- 1Institute of Pharmacology, University of Bern, Bern, Switzerland
- 2Bern Center for Precision Medicine (BCPM), University of Bern, Bern, Switzerland
Aberrant glycosylation is a key feature of malignant transformation. Hypersialylation, the enhanced expression of sialic acid-terminated glycoconjugates on the cell surface, has been linked to immune evasion and metastatic spread, eventually by interaction with sialoglycan-binding lectins, including Siglecs and selectins. The biosynthesis of tumor-associated sialoglycans involves sialyltransferases, which are differentially expressed in cancer cells. In this review article, we provide an overview of the twenty human sialyltransferases and their roles in cancer biology and immunity. A better understanding of the individual contribution of select sialyltransferases to the tumor sialome may lead to more personalized strategies for the treatment of cancer.
Introduction
Cancer remains one of the leading cause of death worldwide (1). During their development, cancer cells undergo important genetic and structural modifications (2). A well-known feature of malignant transformation is aberrant glycosylation (3, 4). Altered tumor glycosylation was initially described in the mid-twentieth century (5–7), and has since been studied in-depth with regard to its role in tumor progression. Tumor-specific glycosylation has been linked to many processes involved in oncogenesis, such as tumor growth and progression, invasion, metastasis, angiogenesis, chemoresistance and tumor immunity (3, 4, 8–12).
Commonly found glycosylation changes in cancer cells include hypersialylation, incomplete synthesis, truncation of O- and N-glycans, altered branching, and even xenoglycosylation (3, 13). Hypersialylation, referring to the increased density of sialic acid-containing glycans (sialoglycans), is one of the most common features of altered tumor glycosylation (3). Overexpressed sialoglycans include sialylated derivatives of Lewis antigens (sialyl-Lewis X [sLeX]), sialyl-Lewis A [sLeA]), which as ligands of selectins are long known to promote tumor metastasis (3, 14). Accumulating evidence suggests that distinct sialoglycans act as glycoimmune checkpoints that suppress anti-tumor immune reactivity by engagement of immunoregulatory Siglec receptors on myeloid and lymphoid immune cells (12, 15–17). Indeed, ligands of Siglecs are broadly expressed on primary human cancer cells and cell lines of different origin (18).
In humans, twenty different sialyltransferases (SiaTs) are involved in the biosynthesis of glycans and each exhibits distinct characteristics and preferences such as for substrates and glycosidic linkages. The expression levels of individual SiaTs varies significantly between different types of tumors (19), but also within tumors of the same origin (20). While the overexpression of certain sialyltransferases in cancer is associated with tumor hypersialylation and adverse outcome, such positive correlation is not found for all sialyltransferases and may also depend on the type of tumor (see below). Given the significance of distinct sialylation patterns for cancer biology and immunity, in this review article we provide an overview on expression and roles of individual sialyltransferases in cancer.
Sialic Acids and Sialyltransferases
Sialic acids (neuraminic acids) are nine-carbon (C1-9) monosaccharides most commonly found at a terminal position on the outer end of glycoconjugates on many glycoproteins and glycolipids synthesized by living cells (21). Their prominent position on the cell surface glycans of mammalian cells keeps them at the forefront of cellular processes in health, but also in cancer biology and immunity (22–25).
The most prevalent sialic acids in mammals comprise N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) monosaccharides (Figure 1A). 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid (Kdn) sialic acids are more widespread in lower vertebrates (26) (Figure 1B). When one or more hydroxyl groups of Neu5Ac, Neu5Gc or deaminated neuraminic acid (Kdn) are substituted with acetyl, methyl or sulfate residues, more than 50 derivatives with a high diversity are formed (21, 27). As opposed to most mammals, humans do not naturally express Neu5Gc due to the deletion of the CMAH (Cytidine monophospho-N-acetylneuraminic acid hydroxylase) gene, which is responsible for the conversion of Neu5Ac into Neu5Gc (28) (Figure 1A). It is thought that the deletion of this gene could have provided selective advantages during human evolution and eventually played a role in brain development and running endurance in humans (29, 30). Remarkably, Neu5Gc is often expressed in glycoconjugates of human tumors (13, 31, 32). Due to altered metabolic pathways tumor cells are able to incorporate non-human Neu5Gc (3, 13, 33, 34), which humans can retrieve from foods such as red meat (35, 36).
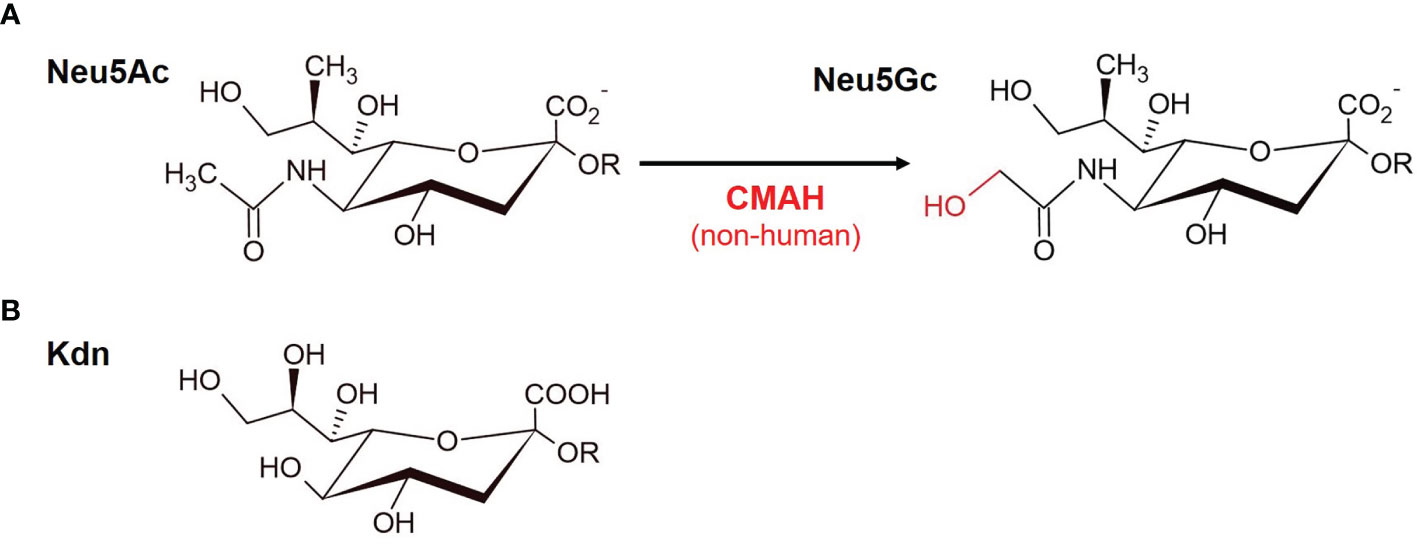
Figure 1 Sialic acids. Sialic acids are nine-carbon monosaccharides. (A) The two main mammalian sialic acids N-acetyl neuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) are shown. Neu5Gc is derived from Neu5Ac and differs by one oxygen atom in the N-glycolyl group, which is added by the enzyme cytidine monophosphate N-acetylneuraminic acid hydroxylase (CMAH) in the cytosol. Humans have an inactivating mutation of the CMAH gene and therefore they lack this enzymatic activity. (B) Kdn (2-keto-3-deoxy-D-glycero-D-galacto-nononic acid), which is more common among lower vertebrates and bacteria (see text).
The sialic acid metabolism involves enzymes that catalyze the biosynthesis and transfer of sialic acid to a glycoconjugate, as well as the removal and degradation of sialic acid (37) (Figure 2). Sialic acid biosynthesis starts with UDP-GlcNAc (uridine diphosphate N-acetylglucosamine) produced via the hexosamine pathway, which is converted to ManNAC-6-P (N-Acetyl-mannosamine 6-phosphate) by UDP-GlcNAc 2-epimerase/ManNAc-6 (GNE) in a two-step process (38). Then, Neu5Ac synthase (NANS) generates 9-phosphorylated forms of sialic acid (Neu5Ac-9-P), which is then dephosphorylated by Neu5Ac-P-phosphatase (NANP) to generate free sialic acid (Neu5Ac) in the cytoplasm (39). Next, cytosolic Neu5Ac enters the nucleus and is activated by coupling cytidine monophosphate (CMP) via the action of cytosine 5’-monophosphate N-acetylneuraminic acid synthetase (CMAS) to produce CMP-Neu5Ac (40). CMP-Neu5Ac is used by sialyltransferases in the Golgi apparatus for sialylation of glycoconjugates. Finally, sialylated glycoproteins and glycolipids are exported to the cell membrane or secreted.
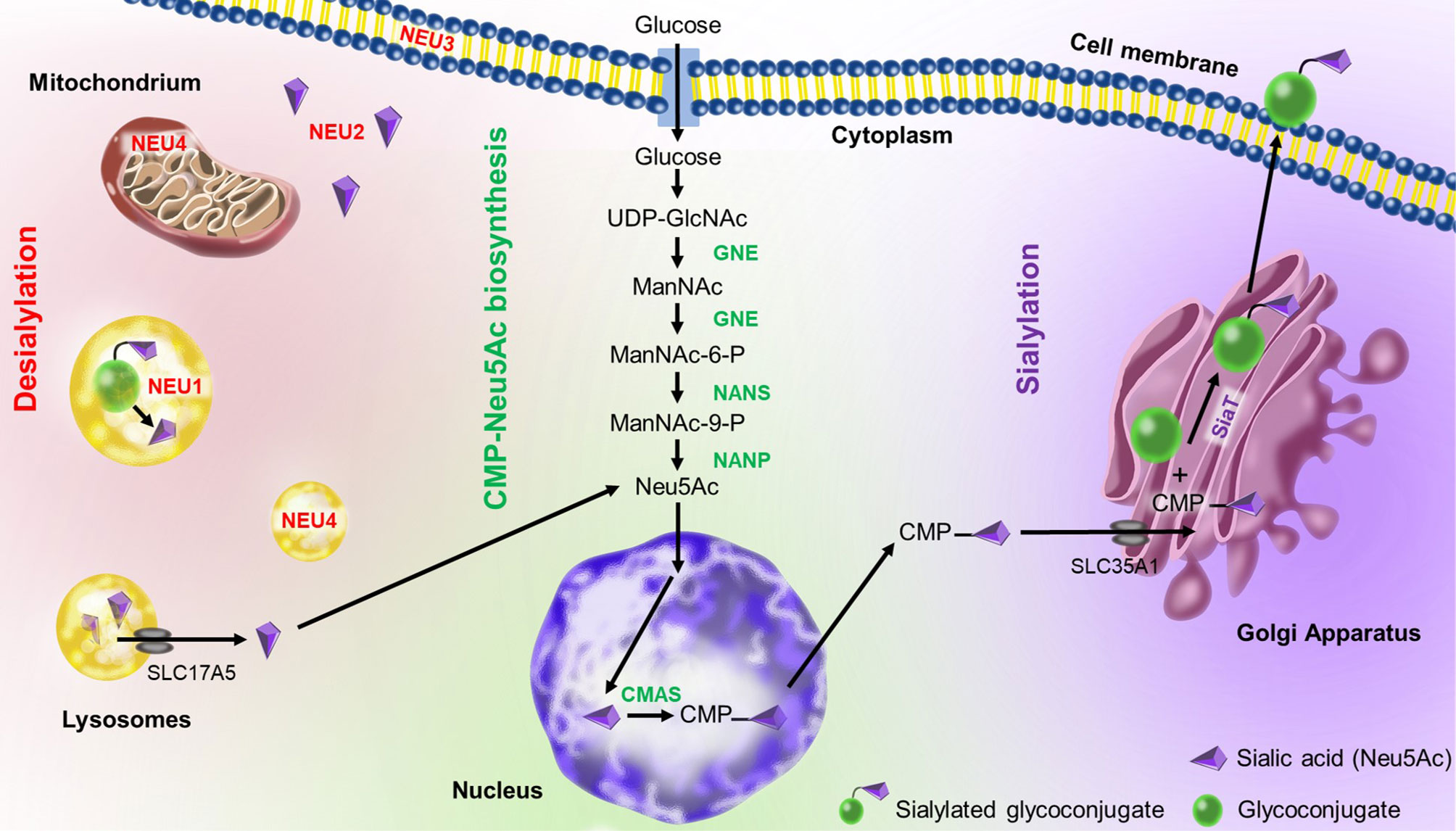
Figure 2 Sialic acid metabolism in humans. CMP-Neu5Ac mostly occurs in the cytoplasm except of the CMP-sialic acid synthase (CMAS)-mediated reaction which takes place in the nucleus. UDP-GlcNAc-2 epimerase (GNE) synthesizes N-acetylmannosamine (ManNAc) in two steps. Then, Neu5Ac synthase (NANS) generates ManNAc-9-P, which is then dephosphorylated by Neu5Ac-P-phosphatase (NANP) to generate free sialic acid in the cytoplasm. The free sialic acid can enter the nucleus to be linked to CMP (cytidine-5’-monophosphate). The CMP-Neu5Ac is transferred to the Golgi apparatus via SLC35A1 transporter (solute carrier family 35 member A1), where it is used as a substrate for sialylation by different sialyltransferases (SiaT). Sialylated glycoconjugates are then exported to the cellular membrane or secreted. They can also be broken down by various neuraminidases (NEU1-4) present in different cellular localizations. The released sialic acid can reenter the biosynthesis pathway. Illustration by Aldona von Gunten.
On the other hand, sialic acid can also be released by neuraminidase (also called sialidase) from sialylated glycoconjugates (40). There are 4 mammalian neuraminidases with different cellular localizations: the lysosomal neuraminidase NEU1 (41), the cytosolic neuraminidase NEU2 (42), the plasma membrane-associated neuraminidase NEU3 (43) and the lysosomal or mitochondrial membrane-associated neuraminidase NEU4 (44). The released sialic acids can be reutilized in the biosynthesis pathway (40). Hypersialylation, as occurring in malignancy, is closely associated to an imbalance between sialic acid biosynthesis and desialylation (45).
Human SiaTs comprise a set of 20 glycosyltransferases which all use cytidine monophosphate N-acetylneuraminic acid (CMP-Neu5Ac) as an activated sugar donor for the transfer of sialic acids to the terminal glycosyl group of glycoproteins and glycolipids as acceptor molecules (46). SiaTs catalyze the formation of different glycosidic linkages, α2,3-, α2,6-, or α2,8-linkage, and also vary in their acceptor specificities. Accordingly, SiaTs can be grouped into four different families: ST3Gal, ST6Gal, ST6GalNAc, and the ST8Sia (Figure 3). Even though SiaTs share the same sugar donors, they present specific substrate specificity, although with some degree of redundancies. Indeed, enzymatic analysis conducted in vitro with recombinant enzymes revealed that one linkage can be synthesized by multiple enzymes (47, 48). SiaTs share conserved sialylmotifs, including ‘L’- (for long), ‘S’- (for short), ‘III’ (for being third position in sequence), and ‘VS’- (for very small) motifs (49). The L-motif is thought to mediate the binding of the donor substrate, the III- and VS-motifs bind the acceptor substrate, and the S-sialylmotif contributes to both binding of donor and acceptor substrates (49). A disulfide bond between the L- and S-motifs bring all sialylmotifs closer together to facilitate interactions with substrates (49).
