnjak B, Stahl K, David S, Wiesner O, et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine (2020) 57:102885. doi: 10.1016/j.ebiom.2020.102885
110. Kalyan S, Kabelitz D. Defining the nature of human γδ T cells: a biographical sketch of the highly empathetic. Cell Mol Immunol (2013) 10(1):21–9. doi: 10.1038/cmi.2012.44
111. Soriano-Sarabia N, Archin NM, Bateson R, Dahl NP, Crooks AM, Kuruc JD, et al. Peripheral Vγ9Vδ2 T cells are a novel reservoir of latent HIV infection. PloS Pathog (2015) 11(10):e1005201.
112. Lusso P, et al. Infection of gamma/delta T lymphocytes by human herpesvirus 6: transcriptional induction of CD4 and susceptibility to HIV infection. J Exp Med (1995) 181(4):1303–10. doi: 10.1084/jem.181.4.1303
113. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med (2020) 26(6):842–4. doi: 10.1038/s41591-020-0901-9
114. Bouadma L, Wiedemann A, Patrier J, Surénaud M, Wicky PH, Foucat E, et al. Immune alterations in a patient with SARS-CoV-2-Related acute respiratory distress syndrome. J Clin Immunol (2020) 40(8):1082–92. doi: 10.1007/s10875-020-00839-x
115. Ryan PL, Sumaria N, Holland CJ, Bradford CM, Izotova N, Grandjean CL, et al. Heterogeneous yet stable Vδ2(+) T-cell profiles define distinct cytotoxic effector potentials in healthy human individuals. Proc Natl Acad Sci U.S.A. (2016) 113(50):14378–83. doi: 10.1073/pnas.1611098113
116. Yu Y, Wang M, Zhang X, Li S, Lu Q, Zeng H, et al. Antibody-dependent cellular cytotoxicity response to SARS-CoV-2 in COVID-19 patients. Signal Transduct Target Ther (2021) 6(1):346. doi: 10.1038/s41392-021-00759-1
117. Tso FY, Lidenge SJ, Poppe LK, Peña PB, Privatt SR, Bennett SJ, et al. Presence of antibody-dependent cellular cytotoxicity (ADCC) against SARS-CoV-2 in COVID-19 plasma. PloS One (2021) 16(3):e0247640. doi: 10.1371/journal.pone.0247640
118. Cairo C, Armstrong CL, Cummings JS, Deetz CO, Tan M, Lu C, et al. Impact of age, gender, and race on circulating γδ T cells. Hum Immunol (2010) 71(10):968–75. doi: 10.1016/j.humimm.2010.06.014
119. Caccamo N, et al. Sex-specific phenotypical and functional differences in peripheral human Vgamma9/Vdelta2 T cells. J Leukoc Biol (2006) 79(4):663–6. doi: 10.1189/jlb.1105640
120. Michishita Y, Hirokawa M, Guo YM, Abe Y, Liu J, Ubukawa K, et al. Age-associated alteration of γδ T-cell repertoire and different profiles of activation-induced death of Vδ1 and Vδ2 T cells. Int J Hematol (2011) 94(3):230–40. doi: 10.1007/s12185-011-0907-7
121. Caccamo N, Meraviglia S, Ferlazzo V, Angelini D, Borsellino G, Poccia F, et al. Differential requirements for antigen or homeostatic cytokines for proliferation and differentiation of human Vgamma9Vdelta2 naive, memory and effector T cell subsets. Eur J Immunol (2005) 35(6):1764–72. doi: 10.1002/eji.200525983
122. Fay NS, Larson EC, Jameson JM. Chronic inflammation and γδ T cells. Front Immunol (2016) 7:210. doi: 10.3389/fimmu.2016.00210
123. Costanzo AE, Taylor KR, Dutt S, Han PP, Fujioka K, Jameson JM, et al. Obesity impairs γδ T cell homeostasis and antiviral function in humans. PloS One (2015) 10(3):e0120918.
