- 1Medical Research Center, Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Linhai, Zhejiang, China
- 2School of Clinical Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China
- 3Key Laboratory of Minimally Invasive Techniques & Rapid Rehabilitation of Digestive System Tumour of Zhejiang Province, Linhai, China
Background: Human leukocyte antigen G (HLA-G) is an immune checkpoint molecule with relevance in several cancers. The aim of this study was to evaluate the potential role of soluble HLA-G (sHLA-G), its genetic polymorphisms and its haplotype structure in the susceptibility and prognosis of primary cervical cancer in a Chinese Han population.
Methods: We investigated sHLA-G plasma levels and 3’ untranslated region (3’UTR) polymorphisms through ELISA and direct DNA sequencing, respectively, in cervical cancer patients (120 cases) and healthy control women (96 cases). The data were analyzed for associations using PowerMarker, Haploview, and GraphPad Prism.
Results: In this study, 8 polymorphic sites, 16 haplotypes and 23 diplotypes in the HLA-G 3’UTR were identified in our study population. We observed that each pair of 8 polymorphic sites exhibited linkage disequilibrium. The heterozygote CT genotype at position +3422 (rs17875408) was more common in cervical cancer patients than in healthy women (OR=5.285, P<0.05). Haplotypes UTR-1, UTR-3, and UTR-7 accounted for more than 85% of both groups, but no significant difference was found. The frequency of the UTR-1/UTR-3 diplotype in patients was significantly higher than that in controls (P<0.05). In addition, we further observed that HLA-G 3’UTR polymorphisms may influence the sHLA-G plasma level in patients’ peripheral blood, especially 14 bp Ins/Del (rs371194629) and +3142 C/G (rs1063320). A receiver operating characteristic (ROC) curve analysis showed that the sHLA-G level had good diagnostic performance in differentiating patients with cervical cancer from healthy women (AUC>0.7). Among patients, mean sHLA-G levels increased with increasing FIGO stages but were not related to the overall survival time.
Conclusions: The results of the present study enhance our understanding of how HLA-G 3’UTR polymorphisms can influence the peripheral sHLA-G plasma level and play a key role in cervical carcinogenesis. This study further confirmed that sHLA-G may represent a novel plasma biomarker for the prognosis and potential therapeutic target of cervical cancer.
Background
Cervical cancer is a common malignant tumor in females that originates in the cervix. Persistent infection with human papillomavirus (HPV) leads to almost all cervical cancers and their precursors (1). Human leukocyte antigen G (HLA-G) is an immunosuppressive molecule involved in the escape mechanisms of virus-infected and malignant cells. Our previous studies showed that HLA-G polymorphism was a susceptibility factor for active HPV infection, especially high-risk HPV types (2, 3). We speculate that HPV infection may escape host immunosurveillance by inducing HLA-G expression in cervical epithelial cells (4). An increasing number of studies have confirmed that HLA-G is an immune checkpoint molecule with immunosuppressive and carcinogenic activities (5, 6). HLA-G expression is strongly related to higher tumor grade and worse prognosis for patients with cervical cancer (7–10).
HLA-G is a nonclassical HLA class I molecule, and its coding gene is located on chromosome 6p21.3, which has limited polymorphisms. To date, only 110 alleles and 36 proteins have been identified (http://hla.alleles.org/nomenclature/stats.html, September 2022). HLA-G mRNA is alternatively spliced, resulting in at least four membrane-bound (HLA-G1, -G2, -G3, -G4) and three soluble (HLA-G5, -G6, -G7) protein isoforms (11, 12). Membrane-bound HLA-G1 isoform can be cleaved by metalloproteinases, producing soluble shedding HLA-G1 (sHLA-G1). Soluble HLA-G (sHLA-G) in peripheral blood mainly includes sHLA-G1 and HLA-G5 isoforms, which maybe expressed and released by cancer cells in the tissue microenvironment (10). It has been found that polymorphic sites may affect the expression of HLA-G molecules by changing the affinity of gene-targeted sequences or modifying the stability of HLA-G mRNA or the binding sites of microRNAs (13, 14). Most of the relevant studies have focused on the role of 3’ untranslated region (3’UTR) polymorphisms of the HLA-G gene, particularly 14 bp insertion/deletion (Ins/Del) (5’-ATTTGTTCATGCCT-3’, rs371194629) and +3142 C/G (rs1063320) (2, 3, 15). Our previous studies showed that 14 bp Ins or +3142 G alleles increased the risk of Chinese women acquiring cervical HPV (2, 3). The 14 bp Ins allele produces an additional splice, removing 92 bp, which may affect HLA-G mRNA stability and cause a decrease in sHLA-G levels in plasma (16, 17). The +3142 G allele favors binding miR-148a, miR-148b, and miR-152, causing increased degradation of HLA-G mRNA (18). It was previously reported that the expression of HLA-G in cervical intraepithelial neoplasia (CIN) with HPV16/18 infection was significantly higher than that in CIN without HPV infection, and the sHLA-G plasma level was significantly higher in CIN and SCC patients (8, 9). The above data indicate that the genetic heterogeneity of the HLA-G gene may lead to heterogeneous HLA-G expression in different populations. Therefore, the HLA-G 3’UTR polymorphisms of women’s genetic backgrounds were related to susceptibility to HPV infection, and we speculated that HLA-G 3’UTR polymorphisms may affect the development of cervical carcinogenesis by regulating sHLA-G level. However, the distribution of HLA-G alleles, genotypes, and haplotypes in cervical cancer patients and their possible roles in the expression of HLA-G levels remain unclear. The aim of this study was to investigate the association between HLA-G 3’UTR polymorphisms and sHLA-G plasma levels in Chinese Han cervical cancer patients. In addition, the association between the sHLA-G level and the diagnosis and prognosis of cervical cancer was also investigated.
