- 1Department of Cardiovascular Medicine, The Second Affiliated Hospital of Nanchang University, Nanchang, China
- 2Center for Metabolic Disease Research, Department of Cardiovascular Science, Lewis Kats School of Medicine, Temple University, Philadelphia, PA, United States
- 3Department of Endocrinology, The Second Affiliated Hospital of Nanchang University, Nanchang, China
- 4Department of Cardiovascular Medicine, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 5Key Laboratory of Cardiovascular Disease and Molecular Intervention, Nanjing Medical University, Nanjing, China
- 6Tulane National Primate Research Center, School of Medicine, Tulane University, Covington, LA, United States
Rationale: Inflammatory monocyte (MC) subset differentiation is a major feature in tissue inflammatory and atherosclerosis. The underlying molecular mechanism remains unclear.
Objective: This study aims to explore molecule targets and signaling which determinate immunological features in MC subsets.
Methods and Results: Blood Ly6Chigh and Ly6Clow MC subsets from control and ApoE-/- mice were isolated by flow cytometry sorting and subjected for bulk high-throughput RNA-sequencing. Intensive bioinformatic studies were performed by analyzing transcriptome through four pairs of comparisons: A) Ly6Chigh vs Ly6Clow in control mice; B) Ly6Chigh vs Ly6Clow in ApoE-/- mice; C) ApoE-/- Ly6Chigh vs control Ly6Chigh MC; D) ApoE-/- Ly6Clow vs control Ly6Clow MC. A total of 80 canonical pathways and 16 enriched pathways were recognized by top-down analysis using IPA and GSEA software, and further used for overlapping analysis. Immunological features and signaling were assessed on four selected functional groups, including MHCII, immune checkpoint, cytokine, and transcription factor (TF). Among the total 14578 significantly differentially expressed (SDE) genes identified though above four comparison, 1051 TF and 348 immunological genes were discovered. SDE immunological genes were matched with corresponding upstream SDE TF by IPA upstream analysis. Fourteen potential transcriptional axes were recognized to modulate immunological features in the Ly6C MC subset. Based on an intensive literature search, we found that the identified SDE immune checkpoint genes in Ly6Chigh MC are associated with pro-inflammatory/atherogenic balance function. Immune checkpoint genes GITR, CTLA4, and CD96 were upregulated in Ly6Clow MC from all mice and presented anti-inflammatory/atherogenic features. Six cytokine genes, including Ccl2, Tnfsf14, Il1rn, Cxcl10, Ccl9, and Cxcl2, were upregulated in Ly6Chigh MC from all mice and associated with pro-inflammatory/atherogenic feature. Cytokine receptor gene Il12rb2, Il1r1, Il27ra, Il5ra, Ngfr, Ccr7, and Cxcr5 were upregulated in Ly6Clow MC from all mice and presented anti-inflammatory/atherogenic features. MHCII genes (H2-Oa, H2-DMb2, H2-Ob, H2-Eb2, H2-Eb1, H2-Aa, and Cd74) were elevated in Ly6Clow MC from all mice. ApoE-/- augmented pro-atherogenic/inflammatory and antigen-presenting cells (APC) feature in both subsets due to elevated expression of cytokine genes (Cxcl11, Cntf, Il24, Xcl, Ccr5, Mpl, and Acvr2a) and MHCII gene (H2-Aa and H2-Ea-ps). Finally, we modeled immunological gene expression changes and functional implications in MC differentiation and adaptive immune response for MC subsets from control and ApoE-/- mice.
Conclusions: Ly6Chigh MC presented pro-inflammatory/atherogenic features and lower APC potential. Ly6Clow MC displayed anti-inflammatory/atherogenic features and higher APC potential. ApoE-/- confers upon both subsets with augmented pro-atherogenic/inflammatory function and APC potential.
Introduction
Atherosclerosis is a chronic inflammatory disease of blood vessels and the essential pathological cause of cardiovascular disease, the leading cause of mortality worldwide (1, 2). Innate immune cell monocyte (MC) and macrophage (MΦ) is the major cellular components in the advanced atherosclerosis lesion, which is correlated with increased inflammatory MC differentiation (3–5). Emerging evidence supports the role of the adaptive immune system in atherosclerosis (6). However, the molecular mechanism underlying inflammatory MC differentiation, especially under hyperlipidemia (HL) conditions, and related adaptive immune response remains unknown.
MC circulates in the blood and migrates to inflammatory tissues, but their functions can be either detrimental or beneficial, determined by their subsets. Human CD14++CD16+ intermediate and CD14+CD40+ MC are considered as inflammatory MC subsets, similar to murine Ly6Chigh and Ly6Cmiddle MC (7, 8). Human CD14+CD16++ nonclassical, CD14++CD16- classical, and CD14+CD40- MC are anti-inflammatory MC, similar as mouse Ly6Clow MC (7, 8).
In response to environmental stimulation, MC can be differentiated into different MC/MΦ subsets. Inflammatory MC/MΦ subsets are the major component in inflammatory tissue and advanced atherosclerosis. A recent study supported that the emerging roles of MC as antigen-presenting cells (APC) that enable adaptive immunity (9). Adaptive immunity is critical for disease progression and modulates T cell and APC functions, which was mediated by three concerted signals, including signal 1 antigen recognition, signal 2 immune checkpoint and signal 3 cytokine stimulation (10, 11). We recently proposed an additional novel signal 4, metabolism-associated danger signal recognition, which transmit metabolic risk factor response via metabolic sensor-mediated and pattern recognition leading to MC to APC differentiation and T cell activation (10, 11).
It is believed that inflammation triggers the differentiation of Ly6Chigh MC into APC, mostly microbicidal MΦ or MC-derived dendritic cells (moDCs). Yet, little is known about the molecular mechanism determining Ly6Chigh MC differentiation and their APC potential. Recently, we and others reported that TF NR4A1, CEBPα, CEBPβ, and PU.1 are involved in the process of Ly6Chigh MC differentiation to Ly6Clow MC (3, 12, 13). The activation of STAT3 was described in MC-to-MΦ differentiation and inflammation (14). It was shown that major histocompatibility complex II (MHCII) was elevated in Ly6Chigh MC after infiltrating into the skin or skin-draining lymph nodes (15). Through transcriptome analysis in sorted mouse Ly6C MC subset, we reported that the Ly6Chigh MC displayed enriched inflammatory pathways and favored to be differentiated into MΦ and osteoclast (13). However, molecular mechanism underlying MC subset differentiation, specially under HL condition, remains to be elucidated.
MC/MΦ account for about 65% of CD45+ immune cells in advanced atherosclerotic aorta in human (16). MC/MΦ population is up to about 48.1% of CD45+ cells and the major immune cell populations in lesion of hyperlipidemic ApoE-/- and Ldlr-/- mice, and their combination with hyperhomocysteinemia (HHcy) cystathionine b-synthase gene-deficient (Cbs-/-) (4, 16–18). The myeloid response is accompanied by the infiltration of adaptive immune cells (18). In atherosclerosis-prone ApoE-/-CD11c-YFP+ mice, APC and CD4+ T helper cell interactions were increased in the plaque that resulted in pro-inflammatory cytokine IFN-γ and TNF-α secretion (19). Through transcriptome analysis, we recently revealed that Ly6Clow MC is enriched with genes manifesting T cell activation signaling in wild type and Cbs-/- mice (13). Although the potential role of APC and adaptive immune cells interaction in atherosclerosis is known, the specific molecular mechanism in regulating innate/adaptive immunity in disease conditions remains unclear.
This study aims to identify molecular mechanisms underlying MC subset differentiation, with a focus on HL-induced MC differentiation and innate/adaptive interplay, by analyzing MC subset transcriptome using intensive bioinformatic studies.
Research Design and Methods
We summarized the overall study approaches and strategies in Figure 1.
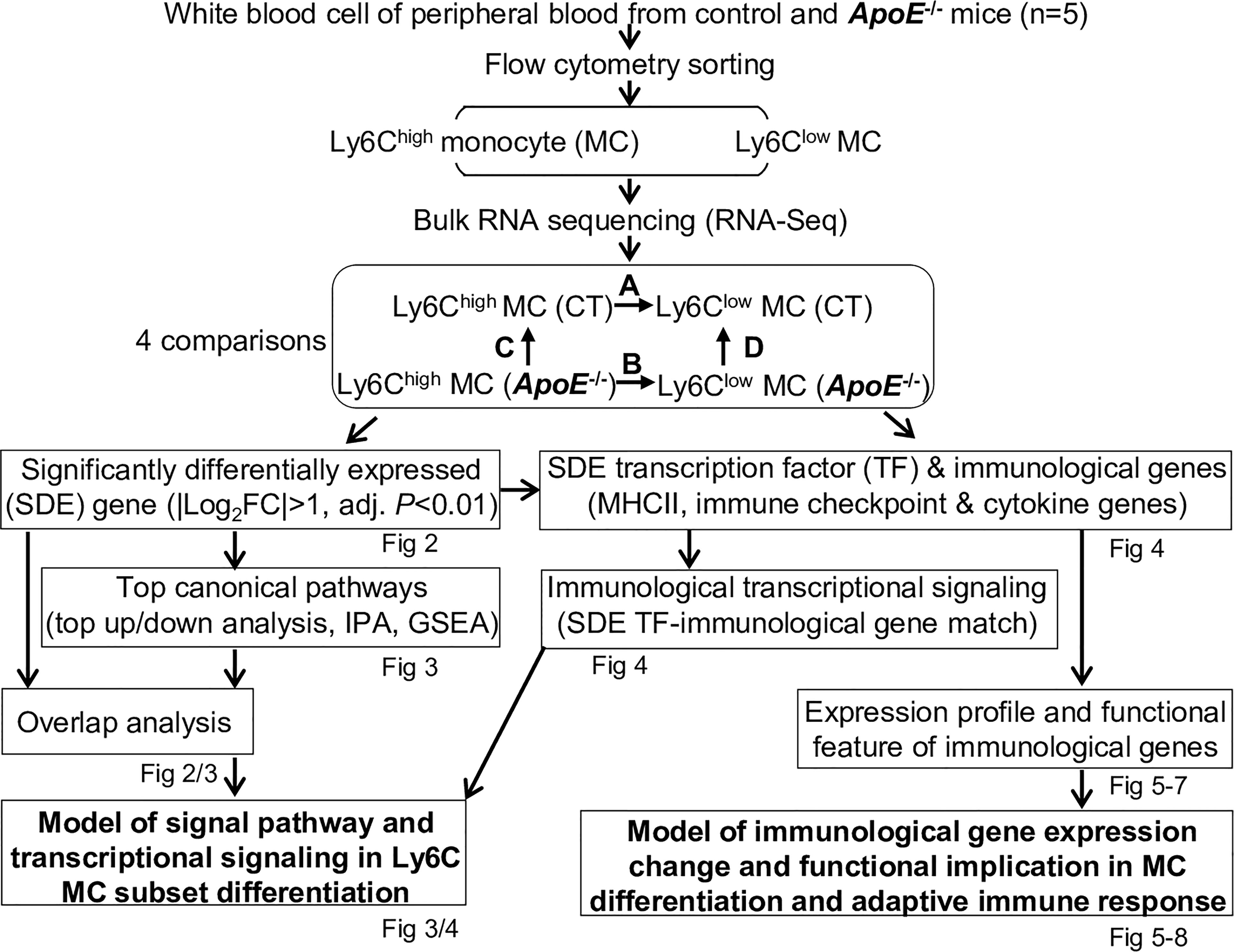
Figure 1 Overall strategy of the identification of molecular signaling in Ly6C MC subset differentiation and adaptive immune response in control and ApoE-/- mice. RNA-Seq were performed in Ly6Chigh (CD11b+Ly6G−Ly6Chigh) and Ly6Clow (CD11b+Ly6G−Ly6Clow) MC isolated by flow cytometry sorting from peripheral blood of C57/BL6 control and ApoE-/- mice. Transcriptome data were analyzed by performing four pairs of comparisons: (A) Ly6Chigh vs Ly6Clow (CT), (B) Ly6Chigh vs Ly6Clow (ApoE-/-), (C) ApoE-/- vs CT (Ly6Chigh), (D) ApoE-/- vs CT (Ly6Clow). SDE genes were identified by using the criteria of |Log2FC| more than 1 (2-FC) and adjusted P value less than 0.01. Top canonical pathways were recognized by top-down analysis using IPA with |Z-score|>2, P value<0.05. Overlapped analysis were performed for canonical pathways between groups. SDE TF and three sets of SDE immunological genes (MHCII, immune checkpoint and cytokine genes) were identified. Immunological SDE genes were matched with corresponding upstream SDE TF by IPA upstream analysis. Models of signal pathway and transcriptional signaling of Ly6C MC subset differentiation were developed. Expression profile and functional feature of immunological SDE genes were characterized. Model of immunological gene expression change and functional implication in MC differentiation and adaptive immune response in MC subsets of both mice were developed. CT, control, ApoE, Apolipoprotein E; FC, fold change; RNA-seq, RNA-sequencing; MC, monocyte; SDE, significant differentially expressed; IPA, Ingenuity Pathway Analysis; GSEA, gene set enrichment analysis; TF, transcription factor.
