- Department of Colorectal Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Tumor immunity is involved in malignant tumor progression. Myeloid-derived suppressor cells (MDSCs) play an irreplaceable role in tumor immunity. MDSCs are composed of immature myeloid cells and exhibit obvious immunomodulatory functions. Exosomes released by MDSCs (MDSCs-Exos) have similar effects to parental MDSCs in regulating tumor immunity. In this review, we provided a comprehensive description of the characteristics, functions and mechanisms of exosomes. We analyzed the immunosuppressive, angiogenesis and metastatic effects of MDSCs-Exos in different tumors through multiple perspectives. Immunotherapy targeting MDSCs-Exos has demonstrated great potential in cancers and non-cancerous diseases.
Introduction
Malignant tumors are considered a major threat to human health (1). Studying the contribution of tumor immunity in tumor progression may improve the extremely narrow therapeutic strategy regarding cancer. Myeloid-derived suppressor cells (MDSCs) are immature cells consisting of myeloid progenitor cells, immature macrophages, immature granulocytes and immature dendritic cells. They are closely related to patients’ poor prognosis due to its powerful effects on tumor immune suppression, tumor angiogenesis, drug resistance, and tumor metastases (2–4). For example, The interaction between MDSCs and macrophages can reduce the production of IL-12 by macrophages and increase the production of IL-10 by MDSCs, which promotes tumor progression (5).
Exosomes are 40- to 100-nm small vesicles that are released by the vast majority of cells and distributed in all body fluids (6, 7). Exosomes derived from different cells perform variable functions. Exosomes carry proteins, DNA, messenger RNAs(mRNAs), noncoding RNAs(ncRNAs), and lipids (8). Exosomes exert cancer-inhibiting or cancer-promoting effects. For example, exosomal miR-19a is delivered to osteoblasts to promote bone metastasis in breast cancer (9). While human umbilical cord mesenchymal stem cell -derived exosomal miR-320a inhibits lung cancer cell growth via SOX4/Wnt/β-catenin axis (10). In addition, emerging studies have shown that exosomes have potential clinical applications as biomarkers for disease diagnosis and prognosis (11–14).
The exosomes derived from MDSCs (MDSCs-Exos) are involved in the function of immunosuppression, promoting tumor angiogenesis, tumor metastasis, drug resistance of malignant tumors (15–17). Here, we summarized linkages and differences between MDSCs-Exos and parental cells, as well as regulatory roles and possible diagnostic and prognostic values in tumor immunity.
MDSCs in Tumor Immunity
MDSCs are a special kind of cells that have important tumor immunomodulatory effects, composed of several immature heterogeneous cells originating from myeloid cells (18). Normally, immature myeloid cells differentiate into mature immediately after entering the peripheral organs (19). Under a variety of pathological conditions, MDSCs expand and can be detected in blood, cancer tissue, inflammatory sites, lymph nodes and spleen (20). In the tumor microenvironment (TME), the differentiation and amplification of MDSCs is mediated by a variety of molecules, such as: granulocyte-macrophage colony–stimulating factor, granulocyte colony–stimulating factor (G-CSF), macrophage colony–stimulating factor, stem cell factor, vascular endothelial growth factor (VEGF), and polyunsaturated fatty acids (21–23). Activation of MDSCs is dependent on the following cytokines: IFN-γ, IL-1β, TNF, IL-4, IL-6, IL-13 and high mobility group box protein 1 which function through NF-κB, STAT1 and STAT6 pathways (24). According to different surface markers, MDSCs are divided into two subtypes: granulocytic MDSCs (G-MDSCs, CD11b+Ly6G+Ly6Clow), termed polymorphonuclear MDSCs (PMN-MDSCs) simultaneously, and monocytic MDSCs (M-MDSCs, CD11b+Ly6G-Ly6Chi) (23, 25, 26). The number of G-MDSCs is far outweighed by that of M-MDSCs, and a majority of current studies have focused on investigating the capabilities of G-MDSCs, while the role of M-MDSCs remains to be further investigated (27). Their immunosuppressive abilities and mechanisms also differ (23, 28). G-MDSCs inhibit T-cell responses mainly through the production of reactive oxygen species (ROS) by antigen-specific methods. M-MDSCs produce large amounts of NO, arginase 1 (Arg-1) and immunosuppressive cytokines, such as IL-10, which inhibit both antigen-specific and non-specific T-cell responses. M-MDSCs have a higher inhibitory activity than G-MDSCs (29–33).
MDSCs exert their immunosuppressive effect by promoting the expansion of regulatory T cells (Tregs) (34–36), promoting the production of helper T cell 17 (Th17) (35), inducing macrophage differentiation into the M2 phenotype (2, 37, 38) and inhibiting of immune response of NK cells (39) and B cells (40). In addition to suppressing the immune response, MDSCs also accelerate tumor progression by remodeling tumor microenvironment (22). MDSCs promote tumor angiogenesis through up-regulating VEGF, matrix metallopeptidase 9 (MMP9) and bombina variegata peptide 8 (Bv8) (41). MDSCs also promote tumor metastases by infiltrating primary tumors and facilitating the formation of premetastatic niches (42).
In conclusion, MDSCs exhibit great prospect in the treatment of cancer by blocking T cells, B cells and NK cells activity and bolstering Tregs expansion and mobilization.
Exosome
Extracellular vesicles (EVs) were initially considered to be “platelet dust” by Peter Wolf in 1967 (43). EVs transport proteins, nucleic acids, lipids, cytokines, metabolites, and enable intercellular information communication (44). Depending on their origin, markers, properties and functions, EVs are classified into two main groups: ectosomes and exosomes (45). Ectosomes are vesicles formed by outward budding of the plasma membrane, whereas exosomes are intraluminal vesicles formed by plasma membrane invagination, the release of the latter involves the fusion of multivesicular bodies with the plasma membrane (46–48). Recent studies suggest that CD63 is the signature exosome-specific protein, while CD9 and CD81 are not specific (49). Statistically, the cargoes that have been identified in exosomes include 9769 proteins, 3408 mRNAs, 2838 miRNAs and 1116 lipids [data from http://www.exocarta.org (a database collecting many studies)]. These components are involved in cellular signaling pathways, regulation of lipid metabolism, tumor progression, recurrence and metastasis (50–53).
After exosomes are released outside the cell, they participate in information transmission with the target cells through membrane fusion, endocytosis and binding to the receptors on the surface of the target cells (54). Currently, it is becoming increasingly evident that exosomes play an essential role in disease, especially in tumors by promoting the reprogramming of receptor cells (55–57). In non-small cell lung cancer(NSCLC), tumor-derived exosomes(TDEs) polarize macrophages to an immunosuppressive phenotype that increases programmed death ligand-1 expression through NF-kB-dependent, glycolysis-dominated metabolic reprogramming, triggering the formation of pre-metastatic niche (58). Exosomal lncARSR propagates sunitinib resistance through competitive binding of miR-34/miR-449 in renal cell carcinoma (59).
In briefly, exosomes participate in the physiopathological processes of coagulation, inflammation, angiogenesis and immune response (12). Exosomes are widely distributed and easy to modulate, can be used as a promising minimally invasive tool for diagnosis and treatment (7, 60–62).
Characterization of Exosomes Derived From MDSCs
It is now known that exosomes carry proteins, DNA, messenger RNAs(mRNAs), noncoding RNAs(ncRNAs), and lipids. MDSCs-Exos exert a unique function due to the specificity of the cargoes carried. MDSCs-Exos are also rich in proteins, RNA and DNA. We next discuss the properties of exosomes in five dimensions.
Protein Differences Between MDSCs and Their Exosomes
It is well known that protein is the material basis of life activities (63). The same protein exists in different structures and performs different biological functions due to post-translational modifications (64). Current studies have shown that the cargoes carried by MDSCs-Exos are mainly involved in the immunosuppressive effect of MDSCs (65). S100A8/A9 (calcium binding protein, with chemotactic activity) is present in both MDCSs and MDSC-EXO. Chronic inflammation increased S100A8/A9 content in MDSCs (66), with insignificant changes in exosomes (65). At present, numerous studies on exosomes in tumor-bearing mice are mainly focused on the differences of cargoes (especially ubiquitination protein (67), glycoprotein (68, 69) and RNA (70, 71) carried by exosomes and parental cells.
A study identified 1726 proteins in MDSCs and their exosomes, of which 58% were identified in MDSCs and their exosomes simultaneously. Regardless of inflammation, 30% of the proteins in MDSCs are enriched in their exosomes, especially those involved in exosome formation and protein sorting as well as proteins that load miRNAs into exosomes. Through this selective sorting mechanism, MDSCs-Exos may mediate some functions different from those of MDSCs (15). Similar to other exosomes, MDSCs-Exos enrich many characteristic components, such as tetraspanins (including CD9, CD177), Hsp70, Hsp90α, Hsp90β, Alix, and the ESCRT complex, which are involved in exosome formation and protein sorting. Compared with parental cells, the abundance of CD9 was 89-fold increased regardless of inflammatory status (15, 65). MDSCs-Exos also contain many other protein cargoes, including many nucleic acid binding proteins, numerous histone variants and several elongation factors. It has been reported that these proteins can bind to nucleic acids and induce changes in nucleic acids expression and the protein spectrum of receptor cells (72). Some chemotactic proteins are enriched in MDSCs-Exos, such as the pro-inflammatory proteins S100A8/9, CD47 and thrombospondin-1. These proteins mediate the aggregation of MDSCs and enhance the immunosuppressive function of MDSCs. The relative abundance of the pro-inflammatory proteins S100A8/9, which are secreted by MDSCs and mediate >90% of the chemotactic effect on MDSCs, are not affected by inflammatory conditions. The cytokine macrophage migration inhibition factor and the chemokine platelet factor-4 are also enriched in exosomes, and these proteins exhibit chemotactic activity on leukocytes (15). Regardless of inflammatory conditions, transforming growth factor-β1(TGF-β1) is 4.3-fold more abundant in the exosomes compared with parental cells (15). TGF-β1 participates in the expansion of T cells and the inhibition of NK cells (73, 74). Immunoglobulins, complement regulatory factor H and C4B-binding proteins(C4B-bp) are enriched in exosomes. They may also be involved in the regulation of immune system by MDSCs and their exosomes (15).
The above results indicated that the cancer-promoting effects of MDCSs are partially achieved by exosomes. Targeting MDSCs-Exos holds a bright future for cancer treatment.