Materials and methods
Study population
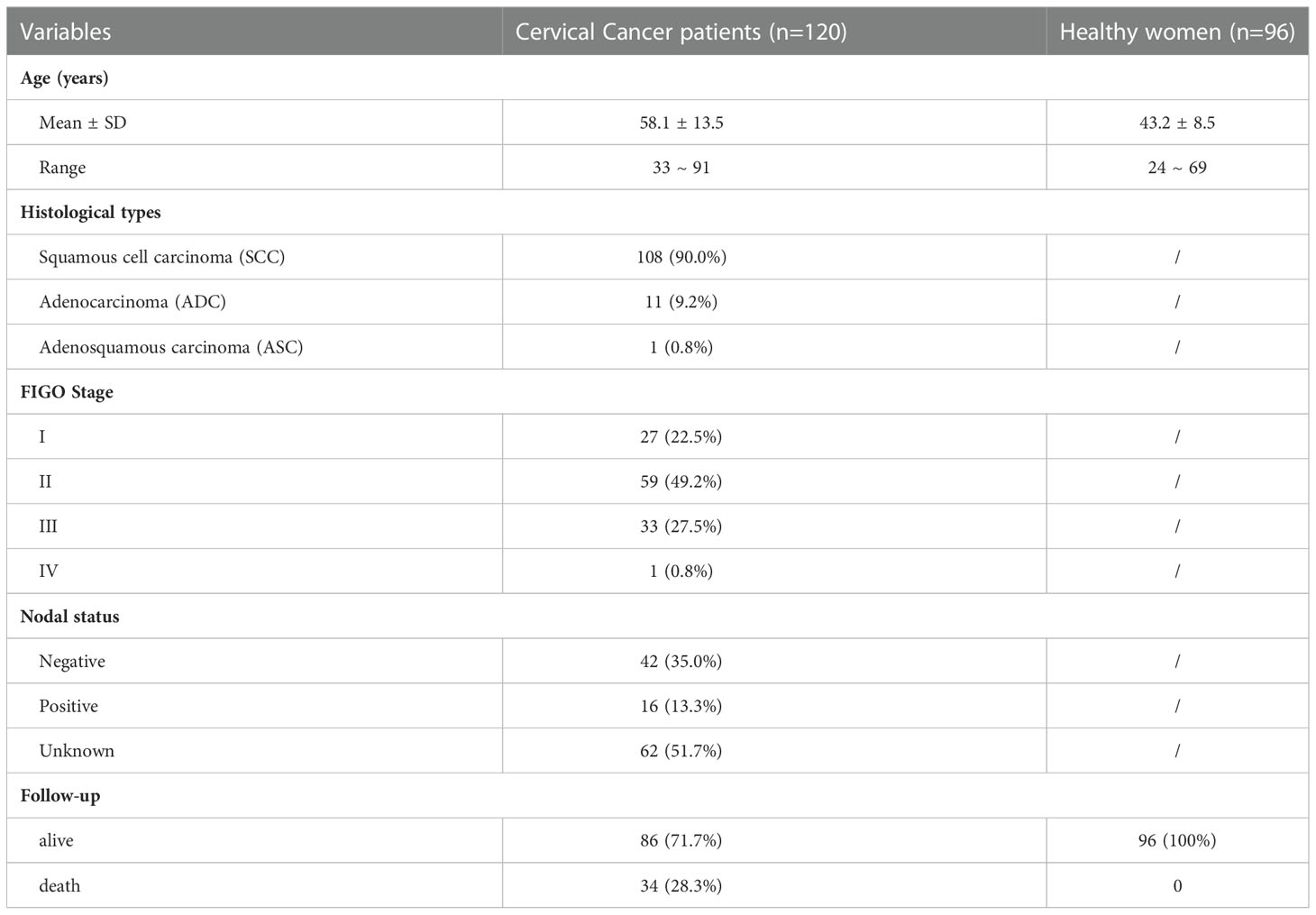
Since 2008, cervical cancer patients have been recruited at Taizhou Hospital, which is affiliated with Wenzhou Medical University, China. In total, 120 patients who were initially diagnosed with cervical cancer were enrolled. The exclusion criteria were radiotherapy, chemotherapy, or other medical interventions before blood collection. Staging of cervical cancer was performed according to the International Federation of Gynaecology and Obstetrics (FIGO) criteria, and patients were classified into Stages I, II, III, and IV. Follow-up was performed on 21 July 2022. In addition, 96 unrelated healthy women who were negative for HPV were also enrolled in this study. All included subjects were of Chinese Han descent.
This study was approved by the Medical Ethics Review Committee of Taizhou Hospital (approval #K20220226), and written informed consent from all study participants was obtained before enrolment.
HLA-G gene 3’UTR analysis
The NCBI reference sequence (RefSeq: NC_000006.12) was used as the standard for primer design and sequence alignment in this study. Genomic DNA was extracted using the commercially available DNA Extraction Kit (#GK0122, GENEray, China). The HLA-G gene 3’UTR was amplified by PCR (forward primer: 5’-TGAGCATGTGATGGGCTGTT-3’ and reverse primer: 5’-GGGAAGAGGTGTAGGGGTCT-3’). The PCR amplification product had a length of 704 bp or 718 bp, encompassing nucleotides +2846 to +3563 with 23 single nucleotide polymorphism (SNP) sites (rs371194629, rs567747015, rs1707, rs1710, rs17179101, rs146339774, rs17179108, rs569057854, rs180827037, rs554784083, rs138249160, rs1063320, rs554076817, rs187320344, rs9380142, rs1610696, rs1233331, rs541542414, rs556033566, rs561554956, rs530204611, rs17875408, rs190871790). In addition, we performed another PCR with primers 14F and 14R to validate the 14 bp Ins/Del polymorphism in each sample as previously described (2).
The PCR mixture included 25 μL 2×PrimeSTAR Max Premix (#R040A, TaKaRa, Japan), 20 μL nuclease-free water, 2 μL forward primer, 2 μL reverse primer, and 1 μL template DNA in a final volume of 50 μL. The PCR conditions were 95°C for 5 min; 35 repeated cycles of 94°C for 45 s, 62°C for 45 s and 72°C for 1 min; and a final extension step at 72°C for 7 min. Subsequently, the PCR amplification products were purified and sequenced using the reverse primer on an ABI 3730XL instrument at BGI Company. HLA-G polymorphic sites were analyzed by interpreting the chromatogram peaks using BioEdit.
sHLA-G measurement
Plasma was separated and stored at -80°C until sHLA-G level determination. The concentration of sHLA-G (U/mL) in the peripheral plasma of cervical cancer patients and healthy women was quantified by ELISA kit (Cat# RD194070100R, sHLA-G kit; BioVendor) using our previously described method (10). In this ELISA kit, combinations monoclonal antibody (mAb) MEM-G/09 (capture) + anti-β2m (detection) were chosen for the simultaneous determination of shed HLA-G1 and soluble HLA-G5.
Statistical analysis
HLA-G allele, genotype, haplotype and diplotype frequencies were compared between patients with cervical cancer and healthy women using the chi-squared (χ2) test. Hardy−Weinberg equilibrium for HLA-G genotypic data was estimated using the χ2 test with 1 df (SPSS 23.0). Linkage disequilibrium (LD) between each pair of polymorphic sites was evaluated using PowerMarker 3.25 and Haploview 4.2. The sHLA-G concentration was analyzed using the Mann−Whitney U test and Spearman rank correlation test (GraphPad Prism 5.0). Survival probabilities were calculated using Kaplan−Meier analysis. A receiver operating characteristic (ROC) curve was used to evaluate the diagnostic performance of sHLA-G concentration to discriminate between patients and healthy women. A two-sided P value less than 0.05 was considered statistically significant.
Results
Characteristics of the study population
A total of 120 patients (58.1 ± 13.5 years; range, 33~91 years) with an initial diagnosis of cervical cancer were included; 108 (90.0%) patients also had squamous cell carcinoma (SCC), 11 (9.2%) patients had adenocarcinoma (ADC) and 1 (0.8%) patient had adenosquamous carcinoma (ASC). The follow-up period was 14 years (range, 2~174 months) or until death. The clinical characteristics of the study population are summarized in Table 1.
