
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 28 October 2022
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1014985
This article is part of the Research Topic Bi-directional communication between the microbiome and the innate immune system View all 7 articles
Poria cocos polysaccharides (PS) have been used as Chinese traditional medicine with various pharmacological effects, including antiviral, anti-oxidative, and immunomodulatory activities. Herein Bombyx mori silkworm was used as a model animal to evaluate the immunomodulatory effects of PS via detecting the changes of innate immune parameters and explore the underlying molecular mechanism of the immunoregulatory effect of PS using Illumina HiSeq Xten platform. The results presented here demonstrated that a hemocoel injection of PS significantly enhanced the cellular immunity of silkworm, including hemocyte phagocytosis, microaggregation, and spreading ability. A total of 335 differentially expressed genes (DEGs) were screened, including 214 upregulated genes and 121 downregulated genes by differential expression analysis. Gene annotation and enrichment analyses showed that many DEGs related to immune signal recognition, detoxification, proPO activation, carbohydrate metabolism, and lipid metabolism were significantly upregulated in the treatment group. The Kyoto Encyclopedia of Genes and Genomes-based Gene Set Enrichment Analysis also revealed that the more highly expressed gene sets in the PS treatment silkworm were mainly related to immune signal transduction pathways and energy metabolism. In addition, the activity of four enzymes related to immunity and energy metabolism—including phenoloxidase, glucose-6-phosphate dehydrogenase, hexokinase, and fatty acid synthetase—were all significantly increased in the larvae injected with PS. We performed qRT-PCR to examine the expression profile of immune and metabolic-related genes, which further verified the reliability of our transcriptome data and suggested that PS can regulate the immunity of silkworm by enhancing the cellular immunity and modulating the expression levels of genes related to immune responses and physiological metabolism. These findings will lay a scientific foundation for the use of PS as an immunomodulator in disease prevention in human beings or animals.
Bombyx mori, belonging to the silkworm moth family of Lepidoptera, is an oligophagous and holometabolous insect that feeds on mulberry leaves, and it is an ideal animal model with a short growth cycle. Compared with many mammal models (e.g., mouse, rat and rabbit), silkworm has many unique advantages, such as low breeding cost, fewer ethical problems, rich genetic mutation resources, clear genetic background, and suitability for large-scale breeding.
Owing to the above-mentioned advantages of silkworm as well as its similarities with mammals in basic life system, energy metabolism, and inheritance patterns, silkworm has gained increasing attention in many fields, including pharmaceutical research (1), human disease investigation (2), and toxicological research (3–5)—for example, researchers evaluated drug toxicity using silkworm as an animal model and found that the silkworm eliminated the chemicals through a common metabolic pathway with the mammals (6). By using the silkworm–baculovirus infection mode, Orihara et al. screened antiviral agents that are effective for treating humans infected with DNA viruses (7). Silkworm models are also widely used for the evaluation of anti-diabetic drugs for both type I diabetes and type II diabetes (8). Our previous report showed that silkworm possessed an immune-priming response, which was similar to the adapted immunity of vertebrates, and the specificity of the priming response was mediated by hemocyte phagocytosis (9). We further investigated the molecular mechanism of specific immune priming in silkworm using transcriptome sequencing and found that many immune-related genes, such as pattern recognition receptors, antimicrobial peptides (AMPs), and detoxification genes, were involved in the enhanced immune response (10). More and more researchers use silkworm as an alternative model of experimental animals, as it is an ideal experimental model in the fields of medicine, genetics, immunology, and toxicological research.
Fungal polysaccharide refers to a high-molecular-weight polymer with bioactive functions and connected by 10 or more monosaccharides in the form of glycosidic bonds. They are not only the main components of cells and tissues but also participate in energy storage and a variety of life activities. Fungal polysaccharides are known as “biological response modifiers” (BRMs). They have attracted much attention due to their biological activities, such as antitumor, antioxidant, anti-inflammatory, antibacterial, and immunoregulatory activities (11–15).
Poria cocos is a saprophytic fungus belonging to the family of Polyporaceae. Its sclerotium, called fu-ling in China, has been used as a Chinese traditional medicine for more than 2000 years. Many studies have shown that P. cocos polysaccharides (PS) possess excellent immunomodulatory effects—for example, a polysaccharide-rich fraction derived from P. cocos could improve the adaptive immune cell activation and modulate the specific immune response of mice (16). PS could exert immunomodulatory effects by activating the Ca2+/PKC/p38/NF-κB signaling pathway in macrophages and also serve to enhance the immune activity against lung cancer via TLR4/TRAF6/NF-κB signaling (17, 18). Although the past results suggested that PS displayed an immunomodulatory activity, the specific molecular mechanisms underlying how PS regulates the host immunity and metabolism system remain to be fully explored.
Insects possess effective immune systems composed of both humoral (secretion of AMPs and activation of proteinase cascades) and cellular (hemocyte phagocytosis, encapsulation and microaggregation) components (19, 20). The blood cavity of insects contains a large amount of fat body, which is the central tissue of metabolic activities such as insect growth, development, metamorphosis, and reproduction. The main functions of the fat body are storing nutrition, detoxification, and providing various biosynthetic metabolites for insect life activities, which are similar to those of the vertebrate liver. Most AMPs are also produced mainly in the fat body.
In this study, we used transcriptome sequencing technology to explore the immune signaling pathways, key metabolic pathways, and genes in the fat body of silkworm affected by PS. Many immune and metabolic-related differentially expressed genes (DEGs) were selected for further analysis by qPCR. Furthermore, we systematically analyzed the changes of innate immune parameters, such as phagocytosis, hemocyte microaggregation, and spreading of silkworm after inoculation with PS. To our knowledge, this is the first study on the molecular mechanisms of immunoregulation as exerted by PS using silkworm as an animal model. The results will lay a scientific foundation for the use of PS as an immunomodulator in disease prevention in human beings or animals.
B. mori strain p50 was reared on mulberry leaves at 25°C with a 12-h light and 12-h dark cycle. Then, 1-day-old fifth-instar larvae were selected for the following experiments. PS (purity >90%) was purchased from Sangon Biotech (Shanghai) Co., Ltd. For the immune parameters and survival assays, four groups of larvae (50 larvae per group) were treated as follows: The first group of larvae was injected with 10 μl of phosphate-buffered saline (PBS: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 in 1,000 ml distilled water, pH 7.2) as control. The other three groups were injected with 10 μl of PBS containing 0.1, 0.2, and 0.4 µg PS, respectively. The preliminary experiments showed that PS had no lethal effect on the silkworm larvae in the range of the concentrations selected. At 24 h after injection, 30 larvae of each group were selected and injected with the lethal doses of (1 × 105 cells/larva) Bacillus thuringiensis (Bt) for the survival assay. The hemolymph was collected from the remaining larvae of each group for microaggregate counts, phagocytosis, and hemocyte spreading assays. For all the treatments, three independent trials were performed.
In detail, 50 µl of hemolymph was diluted into five volumes of ice-cold anticoagulant solution (93 mM NaCl, 100 mM glucose, 30 mM trisodium citrate, 26 mM citric acid, 10 mM Na2EDTA, and a few crystals of phenylthiourea, pH 4.6.) and centrifuged at 800 g for 5 min. The pellet was resuspended in 200 µl of Grace’s tissue culture medium (GIM). After incubation at 28°C for 1 h in a 24-well plate, the hemocyte spreading behavior was observed under a phase-contrast microscope by counting the hemocyte-exhibiting cytoplasmic extension along with pseudopodial growth. Hemocyte spreading was expressed as the percentage of spreading plasmatocytes to the total number of plasmatocytes.
Moreover, 20 µl of hemolymph was diluted into 80 µl PBS with a few phenylthioureas and added to a Neubauer hemocytometer immediately. The number of microaggregates was determined directly by phase-contrast optics. Only hemocyte clusters containing five cells or more were considered as microaggregates. The number of microaggregates was normalized to microaggregates/µl hemolymph.
To prepare fluorescein isothiocyanate (FITC-labeled bacteria, heat-killed Escherichia coli was resuspended in carbonate buffer (0.2 M Na2CO3 and 0.2 M NaHCO3, pH 9.4) containing FITC (0.1 mg/ml) and then incubated in a rotary mixer (200 rpm) for 30 min in the dark at 28°C. After removing the free FITC, the suspension was diluted with GIM to a concentration of 109 cells per milliliter. An in vitro phagocytosis assay was conducted as described in our previous study (9). The phagocytosed FITC-labeled E. coli was detected by fluorescence microscopy, and the average fluorescence intensity of each phagocytic hemocyte was quantified by Image J software.
Two groups (45 larvae per group) of B. mori larvae were selected for the enzyme activity assay. One group was injected with PS (0.2 µg/larva) solution, and the other was injected with PBS and served as the control. At 24 h after injection, the fat body of each larva was isolated and ground with liquid nitrogen using a mortar. Subsequently, 1 ml of Tris-HCL (10 mmol/L, pH 7.0) was added, and the mixture was centrifuged at 10,000 rev/min for 5 min at 4°C. The supernatant was used for enzyme activity assay.
For the PO activity assay, 1.4 ml of PBS was added to 1.50 ml of L-DOPA (0.05 mol/L) and preincubated at 30°C for 30 min. Then, 0.1 ml of enzyme solution was added, and phenoloxidase (PO) activity was measured using a spectrophotometer at 490-nm wavelength. One unit of PO was described as 0.01 absorbance increase at 490 nm per minute. Glucose-6-phosphate dehydrogenase (G6PD), hexokinase (HK), and fatty acid synthetase (FAS) activities were determined using corresponding detection kits (Jiancheng, China). Total protein concentration was determined using the Bradford protein assay, with bovine serum albumin as the standard (Sigma-Aldrich, USA). Each milligram of protein that oxidizes 1 µmol of NADPH per minute was expressed as one unit of FAS activity. One unit of G6PD and HK activities were defined as each milligram of tissue protein that generates 1 nmol of NADPH per minute. Three biological replications were used to determine the average of enzyme activity in all experiments.
A total of 60 B. mori larvae were selected and divided into two groups—one group was injected with 10 μl of PBS containing 0.2 µg PS, and the other group was injected with PBS only as a control. All the larvae were reared on mulberry leaves. After having been fed for 24 h, the larvae were anesthetized on ice, and fat bodies were collected and frozen in liquid nitrogen quickly. Total RNA was extracted from the fat body using TRIzol® Reagent according to the manufacturer’s instructions (Invitrogen), and genomic DNA was removed using DNase I (TaKara). Then, RNA quality was determined by 2100 Bioanalyser (Agilent). Only high-quality RNA sample was used to construct a sequencing library.
The RNA-seq transcriptome library was constructed according to the instructions of TruSeq™ RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Firstly, poly-A tail mRNA was enriched from 1 µg of total RNA by magnetic beads with oligo (DT) and then fragmented by a fragmentation buffer. Secondly, double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA, USA) with random hexamer primers. Then, the synthesized cDNA was subjected to end-repair, phosphorylation, and “A” base addition according to Illumina’s library construction protocol. The libraries were size-selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose, followed by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles. After having been quantified by TBS380, the paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq xten sequencer (2 × 150-bp read length).
