- 1Immunology Division, Walter & Eliza Hall Institute of Medical Research, Melbourne, VIC, Australia
- 2Department of Medical Biology, The University of Melbourne, Melbourne, VIC, Australia
- 3Department of Clinical Immunology & Allergy, Royal Melbourne Hospital, Melbourne, VIC, Australia
1. Introduction
Inborn errors of immunity (IEI) are a group of inherited disorders caused by damaging variants in genes essential for immunity. Cases in which a single gene causes disease provides fundamental insights into how a single protein’s function directly impacts specific components of the immune system. Patients with IEI may present clinically with primary immunodeficiency, autoinflammation, autoimmunity and/or malignancy. IEI research is a rapidly growing field, with the recent advances in genome sequencing leading to 485 currently known monogenetic defects that cause IEI (1).
Mucosal associated invariant T (MAIT) cells are a subset of unconventional T cells that are activated following engagement of their T cell receptor (TCR) with MR1, a major histocompatibility complex (MHC) class I-related molecule that presents vitamin B metabolite antigens (2). However, MAIT cells can also be activated in a TCR-independent manner via cytokines, namely interleukin (IL)-12 and IL-18 (3). MAIT cell effector responses mirror conventional T-helper (Th)1 and Th17 cytokine profiles (4), but can also engage in CD8 T cell-like cytotoxic responses via release of granzymes and perforin (5). Due to this broad activation and effector function potential, MAIT cells have been implicated as key immune players in defense against a range of bacterial and viral infections, in addition to a role in autoimmunity and cancer (6). Despite these insights, the proteins and cells essential to support MAIT cell frequency and function, and the implications for human immunity in the context of dysfunctional MAIT cells, are only just beginning to be uncovered. Recent reports of IEI that include MAIT cell immunophenotyping, and to a limited extent functional analysis, provide an ideal opportunity to discover the fundamental factors that govern MAIT cell biology.
2. IEI with disruptions to MAIT cell compartment
Here, we present a curated review of IEI in which MAIT cells have been assessed for frequency, phenotype and/or function (Table 1). The most striking disruptions reported in IEI are cases that report a complete absence of MAIT cells (Figure 1). Complete MAIT cell deficiency, along with an expansion of γδ T cells was observed in an individual with MR1 deficiency (29). This was the result of a homozygous point mutation in the antigen binding groove of MR1, rendering it unable to present antigen. This resulted in an immune system with a selective loss of MAIT cells. This individual’s infection history included Varicella zoster viral infection (complicated by secondary bacterial pneumonia and subsequent lung scarring) prolonged Campylobacter gastroenteritis with haematochezia (which was initially refractory to treatment), and extensive human papilloma virus (HPV)+ warts refractory to treatment. This case provided direct evidence for the importance of MAIT cells’ antigen-dependent role in controlling human bacterial infections, but also highlighted their antigen-independent role in controlling human viral infections, as had been suggested by previous mouse model (45) and observational human studies (46, 47).

Table 1 Summary of inborn errors of immunity that have assessed MAIT cell frequency and/or function.
