- 1Center of Excellence for Molecular Biology and Genomics of Shrimp, Department of Biochemistry, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 2Marine Biotechnology Research Team, Integrative Aquaculture Biotechnology Research Group, National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), Pathumthani, Thailand
- 3Center of Excellence for Marine Biotechnology, Department of Marine Science, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 4Laboratory of Molecular Immunobiology, Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology (NAIST), Ikoma, Japan
Interferon regulatory factors (IRFs) are transcription factors found in both vertebrates and invertebrates that were recently identified and found to play an important role in antiviral immunity in black tiger shrimp Penaeus monodon. In this study, we investigated the mechanism by which P. monodon IRF (PmIRF) regulates the immune-related genes downstream of the cytosolic DNA sensing pathway. Depletion of PmIRF by double-stranded RNA-mediated gene silencing significantly reduced the mRNA expression levels of the IFN-like factors PmVago1, PmVago4, and PmVago5 and antilipopolysaccharide factor 6 (ALFPm6) in shrimp. In human embryonic kidney (HEK293T) cells transfected with PmIRF or co-transfected with DEAD-box polypeptide (PmDDX41) and simulator of IFN genes (PmSTING) expression plasmids, the promoter activity of IFN-β, nuclear factor (NF-κB), and ALFPm6 was synergistically enhanced following stimulation with the nucleic acid mimics deoxyadenylic–deoxythymidylic acid sodium salt [poly(dA:dT)] and high molecular weight (HMW) polyinosinic–polycytidylic acid [poly(I:C)]. Both nucleic acid mimics also significantly induced PmSTING, PmIRF, and ALFPm6 gene expression. Co-immunoprecipitation experiments showed that PmIRF interacted with PmSTING in cells stimulated with poly(dA:dT). PmSTING, PmIRF, and PmDDX41 were localized in the cytoplasm of unstimulated HEK293T cells and PmIRF and PmDDX41 were translocated to the nucleus upon stimulation with the nucleic acid mimics while PmSTING remained in the cytoplasm. These results indicate that PmIRF transduces the pathogen signal via the PmDDX41–PmSTING DNA sensing pathway to induce downstream production of interferon-like molecules and antimicrobial peptides.
Introduction
The innate immune system is the first line of host defense against invasive pathogens (1). Host pattern recognition receptors (PRRs) play a key role in recognizing nonself pathogen-associated molecular patterns (PAMPs). A number of PRRs have been described, including Toll-like receptor (TLR), nucleotide-binding oligomerization domain-like receptors (NLRs) and retinoic acid-inducible gene (RIG)-I–like receptors (RLRs) (2–4). Intracellular DNA sensors including DEAD-box polypeptide (DDX)41 and cyclic GMP–AMP synthase (cGAS) recognize cytoplasmic or nuclear pathogen-derived DNA (5–9).
DDX41 has been shown to directly bind to double-stranded (ds) DNA and stimulator of IFN genes (STING) protein via the DEAD domain and induce the activation of nuclear factor kappa B (NF-κB) and IFN production in mouse dendritic cells (10). In vertebrates, STING is an adaptor protein of the cytosolic DNA-sensing pathway that mediates the immune response to pathogens. Upon binding to foreign DNA or cyclic (c)GMP–AMP (a second messenger), DNA sensors activate STING (11, 12), leading to activation of TANK-binding kinase (TBK)1, which then phosphorylates STING and the downstream transcription factor IRF3 to induce the expression of type I IFNs (13) in the immune response to DNA viruses and tumors (14–16).
In vertebrates, cGAS activates STING which initiates a downstream signaling pathway leading to induce the expression of IFNs and other cytokines such as TNF-α and IL-6, and thereby trigger the host immune response. After viral and bacterial infection, dsDNA was released. Cytosolic DNA binds and activates cGAS, which catalyzes the synthesis of 2′3′-cGAMP from ATP and GTP. 2′3′-cGAMP binds to the ER adaptor STING, which traffics to the ER and the Golgi apparatus. STING then activates IKK and TBK1. TBK1 phosphorylates STING, which in turn recruits IRF3 for phosphorylation by TBK1. IRF3 then enters the nucleus, where it functions with NF-kB to synthesize the expression of IFNs (6, 17).
In Drosophila, the inhibitor of kappa B kinase (dIKKβ) and Relish genes were found to be induced by viral infection. The Drosophila STING ortholog (dSTING) participates in host defense against infection by picorna-like viruses, acting upstream of dIKKβ to regulate the expression of the antiviral factor Nazo (18). In Pacific white shrimp (Litopenaeus vannamei), LvSTING was shown to contribute to the innate immune response to infection by Vibrio parahaemolyticus (19). Similarly, PmSTING in black tiger shrimp Penaeus monodon which shared high sequence similarity to LvSTING (92%), was important for the antiviral innate response against white spot syndrome virus (WSSV) infection (20).
IRFs are a family of transcription factors involved in the antiviral response (21–23). To date, eleven IRFs (IRF-1 to IRF-11) have been identified in fish, all containing a highly conserved DNA-binding domain in the N-terminal region that recognizes a consensus sequence similar to the IFN-stimulated response element (ISRE) (24). IRF3 and IRF7 are activated by TLR3 and TLR4 signaling pathways, respectively, leading to IFN expression (25). IRF has been identified in L. vannamei (LvIRF) (26) and more recently, in P. monodon (PmIRF) (20). Like their vertebrate counterparts, LvIRF and PmIRF as well as PmDDX41 and PmSTING are activated during virus infection (26,27). It was reported that LvIRF mediated the activation of the ISRE-containing promoters in mammalian cells to regulate the expression of LvVago4 and LvVago5 genes, which encode a virus-activated secreted peptide that blocks virus infection via activation of the Janus kinase (JAK)–STAT signaling pathway (26).
Recently, we identified PmDDX41, PmSTING and PmIRF in shrimp P. monodon. PmDDX41 plays an important role as a cytosolic DNA sensor which interacted with STING and triggered the IFNs and NF-κB signaling pathway to activate the innate immune response (27). While, PmSTING and PmIRF play a key role in protecting shrimp from WSSV infection (20). Moreover, silencing of PmDDX41 caused a decrease expression of PmSTING and PmIRF (20). As the regulation of the cytosolic DNA-sensing pathways in shrimp is not fully understood, in this study, we further characterized the function of PmIRF in antiviral innate immunity by identifying its downstream immune-related target genes. Nucleic acid mimics were used to stimulate the cells and investigate the innate immune response. Poly(dA:dT) and poly(I:C) are the synthetic analog of B form DNA and synthetic dsRNA polymer, respectively, thus representative of a DNA virus and RNA virus. It was found that PmIRF, PmDDX41, and PmSTING synergistically activated the IFN-β, NF-κB, and antilipopolysaccharide factor (ALFPm6) gene promoters following stimulation with DNA mimics and that PmIRF interacts with PmSTING in the cytoplasm and translocates to the nucleus to stimulate the expression and production of IFN-like molecules as part of the antiviral immune response in P. monodon.
Materials and Methods
Shrimp and Sample Preparation
Healthy black tiger shrimps (P. monodon; 3-5 g body weight) were provided by Charoen Pokphand Foods in Chanthaburi province, Thailand, and maintained in aerated seawater (20 ppt) at 28°C for 1 week prior to experiments. Shrimp samples were screened for pathogen-free including WSSV, YHV, EHP and Vibrio parahaemolyticus AHPND by PCR before used in the experiment. To determine the expression level of the PmIRF transcript, intestine from triplicate groups of 3 shrimps each were separately collected as previously described (28). All samples were stored at -80°C until RNA extraction. This study was conducted under the ethical principles and guidelines according to the animal use protocol approved by Chulalongkorn University Animal Care and Use Committee (CU-ACUC).
Total RNA Extraction and Reverse Transcription
Shrimp intestine was homogenized in GENEzol (Geneaid, New Taipei City, Taiwan) and total RNA was isolated according to the manufacturer’s protocol and treated with DNaseI (New England Biolabs, Ipswich, MA, USA) to destroy contaminating DNA. First-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) and stored at -20°C until used for qRT-PCR.
Double-Stranded RNA Preparation
To prepare dsRNA specifically targeting PmIRF, DNA fragments of the PmIRF (614 bp) gene were amplified by PCR using specific primers (PmIRFi-F1 and PmIRFi-R1) designed using the Primer Premier 5 program (PREMIER Biosoft, Palo Alto, CA, USA) (Table 1). In vitro transcription with T7 RNA polymerase was performed to obtain sense and antisense RNA strands. Sense and antisense DNA templates containing the T7 promoter RNA polymerase sequence at the 5’ end of each strand were generated by PCR using oligonucleotide primers containing the sequence at the 5’ end (PmIRFi-T7F1 and PmIRFi-T7R1) (Table 1). For the negative control dsRNA, the GFP gene was amplified from the pEGFP-1 plasmid (Clontech, Mountain View, CA, USA) (29). The T7 RiboMAX Express Large Scale RNA Production System (Promega, Madison, WI, USA) was used to synthesize RNA by in vitro transcription according to the manufacturer’s protocol. The quality of the dsRNA was verified by agarose gel electrophoresis and quantification was performed by spectrophotometry.
Gene Knockdown by RNA Interference (RNAi)
PmIRF or GFP (control) dsRNA was injected into juvenile shrimp (3–5 g, fresh weight) using a 0.5-ml insulin syringe with a 29-gauge needle as previously described (29). Shrimp were injected with 25 ul of PmIRF dsRNA (5 μg/g) diluted in 150mM NaCl. and delivered by intramuscular injection into the third abdominal segment of each shrimp. NaCl (150 mM) was injected as a control for handling- and injection-induced mortality. After 24 h, shrimp intestine was collected for total RNA extraction and first-strand cDNA was synthesized from 200 ng of total RNA as described above.
The efficiency of PmIRF knockdown was analyzed by qRT-PCR using specific primers for PmIRF (Table 1). A fragment of the elongation factor (EF)1-α gene was amplified in a separate tube and served as an internal control for normalization of expression levels. The PCR reactions and thermal cycling conditions were as previously reported (Soponpoong et al., 2008). In brief, the PCR reaction was performed in 10-µl reaction volume, containing 0.5 µl of intestine cDNA, 0.2 µl of specific primer (10µM each), 5 µl of Luna® Universal qPCR Master Mix (New England Biolabs, Ipswich, MA, USA) and 4.3 µl of nuclease-free water. The thermal cycling was performed in triplicate at 94°C for 1 min, followed by 40 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 30 s.
