- 1School of Animal Husbandry and Veterinary Medicine, Jiangsu Vocational College of Agriculture and Forestry, Jurong, China
- 2College of Animal Science and Technology, Chinese Agricultural University, Beijing, China
- 3Key Laboratory of Agricultural Animal Breeding and Healthy Breeding of Tianjin, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China
Abundant microbiota resides in the organs of the body, which utilize the nutrition and form a reciprocal relationship with the host. The composition of these microbiota changes under different pathological conditions, particularly in response to stress and digestive diseases, making the microbial composition and health of the hosts body interdependent. Probiotics are living microorganisms that have demonstrated beneficial effects on physical health and as such are used as supplements to ameliorate symptoms of various digestive diseases by optimizing microbial composition of the gut and restore digestive balance. However, the supplementary effect does not achieve the expected result. Therefore, a targeted screening strategy on probiotic bacteria is crucial, owing to the presence of several bacterial strains. Core bacteria work effectively in maintaining microbiological homeostasis and stabilization in the gastrointestinal tract. Some of the core bacteria can be inherited and acquired from maternal pregnancy and delivery; others can be acquired from contact with the mother, feces, and the environment. Knowing the genera and functions of the core bacteria could be vital in the isolation and selection of probiotic bacteria for supplementation. In addition, other supporting strains of probiotic bacteria are also needed. A comprehensive strategy for mining both core and supporting bacteria before its clinical use is needed. Using metagenomics or other methods of estimation to discern the typically differentiated strains of bacteria is another important strategy to treat dysbiosis. Hence, these two factors are significant to carry out targeted isolation and selection of the functional strains to compose the resulting probiotic preparation for application in both research and clinical use. In conclusion, precise probiotic supplementation, by screening abundant strains of bacteria and isolating specific probiotic strains, could rapidly establish the core microbiota needed to confer resilience, particularly in bacterial dysfunctional diseases. This approach can help identify distinct bacteria which can be used to improve supplementation therapies.
1 Introduction
Probiotics are live bacteria that confer health benefits to the host when administered in adequate quantities (1). Recent studies have demonstrated that the host’s microbiota plays an important role in maintaining overall health, and is thus an attractive target for clinical interventions (2, 3). In humans and animals, the gastrointestinal tract (GIT) contains a dense population of microorganisms that cohesively interplay with the host’s digestion and help to fight against infections (2, 4). Some of the bacterial strains present belong to core genera that may represent a significant proportion of the GIT microbiota or form smaller dominant microbial groups (5). However, many factors, such as alcohol consumption and high-energy diets containing lots of carbohydrates and proteins, can cause dysbiosis of the GIT (6). Significant consequences of nutritional and metabolic diseases are disturbance of the GIT microbiota, especially under stress conditions (7, 8). Treat these diseases involves restoring the microecological environment of the GIT. Thus, as an alternative to administering antibiotic drugs, supplementation with probiotic bacteria has been found to be an effective approach. Scientists have developed a growing interest in assessing the ability of probiotics to enhance the health of humans or animals experiencing microbial dysbiosis (9). Numerous strains of bacteria, from both humans and animals, have been isolated from different organs (10). Furthermore, these isolated probiotic bacteria have been used in products designed and developed for clinical use, both in human medicine and animal husbandry and breeding (6, 11).
Currently the most widely used probiotics such as Bifidobacteria, Propionibacteria, Lactobacillus, Bacillus, Akkermansia muciniphila, and Saccharomyces play important roles when used and administrated in certain strains and approaches. Supplementing these probiotics can help modulate the GIT microbiota, thereby alleviating symptoms of inflammatory bowel diseases. The myriad of benefits associated with probiotic supplementation are shown in Table 1 (12–27). However, it is unclear if probiotic bacteria containing products are effective in treating all cases of dysbacteriosis or if they can function as a nutritional supplement (28). The probiotic bacteria used in clinical did not realize the respected aims even meaningless (29–31). Optimal methods of isolating and identifying suitable probiotic bacterial strains for use in humans and animals are yet to be determined.
The review discusses the intrinsic roles of bacteria in the GIT and typical microbial differences observed in digestive diseases and stress-induced micro dysbiosis. Identifying the unique strains of bacteria associated with intestinal microbial dysfunctional diseases is greatly important for developing effective treatments for the disease. This review also aims to discuss strategies for the precise choice of probiotic bacteria based on how the host’s microbiota needs to be modulated to restore balance, to provide researchers with effective methods to isolate probiotic bacteria and reduce candidate strains to achieve effective targeting and precise supplementation for clinical applications.
2 Causal relationship between dysbacteriosis and diseases
The gut microbiota consists of trillions of microbial cells belonging to many different strains of bacteria. This complex group of microorganisms is established shortly after birth and is subsequently influenced by factors such as diet, geography, genetics, medications, and lifestyle (32). Since the GIT is the site of digestion and absorption of nutrients, the microbial composition is primarily influenced by nutritional changes (33). Under homeostatic conditions, the gut microbiota is in a reciprocal relationship with the host and has important roles in maintaining health in relation to food metabolism, pathogen defense, immune training, production of important metabolites, and neuro-endocrine regulation (34). Most of the constituent strains are the same across different healthy individuals. The diversity and similarity in microbial composition of the GIT can both be caused by or result in digestive disease, especially under stress conditions (8).
Nutritional and metabolic disease threatens human health, and are therefore a focus of studies on the relationship between altered microbial composition and poor physical health (35, 36). Nutritional and metabolic diseases include obesity, gout, and non-alcoholic fatty liver disease. Consuming a diet rich in carbohydrates and proteins usually causes an alteration in the composition of GIT bacteria (37). Obesity is an epidemic phenomenon and is a prime risk factor for type 2 diabetes, gout, and cardiovascular diseases (38). In general, obese individuals show lower microbial diversity and gene richness than do non-obese individuals, and the difference in phylum levels appeared to be significant (39). An increased abundance of the phylum Firmicutes and a decreased abundance of Bacteroidetes have been observed in obese individuals. In mouse models, the caecum was found to normally be dominated by Firmicutes (60–80% of the phylotypes) and Bacteroidetes (20–40% of the phylotypes), but an approximately 50% reduction in the abundance of Bacteroidetes can be observed in obese mice relative to lean mice (40, 41). An increased ratio (5:1 to 6:1) of Firmicutes to Bacteroidetes is regularly be observed in obese humans and animals. According to the conceptual framework of Koch’s postulates, microbial dysbiosis is both a symptom and cause of many diseases (42, 43). Gut microbiota transplantation experiments have revealed a causal correlation between the gut microbiota and the development of obesity and type 2 diabetes (42). When germ-free (GF) mice were colonized with GIT microbiota isolated from obese mice, the GF mice acquired more body fat than mice colonized with microbiota from lean mice, indicating a contributory effect of the microbiota composition to obesity (5, 37).
Obesity, type 2 diabetes, and inflammatory bowel diseases (IBDs), including Crohn’s disease and ulcerative colitis, are chronic inflammatory conditions of the intestinal tract that affect humans and cause significant morbidity and occasional mortality (44, 45). IBD patients tend to have low bacterial diversity as well as lower numbers of Bacteroidetes and Firmicutes. These factors, together, reduced concentrations of microbial-derived butyrate. Butyrate and other short chain fatty acids are thought to have a direct anti-inflammatory effect in the gut (45). IBDs are often accompanied by microbial dysfunction which is not simply caused by a single pathogen. Furthermore, different indices of IBD activities have each been characterized by specific gut mucosa-attached bacteria. Strains of probiotic bacteria have proved effective in reducing IBD symptoms by improving the microbial composition and repairing the mucous membrane (46). However, there are no suitable cures containing probiotics and studies on the use of probiotics in clinical applications have mixed results (47, 48).
3 Distinguishing microbial differences associated with diseases
In nutritional and metabolic diseases, significant microbial differences have been observed at the phylum and genus levels. Similar results were observed for colon cancer, obesity, and non-alcoholic fatty liver disease (49).
Especial in little fatties’ children, with some obesity was inheritable, the overweight caused hyperglycemia, hypertension, and dyspnea. This situation has been popular in high carbohydrate contained diet. The fecal transplanted from the obesity children to germ free mice caused the same syndromes (Figure 1A). The strain of Enterobacter cloacae B29 was proved as the prime culprit to little fatties’ children syndrome. The toxin produced by Enterobacter cloacae B29 can promote the syndrome (43). Toxins such LPS produced through playing pleiotropic role one pathway was recognized by toll like receptor 4 (TLR4) (Figure 1B) to upstream signal pathway to regulate gut permeability. Classic mitogen-activated protein kinases (MAPK), which were divided into 4 subgroups: ERK/p38/JNK and BMK1 (50). The signaling pathways involved include the ERK/p38/JNK and nuclear transcription factor (NF-kB) pathways were enrolled to control adipose tissue metabolism through mediating cannabinoid-driven adipogenesis, which accelerated the syndrome of obesity (50, 51). Cross talk between the endotoxins produced by the strain and the TLR4 of the host is the most upstream and vital molecular event responsible for inducing all the phenotypes of obesity and other digestive diseases (52–54). Overgrowth in the human gut of these nonvirulent endotoxin-producing strains of pathogenic bacteria species, may collectively become a predictive biomarker or serve as a novel therapeutic target for treatment of obesity, non-alcoholic fatty liver disease, and other related metabolic disorders (55).
