- Department of Microbiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
The complex network of microscopic organisms living on and within humans, collectively referred to as the microbiome, produce wide array of biologically active molecules that shape our health. Disruption of the microbiome is associated with susceptibility to a range of diseases such as cancer, diabetes, allergy, obesity, and infection. A new series of next-generation microbiome-based therapies are being developed to treat these diseases by transplanting bacteria or bacterial-derived byproducts into a diseased individual to reset the recipient’s microbiome and restore health. Microbiome transplantation therapy is still in its early stages of being a routine treatment option and, with a few notable exceptions, has had limited success in clinical trials. In this review, we highlight the successes and challenges of implementing these therapies to treat disease with a focus on interactions between the immune system and microbiome-based therapeutics. The immune activation status of the microbiome transplant recipient prior to transplantation has an important role in supporting bacterial engraftment. Following engraftment, microbiome transplant derived signals can modulate immune function to ameliorate disease. As novel microbiome-based therapeutics are developed, consideration of how the transplants will interact with the immune system will be a key factor in determining whether the microbiome-based transplant elicits its intended therapeutic effect.
Section 1: Introduction. Microbiome-based therapy and immune health
Millions of years of co-evolution between microbiome and the mammalian host has resulted in mutualism and homeostasis (1). Microbial diversity is a hallmark feature of a healthy microbiome as the trillions of microbes act in concert to support host health (2, 3). However, this mutualistic relationship can be breached by environmental factors, such as antibiotic use or dietary changes, that lead to decreased bacterial diversity or skewed relative abundance of bacterial species (4–6). Collectively, these changes in microbiome composition are referred to as microbial dysbiosis (7). Consistently, altered or ‘dysbiotic’ intestinal microbial communities are associated with disease states such as cancer, diabetes, allergy, obesity, neurological disorders, and infectious diseases (8–15). These associations have been replicated in animal studies, leading to increased mechanistic understanding of how microbial dysbiosis can drive disease progression. Numerous observations linking dysbiosis and disease have provoked the biomedical community to hypothesize that resetting the microbiome with a composition of bacteria from healthy individuals could restore health (16–18). In support of this theory, numerous animal studies demonstrate resolution of disease following microbiome transplantation directly into the host’s intestine (19–24). In humans, however, the efficacy of microbiome-based therapies to treat disease have been inconsistent (25). The complex challenge of developing successful microbiome-based therapies is exemplified by the multifaceted, bidirectional interactions between commensal bacteria and the host immune system (7).
At steady-state, intestinal bacteria are critical regulators of the host immune system that promote immune education, immunotolerance, as well as resistance to pathogens (15). For example, distinct species of the endogenous microbiome directly impact CD4+ T helper (TH) cell differentiation and function in the intestine. Resident segmented filamentous bacteria (SFB) attach to the small intestinal epithelium and drive CD4+ TH17 cell differentiation that is essential to maintain the integrity of the intestinal barrier (26, 27). SFB colonization induce antigen-specific TH17 cells in the small intestine via IL-22, IL-23, and serum amyloid A protein (SAA) protein and promote host defenses against enteric bacterial pathogens through CD4+ T cell dependent and independent mechanisms (26, 28, 29). Similarly, Bacteroides fragilis contributes to the differentiation of TH1 cells and lymphoid organogenesis by its capsular polysaccharide A (PSA). Colonization of germ-free mice with PSA produced from B. fragilis restores splenic IFN-γ producing CD4+ T cells through antigen presentation in an IL-12/STAT4 specific manner (30). Further, B. fragilis helps to maintain TH1/TH2 balance and directs the generation of the CD4+ regulatory T cells (Treg) via PSA (30, 31). Firmicutes, specifically bacteria belong to the Clostridia class, also promote immune homeostasis at steady-state by promoting Treg cell differentiation in the intestine via production of short chain fatty acids (SCFA) and secondary bile acids (32–39). Insights gained from these studies as well as many others (40, 41) that investigate immune-microbiome interactions at steady-state have laid the foundation to use the microbiome as a therapeutic that can be administered to an individual to recalibrate the immune system and treat disease.
The collective microbiome has been referenced as a distinct organ system, comparable to the kidney or liver organ, due to the capacity of the microbiome to synthesize signaling molecules that shape host physiology (42). A dysbiotic microbiome that fails to function properly is analogous to an organ system failure (43). Likewise, a microbiome-based transplant can be considered a bacterial “organ” transplant meant to replace the dysbiotic “failing organ” with a healthy one. As with kidney transplants, there are complex molecular interactions between the donor microbiome “organ” and the host that will determine whether the transplant successfully engrafts or is rejected. Crosstalk between the microbiome transplant and the host immune system has an important role in determining transplant success or failure and is the focus of this review.
Section 2: Dysbiosis and disease
It is often challenging to determine whether a dysbiotic intestinal microbiome is the cause or the consequence of the disease state. Chronic intestinal dysbiosis is associated with elevated expression of proinflammatory mediators that are hallmarks of various chronic inflammatory diseases (7, 41). For example, the intestinal microbiome in patients with inflammatory bowel disease (IBD) is characterized by a significant decrease in Firmicutes bacteria and an increase in Bacteroidetes and Proteobacteria (44). Instead of a diverse, Firmicutes-enriched microbiome that promotes intestinal Treg cell population and immune tolerance (34, 35, 45), the dysbiotic microbiome in IBD patients supports pathogenic CD4+ type-17 T helper (TH17) cells in the intestine. In contrast to the TH17 cell population induced by commensal bacteria that maintain barrier integrity in the context of a diverse microbiome, TH17 cells that arise under dysbiotic conditions release pro-inflammatory cytokines, causing intestinal damage (46–48). Inflammatory TH17 cells accumulate in IBD patients’ intestinal lesions both in the ileum and colon (49, 50). Several studies support that the dysbiotic intestinal microbiome in IBD is the causative agent that initiates immune changes in the colon. For instance, Britton et al. show that germ-free mice transplanted with the microbiome obtained from IBD patients had increased numbers of TH17 and TH2 cells and reduced RORγt+ Treg cells in their colon compared to germ-free mice that received microbiome from healthy donors (51). Using a T cell transfer model of colitis this group demonstrated the IBD microbiome caused more severe CD4+ T cell-mediated colitis, compared to healthy microbiome (51). Dysbiosis caused by external perturbation, such as antibiotic treatment, can also lead to activation of host inflammatory mediators, that provide feedback to the microbiome and drive expansion of potentially pathogenic intestinal bacteria. Byndloss et al. found that antibiotic-mediated depletion of butyrate-producing bacteria reduced peroxisome proliferator–activated receptor gamma (PPAR-γ) expression in the intestinal epithelium, leading to the induction of NOS2 and increased the nitrate levels in the colon (52). Host-derived nitrate can be used as respiratory electron acceptors by colitogenic Escherichia/Shigella or invasive Samonella bacteria in the intestine (53, 54). In both examples, the antibiotic-induced dysbiotic microbiome led to breakdown of intestinal immune homeostasis that leaves the host more vulnerable to colonization with disease-promoting bacteria.
Reciprocal to dysbiosis driving aberrant immune responses, host inflammation can cause intestinal dysbiosis. Several studies show that excessive intestinal inflammation due to infection or IBD can alter the intestinal lumen microbial population (55, 56). As a consequence of intestinal mucosal inflammation, inflammatory by-products like reactive oxygen species and reactive nitrogen species derivatives can change the intestinal microbiome by providing a growth advantage to inflammation-tolerant colitogenic bacteria (e.g., Gammaproteobacteria) over the inflammation-sensitive (e.g., Bifidobacteria) beneficial bacteria (53, 57, 58). For instance, infection due to Toxoplasma gondii, causes a marked immune-driven intestinal dysbiosis by increasing macrophage-derived NO byproducts in the colon that supports anaerobic respiration in E. coli (59). Likewise, in recurrent Clostridioides difficile infection patients, increased intestinal inflammation is associated with an abundance of Escherichia/Shigella in the intestinal tract (60, 61). Studies using genetically modified mice or mice treated with immunostimulatory agents to initiate immune activation demonstrate that immune activity can result in dysbiosis. Dysbiosis that is caused by immune activation renders the host more susceptible to severe colitis (62–64). Combined, these studies demonstrate the bidirectional influence of immune system and dysbiosis.
In dysbiosis-associated diseases, it is difficult to determine whether the altered microbiome is driving pathogenic immune responses or, conversely, dysregulated immunity promotes microbial dysbiosis. In reality, a reciprocal relationship exists where both the dysbiotic microbiome and aberrant immune system feed forward to reinforce dysbiosis and immunopathology. Microbiome-based therapies seek to break this cycle by resetting the microbiome back to a “healthy” state. The immunologic consequences of such therapies need to be considered for successful implementation of such therapies.
Section 3: Types of microbiome-based therapies
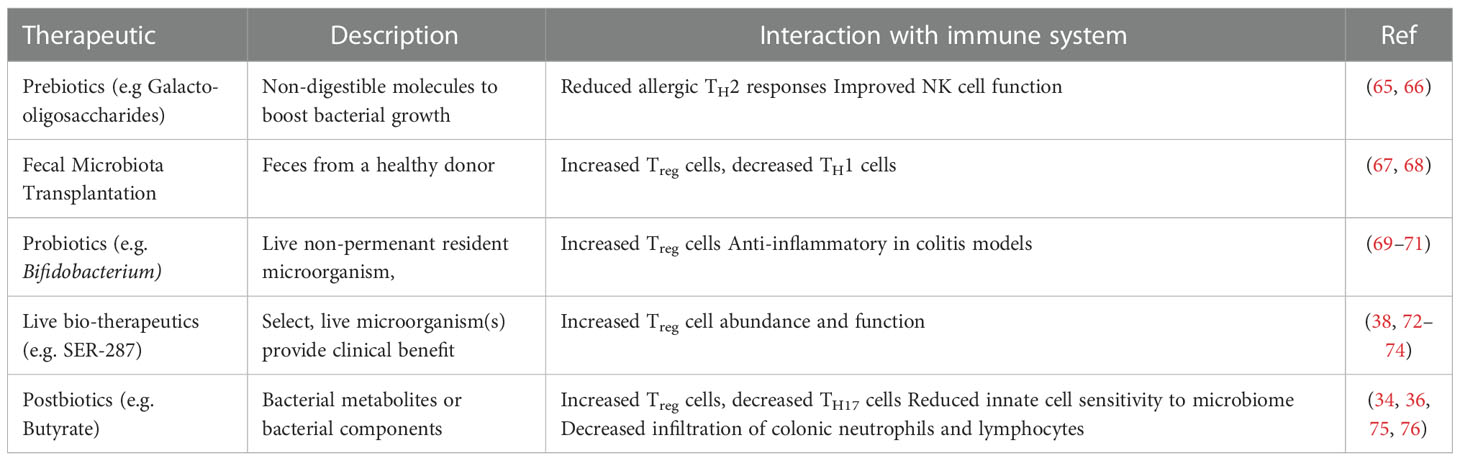
Microbiome-based therapeutics seek to restore the healthy microbial populations that reside on or within the host and the downstream metabolic networks that the microbiome directs. Broadly, microbiome-based therapies can be categorized into five approaches. These are prebiotics, fecal microbiota transplantation (FMT), probiotics, live-biotherapeutics, and postbiotics (Table 1).
3.1 Prebiotics
Prebiotics are non-living food components that are not digestible by the host but beneficially impact the host’s health by selectively stimulating the activity and growth of some genera of microbes in the intestine (65, 77). Current prebiotics are predominantly carbohydrate-based, but other substances, such as polyunsaturated fatty acids and polyphenols, also exert prebiotic effects (65). An ideal prebiotic can resist the actions of acids, bile salts, and other hydrolyzing enzymes in the intestine, is non-absorbable in the upper gastrointestinal tract, and can be fermented by beneficial intestinal microbes Fecal microbiota transplantation (FMT) (77). Among many prebiotics, non-digestible oligosaccharides, such as inulin and its hydrolysis products oligofructose, and (trans) galacto-oligosaccharides fulfill all those criteria (65, 66). Prebiotics prevent pathogen growth by increasing intestinal organic acids thereby reducing luminal pH levels and establishing a stable population of commensal bacteria that compete with invading pathogens for nutrients (78–80). Efficacy of prebiotic therapy, however, is dependent on beneficial microbes already residing within the host, a condition that may not be met in dysbiosis. Therefore, more direct microbiome-based therapeutic modalities may be needed.
