- 1Department of Infectious Diseases and Immunology, College of Veterinary Medicine, University of Florida, Gainesville, Florida, FL, United States
- 2Department of Comparative, Diagnostic and Population Medicine, University of Florida, Gainesville, FL, United States
Helicobacter pylori is a major cause of gastric mucosal inflammation, peptic ulcers, and gastric cancer. Emerging antimicrobial-resistant H. pylori has hampered the effective eradication of frequent chronic infections. Moreover, a safe vaccine is highly demanded due to the absence of effective vaccines against H. pylori. In this study, we employed a new innovative Protective Immunity Enhanced Salmonella Vaccine (PIESV) vector strain to deliver and express multiple H. pylori antigen genes. Immunization of mice with our vaccine delivering the HpaA, Hp-NAP, UreA and UreB antigens, provided sterile protection against H. pylori SS1 infection in 7 out of 10 tested mice. In comparison to the control groups that had received PBS or a PIESV carrying an empty vector, immunized mice exhibited specific and significant cellular recall responses and antigen-specific serum IgG1, IgG2c, total IgG and gastric IgA antibody titers. In conclusion, an improved S. Typhimurium-based live vaccine delivering four antigens shows promise as a safe and effective vaccine against H. pylori infection.
1 Introduction
H. pylori is a Gram-negative bacterium infecting the stomach of more than half of the Earth’s human population and is the main cause of gastric pathologies including peptic ulcers, dyspepsia and gastric cancer (1, 2). Prevalence differs from 10 to >70% (3). H. pylori has been confirmed as a class I carcinogen by the World Health Organization (4). Gastric cancer is still the second leading cause of death by cancer worldwide (5). Eradication of H. pylori has been frequently considered as an efficient approach to cure peptic ulcer disease in addition to gastric cancer (6). No doubt, antibiotics are the first choice in treatment. However, H. pylori infections present numerous challenges to successful antimicrobial therapy, some of which are limited to H. pylori and others are experienced in the treatment of other infections. Challenges arise from the fact that H. pylori colonize the stomach where they are protected by a dense mucus layer and an acidic environment. Additionally, the stomach is continuously secreting acid and discharging its contents such that typical therapy would be diluted and washed out (7). The efficacy of several antimicrobials is greatly reduced at acidic pH and proper pH is needed for them to be effective. H. pylori can aquire resistant genotypes and become multi-drug resistant (MDR). Importantly, a recent study has shown that eradication of toxigenic H. pylori expressing VacA is not achievable using only antibiotics (8). In clinical practice, the quick emergence of resistance raised concerns about the correct management of this bacterial infection (9, 10). Thus, protective and therapeutic vaccines could be an alternative method for antibiotic treatment against H. pylori infection. The importance of CD4+ T cells in protective immunity against H. pylori has been broadly accepted (11). Oral immunization of recombinant Salmonella vectored vaccines could provoke classical Th1-type responses and also induce a significant mucosal SIgA response (12, 13) through transcytosis by micro-fold cells (14) or by direct antigen presentation by interstitial dendritic cells (15). It was reported that a considerable number of foreign antigens, synthesized and delivered by live attenuated Salmonella vector, protected animals against a diversity of pathogens including viruses, bacteria and parasites (16). Oral administration of a live attenuated Salmonella vector vaccine synthesizing and delivering H. pylori protective antigens was therefore early tested as an approach to eradicate H. pylori infections (17–26).
In this study, we employed recently developed much improved PIESVs that specifically synthesize heterologous antigens to enhance the induction of immune responses and protection against viruses and bacteria (27–29). The specific PIESV strain used in this study (29) has just been licensed by APHIS as the first genetically modified bacterial vectored vaccine in commercial distribution. Such mucosal PIESV delivery induces strong mucosal, systemic and cellular immunities against the targeted pathogen (30–35). Since H. pylori colonize and move through a mucosal body surface, the induction of mucosal immunity dependent on the production of secretory antibodies (SIgA/SIgM) as well as cellular immunity offers the first line of defense against infection with such pathogens (36–41). Injectable subunit recombinant protein vaccines are unable to induce such mucosal immunity in contrast to PIESVs that induce mucosal, systemic and cellular immunities that can block infections and preclude disease (27, 42–49). Here, we developed PIESVs using this most recently approved and licensed PIESV vector strain to synthesize and deliver H. pylori protective antigens to prevent H. pylori SS1 infection.
2. Materials and methods
2.1 Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and S. Typhimurium UK-1 derivative strains were routinely cultured in LB broth [19] or on LB agar at 37°C. S. Typhimurium UK-1 mutant strains were supplemented with 50 μg/ml of diaminopimelic acid (DAP), 0.05% arabinose, 0.1% mannose and 0.1% rhamnose when necessary for bacterial growth as described in previous work (29, 52–54). For animal experiments, S. Typhimurium χ12341 was cultured in LB broth with necessary supplements. Overnight cultures were diluted 1:100 into fresh medium and grown with aeration (200 rpm) to an optical density at 600 nm of ~0.85. Bacteria were then centrifuged at 5,000 × g for 15 min at room temperature and resuspended in buffered saline with 0.01% gelatin (BSG) (55). The H. pylori strain SS1 (a kind gift from Prof. James G. Fox, Massachusetts Institute of Technology) was grown on Brucella agar supplemented with 5% sheep blood, 25 μg/ml trimethoprim, 100 μg/ml vancomycin, 3.3 μg/ml polymyxin B, 50 μg/ml amphotericin B, 200 μg/ml bacitracinin and 10 μg/ml nalidixic acid in an anaerobic jar with a microaerophilic gas generating kit (BD, USA) for 5 days at 37°C.
2.2 Plasmids and constructs
The regulated delayed lysis vector pG8R111 (Figure 1A) for synthesis and delivery of antigens has a Ptrc-regulated synthesis of encoded protein antigens for delivery by cell lysis and araC ParaBAD-regulated murA and asdA genes with GTG start codons to lessen translation efficiency. The pG8R111 has a weaker SD AAGGCAA to further reduce the production of AsdA. The P22 PR located with opposite orientation to the transcription of the araC ParaBAD GTG-murA GTG-asdA genes is repressed by the C2 repressor made during the growth of χ12341 with arabinose. C2 concentration decreases due to cell division in vivo to cause PR-directed anti-sense mRNA synthesis to block translation of residual asdA and murA mRNA. Transcription terminators (TT) flank all plasmid domains for regulated lysis, replication, and gene expression so that expression in one domain does not interfere activities of adjacent domain. pG8R114 (Figure 1B) is derived from pG8R111 with a much-improved optimized β-lactmase signal sequence (56), the fusion of molecules to the bla SS in pG8R114 leads to delivery of molecules to the periplasm that results in increased production of outer membrane vesicles (OMVs) and releasing into the supernatant fluid surrounding PIESV cells that enhance induced immune responses (57, 58). We made constructs to synthesize nine putative protective H. pylori antigens according to the antigen gene sequences in H. pylori SS1 (2, 59–67) (Table 1). These constructs were evaluated for synthesis and delivery and protective immunity induction in mice. Antigens included VacA, CagA, UreB, UreA, HpaA, BabA, HopM, Hp-NAP and a chimeric antigen. To design the chimeric antigen with the most antigenic fragments of FliD, UreB, VacA and CagA, bioinformatic tools (65) were used to identify T-cell and B-cell epitopes. Sequences encoding FliD (1–600), UreB (327–385), VacA (744–805) and CagA (51–100) polypeptides were accessed from GenBank. To assist epitope exposure, flexible glycine-serine (GS) linkers were included between the gene segments. Sequences were codon-optimized to have a high-level expression in S. Typhimurium and GC contents adjusted to be closer to that for Salmonella. All genes were synthesized by Biomatik (Cambridge, Ontario, Canada). To detect the synthesis of recombinant proteins, a 6 His-tag sequence was added at the 3′ end of each gene before the stop codon. Based on the presence or absence of a signal peptide encoded in each antigen gene, antigens were divided into two groups. For those antigens without a signal peptide, the synthesized genes were cloned into pG8R111 whereas for those H. pylori antigens with signal sequences, these were removed, and the codon-optimized sequences were inserted into pG8R114 with fusion to the bla SS (Table 1). Considering the results obtained from the initial protection experiments (data not shown), four antigens were selected for further study. The codon-optimized H. pylori genes were fused with Plpp in front of the second gene and then were cloned under control of the Ptrc promoter of pG8R111 or pGR114 (Table 1) (Figure 1). Plasmids carrying genes were finally electroporated into χ12341. To obtain purified recombinant Hp-NAP, UreA, UreB and HpaA, sequences encoding each antigen were cloned into pET28a (+) (Table 1) using NcoI at N-terminal and XbaI at C-terminal sites. pET28a derivatives with inserts were electroporated into E. coli BL21(DE3) (Table 1) for synthesis and purification of gene products.
