- 1State Key Laboratory of Silkworm Genome Biology, Biological Science Research Center, Southwest University, Chongqing, China
- 2Chongqing Key Laboratory of Sericultural Science, Chongqing Engineering and Technology Research Center for Novel Silk Materials, Southwest University, Chongqing, China
Translationally controlled tumor protein (TCTP) is a highly conserved protein possessing numerous biological functions and molecular interactions, ranging from cell growth to immune responses. However, the molecular mechanism by which TCTP regulates immune function is largely unknown. Here, we found that knockdown of Bombyx mori translationally controlled tumor protein (BmTCTP) led to the increased susceptibility of silkworm cells to virus infection, whereas overexpression of BmTCTP significantly decreased the virus replication. We further demonstrated that BmTCTP could be modified by SUMOylation molecular BmSMT3 at the lysine 164 via the conjugating enzyme BmUBC9, and the stable SUMOylation of BmTCTP by expressing BmTCTP-BmSMT3 fusion protein exhibited strong antiviral activity, which confirmed that the SUMOylation of BmTCTP would contribute to its immune responses. Further work indicated that BmTCTP is able to physically interact with interleukin enhancer binding factor (ILF), one immune molecular, involved in antivirus, and also induce the expression of BmILF in response to virus infection, which in turn enhanced antiviral activity of BmTCTP. Altogether, our present study has provided a novel insight into defending against virus via BmTCTP SUMOylation signaling pathway and interacting with key immune molecular in silkworm.
Introduction
Translationally controlled tumor protein (TCTP), also named tumor protein translationally controlled 1 (TPT1), histamine release factor (HRF), fortilin, p23, or Mmi, was originally identified as a protein that undergoes an early and prominent increase upon serum stimulation in murine cell lines (1–4) and thus establishing TCTP as a translationally regulated protein (5). It was subsequently found that TCTP has been an evolutionary conserved protein present in eukaryotic organisms, including mammals, plants, and insects (6–9). TCTP has been characterized as a multifunction protein that participates in multiple biological processes, such as cell growth, cell cycle progression, cell apoptosis, protein synthesis, immune responses, tumor reversion, and tumorigenesis (10–14). Due to the important function in tumorigenesis and upregulated expression in tumors, TCTP has been considered as a promising therapeutic target for cancer prevention and intervention (15, 16).
Given the important roles of TCTP in various species, deciphering the molecular mechanisms underlying TCTP involvement in cellular processes will be critical for understanding its diverse biological functions. One of the important ways is to identify the specific interactions with TCTP, which could provide the potential mechanisms to regulate target processes. Recently, a majority of proteins have been demonstrated to be interacting with TCTP using coimmunoprecipitation (Co-IP) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (17–19). For example, TCTP could interact with cytoskeleton proteins, such as Tubulin and Actin, and participate in regulating microtubule organization and cell morphology (20, 21). TCTP also interacts with ribosomal proteins and translation elongation factors, indicating its role in the translation process (17, 19). By interacting with Bcl-xL and Mcl-1, TCTP increases their stability and promotes antiapoptotic effects (22, 23). TCTP is able to form a complex with p53 and MDM2 and further regulates the degradation of p53 (24–26). The interaction of TCTP with Na,K-ATPase inhibits the pumping activity of Na,K-ATPase and induces Na,K-ATPase-mediated tumorigenic signaling pathways (27, 28).
Posttranslational modification (PTM) of a protein can also determine the target protein activity, localization, and stabilization (29). Another way to understand TCTP function is to investigate its modification processes. It has been demonstrated that human TCTP can be phosphorylated by polo-like kinase, and this phosphorylation inhibits the microtubule-stabilizing activity of TCTP, which in turn regulates the progression of cell cycle (30, 31). Increased phosphorylation of TCTP has also been shown to be associated with a poor clinical response to trastuzumab therapy in human epithelial growth factor receptor-2 (HER2)-positive breast cancer (15). In addition to TCTP phosphorylation, a recent report shows that human TCTP is SUMOylated by small ubiquitin-like modifier (SUMO) at a lysine 164 site. The SUMOylation of TCTP regulates its nuclear transport and promotes antioxidant function of TCTP (32). These modifications on TCTP have shed light on the novel regulation of TCTP activity in organisms.
The demonstrated silkworm Bombyx mori is an economically important insect for silk production and an excellent lepidopteran model for studying gene functions (33–35). The role of Bombyx mori translationally controlled tumor protein (BmTCTP) in the silkworm is emerging in recent years. Clustered regularly interspaced short palindromic repeat/CRISPER-associated protein 9 (CRISPR/Cas9)-mediated depletion of BmTCTP led to influenced cell size, cell proliferation, and differentiation of intestinal epithelial cells and thus delayed development and lethality in the third instar larvae of the silkworm (36). Additionally, transgenic RNA interference (RNAi) knockdown of the midgut BmTCTP expression suppressed the antimicrobial capacity and the intestinal innate immunity of the silkworm during oral microbial challenge (37, 38). All these results have demonstrated the roles of BmTCTP in regulating larval development and antimicrobial activity. However, the molecular mechanism underlying BmTCTP involved in the immunity is largely unknown.
To further understand the possible mechanism of BmTCTP in the immune response, we here focused on the modification of BmTCTP and investigated its role in Bombyx mori nucleopolyhedrovirus (BmNPV) infection, one of the serious diseases in sericulture (39, 40). Our present data confirmed that the silkworm BmTCTP can be SUMOylated at a conserved lysine site, and the SUMO conjugating enzyme BmUBC9-mediated SUMOylation of BmTCTP contributes to its immune response. Furthermore, we showed that BmTCTP may act as antiviral immunity via regulating the expression of interleukin enhancer binding factor (ILF).
Materials and Methods
Plasmids
Full-length cDNAs of BmTCTP and BmILF were amplified from the cDNA library of cultured silkworm BmN cells by using primers listed in Table S1 and further cloned into an NcoI–XhoI site of pENTR11 (Invitrogen, Carlsbad, CA, USA) vector. Plasmids expressing BmTCTP and BmILF with different tags (FLAG, HA, EGFP, and Red) were generated using the gateway technology (Invitrogen, Carlsbad, CA, USA) (41). BmSMT3 and BmUBC9 plasmids were from our previous publication (42). Point mutant of BmTCTP at K164 site was generated by PCR-based mutagenesis (43). To tag BmTCTP with a BmSMT3 chain, the silkworm BmSMT3 gene was cloned into the pENTR11 vector, and the resulting plasmid was further ligated with the PCR product of BmTCTP (43). All plasmids were verified by sequencing.
Cell Lines
The silkworm ovary-derived BmN cells were cultured in TC-100 (Sigma, St. Louis, MO, USA) medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) and penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) at 27°C, respectively.
The expression vectors for FLAG-TCTP and FLAG-TCTP-K164R were inserted into the genome of BmN cells by using piggyBac transposition system according to the previous report, and the stably transformed cells were selected by puromycin (Beyotime, Shanghai, China).
Sequence Analysis
We performed multiple sequence alignments using CLC sequence viewer 7 based on the silkworm BmTCTP and Human HsTCTP. The potential modification of lysine residue was indicated by an asterisk.
RNA Interference
The synthesis of double-stranded RNAs (dsRNAs) for BmTCTP was carried out by T7 RNA polymerase (Promega, Madison, WI, USA) in vitro according to the previous report (44). The dsRNAs for control gene of Red were also synthesized. G5-PAMAM-mediated delivery of dsRNA was used according to the previous protocol (45). G5-PAMAM was purchased from Chenyuan Company (Shandong, China).
PCR
Total RNA samples were prepared from BmN cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and 1 μg of total RNA was used for cDNA synthesis according to the manufacturer’s protocol of the M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with an SYBR Premix ExTaq Kit (Takara, Kyoto, Japan) and a qTower 2.2 Real-time PCR Detection System. The silkworm eukaryotic translation initiation factor 4A (BmeIF-4a), BmActin3, and glyceraldehyde-3-phosphate dehydrogenase (BmGAPDH) genes were used as the internal control. All experiments were independently performed with three biological replicates, and all primers used for PCR were listed in Table S1.
Virus Infection
BmNPV-expressing green fluorescent protein (BmNPV-GFP) was stored in our laboratory of Biological Science Research Center (46). BmN cells infected with BmNPV-GFP at the different time points were collected for different assays.
Virus Proliferation
BmNPV-GFP-infected BmN cells were harvested at the indicated time points and suspended in phosphate-buffered saline (PBS). Total DNA from each sample was prepared by using a TaKaRa MiniBEST Universal Genomic DNA Extraction Kit (Takara, Kusatsu, Japan) according to the manufacturer’s protocol. The viral DNA abundance of BmNPV was examined by the expression of virus GP41 gene (46), and the silkworm GAPDH gene was used as the internal control (Table S1). The viral fluorescence was observed at 72 h post infection (hpi), and the aggregated nuclear DNA was counterstained by 4’,6-diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA, USA). Fluorescence signals were captured by a fluorescent microscope (Z16, Leica, Wetzlar, Germany).
Fluorescence Microscopy
For subcellular localization analysis, cells expressing Red-TCTP, Red-TCTP-K164R, Red-TCTP-SMT3, or EGFP-ILF were cultured on a coverslip, fixed with 3.7% formaldehyde in PBS for 10 min, and the nuclear DNA was counterstained by DAPI. Fluorescence signals were captured by a fluorescence microscope (Z16, Leica, Wetzlar, Germany).
Immunoprecipitation and Immunoblotting
Immunoprecipitation was carried out as described previously (41). Briefly, cells co-expressing proteins were lysed in radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] supplemented with protease inhibitor (Complete, EDTA-free, Roche, New Zealand). The lysates were immunoprecipitated by using an anti-FLAG M2 Affinity Gel (F2426, Sigma, St. Louis, MO, USA), and the eluted proteins were further detected by immunoblotting using anti-HA (AF518, Beyotime, Shanghai, China), anti-FLAG (AH159, Beyotime, Shanghai, China), anti-Smt3 (ab135758, Abcam, Cambridge, UK), and anti-Tubulin (ab7291, Abcam, Cambridge, UK) antibodies.
