- 1General Surgery Center, Beijing YouAn Hospital, Capital Medical University, Beijing, China
- 2Department of Liver Disease Center, Beijing YouAn Hospital, Capital Medical University, Beijing, China
- 3Biomedical Information Center, Beijing YouAn Hospital, Capital Medical University, Beijing, China
Introduction: Although we had identified that the methylation of AHNAK was a good diagnostic marker for hepatopathy, here we speculate that there was also another marker, STAP1, whose methylation also involved in the detection of hepatopathy.
Methods: We investigated the methylation levels of the AHNAK and STAP1 in peripheral blood mononuclear cells of chronic hepatitis B (CHB) patients, compensatory liver cirrhosis (CLC) patients, decompensated liver cirrhosis (DCLC) patients, hepatocellular carcinoma (HCC) patients and healthy controls by methylation-specific PCR. We also evaluated the differences and changes of methylation and expression of AHNAK and STAP1 at different stages of liver disease using the TCGA and GEO public datasets.
Results: Methylation level of STAP1 in PBMC was positively correlated with the course of liver cancer. The combination of AHNAK and STAP1 methylation was able to predict differrent HBV related hepatopathy. The GEO datasets also supported that the methylation of AHNAK and STAP1 was associated with different types of hepatopathy. The TCGA data showed that the levels of methylation and expression of STAP1 were down-regulated in HCC. We also found the STAP1 methylation level in PBMC and T cells was associated with age, gender, alcohol drinking and anti-HBe. Hyper-methylation of STAP1 was correlated with the poor prognosis of patients but its expression had no association.
Conclusion: We concluded that combination of AHNAK and STAP1 methylation in peripheral blood immune cells can be used as a diagnostic marker for HBV related hepatopathy and STAP1 methylation may be a potential prognostic marker for HBV related HCC. Our clinical study registration number was ChiCTR2000039860.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common neoplasm and the third most common cause of cancer-related deaths in the world (1–3). Almost all patients with liver cirrhosis (4) and a large portion of those with chronic liver disease eventually progress to liver fibrosis and HCC (5). Because the majority of people with liver cirrhosis are asymptomatic and because HCC patients frequently come with hepatitis and HCC symptoms at an advanced stage, the effectiveness of treatment is frequently limited (6, 7). Therefore, finding efficient biomarkers for early diagnosis detection, therapy evaluation, and prognosis prediction of liver cancer is therefore very valuable.
Emerging evidence suggests that many genetic and epigenetic abnormalities are involved in the development of HCC (8). DNA methylation is recognized as one of the first epigenetic modifications discovered, which can regulate gene expression by influencing the chromatin structure, DNA conformation, DNA stability, and DNA–protein action mode (9). A growing number of studies have shown that DNA methylation can be used as a diagnostic and prognostic molecular marker for multiple diseases (10–12). Our previous study has reported that the promoter methylation level of AHNAK gene in peripheral blood mononuclear cells (PBMC) was associated with the increase of the severity of liver diseases, and AHNAK can be employed as an early diagnostic marker for HCC (13). However, a few patients did not conform to the AHNAK-based rule. In addition, our previous research also showed that STAP1 methylation in PBMC is also associated with the progression of hepatitis B virus (HBV)-related liver diseases. Therefore, in this study, we collected new patient samples to investigate the correlation between the methylation of another gene, STAP1, as well as AHNAK, and the progression of hepatitis and HCC and to assess the diagnostic effect of the combination using the genes STAP1 and AHNAK.
The protein STAP1 is a substrate of tyrosine-protein kinase Tec, participating in a positive feedback loop by upregulating the activity of tyrosine-protein kinase Tec. Although STAP1 is reported to be involved in B cell receptor signaling (14) and in the prognosis gene signature of lung adenocarcinoma (15), the role of STAP1 in HCC is not clear. We here used methylation-specific polymerase chain reaction (MSP) to study the STAP1 and AHNAK methylation level in hepatitis B patients and its correlation to the progress of HCC from BCLC stage 0 to stage C. We also explored the association of methylation level and expression level of STAP1 and AHNAK to the different stages of HCC and survival of patients using several public datasets. The current study aimed to explore the diagnostic and prognostic value of STAP1 methylation in liver cirrhosis and HCC combined with AHNAK.
Materials and methods
Patients and collection of samples
A total of 302 HBV-infected patients in Beijing YouAn Hospital, Capital Medical University were recruited during June 2014 to August 2020, including 46 cases of chronic hepatitis B (CHB), 46 cases of compensatory liver cirrhosis (CLC), 53 cases of decompensated liver cirrhosis (DCLC), 157 cases of HCC (stage 0 = 34, stage A = 42, stage B =19, and stage C = 62). In total, 22 healthy volunteers were recruited as controls during a routine physical examination, and none of them had hepatobiliary diseases before the peripheral blood sample collection. The whole PBMC suspension was prepared by taking 3–5 ml peripheral blood of the patients into EDTA anticoagulant tubes. T cells were also isolated from the PBMC by using the magnetic activated cell sorter (MACS®) T-cell isolation kit following the manufacturer’s instructions. All the participants signed informed consents, and the study was approved by the medical ethics committee of Beijing YouAn Hospital (BJYACE-201721).
DNA extraction, bisulfite modification, and MSP
Genomic DNA was extracted from PBMC or T cells of patients and healthy controls using the QIAamp DNA mini kit (Qiagen NV, Venlo, The Netherlands) in line with the manufacturer’s protocol. The quality and the quantity of the isolated DNA were measured by NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). The bisulfite treatment of genomic DNA was processed using an EpiTect Fast DNA Bisulfite Kit (Qiagen NV). The bisulfite-treated DNA samples were next kept at -20°C for further use. After analyzing a 2,000-bp region upstream of the transcription start site (AHNAK or STAP1 promoter region), one CpG island was identified (Figure 1A). With bisulfite-treated DNA as the template, the methylation pattern in the CpG island within AHNAK or STAP1 promoter was measured by MSP, and the MSP primers are presented in Supplementary Table S1. The MSP primers were designed according to the principle described previously (16). The reaction condition for MSP was 10 min at 95°C, followed by 40 cycles of 30 s at 94°C and 30 s at 72°C, and then 5 min of extension at 72°C. After having been separated on 1.5% agarose gels, the MSP products were then stained by ethidium bromide and visualized under a UV spectrophotometer. Water blanks served as the negative control. We obtained the methylation levels of AHNAK or STAP1 in PBMC or T cells.
