
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 12 October 2022
Sec. Alloimmunity and Transplantation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1007647
This article is part of the Research TopicRising Stars in Alloimmunity and Transplantation 2021View all 7 articles
 Roberto Littera1,2*†
Roberto Littera1,2*† Andrea Perra2,3*†
Andrea Perra2,3*† Michela Miglianti4
Michela Miglianti4 Ignazio S. Piras5
Ignazio S. Piras5 Stefano Mocci6*†
Stefano Mocci6*† Sara Lai1
Sara Lai1 Maurizio Melis2
Maurizio Melis2 Teresa Zolfino7
Teresa Zolfino7 Cinzia Balestrieri8
Cinzia Balestrieri8 Maria Conti8
Maria Conti8 Giancarlo Serra8
Giancarlo Serra8 Francesco Figorilli7
Francesco Figorilli7 Davide Firinu4
Davide Firinu4 Simona Onali4
Simona Onali4 Laura Matta4
Laura Matta4 Carmen Porcu4
Carmen Porcu4 Francesco Pes4
Francesco Pes4 Daniela Fanni9
Daniela Fanni9 Cristina Manieli10
Cristina Manieli10 Monica Vacca6
Monica Vacca6 Roberto Cusano11
Roberto Cusano11 Marcello Trucas3
Marcello Trucas3 Selene Cipri2
Selene Cipri2 Stefania Tranquilli6
Stefania Tranquilli6 Stefania Rassu1
Stefania Rassu1 Federica Cannas6
Federica Cannas6 Mauro Giovanni Carta4
Mauro Giovanni Carta4 Marta Anna Kowalik3
Marta Anna Kowalik3 Erika Giuressi1
Erika Giuressi1 Gavino Faa9
Gavino Faa9 Luchino Chessa2,4,8*†
Luchino Chessa2,4,8*† Sabrina Giglio1,2,6,12
Sabrina Giglio1,2,6,12The immunomodulatory effects of HLA-G expression and its role in cancers, human liver infections and liver transplantation are well documented, but so far, there are only a few reports addressing autoimmune liver diseases, particularly autoimmune hepatitis (AIH).
Method and materials: We analyzed the genetic and phenotypic characteristics of HLA-G in 205 type 1 AIH patients (AIH-1) and a population of 210 healthy controls from Sardinia (Italy).
Results: Analysis of the HLA-G locus showed no substantial differences in allele frequencies between patients and the healthy control population. The HLA-G UTR-1 haplotype was the most prevalent in both AIH-1 patients and controls (40.24% and 34.29%). Strong linkage was found between the HLA-G UTR-1 haplotype and HLA-DRB1*03:01 in AIH-1 patients but not controls (D’ = 0.92 vs D’ = 0.50 respectively; P = 1.3x10-8). Soluble HLA-G (sHLA-G) levels were significantly lower in AIH-1 patients compared to controls [13.9 (11.6 – 17.4) U/mL vs 21.3 (16.5 – 27.8) U/mL; P = 0.011]. Twenty-four patients with mild or moderate inflammatory involvement, as assessed from liver biopsy, showed much higher sHLA-G levels compared to the 28 patients with severe liver inflammation [33.5 (23.6 – 44.8) U/mL vs 8.8 (6.1 – 14.5) U/mL; P = 0.003]. Finally, immunohistochemistry analysis of 52 liver biopsies from AIH-1 patients did not show expression of HLA-G molecules in the liver parenchyma. However, a percentage of 69.2% (36/52) revealed widespread expression of HLA-G both in the cytoplasm and the membrane of plasma cells labeled with anti-HLA-G monoclonal antibodies.
Conclusion: This study highlights the positive immunomodulatory effect of HLA-G molecules on the clinical course of AIH-1 and how this improvement closely correlates with plasma levels of sHLA-G. However, our results open the debate on the ambiguous role of HLA-G molecules expressed by plasma cells, which are pathognomonic features of AIH-1.
Autoimmune hepatitis (AIH) is a rare autoimmune disease in which the immune system attacks autologous hepatocytes, causing a chronic/relapsing inflammation of the liver that can lead to cirrhosis and liver failure. In the United States of America, the 5-year prevalence rate of AIH is estimated at 31.2 per 100,000 persons with a higher prevalence among females, the elderly (above 65 years), and individuals of Caucasian ethnicity (1). Studies from Europe indicate an incidence ranging from 0.9 to 2 per 100,000 population per year. A similar incidence has recently been reported in South Korea whereas New Zealand would seem to have an incidence of AIH among the highest reported worldwide (2). The highest point prevalence of the disease has been reported in Alaska (42.9/100,000) (3).
Although it is considered a rare disease, the incidence and prevalence of AIH are growing worldwide, and its complications are the principal indication for liver transplantation for up to 3% and 6% of the pediatric and adult population, respectively. Variants of AIH may occur, with features overlapping with primary biliary sclerosis (PBC) and/or primary sclerosing cholangitis (PSC) occurring in up to 10% of cases (4, 5). While more than 40% of patients develop acute hepatitis, sometimes in the form of fulminant hepatitis, some patients remain asymptomatic (6–8).
To date, the precise pathogenesis of AIH has not been clarified yet. There is strong evidence that environmental factors contribute to the disease, interacting with a predisposing genetic background (2, 9). Since AIH is a polygenic disease, the identification of triggers and genetic predisposing factors has proved challenging, although several studies have pointed to HLAs and TCR genes as potentially playing a role (10, 11). Even though the genetic background of AIH remains elusive, similar to other autoimmune diseases, it is characterized by a loss of immune tolerance that leads to chronic and progressive liver damage and inflammation (12). Among the most important tolerogenic systems, the human leukocyte antigen-G (HLA-G) is the best characterized non-classical major histocompatibility complex (MHC) Class Ib molecule (13, 14). The immunosuppressive effects exerted by this molecule on all types of immune cells are well documented and account for its pivotal role in the induction and maintenance of immune tolerance (15). The HLA-G gene maps to the short arm of chromosome 6 in the HLA region (6p21.2-21.3), between the HLA-A and HLA-F genes. Overall, its gene structure is similar to the classical HLA class I genes, bearing seven introns and eight exons encoding the molecule heavy chain (16, 17). The HLA-G gene promoter region comprises a modified enhancer A (enh A), S, and X1 sequence and a few alternative HLA-G transcriptional regulatory elements (18).
Alternative splicing of HLA-G primary transcript generates seven alternative mRNAs encoding membrane-bound (HLA-G1, -G2, -G3, -G4) and soluble (HLA- G5, -G6, -G7) protein isoforms. Soluble HLA-G (sHLA-G) is detectable in both serum and plasma and is mainly produced by monocytes (19, 20). HLA-G expression is regulated by a variety of factors, including genetic variability, post-transcriptional regulation, intracellular and extracellular microenvironmental factors (21). Depending on the cell type and the specific stimulation, some isoforms may be expressed, whereas others may not (22). Compared to the other HLA Class I genes, HLA-G displays a relatively low degree of variability with only 102 alleles (https://www.ebi.ac.uk/ipd/imgt/hla/about/statistics/, April 2022) and 35 proteins. Two non-coding regions have been identified as the polymorphic sites with the strongest influence on HLA-G expression: the 5’ upstream regulatory region (5’URR) and the 3’ untranslated region (3’UTR) of the HLA-G gene. These regions also contain a variety of regulatory elements capable of influencing mRNA stability, turnover, splicing and mobility mechanisms (23). According to the results of a worldwide genetic analysis published in 2014, higher or lower secretion of soluble HLA-G (sHLA-G) seems to be associated with haplotypes mapping in the 3’UTR of HLA-G (24, 25). More specifically, certain polymorphic sites located in the HLA-G 3’UTR (14-base pair insertion/deletion, 3003C/T, 3010C/G, 3027A/C, 3035C/T, 3142C/G, 3187A/G, 3196C/G) generate UTR haplotypes (26) which determine different plasma levels of sHLA-G. The UTR-1 haplotype is classified as the highest producer of sHLA-G, whereas UTR-2, UTR-3, UTR-4, and UTR-6 are classified as medium producers and UTR-5 and UTR-7 as low producers (27).
Alongside its limited degree of polymorphism, HLA-G differs from the classical HLA Class I molecules for its restricted pattern of protein expression. Initially, HLA-G expression was considered to be restricted to extravillous cytotrophoblasts (EVT) at the maternal-fetal interface, possibly suppressing immune response in the placenta to protect the fetus from lysis mediated by natural killer (NK) cells (28). Later, it was established that HLA-G protein in healthy non-fetal subjects is found in the thymic medulla, the cornea, proximal nail matrix, beta cells of the islets of Langerhans, mesenchymal stem cells, and erythroid and endothelial precursors (15, 29–32). It is well known that EVT expressing HLA-G has a tolerogenic function at the maternal-fetal interface, where they protect the fetus from destruction by the maternal immune system (33). As reported in Table 1, the immunosuppressive role of HLA-G molecules has also been widely explored in numerous inflammatory, immune-mediated and infective conditions as well as in the transplantation setting (25, 39–43, 45, 54). In malignancies, HLA-G expression can directly help tumor cells escape immune surveillance through receptor binding, impairment of chemotaxis, and a mechanism known as trogocytosis (34–37, 44). Recent data in the literature indicate that HLA-G also plays an important role in liver homeostasis and immune response to liver injury and/or cancer (46). These studies demonstrate detectable levels of HLA-G expression in the hepatocytes and biliary epithelial cells of patients with chronic hepatitis B. These patients had higher levels of plasma sHLA-G than healthy controls, albeit different levels of sHLA-G were found according to disease stage (48, 49). Similar results were reported for patients with chronic hepatitis C. In these patients, HLA-G was expressed by mast cells that promote the formation of fibrous septa. Additionally, HLA-G was shown to worsen liver fibrosis by creating a shift in host immune response mechanisms in favor of a TH2 cytokine profile (47, 50).
Moreover, high levels of sHLA-G were detected in patients diagnosed with hepatocellular carcinoma (HCC), possibly protecting cancer cells from immune attack by inhibiting the activation of immune cells such as CD8-positive T cells, NK cells, B cells, and dendritic cells (DCs) (38). Hence, in HCC, like in other cancers, expression of the tolerogenic HLA-G molecule correlates with a poor prognosis (51–53).
However, despite the recent significant advances in the understanding of HLA-G biology, there are, to the best of our knowledge, no reports investigating the immunomodulatory effects of HLA-G expression and its role in AIH-1. Starting from the evidence that HLA-G molecules have anti-inflammatory effects both in physiological and pathological conditions, this study aimed at investigating the role of HLA-G molecules in type-1 AIH. In particular, we analyzed the genetic and phenotypic characteristics of HLA-G by comparing them with a control population originating from the same geographical area (Sardinia, Italy). Furthermore, we investigated the distribution of HLA-G molecules in liver tissue.
