
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 22 February 2022
Sec. Vaccines and Molecular Therapeutics
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.819222
This article is part of the Research Topic Nature's Own Host Defense Peptides: From Discovery to Drugs View all 4 articles
Enhancing the synthesis of microbicidal and immunomodulatory host defense peptides (HDP) is a promising host-directed antimicrobial strategy to combat a growing threat of antimicrobial resistance. Here we investigated the effect of several natural cyclooxygenase-2 (COX-2) inhibitors on chicken HDP gene regulation. Our results indicated that phenolic COX-2 inhibitors such as quercetin, resveratrol, epigallocatechin gallate, anacardic acid, and garcinol enhanced HDP gene expression in chicken HTC macrophage cell line and peripheral blood mononuclear cells (PBMCs). Moreover, these natural COX-2 inhibitors showed a strong synergy with butyrate in augmenting the expressions of multiple HDP genes in HTC cells and PBMCs. Additionally, quercetin and butyrate synergistically promoted the expressions of mucin-2 and claudin-1, two major genes involved in barrier function, while suppressing lipopolysaccharide-triggered interleukin-1β expression in HTC macrophages. Mechanistically, we revealed that NF-κB, p38 mitogen-activated protein kinase, and cyclic adenosine monophosphate signaling pathways were all involved in the avian β-defensin 9 gene induction, but histone H4 was not hyperacetylated in response to a combination of butyrate and quercetin. Because of their HDP-inducing, barrier-protective, and antiinflammatory activities, these natural COX-2 inhibitors, when combined with butyrate, may be developed as novel host-directed antimicrobial therapeutics.
The rapid emergence of antibiotic-resistant bacteria has become a major public health concern (1), creating an urgent need for the development of novel antimicrobial therapeutic strategies with a minimum risk to trigger resistance (2). Boosting host innate immunity through induction of endogenous host defense peptide (HDP) synthesis has attracted an increasing attention as a host-directed antiinfective approach (3–5). HDPs, also known as antimicrobial peptides, are a group of small cationic and amphipathic peptides with preferential expression in phagocytes and epithelial cells of the host (6). HDPs consist mainly of the defensin or cathelicidin families in vertebrates. Defensins are categorized by the presence of six cysteine residues, while cathelicidins are comprised of a conserved cathelin domain and a diversified mature peptide sequence (4, 7). A total of 14 defensins (AvBD1–14) and four cathelicidins (CATH1–3 and CATHB1) exist in chickens (7, 8). Chicken HDPs are widely expressed in the respiratory, gastrointestinal, and urogenital tracts as well as in multiple lymphoid cells, and regulated differentially in response to infection and inflammation (7, 8). With antimicrobial, immunomodulatory, and barrier protective activities, HDPs constitute a critically important component of the innate immunity system (6, 9). A range of small-molecule compounds such as short-chain fatty acids, vitamin D3, and histone deacetylase (HDAC) inhibitors have been found to be capable of inducing HDP synthesis and some have been explored for disease control and prevention (3–5, 10, 11).
Butyrate, a short-chain fatty acid fermented from dietary carbohydrates by intestinal bacteria, is a well-known HDP inducer in humans and animals (4, 5). Inhibition of HDAC is a major mechanism by which butyrate promotes HDP expression (12), while mitogen-activated protein kinase (MAPK) signaling pathways are also involved in butyrate-mediated HDP induction (13). Butyrate has been shown to confer protection to infections at least in part via induction of HDPs (14, 15). Synergic augmentation of HDPs has been observed between butyrate and several other HDP-inducing compounds such as vitamin D3 (16), lactose (17, 18), forskolin (19, 20), and wortmannin (21).
Cyclooxygenase (COX) is a key enzyme responsible for the conversion of arachidonic acid to prostanoids (22, 23). COX-1 is a constitutive isoform involved in maintaining barrier integrity, while COX-2 is inducible by inflammatory stimuli and often a target for treating inflammatory diseases. Several natural phenolic compounds such as quercetin, resveratrol, and epigallocatechin gallate (EGCG), anacardic acid, and garcinol are well-known COX-2 inhibitors (22, 23). In addition, a few of these polyphenols such as resveratrol (24), curcumin (25), EGCG (26, 27), and genistein (28) are capable of inducing HDP genes. It is unknown whether these polyphenols induce HDP genes by acting as COX-2 inhibitors. Additionally, a potential HDP-inducing synergy between butyrate and COX-2 inhibitors is yet to be explored.
In the current study, we investigated a potential synergy between butyrate and several natural phenolic COX-2 inhibitors in regulating the expression of various HDP genes in chicken HTC macrophages and peripheral blood mononuclear cells (PBMCs). To confirm these polyphenols synergize with butyrate in HDP induction through COX-2 inhibition, two synthetic COX-2-specific inhibitors namely nimesulide and niflumic acid were also evaluated for their synergy with butyrate in HDP induction in chicken cells. The involvement of histone acetylation as well as the MAPK, nuclear factor-κB (NF-κB), and cyclic adenosine monophosphate (cAMP) signaling pathways in the avian β-defensin 9 (AvBD9) gene induction mediated by butyrate and COX-2 inhibitors was also examined.
Quercetin and garcinol were acquired from Cayman Chemical (Ann Arbor, MI), while sodium butyrate, tributyrin (glyceryl tributyrate), and lipopolysaccharide (LPS) (Escherichia coli O55:B5) were obtained from MilliporeSigma (St. Louis, MO). Resveratrol, anacardic acid, EGCG, nimesulide, niflumic acid, SB203580, SP600125, PD98059, PDTC, MG132, and 2’,5’-dideoxyadenosine (DDA) were procured from Santa Cruz Biotechnology (Dallas, TX). Sodium butyrate was dissolved in Roswell Park Memorial Institute (RPMI) 1640 medium (Hyclone, Logan, UT), and all other chemicals were dissolved in dimethyl sulfoxide (DMSO, Santa Cruz Biotechnology). All chemical stock solutions were stored at –20°C or –80°C. An equal volume of RPMI or DMSO was used in all cell stimulation assays as a negative control.
Chicken HTC macrophage cells (29), kindly provided by Dr. Narayan C. Rath at United States Department of Agriculture–Agricultural Research Service, were cultured in complete RPMI 1640 medium containing 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA), 1% streptomycin-penicillin (Lonza, Walkersville, MD) at 1 × 106 cells/well in 6-well tissue culture plates overnight at 37°C and 5% CO2 prior to 24-h treatment with different concentrations of natural or synthetic COX-2 inhibitors separately or in combination with 2 mM sodium butyrate or 0.25 mM tributyrin (19, 30, 31), followed by total RNA extraction and expression analyses of chicken HDP genes as described below. HTC cells were also stimulated with butyrate, quercetin, or their combination for 24 h, followed by 3-h stimulation with 10 ng/mL LPS and subsequent expression analyses of the genes involved in barrier function and inflammation.
Chicken PBMCs were isolated from the peripheral blood of 2- to 4-week-old male Cobb broilers through density gradient centrifugation using Histopaque®-1077 (MilliporeSigma) following the manufacturer’s instructions. Briefly, EDTA-anticoagulated blood was overlaid onto an equal volume of Histopaque®-1077 and centrifuged at 400 × g for 30 min. The interface containing PBMCs was carefully aspirated and if necessary, red blood cells were hypotonically lysed with 0.2% sodium chloride and reconstituted with 1.6% sodium chloride. After three washes in phosphate buffered saline, PBMCs were suspended in complete RPMI 1640 medium containing 10% fetal bovine serum, 1% streptomycin-penicillin, and 20 mM HEPES (Hyclone). After 2-h seeding at 5 × 107 cells/well in 6-well plates, PBMCs were stimulated with various chemicals individually or in combination for 24 h. In the signaling experiments, PBMCs were pre-treated for 1 h with individual specific inhibitors, including 25 µM SB203580 (p38 MAPK inhibitor), 20 µM SP600125 (JNK inhibitor), 50 µM PD98059 (MEK1/2 inhibitor), 40 µM PDTC (NF-κB inhibitor), 20 µM MG132 (NF-κB inhibitor), or 100 or 500 µM DDA (adenylyl cyclase inhibitor) (19, 31), followed by stimulation with 10 µM quercetin and 2 mM butyrate for 24 h.
Cells were lysed in RNAzol (Molecular Research Center, Cincinnati, OH) for total RNA isolation according to the manufacturer’s protocol. The quantity and quality of total RNA were determined using Nanodrop (Nanodrop Products, Wilmington, DE). Reverse transcription was performed using QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) following the instructions of the manufacturer. Briefly, genomic DNA was removed from total RNA with gDNA Wipeout Buffer provided in the kit prior to reverse transcription at 42°C for 30 min. The resulting cDNA was then diluted with RNase-free water and used in subsequent qPCR analysis with QuantiTect SYBR Green PCR Kit (Qiagen) according to the manufacturer’s instruction. PCR was performed in 10-µL reactions with initial activation at 95°C for 10 min, followed by 40 cycles at 94°C for 15 s, 55°C for 20 s, and 72°C for 30 s in CFX Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The primer sequences for chicken HDP genes, claudin 1 (CLDN1), mucin 2 (MUC2), interleukin-1β (IL-1β), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were previously described (14, 18, 20). The specificity of PCR amplification was confirmed using melt curve analysis, and the CLDN1 and MUC2 amplicons were further verified through direct Sanger sequencing using gene-specific forward primers. Relative fold changes in gene expressions were calculated using the ΔΔCt method (32) normalized against GAPDH as described (14, 18–20).