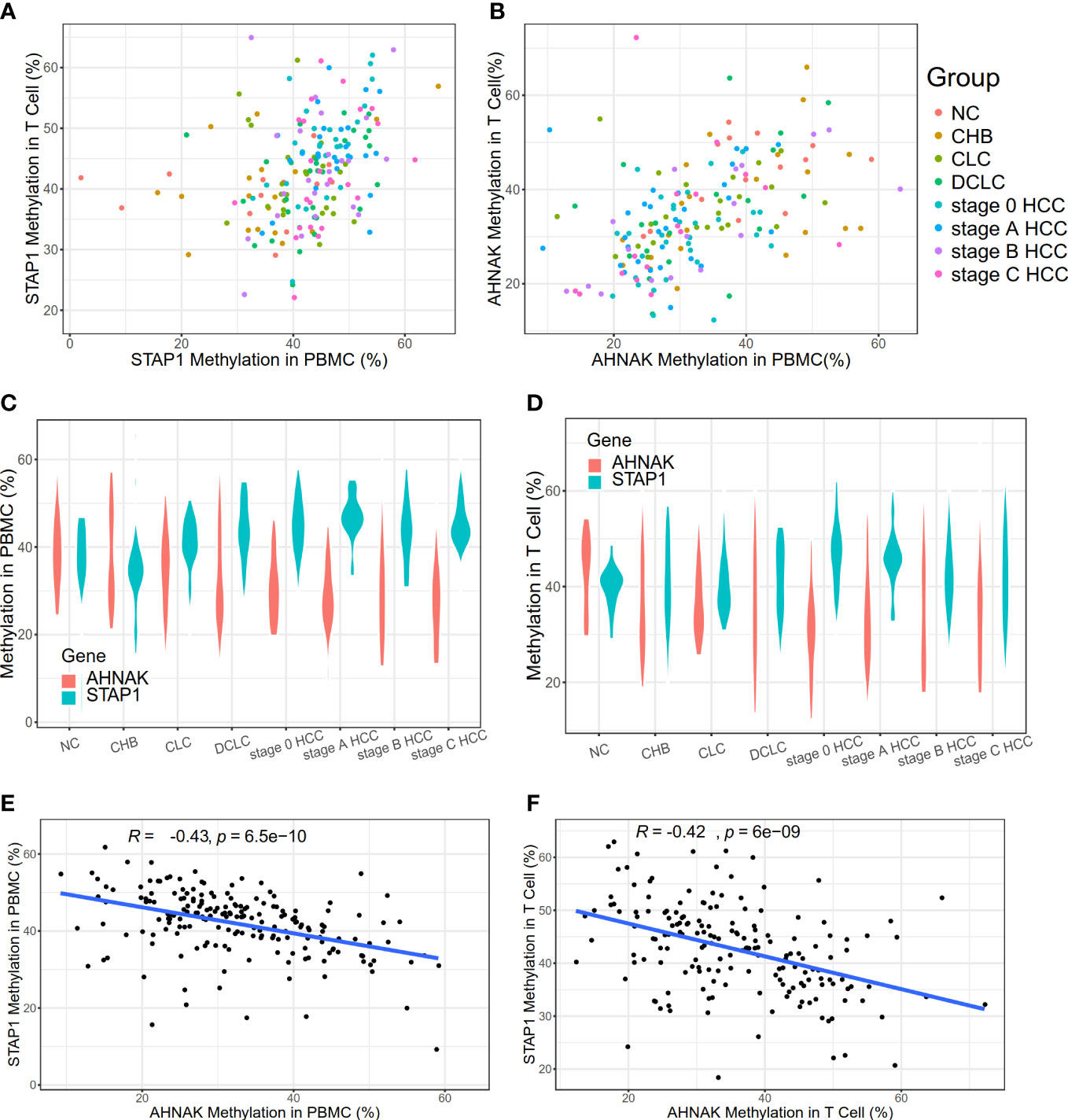
Figure 1 Methylation level of AHNAK and STAP1 in peripheral blood mononuclear cells (PBMC) and T cells in different liver diseases. (A) Methylation levels of STAP1 in PBMC and T cells. (B) Methylation levels of AHNAK in PBMC and T cells. (C) Methylation levels of AHNAK and STAP1 in PBMC at different disease stages. (D) Methylation level of AHNAK and STAP1 in T cells at different disease stages. (E, F) Negative correlation of methylation of AHNAK and STAP1 in PBMC and T cells.
Public datasets used
The expression and methylation data of HCC in the LIHC project in The Cancer Genome Atlas (TCGA) (17) was used to confirm the correlation of AHNAK and STAP1 with the HCC and the overall survival of patients. DNA methylation data from the Gene Expression Omnibus (GEO) database were employed to investigate the methylation profile of AHNAK and STAP1 in the different types of liver cirrhosis or HCC: (1) methylation profiling of 1,204 HCC patients, 392 patients with CHB or liver cirrhosis, and 958 healthy individuals and patients with benign liver lesions (GSE112679); (2) methylation profiling of 34 healthy liver tissues and 122 primary liver disease tissues arising in the setting of chronic HBV or C viral infection, alcoholism (EtOH) (GSE60753); and (3) methylation profiling of 48 HBsAg carriers who developed HCC and 48 HBsAg carriers without HCC during follow-up (GSE78904). The normalized data of those GEO datasets were downloaded through the GEOquery package in R.
Statistical analysis
All 302 HBV-infected patients in Beijing YouAn Hospital were included in the statistical analysis. Student’s t-test and one-way analysis of variance (ANOVA) were used to compare the differences of AHNAK or STAP1 methylation between groups. Spearman’s correlation was carried out to compare the difference of continuous variables. Linear regression was utilized to compare the difference of discrete variables. Correlations between the methylation levels of STAP1 or AHNAK and the clinical characteristics (including ages, sex, alcohol drinking, and anti-HBeAb) were determined via Spearman rank correlation analysis. Using the data of patients with information of overall survival (OS), we investigated the association of methylation or expression of STAP1 as well as AHNAK to the patients’ OS. We grouped the patients into uppers and lowers according to the median of methylation or the expression level of STAP1 or AHNAK. Kaplan–Meier curves with log-rank test were used for the overall survival. Statistical significance was defined as two-tailed P <0.05 for all analyses. To assess the diagnostic value of STAP1 and AHNAK methylation levels as markers for CHB, CLC, DCLC, or HCC, respectively, we performed receiver operating characteristic (ROC) analysis based on their methylation levels and used the area under the curve (AUC) as an assessment of diagnostic accuracy. The ROC curve analysis was performed using SPSS19.0 statistical software. Other data were analyzed using statistical software R3.1.1 or GraphPad Prism version 6.0.
Results
The methylation level of STAP1 in PBMC was positively correlated with the course of liver cancer, while AHNAK had the reverse correlation
We found out that there was no significant difference in the methylation level of STAP1 or AHNAK between the PBMC and T cells (Figures 1A, B). The STAP1 methylation level was positively correlated with the severity of the liver disease not only in the PBMC but also the T cells, while AHNAK showed a negative correlation (Figures 1C, D). Thus, there was a reverse correlation between AHNAK and STAP1 in the PBMC and T cells (Figures 1E, F). The ANOVA test suggested that the methylation levels of AHNAK and STAP1 both showed significant differences among different groups whether in the PBMC or T cells (Table 1). A further two-paired comparison of STAP1 methylation levels demonstrated that there existed significant differences between NC and DCLC and HCC (stage 0, A, B, C), between CHB and DCLC and HCC, and between CLC and DCLC, stage 0 HCC, and stage C HCC in the PBMC. There were significant differences between NC, CHB, CLC, and HCC stage 0 and HCC stage A in the T cells (Supplementary Tables S2–S5).