ApoE-/- Mice and Plasma Lipid Determination
— ApoE-/- mice were fed a normal chow diet and switched to a high-fat diet [21% fat (w/w), 0.15% cholesterol (w/w); Dyets Inc., Bethlehem, PA] at age 8 weeks and maintained on a high-fat diet for 12 weeks. The control (CT) C57BL/6 mice were fed a normal chow diet throughout. Animals were sacrificed at 20-22 weeks of age for blood collection after euthanization. Mouse blood was collected into 1 mM ethylenediaminetetraacetic acid (EDTA)-coated tubes for MC and plasma preparation. The plasma was separated (3,000g for 20 min). Plasma total cholesterol and triglyceride (TG) were analyzed as we previously described (20). All experiments were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and with approval from Temple University School of Medicine Institutional Animal Care and Use Committee.
Flow Cytometry and Cell Sorting
MC from mouse peripheral blood were collected and sorted by flow cytometry for Ly6Chigh (CD11b+Ly6G−Ly6Chigh) and Ly6Clow (CD11b+Ly6G−Ly6Clow) MC from CT and ApoE-/- cells isolation as described (13) and detailed in a supplementary document. The difference of Ly6Chigh and Ly6Clow MC between two groups of CT and ApoE-/- after sorting was shown in the Supplementary Figure 1.
RNA Sequencing and Data Processing
RNA was extracted. Pooled samples (5-6 in each) were run for sequencing analysis in duplication on NextSeq 500 (CT) and Illumina Hiseq 4000 sequencer (ApoE-/-). Overall, we obtained around 40 million reads per sample. The raw RNA-seq data were analyzed using R and RStudio. The raw reads were mapped to the mouse reference transcriptome using Kallisto as described (13) and detailed in a supplementary document.
Bioinformatic Analysis and Model Establishment
Intensive bioinformatic analysis was performed as described (13) and detailed in a supplementary document. Briefly, principle components analysis (PCA) was performed to examine the variance of RNA-seq data. Significantly differentially expressed (SDE) genes were identified by using the Bioconductor suite of Limma packages in RStudio software with the criteria of |fold change (FC)|>2 and adjusted P-value<0.01. Heatmap was generated in RStudio using the “pheatmap” package to present the expression levels of SDE genes. Ingenuity Pathway Analysis (IPA) version 7.1 was used to identify functional pathways and matched SDE TF with their corresponding SDE immunological genes. Gene set enrichment analysis (GSEA) was used for pathway enrichment study. Venn diagrams were displayed to present the overlaps of SDE genes and pathways between comparisons. Finally, we developed models of hypothetic signal pathway and transcriptional signaling of Ly6C MC subset differentiation and immunological functional implication.
Results
RNA-Seq Analysis and SDE Gene Identification From Blood Ly6Chigh and Ly6Clow MC of CT and ApoE-/- Mice
Eleven mice at age of 20 weeks were used. Severe HL were developed in ApoE-/- mice fed an HF diet for 12 weeks (plasma TG 289 mg/dl and cholesterol 587 mg/dl) (Figure 2A). In contrast, C57BL/6 CT animals had plasma TG of 87 mg/dl and cholesterol of 44 mg/dl. RNA-seq was performed in Ly6Chigh (CD11b+Ly6G−Ly6Chigh) and Ly6Clow (CD11b+Ly6G−Ly6Clow) MC isolated by flow cytometry sorting from mouse peripheral blood (Figure 2B). MC (100,000 cells) were sorted by flow cytometry with 43.7% Ly6Chigh, 23.7% Ly6Cmiddle and 32.3% Ly6Clow MC from ApoE-/- mice, and 30.3% Ly6Chigh, 33.1% Ly6Cmiddle and 36% Ly6Clow MC in CT mice (Figure 2C, details in Supplementary Figure 1). ApoE-/- elevated Ly6Chigh and reduced Ly6Cmiddle/low subsets. Principal component (PC) analysis (PCA) incorporated 8 samples from 4 groups of MC subsets [Ly6Chigh (CT), Ly6Clow (CT), Ly6Chigh (ApoE-/-) and Ly6Clow (ApoE-/-)]. PC1, PC2 and PC3 variance is 47.3%, 21.0% and 13.6% (Figure 2D). The PC1 axis separated Ly6Chigh and Ly6Clow MC in both mice, and explains 47.3% of the variance. Although the PC2 axis, which explains 21.0% of the variance, separated the Ly6Chigh MC from the ApoE-/- and CT groups, but not the Ly6Clow MC. However, the hierarchical clustering well separated the MC subsets in both groups (Figure 2E). We identified 1423-upregulated/1641-downregulated SDE genes, and 1410-upregulated/2113-downregulated SDE genes in Ly6Chigh MC from CT and ApoE-/- mice by A/B comparison. HL in ApoE-/- mice with 12-weeks HF diet-induced 1799-upregulated/2155-downregulated SDE gene in Ly6Chigh MC and induced 1792- upregulated/2245-downregulated SDE gene in Ly6Clow MC (Figures 2F, G).
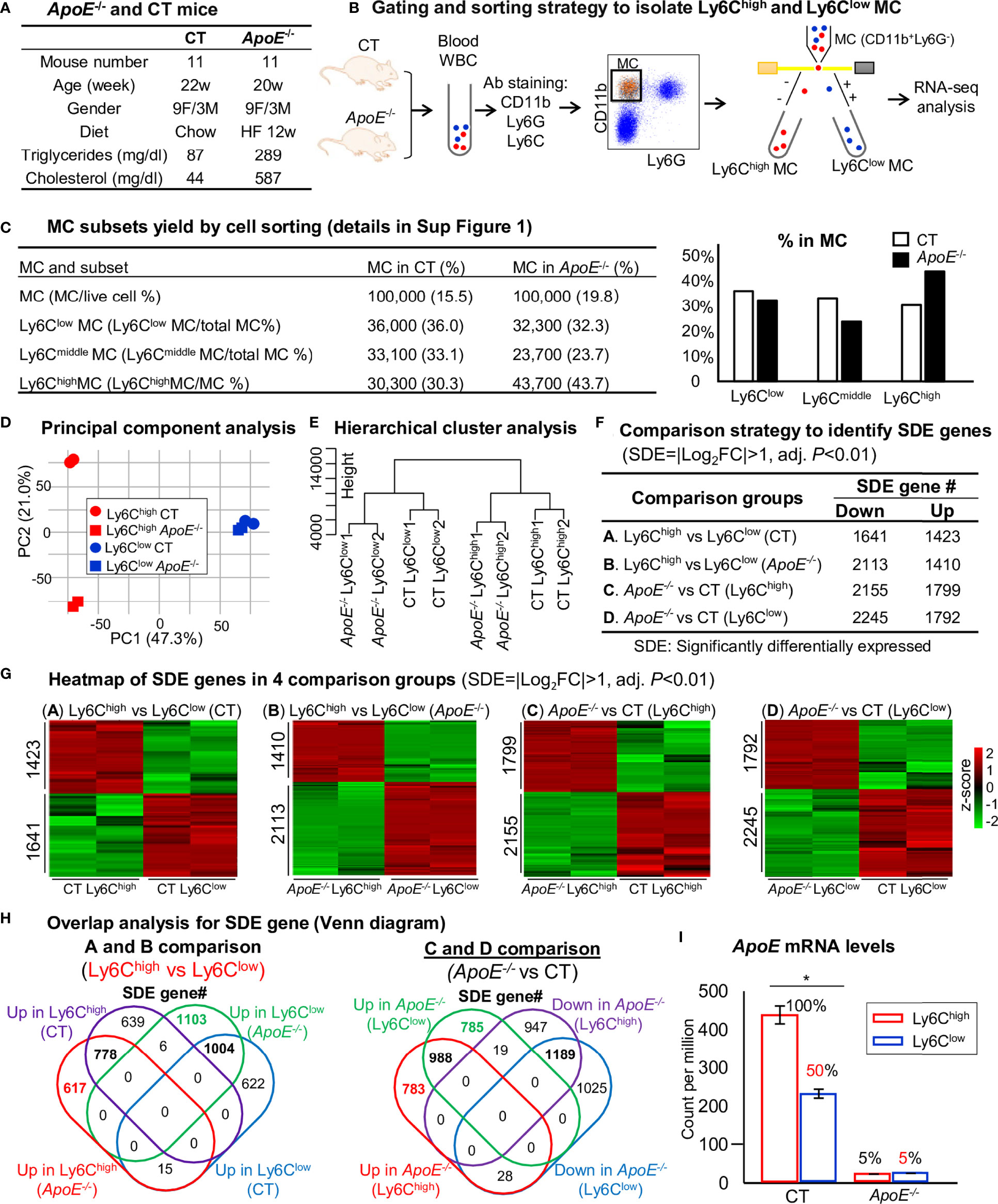
Figure 2 RNA-Seq analysis and SDE gene identification from blood Ly6Chigh and Ly6Clow MC of control and ApoE-/- mice. (A) ApoE-/- and CT mice. Eleven mice at the age of 20 weeks were used in each group. C57/BL6 mice on rodent chaw were used as CT. ApoE-/- mice were fed a high-fat diet for 8 weeks. (B) Gating and sorting strategy to isolate Ly6Chigh and Ly6Clow MC. Mouse white blood cell were prepared from peripheral blood, pooled, stained with antibody against CD11b, Ly6G and Ly6C and subjected for flow cytometry cell sorting. CD11b+Ly6G- cells were characterized as MC. MC subsets (CD11b+Ly6G-Ly6Chigh, and CD11b+Ly6G-Ly6Clow) were sorted and used for bulk RNA-seq analysis. (C) MC subsets yield by cell sorting. MC (100,000 cells) were sorted by flow cytometry with 43.7% Ly6Chigh, 23.7% Ly6Cmiddle and 32.3% Ly6Clow MC from ApoE-/- mice, and 30.3% Ly6Chigh, 33.1% Ly6Cmiddle and 36% Ly6Clow MC in CT mice. The detail of flow cytometry sorting cell subset was presented in Supplementary Figure 1. (D) Principal component analysis. PCA analysis incorporated 8 samples from 4 groups of MC subsets {Ly6Chigh (CT), Ly6Clow (CT), Ly6Chigh (ApoE-/-) and Ly6Clow (ApoE-/-)}. (E) Hierarchical cluster analysis. The similarity of gene expression between different samples is represented by the vertical distances on each branch of the dendrogram. Biological replicates show the highest degree of correlation within samples, represented by short vertical distances. (F) Comparison strategy to identified SDE genes. Four group comparisons (A–D) were performed. Down-regulated and up-regulated SDE genes were identified using the criteria of |Log2FC| more than 1 (2-FC) and adjusted P value less than 0.01. (G) Heatmap of SDE gene in 4 comparison groups. Heatmap shows the expression levels of the SDE gene in Ly6C MC. The color density indicates the average expression of a given gene normalized by z-score. (H) Overlap analysis for SDE gene. Venn diagram summarized the total SDE genes from four pairs of comparisons. Numbers depict the amount of SDE genes. (I) ApoE mRNA levels. MC, monocyte; CT, wild type; ApoE, Apolipoprotein E; FACS, fluorescent-activated cell sorting; PCA, principal component analysis; PC, principal component; SDE, significantly differentially expressed; FC, fold change. *P < 0.05.