Effect of PmIRF Gene Silencing on Immune-Related Gene Expression
The effect of dsRNA-mediated PmIRF gene silencing on the transcript levels of other immune-related genes was examined by qRT-PCR using primers specific to P. monodon antimicrobial peptides (PmPEN3, PmPEN5, ALFPm3, ALFPm6, CrustinPm1, and CrustinPm7) and IFN-like molecules (PmVago1, PmVago2, PmVago3, PmVago4, and PmVago5) (Table 1). EF1-α served as the internal control for normalization.
Gene Expression Profiles in Response to Stimulation With Nucleic Acid Mimic
Changes in PmSTING, PmIRF, and ALFPm6 transcript levels in P. monodon intestine following injection with the nucleic acid mimics poly(dA:dT) and HMW poly(I:C) were evaluated by qRT-PCR. Shrimp (3–5 g) were divided into triplicate groups of 3 shrimps each and 50 µl poly(dA:dT) (2 µg/g) or 50 µl HMW poly(I:C) (2 µg/g) diluted in phosphate-buffered saline [PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4 (pH 7.4)] was injected into the second abdominal segment (50 µl per shrimp). The control group was injected with PBS. The shrimps were reared in seawater tanks and the intestine was randomly collected at 0, 3, 6, 24, and 48 h post injection. Total RNA was extracted and first-strand cDNA was synthesized as described above. The RNA from 3 shrimps per treatment at each time point was pooled. qRT-PCR was performed as previously described (30) using target gene-specific primers (Table 1). EF1-α was amplified as the internal control and reference standard. Three replicates were prepared for each template with 3 independent replicates for each data point. The Ct value at each time point was normalized to PBS-injected samples. A previously established mathematical model (31) was used to determine the relative expression ratio.
Cells and Reagents
HEK293T cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) (Life Technologies, Carlsbad, CA, USA) with 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies) in an incubator at 5% CO2 and 37°C. HMW poly(I:C) and poly(dA:dT) (In vivoGen, San Diego, CA, USA) were separately mixed with Lipofectamine 3000 (Life Technologies) at a 1:1 ratio (µg/µl) in Opti-MEM (Life Technologies) for cell stimulation. Anti-Flag and anti-Myc antibodies were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Plasmid Construction
Full-length PmSTING and PmIRF cDNA sequences were cloned into pFlag-CMV5 (Sigma-Aldrich, St. Louis, MO, USA) and pcDNA3-Myc (Santa Cruz Biotechnology, Santa Cruz, CA, USA) expression plasmids using specific primers (Table 1). The 25-µl amplification reaction mixture contained 1× KOD FX PCR buffer, 4 mM dNTP, 0.3 µM each primer, 1 µl normal shrimp cDNA, and 1 U KOD FX DNA polymerase (Toyobo, Osaka, Japan). The PCR thermal cycling conditions were 94°C for 2 min; 35 cycles of 98°C for 10 s, 60°C for 30 s, and 68°C for 1 min 30 s; and 68°C for 7 min. PCR products were separated by agarose gel electrophoresis and bands of the expected size were excised and purified using a FavorPrep GEL/PCR Purification Kit (Favorgen Biotech, Ping-Tung, Taiwan). The purified fragments were cloned into pFlag-CMV5 and pcDNA3-Myc expression plasmids. To construct the pGL3 promoter plasmid (pGL3) harboring ALFPm6 and ALFPm3 promoter sequences, the sequences were amplified by PCR from gill genomic DNA and inserted into the pGL3 plasmid digested with BglII and NotI restriction enzymes. The reporter plasmids for IFN-β and NF-κB were constructed as previously described (32, 33).
Luciferase Reporter Assay
HEK293T cells (1×105 cell/ml) were cultured in DMEM and seeded in a 24-well plate, then transfected with 100 ng IFN-β or NF-κB reporter plasmid and 500 ng of expression plasmid or empty plasmid using Lipofectamine 3000 in Opti-MEM at a 1:1 ratio (µg/µl). As an internal control, 10 ng of pRL-TK (Promega, Madison, WI, USA) was transfected. After 24 h, cells were stimulated with 1 µg/ml of poly(dA:dT) or HMW poly(I:C) and 6 h later, luciferase activity was detected using the Dual-Glo Luciferase System (Promega, Madison, WI, USA) according to the manufacturer’s instructions, with absorbance measured using a TriStar2 LB 942 Multidetection Microplate Reader (Berthold Technologies, Bad Wildbad, Germany).
Protein Expression and Co-Immunoprecipitation
HEK293T cells (1×106 cell/ml) were seeded in 10-cm cell culture dishes and transfected with 4 µg of Flag- and Myc-tagged expression constructs using Lipofectamine 3000. After 24 h, the cells were stimulated with 1 µg/ml poly(dA:dT) and HMW poly(I:C) for 6 h and then lysed with homo buffer [150 mM NaCl, 5 mM EDTA (pH 8.0), 25 mM Tris-HCl (pH 8.0), and 0.2% Triton X-100] containing protease inhibitor cocktail (Roche, Basel, Switzerland). After sonication, cell lysates were immunoprecipitated overnight at 4°C with mouse anti-Myc antibody diluted 1:500 and then treated for 4 h at 4°C with protein A sepharose beads (GE Healthcare, Little Chalfont, UK). The beads with immunoprecipitates were washed 3 times with PBS buffer. Whole-cell lysates and immunoprecipitates were probed with the appropriate antibodies.
Western Blotting
HEK293T cells cultured in 6-well plates were lysed in homo buffer [150 mM NaCl, 5 mM EDTA (pH 8.0), 25 mM Tris-HCl (pH 8.0), and 0.2% Triton X-100] containing protease inhibitor cocktail (Roche). Following centrifugation, the supernatant was mixed with sodium dodecyl sulfate (SDS) sample buffer and proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to an Immun-Blot polyvinylidene difluoride membrane that was probed with anti-Flag and -Myc antibodies. Protein bands were visualized with horseradish peroxidase (HRP)-conjugated antibodies against mouse, rabbit, or goat IgG (Sigma-Aldrich, St. Louis, MO, USA) using Western Lighting Plus-ECL reagent (Perkin Elmer, Waltham, MA, USA). HRP activity was detected with an LAS 4000 imaging system (Fujitsu Life Sciences, Tokyo, Japan).
Immunofluorescence Analysis and Confocal Microscopy
Cells were cultured on poly-L-lysine–coated coverslips in 24-well plates for 6 h, then transfected with 500 ng of expression plasmid for 16 h and stimulated with 1 µg/ml of poly(dA:dT) and HMW poly(I:C) for 6 h before fixation with 4% paraformaldehyde for 30 min. The cells were washed 3 times with 0.02% Triton X-100 in PBS, permeabilized with PBS containing 100 mM glycine and 0.2% Triton X-100 for 30 min, blocked overnight at 4°C in PBS containing 10% FBS and 0.02% Triton X-100, and probed overnight at 4°C with anti-Flag and/or -Myc antibody diluted 1:100. The coverslips were then washed and incubated for 1 h at room temperature with Alexa Fluor 488- or Alexa Fluor 568-conjugated anti-mouse and/or -rabbit secondary antibody (both from Invitrogen). Nuclei were stained with Hoechst 33342 (Invitrogen). Stained cells were mounted with Fluoro-KEEPER Antifade Reagent (Nacalai Tesque, Kyoto, Japan), and images were acquired with an LSM 700 laser scanning confocal microscope (Carl Zeiss, Wetzlar, Germany).
Statistical Analysis
The experiments were performed in three independent experiments with three technical replicates per experiment. Relative gene expression data were obtained according to the method described by Pfaffl and comparisons between groups were analyzed by one-way analysis of variance followed by Duncan’s multiple comparison tests.
Results
PmIRF Gene Silencing Inhibits the Expression of Shrimp Antimicrobial Peptides and IFN-Like Molecules
IRFs regulate gene expression in both innate and adaptive immunity (34). In order to identify genes that are regulated by PmIRF, we suppressed PmIRF expression by RNA interference (RNAi) and examined the changes in expression of immune-related genes by quantitative real-time (qRT)-PCR. P. monodon (3–5 g) was injected with PmIRF dsRNA (5 μg/g shrimp), control green fluorescent protein (GFP) dsRNA, or 150 mM NaCl. Intestine from triplicate groups of shrimp (n = 3 for each group) were collected and extracted the total RNA. PmIRF transcript was depleted by dsRNA-mediated knockdown, whereas injection of GFP dsRNA or NaCl had no effect on PmIRF expression (Figure 1). We also analyzed the expression of genes encoding shrimp antimicrobial peptides (PmPEN3, PmPEN5, ALFPm3, ALFPm6, CrustinPm1, and CrustinPm7) and IFN-like molecules (PmVago1, PmVago2, PmVago3, PmVago4, and PmVago5) after PmIRF silencing and found that ALFPm6, PmVago1, PmVago4, and PmVago5 were significantly downregulated (p<0.05) compared to the control whereas the expression of other genes (PmPEN3, PmPEN5, ALFPm3, CrustinPm1, CrustinPm7, PmVago2, and PmVago3) was unaffected (Figure 1). The results suggest that ALFPm6, PmVago1, PmVago4, and PmVago5 are possibly regulated by PmIRF.
