- 1State Key Laboratory of Biocontrol, Guangdong Key Laboratory of Pharmaceutical Functional Genes, School of Life Sciences, Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Sun Yat-sen University, Higher Education Mega Center, Guangzhou, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Serum resistance is recognized as one of the most important pathogenic traits of bacterial pathogens, and no control measure is available. Based on our previous discovery that pathogenic Escherichia coli represses glycine, serine, and threonine metabolism to confer serum resistance and that the reactivation of this pathway by exogenous glycine could restore serum sensitivity, we further investigate the mechanism underlying the action of glycine in Vibrio alginolyticus. Thus, V. alginolyticus is treated with glycine, and the proteomic change is profiled with tandem mass tag-based quantitative proteomics. Compared to the control group, glycine treatment influences the expression of a total of 291 proteins. Among them, a trap-type mannitol/chloroaromatic compound transport system with periplasmic component, encoded by N646_0992, is the most significantly increased protein. In combination with the pathway enrichment analysis showing the altered fructose and mannitol metabolism, mannitol has emerged as a possible metabolite in enhancing the serum killing activity. To demonstrate this, exogenous mannitol reduces bacterial viability. This synergistic effect is further confirmed in a V. alginolyticus–Danio rerio infection model. Furthermore, the mechanism underlying mannitol-enabled serum killing is dependent on glycolysis and the pyruvate cycle that increases the deposition of complement components C3b and C5b-9 on the bacterial surface, whereas inhibiting glycolysis or the pyruvate cycle significantly weakened the synergistic effects and complement deposition. These data together suggest that mannitol is a potent metabolite in reversing the serum resistance of V. alginolyticus and has promising use in aquaculture.
Introduction
The complement system plays essential roles in innate immunity, and it consists of more than 30 proteins. Three pathways are known to activate the complement system to clear bacterial pathogens, namely: classical pathway, alternative pathway, and lectin pathway (1–3). By these pathways, the complement system not only functions by directly killing the bacteria via forming a membrane attack complex at the bacterial surface but also opsonizes bacteria for phagocytosis, wherein antigens are presented to B cells for antibody production (4–6). Thus, the complement system plays critical roles in bridging the innate immune system and the adaptive immune system during a bacterial infection.
Accumulating evidence demonstrate that bacterial pathogens have evolved different mechanisms to evade complement-mediated killing (7, 8). Three successful strategies have been recognized. Bacteria produce proteins like complement-regulator-acquiring protein and M protein family to recruit complement factors like factor H, factor H-like protein-1, and C4-binding protein so that the complement system is disabled (9, 10). Bacteria, especially Staphylococcal, express proteins that directly interact with the hub component of the complement system, C3, C5, C3 convertase, or C5 convertase, to inhibit their functions (11, 12) Staphylokinase, Pseudomonas elastase, and 56-kDa protease are known bacterial proteases to degrade C3, C3b, C5a, and IgG (13–15). Although the knowledge on serum resistance is expanding, control measures are still lacking due to the varied mechanisms that are hard to be specifically targeted.
Recently, we proposed that metabolic state determines serum resistance (16–19). By comparing serum-resistant and serum-sensitive bacteria, we find that glycine, serine, and threonine metabolism is repressed in serum-resistant bacteria. Glycine is one of the most decreased metabolites in serum-resistant bacteria. Thus, we supplement serum-resistant bacteria with exogenous glycine that reverses serum resistance and increases serum killing efficacy. This synergistic effect is confirmed both in vivo and in vitro. The underlying mechanism is that glycine depletes intracellular ATP through the inhibition of ATP synthase and downregulates purine metabolism. The reduced ATP, in turn, decreases cAMP/CRP that enhances complement-binding protein expression. However, whether other mechanisms are present in a different species of E. coli is unknown, and the identification of the working mechanism would find a new metabolite in reversing serum resistance in Vibrios.
Results
Glycine enhances fish serum to kill Vibrios
The efficacy of glycine on potentiating serum to kill Vibrio pathogens was investigated with our previously established conditions that pathogens were incubated with serum in the presence or absence of glycine (18). Clinic isolates including V. alginolyticus (ATCC33787 and 12G01), V. parahaemolyticus (ATCC17802 and VP01), V. mimicus vmi01, and V. vulnificus ATCC27562 were investigated. As shown in Figure 1, glycine alone had limited effects on the survival of Vibrios, while serum killed a minor portion of the pathogens. The presence of glycine greatly increased the susceptibility of pathogens to serum killing. The viability of the pathogens decreased 2.2–40.1 folds in the presence of glycine as compared to glycine only, whereas V. alginolyticus 12G01 was the most susceptible strain that was used for further functional study.
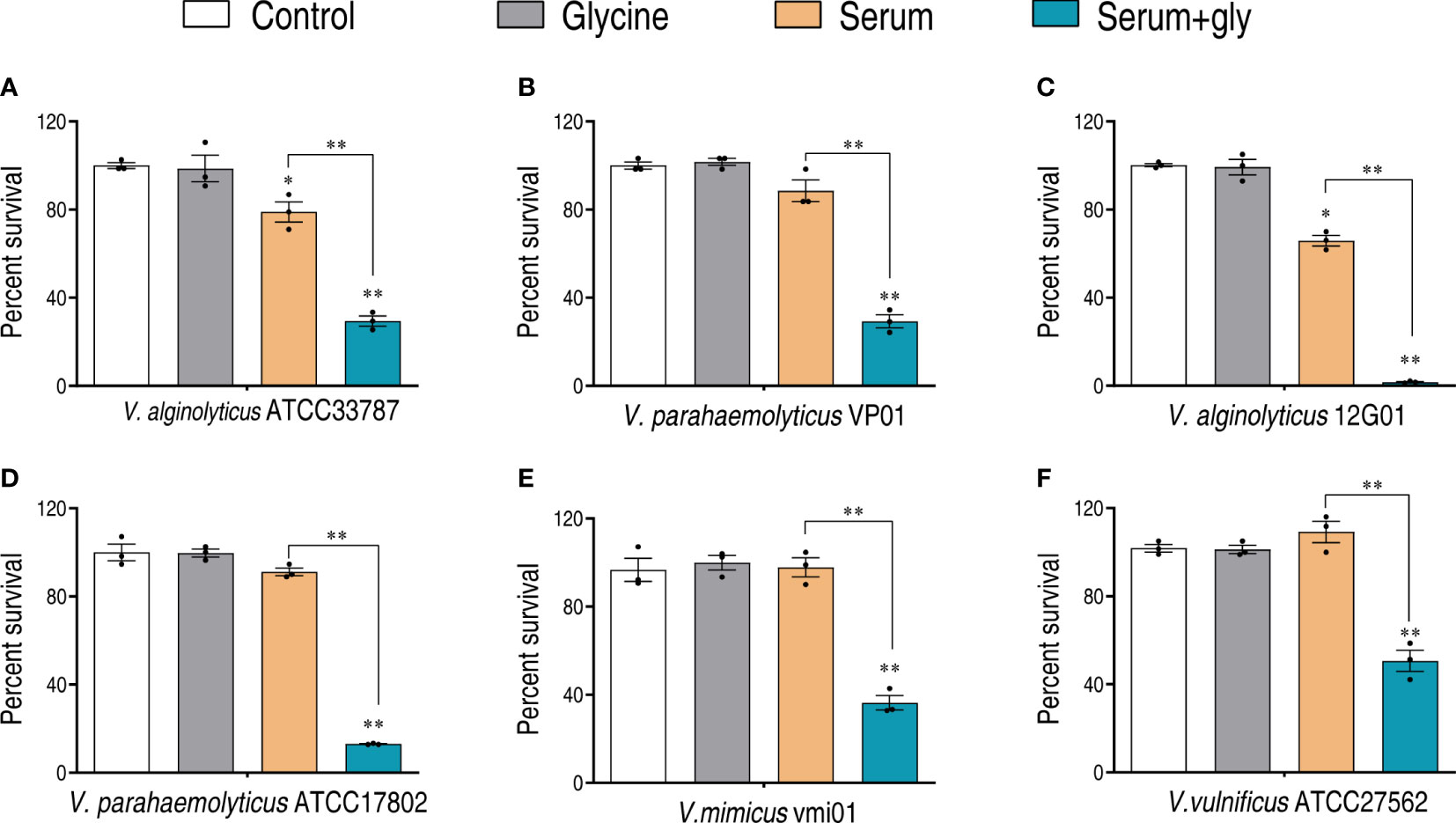
Figure 1 Glycine enhances serum killing to Vibrio. Percent survival of V. alginolyticus ATCC33787 (A), V. parahaemolyticus VP01 (B), V. alginolyticus 12G01 (C), V. parahaemolyticus ATCC17802 (D), Vibrio mimicus vmi01 (E), and V. vulnificu ATCC27562 (F) in the presence of 100 mM glycine or/and 100 μl serum. The results are displayed as means ± SEM, and significant differences are identified (*p < 0.05; **p < 0.01) as determined by Student’s t-test. At least three biological repeats were carried out.
Proteomic analysis of glycine-treated V. alginolyticus 12G01
To better understand how glycine sensitizes Vibrio to fish serum, we adopted quantitative proteomics to investigate the glycine-triggered proteomic change. Thus, V. alginolyticus 12G01 was treated with glycine for 120 min and then processed for proteomic sample preparation. Proteins were labeled with tandem mass tags (TMT) that are isobaric chemical tags, providing multiplexing capabilities for relative quantitative proteomic analysis (20). The labeled proteins were processed for mass spectrometry analysis. Lastly, a total of 2,881 proteins with unique peptides ≥2 were identified, where the corresponding false discovery rate was less than 0.05%. The ratio of the expression level between the glycine-treated group and the non-treated group was calculated, summarized as frequency, and displayed as histograms. The frequency showed a normal distribution, where the median ratio value was -0.029, implying the protein of altered expression was not skewed (Figure 2A). To further demonstrate that glycine induced proteomic change, principal component analysis was adopted, which demonstrated that the control (non-treated bacteria) and the glycine groups (glycine-treated bacteria) were clearly separated from each other (Figure 2B). Proteins of differential abundance were screened by setting the abundance ratio at greater than 1.2 or smaller than 0.83, corresponding to increased expression and decreased expression, respectively Figure 2C. Meanwhile, the number of matching peptides was greater than or equal to 1. Therefore, 291 proteins of altered abundance were identified, including 111 proteins of increased abundance and 180 proteins of decreased abundance Figure 2D. Proteins of differential expression were summarized in Supplementary Table 1.