Liquid Chromatography-Tandem Mass Spectrometry Assay
After digestion of the immunoprecipitated BmTCTP complex, liquid chromatography-tandem mass spectrometry (LC-MS/MS) was carried out to identify the protein interactions according to the previous protocol (47). Briefly, the protein solution after immunoprecipitation was chemically reduced with 10 mM dithiothreitol (DTT) for 1 h at 37°C and then alkylated with 50 mM iodoacetamide for 1 h at room temperature in the dark. After washing with 8 M urea and 50 mM NH4HCO3 in an ultrafiltration tube, proteins were digested with trypsin for 20 h at 37°C. The peptide mixture was acidified by 0.1% formic acid and resolved by using a Thermo Fisher Scientific EASY-nLC 1000 system (Waltham, MA, USA) under the standard parameters.
Data Analysis
Protein identification was analyzed with MaxQuant software (version 1.3.0.1) against an integrated silkworm proteome database. The search parameters for protein identification were set according to the published procedure (48). At least one unique peptide was designated as an identified protein, and the protein information was listed in Table S2. For the functional annotation of the identified proteins, we used the Blast2GO program (https://www.blast2go.com/) (49) to search against the non-redundant protein database (NR, NCBI, https://www.ncbi.nlm.nih.gov/). The WEGO database (http://wego.genomics.org.cn/) was used to analyze the interacting proteins.
Statistical Analysis
Statistical data are presented as the mean ± standard deviation (SD) of three independent biological replicates. The significance (P-value) was analyzed by the Student’s t-test and denoted as follows: *P < 0.05, **P <0.01, and ***P < 0.001.
Results
BmTCTP Was SUMOylated at a Conserved Lysine Site in Silkworm
It has been shown that the human HsTCTP can be SUMOylated at a lysine 164 site (32). In order to identify whether the SUMOylation of TCTP is conserved in other species, we here investigated the potential SUMOylation target in silkworm. A comparison of the TCTP sequences from human and silkworm showed very high similarity in full-length and possessed a conserved lysine at 164 site (Figure 1A). To verify the SUMOylation, we constructed wild-type (WT) and mutant (K164R) overexpression plasmids of FLAG-TCTP-WT and FLAG-TCTP-K164R. These plasmids were respectively co-transfected with SUMOylation molecular HA-SMT3 into the silkworm BmN cells. The results of Co-IP analysis showed that TCTP-WT had an obvious band of SUMOylation compared with TCTP-K164R (Figure 1B). These findings suggested that the lysine residue at 164 was also the SUMOylation accept site of silkworm BmTCTP, which was consistent with the human HsTCTP (32).
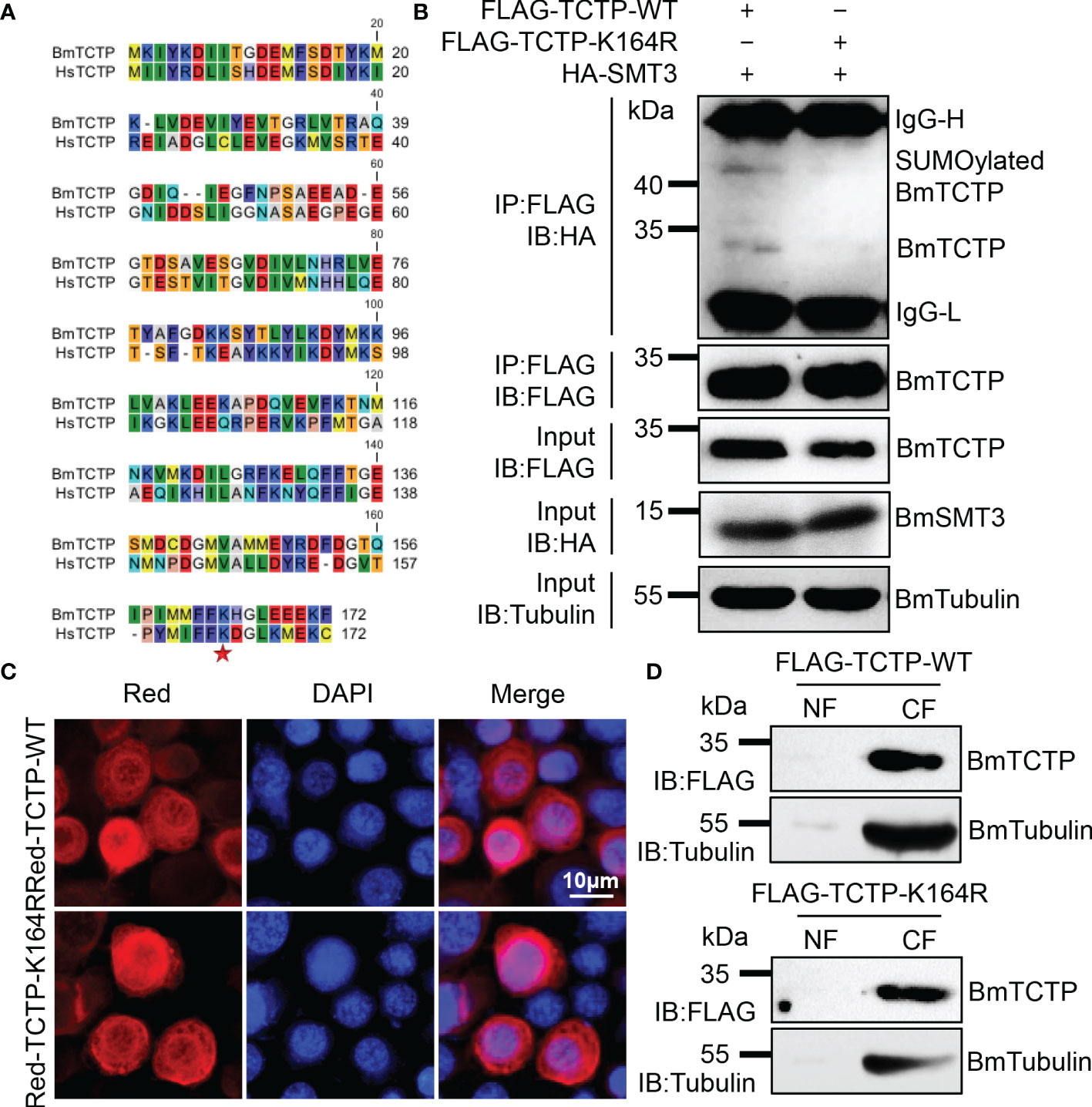
Figure 1 Bombyx mori translationally controlled tumor protein (BmTCTP) was SUMOylated at a conserved lysine site in silkworm. (A) The amino acid sequence alignment was performed using the CLC sequence viewer 7 software on human and silkworm sequences of TCTP. Asterisk indicated residue of the potential SUMOylation site. (B) Plasmids as indicated were co-transfected into the silkworm BmN cells. Lysates from BmN cells were used for immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with indicated antibodies. IgG-L and IgG-H were the light and heavy chains of anti-FLAG antibody. (C) Subcellular localization of Red-TCTP-WT or Red-TCTP-K164R fusion protein in BmN cells was determined by fluorescence (red), and the nuclear DNA was counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 10 μm. (D) Cytoplasm (CF) and nuclear (NF) fractions isolated from BmN cells expressing FLAG-TCTP-WT or FLAG-TCTP-K164R were probed with an anti-FLAG antibody. An anti-Tubulin antibody was used to label CF.
We next analyzed if the SUMOylation mutation of BmTCTP would affect its subcellular localization. Red-tagged TCTP-WT and TCTP-K164R were transiently transfected into the BmN cells, respectively. As shown in Figure 1C, WT BmTCTP was primarily localized in the cytoplasm, indicating that silkworm BmTCTP was a cytoplasmic protein, which further confirmed the previous report (50). Also, K164R mutant BmTCTP showed similar cytoplasm localization as WT, implying that the SUMOylation mutation of BmTCTP did not affect its localization under the normal state of cultured cells. In agreement with the fluorescence signals, isolation of the cytoplasm and nuclear fractions of BmN cells showed that either TCTP-WT or TCTP-K164R was present in the cytoplasm by immunoblotting analysis (Figure 1D). Taken together, these data demonstrated that BmTCTP is SUMOylated at a conserved lysine site in silkworm and that the SUMOylation of BmTCTP did not influence its cytoplasm localization.
BmTCTP Knockdown Promoted Viral Replication
TCTP has been reported to be involved in the resistance against bacteria or viruses, and silencing of BmTCTP expression resulted in a decrease in survival rate of silkworm individuals after bacterial infection (38). To gain further insight into the function of BmTCTP in immune response, we here investigated its immune function in response to BmNPV infection by RNAi-mediated knockdown of BmTCTP. We firstly synthesized specific dsRNAs against ORF and UTR of BmTCTP (referred to as dsTCTP-ORF and dsTCTP-UTR) (Figure 2A). G5-PAMAM system was used to deliver dsRNAs of BmTCTP into the BmN cells to silence BmTCTP expression (45). RT-PCR result showed that both dsTCTP-ORF and dsTCTP-UTR were able to decrease the mRNA transcription of BmTCTP compared with the dsRed control treatment (Figure 2B), and also dsTCTP-ORF could specifically decrease the expression of FLAG-TCTP by immunoblotting (Figure S1), which together revealed that dsRNAs of BmTCTP could efficiently silence its own expression.
