- 1Center for Sepsis Control and Care & Institute of Biochemistry II, University Hospital Jena, Jena, Germany
- 2Université de Nantes, Inserm, TENS, The Enteric Nervous System in Gut and Brain Diseases, IMAD, Nantes, France
- 3Department of Pediatric General and Thoracic Surgery, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 4Department of Pediatrics, University of Cincinnati, Cincinnati, OH, United States
The gut is a tubular organ responsible for nutrient absorption and harbors our intestinal microbiome. This organ is composed of a multitude of specialized cell types arranged in complex barrier-forming crypts and villi covered by a mucosal layer controlling nutrient passage and protecting from invading pathogens. The development and self-renewal of the intestinal epithelium are guided by niche signals controlling the differentiation of specific cell types along the crypt-villus axis in the epithelium. The emergence of microphysiological systems, or organ-on-chips, has paved the way to study the intestinal epithelium within a dynamic and controlled environment. In this review, we describe the use of organ-on-chip technology to control and guide these differentiation processes in vitro. We further discuss current applications and forthcoming strategies to investigate the mechanical processes of intestinal stem cell differentiation, tissue formation, and the interaction of the intestine with the microbiota in the context of gastrointestinal diseases.
Introduction
The gastrointestinal tract is a central organ system that enables the ingestion, digestion, absorption, and the utilization of processed nutrients to fuel the overall body. Within this system, several organs are responsible for digestion and the uptake of nutrients, including the stomach, the small intestine, and the large intestine. The gut forms a tubular structure whose central lumen is enclosed by a protective mucosa composed of a monostratified epithelium forming invaginations (or crypts) and finger-like protrusions (or villi). The epithelium allows nutrient uptake and acts as the first line of protection against invading pathogens (1). A crypt-villus structure is seen in the small intestine, while only crypts are present within the colon. Both the initial development and continuous self-renewal of the intestinal epithelium are guided by niche signals in addition to cell-autonomous processes that initiate the outgrowth and differentiation of intestinal stem cells (ISCs), driving proliferation and differentiation along the crypt-to-villus axis in the small intestine. Transit-amplifying cells derived from ISC further separate into lineages showing absorptive or secretory characteristics that terminally differentiate into specific cell types within the epithelium (2). Enterocytes are the predominant cell type in the epithelium and involved in the absorption and transport of small molecules from the intestinal lumen into the bloodstream. Enterochromaffin cells, a enteroendocrine cell type, are specialized cells of the epithelial tissue that secrete hormones and modulate neural activity by releasing neurotransmitters (3). Microfold cells (M cells) are responsible for the transport of luminal antigens to the lymphoid follicles, thereby initiating an adapted immune response to the microbiota (4). The composition of the microbiota at the epithelium can be modulated by Paneth cells through secretion of antimicrobial peptides. In addition, this cell type contributes to the maintenance of the ISC niche (5). Tuft cells and cup cells belong to the rarer cell types of the intestinal epithelium. For Tuft cells, a contribution to neural signaling has been proposed (6) whereas the function of cup cells still awaits clarification. This complex epithelium composed of several cell types is surrounded by smooth muscle cell layers, the enteric nervous system, and connective tissue that contains arteries and lymphatics and is abundant in fibroblasts and mast cells (7).
The last few decades in biomedical research have led to a number of robust experimental strategies and techniques to study intestinal physiology. Yet, in vitro studies aiming to model the gut mucosa have mostly been limited to static monocultures and co-cultures. In contrast to these older models, recent developments that use microphysiological systems allow scientists to probe both the intestinal epithelium and its environment in a dynamic and physiologically relevant manner (8). In this review, we will emphasize the transitions from immortalized cell lines to stem cells and describe their use in combination with gut-on-chip models. These “on-chip” models are defined as three-dimensional (3D) cell cultures arranged as multiple cell layers actively perfused by microfluidics for medium exchange to enable an improved tissue formation with increased lifespan. We will highlight selected applications of these gut-on-chip models to understand gastrointestinal diseases and forthcoming strategies to investigate human gut physiology.
Cell Sources for Gut-on-Chip Models
The recapitulation of whole embryonic development can be challenging due to a lack of knowledge of all the required biochemical cues driving the differentiation and morphogenic processes. One of the most novel in vitro approaches in intestinal mucosa modeling is the use of microfluidically perfused organ-on-chip seeded with cell lines, primary cells, or stem cells (9–14). A clear benefit of these models is that they allow for better control of the microenvironment. One example includes the addition of flow on Caco-2 cells to differentiate them and express the typical cell markers of essential cell types of the small intestine (Goblet- and Paneth-, enterochromaffin-, and enterocytes) (15). Further, flow conditions stimulated an increased expression of mucin-2 with the development of a thickened mucus layer covering the epithelial tissue, indicating the functional relevance of flow conditions for improved goblet-like cell functionality (9, 16). Perfused organ-on-chip further facilitates physiological cellular crosstalk by aiding in the outgrowth of organotypic microstructures such as villi and crypts (9, 12). In addition, an epithelial cell layer has been combined in a number of studies with endothelial cells and immune cells using various tissue engineering approaches (9, 12, 17). These studies highlight the potential of organ-on-chip as a tool to manipulate the microenvironment by facilitating the long-term growth and co-differentiation of different cell types to replicate some typical microanatomical features of the human intestine.
Control and Guidance of Intestinal Cell Development on-Chip
Immortalized cell lines have been used extensively to study the intestine. For example, the colorectal adenocarcinoma cell line Caco-2 had been originally used to study the intestinal epithelial barrier as these cells differentiate spontaneously into a monolayer of enterocytes when reaching confluence (18). However, given the cancerous background of the Caco-2 cell line data generated with these models, the data should be interpreted with caution as this cell line has several limitations in its differentiation potential to individual intestinal cell types compared to adult stem cells or induced pluripotent stem cells (19). Further, immortalized cell lines such as Caco-2 cannot reflect a patient-specific genetic background to enable a personalized approach to study individual mechanisms of disease-related conditions.
To overcome these limitations, the use of human pluripotent stem cells (hiPSCs) or adult stem cells has been explored as a promising alternative to established cell lines. Human adult ISCs maintain an organ-specific imprint and epithelial maturity and possess a patient-specific background, thus enabling personalized studies (9, 14). Biopsy-derived cells and intestinal organoid models have emerged as powerful tools to mimic tissue complexity and high cellular diversity in vitro (20, 21). ISCs are a valuable cell source to grow organoids with organotypic self-patterning, thereby recreating essential microanatomical features of the gut (22). However, the availability of primary tissue for the isolation of ISCs is limited. Hence, hiPSCs are an exciting alternative as they can generate intestinal cells through directed differentiation (23). The use of hiPSCs allows for the generation of multiple differentiated cell types from a patient-specific background with an unlimited supply of human stem cells stored in biobanks (24). Furthermore, hiPSCs from patients with a genetic background of interest can be obtained by reprogramming from a multitude of easily accessible primary cell sources, including urine, blood, and skin (25). Nevertheless, a significant drawback in using pluripotent stem cells can be found in difficulty producing tissues that reach full maturity.
ISC-derived organoids have been used to experimentally address and dissect the individual spatiotemporal effects of different biochemical gradients on the growth and differentiation of ISCs. However, given the spherical nature of organoids, the stable recreation of physiological conditions over a more extended period of culture has remained challenging (26). Microfluidically perfused organ-on-chip platforms mimic in vivo-like biochemical cues to guide stem cell differentiation with typical microanatomical features such as villi and crypts (Figure 1).
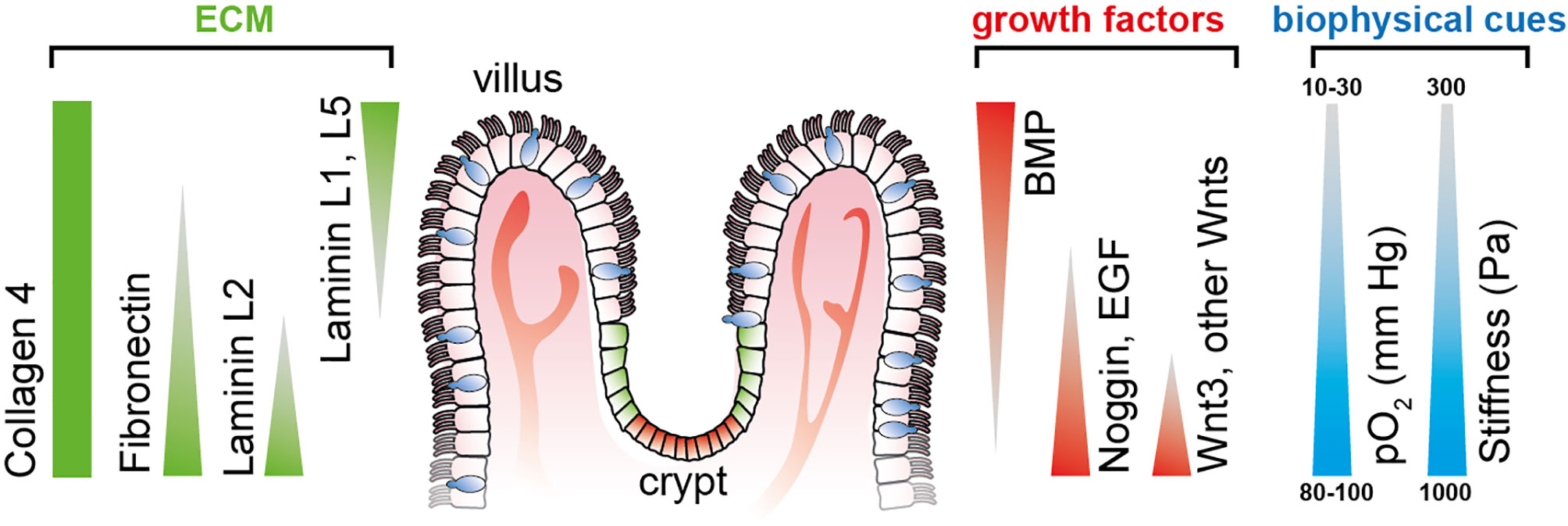
Figure 1 Defined gradients of biochemical and biophysical cues span along the crypt-villus axis in the small intestine, thereby determining cellular stemness and differentiation toward the tips of the villi. Triangles indicate the gradient direction of growth factors and ECM protein. Illustration adapted from (26).
