- Academy of Integrative Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Accumulating evidence suggests that gut microbial dysbiosis is implicated in colorectal cancer (CRC) initiation and progression through interaction with host immune system. Given the intimate relationship between the gut microbiota and the antitumor immune responses, the microbiota has proven to be effective targets in modulating immunotherapy responses of preclinical CRC models. However, the proposed putative mechanisms of how these bacteria affect immune responses and immunotherapy efficacy remains obscure. In this review, we summarize recent findings of clinical gut microbial dysbiosis in CRC patients, the reciprocal interactions between gut microbiota and the innate and/or the adaptive immune system, as well as the effect of gut microbiota on immunotherapy response in CRC. Increased understanding of the gut microbiota-immune system interactions will benefit the rational application of microbiota to the clinical promising biomarker or therapeutic strategy as a cancer immunotherapy adjuvant.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death with more than 1.9 million new cases and 935,000 deaths estimated to occur in 2020 worldwide (1). Although risk factors are recognized as western dietary patterns, excess body weight, and lifestyle factors including heavy alcohol consumption and cigarette smoking, the specific underlying pathogenetic mechanisms are still not fully-elucidated.
The gut microbiota has been discussed widely over the past decades which plays an important role in all the different phases of CRC process from oncogenesis to metastasis, from treatment to prognosis prediction (2, 3). Investigators have identified specific microbial features relevant to CRC, data from cross-sectional epidemiological studies and unbiased microbiome profiling of stools and colorectal tissues have uncovered specific bacterial taxa that contribute to CRC (4). Mechanistic insights into a microbe’s contributions to carcinogenesis support that gut microbiota alterations induce genotoxin production, inflammation, metabolic regulation, and local and systemic immune response, thereby influencing the development of CRC (5). Immune escape or suppression has long been proposed to constitute a critical step in both tumor formation and progression (6). Studies have highlighted that intact mucosal immunity maintains a balanced bacterial composition in the gut, whereas disruption of this immunological circuit, either prior to, or as a consequence of tumor development, accelerates CRC initiation and progression (7, 8). In addition, some specific bacterial taxa are shown to regulate immunotherapy responses in both animal models and human cohorts (9–12). In this sense, researchers are focusing on the deep and complex relationship between microbiota and immune regulation to better understand cancer biology and to formulate novel therapeutic approach. In this review, we outline the clinical findings of changes in gut microbiota composition in CRC patients, highlight potential mechanisms of gut microbiota-modulated immune responses and the possible impacts on responses to immunotherapy in CRC.
Clinical findings of microbial changes in CRC patients
Next-generation sequencing studies have revealed the potential association of microbial compositional changes (dysbiosis) within CRC patients, including increased proportions of Fusobacterium nucleatum, Bacteroides fragilis, Escherichia coli, and Streptococcus, Peptostreptococcus species (13–15). These findings raised the possibilities for clinical applications using gut microbiota analysis as screening, prognostic or predictive biomarkers. Moreover, deciphering key microbiome signatures within different stages of cancer progression may offer possibilities for treatment stratification and metastasis surveillance (16). The gut microbiota changes across all stages of CRC patients were summarized in Table 1. As indicated, dynamic shifts in microbial composition in gut microbiota were observed during multistep CRC progression.
Gut microbiota elicits tumor-promoting inflammation
It is well-known that chronic inflammation is a risk for CRC initiation and development. Overall, 2.2 million new cancer cases were attributable to infections by different etiological agents, including viruses, bacteria and parasites in 2018, representing 13% of all cancer cases (28). The bacterium enterotoxigenic Bacteroides fragilis (ETBF) is a significant source of chronic inflammation and has been implicated as a risk factor for CRC, which can up-regulate spermine oxidase (SMO)-dependent generation of reactive oxygen species (ROS) and induce inflammation, leading to DNA damage in colonic epithelial cells (29). Diverse cytokines like tumor necrosis factor (TNF)α, interlukin (IL)-6, IL-1β, IL-23, and IL-17, can be triggered by microbes or their products, contribute to the progression of intestinal tumorigenesis (30). Upon invading the stroma, bacteria can trigger both innate responses via recognition through pattern recognition receptors (PRRs), eliciting secretion of a repertoire of cytokines and chemokines. Streptococcus gallolyticus has long been associated with colonic pathologies. Although a causal relationship to CRC is not clear, increased production of inflammatory factors, including cyclooxygenase (COX)-2, IL-1 and IL-8, in Streptococcus gallolyticus-bearing tumor tissue might indicate its possible contribution to tumor progression (31). Fusobacterium nucleatum has been reported to drive a pro-inflammatory intestinal microenvironment through metabolite receptor -dependent modulation of IL-17 expression in Apcmin/+ mice (32). Fusobacterium nucleatum colonization leads to increased intestinal short chain fatty acid (SCFA) levels and human CRC tissues harboring Fusobacterium nucleatum are primed to sense these immunomodulatory metabolites, with higher expression of the genes encoding the SCFA receptors FFAR2 and NIACR1. Furthermore, in Ffar2-/- mice, Fusobacterium nucleatum failed to increase Th17 cell frequency, suggesting that Fusobacterium nucleatum shaped Th17 response is FFAR2-dependent (32). The inflammasome NACHT, LRR, and PYD domains-containing protein 3 (NLRP3) is a global sensor of pathogen-associated molecular pattern molecules (PAMPs) and damage-associated molecular patterns (DAMPs) and its activation leads to secretion of both IL-1β and IL-18. Studies have provided evidence to indicate that the gut microbiota serve as critical modulators of inflammasome activity and susceptibility to the development of intestinal inflammation and cancer (33). Indeed, the presence of some of the cytokines (IL-17, IL-6, IL-1β, TNFα) also correlate with poor prognosis in CRC patients (34). The chronic activation of inflammatory signals not only suppresses adaptive immune responses but simultaneously supports tumor growth, via mechanisms such as the increased release of growth and immunomodulatory factors (35). For example, tumor infiltrating myeloid cells release growth factors, such as epidermal growth factor (EGF), hepatocyte growth factor (HGF), and fibroblast growth factor (FGF) (36, 37). These growth factors can support cancer cell proliferation, survival, motility, and invasion, by triggering the activation of MAPK, Wnt/β-catenin or PI3K/AKT/mTOR signaling pathway, thus facilitate CRC progression (38). The continuation of inflammatory responses and tumor progression to malignancy on the one hand, changes the composition of immune cells from immune activators to immune suppressors, on the other hand, potential cytokines and chemokines facilitate the recruitment of immune cells with immunosuppressive functions, such as myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs) (39), which eventually maintains tumor cell survival and immune escape (discussed below). In addition, chronic inflammation is now accepted as a major influence in the outcome of CRC, treatment with anti-inflammatory (such as aspirin) mitigates CRC progression and extends patient survival (40).
