
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 31 October 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1037739
This article is part of the Research Topic The Immune Diversity and its Organization/Regulation in Aquatic Invertebrates View all 8 articles
Molting is one of the most important biological processes of crustacean species, and a number of molecular mechanisms facilitate this complex procedure. However, the understanding of the immune mechanisms underlying crustacean molting cycle remains very limited. This study performed transcriptome sequencing in hemolymph and hepatopancreas of the swimming crab (Portunus trituberculatus) during the four molting stages: post-molt (AB), inter-molt (C), pre-molt (D), and ecdysis (E). The results showed that there were 78,572 unigenes that were obtained in the hemolymph and hepatopancreas of P. trituberculatus. Further analysis showed that 98 DEGs were involved in immunity response of hemolymph and hepatopancreas, and most of the DEGs participated in the process of signal transduction, pattern recognition proteins/receptors, and antioxidative enzymes system. Specifically, the key genes and pathway involved in signal transduction including the GPCR126, beta-integrin, integrin, three genes in mitogen-activated protein kinase (MAPK) signaling cascade (MAPKKK10, MAPKK4, and p38 MAPK), and four genes in Toll pathway (Toll-like receptor, cactus, pelle-like kinase, and NFIL3). For the pattern recognition proteins/receptors, the lowest expression level of 11 genes was found in the E stage, including C-type lectin receptor, C-type lectin domain family 6 member A and SRB3/C in the hemolymph, and hepatopancreatic lectin 4, C-type lectin, SRB, Down syndrome cell adhesion molecule homolog, Down syndrome cell adhesion molecule isoform, and A2M. Moreover, the expression level of copper/zinc superoxide dismutase isoform 4, glutathione peroxidase, glutathione S-transferase, peroxiredoxin, peroxiredoxin 6, and dual oxidase 2 in stage C or stage D significantly higher than that of stage E or stage AB. These results fill in the gap of the continuous transcriptional changes that are evident during the molting cycle of crab and further provided valuable information for elucidating the molecular mechanisms of immune regulation during the molting cycle of crab.
Molting, a phenomenon of shedding the old exoskeleton and re-generating the new one, is one of the most important biological processes of crustacean species, and it is generally divided into three types: larval-metamorphic molts, growth-related molts, and reproductive molts (1, 2). The successful molting of crustacean will contribute to its growth, development, reproduction, and appendage regeneration (3–5). Consequently, the crabs and shrimp undergo dramatic biochemical and physiological changes during the molting stage. Previous studies showed that each molting cycle of crustacean can be normally divided into four recurrent stages, namely, inter-molt (stage C), pre-molt (stage D), ecdysis (stage E), and post-molt (stage AB), according to the morphological differences of epidermis, exoskeleton and the internal setal development of the appendages (6, 7). It has been well known that the crustaceans are too vulnerable to the attacks from other congeners or natural enemy during the period of molting or just finished molting, which leads to their death (8). It can be speculated that the changes in the immune response in crustaceans are closely related to the molting stage.
To date, several studies have shown that crustaceans exhibit different immune responses to pathogens at different molting stages, including white spot syndrome virus (WSSV) and pesticide virus (9, 10). Meanwhile, a few studies have analyzed immune dynamics during the molting cycle of crustaceans using transcriptomic techniques. For example, Xu et al. (11) reported that the expression level of genes related to antimicrobial peptide, Toll signaling pathway factors, phenoloxidase system, and antioxidant enzymes in the hepatopancreas of mud crab (Scylla paramamosain) were significantly upregulated at the post-molt stage and inter-molt stage compared with the pre-molt stage (11). Zhang et al. (12) founded that a significant positive correlation between the expression of two antimicrobial peptide (AMP) genes (SpALF5 and SpCrustin) and the relative abundance of two predominant microorganisms (Halomonas and Shewanella) in hemolymph was observed in the whole molt cycle of S. paramamosain (12). Gao et al. (6) reported that shrimp will upregulate the expression of the antimicrobial peptides in pre-molt stage for generate an immune response (6). Despite these previous findings, the understanding of the immune mechanisms underlying crustacean molting cycle remains very limited.
The swimming crab Portunus trituberculatus (Crustacea, Decapoda, and Brachyura) widely distributed along the coasts of East Asia countries and forms important fisheries in these countries (13–15). Due to its rapid growth rate and higher farming profit, the pond culture of P. trituberculatus has been developing rapidly in China (16, 17). The aquaculture production of P. trituberculatus reached up to 100,895 tonnes in 2020 in China (18). However, the survival rate of pond-reared P. trituberculatus is very low, only 5%, which is one of the important problems hindering the improvement of economic value and aquaculture profit for this species (8, 19). The reason for the low survival rate of pond-reared P. trituberculatus may be related to their higher cannibalism rate and various diseases during the molting period (8, 19, 20). Therefore, in order to sustain the stable swimming crab culture, it is very important to understand its immune response mechanism during molting cycle.
Similar to other crustaceans, the immune response of P. trituberculatus was also mediated by hemolymph and hepatopancreas. To further identify the molecular events altered during the four molting stages, post-molt, inter-molt, pre-molt, and ecdysis, herein, we used RNA-seq to investigate the immune response in the hemolymph and hepatopancreas of P. trituberculatus during the molting cycle. The associated findings in this study will provide an improved understanding of the molecular mechanism for the immune defense in crab and other molting animals.
All crabs were treated in strict accordance with the guidelines for the care and use of experimental animals established by the Administration of Affairs Concerning Experimental Animals of the State Council of the People’s Republic of China and approved by the Committee on Experimental Animal Management of the Jiangsu Ocean University.
The experimental design and sampling procedures are shown in Figure 1. Juvenile P. trituberculatus (body weight: 15–30 g) was collected from outdoor ponds of Qidong scientific research base of Shanghai Fisheries Research Institute, Jiangsu, China and then acclimated in an indoor circulating water system. During the acclimation, each crab was individually housed in culture baskets (Length × Width × Depth = 33 × 27.5 × 35 cm), and all baskets were kept floating in four concrete tanks (Length × Width × Depth = 5.8 m × 2.4 m × 1.8 m) supplied by filtered and treated recirculating seawater. After the crab finished molting, it was used for sampling of different molt cycles.
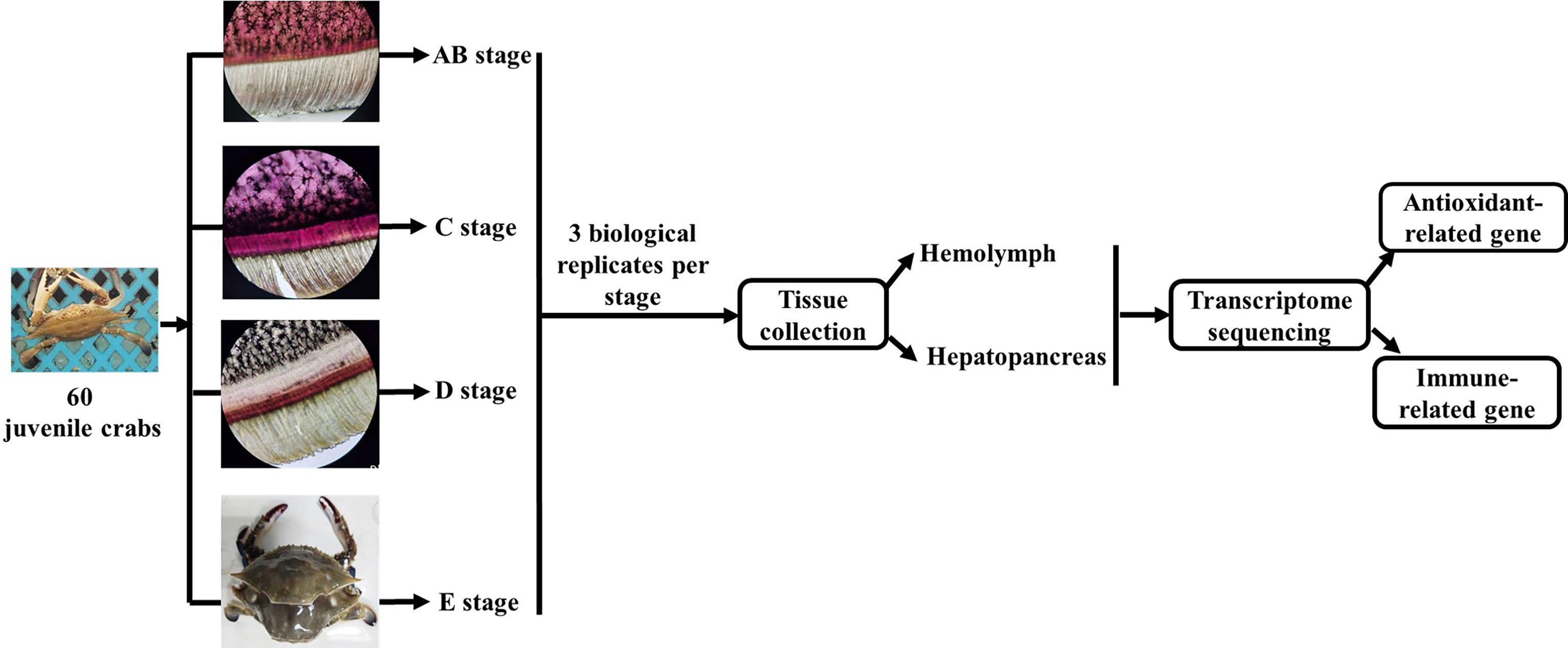
Figure 1 Schematic showing the experimental design and sampling procedures for the transcriptome analysis during the molting cycle of P. trituberculatus. Notes: The molting stage of P. trituberculatus was determined by observing the base of the swimming leg under a microscope before sampling. The characteristics of swimming leg bases in different molting stages of P. trituberculatus are shown in the figure. Stage AB: post-molt; stage C: inter-molt; stage D: per-molt; stage E: ecdysis.
The molt cycle of P. trituberculatus was classified into four stages (post-molt [stage AB], inter-molt [stage C], per-molt [stage D], and ecdysis [stage E]) according to the previously report (21). Specifically, post-molt (AB) stage: P. trituberculatus has higher water content in its body, dorsal carapace edge hard, and the cuticle thickness is between 10 and 35 mm; inter-molt (C) stage: All parts of the body become hard, cuticle thickness is between 35 and 45 mm, and new carapace begins to secrete; per-molt (D) stage: All parts of the body is hard, the secretion of the outer epidermis is completed, and the new carapace is completely separated from the old carapace during dissection in later stage; ecdysis (E) stage: The new shell is clearly visible and the body can further absorb water. Three individual crabs at each stage were sampled, anesthetized on ice and then the hemolymph, and hepatopancreas were removed from the crabs for transcriptome analysis.
Total RNA was isolated from each sample using the mirVana miRNA Isolation Kit (Ambion) according to manufacturer’s protocol. The purity and integrity of total RNA were estimated by a NanoDrop 2000 (Thermo Fisher Scientific, Massachusetts, USA) and 2100 Bioanalyzer (Agilent), respectively. Only high-quality RNA samples (OD260/OD280 ranged 1.8–2.2, RIN ≥ 8.0) were used to construct the sequencing library.
The libraries were constructed using TruSeq Stranded mRNA LTSample Prep Kit (Illumina) according to the manufacturer’s instructions. Then, these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten) and 150 bp paired-end reads were generated. The reads containing ploy-N and the low-quality reads were removed to obtain the clean reads. The clean reads were assembled into expressed sequence tag clusters (contigs) and de novo assembled into transcripts by using Trinityin paired-end method (22).
In order to obtain the annotation information of unigenes, the assembled unigenes were annotated with the NCBI non-redundant protein database (NR), Swiss-Prot protein and Clusters of orthologous groups for eukaryotic complete genomes (KOG) databases using BlastX with a threshold E-value of 1.0 × 10−5. The proteins with the highest hits to the unigenes were used to assign functional annotations thereto. Blast2go (http://www.BLAST2go.org/ ) and WEGO software were used to get the Gene Ontology (GO) annotation and GO functional classification for the unigenes. The unigenes were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to annotate their potential metabolic pathways.
The quantity expression of these unigenes was estimated by calculating FPKM (fragments per kb per million reads) using bowtie2 and eXpress to calculate FPKM. To identify differentially expressed genes (DEGs) across samples, |log2(fold change)| > 1 and P-value < 0.05 were set to be the thresholds for significantly different expression levels. The gene expression profiles were compared among the four stages and the DEGs were then subjected to enrichment analysis of GO functions and KEGG pathways.
To visualize the expression levels between different immune-related genes and molting stages, normalized counts in the form of transcripts per million values were generated. The expression level of immune-related genes was shown as log2 transcript per million plus one (TPM + 1) (23).
Ten important genes involved in immune response during the molting cycle of P. trituberculatus were determined using qRT-PCR to validate the RNA-seq results. First-strand cDNA synthesis was performed with a reverse first strand cDNA synthesis kit (RR036A, Takara Bio, Japan). To normalize the target gene expression, 18S was used as an internal reference gene. Gene-specific primers were designed using Primer Premier 5.0 and listed in Table 1. The reaction system was performed as previously described (24). qRT-PCR was carried with a StepOnePlus™ real-time PCR system using SYBR1Green I (TakaRa, Japan) according to the manufacturer’s instructions. The relative expression level of each gene was calculated using the the 2−ΔΔCt method (25). Data are presented as the mean ± standard error (SE).
After filtration, a total of 384.74 million reads were obtained from the four molting stages of P. trituberculatus transcriptomes, namely, 48.35 (AB-Ha), 47.98 (C-Ha), 48.85 (D-Ha), and 47.93 (E-Ha) million reads from the hemolymph libraries and 46.96 (AB-H), 49.01 (C-H), 48.38 (D-H), and 47.28 (E-H) million reads from the hepatopancreas libraries (Table 2). Finally, 78,572 unigenes were obtained after combining the transcripts, with an average length of 2654 bp, and the N50 was 4407 bp. Among the hemolymph unigenes, 38,570 (85.33%) unigenes could be matched to the NR database, 27 517 (60.88%) unigenes matched in the eggNOG database, whereas 10,978 (24.29%) unigenes matched in the KEGG (Table 3). For the hepatopancreas, 36,224 (86.62%) unigenes could be matched to the NR database, 26,834 (64.16%) unigenes were annotated in the eggNOG database, whereas 11,164 (26.69%) unigenes could be fully annotated with the KEGG (Table 3). The ILLUMINA data were submitted to the National Center for Biotechnology Information (NCBI, PRJNA855992).
