
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 05 January 2023
Sec. Immunological Tolerance and Regulation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1079294
This article is part of the Research Topic Immune Responses and Immunotolerance in the Process of Reproduction View all 16 articles
 Xu Liu1,2†
Xu Liu1,2† Guolin He1,3†
Guolin He1,3† Yue Lan1
Yue Lan1 Weijie Guo1
Weijie Guo1 Xuyuan Liu1
Xuyuan Liu1 Jing Li1
Jing Li1 Anqing Liu4
Anqing Liu4 Miao He4
Miao He4 Xinhui Liu1,3
Xinhui Liu1,3 Zhenxin Fan1,2*
Zhenxin Fan1,2* Yaoyao Zhang1,3*
Yaoyao Zhang1,3*Studies have shown that fetal immune cell activation may result from potential exposure to microbes, although the presence of microbes in fetus has been a controversial topic. Here, we combined metagenomic and virome techniques to investigate the presence of bacteria and viruses in fetal tissues (small intestine, cecum, and rectum). We found that the fetal gut is not a sterile environment and has a low abundance but metabolically rich microbiome. Specifically, Proteobacteria and Actinobacteria were the dominant bacteria phyla of fetal gut. In total, 700 species viruses were detected, and Human betaherpesvirus 5 was the most abundant eukaryotic viruses. Especially, we first identified Methanobrevibacter smithii in fetal gut. Through the comparison with adults’ gut microbiota we found that Firmicutes and Bacteroidetes gradually became the main force of gut microbiota during the process of growth and development. Interestingly, 6 antibiotic resistance genes were shared by the fetus and adults. Our results indicate the presence of microbes in the fetal gut and demonstrate the diversity of bacteria, archaea and viruses, which provide support for the studies related to early fetal immunity. This study further explores the specific composition of viruses in the fetal gut and the similarities between fetal and adults’ gut microbiota, which is valuable for understanding human fetal immunity development during gestation.
Immune system is crucial to recognize and exclude antigenic foreign particles and maintain the stability of the internal environment. The human immune system begins to develop early in fetal development and has obvious sensitivity to external antigens (1–3). The fetal immunity is likely influenced by fragments and metabolites of maternal gut microbes, whereas the presence of the microbiome in utero has been a controversial topic (4). Earlier reports have shown that the amniotic cavity and placenta are sterile (5–10). The superior defenses of placenta mean it’s extremely difficult for microbes to enter the uterine environment.
It is widely known that the placenta is an essential organ for material exchange and the primary barrier between the mother and the fetus during human pregnancy. The placenta composed of amnion, villous trees and decidua basalis, has the function of defense, synthesis and immunity. Anchoring villi, an integral part of the villous tree, are attached to decidua basalis by extravillous trophoblasts (EVTs). Fetal blood passes through the umbilical artery to the villous capillaries and exchanges material with maternal blood in the intervillous space, but fetal blood and maternal blood are not directly connected. The villous trees of placenta at full term are covered by syncytiotrophoblast, and there is a layer of cytotrophoblasts below which is discontinuous. The inner layer of cytotrophoblasts layer is the basement membrane, which acts as the placental barrier (11). In addition, syncytiotrophoblasts and cytotrophoblasts both provide effective protection against viral and non-viral pathogens. Among them, the surface of syncytiotrophoblast has unique physical properties, and the physical barrier formed by syncytiotrophoblast limits the vertical transmission of pathogens at multiple stages of pregnancy (12–16).
However, with the development of microbial detection technology, more and more evidence suggest the presence of microbes in human placenta and fetus (17–26). Studies supporting the sterile womb hypothesis suggest that the microbial signals detected in the womb are actually due to contamination of samples and the DNA purification kits (21, 27). Researches supporting the presence of a low biomass placental microbiome suggest that after filtering out contaminants and low-quality sequences according to negative controls, some microbial signals still exist (26, 28, 29). Excitingly, a recent study showed that microbial exposure reduces fetal immune cells early in human development. This study demonstrated the presence of microbes in fetal organs by inoculating fetal tissue in culture media and visualizing fetal guts, and suggested that these bacteria induce the activation of syngeneic memory T cells in fetal mLN T cells. And the question of how do microbes get into the uterine environment, has it been shown that pathogens (Zika virus, Toxoplasma gondii, HIV, Cytomegalovirus, etc.) might target multiple cells in the decidua to reach the extravillous trophoblasts (EVTs) layer and eventually bypass the syncytial layer (13, 30, 31). Microbes might use this mechanism to breach the placental barrier, but are more likely to evolve distinct strategies at different stages of pregnancy (11).
Most of the recent studies on fetal microbes are based on 16S rRNA gene amplicon sequencing and metagenomic sequencing (10, 26, 32, 33). Metagenomic data includes bacterial, archaear, protozoa, virus, fungus and host genomes. Compared to 16S rRNA sequencing, the DNA used for metagenomic sequencing is not amplified by PCR, and the metagenomic results are relatively unbiased. Besides, the composition, abundance and function of microbiota can be obtained by metagenomic sequencing (34). However, due to the low biomass of fetal samples, it is difficult to detect archaea and virus signals using 16S rRNA and metagenomic sequencing. Virome is a combination of metagenomic theory and existing virus molecular biological detection technology, mainly used for the studies of all viruses genetic material in the environment (35). Therefore, in order to detect as many microbial signals as possible, virome sequencing were used in this study to explore the fetal gut microbiome based on the metagenomic results.
Human fetal tissues were obtained in accordance from West China 2nd University Hospital with ethic approval of Ethics Committees of West China 2nd University Hospital. All women gave written consent to the use of fetal tissues according to internationally recognized guidelines (36). All fetal tissues (gut) were obtained from 2nd trimester (12-22 weeks) elective pregnancy terminations. The fetus was considered structurally normal on ultrasound examination prior to termination and by gross morphological examination following termination. Fetal tissues from 2nd trimester of gestation were used for this study. The participant (or mother, in the case of fetal samples) gave written informed consent. Mid-trimester terminations were medically induced and the fetus was delivered through the birth canal. Fetal organs were collected under sterile conditions in a tissue culture hood. Aseptic equipment was used for collecting the intestinal contents of fetal small intestine, cecum and rectum. The main experimental route was shown in Figure 1.
Fetal samples were sent to Chengdu Life Baseline Technology Co., Ltd. for metagenomic sequencing. The DNA samples were extracted using Tiangen DNA Stool Mini Kit (TIANGEN Biotech Co., Ltd. China) with the manufacturer’s instructions. In total, 0.2 μg DNA per sample was used for the DNA library preparations after DNA extraction. Sequencing library was generated using NEBNext® UltraTM DNA Library Prep Kit for Illumina (NEB, USA, Catalog #: E7370L). The assessment of library quality and quantity was performed by Agilent 5400 system(Agilent, USA) and QPCR (1.5 nM), respectively. The qualified libraries were sequenced on Illumina NovaSeq 6000 with pair-end 150bp reads.