By overlap analysis, we identified 778 upregulated and 1004 downregulated SDE gene in Ly6Chigh MC in CT and ApoE-/- mice (Figure 2H). HL in ApoE-/- mice induced 988 upregulated and 1189 downregulated SDE gene in both Ly6Chigh and Ly6Clow MC. Interestingly, ApoE mRNA levels were significantly reduced to 50% in Ly6Clow MC compared with that in Ly6Chigh MC in CT mice, and depleted to about 5% in ApoE-/- mice (Figure 2I).
Ly6Clow MC Engages More Adaptive Immune Function
We recognized top 10 up/down canonical pathways that were significantly enriched by top-down analysis using SDE genes identified from four comparison groups by using IPA software (Figures 3A–D). The top 2 up/down enrichment pathways from the GSEA study in each group are presented in Figure 3E. The top 20 upregulated or downregulated GSEA pathways were shown in Supplementary Table 1. The GSEA study showed that OXPHOS pathway activation and antigen-activated B cell receptor to second messengers’ generation pathway suppression were overlapped in Ly6Chigh MC from both CT and ApoE-/- mice. In addition, interferon (IFN) α/β signaling was upregulated and Gap junction assembly signaling was downregulated in Ly6Chigh MC from CT mice. Formation of fibrin clot cascade was upregulated and LAT2/NTAL/LAB on calcium mobilization signaling were downregulated in ApoE-/- Ly6Chigh MC. HL in ApoE-/- mice induced IL-12-stimulated JAK-STAT signaling and Hes hey pathway activation and G2 and ATRBRCA pathway suppression in Ly6Chigh MC. In Ly6Clow MC, ApoE-/- induced IFN-α/β signaling and P53 hypoxia pathway activation, and suppressed Tyrosine metabolism and Rho GTPases-related NADPH oxidase activation.
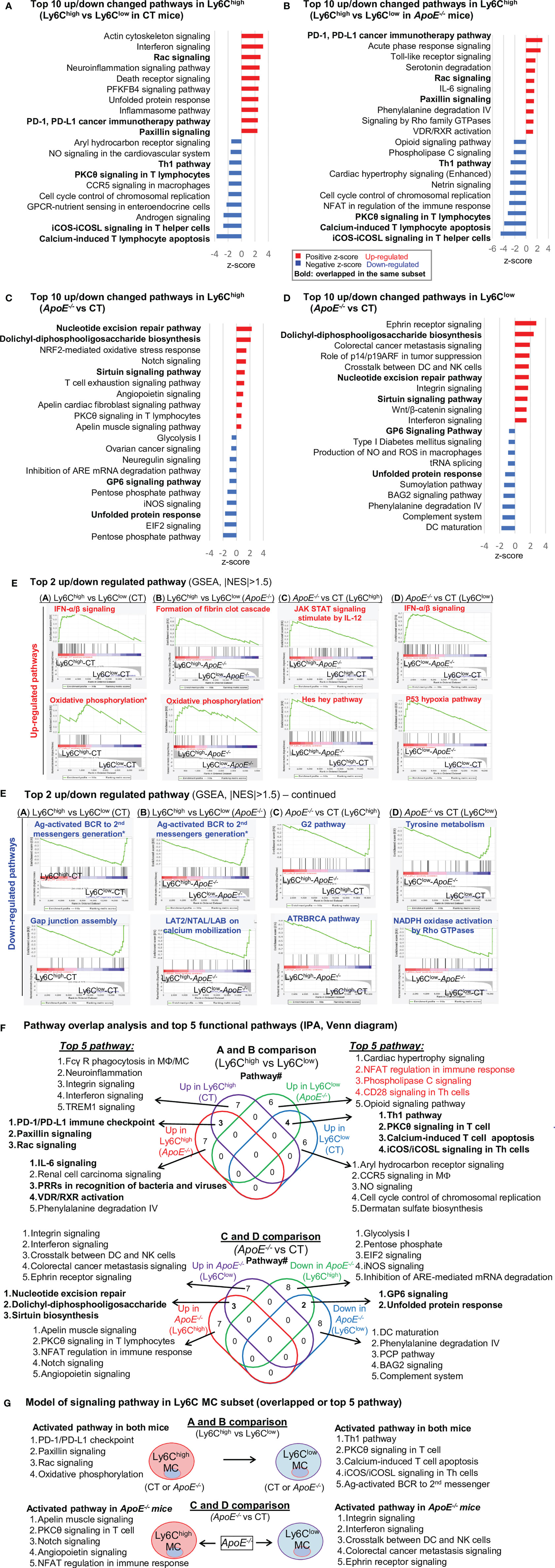
Figure 3 Canonical pathway analysis for SDE genes in four comparison groups (Top 10 up/down changed pathways), (A). CT Ly6Chigh vs CT Ly6Clow; (B). ApoE-/- Ly6Chigh vs ApoE-/- Ly6Clow; (C). Ly6Chigh (ApoE-/- vs CT); (D). Ly6Clow (ApoE-/- vs CT). Top up/down changed canonical pathways were identified by using IPA software with the criteria of adjusted P value<0.05. Blue bar indicates a negative z-score and down-regulated pathways. Red bar indicates a positive z-score and up-regulated pathways. Bold letters indicate overlapped pathway in the same subset. (E) Top 2 up/down regulated pathway. The most enriched significant pathways from GSEA study with threshold of |NES|>1.5 are marked (red=up-regulated, blue=down-regulated). The green curve corresponds to the enriched score. The y-axis indicates the enriched score. The x-axis displaces genes (vertical black lines) represented in their pathway gene set. Rainbow bands represent the corresponding enriched score of the genes (red for positive and blue for negative correlation). The top 20 upregulated or downregulated GSEA pathways were shown in Supplementary Table 1. (F) Pathway overlap analysis and top 5 functional pathways. Venn diagram summarized the overlap of top 10 pathway presented in (A–D) and listed the top 5 pathways in four pairs of comparisons. (G) Model of signaling pathway in Ly6C MC subset (overlapped or top 5 pathway). Four activated pathways in Ly6Chigh MC and 5 activated pathway in Ly6Clow MC were overlapped in A and B comparison. ApoE-/- induced top 5 activated pathway in both Ly6Chigh MC and Ly6Clow MC. MC, monocyte; MΦ, macrophage; TREM1, The triggering receptor expressed on myeloid cells 1; GPCRs, G-protein-coupled receptors; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4; SLE, Systemic Lupus Erythematosus, Th1, T helper 1; PKCθ, Protein Kinase C Theta; IL-7, Interleukin 7; NFAT, Nuclear factor of activated T-cells; CHK, Csk-homologous kinase; nNOS, neuronal nitric oxide synthase; PXR, pregnane X receptor; CAR, constitutive androstane receptor. NO, Nitric Oxide; ROS, Reactive Oxygen Species; PRRs, Pattern Recognition Receptors.
Through IPA pathway overlap analysis (Figure 3F), among the top 10 upregulated pathways from comparison A and B, 3 pathways (PD-1/PD-L1 checkpoint, paxillin, and Rac signaling) were overlapped in Ly6Chigh MC from both CT and ApoE-/- mice, and 7 were only activated in either mouse. ApoE-/- upregulated IL-6, Renal cell carcinoma, PRRs in recognition of bacteria and viruses, VDR/RXR activation, Phenylalanine degradation IV pathways in Ly6Chigh MC.
Among the top 10 downregulated pathways from comparison A and B, 4 pathways (Th1 pathway, PKCθ in T cell, calcium-induced T cell apoptosis and iCOS/iCOSL in Th cells) were overlapped in Ly6Chigh MC from both CT and ApoE-/- mice, and 7 were only suppressed in either mouse. ApoE-/- downregulated NFAT regulation in immune response, Phospholipase C and CD28 (Th cells) pathways in Ly6Clow MC.
From comparison C and D (Figure 3F), we identified ApoE-/- alter pathways in both subsets. ApoE-/- activated 3 pathways (nucleotide excision repair, dolichyl-diphosphooligosaccharide, sirtuin biosynthesis) and suppressed 2 pathways (GP6 and unfolded protein response) in both Ly6Chigh and Ly6Clow MC. In addition, ApoE-/- specifically activated 5 pathways (Apelin, PKCθ, NFAT regulation, notch and angiopoietin), and suppressed 5 pathways (glycolysis I, pentose phosphate, EIF2 and iNOS) only in Ly6Chigh MC. Whereas, ApoE-/- specifically activated 5 pathways (integrin, interferon, crosstalk between DC and NK cells, colorectal cancer metastasis, ephrin receptor) and suppressed 5 pathways (phenylalanine degradation IV, PCP, BAG2 and complement system) only in Ly6Clow MC. Finally, we established a model to describe specific pathways in the Ly6C MC subset based on IPA and GSEA pathway analysis (Figure 3G).
Fourteen Potential Transcriptional Axes Modulate Immunological Feature in Ly6C MC Subset From CT and ApoE-/- Mice
In the efforts to discover transcriptional signal determining immunological feature in Ly6C MC subset, we identified 414-upregulated/637-downregulated TF, 8-upregulated/18-downregulated MHC II, 25-upregulated/49-downregulated checkpoint, 64-upregulated/79-downregulated cytokine ligand, and 51-upregulated/54-downregulated cytokine receptor SDE genes from the 4 comparisons (details in Figure 4A).