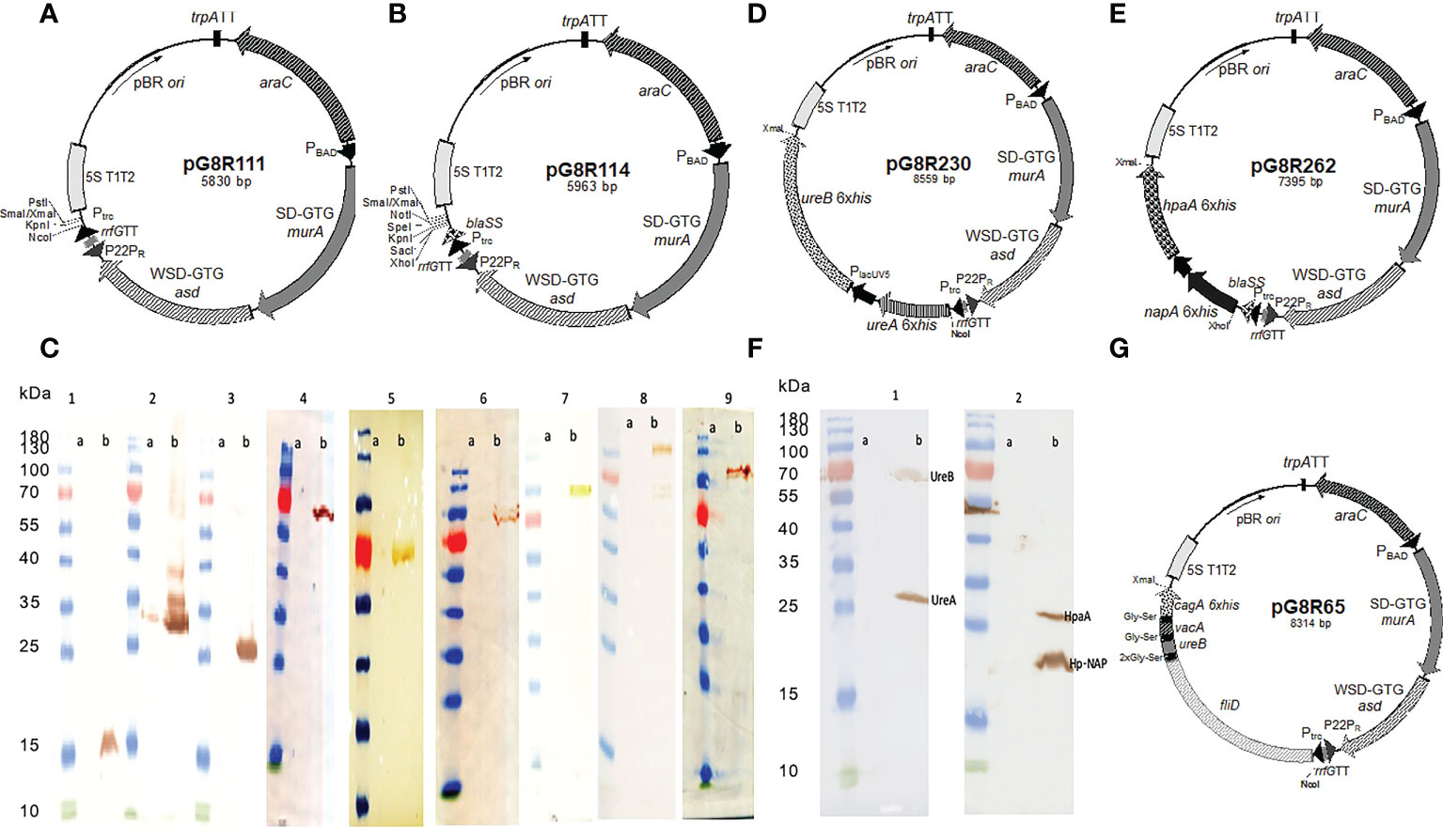
Figure 1 Synthesis of (H) pylori antigens in S. Typhimurium χ12341. Regulated delayed lysis plasmid vectors pG8R111 (A) and pG8R114 with improved bla SS (B). (C) Synthesis of nine putative (H) pylori antigens in S. Typhimurium χ12341. (1) Hp-NAP (17.7 kDa), (2) HpaA (26.9 kDa), (3) UreA (27.5 kDa), (4) UreB (62.3 kDa), (5) HopM (74.6 kDa), (6) BabA (79.6 kDa), (7) Chimeric Protein (90 kDa), (8) CagA (131.8 kDa), (9) VacA(136.3 kDa). (a) Uninduced with IPTG and (b) 2 h after induction with IPTG compared to molecular mass markers. Anti-6xHis monoclonal antibody was used to detect each recombinant protein by western blotting. (D). Regulated delayed lysis plasmid vectors pG8R230 encoding optimized ureA and Plpp ureB (H) pylori genes inserted into pG8R114 with expression controlled by Ptrc promoter and pG8R262 (E) encoding optimized napA and Plpp hpaA (H) pylori genes inserted into pG8R114 with expression controlled by Ptrc promoter. (F) Successful simultaneous synthesis of four protective (H) pylori antigens using two different plasmids in S. Typhimurium χ12341. (1) UreA and UreB encoded on pG8R230 and (2) Hp-NAP and HpaA encoded on pG8R262. (a) Uninduced with IPTG and (b) 2 h after induction with IPTG compared to molecular mass markers. Anti-6xHis monoclonal antibody was used to detect each recombinant protein in western blotting. (G) Regulated delayed lysis plasmid vector pG8R65 encoding the fliD-ureB-vacA-cagA fusion derived from pG8R111.
2.3 Determination of plasmid stability
Measurement of plasmid stability is described previously (55). Briefly, vaccine strains grown overnight (G0) were diluted 1:1000 into pre-warmed fully supplemented LB grown (permissive growth conditions) with aeration for 12 h at 37°C. This process was repeated for approximately 50 generations (G5) and the proportions of cells possessing the Asd+ plasmids were determined for each culture. The percentage of clones holding the plasmids from each culture was obtained by counting the colonies grown on LB agar without and with DAP. The ability of these clones to synthesize H. pylori antigens was also checked after 50 generations using western blotting.
2.4 Recombinant protein synthesis evaluated by SDS-PAGE and western blot analyses
S. Typhimurium strain χ12341 carrying recombinant plasmids (PIESV-Hp) or empty vectors (PIESV-empty vector) were cultured in LB containing 0.05% arabinose, 0.1% mannose and 0.1% rhamnose at 37°C. LB containing kanamycin 50 μg/ml was used to culture E. coli BL21 carrying recombinant plasmids as well. When the bacteria reached an OD600 of 0.6, 1 mM IPTG was added to the cultures to induce heterologous H. pylori protein syntheses. Protein samples from bacterial cultures were separated and analyzed by SDS-polyacrylamide gel electrophoresis with transfer to nitrocellulose membranes as previously described (68). Recombinant proteins were detected by anti-6xHis peroxidase (Roche, Basel, Switzerland) (1/40,000 dilution) for 2 h. E. coli BL21carrying pG8R289 (ureA-6xHis), pG8R290 (ureB-6xHis), pG8R291 (hpaA-6xHis) or pG8R292 (napA-6xHis) were grown overnight at 37°C in LB broth supplemented with 50 μg/ml kanamycin. The procedures for protein synthesis and purification have been described in our previous study (4). Prestained Protein Ladder, 10 to 180 kDa (Thermofisher, USA) was used as the protein marker.
2.5 Immunization of mice
All animal experiments were approved by the University of Florida Institutional Animal Care and Use Committee. Six-to eight-week-old SPF female C57BL/6 mice (n=10/group), were purchased from Charles River Laboratories (Wilmington, MA). Mice were acclimated for one week after arrival before starting the experiments.
Immunization procedures followed the previous description (4, 68, 69). Briefly, food and water were not given to mice for 6 h prior to the immunization and re-supplied 30 min later. Mice were orally immunized with 20 μl BSG containing 109 CFU of each vaccine strain, combination of strains, or with 20 μl BSG alone as the negative control on day 0 and boosted on days 14 and 28. Blood samples were collected individually on days 0, 14, 28, 42 and 72 and serum collected individually for analysis of antibody responses. Mice (10 mice) were immunized with 109 CFU of each PIESV synthesizing and delivering different antigens. A mixture of PIESV strains delivering pG8R111 and pG8R114 (PIESV-empty vectors) were used as controls.
2.6 Protection experiment
To assess whether the immunization of mice with Salmonella delivering H. pylori antigens was able to reduce bacterial burden of H. pylori in the stomachs of infected mice, mice were infected with 5×108 H. pylori SS1 thrice at one day intervals two weeks after the last immunization. Four weeks post infection, mice were euthanized and their stomachs removed and H. pylori CFUs quantified. To gain CFUs, the quantitative bacterial culture of mouse stomach was used. Briefly, a half section of the stomach from each euthanized mouse was completely homogenized, serially diluted, and then plated on selective medium as described above.
2.7 Evaluation of serum and mucosal antibody responses by ELISA
An enzyme-linked immunosorbent assay (ELISA) was used to specifically evaluate serum total IgG, IgG1 and IgG2c titers in immunized mice. 96-well polystyrene plates (Nalge Nunc. Rochester, NY, USA) were coated with purified recombinant HpaA, UreA, UreB and Hp-NAP (5 mg/ml). After over-night incubation, the plates were washed three times with TBST buffer (Tris-buffered saline, pH 7.4, containing 0.05% Tween 20) and followed by blocking with 300 µl PBS containing 10% FBS for 2 h at 37°C. After adding serial dilutions of mouse sera to the plates, they were incubated for 2 h at room temperature and then washed with TBST. HRP-conjugated goat-anti-mouse IgG1, IgG2c or total IgG (Southernbiotech, Birmingham, AL, USA) were added to the wells and incubated for 90 min at 37°C. Following the last washing step, specific reactivity was calculated by the addition of 50 µl/well of the TMB substrate (Thermofisher, USA). The reaction was stopped by adding 15 µl of 2 M H2SO4. Next, the absorbance at 450 nm was measured. To define a cut-off value for the test, the mean OD plus three-fold SD of sera from mice immunized with BSG was calculated at a 1:100 dilution. ELISA titers were calculated as the reciprocal of the last serum dilution providing an OD higher than the cut-off (70). To assess stomach mucosal IgA production, the secretory IgA was extracted by incubation of mouse stomachs in PBS containing 5% non-fat dry milk, 4-(2-aminoethyl)-benzenesulphonyl fluorid (Calbiochem), 1 mg/ml 3.25mM aprotinin, 10 mM leupeptin (Sigma), and bestatin (Boehringer-Mannheim Biochemicals). After extensive vortexing and centrifugation (16,000 g for 10 min), the supernatants were used for the determination of antibody titers. An ELISA was used as described above.
2.8 T-cell activation cell and cytokine profiling
2.8.1 Isolation of DCs from mouse bone marrow
C57BL/6 mice were euthanized, femurs were removed under sterile conditions, and then soaked in 70% alcohol for a minute. Both epiphyses were cut off with scissors, and the needle of a 1-mL syringe was inserted into the bone cavity to rinse the bone marrow out of the cavity into a sterile culture dish with RPMI 1640 supplemented with 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin and 10% heat-inactivated fetal bovine serum. The cell suspension in the dish was collected and counted. The cells suspended were distributed in plates at a density of 2×106 cells/per plate. Subsequently, GM-CSF was added into the medium to a final concentration of 20 ng/mL. The cells were cultured at 37°C in an incubator containing 5% CO2. The culture medium was replaced 72 h later to remove the unattached cells and cell debris, afterwards GM-CSF was added to the fresh medium. On day 10, the semi-suspended cells and loosely attached cells were collected by gently pipetting the medium against the plate. The cells were pulsed with recombinant proteins (HpaA, Hp-NAP, UreA and UreB) and incubated in complete medium overnight before co-culturing with T cells.
2.8.2 Antigen-specific T-cell assay
Mouse spleens were obtained from mice immunized with PIESV-Hp (hpaA, napA, ureA, ureB), PIESV-empty vector or PBS, at 45 days post the first immunization. T cells were isolated and counted using EasySep™ Mouse T Cell Isolation Kit (STEMCELL, Cambridge, MA.USA). T cells were then stained with CellTrace (Thermofisher, USA) to trace the propagation of T cells after re-stimulation by flow cytometry. After that, T cells and DCs already pulsed with recombinant proteins (HpaA, UreA, UreB and Hp-NAP) were co-cultured 10 to 1, (T cells to DCs, respectively) and incubated for 7 days. Supernatants were collected and cells were stained by antibodies against CD3, CD4, and CD8. Using Flow cytometer, T-cell propagation against each antigen in addition to different types of T cells were investigated.
Cytokine assay: The concentration of ten different cytokines including IFN-γ, TNF-α, IL-2, IL-4, IL-5, IL-6, IL-10, IL-17A, IL-22, and IL-23 were measured in both sera and supernatants described in the previous section. We used Multiplex Assays Using Luminex (Millipore-Sigma, USA) to evaluate the concentration of cytokines in the samples. In this method, the sample is added to a mixture of color-coded beads, pre-coated with different cytokine-specific capture antibodies. The antibodies finally bind to the interested cytokines. Specific detection antibodies of cytokines of interest conjugated with biotin are added and form an antibody-antigen sandwich. PE-conjugated streptavidin is then added. It attaches to the biotinylated detection antibodies. Polystyrene beads are read on a dual-laser flow-based detection set, using Luminex 200™. One laser categorizes the bead and defines the cytokine that is being detected. The second laser defines the greatness of the PE-derived signal, which is in direct quantity to the amount of cytokine bound.