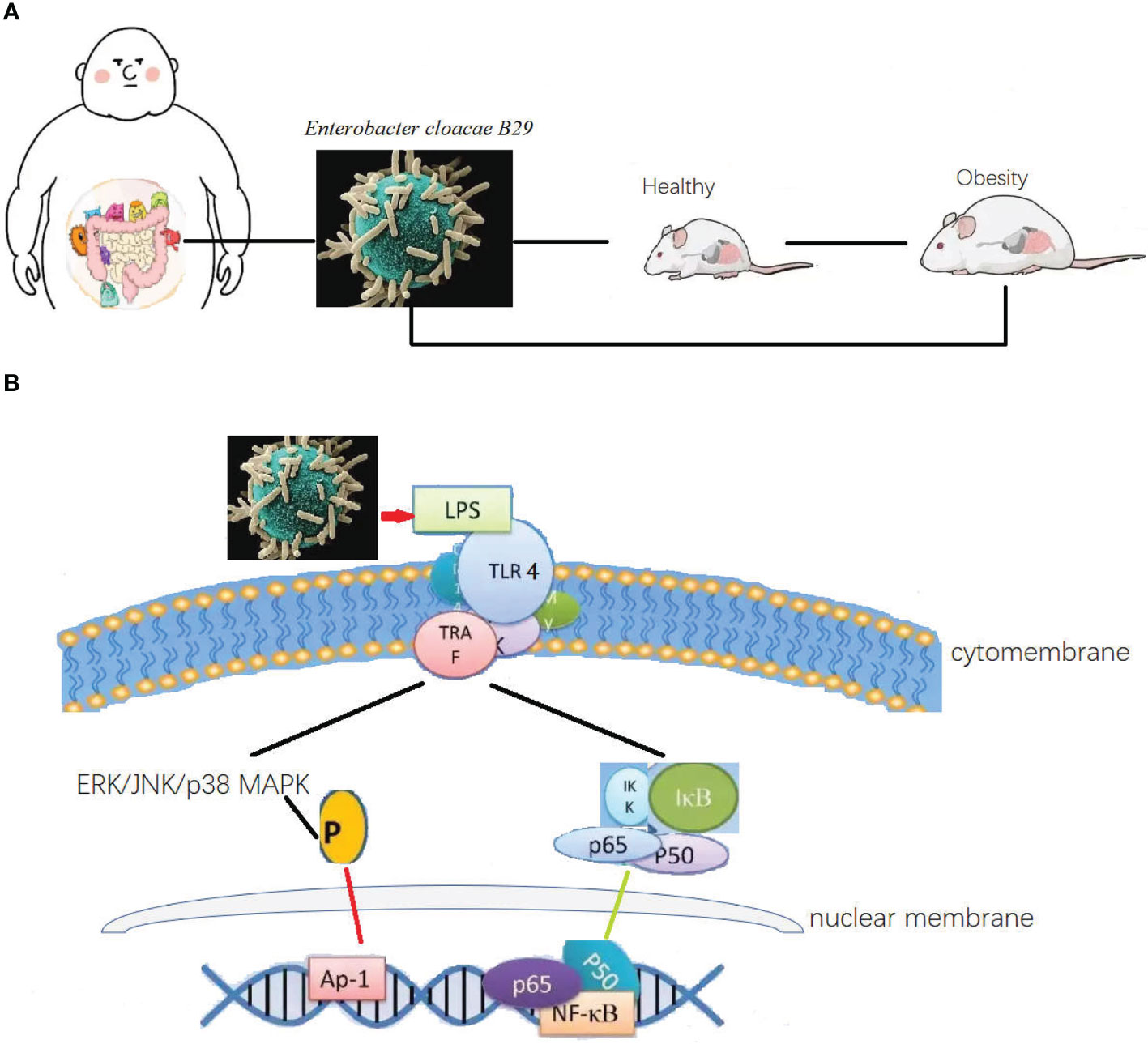
Figure 1 The differential bacteria in obesity and as a causation to obese. (A) Intestinal bacteria was both the cause and syndrome for the obesity. Causation relationship between the disease and organic bacteria. Microbiota transplanted from the obesity children to germ free mice caused the same syndromes. The strain of Enterobacter cloacae B29 was proved as the prime culprit to little fatties’ children syndrome. Toxins such LPS produced through playing pleiotropic role one pathway was recognized by toll like receptor 4 to upstream signal pathway to regulate gut permeability, and control adipose tissue metabolism through mediating cannabinoid-driven adipogenesis, which accelerated the syndrome of obesity. (B) Then possible molecular signal pathway of Enterobacter cloacae B29 regulated the adipose tissue metabolism. The toxin produced by Enterobacter cloacae B29 can promote the syndrome. Toxins such LPS produced through playing pleiotropic role one pathway was recognized by toll like receptor 4 to upstream signal pathway to regulate gut permeability. The signaling pathways involved include the ERK/p38/JNK and nuclear transcription factor pathways were enrolled to control adipose tissue metabolism through mediating cannabinoid-driven adipogenesis, which accelerated the syndrome of obesity.
The commensal bacterial species Bacteroides fragilis, Fusobacterium nucleatum, and Escherichia coli (E. coli) seem to emerge as pathogens and contribute to colorectal carcinogenesis through their inflammatory and oncogenic properties (53). Additionally, Bacteroides fragilis has been shown to be enriched in the gut microbiota of patients with colorectal cancer (54). Along with an increased abundance of Bacteroides fragilis, a decreased population of Bacteroides vulgatus and Bacteroides stercoris has also been observed in the guts of patients with human colorectal cancer (52, 55, 56). Studies on specific bacterial species associated with obesity and non-alcoholic fatty liver disease, and their molecular cross talk with the host, have suggested that overgrowth of nonvirulent endotoxin-producing strains of pathogenic bacteria, such as Enterobacter cloacae B29, E. coli PY102, and Klebsiella pneumoniae A7, in the gut of obese humans can act as causative agents for non-alcoholic fatty liver disease (54).
The microbial composition of the GIT is affected by diet and disease, and is also a typical symptom of many diseases. Further, microbial translocation to the rest of the body can make them causative agents of disease (57). All these results suggest that the microbial composition is not only the causative agent, but also an outcome of the development of various diseases.
Diarrhea and common infectious diseases of the GIT are mainly caused by infection-causing pathogens or endogenous opportunistic pathogens, accompanied with other typical clinical symptoms (58). In patients with diarrhea, the water content of the excreta increases more than that under healthy conditions. The microbial composition is disturbed by endogenous opportunistic pathogens. The functioning of the intestinal barrier is reduced and water filters more readily through the mucous membrane to reach the inner lumen of the intestine. Additionally, an increase in the abundance of endogenous E. coli or Salmonella typhi results in further deterioration of symptoms (59–62). The microbial composition of the gut can be both a causative agent and result of diarrhea and infectious gastrointestinal diseases. Toxins and endotoxins produced by pathogens or overgrowth of opportunistic pathogens act to impair and destroy the mucous membrane through a cascade of molecular signals, which is recognized by TLR4 (63), which presents the signal to MyD88, thereby stimulating the downstream expression of interleukin-1β (IL-1β) and tumor necrosis factor α (TNF-α). The signaling pathways involved include the classic nuclear transcription factor (NF-kB) or c-Jun N-terminal kinase pathways (Figure 1B) which act to up-regulate inflammation and suppress the activity of T regulatory cells (Tregs) (64), which were partly agreed with the obesity caused by Enterobacter cloacae B29. Under unfavorable environmental conditions, such as high ammonia concentration, heat, cold, and transportation, animals can suffer abnormal bodily stress. Stress can induce alterations in the microbiota of the digestive and respiratory tracts. Furthermore, the altered microbiota can negatively impact the health of the animal through microbial translocation.
4 Common supplementary methods of probiotic bacteria
Considering the relationship between organs of the body and microbiota, the restoration of microbiota should be the first step in the treatment of dysbiosis-related diseases. Resilience of dominant bacteria in the GIT of patients can be the most meaningful strategy for in restoring digestive balance (52, 65, 66). Fecal microbiota transplantation (FMT), which involves preparation and administration of distal gut microbiota-containing fecal material from healthy donors to a patient with a disrupted gut microbiota is, a promising strategy in the treatment of microbiota dysbiosis diseases. FMT acts to directly interfere with gut microbiota receptors, thus normalizing the microbial composition and producing therapeutic effects (67, 68). FMT has widely employed since 2013, when the United States Food and Drug Administration approved FMT for treatment of Clostridium difficile infection (69, 70). Since then, the range of its applications extended rapidly and broadly, not only for the treatment of gastrointestinal disorders, but also for extra-gastrointestinal disease, such as obesity, type 2 diabetes, and even cancer (71). Patients with Crohn’s disease, obesity, type 2 diabetes, and chronic IBD who received FMT treatment experienced therapeutic benefits (72, 73). Thus, the restoration of the microbial community is important to overcoming disease.
Reduce the over-growth of bacteria and conditioned bacteria, eliminate the pathogen. For the treatment of diarrhea and infectious gastrointestinal diseases, physicians often prescribe tablets containing strains of probiotic bacteria, as an alternative to antibiotic drugs, owing to the numerous side effects of antibiotics, particularly antibiotic resistance. Many strains of drug resistant bacteria, such as carbapenem-resistant Enterobacteriaceae, vancomycin-resistant Enterococcus, and extended spectrum beta-lactamase carrying strains, represent a major public health concern, as they are potential pathogens associated with a high mortality rate (74). Thus, the common genera of probiotic bacteria, including Bacillus, Bifidobacterium, Akkermansia, and Clostridium have often been used to inhibit E. coli or Salmonella growth (71, 75, 76). However, if the microbial composition of the patient is unknown, a generalized use of probiotic bacteria may not provide the desired effect.
An alternative means to prevent diarrhea and improve body health is dietary supplementation with probiotic bacteria or with fermented food containing probiotic bacteria, namely those in the genera Lactobacillus, Bacillus, Saccharomyces, and Bifidobacterium (77, 78). Effects of probiotic supplementation include improving intestinal secreting mucins, enhancing mucosal barrier function, increasing tight junctions in mucous cells, providing colonization resistance, increasing production of secretory lgA, producing bacteriocins, producing balanced T-helper cell response, and increasing production of IL-10 and Tregs (79, 80). However, the origin of some used probiotic bacteria was not clear, and the physiological characters of bacteria was lack of sufficient studied. In clinical use, the diagnosis effect maybe off-target and not achieved. Many probiotic bacteria were overused. Analyze the composition of microbiota in GIT of patient, provided the precise supplementary strategies could be needed.
5 Precise supplementary strategies
5.1 Core bacteria and its function
Following the common supplementary pathway, supplementation with the scarcer bacteria of the gut is not an ideal strategy. It is hypothesized that the core bacterial community ecology (showing in Figure 2) plays an important role in maintaining the dynamic equilibrium of microbiota in the organs (81). Core bacteria are defined as one or several functional strains, that play important roles in maintaining the physiological wellness (82). Although the number of core bacteria is small, they serve various key functions, and several core strains are inherited in different breeds of the same animal (83). The core bacteria may also be cross-inherited from an ancestral generation or from interactions with the outer environment (84–86), shown in Figure 3. There is evidence that some of the microbial taxa found in the placenta, such as those in the genus Lactobacillus, and the common pathogen Streptococcus agalactiae, may have an oral origin (87–89). It is also known that vaginal microbiota mainly consists of Lactobacillus, namely Lactobacillus crispatus, Lactobacillus iners, and Lactobacillus jensenii, as well as Gardnerella vaginalis. The abundance and diversity of vaginal microbiota vary across different periods of pregnancy. Such data further showed that the housekeeping bacteria were part of the core bacteria can be inherited from matrix and play a key role in establishing a healthy bacterial community (2).