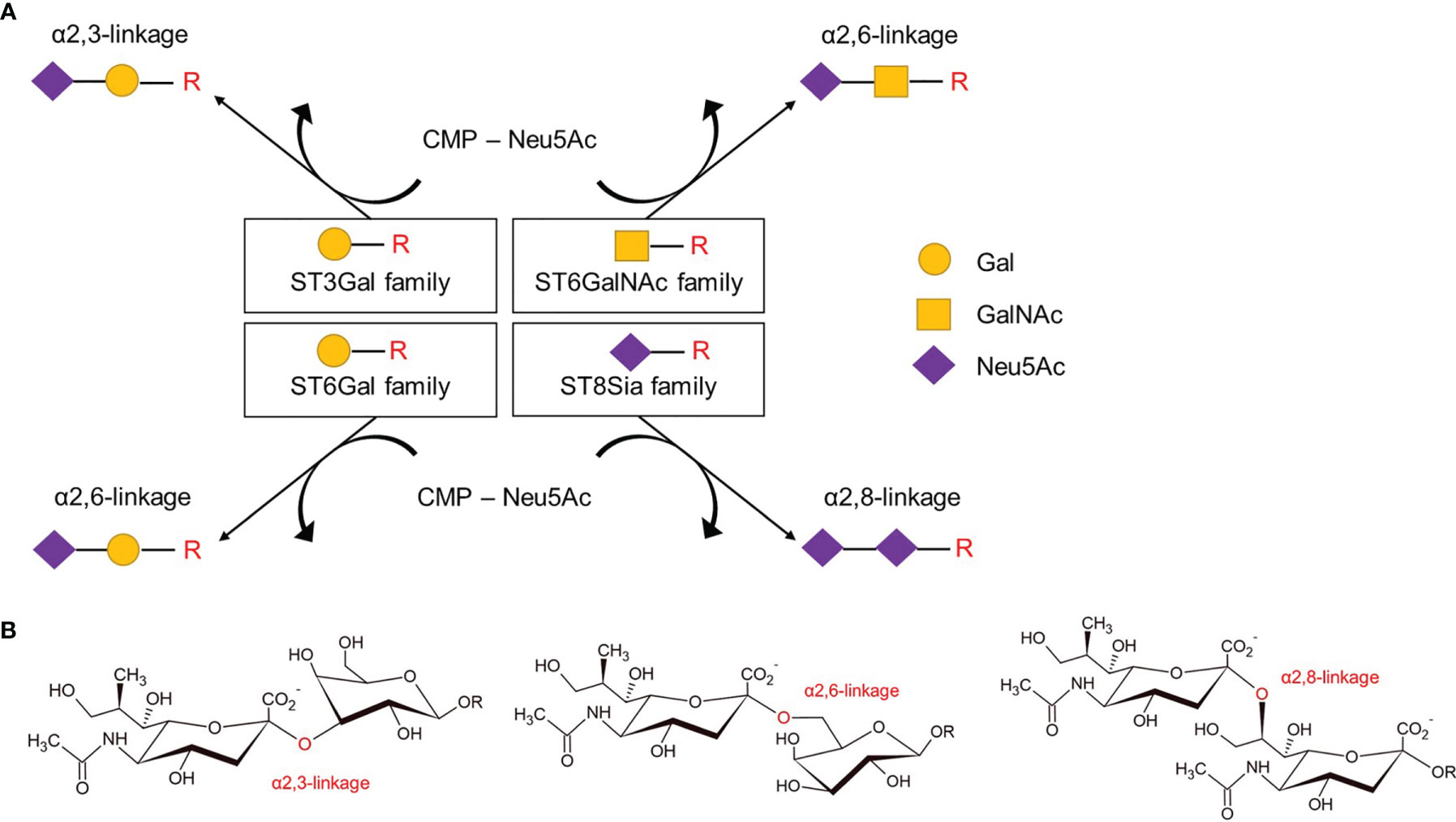
Figure 3 Members of the four families of sialyltransferases catalyze different glycosidic linkages. (A) The four families of sialyltransferases as categorized according to restricted glycosidic linkage and acceptor specificity. Indicated are the transfer of activated CMP-Neu5Ac onto Gal, GalNAc or Neu5Ac moieties of carbohydrate chains (-R), such as on glycoproteins or glycolipids. (B) Examples of glycosidic α2,3, α2,6, and α2–8 -linkages involving the hydroxyl group at carbon atom 2 of Neu5Ac sialic acid with galactose (left, middle) or another sialic acid (right). CMP, cytidine monophosphate; Neu5Ac, N-acetylneuraminic acid; Gal, galactose; GalNAc, N-acetylgalactosamine.
SiaTs have been shown to be primarily restricted to medial- and trans-cisternae of the Golgi apparatus, with some being present in the trans-Golgi network (50), but some SiaTs are also expressed as post-Golgi and secreted enzymes (51, 52), and SiaT activity was also reported to occur at the cell surface of monocyte-derived dendritic cells (53). Their expression pattern among tissues is diverse, but some SiaTs are preferentially expressed at distinct sites. For specific protein expression of SiaTs the Human Protein Atlas (54) can be consulted (proteinatlas.org).
Increased activity or expression of SiaTs leads to the hypersialylation of cell surfaces which is one of the most common glycosylation changes that occurs in tumors; it entails the enhanced expression of sialic acid-terminated glycoconjugates (3). Many studies show elevated levels of SiaTs in the plasma of cancer patients (55–58). The relative diversity and complexity of sialylation patterns in tumors represents a promising area of research, knowing that each SiaT is involved in the synthesis of various structures, therefore, broadly impacting cancer development in various ways, which will be discussed in the following sections.
ST3Gal Family
Six β-galactoside α2,3-sialyltransferases belong to the ST3Gal family in humans and these enzymes transfer sialic acid residue in an α2,3-linkage to terminal galactose (Gal) residues present on glycolipids or glycoproteins (59, 60). Members of this family are involved in the synthesis of gangliosides (ST3Gal2 and 5), and the tumor-associated sialyl-T (ST) (ST3Gal1) and sialyl-Lewis (ST3Gal3, 4, and 6) antigens (Figure 4).
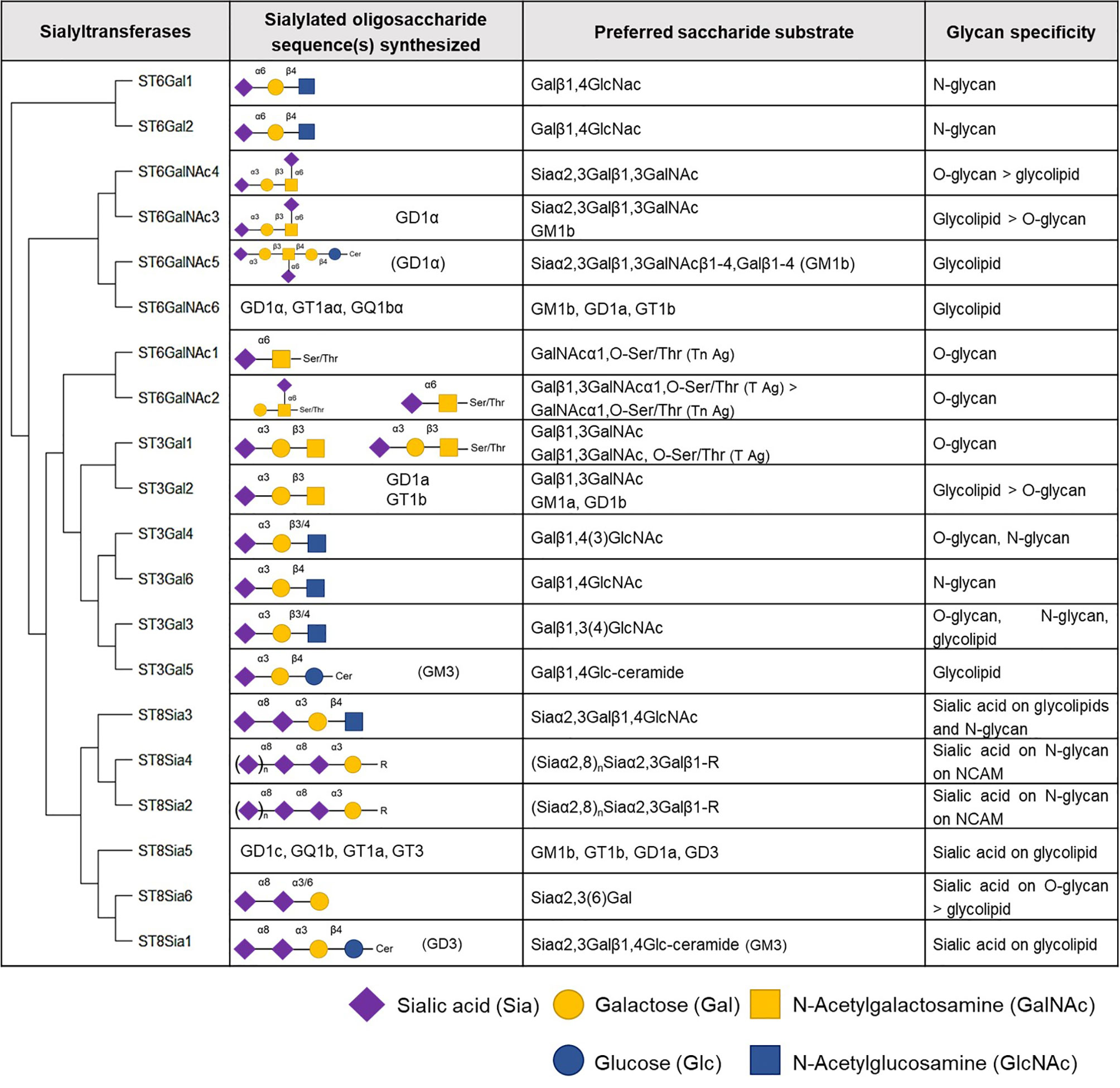
Figure 4 Human sialyltransferases. The twenty human sialyltransferases listed according to their homology (60). Select generated oligosaccharides, preferred substrates and glycan specificities of individual sialyltransferases are shown.
ST3Gal1
ST3Gal1 is known as the major human SiaT to synthesize sialyl-T (ST) antigen from the T antigen Galβ1-3GalNAc. While the T and ST antigens are found on normal O-glycans such as in hematopoietic cells (4), ST3Gal1 overexpression is found in different types of malignancies (61–65), and has been linked to poor prognosis (65, 66). MUC1-ST, a glycoform of the mucin MUC1 carrying the ST antigen found in breast cancer patient serum (67), through Siglec-9 engagement, triggers the differentiation of a unique tumor-associated macrophage (TAM) subtype that has been associated with poor prognosis in breast cancer (68). Recently, Rodriguez et al. identified ST3Gal1 as a main contributor to the synthesis of Siglec-7 ligands in pancreatic cancer cells, which by engagement of the sialic acid-Siglec axis may shift TAM differentiation towards a more suppressive phenotype (69). Overexpression of ST3Gal1 has been shown to promote tumor cell migration and metastasis (65, 70–72), which may involve epidermal growth factor receptor (EGFR) signaling (72), or receptor tyrosine kinase AXL dimerization/activation (71). Moreover, ST3Gal1 seems to also play a role in TGF-β1-induced epithelial-mesenchymal transition (EMT) in ovarian cancer cells (70). ST3Gal1 is also enrolled in promoting resistance to anti-cancer effects of agents, such as of adriamycin directed against chronic myeloid leukemia (CML) cell lines (73), paclitaxel against ovarian cancer cells (70), and tamoxifen and/or vandetanib against breast cancer cells (66). The exact mechanisms of ST3Gal1-mediated resistance to chemotherapeutic drugs remain to be deciphered.
ST3Gal2
In vivo genetic experiments showed that ST3Gal2 is a key enzyme mediating α2,3 sialylation of gangliosides in the brain of mice, in particular of GD1a and GT1b, eventually with support of ST3Gal3 (74). ST3GAL2 mRNA expression was found to be associated with advanced stage and poor clinical outcome in cancer (75, 76). Increased mRNA expression of ST3GAL2, as well as ST3GAL5 and ST8SIA1, was also observed in breast cancer stem cells which is eventually linked to increased expression of gangliosides in these cells (77). ST3Gal2 is a rate-limiting enzyme for SSEA-4 (sialyl-glycolipid stage-specific embryonic antigen 4) synthesis (78), which was shown to be limited in normal tissues but highly expressed in glioblastoma cells (79) and has been associated with epithelial-mesenchymal transition (EMT) (76), loss of cell-cell interactions and adaptation of a migratory phenotype (80). Furthermore, a positive correlation between SSEA4 and chemoresistance was reported (76). Notably, gangliosides are differentially recognized by the immunoregulatory receptors Siglec-7 and -9 receptors (81, 82).
ST3Gal3
ST3Gal3 is involved in the synthesis of sLeA (also known as carbohydrate antigen 19-9 [CA19-9]) and sLeX, which are expressed in different types of cancer (83–87), and have been linked to cancer progression and poor prognosis (88), eventually by selectin-mediated invasion and metastasis of tumor cells (14, 89). Indeed, the expression of ST3GAL3 in breast cancer was found to be associated with poor prognosis (90). ST3Gal3 has also been associated with paclitaxel and cisplatin resistance in ovarian cancer cells (91, 92).
ST3Gal4
ST3Gal4 is involved in the biosynthesis of the tumor-associated antigen sLeX (89, 93). ST3GAL4 expression correlates with enhanced metastatic potential and poor prognosis in some types of cancer, including pancreatic and gastric cancer (94, 95), which may involve selectin-dependent adhesion through sLeX (87). Recently, ST3Gal4 was found to be responsible for the generation of ligands for the immunoregulatory receptor Siglec-9 in pancreatic cancer cell lines (69), and Siglec-7 and -9 ligand in HEK293 cells (96), indicating its potential role in the generation of glyco-immune checkpoints. However, overexpression of ST3GAL4 appears not to be a universal feature of malignancy as downregulation of the enzyme or specific variants has been found for instance in premalignant and malignant cervical tissues (97) and renal cell carcinoma (98). Tissue-specific transcriptional regulation involving alternative splicing and promoter utilization has been described for alpha2,3-sialyltransferases (99), and may explain the differential expression in various types of malignancies.
ST3Gal5/GM3 Synthase
ST3Gal5 initiates the biosynthesis of many downstream gangliosides (100), and is also known by the name “GM3 synthase”. GM3, the simplest ganglioside, is involved in various processes such as transmembrane signaling through the regulation of growth receptor activities and in integrin-mediated cell adhesion and motility (101, 102). Furthermore, GM3 has been shown to be recognized by inhibitory Siglec-9 (103). However, ST3Gal5 also mediates the synthesis of GM4 (104). In a breast cancer model, GM3 synthase knockout mice exhibited enhanced tumor growth and angiogenesis (105). In bladder cancer, the downregulation of ST3Gal5 was associated with reduced patient survival (106). Such experimental evidence suggests a beneficial role of GM3 synthase and certain products, such as distinct a-, b- and c-series gangliosides eventually, in at least some tumors. However, given that GM3 synthase acts at an early stage of ganglioside biosynthesis, it remains unclear which ganglioside products and derivatives are effective in such experimental models and differences may exist among different types of tumors.
ST3Gal6
Like ST3Gal3 and ST3Gal4, ST3Gal6 mediates the sialylation of LeX antigen (83). The resulting sLeX antigen interacts with selectins, such as during the initial tethering before extravasation of cells (107). Indeed, ST3Gal6 was shown to have a crucial role in the generation of selectin ligands in mice (108). High expression of ST3Gal6 in multiple myeloma (MM) patients is associated with poor prognosis (109). Knockdown of ST3GAL6 resulted in a reduced surface expression of α-2,3-linked sialic acid and sLeX on MM cell lines and also reduced the homing and engraftment of malignant cells to the bone marrow niche in vivo (109). Furthermore, mice injected with ST3GAL6 knockdown MM cells demonstrated a decreased tumor burden and prolonged survival. Higher expression of Lewis antigens in neuroblastoma MYCN-amplified cell lines and patient samples could be a consequence of the overexpression of SiaTs, including ST3Gal3/4/6, compared to MYCN-non-amplified counterparts (110). Furthermore, high-grade glioma cell lines exhibit higher expression of terminal sLeX and of the SiaTs ST3Gal3/4/6 compared to low-grade glioma cells (111). ST3Gal6 is also upregulated in human hepatocellular carcinoma (HCC) tissues, and correlates with cell proliferation, migration and invasion ability in HCC cell lines (112). Similar observations were made in urinary bladder cancer with a positive correlation between increased ST3GAL6 expression and tumor stage, grade as well as poor outcome (113).
ST6Gal Family
ST6Gals preferentially link sialic acids in an α2-6 linkage to galactose residues of Galβ1-4GlcNAc-R on N-glycans (59, 60). This family contains two enzymes ST6Gal1 and ST6Gal2, and is thus the smallest SiaT family.
ST6Gal1
ST6Gal1 is the main sialyltransferase contributing to the addition of α-2,6-linked sialic acid to Galβ4GlcNAc chains, usually present in N-linked chains (59). ST6Gal1 is frequently overexpressed in many solid tumors, such as pancreatic, gastric, cervical, ovarian, brain and colorectal cancers and cancer cell lines (114–120). Indeed, this enzyme has been extensively investigated in regard to cancer research [for a review see (121)]. High expression of ST6GAL1 in cancer correlates with worse tumor grade (90, 122), advanced stage of disease (120), and poor prognosis (119, 120, 122). While a greater number of experimental studies support an oncogenic role of ST6Gal1 (discussed below), few reports propose an inverse role of this enzyme based on evidence from select in vitro and in vivo experimental models (123–125). Interestingly, while ST6Gal1 mRNA expression was found to be increased in papillary non-invasive bladder tumors, expression of this enzyme was found to be decreased in muscle-invasive bladder cancer due to epigenetic inactivation of ST6GAL1 by promoter methylation (126).
Interestingly, ST6Gal1 was shown to protect tumor cells from hypoxic stress, eventually by enhancing the expression of hypoxia-inducible factor-1α (HIF-1α) (127). ST6Gal1 activity has been shown to promote EMT in cell lines of different histological origin (128–130), eventually involving E-cadherin transcription and turnover, as well as PI3K/Akt signaling (128). Silencing of ST6Gal1 in prostate cancer cell lines resulted in decreased expression of components of the PI3K/Akt and β-catenin signaling pathways, resulting in reduced proliferation, migration and invasion (122). Furthermore, ST6Gal1 expression is associated with nonmalignant stem and progenitor cells, but also with stemness in cancer and may drive cancer stem cell (CSC)-like characteristics (131–136). Furthermore, high expression of ST6GAL1 in CSCs could eventually promote chemo-resistance (137). Indeed, ST6Gal1 has been linked to resistance to a number of agents including gemcitabine (138), cisplatin (139), trastuzumab (140, 141) or gefitinib (142), latter of which appears to involve sialylation and activation of EGFR (142).
