- 1Department of Cardiothoracic Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Thoracic Surgery, Army Medical Center of People’s Liberation Army of China (PLA), Chongqing, China
Background: Genetic association studies have elucidated the link of variants in the interleukin 17 (IL-17) family genes with susceptibility to human diseases, yet have obtained controversial outcomes. Therefore, we sought to update comprehensive synopsis of variants in the IL-17 family genes with susceptibility to human diseases.
Methods: Our study screened the Pubmed and Web of Science to enroll eligible articles and performed a meta-analysis, then graded the cumulative evidence of significant association using Venice criteria and false-positive report probability test, and finally assessed the function of variants with strong evidence.
Results: Seven variants in IL-17 family genes had significant relationships with susceptibility to 18 human diseases identified by meta-analyses. Strong evidence was assigned to 4 variants (IL-17A rs2275913, IL-17A rs8193037, IL-17F rs1889570, IL-17F rs763780) with susceptibility to 6 human diseases (lung and cervical cancer, spondyloarthritis, asthma, multiple sclerosis, rheumatoid arthritis), moderate to 2 variants with risk of 5 diseases, weak to 5 variants with risk of 10 diseases. Bioinformatics analysis suggested that the variants with strong evidence might fall in putative functional regions. Additionally, positive relationships for 5 variants with risk of 4 diseases (based on two datasets) and 14 variants with risk of 21 diseases (based on one dataset) were considered noteworthy.
Conclusions: This study offers updated and comprehensive clues that variants in the IL-17 family genes are significantly linked with susceptibility to cervical, lung cancer, asthma, multiple sclerosis, rheumatoid arthritis and spondyloarthritis, and elucidates the crucial role of the IL-17 regions in the genetic predisposition to cancer or noncancerous diseases.
Introduction
Interleukin (IL) 17 (IL-17), a homodimeric glycoprotein composed of 155 amino acids, remains a pro-inflammatory and its family genes contain six groups (IL-17A to F) (1). The IL-17 signaling system has a crucial impact on different tissues such as lung, skin, kidney, brain, bone, articular cartilage, meniscus and hematopoietic tissue (1); this system mediated by the binding to IL-17 receptors can active multiple cell types (such as fibroblasts, endothelial cells, epithelial cells, keratinocytes and macrophages) (2). It could be activated which produces cell subsets of IL-17 that play a crucial role in multiple essential biological activities and accelerating occurrences of human diseases, involving novel coronavirus disease 2019 (COVID-19) (3).
As early as 2006, Hizawa et al. found five single nucleotide polymorphisms (SNPs) in IL-17F and found that rs763780 {His-to-Arg substitution at amino acid 161 (H161R)} in the third exon of the IL-17F gene influenced the susceptibility to asthma and chronic obstructive pulmonary disease (COPD) in the Japanese population (4, 5). Since then, a range of genetic association studies found that SNPs in IL-17 family genes have been shown to be linked with the risk of multiple diseases. In 2007, Arisawa et al. identified that IL-17F rs763780 and IL-17A rs2275913 in Japanese population had been shown to be linked with the risk of ulcerative colitis (UC) (6). Subsequently, studies also found that IL-17 family genes are linked with multiple cancers risk, including ovarian (7), breast (8), hepatocellular (9), esophageal (10), gastric (11) and lung cancer (12). In 2014, two researchers independently performed a meta-analysis and attempted to elucidate the relationship between IL-17A rs2275913 and IL-17F rs763780 and cancer risk in Asians (13, 14). Interestingly, the outcomes of the two studies were inconsistent. Recently, in an updated meta-analysis conducted in multiple countries from the Asian ancestry, SNP rs2275913 and SNP rs763780 associated with 31,234 subjects were tested. Then it was discovered that IL-17A rs2275913 acted as risk factors for gastric, cervical, colorectal and oral cancer, and IL-17F rs763780 for cervical and oral cancer (15).
Even though previous studies evaluate the relationship between SNPs in IL-17 family genes and the risk of diseases, the outcomes are controversial. In addition, an updated research synopsis with comprehensive functional annotation had not been conducted to assess the epidemiological evidence of associations with IL-17 family genes and risk of all human diseases thus far. Therefore, we carried out meta-analysis to elucidate the relationships of SNPs in the IL-17 genes with susceptibility to disease, offered the epidemiological evidence for variants with significant relationships, and evaluated the functions of significant variants using public sources.
Materials and methods
Our research followed the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Statement (PRISMA) and the Human Genome Epidemiology Network for systematic review of genetic association studies (16, 17).
We screened genetic association studies from Pubmed and Web of science up to 30 Apr 2022 using “{interleukin-17} OR {IL-17} OR {IL-17}” AND “{variant} OR {variation} OR {polymorphism} OR {genotype} OR {single nucleotide polymorphism} OR {SNP}”. We also collected additional articles by retrieving published reviews, meta-analyses studies, etc.
The inclusion criteria are as follows: (i) they were concentrated on the relationships between SNPs in IL-17 family genes and susceptibility to human cancers or non-neoplastic diseases performed in case-control, cohort or cross-sectional studies (ii) they could provide the genotype data to calculate the odds ratios (ORs) and corresponding 95% confidence intervals (95% CIs) under additive genetic model, (iii) they were published in English by form of full-text. The exclusion criteria are presented, (i) they lacked sufficient information (especially the quantity of genotype and/or allelic distributions), (ii) the study was not focused on SNP in IL-17 family genes, (iii) they were not published as full reports, such as conference abstracts and letters to editors, (iv) they concentrated on cancer mortality.
Data extraction
Two authors extracted the data independently using a predesigned collection sheet and any disagreement could be solved with the corresponding author by discussion together. The extracted data were as follows: first author, publishing year, study design, country or region, ethnicity, gene name, variant, cases and controls, genotype counts, minor allelic frequency (MAF). When previous articles studied on the same or overlapping data, we only extracted data from papers with largest sample size and most detailed information.