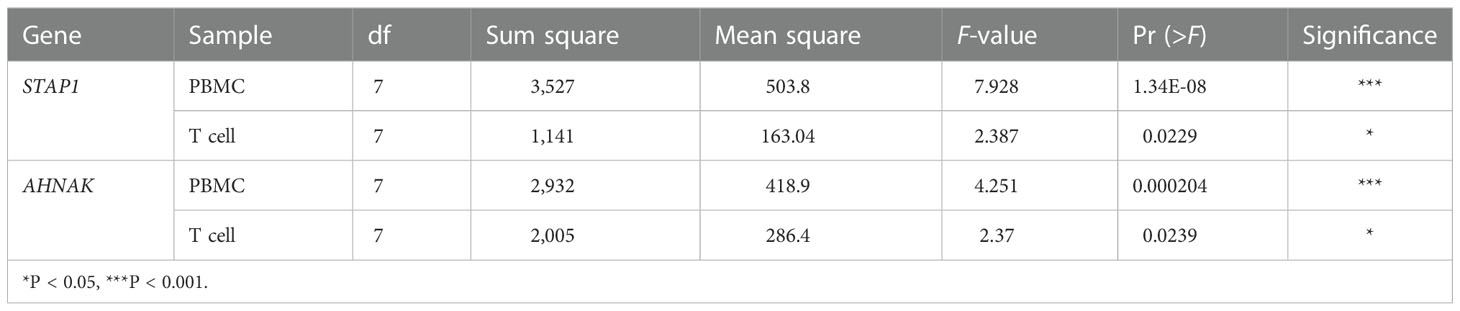
Table 1 AHNAK and STAP1 methylation level among different groups in peripheral blood mononuclear cells (PBMC) and T cells.
Diagnostic value of STAP1 and AHNAK methylation level in patients with different liver diseases
To further verify whether STAP1 or AHNAK methylation in PBMC can be used as an indicator for the diagnosis of liver diseases, the ROC curve was drawn for the analysis (Figure 2). The STAP1 and AHNAK methylation levels were not highly specific for identifying CHB patients. STAP1 methylation was also not specific for predicting patients with CLC (AUC = 0.65 in PBMC and AUC = 0.54 in T cells); however, AHNAK methylation in T cells was predictive of CLC (AUC = 0.7). The methylation of either STAP1 or AHNAK in PBMC may be predictive of DCLC, with AUCs of 0.74 and 0.70, respectively. Either STAP1 or AHNAK demonstrated pretty high specificity in predicting HCC, where the AUC of methylation of either STAP1 or AHNAK in PBMC was mostly greater than 0.70.
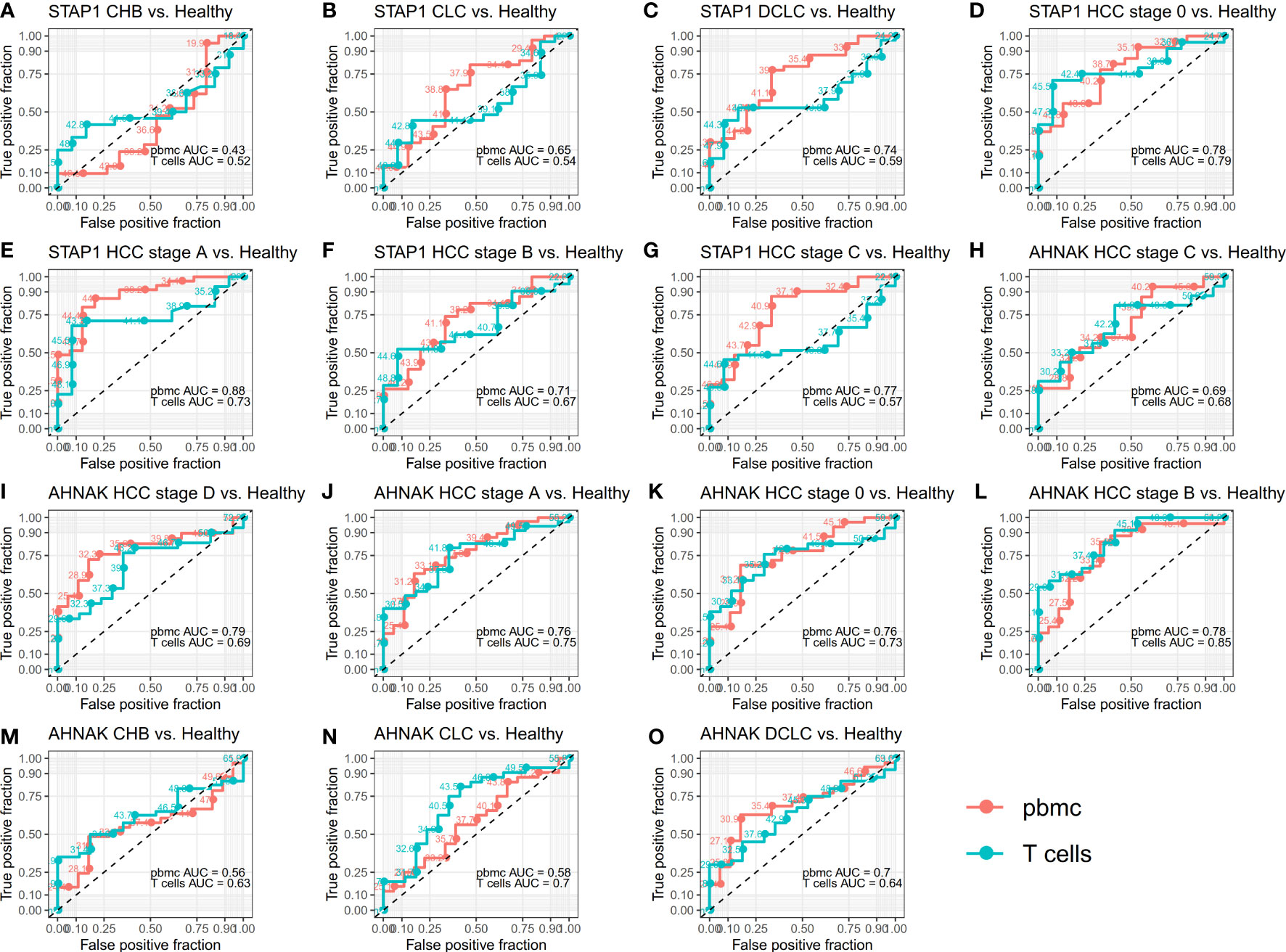
Figure 2 Receiver operating characteristic (ROC) curves for whether STAP1 or AHNAK methylation in peripheral blood mononuclear cells (PBMC) can be used as an indicator for the diagnosis of liver disease. (A–G) ROC curves for methylation of STAP1 in PBMC and T cells. (H-O), ROC curves for methylation of AHNAK in PBMC and T cells.
Methylation level of AHNAK and STAP1 also showed significant differences among different types of liver diseases in the public datasets
In order to confirm the association of STAP1 and AHNAK with liver diseases, we further analyzed the AHNAK and STAP1 methylation levels in the different types of liver cirrhosis or HCC using the GEO datasets. In the GSE60753 data, there was a significant difference in STAP1 methylation level among liver cirrhosis and HCC (Kruskal−Wallis test, p = 1.2e−06) (Figure 3A). In the GSE112679 data, the methylation level of STAP1 was also significantly higher than in healthy controls and CHB and HCC patients (Figure 3B). In the GSE112679 data, AHNAK also showed a significantly high methylation level in liver cirrhosis than other types, and it was significantly higher in HCC compared with benign liver lesions and CHB (Figure 3C).