Several investigators observed that α2-6 sialylation by ST6Gal1 activity may protect cells from cell death, and eventually block homeostatic epithelial cell apoptosis in cancer (133). ST6Gal1-mediated sialylation prevents apoptosis induced by tumor necrosis factor receptor 1 (TNFR1) (143), eventually by restraining the receptor on the cell surface (144). Similarily, α2-6 sialylation of the death receptor FAS by ST6Gal1 prevents receptor activation by blocking its internalization and the subsequent formation of death-inducing signaling complex and activation of apoptotic caspase-dependent signaling pathways (145). Furthermore, sialylation of β1 integrins by ST6Gal1 conferred protection against galectin-3-induced apoptosis in a cancer cell line (146).
Recently, using gene engineered HEK293 cells, ST6Gal1 was found to be partially responsible for the generation of ligands for the immunoregulatory receptor Siglec-7 (96), indicating its potential role in the generation of glyco-immune checkpoints.
ST6Gal2
ST6GAL2 is predominantly expressed in the adult brain and fetal tissues, and to a lesser extent in the thyroid gland, small intestine, colon, and testis (147, 148). While relatively few studies have investigated the expression and role of ST6GAL2 in tumors, overexpression of this enzyme was found in select types of cancer, including breast cancer (149) and follicular thyroid carcinoma (FTC) (150). In breast cancer ST6GAL2 expression associated with poor prognosis for patients (149). Moreover, silencing of ST6GAL2 in breast cancer cells resulted in reduced xenograft tumor growth in vivo (149). Furthermore, this study revealed that ST6GAL2 silenced cell lines exhibited reduced adhesion and invasion properties in vitro, with downregulation of several focal adhesion molecules (ICAM-1, VCAM-1) and metastasis pathways proteins (MMP2, CXCR4). Similarly, silencing of ST6GAL2 in FTC reduced tumor growth in an in vivo model (150). Findings from this study suggest that the overexpression of ST6GAL2 leads to the suppression of the Hippo signaling pathway, a tumor suppressor pathway that regulates cellular differentiation and proliferation by inhibiting YAP and TAZ transcription co-activators (151–153).
ST6GalNAc Family
The six SiaTs of the ST6GalNAc family catalyze the glycosidic linkage of sialic acids to N-galactosamine (GalNAc) residues found on O-glycosylated proteins or glycolipids in an α2-6 linkage.
ST6GalNAc1
ST6GalNAc1 catalyzes the generation of sialyl-Tn (sTn) antigen from Tn antigen (154). sTn is a well-known tumor-associated carbohydrate antigen (TACA) overexpressed in multiple cancers (155–157), and has been linked to poor prognosis (158–160). Expression of the biosynthetic enzyme ST6GalNAc1 has also been directly associated with poor prognosis (161). Indeed, overexpression of ST6GalNAc1 in gastric, breast, prostate and ovarian cancer cell lines and tissues has shown to induce the expression of the sTn antigen (155, 157, 161–166). The expression of ST6GALNAC1 can also be induced by cytokines, such as IL-13 and CCL17 secreted by M2 macrophages co-cultured with colon cancer cells, which may result in higher expression of sTn antigen including on MUC1 (167).
Downregulation of ST6GALNAC1 via hyper-methylation and loss of heterozygosity (LOH) was observed in esophageal carcinoma in tylosis, an inherited epithelial disorder (168). Interestingly, in prostate cancer a splice variant of ST6GalNAc1 is induced by androgens, which consists of a shorter isoform that exhibits sialyltransferase activity yet with slightly different properties (157).
In experimental models, overexpression of ST6GalNac1 reduced cell-cell aggregation and increased extracellular matrix (ECM) adhesion, migration and invasion in vitro (163, 166), and promoted tumor growth and metastasis in vivo (163, 164) (165). Furthermore, ST6GalNAc1 activity might foster cancer cell stemness, as expression of CSC markers and tumor sphere formation capability were increased in ST6GalNAc1 overexpressing colorectal or ovarian cancer cell lines (161, 164). Stemness through the generation of sTn seems to involve Akt pathway signaling (161, 164), eventually in cooperation with Galectin-3 (161).
The immunoreceptor Siglec-15 was shown to recognize sTn antigen (169, 170), and to depend on ST6GalNac1-mediated biosynthesis (170). Engagement of Siglec-15 by binding to tumor-associated sTn antigen resulted in enhanced TGF-β secretion from monocytes/macrophages following DAP12-Syk signaling (171). Notably, a recent study showed that macrophage-associated Siglec-15 suppressed T cell responses in vitro and in vivo, eventually establishing a mechanism for immune evasion in the TME (172).
ST6GalNAc2
ST6GalNac2 synthesizes sialyl-6-T antigen from T antigen, and to a lesser extent it sialylates the Tn antigen (154, 166). High transcriptional expression of ST6GALNAC2 correlated with poor prognosis in colorectal cancer (173), and was found to be associated with higher histological tumor grade, lymph node metastasis, and advanced clinical stage in FTC (174). ST6GalNAc2 has been proposed to enhance invasive properties of cancer cell lines via PI3K/Akt pathway signaling (174, 175). However, the role of ST6GalNac2 in cancer appears not to be unequivocally detrimental as Murugaesu and colleagues identified ST6GalNAc2 as a novel metastasis suppressor in mouse and human breast cancer models (176). Indeed, high levels of ST6GALNAC2 expression correlated with increased survival in patients with breast cancer (176). The authors showed that silencing of ST6GALNAC2 modified the cell surface O-glycome resulting in an increase in unmodified T antigen/core 1 antigen and a reduction in the disialyl core 1 antigen. Such altered glycosylation facilitated the binding of the soluble lectin galectin-3 and resulted in increased tumor cell aggregation, pulmonary tumor cell retention and metastatic burden in vitro or in vivo.
ST6GalNAc3
ST6GalNAc3 uses α2,3-sialylated ganglioside GM1b as a substrate to synthesize the ganglioside GD1α. In healthy individuals, this enzyme is highly expressed in brain and kidney (177). Aberrant promoter hypermethylation of ST6GALNAC3 was found in prostate cancer tissue samples (178), but it remains to be shown whether transcriptional silencing of this gene influences the development or progression of prostate cancer. However, ST6GalNAc3 seems to promote the proliferation of A549 non-small cell lung cancer cells through enhanced expression of transferrin receptor protein 1 (TFR1) (179), which is important for cell proliferation and survival (180).
ST6GalNAc4
ST6GalNAc4 mediates the synthesis of disialyl-T antigen from sialyl-T antigen (O-glycan), and also generates the disialyl-lactotetraosyl-ceramide GD1α from sialyl-lactotetraosyl-ceramide GM1b (gangliosides) yet to a lesser degree than ST6GalNAc3 (181, 182). Upregulation of ST6GalNAc4 and downregulation of the core 2 N-acetylglucosaminyltransferase C2GnT2 (Gcnt3) were shown to be key in conferring tumor cell glycosylation changes that contribute to metastatic activity in a primary lung cancer model, eventually by preserving presentation of the T-antigen and adherence to galectin 3 (183). In another study, higher expression of ST6GALNAC4 was observed in FTC tissues compared to transitional tissues and silencing of this enzyme led to decreased invasive ability in vitro and in vivo (184).
ST6GalNAc5/GD1α Synthase
ST6GalNAc5 transfers a sialic acid residue onto GM1b to form GD1α (185) and this enzyme is also referred to as GD1α synthase (186). Indeed, transfection of the human ST6GalNAc5 cDNA into a breast cancer cell line resulted in the expression of GD1α (187). A study investigating germline single-nucleotide polymorphisms indicates that specific SNPs of ST6GALNAC5 determine susceptibility for colorectal brain metastasis and overall survival (188). Silencing of ST6GALNAC5 in breast cancer cells led to decreased metastasis in a murine model in vivo, and in an in vitro model using human umbilical vein endothelial cells (HUVEC) silenced cells exhibited reduced blood brain barrier (BBB) transmigration activity (189). As opposed, a more recent study showed that ST6GalNac5 overexpression in breast cancer cells leads to a decreased adhesion and no change in transmigration compared to controls in a human BBB model using CD34+ hematopoietic stem cell derived endothelial cells co-cultivated with brain pericytes (190),. The authors of this study suggested that differences in the used BBB models may account for these divergent observations.
ST6GalNAc6
ST6GalNAc6 catalyzes the synthesis of α-series gangliosides, including GD1α, GT1aα and GQ1bα (191), globo-series glycosphingolipids (GSL) (192, 193), and disialyl LeA (194, 195). In humans, ST6GalNAc6 is widely expressed in different organs (193). In human colon cancer ST6GalNAc6 is downregulated compared to nonmalignant epithelium, which is paralleled by a decrease in disialyl LeA expression and a concomitant increase in sialyl LeA (195). Such downregulation of ST6GalNAc6 occurs already in early-stage colon cancer and has been associated with epigenetic silencing (196). The related glycan change from disialyl LeA to sialyl LeA may increase E-selectin binding activity during metastasis and support inflammation-driven carcinogenesis by reduced binding to immunoregulatory Siglec-7 (195). mRNA levels of ST6GalNAc6 have also been found to be reduced in human kidney tumor lesions as compared to healthy tissue from the same patient (193). However, ST6GalNAc6 may also enhance the metastatic capability of tumor cells, as silencing of ST6GalNAc6 in a renal cell carcinoma (RCC) cell line, expressing lower levels of DSGb5, exhibited decreased migration, but not proliferation, in vitro (192). Siglec-7 binds to the RCC cell line ACHN in a DSGb5-dependent fashion and silencing of ST6GalNAc6 led to reduced surface binding of a Siglec-Fc chimera protein in these cells (197). These ST6GalNAc6 knockdown cells were more susceptible to cytotoxicity mediated by sialidase-treated NK cells in vitro, suggesting that this sialyltransferase has the potential to generate glyco-immune checkpoints at least in some types of tumors.
ST8Sia Family
The ST8Sia family catalyzes the transfer of sialic acid to another sialic acid in an α2,8-linkage (60). Oligosialic acid chains display a chain of 2-7 sialic acids, whereas polysialic acid (polySia) chain exhibit a chain of eight or more polysialic acids (198). ST8Sia2 and 4 are also called polysialyltransferases as they participate in extending linear chains of polysialic acids (60). ST8Sia3 also participate in polysialylation, but with less efficacy than ST8Sia2 and 4 (199). ST8Sia1 (GD3 synthase), ST8Sia3, ST8Sia5 and ST8Sia6 are involved in the synthesis of sialylated glycolipids (60).
ST8Sia1/GD3 Synthase
ST8Sia1 is also known as GD3 synthase (GD3S), as it catalyzes the transfer of a sialic acid residue onto GM3 to give raise to the b-series ganglioside GD3, which can eventually be further processed for the biosynthesis of other b-/c-series gangliosides (59). GD3S expression positively correlates with increasing grades of astrocytomas and is highly expressed in glioblastoma (200). In metastatic melanoma high ST8Sia1 expression is associated with detrimental outcome and higher expression in metastatic lesions, particularly in the brain (201). Recent studies analyzing data from The Cancer Genome Atlas (TCGA) showed an association of high ST8Sia1 expression levels in breast cancer with poor patient survival (202–204), which is eventually linked to epigenetic hypomethylation of the ST8SIA1 gene (204). As opposed, in another study higher expression of ST8Sia1 mRNA in estrogen receptor (ER) positive breast cancer patients has been associated with higher disease free survival, while no significant difference was found in ER negative patients (205). However, a growing body of evidence supports the notion that ST8Sia1 is associated with tumor growth and progression. In a murine model of glioma, ST8Sia1-deficient mice exhibited attenuated glioma progression, lower-grade pathology and prolonged lifespan (206). Furthermore, in a breast cancer xenograft model silencing of ST8Sia1 led to reduced tumor growth and triptolide-mediated downregulation of ST8Sia1 inhibited tumor growth and prolonged survival (207). ST8Sia1 overexpression has been shown to bypass the need of serum for cell growth and to enhance migratory properties of breast cancer and glioma cell lines (208, 209). Inhibition of ST8Sia1 function by shRNA or triptolide affected the initiation and maintenance of EMT and ST8Sia1 expression correlated with activation of the c-Met signaling pathway enhancing stemness and metastatic properties (203). The implication of ST8Sia1 in stemness with c-Met signaling downstream of this enzyme was also found in experimental models of glioblastoma (200). ST8Sia1 activity has also been linked to oncogenic signaling through Wnt/b-catenin or Akt, Erk, and Src kinases (206, 210), which eventually may confer chemoresistance (210). GD3 has been identified as a ligand for Siglec-7 (81, 82), and ST8Sia1-transfected P815 cells with high surface expression of GD3 exhibited resistance to NK cell-mediated cytotoxicity due to Siglec-7-dependent inhibition (211).
ST8Sia2/STX
The polysialyltransferase ST8Sia2, also known as sialyltransferase X (STX) is involved in the synthesis of linear polymers of sialic acid, so-called polysialic acid (polySia) chains (212). Polysialic acids are a form of post-translational modifications on different proteins, including the neural cell adhesion molecule (NCAM). Besides expression in healthy neuronal tissues, ST8Sia2 is expressed in neuronal and non-neuronal tumors and expression levels eventually correlate with advanced stage of disease, poor prognosis and risk of relapse (213–215). In an in vivo model, ST8Sia2-transfected glioma cells with high expression of polySia exhibited increased tumor invasion within the brain of recipient mice (216). Overexpression of ST8SIA2 appears to also enhance invasiveness and metastatic capabilities of small cell lung cancer cells in vitro (217). Cytidine monophosphate (CMP) was reported to competitively inhibit ST8Sia2 and treatment with CMP led to reduced migration of ST8Sia2-expressing but not non-expressing cell lines in 2D migration assays (218). ST8Sia2 was upregulated in a subset of primary human carcinoma-associated fibroblasts (CAFs), and ST8SIA2 silencing in co-cultured CAFs resulted in decreased lung tumor cells invasion in a 3D model (215).
ST8Sia3
ST8Sia3 is highly expressed in brain and testis and mediates the sialylation of a diversity of glycolipids (GM3, GD3 and α2,3-sialylparagloboside) and select glycoproteins, including striatal glycoproteins (199, 219, 220). ST8Sia3 can also transfer polySia to NCAM, but with a lower efficacy than ST8Sia2 and ST8Sia4 (199). ST8Sia3 was shown to promote survival, proliferation, clonogenicity, and migration of glioblastoma cells based on ST8SIA3 knockdown experiments in vitro (221). Moreover, in the same study it was observed that mice xenografted intracranially with human glioblastoma cell line silenced for ST8Sia3 showed a better overall survival and tumors obtained from these mice demonstrated a lower Ki67 proliferation index.
ST8Sia4/PST
ST8Sia4, also known as polysialyltransferase (PST), synthesizes slightly longer polySia chains compared to ST8Sia2, eventually conferring different molecular properties (222). Both polysialyltransferases are thought to contribute to the polysialylation of NCAM in mammalian cells (223). ST8Sia4 was also reported to be overexpressed in human RCC and breast cancer tissues and to promote cancer progression (224, 225). In these studies, silencing of ST8Sia4 by short-hairpin RNA (shRNA) or specific microRNA (miRNA) reduced cancer cell proliferation and invasion in vitro, and decreased tumor growth in vivo. High levels of ST8Sia4 expression was observed in chemoresistant leukemic cells (226–228), which may functionally contribute to chemoresistance, eventually by processes involving PI3K/AKT signaling (226, 227). However, in FTC patient tissues, ST8SIA4 was observed to be downregulated compared to normal thyroid tissue, and ST8Sia4 expression in cell lines inversely correlated with proliferation, migration and invasion in vitro or tumor growth in vivo (229). Specific miRNAs targeting ST8SIA4 were reported to promote proliferation and invasion capabilities of FTC and oral squamous carcinoma cells (229, 230), and to foster epithelial-to-mesenchymal transition (230).
ST8Sia5
ST8Sia5 exhibits transferase activity of sialic acid moieties onto several gangliosides to synthesize GT3, GD1c, GT1a and GQ1b, respectively (231, 232). Decreased expression of ST8SIA5 from TCGA dataset was linked to a poor survival in patients suffering from colon cancer, and decreased ST8SIA5 transcript was also observed in a murine model of colitis-associated cancer (233). The reduced expression of ST8Sia5 was linked to gene regulation by forkhead box O3 (FOXO3), the functional deficiency of which may facilitate inflammation-mediated colon cancer growth (233).