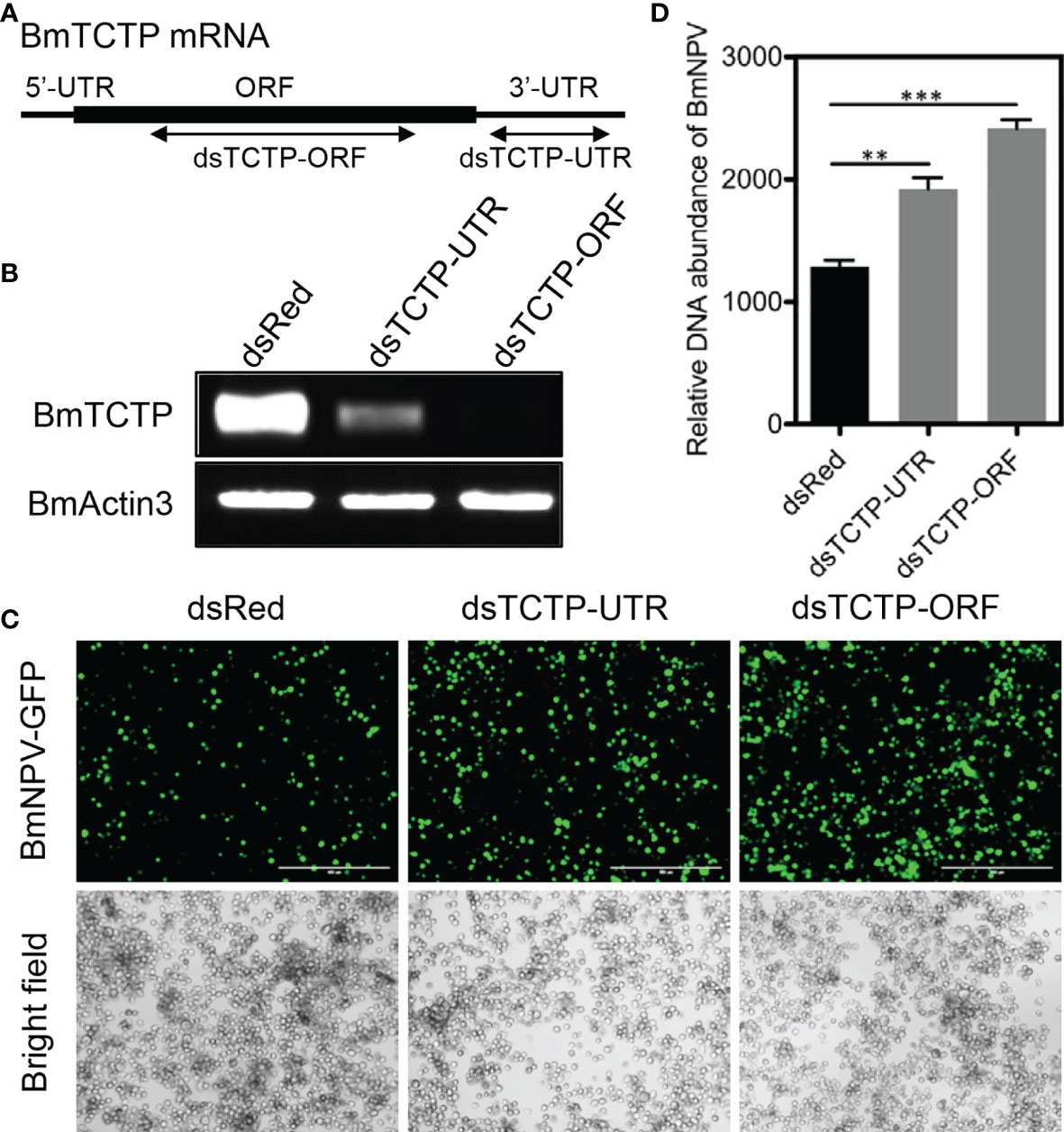
Figure 2 Bombyx mori translationally controlled tumor protein (BmTCTP) knockdown promoted viral replication. (A) Schematic representation of the silkworm BmTCTP gene structure and dsRNA target regions. (B) Semiquantitative RT-PCR was used to evaluate RNAi efficiency of dsRNA targeting TCTP-ORF and TCTP-UTR in cultured silkworm BmN cells. The expression of BmActin3 gene was used as an internal control. (C) Fluorescence microscopy was used to observe the viral amounts when BmN cells were infected with Bombyx mori nucleopolyhedrovirus (BmNPV)-GFP for 72 h in the presence or absence of BmTCTP. Scale bar, 400 μm. (D) The viral DNA contents were detected by quantitative RT-PCR analysis of the BmNPV GP41 gene. The expression of BmGAPDH gene was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: **P < 0.01 and ***P < 0.001.
Then, BmNPV infection in the BmN cells was evaluated after BmTCTP knockdown. BmN cells were infected with BmNPV-GFP when the expression of BmTCTP was silenced. As shown in Figure 2C, fluorescence observation exhibited that the viral fluorescence was obviously stronger in cells of BmTCTP knockdown than that of the control. To further quantify the viral amounts, the viral DNA contents were detected by qRT-PCR analysis of the BmNPV GP41 gene and compared with the control. Low levels of DNA replication were observed in control dsRNA treatment, and knockdown of BmTCTP led to a significant increase of viral amounts (Figure 2D). These data suggested that BmTCTP knockdown promoted the replication of BmNPV, indicating that the silkworm BmTCTP participated in the host immune response against virus infection.
Deficiency of BmTCTP SUMOylation Inhibited Its Antiviral Immunity
To investigate whether SUMOylation of BmTCTP was involved in the immune function to virus infection, we firstly transfected two plasmids expressing FLAG-TCTP-WT and FLAG-TCTP-K164R, respectively, into the BmN cells. After the challenge by BmNPV-GFP, viral fluorescence clearly showed that the expression of mutated TCTP-K164R induced higher BmNPV levels than that of TCTP-WT expression (Figure 3A). Consistent with this observation, qRT-PCR for GP41 gene in TCTP-K164R also exhibited high levels of the viral DNA contents compared with TCTP-WT (Figure 3B). These results may indicate the possibility that the mutant of BmTCTP SUMOylation site affected its own immune response.
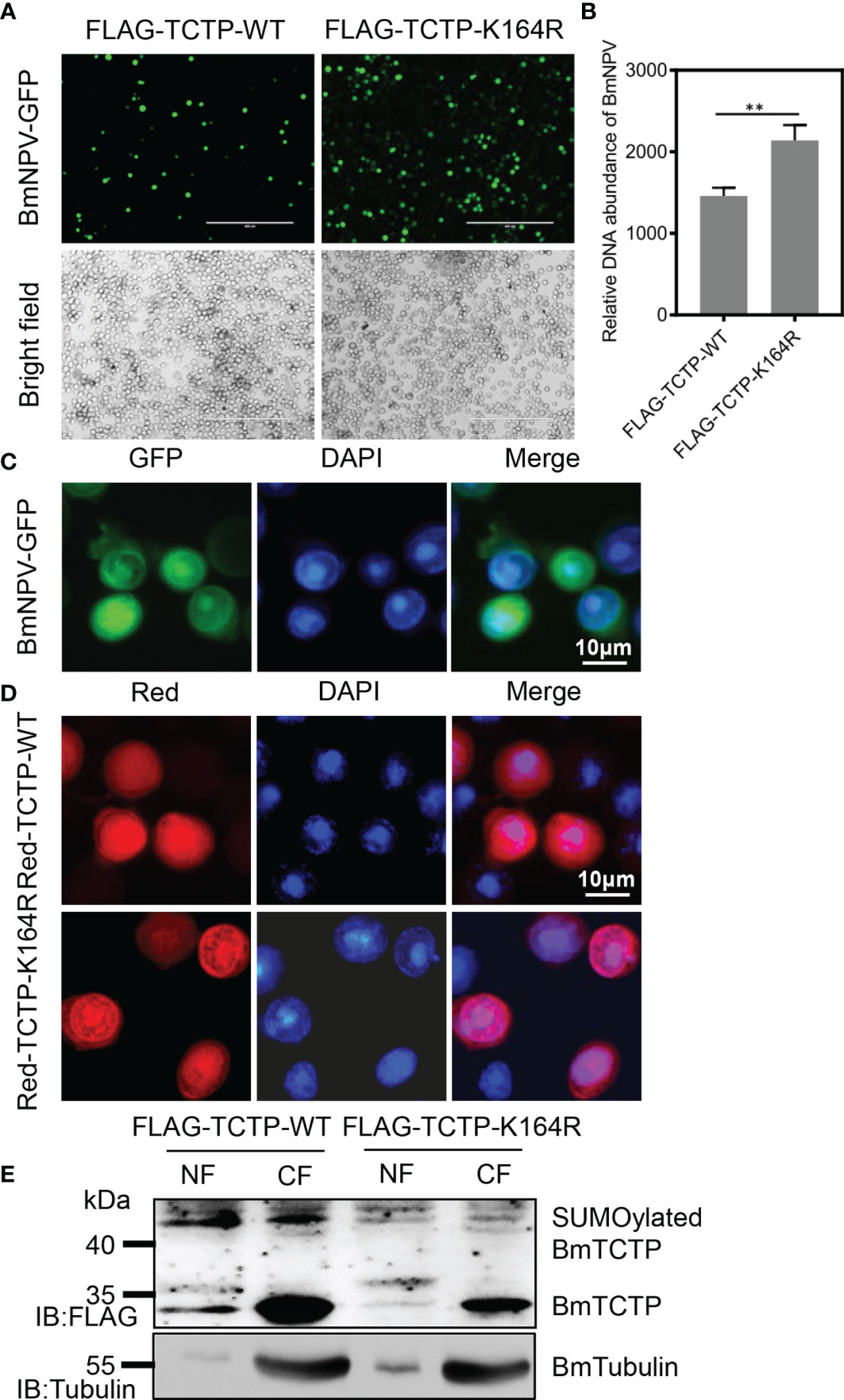
Figure 3 Deficiency of Bombyx mori translationally controlled tumor protein (BmTCTP) SUMOylation affected its antiviral immunity. (A) Plasmids as indicated were transfected into the silkworm BmN cells, and then cells were infected with Bombyx mori nucleopolyhedrovirus (BmNPV)-GFP for 72 h. Fluorescence microscopy was used to observe the viral amounts. Scale bar, 400 μm. (B) The viral DNA contents were detected by qRT-PCR analysis of the BmNPV GP41 gene. The expression of BmGAPDH gene was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: **P < 0.01. (C) BmN cells were infected with BmNPV-GFP for 72 h. BmNPV-positive cells were then determined by fluorescence (green), and the nuclear DNA was counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 10 μm. (D) Red-TCTP-WT or Red-TCTP-K164R plasmid was transfected into the silkworm BmN cells, and then cells were infected with BmNPV-GFP for 72 h. Subcellular localization of Red-TCTP-WT or Red-TCTP-K164R in BmNPV-positive cells were determined by fluorescence (red), and the nuclear DNA was counterstained with DAPI (blue). Scale bar, 10 μm. (E) Cytoplasm (CF) and nuclear (NF) fractions isolated from BmNPV-GFP-infected cells expressing FLAG-TCTP-WT or FLAG-TCTP-K164R were probed with an anti-FLAG antibody. An anti-Tubulin antibody was used to label CF.