It has been long known that many aspects of cell fate are dependent on the geometry and mechanics of their microenvironment, including the induction of apoptosis (27), cellular differentiation (28, 29), and proliferation rate (30). The presence of a flow-through media stream has been demonstrated to be a requirement for forming 3D intestinal structures in cell lines and primary cell-based intestinal models by modulating the biochemical availability of growth factors. The flow-dependent basolateral removal of the Wnt antagonist Dickkopf-1 and the elevated expression of the Frizzled-9 receptor was required in the formation of epithelium villi-like protrusions (31). Our existing knowledge on the individual growth factors driving ISC differentiation provides an excellent basis to engineer more realistic organ models with better accessibility. In this review, we are focusing on the use of the following two principal types of stem cells to differentiate and form intestinal tissue: adult stem cells and induced pluripotent stem cells. Both cell sources possess unique advantages and disadvantages in recapitulating the intestinal tissue (Table 1).
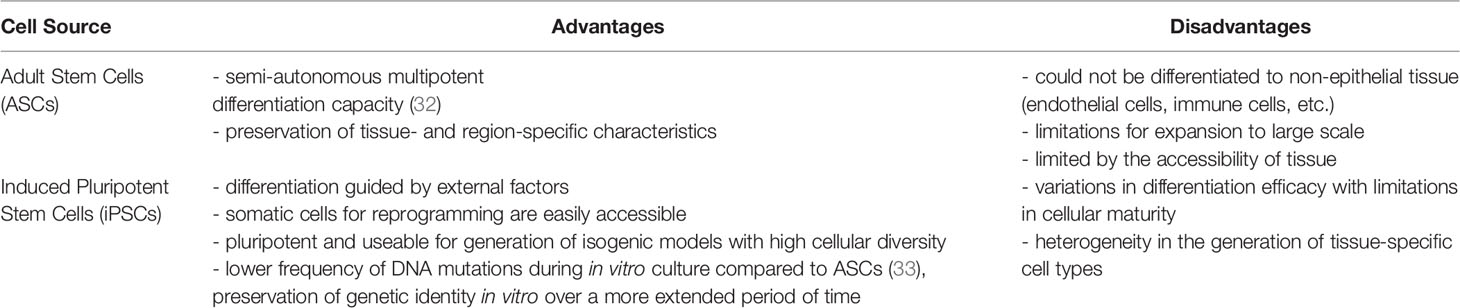
Table 1 Pros and cons of the use of adult stem cells and induced pluripotent stem cells for the generation of intestinal tissue models.
A striking example of the potential of defined growth factor gradients to guide stem cell growth to intestinal tissues has been given by Wang et al. in the Transwell system (34, 35). Arrays of artificial collagen-based crypts and villi structures were created by microfabrication and molding to mimic the topography of the intestine and the colon (34, 35). The scaffold has been used to generate gradients of Wnt-3A, R-spondin, noggin, and gamma-secretase inhibitor. These gradients enabled the establishment and maintenance of a crypt-villus axis composed of adult human ISCs that formed an epithelium containing proliferating progenitor cell compartments. Kasendra et al. recently presented a more complex model of the human duodenum that combines the advantages of organoids and organ-on-chip technologies (9, 36). In this model, the tissue forms a polarized cell architecture under flow conditions and shows improved intestinal barrier function and differentiation of specialized cell subpopulations. This duodenum-on-chip system could exert mechanical stimulation to organoid-derived epithelial cells, resulting in improved tissue architecture. The tissue perfusion significantly improved cell-cell junctions and microvilli density, further improving intestinal barrier function. Remarkably, it was demonstrated that organoid-derived cells in the duodenum chip have an increased similarity in their global gene expression profile to human adult duodenal tissue compared to conventional static organoid culture. Further, in the duodenum-on-chip system, organoid-derived cells show an improved metabolic capacity compared to Caco-2 cells due to an increased induction capability and expression of CYP3A4 (36). The ability to integrate and analyze patient-specific stem cells from different donors within the chip represents a decisive advantage over the use of immortalized cell lines such as Caco-2 or HT-29. Stem cells allow for the reflection of the host’s genetic background and possess the ability to self-organize into an in vivo-like tissue structure with improved functionality. Both aspects are significant assets for studies on drug uptake and drug-drug interaction and its metabolization to predict drug efficacy and safety. Recently, Nikolaev et al. used synthetic hydrogels to guide the growth of adult human ISCs with intrinsic self-organization properties to tubular structures forming crypt- and villus-like domains (37). Furthermore, conventional organoids and 2D cell layers derived from intestinal organoids possess a high regenerative potential when cultured on hydrogels, as demonstrated after stimulation with cytotoxic dextran sodium sulfate (DSS) to model colitis. The perfusion of the system was used to provide a continuous supply of nutrition and the removal of waste products and dead cells, allowing the formation of a tenfold higher cell mass and the emergence of rare cell types such as M cells (37).
Similar approaches have also been demonstrated using hiPSCs. Workman et al. used epithelial cells isolated from hiPSC-derived human intestinal organoids and seeded them in a microfluidic perfused gut-on-chip. In the 3D tissue model, the spontaneous differentiation into epithelial-specific types, such as Paneth cells, goblet cells, enterocytes, enteroendocrine cells, transit-amplifying cells, and Lgr5+ cells similar to in vivo cell populations was observed under flow conditions (38). The immune responsiveness of the intestinal tissue after INFγ stimulation was validated by the measurement of STAT1 phosphorylation after one hour and the upregulation of INFγ−related genes three days after stimulation. Further, combined treatment with INFγ and TNF resulted in increased permeability of the epithelial cell layer. In another study, the same chip platform was used to culture stem cell-derived epithelial cells isolated from human colon organoids (14). Likewise, the OrganoPlate system, a plate-based cell culture system that allows pumpless media movement via a rocking platform, was used for cultivating hiPSC on a gel matrix to form a monolayered epithelial tube (39). The cells expressed the Paneth cell marker Lysozyme, the enterocyte cell marker Villin-1, and the neuroendocrine cell marker Chromogranin A. However, the expression of these cell marker proteins was diffuse and distributed over the entire tissue. Another shortcoming of the model is the absence of organized 3D epithelial structures.
These gut-on-chip systems strikingly demonstrate the vast potential of organ-on-chip platforms to precisely control environmental growth conditions, to guide the differentiation of stem cells, and to form improved intrinsically self-organized tissue with superior properties compared to traditional organoid cultures. Hence, the applications of ISCs include the possibility to investigate intestinal developmental biology and barrier function but also studies related to bacterial colonization, drug discovery, regenerative medicine, and responses to infections in a personalized approach (40–43). The generation and integration of hiPSC-derived immune cells such as T cells, NK cells, and macrophages by use of differentiation protocols that became recently available will aid in the development of isogenic immune-competent intestinal models derived from a single hiPSC line to avoid potential allogenic reactions (44–46).
Gut-on-Chip Models to Study Microbiome-Host Interaction
As a result of coevolution, niches of the human body are inhabited by a variety of commensal, mutualistic, and pathogenic microorganisms, including archaea, bacteria, fungi, phages, protists, and viruses (47–53). The gut provides a variety of niches serving as habitats for different ecological communities. On a macroscopic scale, physiological gradients determine the growth niche of microorganisms along the gut from the small intestine toward the rectum with an increase in the pH and the abundance of bile acids, whereas the availability of oxygen decreases. A change in the environmental conditions thereby determines microbial colonization with 104 CFU/ml bacteria in the small intestine up to 108 CFU/ml microorganisms in the terminal ileum, where elongated passage times and associated enrichment in nutrients provide ideal bacterial growth conditions (54, 55). With that, the number of facultative anaerobic and obligate anaerobic species increases from the small intestine to the colon (56, 57).
Toll-like receptors (TLR) belong to a class of pattern recognition receptors evolutionary highly conserved and centrally involved in mediating host-microbe interactions in epithelial cells (58). Recent studies provide evidence of a defined spatial organization of TLR expression along the gut axis, which seems to be determined already before birth in tissue-resident stem cells and is only partially dependent on the interaction with the commensal gut microbiota (59, 60). TLR dependent pathways have been shown to mediate not only antimicrobial functionality but also directly contribute to the regulation of epithelial cell differentiation, which is of particular importance for the intestine (61). LPS activation of TLR-4 enhances cell differentiation of goblet cell lineages in colonic organoids but inhibits Lgr5+ proliferation with induction of apoptosis in stem cells. Similar observations have been made in intestinal organoids (62). MyD88 is an essential adapter protein to TLR-4 and critically involved in mediating LPS dependent maintenance of mucus production. Consequently, MyD88-/- mice have impaired mucus production with increased bacterial adherence to epithelial cells and loss of barrier function (63). TLR activation has also been demonstrated to contribute to the polarization of IECs depending on the side of stimulation (64). A strict unique spatial distribution pattern of TLRs between the apical or basolateral side of IECs along the gut axis contributes to an adapted interaction with the microbiota as a requirement for the maintenance of barrier functionality (65, 66). The proof of a physiological TLR distribution pattern in IECs cultured in gut-on-chip models remains an open task, but likely represents an important requirement to faithfully recapitulate host-microbiota interaction in vitro. IECs derived from different gut sections cultured in gut-on-chip and precisely stimulated with TLR agonists in a precise spatiotemporal manner by microfluidic perfusion would offer an interesting approach for achieving a physiologically relevant TLR expression pattern in vitro. Recent work already demonstrated the feasibility of a similar approach to achieve intestinal tissue polarization in an immunocompetent gut-on-chip model. Monocyte derived macrophages and dendritic cells here instructed immune tolerance and the maintenance of barrier function depending on the site of LPS stimulation (12).