Differences Roles of the Exosomes Released by G-MDSCs and M-MDSCs in Tumor
As we mentioned earlier, MDSCs are distinguished into two subtypes. Interestingly, the exosomes derived from different subtypes of MDSCs also differ in their impacts on tumor. Rab27a controls exosomes biogenesis (75). The expression of Rab27a was significantly reduced by transfecting siRNA. In a tumor sphere formation assay, after inhibiting of exosome derived from G-MDSCs, the tumor sphere numbers, CD44+ cell percentages and CD133+ cell percentages were decreased. But the CD44+ cell percentages was not decreased when exosomes were inhibited in M-MDSCs (76). This indicated that G-MDSCs-Exos and M-MDSCs-Exos have different effects on cancer cell stemness.
Currently, researchers have mainly focused on investigating the role of G-MDSCs-Exos on tumor progression, and studies on M-MDSCs-Exos are very rare. Although there is an evidence that M-MDSCs-Exos affects tumor immunity. However, researchers have mainly focused on investigating the role of G-MDSCs-Exos on tumor progression, and studies on M-MDSCs-Exos are very rare. To some extent, an insight into the role of M-MDSCs-Exos may lead to new immunotherapeutic approaches. Distinguishing the role differences between M-MDSCs-Exos and G-MDSCs-Exos may be a new research hotspot.
Differences in Proteins Carried by MDSCs-Exos Under Different Inflammatory Conditions
As inflammation increases, an increasing number of MDSCs were identified, and stronger immunosuppressive effect was observed (77). MDSCs play a key role in the control of experimental necrotizing small intestinal colitis in neonatal mice by suppressing T-cell function (78). Immunosuppressive proteins and miRNAs are increased in EVs during chronic inflammation and aging (79). In one study, the researchers identified 412 proteins, of which the abundance of 63 proteins changed greater than 2-fold in an inflammatory environment. It is worth noting that there was no obvious difference in quantity of exosomes shed per MDSC isolated from low-inflammation or high-inflammation environments. Compared with conventional conditions, inflammatory conditions reduced the abundance of 33 proteins, such as C4B-bp, complement C3 and ficolin-1, which participate in the innate immune response. Several cytoskeletal proteins and chemotactic proteins are found to be reduced in an inflammatory environment, which are related to the migration of exosomes. In addition, a highly inflammatory environment increased the abundance of 30 proteins, including Leukocyte elastase inhibitor A, DBF4-type zinc finger-containing protein 2 homologue and Cathepsin G, etc (65).
Reducing inflammation may expand new horizons for cancer treatment by weakening MDCSs in TME. In the future, the majority of cancer patients may be able to benefit from this.
Ubiquitin Proteins and Glycoproteins Carried by MDSCs-Exos
Ubiquitin is a common post-translational modification (80), which affects protein function by influencing protein stability, turnover, cellular localization, and regulating cellular signaling cascade responses (81). The imbalance between ubiquitination and deubiquitination is closely related to the occurrence of human immune diseases, cancer, infection and neuropathy (82). In NSCLC, deubiquitination of PDL-1 promotes immune escape by suppressing CD8+T cell responses (83). NLRC3, a member of the innate immune receptor, impaired CD4+ T cell signaling and metabolism by limiting NF-κB activation, reducing glycolysis and oxidative phosphorylation via decreased K63-linked ubiquitination of TNF-receptor-associated factor 6 (84). Therefore, it is urgently needed to investigate whether MDCSs and MDSCs-Exos carry ubiquitin protein, which will help to develop new treatment strategies based on exosomes. Protein blot analysis demonstrated that the parental cells and their exosomes contained different ubiquitinated protein profiles (85). Initially, 10 ubiquitinated proteins in MDSCs-Exos were identified (65). With the application of mass spectrometry-based bottom-up proteomics technology, scholars isolated and identified 50 ubiquitinated proteins from MDSCs-Exos (86). Specifically, the ubiquitinated nuclear proteins include several histones, ribosomal proteins and nucleic acid binding proteins. The ubiquitinated histones in these exosomes may possess active pro-inflammatory properties (87, 88). Interestingly, the pro-inflammatory high mobility group box protein 1 is ubiquitinated, promotes the accumulation of MDSCs, and enhances the immunosuppressive effect of MDSCs (89). Sorting nexin 13 has been identified to be involved in endosomal transport of ubiquitinated proteins (90). Two ubiquitinated keratins were revealed to play an active role in plasma membrane invagination during the initial phase of EVs formation (91, 92). Other ubiquitinated proteins leucine zipper EF hand-containing transmembrane protein 1 and endoplasmin, which participate in the formation of endosomes and exosomes (65).
Similarly, glycosylation is an important protein modification that determines protein folding and transport and is crucial for mammalian survival (93, 94). Until 2018, 21 N-glycoproteins on the surface of MDCS-Exos exosomes were identified using proteomic methods, including CD44, CD47, CD321, CD157, CD11b, CD97, thrombospondin1 (Tsp1), fibronectin, cytoskeletal krt, fibrinogen, etc (95). Of special interest is CD47, donor CD47 plays an important role in the control of T cell allogeneic response and tolerance induction after hepatocyte transplantation (96, 97). It mediates the chemotaxis and migration of MDSCs by combining with Tsp1 on MDSCs. When CD47 on tumor cells binds to CD172a (signal regulatory protein α or SIRP α), it can prevent macrophages from phagocytosing tumors (98) and maintain acquired immune tolerance (97). Therefore, CD47 is a potential drug target (99). In addition, it is worth noting that MDSCs-Exos may transport immunosuppressive cargoes to T cells through the binding of CD321 to TLFA-1 of T cells (95).
Both ubiquitinated and glycosylated proteins are present in MDSC and its derived exosomes, respectively, which supports the idea that exosomes have an analog to parental cells.
mRNAs and miRNAs Carried by MDSCs-Exos
In addition to carrying protein cargo, MDSCs-Exos also carry a large number of RNAs, including mRNAs and miRNAs (15), similar to the results of previous studies, almost no ribosomal RNA was found (100–102). The mRNAs carried by exosomes is also transferred to recipient cells and translated into functionally active proteins, which produces more lasting effects than proteins. Compared with parental cells, 45% of mRNA transcripts in exosomes exhibited statistically differences in abundance regardless of the inflammatory conditions. The transcripts of these mRNAs took part in several signaling pathways including “calcium signaling pathway”, “cAMP signaling pathway” and “hippo signaling pathway”. In addition, only approximately 3.5% of the mRNA transcripts differ in abundance under inflammatory conditions compared with parental cells. These mRNA transcripts played role in the signaling pathway associated with TGF-β and VEGF. Compared with conventional exosomes, several biological processes were identified enriched in inflammatory exosomes, including “cell-cell signaling”, “macrophage differentiation” (15).
Simultaneously, the study identified approximately 1500 differentially expressed miRNAs in MDSCs-Exos, and approximately half of them exhibited increased abundance in inflammatory exosomes. According to the prediction of these miRNA targets, if these miRNAs are transferred to the target cells and bind to mRNA targets, they will affect the proliferation, differentiation and apoptosis of target cells. These miRNAs can regulate the immune system and tumor microenvironment and thus affect tumor progression and metastasis.
The miRNAs enriched in inflammatory exosomes include miRNA-704, miRNA-5134, miRNA-7022 and miRNA-7062, which bind to the target mRNA taking part in the apoptosis pathway, including Fas. Compared to parental cells, miRNA-690 and miRNA-155 are enriched in exosomes and may be delivered to MDSCs. MiRNA-690 promotes MDSCs expansion through regulating the cell cycle of myeloid cells. MiRNA-155 increases the production of IL-10. IL-10 induces the proliferation of regulatory T cells and causes the transformation of macrophages to tumor growth-promoting M2-Mϕ. Interestingly, miR-146a negatively regulates the activation of the NF-κB pathway and subsequently controls inflammation by targeting the IL-1 receptor-associated kinase 1 and TNF receptor-associated factor 6 mRNAs (15, 103). In contrast to miRNA-690 and miRNA-155, miR-146a suppresses the development of malignant tumors (15).
We have realized that MDSCs-Exos regulate the signaling pathways and biological processes of target cells through the carried proteins and RNAs. At present, it is necessary to further clarify the type and abundance of cargoes contained in MDSCs and their exosomes, and compare the similarities and differences of cargoes carried by MDSCs and exosomes under different conditions. It is helpful to predict the function of MDSCs-Exos according to the existing research on the composition and function of MDSCs.
Effects of MDSCs-Exos on Tumor Immunity in Various Cancers
Tumor immunosuppression is a feature of malignant tumors (104). MDSCs-Exos play an irreplaceable role in tumor immunity, similar to parental MDSCs. Here, we summarized the role of MDSCs-Exos in cancer immunity.
Immune Suppression Induced by MDSCs-Exos in Cancers
MDSCs are one of the components of TME and are involved in tumor progression mainly by suppressing the function of T cells (26). MDSCs-Exos, as the immunosuppressive factor in the TME, carry many bioactive substances from MDCSs. In tumor-bearing mice, MDSCs-Exos were significantly higher in tumor tissue than at the spleen and bone marrow. MDSCs-Exos activate CD8+ T cells and drive them to produce more IFN-γ, but MDSCs-Exos increase ROS production, activate the Fas/FasL pathway in T cells, and trigger so-called activation-induced cell death (AICD) (105, 106). In tumor patients, this process is induced by the high expression of S100A8/9 (20). TDEs-provided membrane-associated Hsp72 triggers the activation of TLR2/MyD88-dependent STAT3 pathway in MDSCs through autocrine IL-6, which triggers significant immunosuppressive activity (107). The miRNAs carried by TDEs are also involved in enhancing the expansion and immunosuppression of MDSCs. For example, hypoxia-inducible miRNA-21 in TDEs enhances MDSC expansion and activation by targeting RORα and PTEN (108).
MDSCs-Exos regulate tumor immunity by carrying differential bioactive contents that mainly act on MDSCs and other target cells in the immune system. The pro-inflammatory S100A8/9 heterodimer carried by MDSCs-Exos is chemotactic for MDSCs and plays the primary role in promoting the aggregation of MDSCs to the tumor tissue and pre-metastatic niche (109). Other chemotactic proteins enriched in MDSCs-Exos include CD47 and TSP1, which mediate the immunosuppressive function of MDSCs together with S100A8/9 (65). Similar to MDSCs, MDSCs-Exos can also transform macrophages into tumor growth-promoting M2 macrophages by reducing the production of IL12 from macrophages (65). TGF-β1 enriched in MDSCs-Exos induces Tregs or Th17 cells and impair the cytotoxicity of natural killer (NK) cells, which enhances the immunosuppressive effect of MDSCs-Exos (15).
Contribution of MDSCs-Exos to Tumor Progression and Metastasis
MDSCs and their exosomes participate in the entire process of tumor progression through immunosuppression, angiogenesis, invasion and metastasis, the formation of a premetastatic niche and the stemness of tumor cells. G-MDSCs gathered in lung cancer tissue secreted a large amount of miRNA-143-3p. MiRNA-143-3p promotes tumor cell proliferation by inhibiting integral membrane protein 2B and activating PI3K/Akt pathway (110).
