
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 10 November 2022
Sec. Mucosal Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1024330
This article is part of the Research TopicMucosal Vaccines for the Induction of Antimicrobial Immunoglobulin AView all 6 articles
Gut microbiota has extensive and tremendous impacts on human physiology and pathology. The regulation of microbiota is therefore a cardinal problem for the mutualistic relationship, as both microbial overgrowth and excessive immune reactions toward them could potentially be detrimental to host homeostasis. Growing evidence suggests that IgA, the most dominant secretory immunoglobulin in the intestine, regulates the colonization of commensal microbiota, and consequently, the microbiota-mediated intestinal and extra-intestinal diseases. In this review, we discuss the interactions between IgA and gut microbiota particularly relevant to human pathophysiology. We review current knowledge about how IgA regulates gut microbiota in humans and about the molecular mechanisms behind this interaction. We further discuss the potential role of IgA in regulating human diseases by extrapolating experimental findings, suggesting that IgA can be a future therapeutic strategy that functionally modulates gut microbiota.
IgA is the most dominant immunoglobulin class in humans. IgA-producing cells are mainly distributed in the mucosa lining tissues such as the intestine and oral and nasal cavities. IgA produced by these cells are usually dimerized within the cells through disulfide bonds of its C-terminus with the joining chain (J chain). Dimeric IgA is bound with polymeric Ig receptor (pIgR) on the basolateral surface of intestinal epithelial cells and endocytosed. During transcytosis through intestinal epithelial cells, the extracellular domain of pIgR is cleaved by endopeptidase in the transport vesicle, and this domain, now called secretary component (SC), is secreted with bound IgA to the intestinal lumen. IgA complexed with SC is designated as secretory IgA (SIgA). SIgA is protected from bacterial proteases thanks to the presence of SC, and thus has a longer half-life than IgA (1).
The targets of IgA are “foreign bodies” for us, such as microbes, dietary antigens, etc. In the past decades, IgA has been studied as a cardinal component of mucosal defense against pathogens such as genera Salmonella and Vibrio based on the assumption that the control of pathogens is the primary role of IgA (2–6). More recently, many studies have also appreciated the role of IgA in regulating gut commensal microbiome (7, 8). Surprisingly, however, the lack of IgA in humans rarely causes severe complications such as infectious diseases (9, 10), in part because of compensation by other immunoglobulin classes such as IgM (11, 12). Nevertheless, growing evidence also links IgA deficiency and altered microbiome in humans (11, 13). Considering the tremendous impact of the human microbiome on health and diseases, ranging from local intestinal diseases to systemic disorders relating to the central nervous system, systemic metabolism, and autoimmunity, it is tempting to presume that regulation of the microbiome by IgA may modulate the susceptibility of these human diseases other than infections. In addition, understanding the biology of IgA may lead to a novel opportunity to artificially modulate the susceptibility for the diseases through microbiome regulation. In this review, we summarize the current knowledge regarding the relationship between IgA and gut microbiome and discuss how IgA controls the microbiome and, as a consequence, our health.
IgA has long been studied in the context of infectious diseases and their pathogens, such as genera Salmonella and Vibrio, their toxins like Cholera toxin. For example, IgA can bind to Salmonella and suppress their motility (14) and invasion (5). IgA also facilitates agglutination, enchaining, and therefore, clearance of Salmonella (4). In addition, IgA can neutralize Cholera toxin and mitigate its pathogenicity (6). IgA not only directly acts on these pathogens to limit their pathogenicity but also facilitates immune reactions against these pathogens through retro transcytosis (15). In this transport mechanism, IgA binds to antigens such as bacteria and then bind to surface receptors of microfold cells (M cells) such as Dectin-1 on the Peyer’s patches (15), although a later study shows that Dectin-1 is dispensable for retro transcytosis of IgA-coated Salmonella (16).
However, these studies largely rely on animal studies. The effects of IgA on pathogens have not been fully elucidated in humans. Selective IgA deficiency, which is defined as undetectable levels of IgA in the serum, stool, etc. while other immunoglobulin classes remain intact, is the most common primary immunodeficiency in humans. It affects one in 200 to 1,000 individuals in Caucasians, although the prevalence is varied among populations (9, 10). Although selective IgA deficiency is frequently complicated with other immune-related diseases such as Graves’ disease, type 1 diabetes, and celiac disease (17, 18), it has been reported that IgA deficiency mainly increases the risk of infectious diseases. For example, Jorgensen et al. have reported that IgA deficiency increases the incidence of upper and lower respiratory tract infection and allergic diseases compared with healthy individuals (19). Aytekin et al. have also analyzed 118 patients with selective IgA deficiency in Turkey and revealed that 83.9% of patients develop some infectious diseases during 7 years (median) of the observation period (20). Recurrent infections are observed in up to one-third of symptomatic patients (21). However, these studies also show that critical infectious diseases are not common in patients with selective IgA deficiency, and it appears that its susceptibility to infectious diseases is not as severe as other primary immunodeficiency diseases such as common variable immunodeficiency (22). This fact may be attributable to compensation by other immunoglobulin classes such as IgM. There are five immunoglobulin classes in humans: IgM, IgG, IgA, IgD, and IgE. Although IgA is the most dominant immunoglobulin in the intestine, IgM and IgG are also responsible for regulating intestinal immunity and infections (23, 24). Several reports have revealed that IgM is highly detected in the stool of IgA deficient patients (25), implying the compensatory role of IgM in regulating the intestinal environment. However, the relationship between enhanced IgM secretion and pathogen control in humans has not been fully understood.
Recent studies also highlight the impact of IgA deficiency on gut commensal bacteria (11, 13, 25). As in mice, human IgA can react to gut commensals (26). In addition, Sterlin et al. have specifically addressed the microbial reactivity of two human IgA subclasses: IgA1 and IgA2 (27). Interestingly, both IgA1 and IgA2 similarly bind to small intestinal bacteria, whereas genera from phylum Bacteroidetes, especially Flavobacterium, are preferentially bound by IgA2. Potentially concordant with this observation, IgA2 is more common, especially in the colon (28). The major difference between two IgA subclass is the length of hinge region, with IgA1 having an extended hinge, which may confer advantage in antigen binding (29). However, the longer hinge region also allows pathogenic bacteria to cleave and impair IgA1 function through their proteolytic enzymes (30). It has also been reported that IgA1 and IgA2 show different glycosylation profiles, with IgA1 having more sialic acids (31). The difference in the glycosylation profiles seems to affect their functional properties, as serum IgA2 shows more proinflammatory profiles than IgA1. Although these studies did not specifically address the difference of IgA1 and IgA2 in the intestine, we can extrapolate these data to speculate that IgA1 and IgA2 may play a different role in regulating microbiota in humans. To support our speculation, it has been reported that IgA2 but not IgA1 is able to bind to M cells on the Peyer’s patches, which play an important role in initiating the production of antigen-specific IgA (32).
In line with the role of IgA in regulating pathogens, IgA deficiency seems to increase the abundance of pathobionts, i.e. members of commensal bacteria potentially harmful to host homeostasis under certain conditions. For example, Fadlallah et al., have reported that Escherichia coli, a known pathobiont and target of IgA binding, are increased in the stool of patients with selective IgA deficiency (11). Similarly, Moll et al. have shown that E. coli is one of the major gut colonizers in healthy individuals and more prominent in patients with selective IgA deficiency (13). Although direct associations have not been elucidated, selective IgA deficiency is associated with the elevation of proinflammatory cytokines such as interleukin (IL)-6 and IL-17, which may be attributable to the increase in pathobionts (11). Again, IgM seems to compensate for part of IgA roles in regulating commensal bacteria; however, Enterobacteriaceae including E. coli is not targeted by IgM (11). In addition, Magri et al., have shown that dual coating of microbes by SIgA and SIgM is a cardinal feature in humans rather than mice (24), suggesting a compensatory role of IgM. However, the extent of compensation by IgM is not fully understood, as IgM appears to be less specific to gut commensals (25).
Interestingly, many studies have linked IgA deficiency to several non-infectious diseases such as autoimmune diseases (17, 18), which have also been associated with the gut commensal microbiota. It also appears that autoimmunity in turn affects the regulation of microbiota since anti-IgA antibodies in selective IgA deficiency are associated with the expansion of pathobionts like E. coli (13). These findings raise the possibility that IgA may modulate the susceptibility of human diseases through the regulation of commensal microbiota, and it encourages researchers to understand the impact of commensal microbiota on our health and how IgA can interact with these bacteria.
We here discuss how IgA interacts with the gut microbiota. Unfortunately, vast evidence in this field is established in mouse experiments. Nevertheless, we think it is possible to extrapolate these experimental findings to human settings, particularly thanks to the recent development of IgA-sequencing (IgA-seq) analysis. Previously, it was necessary to examine the affinity of IgA with microbes one by one, while IgA-seq analysis, which takes advantage of next-generation sequencing techniques, provides an overall picture of IgA bound to commensal microbes and enables researchers to comprehensively understand IgA-microbe interactions in both humans and mice. The detailed molecular mechanisms by which IgA binds to microbial antigens are also discussed elsewhere (7, 8, 33).