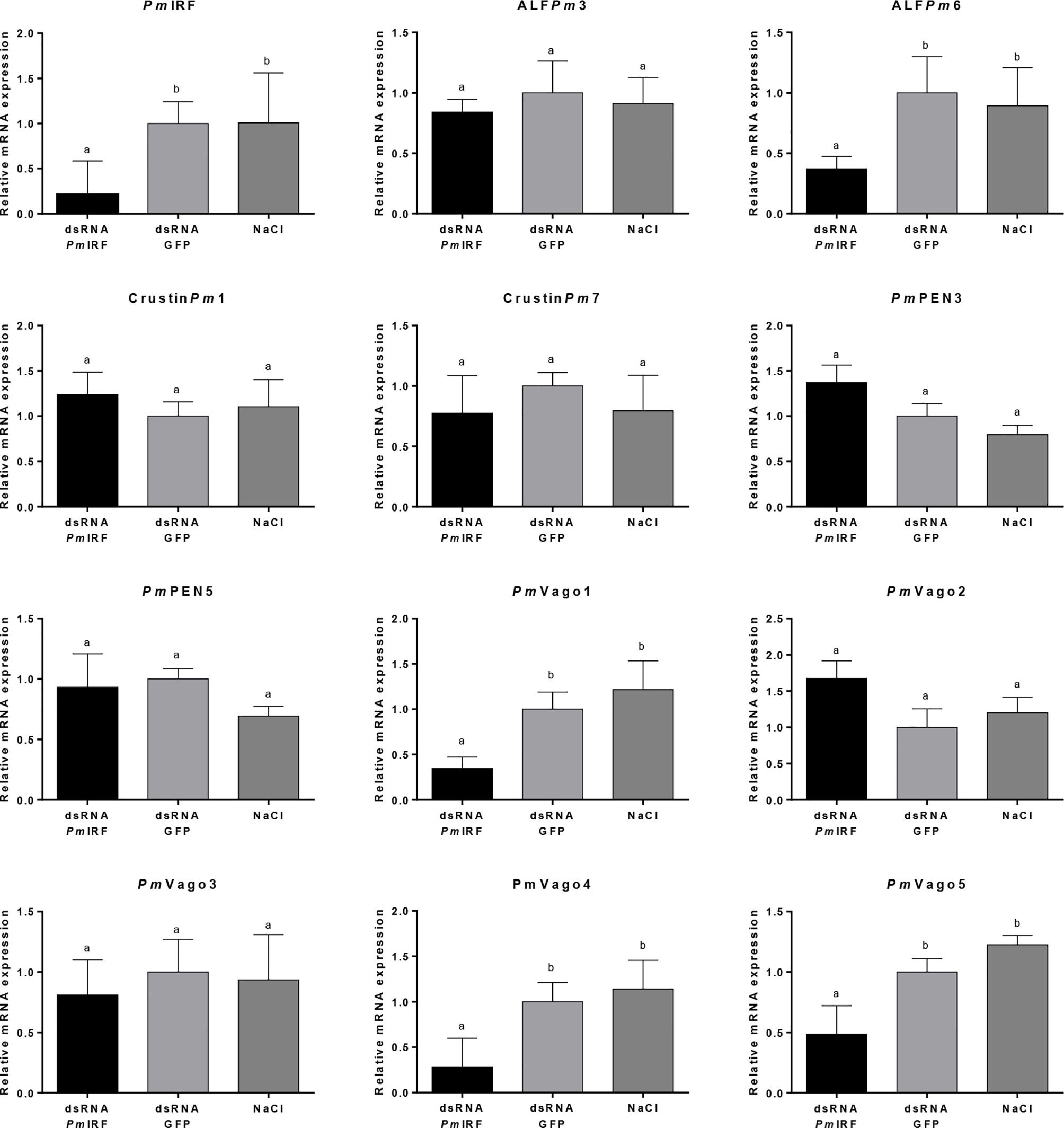
Figure 1 Effect of PmIRF knockdown on the expression levels of antiviral and antimicrobial peptide genes. Shrimps were injected with PmIRF dsRNA, GFP dsRNA, or 150 mM NaCl. After 24h, the intestine was collected for qRT-PCR analysis. EF1-α was used as an internal control. Data represent mean ± SD of 3 independently replicated experiments. Significant difference in the mean relative expressions compared with the control group at the level of P < 0.05 is indicated by a different lowercase letter.
PmIRF Overexpression Activates IFN-β, NF-κB, and ALFPm6 Promoters
To further investigate the function of PmDDX41, PmSTING and PmIRF, recombinant of PmDDX41, PmSTING and PmIRF proteins were produced in HEK293T cells. The cells were transfected with Flag-tagged-PmDDX41 or Myc-tagged-PmSTING or Flag-tagged-PmIRF for 24-72 h. The recombinant protein in HEK293T cells were detected by immunoblotting using anti-Flag or anti-Myc antibody, respectively (Supplement Figure 1).
To identify the immune signaling pathway involved in the activation of PmIRF and expression of immune-related genes, we co-transfected a PmIRF overexpression construct and luciferase reporter plasmid driven by the IFN-β, NF-κB, ALFPm3, or ALFPm6 promoter into human embryonic kidney (HEK293T) cells. In PBS-control condition, PmIRF overexpression increased the activity of the IFN-β, NF-κB, and ALFPm6 promoters 2.29, 1.17 and 2.39 fold, respectively; and in cells stimulated with the nucleic acid mimic deoxyadenylic–deoxythymidylic acid sodium salt [poly(dA:dT)], promoter activity was increased 7.27, 3.69, and 4.52 fold, respectively (Figures 2A–C) while poly(I:C) induced the promoter activity of IFN-β, NF-κB and ALFPm6 to a similar extent to the PBS treatment. ALFPm3 promoter activity was unchanged by stimulation with DNA mimics compared to the control (Figure 2D).
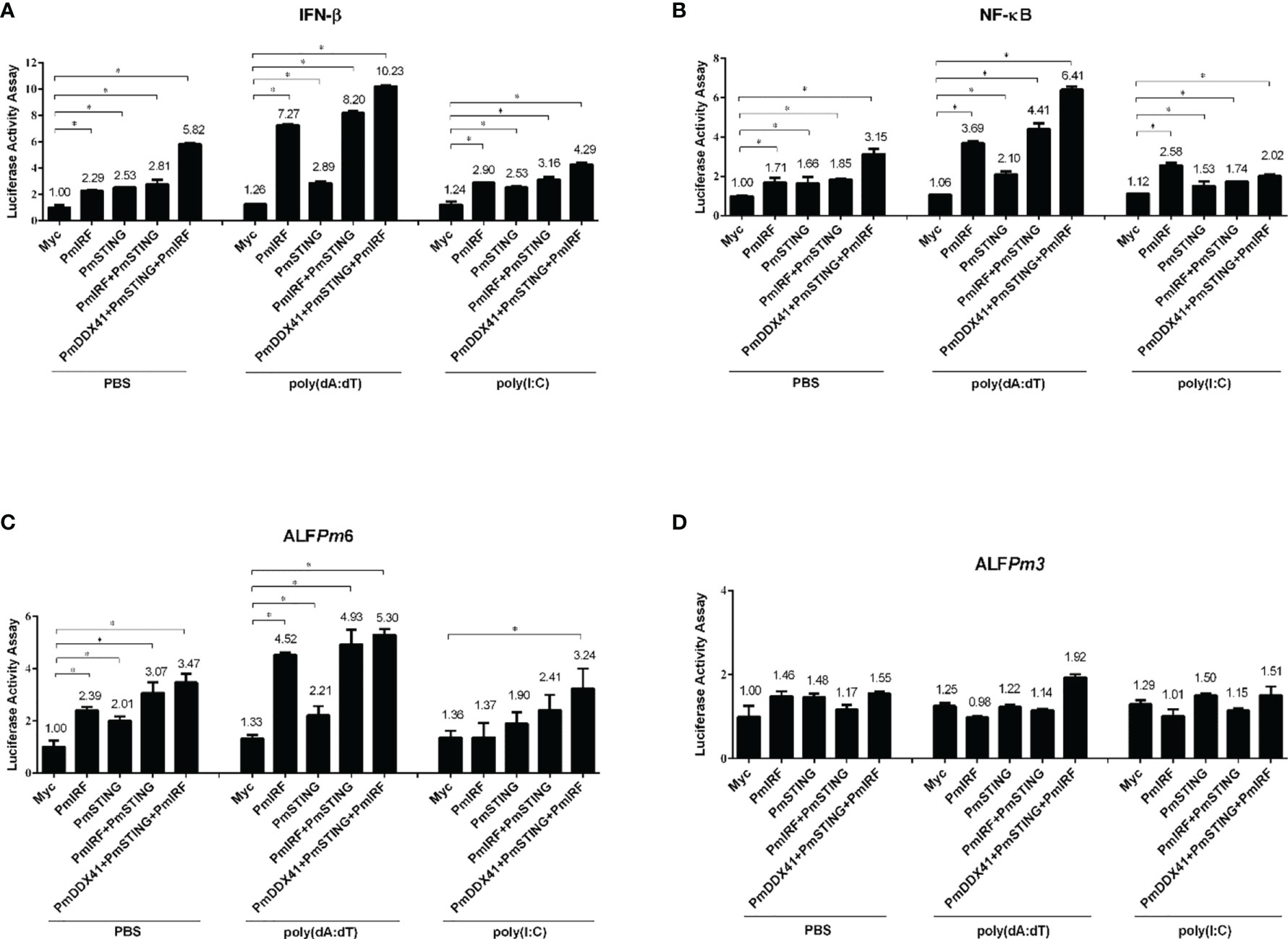
Figure 2 Luciferase assay for promoter activation induced by various immune-related factors in HEK293T cells. (A–D) The activation of IFN-β (A), NF-κB (B), ALFPm6 (C), and ALFPm3 (D) promoters was evaluated. Cells were co-transfected with 0.5 μg of Flag-tagged PmIRF expression plasmid and 0.5 μg of Myc tagged-PmSTING along with IFN-β-Luc, NF-κB-Luc, ALFPm6-Luc, and/or ALFPm3-Luc (all 0.1 μg) plasmid and the Renilla luciferase reporter pRL-TK (0.01 μg) plasmid, then stimulated with poly(dA:dT) or HMW poly(I:C); the luciferase assay was performed after 6 h Data represent mean ± SD of 3 independently replicated experiments. *p < 0.05.