3.2 Fecal microbiota transplantation
FMT, also known as bacteriotherapy, is a procedure to obtain feces from a healthy individual and introduce the fecal slurry into a patient’s intestinal tract. FMT can restore the recipient’s gastrointestinal bacterial diversity and bacterial-derived metabolites termed the metabolome (81, 82). FMT is considered the crudest form of live microbiome based therapies since the transplant inoculum is often an undefined mixture of microbes. The first known description of the therapeutic use of feces was by Ge Hong for the treatment of human diseases, including diarrhea, in the fourth century in China (83). However, it was not until the mid-twentieth century that feces were used to treat a specific disease in a controlled way. In 1958, Eisenmen and colleagues were the first to treat pseudomembranous colitis with FMT (84). More recently, FMT to treat recurrent C. difficile infection is the most widely used application of this microbiome-based therapeutic. In controlled, double-blinded studies, FMT has been proven to be ~89% efficacious in preventing recurrent C. difficile infection (85). During C. difficile infection there is a decrease in SCFAs and secondary bile acids in patients’ colons (60, 86). Following FMT, the clinical improvement in patients is directly associated with engraftment of donor microbes, restoration of SCFAs and secondary bile acids, and resolution of inflammation in the colon (82, 87, 88). Bacteria to bacteria interactions and restoration of commensal bacteria colonization resistance are thought to be the primary FMT mechanism of action for resolution of C. difficile infection. In agreement with this prevailing theory, SCFAs and secondary bile acid metabolites can directly inhibit C. difficile growth (89, 90). However, as described in the following sections, FMT may also work indirectly via signaling with the host immune system.
FMT is also being studied for the treatment of non-infectious intestinal diseases like IBD (91) as well as extraintestinal diseases such as immune checkpoint inhibitor (ICI) cancer immunotherapies (92), metabolic, and neuropsychiatric disorders (93). Clinical trials using FMT to treat these diseases have more varied success compared to the use of FMT to treat recurrent C. difficile infection. In a systematic meta-analysis assessing the efficacy of FMT for the treatment of ulcerative colitis (UC), Narula et al. found that 28% of patients progressed to clinical remission of active UC following FMT. This was a marked improvement over the 9% remission in the placebo groups but still demonstrated that the majority of FMT recipients did not experience a therapeutic benefit from the FMT (94). Both animal and human trials with FMT have demonstrated the feasibility but also potential dangers of using an undefined bacterial consortium as a therapy (95). This concern has led to slow adoption of crude FMT as a standard practice and search for more refined forms of FMT. A clean mixture of purely isolated gut bacteria collected from a healthy donor termed ‘defined gut microbial ecosystem components’- effectively cleared C. difficile from two patients who failed to clear the pathogen with several courses of traditional antibiotics therapy (96, 97). This defined FMT approach may be a safer, more controlled, and more acceptable method. However, future large-scale randomized controlled trials are needed to determine the efficacy of this defined FMT approach. Next generation microbiome-based therapies that are described below seek to further refine and remove donor variability inherent with FMT.
3.3 Probiotics
Probiotics are live microorganisms, mainly Lactobacillus, Bifidobacterium and Saccharomyces, that do not naturally reside in the mammalian host but provide health benefits when administered (98). The discovery that fermented food such as yogurt and fermented milk contain mixtures of bacteria with health benefits led to the development of modern probiotics. These bacteria can be isolated, grown, and administered in controlled doses. Probiotics can provide health benefits through several mechanisms including producing antimicrobial peptides, enhancing mucus production, and regulating mucosal immune functions (65). Several studies show an anti-inflammatory role of probiotics in alleviating colitis in mouse model (69–71). Although promising in animal studies, the therapeutic effect of probiotics in treating inflammatory diseases in patients is not well established. Lactobacillus and Bifidobacterium probiotic treatment failed to reduce TNF-α or increase IL-10 expression in IBD patients (99). However, several clinical studies demonstrate that combinations of probiotics provide some benefit in UC patients (100). A systematic review with a meta-analysis by Derwa et al. that included 22 randomized controlled trials that administered probiotics to IBD patients demonstrated no therapeutic benefit of probiotics at inducing remission of active UC compared to the placebo group (101). In the same meta-analysis, the probiotics effect was similar to 5-aminosalicylic acid (5-ASA), a commonly used anti-inflammatory drug, in preventing UC relapse. In addition, no benefit of probiotics was observed in inducing remission of active Crohn’s disease (CD) as well as in preventing relapse of quiescent CD. In C. difficile infection, administration of the probiotic, Saccharomyces boulardii showed promise in preventing recurrence. McFarland and colleagues (102) found that oral administration of S. boulardii for four weeks with standard antibiotics therapy significantly reduced recurrent C. difficile-associated disease (34.6%), compared to the placebo group (64.7%). In another study (103), S. boulardii was effective in decreasing recurrences (16.7%) only when administered with a high-dose of vancomycin (2 grams/day), compared to placebo with a high-dose-vancomycin group (50%). In both studies, S. boulardii was effective with concurrent antibiotic therapy. The limitation of probiotic therapy could be due to the inability of the probiotic bacteria to colonize and take up residence in the dysbiotic intestinal tract and stably restore microbiome diversity.
3.4 Live bio-therapeutics
Distinct from probiotics, live bio-therapeutics are a single bacterial species or a selected combination of bacteria, that can colonize the intestine and are designed to provide a clinical benefit for a particular disease (104). Live bio-therapeutics are often identified and isolated from fecal bacterial populations. They are considered “next-generation” microbiome therapy products to treat diseases like C. difficile and IBD with more refinement than a heterogeneous fecal transplant mixture. A series of encouraging clinical trials have demonstrated the potential of this more refined microbiome-based therapy approach. A consortium of purified Firmicutes spores developed by Seres Therapeutics was effective in a phase 3 clinical trial to reduce the risk of C. difficile recurrence compared to standard of care antibiotics (105). Further, a defined collection of 8 bacteria, named VE303, developed by Vedanta Biosciences also shows promise in phase 2 clinical trial for the treatment of recurrent C. difficile infection (88). Similarly, SER-287, an oral formulation of Firmicutes spores, showed promising results in a phase 1 clinical trial for the remission of mild to moderate UC (106). Due to the complexity of microbial interactions with the host, identifying a single or small combination of bacterial species that convey comparable therapeutic effects to FMT is challenging. However, the studies described above demonstrate that live bio-therapeutics in development have the capacity to treat a broad range of dysbiosis associated diseases and could be used as a safer alternative to FMT.
3.5 Postbiotics
Postbiotics are soluble components of microbial cells or their derived metabolites that can provide therapeutic benefits (107). SCFAs and secondary bile acids are two examples of well-studied microbial-derived metabolites being actively tested for therapeutic effects. SCFAs consist of a class of molecules produced during bacterial fermentation of complex fibers in the colon (108). Administration of SCFAs reduces colitis in mice (45, 109), however, human IBD trials show inconsistent results (110). Further, secondary bile acids show anti-inflammatory effects on immune cells and alleviate intestinal inflammation in a mouse model of colitis (111). A clinical trial on the therapeutic effect of secondary bile acid, ursodeoxycholic acid in IBD is ongoing (NCT03724175). Notably, high physiologic concentrations of secondary bile acids drive colorectal tumorigenesis through oxidative stress and DNA damage in the epithelium (112, 113). The goal of postbiotics therapy is to identify and isolate downstream effector metabolites produced by the beneficial bacteria of the microbiome and administer these molecules to treat disease without the detrimental side effects. Postbiotic therapeutics offer the potential for chemical modifications to reduce off-target interactions and enhance on-target therapeutic effects.
Section 4: The role of the immune system in shaping the outcome of microbiome-based therapy
The immune status of the recipient prior to microbiome transplantation shapes the engraftment, function and ultimately efficacy of any microbiome therapy to treat disease (Figure 1). The microbiome transplant recipient’s immune status can be impacted by infection, genetics or pre-existing dysbiosis and in turn shape the intestinal microenvironment into a hostile or receptive setting for the microbial consortium attempting to colonize the host. Work from our group has begun to identify the immune components prior to microbiome therapy that determine whether the microbiome transplant is successful. In a mouse model of FMT-mediated resolution of C. difficile infection, FMT recipient Rag1-/- mice, which lack B and T cells, failed to engraft the transplanted bacteria, did not restore cecal secondary bile acid levels, and were not able to resolve C. difficile infection compared to immunocompetent littermate control mice (114). CD4+ T cells, specifically Treg cells, were the critical adaptive immune cell type that supported FMT success as depletion of Treg cells rendered C. difficile infected mice non-responsive to FMT therapy. Notably, uninfected Rag1-/- mice readily engraft the FMT bacterial consortium indicating that immune deficiency in combination with infection-driven inflammation is both needed to inhibit FMT engraftment. The intestinal Treg cell compartment is comprised of heterogenous subsets of thymic-derived and peripherally-induced Treg cells (115–118). These Treg cell subsets promote tissue homeostasis mediated through a diverse repertoire of anti-inflammatory mechanisms, such as secretion of IL-10 and TGF-β, sequestration of IL-2, and expression of co-inhibitory molecules CTLA-4, ICOS and PD-1. In the absence of Treg cell-mediated regulation, the intestinal environment can shift towards an inflammatory setting that results in elevated expression of pro-inflammatory mediators that provide a growth advantage to inflammation-tolerant bacterial species, and may skew microbial engraftment to favor these bacterial species over donor-derived, inflammation-sensitive bacteria (54, 119, 120). While it is known that inflammation can favor selective growth of some bacteria (54) further study is needed to identify which specific molecular pathways can be targeted to enable bacterial species from a microbiome transplant to engraft.
