
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 29 September 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1001255
 Wujuan Zhong1†
Wujuan Zhong1† Avinash Kollipara2†
Avinash Kollipara2† Yutong Liu1
Yutong Liu1 Yuhan Wang1
Yuhan Wang1 Catherine M. O’Connell2
Catherine M. O’Connell2 Taylor B. Poston2
Taylor B. Poston2 Kacy Yount2
Kacy Yount2 Harold C. Wiesenfeld3
Harold C. Wiesenfeld3 Sharon L. Hillier3
Sharon L. Hillier3 Yun Li1,4
Yun Li1,4 Toni Darville2*‡
Toni Darville2*‡ Xiaojing Zheng1,2*‡
Xiaojing Zheng1,2*‡Objectives: Identify genetic loci of enhanced susceptibility to Chlamydial trachomatis (Ct) upper genital tract infection in women.
Methods: We performed an integrated analysis of DNA genotypes and blood-derived mRNA profiles from 200 Ct-exposed women to identify expression quantitative trait loci (eQTL) and determine their association with endometrial chlamydial infection using a mediation test. We further evaluated the effect of a lead eQTL on the expression of CD151 by immune cells from women with genotypes associated with low and high whole blood expression of CD151, respectively.
Results: We identified cis-eQTLs modulating mRNA expression of 81 genes (eGenes) associated with altered risk of ascending infection. In women with endometrial infection, eGenes involved in proinflammatory signaling were upregulated. Downregulated eGenes included genes involved in T cell functions pivotal for chlamydial control. eGenes encoding molecules linked to metabolism of tryptophan, an essential chlamydial nutrient, and formation of epithelial tight junctions were also downregulated in women with endometrial infection. A lead eSNP rs10902226 was identified regulating CD151, a tetrospanin molecule important for immune cell adhesion and migration and T cell proliferation. Further in vitro experiments showed that women with a CC genotype at rs10902226 had reduced rates of endometrial infection with increased CD151 expression in whole blood and T cells when compared to women with a GG genotype.
Conclusions: We discovered genetic variants associated with altered risk for Ct ascension. A lead eSNP for CD151 is a candidate genetic marker for enhanced CD4 T cell function and reduced susceptibility.
Chlamydia trachomatis (Ct) is the most commonly reported bacterial infection in the US. Infection is often asymptomatic and consequently untreated. No vaccine is available. In about 50% of women (1), infection can ascend from the cervix to the upper genital tract (uterus and fallopian tubes), potentially resulting in pelvic inflammatory disease (PID) and leading to devastating long-term sequela including chronic pelvic pain and infertility (2). In previous studies we have identified behavioral, biological, blood transcriptional, cervical cytokine, and Ct antigen-specific T cell and antibody responses, associated with altered risk of endometrial Ct infection (1, 3–6). However, a major research gap that remains is the lack of biomarker(s) that can identify women at elevated risk of disease. DNA variants, such as single nucleotide polymorphisms (SNPs) associated with enhanced risk of endometrial ascension could serve as biomarkers for susceptibility, promoting more effective screening that can be assessed independent of infection.
Several candidate gene and genome wide association studies (GWAS) of infertile women have reported genetic variants associated with altered risk of infertility, and used multiple parameters to link infertility with prior Ct exposure (7–11). However, development of infertility can be multifactorial, and confounded by occurrence of other sexually transmitted infections, behavioral, biological, and/or environmental factors. Although GWAS supports systematic detection of genetic variation when compared to candidate gene studies which are inherently biased, using GWAS to identify SNPs associated with ascending Ct infection would require thousands of women with defined endometrial Ct infection status. Furthermore, most loci identified by GWAS are not mapped to protein-coding regions, so the mechanism underlying their effect can be difficult to interpret.
An intermediate trait such as ascension of Ct, a prerequisite for tubal factor infertility, provides a substantial advantage to gene discovery studies. In this study, we analyzed blood samples from 200 Ct-exposed women with known endometrial Ct infection status with the goal of identifying expression quantitative trait loci (eQTLs) that modulate expression of genes associated with altered risk of Ct ascension. eQTLs are genomic SNPs (eSNPs) that influence expression of eGenes at the level of their regulatory elements. Our analysis focused on identifying SNPs in a cis window spanning a 1Mb region up- and downstream of the transcriptional start site of each gene. In our previous transcriptional profiling of women with symptomatic Ct PID and histologic endometritis, we determined that gene pathways upregulated in whole blood, e.g., type I interferon and myeloid cell activation, paralleled responses detected in endometrial tissue and cervical secretions, making blood eQTLs attractive candidate biomarkers predictive of risk for Ct-induced disease (3).
We found that cis-eQTLs altered the risk of ascending infection by modulating expression of 81 eGenes (FDR<0.2). eGenes involved in innate and adaptive immune responses, cellular trafficking, and metabolic pathways were associated with differential risk of endometrial Ct, suggesting that these eGenes and their regulating cis-eQTLs may serve as candidate genetic biomarkers of Ct ascension risk.
Of particular interest was a cis-eQTL modulating CD151 expression that was significantly linked to risk of ascension. CD151 is a member of the tetraspanin family and has been associated with immune cell migration and adhesion and identifies T cells with hyperproliferative capabilities (12–15). Examination of genotypes for the lead cis-eQTL of CD151 revealed women carrying a CC genotype displayed elevated whole blood and T cell expression of CD151 with reduced risk of endometrial infection, when compared to women with a GG genotype. This CD151-associatead eQTL could serve as a potential biomarker of risk for endometrial Ct infection.
This study complied with the Declaration of Helsinki guidelines and all study participants provided written informed consent prior to initiation of study procedures. The Institutional Review Boards for Human Subjects Research at the University of North Carolina, and the University of Pittsburgh approved the study.
This study used whole blood collected from cis-gender female participants recruited into two cohorts: the Anaerobes and Clearance of Endometritis (ACE) cohort, comprised of women (age 16-40 years) who participated in a clinical trial (NCT01160640) comparing antibiotic regimens for the treatment of clinically diagnosed PID (16), and the T cell Response Against Chlamydia (TRAC) cohort, comprised of women with recognized risk factors for Ct who had cervicitis or were asymptomatic (age 15-35 years) (1). Participants in both cohorts were recruited at the University of Pittsburgh during 2011-2015. Clinical and microbiological data for the ACE and TRAC cohorts, mRNA transcriptional profiling, and DNA genotyping were generated as described previously (1, 3, 11, 16). DNA and mRNA extracted from whole blood samples from women with Ct infection in ACE and from all TRAC participants were used in this study. After quality control procedures, data from 200 women, including 57 uninfected, 71 cervical only infected (Endo-), and 72 with both cervical and endometrial infection (Endo+), were analyzed for eQTL mapping and mediation test for Ct ascension.
Endometrial sampling was obtained transcervically at enrollment after careful preparation of the cervix. The exocervix and endocervical canal were each cleansed with two applications of povidone alcohol. In order to prevent antiseptic solution interfering with microbiologic assays, we then dried the excess antiseptic in the endocervix with two sterile swabs. A sterile suction curette (Unimar Pipelle de Cornier, Cooper Surgical, Shelton, Connecticut) was placed into the endometrial cavity using sterile technique, and a tissue sample was aspirated into the cannula. After removal of the catheter, the tissue specimen was discharged into a sterile Petri dish. Tissue proximal to the sampling portal of the cannula was placed in 10% formalin fixative. In order to further minimize the contamination of the specimen from the cervix, a swab absorbed 5 mm of distal tissue for qualitative nucleic acid amplification testing (Aptima).
Genotyping was performed with the Illumina HumanOmniExpressExome-8 v1.2 BeadChip array. Markers with call rate < 95%, deviation from Hardy-Weinberg equilibrium (P < 1.00E−6), or more than two alleles were removed from subsequent imputation. Samples were also screened for relatedness based on identity by descent, and two samples with subject relatedness (PI_HAT > 0.185) were filtered [Anderson CA, 2010]. We phased and imputed post-QC genotype data using the 1,000 Genomes phase 1 dataset (17), SHAPEIT software (version 2.837) (18, 19), and BEAGLE software (version 4.1) (20). After imputation, variants with poor imputation quality (Beagle r2 < 0.1) and low minor-allele frequency (MAF < 0.1) were filtered.
Total RNA was isolated from blood and analyzed via microarray (Illumina Human HT12 v3.0 expression beadchip) as described previously (3). Expression profiles can be accessed from GEO (Gene Expression Omnibus) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110106). Transcripts were quantile normalized (21) and log2 transformed. Batch effects were measured by guided principal component analysis (22), and corrected using ComBat (23).
We used Plink (version 1.9) (24) (24)[Chang CC, 2015. www.cog-genomics.org/plink/1.9/] to control for population stratification. Population stratification was adjusted using the first 15 principal components, which explained >80% of the total variance. Oral contraceptive use, and gonorrhea co-infection were selected as covariates in the final model (1), and included in downstream analysis. We previously established that these covariates are risk factors for ascending infection by stepwise logistic regression of cross-sectional enrollment data, with the α to enter set at P ≤ 0.2 and the α to be maintained set at P ≤ 0.1.
We applied the probabilistic estimation of expression residuals (PEER) method (25), to infer and account for confounding factors affecting gene expression levels. We set maximum relevant factors in PEER to 50, then used the PEER-processed residuals of gene expression for downstream analyses. PEER factor analysis infers broad variance components in the measurements and is used to correct for observed and hidden confounding factors including bacterial burden (26–28). It has been demonstrated to correct blood cell compositions successfully in a similar, large blood-derived eQTL study (26).
Heritability was calculated via a linear mixed model using GCTA (Genome-wide Complex Trait Analysis) software (29).
eQTLs were defined as cis if eSNPs were located within 1 Mb of the exon boundaries flanking a gene (eGene). We conducted single locus eQTL mapping using a linear regression model with adjustment of covariates and population stratification. Infection status (uninfected vs. infected), and genotype-infection interaction were included in the model to enable the detection of response eQTLs displaying varying effects between uninfected and infected women, and constant eQTLs with consistent effects regardless of infection status. Presence of genotype-infection interaction was examined with a cutoff p-value of 0.05. In the absence of significant genotype-infection interaction, a main effect model was used. Otherwise, a two degree of freedom test was performed. This compared a model in which transcript expression depends on the additive effects of a SNP and infection status together with their interaction term, to a model in which expression depends only on infection status. All analyses were conducted using R. We used all association p-values in the cis region to estimate the false-discovery rate (FDR-qvalue) using the qvalue package in R. LocusZoom (30) was used for the regional visualization of eQTL results on the basis of linkage disequilibrium (LD) ascertained from samples used in this study.
Associations of genotype at the lead cis-eQTL at the CD151 locus and expression of CD151 in uninfected participants and infected participants respectively, were tested using a linear regression model with adjustment of covariates and population stratification.
We developed a generalized multi-SNP mediation intersection-union test to rigorously assess the effects of multiple eSNPs on ascension through their alteration of eGene expression (31). The locations and FDR of significant mediator eGenes were visualized using Circos software (32).
We used extreme phenotype sampling to select samples for evaluation of the genetic effect of the lead SNP on CD151 expression in blood T cells and T cell subsets after Ct stimulation using freshly thawed peripheral blood mononuclear cell (PBMCs). This is a well-established strategy for achieving statistical power to detect genotype-phenotype associations in the face of limited sample sizes with the rationale that the phenotypic extremes are enriched for either deleterious or protective variants (33, 34). For a SNP with an additive effect, the z-extreme sampling design, where extremes are defined based on “residual phenotype” after adjustment for non-genetic covariates, provides better power than random sampling (35). The phenotypic extremes in this study were defined as all women homozygous for the lead eSNP at the CD151 locus with the upper or lower 20% extreme whole blood CD151 expression after adjustment of covariates. We selected 6 women from each of two extreme groups by random sampling. After sampling, all 6 women from the upper extreme had CC genotype, and all 6 women from the lower extreme carried GG genotype.