As shown in Figure 2, there were 2.566 upregulated genes and 2,001 downregulated genes in AB-Ha versus C-Ha, 2,618 upregulated genes and 2,141 genes downregulated in AB-Ha versus D-Ha, 4,453 upregulated genes and 3,876 genes downregulated in AB-Ha versus E-Ha, 2,040 upregulated genes and 2 104 genes downregulated in C-Ha versus D-Ha, 3,167 upregulated genes and 2,968 genes downregulated in C-Ha versus E-Ha, 4,393 upregulated genes and 4,482 genes downregulated in D-Ha versus E-Ha. For the hepatopancreas, 6,146 upregulated genes and 5,153 downregulated genes in AB-H versus C-H, 7,116 upregulated genes and 5,787 genes downregulated in AB-H versus D-H, 10,799 upregulated genes and 11,131genes downregulated in AB-H versus E-H, 2,948 upregulated genes and 2,906 genes downregulated in C-H versus D-H, 9,747 upregulated genes and 12,957 genes downregulated in C-H versus E-H, 9,915 upregulated genes and 13,126 genes downregulated in D-H versus E-H (Figure 2).
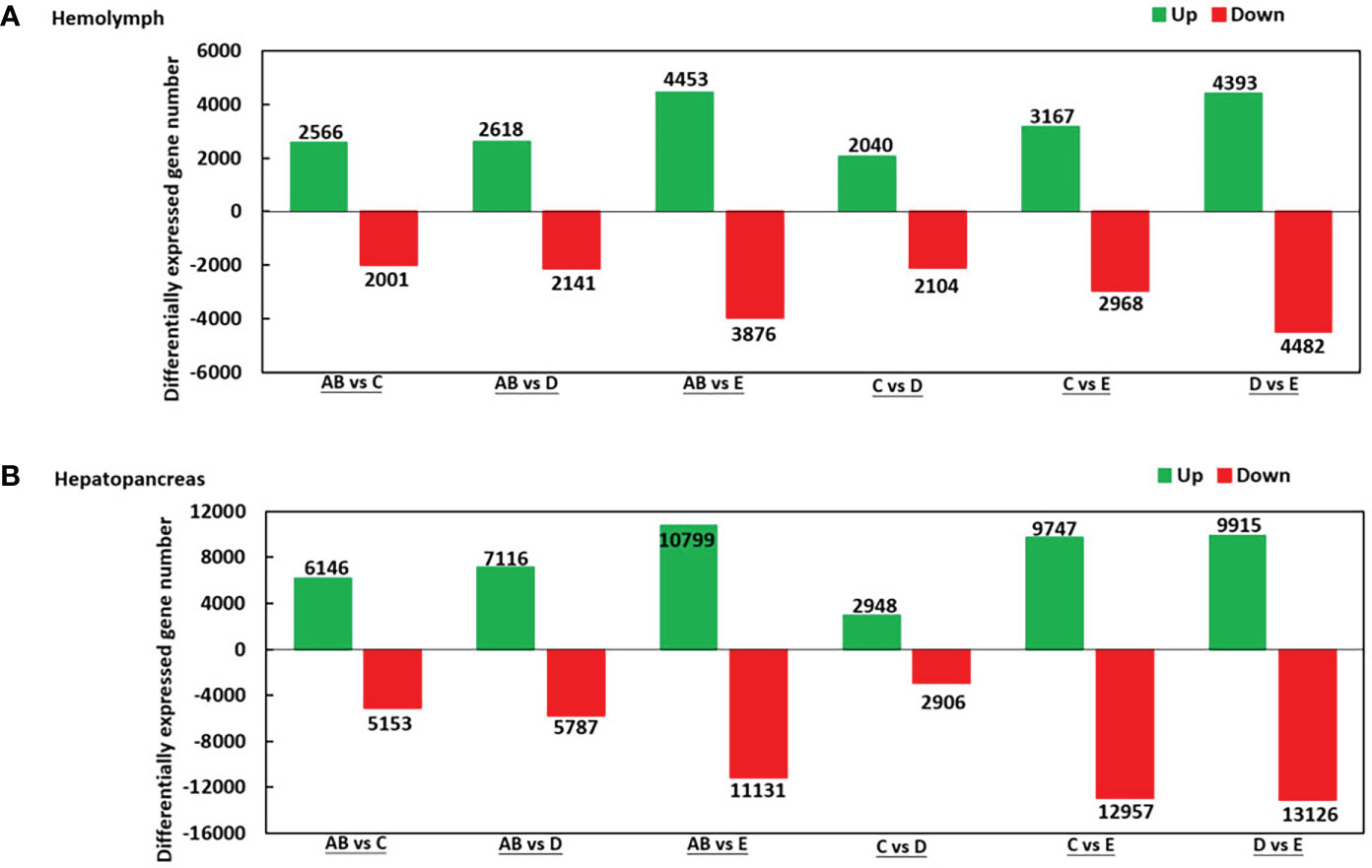
Figure 2 Summary of differentially expressed genes (DEGs) in transcriptome of hemolymph (A) and hepatopancreas (B) of P. trituberculatus. Notes (1): AB: post-molt; C: inter-molt; D: per-molt; E: ecdysis (2). Up- and downregulated genes are shown in green and red, respectively (3). The x-axis shows the comparisons between two stages, whereas the y-axis represents the total number of DEGs for each comparison. The numbers on the columns represent the number of DEGs.
To further assign the putative functions to DEGs, GO and KEGG analyses were performed. GO annotation output showed that the DEGs were mainly classified into three parts: biological process, cellular component, and molecular function. GO annotation of the DEGs in the different molting stage in the hemolymph showed that the DEGs were categorized mainly into cellular process (GO:0009987) and metabolic process (GO:0008152) in biological processes, organelle (GO:0031090) and membrane (GO:0016020) in cellular component, and catalytic activity (GO:0003824) and binding (GO:0005488) in molecular function, respectively (Figures 3, 4). In the hepatopancreas, compared AB stage and E stage, a total of 6,268 DEGs were annotated with GO terms, 7,218 DEGs were annotated with GO terms between the AB stage and D stage, and 12,273 DEGs were annotated with GO terms between the AB stage and E stage. In the comparison of C stage and D stage, 3,073 DEGs were annotated with GO terms. When C stage and E stage were compared, 12,460 DEGs were assigned to 7,571 GO terms (level 2). In the comparison of D stage and E stage, 12,750 DEGs were annotated with GO terms. Overall, the GO assignment result of hepatopancreatic DEGs was similar to the hemolymph (Figures 5, 6). KEGG enrichment results showed that the DEGs in the hemolymph were enriched in 331 specific KEGG metabolic pathways, whereas the DEGs in the hepatopancreas were enriched in 337 specific KEGG metabolic pathways. The top 10 most significantly enriched metabolic pathways in the hemolymph and hepatopancreas are represented in Table 4. In the hemolymph, the significantly enriched pathways included “maturity onset diabetes of the young,” “Jak-STAT signaling pathway,” “Bacterial invasion of epithelial cells.” The DEGs in the hepatopancreas were significantly clustered in “ribosome,” “amino sugar and nucleotide sugar metabolism,” and “collecting duct acid secretion.”
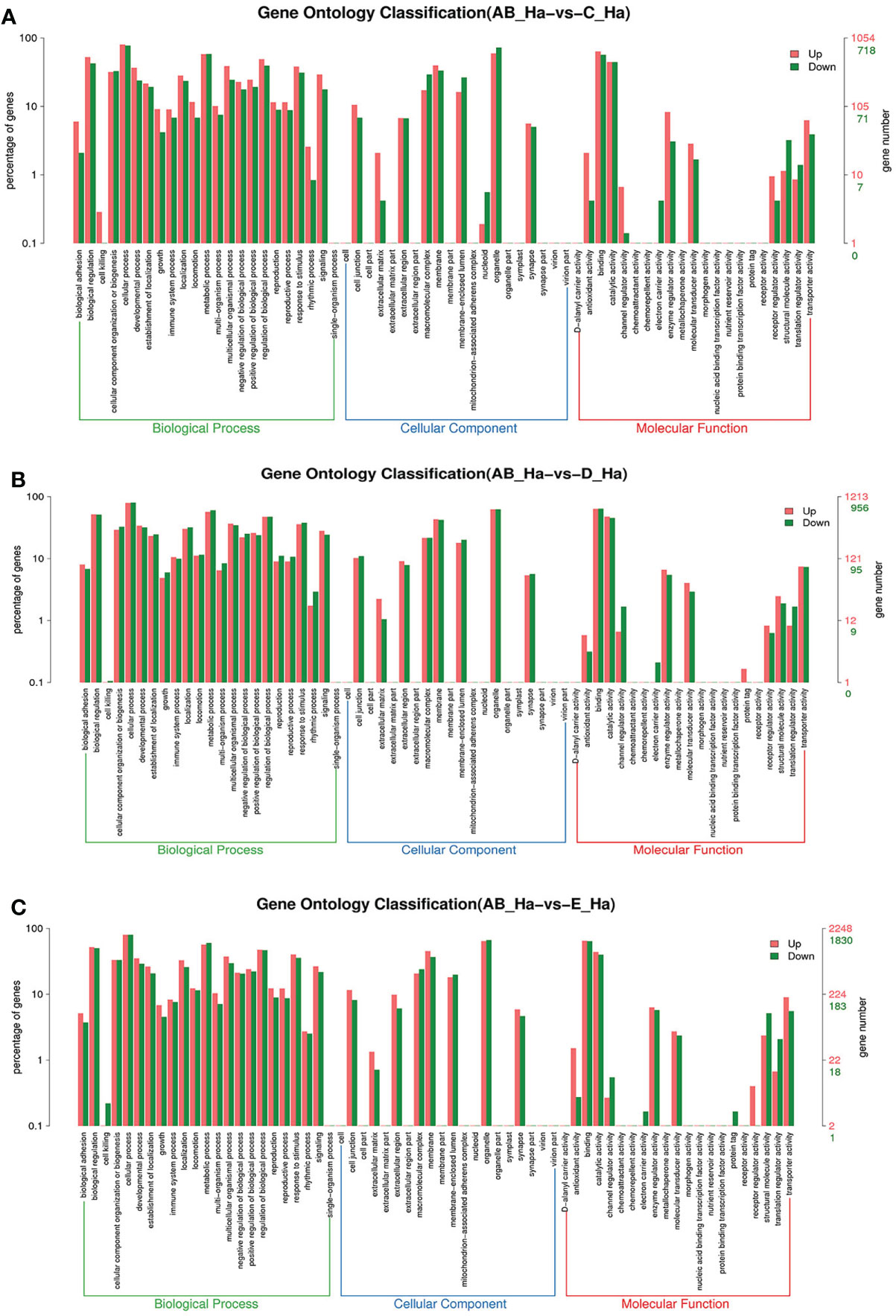
Figure 3 GO enrichment analyses of differentially expressed genes (DEGs) in hemolymph transcriptome. Notes: (A) Comparison between AB-Ha and C-Ha; (B) comparison between AB-Ha and D-Ha. (C) Comparison between AB-Ha and E-Ha. Notes (1): AB-Ha: hemolymph from post-molt; C-Ha: hemolymph from inter-molt; D-Ha: hemolymph from per-molt; E-Ha: hemolymph from ecdysis (2). Red columns indicate GO item entries enriched by upregulated differentially expressed genes; green columns indicate GO item entries enriched by downregulated differentially expressed genes. (3) The x-axis shows the item name between two stages, whereas the y-axis represents the number of genes and their percentage of the corresponding item.
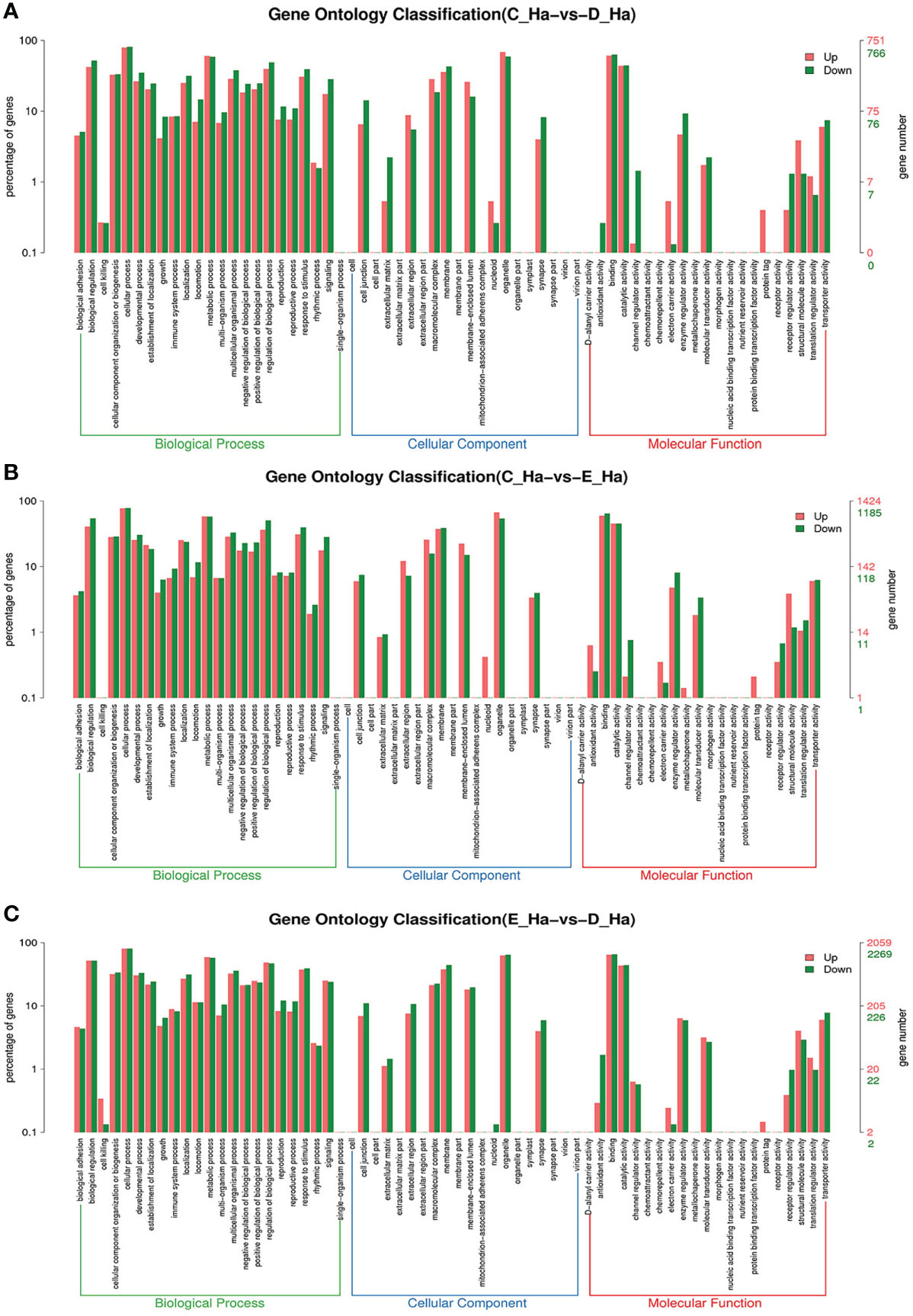
Figure 4 GO enrichment analyses of differentially expressed genes (DEGs) in hemolymph transcriptome. Notes: (A) Comparison between C-Ha and D-Ha; (B) comparison between C-Ha and E-Ha. (C) Comparison between E-Ha and D-Ha. Notes: (1) C-Ha: hemolymph from inter-molt; D-Ha: hemolymph from per-molt; E-Ha: hemolymph from ecdysis. (2) Red columns indicate GO item entries enriched by upregulated differentially expressed genes; green columns indicate GO item entries enriched by downregulated differentially expressed genes (3). The x-axis shows the item name between two stages, whereas the y-axis represents the number of genes and their percentage of the corresponding item.