As cells infected by BmNPV caused the nuclear chromosomes to aggregate (Figure 3C), we next analyzed the distribution of BmTCTP in BmN cells after BmNPV treatment. It was shown that the TCTP-WT could enter the nucleus wherein the fluorescence signals were very strong. When compared with the TCTP-WT, however, the localization of TCTP-K164R mutation in the nucleus was decreased (Figure 3D). To further verify this result, we isolated the cytoplasm and nuclear fractions to determine the localization of BmTCTP. As shown in Figure 3E, we can detect the TCTP-WT protein in both cytoplasm and nucleus; by contrast, only a weak signal of the TCTP-K164R can be detected in the nucleus, suggesting that TCTP-WT indeed entered the nucleus post BmNPV infection. Interestingly, we found very high levels of SUMOylation in the TCTP-WT, but not in the TCTP-K164R, which indicated that BmNPV infection may induce the modification of BmTCTP SUMOylation and the enrichment into the nucleus. Taken together, these data suggested that SUMOylation may contribute to antiviral immunity of BmTCTP.
Stabilization of BmTCTP SUMOylation Enhanced Its Antiviral Immunity
To further confirm that SUMOylation of BmTCTP was involved in the antiviral immunity, we next generated a fusion vector of BmTCTP and BmSMT3 proteins in order to maintain the state of BmTCTP SUMOylation. Expression of FLAG-TCTP-SMT3 was measured by immunoblotting, and the protein size of FLAG-TCTP-SMT3 was larger than that of FLAG-TCTP (Figure 4A), which also supported the SUMOylated BmTCTP in Figure 1B. We then transfected the FLAG-TCTP or FLAG-TCTP-SMT3 into the BmN cells infected with BmNPV-GFP, and post 72 h treatment, the viral fluorescence was observed. It was shown that the expression of BmTCTP-BmSMT3 obviously inhibited BmNPV levels compared with that of BmTCTP itself, although both treatments also reduced viral signals when compared with the control (Figure 4B). In agreement with this, qRT-PCR for GP41 gene in BmTCTP-BmSMT3 also significantly decreased levels of the viral DNA contents compared with BmTCTP and control (Figure 4C). These results indicated that stabilization of BmTCTP SUMOylation indeed enhanced its antiviral function.
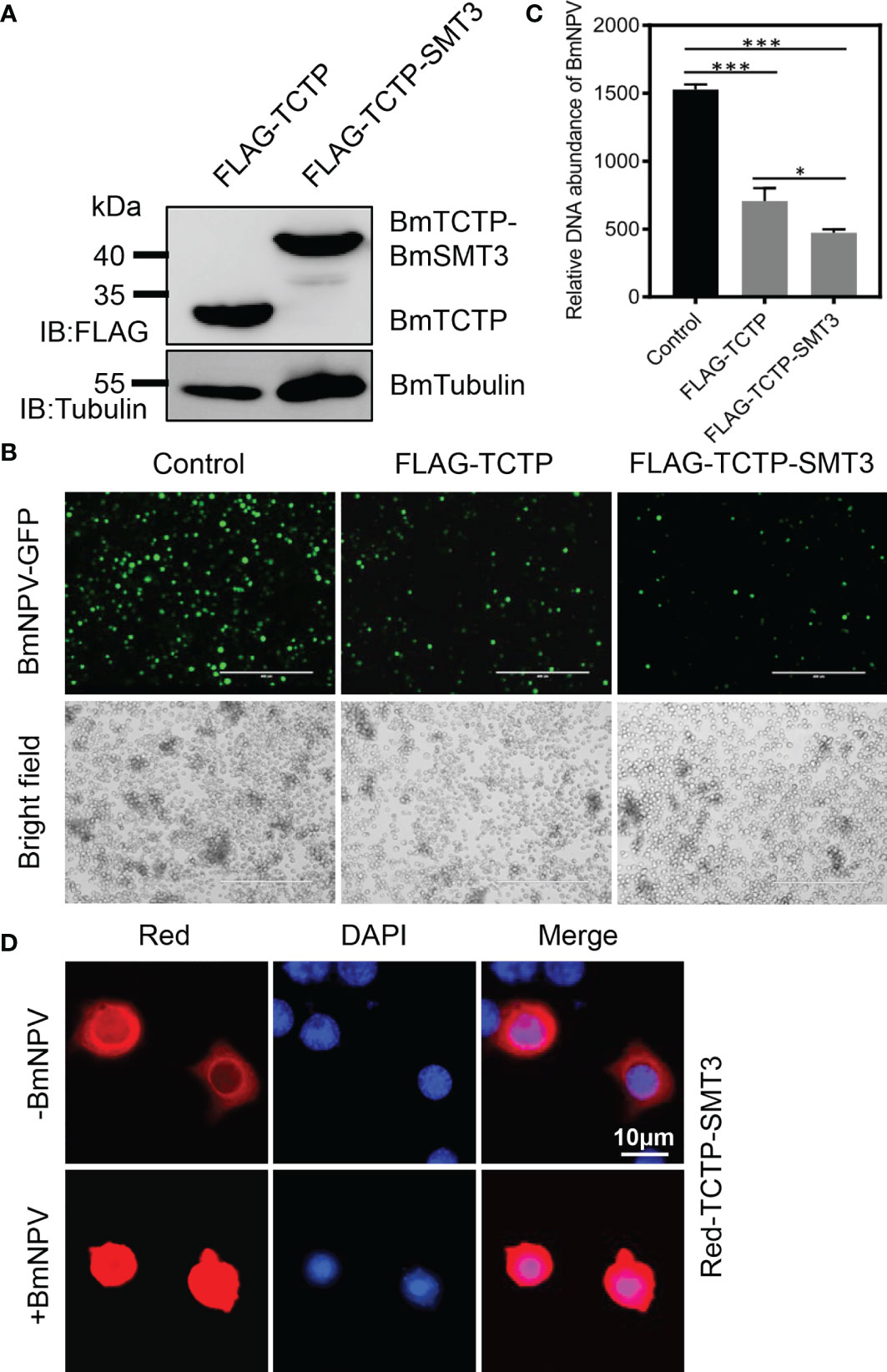
Figure 4 Stabilization of Bombyx mori translationally controlled tumor protein (BmTCTP) SUMOylation enhanced its antiviral immunity. (A) Plasmids expressing FLAG-TCTP and FLAG-TCTP-SMT3 were transfected into the silkworm BmN cells, and then cells were collected after 72 h. Proteins from FLAG-TCTP and FLAG-TCTP-SMT3 were detected by anti-FLAG and anti-Tubulin antibodies. (B) Plasmids as indicated were transfected into the silkworm BmN cells, and then cells were infected with Bombyx mori nucleopolyhedrovirus (BmNPV)-GFP for 72 h. Fluorescence microscopy was used to observe the viral amounts. Scale bar, 400 μm. (C) The viral DNA contents were detected by qRT-PCR analysis of the BmNPV GP41 gene. The expression of BmGAPDH gene was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: *P < 0.05 and ***P < 0.001. (D) Red-TCTP-SMT3 was transfected into the silkworm BmN cells, and then cells were infected with or without BmNPV-GFP for 72 h. Subcellular localization of Red-TCTP-SMT3 was determined by fluorescence (red), and the nuclear DNA was counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 10 μm.
To detect the subcellular localization of BmTCTP-BmSMT3 upon the infection with or without BmNPV, Red-TCTP-SMT3 was transfected into cells to analyze its distribution. In the absence of BmNPV, Red-TCTP-SMT3 was mainly localized in the cytoplasm (Figure 4D), consistent with the WT BmTCTP localization (Figure 1C). By contrast, in the presence of BmNPV, Red-TCTP-SMT3 was able to largely accumulate in the nucleus (Figure 4D). Altogether, these data demonstrated that SUMOylation enabled BmTCTP to promote the immune response against the virus.
SUMO Conjugating Enzyme BmUBC9 Was Required for the SUMOylation of BmTCTP
To address how SUMOylation pathway was involved in the BmTCTP SUMOylation and immune response, we investigated the potential role of BmUBC9, a SUMO conjugating enzyme responsible for all SUMO conjugations in eukaryotic cells (42, 51, 52), in the BmTCTP SUMOylation. We firstly monitored the DNA amounts of BmNPV infection in cells at the indicated time points. It was shown that following the virus infection, BmNPV replications were increased rapidly (Figure 5A). By contrast, the expression of BmTCTP was downregulated after BmNPV infection (Figure 5B). These results suggested that the virus might inhibit the immune function of BmTCTP by downregulating BmTCTP expression levels in the host cells and thus prompted its own replications.