Chicken HTC cells were treated with 2 mM butyrate, 20 μM quercetin, or their combination for 6 or 12 h, followed by lysis in the radioimmunoprecipitation (RIPA) lysis buffer (Santa Cruz Biotechnology). The resulting proteins were quantified using the Bradford Assay (Bio-Rad), followed by loading of 20 µg proteins from each sample in 12.5% SDS-PAGE gels and transferring to Immobilon-P® polyvinylidene difluoride membranes (MilliporeSigma). After overnight blocking in 5% dry skim milk in TTBS (0.05% Tween 20, 20 mM Tris-HCl, 150 mM NaCl, pH 7.5) at 4°C, the membranes were incubated with a primary rabbit antibody against acetyl-histone H4 (Cell Signaling, Danvers, MA; diluted 1:500) or a rabbit antibody against β-Actin (MilliporeSigma; diluted 1:1000) in the blocking buffer for 1 h at room temperature. After three washes in TTBS, the membranes were incubated with an alkaline phosphatase-conjugated goat anti-rabbit IgG antibody (MilliporeSigma; diluted 1:2,000) for 45 min at room temperature, followed by visualization using enhanced chemiluminescence (ThermoFisher Scientific). The band intensity of acetyl-histone H4 was quantified as the area under the curve using ImageJ (https://imagej.nih.gov/ij/) and further normalized against the band intensity of β-Actin for each sample.
Statistical analysis and data visualization were performed using GraphPad Prism (GraphPad Software, La Jolla, CA). The results were expressed as means ± standard error of the mean (SEM) from 2–3 independent experiments. One-way ANOVA and post hoc Tukey’s test were applied to determine statistical significance. The results were considered statistically significant if P < 0.05.
To study whether chicken HDP genes are regulated by quercetin, both dose-response and time-course experiments were conducted with quercetin in chicken HTC macrophage cells. AvBD9, the most inducible chicken HDP genes in response to a variety of small-molecule compounds (14, 18, 31), was gradually augmented in response to increasing concentrations of quercetin, reaching a peak of 881-fold induction at 80 μM (Figure 1A). In addition, almost all HDP genes that are expressed in HTC cells were induced by quercetin, although the magnitude of induction varied greatly among different genes (Figure 1B). For example, AvBD14 was readily induced, while AvBD10 showed minimum alterations. An obvious time-dependent induction of AvBD9 expression was also observed in HTC cells in response to 40 μM quercetin, peaking at 24 h (Figure 1C).
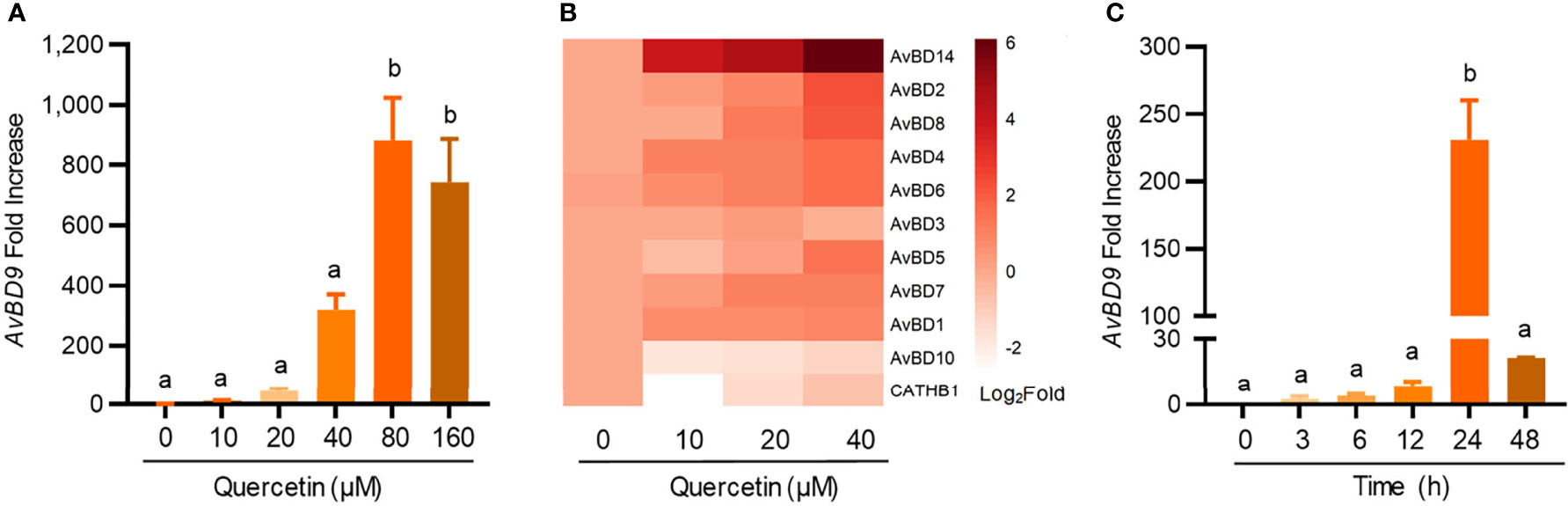
Figure 1 Dose- and time-dependent induction of chicken host defense peptide (HDP) gene expressions by quercetin. Chicken HTC macrophage cells were treated in duplicate with indicated concentrations of quercetin for 24 h, followed by RT-qPCR analysis of avian β-defensin 9 (AvBD9) (A) and other HDP genes (B). (C) HTC cells were also treated with 40 µM quercetin for indicated times, followed by AvBD9 expression analysis. Results are expressed as means ± SEM of three independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
To explore a possible synergy in HDP induction between quercetin and butyrate, chicken HTC cells were treated with different concentrations of quercetin in the presence or absence of 2 mM sodium butyrate for 24 h. A clear synergy was observed with a peak 1000-fold induction when 2 mM butyrate was combined with 20 µM quercetin, while they separately gave a 50- and 41-fold AvBD9 induction, respectively (Figure 2A). To confirm the quercetin/butyrate synergy in HDP induction in a different cell type, chicken PBMCs were treated with butyrate and quercetin individually or in combination. Similar to HTC macrophages, quercetin alone elevated AvBD9 transcription in a dose-dependent fashion and more importantly, synergized dramatically with butyrate (Figure 2B). It is noteworthy that the synergism was diminished when butyrate was combined with higher concentrations of quercetin in both HTC cells (Figure 2A) and PBMCs (Figure 2B). Quercetin also synergized strongly with tributyrin, a triglyceride analog of butyrate (33), in improving AvBD9 expression in chicken PBMCs (Figure 2C). To study the kinetics of the butyrate/quercetin synergy in HDP induction, a time-course experiment was conducted using quercetin and butyrate separately or in combination. The synergy became more pronounced as the treatment time was gradually increased, with a 24-h treatment giving the strongest synergy in inducing AvBD9, while a longer 48-h stimulation caused a diminished synergy in HTC cells (Figure 2D). Besides AvBD9, butyrate and quercetin also synergistically increased AvBD1, AvBD2, AvBD3, AvBD4, AvBD6, and AvBD7, but not AvBD5, AvBD8, AvBD10, AvBD14, or CATHB1, in HTC macrophages (Figure 3). Notably, varying magnitudes of induction were observed for different HDP genes (Figure 3), indicating a gene-specific effect.