2.9 Immunological responses in stomach of immunized mice after infection with H. pylori
2.9.1 Spleen Single-cell Suspension Preparation and Flow Cytometry
To obtain further information about how the immune system responded in the immunized mice after infection with H. pylori, ten days after the challenge (day 55) mouse spleens were collected. Single-cell suspensions from spleens were prepared by pressing spleens through 100 μm cell strainers. Cell suspensions were washed in PBS and resuspended in red blood cell lysis buffer [155 mM NH4Cl; 10 mM KHCO3; 0.1 mM EDTA] for 8 min on ice. Cell suspensions were washed again with PBS before staining. Antibodies were purchased from eBioscience (CD8a, IFN-γ, IL-17A, CD16/32, and TNFα), BioLegend (GranzymeB, Aqua Live/Dead), or TONBO(CD3 and CD4). CD16/32 antibody was used to block nonspecific binding to Fc receptors before all surface staining. For cytokine staining, cells were stimulated with 500 ng/mL ionomycin and 50 ng/mL phorbol-12-myristate-13-acetate (PMA) for 4 h, and Brefeldin A (2 mg/mL) was added 2 h before cell harvest. Dead cells were discriminated by LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Invitrogen). Sample acquisition was performed on FACSCantoII and analyzed with FlowJo (version 10.2, Tree Star).
2.9.2 Quantitative real-time PCR analysis
The expression of CXCL1, CXCL2, CXCL5, Reg3a, Reg3b, Reg3d and Reg3g were investigated using qPCR (71, 72). Briefly, RNA extracted from mouse stomach biopsy specimens obtained ten days after infection were reverse-transcribed to cDNA by PrimeScriptTM RT reagent Kit (ThermoFisher). Real-time PCR was performed on the IQ5 (Bio-Rad) with PowerUpTM SYBERTM Green Master Mix (appliedbiosystems, USA) according to the manufacturer’s specifications. β-actin was used for normalizing the expression. The relative gene expression was expressed as fold change calculated by the ΔΔCt method (73).
2.10 Histochemistry
Mouse stomachs were obtained 75 days post the first immunization. Stomach tissues were fixed in 10% neutral buffered formalin for at least 24 h. The trimmed tissues were processed, paraffin-embedded, sectioned at 5 μm, and stained with hematoxylin and eosin. Selected sections of stomach tissues were also evaluated with Warthin-Starry and Gram stains to identify bacteria. The pathologist was blinded as to the experimental groups and treatments of the study. Histologic samples of both glandular and squamous portions of the stomach were examined and evaluated and scored based on intensity 0 to 3 for criteria including mucosal inflammation and type, submucosal inflammation and type, non-H. pylori bacteria, mucosal ulceration, and hyperkeratosis of squamous stomach.
2.11 Statistical analysis
SPSS computer software, version 17.0. was used to do statistical analysis. The statistical differences between two groups were studied by t-test and results among several groups were obtained by one factor analysis of variance (ANOVA) and Tukey’s post hoc test. The statistical border for accepting significance was P< 0.05. Data were envisaged using the GraphPad Prism 7 program.
3. Results
3.1 Salmonella Strain χ12341 and Plasmid encoding H. pylori antigens
The construction and properties of χ12341 have been recently described (29). Three sugars, arabinose, mannose and rhamnose which are not present in animal tissues, were used to regulate the virulence trait. The ΔPmurA::TT araC ParaBAD murA and ΔasdA::TT araC ParaBAD c2 mutations regulate synthesis of diaminopimelic acid and muramic acid, two essential constituents of peptidoglycan, which enable PIESV lysis in the absence of DAP and inability to synthesize the MurA enzyme without arabinose in vivo (53). The Δpmi ΔwaaL ΔpagL::TT rhaRS PrhaBAD waaL mutations collectively provide regulated delayed attenuation and cause cessation in the synthesis of LPS O-antigen in the absence of the sugars mannose and rhamnose in vivo (52, 74, 75). The deletion Δ(wza-wcaM) enhances complete lysis, blocks the synthesis of cell surface polymers to prevent biofilm formation and enhances induced immunities (76). The ΔrelA::araC ParaBAD lacI TT mutation confers the regulated delayed antigen synthesis phenotype that enhances PIESV colonization and induction of immune responses by the arabinose-dependent regulated LacI synthesis so that Ptrc-regulated genes on plasmid vectors are gradually expressed as LacI is diluted at each cell division during in vivo growth (77–79). The ΔrecF mutation decreases inter- and intra-plasmidic recombination to stabilize PIESVs (80). The ΔsifA mutation enables PIESV escape from the Salmonella-containing vesicles (SCVs) to cause PIESV cells to lyse in the cytosol (81, 82). This enables delivery of protective antigens to the proteasome for class I presentation to generate CD8-, CD17- and NKT-dependent immunities.
Codon-optimized sequences encoding all nine H. pylori antigens were successfully inserted into the plasmid vectors pG8R111 or pG8R114 (Figures 1A, B) to yield the plasmids listed in Table 1. The synthesis of each antigen was confirmed by western blot using a monoclonal antibody against the 6His-tag (Figure 1C) to yield the plasmids listed in Table 1. Results of the plasmid stability experiment showed that all recombinant plasmids encoding antigens except HopM were stable in the Salmonella vaccine strain. However, only 30% of Salmonella cells lost the plasmid specifying HopM synthesis after 50 generations of growth under permissive conditions. Based on results obtained from initial protection experiments, UreA+UreB (pG8R230) and HpaA+Hp-NAP (pG8R262) (Figures 1D, E), the set of antigens providing considerable protection, were selected to be expressed by pG8R111 and pG8R114, respectively. χ 12341(pG8R230) and χ 12341(pG8R262) specified synthesis of the four encoded antigens after IPTG induction (Figure 1F). We also designed and expressed a chimeric antigen including protective parts of four four putative antigenis (FliD, VacA, CagA and UreB) of H. pylori using pG8R111 in PIESV χ 12341 (Figure 1G).
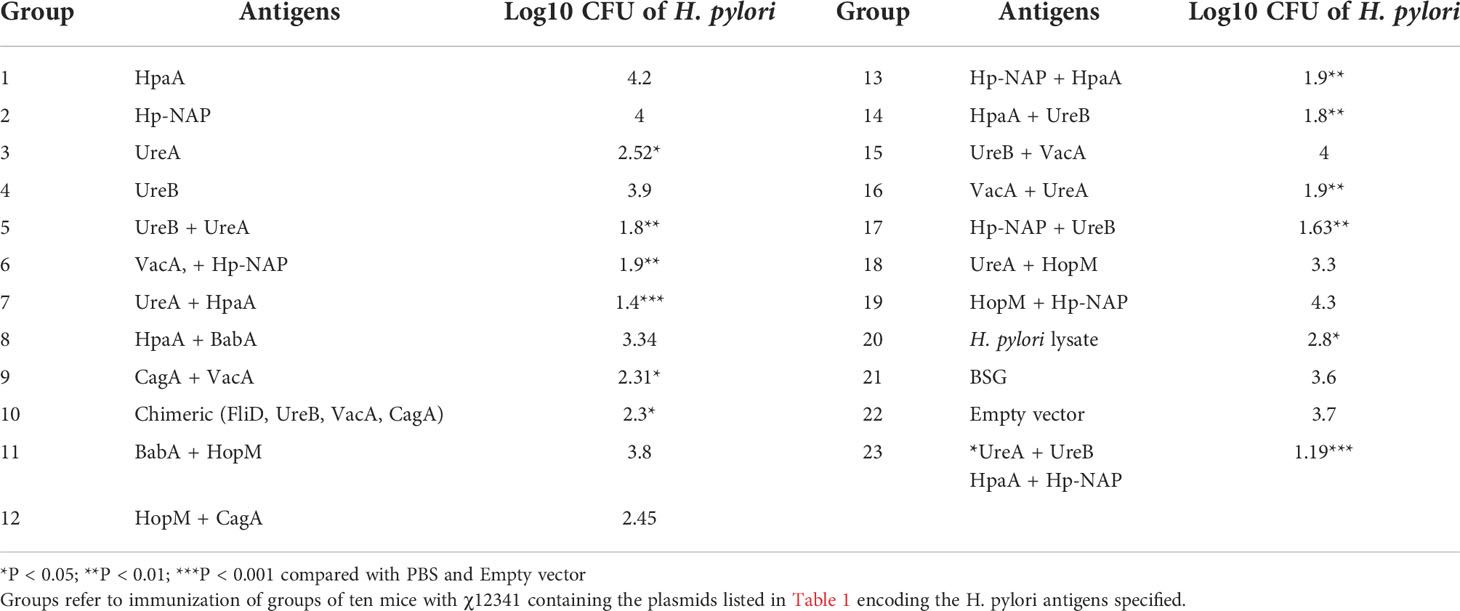
3.2. Immunization of mice with Salmonella carrying pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP) induces significant protection against H. pylori SS1 challenge with strong specific humoral and mucosal immune responses
To determine whether immunization with vaccine candidates lowers the bacterial burden in the stomachs of infected mice, we ascertained CFUs of H. pylori using quantitative bacterial culture procedures. Immunized mice were infected with H. pylori SS1 two weeks after the last immunization. Then, four weeks after challenge, the stomachs of euthanized mice were removed, minced, homogenized, serially diluted and then plated on the selective agar medium. As shown in Table 2; Figure 2A, higher levels of protection were observed in mice immunized with the Salmonella vaccine PIESV-Hp strains carrying pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP). Notably, seven out of 10 immunized mice in this group showed sterile protection and the other three mice showed a significant reduction in a load of bacteria compared to mice receiving BSG or empty vector (Figure 2A).