The reliable monitoring of the microenvironment and its change upon tissue formation or bacterial colonization is key to control and understand the stem cell development and differentiation as well as the interaction of the intestinal tissue with the microbiota. Luminescent-based sensors integrated into organ-on-chip systems have been used to quantify medium dissolved oxygen levels (67, 68), cellular glucose consumption (69), and the changes of the pH by the release of lactate by metabolic active tissues or microorganisms (70–72). To enable the growth of anaerobic bacteria in gut-on-chip, several platforms have been tailored to allow the stable formation of neighboring cell layers perfused with normoxic and hypoxic media streams. The characterization and quantification of microbiota-associated changes in the metabolic profile require reliable and continuous measurement of key metabolites and mediators of microbial colonization. Electrochemical enzyme-based biosensors and microbead-based microfluidic assays expand the ability of organ-on-chip systems to perform detailed monitoring of metabolic parameters, including the formation of glutamine and glutamate (73, 74), the release of reactive oxygen species (75), and the secretion of various cytokines (76, 77). The determination of transepithelial electrical resistance (TEER) is another widely used method to assess barrier function and has been adapted in recent years for organ-on-chip platforms. In particular, the combination of impedance spectroscopy and electrical stimulation has been used to quantify cell layer capacitance as a measure of villi differentiation in gut-on-chip models, which could be applied across different gut-on-chip platforms (78).
Gut-on-chip models have been used in recent years to better understand the complex interactions between the microbiota and their host. These systems allow balancing between robust and predictable approaches and the recreation of physiological-relevant culture conditions (Figure 2). The incremental nature of these models makes it relatively simple for them to be designed as “simple as possible and complex as required,” and they, therefore, offer additional flexibility in identifying contributing factors of host-microbiota interaction. The controlled escalation of biological complexity on the host side as well as in the composition of microbiota-derived factors and live microorganism communities will enable the elaboration and the proof of a complex interaction mechanism in a well-controlled and standardized environment provided by organ-on-chip platforms (Figure 3).
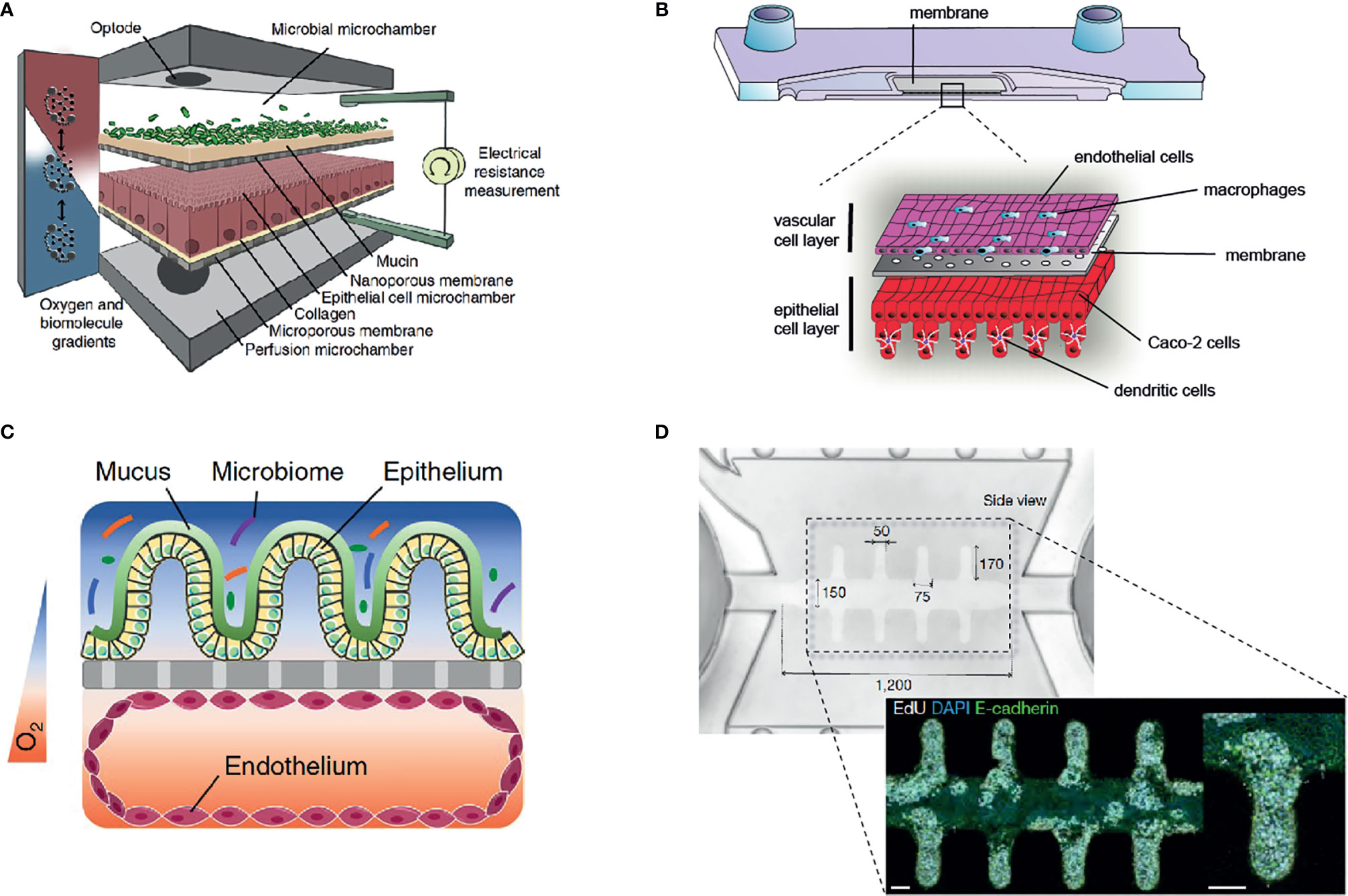
Figure 2 Overview of selected gut-on-chip models to study host-microbiota interaction in vitro. (A) The HuMiX gut-on-chip model allows the co-culture of anaerobic bacteria with an epithelial cell layer (68) (Creative Commons CC BY license). (B) Immunocompetent, multilayered gut-on-chip model comprising endothelial cells, epithelial cells, macrophages, and dendritic cells (12) (Copyright © 2019 Elsevier Ltd. All rights reserved). (C) Gut-on-chip model with oxygen tension gradient for the culture of a complex microbiome up to three days (11) (Copyright © 2019, The Author(s), under exclusive license to Springer Nature Limited). (D) Mini-intestine formed by organoid-derived epithelial cells, guided in growth by a perfused 3D hydrogel scaffold (37) (Copyright © 2020, The Author(s), under exclusive license to Springer Nature Limited).
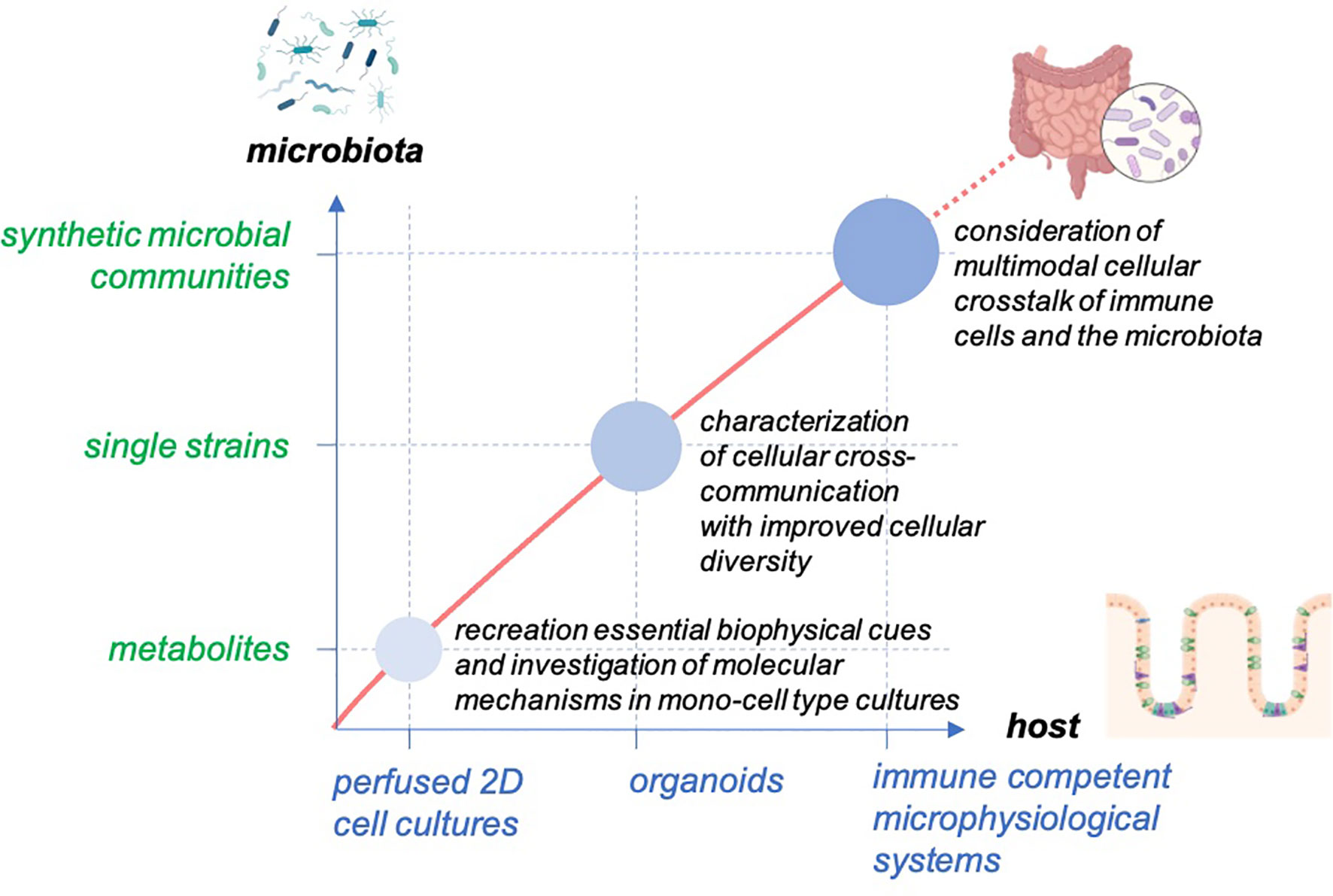
Figure 3 Concept for studying host-microbiome interaction in gut-on-chip. The models enable the balancing of the cellular complexity of host tissue and provide the technical basis to facilitate the outgrowth of host cells to self-organized organoid structures with high cellular diversity. The systems allow the scaling of the biological complexity of the host and the microbiota to study the interaction of the host tissue with the microbiota from microbial metabolites to single microbial strains and diverse microbial communities.