ST8Sia6
ST8Sia6 generates disialic acid structures, eventually by transfer of a sialic acid moiety onto a NeuAcα2,3 (6)Gal disaccharide on acceptor substrates, which include glycolipids, but preferentially O-linked glycoproteins (234). Some investigators suggest that ST8SIA6 Antisense RNA 1 (ST8SIA6-AS1) is associated with poor prognosis and enhances the proliferative and metastatic potential of cancer cells (235–240). Furthermore, ST8Sia6 may increase the chemosensitivity of tumor cells at least to certain drugs (226). However, ST8SIA6 expression was found to be upregulated in several types of cancer and to be associated with a poor prognosis (241). Engineered murine colon and melanoma cancer cell lines expressing ST8Sia6 grew faster and led to a decreased survival in vivo and depending on host Siglec-E (241). Also depending on Siglec-E, ST8SIA6 expression induced an antitumor immune responses characterized by macrophage polarization toward M2 and upregulation of arginase, which required Siglec-E (241). Notably, 2,8-disialic acid structures were shown to be ligands of murine Siglec-E (242), as well as human Siglec-7 and -9 (81, 241), and may thus act as glyco-immune checkpoints in human cancer.
Sialic Acid-Binding Proteins in Cancer
Sialyltransferases are involved in the biosynthesis of tumor-associated sialoglycans, which via recognition by sialic acid-binding proteins, influence tumor progression and the immune response of the host. Siglecs and selectins are among the most intensively studied sialic acid-binding lectins, and their implication in cancer will be briefly discussed in this section.
Siglecs
Sialic acid-binding immunoglobulin-type lectins (Siglecs), are a family of I-type lectins that belong to the immunoglobulin superfamily. Siglecs are cell-surface receptors predominantly expressed on leukocytes in a cell-specific and differentiation-dependent manner (243). On the basis of evolutionary conservation and sequence similarity, they are divided into two subsets: the first comprises sialoadhesin (also known as Siglec-1 and CD169), CD22 (also known as Siglec-2), myelin-associated glycoprotein (MAG; also known as Siglec-4) and Siglec-15 (244), and are quite distantly related (∼25-30% sequence identity) (245). The other group comprises CD33-related Siglecs (Siglec-3 (CD33), Siglec-5, Siglec-6, Siglec-7, Siglec-8, Siglec-9, Siglec-10, Siglec-11, Siglec-14, and Siglec-16), which have ∼50-99% identity and have evolutionary rapidly evolved due to exon shuffling, exon loss, gene conversion and gene duplication (244, 245). Structurally, Siglecs consist of an amino-terminal V-set domain that confers binding specificity for select sialoglycan ligands, which differ across individual family members (246), and between species (245). The V-set domain is followed by a differing number of immunoglobulin-like domains, a transmembrane domain, and the carboxy-terminal cytoplasmic tail that contains inhibitory, or for fewer members activating, signaling motifs (247). It has been proposed that Siglec ligands might serve as self-associated molecular patterns (SAMPs) to avoid autoreactivity of immune cells (248).
Ligands for Siglecs are broadly expressed in different types of human tumors and in a diversity of common cancer cell lines (18). The expression of Siglec-7 and -9 ligands protected tumor cells from NK cell-mediated cytotoxicity in vitro, and in a Siglec humanized in vivo model (18). In a complementary approach, it was shown that tumor cells decorated with synthetic glycopolymers inhibited NK cell cytotoxicity by engagement of Siglec-7 (249). The body of evidence for Siglec-mediated immune checkpoints in cancer is rapidly growing and indicates that the sialic acid-Siglec axis is relevant for the control of both myeloid and lymphoid immune cells within the tumor microenvironment (12, 16, 17). Interestingly, Siglecs have been shown to be up-regulated on subsets of tumor-infiltrating and circulating cytotoxic T cells in cancer patients (20, 250), in particular on functionally potent effector memory and EMRA T cells (20). While a variety of Siglec-based therapeutic strategies for cancer immunotherapy are currently under investigation (17, 251), a better understanding of the identity and expression not only of tumor-associated sialoside ligands, but also of underlying carrier molecule (252, 253), in specific tumors and patients, may allow for more tailored treatment strategies.
Selectins
Selectins are a family of three calcium-dependent (C-type) lectins comprising E-selectin, L-selectin, and P-selectin, named after their expression on endothelial cells, leukocytes and platelets. In contrast to L-selectin that is constitutively expressed on leukocytes and E-selectin in postcapillary venules of the skin and bone marrow (17), however, E- and P-selectin expression on endothelial cells or platelets are mainly induced following cellular activation (254). The main physiological function of all selectins is to mediate the rolling and adhesion of leukocyte during leukocyte recruitment to sites of inflammation or to lymphoid tissues (254). The carbohydrate-recognition domain (CDR) of all selectins has modest affinity to sLeX and its isomer sLeA (254), which are among the best described ligands for selectins (17). The synthesis of these tetrasaccharides occurs due to the integrated action of α2,3-sialyltransferases with α1,3-fucosyltransferases, β1,4-galactosyltranferases, and N-acetyl-β-glucosaminyltransferases (255). As discussed above, ST3Gal3, ST3Gal4 and ST3Gal6 are involved in the synthesis of sLeX, while sLeA is predominantly generated by ST3Gal3.
sLeA and sLeX are known tumor markers and functionally implicated in the malignant behavior of cancer cells (88). Glycosylated proteins carrying sLeX/A moieties, such as PSGL-1, CD24, CD44, ESL-1, and death receptor-3 represent major selectin ligands on cancer cells (14). The overexpression of selectin ligands has been linked to cancer progression and poor prognosis in some cancers (14, 88, 256). In vivo studies using selectin knockout or selectin ligand deficient mice highlighted the importance of selectins in metastasis (3). Selectins seems to contribute to metastasis through heterotypic interactions between tumor cells, leukocytes and endothelial cells (14, 256). These interactions may also foster tumor embolus formation with local activation of endothelial cells and increased transendothelial migration of both tumor cells and leukocytes (3). Recruited leukocytes might further enhance vascular permeability and cancer cell extravasation, and also shape the tumor microenvironment (14). While earlier studies on selectin-targeted therapies focused on cardiovascular disease, positive outcomes from clinical trials have raised the interest in strategies targeting selectin receptor-ligand interactions in cancer.
Conclusion
In the last decade we have witnessed a significant body of discoveries that highlight the importance of sialic acids in cancer biology and immuno-oncology. As biosynthetic enzymes for sialosides, human SiaTs have long been linked to cancer hypersialylation. However, the twenty SiaTs exhibit different characteristics and their roles in cancer are manyfold and complex, and remain to be fully explored. The expression of SiaTs, sialosides and sialic acid interaction partners (e.g. Siglecs), can vary between different types of tumors, between primary tumor and metastatic lesion, and even between patients (19). Furthermore, controversial observations on the role of a select SiaT may be due to its involvement in the synthesis of multiple glycans, eventually generating various ligands for different glycan-binding proteins. Moreover, limitations of methodological approaches need to be considered, such as missing environmental context for in vitro cell cultures or species differences for in vivo studies. Functional redundancy may exist between SiaTs, and while specific small-molecule SiaT inhibitors that bind and block select SiaTs may hold promise for therapeutic and diagnostic use [for recent reviews see (257–259)], combination strategies might be needed in a given context. However, the observation that SiaTs are responsible for the generation of glyco-immune checkpoints has reinvigorated ambitions of researchers to explore the role of individual SiaTs in cancer, which may pave the way for novel immune normalization (260), and more personalized, cancer immunotherapies.
Author Contributions
All authors wrote and approved the manuscript.
Funding
The laboratory of SV is supported by grants from the Swiss National Science Foundation (310030_184757), the Swiss Cancer League/Swiss Cancer Research (KFS-4958-02-2020), and the Bern Center for Precision Medicine (BCPM).
Conflict of Interest
SVG received remuneration for serving on the scientific advisory board of Palleon Pharmaceuticals.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Aldona von Gunten, University of Bern, for support with illustrations. The authors apologize to all colleagues whose work has not been cited due to space limitations.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science (2013) 340:1546–58. doi: 10.1126/science.1235122
3. Boligan KF, Mesa C, Fernandez LE, Von Gunten S. Cancer Intelligence Acquired (CIA): Tumor Glycosylation and Sialylation Codes Dismantling Antitumor Defense. Cell Mol Life Sci (2015) 72:1231–48. doi: 10.1007/s00018-014-1799-5
4. Stowell SR, Ju T, Cummings RD. Protein Glycosylation in Cancer. Annu Rev Pathol Mech Dis (2015) 10:473–510. doi: 10.1146/annurev-pathol-012414-040438
5. Oh-Uti K. Polysaccharides and a Glycidamin in the Tissue of Gastric Cancer. Tohoku J Exp Med (1949) 51:297–304. doi: 10.1620/tjem.51.297
6. Gasic G, Gasic T. Removal and Regeneration of the Cell Coating in Tumour Cells. Nature (1962) 196:170. doi: 10.1038/196170a0
7. Gasic G, Gasic T. Removal of Sialic Acid From the Cell Coat in Tumor Cells and Vascular Endothelium, and its Effects on Metastasis. Proc Natl Acad Sci USA (1962) 48:1172–7. doi: 10.1073/pnas.48.7.1172
8. Munkley J, Elliott DJ. Hallmarks of Glycosylation in Cancer. Oncotarget (2016) 7:35478–89. doi: 10.18632/oncotarget.8155
9. Pinho SS, Reis CA. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat Rev Cancer (2015) 15:540–55. doi: 10.1038/nrc3982
10. Mereiter S, Balmaña M, Campos D, Gomes J, Reis CA. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell (2019) 36:6–16. doi: 10.1016/j.ccell.2019.06.006
11. Fuster MM, Esko JD. The Sweet and Sour of Cancer: Glycans as Novel Therapeutic Targets. Nat Rev Cancer (2005) 5:526–42. doi: 10.1038/nrc1649
12. Rodríguez E, Schetters STT, Van Kooyk Y. The Tumour Glyco-Code as a Novel Immune Checkpoint for Immunotherapy. Nat Rev Immunol (2018) 18:204–11. doi: 10.1038/nri.2018.3
13. Samraj AN, Läubli H, Varki N, Varki A. Involvement of a Non-Human Sialic Acid in Human Cancer. Front Oncol (2014) 4:33. doi: 10.3389/fonc.2014.00033
14. Läubli H, Borsig L. Selectins Promote Tumor Metastasis. Semin Cancer Biol (2010) 20:169–77. doi: 10.1016/j.semcancer.2010.04.005
15. Adams OJ, Stanczak MA, Von Gunten S, Läubli H. Targeting Sialic Acid-Siglec Interactions to Reverse Immune Suppression in Cancer. Glycobiology (2018) 28:640–7. doi: 10.1093/glycob/cwx108
16. Duan S, Paulson JC. Siglecs as Immune Cell Checkpoints in Disease. Annu Rev Immunol (2020) 38:365–95. doi: 10.1146/annurev-immunol-102419-035900
17. Smith BAH, Bertozzi CR. The Clinical Impact of Glycobiology: Targeting Selectins, Siglecs and Mammalian Glycans. Nat Rev Drug Discov (2021) 20:217–43. doi: 10.1038/s41573-020-00093-1
18. Jandus C, Boligan KF, Chijioke O, Liu H, Dahlhaus M, Démoulins T, et al. Interactions Between Siglec-7/9 Receptors and Ligands Influence NK Cell-Dependent Tumor Immunosurveillance. J Clin Invest (2014) 124:1810–20. doi: 10.1172/JCI65899
19. Haas Q, Simillion C. Von Gunten S. A Cartography of Siglecs and Sialyltransferases in Gynecologic Malignancies: Is There a Road Towards a Sweet Future? Front Oncol (2018) 8:68. doi: 10.3389/fonc.2018.00068
20. Haas Q, Boligan KF, Jandus C, Schneider C, Simillion C, Stanczak MA, et al. Siglec-9 Regulates an Effector Memory Cd8β T-Cell Subset That Congregates in the Melanoma Tumor Microenvironment. Cancer Immunol Res (2019) 7:707–18. doi: 10.1158/2326-6066.CIR-18-0505
21. Schauer R, Kamerling JP. Exploration of the Sialic Acid World. Adv Carbohydr Chem Biochem (2018) 75:1–213. doi: 10.1016/bs.accb.2018.09.001
22. Pearce OMT, Läubli H. Sialic Acids in Cancer Biology and Immunity. Glycobiology (2015) 26:111–28. doi: 10.1093/glycob/cwv097
23. Varki A. Sialic Acids in Human Health and Disease. Trends Mol Med (2008) 14:351–60. doi: 10.1016/j.molmed.2008.06.002
24. Büll C, Stoel MA, Den Brok MH, Adema GJ. Sialic Acids Sweeten a Tumor’s Life. Cancer Res (2014) 74:3199–204. doi: 10.1158/0008-5472.CAN-14-0728
25. Varki A. Glycan-Based Interactions Involving Vertebrate Sialic-Acid-Recognizing Proteins. Nature (2007) 446:1023–9. doi: 10.1038/nature05816
26. Inoue S, Kitajima K. KDN (Deaminated Neuraminic Acid): Dreamful Past and Exciting Future of the Newest Member of the Sialic Acid Family. Glycoconj J (2006) 23:277–90. doi: 10.1007/s10719-006-6484-y
27. Angata T, Varki A. Chemical Diversity in the Sialic Acids and Related α-Keto Acids: An Evolutionary Perspective. Chem Rev (2002) 102:439–69. doi: 10.1021/cr000407m
28. Varki A. Uniquely Human Evolution of Sialic Acid Genetics and Biology. Proc Natl Acad Sci USA (2010) 107:8939–46. doi: 10.1073/pnas.0914634107
29. Chou HH, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, et al. Inactivation of CMP-N-Acetylneuraminic Acid Hydroxylase Occurred Prior to Brain Expansion During Human Evolution. Proc Natl Acad Sci USA (2002) 99:11736–41. doi: 10.1073/pnas.182257399
30. Okerblom J, Fletes W, Patel HH, Schenk S, Varki A, Breen EC. Human-Like CMAH Inactivation in Mice Increases Running Endurance and Decreases Muscle Fatigability: Implications for Human Evolution. Proc R Soc B Biol Sci (2018) 285. doi: 10.1098/rspb.2018.1656
31. Malykh YN, Schauer R, Shaw L. N-Glycolylneuraminic Acid in Human Tumours. Biochimie (2001) 83:623–34. doi: 10.1016/S0300-9084(01)01303-7
32. Labrada M, Dorvignit D, Hevia G, Rodríguez-Zhurbenko N, Hernández AM, Vázquez AM, et al. GM3(Neu5Gc) Ganglioside: An Evolution Fixed Neoantigen for Cancer Immunotherapy. Semin Oncol (2018) 45:41–51. doi: 10.1053/j.seminoncol.2018.04.003
33. Dorvignit D, Boligan KF, Relova-Hernández E, Clavell M, López A, Labrada M, et al. Antitumor Effects of the GM3(Neu5Gc) Ganglioside-Specific Humanized Antibody 14f7ht Against Cmah-Transfected Cancer Cells. Sci Rep (2019) 9:1–12. doi: 10.1038/s41598-019-46148-1
34. Yin J, Hashimoto A, Izawa M, Miyazaki K, Chen GY, Takematsu H, et al. Hypoxic Culture Induces Expression of Sialin, a Sialic Acid Transporter, and Cancer-Associated Gangliosides Containing Non-Human Sialic Acid on Human Cancer Cells. Cancer Res (2006) 66:2937–45. doi: 10.1158/0008-5472.CAN-05-2615
35. Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, et al. Human Uptake and Incorporation of an Immunogenic Nonhuman Dietary Sialic Acid. Proc Natl Acad Sci USA (2003) 100:12045–50. doi: 10.1073/pnas.2131556100
36. Bashir S, Fezeu LK, Leviatan Ben-Arye S, Yehuda S, Reuven EM, Szabo De Edelenyi F, et al. Association Between Neu5Gc Carbohydrate and Serum Antibodies Against It Provides the Molecular Link to Cancer: French NutriNet-Santé Study. BMC Med (2020) 18(1):262. doi: 10.1186/s12916-020-01721-8
37. Tanner ME. The Enzymes of Sialic Acid Biosynthesis. Bioorg Chem (2005) 33:216–28. doi: 10.1016/j.bioorg.2005.01.005
38. Stäsche R, Hinderlich S, Weise C, Effertz K, Lucka L, Moormann P, et al. A Bifunctional Enzyme Catalyzes the First Two Steps in N- Acetylneuraminic Acid Biosynthesis of Rat Liver. Molecular Cloning and Functional Expression of UDP-N-Acetyl-Glucosamine 2-Epimerase N- Acetylmannosamine Kinase. J Biol Chem (1997) 272:24319–24. doi: 10.1074/jbc.272.39.24319
39. Li Y, Chen X. Sialic Acid Metabolism and Sialyltransferases: Natural Functions and Applications. Appl Microbiol Biotechnol (2012) 94:887–905. doi: 10.1007/s00253-012-4040-1
40. Verheijen FW. “Sialic Acid,”. In: Laboratory Guide to the Methods in Biochemical Genetics. Springer, Berlin, Heidelberg: Springer (Cold Spring Harbor Laboratory Press. (2017). p. 335–49. doi: 10.1007/978-3-540-76698-8_19
41. Pshezhetsky AV, Richard C, Michaud L, Igdoura S, Wang S, Elsliger MA, et al. Cloning, Expression and Chromosomal Mapping of Human Lysosomal Sialidase and Characterization of Mutations in Sialidosis. Nat Genet (1997) 15:316–20. doi: 10.1038/ng0397-316
42. Monti E, Preti A, Rossi E, Ballabio A, Borsani G. Cloning and Characterization of NEU2, a Human Gene Homologous to Rodent Soluble Sialidases. Genomics (1999) 57:137–43. doi: 10.1006/geno.1999.5749
43. Wada T, Yoshikawa Y, Tokuyama S, Kuwabara M, Akita H, Miyagi T. Cloning, Expression, and Chromosomal Mapping of a Human Ganglioside Sialidase. Biochem Biophys Res Commun (1999) 261:21–7. doi: 10.1006/bbrc.1999.0973
44. Monti E, Bassi MT, Bresciani R, Civini S, Croci GL, Papini N, et al. Molecular Cloning and Characterization of NEU4, the Fourth Member of the Human Sialidase Gene Family. Genomics (2004) 83:445–53. doi: 10.1016/j.ygeno.2003.08.019
45. Vajaria BN, Patel KR, Begum R, Patel PS. Sialylation: An Avenue to Target Cancer Cells. Pathol Oncol Res (2016) 22:443–7. doi: 10.1007/s12253-015-0033-6
46. Paulson JC, Rademacher C. Glycan Terminator. Nat Struct Mol Biol (2009) 16:1121–2. doi: 10.1038/nsmb1109-1121
47. Harduin-Lepers A. Comprehensive Analysis of Sialyltransferases in Vertebrate Genomes. Glycobiol Insights (2010) 2:29–61. doi: 10.4137/gbi.s3123
48. Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The Human Sialyltransferase Family. Biochimie (2001) 83:727–37. doi: 10.1016/S0300-9084(01)01301-3
49. Datta A. Comparative Sequence Analysis in the Sialyltransferase Protein Family: Analysis of Motifs. Curr Drug Targets (2009) 10:483–98. doi: 10.2174/138945009788488422
50. Whitehouse C, Burchell J, Gschmeissner S, Brockhausen I, Lloyd KO, Taylor-Papadimitriou J. A Transfected Sialyltransferase That Is Elevated in Breast Cancer and Localizes to the Medial/Trans-Golgi Apparatus Inhibits the Development of Core-2-Based O-Glycans. J Cell Biol (1997) 137:1229–41. doi: 10.1083/jcb.137.6.1229
51. Burger PC, Lötscher M, Streiff M, Kleene R, Kaissling B, Berger EG. Immunocytochemical Localization of α2,3(N)-Sialyltransferase (ST3Gal III) in Cell Lines and Rat Kidney Tissue Sections: Evidence for Golgi and Post-Golgi Localization. Glycobiology (1998) 8:245–57. doi: 10.1093/glycob/8.3.245
52. Jones MB, Nasirikenari M, Feng L, Migliore MT, Choi KS, Kazim L, et al. Role for Hepatic and Circulatory ST6Gal-1 Sialyltransferase in Regulating Myelopoiesis. J Biol Chem (2010) 285:25009–17. doi: 10.1074/jbc.M110.104406
53. Cabral MG, Piteira AR, Silva Z, Ligeiro D, Brossmer R, Videira PA. Human Dendritic Cells Contain Cell Surface Sialyltransferase Activity. Immunol Lett (2010) 131:89–96. doi: 10.1016/j.imlet.2010.02.009
54. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-Based Map of the Human Proteome. Science (80- ) (2015) 347(6220). doi: 10.1126/science.1260419
55. Berge PG, Wilhelm A, Schriewer H, Wüst G. Serum-Sialyltransferase Activity in Cancer Patients. Klin Wochenschr (1982) 60:445–9. doi: 10.1007/BF01720358
56. Shen ALY, Chou MD, Chi CW, Lee LS. Alterations in Serum Sialyltransferase Activities in Patients With Brain Tumors. Surg Neurol (1984) 22:509–14. doi: 10.1016/0090-3019(84)90313-6