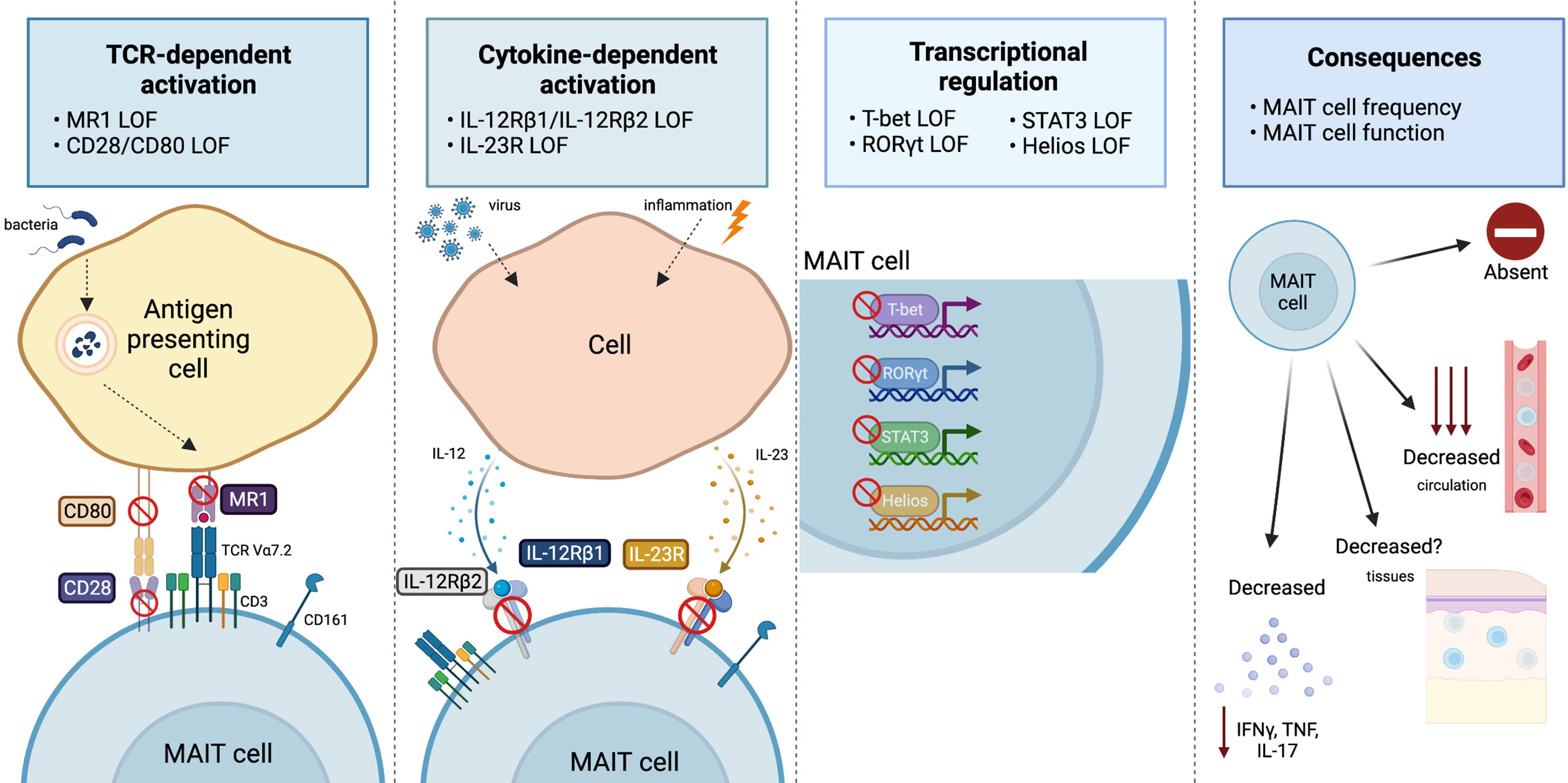
Figure 1 Overview of the range and consequences of MAIT cell activation and signaling pathways disrupted by IEI. MAIT cells can be stimulated via TCR-dependent activation, where microbial-derived vitamin B metabolites are presented on MR1 and recognized by the MAIT cell TCR Vα7.2. Disruptions to MR1 or costimulatory molecules (CD28/CD80) has been shown to impact the MAIT cell compartment. MAIT cells are also activated by viral or inflammatory conditions in which cells produce IL-12 or IL-23 in response. Cases in which IL-12Rβ1, IL-12Rβ2, or IL-23R are deficient alters the MAIT cells compartment. The transcription factors T-bet, RORγT, STAT3 and Helios all play a vital role for MAIT cell development and/or effector function, and cases in which they are deficient report alterations to the MAIT cell compartment. Ultimately, all these pathway disruptions can cause varying consequences to the MAIT cell compartment, that includes: an absence or reduction in MAIT cells in circulation (it is unknown whether this is also the case at tissue sites) or a reduction in pro-inflammatory cytokine production (IFNγ, TNF, or IL-17). Figure created with BioRender.com.