124. Poccia F, Gioia C, Martini F, Sacchi A, Piacentini P, Tempestilli M, et al. Zoledronic acid and interleukin-2 treatment improves immunocompetence in HIV-infected persons by activating Vgamma9Vdelta2 T cells. Aids (2009) 23(5):555–65. doi: 10.1097/QAD.0b013e3283244619
125. Sabbaghi A, Miri SM, Keshavarz M, Mahooti M, Zebardast A, Ghaemi A. Role of γδ T cells in controlling viral infections with a focus on influenza virus: implications for designing novel therapeutic approaches. Virol J (2020) 17(1):174. doi: 10.1186/s12985-020-01449-0
126. Zheng J, Wu WL, Liu Y, Xiang Z, Liu M, Chan KH, et al. The therapeutic effect of pamidronate on lethal avian influenza a H7N9 virus infected humanized mice. PloS One (2015) 10(8):e0135999. doi: 10.1371/journal.pone.0135999
127. Wang T, Gao Y, Scully E, Davis CT, Anderson JF, Welte T, et al. Gamma delta T cells facilitate adaptive immunity against West Nile virus infection in mice. J Immunol (2006) 177(3):1825–32. doi: 10.4049/jimmunol.177.3.1825
128. Agrati C, Castilletti C, Cimini E, Romanelli A, Lapa D, Quartu S, et al. Antiviral activity of human Vδ2 T-cells against WNV includes both cytolytic and non-cytolytic mechanisms. New Microbiol (2016) 39(2):139–42.
129. Agrati C, Alonzi T, De Santis R, Castilletti C, Abbate I, Capobianchi MR, et al. Activation of Vgamma9Vdelta2 T cells by non-peptidic antigens induces the inhibition of subgenomic HCV replication. Int Immunol (2006) 18(1):11–8. doi: 10.1093/intimm/dxh337
130. Cimini E, Bonnafous C, Bordoni V, Lalle E, Sicard H, Sacchi A, et al. Interferon-α improves phosphoantigen-induced Vγ9Vδ2 T-cells interferon-γ production during chronic HCV infection. PloS One (2012) 7(5):e37014.
131. Covián C, ernández-Fierro A, Retamal-Díaz A, Díaz FE, Vasquez AE, Lay MK, et al. BCG-Induced cross-protection and development of trained immunity: Implication for vaccine design. Front Immunol (2019) 10:2806. doi: 10.3389/fimmu.2019.02806
132. Shann F. The non-specific effects of vaccines. Arch Dis Child (2010) 95(9):662–7. doi: 10.1136/adc.2009.157537
133. Morrison AL, et al. Cheap and commonplace: Making the case for BCG and γδ T cells in COVID-19. Front Immunol (2021) 12:743924. doi: 10.3389/fimmu.2021.743924
134. White AD, Sibley L, Sarfas C, Morrison AL, Bewley K, Churchward C, et al. Influence of aerosol delivered BCG vaccination on immunological and disease parameters following SARS-CoV-2 challenge in rhesus macaques. Front Immunol (2021) 12:801799. doi: 10.3389/fimmu.2021.801799
135. Wallace M, Bartz SR, Chang WL, Mackenzie DA, Pauza CD, Malkovsky M, et al. Gamma delta T lymphocyte responses to HIV. Clin Exp Immunol (1996) 103(2):177–84.
136. Berenson JR. Zoledronic acid in cancer patients with bone metastases: results of phase I and II trials. Semin Oncol (2001) 28(2 Suppl 6):25–34. doi: 10.1016/S0093-7754(01)90262-3
137. Kunzmann V, Bauer E, Wilhelm M. Gamma/delta T-cell stimulation by pamidronate. N Engl J Med (1999) 340(9):737–8. doi: 10.1056/NEJM199903043400914
138. Brufsky A, et al. Boning up: amino-bisphophonates as immunostimulants and endosomal disruptors of dendritic cell in SARS-CoV-2 infection. J Transl Med (2020) 18(1):261. doi: 10.1186/s12967-020-02433-6
139. Degli Esposti L, Perrone V, Sangiorgi D, Andretta M, Bartolini F, Cavaliere A, et al. The use of oral amino-bisphosphonates and coronavirus disease 2019 (COVID-19) outcomes. J Bone Miner Res (2021) 36(11):2177–83. doi: 10.1002/jbmr.4419
140. Kabelitz D, et al. Cancer immunotherapy with γδ T cells: many paths ahead of us. Cell Mol Immunol (2020) 17(9):925–39. doi: 10.1038/s41423-020-0504-x
141. Thompson K, Roelofs AJ, Jauhiainen M, Mönkkönen H, Mönkkönen J, Rogers MJ. Activation of γδ T cells by bisphosphonates. Adv Exp Med Biol (2010) 658:11–20. doi: 10.1007/978-1-4419-1050-9_2
142. Davey MS, Lin CY, Roberts GW, Heuston S, Brown AC, Chess JA, et al. Human neutrophil clearance of bacterial pathogens triggers anti-microbial γδ T cell responses in early infection. PloS Pathog (2011) 7(5):e1002040. doi: 10.1371/journal.ppat.1002040
143. Lafont V, Sanchez F, Laprevotte E, Michaud HA, Gros L, Eliaou JF, Bonnefoy N. Plasticity of γδ T cells: Impact on the anti-tumor response. Front Immunol (2014) 5:622. doi: 10.3389/fimmu.2014.00622
144. Thompson K, Rogers MJ. Statins prevent bisphosphonate-induced gamma,delta-t-cell proliferation and activation in vitro. J Bone Miner Res (2004) 19(2):278–88.