Examples of a successful application of this approach have already been provided. Sontheimer-Phelps et al. recently reported a system lined with primary patient-derived colonic epithelial cells (14). A mucus bilayer is built up in the chip by perfused epithelial cells and provides a suitable substrate for colonization with living microbiota to study host-microbiome interactions in the colon in more detail. Similarly, Shin et al. generated an anoxic-oxic interface on a chip inhabited with CaCo 2-cells and implemented Bifidobacterium adolescentis (79). The system is able to co-cultivate bacteria for up to one week without the impairment of epithelial performance. This represents an interesting platform to assess mechanistic questions regarding Bifidobacteria-host interactions in the neonate period in more detail. Similarly, Zhang et al. reported the co-culture of the obligate anaerobic bacterium Faecalibacterium prausnitzii with colon organoid-derived epithelial cells in their physiomimetic platform for up to four days. In the study, the authors were able to uncover the mechanisms of an anti-inflammatory host response related to butyrate release with the downregulation of the gene expression of histone deacetylase complexes, Toll-like receptors, and nuclear factor kB (NF-kB) (80). Different approaches have also been used to reconstitute the microbiome and its function in vitro in a more complex and detailed manner. A top-down approach consists of using the full microbiome derived from human stool samples, which would, in principle mimic the in vivo situation ideally. However, it remains challenging to characterize its full composition in a short period of time before transfer into an in vitro model, even under hypoxic conditions. Jalili-Firoozinezhad and co-workers managed to implement complex microbiomes in their in vivo-like system but pointed out that overgrowth and species shift due to artificial culture conditions might be problematic. Thus, a sophisticated pre-culture step (i.e., in large SHIME bioreactors) is still required to expand a defined microbiota prior to colonization experiments (11). However, these studies illustrate nicely the potential of advanced in vitro models to dissect the individual microbiota in humans.
Although conventional organoid cultures already provide invaluable insight into detailed ISC dynamics, their application for long-term studies or the dissection of host-microbiota interaction has some limitations. The long-term exposure of microbial metabolites or co-culturing living microorganisms in an enclosed lumen of organoids under static conditions favors the rapid overgrowth of bacteria, eventually resulting in the rupture and cell death of the organoids (32, 81). The microfluidic perfusion of gut-on-chip offers the ability of the long-term exposure of the tissue to microbiota-associated metabolites under well-defined conditions. Perfusion media can be supplemented with defined single molecules of microbial metabolites up to filtrates of stool samples, single strain microorganisms, or defined microbial communities. To better recapitulate the in vivo situation, gut-on-chip platforms could be leveraged to mimic the conditions of different sides of the gut by use of adult stem cells derived from specific sections (i.e., intestine, colon, etc.) to generate side-specific tissue (Table 2). Colonization with oxygen-scavenging bacteria in combination with the continuous sensing and regulation of oxygen levels would allow the maintenance of physiological-relevant oxygenation levels and a co-culture with defined anaerobic and facultative-anaerobic bacteria under homeostatic conditions.
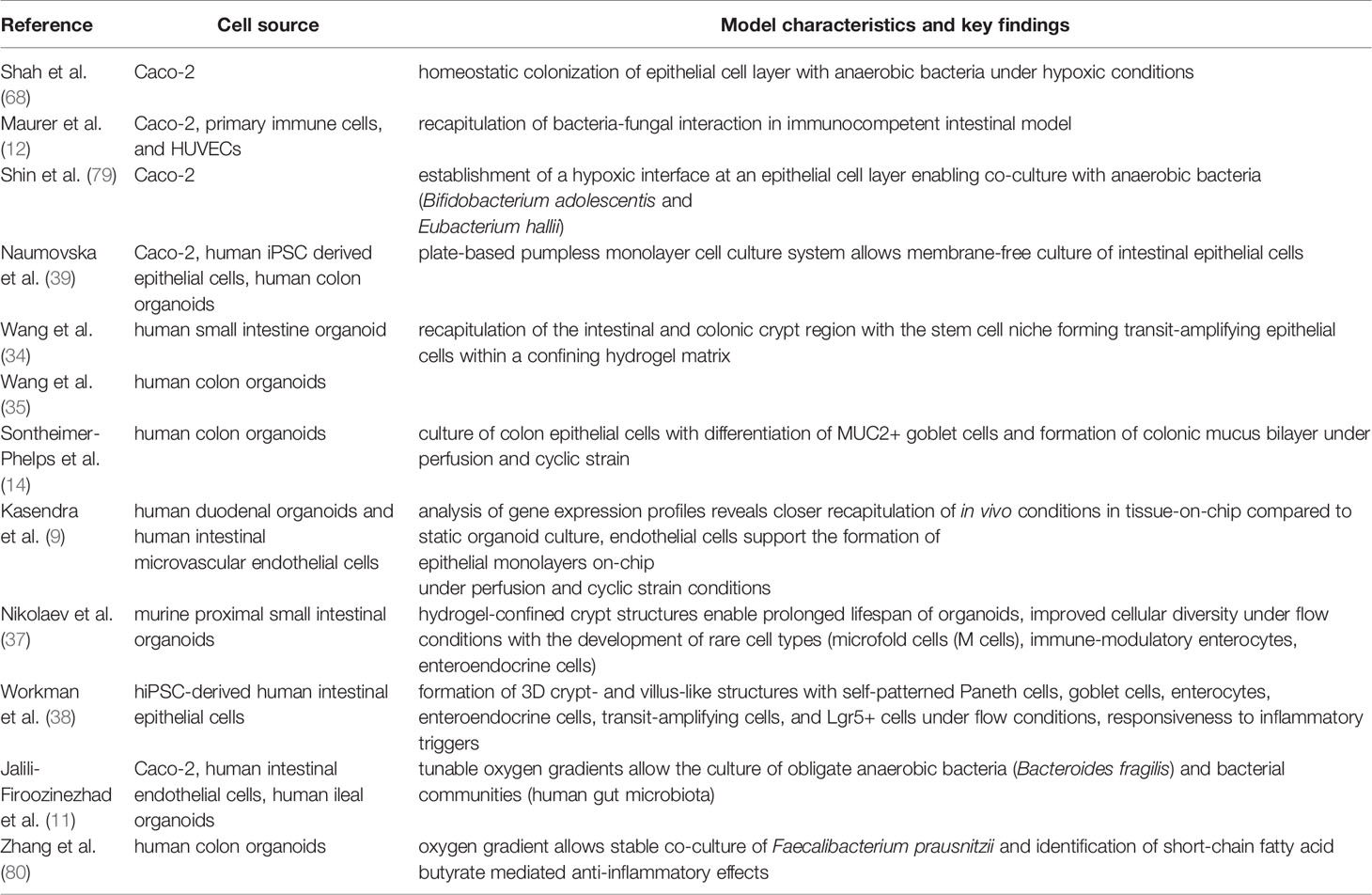
Table 2 Key references of selected gut-on-chip systems with important characteristics to recapitulate the gut environment.