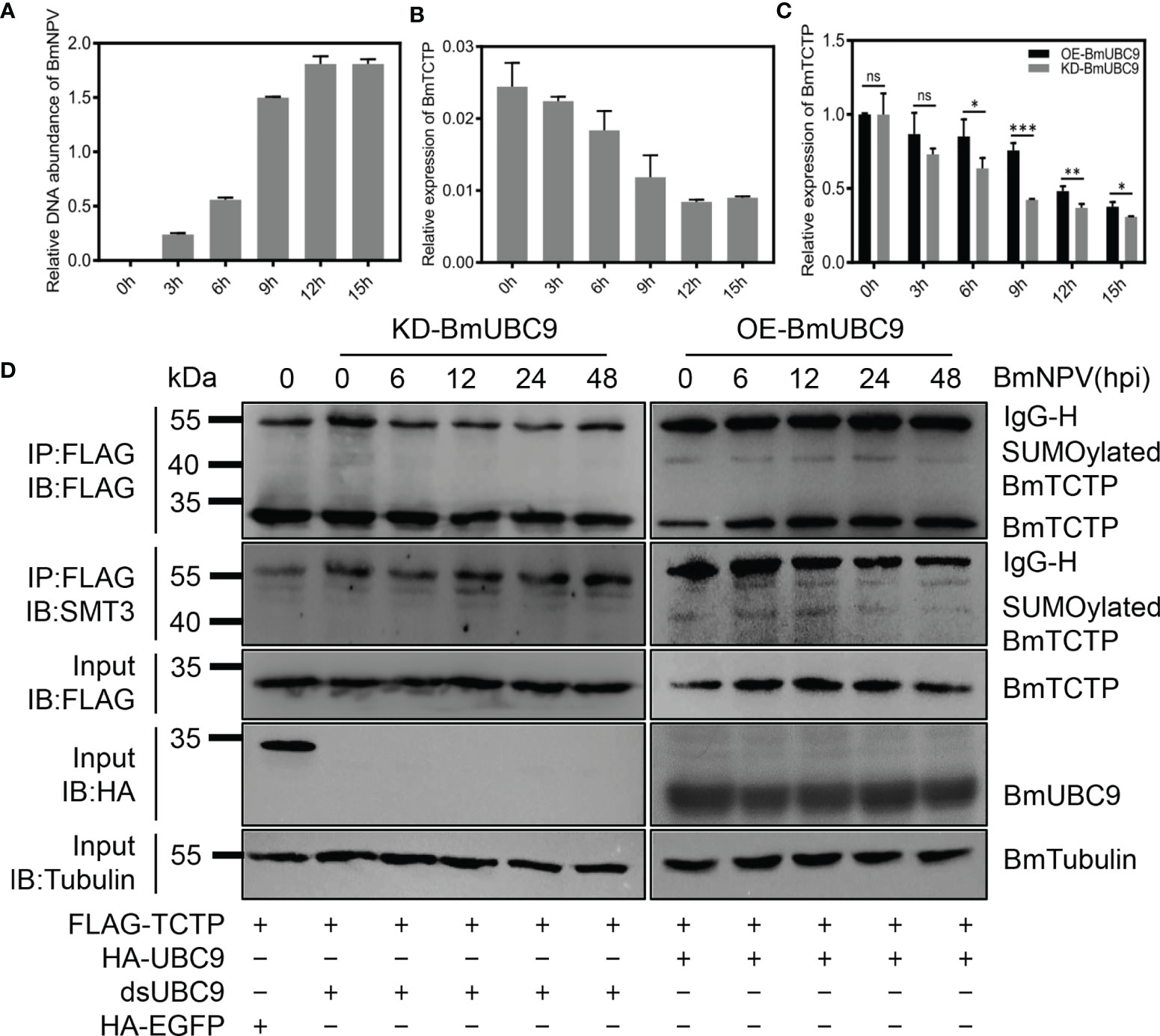
Figure 5 SUMO conjugating enzyme BmUBC9 was required for the SUMOylation of Bombyx mori translationally controlled tumor protein (BmTCTP). (A) BmN cells infected with Bombyx mori nucleopolyhedrovirus (BmNPV) were collected at the indicated time points and used for qRT-PCR analysis of the BmNPV GP41 gene. The expression of BmGAPDH gene was used as an internal control. The data presented are the means ± SD (n = 3). (B) The expression of BmTCTP gene was analyzed by qRT-PCR following the BmNPV infection, and the BmeIF-4a was used as an internal control. The data presented are the means ± SD (n = 3). (C) The expression of BmTCTP gene was analyzed by qRT-PCR in BmUBC9 knockdown or overexpression cells following the BmNPV infection, and the BmeIF-4a was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: *P < 0.05, **P < 0.01, and ***P < 0.001, and ns represents no significance. (D) Plasmids as indicated were co-transfected into the BmN cells infected with BmNPV. Lysates from BmN cells were used for immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with the indicated antibodies. SUMOylated BmTCTP was further determined by an anti-SMT3 antibody. IgG-L and IgG-H were the light and heavy chains of anti-FLAG antibody.
It was interesting that when we knocked down or overexpressed the BmUBC9 in the BmN cells infected with BmNPV, the decrease of BmTCTP expression was delayed in BmUBC9 overexpression cells compared with that in BmUBC9 knockdown cells (Figure 5C), implying the possibility that BmUBC9 could be involved in the immune response of BmTCTP. To test this, we carried out the immunoprecipitation analysis to analyze the SUMOylation levels of BmTCTP in BmUBC9 knockdown or overexpression cells upon BmNPV infection. As shown in Figure 5D, deletion of BmUBC9 clearly disrupted the SUMOylation of BmTCTP, whereas overexpression of BmUBC9 allowed cells to maintain high levels of BmTCTP SUMOylation even in the cells infected with a long period of BmNPV. Hence, the present data indicated that BmUBC9 would be essential for BmTCTP SUMOylation as well as its stabilization.
BmTCTP Interactome Identified the Interaction With an Immune Molecule Interleukin Enhancer Binding Factor
To better understand how BmTCTP participates in the immune process, we next aimed to identify proteins interacting with BmTCTP in BmN cells. We established two cell lines stably expressing FLAG-TCTP-WT and FLAG-TCTP-K164R, respectively. Whole-cell lysates were prepared from both cell lines, and FLAG antibody against FLAG peptide was used for immunoprecipitation in the above cell lysates. The resulting bound or flow proteins were resolved on 10% SDS-polyacrylamide gel electropheresis (SDS-PAGE) gels and probed with anti-FLAG antibody (Figure 6A). The eluate after the elution was further applied to SDS-PAGE, and the peptides were generated via in-gel trypsin digestion and subjected to LC-MS/MS analysis. As a result, we obtained 271 interaction proteins in FLAG-TCTP-WT and 102 proteins in FLAG-TCTP-K164R, and there were 64 common proteins (Figure 6B). These differential interactomes between WT and K164R mutant indicated that the SUMOylation of BmTCTP may contribute to its own interaction with other proteins.
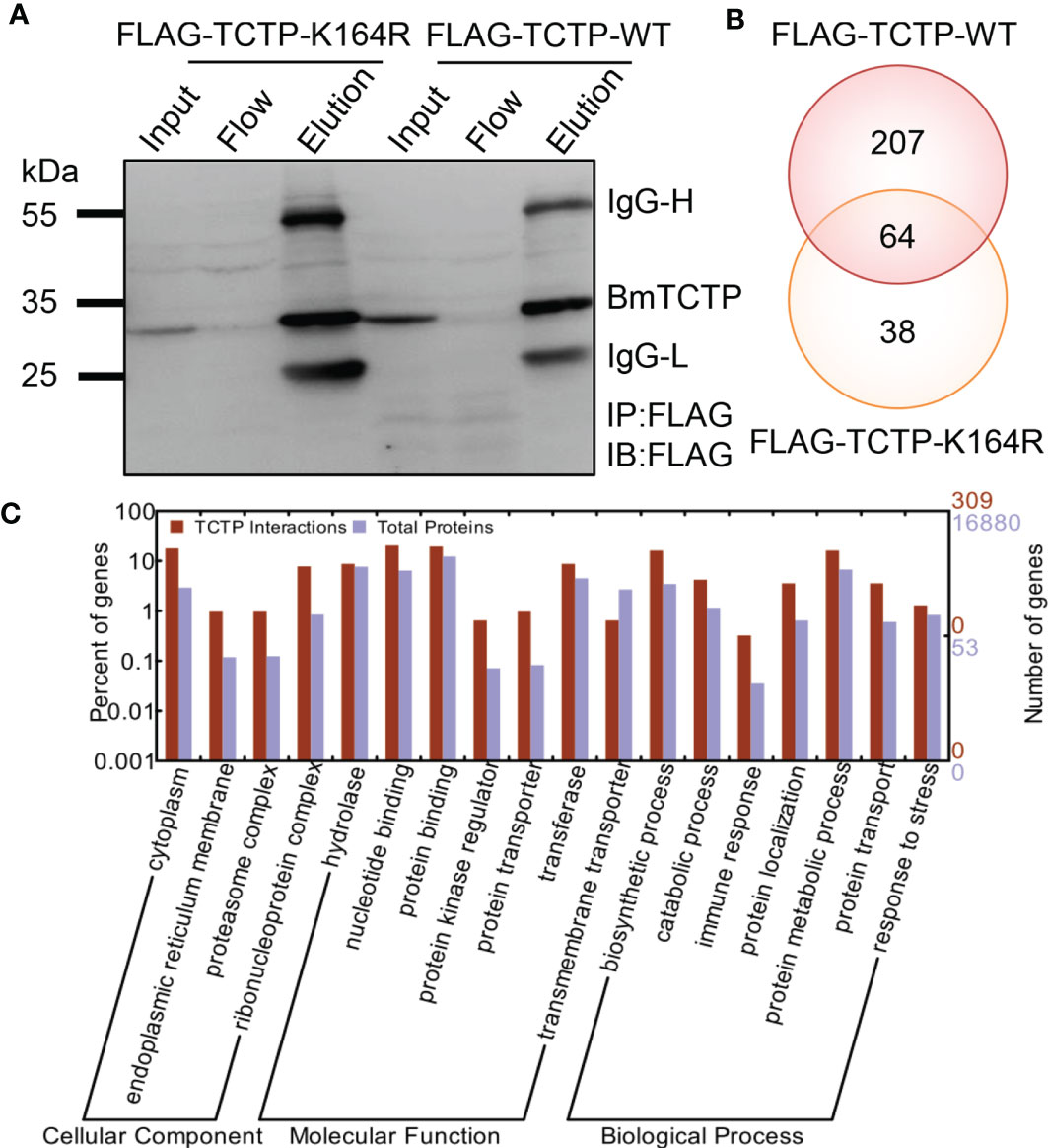
Figure 6 Bombyx mori translationally controlled tumor protein (BmTCTP) interactome identified the interaction with an immune molecule Bombyx mori interleukin enhancer binding factor (BmILF). (A) The efficiency of anti-FLAG antibody-mediated BmTCTP immunoprecipitation. Anti-FLAG antibody was used for both IP and immunoblotting (IB). The input, flow, and elution proteins were resolved on 10% SDS-PAGE gels and probed with anti-FLAG antibody. IgG-L and IgG-H were the light and heavy chains of anti-FLAG antibody. (B) Venn diagram assay of the FLAG-TCTP-WT and FLAG-TCTP-K164R interacting proteins identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The LC-MS/MS data for each expression plasmid were shown in Table S2. (C) Functional categories of the interacting proteins were analyzed by WEGO. Proteins were classified into cellular component, molecular function, and biological process according to their Gene Ontology (GO) signatures, and the total silkworm genes were shown as a reference.