To examine the function of PmIRF in the cytosolic nucleic acid sensing pathway, we co-transfected PmSTING and PmIRF expression plasmids into HEK293T cells. PmSTING overexpression induced the activation of IFN-β, NF-κB, and ALFPm6 promoters 2.53, 1.66, and 2.01-fold, respectively; in the presence of poly(dA:dT), the activity was induced 2.89, 2.10, and 2.21 fold, respectively, as determined with the luciferase assay (all p<0.05). Similarly, co-expression of PmSTING and PmIRF increased IFN-β, NF-κB, and ALFPm6 promoter activity 2.81, 1.85, and 3.07-fold, respectively; and stimulation with poly(dA:dT) enhanced the activity 8.20, 4.41, and 4.93 fold, respectively. Notably, co-expression of PmDDX41, PmSTING, and PmIRF synergistically increased IFN-β, NF-κB, and ALFPm6 promoter activity 5.82, 3.15, and 3.47-fold, respectively, without stimulation and 10.23, 6.41, and 5.30 fold, respectively, in the presence of poly(dA:dT) (Figures 2A–C). These results suggest that PmIRF is involved in the STING-dependent cytosolic DNA sensing pathway leading to interferon and AMPs activation.
DNA Sensing Pathway Genes Are Induced in P. monodon Intestine Following Stimulation With Nucleic Acid Mimic
We investigated whether nucleic acid mimics could also activate DNA sensing pathway-related genes (PmSTING, PmIRF, and ALFPm6) in shrimp intestine by qRT-PCR. After poly(dA:dT) injection, PmSTING expression was significantly upregulated at 3, 24, and 48 h post injection (p<0.05), with the greatest increase (4.78 fold) at 3 h (Figure 3A). HMW poly(I:C) also induced PmSTING expression 3.71 fold at 48 h (p<0.05; Figure 3B). PmIRF transcript was upregulated 2.07, 2.21, 2.31, and 5.06 fold at 3, 6, 24, and 48 h, respectively, after poly(dA:dT) injection (Figure 3C); and HMW poly(I:C) injection induced PmIRF expression 3.51, 2.09, 3.72, and 2.27 fold at 3, 6, 24, and 48 h, respectively (Figure 3D). Moreover, after poly(dA:dT) injection, ALFPm6 expression was increased 3.19, 1.63, and 1.88 fold at 6, 24, and 48 h, respectively (Figure 3E). HMW poly(I:C) injection also enhanced ALFPm6 expression 2.26 and 1.42 fold at 24 and 48 h, respectively (both p<0.05; Figure 3F). Thus, the expression of PmSTING, PmIRF, and ALFPm6 was induced by both nucleic acid mimics.
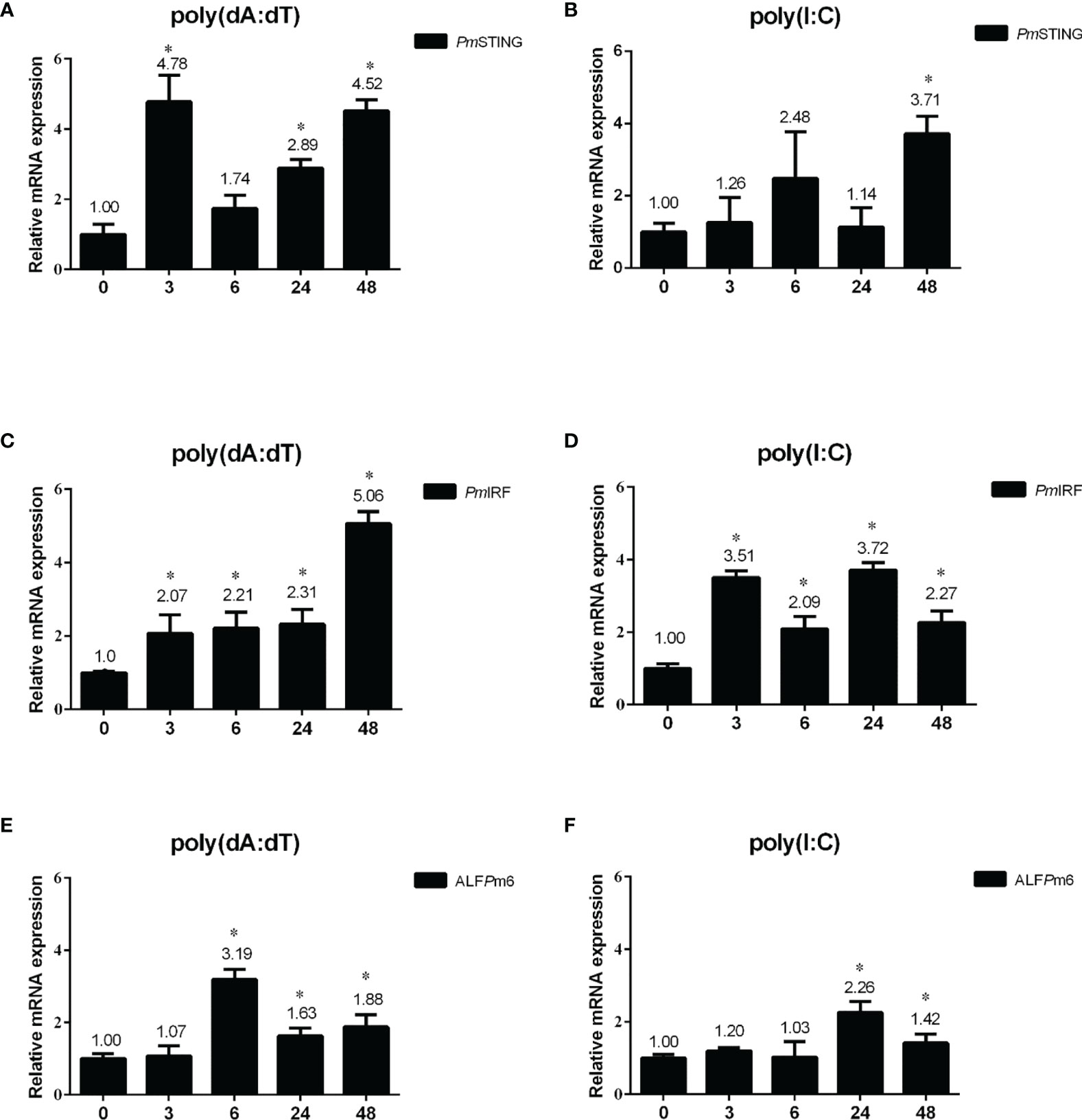
Figure 3 Expression profiles of immune-related genes in P. monodon intestine after injection of nucleic acid mimics. (A–F) Relative expression levels of PmSTING (A, B), PmIRF (C, D), and ALFPm6 (E, F) were evaluated by qRT-PCR at 0, 3, 6, 24, and 48 h after injection of poly(dA:dT) or HMW poly(I:C), with the EF1-α gene serving as an internal control. The expression level at 0 h was set as the baseline (1.0). Data represent mean ± SD of the assay performed with triplicate samples. *p < 0.05.
Interaction of PmIRF and PmSTING in HEK293T Cells
To further clarify the function of PmIRF in the STING-dependent cytosolic DNA sensing pathway, we analyzed the interaction between PmIRF and PmSTING. We co-transfected HEK293T cells with plasmids encoding Flag-tagged PmIRF and Myc-tagged PmSTING; a plasmid encoding Myc-tagged Mus musculus (MmSTING) protein served as a control. The proteins were incubated with anti-Myc antibody conjugated with A-sepharose beads and then detected by western blotting using anti-Flag or -Myc antibody. PmIRF bound PmSTING after stimulation with poly(dA:dT) but not poly(I:C) (Figure 4A).
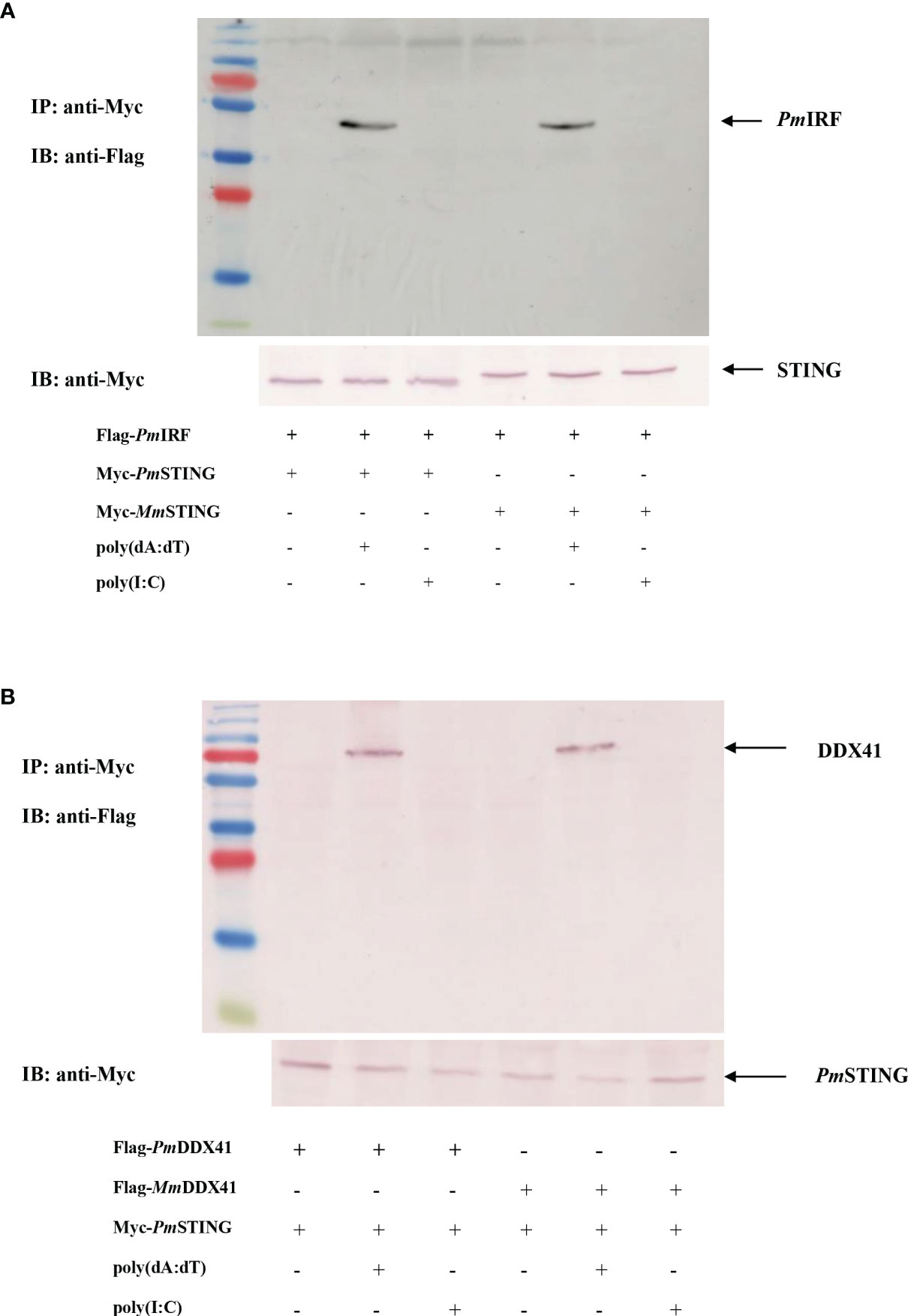
Figure 4 Co-immunoprecipitation of P. monodon proteins in HEK293T cells. (A, B) Cells were co-transfected with Flag-tagged full-length PmIRF (A) or PmDDX41 (B) plasmid, and 24 h later the co-immunoprecipitation of the P. monodon proteins with Myc-tagged PmSTING was detected following stimulation with poly(dA:dT) or HMW poly(I:C) for 6 h using HRP-conjugated anti-Flag and -Myc antibodies; the mouse homologs MmSTING [in panel (A)] and MmDDX41 [in panel (B)] were used as controls.