In this study, we show that the role of HLA-G molecules in type-1 AIH is not univocal: on the one side, the immunomodulatory effect of HLA-G molecules improves the clinical course of AIH-1; on the other side, HLA-G expressed in plasma cells may be correlated with more aggressive AIH-1 clinical forms.
We recruited a cohort of 205 Sardinian outpatients with AIH type I who were referred to the Center for the Study of Liver Diseases, Department of Medical Sciences and Public Health of the University of Cagliari, and from the Division of Gastroenterology of the ARNAS, S. Michele Hospital (Cagliari, Italy). All patients originated from central-southern Sardinia, were recruited between 02/2014 and 12/2020, and were diagnosed according to the International Autoimmune Hepatitis Group (IAIHG) revised scoring system and the American Association for the Study of Liver Diseases (AASLD) Guidelines (55). To avoid bias, patients with type 2 AIH or overlap syndrome were excluded from the study, as well as patients affected by chronic liver disorders induced by drug or alcohol abuse, fatty liver disease, metabolic disorders, genetic disorders, autoimmune cholangitis, primary biliary cirrhosis, and primary sclerosing cholangitis. All enrolled patients were negative for hepatitis B surface antigen (HbsAg), anti-hepatitis A virus IgM antibody, anti-hepatitis C virus IgG antibody, and anti-hepatitis D virus IgG antibody (Figure 1).
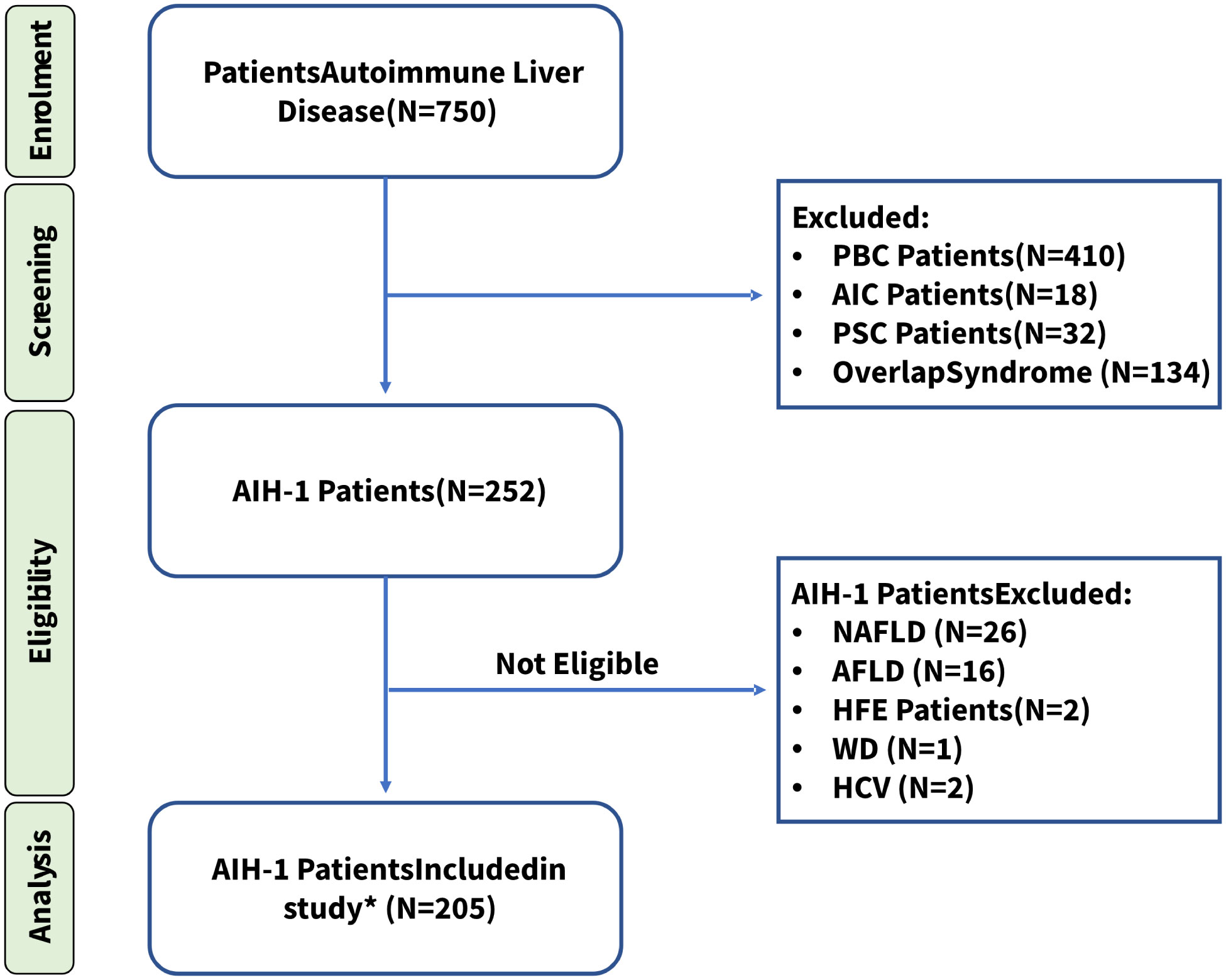
Figure 1 Patient enrollment workflow diagram. * All patients included in the study, were diagnosed according to the International Autoimmune Hepatitis Group (IAIHG) revised scoring system and the American Association for the Study of Liver Diseases (AASLD) Guidelines (55). PBC, Primary biliary cholangitis; AIC, Autoimmune cholangitis; PSC, Primary sclerosing cholangitis; NAFLD, Non-alcoholic fatty liver disease; AFLD, Alcoholic Fatty Liver Disease; HFE, Hereditary hemochromatosis; WD, Wilson Disease; HCV, Hepatitis C. *Fifteen patients (7.3%) tested negative for all AIH-related antibodies but were included in this study because of their high score (≥ 15) according to the IAIHG scoring system at the time of diagnosis.
The healthy control population corresponded to a panel of 210 individuals of Sardinian origin, going back at least two generations, selected from the regional bone marrow donor registry to represent the genetic background of the Sardinian population (56). This population group appropriately represented the male-to-female ratio and genetic profiles of the population in the central-south geographical areas from where the AIH-1 patients were recruited.
HLA-G alleles and 3’UTR haplotypes were compared between patients and healthy controls. Plasma sHLA-G levels were determined in patients and controls and stratified according to the different HLA-G 3’UTR haplotypes that have been described as influencing the expression of HLA-G (27).
Moreover, a subset of 52 AIH-1 patients whose liver biopsy was adequate for additional immunohistochemical staining, were analyzed to detect the presence of HLA-G in hepatic tissue and to evaluate its possible correlation with the level of hepatic inflammation (mild/moderate or severe).
Written informed consent was obtained from each patient or healthy subject included in the study, following the institutional and national ethical standards of the local human research committee. The study protocol, including informed consent procedures, conforms to the ethical guidelines of the Declaration of Helsinki and was approved by the responsible ethics committee (Ethics Committee of the Cagliari University Hospital; date of approval: January 23, 214; protocol number NP/2014/456). Records of written informed consent are kept on file and are included in the clinical record of each patient.
Genomic DNA was extracted from peripheral blood mononuclear cells according to standard methods (57). Patients and controls were typed at high-resolution for the alleles at the HLA-A, -B, -C -DR loci and other 13 HLA loci (including HLA-G) using a Next-Generation Sequencing (NGS) method. AlloSeq Tx17 (CareDx), an innovative NGS high-resolution HLA typing solution based on Hybrid Capture Technology (https://caredx.com/products-and-services/transplant-lab-products/hla-typing-solutions/alloseq-tx17/) was used to type all 415 samples (patients and controls). Data analysis was performed with AlloSeq Assign® software (v.1.0.2). Rare or ambiguous HLA alleles were methodically re-typed according to the Sanger sequencing-based typing (SBT) method using the following SBT kits: AlleleSEQR® HLA for the HLA-A, -B, -C and -DRB1 loci (GenDx&GenDx Products, Utrecht, The Netherlands).
Full-length NGS of the HLA-G gene including the 5’URR, introns, and 3’UTR, was performed using the MiSeq platform (Illumina). The HLA-G gene region extending from positions −1550 to 3404, relative to the start codon, was analyzed using long-range PCR. Specific primers were designed in relation to HLA-G RefSeqGene version NG_029039.1 (NCBI database), as previously described (58). The libraries were prepared starting from 1 ng of PCR product using the Nextera XT DNA Library Preparation Kit. The normalized libraries (4 nM) were pooled and loaded onto V3 flow cells for 600 cycles of bidirectional sequencing in a MiSeq Illumina Sequencer. The automatically generated FASTQ files were then processed with the MiSeq Reporter v2.6 for alignment and variant calling, and VariantStudio Software v3.0 for variant classification (Illumina, Netherlands). The variants were verified individually and then inserted into appropriate electronic spreadsheets for statistical analysis (see below). HLA-G 3’UTR haplotypes were determined by nucleotide sequence variations from +2945 to +3259 nucleotides, using a methodology and a nomenclature described elsewhere (14, 59).
Plasma samples were obtained from 205 patients at the time of diagnosis, before treatment and from 210 controls at the time of enrollment. Levels of sHLA-G were determined by the sHLA-G ELISA assay kit (Exbio, Prague, Czech Republic) according to the manufacturer’s instructions. This kit detects both shedding HLA-G-1 and soluble HLA-G5 molecules. Briefly, plasma samples were immediately frozen after separation and stored at -80°C until use. Fifty µl of each sample were diluted 1:80 in the plasma-specific buffer prior to running the HLA-G assay. A six-point calibration curve was obtained using the human native HLA-G protein supplied with the kit. A microplate reader with a 450 nm filter was used to measure the optical density at the end of the reaction. The limit of sensitivity was 0.6 U/ml. All samples were assayed in duplicate.
A subset of 52 AIH-1 patients whose liver biopsy was adequate for additional immunohistochemical stains, were analyzed to detect the presence of HLA-G in tissue. All biopsy specimens had at least 10 portal spaces and were suitable for immunohistochemical analysis.