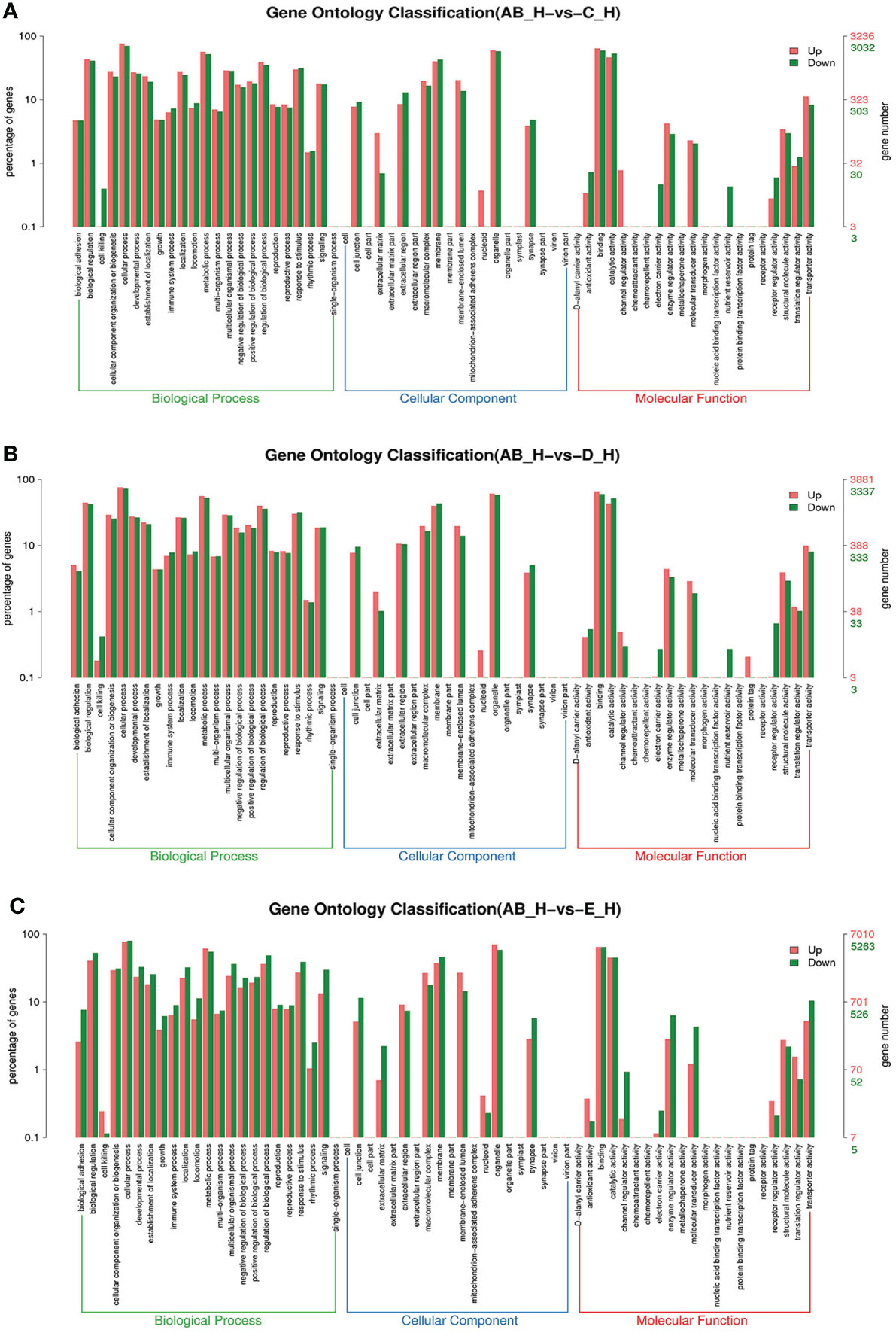
Figure 5 GO enrichment analyses of differentially expressed genes (DEGs) in hepatopancreas transcriptome. Notes: (A) Comparison between AB-H and C-H; (B) comparison between AB-H and D-H. (C) Comparison between AB-H and E-H. Notes: (1) AB-H: hepatopancreas from post-molt; C-H: hepatopancreas from inter-molt; D-H: hepatopancreas from per-molt; E-H: hepatopancreas from ecdysis. (2) Red columns indicate GO item entries enriched by upregulated differentially expressed genes; green columns indicate GO item entries enriched by downregulated differentially expressed genes. (3) The x-axis shows the item name between two stages, whereas the y-axis represents the number of genes and their percentage of the corresponding item.
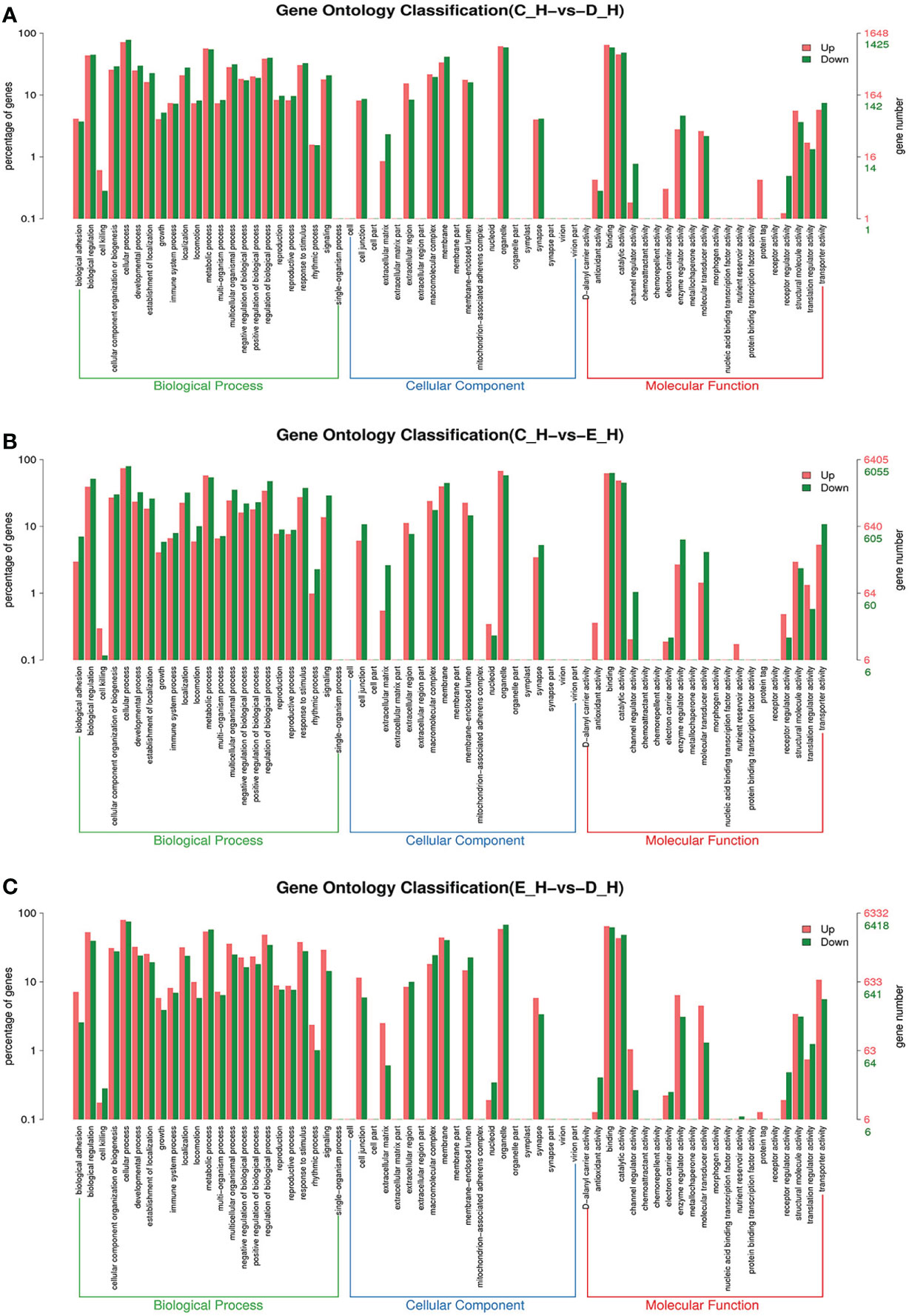
Figure 6 GO enrichment analyses of differentially expressed genes (DEGs) in hepatopancreas transcriptome. Notes: (A) Comparison between C-H and D-H; (B) comparison between C-H and E-H. (C) Comparison between E-H and D-H. Notes: (1) C-H: hepatopancreas from inter-molt; D-H: hepatopancreas from per-molt; E-H: hepatopancreas from ecdysis. (2) Red columns indicate GO item entries enriched by upregulated differentially expressed genes, green columns indicate GO item entries enriched by downregulated differentially expressed genes (3) The x-axis shows the item name between two stages, whereas the y-axis represents the number of genes and their percentage of the corresponding item.
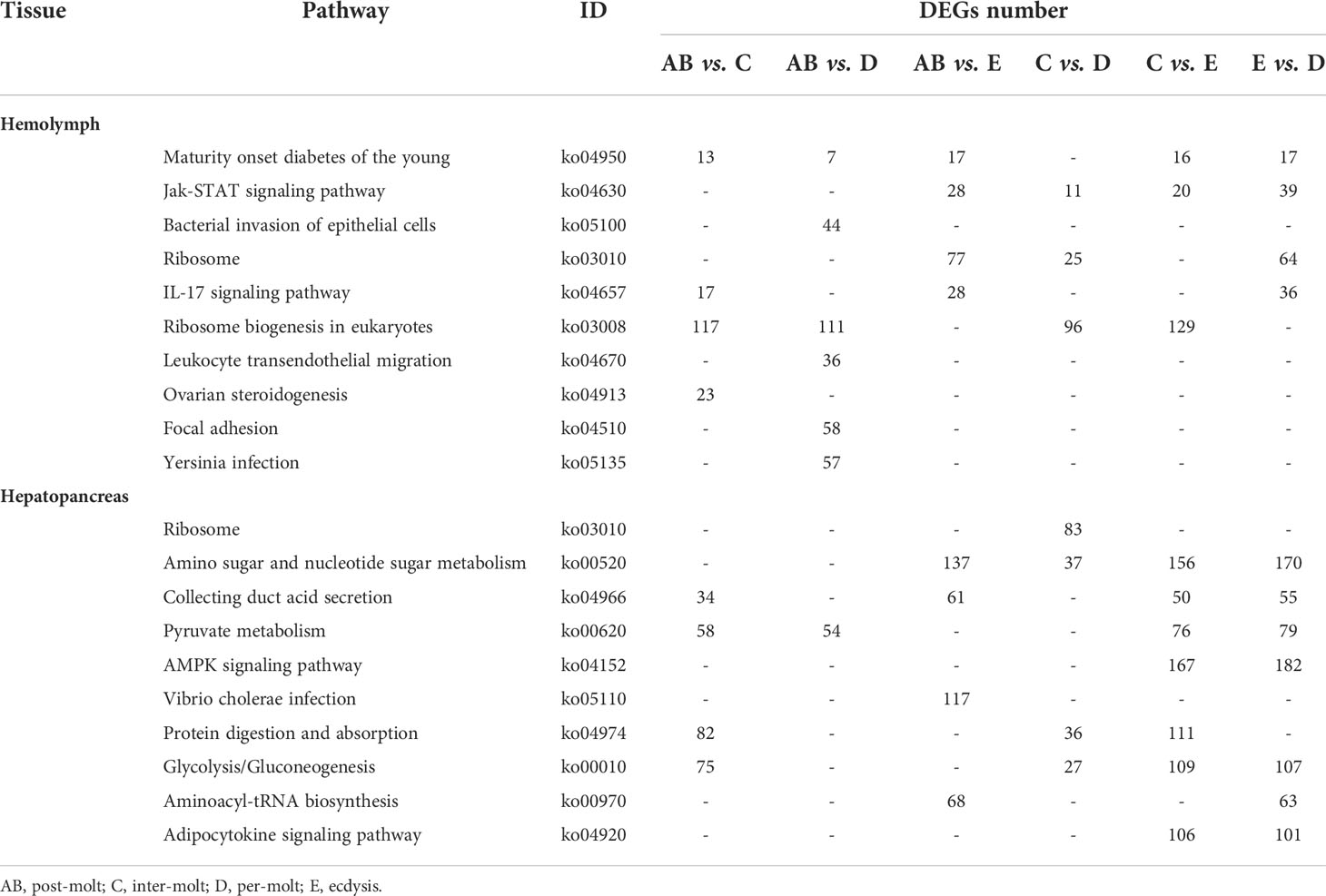
Table 4 Significantly changed pathways obtained with KEGG, using the significantly changed genes from hemolymph and hepatopancreas during the molt cycle of P. trituberculatus.