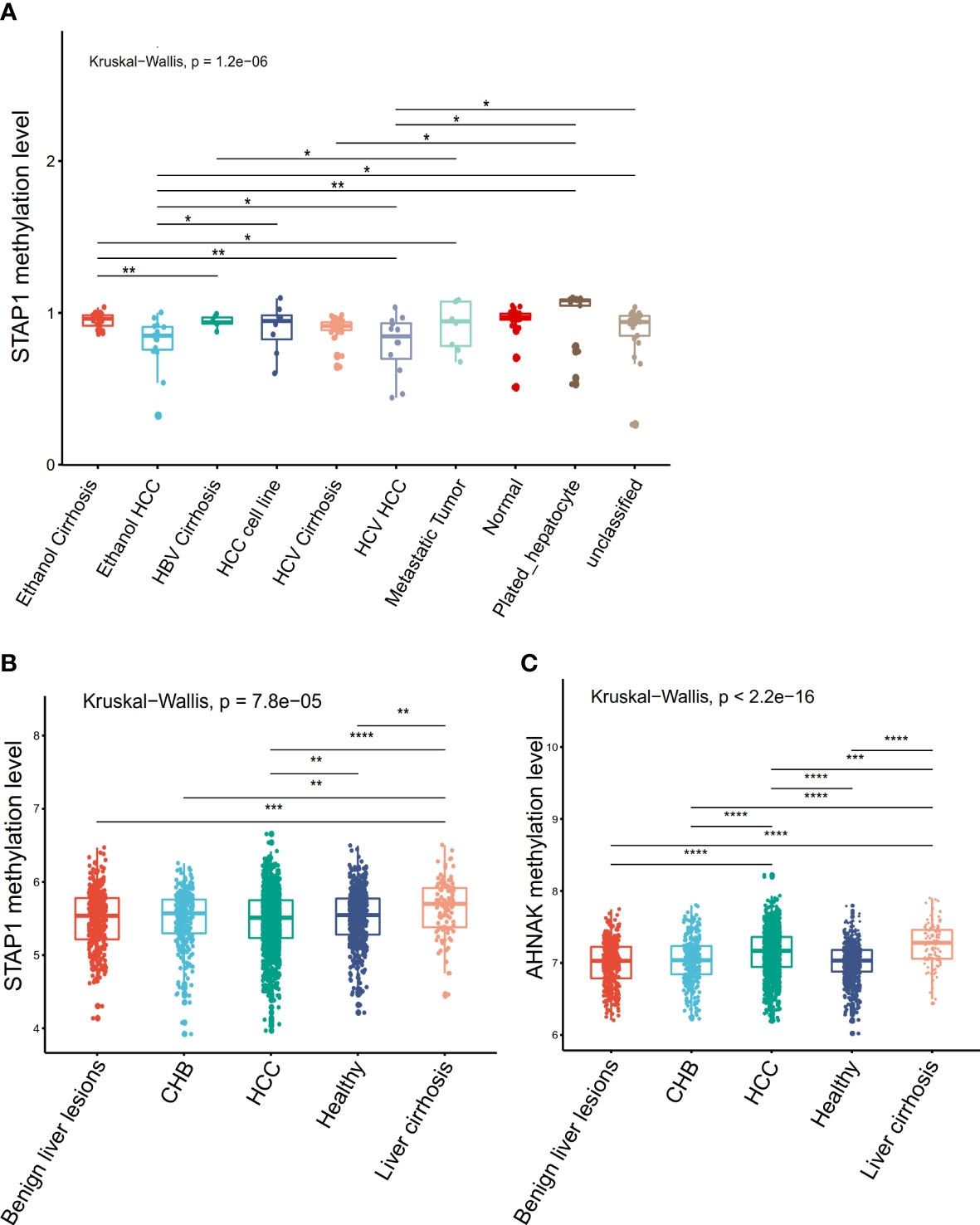
Figure 3 Comparison of the methylation of AHNAK and STAP1 among different types of liver disease in the Gene Expression Omnibus datasets. (A) Comparisons in STAP1 methylation among the different types of liver cirrhosis and hepatocellular carcinoma (HCC) patients (GSE60753 data). (B) Comparisons in STAP1 methylation among the chronic hepatitis B (CHB), liver cirrhosis, and HCC patients and benign liver lesions (GSE112679 data). (C) Comparisons in AHNAK methylation among the CHB, liver cirrhosis, and HCC patients and benign liver lesions (GSE112679 data). *P < 0.05, **P < 0.01, ***p < = 0.001, ****p < = 0.0001.
Lower methylation level and expression level of STAP1 and AHNAK in liver cancer tissues
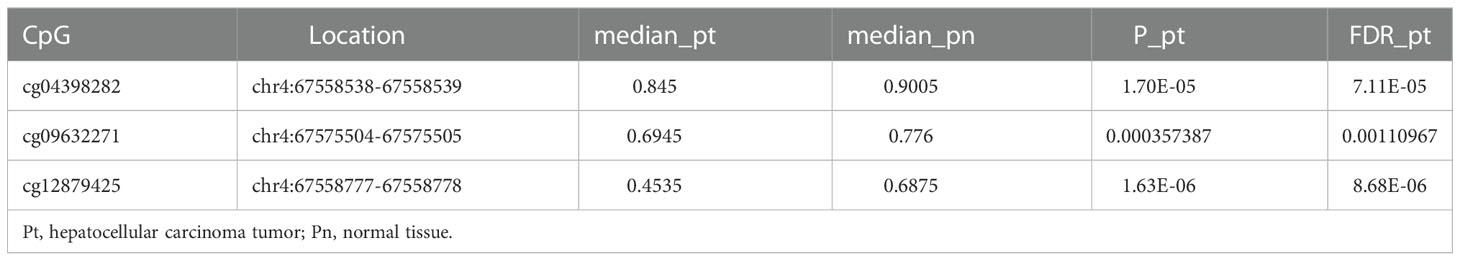
We found that the cancer tissues displayed a lower methylation level of STAP1 than the adjacent tissues (by t-test, P < 0.05) (Table 2). However, there was no significant difference between HBsAg carriers who developed HCC and HBsAg carriers who did not during follow-up in the GSE78904 data (Figure 4A).