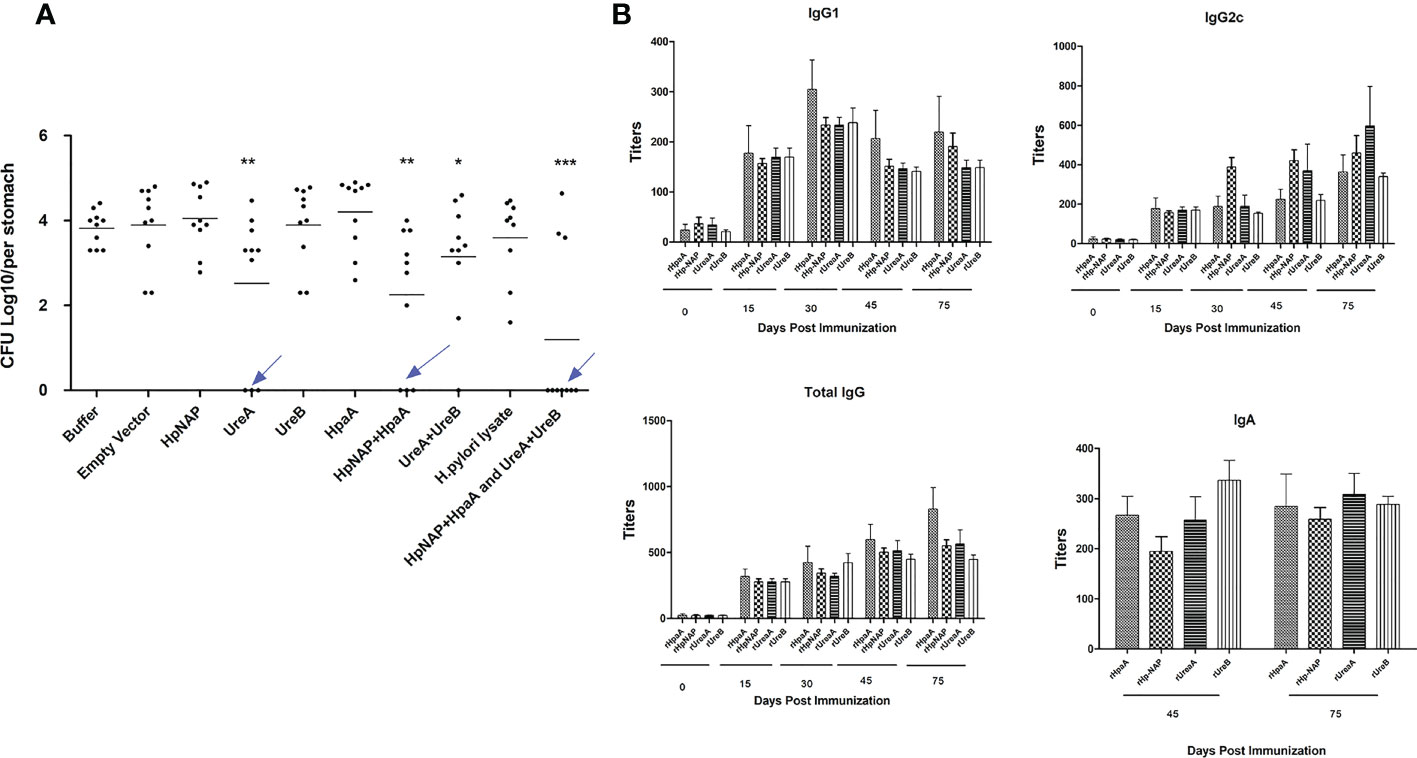
Figure 2 (A) Protection against (H) pylori SS1 challenge infection following oral vaccination of mice with individual and combinations of χ12341 strains harboring plasmids listed in Table 1 synthesizing and delivering specified (H) pylori antigens. (B) Specific antibody responses in immunized animals. Kinetics of specific antibody responses after oral immunization with a cocktail vaccine of χ12341(pG8R230) and χ12341(pG8R262) delivering UreA and UreB and NapA and HpaA, respectively. Mice were bled (submandibular bleeding method) on the indicated days, and specific IgG1 and IgG2c and total IgG antibody titers against recombinant UreA, UreB, NapA and HpaA evaluated by ELISA. Titer values represent the mean ± SD of sera from three analyses of five animals each. Stomach suspensions were obtained 45 days post first immunizationand specific IgA titers were analyzed by ELISA.
To study the humoral response against antigens in mice immunized with PIESV vector strains delivering four antigens, sera were obtained at several time points over three months following initial immunization. Immunization of mice induced strong and specific immunoglobulin G (IgG) responses against each component of the cocktail vaccine including pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP), in which IgG2c (Th1-related isotypes) titers were usually slightly higher than those of the IgG1 subtype (Figure 2B). The total IgG and IgG2c titers against each antigen started to increase during the second week after the first immunization and peaked after 75 days. However, IgG1 titers peaked at 30 days post first immunization and titers fell after 75 days. These results indicated that immunization of mice with Salmonella vaccine vectors carrying pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP) elicit a Th1-biased immune response. To evaluate whether immunization with PIESV strains delivering these four antigens also provoked mucosal immune responses, gastric IgA production was evaluated for each antigen in stomachs of immunized mice. As shown in Figure 2B, the immunized mice with the cocktail of four antigens significantly augmented gastric mucosal IgA titers against each component of the cocktail vaccine. These findings indicate that immunization of mice with a combination of pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP) synthesized and orally delivered by two PIESV vector strains provokes both specific systemic and mucosal humoral immune responses.
3.3. Immunization of mice with Salmonella carrying pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP) induces mixed Th1-, Th2-, and Th17-type immune responses
3.3.1 T-cell propagation
To find out the dominant subset of T cells in the spleen of immunized mice and assess whether such T cells could propagate after re-stimulation with related antigens in vitro, T cells were co-cultured with dendritic cells pulsed with each component of a cocktail vaccine composed of recombinant UreA, UreB, HpA and HpNAP proteins. The percentage of CD4+ and CD8+ cells in the immunized group were significantly increased compared with PBS or PIESV-empty vector immunized mice (Figure 3A). Additionally, CD8+ cells propagated more than CD4+ cells showing a dominant cytotoxic response in immunized mice in response to each antigen (Figure 3A).
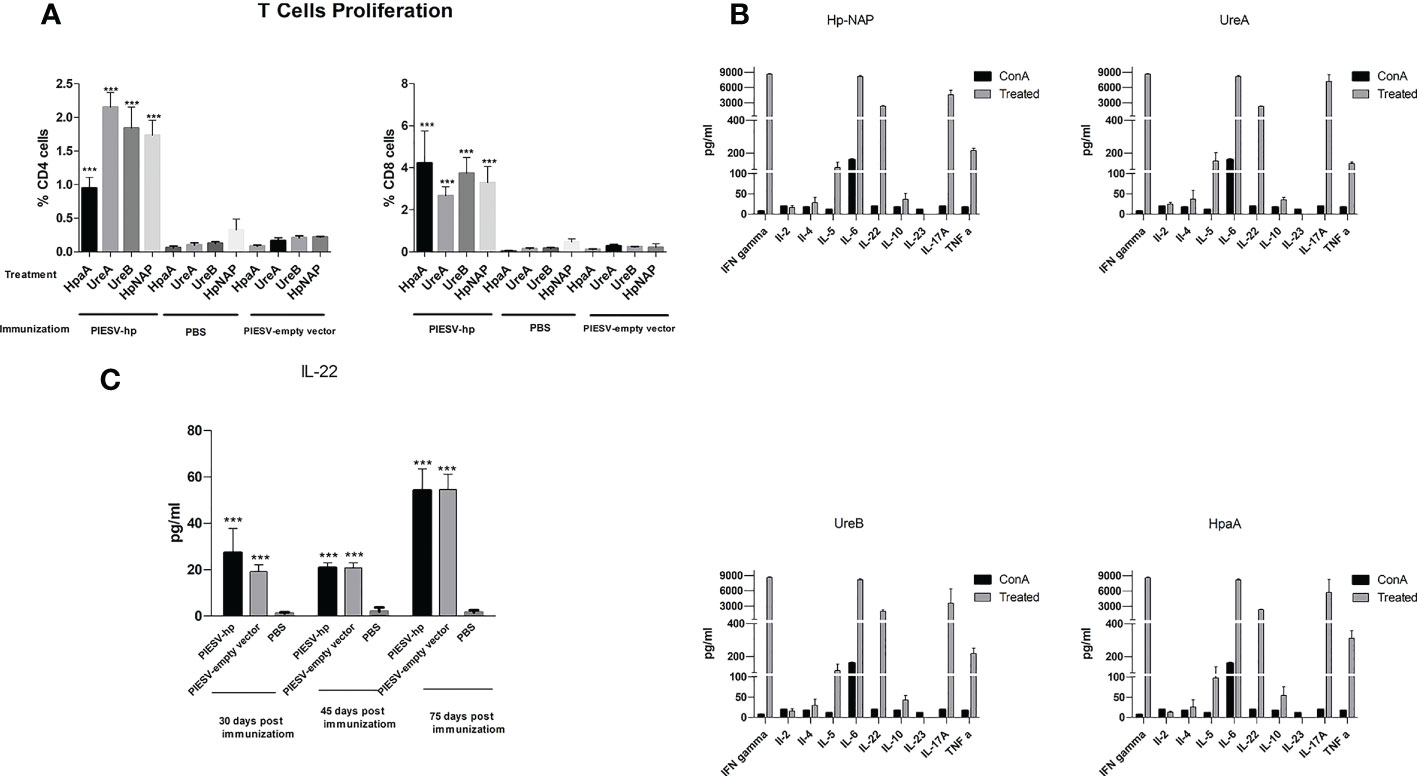
Figure 3 T cell responses (A) Proliferative responses of T cells from mice immunized with the χ12341(pG8R230) and χ12341(pG8R262) mixture, χ12341(pG8R111) = PIESV-empty vector) and PBS. T cells from immunized mice were isolated 45 days post first immunization and co-cultured with dendritic cells treated with 20 µg/ml recombinant NapA, HpaA, UreA and UreB and the proliferative responses were subsequently assayed by CellTrace. T cells were also stained with proper antibodies to be analyzed for the proliferation of CD4+ and CD8+ T cells by flow cytometry. (B) Cytokine production by T cells from mice immunized with the mixture of χ12341(pG8R230) and χ12341(pG8R262). T cells were obtained 45 days post first immunization from immunized mice and co-cultured with dendritic cells already treated with 20 µg/ml recombinant NapA, HpaA, UreA and UreB. Cytokine concentrations in culture supernatants were measured by multiplex ELISA. The data are the mean ± SD of five individual mice from each group. (C). Level of IL-22 increased in mice immunized with the PIESV-Hp mixture. Sera obtained 30, 45 and 75 days after first immunization of mice with PIESV-Hp, PIESV-empty vectors and PBS were analyzed for the presence of IL-22. ***P < 0.001.
3.3.2. Cytokine production
Since cytokines secreted by activated T cells are indicators of the type of Th responses, we also measured the amounts of ten different cytokines in supernatants obtained in T-cell propagation experiments. Cytokine production by the re-stimulated T cells was determined by Multiplex ELISA. In comparison to the controls, T cells of mice immunized with the combination of pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP) synthesized and delivered by two PIESV vector strains secreted significantly higher amounts of IFN-γ, IL-5, IL-6, IL-22, IL-17A and TNF-α in response to each recombinant protein used for immunization (Figure 3B). We could also detect small amounts of IL-2, IL-4 and IL-10 but not IL-23 secreted against each antigen after re-stimulation with related antigens. These results suggest that immunization of mice with a combination of Hp-NAP, UreA, UreB and HpaA synthesized by the PIESV strains induces a mix of Th1, Th2, and Th17 responses. Further study using sera obtained from immunized mice showed that the level of circulating IL-22 cytokine increased following immunization compared to the non-immunized control groups (Figure 3C).
3.4 Immunized mice increase IFN-γ+ CD4 T and CXCL2 after infection with H. pylori SS1
Antimicrobial peptide plays a important role in immune responses to H. pylori (83, 84). To obtain a deeper understanding of the roles of antimicrobial peptides and immunological markers in the protection provided in immunized mice, ten days after infecting the tested mice with H. pylori SS1, mouse stomachs and spleens were obtained. Reg3a, Reg3b, CXCL1, CXCL2, and CXCL5 in stomach tissues were investigated by qPCR. These chemokines and antimicrobial peptides were selected since they have been shown to work in correlation with IL-17 (85, 86). Spleen samples were further analyzed for T-cell markers and cytokines. In the stomachs of immunized mice after infection with H. pylori SS1, the expression of CXCL2 increased two-fold while the expression of the remaining genes did not change (Figure 4A).