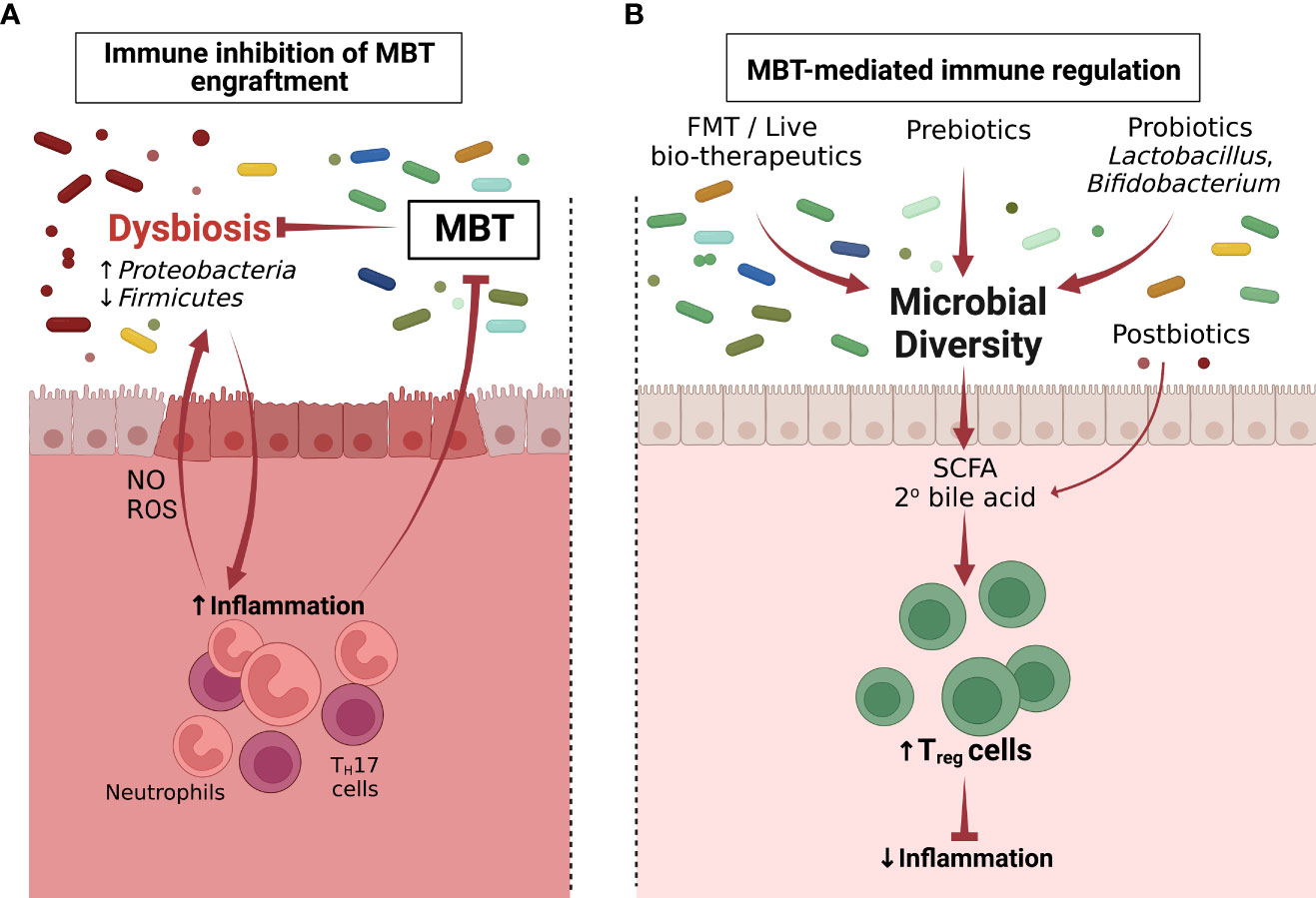
Figure 1 Microbiome-immune cell interactions determine the failure and success of microbiome-based therapies (MBTs). (A) Inflammation associated with dysbiosis renders the intestinal environment refractory to MBT engraftment (Section 4). (B) MBT can promote therapeutic effects through immune regulation following administration (Section 5). Following successful MBT engraftment, the overall microbiome diversity increases. Postbiotics or metabolites produced by incoming bacteria including SCFAs and 2° bile acids can induce CD4+ Treg cells, which reduce inflammation and restore the immune homeostasis of the colon.
Clinical studies assessing the host inflammatory status prior to FMT further highlight the importance of immune status in transplant engraftment and thereby efficacy of such therapies. For example, recurrent C. difficile patients that have higher intestinal inflammation prior FMT, as measured by fecal calprotectin level, are more likely to require repeated FMT administration to resolve infection (121). Further, patients with recurrent C. difficile infection that have underlying IBD experience reduced beneficial effects following FMT therapy (122). Microbiome-based treatment of recurrent C. difficile infection in patients with underlying IBD is a difficult clinical challenge since both diseases have the potential to amplify mucosal barrier breakdown and dysregulate mucosal immunity. In a large retrospective study that used clinical metadata and colonoscopy to diagnosis patients’ IBD status, 74.4% of patients with concomitant IBD resolved C. difficile at 2 months following a single infusion of FMT compared to 92.1% of patients who did not have IBD (122). More evidence that pre-existing immune-driven inflammation impairs FMT efficacy comes from a study that found that active IBD patients on high dose drug regimens to control IBD flares, indicating more aggressive inflammation in this patient subset, had reduced success when administered FMT to treat recurrent C. difficile infection (123). In addition, IBD patients are more likely to experience intestinal flares following FMT, although the precipitating factors for these flares could be the progression of the underlying IBD condition, the recurrence of C. difficile or the FMT itself (124). Data on whether a microbiome transplant recipient’s intestinal inflammatory status is in an active flare phase or in maintenance phase may be crucial in understanding microbiome transplant failure or success. Currently, there are very limited clinical studies considering this factor when reporting trial results. In a small study on patients with active UC, individuals taking immunosuppressants had a stronger response rate following FMT (5 out of 11 patients) compared to UC patients not on immunosuppressants (4 out of 27 patients) (125). In the same study FMT was found to be more effective in patients (75% efficacy) who were recently diagnosed with UC compared to those with chronic disease (18% efficacy) suggesting persistent chronic inflammation may shape the intestinal microenvironment to be refractory to FMT (125).
Section 5: Microbiome-based therapies acting on the immune system
Microbiome-based therapies are designed to reshape the composition of resident microbial communities and restore health. One mechanism through which these therapies elicit their beneficial properties is via stimulation of the immune system. Broadly, the microbiome transplant can promote immune tolerance mechanisms to decrease inflammation (Figure 1) or activate the immune system to enhance effector activity (Figure 2).
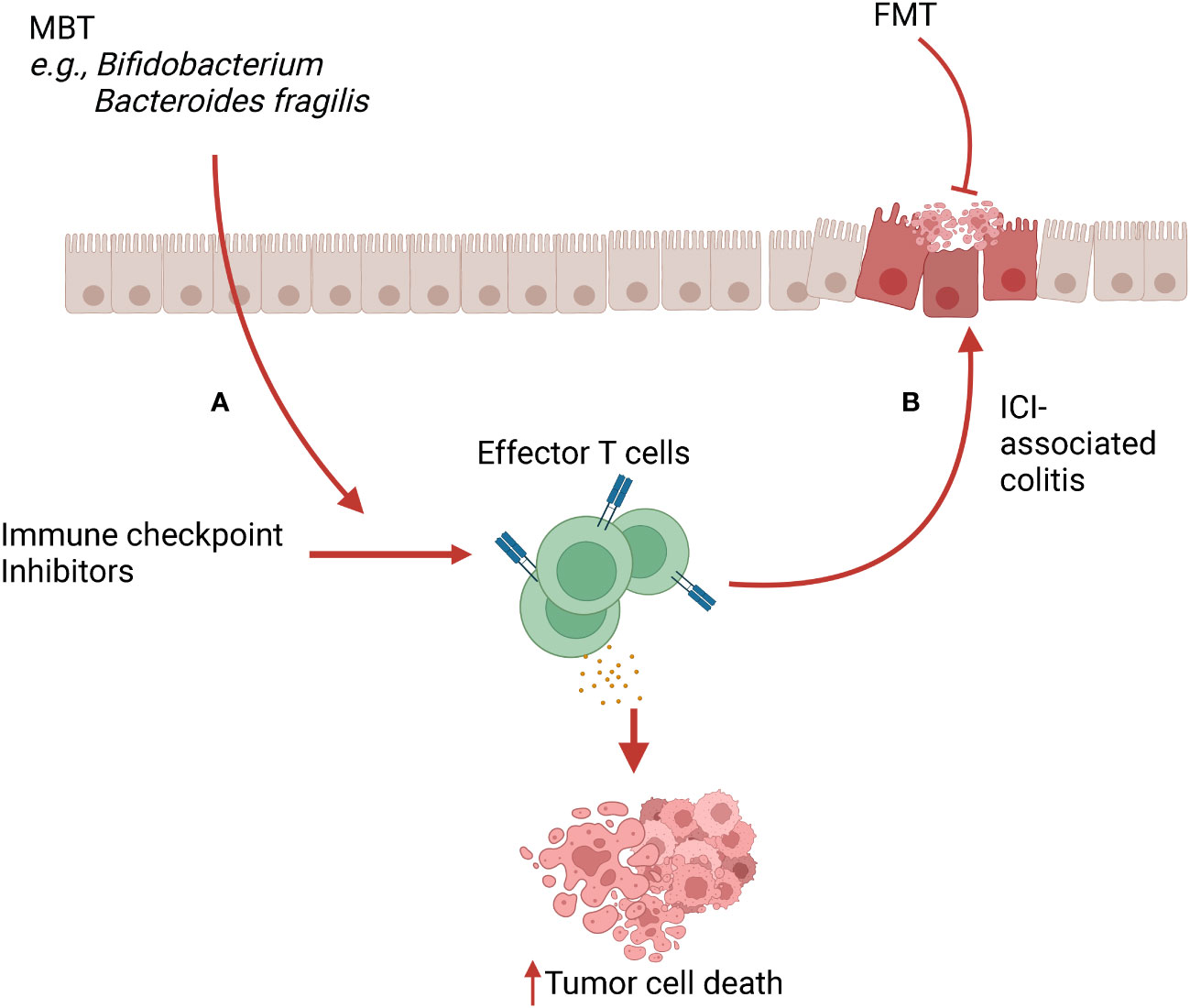
Figure 2 Microbiome-based therapy (MBT) in immune checkpoint inhibitor (ICI) therapy and ICI-associated colitis. (A) MBTs such as Bifidobacterium spp and Bacteroides fragilis cooperate with ICI, leading to increased infiltration and activation of effector T cells in the tumor microenvironment (TME). This mechanism boosts the anti-tumor activity of ICI therapy. (B) Activated T cells and their released cytokines induced by ICI therapy can cause off-target inflammation, leading to ICI-associated colitis. FMT can alleviate this ICI-associated colitis via inducing immunoregulatory properties.
5.1 Microbiome-based therapies promoting immunoregulation in intestinal disorders
5.1.1 IBD
Animal and clinical studies investigating microbiome-based therapies for IBD have focused on assessing whether the recipient's intestinal microbial communities and their derived metabolites are restored following treatment. Studies focused on the impact of microbiome-based therapies on the host immune response are less prevalent. In the dextran sodium sulfate (DSS)-induced colitis mouse model, FMT reduces colonic inflammation and improves mucosal immune health by inducing IL-10 expressing CD4+ T cells and invariant NK T cells in the colon of mice (126). Additionally, FMT reduces macrophage and neutrophil numbers in the colon and dampens antigen presentation capabilities to T cells, which is critical to controlling exaggerated inflammation (126). In agreement, a study by Wei et al. found that in DSS-induced colitis, FMT upregulates the aryl hydrocarbon receptor, IL-10, and transforming growth factor beta (TGF-β) expression in the mouse colon, providing favorable therapeutic effect in limiting DSS-induced colitis (127). Interestingly, contrary to the above studies, Il10-/- mice that develop spontaneous microbiome driven colitis (128) exhibit increased pro-inflammatory cytokines in the colon and exacerbated colitis following ileocolic resection and subsequent FMT administration (129), suggesting the FMT loses it therapeutic effect in the absence of IL-10. These studies suggest the prominent role of IL-10 signaling in the therapeutic effects of FMT against colitis in mice. Identifying specific immune pathways activated by the FMT will be important in determining the therapeutic outcome of the FMT.
Like FMT, live biotherapeutic bacteria have protective properties in mouse colitis models by modulating the immune response in the colon. For example, Round et al. showed that colonization of germ-free mice with B. fragilis, a prominent commensal bacterium, induced Treg cell function and anti-inflammatory cytokine release from the Treg cell population (72). This immunomodulatory effect of B. fragilis was driven by PSA, inducing differentiation of CD4+ T cells into FoxP3+ Treg cells that produce IL-10. B. fragilis-derived PSA administered as a postbiotic therapy alone had an immunomodulatory effect and prevented and ameliorated TNBS-induced colitis in mice. Oral administration of a defined consortia of 17 Clostridia commensal bacteria also alleviated colitis in mice through Treg-mediated immunomodulatory effects (38). Mechanistically, these 17 Clostridia strains produced SCFAs that increased TGF-β concentration in the mouse colon leading to elevated colonic Treg cells that expressed ICOS and produced IL-10. A follow up study further demonstrated the ability of these 17 Clostridia strains to produce SCFAs (73). Another defined consortium of bacteria reported by Zhu et al. also found an IL-10 associated beneficial effect. This probiotic cocktail with an equal mixture of 3 Bifidobacterium and 7 Lactobacillus sub-strains significantly reduced signs and symptoms of DSS-induced colitis and was associated with an increase of IL-10 and decrease of proinflammatory cytokines in the serum (74). The improvement in colitis was also associated with an increased expression of epithelial tight junction proteins, indicating restoration of compromised barrier functions. These observations suggest an IL-10-dependent pathway as a prominent mechanism through which microbiome transplants convey their therapeutic effect in limiting inflammation.