Gut dysbiosis facilitates immune escape in CRC
Here, we outline the potential mechanisms of microbial-modulated immune escape from innate and adaptive immune perspectives and discuss dysbiosis-immune interaction during CRC progression.
Modulating innate immune surveillance against tumors
The immune system has an extraordinary capacity to recognize and respond to a range of microbial patterns and danger signals. The first cells to fight microbes are the myeloid derived innate immune cells (41). Bacterial species that translocate through the epithelial barrier induce recruitment of myeloid cells. Through their PPRs, these cells recognize microbes, thus influencing the type and intensity of innate and adaptive immunity (42). Dysbiosis can enhance gut responsiveness to bacteria and its products, however during chronic infections, it may lead to a miscoordination between inflammation and immune suppression, thus favors tumor growth. In fact, an important feature of tumors is the generation, development, and expansion of myeloid cells with special immunosuppressive properties including tumor-associated neutrophils (TANs), tumor-associated macrophages (TAMs), regulatory dendritic cells (DCs), and MDSCs (43). For example, Fusobacterium promotes the growth of colorectal cancer through the induction of the activity of MDSCs and TAMs (44, 45). Tumor invading pathogen bacteria also trigger the activation of neutrophils that infiltrate into tumor stroma, whereby the earliest tumor-infiltrating neutrophils may serve to inhibit expansion of colon microbiota to limit tumorigenesis and progression (46–48). However, established tumors may evolve a more pro-tumorigenic TAN phenotype and elicit the production of tumor-promoting cytokines (49). Neutrophils can also produce ‘neutrophil extracellular traps’ (NETs), upon activation to ensnare and neutralize pathogens. Recent studies highlight the function of NETs in cancer progression and metastasis, NETs are able to wake up dormant cancer cells promoting cancer relapse, and are able to entrap circulating cancer, thus enhancing metastasis spread (50). Bacteria derivations also play a role in escalating the tumor-associated immune suppression. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in TAMs to suppress anti-tumor immunity (51). Gut microbiota tend to produce butyrate, which in turn can inhibit the DCs’ antigen presentation (52, 53). Beyaz et al. showed that high fat diets (HFDs) resulted in changes in the composition of the gut microbiota (54), and in a Kras-driven mouse model of intestinal cancer, HFD-altered gut microbiome, which, in turn, resulted in reduced major histocompatibility complex (MHC) II expression on DCs and engages in immune evasion (55), suggesting that dietary in association with the gut microbiota, are critical modulators to the development of intestinal cancer. In addition to the aforementioned myeloid cells, another example is the inhibitory effect of Fusobacterium nucleatum on natural killer (NK) cells. Fusobacterium nucleatum produces the Fap2 protein, which binding to the inhibitory receptor T cell immunoglobulin and ITIM domain (TIGIT) on NK cells, thus directly inhibiting cell-killing of tumor cells (56). Yet, we have very little insight into the roles of microbiota on innate immune cell populations, further studies are needed to interrogate the precise functional contributions of gut microbe on these innate immune cell subsets.
Reprogramming adaptive anti-tumoral responses
With improved tools, recent work has suggested two broad categories of bacteria-related tumor escape of adaptive immune attack: 1. Microbes influence anti-tumor effectors directly by serving as antigens which mediate recognition by host T cells (57, 58), 2. Microbes facilitate the resistance of immune attack through the immune suppressive pathways such as inducing immune exhaustion (59, 60).
Intestinal microbiota has been proposed to induce commensal-specific memory T cells that cross-react with tumor-associated antigens. Indeed, memory responses by CD4+ and CD8+ T cells specific for Enterococcus hirae, Bacteroides fragilis, and Akkermansia muciniphila are associated with favorable clinical outcome in cancer patients (11, 57, 58), suggesting that microbe-specific T lymphocytes may contribute to anti-tumoral immune responses. The optimal recognition of the antigen induces a specific activation of T cells, thereby driving T cell activation and differentiation of CD4+T cell subsets into Th1, Th2, and Th17 or Tregs (61, 62). It should be noted that dysbiosis induced T cells are capable of switching their phenotypes, which in turn set the proclivity to inflammatory, immunostimulatory or immunosuppressive reactions depending on tumor context and specific bacteria. The modulation by distinctive microbiome antigens can also consequently affect the activation of cytotoxic CD8+ T cells that limit the direct lysis of cancer cells (63).
In cancer, like in chronic infection, the long exposure to the antigen leads to a dysfunction of T cells, represents the state of “exhaustion” (64, 65). In the early stage of azoxymethane (AOM)/dextran sulfate sodium (DSS) mice model, gut dysbiosis (increased Prevotellaceae and decreased Anaeroplasmataceae) promoted tumorigenesis by stimulating CD8+ T cells activation, durable hyperstimulation of CD8+ T cells resulted in T cell exhaustion, leading to increased tumor susceptibility (66). Microbiota can also provoke sustained expression of the inhibitory molecules, such as cytotoxic T lymphocyte antigen 4 (CTLA-4), T cell immunoglobulin and mucin domain-containing protein 3 (TIM3), programmed cell−death protein 1 (PD−1), or the ligand PD-L1, which are the most prominent examples of immune−checkpoint molecules underlying immune−escape mechanisms (60). The FAS/FAS ligand (FASL) apoptotic pathway is also highly relevant to immune evasion, which induces apoptosis of lymphocytes (67). Histopathological analyses have revealed that FASL is upregulated in metastatic tumor compared the primary tumor in patients with CRC (68). Thus, simultaneous loss or downregulation of FAS and upregulation of FASL on tumor cells might contribute to tumor evasion of immune−mediated cytolysis. The microbiota-derived SCFAs, such as butyrate, can be absorbed across the intestinal epithelium and exert their influence on T cells via G-protein-coupled receptors (GPRs). Butyrate was shown to promote cellular metabolism, enhance memory potential of activated CD8+ T cells through promoting mitochondrial function and cellular metabolism (69). Taken together, these findings reveal a role for the microbiota in the modulation of T cell responses in CRC which may have important implications on immunotherapy.