Statistical analysis
In our study, statistical tests of meta-analysis in the additive genetic association were two-tailed, and a P < 0.05 was significant level unless otherwise stated, which were conducted using Stata, version 15 (Stata, College Station, TX, USA). Meta-analyses were performed for variants with at least three datasets. We used the Cochran’s Q test to evaluate the heterogeneity between studies (18), while I2 statistic was applied to quantify and evaluate the heterogeneity (19). Sensitive analyses were performed to evaluate whether the significant association was lost when excluding a single study (dataset), or the first published study, or studies deviated from the Hardy-Weinberg equilibrium (HWE) in the controls. We investigated the probability of an excess of significant findings for single meta-analysis (20). Begg’s test and Egger’s test were conducted to assess potential publication bias and small-study bias, respectively (21, 22). Moreover, P<0.1 as the significant level in the assessment of heterogeneity, an excess of significant findings, Begg’s test and Egger’s test.
Assessment of epidemiological credibility
Our study graded the epidemiological credibility of significant associations identified by main meta analyses using the Venice guideline (23) and false positive report probability (FPRP) test (24) (see Supplementary Method).
Functional annotation
Our study assessed the potential functional effect of variants on 6p12.2 using data from the Encyclopedia of DNA Elements (ENCODE) tool HaploReg (v4.1) (25) and the UCSC Genome browser (http://genome.ucsc.edu/). We analyzed the regions of promoter or enhancer activity, local histone modification, DNase I hypersensitivity, transcription factor binding motifs and proteins bound to these regulatory sites. In addition, we examined genome-wide cis-eQTL data in multiple tissues from two major eQTL databases: the Genotype-Tissue Expression Project (26) and the Multiple Tissue Human Expression Resource Project (27) to determine whether these genes might explain the observed associations in these loci. We used the data from the Phase 3 of the 1000 Genomes Project to perform linkage disequilibrium (LD) analysis for variants positively associated with susceptibility to cancer and noncancerous diseases in current study (28).
Result
Characteristics of the included studies
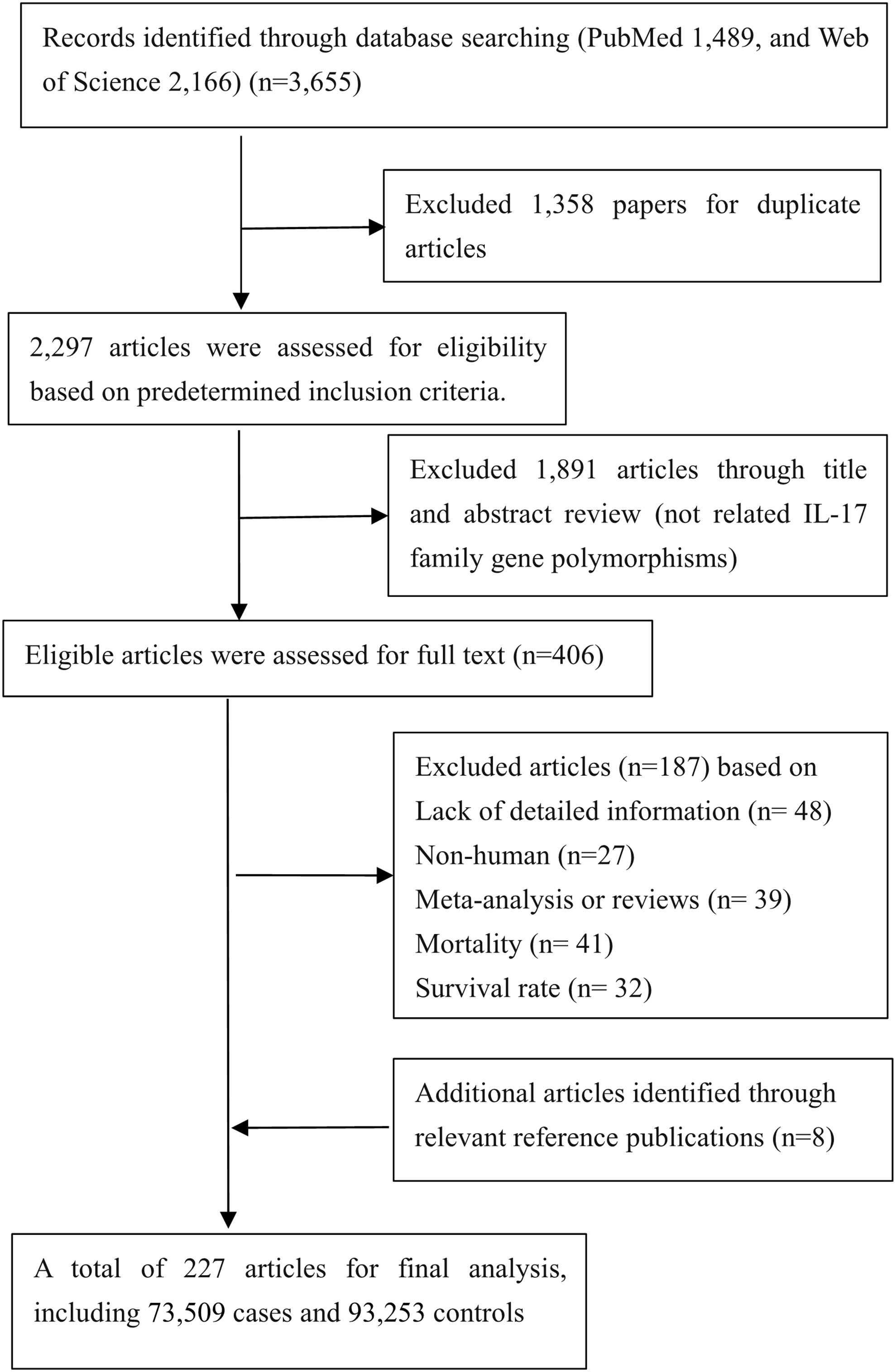
As presented in Figure 1, a total of 227 eligible papers including 73,509 cases and 93,253 controls were enrolled (Supplementary Table 1); 135 papers focused on relationships between 10 variants in IL-17 family genes and 25 diseases (5 cancers as well as 20 noncancerous diseases). The distributions of SNPs (n) with human diseases were presented: asthma (n=5), autoimmune thyroid diseases (AITD) (n=1), cervical cancer (n=3), COPD (n=2), colorectal cancer (n=2), coronary artery disease (CAD) (n=2), functional dyspepsia (FD) (n=3), gastric cancer (n=4), hepatitis B Virus (HBV) infection (n=4), hepatocellular carcinoma (n=1), immune thrombocytopenia (ITP) (n=3), inflammatory bowel disease (IBD) (n=3), leprosy (n=2), lung cancer (n=4), multiple sclerosis (MS) (n=1), osteoarthritis (OA) (n=2), periodontitis (n=2), pre-eclampsia (PE) (n=2), psoriasis (n=1), recurrent miscarriage (RM) (n=1), rheumatoid arthritis (RA) (n=5), spondyloarthritis (SpA) (n=2), systemic lupus erythematosus (SLE) (n=1), tuberculosis (TB) (n=3), Type 1 diabetes mellitus (T1DM) (n=1).