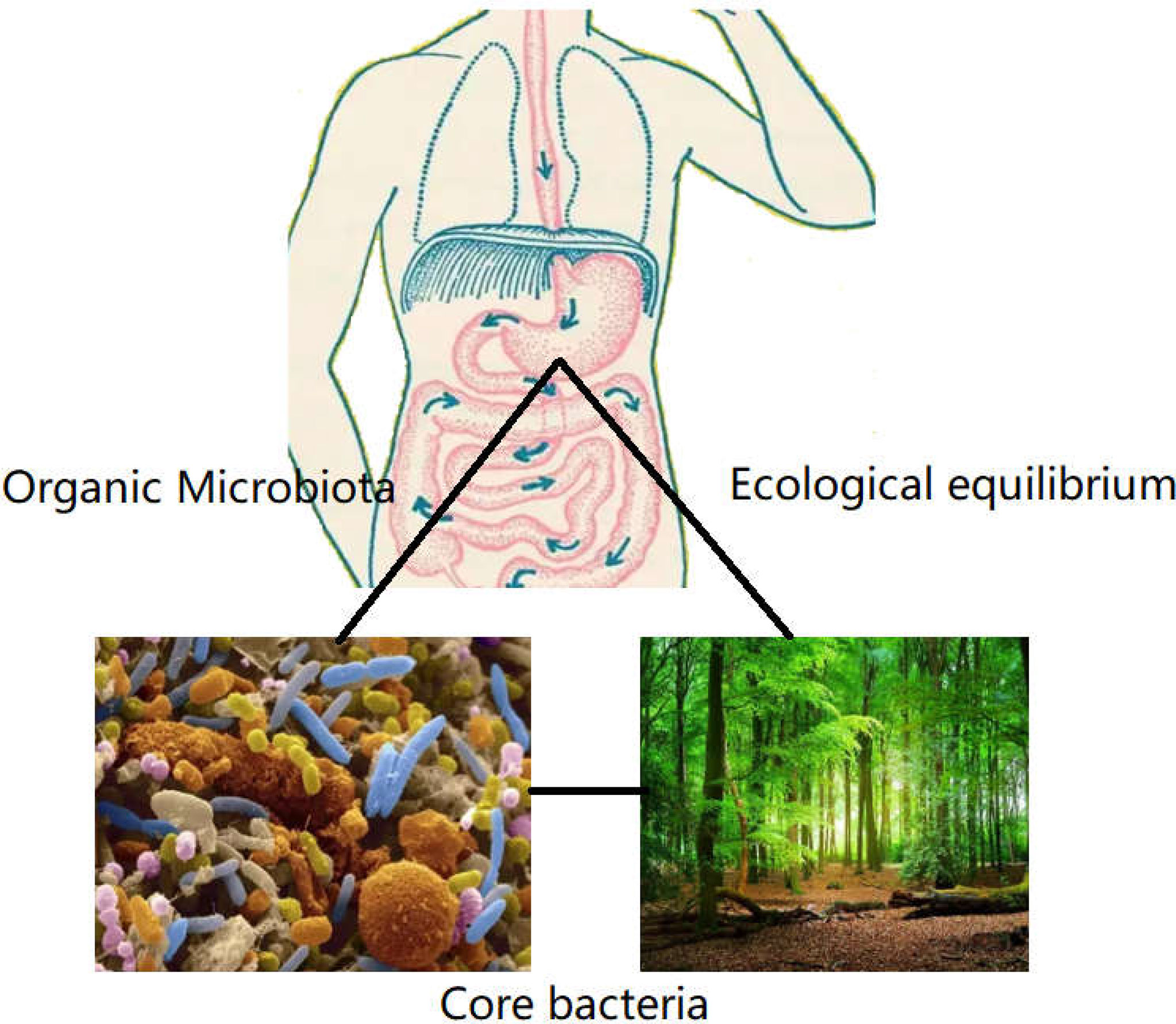
Figure 2 The role of core bacteria in organs. Organs such as mouth, gastrointestinal tract, lung, and genitourinary tract harbored many strains of bacteria, like the virgin forest existing many floras. The core bacteria were the exuberant floras, which presided in the microbiome. Although the number of core bacteria covered a few proportions in total flora. The function was the biggest in defending the equilibrium of microecology, which was essential to establish the dominate bacteria in organs.
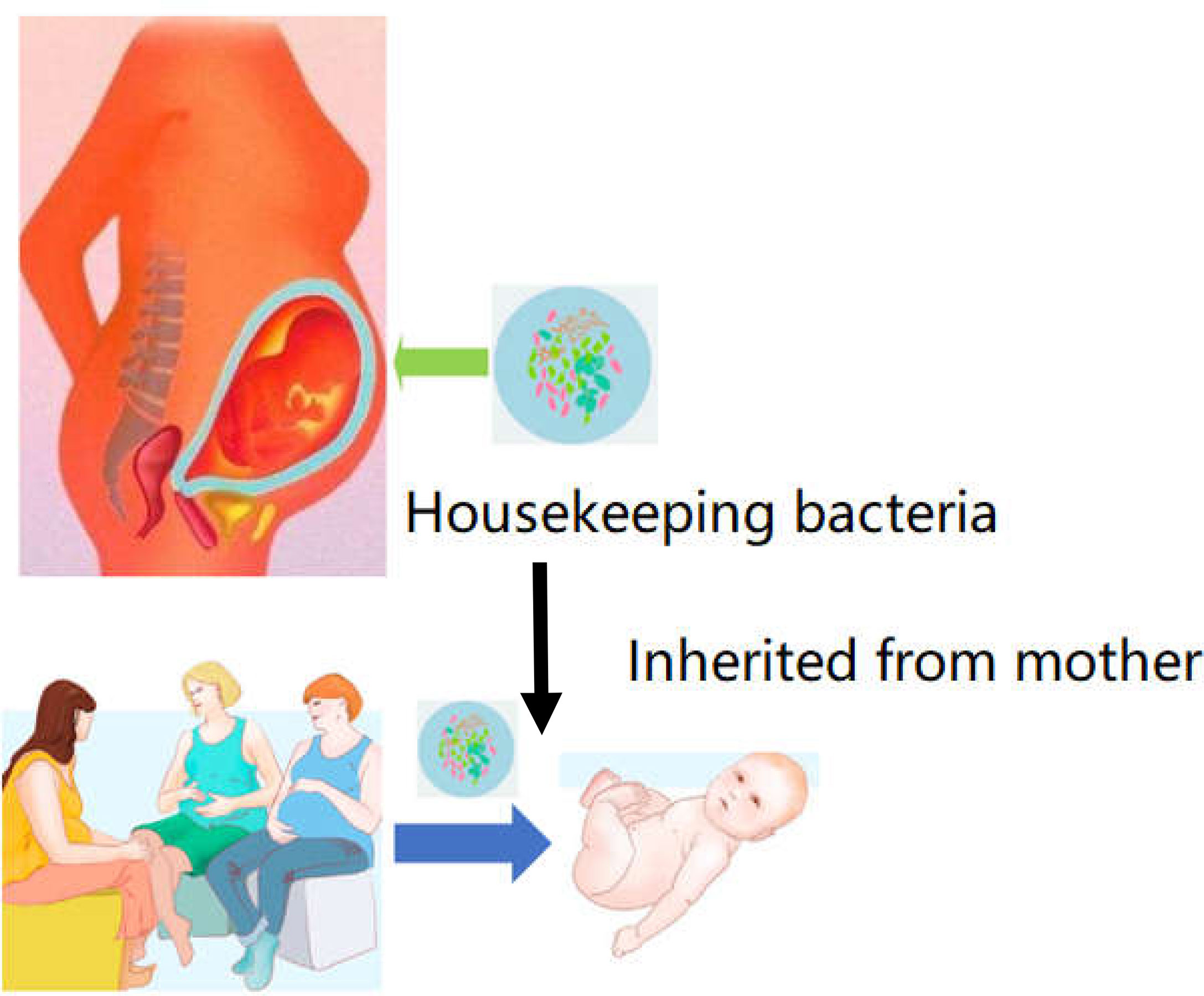
Figure 3 Housekeeper bacteria are the strains can be inheritable from mother to infant. Once these strains of housekeeper bacteria can be acquired from the placenta or amniotic fluid. The bacteria could be founded as a primary microbiome data to cure and prevent the dysbiosis in future GIT dysfunction.
Certain core bacteria were commonly owned by rodent animals. In humans, using 16S rDNA cloned sequences from 17 individuals found seven genera that were common in 50% of the cohort (90). On the other hand, a study investigating metagenomes from 124 European individuals discovered that 90% of the individuals of the cohort share a common core of nine genera, at a 10% sequence coverage threshold through sequenced metagenome (91). The comparative analysis of six mouse gut microbiota datasets (92, 93) showed that the core mouse gut microbiota plateaus were partly overlapped with human.
The genera of Lactobacillus, Akkermansia, Bacillus, Bifidobacterium, Clostridium and Prevotellacea form the core bacteria in the GIT of humans, and their merits in the recovery from various diseases have been documented (2, 4, 94, 95). Oral administration of Akkermansia muciniphila has been used as a treatment to reduce the symptoms of IBD (92). Supplementation with the strains Lactobacillus reuteri and Lactobacillus johnsonii has been useful in optimizing the bacterial composition of both humans and pigs (5, 82). Addition of the strain Clostridium propionate helped to improve body health and overcomes stress (93). Co-correlation network analysis revealed that the genus of Prevotellacea UCG-003 was the key bacterium in microbiota of piglets. Furthermore, changes in bacterial metabolic function between diarrheic piglets and non-diarrheic piglets were estimated by picrust analysis (contained in Metagenomic analysis), which revealed that the dominant functions of fecal microbes were membrane transport, carbohydrate metabolism, amino acid metabolism, and energy metabolism. Also, the 16S rDNA cloned sequence on colostrum of cows found that ten core genera contained Bacillus, Bacteroides, Staphylococcus, Acinetobacter, and Pseudomonas. A part of the found core bacteria belonged to the common intestinal bacteria and were also identified as the core ones.
5.2 Established the domination of core bacteria
Gut microbial diversity and the dominate microbiota changed in disease. It is assumed that the core strains of bacteria play more direct roles in assisting the receptors to restore bodily health (96). From these studies, it can be concluded that the core strains of bacteria should be further identified and their functions in different stages of growth should be determined (97). Replaced with bacteria at the site of microbial translocation by replacement with the core bacterial strains, following supplementation, can be done based on the relationship between the disease and bacterial composition (98). Restore the dominant flora, and repair the equilibrium of microecology (Figure 4). The supplementation of core bacteria is essential to promote resilience of bacterial abundances. The theory of dominant flora in microbiology suggested that the core bacteria inherited form ancestors dominated in all strains of bacteria in gastrointestinal tract. More than 1700 strains of bacteria found in obese volunteers and these accept the dietary intervention with high fibers, suggested that the total of 141 strains of bacteria composed 2 different sets, with one set increased and the other decreased. Otherwise, the situation was reverse after dietary intervention. Nearly, 50 strains of bacteria were composed core bacteria increased after dietary intervention and suppress the remained strains. The 141 strains of bacteria covered less than 10% total bacteria in GIT. Others were owing no ecological network in micro-system (99–101).