Modeling Human Intestinal Disease on-Chip
Inflammatory Diseases
Gut-on-chip models offer a new avenue through which to study the pathophysiological mechanisms of gastrointestinal diseases. For example, patients suffering from Celiac disease (CeD), an autoimmune disease that affects about 1% of the human population, develop a strong inflammatory response causing severe damage to the small intestine upon the uptake of a gluten-containing diet (82). Still, the mechanistic details of the disease await further investigation before effective drugs can be developed. Moerkens et al. recently proposed an immunocompetent organ-on-chip model using both hiPSC and primary epithelial cells to mimic the genetic background and environmental factors more reliably. Using this system, it is now possible to determine the disease onset in CeD patients (83). Similarly, gut-on-chip models have been used to study the disease mechanism of inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC), characterized by chronically relapsing intestinal inflammation and representing a worldwide healthcare problem with continually increasing incidence (84). A 3D gut model based on primary patient-derived colonic epithelial cells forming a mucus layer was used to recapitulate prostaglandin E2-dependent mucus volume expansion via the activation of the Na-K-Cl cotransporter 1 ion channel (14). In a similar study, the pathophysiology of DSS-induced inflammation was also recapitulated in the presence of E. coli (16). DSS cessation quickly induced the recovery of barrier integrity, villus formation, and mucus production, indicating that this model might also be suitable to study processes of mucosal regeneration in IBD. The effects of microbial colonization were also investigated with a mixture of commensal Streptococcus thermophilus, Bifidobacteria, and Lactobacilli (VSL#3) as well as pathogenic enteroinvasive E. coli (EIEC). In this study, the authors were able to replicate inflammation-associated tissue damage and the invasion of EIEC. Furthermore, they proved the growth-limiting effect of VSL#3 on EIEC colonization in vitro (85). Recently, our group has been able to further replicate the protective characteristics of Lactobacillus rhamnosus against the tissue infiltration of the opportunistic yeast Candida albicans in an immunocompetent gut-on-chip model. In addition to peripheral immune cells, this model also contained functional mucosal macrophages and dendritic cells integrated into the epithelial tissue layer (12). In contrast to conventional organoid cultures, chip-based ISC cultures enable long-term studies and perfusion with peripheral immune cells with superior accessibility and the continuous monitoring of key environmental factors to maintain and manipulate disease-related growth conditions. In combination with a homeostatic bacterial co-culture, these systems might serve in future studies as patient-specific tools to develop new therapeutics or probiotics that modulate the mucus barrier.
Infectious Diseases
Gut-on-chip has further been used to study infectious diseases, including Clostridioides difficile infection (CDI), Enterohemorrhagic E. coli (EHEC), and viral infections. Infection with Clostridioides difficile (formerly Clostridium difficile) is the most common cause of hospital-acquired gastrointestinal infections (86, 87). While several animal models have been used to study CDI in vivo, the manipulation of infection processes and the direct assessment of host-bacterial interaction is challenging (88–90). Advanced gut-on-chip models have been described to investigate CDI. This model allows for the co-culture of primary epithelial and endothelial cells, with the addition of a complex microbiome under anaerobic culture conditions for several days (11). This is a promising translational tool to study the CDI infection process and associated damage in humans. As with CDI, infections with EHEC are frequent, with up 100,000 cases per year in the U.S. and are associated with several symptoms such as severe bloody diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome (HUS). Interestingly, the microbiota seems to be a critical determinant of susceptibility to EHEC infection. Studies in mice revealed that not only is a 100,000-fold higher dose of the bacterium required (107 microbes) but that the whole murine microbiome also needs to be depleted to induce symptoms. This pathology is quite different in humans, where as little as 102 microbes are sufficient to cause severe symptoms (91–93). Tovaglieri et al. were able to recapitulate this phenomenon in vitro in a gut-on-chip model by co-culturing EHEC in the context of a murine or a human microbiome. The species-specific tissue damage induced by the human microbiome could be verified. This model was also used to identify several metabolites related to or even causative of the high susceptibility of humans to EHEC (13). Finally, human enteroviruses are responsible for an estimated 10–15 million infections and at least 30,000–50,000 hospitalizations per year (94). Viruses can replicate in the epithelial cells of the gastrointestinal tract and subsequently disseminate to secondary sites of infection, such as the respiratory tract (95). So far, no approved drugs for the treatment of enterovirus infections in humans besides polio exist (95). Villenave et al. used coxsackievirus B1 (CVB1) as a prototype enterovirus strain and established a human infection model based on a human gut-on-a-chip microfluidic device to study replication and infectious virus production in vitro (94). With this model, they were able to follow the infection of epithelial cells, the release of inflammatory cytokines, and the secretion of infectious virions. The model also permitted to reproduce the different susceptibility of epithelial and endothelial cell layers for viral infection by using separate microchannels, thus independently perfusing the vascular and epithelial sides of the polarized gut tissue. Gut-on-chip models have further just recently been used to study infection with human parasites. “Mini-intestines” fully accessible from the luminal side enabled colonization with Cryptosporidium parvum, an obligate parasite causing life-threatening diarrhea in immunocompromised adult hosts and in infants. The tissue model supported the full life cycle of the parasite, thereby allowing for the first time long-term studies in a primary cell-derived in vitro culture system (37).
With the emergence of the SARS-CoV2 virus and the COVID-19 pandemic, the need for sophisticated models to study viral infection in human tissue has become even more urgent. Human small intestinal organoids have proven to be valuable tissue models to study virus docking to angiotensin converting enzyme II and its replication within enterocytes. Combining ISC-derived organoids with the advantages of organ-on-chip would offer additional options to manipulate the viral infection route and provide different parameters to faithfully reflect the intestinal microenvironment, prolong the culture of organoids with the improvement of cellular differentiation, and offer a variety of additional monitoring and readout techniques available for gut-on-chip systems (96).
Colorectal Cancer
The effects of short-chain fatty acids (SCFAs) released by probiotic Lactobacillus rhamnosus GG (LGG) bacteria on the growth of colorectal cancer cells was recently investigated in a version of the HuMiX model, a modular, microfluidics-based gut-on-chip model, which allows the co-culture of human and microbial cells and facilitates anaerobic culture conditions (97). Symbiotic treatment with a high-fiber diet was simulated in this model, and the generation of organics and SCFAs was assessed. Strikingly, it was demonstrated that SCFAs and lactate production was altered by a simulated high-fiber diet compared to a reference diet medium containing only simple sugars. The simulated high-fiber diet caused an increased expression of oncogenes and proinflammatory signaling in the absence of LGG bacteria, whereas, in the presence of LGG, both gene clusters were found to be significantly downregulated and associated with a reduced cell proliferation rate of primary colon-rectal cancer cells. This study illustrates the capability of gut-on-chip systems to precisely dissect the individual aspects of the microbiota-host crosstalk at the metabolic level. Hopefully, similar studies will further expand our insight into this complex relationship by identifying individual metabolic targets with potential as tailored pre- and probiotic-based therapeutic options. Uncovering individual beneficial metabolic profiles by applying hiPSC-based organ models and individualized microbiota surrogates in personalized in vitro models would represent a major step forward in the treatment of IBD and IBD sequelae such as colorectal cancer.
Conclusions and Outlook
The intestinal tissue shows considerably variance in its architecture, cellular and molecular composition along the cephalocaudal axis, thereby creating various defined environmental niches populated with a specifically adapted microbiota (54, 60, 98). These niches are defined by differences in biophysical cues such as luminal oxygen levels or changes in pH and formed by adapted epithelial cells. Future gut-on-chip systems should be able to recapitulate this topography and allow the direct monitoring of environmental changes i.e., induced by changes in the microbiota composition and its impact on drug metabolization. It has been demonstrated that chip-based cultivation of organoid-derived cells could accelerate the formation of a self-organized epithelial microarchitecture, allowing the long-term culture with a living microbiota (11, 37, 79). These models have already proven to produce data similar to in vivo contexts in studies ranging from infectious disease modeling to drug metabolism (12, 13, 36, 41, 42, 94). The benefits and wide range of applications of the gut-on-chip models can be attributed to their versatility and their capability to increment parameters of various types, from multiple cell types to medium gradients, flow mechanics, or global topography. Despite these numerous advances in the complexification of microfluidics systems, many requirements still need to be met in order to obtain models with an in vitro microenvironment that closely mimics its in vivo counterpart.
Gut-on-chip technology offers unique options by providing the technical basis for a reliable and guided differentiation of stem cells to human intestinal tissue, combining the strengths of self-patterned stem-cell-based organoids with the ability to precisely regulated biochemical and biophysical cues in a spatiotemporal manner. The manipulation of the microenvironment by tissue engineering and the guided growth of stem cells derived from patients will create a new angle for the dissection of personalized host-microbiota interaction with scalable levels of complexity for the host and the microbiota. Optimizing fully defined and tunable hydrogel matrices as an alternative to the widely used Matrigel will allow the generation of organoids in a more standardized and reproducible way as a reliable cell source for gut-on-chip systems. Promising approaches have already demonstrated the potential of novel synthetic polymer matrices based on alginate (99), thrombin cross-linked fibrin gels (100) or PEG macromers decorated with maleimide groups (101).
The combination of advanced cell substrates and the precise control of differentiation conditions offered by microfluidic perfusion has great potential for improving stem cell differentiation and maturation to faithfully mimic epithelial cell layers (37). With the ability to monitor and perturb the milieu by manipulating isolated biophysical and biochemical cues in real-time these systems provide a deeper mechanistic insight into stem cell differentiation and effects of changes in the microenvironment on tissue formation its interaction with the microbiota. Further, gut-on-chip will help identify the molecular and cellular targets of disease-related mechanisms for individual patients that cannot be resolved in similar detail in animal models. Improvements of the materials used for chip manufacturing will also increase its relevance for drug testing. Polydimethylsiloxane so far widely used for organ-on-chip is known to non-specifically bind small hydrophobic molecules, which potentially causes bias in drug-absorption and metabolization studies (102, 103). Advanced microfabrication processes using i.e., thermoplastic polymers with more inert properties, will foster the acceptance of gut-on-chip models for drug screening (104).
Clearly, organ-on-chip and in particular complex gut-on-chip models have several limitations. In contrast to conventional organoid cultures, their usability in high-throughput testing is limited.