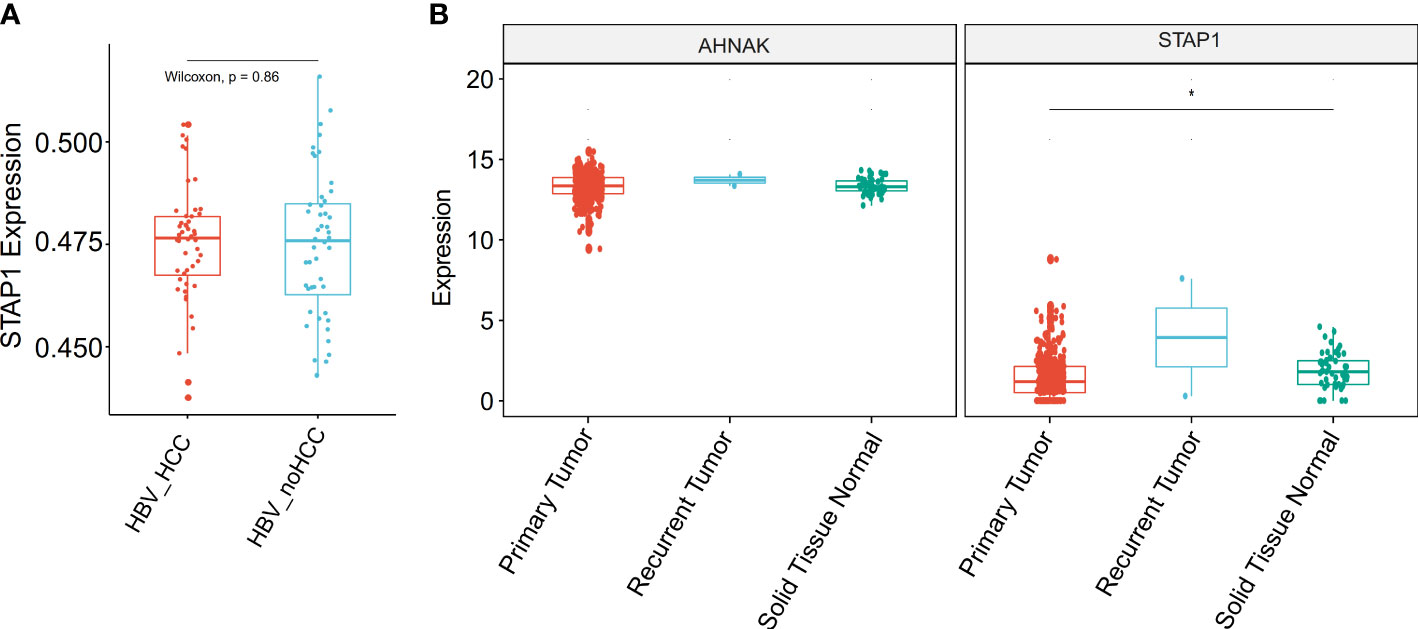
Figure 4 Comparison of the methylation and expression of AHNAK and STAP1 between hepatocellular carcinoma (HCC) and normal patients. (A) Comparison of the methylation level of STAP1 between HBsAg carriers who developed HCC and HBsAg carriers who did not. (B) Comparison of the expression level of AHNAK and STAP1 between tumor and normal tissues in The Cancer Genome Atlas data. *P < 0.05.
As gene methylation is able to regulate the gene expression of itself, we analyzed the expression levels of STAP1 between primary tumor and solid normal tissue in TCGA data. We found that STAP1 had a lower expression in primary tumor, while AHNAK showed no significant difference (Wilcoxon rank sum test, P < 0.05) (Figure 4B).
STAP1 methylation level was pertinent to age, sex, alcohol drinking, and anti-HBe
In our new collection of patients with HBV, we found that the STAP1 methylation level was positively correlated with age not only in the PBMC (R = 0.289, p = 1.9e-5) but also in T cells (R = 0.243, p = 6.2e-4) (Figures 5A, B). The STAP1 methylation level in the PBMC was significantly higher in male than in female patients, while there was no significant difference in T cells (Figure 5C). Similarly, the STAP1 methylation level in the PBMC was significantly higher in the alcohol-drinking patients than in the non-alcoholic patients and higher in the anti-HBe+ patients than in the anti-HBe- patients, but it showed no significant difference in T cells (Figures 5D, E). In the same set of patients, the AHNAK methylation level, as it was negatively correlated with STAP1 methylation, was lower in the older patients and was significantly higher in the female patients, in the non-alcoholic patients, and in non-smoking patients (Supplementary Figure S1).
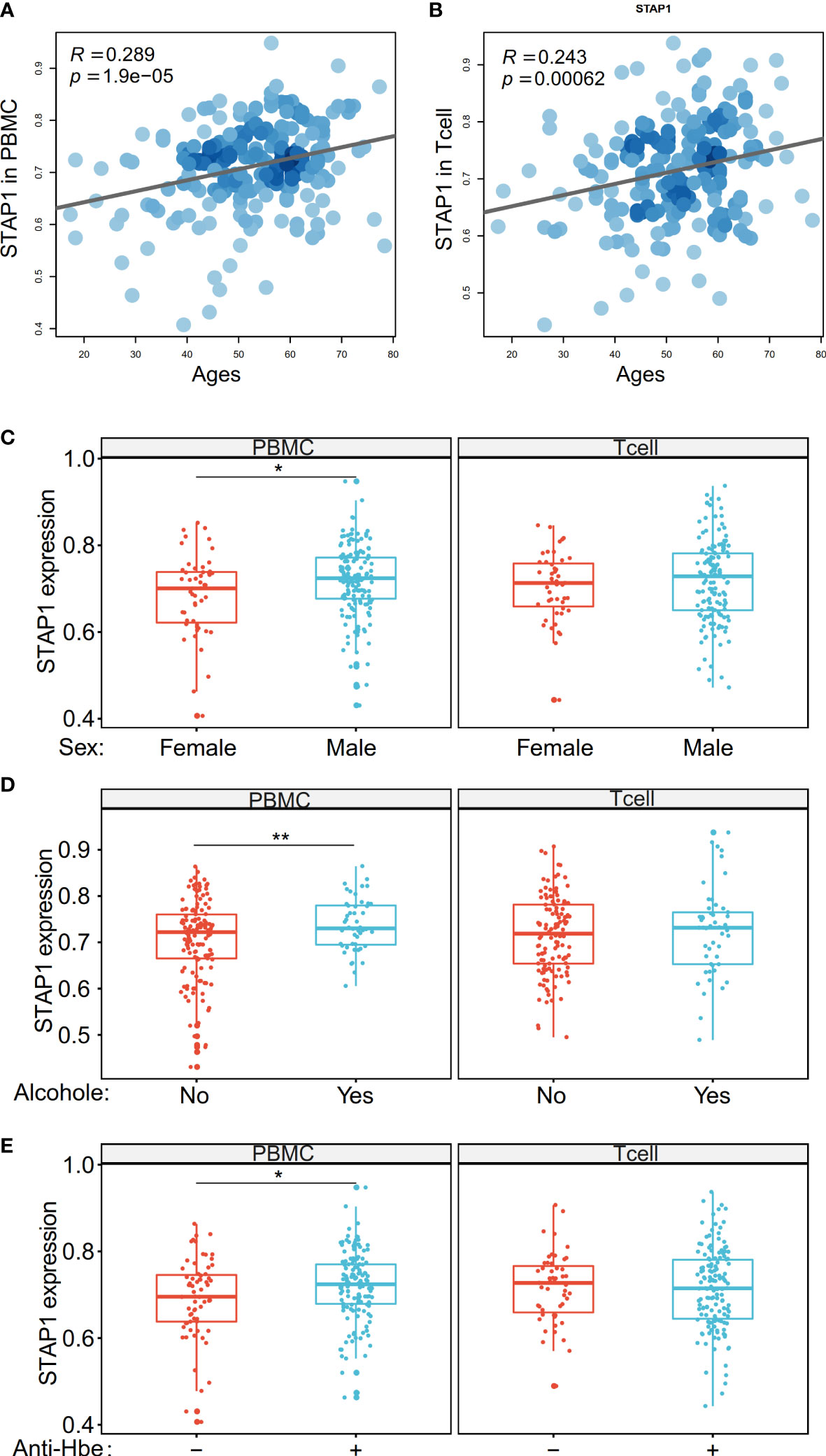
Figure 5 The methylation level of STAP1 was correlated with age, sex, alcohol drinking, and anti-HBe in peripheral blood mononuclear cells (PBMC) and T cells. (A, B) Correlation of STAP1 methylation level with ages in PBMC and T cells. (C) Difference of STAP1 methylation in different genders in PBMC and T cells. (D) Difference of STAP1 methylation between PBMC and T cells relative to alcohol abuse. (E) Difference of STAP1 methylation between anti-HBe+ and anti-HBe- patients in PBMC and T cells. *P < 0.05, **P < 0.01.