Virome sequencing was performed in Chengdu Life Baseline Technology Co., Ltd. To remove debris and cells, samples were centrifuged at 2,500 x g for 5 minutes, and supernate was passed through a 0.45 μm filter after another centrifugation (5,000 x g, 20 min). After the treatment with 2 ul lysozyme (50 mg/ml) at 37 °C for 30 minutes, samples were treated with 0.2x volume chloroform at RT for 10 minutes. Then 10U Tubro DNase I (Ambion), 2 ug RNase A (Roche) or 20 U of RNase I (ThermoFisher Scientific) were added to the new centrifugation supernate (17,000 x g, 10 min) followed by heat inactivation at 65 °C for 10 minutes. VLPs DNA extraction and quantification were performed by Qiagen MinElute virus kit and Qubit dsDNA HS Assay Kit (ThermoFisher Scientific), respectively. After the library preparation, sequencing was performed on an Illumina Nova Seq 6000 platform using pair-end 150bp reads.
To compare the gut microbiota of the fetus and adults, we downloaded 13 gut metagenomic data of healthy adults from public database (https://www.ncbi.nlm.nih.gov/). The raw data obtained from metagenomic and virome sequencing was used for subsequent analysis. Trimmomatic was used to remove the adapters and low-quality reads of raw reads after the sequencing with the setting of average quality per base >20 and minimum length 90 bp (37). The host contamination was removed by Bowtie2 with human reference genome (38). MEGAHIT (39) was used to the de novo assembly (–min-contig-len 300). We performed gene prediction and translation of amino acid sequences by Prodigal (40) and DIAMOND (41), respectively. The taxonomic annotation were assigned by Kraken2 (42) with option “–use-mpa-style”. Functional annotations, including microbial metabolic pathway and ARGs, were assessed by using HUMANn3 (43) and comprehensive antibiotic resistance database (CARD) (44).
Metagenomic sequencing was performed on the contents of the fetal small intestine, cecum and rectum, a total of 486 species of bacteria were detected, including 8 phyla and 238 genera (Figure 2A–C). Among the 486 species, 120 species were shared by small intestine, cecum and rectum, 172 species were the peculiar species of small intestine, and the number of peculiar species in rectum was 55 (Figure 2D). As shown in Table 1, the top 3 phyla in the small intestine and cecum were Proteobacteria, Actinobacteria and Bacteroidetes. However, Firmicutes replaced Bacteroidetes as the third phylum in the rectum. At the genus level, the top 3 of the small intestine (specifically Microbacterium, Burkholderia and Rhizobium) and rectum (specifically Microbacterium, Rhizobium and Burkholderia) were similar. While there were significant changes in cecum, the top 3 genera of cecum were Burkholderia, Microbacterium and Paraburkholderia. At the specie level, the top 3 in relative abundances of the small intestine and rectum were Microbacterium sp. LKL04, Methylorubrum populi and Agrococcus sp. SGAir0287, while the top 3 species of cecum were Agrococcus sp. SGAir0287, Paraburkholderia fungorum and Burkholderia multivorans. In addition, very tiny amounts of viruses and archaea were detected. To further explore the function of fetal gut microbiome, we performed the functional enrichment analysis by HUMANn3, and there was no pathway enriched.
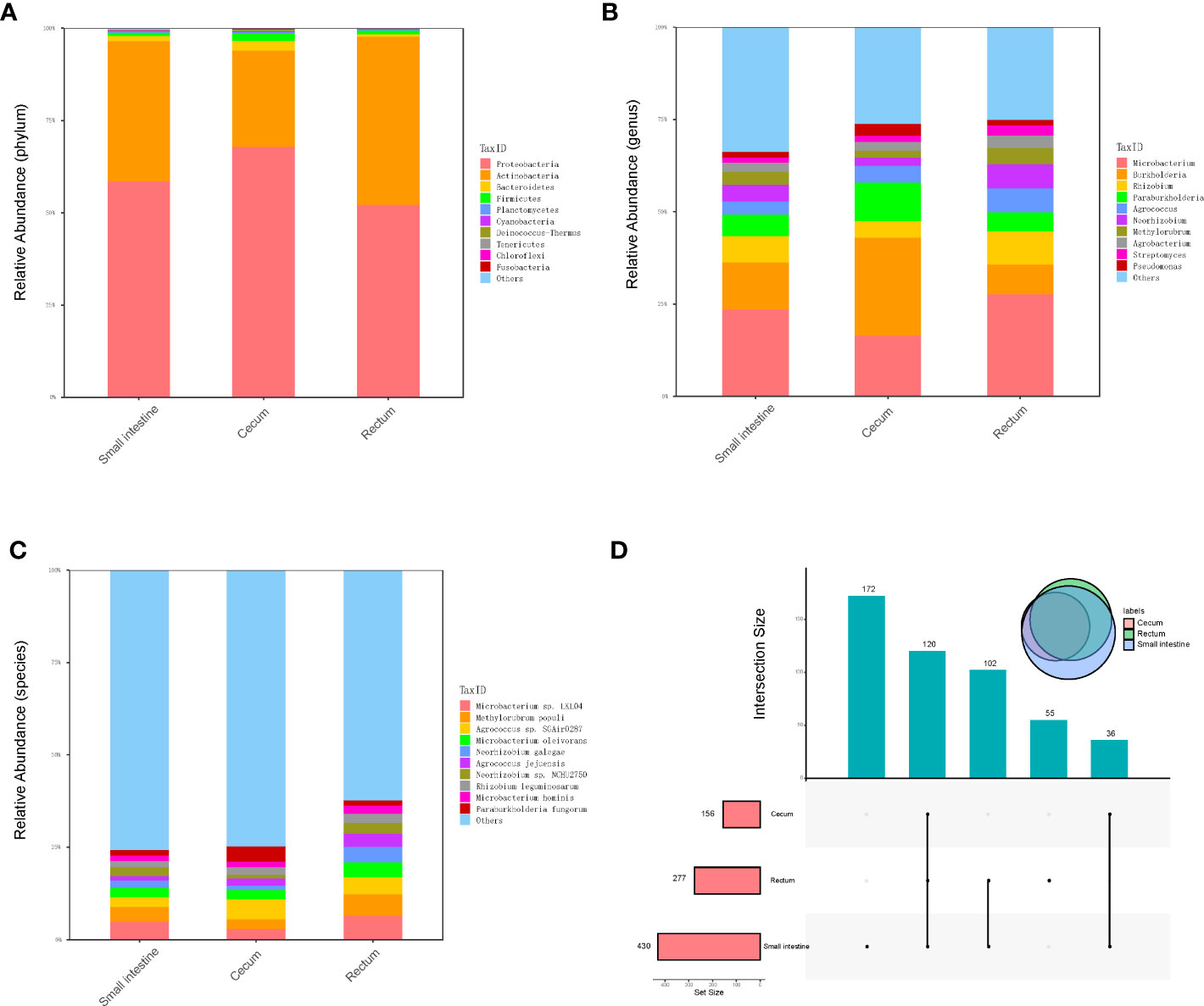
Figure 2 Metagenomics analysis of gut microbiota. (A) The top 10 abundant phyla in three different parts of fetal gut. (B) The top 10 abundant genera in three different parts of fetal gut. (C) The top 10 abundant species in three different parts of fetal gut. (D) Upset plot and venn plot of fetal gut microbiota in species level. The column above indicates the intersection of several samples in the row of which the point is in.