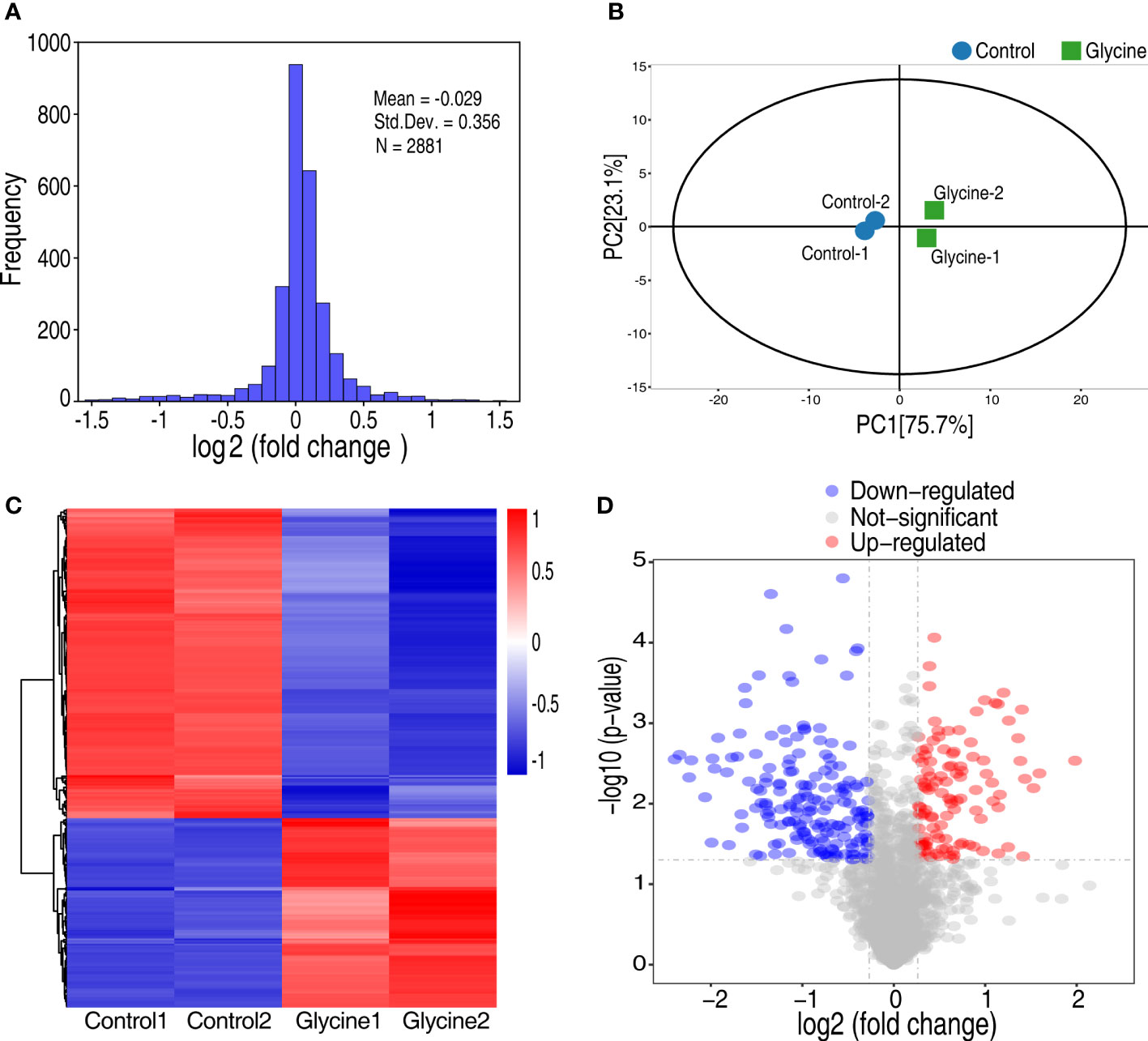
Figure 2 Analysis of glycine on the proteomics of Vibrio alginolyticus. (A) Frequency distribution of protein ratios (log2 scale, glycine vs. control). (B) Principal component analysis between glycine and control. Each dot represents the technique replicas in the plot. (C) Hierarchical cluster analysis of 291 proteins expressed with statistically significant differences (p < 0.05; fold change >1.2 or <0.83) in glycine-treated and untreated (control) samples. (D) Volcano plot of the P-values vs. the log2 protein abundance differences between glycine-treated and control samples. Colors: red, upregulation; blue, down-regulation; gray, non-significant differences.
GO enrichment of differentially expressed proteins upon glycine treatment
To visualize the parts that were influenced by glycine, the differentially expressed proteins were analyzed with Gene Ontology (GO). The terms of biological process, cellular component, and molecular functions were analyzed. A total of 10 biological processes were enriched Figure 3A. Interestingly, all the biological processes were related to metabolism, like biosynthesis of amino acids and carbohydrate metabolism. It is not surprising that glycine triggers a metabolic response since it is itself an amino acid Figure 3A. However, it should be noted that glycine affects transport activity as well as that it might have an important function on regulating serum resistance. Consistent with the altered metabolism, differentially expressed proteins were mainly localized in intracellular parts (Figure 3B). Moreover, the enrichment of glycine on molecular functions demonstrates that glycine greatly impacts the redox system since four out of 11 molecular functions were oxidoreductase activity or were iron–sulfur proteins (Figure 3C).
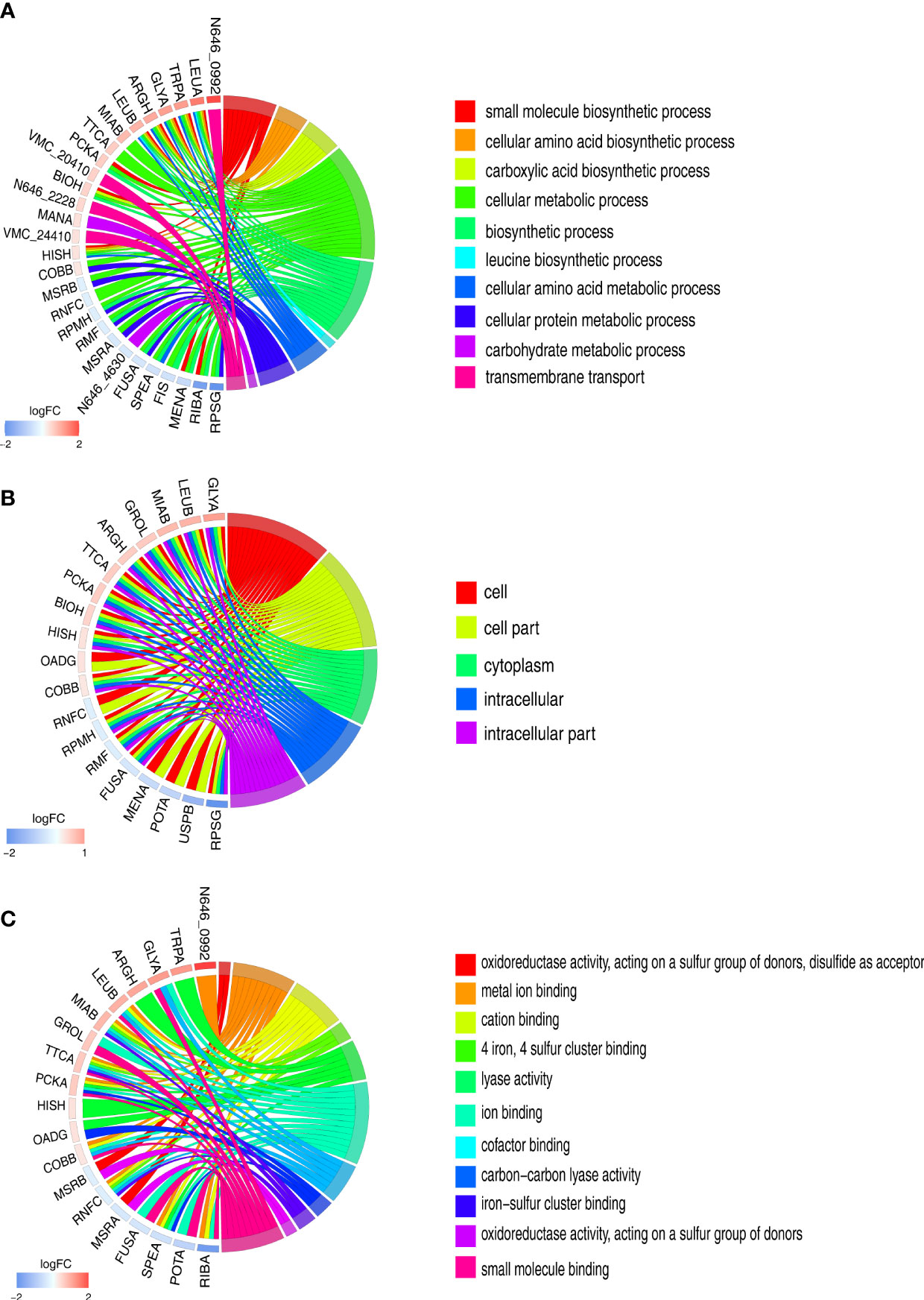
Figure 3 Gene Ontology (GO) term enrichment analysis of differentially expressed proteins. (A) Chord chart analysis of GO annotation in biological processes. (B) Chord chart analysis of GO annotation in cellular component. (C) Chord chart analysis of GO annotation in molecular functions. The left side of the GO Chord displayed whether the gene was up- or down-expression. Red represents up-expression genes and blue represents down-expression. The right side represents different GO terms with different colors. A gene was linked to a certain GO term by the colored bands.