Of the twenty-three SNP sites, only eight were recognized in the study population as 3’UTR polymorphisms of the HLA-G gene. We subsequently analyzed eight polymorphic sites, including 14 bp Ins/Del (rs371194629), +3010 C/G (rs1710), +3027 A/C (rs17179101), +3035 C/T (rs17179108), +3142 C/G (rs1063320), +3187 A/G (rs9380142), +3196 C/G (rs1610696), and +3422 C/T (rs17875408). All HLA-G genotypic frequencies fit Hardy−Weinberg equilibrium (data not shown).
Analysis of 3’UTR polymorphisms in the HLA-G gene
The allelic and genotypic frequencies observed in patients and healthy women are summarized in Table 2. Of the 8 SNP sites, a significant difference in genotypic frequencies was observed only in the heterozygote CT genotype at position +3422 (rs17875408), which was more frequent in cervical cancer patients than in healthy women (OR=5.285, P=0.039).
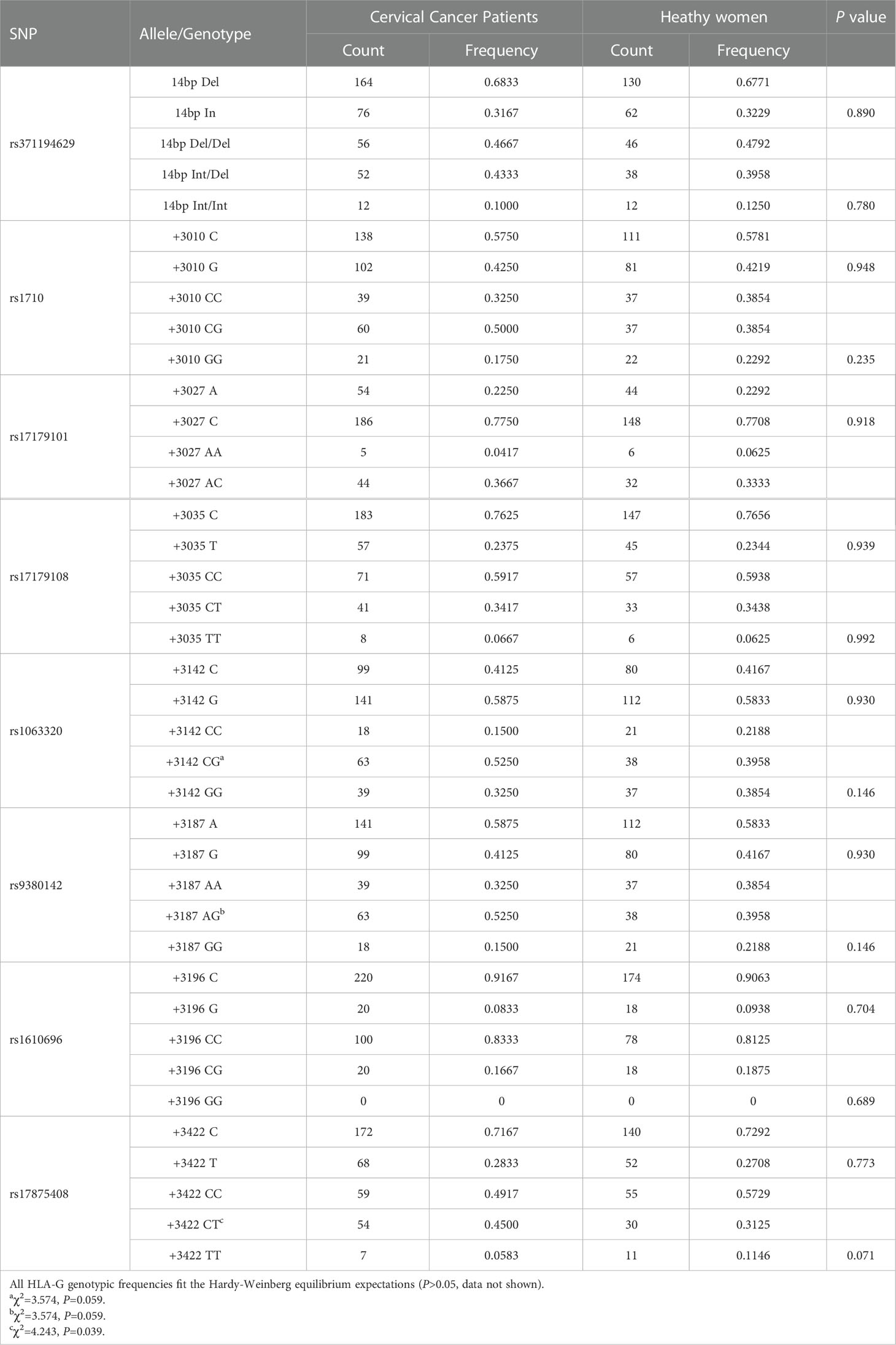
Table 2 Comparisons of the HLA-G 3’UTR allelic and genotypic frequencies between patients and controls.
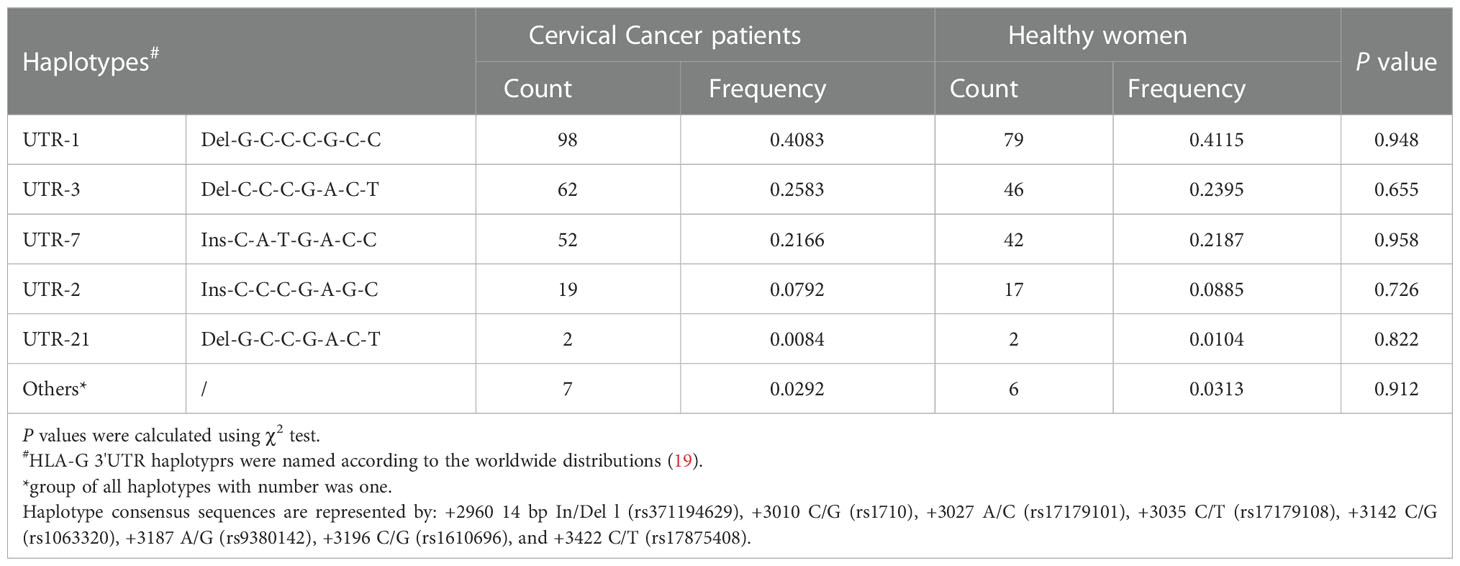
To investigate whether haplotypes are more predictive of cancer susceptibility than single SNP sites, we performed a haplotype linkage analysis on alleles. Among the study population, 16 different haplotypes were generated using PowerMarker software. These haplotypes were named in this study and were consistent with those previously published, according to the worldwide distributions addressing the HLA-G 3’UTR polymorphic sites (19). The results showed that the haplotype frequencies of UTR-1, UTR-3, and UTR-7 accounted for more than 85% of both groups. However, no significant differences were observed between patients and healthy women (P>0.05) (Table 3). As depicted in Figure 1, all eight polymorphic sites exhibited linkage disequilibrium (LD). A high LD was observed, especially among the 14 bp Ins, +3142 G and +3187 A alleles, with rare exceptions. In addition, a nearly perfect LD was observed between +3027 A/C and +3035 C/T alleles, +3010 C/G and +3142 C/G alleles, and +3010 C/G and +3187 A/G alleles (D’=0.98, r2 = 0.92).