According to the gene data, 98 DEGs are involved in various processes of crab immunity response in the hemolymph and hepatopancreas and those DEGs were categorized into nine groups (Table 5). Specifically, 25 key genes were mainly involved in signal transduction, namely, G-protein coupled receptor 126 (GPCR126), G-protein coupled receptor Mth2, mitogen-activated protein kinase kinase 4 (MAPKK4), MAPKKK10, MAPKKK3, MAPKKK12, p38 MAPK, beta-integrin, integrin, protein kinase C and casein kinase substrate in neurons protein 1-like and four genes (Toll-like receptor, cactus, pelle-like kinase, nuclear factor interleukin 3-regulated transcription factor [NFIL3]) in Toll pathway. Sixteen genes related to pattern recognition proteins/receptors were identified in the hemolymph and hepatopancreas, such as lectin 4, C-type lectin, C-type lectin receptor, C-type lectin domain family 6 member A, C-type lectin-like domain-containing protein PtLP, scavenger receptor B (SRB), SRB3, SRC, Down syndrome cell adhesion molecule homolog, Down syndrome cell adhesion molecule isoform, alpha-2 macroglobulin (A2M), tetraspanin, tetraspanin-11, tetraspanin-2A-like, tetraspanin-13-like, and lipopolysaccharide and beta-1,3-glucan binding protein (LBGBP). Ten genes related to other immune molecules were identified in the hemolymph and hepatopancreas, such as metallothionein, metallothionein 2, macrophage migration inhibitory factor, macrophage migration inhibitory factor MIF1, macrophage migration inhibitory factor MIF2, hemocyanin, hemocyanin subunit, hemocyanin subunit 1, hemocyanin subunit 2, and hemocyanin subunit 5. Nine genes related to apoptosis were identified in the hemolymph and hepatopancreas, such as caspase, caspase 2, caspase 3, caspase 4, caspase 8, cathepsin A, cathepsin C, neprilysin-11, and autophagy protein 5. For the prophenoloxidase cascade, seven genes were identified in the hemolymph and hepatopancreas, namely, prophenoloxidase, prophenoloxidase activating factor 1, serine proteinase, serine protease 4, serine protease 5, serine protease 27, and serine proteinase inhibitor 2. For the antimicrobial peptides, six genes were identified in the hemolymph and hepatopancreas, namely, lysozyme, crustin antimicrobial peptide, antimicrobial peptide hyastatin, anti-lipopolysaccharide factor isoform 4 (ALF4), ALF5, and ALF7. For the blood clotting system, the transglutaminase and transglutaminase 2 were found in the hemolymph and hepatopancreas.
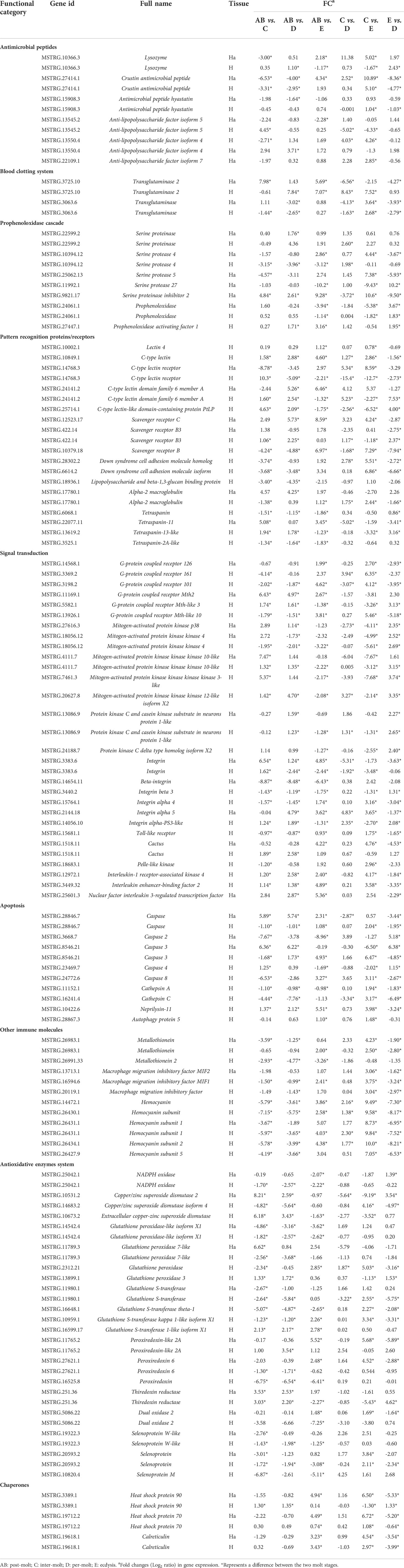
Table 5 Summary of differential expressed genes related to immune in the transcriptome of P. trituberculatus.
In addition, several genes of the antioxidative enzymes system and chaperones also involved in immunity response of P. trituberculatus. Thus, for the antioxidative enzymes system, 20 genes were identified in the hemolymph and hepatopancreas, namely, NADPH oxidase, copper/zinc superoxide dismutase 2 (CuZnSOD-2), CuZnSOD-4, ecCuZnSOD, glutathione peroxidase-like isoform X1, glutathione peroxidase 7-like, glutathione peroxidase, glutathione peroxidase 3, glutathione S-transferase, glutathione S-transferase theta-1, glutathione S-transferase kappa 1-like isoform X1, glutathione S-transferase 1-like isoform X1, peroxiredoxin-like 2A, peroxiredoxin, peroxiredoxin 6, thiredoxin reductase, dual oxidase 2, selenoprotein, selenoprotein W-like, and Selenoprotein M. Moreover, three genes related to chaperones also showed various changes pattern in different molting stages, such as heat shock protein 90 (HSP90), HSP70, and calreticulin.
To better understand the changes of immune-related genes in the hemolymph and hepatopancreas during the molting cycle, the expression level of key genes involved in signal transduction, pattern recognition proteins/receptors, apoptosis, prophenoloxidase cascade, antimicrobial peptides, antioxidative enzymes system, chaperones, and other immune molecules were analyzed (Figures 7, 8). The results showed that the expression levels of p38 MAPK, MAPKK4, MAPKK3, MAPKKK10, and MAPKKK12 decline from stage AB to stage C and then rise from stage D to stage E. However, the expression level of GPCR 126, GPCR 161, and GPCR 101 rises from stage AB to stage C and then decline from stage D to stage E. Moreover, the higher expression level of integrin, integrin alpha-4, and integrin alpha 5 was found in stage AB, whereas the expression level of beta-integrin was lower in stages AB during the molting cycle of P. trituberculatus. Remarkably, the expression level of Toll-like receptor was higher during the molting cycle of P. trituberculatus and reached the highest level in stage D. For pattern recognition proteins/receptors, the lowest expression level of 10 genes was found in the E stage, namely, C-type lectin receptor, SRB3, and SRC in the hemolymph and lectin 4, C-type lectin, SRB, Down syndrome cell adhesion molecule homolog, Down syndrome cell adhesion molecule isoform, and A2M in the hepatopancreas. For the other immune molecules, the expression level of those genes reached the highest level in stage C or stage D and the lowest level in E stage or AB stage, such as metallothionein, metallothionein 2, macrophage migration inhibitory factor, macrophage migration inhibitory factor MIF1, hemocyanin subunit 1, hemocyanin subunit 2, and hemocyanin subunit 5.
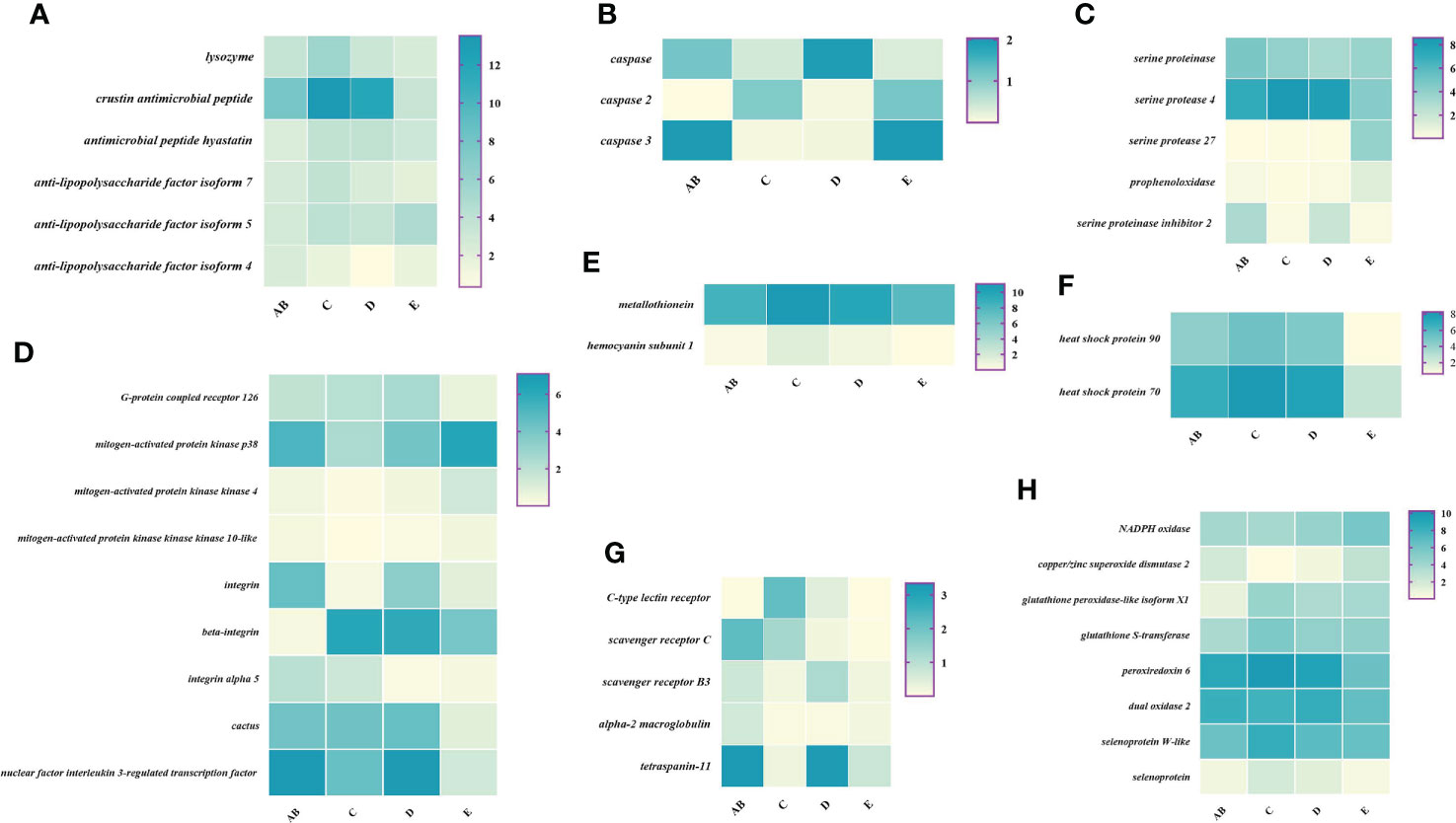
Figure 7 Expression patterns of immune-related genes in the hemolymph during the molting cycle of P. trituberculatus. (A) The genes related to antimicrobial peptides; (B) the genes related to apoptosis; (C) the genes related to prophenoloxidase cascade; (D) the genes related to signal transduction; (E) the genes related to other immune molecules; (F) the genes related to chaperones; (G) the genes related to pattern recognition proteins/receptors; (H) The genes related to antioxidative enzymes system. Notes: (1) The x-axis shows the molting stages, whereas the y-axis represents the name of genes. (2) The color represents the expression level of the gene; the darker the color, the higher gene expression level. (3) The expression level of immune-related genes was shown as log2 transcript per million plus one (TPM + 1).
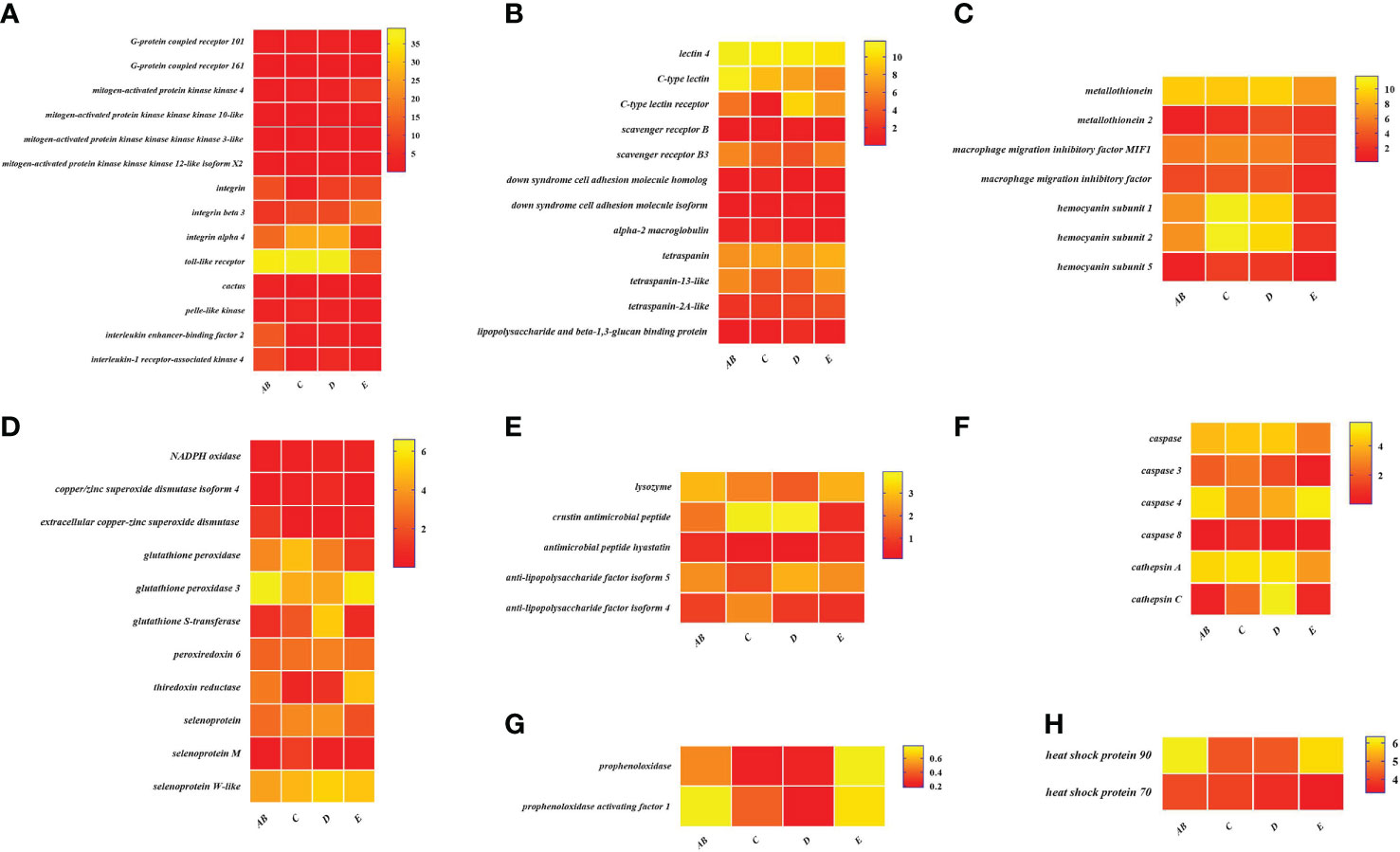
Figure 8 Expression patterns of immune-related genes in the hepatopancreas during the molting cycle of P. trituberculatus. (A) The genes related to signal transduction; (B) the genes related to pattern recognition proteins/receptors; (C) the genes related to other immune molecules; (D) the genes related to antioxidative enzymes system; (E) the genes related to antimicrobial peptides; (F) the genes related to apoptosis; (G) the genes related to prophenoloxidase cascade; (H) the genes related to chaperones. Notes: (1) The x-axis shows the molting stages, whereas the y-axis represents the name of genes. (2) The color represents the expression level of the gene; the lighter the color, the higher the gene expression level. (3) The expression level of immune-related genes was shown as log2 transcript per million plus one (TPM + 1).
