
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 29 September 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1014377
This article is part of the Research TopicSingle Domain Antibody Approaches in Pathogen Biology, Diagnosis, and NeutralizationView all 5 articles
 Qian Qin1,2†
Qian Qin1,2† Hao Liu3†
Hao Liu3† Wenbo He1
Wenbo He1 Yucheng Guo2
Yucheng Guo2 Jiaxin Zhang2
Jiaxin Zhang2 Junjun She1
Junjun She1 Fang Zheng2
Fang Zheng2 Sicai Zhang3*
Sicai Zhang3* Serge Muyldermans4
Serge Muyldermans4 Yurong Wen1,2*
Yurong Wen1,2*Increasing antibiotic resistance to bacterial infections causes a serious threat to human health. Efficient detection and treatment strategies are the keys to preventing and reducing bacterial infections. Due to the high affinity and antigen specificity, antibodies have become an important tool for diagnosis and treatment of various human diseases. In addition to conventional antibodies, a unique class of “heavy-chain-only” antibodies (HCAbs) were found in the serum of camelids and sharks. HCAbs binds to the antigen through only one variable domain Referred to as VHH (variable domain of the heavy chain of HCAbs). The recombinant format of the VHH is also called single domain antibody (sdAb) or nanobody (Nb). Sharks might also have an ancestor HCAb from where SdAbs or V-NAR might be engineered. Compared with traditional Abs, Nbs have several outstanding properties such as small size, high stability, strong antigen-binding affinity, high solubility and low immunogenicity. Furthermore, they are expressed at low cost in microorganisms and amenable to engineering. These superior properties make Nbs a highly desired alternative to conventional antibodies, which are extensively employed in structural biology, unravelling biochemical mechanisms, molecular imaging, diagnosis and treatment of diseases. In this review, we summarized recent progress of nanobody-based approaches in diagnosis and neutralization of bacterial infection and further discussed the challenges of Nbs in these fields.
With the increasing antibiotic resistance, bacterial infection constitutes a serious threat to human health. It can lead to tremendous morbidity and mortality, emphasizing the need for rapid and effective identification and treatment of bacteria pathogens (1). At present, clinical bacterial diagnosis mainly involves bacterial culture, molecular diagnostics and colony formation methods which are time-consuming, labor intensive and requiring expensive equipment, all of which limit the utility, especially in resource limited settings (2–4). Oral and intravenous antibiotics are the most common treatments against bacterial infections; however, they are usually administered against ill-defined pathogens. This abuse of antibiotics plays an important role in the increase of antibiotic resistance (5–8). Therefore, it is important to develop fast, cost-effective, and accurate methods for the detection, identification and treatment of bacterial infections. Antibodies became promising molecules for bacterial detection and treatment due to their high sensitivity and specificity.
Antibodies are essential components of adaptive immunity. Antibody-based diagnosis and therapeutics are the fastest growing classes of drugs on the market. The US FDA has approved over 100 antibodies mainly for treating cancer (45%) and immune-mediated disorders (27%) while only 8% against infectious diseases (9). The high production cost, low stability and large size may be the main obstacles to develop the antibodies for treating infectious diseases (10). Therefore, single domain antibodies (sdAbs), which have low production costs, high stability and small size become a promising alternative to canonical antibodies (11).
In 1990s, scientists found a unique class of “heavy-chain-only” antibodies (HCAbs) in the serum of camelids and sharks. Owing to the absence of light chains, HCAbs binds to the antigen through only one variable region, referred to as VHH or also sdAb or nanobody (Nbs). The antigen-binding domain of shark HCAbs are known as VNAR (12, 13). Their special structure endowed sdAbs with superior properties and enabled them to be extensively employed in structural biology (14–16), unravelling biochemical mechanisms (17), molecular imaging (18–20), diagnosis and treatment of tumors (21, 22) and infection diseases (23–27). As for infectious diseases, sdAb have been widely used in the diagnosis and treatment of a variety of viral infections (28). It is noteworthy that a lot of nanobodies have been generated targeting the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, COVID-19), and there have been excellent reviews to summarize the current research progress (29, 30). This review will focus on the current progress and perspectives of diagnostic and neutralizing sdAbs against bacterial infection (Figure 1).
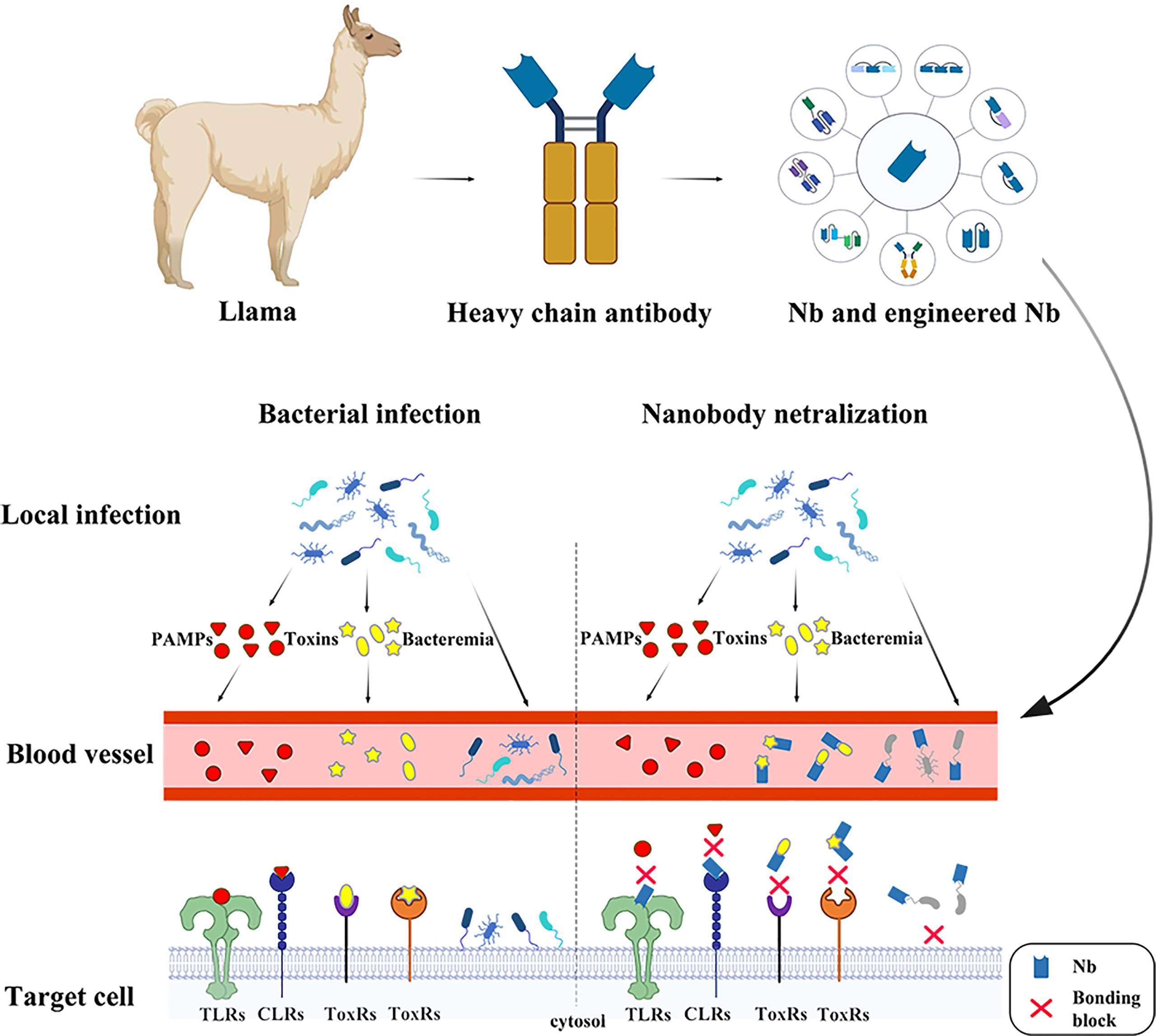
Figure 1 The nanobodies take effects in several ways against bacterial infection. In the early stage of infection, pathogenic microorganisms are confined to the lesion. At this time, the PAMPs and the toxins are released into the bloodstream. The nanobodies binding onto the receptors prevent PAMPs recognition by PRRs, such as Toll-like receptors, Nod-like receptors and C-type lectins such as Clec4f, leading to a series of bodily reactions. The toxins, such as CDT, Tcd and BoNT are the major part in bacterial pathogenicity, which serve in different ways to cause damage in hosts and assist in enlargement of infection foci. The neutralizing nanobodies protect the host through specifically binding the toxins. At late stage of infection, pathogens are released into blood causing bacteremia. The nanobodies recognizing surface antigens, such as pilus and flagellum, bind onto the pathogen surface, preventing bacterial attachment.
SdAbs are the smallest known natural antigen-specific binding functional fragment, with dimensions of 2.5 nm in diameter and 4 nm in length. They consist of approximately 120 amino acids and merely 12~17 kD in weight, which is only one-tenth of canonical antibodies (150kD) (31). Similar to the VH domain of canonical antibodies, sdAbs consist of three hypervariable antigen-binding loops (complementarity determining regions, CDR1-CDR3) and four conserved framework regions (FR1-FR4) (32–34). There are mainly two differences between sdAbs and VHs of canonical antibodies. SdAbs have elongated CDR1 and CDR3, which to some extent compensate the loss of antigen-binding surface contributed by the light chain CDRs. In addition, the elongated CDR3 can adopt larger variety of structures and has a preference to interact with concave shaped antigen surfaces (35). Another notable difference is the conservative hydrophobic amino acids (Val47, Gly49, Leu50, and Trp52) in canonical antibody FR2 substitution by hydrophilic amino acids (Phe42, Glu49, Arg50, and Gly52), increasing solubility and stability of sdAbs (33, 36).