57. Kassel D, Allen J. Elevated Plasma Sialyltransferase in the Cancer Patient. Cancer Res (1975) 35:670–2.
58. Ganzinger U, Deutsch E. Serum Sialyltransferase Levels as a Parameter in the Diagnosis and Follow-Up of Gastrointestinal Tumors. Cancer Res (1980) 40:1300–4.
59. Stanley P, Cummings RD. “Structures Common to Different Glycans,”. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al, editors. Essentials of Glycobiology, 3rd Edition, vol. chapter 14. Cold Spring Harbor New York: Cold Spring Harbor Laboratory Press; 2015-2017 (2017). p. 161–78. doi: 10.1101/glycobiology.3e.014
60. Harduin-Lepers A, Mollicone R, Delannoy P, Oriol R. The Animal Sialyltransferases and Sialyltransferase-Related Genes: A Phylogenetic Approach. Glycobiology (2005) 15:805–17. doi: 10.1093/glycob/cwi063
61. Videira PA, Correia M, Malagolini N, Crespo HJ, Ligeiro D, Calais FM, et al. ST3Gal I Sialyltransferase Relevance in Bladder Cancer Tissues and Cell Lines. BMC Cancer (2009) 9:357. doi: 10.1186/1471-2407-9-357
62. Picco G, Julien S, Brockhausen I, Beatson R, Antonopoulos A, Haslam S, et al. Over-Expression of ST3Gal-I Promotes Mammary Tumorigenesis. Glycobiology (2010) 20:1241–50. doi: 10.1093/glycob/cwq085
63. Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H, et al. An α2,3 Sialyltransferase (ST3Gal I) Is Elevated in Primary Breast Carcinomas. Glycobiology (1999) 9:1307–11. doi: 10.1093/glycob/9.12.1307
64. Chong YK, Sandanaraj E, Koh LWH, Thangaveloo M, Tan MSY, Koh GRH, et al. ST3Gal1-Associated Transcriptomic Program in Glioblastoma Tumor Growth, Invasion, and Prognosis. J Natl Cancer Inst (2016) 108(2). doi: 10.1093/jnci/djv326
65. Wu H, Shi XL, Zhang HJ, Song QJ, Yang XB, Hu WD, et al. Overexpression of ST3Gal-I Promotes Migration and Invasion of HCCLM3 In Vitro and Poor Prognosis in Human Hepatocellular Carcinoma. Onco Targets Ther (2016) 9:2227–36. doi: 10.2147/OTT.S96510
66. Fan T, Yeo HL, Hsu HM, Yu JC, Ho MY, der Lin W, et al. Reciprocal Feedback Regulation of ST3Gal1 and GFRA1 Signaling in Breast Cancer Cells. Cancer Lett (2018) 434:184–95. doi: 10.1016/j.canlet.2018.07.026
67. Storr SJ, Royle L, Chapman CJ, Hamid UMA, Robertson JF, Murray A, et al. The O-Linked Glycosylation of Secretory/Shed MUC1 From an Advanced Breast Cancer Patient’s Serum. Glycobiology (2008) 18:456–62. doi: 10.1093/glycob/cwn022
68. Beatson R, Tajadura-Ortega V, Achkova D, Picco G, Tsourouktsoglou TD, Klausing S, et al. The Mucin MUC1 Modulates the Tumor Immunological Microenvironment Through Engagement of the Lectin Siglec-9. Nat Immunol (2016) 17:1273–81. doi: 10.1038/ni.3552
69. Rodriguez E, Boelaars K, Brown K, Eveline Li RJ, Kruijssen L, Bruijns SCM, et al. Sialic Acids in Pancreatic Cancer Cells Drive Tumour-Associated Macrophage Differentiation via the Siglec Receptors Siglec-7 and Siglec-9. Nat Commun (2021) 12:1270. doi: 10.1038/s41467-021-21550-4
70. Wu X, Zhao J, Ruan Y, Sun L, Xu C, Jiang H. Sialyltransferase ST3GAL1 Promotes Cell Migration, Invasion, and TGF-β1-Induced EMT and Confers Paclitaxel Resistance in Ovarian Cancer. Cell Death Dis (2018) 9. doi: 10.1038/s41419-018-1101-0
71. Pietrobono S, Anichini G, Sala C, Manetti F, Almada LL, Pepe S, et al. ST3GAL1 Is a Target of the SOX2-GLI1 Transcriptional Complex and Promotes Melanoma Metastasis Through AXL. Nat Commun (2020) 11. doi: 10.1038/s41467-020-19575-2
72. Wen KC, Sung PL, Hsieh SL, Chou YT, Lee OKS, Wu CW, et al. α2,3-Sialyltransferase Type I Regulates Migration and Peritoneal Dissemination of Ovarian Cancer Cells. Oncotarget (2017) 8:29013–27. doi: 10.18632/oncotarget.15994
73. Li Y, Luo S, Dong W, Song X, Zhou H, Zhao L, et al. Alpha-2, 3-Sialyltransferases Regulate the Multidrug Resistance of Chronic Myeloid Leukemia Through miR-4701-5p Targeting ST3GAL1. Lab Investig (2016) 96:731–40. doi: 10.1038/labinvest.2016.50
74. Sturgill ER, Aoki K, Lopez PHH, Colacurcio D, Vajn K, Lorenzini I, et al. Biosynthesis of the Major Brain Gangliosides GD1a and GT1b. Glycobiology (2012) 22:1289–301. doi: 10.1093/glycob/cws103
75. Mehta KA, Patel KA, Pandya SJ, Patel PS. Aberrant Sialylation Plays a Significant Role in Oral Squamous Cell Carcinoma Progression. J Oral Pathol Med (2020) 49:253–9. doi: 10.1111/jop.12976
76. Aloia A, Petrova E, Tomiuk S, Bissels U, Déas O, Saini M, et al. The Sialyl-Glycolipid Stage-Specific Embryonic Antigen 4 Marks a Subpopulation of Chemotherapy-Resistant Breast Cancer Cells With Mesenchymal Features. Breast Cancer Res (2015) 17:146. doi: 10.1186/s13058-015-0652-6
77. Liang YJ, Ding Y, Levery SB, Lobaton M, Handa K, Hakomori SI. Differential Expression Profiles of Glycosphingolipids in Human Breast Cancer Stem Cells vs. Cancer Non-Stem Cells. Proc Natl Acad Sci USA (2013) 110:4968–73. doi: 10.1073/pnas.1302825110
78. Saito S, Aoki H, Ito A, Ueno S, Wada T, Mitsuzuka K, et al. Human α2,3-Sialyltransferase (ST3Gal II) Is a Stage-Specific Embryonic Antigen-4 Synthase. J Biol Chem (2003) 278:26474–9. doi: 10.1074/jbc.M213223200
79. Lou YW, Wang PY, Yeh SC, Chuang PK, Li ST, Wu CY, et al. Stage-Specific Embryonic Antigen-4 as a Potential Therapeutic Target in Glioblastoma Multiforme and Other Cancers. Proc Natl Acad Sci USA (2014) 111:2482–7. doi: 10.1073/pnas.1400283111
80. Sivasubramaniyan K, Harichandan A, Schilbach K, Mack AF, Bedke J, Stenzl A, et al. Expression of Stage-Specific Embryonic Antigen-4 (SSEA-4) Defines Spontaneous Loss of Epithelial Phenotype in Human Solid Tumor Cells. Glycobiology (2015) 25:902–17. doi: 10.1093/glycob/cwv032
81. Yamaji T, Teranishi T, Alphey MS, Crocker PR, Hashimoto Y. A Small Region of the Natural Killer Cell Receptor, Siglec-7, Is Responsible for Its Preferred Binding to α2,8-Disialyl and Branched α2,6-Sialyl Residues. A Comparison With Siglec-9. J Biol Chem (2002) 277:6324–32. doi: 10.1074/jbc.M110146200
82. Ito A, Handa K, Withers DA, Satoh M. Hakomori S Itiroh. Binding Specificity of Siglec-7 to Disialogangliosides of Renal Cell Carcinoma: Possible Role of Disialogangliosides in Tumor Progression. FEBS Lett (2001) 504:82–6. doi: 10.1016/S0014-5793(01)02734-X
83. Dall’Olio F, Malagolini N, Trinchera M, Chiricolo M. Sialosignaling: Sialyltransferases as Engines of Self-Fueling Loops in Cancer Progression. Biochim Biophys Acta - Gen Subj (2014) 1840:2752–64. doi: 10.1016/j.bbagen.2014.06.006
84. Carvalho AS, Harduin-Lepers A, Magalhães A, Machado E, Mendes N, Costa LT, et al. Differential Expression of α-2,3-Sialyltransferases and α-1,3/4-Fucosyltransferases Regulates the Levels of Sialyl Lewis a and Sialyl Lewis X in Gastrointestinal Carcinoma Cells. Int J Biochem Cell Biol (2010) 42:80–9. doi: 10.1016/j.biocel.2009.09.010
85. Cui HX, Wang H, Wang Y, Song J, Tian H, Xia C, et al. ST3Gal III Modulates Breast Cancer Cell Adhesion and Invasion by Altering the Expression of Invasion-Related Molecules. Oncol Rep (2016) 36:3317–24. doi: 10.3892/or.2016.5180
86. Pérez-Garay M, Arteta B, Pagés L, de Llorens R, de Bolós C, Vidal-Vanaclocha F, et al. α2,3-Sialyltransferase ST3Gal III Modulates Pancreatic Cancer Cell Motility and Adhesion In Vitro and Enhances Its Metastatic Potential In Vivo. PLoS One (2010) 5:1–11. doi: 10.1371/journal.pone.0012524
87. Guerrero PE, Miró L, Wong BS, Massaguer A, Martínez-Bosch N, de Llorens R, et al. Knockdown of α2,3-Sialyltransferases Impairs Pancreatic Cancer Cell Migration, Invasion and E-Selectin-Dependent Adhesion. Int J Mol Sci (2020) 21:1–24. doi: 10.3390/ijms21176239
88. Kannagi R, Izawa M, Koike T, Miyazaki K, Kimura N. Carbohydrate-Mediated Cell Adhesion in Cancer Metastasis and Angiogenesis. Cancer Sci (2004) 95:377–84. doi: 10.1111/j.1349-7006.2004.tb03219.x
89. Jin F, Wang F. The Physiological and Pathological Roles and Applications of Sialyl Lewis X, A Common Carbohydrate Ligand of the Three Selectins. Glycoconj J (2020) 37:277–91. doi: 10.1007/s10719-020-09912-4
90. Recchi MA, Hebbar M, Hornez L, Harduin-Lepers A, Peyrat JP, Delannoy P. Multiplex Reverse Transcription Polymerase Chain Reaction Assessment of Sialyltransferase Expression in Human Breast Cancer. Cancer Res (1998) 58:4066–70.