As another important adaptive immune cell type, B cells perform immune surveillance as antigen presenting cells (APCs) or function by stimulating immunoglobulins (Ig A) and producing cytokines (IL-10, TGFβ, often terms as regulatory B cells, Bregs) (70). Changes in gut microbiota composition and a diverse role of B cells have been implicated at the mucosal interface. Helicobacter hepaticus colonization has been shown to relieve tumor burden in CRC mice and increase B cells maturation and infiltration (71). Parvimonas micra was also shown to be closely associated with the antigen-presenting HLA-DR (+) B cells in a CRC cohort (72). In another study of familial adenomatous polyposis (FAP), loss of resident memory T cells and γδ T cells, excess IgA antibody secretion and increased IgA+ peripheral B cells were found to accompany intestinal microbial dysbiosis, implicating mucosal immune dysfunction as a contributing factor in the etiology of CRC (73). Above studies highlighted the interactions between microbiota and B cells in CRC, but the underlying mechanism remains largely unresolved. Some recent reports have shed light on the microbial metabolites, SCFAs and microbial tryptophan catabolites to regulate B cell activation and antibody responses (74–76). More recently, Wang et al. found that leucine-tRNA-synthetase-2 (LARS2)-expressing B cell (LARS B) with TGF-β1-dominant feature correlates with shortened survival in CRC, mechanistically, LARS2 programmed mitochondrial nicotinamide adenine dinucleotide (NAD+) regeneration and oxidative metabolism, thus determining the regulatory feature of LARS B cells in which the NAD-dependent protein deacetylase sirtuin-1 (SIRT1) was involved (77).
Overall, the immune impact of gut microbiota on CRC partially depends on shaping innate and adaptive immune responses (e.g., suppressing immunosurveillance, inducing T cell exhaustion and apoptosis, etc.), consequently leading to immune escape (Figure 1). In future, clarification of possible role of microbiota in modulation of populations and functions of innate and adaptive immune cells, as well as the crosstalk between different types of immune cells remain important research areas.
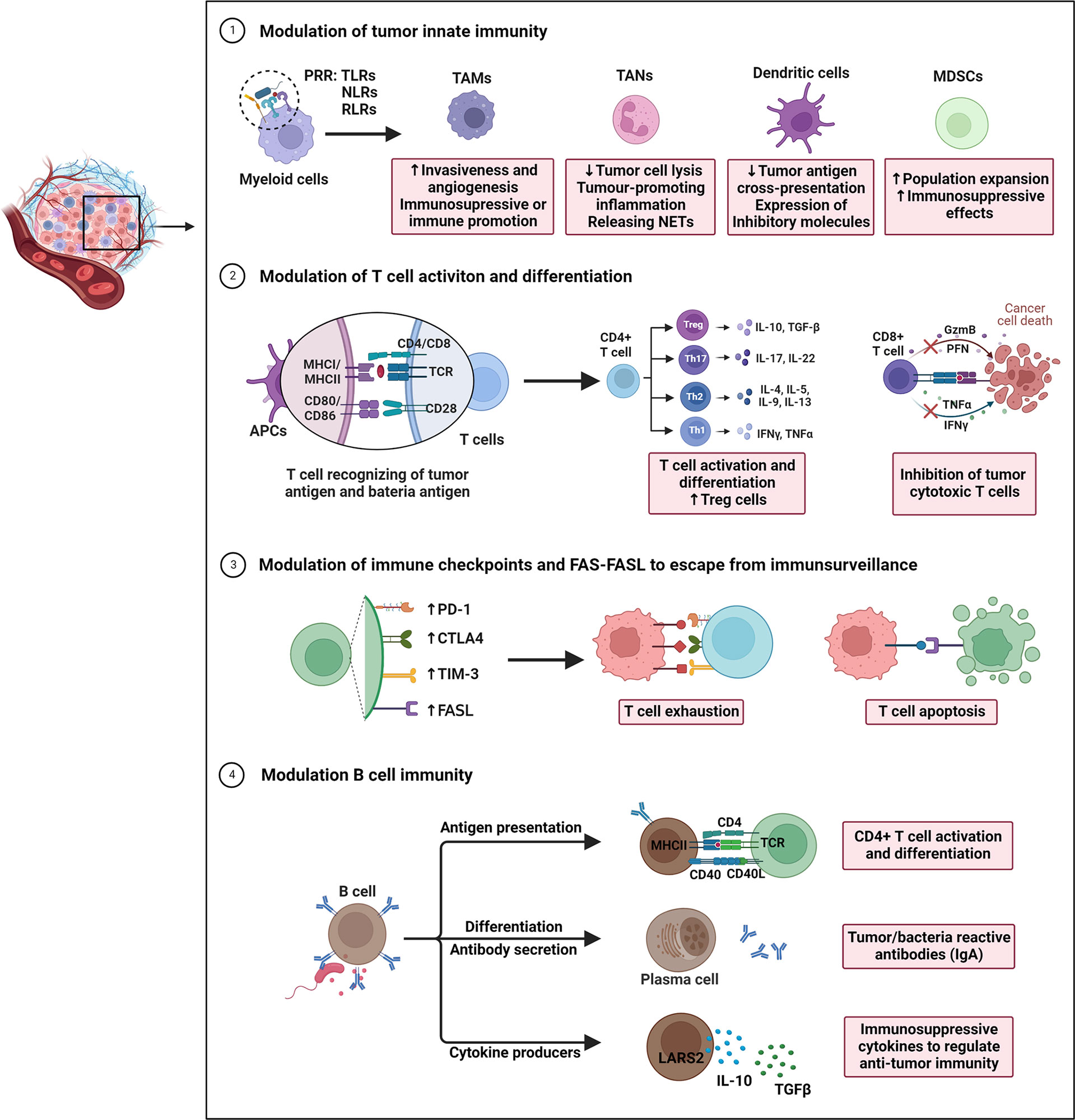
Figure 1 Overview of gut microbiota-modulated immune responses in tumor progression. The underlying actions and mechanisms by which the microbiota affects tumor immune escape are summarized as follows: 1) Under pathogenic conditions (dysbiosis), bacterial species that translocate through the epithelial barrier induce recruitment of myeloid cells. Myeloid cells recognize microbes via their pattern recognition receptors (PPR). During this process, the immunosuppressive mechanisms observed in the tumor microenvironment (TME) are activated underlie chronic infections. 2) Intestinal microbiota and intra-tumoral bacteria can be directly presented by antigen-presenting cells, thereby driving T cell activation and differentiation of CD4+T cell subsets into Th1, Th2, and Th17 or Tregs. Microbiota may also inhibit the dendritic cells’ antigen presentation, consequently decreasing the CD8+T cell response. 3) By modulating T cell immune checkpoint receptor-ligand pairs (e.g., CTLA-4/CD80/CD86 and PD1/PD-L1/PD-L2), they impact T cell exhaustion, and directly impacting efficacy of immune checkpoint inhibitors. Additional mechanisms of immune escape include expression of the apoptotic proteins FAS/FASL to induce immune cell apoptosis. 4) Gut microbiota may also regulate B cell infiltration, development, and polarization. These B cells exert anti/pro-tumor immunity through acting as APCs to reshape T cell responses, secreting tumor/bacteria-specific antibodies (IgA), and producing cytokines (IL-10, TGFβ), all of which are associated with immune processes in CRC.
