
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 12 January 2023
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1060985
This article is part of the Research Topic Women In Viral Immunology View all 5 articles
HIV emerged following cross-species transmissions of simian immunodeficiency viruses (SIVs) that naturally infect non-human primates (NHPs) from Africa. While HIV replication and CD4+ T-cell depletion lead to increased gut permeability, microbial translocation, chronic immune activation, and systemic inflammation, the natural hosts of SIVs generally avoid these deleterious consequences when infected with their species-specific SIVs and do not progress to AIDS despite persistent lifelong high viremia due to long-term coevolution with their SIV pathogens. The benign course of natural SIV infection in the natural hosts is in stark contrast to the experimental SIV infection of Asian macaques, which progresses to simian AIDS. The mechanisms of non-pathogenic SIV infections are studied mainly in African green monkeys, sooty mangabeys, and mandrills, while progressing SIV infection is experimentally modeled in macaques: rhesus macaques, pigtailed macaques, and cynomolgus macaques. Here, we focus on the distinctive features of SIV infection in natural hosts, particularly (1): the superior healing properties of the intestinal mucosa, which enable them to maintain the integrity of the gut barrier and prevent microbial translocation, thus avoiding excessive/pathologic immune activation and inflammation usually perpetrated by the leaking of the microbial products into the circulation; (2) the gut microbiome, the disruption of which is an important factor in some inflammatory diseases, yet not completely understood in the course of lentiviral infection; (3) cell population shifts resulting in target cell restriction (downregulation of CD4 or CCR5 surface molecules that bind to SIV), control of viral replication in the lymph nodes (expansion of natural killer cells), and anti-inflammatory effects in the gut (NKG2a/c+ CD8+ T cells); and (4) the genes and biological pathways that can shape genetic adaptations to viral pathogens and are associated with the non-pathogenic outcome of the natural SIV infection. Deciphering the protective mechanisms against SIV disease progression to immunodeficiency, which have been established through long-term coevolution between the natural hosts and their species-specific SIVs, may prompt the development of novel therapeutic interventions, such as drugs that can control gut inflammation, enhance gut healing capacities, or modulate the gut microbiome. These developments can go beyond HIV infection and open up large avenues for correcting gut damage, which is common in many diseases.
The statement “The origin of AIDS is more ancient than the origin of HIV-1” by Sharp and Hahn (1) emphasizes the long-term presence of the SIVs, the simian ancestors of the HIV pandemic strains, in African non-human primate (NHP) populations (2–7). Species-specific strains of SIV have emerged and spread among African NHPs through the codivergence of SIV lineages with host speciation, e.g., contemporarily, SIVs are naturally infecting over 45 NHP species in Africa, while they are absent in NHPs native to the Asian continent and the Americas (5, 8, 9).
HIV-1 emerged through cross-species transmission of SIV to humans (1), leading to massive CD4+ T-cell loss, inflammation, and chronic immune activation in the gut and systemically, which altogether eventually lead to an exhaustion of the immune system and the development of opportunistic infections and AIDS-associated comorbidities, such as hypercoagulation and other cardiovascular diseases, hyperlipidemia, chronic kidney and hepatic diseases, osteoporosis, endocrine diseases, and cancers (10, 11). While some aspects of AIDS/HIV pathogenesis are reproduced in experimentally infected Asian macaques that are vulnerable to SIVmac-induced immunodeficiency (12, 13), the vast majority of modern-day African NHPs manifest a benign course of natural SIV infection when infected with their species-specific SIV strains (14–20). These natural host species do not progress to immunodeficiency despite life-long viremia and CD4+ T-cell loss in the gut and periphery, e.g., sooty mangabey (SM, Cercocebus atys) (18, 21), African green monkey (AGM, genus Chlorocebus) (22, 23), and mandrills (MND, Mandrillus sphinx) (19, 24).
Although the course of SIV infection in the natural host species is generally benign, several lines of evidence demonstrate that the viruses that naturally infect African NHP hosts do, in fact, retain their virulence and have the capacity to cause pathogenic infections; however, this is controlled by the superior species-specific host defense mechanisms (1). Similar features of SIV infections between natural hosts and progressing hosts (25–27) include early and massive loss of CD4+ T cells in the gut mucosa (23, 28–30) but the ability to preserve healthy levels of Th17 cells, which play a key role in mucosal immunity (promoting the recruitment of neutrophils and expression of antimicrobial products) (31, 32), in contrast to SIV-infected macaques (32) and HIV-infected humans (31) (2). The description of rare cases of AIDS in SIV-infected African NHPs after a long incubation period or in senescing individuals revealing that the pathogenic effects of SIV manifested in a background of a weakening immune system (33, 34): a feral born AGM (aged 12 years) (35), a captive-born SM naturally infected by SIVsmm transmitted in the colony (aged 20 years) (36), a feral-born mandrill naturally infected in the wild (aged 18 years) (37), and a black mangabey experimentally infected with SIVsmm (38) (3). Pathogenicity of species-specific strains from natural hosts to at least some species of Asian macaques upon direct cross-species transmission is routinely leveraged by employing the rhesus macaque (Macaca mulatta), the pigtailed macaque (Macaca nemestrina), and the cynomolgus macaque (Macaca fascicularis) as surrogate models for studying HIV-1 infection in humans (39, 40) (4). Lack of immunodeficiency or other disease in AGMs infected with a pathogenically enhanced HIV-1-like SIVagm (expressing Vpu and Nef proteins) was reported over a 5-year infection/follow-up. Despite being associated with moderately increased levels of chronic immune activation, this infection neither accelerated CD4+ T-cell depletion nor caused overt AIDS (41). Taken together, the general lack of disease progression in the natural hosts is not due to differences in natural history or to a lack of cytopathicity of the virus but rather the result of active protective mechanisms against immunodeficiency resulting from host adaptations to highly replicating virus.
This ‘well-tempered SIV infection’ (42) in the African NHP hosts has been previously linked to several main protective strategies (1): a lack of microbial translocation from the gut, and therefore, a lack of stimulation of inflammation and immune activation (43) (2); anti-inflammatory mechanisms controlling these processes and preventing chronic inflammation (44–46) (3); homeostatic regulation of various immune cell populations, including the sparing of critical immune cell subsets through the downregulation of CD4 receptors (47–50) while preserving some of their functionalities, or downregulation of CCR5 co-receptor on the CD4+ T-cell surface at sites critical for SIV pathogenesis (51) and in immature individuals (52–54) (Figure 1).
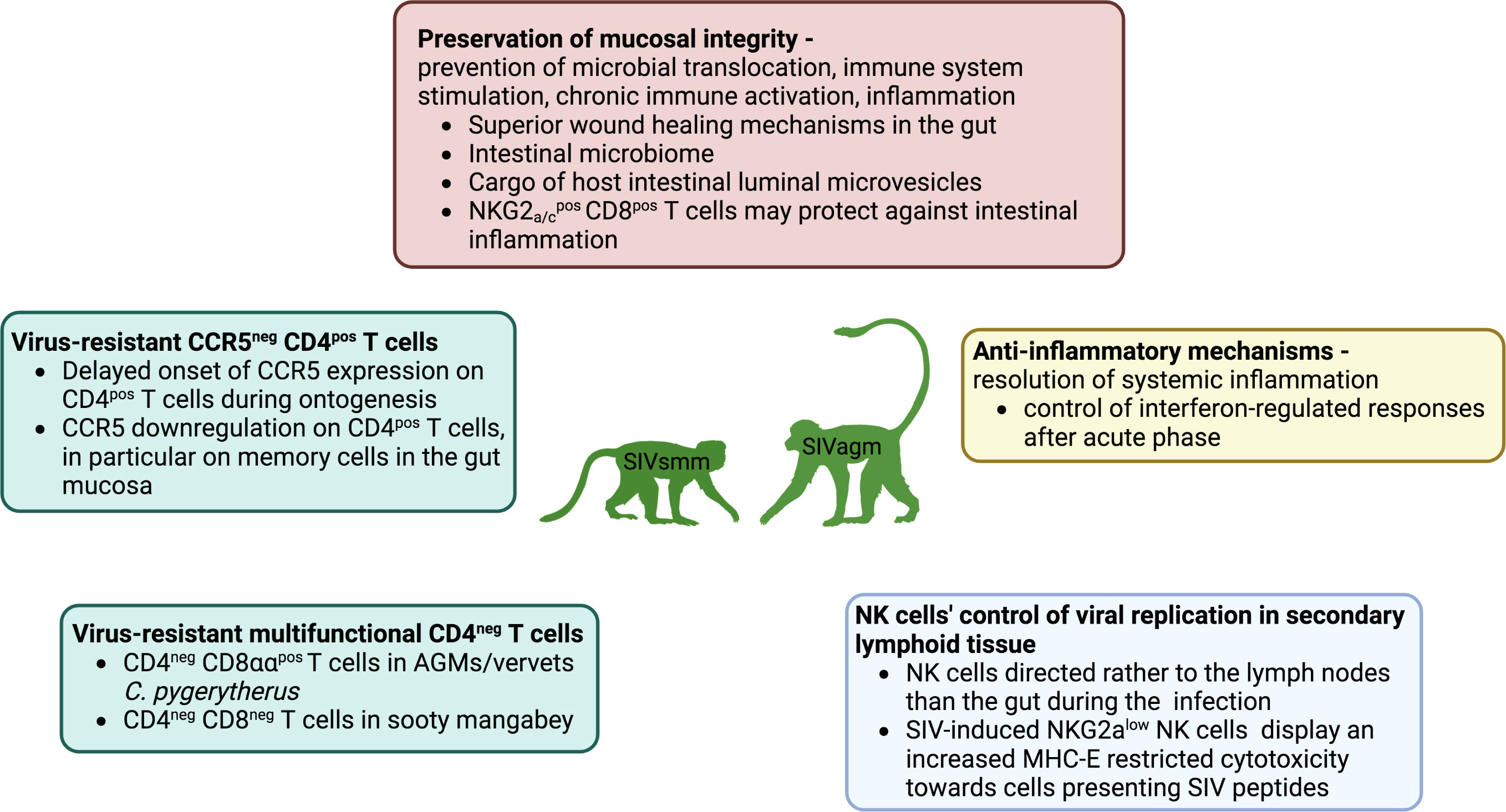
Figure 1 Mechanisms of protection against SIV infections include homeostatic regulation of immune cell populations through target cell restriction by downregulation of CD4 or CCR5 in T-cells (green frame) and NK cell colocalization in secondary lymphoid tissues, expansion and enhanced MHC-E restricted activity (blue frame), preservation of mucosal immunity, and thus prevention of microbial translocation, immune system stimulation and chronic immune activation. Inflammation is undergirded by highly effective non-inflammatory wound healing in the intestinal mucosa, the anti-viral content of epithelial microvesicles in the gut, and a potential anti-inflammatory role of NKG2a/c+CD8+ T cells in the gut, while the role of the intestinal microbiome remains unclear (red frame), and systemic anti-inflammatory mechanisms control the expression of interferon-stimulated genes after the acute phase (yellow).
The identification of critical differences in the host mechanisms counteracting the pathogenic HIV infection in humans (or SIV infection of experimental NHP models of simian AIDS) and non-pathogenic infection in some natural hosts may shed light on the host mechanisms controlling the pathogenicity of lentiviral infection and inspire novel approaches to HIV/AIDS treatment.
SIV is an old pathogen in NHPs, as suggested by the lack of pathogenicity in the host species and demonstrated by two studies that combined molecular phylogeny with biogeography to recalibrate molecular clock analyses. The first study compared SIV sequences originating from the same species collected from Bioko Island and the Equatorial Guinea African mainland, which were once connected via a land bridge but became separated 12,800 years ago at the end of ice age. Recalibration of the molecular clocks to include the biogeography, and thus the fact that the last time when these two strains could have been in contact was before the collapse of the land bridge, led to the conclusion that SIV could have been infecting these NHP species for at least 32,000 years but was likely much more ancient (4).
The second study was performed on vervets from South Africa, which have been postulated to have coevolved with SIV over a long period of evolutionary time, initially estimated to be between 3 million to 100,000 years ago, with limits corresponding to the vervets dispersal across sub-Saharan Africa and mass Plio-Pleistocene migration of various African species. Mutation analysis in SIVs present in the South African vervets showed a striking divergence of SIVagm around the Drakensberg Mountain range, suggesting that the mountains represented an insurmountable geographical barrier (due to the lack of water and food resources) during the vervet and SIVagm spread, the timing of which could accordingly be established. The molecular clock based on the phylogenetic relationships among Chlorocebus SIVs and further geologically calibrated based on the Bioko split of SIVdrl of the drill monkey (Mandrillus leucophaeus) further narrowed down the time of divergence of vervets in South Africa to approximately 200,000 years (6). That allowed time for long-term selective pressure from the SIV pathogen to take place in the African vervets.
Cross-species transmission from NHPs to humans is not a very rare event, as demonstrated by the observation that, during the last century, multiple cross-species transmissions contributed to HIV emergence in humans: two from chimpanzees and two from gorillas, which were responsible for the emergence of the four HIV-1 groups, (M-P) and eight from the sooty mangabeys, which were responsible for the emergence of the eight HIV-2 groups (A-H) (55, 56). In the great apes, SIVcpz emerged following a recombination between SIVrcm from red-capped mangabey (Cercocebus torquatus torquatus) and SIVgsn/mus/mon from one of the species on which chimpanzees prey, i.e., greater spot-nosed monkey (Cercopithecus nictitans), mustached monkey (C. cephus), and mona monkey (C. mona), respectively (57). Such chimeric viruses emerge during superinfections/coinfection with different SIVs from different hosts, which enable recombinations (58–60).
Two out of four chimpanzee subspecies in West-Central and East-Central Africa, Pan troglodytes troglodytes and P. t. schweinfurthii, are infected with SIVcpz (SIVcpzPtt and SIVcpzPts, respectively) at a prevalence of 6%-14%, respectively (60–64), although the distribution of the virus is very uneven between different chimpanzee troops (60). Phylogenetic studies revealed that only SIVcpzPtt is an ancestor of HIV-1 (61, 65). This virus crossed the species barrier to humans in at least two instances, being responsible for the emergence of two HIV-1 groups: M (major), which likely emerged in the first half of the twentieth century (66) and is responsible for the world AIDS pandemic (estimated 98% of HIV infections worldwide) (55); and the non-pandemic HIV1 group N, limited to a small number of infections in Cameroon (61, 67, 68). The HIV-1 groups O and P originated following cross-species transmission of the SIVgor, a virus that naturally infects wild western-lowland gorillas from Cameroon (SIVgor) (69, 70). SIVgor also originated from cross-species transmissions of SIVcpzPtt (69, 71).