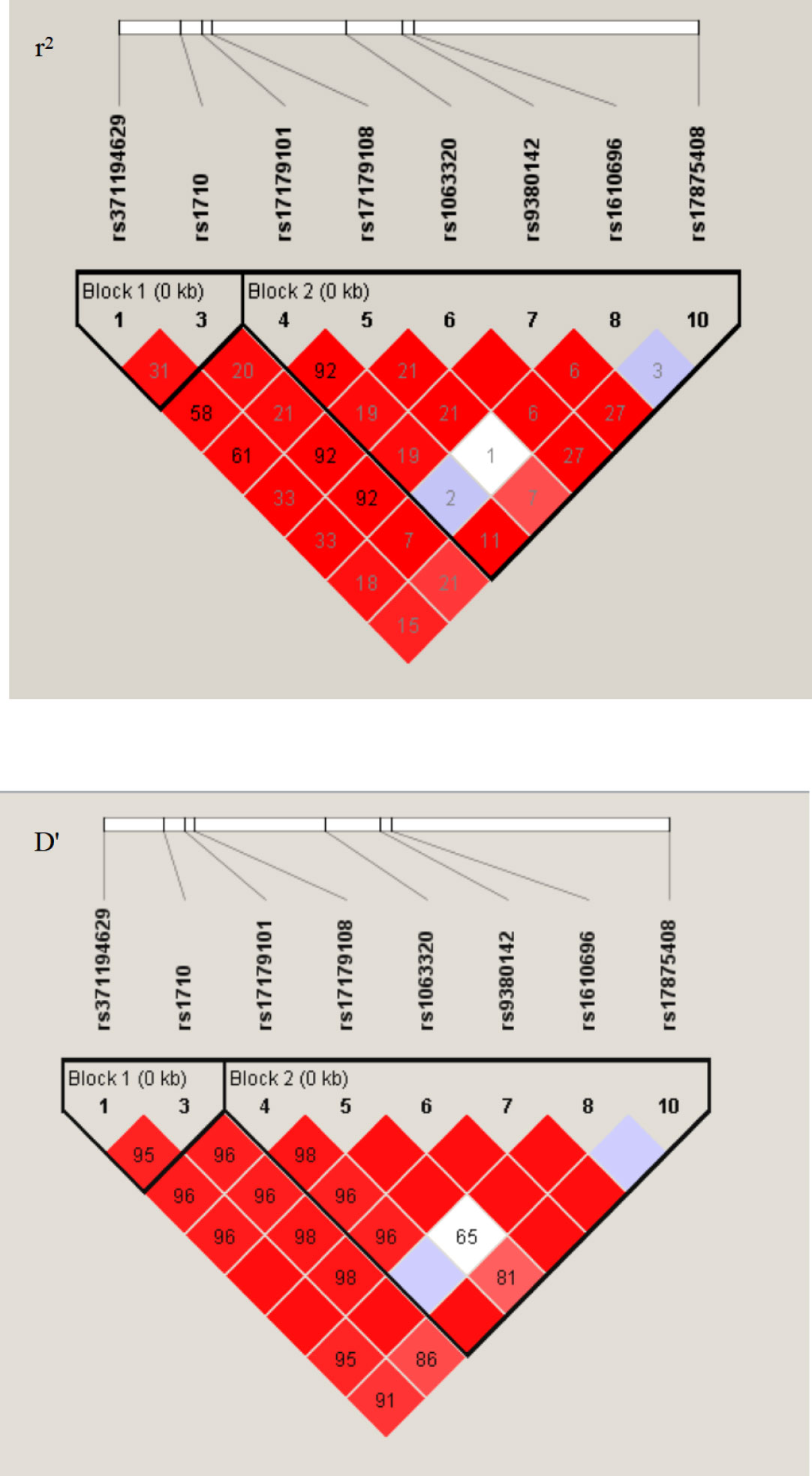
Figure 1 LD patterns between eight pairs polymorphic sites at the 3’UTR region of HLA-G gene. The image was generated in the Haploview 4.2. D’ and r2, pairwise correlation between polymorphic sites. Areas in red indicated strong LD (r2>0.8), shades of pink indicated moderate LD, and white indicates low LD. The r2 values (×100) for the marker pairs are listed in the corresponding boxes.