Associations between IL-17 variants and risk of human diseases
we carried out meta-analyses to investigate correlations of 10 SNPs in IL-17 family genes with 5 cancers as well as 20 non-cancer diseases based on at least 3 datasets, under an additive genetic model. As presented in Table 1 and Supplementary Table 2, 7 polymorphisms (rs1889570, rs2275913, rs2397084, rs3748067, rs763780, rs8193036, rs8193037) were associated with susceptibility to 4 types of carcinoma (colorectal, cervical, lung and gastric cancer) and 14 non-cancer diseases (AITP, asthma, CAD, HBV infection, ITP, IBD, leprosy, MS, OA, psoriasis, RA, SpA, SLE, TB) (28 associations, P<0.05). The cervical cancer susceptibility had positive association with minor allele of rs2275913 in Asians (OR=1.391), rs3748067 (OR=1.493) and rs763780 (OR=1.184). Apart from that, rs2275913 had an elevated susceptibility to colorectal cancer (OR=1.452); the positive association could be found in Asians and mixed populations (OR=1.524, OR=2.038, respectively), rather than in Caucasians. Apart from colorectal cancer, rs2275913 had an increased predisposition to gastric cancer (OR=1.281); the positive association could be found both in Asians and Caucasians (OR=1.264, OR=1.723, respectively); this SNP could increase risk of lung cancer (OR=1.213). Moreover, rs8193037 could elevate lung carcinoma susceptibility (OR=2.129).
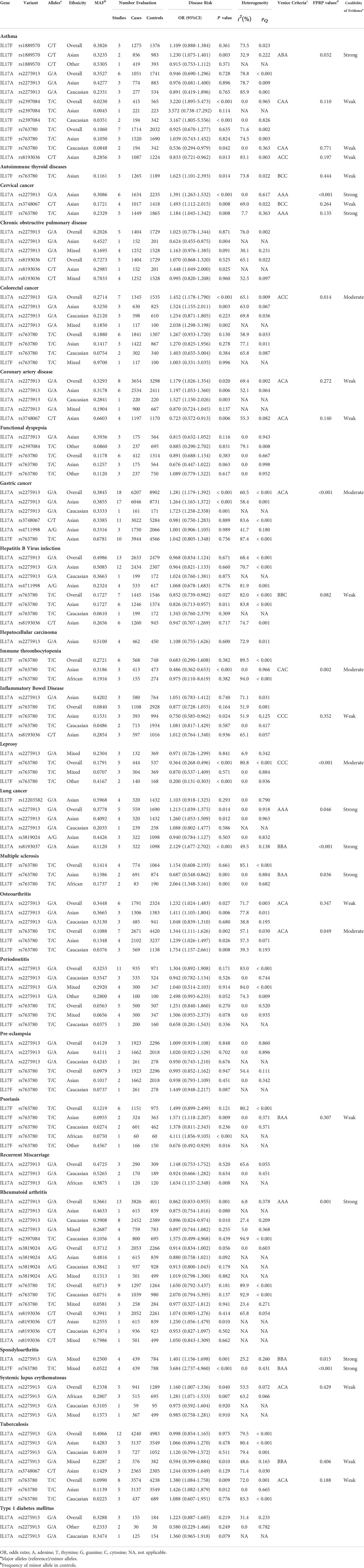
Table 1 Associations between variants in the IL-17 family genes associated with risk of human disease in meta-analysis under additive model (at least 3 datasets).
For non-cancer disease, current results showed that significant relationships with asthma risk were found for rs1889570 in Asians (OR=1.230), rs2397084 (OR=3.220), rs763780 (OR=0.536) and rs8193036 (OR=0.833). In addition, rs763780 had an elevated susceptibility to AITD (OR=1.623) in Asians. For CAD, significant relationships were found for rs2275913 (OR=1.179). Additionally, rs3748067 could reduce risk of CAD in Asians (OR=0.723). For HBV infection, rs763780 had a decreased susceptibility to HBV infection (OR=0.852), especially among Asians (OR=0.826). Apart from that, rs763780 could decrease susceptibility to ITP (OR=0.486) and risk of IBD (OR=0.750) in Asians. Apart from that, rs763780 had a reduced susceptibility to leprosy (OR=0.364) and risk of MS (OR=0.687), respectively. Interestingly, current results showed that rs2275913 could increase risk of OA (OR=1.232), SpA (OR=1.401) and SLE (OR=1.160), whereas decrease susceptibility to RA (OR=0.862) and TB (OR=0.594); rs763780 could increase risk of OA (OR=1.344), psoriasis (OR=1.571), SpA (OR=3.684) and TB (OR=1.380), respectively.
Additionally, 8 SNPs (rs12203582, rs2275913, rs2397084, rs3748067, rs3819024, rs8193036, rs763780 and rs4711998) had no association with susceptibility to 14 human diseases (asthma, COPD, colorectal cancer, FD, gastric cancer, HBV infection, hepatocellular carcinoma, IBD, lung cancer, PE, RM, RA, TB and T1DM) in additive model. Of these, 3 SNPs had no association with 3 diseases (rs3748067 and rs763780 for gastric cancer; rs2275913 for HBV infection; rs3748067 for TB) with at least 2,300 case and 2,300 controls. Also, we calculated the statistical power to confirm whether the bigger sample size confirming these relationships is required in next study (Table 2 and Supplementary Table 3).