HIV-2, comprising eight groups, originated following eight cross-species transmissions of SIVsmm from sooty mangabeys (72–76). Interestingly, SMs naturally infected with SIVsmm were the origin of the major reference strains in macaques (77, 78), following accidental transmissions that occurred during kuru (79) and leprosy (80) experiments carried out in the 1970s in the National Primate Research Centers in the US (81). This accidental experimental transmission of these viruses (which involved serial passages) is probably the reason for their high pathogenicity in macaques (82). Note that direct cross-species transmission of SIVsmm to rhesus macaques may have a very variable pathogenic outcome, ranging from pathogenic infection to virus control (Apetrei, unpublished). Other SIVs are completely controlled when experimentally transmitted to RMs (83–85). Among the different species of macaques, the pigtailed macaques appear to be the most permissive to cross-species infections, as shown by the persistent infections with SIVagm or SIVrcm in PTMs (86–88). The course of progressive SIV infection in macaques recapitulates all aspects of HIV-1 infection in humans, albeit in a more condensed timeframe, and as such, the macaques represent a pathogenic NHP model of simian AIDS (40, 89).
AGM, also called the savanna monkey (vervet), is a term that is broadly applied to members of the major subspecies of the single-species genus Chlorocebus of the Old World monkey superfamily (90). They are highly abundant primates ranging across sub-Saharan Africa, with the exception of tropical forests and deserts, and heavily infected with species-specific SIVagm (SIVsab in C. sabaeus, SIVgri in C. aethiops, SIVtan in C. tantalus, SIVmal-ZMB in C. cynosuros, and SIVver in C. pygerythrus) (6, 52, 91). Extensive laboratory data gathered from experimental SIVsab infection in Caribbean AGMs demonstrates that a benign course of the infection characterized by high peak viremia of 107-108 copies/ml 8-10 days postinfection (dpi) that drops to set point values of 2x105 copies/ml by 28 dpi, a transient CD4+ T-cell decrease in the blood and lymph nodes, and a lack of systemic T-cell activation (22). In AGMs, inflammatory responses (IL-10 and INF-γ) are activated immediately yet transiently, while lagging in the progressing SIVmac251 infection of the RMs (44). The gut is a major replication site for SIVagm, yet virus is detectable across various tissues, including blood and lymph nodes (the virus from these tissues has more restricted growth in human T-cell lines) and brain and CSF (the virus from these samples is capable of infecting macrophage cultures) (16). However, despite markedly high virus levels in the nervous system, no neuropathologies were observed in AGMs (16).
SIV infection in wild AGM populations, in particular, in South African C. pygerythrus (6) and West African C. sabaeus in the Gambia (52) has been broadly studied using minimally invasive procedures to assess the markers of chronic immune activation, microbial translocation, abundances of different cell populations, and gut and genital microbiome (6, 52, 92). A combination of viral load measurements and serological testing (ELISA for gp41 peptide) in the plasma allowed the staging of SIV infection (acute/chronic) in these cross-sectional samples based on the Fiebig criteria, according to which chronic infection is diagnosed based on the moderately high viral loads in the presence of anti-SIV antibodies, and acute infection can be defined based on high viral loads in the absence of anti-SIV antibodies (93). The major findings of these pathogenesis studies in the wild (the only studies ever performed in wild animals) were:
Wild AGMs showed a high prevalence of the SIV infection in adults, with a strong bias towards a higher infection rate in females than males (SIV prevalence in females was 78% in South Africa and 90% in The Gambia, while in the males, the prevalence was significantly lower: 57% in South Africa, and 36% in the Gambia) concordant with unequal male access to females and possibly an increased transmissibility to females (6, 52) [a similar bias to that observed for HIV-1 transmission in humans (94)]. The high frequency of SIV infection was also associated with a very active SIV transmission in the wild populations, as suggested by the high frequency of acute infection (leading to an estimate of SIVver incidence of 4.4% in South Africa) (6).
Interestingly, a large proportion of reproductively active adult females (10-22%) avoid SIV infection through heterosexual transmission (6, 52), yet it remains unknown whether this apparent resistance is transient or permanent. It would be interesting to evaluate what factors (e.g., host genetic variants, gene expression, epigenetic modifications, and/or genital and gut microbiomes are associated with the resistance) are protecting the ESN individuals against SIV acquisition and how they are shaping the key features of viral resistance.
SIV infection is nearly absent in infants and very rare in juveniles, intermediate in young adults, and high in adults, pointing to a very rare MTIT in the wild and suggesting that the vast majority of transmissions occur around the time when monkeys reach sexual maturity, join the reproductive community, and become exposed to the virus via heterosexual contacts and aggressive behavior (6, 52, 95, 96). The very low incidence of SIV MTIT has also been confirmed in captive AGMs, SMs, and MNDs (97–100).
This bias implicates the protection of immature individuals through adaptive biological mechanism(s). CCR5, the main co-receptor of SIV and HIV, shows a striking age-specific expression pattern in CD4+ T cells, rising from nearly absent in young individuals to highly abundant in adults (52). Additionally, infected young individuals tend to show higher CCR5 expression in CD4+ T cells than uninfected ones (52). The age-related maturation of CCR5 expression in CD4+ T cells is associated with a similar age-related increase in the frequency of SIV infection, which points to target cell restriction as a critical defense mechanism against the lentiviral infection of immature individuals (53, 101). This hypothesis was confirmed in experimental mucosal transmission studies in AGMs (51). In experimental studies of breast-feeding transmission of SIVmnd-1 in infant mandrills, low CCR5 expression in CD4+ T cells was reported to be the main determinant of the lack of SIVmnd-1 transmission to the offspring (54).
SIV infection status (infected/uninfected) in wild AGMs in Africa does not impact the following major health-related measurements (6, 52) (1): body mass index (a proxy for clinical phenotypes not available in field conditions) (2); physiological measures, which are biomarkers of mortality and disease progression in HIV/SIV disease in progressing infection, such as sCD14 (a biomarker of macrophage activation) (102), a wide panel of cytokines and chemokines (103), and C-reactive protein (CRP, a negative biomarker of survival time in HIV-infected patients) (104) (3); the blood counts of major immune cell populations (including CD4+ T cells, CD8+ T cells, B cells, NK cells, myeloid cells, and plasmacytoid dendritic cells), which do not differ between SIVsab-infected and uninfected AGMs (52). T-cell activation biomarkers (such as the expression of HLA-DR and Ki-67 in CD4+ and CD8+ T cells) are similarly expressed in SIVsab-infected and uninfected AGMs (52). The three following biomarkers stood out as being associated with SIV infection in African AGMs: D-dimer (a hypercoagulation biomarker), the levels of which are positively correlated with SIV infection, although D-dimer also increases with age and, as such, considering the age-related bias in SIV infection prevalence, it is a reflection of age rather than SIV infection status; IL-6 (an inflammatory cytokine), which is elevated in SIV-infected individuals, but this signal is driven by acute infection, when the high levels of viral replication trigger a transient increase in systemic inflammation in the natural hosts; and CCR5 expression in circulating CD4+ T cells; the increase with age of CD4+ T cells appears to be a very effective mechanism for preventing MTIT in natural hosts of SIVs (6, 52, 101).
In summary, the absence of differences in the levels of biomarkers associated with disease progression and mortality in natural NHP hosts of SIVs demonstrate that in the presence of other pathogens and parasites, even under harsh natural conditions and sometimes with limited water and food resources, African NHPs that are natural hosts of SIVs show resilience to immunodeficiency upon SIV infection (6, 52).
SIVcpz infection is pathogenic and manifests with an increased mortality rate (10-16-fold), lower birth rates, and higher infant mortality in natural chimpanzee populations (62, 105). A high rate of SIVcpz infection (40-50%) was associated with local chimpanzee population decline (62). Some SIVcpz-infected chimpanzees in the wild showed lymphatic tissue destruction, depletion of cortical and paracortical T- and B-lymphocytes, and follicular hyperplasia with prominent germinal centers (105, 106). Full-blown AIDS was reported from necropsies of several SIVcpz-infected wild-living chimpanzees that died as a result of AIDS, conspecific (i.e., within species) aggressive behavior, and injuries. Captive chimpanzees showed a blend of features characteristic of benign and pathogenic lentiviral infections. On the one hand, they showed a lack of increased mortality (some individuals were surviving 25 years post infection), no increase in the plasma levels of the sCD14 marker of microbial translocation during chronic infection (yet no direct assessments of SIVcpz showed whether there is indeed a lack of intestinal damage), a lack of AIDS-like clinical signs, and limited immune activation typical of non-progressing infection in well-adapted natural hosts. Yet, captive SIVcpz-infected chimps also displayed reduced CD4+ T-cell abundance and disruption of secondary lymphoid tissue architecture typically found in pathogenic infection (107). Soluble markers of immune activation (β-2 microglobulin, neopterin, and sTRAIL) and markers of T-cell activation in the peripheral blood (Patr-DR MHC class II and Ki-67) showed a temporary increase during acute infection, but no lasting increase in the chronic phase, while CD69, also a conventional marker of T-cell activation, showed a fluctuating increase during acute and chronic infection (107). These combinations of pathogenic and non-pathogenic features of lentiviral infection in captive chimpanzees infected with SIVcpz may be associated with advantageous/favorable conditions in controlled environments, i.e., reduced exposure to other pathogens, parasites, conspecific aggression and trauma, availability of food resources (107). In the case of SIVcpz infection of chimpanzees, studies in the wild and a long follow-up decisively contributed to the definition of the pathogenic nature of SIV infection.
The immunopathogenicity of SIVcpz was documented in a western chimpanzee (Pan troglodytes verus, a subspecies that does not harbor SIV in the wild) that was experimentally infected with SIVcpz for 20 years. The chimpanzee developed several clinical hallmarks of AIDS (high viremia [105-106 SIVcpz copies/ml of plasma], massive CD4+ T-cell depletion [220 cells/ul], and thrombocytopenia [90,000 platelets/ul]), and was effectively treated with antiretroviral therapy (108).
In conclusion, existing data point to the gradual adaptation of the NHP host to their SIVs, i.e., the longer the time of coevolution between SIV and its host, the more effective the host defense mechanisms are at counteracting SIV pathogenic potential and preventing immunodeficiency.
To avoid pathogenic SIV infection, natural hosts have developed adaptations that control the use of the CCR5-mediated entry pathway (101).
Natural hosts have acquired CCR5 null mutations that, similar to the CCR5-Δ32 loss-of-function allele in human populations that can modulate the risk of HIV transmission (109–111), abrogate CCR5 surface expression and protect null homozygotes from SIV entry through the CCR5 pathway [CCR5-Δ24 with a frequency of 86.6% in RCMs, 4.1% in SMs (112) and CCR5- Δ2 in SMs (113)]. Yet, the SIV pathogen bypasses the lack of functional CCR5 co-receptor in some RCMs and SMs and utilizes the non-CCR5-mediated entry pathway with alternative co-receptors. Thus, SIVrcm in RCMs uses CCR2b as the main entry co-receptor, while SIVsmm in SM can use CXCR6 as an alternative co-receptor in addition to CCR5 (113).
As a result of CCR5 downregulation, SMs and AGMs have significantly lower levels of CD4+ CCR5+ T cells than humans or RMs, both in the blood and at mucosal sites (53). The shift in tropism from CCR5 to CXCR6 may benefit the host, as it directs the virus to a different and putatively more dispensable cell population (113). On the other hand, the loss of CXCR6 co-receptor use is characteristic of the lineage of more pathogenic lentiviruses—SIVcpz in chimpanzees and HIV-1 in humans (114).
Restricted expression of entry receptors eliciting resistance to SIV infection is a mechanism that helps to spare critically important cell populations among the CD4+ T lymphocyte memory pool—the long-lived central memory CD4+ T cells (Tcm). Tcm T cells have stem-like properties, reside primarily in the lymphoid tissue, and have strong proliferation capabilities in response to stimuli. In stark contrast, effector memory CD4+ T cells (Tem) have lower proliferation potential, are located in non-lymphoid tissues, and remain susceptible to SIV infection (5).
In SMs, CD4+ Tcm cells are relatively protected from SIV-mediated depletion through the downregulation of CCR5 (115). CCR5, which is the canonical co-receptor for HIV/SIV infection in humans and RMs, is also expressed in the CD4+ T cell of the natural hosts (SMs, AGMs, mandrills, and sun-tailed monkeys) but at very low levels (53). Instead of CCR5, CXCR6 is the major SIV co-receptor in SMs and AGMs (116–118). As a result, SIV still can infect and replicate in natural hosts but only in the cell populations expressing CXCR6. The targeting of different cell populations by SIV is dependent on the pattern of distribution of the co-receptors CCR5 and CXCR6. In SMs, they are expressed in largely non-overlapping populations of CD4+ T cells, with only 0.3% of CD4+ T cells being dual-positive for CCR5 and CXCR6 (114). In CD4+ T cell memory subsets, CXCR6 was enriched in Tem (5.9%) but less so in naive CD4+ T cells (0.5%) and Tcm (1%) (114). Interestingly, some features of non-progressing SIVsmm infection in SMs—low immune activation despite high viremia and low CCR5 density on CD4+ Tcm cells—were observed in a pediatric HIV non-progressor cohort from South Africa (119). The fine-tuned control of co-receptor expression helps to preserve the CD4+ Tcm population, which is critical for maintaining immune homeostasis. A similar phenomenon of protecting Tcm CD4+ cells from SIV infection is achieved through the downregulation of CD4 in CD4+ Tcm cells in AGMs (48, 120).
Salient features of non-pathogenic SIV infections in natural hosts that differentiate them from pathologic infections include preservation of the mucosal barrier, preservation of the Th-17 cells (the T-cell subset that protects mucosa against bacterial and fungal infection) in the gut and periphery (31, 32), a lack of microbial translocation, and a lack of chronic inflammation and T-cell immune activation (121–123), which suggest that gut health is a key determinant of the pathogenic or non-pathogenic course of infection.
The gut plays a dual role as a structural and immunological barrier between the external and internal environment. It requires a fine-tuned balance between tolerance and sensitivity and the ability to provide rapid yet sustainable long-term protection against pathogens (124, 125). Nearly 70% of the body’s T-cell-generating lymphoid tissue is located in the gut (124), which is also the primary site of SIV/HIV replication (25, 43, 126–128), and intestinal epithelium is highly susceptible to inflammation. Lentiviral infection leads to physical and immunological dysfunction of the intestinal mucosa (129, 130). Damage to the enteric barrier allows microbial and fungal translocation, i.e., the leakage of microbial and fungal products across the breached intestinal epithelium into the blood circulation, and causes the systemic dissemination of microbial biomolecules, such as LPS (bacterial translocation marker), peptidoglycan, bacterial DNA, and viral genomes, which can be found in various tissues even at distant anatomical sites with progressive SIV infection (131) and, in the case of the major fungal cell wall antigen β-d-glucan (BDG) (a biomarker of fungal translocation), at elevated levels in the plasma (132, 133).