Absence of MAIT cells was also reported in seven individuals with RORγT deficiency (35), along with a lack of Th17 and natural killer T (NKT) cells in these patients, who presented with common features of candidiasis and mycobacterial disease. MAIT cells were also reportedly absent in a ZAP70-deficient patient who initially presented with CD8+ T cell lymphopenia and severe viral infections (42). These examples highlight the exceptionally rare instances of individuals with a deficiency of a protein essential for either MAIT cell development or peripheral maintenance. The immunological phenotype and clinical presentation of those with a MAIT cell deficiency were varied, but all involved disturbances to the T cell compartment and frequent, severe, or difficult to treat infections.
By far the most common observation reported across IEI describe a decrease in the proportion (or total number) of circulating MAIT cells. Reduced frequencies of circulating MAIT cells have been reported for a range of different IEI that have a diverse clinical and/or immunological presentation, including: combined immunodeficiency (CID), X-linked agammaglobulinemia (XLA), Mendelian susceptibility to mycobacterial diseases (MSMD), X-linked immunodeficiency with magnesium defect, Epstein-Barr virus infection, and neoplasia (XMEN), and X-linked lymphoproliferative (XLP) syndrome. The majority of which are characterized by altered T and/or B cell compartments. Genes with variants related to a decrease in MAIT cells can range from: costimulatory receptors (e.g. CD28) (12, 13), cell structure proteins (e.g. CARMIL2/RLTPR) (11, 16, 18), cytokine receptors (e.g. IL12RB1/IL12RB2) (24–27), DNA replication proteins (e.g. GINS1) (17, 20) and transcription factors (e.g. TBX21) (19, 22, 30, 31, 40, 43) (see Table 1 for full list).
Interestingly, a single case report described an expansion of MAIT cells in a child with c-Rel deficiency presenting with a history of severe viral, bacterial, fungal, and parasitic infections (34). Vδ1 and innate-lymphoid cells (ILC) were also expanded, and reduced frequencies of natural killer (NK) and regulatory T cells (Tregs), compared to pediatric healthy controls. However, with only a single case, it is difficult to interpret whether this MAIT cell expansion is attributable to the specific IEI, or simply individual variation. Of the IEI studies that measured and reported MAIT cell frequency, six have described frequencies of MAIT cells within a normal range in their patient cohorts (10, 14, 21, 36, 38, 44). Together, these reports demonstrate that reduced frequency of MAIT cells is a common, but not a universal, observation in IEI.
MAIT cell frequency is also impacted by loss-of-function variants in IKZF2, which encodes the T cell transcriptional regulator Helios. Helios deficiency can present as dominant or recessive CID with varying severity. A heterozygous IKZF2 variant was reported in a proband and her father presenting with mild CID characterized by recurrent upper respiratory infections, mucosal ulcers, and chronic lymphadenopathy (22). The immune phenotype was chronic activation and proinflammatory cytokine production by both effector and regulatory T cells, but immune subset frequencies largely remained intact. A homozygous IKZF2 variant in a single case presented with a more severe CID characterized by recurrent lower respiratory tract infections, leading to multiple pneumonias requiring hospitalization (23). The immune phenotype was more pronounced, with reductions in: CD4+ T, B, and NK cells and an absence of NKT cells. Even with differing presentations, both studies reported a decrease or absence of MAIT cells due to the IKZF2 variants. Together, this demonstrates that MAIT cells are particularly susceptible to changes in Helios function, compared to other immune cell subsets.
The Helios deficiency study by Hetemäki et al. (22) extended beyond the typical circulating MAIT cell enumeration to measure tissue resident MAIT cells. MAIT cells are mucosal associated as their name suggests, with a large proportion populating mucosal sites. It is not well understood whether the MAIT cell circulating frequency reflects that of their tissue-associated counterparts. Colon and duodenal biopsies were examined from two individuals with Helios deficiency and a decrease in MAIT cell frequency was observed in all tissues examined when compared to healthy donor tissue (22). Therefore, this reduction in tissue associated MAIT cells suggests a global decrease of MAIT cells, rather than a redistribution to the tissues.
Establishing whether reported changes in frequency are due directly to the inborn error itself, a result of a secondary effect of the IEI on other immune components that interact with MAIT cells, or simply the result of MAIT cells responding to a clinical history of repeated episodes of prolonged infection and inflammation, is challenging. Thus, we will next discuss the strict considerations for reporting on MAIT cells in the context of IEI.