Peripheral blood mononuclear cells (PBMCs) were thawed in T cell media (10% filtered, heat inactivated FBS, 5 mL of 1M HEPES, 5 mL of 200mM L-Glutamine, 5 mL of 100mM Sodium pyruvate, 200 μL of 100 mg/mL vancomycin and 25 mg of gentamicin) containing benzonase (50 units/mL of media; Sigma-Aldrich; Cat # E1014) followed by a wash in plain RPMI media and enumerated. For T cell evaluations, 1.5 x 106 cells were incubated in media alone in a 96 well plate, or wells were coated with 10 μg/mL of anti-CD3 (UCHT1) and 5 μg/mL of anti-CD28 (28.2) or pooled gamma-irradiated Ct EB/RBs from serovars D, F and H (5 μg/mL) and incubated for 7 days at 37°C in a 5% CO2 incubator with media replenishment every 24 hrs. Cells were harvested and stained with 1:100 of Zombie UV fixable viable dye (BioLegend, Cat # 423107) in 1x PBS for 15 min at room temperature. Cells were subsequently washed with cell staining buffer (BioLegend, Cat # 420201) before staining with an optimized concentration of antibody cocktail comprising CD45-BV510 (BioLegend, Clone: HI30, Cat # 304036), CD3-APC (BioLegend, Clone: HIT3a, Cat # 300312), CD4-PerCP-Cy5.5 (BioLegend, Clone: RPA-T4, Cat # 300530), CD8-AF700 (BioLegend, Clone: SK1, Cat # 344724), CD45RA-BV421 (BioLegend, Clone: HI100, Cat # 304130), CCR7-BV605 (BioLegend, Clone: G043H7, Cat # 353224), CD151-PE (BD Biosciences, Clone: 14A2.H1, Cat # 556057) and Ki67-PE/Cy7 (BD Biosciences, Clone: B56, Cat # 561283) for 30 min at 4°C in dark. After staining, cells were washed twice and fixed with 2% PFA. Fixed cells were washed and fluorescence acquired on a BD LSR II, 7 Laser cytometer. Wilcoxon rank-sum test was used to compare percentages of CD151+ T cells and CD151 MFI between two genotype groups (GG vs. CC); paired samples Wilcoxon test was used to compare groups before and after in-vitro stimulation.
The characteristics of participants in this study are summarized in Table 1. Only oral contraceptive pill use and co-infection with Neisseria gonorrhoeae were significantly different among the three groups with rates being higher in women with endometrial infection, consistent with our previous report (1). Consequently, these factors were adjusted as covariates in the models used in this study.
We analyzed genotype data of ∼2.6 million SNPs and the expression profiles of 47,109 probe sets corresponding to 34,540 unique genes. The estimated mean narrow-sense heritability, h2, for variance of gene expression was 0.23 (standard error = 0.098, P = 0.009), suggesting significant genetic effects on gene expression. We identified 105,873 SNPs with effects in cis (<1 Mb) for 6,837 genes (FDR < 0.1, P < 2.17E-4). Table 2 summarizes the identified cis-eQTLs.
We applied the generalized multi-SNP mediation intersection-union test (31) to the identified cis-eQTLs with FDR < 0.1, and determined that expression of 33 eGenes was increased in women with endometrial infection (FDR < 0.2, Table 3), and expression of 48 eGenes was decreased in women with endometrial infection (FDR < 0.2, Table 4). Distribution and size of these 81 eGenes, their positions within the chromosomes and -log 10(FDR) of mediation tests were demonstrated by circos plot (Figure 1). We have highlighted eGenes below that are related to biologic pathways involved in Ct infection and inflammation (Figure 2) and/or T cell mediated control/clearance (Figure 3).
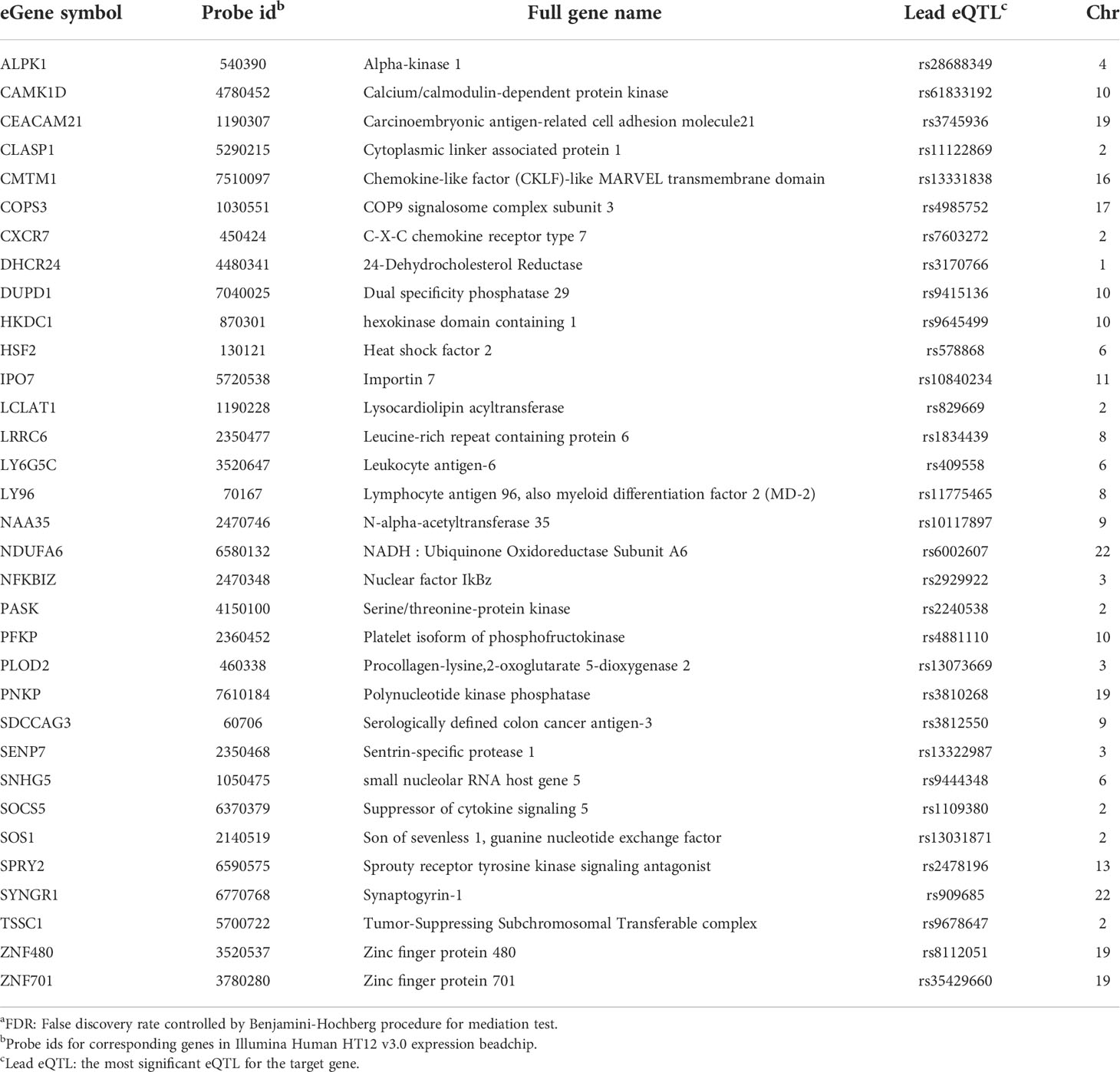
Table 3 eGenes with increased expression in women with endometrial infection, and their corresponding cis-eQTLs and chromosomal locations (mediation test FDRa<0.2).
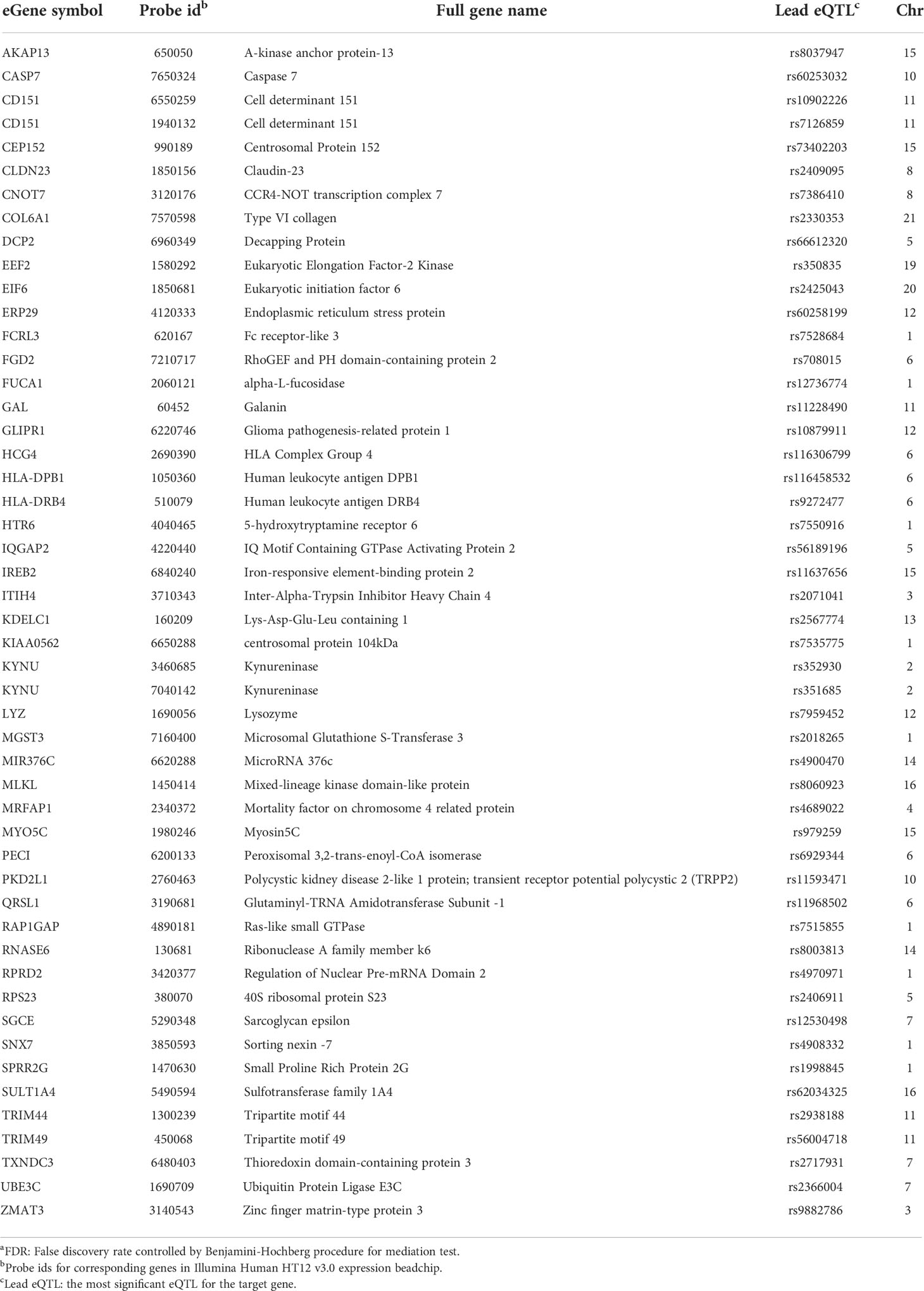
Table 4 eGenes with decreased expression in women with endometrial infection, and their corresponding cis-eQTLs and chromosomal locations (mediation test FDRa<0.2).