Microbial products translocated from the gut strongly stimulate the immune system and the production of cytokines, contributing to persistent local and systemic inflammation and T-cell activation, which further amplify viremia by generating more target cells in both treated and untreated HIV-infected individuals (43). While initially CD4+ T-cell counts or plasma viral loads appeared to be the most accurate biomarkers of disease progression, immune activation that enhances virus replication by providing the virus with activated CD4+ T-cell targets is now considered a more accurate predictor of survival time in advanced HIV-1 disease (134). Note that persistent CD4+ T cell depletion for over 1.5 years did not result in SIV disease progression in AGMs (135). Plasma levels of soluble CD14 (a biomarker of microbial translocation) are an independent predictor of mortality in HIV infection (102). Microbial translocation is characteristic of the disease pathogenesis of not only acquired immunodeficiency (HIV infection) (136) but also primary deficiencies, such as idiopathic CD4+ T-cell lymphocytopenia (ICL) (137); however, the link between microbial translocation and the perturbation of CD4+ T cell homeostasis in ICL is not as obvious as in the case of HIV-1/AIDS. In ICL, sCD14 levels are only modestly increased, and the local architecture of the GI tract is preserved, with normal enterocyte turnover and no apparent loss of Th17 cells, which is in contrast to patients with HIV-1/AIDS (138).
BDG is a biomarker of fungal translocation and is elevated in HIV-infected individuals (139) and is associated with gut damage, immune activation and inflammation (140), and a risk of neurocognitive comorbidities (141). In HIV-infected individuals, fungal products stimulate antigen-presenting cells (monocytes and macrophages) and NK cells, leading to the excessive secretion of proinflammatory cytokines and inflammation (133).
The following mechanisms may be responsible for triggering the functional loss of the gut barrier in progressive HIV/SIV infections (142) (1): loss of the CD4+ T cells, the primary target of HIV-1/SIV infection, undergoing virus-induced cell death, which results in a severe reduction of this cell population in circulation and mucosal sites during acute infection in both progressive and non-progressive hosts (22, 23, 27, 53, 143) (2); preferential infection and loss of Th-17 cells, which play a critical role in protecting against microbial and fungal pathogens and regenerating the gut epithelium, thus being a key contributor to the maintenance of mucosal integrity (144) in the GI tract. Differentiation of Th-17 cells and HIV replication in these cells are regulated by a retinoic acid-related nuclear hormone receptor, RORC2 (145). The long-term survival and proliferation capacities of Th-17 cells make them preferential targets for HIV reservoirs in individuals receiving ART (146, 147). By contrast to progressing infection, Th-17 abundance is well maintained in the natural hosts during non-progressing infection (31, 32) (3); persistent gut cell death and mucosal apoptosis (130, 148, 149). The loss of integrity of the intestinal immune barrier occurs during the acute pathogenic infection and continues through the chronic phase due to incomplete repair of the barrier. These gut breaches permit an influx of microbial toxins into the circulation, which drive chronic T-cell immune activation and systemic inflammation. In contrast to the progressing hosts, natural hosts chronically infected with species-specific SIVs (SIVsmm-infected SMs and SIVagm-infected AGMs) manage to prevent permeabilization of the mucosal barrier (23, 28). As such, gut integrity is maintained in the natural hosts throughout the SIV infection (150, 151). As a result, in spite of the relatively robust viral replication resulting in levels of viremia, which are similar or higher than those observed during pathogenic SIV infection, the natural hosts of SIVs do not display any of the hallmarks of HIV disease progression, i.e., breakdown of the gut epithelial barrier, microbial translocation, and chronic T-cell immune activation and inflammation, CD4+ T-cell decline, or hypercoagulation (6, 28, 52, 122, 130, 151–153).
Alterations of the mucosal barrier of SIV-infected natural hosts drive the otherwise non-pathogenic infection towards disease progression. The intestinal mucosa functions as a physical and immunological barrier and is thus critical for gut and systemic health. All impairments of gut integrity, such as pathogenic lentiviral infections, inflammatory bowel disease (IBD) (154), are associated with chronic systemic inflammation and immune activation (124, 125). Damage to the intestinal mucosa and loss of its barrier function can occur in the absence of lentiviral infection during multiple clinical conditions, including celiac disease, IBD, colon carcinoma, chronic liver disease, type 1 diabetes, and obesity (155) (Figure 2). Experimental destruction of the mucosal barrier of uninfected RMs with dextran sulfate sodium (DSS), which is toxic to enterocytes in the crypts and causes chemically induced colitis in rodents and non-human primates (157), induces gut damage that evokes features of pathogenic lentiviral infection, including neutrophil infiltration of the lamina propria in the colon, microbial translocation to local and distant anatomical sites, local inflammation and activation of local T cells, systemic inflammation (IL-6) and immune activation, and fibrosis in lymphoid tissues, which is a histopathologic hallmark of pathogenic lentiviral infection (157).
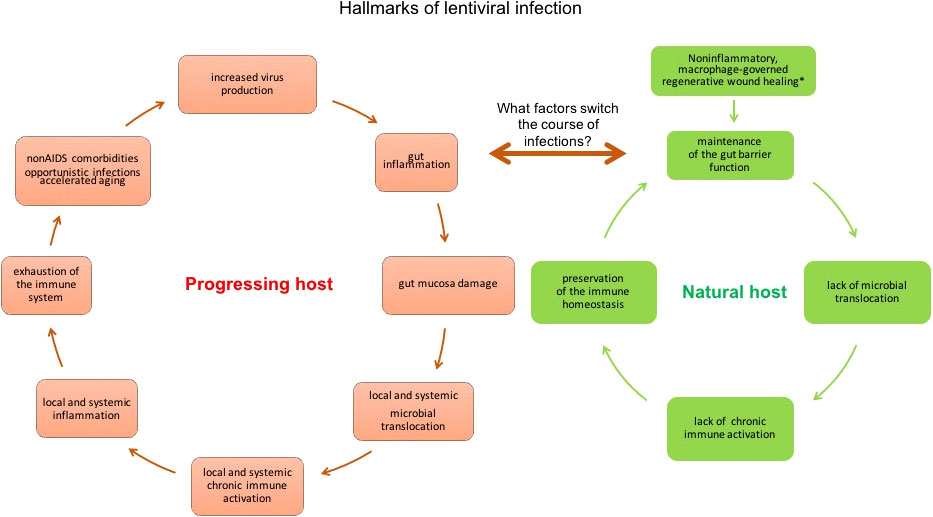
Figure 2 Hallmarks of lentiviral infection in natural non-progressing hosts (vervets/AGMs and SMs) and progressing hosts (humans and RMs). In the acute phase of infection in vervet/AGMs, the immediate activation of macrophage-associated regenerative wound healing bypassing the early inflammatory stage prevents the loss of immune barrier function of the gut (156). However, chemically induced damage of the intestinal mucosa with DSS treatment during acute infection shifts the course of pathogenesis from a non-pathogenic course (controlling microbial translocation and its consequences of persistent activation of the immune system) to a pathogenic course (losing the containment of the luminal microbiota, leading to immune activation and inflammation on a local and systemic scale), demonstrating that dysfunction of the gut barrier can initiate a vicious cycle in SIV-infected non-progressing hosts (157). Whether the DSS-induced hallmarks of the pathogenic course of infection in AGMs are predictive of the development of HIV-like clinical manifestations typical to progressing infection, needs to be determined. Longer DSS administration is needed to determine whether that would be sufficient to evoke progression to AIDS.
Furthermore, the administration of DSS to chronically SIVagm-infected AGMs induces a gut dysfunction (mucosal thickening, redness, ulcerations, and colitis) that can drive the non-pathogenic SIV infection towards a more progressive pattern. This experimentally induced gut damage is associated with elevated levels of the plasma markers of gut barrier dysfunction, microbial translocation, and systemic immune activation (sCD14) and inflammation (CRP) that further increases plasma viral loads and CD4+ T-cell depletion (157). Note, however, that while chemical-induced gut dysfunction clearly has the potential to alter the clinical pattern of SIV infection in natural hosts, studies of longer DSS administration are needed to test whether or not the induced defects of the mucosal barrier are sufficient to trigger progression to AIDS.
Intestinal barrier robustness was tested in a single study that assessed, on serial necropsies, systemic health during the acute phase of non-pathogenic SIVagm infection of AGMs (151). The observations derived from this study were as follows: (1) SIV infection exerted a minimal impact on the major immune populations both in the periphery and at the mucosal sites. Thus, circulating CD4+ T cells were only minimally depleted during acute SIVagm infection, and were already restored to pre-infection levels by the time of transition to chronic infection; (2) very importantly, and in agreement with previous observations (158), CD4+ T-cell loss through bystander mechanisms was not increased in SIV-infected AGMs. Thus, little or no apoptosis in the gut lamina propria and epithelium was observed (based on the caspase-3 biomarker). This minimal loss, in the absence of bystander depletion mechanisms, did not induce a major local inflammation; instead, (3) local inflammation of the gut mucosa was very transient, and likely virus induced, as demonstrated by a temporary increase of MPO-neutrophils that returned to the baseline after the peak of viremia, an increase of MX-1 at the peak of viremia, and a transient increase of Ki-67 expression in the lymph nodes at the peak of viremia during the chronic phase; as a result, (4) the overall integrity of the mucosal barrier was maintained, as demonstrated by the overall preservation of the continuity of colonic epithelium, which was documented by the distribution of claudin-3, a biomarker of the proper sealing of tight junctions, which is critical for regulating the permeability of the epithelial barrier; (5) microbial translocation did not increase with SIV infection, as indicated by biomarker analysis (based on in situ LPS and an E. coli IHS assay), or at best was transiently increased around the peak of virus replication (based on the plasma levels of LPS and sCD14). (6) the absence of SIV-induced fibrosis in the gut or lymph nodes was documented by the lack of increased collagen deposits at these sites and normal plasma levels of hyaluronic acid (a biomarker of liver fibrosis) (151). These findings underscore that the key distinction of the natural host’s response to the lentiviral infection is the capability to withstand SIV infection without damage to gut mucosa integrity, rather than rapidly repair and restore breaks in it (151). This might be because of superior healing mechanisms that enable AGMs to sustain gut integrity without permanent or even transient damage (156).
The issue of how natural hosts prevent gut dysfunction was investigated through comparative transcriptomic analysis of responses to SIVsab and SIVmac in the rectal tissues of AGMs and RMs, respectively, during acute infection (156). Gene expression responses characteristic of the pathogenic infection of RMs were primarily orchestrated by LPS as the main upstream regulator and showed a stronger activation of antiviral and antimicrobial immune responses. By contrast, reactions specific to the non-pathogenic infection in AGMs did not manifest antimicrobial signatures, but appeared to be driven by IFN-γ and prolactin genes (both inflammatory cytokines) as the upstream regulators and were marked by the recruitment of leukocytes and myeloid cells. AGMs activated IFN-driven targets, myeloid cell migration, and the developmental processes of muscle, epithelial, and adipose tissues in the absence of microbe-activated patterns as early as the previremic phase, suggesting that preventive repair mechanisms may be responsible for stopping microbial translocation (156).
The superior ability to maintain the gut barrier function of the gut mucosa throughout acute infection is a critical feature that differentiates pathogenic and non-pathogenic infection (151). Transcriptomic profiles in rectal tissues through the course of non-pathogenic SIVsab infection in AGMs pointed to regenerative wound healing mechanisms, which bypass the early inflammatory processes and rapidly activate the late stage of wound healing, resembling the transcriptional profile that regulates the regeneration process in the aquatic axolotl, with fully functional scar-free repairs, including blood vessel development (angiogenesis). This process in AGMs involved fibronectin as a key connector between collagen and the laminin components of the extracellular matrix in transcriptional networks. Among the predicted upstream regulators of the wound healing networks was EGFB release factor, which was also upregulated during the axolotl wound repair process, suggesting that TGF-β secretion is a regulator of monocyte/macrophage-mediated epithelial cell repair in AGMs (156).
Taken together, while pathogenic infections are characterized by inflammatory tissue damage signatures and microbial pattern recognition receptor and inflammatory signaling, non-pathogenic hosts displayed a strong wound-healing signature, probably regulated by monocytes early in the acute infection. This wound-healing ability prevented infection from taking a pathogenic course and causing microbial translocation and subsequent local and systemic inflammation and chronic immune activation (156).
The development and health of the immune system is shaped by commensal microbiome, and disruption of its homeostasis leads to the impairment of the immune system. The intestinal ecosystem is the site of multidirectional interactions between the host, the commensal microbiome, and the pathogen. Progressive loss of gut immune function (due to loss of CD4+ T cells in the gut mucosa, in particular the loss of the Th-17 CD4+ T cells that are key effectors that maintain immune barrier function and memory cells) contributes to the destabilization of the intestinal microbiome in HIV-infected individuals (159). A shift towards dysbiotic mucosa-associated communities enriched in Proteobacteria and depletion of Bacteroidia species was associated with mucosal immune disruption, T-cell activation, and chronic inflammation in HIV-infected subjects (160). Biomarkers of HIV disease progression, such as levels of mucosal and systemic immune activation and inflammation, CD4+ T-cell counts, and viral loads, were correlated with gut dysbiosis manifesting with the disruption of microbial homeostasis, characterized by an expansion of pathogenic bacteria compared with commensal species (160–162). However, various studies produced diverse results, probably due to variability within and between the study cohorts. Pathogenic infection in RMs led to shifts in the composition of gut microbiota, yet studies lacked consistency, probably due to cohort and experimental variability (163). Nevertheless, the gut microbiome can act as a potential modulator of SIV/HIV disease, probably as one of the main determinants of the resilience to lentiviral-induced destruction of the gut mucosa and microbial translocation and their consequences.
The extent to which the gut microbiome helps natural hosts preserve the integrity of the gut mucosa and prevent translocation of bacterial products from the gut is not fully understood, but there is increasing insight into the natural microbiome composition in wild populations of natural hosts (Figure 3). We anticipated microbiota stability or activation of protective mechanisms in ancient well-adapted hosts (SMs and AGMs) and destabilization of the microbiome in more recent hosts (CPZ).