To classify the identified BmTCTP interacting proteins according to biological processes, molecular functions, or cellular component, we performed a Gene Ontology (GO) analysis to annotate protein functions. The resulting GO terms were used to categorize the proteins using WEGO program. It was shown that the majority of proteins will function in protein binding, as a protein kinase regulator, and as a protein transporter and participate in immune response and protein localization in silkworm (Figure 6C), suggesting that the silkworm BmTCTP would also act as a multifunctional protein through binding directly or indirectly to a number of proteins. Interestingly, among the interaction proteins, we found that only one immune molecule called ILF was just presented in TCTP-WT interactome but not in TCTP-K164R (Table S2), which implied that the BmILF may be involved in the immune function of BmTCTP.
BmTCTP Regulated BmILF Expression and Inhibited Viral Replication
Indeed, previous studies have demonstrated that human interleukin enhancer binding factor 2 (ILF2) is considered as an important host cellular protein, which exerts a negatively regulatory effect on the replication of various viruses, such as human immunodeficiency virus type 1 (HIV-1), porcine reproductive and respiratory syndrome virus (PRRSV), and Japanese encephalitis virus (JEV) (53–57). To further characterize the correlation between BmTCTP and BmILF and whether the silkworm BmILF was able to regulate BmNPV replication, we investigated their functions in the response to BmNPV infection. It has been demonstrated that TCTP-WT was able to interact with BmILF by IP experiment. We further analyzed their interaction by colocalization. As shown in Figure 7A, BmILF was primarily localized in the nucleus, and only a small part of BmILF in the cytoplasm could be colocalized with BmTCTP.
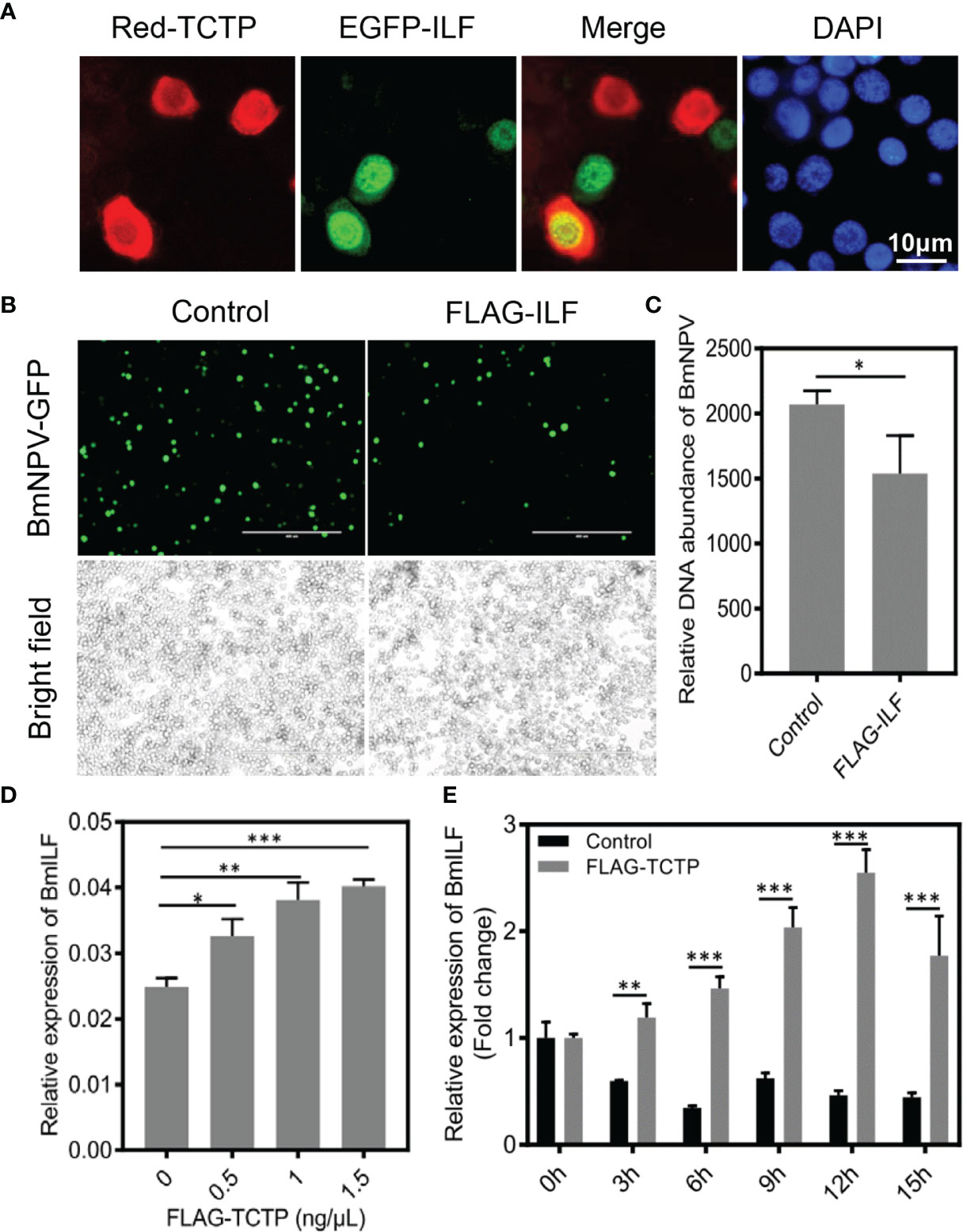
Figure 7 Bombyx mori translationally controlled tumor protein (BmTCTP) regulated Bombyx mori interleukin enhancer binding factor (BmILF) expression and inhibited viral replication. (A) Plasmids encoding Red-TCTP and EGFP-ILF were co-transfected into the BmN cells. After 3 days of transfection, cells were fixed and stained with 4’,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 10 μm. (B) Plasmids as indicated were transfected into the BmN cells, and then cells were infected with Bombyx mori nucleopolyhedrovirus (BmNPV)-GFP for 72 h. Fluorescence microscopy was used to observe the viral amounts. Scale bar, 400 μm. (C) The viral DNA contents were detected by qRT-PCR analysis of the BmNPV GP41 gene. The expression of BmGAPDH gene was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: *P < 0.05. (D) The expression of BmILF gene was analyzed by qRT-PCR in BmTCTP overexpression cells at different concentrations, and the BmeIF-4a was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: *P < 0.05, **P < 0.01, and ***P < 0.001. (E) The fold changes of BmILF expression was analyzed by qRT-PCR in BmTCTP overexpression cells following the BmNPV infection at the indicated time points, and the BmeIF-4a was used as an internal control. The data presented are the means ± SD (n = 3). For the significant analysis: **P < 0.01 and ***P < 0.001.
To study the response of BmILF to the virus infection, we overexpressed BmILF in the BmN cells and infected BmNPV-GFP for 72 h. The viral fluorescence clearly showed that the expression of FLAG-ILF inhibited BmNPV compared with the control (Figure 7B). FLAG-ILF expression also significantly decreased levels of the viral DNA contents compared with the control by qRT-PCR for GP41 gene (Figure 7C). These results implied that the silkworm BmILF was able to negatively regulate BmNPV replication.
Interestingly, we found that the expression of BmILF could be induced by overexpressed BmTCTP; moreover, this increased expression of BmILF was dependent on the levels of BmTCTP (Figure 7D), which suggested a possibility that BmTCTP may regulate BmILF expression as well. Although the expression of BmILF was also decreased post-infection with BmNPV at the indicated time points, overexpression of BmTCTP, however, significantly upregulated BmILF expression (Figure 7E). Therefore, our data demonstrated that BmTCTP will exert antiviral function via regulating BmILF expression and may in turn form an immune complex to inhibit the replication of virus in silkworm.
Discussion
Increasing amount of evidence has proven that TCTP contributes to regulating immune function and plays important roles in diverse biological processes. In eukaryotes, TCTP has been considered as an antibacterial or antiviral factor. For example, RNAi of Drosophila melanogaster TCTP increased the mortality after oral infection with Serratia marcescens (37). The silence of TCTP in Litopenaeus vannamei led to the increase of viral replication of white spot syndrome virus (WSSV), and injection of purified TCTP into WSSV-infected Penaeus monodon increased its survival rate (58). RNAi of the silkworm TCTP inhibited the antimicrobial capacity and the intestinal innate immunity of the silkworm after oral infection of bacteria (38). Although the immune response of TCTP is widely accepted, its regulatory mechanisms are still largely unknown.
In the present study, we established that the silkworm BmTCTP could be modified by SUMOylation molecular BmSMT3 at the lysine 164, and the SUMOylated BmTCTP would contribute to its immune response and promote antiviral activity. Our data showed that BmTCTP RNAi led to increased susceptibility of the silkworm cells to virus infection, and by contrast, overexpression of BmTCTP significantly decreased the virus replication, implying a strong antiviral function of BmTCTP in silkworm. Unexpectedly, this antiviral function of BmTCTP had been attenuated by mutation of BmTCTP SUMOylation site, suggesting that the SUMOylation of BmTCTP would be required for the survival of the silkworm cells upon the virus challenge. Consistent with this, it was interestingly shown that the stabilization of BmTCTP SUMOylation by overexpressing BmTCTP-BmSMT3 fusion protein further suppressed the replication of the virus. Hence, the enhanced immune response of BmTCTP SUMOylation would be fascinating to evaluate the antiviral function in silkworm individuals. Recent work also showed that Drosophila Relish can be SUMOylated, and this SUMOylation prevents cleavage and activation of Relish, thus negatively regulating the immune response (59). All these data suggested that SUMOylation status in organisms would exactly contribute to differential immune response dependent on the different substrate proteins.
Furthermore, our results showed that BmUBC9 can act as a SUMO conjugating enzyme for BmTCTP SUMOylation. After the virus infection, knockdown or overexpression of UBC9 displayed differential SUMOylation status of BmTCTP. It was also shown that the decreased expression of BmTCTP following the virus challenge was partially rescued by overexpression of BmUBC9 rather than its knockdown, which implied that BmUBC9 may regulate BmTCTP expression as well. Future research is needed to improve our understanding of the molecular mechanisms underlying the BmUBC9-mediated regulation of BmTCTP promoter activity. However, it was demonstrated that the BmUBC9-mediated BmTCTP SUMOylation would be essential for antiviral function.