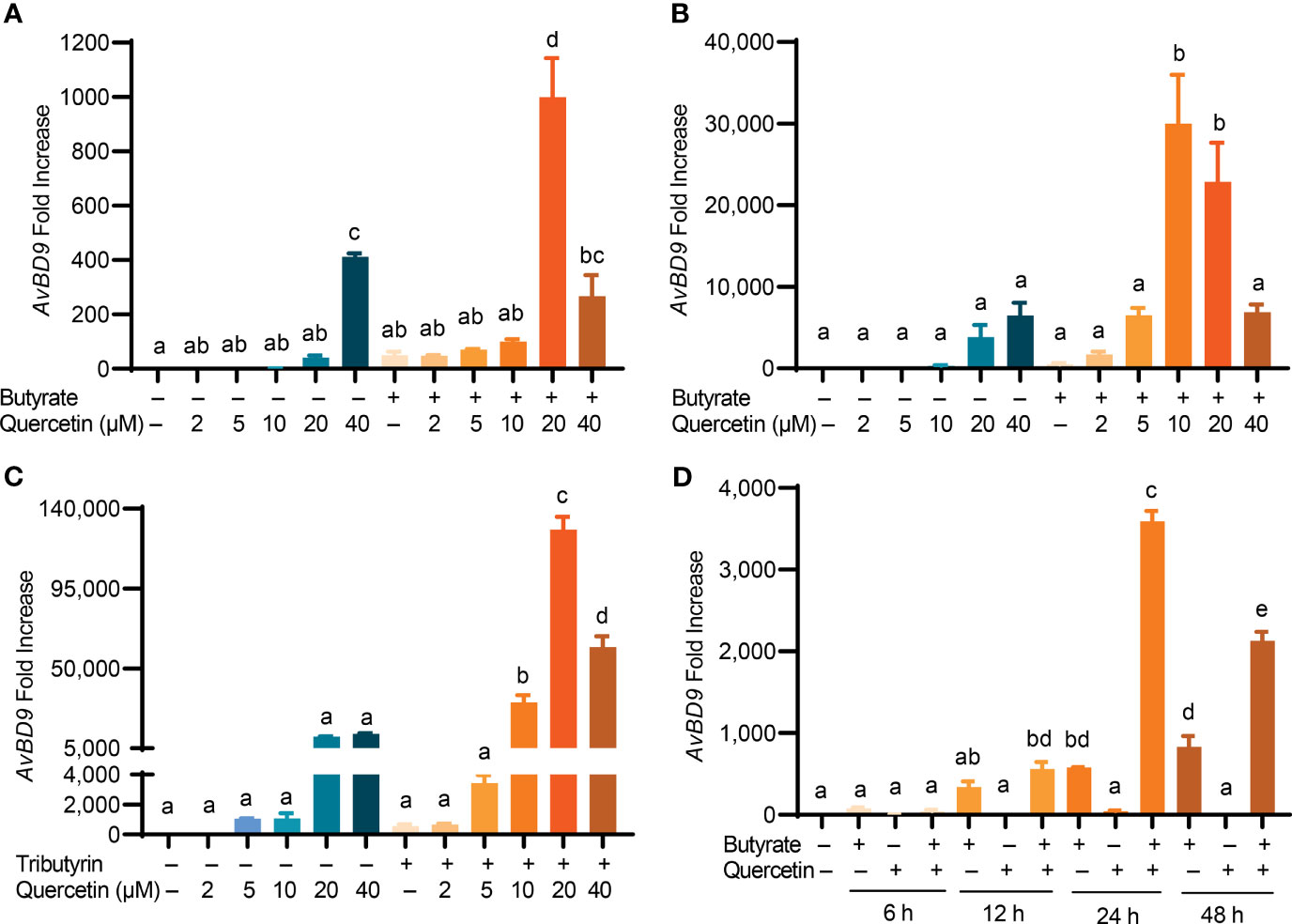
Figure 2 Synergistic induction of AvBD9 gene expression by a combination of quercetin and butyrate or tributyrin. (A) Chicken HTC macrophages or (B) peripheral blood mononuclear cells (PBMCs) were treated in duplicate with indicated concentrations of quercetin with or without 2 mM butyrate for 24 h. (C) Chicken PBMCs were stimulated with indicated concentrations of quercetin with or without 0.25 mM tributyrin for 24 h. (D) Chicken PBMCs were treated in duplicate with 2 mM butyrate and 10 μM quercetin individually or in combination for indicated times. The expression levels of the AvBD9 gene were analyzed by RT-qPCR. Results are presented as means ± SEM of 2–3 independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
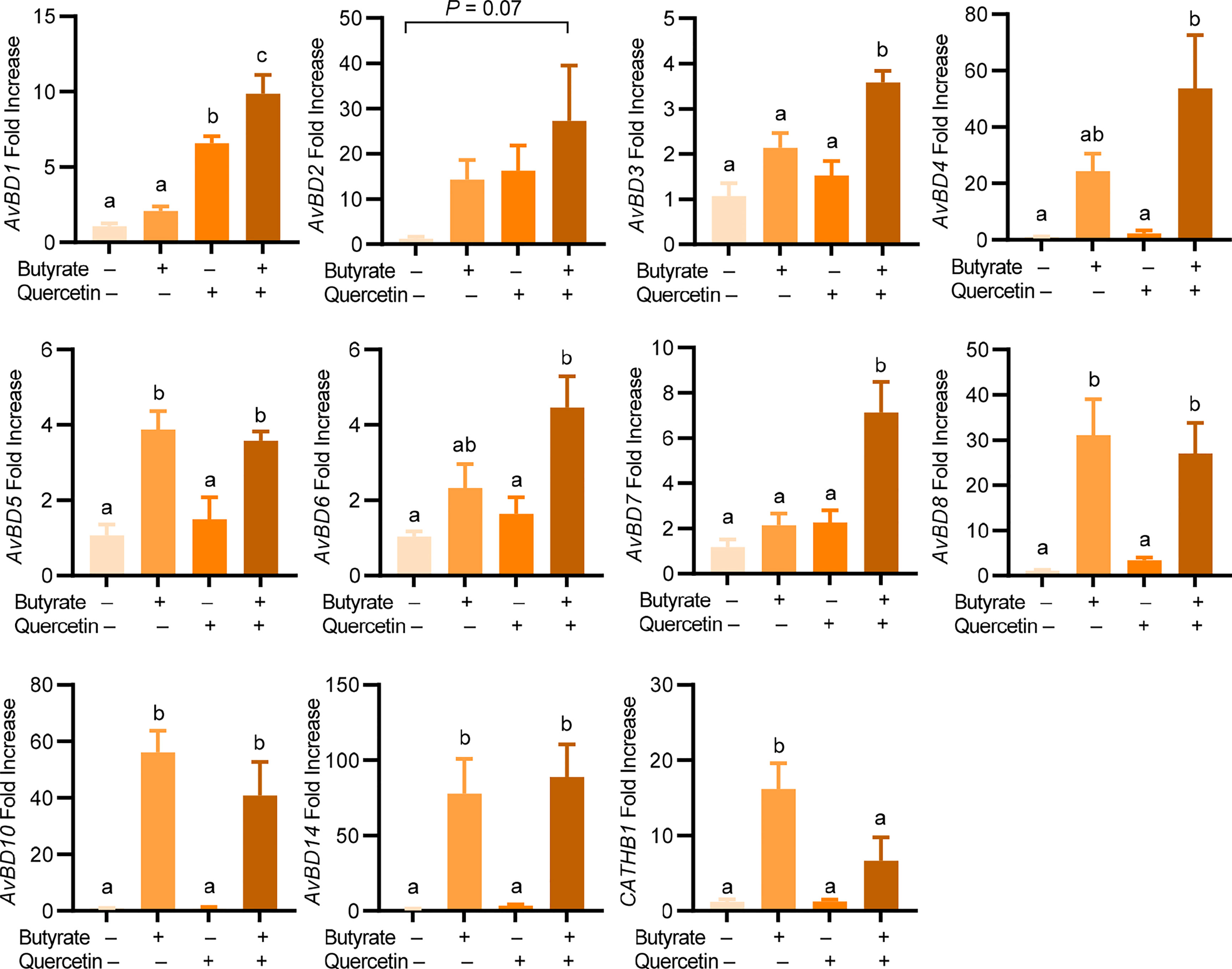
Figure 3 Induction of chicken HDP genes by quercetin and butyrate. Chicken HTC macrophage cells were treated in duplicate with 20 μM quercetin and 2 mM sodium butyrate separately or in combination for 24 h, followed by RT-qPCR analysis of chicken HDP gene expression. Results are presented as means ± SEM of two independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
Butyrate and quercetin are known to enhance barrier function by inducing mucin and tight junction protein expressions while suppressing inflammation (34, 35). CLDN1 is critically involved in tight junction assembly (35), while MUC2 is the most abundant component of the intestinal mucus layer (36). To investigate a potential synergy between butyrate and quercetin in regulating barrier function and inflammation, chicken HTC macrophage cells were treated for 24 h with butyrate and quercetin separately or in combination prior to a 3-h stimulation with LPS. A synergistic induction of AvBD9 by butyrate and quercetin was obviously maintained following LPS stimulation (Figure 4A). While 2 mM butyrate induced CLDN1 gene expression by 875-fold and 20 μM quercetin showed a minimum effect, the butyrate/quercetin combination elicited a synergistic 2,024-fold increase in CLDN1 expression, which was unaffected by LPS (Figure 4B). Moreover, butyrate or quercetin improved the MUC2 mRNA expression individually, and the combination synergistically enhanced MUC2 expression even in the presence of LPS (P < 0.05) (Figure 4C). Additionally, butyrate, quercetin, or the combination had no significant impact on IL-1β expression. As expected, butyrate or quercetin suppressed LPS-induced IL-1β expression in HTC cells (P < 0.05). Desirably, the butyrate/quercetin combination had the strongest effect in reducing LPS-triggered IL-1β expression (Figure 4D). Collectively, quercetin is synergistic with butyrate in boosting the expressions of AvBD9, CLDN1, and MUC2, while suppressing IL-1β during LPS-triggered stimulation. It is noted that LPS had no effect on AvBD9 expression, but triggered a 754-fold induction of IL-1β gene expression (Figures 4A, D).
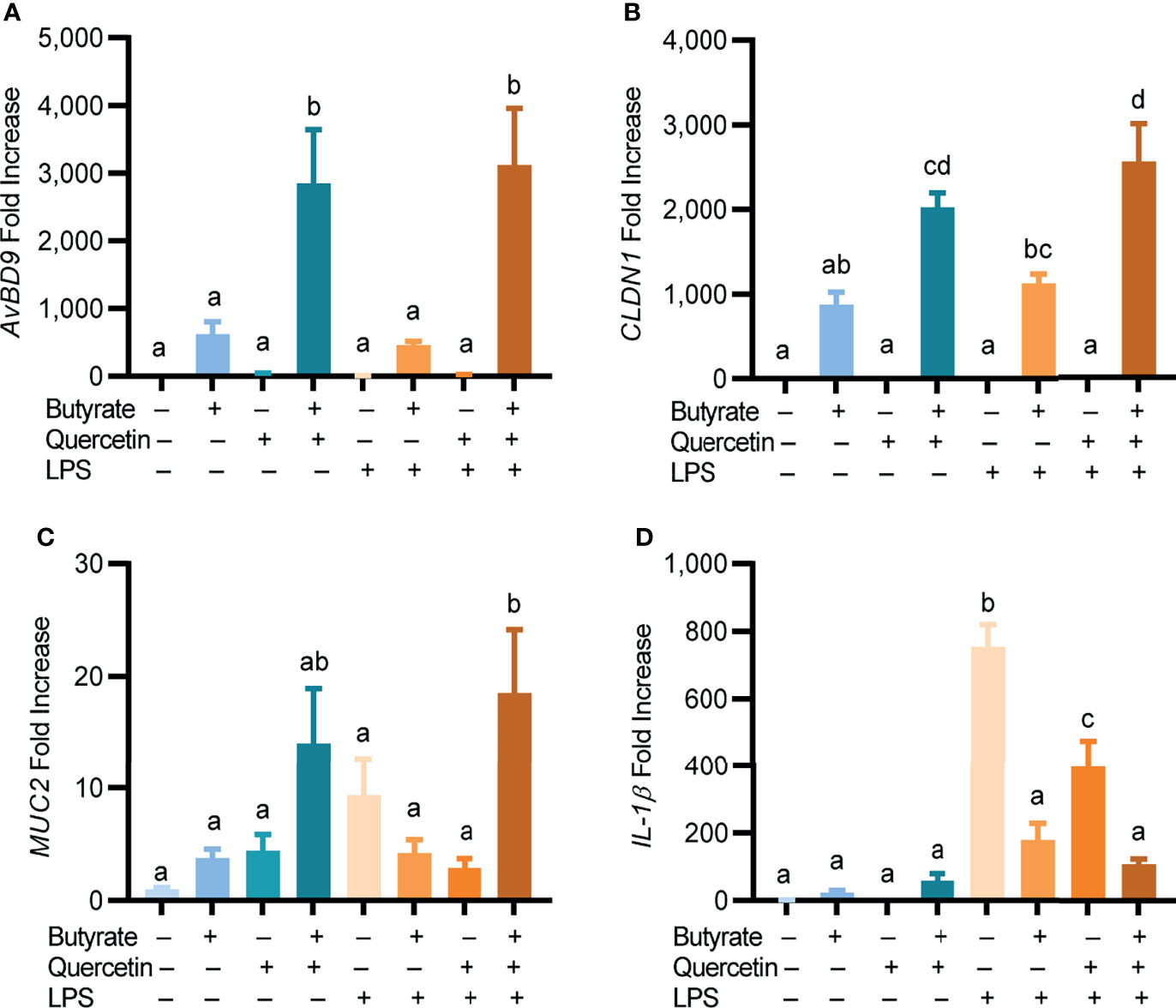
Figure 4 Regulation of HDP, barrier function, and inflammatory cytokine gene expressions by quercetin and butyrate. Chicken HTC macrophage cells were treated in duplicate with 20 μM quercetin and 2 mM sodium butyrate separately or in combination for 24 h, followed by stimulation with 10 ng/mL lipopolysaccharides (LPS) for another 3 h. Gene expressions of AvBD9 (A), claudin 1 (CLDN1) (B), mucin 2 (MUC2) (C), and interleukin 1β (IL-1β) (D) were analyzed by RT-qPCR. Results are presented as means ± SEM of two independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
To further evaluate the synergy between butyrate and other COX-2 inhibitors in chicken HDP gene induction, four additional natural COX-2 inhibitors including resveratrol, anacardic acid, EGCG, and garcinol were tested in chicken PBMCs with or without butyrate. Similar to quercetin, each compound dose-dependently elevated AvBD9 expression and also synergized markedly with butyrate in AvBD9 gene expression (Figure 5). For example, 50 µM resveratrol and 2 mM butyrate induced AvBD9 expression by 43- and 511-fold, respectively, while the combination augmented AvBD9 expression by 2,519-fold (Figure 5A). Similarly, anacardic acid (Figure 5B), EGCG (Figure 5C), and garcinol (Figure 5D) also demonstrated a strong synergy with butyrate in AvBD9 mRNA induction, although garcinol was relatively weak in its ability to synergize with butyrate.