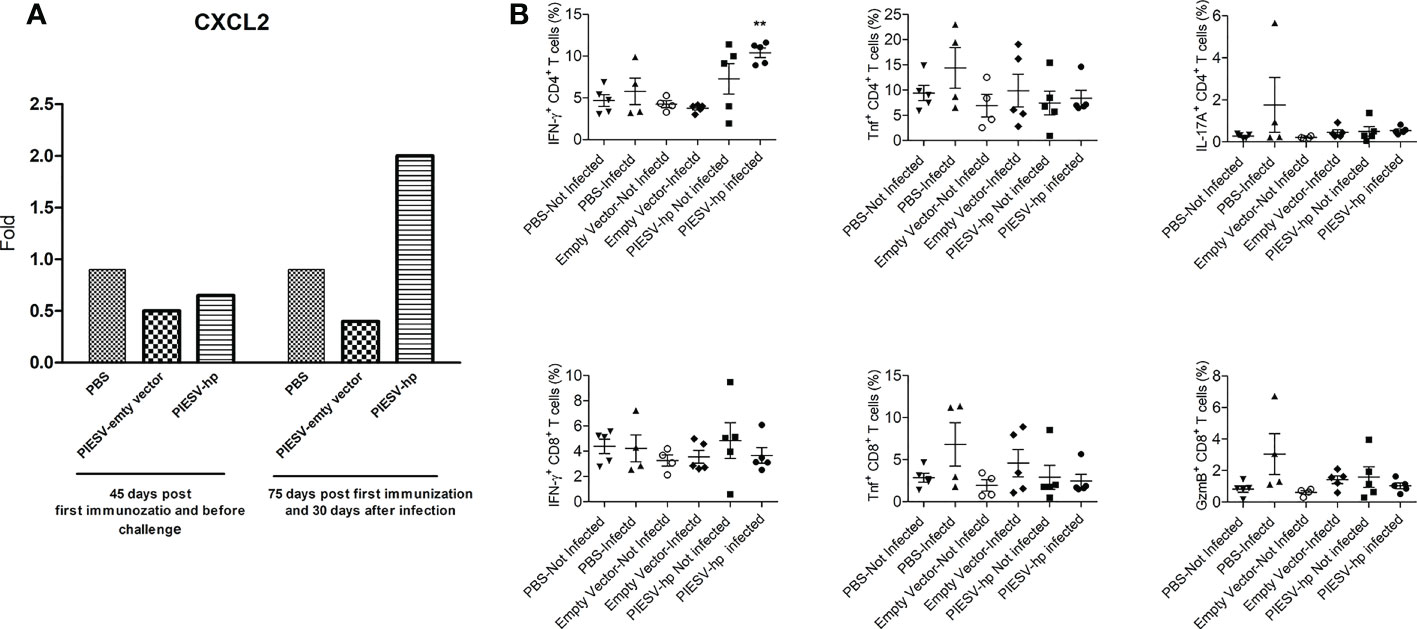
Figure 4 (A) CXCL2 is increased in stomach of (H) pylori SS1-infected mice immunized with PIESV-Hp mixture. CXCL2 mRNA synthesis in stomach of (H) pylori SS1-infected mice immunized with PIESV-Hp, and PIESV (empty vector) and PBS were compared (n = 10). (B) PIESV-Hp mixture immunized mice showing increases in IFN-γ+ CD4 T after challenge infection with (H) pylori SS1. Fifty-five days after the first immunization, mouse spleens were collected. Single-cell suspensions from spleens were isolated and then stained with proper antibodies to be analyzed by flow cytometry. **P < 0.01.
In response to H. pylori SS1 infection, IFN-γ+ CD4 T cells increased in the spleens of immunized mice compared to the controls. Additionally, no significant changes in TNF+ or IL-17A+ CD4 T cells in the spleen for any test groups were seen. There were also no significant changes in IFN-γ+, TNF+ or GranzymeB+ CD8 T cells in the spleen for any test group (Figure 4B).
3.5 Histopathology
The stomach tissues were evaluated and scored based on severity 0 to 3 for criteria including mucosal inflammation and type, submucosal inflammation and type, presence of non-H. pylori bacteria, mucosal ulceration, and hyperkeratosis of squamous portions of the stomach. The scale 0-3 refers to 0=normal, 1=mild, 2=moderate, 3=severe. The inflammation ranged from nominal to severe both in the mucosa and submucosa of both the glandular and squamous portions of the stomach and consisted of lymphocytes, plasma cells with or without smaller numbers of neutrophils. Mice in control group had large amounts of hyperkeratosis in the squamous portions of the stomach which are often associated with moderate to large numbers of large, Gram-positive, rod-shaped bacteria on the surface or within the laminated keratin. The hyperkeratosis and associated bacteria are suggestive of hyporexia or anorexia for a relatively prolonged period of at least 2 days. In anorectic rodents, the build-up of excess keratin is presumably caused by reduced mechanical removal by the passage of food (87). The hyperkeratosis was not seen in a single area as suggested at the limiting ridge, but throughout all of the squamous lined areas of the forestomach.
However, significant weight loss was not seen in any group of mice. Overall, the mice receiving the PIESV-empty vector had relatively higher inflammation and numbers of non-H. pylori bacteria (gram-positive rod-shaped bacilli) compared to immunized mice. These findings might suggest the role of other gram-positive bacteria as well as H. pylori infection in the development of stomach complications. Additionally, these findings demonstrated that the vaccine candidate not only protects against H. pylori SS1 but also against other non-H. pylori flora, which may have a role in stomach complications (Figure 5).
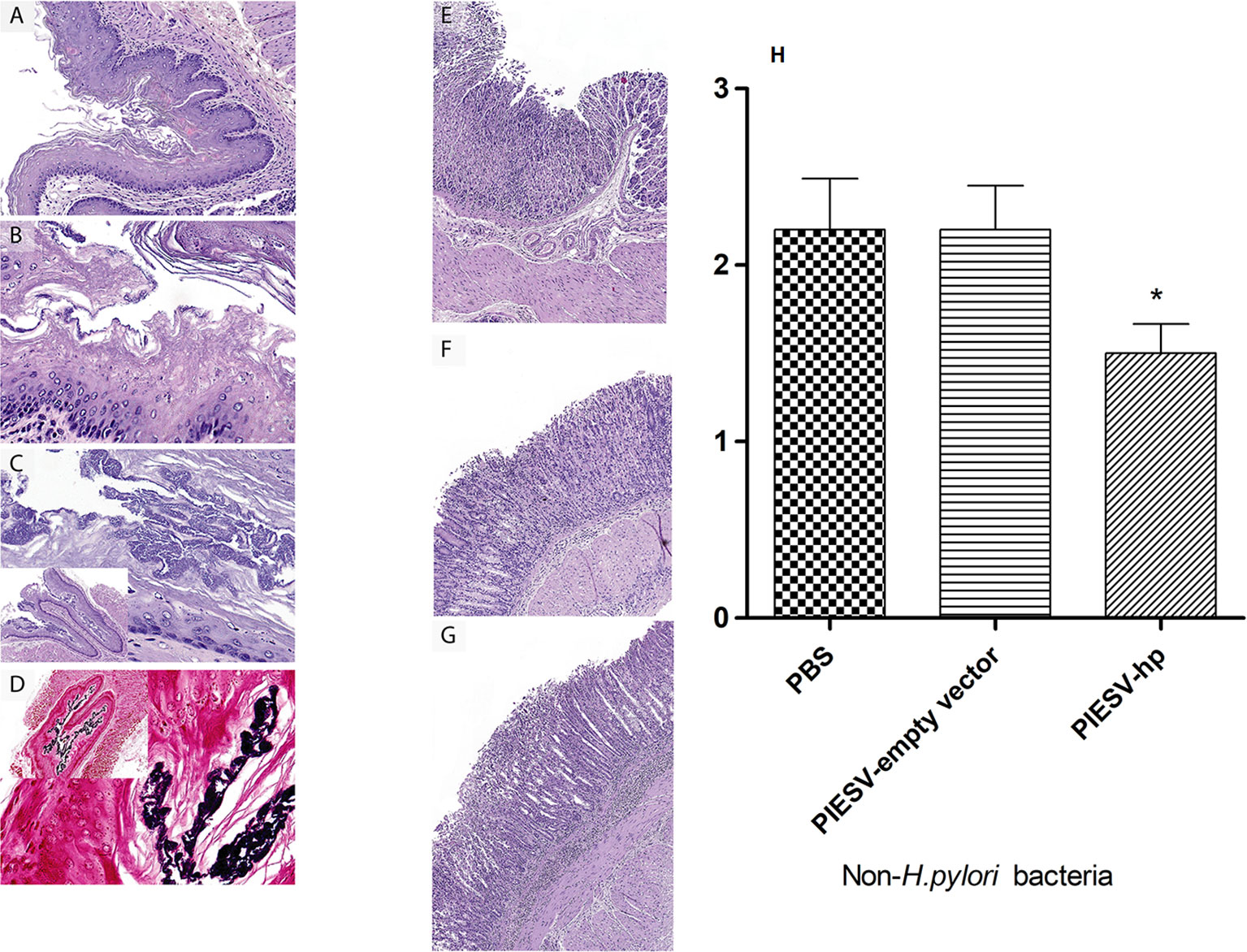
Figure 5 Immunization with the PIESV-Hp mixture reduces inflammation and the number of Gram-positive rod-shaped bacteria in the mouse stomach. Mice immunized with the PIESV-Hp mixture, the PIESV-empty vector and PBS were infected with (H) pylori SS1 two weeks post last immunization. After thirty days, mice were sacrificed, and stomach tissues were obtained for H&E staining. (A) Squamous portion of stomach of mice immunized with the PIESV-Hp mixture showed moderate amounts of hyperkeratosis. (B) Abundant hyperkeratosis with moderate numbers of large rod-shaped bacteria (Gram stain) associated with the keratin layer in mice immunized with PIESV-empty vector. (C) Same as B but with much larger populations of gram-positive bacteria in the keratin layer of mice immunized with PIESV empty vector. (D) Same as C but in mice immunized with PBS. Glandular portion of the stomach with (E) mild, (F) moderate, and (G) severe mucosal and submucosal inflammation in mice immunized with the PIESV-Hp mixture (E & F) or the PIESV empty vector or PBS (G). (H) Reduction in the load of rod-shaped bacteria in immunized mice based of relative frequency scores of 0 (absent), 1 (low), 2 (moderate) and 3 (high). P<0.05.
4 Discussion
Salmonella has been extensively investigated for delivering recombinant protective antigens and DNA vaccine vectors due to its capability to be delivered mucosally, thereby providing needle-free immunization especially against H. pylori (17, 18, 27, 88). Although these vaccines were protective in animal models, they were less effective in humans (17, 21, 89, 90). Therfore, there is a need for an efficient Salmonella to synthesize and deliver protective antigens inside the host to immunize all armes of immune systems efficiently. Live attenuated bacterial vaccines preferably comprise strains that possess two or more stable attenuating mutations dispersed in the bacteria chromosome and plasmids (without antibiotic-resistant markers) into which genes encoding heterologous antigens from microbial pathogens can be introduced. The attenuating mutations should be in genes encoding essential components of bacterial cell structures or biosynthetic pathways for crucial nutrients that are not freely available in any environment that the PIESV may reside in the immunized animal host or outside of the laboratory (Clark-Curtiss and 51).