On the other hand, we verified the relationship between the clinical characteristics and STAP1 and AHNAK in the HCC patients using the TCGA data. The STAP1 methylation level was associated with postoperative rx tx (Figure 6A), gender (Figure 6B), and age (Figure 6C), while the AHNAK methylation level was associated with additional pharmaceutical therapy (Figure 6D) and sex (Figure 6B).
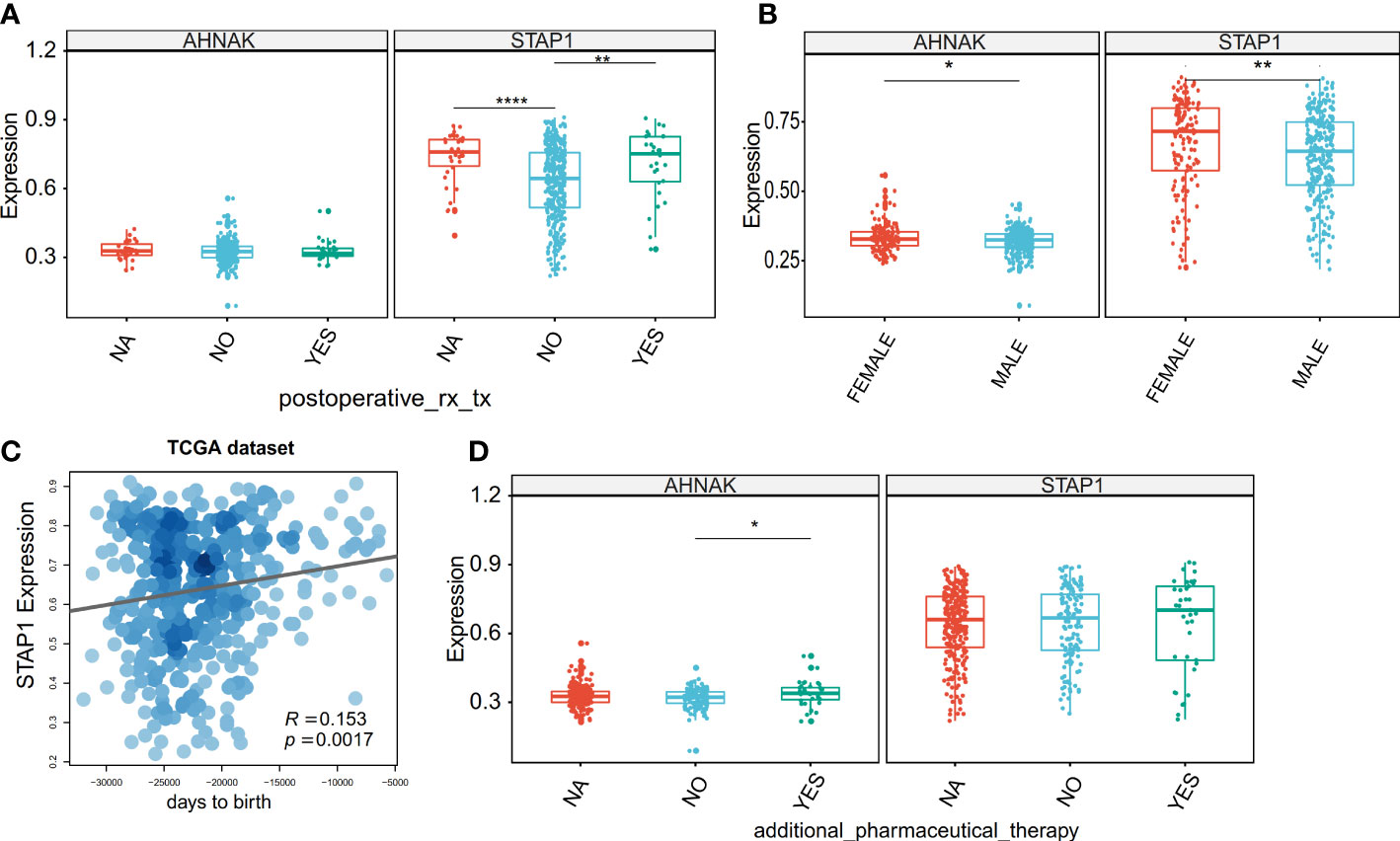
Figure 6 Association between the methylation level of STAP1 or AHNAK and clinical characteristics in The Cancer Genome Atlas data. (A) Difference of STAP1 and AHNAK methylation between whether postoperative rx tx. (B) Difference of STAP1 and AHNAK methylation between different genders. (C) Correlation of STAP1 methylation with ages. (D) Difference of STAP1 and AHNAK methylation between whether additional pharmaceutical therapy was used or not. *P < 0.05, **P < 0.01, ****P < 0.0001.
The combination of AHNAK and STAP1 methylation levels was associated with the overall survival of patients
The result suggested that methylation of STAP1 was significantly associated with OS, and it was a risk factor (HR = 1.49, P = 0.0282), but the expression level of STAP1 was not significantly associated with OS (HR = 0.75, P = 0.0908) (Figures 7A, B). Neither the methylation nor the expression of AHNAK was significantly associated with OS (HR <1, P > 0.05) (Figures 7C, D). However, there was a significant distinction in OS (HR = 0.98, P = 0.0396) when using the combination of methylation of STAP1 and AHNAK with the patients who carried lower AHNAK and upper STAP1 having poor OS (Figure 7E).