Due to the specific structure, sdAbs possess several outstanding characteristics compared to the traditional antibodies. 1) SdAbs represent the smallest naturally derived antigen-binding functional fragments (~15 kD). The small size allows sdAbs to penetrate deeper in dense tissues and might cross the blood-brain barrier(BBB) (37, 38), and to be quickly eliminated via the kidney (39). Besides, the higher isoelectric point makes SdAbs positively charged and easier to penetrate the BBB. Therefore, sdAbs are more suitable for targeting solid tumors (40, 41) and brain diseases (37, 42, 43). 2) Compared with traditional antibodies, the sdAbs have only one domain with disulfide bonds, which folds into a relatively stable structure. Amazingly, sdAbs maintained their antigenic binding ability after being incubated for one week at 37°C. They even can tolerate higher temperatures of 60-80°C (36). In some cases, they can regain antigen-binding activity after thermal denaturation by exposure at high temperatures (90°C) (44).When exposed to chemical denaturing agents and proteases, as well as non-physiological pH (pH range 3.0-9.0), sdAbs can also retain most antigen-binding capabilities (45). These properties permit the use of more demanding chemical and physical conditions during treatments or modifications of sdAbs than other types of antibodies. 3) The longer amino acid sequences of the CDR3 enlarges the antigen binding surface of the sdAbs, increases their structural repertoire, and further expands the binding ability to some hidden antigenic epitopes by creating new finger-like structures. Thus, they are enhancing the recognition ability of concave epitopes as well as binding to such epitope architectures with high affinity (31, 46–51). 4) The hydrophilic amino acids on the side of VHH, corresponding to the VL interface of VH domains, improve the solubility in aqueous solutions and lower the tendency to aggregate (52). 5) The high degree of sequence identity with human VH domains of family-3 of VHHs, their small size, resistance to form aggregates and rapid blood clearance favor a low immunogenicity. VNARs may have a higher immunogenicity due to low sequence identity between VNARs and human VH or VL domains (~ 30% overall) (53). Overall, the immunogenicity risks with VHHs are low (54). In addition, the humanization of sdAbs provides a safe option for long-term treatment (55, 56). 6) SdAbs can be efficiently, easily and economically produced recombinantly in bacteria, mammalian cell lines, yeast and plants at an affordable cost (11, 57), while the production of canonical monoclonal antibodies requires mammalian expression system, which is complex in technology and expensive to maintain. Apart from these outstanding characteristics, sdAbs also have some limitations. The biggest drawback of sdAbs is their inadequate pharmacokinetics. Compared with conventional antibodies, sdAbs have a faster serum clearance rate, which limits their application in the field of therapy. In addition, sdAbs may have some adverse effects and a humanized tetravalent Nb have been reported with hepatotoxicity. The characteristics compared between nanobodies and conventional antibodies are listed in Table 1.
Enterotoxigenic E. coli (ETEC) is one of the most common causes of diarrhea in toddlers, adults in the developing world and in travelers to endemic areas. According to WHO reports, ETEC related diarrhea is one of the leading causes of death in the children under the age of 5 in developing countries (58). In addition, ETEC strains causing severe, watery diarrhea are responsible for significant death and morbidity in neonatal and post-weaned piglets, leading to worldwide tremendous economic losses in pork industry (59).
ETEC is a non-invasive pathogen that mediates small intestine adherence through bacterial surface structures, known as colonization factors (CFs). Once bound to the small intestine, the bacteria produce toxins causing a net flow of water from enterocytes, leading to watery diarrhea (60). ETEC strains can also produce many types of fimbriae that are involved in bacterial attachment. F4 fimbriae are commonly found on ETEC from diarrheic piglets (61, 62). In 2005, Harmsen et al. immunized a llama with F4ac fimbriae from the F4-positive (F4+) ETEC strain CVI-1000 and obtained a few monoclonal VHHs. However, the best monovalent VHH, K609, could not significantly reduce diarrhea and reduced piglet mortality was poor (63). In contrast, orally administration of the linker connected bivalent VHH could enhance the clearance of F4+ ETEC and decrease the number of infected piglets (64). The different in vivo activity of mono- and bivalent VHH suggested that the ability to agglutinate bacteria may have a higher impact on infection, consistent with other studies where only bivalent antibodies showed in vivo protection (65–67). In another study, Moonens et al. (59) fused four different variable domains of llama heavy chain-only antibodies (V1-4), raised against F4ac, to the Fc domain of a porcine immunoglobulin IgA. These four different VHH targeted conserved epitopes of FaeG, a major adhesive subunit of F4. The four VHHs were fused to porcine IgA-Fc and subsequently expressed in Arabidopsis thaliana seeds to feed piglets. The oral feed-based passive immunization strategy protected piglets as demonstrated by (i) the progressive decline in shedding of F4 positive ETEC bacteria, (ii) the significantly lower immune responses of the piglets to F4 fimbriae, which suggest a reduced exposure to the ETEC pathogen, and (iii) a significantly higher body weight in comparison with control piglets (63, 68). The structural study of V1-4 in complex with FaeG indicated that they sterically hindered FaeG associating with the F4 receptor but they did not directly interfere with the carbohydrate binding site (59). Besides F4+ ETEC, four VHHs targeting F18 fimbriae FedF domain were generated by llama immunization and selection as well. They could inhibit F18+ ETEC attaching to piglet enterocytes in vitro, and either sterically hinder or induce conformational changes of the binding surface of FedF (69). In a recent study, Amcheslavsky (60) and his colleagues immunized two male llamas with N-terminal fragments of eight class-5 ETEC adhesins to generate nanobodies with broad cross-reactivity against ETEC adhesins. They identified single nanobodies that show cross protective potency against eleven major pathogenic ETEC strains in vitro and inhibited ETEC colonization in vivo. Molecular docking and mutagenesis analysis revealed that nanobodies recognized a highly conserved epitope within the putative receptor binding region of ETEC adhesins (60).
Shiga toxin-producing Escherichia coli (STEC) are a subset of E. coli pathogens leading to illnesses such as diarrhea, hemolytic uremic syndrome (HUS) and even death. Shiga toxins, the main virulence factors are divided in two groups: Stx1 and Stx2, of which the latter is more frequently associated with severe pathologies in humans and newly weaned pigs (70). Stx2e consists of an enzymatically active A subunit and five B subunits that bind to globotriaosylceramide (Gb3) on host cells (71). Lo et al. reported the discovery and characterization of a VHH, NbStx2e1, isolated from a llama phage display library that confers potent neutralizing capacity against Stx2e toxin. Structural analysis revealed that for each B subunit of Stx2e, one NbStx2e1 is interacting in a head-to-head orientation and directly competing with the glycolipid receptor binding site on the surface of the B subunit. The neutralizing NbStx2e1 can be used to prevent or treat edema disease in the future. Tremblay et al. immunized llama with Stx1 and 2 together and identified a panel of neutralizing VHHs, two of which demonstrated cross activity to Stx1 and 2 (72). A VHH heterodimer consisting of one Stx1-specific VHH/Stx2-specific VHH, and one Stx1/Stx2 cross-specific VHH, significantly improved the survival and reduced the kidney damage of mice challenged with Stx1 or 2. In addition, co-administration of the heterodimeric VHH with an effector Ab that binds to the VHH heterodimer, was effective in preventing all symptoms of intoxication from Stx1 and Stx2. In 2016, Mejías and his colleagues reported the generation of a family of Stx2B-binding VHHs that neutralize Stx2 in vitro at a nanomolar to subnanomolar range. The anti-Stx2B VHH, 2vb27, was selected and two copies were fused to an anti-human serum albumin VHH. This engineered antibody showed increased retention in circulation and was able to neutralize Stx2 in three different mouse models. This novel and simple antitoxin agent should offer new therapeutic options for treating STEC infections to prevent or ameliorate HUS outcome (73). In another study, Navarro et al. described the identification and characterization of a nanobody (Nb113) with the potential to neutralize the Stx2a and Stx2c toxins that are associated with human clinical infections. The crystal structural study revealed that each B subunit in the pentameric B5 ring is associated with a single Nb113 molecule. A detailed analysis of the epitope targeted by Nb113 suggests that this Nb prevents the formation of the Stx2a–Gb3 complex, thereby impeding the subsequent steps of the internalization and enzymatic activity of the Stx2a holotoxin (70).
Besides Stx2-neutralizing VHHs, two VHHs were identified from immunized llama for detection of Stx2 using ELISA, which was even more sensitive than commercial ELISA kits (74). The ELISA was best for the major subtype Stx2a and less sensitive for Stx2f. VHH based ELISA is expected to be more cost effective than IgG ELISA.
Pseudomonas aeruginosa is one of the leading causes of hospital-acquired infections. It is difficult to treat the infections due to the high intrinsic antibiotic resistance and the organism’s capability to occur in biofilms in the host. Adams et al. immunized a llama with P. aeruginosa antigens and identified monoclonal anti-flagellin VHHs. In an in vitro assay, they showed that the anti-flagellin VHHs are capable of inhibiting P. aeruginosa from swimming and that they prevented biofilm formation (75).