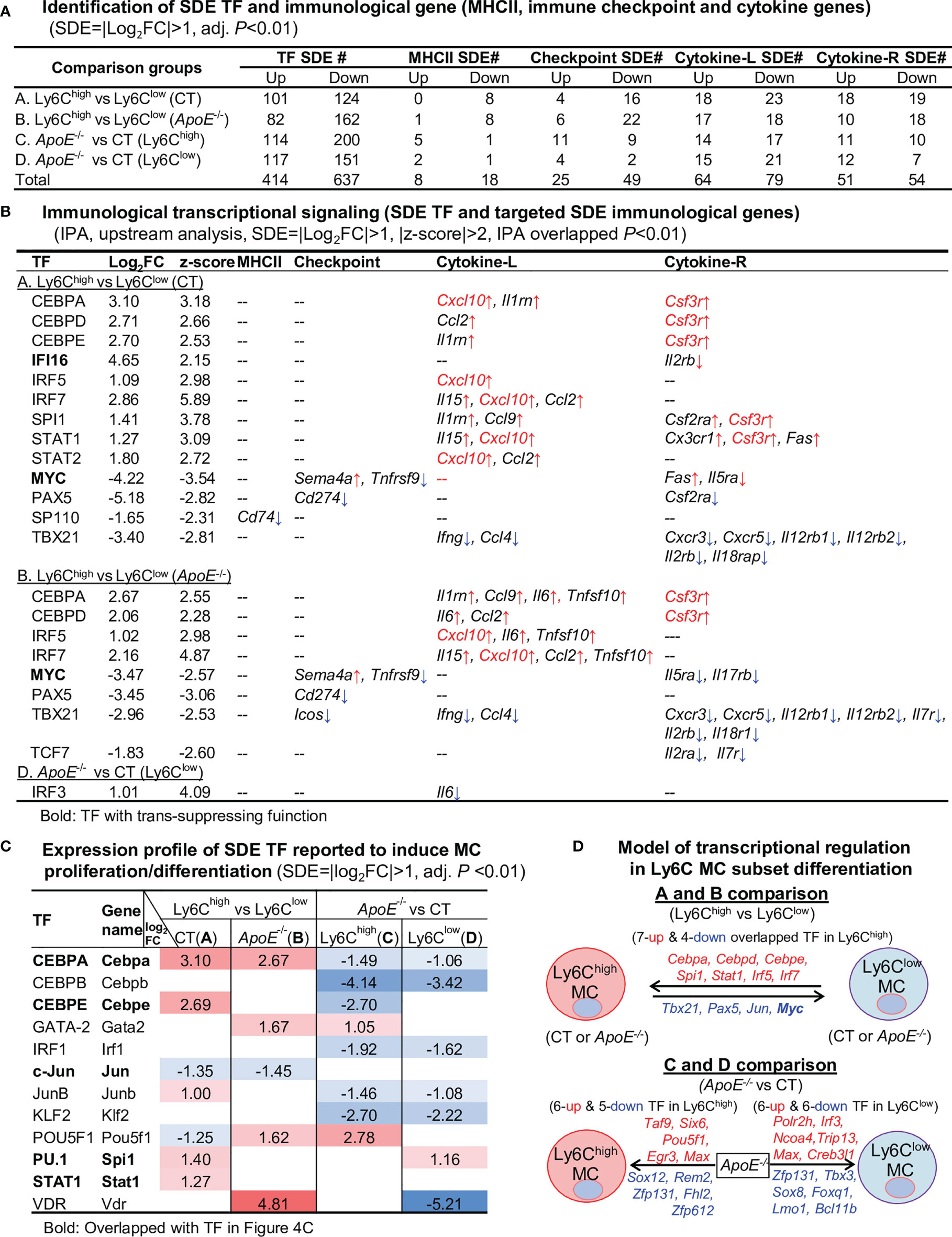
Figure 4 Identification of SDE TF, immunological gene and transcriptional regulatory models. (A) Identification of SDE TF and immunological genes. SDE TF (1150) and MHCII (28), immune checkpoint (82), cytokine ligand (135) and receptor (89) were identified using the criteria of |Log2FC|>1 (2-FC) and adjusted P<0.01. (B) Immunological transcriptional signaling. SDE immunological genes were matched with SDE TF by IPA upstream analysis. Transcriptional regulatory relationship between SDE TF and SDE immunological genes was justified by p-value<0.01 and |z-score|>2. The detailed list of SDE TF matching with the corresponding SDE gene is presented in Supplementary Table 2. ↑, upregulated; ↓, downregulated (C) Expression profile of SDE TF reported to induce MC proliferation/differentiation. Twelve SDE TF involved in MC generation are differentially expressed in four comparison groups in these subsets. Numbers with red-colored background indicate fold change>2 (log2FC>1). Numbers with blue-colored background indicate fold change<0.5 (log2FC<-1). The completed list of TF reported to induce MC proliferation/differentiation is in Supplementary Table 3. (D) Model of transcriptional regulation in Ly6C MC subset differentiation. Model describes potential transcriptional regulatory machinery. In A and B comparison, we identified 7-upregulated and 5-downregulated overlapped TF regulating Ly6C MC differentiation. In comparison C, 11 SDE TF (6 up and 5 down) are identified in ApoE-/- Ly6Chigh MC. While, in comparison D, 12 SDE TF (6 up and 6 down) are identified in ApoE-/- Ly6Clow MC. Red letter highlighted the representative up-regulated gene. Blue letter highlighted down-regulated genes.
By matching the immunological SDE genes with their corresponding upstream SDE TF by using IPA upstream analysis, we identified 12 trans-activating TF (activating transcription factor), one trans-suppressing TF (transcription repressors) IFI16, and one dual-functional TF MYC, which potentially regulate the identified SDE immunological genes transcription (Figure 4B). In A comparison, there are 8 upregulated SDE trans-activating TF (CEBPA, CEBPD, CEBPE, IRF5/7, PU.1 and STAT1/2) corresponding to 9 upregulated cytokine-L/R, and 3 downregulated SDE trans-activating TF (PAX5, SP110 and TBX21) corresponding to 10 downregulated immunological gene in CT Ly6Chigh MC. We also found one upregulated transcription repressors IFI16, which was associated with the downregulation of Il2rb. The downregulated dual functional TF MYC (21, 22) was associated with Tnfrsf9/Il5ra downregulation and Sema4a/Fas upregulation. In B comparison, 4 upregulated SDE trans-activating TF (CEBPA, CEBPD and IRF5/7) was associated with 8 upregulated cytokine/receptors in ApoE-/- Ly6Chigh MC. Three SDE trans-activating TF (PAX5, TBX21 and TCF21) were downregulated and corresponded to 12 downregulated immunological gene in ApoE-/- Ly6Chigh MC. TF MYC downregulation was associated with Tnfrsf9/Il5ra/Il17rb downregulation and Sema4a upregulation. In D comparison, we identified upregulated SDE trans-activating TF IRF3 corresponding to upregulated cytokine-L IL6 in ApoE-/- Ly6Chigh MC. The detailed list of SDE TF matching with the corresponding SDE gene is presented in Supplementary Table 2.
Among the matched SDE TF identified in Figure 4B, 12 were validated with conformed function in regulating MC proliferation/differentiation (Figure 4C, details in Supplementary Table 3). Apparently, these validated SDE TF were mostly upregulated in Ly6Chigh MC, and largely downregulated by ApoE-/- in both Ly6Chigh and Ly6Clow MC. We established models to describe potential transcriptional regulatory machinery in Ly6C MC subset differentiation in Figure 4D.
Immune Checkpoint Genes Present Balanced Pro-Inflammatory/Atherogenic Features in Ly6Chigh MC, and Anti-Inflammatory/Atherogenic Features Ly6Clow MC
To examine the differential role of Ly6C MC subsets in regulating adaptive immunity, we analyzed the expression pattern of immune checkpoint molecules. As depicted in Figure 5A, 27 out of 49 checkpoint pairs displayed differential expression in Ly6C MC subsets. In general, Ly6Chigh MC expressed relatively low levels of both co-stimulatory and co-inhibitory immune checkpoint receptors compared to Ly6Clow in both mice, except for TIM1. ApoE-/- upregulated immune checkpoint receptor GITR, SLAM, TIM2 and CD96 in Ly6Chigh and TIGIT in Ly6Clow MC subsets. We identified a trend of downregulation of some immune checkpoint ligands genes in Ly6Chigh compared to Ly6Clow MC in both mice. ApoE-/- upregulate immune checkpoint ligands SLAM, BTLA, CD160, CD155, PD-L1, PD-L2, CD155 and Galectin9 in Ly6Chigh and CD112, TL1A, PD-L2 and Galectin9 in Ly6Clow MC subsets.
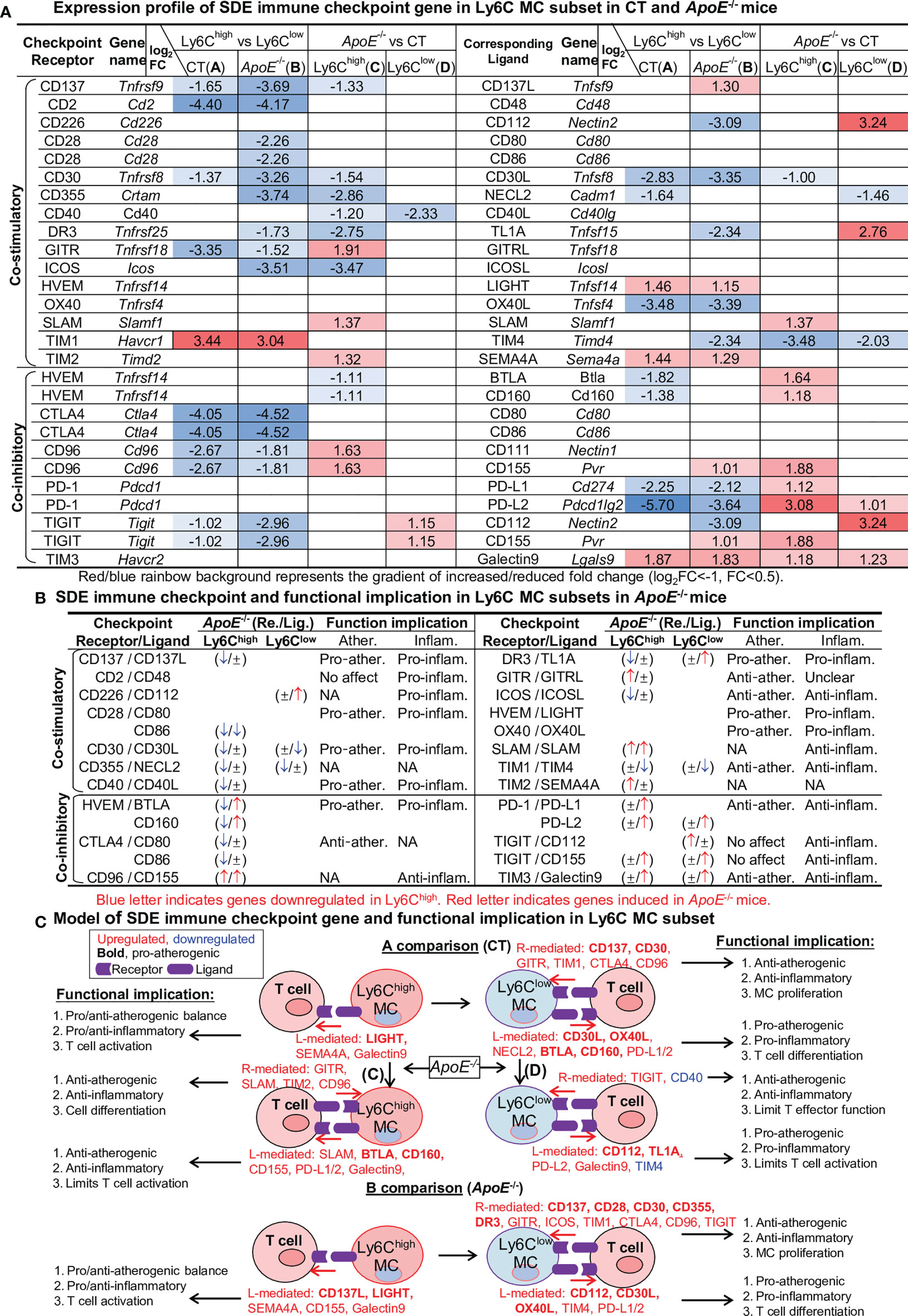
Figure 5 Identification of SDE immune checkpoint gene and function implication in Ly6C MC subset in CT and ApoE-/- mice. (A) Expression profile of SDE immune checkpoint gene. Sixteen pairs of SDE co-stimulatory and 11 pairs of SDE co-inhibitory molecules are identified in four comparison groups. (B) SDE immune checkpoint gene functional implication in Ly6C MC subsets in ApoE-/- mice. ↑ refers to induce expression by ApoE-/-. ↓ refers to reduce expression by ApoE-/-, ± refers to no changes in ApoE-/-. (C) Model of SDE immune checkpoint gene and functional implication. Red letter highlighted the representative up-regulated gene. Blue letter highlighted down-regulated genes. Bold letter emphasized pro-atherogenic function. NA, Not applicable.