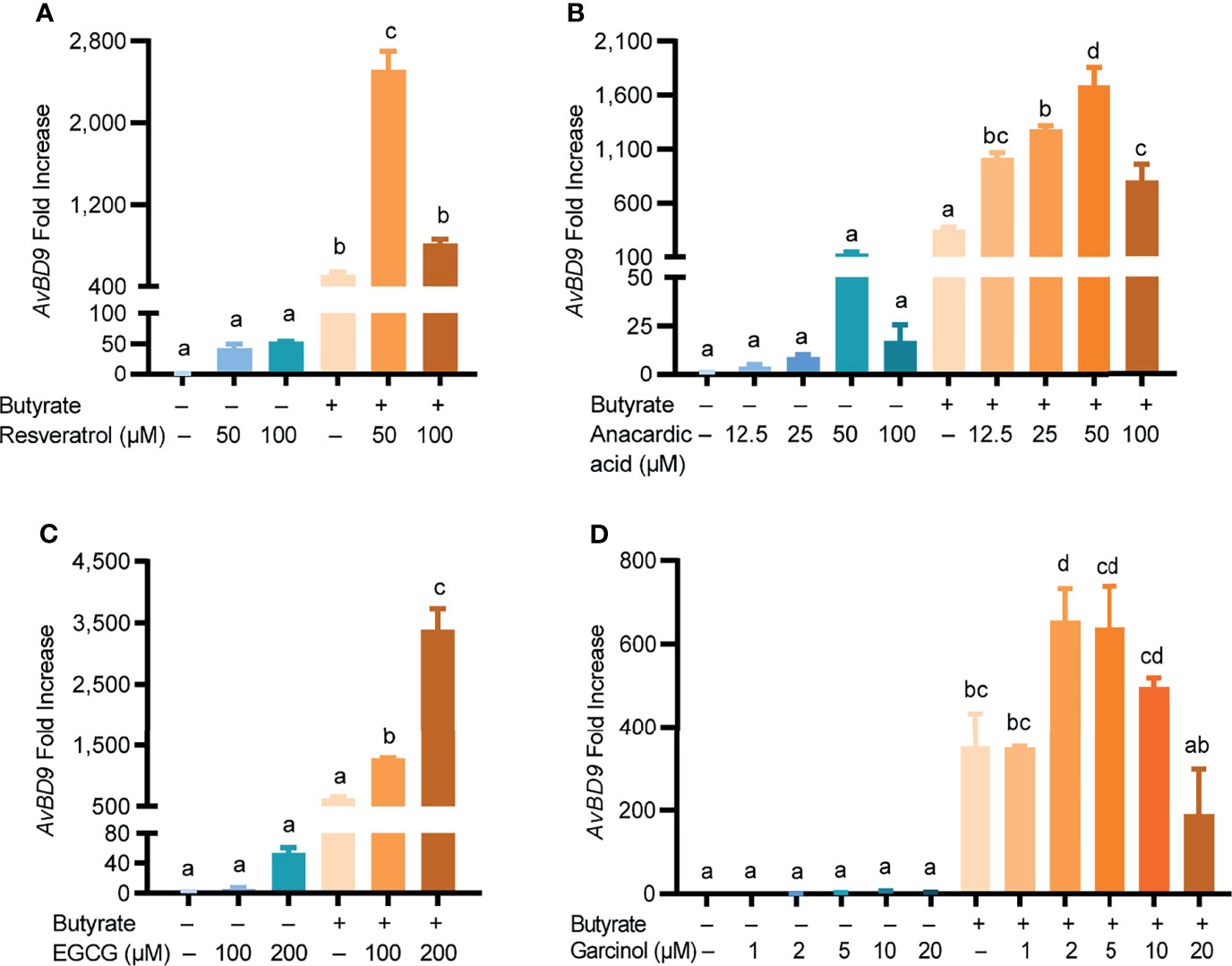
Figure 5 Induction of AvBD9 by natural COX-2 inhibitors and butyrate. Chicken PBMCs were treated in duplicate with indicated concentrations of resveratrol (A), anacardic acid (B), epigallocatechin gallate (EGCG) (C), or garcinol (D), in the presence or absence of 2 mM butyrate for 24 h, followed by AvBD9 expression analysis. Results are presented as means ± SEM of 2–3 independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
In addition to acting as a COX-2 inhibitor, quercetin, resveratrol, anacardic acid, EGCG, and garcinol also exert a range of other effects on host cells (37, 38). To confirm COX-2-specific inhibitors can also synergize with butyrate in HDP induction, two highly selective COX-2 inhibitors namely nimesulide and niflumic acid (39, 40) were separately applied to chicken PBMCs with or without butyrate. Similar to their natural counterparts, both nimesulide and niflumic acid increased AvBD9 gene expression in a dose-dependent manner, further with a marked synergy with butyrate (Figure 6). For example, 250 µM nimesulide and 2 mM butyrate together enhanced AvBD9 gene expression by 10,524-fold, while they individually gave 69- and 349-fold induction, respectively (Figure 6A). A robust synergy also occurred between niflumic acid and butyrate (Figure 6B). Taken together, inhibition of the COX-2 pathway upregulates AvBD9 gene expression and further synergizes with butyrate to promote AvBD9 transcription.
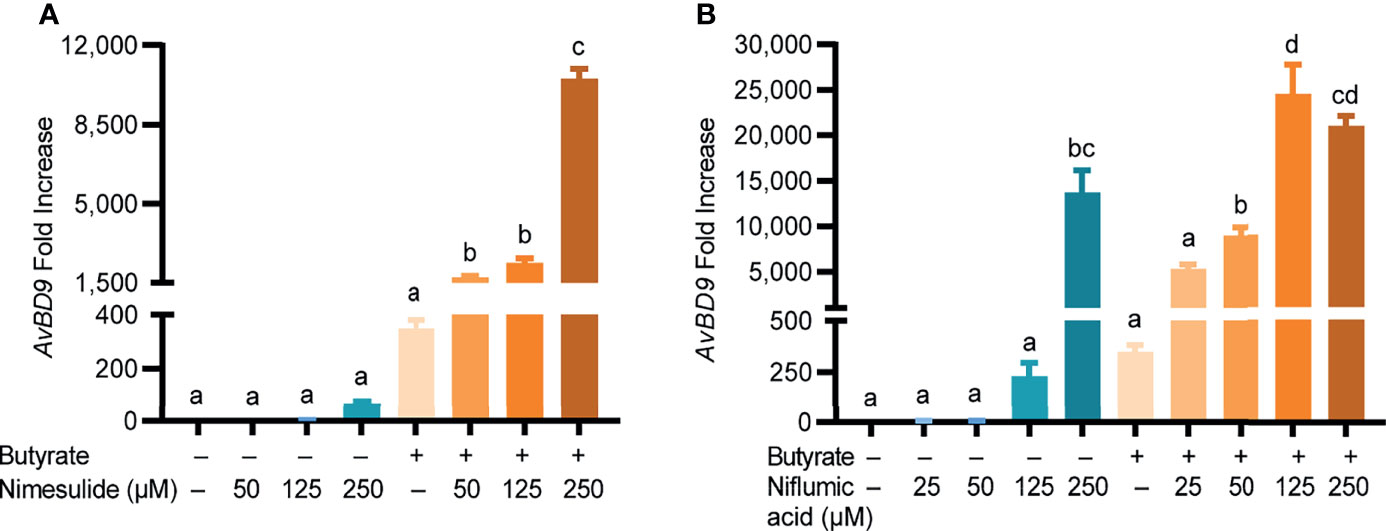
Figure 6 Synergistic induction of AvBD9 gene expression by synthetic COX-2-specific inhibitors and butyrate. Chicken PBMCs were treated in duplicate with indicated concentrations of nimesulide (A) or niflumic acid (B) with or without 2 mM butyrate for 24 h, followed by RT-qPCR analysis of AvBD9 gene expression. Results are presented as means ± SEM of two independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
Butyrate is a well-known HDAC inhibitor to maintain the hyper-acetylation status of histones to facilitate chromatin relaxation and subsequent gene expression (41). To investigate whether histone acetylation is involved in butyrate/quercetin-mediated synergy in AvBD9 induction, chicken HTC cells were treated with butyrate and quercetin separately or in combination for 6 and 12 h, followed by examination of histone 4 (H4) acetylation using Western blotting. Butyrate, but not quercetin, significantly increased H4 acetylation at 6 h and the acetylation was diminished at 12 h (Figures 7A, B). Relative to butyrate, a combination of butyrate and quercetin showed no hyper-acetylation at either time point (Figures 7A, B), suggesting that the HDP-inducing synergy between butyrate and quercetin is not caused by hyper-acetylation of histones in the HDP gene promoters.