Helicobacter pylori infection is associated with gastritis, gastric and duodenal ulcers, and even gastric adenocarcinoma. It is important to seek alternative therapeutic strategies due to the increasing occurrence of antibiotic resistance. Some studies reported the isolation and purification of nanobodies with high affinity against UreC subunit of urease enzyme from H. pylori. These nanobodies could be a novel class of treatments against H. pylori infection (76, 77). The sdAbs employed for diagnosis and neutralization of Gram- negative bacterial infection are listed in Table 2.
Clostridium difficile is an opportunistic pathogen residing in the gastrointestinal tract of humans, causing antibiotic-associated diarrhea and pseudomembranous colitis (78). Antibiotics metronidazole and/or vancomycin are the primary treatment for C. difficile-associated disease (CDI) and surgeries are often required in the case of fulminant CDI (79). Due to the difficulties of treatment and high rates of recurrence, it’s necessary to explore new therapeutic agents (80). The Gram-positive bacterium produces two large clostridial exotoxins, toxin A (TcdA) and toxin B (TcdB), which are the major virulence factors responsible for CDI and are potential targets for CDI therapy (81). TcdA and TcdB are homologous to each other, having a similar domain organization including glucosyltransferase domain (GTD), cysteine protease domain (CPD), delivery and receptor binding domain (RBD) and combined repetitive oligopeptide domain (CROPs) (82, 83). In 2011, Hussack and his colleagues isolated after phage display from an immune sdAb llama library four VHHs specifically targeting partial CROPs of TcdA or TcdB. In vitro assay on fibroblast cells demonstrated potent protection from the cytopathic effects of toxin A by these VHHs. Moreover, the protection efficiency was further enhanced when VHHs were administered in a manner of paired or triplet combinations (81). In another study, they characterized a panel of VHHs against partial RBD and CROPs of TcdB. Unfortunately, none of these VHHs exhibited inhibitory effects against TcdB cytotoxicity in a cell-based assay, given that several VHHs showed high affinity to toxin. This incapability of neutralization is probably due to TcdB accepting multiple proteins as receptors (84–86) and blockage of a single epitope might not be effective inhibition of TcdB toxicity. Nevertheless, when bivalent VHHs fused to the Fc fragment, their neutralization efficiency reached to the level of the recently approved anti-toxin B monoclonal antibody, bezlotoxumab (87). Furthermore, VHHs targeting different vulnerable regions on TcdB were also developed. SdAb named E3, 7F and 5D were demonstrated to bind with GTD, the connecting region between GTD and CPD, and RBD, respectively. Among which, E3 showed the best inhibition of TcdB cytotoxicity (88, 89). Yang and his colleagues created a tetravalent and bispecific antibody called “ABA” which comprised two VHHs against both, TcdA and TcdB. ABA was capable of binding to both toxins simultaneously and neutralizing toxins from clinical C. difficile isolates. Therefore, ABA showed a significantly enhanced neutralizing activity both in vitro and in vivo (90). Schimdt and colleagues constructed a heteromultimeric VHH-based neutralizing agent, which potently neutralized both C.difficile toxins in cell assays and protected animals from CDI to different extents (88). In addition to development of VHHs, strategies to administer VHHs were also explored. For example, adenovirus, engineered Lactobacillus and probiotic Saccharomyces boulardii, expressing different forms of VHHs, were utilized to treat CDI effectively in animal models and proved to be promiscuous for combating the diseases invoked by C. difficile (91–93). Beside TcdA and TcdB, surface layer proteins (SLPs), mediating adherence to host cells, represents an alternative target for CDI treatment. Kandalaft and his colleagues used SLPs isolated from C. difficile hypervirulent strain QCD32g58 (027 ribotype) to immunize a llama and identified a panel of SLP-specific VHHs, which exhibited inhibition of C. difficile QCD32g58 motility in vitro. Therefore, targeting SLPs with VHHs may be a viable therapeutic approach against CDI (94).
Anthrax is a severe and fatal disease caused by the Gram-positive Bacillus anthracis. Anthrax toxin is a mixture of one non-toxic protein, protective antigen (PA) and two toxins, edema factor (EF) and lethal factor (LF). Protective antigen (PA) could bind to anthrax toxin receptors on cell surface forming oligomer pore and translocate the lethal factor (LF) and edema factor (EF) into the cytosol to take effects (95). In 2015, Moayeri and his colleagues identified two classes VHHs (JIK-B8 and JKH-C7) targeting two epitopes of PA from immunized alpacas. The two VHHs were expressed as a heterodimeric VHH-based neutralizing agent (VNA2-PA) and displayed improved neutralizing potency in in vitro and in vivo assays compared with monomeric VHH (96). In another study, they used a gene therapy approach using recombinant replication-incompetent human adenovirus serotype 5 (Ad5) vector to express and secret the VNA (Ad/VNA2-PA) into the serum, and found that it can protect mice against an anthrax toxin challenge and anthrax spore infection (97). Apart from PA, the same group identified a set of 15 VHHs against EF and/or LF. Six of these VHHs were cross-reactive with both, EF and LF N-terminal domain, which is responsible for association with PA. Unlike the other selected VHHs, one LF specific VHH bound the C-terminal of LF inhibiting its enzymatic activity. Two bispecific heterodimers of the selected neutralizing VHHs demonstrated full protection against lethal anthrax spore infection (98).
The cell surface of B. anthracis is covered by a protective surface layer or S-layer, composed of the highly-conserved S-layer protein (Sap). S-layers are proposed to function (i) as exoskeletons, (ii) as protection against harmful environments, (iii) as scaffolding structures for surface-localized enzymes and adhesins, (iv) as molecular sieves for nutrient uptake and (v) as a contact zone with the extracellular environment, including host cells in case of pathogenic bacteria (99). Fioravanti et al. generated Sap self-assembly inhibiting nanobodies, which exhibited disruption of the S-layer and attenuated the bacterial growth. Subcutaneous injection of the Sap inhibiting nanobodies cleared anthrax infection and prevented death in a mouse model of anthrax (100).
Botulinum neurotoxins (BoNTs) are a category of bacterial toxins produced by Clostridium Botulinum and related strains, they are dangerous potential bioterrorism agents (Category A and Tier 1 select agent) (101). BoNTs cause a life-threatening disease called botulism, which develops flaccid paralysis and autonomic dysfunctions. Once infected, patients have to stay in the intensive care unit (ICU) and rely on mechanical ventilation for weeks to months, which is costly and time consuming (102). There are seven known serotypes of BoNTs (BoNT/A to BoNT/G), in which serotypes A, B and E are often associated with human botulism (103). Currently, antitoxins such as equine antitoxin and human botulism immunoglobulin represent the main strategy for treatment. However, adverse reactions, including early anaphylactic shock and late serum sickness, have been reported (103), which poses the necessity for developing new therapeutics to treat botulism. To this end, nanobodies could play an important role in such tasks.
For this purpose, a variety of VHHs against BoNT/A were generated in the past years from phage or yeast display libraries derived from camel, alpaca and llama, respectively. Thanongsaksrikul et al. reported a neutralizing nanobody, VHH17, binding specifically to the catalytic cleft in light chain of BoNT/A via its CDR2 region, which is inaccessible to conventional antibodies due to their large size (104). In a similar study, Dong et al. identified a VHH Aa1 using yeast display. Rather than binding to the catalytic site of BoNT/A, Aa1 targeted the non-catalytic α-exosite binding region and inhibited enzyme activity of the toxin. Besides, Aa1 exhibited extraordinary thermal and reducing stability, which is optimal for therapeutic purposes (105). Tremblay and colleagues identified and characterized two VHHs ALc-B8 and ALc-H7 having affinity up to the nanomolar level to the light chain of BoNT/A. They further confirmed that ALc-B8 was able to inhibit SNAP-25 proteolysis in neuronal cells intoxicated by BoNT/A (106), which demonstrated its potential for therapy. In a recent study, Lam et al. discussed the inhibitory mechanism of VHHs against BoNT/A light chain via structural studies and found that the recognized epitopes of the light chain are quite conserved across different subtypes, laying the foundation for structure-based drug design (107, 108). Besides the protease domain, VHHs such as ciA-C2, specifically recognizing the receptor binding domain of BoNT/A were also identified and proven to exert an inhibitory function (109). Furthermore, various strategies to enhance the efficacy of VHHs neutralization of BoNT/A have been exploited, such as (i) tagging the VHHs for better and faster clearance of bound toxin (110), (ii) fusing the VHHs with human Fc fragment or Glycophorin A on red blood cell surface to increase their circulation half-life (111, 112), or (iii) expressing VHHs in replication-incompetent adenovirus to provide prolonged protection (113). With similar strategies, several VHHs bound to BoNT/E were also produced and characterized. Bakherad et al. selected a VHH, BMR2, specifically targeting the receptor binding domain of BoNT/E, which completely neutralized 3LD50 of BoNT/E in mice (103). Lately, Tremblay et al. identified plenty of BoNT/E-neutralizing VHHs and Lam et al. characterized two of them, JLE-E5 and JLE-E9, targeting the translocation domain of BoNT/E. They confirmed that these two VHHs blocked a structural change of BoNT/E in acidic pH, a process necessary for its biological function, which could hamper toxicity of BoNT/E (114). The pitfall to treat botulism is that no drugs are able entering into neurons to take effect once the toxins are endocytosed. A hallmark application of VHH for treating botulism was to deliver VHHs into neural cells by coupling them to intoxicated BoNTs. Utilizing this strategy, two independent groups successfully delivered VHHs into neurons and provided animals with full recovery from botulism, which opened new avenues of using VHHs to treat diseases (115, 116).