Commensal bacteria-derived metabolites, such as SCFAs, have also been studied for therapeutic potential in murine colitis models. In a TNBS-induced colitis model, Chen et al. found that butyrate administered through drinking water protected mice from severe colitis by suppressing proinflammatory responses in the colon and improving gut barrier integrity (130). This protective effect of butyrate was GPR109a receptor-mediated as GPR109a deficient mice completely lost butyrate-mediated protection. Several other animal studies also demonstrate protective role of SCFA against experimental colitis through boosting Treg cells and downregulating pro-inflammatory cytokines in the colon (131, 132). Similarly, secondary bile acids demonstrate anti-inflammatory effects on immune cells and alleviate intestinal inflammation in a mouse model of colitis (111). Secondary bile acids derivatives, such as 3-oxoLCA and isoalloLCA can promote the differentiation of Treg cells and inhibit TH17 cells (36, 37, 39).
Numerous clinical microbiome transplant studies report changes in transplant recipient’s immune parameters following transplantation in IBD. In a pilot study on active CD by Vaughn et al., a single colonic FMT infusion lead to an increase of Treg cells in the colon at 12 weeks. However, the Treg cell population increased in both FMT recipient patients who responded and failed to respond to the FMT (67). Another prospective, open-label study with 20 active UC patients showed a decrease in Treg and TH1 cells in the colon at four weeks following a single dose of colonoscopy-delivered FMT, with no differences in TH2 and TH17 cells (68). Patients treated with Lactobacillaceae-enriched probiotic, named VSL#3, showed therapeutic benefit against active UC, compared to the placebo group. VSL#3 had an immunomodulatory effect on colonic dendritic cells (DCs). Specifically, immune profiling from rectal biopsies obtained from patients demonstrated a decrease in IL-12 and an increase in IL-10 production from the colonic DCs. Probiotics can modulate DC populations and lower the proinflammatory response in the colon, contributing to clinical improvement of UC patients (133). Lactobacillus casei administered rectally, but not orally, with 5-ASA therapy significantly improved clinical outcomes in UC patients, compared to 5-ASA-treatment alone (134). The clinical improvement was associated with immune changes in the colon. Biopsies from the sigmoid region of the large intestine showed decreased expression of IL-1β and TLR-4 and increased levels of IL-10 in the colon of patients who benefitted from L. casei probiotic. SCFAs have also been tested as a postbiotic therapy to treat IBD in patients. Luhrs and colleagues report that UC patients treated with butyrate have a significant clinical improvement compared to a non-butyrate treated group (135). Histologically, butyrate treated patients had less mucosal inflammation, reduced infiltration of neutrophils and lymphocytes in the lamina propria of the colon, associated with decreased activation of NF-κB signaling in macrophages. Other clinical studies also demonstrate immune mediated beneficial effect of SCFA in UC patients through decreasing the infiltration of neutrophils (75) and suppressing mucosal inflammation in the colon (76).
5.1.2 ICI-colitis
Immune checkpoint inhibitors (ICI) work by removing the intrinsic breaks of the immune system and thereby boosting the effector functions of T cells effector function (136). One serious adverse effect of ICI use is colitis caused by an overactivated immune response, which resembles IBD and can be life-threatening. Recent studies suggest that the development of ICI-associated colitis is correlated with gut microbiome dysbiosis and restoration of the bacterial population can mitigate the ICI-associated colitis in animals and patients (137) (Figure 2).
In a mouse study, anti-CTLA-4 treatment was combined with DSS administration to model severe ICI-associated colitis. Oral administration of Bifidobacterium species rescued mice from colitis in a Treg cell dependent manner without an apparent impact on the antitumor activity of anti-CTLA-4 immunotherapy (138). In a follow up study, this group found that Bifidobacterium enhanced mitochondrial fitness and IL-10 activity of intestinal Treg cells to limit ICI-induced colitis (139). Similarly, Lactobacillus reuteri administration inhibits the progression of ICI-associated colitis in mice administered DSS in their drinking water and treated with anti-CTLA-4 and anti-PD-1 immunotherapy (140). The protective effect of L. reuteri in preventing colitis was associated with decreased infiltration of leukocytes in the intestinal tissue and decreased levels of proinflammatory cytokines such as TNF-α, IFN-γ, and IL-6 in serum, compared to non L. reuteri-administered mice. Additional studies have identified specific bacterial-derived metabolites with therapeutic effects against ICI-associated colitis. Mice orally administered with indole-3-carboxaldehyde (3-IAld), a microbial-derived tryptophan catabolite, had decreased disease severity, weight loss, and colonic damage compared to mice non-treated with 3-IAld (141). This protective effect of 3-IAld was AhR-signaling mediated and dependent on IL-22 cytokine production, which protected mice by inducing Reg3γ expression in the colon.
In humans, a case report of two patients with ICI-associated colitis that were refractory to steroid treatment, found FMT successfully treated the colitis (142). In one patient, the clinical improvement was associated with a substantial reduction in colonic CD8+ T cells with a concomitant increase in Treg cells. The second patient had a decrease of all the T cell subtypes following FMT; however, the CD4+ T cell population was relatively unchanged compared to the CD8+ T cells, which was associated with the persistence of Treg cells in the colonic mucosa. In another recent case study (143), a patient suffering from palatal malignant melanoma developed severe ICI-associated colitis after the third dose of anti-PD-1 treatment. After unsuccessful treatment with corticosteroids, an FMT was administered to successfully resolve colitis symptoms. The patient’s clinical outcome was associated with decreased fecal calprotectin and restoration of the congested, inflamed colonic mucosa to near normal level (143). These mucosal immune changes reported in the case reports suggest that FMT is signaling to reshape intestinal immune cell populations resulting in therapeutic effect against ICI-associated colitis. It is unknown, however, whether FMT-mediated immunoregulation during ICI treatment counteracts the anti-tumor activity of the checkpoint blockade therapy and changes cancer progression. For example, Treg cells contribute to tumor growth by suppressing effector T cell functions (144). In addition, increased levels of Treg cells in the tumor microenvironment are positively correlated with poor prognosis in various cancers (145, 146). As mentioned above (142), increased level of Treg cells in the colon could be one of the potential mechanisms of FMT-mediated amelioration of ICI-associated colitis. Therefore, it will be important for future studies to determine whether there is any detrimental effect of FMT-driven Treg cells on the capacity of ICI to limit cancer progression. Studies investigating the efficacy of microbiome-based therapies to limit ICI-associated colitis have involved a small number of patients with various types of cancers. Large-scale randomized trials are needed. A clinical trial is currently underway on the use of FMT for treating ICI-associated diarrhea or colitis in patients with genitourinary tract cancer (NCT04038619).
5.2 Microbiome-based therapies driving immune activation
Data from animal studies and clinical trials also indicate that the microbiome can enhance the antitumor responses of ICI (Figure 2). In mice, anti-CTLA-4 antibody treatment controls melanoma and colon cancer progression in specific pathogen-free (SPF) mice but not in germ-free mice. Additionally, the anti-CTLA-4 effect was compromised in SPF mice treated with broad-spectrum antibiotics supporting an active role of the bacteria (24). Further animal studies demonstrated the therapeutic role of microbiome transplants in boosting the antitumor effect of ICIs. Routy et al. found that germ-free or antibiotic-treated mice receiving FMT from cancer patients who responded to ICIs, had improved antitumor activity following anti-PD-1 treatment compared to mice receiving FMT from ICI-non-responder (147). This study also showed a correlation between the clinical benefit of ICIs with the relative abundance of Akkermansia muciniphila. Oral supplementation with A. muciniphila restored the efficacy of PD-1 blockade in non-responsive mice by increasing the recruitment of CCR9+ CXCR3+ CD4+ T cells into mouse tumors in an IL-12-dependent manner. This study demonstrates a clear immune-mediated role of a specific bio-therapeutic bacteria species in improving the efficacy of ICIs during cancer therapy. Tanoue et al. showed that a consortium of 11 bacterial strains obtained from healthy human donors can boost the efficacy of anti-PD-1 immunotherapy in suppressing adenocarcinoma and melanoma growth in mice by increasing the infiltration of IFN-γ+ CD8+ T cells in the tumor microenvironment (148). Interestingly, these 11 strains of bacteria are normally present in low abundance in the intestinal microbiome but could have strong potential for ICI-mediated cancer therapy if enabled to expand in the intestinal tract. Another study by Sivan et al. demonstrated that oral administration of Bifidobacterium enhanced the capacity of anti-PD-L1 treatment to control melanoma growth (23). This effect was mediated through activation of antigen presenting cells that enhanced CD8+ T cell infiltration into the tumor microenvironment. Similarly, in another mouse study, B. fragilis was shown to boost anti-CTLA-4 effect against tumors in mice (24). The anti-CTLA-4 anti-tumor response in germ-free or antibiotic-treated mice was rescued by oral administration of B. fragilis, immunization with B. fragilis polysaccharide, or by adoptive transfer of memory B. fragilis-specific TH1 cells. This immunostimulatory effect was mediated by an IL-12 dependent TH1 cell response that controlled the tumor progression. In another study of colorectal cancer and melanoma in mice, oral administration of live Lactobacillus rhamnosus augmented the antitumor activity of anti-PD-1 immunotherapy by triggering IFN-β production in dendritic cells with a concomitant increase of CD8+ T cells in the tumor microenvironment (149). These animal studies demonstrate the promise of microbiome-based therapy in supporting immune checkpoint inhibitor therapy’s antitumor effects.
Combining microbiome-based therapies with immune checkpoint inhibitors to enhance anti-cancer effects is actively being tested in the clinic with several promising results. Baruch et al. investigated the role of FMT in ICI-refractory melanoma. In this Phase I clinical trial, FMT was effective in three out of ten patients at improving the efficacy of anti-PD-1 therapy to treat metastatic melanoma. In responders, treatment with FMT was associated with a favorable immune change in tumor microenvironment, including upregulation of the IFN-γ-mediated signaling pathway, MHC-II protein expression, dendritic cell differentiation, TH1 cell response, and an increased infiltration of CD8+ T cells (150). Another study by Davar and colleagues also showed some promise using FMT for patients with refractory melanoma resistant to ICI (92). In this study fecal samples were obtained from melanoma patients who responded to ICI therapy previously and transferred to non-responsive patients. Following FMT, six out of fifteen patients clinically benefitted from FMT therapy in conjunction to anti-PD-1 treatment. Patients who responded had increased microbiome diversity, increased activation of CD8+ T cells, and decreased IL-8 expressing myeloid cells in tumor microenvironment (92). Beyond the two studies mentioned, several other ongoing clinical trials are investigating FMT in improving ICI therapy against various tumors, including gastrointestinal cancer (NCT04130763), renal cell carcinoma (NCT04758507) and prostate cancer (NCT04116775).
Live biotherapeutics also show promise to improve the efficacy of ICIs in human cancers. Patients with metastatic renal cell carcinoma, administered CBM588 (contained Clostridium butyricum) and both anti-PD-1 and anti-CTLA-4 immunotherapies had improved clinical outcome (151). The clinical improvement following CBM588 therapy was associated with an anti-tumorigenic immune change in patients. Higher level of pro-inflammatory cytokines, such as GM-CSF, IL-8, TNF-α, IFN-γ, MCP-1, IL-1RA and decreased level of Treg cells were observed in peripheral blood of CBM588 plus anti-PD-1 and anti-CTLA4 group, compared to only anti-PD-1 and anti-CTLA-4 treated group. A probiotic, called EDP1503 containing Bifidobacterium spp, developed by Evelo Biosciences is now in Phase I/II trial for the treatment of colorectal cancer and breast cancer with ICIs (NCT03775850). Microbiome-derived SCFAs are also found to be associated with favorable clinical outcome to ICI therapy against human cancer. A study by Nomura et al. found that patients having high SCFA levels in blood and feces were associated with improved efficacy of anti-PD1 therapy against solid tumors (152), while in another study, patients with non-small cell lung carcinoma treated with anti-PD-1 immunotherapy showed a positive correlation between anti-PD-1 response and high levels of fecal propionate and butyrate (153). These studies do not show a direct causative link between administering SCFAs to patients and improved efficacy of ICI nor provide a mechanism of how SCFAs elicit clinical improvement in patients. Future large-scale animal and clinical studies will be needed to determine the role of postbiotics in this aspect.