Protein–protein interaction network analysis
An analysis of protein–protein interaction (PPI) may help uncover more proteomic signatures upon glycine treatment (21, 22). Hence, the differentially expressed proteins triggered by glycine were further analyzed by PPI and pathway enrichment analysis (Figure 4). Seven pathways were enriched, namely: glycine, serine, and threonine metabolism; fructose and mannose metabolism; pyruvate metabolism; metabolic pathways; carbon metabolism; quorum sensing; and biosynthesis of amino acids. It is not surprising that glycine, serine, and threonine metabolism was enriched since glycine directly activates this metabolic pathway. Four altered proteins (ThrB, GlyA, GcvP, and N646_1807) were identified in the proteomic data. Three proteins were involved in carbon metabolism (PckA, N646_2995, and N646_1929), whose abundance was significantly increased, while two altered proteins related with pyruvate metabolism (PdhA and N646_4476) were largely decreased in terms of abundance. Seven proteins involved in the biosynthesis of amino acids were significantly increased. Interestingly, in addition to glycine, serine, and threonine metabolism, fructose and mannose metabolism connects to pyruvate metabolism and carbon metabolism via glycolysis. These data suggest that glycine may impact fructose and mannose metabolism to enhance serum killing.
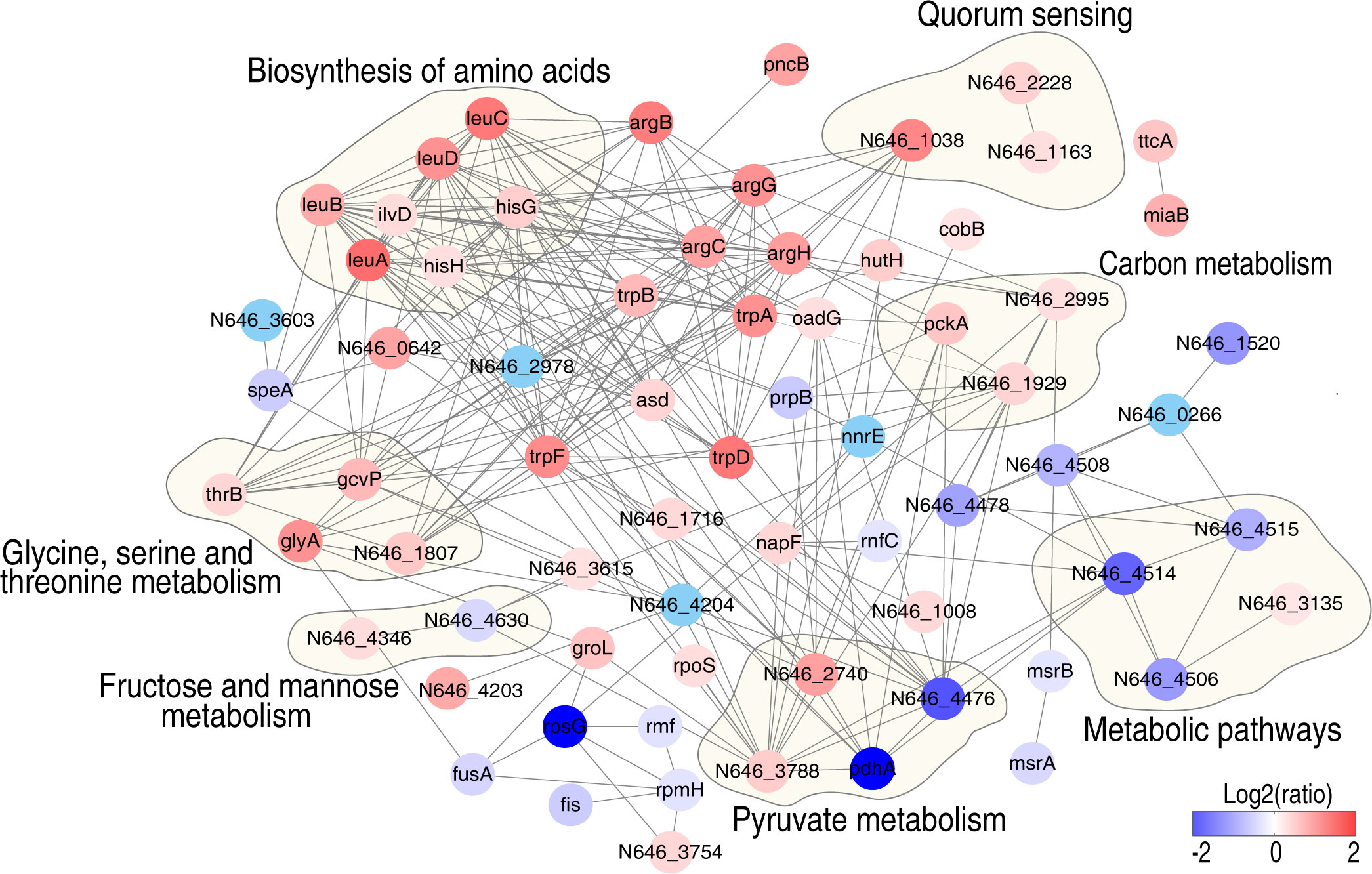
Figure 4 Interaction of differentially expressed proteins in the presence of glycine in V. alginolyticus. Protein–protein interaction prediction using STRING software. The circles indicate the enriched Kyoto Encyclopedia of Genes and Genomes pathways. Differently colored dots indicate the altered proteins, blue dots represent downregulated proteins, and red dots represent upregulated proteins, respectively (see the color scale).
Mannitol enhances serum killing of Vibrios
In combination with the pathway enrichment analysis and differentially expressed proteins, we speculate that mannitol may be a possible metabolite in enhancing the serum killing activity. There are two reasons for this in the trap-type mannitol/chloroaromatic compound transport system: periplasmic component (encoded by N646_0992) was the most significantly increased protein (3.95 folds, Supplementary Table 1), and mannitol could activate fructose and mannose metabolism. Based on this hypothesis, we tested whether mannitol could potentiate serum killing. Mannitol increased the serum killing activity in a dose-dependent manner (Figure 5A), and the best efficacy was obtained when mannitol was used at 5 mM. The mannitol-potentiated effect was confirmed by increasing the serum concentration (Figure 5B), and mannitol enhanced serum killing in a time-dependent manner (Figure 5C).
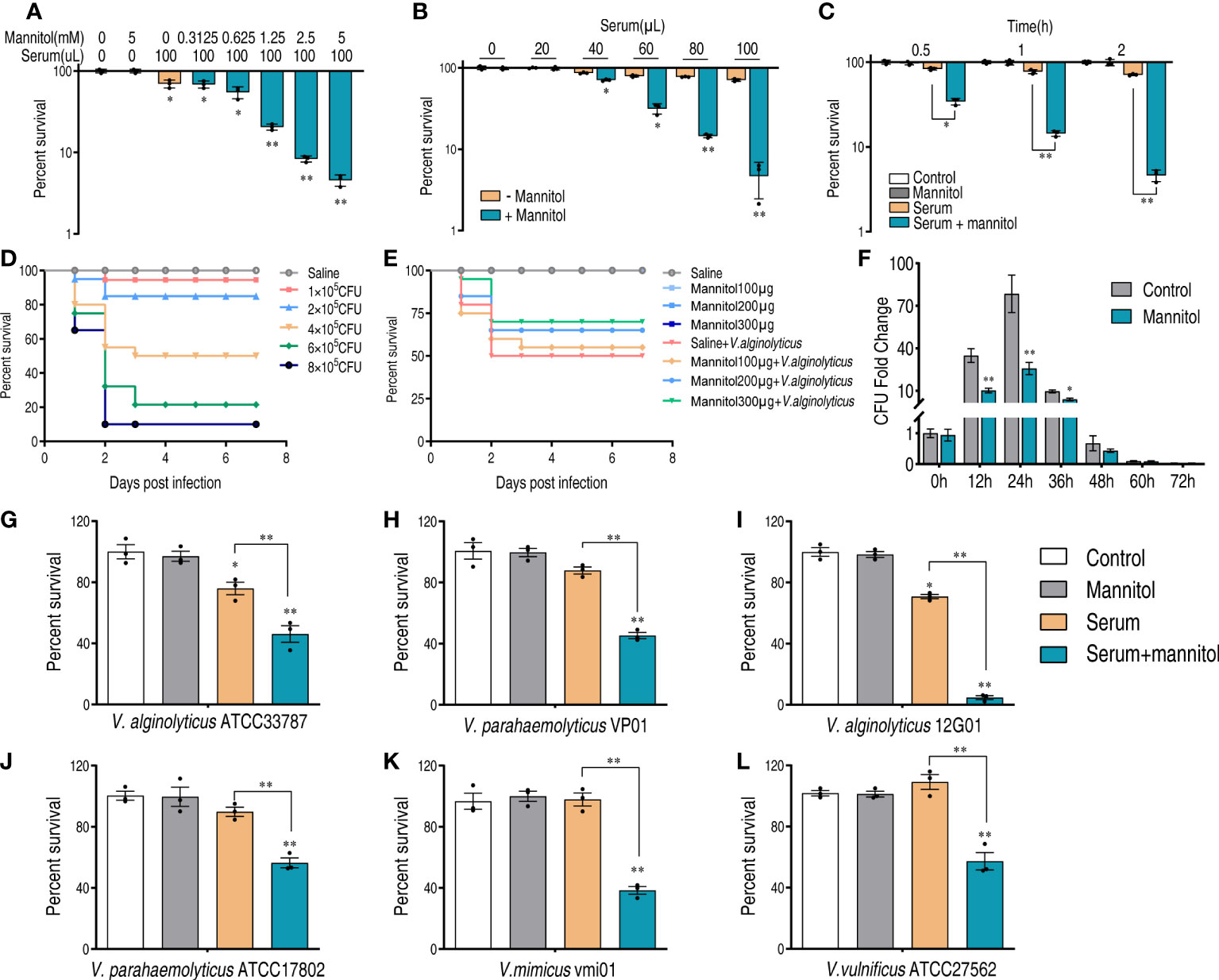
Figure 5 Mannitol promotes the killing effect of serum on Vibrios. (A) Synergetic effects of 100 μl serum and mannitol on the viability of V. alginolyticus 12G01 with the indicated dose of mannitol (0–5 mM). (B) Percent survival of V. alginolyticus 12G01 incubated with 5 mM mannitol plus serum (0–100 μl) or without mannitol. (C) Percent survival of V. alginolyticus 12G01 in the presence of 5 mM mannitol or/and 100 μl serum for the indicated length of time. (D) Determination of LD50 of D. rerio infected with V. alginolyticus. Mortality was monitored for 14 days (only 7 days are shown as no death was observed after 7 days). (E) Percent survival of D. rerio challenged with V. alginolyticus, which received mannitol treatment at 1, 4, 10, and 20 h post-infection. Mortality was monitored for 14 days (only 7 days are shown as no death was observed after 7 days). (F) The amounts of bacteria in each fish after being treated with or without mannitol (300 μg) and infected with a sublethal dose of bacterial challenge. (G–L) Percent survival of V. alginolyticus ATCC33787 (G), V. parahaemolyticus VP01 (H), V. alginolyticus 12G01 (I), V. parahaemolyticus ATCC17802 (J), V. mimicus vmi01 (K), and V. vulnificu ATCC27562 (L) in the presence of 5 mM mannitol or/and 100 μl serum. The results are displayed as means ± SEM, and significant differences are identified (*p < 0.05 and **p < 0.01) as determined by Student’s t-test. At least three biological repeats were carried out.