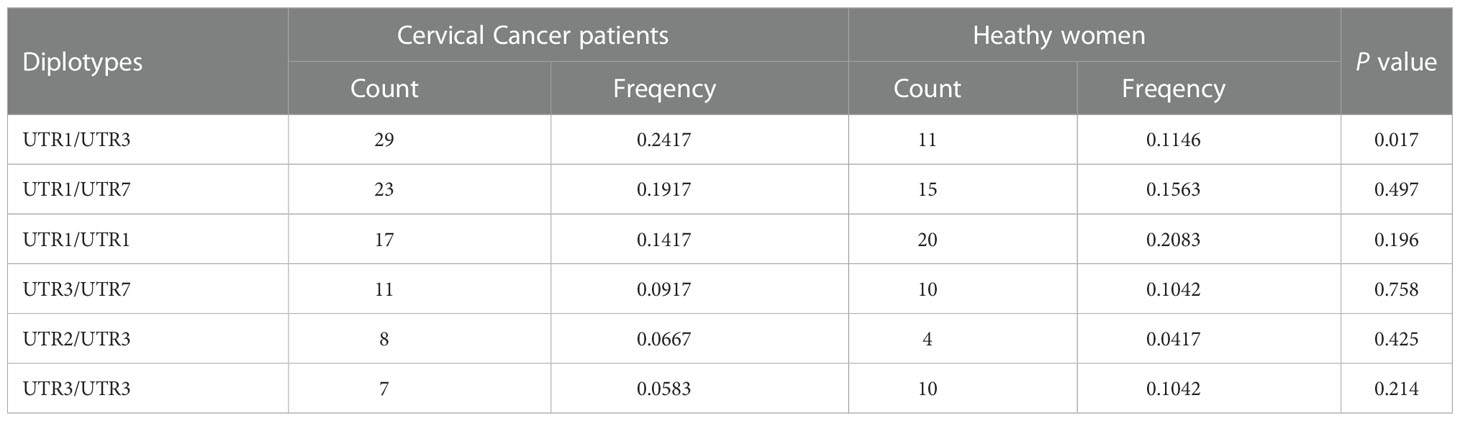
We further analyzed 23 different diplotypes in the study population. The most common diplotypes in cervical cancer patients were UTR-1/UTR-3 (24.2%), UTR-1/UTR-7 (19.2%) and UTR-1/UTR-1 (14.2%), while the most common diplotypes in healthy women were UTR-1/UTR-1 (20.8%), UTR-1/UTR-7 (15.6%) and UTR-1/UTR-3 (11.5%) (Table 4). The frequency of the UTR-1/UTR-3 diplotype in patients was significantly higher than that in healthy women (P<0.05). These findings further confirmed that HLA-G genetic polymorphism was related to the risk of cervical cancer in the Chinese Han population in the Taizhou area, especially in women with diplotype UTR-1/UTR-3, which may favor susceptibility to cervical cancer.
sHLA-G plasma levels in cervical cancer patients and healthy women
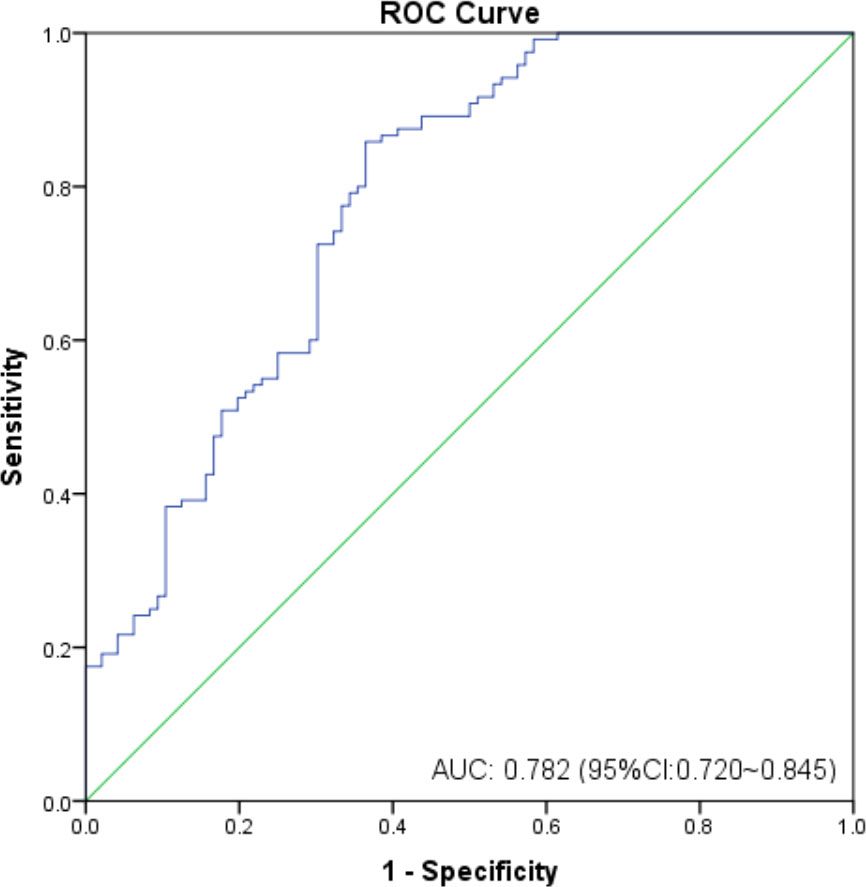
As shown in Figure 2, the mean sHLA-G level in patients’ peripheral blood was almost threefold higher than that in healthy women (P<0.0001, 115.80 ± 177.6 vs. 32.99 ± 26.74 U/mL), regardless of HLA-G polymorphisms. We further analyzed whether sHLA-G levels could differentiate cervical cancer patients from healthy women by ROC analysis (Figure 3). The results showed that sHLA-G had good diagnostic performance in differentiating patients with cervical cancer (AUC=0.782). The ROC curve showed that at a threshold value of 33.75 U/mL, the diagnostic sensitivity and specificity of sHLA-G for cervical cancer were 86% and 64%, respectively. Among patients, the sHLA-G plasma level increased with increasing FIGO stage, but the difference was not significant (P>0.05). Compared with healthy women, the sHLA-G plasma level in the patients with stage I were significantly higher (P<0.001). Therefore, sHLA-G can be also considered a supplementary biomarker for the early diagnosis of cervical cancer.
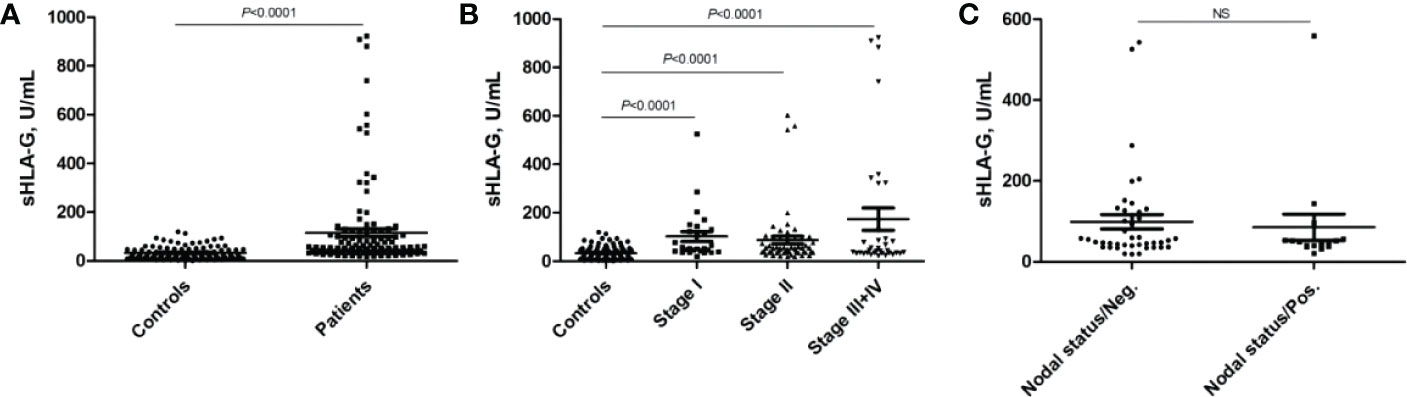
Figure 2 Comparison of sHLA-G plasma levels of cervical cancer patients and healthy women. (A) sHLA-G is significantly (P<0.0001) elevated in patients, (B) sHLA-G levels increase with ascending FIGO stage in patients without reaching significance. (C) sHLA-G levels based on nodal status. Bars indicate mean ± SEM. Statistic was performed by Mann-Whitney test. NS, not significant.