5.3 Microbiome-based therapies tuning immune defenses against infection
5.3.1 C. difficile
As detailed previously, the most successful implementation of FMT is in resolving C. difficile infection. Most animal studies investigating FMT’s mechanism of action have focused on bacteria-bacteria interactions between the FMT and C. difficile rather than the microbiome transplant driving host-immune changes that then lead to resolution of infection. For instance, secondary bile acid producing commensal bacteria like Clostridium scindens directly inhibit the growth of C. difficile through production of these secondary bile acids (90). However, human FMT trials have reported considerable immune changes following transplant. A pilot study found that following successful FMT for recurrent C. difficile infection, there was an upregulation of bile acid driven FXR-FGF signaling in the ileum, which resulted in an increase of fibroblast growth factor (FGF)-19 and decrease in FGF-21 in serum (154). The upregulation of this pathway was associated with the restoration of intestinal microbiome and secondary bile acid profile in the patient’s colon. Both secondary bile acids and FXR signaling have been linked to restoring immune tolerance; secondary bile acids via promotion of Treg cell differentiation and FXR signaling by improving intestinal barrier integrity (36, 37, 39, 155). Therefore, restoration of secondary bile acids following FMT (86), in addition to directly inhibiting C. difficile growth (90, 156), may signal through the host to indirectly support resolution of C. difficile infection. Similarly, another study by the same group showed recurrent C. difficile infection patients have increased specific micro RNAs (miRNA) following FMT compared to the screening control group (157). miRNAs are short noncoding RNA molecules that can bind to complementary sequences in the 3’ UTR (3 prime untranslated region) of messenger RNAs (mRNAs), resulting in translational suppression of target genes (158). The increase of two miRNAs observed in the patient population following FMT, miR-23a and miR-150, was confirmed using a mouse model of FMT and recurrent C. difficile infection (157). Through target scan prediction analysis and over expression of miRNA in epithelial cells, this report found that miR-150 and miR-23a target Il18 and Il12b respectively, two cytokines that promote a type-1 proinflammatory immune response. Thus, FMT driven increases in miRNA that could work to limit deleterious inflammation during C. difficile infection and enable inflammation-sensitive microbes from the FMT to recolonize the intestine. Immunoregulatory changes following FMT were also reported by Marie and colleagues who observed increased IL-25 levels in the colon following FMT (61). IL-25 drives type 2 immunity, which is protective following acute C. difficile infection in mice by skewing the host response away from a pathogenic proinflammatory response (159, 160). In the same study, the FMT suppressed the pro-inflammatory immune response and increased expression of a family of homeobox and laminin genes that promote epithelial development and homeostatic function of the colon. Further, TH17 cells in the peripheral blood were decreased following FMT (61). While the total TH17 cell populations decreased, another study reported that C. difficile toxin B-specific TH17 cells as well as toxin-A and toxin-B-specific IgG and IgA antibodies increased in the blood following successful FMT (161). The authors proposed that the enhanced C. difficile toxin-specific adaptive immune response could be a mechanism through which FMT works. Combined, the reported immune changes following FMT support the concept that the FMT, in addition to direct bacteria-to-bacteria interactions, could boost host immunity to clear C. difficile infection.
5.3.2 Multidrug-resistant organisms
The success of FMT in treating recurrent C. difficile infection has led to the application of microbiome-based therapies to treat other bacterial infections. Multidrug-resistant organisms (MDROs) in the intestinal tract, such as vancomycin-resistant enterococci (VRE), carbapenem-resistant enterococci, methicillin-resistant Staphylococcus aureus (MRSA) and extended-spectrum β-lactamase producing bacteria, are resistant to more than three classes of antibiotics (162, 163). Therefore alternative treatment options are needed to combat infection originating from these organisms. In general, FMT decreases the abundance of antibiotic resistance genes in the intestinal microbiome reservoir (164). Several clinical studies have described the use of FMT for the intestinal decolonization of MDROs (165, 166). In a meta-analysis of 23 studies that included 142 patients undergoing FMT to decolonize MDROs in the intestines around 77.5% of patients cleared MDROs as determined by microbiological assessment of fecal samples and rectal swabs (167). Probiotics are also being used to prevent and treat infections caused by MDROs. For example, oral administration of two Lactobacillus strains (Y74 and HT121) reduced the colonization of VRE in mice colons (168). In another clinical study MRSA colonization of the gut was reduced by oral administration of Lactobacillus rhamnosus (169). In these examples microbiome-based therapies are targeting MDROs via direct bacteria to bacteria interactions that are immune independent. The mechanisms of action for the transplanted bacterial inoculum include competition for resources, production of bacteriocins, or metabolic byproducts that are toxic to the pathogenic bacteria. For example, administering an FMT or a defined bacterial consortium containing Blautia producta and Clostridium bolteae cleared persistent VRE colonization in mice colons (170, 171). Kim et al. identified that a lantibiotic produced by a specific strain of B. producta was necessary for clearance of VRE (172). Of note, this group observed that transplantation with the B. producta and C. bolteae consortium resolved VRE colonization even in severely immunodeficient mice (Rag2-/- Il2rg-/-) further demonstrating the mechanism of action of this microbiome-based therapy was immune independent. This result contrasts with Littman et al. (114) that observed that success or failure FMT therapy was dependent on the host’s immune status. These examples illustrate that immune-independent benefits of microbiome-based therapy, which are not the focus of this review, is a rich field of study to itself (18). Understanding when microbiome-based therapies work through the immune system and when they are immune independent will be critical information as design of microbiome-based therapies become more refined and targeted.
5.3.3 Viral infection
Microbiome-based therapies also can exert trans-kingdom protection against enteric and systemic viral infections. Several probiotic studies show decreased severity or duration of viral infection following probiotic administration (173, 174). Some probiotic effects are via non-immune-mediated mechanisms such as bacteriocin production that also have viricidal properties (175, 176) or inhibition of viral attachment to host cells (177). The microbiome also induces activation of antiviral immune defenses to poise the host to rapidly respond to invading viruses (178–182). Several probiotic formulations have been linked to enhanced type I and II IFN signaling and natural killer cell activity to limit viral infection (183–185). This is further supported by mouse studies that isolated and transplanted live bio-therapeutic bacterial species that activated type I IFN-mediated antiviral defense pathways resulting in improved host survival against enteric or systemic viral infection (181, 182). Steed et al. reported that an FMT could protect microbiome-depleted mice from influenza virus infection and went on to identify that a specific postbiotic, desaminotyrosine, produced by Clostridum orbiscindens through flavonoid metabolism could be administered to enhance type I IFN pathways in the lung and protect against lethal influenza virus challenge (186). Microbiome-based therapies can also enhance adaptive antiviral immunity. Several probiotics can enhance secretory IgA antibody production to limit enteric viral entry (187–189). Meanwhile administration of the postbiotic PSA derived from B. fragilis limited fatal herpes simplex encephalitis. This therapeutic effect was driven by PSA tuning down the T cell response through increasing IL-10 production (190). Indeed, probiotics have even been proposed as part of a treatment regimen to correct the dysbiosis associated with SARS-CoV-2 infection (191, 192).
6 Concluding remarks
The promise of using microbiome-based therapies to treat disease has as much potential to improve human life as when the first heart and liver transplant surgeries were performed in the 1960-70’s. However, the same microbiome transplant inoculum often has a heterogenous clinical outcome when administered to multiple patients (193). Long term engraftment in the recipients can be determined by multiple factors such as diet, genetics, or the pre-existing microbiome (194–197). Therefore, in the same vein as fifty years ago when the rules of organ donor matching and immunosuppression medication were systematically teased apart through careful, basic scientific research, the mechanisms determining microbiome transplantation success need to be identified to harness the potential power of this nascent field.
Author contributions
ZA researched and wrote the manuscript. JM designed the figure and provided edits feedback. MA reviewed the manuscript and provided edits and feedback. All authors contributed to the article and approved the submitted version.
Funding
The authors of this manuscript were supported by NIH NIAID funding (R01 AI158830 to ZA, JM and MA; R21 AI164385, R01 AI130115 to MA).
Acknowledgments
Figures were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
FMT, fecal microbiota transplantation; Treg, regulatory T cells; IBD, Inflammatory Bowel Disease; SCFA, short chain fatty acid; FXR, farnesoid X receptor; ICI, Immune Checkpoint Inhibitor; DSS, dextran sodium sulfate; SPF, specific pathogen free; 5-ASA- 5-aminosalicylic acid; UC, ulcerative colitis; MBT, Microbiome-based therapy.