Role of gut microbiota in shaping an immune-privileged (pre-) metastatic niche
Although studies have observed the presence of microbiota in metastatic liver or lung organs of CRC patients, the underline mechanisms by which microbiota affects CRC metastases formation are only now being uncovered. In the secondary sites, immunosuppressive cell types, such as TAMs and MDSCs populate (pre) metastatic niches, where they help direct metastatic dissemination by creating a niche that is permissive to tumor colonization (78). These cells have been shown to achieve these pro-tumoral functions by (1) generating a proinflammatory milieu (2), remodeling the matrix and creating a pro-angiogenic, pro-invasive environment (3), maintaining an immunosuppressive microenvironment, and (4) secreting growth factors that maintains the growth of metastatic cells. Pathogen Escherichia coli can upregulate Cathepsin K (CTSK) expression which serves as a vital mediator between the imbalance of intestinal microbiota and CRC metastasis (79). CRC-secreted CTSK stimulates CRC progression through accelerating M2 polarization of TAMs in a TLR4-mTOR-dependent pathway. Meanwhile, cytokines (IL-10, IL-17) secreted by activated M2 macrophage, in turn, promote CRC cells invasion and metastasis by activating NF-κB pathway (79). Fusobacterium nucleatum can boost liver metastasis by modulating liver microenvironment featured with accumulation of MDSCs, and reduction of NK and Th17 cells (80, 81). Peptostreptococcus anaerobius was also reported to induce chronic inflammation and modulate tumor microenvironment by recruiting MDSCs, TANs and TAMs (82). More recently, Bertocchi et al. demonstrate that Escherichia coli induces gut vascular barrier (GVB) disruption, which allows bacteria to reach the liver and initiate the recruitment of inflammatory cells, contributing to pre-metastatic niche maturation and favoring metastases formation (83). These results demonstrate that host microbiota acts as a key modulator during CRC metastasis by facilitating (pre-) metastatic niche formation which support cancer cells seeding in secondary organs (Figure 2). Re-education of the metastatic niche, through alterations in metastasis-related bacteria and associated pathways, may have favorable consequences for metastatic CRC therapy.
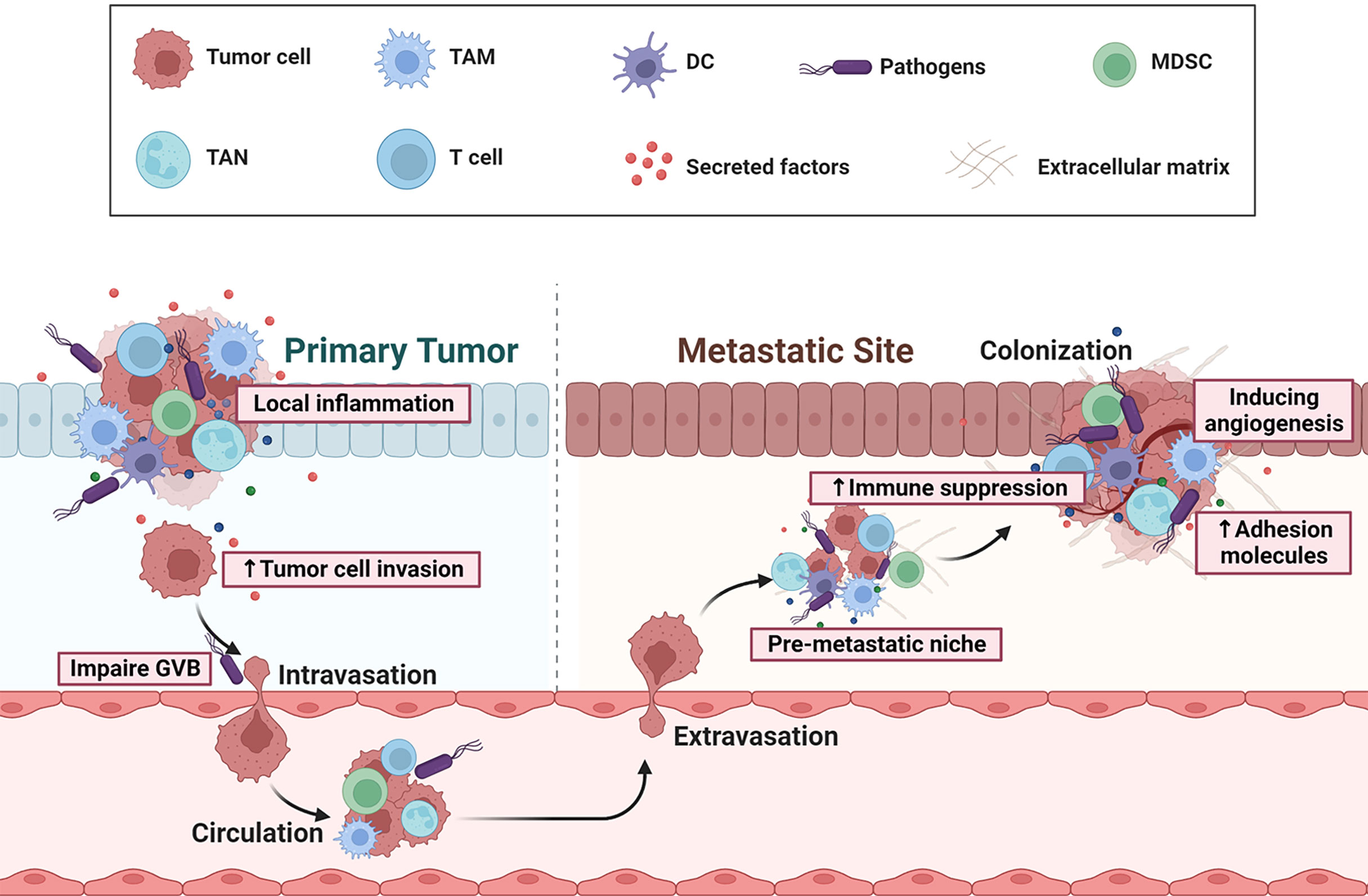
Figure 2 Overview of gut microbial dysbiosis on CRC initiation and metastasis. Dysbiosis can cause a chronic, pro-inflammatory milieu, by eliciting secretion of a repertoire of cytokines or growth factors, and thereby facilitates tumor cell invasion, adhesion, extravasation and survival. Some pathogens (eg: Escherichia coli) can induce gut vascular barrier (GVB) disruption, which allows bacteria to reach the distant organ and initiate the recruitment of immune cells with immunosuppressive functions, such as MDSCs and Tregs. Thus, the microbiota can contribute to the immune escape of distant tumors, the pre-metastatic niche maturation, adhesion, angiogenesis, and eventually the formation of metastasis.