In line with the central role of immunoglobulins, IgA mainly regulates the colonization, invasion, growth, and motility of commensal bacteria. The importance of intestinal IgA in the regulation of commensal microbiota is initially recognized by Fagarasan et al. In their study, they used mice deficient in activation-induced cytidine deaminase (AID), a critical regulator of both class switching and somatic hypermutation of the immunoglobulin locus, and thereby lacking immunoglobulins other than IgM. The authors found an overgrowth of gut microbiota in AID deficient mice (34). AID deficiency particularly expands segmented filamentous bacteria (SFB) colonization in the intestine (35). Later, the same group also reported that a point mutation in AID which leads to loss of somatic hypermutation but not class switching resulted in similar consequences (36), suggesting the importance of affinity maturation in immunoglobulins, particularly IgA. Similarly, it has been reported that PD1-deficient mice show decreased IgA-binding bacteria and dysregulation of gut microbiota, which result from abnormal follicular T-cell function (37). More recently, Nagaishi et al. have specifically addressed the alterations of gut microbiota and intestinal homeostasis in IgA-deficient mice (38). They showed a skewed microbiota composition in IgA-deficient mice, including an expansion of SFB. IgA-deficient mice also exhibited spontaneous inflammation in the ileum, which was canceled by antibiotic treatment. These studies indicate that IgA, especially those produced by the T cell-dependent pathway and therefore characterized by its high affinity, could functionally regulate the growth of intestinal bacteria, especially pathobionts.
With the widespread use of IgA-seq techniques, it has become recognized that the patterns of IgA-binding bacteria in specific diseases and environments are different. Palm et al. first showed in 2014 that the pattern of IgA-binding bacteria is altered in patients with IBD and that colonization of IgA-binding bacteria from IBD patients but not healthy individuals in germ-free mice exacerbates intestinal inflammation in the colitis model (39). Similarly, IgA binding to pathogenic E. coli is increased in the feces of patients with spondyloarthritis (40). IgA-coated bacteria are also altered in multiple sclerosis (41, 42), kwashiorkor (43), obesity after bariatric surgery (44), and certain types of cancer (45). These results suggest that IgA may have the ability to bind to and control potentially harmful commensal bacteria that are involved in human pathogenesis.
It is also important to note that IgA can activate the immune system against gut microbes via retro transcytosis into the Peyer’s patches (15). Although antigens derived from pathogens are transported through M cells scattered in the epithelium covering the Peyer’s patches and recognized by dendritic cells located right beneath M cells, several studies show that M cell-dependent transcytosis also appears important for commensal bacteria such as E. coli and SFB (46–48). Mikulic et al. have experimentally shown that IgA coating of Lactobacillus rhamnosus, a symbiotic gut microbe, plays a role in conditioning dendritic cells, which facilitates tolerogenic responses such as induction of regulatory T cells (49). These findings suggest that IgA not only regulates commensal bacteria through a direct interaction but also affects the gut environment by modulating the intestinal immune system through M cell-mediated uptake of commensal microbes.
Interestingly, IgA can promote the colonization of gut microbes in certain situations. This is especially true for Bacteroides, a major symbiotic genus in the gut. For example, Nakajima et al. have reported that IgA facilitates colonization of B. thetaiotaomicron (50). As a mechanism, the authors have suggested that IgA alters the gene expression of B. thetaiotaomicron related to polysaccharide utilization required for colonization in the mucus layer, which they named Mucus-Associated Functional Factor (MAFF). Another report has shown that IgA fosters stable colonization of B. thetaiotaomicron by altering its epitope expressions and thereby silencing excessive inflammatory responses (51). Donaldson et al. have reported that IgA mediates the adhesion of B. fragilis to intestinal epithelial cells in a capsular polysaccharide-dependent manner (52). IgA is also found to facilitate biofilm formation of E. coli in an in vitro setting (53, 54). The mechanism by which IgA regulates commensal microbes in opposite ways is not fully understood; however, it has been proposed that adhesive molecules produced by the host (e.g., IgA and mucus) and mucus flow could select for and against certain microbes (55). In addition, replication rates of microbes and the bound break of IgA enchainment may also explain this difference (56, 57). In this model, the host produces IgA to all types of bacteria; however, those characterized by higher replication rates may remain enchained even after their division, since their replication rates are faster than SIgA-crosslinking breaks of enchained clusters. This model may explain why SIgA is likely to affect fast-growing bacteria such as pathobionts and eliminate them from the intestine.
It is generally recognized that the synthetic pathways of IgA largely determine its functional characteristics. The T cell-dependent pathway is supported by CD4 T cells that results in differentiation of high-affinity IgA-producing cells, while the T cell-independent pathway is mainly supported by dendritic cells and results in the production of low-affinity IgA (58–60). There is evidence showing that the high affinity IgA resulting from T-cell help and somatic hypermutation is the cardinal feature of IgA function in regulating gut microbiota. For example, as we discussed earlier, a point mutation in AID that impairs somatic hypermutation elicits dysregulation of gut microbiota (36). Kabbert et al. have also analyzed the characteristics of human IgA and reported that somatic hypermutation rather than polyreactivity, another aspect of IgA, is associated with microbial reactivity (26). Furthermore, Okai et al. have reported that oral administration of high-affinity IgA can alter gut microbiota composition (61). In a report by Bunker et al., the authors compared the microbiota composition in wild-type and T cell-deficient mice and showed that IgA produced through the T cell-dependent pathway was capable of binding to microbes that colonize close to epithelial cells, such as SFB and Mucispirillum (62). It has been proposed that although these bacteria are generally considered as pathobionts, their proximity to epithelial cells makes them more likely to be captured by antigen-presenting cells and consequently more likely to be presented their antigens to CD4 T cells, leading to the production of high-affinity IgA toward them in the T cell-dependent pathway (62). Indeed, mice with impaired T-cell function, including those deficient in T-cell receptor beta and delta chains, have altered affinity and specificity of IgA, resulting in changes in the composition of intestinal bacteria (37, 62, 63). Yang et al. have also reported that IgA specificity to commensal microbe is dependent on CD4 T cells (63). We similarly reported that acetate, one of the SCFAs, is potent in enhancing T cell-dependent IgA production and that it has a substantial impact on the composition of mucosa-associated bacteria in an IgA-dependent manner (64). We also showed that acetate facilitates the clearance of E. coli in the mucus layer, which also appears dependent on its IgA coating. On the other hand, the role of IgA produced by the T cell-independent pathway in regulating gut microbiota has not been well elucidated. It is widely considered that T cell-independent IgA generally binds to a wide variety of bacterial antigens.
Meanwhile, an IgA repertoire analysis reveals that V gene usage among IgA-producing B cells in the intestine is restricted and that somatic hypermutation profile is less likely shared among clonally related cells, suggesting that antigen selection and affinity maturation may be uncommon (65). In addition, IgA-coated bacteria are detected in T-cell deficient mice to a similar extent to their wild-type counterpart despite a severe reduction in SIgA (60, 62, 66), suggesting a capability of T cell-independent IgA in binding commensal bacteria. Polyclonal and low affinity antibodies, mainly IgE and IgG1, are also considered to play a role in host defense against certain gut pathogens such as helminth (67). These studies may support the role of low-affinity IgA in regulating gut homeostasis. Nevertheless, the role of T cell-independent IgA in regulating gut bacteria has not been fully elucidated. It has been recognized that T cell substantially impacts microbiota composition through IgA production (68). Furthermore, Grasset et al. showed that IgA in TACI-deficient mice, which lack T cell-independent IgA, did not alter gut microbiota in the colonic tissue and stool contents (60). These studies show that although T cell-independent IgA can bind to gut microbes, its role in regulating microbiota composition, in contrast to T cell-dependent IgA, remains elusive. Although not proven, this may be attributable to the low affinity of T cell-independent IgA, since sufficient affinity to gut bacteria appears required for their discharge from the intestinal lumen (56, 57). It is also important to note that apart from somatic hypermutation, N-linked glycosylation of the variable domain in antibody also affects antigen binding (69). In addition, the affinity of glycan-glycan interactions with bacterial components such as lipopolysaccharides could be sufficiently high (70). Although this is a hypothesis requiring future investigation, glycan-glycan interactions may be another perspective of IgA in regulating microbiota. Taken together, the function of IgA on the gut microbiota is likely to be defined by its mode of induction and structure, and in particular, its affinity appears to have a significant impact.
Another important aspect is the regulation of IgA-microbe interactions by environmental factors. Microbe-derived small molecules such as metabolites are important regulator of intestinal immunity (Figure 1). As we discussed above, microbial metabolites (e.g., SCFA) augment intestinal IgA production and regulate their reactivity with microbes (64, 71, 72). Other dietary components and nutrients such as dietary antigens (73), glutamine (74), vitamin A (75, 76), and dietary fat (77) are also considered important regulators of intestinal IgA. ATP, which could be released during tissue damage and presumably during the invasion of pathogens, is another important environmental stimulus that regulates IgA production and mucosal colonization (78). Nutrient status can also impact gut microbiota and their properties of being bound by IgA. For example, under starvation, Lactobacillus spp. that minimally express surface antigens for IgA binding can selectively expand, resulting in altered IgA-microbe interrelationships at the community level (79).