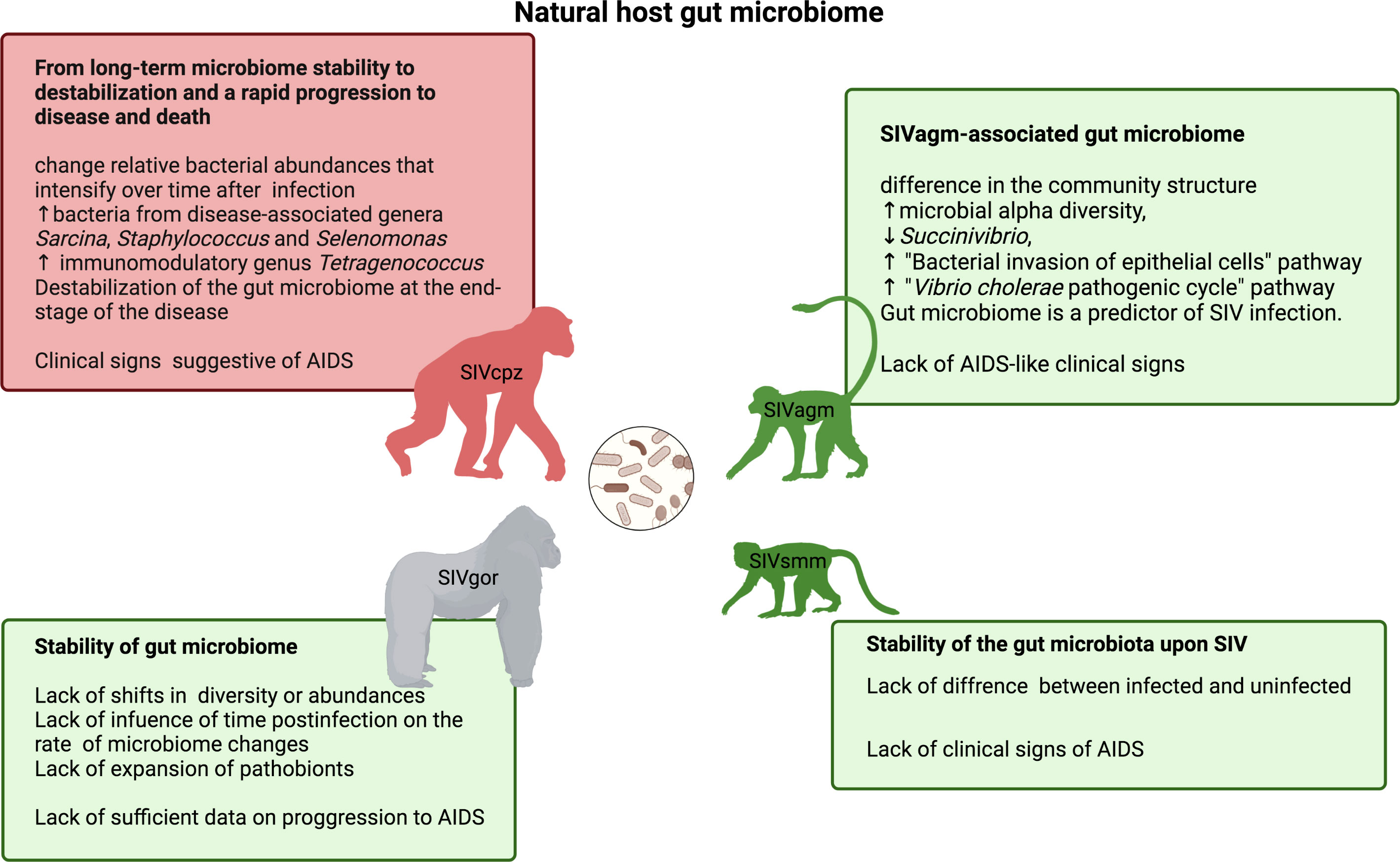
Figure 3 The gut microbiome of natural NHP hosts of SIV infection: chimpanzee, a host displaying mixed clinical signs of progressing and non-progressing SIV infection (top left) (164, 165); gorilla, a host with lacking sufficient data on clinical indicators of AIDS (bottom left) (166); and hosts that typically do not develop immunodeficiency and simian AIDS - vervet/AGM (top right) (92) and sooty mangabey (bottom right) (167).
SIVcpz impact on the gut microbiomes of chimpanzees was studied in wild eastern chimpanzees (Pan troglodytes schweinfurthii) from Gombe National Park, Tanzania that naturally host pathogenic SIVcpzPts (164). Long-term non-invasive monitoring of SIV infection status and AIDS-like symptoms in this chimpanzee population allowed the assessment of changes in the fecal microbiome upon SIVcpzPts infection (164). While the core gut microbiome suggested a composition that was overall stable at the phylum level, with no significant changes in the abundance of individual bacteria upon SIVcpzPts infection, it underwent marked changes in relative microbial abundance that increased with time and an upturn in microbes not observed prior to infection and potentially relevant to immune health, i.e., bacteria from disease-associated genera (Sarcina, Staphylococcus and Selenomonas) and Tetragenococcus (164), known for their immunomodulatory effect (168). Further studies using a combination of metagenomic sequencing and larger-scale bacterial 16SrRNA gene sequencing showed that the chimpanzee gut microbiome is very robust throughout most of the course of SIV infection and that its stability collapses only in the final stage of disease and for the few months preceding AIDS-related death (165). SIVcpz-infected chimpanzees showed a tendency towards enrichment for bacteria from the Prevotellaceae family (165). Chimpanzee stool-associated circular virus (Chi-SCV) and adenovirus (ChAdV), quantified using metagenomic sequencing, did not show differential abundances in SIVcpz-infected chimpanzees (165). The microbial composition in the guts of SIVcpz-infected chimpanzees showed continuous drifts, resulting in gradual changes in response to SIVcpz infection in wild chimpanzees rather than a single shift.
Non-invasive sampling methods investigating SIV infection status and evaluating SIV pathogenicity via microbiome analysis in feces provided insight into the influence of SIVgor infection in the gut microbiome of western lowland gorillas (Gorilla gorilla gorilla). While SIVcpz in chimpanzees and HIV-1 in humans are pathogenic and cause substantial alterations to the composition of the gut microbiome (164, 165), it remains unknown whether the closely related SIVgor leads to pathogenesis and clinical signs of AIDS in its natural host. Despite SIVgor originating from the SIVcpz strain, which destabilizes the gut microbiome in chimpanzees, SIVgor infection in gorillas does not show similarly pathogenic behavior and does not lead to gut microbiome variation in western gorillas from Southern Cameroon. Unlike chimpanzees, the gorilla gut microbiome does not show a progressing shift in composition over time or a tendency towards the emergence of opportunistic pathogens upon infection (166). There were no noticeable differences in diversity and composition between SIVgor-infected and uninfected gorillas or accelerated changes in microbiome composition over time in infected individuals.
Typically, 5-7 taxa are delineated within the genus Chlorocebus, which are characterized by different geographical ranges across tropical and temperate climatic zones in Africa and a massive spread of SIVagm infection (90). However, the majority of controlled experiments are conducted in AGMs originating from a founder isolate population in the Caribbean Islands (St. Kitts and Nevis) that was more recently established from the west African populations and is free of SIV (169, 170). So far, microbiome studies relating to SIV infection have been conducted only in the natural populations of South African AGMs (Chlorocebus pygerythrus) (92). Studies of natural microbiome in wild vervets involved a wide range of ages, from infants to old adults, to cover as much as possible the epidemiological profile of SIV infection/transmission and the course of the diseases (6). While SIV-infected vervets (both in natural and captive populations) do not experience gut dysfunction, microbial translocation, and chronic immune activation and in general do not progress to immunodeficiency, their microbiome displays significant differences in the microbial ecosystems of the gut and genital tract compared with uninfected individuals (92). The gut microbiota in SIV-infected vervets showed (1) increased alpha diversity (2), decreased abundance of the phylum Proteobacteria (particularly the genus Succinivibrio) (3), decreased ‘bacterial invasion of epithelial cells’ and ‘Vibrio cholerae pathogenic cycle’ KEGG pathways in the predicted fecal metagenome of SIV-infected individuals compared with uninfected individuals, and (4) partial control of early SIV-induced alterations during chronic infection (92). The host microbiome differences between SIV-infected and uninfected individuals may represent adaptations to the virus that prevent microbial translocation, persistent immune activation, and the resultant disease progression in vervets. This cross-sectional study showed that SIVver infection in vervets is characterized by a distinct gut microbiome composition and functionality yet did not determine whether these differences are pre-existing or responsive to the infection, a question that could be further addressed through longitudinal studies.
Fecal samples were an accurate predictor of SIV infection status in wild vervets (92). While this observation was obtained from samples collected from the rectum, it seems possible that non-invasively collected samples on the ground may provide a feasible microbial biomarker predictive of immune health in wild primate populations.
Conventional studies of disease correlates in animal models, including AGMs, are typically conducted under well-controlled standardized laboratory conditions in a small number of animals that can be subjected to invasive sampling. Natural populations, on the other hand, allow for more scalable studies (dozens or even hundreds of individuals) in the context of a complex and variable environment (including natural habitat and social groups) using non- or minimally invasive sampling. The natural setting differs from the laboratory as wild primates usually expend more energy and may ingest less nutritional food, which is highly dependent on timing, habitat, geographic location, and seasonality of environment, and aggressive interactions in the group may reduce equal access to foraging. In wild AGMs from South Africa, gut and vaginal microbiome revealed an association with geographic location and environmental variables, such as geographical biomes and temperature (92). The within-genus differences may be significant, as illustrated by differential/distinctive vaginal microbiome composition between South African and Caribbean AGM populations (92). Comparison of the effects of diet between Caribbean-origin AGMs in the wild consuming a natural diet and AGMs fed a typical western diet (TWD) under controlled conditions showed similar microbial richness, yet the composition of their microbiome was distinct, with lower abundances of Firmicutes, Lentisphaerae, Proteobacteria, Tenericutes, and Verrucomicrobia and higher abundances of Bacteroidetes and TM7 in AGMs with a TWD (171). Yet, wild populations more closely reproduce conditions reflecting host-pathogen coevolution and are the preferred setting for studies of natural transmission.
The gut microbiomes of captive SMs did not show significant SIVsmm-associated variation in microbial diversity or community structure, yet there were lower levels of pathobiont bacteria in SMs not infected with SIVsmm than in diet-matched RMs experimentally pathogenically infected with SIVmac239 (in the early chronic phase) (167). The stability of gastrointestinal microbiota was paralleled with the compositional stability of the cargo of the luminal intestinal microvesicles produced by the host enterocytes in chronic SIVsmm infection in SMs, which was in contrast to RMs infected with closely related SIVmac (172). Progressive SIVmac infection in RMs showed shifts in microbial communities concomitant with changes in the content of gut microvesicles compared with uninfected RMs, including a reduced amount of several miRNAs and an increased level of beta-defensin 1 (DEFB1). The microvesicles from the progressive infection hampered the growth of Lactobacillus salivarius, one of the commensal bacteria undergoing microbial translocation, suggesting that their potential role is to control microbial growth and shape microbial translocation (172).
A series of phenotypic adaptations, which are considered to have occurred during the millenary virus-host coevolution as tools of the host adaptation to circumvent the deleterious effects of SIV infection, characterize the immune system of the natural hosts of SIV:
Some natural hosts of SIV (AGMs, SMs, and patas monkeys) harbor an evolutionarily conserved downregulation of the CD4 molecule on the T-cell surface, resulting in low levels of this cell subset in contrast to its abundance in non-natural hosts of SIV/HIV (47, 48). Chlorocebus pygerythrus has large CD4neg CD8αα+ T-cell populations (lacking CD4, and thus spared from SIV infection, and distinct from canonical CD8+ T cells expressing heterodimers with α and β chains) (50). Meanwhile, SMs have abundant populations of double-negative cells (expressing neither CD4 nor CD8) that are critical for preventing massive homeostatic alterations during acute CD4+ T-cell loss (49, 173, 174). These CD4neg cell populations are virus-resistant and preserve some CD4+ T-cell functions (49, 50, 175). In C. pygerythrus, the reduced CD4 expression in CD4+ T cells may be related to the regulation of RUNX3 expression, the master transcription factor regulating CD4/CD8 expression (175). Runx3 is involved in CD4 suppression in murine models; in RMs, its expression is higher in CD8αβ T cells than in CD4+ T cells. Yet, in vervets, RUNX3 expression does not vary among all T cell subsets (including, CD8αα+ T cells, CD4+, and CD8αβ+ T cells), irrespective of their CD4 and CD8 phenotype, suggesting that RUNX3 may be the factor contributing to CD4 downregulation in CD4+ T cells (175).
The CD4neg CD8αα+ T-cell populations of C. pygerythrus also display functions characteristic to classical CD4+ T cells (MHC class II restriction, expression of FoxP3, CD40 ligand, and production of IL-17 and/or IL-2) (120). These cells can avoid SIV infection while performing some CD4+ T cell-like functions in the absence or with very low levels of classical CD4+ T-cells. The conversion process is accelerated by SIV infection in AGMs in vitro and induced both in vitro and in vivo by homeostatic cytokines, such as IL-2 (50, 176). These virus-resistant hyperfunctional T cells arise through post-thymical downregulation of CD4 by CD4+ T cells causing CD4-to-CD8αα conversion (120). This is a developmental process resulting in the increase of the initially low abundance of CD4neg CD8αα+ T cells with age. CD4 downregulation is evoked through epigenetic silencing mechanisms involving CpG methylation in the CD4 promoter (177), which colocalizes with inaccessible chromatin regions in the CD4 gene region, as determined using ATAC-seq (178). Differential chromatin accessibility across the genome attributable to the CD4-to-CD8αα conversion was associated with immune processes and T-cell biology, and binding sites for transcription factors involved in the immune processes. In CD4+ memory T-cells, more accessible regions were enriched for high mobility group (HMG) family binding sites involved in T-cell development and differentiation. In CD8aa+ T cells, open chromatin regions were enriched for binding sites for Runt domain transcription factor and erythroblast transformation specific (ETS) regulating hematopoiesis (178).
Lymph node follicles of the natural host species remain virus-free during SIV infection, in contrast to HIV-infected individuals and SIV-infected NHP progressors, in which they are major viral reservoirs, even in patients on ART (179). This observation raises the following question: what are the protecting mechanisms preventing infection of the cells in the lymph nodes in AIDS-resistant NHPs? The frequencies and phenotypes of NK cells (defined as CD45+ CD3neg CD14neg CD20neg NKG2a/c+) assessed in African and Asian NHPs were similarly anatomically distributed in NHPs with different origins, yet the frequencies of circulating NKG2a/clow NK cells was higher in African species than in the Asian ones (180). It was also reported that SIV infection differentially drives NK distribution in natural and non-natural hosts: in AGMs, contrary to cynomolgus macaque, NK cells did not home to the gut, and their abundance at mucosal sites was lower during SIV infection but increased in lymphoid tissues (180). This was consistent with an effective control of viral replication in the T zone and B cell follicles in secondary lymph nodes mediated by NK cells occurring in AGMs, in contrast to progressing SIV infection in macaques in which these anatomical locations are the major viral reservoir (181). The process of effective control of SIVagm in the lymph nodes of natural hosts involves SIVagm-induced expansion of terminally differentiated NKG2alow NK cells in lymphoid tissues. These expanded cell subsets showed upregulation of genes in the pathways related to lytic activities of NK cells and displayed adaptive transcriptional profiles (based on marker analysis) and increased MHC-E-restricted cytotoxicity towards cells presenting SIV peptides (182). MHC-E-restricted NK cell activity against target cells presenting SIV peptides was enhanced during SIVagm infection in AGMs but decreased with SIV infection in macaques (182).