Among the genes related to apoptosis, the expression of caspase, caspase 3, and caspase 8 in the hepatopancreas reached the highest level in stage C and the lowest level in stage E. The transcript levels of the genes (prophenoloxidase, prophenoloxidase-activating factor 1, and serine protease 27) that involved in activating proPO system increased significantly from stage D to stage E, whereas the transcript levels of serine proteinase inhibitor 2 decreased significantly from stage D to stage E. Among the genes related to antimicrobial peptides, the highest expression level of lysozyme in hemolymph was found in the E stage while the lowest expression level of lysozyme in hepatopancreas was found in the E stage. The expression levels of crustin and antimicrobial peptide hyastatin in the stage C or stage D significantly higher than AB stage or E stage in the hemolymph and hepatopancreas. The expression levels of ALF4, ALF5, and ALF7 in the hemolymph and hepatopancreas showed various changes pattern in different molting stages.
For the antioxidative enzymes system and chaperones, the expression level of copper/zinc superoxide dismutase isoform 4, glutathione peroxidase, glutathione S-transferase, peroxiredoxin, peroxiredoxin 6, and dual oxidase 2 in stage C or stage D significantly higher than that of stage E or stage AB. Moreover, the expression level of selenoprotein (selenoprotein M and selenoprotein W-like) increased significantly from stage AB to stage C and then decline gradually from stage C to stage E. The expression level of chaperones (HSP90 and HSP70) gradually decreased from stage C to stage E.
Ten selected important genes in the molting cycle of P. trituberculatus were determined using qRT-PCR to validate the RNA-seq results. The qPCR results showed that the relative expression patterns of these nine genes were consistent with the RNA-seq results (Figures 9, 10). Notably, the qRT-PCR results showed that the expression level of Toll-like receptor in the hepatopancreas of stages AB to D was higher than that of stage E, and reached the highest level in stage AB. However, the RNA-seq results showed that the expression level of Toll-like receptor in the hepatopancreas of stages AB to D was higher than that of stage E, and reached the highest level in stage D.
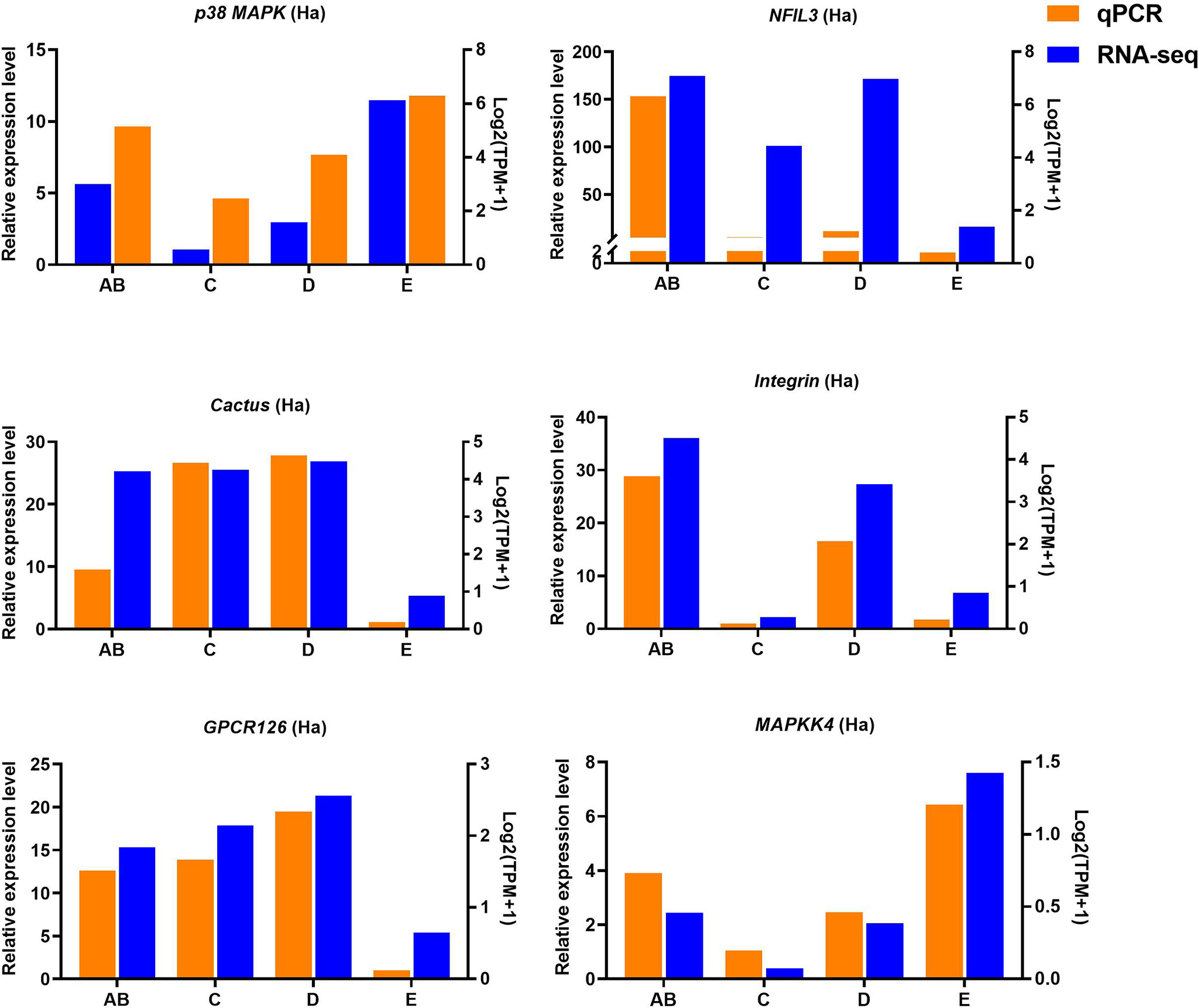
Figure 9 Quantitative expression of six genes was determined by qPCR in hemolymph from different molting stage of P. trituberculatus. p38 MAPK: p38 mitogen-activated protein kinase; NFIL3: nuclear factor interleukin 3–regulated transcription factor; GPCR126: G-protein coupled receptor 126; MAPKK4: mitogen-activated protein kinase kinase 4.
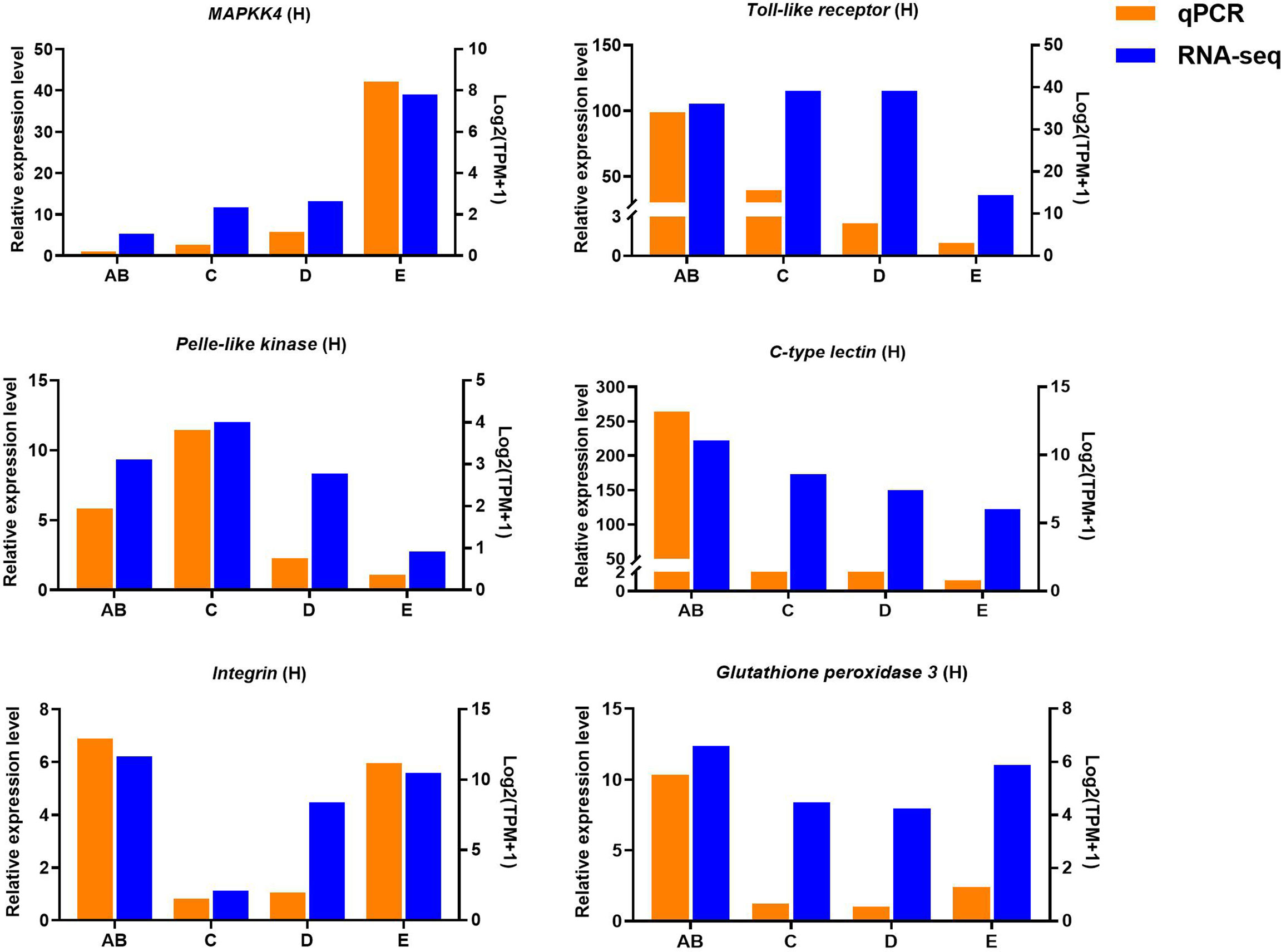
Figure 10 Quantitative expression of six genes was determined by qPCR in hepatopancreas from different molting stage of P. trituberculatus. MAPKK4: mitogen-activated protein kinase kinase 4.
Like other invertebrates, crustaceans rely entirely on the innate immune system to combat invading pathogenic microorganisms, which mainly consists of cellular and humoral immunity (26). Specifically, the cellular immunity involves phagocytosis, apoptosis, nodulation, and encapsulation, whereas the humoral immunity includes antimicrobial peptides (AMPs), prophenoloxidase cascade, clottable proteins, the toll and immune deficiency (IMD) pathway, and so forth (27). Previous studies have shown that the crustacean immune system is a tightly regulated network, and the immune molecules that synthesized or released by hemolymph and hepatopancreas are able to maintain the balance of immune homeostasis under normal physiological conditions (27, 28). As an effective physical barrier, the hard carapace and external cuticle are the crab’s first line of immune defense, protecting the crab from the attachment and penetration of pathogens (28). However, the crabs are too vulnerable to pathogenic bacteria when they are molting or just finished molting. Thus, this study performed a comprehensive transcriptome analysis of the hemolymph and hepatopancreas in different molting stage of P. trituberculatus, aiming to uncover the immune-related to genes or pathway altered during the four molting stages and further improve the understanding of the immune mechanisms applicable to the molting cycle process in crustaceans.
In this study, the significantly changed molecules associated with immune processes during the molting cycle of P. trituberculatus could divided into seven functional categories, namely, antimicrobial peptides (AMPs), blood clotting system, prophenoloxidase (proPO) cascade, pattern recognition receptors (PRRs), signal transduction, apoptosis, and other immune molecules (Table 5). AMP are key components of innate immunity and play key roles in the defense against invading microorganisms, including lysozyme, crustin, and so forth (29). Lysozymes can be classified into six types, and the c-type lysozymes have been reported in shrimp, crabs, and crayfish (30). Pan et al. (31) reported that the c-type lysozymes of P. trituberculatus may be an inducible acute-phase protein and bacterial challenge could decline its mRNA expression (31). Crustin group, the largest family of crustacean AMPs, exhibits broad antimicrobial activity against Gram-negative bacteria (32). Anti-lipopolysaccharide factors (ALFs) are considered to have broad-spectrum antimicrobial activity against Gram-positive bacteria, Gram-negative bacteria, fungi, and viruses (33). The present study showed that the highest expression level of several AMPs (crustin, lysozyme, antimicrobial peptide hyastatin, and anti-lipopolysaccharide factor isoform 7) was found in the stage C hemolymph, indicating that the higher antimicrobial activity was found in this stage. Moreover, the lowest expression level of crustin, antimicrobial peptide hyastatin, and anti-lipopolysaccharide factor isoform 4/5/7 was found in the stage E, which may lead to the lower antimicrobial activity of crab in this stage and susceptible to bacterial or virus infection.
The prophenoloxidase (proPO) cascade system is one of the most essential immune reactions, which involves the participation of proPO, phenoloxidase (PO), serine proteases, and inhibitors (34). The Phenoloxidase (PO) is believed to be derived from its precursor prophenoloxidase (proPO), which is considered as the rate-limiting step of melanin formation (35, 36). Melanization is an important component of the innate immune responses in arthropods and allowing a rapid response to pathogen infection (36). Serine proteases, especially those with one or two clip domains, play critical roles in the activation of prophenoloxidase (proPO) activating system (37). Serine proteinase inhibitors have been functionally characterized as negative regulators of the proPO activation cascade by inhibiting the serine proteases in different crustacean species (35, 38). In the present study, the transcript levels of the genes (prophenoloxidase, prophenoloxidase-activating factor 1, and serine protease 27) that involved in activating proPO system increased significantly from stage D to stage E, suggesting that the proPO cascade system is involved in the immune defense of P. trituberculatus during ecdysis. The transcript levels of serine proteinase inhibitor 2 that inhibiting in activating proPO system decreased significantly from stage D to stage E further confirmed the above speculation.