Tissue samples from liver biopsies were fixed in 10% buffered formalin, routinely processed, and embedded in paraffin. Serial 3-µm-thick sections were obtained from each paraffin block. The serial sections were dewaxed, rehydrated and pretreated for histochemical and immunohistochemical staining. Standard hematoxylin & eosin staining was performed with Mayer hematoxylin and eosin in an aqueous solution. Serial sections were used for HLA-G immunostaining. Briefly, sections were subjected to a 10-min heat-induced epitope retrieval in buffer pH 9.00 (EnVisionTM FLEX Target Retrieval Solution High pH - Dako Denmark A/S, Glostrup, Denmark, Code K8004). Slides were then incubated for 20 min at room temperature with a specific anti-human MHC Class I HLA-G (LifeSpanBioSciences, Inc., Seattle, WA, USA, Code LS-B3734) mouse monoclonal antibody clone 4H84 at 1:100 dilution. Staining procedures were performed with the EnvisionTM FLEX+ (Dako, Code K8002) Detection System and the Autostainer Link 48 instrument according to the manufacturer’s instructions. The immunohistochemical evaluation of HLA-G protein expression in stained liver biopsy specimens was performed by three independent pathologists according to the following scores: 0 points for negative staining (–), 1 for staining <25% of examined cells (+), 2 between 26 and 50% (+), 3 between 51 and 75% (++), 4 between 76 and 100% (+++). The intensity of the staining was not considered for the evaluation of HLA-G expression. The average agreement for the 3 assessed variables among the three pathologists (DF, CM, GF) was 80%, ranging from 70% for Kupffer cells, histiocytes and macrophages to 90% for hepatocytes and plasma cells.
Hepatic inflammation was histologically graded as mild, moderate and severe according to the international grading system of hepatitis activity based on of the presence of portal inflammation, interface hepatitis, and lobular inflammation (60). A correlation between the degree of hepatic inflammation and the HLA-G tissue presence was also investigated.
Clinical and biochemical parameters of the AIH-1 patients were computed using mean and standard deviations (SD) for continuous variables and percentages for categorical data. Ninety-five percent confidence intervals (95% CI) were calculated for all variables. Results for patients were compared by applying the Student’s test and Fisher’s exact test as appropriate.
Comparisons of HLA-G alleles and HLA-G 3’UTR haplotypes between AIH-1 patients and healthy controls were performed using the exact Fisher’s test. P values were adjusted for multiple testing by Bonferroni method accounting for the number of HLA-G 3’UTR haplotypes analyzed. Only corrected P values (Pc) lower than 0.05 were considered to be statistically significant.
Soluble HLA-G plasma levels of AIH-1 patients and healthy controls were expressed using median values, 95% confidence intervals and boxplots. Subgroups of patients and/or controls stratified according to the presence or absence of the UTR-1 haplotype or mild/moderate and severe inflammatory conditions were also evaluated. P values were computed by using the Wilcoxon rank-sum test (61) for data not following a normal distribution. The 95% confidence intervals of the median values were obtained with Hodges-Lehmann statistics (62).
Kendall’s tau rank method (61) was used to evaluate the correlation between sHLA-G levels and other biochemical parameters (ALT, ALP and γ-globulin) in a subset of 52 AIH-1 patients whose liver biopsy was adequate for analysis.
The linkage disequilibrium (LD) between the HLA-G UTR-1 haplotype and the complete or partial Sardinian HLA extended haplotypes was evaluated in AIH-1 patients and healthy controls. The expected frequencies were obtained by multiplying the frequency of the HLA-G UTR-1 haplotype and that of the considered HLA haplotype. The observed and expected frequencies in each sample were compared using the chi-square test.
Linkage disequilibrium was measured by the parameters D (difference between the observed and expected frequencies) and D′ (i.e. D normalized to one: -1 ≤ D′ ≤ 1). D′ was obtained using the normalization formulas proposed by Lewontin (63) for two-loci haplotypes.
In order to compare the linkage disequilibrium in the control and patient group, we computed the P value associated with the Chi-square variable (with two degrees of freedom) given by the difference between the chi-square variables in the two cohorts (with one degree of freedom).
Statistical analysis was performed using R software version 4.2.1 [R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/].
The clinical and immunological characteristics of 205 AIH-1 patients are shown in Table 2. The mean age at diagnosis was 51.7 (SD 13.3) years and 177 patients (86.3%) were females. Median disease duration at the time of evaluation for this study was 6 years (IQR 6 years). Most of the patients had high titers (64.9%) of antinuclear antibodies (ANA), detected either alone or in combination with anti-smooth muscle antibodies (SMA) (42.9%). Liver kidney microsomal type-1 (LKM-1) and type-3 (LKM-3) antibodies were absent in all samples. Fifteen patients (7.3%) tested negative for all AIH-related antibodies but were included in this study because of their high score (≥ 15) according to the IAIHG scoring system at the time of diagnosis. More than one-third of the patients (35.6%) presented one or more associated autoimmune diseases, including Hashimoto’s thyroiditis, rheumatoid arthritis and mixed connectivitis. One hundred and thirty-eight patients (67.3%) tested positive for HLA-DRB1*03 antigens, and 90 patients (43.9%) tested positive for HLA-DRB1*04 antigens. Most patients (66.8%) had been treated with steroids, either alone or in combination with azathioprine (59.5%), and only eleven patients (5.4%) were non-responders or partial responders to therapy. These results are in line with those reported in previous studies on European and North American populations (1, 64, 65).
Out of the 52 patients whose liver biopsy was adequate for analysis, 44 were females (84.6%), and 8 were males (15.4%). Twenty-eight out of 52 patients (53.8%) presented severe inflammation, while in the remaining 24 patients (46.2%), parenchymal inflammation was mild or moderate. Patients who showed severe inflammation in the liver tissue had higher transaminase (AST, ALT), alkaline phosphatase (ALP), and γ-globulin levels compared to the other AIH-1 patients. Furthermore, in this group of 28 patients, AIH-1 was associated with an increased frequency of other autoimmune diseases [53.8% vs 33.0%, P = 0.048]. No other significant differences were found for the remaining clinical and immuno-genetical characteristics (Table 2).
The comparison of HLA-G alleles and 3’UTR haplotype frequencies between 205 AIH-1 patients and 210 healthy controls has been reported in Table 3. Similar to Northern Europe and Northern America populations, in our AIH-1 patients, the HLA-DRB1*03:01 allele was observed with the highest frequency at the DRB1 locus (Supplementary Table S1), reaching a statistically significant difference compared to the control population [38.8% vs 21.7%; OR = 2.29 (95% CI 1.67 – 3.15) P = 1.0·10-7].
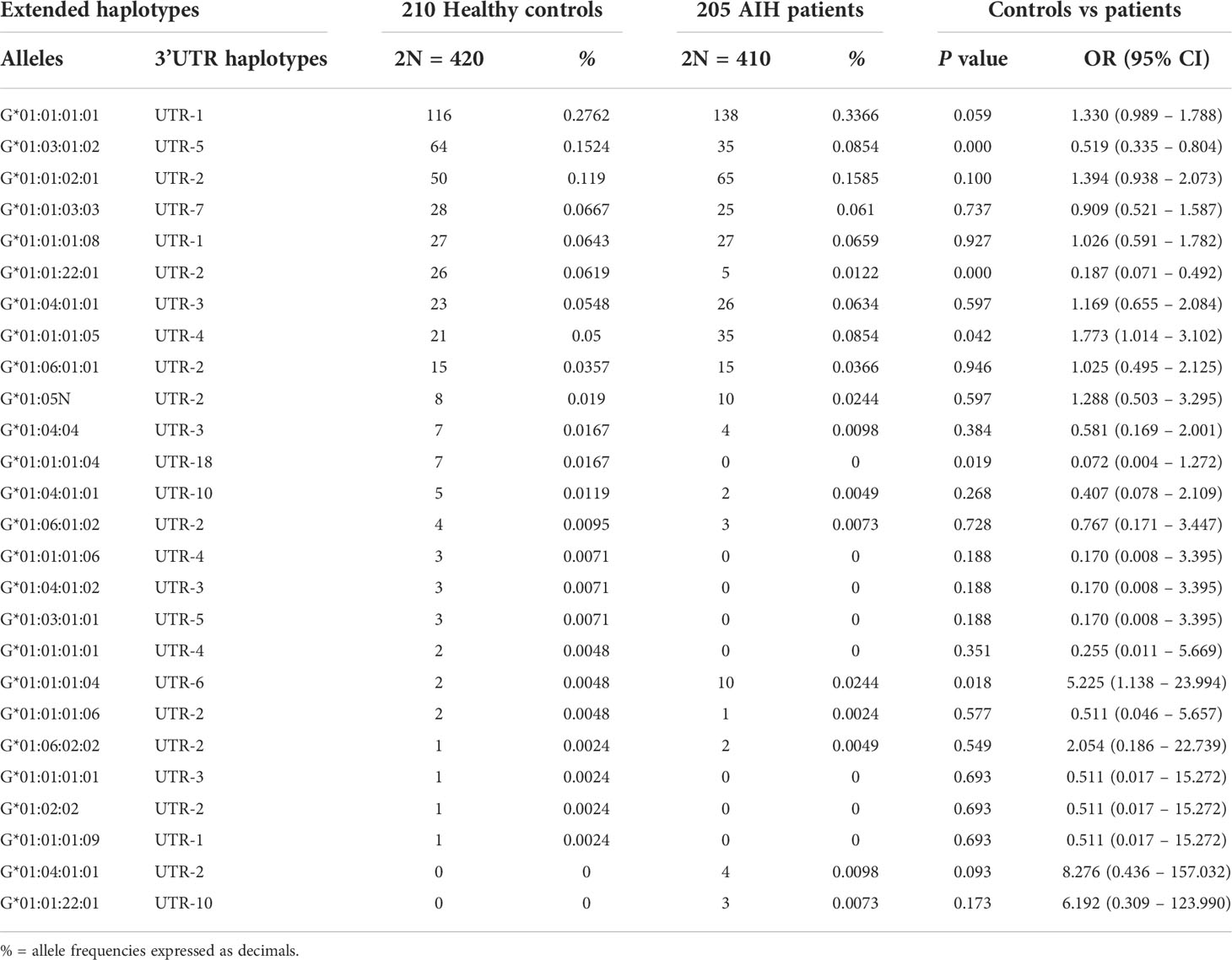
Table 3 Extended haplotypes (HLA-G alleles and 3’UTR haplotypes) frequencies in healthy controls and AIH type 1 patients.
Analysis of the extended haplotypes (HLA-G alleles and 3’UTR haplotypes) showed no substantial difference in frequencies between patients and healthy controls (Table 3). The most prevalent extended haplotypes in both groups were HLA-G*01:01:01:01 with UTR-1, HLA-G*01:01:02:01 with UTR-2, HLA-G*01:03:01:02 with UTR-5 and G*01:01:01:05 with UTR-4 (33.66%, 15.85%, 8.54% and 8.54% in patients vs 27.62%, 11.90%, 15.24% and 5.0% in controls). Only HLA-G*01:03:01:02 with UTR-1 showed a lower frequency in patients [8.54% vs 15.24%; OR = 0.52 (95% CI 0.33 – 0.82) P = 0.004, Pc = NS]. However, the differences between the frequencies of the HLA-G 3’UTR extended haplotypes in the two groups (AIH-1 patients and controls) never exceeded 6.7%, and the P value was not statistically significant after multiple testing adjustments (Pc = NS for all extended haplotypes, except for HLA-G*01:01:22:01 with UTR-2: 1.22% vs 6.19%; P = 1.5·10-4 and Pc = 0.004). HLA-G 3’UTR haplotype frequencies are described in Table 4. The UTR-1 haplotype was the most prevalent in both AIH-1 patients and controls (40.24% vs 34.29% respectively; P = 0.076). A significant difference in frequency was only found for the UTR-5 haplotype [8.54% vs 15.95%, OR = 0.49 (95% CI 0.32 – 0.76); P = 0.001, Pc = 0.008].