PmDDX41 was shown to bind mouse STING protein upon poly(dA:dT) stimulation in HEK293T cells (27). We recently identified a P. monodon homolog of STING (PmSTING) (20). As some amino acid residues of PmSTING are conserved from arthropods to mammals, we speculated that PmSTING could bind to cyclic dinucleotides in a manner similar to mammalian STING (19). To test this hypothesis, we carried out a co-immunoprecipitation assay to analyze the interaction of PmDDX41 and PmSTING in HEK293T cells. The cells were co-transfected with plasmids encoding Flag-tagged PmDDX41 and Myc-tagged PmSTING, with Flag-tagged mouse MmDDX41 recombinant protein used as a control. Cell lysates were precipitated with an anti-Myc antibody conjugated with protein A-sepharose beads and the Flag- or Myc-tagged protein was detected by western blotting using an anti-Flag or -Myc antibody, respectively. PmDDX41 was found to interact with PmSTING after poly(dA:dT) but not HMW poly(I:C) stimulation (Figure 4B). Additionally, MmDDX41 interacted with PmSTING in the presence of poly(dA:dT) (Figure 4B). These results confirm that STING mediates cytosolic DNA sensing in response to a signal from DDX41 that activates IRF, leading to the production of IFN-like molecules.
Subcellular Localization of PmIRF, PmSTING, and PmDDX41 in HEK293T Cells
In the STING-dependent cytosolic DNA sensing pathway, DDX41 interacts with viral dsDNA and STING, leading to activation of TBK1 or IKK and IFN production via IRF. To clarify the mechanism by which PmIRF senses nucleic acids, we performed immunofluorescence microscopy to examine the subcellular localization of PmIRF and PmSTING in HEK293T cells co-transfected with Myc-tagged PmSTING and Flag-tagged PmIRF expression plasmids and stimulated 24 h later with poly(dA:dT) or HMW poly(I:C). PmIRF and PmSTING were localized in the cytoplasm in unstimulated cells (Figure 5). After treatment with poly(dA:dT) and HMW poly(I:C), PmIRF was detected in the cytoplasm and nucleus whereas PmSTING remained exclusively cytoplasmic (Figure 5). In cells co-transfected with Myc-tagged PmDDX41 and Flag-tagged PmIRF plasmids, PmIRF co-localized with PmDDX41 in the cytoplasm. Poly(dA:dT) and HMW poly(I:C) treatment induced the expression of PmIRF and PmDDX41, which were localized in both the cytoplasm and nucleus (Figure 6).
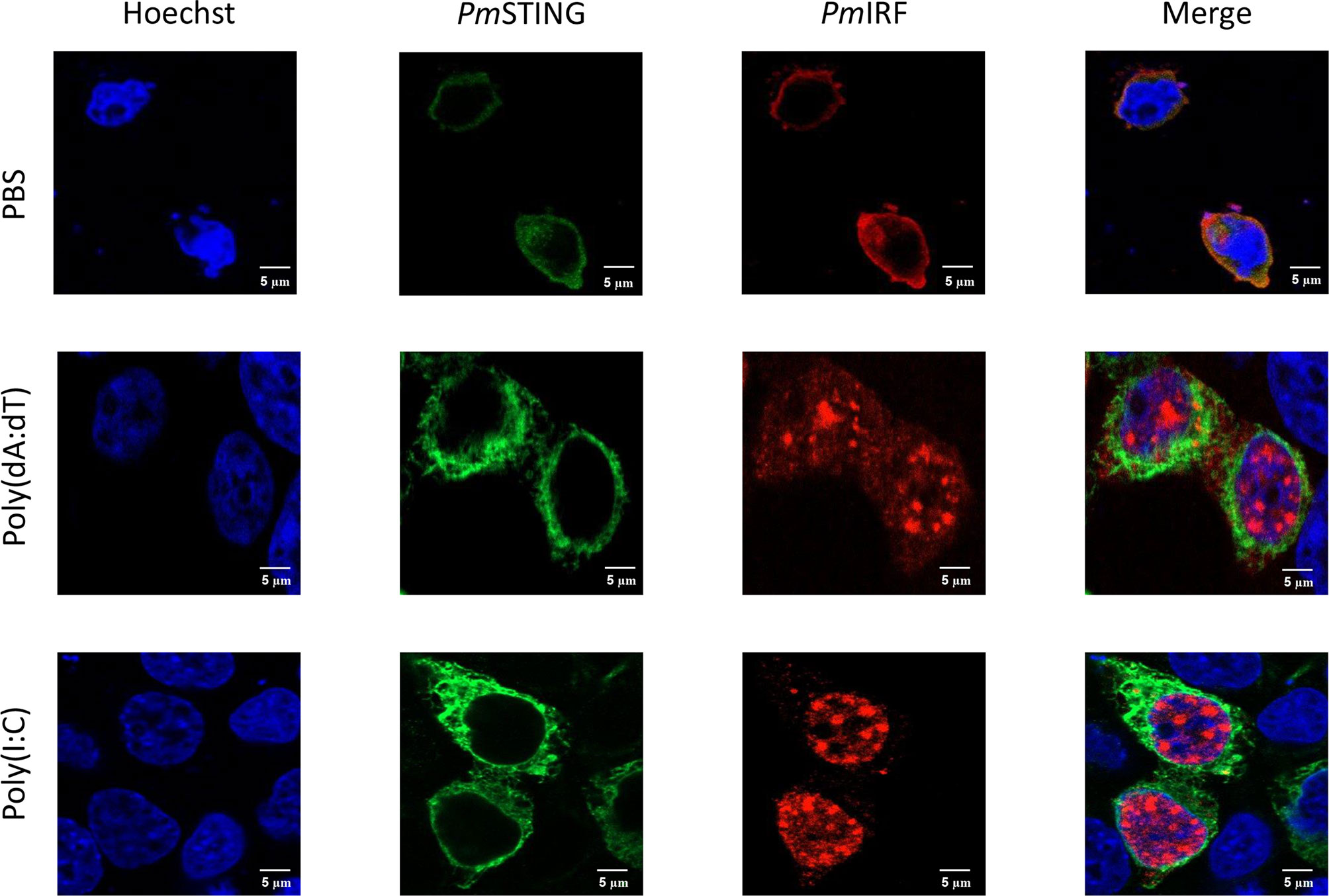
Figure 5 Subcellular localization of PmIRF and PmSTING in HEK293T cells. Cells were co-transfected with Flag-tagged PmIRF and Myc-tagged PmSTING expression plasmids for 24 h, then stimulated with 1 μg/ml poly(dA:dT) or HMW poly(I:C) for 6 h before labeling with an Alexa Fluor 568-conjugated anti-Flag antibody (red) and Alexa Fluor 488-conjugated anti-Myc antibody (green). Nuclei were stained with Hoechst 33342 (blue). Scale bar, 5 μm. Fluorescence was detected by laser scanning confocal microscopy (63× magnification).