3. Considerations for MAIT cell frequency reporting for individuals with IEI
There is no standardized method for reporting MAIT cell frequency, with variation in how they are reported and how they are identified/defined. The most common (and accessible) method for MAIT cell identification is via expression of surrogate surface markers: TCR-Vα7.2 and CD161 (10). However, it is more accurate to define MAIT cells using MR1 tetramers loaded with 5-OP-RU (48). TCR-Vα7.2+ and CD161++ cells overlap 96% with MR1-tetramer+ cells in circulation. While it is an appropriate method for identifying MAIT cells (49), it is important to consider if the IEI impacts expression of CD161, in which case MR1 tetramers should be used for identification instead.
A major issue in reporting circulating MAIT cell frequency in humans is that no standardized frequency or number values have been established. Also, MAIT cell markers/tetramers are not typically included in standard clinical T cell panels. Thus, studies either establish their own standard values, with reference to an internal healthy control group, or patient values are compared to the typical 1–5% of circulating T cells reference range set by earlier studies of MAIT cells (4, 10, 49). Importantly, this simplified reference range does not consider variation of circulating MAIT cells present in different healthy control populations. Previous studies report MAIT cell frequency is significantly impacted by both age and sex (50, 51). MAIT cells steadily increase and peak at 20–29 years of age, before progressively declining during aging (49). Thus, it is important for IEI studies to compare to an age-matched MAIT cell value for each patient, either internally or referencing external age-matched values, to confidently report any alterations to normal frequency.
Another consideration when interpreting data on MAIT cells is the infection status of the patient at the time of analysis. MAIT cells have been shown to dynamically change in frequency during an acute infection (52) with studies in mice suggesting they accumulate and expand at the tissue site of infection (45, 53). Thus, an active infection could cause a decrease in circulating MAIT cells that may be unrelated to the underlying genetic defect. In addition, the use of corticosteroids to treat asthma and chronic obstructive pulmonary disease have also been shown to impact MAIT cell frequency (54, 55). Therefore, detailed clinical history, list of current medications, and the current infection status at the time of sampling should be provided. A more definitive approach to assess circulating MAIT cell frequency is to measure multiple samples over time. This would provide important insight into the stability of any observed change in MAIT cell frequency. This approach would also control for any infection-induced fluctuations when functionally assessing T cell (including MAIT cell) activation, proliferation, and cytotoxicity markers. As these would also be expected to alter with varying infection status.
Examination of tissue biopsies (particularly from areas of inflammation or infection), although challenging to obtain, would also address the question of MAIT cell kinetics in IEI. By directly examining MAIT cell frequency at tissue sites, it could then be correlated back to the proportion of circulating cells. This would provide an understanding of the relationship between circulating and tissue-resident MAIT cell populations, and if disturbances in MAIT cell frequency are directly attributable to the underlying IEI, rather than a consequence of increased inflammation/infection-induced tissue-homing.
4. Limited MAIT cell functional analysis in IEI
A less explored aspect of MAIT cells in IEI is the potential changes in their ability to respond to stimuli. MAIT cells can be activated via TCR-dependent or TCR-independent stimulation (2, 3). Factors that control or influence these separate activation pathways in MAIT cells could be elucidated by studying the functional response of MAIT cells from IEI patients.
Several studies have examined interferon (IFN)γ production by MAIT cells in IEI. MAIT cells from a PD-1 deficient patient produced less IFNγ in response to bacille Calmette-Guérin (BCG) + IL-12 stimulation (32). Also, MAIT cells from a T-bet deficient patient produced less IFNγ in response to phorbol myristate acetate (PMA)/ionomycin stimulation (40). However, in addition to IFNγ, MAIT cells can also produce proinflammatory cytokines TNF and IL-17A, as well as cytotoxic granules and perforin, that should be considered when undertaking functional analysis (4).