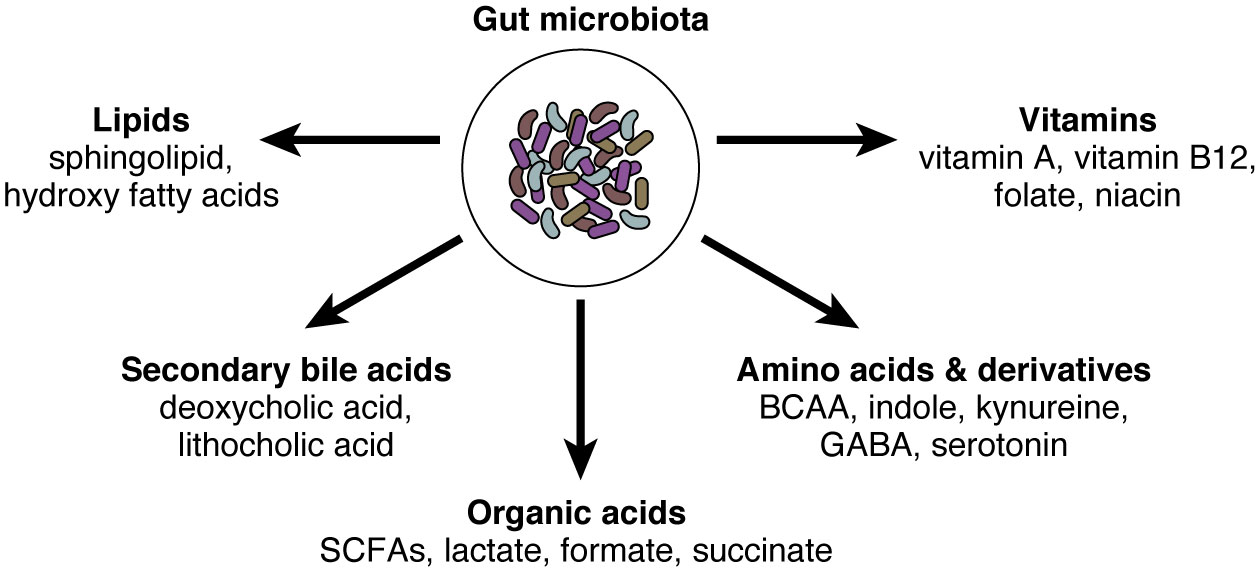
Figure 1 The gut microbiota and their small molecules. The gut microbiota possesses millions of genes and produces thousands of small molecules. Here we summarized representative microbe-derived small molecules, including organic acids, amino acids and their derivatives, vitamins, secondary bile acids, and lipids. SCFAs, short-chain fatty acids; BCAA, branched-chain amino acids; GABA, gamma-aminobutyric acid.
Finally, recent studies also appreciate the commensal fungi as important modulators of the immune system and host diseases such as IBD (80–82). Importantly, IgA appears to regulate the commensal fungi as well. For example, Ost et al. have shown that intestinal IgA can bind to and suppress hyphae of Candida albicans, which can be potentially harmful to intestinal homeostasis (83). Interestingly, IgA reactive with C. albicans induced by a vaccination can reduce the severity of DSS-induced colitis. Similarly, Doron et al. have also reported that SIgA in mice and humans preferentially binds to C. albicans hyphae (84). Notably, SIgA reactive with hyphae-producing antigens such as Sap6 and candidalysin were significantly reduced in the mucosa of Crohn’s disease, implying a protective role of candida-specific IgA in disease pathogenesis.
Considering that gut microbiota has tremendous impacts on human physiology and pathology and that IgA effectively alters the composition of the microbiota, it is tempting to assume that IgA may modulate the pathogenesis of intestinal and extra-intestinal diseases in humans. Understanding the underlying mechanisms by which IgA impacts disease susceptibility may also be important for the clinical application of IgA in future. As most human findings in IgA-microbe interactions are purely associative and correlative, we discuss experimental findings to extrapolate the role of IgA in regulating microbiota-mediated diseases.
As we discussed above, it is not surprising that intestinal diseases such as IBD are more prominently influenced by the gut microbiota. It has been shown that IBD is characterized by overgrowth of pathobionts such as E. coli and Klebsiella in the intestine, which may be functionally involved in the progression of disease activity and complications (39, 40, 85–87). These bacteria seem to directly penetrate the mucosal layer and thereby excessively fuel immune reactions (39). In line with the protective role of IgA against pathobionts, many human studies have revealed that IgA is bound preferentially to pathobionts and/or colitogenic bacteria in IBD (39, 40, 88). A report has also shown that SIgA is increased in IBD (89), implying that IgA is produced in response to the expansion of pathobionts. Furthermore, a recent paper by Shapiro et al., revealed that IgA coating to Oscillospira is linked with a delay in time to surgery in IBD patients, suggesting that IgA-coated bacteria can be clinically applied as an IBD biomarker (88). Together, these human findings warrant the application of IgA to IBD for therapeutic and/or diagnostic purposes. In this regard, Okai et al. have reported that administration of a high affinity and polyreactive IgA, named W27, can bind to a wide variety of gut commensal bacteria including pathobionts and that this high-affinity IgA can mitigate colitis in experimental models (61). Similarly, Xiong et al. have shown that oral administration of W27 ameliorates a colitis model in marginal zone B and B-1 cell-specific protein (MZB1)-deficient mice, which largely impairs the secretion of IgA into the gut lumen (90). The effects of W27 have also been tested in an in vitro model of the human microbiota, with a prominent effect on the growth inhibition of E. coli (91). These findings suggest that limit of bacterial growth and/or faster discharge from the intestinal lumen is facilitated by exogenous IgA administration, and that this may serve as a protection against colitis. However, it is important to note that IgA binding to pathobionts is not always a good thing; for example, in Crohn’s disease, NOD2 mutation facilitates retro transcytosis of IgA-coated bacteria through M cells, which may fuel intestinal inflammation and permeability (92).
As we discussed above, commensalism of other microorganisms such as fungi may also regulate the disease severity and outcome of IBD (83, 84). Virus is yet another pathogen that is controlled by IgA. The regulation of virus by SIgA is particularly important for protection against enteropathogenic virus such as norovirus and rotavirus (93–95). Rotavirus vaccination failure is associated with lower plasma rotavirus-specific IgA (96), suggesting that IgA induction through vaccination functionally protect against rotavirus infection. How IgA regulates commensal virome has not been fully elucidated. Although many studies have suggested altered gut viromes in IBD patients (97–99), one study show that IgA deficiency does not substantially impact viral profile at least in the saliva (100).
In addition to IBD, it has been recognized that irritable bowel syndrome (IBS) is also associated with altered gut microbiota and their metabolites (101, 102), which is characterized by an increase in pathobionts such as the family Enterobacteriaceae (101). Both SIgA and IgA-coated bacteria are increased in IBS patients, especially those of diarrhea-predominant type (IBS-D) (103, 104). It also appears that IgA coating toward the genus Escherichia–Shigella is particularly promoted in patients with IBS-D and is positively correlated with some clinical manifestations of IBS such as anxiety and depression scores (103). These findings suggest that, like IBD, IgA may be produced and secreted upon an expansion of pathobionts in IBS, although a protective role of IgA in the pathogenesis of IBS remains elusive.
Cardiometabolic diseases is another good example of how the regulation of microbiome by IgA can be a potential treatment strategy. For example, IgA deficiency in a mouse model of obesity (e.g., high-fat diet-induced obesity) aggravates the metabolic phenotypes (77). In addition, IgA positively correlated with the improvement of metabolic parameters in the patients who underwent bariatric surgery (77). Interestingly, serum IgA has shown to increase in patients with type 2 diabetes, although fecal IgA has not been studied (105, 106). These human studies raise the possibility that IgA may somehow affect the pathogenesis of cardiometabolic diseases. As we noted above, translocation of microbial components such as LPS and flagellin elicits low-grade inflammation in the liver and adipose tissue, resulting in insulin resistance and obesity (107, 108). In this regard, it has been reported that vaccination with flagellin is potent in regulating the localization of gut microbes through induction of flagellin-specific IgA (109). Furthermore, the vaccination ameliorates colitis and diet-induced obesity. Fujimoto et al. have similarly reported that immunization with a microbial antigen with adjuvants (i.e. CpG and curdlan) shows potent induction of anti-Clostridium ramosum IgA in the intestine, and this specific antibody can ameliorate obesity and other metabolic consequences (110). Since microbe-derived metabolites also play an important role in the development of cardiometabolic diseases, the regulation of microbial transcriptional activity could be another therapeutic strategy. In this regard, several papers reveal that host immune reactions including IgA binding to gut microbes also alter the transcriptional activity and metabolite production by gut microbes (50, 111). As we discussed earlier, T-cell help is important for the production of high-affinity IgA that can regulate microbiota. In this regard, Petersen et al. have revealed that MyD88 deficiency in T cells, which results in impaired T-cell help function in producing intestinal IgA, aggravates obesity in a microbiota-dependent manner (112). Collectively, these findings suggest that IgA is involved in the pathogenesis of metabolic diseases and that IgA can be a useful toolbox to alter the gut microbiota and thereby host metabolism.