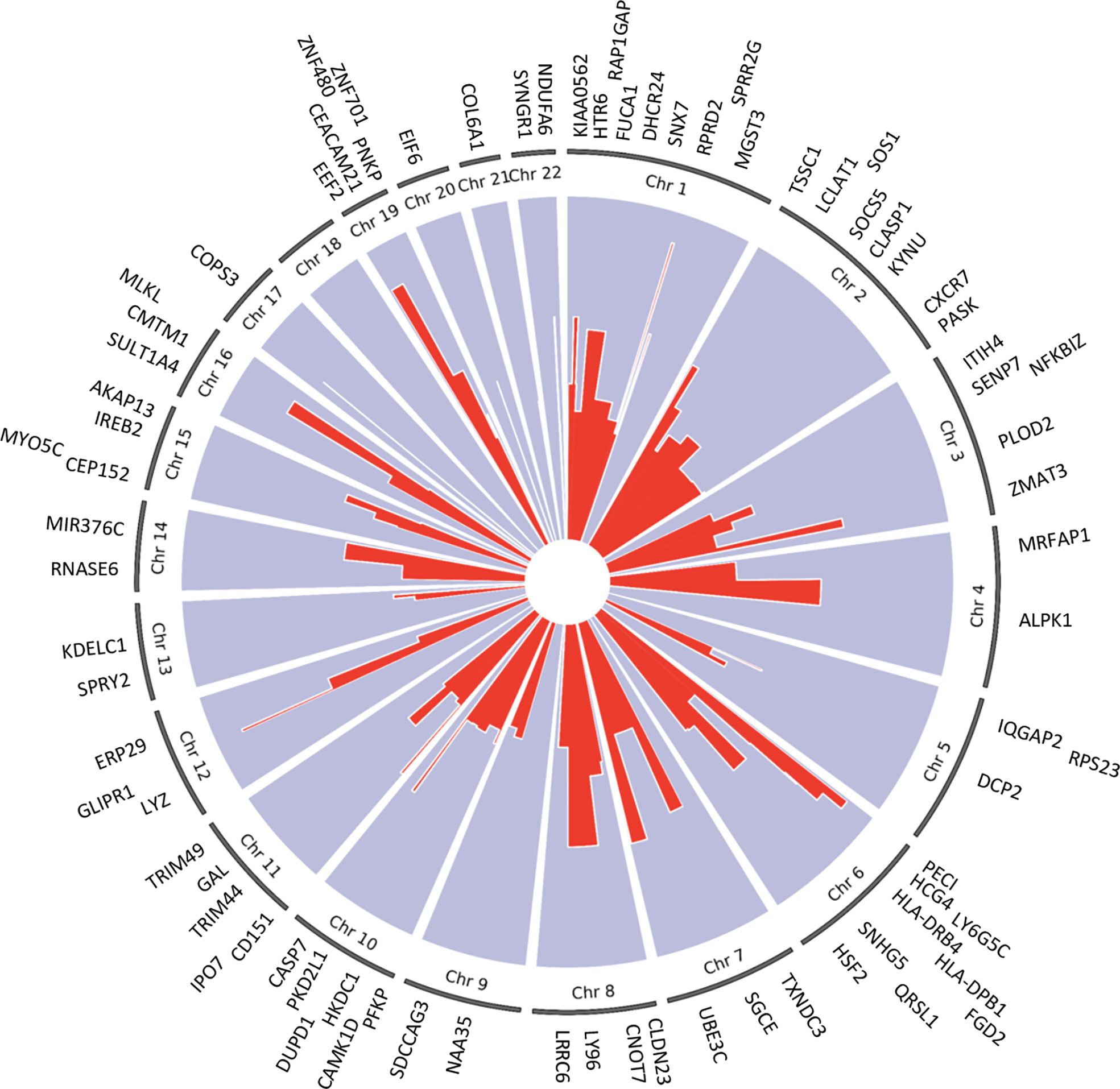
Figure 1 Circos plot of 81 eGenes mediating effects of cis-eQTLs on ascension (FDR <0.2) and their chromosome number. The -log 10(FDR) mediation test results for each eGene are represented by the red lines. The width of the red lines indicates the size of each gene. The eGenes are labeled at their respective chromosome base pair location boundaries.
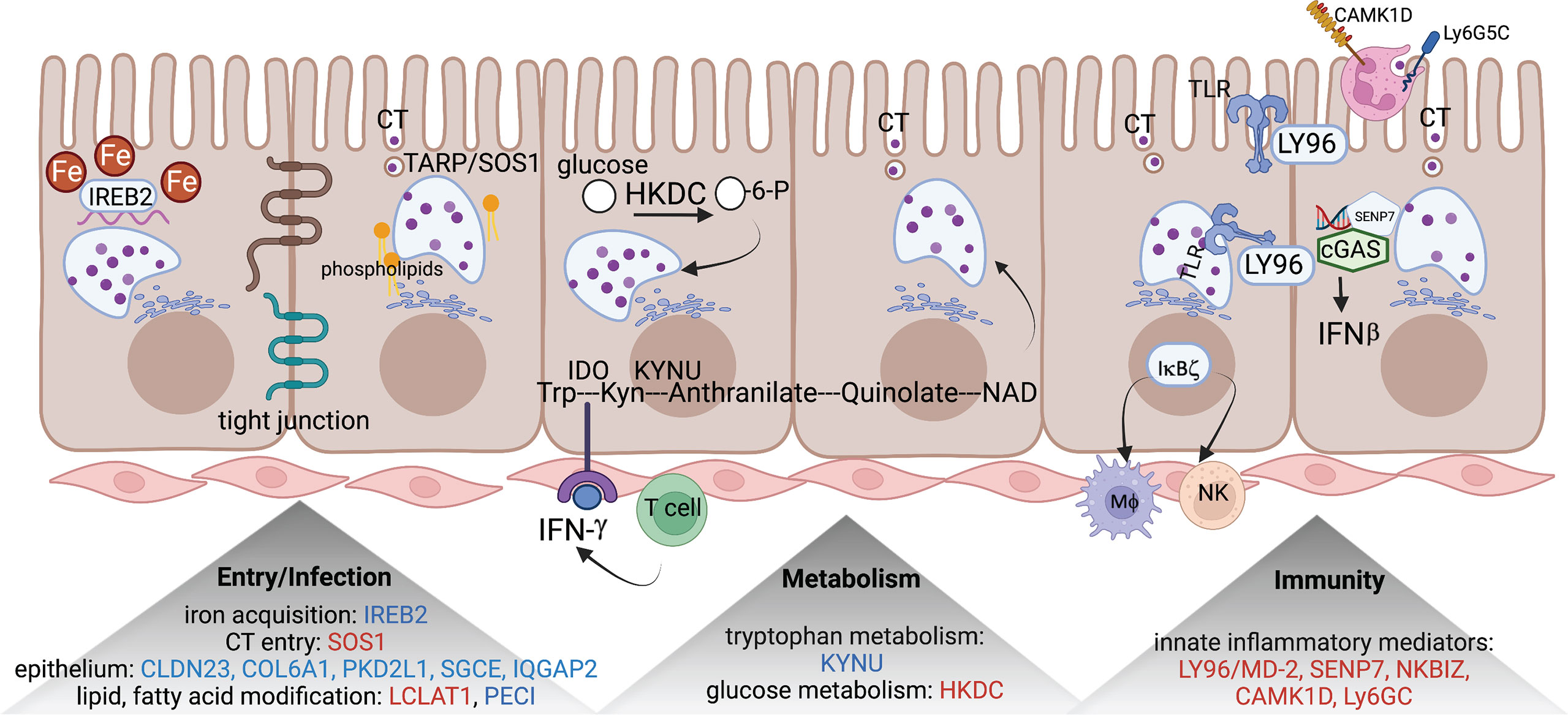
Figure 2 eGenes encoding proteins linked to biologic pathways involved in chlamydial infection. eGenes downregulated in women with endometrial infection are listed in blue and include: IREB2, an iron acquisition protein; multiple molecules involved in formation of tight junctions and epithelial integrity (CLDN23, COL6A1, PKD2L1, SGCE, and IQGAP2); PECI, that modifies fatty acids; and KYNU, important in tryptophan metabolism. eGenes upregulated in women with endometrial infection are listed in red and include SOS1, that encodes a protein that directly interacts with chlamydial TARP to facilitate entry; LCAT1 important for lipid modification; HKDC, glucose metabolism; and genes for multiple innate inflammatory mediators (LY96/MD-2, SENP7, NKBIZ, CAMK1D, and Ly6GC). The figure was created using BioRender.com.
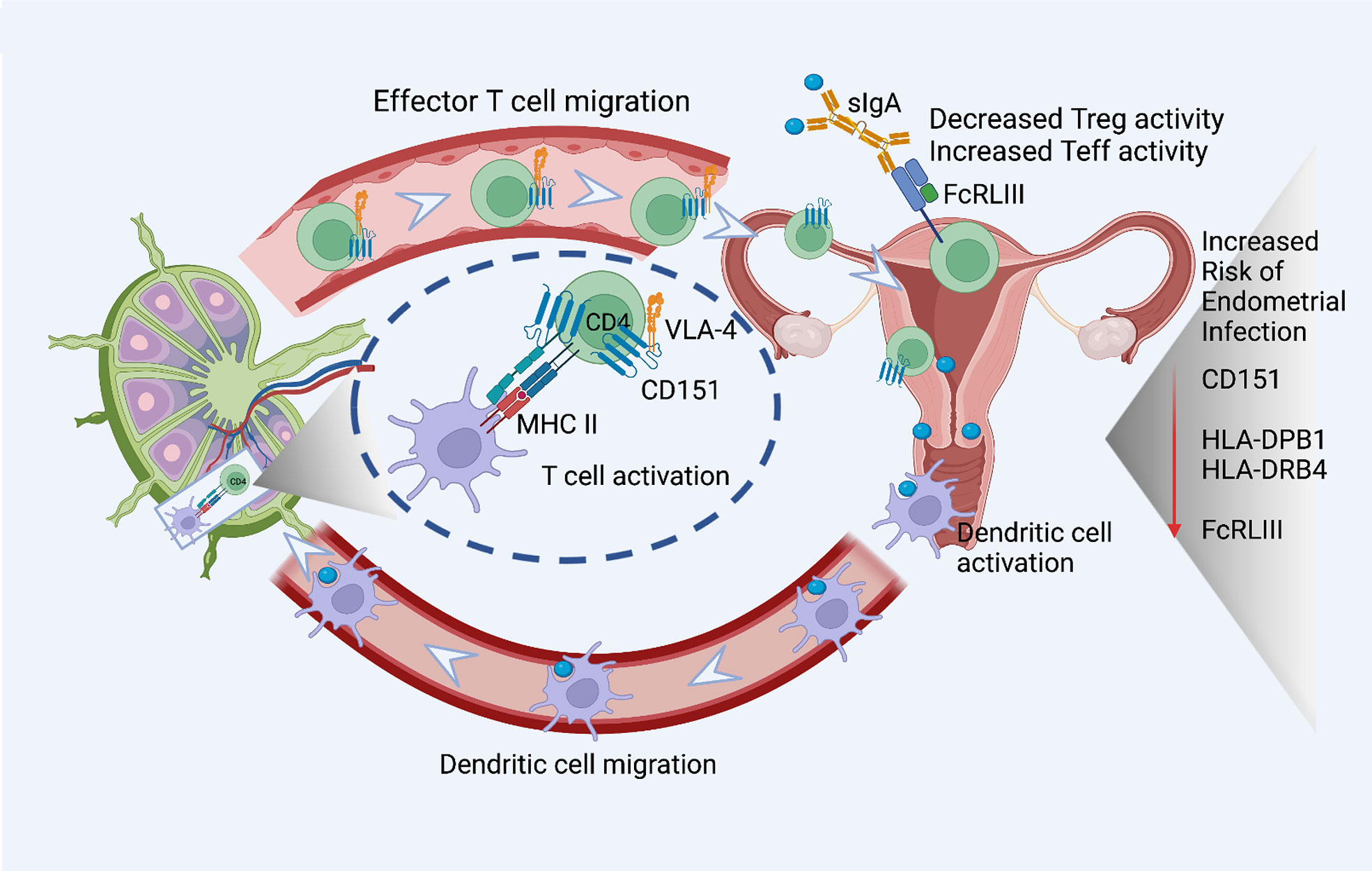
Figure 3 eGenes important for T cell activation, function, and migration downregulated in women with endometrial infection include: CD151, a tetraspanin intimately involved in the T cell/antigen presenting cell synapse that also interacts with integrins to promote T cell migration; MHC Class II molecules important for antigen presentation to CD4 T cells, HLA-DPB1 and HLA-DRB4; and FcRLIII, that binds bacterial-specific sIgA at mucosal surfaces to inhibit T suppressor functions and enhance T effector functions. The figure was created using BioRender.com.