145. Moghadasian MH. Clinical pharmacology of 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors. Life Sci (1999) 65(13):1329–37. doi: 10.1016/S0024-3205(99)00199-X
146. Sun X, Whittaker GR. Role for influenza virus envelope cholesterol in virus entry and infection. J Virol (2003) 77(23):12543–51. doi: 10.1128/JVI.77.23.12543-12551.2003
147. Glende J, Schwegmann-Wessels C, Al-Falah M, Pfefferle S, Qu X, Deng H, et al. Importance of cholesterol-rich membrane microdomains in the interaction of the s protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology (2008) 381(2):215–21. doi: 10.1016/j.virol.2008.08.026
148. Blanco-Colio LM, et al. Anti-inflammatory and immunomodulatory effects of statins. Kidney Int (2003) 63(1):12–23. doi: 10.1046/j.1523-1755.2003.00744.x
149. Overton ET, Sterrett S, Westfall AO, Kahan SM, Burkholder G, Zajac AJ, et al. Effects of atorvastatin and pravastatin on immune activation and T-cell function in antiretroviral therapy-suppressed HIV-1-infected patients. Aids (2014) 28(17):2627–31. doi: 10.1097/QAD.0000000000000475
150. Diaz-Arocutipa C, Melgar-Talavera B, Alvarado-Yarasca Á, Saravia-Bartra MM, Cazorla P, Belzusarri I, et al. Statins reduce mortality in patients with COVID-19: an updated meta-analysis of 147 824 patients. Int J Infect Dis (2021) 110:374–81. doi: 10.1016/j.ijid.2021.08.004
151. Adami S, Bhalla AK, Dorizzi R, Montesanti F, Rosini S, Salvagno G, et al. The acute-phase response after bisphosphonate administration. Calcif Tissue Int (1987) 41(6):326–31. doi: 10.1007/BF02556671
152. Abrahamsen B. Adverse effects of bisphosphonates. Calcif Tissue Int (2010) 86(6):421–35. doi: 10.1007/s00223-010-9364-1
153. Kennel KA, Drake MT. Adverse effects of bisphosphonates: implications for osteoporosis management. Mayo Clin Proc (2009) 84(7):632–7. doi: 10.1016/S0025-6196(11)60752-0
154. Schneiders FL, Huijts CM, Reijm M, Bontkes HJ, Verheul HMW, de Gruijl TD, et al. The effects of systemic treatment with aminobisphosphonates and statins on circulating Vγ9Vδ2-T cells in patients with advanced cancer. Immunobiology (2018) 223(2):171–7. doi: 10.1016/j.imbio.2017.10.029
155. Hewitt RE, Lissina A, Green AE, Slay ES, Price DA, Sewell AK, et al. The bisphosphonate acute phase response: rapid and copious production of proinflammatory cytokines by peripheral blood gd T cells in response to aminobisphosphonates is inhibited by statins. Clin Exp Immunol (2005) 139(1):101–11.
Keywords: gamma delta (γδ) T cells, SARS-CoV-2, innate immunity, aminobisphosphonates COVID-19, COVID-19
Citation: Sanz M, Mann BT, Chitrakar A and Soriano-Sarabia N (2022) Defying convention in the time of COVID-19: Insights into the role of γδ T cells. Front. Immunol. 13:819574. doi: 10.3389/fimmu.2022.819574
Received: 21 November 2021; Accepted: 14 July 2022;
Published: 11 August 2022.
Edited by:
Qingfeng Chen, Institute of Molecular and Cell Biology (A*STAR), SingaporeReviewed by:
Sina Taefehshokr, University of Manitoba, CanadaCopyright © 2022 Sanz, Mann, Chitrakar and Soriano-Sarabia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalia Soriano-Sarabia, bmF0YWxpYXNvcnNhckBnd3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.