Pattern recognition receptors (PRPs) are a group of germ line encoded receptors that recognize common epitopes on pathogen surfaces and then trigger a series of downstream responses (39). In this study, a series of PRRs were identified, namely, lectin 4, C-type lectin, C-type lectin receptor, scavenger receptor B (SRB), SRC, Down syndrome cell adhesion molecule homolog, alpha-2 macroglobulin, lipopolysaccharide and beta-1,3-glucan binding protein, and tetraspanin, indicating that the immune defense mechanism of crab has a very complex recognition network. Lectins are carbohydrate-binding proteins that can be classified into several families and C-type lectins families are the largest and most distributed in crustaceans (40). Previous studies reported that the C-type lectins not only can regulate the expression of antilipopolysaccharide factor and inhibit bacterial growth but also participate in phagocytosis, encapsulation, and the activation and regulation of the proPO system (41–44). The present study showed that the expression level of C-type lectins and lectin 4 higher than that of other PRPs, demonstrating that the lectins play a key role in recognizing invading microorganisms during the molting cycle of P. trituberculatus. Moreover, the results showed that the expression level of C-type lectins and lectin 4 in the hepatopancreas decreased continuously from stage AB to stage E, which is similar to the result of Scylla paramamosain (11). However, the expression level of C-type lectin receptor in the hepatopancreas dramatically increased from stage C to stage E, illustrating that C-type lectin receptor played a key role in trigger other intracellular signal pathways. Moreover, Sanchez-Salgado et al. (45) reported that the receptor of lectin was β-integrin when the lectin involved in the phagocytosis of shrimp Fenneropenaeus chinensis (45). SR comprised nine heterogeneous classes, of which SRB and SRC promote phagocytosis of viruses in crustaceans (46, 47). The transcript of SRB and SRC in the hemolymph and hepatopancreas showed that the opposite expression pattern from the stage AB to stage C may suggest that the different role of two tissues in SR mediates phagocytosis. However, the lowest expression level of SRB and SRC in the hemolymph and hepatopancreas was found in stage E, indicating that the low phagocytosis in this stage.
As an immune effector, Down syndrome cell adhesion molecule (Dscam) can produce thousands of isoforms and specifically binds to different types of bacteria and promotes challenge-specific protection (48, 49). Lipopolysaccharide-and beta-1,3-glucan-binding protein (LGBP) plays a crucial role in immune mechanism by mediating the recognition of invading pathogens to host cells (50). In this study, the expression level of hepatopancreatic Dscam and LGBP in stage C/D significantly higher than that in stage AB/E, demonstrating that the crab in stages C and D are able to make a rapid immune response when invaded by pathogens. Alpha-2-macroglobulin (A2M) is highly upregulated upon microbial infection and involved in several immune pathways (51). The transcript of alpha-2 macroglobulin in the hemolymph and hepatopancreas showed that a decline trend from the stage AB to stage E may suggest that the A2M was not necessarily to participate in the immune defense during the molting cycle of crab. Tetraspanins, belong to the transmembrane 4 superfamily (TM4SF), are highly involved in viral and bacterial infections (52, 53). Previous studies showed that the tetraspanin-3 and tetraspanin 8 play a key role in WSSV and Aeromonas hydrophila infection of shrimp (52, 53). Li et al. (54) reported that tetraspanin-18 may play an important role in the beginning of molting cycle of Eriocheir sinensis (54). The present study showed that tetraspanin, tetraspanin-13-like and tetraspanin-2A-like were identified, and its higher transcript level was detected in the hepatopancreas during the molting cycle of P. trituberculatus. It was speculated that the tetraspanins may involve in the immune defense or other physiological process of P. trituberculatus.
G-protein coupled receptors (GPCRs) are important transmembrane receptors that played a vital role in the innate immune defense of crustacean, including the defense pathogens, melanization, sclerotization, and bacterial infection (55–57). Mitogen-activated protein kinase (MAPK) signaling cascade consists of extracellular signal-regulated kinases (ERKs), C-Jun N-terminal or stress-activated protein kinases (JNKs/SAPKs), and p38 MAPKs pathways, which and are involved in cellular response to inflammatory cytokines, environmental stress, and pathogenic infection (58, 59). Among them, p38 MAPK can be activated by various extracellular stimuli and play a critical role in innate immune responses by induces the expression of interleukin-1 and tumor necrosis factor-alpha and initiates (60, 61). Mitogen-activated protein kinase 4 is a key upstream kinase in the JNK/p38 MAPK pathway that has been reported to play an anti-bacterial role in shrimp by regulate the expression of antimicrobial peptides (62, 63). In this study, the expression level of GPCR 126, GPCR 161, and GPCR 101 rise from stage AB to stage C and then decline from stage D to stage E. However, the expression level of p38 MAPK, MAPKK4 MAPKKK3, MAPKKK10, and MAPKKK12 decline from stage AB to stage C and then rise from stage D to stage E. The opposite expression pattern of GPCR and MAPK family genes showed that the GPCR signaling pathway and MAPK signaling cascade play a critical role in the innate immune responses of stage C/D or stage AB/E, respectively.
Integrins belong to a large family of heterodimeric cell surface receptors that composed of α and β subunits (64). Previous studies have shown that integrin involved in WSSV infection, bacterial infection, proPO activation, and phagocytosis of shrimp and crab (65–69). The present study showed that the higher expression level of integrin, integrin alpha-4, and integrin alpha 5 was found in stage AB, illustrating that integrin-mediated signal transduction has significant function in balanced immune defense of this stage. Moreover, Lin et al. (2013) reported that the expression level of integrin beta in the haemocytes was lower in stages D and higher in stages AB during the molting cycle of white shrimp Litopenaeus vannamei (66). In this study, the expression level of beta-integrin in the haemocytes was lower in stages AB and higher in stages C/D during the molting cycle of P. trituberculatus. The opposite expression pattern of beta-integrin in two animals may be related to species specificity.
Toll-like receptor (TLR) signaling plays an essential role in initiating innate immune responses in crustaceans (70). Specifically, the activation of the Toll pathway is initiated by the Spätzle and then Spätzle binds the N terminus of Toll inducing a conformational change. Subsequently, the trimeric complex MyD88/Tube/TRAF6 is formed then transducing signals to the Dorsal/Cactus complex that regulates the Toll-dependent gene expression of AMPs, and a significant number of innate immune responsive genes (71). In this study, four significantly expressed genes (Toll-like receptor, cactus, pelle-like kinase, and nuclear factor interleukin 3-regulated transcription factor) involve in TLR signaling were identified and the higher expression level of Toll-like receptor was found from stage AB to stage D of P. trituberculatus, further demonstrating that the TLR signalling pathway play an important role of innate immunity of this crab. Notably, the qRT-PCR results showed that the expression level of Toll-like receptor in the hepatopancreas reached the highest level in stage AB, whereas the RNA-seq results showed that the expression level of Toll-like receptor in the hepatopancreas reached the highest level in stage D. We speculate the possible reasons for this difference are the differences in detection methods or samples.
Apoptosis is a fundamental cellular process that plays a critical role in innate immunity, normal development, and tissue homeostasis (72, 73). Caspases are a conserved family of cysteine proteases, which could initiate and execute apoptosis, and the apoptotic caspases are further categorized into initiator caspases (caspase-2, -8, -9, and -10), inflammatory caspase (caspase-1, -4, and -5) and effector caspases (caspase-3, -6, and -7) (74, 75). In crustacean, increasing evidences have indicated that caspase -2, -3, and -8 could suppress WSSV infection via apoptosis induction (76–78). The present study showed that the expression level of caspases (caspase-2, -3, -4, and -8), changed significantly during the molting cycle of P. trituberculatus, indicating that caspase would contribute to apoptosis homeostasis and immune defense of this crab. Hemocyanin, the respiratory protein of arthropods and mollusks, has been linked to key aspects of innate immunity, in particular antibacterial, antiviral and induced the phenoloxidase-like activities (79). The expression level of hemocyanin subunit (1, 2, and 5) in this study increased significantly from stage AB to stage C and then decline gradually from stage C to stage E, illustrating that the activity of hemocyanin-mediated immune defense was highest in the stage C and lowest in the stage E.
During pathogen invasion, innate immunity can produce free radical molecules and their derivatives reactive nitrogen/oxygen species (RNS/ROS) against invading pathogens in shrimps and crabs (80, 81). Previous study has established that the NADPH oxidase play important roles in host defense system by catalyzing the production of superoxide anions (82). In this study, the expression level of NADPH oxidase increased gradually from stage AB to stage E in the hemolymph and hepatopancreas, suggesting that crabs may carry out the immune defense at this stage by increasing the content of superoxide anion to protect themselves. Although ROS are beneficial for host defense against pathogens, their accumulation damages cells and disrupts immune system homeostasis (83). Thus, organisms eliminate excessive ROS through the antioxidant systems composed of some endogenous antioxidant enzymes, such as superoxide dismutase (SOD), glutathione peroxidase (GPx), glutathione S-transferase (GST), and peroxiredoxin (Prx) (84, 85). It has been reported that those antioxidant enzymes can collaboratively contribute to the homeostasis of redox status in organisms, and those enzymes were also closely involved in the crustacean immune responses (81, 86–88). Specifically, the mRNA expressions of copper/zinc superoxide dismutase (Cu/ZnSOD), extracellular copper-zinc superoxide dismutase (ecCu/ZnSOD) and glutathione-s-transferases (GSTs) significantly upregulated when the crustacean challenging with virus infection (89–91). Mu et al. (86) reported that the involvement of peroxiredoxins 6 in responses against bacterial infection (86). In this study, the transcript of three SOD isoforms showed opposite expression trends during the molting cycle of P. trituberculatus, indicating that SOD can protect the body from injury by maintaining ROS homeostasis in crabs. Moreover, the transcript of glutathione peroxidase, glutathione-s-transferases (GSTs) and peroxiredoxin 6 significantly increased from stage AB to stage D and then decreased in stage E, illustrating that those endogenous antioxidant enzymes are important in maintaining redox equilibrium in crab.
As an antioxidant, selenoprotein M and selenoprotein W-like have been reported that involved in alleviating the oxidative stress and apoptosis caused by virus infection (92, 93). The expression level of selenoprotein (selenoprotein M and selenoprotein W-like) in this study increased significantly from stage AB to stage C and then decline gradually from stage C to stage E, indicating that the alleviation effect of selenoprotein on oxidative stress was the highest in the C stage and the lowest in the E stage. Heat shock proteins (HSPs), also known as stress proteins and extrinsic chaperones, are essential for the maintenance of cellular proteostasis (94). In recent years, increasing evidences have reported that HSPs (particularly HSP70) elicit a variety of immune responses to infection and modulate several inflammatory cascades (95–97). The higher expression level of heat shock protein 70 in this study was found from stage AB to stage C and then significantly decline in stage E, indicating that HSP 70 cannot effectively activate the immune pathway in stage E. Moreover, the transcript of HSP 90 in the hemolymph and hepatopancreas showed opposite expression pattern during the molting cycle of P. trituberculatus may relate to the multiple roles of HSP90. It has been reported that the HSP90 participated in the process of lipid droplet biogenesis and lipids homeostasis (98, 99). Except for the immune function, the hepatopancreas of crustaceans is the main organ for lipid metabolism, which provides energy for growth and development (100, 101).
In this study, transcriptome sequencing has explored the expression level of immune-related genes in the hemolymph and hepatopancreas during the molting cycle of P. trituberculatus. The present results showed that the expression levels of immune-related genes changed significantly throughout the molting cycle of P. trituberculatus. For example, genes related to GPCR and MAPK signalling pathways exhibit opposite expression patterns from stage AB to stage E. The expression level of Toll-like receptor was higher from stage AB to stage D of P. trituberculatus. The lowest expression level of ten genes related to pattern recognition proteins/receptors in the hepatopancreas was found in the E stage. The transcript levels of the genes (prophenoloxidase, prophenoloxidase-activating factor 1, serine protease 27) that involved in activating proPO system increased significantly from stage D to stage E. Moreover, the expression level of copper/zinc superoxide dismutase isoform 4, glutathione peroxidase, glutathione S-transferase, peroxiredoxin, peroxiredoxin 6, and dual oxidase 2 in stage C or stage D significantly higher than that of stage E or stage AB. The speculated keys genes and pathway for immune response during the molting cycle of P. trituberculatus based on the transcriptome analysis is summarized in Figure 11. This study results not only provide basic information to discover genes and pathways involved in immune regulation on the molting of swimming crab but also provided important clues for the innovation of crustacean disease prevention and control technology.
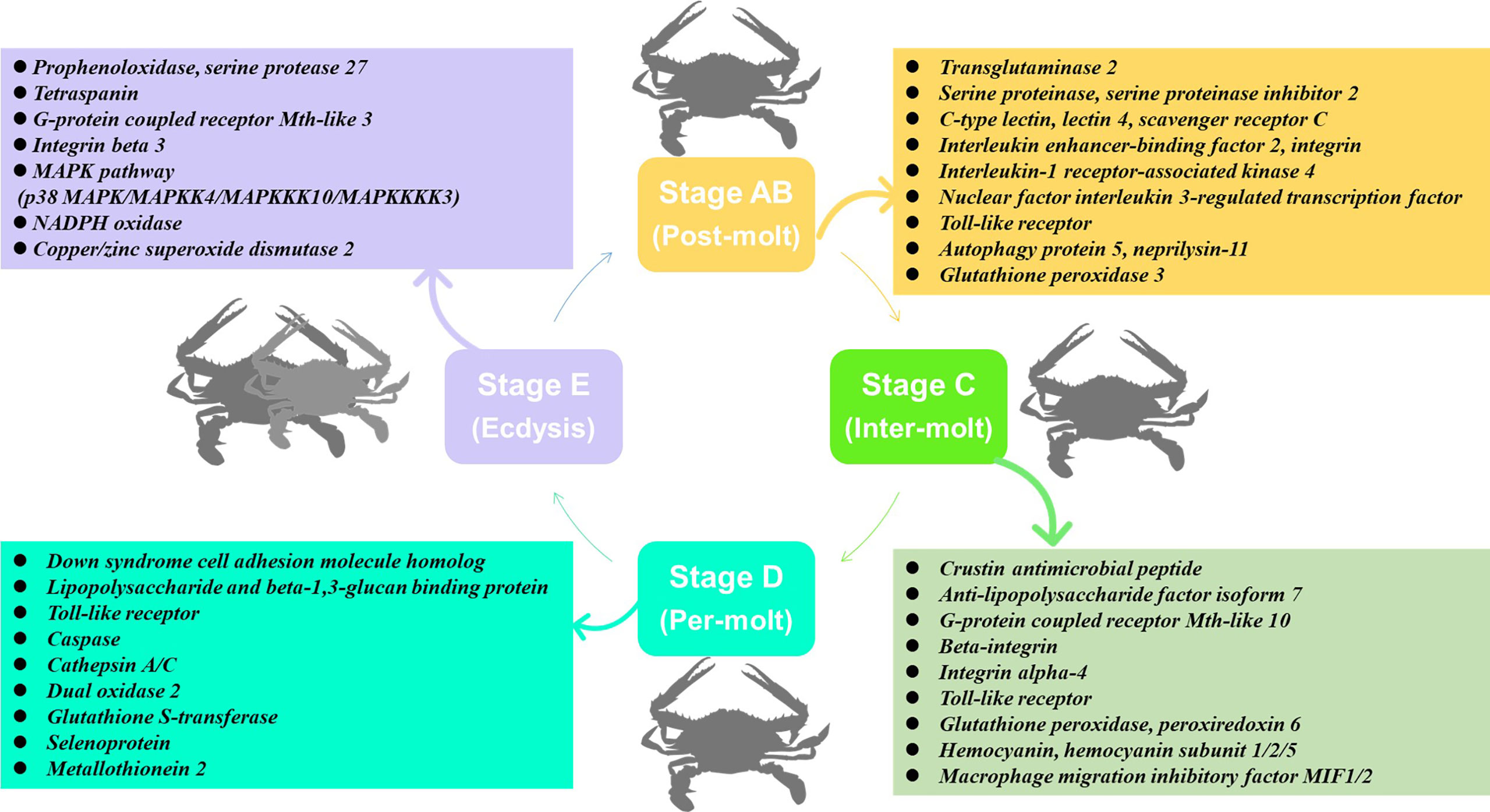
Figure 11 The speculated key genes and pathway for immune response during the molting cycle of P. trituberculatus based on the transcriptome analysis.