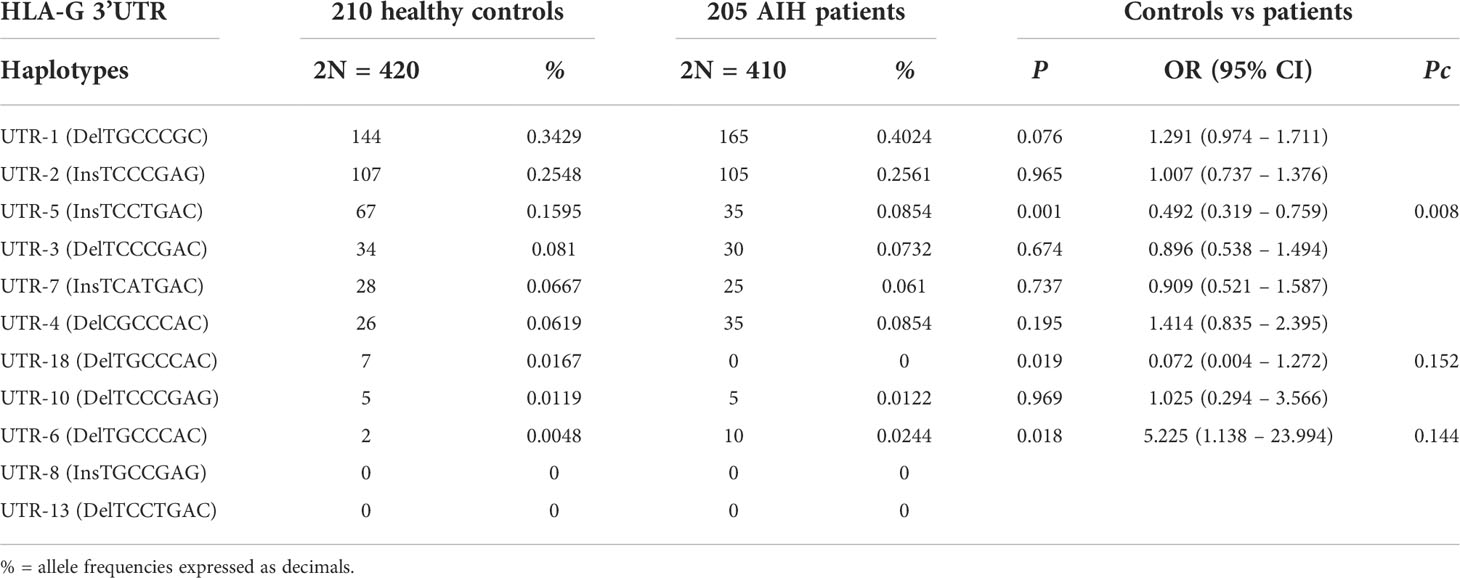
Table 4 Haplotype frequencies observed at the HLA-G 3’UTR polymorphic sites (14bp Ins/Del, 3003C/T, 3010C/G, 3027A/C, 3035C/T, 3142C/G, 3187A/G, 3196C/G) in healthy controls and AIH type 1 patients.
Because a previous study of Sardinian patients indicates a strong association of AIH-1 with a particular HLA extended haplotype (HLA-A*30:02, C*05:01, B*18:01, DRB1*03:01) (66), the presence of linkage disequilibrium between this HLA extended haplotype and the most prevalent HLA-G3’UTR haplotype (UTR-1) was investigated (Table 5). We observed a strong linkage disequilibrium between the UTR-1 haplotype and the aforementioned HLA extended haplotype in patients compared to controls (D = 12.97%, D1 = 1 and D = 6.49%, D1 = 0.96 respectively; P = 0.002). The most significant difference between AIH-1 patients and controls was found for the linkage disequilibrium between UTR-1 and HLA-DRB1*03:01 (D = 7.09%, D’ = 0.92 and D = 46.62%, D’= 0.50 respectively; P = 1.3·10-8).
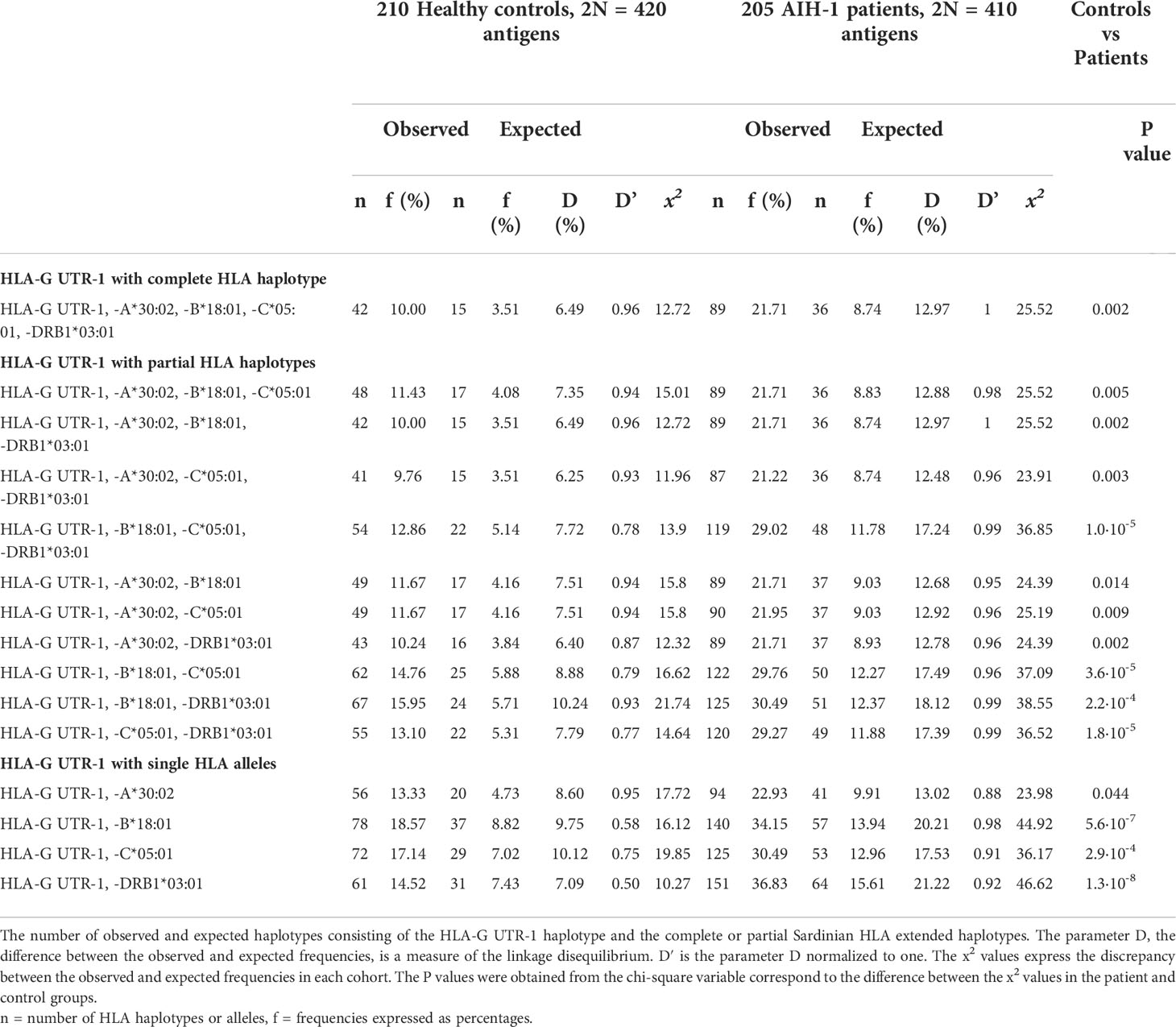
Table 5 Linkage Disequilibrium between the HLA-G UTR-1 haplotype and HLA alleles and haplotypes in the population control and AIH-1 patient groups.
Soluble HLA-G (sHLA-G) levels were measured in patients at the time of diagnosis and in healthy controls at the time of enrollment (Figure 2).