Microbiota influence response of cancer immunotherapy
Cancer immunotherapies, specifically immune checkpoint inhibitors (ICIs) PD-1/PD-L1 and CTLA-4, have become effective strategies for cancer treatment (84). The reciprocal interactions between gut microbiota and cancer immune response raised the possibility that gut microbiota could significantly influence cancer immunotherapy response. In CRC, microbiota-based methods to enhance immunotherapy efficacy has not yet been demonstrated in human cohort. However, in animal models, specific bacterial species have been shown associated with immunotherapy response, including Bifidobacterium spp (9, 85). Bacteroides fragilis (10), Akkermansia muciniphila (11, 86), and Alistipes shahii (12). In Microsatellite Stability (MSS)-type CRC tumor-bearing mice, changes in gut microbiome affected the expression of immune-related cytokines IFN-γ and IL-2 in the tumor microenvironment, resulting in a different therapeutic effect of PD-1 antibody, and Prevotella_sp._CAG:485 and Akkermansia may maintain the normal efficacy of PD-1 antibody (87). Mager et al. investigated the efficacy of ICIs therapy in both AOM/DSS induced colitis-associated cancer and MC38 tumor-bearing models, they found that Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella species—that significantly enhanced efficacy of anti-PD-L1 and anti-CTLA-4 (88). Another study identified that tumors in antibiotic-treated or germ-free mice did not respond to CTLA-4 blockade, oral gavage of Bacteroides thetalotaomicron, Bacteroides fragilis, Burkholderia cepacia, or the combination of Bacteroides fragilis and Burkholderia cepacian recovered the anticancer response to CTLA-4 Ab following antibiotic treatment in mice subcutaneously engrafted with MC38 cells (10). Tanoue et al. isolated a consortium of 11 bacterial strains that is capable of enhancing therapeutic efficacy of ICIs in subcutaneous MC38 tumor models (89). Furthermore, gut microbiota was shown to impact immunotherapy efficacy related to innate responses. Researchers have observed that systemic administration of Bifidobacterium converts the nonresponder mice into responders to anti-CD47 immunotherapy and improves the antigen-presenting capacity of DCs (90). Song et al. found that an engineered LPS-targeting fusion protein significantly boosts anti-PD-L1 therapy against CRC tumors (91), suggesting that anti-LPS treatment may promote anti-PD-L1 immunotherapy for mouse model of CRC. Collectively, these results indicate that the gut microbiota could be used to develop new therapeutic strategies to enhance CRC immunotherapy response. Recently, a human clinical trial (NCT04729322) which studies the effect of fecal microbiota transplant and re-introduction of anti-PD-1 therapy (pembrolizumab or nivolumab) for the treatment of metastatic CRC in anti-PD-1 non-responders is under recruiting. Although still under investigation, we are beginning to exploit the tremendous potential of the gut microbiota to predict immunotherapy response, and to enhance immune surveillance for a more precise immunotherapeutic intervention.