To confirm this result in vivo, we used a V. alginolyticus–Danio rerio infection model. The half lethal dose (LD50) was determined by infecting D. rerio with a series of doses of V. alginolyticus. Lastly, 4 × 105 colony-forming units (CFU) were used as LD50 for the following analysis (Figure 5D). D. rerio was infected with V. alginolyticus, followed by administration with saline or different amounts of mannitol as indicated at 1, 4, 10, and 20 h post-infection. Meanwhile, the same amounts of mannitol were injected into D. rerio to exclude possible toxic effects. In total, 50% of D. rerio without mannitol treatment died, whereas the survival rate was increased in a mannitol dose-dependent manner (Figure 5E). When mannitol concentration was used at 300 μg/fish, the best efficacy that increased survival for 20% was observed. Furthermore, the bacterial loads of the whole fish were quantified. The number of bacteria increased at the first 24 h and then decreased afterwards. At 48 h, the number of bacteria was similar to that at 0 h. This was consistent with the death curve in Figure 5E in that D. rerio mainly died at the first 48 h. It is noticeable that the presence of mannitol decreased the bacterial loads from 12 to 48 h and that these were the same regardless of the application of the mannitol treatment or not (Figure 5F). Furthermore, mannitol exerted a serum-promoting effect to other Vibrio strains including V. alginolyticus ATCC33787, V. parahaemolyticus VP01, V. alginolyticus 12G01, V. parahaemolyticus ATCC17802, V. mimicus vmi01, and V. vulnificus ATCC27562 (Figures 5G–L). These data suggest that mannitol promotes serum killing in vitro and bacterial clearance in vivo.
Mannitol increases the activity of glycolysis and the pyruvate cycle
There are two possibilities that mannitol may enhance the serum killing activity. One is that mannitol directly regulates the serum killing activity, and the other one is that the mannitol metabolism contributes to the serum killing activity. We speculate that the latter is the possibility since mannitol can be phosphorylated to form mannitol-1P by D-mannitol PTS permease during the transport (23, 24) (Figure 6A). To test this possibility, the expression levels of mannitol metabolism were quantified in the control, mannitol only, serum only, and mannitol plus serum groups. Mannitol alone or mannitol plus serum significantly increased the gene expression of glycolysis, especially for pfkA that was repressed by serum (Figure 6B). More importantly, several of the genes (N646_1264, pgk, gpmM, and eno) were decreased by the presence of serum but can be rescued by mannitol, suggesting that serum impaired glycolysis. Glycolysis is upstream of the pyruvate cycle (25–28). Similarly, the expression of the genes (pykA and pykF) that connect glycolysis and the pyruvate cycle was decreased by serum but enhanced by glycine plus serum.
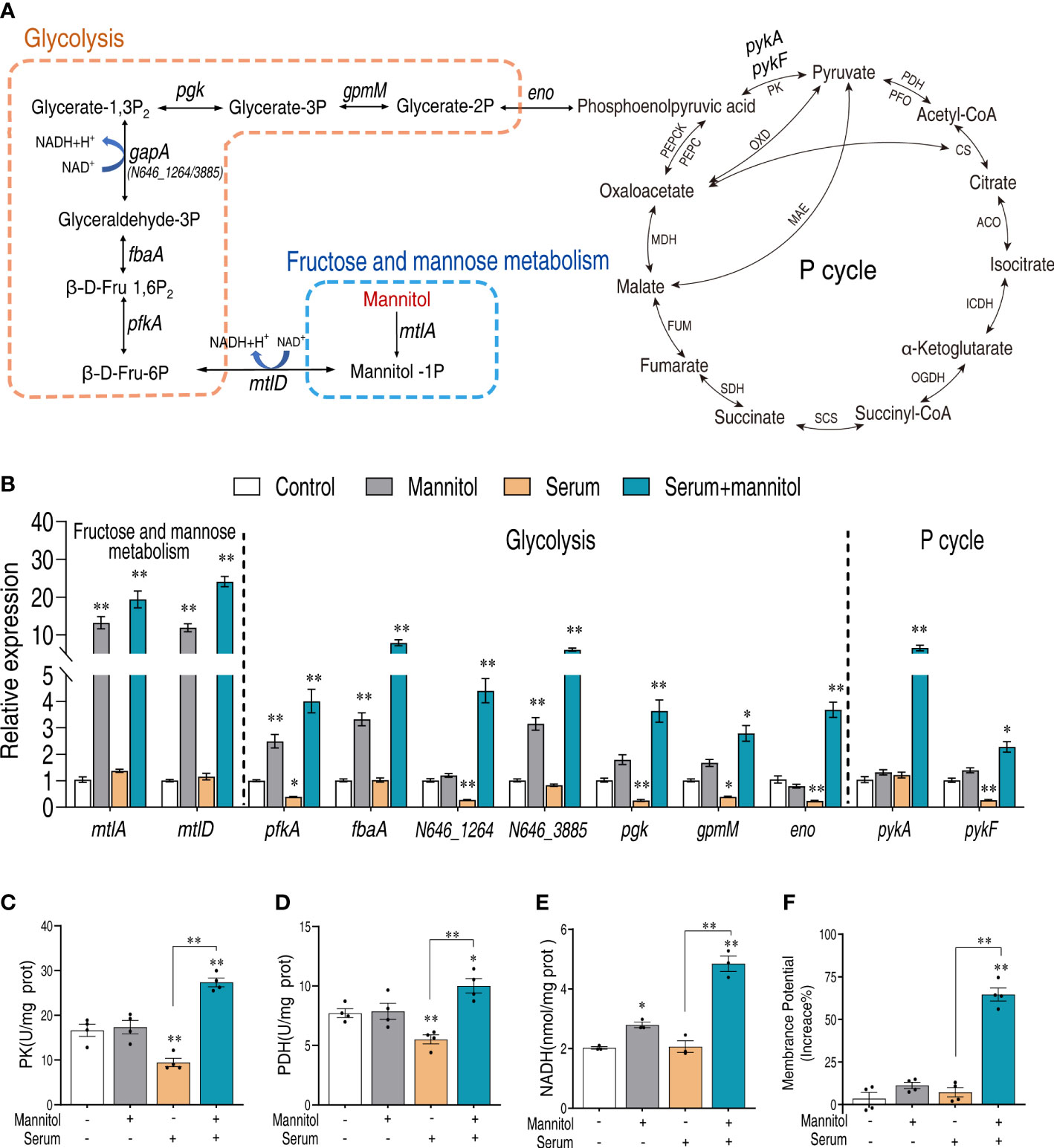
Figure 6 Mannitol promotes glycolysis and the pyruvate cycle. (A) Mannitol metabolism pathway. (B) Real-time quantitative reverse transcription-PCR (qRT-PCR) for the quantification of gene expression of glycolysis and P cycle. (C, D) Activity of pyruvate kinase and pyruvate dehydrogenase in the presence of mannitol or/and serum. (E) Effects of mannitol or/and serum on the content of NADH. (F) Membrane potential of Vibrio alginolyticus treated with 100 μl serum for 2 h in the presence or absence of 5 mM mannitol. The results are displayed as means ± SEM, and significant differences are identified (*p < 0.05 and **p < 0.01) as determined by Student’s t-test. At least three biological repeats were carried out.
To further confirm this result, the enzymatic activities of the pyruvate cycle, pyruvate kinase (PK), and pyruvate dehydrogenase (PDH) were investigated. Serum consistently decreased the activities of the two enzymes, but the presence of mannitol increased their activity (Figures 6C, D). The activated glycolysis and pyruvate cycle, by the presence of mannitol and serum, were confirmed with the content of NADH (Figure 6E) and proton motive force (Figure 6F). These data together suggest that mannitol enhances glycolysis and the pyruvate cycle that enhance serum killing.