To identify differentially expressed genes (DEGs) between the PS treatment and the control group, the expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FPKM) method. RSEM was used to quantify gene abundances. The differential expression level was analyzed by R statistical package software EdgeR. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) (21).
All differentially expressed genes, whether significant or not, were used for Gene Set Enrichment Analysis (GSEA) analysis (http://software.broadinstitute.org/gsea/index.jsp). GSEA was carried out with default algorithm as 1,000 permutations, with a maximum term size of 500 and a minimum term size of 15. The enrichment score (ES) for each gene set is calculated using the entire ranked list, which reflects how the genes for each set are distributed in the ranked list. The ES and normalized ES (NES) were determined for each gene set. The significant enrichment of a gene set was assigned based on nominal p-value <0.05 and false discovery rate <0.25.
All primers of the candidate gene were designed using Primer 5.0 software, and the sequences are provided in Supplementary Table S1. One microgram of total RNA was used for the cDNA synthesis using HiScript Q RT SuperMix (Vazyme). qRT-PCR analysis was performed using ChamQ SYBR Color qPCR Master Mix (Vazyme) and carried out on a ABI7300 apparatus (Applied Biosystems, UK) with the following program: initial denaturation at 95°C for 5 min, followed by 40 cycles of 5 s at 95°C, 30 s at 58°C, and 40 s at 72°C. Each sample was run in triplicate, and the average threshold cycle (Ct) was calculated. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used to normalize the expression levels, and the relative gene expression levels of the target genes were calculated using the 2-ΔΔCT method (22).
In order to assay the immune regulation of PS on the silkworm, we inoculated the larvae with different doses of PS and examined the changes of innate immune parameters. We found that there was no significant change of microaggregate number in the hemolymph of larvae injected with a low dose (0.1 μg/larva) of PS. However, compared with the control, the number of microaggregates has significantly increased in the middle (0.2 μg/larva) and high (0.4 μg/larva) doses of PS treatment group (Figure 1A). Meanwhile, a stronger activation effect of phagocytic and hemocyte spreading abilities could be observed in the low, middle, and high doses of PS treatment groups because they showed significantly higher phagocytic index and hemocyte spreading ability (Figures 1B, C).
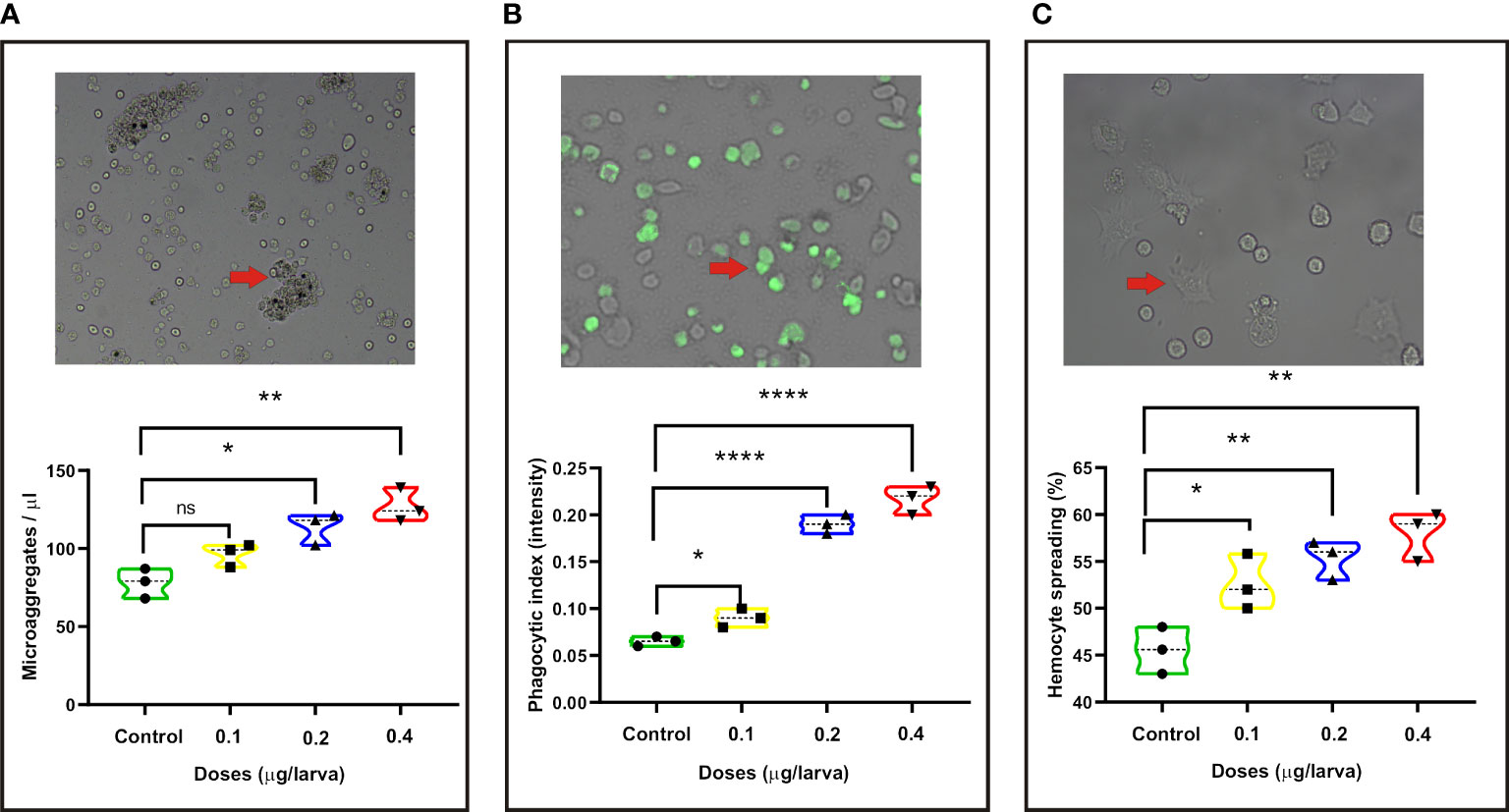
Figure 1 Changes in the following innate immune parameters of silkworm at 24 h after injection with different doses of Poria cocos polysaccharides (PS). (A) Microaggregate numbers. (B) Phagocytic index. (C) Hemocyte spreading. The average values calculated from three independent experiments and the statistical differences between the treatment and the control groups are displayed with an asterisk (*). *P < 0.05, **P < 0.01, ****P ≤ 0.0001.
The larvae primed with high (log-rank Mantel–Cox test, χ2: 14.94, p < 0.001) and middle (log-rank Mantel–Cox test, χ2: 13.77, p < 0.01) doses of PS showed significantly higher survival rates than that of the PBS control when they were infected with the lethal dose of Bt at 24 h after priming, while there was no significant difference between the low dose of PS priming group and the control group (Figure 2).
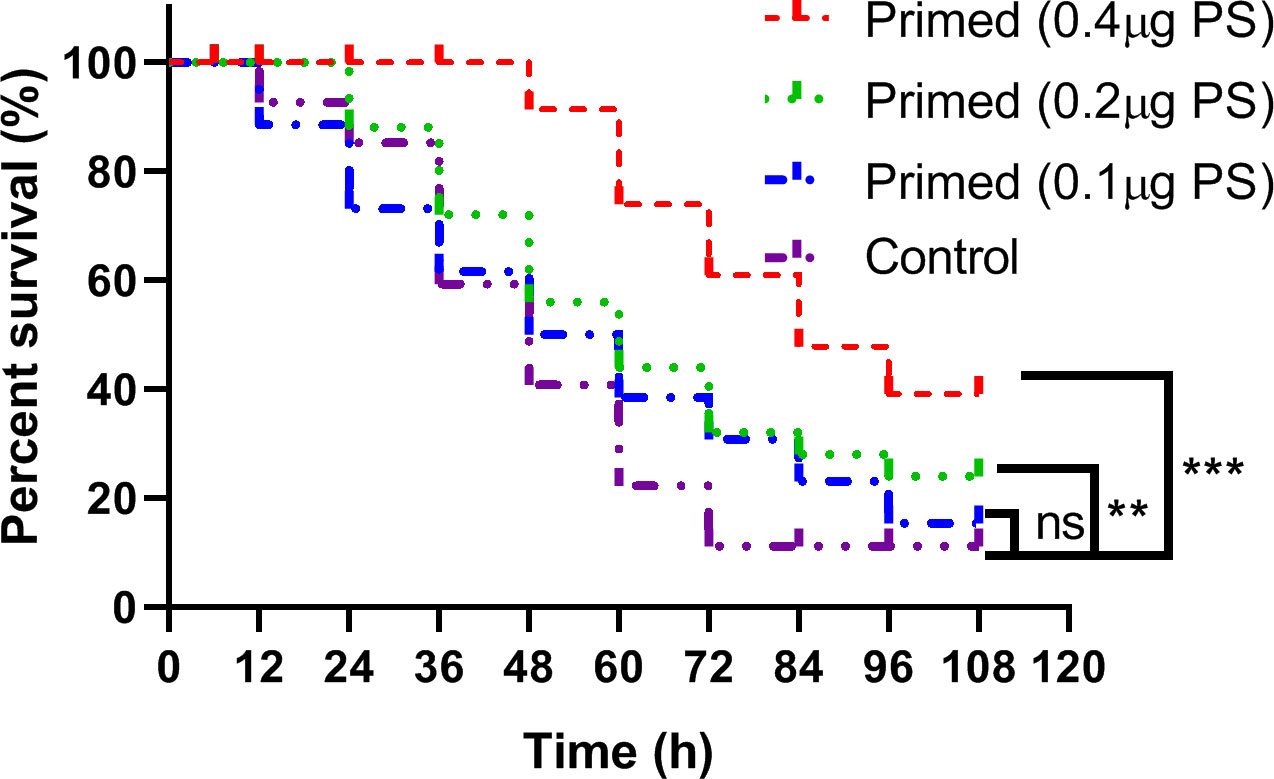
Figure 2 Kaplan–Meier curve analysis of Bombyx mori larvae immune-primed with different doses of Poria cocos polysaccharides followed by infection with a lethal dose of Bacillus thuringiensis at 24 h post-priming. ** , *** and ns indicate significance at P≤0.01, P≤0.001 and non-significant, respectively.
In total, 145,514,334 and 150,376,202 raw reads were gained, respectively, from the PBS (control_1, control_2, and control_3) and PS-treated (PS_1, PS_2, and PS_3) samples using an Illumina HiSeq NovaSeq 6000 platform. After quality filtering and trimming, 45,850,194 to 52,154,744 reads were obtained for six samples with high Q20 and Q30 quality scores, resulting in 87.75–89.73% of the clean reads that were mapped to the B. mori genome (Supplementary Table S2). From these six libraries, 79.54, 77.52, 76.26, 77.15, 76.71, and 78.47% reads were mapped to the CDS regions of the reference genome (Supplementary Figure S1A), and there was no difference in the gene expression levels among all samples (Supplementary Figure S1B). All reads were distributed to varying degrees on all chromosomes. Among them, the most distributed chromosome is BMSK_chr25 (Supplementary Figure S1C). Therefore, the data obtained from this sequencing were reliable and could be used for subsequent analysis. The original data has been uploaded to NCBI and can be found under accession numbers SRR18297772, SRR18297773, SRR18297774, SRR18297778, SRR18297779, and SRR18297780.