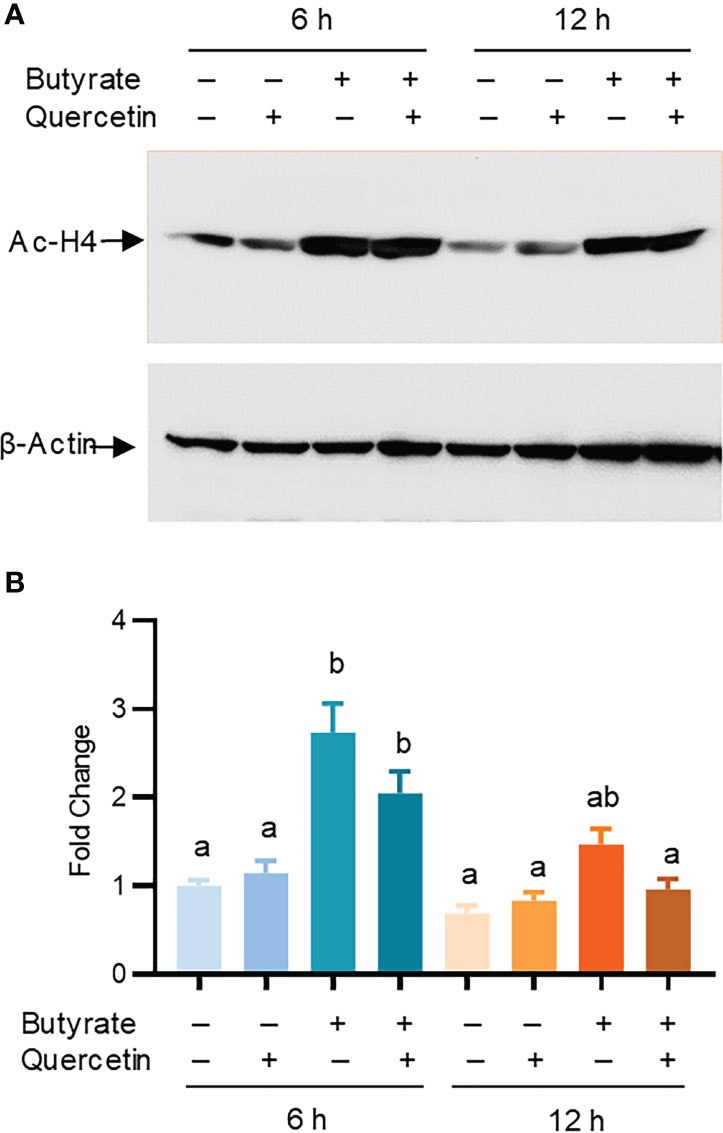
Figure 7 Role of histone acetylation in AvBD9 induction by butyrate and quercetin. Chicken HTC cells were treated with 2 mM sodium butyrate or 20 μM quercetin individually or in combination for 6 or 12 h, followed by western blot analysis of acetyl-histone H4 (Ac-H4) and β-actin. (A) A representative of two independent blots. (B) Fold changes in the acetylation of histone H4 relative to unstimulated cells. Results are presented as means ± SEM of two independent experiments normalized against β-actin. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
To examine the role of MAPK, NF-κB, and cAMP signaling in AvBD9 induction, chicken PBMCs were treated with quercetin and butyrate in the presence or absence of specific inhibitors for three major MAPK pathways as well as NF-κB, and cAMP pathways. To our surprise, inhibition of three MAPK pathways (with SB203580, SP60025, or PD98059) modestly activated the basal expression of the AvBD9 gene (Figure 8), suggesting that blocking MAPK signaling is beneficial for the constitutive expression of AvBD9. However, inhibiting the p38 MAPK pathway with SB203580 gave a substantial reduction in butyrate-induced AvBD9 expression, while blocking the JNK and MEK1/2 MAPK pathways with SP60025 and PD98059, respectively, potentiated AvBD9 expression (Figure 8), implying that p38, but not JNK or MEK1/2 MAPK, is required for butyrate-mediated AvBD9 induction. However, p38 MAPK appeared to have no effect on quercetin-induced AvBD9 expression, while suppressing the JNK and MEK1/2 pathways increased AvBD9 expression. Blocking p38 MAPK substantially reduced AvBD9 gene expression in chicken PBMCs in response to a combination of butyrate and quercetin, while inhibiting the JNK and MEK-ERK pathways had a minimum effect on AvBD9 expression (Figure 8). These results suggest a differential involvement of three canonical MAPK signaling pathways in AvBD9 induction mediated by butyrate, quercetin, or the combination.
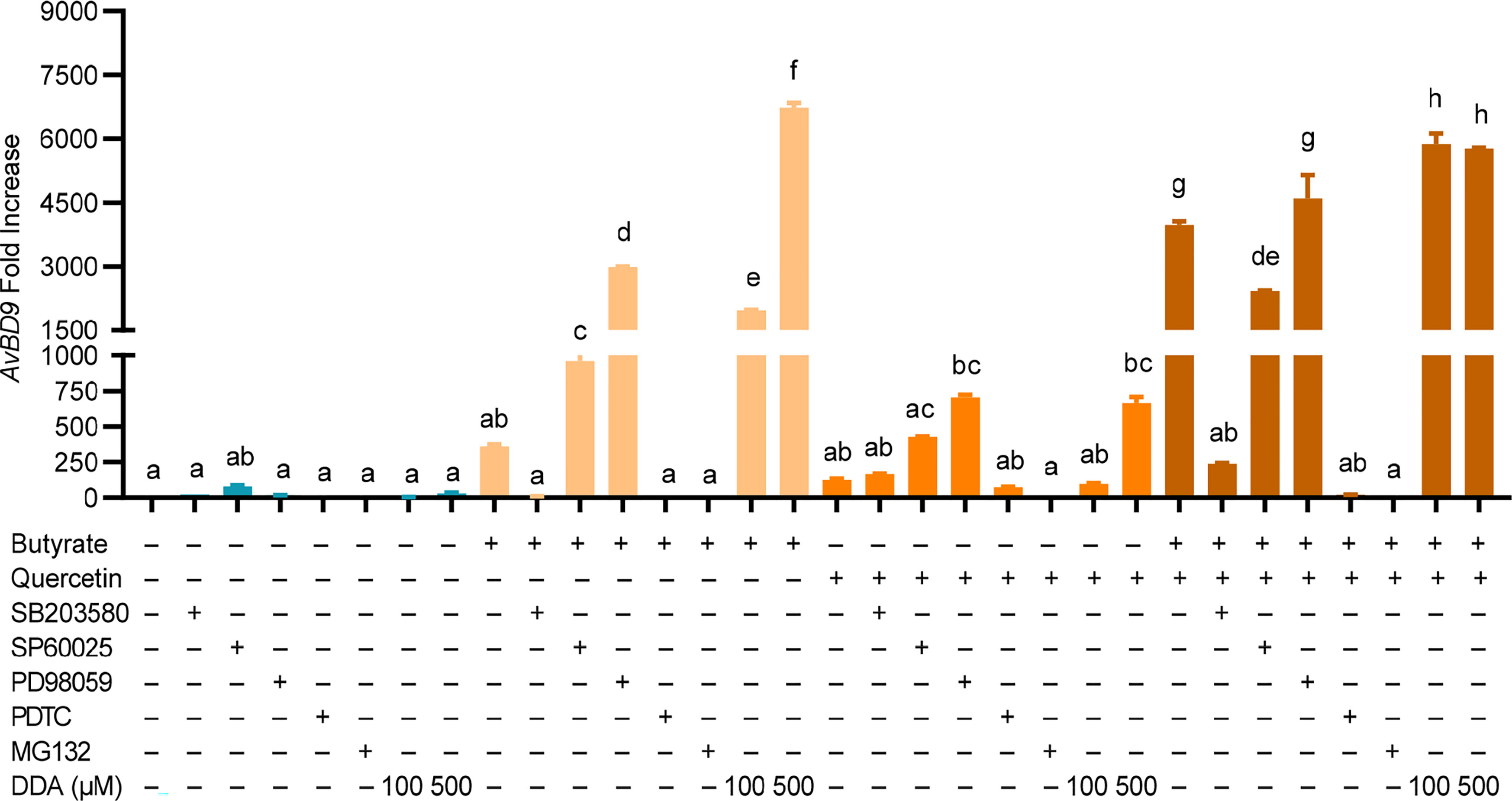
Figure 8 Role of MAPK, NF-κB, and cAMP signaling pathways in AvBD9 induction mediated by quercetin and butyrate. Chicken PBMCs were pretreated for 1 h with or without 25 µM SB203580 (p38 MAPK inhibitor), 20 µM SP60025 (JNK inhibitor), 50 µM PD98059 (MEK1/2 inhibitor), 40 µM PDTC (NF-κB inhibitor), 20 µM MG132 (NF-κB inhibitor), or 100 or 500 µM 2’,5’-dideoxyadenosine (DDA) (cAMP inhibitor), followed by stimulation with 10 µM quercetin with or without 2 mM sodium butyrate for another 24 h. RT-qPCR was performed to determine AvBD9 mRNA expression. Results are presented as means ± SEM of two independent experiments. The bars not sharing common superscript letters are significantly different (P < 0.05) as determined by one-way ANOVA and post hoc Tukey’s test.