SOS1, encoding a nucleotide exchange factor that interacts with chlamydial translocated actin recruiting phosphoprotein (TARP) to facilitate chlamydial entry (36), was upregulated in women with endometrial infection, while several eGenes involved in formation of tight junctions and epithelial cell adhesion (CLDN23 (37), COL6A1 (38)(REFs), PKD2L1 (39), SGCE (40), IQGAP2 (41, 42)) were downregulated in women with endometrial infection (Table 4). Increased Ct entry into epithelial cells and compromised epithelial integrity are plausible mechanisms for increased risk of ascension.
eGenes also mapped to loci involved in phospholipid modification and fatty acid metabolism. LCLAT1 (Lysocardiolipin Acyltransferase 1), exhibiting increased expression in women with endometrial infection (Table 3), encodes a protein that catalyzes the reacylation of lyso-cardiolipin to cardiolipin (43), a key step in cardiolipin remodeling, while PECI (Table 4), decreased in women with endometrial infection, encodes peroxisomal 3,2-trans-enoyl-CoA isomerase which catalyzes an isomerization step leading to beta-oxidation of unsaturated fatty acids (44). Since the chlamydial genome carries all the genes necessary for phospholipid and fatty acid synthesis and biochemical studies have confirmed that bacterial membrane biogenesis is achieved with autonomously synthesized phospholipids (45–47), it seems unlikely that these genes could influence chlamydial replicative success directly. However, host membrane trafficking pathways are hijacked to assemble the expanding inclusion membrane required to accommodate multiplying chlamydial progeny (48). Inhibition of host lipid synthesis/trafficking decreases the yield of Ct (49), and lipid biosynthesis inhibitors lead to premature rupture of the inclusion membrane (50), arguing that these eGenes may be important for maximizing infectious yield. An eGene involved in the regulation of iron homeostasis was also mapped. Down-regulated expression of the gene for iron-responsive element-binding protein 2 (IREB2) (Table 4), which regulates cellular iron homeostasis, might restrict intracellular growth of chlamydiae directly because iron restriction limits chlamydial infection (51) or indirectly through co-regulatory effects modulating tryptophan metabolism (52).
A metabolic pathway eGene with potential to impact Ct replication and development, KYNU which encodes kynureninase, was downregulated in women with endometrial infection (Table 4). Interferon-gamma released from immune cells infiltrating Ct infected tissues activates indoleamine 2,3-dioxygenase (IDO) which catalyzes the breakdown of tryptophan to kynurenine (KYN) (53). Ct lack several enzymes of the tryptophan biosynthetic pathway (54) and are largely dependent on cellular metabolism to access sufficient tryptophan to support replication (55). Kynureninase cleaves KYN to anthranilic acid, which eventually results in production of nicotinamide adenine dinucleotide (NAD). Since dioxygenases are partially regulated by end product inhibition (56), reduced KYN activity could increase tryptophan metabolism and reduce its availability to Ct. Fisher and colleagues determined that the chlamydial ATP/ADP translocase, Npt1, preferentially transports NAD, indicating that the bacteria scavenge this energy substrate from their host, despite their ability to complete de novo synthesis using folate (57). Finally, kynurenine metabolites are potent regulators of immune function (58). Thus, altered expression of KYNU may affect chlamydial growth through changes in the availability of tryptophan or NAD or immune response alterations. Another metabolic gene, encoding HKDC (hexokinase domain containing 1), that catalyzes ATP-dependent phosphorylation of glucose to G6P (59), a primary carbohydrate substrate for Ct (60, 61), was elevated in women with endometrial infection (Table 3).
Myeloid cell and innate inflammatory responses are primarily associated with development of reproductive tract tissue pathology during Ct infection rather than infection resolution (62, 63). LY96, encoding lymphocyte antigen 96, also called MD-2, which associates with TLR2 and TLR4 to increase their responsiveness to lipoproteins and LPS (64, 65), and chlamydial heat shock protein 60 (66), was upregulated in women with endometrial infection (Table 3). Other inflammatory genes that were upregulated in women with endometrial infection included SENP7, CAMK1D, and Ly6G5C. The protease encoded by SENP7 potentiates the DNA sensor cyclic GMP-AMP synthase (cGAS) by relieving small ubiquitin-like-related modifier (SUMO) inhibition of cytosolic DNA sensing (67). cGAS is required for IFN-β expression during chlamydial infection in multiple cell types (68), and murine (69) and human studies (3) indicate IFN-β compromises resolution of chlamydial infection and exacerbates pathology. The calcium/calmodulin-dependent protein kinase, encoded by CAMK1D, regulates activation of neutrophils (70), and the protein encoded by LY6G5C regulates neutrophil migration (71).
A robust CD4 T cell response is key to elimination of Ct infection (72, 73). Women with endometrial infection exhibited downregulation of CD151, which encodes a protein that augments T cell activation, adhesion and migration (12, 13, 74–76). The eGene for FCRL3, which enhances generation of CD4 effector memory T cells at mucosal sites (77), and eGenes for HLA-DPB1, HLA-DRB4, MHC Class II molecules important for activation of CD4 T cells (78, 79) were also down-regulated (Figure 3). Previous work demonstrated that FCRL3 engagement inhibited regulatory T cell suppressive functions, and induced IL-17, IL-26, and IFNγ production, suggesting that FCRL3 engagement mediates a transition of regulatory T cells to a pro-inflammatory Th17-like phenotype. Secretory IgA (SIgA) is a FCRL3-specific ligand, suggesting pathogen-specific sIgA might drive mucosal regulatory T cell plasticity to help control infection (77). The associated decrease in expression of eGene for HLA-DPB1 and HLA-DRB4 could also diminish Class II presentation of chlamydial antigens that would further inhibit protective CD4 T cell responses. Increased expression of the eGene for SOCS5 in women with endometrial infection (Table 3) may reflect inhibition of STAT1 and STAT3 signaling necessary for the differentiation of Th1 and Th17 cells, respectively (80).
Examples of significant cis-eQTL signals at the CD151 and HLA-DPB1 loci are demonstrated in Figure 4. The lead eSNP (strongest eQTL) rs10902226 (Figure 4A) is located 4266 base pairs downstream of CD151 gene and is a TSPAN4 intronic variant. The lead eSNP rs10902226 (Figure 4B) is located 9363 base pairs upstream of HLA-DPB1 gene and is a HLA-DPA1 intronic variant. These two lead eSNPs are highly significantly associated with expression of CD151 and HLA-DPB1 with P<1.00E-11 and P<1.00E-13, respectively. Multiple nearby SNPs in LD (r2>0.6) with the lead eSNPs are also significant cis-eQTLs (P<1.00E-8 for CD151 and P<1.00E-10 for HLA-DPB1, respectively), implying that both lead eSNPs are true.
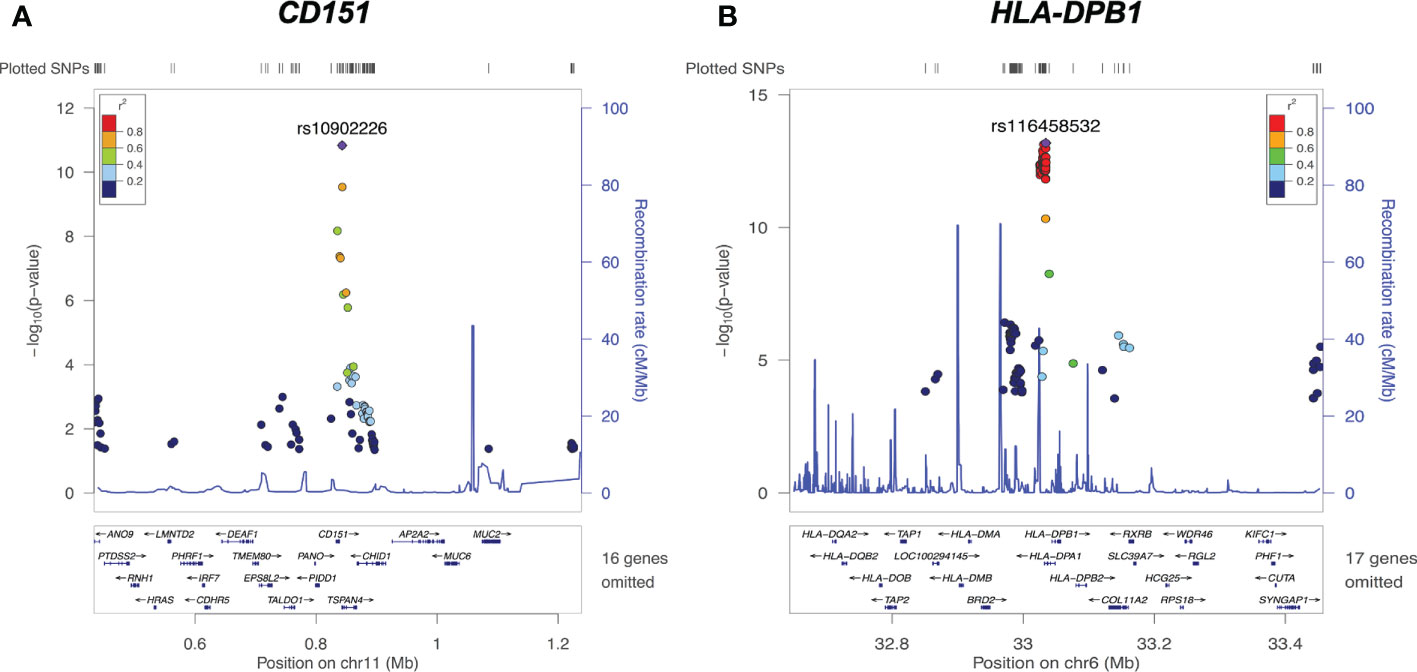
Figure 4 cis-eQTLs of CD151 and HLA-DPB1. (A) Regional plot on chromosome 11 CD151 locus reveals the lead cis-eQTL rs10902226 (purple diamond) is significantly associated with CD151 expression (P<10E-11). (B) Regional plot on chromosome 6 HLA-DPB1 locus reveals the lead cis-eQTL rs116458532 (purple diamond) is significantly associated with HLA-DPB1 expression (P<1.00E-13). For both graphs, each dot represents one SNP. X axis shows the physical location of the SNPs, gene names and flanking region; Y axis indicates the −log10 value (P-value) of the respective SNP. The color for each dot represents the pairwise linkage disequilibrium r2-value against respective lead cis-eQTL.
The effect of genotypes on CD151 expression in uninfected and infected participants are further represented by Figure 5. Figure 5A presents the association of genotype at the lead cis-eQTL rs10902226 at the CD151 locus and expression of CD151 in uninfected participants and Figure 5B for infected participants. In uninfected participants (A), the SNP shows suggestive evidence to be an eQTL (P value = 0.0018), but it is not significant after multiple testing correction. In infected participants (B), the SNP is a strong eQTL (P value = 6.7E-9) with an additive effect. In (C) the effect of the C allele on CD151 expression and its relationship to endometrial infection can be visualized (P=0.012 by mediation test on this single SNP). This graph demonstrates that CC genotype is associated with increased CD151 expression and reduced occurrence of endometrial infection, in contrast GG genotype is associated with lower CD151 expression and increased occurrence of endometrial infection.