Non-pathogenic SIVagm infection in AGMs is characterized by a rapid expansion of NKG2a/c+ CD8+ T cells in circulation and at mucosal sites but not in secondary lymphoid tissues (183). The transcriptomic signatures of these expanded NKG2a/c+ CD8+ T cells are distinctive from other CD8+ T cells, implying a cytotoxic effect and immunoregulatory function, as well gut homing properties of these cells. Such a pattern of cellular dynamics was not observed during the pathogenic SIV infection of RMs. In the progressing model, NKG2a/c+ CD8+ T cell abundancy negatively correlated with the mucosal levels of IL-23, an inflammatory cytokine. This negative correlation between NKG2a/c+ CD8+ T cells with intestinal inflammation in the pathogenic models suggests that these cells protect against intestinal inflammation during SIV infection by controlling immunopathology in the intestine (183).
What genes and pathways underlie a non-progressive course of SIV infection in natural hosts despite chronic virus replication? Genomic analysis enabled an unbiased genome-wide search for host adaptive links against disease progression in SIV-infected NHPs. Genomic studies revealed genetic adaptations in the genes involved in biological processes acting in response to viral infections in AGMs and SMs.
Comprehensive omics approaches aiming for a complete perspective, such as genome-, transcriptome-, and epigenome-wide studies, facilitate an unbiased identification of the host molecular reactions to infection and adaptive responses against the progression to disease. The identification of molecular processes and key genes driving the ability to evade immunodeficiency may set new directions for the development of novel therapeutic approaches.
Genome-wide studies using next generation sequencing revealed the genetic adaptations in the genes involved in biological processes responding to viral infection, including SIVs, in AGMs (184) and SMs (185–188). Genome sequencing of representatives of diverse vervet populations across major Chlorocebus species revealed massive polygenic adaptations to SIV in vervets (6, 184). Genes with strong selection signals were enriched in the GO category ‘viral processes’, vervet orthologs of human genes interacting with HIV, and genes transcriptionally regulated in response to SIV infection in the natural host (AGMs), but not non-natural hosts (RMs). Early expressional responses were enriched for clathrin-mediated endocytosis (a process utilized by HIV to enter cells lacking CD4), autophagosome assembly (autophagy being a key component of host responses to HIV-1 infection), and the regulation of type I IFN production (a key driver of early transcriptomic responses both in natural and non-natural hosts), while expressional changes in the chronic phase were, among others, enriched for the regulation of NK cell activation (184).
Comparative genomic studies contrasting the protein coding regions of SMs with humans and RMs revealed the genes that were most diverged at the amino acid level and in structures between the natural host and pathogenic hosts—intercellular adhesion molecule 2 (ICAM-2) and toll-like receptor-4 (TLR4) (188). Functional studies of the top immunoregulatory proteins displaying major structural differences specific to the natural host demonstrated that sequence differences have significant functional consequences: abrogation of the cell surface expression of ICAM-2 and TLR4 blunted the production of pro-inflammatory cytokines in response to LPS stimulation in vitro, a potential factor contributing to reduced immune activation (188). Genomics combined with functional assays allowed the identification of candidate modifier genes shaping the resistance of the natural host to immunodeficiency.
The benign phenotype of SIV infection in SMs has been associated with genes in pathways contributing to interferon signaling, BRCA1/DNA damage response, PKR/INF induction, and CGALS8 in SMs (189) and point to the leading role of the genes LGALS8 and IL-17RA, which enhance gut barrier function and shape homeostasis with gut microbiota.
Wild NHPs are important models for understanding the host mechanisms involved in the control or pathogenesis of viral infections. Ugandan red colobus (URC) in Kibale National Park are naturally infected with SIVkrc (22.3%) (190). RNA-seq in URC from this wild population pointed to pathways associated with cellular immunity, cell activation, leukocyte activation in infected URC, and inflammatory responses in uninfected URC. SIVkrc infection has been characterized by downregulation of SLAMF6 and CD4, immune genes undergoing downregulation in HIV infection, and upregulation of the immunosuppressive gene CD101 during infection (191). The pattern of temporary upregulation of ISGs observed in AGMs and SMs was not displayed by SIVkrc-infected URC, most likely because in a moderately sized group a vast majority, or all, are in the chronic phase of infection when ISG expression has already normalized (191).
SIV infection has a different influence on DNA methylation patterns in CD4+ T cells purified from circulation and lymphoid tissue in non-pathogenic SIVagm infection of AGMs and pathogenic SIV infection of Chinese RMs (192). Non-pathogenic SIV infection of AGMs is characterized by the enrichment for methylation signals in regulatory proteins, while pathogenic infection in macaques is associated with Th1-signaling and metabolic pathways, suggesting that epigenetic mechanism may contribute to the risk of metabolic diseases during progressing infection (192).
As shown above, studies of natural hosts that remain disease-free despite life-long SIV infection identified several directions towards the development of therapeutic approaches: (1) Ameliorating gut inflammation and damage that leads to aberrant mucosal permeability and translocation of enteric bacteria to the circulation appears to be a promising therapeutic direction. Commensal enteric microbes play an important role in intestinal wound healing and the maintenance of barrier function (193) and can decrease gut permeability associated with low-grade inflammation and normalize the levels of tight junction proteins, such as claudin, in mice (194). (2) Therefore, stabilization of the natural microbiome, preserving and/or enriching beneficial taxa, manipulation of community composition, and metabolic activity is another possible therapeutic direction. Attempts to reduce gut inflammation through modification of the enteric microbiome using selected bacterial strains as probiotics alone or combined with prebiotics, or bacterial metabolic products, such as short-chain fatty acids, brought mixed results (123). Results of the administration of selected microbial strains ranged from a lack of effect on systemic inflammation (195), no effect on immune activation biomarkers (196, 197) through reduced systemic inflammation markers, but not microbial translocation markers (LPS or sCD14) (198), to a significant decrease of enterocyte apoptosis and increase in Th17 cells in the gut (199). The role of luminal vesicle cargo of microbiome-stabilizing molecules (miRNA and defensins) as potential therapeutic tools to control the growth of pathobionts is emerging, given the development of technologies allowing scalable manufacturing and delivery systems (172). (3) Macrophages are key drivers of the wound-healing process in the intestinal mucosa during the non-pathogenic course of SIV infection in the natural host (156) and participants of the wound-healing process in various disease phenotypes stemming from excessive or insufficient healing (aberrant aging, diabetes, and fibrosis) (200). Therefore, they can provide a novel therapeutic target for promoting wound-healing disorders (151, 200). (4) Downregulation or blockade of SIV/HIV co-receptors. An allogenic CCR5-deficient bone marrow transplant carrying a homozygous deletion (CCR5 Δ32/Δ32) led to sustained HIV-1 disease remission (201, 202). However, another leukemia patient who received a stem cell transplant from a CCR5-deficient donor showed that post transplant the HIV virus changed tropism from CCR5 toward an alternative co-receptor, CXCR4 (203). Targeting the interaction of CCR5 with HIV led to development of CCR5-blocking agents that prevented virus entry. Maraviroc (MVC) is a non-peptidic small molecule that prevents viral envelope binding (204, 205). Leronlimab, a CCR5-specific antibody (206), effectively increases CCR5+ CD4+ T cells in circulation in infected humans and macaques (207), suppresses the virus in humans chronically infected with CCR5-tropic HIV-1 and RMs acutely infected with CCR5-tropic SHIV (208), and protects RMs from mucosal SHIV acquisition (209). Downregulation of CD4, which is a natural process in the maturation of CD4+ T cells into CD4neg CD8aa+ T-cells in non-progressing hosts and does not seem to lead to any pathologies, appears to be a potential approach to effectively prevent cell infection regardless of co-receptor usage (178). (5) SIV-related epigenetic modifications in genes associated with immunoregulation and tissue integrity may constitute novel candidates for immunotherapies (192). (6) PD-1-targeting/related therapies for controlling virus in lymphoid tissues (182). Overexpression of programmed death-1 (PD-1) is characteristic of HIV/SIV-induced exhaustion of the immune system (CD4+ and CD8+ T cells) (210). PD-1 blockage was leveraged to reduce viral reservoirs in lymphoid tissues and specifically redirect NK cells to the follicles. It increased the therapeutic benefits of SIV vaccine in pathogenic SIV infection in macaques by enhancing the function and follicular homing of vaccine-induced CD8+ T cells and decreasing viral reservoirs in lymphoid tissue (211). The high expression of PD-1 in the follicular helper CD4+ T cells was harnessed to direct NK-cells expressing a chimeric antigen receptor (CAR) to the follicles and selectively deplete of PD-1high target cells (212).
The last two decades of studies investigating SIV infections in their natural hosts have proven to be critical for driving our understanding of the pathogenicity of SIV infection. Current research points towards therapeutic avenues for controlling the deleterious consequences of pathogenic infections.
AJ, CA, and IP designed, wrote, and edited the manuscript. All authors contributed to the article and approved the submitted version.
IP and CA are supported by grants from the National Institutes of Health, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Heart, Lung, and Blood Institute, and the National Institute of Allergy and Infectious Diseases (R01 DK130481 [IP], R01 DK113919 [IP/CA], R01 DK119936 [CA], R01 DK131476 [CA], RO1 HL117715 [IP], R01 HL123096 [IP], R01 HL154862 [IP], R01 AI119346 [CA]). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med (2011) 1:a006841. doi: 10.1101/cshperspect.a006841
2. VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev (2006) 19:728–62. doi: 10.1128/CMR.00009-06
3. Pandrea I, Apetrei C. Where the wild things are: pathogenesis of SIV infection in African nonhuman primate hosts. Curr HIV/AIDS Rep (2010) 7:28–36. doi: 10.1007/s11904-009-0034-8
4. Worobey M, Telfer P, Souquière S, Hunter M, Coleman CA, Metzger MJ, et al. Island biogeography reveals the deep history of SIV. Science (2010) 329:1487. doi: 10.1126/science.1193550
5. Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science (2012) 335:1188–93. doi: 10.1126/science.1217550
6. Ma D, Jasinska A, Kristoff J, Grobler JP, Turner T, Jung Y, et al. SIVagm infection in wild African green monkeys from south Africa: epidemiology, natural history, and evolutionary considerations. PloS Pathog (2013) 9:e1003011. doi: 10.1371/journal.ppat.1003011
7. Kobayashi T, Takeuchi JS, Ren F, Matsuda K, Sato K, Kimura Y, et al. Characterization of red-capped mangabey tetherin: implication for the co-evolution of primates and their lentiviruses. Sci Rep (2014) 4:5529. doi: 10.1038/srep05529
8. Apetrei C, Robertson DL, Marx PA. The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front Biosci (2004) 9:225–54. doi: 10.2741/1154
9. Bibollet-Ruche F, Bailes E, Gao F, Pourrut X, Barlow KL, Clewley JP, et al. New simian immunodeficiency virus infecting de brazza’s monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade. J Virol (2004) 78:7748–62. doi: 10.1128/JVI.78.14.7748-7762.2004
10. Gallant J, Hsue PY, Shreay S, Meyer N. Comorbidities among US patients with prevalent HIV infection-a trend analysis. J Infect Dis (2017) 216:1525–33. doi: 10.1093/infdis/jix518
11. Gallant J, Hsue P, Budd D, Meyer N. Healthcare utilization and direct costs of non-infectious comorbidities in HIV-infected patients in the USA. Curr Med Res Opin (2018) 34:13–23. doi: 10.1080/03007995.2017.1383889
12. Johnson PR, Hirsch VM. SIV infection of macaques as a model for AIDS pathogenesis. Int Rev Immunol (1992) 8:55–63. doi: 10.3109/08830189209056641
13. Lackner AA, Veazey RS. Current concepts in AIDS pathogenesis: insights from the SIV/macaque model. Annu Rev Med (2007) 58:461–76. doi: 10.1146/annurev.med.58.082405.094316
14. Beer B, Denner J, Brown CR, Norley S, zur Megede J, Coulibaly C, et al. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J Acquir Immune Defic Syndr Hum Retrovirol (1998) 18:210–20. doi: 10.1097/00042560-199807010-00003
15. Rey-Cuillé MA, Berthier JL, Bomsel-Demontoy MC, Chaduc Y, Montagnier L, Hovanessian AG, et al. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J Virol (1998) 72:3872–86. doi: 10.1128/JVI.72.5.3872-3886.1998
16. Broussard SR, Staprans SI, White R, Whitehead EM, Feinberg MB, Allan JS. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J Virol (2001) 75:2262–75. doi: 10.1128/JVI.75.5.2262-2275.2001
17. Onanga R, Kornfeld C, Pandrea I, Estaquier J, Souquière S, Rouquet P, et al. High levels of viral replication contrast with only transient changes in CD4(+) and CD8(+) cell numbers during the early phase of experimental infection with simian immunodeficiency virus SIVmnd-1 in mandrillus sphinx. J Virol (2002) 76:10256–63. doi: 10.1128/JVI.76.20.10256-10263.2002
18. Silvestri G, Sodora DL, Koup RA, Paiardini M, O’Neil SP, McClure HM, et al. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity (2003) 18:441–52. doi: 10.1016/S1074-7613(03)00060-8