Blocking NF-κB with PDTC or MG132 had no impact on the basal expression of AvBD9, but caused nearly a complete abolishment of AvBD9 expression induced by butyrate, quercetin, or the combination (Figure 8), indicating a critical role of NF-κB in AvBD9 gene expression. Inhibition of the cAMP pathway with DDA resulted in a modest AvBD9 induction in PBMCs under the basal condition; however, DDA dose-dependently increased AvBD9 expression induced by butyrate or quercetin and further potentiated AvBD9 expression in response to the butyrate/quercetin combination (Figure 8), implying that inhibiting cAMP signaling is beneficial for AvBD9 induction mediated by butyrate, quercetin, or the combination.
Enhancing HDP synthesis is a promising host-directed approach to antimicrobial therapy (3–5, 11). This study revealed a potent HDP-inducing activity of several natural phenolic compounds with COX-2 inhibitory activity. We further reported a strong synergy between butyrate and these natural COX-2 inhibitors in promoting HDP expression and barrier function while suppressing LPS-mediated inflammatory response in chicken cells. These multifaceted beneficial effects of polyphenols and butyrate make them attractive candidates for further evaluation of their efficacy in disease control and prevention in chickens and possibly other animals.
COX-2 is a membrane-bound enzyme responsible for the production of prostanoids including prostaglandins and thromboxanes from arachidonic acid during inflammation (22, 23). As a result, a variety of cell type-specific prostaglandins are synthesized and in turn lead to inflammation and pain (22, 23). Specific inhibition of COX-2 can be achieved by using nonsteroidal anti-inflammatory drugs (NSAIDs) (22, 23). Many polyphenols have COX-2 inhibitory activity (42). For example, quercetin is capable of suppressing COX-2 gene expression and prostaglandin E2 (PGE2) production (43). Recently, COX-2 signaling was linked to HDP expression. Activation of COX-2 downregulates human β-defensin 1 (DEFB1) mRNA expression in intestinal epithelial cells (44), and PGE2 is known to suppress human β-defensin 2 (DEFB4) expression in gingival epithelial cells (45) and human cathelicidin antimicrobial peptide (CAMP) gene in macrophages (46). Conversely, inhibiting COX-2 increases DEFB4 expression in human gingival epithelial cells (45). Several polyphenols such as resveratrol and EGCG have also been shown to induce HDP gene expression (24, 26, 27). However, it remains unknown whether these polyphenols induce HDP expression by acting as COX-2 inhibitors.
In the current study, several structurally unrelated polyphenols such as quercetin, anacardic acid, and garcinol were tested in parallel with resveratrol and EGCG. All tested polyphenols possess HDP-inducing activity and further synergize with butyrate in HDP induction. We also confirmed two highly selective COX-2 inhibitors including nimesulide and niflumic acid (39, 47) are also capable of inducing HDP expression showing a synergistic activity with butyrate in promoting HDP gene expression, similar to those polyphenols. These results suggest that polyphenols enhance HDP expression at least in part through inhibition of COX-2 activation, perhaps by suppressing PGE2 synthesis, which is known to downregulate HDP gene expression (45, 46).
Histone acetylation plays an important role in modulating HDP gene expression and many compounds with HDAC inhibitory activity have been found to induce HDP expression (5). It is unsurprising to see butyrate, a well-known HDAC inhibitor, to cause acetylation of histone H4 in chicken HTC cells in this study. Although many polyphenols such as resveratrol, quercetin, and EGCG have epigenetic functions by modifying DNA methylation and histone methylation and acetylation (48), quercetin fails to obviously induce histone acetylation showing no cooperative activity with butyrate in hyper-acetylation of histones, suggesting that its synergy with butyrate in HDP induction is unrelated to histone acetylation.
However, we found that blocking the p38 MAPK signaling pathway substantially abolishes AvDB9 gene induction in PBMCs mediated by butyrate or the butyrate/quercetin combination, but not quercetin alone; however, inhibiting the MEK1/2 or JNK pathway instead potentiates AvDB9 gene expression in response to butyrate, quercetin, the combination, or even in the basal quiescent state. Differential involvement of the three canonical MAPK pathways in HDP induction in PBMCs is consistent with earlier observations that three MAPK pathways mediate HDP induction in compound- and cell type-specific manners (11). For example, inhibition of p38 and JNK pathways decreases AvBD9 expression, while suppressing MEK1/2 increases AvBD9 expression in response to butyrate or/and forskolin in chicken HD11 macrophage cells (19). On the other hand, resveratrol induces human CAMP gene expression through p38 MAPK, while blocking the MEK1/2 pathway further enhances resveratrol-induced CAMP expression in human keratinocytes (24).
Phosphorylation of CCAAT-enhancer-binding protein α (C/EBPα) by p38 MAPK is believed to be responsible for resveratrol-mediated CAMP expression (24). Whether C/EBPα is also involved in HDP induction by other polyphenols is currently unknown. Given no influence of p38 MAPK on quercetin-mediated AvBD9 induction, it is likely that different polyphenols induce HDP gene expression differently, but it is also possible that a gene- or cell type-specific HDP regulation pattern exists. One possible reason for HDP induction in response to the MEK1/2 blockage could be due to reduced phosphorylation of cAMP response element-binding protein (CREB), which is a major target of the MEK1/2 pathway (49, 50). Reduced CREB phosphorylation in turn suppresses the synthesis of inducible cAMP early repressor (ICER), which could otherwise compete with CREB for the cAMP response element (CRE) on an HDP gene promoter resulting in reduced gene transcription (51, 52).
NF-κB is also required for AvBD9 induction mediated by butyrate, quercetin, or the combination, which is in agreement with earlier observations on the involvement of NF-κB in human CAMP induction by resveratrol and genistein, where NF-κB is believed to activate p38 MAPK for phosphorylation of C/EBPα and activation of CAMP gene transcription (24, 28). It is noted that NF-κB is critical for the induction of the COX-2 gene, and polyphenols block the COX-2 activity and PGE2 synthesis by suppressing NF-κB action (53). Reduced PGE2 production thus leads to increased HDP gene expression (45, 46).
Similar to the MEK1/2 MAPK pathway, inhibition of the cAMP signaling pathway appears to be beneficial for butyrate- and quercetin-mediated AvBD9 gene induction in chicken PBMCs. Consistently, activation of cAMP signaling suppresses the expression of human CAMP and DEFB1 genes in colonic epithelial cells (44). However, forskolin, an adenylyl cyclase agonist and activator of the cAMP-protein kinase A (PKA) signaling (54), has been found to be a weak inducer of AvBD9 and even synergize with butyrate in AvBD9 induction in chicken HD11 macrophages (18–20). The discrepancy is likely due to a difference in the cell type used in different studies, but it is also possible because of a difference in the strength and duration of the cAMP signaling being activated. Forskolin is beneficial to activate the cAMP-PKA pathway to phosphorylate CREB for enhanced HDP synthesis; however, excessive and prolonged CREB activation may sustain ICER synthesis, creating a negative feedback to turn off HDP synthesis (20, 51). Therefore, it is beneficial to fine-tune the amount and duration of CREB synthesis and phosphorylation to maintain a positive ratio of phosphorylated CREB over phosphorylated ICER and increased HDP gene expression. In fact, diminished HDP induction occurs with high concentrations of butyrate or forskolin or prolonged treatment (14, 19, 55), which is associated with increased ICER transcription (20, 51). Similarly, we also observed a diminished synergic AvBD9 induction in response to a combination of butyrate and high concentrations of natural COX-2 inhibitors in this study. ICER is likely involved in the reduced synergism, but an experimental verification is warranted.
Although HDPs are an important part of the innate immunity, not all HDPs are increasingly transcribed and produced in response to infection or injury. In chicken HTC macrophages, a 3-h LPS treatment triggered a massive IL-1β induction, but AvBD9 gene expression was not altered, which is consistent with earlier reports that AvBD9 was unaffected in the vagina or Sertoli cells of chickens for up to 24 h following LPS treatment (56, 57), although another report appeared to suggest that AvBD9 is upregulated by LPS in chicken bone marrow-derived cells (58). The discrepancy is currently unknown, perhaps it is cell type-dependent.
In the current study, we believe that COX-2 inhibition is at least partially responsible for polyphenol-mediated HDP synthesis; however, we cannot rule out relative contributions of other biological activities of individual polyphenols to HDP induction. Most polyphenols have antioxidative, antiinflammatory, and antiproliferative activities by modulating a multitude of biological processes. Besides COX-2 inhibition, NF-κB, MAPK, and signal transducer and activator of transcription proteins (STATs) are all commonly suppressed by polyphenols (42). Additionally, many polyphenols have been shown to influence DNA and histone methylation beyond histone acetylation (48). Therefore, it will be important to dissect the relative contributions of each of those individual activities of polyphenols to HDP induction. It is likely different polyphenols induce HDP expression through different mechanisms beyond COX-2 inhibition. It will be important to expand the testing of additional structurally and functionally distinct polyphenols as well as other highly specific COX-2 inhibitors such as aspirin, ibuprofen, and coxibs (23) for their HDP-inducing activity. Additionally, it is critical to examine whether the HDP-inducing synergy between butyrate and COX-2 inhibitors similarly occurs in other animal species or humans.