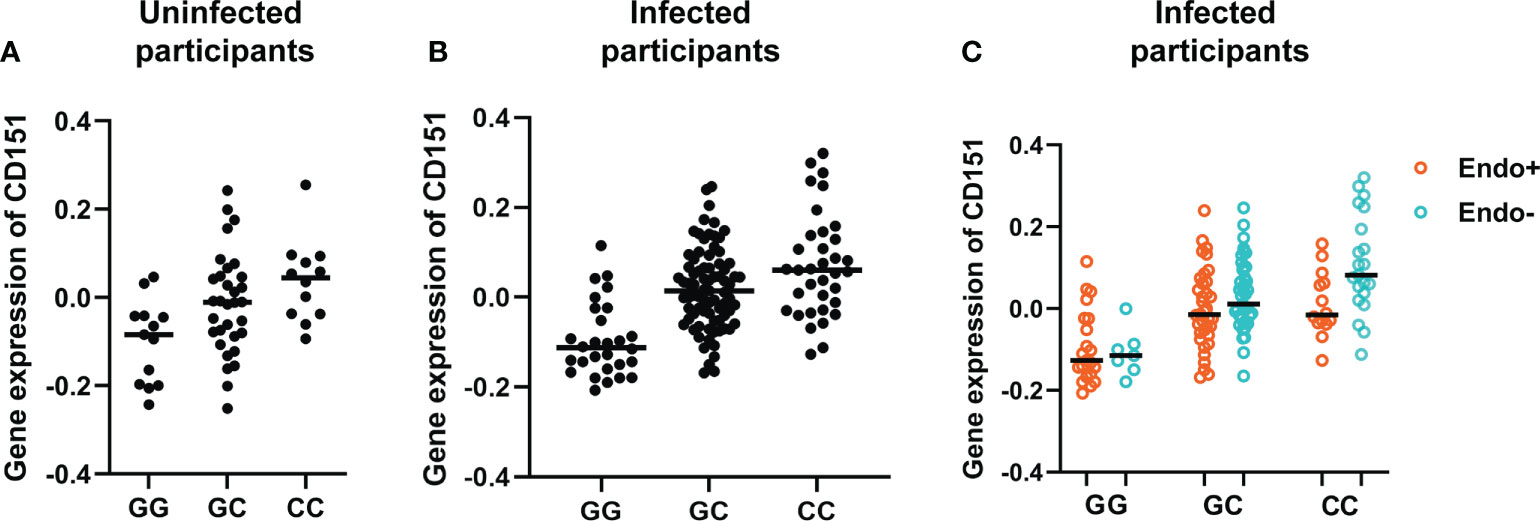
Figure 5 Genotype at the lead cis-eQTL rs10902226 at the CD151 locus is associated with altered expression of CD151 and risk of ascension. Using linear regression, in uninfected participants (A), the SNP shows suggestive evidence to be an eQTL (P value = 0.0018), but it is not significant after multiple testing correction. In infected participants (B), the SNP is a strong eQTL (P value = 6.7E-9) with an additive effect. In infected participants (C), the effect of the C allele on CD151 expression and its relationship to endometrial infection can be visualized (P=0.012 by mediation test on this single SNP). This graph demonstrates that CC genotype is associated with increased CD151 expression and reduced occurrence of endometrial infection. In contrast, GG genotype is associated with lower CD151 expression and increased occurrence of endometrial infection. In all graphs, each dot represents one subject. The black line indicates the median expression of CD151 in each genotype group.
CD151 is a member of the tetraspanin family (TM4SF) associated with immune cell migration and adhesion. It associates with multiple integrins and plays a role in regulating integrin trafficking and/or function (81). CD151 actively changes cell cycle control and cell death processes and identifies T cells with hyperproliferative capabilities (13).
We compared T cell CD151 expression in women with CC and GG genotypes after a 7-day culture of PBMCs in media, media with anti-CD3/CD28, or killed Ct, and determined that frequencies of CD151+CD3+ lymphocytes were increased in in both unstimulated and stimulated cells of women with the CC genotype (Figure 6). Although incubation with Ct led to increases in numbers of CD151+ T cells in both genotype groups, the increases were not statistically significant (Figure 6C). Nevertheless, the mean fluorescent intensities (MFIs) of CD151 expression per cell concatenated from 6 women with CC genotype was higher than ones from women with GG genotype with and without Ct stimulation and increased in both genotype groups after stimulation with Ct (Figures 6D, E), and all these changes were statistically significant when quantified at the individual subject level (Figure 6F). Cell expression of CD151 also increased in both genotype groups with CD3/CD28 stimulation, demonstrating the response also occurs with non-specific stimulation of the T cell receptor (Figures 6D, E). Frequencies of CD4+ (Supplementary Figure 1A), CD8+ (Supplementary Figure 1B), naive (Supplementary Figure 1C), and memory (Supplementary Figure 1D) CD3+ lymphocytes were not different among genotype groups with or without stimulation. However, Ct stimulation led to a downward shift in frequencies of naïve CD3+ lymphocytes and a corresponding increase in memory CD3+ lymphocytes in both genotypes (Supplementary Figures 1C, D, respectively), indicating Ct responsiveness led to memory T cell proliferation in both patient subgroups. Taken together, these data indicate that increased frequencies of CD151+CD3+ lymphocytes in the CC genotype are not due to differences in overall numbers of T cells or alterations in relative frequencies of CD4 or CD8 T cells when compared to the GG genotype (Figure 6).
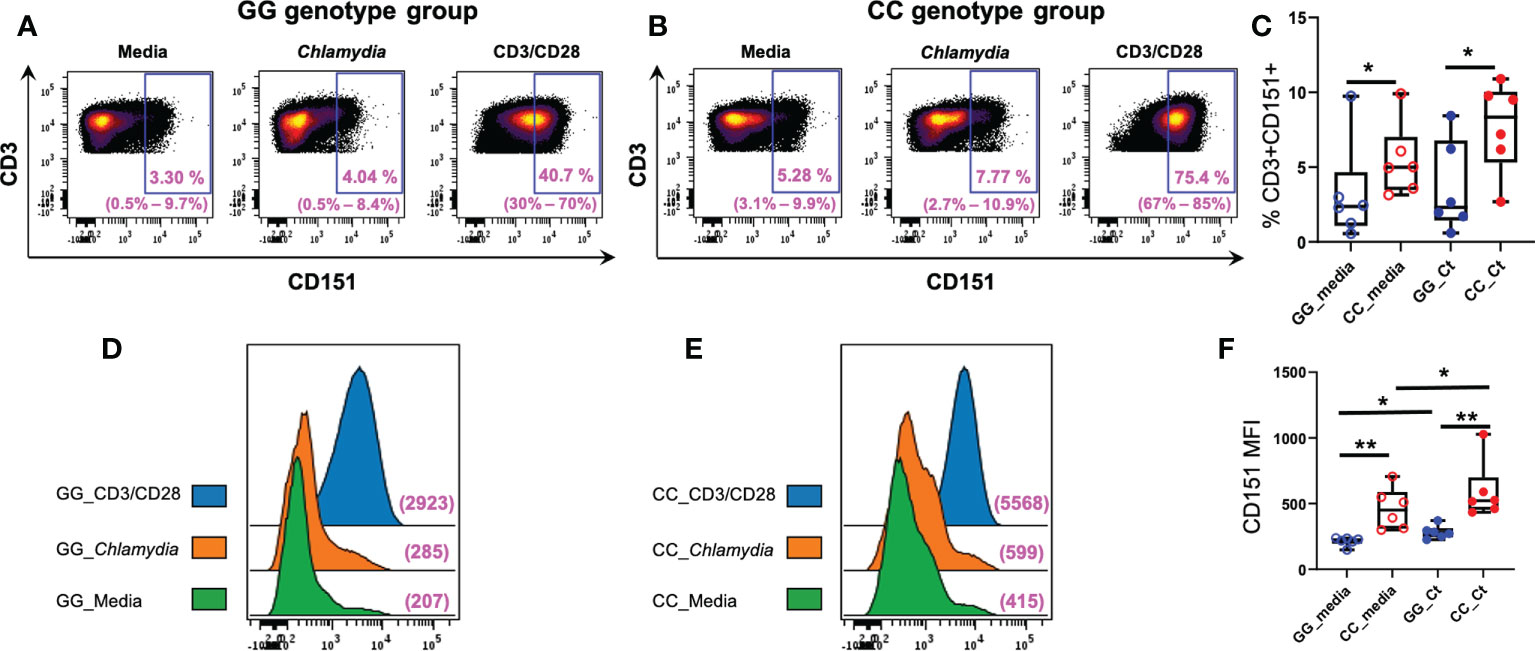
Figure 6 Association of CD151 genotypes with CD3+ lymphocyte CD151 expression in Ct infected women. Each dot plot is a concatenated FCS file consisting of CD3+ cells from six women of (A) GG and (B) CC genotypes, respectively. Range of six individual patients within each group’s condition is in parentheses. (C) Frequencies of CD151+CD3+ cells in GG (n = 6; blue circles) and CC genotype (n = 6; red circles) women with (filled circles) and without (empty circles) Ct stimulation for 7 days. The boundaries of each box indicate the 25th and 75th percentiles; the line within each box indicates the median; whiskers indicate the 0th and 100th percentiles. Each dot represents one subject. The relative frequencies of CD3+CD151+ lymphocytes were significantly increased in the CC group with and without Ct stimulation compared to GG group by Wilcoxon Rank Sum Test. (D) Mean florescence intensities (MFI) of CD151 on CD3+ cells in women with GG genotype compared to women with (E) CC genotype without Ct stimulation (green histograms), after Ct stimulation (orange histograms) and after anti-CD3/CD28 stimulation (blue histograms). The x-axis indicates the fluorescence intensity of CD151. (F) Box and whisker plots summarize the MFIs of CD151 in the different genotype groups with and without Ct stimulation. For MFI, intensities were significantly greater in the CC group compared to GG group with and without stimulation by Wilcoxon Rank Sum Test. Wilcoxon signed rank test indicated Ct stimulation significantly increased the MFI expression of CD151 in both groups, *:P < 0.05; **:P < 0.01.
Finally, we characterized the presence of CD151 expression on T cell subpopulations after stimulation (Figure 7). As reported above, frequencies of CD151+CD3+ cells were increased in CC genotype women and Ct stimulation led to marginal increases in CD151+ cell frequencies. Flow cytometry using T cell specific markers revealed these patterns were consistent across T cell subsets. Frequency of CD151 expression was higher among CD4 T cells compared to CD8 T cells, rare among naïve T cells, but easily detected in memory T cells with the highest expression detected in Ki67+ proliferating T cells. Higher CD151 expression was consistently observed in T cells from women with the CC genotype where endometrial infection was reduced, suggesting that CD151 may play a protective role during Ct infection (Figure 7).