The volcano plots showed that 214 genes were upregulated and that 121 genes were downregulated between the control and PS-treated groups (Figure 3A). All DEGs were presented as FPKM hierarchical clustering heat map, through which we can observe the gene expression patterns between the controls and the PS treatment group (Figure 3B). To obtain more insights into the dynamic changes of DEGs, we performed a cluster analysis of 335 DEGs according to their normalized expression levels. All DEGs were grouped into 10 major clusters. DEGs that belonged to subcluster_1 (40 genes), subcluster_2 (34 genes), subcluster_4 (seven genes), subcluster_5 (29 genes), subcluster_6 (91 genes), and subcluster_9 (13 genes) were significantly induced at 24 h when the B. mori larvae were injected with PS. In contrast, DEGs that belonged to subcluster_3 (14 genes), subcluster_7 (24 genes), subcluster_8 (76 genes), and subcluster_10 (7 genes) showed low expression levels in the treatment group (Figure 3C).
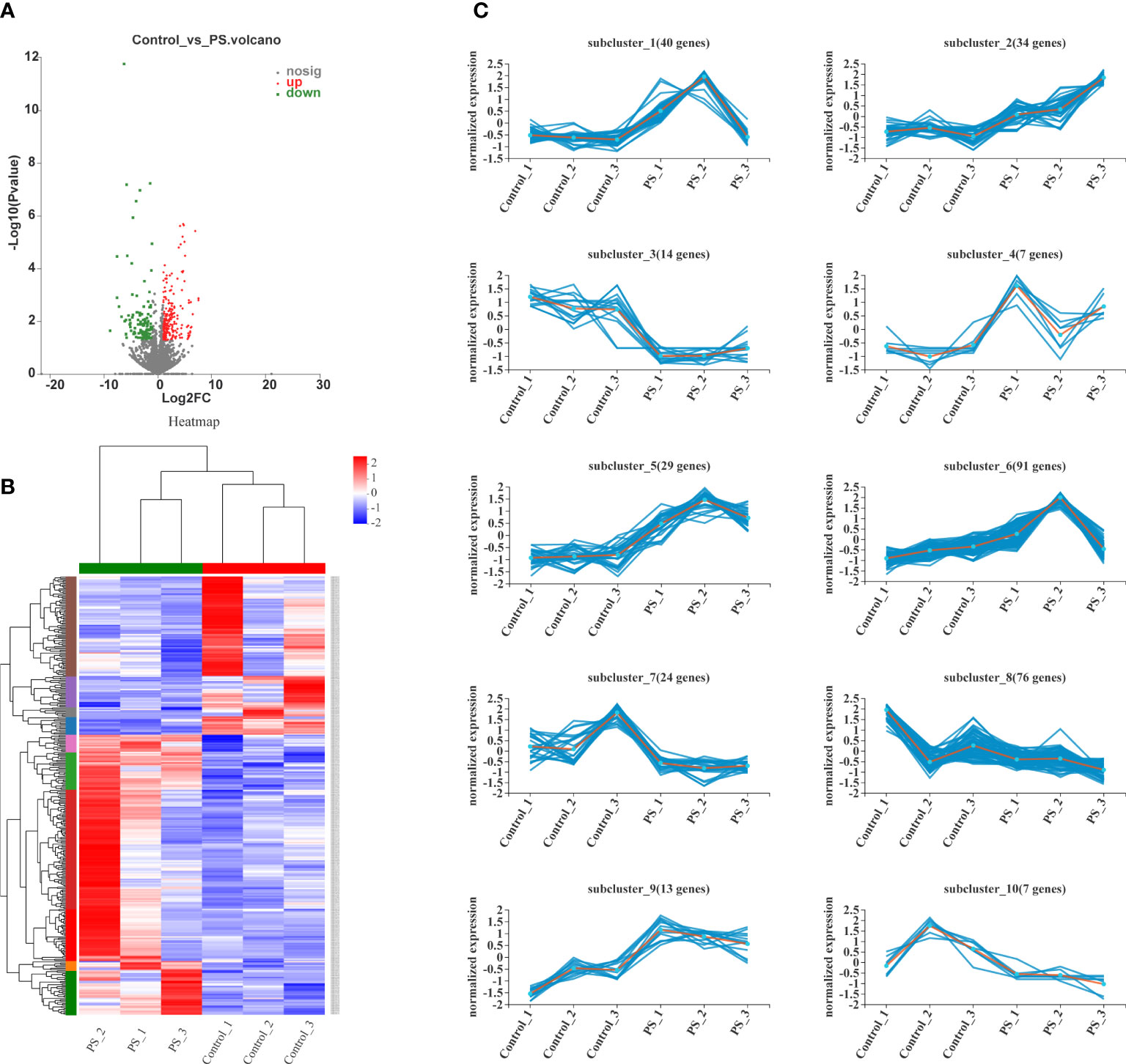
Figure 3 Analysis of differentially expressed genes (DEGs). (A) Volcano plots of the DEGs in control vs. polysaccharides. The red, blue, and gray dots indicate significantly upregulated, significantly downregulated, and non-significantly expressed genes, respectively. (B) Heat map of DEG expression profiles in all samples. The colors ranging from blue to red indicate the genes expressed from low to high levels. (C) Ten different expression patterns analyzed of 335 DEGs.
The functions of DEGs were classified by using the COG, GO, and KEGG databases. In total, 335 DEGs were divided into 19 COG classification categories. Among the 19 COG classifications, “function unknown” represents the largest group, and “posttranslational modification, protein turnover, chaperones”, “lipid transport and metabolism”, and “amino acid transport and metabolism” are next to it (Figure 4A). In terms of GO annotation, all DEGs were annotated to 20 sub-categories and classified into three GO terms: biological processes, cellular components, and molecular function. The top seven subcategories were cell part (135, 40.29%), binding (133, 39.70%), catalytic activity (131, 39.10%), cellular process (122, 36.42%), membrane part (122, 33.73%), biological regulation (98, 29.25%), and metabolic process (94, 28.06%). In addition, many DEGs were annotated to respond to stimulus (40, 11.94%) and immune system process (7, 2.09%) (Figure 4B).
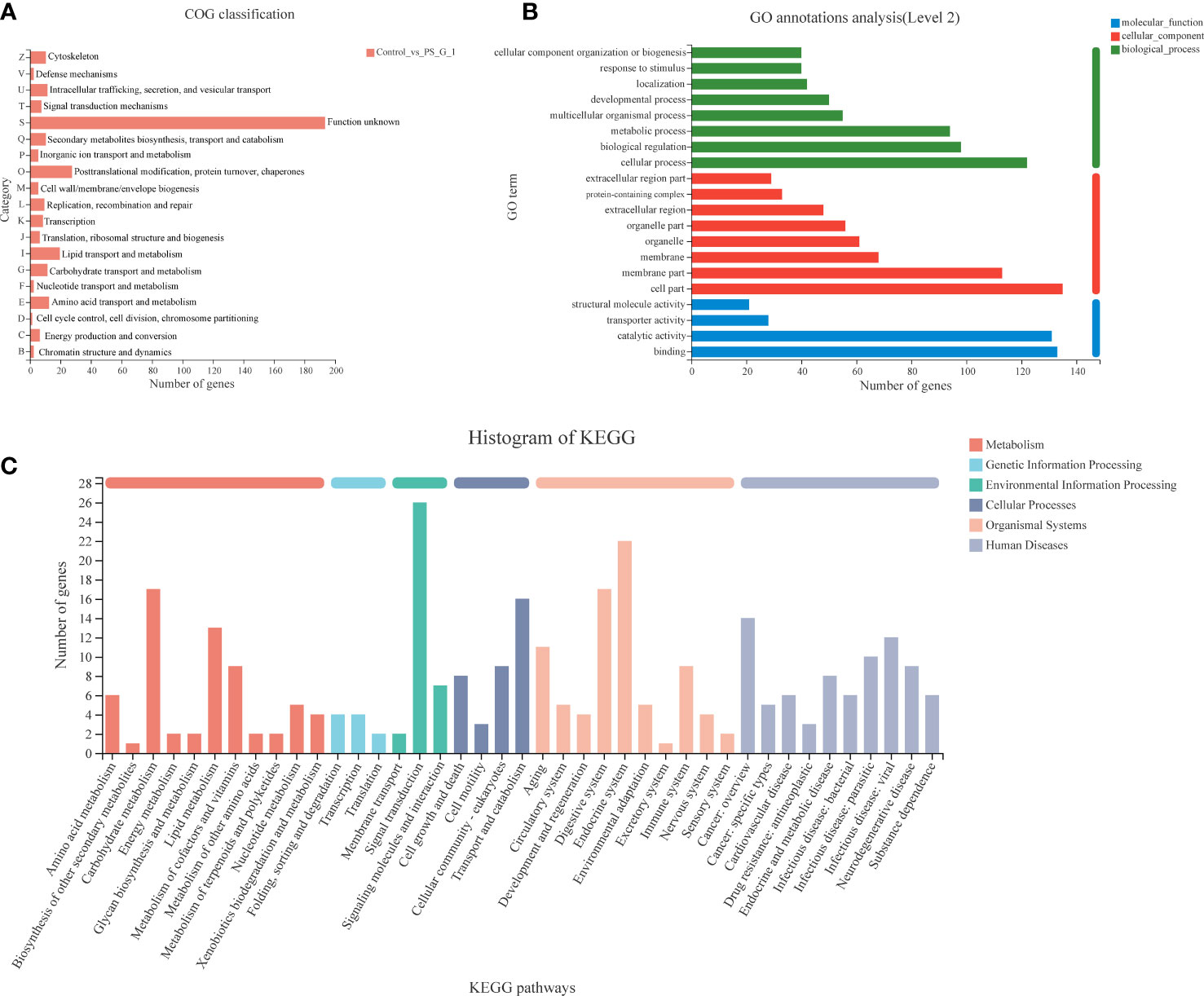
Figure 4 Functional classification of differentially expressed genes (DEGs) between the control and Poria cocos polysaccharides treatment groups. (A) COG, (B) GO, and (C) Kyoto Encyclopedia of Genes and Genomes annotation of DEGs.
We also annotated all the DEGs based on KEGG database. A total of 335 DEGs were clustered in 41 KEGG pathways (level 2), which belonged to six categories, including metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, and human diseases. Among the 41 KEGG pathways, signal transduction (26 DEGs) was attributed with the highest number of DEGs, and the next one was the endocrine system (22 DEGs). Notably, we found that many DEGs were clustered on carbohydrate metabolism (including galactose metabolism: map00052, fructose and mannose metabolism: map00051, pentose and glucuronate interconversions: map00040, TCA cycle: map00020, pyruvate metabolism: map00620, glycolysis/gluconeogenesis: map00010, etc.), lipid metabolism (including fatty acid biosynthesis: map00061, glycerolipid metabolism: map00561, steroid hormone biosynthesis: map00140, etc.), and immune system (including hematopoietic cell lineage: map04640, Toll and Imd signaling pathway: map04624, Fc gamma R-mediated phagocytosis: map04666, NOD-like receptor signaling pathway: map04621, etc.) (Figure 4C).