In this study, we employed a recently much improved PIESV vector strain with superior attributes compared to previously used recombinant attenuated Salmonella vaccine vector strains used by others or by us. Based on the succeed of Salmonlla-C. perfringens vaccine development (29), this PIESV vector strain χ12341 was thus used to deliver and synthesize nine recombinant protective antigens of H. pylori. Mice were immunized with each construct alone or a combination of them. Upon completion of these survey studies, we investigated PIESV constructs delivering combinations of two antigens (Table 1) to determine those providing the most favorable protection. Our findings showed that immunization of mice with pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP) provided the most significant level of protection against H. pylori SS1 infection. H. pylori strains display a high level of genetic heterogeneity (91). However, efforts to find a safe vaccine against H. pylori have resulted in using several protective antigens that are very conserved in H. pylori strains. To form a neutral microenvironment around the bacterium in the acidic environment of the stomach, H. pylori needs to produce large amounts of urease which is formed by two subunits, UreA and UreB (92), which have been broadly used as candidate antigens for the development of vaccines against H. pylori infection (93). HpaA has been identified on the surface of H. pylori and is highly conserved among H. pylori strains (94). Another well conserved antigen is Hp-NAP that is essential for both pathogenesis and immunity (59). Multivalent vaccines may stimulate more efficient and full protection against H. pylori infection. In designing multivalent vaccines, the possible interaction of components with each other should be given special attention such as synergistic and antagonistic effects or competition between components and even epitope suppression, resulting in an inappropriate immune response (93). Importantly, in this study, we observed that combination of UreA, UreB, HpaA and HpNAP when synthesized and delivered by a PIESV vector strain could provide synergistic effects. These results not only emphasize the efficacy of our cocktail vaccine to provide sterile protection but also show the strong applicability of PIESVs, especially the improved plasmids used in this system. To unravel the mechanism of protection provided by our cocktail vaccine, the humoral and cellular immune responses induced by immunization were evaluated and compared to mice immunized with the PIESV-empty vector strain or with PBS.
Though the detailed mechanism by which immunization protects against H. pylori remains to be clarified, the vital role of cell-mediated immunity has been well-known with evidence demonstrating that protection against H. pylori infection can be obtained even in the lack of B cells (95). In this regard, immunological eradication of H. pylori usually is contingent on Th1-type responses and IFN-γ, the Th1 cytokine, playing a main role in the control of H. pylori infection by activating macrophages and changing antibody responses toward protective IgG2c in C57BL/6 mice (96). Our data show very high levels of Th1 (IFN-γ and IL-12) and Th17 (IL-17) cytokines during recall responses of T cells from mice immunized with our cocktail vaccine including pG8R230(UreA+UreB) and pG8R262(HpaA+HpNAP). Consistent with the proliferative response, T cells from mice immunized with our vaccine, produced higher levels of IFN-γ in response to each component of the cocktail than mice that received PBS or a mixture of the empty vector PIESV strains. In addition, T cells of immunized mice produced high levels of IL-6 when re-exposed to the immunizing antigens. IL-6 is deemed one of the mediators directing the transition between innate and adaptive immunity. Its production is mostly targeted by the PAMP-mediated TLR signaling cascade and as a result, IL-6 gene-deficient mice display impaired defense against some bacterial infections (97). Thus, it is possible that this cytokine plays a key role in providing immunity against H. pylori infection. Besides, Th17 cells and their effector cytokines seem to mediate immunity against numerous infections, mainly those caused by extracellular pathogens such as H. pylori (98). Th17 immune responses include neutrophil recruitment, secretion of antimicrobial peptides, and Th1-mediated protection driven by IL-17 (99, 100). In our study, a significant amount of IL-17 was measured after the re-stimulation of T cells with each recombinant antigen in the cocktail vaccine. Our finding showed that the level of circulating IL-22 cytokine spiked after immunization with the PIESV-Hp mixture and the PIESV-empty vectorcompared to the non-immunized control group only receiving PBS. It seems that Salmonella vaccines are able to induce the production of circulating of IL-22 that has been shown to play an important role in protecting the gut tissue integrity and enhance disease lessening through chronic Salmonella Infection (101).
To gain insight further into the underlying mechanism, we evaluated the kinetics of antibody production against each component of the vaccine and found that both antigen-specific IgG1 and IgG2c antibodies were induced. Notably, the IgG1/IgG2c ratio also showed that the response elicited in immunized mice had a strong Th1 bias. Since IFN-γ and IgG2c are markers of the Th1 type of immune response in C57BL/6 mice, and synthesis of IgG1 can be affected by Th2 clones, seeing the results obtained from cytokine assays and these findings, one might conclude that the immune responses elicited by the vaccine was of a mixed Th1/Th17 type (96). However, our cytokine detection was limited to the serum and spleen such that it might not reflect the levels of cytokines produced in the stomach. Further research to evaluate the cytokine production in the stomach with RNA-seq and/or flow cytometry will provide a deeper insight about the dynamics and function of immune cell populations and cytokine production in the stomach.
We then measured the mucosal immune responses produced by immunization. With that goal, the gastric IgA production and immune-mediated inflammation were evaluated in immunized mice. Compared to control immunized mice, immunized mice elicited significant levels of each antigen-specific gastric IgA. These findings indicate that immunization with PIESV strains delivering heterologous antigens efficiently stimulated gastric mucosal immune responses.
We also investigated the responses of other components of the immune system in immunized mice against infection by H. pylori. Thus synthesis of several antimicrobial peptides and chemokines were assessed. Our results only showed that production of CXCL2 is significantly upregulated in the stomach of immunized mice. CXCL2 is also a mighty neutrophil chemoattractant and is involved in numerous immune responses comprising wound healing, cancer metastasis, and angiogenesis (102). This result shows that our vaccine is capable to induce recall innate as well as acquired immunity. Systemic immune responses were also studied in immunized mice. Although in proliferation assays, CD8+ T cells were propagated more than CD4+ T cells in response to re-stimulation, the number of CD4+ T cells but not CD8+ T cells expressing IFN-γ increased after infection with H. pylori SS1 in the spleen of immunized mice. This supports the IgG titer data that indicate that systemic immunity was skewed towards a Th1 response after immunization. However, local immunity in the stomach tissues may be more dependent on CD8+ T cells. Further investigation is therefore needed to clarify the contribution of CD4 versus CD8 T cells in the stomach tissue.
The results of histopathology measurements showed an increased number of gram-positive rod-shaped bacteria and hyperkeratosis resulting in excessive pressure, inflammation or irritation to the stomach mucosa of control groups compared to the immunized group. Although we did not verify the species of this population, this finding shows the role of H. pylori infection in increasing the number of other gram-positive bacilli which might have a role in worsening the stomach complications caused by H. pylori infection. In this regard, it has been indicated that gastric colonization by non-H. pylori bacteria, such as Bacteroides, Actinobacteria, Firmicutes, Fusobacteria, and Proteobacteria, could influence the risk for gastric cancer (103–107). Notably, our vaccine not only provided sterile protection in seven out of 10 of the immunized mice but it also caused a reduction in the titer of a gram-positive rod-shaped bacterium.
To sum up, a S. Typhimurium-based live vaccine including the delivery of four protective antigens of H. pylori confers significantly high protection against H. pylori infection in mice. Additionally, immunological studies showed the induction of both specific acquired immune responses against each antigen and the innate immunity responses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by University of Florida Institutional Animal Care and Use Committee.
Author contributions
The experiments were conceived and designed by AG and RC and performed by AG, SW, BS, and JA. The data were analyzed by AG and RC. The manuscript was written by AG, SW, and RC. All authors contributed to the article and approved the submitted version.
Funding
This work was supported partially by Pilot Award (P0109858) (AG) from the Clinical and Translation Sciences Institute (CTSI) at the University of Florida and by R01 AI60557 (RC) and R21 AI126172 (SW) grants from the (NIH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1034683/full#supplementary-material
References
1. Correa P, Piazuelo MB. Helicobacter pylori infection and gastric adenocarcinoma. US Gastroenterol Hepatol Rev (2011) 7:59–64.
2. Blanchard TG, Czinn SJ. Identification of Helicobacter pylori and the evolution of an efficacious childhood vaccine to protect against gastritis and peptic ulcer disease. Pediatr Res (2017) 81:170–6. doi: 10.1038/pr.2016.199
3. Graham DY, Malaty HM, Evans DG, Evans DJ Jr., Klein PD, Adam E. Epidemiology of Helicobacter pylori in an asymptomatic population in the united states. effect of age, race, and socioeconomic status. Gastroenterology (1991) 100:1495–501. doi: 10.1016/0016-5085(91)90644-Z
4. Ghasemi A, Mohammad N, Mautner J, Taghipour Karsabet M, Amani J, Ardjmand A, et al. Immunization with a recombinant fusion protein protects mice against Helicobacter pylori infection. Vaccine (2018) 36:5124–32. doi: 10.1016/j.vaccine.2018.07.033
5. Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med (2008) 47:1077–83. doi: 10.2169/internalmedicine.47.0975
6. Talebi Bezmin Abadi A. Vaccine against Helicobacter pylori: Inevitable approach. World J Gastroenterol (2016) 22:3150–7. doi: 10.3748/wjg.v22.i11.3150
7. Graham DY, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat Clin Pract Gastroenterol Hepatol (2008) 5:321–31. doi: 10.1038/ncpgasthep1138
8. Capurro MI, Greenfield LK, Prashar A, Xia S, Abdullah M, Wong H, et al. VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Nat Microbiol (2019) 4:1411–23. doi: 10.1038/s41564-019-0441-6
9. Megraud F. H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut (2004) 53:1374–84. doi: 10.1136/gut.2003.022111
10. Sepulveda AR. Helicobacter, inflammation, and gastric cancer. Curr Pathobiol Rep (2013) 1:9–18. doi: 10.1007/s40139-013-0009-8
11. Ermak TH, Giannasca PJ, Nichols R, Myers GA, Nedrud J, Weltzin R, et al. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted responses. J Exp Med (1998) 188:2277–88. doi: 10.1084/jem.188.12.2277
12. Sawai N, Kita M, Kodama T, Tanahashi T, Yamaoka Y, Tagawa Y, et al. Role of gamma interferon in Helicobacter pylori-induced gastric inflammatory responses in a mouse model. Infect Immun (1999) 67:279–85. doi: 10.1128/IAI.67.1.279-285.1999