Conclusions and perspectives
It is important to decipher the specialized roles of gut microbiota in regulating the immune response in cancer, as the current landscape of the gut microbiota-host immune axis has expanded from basic research to clinical development (92). We have gained insights into the gut microbiota dysbiosis in CRC patients (Table 1). However, considerable challenges remain, for example, although multiple studies have identified specific bacteria that are associated with CRC, inconsistency across these studies exists. This may due to diverse life styles, various diet patterns and different disease stages, since gut microbiome varied substantially according to these factors (93). More studies mining of clinical large cohort data, omics, and preclinical models are needed to facilitate consensus for potential characteristics of bacterial alternation and to determine whether such changes are a cause or an effect in CRC development. Additionally, although we have deepened our view on the innate and adaptive immune responses modulated by gut microbiota in CRC (Figure 1), owing to the complex relationship between commensal and pathogenic microbes and host immunity, more detailed studies of the sophisticated network between gut microbiota and host immune system are required. In CRC, the individual heterogeneity between patients in the response to ICIs is largely associated with the gut microbiota composition, suggesting that manipulation of gut microbiota could improve immunotherapy responses (2). Therapeutics that target microbiota is explored in conjunction with cancer immunotherapies such as FMT, prebiotics, probiotics, Chinese traditional medicine, and dietary approaches (94–97). At present, microbial intervention is mainly performed in preclinical studies at the animal level, whereas it is not yet tested with large samples in the context of clinical trials and more clear mechanisms of effective microbiota to enhance immune surveillance and influence immunotherapy responses remain unknown in CRC.
In summary, substantial efforts must be devoted to pursue a deeper understanding of the mechanistic links and to exploit for clinical benefit. The insight gained into the specialized functions of the microbiota on immunity and cancer will help to apply gut microbiota-based strategies into the clinical anti-tumor adjuvant therapies, particularly in the context of conjunction with existing immunotherapies.
Author contributions
XH and LZ conceived the study. XH, ZZ, and JW screened the literatures and wrote the manuscript. LZ reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work is supported by the Shanghai Rising-Star Program (22QA1408700 to LZ), National Natural Science Foundation of China (82104955 to XH) and China Postdoctoral Science Foundation (2022M712154 to XH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol (2019) 16(11):690–704. doi: 10.1038/s41575-019-0209-8
3. Seely KD, Morgan AD, Hagenstein LD, Florey GM, Small JM. Bacterial involvement in progression and metastasis of colorectal neoplasia. Cancers (Basel) (2022) 14(4):1019. doi: 10.3390/cancers14041019
4. Clay SL, Fonseca-Pereira D, Garrett WS. Colorectal cancer: the facts in the case of the microbiota. J Clin Invest (2022) 132(4):e155101. doi: 10.1172/JCI155101
5. Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell (2018) 33(6):954–64. doi: 10.1016/j.ccell.2018.03.004
6. Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discov (2022) 12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059
7. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res (2020) 30(6):492–506. doi: 10.1038/s41422-020-0332-7
8. Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J BioMed Sci (2018) 25(1):79. doi: 10.1186/s12929-018-0483-8
9. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
10. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
11. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
12. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
13. Gao R, Wang Z, Li H, Cao Z, Gao Z, Chen H, et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J Gastroenterol Hepatol (2020) 35(12):2109–21. doi: 10.1111/jgh.15077
14. Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome (2018) 6(1):70. doi: 10.1186/s40168-018-0451-2
15. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med (2019) 25(6):968–76. doi: 10.1038/s41591-019-0458-7
16. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun (2015) 6:8727. doi: 10.1038/ncomms9727
17. Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol (2014) 10:766. doi: 10.15252/msb.20145645
18. Liu X, Cheng Y, Shao L, Ling Z. Alterations of the predominant fecal microbiota and disruption of the gut mucosal barrier in patients with early-stage colorectal cancer. BioMed Res Int (2020) 2020:2948282. doi: 10.1155/2020/2948282
19. Zhang M, Lv Y, Hou S, Liu Y, Wang Y, Wan X. Differential mucosal microbiome profiles across stages of human colorectal cancer. Life (Basel) (2021) 11(8):831. doi: 10.3390/life11080831
20. Zwinsova B, Petrov VA, Hrivnakova M, Smatana S, Micenkova L, Kazdova N, et al. Colorectal tumour mucosa microbiome is enriched in oral pathogens and defines three subtypes that correlate with markers of tumour progression. Cancers (Basel) (2021) 13(19):4799. doi: 10.3390/cancers13194799
21. Kinross J, Mirnezami R, Alexander J, Brown R, Scott A, Galea D, et al. A prospective analysis of mucosal microbiome-metabonome interactions in colorectal cancer using a combined MAS 1HNMR and metataxonomic strategy. Sci Rep (2017) 7(1):8979. doi: 10.1038/s41598-017-08150-3
22. Loftus M, Hassouneh SA, Yooseph S. Bacterial community structure alterations within the colorectal cancer gut microbiome. BMC Microbiol (2021) 21(1):98. doi: 10.1186/s12866-021-02153-x
23. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut (2017) 66(1):70–8. doi: 10.1136/gutjnl-2015-309800
24. Pan HW, Du LT, Li W, Yang YM, Zhang Y, Wang CX. Biodiversity and richness shifts of mucosa-associated gut microbiota with progression of colorectal cancer. Res Microbiol (2020) 171(3-4):107–14. doi: 10.1016/j.resmic.2020.01.001
25. Burns MB, Montassier E, Abrahante J, Priya S, Niccum DE, Khoruts A, et al. Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PLoS Genet (2018) 14(6):e1007376. doi: 10.1371/journal.pgen.1007376