In order to explore the presence of virus in fetal gut and detect as many other microbes as possible, we performed virome sequencing and relaxed the filtering conditions of the virus sequence. Compared with metagenomic data, more viruses, bacteria and archaea were detected. In total, 700 species viruses (including 14 phyla and 432 genera, Figure 3A–C), 267 species of archaea (including 8 phyla and 118 genera, Figure 3D–F) and 5,477 species of bacteria (including 40 phyla and 1,475 genera) were detected (Figure 3G–I).
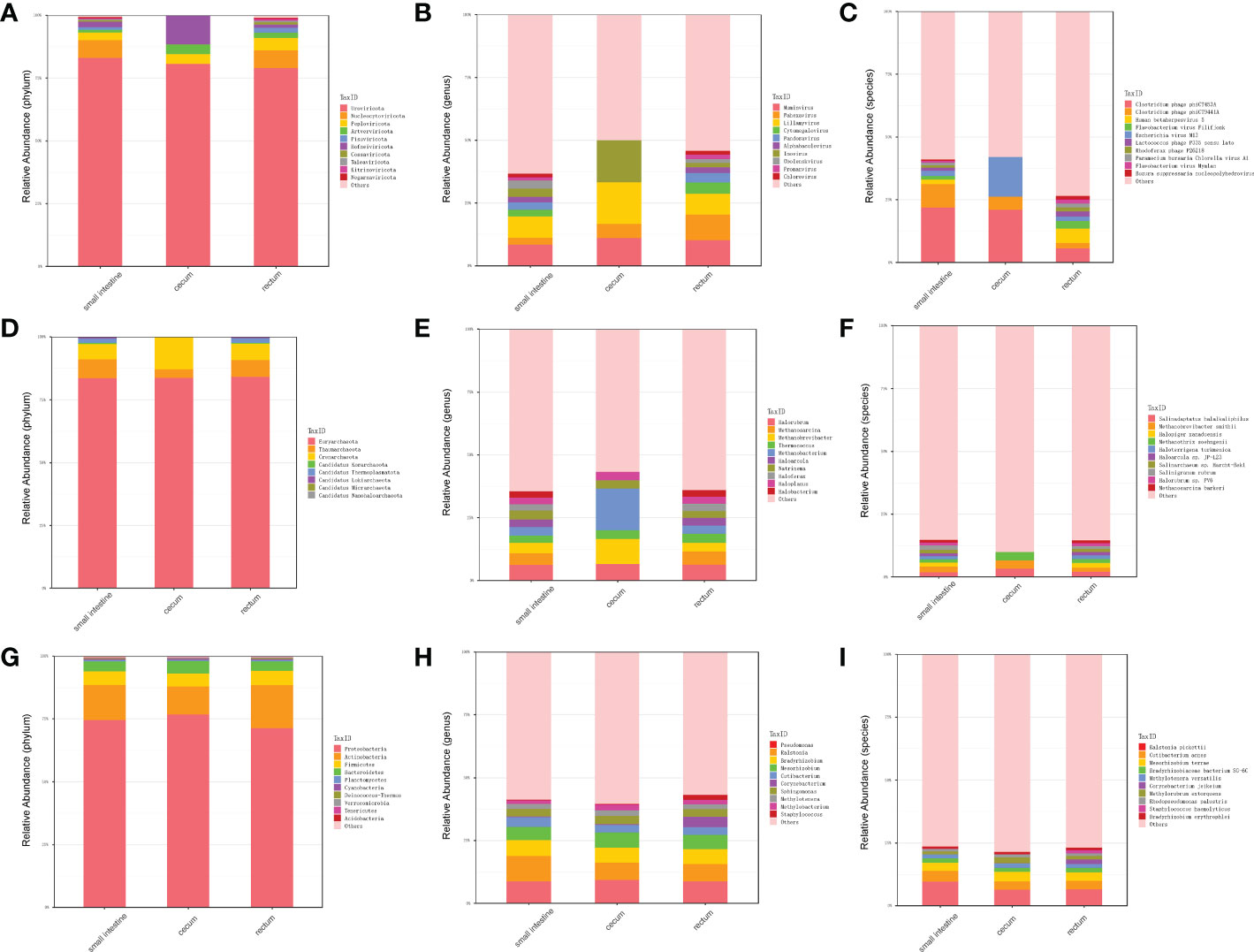
Figure 3 Distribution of microbiota in different parts of fetal gut detected by virome. (A) The top 10 abundant virus phyla in three different parts of fetal gut. (B) The top 10 abundant virus genera in three different parts of fetal gut. (C) The top 10 abundant virus species in three different parts of fetal gut. (D) The top 10 abundant archaea phyla in three different parts of fetal gut. (E) The top 10 abundant archaea genera in three different parts of fetal gut. (F) The top 10 abundant archaea species in three different parts of fetal gut. (G) The top 10 abundant bacteria phyla in three different parts of fetal gut. (H) The top 10 abundant bacteria genera in three different parts of fetal gut. (I) The top 10 abundant bacteria species in three different parts of fetal gut.
In terms of viruses, 11 species were shared by small intestine, cecum and rectum, 189 species were the peculiar species of small intestine, and the number of peculiar species in rectum was 1 (Figure 4A). The top 3 phyla of small intestine and rectum were Uroviricota, Nucleocytoviricota and Peploviricota, while only Uroviricota was detected in cecum. The top 3 genera of small intestine were Lillamyvirus, Muminvirus and Inovirus, while Pahexavirus, Muminvirus and Lillamyvirus were the top 3 genera of rectum. And no viral genera were detected in the cecum, which is consistent with species level. At the specie level, Clostridium phage phiCT453A was the most abundant species in small intestine and rectum, and Human betaherpesvirus 5 was detected in the rectum (Table 2).