The top 20 enrichment GO terms are shown in Figure 5A. Among the DEGs, the most enriched GO terms were multicellular organismal process (GO: 0032501), extracellular region (GO: 0005576), extracellular region part (GO: 0044421), extracellular space (GO: 0005615), and serine-type endopeptidase activity (GO: 0004252). Moreover, we performed a chord plot to analyze the top 10 enrichment GO terms, and the results showed that most of the GO terms with high enrichment were associated with metabolism, such as carbohydrate metabolic process (GO: 0005975), drug metabolic process (GO: 0017144), small molecule metabolic process (GO: 0044281), and terpenoid metabolic process (GO: 0006721) (Figure 5B).
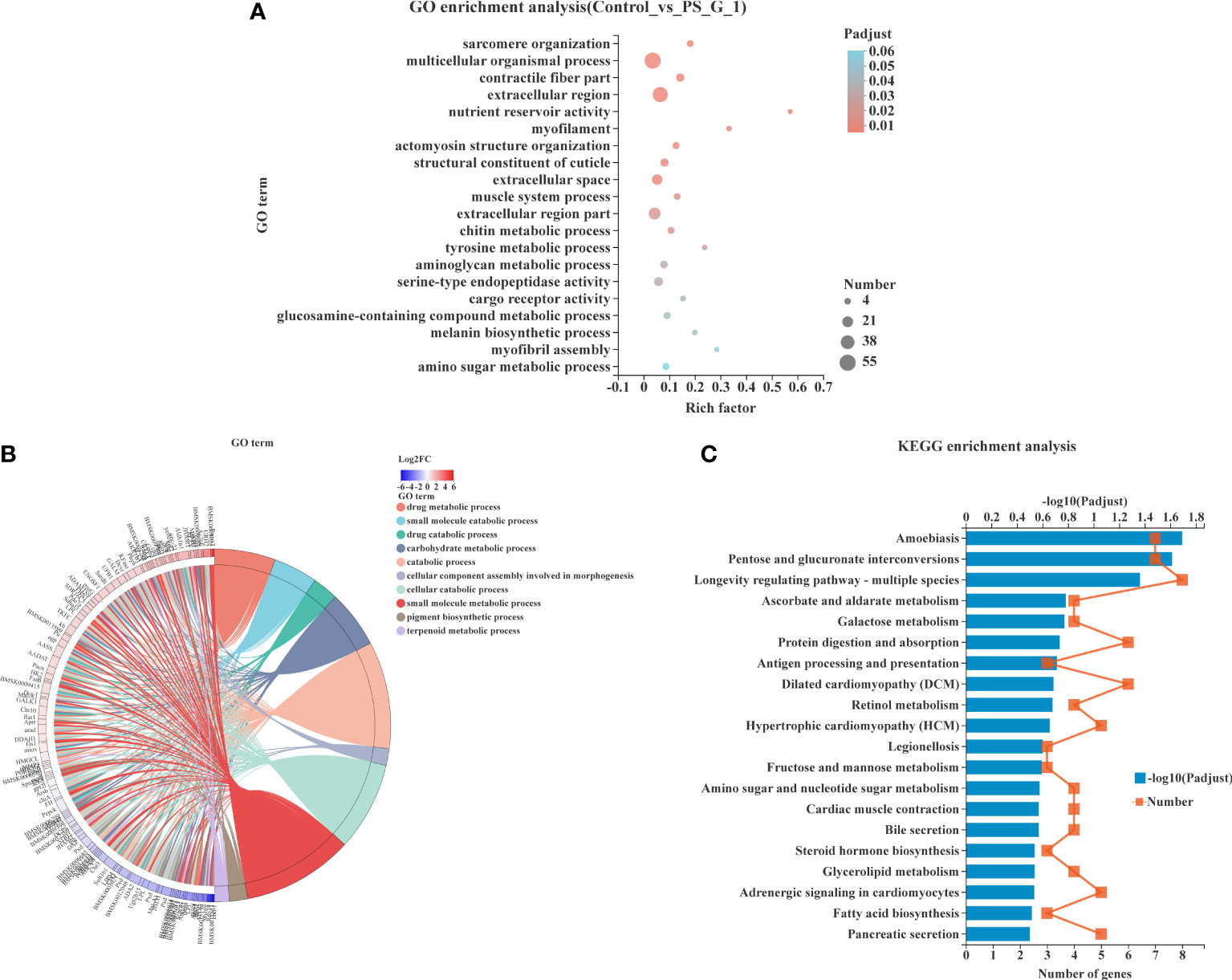
Figure 5 Functional enrichment analysis of differentially expressed genes (DEGs). (A) Gene Ontology (GO) enrichment of DEGs in silkworm exposed to Poria cocos polysaccharides (PS). (B) Chord plot indicating the relationship of the most involved genes and the top 10 terms of GO. (C) Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of DEGs in silkworm exposed to PS.
The KEGG pathway enrichment analysis was also conducted to explore the possible functions of all DEGs. The top 20 enriched KEGG pathways are shown in Figure 5C. We found that amoebiasis (map05146), pentose and glucuronate interconversions (map00040), and longevity-regulating pathway—multiple species (map04213) were significantly enriched pathways with the largest number of DEGs being enriched.
We performed GSEA using the KEGG-based list to enrich the gene sets, and the GSEA enrichment plots of representative pathways are presented in Supplementary Figure S2. The ES, NES, p-value, adjusted p-value and leading edges are detailed in Table 1. The results revealed that many more highly expressed gene sets in the PS-treated group were involved in immune responses and energy metabolism. Among these gene sets, three gene sets related to immune responses, including mitogen-activated protein kinase (MAPK) signaling pathway, PI3K-Akt signaling pathway, and peroxisome, were identified as significantly enriched.
On the other hand, pathways related to glucose and lipid metabolisms, such as fatty acid biosynthesis, glycolysis/gluconeogenesis, galactose metabolism, citrate cycle (TCA cycle), fructose and mannose metabolism, and glycerolipid metabolism, were also significantly enriched and showed significant upregulation of gene sets in the PS-treated group. The results presented here showed that many of the pathways related to the immune system and energy metabolism detected by GSEA-based KEGG were overlapped with that based on DEG.
As shown in Table 2, a big number of genes involved in innate immunity were significantly upregulated in the fat body of silkworm after treatment with PS, such as pattern recognition receptor genes, genes involved in prophenoloxidase-activating system, AMPs genes, and transcription factors genes. Moreover, the expression levels of genes involved in detoxification, including Hsp68, CYP9E2, CYP6B2, Cyp4g15, sod1, and Csp, were also significantly upregulated.
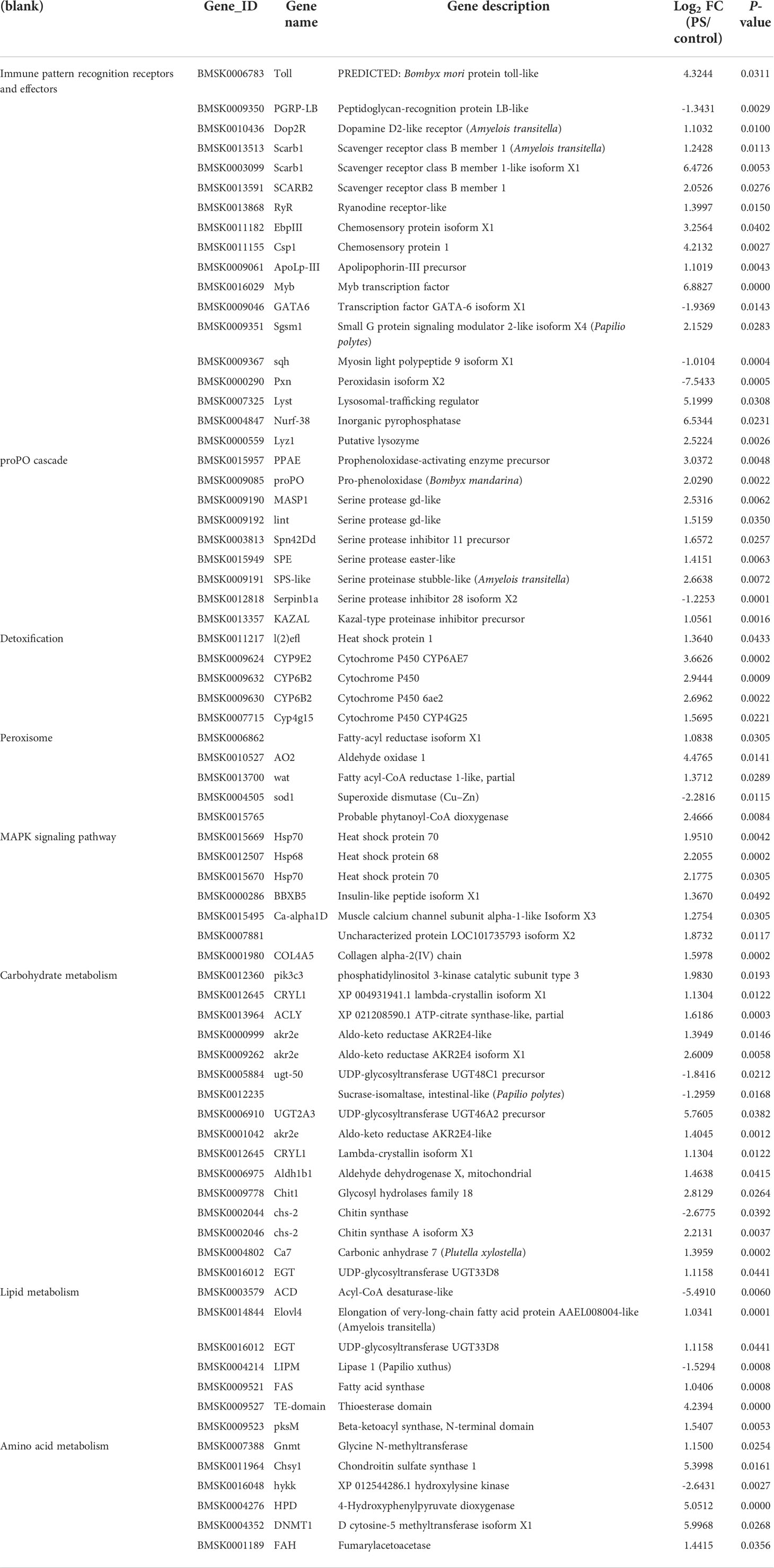
Table 2 Differentially expressed genes related to immunity, detoxification, and metabolic processes.
In addition, we also analyzed the DEGs implicated in energy metabolism. Significant upregulation occurred in many genes encoding enzymes involved in carbohydrate metabolism, lipid metabolism, and amino acid metabolism.
The expression levels of genes involved in Imd and Toll signaling pathways including Toll and Imd receptors, AMP genes, Myd88, and spaetzle were detected in the fat body of silkworm after PS inoculation. Upon treatment with PS, Toll expression increased at 6 h and reached the highest level at 24 h, and this was significantly higher than that in the control group (p < 0.01); the difference lasted until 48 h (Figure 6A). The Imd expression reached the highest level at 6 h (p < 0.01) and subsequently decreased from 12 to 48 h (Figure 6B). Gloverin1 had a similar expression pattern with Imd, while it reached a peak at 12 h post-injection (Figure 6C). The level of the Ceceropin D transcript increased from 6 to 12 h after PS injection and decreased dramatically to a minimum value at 24 h, while it returned to a higher level at 48 h (Figure 6D). Both the Myd88 and spaetzle transcripts reached their peaks at 12 h (p < 0.01) after PS injection (Figures 6E, F).