The most comprehensive functional analysis of MAIT cells in IEI was in individuals with STAT3 loss-of-function (n = 5–7) (24). STAT3-deficient MAIT cells produced normal levels of IFNγ, TNF and granzyme B when stimulated with PMA/ionomycin. However, they showed impaired IL-17A production under these conditions. In addition, STAT3-deficient MAIT cells were unable to produce IL-17A or IL-17F in Th17 culture conditions, suggesting a direct role for STAT3 regulating IL17A/IL17F transcription in MAIT cells. These functional results mirror what was observed for the functional dysregulation of STAT3-deficient CD4+ (Th17) T cells in the same individuals. These observations highlight the importance of assessing polyfunctionality of MAIT cell responses to stimuli in IEI, as it may provide fundamental insights into the key proteins required for differing MAIT cell effector functions.
5. Conclusion
MAIT cells are a particularly interesting immune subset to study in IEI. Given the signaling, activation, and functional pathways shared with NKT, γδ, CD8+ and Th17 T cells, it is not surprising that MAIT cells are often at the intersection of various immune cell effector responses across innate and adaptive immunity. However, it is important when contributing to, and assessing, the literature on MAIT cells in IEI that certain key factors are taken into consideration. It is essential to understand how MAIT cells are defined, and the comparative healthy reference ranges, to make informed interpretations of the impact of IEI on MAIT cell biology. Finally, the infection status at the time of sampling can also impact the strength of conclusions of these studies. In conclusion, MAIT cells are understudied yet play a unique role in human immunity, at the intersection of innate and adaptive responses. Understanding MAIT cells in the context of IEI provides an opportunity to understand their role and potential to contribute to immune dysregulation in IEI.
Author contributions
LJH: conceptualization, writing - original draft preparation. VLB: conceptualization, writing - reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
LJH is supported by a National Health and Medical Research Council of Australia (NHMRC) Investigator Grant (2007884) and a Jack Brockhoff Foundation Early Career Research Grants (JBF 4847-2021). VLB is supported by Sir Clive McPherson Family Research Fellowship and DW Keir Fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human inborn errors of immunity: 2022 update on the classification from the international union of immunological societies expert committee. J Clin Immunol (2022) 42:1473–507. doi: 10.1007/s10875-022-01289-3
2. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L. Et al. MR1 presents microbial vitamin b metabolites to MAIT cells. Nature. (2012) 491(7426):717–23. doi: 10.1038/nature11605
3. Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol (2014) 44(1):195–203. doi: 10.1002/eji.201343509
4. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117(4):1250–9. doi: 10.1182/blood-2010-08-303339
5. Kurioka A, Ussher JE, Cosgrove C, Clough C, Fergusson JR, Smith K, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol (2015) 8(2):429–40. doi: 10.1038/mi.2014.81
6. Howson LJ, Salio M, Cerundolo V. MR1-restricted mucosal-associated invariant T cells and their activation during infectious disease. Front Immunol (2015) 6:303. doi: 10.3389/fimmu.2015.00303
7. Yap JY, Moens L, Lin MW, Kane A, Kelleher A, Toong C, et al. Intrinsic defects in b cell development and differentiation, T cell exhaustion and altered unconventional T cell generation characterize human adenosine deaminase type 2 deficiency. J Clin Immunol (2021) 41(8):1915–35. doi: 10.1007/s10875-021-01141-0
8. Kaleviste E, Rühlemann M, Kärner J, Haljasmägi L, Tserel L, Org E, et al. 22 paucity in APECED is associated with mucosal and microbial alterations in oral cavity. Front Immunol (2020) 11:838. doi: 10.3389/fimmu.2020.00838
9. Garcia-Solis B, Van Den Rym A, Pérez-Caraballo JJ, Al-Ayoubi A, Alazami AM, Lorenzo L, et al. Clinical and immunological features of human BCL10 deficiency. Front Immunol (2021) 12:786572. doi: 10.3389/fimmu.2021.786572
10. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PloS Biol (2009) 7(3):e54. doi: 10.1371/journal.pbio.1000054
11. Wang Y, Ma CS, Ling Y, Bousfiha A, Camcioglu Y, Jacquot S, et al. Dual T cell- and b cell-intrinsic deficiency in humans with biallelic RLTPR mutations. J Exp Med (2016) 213(11):2413–35. doi: 10.1084/jem.20160576
12. Ghosh S, Köstel Bal S, Edwards ESJ, Pillay B, Jiménez Heredia R, Erol Cipe F, et al. Extended clinical and immunological phenotype and transplant outcome in CD27 and CD70 deficiency. Blood. (2020) 136(23):2638–55. doi: 10.1182/blood.2020006738
13. Béziat V, Rapaport F, Hu J, Titeux M, Bonnet des Claustres M, Bourgey M, et al. Humans with inherited T cell CD28 deficiency are susceptible to skin papillomaviruses but are otherwise healthy. Cell. (2021) 184(14):3812–28.e30. doi: 10.1016/j.cell.2021.06.004
14. Bucciol G, Pillay B, Casas-Martin J, Delafontaine S, Proesmans M, Lorent N, et al. Systemic inflammation and myelofibrosis in a patient with takenouchi-kosaki syndrome due to CDC42 Tyr64Cys mutation. J Clin Immunol (2020) 40(4):567–70. doi: 10.1007/s10875-020-00742-5
15. Smith DJ, Hill GR, Bell SC, Reid DW. Reduced mucosal associated invariant T-cells are associated with increased disease severity and pseudomonas aeruginosa infection in cystic fibrosis. PloS One (2014) 9(10):e109891. doi: 10.1371/journal.pone.0109891
16. Moshous D, Martin E, Carpentier W, Lim A, Callebaut I, Canioni D, et al. Whole-exome sequencing identifies coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated b-cell lymphoproliferation. J Allergy Clin Immunol (2013) 131(6):1594–603. doi: 10.1016/j.jaci.2013.01.042
17. Martin E, Minet N, Boschat AC, Sanquer S, Sobrino S, Lenoir C, et al. Impaired lymphocyte function and differentiation in CTPS1-deficient patients result from a hypomorphic homozygous mutation. JCI Insight (2020) 5(5):e133880. doi: 10.1172/jci.insight.133880
18. Pillay BA, Avery DT, Smart JM, Cole T, Choo S, Chan D, et al. Hematopoietic stem cell transplant effectively rescues lymphocyte differentiation and function in DOCK8-deficient patients. JCI Insight (2019) 5(11):e127527. doi: 10.1172/jci.insight.127527
19. Dickinson RE, Milne P, Jardine L, Zandi S, Swierczek SI, McGovern N, et al. The evolution of cellular deficiency in GATA2 mutation. Blood. (2014) 123(6):863–74. doi: 10.1182/blood-2013-07-517151
20. Cottineau J, Kottemann MC, Lach FP, Kang YH, Vély F, Deenick EK, et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. (2017) 127(5):1991–2006. doi: 10.1172/JCI90727
21. Kerner G, Rosain J, Guérin A, Al-Khabaz A, Oleaga-Quintas C, Rapaport F, et al. Inherited human IFN-γ deficiency underlies mycobacterial disease. J Clin Invest. (2020) 130(6):3158–71. doi: 10.1172/JCI135460
22. Hetemäki I, Kaustio M, Kinnunen M, Heikkilä N, Keskitalo S, Nowlan K, et al. Loss-of-function mutation in IKZF2 leads to immunodeficiency with dysregulated germinal center reactions and reduction of MAIT cells. Sci Immunol (2021) 6(65):eabe3454. doi: 10.1126/sciimmunol.abe3454
23. Shahin T, Kuehn HS, Shoeb MR, Gawriyski L, Giuliani S, Repiscak P, et al. Germline biallelic mutation affecting the transcription factor Helios causes pleiotropic defects of immunity. Sci Immunol (2021) 6(65):eabe3981. doi: 10.1126/sciimmunol.abe3981
24. Wilson RP, Ives ML, Rao G, Lau A, Payne K, Kobayashi M, et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med (2015) 212(6):855–64. doi: 10.1084/jem.20141992
25. Martínez-Barricarte R, Markle JG, Ma CS, Deenick EK, Ramírez-Alejo N, Mele F, et al. Human IFN-γ immunity to mycobacteria is governed by both IL-12 and IL-23. Sci Immunol (2018) 3(30):eaau6759. doi: 10.1126/sciimmunol.aau6759
26. Cagdas D, Mayr D, Baris S, Worley L, Langley DB, Metin A, et al. Genomic spectrum and phenotypic heterogeneity of human IL-21 receptor deficiency. J Clin Immunol (2021) 41(6):1272–90. doi: 10.1007/s10875-021-01031-5
27. Béziat V, Tavernier SJ, Chen YH, Ma CS, Materna M, Laurence A, et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J Exp Med (2020) 217(6):e20191804. doi: 10.1084/jem.20191804.