It should be stressed, however, that metabolic phenotypes have not been reported in patients with IgA deficiency. We therefore consider that IgA deficiency itself may minimally contribute to host metabolism in humans, possibly due to compensatory mechanisms by other immunoglobulin classes and/or other factors influencing host metabolism (e.g., recurrent infections). Nevertheless, this does not exclude any possibilities that an IgA-based intervention targeting certain commensal bacteria could effectively alter the microbial community and thereby host metabolic diseases. Needless to mention, more studies are required to prove the concept in future.
Gut microbes can penetrate and translocate into the host body in certain conditions such as obesity and diabetes (113–115). Therefore, the role of IgA as a defense against microbes seems also important in organs other than the intestine. The liver is considered a firewall against bacterial translocation. Moro-Sibilot et al. have revealed that IgA-producing plasma cells are distributed in the human liver, and liver-derived IgA appears to be highly reactive to B. vulgatus, a human commensal species (116). They also show in mice that these IgA-producing cells in the liver are derived from the Peyer’s patches. However, in an alcohol-induced hepatitis model, they report that an increase in liver IgA-producing cells is associated with worse outcomes and that suppressing the migration of IgA-producing cells from the Peyer’s patches to the liver can ameliorate liver injury. Therefore, although IgA produced in the liver seems to react with the commensal bacteria, the functional role in terms of IgA-bacteria interactions, especially in the liver, remains elusive.
IgA is reportedly involved in autoimmune diseases such as multiple sclerosis (MS). However, unlike IBD or cardiometabolic diseases, it seems that IgA-producing cells themselves may affect the pathogenesis of experimental autoimmune encephalomyelitis (EAE), a disease model for MS, by producing immune suppressive cytokines such as IL-10 (117). Interestingly, the authors also showed a reduction of IgA-coated bacteria in severe MS patients, which is also confirmed in another report (41). In addition, they experimentally proved that increase in intestinal and brain IgA by the colonization of Trichomonas musculis, an intestinal protozoan, ameliorated EAE susceptibility independently of T-cell subsets such as Th1 and Th17 (117). Although this paper did not address the IgA reactivity with specific bacteria, another study characterized the bacterial reactivity of IgA in the central nervous system in MS patients (42). In this study, the authors showed that IgA-producing cells were reactive with a diverse array of gut microbiota but not with self-antigens including those of human brains. These findings reveal an intimate interaction between the gut and central nervous system in MS, although the migratory mechanisms have not yet been elucidated. It also remains elusive whether the specificity of IgA to certain commensals affects the disease susceptibility.
Aging is characterized by low-grade inflammatory responses in multiple organs. Since it is also associated with sequential alterations of gut commensal microbiome, many researchers consider that microbes may be a source of inflammation during the aging process. Interestingly, not only the commensals but also IgA-coated bacteria are altered during aging. Sugahara et al. have reported that while Bifidobacteriaceae was reduced in the adult group (35 years old on average), Enterobacteriaceae and Clostridiaceae were increased in the aged group (76 years old on average) (118), implying that consistent with other reports (119), dysbiosis is progressively aggravated during aging. Notably, IgA binding to Enterobacteriaceae and Clostridiaceae was significantly reduced in the aged group despite that the total IgA amount in the stool remained unchanged (118). This may explain why these pathobionts flourished in the aged group. It has been reported that antigen uptake and antigen-specific immune responses in the intestine are impaired with aging (120–123). Although immunosenescence, i.e. age-related impairment of immune cell functions, is generally considered intrinsic to host organs, tissues and cells, diminished antigen-specific responses may also be attributable to the alterations in the gut environment as “beneficial” signals for immune homeostasis from the intestinal lumen such as SCFAs is reduced with aging (124). More studies are required to unveil the interaction between gut microbiome and immunosenescence and how this could impact the host pathophysiology.
Given the promising roles of IgA in regulating and modulating the gut commensal microbiota, it is tempting to assume that IgA can be a novel therapeutic tool for many human diseases. Here we propose four directions for future development of IgA-oriented therapies (Figure 2). First, prebiotics and probiotics can be harnessed to augment intestinal IgA, although it has various effects on the mucosal immune system (125, 126). As we discussed earlier, prebiotics such as dietary fibers that increase intestinal SCFA levels can potently augment intestinal IgA (71), in particular acetate consistently shows this effect as reported in several papers (64, 72). In addition, other prebiotics including fructo- and galacto-oligosaccharides also show robust effects on mucosal IgA response in mice and humans (127–129). This seems relatively easy for application since all we need is to consume these fibers and/or SCFAs. However, a problem would be that prebiotics cannot specify the target microbes of IgA. In addition, many papers show that the effect of dietary fiber is highly variable, probably due to the differences in individual microbiota profiles (130, 131), suggesting that it might be difficult to predict IgA responses after administration of the dietary fibers.
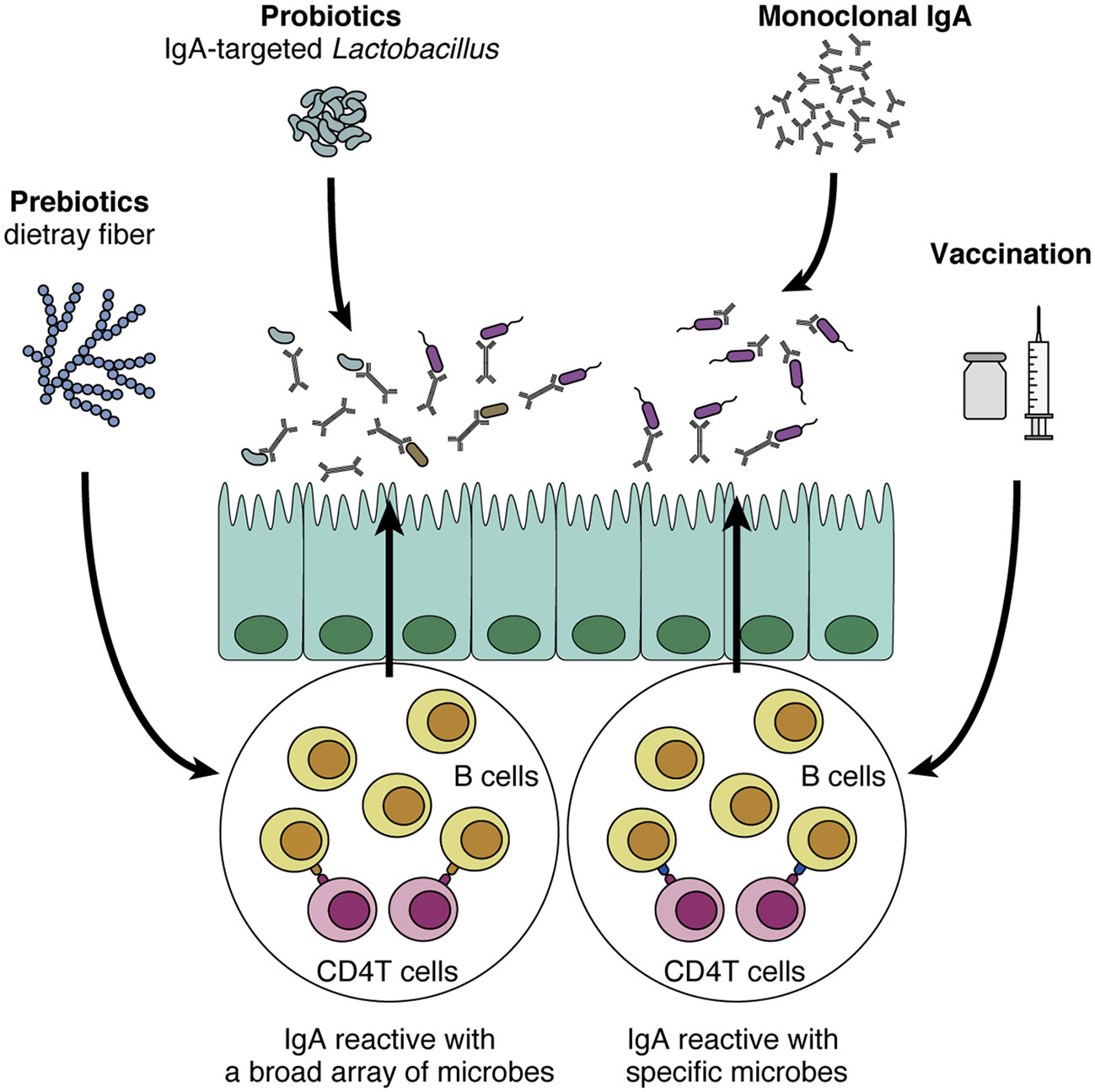
Figure 2 Potential application of IgA to human diseases. IgA-oriented therapies that modulate the gut microbiota could be utilized to control disease susceptibility. First, prebiotics (e.g., dietary fiber) and probiotics increase intestinal IgA production and modulate IgA-coating bacteria. In turn, IgA-coated Lactobacillus could be isolated and harnessed as a probiotic strain that directly interacts with the mucus layer and epithelial cells. Although these methods are relatively easy to implement, their effects on the intestinal immune system are not specific to IgA production. In addition, it is difficult to induce IgA reactive with specific microbes of interest. By contrast, vaccination and monoclonal IgA administration are more sophisticated in the sense that these methods leverage “specificity” toward certain microbes, a cardinal feature of adaptive immunity. As the efficacy of these methods has been shown in mouse models (e.g., colitis and high-fat diet-induced obesity), these approaches that target specific microbes of interest would be promising.