26. Sheng Q, Du H, Cheng X, Cheng X, Tang Y, Pan L, et al. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol Lett (2019) 18(5):4834–44. doi: 10.3892/ol.2019.10841
27. Gao R, Zhu Y, Kong C, Xia K, Li H, Zhu Y, et al. Alterations, interactions, and diagnostic potential of gut bacteria and viruses in colorectal cancer. Front Cell Infect Microbiol (2021) 11:657867. doi: 10.3389/fcimb.2021.657867
28. de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health (2020) 8(2):e180–90. doi: 10.1016/S2214-109X(19)30488-7
29. Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, et al. Polyamine catabolism contributes to enterotoxigenic bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U.S.A. (2011) 108(37):15354–59. doi: 10.1073/pnas.1010203108
30. Lasry A, Zinger A, Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat Immunol (2016) 17(3):230–40. doi: 10.1038/ni.3384
31. Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer (2010) 9:249. doi: 10.1186/1476-4598-9-249
32. Brennan CA, Clay SL, Lavoie SL, Bae S, Lang JK, Fonseca-Pereira D, et al. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut Microbes (2021) 13(1):1987780. doi: 10.1080/19490976.2021.1987780
33. Man SM. Inflammasomes in the gastrointestinal tract: infection, cancer and gut microbiota homeostasis. Nat Rev Gastroenterol Hepatol (2018) 15(12):721–37. doi: 10.1038/s41575-018-0054-1
34. Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity (2016) 44(3):698–711. doi: 10.1016/j.immuni.2016.02.025
35. Trinchieri G. Cancer immunity: Lessons from infectious diseases. J Infect Dis (2015) 212 Suppl 1:S67–73. doi: 10.1093/infdis/jiv070
36. Guc E, Pollard JW. Redefining macrophage and neutrophil biology in the metastatic cascade. Immunity (2021) 54(5):885–902. doi: 10.1016/j.immuni.2021.03.022
37. Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res (2006) 66(2):605–12. doi: 10.1158/0008-5472.CAN-05-4005
38. Stefani C, Miricescu D, Stanescu-Spinu II, Nica RI, Greabu M, Totan AR, et al. Growth factors, PI3K/AKT/mTOR and MAPK signaling pathways in colorectal cancer pathogenesis: Where are we now? Int J Mol Sci (2021) 22(19):10260. doi: 10.3390/ijms221910260
39. Barnet MB, Blinman P, Cooper W, Boyer MJ, Kao S, Goodnow CC. Understanding immune tolerance of cancer: Re-purposing insights from fetal allografts and microbes. Bioessays (2018) 40(8):e1800050. doi: 10.1002/bies.201800050
40. Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA (2009) 302(6):649–58. doi: 10.1001/jama.2009.1112
41. Alvarez R, Oliver L, Valdes A, Mesa C. Cancer-induced systemic myeloid dysfunction: Implications for treatment and a novel nanoparticle approach for its correction. Semin Oncol (2018) 45(1-2):84–94. doi: 10.1053/j.seminoncol.2018.05.001
42. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
43. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol (2013) 14(10):1014–22. doi: 10.1038/ni.2703
44. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1053/j.gastro.2018.04.001
45. Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology (2018) 155(1):33–7.e36. doi: 10.1053/j.gastro.2018.04.001
46. Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, et al. Neutrophil ageing is regulated by the microbiome. Nature (2015) 525(7570):528–32. doi: 10.1038/nm.2278
47. Yang XD, Ai W, Asfaha S, Bhagat G, Friedman RA, Jin G, et al. Histamine deficiency promotes inflammation-associated carcinogenesis through reduced myeloid maturation and accumulation of CD11b+Ly6G+ immature myeloid cells. Nat Med (2011) 17(1):87–95. doi: 10.1038/nm.2278
48. Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer (2016) 16(7):431–46. doi: 10.1038/nrc.2016.52
49. Mizuno R, Kawada K, Itatani Y, Ogawa R, Kiyasu Y, Sakai Y. The role of tumor-associated neutrophils in colorectal cancer. Int J Mol Sci (2019) 20(3):529. doi: 10.3390/ijms20030529
50. Masucci MT, Minopoli M, Del Vecchio S, Carriero MV. The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front Immunol (2020) 11:1749. doi: 10.3389/fimmu.2020.01749
51. Hezaveh K, Shinde RS, Klotgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity (2022) 55(2):324–40.e328. doi: 10.1016/j.immuni.2022.01.006
52. Kaisar MMM, Pelgrom LR, van der Ham AJ, Yazdanbakhsh M, Everts B. Butyrate conditions human dendritic cells to prime type 1 regulatory T cells via both histone deacetylase inhibition and G protein-coupled receptor 109A signaling. Front Immunol (2017) 8:1429. doi: 10.3389/fimmu.2017.01429
53. Liu L, Li L, Min J, Wang J, Wu H, Zeng Y, et al. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell Immunol (2012) 277(1-2):66–73. doi: 10.1016/j.cellimm.2012.05.011
54. Beyaz S, Chung C, Mou H, Bauer-Rowe KE, Xifaras ME, Ergin I, et al. Dietary suppression of MHC class II expression in intestinal epithelial cells enhances intestinal tumorigenesis. Cell Stem Cell (2021) 28(11):1922–35.e1925. doi: 10.1016/j.stem.2021.08.007
55. Schulz MD, Atay C, Heringer J, Romrig FK, Schwitalla S, Aydin B, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature (2014) 514(7523):508–12. doi: 10.1038/nature13398
56. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
57. Zitvogel L, Kroemer G. Cross-reactivity between microbial and tumor antigens. Curr Opin Immunol (2022) 75:102171. doi: 10.1016/j.coi.2022.102171
58. Fluckiger A, Daillère R, Sassi M, Sixt BS, Liu P, Loos F, et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science (2020) 369(6506):936–42. doi: 10.1126/science.aax0701
59. Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell (2021) 39(10):1317–41. doi: 10.1016/j.ccell.2021.08.006
60. Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and anticancer immunosurveillance. Cell (2016) 165(2):276–87. doi: 10.1016/j.cell.2016.03.001
61. De Almeida CV, de Camargo MR, Russo E, Amedei A. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J Gastroenterol (2019) 25(2):151–62. doi: 10.3748/wjg.v25.i2.151
62. Geuking MB, Burkhard R. Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunol (2020) 13(6):855–66. doi: 10.1038/s41385-020-00335-w
63. Oster P, Vaillant L, Riva E, McMillan B, Begka C, Truntzer C, et al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut (2022) 71(3):457–66. doi: 10.1136/gutjnl-2020-323392