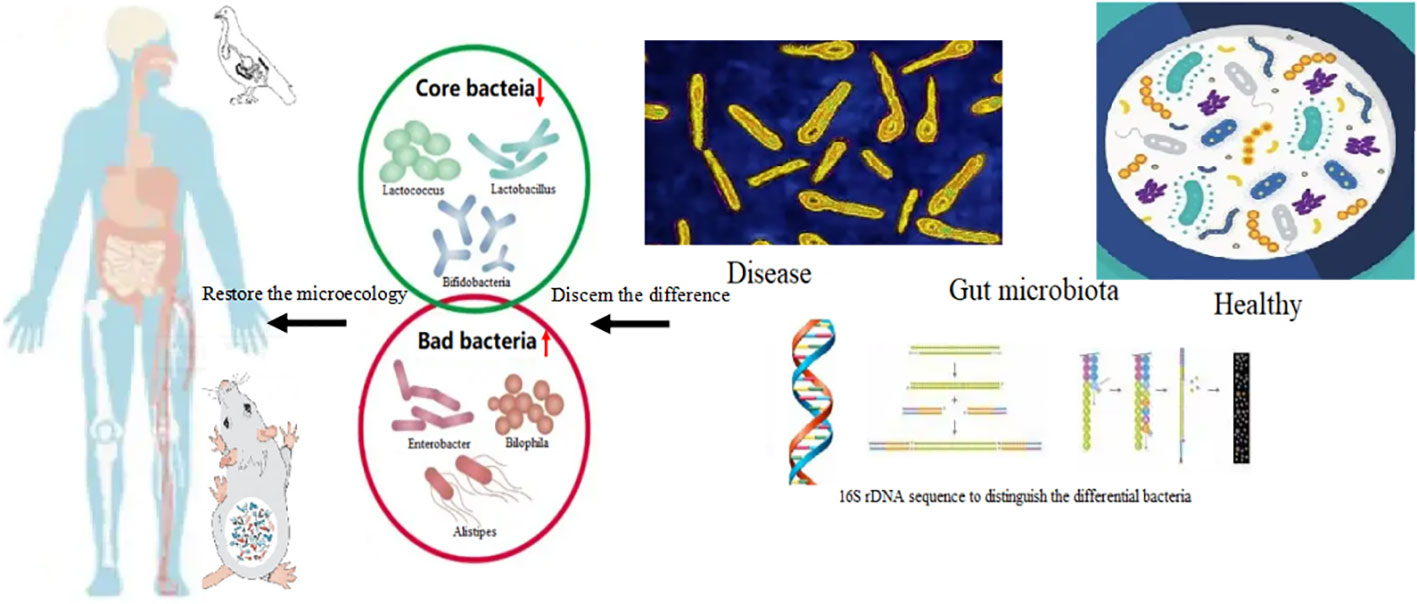
Figure 4 The core bacteria in diseases and restore the microbiota first supplementation with the core bacteria. The core bacteria such as the genus of Lactobacillus, Akkermansia, Bifidobacterium, the abundance was depleted in gut bacteria of patient with obesity and gout. The abnormal increasing of the bacteria namely, Enterobacter, Alistipes, Bilophila, and Bacteroides can be detected. Supplementation with core bacteria such as Lactobacillus reuteri, Lactobacillus salivarius, Akkermansia muciniphila Bifidobacteria infantis, Bifidobacteria longum and Bifidobacteria animalis, Faecalibacterium prausnitzii, restore the dominant flora, and repair the equilibrium of microecology. Storage of core bacterial database in health condition to establish a unique individual sets is necessary for self-aid for whom in disease. The cases in animal model such as poultry and mouse were also found the core bacteria supplementation could alleviate the symptoms. Chicks fed with high crude protein diet induced gout, and supplementation with Lactobacillus reuteri can restore the bacterial composition. Through comparing the differences of bacterial composition of health and diseased conditions, get the common useful strains of core bacteria through culture-independent metagenomic approaches is essential, which would be achieve the targeted supplementation of bacteria.
Supplementation with the reduced abundance of fiber fermenting bacterial strains such as Lactobacillus acidophilus, Lactobacillus casei, Lactococcus lactis, Bifidobacterium bifidum, and Bifidobacterium lactis were helpful in obese patients (102), which can reduce the abundance of Desulfovibrio species and increase in the abundance of Clostridium species, are key features restore the dominate bacteria in obesity model and are seen in humans with age-associated metabolic syndrome (81). Cases in animal model such as poultry and mouse were also found the core bacteria supplementation could alleviate the symptoms. Chicks fed with high crude protein diet induced gout, and supplementation with Lactobacillus reuteri can restore the bacterial composition (99, 103).
By identifying the differences in bacterial composition associated with different diseases, microbiota transplantation can be tailored to supplement the deficient bacterial genera. In unhealthy individuals, outnumbered and short-numbered microbiota can be identified through meta-genome sequencing (104) after which the dominant genera of bacteria in the healthy status must be defined (105, 106). The goal of such research is to develop powerful probiotic regimens that can replace FMT.
However, there is still a lack of sufficient meta-data to distinguish bacterial differences in patients with different disease currently. Storage of core bacterial database in health condition to establish a unique individual sets is necessary for self-aid for whom in disease. Differences in the microbiota profile have been reported to be related to known clinical risk factors for many diseases, such as metabolic diseases, asthma, arthritis, and cancer. In this sense, personalized probiotic therapies aiming to manipulate the host microbiota using specific and different strains have gained considerable interest in recent years. If we continue to learn about other gut microorganisms and their roles in human health, we may obtain a complete rationale for selecting the next generation of probiotics, such as Clostridia clusters IV, XIVa and, XVIII and Faecalibacterium prausnitzii, which have emerged as non-traditional probiotics and studies on their effects on inflammatory diseases have been met with promising results. F. prausnitzii is a commensal bacterial strain that has been reported to be less abundant in colitis patients. However, F. prausnitzii exhibited an important anti-inflammatory response in mouse experimental colitis models. Most of these effects were linked to the high capacity of this strain to produce metabolites with anti-inflammatory effects. Clostridium butyricum is another potential non-traditional candidate probiotic that can produce metabolites with anti-inflammatory effects, such as butyrate (30, 107).
With the development of sequencing technology, microbiome data such as strain-level variation, transcriptomics, proteomics, and metabolomics (108), can be determined. Future avenues and challenges for understanding the interplay between human nutrition, genetics, and microbial genetics, need to be addressed. In addition, integrating microbiome data with human multi-omics data, such as genetics, transcriptomics, epigenetics, and metabolomics, should be considered to develop successful treatments in the future.
In the treatment of these diseases, beneficial microbial functioning decreases, thus, supplementation with core functional probiotics can restore the resilience of the microbial community (5). Second, identification of suitable core probiotics to inhibit the growth of specific pathogenic bacteria in patients seems to be an effective strategy to control deterioration in health caused by disease (65, 109).
5.3 Strategies of probiotic isolation and selection for supplementation
5.3.1 Metagenomic and comparative genomics
Culture-independent metagenomic approaches to characterize the microbiota, enabled by next-generation sequencing, have increased the sensitivity and power of such associative studies by enabling high throughput analysis. Microbiota in dysbiosis-mediate diseases could be analyzed and compared with health condition in many cases of illness (110–114). Such culture independent methods of bacterial species analysis where >99% of microbial species can be unculturable (81). Taxonomic diversity can also be determined from shotgun sequenced metagenomics data sets (84, 85).
The storage of core bacterial database in health condition to establish a unique individual sets is necessary for self-aid for whom in disease. Also, compare the differences between donors or animals to conduct the common useful strains of core bacteria through culture-independent metagenomic approaches is essential (Figure 4). Acknowledged that core bacteria and dominant bacteria, use culture-dependent approaches to acquire the needed bacteria to supplement become urgent. The strategies for targeted choice of probiotic bacteria were suggested.
5.3.2 Homogenous origin
In restoring a healthy microbial composition, the supplementary bacteria must be ascertained to mine the probiotic strains needed to achieve precise supplementation. Currently, the most frequently used genus of bacteria in human and animal probiotic products are Bacillus, Lactobacillus, Bifidobacterium, and Clostridium (115). However, the origin of the strains being used is diverse. Many candidate strains of bacteria can be harvested through various isolation methods (116–118). The strains can be further narrowed down after certain conditional settings. In different animals, the isolation conditions of the core bacteria can be diverse (94, 119–121). Homogeneous isolation involves origination of the probiotic bacterial strains from the organs of used animals (Figure 5A). There are slight differences between the organs in different animal species (122, 123) and the core probiotic strains do not correspond with each other. Thus, the adaption of using bacteria colonized in the organs of animals could be more optimal than the use of non-homogeneous strains of bacteria (124). The mechanism of action is mainly related to the digestive enzymes and their roles in the gastrointestinal microenvironment. For Bacillus isolated from chicken, homologous feeding of probiotic bacteria for the same breed of chick could play more beneficial roles than non-homologous feeding (89, 125). Targeted supplementation in animals also considers the regular pattern of microbial development (126). In addition, supplementation with scarce strains of bacteria seems to be a limiting factor in disease recovery. The growth stage and health status should be considered as additional elimination conditions during isolation of bacteria.