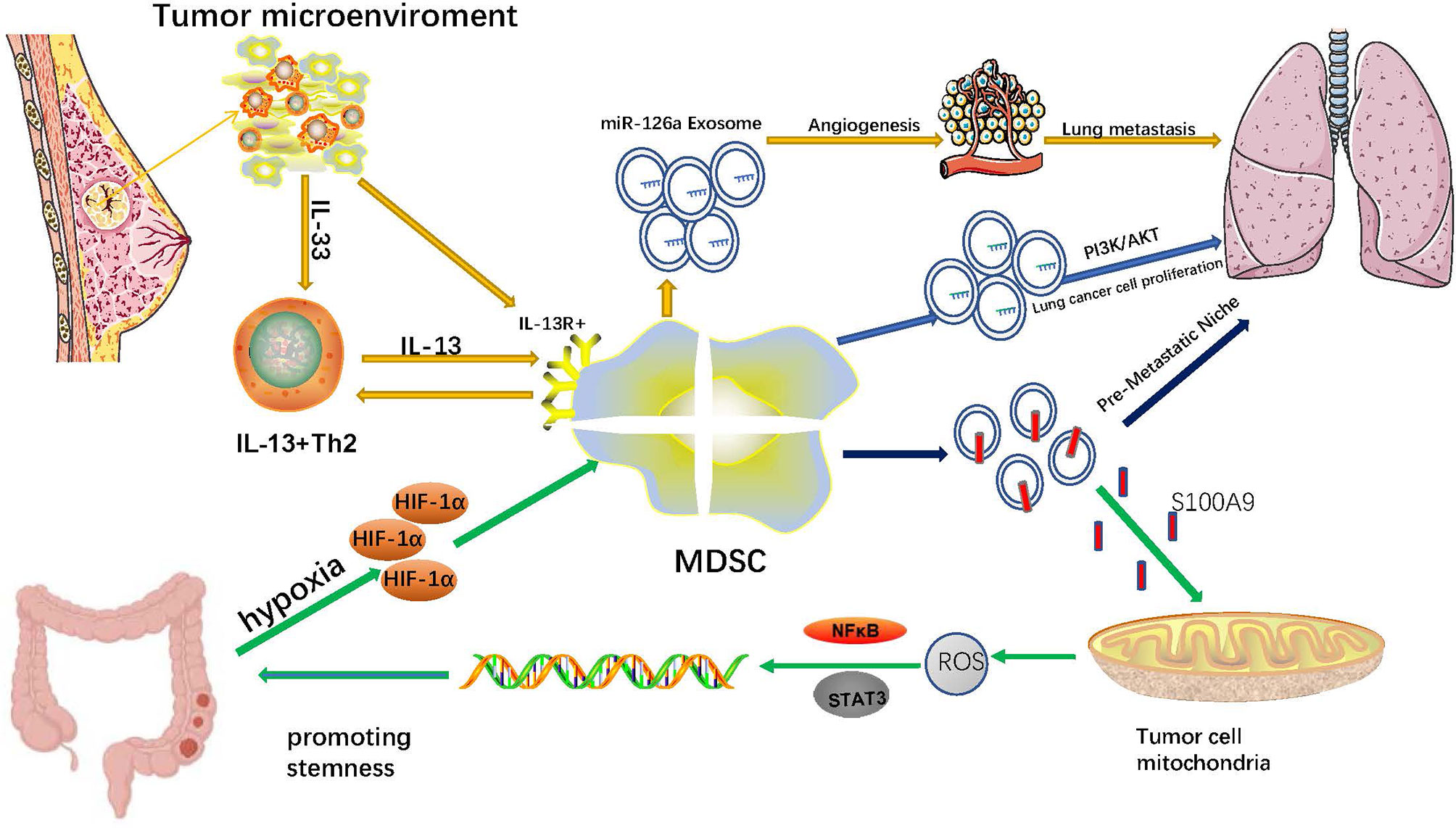
Angiogenesis is fundamental for the growth and metastasis of solid tumors (111). Tumors can induce the upregulation of growth factors, including VEGF, ANG, PDGF, TGF and EGF, which disrupt the balance between proangiogenic and antiangiogenic signals. Growth factors also induce the ‘‘angiogenic switch’’ and subsequently promote the proliferation of vascular endothelial cells and the formation of capillaries (112). In addition, hypoxia in the tumor microenvironment aggravates this process by increasing the expression of proangiogenic factors (113). Several recent studies demonstrated that MDSCs and their exosomes also participate in tumor angiogenesis by recruiting MDSCs to the tumor site with several chemokines. MDSCs can secrete proangiogenic factors, including BV8 (bombina variegata peptide 8), VEGF, and basic fibroblast growth factor, by activating the STAT3 signaling pathway (41). MDSCs can also produce MMP-9, a protease that degrades extracellular matrix, which triggers the release of VEGF deposited in the matrix and increases its bioavailability (114). Moreover, the production of CCL2 in the TME is another important mechanism by which MDSCs promote tumor angiogenesis (115). Notably, splenic MDSCs can differentiate into endothelial progenitor cells that directly participate in tumor angiogenesis (116). It has been reported that miR-126a+MDSCs induced by doxorubicin (DOX) treatment in breast tumor-bearing mice interact with IL-13+Th2 cells in a positive feedback loop manner, increasing the production of Th2 cells and miR-126a+MDSC-Exo. Consequently, the increased level of miR-126a+MDSC-Exo lead to lung metastasis by promoting tumor angiogenesis (17).
One of the negative features of malignant tumors is their unlimited proliferation ability, and cancer stem cells (CSCs) endow them with this ability (117). CSCs have been considered a significant supporter of tumor progression and chemoresistance, and emerging evidence suggests that MDSCs and their exosomes exert crucial influence on the stemness of tumor cells. In patients with ovarian cancer, MDSCs induce ovarian cancer cells to express microRNA101. MicroRNA101 increases the expression of stem cell genes, including OCT3/4, SOX2, and NANOG, via inhibiting the expression of C-terminal binding protein-2 in ovarian cancer cells. As a result, MDSCs promote the stemness of ovarian cancer cells (118). In breast cancer patients, MDSCs promote cancer cell stemness by activating the NO/NOTCH and IL-6/STAT3 signaling pathways (119). New evidence in cervical cancer patients indicates that MDSCs induced by tumor-derived G-CSF enhance the stemness of cancer cells via producing Prostaglandin E2(PGE2) (120). MDSCs infiltrate into PTEN null prostate cancer cells and induce the stemness of prostate cancer cells via producing IL-1Ra and blocking the IL-1α/IL-1R axis (121). In addition, MDSCs promote the stemness and induce mesenchymal characteristics of pancreatic cancer cells by upregulating the levels of p-STAT3 (122). In A549 transplantation tumors treated with endostatin, MDSCs and MDSC-derived TGF-β1 and hypoxia enhanced the stemness of A549 cells and their resistance to endostatin (123). A new study of colorectal cancer revealed that hypoxia can promote G-MDSCs to generate more MDSCs-Exos by up-regulating HIF-1α. MDSCs-Exos aggravate the stemness of colorectal cancer cells through exosomal S100A9. Blocking S100A9 expression in MDSCs-Exos can inhibit the stemness of colorectal cancer cells and prevent the occurrence of colon cancer in mice with colitis (76) (Figure 1).
The Role of MDSCs in Tumor Chemoresistance
Chemotherapy is one of the most important treatments for malignant tumors, but chemoresistance is a crucial obstacle impeding clinical treatment (124). At present, the mechanism underlying chemoresistance has not been well elucidated, but several emerging lines of evidence suggest that MDSCs induce reduced tumor cell sensitivity to chemotherapy. In mice with colorectal cancer, oxaliplatin leads to chemoresistance by restraining the polarization of MDSCs into M1-like macrophages. In addition, MDSCs and their differentiated M2-like macrophages promote immunosuppression, angiogenesis and chemoresistance by producing protumorigenic cytokines (IL-10, TGF-β, VEGF and proteases) and suppressping the function of CD8+ T cells (125).
Tumor-derived G-CSF increases the production of MDSCs and attenuates the spontaneous apoptosis of MDSCs by activating the STAT3 pathway. Then, the increased G-MDSCs induce angiogenesis through Bv8, which leads to chemoresistance in cervical cancer. The study also showed that treating mice with depleting MDSCs enhances the effect of chemotherapy for cervical cancer (126). It has been reported that IL-6 released by drug-resistant hepatocellular cancer promotes the expansion and activity of MDSCs, and the interactions between IL-6 and MDSCs promote the chemoresistance of hepatocellular cancer. The sensitivity to chemotherapy can be enhanced via depleting MDSCs or blocking IL-6 (127). Similarly, PMN-MDSCs promote multiple myeloma survival in response to chemotherapies, such as doxorubicin and melphalan, and the process is mediated by soluble factors, including IL-6 (128). Benzyl butyl phthalate exposure aggravates the resistance of breast cancer to doxorubicin. Mechanically, it promotes MDSCs to infiltrate into tumors and increases the secretion of S100A8/A9 by MDSCs (129). A study shows that MDSCs promote Tregs infiltrate into lung tumors and trigger CD8T cells depletion, which strongly induces immunosuppression and chemoresistance (130).
In numerous studies, MDSCs-Exos exhibit a highly similar role to MDSCs, but whether they play a role in tumor drug resistance has not been explored. In-depth studies on the role of MDSCs-Exos in drug resistance may provide new perspectives for anti-cancer treatment strategies.