64. Botticelli A, Zizzari I, Mazzuca F, Ascierto PA, Putignani L, Marchetti L, et al. Cross-talk between microbiota and immune fitness to steer and control response to anti PD-1/PDL-1 treatment. Oncotarget (2017) 8(5):8890–9. doi: 10.18632/oncotarget.12985
65. Crespo J, Sun H, Welling TH, Tian Z, Zou W. T Cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol (2013) 25(2):214–21. doi: 10.1016/j.coi.2012.12.003
66. Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S, et al. Gut microbiota modulate CD8 T cell responses to influence colitis-associated tumorigenesis. Cell Rep (2020) 31(1):107471. doi: 10.1016/j.celrep.2020.03.035
67. Guégan JP, Ginestier C, Charafe-Jauffret E, Ducret T, Quignard JF, Vacher P, et al. CD95/Fas and metastatic disease: What does not kill you makes you stronger. Semin Cancer Biol (2020) 60:121–31. doi: 10.1016/j.semcancer.2019.06.004
68. Nozoe T, Yasuda M, Honda M, Inutsuka S, Korenaga D. Fas ligand expression is correlated with metastasis in colorectal carcinoma. Oncology (2003) 65(1):83–8. doi: 10.1159/000071208
69. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity (2019) 51(2):285–97.e5. doi: 10.1016/j.immuni.2019.06.002
70. Liu X, Chen Y, Zhang S, Dong L. Gut microbiota-mediated immunomodulation in tumor. J Exp Clin Cancer Res (2021) 40(1):221. doi: 10.1186/s13046-021-01983-x
71. Overacre-Delgoffe AE, Bumgarner HJ, Cillo AR, Burr AHP, Tometich JT, Bhattacharjee A, et al. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity (2021) 54(12):2812–24.e2814. doi: 10.1016/j.immuni.2021.11.003
72. Lowenmark T, Li X, Lofgren-Burstrom A, Zingmark C, Ling A, Kellgren TG, et al. Parvimonas micra is associated with tumour immune profiles in molecular subtypes of colorectal cancer. Cancer Immunol Immunother (2022) 71(10):2565–75. doi: 10.1007/s00262-022-03179-4
73. Noble A, Durant L, Dilke SM, Man R, Martin I, Patel R, et al. Altered mucosal immune-microbiota interactions in familial adenomatous polyposis. Clin Transl Gastroenterol (2022) 13(7):e00428. doi: 10.14309/ctg.0000000000000428
74. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory b cells. Cell Metab (2020) 31(4):837–51.e810. doi: 10.1016/j.cmet.2020.03.003
75. Yang W, Cong Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol Immunol (2021) 18(4):866–77. doi: 10.1038/s41423-021-00661-4
76. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature (2016) 535(7610):75–84. doi: 10.1038/nature18848
77. Wang Z, Lu Z, Lin S, Xia J, Zhong Z, Xie Z, et al. Leucine-tRNA-synthase-2-expressing b cells contribute to colorectal cancer immunoevasion. Immunity (2022) 55(6):1067–81.e8. doi: 10.1016/j.immuni.2022.04.017
78. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med (2013) 19(11):1423–37. doi: 10.1038/nm.3394
79. Li R, Zhou R, Wang H, Li W, Pan M, Yao X, et al. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ (2019) 26(11):2447–63. doi: 10.1038/s41418-019-0312-y
80. Yin H, Miao Z, Wang L, Su B, Liu C, Jin Y, et al. Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging (2022) 14(4):1941–58. doi: 10.18632/aging.203914
81. Sakamoto Y, Mima K, Ishimoto T, Ogata Y, Imai K, Miyamoto Y, et al. Relationship between fusobacterium nucleatum and antitumor immunity in colorectal cancer liver metastasis. Cancer Sci (2021) 112(11):4470–7. doi: 10.1111/cas.15126
82. Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
83. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell (2021) 39(5):708–24.e711. doi: 10.1016/j.ccell.2021.03.004
84. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol (2020) 17(8):807–21. doi: 10.1038/s41423-020-0488-6
85. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
86. Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut (2019) 68(3):385–8. doi: 10.1136/gutjnl-2018-317220
87. Xu X, Lv J, Guo F, Li J, Jia Y, Jiang D, et al. Gut microbiome influences the efficacy of PD-1 antibody immunotherapy on MSS-type colorectal cancer via metabolic pathway. Front Microbiol (2020) 11:814. doi: 10.3389/fmicb.2020.00814
88. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
89. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
90. Fu Y-X, Weichselbaum RR, Chang EB, Lin W, Xue L, Ni K, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med (2020) 217(5):e20192282. doi: 10.1084/jem.20192282
91. Song W, Tiruthani K, Wang Y, Shen L, Hu M, Dorosheva O, et al. Trapping of lipopolysaccharide to promote immunotherapy against colorectal cancer and attenuate liver metastasis. Adv Mater (2018) 30(52):e1805007. doi: 10.1002/adma.201805007
92. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33(4):570–80. doi: 10.1016/j.ccell.2018.03.015
93. Song M, Chan AT. Environmental factors, gut microbiota, and colorectal cancer prevention. Clin Gastroenterol Hepatol (2019) 17(2):275–89. doi: 10.1016/j.cgh.2018.07.012
94. Zhou CB, Zhou YL, Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer (2021) 7(7):647–60. doi: 10.1016/j.trecan.2021.01.010
95. Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology (2021) 160(2):600–13. doi: 10.1053/j.gastro.2020.11.041
96. Xu B, Wang X, Wang H, Cao L, Ge Y, Yuan B, et al. Efficacy and safety of herbal formulas with the function of gut microbiota regulation for gastric and colorectal cancer: A systematic review and meta-analysis. Front Cell Infect Microbiol (2022) 12:875225. doi: 10.3389/fcimb.2022.875225
Keywords: colorectal cancer, gut microbiota, inflammation, immune response, immunotherapy
Citation: Hou X, Zheng Z, Wei J and Zhao L (2022) Effects of gut microbiota on immune responses and immunotherapy in colorectal cancer. Front. Immunol. 13:1030745. doi: 10.3389/fimmu.2022.1030745
Received: 29 August 2022; Accepted: 24 October 2022;
Published: 08 November 2022.
Edited by:
Catherine Sautes-Fridman, INSERM U1138 Centre de Recherche des Cordeliers (CRC), FranceReviewed by:
Sanae Ben Mkaddem, Mohammed VI Polytechnic University, MoroccoRosalba Salcedo, National Institutes of Health (NIH), United States
Copyright © 2022 Hou, Zheng, Wei and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Zhao, bHpoYW9Ac2h1dGNtLmVkdS5jbg==
†These authors have contributed equally to this work
 Xinxin Hou
Xinxin Hou Zongmei Zheng
Zongmei Zheng Jiao Wei
Jiao Wei Ling Zhao
Ling Zhao