The effect of probiotics on IgA production and secretion has also been extensively researched. Lactic acid bacteria such as Lactobacillus and Pediococcus strains have been shown to increase IgA production by augmenting dendritic cells to enhance secretion of IL-6 and/or IL-10, important cytokines for IgA production (132, 133). Another study reveals that oral administration of a heat-killed Lactobacillus strain increases antigen-specific IgA secretion in OVA-immunized mice, presumably through a T cell-dependent manner (134). In addition, oral administration of Lactobacillus and Bifidobacteria strains to formula-fed infants increases their cow milk-specific IgA-producing cells in blood samples (135). These findings suggest that application of probiotic strains may augment IgA secretion and function to protect barrier function. From a different perspective, considering that IgA-seq analysis can distinguish a certain group of microbes, we may be able to isolate an IgA-coated microbe as a next-generation probiotic strain that possesses interesting functions in the intestine. In this regard, Sun et al. have analyzed the differences between IgA-coated and -uncoated Lactobacillus and shown that IgA-coated species can modulate gut barrier function (136).
The adaptive immune system, and more specifically T-cell help, is recognized as a cardinal component for specific IgA production, which is important for the regulation of pathobionts in the intestine. From this perspective, triggering the adaptive immune system with vaccination via either oral or systemic route is another possible strategy for IgA-oriented therapy. For example, immunization of flagellin via intraperitoneal injection is efficacious in inducing anti-flagellin IgA in mice (109). Interestingly, this systemic immunization also alters gut microbiota and several microbiota-associated host pathologies such as high-fat diet-induced obesity (109). In addition, Fujimoto et al. have shown that intramuscular injection of antigens with adjuvants could efficiently induce antigen-specific IgA responses (110). They showed that vaccination with C. ramosum-derived antigens induced anti-C. ramosum-specific IgA in the feces and reduced the abundance of this microbe in the colonic mucosa, while this vaccination did not alter the microbial community structure. Given that mucosal vaccination is believed to efficiently induce IgA especially and effectively protect against respiratory diseases such as influenza virus infection (137), the development of a mucosal vaccination strategy that regulates the microbiota may also be useful.
Monoclonal antibodies targeting specific molecules have been clinically applied to many human diseases such as cancers for many years (138). Likewise, monoclonal IgA antibodies that show potent effects on the gut microbiota could be selectively expanded and administered orally to the patients. This concept was first shown by Okai et al.; as we discussed above, they nicely revealed the efficacy of high-affinity and polyreactive IgA in regulating the gut microbiota and improving colitis after its oral administration (61). Another group has also shown a similar effect of oral monoclonal IgA administration (90). In a Salmonella infection model, oral administration of monoclonal IgA named Sal4 efficiently blocks the invasion of Salmonella into the Peyer’s patches (139). As both vaccination and oral administration are sophisticated strategies leveraging cardinal features of adaptive immunity, i.e. high affinity and specificity especially with pathobionts, it is tempting to anticipate future application to human diseases relating to gut microbiota. For further development, it also seems important to address the safety and stable delivery of vaccines/IgA in humans and to define target antigens/microbes appropriate for vaccines/IgA. Taken together, these pioneering studies point to a promising possibility that IgA can be exploited as an efficient regulator of the gut commensal microbiota.
In this review, we discuss how IgA regulates microbiota in the intestine and how this interrelationship impacts host physiology and pathology, especially in the context of human homeostasis. IgA-based strategies aiming to modulate the gut microbiota such as prebiotics, vaccination, and exogenous administration have been increasingly studied and regarded as a promising therapeutic toolbox for human diseases mediated by the gut microbiota. Nevertheless, much evidence of IgA-microbe interactions has been established based upon experimental findings, emphasizing the necessity to explore the role of IgA in humans. In this regard, a clinical application of IgA-based strategies may serve not only therapeutic purposes but also human evidence to show causality between IgA and the gut microbiota in basic science, which would further accelerate our understanding of human pathophysiology at the system level.
TT and HO conceptualized the work and co-wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by internal research funds from RIKEN.
We thank Drs. Takashi Kanaya, Eiji Miyauchi, and Yumiko Nakanishi for their helpful comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Johansen F-E, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol (2011) 4:598–602. doi: 10.1038/mi.2011.37
2. Michetti P, Mahan MJ, Slauch JM, Mekalanos JJ, Neutra MR. Monoclonal secretory immunoglobulin a protects mice against oral challenge with the invasive pathogen salmonella typhimurium. Infect Immun (1992) 60:1786–92. doi: 10.1128/iai.60.5.1786-1792.1992
3. Wijburg OLC, Uren TK, Simpfendorfer K, Johansen F-E, Brandtzaeg P, Strugnell RA. Innate secretory antibodies protect against natural salmonella typhimurium infection. J Exp Med (2006) 203:21–6. doi: 10.1084/jem.20052093
4. Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature (2017) 544:498–502. doi: 10.1038/nature22058
5. Forbes SJ, Martinelli D, Hsieh C, Ault JG, Marko M, Mannella CA, et al. Association of a protective monoclonal IgA with the O antigen of salmonella enterica serovar typhimurium impacts type 3 secretion and outer membrane integrity. Infect Immun (2012) 80:2454–63. doi: 10.1128/IAI.00018-12
6. Apter FM, Lencer WI, Finkelstein RA, Mekalanos JJ, Neutra MR. Monoclonal immunoglobulin a antibodies directed against cholera toxin prevent the toxin-induced chloride secretory response and block toxin binding to intestinal epithelial cells in vitro. Infect Immun (1993) 61:5271–8. doi: 10.1128/iai.61.12.5271-5278.1993
7. Weis AM, Round JL. Microbiota-antibody interactions that regulate gut homeostasis. Cell Host Microbe (2021) 29:334–46. doi: 10.1016/j.chom.2021.02.009
8. Huus KE, Petersen C, Finlay BB. Diversity and dynamism of IgA–microbiota interactions. Nat Rev Immunol (2021) 21:514–25. doi: 10.1038/s41577-021-00506-1
10. Cunningham-Rundles C. Physiology of IgA and IgA deficiency. J Clin Immunol (2001) 21:303–9. doi: 10.1023/A:1012241117984
11. Fadlallah J, Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med (2018) 10:eaan1217. doi: 10.1126/scitranslmed.aan1217
12. Brandtzaeg P, Karlsson G, Hansson G, Petruson B, Björkander J, Hanson LA. The clinical condition of IgA-deficient patients is related to the proportion of IgD- and IgM-producing cells in their nasal mucosa. Clin Exp Immunol (1987) 67:626–36.