Through an intensive literature search, we established the functional implication relevant to atherosclerosis and inflammation of these differentially expressed checkpoint genes in ApoE-/- Ly6Chigh and Ly6Clow MC (Figure 5B). Finally, we modeled this functional implication in Figure 5C.
Cytokine Expression Profile Endued Ly6Chigh MC With Pro-Inflammatory/Atherogenic Function and Ly6Clow MC With Anti-Atherogenic Function. ApoE-/- Confer Additional Pro-Atherogenic/Inflammatory Function
To test the differential role of Ly6C MC subsets in regulating inflammatory response, we further examined differential expression of cytokine ligand and receptor in MC subsets. The identified 58 SDE cytokine ligand and expression profile are presented in Figure 6A. The expression profile of 52 SDE cytokine receptors and their functional implication established through literature search are exhibited in Figure 6B. We modeled the inflammatory/atherogenic functional implication of cytokine in Ly6C MC subsets in Figure 6C.
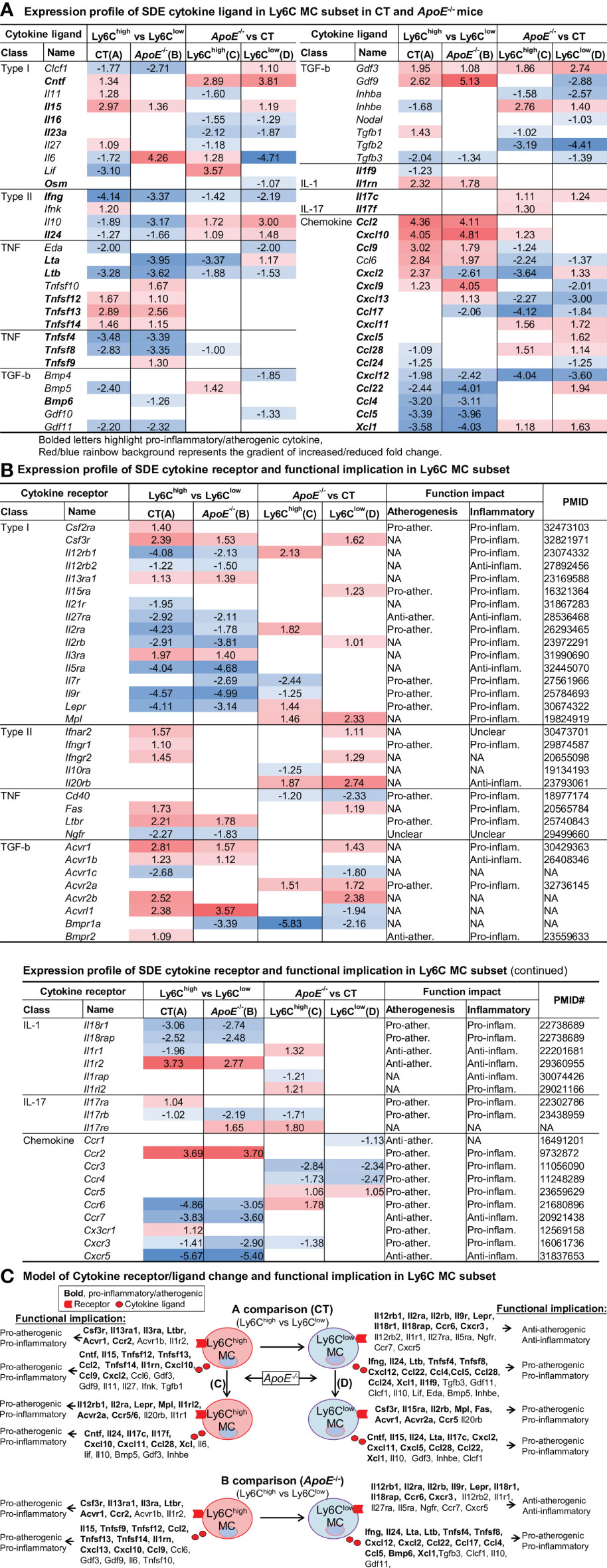
Figure 6 Identification of SDE cytokine gene and function implication in Ly6C MC subset in CT and ApoE-/- mice. (A) Expression profile of SDE cytokine ligand. Fifty-eight SDE cytokine ligand are identified in four comparison groups. (B) Expression profile of SDE cytokine receptor and function implication. Fifty-two SDE cytokine receptor are identified in four comparison groups. There function implication are summarized based on literature search (PMID cited). Red/blue rainbow background represents the gradient of increased/reduced fold change (log2FC>1 and log2FC<-1, respectively). (C) Model of cytokine receptor/ligand change and functional implication. Functional implication is conclude based on cytokine ligand and receptor change. Bold letter emphasized pro-inflammatory and pro-atherogenic function. NA, Not applicable.
We identified that Ly6Chigh MC expressed elevated pro-inflammatory/atherogenic cytokines (Ccl2, Tnfsf14, Il1rn, Cxcl10, Ccl9, Cxcl2), and receptors (Csf3r, Il13ra1, Il3ra, Ltbr, Acvr1 and Ccr2). Whereas Ly6Clow MC expressed high levels of anti-atherogenic function cytokine receptor (Il12rb2, Il1r1, Il27ra, Il5ra, Ngfr, Ccr7, Cxcr5), but produced pro-atherogenic function cytokine (Ifng, Il24, Ltb, Tnfsf4, Tnfsf8, Cxcl12, Ccl22, Ccl4, Ccl5 and Xcl1) in both mice. ApoE-/- induced pro-atherogenic and pro-inflammatory cytokines (Cntf, Il24, Il17c, Cxcl11, Ccl28, Xcl) and receptors (Mpl, Acvr2a, Ccr5) in Ly6Chigh and Ly6Clow MC.
APC Potential Is Higher in Ly6Clow MC and Elevated by ApoE-/- in Ly6Chigh and Ly6Clow MC
Expression profile of 11 SDE MHCII gene are identified through four comparisons (Figure 7A). Ly6Clow MC is more like APC because of their higher levels of MHCII genes (H2-Oa, H2-DMb2, H2-Ob, H2-Eb2, H2-Eb1, H2-Aa, Cd74) compared to Ly6Chigh MC in both mice. ApoE-/- elevated the expression levels of MHCII genes in Ly6Chigh MC (H2-Aa, H2-Ab1, CD74, H2-Dma and H2-Ea-ps) and in Ly6Clow MC (H2-Aa and H2-Ea-ps). APC functional feature of SDE MHCII gene is described in Figure 7B. We modeled APC features in relevant with MHCII gene change in Ly6C MC subsets in Figure 7C.
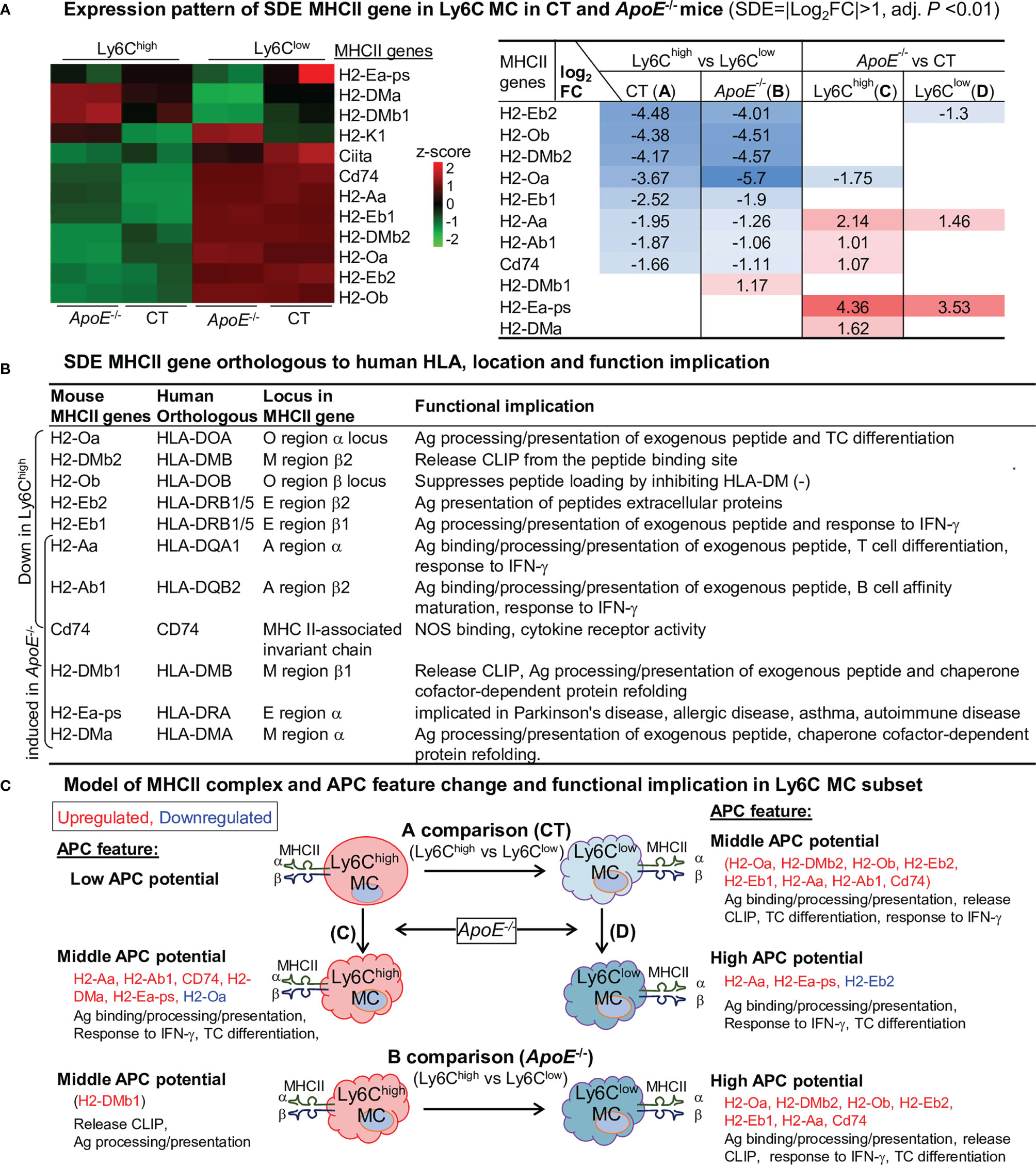
Figure 7 APC potential is higher in Ly6Clow MC and elevated by ApoE-/- in Ly6Clow and Ly6Clow MC. (A) Expression pattern of SDE MHCII gene in Ly6C MC in CT and ApoE-/- mice. Heatmap color density indicates the average expression level of a given gene normalized by z-score. Red/blue rainbow background in the table represents the gradient of increased/reduced fold change (log2FC>1, log2FC<-1). (B) SDE MHCII gene orthologous to human HLA, location and function implication. Identified mouse SDE MHCII gene orthologous to human corresponding HLA gene, gene locus and function implication were search from large database (https://www.alliancegenome.org). (C) Model of MHCII complex and APC feature and functional implication in Ly6C MC subset. Model outlines that eight SDE MHCII molecules were upregulated in Ly6Clow MC in both mice and that ApoE-/- further induced MHCII gene expression. Cloud-like cell shape indicade APC potential. MHCII, major histocompatibility complex class II; Ag, antigen; IFN-γ, Interferon gamma; CLIP, class II-associated invariant chain peptide; NOS, Nitrous Oxide Systems.