References
1. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486:207–14. doi: 10.1038/nature11234
2. Richard ML, Sokol H. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol (2019) 16:331–45. doi: 10.1038/s41575-019-0121-2
3. Ruan W, Engevik MA, Spinler JK, Versalovic J. Healthy human gastrointestinal microbiome: Composition and function after a decade of exploration. Dig Dis Sci (2020) 65:695–705. doi: 10.1007/s10620-020-06118-4
4. Houtman TA, Eckermann HA, Smidt H, de Weerth C. Gut microbiota and BMI throughout childhood: the role of firmicutes, bacteroidetes, and short-chain fatty acid producers. Sci Rep (2022) 12:3140. doi: 10.1038/s41598-022-07176-6
5. Stojanov S, Berlec A, Štrukelj B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms (2020) 8. doi: 10.3390/microorganisms8111715
6. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U.S.A. (2005) 102:11070–5. doi: 10.1073/pnas.0504978102
7. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17:219–32. doi: 10.1038/nri.2017.7
8. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of parkinson’s disease. Cell (2016) 167:1469–1480.e12. doi: 10.1016/j.cell.2016.11.018
9. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature (2011) 474:327–36. doi: 10.1038/nature10213
10. Hartstra AV, Bouter KEC, Bäckhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care (2015) 38:159–65. doi: 10.2337/dc14-0769
11. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33:570–80. doi: 10.1016/j.ccell.2018.03.015
12. Duan M, Wang Y, Zhang Q, Zou R, Guo M, Zheng H. Characteristics of gut microbiota in people with obesity. PloS One (2021) 16:e0255446. doi: 10.1371/journal.pone.0255446
13. Das T, Jayasudha R, Chakravarthy S, Prashanthi GS, Bhargava A, Tyagi M, et al. Alterations in the gut bacterial microbiome in people with type 2 diabetes mellitus and diabetic retinopathy. Sci Rep (2021) 11:2738. doi: 10.1038/s41598-021-82538-0
14. Chen C, Liao J, Xia Y, Liu X, Jones R, Haran J, et al. Gut microbiota regulate alzheimer’s disease pathologies and cognitive disorders via PUFA-associated neuroinflammation. Gut (2022) 71:2233–52. doi: 10.1136/gutjnl-2021-326269
15. Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol (2013) 13:321–35. doi: 10.1038/nri3430
16. Bashiardes S, Tuganbaev T, Federici S, Elinav E. The microbiome in anti-cancer therapy. Semin Immunol (2017) 32:74–81. doi: 10.1016/j.smim.2017.04.001
17. Fitzgibbon G, Mills KHG. The microbiota and immune-mediated diseases: Opportunities for therapeutic intervention. Eur J Immunol (2020) 50:326–37. doi: 10.1002/eji.201948322
18. Sorbara MT, Pamer EG. Microbiome-based therapeutics. Nat Rev Microbiol (2022) 20:365–80. doi: 10.1038/s41579-021-00667-9
19. Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell (2016) 165:1762–75. doi: 10.1016/j.cell.2016.06.001
20. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell (2013) 155:1451–63. doi: 10.1016/j.cell.2013.11.024
21. Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing clostridium difficile disease in mice. PloS Pathog (2012) 8:e1002995. doi: 10.1371/journal.ppat.1002995
22. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. doi: 10.1126/science.1241214
23. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350:1084–9. doi: 10.1126/science.aac4255
24. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350:1079–84. doi: 10.1126/science.aad1329
25. Mimee M, Citorik RJ, Lu TK. Microbiome therapeutics - advances and challenges. Adv Drug Delivery Rev (2016) 105:44–54. doi: 10.1016/j.addr.2016.04.032
26. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
27. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity (2014) 40:594–607. doi: 10.1016/j.immuni.2014.03.005
28. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell (2016) 164:324. doi: 10.1016/j.cell.2015.12.047
29. Woo V, Eshleman EM, Hashimoto-Hill S, Whitt J, Wu S-E, Engleman L, et al. Commensal segmented filamentous bacteria-derived retinoic acid primes host defense to intestinal infection. Cell Host Microbe (2021) 29:1744–1756.e5. doi: 10.1016/j.chom.2021.09.010
30. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell (2005) 122:107–18. doi: 10.1016/j.cell.2005.05.007
31. Troy EB, Kasper DL. Beneficial effects of bacteroides fragilis polysaccharides on the immune system. Front Biosci (Landmark Ed) (2010) 15:25–34. doi: 10.2741/3603
32. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science (2013) 341:569–73. doi: 10.1126/science.1241165
33. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity (2014) 40:128–39. doi: 10.1016/j.immuni.2013.12.007
34. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504:446–50. doi: 10.1038/nature12721
35. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature (2013) 504:451–5. doi: 10.1038/nature12726
36. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control T(H)17 and t(reg) cell differentiation. Nature (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
37. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
38. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature (2013) 500:232–6. doi: 10.1038/nature12331
39. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature (2020) 581(7809):475–9. doi: 10.1038/s41586-020-2193-0
40. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity (2017) 46:562–76. doi: 10.1016/j.immuni.2017.04.008
41. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
42. Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect (2012) 18 Suppl 4:2–4. doi: 10.1111/j.1469-0691.2012.03916.x
43. DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflammation Bowel Dis (2016) 22:1137–50. doi: 10.1097/MIB.0000000000000750
44. Clooney AG, Eckenberger J, Laserna-Mendieta E, Sexton KA, Bernstein MT, Vagianos K, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut (2021) 70:499–510. doi: 10.1136/gutjnl-2020-321106
45. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
46. Reinisch W, Gasché C, Tillinger W, Wyatt J, Lichtenberger C, Willheim M, et al. Clinical relevance of serum interleukin-6 in crohn’s disease: single point measurements, therapy monitoring, and prediction of clinical relapse. Am J Gastroenterol (1999) 94:2156–64. doi: 10.1111/j.1572-0241.1999.01288.x
47. Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, et al. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity (2019) 51:77–89.e6. doi: 10.1016/j.immuni.2019.05.004
48. Lee J-Y, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, et al. Serum amyloid a proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell (2020) 183:2036–9. doi: 10.1016/j.cell.2020.12.008
49. Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut (2003) 52:65–70. doi: 10.1136/gut.52.1.65
50. Rovedatti L, Kudo T, Biancheri P, Sarra M, Knowles CH, Rampton DS, et al. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut (2009) 58:1629–36. doi: 10.1136/gut.2009.182170
51. Britton GJ, Contijoch EJ, Mogno I, Vennaro OH, Llewellyn SR, Ng R, et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt(+) regulatory T cells and exacerbate colitis in mice. Immunity (2019) 50:212–224.e4. doi: 10.1016/j.immuni.2018.12.015
52. Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic enterobacteriaceae expansion. Science (2017) 357:570–5. doi: 10.1126/science.aam9949
53. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of e. coli in the inflamed gut. Science (2013) 339:708–11. doi: 10.1126/science.1232467
54. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for salmonella. Nature (2010) 467:426–9. doi: 10.1038/nature09415
55. Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PloS Biol (2007) 5:2177–89. doi: 10.1371/journal.pbio.0050244
56. Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe (2007) 2:204. doi: 10.1016/j.chom.2007.08.002
57. Chanin RB, Winter MG, Spiga L, Hughes ER, Zhu W, Taylor SJ, et al. Epithelial-derived reactive oxygen species enable AppBCX-mediated aerobic respiration of escherichia coli during intestinal inflammation. Cell Host Microbe (2020) 28:780–788.e5. doi: 10.1016/j.chom.2020.09.005
58. Scales BS, Dickson RP, Huffnagle GB. A tale of two sites: how inflammation can reshape the microbiomes of the gut and lungs. J Leukoc Biol (2016) 100:943–50. doi: 10.1189/jlb.3MR0316-106R
59. Wang S, El-Fahmawi A, Christian DA, Fang Q, Radaelli E, Chen L, et al. Infection-induced intestinal dysbiosis is mediated by macrophage activation and nitrate production. mBio (2019) 10. doi: 10.1128/mBio.00935-19
60. Li X, Gao X, Hu H, Xiao Y, Li D, Yu G, et al. Clinical efficacy and microbiome changes following fecal microbiota transplantation in children with recurrent clostridium difficile infection. Front Microbiol (2018) 9:2622. doi: 10.3389/fmicb.2018.02622
61. Jan N, Hays RA, Oakland DN, Kumar P, Ramakrishnan G, Behm BW, et al. Fecal microbiota transplantation increases colonic IL-25 and dampens tissue inflammation in patients with recurrent clostridioides difficile. mSphere (2021) 6:e0066921. doi: 10.1128/mSphere.00669-21
62. Meisel M, Mayassi T, Fehlner-Peach H, Koval JC, O’Brien SL, Hinterleitner R, et al. Interleukin-15 promotes intestinal dysbiosis with butyrate deficiency associated with increased susceptibility to colitis. ISME J (2017) 11:15–30. doi: 10.1038/ismej.2016.114
63. Kiyohara H, Sujino T, Teratani T, Miyamoto K, Arai MM, Nomura E, et al. Toll-like receptor 7 agonist-induced dermatitis causes severe dextran sulfate sodium colitis by altering the gut microbiome and immune cells. Cell Mol Gastroenterol Hepatol (2019) 7:135–56. doi: 10.1016/j.jcmgh.2018.09.010
64. Darnaud M, Dos Santos A, Gonzalez P, Augui S, Lacoste C, Desterke C, et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology (2018) 154:1009–1023.e14. doi: 10.1053/j.gastro.2017.11.003
65. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol (2019) 16:605–16. doi: 10.1038/s41575-019-0173-3
66. Arnold JW, Roach J, Fabela S, Moorfield E, Ding S, Blue E, et al. The pleiotropic effects of prebiotic galacto-oligosaccharides on the aging gut. Microbiome (2021) 9. doi: 10.1186/s40168-020-00980-0
67. Vaughn BP, Vatanen T, Allegretti JR, Bai A, Xavier RJ, Korzenik J, et al. Increased intestinal microbial diversity following fecal microbiota transplant for active crohn’s disease. Inflammation Bowel Dis (2016) 22:2182–90. doi: 10.1097/MIB.0000000000000893
68. Jacob V, Crawford C, Cohen-Mekelburg S, Viladomiu M, Putzel GG, Schneider Y, et al. Single delivery of high-diversity fecal microbiota preparation by colonoscopy is safe and effective in increasing microbial diversity in active ulcerative colitis. Inflammation Bowel Dis (2017) 23:903–11. doi: 10.1097/MIB.0000000000001132
69. Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. Differential intestinal anti-inflammatory effects of lactobacillus fermentum and lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol Nutr Food Res (2017) 61. doi: 10.1002/mnfr.201700144
70. Jang YJ, Kim W-K, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes (2019) 10:696–711. doi: 10.1080/19490976.2019.1589281
71. Zhou X, Liu H, Zhang J, Mu J, Zalan Z, Hegyi F, et al. Protective effect of lactobacillus fermentum CQPC04 on dextran sulfate sodium-induced colitis in mice is associated with modulation of the nuclear factor-κB signaling pathway. J Dairy Sci (2019) 102:9570–85. doi: 10.3168/jds.2019-16840
72. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U.S.A. (2010) 107:12204–9. doi: 10.1073/pnas.0909122107
73. Narushima S, Sugiura Y, Oshima K, Atarashi K, Hattori M, Suematsu M, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived clostridia. Gut Microbes (2014) 5:333–9. doi: 10.4161/gmic.28572
74. Zhu Y, Xu Y, Wang X, Rao L, Yan X, Gao R, et al. Probiotic cocktail alleviates intestinal inflammation through improving gut microbiota and metabolites in colitis mice. Front Cell Infect Microbiol (2022) 12:886061. doi: 10.3389/fcimb.2022.886061
75. Scheppach W, Müller JG, Boxberger F, Dusel G, Richter F, Bartram HP, et al. Histological changes in the colonic mucosa following irrigation with short-chain fatty acids. Eur J Gastroenterol Hepatol (1997) 9:163–8. doi: 10.1097/00042737-199702000-00010
76. Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology (1992) 103:51–6. doi: 10.1016/0016-5085(92)91094-k
77. Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics- a review. J Food Sci Technol (2015) 52:7577–87. doi: 10.1007/s13197-015-1921-1
78. Fooks LJ, Gibson GR. In vitro investigations of the effect of probiotics and prebiotics on selected human intestinal pathogens. FEMS Microbiol Ecol (2002) 39:67–75. doi: 10.1111/j.1574-6941.2002.tb00907.x
79. Tzortzis G, Baillon M-LA, Gibson GR, Rastall RA. Modulation of anti-pathogenic activity in canine-derived lactobacillus species by carbohydrate growth substrate. J Appl Microbiol (2004) 96:552–9. doi: 10.1111/j.1365-2672.2004.02172.x
80. Gibson GR, McCartney AL, Rastall RA. Prebiotics and resistance to gastrointestinal infections. Br J Nutr (2005) 93 Suppl 1:S31–34. doi: 10.1079/bjn20041343
81. Waller KMJ, Leong RW, Paramsothy S. An update on fecal microbiota transplantation for the treatment of gastrointestinal diseases. J Gastroenterol Hepatol (2022) 37:246–55. doi: 10.1111/jgh.15731
82. Brown JR-M, Flemer B, Joyce SA, Zulquernain A, Sheehan D, Shanahan F, et al. Changes in microbiota composition, bile and fatty acid metabolism, in successful faecal microbiota transplantation for clostridioides difficile infection. BMC Gastroenterol (2018) 18:131. doi: 10.1186/s12876-018-0860-5
83. Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol (2016) 9:229–39. doi: 10.1177/1756283X15607414
84. Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery (1958) 44:854–9.