The data presented in the study are deposited in the Sequence Read Archive (SRA) repository, accession number: PRJNA855992.
ML: Experimental design, Writing - Original Draft, Data Curation. HN: Formal analysis, Data Curation. XZ: Data Curation, Validation. QS: Data Curation, Visualization. XW: Experimental design, Supervision. JH: Writing-Editing, Funding acquisition.
This study was funded by the two projects (No. 42106088 and No. 32172993) from the Natural Science Foundation of China, an open fund from Zhejiang Marine Fisheries Research Institute (2021KF005), a Basic Public Welfare Program of Zhejiang Province (Grant No. LGN22C190005), a Project of Major Agricultural Technology Cooperation Plan of Zhejiang Province (Grant No. 2020XTTGSC03 and No. 2022XTTGSC04), an Open-end Funds (SH20201205) of Jiangsu Key Laboratory of Marine Bioresources and Environment, a practice innovation training program projects (No. 202111641127Y and No. SY202257X) for the Jiangsu College students. Infrastructure costs were partially supported by the Project of Jiangsu Fisheries Science 537 and Technology (SZ-LYG202029).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Promwikorn W, Kirirat P, Thaweethamsewee P. Index of molt staging in the black tiger shrimp (Penaeus monodon). Songklanakarin J Sci Technol (2004) 26(5):765–72.
2. Zhou Z, Gu W, Wang C, Zhou Y, Tu D, Liu Z, et al. Seven transcripts from the chitinase gene family of the mud crab Scylla paramamosain: Their expression profiles during development and moulting and under environmental stresses. Aquac Res (2018) 49(10):3296–308. doi: 10.1111/are.13793
3. Hopkins P. Limb regeneration in the fiddler crab, Uca pugilator: Hormonal and growth factor control. Am Zool (2001) 41(3):389–98. doi: 10.1668/0003-1569
4. Morris S, Postel U, Mrinalini, Turner L, Palmer J, Webster S. The adaptive significance of crustacean hyperglycaemic hormone (CHH) in daily and seasonal migratory activities of the Christmas island red crab Gecarcoidea natalis. J Exp Biol (2010) 213(17):3062–73. doi: 10.1242/jeb.045153
5. Jung H, Lyons R, Hurwood D, Mather P. Genes and growth performance in crustacean species: a review of relevant genomic studies inVrustaceans and other taxa. Rev Aquacult (2013) 5(2):77–110. doi: 10.1111/raq.12005
6. Gao Y, Zhang X, Wei J, Sun X, Yuan J, Li F, et al. Whole transcriptome analysis provides insights into molecular mechanisms for molting in. Litopenaeus Vannamei. PloS One (2015) 10(12):e0144350. doi: 10.1371/journal.pone.0144350
7. Huang S, Wang J, Yue W, Chen J, Gaughan S, Lu W, et al. Transcriptomic variation of hepatopancreas reveals the energy metabolism and biological processes associated with molting in Chinese mitten crab. Eriocheir Sinensis. Sci Rep (2015) 5:14015. doi: 10.1038/srep14015
8. Hou W, Pan G, Long X, Wu X, Zhou W, Cheng Y. Effects of three diets on survival, molting, growth and biochemical composition of pre-adult Portunus trituberculatus. Chin J Zool (2016) 51:642–54. doi: 10.13859/j.cjz.201604016
9. Corteel M, Dantas-Lima J, Wille M, Alday-Sanz V, Pensaert M, Sorgeloos P, et al. Molt stage and cuticle damage influence white spot syndrome virus immersion infection in penaeid shrimp. Vet Microbiol (2009) 137(3-4):209–16. doi: 10.1016/j.vetmic.2009.01.018
10. Tumburu L, Shepard E, Strand A, Browdy C. Effects of endosulfan exposure and taura syndrome virus infection on the survival and molting of the marine penaeid shrimp. Litopenaeus Vannamei. Chemosphere (2012) 86(9):912–8. doi: 10.1016/j.chemosphere.2011.10.057
11. Xu Z, Liu A, Li S, Wang G, Ye H. Hepatopancreas immune response during molt cycle in the mud crab. Scylla Paramamosain. Sci Rep (2020) 10(1):13102. doi: 10.1038/s41598-020-70139-2
12. Zhang M, Zhang X, Tran N, Sun Z, Zhang X, Ye H, et al. Molting alters the microbiome, immune response, and digestive enzyme activity in mud crab (Scylla paramamosain). Msystems (2021) 6(5):e00917–21. doi: 10.1128/mSystems.00917-21
14. Yu C, Song H, Yao G, Lv H. Composition and distribution of economic crab species in the East China Sea. ET Limnol Sin (2006) 37:53–60. doi: 10.3321/j.issn:0029-814X.2006.01.009
15. Dong Z, Mao S, Chen Y, Ge H, Li X, Wu X, et al. Effects of air-exposure stress on the survival rate and physiology of the swimming crab Portunus trituberculatus. Aquaculture (2019) 500(1):429–34. doi: 10.1016/j.aquaculture.2018.10.049
16. Shi H, Xie J, Wang G, Xu W. Preliminary study on a pathogenic kinetoplastid protist in wild-captured Portunus trituberculatus. Mar Fish (2013) 35(3):309–15. doi: 10.3969/j.issn.1004-2490.2013.03.008
17. Duan H, Mao H, Xia Q, Ge H, Liu M, Li W, et al. Comparisons of growth performance, gonadal development and nutritional composition among monosex and mixed-sex culture modes in the swimming cab (Portunus trituberculatus). Aquac Res (2021) 52(7):3403–14. doi: 10.1111/are.15185
18. Management of Fisheries and Fishery Management. China Fisheries statistical yearbook in 2021. Beijing: Chinese Agricultural Press (2021).
19. Duan Q, Mai K, Shentu J, Gao Q, Yang J. Effects of dietary protein and lipid levels on growth and ovary pigmentation in Portunus trituberculatus. J Fish China (2011) 18:809–18. doi: 10.3724/SP.J.1118.2011.00809
20. Ding T, Liu Z. The factors that affecting the survival rate of Portunus trituberculatus during the period of pond culture and countermeasures. Hebei Fish (2009) 8):21–2. doi: 10.3969/j.issn.1004-6755.2009.08.008
21. Shen J, Zhu D, Hu Z, Qi Y, Wang C. Molt staging in the swimming crab portunus trituberculatus. J Fish China (2011) 35(10):1481–7. doi: 10.3724/SP.J.1231
22. Grabherr M, Haas B, Yassour M, Levin J, Thompson D, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol (2011) 29(7):644–U130. doi: 10.1038/nbt.1883
23. Jin Y, Olsen R, Harvey T, Ostensen M, Li K, Santi N, et al. Comparative transcriptomics reveals domestication-associated features of Atlantic salmon lipid metabolism. Mol Ecol (2020) 29(10):1860–72. doi: 10.1111/mec.15446
24. Ni Q, Li W, Liang X, Liu J, Ge H, Dong Z. Gill transcriptome analysis reveals the molecular response to the acute low-salinity stress in. Cyclina sinensis. Aquacult Rep (2021) 19:100564. doi: 10.1016/j.aqrep.2020.100564
25. Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta delta c) method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
26. Huang Y, Ren Q. Research progress in innate immunity of freshwater crustaceans. Dev Comp Immunol (2020) 104:103569. doi: 10.1016/j.dci.2019.103569
27. Li F, Xiang J. Recent advances in researches on the innate immunity of shrimp in China. Dev Comp Immunol (2013) 39(1-2):11–26. doi: 10.1016/j.dci.2012.03.016
28. Chen F, Wang K. Characterization of the innate immunity in the mud crab Scylla paramamosain. Fish Shellfish Immunol (2019) 93:436–48. doi: 10.1016/j.fsi.2019.07.076
29. Selsted M, Ouellette A. Mammalian defensins in the antimicrobial immune response. Nat Immunol (2005) 6(6):551–7. doi: 10.1038/ni1206
30. Smith V, Dyrynda E. Antimicrobial proteins: From old proteins, new tricks. Mol Immunol (2015) 68(2):383–98. doi: 10.1016/j.molimm.2015.08.009
31. Pan L, Yue F, Miao J, Zhang L, Jian L. Molecular cloning and characterization of a novel c-type lysozyme gene in swimming crab Portunus trituberculatus. Fish Shellfish Immunol (2010) 29(2):286–92. doi: 10.1016/j.fsi.2010.04.011
32. Wang H, Zhang J, Wang Y, Fang W, Wang Y, Zhou J, et al. Newly identified type II crustin (SpCrus2) in Scylla paramamosain contains a distinct cysteine distribution pattern exhibiting broad antimicrobial activity. Dev Comp Immunol (2018) 84:1–13. doi: 10.1016/j.dci.2018.01.021
33. Li S, Li F. The anti-lipopolysaccharide factors in crustaceans. Sub-cellular Biochem (2020) 94:63–80. doi: 10.1007/978-3-030-41769-7_3
34. Sirikharin R, Soderhall K, Soderhall I. The n-terminal peptide generated after activation of prophenoloxidase affects crayfish hematopoiesis. Dev Comp Immunol (2020) 108:103687. doi: 10.1016/j.dci.2020.103687
35. Cerenius L, Lee B, Soderhall K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol (2008) 29(6):263–71. doi: 10.1016/j.it.2008.02.009
36. Amparyup P, Charoensapsri W, Tassanakajon A. Prophenoloxidase system and its role in shrimp immune responses against major pathogens. Fish Shellfish Immunol (2013) 34(4):990–1001. doi: 10.1016/j.fsi.2012.08.019
37. Monwan W, Amparyup P, Tassanakajon A. A snake-like serine proteinase (PmSnake) activates prophenoloxidase-activating system in black tiger shrimp Penaeus monodon. Dev Comp Immunol (2017) 67:229–38. doi: 10.1016/j.dci.2016.09.016
38. Li M, Li C, Wang J, Song S. Immune response and gene expression in hemocytes of Portunus trituberculatus inoculated with the parasitic dinoflagellate hematodinium. Mol Immunol (2015) 65(1):113–22. doi: 10.1016/j.molimm.2015.01.002
39. Vazquez L, Alpuche J, Maldonado G, Agundis C, Pereyra-Morales A, Zenteno E. Immunity mechanisms in crustaceans. Innate Immun (2009) 15(3):179–88. doi: 10.1177/1753425909102876
40. Drickamer K, Taylor M. Recent insights into structures and functions of c-type lectins in the immune system. Curr Opin Struct Biol (2015) 34:26–34. doi: 10.1016/j.sbi.2015.06.003
41. Wang X, Xu J, Zhao X, Vasta G, Wang J. A shrimp c-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. J Biol Chem (2014) 289(17):11779–90. doi: 10.1074/jbc.M114.552307
42. Luo T, Yang H, Li F, Zhang X, Xu X. Purification, characterization and cDNA cloning of a novel lipopolysaccharide-binding lectin from the shrimp Penaeus monodon. Dev Comp Immunol (2006) 30(7):607–17. doi: 10.1016/j.dci.2005.10.004
43. Zhang X, Pan L, Yu J, Huang H. One recombinant c-type lectin (LvLec) from white shrimp Litopenaeus vannamei affected the haemocyte immune response in vitro. Fish Shellfish Immunol (2019) 89:35–42. doi: 10.1016/j.fsi.2019.03.029
44. Huang M, Mu C, Wu Y, Ye F, Wang D, Sun C, et al. The functional characterization and comparison of two single CRD containing c-type lectins with novel and typical key motifs from Portunus trituberculatus. Fish Shellfish Immunol (2017) 70:398–407. doi: 10.1016/j.fsi.2017.09.029
45. Sanchez-Salgado J, Pereyra M, Agundis C, Vivanco-Rojas O, Sierra-Castillo C, Alpuche-Osorno J, et al. Participation of lectins in crustacean immune system. Aquac Res (2017) 48(8):4001–11. doi: 10.1111/are.13394
46. Yang M, Shi X, Yang H, Sun J, Xu L, Wang X, et al. Scavenger receptor c mediates phagocytosis of white spot syndrome virus and restricts virus proliferation in shrimp. PloS Pathog (2016) 12(12):e1006127. doi: 10.1371/journal.ppat.1006127
47. Tang M, Li X, Yang L, Wang Q, Li W. A class b scavenger receptor mediates antimicrobial peptide secretion and phagocytosis in Chinese mitten crab (Eriocheir sinensis). Dev Comp Immunol (2020) 103:103496. doi: 10.1016/j.dci.2019.103496
48. Li X, Yang L, Li D, Zhu Y, Wang Q, Li W. Pathogen-specific binding soluble down syndrome cell adhesion molecule (Dscam) regulates phagocytosis &ITvia&IT membrane-bound dscam in crab. Front Immunol (2018) 9:801. doi: 10.3389/fimmu.2018.00801
49. Li W. Dscam in arthropod immune priming: What is known and what remains unknown. Dev Comp Immunol (2021) 125:104231. doi: 10.1016/j.dci.2021.104231
50. Phupet B, Pitakpornpreecha T, Baowubon N, Runsaeng P, Utarabhand P. Lipopolysaccharide- and beta-1,3-glucan-binding protein from Litopenaeus vannamei: Purification, cloning and contribution in shrimp defense immunity via phenoloxidase activation. Dev Comp Immunol (2018) 81:167–79. doi: 10.1016/j.dci.2017.11.016
51. Ponprateep S, Vatanavicharn T, Lo C, Tassanakajon A, Rimphanitchayakit V. Alpha-2-Macroglobulin is a modulator of prophenoloxidase system in pacific white shrimp Litopenaeus vannamai. Fish Shellfish Immunol (2017) 62:68–74. doi: 10.1016/j.fsi.2016.12.028