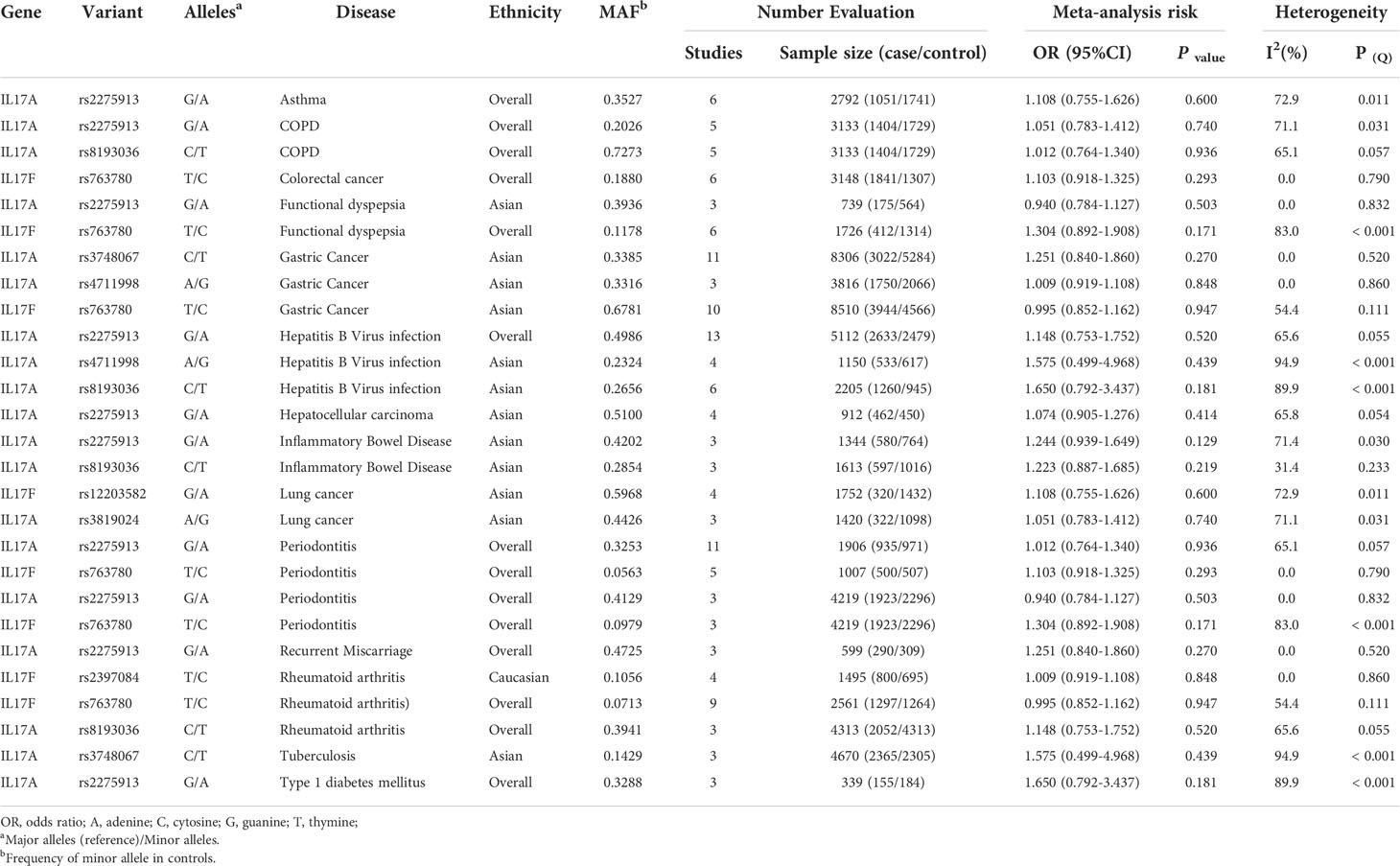
Table 2 Variants in IL-17 family genes showing no relation to risk of disease in meta-analyses in additive model.
Heterogeneity, bias and sensitivity analysis
As shown in Table 1, heterogeneity was investigated for 28 significant associations (7 SNPs for 18 human diseases). Mild heterogeneity (I2 < 25%) was assigned to 3 variants with risk of 2 cancers and 6 noncancerous diseases (10 associations); moderate heterogeneity (25% ≤ I2 ≤ 50%) was assigned to 3 variants with risk of 1 cancer and 3 noncancerous diseases (4 associations); high heterogeneity (I2 > 50%) was assigned to 4 variants with risk of 3 cancers and 9 noncancerous diseases (14 associations). Moreover, the results indicated that publication bias existed (p < 0.10) in associations for rs3748067 and rs2275913 with colorectal cancer risk. Apart from that, sensitivity analyses indicated that some significant summary ORs were lost, including rs8193036 in asthma and AITD, rs763780 in HBV infection (excess of significant findings); rs763780 in AITD and ITP (small study), and in IBD (HWE), and in leprosy (small study).
Cumulative evidence of association
As shown in Table 1, our study firstly used the Venice guideline and FPRP tests to grade epidemiological credibility of 28 significant. In terms of Venice guideline, strong, moderate and weak evidence were assigned to 4, 7 and 17 associations, respectively. Then, the probability for a true association between the 28 positive associations was investigated based on FPRP tests. The FPRP value < 0.05 was observed for 13 associations, FPRP 0.05 to 0.2 for 6 associations, and FPRP > 0.2 was found for 9 associations, respectively. At last, combing Venice guideline and FPRP tests, strong evidence was assigned to 4 variants (IL-17F rs1889570, IL-17A rs2275913, IL-17F rs763780, IL-17A rs8193037) and 2 cancer (cervical and lung cancer) as well as 4 noncancerous diseases (asthma, MS, RA, SpA), moderate to 2 SNPs (IL-17A rs2275913, IL-17F rs763780) and colorectal and gastric cancer as well as 3 noncancerous diseases (ITP, leprosy, OA), weak to 5 SNPs (IL-17F rs2397084, IL-17F rs763780, IL-17A rs8193036, IL-17A rs3748067, IL-17A rs2275913) and 1 cancer (cervical cancer) as well as 9 noncancerous diseases (asthma, AITD, CAD, HBV infection, IBD, OA, psoriasis, SLE, TB).