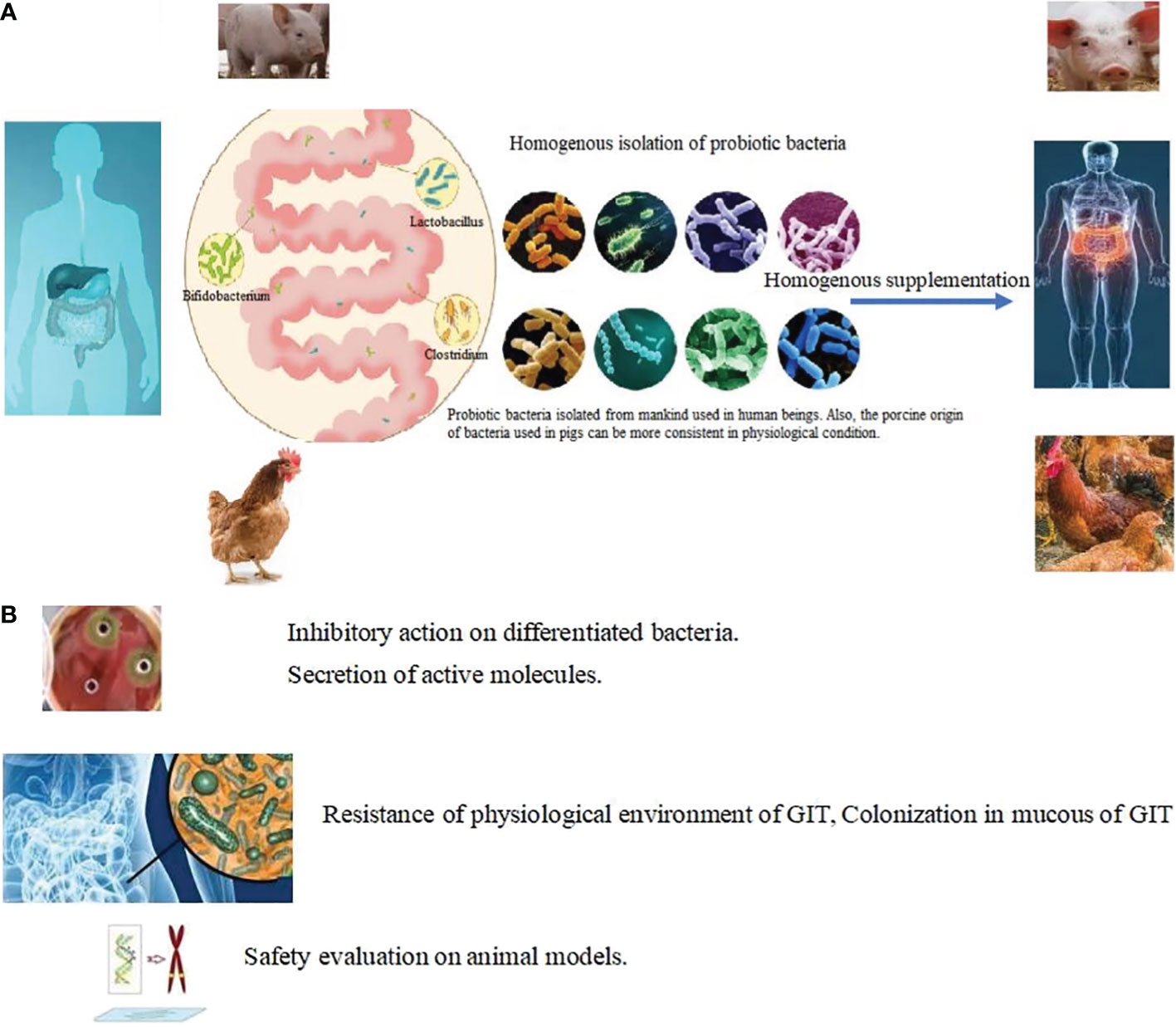
Figure 5 Targeted isolation in choosing probiotic bacteria. (A) Homogeneous isolation: Probiotic bacteria isolated from mankind used in human beings. Also, the porcine origin of bacteria used in pigs can be more consistent in physiological condition. Lactobacillus, Akkermansia, Bacillus, Bifidobacterium, Lactococcus, Streptococcus, Clostridium and Prevotellacea were harvested from organs and supplemented homogenously in same animals. (B) Targeted methods in choosing probiotic bacteria.
5.3.3 Screening conditions
Through selective culturable medium and culturing conditions, the target genus of bacteria could be isolated, such as isolation of Lactobacilli grown in de Man, Rogosa, and Sharpe medium, the use of BBL Trypticase soy agar with sheep blood (TSA II) broth medium for Bifidobacteria isolation, and M17 broth for culturing probiotic lactic streptococci (127).
High temperatures such as 80 °C can be used to isolate heat resistance bacteria and the oxygen content used in the culturing conditions can be adjusted to narrow the scope of isolated bacteria. Other assays of beneficial factors produced by bacteria have also proven useful in minimizing the candidate strains. These include inhibition of pathogens or conditional pathogens (128, 129); secretion of digestive enzymes, lactic acid, and digestive enzymes (130–133), the production of bioactive peptides, and vitamins such as bacteriocins and vitamin B6, K, and gamma amino butyric acid. The ability to perform bile salt degradation, cholesterol lysis (89, 134) and tolerance bile salts, gastric acid, and heat stress (116, 135, 135) are often used as elimination conditions (Figure 5B). Through a series of isolation methods, two or three strains of bacteria can finally be identified for clinical use (136–138). Details of various strategies for isolation and characterization including adhesion and colonization in the mucous membrane of the GIT (27) and safety assessments (139, 140) such as oral acute toxicity, sub-chronic toxicity, chronic toxicity, and reproductive and developmental toxicity (140, 141); antibiotic resistance; and lack of DNase (95, 139), gelatinase, and hemolytic activity (142) are shown in Table 2. Through a series of target conditional choice, the remained strains can be used to monitor the supplementary effect.
5.3.4 Other omics technologies
While the dominant strains of bacteria play a crucial role in overall bodily health, it is also important to isolate undiscovered strains and study their functions, to aid in targeted isolation (147–150). Some bacteria are still difficult to isolate due to being unculturable in aerobic conditions (151–153). For culturable bacteria, there are abundant strains whose physiological characters can be studied to determine their benefits to the body (154, 155). Meanwhile, progress in culture techniques along with high-resolution mass spectrometry have provided an additional sight through which to identify small molecules correlated with disease, setting new direction for more rigorous studies. Multi-omic technologies, such as metagenomics, metabolomics, and culturomics, should be applied in combination to further explore core strains of probiotic bacteria. Culture-independent metagenomic approaches have increased the sensitivity and power of such associative studies by enabling high-throughput analysis (156). By applying shotgun metagenomic sequencing, the bacterial species present in the original fecal samples that grew as distinct colonies under different culture conditions can be compared and profiled. When compared with a comprehensive gene catalogue that was derived from culture-independent methods used to assess the intestinal microbiota of normal healthy individuals, identified genes in the larger database were represented, and derived metagenomic species were detected in the cultured samples (148, 157, 158). Through these methods an increasing number of unidentified microorganisms can be cultured and recognized.
By harvesting the targeted probiotic bacteria, the whole genome can be sequenced and blasted to ascertain the function employed in matrix assisted laser desorption ionization (MALDI) to identify the unknown bacteria in the database.
Although the omics technologies have improved the capacity of bacterial analysis and culture. A total of less than 1% bacteria can be successfully cultured in vitro, a greater number of core bacteria and cross the animals still need to further unveiled to help patient reconstruct the health microbiological flora.
6 Conclusion
Considering the causal relationship on intestinal bacteria and disease, supplementary probiotics to restore the bacterial composition to cure disease is performed in human and animals. In their clinical use, probiotic bacteria were not conforming with the differential scared bacteria owning the symptom, which functioned in healthy condition. Supplemented with core bacteria to re-construct the dominant flora and repair the differential bacteria was the rule. Then repair the conditioned and supper growth-ed bacteria to restore the intestinal bacteria. Skills in choice of bacteria, such as isolated from homogenous animal, conditioned with colonization, temperature, inhibition on certain pathogen, and others could minimize the candidate strains should be focused. Also, research on metagenomics and culturomics can further help to unveil the microbial composition in the organs. Strategies for targeted selection of strains directly corresponding to the deficiencies can be used to tailor precise probiotic bacterial supplementation. Considering the core bacteria in regulation bodily function, get the personal bacterial database in young and health period and make the personal tabs to distinguish the disabled conditions, which can be efficient to achieve targeted supplementation. Our review provided a venue on precise supplementation of probiotic bacteria in clinical use.