91. Zhang X, Yang X, Chen M, Zheng S, Li J, Lin S, et al. ST3Gal3 Confers Paclitaxel-Mediated Chemoresistance in Ovarian Cancer Cells by Attenuating Caspase-8/3 Signaling. Mol Med Rep (2019) 20:4499–506. doi: 10.3892/mmr.2019.10712
92. Wang X, Zhang Y, Lin H, Liu Y, Tan Y, Lin J, et al. Alpha2,3-Sialyltransferase III Knockdown Sensitized Ovarian Cancer Cells to Cisplatin-Induced Apoptosis. Biochem Biophys Res Commun (2017) 482:758–63. doi: 10.1016/j.bbrc.2016.11.107
93. Gomes C, Osório H, Pinto MT, Campos D, Oliveira MJ, Reis CA. Expression of ST3Gal4 Leads to SLex Expression and Induces C-Met Activation and an Invasive Phenotype in Gastric Carcinoma Cells. PLoS One (2013) 8:e66737. doi: 10.1371/journal.pone.0066737
94. Shen L, Luo Z, Wu J, Qiu L, Luo M, Ke Q, et al. Enhanced Expression of α2, 3-Linked Sialic Acids Promotes Gastric Cancer Cell Metastasis and Correlates With Poor Prognosis. Int J Oncol (2017) 50:1201–10. doi: 10.3892/ijo.2017.3882
95. Pérez-Garay M, Arteta B, Llop E, Cobler L, Pagès L, Ortiz R, et al. α2,3-Sialyltransferase ST3Gal IV Promotes Migration and Metastasis in Pancreatic Adenocarcinoma Cells and Tends to be Highly Expressed in Pancreatic Adenocarcinoma Tissues. Int J Biochem Cell Biol (2013) 45:1748–57. doi: 10.1016/j.biocel.2013.05.015
96. Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, et al. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol Cell (2019) 75:394–407.e5. doi: 10.1016/j.molcel.2019.05.017
97. Roa-De la Cruz L, Martínez-Morales P, Morán-Cruz I, Milflores-Flores L, Rosas-Murrieta N, González-Ramírez C, et al. Expression Analysis of ST3Gal4 Transcripts in Cervical Cancer Cells. Mol Med Rep (2018) 18:617–21. doi: 10.3892/mmr.2018.8938
98. Saito S, Yamashita SI, Endoh M, Yamato T, Hoshi S, Ohyama C, et al. Clinical Significance of ST3Gal IV Expression in Human Renal Cell Carcinoma. Oncol Rep (2002) 9:1251–5. doi: 10.3892/or.9.6.1251
99. Taniguchi A. Promoter Structure and Transcriptional Regulation of Human β-Galactoside α2, 3-Sialyltransferase Genes. Curr Drug Targets (2008) 9:310–6. doi: 10.2174/138945008783954998
100. Bowser LE, Young M, Wenger OK, Ammous Z, Brigatti KW, Carson VJ, et al. Recessive GM3 Synthase Deficiency: Natural History, Biochemistry, and Therapeutic Frontier. Mol Genet Metab (2019) 126:475–88. doi: 10.1016/j.ymgme.2019.01.013
101. Inokuchi J, Inamori K, Kabayama K, Nagafuku M, Uemura S, Go S, et al. Biology of GM3 Ganglioside. Prog Mol Biol Transl Sci (2018) 156:151–95. doi: 10.1016/bs.pmbts.2017.10.004
102. Hakomori SI, Handa K. GM3 and Cancer. Glycoconj J (2015) 32:1–8. doi: 10.1007/s10719-014-9572-4
103. Rapoport E, Mikhalyov I, Zhang J, Crocker P, Bovin N. Ganglioside Binding Pattern of CD33-Related Siglecs. Bioorganic Med Chem Lett (2003) 13:675–8. doi: 10.1016/S0960-894X(02)00998-8
104. Uemura S, Go S, Shishido F, Inokuchi JI. Expression Machinery of GM4: The Excess Amounts of GM3/GM4S Synthase (ST3Gal5) Are Necessary for GM4 Synthesis in Mammalian Cells. Glycoconj J (2014) 31:101–8. doi: 10.1007/s10719-013-9499-1
105. Suzuki M, Nagane M, Kato K, Yamauchi A, Shimizu T, Yamashita H, et al. Endothelial Ganglioside GM3 Regulates Angiogenesis in Solid Tumors. Biochem Biophys Res Commun (2021) 569:10–6. doi: 10.1016/j.bbrc.2021.06.063
106. Ouyang S, Liu JH, Ni Z, Ding GF, Wang QZ. Downregulation of ST3Gal5 is Associated With Muscle Invasion, High Grade and a Poor Prognosis in Patients With Bladder Cancer. Oncol Lett (2020) 20:828–40. doi: 10.3892/ol.2020.11597
107. Ley K. The Role of Selectins in Inflammation and Disease. Trends Mol Med (2003) 9:263–8. doi: 10.1016/S1471-4914(03)00071-6
108. Yang WH, Nussbaum C, Grewal PK, Marth JD, Sperandio M. Coordinated Roles of ST3Gal VI and ST3Gal IV Sialyltransferases in the Synthesis of Selectin Ligands. Blood (2012) 120:1015–26. doi: 10.1182/blood-2012-04-424366
109. Glavey SV, Manier S, Natoni A, Sacco A, Moschetta M, Reagan MR, et al. The Sialyltransferase ST3GAL6 Influences Homing and Survival in Multiple Myeloma. Blood (2014) 124:1765–76. doi: 10.1182/blood-2014-03-560862
110. Cuello HA, Segatori VI, Albertó M, Gulino CA, Aschero R, Camarero S, et al. Aberrant O-Glycosylation Modulates Aggressiveness in Neuroblastoma. Oncotarget (2018) 9:34176–88. doi: 10.18632/oncotarget.26169
111. Cuello HA, Ferreira GM, Gulino CA, Toledo AG, Segatori VI, Gabri MR. Terminally Sialylated and Fucosylated Complex N-Glycans Are Involved in the Malignant Behavior of High-Grade Glioma. Oncotarget (2021) 11:4822–35. doi: 10.18632/ONCOTARGET.27850
112. Sun M, Zhao X, Liang L, Pan X, Lv H, Zhao Y. Sialyltransferase ST3Gal6 Mediates the Effect of microRNA-26a on Cell Growth, Migration, and Invasion in Hepatocellular Carcinoma Through the Protein Kinase B/Mammalian Target of Rapamycin Pathway. Cancer Sci (2017) 108:267–76. doi: 10.1111/cas.13128
113. Dalangood S, Zhu Z, Ma Z, Li J, Zeng Q, Yan Y, et al. Identification of Glycogene-Type and Validation of ST3GAL6 as a Biomarker Predicts Clinical Outcome and Cancer Cell Invasion in Urinary Bladder Cancer. Theranostics (2020) 10:10078–91. doi: 10.7150/thno.48711
114. Venturi G, Gomes Ferreira I, Pucci M, Ferracin M, Malagolini N, Chiricolo M, et al. Impact of Sialyltransferase ST6Gal1 Overexpression on Different Colon Cancer Cell Types. Glycobiology (2019) 29:684–95. doi: 10.1093/glycob/cwz053
115. Kaneko Y, Yamamoto H, Kersey DS, Colley KJ, Leestma JE, Moskal JR. The Expression of Galβ1,4glcnac α2,6 Sialyltransferase and α2, 6-Linked Sialoglycoconjugates in Human Brain Tumors. Acta Neuropathol (1996) 91:284–92. doi: 10.1007/s004010050427
116. Olio FD, Malagolini N, di Stefano G, Minni F, Marrano D, Serafini-Cessi F. Increased CMP-NeuAc:Galβ1,4glcnac-R α2,6 Sialyltransferase Activity in Human Colorectal Cancer Tissues. Int J Cancer (1989) 44:434–9. doi: 10.1002/ijc.2910440309
117. Gretschel S, Haensch W, Schlag PM, Kemmner W. Clinical Relevance of Sialyltransferases ST6Gal I and ST3Gal III in Gastric Cancer. Oncology (2003) 65:139–45. doi: 10.1159/000072339
118. Wang PH, Feng Li Y, Juang CM, Lee YR, Chao HT, Tsai YC, et al. Altered mRNA Expression of Sialyltransferase in Squamous Cell Carcinomas of the Cervix. Gynecol Oncol (2001) 83:121–7. doi: 10.1006/gyno.2001.6358
119. Hsieh CC, Shyr YM, Liao WY, Chen TH, Wang SE, Lu PC, et al. Elevation of β-Galactoside α2,6-Sialyltransferase 1 in a Fructose-Responsive Manner Promotes Pancreatic Cancer Metastasis. Oncotarget (2016) 8:7691–709. doi: 10.18632/oncotarget.13845
120. Wichert B, Milde-Langosch K, Galatenko V, Schmalfeldt B, Oliveira-Ferrer L. Prognostic Role of the Sialyltransferase ST6Gal1 in Ovarian Cancer. Glycobiology (2018) 28:898–903. doi: 10.1093/glycob/cwy065
121. Garnham R, Scott E, Livermore KE, Munkley J. ST6Gal1: A Key Player in Cancer. Oncol Lett (2019) 18:983–9. doi: 10.3892/ol.2019.10458
122. Wei A, Fan B, Zhao Y, Zhang H, Wang L, Yu X, et al. ST6Gal I Overexpression Facilitates Prostate Cancer Progression via the PI3K/Akt/GSK-3β/β-Catenin Signaling Pathway. Oncotarget (2016) 7:65374–88. doi: 10.18632/oncotarget.11699
123. Jung YR, Park JJ, Jin YB, Cao YJ, Park MJ, Kim EJ, et al. Silencing of ST6Gal I Enhances Colorectal Cancer Metastasis by Down-Regulating KAI1 via Exosome-Mediated Exportation and Thereby Rescues Integrin Signaling. Carcinogenesis (2016) 37:1089–97. doi: 10.1093/carcin/bgw091
124. Yamamoto H, Kaneko Y, Rebbaa A, Bremer EG, Moskal JR. α2,6-Sialyltransferase Gene Transfection Into a Human Glioma Cell Line (U373 MG) Results in Decreased Invasivity. J Neurochem (1997) 68:2566–76. doi: 10.1046/j.1471-4159.1997.68062566.x
125. Yamamoto H, Oviedo A, Sweeley C, Saito T, Moskal JR. α2, 6-Sialylation of Cell-Surface N-Glycans Inhibits Glioma Formation In Vivo. Cancer Res (2001) 61:6822–9.
126. Antony P, Rose M, Heidenreich A, Knüchel R, Gaisa NT, Dahl E. Epigenetic Inactivation of ST6GAL1 in Human Bladder Cancer. BMC Cancer (2014) 14. doi: 10.1186/1471-2407-14-901
127. Jones RB, Dorsett KA, Hjelmeland AB, Bellis SL. The ST6Gal-I Sialyltransferase Protects Tumor Cells Against Hypoxia by Enhancing HIF-1 Signaling. J Biol Chem (2018) 293:5659–67. doi: 10.1074/jbc.RA117.001194
128. Lu J, Isaji T, Im S, Fukuda T, Hashii N, Takakura D, et al. β-Galactoside α2,6-Sialyltranferase 1 Promotes Transforming Growth Factor-β-Mediated Epithelial-Mesenchymal Transition. J Biol Chem (2014) 289:34627–41. doi: 10.1074/jbc.M114.593392
129. Britain CM, Bhalerao N, Silva AD, Chakraborty A, Buchsbaum DJ, Crowley MR, et al. Glycosyltransferase ST6Gal-I Promotes the Epithelial to Mesenchymal Transition in Pancreatic Cancer Cells. J Biol Chem (2021) 296. doi: 10.1074/jbc.RA120.014126
130. Meng Q, Ren C, Wang L, Zhao Y, Wang S. Knockdown of ST6Gal-I Inhibits the Growth and Invasion of Osteosarcoma MG-63 Cells. BioMed Pharmacother (2015) 72:172–8. doi: 10.1016/j.biopha.2015.04.020
131. Swindall AF, Londoño-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. ST6Gal I Protein Expression Is Upregulated in Human Epithelial Tumors and Correlates With Stem Cell Markers in Normal Tissues and Colon Cancer Cell Lines. Cancer Res (2013) 73:2368–78. doi: 10.1158/0008-5472.CAN-12-3424
132. Dorsett KA, Jones RB, Ankenbauer KE, Hjelmeland AB, Bellis SL. Sox2 Promotes Expression of the ST6Gal-I Glycosyltransferase in Ovarian Cancer Cells. J Ovarian Res (2019) 12:93. doi: 10.1186/s13048-019-0574-5
133. Alexander K, Serrano C, Chakraborty A, Nearing M, Council L, Riquelme A, et al. Modulation of Glycosyltransferase ST6Gal-I in Gastric Cancer-Derived Organoids Disrupts Homeostatic Epithelial Cell Turnover. J Biol Chem (2020) 295:14153–63. doi: 10.1074/JBC.RA120.014887
134. Vergé C, Bouchatal A, Chirat F, Guérardel Y, Maftah A, Petit J. Involvement of ST6Gal I-Mediated α2, 6 Sialylation in Myoblast Proliferation and Differentiation. FEBS Open Bio (2020) 10:56–69. doi: 10.1002/2211-5463.12745
135. Wang YC, Stein JW, Lynch CL, Tran HT, Lee CY, Coleman R, et al. Glycosyltransferase ST6Gal1 Contributes to the Regulation of Pluripotency in Human Pluripotent Stem Cells. Sci Rep (2015) 5:1–13. doi: 10.1038/srep13317
136. Schultz MJ, Holdbrooks AT, Chakraborty A, Grizzle WE, Landen CN, Buchsbaum DJ, et al. The Tumor-Associated Glycosyltransferase ST6Gal-I Regulates Stem Cell Transcription Factors and Confers a Cancer Stem Cell Phenotype. Cancer Res (2016) 76:3978–88. doi: 10.1158/0008-5472.CAN-15-2834
137. Cui H, Yang S, Jiang Y, Li C, Zhao Y, Shi Y, et al. The Glycosyltransferase ST6Gal-I Is Enriched in Cancer Stem-Like Cells in Colorectal Carcinoma and Contributes to Their Chemo-Resistance. Clin Transl Oncol (2018) 20:1175–84. doi: 10.1007/s12094-018-1840-5
138. Chakraborty A, Dorsett KA, Trummell HQ, Yang ES, Oliver PG, Bonner JA, et al. ST6Gal I Sialyltransferase Promotes Chemoresistance in Pancreatic Ductal Adenocarcinoma by Abrogating Gemcitabine-Mediated DNA Damage. J Biol Chem (2018) 293:984–94. doi: 10.1074/jbc.M117.808584
139. Zhang X, Pan C, Zhou L, Cai Z, Zhao S, Yu D. Knockdown of ST6Gal I Increases Cisplatin Sensitivity in Cervical Cancer Cells. BMC Cancer (2016) 16:949. doi: 10.1186/s12885-016-2981-y
140. Liu N, Zhu M, Linhai Y, Song Y, Gui X, Tan G, et al. Increasing HER2 A2, 6 Sialylation Facilitates Gastric Cancer Progression and Resistance via the Akt and ERK Pathways. Oncol Rep (2018) 40:2997–3005. doi: 10.3892/or.2018.6680
141. Duarte HO, Rodrigues JG, Gomes C, Hensbergen PJ, Ederveen ALH, de Ru AH, et al. ST6Gal1 Targets the Ectodomain of ErbB2 in a Site-Specific Manner and Regulates Gastric Cancer Cell Sensitivity to Trastuzumab. Oncogene (2021) 40:3719–33. doi: 10.1038/s41388-021-01801-w
142. Britain CM, Holdbrooks AT, Anderson JC, Willey CD, Bellis SL. Sialylation of EGFR by the ST6Gal I Sialyltransferase Promotes EGFR Activation and Resistance to Gefitinib-Mediated Cell Death. J Ovarian Res (2018) 11:12. doi: 10.1186/s13048-018-0385-0
143. Liu Z, Swindall AF, Kesterson RA, Schoeb TR, Bullard DC, Bellis SL. ST6Gal I Regulates Macrophage Apoptosis via α2-6 Sialylation of the TNFR1 Death Receptor. J Biol Chem (2011) 286:39654–62. doi: 10.1074/jbc.M111.276063
144. Holdbrooks AT, Britain CM, Bellis SL. ST6Gal-I Sialyltransferase Promotes Tumor Necrosis Factor (TNF)-Mediated Cancer Cell Survival via Sialylation of the TNF Receptor 1 (TNFR1) Death Receptor. J Biol Chem (2018) 293:1610–22. doi: 10.1074/jbc.m117.801480
145. Swindall AF, Bellis SL. Sialylation of the Fas Death Receptor by ST6Gal-I Provides Protection Against Fas-Mediated Apoptosis in Colon Carcinoma Cells. J Biol Chem (2011) 286:22982–90. doi: 10.1074/jbc.M110.211375
146. Zhuo Y, Chammas R, Bellis SL. Sialylation of Beta1 Integrins Blocks Cell Adhesion to Galectin-3 and Protects Cells Against Galectin-3-Induced Apoptosis. J Biol Chem (2008) 283:22177–85. doi: 10.1074/jbc.m8000015200
147. Krzewinski-Recchi MA, Julien S, Juliant S, Teintenier-Lelièvre M, Samyn-Petit B, Montiel MD, et al. Identification and Functional Expression of a Second Human β-Galactoside α2, 6-Sialyltransferase, ST6Gal II. Eur J Biochem (2003) 270:950–61. doi: 10.1046/j.1432-1033.2003.03458.x
148. Takashima S, Tsuji S, Tsujimoto M. Characterization of the Second Type of Human β-Galactoside α2, 6-Sialyltransferase (ST6Gal II), Which Sialylates Galβ1, 4glcnac Structures on Oligosaccharides Preferentially: Genomic Analysis of Human Sialyltransferase Genes. J Biol Chem (2002) 277:45719–28. doi: 10.1074/jbc.M206808200
149. Cheng J, Wang R, Zhong G, Chen X, Cheng Y, Li W, et al. ST6GAL2 Downregulation Inhibits Cell Adhesion and Invasion and Is Associated With Improved Patient Survival in Breast Cancer. Onco Targets Ther (2020) 13:903–14. doi: 10.2147/OTT.S230847
150. Xu G, Chen J, Wang G, Xiao J, Zhang N, Chen Y, et al. Resveratrol Inhibits the Tumorigenesis of Follicular Thyroid Cancer via ST6Gal2-Regulated Activation of the Hippo Signaling Pathway. Mol Ther - Oncolytics (2020) 16:124–33. doi: 10.1016/j.omto.2019.12.010
151. Maugeri-Saccà M, De Maria R. The Hippo Pathway in Normal Development and Cancer. Pharmacol Ther (2018) 186:60–72. doi: 10.1016/j.pharmthera.2017.12.011
152. Qiao Y, Li T, Zheng S, Wang H. The Hippo Pathway as a Drug Target in Gastric Cancer. Cancer Lett (2018) 420:14–25. doi: 10.1016/j.canlet.2018.01.062
153. Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell (2015) 163:811–28. doi: 10.1016/j.cell.2015.10.044