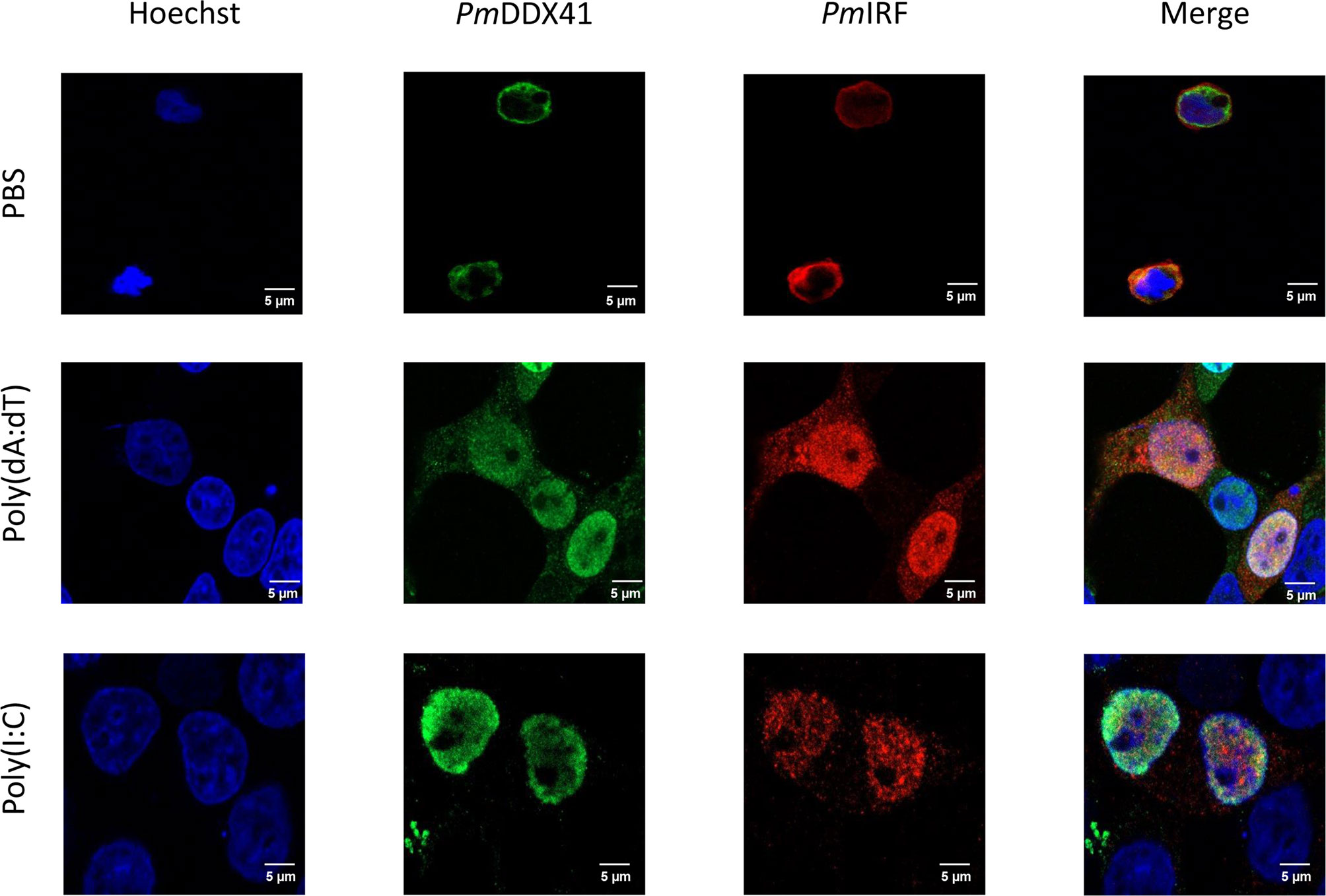
Figure 6 Subcellular localization of PmIRF and PmDDX41 in HEK293T cells. Cells were co-transfected with Myc-tagged PmDDX41 and Flag-tagged PmIRF expression plasmids for 24 h, then stimulated with 1 μg/ml poly((dA:dT) or HMW poly(I:C) for 6 h before labeling with an Alexa Fluor 568-conjugated anti-Flag antibody (red) and Alexa Fluor 488-conjugated anti-Myc antibody (green). Nuclei were stained with Hoechst 33342 (blue). Scale bar, 5 μm. Fluorescence was detected by laser scanning confocal microscopy (63× magnification).
Discussion
IRFs participate in pathogen-induced innate and acquired immunity in both vertebrates and invertebrates by regulating the expression of genes in multiple signaling pathways, especially those involved in antiviral immunity and that control cell differentiation and growth, apoptosis, the DNA damage response, and tumor suppression (22, 34, 35). To date, 9 IRFs have been identified in mammals; these play critical roles in the activation of immune responses (34, 36). The first crustacean IRF-like gene was identified in Pacific white shrimp; subsequent analyses revealed that LvIRF is involved in antiviral immunity, similar to the mammalian homologs. We recently identified PmIRF and PmSTING in P. monodon and demonstrated through RNAi-mediated loss-of-function experiments that they contribute to antiviral defense in shrimp (20).
The cytosolic DNA sensing pathway plays an important role in host defense. Upon infection with a virus, host PRRs sense viral PAMPs and activate an immune response. cGAS is one of the cytosolic DNA sensor in the innate immune system (12). It detects cytosolic DNA from intracellular bacteria, damaged mitochondria, DNA viruses and retroviruses and triggers IFNs response (37, 38). Moreover, DDX41 is also a DNA-binding protein that can detect viral or bacterial DNA and activates intracellular signaling cascades of the innate immune system (10, 39, 40). DDX41 was reported to activate the endoplasmic reticulum (ER) adaptor protein STING (10, 41), and various DNA viruses were shown to induce STING signaling. Recently, PmDDX41, PmSTING, and PmIRF were identified in the shrimp P. monodon and were shown to be involved in the innate immune response against WSSV infection (27). In this study, we further investigated the regulation of PmIRF via the STING-dependent cytosolic DNA sensing pathway. Silencing the PmIRF gene reduced the expression of several antimicrobial peptides and IFN-like molecules in shrimp. In our previous work, depletion of PmDDX41 by RNAi increased the mortality rate of WSSV-infected shrimp and significantly reduced the mRNA expression levels of several immune-related genes (PmIKKβ, PmIKKϵ, PmRelish, PmCactus, PmDorsal, PmPEN3, PmPEN5, and ALFPm6) (42). Moreover, the expression levels of IFN-like molecules (LvVago4 and LvVago5) were significantly decreased in IRF-silenced L. vannamei (26). Vago is a viral infection-inducible peptide first identified in Drosophila that can suppress the viral load of Drosophila C virus in the fat body (43). In mosquito, Vago is a secreted peptide that blocks viral infection by activating JAK–STAT signaling, which is homologous to the mammalian IFN system (44). Thus, ALFPm6, PmVago1, PmVago4, and PmVago5 are likely target genes of PmIRF in the immune signaling pathway of P. monodon.
The results of the promoter activity assay demonstrated that PmIRF activated the IFN-β, NF-κB, and ALFPm6 promoters in HEK293T cells, especially upon stimulation with nucleic acid mimics. DDX41 and STING overexpression synergistically enhanced the activity of the IFN-β promoter in L929 mouse fibroblast cells (10). In P. monodon, co-transfection of PmDDX41 and MmSTING plasmids enhanced the promoter activity of IFN-β and NF-κB (27). In L. vannamei, IRF activated promoters which contain ISRE element to regulate the expression of mammalian type I IFNs and induce an antiviral state in S2 cells (26). Our results indicated that PmIRF is a downstream gene in the PmDDX41–PmSTING DNA sensing pathway that regulates the activity of IFN-β, NF-κB, and ALFPm6 promoters.
We demonstrated that PmDDX41, PmSTING, and PmIRF activated the promoter of the ALFPm6 gene but not the ALFPm3 gene. In a previous study, PmDDX41 knockdown resulted in the downregulation of ALFPm6 expression (42). On the contrary, PmSTING, PmIRF, and ALFPm6 levels was strongly upregulated by stimulation with nucleic acid mimic [poly(dA:dT) and to a lesser extent, HMW poly(I:C)]. PmDDX41, a DNA sensor, was previously shown to be upregulated upon infection with DNA virus and stimulation with nucleic acid mimic (27, 42). In one study, DDX41 recognized a dsDNA virus in vertebrates and acted through the STING–TBK1–IRF3 pathway to directly bind DNA and STING via its DEAD box domain (10). In L. vannamei infected with WSSV or injected with poly(I:C), IRF was shown to be upregulated in the hepatopancreas (26); and PmIRF and ALFPm6 transcripts were upregulated in P. monodon challenged with WSSV (20, 26, 45). Thus, PmSTING, PmIRF, and ALFPm6 respond more specifically to DNA virus or mimic than to RNA virus or mimic, and may be involved in the nucleic acid-induced antiviral immune response in shrimp.
IRFs have a conserved N-terminal region of about 100 amino acid residues, including 5 conserved Tryptophan that mediate DNA binding (46). Here we found that PmIRF interacted with PmSTING in HEK293T cells following stimulation with poly(dA:dT) but not HMW poly(I:C). STING Ser366 participates in IRF3 binding and activation, and its mutation to alanine (S366A) abolished DNA-induced IRF3 activation (47). The amino acid sequence of PmSTING was analyzed and found the ‘PLPLRT/SD’ motif which might also contribute to the interaction between PmSTING and PmIRF (20). However, the investigation of crucial domain responsible for the function of PmSTING will be further performed. We also found that PmIRF co-localized with PmSTING in the cytoplasm but was translocated to the nucleus while PmSTING remained cytoplasmic upon treatment with HMW poly(dA:dT) and poly(I:C). In L. vannamei, LvIRF protein is mainly present in the cytoplasm but is translocated to the nucleus after WSSV infection or poly(I:C) treatment (26). Moreover, PmSTING bound PmDDX41 in the presence of DNA mimic and the two proteins were co-localized in the cytoplasm, but the latter was translocated to the nucleus while PmSTING remained cytoplasmic following the stimulation. Similar results were observed in our previous study in HEK293T cells co-transfected with PmDDX41 and MmSTING plasmids and stimulated with poly(dA:dT) (27), as well as in Danio rerio (48). In vertebrates, the DDX41–STING complex was shown to localize in the cytosol, and poly(dA:dT) stimulation reduced the expression of DDX41 and STING in the ER and mitochondria (10). STING was translocated along with TBK1 from the ER to the endosome in murine embryonic fibroblasts (49). Thus, PmDDX41 may function as a DNA sensor in the cytosol and interacts with PmSTING to form a complex with TBK and PmIRF that enters the nucleus and activates IFN and other genes related to the antiviral response.
TLRs activate the production of type I IFNs through IRFs (2). Signaling through TLRs can be divided into TIR domain-containing adapter-inducing IFN-β (TRIF)- and MyD88-dependent pathways (2, 50). In the latter, IRF4, IRF5, and IRF7 directly interact with MyD88 to regulate the expression of immune-related genes. IRF7 is essential for type I IFN gene induction by TLR7 or TLR9; IRF5 is required for the expression of pro-inflammatory cytokine genes (50–52); and IRF3 plays an essential role in the TRIF-dependent induction of type I IFN genes by TLR3 and TLR4 via TBK1 (2).
Poly(dA:dT) and poly(I:C) are the synthetic compounds which represent as a DNA virus and RNA virus., respectively. Both of them are potent inducers of the innate antiviral response in vertebrates. Poly(dA:dT) is recognized by cytosolic DNA sensors (CDS), including cGAS, AIM2, DAI, DDX41, IFI16, and LRRFIP1, triggers the production of type I interferons (10, 12, 53). Moreover, it is sensed to the cytosolic DNA sensor AIM2 triggers the formation of an inflammasome and the subsequent secretion of IL-1β and IL-18 (54). Poly(I:C) is recognized by TLR3 which mediated the IFNs synthesis (55). These signaling pathways shared the immune-related protein such as STING, IKK, TBK1, and IRF3. After poly(dA:dT) stimulation, PmSTING which composed of c-di-GMP-binding domain (CBD) at C-terminal (20), directly binds to the DNA mimic virus and sends the signal to activate PmIRF leading to IFNs production. PmIRF is the downstream gene in the signaling cascade so, poly(dA:dT) and poly(I:C) might induce PmIRF transcript.