19. Pandrea I, Onanga R, Kornfeld C, Rouquet P, Bourry O, Clifford S, et al. High levels of SIVmnd-1 replication in chronically infected mandrillus sphinx. Virology (2003) 317:119–27. doi: 10.1016/j.virol.2003.08.015
20. Silvestri G, Fedanov A, Germon S, Kozyr N, Kaiser WJ, Garber DA, et al. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J Virol (2005) 79:4043–54. doi: 10.1128/JVI.79.7.4043-4054.2005
21. Apetrei C, Gautam R, Sumpter B, Carter AC, Gaufin T, Staprans SI, et al. Virus subtype-specific features of natural simian immunodeficiency virus SIVsmm infection in sooty mangabeys. J Virol (2007) 81:7913–23. doi: 10.1128/JVI.00281-07
22. Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, et al. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J Virol (2006) 80:4858–67. doi: 10.1128/JVI.80.10.4858-4867.2006
23. Pandrea IV, Gautam R, Ribeiro RM, Brenchley JM, Butler IF, Pattison M, et al. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J Immunol (2007) 179:3035–46. doi: 10.4049/jimmunol.179.5.3035
24. Apetrei C, Sumpter B, Souquiere S, Chahroudi A, Makuwa M, Reed P, et al. Immunovirological analyses of chronically simian immunodeficiency virus SIVmnd-1- and SIVmnd-2-infected mandrills (Mandrillus sphinx). J Virol (2011) 85:13077–87. doi: 10.1128/JVI.05693-11
25. Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol (2003) 77:11708–17. doi: 10.1128/JVI.77.21.11708-11717.2003
26. Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med (2004) 200:761–70. doi: 10.1084/jem.20041196
27. Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med (2004) 200:749–59. doi: 10.1084/jem.20040874
28. Gordon SN, Klatt NR, Bosinger SE, Brenchley JM, Milush JM, Engram JC, et al. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J Immunol (2007) 179:3026–34. doi: 10.4049/jimmunol.179.5.3026
29. Li Q, Duan L, Estes JD, Ma Z-M, Rourke T, Wang Y, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature (2005) 434:1148–52. doi: 10.1038/nature03513
30. Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature (2005) 434:1093–7. doi: 10.1038/nature03501
31. Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood (2008) 112:2826–35. doi: 10.1182/blood-2008-05-159301
32. Favre D, Lederer S, Kanwar B, Ma Z-M, Proll S, Kasakow Z, et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog (2009) 5(2):e1000295 (2009). doi: 10.1371/journal.ppat.1000295
33. Pandrea I, Onanga R, Rouquet P, Bourry O, Ngari P, Wickings EJ, et al. Chronic SIV infection ultimately causes immunodeficiency in African non-human primates. AIDS (2001) 15:2461–2. doi: 10.1097/00002030-200112070-00019
34. Pandrea I, Silvestri G, Apetrei C. AIDS in african nonhuman primate hosts of SIVs: a new paradigm of SIV infection. Curr HIV Res (2009) 7:57–72. doi: 10.2174/157016209787048456
35. Traina-Dorge V, Blanchard J, Martin L, Murphey-Corb M. Immunodeficiency and lymphoproliferative disease in an African green monkey dually infected with SIV and STLV-I. AIDS Res Hum Retroviruses (1992) 8:97–100. doi: 10.1089/aid.1992.8.97
36. Ling B, Telfer P, Reed P, Robertson DL, Marx PA. A link between SIVsm in sooty mangabeys (SM) in wild-living monkeys in Sierra Leone and SIVsm in an American-based SM colony. AIDS Res Hum Retroviruses (2004) 20:1348–51. doi: 10.1089/aid.2004.20.1348
37. Souquière S, Bibollet-Ruche F, Robertson DL, Makuwa M, Apetrei C, Onanga R, et al. Wild mandrillus sphinx are carriers of two types of lentivirus. J Virol (2001) 75:7086–96. doi: 10.1128/JVI.75.15.7086-7096.2001
38. Apetrei C, Gormus B, Pandrea I, Metzger M, ten Haaft P, Martin LN, et al. Direct inoculation of simian immunodeficiency virus from sooty mangabeys in black mangabeys (Lophocebus aterrimus): first evidence of AIDS in a heterologous African species and different pathologic outcomes of experimental infection. J Virol (2004) 78:11506–18. doi: 10.1128/JVI.78.21.11506-11518.2004
39. Hatziioannou T, Evans DT. Animal models for HIV/AIDS research. Nat Rev Microbiol (2012) 10:852–67. doi: 10.1038/nrmicro2911
40. Evans DT, Silvestri G. Nonhuman primate models in AIDS research. Curr Opin HIV AIDS (2013) 8:255–61. doi: 10.1097/COH.0b013e328361cee8
41. Joas S, Parrish EH, Gnanadurai CW, Lump E, Stürzel CM, Parrish NF, et al. Species-specific host factors rather than virus-intrinsic virulence determine primate lentiviral pathogenicity. Nat Commun (2018) 9:1371. doi: 10.1038/s41467-018-03762-3
42. Raehtz K, Pandrea I, Apetrei C. The well-tempered SIV infection: Pathogenesis of SIV infection in natural hosts in the wild, with emphasis on virus transmission and early events post-infection that may contribute to protection from disease progression. Infect Genet Evol (2016) 46:308–23. doi: 10.1016/j.meegid.2016.07.006
43. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med (2006) 12:1365–71. doi: 10.1038/nm1511
44. Kornfeld C, Ploquin MJ-Y, Pandrea I, Faye A, Onanga R, Apetrei C, et al. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J Clin Invest (2005) 115:1082–91. doi: 10.1172/JCI23006
45. Mandl JN, Barry AP, Vanderford TH, Kozyr N, Chavan R, Klucking S, et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med (2008) 14:1077–87. doi: 10.1038/nm.1871
46. Jacquelin B, Mayau V, Targat B, Liovat A-S, Kunkel D, Petitjean G, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest (2009) 119(12):3544–55. doi: 10.1172/JCI40093
47. Apetrei C, Gaufin T, Gautam R, Vinton C, Hirsch V, Lewis M, et al. Pattern of SIVagm infection in patas monkeys suggests that host adaptation to simian immunodeficiency virus infection may result in resistance to infection and virus extinction. J Infect Dis (2010) 202:S371–6. doi: 10.1086/655970
48. Vinton C, Klatt NR, Harris LD, Briant JA, Sanders-Beer BE, Herbert R, et al. CD4-like immunological function by CD4- T cells in multiple natural hosts of simian immunodeficiency virus. J Virol (2011) 85:8702–8. doi: 10.1128/JVI.00332-11
49. Sundaravaradan V, Saleem R, Micci L, Gasper MA, Ortiz AM, Else J, et al. Multifunctional double-negative T cells in sooty mangabeys mediate T-helper functions irrespective of SIV infection. PLoS Pathog (2013) 9(6):e1003441. doi: 10.1371/journal.ppat.1003441
50. Perkins MR, Briant JA, Calantone N, Whitted S, Vinton CL, Klatt NR, et al. Homeostatic cytokines induce CD4 downregulation in African green monkeys independently of antigen exposure to generate simian immunodeficiency virus-resistant CD8αα T cells. J Virol (2014) 88:10714–24. doi: 10.1128/JVI.01331-14
51. Pandrea I, Parrish NF, Raehtz K, Gaufin T, Barbian HJ, Ma D, et al. Mucosal simian immunodeficiency virus transmission in African green monkeys: susceptibility to infection is proportional to target cell availability at mucosal sites. J Virol (2012) 86:4158–68. doi: 10.1128/JVI.07141-11
52. Ma D, Jasinska AJ, Feyertag F, Wijewardana V, Kristoff J, He T, et al. Factors associated with siman immunodeficiency virus transmission in a natural African nonhuman primate host in the wild. J Virol (2014) 88:5687–705. doi: 10.1128/JVI.03606-13
53. Pandrea I, Apetrei C, Gordon S, Barbercheck J, Dufour J, Bohm R, et al. Paucity of CD4+CCR5+ T cells is a typical feature of natural SIV hosts. Blood (2007) 109:1069–76. doi: 10.1182/blood-2006-05-024364
54. Pandrea I, Onanga R, Souquiere S, Mouinga-Ondéme A, Bourry O, Makuwa M, et al. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J Virol (2008) 82:5501–9. doi: 10.1128/JVI.02555-07
55. Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci (2010) 365:2487–94. doi: 10.1098/rstb.2010.0031
56. Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science (2000) 287:607–14. doi: 10.1126/science.287.5453.607
57. Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, et al. Hybrid origin of SIV in chimpanzees. Science (2003) 300:1713. doi: 10.1126/science.1080657
58. Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature (1990) 345:356–9. doi: 10.1038/345356a0
59. Peeters M, Fransen K, Delaporte E, Van den Haesevelde M, Gershy-Damet GM, Kestens L, et al. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS (1992) 6:447–51. doi: 10.1097/00002030-199205000-00002
60. Santiago ML, Rodenburg CM, Kamenya S, Bibollet-Ruche F, Gao F, Bailes E, et al. SIVcpz in wild chimpanzees. Science (2002) 295:465. doi: 10.1126/science.295.5554.465
61. Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science (2006) 313:523–6. doi: 10.1126/science.1126531
62. Rudicell RS, Holland Jones J, Wroblewski EE, Learn GH, Li Y, Robertson JD, et al. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PloS Pathog (2010) 6:e1001116. doi: 10.1371/journal.ppat.1001116
63. Li Y, Ndjango J-B, Learn GH, Ramirez MA, Keele BF, Bibollet-Ruche F, et al. Eastern Chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J Virol (2012) 86:10776–91. doi: 10.1128/JVI.01498-12
64. Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology (2007) 368:155–71. doi: 10.1016/j.virol.2007.06.018
65. Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, et al. Origin of HIV-1 in the chimpanzee pan troglodytes troglodytes. Nature (1999) 397:436–41. doi: 10.1038/17130
66. Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, et al. Timing the ancestor of the HIV-1 pandemic strains. Science (2000) 288:1789–96. doi: 10.1126/science.288.5472.1789
67. Simon F, Mauclère P, Roques P, Loussert-Ajaka I, Müller-Trutwin MC, Saragosti S, et al. Identification of a new human immunodeficiency virus type 1 distinct from group m and group O. Nat Med (1998) 4:1032–7. doi: 10.1038/2017
68. Roques P, Robertson DL, Souquière S, Apetrei C, Nerrienet E, Barré-Sinoussi F, et al. Phylogenetic characteristics of three new HIV-1 N strains and implications for the origin of group n. AIDS (2004) 18:1371–81. doi: 10.1097/01.aids.0000125990.86904.28
69. Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature (2006) 444:164. doi: 10.1038/444164a
70. Plantier J-C, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemée V, et al. A new human immunodeficiency virus derived from gorillas. Nat Med (2009) 15:871–2. doi: 10.1038/nm.2016
71. Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol (2009) 83:1635–48. doi: 10.1128/JVI.02311-08
72. Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, et al. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature (1992) 358:495–9. doi: 10.1038/358495a0
73. Gao F, Yue L, Robertson DL, Hill SC, Hui H, Biggar RJ, et al. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol (1994) 68:7433–47. doi: 10.1128/jvi.68.11.7433-7447.1994
74. Marx PA, Alcabes PG, Drucker E. Serial human passage of simian immunodeficiency virus by unsterile injections and the emergence of epidemic human immunodeficiency virus in Africa. Philos Trans R Soc Lond B Biol Sci (2001) 356:911–20. doi: 10.1098/rstb.2001.0867
75. Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, et al. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the taï forest, côte d’Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol (2005) 79:12515–27. doi: 10.1128/JVI.79.19.12515-12527.2005
76. Damond F, Worobey M, Campa P, Farfara I, Colin G, Matheron S, et al. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res Hum Retroviruses (2004) 20:666–72. doi: 10.1089/0889222041217392
77. Apetrei C, Metzger MJ, Richardson D, Ling B, Telfer PT, Reed P, et al. Detection and partial characterization of simian immunodeficiency virus SIVsm strains from bush meat samples from rural Sierra Leone. J Virol (2005) 79:2631–6. doi: 10.1128/JVI.79.4.2631-2636.2005
78. McCarthy KR, Johnson WE, Kirmaier A. Phylogeny and history of the lost SIV from crab-eating macaques: SIVmfa. PloS One (2016) 11:e0159281. doi: 10.1371/journal.pone.0159281
79. Apetrei C, Lerche NW, Pandrea I, Gormus B, Silvestri G, Kaur A, et al. Kuru experiments triggered the emergence of pathogenic SIVmac. AIDS (2006) 20:317–21. doi: 10.1097/01.aids.0000206498.71041.0e
80. Gormus BJ, Martin LN, Baskin GB. A brief history of the discovery of natural simian immunodeficiency virus (SIV) infections in captive sooty mangabey monkeys. Front Biosci (2004) 9:216–24. doi: 10.2741/1151
81. Apetrei C, Kaur A, Lerche NW, Metzger M, Pandrea I, Hardcastle J, et al. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J Virol (2005) 79:8991–9005. doi: 10.1128/JVI.79.14.8991-9005.2005
82. Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O’Connor S, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PloS Biol (2010) 8:e1000462. doi: 10.1371/journal.pbio.1000462
83. Beer BE, Brown CR, Whitted S, Goldstein S, Goeken R, Plishka R, et al. Immunodeficiency in the absence of high viral load in pig-tailed macaques infected with simian immunodeficiency virus SIVsun or SIVlhoest. J Virol (2005) 79:14044–56. doi: 10.1128/JVI.79.22.14044-14056.2005
84. Hirsch VM, Dapolito GA, Goldstein S, McClure H, Emau P, Fultz PN, et al. A distinct African lentivirus from sykes’ monkeys. J Virol (1993) 67:1517–28. doi: 10.1128/jvi.67.3.1517-1528.1993
85. Pandrea I, Gaufin T, Gautam R, Kristoff J, Mandell D, Montefiori D, et al. Functional cure of SIVagm infection in rhesus macaques results in complete recovery of CD4+ T cells and is reverted by CD8+ cell depletion. PloS Pathog (2011) 7:e1002170. doi: 10.1371/journal.ppat.1002170
86. Goldstein S, Ourmanov I, Brown CR, Plishka R, Buckler-White A, Byrum R, et al. Plateau levels of viremia correlate with the degree of CD4+-t-cell loss in simian immunodeficiency virus SIVagm-infected pigtailed macaques: variable pathogenicity of natural SIVagm isolates. J Virol (2005) 79:5153–62. doi: 10.1128/JVI.79.8.5153-5162.2005
87. Gautam R, Gaufin T, Butler I, Gautam A, Barnes M, Mandell D, et al. Simian immunodeficiency virus SIVrcm, a unique CCR2-tropic virus, selectively depletes memory CD4+ T cells in pigtailed macaques through expanded coreceptor usage. vivo. J Virol (2009) 83:7894–908. doi: 10.1128/JVI.00444-09
88. Mandell DT, Kristoff J, Gaufin T, Gautam R, Ma D, Sandler N, et al. Pathogenic features associated with increased virulence upon simian immunodeficiency virus cross-species transmission from natural hosts. J Virol (2014) 88:6778–92. doi: 10.1128/JVI.03785-13
89. Kleinman AJ, Pandrea I, Apetrei C. So pathogenic or so what?-a brief overview of SIV pathogenesis with an emphasis on cure research. Viruses (2022) 14:135. doi: 10.3390/v14010135
90. Turner TR, Schmitt CA, Cramer JD. Savanna monkeys: The genus chlorocebus. Cambridge Univ Press (2019). doi: 10.1017/9781139019941