Patients were grouped according to the sHLA-G cut-off value, and no significant difference was found in the overall survival time between patients with low sHLA-G levels (< 33.75 U/mL) and those with high sHLA-G levels (> 33.75 U/mL). Survival analysis showed that there was no correlation between the sHLA-G plasma level and the overall survival time of patients.
Analysis of sHLA-G plasma levels based on HLA-G genetic polymorphisms
To further investigate whether the plasma sHLA-G levels were affected by the patients’ genetic background of HLA-G polymorphic sites, we analyzed sHLA-G plasma levels in patients’ peripheral blood according to HLA-G polymorphism. As depicted in Figure 4, we compared the sHLA-G levels between HLA-G 3’UTR diplotypes and observed difference. Patients with the Del-3142C/Del-3142C diplotype had higher sHLA-G production than patients with other diplotypes. We observed that the sHLA-G level increased in patients with the Del-3142C/Del-3142C diplotype (76.63 ± 61.71 U/mL) compared to patients with the Ins-3142G/Ins-3142G diplotype (60.34 ± 48.58 U/mL) without significance. These results confirmed that HLA-G polymorphisms may influence sHLA-G levels in patients’ peripheral blood. None of the HLA-G 3’UTR diplotypes were related to the overall survival time of patients.
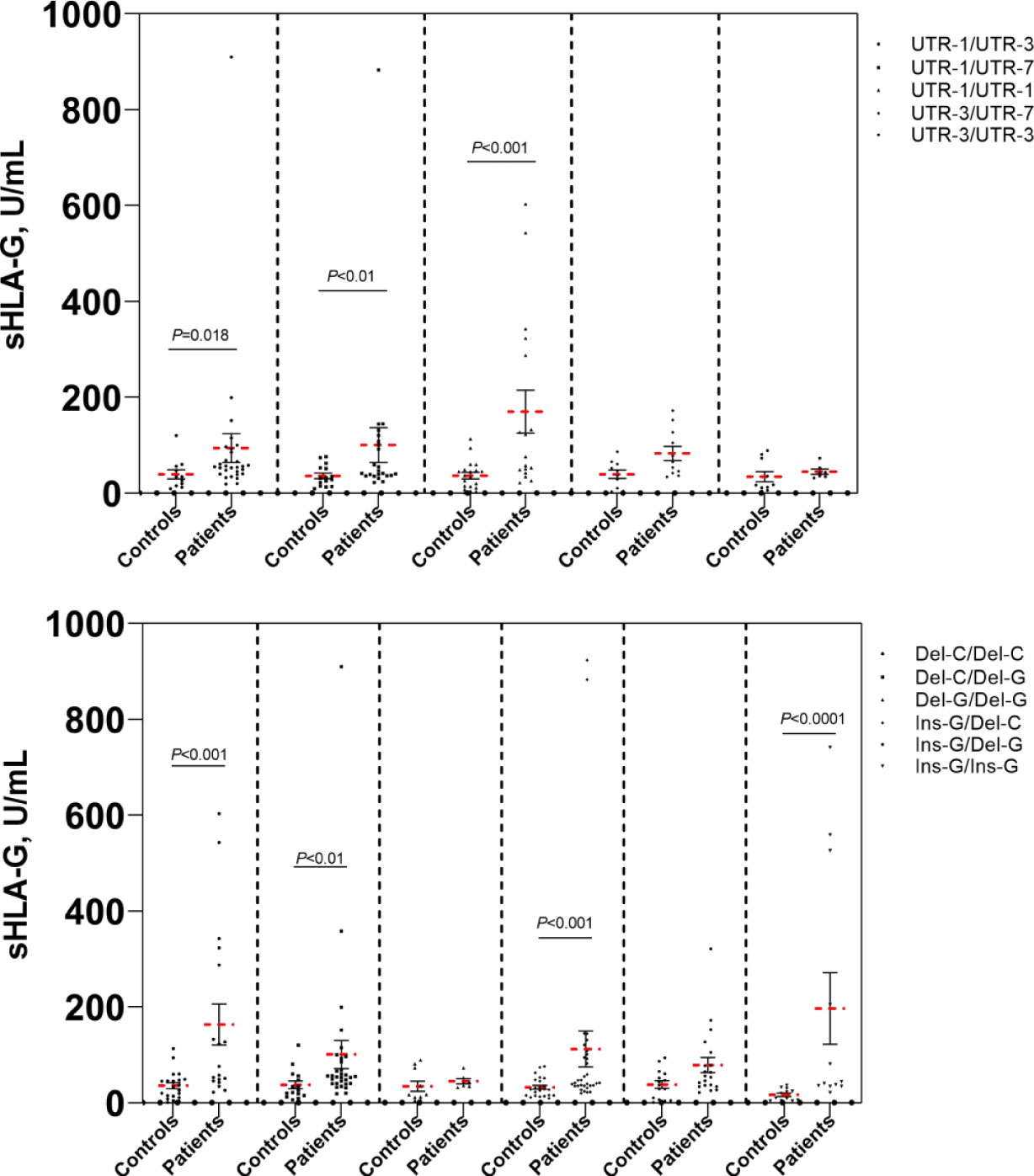
Figure 4 Comparison of sHLA-G levels of cervical cancer patients and healthy controls. Bars indicate mean ± SEM.
Discussion
HLA-G is an inhibitory checkpoint molecule, which is highly expressed in gynecological cancers, such as cervical cancer (3, 4, 7–10), ovarian cancer (20), breast cancer (21, 22), and endometrial cancer (23). It’s known that virus-infected cells and malignant cells escape the host’s immune surveillance by inducing the expression of HLA-G (4–6, 10). The HLA-G 3’UTR contains several SNP sites related to HLA-G mRNA stability that impact HLA-G expression (13–19, 24, 25). It has been reported that HLA-G/sHLA-G is highly expressed in the tissues or peripheral blood of cervical cancer patients, contributing to the immune escape of cancer cells (4, 7–10). Here, we focused on evaluating the possible association of HLA-G 3’UTR polymorphisms with sHLA-G plasma levels in the Chinese Han population and its value in clinical prognostic evaluation and survival outcome prediction in patients with cervical cancer. In the present study, we observed that (i) the frequency of the heterozygote CT genotype at position +3422 (rs17875408) was significantly higher in cervical cancer patients than in healthy women (P<0.05), suggesting that the +3422 C/T genotype may be a highly significant risk factor for cervical cancer; (ii) the frequency of the UTR-1/UTR-3 diplotype in patients was significantly higher than that in healthy women (P<0.05), which may play a key role in cervical carcinogenesis; (iii) HLA-G 3’UTR polymorphic sites may influence the sHLA-G plasma level in patients’ peripheral blood, especially 14 bp Ins/Del (rs371194629) and +3142 C/G (rs1063320); and (iii) the sHLA-G level had good diagnostic performance in differentiating patients with cervical cancer from healthy women, suggesting that sHLA-G can be a supplementary biomarker of cervical cancer.