The Ly6Chigh MC displayed lower APC potential because of relatively lower levels of all MHCII gene expression. Ly6Clow MC had middle APC potential and expressed higher levels of MHCII genes. Most of the MHCII genes in both MC subsets were elevated in ApoE-/- mice. HL promoted Ag binding/processing/presentation, release CLIP in both subsets.
Summarized Functional Feature of the Innate-Adaptive Immunological Interplay of Ly6C MC in CT and ApoE-/- Mice
We described the differential functional feature of Ly6C MC subsets in innate-adaptive immunological interplay as 4 aspects: 1. Ag recognition (signal 1), 2. immune checkpoint (signal 2), 3. cytokine stimulation (signal 3), and 4. receptor-mediated innate immune cell regulation (Figure 8A). We emphasized APC potential for Ag recognition signal, and focused on 3 function aspects for signals 2 and 3 (pro/anti-atherogenic, pro/anti-inflammatory, MC proliferation and T cell activation) modulated by immune checkpoint and cytokine.
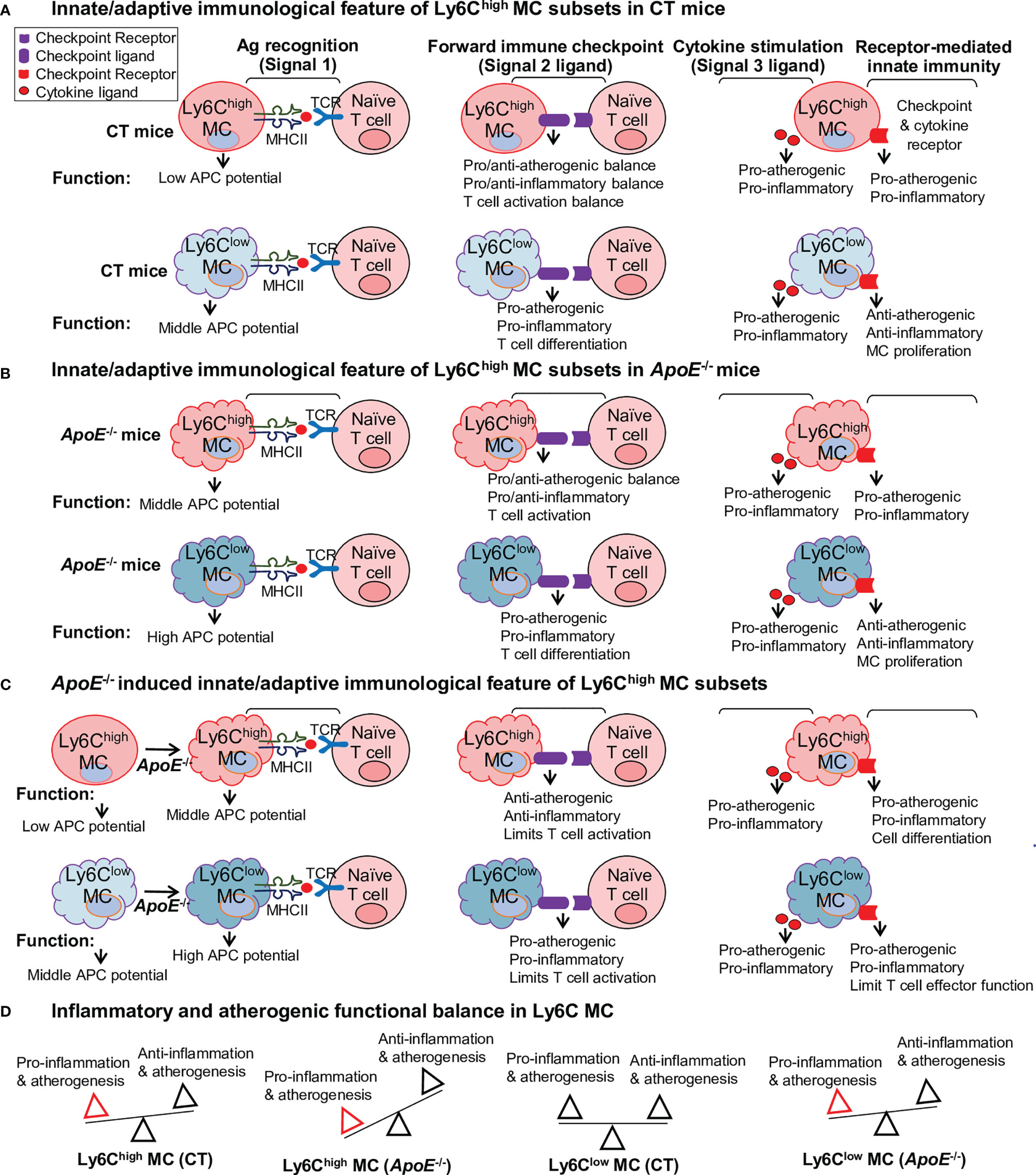
Figure 8 Summarized functional feature of innate-adaptive immunological interplay of Ly6C MC in CT and ApoE-/- mice. Model summarized functional changes derived from molecule signaling presented in Figures 5–7. (A) Innate/adaptive immunological feature of Ly6Chigh MC subsets in CT mice. (B) Innate/adaptive immunological feature of Ly6Chigh MC subsets in ApoE-/- mice. (C) ApoE-/–induced innate/adaptive immunological feature of Ly6Chigh MC subsets. (D) Inflammatory and atherogenic functional balance in Ly6C MC. APC, antigen presenting cell; TCR, T cell receptor.
The CT Ly6Chigh MC presented low APC potential based on their relatively low MHCII molecular expression. Their ligand expression profile implicated a more pro-atherogenic/inflammatory and T cell activation function toward the adaptive immune cells. Their receptor differential expression indicated pro-atherogenic/inflammatory on their own innate immunological function (Figure 8A). The CT Ly6Clow MC displayed middle APC potential based on their relatively higher MHCII molecular expression.
Their ligand expression profile implicated a more pro-atherogenic/inflammatory and T cell differentiation function toward adaptive immune cell. Differently, the receptor differential expression in CT Ly6Clow MC endowed them with anti-atherogenic/inflammatory and MC proliferation function. When comparing Ly6Chigh MC with Ly6Clow MC in the ApoE-/- mice, we found that APC potential was elevated, but other ligand/receptor-determined immunological features were similar as that in CT mice in both MC subsets (Figure 8B).
These innate-adaptive immunological functional features were altered by HL condition. ApoE-/- further enhanced APC potential and pro-inflammatory/atherogenic function in both MC subsets when compared with the same MC subset in CT mice (Figure 8C). Differently, ApoE-/- limited ligand-related T cell activation in Ly6Chigh MC and switched the Ly6Clow MC from an anti-atherogenic/inflammatory to a pro-atherogenic/inflammatory feature. We modeled pro-inflammatory/atherogenic balance in Ly6C MC subset from ApoE-/- and CT mice in Figure 8D. In summary, Ly6Chigh MC has pro-inflammatory/atherogenic in CT mice. This balance is further inclined towards pro-inflammatory/atherogenic direction in ApoE-/- mice. Ly6Clow MC has a balanced inflammatory/atherogenic feature, which is disturbed in ApoE-/- mice and becomes pro-inflammatory/atherogenic.
Discussion
Inflammatory MC subset differentiation is a major feature in tissue inflammation and atherosclerosis. However, the underlying molecular mechanism remains unclear. This study established transcription profiles of flow cytometry sorted Ly6Chigh and Ly6Clow MC subsets from CT and ApoE-/- mice and explored molecule targets and signaling which potentially determinate immunological features in MC subsets by performing intensive bioinformatic analysis and literature integration. We have 6 major findings: 1) Ly6Clow MC engaged more adaptive immune function. 2) Fourteen potential transcriptional axes modulated immunological features in Ly6C MC subset from CT and ApoE-/- mice. 3) Immune checkpoint and cytokine gene profile conferred upon Ly6Chigh MC pro-inflammatory/atherogenic features and Ly6Clow MC anti-inflammatory/atherogenic features. 4) ApoE-/- promoted pro-atherogenic/inflammatory function in both subsets. 5) APC potential is higher in Ly6Clow MC and elevated by ApoE-/- in Ly6Chigh and Ly6Clow MC. 6) We established 4 groups of hypothetic molecular signaling models to summarize the regulation of innate and adaptive immunological features in Ly6C MC from both mice. Our findings provided potential regulatory molecular mechanisms in MC subsets delineated innate-adaptive immune response and important guidance for future investigation in this direction.
Previous studies suggested a group of TF regulating MC differentiation from progenitor cells, such as PU.1, CEBPα/β/ε, IRF1/4/8, c-Jun, JunB, STAT1/3 and VDR (7, 8, 23). Transcriptional mechanism has not been clearly addressed for MC subset differentiation. We recently established transcriptome of flow cytometry-sorted Ly6C MC subsets from CT and HHcy Cbs−/− mice and discovered 9 upregulated TF (Cebpa, Cebpd, Cebpe, Irf5/7, Ifi16, Spi1, and Stat1/2) and 6 downregulated TF (Neurod4, Asb2, Sox5, Pou2af1, Pax5 and Tbx21) in Ly6Chigh MC from CT and HHcy mice (13). We proposed that these 15 TF are potentially involved in Ly6Chigh (9 TF) and Ly6Clow (6 TF) MC differentiation (13). We confirmed that TF CEBPα binds to the Ly6c promoter and that its expression was elevated and synergistically increased in HHcy and Type 2 Diabetes Mellitus mice, supported the note that TF CEBPα transactivates Ly6c gene and mediates inflammatory MC subset differentiation (3). Others evidence also supported the regulatory role of TF Irf8 in Ly6Chigh MC generation using Irf8−/− mice, and TF Cebpb/Irf5 in Ly6Clow MC generation using Cebpb-/- or Irf5−/− mice (24–27).
In this study, we were the first to explore systemic transcriptional regulatory mechanisms in the MC subset in the HL condition. Based on information present in Figure 4, we concluded that 12 transcriptional axes potentially modulate immunological features in Ly6Chigh MC subset. Six trans-activating axis CEBPA-Cxcl10/Il1rn/Csf3r, CEBPD-Ccl2/Csf3r, CEBPE-Il1rn/Csf3r, SPI1-Il1rn/Ccl9/Csf2ra/Csf3r, STAT1-Il15/Cxcl10/Cx3cr1/Csd3r/Fas, IRF5-Cxcl10/Il6/Tnfsf10, IRF7- Il15/Cxcl10/Ccl2, and one trans-suppressing IFI16 -Csf3r may be involved in Ly6Chigh MC differentiation in CT mice. Two trans-activating axis TBX21-Ifng/Ccl4/Cxcr3/Cxcr5/Il12rb1/Il12rb2/Il2rb/Il7r/Il18rap and Pax5-Cd274/Csf2ra, and one dual-functional TF Myc-Sema4a↑/Tnfrsf9↓/Fas↑/Il5ra↓/Il17rb↓ are potentially involved in Ly6Clow MC differentiation. Interestingly, ApoE-/- induced IRF3 by 2-folds which was associated with a 3-folds induction of its downstream target Il6.