In addition, we attempted to pool the ORs and 95%CIs on the basis of two datasets and found that 9 variants (rs1889570, rs2275913, rs3819024, rs4711998, rs4819554, rs6973569, rs763780, rs8193036 and rs8193037) had significantly associated with susceptibility to 4 cancers (bladder, colorectal, papillary thyroid cancer and hepatocellular carcinoma) as well as 10 noncancerous diseases (ankylosing spondylitis, Behcet’s disease, bronchiolitis, brucellosis, chronic chagas cardiomyopathy, CAD, gastro-duodenal ulcer, recurrent miscarriage, silicosis and TB); of these, 5 SNPs (rs4819554, rs8193036, rs8193037, rs2275913, rs763780) and risk of 4 noncancerous diseases (ankylosing spondylitis, CAD, gastro-duodenal ulcer, recurrent miscarriage) were considered noteworthy (P < 0.2 for FPRP) (see Supplementary Table 4). Additionally, we calculated the ORs and 95% CI in the additive model to assess the relationships between 53 variants and susceptibility to 90 diseases (based on one dataset), yielding significant relationships between 22 variants and the risk of 47 types of carcinoma. Apart from that, P value of FPRP for the significant associations also be calculated. Finally, we considered the associations between 14 variants and susceptibility to 21 diseases noteworthy (see Supplementary Table 5).
Functional annotation for variants with strong evidence
As shown in Table 3, we used the Encyclopedia of DNA Elements tool HaploReg v4.1 to assess the potential functional roles for strong evidence (4 variants with risk of 6 human diseases). For functional annotations, rs763780 was annotated as missense. The total 4 SNPs might be located in a region with strong promoter and enhancer activity, and two SNPs in alteration in regulatory motif. Subsequently, as the consequence of the function evaluation using the PolyPhen-2 web server (29), the unique non-synonymous variant rs763780 was qualitatively predicted to be “benign” with a naïve Bayes posterior probability of less than 0.15. As shown in Supplementary Figure 1, the linkage disequilibrium (LD) plots presented that the regions represented by significant SNPs had distinct genetic structures among in European, Asian and African ancestry. The information extracted from the Genotype-Tissue Expression Project shows that rs2275913, rs763780, rs8193037, rs1889570 are eQTLs for the IL-17A, IL-17F, GSTA8P, MCM3. In addition, rs2275913 had an increased expression in GSTA8P, IL-17A genes in testis tissues; rs763780 and rs8193037 had an increased expression in MCM3,IL-17F genes in muscle and esophagus tissues, respectively (Supplementary Table 6). In our study, rs2275913 and rs763780 had significantly associated with susceptibility to cervical cancer and SpA. The Phase 3 of the 1000 Genomes Project (30) (Supplementary Table 7) indicated IL-17A rs2275913 is uncorrelated with IL-17F rs763780 in Europeans, East Asians and Africans (r2< 0.05 for all tests). Moreover, IL-17A rs2275913 and IL-17A rs8193037 had associated with predisposition to lung cancer. We also found that rs2275913 is weak LD with rs8193037 in East Asians (r2 = 0.1) and is uncorrelated with rs8193037 in Europeans and Africans (r2< 0.05).
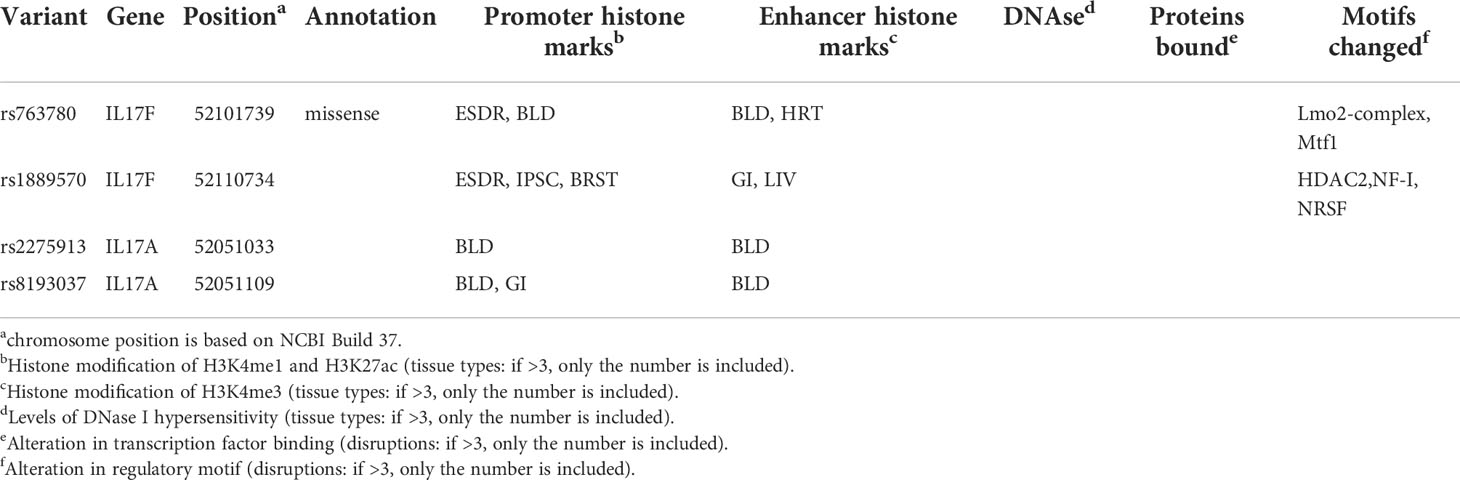
Table 3 Summary of functional annotations for 4 SNPs in 6 human diseases (strong epidemiological credibility).
Discussion
Our study performs a comprehensive research synopsis and meta-analysis, summarizes and updates the associations between SNPs in IL-17 family genes and predisposition to human diseases for the first time, which offers precise results for the SNPs and provides more variants and diseases that never been investigated before. Our study included 227 papers with 73,509 cases and 93,253 controls and performed a meta-analysis using 135 papers with available information to assess relationships of 10 variants with susceptibility to 25 diseases (5 cancers as well as 20 non-cancer diseases); 7 SNPs had positively associated with 18 human disease predisposition. Our study used the Venice guidelines and FPRP tests to grade the cumulative evidence of significant relationships. At last, 4 SNPs were assigned to strong evidence with predisposition to 6 human diseases (9 associations: IL-17F rs1889570 in asthma; IL-17A rs2275913 in lung cancer, cervical cancer, RA, SpA; IL-17F rs763780 in cervical cancer, MS, SpA; IL-17A rs8193037 in lung cancer), moderate to 2 SNPs and 2 cancer as well as 3 noncancerous diseases, weak to 5 SNPs and 1 cancer as well as 9 noncancerous diseases. Moreover, we attempted to construct functional annotations for these 4 variants with strong evidence using data from the Encyclopedia of DNA Elements Project and other public databases and then uncovered that the SNPs with strong evidence might fall in several putative regulatory regions. In summary, this study provides updated evidence that SNPs in the IL-17 family genes had significant associations with predisposition to lung, cervical cancer, asthma, RA, SpA and MS.