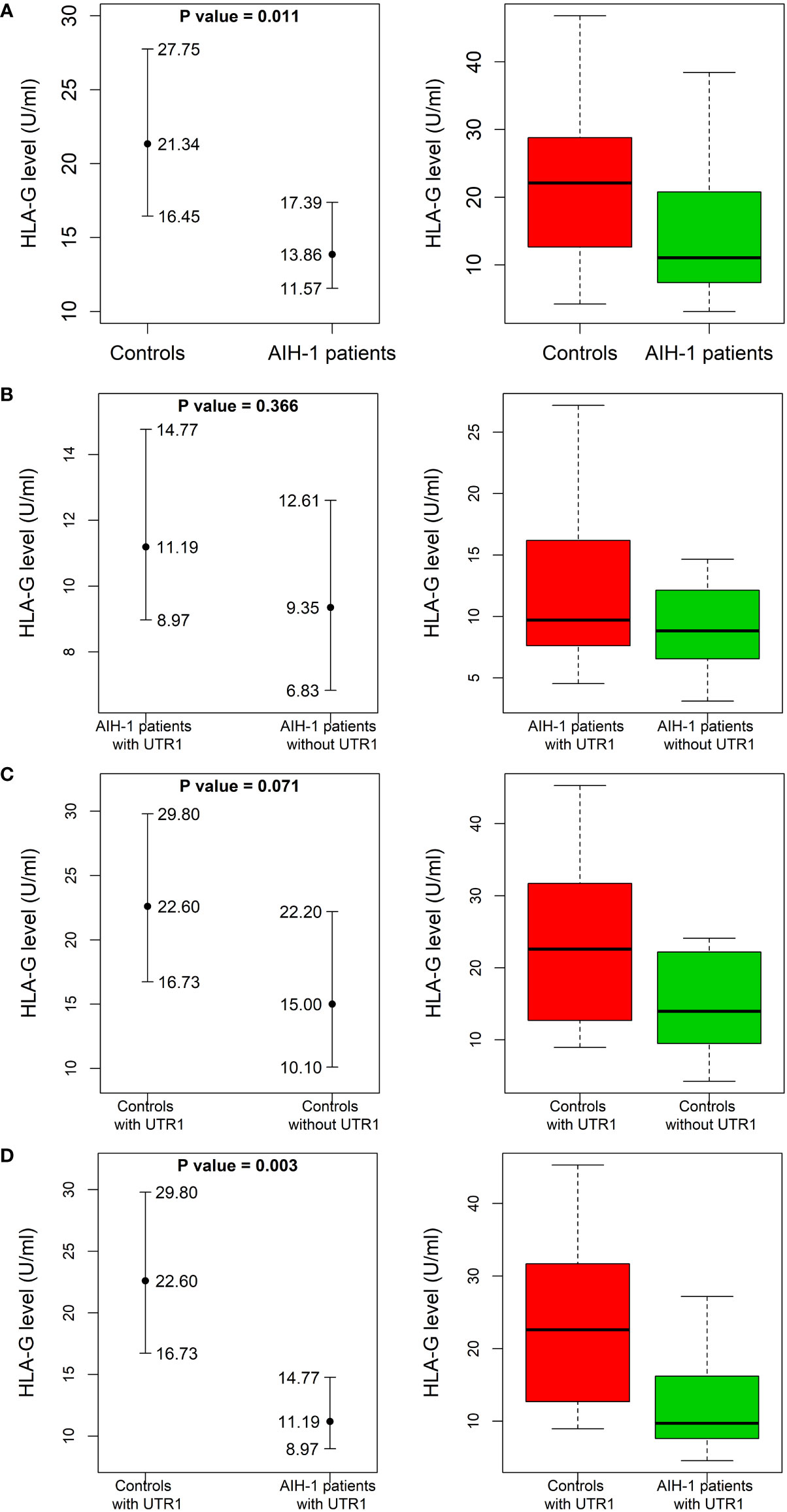
Figure 2 Soluble HLA-G plasma levels in AIH-1 patients and control population. Soluble HLA-G (sHLA-G) plasma levels (U/mL) were compared between AIH-1 patients and the control group (A), two subgroups of the control population stratified according to the presence or absence of the HLA-G UTR-1 haplotype (B), two subgroups of AIH-1 patients stratified according to the presence or absence of the HLA-G UTR-1 haplotype (C) and finally between AIH-1 patients and controls who presented the HLA-G UTR 1 haplotype (D). The P values reported in each figure were computed using the Wilcoxon rank sum test. The vertical bars represent the 95% confidence intervals of the median values (full dots). The boxplots show the spread of the soluble HLA-G plasma levels around the median.
The levels of sHLA-G were significantly lower in AIH-1 patients than controls [13.9 (11.6 – 17.4) U/mL vs 21.3 (16.5 – 27.8) U/mL respectively; P = 0.011] (Figure 2A). According to the literature, the HLA-G UTR-1 haplotype is associated with a higher expression of sHLA-G. This prompted us to compare sHLA-G levels between patients and controls stratified according to the presence or absence of the HLA-G UTR-1 haplotype (Figures 2B–D). The expression of sHLA-G resulted to be significantly different even when the two subgroups of patients and controls carrying the HLA-G UTR-1 haplotype were compared [11.2 (9.0 – 14.8) U/mL vs 22.6 (16.7 – 29.8) U/mL respectively; P = 0.003; Figure 2D].
AIH-1 patients with the UTR-1 haplotype had higher and less variable levels of sHLA-G compared to patients without the UTR-1 haplotype although these differences were not statistically significant [11.2 (9.0 – 14.8) U/mL in UTR-1 positive patients vs 9.4 (6.8 – 12.6) U/mL in UTR-1 negative patients; P = 0.37; Figure 2B]. Similar results were found in the control group [22.6 (16.7 – 29.8) U/mL in UTR-1 positive controls vs 15.0 (10.1 – 22.2) U/mL in negative controls; P = 0.07; Figure 2C]. Basal sHLA-G levels measured at the time of diagnosis in the 52 AIH-1 patients with liver biopsy were evaluated for a possible correlation with the severity of liver inflammation (Figure 3). It is relevant to note that the 24 patients with mild or moderate inflammatory involvement had much higher sHLA-G levels compared to the 28 patients with severe liver inflammation [33.5 (23.6 – 44.8) U/mL vs 8.8 (6.1 – 14.5) U/mL respectively; P = 0.003; Figure 3].
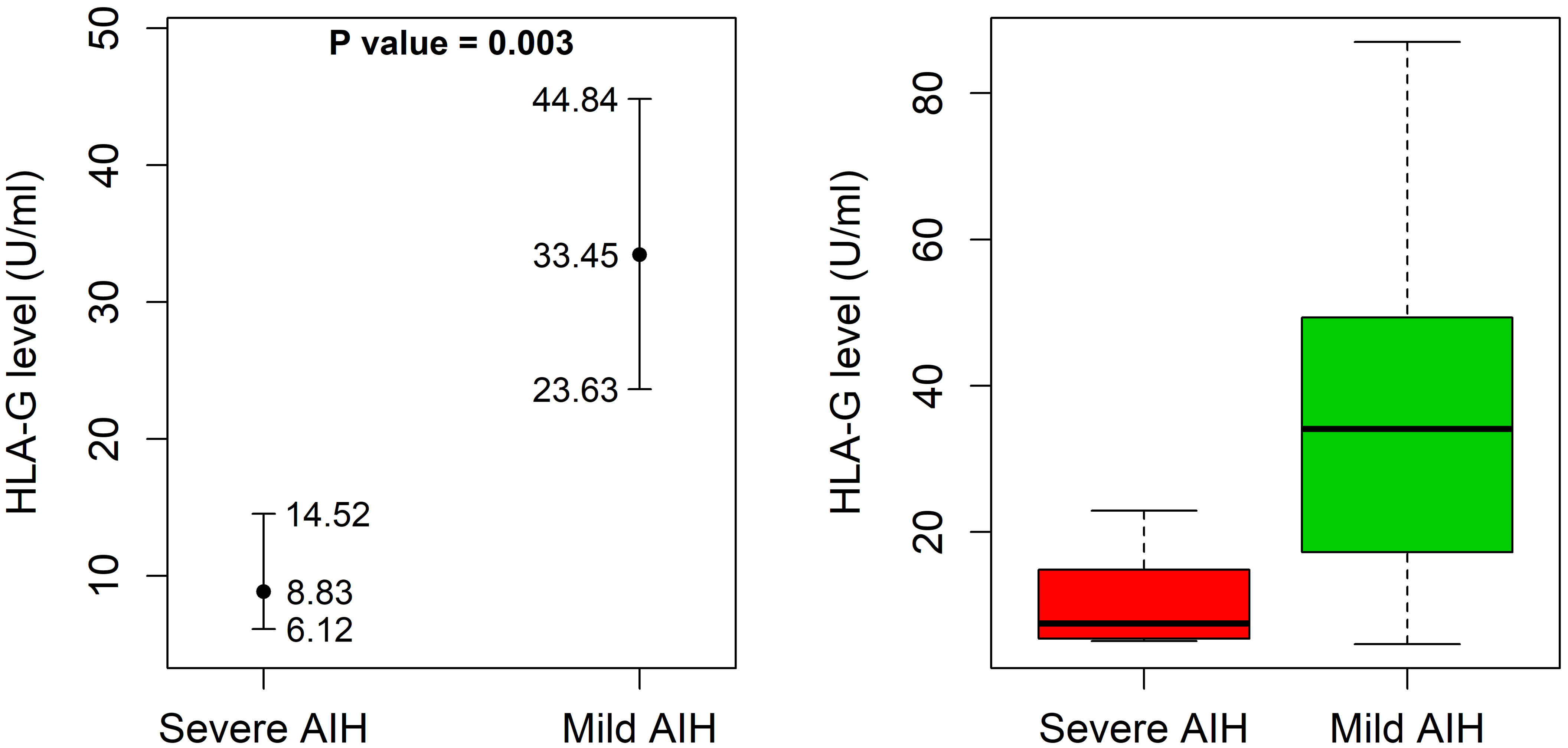
Figure 3 Soluble HLA-G plasma levels in 52 AIH-1 patients for which liver biopsy was available. Soluble HLA-G (sHLA-G) plasma levels (U/mL) were analyzed in 52 AIH-1 patients for which liver biopsy was available, stratified according to the grade of liver inflammation (mild/moderate or severe). The P value reported in the figure was computed using the Wilcoxon rank sum test. The vertical bars represent the 95% confidence intervals of the median values (full dots). The boxplots show the spread of the soluble HLA-G plasma levels around the median.
Interestingly, as opposed to the biochemical markers ALT, ALP, and γ -globulin, the plasma levels of sHLA-G tend to decrease in severe cases of AIH-1 (Table 2). However, correlation analysis based on Kendall’s tau rank did not indicate any statistically significant correlation between sHLA-G levels and ALT, ALP, or globulin levels (τ = -0.17, -0.03, 0.04 and P = 0.10, 0.79, 0.70 respectively).
Analysis of liver biopsies from AIH-1 patients did not reveal the expression of HLA-G molecules in the liver parenchyma: hepatocytes, cholangiocytes, Kupffer cells, and Ito stellate cells (Table 6). One of the most interesting findings stemming from our study was the widespread expression of HLA-G molecules both in the cytoplasm and membrane of plasma cells labeled with anti-HLA-G monoclonal antibodies (Figure 4). This finding was confirmed in the large majority of the examined biopsy specimens [Thirty-six out of 52 samples (69.2%) presented positive plasma cells].