Effects of MDSCs-Exo in Non-Oncologic Diseases
Except in tumors, the massive expansion of MDSCs is always accompanied by non-oncologic diseases, especially in autoimmune diseases. Unlike their role in tumors, MDSCs and MDSCs-Exos can alleviate autoimmune diseases (131). Exosomes derived from MDSCs carry a variety of bioactive contents, including proteins and RNAs, which play a similar role as MDSCs. MDSCs play an irreplaceable role in maternal-fetal tolerance in normal pregnancy. The isolation and identification of exosomes from maternal peripheral blood G-MDSCs revealed that G-MDSCs-Exos inhibited CD4+T cells and CD8+T cells, induced Tregs production, Th2 cell differentiation, and this effect was preserved under frozen conditions (132). (Figure 2B). This is very beneficial for application in clinical treatment. G-MDSCs-Exos attenuated the damage of inflammatory cell infiltration and reduced the activity index of DSS-induced colitis in mice, thus significantly alleviating the severity of the disease. This effect was mainly achieved via repressing the proliferation of Th1 cells, promoting the expansion of Tregs, and reducing the levels of serum IFN-γ and TNF-α in mice (133) (Figure 2A). In the mouse model of autoimmune alopecia areata (AA), MDSCs-Exos reversed the progression of the disease and promoted hair regeneration. MDSCs-Exos accumulated in the draining lymph nodes and cells near residual hair follicles. They are absorbed by T cells, macrophages and NK cells, especially Tregs. As a result, MDSCs-Exos significantly alleviated the disease by amplifying Tregs, weakening the cytotoxic activity of T cells, reducing the proliferation of T helper cells and increasing lymphocyte apoptosis (134) (Figure 2C). In mice with collagen-induced arthritis, G-MDSC-derived exosomes attenuated joint destruction efficiently by reducing the number of Th1 and Th17 cells. Mechanistically, miR-29a-3p carried by G-MDSCs-Exos targets T-bet to suppress the differentiation of Th1 cells, and miR-93-5p carried by G-MDSCs-Exos targets STAT3 to suppress the differentiation of Th17 cells (16). There is no such ability in M-MDSCs exosomes. Under hypoxic conditions, the higher levels of miR-29a-3p and miR-93-5p in G-MDSCs-Exos more effectively inhibited the proliferation of CD4+T cells and thus more effectively attenuate arthropathy (135). In addition, PGE2 in MDSCs-Exos upregulated the phosphorylation levels of GSK-3β and CREB to promote IL-10+ Breg cell production to attenuate CIA in mice. This effect was blocked by celecoxib (136). (Figure 2D)
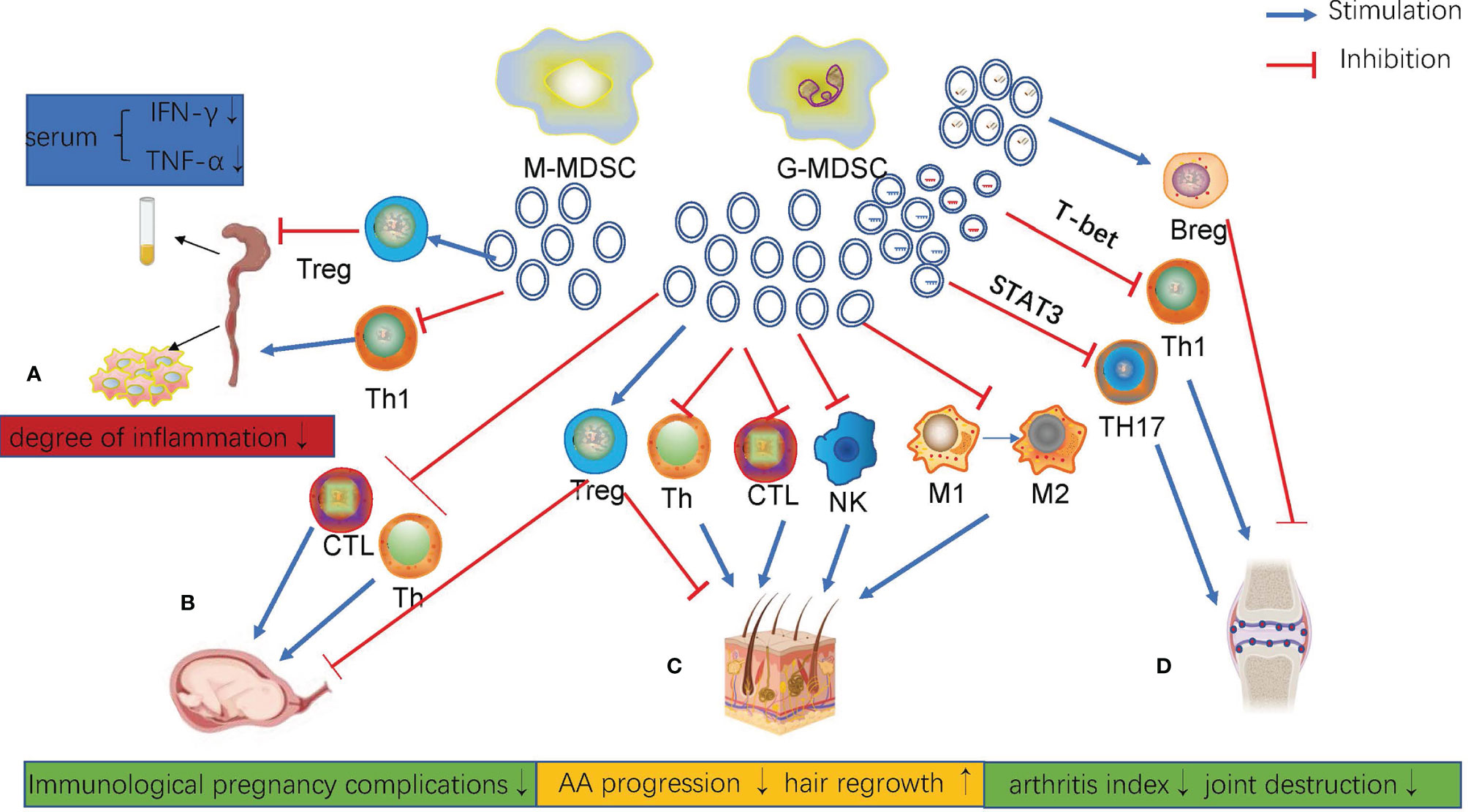
Figure 2 (A) In DSS-induced colitis, G-MDSCs-Exos inhibited the proliferation of Th1 cells, promoted the proliferation of Treg cells, and decreased serum IFN-g and TNF-AIN levels in mice. (B) Maternal peripheral blood G-MDSCs-Exos regulated different subtypes of T cell differentiation and function. (C) MDSCs-Exos inhibited AA progression and promoted hair regrowth. (D) G-MDSCs-Exos attenuated CIA in mice by regulating Th1, Th17 and Breg cells.
Although the understanding of the relationship between MDSCs and autoimmune diseases has become increasingly clear, research on the role of MDSCs-Exos in autoimmune diseases remains limited. Because abundant bioactive molecules overlap in MDSCs and MDSCs-Exos, the function of MDSCs-Exos can be predicted and verified according to known research on MDSCs. Related researches will provide fresh insight into the diagnosis and treatment of autoimmune diseases.
Clinical Applications of MDSCs-Exos
Given the crucial role of MDSC-mediated immunosuppression in tumor progression, several studies have explored a number of therapeutic strategies by targeting MDSCs. These treatments mainly include two aspects. On one hand, the number of MDSCs are reduced by using chemotherapeutic drugs (137), inhibiting the expansion of MDSCs (138) and promoting the differentiation of myeloid cells (139). On the other hand, the functions of MDSCs are suppressed. For example, nitroaspirin is used as an ROS inhibitor (140), and cyclooxygenase 2 inhibitors are used to restrain the production of arginase 1 (141). In addition, phosphodiesterase 5 inhibitors are used to suppress the production of iNOS (142).
In recent years, research on the clinical application of exosomes has become a hot topic. In terms of diagnosis, proteins and ncRNAs expressed in exosomes can be used as markers for early diagnosis, drug sensitivity and prognosis of many cancers (143). In terms of treatment, exosomes are used in tumor immunotherapy and as a new carrier for loading drugs, proteins and ncRNAs (144). When exosomes contact with the extracellular matrix or membrane of the target cells, the exosome contents will be directly transported into the target cells. According to these phenomena, drugs can be loaded into exosomes to target specific areas to treat the disease. At present, there are only a few attempts on the clinical application of MDSCs-Exos. The level of plasma S100A9 expressed in exosomes in patients with colorectal cancer is significantly increased compared with that in normal controls, and the serum level in patients with recurrent tumors is increased compared with that in patients with successful resection of colorectal cancer. Consequently, MDSC-Exo S100A9 can be used as a marker to predict the occurrence and development of colorectal cancer (76). In addition, respiratory hyperoxia inhibits the stemness of colorectal cancer cells by reducing the production of G-MDSCs-Exos, which may be used to assist in the treatment of colorectal cancer. Similarly, breast cancer patients who are resistant to DOX chemotherapy exhibit high levels of circulating miR-126a+MDSCs-Exos in their serum. Therefore, miR-126a+MDSC-Exo can be used as a potential biomarker of chemotherapy resistance to DOX in breast cancer and to guide the use of DOX in the treatment of breast cancer patients. Moreover, the systematic application of miR-126a inhibitor can improve the chemotherapeutic efficiency of DOX against lung metastasis by inhibiting tumor angiogenesis, which provides a basis for targeting MDSCs-Exos (17).
Non-steroidal anti-inflammatory drugs, glucocorticoids, and immunosuppressants have always been the main treatments of autoimmune diseases, and now MDSCs-Exos may be used as a new treatment strategy. Exosomes derived from G-MDSCs relieved collagen-induced arthritis by inhibiting the proliferation of Th1 and Th17 cells (16) and inducing IL10+Breg cells (136). Furthermore, high expression of miR-29a-3p and miR-93-5p induced by hypoxia in exosomes improved the condition (135). In addition, the application of G-MDSC-derived exosomes attenuated DSS-induced colitis by decreasing the percentages of Th1 cells and promoting the expansion of Tregs (133). MDSCs-Exos were also used in the treatment of autoimmune alopecia areata, which reversed the progression of the disease and promoted hair regeneration (134). Relay transfer of MDSCs protected pregnant mice from miscarriage, and exosomes exhibited similar effects, making MDSCs-Exos a possible target for the treatment of immune pregnancy complications. However, its safety as well as reliability still need to be further explored in depth (132).
There are still many blank areas in related research, and it is a demanding task to gain insight. However, future research on the characteristics, mechanism and clinical application of MDSCs-Exos will offer promising information.
Conclusion
In summary, MDSCs-Exos play multiple roles in the tumor immunity. MDSCs-Exos exhibit tumor immunosuppressive, angiogenic and metastatic effects similar to parental cells due to similar substances to parental cells. However, research on functions, mechanisms, and contribution rates of MDSCs-Exos in tumor immunity is limited. Thus, the scientific questions that require urgent exploration include: 1. What other specific substances in MDSCs-Exos are highly effective in affecting tumor immunity? 2. Whether these substances regulate each other and subsequently affect tumor immunity? 3. How to apply MDSCs-Exos to advanced cancer patients as soon as possible?4. How to ensure the safety of MDSCs-Exos in clinical applications?
To conclude, MDSCs-Exos play an essential role in the tumor immunity. Further investigation of MDSCs-Exos in tumor immunity will be beneficial for overcoming tumor progression, recurrence, metastasis and drug resistance, providing potential biomarkers and targets for immunotherapy of cancers. Therefore, people no longer turn pale at the mention of a “cancer” in the future.