In order to decipher the immune function of BmTCTP, we also performed a systematic proteomics study by LC-MS/MS analysis in combination with IP to investigate its interaction proteins. As a result, a total of 309 potential BmTCTP binding proteins have been identified. The protein functional classification system showed that these proteins participate in various functional processes including protein binding, protein kinase regulation, protein transport, immune response, and protein localization, which is consistent with the reports of previous studies (17, 18). It was intriguing that the mutant of TCTP-K164R clearly affected the interaction of BmTCTP with a majority of target proteins, suggesting that the SUMOylation of BmTCTP would be also important for the interactions. Moreover, SUMO-activating enzyme was also specifically pulled down by TCTP-WT rather than by TCTP-K164R, further confirming the presence of SUMOylation in BmTCTP protein, which together provides a novel insight into deciphering the BmTCTP functions by SUMOylation.
Among the BmTCTP interactions, it was shown that BmILF, the only one immune molecule, is able to interact with TCTP-WT but not with TCTP-K164R, which gives us an exciting clue to understand the immunity function of BmTCTP. The human ILF homologue has been demonstrated as an important host cellular protein that exerts a negatively regulatory effect on the replication of various viruses (53, 55, 57). Our result also showed that overexpression of the silkworm BmILF significantly inhibited virus replication. Importantly, BmTCTP could induce the expression of BmILF, especially in the response of BmNPV infection. Given the interaction between BmTCTP and BmILF, and the induced expression of BmILF by BmTCTP, it was interesting to speculate that BmTCTP may regulate the expression of BmILF and further form the complex with BmILF upon virus challenge so as to act antivirus roles. Nevertheless, the molecular mechanisms by which BmTCTP regulates BmILF expression and activity require further investigation.
Taken together, our study unravels a signaling pathway mediated by BmUBC9-mediated SUMOylation of BmTCTP that can inhibit the replication of BmNPV. Further downstream signaling in the antivirus activity of BmTCTP would be ascribed to by the regulation of BmILF interaction with BmTCTP and/or BmILF expression in response to virus infection. It has been shown that diseases caused by viruses, especially by BmNPV, are the greatest challenge to sericulture and usually cause major economic losses of cocoon production every year (39, 40). A better understanding of the silkworm immune response will be helpful for the disease control. Therefore, our present study has provided a novel strategy to defend against a virus via the antiviral capacity of BmTCTP SUMOylation in cultured silkworm cells. It will be of great interest to examine whether the transgenic expression of BmTCTP SUMOylation form in silkworm individuals plays a similar role in replication control of the virus, which would be useful for generating antiviral silkworm lines in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
ZL, QX, and PZ contributed to conception and design of the study. CL and ZL wrote the article. CL, ZL, WZ, HG, WL, and GS performed the research and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32030103), Natural Science Foundation of Chongqing (Nos. cstc2020jcyj-cxttX0001 and cstc2021jsyj-yzysbAX0004), and Fundamental Research Funds for the Central Universities (No. XDJK2019B007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the editor and reviewers for their constructive comments and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.807097/full#supplementary-material
Supplementary Figure 1 | RNAi efficiency of dsRNA for BmTCTP by immunoblotting analysis. Immunoblotting was used to evaluate RNAi efficiency of dsRNA targeting TCTP-ORF and TCTP-UTR in FLAG-TCTP expressing BmN cells. Proteins from different treatments were isolated and detected by anti-FLAG and anti-Tubulin antibodies.
Supplementary Table 2 | Identification of BmTCTP interacting proteins by mass spectrometry in silkworm.
References
1. MacDonald SM, Rafnar T, Langdon J, Lichtenstein LM. Molecular Identification of an IgE-Dependent Histamine-Releasing Factor. Science (1995) 269(5224):688–90. doi: 10.1126/science.7542803
2. Yenofsky R, Bergmann I, Brawerman G. Messenger RNA Species Partially in a Repressed State in Mouse Sarcoma Ascites Cells. Proc Natl Acad Sci USA (1982) 79(19):5876–80. doi: 10.1073/pnas.79.19.5876
3. Gross B, Gaestel M, Bohm H, Bielka H. cDNA Sequence Coding for a Translationally Controlled Human Tumor Protein. Nucleic Acids Res (1989) 17(20):8367. doi: 10.1093/nar/17.20.8367
4. Bommer UA, Borovjagin AV, Greagg MA, Jeffrey IW, Russell P, Laing KG, et al. The mRNA of the Translationally Controlled Tumor Protein P23/TCTP is a Highly Structured RNA, Which Activates the dsRNA-Dependent Protein Kinase PKR. RNA (2002) 8(4):478–96. doi: 10.1017/s1355838202022586
5. Bohm H, Benndorf R, Gaestel M, Gross B, Nurnberg P, Kraft R, et al. The Growth-Related Protein P23 of the Ehrlich Ascites Tumor: Translational Control, Cloning and Primary Structure. Biochem Int (1989) 19(2):277–86.
6. Bommer UA, Thiele BJ. The Translationally Controlled Tumour Protein (TCTP). Int J Biochem Cell Biol (2004) 36(3):379–85. doi: 10.1016/s1357-2725(03)00213-9
7. Amson R, Pece S, Marine JC, Di Fiore PP, Telerman A. TPT1/TCTP-Regulated Pathways in Phenotypic Reprogramming. Trends Cell Biol (2013) 23(1):37–46. doi: 10.1016/j.tcb.2012.10.002
8. Acunzo J, Baylot V, So A, Rocchi P. TCTP as Therapeutic Target in Cancers. Cancer Treat Rev (2014) 40(6):760–9. doi: 10.1016/j.ctrv.2014.02.007
9. Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally Controlled Tumor Protein is a Conserved Mitotic Growth Integrator in Animals and Plants. Proc Natl Acad Sci USA (2010) 107(37):16384–9. doi: 10.1073/pnas.1007926107
10. Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is Essential for Growth and Proliferation Through Regulation of Drheb GTPase. Nature (2007) 445(7129):785–8. doi: 10.1038/nature05528
11. Koziol MJ, Gurdon JB. TCTP in Development and Cancer. Biochem Res Int (2012) 2012:105203. doi: 10.1155/2012/105203
12. Gu X, Yao L, Ma G, Cui L, Li Y, Liang W, et al. TCTP Promotes Glioma Cell Proliferation In Vitro and In Vivo via Enhanced Beta-Catenin/TCF-4 Transcription. Neuro Oncol (2014) 16(2):217–27. doi: 10.1093/neuonc/not194
13. Tuynder M, Fiucci G, Prieur S, Lespagnol A, Geant A, Beaucourt S, et al. Translationally Controlled Tumor Protein is a Target of Tumor Reversion. Proc Natl Acad Sci USA (2004) 101(43):15364–9. doi: 10.1073/pnas.0406776101
14. van de Sande WW, Janse DJ, Hira V, Goedhart H, van der Zee R, Ahmed AO, et al. Translationally Controlled Tumor Protein From Madurella Mycetomatis, a Marker for Tumorous Mycetoma Progression. J Immunol (2006) 177(3):1997–2005. doi: 10.4049/jimmunol.177.3.1997
15. Lucibello M, Adanti S, Antelmi E, Dezi D, Ciafre S, Carcangiu ML, et al. Phospho-TCTP as a Therapeutic Target of Dihydroartemisinin for Aggressive Breast Cancer Cells. Oncotarget (2015) 6(7):5275–91. doi: 10.18632/oncotarget.2971
16. Baylot V, Katsogiannou M, Andrieu C, Taieb D, Acunzo J, Giusiano S, et al. Targeting TCTP as a New Therapeutic Strategy in Castration-Resistant Prostate Cancer. Mol Ther (2012) 20(12):2244–56. doi: 10.1038/mt.2012.155
17. Li S, Chen M, Xiong Q, Zhang J, Cui Z, Ge F. Characterization of the Translationally Controlled Tumor Protein (TCTP) Interactome Reveals Novel Binding Partners in Human Cancer Cells. J Proteome Res (2016) 15(10):3741–51. doi: 10.1021/acs.jproteome.6b00556
18. Li Y, Sun H, Zhang C, Liu J, Zhang H, Fan F, et al. Identification of Translationally Controlled Tumor Protein in Promotion of DNA Homologous Recombination Repair in Cancer Cells by Affinity Proteomics. Oncogene (2017) 36(50):6839–49. doi: 10.1038/onc.2017.289
19. Kwon YV, Zhao B, Xu C, Lee J, Chen CL, Vinayagam A, et al. The Role of Translationally Controlled Tumor Protein in Proliferation of Drosophila Intestinal Stem Cells. Proc Natl Acad Sci USA (2019) 26591–8. doi: 10.1073/pnas.1910850116
20. Tsarova K, Yarmola EG, Bubb MR. Identification of a Cofilin-Like Actin-Binding Site on Translationally Controlled Tumor Protein (TCTP). FEBS Lett (2010) 584(23):4756–60. doi: 10.1016/j.febslet.2010.10.054
21. Tuynder M, Susini L, Prieur S, Besse S, Fiucci G, Amson R, et al. Biological Models and Genes of Tumor Reversion: Cellular Reprogramming Through Tpt1/TCTP and SIAH-1. Proc Natl Acad Sci USA (2002) 99(23):14976–81. doi: 10.1073/pnas.222470799
22. Yang Y, Yang F, Xiong Z, Yan Y, Wang X, Nishino M, et al. An N-Terminal Region of Translationally Controlled Tumor Protein Is Required for Its Antiapoptotic Activity. Oncogene (2005) 24(30):4778–88. doi: 10.1038/sj.onc.1208666
23. Liu H, Peng HW, Cheng YS, Yuan HS, Yang-Yen HF. Stabilization and Enhancement of the Antiapoptotic Activity of Mcl-1 by TCTP. Mol Cell Biol (2005) 25(8):3117–26. doi: 10.1128/MCB.25.8.3117-3126.2005
24. Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, et al. Reciprocal Repression Between P53 and TCTP. Nat Med (2011) 18(1):91–9. doi: 10.1038/nm.2546
25. Rho SB, Lee JH, Park MS, Byun HJ, Kang S, Seo SS, et al. Anti-Apoptotic Protein TCTP Controls the Stability of the Tumor Suppressor P53. FEBS Lett (2011) 585(1):29–35. doi: 10.1016/j.febslet.2010.11.014
26. Funston G, Goh W, Wei SJ, Tng QS, Brown C, Jiah Tong L, et al. Binding of Translationally Controlled Tumour Protein to the N-Terminal Domain of HDM2 is Inhibited by Nutlin-3. PloS One (2012) 7(8):e42642. doi: 10.1371/journal.pone.0042642
27. Jung J, Kim M, Kim MJ, Kim J, Moon J, Lim JS, et al. Translationally Controlled Tumor Protein Interacts With the Third Cytoplasmic Domain of Na,K-ATPase Alpha Subunit and Inhibits the Pump Activity in HeLa Cells. J Biol Chem (2004) 279(48):49868–75. doi: 10.1074/jbc.M400895200
28. Jung J, Ryu S, Ki IA, Woo HA, Lee K. Some Biological Consequences of the Inhibition of Na,K-ATPase by Translationally Controlled Tumor Protein (TCTP). Int J Mol Sci (2018) 19(6):1657. doi: 10.3390/ijms19061657
29. Schlosser A, Klockow B, Manstein DJ, Lehmann WD. Analysis of Post-Translational Modification and Characterization of the Domain Structure of Dynamin A From Dictyostelium Discoideum. J Mass Spectrom (2003) 38(3):277–82. doi: 10.1002/jms.438
30. Yarm FR. Plk Phosphorylation Regulates the Microtubule-Stabilizing Protein TCTP. Mol Cell Biol (2002) 22(17):6209–21. doi: 10.1128/MCB.22.17.6209-6221.2002
31. Cucchi U, Gianellini LM, De Ponti A, Sola F, Alzani R, Patton V, et al. Phosphorylation of TCTP as a Marker for Polo-Like Kinase-1 Activity In Vivo. Anticancer Res (2010) 30(12):4973–85.