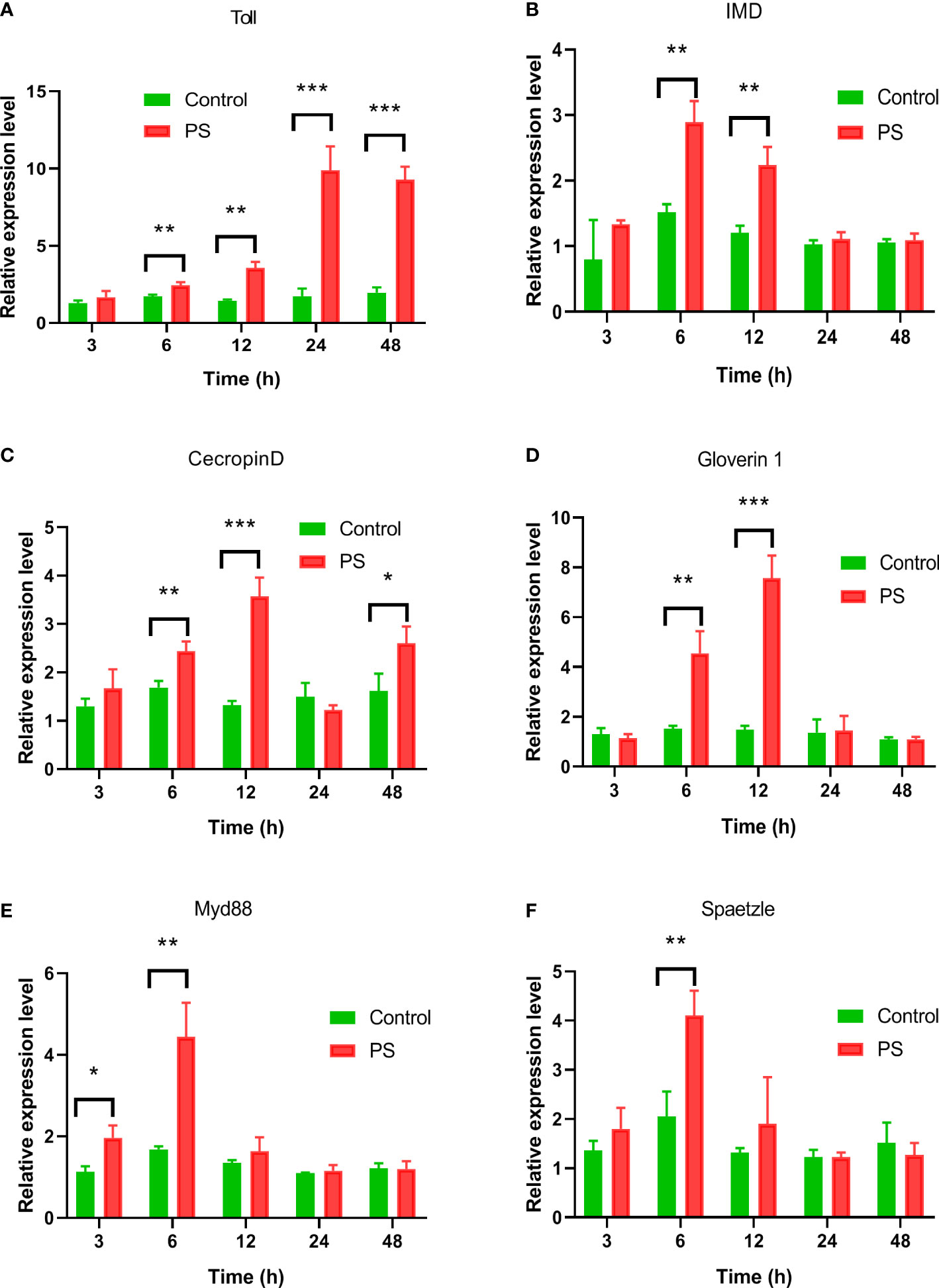
Figure 6 The expression profiles of genes involved in IMD, or Toll pathway induced by Poria cocos polysaccharides (PS). After the hemocoel injection of Bombyx mori larvae with PS (0.2 μg/larva), the mRNA levels of (A) Toll, (B) Imd, (C) Ceceropin D, (D) Gloverin1, (E) Myd88, and (F) spaetzle were detected by qRT-PCR. Two-way ANOVA/Tukey was conducted for statistical analysis. Asterisks indicate the levels of significant differences (*P < 0.05, **P < 0.01, ***P < 0.001).
The activity of four enzymes related to immunity and energy metabolism was also assayed. We found that PS inoculation could significantly enhance the PO (Figure 7A), G6PD (Figure 7B), HK (Figure 7C), and FAS (Figure 7D) activities. They reached levels of up to 0.275 (U/mg protein), 0.151 (U/mg protein), 0.453 (U/g protein), and 25.97 (U/g protein) at 24 h, respectively, and were significantly higher than those of the control group.
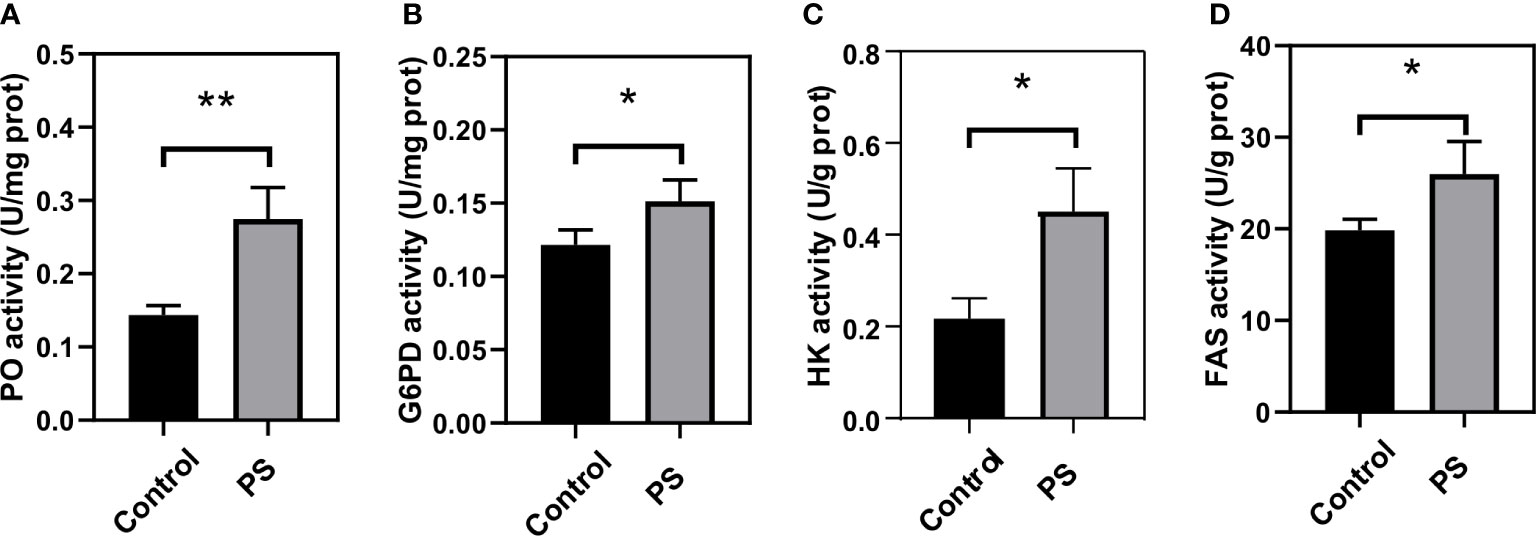
Figure 7 Changes of key enzymes involved in immunity and energy metabolism in the fat body of Bombyx mori larvae at 24 h after injecting with Poria cocos polysaccharides (PS). (A) phenoloxidase, (B) glucose-6-phosphate dehydrogenase, (C) hexokinase, and (D) fatty acid synthetase activities. The data was presented as mean ± SD for three independent experiments (*P < 0.05, **P < 0.01).
To determine the reliability of the transcript data, 14 DEGs related to innate immunity, detoxification, and metabolism were selected upon performing real-time PCR using the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene for normalization. As shown in Figure 8, the relative expression of qRT-PCR exhibited good consistency with the log2FC of the RNA-seq data, which indicated that our findings using RNA-seq were reliable and credible.