The IL-17, a kind of proinflammatory cytokine, plays a crucial role in both innate and acquired immune responses (31). Previous papers demonstrated that IL-17 is activated by microbial products, and may accelerate carcinoma occurrence and development by angiogenic functions (32). The IL-17A gene (Gene ID: 3605) is located at chromosome 6p12.2, and the encoded protein is a proinflammatory cytokine produced by activated T cells, which might involve in the development of human diseases (31). In the previous paper, it was pointed out that IL-17A rs2275913 acted as risk factor for multiple cancers (gastric, cervical, colorectal and oral carcinoma) (15) and non-cancerous diseases (RA) (33). Consistent with our meta-analysis, strong evidence was assigned to IL-17A rs2275913 in lung cancer, cervical cancer, RA, SpA, and IL-17A rs8193037 in lung cancer. LD analysis indicates that IL-17A rs2275913 and IL-17A rs8193037 were associated with susceptibility to lung cancer. We also found that rs2275913 is weak LD with rs8193037 in East Asians and is uncorrelated with rs8193037 in Europeans and Africans, indicating that the functional mechanisms of the two variants associated with lung cancer risk may be distinct in different ethnic groups and partly explain why some variants are found to be associated with a cancer site in one ethnic group but not in others. Current evidence presents that high expression of IL-17A is linked with the development and progression of cancers, and IL-17A could be regulated at the transcriptional level (34). IL-17A rs2275913 could influence the expression of the IL-17A protein and trigger cell transformation and maintain the autonomous proliferation of the transformed cells, and thus increase the susceptibility of cervical cancer, especially in HPV infection individuals (35). IL-17A could influence the transcriptional activity of NFAT and trigger the stimulation of T lymphocytes cells, which might increase risk of lung cancer (36). Moreover, a recent study indicated that the G allele polymorphism of IL-17A rs2275913 (a change from glutamic acid to lysine) was protective in RA individuals (37), which is consistent with our results; IL-17A and TNF-α had been considered as a predictor of a poor outcome in RA individuals; interestingly, previous study concluded that therapies targeting IL-17 in autoimmune diseases ameliorated the inadequate response to anti-TNF-α therapy (38), which indicated that SNP rs2275913 could be considered as a novel target for gene therapy of RA and promote drug developments against RA. Moreover, the SNP rs2275913 A allele is linked with high IL-17 expression, which has an elevated susceptibility to autoimmune and inflammatory diseases, including SpA (39). Moreover, previous papers found that drugs target other molecules of the immune system, such as anti-IL-17A (ixekizumab and secukinumab) and anti-IL-17A receptor (brodalumab). The efficacy of anti-IL-17R and anti-IL-17 agents has been shown in Phase II (40) and III trials (41, 42), indicating that IL-17A might have impact on the pathogenesis of psoriasis.
The IL-17F gene (Gene ID:112744) is located at chromosome 6p12.2, and the protein encoded by IL-17F gene is a cytokine activated by T cells. It could stimulate the production of other cytokines, such as IL-6, IL-8, and CSF2/GM_CSF (31, 43). These cytokines might have similar synergistic effects on risk of human diseases (44). In the previous paper, it was pointed out that IL-17F rs763780 might trigger the development of cervical and oral carcinoma (15) and non-cancerous diseases (such as asthma) (45). Consistent with our meta-analysis, strong evidence was assigned to IL-17F rs1889570 in asthma, and IL-17F rs763780 in cervical cancer, MS, SpA. In our study, rs2275913 and rs763780 had positive association with susceptibility to SpA and cervical cancer. LD analysis indicates that IL-17A rs2275913 is uncorrelated with IL-17F rs763780 in Europeans, East Asians and Africans, demonstrating that there might be different causal variants and functional mechanisms involved in relationships of variants in the IL-17A-IL-17F genes with risk of cervical cancer and SpA. Current evidence presents that IL-17F rs1889570 could increase risk of asthma by influencing the expression of proinflammatory cytokines, chemokines, and growth factors associated with leukocyte activation and airway remodeling (46). Additionally, a previous study found that the IL-17F rs763780, a missense located in the IL-17F exon3 region, could trigger high IL-17 expression which influenced cervical cancer cell growth, and thus increased risk of cervical cancer (47). Moreover, the SNP rs763780 C allele is linked with high IL-17 expression, which has proved to increase risk of SpA (39).
Additionally, we calculated the ORs and 95% CI in the additive model to assess the relationships between 53 variants and susceptibility to 90 diseases (based on one dataset), yielding significant relationships between 22 variants and the risk of 47 types of carcinoma. For example, our results found that some non-cancerous diseases including autoimmune diseases (such as autoimmune thyroid diseases), alopecia areata, and some type of autoimmune blistering diseases (such as bullous pemphigoid), which have presented in our supplementary Tables (see Supplementary Table 1). Moreover, in our study, we performed meta-analysis based on at least three datasets. However, we found that alopecia areata and bullous pemphigoid only contained 1 dataset for each SNP, which could not be assessed by meta-analysis. Therefore, we just presented these information in our supplementary files (see Supplementary Table 1 and Supplementary Table 5). Finally, we hope to attempt to collect more information in order to solve this issue in our study in the future. Apart from that, P value of FPRP for the significant associations also be calculated. In summary, we considered the associations between 14 variants and susceptibility to 21 diseases noteworthy (see Supplementary Table 5). Further, well-designed studies are recommended to clarify the association with multiple diseases for these variants.
Additionally, 8 SNPs had no association with susceptibility to 14 human diseases in additive model. Of these, 3 SNPs had no association with 3 diseases (rs3748067 and rs763780 for gastric cancer; rs2275913 for HBV infection; rs3748067 for TB) with at least 2,300 cases and 2,300 controls, which presented over 80% statistical power to detect an OR of 1.15 for a variant with MAF 0.20 (Type 1 error 0.05). Further study less than current individuals on these 3 variants for these 3 diseases will not yield fruitful results (Supplementary Table 3). Apart from that, our study identified that significant relationships for 5 variants with risk of 4 diseases (based on two datasets) and 14 variants with risk of 21 diseases (based on one dataset) were considered noteworthy, which might be required to confirm or refute these associations by large-scale studies in the future.