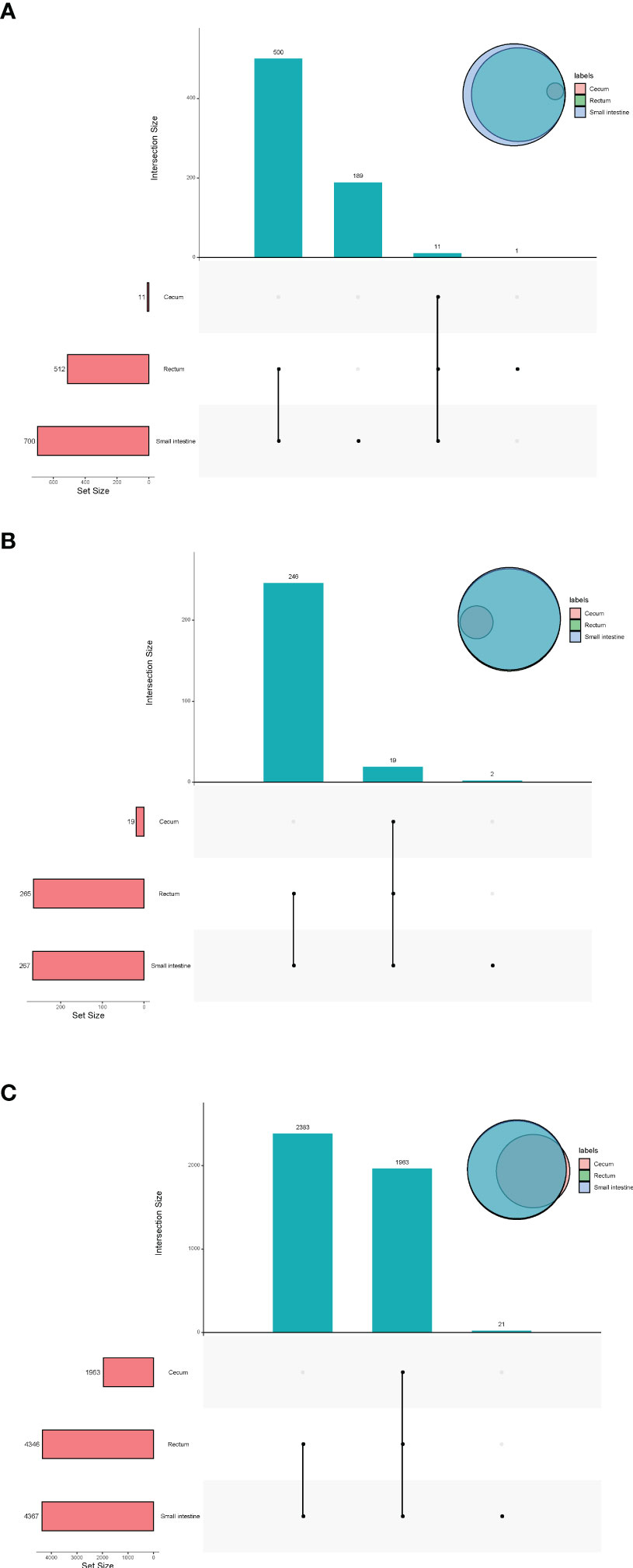
Figure 4 Upset plot and venn plot of fetal gut microbiota detected by virome (species level). (A) Upset plot and venn plot of fetal gut virus in three different parts. (B) Upset plot and venn plot of fetal gut archaea in three different parts. (C) Upset plot and venn plot of fetal gut bacteria in three different parts.
As shown in Figure 4B, in terms of archaea, 19 species were shared by small intestine, cecum and rectum, 2 species were the peculiar species of small intestine. The top 3 phyla of small intestine, cecum and rectum were roughly the same, mainly included Euryarchaeota, Thaumarchaeota and Crenarchaeota. At the genus level, Halorubrum, Methanosarcina and Methanobrevibacter were the top 3 of small intestine, Halorubrum, Methanosarcina and Thermococcus were the top 3 of rectum, only Halovivax And Methanobacterium were detected in cecum. At the specie level, Methanobrevibacter smithii, Salinadaptatus halalkaliphilus and Salinigranum rubrum were the top 3 of small intestine, Salinadaptatus halalkaliphilus, Halopiger xanaduensis and Methanobrevibacter smithii were the top 3 of rectum, Halovivax ruber was the only specie detected in cecum (Table 3).
In terms of bacteria, 1,963 species were shared by small intestine, cecum and rectum, 21 species were the peculiar species of small intestine (Figure 4C). Proteobacteria, Actinobacteria and Firmicutes were the predominant phyla in three different parts of fetal gut. The top 3 genera in relative abundances of small intestine and rectum were Ralstonia, Pseudomonas and Bradyrhizobium. Pseudomonas, Ralstonia and Mesorhizobium were the top 3 of cecum. At the specie level, the top 3 of small intestine, cecum and rectum were all composed of Ralstonia pickettii, Cutibacterium acnes and Mesorhizobium terrae (Table 4).
Indeed, the results of functional enrichment analysis showed that the microbes of small intestine and rectum are mainly enriched in the synthesis and metabolism pathways of amino acids and energy metabolism pathways, such as L-valine biosynthesis, ureide biosynthesis, superpathway of glyoxylate bypass and TCA, L-tyrosine degradation I and TCA cycle I prokaryotic (Figures 5A, B). Moreover, the identification results of antibiotic resistance genes (ARGs) showed that a total of 25 ARGs were detected, including 8 in small intestine and 24 in rectum, while no ARG was detected in cecum (Figures 5C, D).
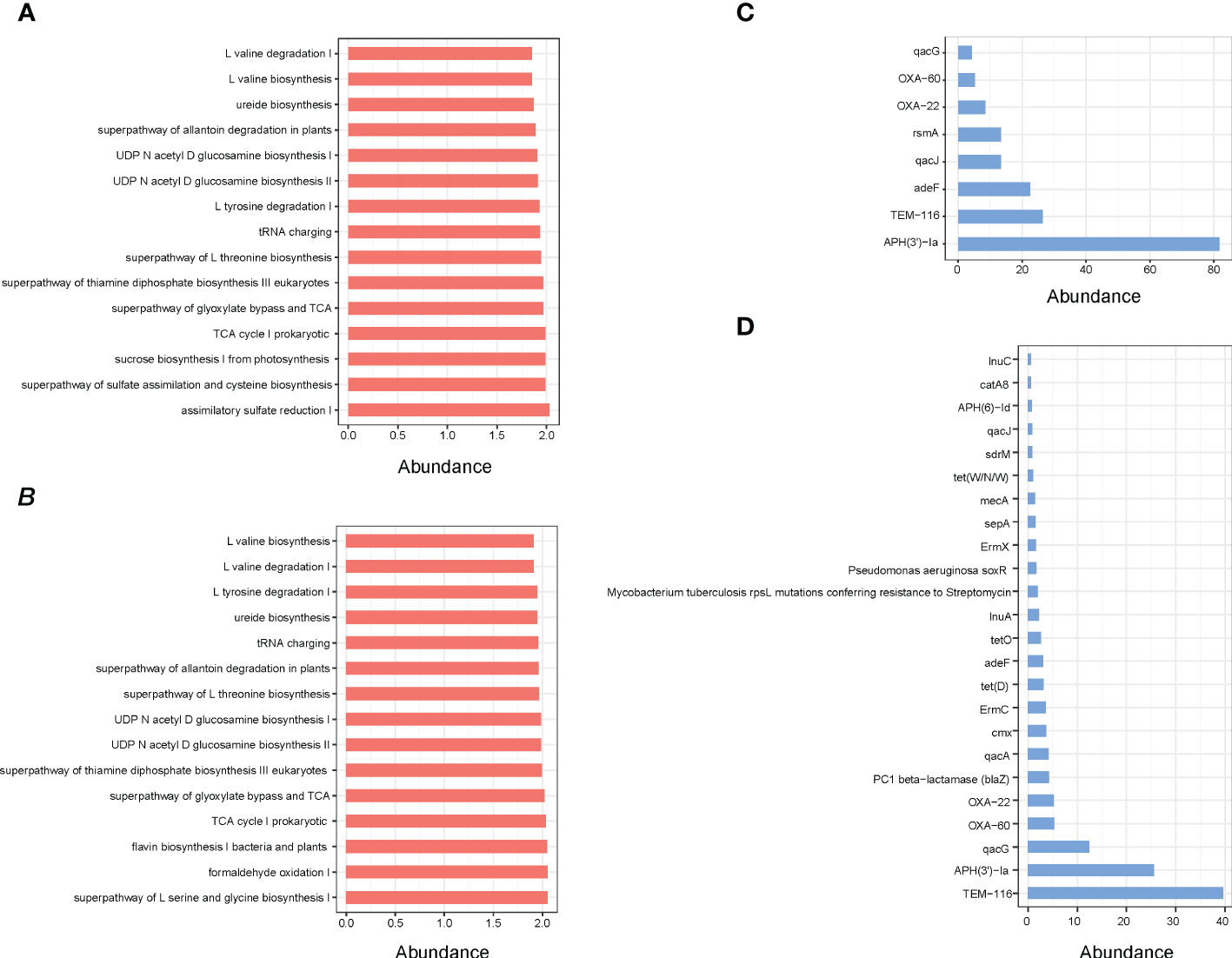
Figure 5 (A) Pathway enrichment analyses of small intestine microbiota. (B) Pathway enrichment analyses of rectum microbiota. (C) ARGs analysis of small intestine microbial. (D) ARGs analysis of rectum microbial.