Together with previous findings on the components of the cytosolic DNA sensing pathway in shrimp (PmDDX41, PmSTING, PmIKKs, PmIRF, and PmVago) (20, 42, 56), we propose a model of how these proteins interact in the cytosolic DNA sensing pathway to activate the antiviral immune response in P. monodon (Figure 7). Possibly during infection with the DNA virus WSSV, dsDNA is detected and bound by the DNA sensor PmDDX41, which forms a complex with PmSTING that acts via TBK–IKK–IRF3 to induce the IFN response. This research extends our knowledge of the regulatory role PmIRF in the antiviral response of crustaceans, and provides insight into the molecular mechanism of the cytosolic DNA sensing pathway in P. monodon.
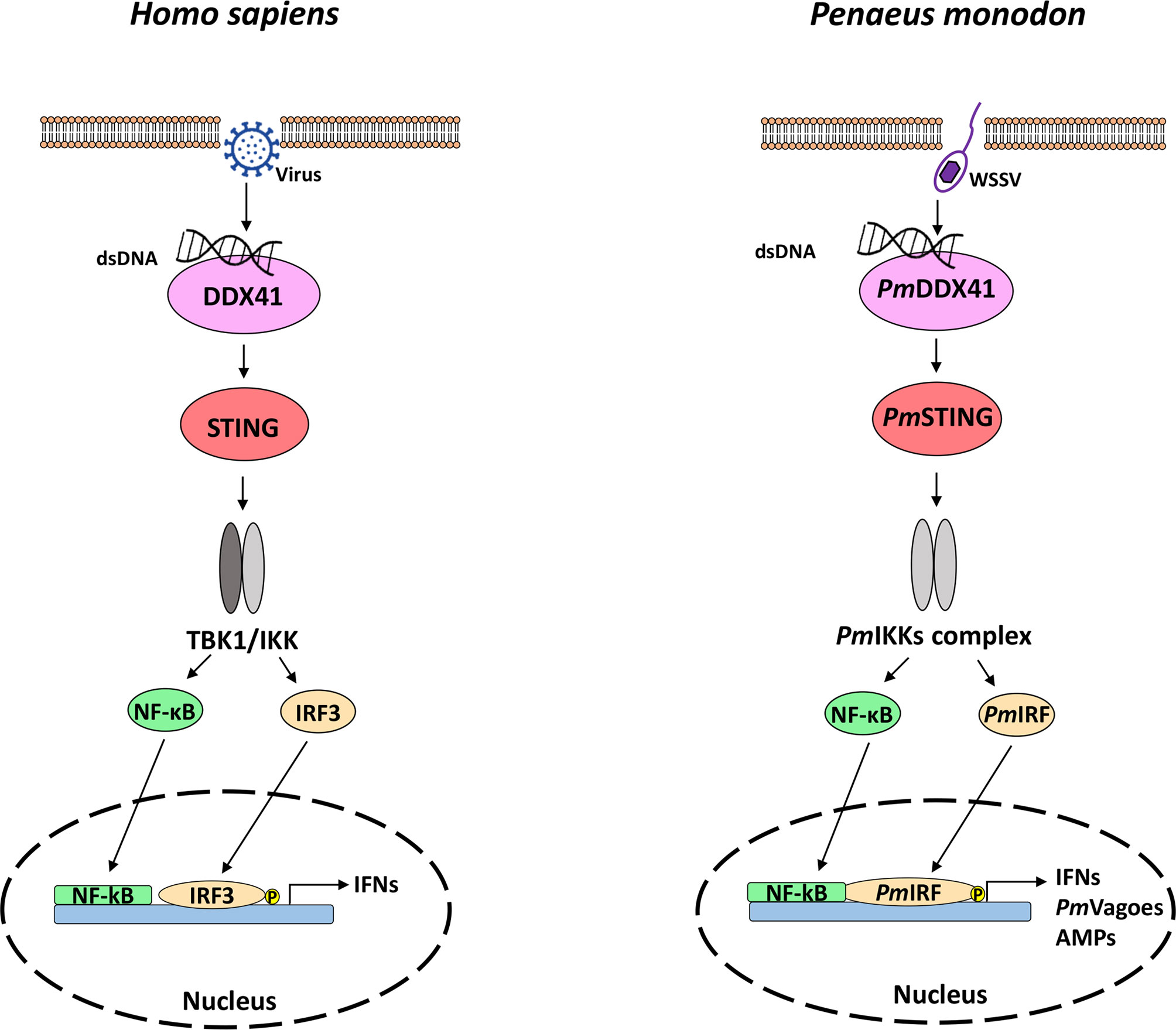
Figure 7 The DNA sensing pathway in mammals (left) and shrimp (right). The homologs of mammalian genes were found in shrimp, indicating that the signaling pathway is conserved from crustaceans to mammals. In mammals, dsDNA is recognized by DDX41, which induces IFN production [adapted from ref. (57)]. In the present study, we showed that dsDNA released from a viral pathogen such as WSSV interacts with the cytosolic DNA sensor PmDDX41 to induce PmSTING, thereby promoting the translocation of PmIRF to the nucleus, resulting in the production of IFNs and antimicrobial peptides that eliminate the viral infection.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
AT and PA contributed to the experimental design and helped to obtain funding. TK conceived the study with HEK293T cells. SS performed the experiments and wrote the manuscript. AT, TK, and PA reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the Thailand Research Fund (International Research Network) (Scholar no. IRN61W0001 to AT) and The Second Century Fund (C2F), Chulalongkorn University (to SS). We also gratefully acknowledge an additional support from the Ratchadaphisek Somphot Endowment Fund, Chulalongkorn University to the Center of Excellence for Molecular Biology and Genomics of Shrimp.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr.Warunthorn Monwan for technical assistance with cell cultures.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.818267/full#supplementary-material
References
1. Kumar H, Kawai T, Akira S. Pathogen Recognition by the Innate Immune System. Int Rev Immunol (2011) 30(1):16–34. doi: 10.3109/08830185.2010.529976
2. Kawai T, Akira S. The Role of Pattern-Recognition Receptors in Innate Immunity: Update on Toll-Like Receptors. Nat Immunol (2010) 11(5):373–84. doi: 10.1038/ni.1863
3. Kawasaki T, Kawai T, Akira S. Recognition of Nucleic Acids by Pattern-Recognition Receptors and its Relevance in Autoimmunity. Immunol Rev (2011) 243(1):61–73. doi: 10.1111/j.1600-065X.2011.01048.x
4. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu Rev Immunol (2015) 33:257–90. doi: 10.1146/annurev-immunol-032414-112240
5. Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA Sensor for IRF-3-Dependent Innate Immunity. Elife (2012) 1:e00047. doi: 10.7554/eLife.00047
6. Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, et al. cGAS Produces a 2'-5'-Linked Cyclic Dinucleotide Second Messenger That Activates STING. Nature (2013) 498(7454):380–4. doi: 10.1038/nature12306
7. Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, et al. DNA Damage Sensor MRE11 Recognizes Cytosolic Double-Stranded DNA and Induces Type I Interferon by Regulating STING Trafficking. Proc Natl Acad Sci USA (2013) 110(8):2969–74. doi: 10.1073/pnas.1222694110
8. Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, et al. Interferon Gamma-Inducible Protein (IFI) 16 Transcriptionally Regulates Type I Interferons and Other Interferon-Stimulated Genes and Controls the Interferon Response to Both DNA and RNA Viruses. J Biol Chem (2014) 289(34):23568–81. doi: 10.1074/jbc.M114.554147
9. Dempsey A, Bowie AG. Innate Immune Recognition of DNA: A Recent History. Virology (2015) 479-480:146–52. doi: 10.1016/j.virol.2015.03.013
10. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The Helicase DDX41 Senses Intracellular DNA Mediated by the Adaptor STING in Dendritic Cells. Nat Immunol (2011) 12(10):959–65. doi: 10.1038/ni.2091
11. Paludan SR, Bowie AG. Immune Sensing of DNA. Immunity (2013) 38(5):870–80. doi: 10.1016/j.immuni.2013.05.004
12. Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science (2013) 339(6121):826–30. doi: 10.1126/science.1229963
13. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of Innate Immune Adaptor Proteins MAVS, STING, and TRIF Induces IRF3 Activation. Science (2015) 347(6227):aaa2630. doi: 10.1126/science.aaa2630
14. Motwani M, Pesiridis S, Fitzgerald KA. DNA Sensing by the cGAS-STING Pathway in Health and Disease. Nat Rev Genet (2019) 20(11):657–74. doi: 10.1038/s41576-019-0151-1
15. Kargbo RB. Targeting Stimulator of Interferon Genes (STING) for the Potential Treatment of Cancer and Infectious Diseases. ACS Med Chem Lett (2021) 12(3):328–30. doi: 10.1021/acsmedchemlett.1c00085
16. Yum S, Li M, Fang Y, Chen ZJ. TBK1 Recruitment to STING Activates Both IRF3 and NF-kappaB That Mediate Immune Defense Against Tumors and Viral Infections. Proc Natl Acad Sci USA (2021) 118(14):1–9. doi: 10.1073/pnas.2100225118
17. Margolis SR, Wilson SC, Vance RE. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol (2017) 38(10):733–43. doi: 10.1016/j.it.2017.03.004
18. Goto A, Okado K, Martins N, Cai H, Barbier V, Lamiable O, et al. The Kinase IKKbeta Regulates a STING- and NF-kappaB-Dependent Antiviral Response Pathway in Drosophila. Immunity (2018) 49(2):225–234 e224. doi: 10.1016/j.immuni.2018.07.013
19. Li H, Wang S, Lu K, Yin B, Xiao B, Li S, et al. An Invertebrate STING From Shrimp Activates an Innate Immune Defense Against Bacterial Infection. FEBS Lett (2017) 591(7):1010–7. doi: 10.1002/1873-3468.12607
20. Amparyup P, Charoensapsri W, Soponpong S, Jearaphunt M, Wongpanya R, Tassanakajon A. Stimulator of Interferon Gene (STING) and Interferon Regulatory Factor (IRF) are Crucial for Shrimp Antiviral Defense Against WSSV Infection. Fish Shellfish Immunol (2021) 117:240–7. doi: 10.1016/j.fsi.2021.08.016
21. Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, et al. Regulated Expression of a Gene Encoding a Nuclear Factor, IRF-1, That Specifically Binds to IFN-Beta Gene Regulatory Elements. Cell (1988) 54(6):903–13. doi: 10.1016/s0092-8674(88)91307-4
22. Barnes B, Lubyova B, Pitha PM. On the Role of IRF in Host Defense. J Interferon Cytokine Res (2002) 22(1):59–71. doi: 10.1089/107999002753452665
23. Nehyba J, Hrdlickova R, Bose HR. Dynamic Evolution of Immune System Regulators: The History of the Interferon Regulatory Factor Family. Mol Biol Evol (2009) 26(11):2539–50. doi: 10.1093/molbev/msp167
24. Sun F, Zhang YB, Liu TK, Gan L, Yu FF, Liu Y, et al. Characterization of Fish IRF3 as an IFN-Inducible Protein Reveals Evolving Regulation of IFN Response in Vertebrates. J Immunol (2010) 185(12):7573–82. doi: 10.4049/jimmunol.1002401
25. Sun F, Zhang YB, Liu TK, Shi J, Wang B, Gui JF. Fish MITA Serves as a Mediator for Distinct Fish IFN Gene Activation Dependent on IRF3 or IRF7. J Immunol (2011) 187(5):2531–9. doi: 10.4049/jimmunol.1100642