154. Marcos NT, Pinho S, Grandela C, Cruz A, Samyn-Petit B, Harduin-Lepers A, et al. Role of the Human ST6GalNAc I and ST6GalNAc II in the Synthesis of the Cancer-Associated Sialyl-Tn Antigen. Cancer Res (2004) 64:7050–7. doi: 10.1158/0008-5472.CAN-04-1921
155. Munkley J. The Role of Sialyl-Tn in Cancer. Int J Mol Sci (2016) 17(3):275. doi: 10.3390/ijms17030275
156. Schmitt FC, Figueiredo P, Lacerda M. Simple Mucin-Type Carbohydrate Antigens (T, Sialosyl-T, Tn and Sialosyl-Tn) in Breast Carcinogenesis. Virchows Arch (1995) 427:251–8. doi: 10.1007/BF00203391
157. Munkley J, Oltean S, Vodák D, Wilson BT, Livermore KE, Zhou Y, et al. The Androgen Receptor Controls Expression of the Cancer Associated sTn Antigen and Cell Adhesion Through Induction of ST6GalNAc1 in Prostate Cancer. Oncotarget (2015) 6:34358–74. doi: 10.18632/oncotarget.6024
158. Victorzon M, Nordling S, Nilsson O, Roberts PJ, Haglund C. Sialyl Tn Antigen Is an Independent Predictor of Outcome in Patients With Gastric Cancer. Int J Cancer (1996) 65:295–300. doi: 10.1002/(SICI)1097-0215(19960126)65:3<295::AID-IJC3>3.0.CO;2-V
159. Leivonen M, Nordling S, Lundin J, Von Boguslawski K, Haglund C. STn and Prognosis in Breast Cancer. Oncology (2001) 61:299–305. doi: 10.1159/000055337
160. Costa C, Pereira S, Lima L, Peixoto A, Fernandes E, Neves D, et al. Abnormal Protein Glycosylation and Activated PI3K/Akt/mTOR Pathway: Role in Bladder Cancer Prognosis and Targeted Therapeutics. PLoS One (2015) 10:e0141253. doi: 10.1371/journal.pone.0141253
161. Ogawa T, Hirohashi Y, Murai A, Nishidate T, Okita K, Wang L, et al. ST6GalNAc1 Plays Important Roles in Enhancing Cancer Stem Phenotypes of Colorectal Cancer via the Akt Pathway. Oncotarget (2017) 8:112550–64. doi: 10.18632/oncotarget.22545
162. Sewell R, Bäckström M, Dalziel M, Gschmeissner S, Karlsson H, Noll T, et al. The ST6GalNAc I Sialyltransferase Localizes Throughout the Golgi and Is Responsible for the Synthesis of the Tumor-Associated Sialyl-Tn O-Glycan in Human Breast Cancer. J Biol Chem (2006) 281:3586–94. doi: 10.1074/jbc.M511826200
163. Julien S, Adriaenssens E, Ottenberg K, Furlan A, Courtand G, Vercoutter-Edouart AS, et al. ST6GalNAc I Expression in MDA-MB-231 Breast Cancer Cells Greatly Modifies Their O-Glycosylation Pattern and Enhances Their Tumourigenicity. Glycobiology (2006) 16:54–64. doi: 10.1093/glycob/cwj033
164. Wang WY, Cao YX, Zhou X, Wei B, Zhan L, Sun SY. Stimulative Role of ST6GalNAc1 in Proliferation, Migration and Invasion of Ovarian Cancer Stem Cells via the Akt Signaling Pathway. Cancer Cell Int (2019) 19:86. doi: 10.1186/s12935-019-0780-7
165. Ozaki H, Matsuzaki H, Ando H, Kaji H, Nakanishi H, Ikehara Y, et al. Enhancement of Metastatic Ability by Ectopic Expression of ST6GalNAc I on a Gastric Cancer Cell Line in a Mouse Model. Clin Exp Metastasis (2012) 29:229–38. doi: 10.1007/s10585-011-9445-1
166. Pinho S, Marcos NT, Ferreira B, Carvalho AS, Oliveira MJ, Santos-Silva F, et al. Biological Significance of Cancer-Associated Sialyl-Tn Antigen: Modulation of Malignant Phenotype in Gastric Carcinoma Cells. Cancer Lett (2007) 249:157–70. doi: 10.1016/j.canlet.2006.08.010
167. Kvorjak M, Ahmed Y, Miller ML, Sriram R, Coronnello C, Hashash JG, et al. Cross-Talk Between Colon Cells and Macrophages Increases ST6GALNAC1 and MUC1-sTn Expression in Ulcerative Colitis and Colitis-Associated Colon Cancer. Cancer Immunol Res (2020) 8:167–78. doi: 10.1158/2326-6066.CIR-19-0514
168. Iwaya T, Sawada G, Amano S, Kume K, Ito C, Endo F, et al. Downregulation of ST6GalNAc1 Is Associated With Esophageal Squamous Cell Carcinoma Development. Int J Oncol (2017) 50:441–7. doi: 10.3892/ijo.2016.3817
169. Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: An Immune System Siglec Conserved Throughout Vertebrate Evolution. Glycobiology (2007) 17:838–46. doi: 10.1093/glycob/cwm049
170. Büll C, Nason R, Sun L, Van Coillie J, Sørensen DM, Moons SJ, et al. Probing the Binding Specificities of Human Siglecs by Cell-Based Glycan Arrays. Proc Natl Acad Sci USA (2021) 118:1–12. doi: 10.1073/pnas.2026102118
171. Takamiya R, Ohtsubo K, Takamatsu S, Taniguchi N, Angata T. The Interaction Between Siglec-15 and Tumor-Associated Sialyl-Tn Antigen Enhances TGF-β Secretion From Monocytes/Macrophages Through the DAP12–Syk Pathway. Glycobiology (2013) 23:178–87. doi: 10.1093/GLYCOB/CWS139
172. Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, et al. Siglec-15 as an Immune Suppressor and Potential Target for Normalization Cancer Immunotherapy. Nat Med (2019) 25:656. doi: 10.1038/S41591-019-0374-X
173. Schneider F, Kemmner W, Haensch W, Franke G, Gretschel S, Karsten U, et al. Overexpression of Sialyltransferase CMP-Sialic Acid: Galbeta1,3GalNAc-R Alpha6-Sialyltransferase Is Related to Poor Patient Survival in Human Colorectal Carcinomas. Cancer Res (2001) 61:4605–11.
174. Miao X, Zhao Y. ST6GalNAc II Mediates Tumor Invasion Through PI3K/Akt/NFB Signaling Pathway in Follicular Thyroid Carcinoma. Oncol Rep (2016) 35:2131–40. doi: 10.3892/or.2016.4590
175. Ren D, Jia L, Li Y, Gong Y, Liu C, Zhang X, et al. ST6GalNAc II Mediates the Invasive Properties of Breast Carcinoma Through PI3K/Akt/NF-κb Signaling Pathway. IUBMB Life (2014) 66:300–8. doi: 10.1002/iub.1268
176. Murugaesu N, Iravani M, Van Weverwijk A, Ivetic A, Johnson DA, Antonopoulos A, et al. An In Vivo Functional Screen Identifies ST6GalNAc2 Sialyltransferase as a Breast Cancer Metastasis Suppressor. Cancer Discovery (2014) 4:304–17. doi: 10.1158/2159-8290.CD-13-0287
177. Tsuchida A, Ogiso M, Nakamura Y, Kiso M, Furukawa KK, Furukawa KK. Molecular Cloning and Expression of Human ST6GalNAc III: Restricted Tissue Distribution and Substrate Specificity. J Biochem (2005) 138:237–43. doi: 10.1093/jb/mvi124
178. Haldrup C, Pedersen AL, Øgaard N, Strand SH, Høyer S, Borre M, et al. Biomarker Potential of ST6GalNAc3 and ZNF660 Promoter Hypermethylation in Prostate Cancer Tissue and Liquid Biopsies. Mol Oncol (2018) 12:545–60. doi: 10.1002/1878-0261.12183
179. Jung S-Y, Lee HK, Kim H, Kim S, Kim JS, Kang JG, et al. Depletion of ST6GALNACIII Retards A549 Non-Small Cell Lung Cancer Cell Proliferation by Downregulating Transferrin Receptor Protein 1 Expression. Biochem Biophys Res Commun (2021) 575:78–84. doi: 10.1016/j.bbrc.2021.08.055
180. Hentze MW, Muckenthaler MU, Andrews NC. Balancing Acts: Molecular Control of Mammalian Iron Metabolism. Cell (2004) 117:285–97. doi: 10.1016/S0092-8674(04)00343-5
181. Harduin-Lepers A, Stokes DC, Steelant WFA, Samyn-Petit B, Krzewinski-Recchi MA, Vallejo-Ruiz V, et al. Cloning, Expression and Gene Organization of a Human Neu5Acα2-3galβ1-3galnac α2,6-Sialyltransferase: Hst6galnac IV. Biochem J (2000) 352:37–48. doi: 10.1042/0264-6021:3520037
182. Lee YC, Kaufmann M, Kitazume-Kawaguchi S, Kono M, Takashima S, Kurosawa N, et al. Molecular Cloning and Functional Expression of Two Members of Mouse Neuacα2,3galβ1,3galnac Galnacα2,6-Sialyltransferase Family, ST6GalNAc III and Iv. J Biol Chem (1999) 274:11958–67. doi: 10.1074/jbc.274.17.11958
183. Reticker-Flynn NE, Bhatia SN. Aberrant Glycosylation Promotes Lung Cancer Metastasis Through Adhesion to Galectins in the Metastatic Niche. Cancer Discov (2015) 5:168–81. doi: 10.1158/2159-8290.CD-13-0760
184. Miao X, Jia L, Zhou H, Song X, Zhou M, Xu J, et al. MiR-4299 Mediates the Invasive Properties and Tumorigenicity of Human Follicular Thyroid Carcinoma by Targeting ST6GALNAC4. IUBMB Life (2016) 68:136–44. doi: 10.1002/iub.1467
185. Ikehara Y, Shimizu N, Kono M, Nishihara S, Nakanishi H, Kitamura T, et al. A Novel Glycosyltransferase With a Polyglutamine Repeat; A New Candidate for GD1α Synthase (ST6GalNAc V). FEBS Lett (1999) 463:92–6. doi: 10.1016/S0014-5793(99)01605-1
186. Okajima T, Fukumoto S, Ito H, Kiso M, Hirabayashi Y, Urano T, et al. Molecular Cloning of Brain-Specific GD1α Synthase (ST6GalNAc V) Containing CAG/glutamine Repeats. J Biol Chem (1999) 274:30557–62. doi: 10.1074/jbc.274.43.30557
187. Vandermeersch S, Vanbeselaere J, Delannoy CP, Drolez A, Mysiorek C, Guérardel Y, et al. Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V. Molecules (2015) 20:6913–24. doi: 10.3390/molecules20046913
188. Stremitzer S, Berghoff AS, Volz NB, Zhang W, Yang D, Stintzing S, et al. Genetic Variants Associated With Colorectal Brain Metastases Susceptibility and Survival. Pharmacogenomics J (2017) 17:29–35. doi: 10.1038/tpj.2015.86
189. Bos PD, Zhang XHF, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes That Mediate Breast Cancer Metastasis to the Brain. Nature (2009) 459:1005–9. doi: 10.1038/nature08021
190. Drolez A, Vandenhaute E, Delannoy CP, Dewald JH, Gosselet F, Cecchelli R, et al. ST6GalNAc5 Expression Decreases the Interactions Between Breast Cancer Cells and the Human Blood-Brain Barrier. Int J Mol Sci (2016) 17:1309. doi: 10.3390/ijms17081309
191. Okajima T, Chen HH, Ito H, Kiso M, Tai T, Furukawa K, et al. Molecular Cloning and Expression of Mouse GD1α/Gtiaα/Gq1bα Synthase (ST6GalNAc VI) Gene. J Biol Chem (2000) 275:6717–23. doi: 10.1074/jbc.275.10.6717
192. Kawasaki Y, Ito A, Kakoi N, Shimada S, Itoh J, Mitsuzuka K, et al. Ganglioside, Disialosyl Globopentaosylceramide (DSGB5), Enhances the Migration of Renal Cell Carcinoma Cells. Tohoku J Exp Med (2015) 236:1–7. doi: 10.1620/tjem.236.1
193. Senda M, Ito A, Tsuchida A, Hagiwara T, Kaneda T, Nakamura Y, et al. Identification and Expression of a Sialyltransferase Responsible for the Synthesis of Disialylgalactosylgloboside in Normal and Malignant Kidney Cells: Downregulation of ST6GalNAc VI in Renal Cancers. Biochem J (2007) 402:459–70. doi: 10.1042/BJ20061118
194. Tsuchida A, Okajima T, Furukawa K, Ando T, Ishida H, Yoshida A, et al. Synthesis of Disialyl Lewis a (Lea) Structure in Colon Cancer Cell Lines by a Sialyltransferase, ST6GalNAc VI, Responsible for the Synthesis of α-Series Gangliosides. J Biol Chem (2003) 278:22787–94. doi: 10.1074/jbc.M211034200
195. Miyazaki K, Ohmori K, Izawa M, Koike T, Kumamoto K, Furukawa K, et al. Loss of Disialyl Lewisa, the Ligand for Lymphocyte Inhibitory Receptor Sialic Acid-Binding Immunoglobulin-Like Lectin-7 (Siglec-7) Associated With Increased Sialyl Lewisa Expression on Human Colon Cancers. Cancer Res (2004) 64:4498–505. doi: 10.1158/0008-5472.CAN-03-3614
196. Huang HC, Chao CC, Wu PH, Chung HY, Lee HY, Suen CS, et al. Epigenetic Silencing of the Synthesis of Immunosuppressive Siglec Ligand Glycans by NF-κb/EZH2/YY1 Axis in Early-Stage Colon Cancers. Biochim Biophys Acta - Gene Regul Mech (2019) 1862:173–83. doi: 10.1016/j.bbagrm.2019.01.002
197. Kawasaki Y, Ito A, Withers DA, Taima T, Kakoi N, Saito S, et al. Ganglioside DSGb5, Preferred Ligand for Siglec-7, Inhibits NK Cell Cytotoxicity Against Renal Cell Carcinoma Cells. Glycobiology (2010) 20:1373–9. doi: 10.1093/glycob/cwq116
198. Sato C, Kitajima K. Polysialylation and Disease. Mol Aspects Med (2021) 79:100892. doi: 10.1016/J.MAM.2020.100892
199. Angata K, Suzuki M, McAuliffe J, Ding Y, Hindsgaul O, Fukuda M. Differential Biosynthesis of Polysialic Acid on Neural Cell Adhesion Molecule (NCAM) and Oligosaccharide Acceptors by Three Distinct α2,8- Sialyltransferases, ST8Sia IV (PST), ST8Sia II (STX), and ST8Sia III. J Biol Chem (2000) 275:18594–601. doi: 10.1074/jbc.M910204199
200. Yeh SC, Wang PY, Lou YW, Khoo KH, Hsiao M, Hsu TL, et al. Glycolipid GD3 and GD3 Synthase Are Key Drivers for Glioblastoma Stem Cells and Tumorigenicity. Proc Natl Acad Sci USA (2016) 113:5592–7. doi: 10.1073/pnas.1604721113
201. Ramos RI, Bustos MA, Wu J, Jones P, Chang SC, Kiyohara E, et al. Upregulation of Cell Surface GD3 Ganglioside Phenotype Is Associated With Human Melanoma Brain Metastasis. Mol Oncol (2020) 14:1760–78. doi: 10.1002/1878-0261.12702
202. Kan JY, Moi SH, Hung WC, Hou MF, Chen FM, Shih SL, et al. Comprehensive Transcriptomic Analysis Identifies ST8Sia1 as a Survival-Related Sialyltransferase Gene in Breast Cancer. Genes (Basel) (2020) 11:1–15. doi: 10.3390/genes11121436
203. Sarkar TR, Battula VL, Werden SJ, Vijay GV, Ramirez-Peña EQ, Taube JH, et al. GD3 Synthase Regulates Epithelial-Mesenchymal Transition and Metastasis in Breast Cancer. Oncogene (2015) 34:2958–67. doi: 10.1038/onc.2014.245
204. Li W, Zheng X, Ren L, Fu W, Liu J, Xv J, et al. Epigenetic Hypomethylation and Upregulation of GD3s in Triple Negative Breast Cancer. Ann Transl Med (2019) 7:723–3. doi: 10.21037/atm.2019.12.23
205. Ruckhäberle E, Karn T, Rody A, Hanker L, Gätje R, Metzler D, et al. Gene Expression of Ceramide Kinase, Galactosyl Ceramide Synthase and Ganglioside GD3 Synthase is Associated With Prognosis in Breast Cancer. J Cancer Res Clin Oncol (2009) 135:1005–13. doi: 10.1007/s00432-008-0536-6
207. Battula VL, Shi Y, Evans KW, Wang RY, Spaeth EL, Jacamo RO, et al. Ganglioside GD2 Identifies Breast Cancer Stem Cells and Promotes Tumorigenesis. J Clin Invest (2012) 122:2066–78. doi: 10.1172/JCI59735
208. Iwasawa T, Zhang P, Ohkawa Y, Momota H, Wakabayashi T, Ohmi Y, et al. Enhancement of Malignant Properties of Human Glioma Cells by Ganglioside GD3/GD2. Int J Oncol (2018) 52:1255–66. doi: 10.3892/ijo.2018.4266
209. Cazet A, Groux-Degroote S, Teylaert B, Kwon KM, Lehoux S, Slomianny C, et al. GD3 Synthase Overexpression Enhances Proliferation and Migration of MDA-MB-231 Breast Cancer Cells. Biol Chem (2009) 390:601–9. doi: 10.1515/BC.2009.054
210. Wan H, Li Z, Wang H, Cai F, Wang L. ST8SIA1 Inhibition Sensitizes Triple Negative Breast Cancer to Chemotherapy via Suppressing Wnt/β-Catenin and FAK/Akt/mTOR. Clin Transl Oncol (2021) 23:902–10. doi: 10.1007/s12094-020-02484-7
211. Nicoll G, Avril T, Lock K, Furukawa K, Bovin N, Crocker PR. Ganglioside GD3 Expression on Target Cells Can Modulate NK Cell Cytotoxicity via Siglec-7-Dependent and -Independent Mechanisms. Eur J Immunol (2003) 33:1642–8. doi: 10.1002/eji.200323693
212. Scheidegger EP, Sternberg LR, Roth J, Lowe JB. A Human STX cDNA Confers Polysialic Acid Expression in Mammalian Cells. J Biol Chem (1995) 270:22685–8. doi: 10.1074/jbc.270.39.22685
213. Tanaka F, Otake Y, Nakagawa T, Kawano Y, Miyahara R, Li M, et al. Expression of Polysialic Acid and STX, a Human Polysialyltransferase, Is Correlated With Tumor Progression in Non-Small Cell Lung Cancer. Cancer Res (2000) 60:3072–80.