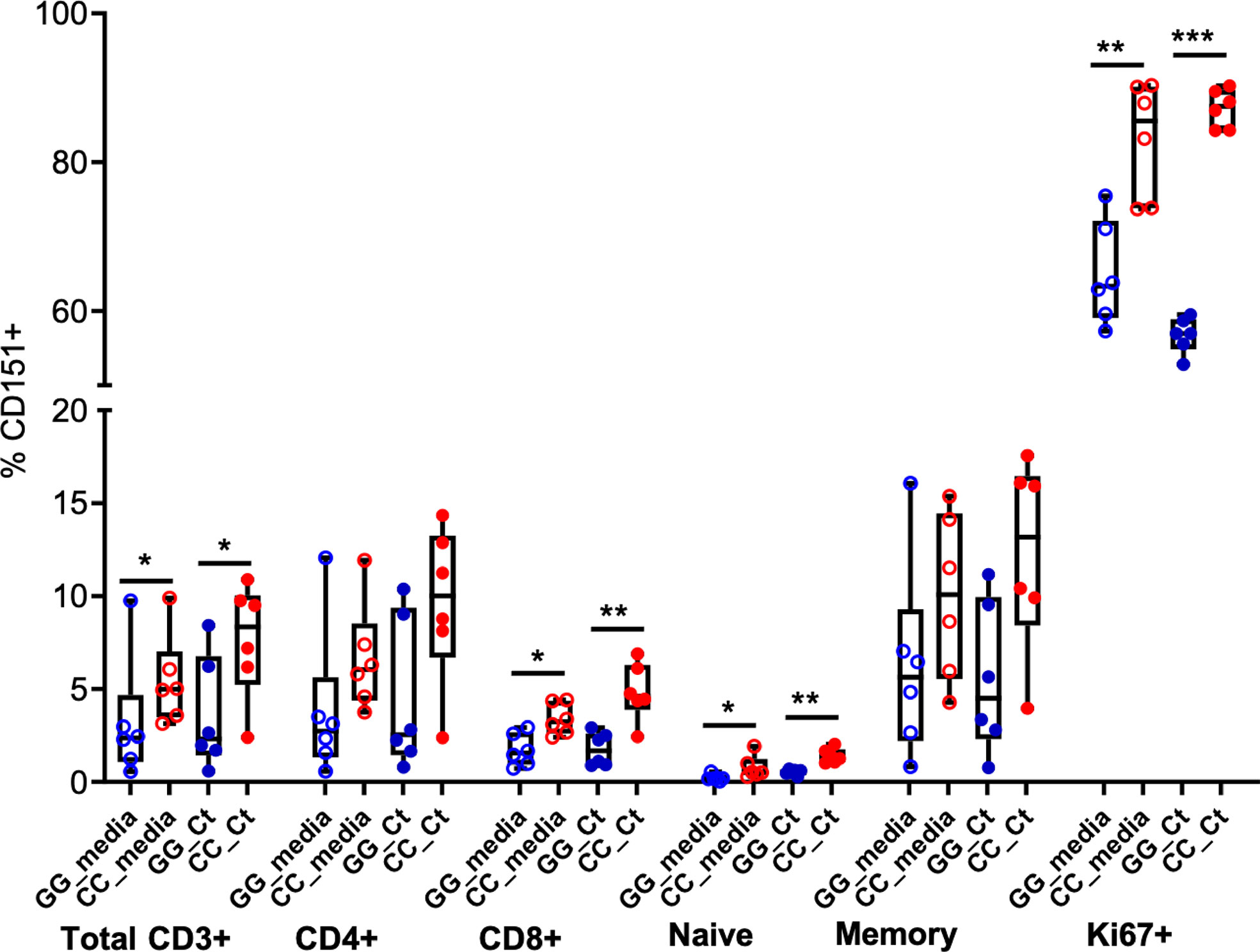
Figure 7 CD151 is enriched in memory and Ki67+ T cells. Box and whisker plots indicate frequencies of CD151+ cells among total CD3+ cells, CD4, CD8, naïve, memory and Ki67+ T cells in GG (n = 6; blue circles) and CC (n = 6; red circles) genotype groups, without (open circles) and with (closed circles) Ct stimulation for seven days. The boundaries of the box indicate the 25th and 75th percentiles; the line within the box indicates the median; whiskers indicate the 10th and 90th percentiles. Each dot represents one subject. The relative frequencies of CD151+ lymphocytes were significantly increased in the CC group with and without Ct stimulation compared to GG group among total CD3+ cells, CD8, naïve and Ki67+ T cells by Wilcoxon Rank Sum Test. *:P < 0.05; **:P < 0.01; ***:P < 0.005. No statistical differences were detected between media and Ct-stimulated groups for any cell type.
We performed eQTL mapping and mediation tests to link gene expression-associated loci with presence of endometrial infection and provided evidence for 81 eGenes mediating the cis-eQTL effects on ascending infection (FDR < 0.2), inherently associated with altered disease risk.
Expression of multiple eGenes important for formation of epithelial tight junctions was downregulated in women with endometrial infection. Compromised epithelial integrity may lead to enhanced Ct spread directly, and accompanying decreases in expression of molecules engaged in actin cytoskeleton activities, IQGAP2, and cell-cell matrix interactions, such as SGCE, could result in dysregulated immune cell adhesion and migration, further enhancing spread of infection. The intracellular niche of Ct imposes the need and/or opportunity for the bacterium to utilize host cell metabolic pathways for its benefit. Differential expression of eGenes involved in cellular iron acquisition, lipid and fatty acid modification, tryptophan and glucose metabolism, which could directly affect chlamydial fitness, growth, and replication, were associated with altered risk of Ct ascension (Figure 2). Changes in tryptophan metabolism could also exert indirect effects on Ct ascension. Tryptophan catabolites inhibit both T cell and natural killer (NK) cell proliferation and activation (58), immune cell subsets important in Ct infection resolution. Alterations in glucose metabolism could also affect Ct infection indirectly via effects on T cell functionality (82).
eGenes involved in innate immune responses, neutrophil migration and activation, and type I interferon signaling were upregulated in women with endometrial infection, suggesting that activation of these pathways fail to prevent Ct ascension (Figure 2). Prior work in mice and humans indicate that enhanced TLR activation (83–85), phagocyte activation (71, 86, 87), and type I interferons (3, 69) are associated with increased disease development, rather than protection. Thus, host genetics may play an important role in driving enhanced inflammatory responses that are ineffective for host defense against Ct and lead to disease.
Endometrial infection was associated with decreased expression of eGenes involved in generation of adaptive T cell immunity. This correlation agrees with the central role of T cells, particularly Ct-specific CD4 T cells, in resolution and protection from Ct infection. Women with endometrial infection had lower expression of genes for MHC Class II molecules, HLA-DRB1 and HLA-DPB1. A recent study reported that a variety of HLA-DRB1 alleles/haplotypes were associated with altered Ct infection risk, and differential rate of clearance in a cohort of Columbian women (88); HLA-DPB1 alleles were not examined.
The finding of decreased expression of eGene for FcRL3 in women with endometrial infection is particularly interesting given its role in mucosal immunity through binding of secretory IgA. Agarwal et al. (77) demonstrated the importance of pathogen-specific secretory IgA binding of FcRL3 on T cells to inhibit their regulatory function and drive their conversion to proinflammatory Th17-like cells that could combat pathogens breaching the mucosa.
We determined that women with the GG genotype for a lead eSNP for CD151 exhibited significantly decreased whole blood and T cell CD151 expression compared to women with GC and CC genotypes that associated with presence of endometrial infection. We experimentally confirmed that the cis effect of the lead eSNP of CD151 on expression of CD151 by T cells was determined by genotype both before and after in-vitro stimulation with Ct or anti-CD3/CD28, suggesting this cis eQTL can be considered as a candidate biomarker for ascension.
In cooperation with other tetraspanin molecules in tetraspanin-enriched microdomains (TEMs), CD151, regulates cell adhesion and migration through physical and functional interactions with integrins (89). CD151 strongly associates with LFA-1 and the β1 integrin family integrins (75). Complexes form between CD151 and αLβ2 (LFA-1) and between CD151 and α4β1 integrin (VLA-4). LFA-1 has an essential role in mediating firm adhesion and transendothelial migration of T cells from the bloodstream into tissues. T cell expression of CD151 has been shown to be important in T cell trafficking towards chemokine gradients and to inflamed mucosal sites in vivo. In addition to their role in T cell migration, CD151-enriched microdomains stabilize the immune synapse at the T cell/antigen presenting cell (APC) interface (76). Consistent with its interactions with integrins, silencing of CD151 expression diminishes relocalization of α4β1 to the IS, resulting in reduced phosphorylation of integrin targets FAK and ERK1/2, and blunting of IL-2 secretion (76). We determined that not only were the frequencies of CD151+ T cells lower in women with the GG genotype, CD151 expression levels per T cell were also reduced, which would further decrease migratory and T cell activation functions augmented by CD151.
Seu et al. compared human CD151+ and CD151- T cells using an in vitro kinome array and determined that CD151 actively changes cell cycle control and cell death process motifs, leading to a hyperresponsive proliferation phenotype (13). Consistent with this report, we observed marked enrichment of CD151 in proliferating Ki67+ T cells after a 7 day culture of PBMCs in media alone or media containing inactivated Ct, regardless of genotype. However, significant differences in CD151 expression were observed across all T cell subsets according to genotype, and especially among frequencies of Ki67+CD151+ T cells, with significantly higher frequencies of double positive T cells being present in CC genotype women who had a reduced risk of endometrial infection. We also observed CD151 expression frequency was higher in memory T cells versus naïve T cells, as reported by Seu et al. (13). Our study examined PBMCs from Ct-infected women after 7 days in culture with media or inactivated Ct, and found that CD151 expression was higher in CD4 T cells compared to CD8 T cells, particularly for women with the CC genotype, where Ct stimulation led to further increases in CD151 expression (Figure 6). Perez et al. (12) and Seu et al. (13) reported CD151 expression frequencies were generally higher on CD8 T cells than CD4 T cells when freshly thawed PBMCs from healthy human donors were analyzed. We determined that frequencies of Ki67+ T cells were 30% and 36% greater for CD4 versus CD8 T cells from women with GG and CC genotypes, respectively on day 7 of culture with inactivated Ct (data not shown). The frequent coexpression of Ki67 and CD151 combined with enhanced proliferation of CD4 T cells after Ct stimulation likely explains our detection of CD151 enrichment in the CD4 T cell subset.
In summary, literature supports CD151 is important in CD4 T cell migration, immune synapse stabilization, and T cell proliferation. Our data demonstrate CD151 allelic differences determined by the lead cis eQTL impact whole blood CD151 expression, and this correlates with risk of endometrial infection. Furthermore, we detected differences in CD4 and CD8 T cell expression of CD151 that could alter proliferative capability, particularly that of memory T cells where CD151 expression is enriched. Enhanced T cell responses may reduce risk of ascending infection in a host genotype dependent manner. These results are highly consistent with our previous blood transcriptomic network analysis where we determined MHC antigen presentation and T cell signaling pathways were upregulated in women without CT induced endometritis and symptomatic pelvic inflammatory disease (3).
We previously determined candidate infertility loci by conducting GWAS in Ct-seropositive women with defined fertility status and investigated their concurrence with SNPs associated with Ct ascension in an independent Ct-infected cohort with biopsy-diagnosed endometrial infection status (11). This approach enabled functional annotation of SNPs to response pathways that alter risk for ascension. Innate immune response pathways, including type I IFN production, pathways important for T cell activation and function, and tryptophan metabolism were identified, as observed in the current study. A non-overlapping gene set unique to the infertility study included genes specifically related to female reproductive tract health, which were not detected in our current eQTL study of acute Ct infection. Unique to the current analysis was a determination of genes involved in immune cell migration. These findings suggest that using Ct ascension as an intermediate trait increases the power to identify genetic loci for responses that lead to control of Ct infection, which may be underestimated by infertility GWAS.
Another strength of this study is the availability of comprehensive clinical data, multi-omics profiles and biopsy-diagnosed endometrial Ct infection in study participants. Furthermore, we conducted eQTL analysis and adopted a statistically rigorous mediation test to link the eQTLs with Ct ascension. We identified blood eQTLs associated with ascension, which provide candidate biomarkers for targeted screening and vaccine development. However, identification of genital tract tissue eQTLs is warranted for improving understanding of disease mechanisms.
This study has potential limitations. Endometrial infection was determined by transcervical sampling which may lead to inadvertent contamination of the specimen by microorganisms in the cervix. We minimized contamination by carefully sterilizing the endocervical canal through which a sterile endometrial sampler was placed. Our prior work has demonstrated higher rates of endometrial Ct and N. gonorrhoeae among women with acute and subclinical PID (90), determined by histological endometritis, compared to women without PID, lending support to endometrial sampling as a valid tool to detect true upper genital tract infection.
Sample size is also limited due to the difficulty in obtaining endometrial biopsy samples. It has been reported that eGenes whose causal SNPs had small allele frequencies using small sample sizes (e.g. frequency <10% in 100 samples) would have inflated FDR (91). We therefore filtered SNPs with minor allele frequency (MAF) < 10% from further analysis in this study. In addition, our mediation test examined the joint effects of multiple SNPs on ascension rather than effects of individual SNPs. The SNPs are not expected to be eQTLs. eQTL screening was used to prioritize the SNPs and reduce multiple testing. Although the lead eSNP for CD151 is a very common SNP, it may not necessarily be a causal SNP. The association between this eSNP and ascension could result from a causal variant in linkage disequilibrium with this eSNP. A large independent cohort to validate the findings of this study is warranted and the new method of regulatory element-sequencing may be used to determine functional SNPs (92).