85. Hui W, Li T, Liu W, Zhou C, Gao F. Fecal microbiota transplantation for treatment of recurrent c. difficile infection: An updated randomized controlled trial meta-analysis. PloS One (2019) 14:e0210016. doi: 10.1371/journal.pone.0210016
86. Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol (2014) 306:G310–319. doi: 10.1152/ajpgi.00282.2013
87. Baunwall SMD, Lee MM, Eriksen MK, Mullish BH, Marchesi JR, Dahlerup JF, et al. Faecal microbiota transplantation for recurrent clostridioides difficile infection: An updated systematic review and meta-analysis. EClinicalMedicine (2020) 29–30:100642. doi: 10.1016/j.eclinm.2020.100642
88. Gilbert JA. Microbiome therapy for recurrent clostridioides difficile. Lancet Microbe (2022) 3:e334. doi: 10.1016/S2666-5247(22)00096-9
89. Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, et al. Microbiota-accessible carbohydrates suppress clostridium difficile infection in a murine model. Nat Microbiol (2018) 3:662–9. doi: 10.1038/s41564-018-0150-6
90. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to clostridium difficile. Nature (2015) 517:205–8. doi: 10.1038/nature13828
91. Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: A randomized clinical trial. JAMA (2019) 321:156–64. doi: 10.1001/jama.2018.20046
92. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science (2021) 371:595–602. doi: 10.1126/science.abf3363
93. Xu M-Q, Cao H-L, Wang W-Q, Wang S, Cao X-C, Yan F, et al. Fecal microbiota transplantation broadening its application beyond intestinal disorders. World J Gastroenterol (2015) 21:102–11. doi: 10.3748/wjg.v21.i1.102
94. Narula N, Kassam Z, Yuan Y, Colombel J-F, Ponsioen C, Reinisch W, et al. Systematic review and meta-analysis: Fecal microbiota transplantation for treatment of active ulcerative colitis. Inflammation Bowel Dis (2017) 23:1702–9. doi: 10.1097/MIB.0000000000001228
95. DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, et al. Drug-resistant e. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med (2019) 381:2043–50. doi: 10.1056/NEJMoa1910437
96. Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, et al. Stool substitute transplant therapy for the eradication of clostridium difficile infection: “RePOOPulating” the gut. Microbiome (2013) 1:3. doi: 10.1186/2049-2618-1-3
97. Carlucci C, Jones CS, Oliphant K, Yen S, Daigneault M, Carriero C, et al. Effects of defined gut microbial ecosystem components on virulence determinants of clostridioides difficile. Sci Rep (2019) 9:885. doi: 10.1038/s41598-018-37547-x
98. Kerry RG, Patra JK, Gouda S, Park Y, Shin H-S, Das G. Benefaction of probiotics for human health: A review. J Food Drug Anal (2018) 26:927–39. doi: 10.1016/j.jfda.2018.01.002
99. Leccese G, Bibi A, Mazza S, Facciotti F, Caprioli F, Landini P, et al. Probiotic lactobacillus and bifidobacterium strains counteract adherent-invasive escherichia coli (AIEC) virulence and hamper IL-23/Th17 axis in ulcerative colitis, but not in crohn’s disease. Cells (2020) 9. doi: 10.3390/cells9081824
100. Coqueiro AY, Raizel R, Bonvini A, Tirapegui J, Rogero MM. Probiotics for inflammatory bowel diseases: a promising adjuvant treatment. Int J Food Sci Nutr (2019) 70:20–9. doi: 10.1080/09637486.2018.1477123
101. Derwa Y, Gracie DJ, Hamlin PJ, Ford AC. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther (2017) 46:389–400. doi: 10.1111/apt.14203
102. McFarland LV, Surawicz CM, Greenberg RN, Fekety R, Elmer GW, Moyer KA, et al. A randomized placebo-controlled trial of saccharomyces boulardii in combination with standard antibiotics for clostridium difficile disease. JAMA (1994) 271:1913–8.
103. Surawicz CM, McFarland LV, Greenberg RN, Rubin M, Fekety R, Mulligan ME, et al. The search for a better treatment for recurrent clostridium difficile disease: use of high-dose vancomycin combined with saccharomyces boulardii. Clin Infect Dis (2000) 31:1012–7. doi: 10.1086/318130
104. O’Toole PW, Marchesi JR, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol (2017) 2:17057. doi: 10.1038/nmicrobiol.2017.57
105. Feuerstadt P, Louie TJ, Lashner B, Wang EEL, Diao L, Bryant JA, et al. SER-109, an oral microbiome therapy for recurrent clostridioides difficile infection. N Engl J Med (2022) 386:220–9. doi: 10.1056/NEJMoa2106516
106. Henn MR, O’Brien EJ, Diao L, Feagan BG, Sandborn WJ, Huttenhower C, et al. A phase 1b safety study of SER-287, a spore-based microbiome therapeutic, for active mild to moderate ulcerative colitis. Gastroenterology (2021) 160:115–127.e30. doi: 10.1053/j.gastro.2020.07.048
107. Homayouni Rad A, Aghebati Maleki L, Samadi Kafil H, Abbasi A. Postbiotics: A novel strategy in food allergy treatment. Crit Rev Food Sci Nutr (2021) 61:492–9. doi: 10.1080/10408398.2020.1738333
108. Mallott EK, Amato KR. Butyrate production pathway abundances are similar in human and nonhuman primate gut microbiomes. Mol Biol Evol (2022) 39. doi: 10.1093/molbev/msab279
109. Zhang Z, Zhang H, Chen T, Shi L, Wang D, Tang D. Regulatory role of short-chain fatty acids in inflammatory bowel disease. Cell Commun Signal (2022) 20. doi: 10.1186/s12964-022-00869-5
110. Russo E, Giudici F, Fiorindi C, Ficari F, Scaringi S, Amedei A. Immunomodulating activity and therapeutic effects of short chain fatty acids and tryptophan post-biotics in inflammatory bowel disease. Front Immunol (2019) 10:2754. doi: 10.3389/fimmu.2019.02754
111. Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe (2020) 27:659–670.e5. doi: 10.1016/j.chom.2020.01.021
112. Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res (2005) 589:47–65. doi: 10.1016/j.mrrev.2004.08.001
113. Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol (2014) 12:164. doi: 10.1186/1477-7819-12-164
114. Littmann ER, Lee J-J, Denny JE, Alam Z, Maslanka JR, Zarin I, et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of clostridioides difficile infection. Nat Commun (2021) 12:755. doi: 10.1038/s41467-020-20793-x
115. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature (2010) 463:808–12. doi: 10.1038/nature08750
116. Whibley N, Tucci A, Powrie F. Regulatory T cell adaptation in the intestine and skin. Nat Immunol (2019) 20:386–96. doi: 10.1038/s41590-019-0351-z
117. Luu M, Steinhoff U, Visekruna A. Functional heterogeneity of gut-resident regulatory T cells. Clin Transl Immunol (2017) 6:e156. doi: 10.1038/cti.2017.39
118. Singh K, Hjort M, Thorvaldson L, Sandler S. Concomitant analysis of Helios and neuropilin-1 as a marker to detect thymic derived regulatory T cells in naïve mice. Sci Rep (2015) 5:7767. doi: 10.1038/srep07767
119. Zhang S, Morgan X, Dogan B, Martin F-P, Strickler S, Oka A, et al. Mucosal metabolites fuel the growth and virulence of e. coli linked to crohn’s disease. JCI Insight (2022) 7. doi: 10.1172/jci.insight.157013
120. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, et al. Intestinal inflammation allows salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U.S.A. (2011) 108:17480–5. doi: 10.1073/pnas.1107857108
121. Gallo A, Cancelli C, Ceron E, Covino M, Capoluongo E, Pocino K, et al. Fecal calprotectin and need of multiple microbiota trasplantation infusions in clostridium difficile infection. J Gastroenterol Hepatol (2020) 35:1909–15. doi: 10.1111/jgh.15072
122. Khoruts A, Rank KM, Newman KM, Viskocil K, Vaughn BP, Hamilton MJ, et al. Inflammatory bowel disease affects the outcome of fecal microbiota transplantation for recurrent clostridium difficile infection. Clin Gastroenterol Hepatol (2016) 14:1433–8. doi: 10.1016/j.cgh.2016.02.018
123. Hirten RP, Grinspan A, Fu S-C, Luo Y, Suarez-Farinas M, Rowland J, et al. Microbial engraftment and efficacy of fecal microbiota transplant for clostridium difficile in patients with and without inflammatory bowel disease. Inflammation Bowel Dis (2019) 25:969–79. doi: 10.1093/ibd/izy398
124. Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, et al. Fecal microbiota transplant for treatment of clostridium difficile infection in immunocompromised patients. Am J Gastroenterol (2014) 109:1065–71. doi: 10.1038/ajg.2014.133
125. Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology (2015) 149:102–109.e6. doi: 10.1053/j.gastro.2015.04.001
126. Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun (2018) 9:5184. doi: 10.1038/s41467-018-07359-8
127. Wei Y-L, Chen Y-Q, Gong H, Li N, Wu K-Q, Hu W, et al. Fecal microbiota transplantation ameliorates experimentally induced colitis in mice by upregulating AhR. Front Microbiol (2018) 9:1921. doi: 10.3389/fmicb.2018.01921
128. Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell (1993) 75:263–74. doi: 10.1016/0092-8674(93)80068-p
129. Perry T, Jovel J, Patterson J, Wong G, Fedorak RN, Thiesen A, et al. Fecal microbial transplant after ileocolic resection reduces ileitis but restores colitis in IL-10-/- mice. Inflamm Bowel Dis (2015) 21:1479–90. doi: 10.1097/MIB.0000000000000383
130. Chen G, Ran X, Li B, Li Y, He D, Huang B, et al. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine (2018) 30:317–25. doi: 10.1016/j.ebiom.2018.03.030
131. Pacheco RG, Esposito CC, Müller LCM, Castelo-Branco MTL, Quintella LP, Chagas VLA, et al. Use of butyrate or glutamine in enema solution reduces inflammation and fibrosis in experimental diversion colitis. World J Gastroenterol (2012) 18:4278–87. doi: 10.3748/wjg.v18.i32.4278
132. Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, et al. Butyrate inhibits interleukin-17 and generates tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol (2016) 16:84. doi: 10.1186/s12876-016-0500-x
133. Ng SC, Plamondon S, Kamm MA, Hart AL, Al-Hassi HO, Guenther T, et al. Immunosuppressive effects via human intestinal dendritic cells of probiotic bacteria and steroids in the treatment of acute ulcerative colitis. Inflammation Bowel Dis (2010) 16:1286–98. doi: 10.1002/ibd.21222
134. D’Incà R, Barollo M, Scarpa M, Grillo AR, Brun P, Vettorato MG, et al. Rectal administration of lactobacillus casei DG modifies flora composition and toll-like receptor expression in colonic mucosa of patients with mild ulcerative colitis. Dig Dis Sci (2011) 56:1178–87. doi: 10.1007/s10620-010-1384-1
135. Lührs H, Gerke T, Müller JG, Melcher R, Schauber J, Boxberge F, et al. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol (2002) 37:458–66. doi: 10.1080/003655202317316105
136. Banday AH, Abdalla M. Immune checkpoint inhibitors: Recent clinical advances and future prospects. Curr Med Chem (2022) 30. doi: 10.2174/0929867329666220819115849
137. Pezo RC, Wong M, Martin A. Impact of the gut microbiota on immune checkpoint inhibitor-associated toxicities. Therap Adv Gastroenterol (2019) 12:1756284819870911. doi: 10.1177/1756284819870911
138. Wang F, Yin Q, Chen L, Davis MM. Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc Natl Acad Sci U.S.A. (2018) 115:157–61. doi: 10.1073/pnas.1712901115
139. Sun S, Luo L, Liang W, Yin Q, Guo J, Rush AM, et al. Bifidobacterium alters the gut microbiota and modulates the functional metabolism of T regulatory cells in the context of immune checkpoint blockade. Proc Natl Acad Sci U.S.A. (2020) 117:27509–15. doi: 10.1073/pnas.1921223117
140. Wang T, Zheng N, Luo Q, Jiang L, He B, Yuan X, et al. Probiotics lactobacillus reuteri abrogates immune checkpoint blockade-associated colitis by inhibiting group 3 innate lymphoid cells. Front Immunol (2019) 10:1235. doi: 10.3389/fimmu.2019.01235
141. Renga G, Nunzi E, Pariano M, Puccetti M, Bellet MM, Pieraccini G, et al. Optimizing therapeutic outcomes of immune checkpoint blockade by a microbial tryptophan metabolite. J Immunother Cancer (2022) 10. doi: 10.1136/jitc-2021-003725
142. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med (2018) 24:1804–8. doi: 10.1038/s41591-018-0238-9