Author Contributions
ZS and WY provided direction and guidance throughout the preparation of this manuscript. ZC and RY wrote and edited the manuscript. SH reviewed and made significant revisions to the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by The National Natural Science Foundation of China (82173055, 81972663, U2004112), Key Scientific Research Projects of Institutions of Higher Education in Henan Province (19A310024) and The National Natural Science Foundation of Henan Province (212300410074).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
Bv8, bombina variegata peptide 8; C4B-bp, C4B-binding proteins; CSCs, cancer stem cells; MDSCs, Myeloid-derived suppressor cells; MDSCs-Exos, exosomes released by MDSCs; G-MDSCs, granulocytic MDSCs; M-MDSCs, monocytic MDSCs; mRNAs, messenger RNAs; ncRNAs, noncoding RNAs; MIF, migration inhibition factor; PF-4, platelet factor-4; STAT, signal transducer and activator of transcription; TDEs, tumor-derived exosomes; PGE2, Prostaglandin E2; TGF-β1, transforming growth factor-β1; Th17, helper T cell 17; TME, tumor microenvironment; Tregs, regulatory T cells; NSCLC, in non-small cell lung cancer; G-CSF, granulocyte colony–stimulating factor; VEGF, vascular endothelial growth factor; Tsp1, thrombospondin1.
References
1. Fan CM, Wang JP, Tang YY, Zhao J, He SY, Xiong F, et al. Circman1a2 Could Serve as a Novel Serum Biomarker for Malignant Tumors. Cancer Sci (2019) 110(7):2180–8. doi: 10.1111/cas.14034
2. Gabrilovich DI, Nagaraj S. Myeloid-Derived Suppressor Cells as Regulators of the Immune System. Nat Rev Immunol (2009) 9(3):162–74. doi: 10.1038/nri2506
3. Zhao Y, Shao Q, Zhu H, Xu H, Long W, Yu B, et al. Resveratrol Ameliorates Lewis Lung Carcinoma-Bearing Mice Development, Decreases Granulocytic Myeloid-Derived Suppressor Cell Accumulation and Impairs Its Suppressive Ability. Cancer Sci (2018) 109(9):2677–86. doi: 10.1111/cas.13720
4. Safarzadeh E, Hashemzadeh S, Duijf PHG, Mansoori B, Khaze V, Mohammadi A, et al. Circulating Myeloid-Derived Suppressor Cells: An Independent Prognostic Factor in Patients With Breast Cancer. J Cell Physiol (2019) 234(4):3515–25. doi: 10.1002/jcp.26896
5. Tan Z, Liu L, Chiu MS, Cheung KW, Yan CW, Yu Z, et al. Virotherapy-Recruited PMN-MDSC Infiltration of Mesothelioma Blocks Antitumor CTL by IL-10-Mediated Dendritic Cell Suppression. Oncoimmunology (2019) 8(1):e1518672. doi: 10.1080/2162402X.2018.1518672
6. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current Knowledge of Their Composition, Biological Functions, and Diagnostic and Therapeutic Potentials. Biochim Biophys Acta (2012) 1820(7):940–8. doi: 10.1016/j.bbagen.2012.03.017
7. Boukouris S, Mathivanan S. Exosomes in Bodily Fluids Are a Highly Stable Resource of Disease Biomarkers. Proteomics Clin Appl (2015) 9(3-4):358–67. doi: 10.1002/prca.201400114
8. Morini M, Cangelosi D, Segalerba D, Marimpietri D, Raggi F, Castellano A, et al. Exosomal microRNAs From Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study. Cancers (Basel) (2019) 11(10):1476. doi: 10.3390/cancers11101476
9. Wu K, Feng J, Lyu F, Xing F, Sharma S, Liu Y, et al. Exosomal miR-19a and IBSP Cooperate to Induce Osteolytic Bone Metastasis of Estrogen Receptor-Positive Breast Cancer. Nat Commun (2021) 12(1):5196. doi: 10.1038/s41467-021-25473-y
10. Xie H, Wang J. MicroRNA-320a-Containing Exosomes From Human Umbilical Cord Mesenchymal Stem Cells Curtail Proliferation and Metastasis in Lung Cancer by Binding to SOX4. J Recept Signal Transduct Res (2021), 1–11. doi: 10.1080/10799893.2021.1918166
11. Kourembanas S. Exosomes: Vehicles of Intercellular Signaling, Biomarkers, and Vectors of Cell Therapy. Annu Rev Physiol (2015) 77:13–27. doi: 10.1146/annurev-physiol-021014-071641
12. Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: Novel Biomarkers for Clinical Diagnosis. Sci World J (2015) 2015:657086. doi: 10.1155/2015/657086
13. Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, et al. Circulating Exosomal miR-125a-3p as a Novel Biomarker for Early-Stage Colon Cancer. Sci Rep (2017) 7(1):4150. doi: 10.1038/s41598-017-04386-1
14. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, et al. Exosomal Long Noncoding RNA HOTTIP as Potential Novel Diagnostic and Prognostic Biomarker Test for Gastric Cancer. Mol Cancer (2018) 17(1):68. doi: 10.1186/s12943-018-0817-x
15. Geis-Asteggiante L, Belew AT, Clements VK, Edwards NJ, Ostrand-Rosenberg S, El-Sayed NM, et al. Differential Content of Proteins, mRNAs, and miRNAs Suggests That MDSC and Their Exosomes May Mediate Distinct Immune Suppressive Functions. J Proteome Res (2018) 17(1):486–98. doi: 10.1021/acs.jproteome.7b00646
16. Zhu D, Tian J, Wu X, Li M, Tang X, Rui K, et al. G-MDSC-Derived Exosomes Attenuate Collagen-Induced Arthritis by Impairing Th1 and Th17 Cell Responses. Biochim Biophys Acta Mol Basis Dis (2019) 1865(12):165540. doi: 10.1016/j.bbadis.2019.165540
17. Deng Z, Rong Y, Teng Y, Zhuang X, Samykutty A, Mu J, et al. Exosomes miR-126a Released From MDSC Induced by DOX Treatment Promotes Lung Metastasis. Oncogene (2017) 36(5):639–51. doi: 10.1038/onc.2016.229
18. Solito S, Falisi E, Diaz-Montero CM, Doni A, Pinton L, Rosato A, et al. A Human Promyelocytic-Like Population Is Responsible for the Immune Suppression Mediated by Myeloid-Derived Suppressor Cells. Blood (2011) 118(8):2254–65. doi: 10.1182/blood-2010-12-325753
19. Trac NT, Chung EJ. Peptide-Based Targeting of Immunosuppressive Cells in Cancer. Bioact Mater (2020) 5(1):92–101. doi: 10.1016/j.bioactmat.2020.01.006
20. Condamine T, Gabrilovich DI. Molecular Mechanisms Regulating Myeloid-Derived Suppressor Cell Differentiation and Function. Trends Immunol (2011) 32(1):19–25. doi: 10.1016/j.it.2010.10.002
21. Zhao Y, Wu T, Shao S, Shi B, Zhao Y. Phenotype, Development, and Biological Function of Myeloid-Derived Suppressor Cells. Oncoimmunology (2016) 5(2):e1004983. doi: 10.1080/2162402X.2015.1004983
22. Veglia F, Perego M, Gabrilovich D. Myeloid-Derived Suppressor Cells Coming of Age. Nat Immunol (2018) 19(2):108–19. doi: 10.1038/s41590-017-0022-x
23. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for Myeloid-Derived Suppressor Cell Nomenclature and Characterization Standards. Nat Commun (2016) 7:12150. doi: 10.1038/ncomms12150
24. Jin S, Yang Z, Hao X, Tang W, Ma W, Zong H. Roles of HMGB1 in Regulating Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. biomark Res (2020) 8:21. doi: 10.1186/s40364-020-00201-8
25. Zhang Y, Xu J, Zhang N, Chen M, Wang H, Zhu D. Targeting the Tumour Immune Microenvironment for Cancer Therapy in Human Gastrointestinal Malignancies. Cancer Lett (2019) 458:123–35. doi: 10.1016/j.canlet.2019.05.017
26. Yang Z, Guo J, Weng L, Tang W, Jin S, Ma W. Myeloid-Derived Suppressor Cells-New and Exciting Players in Lung Cancer. J Hematol Oncol (2020) 13(1):10. doi: 10.1186/s13045-020-0843-1
27. Kostlin N, Kugel H, Spring B, Leiber A, Marme A, Henes M, et al. Granulocytic Myeloid Derived Suppressor Cells Expand in Human Pregnancy and Modulate T-Cell Responses. Eur J Immunol (2014) 44(9):2582–91. doi: 10.1002/eji.201344200
28. Damuzzo V, Pinton L, Desantis G, Solito S, Marigo I, Bronte V, et al. Complexity and Challenges in Defining Myeloid-Derived Suppressor Cells. Cytometry B Clin Cytom (2015) 88(2):77–91. doi: 10.1002/cyto.b.21206
29. Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. J Immunol (2008) 181(8):5791–802. doi: 10.4049/jimmunol.181.8.5791
30. Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, et al. Identification of Discrete Tumor-Induced Myeloid-Derived Suppressor Cell Subpopulations With Distinct T Cell-Suppressive Activity. Blood (2008) 111(8):4233–44. doi: 10.1182/blood-2007-07-099226
31. Safarzadeh E, Orangi M, Mohammadi H, Babaie F, Baradaran B. Myeloid-Derived Suppressor Cells: Important Contributors to Tumor Progression and Metastasis. J Cell Physiol (2018) 233(4):3024–36. doi: 10.1002/jcp.26075
32. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol (2016) 37(3):208–20. doi: 10.1016/j.it.2016.01.004
33. Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, et al. Long Non-Coding RNA HOXA Transcript Antisense RNA Myeloid-Specific 1-HOXA1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front Immunol (2018) 9:473. doi: 10.3389/fimmu.2018.00473
34. Shu CC, Pan SW, Feng JY, Wang JY, Chan YJ, Yu CJ, et al. The Clinical Significance of Programmed Death-1, Regulatory T Cells and Myeloid Derived Suppressor Cells in Patients With Nontuberculous Mycobacteria-Lung Disease. J Clin Med (2019) 8(5):736. doi: 10.3390/jcm8050736