In addition to the bacteria mentioned above, nanobodies also play an important role in the diagnosis and therapy of other bacteria. Nanobodies can also be used to establish immuno-assays to uncover bacteria contaminations in foods. Staphylococcus aureus is one of the most common food-borne pathogens. Hu et al. selected a specific nanobody Nb147 to develop an immuno-assay detecting S. aureus in milk (117). Staphylococcal enterotoxins (SEs) are the major causes of staphylococcal food poisoning (SFP) and various other diseases. Ji et al. developed a double nanobody-based sandwich immunoassay for the detection of staphylococcal enterotoxin C in dairy products (118) while Zanganeh et al. developed a rapid and sensitive detection of staphylococcal enterotoxin B by recombinant nanobodies (119). Listeria monocytogenes (LM) causes listeriosis, a potentially fatal food-borne disease especially harmful to pregnant women. Tu and his colleagues developed an ELISA using the VHH clone L5-79 and a monoclonal antibody to detect LM in pasteurized milk (120). King et al. identified a group of VHHs targeting internalin B (InlB) of LM which were competitive inhibitors preventing bacterial invasion. These results point to the potential of VHH as a novel class of therapeutics for the prevention of listeriosis (121). The sdAbs applications to diagnose and neutralize Gram- positive bacterial infection are overviewed in Table 3.
Pattern recognition receptors (PRRs) are a class of receptors that play crucial roles in detecting conserved pathogen associated molecular patterns (PAMPs) shared among many microorganisms or endogenous damage-associated molecular patterns (DAMPs) to initiate downstream signaling (123–126). PRRs have been identified and are notably classified into the following families: Toll-like receptors (TLRs), the Ctype lectin receptors(CLRs), the nucleotide-binding oligomerisation (NOD)-like receptors (NLRs), the RIG-I-like receptors, the absent in melanoma 2 (AIM2)-like receptors and the OAS like receptors (127–130). PRRs connect PAMPs or DAMPs to trigger a variety of signal pathways, eventually activating interferon regulatory factor (IRFs), nuclear factor-kappa B (NF-κ B), mitogen-activated protein kinase (MAPKs) and etc., which promotes the expression of pro-inflammatory cytokines (131–133). The sdAbs against Pattern Recognition Receptor are listed in Table 4.
Toll-like receptor 4 (TLR4) is a member of the TLR family, which participates in innate immunity and mediates inflammation by recognizing lipopolysaccharide (LPS) or bacterial endotoxin (125, 136, 137). Overactivation of TLR4 can trigger the production of various inflammatory factors, which are related to the occurrence and development of a series of diseases including sepsis (138), endotoxemia, pregnancy-related disorders (139, 140), cardiovascular disease (141, 142), intestinal inflammation (143), rheumatoid arthritis (144), acute kidney injury (AKI) (145, 146), and acute lung injury (147). Therefore, the drug design and development for this target have high therapeutic potential and the anti-inflammatory effect of TLR4 inhibitors has been confirmed by several studies (148–150). Liao and his colleagues (134) identified an anti-TLR4 intermediate and C-terminal domain-recognizing nanobodies using phage display. Then, through in vitro and in vivo experiments, they confirmed that the anti-TLR4 nanobody can effectively reduce the release of inflammatory factors and improve the animal survival rate. The effect is even more pronounced when two different nanobodies are combined.
C-type lectins can recognize a variety of ligands and play an important role in a variety of physiological functions. Particularly, C-type lectins contribute to innate and adaptive antibacterial immune responses by recognizing surface polysaccharides of specific pathogens (151). Clec4f is a member of the type II C-type lectin family and is only expressed by Kupffer cells (152–154). In addition, studies have shown that Clec4f is involved in α-galactose ceramide presentation and Listeria monocytogenes infection in mouse liver (155). Zheng et al. developed a series of nanobodies from an alpaca immunized with recombinant mouse Kupffer cell receptor Clec4F by using a phage display. After bio-panning selections, they obtained 14 different nanobodies against Clec4F with an affinity ranging from 0.2 to 2 nM. Furthermore, they have characterized the structure of two Clec4F nanobodies, Nb1.46 and Nb2.22, with different CDR2 and CDR3 sequence features. These works may contribute to the study of Clec4F structure and function as well as its use as a molecular imaging agent and therapeutic agent (135). In another study, they indicated that Clec4F nanobodies could be used to track changes in Kupffer cell (KCs) dynamics in mice via non-invasive imaging (153).
As bacterial antibiotic resistance is developed at increasing pace, there is a great urgency to develop a non-antibiotic approach to treat bacterial infections. SdAbs are versatile molecules with favorable properties representing an alternative tactic for both therapeutic and diagnostic applications in bacterial infections. SdAbs are characterized by minimal size, high stability, strong affinity, good solubility, and low immunogenicity which open pathways to target antigens that were previously inaccessible during bacterial infection. Therapeutic nanobodies are still in early phase development, however they have a promising future. The first therapeutic nanobody-based drug, Caplicizumab (Cablivi), was approved by EMA in August 2018 and by FDA in March 2019 for the treatment of blood clotting disorder. Since then, Ciltacabtagene autoleucel (Carvykti) a nanobody based Chimeric Antigen Receptor T cell (CAR-T)-based medication against relapsed or refractory multiple myeloma was approved by FDA and EMA (February and May, 2022) and Envafolimab, a subcutaneous injectable sdAb directed against PD-L1 (approved in November 2021) by the Chinese National Medical Products Administration (NMPA) for adult patients with microsatellite instability-high or mismatch repair deficient advanced solid tumors followed soon after. These successes demonstrate the flexibility in engineering and administration of sdAbs as well as the variety of diseases that can be tackled. It will probably not take long before sdAbs with their considerable potential as a diagnostic and therapeutic agent will enter the market for bacterial infectious diseases and will contribute to public health.
QQ, HL wrote the review under the supervision of YW and SZ. WH, YG, JZ, FZ, JS and SM made the figure, tables and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 31870132, No. 82072237), Shaanxi Province Natural Science Funding, and Institutional Foundation of the First Affiliated Hospital of Xi’an Jiaotong University. SZ was supported by Northwest A&F University Star-up Funding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Deusenbery C, Wang YY, Shukla A. Recent innovations in bacterial infection detection and treatment. ACS Infect Dis (2021) 7(4):695–720. doi: 10.1021/acsinfecdis.0c00890
2. Lagier JC, Edouard S, Pagnier I, Mediannikov O, Drancourt M, Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev (2015) 28(1):208–36. doi: 10.1128/Cmr.00110-14
3. Nagy E, Boyanova L, Justesen US, Infe ESGA. How to isolate, identify and determine antimicrobial susceptibility of anaerobic bacteria in routine laboratories. Clin Microbiol Infect (2018) 24(11):1139–48. doi: 10.1016/j.cmi.2018.02.008
4. Abram TJ, Cherukury H, Ou CY, Vu T, Toledano M, Li YY, et al. Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital pcr. Lab Chip (2020) 20(3):477–89. doi: 10.1039/c9lc01212e
5. Solomon SL, Oliver KB. Antibiotic resistance threats in the united states: Stepping back from the brink. Am Fam Phys (2014) 89(12):938–41.