For adults’ microbiota, a total of 5385 species of bacteria (40 phyla and 1471 genera), 47 species of viruses (4 phyla and 20 genera) and 258 species of archaea were detected (6 phyla and 112 genera, Figure 6A). As shown in Table 1, the top 3 phyla were Firmicutes, Bacteroidetes and Actinobacteria. At the genus level, the top 3 were Bacteroides, Phocaeicola and Faecalibacterium. The top 3 species were Phocaeicola vulgatus, Faecalibacterium prausnitzii and Bacteroides uniformi. Since very tiny amounts of viruses and archaea were detected in fetal metagenomic sequencing, we compared the archaea and viruses detected in adults’ metagenomic sequencing to the virome data. In terms of viruses, the top 3 phyla were Uroviricota, Hofneiviricota and Nucleocytoviricota. The top 3 genus were Toutatisvirus, Brigitvirus and Oengusvirus. At the species level, the top 3 were Faecalibacterium virus Toutatis, Faecalibacterium virus Brigit and Faecalibacterium virus Oengus (Table 2). As shown in Table 3, Euryarchaeota, Crenarchaeota and Candidatus Thermoplasmatota were the top 3 phyla. Methanosarcina, Methanobrevibacter and Thermococcus were the top 3 genera in relative abundances of adults’ microbiota. The top 3 species were Methanobrevibacter smithii, Methanosalsum zhilinae and Methanococcus maripaludis. Moreover, we characterized the global function of adults’ gut microbiota by using HUMANn3. As shown in the Figure 6B, the metabolic pathways of adults’ gut microbiota were mainly enriched in sucrose biosynthesis II, glycolysis IV, dTDP β L-rhamnose biosynthesis and L-valine biosynthesis pathways.
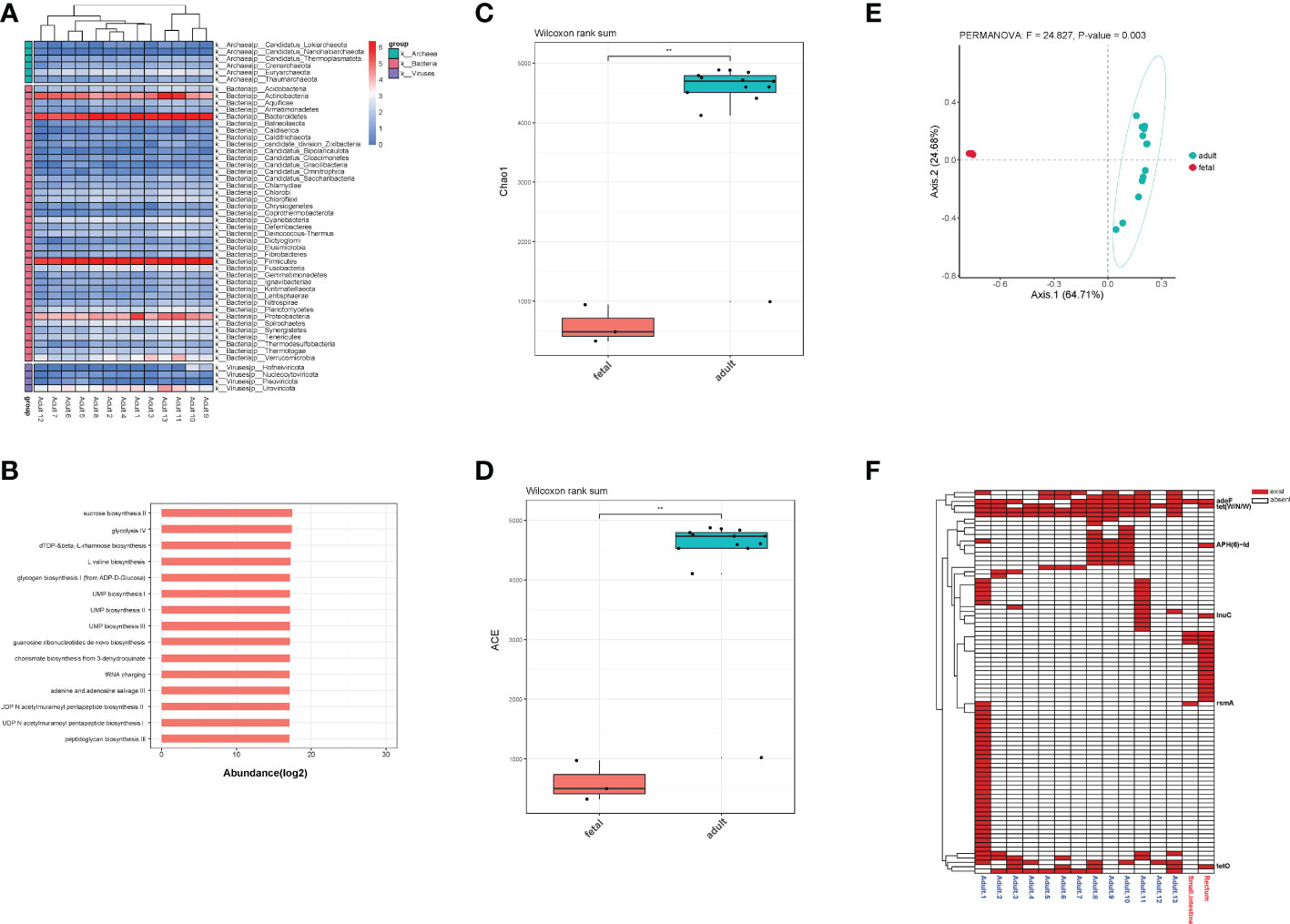
Figure 6 (A) The distribution of bacteria, archaea and viruses in each adult sample at phylum level. (B) Pathway enrichment analyses of adults’ gut microbiota. (C). Alpha diversity (Chao1 index) estimates between fetal and adult groups. (D) Alpha diversity (ACE index) estimates between fetal and adult groups. (E) The PCoA plot based on Bray-Curtis distance. (F) The distribution of all ARGs in fetal and adult groups. ** means p<0.05, and we chose p value cutoff of 0.05 as the significance level.