Mannitol enhances complement deposition via glycolysis and the pyruvate cycle
As suggested by the abovementioned results that glycolysis is critical for the function of mannitol, we treated bacteria with inhibitors shikonin and bromopyruvate that target pyruvate kinase and pyruvate dehydrogenase, respectively. Either shikonin or bromopyruvate alone had a limited effect on bacterial survival. The killing of bacteria by mannitol and serum was blocked by either treatment and showed a dose-dependent manner (Figures 7A, B). The deposition of complement to the bacterial surface is believed to be the key part in enhancing serum lytic activity. Mannitol increased the amount of complements C3b and C5b-9 on the bacterial surface (Figures 7C, D). However, shikonin or bromopyruvate decreased the amount of deposited complement components. Moreover, we tested the synergistic effects of glycine and mannitol on serum killing. Mannitol had a limited effect on glycine-enabled serum killing (Figure 7E), which was the same as the effect of glycine on mannitol (Figure 7F). Consistently, mannitol and glycine do not mutually promote each other’s ability on C3b and C5b-9 deposition (Figures 7G, H). Thus, these data suggest that mannitol is downstream of glycine whose action is dependent on glycolysis and the pyruvate cycle to enhance complement deposition to promote killing.
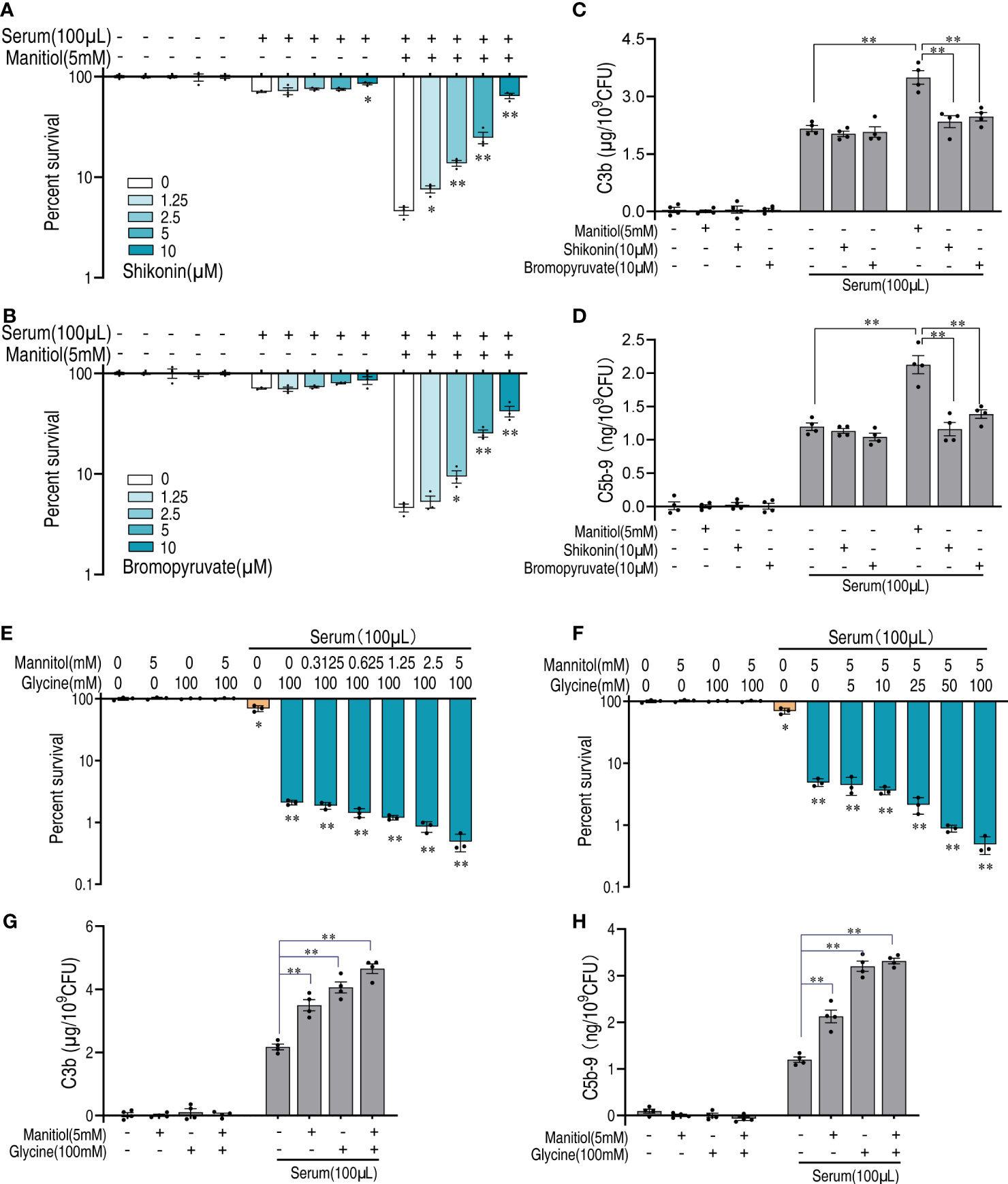
Figure 7 Mannitol increases serum sensitivity via glycolysis and the pyruvate cycle. (A, B) Percent survival of V. alginolyticus in the presence or absence of increasing doses of shikonin (A) or bromopyruvate (B) and in the presence of serum and/or mannitol. (C, D) Quantification of C3b (C) and C5b-9 (D) incubated with 10 μM shikonin or 10 μM bromopyruvate in the presence or absence of 100 μl serum alone or in the presence of or without mannitol (5 mM). (E) Percent survival of V. alginolyticus 12G01 incubated with 100 mM glycine plus serum (100 μl) in a mannitol dose-dependent manner (0–5 mM). (F) Percent survival of V. alginolyticus 12G01 incubated with 5 mM mannitol plus serum (100 μl) in a glycine dose-dependent manner (0–100 mM). (G, H) Quantification of C3b (G) and C5b-9 (H) incubated with 5 mM mannitol and/or 100 mM glycine in the presence or absence of 100 μl serum. The results are displayed as means ± SEM, and significant differences are identified (*p < 0.05 and **p < 0.01) as determined by Student’s t-test. At least three biological repeats were carried out.
Discussion
Managing a bacterial infection is critical for sustainable agriculture and human health. Antibiotics are the major choice for the control of a bacterial infection. However, this option is challenged in recent years because of the spread of antibiotic-resistant bacteria (29). Policies have been issued to ban the use of antibiotics as feed additive in agriculture, which presents a severe threat for the production of food animals, in many countries (30, 31). Therefore, alternatives to control a bacterial infection is urgently needed.
Harness host’s immune system has been a hot spot for identifying potential targets for the control of a bacterial infection. The complement system is a key part in defending against bacterial pathogens. However, serum resistance is a common virulent trait for different pathogens (32, 33). Therefore, reversing serum resistance is a potential way to reduce bacterial loads to a level that can be eliminated by other parts of the immune system. One challenge for such study is the diversified mechanisms utilized by bacterial pathogens to escape serum killing. Different bacteria use different virulent factors to disrupt the normal function of the complement system (34). This is the possible reason that there is still lack of proper control measures. This challenge is addressed by a recent report stating that metabolic reprogramming of serum-resistant bacteria can be a potential way to enable serum killing. Glycine was identified to be a metabolite to reverse serum resistance, but its efficacy varied from pathogen to pathogen. Thus, identifying the underlying mechanism would be a way to find other metabolites that can reverse in a different pathogen.
Deposition of complement components to the bacterial surface is crucial for the action of a complement system that either directly lyse bacteria via membrane-attached complex or opsonized bacteria toward immune cells for phagocytosis or triggering an adaptive immune response (35). As such, promoting complement deposition is a strategy to overcome complement resistance. In this study, we quantify how mannitol promotes C3b and C5b-9 deposition, which are the terminal complex for complement activity. C3 is located at the hub where all three complement pathways—classic pathway, lectin pathway, and alternative pathway—converge and activate C3 by proteolytic cleavage that form C3b. C3b covalently attaches to the bacterial surface for the following immune activation. C3b also activates C5 by splitting which generates C5b to form membrane-attached complexes with C6, C7, C8, and C9 at the bacterial surface (36). Thus, the deficiency of C3 leads to severe a bacterial infection, including those caused by meningococci and pneumococci (37). The exogenous administration of complement component C3 and the active form C3a in Japanese flounder increased its survival against bacterial infection by inducing chemotaxis to peripheral blood leukocytes (38). Given the importance of C3 and C5b-9, enhancing C3 and C5-9 deposition is key to boost the complement killing activity.
Taking advantage of proteomics, we find that the expression of transporters for mannitol are increased, which motivates us to investigate the potential function of mannitol in reversing serum resistance. Interestingly, mannitol promotes serum killing effects, and we find that this effect is dependent on glycolysis and the pyruvate cycle (25, 26, 28). We have previously shown that the pyruvate cycle regulates the TCA cycle in Escherichia coli, Edwardsiella tarda, and V. alginolyticus (28, 39). Thus, V. alginolyticus contained full sets of glycolytic genes and pyruvate cycle genes. Both of glycolysis and the pyruvate cycle are energy metabolism that provides energy for their growth and virulence. Glycolysis is critical for Vibrio growth as they predicated highly expressed genes to support their fast growth (40). Modulation of such energy metabolism is crucial for the bactericidal activity disinfectant. Slightly acidic electrolyzed water, acidic electrolyzed water, and sodium hypochlorite, commonly used to inactivate the food-born pathogen V. parahaemolyticus, decreased the expression of adenylate kinase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and enolase to exert an antibacterial activity (41). Moreover, central carbon metabolism is strongly associated with antibiotic resistance in Vibrio. Decreased glycolytic activity and pyruvate cycle activity is associated with levofloxacin resistance, gentamycin resistance, and colistin resistance (25, 42). The mechanism is that the downregulation of glycolysis and the pyruvate cycle reduced the intracellular generation of reactive oxygen species, whereas reactivating glycolysis and the pyruvate cycle, e.g., glucose, re-sensitizes antibiotic-resistant bacteria to antibiotics (25, 43). Recent studies also show that the glycolytic pathway and the pyruvate cycle are subjected to the regulation of acetylation and succinylation, indicating that protein post-translation modification may be involved in serum resistance (44–46). Actually, our previous study demonstrated that the activity of the pyruvate cycle was decreased by serum (17). Here we complement this finding that it is the serum that decreased the glycolytic pathway to repress glycolysis by withdrawing fuel sources.