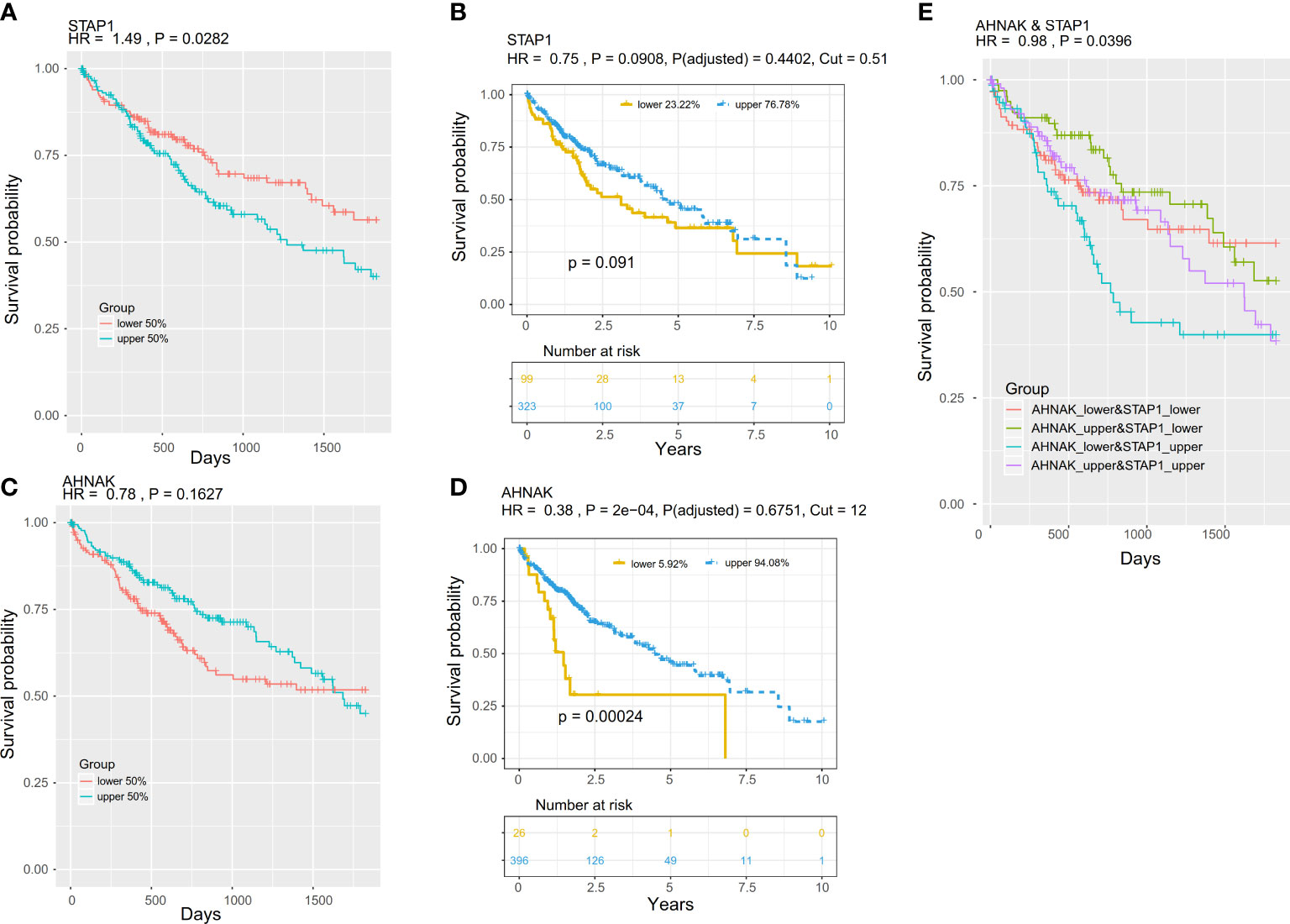
Figure 7 Prognostic value of the methylation and expression of AHNAK and STAP1. The K–M survival curves show the overall survival of patients in The Cancer Genome Atlas project. The patients were stratified (upper and lower) according to the median of the methylation or expression level. (A, B) K–M survival curves for STAP1 methylation (A) and expression (B). (C, D) K–M survival curves for the AHNAK methylation (C) and expression (D). K–M survival curves for the combination of methylation of AHNAK and STAP1. (E) OS of different AHNAK &STAP1 combined groups.
Discussion
AHNAK, as a good diagnostic marker [with AUC as 0.883 (P < 0.001) in the diagnosis of CHB, 0.885 (P < 0.001) in the diagnosis of compensatory liver cirrhosis, 0.955 (P < 0.001) in the diagnosis of decompensated liver cirrhosis, and 0.981 (P < 0.001) in the diagnosis of hepatocellular carcinoma], has been identified in our previous research (13). We here employed a new set of patients to investigate the methylation level of a novel marker, STAP1, as well as AHNAK, in the PBMC and T cells also in different course samples of hepatitis B. We obtained a similar result for AHNAK. In this study, we investigated the methylation of STAP1 and found that the methylation of STAP1 was negatively correlated with the methylation of AHNAK. Although AHNAK can distinguish hepatitis B patients to a certain extent in the methylation, the combination of these two markers can achieve better performance. It is reported that STAP1 was associated with hypercholesterolemia (18–20). However, there were a few reports about STAP1 in cancer (14, 15) and no publication for hepatopathy. Therefore, our study is the first to discover the relationship of methylation of STAP1 with hepatopathy and HCC. The combination of methylation of STAP1 and AHNAK indeed achieved a better performance in distinguishing the different types of hepatopathy patients, which suggests the diagnostic value of STAP1.
On the other hand, using the expression data and the methylation data of public datasets, we investigate the relationship between the methylation and the expression of STAP1 and AHNAK. For AHNAK, there was hypo-methylation shown in the HCC than in adjacent tissues in TCGA dataset but conversely in the GEO data. In our data, we found a negative correlation of the methylation of AHNAK and STAP1 in the PBMC. However, in the public data, there was hyper-methylation of both AHNAK and STAP1 in liver cirrhosis and hypo-methylation both in the HCC and adjacent tissues. The expression of the gene can be downregulated by the hyper-methylation of its promoter region through inhibition of the binding of transcription factors or recruitment of methyl-CpG-binding proteins to silence the gene (21, 22). However, with the data including expression and methylation simultaneously in TCGA, we found decreases in both the methylation and the expression levels of STAP1 in HCC. We think that the contradiction mentioned above is due to the difference between tissue and blood or the difference between the detected positions of methylation.
Similar to AHNAK, STAP1 methylation was also related to alcohol drinking, which suggests that these bad habits do affect the occurrence of cancer. Liver cancer is often diagnosed at an advanced stage and has a high mortality rate (23, 24). Therefore, early diagnosis is very important. The combination of these two genes can be used in the early screening and prognostic judgments and provide a basis for the formulation of treatment strategies.