In this study, we identified eight alleles, sixteen haplotypes and twenty-three diplotypes in the HLA-G gene 3’UTR in our Taizhou population. Each of these different haplotypes was in LD with specific HLA-G 3’UTR alleles. A high LD among these alleles was detected, especially among 14 bp Ins, +3142 G and +3187 A. These alleles have previously been reported to have lower levels of HLA-G mRNA, indicating that their effects are not independent, thereby leading to decreased HLA-G expression (16–18, 24, 25). Our results were consistent with this conclusion; we observed that the 14 bp Ins allele was related to low sHLA-G production and that the 14 bp Del allele was related to high sHLA-G production (Figure 4). Previous studies have shown that elevation of plasma sHLA-G levels is common in patients with various kinds of tumors (10, 20, 24, 26, 27). Notably, sHLA-G plasma levels decreased significantly within 30 days after radical hysterectomy in our previous study (10). This findings confirmed that the source of plasma sHLA-G was mainly expressed and released by cancer cells in the tissue microenvironment. Therefore, we proposed that the increase in sHLA-G was regulated not only by the immune microenvironment of patients but also by their own genetic background. In addition, several studies have debated the regulation of sHLA-G levels by HLA-G 3’UTR polymorphisms. These disputes may vary among different populations with different genetic backgrounds (17, 28, 29).
UTR-1 was the most common haplotype (41.2%), followed by UTR-3 (24.0%), UTR-7 (21.9%), UTR-2 (8.9%) and UTR-21 (1.0%) in patients from the Taizhou area, Southeast China. The order of haplotype frequencies in our Taizhou population was almost consistent with the frequencies previously reported in populations from South China, which were UTR-1 (43.0%), UTR-3 (26.5%), UTR-7 (22.0%), UTR-2 (6.5%) and UTR-4 (2.0%) (19). In the Taizhou population, UTR-1 and UTR-3 with the 14 bp Del allele were the most common haplotypes, which were theoretically considered to exhibit greater sHLA-G production than those with the 14 bp Ins allele. As shown in Figure 4, the mean sHLA-G level in patients with the UTR-1/UTR-1 diplotype was almost twofold higher than that in healthy women with the UTR-1/UTR-1 diplotype. The results suggested that the peripheral sHLA-G level of cancer patients was also regulated by other clinical factors. Our previous study showed that sHLA-G expression may be induced by cytokines in the peripheral blood circulation (10). UTR-2 and UTR-7 with 14 bp Ins, +3142 G and +3187 A alleles may be related to lower sHLA-G levels (17, 19, 30–32). Of note, our results showed that the frequency of the UTR-1/UTR-7 diplotype in patients was significantly higher than that in healthy women (P<0.05). HPV-infected women with the UTR-1/UTR-7 diplotype may be more susceptible to cervical cancer.
In addition, this study investigated the performance of the sHLA-G level in the diagnosis of cervical cancer patients and determined that it has a good diagnostic performance (AU-ROC > 0.7). sHLA-G can be considered a supplementary biomarker of cervical cancer. When the patients were grouped according to the sHLA-G cut-off value, no significant difference was found in the overall survival time between patients with low sHLA-G levels (< 33.75 U/mL) and those with high sHLA-G levels (> 33.75 U/mL). Survival analysis showed that there was no correlation between the sHLA-G plasma level and the overall survival time of patients.
The limitations of this study are as follows: (i) lack of identification of HLA-G dimers and monomers in the peripheral plasma of cervical cancer patients; such analysis might help to corroborate the function of different HLA-G polymers; and (ii) lack of identification of shed HLA-G1 and soluble HLA-G5 in sHLA-G; assessing different sHLA-G isoforms might help for development of immunotherapy for cervical cancer.
In conclusion, our study provides evidence that certain HLA-G 3’UTR polymorphisms can influence sHLA-G levels in cancer patients’ peripheral blood, especially 14 bp Ins/Del (rs371194629) and 3142 C/G (rs1063320). Our previous and current findings enhance our understanding of HLA-G polymorphisms, which are not only associated with susceptibility to HPV infection but also involved in the susceptibility to and progression of cervical cancer by affecting sHLA-G levels. Particular HLA-G diplotypes are related to the cervical cancer outcome. In addition, the present study further confirmed that sHLA-G may represent a novel plasma biomarker for the prognosis and potential therapeutic target of cervical cancer.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
Ethics statement
This study was approved by the Medical Ethics Review Committee of Taizhou Hospital (approval #K20220226), and written informed consent from all study participants was obtained before enrolment.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by grants from National Natural Science Foundation of China (81901625), Natural Science Foundation of Zhejiang province (LY20H100004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yuan Y, Cai X, Shen F, Ma F. HPV post-infection microenvironment and cervical cancer. Cancer Lett (2021) 497:243–4. doi: 10.1016/j.canlet.2020.10.034
2. Xu HH, Shi WW, Lin A, Yan WH. HLA-G 3’ untranslated region polymorphisms influence the susceptibility for human papillomavirus infection. Tissue Antigens (2014) 84(2):216–22. doi: 10.1111/tan.12359
3. Xu HH, Zhang X, Zheng HH, Han QY, Lin AF, Yan WH. Association of HLA-G 3’ UTR polymorphism and expression with the progression of cervical lesions in human papillomavirus 18 infections. Infect Agent Cancer (2018) 13:42. doi: 10.1186/s13027-018-0217-2
4. Xu HH, Yan WH, Lin A. The role of HLA-G in human papillomavirus infections and cervical carcinogenesis. Front Immunol (2020) 11:1349. doi: 10.3389/fimmu.2020.01349
5. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: An immune checkpoint molecule. Adv Immunol (2015) 127:33–144. doi: 10.1016/bs.ai.2015.04.001
6. Lin A, Yan WH. HLA-G as an inhibitor of immune responses. Methods Mol Biol (2016) 1371:3–9. doi: 10.1007/978-1-4939-3139-2_1
7. Li XJ, Zhang X, Lin A, Ruan YY, Yan WH. Human leukocyte antigen-G (HLA-G) expression in cervical cancer lesions is associated with disease progression. Hum Immunol (2012) 73(9):946–9. doi: 10.1016/j.humimm.2012.07.041