Although there is no direct evidence to demonstrate how serum modulates the bacterial physiology, we postulate two possibilities. First, serum contains certain metabolite/factors that can be utilized by bacteria to remodel their metabolism to prevent complement killing—for example, the recent proposal of microbial endocrinology suggests that signals of the host endocrine system impact the microbial physiology. Epinephrine, norepinephrine, dopamine, estriol, estradiol, estrogen, and progesterone influence bacterial growth, bacterial resistance to antimicrobial peptides, quorum sensing, biofilm formation, and virulent gene expression such as E. coli adhering to the intestinal epithelial cells in pigs (47–56). It is highly possible that such signal may participate in serum resistance, which requires further investigation. Second, serum induces a stress response that causes a global metabolomic shift. Our previous study showed that glycine downregulates cAMP/CRP transcriptional factors. Since this transcription factor is involved in both metabolism and stress response, it is not surprising that bacteria activate cAMP/CRP to defend against serum killing but decrease metabolism as well. Thus, it makes sense that enhancing metabolism by glycine or mannitol reverses this phenotype.
In summary, we demonstrate that mannitol is a downstream metabolite effector induced by glycine. Mannitol flux into glycolysis to drive the activation of the pyruvate cycle, which is critical to enhance the serum killing activity. This effect was confirmed both in vitro and in vivo. Our study paves a possible way to develop an eco-friendly compound in aquaculture to treat bacterial infections.
Materials and methods
Ethics statement
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (approval no. SYSU-IACUC-2020-B126716).
Bacterial strain
Bacterial strains V. alginolyticus ATCC33787, V. parahaemolyticus ATCC17802, and V. vulnificus ATCC27562 and clinical isolates including Vibrio alginolyticus 12G01, V. parahaemolyticus VP01, and V. mimicus vmi01 (42, 57) were among the collection of our laboratory. Bacteria were grown in standard Luria–Bertani broth medium (1% bacterial peptone and 0.5% yeast extract) plus 3% NaCl and were propagated at 30°C. The bacterial culture grown overnight was diluted to 1:100 using fresh medium and grown until the optical density was 0.5. Bacterial cells were collected by centrifugation and were washed three times with saline solution.
Fish and rearing conditions
Nile tilapia (Oreochromis niloticus) with an average weight of 500 ± 10 g was obtained from a local fish company (Guangdong Tilapia Breeding Farm, Guangzhou, China). The tilapia was maintained in 180-L tanks whose parameters were set as follows: water temperature at 28°C, pH value of 7.0–7.5, carbon dioxide <10 mg/L, oxygen content of 6–7 mg/L, and nitrogen content of 1–2 mg/L. The tilapia was acclimated for at least 2 weeks before the experimental manipulation (58–60).
Zebrafish with an average body weight as 0.22 ± 0.03g was obtained from a local zebrafish corporation (Guangzhou, China). Before the experiment, the fish was tested by microbiological detection to ensure that it is free from Vibrio species infection. The zebrafish was cultured in a 25-L open-circuit filtered water tanks at room temperature with aeration (39, 61, 62). The animals were maintained in the condition stated above for 2 weeks before the experiment. The zebrafish was fed twice per day with commercial blood worm.
Protein sample preparation and TMT labeling
Bacterial colonies were inoculated in a 5-ml medium and cultured at 30°C. The overnight cultures were re-inoculated into 100 ml medium with 1:100 ratio, followed by growth until OD600 was 0.5. Bacterial cells were collected by centrifugation, washed three times with saline solution, and resuspended until OD600 was 1.0. An aliquot of 3 ml bacteria was collected in a 5-ml centrifugation tube, and glycine dissolved in saline solution was added to the bacterial solution to a final concentration of 100 mM. Bacteria treated with saline solution only was established as the control group. Afterwards, after 2 h of incubation, bacteria were collected for protein labeling. For each sample, two biological replicates were performed. The collected cells were lysed with lysis buffer. Trypsin (sequencing grade, Promega, Madison, WI, USA) was used to digest the proteins. Then, the generated peptide mixture was labeled as constructed by TMT kit (Thermo Fisher Scientific, USA). The labeled peptides were desalted with C18 desalting column (Sigma Aldrich, USA). The polypeptide sample of 100 μg was separated by reverse-phase ultra-performance liquid chromatography under alkaline conditions and then identified using mass spectrometry (63).
Proteomics analysis
For each fraction, 1 μg peptide was separated with a nano-UPLC (EASYnLC1200) coupled to a Q Exactive HFX Orbitrap instrument (Thermo Fisher Scientific) that is equipped with a nano-electrospray ion source. The peptide mixture was separated by a reverse-phase column (100 μm ID × 15 cm, ReprosilPur120 C18AQ, 1.9 μm, Dr. Math). The mobile phases contained 0.1% FA and 2% ACN (phase A) as well as 90% CAN and 0.1% FA (phase B). Separation was carried out for a total of 90 min, with a flow rate of 300 nl/min. Gradient B was performed as follows: 2% to 5% for 2 min, 5% to 22% for 68 min, 22% to 45% for 16 min, 45% to 95% for 2 min, and 95% for 2 min. Raw MS files were exported and analyzed with Max Quant software (version 1.6.15.0). Carbamidomethyl [C] was set as a fixed modification, while oxidation (M), acetyl (protein N term), deamidation (N), and deamidation (NQ) were set as variable modifications (the maximum number of variable modifications was five per peptide). The false discovery rate was set to 0.01 for both protein and peptide, where the minimum length for a peptide was seven amino acids. Proteins of altered expression were selected if their folds of change <0.83 or >1.2) (64) and with a P-value ≤0.05 (65, 66).
Sample preparation and serum killing
Fish serum was collected from 50 fish with equal numbers of male and female individuals. Serum was collected via a vein puncture and was isolated by centrifugation at 3,000 × g for 10 min at 4°C. The sera were aliquoted to a small volume and stored at −80°C (17). The sera were used only one time to avoid the thaw–frozen cycle. To perform serum killing, bacteria were resuspended in saline solution to OD600 of 1.0. Then, 3 ml of the bacteria was collected by centrifugation and resuspended in 100 μl serum or/and metabolites. The bacteria were incubated at 30°C for 2 h with shaking. After that, bacteria were collected to determine colony-forming units by plating.
Quantitative real-time PCR
qRT-PCR was performed as previously described (67, 68). Total RNA was extracted from V. alginolyticus with TRIzol reagent (Invitrogen, USA). The quality and the purity of RNA were checked by electrophoresis and Nanodrop. Reverse transcription was performed with EvoM-MLV RT kit with gDNA clean for qPCR (AG11705; Accurate Biology). The reaction for qRT-PCR was conducted in a 10-μl reaction including 5 μl 2× SYBR green premix pro Taq HS qPCR kit (AG11701; Accurate Biology), 2.6 μl H2O, 2 μl cDNA template, and 0.2 μl forward and reverse primers. Three technical replicas were included for each sample, and the reaction was run on a CFX384 Touch (Bio-Rad, USA).
Bacterial infection
Overnight bacteria were diluted 1:100 in growth medium until OD600 was 1.0 at 30°C (69–71). Bacteria were collected for bacterial challenge. The zebrafish were randomly grouped and injected intramuscularly with 5 μl of 1 × 105 CFU, 2 × 105 CFU, 4 × 105 CFU, 6 × 105 CFU, or 8 × 105 CFU bacteria or saline solution (n = 30 for each treatment). Death of fish was checked twice a day for a total of 14 days to obtain the accumulative death.
Measurement of NAD+/NADH
NAD+/NADH measurement was performed as previously reported (26, 72, 73). Bacteria were suspended in saline solution to 1.0 of OD600. Furthermore, 2 ml of bacteria was resuspended in NADH/NAD+ extraction buffer and incubated in 60°C water for 5 min, and then the assay buffer was added. After vertexing and centrifugation, the supernatant was collected and read according to the protocol (EnzyChrom NAD+/NADH Assay Kit, BioAssay Systems).
Measurement of membrane potential
Membrane potential was measured as reported (73, 74). Briefly, the bacteria (1 × 106 CFU) were stained with 3,3′-diethyloxacarbocyanine iodide for 30 min. The reading was performed by flow cytometry (FACSCalibur flow cytometer, Becton Dickinson, San Jose, CA, USA).
Measurement of enzyme activity
The activity of PDH was measured as reported (75–77), and the PK enzymatic activity was assessed using a commercial assay kit (BC0545, Beijing Solarbio Science and Technology Co., Ltd.). The bacteria were treated as stated above. After incubation with serum or/and mannitol, the bacteria were collected and adjusted to OD600 of 1.0. Moreover, 20 ml of OD600 = 1.0 bacterial solution was collected in a 1.5-ml centrifugation tube, resuspended with 1 ml sterile saline, and disrupted by sonic oscillation (200 W total power with 35% output, 2 s pulse, and 3 s pause over ice) for 3 min. After centrifugation, the supernatants of the enzyme activity assay were collected. Protein concentration was measured with enhanced BCA Protein Assay Kit (P0010, Beyotime, Shanghai, China).
Quantification of C3b/C5b-9 deposited on the bacterial outer membrane
Commercial assay kits (FS-E63210-96T and A119939-96T, Shanghai Fusheng Industrial Co., Ltd.), were used to quantify the C3b/C5b-9 on the bacterial outer membrane. The bacteria were treated as described above and were processed for ELISA.
Data availability statement
The data presented in the study are deposited in ProteomeXchange Consortium via the iProX partner repository(78), accession number PXD035517.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Sun Yat-sen University.
Author contributions
T-sK and J-hW conducted the experiments. T-sK, J-hW, and X-wC performed data analysis. T-sK, X-wC, and BP interpreted the data. BP wrote the manuscript. BP conceptualized and designed the project. All the authors reviewed the manuscript and acknowledged the contributions. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by grants from the National Natural Science Foundation of China (NSFC) project [32061133007, 31872602], Project supported by Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) [No. 311020006], and The Youth Talent Support Program of Guangdong Province [2017GC010617] (to B. P.).