35. Ji J, Xu J, Zhao S, Liu F, Qi J, Song Y, et al. Myeloid-Derived Suppressor Cells Contribute to Systemic Lupus Erythaematosus by Regulating Differentiation of Th17 Cells and Tregs. Clin Sci (Lond) (2016) 130(16):1453–67. doi: 10.1042/CS20160311
36. Park MJ, Lee SH, Kim EK, Lee EJ, Baek JA, Park SH, et al. Interleukin-10 Produced by Myeloid-Derived Suppressor Cells Is Critical for the Induction of Tregs and Attenuation of Rheumatoid Inflammation in Mice. Sci Rep (2018) 8(1):3753. doi: 10.1038/s41598-018-21856-2
37. Han X, Shi H, Sun Y, Shang C, Luan T, Wang D, et al. CXCR2 Expression on Granulocyte and Macrophage Progenitors Under Tumor Conditions Contributes to Mo-MDSC Generation via SAP18/ERK/Stat3. Cell Death Dis (2019) 10(8):598. doi: 10.1038/s41419-019-1837-1
38. Kumar V, Cheng P, Condamine T, Mony S, Languino LR, McCaffrey JC, et al. CD45 Phosphatase Inhibits STAT3 Transcription Factor Activity in Myeloid Cells and Promotes Tumor-Associated Macrophage Differentiation. Immunity (2016) 44(2):303–15. doi: 10.1016/j.immuni.2016.01.014
39. Bruno A, Mortara L, Baci D, Noonan DM, Albini A. Myeloid Derived Suppressor Cells Interactions With Natural Killer Cells and Pro-Angiogenic Activities: Roles in Tumor Progression. Front Immunol (2019) 10:771. doi: 10.3389/fimmu.2019.00771
40. Wang Y, Schafer CC, Hough KP, Tousif S, Duncan SR, Kearney JF, et al. Myeloid-Derived Suppressor Cells Impair B Cell Responses in Lung Cancer Through IL-7 and STAT5. J Immunol (2018) 201(1):278–95. doi: 10.4049/jimmunol.1701069
41. Qu X, Zhuang G, Yu L, Meng G, Ferrara N. Induction of Bv8 Expression by Granulocyte Colony-Stimulating Factor in CD11b+Gr1+ Cells: Key Role of Stat3 Signaling. J Biol Chem (2012) 287(23):19574–84. doi: 10.1074/jbc.M111.326801
42. Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell (2012) 21(3):309–22. doi: 10.1016/j.ccr.2012.02.022
43. Wolf P. The Nature and Significance of Platelet Products in Human Plasma. Br J Haematol (1967) 13(3):269–88. doi: 10.1111/j.1365-2141.1967.tb08741.x
44. S ELA, Mager I, Breakefield XO, Wood MJ. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat Rev Drug Discov (2013) 12(5):347–57. doi: 10.1038/nrd3978
45. Weng Z, Zhang B, Wu C, Yu F, Han B, Li B, et al. Therapeutic Roles of Mesenchymal Stem Cell-Derived Extracellular Vesicles in Cancer. J Hematol Oncol (2021) 14(1):136. doi: 10.1186/s13045-021-01141-y
46. Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J Neurooncol (2013) 113(1):1–11. doi: 10.1007/s11060-013-1084-8
47. Colombo M, Raposo G, Thery C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol (2014) 30:255–89. doi: 10.1146/annurev-cellbio-101512-122326
48. van Niel G, D’Angelo G, Raposo G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat Rev Mol Cell Biol (2018) 19(4):213–28. doi: 10.1038/nrm.2017.125
49. Mathieu M, Nevo N, Jouve M, Valenzuela JI, Maurin M, Verweij FJ, et al. Specificities of Exosome Versus Small Ectosome Secretion Revealed by Live Intracellular Tracking of CD63 and CD9. Nat Commun (2021) 12(1):4389. doi: 10.1038/s41467-021-24384-2
50. Choi DS, Yang JS, Choi EJ, Jang SC, Park S, Kim OY, et al. The Protein Interaction Network of Extracellular Vesicles Derived From Human Colorectal Cancer Cells. J Proteome Res (2012) 11(2):1144–51. doi: 10.1021/pr200842h
51. Rana S, Yue S, Stadel D, Zoller M. Toward Tailored Exosomes: The Exosomal Tetraspanin Web Contributes to Target Cell Selection. Int J Biochem Cell Biol (2012) 44(9):1574–84. doi: 10.1016/j.biocel.2012.06.018
52. Skotland T, Sandvig K, Llorente A. Lipids in Exosomes: Current Knowledge and the Way Forward. Prog Lipid Res (2017) 66:30–41. doi: 10.1016/j.plipres.2017.03.001
53. Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The Fusion of Two Worlds: Non-Coding RNAs and Extracellular Vesicles–Diagnostic and Therapeutic Implications (Review). Int J Oncol (2015) 46(1):17–27. doi: 10.3892/ijo.2014.2712
54. Tian T, Zhu YL, Hu FH, Wang YY, Huang NP, Xiao ZD. Dynamics of Exosome Internalization and Trafficking. J Cell Physiol (2013) 228(7):1487–95. doi: 10.1002/jcp.24304
55. Lo Cicero A, Stahl PD, Raposo G. Extracellular Vesicles Shuffling Intercellular Messages: For Good or for Bad. Curr Opin Cell Biol (2015) 35:69–77. doi: 10.1016/j.ceb.2015.04.013
56. Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, et al. Neoplastic Reprogramming of Patient-Derived Adipose Stem Cells by Prostate Cancer Cell-Associated Exosomes. Stem Cells (2014) 32(4):983–97. doi: 10.1002/stem.1619
57. Subramanian A, Gupta V, Sarkar S, Maity G, Banerjee S, Ghosh A, et al. Exosomes in Carcinogenesis: Molecular Palkis Carry Signals for the Regulation of Cancer Progression and Metastasis. J Cell Commun Signal (2016) 10(3):241–9. doi: 10.1007/s12079-016-0338-6
58. Morrissey SM, Zhang F, Ding C, Montoya-Durango DE, Hu X, Yang C, et al. Tumor-Derived Exosomes Drive Immunosuppressive Macrophages in a Pre-Metastatic Niche Through Glycolytic Dominant Metabolic Reprogramming. Cell Metab (2021) 33(10):2040–58.e10. doi: 10.1016/j.cmet.2021.09.002
59. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell (2016) 29(5):653–68. doi: 10.1016/j.ccell.2016.03.004
60. Jia S, Zocco D, Samuels ML, Chou MF, Chammas R, Skog J, et al. Emerging Technologies in Extracellular Vesicle-Based Molecular Diagnostics. Expert Rev Mol Diagn (2014) 14(3):307–21. doi: 10.1586/14737159.2014.893828
61. Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A Comprehensive Overview of Exosomes as Drug Delivery Vehicles - Endogenous Nanocarriers for Targeted Cancer Therapy. Biochim Biophys Acta (2014) 1846(1):75–87. doi: 10.1016/j.bbcan.2014.04.005
62. Kotmakci M, Bozok Cetintas V. Extracellular Vesicles as Natural Nanosized Delivery Systems for Small-Molecule Drugs and Genetic Material: Steps Towards the Future Nanomedicines. J Pharm Pharm Sci (2015) 18(3):396–413. doi: 10.18433/j36w3x
63. Jin S, Zeng X, Xia F, Huang W, Liu X. Application of Deep Learning Methods in Biological Networks. Brief Bioinform (2021) 22(2):1902–17. doi: 10.1093/bib/bbaa043
64. Liu Y, Cheng A, Wang M, Mao S, Ou X, Yang Q, et al. Duck Hepatitis A Virus Type 1 Induces Eif2alpha Phosphorylation-Dependent Cellular Translation Shutoff via PERK/Gcn2. Front Microbiol (2021) 12:624540. doi: 10.3389/fmicb.2021.624540
65. Burke M, Choksawangkarn W, Edwards N, Ostrand-Rosenberg S, Fenselau C. Exosomes From Myeloid-Derived Suppressor Cells Carry Biologically Active Proteins. J Proteome Res (2014) 13(2):836–43. doi: 10.1021/pr400879c
66. Choksawangkarn W, Graham LM, Burke M, Lee SB, Ostrand-Rosenberg S, Fenselau C, et al. Peptide-Based Systems Analysis of Inflammation Induced Myeloid-Derived Suppressor Cells Reveals Diverse Signaling Pathways. Proteomics (2016) 16(13):1881–8. doi: 10.1002/pmic.201500102
67. Lee HD, Kim YH, Kim DS. Exosomes Derived From Human Macrophages Suppress Endothelial Cell Migration by Controlling Integrin Trafficking. Eur J Immunol (2014) 44(4):1156–69. doi: 10.1002/eji.201343660
68. Batista BS, Eng WS, Pilobello KT, Hendricks-Munoz KD, Mahal LK. Identification of a Conserved Glycan Signature for Microvesicles. J Proteome Res (2011) 10(10):4624–33. doi: 10.1021/pr200434y
69. Shimoda A, Tahara Y, Sawada SI, Sasaki Y, Akiyoshi K. Glycan Profiling Analysis Using Evanescent-Field Fluorescence-Assisted Lectin Array: Importance of Sugar Recognition for Cellular Uptake of Exosomes From Mesenchymal Stem Cells. Biochem Biophys Res Commun (2017) 491(3):701–7. doi: 10.1016/j.bbrc.2017.07.126
70. Hegde VL, Tomar S, Jackson A, Rao R, Yang X, Singh UP, et al. Distinct microRNA Expression Profile and Targeted Biological Pathways in Functional Myeloid-Derived Suppressor Cells Induced by Delta9-Tetrahydrocannabinol In Vivo: Regulation of CCAAT/enhancer-Binding Protein Alpha by microRNA-690. J Biol Chem (2013) 288(52):36810–26. doi: 10.1074/jbc.M113.503037
71. Chen S, Zhang Y, Kuzel TM, Zhang B. Regulating Tumor Myeloid-Derived Suppressor Cells by MicroRNAs. Cancer Cell Microenviron (2015) 2(1):e637. doi: 10.14800/ccm.637
72. Geis-Asteggiante L, Dhabaria A, Edwards N, Ostrand-Rosenberg S, Fenselau C. Top-Down Analysis of Low Mass Proteins in Exosomes Shed by Murine Myeloid-Derived Suppressor Cells. Int J Mass Spectrom (2015) 378:264–9. doi: 10.1016/j.ijms.2014.08.035
73. Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv Cancer Res (2015) 128:95–139. doi: 10.1016/bs.acr.2015.04.002
74. Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-Expanded Myeloid-Derived Suppressor Cells Induce Anergy of NK Cells Through Membrane-Bound TGF-Beta 1. J Immunol (2009) 182(1):240–9. doi: 10.4049/jimmunol.182.1.240