ApoE-/- mice develop HL similar as that in humans and are an ideal model for atherosclerosis research. We and others reported that inflammatory Ly6Cmiddle+high MC subsets were elevated in early and advanced atherosclerosis lesion, which promoted vascular inflammation (3, 4, 17). Transcriptional molecular processes identified in this study provide models for future investigation.
Very limited information is available to review mechanisms underlying MC subset differentiation. It was suggested that PKCθ may suppress Ly6Clow MC in ApoE-/- mice (28), and that cell-intrinsic Notch2 and TLR7-Myd88 pathways may promote Ly6Clow MC development from Ly6Chigh MC under inflammatory conditions (29). Our laboratory is the first to systemically investigate signal pathways mediating MC subset differentiation. In this study, we established a model to summarize top canonical pathways involved in Ly6C MC subset differentiation and responded to ApoE-/- identified by using two bioinformatics tools (IPA and GSEA) (Figure 3G). Four pathways (PD-1/PD-L1 checkpoint, paxillin, Rac and OXPHOS) are activated in Ly6Chigh MC from both ApoE-/- and CT mice, and are potentially involved in Ly6Chigh MC differentiation. Five pathways (Th1, PKCθ, calcium-induced T cell apoptosis, iCOS/iCOSL signaling in Th cells and Ag-activated B cell receptor to 2nd messenger) are activated in Ly6Clow MC from both mice, and may contribute to Ly6Clow MC differentiation. In addition, ApoE-/- activated 5 pathways (apelin muscle, PKCθ in T cell, notch, angiopoietin signaling and NFAT regulation) in Ly6Chigh MC, and 5 pathways (integrin, interferon, crosstalk between DC and NK cells, colorectal cancer metastasis and ephrin receptor) in ly6Clow MC, which may be responsible for subsequent function changes.
Recently, through similarly transcriptome analysis approaches, we demonstrated that interferon, inflammasome and PD-1/PD-L1 checkpoint pathways were activated in Ly6Chigh MC, and calcium-induced T cell apoptosis and iCOS/iCOSL signaling in Th cells were activated in Ly6Clow MC from both CT and HHcy Cbs-/- mice, and that Cbs-/- activated NK cell signaling in Ly6Chigh and suppressed xenobiotic metabolism and melatonin degradation in Ly6Clow MC (13). Thus far, we identified 3 canonical pathways shared by Ly6C MC subset from CT, ApoE-/-, and Cbs-/- mice. These include PD-1/PD-L1 checkpoint pathway activation in Ly6Chigh MC, calcium-induced T cell apoptosis and iCOS/iCOSL signaling activation in Ly6Clow MC from all three mouse lines.
Interestingly, we found that IFN-α/β signaling was activated in Ly6Chigh MC from CT mice, and also activated by ApoE-/- in Ly6Clow MC suggesting that IFN-α/β signaling represent inflammatory feature in CT Ly6Chigh and ApoE-/- Ly6Clow MC. We also observed a downregulated Ag-activated B cell receptor to 2nd messenger pathway in Ly6Chigh MC from CT and ApoE-/- mice, supporting the hypothesis that Ly6Chigh MC has a lower potential for adaptive immune cell activation.
It is known that MC enables adaptive immunity through its APC potential (9). It was reported that circulating Ly6Chigh MC, and not LY6Clow MC, can enter tissues and differentiate into MHCII+ MC through interaction with the endothelium (15). Enhanced expression of MHC class II in the Ly6Chigh MC was associated with inflammatory MC differentiation into mature APC that can activate T cells (30). In the experimental model of multiple sclerosis, Ly6Chigh MC migrated into the central nervous system and further differentiated into APC during disease progression (31). Despite the above-mentioned studies, the APC potential and regulatory mechanism of MC subsets have not been systemically investigated. This study, for the first time, demonstrated that the Ly6Chigh MC displayed lower APC potential because of relatively lower levels of all MHCII gene expression, that Ly6Clow MC had middle APC potential and expressed higher levels of MHCII genes, and that ApoE-/- elevated most of MHCII genes in both MC subsets. We hypothesize that the higher APC potential in Ly6Clow MC may ignite protective adaptive immune response and that ApoE-/- elevated APC potential in both MC subsets may be related to inflammatory response.
The immune checkpoint is a delicate process that controls the direction towards immune suppression or activation. It was reported that LIGHT enhanced proliferating Ly6Chigh MC and increased atherosclerosis lesion size in ApoE-/-Irs2+/-HL-/- mice (32). Our current study displayed a systemic expression profile of immune checkpoints in the MC subset and explored their potential role in regulating atherosclerosis based on literature information. We proposed that elevated co-stimulatory checkpoint receptor LIGHT expression in Ly6Chigh MC may lead to pro-inflammatory/atherogenic reaction, that elevated co-stimulatory checkpoint ligand (CD30L and OX40L), co-inhibitory checkpoint ligand (BTLA and CD160) in Ly6Clow MC may induce pro-atherogenic function, and that elevated co-stimulatory checkpoint receptor (GITR and TIM1) and co-inhibitory (CTLA4 and CD96) may contribute to anti-atherogenic function in Ly6Clow MC.
MC and MΦ can secrete inflammatory or anti-inflammatory cytokines which play a critical role in the pathogenesis. We found that elevated cytokines (Cxcl10, Ccl2, Tnfsf14, Il1rn, Ccl9, and Cxcl2), and cytokine receptors (Ccr2, Csf3r, Il13ra1, Il3ra, Ltbr, and Acvr1) expression in Ly6Chigh MC may be associated with their pro-inflammatory/atherogenic function. In contrast, in Ly6Clow MC, elevated cytokine receptors (Il12rb2, Il1r1, Il27ra, Il5ra, Ngfr, Ccr7 and Cxcr5) may be related to their anti-inflammatory/atherogenic function toward MC. In addition, elevated expression of pro-inflammatory/atherogenic cytokine ligand (Ifng, Il24, Ltb, Tnfsf4, Tnfsf8, Cxcl12, Ccl22, Ccl4, Ccl5 and Xcl1) in Ly6Clow MC may influence immunological balance in other cells. Consistent with our finding, CXCL10 was shown essential for MC pro-inflammatory function (33). ApoE-/-Cxcl10-/- mice presented increased aortic size and a higher incidence of death due to aortic rupture (34). Many studies in both humans and animals have shown the importance of MC chemoattractant protein-1 (MCP-1, also named CCL2) and its receptor CCR2 in pathologies, such as atherosclerosis (35). the CC genotype of MCP-1 SNP rs2857656, independently or in combination with CCR2 V64I genotype, is associated with a high prevalence of carotid artery plaque (36).
Interestingly, we found that ApoE-/- elevated pro-inflammatory Ly6Chigh MC and reduced anti-inflammatory Ly6Clow MC subsets, and ApoE-/- promoted Ly6Chigh and Ly6Clow MC to express higher levels of additional pro-atherogenic and pro-inflammatory cytokines (Cxcl11, Cntf, Il24, and Xcl1) and receptor (Ccr5, Mpl, and Acvr2a). These might be the molecular bases underlying HL-induced atherosclerosis. Pro-inflammatory chemokines CXCL11 was increased in patients with severe transplant coronary artery disease (37). CCR5 has been suggested as a predictor of atherosclerosis progression (38). A CCR5 antagonist (111In-DOTA-DAPTA) was tested as an inflammation imaging tracer for atherosclerosis in mice (39). Bone morphogenetic proteins receptor Acvr2a was reported to induce osteogenic differentiation and MC infiltration in atherosclerosis (40). Mouse MC subset cytokine transcriptome profile provided systemic molecular details and a potential mechanism for MC-related inflammatory reaction.
The current study provided multiple insightful molecular signaling model system for immunological features responsible for pro-inflammatory/atherogenic or anti-inflammatory function of different MC subset. However, future studies are needed to validate these molecular signature and signal pathway changes and to confirm their role in disease condition. The experimental condition of using 20 weeks old ApoE-/- mice fed a high-fat diet for 12 weeks may middle stage of atherosclerotic condition. Immunological molecular change may alter in earlier or late stage of atherosclerosis advanced lesion at different time points.
Conclusion
Ly6Chigh MC presented pro-inflammatory/atherogenic features and lower APC potential. Ly6Clow MC displayed anti-inflammatory/atherogenic features and higher APC potential. ApoE-/- confers upon both subsets with augmented pro-atherogenic/inflammatory function and APC potential. Our study presented the expression profile of immunological genes in MC subsets, and shaded light on new strategies for the blocking of key immunological genes and signaling as therapeutic targets for treating inflammatory diseases, including atherosclerosis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/GSE189097.
Ethics Statement
The animal study was reviewed and approved by Temple University, Institutional Animal Care & Use Committee.