Acknowledgments
We also thank Dr. D.X. Yang and Mr. Y.C. Cao for their assistance in the collection of fish serum.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1010526/full#supplementary-material
References
1. Recheis B, Rumpler H, Schneider WJ, Nimpf J. Receptor-mediated transport and deposition of complement component C3 into developing chicken oocytes. Cell Mol Life Sci (2005) 62(16):1871–80. doi: 10.1007/s00018-005-5193-1
2. Thurman JM, Holers VM. The central role of the alternative complement pathway in human disease. J Immunol (2006) 176(3):1305–10. doi: 10.4049/jimmunol.176.3.1305
3. Wallis R, Mitchell DA, Schmid R, Schwaeble WJ, Keeble AH. Paths reunited: Initiation of the classical and lectin pathways of complement activation. Immunobiology (2010) 215(1):1–11. doi: 10.1016/j.imbio.2009.08.006
4. Morgan BP, Boyd C, Bubeck D. Molecular cell biology of complement membrane attack. Semin Cell Dev Biol (2017) 72:124–32. doi: 10.1016/j.semcdb.2017.06.009
5. Heesterbeek DA, Bardoel BW, Parsons ES, Bennett I, Ruyken M, Doorduijn DJ, et al. Bacterial killing by complement requires membrane attack complex formation Via surface-bound C5 convertases. EMBO J (2019) 38:e99852. doi: 10.15252/embj.201899852
6. Doorduijn DJ, Bardoel BW, Heesterbeek DAC, Ruyken M, Benn G, Parsons ES, et al. Bacterial killing by complement requires direct anchoring of membrane attack complex precursor C5b-7. PloS Pathog (2020) 16(6):e1008606. doi: 10.1371/journal.ppat.1008606
7. Thomassin JL, Brannon JR, Kaiser J, Gruenheid S, Le Moual H. Enterohemorrhagic and enteropathogenic Escherichia coli evolved different strategies to resist antimicrobial peptides. Gut Microbes (2012) 3(6):556–61. doi: 10.4161/gmic.21656
8. Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect (2015) 17(3):173–83. doi: 10.1016/j.micinf.2015.01.004
9. Kraiczy P, Stevenson B. Complement regulator-acquiring surface proteins of borrelia burgdorferi: Structure, function and regulation of gene expression. Ticks Tick Borne Dis (2013) 4(1-2):26–34. doi: 10.1016/j.ttbdis.2012.10.039
10. Kraiczy P, Skerka C, Brade V, Zipfel PF. Further characterization of complement regulator-acquiring surface proteins of borrelia burgdorferi. Infect Immun (2001) 69(12):7800–9. doi: 10.1128/IAI.69.12.7800-7809.2001
11. Armbruster CR, Wolter DJ, Mishra M, Hayden HS, Radey MC, Merrihew G, et al. Staphylococcus aureus protein a mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio (2016) 7(3):e00538–16. doi: 10.1128/mBio.00538-16
12. Pietrocola G, Nobile G, Rindi S, Speziale P. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front Cell Infect Microbiol (2017) 7:166. doi: 10.3389/fcimb.2017.00166
13. Kharazmi A, Doring G, Hoiby N, Valerius NH. Interaction of Pseudomonas aeruginosa alkaline protease and elastase with human polymorphonuclear leukocytes in vitro. Infect Immun (1984) 43(1):161–5. doi: 10.1128/iai.43.1.161-165.1984
14. Kessler E, Safrin M, Gustin JK, Ohman DE. Elastase and the lasa protease of Pseudomonas aeruginosa are secreted with their propeptides. J Biol Chem (1998) 273(46):30225–31. doi: 10.1074/jbc.273.46.30225
15. Bokarewa MI, Jin T, Tarkowski A. Staphylococcus aureus: Staphylokinase. Int J Biochem Cell Biol (2006) 38(4):504–9. doi: 10.1016/j.biocel.2005.07.005
16. Wang Z, Li MY, Peng B, Cheng ZX, Li H, Peng XX. GC-MS-Based metabolome and metabolite regulation in serum-resistant Streptococcus agalactiae. J Proteome Res (2016) 15(7):2246–53. doi: 10.1021/acs.jproteome.6b00215
17. Cheng ZX, Gong QY, Wang Z, Chen ZG, Ye JZ, Li J, et al. Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front Immunol (2017) 8:1706. doi: 10.3389/fimmu.2017.01706
18. Cheng ZX, Guo C, Chen ZG, Yang TC, Zhang JY, Wang J, et al. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat Commun (2019) 10(1):3325. doi: 10.1038/s41467-019-11129-5
19. Jiang M, Chen ZG, Li H, Zhang TT, Yang MJ, Peng XX, et al. Succinate and inosine coordinate innate immune response to bacterial infection. PloS Pathog (2022) 18(8):e1010796. doi: 10.1371/journal.ppat.1010796
20. Hung CW, Tholey A. Tandem mass tag protein labeling for top-down identification and quantification. Anal Chem (2012) 84(1):161–70. doi: 10.1021/ac202243r
21. von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. String: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res (2005) 33(Database issue):D433–7. doi: 10.1093/nar/gki005
22. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. String V11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res (2019) 47(D1):D607–D13. doi: 10.1093/nar/gky1131
23. Martinez-Miranda JG, Chairez I, Duran-Paramo E. Mannitol production by heterofermentative lactic acid bacteria: A review. Appl Biochem Biotechnol (2022) 194(6):2762–95. doi: 10.1007/s12010-022-03836-5
24. Saha BC, Racine FM. Biotechnological production of mannitol and its applications. Appl Microbiol Biotechnol (2011) 89(4):879–91. doi: 10.1007/s00253-010-2979-3
25. Zhang S, Yang MJ, Peng B, Peng XX, Li H. Reduced ros-mediated antibiotic resistance and its reverting by glucose in Vibrio alginolyticus. Environ Microbiol (2020) 22(10):4367–80. doi: 10.1111/1462-2920.15085
26. Peng B, Su YB, Li H, Han Y, Guo C, Tian YM, et al. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab (2015) 21(2):249–62. doi: 10.1016/j.cmet.2015.01.008
27. Su YB, Peng B, Han Y, Li H, Peng XX. Fructose restores susceptibility of multidrug-resistant Edwardsiella tarda to kanamycin. J Proteome Res (2015) 14(3):1612–20. doi: 10.1021/pr501285f
28. Su YB, Peng B, Li H, Cheng ZX, Zhang TT, Zhu JX, et al. Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc Natl Acad Sci U.S.A. (2018) 115(7):E1578–E87. doi: 10.1073/pnas.1714645115
29. Zhao XL, Chen ZG, Yang TC, Jiang M, Wang J, Cheng ZX, et al. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci Transl Med (2021) 13(625):eabj0716. doi: 10.1126/scitranslmed.abj0716
30. Pugh DM. The eu precautionary bans of animal feed additive antibiotics. Toxicol Lett (2002) 128(1-3):35–44. doi: 10.1016/s0378-4274(01)00531-8
31. Chattopadhyay MK. Use of antibiotics as feed additives: A burning question. Front Microbiol (2014) 5:334. doi: 10.3389/fmicb.2014.00334
32. Coggon CF, Jiang A, Goh KGK, Henderson IR, Schembri MA, Wells TJ. A novel method of serum resistance by Escherichia coli that causes urosepsis. mBio (2018) 9(3):e00920–18. doi: 10.1128/mBio.00920-18
33. Moore SR, Menon SS, Cortes C, Ferreira VP. Hijacking factor h for complement immune evasion. Front Immunol (2021) 12:602277. doi: 10.3389/fimmu.2021.602277
34. Wu HJ, Wang AH, Jennings MP. Discovery of virulence factors of pathogenic bacteria. Curr Opin Chem Biol (2008) 12(1):93–101. doi: 10.1016/j.cbpa.2008.01.023
35. Haapasalo K, Meri S. Regulation of the complement system by pentraxins. Front Immunol (2019) 10:1750. doi: 10.3389/fimmu.2019.01750
36. Heesterbeek DAC, Angelier ML, Harrison RA, Rooijakkers SHM. Complement and bacterial infections: From molecular mechanisms to therapeutic applications. J Innate Immun (2018) 10(5-6):455–64. doi: 10.1159/000491439
37. Ram S, Lewis LA, Rice PA. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin Microbiol Rev (2010) 23(4):740–80. doi: 10.1128/CMR.00048-09
38. Wu M, Jia BB, Li MF. Complement C3 and activated fragment C3a are involved in complement activation and anti-bacterial immunity. Front Immunol (2022) 13:813173. doi: 10.3389/fimmu.2022.813173
39. Yang J, Yang XL, Su YB, Peng XX, Li H. Activation of the tca cycle to provide immune protection in zebrafish immunized by high magnesium-prepared Vibrio alginolyticus vaccine. Front Immunol (2021) 12:739591. doi: 10.3389/fimmu.2021.739591
40. Karlin S, Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J Bacteriol (2000) 182(18):5238–50. doi: 10.1128/JB.182.18.5238-5250.2000
41. Chen TY, Kuo SH, Chen ST, Hwang DF. Differential proteomics to explore the inhibitory effects of acidic, slightly acidic electrolysed water and sodium hypochlorite solution on Vibrio parahaemolyticus. Food Chem (2016) 194:529–37. doi: 10.1016/j.foodchem.2015.08.019
42. Li L, Su YB, Peng B, Peng XX, Li H. Metabolic mechanism of colistin resistance and its reverting in Vibrio alginolyticus. Environ Microbiol (2020) 22(10):4295–313. doi: 10.1111/1462-2920.15021
43. Zhang S, Wang J, Jiang M, Xu D, Peng B, Peng XX, et al. Reduced redox-dependent mechanism and glucose-mediated reversal in gentamicin-resistant Vibrio alginolyticus. Environ Microbiol (2019) 21(12):4724–39. doi: 10.1111/1462-2920.14811
44. Wang J, Pang H, Yin L, Zeng F, Wang N, Hoare R, et al. A comprehensive analysis of the lysine acetylome in the aquatic animals pathogenic bacterium Vibrio mimicus. Front Microbiol (2022) 13:816968. doi: 10.3389/fmicb.2022.816968
45. Xu Z, Wang L, Wang X, Wan M, Tang M, Ding Y. Characterizing the effect of the lysine deacetylation modification on enzyme activity of pyruvate kinase I and pathogenicity of Vibrio alginolyticus. Front Vet Sci (2022) 9:877067. doi: 10.3389/fvets.2022.877067
46. Zeng F, Pang H, Chen Y, Zheng H, Li W, Ramanathan S, et al. First succinylome profiling of Vibrio alginolyticus reveals key role of lysine succinylation in cellular metabolism and virulence. Front Cell Infect Microbiol (2020) 10:626574. doi: 10.3389/fcimb.2020.626574
47. Lyte JM, Lyte M. Review: Microbial endocrinology: Intersection of microbiology and neurobiology matters to swine health from infection to behavior. Animal (2019) 13(11):2689–98. doi: 10.1017/S1751731119000284
48. Neuman H, Debelius JW, Knight R, Koren O. Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiol Rev (2015) 39(4):509–21. doi: 10.1093/femsre/fuu010
49. Lyte M, Ernst S. Catecholamine induced growth of gram negative bacteria. Life Sci (1992) 50(3):203–12. doi: 10.1016/0024-3205(92)90273-r
50. Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: The language of hormones. Proc Natl Acad Sci U.S.A. (2003) 100(15):8951–6. doi: 10.1073/pnas.1537100100
51. Freestone PP, Lyte M. Microbial endocrinology: Experimental design issues in the study of interkingdom signalling in infectious disease. Adv Appl Microbiol (2008) 64:75–105. doi: 10.1016/S0065-2164(08)00402-4
52. Hegde M, Wood TK, Jayaraman A. The neuroendocrine hormone norepinephrine increases Pseudomonas aeruginosa Pa14 virulence through the las quorum-sensing pathway. Appl Microbiol Biotechnol (2009) 84(4):763–76. doi: 10.1007/s00253-009-2045-1
53. Lyte M, Freestone PP, Neal CP, Olson BA, Haigh RD, Bayston R, et al. Stimulation of Staphylococcus epidermidis growth and biofilm formation by catecholamine inotropes. Lancet (2003) 361(9352):130–5. doi: 10.1016/S0140-6736(03)12231-3
54. Kornman KS, Loesche WJ. Effects of estradiol and progesterone on Bacteroides melaninogenicus and Bacteroides gingivalis. Infect Immun (1982) 35(1):256–63. doi: 10.1128/iai.35.1.256-263.1982
55. Angel D, Baizabal-Aguirre V, Bravo-Patiño A, Valdez-Alarcón JJ. Microbial endocrinology: Target for new antimicrobial strategies to control infectious diseases. In: Mendez-Vilas A, editor. The battle against microbial pathogens: technological advances and educational programs. Fomatex Research Center (2015), 938–946.