To further explore the differences in the microbiota diversity and composition of adults and the fetus, we performed α-diversity and β-diversity analysis. Our results show that there was significant difference in Chao1 and ACE indexes (p<0.05, Figures 6C, D). As the principal co-ordinates analysis (PCoA) shown, fetal samples and adult samples were significant separated (p<0.05, Figure 6E). Besides, total 71 ARGs were detected in adults’ gut microbiota. Among them, rsmA, tet(W/N/W), adeF, tetO, lnuC and APH(6)-Id were shared by the fetus and adults (Figure 6F).
The presence of microbes in fetal gut has long been controversial. After strict experimental conditions and environmental control settings, our results showed that there were indeed microbes in fetal gut, including bacteria, archaea and viruses.
The integration results of metagenomics and virome showed that Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes were the dominant phyla of fetal gut at the phylum level. At the genus level, Pseudomonas, Bradyrhizobium, Microbacterium, Burkholderia and Ralstonia were the dominant genera of fetal gut, of which Ralstonia and Burkholderia were detected in the environmental control groups in recent studies (26). However, Ralstonia insidiosa, as a member of Ralstonia, was a resident at the maternal-fetal interface in another research (45). Indeed, we also detected a high abundance of Ralstonia pickettii and Ralstonia insidiosa in fetal gut. Our results confirm that a recent study by Mishra et al. analyzed fetal microbes and found that Pseudomonas and Bradyrhizobium were enriched in fetal samples (26).
Besides, we detected 700 species viruses, which together with other gut microbial communities maintain the dynamic balance of gut and are key players in the regulation of intestinal homeostasis and inflammation, including 130 species of bacterial viruses (bacteriophages) and 570 species of eukaryotic viruses in fetal gut. (46, 47). Among the 570 species of eukaryotic viruses, Human betaherpesvirus 5 (also termed human cytomegalovirus) was the most abundant, which is a common cause of congenital viral infection in fetuses and neonates and a major non-genetic cause of congenital sensorineural hearing loss and neurological disability (48, 49). Humans are the only host of human cytomegalovirus (HCMV), which can replicate in most types of cells. HCMV can lead to infection in the developing fetus through vertical transmission during maternal infection (50, 51), and the transmission rate in the first, second, and third trimesters are 26%, 28%, and 65% (52–55). Among the 198 species of bacteriophages that can interact with bacteria to regulate bacterial composition, phages of Clostridium, Escherichia and Flavobacterium were the predominant species. Moreover, we detected crAssphage, which is not only the most abundant virus known to exist in humans but also almost ubiquitous (56), suggesting that crAssphage was acquired in early life. This was contrary to the research from Lim et al. (57).
Most studies currently focused on bacteria, fungi or virus, while archaea are often overlooked. However, the interaction between archaea and host can affect the host in many ways, as archaea were proposed to use for the prevention of trimethylaminuria and cardiovascular disease (58); archaea found on human skin may be related to age and skin physiology (59); archaea participate in the pro-inflammatory process (60). Methane-producing archaea were the predominant component of the archaeome, including Methanobacteriales and Methanomassiliicoccales (61). Methanobrevibacter smithii is the most abundant methanogen in the human gut and was isolated as the first representative nearly 40 years ago (62). Take the advantage of virome sequencing, 262 archaeal species (including 8 phyla and 117 genera) were detected in fetal gut. Euryarchaeota was the most abundant phylum, which contained most of the species of archaea (Methanogens, halophiles and Thermophiles). Methanobrevibacter smithii was the most abundant specie, and this is the first ever detection in fetal gut of Methanobrevibacter smithii, which confirmed the hypothesis from Sereme et al. that Methanobrevibacter smithii was an in-utero member of gut microbiota (63). Our results opposed the hypothesis that breast milk is the source of Methanobrevibacter smithii in premature neonates (64). The study from Grine et al. showed that Methanobrevibacter smithii was detected in vaginal fluid only in cases of vaginal disease (65), although the fetus passed through the vagina, the mother of our study without bacterial vaginosis. Besides, more methanogens such as Methanobrevibacter millerae, Methanobrevibacter olleyae, Methanobrevibacter ruminantium and Methanosphaera stadtmanae were detected in fetal gut.
Both metagenomic and virome sequencing results showed that rectal microbial species were higher than those of small intestine and cecum in different classification levels (phylum, genus and specie). Besides, the composition and functional pathway of the small intestine and rectum were more similar. The difference between the cecum and the other two sites might be due to the host contamination of the cecum was more severe than that of the small intestine and rectum.
We compared the fetal gut microbiota to the adults’ and found that the diversity of adults’ gut microbiota was significantly higher than that in fetal group. The composition of gut microbiota between two groups was significantly different. Specifically, Proteobacteria and Actinobacteria were the dominant bacteria phyla in fetal gut. While the dominant bacteria phyla in adults’ gut were Firmicutes and Bacteroidetes, which indicated that Firmicutes and Bacteroidetes gradually replaced Proteobacteria and Actinobacteria as the main force of gut microbiota during the process of growth and development. For viruses and archaea, the dominant phyla in both groups were similar. Besides, we found 6 ARGs in fetal group were consistent with adult group, suggesting that microbes carried ARGs might transmit vertically during pregnancy.
In conclusion, we detected a variety of microbes in fetal gut through metagenomic and virome sequencing, including bacteria, virus and archaea. Especially, we first identified Methanobrevibacter smithii in fetal gut, which was the most prevalent and abundant methanogen. In addition, by comparing the fetal and adults’ gut microbiota, we found that gut bacterial composition changed greatly during the growth and development process and the same ARGs existed in fetuses and adults. Thus, we suggest the fetal gut is not a sterile environment and has a low abundance but metabolically rich microbiome. Our study provided valuable resource for understanding human fetal immunity development during gestation.
The data presented in the study are deposited in the Genome Sequence Archive for Human repository, accession number: HRA003676.