28. Klinken EM, Gray PE, Pillay B, Worley L, Edwards ESJ, Payne K, et al. Diversity of XMEN disease: Description of 2 novel variants and analysis of the lymphocyte phenotype. J Clin Immunol (2020) 40(2):299–309. doi: 10.1007/s10875-019-00732-2
29. Howson LJ, Awad W, von Borstel A, Lim HJ, McWilliam HEG, Sandoval-Romero ML, et al. Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1. Sci Immunol (2020) 5(49). doi: 10.1126/sciimmunol.abc9492
30. Gonzalez-Granado LI, Ruiz-García R, Blas-Espada J, Moreno-Villares JM, Germán-Diaz M, López-Nevado M, et al. Acquired and innate immunity impairment and severe disseminated mycobacterium genavense infection in a patient with a NF-κB1 deficiency. Front Immunol (2018) 9:3148. doi: 10.3389/fimmu.2018.03148
31. Klemann C, Camacho-Ordonez N, Yang L, Eskandarian Z, Rojas-Restrepo JL, Frede N, et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol (2019) 10:297. doi: 10.3389/fimmu.2019.00297
32. Ogishi M, Yang R, Aytekin C, Langlais D, Bourgey M, Khan T, et al. Inherited PD-1 deficiency underlies tuberculosis and autoimmunity in a child. Nat Med (2021) 27(9):1646–54. doi: 10.1038/s41591-021-01388-5
33. Winter S, Martin E, Boutboul D, Lenoir C, Boudjemaa S, Petit A, et al. Loss of RASGRP1 in humans impairs T-cell expansion leading to Epstein-Barr virus susceptibility. EMBO Mol Med (2018) 10(2):188–99. doi: 10.15252/emmm.201708292
34. Lévy R, Langlais D, Béziat V, Rapaport F, Rao G, Lazarov T, et al. Inherited human c-rel deficiency disrupts myeloid and lymphoid immunity to multiple infectious agents. J Clin Invest (2021) 131(17). doi: 10.1172/JCI150143
35. Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, et al. IMMUNODEFICIENCIES. impairment of immunity to candida and mycobacterium in humans with bi-allelic RORC mutations. Science. (2015) 349(6248):606–13. doi: 10.1126/science.aaa4282
36. Rigaud S, Fondanèche MC, Lambert N, Pasquier B, Mateo V, Soulas P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. (2006) 444(7115):110–4. doi: 10.1038/nature05257
37. Delmonte OM, Bergerson JRE, Kawai T, Kuehn HS, McDermott DH, Cortese I, et al. SASH3 variants cause a novel form of X-linked combined immunodeficiency with immune dysregulation. Blood. (2021) 138(12):1019–33. doi: 10.1182/blood.2020008629
38. Kong XF, Martinez-Barricarte R, Kennedy J, Mele F, Lazarov T, Deenick EK, et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol (2018) 19(9):973–85. doi: 10.1038/s41590-018-0178-z
39. Schaballie H, Rodriguez R, Martin E, Moens L, Frans G, Lenoir C, et al. A novel hypomorphic mutation in STIM1 results in a late-onset immunodeficiency. J Allergy Clin Immunol (2015) 136(3):816–9.e4. doi: 10.1016/j.jaci.2015.03.009
40. Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, et al. Human T-bet governs innate and innate-like adaptive IFN-γ immunity against mycobacteria. Cell. (2020) 183(7):1826–47.e31. doi: 10.1016/j.cell.2020.10.046
41. Martin-Fernandez M, Buta S, Le Voyer T, Li Z, Dynesen LT, Vuillier F, et al. A partial form of inherited human USP18 deficiency underlies infection and inflammation. J Exp Med (2022) 219(4). doi: 10.1084/jem.20211273
42. Hauck F, Blumenthal B, Fuchs S, Lenoir C, Martin E, Speckmann C, et al. SYK expression endows human ZAP70-deficient CD8 T cells with residual TCR signaling. Clin Immunol (2015) 161(2):103–9. doi: 10.1016/j.clim.2015.07.002