Author Contributions
PY analyzed the data, conceived all figures, and prepared manuscript. QW afforded strong intellectual, data analysis and manuscript support. LS participated in some part of data analysis and manuscript preparation. PF isolated MC subsets from mice, designed RNA-Seq analysis, and provided editing assistance. LL conducted the bioinformatics analyses. J-YP and XQ participated in some of data analysis and provided editing assistance. XY provided strong intellectual and data analysis support. HW designed the study, supervised the project, and prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Institutes of Health (NIH) grants HL82774, HL-110764, HL130233, HL131460, DK104114, DK113775, and HL131460 to HW.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.809208/full#supplementary-material
References
1. Hansson GK, Hermansson A. The Immune System in Atherosclerosis. Nat Immunol (2011) 12:204–12. doi: 10.1038/ni.2001
2. Zhang J, Zu Y, Dhanasekara CS, Li J, Wu D, Fan Z, et al. Detection and Treatment of Atherosclerosis Using Nanoparticles. Wiley Interdisciplinary Reviews. Nanomed Nanobiotechnol (2017) 9:10.1002/wnan.1412. doi: 10.1002/wnan.1412
3. Fang P, Li X, Shan H, Saredy JJ, Cueto R, Xia J, et al. Ly6c(+) Inflammatory Monocyte Differentiation Partially Mediates Hyperhomocysteinemia-Induced Vascular Dysfunction in Type 2 Diabetic Db/Db Mice. Arterioscler Thromb Vasc Biol (2019) 39:2097–119. doi: 10.1161/ATVBAHA.119.313138
4. Zhang D, Jiang X, Fang P, Yan Y, Song J, Gupta S, et al. Hyperhomocysteinemia Promotes Inflammatory Monocyte Generation and Accelerates Atherosclerosis in Transgenic Cystathionine Beta-Synthase-Deficient Mice. Circulation (2009) 120:1893–902. doi: 10.1161/CIRCULATIONAHA.109.866889
5. Sun L, Yang X, Yuan Z, Wang H. Metabolic Reprogramming in Immune Response and Tissue Inflammation. Arterioscler Thromb Vasc Biol (2020) 40:1990–2001. doi: 10.1161/ATVBAHA.120.314037
6. Pastrana JL, Sha X, Virtue A, Mai J, Cueto R, Lee IA, et al. Regulatory T Cells and Atherosclerosis. J Clin Exp Cardiol (2012) 2012:2. doi: 10.4172/2155-9880.S12-002
7. Fang P, Li X, Dai J, Cole L, Camacho JA, Zhang Y, et al. Immune Cell Subset Differentiation and Tissue Inflammation. J Hematol Oncol (2018) 11:97. doi: 10.1186/s13045-018-0637-x
8. Yang J, Zhang L, Yu C, Yang XF, Wang H. Monocyte and Macrophage Differentiation: Circulation Inflammatory Monocyte as Biomarker for Inflammatory Diseases. Biomark Res (2014) 2:1. doi: 10.1186/2050-7771-2-1
9. Jakubzick CV, Randolph GJ, Henson PM. Monocyte Differentiation and Antigen-Presenting Functions. Nat Rev Immunol (2017) 17:349–62. doi: 10.1038/nri.2017.28
10. Dai J, Fang P, Saredy J, Xi H, Ramon C, Yang W, et al. Metabolism-Associated Danger Signal-Induced Immune Response and Reverse Immune Checkpoint-Activated Cd40(+) Monocyte Differentiation. J Hematol Oncol (2017) 10:141. doi: 10.1186/s13045-017-0504-1
11. Tang T, Cheng X, Truong B, Sun L, Yang X, Wang H. Molecular Basis and Therapeutic Implications of Cd40/Cd40l Immune Checkpoint. Pharmacol Ther (2021) 219:107709. doi: 10.1016/j.pharmthera.2020.107709
12. Iwasaki H, Akashi K. Myeloid Lineage Commitment From the Hematopoietic Stem Cell. Immunity (2007) 26:726–40. doi: 10.1016/j.immuni.2007.06.004
13. Yang P, Liu L, Sun L, Fang P, Snyder N, Saredy J, et al. Immunological Feature and Transcriptional Signaling of Ly6c Monocyte Subsets From Transcriptome Analysis in Control and Hyperhomocysteinemic Mice. Front Immunol (2021) 12:632333. doi: 10.3389/fimmu.2021.632333
14. Vasamsetti SB, Karnewar S, Kanugula AK, Thatipalli AR, Kumar JM, Kotamraju S. Metformin Inhibits Monocyte-to-Macrophage Differentiation via Ampk-Mediated Inhibition of Stat3 Activation: Potential Role in Atherosclerosis. Diabetes (2015) 64:2028–41. doi: 10.2337/db14-1225
15. Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal Differentiation of Classical Monocytes as They Survey Steady-State Tissues and Transport Antigen to Lymph Nodes. Immunity (2013) 39:599–610. doi: 10.1016/j.immuni.2013.08.007
16. Sun L, Wang X, Saredy J, Yuan Z, Yang X, Wang H. Innate-Adaptive Immunity Interplay and Redox Regulation in Immune Response. Redox Biol (2020) 37:101759. doi: 10.1016/j.redox.2020.101759
17. Zhang D, Fang P, Jiang X, Nelson J, Moore JK, Kruger WD, et al. Severe Hyperhomocysteinemia Promotes Bone Marrow-Derived and Resident Inflammatory Monocyte Differentiation and Atherosclerosis in Ldlr/Cbs-Deficient Mice. Circ Res (2012) 111:37–49. doi: 10.1161/CIRCRESAHA.112.269472
18. Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, et al. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ Res (2018) 122:1675–88. doi: 10.1161/CIRCRESAHA.117.312513
19. Koltsova EK, Garcia Z, Chodaczek G, Landau M, McArdle S, Scott SR, et al. Dynamic T Cell-Apc Interactions Sustain Chronic Inflammation in Atherosclerosis. J Clin Invest (2012) 122:3114–26. doi: 10.1172/JCI61758
20. Fang P, Zhang D, Cheng Z, Yan C, Jiang X, Kruger WD, et al. Hyperhomocysteinemia Potentiates Hyperglycemia-Induced Inflammatory Monocyte Differentiation and Atherosclerosis. Diabetes (2014) 63:4275–90. doi: 10.2337/db14-0809
21. Nguyen HV, Vandenberg CJ, Ng AP, Robati MR, Anstee NS, Rimes J, et al. Development and Survival of Myc-Driven Lymphomas Require the Myc Antagonist Mnt to Curb Myc-Induced Apoptosis. Blood (2020) 135:1019–31. doi: 10.1182/blood.2019003014
22. Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, Eisenman RN. Genomic Binding and Transcriptional Regulation by the Drosophila Myc and Mnt Transcription Factors. Cold Spring Harbor Symp Quant Biol (2005) 70:299–307. doi: 10.1101/sqb.2005.70.019
23. Huber R, Pietsch D, Gunther J, Welz B, Vogt N, Brand K. Regulation of Monocyte Differentiation by Specific Signaling Modules and Associated Transcription Factor Networks. Cell Mol Life Sci CMLS (2014) 71:63–92. doi: 10.1007/s00018-013-1322-4
24. Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, et al. Essential Role of the Irf8-Klf4 Transcription Factor Cascade in Murine Monocyte Differentiation. Blood (2013) 121:1839–49. doi: 10.1182/blood-2012-06-437863
25. Corbin AL, Gomez-Vazquez M, Berthold DL, Attar M, Arnold IC, Powrie FM, et al. Irf5 Guides Monocytes Toward an Inflammatory Cd11c(+) Macrophage Phenotype and Promotes Intestinal Inflammation. Sci Immunol (2020) 5:eaax6085. doi: 10.1126/sciimmunol.aax6085
26. Mildner A, Schonheit J, Giladi A, David E, Lara-Astiaso D, Lorenzo-Vivas E, et al. Genomic Characterization of Murine Monocytes Reveals C/Ebpbeta Transcription Factor Dependence of Ly6c(-) Cells. Immunity (2017) 46:849–862 e847. doi: 10.1016/j.immuni.2017.04.018
27. Tamura A, Hirai H, Yokota A, Kamio N, Sato A, Shoji T, et al. C/ebpbeta Is Required for Survival of Ly6c(-) Monocytes. Blood (2017) 130:1809–18. doi: 10.1182/blood-2017-03-772962
28. Raghavan S, Singh NK, Gali S, Mani AM, Rao GN. Protein Kinase Ctheta via Activating Transcription Factor 2-Mediated Cd36 Expression and Foam Cell Formation of Ly6c(Hi) Cells Contributes to Atherosclerosis. Circulation (2018) 138:2395–412. doi: 10.1161/CIRCULATIONAHA.118.034083
29. Gamrekelashvili J, Kapanadze T, Sablotny S, Ratiu C, Dastagir K, Lochner M, et al. Notch and Tlr Signaling Coordinate Monocyte Cell Fate and Inflammation. eLife (2020) 9:e57007. doi: 10.7554/eLife.57007
30. Matsuoka Y, Kawauchi Y, Kuroda Y, Kawauchi K, Kojima N. In Vitro Uptake of Oligomannose-Coated Liposomes Leads to Differentiation of Inflammatory Monocytes Into Mature Antigen-Presenting Cells That Can Activate T Cells. Int Immunopharmacol (2018) 57:102–11. doi: 10.1016/j.intimp.2018.02.013
31. Monaghan KL, Zheng W, Hu G, Wan ECK. Monocytes and Monocyte-Derived Antigen-Presenting Cells Have Distinct Gene Signatures in Experimental Model of Multiple Sclerosis. Front Immunol (2019) 10:2779. doi: 10.3389/fimmu.2019.02779
32. Andres-Blasco I, Vinue A, Herrero-Cervera A, Martinez-Hervas S, Nunez L, Piqueras L, et al. Hepatic Lipase Inactivation Decreases Atherosclerosis in Insulin Resistance by Reducing Light/Lymphotoxin Beta-Receptor Pathway. Thromb Haemostasis (2016) 116:379–93. doi: 10.1160/TH15-10-0773
33. Zhao Q, Kim T, Pang J, Sun W, Yang X, Wang J, et al. A Novel Function of Cxcl10 in Mediating Monocyte Production of Proinflammatory Cytokines. J Leuk Biol (2017) 102:1271–80. doi: 10.1189/jlb.5A0717-302
34. King VL, Lin AY, Kristo F, Anderson TJ, Ahluwalia N, Hardy GJ, et al. Interferon-Gamma and the Interferon-Inducible Chemokine Cxcl10 Protect Against Aneurysm Formation and Rupture. Circulation (2009) 119:426–35. doi: 10.1161/CIRCULATIONAHA.108.785949
35. Franca CN, Izar MCO, Hortencio MNS, do Amaral JB, Ferreira CES, Tuleta ID, et al. Monocyte Subtypes and the Ccr2 Chemokine Receptor in Cardiovascular Disease. Clin Sci (2017) 131:1215–24. doi: 10.1042/CS20170009
36. Nyquist PA, Winkler CA, McKenzie LM, Yanek LR, Becker LC, Becker DM. Single Nucleotide Polymorphisms in Monocyte Chemoattractant Protein-1 and its Receptor Act Synergistically to Increase the Risk of Carotid Atherosclerosis. Cerebrovasc Dis (2009) 28:124–30. doi: 10.1159/000223437
37. Kao J, Kobashigawa J, Fishbein MC, MacLellan WR, Burdick MD, Belperio JA, et al. Elevated Serum Levels of the Cxcr3 Chemokine Itac Are Associated With the Development of Transplant Coronary Artery Disease. Circulation (2003) 107:1958–61. doi: 10.1161/01.CIR.0000069270.16498.75
38. Fernandez-Sender L, Alonso-Villaverde C, Rull A, Rodriguez-Gallego E, Riera-Borrull M, Hernandez-Aguilera A, et al. A Possible Role for Ccr5 in the Progression of Atherosclerosis in Hiv-Infected Patients: A Cross-Sectional Study. AIDS Res Ther (2013) 10:11. doi: 10.1186/1742-6405-10-11
39. Wei L, Petryk J, Gaudet C, Kamkar M, Gan W, Duan Y, et al. Development of an Inflammation Imaging Tracer, (111)in-Dota-Dapta, Targeting Chemokine Receptor Ccr5 and Preliminary Evaluation in an Apoe(-/-) Atherosclerosis Mouse Model. J Nucl Cardiol Off Publ Am Soc Nucl Cardiol (2019) 26:1169–78. doi: 10.1007/s12350-018-1203-1
Keywords: atherosclerosis, Ly6C MC, adaptive immune response, inflammatory, ApoE
Citation: Yang P, Wu Q, Sun L, Fang P, Liu L, Ji Y, Park J-Y, Qin X, Yang X and Wang H (2021) Adaptive Immune Response Signaling Is Suppressed in Ly6Chigh Monocyte but Upregulated in Monocyte Subsets of ApoE-/- Mice — Functional Implication in Atherosclerosis. Front. Immunol. 12:809208. doi: 10.3389/fimmu.2021.809208
Received: 04 November 2021; Accepted: 23 November 2021;
Published: 20 December 2021.
Edited by:
Liwu Li, Virginia Tech, United StatesReviewed by:
Rongxue Wu, University of Chicago, United StatesLiya Yin, Northeast Ohio Medical University, United States
Copyright © 2021 Yang, Wu, Sun, Fang, Liu, Ji, Park, Qin, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Wang, aG9uZy53YW5nQHRlbXBsZS5lZHU=
†These authors have contributed equally to this work
 Pingping Yang
Pingping Yang Qinghua Wu1†
Qinghua Wu1† Lizhe Sun
Lizhe Sun Pu Fang
Pu Fang Yong Ji
Yong Ji Joon-Young Park
Joon-Young Park Xiaofeng Yang
Xiaofeng Yang Hong Wang
Hong Wang