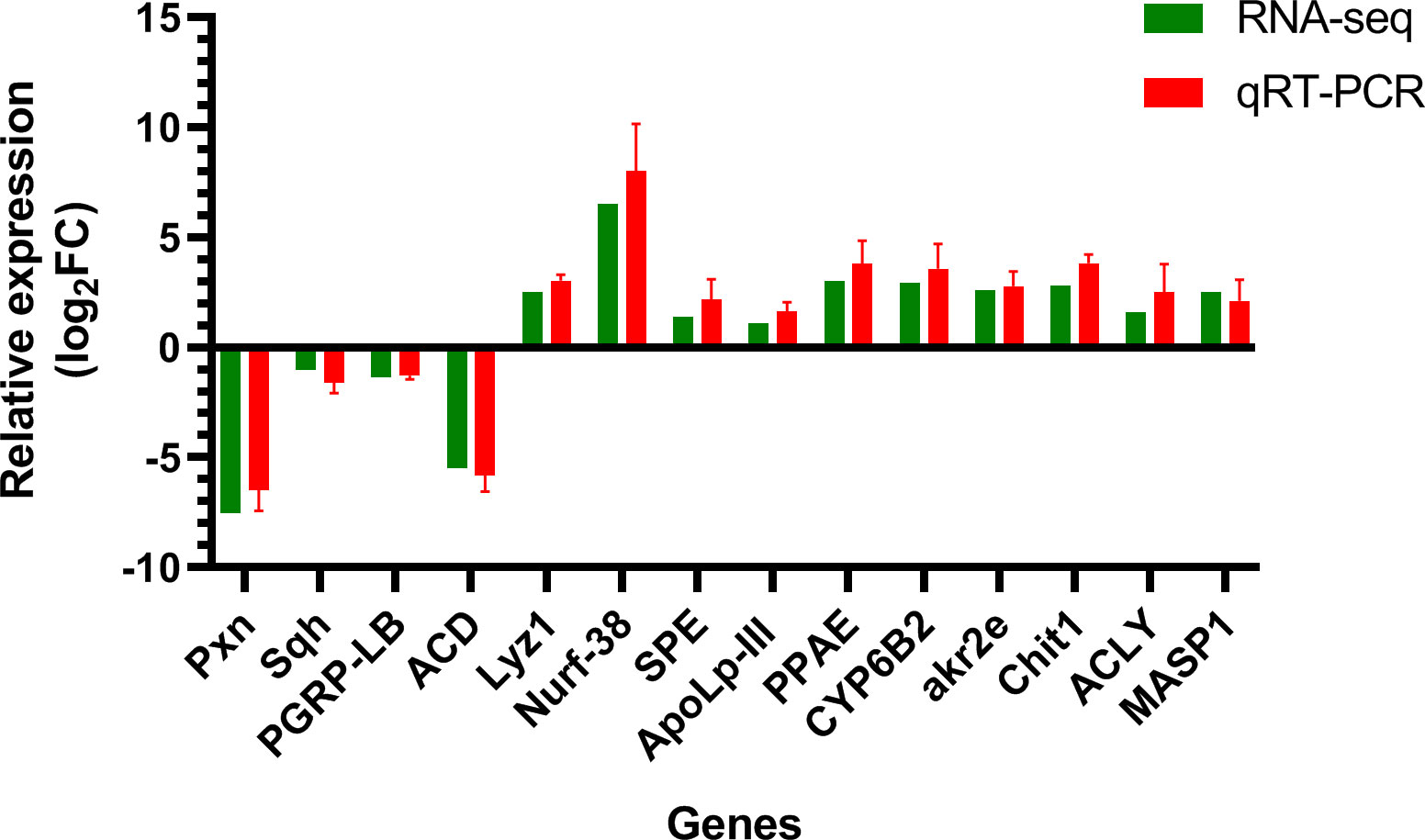
Figure 8 Validation results of RNA-seq profiles by real-time PCR. The y-axis indicates the value of relative expression level (2-ΔΔCt) by qRT-PCR and log2 ratio of PS/control RNA-seq. Pxn, peroxidasin isoform X2; Sqh, myosin light polypeptide 9 isoform X1; Lyz1, putative lysozyme; Nurf-38, inorganic pyrophosphatase; SPE, serine protease easter-like; ApoLp-III, apolipophorin-III precursor; PPAE, prophenoloxidase activating enzyme precursor; CYP6B2, cytochrome P450; PGRP-LB, peptidoglycan-recognition protein LB-like; akr2e, aldo-keto reductase AKR2E4-like; Chit1, Glycosyl hydrolases family 18; ACLY, ATP-citrate synthase-like; MASP1, serine protease gd-like; ACD, acyl-CoA desaturase-like; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To reveal the mechanisms underlying the immune regulation on silkworm by PS, the GO terms and KEGG pathways corresponding to DEGs in the control vs. PS were analyzed. According to the results, a lot of DEGs were functionally annotated as “immune system process”, “response to stimulus”, and “metabolic process”. Meanwhile, many DEGs were annotated as “metabolism” category, including “carbohydrate metabolism”, “lipid metabolism”, and “amino acid metabolism” subcategories, in the KEGG database. Our finding indicated that PS could significantly enhance the immune system and altera the physiology metabolism of silkworm. Our results are consistent with previous transcriptome studies, which showed that Atractylodis macrocephalae Koidz polysaccharides exert its immunoregulation in lymphocyte by modulating the immune signaling pathway (23) and that dietary Astragalus polysaccharides could positively influence the innate immune response of grass carp (24).
It is known that the immune system of insects consists of humoral and cellular immunity. Induction of the Toll or Imd pathway by pathogen-associated molecular patterns (PAMPs) leads to the activation of humoral immunity and synthesis of certain AMPs. A total of 11 putative Toll-related receptors and two Toll analogs have been identified in the silkworm genome by Chen et al. (25), and they speculated that these receptors may play a key role in immune defense or other biological functions. Our transcriptome data showed that genes encoding pattern recognition receptor (PRR), including Toll and scavenger receptor (Scarb1 and SCARB2), were significantly increased in the fat body of silkworm after injection with PS. Scavenger receptor plays a critical role in activating hemocyte phagocytosis and inducing the expression of AMPs (26–28), so the upregulation of Scarb1 and SCARB2 genes induced by PS revealed a positive significance in antimicrobial, antiviral, and phagocytic functions for the host. The significant increases in the phagocytic activity of hemocytes in the silkworm after PS injection support this opinion. Moreover, similar results were found in RAW264.7 cells, such that polysaccharides can exert their immunomodulatory effect by enhancing the phagocytic activity (29). It is well known that Spaetzle is a key gene in the Toll pathway. Spaetzle protein can activate the Toll receptor and recruit the downstream protein, including MyD88, Tube, and the kinase Pell, to form a complex, which initiates downstream signal cascades (30). In this study, after PS exposure, the transcriptional levels of Toll and Imd signaling pathway genes, such as Toll, Imd, Myd88, and Spaetzle, were significantly upregulated, which was consistent with the upregulation of Cecropin D and gloverin 1 genes. These results suggested that PS can activate AMP expression through the Toll or Imd signal pathway in silkworms. Noticeably, numerous research have also demonstrated that polysaccharides have the function of regulating host immunity and activating the immune cells effectively in vertebrates. They not only can act directly on natural killer cells, dendritic cells, and macrophages but also effectively activate T/B lymphocytes or regulate the secretion of cell cytokines (31, 32). However, a more detailed experiment is needed in the future to elucidate the underlying mechanism of the signal transduction pathway triggered by PS in silkworm.
We detected a high transcriptional level of ryanodine receptor genes in the fat body of PS injection silkworm, which have been demonstrated to be involved in the cellular immune defenses of insects, such as phagocytosis, hemocyte spreading, and encapsulation (33, 34). Our results suggested that PS can regulate the cellular immunity of silkworm, and they were further confirmed by the significant increases in hemocyte spreading ability of the silkworm after PS treatment. Differential gene expression analysis also showed that many genes related to the protophenol oxidase system were upregulated, such as PPAE, proPO, MASP1 and SPE, lint, Spn42Dd, and SPS-like. These genes are the members of a proPO cascade, which has been demonstrated to be one of the key immune defense mechanisms in silkworms (35). In this cascade, proPO is first cleaved by PPAE, which also exists in silkworm tissue in the form of inactive zymogen (pro-PPAE). The proteolytic processing both of pro-PPAE and proPO leads to the formation of a large number of immune effector molecules such as AMPs, opsonic, and agglutinin and to the promotion of hemocyte differentiation (36). A previous study reported that PPAE transcripts were expressed in the hemocytes, integument, and salivary glands but not in the mid-gut or fat body of silkworm (37). Another study showed that the transcript abundance of SPE in Anopheles gambiae increased significantly following a bacterial infection (38). It has also been demonstrated that serine protease inhibitor (serpin), such as Spn28D in D. melanogaster and Spn 40, Spn 55, and Spn 48 in Tenebrio molitor, revealed upregulated transcript levels and participated in modulating the melanization reaction during a pathogenic infection (39, 40). Combined with the results of the above-mentioned studies and our RNA-seq data, we can conclude that PS can trigger the proPO cascade and then convert proPO to the active form of phenoloxidase which is crucial for the formation of melanin and in the killing of invading pathogens. In fact, the significant increase of the PO activity detected in the fat body of PS-inoculated silkworm further confirmed the above-mentioned conclusion.
ApoLp-III acts as a PRR in insect and regulates the humoral and cellular immune responses, such as the synthesis of AMPs and immune-related proteins, nodule formation, and opsonization (41). It has been reported that the upregulation of BmApoLp-III in silkworm is beneficial for fighting against Beauveria bassiana (42). BmApoLp-III protein may be involved in the activation of the Toll pathway to enhance the defense ability of the silkworm against invaders. The synergistic action of lysozyme and apoLp-III against selected Gram-negative and Gram-positive bacteria was documented in G. mellonella larvae (43). In silkworm, we found that both lysozyme and apoLp-III were similarly upregulated, indicating that the host immune defenses were positively regulated by exogenous PS. In addition, we also observed that a significant decrease in PGRP-LB mRNA occurred in the fat body of PS treatment larvae. These results further confirmed that PGRP-LB played an important negative regulatory role in systemic immunity and intestinal epithelial immunity (44).
The GSEA showed that the PS treatment larvae exhibited highly enriched signatures for many immune signaling pathways, such as MAPK, PI3K-Akt, and peroxisome pathway compared with the control. It is well known that the MAPK signaling pathways, including p38, JNK, and ERK, functions in triggering host innate immune responses. Polysaccharide can activate the MAPK and NF-κB signal through triggering specific receptors, including TLR-4, scavenger receptor, complement receptor 3, and Dectin-1, and induce the expression of inflammation factors in macrophages (45). In humans, the MAPK can regulate the production of molecules such as IL-6, TNF-α, and nitric oxide (NO), which play an important role in immune response and inflammation (46). Furthermore, a previous study revealed that activating the MAPK signaling cascade is necessary for diamondback moth, Plutella xylostella, to overcome the toxic action of Bacillus thuringiensis (47), and there is also evidence to suggest that activation of MAPK signaling in Drosophila has contributed to restricting the strength of IMD signaling, which could prevent the occurrence of an excessive immune response (48). When insects initiate the immune system to fight against invaders, a huge number of reactive oxygen species (ROS) will be generated by the immune cells. An appropriate amount of ROS is conducive to the killing of invaders, but excessive levels of ROS are toxic. The peroxisome pathway is involved in the detoxification of ROS and also mediate the cytoskeletal rearrangement required for phagocytosis (49). The PI3K-Akt pathway plays an essential role in several biological processes such as cell apoptosis, proliferation, and differentiation (50). To summarize, the GSEA in this study displayed an overview of various regulated genes, suggesting that PS are capable of regulating the immune functions of silkworm through activating the MAPK, PI3K-Akt, and peroxisome signaling pathways.
In this study, we found that many DEGs participated in carbohydrate metabolism (e.g., pentose and glucuronate interconversions, ascorbate and aldarate metabolism, galactose metabolism, and amino sugar and nucleotide sugar metabolism) and lipid metabolism (e.g., steroid hormone biosynthesis, glycerolipid metabolism, and fatty acid biosynthesis). Most of the DEGs in these pathways were upregulated in the experimental group, such as pik3c3, UGT2A3, CRYL1, Chit1, Elovl4, EGT, FAS, TE-domain, and pksM. These results indicated that PS injection enhanced the level of carbohydrate and lipid metabolism in the fat body of silkworm. The increased enzyme activities, including G6PD, HK, and FAS, related to carbohydrate metabolism or lipid metabolism in the fat body of PS treatment silkworm further confirmed the above-mentioned opinion. G6PD is an important enzyme for NADPH biosynthesis, and a high level of G6PD activity could extend the lifespan of transgenic Drosophila melanogaster by improving the oxidative stress (51). HK is a key enzyme that controls glycolysis because it can phosphorylate glucose, fructose, and mannose to the corresponding hexose 6-phosphates (52). It also acts as a PRR for the recognition of various PAMPs and activation of an anti-bacterial immune response (53). FAS catalyzes acetyl coenzyme A (acetyl-CoA) and malonyl-CoA to produce palmitate, which plays important roles in adenosine triphosphate (ATP) production (54). Large amounts of ATP are needed for insects to activate their innate immune responses.