Author contributions
JY carried out the literature study and JY drafted the manuscript. SQ critically evaluated the manuscript. HZ reviewed the manuscript. All authors approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by fund of Tibet Major Science and Technology Project (XZ202101ZD0005N), Start-up for Scientific Research of High-level Talents of Jiangsu Vocational College of Agriculture and Forestry, Youth support project of Jiangsu Vocational College of Agriculture and Forestry (2021kj19), China Agriculture Research System (CARS-40), Key projects of Natural Science Foundation of Tianjin (20JCZDJC00170).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature reviews. Gastroenterol Hepatol (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
2. Yang J, Qian K, Wang C, Wu Y. Roles of probiotic Lactobacilli inclusion in helping piglets establish healthy intestinal inter-environment for pathogen defense. Probiotics Antimicrob Proteins (2018) 10:243–50. doi: 10.1007/s12602-017-9273-y
3. Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy JM, Dequenne I, et al. How probiotics affect the microbiota. Front Cell Infect Microbiol (2019) 9:454. doi: 10.3389/fcimb.2019.00454
4. Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microbial Pathog (2017) 106:171–81. doi: 10.1016/j.micpath.2016.02.005
5. Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol (2009) 587:4153–8. doi: 10.1113/jphysiol.2009.174136
6. Yang J, Huang K, Wang J, Wu D, Liu Z, Yu P, et al. Combined use of bacillus subtilis yb-114,246 and bacillus licheniformis yb-214,245 improves body growth performance of Chinese huainan partridge shank chickens by enhancing intestinal digestive profiles. Probiotics Antimicrob Proteins (2021) 13:327–42. doi: 10.1007/s12602-020-09691-2
7. Cox AJ, West NP, Cripps AW. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol (2015) 3:207–15. doi: 10.1016/S2213-8587(14)70134-2
8. Garrett WS. The gut microbiota and colon cancer. Science (2019) 364:1133–5. doi: 10.1126/science.aaw2367
9. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol (2004) 54:1469–76. doi: 10.1099/ijs.0.02873-0
10. Yang J, Wang C, Huang K, Zhang M, Wang J, Pan X. Compound lactobacillus sp. administration ameliorates stress and body growth through gut microbiota optimization on weaning piglets. Appl Microbiol Biotechnol (2020) 104:6749–65. doi: 10.1007/s00253-020-10727-4
11. García-Hernández Y, Pérez-Sánchez T, Boucourt R, Balcázar JL, Nicoli JR, Moreira-Silva J, et al. Isolation, characterization, and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res Vet Sci (2016) 108:125–32. doi: 10.1016/j.rvsc.2016.08.009
12. Groeger D, O’Mahony L, Murphy EF, Bourke JF, Dinan TG, Kiely B, et al. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes (2013) 4:325–39. doi: 10.4161/gmic.25487
13. Guglielmetti S, Mora D, Gschwender M, Popp K. Randomized clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life – A double-blind, placebo-controlled study. Alimentary Pharmacol Ther (2011) 33:1123–32. doi: 10.1111/j.1365-2036.2011.04633.x
14. Bomhof MR, Saha DC, Reid DT, Paul HA, Reimer RA. Combined effects of oligofructose and bifidobacterium animalis on gut microbiota and glycemia in obese rats. Obes (Silver Spring) (2013) 22:763–71. doi: 10.1002/oby.20632
15. Lazarenko L, Babenko L, Sichel LS, Pidgorskyi V, Mokrozub V, Voronkova O, et al. Antagonistic action of lactobacilli and bifidobacteria in relation to staphylococcus aureus and their influence on the immune response in cases of intravaginal staphylococcosis in mice. Probiotics Antimicrob Proteins (2012) 4:78–89. doi: 10.1007/s12602-012-9093-z
16. Zárate G, Chaia AP. Influence of lactose and lactate on growth and b-galactosidase activity of potential probiotic propionibacterium acidipropionici. Anaerobe (2012) 18:25–30. doi: 10.1016/j.anaerobe.2011.12.005
17. Zárate G, Chaia AP. Feeding with dairy propionibacterium acidipropionici CRL 1198 reduces the incidence of concanavalin-a induced alterations in mouse small intestinal epithelium. Food Res Int (2012) 47:3–22. doi: 10.1016/j.foodres.2012.01.005
18. Oksaharju A, Kooistra T, Kleemann R, Lindstedt KA, Kovanen PT, Korpela R, et al. Effects of probiotic lactobacillus rhamnosus GG and propionibacterium freudenreichii ssp. shermanii JS supplementation on intestinal and systemic markers of inflammation in ApoE*3Leiden mice consuming a high-fat diet. Br J Nutr (2013) 110:77–85. doi: 10.1017/S0007114512004801
19. Varankovich NV, Nickerson MT, Korber DR. Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front Microbiol (2015) 6:685. doi: 10.3389/fmicb.2015.00685
20. Yang J, Wang C, Liu L, Zhang M. Lactobacillus reuteri KT260178 supplementation reduced morbidity of piglets through its targeted colonization, improvement of cecal microbiota profile, and immune functions. Probiotics Antimicrob Proteins (2020) 12:194–203. doi: 10.1007/s12602-019-9514-3
21. Abhari K, Shekarforoush SS, Hosseinzadeh S, Nazifi S, Sajedianfard J, Eskandari MH. The effects of orally administered bacillus coagulans and inulin on prevention and progression of rheumatoid arthritis in rats. Food Nutr Res (2016) 60:30876. doi: 10.3402/fnr.v60.30876
22. Manhar AK, Bashir Y, Saikia D, Nath D, Gupta K, Konwar BK, et al. Cellulolytic potential of probiotic bacillus subtilis AMS6 isolated from traditional fermented soybean (Churpi): An in-vitro study with regards to application as an animal feed additive. Microbiol Res (2016) 186-187:62–70. doi: 10.1016/j.micres.2016.03.004
23. Vieira AT, Fukumori C, Ferreira CM. New insights into therapeutic strategies for gut microbiota modulation in inflammatory diseases. Clin Trans Immunol (2016) 5:e87. doi: 10.1038/cti.2016.38
24. Yang J, Huang K, Qin S, Wu X, Zhao Z, Chen F. Antibacterial action of selenium-enriched probiotics against pathogenic Escherichia coli. Digestive Dis Sci (2009) 54:246–54. doi: 10.1007/s10620-008-0361-4
25. Corrêa NB, Penna FJ, Lima FM, Nicoli JR, Filho LA. Treatment of acute diarrhea with saccharomyces boulardii in infants. J Pediatr Gastroenterol Nutr (2011) 53:497–501. doi: 10.1097/MPG.0b013e31822b7ab0
26. Thomas S, Metzke D, Schmitz J, Dörffel Y, Baumgart DC. Anti-inflammatory effects of saccharomyces boulardii mediated by myeloid dendritic cells from patients with crohn’s disease and ulcerative colitis. Am J Physiology-Gastroint Liver Physiol (2011) 301:G1083–1092. doi: 10.1152/ajpgi.00217.2011
27. Yang J, Wang J, Huang K, Zhu M, Liu Q, Liu G, et al. Selenium enriched bacillus subtilis yb-1114246 activated the TLR2-NF-κB1 signaling pathway to regulate chicken intestinal β-defensin 1 expression. Food Funct (2021) 12:5913–26. doi: 10.1039/D1FO01158H
28. Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr (2019) 73:24–37. doi: 10.1038/s41430-018-0135-9
29. Choi SH, Yoon TJ. Non-specific immune response of rainbow trout (Oncorhynchus mykiss) by dietary heat-inactivated potential probiotics. Immune Netw (2008) 8:67–74. doi: 10.4110/in.2008.8.3.67
30. Wang H, Ni X, Liu L, Zeng D, Lai J, Qing X, et al. Controlling of growth performance, lipid deposits and fatty acid composition of chicken meat through a probiotic, lactobacillus johnsonii during subclinical clostridium perfringens infection. Lipids Health Dis (2017) 16:38. doi: 10.1186/s12944-017-0408-7
31. Al-Khalaifa H, Al-Nasser A, Al-Surayee T, Al-Kandari S, Al-Enzi N, Al-Sharrah T, et al. Effect of dietary probiotics and prebiotics on the performance of broiler chickens. Poultry Sci (2019) 98:4465–79. doi: 10.3382/ps/pez282
32. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science (2009) 326:1694–7. doi: 10.1126/science.1177486
33. Alves J, Peres S, Gonçalves E, Mansinho K. Anaerobic bacteria with clinical relevance: morphologic and taxonomic classification, distribution among human microbiota and microbiologic diagnosis. Acta Med Portuguesa. (2017) 30:409–17. doi: 10.20344/amp.8098
34. de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Reviwes (2012) 70:S45–56. doi: 10.1111/j.1753-4887.2012.00505.x
35. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. doi: 10.1126/science.1241214
36. Raisch J, Dalmasso G, Bonnet R, Barnich N, Bonnet M, Bringer MA. How some commensal bacteria would exacerbate colorectal carcinogenesis? Médecine Sciences: M/S (2016) 32:175–82. doi: 10.1051/medsci/20163202011
37. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature (2013) 498:99–103. doi: 10.1038/nature12198
38. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490:55–60. doi: 10.1038/nature11450
39. Ning Y, Yang G, Chen Y, Zhao X, Qian H, Liu Y, et al. Characteristics of the urinary microbiome from patients with gout: A prospective study. Front Endocrinol (2020) 11:272. doi: 10.3389/fendo.2020.00272
40. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature (2013) 500:585–8. doi: 10.1038/nature12480
41. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature (2013) 500:541–6. doi: 10.1038/nature12506
42. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature (2006) 444:1022–3. doi: 10.1038/4441022a
43. Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J (2012) 6:1848–57. doi: 10.1038/ismej.2012.27
44. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA (2007) 104:13780–5. doi: 10.1073/pnas.0706625104
45. Doherty G, Bennett G, Patil S, Cheifetz A, Moss AC. Interventions for prevention of post-operative recurrence of crohn's disease. Cochrane Database syst Rev (2009) 7:CD006873. doi: 10.1002/14651858.CD006873.pub2
46. Veerappan GR, Betteridge J, Young PE. Probiotics for the treatment of inflammatory bowel disease. Curr Gastroenterol Rep (2012) 14:324–33. doi: 10.1007/s11894-012-0265-5
47. Shen B, Brzezinski A, Fazio VW, Remzi FH, Achkar JP, Bennett AE, et al. Maintenance therapy with a probiotic in antibiotic-dependent pouchitis: experience in clinical practice. Alimentary Pharmacol Ther (2005) 22:721–8. doi: 10.1111/j.1365-2036.2005.02642.x
48. Chen M, Feng Y, Liu W. Efficacy and safety of probiotics in the induction and maintenance of inflammatory bowel disease remission: a systematic review and meta-analysis. Ann Palliative Med (2021) 10:11821–9. doi: 10.21037/apm-21-2996
49. Fei N, Bruneau A, Zhang X, Wang R, Wang J, Rabot S, et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio (2020) 11:e03263–19. doi: 10.1128/mBio.03263-19
50. Zhao L. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol (2013) 11:639–47. doi: 10.1038/nrmicro3089
51. Muccioli GG, Naslain D, Bäckhed F, Reigstad CS, Lambert DM, Delzenne NM, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol (2010) 6:392. doi: 10.1038/msb.2010.46
52. Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (2012) 338:120–3. doi: 10.1126/science.1224820
53. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J (2012) 6:320–9. doi: 10.1038/ismej.2011.109
54. Yan H, Fei N, Wu G, Zhang C, Zhao L, Zhang M. Regulated inflammation and lipid metabolism in colon mRNA expressions of obese germfree mice responding to Enterobacter cloacae B29 combined with the high fat diet. Front Microbiol (2016) 7:1786. doi: 10.3389/fmicb.2016.01786
55. de Clercq NC, Groen AK, Romijn JA, Nieuwdorp M. Gut microbiota in obesity and undernutrition. Adv Nutr (2016) 7:1080–9. doi: 10.3945/an.116.012914
56. Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J (2013) 7:880–4. doi: 10.1038/ismej.2012.153
57. Olsson LM, Poitou C, Tremaroli V, Coupaye M, Aron-Wisnewsky J, Bäckhed F, et al. Gut microbiota of obese subjects with prader–willi syndrome is linked to metabolic health. Gut (2020) 69:1229–38. doi: 10.1136/gutjnl-2019-319322
58. Zhang M, Zhang M, Zhang C, Du H, Wei G, Pang X, et al. Pattern extraction of structural responses of gut microbiota to rotavirus infection via multivariate statistical analysis of clone library data. FEMS Microbiol Ecol (2009) 70:21–9. doi: 10.1111/j.1574-6941.2008.00604.x
59. Wagner PL, Acheson DW, Waldor MK. Human neutrophils and their products induce shiga toxin production by enterohemorrhagic Escherichia coli. Infect Immun (2001) 69:1934–7. doi: 10.1128/IAI.69.3.1934-1937.2001
60. Fazelnia K, Fakhraei J, Yarahmadi HM, Amini K. Dietary supplementation of potential probiotics bacillus subtilis, bacillus licheniformis, and Saccharomyces cerevisiae and synbiotic improves growth performance and immune responses by modulation in intestinal system in broiler chicks challenged with salmonella typhimurium. Probiotics Antimicrob Proteins (2021) 13:1081–92. doi: 10.1007/s12602-020-09737-5
61. Fakruddin M, Hossain MN, Ahmed MM. Evaluation of microplate immunocapture method for detection of vibrio cholerae, salmonella typhi and shigella flexneri from food. BMC Microbiol (2017) 17:189. doi: 10.1186/s12866-017-1099-y
62. Amin N, Liu P, Foster T, Rahman M, Miah MR, Ahmed GB, et al. Pathogen flows from on-site sanitation systems in low-income urban neighborhoods, Dhaka: A quantitative environmental assessment. Int J Hygiene Environ Health (2020) 230:113619. doi: 10.1016/j.ijheh.2020.113619
63. Zhang SS, Liu M, Liu DN, Shang YF, Wang YH, Du GH. ST2825, a small molecule inhibitor of MyD88, suppresses NF-κB activation and the ROS/NLRP3/Cleaved caspase-1 signaling pathway to attenuate lipopolysaccharide-stimulated neuroinflammation. Molecules (Basel Switzerland) (2022) 27:2990. doi: 10.3390/molecules27092990
64. Suryavanshi SV, Zaiachuk M, Pryimak N, Kovalchuk I, Kovalchuk O. Cannabinoids alleviate the LPS-induced cytokine storm via attenuating NLRP3 inflammasome signaling and TYK2-mediated STAT3 signaling pathways In vitro. . Cells (2022) 11:1391. doi: 10.3390/cells11091391
65. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature (2009) 457:480–4. doi: 10.1038/nature07540
66. Brunkwall L, Orho-Melander M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia (2017) 60:943–51. doi: 10.1007/s00125-017-4278-3
67. Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology (2013) 145:946–53. doi: 10.1053/j.gastro.2013.08.058
68. Lo Vecchio A, Cohen MB. Fecal microbiota transplantation for clostridium difficile infection: benefits and barriers. Curr Opin Gastroenterol (2014) 30:47–53. doi: 10.1097/MOG.0000000000000023
69. van Nood E, Speelman P, Nieuwdorp M, Keller J. Fecal microbiota transplantation: facts and controversies. Curr Opin Gastroenterol (2014) 30:34–9. doi: 10.1097/MOG.0000000000000024
70. Aron-Wisnewsky J, Clément K, Nieuwdorp M. Fecal microbiota transplantation: A future therapeutic option for obesity/diabetes? Curr Diabetes Rep (2019) 19:51. doi: 10.1007/s11892-019-1180-z
71. Zhang F, Cui B, He X, Nie Y, Wu K, Fan D, et al. Microbiota transplantation: Concept, methodology and strategy for its modernization. Protein Cell (2018) 9:462–73. doi: 10.1007/s13238-018-0541-8
72. Wei Y, Yang J, Wang J, Yang Y, Huang J, Gong H, et al. Successful treatment with fecal microbiota transplantation in patients with multiple organ dysfunction syndrome and diarrhea following severe sepsis. Crit Care (2016) 20:332. doi: 10.1186/s13054-016-1491-2
73. Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: current status and perspectives. Int J Cancer (2019) 145:2021–31. doi: 10.1002/ijc.32003
74. Caballero S, Carter R, Ke X, Sušac B, Leiner IM, Kim GJ, et al. Distinct but spatially overlapping intestinal niches for vancomycin-resistant Enterococcus faecium and carbapenem-resistant klebsiella pneumoniae. PloS Pathog (2015) 11:e1005132. doi: 10.1371/journal.ppat.1005132
75. Lai HH, Chiu CH, Kong MS, Chang CJ, Chen CC. Probiotic lactobacillus casei: effective for managing childhood diarrhea by altering gut microbiota and attenuating fecal inflammatory markers. Nutrients (2019) 11:1150. doi: 10.3390/nu11051150
76. Zhai R, Xue X, Zhang L, Yang X, Zhao L, Zhang C. Strain-specific anti-inflammatory properties of two Akkermansia muciniphila strains on chronic colitis in mice. Front Cell Infect Microbiol (2019) 9:239. doi: 10.3389/fcimb.2019.00239
77. Meng QW, Yan L, Ao X, Zhou TX, Wang JP, Lee JH, et al. Influence of probiotics in different energy and nutrient density diets on growth performance, nutrient digestibility, meat quality, and blood characteristics in growing-finishing pigs. J Anim Sci (2010) 88:3320–6. doi: 10.2527/jas.2009-2308
78. Friedman G. Preface: clinical applications of probiotics in gastroenterology: questions and answers. Gastroenterol Clin North Am (2012) 41:763–79. doi: 10.1016/j.gtc.2012.08.002
79. Hu S, Wang L, Jiang Z. Dietary additive probiotics modulation of the intestinal microbiota. Protein Pept Lett (2017) 24:382–7. doi: 10.2174/0929866524666170223143615
80. Hou Q, Zhao F, Liu W, Lv R, Khine WWT, Han J, et al. Probiotic-directed modulation of gut microbiota is basal microbiome dependent. Gut Microbes (2020) 12:1736974. doi: 10.1080/19490976.2020.1736974
81. Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, et al. The infant gut microbiome as a microbial organ influencing host well-being. Ital J Pediatr (2020) 46:16. doi: 10.1186/s13052-020-0781-0
82. Buhnik-Rosenblau K, Danin-Poleg Y, Kashi Y. Predominant effect of host genetics on levels of lactobacillus johnsonii bacteria in the mouse gut. Appl Environ Microbiol (2011) 77:6531–8. doi: 10.1128/AEM.00324-11
83. Younes JA, Lievens E, Hummelen R, van der Westen R, Reid G, Petrova MI. Women and their microbes: the unexpected friendship. Trends Microbiol (2018) 26:16–32. doi: 10.1016/j.tim.2017.07.008
84. Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. Q Rev Biol (2012) 87:325–41. doi: 10.1086/668166
85. Duron O, Hurst GD. Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol (2013) 11:45. doi: 10.1186/1741-7007-11-45
86. Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. mBio (2016) 7:e01395. doi: 10.1128/mBio.01395-15
87. de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature (2019) 572:329–34. doi: 10.1038/s41586-019-1451-5
88. Zakis DR, Paulissen E, Kornete L, Kaan AMM, Nicu EA, Zaura E. The evidence for placental microbiome and its composition in healthy pregnancies: A systematic review. J Reprod Immunol (2022) 149:103455. doi: 10.1016/j.jri.2021.103455
89. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep (2016) 6:23129. doi: 10.1038/srep23129
90. Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol (2009) 11:2574–284. doi: 10.1111/j.1462-2920.2009.01982.x
91. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
92. Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, et al. Intestinal microbiota containing barnesiella species cures vancomycin-resistant enterococcus faecium colonization. Infect Immun (2013) 81:965–73. doi: 10.1128/IAI.01197-12
93. Hang BPT, Wredle E, Dicksved J. Analysis of the developing gut microbiota in young dairy calves-impact of colostrum microbiota and gut disturbances. Trop Anim Health Prod (2020) 53:50. doi: 10.1007/s11250-020-02535-9
94. Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J (2017) 11:2834–47. doi: 10.1038/ismej.2017.138
95. Martínez JE, Vargas A, Pérez-Sánchez T, Encío IJ, Cabello-Olmo M, Barajas M. Human microbiota network: unveiling potential crosstalk between the different microbiota ecosystems and their role in health and disease. Nutrients (2021) 13:2905. doi: 10.3390/nu13092905
96. Guo Q, Goldenberg JZ, Humphrey C, El Dib R, Johnston BC. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev (2019) 4:CD004827. doi: 10.1002/14651858.CD004827.pub5
97. Hagihara M, Kuroki Y, Ariyoshi T, Higashi S, Fukuda K, Yamashita R, et al. Clostridium butyricum modulates the microbiome to protect intestinal barrier function in mice with antibiotic-induced dysbiosis. iScience (2020) 23:100772. doi: 10.1016/j.isci.2019.100772
98. Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, et al. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci CMLS (2018) 75:103–18. doi: 10.1007/s00018-017-2672-0
99. Yuan J, Qin S, Hu S, Liu Z, Song Y, Li L. Restoration of cefixime-induced gut microbiota changes by a prebiotic blend in a mouse model. Appl Microbiol Biotechnol (2022) 106(13-16):5197–209. doi: 10.1007/s00253-022-12044-4
100. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science (2018) 359(6380):1151–6. doi: 10.1126/science.aao5774
101. Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in bifidobacterium animalis and methanobrevibacter smithii. Int J Obes (Lond) (2012) 36:817–25. doi: 10.1038/ijo.2011.153
102. Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microbial Biotechnol (2019) 12:1109–25. doi: 10.1111/1751-7915.13410
103. Xi Y, Huang Y, Li Y, Huang Y, Yan J, Shi Z. The effects of dietary protein and fiber levels on growth performance, gout occurrence, intestinal microbial communities, and immunoregulation in the gut-kidney axis of goslings. Poultry Sci (2022) 101:101780. doi: 10.1016/j.psj.2022.101780
104. Claesson MJ, Wang QO, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res (2010) 38:e200. doi: 10.1093/nar/gkq873
105. Fodor AA, DeSantis TZ, Wylie KM, Badger JH, Ye Y, Hepburn T, et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PloS One (2012) 7:e41294. doi: 10.1371/journal.pone.0041294
106. Bottacini F, van Sinderen D, Ventura M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem J (2017) 474:4137–52. doi: 10.1042/BCJ20160756
107. Choi Y, Bose S, Seo J, Shin JH, Lee D, Kim Y, et al. Effects of live and pasteurized forms of akkermansia from the human gut on obesity and metabolic dysregulation. Microorganisms (2021) 9:2039. doi: 10.3390/microorganisms9102039