143. Chen M, Liu M, Li C, Peng S, Li Y, Xu X, et al. Fecal microbiota transplantation effectively cures a patient with severe bleeding immune checkpoint inhibitor-associated colitis and a short review. Front Oncol (2022) 12:913217. doi: 10.3389/fonc.2022.913217
144. Saleh R, Elkord E. FoxP3(+) T regulatory cells in cancer: Prognostic biomarkers and therapeutic targets. Cancer Lett (2020) 490:174–85. doi: 10.1016/j.canlet.2020.07.022
145. Suzuki H, Chikazawa N, Tasaka T, Wada J, Yamasaki A, Kitaura Y, et al. Intratumoral CD8(+) T/FOXP3 (+) cell ratio is a predictive marker for survival in patients with colorectal cancer. Cancer Immunol Immunother (2010) 59:653–61. doi: 10.1007/s00262-009-0781-9
146. Griffiths RW, Elkord E, Gilham DE, Ramani V, Clarke N, Stern PL, et al. Frequency of regulatory T cells in renal cell carcinoma patients and investigation of correlation with survival. Cancer Immunol Immunother (2007) 56:1743–53. doi: 10.1007/s00262-007-0318-z
147. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359:91–7. doi: 10.1126/science.aan3706
148. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature (2019) 565:600–5. doi: 10.1038/s41586-019-0878-z
149. Si W, Liang H, Bugno J, Xu Q, Ding X, Yang K, et al. Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut (2022) 71:521–33. doi: 10.1136/gutjnl-2020-323426
150. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science (2021) 371:602–9. doi: 10.1126/science.abb5920
151. Dizman N, Meza L, Bergerot P, Alcantara M, Dorff T, Lyou Y, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat Med (2022) 28:704–12. doi: 10.1038/s41591-022-01694-6
152. Nomura M, Nagatomo R, Doi K, Shimizu J, Baba K, Saito T, et al. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw Open (2020) 3:e202895. doi: 10.1001/jamanetworkopen.2020.2895
153. Botticelli A, Vernocchi P, Marini F, Quagliariello A, Cerbelli B, Reddel S, et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J Transl Med (2020) 18:49. doi: 10.1186/s12967-020-02231-0
154. Monaghan T, Mullish BH, Patterson J, Wong GK, Marchesi JR, Xu H, et al. Effective fecal microbiota transplantation for recurrent clostridioides difficile infection in humans is associated with increased signalling in the bile acid-farnesoid X receptor-fibroblast growth factor pathway. Gut Microbes (2019) 10:142–8. doi: 10.1080/19490976.2018.1506667
155. Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen ECL, Renooij W, Murzilli S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut (2011) 60:463–72. doi: 10.1136/gut.2010.212159
156. Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for clostridium difficile spore germination and outgrowth in the Large intestine. mSphere (2016) 1. doi: 10.1128/mSphere.00045-15
157. Monaghan TM, Seekatz AM, Markham NO, Yau TO, Hatziapostolou M, Jilani T, et al. Fecal microbiota transplantation for recurrent clostridioides difficile infection associates with functional alterations in circulating microRNAs. Gastroenterology (2021) 161:255–270.e4. doi: 10.1053/j.gastro.2021.03.050
158. Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol (2019) 20:21–37. doi: 10.1038/s41580-018-0045-7
159. Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, et al. The binary toxin CDT enhances clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol (2016) 1:16108. doi: 10.1038/nmicrobiol.2016.108
160. Buonomo EL, Cowardin CA, Wilson MG, Saleh MM, Pramoonjago P, Petri WAJ. Microbiota-regulated IL-25 increases eosinophil number to provide protection during clostridium difficile infection. Cell Rep (2016) 16:432–43. doi: 10.1016/j.celrep.2016.06.007
161. Cook L, Rees WD, Wong MQ, Peters H, Levings MK, Steiner TS. Fecal microbiota transplantation for recurrent clostridioides difficile infection enhances adaptive immunity to c difficile toxin b. Gastroenterology (2021) 160:2155–2158.e4. doi: 10.1053/j.gastro.2021.01.009
162. van Schaik W. The human gut resistome. Philos Trans R Soc Lond B Biol Sci (2015) 370:20140087. doi: 10.1098/rstb.2014.0087
163. Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
164. Jouhten H, Mattila E, Arkkila P, Satokari R. Reduction of antibiotic resistance genes in intestinal microbiota of patients with recurrent clostridium difficile infection after fecal microbiota transplantation. Clin Infect Dis (2016) 63:710–1. doi: 10.1093/cid/ciw390
165. Gopalsamy SN, Woodworth MH, Wang T, Carpentieri CT, Mehta N, Friedman-Moraco RJ, et al. The use of microbiome restoration therapeutics to eliminate intestinal colonization with multidrug-resistant organisms. Am J Med Sci (2018) 356:433–40. doi: 10.1016/j.amjms.2018.08.015
166. Mahieu R, Cassisa V, Hilliquin D, Coron N, Pailhoriès H, Kempf M, et al. Impact of faecal microbiota transplantation on mouse digestive colonization with two extensively resistant bacteria. J Infect (2017) 75:75–7. doi: 10.1016/j.jinf.2017.04.008
167. Gargiullo L, Del Chierico F, D’Argenio P, Putignani L. Gut microbiota modulation for multidrug-resistant organism decolonization: Present and future perspectives. Front Microbiol (2019) 10:1704. doi: 10.3389/fmicb.2019.01704
168. Li X, Song L, Zhu S, Xiao Y, Huang Y, Hua Y, et al. Two strains of lactobacilli effectively decrease the colonization of VRE in a mouse model. Front Cell Infect Microbiol (2019) 9:6. doi: 10.3389/fcimb.2019.00006
169. Eggers S, Barker AK, Valentine S, Hess T, Duster M, Safdar N. Effect of lactobacillus rhamnosus HN001 on carriage of staphylococcus aureus: results of the impact of probiotics for reducing infections in veterans (IMPROVE) study. BMC Infect Dis (2018) 18:129. doi: 10.1186/s12879-018-3028-6
170. Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, et al. Cooperating commensals restore colonization resistance to vancomycin-resistant enterococcus faecium. Cell Host Microbe (2017) 21:592–602.e4. doi: 10.1016/j.chom.2017.04.002
171. Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, et al. Intestinal microbiota containing barnesiella species cures vancomycin-resistant enterococcus faecium colonization. Infect Immun (2013) 81:965–73. doi: 10.1128/IAI.01197-12
172. Kim SG, Becattini S, Moody TU, Shliaha PV, Littmann ER, Seok R, et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant enterococcus. Nature (2019) 572:665–9. doi: 10.1038/s41586-019-1501-z
173. Sazawal S, Hiremath G, Dhingra U, Malik P, Deb S, Black RE. Efficacy of probiotics in prevention of acute diarrhoea: a meta-analysis of masked, randomised, placebo-controlled trials. Lancet Infect Dis (2006) 6:374–82. doi: 10.1016/S1473-3099(06)70495-9
174. Harper A, Vijayakumar V, Ouwehand AC, Ter Haar J, Obis D, Espadaler J, et al. Viral infections, the microbiome, and probiotics. Front Cell Infect Microbiol (2020) 10:596166. doi: 10.3389/fcimb.2020.596166
175. Botić T, Klingberg TD, Weingartl H, Cencic A. A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int J Food Microbiol (2007) 115:227–34. doi: 10.1016/j.ijfoodmicro.2006.10.044
176. Quintana VM, Torres NI, Wachsman MB, Sinko PJ, Castilla V, Chikindas M. Antiherpes simplex virus type 2 activity of the antimicrobial peptide subtilosin. J Appl Microbiol (2014) 117:1253–9. doi: 10.1111/jam.12618
177. Su Y, Zhang B, Su L. CD4 detected from lactobacillus helps understand the interaction between lactobacillus and HIV. Microbiol Res (2013) 168:273–7. doi: 10.1016/j.micres.2012.12.004
178. Wu S, Jiang Z-Y, Sun Y-F, Yu B, Chen J, Dai C-Q, et al. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza a virus infection. Curr Microbiol (2013) 67:414–22. doi: 10.1007/s00284-013-0380-z
179. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity (2012) 37:171–86. doi: 10.1016/j.immuni.2012.05.020
180. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity (2012) 37:158–70. doi: 10.1016/j.immuni.2012.04.011
181. Winkler ES, Shrihari S, Hykes BLJ, Handley SA, Andhey PS, Huang Y-JS, et al. The intestinal microbiome restricts alphavirus infection and dissemination through a bile acid-type I IFN signaling axis. Cell (2020) 182:901–918.e18. doi: 10.1016/j.cell.2020.06.029
182. Yang X-L, Wang G, Xie J-Y, Li H, Chen S-X, Liu W, et al. The intestinal microbiome primes host innate immunity against enteric virus systemic infection through type I interferon. mBio (2021) 12. doi: 10.1128/mBio.00366-21
183. Aziz N, Bonavida B. Activation of natural killer cells by probiotics. For Immunopathol Dis Therap (2016) 7:41–55. doi: 10.1615/ForumImmunDisTher.2016017095
184. Park M-K, Ngo V, Kwon Y-M, Lee Y-T, Yoo S, Cho Y-H, et al. Lactobacillus plantarum DK119 as a probiotic confers protection against influenza virus by modulating innate immunity. PloS One (2013) 8:e75368. doi: 10.1371/journal.pone.0075368
185. Kumova OK, Fike AJ, Thayer JL, Nguyen LT, Mell JC, Pascasio J, et al. Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal lactobacillus rhamnosus GG. PloS Pathog (2019) 15:e1008072. doi: 10.1371/journal.ppat.1008072
186. Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science (2017) 357:498–502. doi: 10.1126/science.aam5336
187. Wang Y, Liu L, Moore DJ, Shen X, Peek RM, Acra SA, et al. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol (2017) 10:373–84. doi: 10.1038/mi.2016.57
188. Song JA, Kim HJ, Hong SK, Lee DH, Lee SW, Song CS, et al. Oral intake of lactobacillus rhamnosus M21 enhances the survival rate of mice lethally infected with influenza virus. J Microbiol Immunol Infect (2016) 49:16–23. doi: 10.1016/j.jmii.2014.07.011
189. Holscher HD, Czerkies LA, Cekola P, Litov R, Benbow M, Santema S, et al. Bifidobacterium lactis Bb12 enhances intestinal antibody response in formula-fed infants: a randomized, double-blind, controlled trial. JPEN J Parenter Enteral Nutr (2012) 36:106S–17S. doi: 10.1177/0148607111430817
190. Ramakrishna C, Kujawski M, Chu H, Li L, Mazmanian SK, Cantin EM. Bacteroides fragilis polysaccharide a induces IL-10 secreting b and T cells that prevent viral encephalitis. Nat Commun (2019) 10:2153. doi: 10.1038/s41467-019-09884-6
191. Nguyen QV, Chong LC, Hor Y-Y, Lew L-C, Rather IA, Choi S-B. Role of probiotics in the management of COVID-19: A computational perspective. Nutrients (2022) 14. doi: 10.3390/nu14020274
192. Bottari B, Castellone V, Neviani E. Probiotics and covid-19. Int J Food Sci Nutr (2021) 72:293–9. doi: 10.1080/09637486.2020.1807475
193. Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science (2016) 352:586–9. doi: 10.1126/science.aad8852
194. Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe (2018) 23:229–240.e5. doi: 10.1016/j.chom.2018.01.003
195. Moss EL, Falconer SB, Tkachenko E, Wang M, Systrom H, Mahabamunuge J, et al. Long-term taxonomic and functional divergence from donor bacterial strains following fecal microbiota transplantation in immunocompromised patients. PloS One (2017) 12:e0182585. doi: 10.1371/journal.pone.0182585
196. Kootte RS, Levin E, Salojärvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab (2017) 26:611–619.e6. doi: 10.1016/j.cmet.2017.09.008
Keywords: mucosal immunity, regulatory T cells, fecal microbiota transplantation, probiotics, live biotherapeutic products, C. difficile, microbiome-based therapeutic, immune checkpoint inhibitors
Citation: Alam MZ, Maslanka JR and Abt MC (2023) Immunological consequences of microbiome-based therapeutics. Front. Immunol. 13:1046472. doi: 10.3389/fimmu.2022.1046472
Received: 16 September 2022; Accepted: 16 December 2022;
Published: 12 January 2023.
Edited by:
Kevin Maloy, University of Glasgow, United KingdomReviewed by:
Clarissa Campbell, Austrian Academy of Sciences (OeAW), AustriaKatie L. Alexander, University of Alabama at Birmingham, United States
Copyright © 2023 Alam, Maslanka and Abt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael C. Abt, TWljaGFlbC5hYnRAcGVubm1lZGljaW5lLnVwZW5uLmVkdQ==
 Md Zahidul Alam
Md Zahidul Alam Jeffrey R. Maslanka
Jeffrey R. Maslanka Michael C. Abt
Michael C. Abt