The KEGG-based GSEA also revealed that glycolysis, TCA cycle, glycerolipid metabolism, and fatty acid biosynthesis were significantly enriched gene sets, with higher expression levels in the PS treatment group compared with the control group. These changes enabled the immune cells to respond effectively during a pathogen infection. In mammals, this similar phenomenon is called Warburg effect (55). The TCA cycle is the central metabolic hub of sugars, lipids, and amino acids and finally produces a large amount of ATP through oxidative phosphorylation (56). As we all know, the insect fat body is a key nutrient metabolic organ which plays an important role in storing energy and providing energy to the rest of the organism, but it is also an organ important for exerting immune responses. The upregulated transcriptions of genes involved in Toll or Imd signal pathway and the increase in cellular immune parameters in silkworm after a PS injection indicated that PS could activate the innate immune responses of silkworm. However, the activation of the immune system leads to a high level of energy consumption (57); therefore, the organism will undergo a metabolic switch or metabolic reprogramming, which supplies adequate energy to synthesize immune effector molecules. Metabolic reprogramming, including glucose consumption or glycolysis, has been well documented in mammalian cells after immune stimulation (57). However, such changes in energy metabolism have received little attention in insects. It has been demonstrated that mounting humoral (synthesis of AMPs and activation of proPO system) and cellular (phagocytosis, encapsulation, and nodulation) immune responses are energy-consuming processes; the energy requirements of immune cells may rise from 10% to almost 30% of the whole energy consumption (58). The increase in energy consumption and the rapid production of new biomolecules lead to the metabolic conversion of immune cells. In other words, carbohydrate and lipid metabolism must be synchronized with these changes in energy requirements. The mRNA levels of genes encoding ACLY, akr2e, and Aldh1b1, which participate in ATP synthesis, were also significantly increased, further indicating that energy metabolism and ATP production were enhanced in the PS-treated silkworm. Our results suggested that PS can enhance the energy metabolism of B. mori larvae by activating carbohydrate and lipid metabolism, especially glycolysis and TCA cycle, so as to meet the needs of high-intensity immune ability. This is supported by a previous study which examined an increase in ATP production due to the energy requirements of mounting an immune response in LPS-challenged Gambusia holbrooki (59). PS could also rescue the acetaminophen-induced liver injury by suppressing the excessive inflammatory response and apoptosis (60).
The COG annotation analysis revealed that a lot of DEGs were annotated to amino acid transport and metabolism. Many DEGs involved in amino acid metabolism, such as Gnmt, HPD, Chsy1, DNMT1, and FAH, were upregulated in the PS treatment group. Amino acids and their metabolites participate in the activation of cellular immunity, including cell proliferation, differentiation, phagocytosis, nodulation, and encapsulation, as well as in the synthesis of immune-related molecules and regulation of immune signaling pathways and oxidative stress—for instance, arginine (Arg) can be converted into nitric oxide NO, which functions as a regulator of many immune cells (61). Dietary Trp improved the hemocyte phagocytic activity and increased the total hemocyte count, hemocyanin, acid phosphatase, and alkaline phosphatase activity of Eriocheir sinensis (62). Therefore, combined with our results, we can conclude that PS may improve the immune system by regulating the amino acid metabolism.
The transcriptome data revealed that many DEGs involved in energy metabolism, including carbohydrate metabolism, lipid metabolism, and amino acid metabolism, were significantly upregulated in the fat body of silkworm after an injection with PS. The gene expression profiles, innate immune parameters, and enzyme activity assays demonstrated that PS could enhance the immunity of silkworm by increasing hemocyte phagocytosis, microaggregation, and spreading ability as well as upregulating the expression levels of genes related to immune signal recognition, detoxification, and proPO activation. These findings provided us valuable insights into how PS regulated the host immune and physiology systems and broadened our understanding of the application of polysaccharides as drugs and vaccine adjuvant.
The data presented in the study are deposited in the SRA repository, accession numbers SRR18297772, SRR18297773, SRR18297774, SRR18297778, SRR18297779, and SRR18297780.
Wrote the paper: GW. Performed the experiments: JL and WH. Provided suggestions for the experiments and manuscript: YX, ML and YL. Designed the experiments: YY and GW.
This study was supported by the National Natural Science Foundation of China (31701852, 31802135), Characteristic Innovation Projects of Colleges and Universities in Guangdong Province in 2019 (2019KTSCX074), Major Scientific Research Projects of Social Public Welfare in Zhongshan City (2019B2013), Engineering Technology Research Center of Gene and Bioinformatics (Zhongshan Institute) in Zhongshan City (170702100193188), and Gene and Protein Engineering Technology Research Center of Guangdong Province (418S43).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1014985/full#supplementary-material
Supplementary Table 1 | List of immune and metabolism-related DEGs primers used for qPCR validation.
Supplementary Table 2 | Statistical analysis and quality evaluation of RNA-seq data.
Supplementary Figure 1 | Quality assessment of the transcriptome data. (A) Pie chart of reads distribution in different genomic regions. (B) Violin plot of the expression distribution of different samples. The bulky part of each sample represents the region with the most concentrated gene expression in the whole sample, the black rectangle encompasses the lower and upper quartiles, and the white dot represents the median. (C) The reads distribution in chromosomes for different samples. The abscissa represents different chromosomes; the ordinate represents the number of reads that mapped to the corresponding chromosome.
Supplementary Figure 2 | Gene Set Enrichment Analysis (GSEA) enrichment plot of significantly enriched pathways related to immune response and metabolism as identified by GSEA-based Kyoto Encyclopedia of Genes and Genomes analysis. (A) MAPK signaling pathway, (B) peroxisome, (C) PI3K-Akt signaling pathway, (D) fructose and mannose metabolism, (E) galactose metabolism, (F) citrate cycle (TCA cycle), (G) glycolysis/gluconeogenesis, (H) fatty acid biosynthesis, and (I) glycerolipid metabolism.
1. Chen M, Song J-b, Li Z-q, Tang D-m, Tong X-. [Progress and perspective of silkworm as a model of human diseases for drug screening]. Yao xue xue bao = Acta Pharm Sin (2016) 51:690–700. doi: 10.16438/j.0513-4870.2015-0717
2. Hiroko T, Hidemasa B, Katsuhiko I, Takeshi Y. Can the silkworm (Bombyx mori) be used as a human disease model? Drug Discov Ther (2016) 10:3. doi: 10.5582/ddt.2016.01011
3. Ma L, Andoh V, Adjei MO, Liu H, Shen Z, Li L, et al. In vivo toxicity evaluation of boron nitride nanosheets in bombyx mori silkworm model. Chemosphere (2020) 247:125877. doi: 10.1016/j.chemosphere.2020.125877
4. Gao K, Li B, Chen R, Qian P, Dong J, Xue C, et al. A feasibility study of using silkworm larvae as a novel in vivo model to evaluate the biotoxicity of ionic liquids. Ecotoxicol Environ Saf (2021) 209:111759. doi: 10.1016/j.ecoenv.2020.111759
5. Abdelli N, Peng L, Keping C. Silkworm, bombyx mori, as an alternative model organism in toxicological research. Environ Sci pollut Res Int (2018) 25:35048–35054. doi: 10.1007/s11356-018-3442-8
6. Hamamoto H, Tonoike A, Narushima K, Horie R, Sekimizu K. Silkworm as a model animal to evaluate drug candidate toxicity and metabolism. Comp Biochem Physiol C-Toxicol Pharmacol (2009) 149:334–9. doi: 10.1016/j.cbpc.2008.08.008
7. Orihara Y, Hamamoto H, Kasuga H, Shimada T, Kawaguchi Y, Sekimizu K. A silkworm baculovirus model for assessing the therapeutic effects of antiviral compounds: Characterization and application to the isolation of antivirals from traditional medicines. J Gen Virol (2008) 89:188–94. doi: 10.1099/vir.0.83208-0
8. Yasuhiko M, Kazuhisa S. Evaluation of anti-diabetic drugs by using silkworm, bombyx mori. Drug Discov Ther (2016) 10:19. doi: 10.5582/ddt.2016.01017
9. Wu GQ, Li M, Liu Y, Ding Y, Yi YH. The specificity of immune priming in silkworm, bombyx mori, is mediated by the phagocytic ability of granular cells. J Insect Physiol (2015) 81:60–8. doi: 10.1016/j.jinsphys.2015.07.004
10. Yi Y, Xu H, Li M, Wu G. RNA-Seq profiles of putative genes involved in specific immune priming in bombyx mori haemocytes. Infect Genet Evol (2019) 74:103921. doi: 10.1016/j.meegid.2019.103921
11. Sun Y. Biological activities and potential health benefits of polysaccharides from poria cocos and their derivatives. Int J Biol Macromol (2014) 68:131–4. doi: 10.1016/j.ijbiomac.2014.04.010
12. He Y, Zhang L, Wang H. The biological activities of the antitumor drug grifola frondosa polysaccharide. Prog Mol Biol Transl Sci (2019) 163:221–61. doi: 10.1016/bs.pmbts.2019.02.010
13. Zhu W, Zhou S, Liu J, McLean RJC, Chu W. Prebiotic, immuno-stimulating and gut microbiota-modulating effects of lycium barbarum polysaccharide. BioMed Pharmacother (2020) 121:109591. doi: 10.1016/j.biopha.2019.109591
14. Abdala Diaz RT, Casas Arrojo V, Arrojo Agudo MA, Cardenas C, Dobretsov S, Figueroa FL. Immunomodulatory and antioxidant activities of sulfated polysaccharides from laminaria ochroleuca, porphyra umbilicalis, and gelidium corneum. Mar Biotechnol (NY) (2019) 21:577–87. doi: 10.1007/s10126-019-09905-x
15. Li H, Xie W, Sun H, Cao K, Yang X. Effect of the structural characterization of the fungal polysaccharides on their immunomodulatory activity. Int J Biol Macromol (2020) 164:3603–10. doi: 10.1016/j.ijbiomac.2020.08.189
16. Ma CY, Chang WC, Chang HM, Wu SB. Immunomodulatory effect of the polysaccharide-rich fraction from sclerotium of medicinal mushroom poria cocos F.A. wolf (Aphyllophoromycetideae) on balb/c mice. Int J Med Mushrooms (2010) 12:111–21. doi: 10.1615/IntJMedMushr.v12.i2.10