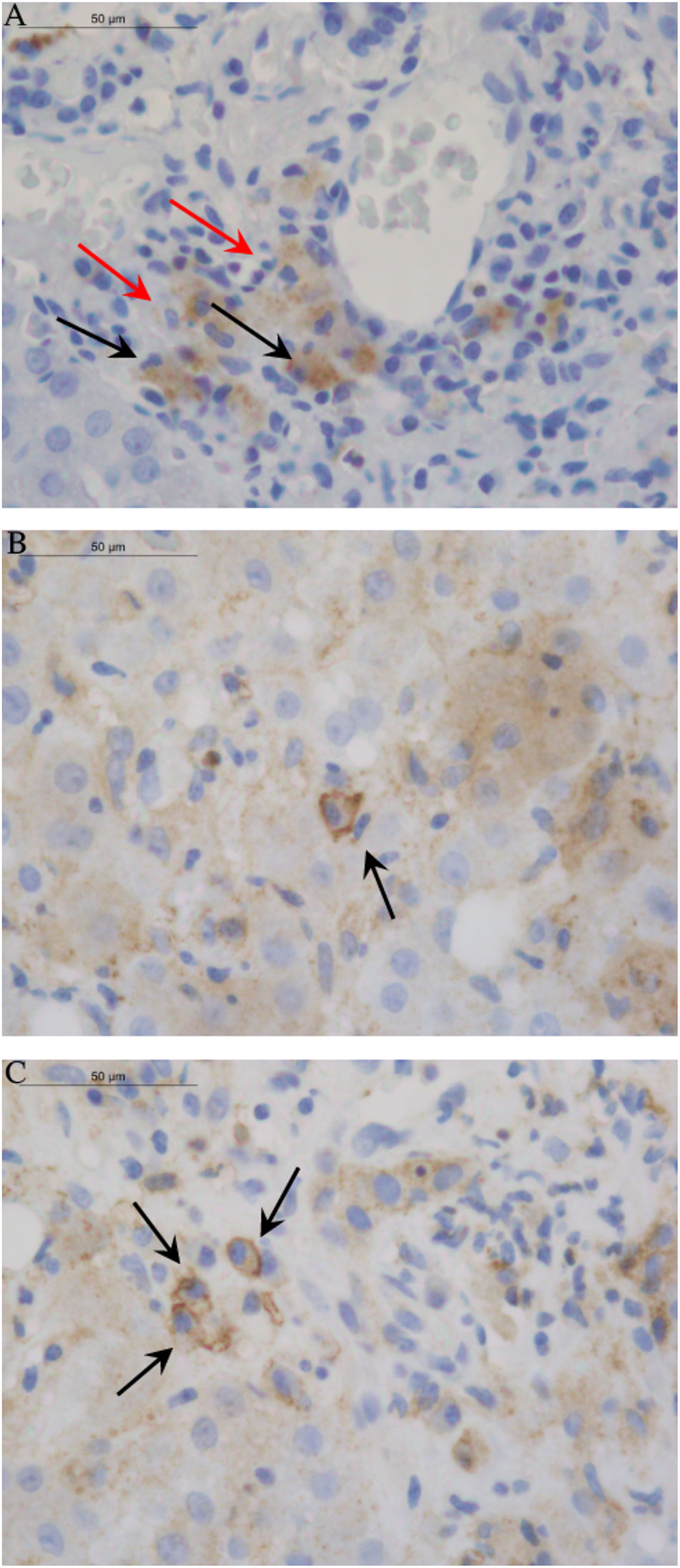
Figure 4 Images of HLA-G distribution in liver tissue. The 4H84 mAb were used to detect HLA-G molecules in different cell types of liver biopsy from AIH-1 patients. (A) The picture displayed the absence of HLA-G expression in hepatocytes, cholangiocytes, Kupffer cells and Ito stellate cells. Only Histiocytes/Macrophages (marked by red arrow) and Plasma cells (marked by black arrow) showed widespread HLA-G expression in the cytoplasm. (B, C) Single and clustered Plasma cells of AIH-1 patients with severe clinical manifestations showed expression of HLA-G molecules both in the cytoplasm and in the membrane (sHLA-G ≥ 50% and score ≥ ++; see ).
Another important finding was the greater expression of HLA-G molecules in the plasma cells of patients with severe liver inflammation and serious clinical manifestations of AIH-1 (sHLA-G ≥ 50% and score ≥ ++, except for two patients). On the contrary, no AIH-1 patients with mild hepatic inflammation had plasma cells characterized by the presence of cytoplasmic and/or membrane HLA-G molecules. No other element of the typical inflammatory infiltrate was positive for anti-HLA-G antibodies.
In this study, we investigated the role of HLA-G molecules in type-1 AIH. Specifically, we analyzed the genetic and phenotypic characteristics of HLA-G comparing a group of AIH-1 patients with a control population originating from the same geographical area (Sardinia, Italy). Additionally, we investigated the distribution of HLA-G molecules in liver tissue in a subset of patients who underwent liver biopsy. Previous studies have described the pathways of HLA-G-induced tolerance and the role that these molecules play in autoimmune diseases (67, 68), such as rheumatoid arthritis (69, 70) and systemic lupus erythematosus (71, 72), but so far, no studies have investigated the impact of HLA-G expression on autoimmune hepatitis. This may partly be due to the difficulty of recruiting a large enough cohort of AIH-1 patients, as well as a genetically homogeneous control population. Even today, the homogeneity of the Sardinian Island population continues to offer an ideal study ground for the analysis of both complex genetic traits and diseases, providing information that is difficult to obtain in other populations.
First of all, this study demonstrates a considerable overlap between the clinical and immuno-genetical parameters observed in Sardinian AIH-1 patients and others from mainland Italy. Moreover, similar results have been reported for patients from North America (1, 64).
In particular, as described in our AIH-1 patients, the HLA-DRB1*03 (*03:01) allele is the main susceptibility factor for AIH-1 in white northern Europeans and North Americans, although a high frequency has also been reported for the HLA-DRB1*04 allele (65).
It is important to note that in our group of AIH-1 patients, the HLA-DRB1*04 susceptibility antigen was represented by HLA-DRB1*04:05 instead of HLA-DRB1*04:01 observed in white North American and Northern European AIH-1 patients (65).
Another interesting observation is that the Sardinian population showed similar frequencies of HLA-G alleles and 3’UTR haplotypes between AIH-1 patients and controls.
Although the HLA-G UTR-1 haplotype was more frequent in AIH-1 patients, it did not reach statistical significance in comparison with controls. Interestingly, this haplotype was in strong linkage disequilibrium with the extended haplotype HLA-A*30:02, -C*05:01, -B*18:01, -DRB1*03:01 (Table 4), and even more so with the three-loci haplotype HLA-C*05:01, -B*18:01, -DRB1*03:01 (P = 1.0 x 10-5) which we found strongly associated with AIH-1 patients in a previous study (66). HLA-C*05:01, -B*18:01, -DRB1*03:01 is an ancestral extended haplotype probably deriving from the paleo-Mediterranean population that lived in the western regions of the Mediterranean up until the Mesolithic period (56). Infectious diseases such as brucellosis (73, 74), genetic drift, besides the peculiar structure of this haplotype (75), are likely to account for the high frequency of this haplotype in the Sardinian population. However, the high ability of this extended haplotype to activate the immune system has the disadvantage of increasing the risk of serious autoimmune disease in carriers of this haplotype (66, 75). Most probably, the strong LD between the HLA-G UTR-1 polymorphism and the HLA-C*05:01, -B*18:01, -DRB1*03:01 extended haplotype has the function of counterbalancing the reactivity of the immune system triggered by this HLA extended haplotype, by promoting expression of HLA-G molecules with a tolerogenic function.
Similar to healthy controls, we found that sHLA-G levels were higher in AIH-1 patients who express UTR-1 (Figures 2B, C).
Overall, AIH-1 patients exhibited sHLA-G levels significantly lower than those observed in the control population (P = 0.011), which is similar to what has been reported for other autoimmune diseases, such as Systemic Lupus Erythematosus (76), Multiple Sclerosis (77), and rheumatoid arthritis (69). Expression of these tolerogenic HLA-G molecules seems to exert a beneficial tolerogenic effect on AIH-1, as evidenced by higher levels of sHLA-G (Figure 3) found in the plasma of patients with mild/moderate hepatic inflammation compared to severe forms. Also, the histological picture of patients with high sHLA-G levels was much less severe than that of patients with low plasma levels of sHLA-G, whose biopsies revealed strong inflammatory activity with marked lymphocytic infiltrate.
It is interesting to note that sHLA-G levels show an opposite trend to the biochemical markers ALT, ALP and γ-globulin which tend to increase in severe inflammatory forms of AIH-1.
However, the correlation analysis did not show any statistically significant correlation between sHLA-G levels and ALT, ALP and γ-globulin levels (τ = -0.17, -0.03, 0.04). Therefore, this seems to indicate that sHLA-G levels are independent of these biochemical markers, thus making it useful for discriminating between mild and severe forms of AIH-1. Future studies involving large case series are needed to confirm this result, which was obtained by analyzing a limited number of biopsy samples.
Analysis of liver biopsies from AIH-1 patients did not reveal expression of HLA-G molecules in the liver parenchyma (hepatocytes, cholangiocytes, Kupffer cells, Ito stellate cells). This finding is similar to what has already been described in the literature for the liver of healthy subjects (www.proteinatlas.org). The presence of plasma cells in liver parenchyma represents the hallmark of diagnosis in AIH-1. One of the most interesting findings emerging from our study was the positivity of plasma cells with anti-HLA-G monoclonal antibodies, observed in the large majority of the examined bioptic specimens. Interestingly, no other element of the typical inflammatory infiltrate was positive for anti-HLA-G antibodies (Figure 4). Diffuse expression of sHLA-G in the cytoplasm of the plasma cells suggests that these cells not only produce the autoantibodies leading to the inflammatory reaction in AIH-1, but contemporarily produce sHLA-G, probably to protect themselves against the immune cell attacks for which they are responsible. This hypothesis is supported by the fact that in the examined histological samples, the increased expression of HLA-G molecules by plasma cells is directly associated with more severe clinical manifestations (Table 6) and a marked inflammatory infiltrate in the hepatic parenchyma.
In conclusion, this is the first study that thoroughly investigated the role of HLA-G molecules in type-1 autoimmune hepatitis. In particular, our results demonstrate how the immunomodulatory effect of HLA-G molecules has a positive impact on the clinical course of AIH-1 and how this improvement closely correlates with plasma levels of sHLA-G. We highly recommend similar and larger studies on genetically different populations to confirm the positive prognostic value of plasma levels of sHLA-G in AIH-1. Based on our findings, it should be possible to hypothesize the use of these molecules in the near future for therapy in autoimmune disorders, particularly considering that these molecules are already considered promising and relevant targets for cancer immunotherapy (78). Meanwhile, our results open the debate on the role of HLA-G molecules expressed by plasma cells, that are ‘pathognomonic’ of AIH-1. In myeloma patients, HLA-G transfer from tumor plasma cells to T cells via trogocytosis was associated with a poor clinical outcome (36). It is possible that also in AIH-1, the plasma cells utilize the transfer of HLA-G molecules through the release of exosomes and/or via trogocytosis to counteract the immune cell activity of the surrounding microenvironment. The effect of HLA-G plasma cell expression on AIH-1 clinical outcome might therefore be highly dependent on the expression pattern of HLA-G isoforms. Perhaps HLA-G expressed in plasma cells may reflect a high level of epigenetic deregulation and/or genetic mutation, correlating with the more aggressive clinical forms of AIH-1.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA859661.
This study was reviewed and approved by Ethics Committee of the Cagliari University Hospital. The patients/participants provided their written informed consent to participate in this study.
All authors contributed to the article and approved the submitted version.