In conclusion, we have successfully identified cis-eQTLs associated with ascending infection. Our study highlights the power of a systematic genetics approach and mediation test in dissecting a complex disease and identifying potential genetic biomarkers and pathways. Recruitment of an independent cohort, replicating the recruitment strategy and sample collection of this cohort is ongoing and will enable reexamination of these ascension associated cis-eQTLs for validation and extend our ability to further investigate CD151 as a potential genetic biomarker of Ct risk.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by The Institutional Review Boards for Human Subjects Research at the University of North Carolina, and the University of Pittsburgh approved the study. The patients/participants provided their written informed consent to participate in this study.
Conceived and designed the study: XZ and TD. Analyzed the data: WZ, XZ, AK, YL, YW, and YTL. Performed the experiments: AK. Wrote the paper: XZ, TD, WZ and AK. Data collection and interpretation: CO’C, TP, KY, HW, and SH. All authors reviewed the results and the manuscript and approved the final version of the manuscript.
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health through R01 AI119164 and U19 AI084024 to TD, and U19 AI144181 to TD, CO’C, and XZ.
The authors thank the women who agreed to participate in this study; Ingrid Macio, Melinda Petrina, Carol Priest, Abi Jett, and Lorna Rabe for their efforts in the clinic and the microbiology laboratory; the staff at the Allegheny County Health Department Sexually Transmitted Disease Clinic for their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1001255/full#supplementary-material
1. Russell AN, Zheng X, O'Connell CM, Taylor BD, Wiesenfeld HC, Hillier SL, et al. Analysis of factors driving incident and ascending infection and the role of serum antibody in chlamydia trachomatis genital tract infection. J Infect Dis (2016) 213:523–31. doi: 10.1093/infdis/jiv438
2. Brunham RC, Gottlieb SL, Paavonen J. Pelvic inflammatory disease. N Engl J Med (2015) 372:2039–48. doi: 10.1056/NEJMra1411426
3. Zheng X, O'Connell CM, Zhong W, Nagarajan UM, Tripathy M, Lee D, et al. Discovery of blood transcriptional endotypes in women with pelvic inflammatory disease. J Immunol (2018) 200:2941–56. doi: 10.4049/jimmunol.1701658
4. Poston TB, Lee DE, Darville T, Zhong W, Dong L, O'Connell CM, et al. Cervical cytokines associated with chlamydia trachomatis susceptibility and protection. J Infect Dis (2019) 220:330–9. doi: 10.1093/infdis/jiz087
5. Russell AN, Zheng X, O'Connell CM, Wiesenfeld HC, Hillier SL, Taylor BD, et al. Identification of chlamydia trachomatis antigens recognized by T cells from highly exposed women who limit or resist genital tract infection. J Infect Dis (2016) 214:1884–92. doi: 10.1093/infdis/jiw485
6. Liu C, Hufnagel K, O'Connell CM, Goonetilleke N, Mokashi N, Waterboer T, et al. Reduced endometrial ascension and enhanced reinfection associated with IgG antibodies to specific chlamydia trachomatis proteins in women at risk for chlamydia. J Infect Dis (2021) 225(5):846–55. doi: 10.1093/infdis/jiab496
7. den Hartog JE, Ouburg S, Land JA, Lyons JM, Ito JI, Pena AS, et al. Do host genetic traits in the bacterial sensing system play a role in the development of chlamydia trachomatis-associated tubal pathology in subfertile women? BMC Infect Dis (2006) 6:122. doi: 10.1186/1471-2334-6-122
8. Barr EL, Ouburg S, Igietseme JU, Morre SA, Okwandu E, Eko FO, et al. Host inflammatory response and development of complications of chlamydia trachomatis genital infection in CCR5-deficient mice and subfertile women with the CCR5delta32 gene deletion. J Microbiol Immunol Infect (2005) 38:244–54.
9. Ouburg S, Spaargaren J, den Hartog JE, Land JA, Fennema JS, Pleijster J, et al. The CD14 functional gene polymorphism -260 C>T is not involved in either the susceptibility to chlamydia trachomatis infection or the development of tubal pathology. BMC Infect Dis (2005) 5:114. doi: 10.1186/1471-2334-5-114
10. Morre SA, Murillo LS, Bruggeman CA, Pena AS. The role that the functional Asp299Gly polymorphism in the toll-like receptor-4 gene plays in susceptibility to chlamydia trachomatis-associated tubal infertility. J Infect Dis (2003) 187:341–2. doi: 10.1086/346044
11. Zheng X, Zhong W, O'Connell CM, Liu Y, Haggerty CL, Geisler WM, et al. Host genetic risk factors for chlamydia trachomatis-related infertility in women. J Infect Dis (2021) 224:S64–s71. doi: 10.1093/infdis/jiab149
12. Perez MD, Seu L, Lowman KE, Moylan DC, Tidwell C, Samuel S, et al. The tetraspanin CD151 marks a unique population of activated human T cells. Sci Rep (2020) 10:15748. doi: 10.1038/s41598-020-72719-8
13. Seu L, Tidwell C, Timares L, Duverger A, Wagner FH, Goepfert PA, et al. CD151 expression is associated with a hyperproliferative T cell phenotype. J Immunol (2017) 199:3336–47. doi: 10.4049/jimmunol.1700648
14. Gao C, Jia W, Xu W, Wu Q, Wu J. Downregulation of CD151 restricts VCAM-1 mediated leukocyte infiltration to reduce neurobiological injuries after experimental stroke. J Neuroinflamm (2021) 18:118. doi: 10.1186/s12974-021-02171-6
15. Te Molder L, Juksar J, Harkes R, Wang W, Kreft M, Sonnenberg A. Tetraspanin CD151 and integrin alpha3beta1 contribute to the stabilization of integrin alpha6beta4-containing cell-matrix adhesions. J Cell Sci (2019) 132(19):jcs235366. doi: 10.1242/jcs.235366
16. Wiesenfeld HC, Meyn LA, Darville T, Macio IS, Hillier SL. A randomized controlled trial of ceftriaxone and doxycycline, with or without metronidazole, for the treatment of acute pelvic inflammatory disease. Clin Infect Dis (2021) 72:1181–9. doi: 10.1093/cid/ciaa101
17. Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature (2015) 526:68–74. doi: 10.1038/nature15393
18. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods (2011) 9:179–81. doi: 10.1038/nmeth.1785
19. Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods (2013) 10:5–6. doi: 10.1038/nmeth.2307
20. Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am J Hum Genet (2016) 98:116–26. doi: 10.1016/j.ajhg.2015.11.020
21. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (2003) 4:249–64. doi: 10.1093/biostatistics/4.2.249
22. Reese SE, Archer KJ, Therneau TM, Atkinson EJ, Vachon CM, de Andrade M, et al. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics (2013) 29:2877–83. doi: 10.1093/bioinformatics/btt480
23. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics (2007) 8:118–27. doi: 10.1093/biostatistics/kxj037
24. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, et al. Rising to the challenge of larger and richer datasets. Gigascience (2015) 4:7. doi: 10.1186/s13742-015-0047-8
25. Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc (2012) 7:500–7. doi: 10.1038/nprot.2011.457
26. Lloyd-Jones LR, Holloway A, McRae A, Yang J, Small K, Zhao J, et al. The genetic architecture of gene expression in peripheral blood. Am J Hum Genet (2017) 100:228–37. doi: 10.1016/j.ajhg.2016.12.008
27. Peters JE, Lyons PA, Lee JC, Richard AC, Fortune MD, Newcombe PJ, et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet (2016) 12:e1005908. doi: 10.1371/journal.pgen.1005908
28. Yao C, Chen BH, Joehanes R, Otlu B, Zhang X, Liu C, et al. Integromic analysis of genetic variation and gene expression identifies networks for cardiovascular disease phenotypes. Circulation (2015) 131:536–49. doi: 10.1161/CIRCULATIONAHA.114.010696
29. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet (2011) 88:76–82. doi: 10.1016/j.ajhg.2010.11.011
30. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics (2010) 26:2336–7. doi: 10.1093/bioinformatics/btq419
31. Zhong W, Darville T, Zheng X, Fine J, Li Y. Generalized multi-SNP mediation intersection–union test. Biometrics n/a (2020) 78(1):364–75. doi: 10.1101/780767
32. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res (2009) 19:1639–45. doi: 10.1101/gr.092759.109
33. Li Y, Levran O, Kim J, Zhang T, Chen X, Suo C. Extreme sampling design in genetic association mapping of quantitative trait loci using balanced and unbalanced case-control samples. Sci Rep (2019) 9:15504. doi: 10.1038/s41598-019-51790-w
34. Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet Epidemiol (2011) 35:790–9. doi: 10.1002/gepi.20628
35. Bjornland T, Bye A, Ryeng E, Wisloff U, Langaas M. Powerful extreme phenotype sampling designs and score tests for genetic association studies. Stat Med (2018) 37:4234–51. doi: 10.1002/sim.7914
36. Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog (2008) 4:e1000014. doi: 10.1371/journal.ppat.1000014
37. Paradis T, Begue H, Basmaciyan L, Dalle F, Bon F. Tight junctions as a key for pathogens invasion in intestinal epithelial cells. Int J Mol Sci 22 (2021) 22(5):2506–26. doi: 10.3390/ijms22052506
38. Williams L, Layton T, Yang N, Feldmann M, Nanchahal J. Collagen VI as a driver and disease biomarker in human fibrosis. FEBS J (2021) 289(13):3603–29. doi: 10.1111/febs.16039
39. Cai Y, Maeda Y, Cedzich A, Torres VE, Wu G, Hayashi T, et al. Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem (1999) 274:28557–65. doi: 10.1074/jbc.274.40.28557
40. Kessler M, Hoffmann K, Fritsche K, Brinkmann V, Mollenkopf HJ, Thieck O, et al. Chronic chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat Commun (2019) 10:1194. doi: 10.1038/s41467-019-09144-7
41. Kumar D, Patel SA, Hassan MK, Mohapatra N, Pattanaik N, Dixit M. Reduced IQGAP2 expression promotes EMT and inhibits apoptosis by modulating the MEK-ERK and p38 signaling in breast cancer irrespective of ER status. Cell Death Dis (2021) 12:389. doi: 10.1038/s41419-021-03673-0
42. Yamashiro S, Abe H, Mabuchi I. IQGAP2 is required for the cadherin-mediated cell-to-cell adhesion in xenopus laevis embryos. Dev Biol (2007) 308:485–93. doi: 10.1016/j.ydbio.2007.06.001
43. Agarwal AK, Barnes RI, Garg A. Functional characterization of human 1-acylglycerol-3-phosphate acyltransferase isoform 8: cloning, tissue distribution, gene structure, and enzymatic activity. Arch Biochem Biophys (2006) 449:64–76. doi: 10.1016/j.abb.2006.03.014
44. Engeland K, Kindl H. Evidence for a peroxisomal fatty acid beta-oxidation involving d-3-hydroxyacyl-CoAs. Characterization of two forms of hydro-lyase that convert d-(-)-3-hydroxyacyl-CoA into 2-trans-enoyl-CoA. Eur J Biochem (1991) 200:171–8. doi: 10.1111/j.1432-1033.1991.tb21064.x
45. Yao J, Abdelrahman YM, Robertson RM, Cox JV, Belland RJ, White SW, et al. Type II fatty acid synthesis is essential for the replication of chlamydia trachomatis. J Biol Chem (2014) 289:22365–76. doi: 10.1074/jbc.M114.584185
46. Yao J, Dodson VJ, Frank MW, Rock CO. Chlamydia trachomatis scavenges host fatty acids for phospholipid synthesis via an acyl-acyl carrier protein synthetase. J Biol Chem (2015) 290:22163–73. doi: 10.1074/jbc.M115.671008