32. Munirathinam G, Ramaswamy K. Sumoylation of Human Translationally Controlled Tumor Protein is Important for its Nuclear Transport. Biochem Res Int (2012) 2012:831940. doi: 10.1155/2012/831940
33. Goldsmith MR, Shimada T, Abe H. The Genetics and Genomics of the Silkworm, Bombyx Mori. Annu Rev Entomol (2005) 50:71–100. doi: 10.1146/annurev.ento.50.071803.130456
34. International Silkworm Genome C. The Genome of a Lepidopteran Model Insect, the Silkworm Bombyx Mori. Insect Biochem Mol Biol (2008) 38(12):1036–45. doi: 10.1016/j.ibmb.2008.11.004
35. Xia Q, Li S, Feng Q. Advances in Silkworm Studies Accelerated by the Genome Sequencing of Bombyx Mori. Annu Rev Entomol (2014) 59:513–36. doi: 10.1146/annurev-ento-011613-161940
36. Liu ZL, Xu J, Ling L, Zhang R, Shang P, Huang YP. CRISPR Disruption of TCTP Gene Impaired Normal Development in the Silkworm Bombyx Mori. Insect Sci (2019) 26(6):973–82. doi: 10.1111/1744-7917.12567
37. Wang F, Hu C, Hua X, Song L, Xia Q. Translationally Controlled Tumor Protein, a Dual Functional Protein Involved in the Immune Response of the Silkworm, Bombyx Mori. PloS One (2013) 8(7):e69284. doi: 10.1371/journal.pone.0069284
38. Hu C, Wang F, Ma S, Li X, Song L, Hua X, et al. Suppression of Intestinal Immunity Through Silencing of TCTP by RNAi in Transgenic Silkworm, Bombyx Mori. Gene (2015) 574(1):82–7. doi: 10.1016/j.gene.2015.07.089
39. Jiang L, Xia Q. The Progress and Future of Enhancing Antiviral Capacity by Transgenic Technology in the Silkworm Bombyx Mori. Insect Biochem Mol Biol (2014) 48:1–7. doi: 10.1016/j.ibmb.2014.02.003
40. Jiang L, Goldsmith MR, Xia Q. Advances in the Arms Race Between Silkworm and Baculovirus. Front Immunol (2021) 12:628151. doi: 10.3389/fimmu.2021.628151
41. Li Z, Cheng D, Mon H, Tatsuke T, Zhu L, Xu J, et al. Genome-Wide Identification of Polycomb Target Genes Reveals a Functional Association of Pho With Scm in Bombyx Mori. PloS One (2012) 7(4):e34330. doi: 10.1371/journal.pone.0034330
42. Li Z, Mon H, Xu J, Zhu L, Lee JM, Kusakabe T. A Conserved SUMOylation Signaling for Cell Cycle Control in a Holocentric Species Bombyx Mori. Insect Biochem Mol Biol (2014) 51:71–9. doi: 10.1016/j.ibmb.2014.05.008
43. Li Z, Cui Q, Xu J, Cheng D, Wang X, Li B, et al. SUMOylation Regulates the Localization and Activity of Polo-Like Kinase 1 During Cell Cycle in the Silkworm, Bombyx Mori. Sci Rep (2017) 7(1):15536. doi: 10.1038/s41598-017-15884-7
44. Li Z, Tatsuke T, Sakashita K, Zhu L, Xu J, Mon H, et al. Identification and Characterization of Polycomb Group Genes in the Silkworm, Bombyx Mori. Mol Biol Rep (2012) 39(5):5575–88. doi: 10.1007/s11033-011-1362-5
45. Lu C, Li Z, Chang L, Dong Z, Guo P, Shen G, et al. Efficient Delivery of dsRNA and DNA in Cultured Silkworm Cells for Gene Function Analysis Using PAMAM Dendrimers System. Insects (2019) 11(1):12. doi: 10.3390/insects11010012
46. Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, et al. Distinct Functions of Bombyx Mori Peptidoglycan Recognition Protein 2 in Immune Responses to Bacteria and Viruses. Front Immunol (2019) 10:776. doi: 10.3389/fimmu.2019.00776
47. Li Z, Cui Q, Wang X, Li B, Zhao D, Xia Q, et al. Functions and Substrates of NEDDylation During Cell Cycle in the Silkworm, Bombyx Mori. Insect Biochem Mol Biol (2017) 90:101–12. doi: 10.1016/j.ibmb.2017.09.013
48. Dong Z, Zhang W, Zhang Y, Zhang X, Zhao P, Xia Q. Identification and Characterization of Novel Chitin-Binding Proteins From the Larval Cuticle of Silkworm, Bombyx Mori. J Proteome Res (2016) 15(5):1435–45. doi: 10.1021/acs.jproteome.5b00943
49. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics (2005) 21(18):3674–6. doi: 10.1093/bioinformatics/bti610
50. Nie Z, Lv Z, Qian J, Chen J, Li S, Sheng Q, et al. Molecular Cloning and Expression Characterization of Translationally Controlled Tumor Protein in Silkworm Pupae. Mol Biol Rep (2010) 37(6):2621–8. doi: 10.1007/s11033-009-9787-9
51. Johnson ES, Blobel G. Ubc9p Is the Conjugating Enzyme for the Ubiquitin-Like Protein Smt3p. J Biol Chem (1997) 272(43):26799–802. doi: 10.1074/jbc.272.43.26799
52. Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The Ubiquitin-Like Proteins SMT3 and SUMO-1 Are Conjugated by the UBC9 E2 Enzyme. Proc Natl Acad Sci USA (1998) 95(2):560–4. doi: 10.1073/pnas.95.2.560
53. Cui X, Qian P, Rao T, Wei Y, Zhao F, Zhang H, et al. Cellular Interleukin Enhancer-Binding Factor 2, ILF2, Inhibits Japanese Encephalitis Virus Replication In Vitro. Viruses (2019) 11(6):559. doi: 10.3390/v11060559
54. Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, et al. Nuclear Factors Are Involved in Hepatitis C Virus RNA Replication. RNA (2007) 13(10):1675–92. doi: 10.1261/rna.594207
55. Li Y, Belshan M. NF45 and NF90 Bind HIV-1 RNA and Modulate HIV Gene Expression. Viruses (2016) 8(2):47. doi: 10.3390/v8020047
56. Stricker RL, Behrens SE, Mundt E. Nuclear Factor NF45 Interacts With Viral Proteins of Infectious Bursal Disease Virus and Inhibits Viral Replication. J Virol (2010) 84(20):10592–605. doi: 10.1128/JVI.02506-09
57. Wen X, Bian T, Zhang Z, Zhou L, Ge X, Han J, et al. Interleukin-2 Enhancer Binding Factor 2 Interacts With the Nsp9 or Nsp2 of Porcine Reproductive and Respiratory Syndrome Virus and Exerts Negatively Regulatory Effect on the Viral Replication. Virol J (2017) 14(1):125. doi: 10.1186/s12985-017-0794-5
58. Wu W, Wu B, Ye T, Huang H, Dai C, Yuan J, et al. TCTP Is a Critical Factor in Shrimp Immune Response to Virus Infection. PloS One (2013) 8(9):e74460. doi: 10.1371/journal.pone.0074460
Keywords: SUMOylation, BmNPV, Bombyx mori, TCTP, antivirus
Citation: Lu C, Li Z, Zhang W, Guo H, Lan W, Shen G, Xia Q and Zhao P (2022) SUMOylation of Translationally Regulated Tumor Protein Modulates Its Immune Function. Front. Immunol. 13:807097. doi: 10.3389/fimmu.2022.807097
Received: 08 November 2021; Accepted: 13 January 2022;
Published: 07 February 2022.
Edited by:
Erjun Ling, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Wuren Huang, Shanghai Institutes for Biological Sciences (CAS), ChinaYusong Xu, Zhejiang University, China
Copyright © 2022 Lu, Li, Zhang, Guo, Lan, Shen, Xia and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqing Li, lizhiqing@swu.edu.cn
 Chenchen Lu1,2
Chenchen Lu1,2 Zhiqing Li
Zhiqing Li Ping Zhao
Ping Zhao