Some limitations apply to this research: (i) even though we conducted a comprehensive search to screen eligible papers, some articles may have been missed. Also, some malignancies and non-cancer diseases could not be completely assessed by meta-analysis owing to insufficient information (for example, lack of genotype amount, fewer than 3 datasets in some associations); (ii) only ethnicity was assessed by subgroup, other factors (such as pathological/clinical type, gene-gene or gene-environment associations and interactions) might be required to confirm or refute the relationships with susceptibility to disease; (iii) the unreasonable data, like errors in genotype, could not be investigated, and (iv) moderate and weak evidence should be explained with caution.
In summary, this large-scale meta-analysis identified that 4 SNPs in the IL-17 family genes were graded as demonstrating strong association to 2 cancer and 4 non-cancer disease risk. Apart from that, these findings provide a foundation for further demonstrating the variations in the IL-17 family genes are positively linked with susceptibility to cervical cancer, lung cancer, asthma, MS, RA, SpA, and highlight that the variants in IL-17 family genes might become a valuable genetic tool to investigate the pharmacological targeting potential of IL-17 family genes. We should further understand its biological pathway and apply these clues to clinical practice and public health for risk assessment and management.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
TL, LY, and HC designed this work. TL and LY integrated and analyzed the data. TL, LY, and HC wrote this manuscript. TL, LY, XL, CZ, CJ, ZY, CF, and HC finished the related Tables and Figures. TL, LY, and HC edited and revised the manuscript. All authors approved this manuscript.
Funding
This study was supported by funding from the Chongqing Natural Science Foundation (grant No. cstc2020jcyj-msxmX0257).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1008184/full#supplementary-material
References
1. Moseley TA, Haudenschild DR, Rose L, Reddi AH. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev (2003) 14(2):155–74. doi: 10.1016/s1359-6101(03)00002-9
2. Furue M, Furue K, Nakahara G. Interleukin-17A and keratinocytes in psoriasis. Int J Mol Sci (2020) 21(4):1275. doi: 10.3390/ijms21041275
3. Batur LK, Hekim N. Correlation between interleukin gene polymorphisms and current prevalence and mortality rates due to novel coronavirus disease 2019 (COVID-2019) in 23 countries. J Med Virol (2021) 93(10):5853–63. doi: 10.1002/jmv.27127
4. Zou WP, Restifo NP. T(H)17 cells in tumour immunity and immunotherapy. Nat Rev Immunol (2010) 10(4):248–56. doi: 10.1038/nri2742
5. Hizawa N, Kawaguchi M, Huang SK, Nishimura M. Role of interleukin-17F in chronic inflammatory and allergic lung disease. Clin Exp Allergy (2006) 36(9):1109–14. doi: 10.1111/j.1365-2222.2006.02550.x
6. Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol (2008) 28(1):44–9. doi: 10.1007/s10875-007-9125-8
7. Miyahara Y, Odunsi K, Chen WH, Peng GY, Matsuzaki J, Wang RF. Generation and regulation of human CD4+ IL-17-producing T cells in ovarian cancer. Proc Natl Acad Sci USA (2008) 105(40):15505–10. doi: 10.1073/pnas.0710686105
8. Zhu XW, Mulcahy LA, Mohammed RAA, Lee AHS, Franks HA, Kilpatrick L, et al. IL-17 expression by breast-cancer-associated macrophages: IL-17 promotes invasiveness of breast cancer cell lines. Breast Cancer Res (2008) 10(6):R95. doi: 10.1186/bcr2195
9. Kuang DM, Peng C, Zhao QY, Wu Y, Chen MS, Zheng LM. Activated monocytes in peritumoral stroma of hepatocellular carcinoma promote expansion of memory T helper 17 cells. Hepatology (2010) 51(1):154–64. doi: 10.1002/hep.23291
10. Wang B, Li L, Liao Y, Li JQ, Yu XJ, Zhang Y, et al. Mast cells expressing interleukin 17 in the muscularis propria predict a favorable prognosis in esophageal squamous cell carcinoma. Cancer Immunol Immunother (2013) 62(10):1575–85. doi: 10.1007/s00262-013-1460-4
11. Wu XQ, Zeng ZR, Chen B, Yu J, Xue L, MH YTC, et al. Association between polymorphisms in interleukin-17A and interleukin-17F genes and risks of gastric cancer. Int J Cancer (2010) 127(1):86–92. doi: 10.1002/ijc.25027
12. Ma QY, Chen J, Wang SH, Wu N, Hao ZH, Chen XF. Interleukin 17A genetic variations and susceptibility to non-small cell lung cancer. APMIS (2015) 123(3):194–8. doi: 10.1111/apm.12341
13. Dai W, Zhou Q, Tan XX, Sun CF. IL-17A (-197G/A) and IL-17F (7488T/C) gene polymorphisms and cancer risk in Asian population: A meta-analysis. Onco Targets Ther (2014) 7:703–11. doi: 10.2147/OTT.S62781
14. Zhao HY, Wang R, Ma W. IL-17A G197A and IL-17F T7488C polymorphisms and cancer risk in Asian populations: A meta-analysis. J BUON (2014) 19(2):562–6.