47. Yao J, Cherian PT, Frank MW, Rock CO. Chlamydia trachomatis relies on autonomous phospholipid synthesis for membrane biogenesis. J Biol Chem (2015) 290:18874–88. doi: 10.1074/jbc.M115.657148
48. Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the chlamydia trachomatis inclusion. Proc Natl Acad Sci U S A (2003) 100:6771–6. doi: 10.1073/pnas.1131289100
49. Cox JV, Naher N, Abdelrahman YM, Belland RJ. Host HDL biogenesis machinery is recruited to the inclusion of chlamydia trachomatis-infected cells and regulates chlamydial growth. Cell Microbiol (2012) 14:1497–512. doi: 10.1111/j.1462-5822.2012.01823.x
50. Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, et al. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog (2011) 7:e1002198. doi: 10.1371/journal.ppat.1002198
51. Raulston JE. Response of chlamydia trachomatis serovar e to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infection Immun (1997) 65:4539–47. doi: 10.1128/iai.65.11.4539-4547.1997
52. Pokorzynski ND, Brinkworth AJ, Carabeo R. A bipartite iron-dependent transcriptional regulation of the tryptophan salvage pathway in chlamydia trachomatis. Elife 8 (2019) 8:e42295. doi: 10.7554/eLife.42295
53. Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infection Immun (1994) 62:3705–11. doi: 10.1128/iai.62.9.3705-3711.1994
54. Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, et al. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc Natl Acad Sci U S A (2005) 102:10658–63. doi: 10.1073/pnas.0504198102
55. Wood H, Fehlner-Gardner C, Berry J, Fischer E, Graham B, Hackstadt T, et al. Regulation of tryptophan synthase gene expression in chlamydia trachomatis. Mol Microbiol (2003) 49:1347–59. doi: 10.1046/j.1365-2958.2003.03638.x
56. Badawy AA. Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int J Tryptophan Res (2017) 10:1178646917691938. doi: 10.1177/1178646917691938
57. Fisher DJ, Fernandez RE, Maurelli AT. Chlamydia trachomatis transports NAD via the Npt1 ATP/ADP translocase. J Bacteriol (2013) 195:3381–6. doi: 10.1128/JB.00433-13
58. Frumento G, Rotondo R, Tonetti M, Damonte G, Benatti U, Ferrara GB. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J Exp Med (2002) 196:459–68. doi: 10.1084/jem.20020121
59. Irwin DM, Tan H. Molecular evolution of the vertebrate hexokinase gene family: Identification of a conserved fifth vertebrate hexokinase gene. Comp Biochem Physiol Part D Genomics Proteomics (2008) 3:96–107. doi: 10.1016/j.cbd.2007.11.002
60. Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. Developmental stage-specific metabolic and transcriptional activity of chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A (2012) 109:19781–5. doi: 10.1073/pnas.1212831109
61. Grieshaber S, Grieshaber N, Yang H, Baxter B, Hackstadt T, Omsland A. The impact of active metabolism on chlamydia trachomatis elementary body transcript profile and infectivity. J Bacteriol (2018) 200(14):e00065–18. doi: 10.1128/JB.00065-18
62. Darville T, Hiltke TJ. Pathogenesis of genital tract disease due to chlamydia trachomatis. J Infect Dis (2010) 201 Suppl 2:S114–25. doi: 10.1086/652397
63. Darville T. Pelvic inflammatory disease due to neisseria gonorrhoeae and chlamydia trachomatis: Immune evasion mechanisms and pathogenic disease pathways. J Infect Dis (2021) 224:S39–46. doi: 10.1093/infdis/jiab031
64. Dziarski R, Wang Q, Miyake K, Kirschning CJ, Gupta D. MD-2 enables toll-like receptor 2 (TLR2)-mediated responses to lipopolysaccharide and enhances TLR2-mediated responses to gram-positive and gram-negative bacteria and their cell wall components. J Immunol (2001) 166:1938–44. doi: 10.4049/jimmunol.166.3.1938
65. Visintin A, Mazzoni A, Spitzer JA, Segal DM. Secreted MD-2 is a large polymeric protein that efficiently confers lipopolysaccharide sensitivity to toll-like receptor 4. Proc Natl Acad Sci U S A (2001) 98:12156–61. doi: 10.1073/pnas.211445098
66. Bulut Y, Faure E, Thomas L, Karahashi H, Michelsen KS, Equils O, et al. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J Immunol (2002) 168:1435–40. doi: 10.4049/jimmunol.168.3.1435
67. Cui Y, Yu H, Zheng X, Peng R, Wang Q, Zhou Y, et al. SENP7 potentiates cGAS activation by relieving SUMO-mediated inhibition of cytosolic DNA sensing. PLoS Pathog (2017) 13:e1006156. doi: 10.1371/journal.ppat.1006156
68. Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, et al. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during chlamydia trachomatis infection. J Immunol (2014) 193:2394–404. doi: 10.4049/jimmunol.1302718
69. Nagarajan UM, Prantner D, Sikes JD, Andrews CW Jr., Goodwin AM, Nagarajan S, et al. Type I interferon signaling exacerbates chlamydia muridarum genital infection in a murine model. Infect Immun (2008) 76:4642–8. doi: 10.1128/IAI.00629-08
70. Verploegen S, Ulfman L, van Deutekom HW, van Aalst C, Honing H, Lammers JW, et al. Characterization of the role of CaMKI-like kinase (CKLiK) in human granulocyte function. Blood (2005) 106:1076–83. doi: 10.1182/blood-2004-09-3755
71. Frazer LC, O'Connell CM, Andrews CW Jr., Zurenski MA, Darville T. Enhanced neutrophil longevity and recruitment contribute to the severity of oviduct pathology during chlamydia muridarum infection. Infect Immun (2011) 79:4029–41. doi: 10.1128/IAI.05535-11
72. Rank RG, Whittum-Hudson JA. Protective immunity to chlamydial genital infection: Evidence from animal studies. J Infect Dis (2010) 201 Suppl 2:S168–77. doi: 10.1086/652399
73. Igietseme JU, Ramsey KH, Magee DM, Williams DM, Kincy TJ, Rank RG. Resolution of murine chlamydial genital infection by the adoptive transfer of a biovar-specific TH1 lymphocyte clone. Regional Immunol (1993) 5:317–24.
74. Wadkin JCR, Patten DA, Kamarajah SK, Shepherd EL, Novitskaya V, Berditchevski F, et al. CD151 supports VCAM-1-mediated lymphocyte adhesion to liver endothelium and is upregulated in chronic liver disease and hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol (2017) 313:G138–49. doi: 10.1152/ajpgi.00411.2016
75. Zelman-Toister E, Bakos E, Cohen S, Zigmond E, Shezen E, Grabovsky V, et al. CD151 regulates T-cell migration in health and inflammatory bowel disease. Inflammation Bowel Dis (2016) 22:257–67. doi: 10.1097/MIB.0000000000000621
76. Rocha-Perugini V, Gonzalez-Granado JM, Tejera E, Lopez-Martin S, Yanez-Mo M, Sanchez-Madrid F. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur J Immunol (2014) 44:1967–75. doi: 10.1002/eji.201344235
77. Agarwal S, Kraus Z, Dement-Brown J, Alabi O, Starost K, Tolnay M. Human fc receptor-like 3 inhibits regulatory T cell function and binds secretory IgA. Cell Rep (2020) 30:1292–9.e3. doi: 10.1016/j.celrep.2019.12.099
78. Stern LJ, Calvo-Calle JM. HLA-DR: molecular insights and vaccine design. Curr Pharm design (2009) 15:3249–61. doi: 10.2174/138161209789105171
79. Hermann E, Mayet WJ, Thomssen H, Sieper J, Poralla T, Meyer zum Buschenfelde KH, et al. HLA-DP restricted chlamydia trachomatis specific synovial fluid T cell clones in chlamydia induced reiter's disease. J Rheumatol (1992) 19:1243–6.
80. Jiang S, Li C, McRae G, Lykken E, Sevilla J, Liu SQ, et al. MeCP2 reinforces STAT3 signaling and the generation of effector CD4+ T cells by promoting miR-124-mediated suppression of SOCS5. Sci Signal 7 (2014) 7(316):ra25. doi: 10.1126/scisignal.2004824
81. Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK. PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci (1999) 112(Pt 6):833–44. doi: 10.1242/jcs.112.6.833
82. Palmer CS, Ostrowski M, Balderson B, Christian N, Crowe SM. Glucose metabolism regulates T cell activation, differentiation, and functions. Front Immunol (2015) 6:1. doi: 10.3389/fimmu.2015.00001
83. Darville T, O'Neill JM, Andrews CW Jr., Nagarajan UM, Stahl L, Ojcius DM. Toll-like receptor-2, but not toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol (2003) 171:6187–97. doi: 10.4049/jimmunol.171.11.6187
84. Taylor BD, Darville T, Ferrell RE, Ness RB, Haggerty CL. Racial variation in toll-like receptor variants among women with pelvic inflammatory disease. J Infect Dis (2013) 207:940–6. doi: 10.1093/infdis/jis922
85. Taylor BD, Darville T, Ferrell RE, Kammerer CM, Ness RB, Haggerty CL. Variants in toll-like receptor 1 and 4 genes are associated with chlamydia trachomatis among women with pelvic inflammatory disease. J Infect Dis (2012) 205:603–9. doi: 10.1093/infdis/jir822
86. Lee HY, Schripsema JH, Sigar IM, Murray CM, Lacy SR, Ramsey KH. A link between neutrophils and chronic disease manifestations of chlamydia muridarum urogenital infection of mice. FEMS Immunol Med Microbiol (2010) 59:108–16. doi: 10.1111/j.1574-695X.2010.00668.x
87. Wiesenfeld HC, Heine RP, Krohn MA, Hillier SL, Amortegui AA, Nicolazzo M, et al. Association between elevated neutrophil defensin levels and endometritis. J Infect Dis (2002) 186:792–7. doi: 10.1086/342417
88. Pedraza L, Camargo M, Moreno-Perez DA, Sanchez R, Del Rio-Ospina L, Baez-Murcia IM, et al. Identifying HLA DRB1-DQB1 alleles associated with chlamydia trachomatis infection and in silico prediction of potentially-related peptides. Sci Rep (2021) 11:12837. doi: 10.1038/s41598-021-92294-w
89. Chattopadhyay N, Wang Z, Ashman LK, Brady-Kalnay SM, Kreidberg JA. alpha3beta1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J Cell Biol (2003) 163:1351–62. doi: 10.1083/jcb.200306067
90. Wiesenfeld HC, Sweet RL, Ness RB, Krohn MA, Amortegui AJ, Hillier SL. Comparison of acute and subclinical pelvic inflammatory disease. Sex Transm Dis (2005) 32:400–5. doi: 10.1097/01.olq.0000154508.26532.6a
91. Huang QQ, Ritchie SC, Brozynska M, Inouye M. Power, false discovery rate and winner's curse in eQTL studies. Nucleic Acids Res (2018) 46:e133. doi: 10.1093/nar/gky780
Keywords: Chlamydial trachomatis, upper genital tract infection in women, genetic marker for host susceptibility, cis-eQTLs, blood T cells and T cell subsets, CD151, integrated analysis of DNA genotypes and human blood transcriptome
Citation: Zhong W, Kollipara A, Liu Y, Wang Y, O’Connell CM, Poston TB, Yount K, Wiesenfeld HC, Hillier SL, Li Y, Darville T and Zheng X (2022) Genetic susceptibility loci for Chlamydia trachomatis endometrial infection influence expression of genes involved in T cell function, tryptophan metabolism and epithelial integrity. Front. Immunol. 13:1001255. doi: 10.3389/fimmu.2022.1001255
Received: 23 July 2022; Accepted: 09 September 2022;
Published: 29 September 2022.
Edited by:
Amy Rasley, Lawrence Livermore National Laboratory (DOE), United StatesReviewed by:
Wilhelmina May Huston, University of Technology Sydney, AustraliaCopyright © 2022 Zhong, Kollipara, Liu, Wang, O’Connell, Poston, Yount, Wiesenfeld, Hillier, Li, Darville and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojing Zheng, eGlhb2ppbnpAZW1haWwudW5jLmVkdQ==; Toni Darville, dG9uaS5kYXJ2aWxsZUB1bmMuZWR1
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.