75. Ramadass M, Johnson JL, Catz SD. Rab27a Regulates GM-CSF-Dependent Priming of Neutrophil Exocytosis. J Leukoc Biol (2017) 101(3):693–702. doi: 10.1189/jlb.3AB0416-189RR
76. Wang Y, Yin K, Tian J, Xia X, Ma J, Tang X, et al. Granulocytic Myeloid-Derived Suppressor Cells Promote the Stemness of Colorectal Cancer Cells Through Exosomal S100a9. Adv Sci (Weinh) (2019) 6(18):1901278. doi: 10.1002/advs.201901278
77. Zhou L, Miao K, Yin B, Li H, Fan J, Zhu Y, et al. Cardioprotective Role of Myeloid-Derived Suppressor Cells in Heart Failure. Circulation (2018) 138(2):181–97. doi: 10.1161/CIRCULATIONAHA.117.030811
78. He YM, Li X, Perego M, Nefedova Y, Kossenkov AV, Jensen EA, et al. Transitory Presence of Myeloid-Derived Suppressor Cells in Neonates Is Critical for Control of Inflammation. Nat Med (2018) 24(2):224–31. doi: 10.1038/nm.4467
79. Salminen A, Kaarniranta K, Kauppinen A. Exosomal Vesicles Enhance Immunosuppression in Chronic Inflammation: Impact in Cellular Senescence and the Aging Process. Cell Signal (2020) 75:109771. doi: 10.1016/j.cellsig.2020.109771
80. Sun T, Liu Z, Yang Q. The Role of Ubiquitination and Deubiquitination in Cancer Metabolism. Mol Cancer (2020) 19(1):146. doi: 10.1186/s12943-020-01262-x
81. Zhang Y, Chen C, Yu T, Chen T. Proteomic Analysis of Protein Ubiquitination Events in Human Primary and Metastatic Colon Adenocarcinoma Tissues. Front Oncol (2020) 10:1684. doi: 10.3389/fonc.2020.01684
82. Popovic D, Vucic D, Dikic I. Ubiquitination in Disease Pathogenesis and Treatment. Nat Med (2014) 20(11):1242–53. doi: 10.1038/nm.3739
83. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-Methyladenosine-Modified Circigf2bp3 Inhibits CD8(+) T-Cell Responses to Facilitate Tumor Immune Evasion by Promoting the Deubiquitination of PD-L1 in Non-Small Cell Lung Cancer. Mol Cancer (2021) 20(1):105. doi: 10.1186/s12943-021-01398-4
84. Uchimura T, Oyama Y, Deng M, Guo H, Wilson JE, Rampanelli E, et al. The Innate Immune Sensor NLRC3 Acts as a Rheostat That Fine-Tunes T Cell Responses in Infection and Autoimmunity. Immunity (2018) 49(6):1049–1061 e6. doi: 10.1016/j.immuni.2018.10.008
85. Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes Contain Ubiquitinated Proteins. Blood Cells Mol Dis (2005) 35(3):398–403. doi: 10.1016/j.bcmd.2005.08.005
86. Burke MC, Oei MS, Edwards NJ, Ostrand-Rosenberg S, Fenselau C. Ubiquitinated Proteins in Exosomes Secreted by Myeloid-Derived Suppressor Cells. J Proteome Res (2014) 13(12):5965–72. doi: 10.1021/pr500854x
87. Vidal M, Mangeat P, Hoekstra D. Aggregation Reroutes Molecules From a Recycling to a Vesicle-Mediated Secretion Pathway During Reticulocyte Maturation. J Cell Sci (1997) 110(Pt 16):1867–77. doi: 10.1242/jcs.110.16.1867
88. Janig E, Stumptner C, Fuchsbichler A, Denk H, Zatloukal K. Interaction of Stress Proteins With Misfolded Keratins. Eur J Cell Biol (2005) 84(2-3):329–39. doi: 10.1016/j.ejcb.2004.12.018
89. Parker KH, Sinha P, Horn LA, Clements VK, Yang H, Li J, et al. HMGB1 Enhances Immune Suppression by Facilitating the Differentiation and Suppressive Activity of Myeloid-Derived Suppressor Cells. Cancer Res (2014) 74(20):5723–33. doi: 10.1158/0008-5472.CAN-13-2347
90. Gilthorpe JD, Oozeer F, Nash J, Calvo M, Bennett DL, Lumsden A, et al. Extracellular Histone H1 Is Neurotoxic and Drives a Pro-Inflammatory Response in Microglia. F1000Res (2013) 2:148. doi: 10.12688/f1000research.2-148.v1
91. Wen Z, Liu Y, Li F, Ren F, Chen D, Li X, et al. Circulating Histones Exacerbate Inflammation in Mice With Acute Liver Failure. J Cell Biochem (2013) 114(10):2384–91. doi: 10.1002/jcb.24588
92. Allam R, Kumar SV, Darisipudi MN, Anders HJ. Extracellular Histones in Tissue Injury and Inflammation. J Mol Med (Berl) (2014) 92(5):465–72. doi: 10.1007/s00109-014-1148-z
93. Gandini R, Reichenbach T, Tan TC, Divne C. Structural Basis for Dolichylphosphate Mannose Biosynthesis. Nat Commun (2017) 8(1):120. doi: 10.1038/s41467-017-00187-2
94. Xiao H, Suttapitugsakul S, Sun F, Wu R. Mass Spectrometry-Based Chemical and Enzymatic Methods for Global Analysis of Protein Glycosylation. Acc Chem Res (2018) 51(8):1796–806. doi: 10.1021/acs.accounts.8b00200
95. Chauhan S, Danielson S, Clements V, Edwards N, Ostrand-Rosenberg S, Fenselau C. Surface Glycoproteins of Exosomes Shed by Myeloid-Derived Suppressor Cells Contribute to Function. J Proteome Res (2017) 16(1):238–46. doi: 10.1021/acs.jproteome.6b00811
96. Zhang M, Wang H, Tan S, Navarro-Alvarez N, Zheng Y, Yang YG. Donor CD47 Controls T Cell Alloresponses and Is Required for Tolerance Induction Following Hepatocyte Allotransplantation. Sci Rep (2016) 6:26839. doi: 10.1038/srep26839
97. Pengam S, Durand J, Usal C, Gauttier V, Dilek N, Martinet B, et al. SIRPalpha/CD47 Axis Controls the Maintenance of Transplant Tolerance Sustained by Myeloid-Derived Suppressor Cells. Am J Transplant (2019) 19(12):3263–75. doi: 10.1111/ajt.15497
98. Lv Z, Bian Z, Shi L, Niu S, Ha B, Tremblay A, et al. Loss of Cell Surface CD47 Clustering Formation and Binding Avidity to SIRPalpha Facilitate Apoptotic Cell Clearance by Macrophages. J Immunol (2015) 195(2):661–71. doi: 10.4049/jimmunol.1401719
99. Tseng D, Volkmer JP, Willingham SB, Contreras-Trujillo H, Fathman JW, Fernhoff NB, et al. Anti-CD47 Antibody-Mediated Phagocytosis of Cancer by Macrophages Primes an Effective Antitumor T-Cell Response. Proc Natl Acad Sci USA (2013) 110(27):11103–8. doi: 10.1073/pnas.1305569110
100. Ekstrom K, Valadi H, Sjostrand M, Malmhall C, Bossios A, Eldh M, et al. Characterization of mRNA and microRNA in Human Mast Cell-Derived Exosomes and Their Transfer to Other Mast Cells and Blood CD34 Progenitor Cells. J Extracell Vesicles (2012) 1. doi: 10.3402/jev.v1i0.18389
101. Ji H, Chen M, Greening DW, He W, Rai A, Zhang W, et al. Deep Sequencing of RNA From Three Different Extracellular Vesicle (EV) Subtypes Released From the Human LIM1863 Colon Cancer Cell Line Uncovers Distinct miRNA-Enrichment Signatures. PloS One (2014) 9(10):e110314. doi: 10.1371/journal.pone.0110314
102. Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, et al. Identifying mRNA, microRNA and Protein Profiles of Melanoma Exosomes. PloS One (2012) 7(10):e46874. doi: 10.1371/journal.pone.0046874
103. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-Dependent Induction of microRNA miR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses. Proc Natl Acad Sci USA (2006) 103(33):12481–6. doi: 10.1073/pnas.0605298103
104. Feng J, Byrne NM, Al Jamal W, Coulter JA. Exploiting Current Understanding of Hypoxia Mediated Tumour Progression for Nanotherapeutic Development. Cancers (Basel) (2019) 11(12):1989. doi: 10.3390/cancers11121989
105. Hildeman DA, Mitchell T, Kappler J, Marrack P. T Cell Apoptosis and Reactive Oxygen Species. J Clin Invest (2003) 111(5):575–81. doi: 10.1172/JCI18007
106. Rashid MH, Borin TF, Ara R, Piranlioglu R, Achyut BR, Korkaya H, et al. Critical Immunosuppressive Effect of MDSCderived Exosomes in the Tumor Microenvironment. Oncol Rep (2021) 45(3):1171–81. doi: 10.3892/or.2021.7936
107. Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, et al. Membrane-Associated Hsp72 From Tumor-Derived Exosomes Mediates STAT3-Dependent Immunosuppressive Function of Mouse and Human Myeloid-Derived Suppressor Cells. J Clin Invest (2010) 120(2):457–71. doi: 10.1172/jci40483
108. Guo X, Qiu W, Liu Q, Qian M, Wang S, Zhang Z, et al. Immunosuppressive Effects of Hypoxia-Induced Glioma Exosomes Through Myeloid-Derived Suppressor Cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene (2018) 37(31):4239–59. doi: 10.1038/s41388-018-0261-9
109. Eisenblaetter M, Flores-Borja F, Lee JJ, Wefers C, Smith H, Hueting R, et al. Visualization of Tumor-Immune Interaction - Target-Specific Imaging of S100A8/A9 Reveals Pre-Metastatic Niche Establishment. Theranostics (2017) 7(9):2392–401. doi: 10.7150/thno.17138
110. Zhou JH, Yao ZX, Zheng Z, Yang J, Wang R, Fu SJ, et al. G-MDSCs-Derived Exosomal miRNA-143-3p Promotes Proliferation via Targeting of ITM2B in Lung Cancer. Onco Targets Ther (2020) 13:9701–19. doi: 10.2147/ott.s256378
111. Li X, Li Y, Lu W, Chen M, Ye W, Zhang D. The Tumor Vessel Targeting Strategy: A Double-Edged Sword in Tumor Metastasis. Cells (2019) 8(12):1602. doi: 10.3390/cells8121602
112. Qiang W, Zhou T, Lan X, Zhang X, Guo Y, Noman M, et al. A New Nanoscale Transdermal Drug Delivery System: Oil Body-Linked oleosin-hEGF Improves Skin Regeneration to Accelerate Wound Healing. J Nanobiotechnol (2018) 16(1):62. doi: 10.1186/s12951-018-0387-5
113. Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of Tumor Metastasis by Myeloid-Derived Suppressor Cells. Annu Rev Med (2015) 66:97–110. doi: 10.1146/annurev-med-051013-052304
114. Motallebnezhad M, Jadidi-Niaragh F, Qamsari ES, Bagheri S, Gharibi T, Yousefi M. The Immunobiology of Myeloid-Derived Suppressor Cells in Cancer. Tumour Biol (2016) 37(2):1387–406. doi: 10.1007/s13277-015-4477-9
115. Boelte KC, Gordy LE, Joyce S, Thompson MA, Yang L, Lin PC. Rgs2 Mediates Pro-Angiogenic Function of Myeloid Derived Suppressor Cells in the Tumor Microenvironment via Upregulation of MCP-1. PloS One (2011) 6(4):e18534. doi: 10.1371/journal.pone.0018534
116. Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of Myeloid Immune Suppressor Gr+CD11b+ Cells in Tumor-Bearing Host Directly Promotes Tumor Angiogenesis. Cancer Cell (2004) 6(4):409–21. doi: 10.1016/j.ccr.2004.08.031
117. Zhang Z, Zhang C, Miao J, Wang Z, Wang Z, Cheng Z, et al. A Tumor-Targeted Replicating Oncolytic Adenovirus Ad-TD-Nsil12 as a Promising Therapeutic Agent for Human Esophageal Squamous Cell Carcinoma. Cells (2020) 9(11):2438. doi: 10.3390/cells9112438
118. Cui TX, Kryczek I, Zhao L, Zhao E, Kuick R, Roh MH, et al. Myeloid-Derived Suppressor Cells Enhance Stemness of Cancer Cells by Inducing Microrna101 and Suppressing the Corepressor Ctbp2. Immunity (2013) 39(3):611–21. doi: 10.1016/j.immuni.2013.08.025
119. Peng D, Tanikawa T, Li W, Zhao L, Vatan L, Szeliga W, et al. Myeloid-Derived Suppressor Cells Endow Stem-Like Qualities to Breast Cancer Cells Through IL6/STAT3 and NO/NOTCH Cross-Talk Signaling. Cancer Res (2016) 76(11):3156–65. doi: 10.1158/0008-5472.CAN-15-2528
120. Kuroda H, Mabuchi S, Yokoi E, Komura N, Kozasa K, Matsumoto Y, et al. Prostaglandin E2 Produced by Myeloid-Derived Suppressive Cells Induces Cancer Stem Cells in Uterine Cervical Cancer. Oncotarget (2018) 9(91):36317–30. doi: 10.18632/oncotarget.26347
121. Di Mitri D, Toso A, Chen JJ, Sarti M, Pinton S, Jost TR, et al. Tumour-Infiltrating Gr-1+ Myeloid Cells Antagonize Senescence in Cancer. Nature (2014) 515(7525):134–7. doi: 10.1038/nature13638
122. Panni RZ, Sanford DE, Belt BA, Mitchem JB, Worley LA, Goetz BD, et al. Tumor-Induced STAT3 Activation in Monocytic Myeloid-Derived Suppressor Cells Enhances Stemness and Mesenchymal Properties in Human Pancreatic Cancer. Cancer Immunol Immunother (2014) 63(5):513–28. doi: 10.1007/s00262-014-1527-x
123. Wang Y, Jiang M, Li Z, Wang J, Du C, Yanyang L, et al. Hypoxia and TGF-Beta1 Lead to Endostatin Resistance by Cooperatively Increasing Cancer Stem Cells in A549 Transplantation Tumors. Cell Biosci (2015) 5:72. doi: 10.1186/s13578-015-0064-4
124. Shen Q, Xu Z, Xu S. Long Noncoding RNA LUCAT1 Contributes to Cisplatin Resistance by Regulating the Mir514a3p/ULK1 Axis in Human Nonsmall Cell Lung Cancer. Int J Oncol (2020) 57(4):967–79. doi: 10.3892/ijo.2020.5106
125. Liu Z, Xie Y, Xiong Y, Liu S, Qiu C, Zhu Z, et al. TLR 7/8 Agonist Reverses Oxaliplatin Resistance in Colorectal Cancer via Directing the Myeloid-Derived Suppressor Cells to Tumoricidal M1-Macrophages. Cancer Lett (2020) 469:173–85. doi: 10.1016/j.canlet.2019.10.020
126. Kawano M, Mabuchi S, Matsumoto Y, Sasano T, Takahashi R, Kuroda H, et al. The Significance of G-CSF Expression and Myeloid-Derived Suppressor Cells in the Chemoresistance of Uterine Cervical Cancer. Sci Rep (2015) 5:18217. doi: 10.1038/srep18217
127. Xu M, Zhao Z, Song J, Lan X, Lu S, Chen M, et al. Interactions Between Interleukin-6 and Myeloid-Derived Suppressor Cells Drive the Chemoresistant Phenotype of Hepatocellular Cancer. Exp Cell Res (2017) 351(2):142–9. doi: 10.1016/j.yexcr.2017.01.008
128. Ramachandran IR, Condamine T, Lin C, Herlihy SE, Garfall A, Vogl DT, et al. Bone Marrow PMN-MDSCs and Neutrophils Are Functionally Similar in Protection of Multiple Myeloma From Chemotherapy. Cancer Lett (2016) 371(1):117–24. doi: 10.1016/j.canlet.2015.10.040
129. Hsu YL, Hung JY, Tsai EM, Wu CY, Ho YW, Jian SF, et al. Benzyl Butyl Phthalate Increases the Chemoresistance to Doxorubicin/Cyclophosphamide by Increasing Breast Cancer-Associated Dendritic Cell-Derived CXCL1/GROalpha and S100A8/A9. Oncol Rep (2015) 34(6):2889–900. doi: 10.3892/or.2015.4307
130. Anestakis D, Petanidis S, Domvri K, Tsavlis D, Zarogoulidis P, Katopodi T. Carboplatin Chemoresistance Is Associated With CD11b(+)/Ly6C(+) Myeloid Release and Upregulation of TIGIT and LAG3/CD160 Exhausted T Cells. Mol Immunol (2020) 118:99–109. doi: 10.1016/j.molimm.2019.11.008
131. Cripps JG, Gorham JD. MDSC in Autoimmunity. Int Immunopharmacol (2011) 11(7):789–93. doi: 10.1016/j.intimp.2011.01.026
132. Dietz S, Schwarz J, Ruhle J, Schaller M, Fehrenbacher B, Marme A, et al. Extracellular Vesicles Released by Myeloid-Derived Suppressor Cells From Pregnant Women Modulate Adaptive Immune Responses. Cell Immunol (2021) 361:104276. doi: 10.1016/j.cellimm.2020.104276
133. Wang S, Tang X. Exosomes Released by Granulocytic Myeloid-Derived Suppressor Cells Attenuate DSS-Induced Colitis in Mice. Oncotarget (2015) 7(13):15356–68. doi: 10.18632/oncotarget.7324
134. Zoller M, Zhao K, Kutlu N, Bauer N, Provaznik J, Hackert T, et al. Immunoregulatory Effects of Myeloid-Derived Suppressor Cell Exosomes in Mouse Model of Autoimmune Alopecia Areata. Front Immunol (2018) 9:1279. doi: 10.3389/fimmu.2018.01279
135. Chu X, Chen L, Zhang Y. Exosomes Derived From PMN-MDSCs Preconditioned by Hypoxia Attenuate Arthropathy of Collagen-Induced Arthritis Mice. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi (2021) 37(8):728–35.
136. Wu X, Zhu D, Tian J, Tang X, Guo H, Ma J, et al. Granulocytic Myeloid-Derived Suppressor Cell Exosomal Prostaglandin E2 Ameliorates Collagen-Induced Arthritis by Enhancing IL-10(+) B Cells. Front Immunol (2020) 11:588500:588500. doi: 10.3389/fimmu.2020.588500
137. Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM. Gemcitabine Selectively Eliminates Splenic Gr-1+/CD11b+ Myeloid Suppressor Cells in Tumor-Bearing Animals and Enhances Antitumor Immune Activity. Clin Cancer Res (2005) 11(18):6713–21. doi: 10.1158/1078-0432.ccr-05-0883
138. Melani C, Sangaletti S, Barazzetta FM, Werb Z, Colombo MP. Amino-Biphosphonate-Mediated MMP-9 Inhibition Breaks the Tumor-Bone Marrow Axis Responsible for Myeloid-Derived Suppressor Cell Expansion and Macrophage Infiltration in Tumor Stroma. Cancer Res (2007) 67(23):11438–46. doi: 10.1158/0008-5472.can-07-1882
139. Hengesbach LM, Hoag KA. Physiological Concentrations of Retinoic Acid Favor Myeloid Dendritic Cell Development Over Granulocyte Development in Cultures of Bone Marrow Cells From Mice. J Nutr (2004) 134(10):2653–9. doi: 10.1093/jn/134.10.2653
140. De Santo C, Serafini P, Marigo I, Dolcetti L, Bolla M, Del Soldato P, et al. Nitroaspirin Corrects Immune Dysfunction in Tumor-Bearing Hosts and Promotes Tumor Eradication by Cancer Vaccination. Proc Natl Acad Sci USA (2005) 102(11):4185–90. doi: 10.1073/pnas.0409783102
141. Talmadge JE, Hood KC, Zobel LC, Shafer LR, Coles M, Toth B. Chemoprevention by Cyclooxygenase-2 Inhibition Reduces Immature Myeloid Suppressor Cell Expansion. Int Immunopharmacol (2007) 7(2):140–51. doi: 10.1016/j.intimp.2006.09.021
142. Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, et al. Phosphodiesterase-5 Inhibition Augments Endogenous Antitumor Immunity by Reducing Myeloid-Derived Suppressor Cell Function. J Exp Med (2006) 203(12):2691–702. doi: 10.1084/jem.20061104
143. Li M, Xia B, Wang Y, You MJ, Zhang Y. Potential Therapeutic Roles of Exosomes in Multiple Myeloma: A Systematic Review. J Cancer (2019) 10(24):6154–60. doi: 10.7150/jca.31752
Keywords: myeloid-derived suppressor cells, exosome, tumor immunity, cancer, immune escape
Citation: Chen Z, Yuan R, Hu S, Yuan W and Sun Z (2022) Roles of the Exosomes Derived From Myeloid-Derived Suppressor Cells in Tumor Immunity and Cancer Progression. Front. Immunol. 13:817942. doi: 10.3389/fimmu.2022.817942
Received: 18 November 2021; Accepted: 12 January 2022;
Published: 27 January 2022.
Edited by:
Nicolas Larmonier, Université de Bordeaux, FranceReviewed by:
Serena Meraviglia, University of Palermo, ItalyTomer Cooks, Ben-Gurion University of the Negev, Israel
Copyright © 2022 Chen, Yuan, Hu, Yuan and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenqiang Sun, ZmNjc3VuenFAenp1LmVkdS5jbg==; Weitang Yuan, eXVhbndlaXRhbmdAenp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Zhuang Chen†
Zhuang Chen† Zhenqiang Sun
Zhenqiang Sun