The research performed in this report falls within the institutional responsibilities of the investigators of the participating centers, all of which pertain to the Italian National Public Health Service. The authors received no specific funding for this work. Grant funding (2021-290) was received from the “Fondazione di Sardegna”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to the Fondazione di Sardegna for supplying part of the reagents and services necessary for this study. We wish to thank Anna Maria Koopmans for her help in coordinating research activities, professional writing assistance and preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1007647/full#supplementary-material
1. Tunio NA, Mansoor E, Sheriff MZ, Cooper GS, Sclair SN, Cohen SM. Epidemiology of autoimmune hepatitis (AIH) in the united states between 2014 and 2019: A population-based national study. J Clin Gastroenterol (2021) 55(10):903–10. doi: 10.1097/MCG.0000000000001449
2. Heneghan MA, Yeoman AD, Verma S, Smith AD, Longhi MS. Autoimmune hepatitis. Lancet (2013) 382(9902):1433–44. doi: 10.1016/S0140-6736(12)62163-1
3. Hurlburt KJ, McMahon BJ, Deubner H, Hsu-Trawinski B, Williams JL, Kowdley KV. Prevalence of autoimmune liver disease in Alaska natives. Am J Gastroenterol (2002) 97(9):2402–7. doi: 10.1111/j.1572-0241.2002.06019.x
4. Nguyen HH, Shaheen AA, Baeza N, Lytvyak E, Urbanski SJ, Mason AL, et al. Evaluation of classical and novel autoantibodies for the diagnosis of primary biliary cholangitis-autoimmune hepatitis overlap syndrome (PBC-AIH OS). PLoS One (2018) 13(3):e0193960. doi: 10.1371/journal.pone.0193960
5. Bonder A, Retana A, Winston DM, Leung J, Kaplan MM. Prevalence of primary biliary cirrhosis-autoimmune hepatitis overlap syndrome. Clin Gastroenterol Hepatol (2011) 9(7):609–12. doi: 10.1016/j.cgh.2011.03.019
6. Trivedi PJ, Hubscher SG, Heneghan M, Gleeson D, Hirschfield GM. Grand round: Autoimmune hepatitis. J Hepatol (2019) 70(4):773–84. doi: 10.1016/j.jhep.2018.11.006
7. Kirstein MM, Metzler F, Geiger E, Heinrich E, Hallensleben M, Manns MP, et al. Prediction of short- and long-term outcome in patients with autoimmune hepatitis. Hepatology (2015) 62(5):1524–35. doi: 10.1002/hep.27983
8. Muratori P, Granito A, Lenzi M, Muratori L. Limitation of the simplified scoring system for the diagnosis of autoimmune hepatitis with acute onset. Liver Int (2021) 41(3):529–34. doi: 10.1111/liv.14778
9. Floreani A, Restrepo-Jiménez P, Secchi MF, De Martin S, Leung PSC, Krawitt E, et al. Etiopathogenesis of autoimmune hepatitis. J Autoimmun (2018) 95:133–43. doi: 10.1016/j.jaut.2018.10.020
10. Oliveira LC, Porta G, Marin ML, Bittencourt PL, Kalil J, Goldberg AC. Autoimmune hepatitis, HLA and extended haplotypes. Autoimmun Rev (2011) 10(4):189–93. doi: 10.1016/j.autrev.2010.09.024
11. Longhi MS, Ma Y, Mitry RR, Bogdanos DP, Heneghan M, Cheeseman P, et al. Effect of CD4+ CD25+ regulatory T-cells on CD8 T-cell function in patients with autoimmune hepatitis. J Autoimmun (2005) 25(1):63–71. doi: 10.1016/j.jaut.2005.05.001
12. Ferri S, Longhi MS, De Molo C, Lalanne C, Muratori P, Granito A, et al. A multifaceted imbalance of T cells with regulatory function characterizes type 1 autoimmune hepatitis. Hepatology (2010) 52(3):999–1007. doi: 10.1002/hep.23792
13. Alegre E, Rizzo R, Bortolotti D, Fernandez-Landázuri S, Fainardi E, González A. Some basic aspects of HLA-G biology. J Immunol Res (2014) 2014:657625. doi: 10.1155/2014/657625
14. Castelli EC, Ramalho J, Porto IO, Lima TH, Felício LP, Sabbagh A, et al. Insights into HLA-G genetics provided by worldwide haplotype diversity. Front Immunol (2014) 5:476. doi: 10.3389/fimmu.2014.00476
15. Contini P, Murdaca G, Puppo F, Negrini S. HLA-G expressing immune cells in immune mediated diseases. Front Immunol (2020) 11:1613. doi: 10.3389/fimmu.2020.01613
16. Geraghty DE, Koller BH, Orr HT. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proc Natl Acad Sci U.S.A. (1987) 84(24):9145–9. doi: 10.1073/pnas.84.24.9145
17. Koller BH, Geraghty DE, DeMars R, Duvick L, Rich SS, Orr HT. Chromosomal organization of the human major histocompatibility complex class I gene family. J Exp Med (1989) 169(2):469–80. doi: 10.1084/jem.169.2.469
18. Gobin SJ, van den Elsen PJ. Transcriptional regulation of the MHC class ib genes HLA-e, HLA-f, and HLA-G. HumImmunol (2000) 61(11):1102–7. doi: 10.1016/s0198-8859(00)00198-1
19. LeMaoult J, Krawice-Radanne I, Dausset J, Carosella ED. HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc Natl Acad Sci U S A (2004) 101(18):7064–9. doi: 10.1073/pnas.0401922101
20. Le Rond S, Le Maoult J, Créput C, Menier C, Deschamps M, Le Friec G, et al. Alloreactive CD4+ and CD8+ T cells express the immunotolerant HLA-G molecule in mixed lymphocyte reactions: in vivo implications in transplanted patients. Eur J Immunol (2004) 34(3):649–60. doi: 10.1002/eji.200324266
21. LeMaoult J, Yan WH. Editorial: The biological and clinical aspects of HLA-G. Front Immunol (2021) 12:649344. doi: 10.3389/fimmu.2021.649344
22. LeMaoult J, Le Discorde M, Rouas-Freiss N, Moreau P, Menier C, McCluskey J, et al. Biology and functions of human leukocyte antigen-G in health and sickness. Tissue Antigens (2003) 62(4):273–84. doi: 10.1034/j.1399-0039.2003.00143.x
23. Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci (2011) 68(3):369–95. doi: 10.1007/s00018-010-0580-7
24. Sabbagh A, Luisi P, Castelli EC, Gineau L, Courtin D, Milet J, et al. Worldwide genetic variation at the 3' untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. GenesImmun (2014) 15(2):95–106. doi: 10.1038/gene.2013.67
25. Rizzo R, Bortolotti D, Bolzani S, Fainardi E. HLA-G molecules in autoimmune diseases and infections. Front Immunol (2014) 5:592. doi: 10.3389/fimmu.2014.00592
26. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res (2014) 2014:734068. doi: 10.1155/2014/734068
27. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PLoS One (2013) 8(10):e71742. doi: 10.1371/journal.pone.0071742
28. Xu X, Zhou Y, Wei H. Roles of HLA-G in the maternal-fetal immune microenvironment. Front Immunol (2020) 11:592010. doi: 10.3389/fimmu.2020.592010
29. Melo-Lima BL, Poras I, Passos GA, Carosella ED, Donadi EA, Moreau P. The autoimmune regulator (Aire) transactivates HLA-G gene expression in thymic epithelial cells. Immunology (2019) 158(2):121–35. doi: 10.1111/imm.13099
30. Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. HumImmunol (2003) 64(11):1039–44. doi: 10.1016/j.humimm.2003.08.346
31. Amodio G, Gregori S. HLA-G Genotype/Expression/Disease association studies: Success, hurdles, and perspectives. Front Immunol (2020) 11:1178. doi: 10.3389/fimmu.2020.01178
32. Cirulli V, Zalatan J, McMaster M, Prinsen R, Salomon DR, Ricordi C, et al. The class I HLA repertoire of pancreatic islets comprises the nonclassical class ib antigen HLA-G. Diabetes (2006) 55(5):1214–22. doi: 10.2337/db05-0731
33. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science (1990) 248(4952):220–3. doi: 10.1126/science.2326636
34. Caocci G, Greco M, Fanni D, Senes G, Littera R, Lai S, et al. HLA-G expression and role in advanced-stage classical Hodgkin lymphoma. Eur J Histochem (2016) 60(2):2606. doi: 10.4081/ejh.2016.2606
35. Lin A, Yan WH. Heterogeneity of HLA-G expression in cancers: Facing the challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
36. Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, et al. CD86+ or HLA-g+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood (2012) 120(10):2055–63. doi: 10.1182/blood-2012-03-416792
37. Peng Y, Xiao J, Li W, Li S, Xie B, He J, et al. Prognostic and clinicopathological value of human leukocyte antigen G in gastrointestinal cancers: A meta-analysis. Front Oncol (2021) 11:642902. doi: 10.3389/fonc.2021.642902
38. Liu L, Wang L, Zhao L, He C, Wang G. The role of HLA-G in tumor escape: Manipulating the phenotype and function of immune cells. Front Oncol (2020) 10:597468. doi: 10.3389/fonc.2020.597468
39. Bortolotti D, Gentili V, Rotola A, Potena L, Rizzo R. Soluble HLA-G pre-transplant levels to identify the risk for development of infection in heart transplant recipients. Hum Immunol (2020) 81(4):147–50. doi: 10.1016/j.humimm.2019.10.003
40. Ajith A, Portik-Dobos V, Horuzsko DD, Kapoor R, Mulloy LL, Horuzsko A. HLA-G and humanized mouse models as a novel therapeutic approach in transplantation. Hum Immunol (2020) 81(4):178–85. doi: 10.1016/j.humimm.2020.02.006
41. Kang SW, Oh E, Cho W, Kim M, Park EJ, Kwack KH, et al. HLA-G 14bp Ins/Del polymorphism in the 3'UTR region and acute rejection in kidney transplant recipients: An updated meta-analysis. Medicina (Kaunas) (2021) 57(10):1007. doi: 10.3390/medicina57101007
42. Piancatelli D, Maccarone D, Colanardi A, Sebastiani P, Clemente K, Iesari S, et al. HLA-G14bp ins/del polymorphism and post-transplant weight gain in kidney transplantation: potential implications beyond tolerance. BMCNephrol (2020) 21(1):109. doi: 10.1186/s12882-020-01752-6