6. Lupo A, Coyne S, Berendonk TU. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front Microbiol (2012) 3:18. doi: 10.3389/fmicb.2012.00018
7. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol R (2010) 74(3):417–33. doi: 10.1128/Mmbr.00016-10
8. Jernigan JA, Hatfield KM, Wolford H, Nelson RE, Olubajo B, Reddy SC, et al. Multidrug-resistant bacterial infections in us hospitalized patients, 2012-2017. New Engl J Med (2020) 382(14):1309–19. doi: 10.1056/NEJMoa1914433
9. Kaplon H, Chenoweth A, Crescioli S, Reichert JM. Antibodies to watch in 2022. MAbs (2022) 14(1):2014296. doi: 10.1080/19420862.2021.2014296
10. Samaranayake H, Wirth T, Schenkwein D, Raty JK, Yla-Herttuala S. Challenges in monoclonal antibody-based therapies. Ann Med (2009) 41(5):322–31. doi: 10.1080/07853890802698842
11. Ingram JR, Schmidt FI, Ploegh HL. Exploiting nanobodies' singular traits. Annu Rev Immunol (2018) 36:695–715. doi: 10.1146/annurev-immunol-042617-053327
12. Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature (1993) 363(6428):446–8. doi: 10.1038/363446a0
13. Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature (1995) 374(6518):168–73. doi: 10.1038/374168a0
14. Uchanski T, Pardon E, Steyaert J. Nanobodies to study protein conformational states. Curr Opin Struct Biol (2020) 60:117–23. doi: 10.1016/j.sbi.2020.01.003
15. Koehl A, Hu H, Feng D, Sun B, Zhang Y, Robertson MJ, et al. Structural insights into the activation of metabotropic glutamate receptors. Nature (2019) 566(7742):79–84. doi: 10.1038/s41586-019-0881-4
16. Warne T, Edwards PC, Dore AS, Leslie AGW, Tate CG. Molecular basis for high-affinity agonist binding in gpcrs. Science (2019) 364(6442):775–8. doi: 10.1126/science.aau5595
17. Drees C, Raj AN, Kurre R, Busch KB, Haase M, Piehler J. Engineered upconversion nanoparticles for resolving protein interactions inside living cells. Angew Chem Int Ed Engl (2016) 55(38):11668–72. doi: 10.1002/anie.201603028
18. Traenkle B, Rothbauer U. Under the microscope: Single-domain antibodies for live-cell imaging and super-resolution microscopy. Front Immunol (2017) 8:1030. doi: 10.3389/fimmu.2017.01030
19. Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, et al. Cortical constriction during abscission involves helices of escrt-Iii-Dependent filaments. Science (2011) 331(6024):1616–20. doi: 10.1126/science.1201847
20. Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for gfp-based super-resolution microscopy Via nanobodies. Nat Methods (2012) 9(6):582–4. doi: 10.1038/nmeth.1991
21. Feng Y, Zhou Z, McDougald D, Meshaw RL, Vaidyanathan G, Zalutsky MR. Site-specific radioiodination of an anti-Her2 single domain antibody fragment with a residualizing prosthetic agent. Nucl Med Biol (2021) 92:171–83. doi: 10.1016/j.nucmedbio.2020.05.002
22. Farasat A, Rahbarizadeh F, Ahmadvand D, Ranjbar S, Khoshtinat Nikkhoi S. Effective suppression of tumour cells by oligoclonal Her2-targeted delivery of liposomal doxorubicin. J Liposome Res (2019) 29(1):53–65. doi: 10.1080/08982104.2018.1430829
23. Li T, Cai H, Yao H, Zhou B, Zhang N, van Vlissingen MF, et al. A synthetic nanobody targeting rbd protects hamsters from sars-Cov-2 infection. Nat Commun (2021) 12(1):4635. doi: 10.1038/s41467-021-24905-z
24. Gaiotto T, Ramage W, Ball C, Risley P, Carnell GW, Temperton N, et al. Nanobodies mapped to cross-reactive and divergent epitopes on a(H7n9) influenza hemagglutinin using yeast display. Sci Rep (2021) 11(1):3126. doi: 10.1038/s41598-021-82356-4
25. Weiss RA, Verrips CT. Nanobodies that neutralize hiv. Vaccines (Basel) (2019) 7(3):77. doi: 10.3390/vaccines7030077
26. Caljon G, Hussain S, Vermeiren L, Van Den Abbeele J. Description of a nanobody-based competitive immunoassay to detect tsetse fly exposure. PloS Negl Trop Dis (2015) 9(2):e0003456. doi: 10.1371/journal.pntd.0003456
27. Tremblay JM, Vazquez-Cintron E, Lam KH, Mukherjee J, Bedenice D, Ondeck CA, et al. Camelid vhh antibodies that neutralize botulinum neurotoxin serotype e intoxication or protease function. Toxins (Basel) (2020) 12(10):611. doi: 10.3390/toxins12100611
28. Sroga P, Safronetz D, Stein DR. Nanobodies: A new approach for the diagnosis and treatment of viral infectious diseases. Future Virol (2020) 15(3):195–205. doi: 10.2217/fvl-2019-0167
29. Chen F, Liu Z, Jiang F. Prospects of neutralizing nanobodies against sars-Cov-2. Front Immunol (2021) 12:690742. doi: 10.3389/fimmu.2021.690742
30. Wu YL, Jiang SB, Ying TL. Single-domain antibodies as therapeutics against human viral diseases. Front Immunol (2017) 8:1802. doi: 10.3389/fimmu.2017.01802
31. Salvador JP, Vilaplana L, Marco MP. Nanobody: Outstanding features for diagnostic and therapeutic applications. Anal Bioanal Chem (2019) 411(9):1703–13. doi: 10.1007/s00216-019-01633-4
32. Li C, Tang ZR, Hu ZX, Wang YW, Yang XM, Mo FZ, et al. Natural single-domain antibody-nanobody: A novel concept in the antibody field. J BioMed Nanotechnol (2018) 14(1):1–19. doi: 10.1166/jbn.2018.2463
33. Wang YZ, Fan Z, Shao L, Kong XW, Hou XJ, Tian DR, et al. Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. Int J Nanomed (2016) 11:3287–302. doi: 10.2147/Ijn.S107194
34. Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, et al. Single domain antibodies: Promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immun (2009) 198(3):157–74. doi: 10.1007/s00430-009-0116-7
35. Mitchell LS, Colwell LJ. Analysis of nanobody paratopes reveals greater diversity than classical antibodies. Protein Eng Des Sel (2018) 31(7-8):267–75. doi: 10.1093/protein/gzy017
36. Jindal V, Khoury J, Gupta R, Jaiyesimi I. Current status of chimeric antigen receptor T-cell therapy in multiple myeloma. Am J Clin Oncol-Canc (2020) 43(5):371–7. doi: 10.1097/Coc.0000000000000669
37. Li T, Bourgeois JP, Celli S, Glacial F, Le Sourd AM, Mecheri S, et al. Cell-penetrating anti-gfap vhh and corresponding fluorescent fusion protein vhh-gfp spontaneously cross the blood-brain barrier and specifically recognize astrocytes: Application to brain imaging. FASEB J (2012) 26(10):3969–79. doi: 10.1096/fj.11-201384
38. Abulrob A, Sprong H, Van Bergen en Henegouwen P, Stanimirovic D. The blood-brain barrier transmigrating single domain antibody: Mechanisms of transport and antigenic epitopes in human brain endothelial cells. J Neurochem (2005) 95(4):1201–14. doi: 10.1111/j.1471-4159.2005.03463.x
39. Hoefman S, Ottevaere I, Baumeister J, Sargentini-Maier M. Pre-clinical intravenous serum pharmacokinetics of albumin binding and non-Half-Life extended nanobodies®. Antibodies (2015) 4(3):141–56. doi: 10.3390/antib4030141
40. Kijanka M, Dorresteijn B, Oliveira S, Henegouwen PMPVE. Nanobody-based cancer therapy of solid tumors. Nanomedicine-Uk (2015) 10(1):161–74. doi: 10.2217/nnm.14.178
41. Wang L, Zhang GY, Qin L, Ye HL, Wang Y, Long B, et al. Anti-egfr binding nanobody delivery system to improve the diagnosis and treatment of solid tumours. Recent Pat Anti-Canc (2020) 15(3):200–11. doi: 10.2174/1574892815666200904111728
42. Belanger K, Iqbal U, Tanha J, MacKenzie R, Moreno M, Stanimirovic D. Single-domain antibodies as therapeutic and imaging agents for the treatment of cns diseases. Antibodies (2019) 8(2):1. doi: 10.3390/antib8020027
43. Pothin E, Lesuisse D, Lafaye P. Brain delivery of single-domain antibodies: A focus on vhh and vnar. Pharmaceutics (2020) 12(10):937. doi: 10.3390/pharmaceutics12100937
44. van der Linden RHJ, Frenken LGJ, de Geus B, Harmsen MM, Ruuls RC, Stok W, et al. Comparison of physical chemical properties of llama V-hh antibody fragments and mouse monoclonal antibodies. Bba-Protein Struct M (1999) 1431(1):37–46. doi: 10.1016/S0167-4838(99)00030-8
45. De Vos J, Devoogdt N, Lahoutte T, Muyldermans S. Camelid single-domain antibody-fragment engineering for (Pre)Clinical in vivo molecular imaging applications: Adjusting the bullet to its target. Expert Opin Biol Th (2013) 13(8):1149–60. doi: 10.1517/14712598.2013.800478
46. Oliveira S, Heukers R, Sornkom J, Kok RJ, Henegouwen PMPVE. Targeting tumors with nanobodies for cancer imaging and therapy. J Controlled Release (2013) 172(3):607–17. doi: 10.1016/j.jconrel.2013.08.298