52. Wang B, Li F, Xiang J, Gui L, Luo Z, Yan H. Three tetraspanins from Chinese shrimp, Fenneropenaeus chinensis, may play important roles in WSSV infection. J Fish Dis (2010) 33(1):15–29. doi: 10.1111/j.1365-2761.2009.01079.x
53. Zhu X, Yang X, He W, Xiong Y, Liu J, Dai Z. Involvement of tetraspanin 8 in the innate immune response of the giant prawn, Macrobrachium rosenbergii. Fish Shellfish Immunol (2019) 86:459–64. doi: 10.1016/j.fsi.2018.11.055
54. Li J, Sun J, Dong X, Geng X, Qiu G. Transcriptomic analysis of gills provides insights into the molecular basis of molting in Chinese mitten crab (Eriocheir sinensis). Peerj (2019) 7:e7182. doi: 10.7717/peerj.7182
55. Dong C, Zhang P. A putative G protein-coupled receptor involved in innate immune defense of Procambarus clarkii against bacterial infection. Comp Biochem Phys A Mol Integr Physiol (2012) 161(2):95–101. doi: 10.1016/j.cbpa.2011.09.006
56. Qin X, Jin X, Zhou K, Li H, Wang Q, Li W, et al. EsGPCR89 regulates cerebral antimicrobial peptides through hemocytes in Eriocheir sinensis. Fish Shellfish Immunol (2019) 95:151–62. doi: 10.1016/j.fsi.2019.10.015
57. Cao X, Zhang CC, Zhang R, Wang K, Dai X, Huang X, et al. Leucine-rich repeat-containing G-protein-coupled receptor 2 (LGR2) can regulate PO activity and AMP genes expression in Macrobrachium nipponense. Mol Immunol (2020) 126:14–24. doi: 10.1016/j.molimm.2020.07.013
58. Raman M, Chen W, Cobb M. Differential regulation and properties of MAPKs. Oncogene (2007) 26(22):3100–12. doi: 10.1038/sj.onc.1210392
59. He Y, Yao W, Liu P, Li J, Wang Q. Expression profiles of the p38 MAPK signaling pathway from Chinese shrimp I Fenneropenaeus chinensis in response to viral and bacterial infections. Gene (2018) 642:381–8. doi: 10.1016/j.gene.2017.11.050
60. Hansen T, Jorgensen J. Cloning and characterisation of p38 MAP kinase from Atlantic salmon - a kinase important for regulating salmon TNF-2 and IL-1 beta expression. Mol Immunol (2007) 44(12):3137–46. doi: 10.1016/j.molimm.2007.02.006
61. He S, Qian Z, Yang J, Wang X, Mi X, Liu Y, et al. Molecular characterization of a p38 MAPK from Litopenaeus vannamei and its expression during the molt cycle and following pathogen infection. Dev Comp Immunol (2013) 41(2):217–21. doi: 10.1016/j.dci.2013.05.010
62. Wang S, Yin B, Li H, Xiao B, Lu K, Feng C, et al. MKK4 from Litopenaeus vannamei is a regulator of p38 MAPK kinase and involved in anti-bacterial response. Dev Comp Immunol (2018) 78:61–70. doi: 10.1016/j.dci.2017.09.015
63. Zhang R, Wan K, Liu Y, Wang Z, Zhang D, Yin H. Expression pattern of mitogen-activated protein kinase kinase 4 and regulation to antibacterial factor ABF-1/2 in response to bacterial challenge from artemia parthenogenetica. Fish Shellfish Immunol (2021) 115:35–42. doi: 10.1016/j.fsi.2021.03.008
64. Humphries J, Byron A, Humphries M. Integrin ligand at a glance. J Cell Sci (2006) 119(19):3901–3. doi: 10.1242/jcs.03098
65. Huang Y, Zhao L, Feng J, Zhu H, Huang X, Ren Q, et al. A novel integrin function in innate immunity from Chinese mitten crab (Eriocheir sinensis). Dev Comp Immunol (2015) 52(2):155–65. doi: 10.1016/j.dci.2015.05.005
66. Lin Y, Chen J, Chen Y, Liu C, Cheng W, Hsu C, et al. Characterization of white shrimp Litopenaeus vannamei integrin beta and its role in immunomodulation by dsRNA-mediated gene silencing. Dev Comp Immunol (2013) 40(2):167–79. doi: 10.1016/j.dci.2013.01.001
67. Tang X, Wang X, Zhan W. An integrin β subunit of Chinese shrimp fenneropenaeus chinensis involved in WSSV infection. Aquaculture (2012) 368-369:1–9. doi: 10.1016/j.aquaculture.2012.08.004
68. Li D, Zhang M, Yang H, Zhu Y, Xu X. Beta-integrin mediates WSSV infection. Virology (2007) 368(1):122–32. doi: 10.1016/j.virol.2007.06.027
69. Escobedo-Bonilla C, Alday-Sanz V, Wille M, Sorgeloos P, Pensaert M, Nauwynck H. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J Fish Dis (2008) 31(1):1–18. doi: 10.1111/j.1365-2761.2007.00877.x
70. Yu A, Jin X, Guo X, Li S, Wu M, Li W, et al. Two novel toll genes (EsToll1 and EsToll2) from Eriocheir sinensis are differentially induced by lipopolysaccharide, peptidoglycan and zymosan. Fish Shellfish Immunol (2013) 35(4):1282–92. doi: 10.1016/j.fsi.2013.07.044
71. Sanchez-Paz A, Muhlia-Almazan A. Uncovering and defragmenting the role of the toll pathway in the innate immune responses of cultured crustaceans against viral pathogens. Rev Aquacult (2020) 12(3):1818–35. doi: 10.1111/raq.12411
72. Phongdara A, Wanna W, Chotigeat W. Molecular cloning and expression of caspase from white shrimp penaeus merguiensis. Aquaculture (2006) 252(2-4):114–20. doi: 10.1016/j.aquaculture.2005.07.024
73. Voll R, Herrmann M, Roth E, Stach C, Kalden J, Girkontaite I. Immunosuppressive effects of apoptotic cells. Nature (1997) 390(6658):350–1. doi: 10.1038/37022
74. Takle H, Andersen O. Caspases and apoptosis in fish. J Fish Biol (2007) 71:326–49. doi: 10.1111/j.1095-8649.2007.01665.x
75. Jin Z, El-Deiry W. Overview of cell death signaling pathways. Cancer Biol Ther (2005) 4(2):139–63. doi: 10.4161/cbt.4.2.1508
76. Gong Y, Chen J, Cui Y, Li S. MiR-2 montributes to WSSV infection by targeting caspase 2 in mud crab (Scylla paramamosain). Dev Comp Immunol (2021) 125:104217. doi: 10.1016/j.dci.2021.104217
77. Li Y, Xie X, Ma X, Liu H. Identification of a CqCaspase gene with antiviral activity from red claw crayfish Cherax quadricarinatus. Dev Comp Immunol (2019) 91:101–7. doi: 10.1016/j.dci.2018.10.012
78. Wang L, Zhi B, Wu W, Zhang X. Requirement for shrimp caspase in apoptosis against virus infection. Dev Comp Immunol (2008) 32(6):706–15. doi: 10.1016/j.dci.2007.10.010
79. Coates C, Nairn J. Diverse immune functions of hemocyanins. Dev Comp Immunol (2014) 45(1):43–55. doi: 10.1016/j.dci.2014.01.021
80. Kiruthiga C, Rajesh S, Rashika V, Priya R, Narayanan R. Molecular cloning, expression analysis and characterization of peroxiredoxin during WSSV infection in shrimp Fenneropenaeus indicus. J Invertebr Pathol (2012) 109(1):52–8. doi: 10.1016/j.jip.2011.09.006
81. Li Y, Zhan F, Li F, Lu Z, Xu Z, Yang Y, et al. Molecular and functional characterization of mitochondrial manganese superoxide dismutase from Macrobrachium rosenbergii during bacterial infection. Fish Shellfish Immunol (2021) 118:94–101. doi: 10.1016/j.fsi.2021.08.025
82. Leto T, Geiszt M. Role of nox family NADPH oxidases in host defense. Antioxid Redox Signal (2006) 8(9-10):1549–61. doi: 10.1089/ars.2006.8.1549
83. Bogdan C, Rollinghoff M, Diefenbach A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr Opin Immunol (2000) 12(1):64–76. doi: 10.1016/s0952-7915(99)00052-7
84. Chainy G, Paital B, Dandapat J. An overview of seasonal changes in oxidative stress and antioxidant defence parameters in some invertebrate and vertebrate species. Scientifica (2016) 2016:6126570. doi: 10.1155/2016/6126570
85. Li M, Zheng Y, Liang H, Zou L, Sun J, Zhang Y, et al. Molecular cloning and characterization of cat, Gpx1 and Cu/Zn-sod genes in pengze crucian carp (Carassius auratus var. pengze) and antioxidant enzyme modulation induced by hexavalent chromium in juveniles. Comp Biochem Physiol C Toxicol Pharmacol (2013) 157(3):310–21. doi: 10.1016/j.cbpc.2013.02.003
86. Mu C, Zhao J, Wang L, Song L, Zhang H, Li C, et al. Molecular cloning and characterization of peroxiredoxin 6 from Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol (2009) 26(6):821–7. doi: 10.1016/j.fsi.2008.10.006
87. Yang Y, Bazhin A, Werner J, Karakhanova S. Reactive oxygen species in the immune system. Int Rev Immunol (2013) 32(3):249–70. doi: 10.3109/08830185.2012.755176
88. Cheng C, Ma H, Deng Y, Feng J, Chen X, Guo Z. Glutathione peroxidase 3 in the mud crab Scylla paramamosain: Characterization and regulation under nitrite stress. Comp Biochem Physiol C Toxicol Pharmacol (2020) 229:108673. doi: 10.1016/j.cbpc.2019.108673
89. Xiu Y, Wu T, Du J, Yao W, Li W, Ding Z, et al. Molecular characterization and expression analysis of extracellular copper/zinc superoxide dismutase (ecCuZnSOD) from oriental river prawn, Macrobrachium nipponense. Aquaculture (2013) 380:23–8. doi: 10.1016/j.aquaculture.2012.07.014
90. Hung M, Shiomi R, Nozaki R, Kondo H, Hirono I. Identification of novel copper/zinc superoxide dismutase (Cu/ZnSOD) genes in kuruma shrimp Marsupenaeus japonicus. Fish Shellfish Immunol (2014) 40(2):472–7. doi: 10.1016/j.fsi.2014.07.030
91. Zhao D, Chen L, Qin C, Zhang H, Wu P, Zhang F. A delta-class glutathione transferase from the Chinese mitten crab Eriocheir sinensis: cDNA cloning, characterization and mRNA expression. Fish Shellfish Immunol (2010) 29(4):698–703. doi: 10.1016/j.fsi.2010.06.002
92. Chintapitaksakul L, Udomkit A, Smith D, Panyim S, Sonthayanon B. Expression analysis of selected haemocyte transcripts from black tiger shrimp infected with yellow head virus. Scienceasia (2008) 34(3):327–33. doi: 10.2306/scienceasia1513-1874.2008.34.327
93. Clavero-Salas A, Sotelo-Mundo R, Gollas-Galvan T, Hernandez-Lopez J, Peregrino-Uriarte A, Muhlia-Almazan A, et al. Transcriptome analysis of gills from the white shrimp Litopenaeus vannamei infected with white spot syndrome virus. Fish Shellfish Immunol (2007) 23(2):459–72. doi: 10.1016/j.fsi.2007.01.010
94. Junprung W, Supungul P, Tassanakajon A. Structure, gene expression, and putative functions of crustacean heat shock proteins in innate immunity. Dev Comp Immunol (2021) 115:103875. doi: 10.1016/j.dci.2020.103875
95. Wang Q, Huang C, Liu K, Lu M, Dan S, Xu Y, et al. Cloning and expression of three heat shock protein genes in the gills of Cherax quadricarinatus responding to bacterial challenge. Microb Pathogenesis (2020) 142:104043. doi: 10.1016/j.micpath.2020.104043
96. Junprung W, Supungul P, Tassanakajon A. HSP70 and HSP90 are involved in shrimp Penaeus vannamei tolerance to AHPND-causing strain of vibrio parahaemolyticus after non-lethal heat shock. Fish Shellfish Immunol (2017) 60:237–46. doi: 10.1016/j.fsi.2016.11.049
97. Yuan K, Yuan F, He H, Bi H, Weng S, He J, et al. Heat shock 70 kDa protein cognate 5 involved in WSSV toleration of Litopenaeus vannamei. Dev Comp Immunol (2017) 72:9–20. doi: 10.1016/j.dci.2017.02.003
98. Horvath I, Balogh G, Peter M, Gombos I, Glatz A, Tim C, et al. Interrelationship between lipid droplet (LD) biogenesis and heat shock protein (Hsp) response. Chem Phys Lipids (2010) 163:S60–0. doi: 10.1016/j.chemphyslip.2010.05.179
99. Kuan Y, Hashidume T, Shibata T, Uchida K, Shimizu M, Inoue J, et al. Heat shock protein 90 modulates lipid homeostasis by regulating the stability and function of sterol regulatory element-binding protein (SREBP) and SREBP cleavage-activating protein. J Biol Chem (2017) 292(7):3016–28. doi: 10.1074/jbc.M116.767277
100. Vogt G. Life-cycle and functional cytology of the hepatopancreatic cells of Astacus astacus (Crustacea, decapoda). Zoomorphology (1994) 114:83–101. doi: 10.1007/BF00396642
Keywords: Portunus trituberculatus, molting cycle, immunity, antioxidative enzyme system, comparative transcriptome
Citation: Liu M, Ni H, Zhang X, Sun Q, Wu X and He J (2022) Comparative transcriptomics reveals the immune dynamics during the molting cycle of swimming crab Portunus trituberculatus. Front. Immunol. 13:1037739. doi: 10.3389/fimmu.2022.1037739
Received: 06 September 2022; Accepted: 10 October 2022;
Published: 31 October 2022.
Edited by:
Linlin Zhang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Liu, Ni, Zhang, Sun, Wu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie He, aGVfMDkwMkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.