13. Lee JS, Jung ID, Lee CM, Park JW, Chun SH, Jeong SK, et al. Outer membrane protein a of Salmonella enterica serovar Typhimurium activates dendritic cells and enhances Th1 polarization. BMC Microbiol (2010) 10:263. doi: 10.1186/1471-2180-10-263
14. Hase K, Kawano K, Nochi T, Pontes GS, Fukuda S, Ebisawa M, et al. Uptake through glycoprotein 2 of FimH(+) bacteria by m cells initiates mucosal immune response. Nature (2009) 462:226–30. doi: 10.1038/nature08529
15. Sirard JC, Niedergang F, Kraehenbuhl JP. Live attenuated Salmonella: a paradigm of mucosal vaccines. Immunol Rev (1999) 171:5–26. doi: 10.1111/j.1600-065X.1999.tb01340.x
16. Chatfield SN, Strugnell RA, Dougan G. Live Salmonella as vaccines and carriers of foreign antigenic determinants. Vaccine (1989) 7:495–8. doi: 10.1016/0264-410X(89)90271-5
17. Corthésy-Theulaz IE, Hopkins S, Bachmann D, Saldinger PF, Porta N, Haas R, et al. Mice are protected from Helicobacter pylori infection by nasal immunization with attenuated Salmonella Typhimurium phoPc expressing urease a and b subunits. Infect Immun (1998) 66:581–6. doi: 10.1128/IAI.66.2.581-586.1998
18. Gómez-Duarte OG, Lucas B, Yan ZX, Panthel K, Haas R, Meyer TF. Protection of mice against gastric colonization by Helicobacter pylori by single oral dose immunization with attenuated Salmonella Typhimurium producing urease subunits a and b. Vaccine (1998) 16:460–71. doi: 10.1016/S0264-410X(97)00247-8
19. Dipetrillo MD, Tibbetts T, Kleanthous H, Killeen KP, Hohmann EL. Safety and immunogenicity of phoP/phoQ-deleted Salmonella Typhi expressing Helicobacter pylori urease in adult volunteers. Vaccine (1999) 18:449–59. doi: 10.1016/S0264-410X(99)00246-7
20. Gómez-Duarte OG, Bumann D, Meyer TF. The attenuated Salmonella vaccine approach for the control of Helicobacter pylori-related diseases. Vaccine (1999) 17:1667–73. doi: 10.1016/S0264-410X(98)00436-8
21. Bumann D, Metzger WG, Mansouri E, Palme O, Wendland M, Hurwitz R, et al. Safety and immunogenicity of live recombinant Salmonella enterica serovar Typhi Ty21a expressing urease A and B from Helicobacter pylori in human volunteers. Vaccine (2001) 20:845–52. doi: 10.1016/S0264-410X(01)00391-7
22. Koesling J, Lucas B, Develioglou L, Aebischer T, Meyer TF. Vaccination of mice with live recombinant Salmonella Typhimurium aroA against H. pylori: parameters associated with prophylactic and therapeutic vaccine efficacy. Vaccine (2001) 20:413–20. doi: 10.1016/S0264-410X(01)00355-3
23. Londoño-Arcila P, Freeman D, Kleanthous H, O'dowd AM, Lewis S, Turner AK, et al. Attenuated Salmonella enterica serovar Typhi expressing urease effectively immunizes mice against Helicobacter pylori challenge as part of a heterologous mucosal priming-parenteral boosting vaccination regimen. Infect Immun (2002) 70:5096–106. doi: 10.1128/IAI.70.9.5096-5106.2002
24. Chen M, Chen J, Liao W, Zhu S, Yu J, Leung WK, et al. Immunization with attenuated Salmonella Typhimurium producing catalase in protection against gastric Helicobacter pylori infection in mice. Helicobacter (2003) 8:613–25. doi: 10.1111/j.1523-5378.2003.00182.x
25. Rizos K, Lattemann CT, Bumann D, Meyer TF, Aebischer T. Autodisplay: efficacious surface exposure of antigenic UreA fragments from Helicobacter pylori in Salmonella vaccine strains. Infect Immun (2003) 71:6320–8. doi: 10.1128/IAI.71.11.6320-6328.2003
26. Metzger WG, Mansouri E, Kronawitter M, Diescher S, Soerensen M, Hurwitz R, et al. Impact of vector-priming on the immunogenicity of a live recombinant Salmonella enterica serovar Typhi Ty21a vaccine expressing urease A and B from Helicobacter pylori in human volunteers. Vaccine (2004) 22:2273–7. doi: 10.1016/j.vaccine.2003.11.020
27. Clark-Curtiss JE, Curtiss R III. Salmonella vaccines: Conduits for protective antigens. J Immunol (2018) 200:39–48. doi: 10.4049/jimmunol.1600608
28. Kong W, Wang X, Fields E, Okon B, Jenkins MC, Wilkins G, et al. Mucosal delivery of a self-destructing Salmonella-based vaccine inducing immunity against Eimeria. Avian Dis (2020) 64:254–68. doi: 10.1637/aviandiseases-D-19-00159
29. Wang S, Hofacre CL, Wanda SY, Zhou J, Callum RA, Nordgren B, et al. A triple-sugar regulated Salmonella vaccine protects against Clostridium perfringens-induced necrotic enteritis in broiler chickens. Poult Sci (2022) 101:101592. doi: 10.1016/j.psj.2021.101592
30. Cardenas L, Clements JD. Oral immunization using live attenuated Salmonella spp as carriers of foreign antigens. Clin Microbiol Rev (1992) 5:328–42. doi: 10.1128/CMR.5.3.328
31. Curtiss R III. Bacterial infectious disease control by vaccine development. J Clin Invest. (2002) 110:1061–6. doi: 10.1172/JCI0216941
32. Atkins HS, Morton M, Griffin KF, Stokes MG, Nataro JP, Titball RW. Recombinant Salmonella vaccines for biodefence. Vaccine (2006) 24:2710–7. doi: 10.1016/j.vaccine.2005.12.046
33. Cheminay C, Hensel M. Rational design of Salmonella recombinant vaccines. Int J Med Microbiol (2008) 298:87–98. doi: 10.1016/j.ijmm.2007.08.006
34. Curtiss R III, Xin W, Li Y, Kong W, Wanda SY, Gunn B, et al. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit Rev Immunol (2010) 30:255–70. doi: 10.1615/CritRevImmunol.v30.i3.30
35. Wang S, Kong Q, Curtiss R III.New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microbial Pathogenesis (2013) 58:17–28. doi: 10.1016/j.micpath.2012.10.006
36. Brandtzaeg P, Farstad IN, Haraldsen G. Regional specialization in the mucosal immune system: primed cells do not always home along the same track. Immunol Today (1999) 20:267–77. doi: 10.1016/S0167-5699(99)01468-1
37. Brandtzaeg P, Farstad IN, Johansen FE, Morton HC, Norderhaug IN, Yamanaka T. The b-cell system of human mucosae and exocrine glands. Immunol Rev (1999) 171:45–87. doi: 10.1111/j.1600-065X.1999.tb01342.x
39. Brandtzaeg P. Role of secretory antibodies in the defence against infections. Int J Med Microbiol (2003) 293:3–15. doi: 10.1078/1438-4221-00241
40. Woof JM, Mestecky J. Mucosal immunoglobulins. Immunol Rev (2005) 206:64–82. doi: 10.1111/j.0105-2896.2005.00290.x
41. Mantis NJ, Forbes SJ. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol Invest (2010) 39:383–406. doi: 10.3109/08820131003622635
42. Hopkins S, Kraehenbuhl JP, Schodel F, Potts A, Peterson D, De Grandi P, et al. A recombinant Salmonella Typhimurium vaccine induces local immunity by four different routes of immunization. Infect Immun (1995) 63:3279–86. doi: 10.1128/iai.63.9.3279-3286.1995
43. Srinivasan J, Tinge S, Wright R, Herr JC, Curtiss R III. Oral immunization with attenuated Salmonella expressing human sperm antigen induces antibodies in serum and the reproductive tract. Biol Reprod (1995) 53:462–71. doi: 10.1095/biolreprod53.2.462
44. Srinivasan A, Mcsorley SJ. Activation of Salmonella-specific immune responses in the intestinal mucosa. Arch Immunol Ther Exp (Warsz) (2006) 54:25–31. doi: 10.1007/s00005-006-0003-5
45. Tam MA, Rydstrom A, Sundquist M, Wick MJ. Early cellular responses to Salmonella infection: dendritic cells, monocytes, and more. Immunol Rev (2008) 225:140–62. doi: 10.1111/j.1600-065X.2008.00679.x
46. Moon JJ, Mcsorley SJ. Tracking the dynamics of Salmonella specific T cell responses. Curr Top Microbiol Immunol (2009) 334:179–98. doi: 10.1007/978-3-540-93864-4_8
47. Shi H, Wang S, Roland KL, Gunn BM, Curtiss R III. Immunogenicity of a live recombinant Salmonella enterica serovar Typhimurium vaccine expressing pspA in neonates and infant mice born from naive and immunized mothers. Clin Vaccine Immunol (2010) 17:363–71. doi: 10.1128/CVI.00413-09
48. Griffin AJ, Mcsorley SJ. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol (2011) 4:371–82. doi: 10.1038/mi.2011.2
49. Broz P, Ohlson MB, Monack DM. Innate immune response to Salmonella Typhimurium, a model enteric pathogen. Gut Microbes (2012) 3:62–70. doi: 10.4161/gmic.19141
50. Nakayama K, Kelly SM, Curtiss R III. Construction of an ASD+ expression-cloning vector: Stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotech (1988) 6:693–7. doi: 10.1038/nbt0688-693
51. Curtiss R III.Protective immunity enhanced Salmonella vaccine (PIESV) against brucella spp. US patent Appl 17 / 273,801 (2018).
52. Collins LV, Attridge S, Hackett J. Mutations at rfc or pmi attenuate Salmonella Typhimurium virulence for mice. Infect Immun (1991) 59:1079–85. doi: 10.1128/iai.59.3.1079-1085.1991
53. Kong W, Wanda SY, Zhang X, Bollen W, Tinge SA, Roland KL, et al. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc Natl Acad Sci U.S.A. (2008) 105:9361–6. doi: 10.1073/pnas.0803801105
54. Curtiss R III., Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect Immun (2009) 77:1071–82. doi: 10.1128/IAI.00693-08
55. Sanapala S, Rahav H, Patel H, Sun W, Curtiss R III.Multiple antigens of yersinia pestis delivered by live recombinant attenuated Salmonella vaccine strains elicit protective immunity against plague. Vaccine (2016) 34:2410–6. doi: 10.1016/j.vaccine.2016.03.094
56. Jiang Y, Mo H, Willingham C, Wang S, Park JY, Kong W, et al. Protection against necrotic enteritis in broiler chickens by regulated delayed lysis Salmonella vaccines. Avian Dis (2015) 59:475–85. doi: 10.1637/11094-041715-Reg
57. Kang HY, Srinivasan J, Curtiss R III.Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun (2002) 70:1739–49. doi: 10.1128/IAI.70.4.1739-1749.2002
58. Muralinath M, Kuehn MJ, Roland KL, Curtiss R III. Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect Immun (2011) 79:887–94. doi: 10.1128/IAI.00950-10
59. Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, et al. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J Exp Med (2000) 191:1467–76. doi: 10.1084/jem.191.9.1467
60. Carlsohn E, Nystrom J, Bolin I, Nilsson CL, Svennerholm AM. HpaA is essential for Helicobacter pylori colonization in mice. Infect Immun (2006) 74:920–6. doi: 10.1128/IAI.74.2.920-926.2006
61. Del Giudice G, Malfertheiner P, Rappuoli R. Development of vaccines against Helicobacter pylori. Expert Rev Vaccines (2009) 8:1037–49. doi: 10.1586/erv.09.62
62. Oleastro M, Menard A. The role of Helicobacter pylori outer membrane proteins in adherence and pathogenesis. Biol (Basel) (2013) 2:1110–34. doi: 10.3390/biology2031110
63. Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe (2014) 15:306–16. doi: 10.1016/j.chom.2014.02.008
64. Nell S, Kennemann L, Schwarz S, Josenhans C, Suerbaum S. Dynamics of Lewis b binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans. MBio (2014) 5:e02281–02214. doi: 10.1128/mBio.02281-14
65. Mohammad N, Karsabet MT, Amani J, Ardjmand A, Zadeh MR, Gholi MK, et al. In silico design of a chimeric protein containing antigenic fragments of Helicobacter pylori; a bioinformatic approach. Open Microbiol J (2016) 10:97–112. doi: 10.2174/1874285801610010097
66. Moyat M, Velin D. Use of VacA as a vaccine antigen. Toxins (Basel) (2016) 8:181. doi: 10.3390/toxins8060181
67. Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel JP. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol (2017) 15:109–28. doi: 10.1038/nrmicro.2016.171
68. Ghasemi A, Mohammad N, Mautner J, Karsabet MT, Ardjmand A, Moniri R. Immunization with recombinant FliD confers protection against Helicobacter pylori infection in mice. Mol Immunol (2018) 94:176–82. doi: 10.1016/j.molimm.2018.01.001
69. Branger CG, Sun W, Torres-Escobar A, Perry R, Roland KL, Fetherston J, et al. Evaluation of Psn, HmuR and a modified LcrV protein delivered to mice by live attenuated Salmonella as a vaccine against bubonic and pneumonic Yersinia pestis challenge. Vaccine (2010) 29:274–82. doi: 10.1016/j.vaccine.2010.10.033
70. Ghasemi A, Jeddi-Tehrani M, Mautner J, Salari MH, Zarnani AH. Simultaneous immunization of mice with Omp31 and TF provides protection against Brucella melitensis infection. Vaccine (2015) 33:5532–8. doi: 10.1016/j.vaccine.2015.09.013
71. Shi Y, Yang Z, Zhang T, Shen L, Li Y, Ding S. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer. Cell Death Dis (2019) 10:625. doi: 10.1038/s41419-019-1859-8
72. Teng YS, Liu YG, Chen XH, Wang TT, Cheng P, Lv YP, et al. Decreased IL-17RB expression impairs CD11b. Cell Death Dis (2019) 10:79. doi: 10.1038/s41419-019-1312-z
73. Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath (2013) 3:71–85.