The ORCHID consortium thus elaborated recommendations for the development and use of future organ-on-chip models as part of a European Roadmap (105). End users need to be provided with customizable platforms “fit-for-purpose” and provided with tailored training programs. ISC-based gut-on-chip as well as ISC-derived organoids represent reductionistic models, which do not reflect the full physiology offered by animal models. Currently available systems thus need to be selected based on the scientific context of the study i.e., the pathogen that is studied or its targeted cell type. The use of standardized and open platform technology will benefit in the future by the establishment of independent testing centers for the qualification and characterization of organ-on-chip models (105).
In the case of organ-on-chip modeling of the gastrointestinal tract, a more “complete” model would need to be composed of adjacent channels with human microvascular endothelium, tissue-resident immune cells, commensal or pathogenic bacteria, and the application of cyclic mechanical forces that mimic peristalsis-like deformations. The additional presence of muscle or nervous system cells could also contribute to create such a faithful model. To this date, there is no system available which is capable to recapitulate every aspect of the gut, considering biophysical, biochemical, immunological and microbiota derived cues. However, the combination of stem cell-based organoid culture and organ-on-chip will help pave the way to significantly expand our knowledge in the development and maintenance of intestinal tissue and its role in the onset of related diseases. Several challenges, such as the uniformization of the systems and its standardization, need to be overcome to allow researchers and non-organ-on-chip specialists easy access to this technology and to allow its routine use in biomedical research.
Author Contributions
FS, EL, MK, HK, MM, and AM contributed to the writing of the manuscript. MM and AM conceptualized, supervised, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Deutsche Forschungsgemeinschaft through the Cluster of Excellence “Balance of the Microverse” under Germany’s Excellence Strategy - EXC 2051 - Project-ID 690 390713860 and the European Commission through the Marie Skłodowska-Curie Actions (MSCA) Innovative Training Network EUROoC (Grant no. 812954) to AM. This work was also supported by the Agence Nationale de la Recherche ANR-17-CE14-0021 (SyNEDI to MM) and a “New Team” grant (BOGUS to MM) from the Bioregate Regenerative Medicine Cluster, University of Nantes and Région Pays de la Loire.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Otani S, Coopersmith CM. Gut Integrity in Critical Illness. J Intensive Care (2019) 7:17. doi: 10.1186/s40560-019-0372-6
2. Clevers H. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell (2013) 154:274–84. doi: 10.1016/j.cell.2013.07.004
3. Guo X, Lv J, Xi R. The Specification and Function of Enteroendocrine Cells in Drosophila and Mammals: A Comparative Review. FEBS J (2021). doi: 10.1111/febs.16067
4. Kobayashi N, Takahashi D, Takano S, Kimura S, Hase K. The Roles of Peyer's Patches and Microfold Cells in the Gut Immune System: Relevance to Autoimmune Diseases. Front Immunol (2019) 10:2345. doi: 10.3389/fimmu.2019.02345
5. Cray P, Sheahan BJ, Dekaney CM. Secretory Sorcery: Paneth Cell Control of Intestinal Repair and Homeostasis. Cell Mol Gastroenterol Hepatol (2021) 12:1239–50. doi: 10.1016/j.jcmgh.2021.06.006
6. Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, et al. Long-Lived Intestinal Tuft Cells Serve as Colon Cancer-Initiating Cells. J Clin Invest (2014) 124:1283–95. doi: 10.1172/JCI73434
7. Mowat AM. Anatomical Basis of Tolerance and Immunity to Intestinal Antigens. Nat Rev Immunol (2003) 3:331–41. doi: 10.1038/nri1057
8. Mosig AS. Organ-On-Chip Models: New Opportunities for Biomedical Research. Future Sci OA (2017) 3:FSO130. doi: 10.4155/fsoa-2016-0038
9. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, et al. Development of a Primary Human Small Intestine-On-a-Chip Using Biopsy-Derived Organoids. Sci Rep (2018) 8:2871. doi: 10.1038/s41598-018-21201-7
10. Tan HY, Trier S, Rahbek UL, Dufva M, Kutter JP, Andresen TL. A Multi-Chamber Microfluidic Intestinal Barrier Model Using Caco-2 Cells for Drug Transport Studies. PloS One (2018) 13:e0197101. doi: 10.1371/journal.pone.0197101
11. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, et al. A Complex Human Gut Microbiome Cultured in an Anaerobic Intestine-on-a-Chip. Nat BioMed Eng (2019) 3:520–31. doi: 10.1038/s41551-019-0397-0
12. Maurer M, Gresnigt MS, Last A, Wollny T, Berlinghof F, Pospich R, et al. A Three-Dimensional Immunocompetent Intestine-on-Chip Model as In Vitro Platform for Functional and Microbial Interaction Studies. Biomaterials (2019) 220:119396. doi: 10.1016/j.biomaterials.2019.119396
13. Tovaglieri A, Sontheimer-Phelps A, Geirnaert A, Prantil-Baun R, Camacho DM, Chou DB, et al. Species-Specific Enhancement of Enterohemorrhagic E. Coli Pathogenesis Mediated by Microbiome Metabolites. Microbiome (2019) 7:43. doi: 10.1186/s40168-019-0650-5
14. Sontheimer-Phelps A, Chou DB, Tovaglieri A, Ferrante TC, Duckworth T, Fadel C, et al. Human Colon-On-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cell Mol Gastroenterol Hepatol (2020) 9:507–26. doi: 10.1016/j.jcmgh.2019.11.008
15. Kim HJ, Ingber DE. Gut-On-a-Chip Microenvironment Induces Human Intestinal Cells to Undergo Villus Differentiation. Integr Biol (Camb) (2013) 5:1130–40. doi: 10.1039/c3ib40126j
16. Shin W, Kim HJ. Intestinal Barrier Dysfunction Orchestrates the Onset of Inflammatory Host-Microbiome Cross-Talk in a Human Gut Inflammation-on-a-Chip. Proc Natl Acad Sci USA (2018) 115:E10539–47. doi: 10.1073/pnas.1810819115
17. Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A, et al. Microfabrication of Human Organs-on-Chips. Nat Protoc (2013) 8:2135–57. doi: 10.1038/nprot.2013.137
18. Sun H, Chow EC, Liu S, Du Y, Pang KS. The Caco-2 Cell Monolayer: Usefulness and Limitations. Expert Opin Drug Metab Toxicol (2008) 4:395–411. doi: 10.1517/17425255.4.4.395
19. Richmond CA, Breault DT. Move Over Caco-2 Cells: Human-Induced Organoids Meet Gut-On-a-Chip. Cell Mol Gastroenterol Hepatol (2018) 5:634–5. doi: 10.1016/j.jcmgh.2018.01.016
20. Clevers H. Modeling Development and Disease With Organoids. Cell (2016) 165:1586–97. doi: 10.1016/j.cell.2016.05.082
21. Beumer J, Clevers H. Cell Fate Specification and Differentiation in the Adult Mammalian Intestine. Nat Rev Mol Cell Biol (2021) 22:39–53. doi: 10.1038/s41580-020-0278-0
22. Clevers HC. Organoids: Avatars for Personalized Medicine. Keio J Med (2019) 68:95. doi: 10.2302/kjm.68-006-ABST
23. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed Differentiation of Human Pluripotent Stem Cells Into Intestinal Tissue In Vitro. Nature (2011) 470:105–9. doi: 10.1038/nature09691
24. Huang CY, Liu CL, Ting CY, Chiu YT, Cheng YC, Nicholson MW, et al. Human iPSC Banking: Barriers and Opportunities. J BioMed Sci (2019) 26:87. doi: 10.1186/s12929-019-0578-x
25. Takahashi K, Yamanaka S. A Decade of Transcription Factor-Mediated Reprogramming to Pluripotency. Nat Rev Mol Cell Biol (2016) 17:183–93. doi: 10.1038/nrm.2016.8
26. Wang Y, Kim R, Hinman SS, Zwarycz B, Magness ST, Allbritton NL. Bioengineered Systems and Designer Matrices That Recapitulate the Intestinal Stem Cell Niche. Cell Mol Gastroenterol Hepatol (2018) 5:440–453 e441. doi: 10.1016/j.jcmgh.2018.01.008
27. Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric Control of Cell Life and Death. Science (1997) 276:1425–8. doi: 10.1126/science.276.5317.1425
28. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell (2006) 126:677–89. doi: 10.1016/j.cell.2006.06.044
29. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in Mechanotransduction. Nature (2011) 474:179–83. doi: 10.1038/nature10137
30. Murphy WL, Mcdevitt TC, Engler AJ. Materials as Stem Cell Regulators. Nat Mater (2014) 13:547–57. doi: 10.1038/nmat3937
31. Shin W, Hinojosa CD, Ingber DE, Kim HJ. Human Intestinal Morphogenesis Controlled by Transepithelial Morphogen Gradient and Flow-Dependent Physical Cues in a Microengineered Gut-On-a-Chip. iScience (2019) 15:391–406. doi: 10.1016/j.isci.2019.04.037