We have revealed that several structurally different polyphenols are potent HDP inducers likely by acting as COX-2 inhibitors. Furthermore, these natural COX-2 inhibitors are synergistic with butyrate in inducing HDP and barrier function gene expression without eliciting inflammation, suggesting their potential for further exploration as a novel host-directed approach to antimicrobial therapy in chickens and possibly other animals, although additional in vivo studies are warranted.
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
QY, AB, LS, and KX conducted the experiments. QY, AB, and LS performed data analysis. QY and AB drafted the manuscript. GZ conceived the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the USDA National Institute of Food and Agriculture (grant no. 2018-68003-27462 and 2020-67016-31619), Oklahoma Center for the Advancement of Science and Technology (grant no. AR19-027), the Ralph F. and Leila W. Boulware Endowment Fund, and Oklahoma Agricultural Experiment Station Project H-3112.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Narayan C. Rath at Poultry Production and Product Safety Research Unit of USDA- Agricultural Research Services for kindly providing chicken HTC macrophages.
AvBD9, avian β-defensin 9; C/EBPα, CCAAT-enhancer-binding protein α; CAMP, cathelicidin antimicrobial peptide; cAMP, cyclic adenosine monophosphate; CLDN1, claudin 1; COX-2, cyclooxygenase-2; CRE, cAMP response element; CREB, cAMP response element-binding protein; DDA, 2’,5’-dideoxyadenosine; DEFB1, human β-defensin 1; DEFB4, human β-defensin 2; EGCG, epigallocatechin gallate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDAC, histone deacetylase; HDPs, host defense peptides; ICER, inducible cAMP early repressor; IL-1β, interleukin-1β; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MUC2, mucin 2; NF-κB, nuclear factor-κB; NSAIDs, nonsteroidal anti-inflammatory drugs; PBMCs, peripheral blood mononuclear cells; PGE2, prostaglandin E2; PKA, protein kinase A; RIPA, radioimmunoprecipitation; RPMI, Roswell Park Memorial Institute; RT-qPCR, reverse transcriptase-quantitative PCR; SEM, standard error of the mean; STATs, signal transducer and activator of transcription proteins.
1. Majumder MAA, Rahman S, Cohall D, Bharatha A, Singh K, Haque M, et al. Antimicrobial Stewardship: Fighting Antimicrobial Resistance and Protecting Global Public Health. Infect Drug Resist (2020) 13:4713–38. doi: 10.2147/idr.s290835
2. Schrader SM, Vaubourgeix J, Nathan C. Biology of Antimicrobial Resistance and Approaches to Combat It. Sci Transl Med (2020) 12(549):eaaz6992. doi: 10.1126/scitranslmed.aaz6992
3. Bergman P, Raqib R, Rekha RS, Agerberth B, Gudmundsson GH. Host Directed Therapy Against Infection by Boosting Innate Immunity. Front Immunol (2020) 11:1209. doi: 10.3389/fimmu.2020.01209
4. Robinson K, Ma X, Liu Y, Qiao S, Hou Y, Zhang G. Dietary Modulation of Endogenous Host Defense Peptide Synthesis as an Alternative Approach to in-Feed Antibiotics. Anim Nutr (2018) 4(2):160–9. doi: 10.1016/j.aninu.2018.01.003
5. Rodriguez-Carlos A, Jacobo-Delgado YM, Santos-Mena AO, Rivas-Santiago B. Modulation of Cathelicidin and Defensins by Histone Deacetylase Inhibitors: A Potential Treatment for Multi-Drug Resistant Infectious Diseases. Peptides (2021) 140:170527. doi: 10.1016/j.peptides.2021.170527
6. Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A, et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect Dis (2020) 20(9):e216–30. doi: 10.1016/S1473-3099(20)30327-3
7. Cuperus T, Coorens M, van Dijk A, Haagsman HP. Avian Host Defense Peptides. Dev Comp Immunol (2013) 41(3):352–69. doi: 10.1016/j.dci.2013.04.019
8. Zhang G, Sunkara LT. Avian Antimicrobial Host Defense Peptides: From Biology to Therapeutic Applications. Pharmaceuticals (Basel) (2014) 7(3):220–47. doi: 10.3390/ph7030220
9. Robinson K, Deng Z, Hou Y, Zhang G. Regulation of the Intestinal Barrier Function by Host Defense Peptides. Front Vet Sci (2015) 2:57. doi: 10.3389/fvets.2015.00057
10. Lyu W, Curtis AR, Sunkara LT, Zhang G. Transcriptional Regulation of Antimicrobial Host Defense Peptides. Curr Protein Pept Sci (2015) 16(7):672–9. doi: 10.2174/1389203716666150630133432
11. Chen J, Zhai Z, Long H, Yang G, Deng B, Deng J. Inducible Expression of Defensins and Cathelicidins by Nutrients and Associated Regulatory Mechanisms. Peptides (2020) 123:170177. doi: 10.1016/j.peptides.2019.170177
12. Kida Y, Shimizu T, Kuwano K. Sodium Butyrate Up-Regulates Cathelicidin Gene Expression via Activator Protein-1 and Histone Acetylation at the Promoter Region in a Human Lung Epithelial Cell Line, EBC-1. Mol Immunol (2006) 43(12):1972–81. doi: 10.1016/j.molimm.2005.11.014
13. Schauber J, Svanholm C, Termen S, Iffland K, Menzel T, Scheppach W, et al. Expression of the Cathelicidin LL-37 Is Modulated by Short Chain Fatty Acids in Colonocytes: Relevance of Signalling Pathways. Gut (2003) 52(5):735–41. doi: 10.1136/gut.52.5.735
14. Sunkara LT, Achanta M, Schreiber NB, Bommineni YR, Dai G, Jiang W, et al. Butyrate Enhances Disease Resistance of Chickens by Inducing Antimicrobial Host Defense Peptide Gene Expression. PloS One (2011) 6(11):e27225. doi: 10.1371/journal.pone.0027225
15. Xiong H, Guo B, Gan Z, Song D, Lu Z, Yi H, et al. Butyrate Upregulates Endogenous Host Defense Peptides to Enhance Disease Resistance in Piglets via Histone Deacetylase Inhibition. Sci Rep (2016) 6:27070. doi: 10.1038/srep27070
16. Wang TT, Nestel FP, Bourdeau V, Nagai Y, Wang Q, Liao J, et al. Cutting Edge: 1,25-Dihydroxyvitamin D3 Is a Direct Inducer of Antimicrobial Peptide Gene Expression. J Immunol (2004) 173(5):2909–12. doi: 10.4049/jimmunol.173.5.2909
17. Cederlund A, Kai-Larsen Y, Printz G, Yoshio H, Alvelius G, Lagercrantz H, et al. Lactose in Human Breast Milk an Inducer of Innate Immunity With Implications for a Role in Intestinal Homeostasis. PloS One (2013) 8(1):e53876. doi: 10.1371/journal.pone.0053876
18. Yang Q, Whitmore MA, Robinson K, Lyu W, Zhang G. Butyrate, Forskolin, and Lactose Synergistically Enhance Disease Resistance by Inducing the Expression of the Genes Involved in Innate Host Defense and Barrier Function. Antibiotics (Basel) (2021) 10(10):1175. doi: 10.3390/antibiotics10101175
19. Sunkara LT, Zeng X, Curtis AR, Zhang G. Cyclic AMP Synergizes With Butyrate in Promoting Beta-Defensin 9 Expression in Chickens. Mol Immunol (2014) 57(2):171–80. doi: 10.1016/j.molimm.2013.09.003
20. Robinson K, Yang Q, Li H, Zhang L, Aylward B, Arsenault RJ, et al. Butyrate and Forskolin Augment Host Defense, Barrier Function, and Disease Resistance Without Eliciting Inflammation. Front Nutr (2021) 8:778424. doi: 10.3389/fnut.2021.778424
21. Lyu W, Deng Z, Sunkara LT, Becker S, Robinson K, Matts R, et al. High Throughput Screening for Natural Host Defense Peptide-Inducing Compounds as Novel Alternatives to Antibiotics. Front Cell Infect Microbiol (2018) 8:191. doi: 10.3389/fcimb.2018.00191
22. Ambati GG, Jachak SM. Natural Product Inhibitors of Cyclooxygenase (COX) Enzyme: A Review on Current Status and Future Perspectives. Curr Med Chem (2021) 28(10):1877–905. doi: 10.2174/0929867327666200602131100
23. Ferrer MD, Busquets-Cortes C, Capo X, Tejada S, Tur JA, Pons A, et al. Cyclooxygenase-2 Inhibitors as a Therapeutic Target in Inflammatory Diseases. Curr Med Chem (2019) 26(18):3225–41. doi: 10.2174/0929867325666180514112124
24. Park K, Elias PM, Hupe M, Borkowski AW, Gallo RL, Shin KO, et al. Resveratrol Stimulates Sphingosine-1-Phosphate Signaling of Cathelicidin Production. J Invest Dermatol (2013) 133(8):1942–9. doi: 10.1038/jid.2013.133
25. Guo C, Rosoha E, Lowry MB, Borregaard N, Gombart AF. Curcumin Induces Human Cathelicidin Antimicrobial Peptide Gene Expression Through a Vitamin D Receptor-Independent Pathway [Research Support, N.I.H., Extramural]. J Nutr Biochem (2013) 24(5):754–9. doi: 10.1016/j.jnutbio.2012.04.002