13. Moll JM, Myers PN, Zhang C, Eriksen C, Wolf J, Appelberg KS, et al. Gut microbiota perturbation in IgA deficiency is influenced by IgA-autoantibody status. Gastroenterology (2021) 160:2423–34.e5. doi: 10.1053/j.gastro.2021.02.053
14. Forbes SJ, Eschmann M, Mantis NJ. Inhibition of salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin a antibody. Infect Immun (2008) 76:4137–44. doi: 10.1128/IAI.00416-08
15. Rochereau N, Drocourt D, Perouzel E, Pavot V, Redelinghuys P, Brown GD, et al. Dectin-1 is essential for reverse transcytosis of glycosylated SIgA-antigen complexes by intestinal m cells. PLoS Biol (2013) 11:e1001658. doi: 10.1371/journal.pbio.1001658
16. Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, el Aidy S, et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity (2015) 43:527–40. doi: 10.1016/j.immuni.2015.08.011
17. Wang N, Shen N, Vyse TJ, Anand V, Gunnarson I, Sturfelt G, et al. Selective IgA deficiency in autoimmune diseases. Mol Med (2011) 17:1383–96. doi: 10.2119/molmed.2011.00195
18. Ludvigsson JF, Neovius M, Hammarström L. Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol (2014) 34:444–51. doi: 10.1007/s10875-014-0009-4
19. Jorgensen GH, Gardulf A, Sigurdsson MI, Sigurdardottir ST, Thorsteinsdottir I, Gudmundsson S, et al. Clinical symptoms in adults with selective IgA deficiency: a case-control study. J Clin Immunol (2013) 33:742–7. doi: 10.1007/s10875-012-9858-x
20. Aytekin C, Tuygun N, Gokce S, Dogu F, Ikinciogullari A. Selective IgA deficiency: clinical and laboratory features of 118 children in Turkey. J Clin Immunol (2012) 32:961–6. doi: 10.1007/s10875-012-9702-3
21. Bonilla FA, Khan DA, Ballas ZK, Chinen J, Frank MM, Hsu JT, et al. Practice parameter for the diagnosis and management of primary immunodeficiency. J Allergy Clin Immunol (2015) 136:1186–205. doi: 10.1016/j.jaci.2015.04.049
22. Hammarström L, Vorechovsky I, Webster D. Selective IgA deficiency (SIgAD) and common variable immunodeficiency (CVID). Clin Exp Immunol (2000) 120:225–31. doi: 10.1046/j.1365-2249.2000.01131.x
23. Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity (2016) 44:647–58. doi: 10.1016/j.immuni.2016.02.006
24. Magri G, Comerma L, Pybus M, Sintes J, Lligé D, Segura-Garzón D, et al. Human secretory IgM emerges from plasma cells clonally related to gut memory b cells and targets highly diverse commensals. Immunity (2017) 47:118–34.e8. doi: 10.1016/j.immuni.2017.06.013
25. Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A, et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep (2019) 9:13574. doi: 10.1038/s41598-019-49923-2
26. Kabbert J, Benckert J, Rollenske T, Hitch TCA, Clavel T, Cerovic V, et al. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J Exp Med (2020) 217:e20200275. doi: 10.1084/jem.20200275
27. Sterlin D, Fadlallah J, Adams O, Fieschi C, Parizot C, Dorgham K, et al. Human IgA binds a diverse array of commensal bacteria. J Exp Med (2019) 217:e20181635. doi: 10.1084/jem.20181635
28. He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity (2007) 26:812–26. doi: 10.1016/j.immuni.2007.04.014
29. Woof JM, Kerr MA. IgA function–variations on a theme. Immunology (2004) 113:175–7. doi: 10.1111/j.1365-2567.2004.01958.x
30. Pleass RJ, Areschoug T, Lindahl G, Woof JM. Streptococcal IgA-binding proteins bind in the calpha 2-calpha 3 interdomain region and inhibit binding of IgA to human CD89. J Biol Chem (2001) 276:8197–204. doi: 10.1074/jbc.M009396200
31. Steffen U, Koeleman CA, Sokolova M v, Bang H, Kleyer A, Rech J, et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat Commun (2020) 11:120. doi: 10.1038/s41467-019-13992-8
32. Mantis NJ, Cheung MC, Chintalacharuvu KR, Rey J, Corthésy B, Neutra MR. Selective adherence of IgA to murine peyer’s patch m cells: evidence for a novel IgA receptor. J Immunol (2002) 169:1844–51. doi: 10.4049/jimmunol.169.4.1844
33. Takeuchi T, Ohno H. Reciprocal regulation of IgA and the gut microbiota: a key mutualism in the intestine. Int Immunol (2021) 33:781–6. doi: 10.1093/intimm/dxab049
34. Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Sci (1979) (2002) 298:1424–7. doi: 10.1126/science.1077336
35. Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci (2004) 101:1981–6. doi: 10.1073/pnas.0307317101
36. Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol (2011) 12:264–70. doi: 10.1038/ni.1991
37. Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y. The inhibitory receptor PD-1. Sci (1979) (2012) 821:485–9. doi: 10.1126/science.1217718
38. Nagaishi T, Watabe T, Kotake K, Kumazawa T, Aida T, Tanaka K, et al. Immunoglobulin a-specific deficiency induces spontaneous inflammation specifically in the ileum. Gut (2022) 71:487–96. doi: 10.1136/gutjnl-2020-322873
39. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell (2014) 158:1000–10. doi: 10.1016/j.cell.2014.08.006
40. Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, et al. IgA-coated e. coli enriched in crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med (2017) 9:eaaf9655. doi: 10.1126/scitranslmed.aaf9655
41. Sterlin D, Larsen M, Fadlallah J, Parizot C, Vignes M, Autaa G, et al. Perturbed Microbiota/Immune homeostasis in multiple sclerosis. Neurol(R) Neuroimmunol Neuroinflamm (2021) 8(4):e997. doi: 10.1212/NXI.0000000000000997
42. Pröbstel A-K, Zhou X, Baumann R, Wischnewski S, Kutza M, Rojas OL, et al. Gut microbiota–specific IgA<sup<+</sup< b cells traffic to the CNS in active multiple sclerosis. Sci Immunol (2020) 5:eabc7191. doi: 10.1126/sciimmunol.abc7191
43. Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med (2015) 7:276ra24. doi: 10.1126/scitranslmed.aaa4877
44. Scheithauer TPM, Davids M, Winkelmeijer M, Verdoes X, Aydin Ö, de Brauw M, et al. Compensatory intestinal antibody response against pro-inflammatory microbiota after bariatric surgery. Gut Microbes (2022) 14:2031696. doi: 10.1080/19490976.2022.2031696
45. Goedert JJ, Hua X, Bielecka A, Okayasu I, Milne GL, Jones GS, et al. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br J Cancer (2018) 118:471–9. doi: 10.1038/bjc.2017.435
46. Hase K, Kawano K, Nochi T, Pontes GS, Fukuda S, Ebisawa M, et al. Uptake through glycoprotein 2 of FimH + bacteria by m cells initiates mucosal immune response. Nature (2009) 462:226–30. doi: 10.1038/nature08529
47. Nagashima K, Sawa S, Nitta T, Tsutsumi M, Okamura T, Penninger JM, et al. Identification of subepithelial mesenchymal cells that induce IgA and diversify gut microbiota. Nat Immunol (2017) 18:675–82. doi: 10.1038/ni.3732
48. Kunimura K, Sakata D, Tun X, Uruno T, Ushijima M, Katakai T, et al. S100A4 protein is essential for the development of mature microfold cells in peyer’s patches. Cell Rep (2019) 29:2823–34.e7. doi: 10.1016/j.celrep.2019.10.091
49. Mikulic J, Longet S, Favre L, Benyacoub J, Corthesy B. Secretory IgA in complex with lactobacillus rhamnosus potentiates mucosal dendritic cell-mediated treg cell differentiation via TLR regulatory proteins, RALDH2 and secretion of IL-10 and TGF-β. Cell Mol Immunol (2017) 14:546–56. doi: 10.1038/cmi.2015.110
50. Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med (2018) 215:2019–34. doi: 10.1084/jem.20180427
51. Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe (2007) 2:328–39. doi: 10.1016/j.chom.2007.09.013
52. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin a for mucosal colonization. Sci (1979) (2018) 0926:eaaq0926. doi: 10.1126/science.aaq0926
53. Bollinger RR, Everett Ml, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin a may contribute to biofilm formation in the gut. Immunology (2003) 109:580–7. doi: 10.1046/j.1365-2567.2003.01700.x
54. Bollinger RR, Everett Ml, Wahl SD, Lee Y-H, Orndorff PE, Parker W. Secretory IgA and mucin-mediated biofilm formation by environmental strains of escherichia coli: role of type 1 pili. Mol Immunol (2006) 43:378–87. doi: 10.1016/j.molimm.2005.02.013
55. McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe (2016) 19:550–9. doi: 10.1016/j.chom.2016.02.021
56. Bansept F, Schumann-Moor K, Diard M, Hardt W-D, Slack E, Loverdo C. Enchained growth and cluster dislocation: A possible mechanism for microbiota homeostasis. PloS Comput Biol (2019) 15:e1006986. doi: 10.1371/journal.pcbi.1006986
57. Hoces D, Arnoldini M, Diard M, Loverdo C, Slack E. Growing, evolving and sticking in a flowing environment: understanding IgA interactions with bacteria in the gut. Immunology (2020) 159:52–62. doi: 10.1111/imm.13156