32. Kim J, Koo BK, Knoblich JA. Human Organoids: Model Systems for Human Biology and Medicine. Nat Rev Mol Cell Biol (2020) 21:571–84. doi: 10.1038/s41580-020-0259-3
33. Kuijk E, Jager M, van der Roest B, Locati MD, Van Hoeck A, Korzelius J, et al. The Mutational Impact of Culturing Human Pluripotent and Adult Stem Cells. Nat Commun (2020) 11:2493. doi: 10.1038/s41467-020-16323-4
34. Wang Y, Gunasekara DB, Reed MI, Disalvo M, Bultman SJ, Sims CE, et al. A Microengineered Collagen Scaffold for Generating a Polarized Crypt-Villus Architecture of Human Small Intestinal Epithelium. Biomaterials (2017) 128:44–55. doi: 10.1016/j.biomaterials.2017.03.005
35. Wang Y, Kim R, Gunasekara DB, Reed MI, Disalvo M, Nguyen DL, et al. Formation of Human Colonic Crypt Array by Application of Chemical Gradients Across a Shaped Epithelial Monolayer. Cell Mol Gastroenterol Hepatol (2018) 5:113–30. doi: 10.1016/j.jcmgh.2017.10.007
36. Kasendra M, Luc R, Yin J, Manatakis DV, Kulkarni G, Lucchesi C, et al. Duodenum Intestine-Chip for Preclinical Drug Assessment in a Human Relevant Model. Elife (2020) 9. doi: 10.7554/eLife.50135
37. Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, et al. Homeostatic Mini-Intestines Through Scaffold-Guided Organoid Morphogenesis. Nature (2020) 585:574–8. doi: 10.1038/s41586-020-2724-8
38. Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, et al. Enhanced Utilization of Induced Pluripotent Stem Cell-Derived Human Intestinal Organoids Using Microengineered Chips. Cell Mol Gastroenterol Hepatol (2018) 5:669–77.e662. doi: 10.1016/j.jcmgh.2017.12.008
39. Naumovska E, Aalderink G, Wong Valencia C, Kosim K, Nicolas A, Brown S, et al. Direct On-Chip Differentiation of Intestinal Tubules From Induced Pluripotent Stem Cells. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21144964
40. Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, et al. Persistence and Toxin Production by Clostridium Difficile Within Human Intestinal Organoids Result in Disruption of Epithelial Paracellular Barrier Function. Infect Immun (2015) 83:138–45. doi: 10.1128/IAI.02561-14
41. Crespo M, Vilar E, Tsai SY, Chang K, Amin S, Srinivasan T, et al. Colonic Organoids Derived From Human Induced Pluripotent Stem Cells for Modeling Colorectal Cancer and Drug Testing. Nat Med (2017) 23:878–84. doi: 10.1038/nm.4355
42. Negoro R, Takayama K, Kawai K, Harada K, Sakurai F, Hirata K, et al. Efficient Generation of Small Intestinal Epithelial-Like Cells From Human iPSCs for Drug Absorption and Metabolism Studies. Stem Cell Rep (2018) 11:1539–50. doi: 10.1016/j.stemcr.2018.10.019
43. Reiner O, Sapir T, Parichha A. Using Multi-Organ Culture Systems to Study Parkinson's Disease. Mol Psychiatry (2020) 26(3):725–35. doi: 10.1038/s41380-020-00936-8
44. Bernareggi D, Pouyanfard S, Kaufman DS. Development of Innate Immune Cells From Human Pluripotent Stem Cells. Exp Hematol (2019) 71:13–23. doi: 10.1016/j.exphem.2018.12.005
45. Monkley S, Krishnaswamy JK, Goransson M, Clausen M, Meuller J, Thorn K, et al. Optimised Generation of iPSC-Derived Macrophages and Dendritic Cells That Are Functionally and Transcriptionally Similar to Their Primary Counterparts. PloS One (2020) 15:e0243807. doi: 10.1371/journal.pone.0243807
46. Iriguchi S, Yasui Y, Kawai Y, Arima S, Kunitomo M, Sato T, et al. A Clinically Applicable and Scalable Method to Regenerate T-Cells From iPSCs for Off-the-Shelf T-Cell Immunotherapy. Nat Commun (2021) 12:430. doi: 10.1038/s41467-020-20658-3
47. Campion GV, Mccrae F, Alwan W, Watt I, Bradfield J, Dieppe PA. Idiopathic Destructive Arthritis of the Shoulder. Semin Arthritis Rheum (1988) 17:232–45. doi: 10.1016/0049-0172(88)90009-1
48. Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of Mammals and Their Gut Microbes. Science (2008) 320:1647–51. doi: 10.1126/science.1155725
49. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
50. Parfrey LW, Walters WA, Knight R. Microbial Eukaryotes in the Human Microbiome: Ecology, Evolution, and Future Directions. Front Microbiol (2011) 2:153. doi: 10.3389/fmicb.2011.00153
51. Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early Life Dynamics of the Human Gut Virome and Bacterial Microbiome in Infants. Nat Med (2015) 21:1228–34. doi: 10.1038/nm.3950
52. Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, Functions and Dynamics in the Expanded Human Microbiome Project. Nature (2017) 550:61–6. doi: 10.1038/nature23889
53. Kapitan M, Niemiec MJ, Steimle A, Frick JS, Jacobsen ID. Fungi as Part of the Microbiota and Interactions With Intestinal Bacteria. Curr Top Microbiol Immunol (2019) 422:265–301. doi: 10.1007/82_2018_117
54. Donaldson GP, Lee SM, Mazmanian SK. Gut Biogeography of the Bacterial Microbiota. Nat Rev Microbiol (2016) 14:20–32. doi: 10.1038/nrmicro3552
55. Kastl AJ Jr, Terry NA, Wu GD, Albenberg LG. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell Mol Gastroenterol Hepatol (2020) 9:33–45. doi: 10.1016/j.jcmgh.2019.07.006
56. Thadepalli H, Lou MA, Bach VT, Matsui TK, Mandal AK. Microflora of the Human Small Intestine. Am J Surg (1979) 138:845–50. doi: 10.1016/0002-9610(79)90309-X
57. Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular Analysis of Jejunal, Ileal, Caecal and Recto-Sigmoidal Human Colonic Microbiota Using 16S rRNA Gene Libraries and Terminal Restriction Fragment Length Polymorphism. J Med Microbiol (2005) 54:1093–101. doi: 10.1099/jmm.0.45935-0
58. Mogensen TH. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin Microbiol Rev (2009) 22:240–273, Table of Contents. doi: 10.1128/CMR.00046-08
59. Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, et al. A Map of Toll-Like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity (2018) 49:560–575 e566. doi: 10.1016/j.immuni.2018.07.016
60. Kayisoglu O, Weiss F, Niklas C, Pierotti I, Pompaiah M, Wallaschek N, et al. Location-Specific Cell Identity Rather Than Exposure to GI Microbiota Defines Many Innate Immune Signalling Cascades in the Gut Epithelium. Gut (2021) 70:687–97. doi: 10.1136/gutjnl-2019-319919
61. Burgueno JF, Abreu MT. Epithelial Toll-Like Receptors and Their Role in Gut Homeostasis and Disease. Nat Rev Gastroenterol Hepatol (2020) 17:263–78. doi: 10.1038/s41575-019-0261-4
62. Ferguson M, Foley E. Microbial Recognition Regulates Intestinal Epithelial Growth in Homeostasis and Disease. FEBS J (2021). doi: 10.1111/febs.15910
63. Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, Fenton LA, et al. Targeted Deletion of MyD88 in Intestinal Epithelial Cells Results in Compromised Antibacterial Immunity Associated With Downregulation of Polymeric Immunoglobulin Receptor, Mucin-2, and Antibacterial Peptides. Mucosal Immunol (2012) 5:501–12. doi: 10.1038/mi.2012.23
64. De Kivit S, Van Hoffen E, Korthagen N, Garssen J, Willemsen LE. Apical TLR Ligation of Intestinal Epithelial Cells Drives a Th1-Polarized Regulatory or Inflammatory Type Effector Response In Vitro. Immunobiology (2011) 216:518–27. doi: 10.1016/j.imbio.2010.08.005
65. Yu S, Gao N. Compartmentalizing Intestinal Epithelial Cell Toll-Like Receptors for Immune Surveillance. Cell Mol Life Sci (2015) 72:3343–53. doi: 10.1007/s00018-015-1931-1
66. Semin I, Ninnemann J, Bondareva M, Gimaev I, Kruglov AA. Interplay Between Microbiota, Toll-Like Receptors and Cytokines for the Maintenance of Epithelial Barrier Integrity. Front Med (Lausanne) (2021) 8:644333. doi: 10.3389/fmed.2021.644333
67. Rennert K, Steinborn S, Groger M, Ungerbock B, Jank AM, Ehgartner J, et al. A Microfluidically Perfused Three Dimensional Human Liver Model. Biomaterials (2015) 71:119–31. doi: 10.1016/j.biomaterials.2015.08.043
68. Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A Microfluidics-Based In Vitro Model of the Gastrointestinal Human-Microbe Interface. Nat Commun (2016) 7:11535. doi: 10.1038/ncomms11535
69. Lin Z, Cherng-Wen T, Roy P, Trau D. In-Situ Measurement of Cellular Microenvironments in a Microfluidic Device. Lab Chip (2009) 9:257–62. doi: 10.1039/B806907G
70. Mousavi Shaegh SA, De Ferrari F, Zhang YS, Nabavinia M, Binth Mohammad N, Ryan J, et al. A Microfluidic Optical Platform for Real-Time Monitoring of pH and Oxygen in Microfluidic Bioreactors and Organ-on-Chip Devices. Biomicrofluidics (2016) 10:044111. doi: 10.1063/1.4955155