56. Roshchina VV. Evolutionary considerations of neurotransmitters in microbial, plant, and animal cells. In: Lyte M, Freestone PPE, editors. Microbial endocrinology: Interkingdom signaling in infectious disease and health. New York, NY: Springer New York (2010). p. 17–52.
57. Kuang SF, Chen YT, Chen JJ, Peng XX, Chen ZG, Li H. Synergy of alanine and gentamicin to reduce nitric oxide for elevating killing efficacy to antibiotic-resistant Vibrio alginolyticus. Virulence (2021) 12(1):1737–53. doi: 10.1080/21505594.2021.1947447
58. Yang DX, Yang MJ, Yin Y, Kou TS, Peng LT, Chen ZG, et al. Serine metabolism tunes immune responses to promote oreochromis niloticus survival upon Edwardsiella tarda infection. mSystems (2021) 6(4):e0042621. doi: 10.1128/mSystems.00426-21
59. Yang DX, Yang H, Cao YC, Jiang M, Zheng J, Peng B. Succinate promotes phagocytosis of Monocytes/Macrophages in teleost fish. Front Mol Biosci (2021) 8:644957. doi: 10.3389/fmolb.2021.644957
60. Peng LT, Li DL, Yang DX, Peng B. Taurine promotes Oreochromis niloticus survival against Edwardsiella tarda infection. Fish Shellfish Immunol (2022) 129:137–144. doi: 10.1016/j.fsi.2022.08.065
61. Li L, Song M, Peng B, Peng XX, Li H. Identification and innate immunity mechanism of protective immunogens from extracellular proteins of Edwardsiella tarda. Fish Shellfish Immunol (2020) 97:41–5. doi: 10.1016/j.fsi.2019.12.020
62. Gong QY, Yang MJ, Yang LF, Chen ZG, Jiang M, Peng B. Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb Biotechnol (2020) 13(3):796–812. doi: 10.1111/1751-7915.13553
63. Muller JB, Geyer PE, Colaco AR, Treit PV, Strauss MT, Oroshi M, et al. The proteome landscape of the kingdoms of life. Nature (2020) 582(7813):592–6. doi: 10.1038/s41586-020-2402-x
64. Liu X, Wang J, Gao L, Liu H, Liu C. iTRAQ-based proteomic analysis of neonatal kidney from offspring of protein restricted rats reveals abnormalities in intraflagellar transport proteins. Cell Physiol Biochem (2017) 44(1):185–99. doi: 10.1159/000484626
65. Sinitcyn P, Tiwary S, Rudolph J, Gutenbrunner P, Wichmann C, Yilmaz S, et al. Maxquant goes Linux. Nat Methods (2018) 15(6):401. doi: 10.1038/s41592-018-0018-y
66. Tyanova S, Temu T, Cox J. The maxquant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc (2016) 11(12):2301–19. doi: 10.1038/nprot.2016.136
67. Jiang M, Kuang SF, Lai SS, Zhang S, Yang J, Peng B, et al. Na(+)-NQR confers aminoglycoside resistance via the regulation of l-alanine metabolism. mBio (2020) 11(6):e02086–20. doi: 10.1128/mBio.02086-20
68. Xu D, Wang J, Guo C, Peng XX, Li H. Elevated biosynthesis of palmitic acid is required for zebrafish against Edwardsiella tarda infection. Fish Shellfish Immunol (2019) 92:508–18. doi: 10.1016/j.fsi.2019.06.041
69. Cao YC, Kou TS, Peng LT, Munang’andu HM, Peng B. Fructose promotes crucian carp survival against Aeromonas hydrophila infection. Front Immunol (2022) 13:865560. doi: 10.3389/fimmu.2022.865560
70. Yang MJ, Cheng ZX, Jiang M, Zeng ZH, Peng B, Peng XX, et al. Boosted tca cycle enhances survival of zebrafish to Vibrio alginolyticus infection. Virulence (2018) 9(1):634–44. doi: 10.1080/21505594.2017.1423188
71. Jiang M, Yang LF, Zheng J, Chen ZG, Peng B. Maltose promotes crucian carp survival against Aeromonas sobrial infection at high temperature. Virulence (2020) 11(1):877–88. doi: 10.1080/21505594.2020.1787604
72. Yang J, Zeng ZH, Yang MJ, Cheng ZX, Peng XX, Li H. Nacl promotes antibiotic resistance by reducing redox states in Vibrio alginolyticus. Environ Microbiol (2018) 20(11):4022–36. doi: 10.1111/1462-2920.14443
73. Su YB, Kuang SF, Ye JZ, Tao JJ, Li H, Peng XX, et al. Enhanced biosynthesis of fatty acids is associated with the acquisition of ciprofloxacin resistance in Edwardsiella tarda. mSystems (2021) 6(4):e0069421. doi: 10.1128/mSystems.00694-21
74. Cheng ZX, Yang MJ, Peng B, Peng XX, Lin XM, Li H. The depressed central carbon and energy metabolisms is associated to the acquisition of levofloxacin resistance in Vibrio alginolyticus. J Proteomics (2018) 181:83–91. doi: 10.1016/j.jprot.2018.04.002
75. Kuang SF, Feng DY, Chen ZG, Liang ZZ, Xiang JJ, Li H, et al. Inactivation of nitrite-dependent nitric oxide biosynthesis is responsible for overlapped antibiotic resistance between naturally and artificially evolved Pseudomonas aeruginosa. mSystems (2021) 6(5):e0073221. doi: 10.1128/mSystems.00732-21
76. Ye JZ, Lin XM, Cheng ZX, Su YB, Li WX, Ali FM, et al. Identification and efficacy of glycine, serine and threonine metabolism in potentiating kanamycin-mediated killing of Edwardsiella piscicida. J Proteomics (2018) 183:34–44. doi: 10.1016/j.jprot.2018.05.006
77. Yin Y, Yin Y, Yang H, Chen Z, Zheng J, Peng B. Vibrio alginolyticus survives from ofloxacin stress by metabolic adjustment. Front Microbiol (2022) 13:818923. doi: 10.3389/fmicb.2022.818923
Keywords: mannitol, Vibrios, serum, proteomics, glycolysis, pyruvate cycle
Citation: Kou T-s, Wu J-h, Chen X-w and Peng B (2022) Functional proteomics identify mannitol metabolism in serum resistance and therapeutic implications in Vibrio alginolyticus. Front. Immunol. 13:1010526. doi: 10.3389/fimmu.2022.1010526
Received: 03 August 2022; Accepted: 07 October 2022;
Published: 31 October 2022.
Edited by:
Gee W. Lau, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Fei Zhu, Zhejiang Agriculture and Forestry University, ChinaJuan José Valdez Alarcón, Universidad Michoacana de San Nicolás de Hidalgo, Mexico
Copyright © 2022 Kou, Wu, Chen and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Peng, cGVuZ2IyNkBtYWlsLnN5c3UuZWR1LmNu
 Tian-shun Kou
Tian-shun Kou Jia-han Wu
Jia-han Wu Xuan-wei Chen
Xuan-wei Chen Bo Peng
Bo Peng