91. Carr M, Kawaguchi A, Sasaki M, Gonzalez G, Ito K, Thomas Y, et al. Isolation of a simian immunodeficiency virus from a malbrouck (Chlorocebus cynosuros). Arch Virol (2017) 162:543–8. doi: 10.1007/s00705-016-3129-8
92. Jasinska AJ, Dong TS, Lagishetty V, Katzka W, Jacobs JP, Schmitt CA, et al. Shifts in microbial diversity, composition, and functionality in the gut and genital microbiome during a natural SIV infection in vervet monkeys. Microbiome (2020) 8:154. doi: 10.1186/s40168-020-00928-4
93. Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS (2003) 17:1871. doi: 10.1097/00002030-200309050-00005
94. British Medical Journal Publishing Group. Comparison of female to male and male to female transmission of HIV in 563 stable couples. European study group on heterosexual transmission of HIV. BMJ (1992) 304:809–13. doi: 10.1136/bmj.304.6830.809
95. Phillips-Conroy JE, Jolly CJ, Petros B, Allan JS, Desrosiers RC. Sexual transmission of SIVagm in wild grivet monkeys. J Med Primatol (1994) 23:1–7. doi: 10.1111/j.1600-0684.1994.tb00088.x
96. Nerrienet E, Amouretti X, Müller-Trutwin MC, Poaty-Mavoungou V, Bedjebaga I, Nguyen HT, et al. Phylogenetic analysis of SIV and STLV type I in mandrills (Mandrillus sphinx): indications that intracolony transmissions are predominantly the result of male-to-male aggressive contacts. AIDS Res Hum Retroviruses (1998) 14:785–96. doi: 10.1089/aid.1998.14.785
97. Estaquier J, Peeters M, Bedjabaga L, Honoré C, Bussi P, Dixson A, et al. Prevalence and transmission of simian immunodeficiency virus and simian T-cell leukemia virus in a semi-free-range breeding colony of mandrills in Gabon. AIDS (1991) 5:1385–6. doi: 10.1097/00002030-199111000-00018
98. Otsyula MG, Gettie A, Suleman M, Tarara R, Mohamed I, Marx P. Apparent lack of vertical transmission of simian immunodeficiency virus (SIV) in naturally infected African green monkeys, cercopithecus aethiops. Ann Trop Med Parasitol (1995) 89:573–6. doi: 10.1080/00034983.1995.11812990
99. Georges-Courbot MC, Moisson P, Leroy E, Pingard AM, Nerrienet E, Dubreuil G, et al. Occurrence and frequency of transmission of naturally occurring simian retroviral infections (SIV, STLV, and SRV) at the CIRMF primate center, Gabon. J Med Primatol (1996) 25:313–26. doi: 10.1111/j.1600-0684.1996.tb00023.x
100. Chahroudi A, Meeker T, Lawson B, Ratcliffe S, Else J, Silvestri G. Mother-to-infant transmission of simian immunodeficiency virus is rare in sooty mangabeys and is associated with low viremia. J Virol (2011) 85:5757–63. doi: 10.1128/JVI.02690-10
101. Jasinska AJ, Pandrea I, Apetrei C. CCR5 as a coreceptor for human immunodeficiency virus and simian immunodeficiency viruses: A prototypic love-hate affair. Front Immunol (2022) 13:835994. doi: 10.3389/fimmu.2022.835994
102. Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis (2011) 203:780–90. doi: 10.1093/infdis/jiq118
103. Nixon DE, Landay AL. Biomarkers of immune dysfunction in HIV. Curr Opin HIV AIDS (2010) 5:498–503. doi: 10.1097/COH.0b013e32833ed6f4
104. Chaisson LH, Semitala FC, Asege L, Mwebe S, Katende J, Nakaye M, et al. Point-of-care c-reactive protein and risk of early mortality among adults initiating antiretroviral therapy. AIDS (2019) 33:895–902. doi: 10.1097/QAD.0000000000002130
105. Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature (2009) 460:515–9. doi: 10.1038/nature08200
106. Terio KA, Kinsel MJ, Raphael J, Mlengeya T, Lipende I, Kirchhoff CA, et al. Pathologic lesions in chimpanzees (Pan trogylodytes schweinfurthii) from gombe national park, Tanzania, 2004–2010. J Zoo Wildlife Med (2011) 42:597–607. doi: 10.1638/2010-0237.1
107. Greenwood EJD, Schmidt F, Kondova I, Niphuis H, Hodara VL, Clissold L, et al. Simian immunodeficiency virus infection of chimpanzees (Pan troglodytes) shares features of both pathogenic and non-pathogenic lentiviral infections. PloS Pathog (2015) 11:e1005146. doi: 10.1371/journal.ppat.1005146
108. Barbian HJ, Jackson-Jewett R, Brown CS, Bibollet-Ruche F, Learn GH, Decker T, et al. Effective treatment of SIVcpz-induced immunodeficiency in a captive western chimpanzee. Retrovirology (2017) 14:35. doi: 10.1186/s12977-017-0359-0
109. Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene Hemophilia growth and development study, multicenter AIDS cohort study, multicenter hemophilia cohort study, San Francisco city cohort, ALIVE study. Science (1996) 273:1856–62. doi: 10.1126/science.273.5283.1856
110. Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell (1996) 86:367–77. doi: 10.1016/S0092-8674(00)80110-5
111. Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature (1996) 382:722–5. doi: 10.1038/382722a0
112. Chen Z, Kwon D, Jin Z, Monard S, Telfer P, Jones MS, et al. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J Exp Med (1998) 188:2057–65. doi: 10.1084/jem.188.11.2057
113. Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, et al. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use. vivo. PloS Pathog (2010) 6:e1001064. doi: 10.1371/journal.ppat.1001064
114. Wetzel KS, Yi Y, Yadav A, Bauer AM, Bello EA, Romero DC, et al. Loss of CXCR6 coreceptor usage characterizes pathogenic lentiviruses. PloS Pathog (2018) 14:e1007003. doi: 10.1371/journal.ppat.1007003
115. Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, et al. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nat Med (2011) 17:830–6. doi: 10.1038/nm.2395
116. Elliott STC, Wetzel KS, Francella N, Bryan S, Romero DC, Riddick NE, et al. Dualtropic CXCR6/CCR5 simian immunodeficiency virus (SIV) infection of sooty mangabey primary lymphocytes: Distinct coreceptor use in natural versus pathogenic hosts of SIV. J Virol (2015) 89:9252–61. doi: 10.1128/JVI.01236-15
117. Elliott STC, Riddick NE, Francella N, Paiardini M, Vanderford TH, Li B, et al. Cloning and analysis of sooty mangabey alternative coreceptors that support simian immunodeficiency virus SIVsmm entry independently of CCR5. J Virol (2012) 86:898–908. doi: 10.1128/jvi.06415-11
118. Wetzel KS, Yi Y, Elliott STC, Romero D, Jacquelin B, Hahn BH, et al. CXCR6-mediated simian immunodeficiency virus SIVagmSab entry into sabaeus African green monkey lymphocytes implicates widespread use of non-CCR5 pathways in natural host infections. J Virol (2017) 91(4):e01626–16. doi: 10.1128/JVI.01626-16
119. Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, et al. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci Transl Med (2016) 8:358ra125. doi: 10.1126/scitranslmed.aag1048
120. Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, et al. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat Med (2009) 15:879–85. doi: 10.1038/nm.1970
121. Brenchley JM, Silvestri G, Douek DC. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity (2010) 32:737–42. doi: 10.1016/j.immuni.2010.06.004
122. Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. Understanding the benign nature of SIV infection in natural hosts. J Clin Invest (2007) 117:3148–54. doi: 10.1172/JCI33034
123. Pandrea I, Brooks K, Desai RP, Tare M, Brenchley JM, Apetrei C. I’ve looked at gut from both sides now: Gastrointestinal tract involvement in the pathogenesis of SARS-CoV-2 and HIV/SIV infections. Front Immunol (2022) 13:899559. doi: 10.3389/fimmu.2022.899559
124. Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol (2009) 9:799–809. doi: 10.1038/nri2653
125. Martel J, Chang S-H, Ko Y-F, Hwang T-L, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends Endocrinol Metab (2022) 33:247–65. doi: 10.1016/j.tem.2022.01.002
126. Veazey RS, Tham IC, Mansfield KG, DeMaria M, Forand AE, Shvetz DE, et al. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4(+) T cells are rapidly eliminated in early SIV infection. vivo. J Virol (2000) 74:57–64. doi: 10.1128/JVI.74.1.57-64.2000
127. Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev (2013) 254:54–64. doi: 10.1111/imr.12066
128. Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev (2013) 26:2–18. doi: 10.1128/CMR.00050-12
129. Brenchley JM. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol (2013) 6:657–65. doi: 10.1038/mi.2013.15
130. Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PloS Pathog (2010) 6:e1001052. doi: 10.1371/journal.ppat.1001052
131. Klase Z, Ortiz A, Deleage C, Mudd JC, Quiñones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol (2015) 8:1009–20. doi: 10.1038/mi.2014.128
132. Isnard S, Fombuena B, Sadouni M, Lin J, Richard C, Routy B, et al. Circulating β-d-Glucan as a marker of subclinical coronary plaque in antiretroviral therapy-treated people with human immunodeficiency virus. Open Forum Infect Dis (2021) 8:ofab109. doi: 10.1093/ofid/ofab109
133. Isnard S, Lin J, Bu S, Fombuena B, Royston L, Routy J-P. Gut leakage of fungal-related products: Turning up the heat for HIV infection. Front Immunol (2021) 12:656414. doi: 10.3389/fimmu.2021.656414
134. Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis (1999) 179:859–70. doi: 10.1086/314660
135. Le Hingrat Q, Sette P, Xu C, Rahmberg AR, Tarnus L, Annapureddy H, et al. Prolonged antibody-mediated CD4+ T-cell depletion does not cause disease progression in nonprogressively SIV-infected African green monkeys. Nat Commun (2022) 13 in press.
136. Marchetti G, Bellistrì GM, Borghi E, Tincati C, Ferramosca S, La Francesca M, et al. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS (2008) 22:2035–8. doi: 10.1097/QAD.0b013e3283112d29
137. Lee PI, Ciccone EJ, Read SW, Asher A, Pitts R, Douek DC, et al. Evidence for translocation of microbial products in patients with idiopathic CD4 lymphocytopenia. J Infect Dis (2009) 199:1664–70. doi: 10.1086/598953
138. Kovacs SB, Sheikh V, Thompson WL, Morcock DR, Perez-Diez A, Yao MD, et al. T-Cell depletion in the colonic mucosa of patients with idiopathic CD4+ lymphopenia. J Infect Dis (2015) 212:1579–87. doi: 10.1093/infdis/jiv282
139. Morris A, Hillenbrand M, Finkelman M, George MP, Singh V, Kessinger C, et al. Serum (1→3)-β-D-glucan levels in HIV-infected individuals are associated with immunosuppression, inflammation, and cardiopulmonary function. J Acquir Immune Defic Syndr (2012) 61:462–8. doi: 10.1097/QAI.0b013e318271799b
140. Weiner LD, Retuerto M, Hager CL, El Kamari V, Shan L, Sattar A, et al. Fungal translocation is associated with immune activation and systemic inflammation in treated HIV. AIDS Res Hum Retroviruses (2019) 35:461–72. doi: 10.1089/aid.2018.0252
141. Gianella S, Letendre SL, Iudicello J, Franklin D, Gaufin T, Zhang Y, et al. Plasma (1 → 3)-β-D-glucan and suPAR levels correlate with neurocognitive performance in people living with HIV on antiretroviral therapy: a CHARTER analysis. J Neurovirol (2019) 25:837–43. doi: 10.1007/s13365-019-00775-6
142. Le Hingrat Q, Sereti I, Landay AL, Pandrea I, Apetrei C. The hitchhiker guide to CD4+ T-cell depletion in lentiviral infection. a critical review of the dynamics of the CD4+ T cells in SIV and HIV infection. Front Immunol (2021) 12:695674. doi: 10.3389/fimmu.2021.695674
143. Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science (1998) 280:427–31. doi: 10.1126/science.280.5362.427
144. Klatt NR, Brenchley JM. Th17 cell dynamics in HIV infection. Curr Opin HIV AIDS (2010) 5:135–40. doi: 10.1097/COH.0b013e3283364846
145. Wiche Salinas TR, Zhang Y, Sarnello D, Zhyvoloup A, Marchand LR, Fert A, et al. Th17 cell master transcription factor RORC2 regulates HIV-1 gene expression and viral outgrowth. Proc Natl Acad Sci U.S.A. (2021) 118(48):e2105927118. doi: 10.1073/pnas.2105927118
146. Renault C, Veyrenche N, Mennechet F, Bedin A-S, Routy J-P, Van de Perre P, et al. Th17 CD4+ T-cell as a preferential target for HIV reservoirs. Front Immunol (2022) 13:822576. doi: 10.3389/fimmu.2022.822576
147. Fert A, Raymond Marchand L, Wiche Salinas TR, Ancuta P. Targeting Th17 cells in HIV-1 remission/cure interventions. Trends Immunol (2022) 43:580–94. doi: 10.1016/j.it.2022.04.013
148. Li Q, Estes JD, Duan L, Jessurun J, Pambuccian S, Forster C, et al. Simian immunodeficiency virus-induced intestinal cell apoptosis is the underlying mechanism of the regenerative enteropathy of early infection. J Infect Dis (2008) 197:420–9. doi: 10.1086/525046
149. Epple H-J, Allers K, Tröger H, Kühl A, Erben U, Fromm M, et al. Acute HIV infection induces mucosal infiltration with CD4+ and CD8+ T cells, epithelial apoptosis, and a mucosal barrier defect. Gastroenterology (2010) 139:1289–300. doi: 10.1053/j.gastro.2010.06.065
150. Estes JD, Gordon SN, Zeng M, Chahroudi AM, Dunham RM, Staprans SI, et al. Early resolution of acute immune activation and induction of PD-1 in SIV-infected sooty mangabeys distinguishes nonpathogenic from pathogenic infection in rhesus macaques. J Immunol (2008) 180:6798–807. doi: 10.4049/jimmunol.180.10.6798
151. Raehtz KD, Barrenäs F, Xu C, Busman-Sahay K, Valentine A, Law L, et al. African Green monkeys avoid SIV disease progression by preventing intestinal dysfunction and maintaining mucosal barrier integrity. PloS Pathog (2020) 16:e1008333. doi: 10.1371/journal.ppat.1008333
152. Kaur A, Grant RM, Means RE, McClure H, Feinberg M, Johnson RP. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J Virol (1998) 72:9597–611. doi: 10.1128/JVI.72.12.9597-9611.1998
153. Pandrea I, Cornell E, Wilson C, Ribeiro RM, Ma D, Kristoff J, et al. Coagulation biomarkers predict disease progression in SIV-infected nonhuman primates. Blood (2012) 120:1357–66. doi: 10.1182/blood-2012-03-414706
154. Antoni L, Nuding S, Wehkamp J, Stange EF. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol (2014) 20:1165–79. doi: 10.3748/wjg.v20.i5.1165
155. Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol (2017) 11:821–34. doi: 10.1080/17474124.2017.1343143
156. Barrenas F, Raehtz K, Xu C, Law L, Green RR, Silvestri G, et al. Macrophage-associated wound healing contributes to African green monkey SIV pathogenesis control. Nat Commun (2019) 10:5101. doi: 10.1038/s41467-019-12987-9
157. Hao XP, Lucero CM, Turkbey B, Bernardo ML, Morcock DR, Deleage C, et al. Experimental colitis in SIV-uninfected rhesus macaques recapitulates important features of pathogenic SIV infection. Nat Commun (2015) 6:8020. doi: 10.1038/ncomms9020
158. Estaquier J, Idziorek T, de Bels F, Barré-Sinoussi F, Hurtrel B, Aubertin AM, et al. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc Natl Acad Sci U.S.A. (1994) 91:9431–5.
159. Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe (2013) 14:329–39. doi: 10.1016/j.chom.2013.08.006
160. Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med (2013) 5:193ra91. doi: 10.1126/scitranslmed.3006438
161. Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol (2014) 7:983–94. doi: 10.1038/mi.2013.116
162. Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe (2016) 19:311–22. doi: 10.1016/j.chom.2016.02.011
163. Brenchley JM, Ortiz AM. Microbiome studies in non-human primates. Curr HIV/AIDS Rep (2021) 18:527–37. doi: 10.1007/s11904-021-00584-9
164. Moeller AH, Shilts M, Li Y, Rudicell RS, Lonsdorf EV, Pusey AE, et al. SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe (2013) 14:340–5. doi: 10.1016/j.chom.2013.08.005
165. Barbian HJ, Li Y, Ramirez M, Klase Z, Lipende I, Mjungu D, et al. Destabilization of the gut microbiome marks the end-stage of simian immunodeficiency virus infection in wild chimpanzees. Am J Primatol (2018) 80:e22515. doi: 10.1002/ajp.22515
166. Moeller AH, Peeters M, Ayouba A, Ngole EM, Esteban A, Hahn BH, et al. Stability of the gorilla microbiome despite simian immunodeficiency virus infection. Mol Ecol (2015) 24:690–7. doi: 10.1111/mec.13057
167. Bochart RM, Tharp G, Jean S, Upadhyay AA, Crane MM, Vanderford TH, et al. Microbiome stability with chronic SIV infection in AIDS-resistant sooty mangabeys. bioRxiv (2019) 780825. doi: 10.1101/780825
168. Islam SMS, Ryu H-M, Sohn S. Tetragenococcus halophilus alleviates intestinal inflammation in mice by altering gut microbiota and regulating dendritic cell activation via CD83. Cells (2022) 11:1903. doi: 10.3390/cells11121903
169. Jasinska AJ, Schmitt CA, Service SK, Cantor RM, Dewar K, Jentsch JD, et al. Systems biology of the vervet monkey. ILAR J (2013) 54:122–43. doi: 10.1093/ilar/ilt049
170. Jasinska AJ. Resources for functional genomic studies of health and development in nonhuman primates. Am J Phys Anthropol (2020) 171 Suppl 70:174–94. doi: 10.1002/ajpa.24051
171. Amato KR, Yeoman CJ, Cerda G, Schmitt CA, Cramer JD, Miller MEB, et al. Variable responses of human and non-human primate gut microbiomes to a Western diet. Microbiome (2015) 3:53. doi: 10.1186/s40168-015-0120-7
172. Flynn JK, Langner CA, Karmele EP, Baker PJ, Pei L, Gorfu EG, et al. Luminal microvesicles uniquely influence translocating bacteria after SIV infection. Mucosal Immunol (2021) 14:937–48. doi: 10.1038/s41385-021-00393-8
173. Milush JM, Reeves JD, Gordon SN, Zhou D, Muthukumar A, Kosub DA, et al. Virally induced CD4+ T cell depletion is not sufficient to induce AIDS in a natural host. J Immunol (2007) 179:3047–56. doi: 10.4049/jimmunol.179.5.3047
174. Milush JM, Mir KD, Sundaravaradan V, Gordon SN, Engram J, Cano CA, et al. Lack of clinical AIDS in SIV-infected sooty mangabeys with significant CD4+ T cell loss is associated with double-negative T cells. J Clin Invest (2011) 121:1102–10. doi: 10.1172/JCI44876
175. Vinton CL, Ortiz AM, Calantone N, Mudd JC, Deleage C, Morcock DR, et al. Cytotoxic T cell functions accumulate when CD4 is downregulated by CD4+ T cells in African green monkeys. J Immunol (2017) 198:4403–12. doi: 10.4049/jimmunol.1700136
176. Mudd JC, Perkins MR, DiNapoli SR, Hirsch VM, Brenchley JM. Interleukin-2 therapy induces CD4 downregulation, which decreases circulating CD4 T cell counts, in African green monkeys. J Virol (2016) 90:5750–8. doi: 10.1128/jvi.00057-16
177. Mudd JC, Lai S, Shah S, Rahmberg A, Flynn JK, Starke CE, et al. Epigenetic silencing of CD4 expression in nonpathogenic SIV infection in African green monkeys. JCI Insight (2020) 5:e139043. doi: 10.1172/jci.insight.139043
178. Rahmberg AR, Markowitz TE, Mudd JC, Hirsch V, Brenchley JM. Epigenetic reprogramming leads to downregulation of CD4 and functional changes in African green monkey memory CD4+ T cells. J Immunol (2022) 209:337–45. doi: 10.4049/jimmunol.2200109
179. Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med (2017) 23:1271–6. doi: 10.1038/nm.4411
180. Huot N, Rascle P, Petitdemange C, Contreras V, Palgen J-L, Stahl-Hennig C, et al. Non-human primate determinants of natural killer cells in tissues at steady-state and during simian immunodeficiency virus infection. Front Immunol (2020) 11:2134. doi: 10.3389/fimmu.2020.02134
181. Huot N, Jacquelin B, Garcia-Tellez T, Rascle P, Ploquin MJ, Madec Y, et al. Natural killer cells migrate into and control simian immunodeficiency virus replication in lymph node follicles in African green monkeys. Nat Med (2017) 23:1277–86. doi: 10.1038/nm.4421
182. Huot N, Rascle P, Petitdemange C, Contreras V, Stürzel CM, Baquero E, et al. SIV-induced terminally differentiated adaptive NK cells in lymph nodes associated with enhanced MHC-e restricted activity. Nat Commun (2021) 12:1282. doi: 10.1038/s41467-021-21402-1
183. Huot N, Rascle P, Tchitchek N, Wimmer B, Passaes C. Role of NKG2a/c+ CD8+ T cells in pathogenic versus non-pathogenic SIV infections. Iscience (2021) 24:102314. doi: 10.1016/j.isci.2021.102314
184. Svardal H, Jasinska AJ, Apetrei C, Coppola G, Huang Y, Schmitt CA, et al. Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat Genet (2017) 49:1705–13. doi: 10.1038/ng.3980
185. Bosinger SE, Li Q, Gordon SN, Klatt NR, Duan L, Xu L, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest (2009) 119:3556–72. doi: 10.1172/JCI40115
186. Bosinger SE. Nonpathogenic SIV infection of sooty mangabeys. In: Hope T.J, Richman D.D., Stevenson M. (eds) Encyclopedia of AIDS. New York, NY: Springer. pp: 1555–65. doi: 10.1007/978-1-4939-7101-5_205
187. Graham D. The sooty mangabey’s sequence. Lab Anim (2018) 47:62–2. doi: 10.1038/s41684-018-0018-7
188. Palesch D, Bosinger SE, Tharp GK, Vanderford TH, Paiardini M, Chahroudi A, et al. Sooty mangabey genome sequence provides insight into AIDS resistance in a natural SIV host. Nature (2018) 553:77–81. doi: 10.1038/nature25140
189. Yang Z-W, Jiang Y-H, Ma C, Silvestri G, Bosinger SE, Li B-L, et al. Coexpression network analysis of benign and malignant phenotypes of SIV-infected sooty mangabey and rhesus macaque. PloS One (2016) 11:e0156170. doi: 10.1371/journal.pone.0156170
190. Goldberg TL, Sintasath DM, Chapman CA, Cameron KM, Karesh WB, Tang S, et al. Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. J Virol (2009) 83:11318–29. doi: 10.1128/JVI.02616-08
191. Simons ND, Eick GN, Ruiz-Lopez MJ, Hyeroba D, Omeja PA, Weny G, et al. Genome-wide patterns of gene expression in a wild primate indicate species-specific mechanisms associated with tolerance to natural simian immunodeficiency virus infection. Genome Biol Evol (2019) 11:1630–43. doi: 10.1093/gbe/evz099
192. Jochems SP, Jacquelin B, Tchitchek N, Busato F, Pichon F, Huot N, et al. DNA Methylation changes in metabolic and immune-regulatory pathways in blood and lymph node CD4 T cells in response to SIV infections. Clin Epigenet (2020) 12:188. doi: 10.1186/s13148-020-00971-w
193. Alam A, Neish A. Role of gut microbiota in intestinal wound healing and barrier function. Tissue Barriers (2018) 6:1539595. doi: 10.1080/21688370.2018.1539595
194. Martín R, Laval L, Chain F, Miquel S, Natividad J, Cherbuy C, et al. Bifidobacterium animalis ssp. lactis CNCM-I2494 restores gut barrier permeability in chronically low-grade inflamed mice. Front Microbiol (2016) 7:608. doi: 10.3389/fmicb.2016.00608
195. Rousseau RK, Walmsley SL, Lee T, Rosenes R, Reinhard RJ, Malazogu F, et al. Randomized, blinded, placebo-controlled trial of de Simone formulation probiotic during HIV-associated suboptimal CD4+ T cell recovery. J Acquir Immune Defic Syndr (2022) 89:199–207. doi: 10.1097/QAI.0000000000002840
196. Yang OO, Kelesidis T, Cordova R, Khanlou H. Immunomodulation of antiretroviral drug-suppressed chronic HIV-1 infection in an oral probiotic double-blind placebo-controlled trial. AIDS Res Hum Retroviruses (2014) 30:988–95. doi: 10.1089/aid.2014.0181
197. Tenore S de B, Avelino-Silva VI, Costa PR, Franco LM, Sabino EC, Kalil J, et al. Immune effects of lactobacillus casei shirota in treated HIV-infected patients with poor CD4+ T-cell recovery. AIDS (2020) 34:381–9. doi: 10.1097/QAD.0000000000002420
198. Stiksrud B, Nowak P, Nwosu FC, Kvale D, Thalme A, Sonnerborg A, et al. Reduced levels of d-dimer and changes in gut microbiota composition after probiotic intervention in HIV-infected individuals on stable ART. J Acquir Immune Defic Syndr (2015) 70:329–37. doi: 10.1097/QAI.0000000000000784
199. d’Ettorre G, Rossi G, Scagnolari C, Andreotti M, Giustini N, Serafino S, et al. Probiotic supplementation promotes a reduction in T-cell activation, an increase in Th17 frequencies, and a recovery of intestinal epithelium integrity and mitochondrial morphology in ART-treated HIV-1-positive patients. Immun Inflammation Dis (2017) 5:244–60. doi: 10.1002/iid3.160
200. Kim SY, Nair MG. Macrophages in wound healing: activation and plasticity. Immunol Cell Biol (2019) 97:258–67. doi: 10.1111/imcb.12236
201. Gupta RK, Abdul-Jawad S, McCoy LE, Mok HP, Peppa D, Salgado M, et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature (2019) 568:244–8. doi: 10.1038/s41586-019-1027-4
202. Gupta RK, Peppa D, Hill AL, Gálvez C, Salgado M, Pace M, et al. Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet HIV (2020) 7:e340–7. doi: 10.1016/S2352-3018(20)30069-2
203. Kordelas L, Verheyen J, Beelen DW, Horn PA, Heinold A, Kaiser R, et al. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med (2014) 371:880–2. doi: 10.1056/NEJMc1405805
204. Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. New Engl J Med (2008) 359:1429–41. doi: 10.1056/nejmoa0803152
205. Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science (2013) 341:1387–90. doi: 10.1126/science.1241475
206. Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother (2005) 49:4721–32. doi: 10.1128/AAC.49.11.4721-4732.2005
207. Chang XL, Wu HL, Webb GM, Tiwary M, Hughes C, Reed JS, et al. CCR5 receptor occupancy analysis reveals increased peripheral blood CCR5+CD4+ T cells following treatment with the anti-CCR5 antibody leronlimab. Front Immunol (2021) 12:794638. doi: 10.3389/fimmu.2021.794638
208. Chang XL, Reed JS, Webb GM, Wu HL, Le J, Bateman KB, et al. Suppression of human and simian immunodeficiency virus replication with the CCR5-specific antibody leronlimab in two species. PloS Pathog (2022) 18:e1010396. doi: 10.1371/journal.ppat.1010396
209. Chang XL, Webb GM, Wu HL, Greene JM, Abdulhaqq S, Bateman KB, et al. Antibody-based CCR5 blockade protects macaques from mucosal SHIV transmission. Nat Commun (2021) 12:3343. doi: 10.1038/s41467-021-23697-6
210. Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature (2006) 443:350–4. doi: 10.1038/nature05115
211. Rahman SA, Yagnik B, Bally AP, Morrow KN, Wang S, Vanderford TH, et al. PD-1 blockade and vaccination provide therapeutic benefit against SIV by inducing broad and functional CD8+ T cells in lymphoid tissue. Sci Immunol (2021) 6:eabh3034. doi: 10.1126/sciimmunol.abh3034
Keywords: AIDS - acquired immunodeficiency syndrome, HIV - human immunodeficiency virus, nonhuman primate (NHP), African green monkey (AGM) (Chlorocebus aethiops), mucosal immune barrier, inflammation, microbiome, immune activation
Citation: Jasinska AJ, Apetrei C and Pandrea I (2023) Walk on the wild side: SIV infection in African non-human primate hosts—from the field to the laboratory. Front. Immunol. 13:1060985. doi: 10.3389/fimmu.2022.1060985
Received: 04 October 2022; Accepted: 15 December 2022;
Published: 12 January 2023.
Edited by:
Susan Prockop, Boston Children’s Hospital and Harvard Medical School, United StatesReviewed by:
Jean-Pierre Routy, McGill University, CanadaCopyright © 2023 Jasinska, Apetrei and Pandrea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivona Pandrea, cGFuZHJlYUBwaXR0LmVkdQ==
‡ORCID: Anna J Jasinska, orcid.org/0000-0002-1897-4570
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.