To date, little is known about the role of STAP1 in hepatocarcinogenesis. There are several intracellular signaling pathways that derive from the STAP-1 and STAP-2 members of the signal-transducing adaptor protein (STAP) family. These proteins have Pleckstrin homology in their N-terminal sections and SRC homology 2 domains in their middle regions, which are common architectures for adaptor proteins. During carcinogenesis and inflammatory/immune reactions, STAP proteins interact with the inhibitor of nf-κb kinase complex and activator of transcription 3. In hepatocellular carcinoma, the aberrant methylation of STAP1 may also contribute to hepatocarcinogenesis via these signaling pathways. However, there are some limitations in the present study. First, only PBMC and T cell specimens were used in this study, although the methylation and the expression status in liver tissues were investigated using the public data. The finding in the PBMC and T cell for those two genes was not overall consistent with that in the tissues, and even the different datasets showed different status. Thereby, the intrahepatic methylation of AHNAK and STAP1 still needs to be studied in the future. However, the main purpose of our study was to identify the potential clinical diagnostic marker for HBV-related patients, and it is beneficial to use peripheral blood immune cells in the clinical application. In future research, we should investigate the methylation status of STAP1 in different tissues. Second, the MSP approach that we used here only tests whether methylation occurs; other approaches such as gene sequencing would be more helpful. However, it is undeniable that MSP is of excellent specificity, sensitivity, and operability in frequent detection. Finally, the mechanisms underlying STAP1 and AHNAK have not been studied here. Especially for STAP1, little is known about its function, and thus in the future, an experimental investigation about it should be carried out to understand its functional role in HBV-related hepatopathy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving human participants were reviewed and approved by the medical ethics committee of Beijing YouAn Hospital (BJYACE-201721). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
LS contributed to sample and data acquisition and manuscript drafting. JL, KL, HZ, and XZ provided sample and data acquisition and technical support. GL and NL made substantial contributions to the conception and design, funding, and supervision of the study. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from Beijing Natural Science Foundation (7222096), Key Medical Professional Development Plan of Beijing Hospital Authority (ZYLX202124), and Chenxiaoping Technology Development Grant (CXPJJH120008-08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor WY declared a shared affiliation with the authors at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1091103/full#supplementary-material
References
1. Zhang F, Lee J, Liang S, Shum CK. Cyanobacteria blooms and non-alcoholic liver disease: evidence from a county level ecological study in the united states. Environ Health (2015) 14:41. doi: 10.1186/s12940-015-0026-7
2. Wan M, El-Nezami H. Targeting gut microbiota in hepatocellular carcinoma: Probiotics as a novel therapy. Hepatobiliary Surg Nutr (2018) 7(1):11–20. doi: 10.21037/hbsn.2017.12.07
3. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y
4. Takano S, Yokosuka O, Imazeki F, Tagawa M, Omata M. Incidence of hepatocellular carcinoma in chronic hepatitis b and c: A prospective study of 251 patients. Hepatology (1995) 21(3):650–5. doi: 10.1002/hep.1840210308
5. Manos MM, Leyden WA, Murphy RC, Terrault NA, Bell BP. Limitations of conventionally derived chronic liver disease mortality rates: Results of a comprehensive assessment. Hepatology (2008) 47(4):1150–7. doi: 10.1002/hep.22181
6. Obi S, Yoshida H, Toune R, Unuma T, Kanda M, Sato S, et al. Combination therapy of intraarterial 5-fluorouracil and systemic interferon-alpha for advanced hepatocellular carcinoma with portal venous invasion. Cancer (2006) 106(9):1990–7. doi: 10.1002/cncr.21832
7. Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology (2016) 150(4):835–53. doi: 10.1053/j.gastro.2015.12.041
8. Liu WR, Shi YH, Peng YF, Fan J. Epigenetics of hepatocellular carcinoma: a new horizon. Chin Med J (Engl) (2012) 125(13):2349–60. doi: 10.3760/cma.j.issn.0366-6999.2012.13.020
9. Kumar S, Karmakar BC, Nagarajan D, Mukhopadhyay AK, Morgan RD, Rao DN. N4-cytosine DNA methylation regulates transcription and pathogenesis in helicobacter pylori. Nucleic Acids Res (2018) 46(7):3815. doi: 10.1093/nar/gky126
10. Borssen M, Nordlund J, Haider Z, Landfors M, Larsson P, Kanerva J, et al. DNA Methylation holds prognostic information in relapsed precursor b-cell acute lymphoblastic leukemia. Clin Epigenet (2018) 10:31. doi: 10.1186/s13148-018-0466-3
11. Fontecha-Barriuso M, Martin-Sanchez D, Ruiz-Andres O, Poveda J, Sanchez-Nino MD, Valino-Rivas L, et al. Targeting epigenetic DNA and histone modifications to treat kidney disease. Nephrol Dial Transplant (2018) 33(11):1875–86. doi: 10.1093/ndt/gfy009
12. Kato M, Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol (2019) 15(6):327–45. doi: 10.1038/s41581-019-0135-6
13. Sun L, Li K, Liu G, Xu Y, Zhang A, Lin D, et al. Distinctive pattern of AHNAK methylation level in peripheral blood mononuclear cells and the association with HBV-related liver diseases. Cancer Med (2018) 7(10):5178–86. doi: 10.1002/cam4.1778
14. Steeghs E, Bakker M, Hoogkamer AQ, Boer JM, Hartman QJ, Stalpers F, et al. High STAP1 expression in DUX4-rearranged cases is not suitable as therapeutic target in pediatric b-cell precursor acute lymphoblastic leukemia. Sci Rep (2018) 8(1):693. doi: 10.1038/s41598-017-17704-4
15. Ma C, Luo H, Cao J, Zheng X, Zhang J, Zhang Y, et al. Identification of a novel tumor microenvironment-associated eight-gene signature for prognosis prediction in lung adenocarcinoma. Front Mol Biosci (2020) 7:571641. doi: 10.3389/fmolb.2020.571641
16. Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics (2002) 18(11):1427–31. doi: 10.1093/bioinformatics/18.11.1427
17. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet (2013) 45(10):1113–20. doi: 10.1038/ng.2764
18. Fouchier SW, Dallinga-Thie GM, Meijers JC, Zelcer N, Kastelein JJ, Defesche JC, et al. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ Res (2014) 115(6):552–5. doi: 10.1161/CIRCRESAHA.115.304660
19. Blanco-Vaca F, Martin-Campos JM, Perez A, Fuentes-Prior P. A rare STAP1 mutation incompletely associated with familial hypercholesterolemia. Clin Chim Acta (2018) 487:270–4. doi: 10.1016/j.cca.2018.10.014
20. Amor-Salamanca A, Castillo S, Gonzalez-Vioque E, Dominguez F, Quintana L, Lluis-Ganella C, et al. Genetically confirmed familial hypercholesterolemia in patients with acute coronary syndrome. J Am Coll Cardiol (2017) 70(14):1732–40. doi: 10.1016/j.jacc.2017.08.009
21. Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology (2011) 140(5):1501–12.e2. doi: 10.1053/j.gastro.2011.02.006
22. Maemura K, Yoshikawa H, Yokoyama K, Ueno T, Kurose H, Uchiyama K, et al. Delta-like 3 is silenced by methylation and induces apoptosis in human hepatocellular carcinoma. Int J Oncol (2013) 42(3):817–22. doi: 10.3892/ijo.2013.1778
23. Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci (2019) 62(5):640–7. doi: 10.1007/s11427-018-9461-5
Keywords: STAP1, AHNAK, DNA methylation, hepatopathy, diagnosis, prognosis, HBV
Citation: Sun L, Lu J, Li K, Zhang H, Zhao X, Li G and Li N (2023) Diagnostic and prognostic value of STAP1 and AHNAK methylation in peripheral blood immune cells for HBV-related hepatopathy. Front. Immunol. 13:1091103. doi: 10.3389/fimmu.2022.1091103
Received: 06 November 2022; Accepted: 19 December 2022;
Published: 13 January 2023.
Edited by:
Wei Yi, Beijing Ditan Hospital, Capital Medical University, ChinaCopyright © 2023 Sun, Lu, Li, Zhang, Zhao, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangming Li, bGlndWFuZ21pbmdAY2NtdS5lZHUuY24=; Ning Li, bGluaW5neWFAY2NtdS5lZHUuY24=
 Libo Sun
Libo Sun Junfeng Lu
Junfeng Lu Kang Li
Kang Li Haitao Zhang1
Haitao Zhang1 Guangming Li
Guangming Li