8. Dong DD, Yang H, Li K, Xu G, Song LH, Fan XL, et al. Human leukocyte antigen-G (HLA-G) expression in cervical lesions: association with cancer progression, HPV 16/18 infection, and host immune response. Reprod Sci (2010) 17(8):718–23. doi: 10.1177/1933719110369183
9. Zheng N, Wang CX, Zhang X, Du LT, Zhang J, Kan SF, et al. Up-regulation of HLA-G expression in cervical premalignant and malignant lesions. Tissue Antigens (2011) 77(3):218–24. doi: 10.1111/j.1399-0039.2010.01607.x
10. Xu HH, Xie YY, Jun-Gan, Yang Z, Han QY. Dynamic changes of soluble HLA-G and cytokine plasma levels in cervical cancer patients: potential role in cancer progression and immunotherapy. J Cancer Res Clin Oncol (2022). doi: 10.1007/s00432-022-04331-4
11. Ishitani A, Geraghty DE. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc Natl Acad Sci U.S.A. (1992) 89(9):3947–51. doi: 10.1073/pnas.89.9.3947
12. Tronik-Le Roux D, Daouya M, Jacquier A, Schenowitz C, Desgrandchamps F, Rouas-Freiss N, et al. The HLA-G immune checkpoint: a new immuno-stimulatory role for the α1-domain-deleted isoform. Cell Mol Life Sci (2022) 79(6):310. doi: 10.1007/s00018-022-04359-2
13. Dias FC, Castelli EC, Collares CV, Moreau P, Donadi EA. The role of HLA-G molecule and HLA-G gene polymorphisms in tumours, viral hepatitis, and parasitic diseases. Front Immunol (2015) 6:9. doi: 10.3389/fimmu.2015.00009
14. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res (2014) 2014:734068. doi: 10.1155/2014/734068
15. Bortolotti D, Gentili V, Rotola A, Di Luca D, Rizzo R. Implication of HLA-G 3’ untranslated region polymorphisms in human papillomavirus infection. Tissue Antigens (2014) 83(2):113–8. doi: 10.1111/tan.12281
16. Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P. The 14 bp deletion-insertion polymorphism in the 3’ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol (2003) 64(11):1005–10. doi: 10.1016/j.humimm.2003.08.347
17. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3’ untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PloS One (2013) 8(10):e71742. doi: 10.1371/journal.pone.0071742
18. Castelli EC, Moreau P, Oya e Chiromatzo A, Mendes-Junior CT, Veiga-Castelli LC, Yaghi L, et al. In silico analysis of microRNAS targeting the HLA-G 3’ untranslated region alleles and haplotypes. Hum Immunol (2009) 70(12):1020–5. doi: 10.1016/j.humimm.2009.07.028
19. Sabbagh A, Luisi P, Castelli EC, Gineau L, Courtin D, Milet J, et al. Worldwide genetic variation at the 3’ untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. Genes Immun (2014) 15(2):95–106. doi: 10.1038/gene.2013.67
20. Babay W, Boujelbene N, Ben Yahia H, Bortolotti D, Zemni I, Ouzari HI, et al. Prognostic significance of high circulating sHLA-G in ovarian carcinoma. HLA (2021) 98(4):357–65. doi: 10.1111/tan.14374
21. Rebmann V, Schwich E, Michita RT, Grüntkemeier L, Bittner AK, Rohn H, et al. Systematic evaluation of HLA-G 3’Untranslated region variants in locally advanced, non-metastatic breast cancer patients: UTR-1, 2 or UTR-4 are predictors for therapy and disease outcome. Front Immunol (2022) 12:817132. doi: 10.3389/fimmu.2021.817132
22. Zheng G, Jia L, Yang AG. Roles of HLA-G/KIR2DL4 in breast cancer immune microenvironment. Front Immunol (2022) 13:791975. doi: 10.3389/fimmu.2022.791975
23. Ben Yahia H, Babay W, Bortolotti D, Boujelbene N, Laaribi AB, Zidi N, et al. Increased plasmatic soluble HLA-G levels in endometrial cancer. Mol Immunol (2018) 99:82–6. doi: 10.1016/j.molimm.2018.04.007
24. Yie SM, Li LH, Xiao R, Librach CL. A single base-pair mutation in the 3’-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol Hum Reprod (2008) 14(11):649–53. doi: 10.1093/molehr/gan059
25. Chan JJ, Zhang B, Chew XH, Salhi A, Kwok ZH, Lim CY, et al. Pan-cancer pervasive upregulation of 3’ UTR splicing drives tumourigenesis. Nat Cell Biol (2022) 24(6):928–39. doi: 10.1038/s41556-022-00913-z
26. Heidari MH, Movafagh A, Abdollahifar MA, Abdi S, Barez MM, Azimi H, et al. Evaluation of sHLA-G levels in serum of patients with prostate cancer identify as a potential of tumor marker. Anat Cell Biol (2017) 50(1):69–72. doi: 10.5115/acb.2017.50.1.69
27. Kluckova K, Durmanova V, Bucova M. Soluble HLA-G, its diagnostic and prognostic value and potential target molecule for future therapy in cancer. Bratisl Lek Listy (2021) 122(9):60–617. doi: 10.4149/BLL_2021_2097
28. Tizaoui K, Jalouli M, Boujelbene N, Harrath AH, Ouzari HI, Rizzo R, et al. The relationship of 3’UTR HLA-G 14-bp insertion/deletion and +3142 C/G polymorphisms and soluble HLA-G expression with gynecological cancers: An updated meta-analysis. Immun Inflammation Dis (2022) 10(7):e645. doi: 10.1002/iid3.645
29. Lucena-Silva N, Monteiro AR, de Albuquerque RS, Gomes RG, Mendes-Junior CT, Castelli EC, et al. Haplotype frequencies based on eight polymorphic sites at the 3’ untranslated region of the HLA-G gene in individuals from two different geographical regions of Brazil. Tissue Antigens (2012) 79(4):272–8. doi: 10.1111/j.1399-0039.2012.01842.x
30. Durmanova V, Bandzuchova H, Zilinska Z, Tirpakova J, Kuba D, Buc M, et al. Association of HLA-G polymorphisms in the 3’UTR region and soluble HLA-G with kidney graft outcome. Immunol Invest (2019) 48(6):644–58. doi: 10.1080/08820139.2019.1610888
31. Poras I, Yaghi L, Martelli-Palomino G, Mendes-Junior CT, Muniz YC, Cagnin NF, et al. Haplotypes of the HLA-G 3’ untranslated region respond to endogenous factors of HLA-g+ and HLA-g- cell lines differentially. PloS One (2017) 12(1):e0169032. doi: 10.1371/journal.pone.0169032
Keywords: HLA-G, 3’UTR, diplotypes, sHLA-G, cervical cancer
Citation: Gan J, Di X-H, Yan Z-Y, Gao Y-F and Xu H-H (2022) HLA-G 3’UTR polymorphism diplotypes and soluble HLA-G plasma levels impact cervical cancer susceptibility and prognosis. Front. Immunol. 13:1076040. doi: 10.3389/fimmu.2022.1076040
Received: 21 October 2022; Accepted: 06 December 2022;
Published: 21 December 2022.
Edited by:
Ricardo Pujol Borrell, Universitat Autònoma de Barcelona, SpainReviewed by:
Ashwin Ajith, Augusta University, United StatesInes Zidi, Tunis El Manar University, Tunisia
Copyright © 2022 Gan, Di, Yan, Gao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui-Hui Xu, aHVpNzM5QDE2My5jb20=
 Jun Gan1
Jun Gan1 Hui-Hui Xu
Hui-Hui Xu