71. Weltin A, Hammer S, Noor F, Kaminski Y, Kieninger J, Urban GA. Accessing 3D Microtissue Metabolism: Lactate and Oxygen Monitoring in Hepatocyte Spheroids. Biosens Bioelectron (2017) 87:941–8. doi: 10.1016/j.bios.2016.07.094
72. Kieninger J, Weltin A, Flamm H, Urban GA. Microsensor Systems for Cell Metabolism - From 2D Culture to Organ-on-Chip. Lab Chip (2018) 18:1274–91. doi: 10.1039/C7LC00942A
73. Backer M, Rakowski D, Poghossian A, Biselli M, Wagner P, Schoning MJ. Chip-Based Amperometric Enzyme Sensor System for Monitoring of Bioprocesses by Flow-Injection Analysis. J Biotechnol (2013) 163:371–6. doi: 10.1016/j.jbiotec.2012.03.014
74. Tolosa VM, Wassum KM, Maidment NT, Monbouquette HG. Electrochemically Deposited Iridium Oxide Reference Electrode Integrated With an Electroenzymatic Glutamate Sensor on a Multi-Electrode Array Microprobe. Biosens Bioelectron (2013) 42:256–60. doi: 10.1016/j.bios.2012.10.061
75. Enomoto J, Matharu Z, Revzin A. Electrochemical Biosensors for on-Chip Detection of Oxidative Stress From Cells. Methods Enzymol (2013) 526:107–21. doi: 10.1016/B978-0-12-405883-5.00006-5
76. Usuba R, Yokokawa M, Ackermann TN, Llobera A, Fukunaga K, Murata S, et al. Photonic Lab-On-a-Chip for Rapid Cytokine Detection. ACS Sensors (2016) 1:979–86. doi: 10.1021/acssensors.6b00193
77. Cui X, Liu Y, Hu D, Qian W, Tin C, Sun D, et al. A Fluorescent Microbead-Based Microfluidic Immunoassay Chip for Immune Cell Cytokine Secretion Quantification. Lab Chip (2018) 18:522–31. doi: 10.1039/C7LC01183K
78. Van Der Helm MW, Henry OYF, Bein A, Hamkins-Indik T, Cronce MJ, Leineweber WD, et al. Non-Invasive Sensing of Transepithelial Barrier Function and Tissue Differentiation in Organs-on-Chips Using Impedance Spectroscopy. Lab Chip (2019) 19:452–63. doi: 10.1039/C8LC00129D
79. Shin W, Wu A, Massidda MW, Foster C, Thomas N, Lee DW, et al. A Robust Longitudinal Co-Culture of Obligate Anaerobic Gut Microbiome With Human Intestinal Epithelium in an Anoxic-Oxic Interface-On-a-Chip. Front Bioeng Biotechnol (2019) 7:13. doi: 10.3389/fbioe.2019.00013
80. Zhang J, Huang YJ, Yoon JY, Kemmitt J, Wright C, Schneider K, et al. Primary Human Colonic Mucosal Barrier Crosstalk With Super Oxygen-Sensitive Faecalibacterium Prausnitzii in Continuous Culture. Med (N Y) (2021) 2:74–98 e79. doi: 10.1016/j.medj.2020.07.001
81. Dutta D, Clevers H. Organoid Culture Systems to Study Host-Pathogen Interactions. Curr Opin Immunol (2017) 48:15–22. doi: 10.1016/j.coi.2017.07.012
82. Caio G, Volta U, Sapone A, Leffler DA, De Giorgio R, Catassi C, et al. Celiac Disease: A Comprehensive Current Review. BMC Med (2019) 17:142. doi: 10.1186/s12916-019-1380-z
83. Moerkens R, Mooiweer J, Withoff S, Wijmenga C. Celiac Disease-on-Chip: Modeling a Multifactorial Disease In Vitro. United Eur Gastroenterol J (2019) 7:467–76. doi: 10.1177/2050640619836057
84. Xavier RJ, Podolsky DK. Unravelling the Pathogenesis of Inflammatory Bowel Disease. Nature (2007) 448:427–34. doi: 10.1038/nature06005
85. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of Microbiome and Mechanical Deformation to Intestinal Bacterial Overgrowth and Inflammation in a Human Gut-on-a-Chip. Proc Natl Acad Sci USA (2016) 113:E7–15. doi: 10.1073/pnas.1522193112
86. Czepiel J, Drozdz M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, et al. Clostridium Difficile Infection: Review. Eur J Clin Microbiol Infect Dis (2019) 38:1211–21. doi: 10.1007/s10096-019-03539-6
87. Abbas A, Zackular JP. Microbe-Microbe Interactions During Clostridioides Difficile Infection. Curr Opin Microbiol (2020) 53:19–25. doi: 10.1016/j.mib.2020.01.016
88. Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, et al. A Mouse Model of Clostridium Difficile-Associated Disease. Gastroenterology (2008) 135:1984–92. doi: 10.1053/j.gastro.2008.09.002
89. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The Role of Toxin A and Toxin B in Clostridium Difficile Infection. Nature (2010) 467:711–3. doi: 10.1038/nature09397
90. Darkoh C, Odo C, Dupont HL. Accessory Gene Regulator-1 Locus Is Essential for Virulence and Pathogenesis of Clostridium Difficile. eLocator (2016) 7:e01237-16. doi: 10.1128/mBio.01237-16
91. Ikeda M, Ito S, Honda M. Hemolytic Uremic Syndrome Induced by Lipopolysaccharide and Shiga-Like Toxin. Pediatr Nephrol (2004) 19:485–9. doi: 10.1007/s00467-003-1395-7
92. Keepers TR, Psotka MA, Gross LK, Obrig TG. A Murine Model of HUS: Shiga Toxin With Lipopolysaccharide Mimics the Renal Damage and Physiologic Response of Human Disease. J Am Soc Nephrol (2006) 17:3404–14. doi: 10.1681/ASN.2006050419
93. Mohawk KL, O'brien AD. Mouse Models of Escherichia Coli O157:H7 Infection and Shiga Toxin Injection. J BioMed Biotechnol (2011) 2011:258185. doi: 10.1155/2011/258185
94. Villenave R, Wales SQ, Hamkins-Indik T, Papafragkou E, Weaver JC, Ferrante TC, et al. Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro. PloS One (2017) 12:e0169412. doi: 10.1371/journal.pone.0169412
95. Racaniello VR. One Hundred Years of Poliovirus Pathogenesis. Virology (2006) 344:9–16. doi: 10.1016/j.virol.2005.09.015
96. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science (2020) 369:50–4. doi: 10.1126/science.abc1669
97. Greenhalgh K, Ramiro-Garcia J, Heinken A, Ullmann P, Bintener T, Pacheco MP, et al. Integrated In Vitro and In Silico Modeling Delineates the Molecular Effects of a Synbiotic Regimen on Colorectal-Cancer-Derived Cells. Cell Rep (2019) 27:1621–32.e1629. doi: 10.1016/j.celrep.2019.04.001
98. Puschhof J, Pleguezuelos-Manzano C, Clevers H. Organoids and Organs-on-Chips: Insights Into Human Gut-Microbe Interactions. Cell Host Microbe (2021) 29:867–78. doi: 10.1016/j.chom.2021.04.002
99. Capeling MM, Czerwinski M, Huang S, Tsai YH, Wu A, Nagy MS, et al. Nonadhesive Alginate Hydrogels Support Growth of Pluripotent Stem Cell-Derived Intestinal Organoids. Stem Cell Rep (2019) 12:381–94. doi: 10.1016/j.stemcr.2018.12.001
100. Broguiere N, Isenmann L, Hirt C, Ringel T, Placzek S, Cavalli E, et al. Growth of Epithelial Organoids in a Defined Hydrogel. Adv Mater (2018) 30:e1801621. doi: 10.1002/adma.201801621
101. Cruz-Acuna R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, et al. Synthetic Hydrogels for Human Intestinal Organoid Generation and Colonic Wound Repair. Nat Cell Biol (2017) 19:1326–35. doi: 10.1038/ncb3632
102. Toepke MW, Beebe DJ. PDMS Absorption of Small Molecules and Consequences in Microfluidic Applications. Lab Chip (2006) 6:1484–6. doi: 10.1039/b612140c
103. Radisic M, Loskill P. Beyond PDMS and Membranes: New Materials for Organ-On-a-Chip Devices. ACS Biomater Sci Eng (2021) 7:2861–3. doi: 10.1021/acsbiomaterials.1c00831
104. Guttenplan APM, Tahmasebi Birgani Z, Giselbrecht S, Truckenmuller RK, Habibovic P. Chips for Biomaterials and Biomaterials for Chips: Recent Advances at the Interface Between Microfabrication and Biomaterials Research. Adv Healthc Mater (2021) 10:e2100371. doi: 10.1002/adhm.202100371
Keywords: stem cell, gut-on-chip, microbiota, intestine, host-microbiota interaction, in vitro model
Citation: Siwczak F, Loffet E, Kaminska M, Koceva H, Mahe MM and Mosig AS (2021) Intestinal Stem Cell-on-Chip to Study Human Host-Microbiota Interaction. Front. Immunol. 12:798552. doi: 10.3389/fimmu.2021.798552
Received: 20 October 2021; Accepted: 19 November 2021;
Published: 06 December 2021.
Edited by:
Maria José Rodríguez Lagunas, University of Barcelona, SpainsReviewed by:
Toshio Takahashi, Suntory Foundation for Life Sciences, JapanMichal Mokry, University Medical Center Utrecht, Netherlands
Eva Rath, Technical University of Munich, Germany
Copyright © 2021 Siwczak, Loffet, Kaminska, Koceva, Mahe and Mosig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maxime M. Mahe, bWF4aW1lLm1haGVAaW5zZXJtLmZy; Alexander S. Mosig, YWxleGFuZGVyLm1vc2lnQG1lZC51bmktamVuYS5kZQ==
†These authors have contributed equally to this work
 Fatina Siwczak
Fatina Siwczak Elise Loffet
Elise Loffet Mathilda Kaminska
Mathilda Kaminska Hristina Koceva
Hristina Koceva Maxime M. Mahe
Maxime M. Mahe Alexander S. Mosig
Alexander S. Mosig