26. Lombardo Bedran TB, Feghali K, Zhao L, Palomari Spolidorio DM, Grenier D. Green Tea Extract and its Major Constituent, Epigallocatechin-3-Gallate, Induce Epithelial Beta-Defensin Secretion and Prevent Beta-Defensin Degradation by Porphyromonas Gingivalis. J Periodontal Res (2014) 49(5):615–23. doi: 10.1111/jre.12142
27. Wan ML, Ling KH, Wang MF, El-Nezami H. Green Tea Polyphenol Epigallocatechin-3-Gallate Improves Epithelial Barrier Function by Inducing the Production of Antimicrobial Peptide pBD-1 and pBD-2 in Monolayers of Porcine Intestinal Epithelial IPEC-J2 Cells. Mol Nutr Food Res (2016) 60(5):1048–58. doi: 10.1002/mnfr.201500992
28. Park K, Kim YI, Shin KO, Seo HS, Kim JY, Mann T, et al. The Dietary Ingredient, Genistein, Stimulates Cathelicidin Antimicrobial Peptide Expression Through a Novel S1P-Dependent Mechanism. J Nutr Biochem (2014) 25(7):734–40. doi: 10.1016/j.jnutbio.2014.03.005
29. Rath NC, Parcells MS, Xie H, Santin E. Characterization of a Spontaneously Transformed Chicken Mononuclear Cell Line. Vet Immunol Immunopathol (2003) 96(1-2):93–104. doi: 10.1016/s0165-2427(03)00143-0
30. Jiang W, Sunkara LT, Zeng X, Deng Z, Myers SM, Zhang G. Differential Regulation of Human Cathelicidin LL-37 by Free Fatty Acids and Their Analogs. Peptides (2013) 50:129–38. doi: 10.1016/j.peptides.2013.10.008
31. Yang Q, Fong L-A, Lyu W, Sunkara LT, Xiao K, Zhang G. Synergistic Induction of Chicken Antimicrobial Host Defense Peptide Gene Expression by Butyrate and Sugars. Front Microbiol (2021) 12:781649. doi: 10.3389/fmicb.2021.781649
32. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
33. Heidor R, Ortega JF, de Conti A, Ong TP, Moreno FS. Anticarcinogenic Actions of Tributyrin, a Butyric Acid Prodrug. Curr Drug Targets (2012) 13(14):1720–9. doi: 10.2174/138945012804545443
34. Salvi PS, Cowles RA. Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells (2021) 10(7):1775. doi: 10.3390/cells10071775
35. Lee B, Moon KM, Kim CY. Tight Junction in the Intestinal Epithelium: Its Association With Diseases and Regulation by Phytochemicals. J Immunol Res (2018) 2018:2645465. doi: 10.1155/2018/2645465
36. Grondin JA, Kwon YH, Far PM, Haq S, Khan WI. Mucins in Intestinal Mucosal Defense and Inflammation: Learning From Clinical and Experimental Studies. Front Immunol (2020) 11:2054. doi: 10.3389/fimmu.2020.02054
37. Fraga CG, Croft KD, Kennedy DO, Tomas-Barberan FA. The Effects of Polyphenols and Other Bioactives on Human Health. Food Funct (2019) 10(2):514–28. doi: 10.1039/c8fo01997e
38. Yahfoufi N, Alsadi N, Jambi M, Matar C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients (2018) 10(11):1618. doi: 10.3390/nu10111618
39. Suleyman H, Cadirci E, Albayrak A, Halici Z. Nimesulide is a Selective COX-2 Inhibitory, Atypical Non-Steroidal Anti-Inflammatory Drug. Curr Med Chem (2008) 15(3):278–83. doi: 10.2174/092986708783497247
40. Kim BM, Maeng K, Lee KH, Hong SH. Combined Treatment With the Cox-2 Inhibitor Niflumic Acid and Pparγ Ligand Ciglitazone Induces ER Stress/Caspase-8-Mediated Apoptosis in Human Lung Cancer Cells. Cancer Lett (2011) 300(2):134–44. doi: 10.1016/j.canlet.2010.09.014
41. Liu H, Wang J, He T, Becker S, Zhang G, Li D, et al. Butyrate: A Double-Edged Sword for Health? Adv Nutr (2018) 9(1):21–9. doi: 10.1093/advances/nmx009
42. Owczarek K, Lewandowska U. The Impact of Dietary Polyphenols on COX-2 Expression in Colorectal Cancer. Nutr Cancer (2017) 69(8):1105–18. doi: 10.1080/01635581.2017.1367940
43. Xiao X, Shi D, Liu L, Wang J, Xie X, Kang T, et al. Quercetin Suppresses Cyclooxygenase-2 Expression and Angiogenesis Through Inactivation of P300 Signaling. PloS One (2011) 6(8):e22934. doi: 10.1371/journal.pone.0022934
44. Chakraborty K, Ghosh S, Koley H, Mukhopadhyay AK, Ramamurthy T, Saha DR, et al. Bacterial Exotoxins Downregulate Cathelicidin (hCAP-18/LL-37) and Human Beta-Defensin 1 (HBD-1) Expression in the Intestinal Epithelial Cells. Cell Microbiol (2008) 10(12):2520–37. doi: 10.1111/j.1462-5822.2008.01227.x
45. Noguchi T, Shiba H, Komatsuzawa H, Mizuno N, Uchida Y, Ouhara K, et al. Syntheses of Prostaglandin E2 and E-Cadherin and Gene Expression of Beta-Defensin-2 by Human Gingival Epithelial Cells in Response to Actinobacillus Actinomycetemcomitans. Inflammation (2003) 27(6):341–9. doi: 10.1023/b:ifla.0000006702.27906.e9
46. Wan M, Tang X, Rekha RS, Muvva S, Brighenti S, Agerberth B, et al. Prostaglandin E2 Suppresses Hcap18/LL-37 Expression in Human Macrophages via EP2/EP4: Implications for Treatment of Mycobacterium Tuberculosis Infection. FASEB J (2018) 32(5):2827–40. doi: 10.1096/fj.201701308
47. Rossokhin AV, Sharonova IN, Dvorzhak A, Bukanova JV, Skrebitsky VG. The Mechanisms of Potentiation and Inhibition of GABAA Receptors by non-Steroidal Anti-Inflammatory Drugs, Mefenamic and Niflumic Acids. Neuropharmacology (2019) 160:107795. doi: 10.1016/j.neuropharm.2019.107795
48. Carlos-Reyes A, Lopez-Gonzalez JS, Meneses-Flores M, Gallardo-Rincon D, Ruiz-Garcia E, Marchat LA, et al. Dietary Compounds as Epigenetic Modulating Agents in Cancer. Front Genet (2019) 10:79. doi: 10.3389/fgene.2019.00079
49. Steven A, Friedrich M, Jank P, Heimer N, Budczies J, Denkert C, et al. What Turns CREB on? And Off? And Why Does it Matter? Cell Mol Life Sci (2020) 77(20):4049–67. doi: 10.1007/s00018-020-03525-8
50. Wang H, Xu J, Lazarovici P, Quirion R, Zheng W. cAMP Response Element-Binding Protein (CREB): A Possible Signaling Molecule Link in the Pathophysiology of Schizophrenia. Front Mol Neurosci (2018) 11:255. doi: 10.3389/fnmol.2018.00255
51. Chakraborty K, Maity PC, Sil AK, Takeda Y, Das S. cAMP Stringently Regulates Human Cathelicidin Antimicrobial Peptide Expression in the Mucosal Epithelial Cells by Activating cAMP-Response Element-Binding Protein, AP-1, and Inducible cAMP Early Repressor. J Biol Chem (2009) 284(33):21810–27. doi: 10.1074/jbc.M109.001180
52. Rezen T, Zmrzljak UP, Bensa T, Tomas TC, Cirnski K, Stojan J, et al. Novel Insights Into Biological Roles of Inducible cAMP Early Repressor ICER. Biochem Biophys Res Commun (2020) 530(2):396–401. doi: 10.1016/j.bbrc.2020.05.017
53. Surh YJ, Chun KS, Cha HH, Han SS, Keum YS, Park KK, et al. Molecular Mechanisms Underlying Chemopreventive Activities of Anti-Inflammatory Phytochemicals: Down-Regulation of COX-2 and iNOS Through Suppression of NF-Kappa B Activation. Mutat Res (2001) 480-481:243–68. doi: 10.1016/s0027-5107(01)00183-x
54. Sapio L, Gallo M, Illiano M, Chiosi E, Naviglio D, Spina A, et al. The Natural cAMP Elevating Compound Forskolin in Cancer Therapy: Is It Time? J Cell Physiol (2017) 232(5):922–7. doi: 10.1002/jcp.25650
55. Zeng X, Sunkara LT, Jiang W, Bible M, Carter S, Ma X, et al. Induction of Porcine Host Defense Peptide Gene Expression by Short-Chain Fatty Acids and Their Analogs. PloS One (2013) 8(8):e72922. doi: 10.1371/journal.pone.0072922
56. Mageed AM, Isobe N, Yoshimura Y. Expression of Avian Beta-Defensins in the Oviduct and Effects of Lipopolysaccharide on Their Expression in the Vagina of Hens. Poult Sci (2008) 87(5):979–84. doi: 10.3382/ps.2007-00283
57. Michailidis G, Anastasiadou M, Guibert E, Froment P. Activation of Innate Immune System in Response to Lipopolysaccharide in Chicken Sertoli Cells. Reproduction (2014) 148(3):259–70. doi: 10.1530/REP-14-0064
Keywords: cyclooxygenase-2 inhibitors, host defense peptides, antimicrobial peptides, antibiotic alternatives, polyphenols, antimicrobial resistance
Citation: Yang Q, Burkardt AC, Sunkara LT, Xiao K and Zhang G (2022) Natural Cyclooxygenase-2 Inhibitors Synergize With Butyrate to Augment Chicken Host Defense Peptide Gene Expression. Front. Immunol. 13:819222. doi: 10.3389/fimmu.2022.819222
Received: 21 November 2021; Accepted: 31 January 2022;
Published: 22 February 2022.
Edited by:
Rosa Gaglione, University of Naples Federico II, ItalyReviewed by:
Philip Brandon Busbee, University of South Carolina, United StatesCopyright © 2022 Yang, Burkardt, Sunkara, Xiao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guolong Zhang, emd1b2xvbkBva3N0YXRlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.