47. Omidfar K, Zanjani FSA, Hagh AG, Azizi MD, Rasouli SJ, Kashanian S. Efficient growth inhibition of egfr over-expressing tumor cells by an anti-egfr nanobody. Mol Biol Rep (2013) 40(12):6737–45. doi: 10.1007/s11033-013-2790-1
48. Hu YZ, Liu CX, Muyldermans S. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol (2017) 8:1442. doi: 10.3389/fimmu.2017.01442
49. Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biot (2007) 77(1):13–22. doi: 10.1007/s00253-007-1142-2
50. Bannas P, Hambach J, Koch-Nolte F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol (2017) 8:1603. doi: 10.3389/fimmu.2017.01603
51. Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol (2011) 21(4):567–72. doi: 10.1016/j.sbi.2011.06.011
52. Teplyakov A, Obmolova G, Malia TJ, Luo JQ, Muzammil S, Sweet R, et al. Structural diversity in a human antibody germline library. Mabs (2016) 8(6):1045–63. doi: 10.1080/19420862.2016.1190060
53. Kovalenko OV, Olland A, Piche-Nicholas N, Godbole A, King D, Svenson K, et al. Atypical antigen recognition mode of a shark immunoglobulin new antigen receptor (Ignar) variable domain characterized by humanization and structural analysis. J Biol Chem (2013) 288(24):17408–19. doi: 10.1074/jbc.M112.435289
54. Ackaert C, Smiejkowska N, Xavier C, Sterckx YGJ, Denies S, Stijlemans B, et al. Immunogenicity risk profile of nanobodies. Front Immunol (2021) 12:632687. doi: 10.3389/fimmu.2021.632687
55. Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem (2009) 284(5):3273–84. doi: 10.1074/jbc.M806889200
56. Sang Z, Xiang Y, Bahar I, Shi Y. Llamanade: An open-source computational pipeline for robust nanobody humanization. Structure (2022) 30(3):418–29 e3. doi: 10.1016/j.str.2021.11.006
57. Muyldermans S. Nanobodies: Natural single-domain antibodies. Annu Rev Biochem (2013) 82:775–97. doi: 10.1146/annurev-biochem-063011-092449
58. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, gems): A prospective, case-control study. Lancet (2013) 382(9888):209–22. doi: 10.1016/s0140-6736(13)60844-2
59. Moonens K, Van den Broeck I, Okello E, Pardon E, De Kerpel M, Remaut H, et al. Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic escherichia coli by llama single domain antibodies. Vet Res (2015) 46:14. doi: 10.1186/s13567-015-0151-x
60. Amcheslavsky A, Wallace AL, Ejemel M, Li Q, McMahon CT, Stoppato M, et al. Anti-cfae nanobodies provide broad cross-protection against major pathogenic enterotoxigenic escherichia coli strains, with implications for vaccine design. Sci Rep (2021) 11(1):2751. doi: 10.1038/s41598-021-81895-0
61. Chen X, Gao S, Jiao X, Liu XF. Prevalence of serogroups and virulence factors of escherichia coli strains isolated from pigs with postweaning diarrhoea in Eastern China. Vet Microbiol (2004) 103(1-2):13–20. doi: 10.1016/j.vetmic.2004.06.014
62. Osek J. Prevalence of virulence factors of escherichia coli strains isolated from diarrheic and healthy piglets after weaning. Vet Microbiol (1999) 68(3-4):209–17. doi: 10.1016/s0378-1135(99)00109-1
63. Harmsen MM, van Solt CB, Hoogendoorn A, van Zijderveld FG, Niewold TA, van der Meulen J. Escherichia coli F4 fimbriae specific llama single-domain antibody fragments effectively inhibit bacterial adhesion in vitro but poorly protect against diarrhoea. Vet Microbiol (2005) 111(1-2):89–98. doi: 10.1016/j.vetmic.2005.09.005
64. Fiil BK, Thrane SW, Pichler M, Kittila T, Ledsgaard L, Ahmadi S, et al. Orally active bivalent vhh construct prevents proliferation of F4(+) enterotoxigenic escherichia coli in weaned piglets. iScience (2022) 25(4):104003. doi: 10.1016/j.isci.2022.104003
65. de Geus B, Harmsen M, van Zijderveld F. Prevention of diarrhoea using pathogen specific monoclonal antibodies in an experimental enterotoxigenice. coliinfection in germfree piglets. Vet Q (1998) 20(sup3):87–9. doi: 10.1080/01652176.1998.9694978
66. Ma JKC, Hunjan M, Smith R, Kelly C, Lehner T. An investigation into the mechanism of protection by local passive-immunization with monoclonal-antibodies against streptococcus-mutans. Infect Immun (1990) 58(10):3407–14. doi: 10.1128/Iai.58.10.3407-3414.1990
67. van Zijderveld FG, van Zijderveld-van Bemmel AM, Bakker D. The F41 adhesin of enterotoxigenic escherichia coli: Inhibition of adhesion by monoclonal antibodies. Vet Q (1998) 20 Suppl 3:S73–8. doi: 10.1080/01652176.1998.9694974
68. Virdi V, Coddens A, De Buck S, Millet S, Goddeeris BM, Cox E, et al. Orally fed seeds producing designer igas protect weaned piglets against enterotoxigenic escherichia coli infection. Proc Natl Acad Sci U.S.A. (2013) 110(29):11809–14. doi: 10.1073/pnas.1301975110
69. Moonens K, De Kerpel M, Coddens A, Cox E, Pardon E, Remaut H, et al. Nanobody mediated inhibition of attachment of F18 fimbriae expressing escherichia coli. PloS One (2014) 9(12):e114691. doi: 10.1371/journal.pone.0114691
70. Bernedo-Navarro RA, Romao E, Yano T, Pinto J, De Greve H, Sterckx YG, et al. Structural basis for the specific neutralization of Stx2a with a camelid single domain antibody fragment. Toxins (Basel) (2018) 10(3):108. doi: 10.3390/toxins10030108
71. Lo AW, Moonens K, De Kerpel M, Brys L, Pardon E, Remaut H, et al. The molecular mechanism of shiga toxin Stx2e neutralization by a single-domain antibody targeting the cell receptor-binding domain. J Biol Chem (2014) 289(36):25374–81. doi: 10.1074/jbc.M114.566257
72. Tremblay JM, Mukherjee J, Leysath CE, Debatis M, Ofori K, Baldwin K, et al. A single vhh-based toxin-neutralizing agent and an effector antibody protect mice against challenge with shiga toxins 1 and 2. Infect Immun (2013) 81(12):4592–603. doi: 10.1128/IAI.01033-13
73. Mejias MP, Hiriart Y, Lauche C, Fernandez-Brando RJ, Pardo R, Bruballa A, et al. Development of camelid single chain antibodies against shiga toxin type 2 (Stx2) with therapeutic potential against hemolytic uremic syndrome (Hus). Sci Rep (2016) 6:24913. doi: 10.1038/srep24913
74. Melli LJ, Zylberman V, Hiriart Y, Lauche CE, Baschkier A, Pardo R, et al. Development and evaluation of a novel vhh-based immunocapture assay for high-sensitivity detection of shiga toxin type 2 (Stx2) in stool samples. J Clin Microbiol (2020) 58(3):e01566-19. doi: 10.1128/JCM.01566-19
75. Adams H, Horrevoets WM, Adema SM, Carr HE, van Woerden RE, Koster M, et al. Inhibition of biofilm formation by camelid single-domain antibodies against the flagellum of pseudomonas aeruginosa. J Biotechnol (2014) 186:66–73. doi: 10.1016/j.jbiotec.2014.06.029
76. Ardekani LS, Gargari SL, Rasooli I, Bazl MR, Mohammadi M, Ebrahimizadeh W, et al. A novel nanobody against urease activity of helicobacter pylori. Int J Infect Dis (2013) 17(9):e723–8. doi: 10.1016/j.ijid.2013.02.015
77. Hoseinpoor R, Mousavi Gargari SL, Rasooli I, Rajabibazl M, Shahi B. Functional mutations in and characterization of vhh against helicobacter pylori urease. Appl Biochem Biotechnol (2014) 172(6):3079–91. doi: 10.1007/s12010-014-0750-4
78. Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat Rev Microbiol (2009) 7(7):526–36. doi: 10.1038/nrmicro2164
79. Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers (2016) 2:16020. doi: 10.1038/nrdp.2016.20
80. Huang H, Fang H, Weintraub A, Nord CE. Distinct ribotypes and rates of antimicrobial drug resistance in clostridium difficile from shanghai and Stockholm. Clin Microbiol Infect (2009) 15(12):1170–3. doi: 10.1111/j.1469-0691.2009.02992.x
81. Hussack G, Arbabi-Ghahroudi M, van Faassen H, Songer JG, Ng KK, MacKenzie R, et al. Neutralization of clostridium difficile toxin a with single-domain antibodies targeting the cell receptor binding domain. J Biol Chem (2011) 286(11):8961–76. doi: 10.1074/jbc.M110.198754
82. Chumbler NM, Rutherford SA, Zhang Z, Farrow MA, Lisher JP, Farquhar E, et al. Crystal structure of clostridium difficile toxin a. Nat Microbiol (2016) 1(1):15002. doi: 10.1038/nmicrobiol.2015.2
83. Chen P, Tao L, Wang TY, Zhang J, He AN, Lam KH, et al. Structural basis for recognition of frizzled proteins by clostridium difficile toxin b. Science (2018) 360(6389):664–9. doi: 10.1126/science.aar1999
84. Tao L, Zhang J, Meraner P, Tovaglieri A, Wu X, Gerhard R, et al. Frizzled proteins are colonic epithelial receptors for c. Difficile Toxin B Nat (2016) 538(7625):350–5. doi: 10.1038/nature19799
85. Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, et al. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for clostridium difficile toxin b. Cell Res (2015) 25(2):157–68. doi: 10.1038/cr.2014.169
86. LaFrance ME, Farrow MA, Chandrasekaran R, Sheng J, Rubin DH, Lacy DB. Identification of an epithelial cell receptor responsible for clostridium difficile tcdb-induced cytotoxicity. Proc Natl Acad Sci U.S.A. (2015) 112(22):7073–8. doi: 10.1073/pnas.1500791112
87. Hussack G, Ryan S, van Faassen H, Rossotti M, MacKenzie CR, Tanha J. Neutralization of clostridium difficile toxin b with vhh-fc fusions targeting the delivery and crops domains. PloS One (2018) 13(12):e0208978. doi: 10.1371/journal.pone.0208978
88. Schmidt DJ, Beamer G, Tremblay JM, Steele JA, Kim HB, Wang Y, et al. A tetraspecific vhh-based neutralizing antibody modifies disease outcome in three animal models of clostridium difficile infection. Clin Vaccine Immunol (2016) 23(9):774–84. doi: 10.1128/CVI.00730-15
89. Chen P, Lam KH, Liu Z, Mindlin FA, Chen B, Gutierrez CB, et al. Structure of the full-length clostridium difficile toxin b. Nat Struct Mol Biol (2019) 26(8):712–9. doi: 10.1038/s41594-019-0268-0
90. Yang Z, Schmidt D, Liu W, Li S, Shi L, Sheng J, et al. A novel multivalent, single-domain antibody targeting tcda and tcdb prevents fulminant clostridium difficile infection in mice. J Infect Dis (2014) 210(6):964–72. doi: 10.1093/infdis/jiu196
91. Yang Z, Shi L, Yu H, Zhang Y, Chen K, Saint Fleur A, et al. Intravenous adenovirus expressing a multi-specific, single-domain antibody neutralizing tcda and tcdb protects mice from clostridium difficile infection. Pathog Dis (2016) 74(7):ftw078. doi: 10.1093/femspd/ftw078
92. Andersen KK, Strokappe NM, Hultberg A, Truusalu K, Smidt I, Mikelsaar RH, et al. Neutralization of clostridium difficile toxin b mediated by engineered lactobacilli that produce single-domain antibodies. Infect Immun (2016) 84(2):395–406. doi: 10.1128/IAI.00870-15
93. Chen KV, Zhu YX, Zhang YR, Hamza T, Yu H, Saint Fleur A, et al. A probiotic yeast-based immunotherapy against clostridioides difficile infection. Sci Transl Med (2020) 12(567):1. doi: 10.1126/scitranslmed.aax4905
94. Kandalaft H, Hussack G, Aubry A, van Faassen H, Guan Y, Arbabi-Ghahroudi M, et al. Targeting surface-layer proteins with single-domain antibodies: A potential therapeutic approach against clostridium difficile-associated disease. Appl Microbiol Biotechnol (2015) 99(20):8549–62. doi: 10.1007/s00253-015-6594-1
95. Shali A, Hasannia S, Gashtasbi F, Abdous M, Shahangian SS, Jalili S. Generation and screening of efficient neutralizing single domain antibodies (Vhhs) against the critical functional domain of anthrax protective antigen (Pa). Int J Biol Macromol (2018) 114:1267–78. doi: 10.1016/j.ijbiomac.2018.03.034
96. Moayeri M, Leysath CE, Tremblay JM, Vrentas C, Crown D, Leppla SH, et al. A heterodimer of a vhh (Variable domains of camelid heavy chain-only) antibody that inhibits anthrax toxin cell binding linked to a vhh antibody that blocks oligomer formation is highly protective in an anthrax spore challenge model. J Biol Chem (2015) 290(10):6584–95. doi: 10.1074/jbc.M114.627943
97. Moayeri M, Tremblay JM, Debatis M, Dmitriev IP, Kashentseva EA, Yeh AJ, et al. Adenoviral expression of a bispecific vhh-based neutralizing agent that targets protective antigen provides prophylactic protection from anthrax in mice. Clin Vaccine Immunol (2016) 23(3):213–8. doi: 10.1128/CVI.00611-15
98. Vrentas CE, Moayeri M, Keefer AB, Greaney AJ, Tremblay J, O'Mard D, et al. A diverse set of single-domain antibodies (Vhhs) against the anthrax toxin lethal and edema factors provides a basis for construction of a bispecific agent that protects against anthrax infection. J Biol Chem (2016) 291(41):21596–606. doi: 10.1074/jbc.M116.749184
99. Gerbino E, Carasi P, Mobili P, Serradell MA, Gomez-Zavaglia A. Role of s-layer proteins in bacteria. World J Microbiol Biotechnol (2015) 31(12):1877–87. doi: 10.1007/s11274-015-1952-9
100. Fioravanti A, Van Hauwermeiren F, van der Verren SE, Jonckheere W, Goncalves A, Pardon E, et al. Structure of s-layer protein sap reveals a mechanism for therapeutic intervention in anthrax. Nat Microbiol (2019) 4(11):1805–14. doi: 10.1038/s41564-019-0499-1
101. Rossetto O, Pirazzini M, Montecucco C. Botulinum neurotoxins: Genetic, structural and mechanistic insights. Nat Rev Microbiol (2014) 12(8):535–49. doi: 10.1038/nrmicro3295
102. Arnon SS. Botulinum toxin as a biological weapon: Medical and public health management. Jama-J Am Med Assoc (2001) 285(16):2081–.1. doi: 10.1001/jama.285.8.1059
103. Bakherad H, Mousavi Gargari SL, Rasooli I, Rajabibazl M, Mohammadi M, Ebrahimizadeh W, et al. In vivo neutralization of botulinum neurotoxins serotype e with heavy-chain camelid antibodies (Vhh). Mol Biotechnol (2013) 55(2):159–67. doi: 10.1007/s12033-013-9669-1
104. Thanongsaksrikul J, Srimanote P, Maneewatch S, Choowongkomon K, Tapchaisri P, Makino SI, et al. A V h h that neutralizes the zinc metalloproteinase activity of botulinum neurotoxin type a. J Biol Chem (2010) 285(13):9657–66. doi: 10.1074/jbc.M109.073163
105. Dong J, Thompson AA, Fan Y, Lou J, Conrad F, Ho M, et al. A single-domain llama antibody potently inhibits the enzymatic activity of botulinum neurotoxin by binding to the non-catalytic alpha-exosite binding region. J Mol Biol (2010) 397(4):1106–18. doi: 10.1016/j.jmb.2010.01.070
106. Tremblay JM, Kuo CL, Abeijon C, Sepulveda J, Oyler G, Hu X, et al. Camelid single domain antibodies (Vhhs) as neuronal cell intrabody binding agents and inhibitors of clostridium botulinum neurotoxin (Bont) proteases. Toxicon (2010) 56(6):990–8. doi: 10.1016/j.toxicon.2010.07.003
107. Lam KH, Tremblay JM, Perry K, Ichtchenko K, Shoemaker CB, Jin R. Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies. PloS Pathog (2022) 18(1):e1010169. doi: 10.1371/journal.ppat.1010169
108. Lam KH, Tremblay JM, Vazquez-Cintron E, Perry K, Ondeck C, Webb RP, et al. Structural insights into rational design of single-domain antibody-based antitoxins against botulinum neurotoxins. Cell Rep (2020) 30(8):2526–39 e6. doi: 10.1016/j.celrep.2020.01.107
109. Yao G, Lam KH, Weisemann J, Peng L, Krez N, Perry K, et al. A camelid single-domain antibody neutralizes botulinum neurotoxin a by blocking host receptor binding. Sci Rep (2017) 7(1):7438. doi: 10.1038/s41598-017-07457-5
110. Kuo CL, Oyler GA, Shoemaker CB. Accelerated neuronal cell recovery from botulinum neurotoxin intoxication by targeted ubiquitination. PloS One (2011) 6(5):e20352. doi: 10.1371/journal.pone.0020352
111. Godakova SA, Noskov AN, Vinogradova ID, Ugriumova GA, Solovyev AI, Esmagambetov IB, et al. Camelid vhhs fused to human fc fragments provide long term protection against botulinum neurotoxin a in mice. Toxins (Basel) (2019) 11(8):464. doi: 10.3390/toxins11080464
112. Huang NJ, Pishesha N, Mukherjee J, Zhang S, Deshycka R, Sudaryo V, et al. Genetically engineered red cells expressing single domain camelid antibodies confer long-term protection against botulinum neurotoxin. Nat Commun (2017) 8(1):423. doi: 10.1038/s41467-017-00448-0
113. Mukherjee J, Dmitriev I, Debatis M, Tremblay JM, Beamer G, Kashentseva EA, et al. Prolonged prophylactic protection from botulism with a single adenovirus treatment promoting serum expression of a vhh-based antitoxin protein. PloS One (2014) 9(8):e106422. doi: 10.1371/journal.pone.0106422
114. Lam KH, Perry K, Shoemaker CB, Jin R. Two vhh antibodies neutralize botulinum neurotoxin E1 by blocking its membrane translocation in host cells. Toxins (Basel) (2020) 12(10):616. doi: 10.3390/toxins12100616
115. Miyashita SI, Zhang J, Zhang SC, Shoemaker CB, Dong M. Delivery of single-domain antibodies into neurons using a chimeric toxin-based platform is therapeutic in mouse models of botulism. Sci Transl Med (2021) 13(575):eaaz4197. doi: 10.1126/scitranslmed.aaz4197
116. McNutt PM, Vazquez-Cintron EJ, Tenezaca L, Ondeck CA, Kelly KE, Mangkhalakhili M, et al. Neuronal delivery of antibodies has therapeutic effects in animal models of botulism. Sci Transl Med (2021) 13(575):eabd7789. doi: 10.1126/scitranslmed.abd7789