43. Littera R, Piredda G, Pani A, Frongia M, Onano B, Michittu MB, et al. Role of human leukocyte antigen-G 14-base pair polymorphism in kidney transplantation outcomes. J Nephrol (2013) 26(6):1170–8. doi: 10.5301/jn.5000252
44. González A, Rebmann V, LeMaoult J, Horn PA, Carosella ED, Alegre E. The immunosuppressive molecule HLA-G and its clinical implications. Crit Rev Clin Lab Sci (2012) 49(3):63–84. doi: 10.3109/10408363.2012.677947
45. Celsi F, Catamo E, Kleiner G, Tricarico PM, Vuch J, Crovella S. HLA-G/C, miRNAs, and their role in HIV infection and replication. BioMed Res Int (2013) 2013:693643. doi: 10.1155/2013/693643
46. Amiot L, Vu N, Samson M. Biology of the immunomodulatory molecule HLA-G in human liver diseases. J Hepatol (2015) 62(6):1430–7. doi: 10.1016/j.jhep.2015.03.007
47. Amiot L, Vu N, Rauch M, L'Helgoualc'h A, Chalmel F, Gascan H, et al. Expression of HLA-G by mast cells is associated with hepatitis c virus-induced liver fibrosis. J Hepatol (2014) 60(2):245–52. doi: 10.1016/j.jhep.2013.09.006
48. Souto FJ, Crispim JC, Ferreira SC, da Silva AS, Bassi CL, Soares CP, et al. Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis b virus infection. J ViralHepat (2011) 18(2):102–5. doi: 10.1111/j.1365-2893.2010.01286.x
49. Shi WW, Lin A, Xu DP, Bao WG, Zhang JG, Chen SY, et al. Plasma soluble human leukocyte antigen-G expression is a potential clinical biomarker in patients with hepatitis b virus infection. Hum Immunol (2011) 72(11):1068–73. doi: 10.1016/j.humimm.2011.06.012
50. Weng PJ, Fu YM, Ding SX, Xu DP, Lin A, Yan WH. Elevation of plasma soluble human leukocyte antigen-G in patients with chronic hepatitis c virus infection. Hum Immunol (2011) 72(5):406–11. doi: 10.1016/j.humimm.2011.02.008
51. Lin A, Chen HX, Zhu CC, Zhang X, Xu HH, Zhang JG, et al. Aberrant human leucocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J Cell Mol Med (2010) 14(8):2162–71. doi: 10.1111/j.1582-4934.2009.00917.x
52. Wang XK, Liao XW, Yang CK, Yu TD, Liu ZQ, Gong YZ, et al. Diagnostic and prognostic biomarkers of human leukocyte antigen complex for hepatitis b virus-related hepatocellular carcinoma. J Cancer (2019) 10(21):5173–90. doi: 10.7150/jca.29655
53. Wang Y, Ye Z, Meng XQ, Zheng SS. Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int (2011) 10(2):158–63. doi: 10.1016/s1499-3872(11)60025-8
54. Rashidi S, Farhadi L, Ghasemi F, Sheikhesmaeili F, Mohammadi A. The potential role of HLA-G in the pathogenesis of HBV infection: Immunosuppressive or immunoprotective? Infect Genet Evol (2020) 85:104580. doi: 10.1016/j.meegid.2020.104580
55. Mack CL, Adams D, Assis DN, Kerkar N, Manns MP, Mayo MJ, et al. Diagnosis and management of autoimmune hepatitis in adults and children: 2019 practice guidance and guidelines from the American association for the study of liver diseases. Hepatology (2020) 72(2):671–722. doi: 10.1002/hep.31065
56. Contu L, Arras M, Carcassi C, La Nasa G, Mulargia M. HLA structure of the sardinian population: a haplotype study of 551 families. Tissue Antigens (1992) 40(4):165–74. doi: 10.1111/j.1399-0039.1992.tb02041.x
57. QIAGEN. QIAamp DNA mini blood mini handbook (2022). Available at: https://www.qiagen.com/gb/shop/sample-technologies/dna/qiaamp-dna-mini-kit/#resources.
58. Nilsson LL, Funck T, Kjersgaard ND, Hviid TVF. Next-generation sequencing of HLA-G based on long-range polymerase chain reaction. (2018) HLA 92(3):144–53. doi: 10.1111/tan.13342
59. Castelli EC, Mendes-Junior CT, Deghaide NH, de Albuquerque RS, Muniz YC, Simões RT, et al. The genetic structure of 3'untranslated region of the HLA-G gene: polymorphisms and haplotypes. GenesImmun (2010) 11(2):134–41. doi: 10.1038/gene.2009.74
60. Guido M, Mangia A, Faa G. Gruppo italiano patologi apparato digerente (GIPAD); società italiana di anatomia patologica e citopatologia Diagnostica/International academy of pathology, Italian division (SIAPEC/IAP). Chronic Viral hepatitis: Histol Rep Dig Liver Dis (2011) 43(Suppl4):S331–43. doi: 10.1016/S1590-8658(11)60589-6
61. Kirkwood BR, Sterne JAC. Essential medical statistics. 2nd edition. Oxford UK: Blackwell Scientific Publications (2003).
62. Hodges JL, Lehmann EL. Estimates of location based on rank tests. Ann Math Stat (1963) 34:598–611. doi: 10.1214/aoms/1177704172
63. Lewontin RC. The interaction of selection and linkage. i. general considerations; heterotic models. Genetics (1964) 49(1):49–67. doi: 10.1093/genetics/49.1.49
64. Muratori P, Czaja AJ, Muratori L, Pappas G, Maccariello S, Cassani F, et al. Genetic distinctions between autoimmune hepatitis in Italy and north America. World J Gastroenterol (2005) 11(12):1862–6. doi: 10.3748/wjg.v11.i12.1862
65. van Gerven NM, de Boer YS, Zwiers A, Verwer BJ, Drenth JP, van Hoek B, et al. HLA-DRB1*03:01 and HLA-DRB1*04:01 modify the presentation and outcome in autoimmune hepatitis type-1. GenesImmun (2015) 16(4):247–52. doi: 10.1038/gene.2014.82
66. Littera R, Chessa L, Onali S, Figorilli F, Lai S, Secci L, et al. Exploring the role of killer cell immunoglobulin-like receptors and their HLA class I ligands in autoimmune hepatitis. PLoS One (2016) 11(1):e0146086. doi: 10.1371/journal.pone.0146086
67. Martín-Villa JM, Vaquero-Yuste C, Molina-Alejandre M, Juarez I, Suárez-Trujillo F, López-Nares A, et al. HLA-G: Too much or too little? role in cancer and autoimmune disease. Front Immunol (2022) 13:796054. doi: 10.3389/fimmu.2022.796054
68. Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V. Recent advances in our understanding of HLA-G biology: Lessons from a wide spectrum of human diseases. J Immunol Res (2016) 2016:4326495. doi: 10.1155/2016/4326495
69. Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: A review. JAMA (2018) 320(13):1360–72. doi: 10.1001/jama.2018.13103
70. Gautam S, Kumar U, Kumar M, Kanga U, Dada R. Association of HLA-g 3'UTR polymorphisms with soluble HLA-G levels and disease activity in patients with rheumatoid arthritis: A case-control study. Immunol Invest (2020) 49(1-2):88–105. doi: 10.1080/08820139.2019.1657146
71. Veit TD, Cordero EA, Mucenic T, Monticielo OA, Brenol JC, Xavier RM, et al. Association of the HLA-G 14 bp polymorphism with systemic lupus erythematosus. Lupus (2009) 18(5):424–30. doi: 10.1177/0961203308098187
72. Consiglio CR, Veit TD, Monticielo OA, Mucenic T, Xavier RM, Brenol JC, et al. Association of the HLA-G gene +3142C>G polymorphism with systemic lupus erythematosus. TissueAntigens (2011) 77(6):540–5. doi: 10.1111/j.1399-0039.2011.01635.x
73. Piazza A, Mayr WR, Contu L, Amoroso A, Borelli I, Curtoni ES, et al. Genetic and population structure of four sardinian villages. Ann Hum Genet (1985) 49(1):47–63. doi: 10.1111/j.1469-1809.1985.tb01675.x
74. Piazza A, Morton NE. A formal genetic analysis of the HL-a system. Am J Hum Genet (1973) 25(2):119–33.
75. Contu L, Carcassi C, Dausset J. The "Sardinian" HLA-A30,B18,DR3,DQw2 haplotype constantly lacks the 21-OHA and C4B genes. is it an ancestral haplotype without duplication? Immunogenetics (1989) 30(1):13–7. doi: 10.1007/BF02421464
76. Rizzo R, Hviid TV, Govoni M, Padovan M, Rubini M, Melchiorri L, et al. HLA-G genotype and HLA-G expression in systemic lupus erythematosus: HLA-G as a putative susceptibility gene in systemic lupus erythematosus. TissueAntigens (2008) 71(6):520–9. doi: 10.1111/j.1399-0039.2008.01037.x
77. Mohammadi N, Adib M, Alsahebfosoul F, Kazemi M, Etemadifar M. An investigation into the association between HLA-G 14 bp insertion/deletion polymorphism and multiple sclerosis susceptibility. JNeuroimmunol (2016) 290:115–8. doi: 10.1016/j.jneuroim.2015.11.019
Keywords: type 1 autoimmune hepatitis, Sardinian population, human leukocyte antigen, HLA-G alleles, soluble HLA-G, plasma cell, anti-HLA-G monoclonal antibodies, HLA-G 3’UTR haplotypes
Citation: Littera R, Perra A, Miglianti M, Piras IS, Mocci S, Lai S, Melis M, Zolfino T, Balestrieri C, Conti M, Serra G, Figorilli F, Firinu D, Onali S, Matta L, Porcu C, Pes F, Fanni D, Manieli C, Vacca M, Cusano R, Trucas M, Cipri S, Tranquilli S, Rassu S, Cannas F, Carta MG, Kowalik MA, Giuressi E, Faa G, Chessa L and Giglio S (2022) The double-sided of human leukocyte antigen-G molecules in type 1 autoimmune hepatitis. Front. Immunol. 13:1007647. doi: 10.3389/fimmu.2022.1007647
Received: 04 August 2022; Accepted: 27 September 2022;
Published: 12 October 2022.
Edited by:
Mahzad Akbarpour, University of Chicago Medicine, United StatesReviewed by:
Cigdem Kekik, Istanbul University, TurkeyCopyright © 2022 Littera, Perra, Miglianti, Piras, Mocci, Lai, Melis, Zolfino, Balestrieri, Conti, Serra, Figorilli, Firinu, Onali, Matta, Porcu, Pes, Fanni, Manieli, Vacca, Cusano, Trucas, Cipri, Tranquilli, Rassu, Cannas, Carta, Kowalik, Giuressi, Faa, Chessa and Giglio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Littera, cm9ieS5saXR0ZXJAZ21haWwuY29t; Andrea Perra, YW5kcmVhLnBlcnJhQHVuaWNhLml0; Stefano Mocci, c3RlZmFuby5tb2NjaS45QGdtYWlsLmNvbQ==; Luchino Chessa, bHVjaGluby5jaGVzc2FAdW5pY2EuaXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.