43. Béziat V, Li J, Lin JX, Ma CS, Li P, Bousfiha A, et al. A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci Immunol (2018) 3(24):eaat4956. doi: 10.1126/sciimmunol.aat4956
44. Le Voyer T, Neehus AL, Yang R, Ogishi M, Rosain J, Alroqi F, et al. Inherited deficiency of stress granule ZNFX1 in patients with monocytosis and mycobacterial disease. Proc Natl Acad Sci U.S.A. (2021) 118(15). doi: 10.1073/pnas.2102804118
45. van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun (2018) 9(1):4706. doi: 10.1038/s41467-018-07207-9
46. van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nat Commun (2016) 7:11653. doi: 10.1038/ncomms11653
47. Flament H, Rouland M, Beaudoin L, Toubal A, Bertrand L, Lebourgeois S, et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat Immunol (2021) 22(3):322–35. doi: 10.1038/s41590-021-00870-z
48. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-Cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509(7500):361–5. doi: 10.1038/nature13160
49. Gherardin NA, Souter MN, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol (2018) 96(5):507–25. doi: 10.1111/imcb.12021
50. Novak J, Dobrovolny J, Novakova L, Kozak T. The decrease in number and change in phenotype of mucosal-associated invariant T cells in the elderly and differences in men and women of reproductive age. Scand J Immunol (2014) 80(4):271–5. doi: 10.1111/sji.12193
51. Lee OJ, Cho YN, Kee SJ, Kim MJ, Jin HM, Lee SJ, et al. Circulating mucosal-associated invariant T cell levels and their cytokine levels in healthy adults. Exp Gerontol. (2014) 49:47–54. doi: 10.1016/j.exger.2013.11.003
52. Howson LJ, Napolitani G, Shepherd D, Ghadbane H, Kurupati P, Preciado-Llanes L, et al. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with salmonella paratyphi a. Nat Commun (2018) 9(1):253. doi: 10.1038/s41467-017-02540-x
53. Wang H, D'Souza C, Lim XY, Kostenko L, Pediongco TJ, Eckle SBG, et al. MAIT cells protect against pulmonary legionella longbeachae infection. Nat Commun (2018) 9(1):3350. doi: 10.1038/s41467-018-05202-8
54. Hinks TS, Zhou X, Staples KJ, Dimitrov BD, Manta A, Petrossian T, et al. Innate and adaptive T cells in asthmatic patients: Relationship to severity and disease mechanisms. J Allergy Clin Immunol (2015) 136(2):323–33. doi: 10.1016/j.jaci.2015.01.014
55. Hinks TS, Wallington JC, Williams AP, Djukanović R, Staples KJ, Wilkinson TM. Steroid-induced deficiency of mucosal-associated invariant T cells in the chronic obstructive pulmonary disease lung. implications for nontypeable haemophilus influenzae infection. Am J Respir Crit Care Med (2016) 194(10):1208–18. doi: 10.1164/rccm.201601-0002OC
Keywords: MAIT cells, immunodeficiency - primary, autoimmunity, immunogenetics, infection, inborn errors of immunity
Citation: Howson LJ and Bryant VL (2022) Insights into mucosal associated invariant T cell biology from human inborn errors of immunity. Front. Immunol. 13:1107609. doi: 10.3389/fimmu.2022.1107609
Received: 25 November 2022; Accepted: 09 December 2022;
Published: 22 December 2022.
Edited by:
Timothy Stopford Christopher Hinks, University of Oxford, United KingdomReviewed by:
Chie Sugimoto, Dokkyo Medical University, JapanCopyright © 2022 Howson and Bryant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren J. Howson, aG93c29uLmxAd2VoaS5lZHUuYXU=
 Lauren J. Howson
Lauren J. Howson Vanessa L. Bryant
Vanessa L. Bryant