108. Rodriguez J, Hiel S, Neyrinck AM, Le Roy T, Pötgens SA, Leyrolle Q, et al. Discovery of the gut microbial signature driving the efficacy of prebiotic intervention in obese patients. Gut (2020) 69:1975–87. doi: 10.1136/gutjnl-2019-319726
109. Petersen C, Bell R, Klag KA, Lee SH, Soto R, Ghazaryan A, et al. T Cell-mediated regulation of the microbiota protects against obesity. Science (2019) 365:eaat9351. doi: 10.1126/science.aat9351
110. Gomes AC, ffmann C, Mota JF. Gut microbiota is associated with adiposity markers and probiotics may impact specific genera. Eur J Nutr (2020) 59:1751–62. doi: 10.1007/s00394-019-02034-0
111. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature (2015) 528:262–6. doi: 10.1038/nature15766
112. Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG 3rd, Pudlo NA, et al. Neonatal acquisition of clostridia species protects against colonization by bacterial pathogens. Science (2017) 356:315–9. doi: 10.1126/science.aag2029
113. Vacca I. Microbiota: Clostridia protect from gut infections in early life. Nat Rev Microbiol (2017) 15:321. doi: 10.1038/nrmicro.2017.56
114. Vogt SL, Finlay BB. Gut microbiota-mediated protection against diarrheal infections. J Travel Med (2017) 24:S39–43. doi: 10.1093/jtm/taw086
115. Rodríguez JM. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr (2014) 5:779–84. doi: 10.3945/an.114.007229
116. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome (2016) 4:1. doi: 10.1186/s40168-015-0145-y
117. Shokryazdan P, Faseleh Jahromi M, Liang JB, Ho YW. Probiotics: from isolation to application. J Am Coll Nutr (2017) 36:666–76. doi: 10.1080/07315724.2017.1337529
118. Poinsot P, Penhoat A, Mitchell M, Sauvinet V, Meiller L, Louche-Pélissier C, et al. Probiotic from human breast milk, lactobacillus fermentum, promotes growth in animal model of chronic malnutrition. Pediatr Res (2020) 88:374–81. doi: 10.1038/s41390-020-0774-0
119. Walker AW, Duncan SH, Louis P, Flint HJ. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol (2014) 22:267–74. doi: 10.1016/j.tim.2014.03.001
120. Lagier JC, Cadoret F, Raoult D. Critical microbiological view of SER-109. J Infect Dis (2017) 215:161–2. doi: 10.1093/infdis/jiw489
121. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev: MMBR (2017) 81:e00036–17. doi: 10.1128/MMBR.00036-17
122. McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, et al. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA (2013) 110:E2390–9. doi: 10.1073/pnas.1219809110
123. Lugli GA, Milani C, Mancabelli L, Turroni F, Ferrario C, Duranti S, et al. Ancient bacteria of the otzi’s microbiome: a genomic tale from the copper age. Microbiome (2017) 5:5. doi: 10.1186/s40168-016-0221-y
124. Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PloS One (2013) 8:e78331. doi: 10.1371/journal.pone.0078331
125. Dong XD, Li XR, Luan JJ, Liu XF, Peng J, Luo YY, et al. Bacterial communities in neonatal feces are similar to mothers’ placentae. Can J Infect Dis Med Microbiol (2015) 26:90–4. doi: 10.1155/2015/737294
126. Moles L, Gómez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PloS One (2013) 8:e66986. doi: 10.1371/journal.pone.0066986
127. Denys GA, Renzi PB, Koch KM, Wissel CM. Three-way comparison of BBL CHROMagar MRSA II, MRSASelect, and spectra MRSA for detection of methicillin-resistant staphylococcus aureus isolates in nasal surveillance cultures. J Clin Microbiol (2013) 51:202–5. doi: 10.1128/JCM.02022-12
128. Wang J, Zeng Y, Wang S, Liu H, Zhang D, Zhang W, et al. Swine-derived probiotic lactobacillus plantarum inhibits growth and adhesion of enterotoxigenic escherichia coli and mediates host defense. Front Microbiol (2018) 9:1364. doi: 10.3389/fmicb.2018.01364
129. Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: status, concerns, and future perspectives. Front Microbiol (2017) 8:1490. doi: 10.3389/fmicb.2017.01490
130. Ayyash MM, Abdalla AK, AlKalbani NS, Baig MA, Turner MS, Liu SQ, et al. Invited review: Characterization of new probiotics from dairy and nondairy products-insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J Dairy Scie (2021) 104:8363–79. doi: 10.3168/jds.2021-20398
131. Hassan MU, Nayab H, Shafique F, Williamson MP, Almansouri TS, Asim N, et al. Probiotic properties of Lactobacillus helveticus and Lactobacillus plantarum isolated from traditional Pakistani yoghurt. BioMed Res Int (2020) 2020:8889198. doi: 10.1155/2020/8889198
132. Farid W, Masud T, Sohail A, Ahmad N, Naqvi SMS, Khan S, et al. Gastrointestinal transit tolerance, cell surface hydrophobicity, and functional attributes of Lactobacillus acidophilus strains isolated from indigenous dahi. Food Sci Nutr (2021) 9:5092–102. doi: 10.1002/fsn3.2468
133. Lim SM, Lee NK, Kim KT, Paik HD. Probiotic lactobacillus fermentum KU200060 isolated from watery kimchi and its application in probiotic yogurt for oral health. Microbial Pathog (2020) 147:104430. doi: 10.1016/j.micpath.2020.104430
134. Lye HS, Rusul G, Liong MT. Removal of cholesterol by lactobacilli via incorporation and conversion to coprostanol. J Dairy Sci (2010) 93:1383–92. doi: 10.3168/jds.2009-2574
135. Wang J, Zhang W, Wang S, Liu H, Zhang D, Wang Y, et al. Swine-derived probiotic lactobacillus plantarum modulates porcine intestinal endogenous host defense peptide synthesis through tlr2/mapk/ap-1 signaling pathway. Front Immunol (2019) 10:2691. doi: 10.3389/fimmu.2019.02691
136. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science (2012) 336:1262–7. doi: 10.1126/science.1223813
137. Vemuri R, Gundamaraju R, Shinde T, Perera AP, Basheer W, Southam B, et al. Lactobacillus acidophilus DDS-1 modulates intestinal-specific microbiota, short-chain fatty acid and immunological profiles in aging mice. Nutrients (2019) 11:1297. doi: 10.3390/nu11061297
138. Wang X, Tsai T, Deng F, Wei X, Chai J, Knapp J, et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome (2019) 7:109. doi: 10.1186/s40168-019-0721-7
139. Sanders ME, Akkermans LM, Haller D, Hammerman C, Heimbach J, Hörmannsperger G, et al. Safety assessment of probiotics for human use. Gut Microbes (2010) 1:164–85. doi: 10.4161/gmic.1.3.12127
140. Choudhary J, Dubey RC, Sengar G, Dheeman S. Evaluation of probiotic potential and safety assessment of lactobacillus pentosus MMP4 isolated from mare's lactation. Probiotics Antimicrobial Proteins (2019) 11:403–12. doi: 10.1007/s12602-018-9431-x
141. Vesterlund S, Vankerckhoven V, Saxelin M, Goossens H, Salminen S, Ouwehand AC. Safety assessment of lactobacillus strains: presence of putative risk factors in faecal, blood and probiotic isolates. Int J Food Microbiol (2007) 116:325–31. doi: 10.1016/j.ijfoodmicro.2007.02.002
142. Cizeikiene D, Jagelaviciute J. Investigation of antibacterial activity and probiotic properties of strains belonging to lactobacillus and bifidobacterium genera for their potential application in functional food and feed products. Probiotics Antimicrob Proteins. (2021) 13:1387–403. doi: 10.1007/s12602-021-09777-5
143. Ahire JJ, Mokashe NU, Chaudhari BL. Effect of dietary probiotic lactobacillus helveticus on growth performance, antioxidant levels, and absorption of essential trace elements in goldfish (Carassius auratus). Probiotics Antimicrob Proteins. (2019) 11:559–68. doi: 10.1007/s12602-018-9428-5
144. Lim TJ, Lim S, Yoon JH, Chung MJ. Effects of multi-species probiotic supplementation on alcohol metabolism in rats. J Microbiol (2021) 59:417–25. doi: 10.1007/s12275-021-0573-2
145. Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol (2001) 67:1628–35. doi: 10.1128/AEM.67.4.1628-1635.2001
146. Spears JL, Kramer R, Nikiforov AI, Rihner MO, Lambert EA. Safety assessment of bacillus subtilis MB40 for use in foods and dietary supplements. Nutrients (2021) 13:733. doi: 10.3390/nu13030733
147. Pan DD, Zeng XQ, Yan YT. Characterization of lactobacillus fermentum SM-7 isolated from koumiss, a potential probiotic bacterium with cholesterol-lowering effects. J Sci Food Agric (2011) 91:512–8. doi: 10.1002/jsfa.4214
148. Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol (2016) 1:16203. doi: 10.1038/nmicrobiol.2016.203
149. Ghimire S, Roy C, Wongkuna S, Antony L, Maji A, Keena MC, et al. Identification of Clostridioides difficile-inhibiting gut commensals using culturomics, phenotyping, and combinatorial community assembly. mSystems (2020) 5:e00620-19. doi: 10.1128/mSystems.00620-19
150. Pereira AC, Cunha MV. An effective culturomics approach to study the gut microbiota of mammals. Res Microbiol (2020) 171:290–300. doi: 10.1016/j.resmic.2020.09.001
151. Amrane S, Raoult D, Lagier JC. Metagenomics, culturomics, and the human gut microbiota. Expert Rev Anti-Infect Ther (2018) 16:373–5. doi: 10.1080/14787210.2018.1467268
152. Chong PP, Chin VK, Looi CY, Wong WF, Madhavan P. Yong VC the microbiome and irritable bowel syndrome - A review on the pathophysiology, current research, and future therapy. Front Microbiol (2019) 10:1136. doi: 10.3389/fmicb.2019.01136
153. Martellacci L, Quaranta G, Patini R, Isola G, Gallenzi P, Masucci L. A literature review of metagenomics and culturomics of the peri-implant microbiome: current evidence and future perspectives. Materials (2019) 12:3010. doi: 10.3390/ma12183010
154. Abdallah RA, Beye M, Diop A, Bakour S, Raoult D, Fournier PE. The impact of culturomics on taxonomy in clinical microbiology. Antonie van Leeuwenhoek (2017) 110:1327–37. doi: 10.1007/s10482-017-0871-1
155. Suez J, Zmora N, Elinav E. Probiotics in the next-generation sequencing era. Gut Microbes (2020) 11:77–93. doi: 10.1080/19490976.2019.1586039
156. Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature (2016) 533:543–6. doi: 10.1038/nature17645
157. Forster SC, Browne HP, Kumar N, Hunt M, Denise H, Mitchell A, et al. HPMCD: the database of human microbial communities from metagenomic datasets and microbial reference genomes. Nucleic Acids Res (2016) 44:D604–9. doi: 10.1093/nar/gkv1216
Keywords: probiotics, core bacteria, precise supplementation, target isolation, gut abnormalities
Citation: Yang J, Qin S and Zhang H (2022) Precise strategies for selecting probiotic bacteria in treatment of intestinal bacterial dysfunctional diseases. Front. Immunol. 13:1034727. doi: 10.3389/fimmu.2022.1034727
Received: 03 September 2022; Accepted: 07 October 2022;
Published: 20 October 2022.
Edited by:
Zhihong Sun, Inner Mongolia Agricultural University, ChinaReviewed by:
Rajesh Pandey, CSIR-Institute of Genomics and Integrative Biology (CSIR-IGIB), IndiaBenedetta Mattorre, San Raffaele Hospital (IRCCS), Italy
Copyright © 2022 Yang, Qin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Zhang, emhhbmdoYW84MjdAMTYzLmNvbQ==
 Jiajun Yang
Jiajun Yang Shunyi Qin
Shunyi Qin Hao Zhang
Hao Zhang