17. Pu Y, Liu Z, Tian H, Bao Y. The immunomodulatory effect of poria cocos polysaccharides is mediated by the Ca2+/PKC/p38/NF-κB signaling pathway in macrophages. Int Immunopharmacol (2019) 72:252–7. doi: 10.1016/j.intimp.2019.04.017
18. Tian H, Liu Z, Pu Y, Bao Y. Immunomodulatory effects exerted by poria Cocos polysaccharides via TLR4/TRAF6/NF-kappaB signaling in vitro and in vivo. BioMed Pharmacother (2019) 112:108709. doi: 10.1016/j.biopha.2019.108709
19. Hoffmann D, Hoffmann JA. Cellular and molecular aspects of insect immunity. Res Immunol (1990) 141:895–6. doi: 10.1016/0923-2494(90)90045-Z
20. Hillyer JF. Insect immunology and hematopoiesis. Dev Comp Immunol (2016) 58:102–18. doi: 10.1016/j.dci.2015.12.006
21. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res (2011) 39:W316–22. doi: 10.1093/nar/gkr483
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta delta c) method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
23. Xu W, Du A, Hu S. Transcriptome analysis of bovine lymphocytes stimulated by atractylodis macrocephalae koidz. polysaccharides in vitro. Vet Immunol Immunopathol (2018) 196:30–4. doi: 10.1016/j.vetimm.2017.12.005
24. Shi F, Lu ZJ, Yang MX, Li F, Zhan FB, Zhao LJ, et al. Astragalus polysaccharides mediate the immune response and intestinal microbiota in grass carp (Ctenopharyngodon idellus). Aquaculture (2021) 534:736205. doi: 10.1016/j.aquaculture.2020.736205
25. Cheng TC, Zhang YL, Liu C, Xu PZ, Gao ZH, Xia QY, et al. Identification and analysis of toll-related genes in the domesticated silkworm, bombyx mori. Dev Comp Immunol (2008) 32:464–75. doi: 10.1016/j.dci.2007.03.010
26. Bi WJ, Li DX, Xu YH, Xu S, Li J, Zhao XF, et al. Scavenger receptor b protects shrimp from bacteria by enhancing phagocytosis and regulating expression of antimicrobial peptides. Dev Comp Immunol (2015) 51:10–21. doi: 10.1016/j.dci.2015.02.001
27. Yang MC, Shi XZ, Yang HT, Sun JJ, Xu L, Wang XW, et al. Scavenger receptor c mediates phagocytosis of white spot syndrome virus and restricts virus proliferation in shrimp. PloS Pathog (2016) 12:e1006127. doi: 10.1371/journal.ppat.1006127
28. Ramet M, Pearson A, Manfruelli P, Li XH, Koziel H, Gobel V, et al. Drosophila scavenger receptor cl is a pattern recognition receptor for bacteria. Immunity (2001) 15:1027–38. doi: 10.1016/S1074-7613(01)00249-7
29. Bao Z, Yao L, Zhang X, Lin S. Isolation, purification, characterization, and immunomodulatory effects of polysaccharide from auricularia auricula on RAW264.7 macrophages. J Food Biochem (2020) 44:e13516. doi: 10.1111/jfbc.13516
30. Vidya MK, Kumar VG, Sejian V, Bagath M, Krishnan G, Bhatta R. Toll-like receptors: Significance, ligands, signaling pathways, and functions in mammals. Int Rev Immunol (2018) 37:20–36. doi: 10.1080/08830185.2017.1380200
31. Jiang MH, Zhu LA, Jiang JG. Immunoregulatory actions of polysaccharides from Chinese herbal medicine. Expert Opin Ther Targets (2010) 14:1367–402. doi: 10.1517/14728222.2010.531010
32. Estrada A, Yun CH, Van Kessel A, Li B, Hauta S, Laarveld B. Immunomodulatory activities of oat beta-glucan in vitro and in vivo. Microbiol Immunol (1997) 41:991–8. doi: 10.1111/j.1348-0421.1997.tb01959.x
33. Cuttell L, Vaughan A, Silva E, Escaron CJ, Lavine M, Van Goethem E, et al. Undertaker, a drosophila junctophilin, links draper-mediated phagocytosis and calcium homeostasis. Cell (2008) 135:524–34. doi: 10.1016/j.cell.2008.08.033
34. Wu SF, Wang F, Huang J, Fang Q, Shen ZC, Ye GY. Molecular and cellular analyses of a ryanodine receptor from hemocytes of pieris rapae. Dev Comp Immunol (2013) 41:1–10. doi: 10.1016/j.dci.2013.04.006
35. Zou FM, Lee KS, Kim BY, Kim HJ, Gui ZZ, Zhang GZ, et al. Differential and spatial regulation of the prophenoloxidase (proPO) and proPO-activating enzyme in cuticular melanization and innate immunity in bombyx mori pupae. J Asia-Pacific Entomol (2015) 18:757–64. doi: 10.1016/j.aspen.2015.09.007
36. Sirikharin R, Soderhall K, Soderhall I. The n-terminal peptide generated after activation of prophenoloxidase affects crayfish hematopoiesis. Dev Comp Immunol (2020) 108:103687. doi: 10.1016/j.dci.2020.103687
37. Satoh D, Horii A, Ochiai M, Ashida M. Prophenoloxidase-activating enzyme of the silkworm, bombyx mori. purification, characterization, and cDNA cloning. J Biol Chem (1999) 274:7441–53. doi: 10.1074/jbc.274.11.7441
38. Paskewitz SM, Reese-Stardy S, Gorman MJ. An easter-like serine protease from anopheles gambiae exhibits changes in transcript abundance following immune challenge. Insect Mol Biol (1999) 8:329–37. doi: 10.1046/j.1365-2583.1999.83124.x
39. Scherfer C, Tang H, Kambris Z, Lhocine N, Hashimoto C, Lemaitre B. Drosophila serpin-28D regulates hemolymph phenoloxidase activity and adult pigmentation. Dev Biol (2008) 323:189–96. doi: 10.1016/j.ydbio.2008.08.030
40. Jiang R, Kim EH, Gong JH, Kwon HM, Kim CH, Ryu KH, et al. Three pairs of protease-serpin complexes cooperatively regulate the insect innate immune responses. J Biol Chem (2009) 284:35652–8. doi: 10.1074/jbc.M109.071001
41. Wiesner A, Losen S, Kopacek P, Weise C, Gotz P. Isolated apolipophorin III from galleria mellonella stimulates the immune reactions of this insect. J Insect Physiol (1997) 43:383–91. doi: 10.1016/S0022-1910(96)00113-8
42. Wu W, Lin S, Zhao Z, Su Y, Li R, Zhang Z, et al. Bombyx mori apolipophorin-III inhibits beauveria bassiana directly and through regulating expression of genes relevant to immune signaling pathways. J Invertebr Pathol (2021) 184:107647. doi: 10.1016/j.jip.2021.107647
43. Zdybicka-Barabas A, Staczek S, Mak P, Skrzypiec K, Mendyk E, Cytrynska M. Synergistic action of galleria mellonella apolipophorin III and lysozyme against gram-negative bacteria. Biochim Biophys Acta (2013) 1828:1449–56. doi: 10.1016/j.bbamem.2013.02.004
44. Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the drosophila antibacterial response and protects the fly from innocuous infection. Immunity (2011) 35:770–9. doi: 10.1016/j.immuni.2011.09.018
45. Schepetkin IA, Quinn MT. Botanical polysaccharides: Macrophage immunomodulation and therapeutic potential. Int Immunopharmacol (2006) 6:317–33. doi: 10.1016/j.intimp.2005.10.005
46. Yu Q, Zeng K, Ma X, Song F, Jiang Y, Tu P, et al. Resokaempferol-mediated anti-inflammatory effects on activated macrophages via the inhibition of JAK2/STAT3, NF-kappaB and JNK/p38 MAPK signaling pathways. Int Immunopharmacol (2016) 38:104–14. doi: 10.1016/j.intimp.2016.05.010
47. Guo Z, Kang S, Wu Q, Wang S, Crickmore N, Zhou X, et al. The regulation landscape of MAPK signaling cascade for thwarting bacillus thuringiensis infection in an insect host. PloS Pathog (2021) 17:e1009917. doi: 10.1371/journal.ppat.1009917
48. Ragab A, Buechling T, Gesellchen V, Spirohn K, Boettcher AL, Boutros M. Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J (2011) 30:1123–36. doi: 10.1038/emboj.2011.4
49. Di Cara F, Sheshachalam A, Braverman NE, Rachubinski RA, Simmonds AJ. Peroxisome-mediated metabolism is required for immune response to microbial infection. Immunity (2018) 48:832–3. doi: 10.1016/j.immuni.2018.03.035
50. Feng XJ, Liu SX, Wu C, Kang PP, Liu QJ, Hao J, et al. Et al: The PTEN/PI3K/Akt signaling pathway mediates HMGB1-induced cell proliferation by regulating the NF-kappaB/cyclin D1 pathway in mouse mesangial cells. Am J Physiol Cell Physiol (2014) 306:C1119–1128. doi: 10.1152/ajpcell.00385.2013
51. Zhang ZY, Zhang YY, Liew CW, Handy D, Hu J, Zhu B, et al. High glucose-induced decrease in catalase activity in bovine aortic endothelial cells is rescued by overexpression of glucose 6-phosphate dehydrogenase (G6PD). Diabetes (2008) 57:A199–200.
52. Wilson JE. An introduction to the isoenzymes of mammalian hexokinase types I-III. Biochem Soc Trans (1997) 25:103–7. doi: 10.1042/bst0250103
53. Chen XW, Liu ZQ, Gu YF, Zhang YK, Liu Y, Wang LL, et al. A hexokinase from the oyster crassostrea gigas is involved in immune recognition as a pattern recognition receptor. Dev Comp Immunol (2021) 122:104083. doi: 10.1016/j.dci.2021.104083
54. Liu H, Liu JY, Wu X, Zhang JT. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int J Biochem Mol Biol (2010) 1:69–89.
55. Mills CD. Anatomy of a discovery: m1 and m2 macrophages. Front Immunol (2015) 6:212. doi: 10.3389/fimmu.2015.00212
56. Van D, Sedensky MM, Morgan PG. Cell biology of the mitochondrion. Genetics (2018) 208:1673. doi: 10.1534/genetics.118.300816
57. Dolezal T, Krejcova G, Bajgar A, Nedbalova P, Strasser P. Molecular regulations of metabolism during immune response in insects. Insect Biochem Mol Biol (2019) 109:31–42. doi: 10.1016/j.ibmb.2019.04.005
58. Bajgar A, Kucerova K, Jonatova L, Tomcala A, Schneedorferova I, Okrouhlik J, et al. Extracellular adenosine mediates a systemic metabolic switch during immune response. PloS Biol (2015) 13:e1002135. doi: 10.1371/journal.pbio.1002135
59. Bonneaud C, Wilson RS, Seebacher F. Immune-challenged fish up-regulate their metabolic scope to support locomotion. PloS One (2016) 11:e0166028. doi: 10.1371/journal.pone.0166028
60. Wu K, Fan JL, Huang XY, Wu XM, Guo C. Hepatoprotective effects exerted by poria Cocos polysaccharides against acetaminophen-induced liver injury in mice. Int J Biol Macromol (2018) 114:137–42. doi: 10.1016/j.ijbiomac.2018.03.107
61. Huang Z, Aweya JJ, Zhu C, Tran NT, Hong Y, Li S, et al. Modulation of crustacean innate immune response by amino acids and their metabolites: Inferences from other species. Front Immunol (2020) 11:574721. doi: 10.3389/fimmu.2020.574721
Keywords: silkworm, immunoregulation, transcriptome analysis, metabolic alterations, innate immune parameters
Citation: Liu J, Hong W, Li M, Xiao Y, Yi Y, Liu Y and Wu G (2022) Transcriptome analysis reveals immune and metabolic regulation effects of Poria cocos polysaccharides on Bombyx mori larvae. Front. Immunol. 13:1014985. doi: 10.3389/fimmu.2022.1014985
Received: 09 August 2022; Accepted: 30 September 2022;
Published: 28 October 2022.
Edited by:
Simone Becattini, Université de Genève, SwitzerlandReviewed by:
Tomas Dolezal, University of South Bohemia in České Budějovice, CzechiaCopyright © 2022 Liu, Hong, Li, Xiao, Yi, Liu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gongqing Wu, Z29uZ3Fpbmd3dUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.