214. Cheung IY, Vickers A, Cheung N-KV. Sialyltransferase STX (ST8SiaII): A Novel Molecular Marker of Metastatic Neuroblastoma. Int J Cancer (2006) 119:152–6. doi: 10.1002/IJC.21789
215. Hao J, Zeltz C, Pintilie M, Li Q, Sakashita S, Wang T, et al. Characterization of Distinct Populations of Carcinoma-Associated Fibroblasts From Non–Small Cell Lung Carcinoma Reveals a Role for ST8SIA2 in Cancer Cell Invasion. Neoplasia (United States) (2019) 21:482–93. doi: 10.1016/j.neo.2019.03.009
216. Suzuki MM, Suzuki MM, Nakayama J, Suzuki A, Angata K, Chen S, et al. Polysialic Acid Facilitates Tumor Invasion by Glioma Cells. Glycobiology (2005) 15:887–94. doi: 10.1093/glycob/cwi071
217. Gong L, Zhou X, Yang J, Jiang Y, Yang H. Effects of the Regulation of Polysialyltransferase ST8SiaII on the Invasiveness and Metastasis of Small Cell Lung Cancer Cells. Oncol Rep (2017) 37:131–8. doi: 10.3892/or.2016.5279
218. Al-Saraireh YMJ, Sutherland M, Springett BR, Freiberger F, Ribeiro Morais G, Loadman PM, et al. Pharmacological Inhibition of Polysialyltransferase ST8SiaII Modulates Tumour Cell Migration. PLoS One (2013) 8:73366. doi: 10.1371/journal.pone.0073366
219. Yoshida Y, Kojima N, Kurosawa N, Hamamoto T, Tsuji S. Molecular Cloning of Siaα2,3galβ1,4glcnac α2, 8-Sialyltransferase From Mouse Brain. J Biol Chem (1995) 270:14628–33. doi: 10.1074/jbc.270.24.14628
220. Lin CY, Lai HL, Chen HM, Siew JJ, Hsiao CT, Chang HC, et al. Functional Roles of ST8SIA3-Mediated Sialylation of Striatal Dopamine D2 and Adenosine A2A Receptors. Transl Psychiatry (2019) 9:209. doi: 10.1038/s41398-019-0529-z
221. Baeza-Kallee N, Bergès R, Soubéran A, Colin C, Denicolaï E, Appay R, et al. Glycolipids Recognized by A2B5 Antibody Promote Proliferation, Migration, and Clonogenicity in Glioblastoma Cells. Cancers (Basel) (2019) 11(9):1267. doi: 10.3390/cancers11091267
222. Mori A, Hane M, Niimi Y, Kitajima K, Sato C. Different Properties of Polysialic Acids Synthesized by the Polysialyltransferases ST8Sia2 and ST8Sia4. Glycobiology (2017) 27:834–46. doi: 10.1093/glycob/cwx057
223. Foley DA, Swartzentruber KG, Colley KJ. Identification of Sequences in the Polysialyltransferases ST8Sia II and ST8Sia IV That Are Required for the Protein-Specificpolysialylation of the Neural Cell Adhesion Molecule, NCAM. J Biol Chem (2009) 284:15505–16. doi: 10.1074/jbc.M809696200
224. Ma X, Dong W, Su Z, Zhao L, Miao Y, Li N, et al. Functional Roles of Sialylation in Breast Cancer Progression Through miR-26a/26b Targeting ST8SIA4. Cell Death Dis (2016) 7:e2561–1. doi: 10.1038/cddis.2016.427
225. Pan Y, Wu Y, Hu J, Shan Y, Ma J, Ma H, et al. Long Noncoding RNA HOTAIR Promotes Renal Cell Carcinoma Malignancy Through Alpha-2, 8-Sialyltransferase 4 by Sponging microRNA-124. Cell Prolif (2018) 51:e12507. doi: 10.1111/cpr.12507
226. Zhang X, Dong W, Zhou H, Li H, Wang N, Miao X, et al. α-2,8-Sialyltransferase Is Involved in the Development of Multidrug Resistance via PI3K/Akt Pathway in Human Chronic Myeloid Leukemia. IUBMB Life (2015) 67:77–87. doi: 10.1002/iub.1351
227. Ma H, Zhou H, Song X, Shi S, Zhang J, Jia L. Modification of Sialylation Is Associated With Multidrug Resistance in Human Acute Myeloid Leukemia. Oncogene (2015) 34:726–40. doi: 10.1038/onc.2014.7
228. Zhang Z, Zhao Y, Jiang L, Miao X, Zhou H, Jia L. Glycomic Alterations are Associated With Multidrug Resistance in Human Leukemia. Int J Biochem Cell Biol (2012) 44:1244–53. doi: 10.1016/j.biocel.2012.04.026
229. Ma W, Zhao X, Liang L, Wang G, Li Y, Miao X, et al. miR-146a and miR-146b Promote Proliferation, Migration and Invasion of Follicular Thyroid Carcinoma via Inhibition of ST8SIA4. Oncotarget (2017) 8:28028–41. doi: 10.18632/oncotarget.15885
230. Wang F, Ye LJ, Wang FJ, Liu HF, Wang XL. miR-146a Promotes Proliferation, Invasion, and Epithelial-to-Mesenchymal Transition in Oral Squamous Carcinoma Cells. Environ Toxicol (2020) 35:1050–7. doi: 10.1002/tox.22941
231. Kim YJ, Kim KS, Il D, CH K, SK K, Lee YC. Molecular Cloning and Expression of Human α2,8-Sialyltransferase (Hst8sia V). Biochem Biophys Res Commun (1997) 235:327–30. doi: 10.1006/bbrc.1997.6725
232. Kono M, Yoshida Y, Kojima N, Tsuji S. Molecular Cloning and Expression of a Fifth Type of α2,8- Sialyltransferase (ST8Sia V): Its Substrate Specificity is Similar to That of SAT-V/III, Which Synthesize G(D1c), G(T1a), G(Q1b) and G(T3). J Biol Chem (1996) 271:29366–71. doi: 10.1074/jbc.271.46.29366
233. Penrose HM, Cable C, Heller S, Ungerleider N, Nakhoul H, Baddoo M, et al. Loss of Forkhead Box O3 Facilitates Inflammatory Colon Cancer: Transcriptome Profiling of the Immune Landscape and Novel Targets. Cmgh (2019) 7:391–408. doi: 10.1016/j.jcmgh.2018.10.003
234. Takashima S, Ishida HK, Inazu T, Ando T, Ishida HK, Kiso M, et al. Molecular Cloning and Expression of a Sixth Type of α2, 8-Sialyltransferase (ST8Sia VI) That Sialylates O-Glycans. J Biol Chem (2002) 277:24030–8. doi: 10.1074/jbc.M112367200
235. Luo ML, Li J, Shen L, Chu J, Guo Q, Liang G, et al. The Role of APAL/ST8SIA6-AS1 lncRNA in PLK1 Activation and Mitotic Catastrophe of Tumor Cells. J Natl Cancer Inst (2020) 112:356–68. doi: 10.1093/jnci/djz134
236. Zhang B, Liu Z, Liu J, Cao K, Shan W, Wen Q, et al. Long Non−Coding RNA ST8SIA6−AS1 Promotes the Migration and Invasion of Hypoxia−Treated Hepatocellular Carcinoma Cells Through the Mir−338/MEPCE Axis. Oncol Rep (2020) 45:73–82. doi: 10.3892/or.2020.7864
237. Zhang X, Xu S, Hu C, Fang K, Zhou J, Guo Z, et al. LncRNA ST8SIA6-AS1 Promotes Hepatocellular Carcinoma Progression by Regulating MAGEA3 and DCAF4L2 Expression. Biochem Biophys Res Commun (2020) 533:1039–47. doi: 10.1016/j.bbrc.2020.09.115
238. Cao Q, Yang W, Ji X, Wang W. Long non-Coding RNA ST8SIA6-AS1 Promotes Lung Adenocarcinoma Progression Through Sponging miR-125a-3p. Front Genet (2020) 11:597795. doi: 10.3389/fgene.2020.597795
239. Huang CM, Cao GY, Yang CX, Chen Y, Liu GD, Xu BW, et al. LncRNA ST8SIA6-AS1 Promotes Colorectal Cancer Cell Proliferation, Migration and Invasion by Regulating the miR-5195/PCBP2 Axis. Eur Rev Med Pharmacol Sci (2020) 24:4203–11. doi: 10.26355/eurrev_202004_21000
240. Fang K, Hu C, Zhang X, Hou Y, Gao D, Guo Z, et al. LncRNA ST8Sia6-AS1 Promotes Proliferation, Migration and Invasion in Breast Cancer Through the P38 MAPK Signalling Pathway. Carcinogenesis (2020) 41:1273–81. doi: 10.1093/carcin/bgz197
241. Friedman DJ, Crotts SB, Shapiro MJ, Rajcula M, McCue S, Liu X, et al. ST8Sia6 Promotes Tumor Growth in Mice by Inhibiting Immune Responses. Cancer Immunol Res (2021) 9:952–66. doi: 10.1158/2326-6066.CIR-20-0834
242. Belmonte PJ, Shapiro MJ, Rajcula MJ, McCue SA, Shapiro VS. ST8Sia6 Generated Alpha-2, 8-Disialic Acids Mitigate Hyperglycemia in Multiple Low Dose Streptozotocin-Induced Diabetes. J Immunol (2020) 204:3071. doi: 10.4049/JIMMUNOL.2000023
243. Von Gunten S, Bochner BS. Basic and Clinical Immunology of Siglecs. Ann N Y Acad Sci (2008) 1143:61–82. doi: 10.1196/annals.1443.011
244. Angata T, Margulies EH, Green ED, Varki A. Large-Scale Sequencing of the CD33-Related Siglec Gene Cluster in Five Mammalian Species Reveals Rapid Evolution by Multiple Mechanisms. Proc Natl Acad Sci USA (2004) 101:13251–6. doi: 10.1073/pnas.0404833101
245. Crocker PR, Paulson JC, Varki A. Siglecs and Their Roles in the Immune System. Nat Rev Immunol (2007) 7:255–66. doi: 10.1038/nri2056
246. Gonzalez-Gil A, Schnaar RL. Siglec Ligands. Cells (2021) 10(5):1260. doi: 10.3390/cells10051260
247. Jandus C, Simon HU, Von Gunten S. Targeting Siglecs-A Novel Pharmacological Strategy for Immuno- and Glycotherapy. Biochem Pharmacol (2011) 82:323–32. doi: 10.1016/j.bcp.2011.05.018
248. Varki A. Letter to the Glyco-Forum: Since There are PAMPs and DAMPs, There Must be SAMPs? Glycan “Self-Associated Molecular Patterns” Dampen Innate Immunity, But Pathogens Can Mimic Them. Glycobiology (2011) 21:1121–4. doi: 10.1093/glycob/cwr087
249. Hudak JE, Canham SM, Bertozzi CR. Glycocalyx Engineering Reveals a Siglec-Based Mechanism for NK Cell Immunoevasion. Nat Chem Biol (2014) 10:69–75. doi: 10.1038/nchembio.1388
250. Stanczak MA, Siddiqui SS, Trefny MP, Thommen DS, Boligan KF, Von Gunten S, et al. Self-Associated Molecular Patterns Mediate Cancer Immune Evasion by Engaging Siglecs on T Cells. J Clin Invest (2018) 128:4912–23. doi: 10.1172/JCI120612
251. Murugesan G, Weigle B, Crocker PR. Siglec and Anti-Siglec Therapies. Curr Opin Chem Biol (2021) 62:34–42. doi: 10.1016/j.cbpa.2021.01.001
252. Wisnovsky S, Möckl L, Malaker SA, Pedram K, Hess GT, Riley NM, et al. Genome-Wide CRISPR Screens Reveal a Specific Ligand for the Glycan-Binding Immune Checkpoint Receptor Siglec-7. Proc Natl Acad Sci USA (2021) 118(5):e2015024118. doi: 10.1073/pnas.2015024118
253. Yoshimura A, Asahina Y, Chang LY, Angata T, Tanaka H, Kitajima K, et al. Identification and Functional Characterization of a Siglec-7 Counter-Receptor on K562 Cells. J Biol Chem (2021) 296. doi: 10.1016/j.jbc.2021.100477
254. Cummings RD, McEver RP. “C-Type Lectins”. In: Varki A, Cummings CRD, Esko JD, Hart GW, Aebi M, Darvill AGD, Kinoshita T, Packer NH, Prestegard JH editors. Essentials of Glycobiology, 3rd Edition, vol. chapter 34. Cold Spring Harbor New York: Cold Spring Harbor Laboratory Press (2017). p. 675–83. doi: 10.1007/978-4-431-54841-6_138
255. Borsig L. Selectins in Cancer Immunity. Glycobiology (2018) 28:648–55. doi: 10.1093/glycob/cwx105
256. Häuselmann I, Borsig L. Altered Tumor-Cell Glycosylation Promotes Metastasis. Front Oncol (2014) 4:28. doi: 10.3389/fonc.2014.00028
257. Szabo R, Skropeta D. Advancement of Sialyltransferase Inhibitors: Therapeutic Challenges and Opportunities. Med Res Rev (2017) 37:219–70. doi: 10.1002/med.21407
258. Bowles WHD, Gloster TM. Sialidase and Sialyltransferase Inhibitors: Targeting Pathogenicity and Disease. Front Mol Biosci (2021) 0:705133. doi: 10.3389/FMOLB.2021.705133
259. Perez SJLP, Fu CW, Li WS. Sialyltransferase Inhibitors for the Treatment of Cancer Metastasis: Current Challenges and Future Perspectives. Molecules (2021) 26:18. doi: 10.3390/molecules26185673
Keywords: tumor glycosylation, sialyltransferases, sialic acid, cancer, tumor immunology
Citation: Hugonnet M, Singh P, Haas Q and von Gunten S (2021) The Distinct Roles of Sialyltransferases in Cancer Biology and Onco-Immunology. Front. Immunol. 12:799861. doi: 10.3389/fimmu.2021.799861
Received: 22 October 2021; Accepted: 02 December 2021;
Published: 17 December 2021.
Edited by:
Heinz Laubli, University Hospital of Basel, SwitzerlandReviewed by:
Behjatolah Monzavi-Karbassi, University of Arkansas for Medical Sciences, United StatesValeria I. Segatori, Universidad Nacional de Quilmes (UNQ), Argentina
Vered Padler-Karavani, Tel Aviv University, Israel
Thomas Boltje, Radboud University Nijmegen, Netherlands
Copyright © 2021 Hugonnet, Singh, Haas and von Gunten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephan von Gunten, c3RlcGhhbi52b25ndW50ZW5AcGtpLnVuaWJlLmNo
 Marjolaine Hugonnet
Marjolaine Hugonnet Pushpita Singh
Pushpita Singh Quentin Haas1
Quentin Haas1 Stephan von Gunten
Stephan von Gunten