15. Hu YT, Xu DD, Xia HR, Zhang M, Liang CZ. Associations of IL-17A -197G/A and IL-17F 7488T/C polymorphisms with cancer risk in asians: An updated meta-analysis from 43 studies. Gene (2021) 804:145901. doi: 10.1016/j.gene.2021.145901
16. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PloS Med (2009) 6(7):e1000097. doi: 10.1371/journal.pmed.1000097
17. Sagoo GS, Little J, Higgins JPT. Systematic reviews of genetic association studies. Hum Genome Epidemiol Netw PloS Med (2009) 6(3):e28. doi: 10.1371/journal.pmed.1000028
18. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med (1997) 127(9):820–6. doi: 10.7326/0003-4819-127-9-199711010-00008
19. Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med (2002) 21(11):1539–58. doi: 10.1002/sim.1186
20. Ioannidis JPA, Trikalinos TA. An exploratory test for an excess of significant findings. Clin Trials (2007) 4(3):245–53. doi: 10.1177/1740774507079441
21. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics (1994) 50(4):1088–101. doi: 10.2307/2533446
22. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (1997) 315(7109):629–34. doi: 10.1136/bmj.315.7109.629
23. Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: Interim guidelines. Int J Epidemiol (2008) 37(1):120–32. doi: 10.1093/ije/dym159
24. Wacholder S, Chanock S, Garcia-Closas M, Ghormli LE, Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer Inst (2004) 96(6):434–42. doi: 10.1093/jnci/djh075
25. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res (2016) 44(D1):D877–81. doi: 10.1093/nar/gkv1340
26. GTEx consortium. human genomics. the genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (2015) 348(6235):648–60. doi: 10.1126/science.1262110
27. Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet (2012) 44:1084–9. doi: 10.1038/ng.2394
28. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics (2015) 31(21):3555–7. doi: 10.1093/bioinformatics/btv402
29. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods (2010) 7(4):248–9. doi: 10.1038/nmeth0410-248
30. Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics (2015) 31(21):3555–7. doi: 10.1093/bioinformatics/btv402
31. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
32. Shime H, Yabu M, Akazawa T, Kodama K, Matsumoto M, Seya T, et al. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J Immunol (2008) 180(11):7175–83. doi: 10.4049/jimmunol.180.11.7175
33. Chen P, Li YH, Li LL, Zhang GX, Zhang F, Tang Y, et al. Association between the interleukin (IL)-17A rs2275913 polymorphism and rheumatoid arthritis susceptibility: A meta-analysis and trial sequential analysis. J Int Med Res (2021) 49(10):3000605211053233. doi: 10.1177/03000605211053233
34. Chen XM, Wang JF, Wang R, Su QH, Luan JW, Huang HY, et al. Th1-, Th2-, and Th17-associated cytokine expression in hypopharyngeal carcinoma and clinical significance. Eur Arch Otorhinolaryngol (2016) 273(2):431–8. doi: 10.1007/s00405-015-3779-2
35. Niu AQ, Cao YH, Wang H, Zhang X, Zhu B, Li ZH. Role of IL17A rs2275913 and rs3748067 polymorphisms in the risk cervical cancer. Genet Mol Res (2017) 16(3). doi: 10.4238/gmr16038826
36. Kaabachi W, Amor AB, Kaabachi S, Rafrafi A, Tizaoui K, Hamzaoui K. Interleukin-17A and -17F genes polymorphisms in lung cancer. Cytokine (2014) 66(1):23–9. doi: 10.1016/j.cyto.2013.12.012
37. Amin A, Sheikh N, Mukhtar M, Saleem T, Akhtar T, Fatima T, et al. Association of interleukin-17 gene polymorphisms with the onset of rheumatoid arthritis. Immunobiology (2021) 226(1):152045. doi: 10.1016/j.imbio.2020.152045
38. Liang Y, Pan HF, Ye DQ. IL-17A-producing CD8(+)T cells as therapeutic targets in autoimmunity. Expert Opin Ther Targets (2015) 19(5):651–61. doi: 10.1517/14728222.2014.997710
39. Ferreira Neves JS, Laguila Visentainer JE, da Silva Reis DM, Rocha Loures MA, Alves HV, Valentini Zacarias JM, et al. IL17F: A possible risk marker for spondyloarthritis in HLA-B*27 negative Brazilian patients. J Pers Med (2021) 11(6):520. doi: 10.3390/jpm11060520
40. Newsom M, Bashyam AM, Balogh EA, Feldman SR, Strowd LC. New and emerging systemic treatments for atopic dermatitis. Drugs (2020) 80(11):1041–52. doi: 10.1007/s40265-020-01335-7
41. Paul C, Lacour JP, Tedremets L, Kreutzer K, Jazayeri S, Adams S, et al. Efficacy, safety and usability of secukinumab administration by autoinjector/pen in psoriasis: A randomized, controlled trial (JUNCTURE). J Eur Acad Dermatol Venereol (2015) 29(6):1082–90. doi: 10.1111/jdv.12751
42. Blauvelt A, Prinz JC, Gottlieb AB, Kingo K, Sofen H, Ruer-Mulard M, et al. Secukinumab administration by pre-filled syringe: Efficacy, safety and usability results from a randomized controlled trial in psoriasis (FEATURE). Br J Dermatol (2015) 172(2):484–93. doi: 10.1111/bjd.13348
43. Moghari MH, Geva T, Powell AJ. Prospective heart tracking for whole-heart magnetic resonance angiography. Magn Reson Med (2017) 77(2):759–65. doi: 10.1002/mrm.26117
44. Su S, Katopodi XK, Pita-Juarez YH, Maverakis E, Vlachos IS, Adamopoulos IE. Serine and arginine rich splicing factor 1 deficiency alters pathways involved in IL-17A expression and is implicated in human psoriasis. Clin Immunol (2022) 240:109041. doi: 10.1016/j.clim.2022.109041
45. Holster A, Teräsjärvi J, Barkoff AM, Lauhkonen E, Törmänen S, Helminen M, et al. IL17F rs763780 single nucleotide polymorphism is associated with asthma after bronchiolitis in infancy. Acta Paediatr (2021) 110(1):222–7. doi: 10.1111/apa.15390
46. Raeiszadeh Jahromi S, Mahesh PA, Jayaraj BS, Holla AD, Vishweswaraiah S, Ramachandra NB. IL-10 and IL-17F promoter single nucleotide polymorphism and asthma: A case-control study in south India. Lung (2015) 193(5):739–47. doi: 10.1007/s00408-015-9753-3
Keywords: interleukin 17 family gene, variant, cancer, noncancerous diseases, susceptibility
Citation: Liu T, Yang L, Lv X, Zuo C, Jia C, Yang Z, Fan C and Chen H (2022) Cumulative evidence for associations between genetic variants in interleukin 17 family gene and risk of human diseases. Front. Immunol. 13:1008184. doi: 10.3389/fimmu.2022.1008184
Received: 31 July 2022; Accepted: 20 September 2022;
Published: 10 October 2022.
Edited by:
Simone Mader, Ludwig Maximilian University of Munich, GermanyReviewed by:
Reza Akbarzadeh, University of Lübeck, GermanyXue Jun Zhang, Anhui Medical University, China
Copyright © 2022 Liu, Yang, Lv, Zuo, Jia, Yang, Fan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanwen Chen, Y29vbHN0YXJjaHc5NTI3QDE2My5jb20=
†These authors have contributed equally to this work
 Tianyu Liu
Tianyu Liu Lei Yang1†
Lei Yang1† Huanwen Chen
Huanwen Chen