74. Kaniuk NA, Vinogradov E, Whitfield C. Investigation of the structural requirements in the lipopolysaccharide core acceptor for ligation of O antigens in the genus Salmonella: WaaL "ligase" is not the sole determinant of acceptor specificity. J Biol Chem (2004) 279:36470–80. doi: 10.1074/jbc.M401366200
75. Kong Q, Liu Q, Jansen AM, Curtiss R III. Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine (2010) 28:6094–103. doi: 10.1016/j.vaccine.2010.06.074
76. Wang S, Shi H, Li Y, Shi Z, Zhang X, Baek CH, et al. A colanic acid operon deletion mutation enhances induction of early antibody responses by live attenuated Salmonella vaccine strains. Infect Immun (2013) 81:3148–62. doi: 10.1128/IAI.00097-13
77. Xin W, Wanda SY, Li Y, Wang S, Mo H, Curtiss R III.Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect Immun (2008) 76:3241–54. doi: 10.1128/IAI.01623-07
78. Wang S, Li Y, Scarpellini G, Kong W, Shi H, Baek CH, et al. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect Immun (2010) 78:3969–80. doi: 10.1128/IAI.00444-10
79. Juárez-Rodríguez MD, Yang J, Kader R, Alamuri P, Curtiss R III, Clark-Curtiss JE. Live attenuated Salmonella vaccines displaying regulated delayed lysis and delayed antigen synthesis to confer protection against Mycobacterium tuberculosis. Infect Immun (2012) 80:815–31. doi: 10.1128/IAI.05526-11
80. Zhang X, Wanda SY, Brenneman K, Kong W, Roland K, Curtiss R III. Improving Salmonella vector with rec mutation to stabilize the DNA cargoes. BMC Microbiol (2011) 11:31. doi: 10.1186/1471-2180-11-31
81. Ashraf S, Kong W, Wang S, Yang J, Curtiss R III. Protective cellular responses elicited by vaccination with influenza nucleoprotein delivered by a live recombinant attenuated Salmonella vaccine. Vaccine (2011) 29:3990–4002. doi: 10.1016/j.vaccine.2011.03.066
82. Kong W, Brovold M, Koeneman BA, Clark-Curtiss J, Curtiss R III. Turning self-destructing Salmonella into a universal DNA vaccine delivery platform. Proc Natl Acad Sci U.S.A. (2012) 109:19414–9. doi: 10.1073/pnas.1217554109
83. Gopal R, Jeong E, Seo CH, Park Y. Role of antimicrobial peptides expressed by host cells upon infection by Helicobacter pylori. Protein Pept Lett (2014) 21:1057–64. doi: 10.2174/0929866521666140708092032
84. Padra M, Benktander J, Robinson K, Linden SK. Carbohydrate-dependent and antimicrobial peptide defence mechanisms against Helicobacter pylori infections. Curr Top Microbiol Immunol (2019) 421:179–207. doi: 10.1007/978-3-030-15138-6_8
85. Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest (2012) 122:974–86. doi: 10.1172/JCI60588
86. Teng YS, Liu YG, Chen XH, Wang TT, Cheng P, Lv YP, et al. Decreased IL-17RB expression impairs CD11b+CD11c- myeloid cell accumulation in gastric mucosa and host defense during the early-phase of Helicobacter pylori infection. Cell Death Dis (2019) 10:79. doi: 10.1038/s41419-019-1312-z
87. Frantz Jd BG, Cartwright ME, Crissman JW, Macklin AW, Moronpot RR. Proliferative lesions of the non-glandular and glandular stomach in rats, GI-3. In: Guides for toxologic pathology, STP/ARP/AFIP. Whashington, DC (1991).
88. Liu KY, Shi Y, Luo P, Yu S, Chen L, Zhao Z, et al. Therapeutic efficacy of oral immunization with attenuated Salmonella Typhimurium expressing Helicobacter pylori CagA, VacA and UreB fusion proteins in mice model. Vaccine (2011) 29:6679–85. doi: 10.1016/j.vaccine.2011.06.099
89. Angelakopoulos H, Hohmann EL. Pilot study of phoP/phoQ-deleted Salmonella enterica serovar Typhimurium expressing Helicobacter pylori urease in adult volunteers. Infect Immun (2000) 68:2135–41. doi: 10.1128/IAI.68.4.2135-2141.2000
90. Amanna IJ, Slifka MK. Contributions of humoral and cellular immunity to vaccine-induced protection in humans. Virology (2011) 411:206–15. doi: 10.1016/j.virol.2010.12.016
91. Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun (2000) 68:4155–68. doi: 10.1128/IAI.68.7.4155-4168.2000
92. Strugatsky D, Mcnulty R, Munson K, Chen CK, Soltis SM, Sachs G, et al. Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature (2013) 493:255–8. doi: 10.1038/nature11684
93. Pan X, Ke H, Niu X, Li S, Lv J, Pan L. Protection against Helicobacter pylori infection in BALB/c mouse model by oral administration of multivalent epitope-based vaccine of cholera toxin b subunit-HUUC. Front Immunol (2018) 91003 doi: 10.3389/fimmu.2018.01003
94. Lundström AM, Blom K, Sundaeus V, Bolin I. HpaA shows variable surface localization but the gene expression is similar in different Helicobacter pylori strains. Microb Pathog (2001) 31:243–53. doi: 10.1006/mpat.2001.0466
95. Ernst PB, Pappo J. T-Cell-mediated mucosal immunity in the absence of antibody: lessons from Helicobacter pylori infection. Acta Odontol Scand (2001) 59:216–21. doi: 10.1080/00016350152509238
96. Rad R, Brenner L, Bauer S, Schwendy S, Layland L, Da Costa CP, et al. CD25+/Foxp3+ T cells regulate gastric inflammation and Helicobacter pylori colonization in vivo. Gastroenterology (2006) 131:525–37. doi: 10.1053/j.gastro.2006.05.001
97. Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med (2008) 14:109–19. doi: 10.1016/j.molmed.2007.12.007
98. Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity (2008) 28:454–67. doi: 10.1016/j.immuni.2008.03.004
99. Chung DR, Kasper DL, Panzo RJ, Chitnis T, Grusby MJ, Sayegh MH, et al. CD4+ T cells mediate abscess formation in intra-abdominal sepsis by an IL-17-dependent mechanism. J Immunol (2003) 170:1958–63. doi: 10.4049/jimmunol.170.4.1958
100. Kumar P, Chen K, Kolls JK. Th17 cell based vaccines in mucosal immunity. Curr Opin Immunol (2013) 25:373–80. doi: 10.1016/j.coi.2013.03.011
101. Lo BC, Shin SB, Canals Hernaez D, Refaeli I, Yu HB, Goebeler V, et al. IL-22 preserves gut epithelial integrity and promotes disease remission during chronic. J Immunol (2019) 202:956–65. doi: 10.4049/jimmunol.1801308
102. Al-Alwan LA, Chang Y, Mogas A, Halayko AJ, Baglole CJ, Martin JG, et al. Differential roles of CXCL2 and CXCL3 and their receptors in regulating normal and asthmatic airway smooth muscle cell migration. J Immunol (2013) 191:2731–41. doi: 10.4049/jimmunol.1203421
103. Roos S, Engstrand L, Jonsson H. Lactobacillus gastricus sp. nov., lactobacillus antri sp. nov., lactobacillus kalixensis sp. nov. and Lactobacillus ultunensis sp. nov., isolated from human stomach mucosa. Int J Syst Evol Microbiol (2005) 55:77–82. doi: 10.1099/ijs.0.63083-0
104. Ryan KA, Jayaraman T, Daly P, Canchaya C, Curran S, Fang F, et al. Isolation of lactobacilli with probiotic properties from the human stomach. Lett Appl Microbiol (2008) 47:269–74. doi: 10.1111/j.1472-765X.2008.02416.x
105. Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J (2011) 5:574–9. doi: 10.1038/ismej.2010.149
106. Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, et al. Bacterial biogeography of the human digestive tract. Sci Rep (2011) 1:170. doi: 10.1038/srep00170
107. Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut (2014) 63:54–63. doi: 10.1136/gutjnl-2013-305178
Keywords: recombinant protein, cytokines, vaccine, protection, Helicobacter pylori, Salmonella
Citation: Ghasemi A, Wang S, Sahay B, Abbott JR and Curtiss R III (2022) Protective immunity enhanced Salmonella vaccine vectors delivering Helicobacter pylori antigens reduce H. pylori stomach colonization in mice. Front. Immunol. 13:1034683. doi: 10.3389/fimmu.2022.1034683
Received: 01 September 2022; Accepted: 31 October 2022;
Published: 18 November 2022.
Edited by:
Daniela F Hozbor, Universidad Nacional de La Plata, ArgentinaReviewed by:
Holly Algood, Vanderbilt University, United StatesJingbo Zhai, Inner Mongolia Minzu University, China
Copyright © 2022 Ghasemi, Wang, Sahay, Abbott and Curtiss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shifeng Wang, c2hpZmVuZ3dhbmdAdWZsLmVkdQ==; Roy Curtiss III, cmN1cnRpc3NAdWZsLmVkdQ==
 Amir Ghasemi
Amir Ghasemi Shifeng Wang
Shifeng Wang Bikash Sahay
Bikash Sahay Jeffrey R. Abbott2
Jeffrey R. Abbott2