26. Li C, Li H, Chen Y, Chen Y, Wang S, Weng SP, et al. Activation of Vago by Interferon Regulatory Factor (IRF) Suggests an Interferon System-Like Antiviral Mechanism in Shrimp. Sci Rep (2015) 5:15078. doi: 10.1038/srep15078
27. Soponpong S, Amparyup P, Kawai T, Tassanakajon A. A Cytosolic Sensor, PmDDX41, Binds Double Stranded-DNA and Triggers the Activation of an Innate Antiviral Response in the Shrimp Penaeus Monodon via the STING-Dependent Signaling Pathway. Front Immunol (2019) 10:2069. doi: 10.3389/fimmu.2019.02069
28. Amparyup P, Wiriyaukaradecha K, Charoensapsri W, Tassanakajon A. A Clip Domain Serine Proteinase Plays a Role in Antibacterial Defense But is Not Required for Prophenoloxidase Activation in Shrimp. Dev Comp Immunol (2010) 34(2):168–76. doi: 10.1016/j.dci.2009.09.004
29. Amparyup P, Charoensapsri W, Tassanakajon A. Two Prophenoloxidases are Important for the Survival of Vibrio Harveyi Challenged Shrimp Penaeus Monodon. Dev Comp Immunol (2009) 33(2):247–56. doi: 10.1016/j.dci.2008.09.003
30. Amparyup P, Jitvaropas R, Pulsook N, Tassanakajon A. Molecular Cloning, Characterization and Expression of a Masquerade-Like Serine Proteinase Homologue From Black Tiger Shrimp Penaeus Monodon. Fish Shellfish Immunol (2007) 22(5):535–46. doi: 10.1016/j.fsi.2006.07.004
31. Pfaffl MW. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res (2001) 29(9):e45. doi: 10.1093/nar/29.9.e45
32. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat Immunol (2005) 6(10):981–8. doi: 10.1038/ni1243
33. Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The Ubiquitin Ligase TRIM56 Regulates Innate Immune Responses to Intracellular Double-Stranded DNA. Immunity (2010) 33(5):765–76. doi: 10.1016/j.immuni.2010.10.013
34. Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF Family of Transcription Factors as Regulators of Host Defense. Annu Rev Immunol (2001) 19:623–55. doi: 10.1146/annurev.immunol.19.1.623
35. Yanai H, Negishi H, Taniguchi T. The IRF Family of Transcription Factors: Inception, Impact and Implications in Oncogenesis. Oncoimmunology (2012) 1(8):1376–86. doi: 10.4161/onci.22475
36. Zhang XJ, Jiang DS, Li H. The Interferon Regulatory Factors as Novel Potential Targets in the Treatment of Cardiovascular Diseases. Br J Pharmacol (2015) 172(23):5457–76. doi: 10.1111/bph.12881
37. Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, et al. Cyclic GMP-AMP Synthase is an Innate Immune Sensor of HIV and Other Retroviruses. Science (2013) 341(6148):903–6. doi: 10.1126/science.1240933
38. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA Stress Primes the Antiviral Innate Immune Response. Nature (2015) 520(7548):553–7. doi: 10.1038/nature14156
39. Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, et al. The Helicase DDX41 Recognizes the Bacterial Secondary Messengers Cyclic Di-GMP and Cyclic Di-AMP to Activate a Type I Interferon Immune Response. Nat Immunol (2012) 13(12):1155–61. doi: 10.1038/ni.2460
40. Ablasser A, Chen ZJ. cGAS in Action: Expanding Roles in Immunity and Inflammation. Science (2019) 363(6431):1–9. doi: 10.1126/science.aat8657
41. Zahid A, Ismail H, Li B, Jin T. Molecular and Structural Basis of DNA Sensors in Antiviral Innate Immunity. Front Immunol (2020) 11:613039. doi: 10.3389/fimmu.2020.613039
42. Soponpong S, Amparyup P, Tassanakajon A. A Cytosolic Sensor, PmDDX41, Mediates Antiviral Immune Response in Black Tiger Shrimp Penaeus Monodon. Dev Comp Immunol (2018) 81:291–302. doi: 10.1016/j.dci.2017.12.013
43. Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, et al. The DExD/H-Box Helicase Dicer-2 Mediates the Induction of Antiviral Activity in Drosophila. Nat Immunol (2008) 9(12):1425–32. doi: 10.1038/ni.1664
44. Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ. Secreted Vago Restricts West Nile Virus Infection in Culex Mosquito Cells by Activating the Jak-STAT Pathway. Proc Natl Acad Sci USA (2012) 109(46):18915–20. doi: 10.1073/pnas.1205231109
45. Ponprateep S, Tharntada S, Somboonwiwat K, Tassanakajon A. Gene Silencing Reveals a Crucial Role for Anti-Lipopolysaccharide Factors From Penaeus Monodon in the Protection Against Microbial Infections. Fish Shellfish Immunol (2012) 32(1):26–34. doi: 10.1016/j.fsi.2011.10.010
46. Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 With Bound DNA Reveals Determinants of Interferon Regulation. Nature (1998) 391(6662):103–6. doi: 10.1038/34224
47. Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci Signal (2012) 5(214):ra20. doi: 10.1126/scisignal.2002521
48. Ma JX, Li JY, Fan DD, Feng W, Lin AF, Xiang LX, et al. Identification of DEAD-Box RNA Helicase DDX41 as a Trafficking Protein That Involves in Multiple Innate Immune Signaling Pathways in a Zebrafish Model. Front Immunol (2018) 9:1327. doi: 10.3389/fimmu.2018.01327
49. Ishikawa H, Ma Z, Barber GN. STING Regulates Intracellular DNA-Mediated, Type I Interferon-Dependent Innate Immunity. Nature (2009) 461(7265:788–92. doi: 10.1038/nature08476
50. Honda K, Taniguchi T. IRFs: Master Regulators of Signalling by Toll-Like Receptors and Cytosolic Pattern-Recognition Receptors. Nat Rev Immunol (2006) 6(9):644–58. doi: 10.1038/nri1900
51. Balkhi MY, Fitzgerald KA, Pitha PM. Functional Regulation of MyD88-Activated Interferon Regulatory Factor 5 by K63-Linked Polyubiquitination. Mol Cell Biol (2008) 28(24):7296–308. doi: 10.1128/MCB.00662-08
52. Ikushima H, Negishi H, Taniguchi T. The IRF Family Transcription Factors at the Interface of Innate and Adaptive Immune Responses. Cold Spring Harb Symp Quant Biol (2013) 78:105–16. doi: 10.1101/sqb.2013.78.020321
53. Unterholzner L. The Interferon Response to Intracellular DNA: Why So Many Receptors? Immunobiology (2013) 218(11):1312–21. doi: 10.1016/j.imbio.2013.07.007
54. Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, et al. Absent in Melanoma 2 is Required for Innate Immune Recognition of Francisella Tularensis. Proc Natl Acad Sci USA (2010) 107(21):9771–6. doi: 10.1073/pnas.1003738107
55. Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, et al. Cell Type-Specific Involvement of RIG-I in Antiviral Response. Immunity (2005) 23(1):19–28. doi: 10.1016/j.immuni.2005.04.010
56. Nhnhkorn Z, Amparyup P, Kawai T, Tassanakajon A. Penaeus Monodon IKKs Participate in Regulation of Cytokine-Like System and Antiviral Responses of Innate Immune System. Front Immunol (2019) 10:1430. doi: 10.3389/fimmu.2019.01430
Keywords: IRF, STING, DNA sensing pathway, antiviral response, interferon, shrimp immunity
Citation: Soponpong S, Amparyup P, Kawai T and Tassanakajon A (2022) Penaeus monodon Interferon Regulatory Factor (PmIRF) Activates IFNs and Antimicrobial Peptide Expression via a STING-Dependent DNA Sensing Pathway. Front. Immunol. 12:818267. doi: 10.3389/fimmu.2021.818267
Received: 19 November 2021; Accepted: 20 December 2021;
Published: 10 January 2022.
Edited by:
Stephanie DeWitte-Orr, Wilfrid Laurier University, CanadaReviewed by:
Chaozheng Li, Sun Yat-sen University, ChinaJeswin Joseph, Cochin University of Science and Technology, India
Jianguo He, Sun Yat-sen University, China
Copyright © 2022 Soponpong, Amparyup, Kawai and Tassanakajon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchalee Tassanakajon, QW5jaGFsZWUua0BjaHVsYS5hYy50aA==
 Suthinee Soponpong
Suthinee Soponpong Piti Amparyup
Piti Amparyup Taro Kawai
Taro Kawai Anchalee Tassanakajon
Anchalee Tassanakajon