The studies involving human participants were reviewed and approved by West china 2nd university hospital. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Experimental work: GH, AL, JL and MH. Data analysis: YL, XYL and WG. Writing and editing: XL, XHL, ZF and YZ. All authors contributed to the article and approved the submitted version.
This work was supported by Project of Chengdu Science and Technology Bureau, (YZ, Grant No. 2021-YF05-02110-SN), China Postdoctoral Science Foundation (YZ, Grant No. 2020M680149, 2020T130087ZX), Sichuan Science and Technology Program (GH, Grant No. 2020YFS0127).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Tieppo P, Papadopoulou M, Gatti D, Mcgovern N, Chan JKY, Gosselin F, et al. The human fetal thymus generates invariant effector γδ T cells. J Exp Med (2019) 217:e20190580. doi: 10.1084/jem.20190580
2. Park JE, Botting RA, Domínguez Conde C, Popescu DM, Lavaert M, Kunz DJ, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science (2020) 367. doi: 10.1126/science.aay3224
3. Rackaityte E, Halkias J. Mechanisms of fetal T cell tolerance and immune regulation. Front Immunol (2020) 11:588. doi: 10.3389/fimmu.2020.00588
4. Jain N. The early life education of the immune system: Moms, microbes and (missed) opportunities. Gut Microbes (2020) 12:1824564. doi: 10.1080/19490976.2020.1824564
5. Digiulio DB, Gervasi M, Romero R, Mazaki-Tovi S, Vaisbuch E, Kusanovic JP, et al. Microbial invasion of the amniotic cavity in preeclampsia as assessed by cultivation and sequence-based methods. J Perinat Med (2010) 38:503–13. doi: 10.1515/jpm.2010.078
6. Gervasi MT, Romero R, Bracalente G, Chaiworapongsa T, Erez O, Dong Z, et al. Viral invasion of the amniotic cavity (VIAC) in the midtrimester of pregnancy. J Matern Fetal Neonatal Med (2012) 25:2002–13. doi: 10.3109/14767058.2012.683899
7. Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, et al. A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reprod Immunol (2014) 71:330–58. doi: 10.1111/aji.12189
8. Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, et al. Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med (2015) 28:1394–409. doi: 10.3109/14767058.2014.958463
9. Shin H, Pei Z, Martinez KA 2nd, Rivera-Vinas JI, Mendez K, Cavallin H, et al. The first microbial environment of infants born by c-section: the operating room microbes. Microbiome (2015) 3:59. doi: 10.1186/s40168-015-0126-1
10. Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome (2016) 4:29. doi: 10.1186/s40168-016-0172-3
11. Arora N, Sadovsky Y, Dermody TS, Coyne CB. Microbial vertical transmission during human pregnancy. Cell Host Microbe (2017) 21:561–7. doi: 10.1016/j.chom.2017.04.007
12. Maidji E, Nigro G, Tabata T, Mcdonagh S, Nozawa N, Shiboski S, et al. Antibody treatment promotes compensation for human cytomegalovirus-induced pathogenesis and a hypoxia-like condition in placentas with congenital infection. Am J Pathol (2010) 177:1298–310. doi: 10.2353/ajpath.2010.091210
13. Robbins JR, Skrzypczynska KM, Zeldovich VB, Kapidzic M, Bakardjiev AI. Placental syncytiotrophoblast constitutes a major barrier to vertical transmission of listeria monocytogenes. PloS Pathog (2010) 6:e1000732. doi: 10.1371/journal.ppat.1000732
14. Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci U.S.A. (2013) 110:12048–53. doi: 10.1073/PNAS.1304718110
15. Bayer A, Delorme-Axford E, Sleigher C, Frey TK, Trobaugh DW, Klimstra WB, et al. Human trophoblasts confer resistance to viruses implicated in perinatal infection. Am J Obstet Gynecol (2015) 212:71.e71–8. doi: 10.1016/j.ajog.2014.07.060
16. Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET Jr., et al. Type III interferons produced by human placental trophoblasts confer protection against zika virus infection. Cell Host Microbe (2016) 19:705–12. doi: 10.1016/j.chom.2016.03.008
17. Diaz Heijtz R. Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Semin Fetal Neonatal Med (2016) 21:410–7. doi: 10.1016/j.siny.2016.04.012
18. Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: The evolving inner self. Cell (2017) 171:1481–93. doi: 10.1016/j.cell.2017.11.024
19. Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the "sterile womb" and "in utero colonization" hypotheses: implications for research on the pioneer infant microbiome. Microbiome (2017) 5:48. doi: 10.1186/s40168-017-0268-4
20. Willyard C. Could baby's first bacteria take root before birth? Nature (2018) 553:264–6. doi: 10.1038/d41586-018-00664-8
21. De Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature (2019) 572:329–34. doi: 10.1038/s41586-019-1451-5
22. Seferovic MD, Pace RM, Carroll M, Belfort B, Major AM, Chu DM, et al. Visualization of microbes by 16S in situ hybridization in term and preterm placentas without intraamniotic infection. Am J Obstet Gynecol (2019) 221:146.e141–146.e123. doi: 10.1016/j.ajog.2019.04.036
23. Younge N, Mccann JR, Ballard J, Plunkett C, Akhtar S, Araújo-Pérez F, et al. Fetal exposure to the maternal microbiota in humans and mice. JCI Insight (2019) 4. doi: 10.1172/jci.insight.127806
24. Al Alam D, Danopoulos S, Grubbs B, Ali N, Macaogain M, Chotirmall SH, et al. Human fetal lungs harbor a microbiome signature. Am J Respir Crit Care Med (2020) 201:1002–6. doi: 10.1164/rccm.201911-2127LE
25. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med (2020) 26:599–607. doi: 10.1038/s41591-020-0761-3
26. Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, et al. Microbial exposure during early human development primes fetal immune cells. Cell (2021) 184:3394–3409.e3320. doi: 10.1016/j.cell.2021.04.039
27. Sterpu I, Fransson E, Hugerth LW, Du J, Pereira M, Cheng L, et al. No evidence for a placental microbiome in human pregnancies at term. Am J Obstet Gynecol (2021) 224:296.e291–296.e223. doi: 10.1016/j.ajog.2020.08.103
28. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6:237ra265. doi: 10.1126/scitranslmed.3008599
29. Leon LJ, Doyle R, Diez-Benavente E, Clark TG, Klein N, Stanier P, et al. Enrichment of clinically relevant organisms in spontaneous preterm-delivered placentas and reagent contamination across all clinical groups in a Large pregnancy cohort in the united kingdom. Appl Environ Microbiol (2018) 84. doi: 10.1128/AEM.00483-18
30. Weisblum Y, Panet A, Zakay-Rones Z, Haimov-Kochman R, Goldman-Wohl D, Ariel I, et al. Modeling of human cytomegalovirus maternal-fetal transmission in a novel decidual organ culture. J Virol (2011) 85:13204–13. doi: 10.1128/JVI.05749-11
31. Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, et al. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell Host Microbe (2016) 20:155–66. doi: 10.1016/j.chom.2016.07.002
32. Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, et al. Does the human placenta delivered at term have a microbiota? results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol (2019) 220:267.e261–267.e239. doi: 10.1016/j.ajog.2018.10.018
33. Bi Y, Tu Y, Zhang N, Wang S, Zhang F, Suen G, et al. Multiomics analysis reveals the presence of a microbiome in the gut of fetal lambs. Gut (2021) 70:853–64. doi: 10.1136/gutjnl-2020-320951
34. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
35. Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe (2019) 26:527–541.e525. doi: 10.1016/j.chom.2019.09.009
36. Polkinghorne DE. Phenomenological research methods. In: Valle RS, Halling S, editors. Existential-phenomenological perspectives in psychology: Exploring the breadth of human experience. Boston, MA: Springer US (1989). p. 41–60.
37. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
38. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods (2012) 9:357–9. doi: 10.1038/nmeth.1923
39. Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics (2015) 31:1674–6. doi: 10.1093/bioinformatics/btv033
40. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf (2010) 11:119. doi: 10.1186/1471-2105-11-119
41. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods (2015) 12:59–60. doi: 10.1038/nmeth.3176
42. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol (2014) 15:R46. doi: 10.1186/gb-2014-15-3-r46
43. Franzosa EA, Mciver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods (2018) 15:962–8. doi: 10.1038/s41592-018-0176-y
44. Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res (2020) 48:D517–d525. doi: 10.1093/nar/gkz935
45. Parnell LA, Willsey GG, Joshi CS, Yin Y, Wargo MJ, Mysorekar IU. Functional characterization of ralstonia insidiosa, a bona fide resident at the maternal-fetal interface. bioRxiv (2019), 721977. doi: 10.1101/721977
46. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science (2010) 330:1768–73. doi: 10.1126/science.1195568
47. Norman JM, Handley SA, Virgin HW. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology (2014) 146:1459–69. doi: 10.1053/j.gastro.2014.02.001
48. Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol (2007) 17:253–76. doi: 10.1002/rmv.535
49. Wang C, Zhang X, Bialek S, Cannon MJ. Attribution of congenital cytomegalovirus infection to primary versus non-primary maternal infection. Clin Infect Dis (2011) 52:e11–13. doi: 10.1093/cid/ciq085
50. Fowler KB, Stagno S, Pass RF. Maternal immunity and prevention of congenital cytomegalovirus infection. Jama (2003) 289:1008–11. doi: 10.1001/jama.289.8.1008
51. Yamamoto AY, Mussi-Pinhata MM, Boppana SB, Novak Z, Wagatsuma VM, Oliveira Pde F, et al. Human cytomegalovirus reinfection is associated with intrauterine transmission in a highly cytomegalovirus-immune maternal population. Am J Obstet Gynecol (2010) 202:297.e291–298. doi: 10.1016/j.ajog.2009.11.018
52. Bodéus M, Hubinont C, Goubau P. Increased risk of cytomegalovirus transmission in utero during late gestation. Obstet Gynecol (1999) 93:658–60.
53. Enders G, Daiminger A, Bäder U, Exler S, Enders M. Intrauterine transmission and clinical outcome of 248 pregnancies with primary cytomegalovirus infection in relation to gestational age. J Clin Virol (2011) 52:244–6. doi: 10.1016/j.jcv.2011.07.005
54. Feldman B, Yinon Y, Tepperberg Oikawa M, Yoeli R, Schiff E, Lipitz S. Pregestational, periconceptional, and gestational primary maternal cytomegalovirus infection: prenatal diagnosis in 508 pregnancies. Am J Obstet Gynecol (2011) 205:342.e341–346. doi: 10.1016/j.ajog.2011.05.030
55. Leruez-Ville M, Foulon I, Pass R, Ville Y. Cytomegalovirus infection during pregnancy: state of the science. Am J Obstet Gynecol (2020) 223:330–49. doi: 10.1016/j.ajog.2020.02.018
56. Dutilh BE, Cassman N, Mcnair K, Sanchez SE, Silva GG, Boling L, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun (2014) 5:4498. doi: 10.1038/ncomms5498
57. Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med (2015) 21:1228–34. doi: 10.1038/nm.3950
58. Brugère JF, Borrel G, Gaci N, Tottey W, O'toole PW, Malpuech-Brugère C. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes (2014) 5:5–10. doi: 10.4161/gmic.26749
59. Probst AJ, Auerbach AK, Moissl-Eichinger C. Archaea on human skin. PloS One (2013) 8:e65388. doi: 10.1371/journal.pone.0065388
60. Bang C, Vierbuchen T, Gutsmann T, Heine H, Schmitz RA. Immunogenic properties of the human gut-associated archaeon methanomassiliicoccus luminyensis and its susceptibility to antimicrobial peptides. PloS One (2017) 12:e0185919. doi: 10.1371/journal.pone.0185919
61. Borrel G, Brugère JF, Gribaldo S, Schmitz RA, Moissl-Eichinger C. The host-associated archaeome. Nat Rev Microbiol (2020) 18:622–36. doi: 10.1038/s41579-020-0407-y
62. Miller TL, Wolin MJ, Conway De Macario E, Macario AJ. Isolation of methanobrevibacter smithii from human feces. Appl Environ Microbiol (1982) 43:227–32. doi: 10.1128/aem.43.1.227-232.1982
63. Sereme Y, Guindo CO, Filleron A, Corbeau P, Tran TA, Drancourt M, et al. Meconial methanobrevibacter smithii suggests intrauterine methanogen colonization in preterm neonates. Curr Res Microb Sci (2021) 2:100034.doi: 10.1016/j.crmicr.2021.100034
64. Togo AH, Grine G, Khelaifia S, Des Robert C, Brevaut V, Caputo A, et al. Culture of methanogenic archaea from human colostrum and milk. Sci Rep (2019) 9:18653. doi: 10.1038/s41598-019-54759-x
Keywords: fetal gut microbiota, metagenomics, virome, archaea, immunity, gestation
Citation: Liu X, He G, Lan Y, Guo W, Liu X, Li J, Liu A, He M, Liu X, Fan Z and Zhang Y (2023) Virome and metagenomic analysis reveal the distinct distribution of microbiota in human fetal gut during gestation. Front. Immunol. 13:1079294. doi: 10.3389/fimmu.2022.1079294
Received: 25 October 2022; Accepted: 14 December 2022;
Published: 05 January 2023.
Edited by:
Yin Tailang, Reproductive Medicine Center, Wuhan University, ChinaReviewed by:
Binghua Sun, Anhui University, ChinaCopyright © 2023 Liu, He, Lan, Guo, Liu, Li, Liu, He, Liu, Fan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoyao Zhang, eWFveWFvemhhbmdAc2N1LmVkdS5jbg==; Zhenxin Fan, enhmYW5Ac2N1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.