58. Bergqvist P, Gärdby E, Stensson A, Bemark M, Lycke NY. Gut IgA class switch recombination in the absence of CD40 does not occur in the lamina propria and is independent of germinal centers. J Immunol (2006) 177:7772–83. doi: 10.4049/jimmunol.177.11.7772
59. Bergqvist P, Stensson A, Lycke NY, Bemark M. T Cell-independent IgA class switch recombination is restricted to the GALT and occurs prior to manifest germinal center formation. J Immunol (2010) 184:3545–53. doi: 10.4049/jimmunol.0901895
60. Grasset EK, Chorny A, Casas-Recasens S, Gutzeit C, Bongers G, Thomsen I, et al. Gut T cell–independent IgA responses to commensal bacteria require engagement of the TACI receptor on b cells. Sci Immunol (2020) 5:eaat7117. doi: 10.1126/sciimmunol.aat7117
61. Okai S, Usui F, Yokota S, Hori-I Y, Hasegawa M, Nakamura T, et al. High-affinity monoclonal IgA regulates gut microbiota and prevents colitis in mice. Nat Microbiol (2016) 1:16103. doi: 10.1038/nmicrobiol.2016.103
62. Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin a. Immunity (2015) 43:541–53. doi: 10.1016/j.immuni.2015.08.007
63. Yang C, Chen-Liaw A, Spindler MP, Tortorella D, Moran TM, Cerutti A, et al. Immunoglobulin a antibody composition is sculpted to bind the self gut microbiome. Sci Immunol (2022) 7:eabg3208. doi: 10.1126/sciimmunol.abg3208
64. Takeuchi T, Miyauchi E, Kanaya T, Kato T, Nakanishi Y, Watanabe T, et al. Acetate differentially regulates IgA reactivity to commensal bacteria. Nature (2021) 595:560–4. doi: 10.1038/s41586-021-03727-5
65. Stoel M, Jiang H-Q, van Diemen CC, Bun JCAM, Dammers PM, Thurnheer MC, et al. Restricted IgA repertoire in both b-1 and b-2 cell-derived gut plasmablasts. J Immunol (2005) 174:1046–54. doi: 10.4049/jimmunol.174.2.1046
66. Zheng M, Mao K, Fang D, Li D, Lyu J, Peng D, et al. B cell residency but not T cell-independent IgA switching in the gut requires innate lymphoid cells. Proc Natl Acad Sci U S A (2021) 118(27):e2106754118. doi: 10.1073/pnas.2106754118
67. McCoy KD, Stoel M, Stettler R, Merky P, Fink K, Senn BM, et al. Polyclonal and specific antibodies mediate protective immunity against enteric helminth infection. Cell Host Microbe (2008) 4:362–73. doi: 10.1016/j.chom.2008.08.014
68. Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity (2014) 41:152–65. doi: 10.1016/j.immuni.2014.05.016
69. van de Bovenkamp FS, Derksen NIL, Ooijevaar-de Heer P, van Schie KA, Kruithof S, Berkowska MA, et al. Adaptive antibody diversification through n-linked glycosylation of the immunoglobulin variable region. Proc Natl Acad Sci U S A (2018) 115:1901–6. doi: 10.1073/pnas.1711720115
70. Day CJ, Tran EN, Semchenko EA, Tram G, Hartley-Tassell LE, Ng PSK, et al. Glycan:glycan interactions: High affinity biomolecular interactions that can mediate binding of pathogenic bacteria to host cells. Proc Natl Acad Sci U S A (2015) 112:E7266–75. doi: 10.1073/pnas.1421082112
71. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe (2016) 20:202–14. doi: 10.1016/j.chom.2016.07.001
72. Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol (2017) 10:946–56. doi: 10.1038/mi.2016.114
73. Hara S, Sasaki T, Satoh-Takayama N, Kanaya T, Kato T, Takikawa Y, et al. Dietary antigens induce germinal center responses in peyer’s patches and antigen-specific IgA production. Front Immunol (2019) 10:2432. doi: 10.3389/fimmu.2019.02432
74. Ren W, Wang K, Yin J, Chen S, Liu G, Tan B, et al. Glutamine-induced secretion of intestinal secretory immunoglobulin a: A mechanistic perspective. Front Immunol (2016) 7:503. doi: 10.3389/fimmu.2016.00503
75. Mora JR, Iwata M, Eksteen B, Song SY, Junt T, Senman B, et al. Generation of gut-homing IgA-secreting b cells by intestinal dendritic cells. Sci (1979) (2006) 314:1157–60. doi: 10.1126/science.1132742
76. Goverse G, Molenaar R, Macia L, Tan J, Erkelens MN, Konijn T, et al. Diet-derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J Immunol (2017) 198:2172–81. doi: 10.4049/jimmunol.1600165
77. Luck H, Khan S, Kim JH, Copeland JK, Revelo XS, Tsai S, et al. Gut-associated IgA+ immune cells regulate obesity-related insulin resistance. Nat Commun (2019) 10:3650. doi: 10.1038/s41467-019-11370-y
78. Proietti M, Cornacchione V, RezzonicoJost T, Romagnani A, Faliti CE, Perruzza L, et al. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in peyer’s patches to promote host-microbiota mutualism. Immunity (2014) 41:789–801. doi: 10.1016/j.immuni.2014.10.010
79. Huus KE, Bauer KC, Brown EM, Bozorgmehr T, Woodward SE, Serapio-Palacios A, et al. Commensal bacteria modulate immunoglobulin a binding in response to host nutrition. Cell Host Microbe (2020) 27:909–21.e5. doi: 10.1016/j.chom.2020.03.012
80. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the c-type lectin receptor dectin-1 influence colitis. Science (2012) 336:1314–7. doi: 10.1126/science.1221789
81. Jain U, ver Heul AM, Xiong S, Gregory MH, Demers EG, Kern JT, et al. Debaryomyces is enriched in crohn’s disease intestinal tissue and impairs healing in mice. Science (2021) 371:1154–9. doi: 10.1126/science.abd0919
82. Li X v, Leonardi I, Putzel GG, Semon A, Fiers WD, Kusakabe T, et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature (2022) 603:672–8. doi: 10.1038/s41586-022-04502-w
83. Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature (2021) 596:114–8. doi: 10.1038/s41586-021-03722-w
84. Doron I, Mesko M, Li X v, Kusakabe T, Leonardi I, Shaw DG, et al. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in crohn’s disease. Nat Microbiol (2021) 6:1493–504. doi: 10.1038/s41564-021-00983-z
85. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol (2009) 9:313–23. doi: 10.1038/nri2515
86. Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol (2011) 23:473–80. doi: 10.1016/j.coi.2011.07.010
87. Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity (2014) 40:843–54. doi: 10.1016/j.immuni.2014.05.013
88. Shapiro JM, de Zoete MR, Palm NW, Laenen Y, Bright R, Mallette M, et al. Immunoglobulin a targets a unique subset of the microbiota in inflammatory bowel disease. Cell Host Microbe (2021) 29:83–93.e3. doi: 10.1016/j.chom.2020.12.003
89. Lin R, Chen H, Shu W, Sun M, Fang L, Shi Y, et al. Clinical significance of soluble immunoglobulins a and G and their coated bacteria in feces of patients with inflammatory bowel disease. J Transl Med (2018) 16:359. doi: 10.1186/s12967-018-1723-0
90. Xiong E, Li Y, Min Q, Cui C, Liu J, Hong R, et al. MZB1 promotes the secretion of J-chain-containing dimeric IgA and is critical for the suppression of gut inflammation. Proc Natl Acad Sci U S A (2019) 116:13480–9. doi: 10.1073/pnas.1904204116
91. Sasaki K, Mori T, Hoshi N, Sasaki D, Inoue J, Shinkura R, et al. W27 IgA suppresses growth of escherichia in an in vitro model of the human intestinal microbiota. Sci Rep (2021) 11:14627. doi: 10.1038/s41598-021-94210-8
92. Rochereau N, Roblin X, Michaud E, Gayet R, Chanut B, Jospin F, et al. NOD2 deficiency increases retrograde transport of secretory IgA complexes in crohn’s disease. Nat Commun (2021) 12:261. doi: 10.1038/s41467-020-20348-0
93. Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med (2003) 9:548–53. doi: 10.1038/nm860
94. Youngman KR, Franco MA, Kuklin NA, Rott LS, Butcher EC, Greenberg HB. Correlation of tissue distribution, developmental phenotype, and intestinal homing receptor expression of antigen-specific b cells during the murine anti-rotavirus immune response. J Immunol (2002) 168:2173–81. doi: 10.4049/jimmunol.168.5.2173
95. Blutt SE, Miller AD, Salmon SL, Metzger DW, Conner ME. IgA is important for clearance and critical for protection from rotavirus infection. Mucosal Immunol (2012) 5:712–9. doi: 10.1038/mi.2012.51
96. Pollock L, Bennett A, Jere KC, Mandolo J, Dube Q, Bar-Zeev N, et al. Plasma rotavirus-specific IgA and risk of rotavirus vaccine failure in infants in Malawi. Clin Infect Dis (2022) 75:41–6. doi: 10.1093/cid/ciab895
97. Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell (2015) 160:447–60. doi: 10.1016/j.cell.2015.01.002
98. Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe (2019) 26:764–78.e5. doi: 10.1016/j.chom.2019.10.009
99. Zuo T, Lu X-J, Zhang Y, Cheung CP, Lam S, Zhang F, et al. Gut mucosal virome alterations in ulcerative colitis. Gut (2019) 68:1169–79. doi: 10.1136/gutjnl-2018-318131
100. de la Cruz Peña MJ, Gonzalez-Granado LI, Garcia-Heredia I, Carballa LM, Martinez-Garcia M. Minimal-moderate variation of human oral virome and microbiome in IgA deficiency. Sci Rep (2021) 11:14913. doi: 10.1038/s41598-021-94507-8
101. Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology (2019) 157:97–108. doi: 10.1053/j.gastro.2019.03.049
102. Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell (2020) 182:1460–73.e17. doi: 10.1016/j.cell.2020.08.007
103. Liu Y, Yuan X, Li L, Lin L, Zuo X, Cong Y, et al. Increased ileal immunoglobulin a production and immunoglobulin a-coated bacteria in diarrhea-predominant irritable bowel syndrome. Clin Transl Gastroenterol (2020) 11:e00146. doi: 10.14309/ctg.0000000000000146
104. Rengarajan S, Knoop KA, Rengarajan A, Chai JN, Grajales-Reyes JG, Samineni VK, et al. A potential role for stress-induced microbial alterations in IgA-associated irritable bowel syndrome with diarrhea. Cell Rep Med (2020) 1(7):100124. doi: 10.1016/j.xcrm.2020.100124
105. Guo X, Meng G, Liu F, Zhang Q, Liu L, Wu H, et al. Serum levels of immunoglobulins in an adult population and their relationship with type 2 diabetes. Diabetes Res Clin Pract (2016) 115:76–82. doi: 10.1016/j.diabres.2016.03.007
106. Asare-Anane H, Botchey CPK, Ofori EK, Boamah I, Crabbe S, Asamoah-Kusi K. Altered immunoglobulins (A and G) in ghanaian patients with type 2 diabetes. SAGE Open Med (2018) 6:2050312118762042. doi: 10.1177/2050312118762042
107. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes (2007) 56:1761–72. doi: 10.2337/db06-1491.P.D.C
108. Cani PD, Bibiloni R, Knauf C, Neyrinck AM, Delzenne NM. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes (2008) 57:1470–81. doi: 10.2337/db07-1403.Additional
109. Tran HQ, Ley RE, Gewirtz AT, Chassaing B. Flagellin-elicited adaptive immunity suppresses flagellated microbiota and vaccinates against chronic inflammatory diseases. Nat Commun (2019) 10:5650. doi: 10.1038/s41467-019-13538-y
110. Fujimoto K, Kawaguchi Y, Shimohigoshi M, Gotoh Y, Nakano Y, Usui Y, et al. Antigen-specific mucosal immunity regulates development of intestinal bacteria-mediated diseases. Gastroenterology (2019) 157:1530–43.e4. doi: 10.1053/j.gastro.2019.08.021
111. Becattini S, Sorbara MT, Kim SG, Littmann EL, Dong Q, Walsh G, et al. Rapid transcriptional and metabolic adaptation of intestinal microbes to host immune activation. Cell Host Microbe (2021) 29:378–93.e5. doi: 10.1016/j.chom.2021.01.003
112. Petersen C, Bell R, Klag KA, Lee S-H, Soto R, Ghazaryan A, et al. T Cell-mediated regulation of the microbiota protects against obesity. Science (2019) 365(6451):eaat9351. doi: 10.1126/science.aat9351
113. Sato J, Kanazawa A, Ikeda F, Yoshihara T, Goto H, Abe H, et al. Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care (2014) 37:2343–50. doi: 10.2337/dc13-2817
114. Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med (2011) 3:559–72. doi: 10.1002/emmm.201100159
115. Sato J, Kanazawa A, Azuma K, Ikeda F, Goto H, Komiya K, et al. Probiotic reduces bacterial translocation in type 2 diabetes mellitus: A randomised controlled study. Sci Rep (2017) 7:12115. doi: 10.1038/s41598-017-12535-9
116. Moro-Sibilot L, Blanc P, Taillardet M, Bardel E, Couillault C, Boschetti G, et al. Mouse and human liver contain immunoglobulin a-secreting cells originating from peyer’s patches and directed against intestinal antigens. Gastroenterology (2016) 151:311–23. doi: 10.1053/j.gastro.2016.04.014
117. Rojas OL, Pröbstel A-K, Porfilio EA, Wang AA, Charabati M, Sun T, et al. Recirculating intestinal IgA-producing cells regulate neuroinflammation via IL-10. Cell (2019) 176:610–24.e18. doi: 10.1016/j.cell.2018.11.035
118. Sugahara H, Okai S, Odamaki T, Wong CB, Kato K, Mitsuyama E, et al. Decreased taxon-specific IgA response in relation to the changes of gut microbiota composition in the elderly. Front Microbiol (2017) 8:1757. doi: 10.3389/fmicb.2017.01757
119. Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe (2017) 21:455–66.e4. doi: 10.1016/j.chom.2017.03.002
120. Kobayashi A, Donaldson DS, Erridge C, Kanaya T, Williams IR, Ohno H, et al. The functional maturation of m cells is dramatically reduced in the peyer’s patches of aged mice. Mucosal Immunol (2013) 6:1027–37. doi: 10.1038/mi.2012.141
121. Fujihashi K, Koga T, McGhee JR. Mucosal vaccination and immune responses in the elderly. Vaccine (2000) 18:1675–80. doi: 10.1016/s0264-410x(99)00505-8
122. Fujihashi K, Kiyono H. Mucosal immunosenescence: new developments and vaccines to control infectious diseases. Trends Immunol (2009) 30:334–43. doi: 10.1016/j.it.2009.04.004
123. Mabbott NA, Kobayashi A, Sehgal A, Bradford BM, Pattison M, Donaldson DS. Aging and the mucosal immune system in the intestine. Biogerontology (2015) 16:133–45. doi: 10.1007/s10522-014-9498-z
124. Ragonnaud E, Biragyn A. Gut microbiota as the key controllers of “healthy” aging of elderly people. Immun Ageing (2021) 18:2. doi: 10.1186/s12979-020-00213-w
125. Lomax AR, Calder PC. Prebiotics, immune function, infection and inflammation: a review of the evidence. Br J Nutr (2009) 101:633–58. doi: 10.1017/S0007114508055608
126. Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, prebiotics and immunomodulation of gut mucosal defences: homeostasis and immunopathology. Nutrients (2013) 5:1869–912. doi: 10.3390/nu5061869
127. Hosono A, Ozawa A, Kato R, Ohnishi Y, Nakanishi Y, Kimura T, et al. Dietary fructooligosaccharides induce immunoregulation of intestinal IgA secretion by murine peyer’s patch cells. Biosci Biotechnol Biochem (2003) 67:758–64. doi: 10.1271/bbb.67.758
128. Kato T, Fukuda S, Fujiwara A, Suda W, Hattori M, Kikuchi J, et al. Multiple omics uncovers host-gut microbial mutualism during prebiotic fructooligosaccharide supplementation. DNA Res (2014) 21:469–80. doi: 10.1093/dnares/dsu013
129. Vulevic J, Juric A, Tzortzis G, Gibson GR. A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr (2013) 143:324–31. doi: 10.3945/jn.112.166132
130. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, de Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab (2015) 22:971–82. doi: 10.1016/j.cmet.2015.10.001
131. Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, et al. Gut-microbiota-targeted diets modulate human immune status. Cell (2021) 184:4137–53.e14. doi: 10.1016/j.cell.2021.06.019
132. Kotani Y, Kunisawa J, Suzuki Y, Sato I, Saito T, Toba M, et al. Role of lactobacillus pentosus strain b240 and the toll-like receptor 2 axis in peyer’s patch dendritic cell-mediated immunoglobulin a enhancement. PloS One (2014) 9:e91857. doi: 10.1371/journal.pone.0091857
133. Kawashima T, Ikari N, Kouchi T, Kowatari Y, Kubota Y, Shimojo N, et al. The molecular mechanism for activating IgA production by pediococcus acidilactici K15 and the clinical impact in a randomized trial. Sci Rep (2018) 8:5065. doi: 10.1038/s41598-018-23404-4
134. Arai S, Iwabuchi N, Takahashi S, Xiao J-Z, Abe F, Hachimura S. Orally administered heat-killed lactobacillus paracasei MCC1849 enhances antigen-specific IgA secretion and induces follicular helper T cells in mice. PLoS One (2018) 13:e0199018. doi: 10.1371/journal.pone.0199018
135. Rautava S, Arvilommi H, Isolauri E. Specific probiotics in enhancing maturation of IgA responses in formula-fed infants. Pediatr Res (2006) 60:221–4. doi: 10.1203/01.pdr.0000228317.72933.db
136. Sun J, Qi C, Zhu H, Zhou Q, Xiao H, Le G, et al. IgA-targeted lactobacillus jensenii modulated gut barrier and microbiota in high-fat diet-fed mice. Front Microbiol (2019) 10:1179. doi: 10.3389/fmicb.2019.01179
137. Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat Rev Immunol (2012) 12:592–605. doi: 10.1038/nri3251
138. Melero I, Hervas-Stubbs S, Glennie M, Pardoll DM, Chen L. Immunostimulatory monoclonal antibodies for cancer therapy. Nat Rev Cancer (2007) 7:95–106. doi: 10.1038/nrc2051
Keywords: immunoglobulin A, gut microbiota, microbial metabolite, human immunology, adaptive immunity
Citation: Takeuchi T and Ohno H (2022) IgA in human health and diseases: Potential regulator of commensal microbiota. Front. Immunol. 13:1024330. doi: 10.3389/fimmu.2022.1024330
Received: 21 August 2022; Accepted: 27 October 2022;
Published: 10 November 2022.
Edited by:
Marcela Pasetti, University of Maryland, United StatesReviewed by:
Nicolaas Adrianus Bos, University Medical Center Groningen, NetherlandsCopyright © 2022 Takeuchi and Ohno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroshi Ohno, aGlyb3NoaS5vaG5vQHJpa2VuLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.