117. Hu Y, Sun Y, Gu J, Yang F, Wu S, Zhang C, et al. Selection of specific nanobodies to develop an immuno-assay detecting staphylococcus aureus in milk. Food Chem (2021) 353:129481. doi: 10.1016/j.foodchem.2021.129481
118. Ji Y, Chen L, Wang Y, Zhang K, Wu H, Liu Y, et al. Development of a double nanobody-based sandwich immunoassay for the detecting staphylococcal enterotoxin c in dairy products. Foods (2021) 10(10):2426. doi: 10.3390/foods10102426
119. Zanganeh S, Rouhani Nejad H, Mehrabadi JF, Hosseini R, Shahi B, Tavassoli Z, et al. Rapid and sensitive detection of staphylococcal enterotoxin b by recombinant nanobody using phage display technology. Appl Biochem Biotechnol (2019) 187(2):493–505. doi: 10.1007/s12010-018-2762-y
120. Tu Z, Chen Q, Li Y, Xiong Y, Xu Y, Hu N, et al. Identification and characterization of species-specific nanobodies for the detection of listeria monocytogenes in milk. Anal Biochem (2016) 493:1–7. doi: 10.1016/j.ab.2015.09.023
121. King MT, Huh I, Shenai A, Brooks TM, Brooks CL. Structural basis of vhh-mediated neutralization of the food-borne pathogen listeria monocytogenes. J Biol Chem (2018) 293(35):13626–35. doi: 10.1074/jbc.RA118.003888
122. Mukherjee J, Tremblay JM, Leysath CE, Ofori K, Baldwin K, Feng X, et al. A novel strategy for development of recombinant antitoxin therapeutics tested in a mouse botulism model. PloS One (2012) 7(1):e29941. doi: 10.1371/journal.pone.0029941
123. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell (2006) 124(4):783–801. doi: 10.1016/j.cell.2006.02.015
124. Boltana S, Roher N, Goetz FW, MacKenzie SA. Pamps, prrs and the genomics of gram negative bacterial recognition in fish. Dev Comp Immunol (2011) 35(12):1195–203. doi: 10.1016/j.dci.2011.02.010
125. Fawkner-Corbett D, Simmons A, Parikh K. Microbiome, pattern recognition receptor function in health and inflammation. Best Pract Res Cl Ga (2017) 31(6):683–91. doi: 10.1016/j.bpg.2017.11.001
126. Liao ZW, Su JG. Progresses on three pattern recognition receptor families (Tlrs, rlrs and nlrs) in teleost. Dev Comp Immunol (2021) 122:104131. doi: 10.1016/j.dci.2021.104131
127. Kirk P, Bazan JF. Pathogen recognition: Tlrs throw us a curve. Immunity (2005) 23(4):347–50. doi: 10.1016/j.immuni.2005.09.008
128. Loo YM, Gale M. Immune signaling by rig-I-Like receptors. Immunity (2011) 34(5):680–92. doi: 10.1016/j.immuni.2011.05.003
129. Motta V, Soares F, Sun T, Philpott DJ. Nod-like receptors: Versatile cytosolic sentinels. Physiol Rev (2015) 95(1):149–78. doi: 10.1152/physrev.00009.2014
130. Thaiss CA, Levy M, Itav S, Elinav E. Integration of innate immune signaling. Trends Immunol (2016) 37(2):84–101. doi: 10.1016/j.it.2015.12.003
131. Benko S, Magyarics Z, Szabo A, Rajnavolgyi E. Dendritic cell subtypes as primary targets of vaccines: The emerging role and cross-talk of pattern recognition receptors. Biol Chem (2008) 389(5):469–85. doi: 10.1515/Bc.2008.054
132. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol (2008) 8(12):911–22. doi: 10.1038/nri2436
133. Wills-Karp M. Allergen-specific pattern recognition receptor pathways. Curr Opin Immunol (2010) 22(6):777–82. doi: 10.1016/j.coi.2010.10.011
134. Liao S, Liu S, Zhang Y. Preparation of anti toll-like receptor-4 nano-antibody and its effect on gram negative sepsis. J Nanosci Nanotechnol (2021) 21(2):1048–53. doi: 10.1166/jnn.2021.18664
135. Zheng F, Zhou J, Ouyang Z, Zhang J, Wang X, Muyldermans S, et al. Development and characterization of nanobodies targeting the kupffer cell. Front Immunol (2021) 12:641819. doi: 10.3389/fimmu.2021.641819
136. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat Immunol (2010) 11(5):373–84. doi: 10.1038/ni.1863
137. O'Neill LAJ, Golenbock D, Bowie AG. The history of toll-like receptors - redefining innate immunity. Nat Rev Immunol (2013) 13(6):453–60. doi: 10.1038/nri3446
138. Deutschman CS, Tracey KJ. Sepsis: Current dogma and new perspectives. Immunity (2014) 40(4):464–76. doi: 10.1016/j.immuni.2014.04.001
139. Firmal P, Shah VK, Chattopadhyay S. Insight into Tlr4-mediated immunomodulation in normal pregnancy and related disorders. Front Immunol (2020) 11:807. doi: 10.3389/fimmu.2020.00807
140. Robertson SA, Hutchinson MR, Rice KC, Chin PY, Moldenhauer LM, Stark MJ, et al. Targeting toll-like receptor-4 to tackle preterm birth and fetal inflammatory injury. Clin Transl Immunol (2020) 9(4):1. doi: 10.1002/cti2.1121
141. Xiao Z, Kong B, Yang HJ, Dai C, Fang J, Qin TY, et al. Key player in cardiac hypertrophy, emphasizing the role of toll-like receptor 4. Front Cardiovasc Med (2020) 7:579036. doi: 10.3389/fcvm.2020.579036
142. Wang XP, Guo DQ, Li WL, Zhang Q, Jiang YY, Wang QY, et al. Danshen (Salvia miltiorrhiza) restricts Md2/Tlr4-Myd88 complex formation and signalling in acute myocardial infarction-induced heart failure. J Cell Mol Med (2020) 24(18):10677–92. doi: 10.1111/jcmm.15688
143. Tam JSY, Coller JK, Hughes PA, Prestidge CA, Bowen JM. Toll-like receptor 4 (Tlr4) antagonists as potential therapeutics for intestinal inflammation. Indian J Gastroenter (2021) 40(1):5–21. doi: 10.1007/s12664-020-01114-y
144. Unterberger S, Davies KA, Rambhatla SB, Sacre S. Contribution of toll-like receptors and the Nlrp3 inflammasome in rheumatoid arthritis pathophysiology. Immunotargets Ther (2021) 10:285–98.1. doi: 10.2147/Itt.S288547
145. Habib R. Multifaceted roles of toll-like receptors in acute kidney injury. Heliyon (2021) 7(3):1. doi: 10.1016/j.heliyon.2021.e06441
146. Vazquez-Carballo C, Guerrero-Hue M, Garcia-Caballero C, Rayego-Mateos S, Opazo-Rios L, Morgado-Pascual JL, et al. Toll-like receptors in acute kidney injury. Int J Mol Sci (2021) 22(2):1. doi: 10.3390/ijms22020816
147. Jiang DH, Liang JR, Fan J, Yu S, Chen SP, Luo Y, et al. Regulation of lung injury and repair by toll-like receptors and hyaluronan. Nat Med (2005) 11(11):1173–9. doi: 10.1038/nm1315
148. Kawata T, Bristol JR, Rossignol DP, Rose JR, Kobayashi S, Yokohama H, et al. E5531, a synthetic non-toxic lipid a derivative blocks the immunobiological activities of lipopolysaccharide. Br J Pharmacol (1999) 127(4):853–62. doi: 10.1038/sj.bjp.0702596
149. Rossignol DP, Lynn M. Antagonism of in vivo and ex vivo response to endotoxin by E5564, a synthetic lipid a analogue. J Endotoxin Res (2002) 8(6):483–8. doi: 10.1179/096805102125001127
150. Sova M, Rozman K, Svajger U, Rozman P, Gobec S. Synthesis and biological evaluation of n-Aryl-N '-(5-(2-Hydroxybenzoyl)Pyrimidin-2-Yl)Guanidines as toll-like receptor 4 antagonists. Med Chem (2016) 12(8):742–50. doi: 10.2174/1573406412666160314151900
151. Brown GD, Willment JA, Whitehead L. C-type lectins in immunity and homeostasis. Nat Rev Immunol (2018) 18(6):374–89. doi: 10.1038/s41577-018-0004-8
152. Scott C, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, et al. Bone marrow derived monocytes give rise to self-renewing and fully differentiated kupffer cells. Eur J Immunol (2016) 46:1002–. doi: 10.1038/ncomms10321
153. Zheng F, Sparkes A, De Baetselier P, Schoonooghe S, Stijlemans B, Muyldermans S, et al. Molecular imaging with kupffer cell-targeting nanobodies for diagnosis and prognosis in mouse models of liver pathogenesis. Mol Imaging Biol (2017) 19(1):49–58. doi: 10.1007/s11307-016-0976-3
154. Ouyang ZL, Felix J, Zhou JH, Pei YM, Ma BH, Hwang PM, et al. Trimeric structure of the mouse kupffer cell c-type lectin receptor Clec4f. FEBS Lett (2020) 594(1):189–98. doi: 10.1002/1873-3468.13565
Keywords: single domain antibody, nanobody, bacterial infection, diagnosis, neutralization
Citation: Qin Q, Liu H, He W, Guo Y, Zhang J, She J, Zheng F, Zhang S, Muyldermans S and Wen Y (2022) Single Domain Antibody application in bacterial infection diagnosis and neutralization. Front. Immunol. 13:1014377. doi: 10.3389/fimmu.2022.1014377
Received: 08 August 2022; Accepted: 15 September 2022;
Published: 29 September 2022.
Edited by:
Allen D. Smith, Agricultural Research Service (USDA), United StatesReviewed by:
Mahdi Habibi-Anbouhi, Pasteur Institute of Iran, IranCopyright © 2022 Qin, Liu, He, Guo, Zhang, She, Zheng, Zhang, Muyldermans and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yurong Wen, eXVyb25nLndlbkB4anR1LmVkdS5jbg==; Sicai Zhang, c2ljYWkuemhhbmdAbndhZnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.